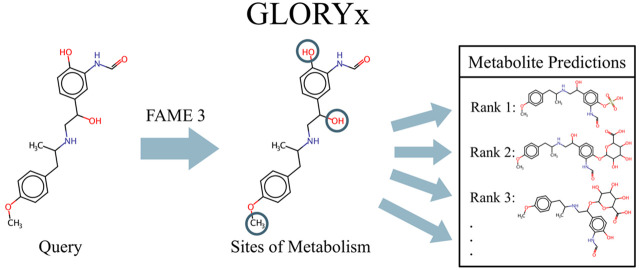
Abstract
Predicting the structures of metabolites formed in humans can provide advantageous insights for the development of drugs and other compounds. Here we present GLORYx, which integrates machine learning-based site of metabolism (SoM) prediction with reaction rule sets to predict and rank the structures of metabolites that could potentially be formed by phase 1 and/or phase 2 metabolism. GLORYx extends the approach from our previously developed tool GLORY, which predicted metabolite structures for cytochrome P450-mediated metabolism only. A robust approach to ranking the predicted metabolites is attained by using the SoM probabilities predicted by the FAME 3 machine learning models to score the predicted metabolites. On a manually curated test data set containing both phase 1 and phase 2 metabolites, GLORYx achieves a recall of 77% and an area under the receiver operating characteristic curve (AUC) of 0.79. Separate analysis of performance on a large amount of freely available phase 1 and phase 2 metabolite data indicates that achieving a meaningful ranking of predicted metabolites is more difficult for phase 2 than for phase 1 metabolites. GLORYx is freely available as a web server at https://nerdd.zbh.uni-hamburg.de/ and is also provided as a software package upon request. The data sets as well as all the reaction rules from this work are also made freely available.
Introduction
Metabolism has a large impact on the safety and efficacy of the xenobiotics that enter the human body, from drugs to cosmetics and agrochemicals, because metabolic reactions can change these compounds into metabolites with different physicochemical and pharmacological properties.1 Computational approaches can be useful for predicting how drugs and other xenobiotics will be metabolized in humans, allowing, for example, the focusing of the drug development process on the most promising compounds in order to save time and reduce costs.
Human xenobiotic metabolism is generally separated into two phases, phase 1 and phase 2, based on the type of reaction (note that the nomenclature does not indicate that a phase 1 reaction must occur before a phase 2 reaction can take place). Phase 1 metabolism consists of oxidation, reduction, and hydrolysis reactions that generally result in increased polarity of the metabolite compared to the parent molecule by creating or unmasking polar functional groups. The main enzyme family responsible for phase 1 metabolism is the cytochrome P450 (CYP) enzyme family, which is responsible for the formation of approximately 60% of first-generation metabolites but only for approximately 40% of metabolites overall (all percentages presented here are based on the current version2 of the MetaQSAR database,3 which contains over 4000 parent molecules, including drugs and other xenobiotics, along with their first-, second-, and third-generation metabolites produced by mammalian metabolic enzymes in vitro and/or in vivo). CYPs are also the cause of a large portion of toxic metabolites and drug–drug interactions.4 Of the non-CYP phase 1 enzymes, the ones with the highest impact on metabolite formation are hydrolases and flavin-containing monooxygenases (FMOs), which account for approximately 9% and approximately 4% of all metabolites, respectively.
Phase 2 metabolism consists of conjugation reactions that, like phase 1 metabolic reactions, tend to modify compounds to more excretable forms. In all, phase 2 metabolism accounts for approximately 30% percent of all metabolites.2 The enzymes responsible for phase 2 drug metabolism belong primarily to five enzyme families: UDP-glucuronosyltransferases (UGTs), glutathione S-transferases (GSTs), sulfotransferases (SULTs), N-acetyltransferases (NATs), and methyltransferases (MTs).5 These five enzyme families are responsible for nearly 90% of all phase 2 metabolites.4
Computational prediction of xenobiotic metabolism encompasses several aspects, including the prediction of the metabolically labile atom positions in molecules, which are known as sites of metabolism (SoM), and prediction of metabolite structures.1,6 Although SoM prediction can provide valuable information and often allows the structure of the resulting metabolite to be inferred by a chemist, it is also of interest to directly predict the metabolites themselves. There are many freely available and commercial methods for metabolite structure prediction, though most focus exclusively on CYP-mediated metabolism. Commercial tools that offer comprehensive prediction of metabolites for both phase 1 and phase 2 metabolism include Meteor Nexus (Lhasa Ltd.),7 TIMES (LMC),8 and MetabolExpert (CompuDrug Ltd.).9
In terms of freely available metabolite structure predictors, a popular and relatively long-lived, open-source tool is SyGMa.10 SyGMa generates and ranks metabolites based on reaction rules and their occurrence ratios derived from the Metabolite database.11 The current version of SyGMa predicts phase 1 and phase 2 metabolites using a set of 145 knowledge-based reaction rules (118 phase 1 rules and 27 phase 2 rules). Using the combined set of reaction rules, SyGMa was able to predict 68% of the metabolites in a test set consisting of 175 parent compounds and 385 reactions (taken from a later release of the Metabolite database compared to the training data).10 The predictor was able to rank 45% of the metabolites in the test set within the top 10 predictions for their corresponding parent molecules. Unfortunately, the Metabolite database, which was used to develop the reaction rules and the occurrence ratios for the scoring, has been discontinued.
Another freely available tool for metabolite prediction is BioTransformer,12 an open-source, comprehensive program that predicts metabolite structures for human CYP and phase 2 metabolism as well as gut microbial, environmental microbial, and human “enzyme commission (EC)-based” metabolism. BioTransformer has 163 CYP rules and 74 phase 2 rules as well as additional constraints regarding molecule types that various rules are allowed to be applied to. Using a combination of the CYP, phase 2, and EC-based modules (408 rules), BioTransformer achieved a recall of 88% on its test set of 40 pharmaceuticals and pesticides, with a precision of 0.49 (188 true positive predictions and 198 putative false positive predictions). BioTransformer does not currently rank its predictions.
A further freely available tool, MetaTox,13,14 predicts metabolites by separately predicting the probability that each potential reaction class is relevant to the given molecule and also predicting the probability of a reaction occurring at each possible reaction center given each possible reaction class. The probability that the resulting metabolite is formed is calculated by combining both of these probabilities, which can then be used to rank the predictions. The reaction types include both phase 1 and phase 2 reaction types, though it is unclear how many reaction rules there are in total. During leave-one-out cross-validation, MetaTox obtained invariant accuracy prediction (IAP, a metric related to area under the receiver operating characteristic curve (AUC)) values between 0.79 and 0.95 for the prediction of the correct reaction class and IAP values between 0.86 and 0.99 for the prediction of the reacting atoms for each of the biotransformation classes.14
We recently reported on the development of a tool called GLORY that predicts the structures of metabolites formed by the CYP enzyme family.15 GLORY includes a new set of reaction rules for CYP-mediated metabolism, whereby common reaction types are distinguished from more unusual reactions. Importantly, GLORY explored how SoM prediction could be effectively employed within the context of metabolite structure prediction. We were able to demonstrate that using the predicted SoM probabilities for each atom in a molecule to score the predicted metabolites, resulting from reactions taking place at those atom positions, led to a meaningful ranking of the predictions.
The software for SoM prediction that was used in GLORY was FAME 2,16 a machine learning-based SoM prediction program that uses extremely randomized trees classifiers combined with two-dimensional (2D) circular descriptors to predict SoMs for CYP-mediated metabolism. Since the development of GLORY, a successor to FAME 2 has become available. FAME 317 continues to use the concept of extra trees classifiers and 2D circular descriptors developed in FAME 2 and applies this approach to generate comprehensive SoM prediction models for both phase 1 and phase 2 metabolism.
There are several other metabolite prediction tools, both commercial (e.g., ADMET Predictor18 from SimulationsPlus, StarDrop19 from Optibrium, and MetaSite from Molecular Discovery20) and freely available (e.g., MetaTox13,14), that incorporate SoM prediction into their metabolite prediction approaches. These tools either focus solely on CYP-mediated metabolism, have not been published, or, as described above for MetaTox, have been evaluated in such a way that makes it difficult to determine how well the metabolite structures themselves were predicted. Thus, although the concept of combining SoM prediction with reaction rules for comprehensive metabolite prediction has been applied in various ways, a systematic analysis of the performance for both phase 1 and phase 2 metabolism has not yet been published.
We have extended the approach developed in GLORY to create a new tool, called GLORYx, that combines SoM prediction with a set of reaction rules to predict metabolites for both phase 1 and phase 2 metabolism. GLORYx employs FAME 3 for SoM prediction, the results of which are used to score and rank the predicted metabolites. Compared to GLORY, GLORYx requires more reaction rules in order to cover non-CYP phase 1 metabolic reactions as well as phase 2 metabolic reactions. GLORYx is freely available via a web server at https://nerdd.zbh.uni-hamburg.de/.
Methods
Reference Data Set
A reference data set of compound-metabolite pairs was compiled from the freely available metabolism data in the DrugBank (drug group “All”)21,22 and MetXBioDB23 databases to serve as a basis for evaluation of the method during the development of GLORYx. For each metabolic reaction in either database, the reactant was considered to be the parent molecule, and the product was considered to be the metabolite. The reference data set is therefore in the format of a map of each parent molecule to its first-generation metabolites, regardless of whether the parent molecule is itself the metabolite of another molecule.
The extraction of the data from DrugBank and MetXBioDB is consistent with the method used in GLORY (see ref (15)). The differences in the preprocessing of the data (i.e., assigning a phase and removing the minor component of salts; see below) arise from considering both phase 1 and phase 2 metabolism rather than just CYP metabolism.
The preprocessing procedure is as follows:
-
(1)
Structural information for both the parent and the metabolite was required in order for a reaction to be included. For DrugBank, the structures are provided in SD file format. In MetXBioDB, only InChIs and InChIKeys are provided, so the InChI was used to generate the structure. Note that stereochemistry information was ignored for parent molecules as well as metabolites, so stereoisomers were thereby condensed.
-
(2)
The multicomponent parent molecules in the DrugBank database had to be handled (no multicomponent parent molecules were found in MetXBioDB). The minor component of each salt was removed (e.g., K+, Ca2+). There was one multicomponent compound (DrugBank ID: DB09327) in which the main component could not be automatically determined, so this compound was excluded from the reference data set. Note that multicomponent metabolites, on the other hand, are simply separated into their individual components and each is considered a separate metabolite.
-
(3)
Any metabolite that contained only one heavy atom (six cases consisting of metal ions, SeH2, and a water molecule; DrugBank only) or had the same InChI, ignoring stereochemistry, as its parent molecule, was excluded.
-
(4)
The metabolites were classified as either phase 1 or phase 2 metabolites, according to the enzyme or biotransformation type annotation (see subsection below for details). If a metabolite could not be assigned a phase, the metabolite was ignored.
-
(5)
Parent molecules with no remaining valid metabolites after applying the above criteria were removed.
-
(6)
The metabolism data corresponding to all parent molecules that overlap with a manually curated test data set (described below) were removed from the reference data set. The removal of the overlap with the test data set affected 15 parent molecules from DrugBank and 9 from MetXBioDB.
The DrugBank and MetXBioDB data were combined in a straightforward manner using InChIs generated without stereochemistry information to compare molecules. If the same parent molecule was present in both DrugBank and MetXBioDB, then the metabolites from both sources were combined, disregarding stereochemistry, into one set.
Assigning a Metabolism Phase to Metabolites in the Data Set
To enable separate evaluation for phase 1 and phase 2 metabolite prediction, we assigned each metabolite in the reference data set to a phase based on the relevant information in DrugBank and MetXBioDB. This allowed the creation of two distinct subsets of the reference data set. The phase 1 and phase 2 subsets of the reference data set represent only phase 1 and phase 2 biotransformations, respectively. If a parent compound has no relevant metabolites for the given phase, then it was excluded from the corresponding subset of the data set.
For the DrugBank data, the metabolites were assigned to a metabolism phase based on the enzyme annotation of the reaction. Some enzymes were omitted completely because they are not enzymes typically associated with human xenobiotic metabolism (e.g., hemoglobin, serum albumin, and lyases). See Table S1 in the Supporting Information for a list of all enzymes that were excluded. This criterion resulted in the exclusion of only 17 metabolites from 11 parent compounds.
For the MetXBioDB data, the appropriate phase for each metabolite was determined based on the “Biotransformation type” annotation in the database. The reactions annotated “Human Phase 1” or “Human Phase 2” were classified as phase 1 or phase 2, respectively.
Manually Curated Test Data Set
The test data set was manually assembled from the scientific literature. We wanted to include all known metabolites of the parent compounds (i.e., all metabolites which have been experimentally observed and reported in the scientific literature), so we chose to structure the data as metabolic trees, including all generations of metabolites that were found in the literature.
The selection of parent molecules for the test data set was based on the top 100 best-selling drugs from 2018.24,25 For all the small-molecule drugs within these 100 drugs that are made up of only the atoms H, C, N, S, O, F, Cl, Br, I, and P, we searched the scientific literature for relevant metabolism information, specifically the structures of human metabolites and preferably a scheme depicting the metabolic tree (see below for more detail). For the listed pharmaceutical products that are a combination of two or more named drugs (e.g., Mavyret is composed of glecaprevir and pibrentasvir), a separate literature search was undertaken for each drug component. For sources of metabolite information, we considered all scientific journal publications that could be found online with Google.
The basic criteria for inclusion in the data set were as follows:
-
(1)
The metabolites must be clearly indicated to be found in humans (either in vivo or in vitro using human hepatocytes, human liver microsomes, or human liver S9 fractions).
-
(2)
Structures of metabolites must be provided. In cases in which a metabolism scheme is not shown, it must be clear, based on chemistry knowledge, that the depicted metabolites are not metabolites of each other, that is, that the depicted metabolites are all first-generation metabolites of the parent drug.
-
(3)
Only fully defined metabolite structures (i.e., the exact position of the added functional group is shown) are included in the data set. The branches of the metabolic tree are followed, and the metabolites included and annotated with the corresponding generation, until a not-fully defined structure is reached. Any further metabolites derived from such a not-fully defined structure are ignored. The maximum metabolite generation included in the data set was generation five, which occurred for only two parent molecules.
-
(4)
Intermediates designated as such in the scheme are not included in the data set.
-
(5)
Some metabolites could not be considered first-generation (based on chemistry knowledge and additional information from the text of the publication), even though the visual scheme indicated that their precursors were only intermediates. Such cases were also removed.
-
(6)
Fatty acid conjugation was not considered.
-
(7)
In the case of one prodrug (abiraterone acetate), we used the drug itself (abiraterone) as the parent molecule in the data set because it had more (first-generation) metabolites shown in the scheme.
The data set was assembled by extracting the SMILES for the parent compounds from the ChEMBL Database26,27 by looking up each drug name. These structures were manually verified for correctness before proceeding. The SMILES for the metabolites were generated with MarvinSketch28 by modifying the parent molecules to create the metabolites and saving them in SMILES format. Metabolite stereoisomers were combined, resulting in a structure with unspecified stereochemistry at the relevant stereocenter.
Data Set Structure
The final data set contains 37 parent molecules and is provided as a JSON file (see Notes). There are 136 first-generation metabolites in total.
The JSON file is structured to represent the metabolic trees, which include multiple generations of metabolites, whenever relevant, following the procedure explained above. For each parent compound, the DOI or PMID of the reference paper(s) is provided, along with the drug name, SMILES, and metabolites. For each metabolite, the name it was given in the publication is provided for reference (this name is often something like “M1”) along with the metabolism generation number and the SMILES. Due to the JSON file format, it is always clear for the second, third, and subsequent generations of metabolites which first-generation metabolites were their precursor, and so on.
No distinction between phase 1 and phase 2 metabolites is made, and enzyme annotations are not included, as this information was only rarely provided in the original literature used to assemble the data set.
Analysis of the Metabolite Data from MetXBioDB and DrugBank
The data from MetXBioDB and DrugBank were considered separately. The data were extracted and preprocessed from each source as described in the Reference Data Set subsection, except that the parent molecules that overlap with the test data set were not removed. For the analysis described here, only the properties of the parent molecules were considered.
Calculations of molecular weight and log P in order to plot the distributions were performed using RDKit.29 The molecular weight calculated was the average molecular weight including hydrogens. One molecule in MetXBioDB was not considered a valid molecule by RDKit (explicit valence greater than permitted) and was therefore excluded from all analysis described in this section.
Principal component analysis (PCA) was performed with scikit-learn30,31 using 44 physicochemical descriptors calculated with the Molecular Operating Environment (MOE).32 A full list of the descriptors, as well as a brief description of each, can be found in Table S2.
Reaction Rules
The metabolic reaction rules used in GLORYx are encoded as SMIRKS.33 Three sets of reaction rules were used: (1) all of SyGMa’s reaction rules, which include both phase 1 and phase 2 rules; (2) GLORY’s reaction rules, covering only CYP metabolism; and (3) a newly developed set of GSH conjugation rules to augment SyGMa’s phase 2 rules, which are missing reactions of this type. The reaction rules from GLORY were used unchanged.
Implementation of SyGMa Reaction Rules
Because the so-called SMIRKS provided in SyGMa’s open-source python package34 are actually in the format of RDKit’s reaction SMARTS, it was necessary to convert them to proper SMIRKS in order to implement them in our software. This conversion was performed manually, with care being taken to preserve the chemical meaning of the reaction.
In one case, namely that of oxidative deamination, additional SMIRKS strings were necessary to achieve the same result with the SMIRKS that SyGMa achieved with its reaction SMARTS. The reason is that double bonds in an aromatic ring are not automatically shifted during transformation in GLORYx. We therefore added two additional SMIRKS in order to explicitly shift the double bonds for 6-rings and 5-rings. Any invalid products generated by the SMIRKS for this reaction are ignored because the molecule validity checker in GLORYx discards transformation products with a carbon atom of invalid atom type (in this case, a valence >4 due to incorrect bond placement).
Development of Reaction Rules for Glutathione Conjugation
The scientific literature indicates that glutathione (GSH) conjugation by the GST enzyme family occurs mainly at the following functional groups: epoxides, α,β-unsaturated carbonyls, quinones, nucleophilic substitution (aliphatic and aromatic), isocyanates (and isothiocyanates), and nitriles.35−40 The SMIRKS for these cases were developed based on the reaction descriptions and example reactions present in the referenced literature.
Metabolite Prediction Program GLORYx
GLORYx applies the reaction rules to all appropriate positions in the molecule, determined by where each reaction rule SMIRKS matches, if it matches at all. Within the program, SoMs are predicted with FAME 3,17 and the predicted SoM probabilities are used to score and rank the predicted metabolites. The software is written in Java and uses CDK version 2.0.41,42
GLORYx performs an initial preprocessing step for all input molecules to check that the input molecule can be successfully parsed by CDK, does not have multiple components, and contains no element types other than C, N, S, O, H, F, Cl, Br, I, P, B, and Si (FAME 3’s allowed element types; note that FAME 3 does not make predictions for B and Si due to a lack of training data, and for this reason the test set was chosen to not include any molecules with a B or Si atom). If any of these checks fail, no predictions are made for the input molecule. Further preprocessing steps that occur within the context of SoM prediction and the application of the reaction rules are described in the following subsections.
SoM Prediction
The SoM prediction in GLORYx is performed using FAME 3.17 FAME 3 was trained on the SoM data from the MetaQSAR database3,43 and offers three SoM prediction models: the P1 model predicts SoMs corresponding to phase 1 metabolic reactions, the P2 model predicts SoMs corresponding to phase 2 metabolic reactions, and the P1 + P2 model predicts SoMs corresponding to both metabolism phases.
The FAME 3 code includes preprocessing of the input molecules, involving the standardization of nitro groups, aromaticity detection, and automatic addition of hydrogens if the hydrogens of the input molecule are not explicitly specified. Because the SoM prediction step comes before the application of the reaction rules within the GLORYx program, the standardization of the molecules described here remains in place for the subsequent transformation step described below.
FAME 3 uses circular descriptors that incorporate 15 basic 2D CDK descriptors and circular atom-type fingerprints (see ref (17) for details). During the development of FAME 3, the effect of the bond depth of the circular descriptors was examined, and a bond depth of five was chosen as the default bond depth for the descriptors. GLORYx uses the default models with a descriptor depth of five.
In order to improve GLORYx’s ability to rank its predictions of phase 2 metabolites (see Results for details), we used previously unpublished reaction class-specific individual phase 2 SoM models from FAME 3. These models were created using the identical modeling procedure described in the FAME 3 paper (see ref (17)), but with each model trained on only a subset of the data. The SoM data from the reaction classes in the MetaQSAR database43 corresponding approximately to the five main enzyme families of phase 2 xenobiotic metabolism (UGTs, GSTs, SULTs, MTs, and NATs) were selected, and a separate model was created for each subset. The reaction class types and the number of molecules used to train the models are described in Table S3. Note that the data from two classes of glucuronidation reactions in MetaQSAR were combined to create a single glucuronidation model.
For the reaction class-specific phase 2 SoM models, GLORYx again uses the models with a descriptor depth of five, for consistency.
Transformation of Molecules According to Reaction Rules
The reaction rules were applied using Ambit-SMIRKS.44,45 As for GLORY, any product containing fewer than three heavy atoms is not included in the set of predicted metabolites.
In order to apply the reaction rules correctly, that is, to achieve the same predicted metabolites as SyGMa while using the same rules, it was necessary to use an aromaticity model that could recognize aromaticity in rings with exocyclic heteroatoms. To achieve this, we chose an aromaticity model in CDK that uses the ElectronDonation.daylight() electron donation model. In order to allow for better ring recognition in molecules with more than three rings, we set the cycles portion of the aromaticity model to Cycles.or(Cycles.all(), Cycles.relevant()), indicating that all cycles would be used whenever possible and the “relevant” cycles would be used in cases in which the molecule contained too many cycles for all of them to be considered. This new aromaticity model is applied directly before reaction rule-based transformation using all reaction rules, not just the rules sourced from SyGMa, and does not affect the aromaticity recognition used for SoM prediction with FAME 3.
There is one noticeable remaining discrepancy related to aromaticity, as determined by comparison on the reference data set, between GLORYx’s predictions and SyGMa’s predictions for the “same” reaction rules. Tetrazoles appear to be recognized as aromatic by GLORYx but not by SyGMa, as indicated by an aromatic glucuronidation reaction being successfully applied by GLORYx but not by SyGMa. This discrepancy affects only three parent molecules in the phase 2 subset of the reference data set.
Scoring
Each predicted metabolite is assigned a priority score in order to rank the predictions. The priority score has two components. The first component is the predicted SoM probability from the FAME 3 model used to make the prediction. The maximum SoM probability among the atoms in the mapping of the reaction rule’s SMIRKs onto the parent molecule is used.
The second component is a reaction rule weighting factor based on a simple designation of either “common” or “uncommon” for each reaction type. This designation, which we previously introduced for CYP reactions in ref (15), was based primarily on a detailed review of CYP-mediated reaction types that described both common and uncommon types of reactions.46 In this work, we use the common–uncommon designation more loosely, as a simple differentiation in reaction type prioritization that allows a binary weighting of the reaction rules. A weighting factor corresponding to the common–uncommon classification is multiplied with the maximum SoM probability mentioned above in order to calculate the priority score for the predicted metabolite. In GLORYx, a weighting factor of 1 is used for reaction rules designated “common”, and a weighting factor of 0.2 is used for reaction rules designated “uncommon”. These weighting factors thereby maintain the same ratio of 5:1 as described previously in ref (15) but are scaled such that the final priority score more reflects a probability-like concept, with values ranging from 0 to 1.
The final priority score of a predicted metabolite is thereby the product of the maximum SoM probability and the weighting factor corresponding to the priority level, common or uncommon, of the reaction type.
The final assignment of a priority level to the reaction rules was determined rationally. The priority levels of the CYP metabolism-based rules from GLORY were not changed. All of the phase 1 rules from SyGMa were designated uncommon, which does not affect the higher priority given to common CYP-mediated reaction types in the case of duplicate reaction types in the SyGMa and GLORY rule sets. The phase 2 rules corresponding to the five main phase 2 enzyme families were designated common, while the others (glycination, phosphorylation, and dephosphorylation) were designated uncommon.
Validation
Predicted metabolites were compared to the known metabolites from either the reference data set or the test data set using InChIs that were generated without stereochemistry information.
Special Consideration for CYP Reactions
Spontaneous oxidation from an aldehyde to a carboxylic acid was considered during the evaluation process, as in GLORY (see ref (15)), but only for predicted metabolites that were the product of a phase 1 reaction rule. It was intended that this consideration only apply to CYP reactions, but SyGMa’s phase 1 reaction rules do not distinguish between CYP and non-CYP, so this step was applied to all phase 1 products. Note that this applies only to the validation and does not affect the predicted metabolites that are provided to the users of GLORYx.
Comparison to SyGMa
The comparison of GLORYx to SyGMa was performed using SyGMa34 with RDKit.29 One change to the standard usage of SyGMa was required, in the case where both phase 1 and phase 2 metabolite predictions were desired. When SyGMa is run with a single metabolism Scenario object specifying both phase 1 and phase 2, the rule sets for the phases are applied sequentially, that is, the first rule set listed (phase 1) is applied first, and then the second rule set (phase 2) is applied to the parent compound as well as the products of the first rule set. This behavior corresponds to a different research question than the one posed in our evaluation, so SyGMa was instead run twice for each molecule in the test set, once using only the phase 1 rules and then separately using only the phase 2 rules. The predictions from both runs were combined.
In addition, any predicted metabolite with the same InChI as the parent compound was ignored, and, for the sake of comparison, a filter to remove all predicted metabolites with fewer than three heavy atoms was added (SyGMa’s built-in percentage-based size filter was turned off). Implicit hydrogens were also added to SyGMa’s output SMILES before generating the InChIs for comparison with GLORYx’s predictions.
Results
The concept of GLORYx is that SoMs, or rather the probability of each heavy atom being a SoM, are predicted with FAME 3, and, building on these predictions, a set of reaction rules is applied in order to generate the structures of predicted metabolites for both phase 1 and phase 2 metabolism. We have previously determined, for our earlier CYP-focused metabolite prediction tool GLORY, that using the predicted SoM probabilities as a hard cutoff to determine whether or not to apply a reaction rule at a given position is not a particularly effective approach, except if the goal is to simply reduce the number of predictions.15 Instead, we found that using the predicted SoM probabilities to score and rank the predicted metabolites enabled a reasonable ranking of the predicted metabolites while retaining a high recall of known metabolites. Therefore, we again use the predicted SoM probabilities to rank the metabolites predicted by GLORYx. For GLORYx, we also have the capability of using a different FAME 3 SoM prediction model depending on which phase of metabolism is being predicted.
GLORYx was developed and analyzed using a large reference data set containing metabolism data from DrugBank and MetXBioDB. This reference data set was used to examine phase 1 and phase 2 metabolism separately to make sure each phase could be handled satisfactorily on its own as well as to determine how to best combine predictions for both phases. The final validation of GLORYx was subsequently performed on a manually curated test data set.
Analysis of the Approach Using a Large Reference Data Set
A reference data set for the development of the GLORYx method was created by combining the freely available metabolism data from DrugBank and MetXBioDB (see Methods for details). Considering both phase 1 and phase 2 metabolism, and using the data preparation process described in Methods, we collected metabolite data for 560 parent molecules from DrugBank and 1188 parent molecules from MetXBioDB. Of these parent molecules, 310 are identical, not considering stereochemistry, meaning there are 1438 parent molecules total from both sources combined. The metabolites for the overlapping parent molecules were consolidated when forming the reference data set. Within this overlap, 555 of 868 metabolites were present in both data sets. Of the rest, 135 were from DrugBank and 178 from MetXBioDB.
It is relevant to mention here that DrugBank does not contain species annotations for the metabolism data, while MetXBioDB specifies “Human Phase 1” and “Human Phase 2” metabolic reactions. Neither data source includes annotations regarding whether any given metabolite data were collected in an in vivo or an in vitro study.
Beyond noting the amount of overlap between the two data sources, we wanted to examine the chemical space covered by each, in terms of the parent molecules. To the best of our knowledge, such an analysis has not yet been done for MetXBioDB. For DrugBank, an analysis focused specifically on the compounds for which there is metabolite data has also not yet been undertaken. When performing this analysis, we retained the overlapping parent molecules in both data sets.
In terms of molecule size, we observe a narrower distribution among the parent molecules of MetXBioDB than among those of DrugBank, as seen in Figure 1A for molecular weight. In addition, we noted a shift in the distributions, whereby DrugBank has a median molecular weight of 322 while MetXBioDB has a median of only 282. The mean values are not compared due to the presence of an outlier with a molecular weight of 4114 Da (semaglutide) in the DrugBank data. For calculated log P (clog P) values as well, a narrower distribution is observed for MetXBioDB (Figure 1B). However, for clog P the median values of the two distributions are very similar, at 3.04 for DrugBank and 3.05 for MetXBioDB.
Figure 1.
Comparison of the metabolite data from MetXBioDB and DrugBank, in terms of parent molecules. (A) Distribution of molecular weight. (B) Distribution of clog P. (C) Histogram of the number of metabolites per parent molecule in terms of percentage of parent molecules. (D) Comparison of the chemical space of the parent molecules from MetXBioDB and DrugBank using PCA calculated using 44 physicochemical descriptors. The percentage of the total variance explained by each of the first two principal components is included in the axis labels.
In the context of metabolite prediction, it is especially interesting to compare the ratio of parent molecules and metabolites recorded in a data set as this ratio can give an indication of the comprehensiveness of the metabolism data (metabolism data are generally incomplete; more metabolites are typically known for compounds of high relevance, in particular approved drugs). In the case of the DrugBank and MetXBioDB data, the distributions of the number of metabolites per parent molecule are quite similar (Figure 1C). In both cases, the majority of parent molecules have only one known metabolite. At the same time, over 40% of the parent molecules from each data source have multiple metabolites.
Finally, to achieve a visual comparison that takes into account multiple physicochemical properties of the parent molecules, we performed principal component analysis (PCA) on each set of parent molecules using 44 physicochemical descriptors (Figure 1D; see Methods for details). From the PCA we see that there is a large amount of overlap between the two data sets, which is unsurprising given that most of the molecules in the DrugBank data set are also included in the MetXBioDB data set. However, we also see that there are portions of the chemical space populated by parent molecules from DrugBank but not from MetXBioDB, which is consistent with the results from the comparison of the distributions of molecular weight and clog P. Inspection of the PCA loading plot (Figure S1) shows that molecule size and polarity seem to play a large role in the variance in the PCA plot. In particular, molecule size seems to influence the first principal component, while polarity seems to influence the second principal component. Interestingly, the five data points (two from DrugBank, three from MetXBioDB) in the far right portion of the PCA plot correspond to the five largest molecules included in the calculation, all of which have a molecular weight between 1000 and 1300 Da (the outlier with a molecular weight of over 4000 Da was not included in the PCA). These five molecules consist of five macrocyclic peptides (including cyclosporine) and one nonmacrocyclic peptide (angiotensin II).
Whereas the above chemical space analysis included all valid metabolite data from DrugBank and MetXBioDB, a further data preprocessing step was performed for the formation of the final reference data set used for the evaluation of the metabolite structure prediction approach. All metabolism data corresponding to parent molecules contained in the test set were removed from the reference data set. This removal resulted in a final reference data set containing 1420 parent molecules and a total of 2453 metabolites.
The reference data set was further separated into two subsets, corresponding to phase 1 and phase 2 metabolism. The phase 1 subset contains 944 parent molecules and 1763 metabolites, and the phase 2 subset contains 582 parent molecules and their 690 metabolites (Table 1). Most of the phase 1 metabolites are CYP metabolites, and most of the phase 2 metabolites are UGT metabolites (Table 1). Note that some of the phase 2 metabolites do not correspond to any of the listed enzyme families, just as some of the phase 1 metabolites are not formed by CYPs.
Table 1. Composition of the Reference Data Seta in Terms of Metabolism Phase and Enzyme Family.
| number of metabolitesb | number of parent molecules | |
|---|---|---|
| phase 1, all | 1763 | 944 |
| CYP | 1640 | – |
| phase 2, all | 690 | 582 |
| UGT | 480 | – |
| SULT | 92 | – |
| GST | 46 | – |
| NAT | 34 | – |
| MT | 17 | – |
| phase 1 + phase 2 | 2453 | 1420 |
The reference data set was created by combining the DrugBank and MetXBioDB metabolism data and removing the data for all parent molecules contained in the test set.
Note that the total numbers of phase 1 and phase 2 metabolites do not equal the sum of the metabolites from the listed enzyme families, because not all metabolites in the data set correspond to these main enzyme families.
The two separate subsets of the reference data set were used to analyze the performance of GLORYx for phase 1 and phase 2 individually, because there are slightly different considerations for each metabolism phase. In addition, the entire reference data set was used to analyze the combined prediction of both phase 1 and phase 2 metabolites.
Note that GLORYx is unable to process two parent molecules in the phase 1 subset of the reference data set and one parent molecule in the phase 2 subset. Both of the phase 1 parent molecules contain a Se atom, which FAME 3 cannot handle (partial charges cannot be calculated; see Methods for a list of allowed element types). Because no SoM predictions can be made, no metabolites are predicted. The parent molecule in the phase 2 subset is unable to be processed because it contains a nitrogen atom with a state that FAME 3 does not recognize. This is the case regardless of which FAME 3 model is used.
Phase 1 Metabolism
The fundamental concept of our approach to predicting metabolites is to integrate machine learning-based SoM prediction in order to score the predicted metabolites. Therefore, the first thing we wanted to know is how GLORYx’s SoM probability-based scoring approach compares to the scoring approach used by the state-of-the-art, open source, comprehensive metabolite prediction tool SyGMa.
To compare the scoring approaches, GLORYx was initially implemented using only the phase 1 reaction rules sourced from SyGMa. The phase 1-specific FAME 3 SoM prediction model (model P1) was used to predict SoMs. The predicted metabolites were scored using the maximum SoM probability predicted among all heavy atoms in the mapping onto the parent molecule of the reaction rule that led to the particular predicted metabolite. In this case, the score was therefore equal to this SoM probability; no weighting based on reaction type was used. SyGMa, on the other hand, ranks its predictions based on probability scores that are calculated using the occurrence ratios of each reaction rule in the Metabolite database. Each of SyGMa’s predicted metabolites is assigned a probability score corresponding to the reaction rule that formed the predicted metabolite.
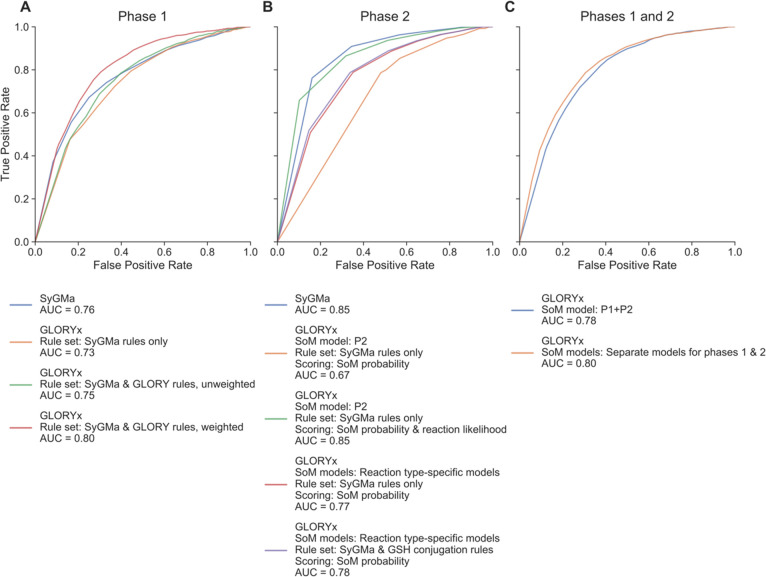
Given the same reaction rules, SyGMa with its reaction probability score-based ranking performed slightly better than our SoM probability-based ranking, with an AUC of 0.76 compared to 0.73, respectively, as shown in Figure 2A. This result is reasonable if we suppose that the Metabolite database, which was used to calculate the occurrence ratios for SyGMa’s reaction types, was so exhaustive even in its 2001 version (the version used to develop SyGMa) that it contained most of the contents of the current versions of DrugBank and MetXBioDB. This supposition is consistent with the observation in 2013 by Kirchmair et al. that nearly all of the Approved Drugs in DrugBank at the time (1341 out of 1391) were found in the 2011 version of the Metabolite database as top-level substrates.47 Unfortunately, without access to the Metabolite database, which is currently unavailable, we are unable to perform a comparison ourselves. Nevertheless, it appears that GLORYx achieves a comparable ranking performance to SyGMa.
Figure 2.
Rank-based ROC curves for the evaluation of metabolite prediction performance on the reference data set. The ranks are calculated based on the priority scores of the predicted metabolites for each parent molecule. (A) Comparison of GLORYx, which scores its predicted metabolites based on predicted SoM probabilities, to SyGMa, which uses reaction probability scores, for phase 1 metabolite prediction. Weighted rules refer to the weighting of the SoM probability-based score based on whether the reaction type is designated common or uncommon. (B) Comparison of the ranking performance of GLORYx with different scoring approaches and rule sets as well as direct comparison to SyGMa’s performance, for phase 2 metabolite prediction. The scoring approach that is based on both SoM probability and reaction probability is achieved by a simple multiplication of the two components. (C) Comparison of the ranking performance of GLORYx for combined prediction of metabolites for phases 1 and 2 metabolism, using different SoM prediction approaches to score the predicted metabolites. In both cases, the score is based on predicted SoM probability with weighting according to reaction type, and the rule set is made up of the final phase 1 rule set (SyGMa and GLORY rules) and final phase 2 rule set (SyGMa and GSH conjugation rules).
Phase 1 Metabolism: Combination of Reaction Rules from SyGMa and GLORY
For GLORY, we had developed a set of reaction rules specific to CYP-mediated metabolism.15 These reaction rules were manually created based on the scientific literature on CYP-mediated reaction types and mechanisms, and each reaction rule received a designation of either common or uncommon reaction type, also according to the literature. SyGMa’s phase 1 reaction rules are not separated into CYP and non-CYP rules, so it was of interest to determine whether adding these CYP-specific rules to the phase 1 rules sourced from SyGMa would result in any gains in performance for GLORYx.
When combining the rule sets, the overlap of the rules from the two different sources is handled in a straightforward manner. Duplicate metabolite predictions are combined by retaining the highest priority score. The addition of the CYP reaction rules from GLORY resulted in a substantial jump in recall (portion of known metabolites that were successfully predicted, also known as sensitivity) from 0.72 to 0.84 (Table 2). The precision (portion of predictions that match known metabolites), on the other hand, was halved, as the number of total predicted metabolites more than doubled, from over 10,000 to nearly 25,000. Note that only a fraction of the metabolites generated by organisms is experimentally observed and reported in the scientific literature and databases, for a number of reasons (e.g., lack of chemical stability, low concentrations of metabolites, limitations of the in vitro system, research interest focused on a specific metabolic enzyme or reaction or metabolite). Therefore, any predicted metabolites that are not “known” should more correctly be considered as putative false positive predictions. Nevertheless, the number of predicted metabolites is enormous, so it is crucial that metabolite prediction methods are able to rank their predictions in a meaningful way.
Table 2. Performance of GLORYx on Predicting Phase 1 Metabolites.
| GLORYx using reaction rules from both SyGMa and GLORY | GLORYx using reaction rules from SyGMa only | |
|---|---|---|
| recall | 0.84 | 0.72 |
| precision | 0.060 | 0.12 |
| total number of predictions | 24,906 | 10,550 |
| number of true positive predictions | 1487 | 1262 |
| AUC (rank-based) | 0.80 | 0.73 |
To examine the ranking performance of GLORYx using the combined rule set, we first used only the SoM probability to score and rank the predicted metabolites, as described above. This nonweighted scoring approach resulted in an AUC of 0.75 (Figure 2A), which was close to SyGMa’s AUC of 0.76. Note that even though the sets of predicted metabolites are different in this case, the ranking ability of each approach can still be compared using the ROC curves and AUC. We then applied the concept of weighting reaction rules that we first developed for GLORY, namely applying a simple common vs uncommon distinction between reaction types and generating the priority score for a predicted metabolite by multiplying the SoM probability by a factor corresponding to whether or not the reaction that led to that particular predicted metabolite was designated common (see ref (15) for details). The common–uncommon designations of the reaction rules from GLORY were used unchanged. Then we simply designated all of SyGMa’s phase 1 reaction rules as uncommon, based on the following logic: The CYP enzyme family is the most prevalent enzyme family involved in phase 1 metabolism,4 SyGMa’s phase 1 reaction rules contain rules for both CYP- and non-CYP-mediated reactions, and our process of combining duplicate predictions by keeping the highest score ensures that any CYP rules from SyGMa that are also “common” rules from GLORY will be in effect scored appropriately as being “common”. The result of this weighting of the rule sets was a jump in AUC to 0.80 (Figure 2A).
A similar trend in AUCs for GLORYx in terms of the weighting approach is observed when the ROC curves are calculated based on score rather than rank (Figure S2A). This means that predicted metabolites are compared across different parent molecules in the reference data set in terms of their priority scores. Here, it is important to note that the original publication of SyGMa implied that its score was only intended to be used to compare likelihoods of predicted metabolites of the same parent molecule, and the evaluation in that publication only considered the ranking per parent molecule.10 This consideration should be kept in mind when viewing all score-based ROC curves for SyGMa throughout this manuscript, which are included for the sake of completeness, especially since the score-based ROC curves for GLORYx tend to yield a higher AUC than the rank-based curves, yet the opposite is true for SyGMa (Figure S2 and Figure 2).
It is also relevant to note that the phase 1 subset of our reference data set is heavily biased toward CYP-mediated metabolism, with over 90% of the metabolites in the data set being CYP metabolites (Table 1). Although CYPs are widely considered the most relevant enzyme family for phase 1 human xenobiotic metabolism, the available data are perhaps even more skewed toward CYP data than would be realistic in humans. Due to the composition of this phase 1 reference data set, it is reasonable that the addition of the CYP-specific rules from GLORY leads to improved performance.
Phase 2 Metabolism
For phase 2 metabolite structure prediction, we again examined the question of how scoring the predicted metabolites based on the SoM probability predicted by FAME 3 compares to SyGMa’s scoring approach. Similarly to the phase 1 protocol, the initial comparison was carried out using only the phase 2 reaction rules from SyGMa, along with the general phase 2 SoM prediction model from FAME 3 (model P2), and scoring the predicted metabolites using only the SoM probability predicted by the SoM model. This comparison showed a large difference in ranking performance between SyGMa and our approach (Figure 2B). SyGMa achieved an AUC of 0.85, while our approach, which used the SoM probabilities predicted by the FAME 3 P2 model to rank the predicted metabolites, achieved an AUC of only 0.67.
It therefore appears that SoM probabilities are a surprisingly poor indication of the likelihood of phase 2 metabolism occurring. We know, however, that FAME 3 predicts SoMs corresponding to phase 2 metabolic reactions very well (AUC of 0.97 on a holdout data set consisting of 157 randomly selected compounds with a total of 3476 annotated atoms).17 The reason for this discrepancy is that multiple predicted metabolites, corresponding to different reaction types, receive the same score because they correspond to the same predicted SoM. Phase 2 metabolic reactions are more specific in terms of functional groups at which they can occur than, for example, CYP-mediated reactions, which makes it easier to predict SoMs but more difficult to predict which reaction type would be more likely to actually occur at a given location. To illustrate this point, consider the case of a hydroxyl group. A hydroxyl group that is a phase 2 SoM could be glucuronidated, sulfated, methylated, or phosphorylated. Another difficult case would be an amine group, which, if it is a phase 2 SoM, could be glucuronidated or N-acetylated. These observations combined with the poor ranking performance indicate that, so far, GLORYx struggles to discriminate between phase 2 reaction types.
In light of this observation and to further investigate the relationship between the predictive capabilities of SoM probabilities and reaction probabilities, we attempted to combine the two scores, since in theory both the SoM and the likelihood of a particular reaction rule compared to other reaction rules that could be applied at a given location are both relevant to the likelihood of the predicted metabolite. We tried two combination approaches: multiplying the reaction probability with the SoM probability and calculating a weighted average. Despite trying various weights (Table S4), a combination score was unable to do better than SyGMa’s reaction probability-based scoring approach alone at ranking the predictions. In addition, by varying the weights, it became clear that the more highly the predicted SoM probability was weighted compared to the reaction probability, the worse the ranking performance was (Table S4). The weighted average score combination, using weights up to 10:1, achieved a maximum rank-based AUC of 0.83 (Table S4), whereas multiplying the SoM probability by the reaction probability resulted in a rank-based AUC of 0.85 (Table S4, Figure 2B), which is the same as for SyGMa’s reaction probability score alone (however, the shape of the ROC curve is slightly different). These results indicated that the SoM probabilities predicted in this way could not compete with SyGMa’s reaction probability scores when it comes to ranking performance.
SyGMa’s good ranking performance was to be expected, for the same reasons discussed in the above section on phase 1 metabolism regarding the use of the Metabolite database to develop SyGMa’s reaction probability scores. Meanwhile, the poor showing by the SoM probability scoring approach indicates that reactivity is not sufficient to discriminate between the different types of phase 2 reactions, especially not when compared to the data-derived likelihoods of each reaction type. We therefore examined how we could use SoM prediction to achieve a distinction between different reaction types without resorting to precomputed occurrence ratios for the reaction rules.
Phase 2 Metabolism: Reaction Type-Specific SoM Prediction Models
In order to attempt to better predict which reaction type would be more likely at a given SoM, without using SyGMa’s reaction probabilities, we developed FAME 3 reaction type-specific SoM prediction models that roughly correspond to the five main phase 2 enzyme families: UGTs, GSTs, SULTs, MTs, and NATs. These models were created using the same training protocol as for the previously published FAME 3 models. Each model was trained on only a subset of the FAME 3 data set, whereby the subsets were selected based on the reaction class annotation in the MetaQSAR database. The reaction classes and the number of molecules used to train each model are provided in Table S3. The 10-fold cross-validation performance of these models was high across the board, with average AUCs all above 0.95 and the average percentage of molecules in which a correct SoM was predicted among the top two atom positions with the highest SoM probabilities (top 2 metric) all above 0.87 (Table 3), despite the relatively small number of molecules used for training in each case. Note, however, that these models are trained on atoms, not molecules, so the number of training instances (although not entirely independent from each other) is much larger than the number of molecules.
Table 3. Average SoM Prediction Performance of the FAME 3 Reaction Class-Specific Models During Cross-Validation.
| reaction class | average top 2 | average AUC |
|---|---|---|
| glucuronidations and glycosylations | 0.957 | 0.988 |
| GSH and RSHa conjugations | 0.874 | 0.950 |
| sulfonations | 0.966 | 0.992 |
| methylations | 0.877 | 0.968 |
| acetylations and acylations | 0.956 | 0.992 |
RSH = protein thiol.
Because these reaction type-specific SoM models were each trained on only a subset of the molecules that were used to train the general phase 2 SoM model, not all atom types (i.e., Sybyl atom types) were represented in the training data for each individual model, which can then not make predictions for molecules containing these unrepresented atom types. Therefore, these individual reaction type-specific SoM models were used to overrule the predicted SoM probabilities from the general P2 model for the molecules to which they apply rather than as a complete substitute for the general model.
There are a few phase 2 reaction rules in SyGMa that do not correspond to any of the five main phase 2 enzyme families. These rules are simply designated “uncommon”, while all other phase 2 reaction rules are designated “common”, and the general P2 SoM prediction model is always used to score the products of these uncommon reaction rules.
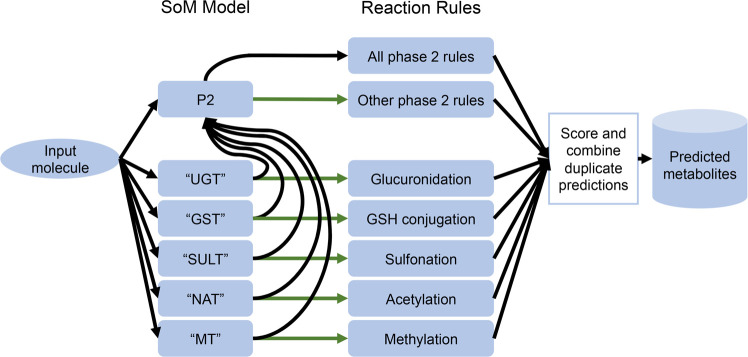
The general P2 model is also used to score the predicted metabolites corresponding to the individual reaction type-specific SoM models that can not make predictions for a given input molecule. For example, if the SoM model for sulfonation reactions could not make predictions, the predicted metabolites resulting from sulfonation reaction rules are scored using the predicted SoM probabilities from the general P2 model. An illustration of the workflow for predicting phase 2 metabolites using the reaction type-specific SoM models for scoring is shown in Figure 3.
Figure 3.
Workflow of phase 2 metabolite prediction using reaction type-specific SoM models to score and rank the predicted metabolites. The reaction type-specific SoM models (“UGT”, “GST”, “SULT”, “NAT”, “MT”) are used instead of the general phase 2 SoM model (P2) to score the products of the relevant reactions for all molecules in which all of the reaction type-specific models are able to make a prediction. The green arrows indicate the molecules that were predicted successfully by the relevant reaction type-specific SoM model. If one or more of the reaction type-specific models cannot make predictions for a given molecule, then that molecule additionally follows the path of the black arrows, followed by a deduplication of predictions. The “UGT” model covers glucuronidation and glycosylation reactions, the “GST” model covers GSH and RSH conjugations, the “SULT” model covers sulfonations, the “NAT” model covers acetylations and acylations, and the “MT” model covers methylation reactions. The “other phase 2 rules” refer to the rules that are neither glucuronidation, GSH conjugation, sulfonation, acetylation, or methylation rules.
In this way, the same metabolites are predicted as if only the general P2 model was used, but the reaction type-specific scoring approach results in different ranks of the metabolites and, perhaps most importantly, a drastic reduction in the number of tied ranks for predicted metabolites of a single parent molecule.
Using the individual reaction type-specific phase 2 SoM models to score the predicted metabolites resulted in a large improvement in the ranking, with an AUC of 0.77 compared to an AUC of 0.67 using the general P2 model (Figure 2B) and only the reaction rules sourced from SyGMa for comparison. Similarly, the score-based AUC increased from 0.66 to 0.79 upon implementation of the reaction type-specific SoM models (Figure S2). Unfortunately, even using the reaction type-specific SoM prediction models resulted in a ranking performance that was worse than SyGMa’s (AUC of 0.77 compared to 0.85, respectively). However, as discussed for phase 1 metabolism above, SyGMa’s approach has the advantage of having derived its scoring approach directly based on, in effect, all available metabolism data from a comprehensive but not freely available database. Meanwhile, the difficulty of using SoM prediction for phase 2 metabolism appears to be that there are relatively few potential SoMs, but that the atom environments may not be specific enough to differentiate between different types of reactions.
Based on these results, we therefore use the individual reaction type-specific phase 2 SoM models to score the phase 2 metabolites predicted in all subsequent sections of this manuscript.
Phase 2 Metabolism: Addition of GSH Conjugation Reaction Rules
We found that the reaction rules sourced from SyGMa do not contain any GSH conjugation reactions, which correspond to the GST enzyme family, one of the five main enzyme families for phase 2 xenobiotic metabolism. We therefore developed a set of GSH conjugation reaction rules based on descriptions of GSH conjugation metabolic reactions in the scientific literature. This resulted in nine new reaction rules.
When only the reaction rules sourced from SyGMa were implemented, GLORYx achieved a recall of 0.78 and a precision of 0.21 (Table 4). When the new GSH conjugation reaction rules were added, GLORYx achieved a recall of 0.80 with the same precision, because only 141 more metabolites were predicted in total (Table 4). Though we do not see a very large improvement in performance on the reference data set after adding these GSH conjugation reaction rules to GLORYx, we believe that this addition is actually meaningful for the purpose of metabolite structure prediction in the real world, because GSTs are actually the second most relevant phase 2 enzyme family for xenobiotic metabolism in terms of number of metabolites formed.4
Table 4. Performance of GLORYx on Predicting Phase 2 Metabolites.
| GLORYx using SyGMa rules only | GLORYx using SyGMa rules plus GSH conjugation rules | |
|---|---|---|
| recall | 0.78 | 0.80 |
| precision | 0.21 | 0.21 |
| total number of predictions | 2509 | 2650 |
| number of true positive predictions | 539 | 555 |
| AUC (rank-based) | 0.77 | 0.78 |
As was expected based on the relatively low number of GST-mediated metabolites in the reference data set (Table 1), the ranking performance remained similar upon the addition of GSH conjugation reaction rules (AUC of 0.77 and 0.78, respectively; Figure 2B). This comparable performance seems to suggest that the products of the new GSH conjugation reaction rules are scored in a meaningful way based on the corresponding reaction type-specific SoM prediction model.
Combined Phase 1 and Phase 2
A general use case of predicting “all” possible metabolites at once was also considered. FAME 3 provides one model, “P1 + P2”, that predicts all SoMs from both phases of metabolism. For this use case, we therefore examined whether it makes sense to use the P1 + P2 FAME 3 model’s predictions to score the predicted metabolites or to use the separate models, as determined separately for phase 1 and phase 2 (see sections Phase 1 Metabolism and Phase 2 Metabolism), and combine the predictions. The predicted metabolite structures are the same in both cases; what changes is their scores, since those are based on the predicted SoM probabilities.
Using separate SoM prediction models for the two phases did provide a slight advantage in terms of the ranking performance, with an improvement in AUC from 0.78 to 0.80 compared to using the P1 + P2 SoM model, as shown in Figure 2C. An improvement of the same amount is seen in the AUCs of the score-based ROC curves (AUC increased from 0.79 to 0.81; Figure S2C). Although this advantage appears small at first glance, it is important to recall the composition of the reference data set. This data set contains more than twice as much phase 1 data as phase 2 data, in terms of number of known metabolites, which may cause the benefit of using separate SoM prediction models for the two phases to be underrepresented by this analysis. Based on these considerations along with the ROC curves, we conclude that the multimodel approach should be used for optimal performance, and we use this approach in the validation on the test data set (see section Performance on a Manually Curated Test Data Set).
Performance on a Manually Curated Test Data Set
The performance of the final version of GLORYx was evaluated on a manually curated test data set consisting of 37 parent molecules that were among the top 100 best-selling drugs in 2018. For these parent molecules, the data set contains a total of 136 first-generation metabolites, which equates to an average of 3.7 known metabolites per parent molecule. This test data set does not contain enzyme or metabolism phase annotations, so the evaluation was carried out from the perspective of predicting all possible metabolites, from both phase 1 and phase 2 metabolism.
GLORYx was able to predict 77% of the known metabolites in the test data set, which is higher than SyGMa’s recall of 68% (Table 5). In conjunction with this higher recall, GLORYx had a lower precision than SyGMa (0.061 compared to 0.12, respectively), which is unsurprising given that GLORYx contains many more reaction rules than SyGMa due to the addition of the CYP metabolism rules from GLORY and the new GSH conjugation rules. The total number of metabolites predicted by GLORYx was nearly double the number predicted by SyGMa. However, SyGMa’s precision of 0.12 was also very low, due to a relatively large number of predictions (800 total). Another potential contribution to the low precision of both tools is that experimentally determined metabolites whose structures have not been fully defined were not included in the test data set. It is possible that this aspect of the data set has an effect on the number of false positive predictions, which would have an effect on the precision as well.
Table 5. Performance of GLORYx and SyGMa on the Test Data Set of 37 Parent Compounds and Their 136 Metabolites.
| GLORYx | SyGMa | |
|---|---|---|
| recall | 0.77 | 0.68 |
| precision | 0.061 | 0.12 |
| total number of predictions | 1724 | 800 |
| number of true positives (out of 136) | 105 | 93 |
| AUC (rank-based) | 0.79 | 0.74 |
The relatively large number of predictions made by both SyGMa and GLORYx is a general problem that is shared by all available metabolite structure prediction approaches.48 This phenomenon clearly underlines the need to have a meaningful way to rank the predicted metabolites. In our case in particular, neither SyGMa nor GLORYx has sufficiently high precision to be used without ranking the predicted metabolites.
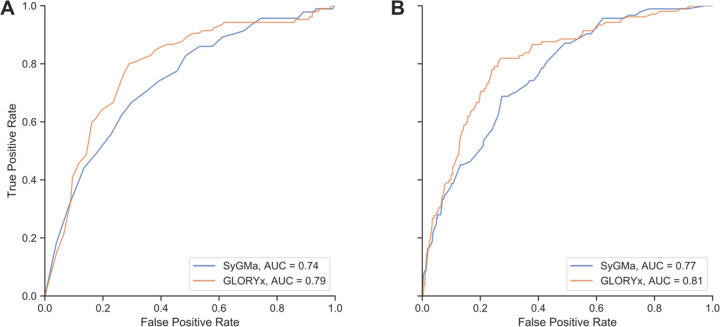
In terms of the ability to rank the predicted metabolites, GLORYx showed better performance than SyGMa, as indicated by the ROC curves shown in Figure 4. The AUC of the rank-based ROC curve was 0.79, compared to 0.74 for SyGMa. In addition, GLORYx’s priority score seems to be a meaningful score in and of itself, not just for ranking the predictions for individual parent molecules separately, because the ROC curve and AUC were actually slightly better using the score than they were using the rank (0.81 compared to 0.79, respectively; Figure 4). For SyGMa, the AUC of the score-based approach was also higher than that of the rank-based approach (0.77 compared to 0.74); however, it is important to note that SyGMa’s score was most likely not intended to be used to compare predicted metabolites from different parent compounds (see section Phase 1 Metabolism: Combination of Reaction Rules from SyGMa and GLORY).
Figure 4.
ROC curves for GLORYx and SyGMa representing ranking performance on the test set based on the (A) ranks and (B) scores of the predicted metabolites.
To get an idea of the variability in the ranking performance on the test data set, we calculated the AUCs while systematically removing one parent molecule at a time from the data set. This resulted in 37 different AUCs for each tool and AUC type (rank-based or score-based), which are plotted in Figure S3. From this analysis, we observed a similar amount of variability in the AUCs between the two tools with the different metrics (score based and rank based). In all cases, the median AUCs from this analysis were within 0.01 of the AUCs reported in Figure 4. From the outliers observed in Figure S3, we learn that two to three molecules are particularly challenging for each tool. In the case of GLORYx, the two outliers correspond to having excluded everolimus (the furthest outlier) and budesonide for both the score-based and rank-based AUCs. For SyGMa, the furthest outlier is also caused by the exclusion of everolimus, while the second-furthest outlier corresponds to having excluded darunavir. Everolimus is a macrocycle with 12 known metabolites in the test data set, while budesonide and darunavir each have 6 known metabolites.
Overall, a clear difference in performance is observed between the two tools, with GLORYx outperforming SyGMa in both cases. The improvement in ranking performance seems to indicate that combining predicted SoM probabilities with reaction rules to score the predicted metabolites, whereby the SoM model and the reaction rules correspond to the same type(s) of reactions, provides very valuable information. This approach also has the benefit of not relying on reaction rule occurrence ratios based on existing metabolism data to score and rank the predictions. Our reference data set was used to measure performance during development but was not used to develop reaction rules or calculate occurrence ratios. This difference could potentially make GLORYx more flexible with regard to never-before-seen input molecules.
Conclusion
GLORYx is a new tool for predicting the structures of metabolites formed by both phase 1 and phase 2 metabolic reactions in humans. The tool utilizes FAME 3 to predict, for all atom positions in a molecule, the likelihood of a biotransformation to take place at this position and, based on these predictions, applies a set of reaction rules to generate and rank likely metabolites.
In conjunction with a high recall of known metabolites (77% on the test data set), GLORYx ranked the predicted metabolites with an AUC of 0.79 on the manually curated test data set. This recall and ranking performance is better than we observed for the established, freely available tool SyGMa on the same data set. However, when considering only phase 2 metabolite prediction, SyGMa’s ranking performance was better than that of GLORYx.
We have observed that it is difficult to predict phase 2 metabolites, that is, difficult to rank the predicted metabolites in a meaningful way, based on predicted SoM probabilities despite high performance of the SoM prediction models themselves. We have concluded that the cause of this difficulty is that reactivity is an insufficient metric for determining which type of conjugation reaction would be more likely to occur at a particular atom position. We were able to mitigate this problem substantially by using individual reaction type-specific SoM prediction models corresponding roughly to the five main phase 2 enzyme families.
During each run of GLORYx, the algorithm generates and ranks one generation of metabolites based on the parent compound(s) provided. Users may of course provide (predicted) metabolites as input to GLORYx, hence enabling multigeneration metabolite prediction.
Given the scarcity of the available high-quality data on small-molecule metabolism, it is difficult to provide a robust definition of the applicability domain of GLORYx. However, we know from thorough analyses of FAME 3 that the metabolic properties of the atoms in a molecule are first and foremost determined by the proximate atom environment, and these environments are much more redundant across the chemical space than the overall (global) structure of molecules. Considering also that the reaction rules implemented in GLORYx are based on only a few connected atoms, GLORYx is expected to provide reliable results for a wide range of synthetic compounds and natural products alike.
GLORYx is freely available as a web server at https://nerdd.zbh.uni-hamburg.de/ and is also provided as a software package upon request. Note that GLORYx should be considered an extension of GLORY rather than a replacement. Hence both tools are available on the Web site, to enable users to choose between CYP-specific metabolite structure prediction with GLORY and comprehensive phase 1 and phase 2 metabolite structure prediction with GLORYx.
Acknowledgments
We would like to thank Nina Jeliazkova and Nikolay Kochev from Idea Consult, Ltd. of Sofia, Bulgaria for discussions about using Ambit-SMIRKS for metabolite structure prediction. We also thank Conrad Stork from the Center of Bioinformatics (ZBH) of the Universität Hamburg for technical support with the Web service. We further acknowledge the authors of BioTransformer for their efforts in assembling the MetXBioDB, which we used as one of the data sources for the compilation of our reference dataset.
Glossary
Abbreviations
- AUC
area under the receiver operating characteristic curve
- clog P
calculated log P
- CYP
cytochrome P450
- EC
Enzyme Commission
- GSH
glutathione
- GST
glutathione S-transferase
- MT
methyltransferase
- NAT
N-acetyltransferase
- ROC
receiver operating characteristic
- SoM
site of metabolism
- SULT
sulfotransferase
- UGT
UDP-glucuronosyltransferase
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.chemrestox.0c00224.
Additional tables and figures (PDF)
C.d.B.K. is funded by a Ph.D. completion scholarship from the Universität Hamburg. M.S. was supported from the Ministry of Education, Youth and Sports of the Czech Republic, project number LM2018130.
The authors declare no competing financial interest.
Notes
The reference and test data sets as well as all the reaction rules from this work are freely available at https://github.com/christinadebruynkops/GLORYx.
Supplementary Material
References
- Kirchmair J.; Göller A. H.; Lang D.; Kunze J.; Testa B.; Wilson I. D.; Glen R. C.; Schneider G. (2015) Predicting Drug Metabolism: Experiment and/or Computation?. Nat. Rev. Drug Discovery 14 (6), 387–404. 10.1038/nrd4581. [DOI] [PubMed] [Google Scholar]
- Pedretti A., Mazzolari A., Vistoli G., and Testa B. (2020) MetaQSAR Database, version MetaQSAR_05_2020.
- Pedretti A.; Mazzolari A.; Vistoli G.; Testa B. (2018) MetaQSAR: An Integrated Database Engine to Manage and Analyze Metabolic Data. J. Med. Chem. 61 (3), 1019–1030. 10.1021/acs.jmedchem.7b01473. [DOI] [PubMed] [Google Scholar]
- Testa B.; Pedretti A.; Vistoli G. (2012) Reactions and Enzymes in the Metabolism of Drugs and Other Xenobiotics. Drug Discovery Today 17 (11–12), 549–560. 10.1016/j.drudis.2012.01.017. [DOI] [PubMed] [Google Scholar]
- Jancova P.; Anzenbacher P.; Anzenbacherova E. (2010) Phase II Drug Metabolizing Enzymes. Biomed. Pap. 154 (2), 103–116. 10.5507/bp.2010.017. [DOI] [PubMed] [Google Scholar]
- Kirchmair J.; Williamson M. J.; Tyzack J. D.; Tan L.; Bond P. J.; Bender A.; Glen R. C. (2012) Computational Prediction of Metabolism: Sites, Products, SAR, P450 Enzyme Dynamics, and Mechanisms. J. Chem. Inf. Model. 52 (3), 617–648. 10.1021/ci200542m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant C. A.; Briggs K. A.; Long A. (2008) In Silico Tools for Sharing Data and Knowledge on Toxicity and Metabolism: Derek for Windows, Meteor, and Vitic. Toxicol. Mech. Methods 18 (2–3), 177–187. 10.1080/15376510701857320. [DOI] [PubMed] [Google Scholar]
- Mekenyan O. G.; Dimitrov S. D.; Pavlov T. S.; Veith G. D. (2004) A Systematic Approach to Simulating Metabolism in Computational Toxicology. I. The TIMES Heuristic Modelling Framework. Curr. Pharm. Des. 10 (11), 1273–1293. 10.2174/1381612043452596. [DOI] [PubMed] [Google Scholar]
- Darvas F. (1987) Metabolexpert: An Expert System for Predicting Metabolism of Substances. In QSAR in Environmental Toxicology - II (Kaiser K. L., Ed.), pp 71–81, Springer, Dordrecht. [Google Scholar]
- Ridder L.; Wagener M. (2008) SyGMa: Combining Expert Knowledge and Empirical Scoring in the Prediction of Metabolites. ChemMedChem 3 (5), 821–832. 10.1002/cmdc.200700312. [DOI] [PubMed] [Google Scholar]
- (2001) MDL Metabolite Database, Elsevier, Amsterdam.
- Djoumbou-Feunang Y.; Fiamoncini J.; Gil-de-la-Fuente A.; Greiner R.; Manach C.; Wishart D. S. (2019) BioTransformer: A Comprehensive Computational Tool for Small Molecule Metabolism Prediction and Metabolite Identification. J. Cheminf. 11 (1), 2. 10.1186/s13321-018-0324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudik A. V.; Bezhentsev V. M.; Dmitriev A. V.; Druzhilovskiy D. S.; Lagunin A. A.; Filimonov D. A.; Poroikov V. V. (2017) MetaTox: Web Application for Predicting Structure and Toxicity of Xenobiotics’ Metabolites. J. Chem. Inf. Model. 57 (4), 638–642. 10.1021/acs.jcim.6b00662. [DOI] [PubMed] [Google Scholar]
- Rudik A.; Bezhentsev V.; Dmitriev A.; Lagunin A.; Filimonov D.; Poroikov V. (2019) Metatox - Web Application for Generation of Metabolic Pathways and Toxicity Estimation. J. Bioinf. Comput. Biol. 17 (1), 1940001. 10.1142/S0219720019400018. [DOI] [PubMed] [Google Scholar]
- de Bruyn Kops C.; Stork C.; Šícho M.; Kochev N.; Svozil D.; Jeliazkova N.; Kirchmair J. (2019) GLORY: Generator of the Structures of Likely Cytochrome P450 Metabolites Based on Predicted Sites of Metabolism. Front. Chem. 7, 402. 10.3389/fchem.2019.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šícho M.; de Bruyn Kops C.; Stork C.; Svozil D.; Kirchmair J. (2017) FAME 2: Simple and Effective Machine Learning Model of Cytochrome P450 Regioselectivity. J. Chem. Inf. Model. 57 (8), 1832–1846. 10.1021/acs.jcim.7b00250. [DOI] [PubMed] [Google Scholar]
- Šícho M.; Stork C.; Mazzolari A.; de Bruyn Kops C.; Pedretti A.; Testa B.; Vistoli G.; Svozil D.; Kirchmair J. (2019) FAME 3: Predicting the Sites of Metabolism in Synthetic Compounds and Natural Products for Phase 1 and Phase 2 Metabolic Enzymes. J. Chem. Inf. Model. 59 (8), 3400–3412. 10.1021/acs.jcim.9b00376. [DOI] [PubMed] [Google Scholar]
- ADMET Predictor Metabolism Module, SimulationsPlus. Simulations-Plus.com/software/admetpredictor/metabolism/ (accessed 2020/05/07).
- Tyzack J. D.; Hunt P. A.; Segall M. D. (2016) Predicting Regioselectivity and Lability of Cytochrome P450 Metabolism Using Quantum Mechanical Simulations. J. Chem. Inf. Model. 56 (11), 2180–2193. 10.1021/acs.jcim.6b00233. [DOI] [PubMed] [Google Scholar]
- Cruciani G.; Carosati E.; De Boeck B.; Ethirajulu K.; Mackie C.; Howe T.; Vianello R. (2005) MetaSite: Understanding Metabolism in Human Cytochromes from the Perspective of the Chemist. J. Med. Chem. 48 (22), 6970–6979. 10.1021/jm050529c. [DOI] [PubMed] [Google Scholar]
- DrugBank, version 5.1.4. https://www.drugbank.ca/ (accessed 2019/09/26).
- Wishart D. S.; Feunang Y. D.; Guo A. C.; Lo E. J.; Marcu A.; Grant J. R.; Sajed T.; Johnson D.; Li C.; Sayeeda Z.; Assempour N.; Iynkkaran I.; Liu Y.; Maciejewski A.; Gale N.; Wilson A.; Chin L.; Cummings R.; Le D.; Pon A.; Knox C.; Wilson M. (2018) DrugBank 5.0: A Major Update to the DrugBank Database for 2018. Nucleic Acids Res. 46 (D1), D1074–D1082. 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MetXBioDB, version 1.0, https://bitbucket.org/djoumbou/biotransformer/src/master/ (accessed 2019/01/11).
- Top 200 Brand Name Drugs by Retail Sales in 2018, The Njardarson Group at the University of Arizona, Tucson, AZ.https://njardarson.lab.arizona.edu/content/top-Pharmaceuticals-Poster (accessed 2019/09/12).
- McGrath N. A.; Brichacek M.; Njardarson J. T. (2010) A Graphical Journey of Innovative Organic Architectures That Have Improved Our Lives. J. Chem. Educ. 87 (12), 1348–1349. 10.1021/ed1003806. [DOI] [Google Scholar]
- Gaulton A.; Hersey A.; Nowotka M.; Bento A. P.; Chambers J.; Mendez D.; Mutowo P.; Atkinson F.; Bellis L. J.; Cibrián-Uhalte E.; Davies M.; Dedman N.; Karlsson A.; Magariños M. P.; Overington J. P.; Papadatos G.; Smit I.; Leach A. R. (2017) The ChEMBL Database in 2017. Nucleic Acids Res. 45 (D1), D945–D954. 10.1093/nar/gkw1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ChEMBL Database. https://www.ebi.ac.uk/chembl/ (accessed 2020/02/21).
- (2017) MarvinSketch, version 17.1.9, ChemAxon, Budapest, Hungary. http://www.chemaxon.com (accessed 2017/10/01).
- (2018) RDKit: Open-Source Cheminformatics, version 2018.09.1.
- Pedregosa F.; Varoquaux G.; Gramfort A.; Michel V.; Thirion B.; Grisel O.; Blondel M.; Prettenhofer P.; Weiss R.; Dubourg V.; Vanderplas J.; Passos A.; Cournapeau D.; Brucher M.; Perrot M.; Duchesnay E. (2011) Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 12, 2825–2830. [Google Scholar]
- Scikit-Learn: Machine Learning in Python, version 0.20.1.
- (2018) Molecular Operating Environment (MOE), version 2018.01, Chemical Computing Group, Montreal, QC, Canada.
- SMIRKS - A Reaction Transform Language, Daylight, Laguna Niguel, CA. https://www.daylight.com/dayhtml/doc/theory/theory.smirks.html (accessed 2020/05/08).
- SyGMa, version 1.1.0.
- Allocati N.; Masulli M.; Di Ilio C.; Federici L. (2018) Glutathione Transferases: Substrates, Inhibitors and pro-Drugs in Cancer and Neurodegenerative Diseases. Oncogenesis 7 (1), 8. 10.1038/s41389-017-0025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bladeren P. J. (2000) Glutathione Conjugation as a Bioactivation Reaction. Chem.-Biol. Interact. 129 (1–2), 61–76. 10.1016/S0009-2797(00)00214-3. [DOI] [PubMed] [Google Scholar]
- Awasthi Y. C. (2006) Toxicology of Glutathione Transferases, CRC Press, Boca Raton, FL. [Google Scholar]
- Armstrong R. N. (1991) Glutathione S-Transferases: Reaction Mechanism, Structure, and Function. Chem. Res. Toxicol. 4 (2), 131–140. 10.1021/tx00020a001. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; den Braver-Sewradj S. P.; den Braver M. W.; Hiemstra S.; Vermeulen N. P. E.; van de Water B.; Commandeur J. N. M.; Vos J. C. (2018) Glutathione S-Transferase P1 Protects Against Amodiaquine Quinoneimines-Induced Cytotoxicity but Does Not Prevent Activation of Endoplasmic Reticulum Stress in HepG2 Cells. Front. Pharmacol. 9, 388. 10.3389/fphar.2018.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFaul P. A.; Morley A. D.; Crawford J. J. (2009) A Simple in Vitro Assay for Assessing the Reactivity of Nitrile Containing Compounds. Bioorg. Med. Chem. Lett. 19 (4), 1136–1138. 10.1016/j.bmcl.2008.12.105. [DOI] [PubMed] [Google Scholar]
- Willighagen E. L.; Mayfield J. W.; Alvarsson J.; Berg A.; Carlsson L.; Jeliazkova N.; Kuhn S.; Pluskal T.; Rojas-Chertó M.; Spjuth O.; Torrance G.; Evelo C. T.; Guha R.; Steinbeck C. (2017) The Chemistry Development Kit (CDK) v2.0: Atom Typing, Depiction, Molecular Formulas, and Substructure Searching. J. Cheminf. 9 (1), 33. 10.1186/s13321-017-0220-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemistry Development Kit, version 2.0.
- Pedretti A., Mazzolari A., Vistoli G., and Testa B. (2018) MetaQSAR Database (snapshot from March 15, 2018).
- Ambit-SMARTS Java Library, version 3.1.0.
- Kochev N.; Avramova S.; Jeliazkova N. (2018) Ambit-SMIRKS: A Software Module for Reaction Representation, Reaction Search and Structure Transformation. J. Cheminf. 10 (1), 42. 10.1186/s13321-018-0295-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich F. P. (2001) Common and Uncommon Cytochrome P450 Reactions Related to Metabolism and Chemical Toxicity. Chem. Res. Toxicol. 14 (6), 611–650. 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- Kirchmair J.; Howlett A.; Peironcely J. E.; Murrell D. S.; Williamson M. J.; Adams S. E.; Hankemeier T.; van Buren L.; Duchateau G.; Klaffke W.; Glen R. C. (2013) How Do Metabolites Differ from Their Parent Molecules and How Are They Excreted?. J. Chem. Inf. Model. 53 (2), 354–367. 10.1021/ci300487z. [DOI] [PubMed] [Google Scholar]
- Judson P. N. (2014) Knowledge-Based Approaches for Predicting the Sites and Products of Metabolism. In Drug Metabolism Prediction; Kirchmair J., Ed.; Wiley-VCH, Weinheim, Germany; pp 293−318. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- (2001) MDL Metabolite Database, Elsevier, Amsterdam.
- DrugBank, version 5.1.4. https://www.drugbank.ca/ (accessed 2019/09/26).
- MetXBioDB, version 1.0, https://bitbucket.org/djoumbou/biotransformer/src/master/ (accessed 2019/01/11).
- ChEMBL Database. https://www.ebi.ac.uk/chembl/ (accessed 2020/02/21).