Abstract
Plant biostimulants are compounds, living microorganisms, or their constituent parts that alter plant development programs. The impact of biostimulants is manifested in several ways: via morphological, physiological, biochemical, epigenomic, proteomic, and transcriptomic changes. For each of these, a response and alteration occur, and these alterations in turn improve metabolic and adaptive performance in the environment. Many studies have been conducted on the effects of different biotic and abiotic stimulants on plants, including many crop species. However, as far as we know, there are no reviews available that describe the impact of biostimulants for a specific field such as transcriptomics, which is the objective of this review. For the commercial registration process of products for agricultural use, it is necessary to distinguish the specific impact of biostimulants from that of other legal categories of products used in agriculture, such as fertilizers and plant hormones. For the chemical or biological classification of biostimulants, the classification is seen as a complex issue, given the great diversity of compounds and organisms that cause biostimulation. However, with an approach focused on the impact on a particular field such as transcriptomics, it is perhaps possible to obtain a criterion that allows biostimulants to be grouped considering their effects on living systems, as well as the overlap of the impact on metabolism, physiology, and morphology occurring between fertilizers, hormones, and biostimulants.
Keywords: gene expression, PGPRs, macroalgae, peptides, humic acid, chitosan, selenium, silicon
Introduction
Biostimulation has been described as a general biological phenomenon dependent on the interactions between the cell’s molecular structures and the external physical, chemical and biological stimuli, or impulses. The biostimulation results in the alteration of metabolic processes that allows the most efficient use of environmental resources, substantially increased growth or yield, and increased tolerance to adverse environmental factors (Juárez-Maldonado et al., 2019). It would be expected that starting from the definition of biostimulation that the definition of biostimulant would be derived; but in reality, that of biostimulant preceded that of biostimulation as a result of the need for regulation in the agricultural sector, which has found a very relevant niche in biostimulants (du Jardin, 2015).
Indeed, at present, biostimulants have gained importance in agriculture from ecological and commercial perspectives. Biostimulants are labeled as ecologically benign since most are considered biodegradable, non-toxic, non-polluting, and non-hazardous for various organisms (Yakhin et al., 2017).
Currently, there is no agreed-upon legal or academic definition of biostimulant; thus, there are several definitions of this term. One academic definition of a plant biostimulant refers to any substance or microorganism applied to plants to improve nutrient use efficiency, stress tolerance, and/or quality traits of plants, regardless of their nutrient content (du Jardin, 2015). Another definition defines biostimulants as materials that, when applied in small amounts, are capable of promoting growth in plants (Juárez-Maldonado et al., 2019).
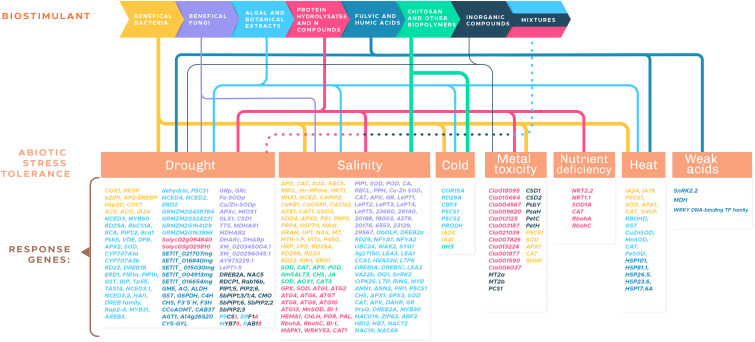
It should be noted that, from the biological point of view, biostimulants are all those biotic and abiotic factors that induce biostimulation (including physical factors such as UV radiation, physicochemical factors such as nanomaterials, and biological factors such as pests, pathogens, competitors, and beneficial organisms and microorganisms) (Juárez-Maldonado et al., 2019). However, for purposes of developing this review with a focus in agriculture, we start from the biostimulants definition and categorization developed by du Jardin (2015). In his paper, the author classified biostimulants into seven groups: (1) beneficial bacteria, (2) beneficial fungi, (3) algae and botanical extracts, (4) protein hydrolysates and other nitrogen-containing compounds, (5) humic acid and fulvic acid, (6) chitosan and other biopolymers, and (7) inorganic compounds.
Currently, the advancement of omics sciences through the development of genome sequencing technologies, as well as the reduction in the costs of these techniques, have revolutionized the understanding of the identification of metabolic pathways in plants (Owen et al., 2017). The review is based on the supposition that through transcriptomic studies, it is possible to identify molecular markers (Harper et al., 2016) that are associated with common responses in a wide range of crop species; these molecular markers can, in turn, be used to understand the mode of action of biostimulants, their interaction with the environment, and the genotype of plants (De Palma et al., 2019). The present review contributes to the description, in terms of transcriptomics, of the impact of the different categories of biostimulants on agricultural crop species, with an emphasis on corn, rice, wheat, tomato, and Arabidopsis. It is intended to describe the transcriptomic landscape induced by biostimulants in plants under abiotic stress. Abiotic stress, such as salinity, drought or temperature variation, can decrease productivity and cause considerable losses in crop yields by more than 60% (Singhal et al., 2016). The following sections are included in this manuscript: (1) beneficial bacteria; (2) beneficial fungi; (3) algae and botanical extracts; (4) protein hydrolysates and other nitrogen-containing compounds; (5) humic acid and fulvic acid; (6) chitosan and other biopolymers; and (7) inorganic compounds. In each section, we present works that describe the genes that whose transcript level changes in plants in response to the application of a biostimulant (in a specific category) in plants under abiotic stress. In total, 56 recent works (published from 2010 to the present) from the seven biostimulant category groups were included.
Beneficial Bacteria
Microorganisms are widely used to produce biostimulants (Xavier and Boyetchko, 2002; Sofo et al., 2014; Colla et al., 2015; Matyjaszczyk, 2015). Biostimulants based on microorganisms are preparations that include living and/or non-living microorganisms and their metabolites. Within this category, the most studied microorganisms are those defined as plant growth-promoting bacteria (PGPBs) (Calvo et al., 2014). Among the PGPBs, the genres most studied for use as biostimulants include Bacillus (Adesemoye et al., 2010), Azospirillum, Pseudomonas, Streptomyces, Achromobacter, and Rhizobium (Calvo et al., 2014). The plant-promoting effects of PGPBs are mostly explained by the release of metabolites directly stimulating growth. The direct mechanism includes those in which the bacteria produce growth regulators [auxins, gibberellins, cytokinins, ethylene, and abscisic acid (ABA)] that are ultimately incorporated into the plant system and thus affect the balance of plant growth regulators, or they act as a sink for plant-released hormones that induce changes in plant metabolism, promoting the overall growth of the plant (Govindasamy et al., 2010). The indirect mechanisms include biological nitrogen fixation, phosphate solubilization, production of siderophores, antibiotics, and other metabolites, production of defense enzymes and modulation of plant stress markers, induction of systemic resistance (ISR) and competition within the rhizosphere (Kumar et al., 2020).
Genes and their function are described below using the results of 10 studies in which beneficial bacteria-based biostimulants were applied to plants under abiotic stress (Table 1).
TABLE 1.
Description of genes whose transcript level changed in response to the application of bacteria-based biostimulants in plants under abiotic stress.
| Gene/gene expression time | Biostimulant type | Abiotic stress type | Crop/gene expression organ | References |
| COX1, PKDP, bZIP1, AP2-EREBP, Hsp20, COC1/65 days old | Pseudomonas fluorescens | Drought | Rice/leaves | Saakre et al., 2017 |
| ACS, AC/45 days after stress | Ochrobactrum pseudogrignonense, Pseudomonas sp., and Bacillus subtilis | Drought | Vigna mungo and Pisum sativum L./leaves and roots | Saikia et al., 2018 |
| OsNAM, OsGRAM/not specified | Bacillus amyloliquefaciens | Salinity | Rice/roots | Chauhan et al., 2019 |
| UP1, NAS, MT, MTR-1-P, VITs, P450, HRP, UP2/not specified | Arthrobacter nitroguajacolicus | Salinity | Wheat/roots | Safdarian et al., 2019 |
| RD29A, RD29B, RD20, RD22, KIN1, ERD1/not specified | Bacillus oryzicola | Salinity | Arabidopsis/shoots and roots | Baek et al., 2020 |
| IA24, IA41, IA19/after 6 h at heat stress, after 6 h at cold stress, after 5 days without water | Bacillus amyloliquefaciens subsp. plantarum | Heat, cold, drought | Wheat/leaves | Abd El-Daim et al., 2018 |
| APX, CAT, SOD, RBCS, RBCL, H+-PPase, HKT1, NHX1, NCED/24 days after transplantation | Serratia liquefaciens | Salinity | Maize/leaves | El-Esawi et al., 2018b |
| P5CS1, SOD, APX1, CAT, SHSP/45 days from sowing | Bacillus cereus, Providencia rettgeri and Myroides odoratimimus | Chromium and heat stresses | Sorghum/leaves | Bruno et al., 2020 |
| CAPIP2, CaKR1, CaOSM1, CAChi2/7 days after inoculation | Bacillus fortis | Salinity | Capsicum/leaves | Yasin et al., 2018 |
| APX1, CAT1, SOD2, SOD4, APX2, PR1, PRP2, PRP4, HSP70/32 days after transplant | Azospirillum brasilense and Rhizobium tropici | Salinity | Maize/leaves and roots | Fukami et al., 2018 |
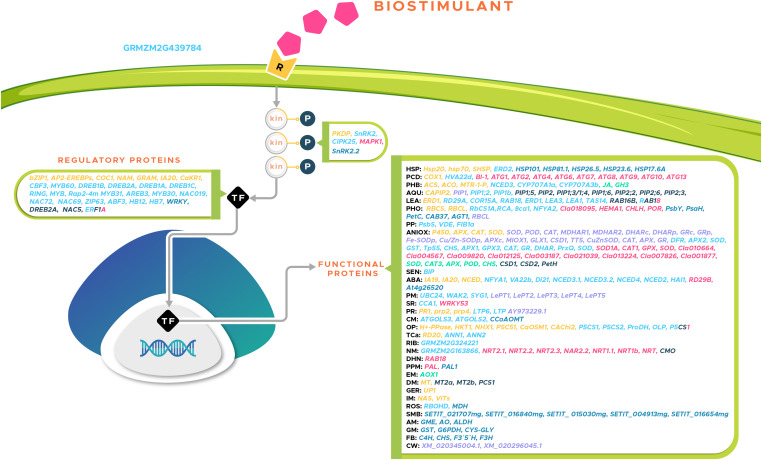
The effect of a biostimulant from Pseudomonas fluorescens applied to reproductive-stage rice plants under drought stress (water restriction for 15 days) was analyzed (Saakre et al., 2017), and six differentially expressed genes (DEGs), COX1, PKDP, bZIP1, AP2-EREBP, Hsp20, and COC1, were reported in leaves of 65-day-old plants. Differentially expressed transcripts correspond to the cytochrome oxidase subunit 1 (COX1) gene, protein kinase domain protein (PKDP), bZIP protein family (bZIP1) members, APETALA2-ethylene-responsive element-binding protein (AP2-EREBP), heat-shock protein 20 (Hsp20), and the circadian oscillator component (COC1). bZIP1, AP2-EREBP, COC1, and PKDP genes are considered regulatory proteins. bZIP1, AP2-EREBPs, and COC1 genes are considered stress-induced transcription factors (TFs) involved in the ABA-dependent signaling pathway (Reeves et al., 2011; Seung et al., 2012). ABA is generally defined as a stress hormone since it intervenes in the regulation of the response to biotic and abiotic stress (Vishwakarma et al., 2017); it also plays a vital role in physiological processes such as seed germination, dormancy, and stomatal closure (Akpinar et al., 2012). In the ABA-dependent signaling pathway, protein receptors (PYR/PIL/RCAR) bind to ABA by inhibiting PP2C activity, allowing SnRK2 activation by autophosphorylation. Subsequently, active SnRK2 can phosphorylate TFs such as AREB/ABF, DREB2A/2B, RD22BP1, and MYC/MYB TFs, which bind to their corresponding cis-elements and modulate the expression of response genes (de Zelicourt et al., 2016; Bulgakov and Wu, 2019). The PKDP gene encodes an enzyme with kinase activity that participates in phosphorylation reactions; this enzyme is a key component of signal transduction (Mohanta et al., 2015) and is involved in multiple cellular processes in plants such as growth and development, carbon and nitrogen metabolism, the formation of the cytoskeleton, senescence and cell death, hormone signal transduction, regulation of ion channels, and the defense response (Lei et al., 2007). Hsp20 and COX1 genes are considered functional proteins. The Hsp20 gene encodes a small (12–42 kDa) heat-shock protein (sHsp) with chaperone activity; its function is to degrade denatured proteins, and it has an ATP-dependent chaperonin function. These proteins can bind partially folded or denatured proteins and avoid permanent unfolding or aggregation (Reddy et al., 2015; Nagaraju et al., 2020). The COX1 gene encodes an enzyme with cytochrome oxidase activity, which is related to the process of apoptosis, a type of programmed cell death (PCD) in plants. PCD is triggered in plants during biotic and abiotic stress since recycling and mobilizing nutrients are carried out under adverse conditions; this process can be triggered by reactive oxygen species (ROS) (Foyer and Noctor, 2005). Moreover, autophagy (another type of PCD) is related to protein turnover and removal of damaged organelle proteins, which can lead to changes in cell behavior (Akpinar et al., 2012).
As a strategy to improve Vigna mungo L. and Pisum sativum L. plants under conditions of water deficit (polyethylene glycol-6000), (Saikia et al., 2018) a PGPB consortium (Ochrobactrum pseudogrignonense, Pseudomonas spp., and Bacillus subtilis) was applied. The plants treated with the consortium presented, in addition to increased growth, a better adaptation to resist stress due to downregulation of the ACS and ACO genes at 45 days after stress. In the plants that had only stress, the opposite occurred, and the expression of these genes was increased. The ACS gene encodes the ACC synthase enzyme, and the ACO gene encodes the ACC oxidase enzyme. ACC synthase is responsible for producing ACC, and ACC oxidase is involved in the production of ethylene (Li et al., 2020b). Ethylene is a hormone related to plant growth and development as well as processes such as fruit ripening and abscission, leaf senescence, seed germination, and organogenesis (Trujillo-Moya and Gisbert, 2012); however, a sudden increase in ethylene concentration in the plant during stress causes negative effects and leads to senescence (Czarny et al., 2006). The mechanism by which the expression of the ACS and ACO genes is reduced is triggered because the consortium of bacteria are producers of hormones, specifically indoleacetic acid (IAA), which triggers the production of relatively high concentrations of ACC and, subsequently, feedback inhibition of IAA synthesis (Glick, 2012). Additionally, some rhizobial strains produce the enzyme ACC deaminase, which removes some of the ACC (the immediate precursor to ethylene) before it can be converted to ethylene (Ma et al., 2002).
In rice plants under salinity stress (100 mM NaCl), the biostimulant effect of Bacillus amyloliquefaciens-SN13 was evaluated (Chauhan et al., 2019). During stress, B. amyloliquefaciens increased the relative water content, biomass, proline content and total soluble sugar content in plants while decreasing the electrolyte leakage and lipid peroxidation. Alterations in gene expression were also observed in the transcriptome of rice roots under salt stress with B. amyloliquefaciens. Changes in the expression of a considerable number of genes related to the stress response, hormones, photosynthesis, lipid metabolism and cell wall were induced. Additionally, DEGs in functional categories such as the stress response (Os11g26750, Os10g38600, Os05g28740, and Os05g45480), metabolism (Os03g08860, Os04g27060, Os01g43750, Os05g41460, Os04g47360, and Os03g12510), transporters (Os07g15460), and regulation (Os04g56990, Os06g41770, Os09g39650, Os09g31031, Os03g14590, Os11g03370, and Os05g10620) were quantified. The functional validation in Saccharomyces cerevisiae of the OsNAM and OsGRAM genes with 5- and 3-fold induction, respectively, in transcriptome of rice roots with B. amyloliquefaciens under salinity was analyzed. Transformed yeast cells expressing OsNAM and OsGRAM showed enhanced tolerance under osmotic stress; moreover, the transformed cells were able to survive at relatively high temperatures, and they displayed increase tolerance to arsenite and arsenate. OsNAM belongs to the NAC family of TFs, which is a large plant-specific gene family that is related to the regulation of tissue development and the response to abiotic stress (Shan et al., 2020). OsGRAM is important for the ABA response, hormone signaling under stress conditions, and the perception of environmental stimuli and regulation of the response to those stimuli (Mauri et al., 2016).
The effect of Arthrobacter nitroguajacolicus (seeds inoculated with 108 CFU mL–1) on wheat tolerance to salinity stress (200 mM NaCl) has been studied (Safdarian et al., 2019). Transcriptomic analysis revealed upregulation of 152 genes, and 5 genes were significantly downregulated. Inoculated roots of plants under salt stress presented differential expression of many genes involved in stilbenoids, diarylheptanoid metabolism, phenylpropanoids, flavonoids, terpenoid, porphyrin, and chlorophyll metabolism. RNA-seq results, 11 DEGs (including UP1, NAS, MT, MTR-1-P, VITs, P450, UP2) were quantified. UP1 is a functional cis-element responsible for germination-associated gene expression (Tatematsu et al., 2008). NAS is induced by salt stress and is related to niacinamide, the synthetic precursor of plant iron (Johnson et al., 2011). MT gene encodes a metal chelatin protein involved in the detoxification of heavy metals and in the homeostasis of intracellular metal ions (Ahn et al., 2012). MTR-1-P encodes a 5-methylthioribose 1-phosphate related to ethylene biosynthesis (Arraes et al., 2015). VIT proteins are involved in the transport and storage of iron, can maintain iron within the optimal physiological range and prevent cellular toxicity (Cao, 2019). Cytochrome P450 proteins are related to signals for growth and development, are responsible for protecting plants from different stresses, and are involved in in redox reactions and a large number of biosynthetic pathways (Bak et al., 2011; Safdarian et al., 2019). UP2 encodes an uncharacterized conserved protein and is related to cell structure (Duan et al., 2017).
In Arabidopsis thaliana plants under salt stress (80 and 100 mM NaCl), the biostimulant effect of Bacillus oryzicola was studied (Baek et al., 2020). Compared with control plants, plants with B. oryzicola while under salinity showed increased numbers of lateral roots, increased fresh weight, and increased chlorophyll content, and they accumulated less salt-induced malondialdehyde and Na+. Additionally, plants with B. oryzicola while under salinity presented enhanced transcription of the RD29A, RD29B, RD20, RD22, KIN1, and ERD1 genes in the shoots and roots. The RD29A (which is responsive to desiccation) gene encodes a protein strongly related to the Brassicaceae family. Its function is similar to that of LEA proteins (Kaur and Gupta, 2005). RD29B encodes a protein that is induced in response to water deprivation, low temperature, salinity, and desiccation, and its response is mediated by ABA (Bihmidine et al., 2013). RD20 is a stress-inducible gene that belongs to the caleosin family and is dependent on ABA signaling. RD20 plays a role in drought tolerance through stomatal control under water-deficit conditions (Aubert et al., 2010). The RD22 (which is responsive to desiccation) gene can be induced by stress caused by drought and salinity and by exogenous applications of ABA, but its role in the stress response is unknown (Wang et al., 2012). The KIN1 gene encodes a protein induced in response to cold, dehydration, osmotica and ABA, possibly functioning as an antifreeze protein (Wang et al., 1995). The ERD1 (early responsive to dehydration stress 1) protein functions in chloroplasts by degrading unassembled or misfolded proteins (Peltier et al., 2001).
Using wheat plants under heat (6 h at 45°C), cold (6 h at −5°C) and drought stress (5 days without water), the effect of B. amyloliquefaciens subsp. plantarum was studied (Abd El-Daim et al., 2018). cDNA-AFLP analysis revealed differential expression of more than 200 transcript-derived fragments (TDFs) in wheat leaves. Five TDFs (IA20, IA24, IA35, IA41, IA19) were selected for RT-PCR analysis. All the TDFs changed in expression pattern in plants treated with B. amyloliquefaciens subsp. plantarum and with the different stresses. IA19, IA20, and IA24 are ABA responsive homologous. IA20 encodes to TF WRKY20, this TF enhance drought tolerance regulating ABA signaling (Luo et al., 2013).
The effect of Serratia liquefaciens on maize plants under salinity stress (0, 80, and 160 mM NaCl) was studied (El-Esawi et al., 2018b). S. liquefaciens inoculation significantly reduced oxidative stress markers but increased the maize growth and biomass production along with better antioxidant defense system, leaf gas exchange, osmoregulation, and nutrient uptake under salinity. Also, it was found the upregulation of stress-related genes (APX, CAT, SOD, RBCS, RBCL, H+-PPase, HKT1, NHX1) and downregulation NCED gene. The APX gene encodes ascorbate peroxidase, which catalyzes the reduction of H2O2 and the oxidation of ascorbate, generating monodehydroascorbate (Eltelib et al., 2012). The CAT gene encodes catalase, and this enzyme is important in the removal of H2O2 generated in peroxisomes by oxidases involved in the β-oxidation of fatty acids, photorespiration and purine catabolism (Gill and Tuteja, 2010). The SOD gene encodes an enzyme that is crucial in the first line of antioxidant defense since it can convert highly reactive O2– radicals into H2O2 and O2 in response to oxidative stress (Wang W. et al., 2016). The RBCS gene encodes the small Rubisco subunit and is part of a multigenic family. The Rubisco enzyme catalyzes the assimilation of CO2 in plants and is related to the photosynthesis process (Atkinson et al., 2017). RBCL is involved in photosynthesis, encodes the large subunit of the primary CO2 fixation enzyme Rubisco. This enzyme serves as the primary engine of carbon assimilation being the most abundant protein on earth (Igamberdiev, 2015). The NCED3 (nine-cis-epoxycarotenoid dioxygenase 3) gene is related to the pathways of ABA biosynthesis and is highly expressed at the root level (Li et al., 2020a). H+-PPase ecodes H+-pumping pyrophosphate and HKT1 encodes high-affinity K+ transporter 1 related with ion balance regulators. NHX1 encodes Na+/H+ antiporter, which participates in Na+ sequestration, export and recirculation (Chen et al., 2016). Using sorghum plants under chromium (200 mg K2Cr2O7 kg–1 in soil) and heat stress (42°C during day and 28°C at night), the effect of Bacillus cereus, Providencia rettgeri, and Myroides odoratimimus (chromium reducing-thermotolerant CRT-PGPB) was studied (Bruno et al., 2020). Inoculation with CRT-PGPB increased plant growth, antioxidant status and decreased malondialdehyde and proline contents in plants under stress. Also, gene expression studies down-regulated the expression of P5CS1 gene, and up-regulated the expression of SOD, APX1, CAT and SHSP. All genes were already mentioned.
The effect of Bacillus fortis (halotolerant HPGPR) on capsicum plants under salt stress (1 and 2 g NaCl kg–1 soil) was studied (Yasin et al., 2018). HPGPR promote growth attributes, chlorophyll, protein content and water use efficiency on capsicum plants under salinity. Also, up-regulated the expression profiles of stress related genes including CAPIP2, CaKR1, CaOSM1, and CAChi2. CAPIP2 encodes plasma membrane intrinsic protein related in transportation of smaller neutral solutes and water (already mentioned). CaKR1 is a member of ankyrin repeat zinc finger protein TF family, the over-expressed of this TF exhibited enhanced antioxidant metabolism (Seong et al., 2007). CaOSM1 is a osmotin gene related with resistance against biotic and abiotic stress (Choi et al., 2013). CAChi2 encodes chitinase class II is related with osmotic stress tolerance (Hong and Hwang, 2006).
Using maize plants under salinity stress (170 mM NaCl), the effect of Azospirillum brasilense and Rhizobium tropici inoculation was studied (Fukami et al., 2018). Inoculation affected antioxidant enzymes, proline, and MDA contents in leaves and roots. The expression of genes related to antioxidant activity were up-regulated (APX1, CAT1, SOD2, SOD4 in leaves, and APX2 in roots), while the expression of pathogenesis-related genes PR1, prp2, prp4i and hsp70 were down-regulated in leaves and roots. Antioxidant genes and were already mentioned. PR1 (a member of a multigene family) is a salicylic acid inducible marker gene for systemic acquired resistance (SAR) (Hussain et al., 2018). The gene prp2 encodes β-1, 3-glucanase, and prp4 encodes chitinase family (Fukami et al., 2017). The gene hsp70 were already mentioned.
Examination of Table 1 allows determining the absence of transcriptomic responses that were common among the 10 studies. One possible explanation, in addition to the small number of studies available, is that the interactions between the plant–bacteria are specific. There were no analogous responses between the different species studied. If to the above it is added that not all the studies were carried out under the same type of stress, it can be concluded that the possibility of demonstrating that a certain class of biostimulant can induce common responses in plants depends on carrying out studies with several species of plants treated with the same type of biostimulant under the same stress condition. However, common genes in some studies were related to ABA, TFs, and antioxidants.
Beneficial Fungi
This category includes fungi and arbuscular mycorrhizal fungi (AMFs), which are considered microbial inoculants along with bacteria (Dodd and Ruiz-Lozano, 2012). Among AMFs, those of the Glomus genus have been widely studied (Calvo et al., 2014). Fungi of the genera Neotyphodium, Curvularia, Colletotrichum, Fusarium, Alternaria, and Trichoderma have been studied as biostimulants in plants, with Trichoderma being the most studied (Calvo et al., 2014; Jardin, 2015; De Palma et al., 2019).
Genes and their functions are described below using the results of eight studies in which beneficial fungi and AMFs were applied to plants under abiotic stress (Table 2).
TABLE 2.
Description of genes whose transcript level changed in response to the application of fungi and AMF-based biostimulants based to plants under abiotic stress.
| Gene/gene expression time | Biostimulant type | Abiotic stress type | Crop/gene expression organ | References |
| LsPIP1/60 days after transplant | Glomus intraradices (AMF) | Salinity | Lettuce/roots | Jahromi et al., 2008 |
| MIOX1, GLX1, CSD1, TT5/after 0, 1, 3, and 6 days of stress | Glomus mosseae (AMF) | Drought | Poncirus trifoliata/leaves | Fan and Liu, 2011 |
| SOD, POD, CAT/15 days of NaCl treatment | Trichoderma longibrachiatum (fungi) | Salinity | Wheat/leaves | Zhang et al., 2016 |
| MDHAR1, MDHAR2, DHARc, DHARp, GRc, GRp, Fe-SODp, Cu/Zn-SODp, APXc/3 days after 50% of seeds germinated | Trichoderma harzianum (fungi) | Drought | Tomato/shoots and roots | Mastouri et al., 2012 |
| RBCL, PPH, Cu-Zn SOD, CAT, APX, GR/6 days after stress | Funneliformis mosseae (AMF) | Salinity-alkalinity | Watermelon/leaves | Ye et al., 2019 |
| LePT1, LePT2, LePT3, LePT4, LePT5/not specified | F. mosseae and Rhizophagus intraradices (AMF) | Drought | Tomato/roots | Volpe et al., 2018 |
| XM_020345004.1, XM_020296045.1, AY973229.1/8 weeks after transplanting germinated seeds | F. mosseae (AMF) | Drought | Wheat/roots | Moradi Tarnabi et al., 2020 |
| 23660, 26140, 20188, 18003, 4379, 20176, 6553, 23129, 29567/10 days after stress | Rhizophagus irregularis (AMF) | Salinity | Asparagus/leaves | Zhang et al., 2019 |
In lettuce plants (Jahromi et al., 2008), researchers studied the effect of applying Glomus intraradices AMFs under salinity stress at two levels (50 and 100 mM NaCl). Compared with plants without the application of AMFs while with stress, plants treated with G. intraradices while under salinity stress presented increased growth and an increase in the relative water content. Additionally, the expression of the gene of an intrinsic plasma membrane protein (LsPIP1) increased 60 days after transplanting; LsPIP1 encodes a type of aquaporin in the plasma membrane and is related to the transport of water and small solutes with no charge (Wang et al., 2019).
Trichoderma longibrachiatum inoculated (108 CFU mL–1) onto wheat seedlings under salt stress (150 mM NaCl) (Zhang et al., 2016) can enhance the plant stress tolerance. The relative water content in the leaves and roots, the chlorophyll content, the proline content and the root activity were increased, but the content of leaf malondialdehyde under salinity stress was decreased. The antioxidant enzymes SOD, POD, and CAT were increased, and the relative expression of the SOD, POD, and CAT genes was upregulated. The possible mechanisms by which salinity suppresses the negative effect on wheat may be due to the improvement of the antioxidant defense system. SOD and CAT are discussed above. The POD gene encodes a peroxidase with a variety of biological functions, including hydrogen peroxide detoxification, hormone signaling, lignin biosynthesis and stress responses (Gao et al., 2010).
In tomato seedlings under water deficit, inoculation of a Trichoderma harzianum biostimulant was evaluated (Mastouri et al., 2012). The enhanced redox state of inoculated plants could be explained by the increased activity of antioxidant enzymes. Additionally, T. harzianum modulated the expression of genes encoding antioxidant enzymes. The MDHAR1, MDHAR2, DHARc, DHARp, GRc, GRp, Fe-SODp, Cu/Zn-SODp, and APXc genes showed increased expression in the shoots, whereas only the APXc gene showed increased expression in the roots. The MDHAR gene encodes a monodehydroascorbate reductase, and together with DHAR (dehydroascorbate reductase) and GR (glutathione reductase), it participates in the ascorbate-glutathione cycle. This cycle plays an important role in the efficient removal of excess ROS. APX and SOD are discussed above.
The effect of Glomus mosseae (AMF) inoculation on the drought (3 days of water depletion) tolerance of Poncirus trifoliata seedlings was studied (Fan and Liu, 2011). Plants inoculated with G. mosseae showed an increase in growth and increased relative water and chlorophyll contents. Under drought, the inoculated plants showed increased levels of proline and increased activity of antioxidant enzymes (SOD, POD). Additionally, four genes (MIOX1, GLX1, CSD1, TT5) involved in ROS homeostasis and counteracting oxidative stress presented increased expression in inoculated plants. The MIOX1 gene encodes myo-inositol oxygenase, which is involved in the biosynthesis of ascorbic acid; ascorbic acid has been shown to be an important antioxidant protecting plants against oxidative damage (Fan and Liu, 2011). The GLX1 gene encodes glyoxalase I, a key enzyme involved in the glutathione-based detoxification of methylglyoxal, a product of lipid and carbohydrate metabolism. The CSD1 gene encodes a copper/zinc SOD, and this gene is discussed above. TT5 (transparent testa 5) encodes a chalcone isomerase involved in flavonoid synthesis. Flavonoids have an important role in the modulation of ROS levels (Fan and Liu, 2011).
Using watermelon plants under salinity-alkalinity stresses (irrigation with 400 mL 60 mM salinity-alkalinity solution [NaCl, Na2SO4, NaHCO3, Na2CO3]), the effect of Funneliformis mosseae (AMF) was studied (Ye et al., 2019). The photosynthesis related parameters were alleviated after incubation of AMF. Under stress, the relative expression level of RBCL, Cu-Zn SOD, CAT, APX, GR were increased after AMF treatment. PPH was reduced after AMF treatment. RBCL, Cu-Zn SOD, CAT, APX, GR genes are involved in antioxidant metabolism and are discussed above.
The effect of F. mosseae and R. intraradices (AMF) on tomato plants under water stress (leaf water potential of about −0.9 and −1.0 MPa) was studied (Volpe et al., 2018). Gene expression analysis involved in inorganic phosphate uptake and transport was made. LePT1, LePT2, LePT3, LePT4, and LePT4 changed their expression under water deficit and AMF treatment. P uptake, transfer and delivery are improved in AM roots. P fertilization can increase stress tolerance and productivity in several plant species (Sawers et al., 2017).
Using wheat plants under water deficit stress (each pot three times weekly irrigating, 25 ml per time), the effect of F. mosseae (AMF) was studied (Moradi Tarnabi et al., 2020). The results showed that symbiotic association between plant and AMF and irrigation not only affect transcription profile of the plant growth, but also membrane components and cell wall. The most DEGs were observed in lipid and carbohydrate metabolic process, membrane transports, cellulose synthase activity, chitinase activity, and nitrogen compound metabolic process related genes. The expression of three randomly selected DEGs were examined. The selected genes were associated to chitinase activity (AY973229.1), cellulose biosynthetic process (XM_020345004.1), and beta-glucosidase BoGH3B-like (XM_020296045.1). Chitin-related genes such as chitinase detect chitin molecules of fungus as a signal to trigger a defense response an increase the plant tolerance (Moradi Tarnabi et al., 2020). A stress-response related role can be considered for cellulose, since cellulose microfibrils and the other factor that lead the direction of cell growth can be regulated by water availability (Wang T. et al., 2016). Beta-glucosidase have important in cell wall biogenesis, which strongly provides protection against biotic and abiotic stress (Moradi Tarnabi et al., 2020).
The effect of Rhizophagus irregularis (AMF) on asparagus plants under salinity stress (100 mM NaCl) was studied (Zhang et al., 2019). The authors conducted a transcriptome analysis on leaves of garden asparagus to identify gene expression under salinity stress. 455 DEGs were identified in plants with salinity to plants with AMF and salinity. The expression profiles of 9 DEGs (23660, 26140, 20188, 18003, 4379, 20176, 6553, 23129, 29567) were by qRT-PCR. Their putative functions involved in nitrogen metabolism, synthesis of secondary metabolites, ion homeostasis, osmotic adjustment, and scavenging ROS, among others.
The review of Table 2 again, as in Table 1, reveals a small number of studies available. Except for the expression of antioxidant enzymes, where there were analogous responses between the different species studied, more studies are needed with the same species of beneficial fungus in different species of plants, under the same stress condition, to effectively demonstrate that the determination of the transcriptomic landscape will contribute to the omic definition of biostimulant.
Algal and Botanical Extracts
Recently, marine algal extracts have been used as biostimulants, as their use in agriculture had previously been limited to being a fertilizer or a source of organic matter (Jardin, 2015). Marine macroalgae are divided into three broad groups (brown, red, and green), of which brown algae are the most widely used in agriculture; brown algae include species such as Fucus spp., Sargassum spp., Laminaria spp., Turbinaria spp., and Ascophyllum nodosum, this last of which is the most studied (Blunden and Gordon, 1986; Ugarte et al., 2006; Hong et al., 2007). The biostimulant function of algal extracts is commonly associated with the content of hormones such as cytokinins, auxins, abscisic acid, gibberellins, and other classes of hormone-like compounds such as sterols and polyamines (Craigie, 2011; Wally et al., 2013). However, they also contain compounds such as polysaccharides (laminarin), alginates, carrageenans, macro- and micronutrients, and nitrogenous compounds such as betaines (Khan et al., 2009; Craigie, 2011).
In the case of botanical extracts, substances extracted from plants (seeds, leaves, roots, and exudates) of various families have generally been used in agriculture as pesticides; in terms their functions as biostimulants, there is little research, thus representing an important area of opportunity (Ertani et al., 2013; Ziosi et al., 2013; Yakhin et al., 2017).
Genes and their functions are described below for 14 studies where biostimulants based on extracts of algae and botanicals were applied to plants under abiotic stress (Table 3).
TABLE 3.
Description of genes whose transcript level changed in response to the application of biostimulants based on extracts of algae and botanicals to plants under abiotic stress.
| Gene/gene expression time | Biostimulant type | Abiotic stress type | Crop/gene expression organ | References |
| AtDREB2a, AtRD29, AtNFYA1, AtNFYA2, AtUBC24, AtWAK2, AtSYG1, At3g27150/6 and 12 h after treatment | Extract of A. nodosum | Salinity | Arabidopsis/whole plant | Shukla et al., 2018 |
| LEA3, LEA1, CCA1, HVA22d, LTP6, DREB1A, DREB1C, LEA2, VA22b, Di21, SnRK2, CIPK25, LTP, RING, MYB/1 and 5 days after treatment | Extract of A. nodosum | Salinity | Arabidopsis/whole plant | Jithesh et al., 2018 |
| ANN1, ANN2, PIP1, P5CS1, CHS, APX1, GPX3/not specified | Extract of A. nodosum and 5-aminolevulinic acid (foliar application) | Salinity | Asparagus aethiopicus/leaves | Al-Ghamdi and Elansary, 2018 |
| COR15A, RD29A, CBF3/when plants were exposed at −2°C | Extract of Ascophyllum nodosum | Cold | Arabidopsis/leaves | Rayirath et al., 2009 |
| P5CS1, P5CS2, PRODH/not specified | A. nodosum lipophilic components | Freezing | Arabidopsis/leaves | Nair et al., 2012 |
| NCED3, MYB60, RAB18, RD29A, RbCS1A, RCA, PIP1;2, B ca1, PsbS, VDE, DFR, APX2, SOD/From the moment of stress until the fourth day | Extract of A. nodosum | Drought | Arabidopsis/leaves | Santaniello et al., 2017 |
| GmCYP707A1a, GmCYP707A3b, GmRD22, GmDREB1B, GmERD1, FIB1a, GmPIP1b, GmGST, GmBIP, GmTp55/75 h and 59 h after treatment | Extract of A. nodosum | Drought | Soybean/leaves | Shukla et al., 2017 |
| TAS14/7th day of stress | Extract of A. nodosum (foliar application) | Drought | Tomato/leaves | Goñi et al., 2018 |
| TaNCED3.1, TaNCED3.2, TaHAI1, DREB family, TaRap2-4, TaMYB31, TaAREB3, dehydrin, TaP5CS1, TaNCED4, TaNCED2, TaERD2/6 days after stress | Extract of Gracilaria dura | Drought | Wheat/leaves | Sharma et al., 2019 |
| GRMZM2G439784, GRMZM2G324221, GRMZM2G164129, GRMZM2G163866/10 days after treatment | Extract of Kappaphycus alvarezii | Drought | Maize/roots | Kumar et al., 2019 |
| SOD, CAT, APX, DAHR, GR, PrxQ/not specified | Licorice root extract | Salinity | Pea (Pisum sativum L.)/leaves | Desoky et al., 2019 |
| DtDREB2A, DtMYB30, DtNAC019, DtNAC72, DtNAC19, DtNAC69, DtZIP63, DtABF3, DtHB12, DtHB7/2, 4, 6, 9, and 24 h from exposure to stress | Extract of borage | Salinity | Wild rocket/leaves | Franzoni et al., 2019 |
| CuZnSOD, MnSOD, CAT, FeSOD/24 and 48 h after stress | Lignin derivatives, plant-derived aminoacids and molybdenum | Heat | Cucumber/seeds | Campobenedetto et al., 2020 |
| ObOLP/3-months old plants | Moringa oleifera extract | Salinity | Ocimum basilicum/leaves | Alkuwayti et al., 2020 |
In A. thaliana plants under freezing stress (−2°C), the effect of extracts (lipophilic fraction) of the brown macroalga species A. nodosum (1 g L–1) was studied (Rayirath et al., 2009), where the expression of three cold-responsive genes (CBF3, RD29A, and COR15A) increased when the plants were at −2°C. The authors pointed out that in response to the application of the extract of A. nodosum to seedlings of A. thaliana, freezing tolerance was obtained by protecting the integrity of the membrane; also, compared with the untreated seedlings, the treated seedlings presented a smaller decrease in the concentration of chlorophyll as a result of stress. The CBF3 (C-repeat-binding factor) gene is a TF that plays an important role in tolerance to low temperatures in Arabidopsis (Takuhara et al., 2011). CBF3 is activated in response to low temperatures and dehydration and is independent of ABA; CBF3 binds to the cis-element DRE-CRT (C-repeat/dehydration-responsive element) that is present in the promoter region of the RD29A and COR15A genes in Arabidopsis, the WCS120 gene in wheat and the BN115 gene in Brassica napus (Medina et al., 2011). Moreover, there are indications that CBF3 regulation is carried out through responses to light quality and the circadian rhythm (Fowler et al., 2005; Franklin and Whitelam, 2007). The RD29A gene is discussed above. The COR15A (cold-regulated) gene regulates cold tolerance by stabilizing chloroplast membranes (Thalhammer et al., 2014).
In A. thaliana plants to which lipophilic components of an extract of A. nodosum (ANE), the effect of tolerance to stress by freezing (−2°C for 24 h) was studied (Nair et al., 2012). Expression of the P5CS1, P5CS2, and ProDH genes was detected; the expression of P5CS1 and P5CS2 increased, while that of ProDH decreased. These genes are involved in proline synthesis (P5CS1 and P5CS2) and degradation (ProDH). P5CS1 and P5CS2 encode delta1-pyrroline-5-carboxylate synthase enzymes that regulates the rate of proline biosynthesis. This gene is expressed in some tissues under normal conditions and throughout the plant under conditions of water deficit, in addition to being induced by ABA and salt stress (Kesari et al., 2012). ProDH catalyzes the degradation of proline to produce glutamic acid; this gene is related to decreasing the oxidative burst and cell death associated with the hypersensitive response (Cecchini et al., 2011).
In A. thaliana plants under drought stress (absence of a hydroponic solution for 4 days), the effect of an extract of A. nodosum was evaluated (Santaniello et al., 2017); the extract positively influenced the survival of the plants. The plants under stress and treated with the extract presented increased expression of the NCED3, MYB60, RAB18, RD29A, RbCS1A, RCA, PIP1;2, βCA1, PsbS, VDE, DFR, APX2, and SOD genes 4 days after stress. These genes are involved in the pathways of the antioxidant system and are ABA dependent. The MYB60 gene is related to the regulation of stomatal movement, and the expression of this gene increases with low levels of ABA; additionally, in an initial drought state, this gene can induce root growth. In contrast, in a severe state of drought, its expression is inhibited, resulting in stomatal closure and a decrease in root growth (Oh et al., 2011). The RAB18 gene (which is sensitive to ABA) encodes a glycine-rich hydrophilic protein (Hoque et al., 2012) that belongs to the LEA family of proteins and has a dehydrin function (Hernández-Sánchez et al., 2019). RAB18 protects membranes under dehydration conditions by binding to anionic phospholipids through electrostatic forces (Eriksson and Harryson, 2011), in addition to binding to other proteins to prevent their denaturation (Graether and Boddington, 2014). The RD29A gene is discussed above. Similarly, the RCA gene encodes the Rubisco activase enzyme, which is a chloroplastic enzyme encoded in the nucleus and participates in the activation of Rubisco (Hasse et al., 2015); this gene is also related to jasmonate-induced leaf senescence (Elizabete Carmo-Silva and Salvucci, 2013). Two genes related to the regulation of mesophilic diffusion restriction include PIP1;2 (discussed above) and βCA1 (β-carbonic anhydrase 1), both of which participate in carboxylation or decarboxylation reactions related to photosynthesis and respiration; as such, they play an important role in the catalysis of CO2 and water to form protons and bicarbonate (Hu et al., 2015). The PsbS (photosystem II subunit S) and VDE (violaxanthin depoxidase) genes are involved in the photoprotection mechanism to avoid damage caused by oxidative stress in plants due to excess energy from sunlight (Fufezan et al., 2012; Ciszak et al., 2015). The DFR (dihydroflavonol reductase) gene is involved in the biosynthesis of anthocyanins, which protect plants from stress through their activity of ROS detoxification (Cui et al., 2014). The APX2 and SOD genes are discussed above.
Researchers (Shukla et al., 2017) studied the effect of an extract of A. nodosum on soybean plants under drought stress (without irrigation). Compared with the untreated plants, the treated plants had higher relative water content, antioxidant activity, and stomatal conductance under drought stress. In addition, there were changes in the expression of the GmCYP707A1a, GmCYP707A3b, GmRD22, GmDREB1B, GmERD1, GmNFYA3, FIB1a, GmPIP1b, GmGST, GmBIP, and GmTp55 genes at 75 h (stress) and 89 h (recovery) after treatment. The GmCYP707A1a and GmCYP707A3b genes encode ABA 8′-hydroxylases, which participate in the regulation of ABA levels during dehydration and rehydration (Umezawa et al., 2006; Zheng et al., 2012a). GmRD22 is discussed above. The GmDREB1B (dehydration response element-binding) gene belongs to a family of TFs induced by drought and salinity, and this gene is ABA dependent. This TF binds to the cis-element DRE-CRT, which is present in the promoter of the COR and RD29A genes, both of which are related to the abiotic stress response (Tuteja, 2007). GmERD1 is discussed above. The FIB1a gene improves the phototolerance of photosystem II (PSII) (Mutava et al., 2015), and the GmPIP1b gene is also discussed above. The GmGST gene, via its glutathione reduction potential, detoxifies ROS and protects cells from oxidative damage (Mcgonigle et al., 2000). The GmBIP gene encodes a chaperone protein that is related to delayed senescence in leaves and therefore increases tolerance to drought stress (Carvalho et al., 2014). The GmTp55 gene encodes an aldehyde dehydrogenase enzyme that reduces reactive aldehydes derived from lipid peroxidation under oxidative stress (Wang et al., 2017).
Using tomato plants under drought stress, researchers (Goñi et al., 2018) studied the effect of several commercial products based on extracts of A. nosodum. All of the ANEs affected drought stress tolerance but to different degrees. Regulation of the TAS14 gene was reported 7 days after stress, and this gene was differentially overexpressed in response to applications of all extracts. TAS14 encodes a group 2 LEA protein called dehydrin, which is induced by osmotic stress and ABA (Godoy et al., 1994). When this gene is overexpressed, long-term tolerance to drought and salinity is achieved through the reduction in osmotic potential and the accumulation of sugars and potassium (Muñoz-Mayor et al., 2012).
Using wheat plants under water stress (no water for 10 days), researchers (Sharma et al., 2019) evaluated foliar applications of Gracilaria dura (red algae) sap. The expression levels of the TaNCED3.1, TaNCED3.2, TaHAI1, DREB, TaRap2-4, TaMYB31, TaAREB3, dehydrin, TaP5CS1, TaNCED4, TaNCED2, and TaCla013224 genes were analyzed, which increased on the sixth day of the onset of stress. TaNCED3.1, TaNCED3.2, TaNCED4, and TaNCED2 belong to the NCED family of genes, which encode 9-cis-epoxycarotenoid dioxygenases; these are key enzymes involved in ABA biosynthesis and are regulated in response to drought and salinity (Behnam et al., 2013). TaHAI1 encodes a member of the PP2C family, whose members includes class A and type 2C phosphatase proteins and are related to the downregulation of osmotic stress and ABA signaling (Nguyen et al., 2019). DREBs are discussed above. TaRap2-4 encodes a DREB-subfamily TF related to light mediation and ethylene signaling (Lin et al., 2008). TaMYB31 belongs to a subfamily of TFs, is ABA dependent and plays an important role in the development of and defense response in plants (Yanhui et al., 2006; Tuteja, 2007). TaAREB3 is also a TF that is sensitive to ABA and is related to stomatal movement and ROS generation in response to ABA (Wang et al., 2013). Dehydrins are LEA-like proteins (as mentioned above), and TaP5CS1 participates in proline biosynthesis (as mentioned above). TaERD2 encodes an HSP70-type chaperonin that is synthesized in response to stress and is the main chaperone in maintaining protein homeostasis (Rowarth et al., 2019).
The effect of Kappaphycus alvarezii extract applied to the soil was analyzed on maize plants under drought stress (no irrigation for 10 days) (Kumar et al., 2019). To obtain a global view of the effect, the authors analyzed the transcriptome of the roots of the plants 10 days after the application of the treatment. A total of 896 upregulated genes and 533 downregulated genes were differentially expressed. However, only 4 genes were overexpressed in response to the application of the extract and stress compared to the application of the extract only, and 18 genes were repressed. The overexpressed genes included GRMZM2G439784, GRMZM2G324221, GRMZM2G164129, and GRMZM2G163866. The GRMZM2G439784 gene encodes an LRR-type kinase, which belong to the receptor kinase subfamily, and its function lies in communication between cells to transmit signals during development and before environmental stimuli to activate defense; it is of the utmost importance in resistance against pathogens (Antolín-Llovera et al., 2014; Dievart et al., 2015). GRMZM2G324221 encodes a structural protein of the small ribosomal subunit (40S) and participates in the regulation of response to virus infections (Li, 2019). The function of GRMZM2G164129 has not been characterized, whereas GRMZM2G163866 is a high-affinity nitrate transporter; overexpression of the latter could translate into an improvement in nitrogen metabolism (Liu X. et al., 2014).
Using A. thaliana plants under saline conditions (150 mM NaCl), researchers (Shukla et al., 2018) studied the effects of an extract of A. nodosum by measuring the expression of several microRNAs (miRNAs) and their target genes. The miRNA miR396a-5p was downregulated, which repressed the expression of the AtGRF7 gene. In turn, this repression positively regulated the expression of AtDREB2a and AtRD29; the greater expression of the AtDREB2a and AtRD29 genes resulted in tolerance to salt stress. AtDREB2a and AtRD29 are discussed above. The expression of the miRNA ath-miR169g-5P also increased, and as a result, the expression of the target nuclear factors AtNFYA1 and AtNFYA2 increased. AtNFYA1 is associated with hypersensitivity to salt stress and ABA during the early stages of post-germination growth (Li et al., 2013). AtNFYA2 participates in nitrogen metabolism, regulation of light signaling, and chloroplast biogenesis (Laloum et al., 2013; Petroni et al., 2013). The expression of the miRNAs ath-miR399, ath-miR827, and ath-miR2111b as well as their target genes AtUBC24, AtWAK2, AtSYG1, and At3g27150 was also altered, suggesting a role of the extract of A. nodosum in phosphate homeostasis (Liu et al., 2012; Shukla et al., 2018).
Using A. thaliana plants under salinity stress (150 mM NaCl), researchers (Jithesh et al., 2018) studied the effect of extracts of A. nodosum subfractionated with ethyl acetate. The transcriptome of the plants was analyzed under salinity and in response to the application of the extract on the first and fifth days after salinity. On the first day, the expression of the LEA3, CCA1, LEA1, HVA22d, LTP6, ATGOLS3, ATGOLS2, DREB1A, and DREB1C genes increased. The products of LEA genes belong to a family of hydrophilic proteins with a protective function of functional proteins (Tunnacliffe and Wise, 2007) to prevent their aggregation (Chakrabortee et al., 2007). The LEA3 gene is induced by drought and salinity (Du et al., 2016), and LEA1 is induced by wounds and mild stress (Dunaeva and Adamska, 2001). CCA1 has a regulatory function in the circadian rhythm and controls various processes, such as the stomatal opening (Hassidim et al., 2017). HVA22d is induced by ABA and by stress and is related to the regulation of autophagy (Chen et al., 2002, 2009). LTP6 is predicted to encode a protein related to pathogenesis (PR) (Le et al., 2014). ATGOLS3 and ATGOLS2 encode the enzyme galactinol synthase, which is key to the biosynthesis of the oligosaccharides of the raffinose family and are related to tolerance to drought, salinity, and cold stress (Taji et al., 2002; Nishizawa et al., 2008). DREB1A and DREB1C are TFs of the DREB family, which is discussed above. On the fifth day after salinity, the expression of the LEA1, LEA2, VA22b, Di21, SnRK2, CIPK25, LTP, RING, and MYB genes increased. VA22b and Di21 regulate stress and are induced by ABA (Jithesh et al., 2018). SnRK2 is discussed above. CIPK25 is a serine-threonine protein kinase that interacts with calcineurin (a calcium sensor); both of these are related to signal transduction in response to environmental stress (Kanwar et al., 2014). LTP is discussed above. RING and MYBs are TFs; RING acts as a ligase and participates in the regulation of gene expression in response to environmental or hormone signals (Qin et al., 2008), and MYBs are discussed above.
Using Asparagus aethiopicus plants subjected to salinity stress (2,000 and 4,000 mg L–1 NaCl), researchers (Al-Ghamdi and Elansary, 2018) studied the synergistic effects of the application of a commercial product based on A. nodosum and 5-aminolevulinic acid (ALA) applied via foliar treatment. ALA is a precursor of porphyrins, so it has effects on the photosynthetic apparatus, thus stimulating the defense system in plants (Wu et al., 2018). The application of both products caused a synergistic effect on plant growth associated with increased expression of the ANN1, ANN2, and PIP1 genes, which are associated with the transport of water and Ca+2; the P5CS1 and CHS genes, which are related to the production of secondary metabolites; and the APX1 and GPX3 genes, which are associated with antioxidant metabolism in plants. ANN1 and ANN2 encode a family of proteins called annexins, which are Ca+2 transporter permeases attached to the plasma membrane. They participate in processes such as stomatal closure, adaptation to stress, and cell signaling (Laohavisit et al., 2012; Wang J. et al., 2018). PIP1 and P5CS1 are discussed above. CHS encodes a chalcone synthase, which is a key enzyme involved in flavonoid biosynthesis and is also involved in auxin transport (Brown et al., 2001; Dana et al., 2006). APX1 and GPX3 play a role in antioxidant metabolism, and GPX3 acts as a redox transducer whose function is similar to that of APX1 in H2O2 homeostasis and is related to ABA signal transduction during stress (Miao et al., 2006).
Using pea plants under salinity stress (150 mM NaCl for 2 weeks), researchers (Desoky et al., 2019) studied the effect of applying licorice (Glycyrrhiza glabra) root extract on pea seeds. Saline stress reduced seedling growth and increased oxidative stress; however, in pretreated seedlings, mitigation of these effects was observed. Treatment with the extract also increased the transcription of the CAT, SOD, APX, GR, DHAR, and PrxQ genes, decreasing oxidative stress. CAT, SOD, APX, GR, DHAR, and PrxQ (peroxiredoxin) participate in the cellular antioxidant system, which maintains ROS homeostasis to mitigate oxidative damage. However, ROS are essential for maintaining metabolic flow and activating acclimation responses to stress through systemic signaling (Ahmad et al., 2010; Suzuki et al., 2012).
Using wild rocket (Diplotaxis tenuifolia L.) plants under salinity stress (200 mM NaCl), researchers (Franzoni et al., 2019) studied the effect of a foliar application of a borage (Borago officinalis) extract. The expression of several TFs related to salinity stress was studied at 2, 4, 6, 9, and 24 h after exposure to stress. TFs such as DtDREB2A, DtMYB30, DtNAC019, DtNAC72, DtNAC19, DtNAC69, DtZIP63, DtABF3, DtHB12, and DtHB7 presented positive regulation. DtDREB2A is discussed above. DtMYB30 is an ABA-sensitive TF that participates in processes such as germination and response to stress (Zheng et al., 2012b). DtNAC019, DtNAC72, DtNAC19, and DtNAC69 compose a family of TFs related to development and stress responses in plants, respond to ABA, and promote the antioxidant system (Xu et al., 2015). DtZIP63 is a TF that regulates the circadian cycle through a low-energy response and is activated by a kinase (SnRK1) (Frank et al., 2018). DtABF3 is a TF induced by ABA and osmotic stress (Bogamuwa and Jang, 2014), and DtHB12 and DtHB7 are ABA-dependent TFs and act by mediating the growth response to water stress (Olsson et al., 2004).
Using cucumber plants (Cucumis sativus L.) subjected to heat stress (35°C), researchers (Campobenedetto et al., 2020) studied the effect of a seed application of a biostimulant based on lignin derivatives and containing plant-derived amino acids and molybdenum (KIEM®). The application of the biostimulant increased the percent germination, fresh biomass, and increased in expression levels RBOHD, CuZnSOD, MnSOD, CAT, and GST genes, while FeSOD gene was decreased. CuZnSOD, MnSOD, CAT, GST, and FeSOD are ROS-scavenging enzymes and are discussed above. RBOHD is a ROS-producing enzyme, H2O2 is a relatively long-lived ROS, and its accumulation is caused by the induction of membrane-bound respiratory burst oxidase homolog proteins (Rboh), which are important players in abiotic stress responses (Huang et al., 2014).
The effect of Moringa oleifera leaves extract applied on Ocimum basilicum plants under salt stress (1,000 mg L–1) was studied (Alkuwayti et al., 2020). The application of M. oleifera extract altered the expression of ObOLP gene and was positively correlated with the plant growth and yield enhancement. ObOLP is a osmotin-like proteins, members of the pathogenesis-related protein 5 (PR-5), which are produced in plants under different abiotic and biotic stresses (Mayer Weber et al., 2014). Under salinity and drought stress, OLP maintains cellular osmolarity by compartmentalization of solutes or by structural and metabolic alterations (Chowdhury et al., 2017).
Examination of Table 3 allows determining a few of transcriptomic responses that were common among the 14 studies. Common genes in some studies were related to ABA, TFs, antioxidants, and LEA proteins. Most of the studies were carried out in A. nodosum in different types of stress. Regarding algae extracts, most of the studies were carried out on A. nodosum under different types of stress. However, in the case of botanical extracts, there are few related studies. More studies are needed with extracts of the same species of algal and botanical in different species of plants, under the same stress condition.
Protein Hydrolysates and Other Nitrogen-Containing Compounds
The two main categories of protein-based products are divided into: (1) protein hydrolysates consisting of mixtures of peptides and amino acids that can be of animal or plant origin and (2) individual amino acids such as proline, glutamate, glutamine, and glycine betaine (Calvo et al., 2014). Mixtures of amino acids and peptides can be obtained by chemical, enzymatic, or thermal hydrolysis from byproducts of plant and animal origin (Calvo et al., 2014; Jardin, 2015). Chemical synthesis can be used to produce amino acids or mixtures of these. This category also includes other nitrogenous molecules, such as polyamines, betaines, and non-protein amino acids (Vranova et al., 2011), which are diverse in higher plants but are poorly characterized in terms of their physiological and ecological functions (Vranova et al., 2011).
Genes and their function are described below based on the results of seven studies in which biostimulants based on protein hydrolysates and other nitrogen-containing compounds were applied to plants under abiotic stress (Table 4).
TABLE 4.
Description of genes whose transcript level changed in response to the application of biostimulants based on protein hydrolysates and other nitrogen-containing compounds to plants under abiotic stress.
| Gene/gene expression time | Biostimulant type | Abiotic stress type | Crop/gene expression organ | References |
| Solyc02g084840, Solyc03g025810/3 to 11 days after application of treatment | A mixture of amino acids, proteins, vitamins, and betaines | Drought | Tomato/leaves | Petrozza et al., 2014a |
| ZmPAL/12 days after stress | Hydrolysate of alfalfa plants, triacontanol, and indole 3-acetic acid | Salinity | Maize/leaves | Ertani and Schiavon, 2013 |
| ZmNRT2.1, ZmNRT2.2, ZmNRT2.3, ZmNAR2.2, ZmNRT1.1, ZmNRT1b, ZmNRT, ZmSOD1A, ZmRbohA, ZmRbohC/3 days after stress | Protein hydrolysate | Hypoxia, salinity, nutrient deficiency | Maize/roots | Trevisan et al., 2019 |
| BI-1, MAPK1, WRKY53, CAT1, GPX, SOD, ATG1, ATG2, ATG4, ATG6, ATG7, ATG8, ATG9, ATG10, ATG13/not specified | Panchagavya | Salinity | Rice/leaves | Khan et al., 2018 |
| CAT, MnSOD, WRKY53, BI-1/not specified | Panchagavya | Salinity | Rice/leaves | Khan et al., 2017 |
| Cla018095, Cla010664, Cla004567, Cla009820, Cla012125, Cla003187, Cla021039, Cla007826, Cla013224, Cla001877, Cla001590, Cla006037/not specified | Melatonin | Vanadium toxicity | Watermelon/roots | Nawaz et al., 2018 |
| HEMA1, CHLH, POR/10 days after treatment | 5-Aminolevulinic acid | Salinity | Cucumber/roots | Wu et al., 2018 |
Using tomato plants under drought stress (no water after the development of the third or fourth leaf), researchers (Petrozza et al., 2014b) evaluated the foliar application of a commercial product based on a complex mixture of vitamins, amino acids, proteins, and betaines. The results indicated that, compared with the untreated plants, the plants treated with the product while under stress had higher growth and stress tolerance. Additionally, the expression of the Solyc02g084840 and Solyc03g025810 genes was studied for 14 days after treatment; these genes were expressed between days 3 and 11. The Solyc02g084840 gene is an ortholog of the Arabidopsis RAB18 gene that is discussed above. Solyc03g025810 is an ortholog of the RD29B gene, which encodes a protein that is induced in response to water deprivation, low temperature, salinity, and desiccation, and its response is mediated by ABA (Bihmidine et al., 2013).
Using maize plants under salinity stress (25, 75, and 150 mM NaCl), researchers (Ertani and Schiavon, 2013) evaluated a biostimulant based on a hydrolysate of alfalfa (Medicago sativa L.) plants, triacontanol (TRIA), and indole-3-acetic acid (IAA) added to the irrigation water for 48 h. Compared with untreated plants under stress, the treated plants under stress had greater biomass and presented greater activity of enzymes related to nitrogen metabolism. Plants treated with the biostimulant while under one of 3 levels of salinity stress presented increased expression of the ZmPAL gene at 12 days after stress. The ZmPAL gene encodes the enzyme phenylalanine ammonium lyase (PAL), which is a key enzyme that catalyzes the first step of the phenylpropanoid pathway, producing precursors of a wide variety of vital secondary metabolites related to plant defense, such as lignin, flavonoids, isoflavonoids, coumarin, and stilbenes (Huang et al., 2010).
Using maize plants treated with a commercial biostimulant based on a protein hydrolysate added to the hydroponic solution, researchers (Trevisan et al., 2019) evaluated the tolerance to three types of abiotic stress: hypoxia (deprivation of air bubbles in the liquid hydroponic solution), salinity (25 mM NaCl), nutrient deficiency (only distilled water was supplied in the hydroponic solution) and the combination of these. The treated plants had increased root and shoot growth and increased tolerance to single and combined stress conditions. Additionally, genes related to nitrate transport (ZmNRT2.1, ZmNRT2.2, ZmNRT2.3, ZmNAR2.2, ZmNRT1.1, ZmNRT1b, and ZmNRT) and ROS metabolism (ZmSOD1A) were expressed. The ZmNRT2.1, ZmNRT2.2, ZmNRT2.3, and ZmNAR2.2 genes belong to the high-affinity nitrate transport system, and the ZmNRT1.1, ZmNRT1b, ZmNRT genes belong to the low-affinity nitrate transport system. An increase in the expression of these genes occurs since the application of protein hydrolysates in plants can modulate the expression of critical genes involved in the assimilation of nitrogen (transporters) (Sestili et al., 2018). ZmSOD1A is discussed above.
Using two rice cultivars (one susceptible and one tolerant to salinity) under salinity stress (100 mM NaCl), researchers (Khan et al., 2018) studied the effects of a natural biostimulant called panchagavya (a mixture of milk, butter, curd, urine, and cow dung) applied as a soil drench. The results indicated that the treated plants under salinity stress showed an improvement in the physiological and biochemical characteristics and presented increased expression of several genes: the expression of BI-1, MAPK1, and WRKY53 increased in the tolerant variety; the expression of CAT1, GPX, and SOD increased in the susceptible variety; and the expression of ATG1, ATG2, ATG4, ATG6, ATG7, ATG8, ATG9, ATG10, and ATG13 increased in both varieties. The BI-1 gene is expressed during senescence and under various stress conditions, and its expression gradually decreases throughout the cell death process (Ishikawa et al., 2015). MAPK1 belongs to a family of kinases (mitogen-activated protein kinases) that regulate the response to abiotic and biotic stress via signaling cascades (Neupane et al., 2019). WRKY53 is an early response factor to water deficit; its expression regulates the stomatal response (Sun and Yu, 2015). The CAT1, GPX, and SOD genes are discussed above. The ATG1, ATG2, ATG4, ATG6, ATG7, ATG8, ATG9, ATG10, and ATG13 genes are related to the autophagy process; together with PCD, this process regulates responses to stress since it is essential to degrade oxidized proteins during oxidative stress (Xiong et al., 2007).
The effect of the panchagavya biostimulant amendment to the soil drench applied to rice plants under salinity stress (100 mM NaCl) has been studied previously (Khan et al., 2017), and the results showed that stressed and treated plants presented upregulated CAT, MnSOD, WRKY53, and BI-1 gene expression. All these genes are discussed above.
In watermelon plants under stress from exposure to vanadium (V) (50 mg L–1), the application of melatonin added to the irrigation solution was studied (Nawaz et al., 2018). The results indicated that treatment with melatonin reduced the concentrations of V in the leaves and stems and reduced the concentrations of H2O2 and malondialdehyde (MDA). The expression of the Cla018095, Cla010664, Cla004567, Cla009820, Cla012125, Cla003187, Cla021039, Cla007826, Cla013224, Cla001877, and Cla001590 genes also increased. The Cla018095 gene is related to chlorophyll biosynthesis. Cla010664 encodes an O-methyl transferase, and Cla004567 encodes an S-methyl transferase, which participates in the biosynthesis of melatonin. Cla010664 can eliminate ROS or activate antioxidant enzymes such as SOD and CAT (Manchester et al., 2015). The Cla009820 (superoxide dismutase), Cla012125 (superoxide dismutase), Cla003187 (peroxidase), Cla021039 (glutathione peroxidase), Cla013224 and Cla007826 (glutathione S-transferase) genes encode enzymes with antioxidant activity. Cla001877 is a respiratory burst oxidase and can integrate Ca+2 signaling and protein phosphorylation with ROS production, the last being key to the regulation of growth, development, responses to environmental stimuli, and cell death (Suzuki et al., 2011). Cla001590 encodes a V-dependent haloperoxidase that may be related to the absorption of inorganic forms of iodine in plants, although its function has not been fully defined (Smoleñ et al., 2019).
Using cucumber plants under salinity stress (50 mmol L–1 NaCl), researches (Wu et al., 2018) evaluated a biostimulant based on ALA applied via foliar sprays. Plants exhibited increased photosynthesis (increased plant height and leaf area, increased gas exchange capacity, increased the use of light by photosystem II and improved chlorophyll biosynthesis) in response to the application of ALA and low stress conditions. There was an increase in the expression of the HEMA1, CHLH, and POR genes at 10 days after treatment. HEMA1 encodes glutamyl-tRNA reductase, CHLH encodes Mg-chelatase, and POR encodes protochlorophyllide oxidoreductase. All three genes are involved in chlorophyll biosynthesis (Stephenson and Terry, 2008). As the chlorophyll content increased in response to the application of ALA, the tolerance of cucumber plants to stress also increased.
The review of Table 4 reveals a small number of studies available (7). Except for the expression of antioxidant enzymes, where there were analogous responses between the different species studied, more studies are needed with the same proteins, hydrolysates, and other nitrogen-containing compounds in different species of plants, under the same stress condition.
Humic Acid and Fulvic Acid
Humic substances are natural components of soil organic matter resulting from the decomposition of animal, plant, and microbial waste (Jardin, 2015). They are considered the main components of soil organic matter and are the most abundant natural organic compounds on Earth (Calvo et al., 2014). Humic substances are heterogeneous compounds classified by their molecular weight and solubility in (1) humic acids, which are soluble in basic media, (2) fulvic acids, which are soluble in alkaline and acidic media, and (3) humins, which are not extractable from the soil (Berbara and García, 2014). In addition to their use as biostimulants, humic substances are related to key processes in the soil and plants, such as carbon and oxygen exchange between the soil and the atmosphere, availability of nutrients, and the detoxification and transport of toxic substances (Piccolo and Spiteller, 2003).
In this category, six studies related to humic and fulvic acid substances was found, where gene expression was analyzed (Table 5).
TABLE 5.
Description of genes whose transcript level changed in response to the application of biostimulants based on humic acids and chitosan and other biopolymers to plants under abiotic stress.
| Gene/gene expression time | Biostimulant type | Abiotic stress type | Crop/gene expression organ | References |
| ZmPAL1/48 h after treatment | Hummus of Nicodrilus caliginosus | None | Maize/leaves | Schiavon et al., 2010 |
| HSP101, HSP81.1, HSP26.5, HSP23.6, HSP17.6A/9h after application of humic acid | Humic acids | Heat | Arabidopsis/not specified | Cha et al., 2020 |
| SnRK2.2, MDH, WRKY DNA-binding transcription factors family/6 days after stress | Humic acids (vermicompost) | Weak acids | Maize/root | Baía et al., 2020 |
| SETIT_021707mg, SETIT_016840mg, SETIT_ 015030mg, SETIT_004913mg, SETIT_016654mg/5 days after stress | Humic acid | Drought | Foxtail millet/leaves | Shen et al., 2020 |
| GME, AO, ALDH, GST, G6PDH, CYS-GYL, C4H, CHS, F3′5′H, F3H/4 and 8 days under stress | Fulvic acid | Drought | Camellia sinensis/shoots | Sun et al., 2020 |
| CCoAOMT, CAB37, AGT1, At4g26520/0, 4, 8, and 12 days after stress | Fulvic acid | Drought | Paeonia ostii/leaves | Fang et al., 2020 |
| SOD, JA/48 h after treatment | nCu-chitosan-PVA and chitosan-PVA complex | Salinity | Tomato/leaves | Hernández-Hernández et al., 2018 |
| AOX1/3 days after treatment | Chitosan | Salinity | Maize/leaves | Turk, 2019 |
| CAT3/24 h after treatment | Xyloglucan oligosaccharides | Salinity | Arabidopsis/leaves | González-Pérez et al., 2018 |
| GH3/24 h after treatment | Chitosan oligosaccharide | Cold | Camellia sinensis/leaves | Li et al., 2020e |
| CAT, APX, POD, SOD, GmSALT3, CHS/12 days of growth | Chitosan modified biochar | Salinity | Soybean/plant tissue | Mehmood et al., 2020 |
Using maize plants, researchers (Schiavon et al., 2010) evaluated a high-molecular-weight (>3,500 Da) humic fraction from Nicodrilus caliginosus feces, which was added for 48 h hydroponically. The effect on phenylpropanoid metabolism was subsequently evaluated, where the expression of the ZmPAL1 gene increased considerably in plants treated with the humic fraction at three different concentrations. The ZmPAL1 gene is discussed above.
Using Arabidopsis plants under heat stress (45°C), the application of humic acid (commercial product) was studied (Cha et al., 2020). The authors performed a transcriptomic analysis to identify the HA-prompted molecular mechanisms. Gene ontology analysis indicated that humic acid up-regulates diverse genes related in the response to stress. Heat stress causes induction in gene families such as heat-shock protein (HSP), coding genes including HSP101, HSP81.1, HSP26.5, HSP23.6, and HSP17.6A. HSPs function as molecular chaperones to protect against thermal denaturation of substrates and stimulate refolding of denatured substrates, also play an important role in maintaining cell membrane integrity, ROS scavenging and production of antioxidants, osmolytes (Khan and Shahwar, 2020).
Using maize seedlings under weak acids stress (acetic and salicylic acids), the application of humic acids extracted from vermicompost produced with cattle manure was studied (Baía et al., 2020). Humic acids decrease the intracellular pH and produce high level of SnRK2.2 and MDH genes, and low level of WRKY TFs family. SnRK2.2 is discussed above. MDH encodes malate dehydrogenase, this enzyme is critical in malate metabolism and is related in ROS producing genes (Akbar et al., 2020). WRKY TF family are associated to transduction of stress signaling and play a major role in plant defense to abiotic and biotic stress (Li et al., 2020c).
The effect of humic acid in foxtail millet plants under drought conditions (natural simulation conditions) was studied (Shen et al., 2020). Transcriptome sequencing and RT-qPCR was performed on plants. Humic acid caused a significant increase in the yield, dry weight and root-shoot ratio. SETIT_021707mg, SETIT_016840mg, and SETIT_ 015030mg genes were significantly up-regulated, while SETIT_004913mg and SETIT_016654mg genes were significantly down-regulated in the plants treated with humic acid and drought. These genes are related with metabolic pathways, secondary metabolite biosynthesis and starch and sucrose metabolism.
The effect of fulvic acid in tea plants (Camellia sinensis) under drought stress was studied (Sun et al., 2020). The authors examined the transcriptomics and metabolomics profiles, 604 and 3331 differentially expressed metabolite genes (DEGs) were found in plants at 4 and 8 days under drought respectively. DEGs are related in diverse biological processes such as ascorbate metabolism (GME, AO, ALDH), glutathione metabolism (GST, G6PDH, CYS-GYL), and flavonoids biosynthesis (C4H, CHS, F3′5′H, F3H). Ascorbic acid functions as an enzymatic cofactor and antioxidant plays roles in maintenance of ROS homeostasis (Conklin and Barth, 2004). Glutathione is one of the important endogenous antioxidants in plants, which functions as a substrate in antioxidative defense mechanisms by scavenging free radicals, conjugating to toxic electrophilic compounds, and reducing peroxides (Anderson and Davis, 2004). Flavonoids are polyphenol compounds with antioxidant activities, the accumulation of flavonoids could be a key step in development of plant tolerance to different stresses (Wang P. et al., 2018).
Using Paeonia ostii plants under natural drought stress (it was mainly characterized by low soil water content, and the roots of plants cannot absorb enough water to compensate for the consumption of transpiration) the effect of fulvic acid was studied (Fang et al., 2020). The fulvic acid treatment increased the leaf water content and antioxidant enzyme activities and decrease proline content, ROS accumulation, and relative electrical conductivity. Also, increased the expression level of drought-tolerant genes, like CCoAOMT, CAB37, AGT1, At4g26520. CCoAOMT ecodes caffeoyl-coenzyme A O-methyltransferase, this enzyme is involved in monolignol synthesis that affects the efficiency of lignification and lignin composition (Rakoczy et al., 2018), and the changes of lignin composition may serve in stress resistance. CAB37 is a important regulatory site of photosynthesis under drought stress (Li et al., 2020d). AGT1 is related to photosynthetic processing and photorespiration (Fang et al., 2020). At4g26520 is ABA dependent signaling pathway in drought response (Fang et al., 2020).
Examination of Table 5 allows determining the absence of transcriptomic responses that were common among the six studies of humic and fulvic acids. One possible explanation, in addition to the small number of studies available, is the presence of chemical or physicochemical differences between the humic substances, or the fact that not all the studies were carried out under the same type of stress.
Chitosan and Other Biopolymers
Chitosan is a polymer obtained by the deacetylation of chitin extracted from crustaceans, fungi, and insects. Chitosan is composed of N-acetyl-D-glucosamine and D-glucosamine units that have different degrees of deacetylation (Riaz Rajoka et al., 2020). The physiological effects of chitosan in plants are related to the ability of this polycationic compound to bind to a wide range of cellular components, such as the plasma membrane, cell wall components, and DNA, in addition to binding to specific receptors involved in plant defense (Katiyar et al., 2015). Other natural and synthetic polymers can be used in agriculture, including polyacrylates, polyacrylamides, and polysaccharides (Ekebafe et al., 2011).
Genes and their function are described below on the basis of the results of five studies in which chitosan-based biostimulants and other biopolymers were applied to plants under abiotic stress (Table 5).
Using tomato plants, researches (Hernández-Hernández et al., 2018) studied the application of hydrogels of nCu-chitosan-PVA and chitosan-PVA as promoters of tolerance to salt stress (100 mM NaCl). The treated and stressed plants presented improved growth and increased expression of JA and SOD genes at 48 h after stress. The JA gene was related to the biosynthesis of jasmonic acid, which has been shown to improve tolerance to osmotic and oxidative stress in plants under salinity stress (Zhao et al., 2014). SOD is discussed above.
Using maize plants under salinity stress (100 mM NaCl), researchers (Turk, 2019) studied the ability of foliar applications of chitosan (0.1%) to mitigate this stress. The treated plants presented increased growth and higher expression of the AOX1 gene on the third day after the treatment; the AOX1 gene encodes the mitochondrial alternative oxidase enzyme. This enzyme is involved in the alternative dissipative flow of the electron transport chain, in addition to optimizing the metabolism of respiration under normal and stress conditions (Erdal and Turk, 2016). AOX1 also plays an essential role in modulating the balance between carbon and nitrogen (Hu et al., 2017).
Using A. thaliana seedlings grown in vitro under salinity stress (100 mM NaCl), researchers (González-Pérez et al., 2018) studied the effect of a biostimulant based on xyloglucan oligosaccharides extracted from Tamarindus indica L. (0.1 mg L–1) applied to the growth media. An increase in the expression of the CAT3 gene was reported 24 h after stress, and the CAT gene is discussed above.
The effect of chitosan oligosaccharide (COS) on tea plants (C. sinensis) under cold stress (−4°C) was studied (Li et al., 2020e). The activity of SOD and POD, content of soluble sugar and chlorophyll in COS-treated tea plant were increased. The tea plants also were analyzed by transcriptomics with RNA-sequencing. There were identified 4503 DEGs between the control and COS under cold stress. By RT-qPCR, the GH3 gene expression was significantly higher in COS-treated plants (under stress). GH3 (indole-3-acetic acid) is an important response gene of auxin-responsive protein, encode a class of IAA-amido synthetase related for balancing endogenous free IAA content, and play an important role in plant growth and development (Feng et al., 2015).
Using soybean under salt-stress, the effect of chitosan modified biochar (CMB) was studied (Mehmood et al., 2020). CMB treatment with salt-stress increased plant growth, root architecture characteristics, biomass yield, nutrients acquisition, chlorophyll, soluble protein and sugar contents. The gene expression levels triggered by salinity but with the application of CMB significantly increased the expression profile of CAT, APX, POD, SOD, GmSALT3, and CHS genes. CAT, APX, POD, and SOD encodes antioxidant enzyme and are discussed above. GmSALT3 (salt tolerance-associated gene on chromosome 3) is associated with limiting the accumulation of sodium ions in shoots and a substantial enhancement in salt tolerance in soybean (Guan et al., 2014). CHS encode a chalcone synthase, this enzyme is the first key enzyme in the biosynthesis of flavonoids (El-Esawi et al., 2018a).
The review of Table 5 allows determining the absence of transcriptomic responses that were common among the five studies of chitosan and other biopolymers, except for the expression of antioxidant enzymes. More studies are needed with the same chitosan and others biopolymers in different species of plants, under the same stress condition.
Inorganic Compounds
Beneficial elements are chemical elements that are not essential for all plants and are related to growth promotion (Pilon-Smits et al., 2009). The main beneficial elements include Si, Se, Co, Al, and Na. These are present in soils and in plants as different inorganic salts and as insoluble forms (Jardin, 2015).
The following describes the genes and their function obtained from the results of six studies in which biostimulants based on inorganic compounds and mixtures with other types of biostimulants were applied to plants under abiotic stress (Table 6).
TABLE 6.
Description of genes whose transcript level changed in response to the application of biostimulants based on inorganic compounds and mixtures to plants under abiotic stress.
| Gene/gene expression time | Biostimulant type | Abiotic stress type | Crop/gene expression organ | References |
| OsDREB2A, OsNAC5, OsRDCP1, OsCMO, OsRab16b/not specified | Selenium and silicon | Drought | Rice/seedlings | Khattab et al., 2014 |
| SlPIP1;5, SlPIP2;6/3 and 5 days after treatment and stress | Silicon | Drought | Tomato/roots | Shi et al., 2016 |
| SbPIP1;3/1;4, SbPIP1;6, SbPIP2;2, SbPIP2;6, SbPIP2;3/4 and 24 h after stress | Silicon | Drought | Sorghum/roots | Liu P. et al., 2014 |
| MT2a, MT2b, PCS1, CSD1, CSD2/3 days after treatment | Silicon | Copper toxicity | Arabidopsis/leaves | Khandekar and Leisner, 2011 |
| Os08g02630 (PsbY), Os05g48630 (PsaH), Os07g37030 (PetC), Os03g57120 (PetH), Os09g26810, Os04g38410/72 h after treatment | Silicon | Zinc toxicity | Rice/leaves | Song et al., 2014 |
| RAB18, P5CS1, ERF1A, MYB75/field capacity 0.1 (10%) | Three different biostimulants (A. nodosum extract, amino acids, and potassium phosphite) | Drought | Arabidopsis/leaves | Fleming et al., 2019 |
Using two rice cultivars under drought stress (no irrigation for 10 days), researchers (Khattab et al., 2014) studied the application of sodium selenate (0.03 mM) and potassium silicate (1.5 mM) to the seeds to promote stress tolerance. Increased expression of genes such as DREB2A, NAC5, RDCP1, CMO, and RAB16b was reported. DREB2A and NAC5 genes encode TFs. NAC5 TFs activate genes related to the production of osmolytes, redox homeostasis, detoxification, and formation of macromolecules (Hu et al., 2008). RDCP1 participates in a set of physiological responses to counteract dehydration stress (Bae et al., 2011). CMO encodes a choline monooxygenase that participates in glycine betaine biosynthesis, resulting in increased tolerance to salinity stress (Luo et al., 2012). RAB16B encodes the dehydrin-like LEA protein, whose expression which is induced in response to abiotic stress, and this protein is ABA dependent (Bies-Ethève et al., 2008).
Using tomato plants under drought stress (induced with 10% polyethylene glycol-6,000 added to the irrigation solution), researchers (Shi et al., 2016) studied the effect of potassium silicate (2.5 mM) added to the irrigation solution. The results showed that Si improved the growth, photosynthesis, and water status of plants under stress. The expression of the SIPIP1;5 and SIPIP2;6 genes increased at 5 and 6 days, respectively. Both genes are discussed above.
Using sorghum plants, researchers (Liu P. et al., 2014) studied the effect of applying sodium silicate (1.67 mM) to counteract water-deficit stress (10% PEG-6000). Increased expression of SbPIP1;3/1;4, SbPIP1;6, SbPIP2;2, SbPIP2;6, and SbPIP2;3 was reported at 4 and 24 h after stress, and this group of genes encodes aquaporins and is discussed above.
Using A. thaliana plants under stress caused by copper toxicity (30 μM), researchers (Khandekar and Leisner, 2011) evaluated the effect of potassium metasilicate (1.5 mM). The MT2a, MT2b, PCS1, CSD1, and CSD2 genes increased at 3 days after treatment. The MT2a and MT2b genes encode metallothioneins, which are proteins that bind to Cu to regulate its concentration in the cell (Guo et al., 2008). PCS1 encodes a phytochelatin, which, like metallothioneins, binds to heavy metals and transports them to the vacuole (Adams et al., 2011). CSD1 and CSD2 encode the Cu/Zn SOD enzymes and are discussed above.
Using rice plants, researchers (Song et al., 2014) studied the effect of silicic acid (1.5 mM) added to the irrigation solution to increase stress tolerance caused by Zn toxicity (2 mM irrigation solution). The levels of the genes Os08g02630 (PsbY), Os05g48630 (PsaH), Os07g37030 (PetC), Os03g57120 (PetH), Os09g26810, and Os04g38410 increased in response to the application of Si and stress at 72 h after the start of the treatments. The Os08g02630 (PsbY) gene encodes a photosystem II polyprotein, which, when its expression increases, subsequently increases both the activity of photosystem II and the amount of chlorophyll (Von Sydow et al., 2016). Os05g48630 (PsaH) encodes a photosystem I subunit (Zhang and Scheller, 2004), and Os07g37030 (PetC) encodes a polypeptide that binds to the Rieske FeS center of cytochrome bf and regulates the electron transport of the b6f complex during photosynthesis (C Breyton et al., 1994). Os03g57120 (PetH) encodes a ferredoxin-NADP+ reductase, which regulates the level of reduced glutathione in the cell (Song et al., 2014).
Using Arabidopsis plants under drought stress, researchers (Fleming et al., 2019) evaluated three commercial products based on A. nodosum, amino acids, and potassium phosphite. An increase in the expression of the RAB18, P5CS1, ERF1A, and MYB75 genes was reported in response to the application of the products and at a field capacity of 0.1 (10%). The ERF1A gene encodes a TF of the ERF (ethylene response factor) family, with ethylene being a highly important hormone in plants (Trujillo-Moya and Gisbert, 2012). The other genes are discussed above.
Examination of Table 6 allows determining a few transcriptomic responses that were common among the six studies. Common genes in some studies were related to TFs, antioxidants and aquaporins. More studies are needed with the same inorganic compound in different species of plants, under the same stress condition.
As a summary of the collection of all the genes differentially expressed in response to the application of biostimulants to plants under abiotic stress, Figure 1 shows an outlook where the different types of biostimulants are presented according to the categorization described in du Jardin (2015). In addition to the 7 categories, mixtures based on 2 or more different types of biostimulants were added. Subsequently, the 7 different types of abiotic stress described in this review are shown. Finally, under each type of stress, DEGs are listed that may be part of the transcriptional modification of plants and may be involved in abiotic stress tolerance. Biostimulants based on algal and botanical extracts have been extensively studied in plants under different types of abiotic stress, such as drought, salinity, cold and heat. On the other hand, biostimulants based on protein hydrolysates and compounds with N have been studied in response to only various types of abiotic stress, such as drought, salinity, metal toxicity, and nutrient deficiency. The most studied is drought stress, followed by salinity, metal toxicity, low and high temperature, weak acids, and nutrient deficiency. However, considering that high-temperature stress (in the form of heatwaves) has become increasingly common in tropical and subtropical regions as climate change progresses, it is necessary to carry out additional studies to verify the usefulness of different biostimulants under extreme temperature conditions.
FIGURE 1.
Global view of the differentially expressed genes in plants treated with different types of biostimulants that are involved in the tolerance of various types of abiotic stress.
In the present review, emphasis was placed on presenting DEGs in plants treated with biostimulants, as well as presenting the cellular function of these genes to try to decipher the transcriptional mechanism underlying the tolerance to abiotic stress. Figure 2 depicts how biostimulants are perceived in cells through receptors. Such recognition triggers a series of downstream events where kinase-like proteins and TFs (regulatory proteins) drive signal transduction, after which specific response genes are ultimately activated to produce functional proteins to counteract the effects caused by abiotic stress. In the genes presented in Figure 2, overlap in the functional categories of functional-type proteins can be observed, in which some genes are homologous since they have the same function in different plant species. Some orthologous genes are also presented in different plant species with similar functions due to a common ancestor. However, to support this information, it is necessary to carry out a phylogenetic analysis of the sequences. Among the functional categories that present the most DEGs are proteins with a role in antioxidant metabolism, followed by functions involving photosynthesis, biosynthesis and signaling of ABA, PCD, aquaporins, osmoprotectants, LEA-like proteins, nitrogen metabolism, heat-shock proteins, biosynthesis of phytohormones, phosphorus metabolism, PR proteins, secondary metabolite biosynthesis, metal detoxification, flavonoids metabolism, photoprotection, carbohydrate metabolism, Ca+2 transport, ascorbate metabolism, glutathione metabolism, stomatal regulation, metabolism of phenylpropanoids, iron metabolism, ROS, cell wall, ribosomes, dehydrins, energy metabolism, senescence, and germination. It is difficult to know if the abovementioned categories correspond to the spectrum of biological responses induced by biostimulants or if the spectrum is an effect of the sampling of the works used in this study (a result of the choice of plant species, type of stress, and the genes chosen in the different works). More studies of specific metabolic pathways that represent plant status against stresses, for example, antioxidant metabolism, biosynthesis and signaling of ABA, PCD, photosynthesis, water transport, and C or N metabolism, are needed. A massive study of gene expression is also proposed through the analysis of transcriptomes or complete proteomes.
FIGURE 2.
Cellular representation of the functional categories of genes differentially expressed in plants in response to the application of different types of biostimulants. R: receptor, kin: kinases, P: phosphate groups, TF: transcription factor, HSP: heat-shock protein, PCD: programmed cell death, PHB: phytohormone biosynthesis, AQU: aquaporin, LEA: late embryogenesis abundant, PHO: photosynthesis, PP: photoprotection, ANTIOX: antioxidant metabolism, SEN: senescence, ABA: biosynthesis and signaling of ABA, FOS: phosphorous metabolism, SR: stomatal regulation, PR: PR protein, CM: carbohydrate metabolism, OP: osmoprotectant, TCa: CaC2 transport, RIB: ribosome, NM: nitrogen metabolism, DHN: dehydrin, PPM: phenylpropanoid metabolism, EM: energy metabolism, DM: metal detoxification, GER: germination, IM: iron metabolism, ROS: reactive oxygen species, SMB: secondary metabolite biosynthesis, AM: ascorbate metabolism, GM: glutathione metabolism, FB: flavonoids metabolism, CW: cell wall. Gene colors: beneficial bacteria, beneficial fungi, algal and botanical extracts, protein hydrolysates and N compounds, fulvic acids and humic acids, chitosan and other biopolymers, inorganic compounds, biostimulant mixtures.
With the information analyzed in this review, in Table 7 some putative molecular markers are proposed that can be used to test the potential of different types of biostimulants under different stress conditions. The genes selected as molecular markers are functional proteins that could be associated with tolerance to abiotic stress in plants.
TABLE 7.
Putative molecular markers for different potential biostimulants in varying abiotic stress conditions.
| Molecular markers | Function | Biostimulant type | Abiotic stress type |
| CAT, SOD, POD, APX, GPX, GR, CHS, CSD1, DHAR | Antioxidant metabolism | Beneficial bacteria, beneficial fungi, algal and botanical extracts, protein hydrolysates and N compounds, chitosan and other biopolymers, inorganic compounds | Drought, salinity, metal toxicity, nutrient deficiency, heat |
| HSP70 | Heat-shock protein (HSP) | Beneficial bacteria, algal and botanical extracts | Drought, salinity |
| PIP1, PIP2 | Aquaporin | Beneficial bacteria, beneficial fungi, algal and botanical extracts, inorganic compounds | Drought, salinity |
| ERD1, RAB18 | Late embryogenesis abundant (LEA) | Beneficial bacteria, algal and botanical extracts, mixtures of biostimulants | Drought, salinity |
| NCED | Biosynthesis and signaling of ABA | Beneficial bacteria, algal and botanical extracts | Drought, salinity |
| P5CS1 | Osmoprotectant | Beneficial bacteria, algal and botanical extracts, mixtures of biostimulants | Drought, salinity, cold, metal toxicity, heat |
| PAL | Phenylalanine ammonium lyase (PAL) | Protein hydrolysates and N compounds, fulvic and humic acids | Salinity |
Conclusion and Perspectives
Given the information described above, genes that can act as molecular markers in abiotic stress tolerance can be globally visualized by applying biostimulants. The genes described encode functional proteins as well as regulatory proteins.
Most related studies have focused on the use of algal extracts, especially those of A. nodosum, and the effects of drought and salinity stress have been the most explored. Thus, there is an area of opportunity to study the different types of biostimulants and abiotic stresses that have been studied little.
However, to focus efforts of future research on the analysis of DEGs in plants in response to the application of biostimulants (those with greater accessibility to producers such as humic substances, vegetable extracts, hydrolysates, and elements such as silicon), it is necessary to take into account several factors, such as the type of biological model (study species with high nutritional importance, such as potato, wheat, maize, rice and tomato), the class of biostimulant, the concentration of the biostimulant (focus on commercial recommendations), the type of application (depending on the crop species and the type of biostimulant), the study organ (include leaves and, if possible, the organ of commercial interest, such as the roots, tubers and fruits), and the time of analysis of gene expression (establish comparable standards of sampling times at 12, 24, and 36 h after the application of the biostimulant).
Author Contributions
AB-M: conceptualization. SG-M: writing of the original draft and editing. SG-M, SS-G, MV-C, AJ-M, AL-T, and AB-M: responsible for processing and organizing information. All authors have read and approved the final manuscript.
Conflict of Interest
SS-G and MV-C were employed by company UPL Mexico. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the UPL for funding to publish this review.
Footnotes
Funding. This research received external funding from the UPL Limited.
References
- Abd El-Daim I. A., Bejai S., Fridborg I., Meijer J. (2018). Identifying potential molecular factors involved in Bacillus amyloliquefaciens 5113 mediated abiotic stress tolerance in wheat. Plant Biol. 20 271–279. 10.1111/plb.12680 [DOI] [PubMed] [Google Scholar]
- Adams J. P., Adeli A., Hsu C.-Y., Harkess R. L., Page G. P., Depamphilis C. W., et al. (2011). Poplar maintains zinc homeostasis with heavy metal genes HMA4 and PCS1. J. Exp. Bot. 62 3737–3752. 10.1093/jxb/err025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adesemoye A., Torbert H., Ecology J. K.-A. S. (2010). Increased plant uptake of nitrogen from 15N-depleted fertilizer using plant growth-promoting rhizobacteria. Appl. Soil Ecol. 46 54–58. [Google Scholar]
- Ahmad P., Jaleel C. A., Salem M. A., Nabi G., Sharma S. (2010). Roles of enzymatic and nonenzymatic antioxidants in plants during abiotic stress. Crit. Rev. Biotechnol. 30 161–175. 10.3109/07388550903524243 [DOI] [PubMed] [Google Scholar]
- Ahn Y. O., Kim S. H., Lee J., Ran Kim H., Lee H. S., Kwak S. S. (2012). Three Brassica rapa metallothionein genes are differentially regulated under various stress conditions. Mol. Biol. Rep. 39 2059–2067. 10.1007/s11033-011-0953-5 [DOI] [PubMed] [Google Scholar]
- Akbar S., Wei Y., Yuan Y., Khan M. T., Qin L., Powell C. A., et al. (2020). Gene expression profiling of reactive oxygen species (ROS) and antioxidant defense system following Sugarcane mosaic virus (SCMV) infection. BMC Plant Biol. 20:532 10.1186/s12870-020-02737-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akpinar B. A., Avsar B., Lucas S. J., Budak H. (2012). Plant abiotic stress signaling. Plant Signal. Behav. 7 1450–1455. 10.4161/psb.21894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Ghamdi A. A., Elansary H. O. (2018). Synergetic effects of 5-aminolevulinic acid and Ascophyllum nodosum seaweed extracts on Asparagus phenolics and stress related genes under saline irrigation. Plant Physiol. Biochem. 129 273–284. 10.1016/j.plaphy.2018.06.008 [DOI] [PubMed] [Google Scholar]
- Alkuwayti M. A., El-Sherif F., Yap Y. K., Khattab S. (2020). Foliar application of Moringa oleifera leaves extract altered stress-responsive gene expression and enhanced bioactive compounds composition in Ocimum basilicum. S. Afr. J. Bot. 129 291–298. 10.1016/j.sajb.2019.08.001 [DOI] [Google Scholar]
- Anderson J. V., Davis D. G. (2004). Abiotic stress alters transcript profiles and activity of glutathione S-transferase, glutathione peroxidase, and glutathione reductase in Euphorbia esula. Physiol. Plant. 120 421–433. 10.1111/j.0031-9317.2004.00249.x [DOI] [PubMed] [Google Scholar]
- Antolín-Llovera M., Petutsching E. K., Ried M. K., Lipka V., Nürnberger T., Robatzek S., et al. (2014). Knowing your friends and foes – plant receptor-like kinases as initiators of symbiosis or defence. New Phytol. 204 791–802. [DOI] [PubMed] [Google Scholar]
- Arraes F. B. M., Beneventi M. A., Lisei de Sa M. E., Paixao J. F. R., Albuquerque E. V. S., Marin S. R. R., et al. (2015). Implications of ethylene biosynthesis and signaling in soybean drought stress tolerance. BMC Plant Biol. 15:213. 10.1186/s12870-015-0597-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson N., Leitão N., Orr D. J., Meyer M. T., Carmo-Silva E., Griffiths H., et al. (2017). Rubisco small subunits from the unicellular green alga Chlamydomonas complement Rubisco-deficient mutants of Arabidopsis. New Phytol. 214 655–667. 10.1111/nph.14414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert Y., Vile D., Pervent M., Aldon D., Ranty B., Simonneau T., et al. (2010). RD20, a stress-inducible caleosin, participates in stomatal control, transpiration and drought tolerance in Arabidopsis thaliana. Plant Cell Physiol. 51 1975–1987. 10.1093/PCP/PCQ155 [DOI] [PubMed] [Google Scholar]
- Bae H., Kim S. K., Cho S. K., Kang B. G., Kim W. T. (2011). Overexpression of OsRDCP1, a rice RING domain-containing E3 ubiquitin ligase, increased tolerance to drought stress in rice (Oryza sativa L.). Plant Sci. 180 775–782. 10.1016/j.plantsci.2011.02.008 [DOI] [PubMed] [Google Scholar]
- Baek D., Rokibuzzaman M., Khan A., Kim M. C., Park H. J., Yun D., et al. (2020). Plant-growth promoting Bacillus oryzicola YC7007 modulates stress-response gene expression and provides protection from salt stress. Front. Plant Sci. 10:1646. 10.3389/fpls.2019.01646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baía D. C., Olivares F. L., Zandonadi D. B., de Paula Soares C., Spaccini R., Canellas L. P. (2020). Humic acids trigger the weak acids stress response in maize seedlings. Chem. Biol. Technol. Agric. 7:31 10.1186/s40538-020-00193-5 [DOI] [Google Scholar]
- Bak S., Beisson F., Bishop G., Hamberger B., Höfer R., Paquette S., et al. (2011). Cytochromes P450. Arab. B 9:e0144. 10.1199/tab.0144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnam B., Iuchi S., Fujita M., Fujita Y., Takasaki H., Osakabe Y., et al. (2013). Characterization of the promoter region of an Arabidopsis gene for 9-cis-epoxycarotenoid dioxygenase involved in dehydration-inducible transcription. DNA Res. 20 315–324. 10.1093/dnares/dst012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbara R. L. L., García A. C. (2014). “Humic substances and plant defense metabolism,” in Physiological Mechanisms and Adaptation Strategies in Plants Under Changing Environment, eds Ahmad P., Wani M. R. (New York, NY: Springer; ), 297–319. [Google Scholar]
- Bies-Ethève N., Gaubier-Comella P., Debures A., Lasserre E., Jobet E., Raynal M., et al. (2008). Inventory, evolution and expression profiling diversity of the LEA (late embryogenesis abundant) protein gene family in Arabidopsis thaliana. Plant Mol. Biol. 67 107–124. 10.1007/s11103-008-9304-x [DOI] [PubMed] [Google Scholar]
- Bihmidine S., Lin J., Stone J. M., Awada T., Specht J. E., Clemente T. E. (2013). Activity of the Arabidopsis RD29A and RD29B promoter elements in soybean under water stress. Planta 237 55–64. 10.1007/s00425-012-1740-9 [DOI] [PubMed] [Google Scholar]
- Blunden G., Gordon S. M. (1986). Betaines and their sulphonio analogues in marine algae. Prog. Phycol. Res. 4 39–80. [Google Scholar]
- Bogamuwa S. P., Jang J.-C. (2014). Tandem CCCH zinc finger proteins in plant growth, development and stress response. Plant Cell Physiol. 55 1367–1375. 10.1093/pcp/pcu074 [DOI] [PubMed] [Google Scholar]
- Breyton C., de Vitry C., Popot J. L. (1994). Membrane association of cytochrome b6f subunits. The Rieske iron-sulfur protein from Chlamydomonas reinhardtii is an extrinsic protein. J. Biol. Chem. 269 7597–7602. [PubMed] [Google Scholar]
- Brown D. E., Rashotte A. M., Murphy A. S., Normanly J., Tague B. W., Peer W. A., et al. (2001). Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol. 126 524–535. 10.1104/pp.126.2.524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno L. B., Karthik C., Ma Y., Kadirvelu K., Freitas H., Rajkumar M. (2020). Amelioration of chromium and heat stresses in Sorghum bicolor by Cr6+ reducing-thermotolerant plant growth promoting bacteria. Chemosphere 244:125521. 10.1016/j.chemosphere.2019.125521 [DOI] [PubMed] [Google Scholar]
- Bulgakov V., Wu H.-C. (2019). Coordination of ABA and chaperone signaling in plant stress responses. Trends Plant Sci. 24 636–651. 10.1016/j.tplants.2019.04.004 [DOI] [PubMed] [Google Scholar]
- Calvo P., Nelson L., Kloepper J. W. (2014). Agricultural uses of plant biostimulants. Plant Soil 383 3–41. 10.1007/s11104-014-2131-8 [DOI] [Google Scholar]
- Campobenedetto C., Grange E., Mannino G., van Arkel J., Beekwilder J., Karlova R., et al. (2020). A biostimulant seed treatment improved heat stress tolerance during cucumber seed germination by acting on the antioxidant system and glyoxylate cycle. Front. Plant Sci. 11:836. 10.3389/fpls.2020.00836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J. (2019). Molecular evolution of the vacuolar iron transporter (VIT) family genes in 14 plant species. Genes 10:144. 10.3390/genes10020144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho H. H., Brustolini O. J. B., Pimenta M. R., Mendes G. C., Gouveia B. C., Silva P. A., et al. (2014). The molecular chaperone binding protein BiP prevents leaf dehydration-induced cellular homeostasis disruption. PLoS One 9:e86661. 10.1371/journal.pone.0086661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini N. M., Monteoliva M. I., Alvarez M. E. (2011). Proline dehydrogenase is a positive regulator of cell death in different kingdoms. Plant Signal. Behav. 6 1195–1197. 10.4161/psb.6.8.15791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha J., Kang S., Ali I., Lee S., Ji M., Reports S. J.-S., et al. (2020). Humic acid enhances heat stress tolerance via transcriptional activation of heat-shock proteins in Arabidopsis. Sci. Rep. 10 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabortee S., Boschetti C., Walton L. J., Sarkar S., Rubinsztein D. C., Tunnacliffe A. (2007). Hydrophilic protein associated with desiccation tolerance exhibits broad protein stabilization function. Proc. Natl. Acad. Sci. U.S.A. 104 18073–18078. 10.1073/pnas.0706964104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan P. S., Lata C., Tiwari S., Chauhan A. S., Mishra S. K., Agrawal L., et al. (2019). Transcriptional alterations reveal Bacillus amyloliquefaciens-rice cooperation under salt stress. Sci. Rep. 9 1–13. 10.1038/s41598-019-48309-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. N., Chu C. C., Zentella R., Pan S. M., Ho T. H. D. (2002). AtHVA22 gene family in Arabidopsis: phylogenetic relationship, ABA and stress regulation, and tissue-specific expression. Plant Mol. Biol. 49 633–644. 10.1023/A:1015593715144 [DOI] [PubMed] [Google Scholar]
- Chen C. N. N., Chen H. R., Yeh S. Y., Vittore G., Ho T. H. D. (2009). Autophagy is enhanced and floral development is impaired in AtHVA22d RNA interference Arabidopsis. Plant Physiol. 149 1679–1689. 10.1104/pp.108.131490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Liu Y., Wu G., Veronican Njeri K., Shen Q., Zhang N., et al. (2016). Induced maize salt tolerance by rhizosphere inoculation of Bacillus amyloliquefaciens SQR9. Physiol. Plant. 158 34–44. 10.1111/ppl.12441 [DOI] [PubMed] [Google Scholar]
- Choi D. S., Hong J. K., Hwang B. K. (2013). Pepper osmotin-like protein 1 (CaOSM1) is an essential component for defense response, cell death, and oxidative burst in plants. Planta 238 1113–1124. 10.1007/s00425-013-1956-3 [DOI] [PubMed] [Google Scholar]
- Chowdhury S., Basu A., Kundu S. (2017). Overexpression of a new osmotin-like protein gene (SindOLP) confers tolerance against biotic and abiotic stresses in sesame. Front. Plant Sci. 8:410. 10.3389/fpls.2017.00410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciszak K., Kulasek M., Barczak A., Grzelak J., Maækowski S., Karpiñski S. (2015). PsbS is required for systemic acquired acclimation and post-excess-light-stress optimization of chlorophyll fluorescence decay times in Arabidopsis. Plant Signal. Behav. 10:e982018. 10.4161/15592324.2014.982018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colla G., Rouphael Y., Di Mattia E., El-Nakhel C., Cardarelli M. (2015). Co-inoculation of Glomus intraradices and Trichoderma atroviride acts as a biostimulant to promote growth, yield and nutrient uptake of vegetable crops. J. Sci. Food Agric. 95 1706–1715. 10.1002/jsfa.6875 [DOI] [PubMed] [Google Scholar]
- Conklin P. L., Barth C. (2004). Ascorbic acid, a familiar small molecule intertwined in the response of plants to ozone, pathogens, and the onset of senescence. Plant Cell Environ. 27 959–970. 10.1111/j.1365-3040.2004.01203.x [DOI] [Google Scholar]
- Craigie J. S. (2011). Seaweed extract stimuli in plant science and agriculture. J. Appl. Phycol. 23 371–393. 10.1007/s10811-010-9560-4 [DOI] [Google Scholar]
- Cui L. G., Shan J. X., Shi M., Gao J. P., Lin H. X. (2014). The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 80 1108–1117. 10.1111/tpj.12712 [DOI] [PubMed] [Google Scholar]
- Czarny J. C., Grichko V. P., Glick B. R. (2006). Genetic modulation of ethylene biosynthesis and signaling in plants. Biotechnol. Adv. 24 410–419. 10.1016/j.biotechadv.2006.01.003 [DOI] [PubMed] [Google Scholar]
- Dana C. D., Bevan D. R., Winkel B. S. J. (2006). Molecular modeling of the effects of mutant alleles on chalcone synthase protein structure. J. Mol. Model. 12 905–914. 10.1007/s00894-005-0071-1 [DOI] [PubMed] [Google Scholar]
- De Palma M., Salzano M., Villano C., Aversano R., Lorito M., Ruocco M., et al. (2019). Transcriptome reprogramming, epigenetic modifications and alternative splicing orchestrate the tomato root response to the beneficial fungus Trichoderma harzianum. Hortic. Res. 6 1–15. 10.1038/s41438-018-0079-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zelicourt A., Colcombet J., Hirt H. (2016). The role of MAPK modules and ABA during abiotic stress signaling. Trends Plant Sci. 21 677–685. 10.1016/j.tplants.2016.04.004 [DOI] [PubMed] [Google Scholar]
- Desoky E. S. M., ElSayed A. I., Merwad A. R. M. A., Rady M. M. (2019). Stimulating antioxidant defenses, antioxidant gene expression, and salt tolerance in Pisum sativum seedling by pretreatment using licorice root extract (LRE) as an organic biostimulant. Plant Physiol. Biochem. 142 292–302. 10.1016/j.plaphy.2019.07.020 [DOI] [PubMed] [Google Scholar]
- Dievart A., Perin C., Hirsch J., Bettembourg M., Lanau N., Artus F., et al. (2015). The phenome analysis of mutant alleles in Leucine-Rich Repeat Receptor-Like Kinase genes in rice reveals new potential targets for stress tolerant cereals. Plant Sci. 242 240–249. 10.1016/j.plantsci.2015.06.019 [DOI] [PubMed] [Google Scholar]
- Dodd I. C., Ruiz-Lozano J. M. (2012). Microbial enhancement of crop resource use efficiency. Curr. Opin. Biotechnol. 23 236–242. 10.1016/j.copbio.2011.09.005 [DOI] [PubMed] [Google Scholar]
- Du J., Wang L., Zhang X., Xiao X., Wang F., Lin P., et al. (2016). Heterologous expression of two Physcomitrella patens group 3 late embryogenesis abundant protein (LEA3) genes confers salinity tolerance in Arabidopsis. J. Plant Biol. 59 182–193. 10.1007/s12374-016-0565-7 [DOI] [Google Scholar]
- du Jardin P. (2015). Plant biostimulants: definition, concept, main categories and regulation. Sci. Hortic. 196 3–14. 10.1016/j.scienta.2015.09.021 [DOI] [Google Scholar]
- Duan M., Wang J., Zhang X., Yang H., Wang H., Qiu Y., et al. (2017). Identification of optimal reference genes for expression analysis in radish (Raphanus sativus L.) and its relatives based on expression stability. Front. Plant Sci. 8:1605. 10.3389/fpls.2017.01605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunaeva M., Adamska I. (2001). Identification of genes expressed in response to light stress in leaves of Arabidopsis thaliana using RNA differential display. Eur. J. Biochem. 268 5521–5529. 10.1046/j.1432-1033.2001.02471.x [DOI] [PubMed] [Google Scholar]
- Ekebafe L. O., Ogbeifun D. E., Okieimen F. (2011). Polymer applications in agriculture. Biokemistri 23 81–89. [Google Scholar]
- El-Esawi M. A., Alaraidh I. A., Alsahli A. A., Alamri S. A., Ali H. M., Alayafi A. A. (2018a). Bacillus firmus (SW5) augments salt tolerance in soybean (Glycine max L.) by modulating root system architecture, antioxidant defense systems and stress-responsive genes expression. Plant Physiol. Biochem. 132 375–384. 10.1016/j.plaphy.2018.09.026 [DOI] [PubMed] [Google Scholar]
- El-Esawi M. A., Alaraidh I. A., Alsahli A. A., Alzahrani S. M., Ali H. M., Alayafi A. A., et al. (2018b). Serratia liquefaciens KM4 improves salt stress tolerance in maize by regulating redox potential, ion homeostasis, leaf gas exchange and stress-related gene expression. Int. J. Mol. Sci. 19:3310. 10.3390/ijms19113310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elizabete Carmo-Silva A., Salvucci M. E. (2013). The regulatory properties of rubisco activase differ among species and affect photosynthetic induction during light transitions. Plant Physiol. 161 1645–1655. 10.1104/pp.112.213348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltelib H. A., Fujikawa Y., Esaka M. (2012). Overexpression of the acerola (Malpighia glabra) monodehydroascorbate reductase gene in transgenic tobacco plants results in increased ascorbate levels and enhanced tolerance to salt stress. S. Afr. J. Bot. 78 295–301. 10.1016/j.sajb.2011.08.005 [DOI] [Google Scholar]
- Erdal S., Turk H. (2016). Cysteine-induced upregulation of nitrogen metabolism-related genes and enzyme activities enhance tolerance of maize seedlings to cadmium stress. Environ. Exp. Bot. 132 92–99. 10.1016/j.envexpbot.2016.08.014 [DOI] [Google Scholar]
- Eriksson S. K., Harryson P. (2011). Dehydrins: Molecular Biology, Structure and Function. Berlin: Springer, 289–305. [Google Scholar]
- Ertani A., Schiavon M. (2013). Alfalfa plant-derived biostimulant stimulate short-term growth of salt stressed Zea mays L. Plants 364 145–158. 10.1007/s11104-012-1335-z [DOI] [Google Scholar]
- Ertani A., Schiavon M., Muscolo A., Nardi S. (2013). Alfalfa plant-derived biostimulant stimulate short-term growth of salt stressed Zea mays L. plants. Plant Soil 364 145–158. [Google Scholar]
- Fan Q. J., Liu J. H. (2011). Colonization with arbuscular mycorrhizal fungus affects growth, drought tolerance and expression of stress-responsive genes in Poncirus trifoliata. Acta Physiol. Plant. 33 1533–1542. 10.1007/s11738-011-0789-6 [DOI] [Google Scholar]
- Fang Z., Wang X., Zhang X., Zhao D., Tao J. (2020). Effects of fulvic acid on the photosynthetic and physiological characteristics of Paeonia ostii under drought stress. Plant Signal. Behav. 15:1774714. 10.1080/15592324.2020.1774714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Yue R., Tao S., Yang Y., Zhang L., Xu M., et al. (2015). Genome-wide identification, expression analysis of auxin-responsive GH3 family genes in maize (Zea mays L.) under abiotic stresses. J. Integr. Plant Biol. 57 783–795. 10.1111/jipb.12327 [DOI] [PubMed] [Google Scholar]
- Fleming T. R., Fleming C. C., Levy C. C. B., Repiso C., Hennequart F., Nolasco J. B., et al. (2019). Biostimulants enhance growth and drought tolerance in Arabidopsis thaliana and exhibit chemical priming action. Ann. Appl. Biol. 174 153–165. 10.1111/aab.12482 [DOI] [Google Scholar]
- Fowler S. G., Cook D., Thomashow M. F. (2005). Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 137 961–968. 10.1104/pp.104.058354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer C. H., Noctor G. (2005). Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17 1866–1875. 10.1105/tpc.105.033589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank A., Matiolli C. C., Viana A. J. C., Hearn T. J., Kusakina J., Belbin F. E., et al. (2018). Circadian entrainment in Arabidopsis by the sugar-responsive transcription factor bZIP63. Curr. Biol. 28 2597.e–2606.e. 10.1016/j.cub.2018.05.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin K. A., Whitelam G. C. (2007). Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat. Genet. 39 1410–1413. 10.1038/ng.2007.3 [DOI] [PubMed] [Google Scholar]
- Franzoni G., Cocetta G., Trivellini A., Ferrante A. (2019). Transcriptional regulation in rocket leaves as affected by salinity. Plants 9:20. 10.3390/plants9010020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fufezan C., Simionato D., Morosinotto T. (2012). Identification of key residues for pH dependent activation of violaxanthin de-epoxidase from Arabidopsis thaliana. PLoS One 7:e35669. 10.1371/journal.pone.0035669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami J., de la Osa C., Ollero F. J., Megías M., Hungria M. (2018). Co-inoculation of maize with Azospirillum brasilense and Rhizobium tropici as a strategy to mitigate salinity stress. Funct. Plant Biol. 45:328. 10.1071/FP17167 [DOI] [PubMed] [Google Scholar]
- Fukami J., Ollero F. J., Megías M., Hungria M. (2017). Phytohormones and induction of plant-stress tolerance and defense genes by seed and foliar inoculation with Azospirillum brasilense cells and metabolites promote maize growth. AMB Express 7 1–13. 10.1186/s13568-017-0453-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C., Wang Y., Liu G., Wang C., Jiang J., Yang C. (2010). Cloning of ten peroxidase (POD) genes from Tamarix Hispida and characterization of their responses to abiotic stress. Plant Mol. Biol. Report. 28 77–89. 10.1007/s11105-009-0129-9 [DOI] [Google Scholar]
- Gill S. S., Tuteja N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48 909–930. 10.1016/j.plaphy.2010.08.016 [DOI] [PubMed] [Google Scholar]
- Glick B. R. (2012). Plant growth-promoting bacteria: mechanisms and applications. Scientifica 2012 1–15. 10.6064/2012/963401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy J. A., Lunar R., Torres-Schumann S., Moreno J., Rodrigo R. M., Pintor-Toro J. A. (1994). Expression, tissue distribution and subcellular localization of dehydrin TAS14 in salt-stressed tomato plants. Plant Mol. Biol. 26 1921–1934. 10.1007/BF00019503 [DOI] [PubMed] [Google Scholar]
- Goñi O., Quille P., O’Connell S. (2018). Ascophyllum nodosum extract biostimulants and their role in enhancing tolerance to drought stress in tomato plants. Plant Physiol. Biochem. 126 63–73. 10.1016/j.plaphy.2018.02.024 [DOI] [PubMed] [Google Scholar]
- González-Pérez L., Páez-Watson T., Álvarez-Suarez J. M., Obando-Rojas M. C., Bonifaz-Arcos E., Viteri G., et al. (2018). Application of exogenous xyloglucan oligosaccharides affects molecular responses to salt stress in Arabidopsis thaliana seedlings. J. Soil Sci. Plant Nutr. 18 1187–1205. 10.4067/S0718-95162018005003301 27315006 [DOI] [Google Scholar]
- Govindasamy V., Senthilkumar M., Magheshwaran V., Kumar U., Bose P., Sharma V., et al. (2010). Bacillus and Paenibacillus spp.: Potential PGPR for Sustainable Agriculture. Berlin: Springer, 333–364. 10.1007/978-3-642-13612-2_15 [DOI] [Google Scholar]
- Graether S. P., Boddington K. F. (2014). Disorder and function: a review of the dehydrin protein family. Front. Plant Sci. 5:576. 10.3389/fpls.2014.00576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan R., Qu Y., Guo Y., Yu L., Liu Y., Jiang J., et al. (2014). Salinity tolerance in soybean is modulated by natural variation in GmSALT3. Plant J. 80 937–950. 10.1111/tpj.12695 [DOI] [PubMed] [Google Scholar]
- Guo W. J., Meetam M., Goldsbrough P. B. (2008). Examining the specific contributions of individual Arabidopsis metallothioneins to copper distribution and metal tolerance. Plant Physiol. 146 1697–1706. 10.1104/pp.108.115782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper A. L., McKinney L. V., Nielsen L. R., Havlickova L., Li Y., Trick M., et al. (2016). Molecular markers for tolerance of European ash (Fraxinus excelsior) to dieback disease identified using Associative Transcriptomics. Sci. Rep. 6 19335. 10.1038/srep19335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasse D., Larsson A. M., Andersson I. (2015). Structure of Arabidopsis thaliana Rubisco activase. Acta Crystallogr. Sect. D Biol. Crystallogr. 71 800–808. 10.1107/S1399004715001182 [DOI] [PubMed] [Google Scholar]
- Hassidim M., Dakhiya Y., Turjeman A., Hussien D., Shor E., Anidjar A., et al. (2017). CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and the circadian control of stomatal aperture. Plant Physiol. 175 1864–1877. 10.1104/pp.17.01214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Hernández H., Juárez-Maldonado A., Benavides-Mendoza A., Ortega-Ortiz H., Cadenas-Pliego G., Sánchez-Aspeytia D., et al. (2018). Chitosan-PVA and copper nanoparticles improve growth and overexpress the SOD and JA genes in tomato plants under salt stress. Agronomy 8:175 10.3390/agronomy8090175 [DOI] [Google Scholar]
- Hernández-Sánchez I. E., Maruri-López I., Molphe-Balch E. P., Becerra-Flora A., Jaimes-Miranda F., Jiménez-Bremont J. F. (2019). Evidence for in vivo interactions between dehydrins and the aquaporin AtPIP2B. Biochem. Biophys. Res. Commun. 510 545–550. 10.1016/j.bbrc.2019.01.095 [DOI] [PubMed] [Google Scholar]
- Hong D. D., Hien H. M., Son P. N. (2007). Seaweeds from Vietnam used for functional food, medicine and biofertilizer. J. Appl. Phycol. 19 817–826. 10.1007/s10811-007-9228-x [DOI] [Google Scholar]
- Hong J., Hwang B. (2006). Promoter activation of pepper class II basic chitinase gene, CAChi2, and enhanced bacterial disease resistance and osmotic stress tolerance in the CAChi2. Planta 223:433. [DOI] [PubMed] [Google Scholar]
- Hoque T. S., Uraji M., Tuya A., Nakamura Y., Murata Y. (2012). Methylglyoxal inhibits seed germination and root elongation and up-regulates transcription of stress-responsive genes in ABA-dependent pathway in Arabidopsis. Plant Biol. 14 854–858. 10.1111/j.1438-8677.2012.00607.x [DOI] [PubMed] [Google Scholar]
- Hu H., Rappel W. J., Occhipinti R., Ries A., Böhmer M., You L., et al. (2015). Distinct cellular locations of carbonic anhydrases mediate carbon dioxide control of stomatal movements. Plant Physiol. 169 1168–1178. 10.1104/pp.15.00646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., You J., Fang Y., Zhu X., Qi Z., Xiong L. (2008). Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol. Biol. 67 169–181. 10.1007/s11103-008-9309-5 [DOI] [PubMed] [Google Scholar]
- Hu W. H., Yan X. H., Yu J. Q. (2017). Importance of the mitochondrial alternative oxidase (AOX) pathway in alleviating photoinhibition in cucumber leaves under chilling injury and subsequent recovery when leaves are subjected to high light intensity. J. Hortic. Sci. Biotechnol. 92 31–38. 10.1080/14620316.2016.1219239 [DOI] [Google Scholar]
- Huang J., Gu M., Lai Z., Fan B., Shi K., Zhou Y. H., et al. (2010). Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 153 1526–1538. 10.1104/pp.110.157370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. W., Zhou Z. Q., Yang H. X., Wei C. X., Wan Y. Y., Wang X. J., et al. (2014). Glucose application protects chloroplast ultrastructure in heat-stressed cucumber leaves through modifying antioxidant enzyme activity. Biol. Plant. 59 131–138. 10.1007/s10535-014-0470-1 [DOI] [Google Scholar]
- Hussain R. M. F., Sheikh A. H., Haider I., Quareshy M., Linthorst H. J. M. (2018). Arabidopsis WRKY50 and TGA transcription factors synergistically activate expression of PR1. Front. Plant Sci. 9:930. 10.3389/fpls.2018.00930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igamberdiev A. U. (2015). Control of Rubisco function via homeostatic equilibration of CO2 supply. Front. Plant Sci. 6:106. 10.3389/fpls.2015.00106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T., Aki T., Yanagisawa S., Uchimiya H., Kawai-Yamada M. (2015). Overexpression of BAX INHIBITOR-1 links plasma membrane microdomain proteins to stress. Plant Physiol. 169 1333–1343. 10.1104/pp.15.00445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahromi F., Aroca R., Porcel R., Ruiz-Lozano J. M. (2008). Influence of salinity on the in vitro development of Glomus intraradices and on the in vivo physiological and molecular responses of mycorrhizal lettuce plants. Microb. Ecol. 55 45–53. 10.1007/s00248-007-9249-7 [DOI] [PubMed] [Google Scholar]
- Jardin P. (2015). Scientia horticulturae plant biostimulants?: definition, concept, main categories and regulation. Sci. Hortic. 196 3–14. [Google Scholar]
- Jithesh M. N., Shukla P. S., Kant P., Joshi J., Critchley A. T., Prithiviraj B. (2018). Physiological and transcriptomics analyses reveal that ascophyllum nodosum extracts induce salinity tolerance in Arabidopsis by regulating the expression of stress responsive genes. J. Plant Growth Regul. 38 463–478. 10.1007/s00344-018-9861-4 [DOI] [Google Scholar]
- Johnson A. A. T., Kyriacou B., Callahan D. L., Carruthers L., Stangoulis J., Lombi E., et al. (2011). Constitutive overexpression of the OsNAS gene family reveals single-gene strategies for effective iron- and zinc-biofortification of rice endosperm. PLoS One 6:e0024476. 10.1371/journal.pone.0024476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juárez-Maldonado A., Ortega-Ortíz H., Morales-Díaz A. B., González-Morales S., Morelos-Moreno Á, Cabrera-De la Fuente M., et al. (2019). Nanoparticles and nanomaterials as plant biostimulants. Int. J. Mol. Sci. 20:162. 10.3390/ijms20010162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwar P., Sanyal S. K., Tokas I., Yadav A. K., Pandey A., Kapoor S., et al. (2014). Comprehensive structural, interaction and expression analysis of CBL and CIPK complement during abiotic stresses and development in rice. Cell Calcium 56 81–95. 10.1016/j.ceca.2014.05.003 [DOI] [PubMed] [Google Scholar]
- Katiyar D., Hemantaranjan A., Singh B. (2015). Chitosan as a promising natural compound to enhance potential physiological responses in plant: a review. Indian J. Plant Physiol. 20 1–9. 10.1007/s40502-015-0139-6 [DOI] [Google Scholar]
- Kaur N., Gupta A. K. (2005). Signal transduction pathways under abiotic stresses in plants. Curr. Sci. 88 1771–1780. 10.2307/24110354 [DOI] [Google Scholar]
- Kesari R., Lasky J. R., Villamor J. G., Des Marais D. L., Chen Y. J. C., Liu T. W., et al. (2012). Intron-mediated alternative splicing of Arabidopsis P5CS1 and its association with natural variation in proline and climate adaptation. Proc. Natl. Acad. Sci. U.S.A. 109 9197–9202. 10.1073/pnas.1203433109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. S., Akther T., Hemalatha S. (2017). Impact of panchagavya on Oryza sativa L. grown under saline stress. J. Plant Growth Regul. 36 702–713. 10.1007/s00344-017-9674-x [DOI] [Google Scholar]
- Khan M. S., Pandey M. K., Hemalatha S. (2018). Comparative studies on the role of organic biostimulant in resistant and susceptible cultivars of rice grown under saline stress - organic biostimulant alleviate saline stress in tolerant and susceptible cultivars of rice. J. Crop Sci. Biotechnol. 21 459–467. 10.1007/s12892-018-0089-0 [DOI] [Google Scholar]
- Khan W., Rayirath U. P., Subramanian S., Jithesh M. N., Rayorath P., Hodges D. M., et al. (2009). Seaweed extracts as biostimulants of plant growth and development. J. Plant Growth Regul. 28 386–399. 10.1007/s00344-009-9103-x [DOI] [Google Scholar]
- Khan Z., Shahwar D. (2020). “Role of heat shock proteins (HSPs) and heat stress tolerance in crop plants,” in Sustainable Agriculture in the Era of Climate Change, eds Hasanuzzaman M., Roychowdhury R., Choudhury S., Srivastava S. (Berlin: Springer International Publishing; ), 211–234. [Google Scholar]
- Khandekar S., Leisner S. (2011). Soluble silicon modulates expression of Arabidopsis thaliana genes involved in copper stress. J. Plant Physiol. 168 699–705. 10.1016/j.jplph.2010.09.009 [DOI] [PubMed] [Google Scholar]
- Khattab H. I., Emam M. A., Emam M. M., Helal N. M., Mohamed M. R. (2014). Effect of selenium and silicon on transcription factors NAC5 and DREB2A involved in drought-responsive gene expression in rice. Biol. Plant. 58 265–273. 10.1007/s10535-014-0391-z [DOI] [Google Scholar]
- Kumar A., Kumar R., Kumari M., Goldar S. (2020). Enhancement of plant growth by using PGPR for a sustainable agriculture: a review. Int. J. Curr. Microbiol. App. Sci. 9 152–165. 10.20546/ijcmas.2020.902.019 [DOI] [Google Scholar]
- Kumar R., Trivedi K., Anand K. G. V., Ghosh A. (2019). Science behind biostimulant action of seaweed extract on growth and crop yield: insights into transcriptional changes in roots of maize treated with Kappaphycus alvarezii seaweed extract under soil moisture stressed conditions. J. Appl. Phycol. 32 599–613. 10.1007/s10811-019-01938-y [DOI] [Google Scholar]
- Laloum T., De Mita S., Gamas P., Baudin M., Niebel A. (2013). CCAAT-box binding transcription factors in plants: y so many? Trends Plant Sci. 18 157–166. 10.1016/j.tplants.2012.07.004 [DOI] [PubMed] [Google Scholar]
- Laohavisit A., Shang Z., Rubio L., Cuin T. A., Véry A. A., Wang A., et al. (2012). Arabidopsis annexin1 mediates the radical-activated plasma membrane Ca2+-and K+-permeable conductance in root cells. Plant Cell 24 1522–1533. 10.1105/tpc.112.097881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le M. H., Cao Y., Zhang X. C., Stacey G. (2014). LIK1, a CERK1-interacting kinase, regulates plant immune responses in Arabidopsis. PLoS One 9:e102245. 10.1371/journal.pone.0102245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Z., Chen J., Technol X. C.-J. (2007). The Physiological Functions of Calcium-Dependent Protein Kinases in Plant Calcium Signal Transduction. en.cnki.com.cn. Available online at: http://en.cnki.com.cn/Article_en/CJFDTotal-FJLK200703061.htm (accessed July 30, 2019). [Google Scholar]
- Li H., Gao Z., Chen Q., Li Q., Luo M., Wang J., et al. (2020a). Grapevine ABA receptor VvPYL1 regulates root hair development in transgenic Arabidopsis. Plant Physiol. Biochem. 149 190–200. 10.1016/j.plaphy.2020.02.008 [DOI] [PubMed] [Google Scholar]
- Li L., Shuai L., Sun J., Li C., Yi P., Zhou Z., et al. (2020b). The Role of 1-Methylcyclopropene in the regulation of ethylene biosynthesis and ethylene receptor gene expression in Mangifera indica L. (Mango Fruit). Food Sci. Nutr. 8 1284–1294. 10.1002/fsn3.1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. (2019). Regulation of ribosomal proteins on viral infection. Cells 8:508. 10.3390/cells8050508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Pang S., Lu Z., Jin B. (2020c). Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plants 9:1515. 10.3390/plants9111515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhu Y., Chen C., Geng Z., Li X., Reports T. Y.-S., et al. (2020d). cloning and characterization of two chlorophyll A/B binding protein genes and analysis of their gene family in Camellia sinensis. Sci. Rep. 10 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. J., Fang Y., Fu Y. R., Huang J. G., Wu C. A., Zheng C. C. (2013). NFYA1 is involved in regulation of postgermination growth arrest under salt stress in Arabidopsis. PLoS One 8:e0061289. 10.1371/journal.pone.0061289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang Q., Ou L., Ji D., Liu T., Lan R., et al. (2020e). Response to the cold stress signaling of the tea plant (Camellia sinensis) elicited by chitosan oligosaccharide. Agronomy 10:915 10.3390/agronomy10060915 [DOI] [Google Scholar]
- Lin R.-C., Park H.-J., Wang H.-Y. (2008). Role of Arabidopsis RAP2.4 in regulating light- and ethylene-mediated developmental processes and drought stress tolerance. Mol. Plant 1 42–57. 10.1093/mp/ssm004 [DOI] [PubMed] [Google Scholar]
- Liu P., Yin L., Deng X., Wang S., Tanaka K., Zhang S. (2014). Aquaporin-mediated increase in root hydraulic conductance is involved in silicon-induced improved root water uptake under osmotic stress in Sorghum bicolor L. J. Exp. Bot. 65 4747–4756. 10.1093/jxb/eru220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. Y., Huang T. K., Tseng C. Y., Lai Y. S., Lin S. I., Lin W. Y., et al. (2012). PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24 2168–2183. 10.1105/tpc.112.096636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Huang D., Tao J., Miller A. J., Fan X., Xu G. (2014). Identification and functional assay of the interaction motifs in the partner protein OsNAR2.1 of the two-component system for high-affinity nitrate transport. New Phytol. 204 74–80. 10.1111/nph.12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D., Niu X., Yu J., Yan J., Gou X., Lu B. R., et al. (2012). Rice choline monooxygenase (OsCMO) protein functions in enhancing glycine betaine biosynthesis in transgenic tobacco but does not accumulate in rice (Oryza sativa L. ssp. japonica). Plant Cell Rep. 31 1625–1635. 10.1007/s00299-012-1276-2 [DOI] [PubMed] [Google Scholar]
- Luo X., Bai X., Sun X., Zhu D., Liu B., Ji W., et al. (2013). Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling. J. Exp. Bot. 64 2155–2169. 10.1093/jxb/ert073 [DOI] [PubMed] [Google Scholar]
- Ma W., Penrose D. M., Glick B. R. (2002). Strategies used by rhizobia to lower plant ethylene levels and increase nodulation. Can. J. Microbiol. 48 947–954. 10.1139/w02-100 [DOI] [PubMed] [Google Scholar]
- Manchester L. C., Coto-Montes A., Boga J. A., Andersen L. P. H., Zhou Z., Galano A., et al. (2015). Melatonin: an ancient molecule that makes oxygen metabolically tolerable. J. Pineal Res. 59 403–419. 10.1111/jpi.12267 [DOI] [PubMed] [Google Scholar]
- Mastouri F., Björkman T., Harman G. E. (2012). Trichoderma harzianum enhances antioxidant defense of tomato seedlings and resistance to water deficit. Mol. Plant Microbe Interact. 25 1264–1271. 10.1094/MPMI [DOI] [PubMed] [Google Scholar]
- Matyjaszczyk E. (2015). Products containing microorganisms as a tool in integrated pest management and the rules of their market placement in the European Union. Pest Manag. Sci. 71 1201–1206. 10.1002/ps.3986 [DOI] [PubMed] [Google Scholar]
- Mauri N., Fernández-Marcos M., Costas C., Desvoyes B., Pichel A., Caro E., et al. (2016). GEM, a member of the GRAM domain family of proteins, is part of the ABA signaling pathway. Sci. Rep. 6 1–11. 10.1038/srep22660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer Weber R. L., Wiebke-Strohm B., Bredemeier C., Margis-Pinheiro M., de Brito G. G., Rechenmacher C., et al. (2014). Expression of an osmotin-like protein from Solanum nigrum confers drought tolerance in transgenic soybean. BMC Plant Biol. 14:343. 10.1186/s12870-014-0343-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcgonigle B., Keeler S. J., Lau S.-M. C., Koeppe M. K., O’keefe D. P. (2000). A Genomics Approach to the Comprehensive Analysis of the Glutathione S-Transferase Gene Family in Soybean and Maize. Available online at: www.plantphysiol.org (accessed February 20, 2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina J., Catalá R., Salinas J. (2011). The CBFs: three Arabidopsis transcription factors to cold acclimate. Plant Sci. 180 3–11. 10.1016/j.plantsci.2010.06.019 [DOI] [PubMed] [Google Scholar]
- Mehmood S., Ahmed W., Ikram M., Imtiaz M., Mahmood S., Tu S., et al. (2020). Chitosan modified biochar increases soybean (Glycine max L.) resistance to salt-stress by augmenting root morphology, antioxidant defense mechanisms and the expression of stress-responsive genes. Plants 9:1173. 10.3390/plants9091173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Lv D., Wang P., Wang X. C., Chen J., Miao C., et al. (2006). An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 18 2749–2766. 10.1105/tpc.106.044230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanta T. K., Arora P. K., Mohanta N., Parida P., Bae H. (2015). Identification of new members of the MAPK gene family in plants shows diverse conserved domains and novel activation loop variants. BMC Genomics 16:58. 10.1186/s12864-015-1244-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradi Tarnabi Z., Iranbakhsh A., Mehregan I., Ahmadvand R. (2020). Impact of arbuscular mycorrhizal fungi (AMF) on gene expression of some cell wall and membrane elements of wheat (Triticum aestivum L.) under water deficit using transcriptome analysis. Physiol. Mol. Biol. Plants 26 143–162. 10.1007/s12298-019-00727-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Mayor A., Pineda B., Garcia-Abellán J. O., Antón T., Garcia-Sogo B., Sanchez-Bel P., et al. (2012). Overexpression of dehydrin tas14 gene improves the osmotic stress imposed by drought and salinity in tomato. J. Plant Physiol. 169 459–468. 10.1016/j.jplph.2011.11.018 [DOI] [PubMed] [Google Scholar]
- Mutava R. N., Prince S. J. K., Syed N. H., Song L., Valliyodan B., Chen W., et al. (2015). Understanding abiotic stress tolerance mechanisms in soybean: a comparative evaluation of soybean response to drought and flooding stress. Plant Physiol. Biochem. 86 109–120. 10.1016/j.plaphy.2014.11.010 [DOI] [PubMed] [Google Scholar]
- Nagaraju M., Reddy P. S., Kumar S. A., Kumar A., Rajasheker G., Rao D. M., et al. (2020). Genome-wide identification and transcriptional profiling of small heat shock protein gene family under diverse abiotic stress conditions in Sorghum bicolor (L.). Int. J. Biol. Macromol. 142 822–834. 10.1016/j.ijbiomac.2019.10.023 [DOI] [PubMed] [Google Scholar]
- Nair P., Kandasamy S., Zhang J., Ji X., Kirby C., Benkel B., et al. (2012). Transcriptional and metabolomic analysis of Ascophyllum nodosum mediated freezing tolerance in Arabidopsis thaliana. BMC Genomics 13:643. 10.1186/1471-2164-13-643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz M. A., Jiao Y., Chen C., Shireen F., Zheng Z., Imtiaz M., et al. (2018). Melatonin pretreatment improves vanadium stress tolerance of watermelon seedlings by reducing vanadium concentration in the leaves and regulating melatonin biosynthesis and antioxidant-related gene expression. J. Plant Physiol. 220 115–127. 10.1016/j.jplph.2017.11.003 [DOI] [PubMed] [Google Scholar]
- Neupane S., Schweitzer S., Neupane A., Andersen E., Fennell A., Zhou R., et al. (2019). Identification and characterization of mitogen-activated protein kinase (MAPK) genes in sunflower (Helianthus annuus L.). Plants 8:28. 10.3390/plants8020028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen N. H., Jung C., Cheong J. J. (2019). Chromatin remodeling for the transcription of type 2C protein phosphatase genes in response to salt stress. Plant Physiol. Biochem. 141 325–331. 10.1016/j.plaphy.2019.06.012 [DOI] [PubMed] [Google Scholar]
- Nishizawa A., Yabuta Y., Shigeoka S. (2008). Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol. 147 1251–1263. 10.1104/pp.108.122465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J. E., Kwon Y., Kim J. H., Noh H., Hong S. W., Lee H. (2011). A dual role for MYB60 in stomatal regulation and root growth of Arabidopsis thaliana under drought stress. Plant Mol. Biol. 77 91–103. 10.1007/s11103-011-9796-7 [DOI] [PubMed] [Google Scholar]
- Olsson A. S. B., Engström P., Söderman E. (2004). The homeobox genes ATHB12 and ATHB7 encode potential regulators of growth in response to water deficit in Arabidopsis. Plant Mol. Biol. 55 663–677. 10.1007/s11103-004-1581-4 [DOI] [PubMed] [Google Scholar]
- Owen C., Patron N. J., Huang A., Osbourn A. (2017). Harnessing plant metabolic diversity. Curr. Opin. Chem. Biol. 40 24–30. 10.1016/j.cbpa.2017.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier J. B., Ytterberg J., Liberles D. A., Roepstorff P., Van Wijk K. J. (2001). Identification of a 350-kDa ClpP protease complex with 10 different Clp isoforms in chloroplasts of Arabidopsis thaliana. J. Biol. Chem. 276 16318–16327. 10.1074/jbc.M010503200 [DOI] [PubMed] [Google Scholar]
- Petroni K., Kumimoto R. W., Gnesutta N., Calvenzani V., Fornari M., Tonelli C., et al. (2013). The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 24 4777–4792. 10.1105/tpc.112.105734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrozza A., Santaniello A., Summerer S., Di Tommaso G., Di Tommaso D., Paparelli E., et al. (2014a). Physiological Responses to Megafol® Treatments in Tomato Plants Under Drought Stress: A Phenomic and Molecular Approach. Amsterdam: Elsevier. [Google Scholar]
- Petrozza A., Santaniello A., Summerer S., Di Tommaso G., Di Tommaso D., Paparelli E., et al. (2014b). Physiological responses to Megafol® treatments in tomato plants under drought stress: a phenomic and molecular approach. Sci. Hortic. 174 185–192. 10.1016/j.scienta.2014.05.023 [DOI] [Google Scholar]
- Piccolo A., Spiteller M. (2003). Electrospray ionization mass spectrometry of terrestrial humic substances and their size fractions. Anal. Bioanal. Chem. 377 1047–1059. 10.1007/s00216-003-2186-5 [DOI] [PubMed] [Google Scholar]
- Pilon-Smits E. A., Quinn C. F., Tapken W., Malagoli M., Schiavon M. (2009). Physiological functions of beneficial elements. Curr. Opin. Plant Biol. 12 267–274. 10.1016/j.pbi.2009.04.009 [DOI] [PubMed] [Google Scholar]
- Qin F., Sakuma Y., Tran L. S. P., Maruyama K., Kidokoro S., Fujita Y., et al. (2008). Arabidopsis DREB2A-interacting proteins function as Ring E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20 1693–1707. 10.1105/tpc.107.057380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoczy M., Femiak I., Alejska M., Figlerowicz M., Podkowinski J. (2018). Sorghum CCoAOMT and CCoAOMT-like gene evolution, structure, expression and the role of conserved amino acids in protein activity. Mol. Genet. Genomics 293 1077–1089. 10.1007/s00438-018-1441-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayirath P., Benkel B., Mark Hodges D., Allan-Wojtas P., MacKinnon S., Critchley A. T., et al. (2009). Lipophilic components of the brown seaweed, Ascophyllum nodosum, enhance freezing tolerance in Arabidopsis thaliana. Planta 230 135–147. 10.1007/s00425-009-0920-8 [DOI] [PubMed] [Google Scholar]
- Reddy P. S., Sharma K. K., Vadez V., Reddy M. K. (2015). Molecular cloning and differential expression of cytosolic class I small Hsp gene family in Pennisetum glaucum (L.). Appl. Biochem. Biotechnol. 176 598–612. 10.1007/s12010-015-1598-y [DOI] [PubMed] [Google Scholar]
- Reeves W. M., Lynch T. J., Mobin R., Finkelstein R. R. (2011). Direct targets of the transcription factors ABA-insensitive(ABI)4 and ABI5 reveal synergistic action by ABI4 and several bZIP ABA response factors. Plant Mol. Biol. 75 347–363. 10.1007/s11103-011-9733-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riaz Rajoka M. S., Mehwish H. M., Wu Y., Zhao L., Arfat Y., Majeed K., et al. (2020). Chitin/chitosan derivatives and their interactions with microorganisms: a comprehensive review and future perspectives. Crit. Rev. Biotechnol. 40 365–379. 10.1080/07388551.2020.1713719 [DOI] [PubMed] [Google Scholar]
- Rowarth N. M., Dauphinee A. N., Denbigh G. L., Gunawardena A. H. (2019). Hsp70 plays a role in programmed cell death during the remodelling of leaves of the lace plant (Aponogeton madagascariensis). J. Exp. Bot. 71 907–918. 10.1093/jxb/erz447 [DOI] [PubMed] [Google Scholar]
- Saakre M., Baburao T. M., Salim A. P., Ffancies R. M., Achuthan V. P., Thomas G., et al. (2017). Identification and characterization of genes responsible for drought tolerance in rice mediated by Pseudomonas fluorescens. Rice Sci. 24 291–298. 10.1016/j.rsci.2017.04.005 [DOI] [Google Scholar]
- Safdarian M., Askari H., Shariati J. V., Nematzadeh G. (2019). Transcriptional responses of wheat roots inoculated with Arthrobacter nitroguajacolicus to salt stress. Sci. Rep. 9 1–12. 10.1038/s41598-018-38398-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia J., Sarma R. K., Dhandia R., Yadav A., Bharali R., Gupta V. K., et al. (2018). Alleviation of drought stress in pulse crops with ACC deaminase producing rhizobacteria isolated from acidic soil of Northeast India. Sci. Rep. 8 1–16. 10.1038/s41598-018-21921-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaniello A., Scartazza A., Gresta F., Loreti E., Biasone A., Di Tommaso D., et al. (2017). Ascophyllum nodosum seaweed extract alleviates drought stress in Arabidopsis by affecting photosynthetic performance and related gene expression. Front. Plant Sci. 8:1362. 10.3389/fpls.2017.01362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawers R. J. H., Svane S. F., Quan C., Grønlund M., Wozniak B., Gebreselassie M. N., et al. (2017). Phosphorus acquisition efficiency in arbuscular mycorrhizal maize is correlated with the abundance of root-external hyphae and the accumulation of transcripts encoding PHT1 phosphate transporters. New Phytol. 214 632–643. 10.1111/nph.14403 [DOI] [PubMed] [Google Scholar]
- Schiavon M., Pizzeghello D., Muscolo A. (2010). High molecular size humic substances enhance phenylpropanoid metabolism in maize (Zea mays L.). J. Chem. Ecol. 36 662–669. 10.1007/s10886-010-9790-6 [DOI] [PubMed] [Google Scholar]
- Seong E. S., Cho H. S., Choi D., Joung Y. H., Lim C. K., Hur J. H., et al. (2007). Tomato plants overexpressing CaKR1 enhanced tolerance to salt and oxidative stress. Biochem. Biophys. Res. Commun. 363 983–988. 10.1016/j.bbrc.2007.09.104 [DOI] [PubMed] [Google Scholar]
- Sestili F., Rouphael Y., Cardarelli M., Pucci A., Bonini P., Canaguier R., et al. (2018). Protein hydrolysate stimulates growth in tomato coupled with N-dependent gene expression involved in N assimilation. Front. Plant Sci. 9:1233. 10.3389/fpls.2018.01233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seung D., Risopatron J. P. M., Jones B. J., Marc J. (2012). Circadian clock-dependent gating in ABA signalling networks. Protoplasma 249 445–457. 10.1007/s00709-011-0304-3 [DOI] [PubMed] [Google Scholar]
- Shan Z., Jiang Y., Li H., Guo J., Dong M., Zhang J., et al. (2020). Genome-wide analysis of the NAC transcription factor family in broomcorn millet (Panicum miliaceum L.) and expression analysis under drought stress. BMC Genomics 21:96 10.1186/s12864-020-6479-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Chen C., Khatri K., Rathore M. S., Pandey S. P. (2019). Gracilaria dura extract confers drought tolerance in wheat by modulating abscisic acid homeostasis. Plant Physiol. Biochem. 136 143–154. 10.1016/j.plaphy.2019.01.015 [DOI] [PubMed] [Google Scholar]
- Shen J., Guo M., Wang Y., Yuan X., Dong S., Song X., et al. (2020). An investigation into the beneficial effects and molecular mechanisms of humic acid on foxtail millet under drought conditions. PLoS One 15:e0234029. 10.1371/journal.pone.0234029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Zhang Y., Han W., Feng R., Hu Y., Guo J., et al. (2016). Silicon enhances water stress tolerance by improving root hydraulic conductance in Solanum lycopersicum L. Front. Plant Sci. 7:196. 10.3389/fpls.2016.00196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla P. S., Borza T., Critchley A. T., Hiltz D., Norrie J., Prithiviraj B. (2018). Ascophyllum nodosum extract mitigates salinity stress in Arabidopsis thaliana by modulating the expression of miRNA involved in stress tolerance and nutrient acquisition. PLoS One 13:e0206221. 10.1371/journal.pone.0206221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla P. S., Shotton K., Norman E., Neily W., Critchley A. T., Prithiviraj B. (2017). Seaweed extract improve drought tolerance of soybean by regulating stress-response genes. AoB Plants 10:plx051. 10.1093/aobpla/plx051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal P., Jan A. T., Azam M., Haq Q. M. R. (2016). Plant abiotic stress: a prospective strategy of exploiting promoters as alternative to overcome the escalating burden. Front. Life Sci. 9:52–63. 10.1080/21553769.2015.1077478 [DOI] [Google Scholar]
- Smoleñ S., Kowalska I., Halka M., Ledwożyw-Smoleñ I., Grzanka M., Skoczylas Ł., et al. (2019). Selected aspects of iodate and iodosalicylate metabolism in lettuce including the activity of vanadium dependent haloperoxidases as affected by exogenous vanadium. Agronomy 10:1 10.3390/agronomy10010001 [DOI] [Google Scholar]
- Sofo A., Nuzzaci M., Vitti A., Tataranni G., Scopa A. (2014). “Control of biotic and abiotic stresses in cultivated plants by the use of biostimulant microorganisms,” in Improvement of Crops in the Era of Climatic Changes, eds Ahmad P., Wani M., Azooz M., Tran L. S. (New York, NY: Springer; ). [Google Scholar]
- Song A., Li P., Fan F., Li Z., Liang Y. (2014). The effect of silicon on photosynthesis and expression of its relevant genes in rice (Oryza sativa L.) under high-zinc stress. PLoS One 9:e0113782. 10.1371/journal.pone.0113782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson P. G., Terry M. J. (2008). Light signalling pathways regulating the Mg-chelatase branchpoint of chlorophyll synthesis during de-etiolation in Arabidopsis thaliana. Photochem. Photobiol. Sci. 7 1243–1252. 10.1039/b802596g [DOI] [PubMed] [Google Scholar]
- Sun J., Qiu C., Ding Y., Wang Y., Sun L., Sun L., et al. (2020). Fulvic acid ameliorates drought stress-induced damage in tea plants by regulating the ascorbate metabolism and flavonoids biosynthesis. BMC Genomics 21:411. 10.1186/s12864-020-06815-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Yu D. (2015). Activated expression of AtWRKY53 negatively regulates drought tolerance by mediating stomatal movement. Plant Cell Rep. 34 1295–1306. 10.1007/s00299-015-1787-8 [DOI] [PubMed] [Google Scholar]
- Suzuki N., Koussevitzky S., Mittler R., Miller G. (2012). ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 35 259–270. 10.1111/j.1365-3040.2011.02336.x [DOI] [PubMed] [Google Scholar]
- Suzuki N., Miller G., Morales J., Shulaev V., Torres M. A., Mittler R. (2011). Respiratory burst oxidases: the engines of ROS signaling. Curr. Opin. Plant Biol. 14 691–699. 10.1016/j.pbi.2011.07.014 [DOI] [PubMed] [Google Scholar]
- Taji T., Ohsumi C., Iuchi S., Seki M., Kasuga M., Kobayashi M., et al. (2002). Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J. 29 417–426. 10.1046/j.0960-7412.2001.01227.x [DOI] [PubMed] [Google Scholar]
- Takuhara Y., Kobayashi M., Suzuki S. (2011). Low-temperature-induced transcription factors in grapevine enhance cold tolerance in transgenic Arabidopsis plants. J. Plant Physiol. 168 967–975. 10.1016/j.jplph.2010.11.008 [DOI] [PubMed] [Google Scholar]
- Tatematsu K., Kamiya Y., Nambara E. (2008). Co-regulation of ribosomal protein genes as an indicator of growth status. Plant Signal. Behav. 3 450–452. 10.4161/psb.3.7.5577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalhammer A., Bryant G., Sulpice R., Hincha D. K. (2014). Disordered cold regulated15 proteins protect chloroplast membranes during freezing through binding and folding, But do not stabilize chloroplast enzymes in vivo. Plant Physiol. 166 190–201. 10.1104/pp.114.245399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevisan S., Manoli A., Quaggiotti S. (2019). A novel biostimulant, belonging to protein hydrolysates, mitigates abiotic stress effects on maize seedlings grown in hydroponics. Agronomy 9:28 10.3390/agronomy9010028 [DOI] [Google Scholar]
- Trujillo-Moya C., Gisbert C. (2012). The influence of ethylene and ethylene modulators on shoot organogenesis in tomato. Plant Cell. Tissue Organ Cult. 111 41–48. 10.1007/s11240-012-0168-z [DOI] [Google Scholar]
- Tunnacliffe A., Wise M. J. (2007). The continuing conundrum of the LEA proteins. Naturwissenschaften 94 791–812. 10.1007/s00114-007-0254-y [DOI] [PubMed] [Google Scholar]
- Turk H. (2019). Chitosan-induced enhanced expression and activation of alternative oxidase confer tolerance to salt stress in maize seedlings. Artic. Plant Physiol. Biochem. 141 415–422. 10.1016/j.plaphy.2019.06.025 [DOI] [PubMed] [Google Scholar]
- Tuteja N. (2007). Abscisic acid and abiotic stress signaling. Plant Signal. Behav. 2 135–138. 10.4161/psb.2.3.4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugarte R. A., Sharp G., Moore B. (2006). “Changes in the brown seaweed Ascophyllum nodosum (L.) Le Jol. plant morphology and biomass produced by cutter rake harvests in southern New Brunswick, Canada,” in Proceedings of the Eighteenth International Seaweed Symposium, (Dordrecht: Springer; ), 125–133. [Google Scholar]
- Umezawa T., Fujita M., Fujita Y., Yamaguchi-Shinozaki K., Shinozaki K. (2006). Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Curr. Opin. Biotechnol. 17 113–122. 10.1016/j.copbio.2006.02.002 [DOI] [PubMed] [Google Scholar]
- Vishwakarma K., Upadhyay N., Kumar N., Yadav G., Singh J., Mishra R. K., et al. (2017). Abscisic acid signaling and abiotic stress tolerance in plants: a review on current knowledge and future prospects. Front. Plant Sci. 8:161. 10.3389/fpls.2017.00161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe V., Chitarra W., Cascone P., Volpe M. G., Bartolini P., Moneti G., et al. (2018). The association with two different arbuscular mycorrhizal fungi differently affects water stress tolerance in tomato. Front. Plant Sci. 9:1480. 10.3389/fpls.2018.01480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Sydow L., Schwenkert S., Meurer J., Funk C., Mamedov F., Schröder W. P. (2016). The PsbY protein of Arabidopsis photosystem II is important for the redox control of cytochrome b559. Biochim. Biophys. Acta Bioenerg. 1857 1524–1533. 10.1016/j.bbabio.2016.05.004 [DOI] [PubMed] [Google Scholar]
- Vranova V., Rejsek K., Skene K. R., Formanek P. (2011). Non-protein amino acids: plant, soil and ecosystem interactions. Plant Soil 342 31–48. 10.1007/s11104-010-0673-y [DOI] [Google Scholar]
- Wally O. S. D., Critchley A. T., Hiltz D., Craigie J. S., Han X., Zaharia L. I., et al. (2013). Regulation of phytohormone biosynthesis and accumulation in Arabidopsis following treatment with commercial extract from the marine macroalga Ascophyllum nodosum. J. Plant Growth Regul. 32 324–339. 10.1007/s00344-012-9301-9 [DOI] [Google Scholar]
- Wang H., Datla R., Georges F., Loewen M., Cutler A. J. (1995). Promoters from kin1 and cor6.6, two homologous Arabidopsis thaliana genes: transcriptional regulation and gene expression induced by low temperature, ABA, osmoticum and dehydration. Plant Mol. Biol. 28 605–617. 10.1007/BF00021187 [DOI] [PubMed] [Google Scholar]
- Wang H., Zhou L., Fu Y., Cheung M.-Y., Wong F.-L., Phang T.-H., et al. (2012). Expression of an apoplast-localized BURP-domain protein from soybean (GmRD22) enhances tolerance towards abiotic stress. Plant. Cell Environ. 35 1932–1947. 10.1111/j.1365-3040.2012.02526.x [DOI] [PubMed] [Google Scholar]
- Wang J., Song J., Clark G., Roux S. J. (2018). ANN1 and ANN2 function in post-phloem sugar transport in root tips to affect primary root growth. Plant Physiol. 178 390–401. 10.1104/pp.18.00713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Xue L., Batelli G., Lee S., Hou Y. J., Van Oosten M. J., et al. (2013). Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. U.S.A. 110 11205–11210. 10.1073/pnas.1308974110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Zhang L., Jiang X., Dai X., Xu L., Li T., et al. (2018). Evolutionary and functional characterization of leucoanthocyanidin reductases from Camellia sinensis. Planta 247 139–154. 10.1007/s00425-017-2771-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., McFarlane H. E., Persson S. (2016). The impact of abiotic factors on cellulose synthesis. J. Exp. Bot. 67 543–552. 10.1093/jxb/erv488 [DOI] [PubMed] [Google Scholar]
- Wang W., Jiang W., Liu J., Li Y., Gai J., Li Y. (2017). Genome-wide characterization of the aldehyde dehydrogenase gene superfamily in soybean and its potential role in drought stress response. BMC Genomics 18:518. 10.1186/s12864-017-3908-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xia M. X., Chen J., Yuan R., Deng F. N., Shen F. F. (2016). Gene expression characteristics and regulation mechanisms of superoxide dismutase and its physiological roles in plants under stress. Biochemistry 81 465–480. 10.1134/S0006297916050047 [DOI] [PubMed] [Google Scholar]
- Wang X., Gao F., Bing J., Sun W., Feng X., Ma X., et al. (2019). Overexpression of the jojoba aquaporin gene, ScPIP1, enhances drought and salt tolerance in transgenic Arabidopsis. Int. J. Mol. Sci. 20:153. 10.3390/ijms20010153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Jin X., Liao W., Hu L., Dawuda M. M., Zhao X., et al. (2018). 5-aminolevulinic acid (ALA) alleviated salinity stress in cucumber seedlings by enhancing chlorophyll synthesis pathway. Front. Plant Sci. 9:635. 10.3389/fpls.2018.00635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier L. J. C., Boyetchko S. M. (2002). Arbuscular mycorrhizal fungi as biostimulants and bioprotectants of crops. Appl. Mycol. Biotechnol. 2 311–340. [Google Scholar]
- Xiong Y., Contento A. L., Nguyen P. Q., Bassham D. C. (2007). Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 143 291–299. 10.1104/pp.106.092106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Gongbuzhaxi, Wang C., Xue F., Zhang H., Ji W. (2015). Wheat NAC transcription factor TaNAC29 is involved in response to salt stress. Plant Physiol. Biochem. 96 356–363. 10.1016/j.plaphy.2015.08.013 [DOI] [PubMed] [Google Scholar]
- Yakhin O. I., Lubyanov A. A., Yakhin I. A., Brown P. H. (2017). Biostimulants in plant science: a global perspective. Front. Plant Sci. 7:2049. 10.3389/fpls.2016.02049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanhui C., Xiaoyuan Y., Kun H., Meihua L., Jigang L., Zhaofeng G., et al. (2006). The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 60 107–124. 10.1007/s11103-005-2910-y [DOI] [PubMed] [Google Scholar]
- Yasin N. A., Akram W., Khan W. U., Ahmad S. R., Ahmad A., Ali A. (2018). Halotolerant plant-growth promoting rhizobacteria modulate gene expression and osmolyte production to improve salinity tolerance and growth in Capsicum annum L. Environ. Sci. Pollut. Res. 25 23236–23250. 10.1007/s11356-018-2381-8 [DOI] [PubMed] [Google Scholar]
- Ye L., Zhao X., Bao E., Cao K., Zou Z. (2019). Effects of arbuscular mycorrhizal fungi on watermelon growth, elemental uptake, antioxidant, and photosystem ii activities and stress-response gene expressions under salinity-alkalinity stresses. Front. Plant Sci. 10:863. 10.3389/fpls.2019.00863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Gan Y., Xu B. (2016). Application of plant-growth-promoting fungi Trichoderma longibrachiatum T6 enhances tolerance of wheat to salt stress through improvement of antioxidative defense system and gene expression. Front. Plant Sci. 07:1405. 10.3389/fpls.2016.01405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Scheller H. V. (2004). Light-harvesting complex II binds to several small subunits of photosystem I. J. Biol. Chem. 279 3180–3187. 10.1074/jbc.M311640200 [DOI] [PubMed] [Google Scholar]
- Zhang X., Han C., Gao H., Cao Y. (2019). Comparative transcriptome analysis of the garden asparagus (Asparagus officinalis L.)reveals the molecular mechanism for growth with arbuscular mycorrhizal fungi under salinity stress. Plant Physiol. Biochem. 141 20–29. 10.1016/j.plaphy.2019.05.013 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Dong W., Zhang N., Ai X., Wang M., Huang Z., et al. (2014). A wheat allene oxide cyclase gene enhances salinity tolerance via jasmonate signaling. Plant Physiol. 164 1068–1076. 10.1104/pp.113.227595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Huang Y., Xian W., Wang J., Liao H. (2012a). Identification and expression analysis of the Glycine max CYP707A gene family in response to drought and salt stresses. Ann. Bot. 110 743–756. 10.1093/aob/mcs133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Schumaker K. S., Guo Y. (2012b). Sumoylation of transcription factor MYB30 by the small ubiquitin-like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 109 12822–12827. 10.1073/pnas.1202630109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziosi V., Zandoli R., Di Nardo A., Biondi S., Antognoni F., Calandriello F. (2013). Biological activity of different botanical extracts as evaluated by means of an array of in vitro and in vivo bioassays. Acta Hortic. 1009 61–66. 10.17660/ActaHortic.2013.1009.5 [DOI] [Google Scholar]