Highlights
-
•
Cross-platform comparisons were conducted across five leading immunoassay platforms.
-
•
Plasma and serum were obtained from healthy controls and clinical populations.
-
•
Analytic parameters included sensitivity, precision, and performance correlation.
-
•
Platform performance was highly variable, particularly for low-abundant cytokines.
-
•
Findings highlight certain immunoassays should be prioritized in future research.
Keywords: Post-traumatic stress disorder, Parkinson’s disease, Immunoassay, Biomarker, Cytokine, Ultrasensitive technologies
Abbreviations: BLQ, below limit of quantification; CV, coefficient of variance; FEAD, frequency of endogenous analyte detection; IFN-γ, interferon-γ; IL-1β, interleukin-1β; IL-6, interleukin-6; IUGB, Indiana University Genetics Biobank; LLOD, lower limit of detection; LLOQ, lower limit of quantification; MSD, Mesoscale Discovery; PBMC, peripheral blood mononuclear cells; PD, Parkinson’s disease; PMA, phorbol myristate acetate; PTSD, post-traumatic stress disorder; TNF-α, tumor necrosis factor-α; ULOD, upper limit of detection; ULOQ, upper limit of quantification
Abstract
There is mounting evidence of systemic inflammation in post-traumatic stress disorder (PTSD) and Parkinson’s disease (PD), yet inconsistency and a lack of replicability in findings of putative biological markers have delayed progress in this space. Variability in performance between platforms may contribute to the lack of consensus in the biomarker literature, as has been seen for a number of psychiatric disorders, including PTSD. Thus, there is a need for high-performance, scalable, and validated platforms for the discovery and development of biomarkers of inflammation for use in drug development and as clinical diagnostics. To identify the best platform for use in future biomarker discovery efforts, we conducted a comprehensive cross-platform and cross-assay evaluation across five leading platform technologies. This initial assessment focused on four cytokines that have been implicated PTSD – interleukin (IL)-1β, IL-6, tumor necrosis factor (TNF)-α, and interferon (IFN)-γ. To assess platform performance and understand likely measurements in individuals with brain disorders, serum and plasma samples were obtained from individuals with PTSD (n = 13) or Parkinson’s Disease (n = 14) as well as healthy controls (n = 5). We compared platform performance across a number of common analytic parameters, including assay precision, sensitivity, frequency of endogenous analyte detection (FEAD), correlation between platforms, and parallelism in measurement of cytokines using a serial dilution series. The single molecule array (Simoa™) ultra-sensitive platform (Quanterix), MESO V-Plex (Mesoscale Discovery), and Luminex xMAP® (Myriad) were conducted by their respective vendors, while Luminex® and Quantikine® high-sensitivity ELISA assays were evaluated by R&D System’s Biomarker Testing Services. The assay with the highest sensitivity in detecting endogenous analytes across all analytes and clinical populations (i.e. the highest FEAD), was the Simoa™ platform. In contrast, more variable performance was observed for MESO V-plex, R&D Luminex® and Quantikine®, while Myriad’s Luminex xMAP® exhibited low FEAD across all analytes and samples. Simoa™ also demonstrated high precision in detecting endogenous cytokines, as reflected in < 20 percent coefficient of variance (%CV) across replicate runs for samples from the healthy controls, PTSD patients, and PD patients. In contrast, MESO V-Plex, R&D Luminex® and Quantikine® had variable performance in terms of precision across cytokines. Myriad Luminex xMAP® could not be included in precision estimates because the vendor did not run samples in duplicate. For cross-platform performance comparisons, the highest cross-platform correlations were observed for IL-6 such that all platforms – except for Myriad’s Luminex xMAP® – had strong correlations with one another in measurements of IL-6 (r range = 0.59 – 0.86). For the other cytokines, there was low to no correlation across platforms, such that reported measurements of IL-1β, TNF-α, and IFN-γ varied across assays. Taken together, these findings provide novel evidence that the choice of immunoassay could greatly impact reported cytokine findings. The current study provides crucial information on the variability in performance between platforms and across immunoassays that may help inform the selection of assay in future research studies. Further, the results emphasize the need for performing comparative evaluations of immunoassays as new technologies emerge over time, particularly given the lack of reference standards for the quantitative assessments of cytokines.
1. Introduction
Post-traumatic stress disorder (PTSD) is a common, debilitating condition that affects ~ 8% of the U.S. population [1], [2]. Current clinical diagnostics for PTSD are subjective and based solely on symptoms experienced by the individual and assessed through interview. Even with specified diagnostic criteria, there is enormous heterogeneity in the PTSD population given that over 636,000 symptom combinations meet current diagnostic criteria [3]. Although extensive research has explored the utility of physiological markers as discrete biomarkers of PTSD, there are no objective biomarkers of this highly complex and heterogenous condition to aid in diagnosis, predict symptom trajectory, or help stratify the patient population [4], [5], [6].
Further, while biomarker development may be a time-consuming and resource-intensive process, biomarkers play an essential role in drug development and can help increase success in clinical trials, decrease costs, and ultimately improve patient outcomes. However, the identification of biomarkers is limited by the sensitivity, accuracy, and precision of available methods of detection. To this end, there is a need to evaluate existing bioassays and technologies around prospective targets such as cytokines to identify high-performing, scalable platforms that can aid in the development of biomarkers for use in drug development and as clinical diagnostics.
There is mounting evidence of systemic inflammation characterized by elevated cytokines in PTSD [7]. For instance, evidence from recent reviews and meta-analyses suggests that a panel of inflammatory cytokines, including interleukin (IL)-6, IL-1β, tumor necrosis factor (TNF)-α, and interferon (IFN)-γ, are elevated in PTSD patients as compared to healthy controls [8], [9], [10], [11], [12]. Similarly, inflammation is suggested to play a role in Parkinson’s disease (PD) pathologic features and symptoms, such that neuroinflammation may be a potential therapeutic target [13], [14], [15], [16]. For example, data extracted from 25 studies and 2654 participants showed that PD patients exhibit heightened levels of peripheral IL-6, TNF-α, IL-1β, IL-2, IL-10, C-reactive protein, and RANTES (regulated on activation, normal T-expressed, and presumably secreted) in comparison to healthy controls [17]. Together, the above studies strongly suggest the utility of an inflammation panel to help diagnose or stratify patients with PTSD or PD, but more work is needed to discover, validate, and develop these as biomarkers.
To that end, validation of platforms for measuring clinical samples is urgently needed. Although many assays are available for the measurement of inflammatory markers, assays vary in performance and sensitivity [18]. The assessment of endogenous cytokines is challenging given these are typically at very low levels in blood samples (pg/mL to sub-pg/mL) and require highly sensitive technologies to ensure biomarker detection, thereby impacting interpretation of biomarker findings [18], [19]. For instance, recent platform comparison studies have shown that IL-6 and TNF-α are present at moderate levels in human blood and can be detected by most commercial assays [20], [21]. However, IL-1β and IFN- γ were not reliably detected using high-sensitivity cytokine multiplex assays on a Luminex or electrochemiluminescence platform and may necessitate use of more sensitive techniques [19], [22].
To the best of our knowledge, the current large-scale cross-platform comparison is the first to compare inflammatory platforms in patients with PTSD and PD. While multiple studies have conducted cross-platform comparisons using pooled control samples and samples with known cytokine concentrations, few have done this in endogenous samples from clinical populations [18], [19], [22], [23], [24], [25], [26]. Hence, a comprehensive cross-platform and cross-assay evaluation was conducted using five leading immunoassay platforms to identify best-in-class platforms for assessing inflammatory markers. The goal of this study was to evaluate both technical performance and dynamic range in samples from healthy controls and individuals with brain disorders to inform platform selection for future studies that would be powered to make comparison between these populations. Thus, while evaluating the inflammatory signature of psychiatric diseases is beyond the scope of the current study, future research can optimize the rigor and robustness of their cytokine measurements based on findings presented in the current publication.
The current manuscript focuses on four cytokines of particular interest in both PTSD and PD (IL-1β, IL-6, TNF-α, and IFN-γ), which have been extensively studied across a variety of technologies and platforms. A total of twelve cytokines were evaluated in the platform comparison, based on cytokines implicated in PTSD [8], [9], [10], [11], [12] as well those based on recommendations by the Michael J. Fox Foundation to ensure coverage of PD inflammatory markers. Comprehensive assessment of all twelve cytokines is beyond the scope of detail possible through manuscript publication and will be assessed, with results available in a subsequent white paper. To enable the research community to optimize measurement of cytokines of interest, all curated data is found in the Supplementary materials and is publicly available on the Brain Commons, a cloud-based platform for computational discovery designed for the brain health community (https://data.braincommons.org/dashboard/Public/publication-page/cytokineplatformcomparison/index.html).
2. Methods
2.1. Platform performance design
Both serum and plasma samples were included in the cross-platform comparison given the frequent use of both biofluids in psychiatric research. Serum and plasma samples and technical controls were evaluated using commercially available kits provided by each vendor, with processing done at the vendor site when possible to ensure optimal technical performance of each assay. In total, 72 samples were run on each platform, including PTSD (n = 13) and PD (n = 14) plasma samples and both plasma and serum samples from healthy controls (n = 5). All other samples were technical samples, including a dilution curve and stimulated serum culture. Each vendor received identical sets of plasma and serum samples that had undergone the same number of freeze–thaw cycles. The evaluated technology platforms included plate- and bead-based assays with singleplex and/or multiplex capabilities, and signal detection using fluorescence, electrochemiluminescence, and chromogenic systems for single molecule counting outputs (Table 1).
Table 1.
Technology Platform and Assay Description.
| Platform (Vendor Name) | Simoa (Quanterix) | MESO V-Plex (Mesoscale Discovery) | Luminex xMAP (Myriad) | Luminex (R&D Systems) |
Quantikine (R&D Systems) |
|---|---|---|---|---|---|
| Instrument required | Simoa HD-1 Analyzer and associated assay kits | MESO QuickPlex SQ - Custom | Luminex 100/200™ instrument | Luminex MAGPIX | Quantikine High Sensitivity ELISA Kit |
| Assay format | singleplex; multiplex capabilities | multiplex | multiplex | multiplex | singleplex |
| Assay Descriptions | Automated paramagnetic bead-based ligand binding assay used to concentrate a diluted solution of molecules; enables digital detection of fluorescence product molecules confined to wells of very small volumes (40 fl). | Custom multiplex plates w/ 12 analyte capture deposited on a carbon sensor surface. Utilizes electrochemiluminescence labels for detection of antibodies. | Simultaneously measures multiple biochemical markers; assays are multiplexed into analyte-specific microspheres that are passed single-file through laser beams to attain fluorescent signatures. | Simultaneously measures multiple biochemical markers; assays are multiplexed into analyte-specific microspheres that are passed single-file through laser beam; uses magnetic antibody-coated beads. | Singleplex plate-based ligand binding assays based on a two-site sandwich principle in which two highly specific antibodies detect target analyte, with tyramide-signal amplification and chromogenic detection. |
| Volume required | 220–350 ul/well | 350 ul/well | 100 ul/well | 125 ul/well or 75 ul/well | 50–250 ul/well |
Platform performance was evaluated based on analytical performance standards, including sensitivity, precision, reproducibility, and parallelism. Sample concentrations were compared between platforms for each analyte to confirm high-performing platforms correlate with one another. Lastly, relative performance in plasma and serum was explored to understand (i) likely measurements from individuals who have low levels of cytokines and chemokines and (ii) whether platform performance differs between biofluid modalities.
For platform comparisons in clinical populations, the measured dynamic range in PTSD and PD for each platform was assessed to determine whether platforms have adequate sensitivity and linear range to detect cytokines in these representative patient samples and, therefore, inform biomarker discovery. This was evaluated by calculating the frequency of endogenous analyte detection (FEAD) in samples from clinical populations and healthy controls.
For further technical comparisons, isolated peripheral blood mononuclear cells (PBMC), which consist of lymphocytes and monocytes, were added to serum samples and then stimulated with phorbol myristate acetate (PMA)/ionomycin to provoke an immune response. This stimulated blood culture enabled testing of samples with elevated levels of endogenous cytokines and chemokines indicative of an activated immune response. The stimulated serum was also evaluated across a serial dilution to assess the parallel behavior of each assay across a range of endogenous cytokines levels and ensure that sample dilution does not result in biased measurements (trending up or down) of the analyte concentration.
2.2. Plasma and serum samples
Plasma and serum samples from healthy donors were purchased from a commercial vendor (n = 5; BioreclamationIVT [BioIVT], Westbury, NY). Clinical samples were obtained for (i) PTSD from the Translational Research Center for TBI and Stress Disorders (TRACTS) cohort and PrecisionMED (n = 13; TRACTS, VA Central Repository, Boston, MA; PrecisionMED, Solana Beach, CA) and (ii) PD from the LRRK2 cohort consortium (n = 14; Michael J. Fox Foundation, New York, NY). For the PTSD populations from the TRACTS study and PrecisionMED, diagnostic criterion was based on the Clinician-Administered PTSD Scale (CAPS) for DSM-IV and CAPS-5, respectively. For simplicity sake, we labeled the TRACTS participants as PTSD 1 to PTSD 8. In fact, all of these participants also sustained a military mild TBI, as assessed with the Boston Assessment of TBI-Lifetime (BAT-L), assessed in Fortier et al. [27]. Information about the TRACTS longitudinal study has been previously reported by McGlinchey et al. [28]. For the PD population from the LRRK2 cohort, diagnostic instruments included the Montreal Cognitive Assessment (MoCA), the Unified Parkinson's Disease Rating Scale (UPDRS), the Geriatric Depression Scale (GDS) and the Non-Motor Symptoms (NMS) questionnaire, as was previously reported [29]. For all participants, informed consent and institutional review board approval was obtained at each site (TRACTS: VA Boston Institution Review Board #2345; PrecisionMED: Western Institutional Review Board® #2900; LRRK2: Tel Aviv Medical Center Ethical Committee; BioIVT: Western Institutional Review Board® #2010-017).
Because the goal of this study is to enable comparison of vendor performance across representative clinical samples, and given both the value of and limited access to large volumes of clinical blood samples, some aliquots for the PTSD population consisted of plasma obtained from two diagnosed individuals from either TRACTS and/or PrecisionMED. Similarly, healthy control serum and plasma aliquots consisted of samples pooled from several healthy donors. This was done to ensure sufficient sample volume for enabling distribution of identical sets of samples to the 5 vendors. Demographic characteristics and pooling details are provided in Supplementary Appendix (Table A.1). Demographic details are provided for informational use only and not to inform population comparisons, which are beyond the scope of the current study.
For the healthy donors, serum samples were obtained by drawing whole blood into serum tubes (SSTs) mixed pursuant to specifications by BioIVT. Following collection, 60 min was allowed for the clot to retract in the tube at room temperature, after which the tube was spun to obtain serum at 1,300 × g for 20 min in a refrigerated centrifuge (5 °C). Plasma was collected by BioIVT by drawing whole blood into a collection bag containing anticoagulant. Contents were mixed gently for 15 s then placed into a cold sterilizing water bath. Within 15 min the whole blood was spun to obtain plasma at 2,800 × g for 20 min in a refrigerated centrifuge (5 °C). For clinical samples, blood was drawn into 10 mL EDTA plasma tubes that were gently inverted 8–10 times and then spun within one hour of collection at either 1200 × g for 10 min at room temperature (PTSD patients from PrecisionMED), 2000 × g for 15 min at 4 °C (PTSD patients from TRACTS), or 1500 × g for 15 min at 4 °C (PD patients from MJFF), reflecting local optimized conditions for each representative clinical cohort.
All samples were shipped on dry ice to Indiana University Genetics Biobank (IUGB, Indianapolis, IN), where they were stored at −80 °C until processing. Clinical and healthy control samples were all obtained and sent to IUGB between March 2018 and June 2018. IUGB purchased all reagents and stimulant products at the direction of Cohen Veterans Bioscience. Commercially-purchased plasma and serum from the healthy donors were subaliquoted, with subaliquots shipped to Quest Diagnostics (Secaucus, NJ) to measure cytokines in technical control samples (described below). The resulting data were returned to Cohen Veterans Bioscience, who calculated and mapped serial dilution factors for final processing. As described above, subaliquots from donors were pooled, then spiked with the stimulated aliquot and serially diluted, aliquoted to the volumes required for each assay, and labeled with randomized barcodes to ensure sample blinding. IUGB then sorted all samples in pre-defined box locations based on plating strategy provided by Cohen Veterans Bioscience to ensure that variation within each assay would be represented in technical replicates. All samples went through identical freeze–thaw cycles before shipment to the vendors, e.g. each sample went through 1 freeze/thaw cycle at IUGB, with the exception of samples in the plasma and serum super pools, which went through 2 freeze/thaw cycles to enable creation of the serial dilution series.
Plates containing the plasma and serum samples were then sent in a blinded manner to each vendor for evaluation in July 2018. Each vendor received samples organized in an identical manner along with a sample manifest that provided information regarding sample bar code, box name, sample position and specimen type (plasma, serum). No information was provided regarding clinical population or technical details of the sample preparation. All vendors but Myriad performed analyses in duplicate per the manufacturer’s protocols, and data from each vendor was returned to Cohen Veterans Bioscience by September 2018.
2.3. Cytokine assays and technology platforms
Simoa (Quanterix), MESO V-Plex (Mesoscale Discovery), and Luminex xMAP® (Myriad) were assessed by vendors at their respective laboratories using optimized protocols, while Luminex® and high-sensitivity Quantikine® enzyme-linked immunosorbent assay (ELISA) assays were evaluated by R&D System’s Biomarker Testing Services based on recommendations by the latter two vendors for external testing of their assay kits. Four cytokines (IL-1β, IL-6, TNF-α, and IFN-γ) were evaluated using commercially available kits optimized by each vendor using either singleplex or multiplex assay formats: Quanterix: Cytokine 3-Plex A for IL-10, IL-6, TNF-α (101160), IL-1b Simoa 2.0 Assay Kit (101605), IFN-g Simoa 2.0 Assay Kit (100200); Mesoscale Discovery: Custom Sample Testing (SU1CD-1); Myriad: Human CustomMAP® (HMPCore 1, HMPCore 2); R&D Systems Luminex: Luminex Panel 1 (FCSTM09-09), Luminex Panel 2 (FCSTM18-02); R&D Systems Quantikine: Human IL-1β/IL-1F2 High Sensitivity Quantikine ELISA Kit (HSLB00D), Human IL-6 High Sensitivity Quantikine ELISA Kit (HS600B), Human IFN-γ Quantikine ELISA Kit (DIF50), Human TNF-α High Sensitivity Quantikine ELISA Kit (HSTA00E).
A brief description of each technology is described in Table 1. The lower limit of quantification (LLOQ) and upper limit of quantification (ULOQ) were provided by each vendor and are listed in Table 2, along with the units of measure and dilution factor necessary for assessing samples on each platform.
Table 2.
Vendor-provided information on units of measure, dilution factor necessary for assessing the sample on each platform, and lower- and upper-limit of quantification per platform and each cytokine (IL-Iß, IL-6, IFN-γ and TNF-α).
| Analyte | Measurement | Simoa (Quanterix) | MESO V-Plex (Mesoscale Discovery) | Luminex (Myriad) | Luminex (R&D Systems) |
Quantikine (R&D Systems) |
|---|---|---|---|---|---|---|
| IL-1ß | Units | pg/mL | pg/mL | pg/mL | pg/mL | pg/mL |
| Dilution | 4 | 2 | 5 | NA | NA | |
| LLOQ | 0.166 | 2.14 | 5.6 | 0.64 | 0.125 | |
| ULOQ | 48 | 375 | NR | 2600 | 8 | |
| IL-6 | Units | pg/mL | pg/mL | pg/mL | pg/mL | pg/mL |
| Dilution | 4 | 2 | 5 | NA | NA | |
| LLOQ | 0.044 | 1.72 | 4.1 | 1.66 | 0.156 | |
| ULOQ | 94.4 | 531 | NR | 6800 | 10 | |
| IFN-γ | Units | pg/mL | pg/mL | pg/mL | pg/mL | pg/mL |
| Dilution | 4 | 2 | 5 | NA | NA | |
| LLOQ | 0.306 | 8.67 | 5.7 | 8.46 | 15.6 | |
| ULOQ | 800 | 1088 | NR | 34,650 | 1000 | |
| TNF-α | Units | pg/mL | pg/mL | pg/mL | pg/mL | pg/mL |
| Dilution | 4 | 2 | 5 | NA | NA | |
| LLOQ | 0.204 | 0.858 | 62 | 1.52 | 0.156 | |
| ULOQ | 170 | 308 | NR | 6200 | 10 |
Note: NA indicates not applicable, NR indicates not reported.
2.4. Preparation of stimulated endogenous cytokines dilutional series
Stimulated samples with elevated levels of endogenous cytokines were generated by BioVT by isolating peripheral blood mononuclear cells (PBMCs) then resuspending 1 million cells/mL of pooled serum and stimulating this with PMA (50 ng/mL)/Ionomycin (1ug/mL). Finally, 25 mL of stimulated serum was collected. A set of cytokines (IL-1β, IL-6, and TNF-α) were then measured in stimulated serum using the V-plex assay (MSD) by Quest Diagnostics (Secaucus, NJ) to guide preparation of the endogenous serial dilution series. Concentrations of 3108, 4898, and 3329 pg/ml were reported for IL-1β, IL-6, and TNF-α, respectively, indicating the stimulated serum aliquot should be diluted by a factor of 16 using the “serum super pool” to generate 4 serial dilutions plus the original stimulated sample. The predicted concentration of cytokines across the serial dilution is shown in Table 3. Note that the decision to measure this subset of cytokines in the stimulated samples was made a priori and stemmed from technical limitations. While IFN-γ was not measured in the stimulated samples, exclusion of IFN-γ did not impact the outcome of dilutional linearity assessment.
Table 3.
Stimulated Serum Dilution Series: Description of endogenous cytokines elevated via stimulated PBMCs and then serially diluted using serum from the healthy controls plus predicted concentrations of cytokines in the dilution series.
| Type | Measured Cytokines | Dilution Factor | Diluent Type | ||
|---|---|---|---|---|---|
| Elevated Endogenous Cytokines | IL-1β, IL-6, TNF-α | 16 | Serum from healthy controls | ||
| Cytokine | Dilution 0 (pg/ml) | Dilution 1 (pg/ml) | Dilution 2 (pg/ml) | Dilution 3 (pg/ml) | Dilution 4 (pg/ml) |
| IL-1β | 3108 | 194 | 12 | 0.8 | 0.05 |
| IL-6 | 4898 | 306 | 19 | 1.2 | 0.07 |
| TNF-α | 3329 | 211 | 16 | 3.5 | 2.75 |
2.5. Platform performance parameters
2.5.1. Measured range of cytokines
Vendors provided reports that included the absolute concentration of each cytokine in all samples. Measurements were synthesized to calculate the median (standard deviation) and measured range (min–max) of cytokine concentrations for the current study and as reported by each platform.
2.5.2. Assay precision (inter-assay and intra-assay)
Precision of cytokine measurement on each platform was determined by calculating the coefficient of variance (%CV) between replicate runs, such that lower %CV was indicative of better performance. This included both %CV of duplicate runs in the clinical samples (e.g., healthy controls, PTSD patients, and PD patients) as well as between paired technical samples that were either plated at different positions within one plate (intra-assay precision or repeatability) or were plated across the two plates (inter-assay precision or platform stability).
2.5.3. Frequency of endogenous analyte detection (FEAD)
FEAD was assessed by analyzing a set of individual samples on each assay/technology platform and calculating the percentage of samples that did not contain detectable endogenous analyte concentrations, e.g. the percentage of samples that were below the limit of quantification (BLQ%). This was calculated for the plasma samples from the healthy controls, PTSD patients, and PD patients as well as the serum samples from the healthy controls.
2.5.4. Platform parallelism
Parallelism was evaluated across the different platforms for the Stimulated Serum Dilution Series (Table 3). A repeated measures general linear model was used to assess the relationships between vendor measurements for each cytokine, with the expected concentration as the between-subjects factor and measurements from the five platforms as the within-subjects factor (SPSS IBM® v. 24). All measured analyte concentrations were log transformed to achieve Poisson distribution. Assumptions related to sphericity were validated using Mauchly’s test of sphericity (all tests > 0.05). Sample measurements that were below the level of quantification were analyzed using a data-driven half minimum (HM) replacement approach [30], [31]. To apply this approach in the dilution curve samples, cytokine values below the threshold of detection were replaced with half of the minimum of the lowest non-missing value reported for that cytokine, within each vendor. This approach is recommended for values missing due to limits of quantification when it is not possible to impute values from a larger distribution of data [30]. Information on measurements that fell below the level of quantification for all analyses and platforms are summarized in the Supplementary Appendix (Table A.4).
2.5.5. Cross-Platform correlation
Cross-platform correlations were calculated using a subset of 33 plasma samples that were available for analysis (corresponding to 32 independent subjects plus one technical replicate) across the 60 assays conducted in the current study (corresponding to 5 technologies X 12 proteins). Note that one sample from a control subject had outlier values across all assays, as confirmed by principal component analysis, which necessitated its removal from all analyses. For downstream analyses, values have been quantile normalized. Results were analyzed from assays that reported values for a minimum of one third of the samples (i.e., at least 11 samples with values). Pearson correlations were calculated to assess the between-platform correlations for the each of the four cytokines. Group comparisons of variability in measurements (i.e., PD vs. healthy controls and PTSD vs. healthy controls) were conducted with limma (http://bioinf.wehi.edu.au/limma), a linear regression-based package in R, controlling for the existence of the replicate sample [32]. In addition, Bland-Altman plots were used to examine differences between measurements of two assays for each cytokine in the clinical samples.
2.5.6. Plasma vs. Serum comparison
To evaluate the effect of biofluid modality, cytokines measurements were compared for plasma vs serum samples obtained from the same subject. For the healthy control subjects (5 out of the 32 independent subjects), both plasma and serum samples were collected at the same visit, with observed data across 60 assays (corresponding to 5 technologies X 12 proteins). For the control subject with duplicate samples (e.g., there were two different samples from one healthy control in different positions in the plate for both plasma and serum), the max values of the two duplicates were used. Pearson correlation tests were conducted for assays that reported values for at least 2 independent subjects for both biofluids to assess the relationship between plasma and serum for the same cytokine.
3. Results
Five analytical platforms were evaluated for their ability to detect endogenous cytokines in healthy controls and clinical populations consisting of PTSD and PD patients. Given that recent meta-analyses and reviews have indicated IL-1β, IL-6, IFN-γ, and TNF-α are elevated in patients with PTSD and other trauma-related brain disorders [8], [9], [10], [11], [12] – and that three of these have also been implicated in PD symptomatology [13] – these cytokines were selected for the focus of this publication. To evaluate platform performance for all cytokines measures by the platform assays included, curated data is available in the Supplementary materials and is accessible on the Brain Commons, a cloud-based platform for computational discovery designed for the brain health community (https://data.braincommons.org/dashboard/Public/publication-page/cytokineplatformcomparison/index.html).
3.1. Evaluation of measured cytokine range
Information on the measured range of cytokine concentration, as well as the % BLQ, is provided for the plasma samples obtained from each of the populations (healthy controls, PTSD patients, and PD patients) and across each platform in Table 4 and for the serum samples obtained from the healthy controls in Table 5. Given that the measured cytokine range differs substantially between platforms, head-to-head comparisons across platforms is not possible, necessitating the use of the following analytical measures to assess immunoassay performance.
Table 4.
Statistical parameters of the measured cytokine concentrations, %CV between replicate runs, and the FEAD (BLQ%) across the technology platforms for plasma samples derived from the clinical population*
| Simoa (Quanterix) |
MESO V-Plex (Mesoscale Discovery) |
Luminex xMAP (Myriad) |
Luminex (R&D Systems) |
Quantikine (R&D Systems) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cyto-kine | Measure | HCs | PTSD | PD | HCs | PTSD | PD | HCs | PTSD | PD | HCs | PTSD | PD | HCs | PTSD | PD |
| IL-1ß | Median (St Dev) | 0.11 (0.09) | 0.1 (0.1) | 0.7 (0.4) | 0.80 (0.82) | 0.31 (0.75) | 0.29 (1.47) | <BLD | 3.4 (2.92) | <BLD | 0.24 (0.18) | 0.29 (1.15) | 0.41 (0.28) | 0.15 (0.03) | 0.10 (0.05) | 0.13 (0.05) |
| Min-Max | 0.04 – 0.15 | 0.05 – 0.27 | 0.05–0.27 | 0.009–2.61 | 0.031–73.6 | 0.002–5.39 | <BLD | 2.9–9.9 | <BLD | 0.14–0.64 | 0.06–4.19 | 0.22–1.22 | 0.06–0.17 | 0.02–1.95 | 0.05–0.20 | |
|
%CV (St Dev) |
7% (9.4%) | 20% (14.3%) | 25% (12.3%) | 50% (41%) | 50% (34%) | 46% (35%) | NA | NA | NA | 4% (3%) | 4% (6%) | 7% (10%) | 9% (15%) | 25% (26%) | 26% (20%) | |
| BLQ % | 40% | 0% | 0% | 100% | 92% | 92% | 100% | 92% | 100% | 100% | 62% | 86% | 33% | 62% | 50% | |
| IL-6 | Median (St Dev) | 1.54 (0.48) | 2.45 (3.58) | 2.47 (4.11) | 0.87 (0.20) | 0.92 (4.20) | 0.91 (1.35) | <BLD | 3.35 (1.05) | 3.00 (2.23) | 1.96 (0.48) | 2.71 (2.10) | 3.16 (2.56) | 2.20 (0.59) | 2.71 (2.10) | 2.15 (4.89) |
| Min-Max | 0.84–2.36 | 0.61–12.10 | 0.84–16.2 | 0.69–1.19 | 0.13–2.91 | 0.71–5.7 | <BLD | 2.3–4.4 | 2.0–7.6 | 1.66–2.82 | 0.59–8.78 | 2.24–12.4 | 1.07–2.68 | 0.56–8.78 | 1.03 - >10.00 | |
|
%CV (St Dev) |
5% (4%) | 6% (5%) |
7% (5%) |
12% (12%) | 12% (16%) | 18% (20%) | NA | NA | NA | 2% (3%) | 5% (8%) | 3% (3%) | 4% (2%) | 5% (8%) | 5% (3%) | |
| BLQ % | 0% | 0% | 50% | 100% | 92% | 100% | 100% | 92% | 100% | 17% | 23% | 0% | 0% | 0% | 0% | |
| IFN-γ | Median (St Dev) | 0.27 (0.24) | 0.24 (0.40) | 0.30 (0.18) | 5.85 (2.25) | 2.41 (5.60) | 4.19 (12.39) | BLD | 2.25 (0.61) | 2.50 (0.62) | BLQ | BLQ | BLQ | 1.00 (0.93) | 1.20 (1.73) | 1.30 (2.08) |
| Min-Max | 0.07–0.71 | 0.08 (1.58) | 0.16–4.89 | 1.52–7.28 | 0.71–21.4 | 1.05–44.75 | BLD | 2.0–3.5 | 2.0–3.5 | NA | NA | NA | 0.04–2.6 | 0.2–4.9 | 0.1–5.0 | |
|
%CV (St Dev) |
9% (7%) | 18% (19%) | 15% (17%) | 39% (78%) | 27% (33%) | 43% (45%) | NA | NA | NA | NA | NA | NA | NA | NA | NA | |
| BLQ % | 50% | 54% | 57% | 100% | 92% | 78% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | |
| TNF-α | Median (St Dev) | 1.92 (0.31) | 2.27 (0.96) | 2.51 (0.97) | 2.41 (0.48) | 2.27 (0.85) | 2.93 (0.68) | 30.00 (21.6) | 30.00 (24.02) | 17.00 (4.55) | 6.82 (2.16) | 5.54 (3.25) | 6.26 (2.15) | 1.28 (0.30) | 1.08 (0.45) | 1.31 (0.41) |
| Min-Max | 1.79–2.58 | 1.15–4.61 | 1.10–4.57 | 1.72–2.85 | 1.27–4.02 | 1.81–4.30 | 17–30 | 17–43 | 17–30 | 4.5–10.62 | 2.28–11.81 | 3.03–11.27 | 0.27–1.37 | 0.56–2.08 | 0.40–5.40 | |
|
%CV (St Dev) |
9% (7%) | 5% (4%) |
6% (5%) |
4% (2%) |
3% (2%) |
3% (3%) |
NA | NA | NA | 2% (2%) | 2% (2%) |
2% (2.5%) | 2% (1%) | 2% (1.5%) | 2% (1.4%) | |
| BLQ % | 0% | 0% | 0% | 17% | 15% | 0% | 100% | 92% | 100% | 0% | 0% | 0% | 0% | 0% | 7% | |
High: Precision, CV ≤ 20%; FEAD (BLQ%) < 50%.
Intermediate: Precision, CV 20–75%; FEAD (BLQ%) 50–75%.
Low: Precision, CV > 75%; FEAD (BLQ%) > 75%.
Assay Performance was ranked as follows.
Table 5.
Statistical parameters of the measured cytokine concentrations, %CV between replicate runs, and the FEAD (BLQ%) across the technology platforms for serum samples derived from the healthy controls*.
| Simoa (Quanterix) | MESO V-Plex (Mesoscale Discovery) | Luminex xMAP (Myriad) |
Luminex (R&D Systems) |
Quantikine (R&D Systems) |
||
|---|---|---|---|---|---|---|
| Cytokine | Measure | HCs | HCs | HCs | HCs | HCs |
| IL-1ß | Median (St Dev) | 0.5 (0.05) | 0.81 (1.51) | 3.23 (0.47) | 0.23 (0.47) | 0.07 (0.02) |
| Min-Max | 0.04 – 0.07 | 33.2–136.2 | 2.9–3.9 | 0–6.15 | 0.02–0.095 | |
| %CV (St Dev) | 25 (10.8) | 85.1 (50.0) | NA | 0.0 (2.9) | 20.6 (15.5) | |
| BLQ % | 40% | 80% | 100% | 100% | 100% | |
| IL-6 | Median (St Dev) | 1.59 (0.80) | 1.0 (0.41) | 2.30 (0.0) | 2.63 (0.70) | 3.03 (4.60) |
| Min-Max | 0.92–3.05 | 0.53–1.65 | 2.3 | 1.27–3.20 | 1.36 (3.58) | |
| %CV (St Dev) | 0.3 (0.1) | 4.5 (4.0) | NA | 0.0 (2.1) | 1.0 (2.1) | |
| BLQ % | 0% | 100% | 100% | 20% | 0% | |
| IFN-γ | Median (St Dev) | 0.18 (0.11) | 2.76 (0.66) | 2.0 (0.0) | NA | 3.2 (2.10) |
| Min-Max | 0.07–0.35 | 3.55 (0.85) | 2 | NA | 0.5–5.6 | |
| %CV (St Dev) | 7.3 (11.2) | 12.4 (39.0) | NA | 2.7 (3.3) | NA | |
| BLQ % | 80% | 100% | 100% | 100% | 100% | |
| TNF-α | Median (St Dev) | 0.95 (0.60) | 1.07 (0.69) | 21.33 (10.66) | 5.94 (3.33) | 0.35 (0.32) |
| Min-Max | 0.91–2.3 | 0.54–2.6 | 11.0–36.0 | 0.8–10.62 | 0.20–1.10 | |
| %CV (St Dev) | 13.9 (3.7) | 2.0 (3.3) | NA | 1.9 (1.6) | 2.1 (6.4) | |
| BLQ % | 0% | 80% | 100% | 20% | 0% |
High: Precision, CV ≤ 20%; FEAD (BLQ%) < 50%.
Intermediate: Precision, CV 20–75%; FEAD (BLQ%) 50–75%.
Low: Precision, CV > 75%; FEAD (BLQ%) > 75.
Assay performance was ranked as follows.
3.2. Evaluation of assay precision
Assay precision was evaluated for the detection of each cytokine by each platform (Table 4). Precision for each cytokine varied widely based on both the specific cytokine being measured and the platform/assay. Simoa (Quanterix) demonstrated high precision (e.g., %CV < 20) in detecting IL-6, IFN-γ, and TNF-α in samples from the healthy controls, PTSD patients, and PD patients (%CV range = 5% − 19%). Only for IL-1ß in the PTSD and PD patients did Simoa (Quanterix) have “intermediate” performance (PTSD: %CV = 21%, PD: %CV = 25%). In contrast, MESO V-Plex (MSD), Luminex (R&D Systems), and Quantikine (R&D Systems) had variable performance across cytokines. For IFN-γ, precision was low to intermediate for MESO V-Plex (%CV range = 27–43%), and the %CV was not calculated for R&D Luminex or Quantikine as all samples were below the limit of detection. For IL-1ß, IL-6, and TNF-α, precision for these three platforms varied from intermediate to high, with generally higher precision seen using R&D’s Luminex and Quantikine platforms (%CV range: IL-1ß; MESO V-Plex MSD = 46–50%, R&D Luminex = 4–7%, R&D Quantikine = 9–26%; IL-6; MSD = 12–18%, R&D Luminex = 2–5%%, R&D Quantikine = 4–5%; TNF-α; MESO V-Plex = 3–4%, R&D Luminex = 2%, R&D Quantikine = 1.5–2%). The Luminex xMAP® (Myriad) assay was conducted by the vendor per their specifications, and samples were not run in duplicate, such that the Luminex assay could not be included in precision estimates.
For the serum samples, a similar pattern of findings was obtained from the healthy controls (Table 5). Precision was high across all four cytokines for R&D Luminex (%CV range = 0–2%) and ranged from intermediate to high for Quanterix’ Simoa platform (%CV range = 0.3–24.6%). Precision by MESO V-Plex and R&D Quantikine ranged from low to high, depending on the cytokine: (%CV range: IL-1ß; MESO V-Plex = 85%, R&D Quantikine = 21%; IL-6; MESO V-Plex = 5%, R&D Quantikine = 0%; IFN-γ; MESO V-Plex = 12%, R&D Quantikine = NA%; TNF-α; MESO V-Plex = 2%, R&D Quantikine = 2%).
The Supplementary Appendix (Figs. A.2 and A.3) provides intra-assay and inter-assay precision based on paired technical samples that were plated in different positions on the same plate (intra-assay precision for an estimate of repeatability) or were on both plates 1 and 2 (inter-plate precision for an estimate of platform stability).
3.3. Evaluation of frequency of endogenous analyte detection (FEAD)
The ability of the platforms to detect the presence of endogenous cytokines was evaluated by calculating the FEAD, or the percentage of samples from each population (healthy controls, PTSD patients, and PD patients) that had analytes below the limit of quantification (BLQ%), such that lower BLQ% was indicative of better performance (Table 4, Fig. 1). Assays were ranked as having high, intermediate, and low performance if the corresponding BLQ% were < 50%, 50–75%, >75%, respectively. There was intermediate to high performance for the more abundant cytokines, IL-6 and TNF-α, across most platforms, with the notable exceptions of MESO V-Plex and Myriad Luminex: both MESO V-Plex and Myriad Luminex failed to detect IL-6 in the majority of samples and Myriad Luminex additionally failed to detect TNF-α (BLQ% range: IL-6, MESO V-Plex = 92–100%, Myriad Luminex = 92–100%; TNF-α, Myriad Luminex = 92–100%). For the less abundant cytokines, IL-1ß and IFN-γ, only the Simoa (Quanterix) platform consistently detected these analytes in the clinical populations with intermediate to high performance. In contrast, MESO V-Plex, Myriad Luminex, and R&D Luminex assay had very low FEAD for both IL-1ß and IFN-γ; in addition, R&D Quantikine performed poorly in detecting IFN-γ (BLQ% range: IL-1ß, MESO V-Plex = 92–100%, Myriad Luminex = 92–100%, R&D Luminex = 62–100%; IFN-γ, MESO V-Plex = 78–100%, Myriad Luminex = 100%, R&D Luminex = 100%; R&D Quantikine = 100%).
Fig. 1.
Comparison of measured cytokine concentrations for (A) IL-Iß, (B) IL-6, (C) IFN-γ and (D) TNF-α assessed in plasma obtained from the clinical samples and healthy controls. Statistical comparisons were conducted for PTSD and PD groups versus healthy controls (*p < 0.05). Black lines indicate the median concentration detected in each sample, while the blue and red dotted lines indicate LLOQ and LLOD, respectively.
A similar pattern of findings was observed in detecting cytokines in serum samples from the healthy controls (Table 5). Quanterix’s Simoa platform exhibited high performance, detecting all cytokines in the majority of serum samples, with the exception of IFN-γ (BLQ% = 80%). Both R&D’s Luminex and Quantikine platforms had excellent performance in terms of detecting IL-6 and TNF-α, but were unable to detect IL-1ß and IFN-γ at levels above the limit of quantification in 100% of the samples. In contrast, MESO V-Plex and Myriad Luminex platforms had poor performance across cytokines, failing to detect IL-1β, IL-6, IFN-γ, and TNF-α in most samples (BLQ% range: MESO V-Plex = 80–100%; Myriad Luminex = 100%).
3.4. Evaluation of parallelism for the stimulated serum dilution series
To further explore the ability of each platform to detect cytokines in endogenous blood samples, the stimulated serum was serially diluted using serum from healthy controls to create a serial dilution series. To test whether the relationship between the expected and measured analyte concentration varied between platforms and across the dilution curve, the degree of parallelism was determined by comparing the concentration of each analyte of interest (IL-1ß, IL-6, and TNF-α) as measured by each platform to the expected concentration curve in the serial dilution (Fig. 2). IFN-γ was not included in this analysis due to technical limitations; however, exclusion of IFN-γ did not impact the overall outcome of dilutional linearity assessment.
Fig. 2.
Stimulated Serum Dilution Series – Parallelism Assessment: Comparison of cytokine measurement across platform to the calculated concentration curve.
Parallelism was lowest for stimulated endogenous levels of IL-1ß and TNF-α, as the relationship between actual and expected measurements for both cytokines varied by platform (Platform Main Effect IL- ß: F(1,4) = 3.32, p = 0.047; TNF-α: F(1,4) = 7.35, p = 0.003). For both IL-1ß and TNF-α, platform performance was affected by the analyte concentration across the dilution curve (Platform × Calculated Concentration Interaction Effect IL- ß: F(1,4) = 5.36, p = 0.010; TNF-α: F(1,4) = 6.16, p = 0.006). For stimulated endogenous levels of IL-6, an overall effect of platform was not detected, but the efficacy of all platforms was affected by the analyte concentration across the serial dilution (Platform Main Effect: F(1,4) = 1.27, p = 0.335; Platform × Calculated Concentration Interaction Effect: F(1,4) = 15.58, p < 0.001).
For the assessment of stimulated IL-1ß levels within platforms, measured concentration accounted for between 76.5% and 99.9% of the reported concentration across the dilution curve, such that up to 23.5% of reported concentration was due to unaccounted for variation (R2: RD Luminex, 99.9%; Myriad, 94.1%; MSD, 98.0%; Quanterix, 87.6%; RD Quantikine, 76.5%). The starting IL-1ß concentration predicted the reported IL-1ß concentration for all vendors (p < 0.05), except RD Quantikine (F1,5 = 9.76, p = 0.052). For a single unit change in calculated concentration, unit change in reported concentration ranged from 1.06 to 1.99 (b1). Thus, most platforms were hypersensitive to change in IL-1ß across the dilution curve.
For stimulated IL-6 levels within platforms, calculated concentrations in the dilution curve accounted for 79.1% to 97.6% of reported concentrations, such that up to 20.9% of reported values resulted from unaccounted for variation (R2: RD Luminex, 97.6%; Myriad, 96.8%; MSD, 95.3%; Quanterix, 89.7%; RD Quantikine, 79.1%). Starting levels of IL-6 predicted reported levels for all vendors (p < 0.05). For a single unit change in the actual concentration, the unit change in the measured concentration ranged from 1.22 to 3.97 (b1). Thus, all platforms were hypersensitive to changes in IL-6, to the degree that a platform reported a single-unit change as nearly a four-unit change (RD Quantikine).
For stimulated TNF-α levels within platforms, calculated concentrations across the dilution curve accounted for between 85.9% and 99.9% of the reported concentration, such that up to 14.1% of the reported concentration was unaccounted for variation (R2: RD Luminex, 99.8%; Myriad, 99.5%; MSD, 99.3%; Quanterix, 89.3%; RD Quantikine, 85.9%). Starting TNF-α concentration predicted the reported concentration for all vendors (p < 0.05). For a single unit change in the actual TNF-α concentration, the unit change in the reported concentration ranged from 0.85 to 1.44 (b1), indicating that platforms varied in hypo- or hyper-sensitivity to changes in TNF-α levels.
3.5. Cross-platform correlation of cytokine results
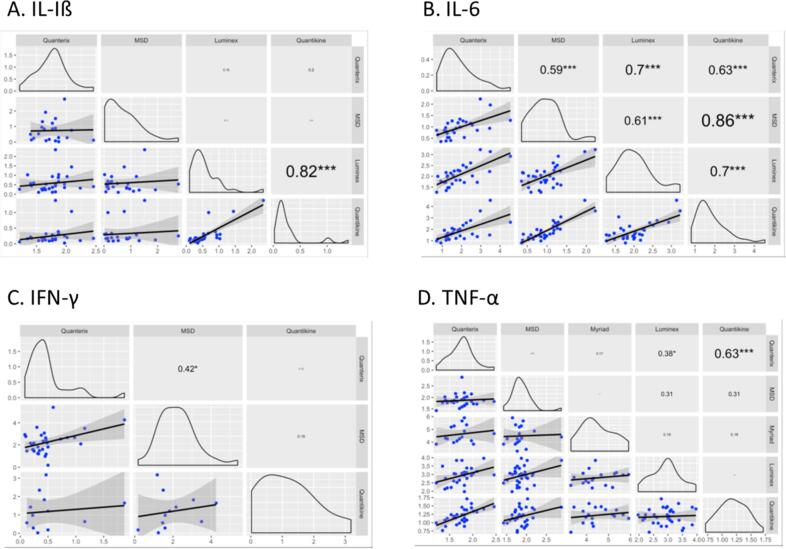
Correlation between platforms is shown in Fig. 3 along with scatter plots depicting the degree of agreement between platforms for each cytokine. The highest platform correlations were observed for IL-6 such that all platforms except for Myriad Luminex had strong correlations with one another (r range = 0.59 – 0.086). For the other cytokines, there was no specific pattern of platform correlation and there was generally low to no correlation for most assays. Platforms with significant correlations for specific cytokines included Simoa and R&D Quantikine (TNF-α: r = 0.63), Simoa and R&D Luminex (TNF-α: r = 0.38), Simoa and MESO V-Plex (IFN-γ: r = 0.42), and R&D’s Luminex and Quantikine platforms (IL-1β: r = 0.82).
Fig. 3.
Platform comparisons for (A) IL-Iß, (B) IL-6, (C) IFN-γ and (D) TNF-α, with Pearson’s correlations conducted for all pair-wise comparisons for assays that had results in >33.33% of the samples (*p < 0.05, **p < 0.01, ***p < 0.001).
3.6. Cross-platform comparison to detect cytokines in plasma vs. serum
To identify whether plasma or serum samples might provide a better matrix for assessing inflammatory markers, Pearson correlations were conducted to assess the plasma-serum correlations for the same protein across the five platforms (Fig. 4). Perhaps due to the small number of individuals represented by both biofluids (e.g. corresponding plasma and serum samples were obtained from only 5 healthy controls), correlations across biofluid measures of IL-Iß, IL-6, TNF-α in plasma vs. serum were significant for only some of the platforms (see Fig. 4A, B, D), and no significant correlations were detected for IFN-γ (Fig. 4C).
Fig. 4.
Plasma vs. Serum Assessment: Comparison of cytokine measurement across platforms for (A) IL-Iß, (B) IL-6, (C) IFN-γ and (D) TNF-α, with Pearson’s correlations conducted for all pair-wise comparisons (*p < 0.05, **p < 0.01, ***p < 0.001).
4. Discussion
This is the first cross-platform comparison study to assess cytokine measurement in PTSD and PD clinical samples across five high-performing technologies, including the single molecule array Simoa™ platform (Quanterix), MESO V-Plex (Mesoscale Discovery), Luminex xMAP® (Myriad), Luminex®(R&D Systems), and Quantikine® (R&D Systems). The goals of this study were to inform platform selection for future studies and to facilitate the interpretation of previous research findings within the context of immunoassays employed for measuring endogenous cytokines.
Sensitivity, one of the most critical analytic parameters for comparing assay performance, was measured as the percentage of samples that had detectable endogenous analyte concentrations for each cytokine. This assessment reflects the relative sensitivity of the assay to detect endogenous cytokines as the absolute concentration of cytokines differed significantly between platforms. Simoa (Quanterix) had the best sensitivity across all four cytokines for both the plasma and serum samples. More variable performance was observed across MESO V-Plex (Mesoscale Discovery), Luminex (R&D Systems), and Quantikine (R&D Systems), particularly for IL-1β and IFN-γ. In contrast, Luminex (Myriad) did not detect even the higher-abundant analytes in any of the endogenous blood samples, inclusive of both plasma and serum, and was only capable of measuring cytokines in the stimulated serum, as described below.
These results merit careful consideration because a number of factors can impact relative sensitivity as assessed by FEAD: (1) FEAD values depend on the individual vendor’s reported threshold of analytical sensitivity, (2) endogenous analyte measurements may not be specific, as seen with multiplexed assays, given that non-specific background interactions can produce high FEAD results not representative of the analyte of interest, and (3) procedures employed to amplify the signal from an analyte may significantly inflate FEAD results stemming from non-specific interactions [18], [33]. Hence, it is necessary to consider additional validation measures to ensure that platforms are detecting the actual analytes of interest and that reported concentrations can be reliably measured over time.
Precision was measured by %CVs between duplicate runs in the clinical samples as well as between technical samples plated at different positions within one plate (intra-assay precision) or across the two plates (inter-assay). Across platforms, precision was designated as high when %CV was > 20%, or low when %CV was > 75%. As anticipated, most platforms exhibited intermediate to high performance for the more abundant cytokines, IL-6 and TNF-α, with lower performance for the less abundant cytokines, IL-1β and IFN-γ. A similar pattern of performance on the precision metric was observed across the five platforms as was reported for assay sensitivity: Quanterix exhibited the highest inter- and intra-assay precision, and more variable precision was observed across MESO V-Plex (Mesoscale Discovery), Luminex (R&D Systems), and Quantikine (R&D Systems).
We also assessed parallelism across the five vendors for the Stimulated Serum Dilution Series, with only IL-6, TNF-α, and IL-1β included in this analysis. In general, platform performance declined on either end of the dilution spectrum based on the measured vs. expected curve, and impact on platform performance was significant for all the measured cytokines. Nevertheless, parallelism was generally high across all platforms and cytokines, particularly for Luminex (Myriad), R&D Luminex, and MESO V-Plex (Mesoscale Discovery). The starting concentration of IL-6, TNF-α, and IL-1β (e.g. Dilution 1 – no dilution) predicted the measured concentration for all vendors (p < 0.05), except for R&D Quantikine for IL-1β. Thus, most platforms were sensitive to change in each cytokine across the dilution curve.
Because a “gold standard” does not exist for the accurate quantitation of inflammatory markers, cross-platform correlations were conducted as exploratory analyses to assess between-platform variation. This analysis assumes that trends in measurements should correlate across high-performing platforms, independent of the specific assay/technology used. Cross-platform correlations were highly variable across the four analytes, and none of the platforms exhibited strong correlations for IFN-γ. The most significant cross-platform comparisons (i.e., the greatest concordance between platforms) were observed for the high-abundant cytokine, IL-6, which is not surprising given most platforms detected this analyte at levels above the limit of quantification. Significant correlations were reported for (1) Simoa (Quanterix) and MESO V-Plex (Mesoscale Discovery), Luminex, and Quantikine and (2) MESO V-Plex (Mesoscale Discovery) and Luminex (R&D Systems) and Quantikine (R&D Systems). To note, the lack of comparable performance between platforms may be explained by differences in the epitopes of the capture and detection antibodies employed for these analytes in the immunoassays. However, this information was not made available by the vendors and is typically not disclosed, and thus warrants further exploration.
These findings were recapitulated when comparing the relative concentration of cytokines between plasma and serum samples obtained from healthy controls in that significant positive correlations were observed for various platforms for all analytes, with the notable exception of IFN-γ. This suggests that the evaluated platforms will have similar performance in plasma and serum for measuring inflammatory markers.
Lastly, to provide an overview of platform performance, the five technology platforms were ranked based on precision (%CV) and relative sensitivity (BLQ%) in the plasma samples obtained from the clinical populations (Table 6) and on precision and relative sensitivity in the serum samples obtained from the healthy controls (Table 7), with relative performance signified by color (high: green, intermediate: orange, low: red). In summary, Simoa (Quanterix) had the highest sensitivity and precision across all four cytokines (IL-1β, IL-6, TNF-α, and IFN-γ). Luminex (R&D Systems) and Quantikine (R&D Systems) had generally high performance for these analytical parameters for all cytokines with the exception of IFN-γ. In contrast, MESO V-Plex (Mesoscale Discovery) performed at a poor to intermediate level for IFN-γ and IL-1β, while performing well for the higher abundant cytokines, IL-6 and TNFα. Luminex (Myriad) failed to detect cytokines of interest in any of the blood samples, indicating this platform may not be appropriate for measuring cytokines known to be at low, sub-pg/mL levels, which may necessitate further investigation.
Table 6.
Summary of platform performance across the five technology platforms based on precision and FEAD in the plasma samples from the clinical populations and healthy controls.
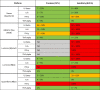 |
*Assay Performance was ranked as follows:
High: Precision, CV ≤ 20%; FEAD (BLQ%) < 50%.
Intermediate: Precision, CV 20–75%; FEAD (BLQ%) 50–75%.
Low: Precision, CV > 75%; FEAD (BLQ%) > 75%.
Table 7.
Summary of platform performance across the five technology platforms based on precision, and FEAD in the serum samples from the healthy controls.
 |
*Assay Performance was ranked as follows:
High: Precision, CV ≤ 20%; FEAD (BLQ%) < 50%.
Intermediate: Precision, CV 20–75%, FEAD (BLQ%) 50–75%.
Low: Precision, CV > 75%; FEAD (BLQ%).
These findings are of high interest given the extensive body of literature evaluating the relationship between elevation in pro-inflammatory cytokines in PTSD. There is mounting evidence of systemic inflammation in PTSD, which may underpin enhanced risk for chronic diseases and adverse health outcomes in PTSD populations, as reviewed by Speer et al. [7]. However, studies investigating inflammatory markers in patients with PTSD have yielded inconsistent findings, with a recent meta-analysis and meta-regression analysis identifying immunoassay as one source of heterogeneity [6], [7], [8], [10].
Consistent with this study, findings from our cross-platform comparison indicate that choice of platform and assay can have a dramatic impact on cytokine measurement and must be carefully considered when designing future studies. Further, it supports the relative comparison of cytokines concentrations across platforms, particularly given the lack of “gold standards” for the quantitative assessment of inflammatory marker and highlights the need to develop reference methods for these analytes.
A recent study by Yeung et al. [18] utilized samples from healthy controls and multiple sclerosis patients to conduct a comprehensive cross-platform and cross-assay evaluation and identified similar outcomes with respect to the analytical performance of the selected platforms. The platforms evaluated in the current study were selected based on availability and use in the literature. These findings will allow for the development of specific recommendations for their use in clinical studies, which must also factor in variables such as cost, availability, and throughput and, ultimately, the opportunity for additional validation for potential clinical use. Future studies should also take into consideration the robustness of these platforms across multiple sites. For example, a cross-site evaluation of the Quanterix IL-6 biomarker kit was recently conducted, which found excellent inter-site reproducibility [34].
It is important to note that although clinical samples were included in this platform comparison, this study was not powered to detect differences in cytokines between these populations, but rather to inform the likely levels of inflammation markers observed in such individuals. Given the small number of samples, demographic variables were not included as covariates in our analysis, and future studies must examine the impact of factors like patient age, gender, race/ethnicity, and socioeconomic status on inflammation markers.
Additionally, considerations around pre-analytical handling and stability of samples should also be taken into account in prospective studies. In this study, individual samples were obtained from existing cohort efforts across different organizations, which vary in procedures for sample collection, handling, processing, and shipping. Such differences could impact cytokine measurements and should be minimized when possible in subsequent large-scale studies.
Overall, the single molecule array (Simoa) ultra-sensitive platform developed by Quanterix exhibited the best performance across all cytokines and should be recommended for use when highly sensitive and precise immunoassays are required. For MESO Vplex (Mesoscale Discovery), Luminex (R&D Systems), and Quantikine (R&D Systems), despite the more variable sensitivity for detection of low abundant cytokines (IFN-γ and IL-1β), these platforms performed well in measuring higher abundant cytokines (IL-6 and TNF-α) and should be considered for use when assessing cytokines at low to high pg/mL levels. Further consideration when choosing the best assay for use may also include factors beyond performance parameter, such as cost, ease of use (e.g. manual vs. automated platforms), technical expertise or equipment required, access and availability, and scalability of the assay.
5. Conclusions
The current study is one of few to compare platform performance in clinical populations and the first, to the best of our knowledge, to do so in PTSD and PD patients [18], [19], [35], [36]. Our findings contribute to the field’s understanding of whether a given assay offers the necessary sensitivity and linear range to evaluate endogenous cytokines in both healthy controls and in populations with a potentially heightened inflammatory state. Thus, the current results can inform future case-control studies that are sufficiently powered for the discovery of a clinical biomarker signature. Additionally, our study demonstrates that immunoassays yield different results across commonly assessed cytokines, which (i) emphasizes the importance of performing such comparative evaluations to guide large-scale biomarker discovery and replication studies, particularly as new technologies emerge over time and (ii) provides insights into which immunoassay is best suited for a particular research endeavor. Moreover, it highlights the need to generate reference materials to allow for commutability across assay platforms.
CRediT authorship contribution statement
Heather C. Lasseter: Conceptualization, Data curation, Formal analysis, Project administration, Visualization, Writing - original draft. Allison C. Provost: Conceptualization, Funding acquisition, Methodology, Project administration, Writing - review & editing. Lauren E. Chaby: Formal analysis, Writing - original draft, Writing - review & editing. Nikolaos P. Daskalakis: Formal analysis, Methodology, Visualization, Writing - review & editing. Magali Haas: Conceptualization, Funding acquisition, Supervision. Andreas Jeromin: Conceptualization, Funding acquisition, Methodology, Supervision.
Declaration of Competing Interest
AJ has been an advisor to Quanterix, Inc and holds stock options. The other authors (HCL, ACP, LC, NPD, and MH) declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
This work was supported primarily by Cohen Veterans Bioscience and generous grants COH-0013 and COH-0003 from Steven A. Cohen for the RAPID-DX program. Additional funding was provided by the Michael J. Fox Foundation (MJFF). Cohen Veterans Bioscience would also like to acknowledge the MJFF for use of Parkinson’s disease samples. Parkinson’s disease data used in the preparation of this article were obtained from the MJFF-sponsored LRRK2 Cohort Consortium (LCC). For up-to-date information on the study, visit www.michaeljfox.org/lcc. The LRRK2 Cohort Consortium is coordinated and funded by The Michael J. Fox Foundation for Parkinson’s Research. In addition, PTSD biospecimens were donated from the longitudinal cohort study of the Translational Research Center for TBI and Stress Disorders (TRACTS), a VA Rehabilitation Research and Development Traumatic Brain Injury Center of Excellence (B3001-C).
In addition, we are immensely grateful to the Indiana University Genetic Biobank for their expertise in the processing and plating of all samples as well as the technology vendors for participating in this cross-platform comparison and providing their technical expertise in processing immunoassays.
HCL, ACP, LC, NPD, MH, and AJ are employed by Cohen Veterans Bioscience Inc., a non-profit public charity research organization.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cytox.2020.100027.
Appendix.
(See Table A1, Table A2, Table A3, Table A4)
Table A1.
Demographic Characteristics and Pooling Information.
| Aliquot | Cohort | Age | Gender | Ethnicity |
|---|---|---|---|---|
| Post-traumatic Stress Disorder | ||||
| PTSD 1 | TRACTS | 23 | Male | White |
| TRACTS | 24 | Male | White | |
| PTSD 2 | TRACTS | 24 | Male | White |
| TRACTS | 28 | Male | Hispanic | |
| PTSD 3 | TRACTS | 29 | Male | White |
| TRACTS | 29 | Male | White | |
| PTSD 4 | TRACTS | 35 | Male | Hispanic |
| TRACTS | 35 | Male | White | |
| PTSD 5 | TRACTS | 35 | Male | White |
| TRACTS | 42 | Male | African American | |
| PTSD 6 | TRACTS | 42 | Male | White |
| TRACTS | 42 | Male | African American | |
| PTSD 7 | TRACTS | 21 | Male | Hispanic |
| TRACTS | 29 | Male | White | |
| PTSD 8 | TRACTS | 27 | Male | Hispanic |
| PrecisionMed | 25 | Male | White | |
| PTSD 9 | PrecisionMed | 31 | Female | White |
| PTSD 10 | PrecisionMed | 31 | Male | Hispanic |
| PTSD 11 | PrecisionMed | 31 | Male | Asian |
| PTSD 12 | PrecisionMed | 33 | Female | White |
| PTSD 13 | PrecisionMed | 61 | Female | White |
| Parkinson's Disease | ||||
| PD 1 | MJFF | 60 | Female | NR |
| PD 2 | MJFF | 62 | Female | NR |
| PD 3 | MJFF | 58 | Female | NR |
| PD 4 | MJFF | 72 | Male | NR |
| PD 5 | MJFF | 67 | Female | NR |
| PD 6 | MJFF | NA | Male | NR |
| PD 7 | MJFF | 67 | Female | NR |
| PD 8 | MJFF | 62 | Male | NR |
| PD 9 | MJFF | 67 | Female | NR |
| PD 10 | MJFF | 67 | Female | NR |
| PD 11 | MJFF | 70 | Female | NR |
| PD 12 | MJFF | 65 | Male | NR |
| PD 13 | MJFF | 78 | Male | NR |
| PD 14 | MJFF | 63 | Female | NR |
| Healthy Controls | ||||
| HC 1 | BioIVT | 52 | Female | Black |
| BioIVT | 47 | Female | Hispanic | |
| HC 2 | BioIVT | NR (multiple donors) | NR | NR |
| HC 3 | BioIVT | NR (multiple donors) | NR | NR |
| HC 4 | BioIVT | NR (multiple donors) | NR | NR |
| HC 5 | BioIVT | 59 | Male | Black |
| BioIVT | 58 | Male | Hispanic | |
| BioIVT | 60 | Male | Hispanic | |
NR indicates not reported.
Table A2.
Intra-assay precision (%CV) across the technology platforms for paired technical replicates of healthy control samples plated at different positions within a plate.
| Cytokine | Replicate Type | Simoa (Quanterix) | MESO V-Plex (Mesoscale Discovery) | Luminex xMAP (Myriad) | Luminex (R&D Systems) | Quantikine (R&D Systems) |
|---|---|---|---|---|---|---|
| IL-1beta | Plasma Pool 1 | 10.5 | NA | NA | 51.2 | 23.4 |
| Plasma Pool 3 | 3.9 | 111.0 | NA | 8.5 | 57.6 | |
| Healthy Control 5 | 4.8 | 95.7 | NA | 0.0 | 9.9 | |
| IL-6 | Plasma Pool 1 | 4.6 | 2.5 | NA | 7.1 | 0.2 |
| Plasma Pool 3 | 3.8 | 20.3 | NA | 5.3 | 0.9 | |
| Healthy Control 5 | 4.6 | 12.4 | NA | 4.9 | 11.0 | |
| IFN-γ | Plasma Pool 1 | 9.5 | NA | NA | NA | |
| Plasma Pool 3 | 1.2 | 57.9 | 33.3 | NA | NA | |
| Healthy Control 5 | 16.2 | 3.3 | NA | NA | NA | |
| TNF-α | Plasma Pool 1 | 7.4 | 9.0 | NA | 5.4 | 2.1 |
| Plasma Pool 3 | 2.8 | 1.9 | 43.3 | 3.2 | 3.6 | |
| Healthy Control 5 | 14.2 | 2.7 | 0.0 | 1.5 | 1.4 |
Table A3.
Inter-assay precision (%CV) across the technology platforms for paired technical replicates of healthy control samples on plate 1 vs plate 2.
| Cytokine | Replicate Type | Simoa (Quanterix) | MESO V-Plex (Mesoscale Discovery) | Luminex xMAP (Myriad) | Luminex (R&D Systems) | Quantikine (R&D Systems) |
|---|---|---|---|---|---|---|
| IL-1beta | Plasma Pool 1 | 0.1 | 129.3 | NA | 23.6 | 18.1 |
| Plasma Pool 2 | 10.7 | NA | NA | 19.3 | 35.9 | |
| Serum Pool 1 | 23.0 | NA | NA | 12.5 | BLD | |
| IL-6 | Plasma Pool 1 | 1.2 | 8.5 | NA | 3.9 | 1.0 |
| Plasma Pool 2 | 7.0 | 1.8 | NA | 9.0 | 1.0 | |
| Serum Pool 1 | 10.2 | 1.6 | NA | 34.5 | 1.9 | |
| IFN-γ | Plasma Pool 1 | 2.1 | 83.7 | NA | NA | 14.3 |
| Plasma Pool 2 | 7.5 | 53.4 | NA | NA | 75.0 | |
| Serum Pool 1 | 13.3 | 16.4 | 0.0 | NA | NA | |
| TNF-α | Plasma Pool 1 | 0.1 | 6.2 | NA | 2.7 | 2.8 |
| Plasma Pool 2 | 7.5 | 0.8 | NA | 8.4 | 0.4 | |
| Serum Pool 1 | 23.0 | 12.2 | 61.3 | 2.5 | 5.2 |
*Assay Performance was ranked as follows:
High: Precision, CV < 20%.
Intermediate: Precision, CV 20–75%.
Low: Precision, CV > 75%.
Table A4.
Analyte measures below the level of quantification in the stimulated serum dilution series.
| Platform Name | Cytokine | LLOD (pg/ug) | Number of Samples BLQ | Lowest Calculated Analyte Concentration Reported (pg/ug) |
|---|---|---|---|---|
| Simoa (Quanterix) | IL-1beta | 0.166 | 0 | 0.19 |
| IL-6 | 0.044 | 0 | 2.25 | |
| TNF-α | 0.204 | 0 | 0.89 | |
| MESO V-Plex (MSD) | IL-1beta | 2.14 | 0 | 0.224 |
| IL-6 | 1.72 | 0 | 1.11 | |
| TNF-α | 0.858 | 0 | 0.83 | |
| Luminex (Myriad) | IL-1beta | 5.6 | 0 | 2.9 |
| IL-6 | 4.1 | 1 | 2.3 | |
| TNF-α | 62 | 1 | 17.0 | |
| Luminex (R&D) | IL-1beta | 0.64 | 0 | 0.44 |
| IL-6 | 1.66 | 0 | 2.06 | |
| TNF-α | 1.52 | 0 | 4.33 | |
| Quantikine (R&D) | IL-1beta | 0.125 | 0 | 0.12 |
| IL-6 | 0.156 | 0 | 2.96 | |
| TNF-α | 0.156 | 0 | 0.48 |
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Pietrzak R.H., Goldstein R.B., Southwick S.M., Grant B.F. Psychiatric comorbidity of full and partial posttraumatic stress disorder among older adults in the United States: results from wave 2 of the national epidemiologic survey on alcohol and related conditions. Am. J. Geriatr. Psychiatry. 2012;20(5):380–390. doi: 10.1097/JGP.0b013e31820d92e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kilpatrick D.G. National estimates of exposure to traumatic events and PTSD prevalence using DSM-IV and DSM-5 criteria. J. Trauma. Stress. 2013;26(5):537–547. doi: 10.1002/jts.21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galatzer-Levy I.R., Bryant R.A. 636,120 ways to have posttraumatic stress disorder. Perspect. Psychol. Sci. 2013;8(6):651–662. doi: 10.1177/1745691613504115. [DOI] [PubMed] [Google Scholar]
- 4.Pitman R.K. Biological studies of post-traumatic stress disorder. Nat. Rev. Neurosci. 2012;13(11):769–787. doi: 10.1038/nrn3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zoladz P.R., Diamond D.M. Current status on behavioral and biological markers of PTSD: a search for clarity in a conflicting literature. Neurosci. Biobehav. Rev. 2013;37(5):860–895. doi: 10.1016/j.neubiorev.2013.03.024. [DOI] [PubMed] [Google Scholar]
- 6.Michopoulos V., Norrholm S.D., Jovanovic T. Diagnostic biomarkers for posttraumatic stress disorder: promising horizons from translational neuroscience research. Biol. Psychiatry. 2015;78(5):344–353. doi: 10.1016/j.biopsych.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Speer K., Upton D., Semple S., McKune A. Systemic low-grade inflammation in post-traumatic stress disorder: a systematic review. J. Inflamm. Res. 2018;11:111–121. doi: 10.2147/JIR.S155903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Passos I.C. Inflammatory markers in post-traumatic stress disorder: a systematic review, meta-analysis, and meta-regression. Lancet Psychiatry. 2015;2(11):1002–1012. doi: 10.1016/S2215-0366(15)00309-0. [DOI] [PubMed] [Google Scholar]
- 9.Black C., Miller B.J. Meta-analysis of cytokines and chemokines in suicidality: distinguishing suicidal versus nonsuicidal patients. Biol. Psychiatry. 2015;78(1):28–37. doi: 10.1016/j.biopsych.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 10.Tursich M. Association of trauma exposure with proinflammatory activity: a transdiagnostic meta-analysis. Transl. Psychiatry. 2014;4 doi: 10.1038/tp.2014.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohler C.A. Peripheral cytokine and chemokine alterations in depression: a meta-analysis of 82 studies. Acta Psychiatr. Scand. 2017;135(5):373–387. doi: 10.1111/acps.12698. [DOI] [PubMed] [Google Scholar]
- 12.Haapakoski R. Cumulative meta-analysis of interleukins 6 and 1beta, tumour necrosis factor alpha and C-reactive protein in patients with major depressive disorder. Brain Behav. Immun. 2015;49:206–215. doi: 10.1016/j.bbi.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGeer P.L., McGeer E.G. Inflammation and neurodegeneration in Parkinson's disease. Parkinsonism Relat. Disord. 2004;10(Suppl 1):S3–S7. doi: 10.1016/j.parkreldis.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 14.Wang Q., Liu Y., Zhou J. Neuroinflammation in Parkinson's disease and its potential as therapeutic target. Transl. Neurodegener. 2015;4:19. doi: 10.1186/s40035-015-0042-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitton P.S. Neuroinflammation and the prospects for anti-inflammatory treatment of Parkinson's disease. Curr. Opin. Invest. Drugs. 2010;11(7):788–794. [PubMed] [Google Scholar]
- 16.Whitton P.S. Inflammation as a causative factor in the aetiology of Parkinson's disease. Br. J. Pharmacol. 2007;150(8):963–976. doi: 10.1038/sj.bjp.0707167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin X.Y. Aberrations in peripheral inflammatory cytokine levels in parkinson disease: a systematic review and meta-analysis. JAMA Neurol. 2016;73(11):1316–1324. doi: 10.1001/jamaneurol.2016.2742. [DOI] [PubMed] [Google Scholar]
- 18.Yeung D. Evaluation of highly sensitive immunoassay technologies for quantitative measurements of sub-pg/mL levels of cytokines in human serum. J. Immunol. Methods. 2016;437:53–63. doi: 10.1016/j.jim.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 19.Casaletto K.B. A comparison of biofluid cytokine markers across platform technologies: correspondence or divergence? Cytokine. 2018;111:481–489. doi: 10.1016/j.cyto.2018.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Locksley R.M., Killeen N., Lenardo M.J. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell. 2001;104(4):487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 21.Wolf J., Rose-John S., Garbers C. Interleukin-6 and its receptors: a highly regulated and dynamic system. Cytokine. 2014;70(1):11–20. doi: 10.1016/j.cyto.2014.05.024. [DOI] [PubMed] [Google Scholar]
- 22.Breen E.C. Multisite comparison of high-sensitivity multiplex cytokine assays. Clin. Vaccine Immunol. 2011;18(8):1229–1242. doi: 10.1128/CVI.05032-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masliah E. Altered expression of synaptic proteins occurs early during progression of Alzheimer's disease. Neurology. 2001;56(1):127–129. doi: 10.1212/wnl.56.1.127. [DOI] [PubMed] [Google Scholar]
- 24.Fu Q., Zhu J., Van Eyk J.E. Comparison of multiplex immunoassay platforms. Clin. Chem. 2010;56(2):314–318. doi: 10.1373/clinchem.2009.135087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malekzadeh A. Comparison of multiplex platforms for cytokine assessments and their potential use for biomarker profiling in multiple sclerosis. Cytokine. 2017;91:145–152. doi: 10.1016/j.cyto.2016.12.021. [DOI] [PubMed] [Google Scholar]
- 26.Chowdhury F., Williams A., Johnson P. Validation and comparison of two multiplex technologies, luminex and mesoscale discovery, for human cytokine profiling. J. Immunol. Methods. 2009;340(1):55–64. doi: 10.1016/j.jim.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Fortier C.B. The Boston Assessment of Traumatic Brain Injury-Lifetime (BAT-L) semistructured interview: evidence of research utility and validity. J. Head Trauma Rehabil. 2014;29(1):89–98. doi: 10.1097/HTR.0b013e3182865859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGlinchey, R.E., W.P. Milberg, J.R. Fonda and C.B. Fortier. A methodology for assessing deployment trauma and its consequences in OEF/OIF/OND veterans: The TRACTS longitudinal prospective cohort study. Int J Methods Psychiatr Res, 2017. 26(3).DOI: 10.1002/mpr.1556. [DOI] [PMC free article] [PubMed]
- 29.San Luciano M. Sex differences in LRRK2 G2019S and idiopathic Parkinson's Disease. Ann. Clin. Transl. Neurol. 2017;4(11):801–810. doi: 10.1002/acn3.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei R. Missing value imputation approach for mass spectrometry-based metabolomics data. Sci. Rep. 2018;8(1):663. doi: 10.1038/s41598-017-19120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brereton, R.G., Chemometrics for Pattern Recognition. 2009: John Wiley & Sons, Ltd.
- 32.Ritchie M.E. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7) doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Koning L, L.C., Shkreta A, Bradwin G, Hu FB, Pradhan AD, Rifai N, Kellogg MD. A multiplex immunoassay gives different results than singleplex immunoassays which may bias epidemiologic associations. Clin Biochem, 2012. 45(10-11): 848-851.DOI: 10.1016/j.clinbiochem.2012.04.006. [DOI] [PubMed]
- 34.Chunyk A.G. A Multi-site In-depth Evaluation of the Quanterix Simoa from a User's Perspective. AAPS J. 2017;20(1):10. doi: 10.1208/s12248-017-0156-7. [DOI] [PubMed] [Google Scholar]
- 35.O'Bryant S.E. Comparing biological markers of Alzheimer's disease across blood fraction and platforms: comparing apples to oranges. Alzheimers Dement (Amst) 2016;3:27–34. doi: 10.1016/j.dadm.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dabitao D., Margolick J.B., Lopez J., Bream J.H. Multiplex measurement of proinflammatory cytokines in human serum: comparison of the Meso Scale Discovery electrochemiluminescence assay and the Cytometric Bead Array. J. Immunol. Methods. 2011;372(1–2):71–77. doi: 10.1016/j.jim.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.