Abstract
Breast cancer (BC) risk for BRCA1 and BRCA2 mutation carriers varies by genetic and familial factors. About 50 common variants have been shown to modify BC risk for mutation carriers. All but three, were identified in general population studies. Other mutation carrier-specific susceptibility variants may exist but studies of mutation carriers have so far been underpowered. We conduct a novel case-only genome-wide association study comparing genotype frequencies between 60,212 general population BC cases and 13,007 cases with BRCA1 or BRCA2 mutations. We identify robust novel associations for 2 variants with BC for BRCA1 and 3 for BRCA2 mutation carriers, P < 10−8, at 5 loci, which are not associated with risk in the general population. They include rs60882887 at 11p11.2 where MADD, SP11 and EIF1, genes previously implicated in BC biology, are predicted as potential targets. These findings will contribute towards customising BC polygenic risk scores for BRCA1 and BRCA2 mutation carriers.
Subject terms: Breast cancer, Cancer epidemiology, Cancer genetics, Risk factors
Breast cancer risk for BRCA1/BRCA2 mutation carriers varies depending on other genetic factors. Here, the authors perform a case-only genome-wide association study and highlight novel loci associated with breast cancer risk for BRCA1/BRCA2 mutation carriers.
Introduction
Breast cancer (BC) is the most common cancer in women worldwide1 and BC family history is one of the most important risk factors for the disease. Women with a history of BC in a first-degree relative are about two times more likely to develop BC than women without a family history2. Around 15–20% of the familial risk of BC can be explained by rare mutations in the BRCA1 or BRCA2 genes3. A recent prospective cohort study estimated the cumulative risk of BC by 80 years to be 72% for BRCA1 mutation carriers and 69% for BRCA2 mutation carriers4. This study also demonstrated that BC risk for mutation carriers varies by family history of BC in first and second degree relatives, suggesting the existence of other genetic factors that modify BC risks4.
A total of 179 common BC susceptibility single nucleotide polymorphisms (SNPs) or small insertions or deletions (INDELs) have been identified through genome-wide association studies (GWAS) in the general population1,5–35. Although risk alleles at individual SNPs (hereafter used as a generic term to refer to common variants, which also includes the small INDELs) are associated with modest increases in BC risk, it has been shown that they combine multiplicatively on risk, resulting in substantial levels of BC risk stratification in the population36–38. Similarly, more than 50 of the common genetic BC susceptibility variants have also been shown to be associated with BC for BRCA1 and BRCA2 mutation carriers5,6,15,18,20,39–48 and their joint effects, summarised as polygenic risk scores (PRS), result in large differences in the absolute risks of developing BC for mutation carriers at the extremes of the PRS distribution49. BC GWAS for BRCA1 and BRCA2 mutation carriers have been carried out through the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA)50. However, despite the large number of BRCA1 and BRCA2 mutation carriers included, the power to detect genetic modifiers of risk remains limited in comparison to that available in the general population7. To date, no variants specifically associated with BC risk for BRCA1 and BRCA2 carriers have been identified.
Here, we apply a novel strategy using a case-only GWAS design51,52, in which SNP genotype frequencies in 7,257 BRCA1 and 5,097 BRCA2 mutation carrier BC cases are compared to those in 60,212 BC cases from the Breast Cancer Association Consortium (BCAC), unselected for mutation status. We aim (1) to identify novel SNPs that modify BC risk for BRCA1 or BRCA2 mutation carriers but are not associated with risk in the general population and (2) for the known 179 BC susceptibility SNPs, assess whether there is evidence of an interaction between the SNPs and BRCA1 or BRCA2 mutations and therefore evaluate whether the SNP effect size estimates applicable to mutation carriers are different.
We identify robust novel associations for 2 variants with BC for BRCA1 and 3 for BRCA2 mutation carriers, P < 10−8, at 5 loci, which are not associated with risk in the general population. They include rs60882887 in 11p11.2 where MADD, SP11 and EIF1, genes previously implicated in BC biology, are predicted as potential targets. These findings will contribute towards customising BC PRS for BRCA1 and BRCA2 mutation carriers.
Results
Sample characteristics
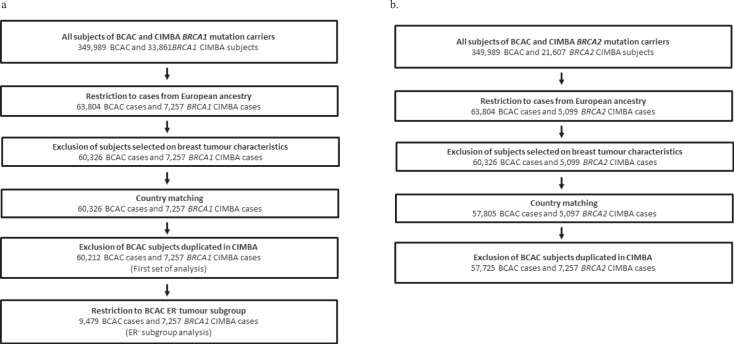
A total of 60,212 BCAC cases and 7,257 BRCA1 mutation carrier cases were available for the BRCA1 case-only analyses and 57,725 BCAC cases and 5,097 BRCA2 mutation carrier cases were available for the BRCA2 case-only analyses (Fig. 1). A total of 45,881 BCAC controls and 5,750 unaffected BRCA1 mutation carriers were available for the BRCA1 control-only analyses and 43,549 BCAC controls and 4,456 unaffected BRCA2 mutation carriers for the BRCA2 control-only analyses (see Fig. 2). Only women of European ancestry were included with 60.9% samples from European countries, 31.1% from the USA, 6.1% from Australia and 1.7% from Israel (Supplementary Tables 1–4). The mean age at BC diagnosis for mutation carrier cases in CIMBA was 42.5 years (40.9 for BRCA1 mutation carriers; 44.1 for BRCA2 mutation carriers) and 58.4 years for cases in BCAC.
Fig. 1. Case-only sample selection.
Sample selection for a BRCA1 and b BRCA2 case-only analysis. *Four studies were excluded because they were included in clinical trials based on breast tumour characteristics as HER-2 receptor status (see Supplementary Table 2).
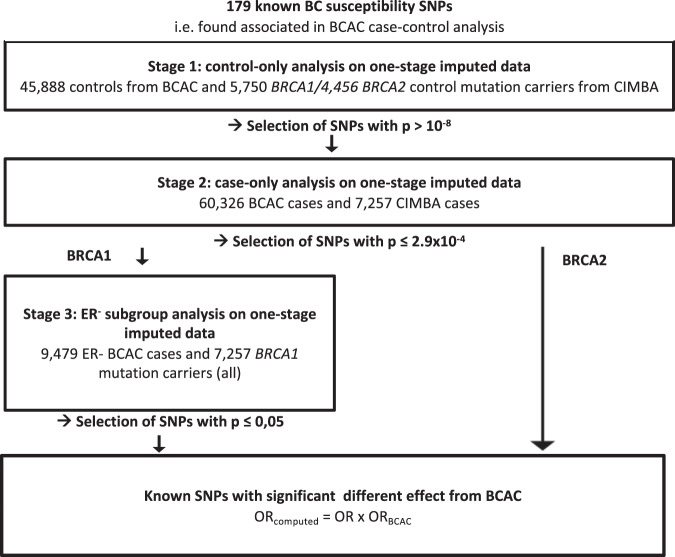
Fig. 2. Control-only sample selection.
Sample selection for a BRCA1 and b BRCA2 control-only analysis.
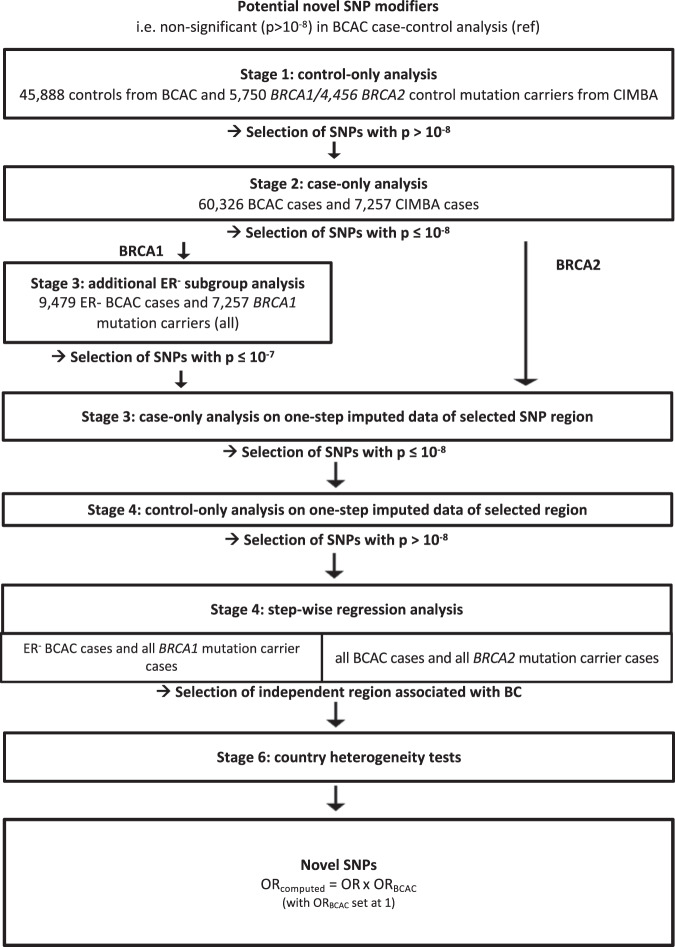
The analytical process for assessing interactions with known BC susceptibility SNP is summarised in Fig. 3 and for the detection of novel modifiers in Fig. 4.
Fig. 3. Analytical process for known BC susceptibility SNPs.
Strategy followed for analysing the associations for the 179 known BC susceptibility SNPs.
Fig. 4. Analytical process for identifying novel modifiers.
Strategy followed for identifying potentially novel SNP modifier.
Independence of SNP frequency with mutation carrier status
Under a case-only study design, it is important to establish independence between the SNPs and BRCA1 or BRCA2 mutation carrier status53. This was assessed a genome-wide level using a control-only analysis which included controls from BCAC and unaffected mutation carriers from CIMBA with SNP data imputed based on the 1,000 genomes project. Genotypes had been imputed separately by each consortium7,50. In the analysis of BRCA1 mutation carriers, 2,164 SNPs were excluded because they were located in or within 500 kb of BRCA1. 2,070 SNPs were excluded from further analyses because they showed associations at p < 10−8 with BRCA1 mutation carrier status in the control-only analysis (2,012 SNPs located on chromosome 17 and 58 on other chromosomes). In the analysis of BRCA2 mutation carriers, 2,947 SNPs were excluded because they were located in or within 500 kb of BRCA2. A further 626 SNPs were excluded from further analyses because they were found to be associated with BRCA2 mutation carrier status in the control-only analysis (566 SNPs on chromosome 13, and 60 on other chromosomes). A total of 9,068,301 SNPs remained for the BRCA1case-only association analysis and 9,043,830 SNPs for the BRCA2case-only analysis.
Interactions with known BC susceptibility SNPs
Based on published data, 179 SNPs were considered as established BC susceptibility SNPs (Fig. 3); 158 SNPs were associated with overall BC risk35 and 21 additional SNPs were found to be associated through studies in ER-negative breast cancer48 (see Supplementary Table 11 in Milne et al.48). One of the 158 SNPs, rs11571833 located within BRCA2 was excluded from the BRCA2 analysis. The detailed results are shown in Supplementary Data 1–3.
For BRCA1 mutation carriers, previous studies have demonstrated heterogeneity in the associations of the SNPs with ER-positive and ER-negative breast cancer35. Since BRCA1 mutation carriers develop primarily ER-negative BC, to comprehensively assess the evidence of interaction with BRCA1 mutation status, we followed a two-step process; we first assessed the associations using all BC cases from BCAC and then we restricted the comparison to BCAC ER-negative BC cases. Of the 158 SNPs35, 59 were associated with BRCA1 mutation carrier status when compared to all BC cases (P < 0.05, Supplementary Data 1). However, after adjusting for multiple testing, only four of these SNPs were associated (P < 2.7 × 10−4) and also showed evidence of association (P < 0.05) when compared with ER-negative BC cases (Table 1). Two additional SNPs on chromosome 1 and 6 (chr1_10566215_A_G and rs17529111) were associated at P < 2.7 × 10−4 with BRCA1 mutation status only when compared with ER-negative BCAC cases. The OR estimates for association with BRCA1 mutation status for these six SNPs were similar under both case-only analyses (all BC and ER-negative BC cases analyses) and varied from 0.85 to 1.07, suggesting that the magnitude of their associations with BC risk for BRCA1 mutation carriers differs from that observed in the general population. For the other 152 SNPs, there was no evidence of association with BRCA1 mutation status when compared against the ER-negative BC cases from BCAC (Supplementary Data 1), suggesting that the OR estimated using case-control data from BCAC are also applicable to BRCA1 mutation carriers.
Table 1.
Known BC susceptibility SNPs demonstrating associations in the BRCA1 case-only analysis.
| Location | SNP namea | Chrb | Positionc | Nearest gene | Estimated effect allele | Referent allele | Frequencyd | r2 | ORe | Pf | ORER¯g | PER-h | ORBCACi | PBCACj | ORcomputedk | Variation in riskl |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All BC SNPs | ||||||||||||||||
| 1p22.3 | rs17426269 | 1 | 88156923 | – | A | G | 0.16 | 1 | 0.90 | 2.70e−04 | 0.92 | 4.22e−02 | 1.05 | 1.70e−04 | 0.95 | IOD |
| 8q24.21 | rs13281615 | 8 | 128355618 | – | G | A | 0.43 | 1 | 0.91 | 1.20e−05 | 0.94 | 4.14e−02 | 1.11 | 5.00e−28 | 1.01 | TT1 |
| 10q22.3 | chr10_80841148_C_T | 10 | 80841148 | ZMZ1 | T | C | 0.40 | 1 | 0.91 | 2.20e−06 | 0.91 | 1.01e−03 | 0.93 | 1.10e−14 | 0.84 | ISD |
| 16q12.1 | chr16_52599188_C_T | 16 | 52599188 | TOX3 | T | C | 0.29 | 1 | 0.85 | 1.80e−13 | 0.91 | 2.80e−03 | 1.23 | 7.00e−88 | 1.04 | TT1 |
| ER- BC SNPs | ||||||||||||||||
| 1p36.22 | chr1_10566215_A_G | 1 | 10566215 | PEX14 | G | A | 0.32 | 1 | 1.07 | 1.30e−03 | 1.12 | 1.10e−04 | 0.94 | 1.80e−09 | 1.05 | TT1 |
| 6q14.1 | rs17529111 | 6 | 82128386 | – | C | T | 0.23 | 0.96 | 0.92 | 7.70e−04 | 0.86 | 1.96e−05 | 1.02 | 4.20e−02 | 0.88 | IOD |
| 8p23.3 | rs66823261 | 8 | 170692 | RPL23AP53 | C | T | 0.23 | 0.92 | – | – | 0.88 | 2.37e−04 | 1.09 | 5.09e−09 | 0.96 | TT1 |
Considering SNPs with known BC (Michailidou et al.)35 and ER-negative-specific BC (Milne et al.)48 associations in the general population.
All BC SNPs: SNPs associated with all BC in the general population.
N = 67,469 breast cancer cases (60,212 BCAC cases and 7,257 BRCA1 mutation carrier cases).
ER- BC SNPs: SNPs associated with ER-negative BC in the general population.
N = 16,736 breast cancer cases (9,479 BCAC ER- cases and 7,257 BRCA1 mutation carrier cases).
TT1 tends to 1, ISD increase in same direction, IOD increase in opposite direction.
aAfter allowing for multiple testing, α* = 2.7 × 10−4.
bChromosome.
cBuild 37 positions.
dFrequency of the allele for which effect is estimated in BCAC cases (OncoArray dataset).
ePer allele odds ratio estimated in the case-only analysis.
fp-value in the case-only analysis (after allowing for multiple testing, p = 2.7 × 10−4).
gPer allele odds-ratio estimated in the case-only ER-negative subgroup analysis. OR values were computed from a two sided logistic regression using a 1 degree freedom likelihood ratio test (1 df lrtest) adjusted for age at BC diagnosis, country and the first four principal components.
hp-value in the case-only ER-negative subgroup analysis.
iPer allele odds-ratio estimated in BCAC (Michailidou et al.)35, except for * (Milne et al.)48.
jp-value in BCAC (Michailidou et al.)35, except for * (Milne et al.)48. For SNPs with PBCAC > 10−8, significance was attained in merging data of Oncoarray, iCOGS and 11 different breast cancer GWAS in Michailidou et al.35 or Milne et al.48.
kPer allele computed odds-ratio (OR × ORBCAC).
lCompared with Michailidou et al’s OR estimates35.
Among the 21 ER-negative SNPs reported in Milne et al.48, only one (rs66823261) demonstrated significant evidence of association in the ER-negative case-only analysis (OR = 0.88, p < 2.7 × 10−4) (Table 1 and Supplementary Data 2). For the 20 other showing no association, the ORs estimated in Milne et al.48 would be applicable to BRCA1 mutation carriers.
To estimate the association of the seven significant SNPs with BC for BRCA1 mutation carriers (ORcomputed), the OR estimated using case-control data from BCAC (ORBCAC) was multiplied by the OR estimated using the case-only analysis (OR). For three SNPs, rs17426269, chr10_80841148_C_T and rs17529111, the magnitude of the association with BC for BRCA1 mutation carriers was greater than that in the general population (ORBCAC) and for two of these three, the ORcomputed was in the opposite direction than the ORBCAC (Table 1). For the four other SNPs (rs13281615, chr16_52599188_C_T, chr1_10566215_A_G and rs66823261), the estimated interaction OR resulted in the OR for associations with BRCA1 BC risk being closer to 1 (Table 1).
Among the remaining 172 SNPs (152 + 20) that showed no associations with BRCA1 mutation status, the estimated ORcomputed was smaller (i.e., closer to 1) than those estimated in the general population (ORBCAC) for 146 SNPs (85%, Supplementary Data 1 and 2). Based on the analysis of ER− tumours, the proportion of SNPs for which the ORcomputed was closer to 1 than the ORBCAC estimates was 59% (Supplementary Data 1 and 2).
For BRCA2 mutation carriers, among the 157 SNPs known to be associated with BC risk in the general population, 43 were associated with BRCA2 mutation carrier status at P < 0.05 in the case-only analysis that included all BCAC BC cases (Supplementary Data 3). However, only three SNPs (rs62355902, rs10759243 and chr22_40876234_C_T) showed associations after adjusting for multiple testing (P < 2.7 × 10−4) with OR estimates in the range of 0.88 to 0.89 (Table 2).
Table 2.
Known BC susceptibility SNPs demonstrating associations in the BRCA2 case-only analysis.
| Location | SNP namea | Chrb | Positionc | Nearest gene | Estimated effect allele | Referent allele | Frequencyd | r2 | ORe | Pf | ORBCACg | PBCACh | ORcomputedi | Variation in riskj |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5q11.2 | rs62355902 | 5 | 56053723 | MAP3K1 | T | A | 0.18 | 0.98 | 0.89 | 1.10e−04 | 1.18 | 8.50e−42 | 1.05 | TT1 |
| 9q31.2 | rs10759243 | 9 | 110306115 | RP11-438P9.2 | A | C | 0.31 | 1 | 0.89 | 4.60e−06 | 1.06 | 4.20e−10 | 0.95 | TT1 |
| 22q13.1 | chr22_40876234_C_T | 22 | 40876234 | MKL1 | C | T | 0.11 | 1 | 0.88 | 2.8e−04 | 1.12 | 5.70e−16 | 0.98 | TT1 |
N = 62,822 breast cancer cases (57,725 BCAC cases and 5,097 BRCA2 mutation carrier cases).
Considering SNPs with known BC (Michailidou et al.)35 associations in the general population.
TT1 tends to 1, ISD increase in same direction, IOD increase in opposite direction.
aAfter allowing for multiple testing, α* = 2.7 × 10−4.
bChromosome.
cBuild 37 position.
dFrequency of the allele for which effect is estimated in BCAC cases (OncoArray dataset).
ePer allele odds ratio estimated in the case-only analysis. OR values were computed from a two sided logistic regression using a 1 df lrtest adjusted for age at BC diagnosis, country and the first four principal components.
fp-value in the case-only analysis (after allowing for multiple testing, p* = 2.7 × 10−4).
gPer allele odds-ratio estimated in BCAC (Michailidou et al.)35.
hp-value in BCAC (Michailidou et al.)35. For SNPs with PBCAC > 10−8, significance was attained in merging data of Oncoarray, iCOGS and 11 different breast cancer GWAS in Michailidou et al.35.
iPer allele computed odds-ratio (OR × ORBCAC).
jCompared with Michailidou et al’s OR estimates35.
For these three SNPs, the observed interaction resulted in the magnitude of association with BC risk for BRCA2 mutation carriers (ORcomputed) being closer to 1 (Table 2).
Of the 154 SNPs that showed no significant associations with BRCA2 mutation status, 79% had ORs of BC for BRCA2 mutation carriers (ORcomputed) that were closer to 1 when compared to the ORs estimated using data in the general population (ORBCAC) (Supplementary Data 3).
Novel SNP modifiers
To identify novel SNPs that modify BC risks for BRCA1 and BRCA2 mutation carriers, we used a case-only design to investigate the associations of SNPs that had not been previously shown to be associated with BC in the general population (Fig. 4).
For BRCA1 mutation carriers, a total of 924 SNPs showed associations at P < 10−8 in all BC case-only analysis. To ensure that none of these associations are driven by differences in the distribution of ER-positive and ER-negative tumours in BCAC cases, an intermediate step was applied, in which we re-analysed the associations after restricting the BCAC data to only ER-negative cases. 220 of these SNPs remained significant at P < 10−7 located in 11 distinct genomic regions. SNPs were considered to belong to the same region if they were located within 500 kb of each other.
To ensure that none of these associations was driven by differences in the genotype imputation in the BCAC and CIMBA data (which had been carried out separately), all the SNPs in these 11 distinct genomic regions were re-imputed in the BCAC and CIMBA samples jointly and the associations for all SNPs in the regions were re-assessed in the control-only and case-only analyses. After the exclusion of 614 SNPs (613 on chromosome 17) that showed associations in the control-only analysis, 71 SNPs in two regions remained significant at P < 10–8 (Supplementary Data 4) in the case-only analyses including all BCAC cases. None of these SNPs had been previously reported in GWAS in the general population (p-values of association ranged from 0.51 to 5.9 × 10−5 with effect sizes in the range 0.96–1.04 in BCAC case-control analyses)35,48. A forward step-wise regression analysis within each of these two regions (restricted to the SNPs exhibiting associations at p < 10−8) starting with the most significant SNP and adding sequentially the other SNPs, identified a set of four conditionally independent SNPs (top SNPs) (Table 3): all SNPs were imputed, with r2 > 0.5, and had minor allele frequency (MAF) > 10%. Three of the top SNPs are located in 17q21.2. rs58117746 is an insertion of 16 bp within an exon of KRTAP4-5 leading to a frameshift of the amino acid sequence. rs5820435 and rs11079012 are both intronic and located in LEPREL4 (also named P3H4) and JUP, respectively, while rs80221606 is intronic and located in 11p11.2, within CELF1. The OR estimates of these four top SNPs ranged from 0.78 to 1.22. All showed evidence of heterogeneity in the OR by country (P < 0.05) (Table 3); however, in a leave-one-out analysis, in which each country was left out in turn, the overall associations remained similar (Supplementary Fig. 1 and 2) suggesting that no individual country had a big impact on the observed associations.
Table 3.
List of potential novel SNP modifiers associated in the case-only analysis for BRCA1 mutation carriers.
| Location | SNP namea | Chrb | Positionc | Nearest gene | Localisation | Estimated effect allele | Referent allele | r² d | Frequencye | ORf | Pg | ORERh | PER¯i | HRCIMBAj | PCIMBAk | ORBCACl | PBCACm | Phetn | Target geneo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11p11.2 | rs80221606 | 11 | 47560211 | CELF1 | Intronic | AT | A | 0.76 | 0.10 | 0.78 | 1.12e−10 | 0.76 | 6.36e−07 | 0.98 | 7.60e−01 | 1.04 | 0.01 | 1.39e−03 | Level 2 |
| 17q21.2 | rs58117746 | 17 | 39305775 | KRTAP4-5 | Pepshift | TGGCAGCAGCTGGGGC | T | 0.60 | 0.39 | 1.18 | 4.33e−10 | 1.15 | 7.71e−05 | 1.05 | 2.20e−02 | 1.02 | 0.26 | 4.60e−04 | – |
| 17q21.2 | rs5820435 | 17 | 39961558 | LEPREL4 | Intronic | A | C | 0.51 | 0.45 | 0.82 | 9.55e−12 | 0.85 | 7.71e−05 | 1.01 | 9.00e−01 | 1.02 | 0.07 | 1.06e−08 | – |
| 17q21.2 | rs11079012 | 17 | 39912880 | JUP | Intronic | G | C | 0.66 | 0.31 | 1.17 | 7.06e−09 | 1.18 | 2.35e−05 | 0.98 | 3.10e−01 | 1.01 | 0.51 | 1.15e−07 | Level 2 |
N = 67,469 breast cancer cases (60,212 BCAC cases and 7,257 BRCA1 mutation carrier cases)
aThe most significant SNP of each region after allowing for multiple testing, α* = 10−8
bChromosome.
cBuild 37 position.
dImputation accuracy.
eFrequency of the allele for which effect is estimated in BCAC cases (OncoArray dataset).
fPer allele odds ratio estimated in the case-only analysis. OR values were computed from a two sided logistic regression using a 1 df lrtest adjusted for age at BC diagnosis, country and the first four principal components.
gp-value in the case-only analysis.
hPer allele odds-ratio estimated in the case-only ER-negative subgroup analysis.
ip-value in the case-only ER-negative subgroup analysis.
jPer allele hazard ratio estimated in CIMBA cohort analysis.
kp-value found in CIMBA cohort analysis.
lPer allele odds-ratio estimated in BCAC (Michailidou et al.)35.
mp-value in BCAC (Michailidou et al.)35. For SNPs with PBCAC > 10−8, significance was attained in merging data of Oncoarray, iCOGS and 11 different breast cancer GWAS in Michailidou et al.35.
np-value of the heterogeneity test by country.
oINQUISIT score level: 1 = most functional evidence supporting a potential link between CCVs and target gene.
For BRCA2 mutation carriers, the case-only analysis identified 273 SNPs, located across 22 regions, with evidence of association at P < 10−8. After the joint re-imputation of the SNPs in these 22 regions, only 102 SNPs located in four regions (2p14, 13q13.1, and 13q13.2) remained associated at P < 10–8 (Supplementary Data 5). The step-wise regression analysis suggested that associations in each of the four regions were driven by a single variant (top SNPs) (Table 4). All four variants were imputed (with r2 > 0.5) and had MAF higher than 5%. At 2p14, rs12470785 (r2 = 0.98) is within an intron of ETAA1. At 13q13.1, rs79183898 (r2 = 0.84) is located between B3GALTL and RXFP2 and rs736596 (r2 = 0.66) is within an intron of STARD13. At 13q13.2, rs4943263 (r2 = 0.99) is located between RP11-266E6.3 and RP11-307O13.1. None of these SNPs had been previously reported to be associated with BC risk in BCAC studies in the general population (p-values from 0.01 to 0.90 in BCAC case-control analyses)35,48. The OR estimates of these four SNPs ranged from 0.85 to 1.37. All showed evidence of heterogeneity in the OR by country at p = 0.05 (Table 4). In the leave-one-country-out sensitivity analysis the two intergenic SNPs, rs79183898 and rs736596 were no longer significant at P < 10−4 when studies from the USA were excluded from the analysis and the OR estimates were substantially attenuated (Supplementary Figs. 3 and 4).
Table 4.
List of potential novel SNP modifiers associated in the case-only analysis for BRCA2 mutation carriers.
| Location | SNP namea | Chrb | Positionc | Nearest gene | Localisation | Estimated effect allele | Referent allele | r² d | Frequencye | ORf | Pg | HRCIMBAh | PCIMBAi | ORBCACj | PBCACk | PHETl | Target genem |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2p14 | rs12470785 | 2 | 67634003 | ETAA1 | Intron | G | A | 0.98 | 0.30 | 0.84 | 2.83e−11 | 0.89 | 1.69e−05 | 0.98 | 0.03 | 2.18e−07 | Level 2 |
| 13q13.1 | rs79183898 | 13 | 32221794 | B3GALTL - RXFP2 | Intergenic | A | T | 0.84 | 0.07 | 1.33 | 2.88e−10 | 1.04 | 3.55e−01 | 1.01 | 0.54 | 1.12e−08 | – |
| 13q13.1 | rs736596 | 13 | 33776506 | STARD13 | Intron | T | G | 0.66 | 0.09 | 1.37 | 3.44e−12 | 0.94 | 2.54e−01 | 0.98 | 0.45 | 4.99e−11 | Level 1 |
| 13q13.2 | rs4943263 | 13 | 35376357 | RP11-266E6.3 - RP11-307O13.1 | Intergenic | T | C | 0.99 | 0.27 | 1.17 | 8.33e−11 | 1.01 | 9.83e−01 | 1.00 | 0.47 | 6.94e−03 | Level 2 |
N = 62,822 breast cancer cases (57,725 BCAC cases and 5,097 BRCA2 mutation carrier cases).
aThe most significant SNP of each region after allowing for multiple testing, α* = 10−8
bChromosome.
cBuild 37 position.
dImputation accuracy.
eFrequency of the allele for which effect is estimated in BCAC cases (OncoArray dataset).
fPer allele odds ratio estimated in the case-only analysis. OR values were computed from a two sided logistic regression using a 1 df lrtest adjusted for age at BC diagnosis, country and the first four principal components.
gp-value in the case-only analysis.
hPer allele hazard ratio estimated in CIMBA cohort analysis.
ip-value found in CIMBA cohort analysis.
jPer allele odds-ratio estimated in BCAC (Michailidou et al.)35.
kp-value in BCAC (Michailidou et al. 2017). For SNPs with PBCAC > 10−8, significance was attained in merging data of Oncoarray, iCOGS and 11 different breast cancer GWAS in Michailidou et al.35.
lp-value of the heterogeneity test by country.
mINQUISIT score level: 1 = most functional evidence supporting potential link between CCVs and target gene.
In silico analyses on credible causal variants (CCV)
In order to determine the likely target genes of each region of the eight novel mutation carriers’ BC risk-associated SNPs, we first defined credible set of SNPs candidates to be causal (credible causal variants [CCVs]) (see “Methods”).
Sets of CCVs were sought for the two regions found in the previous step-wise analyses to be associated with risk in BRCA1 mutation carriers. In the region located at 11p11.2, only one signal composed of 74 CCVs was found (Table 5). All these 74 CCVs were imputed with a r2 higher than 0.92 (Supplementary Data 6). In the region located in 17q21.2, we found nine signals which contained from one to 13 CCVs (Table 5). Two of these CCVs were genotyped and the others had an r2 between 0.50 and 0.98 (Supplementary Data 6).
Table 5.
List of most significant SNPs in the CCV analysis for BRCA1 mutation carriers.
| Fine mapping regiona | Signalb | #CCVc | Location | SNP named | Chre | Positionf | Nearest gene | Localisation | Estimated effect allele | Referent Allele | Frequency g | r² h | Pi | ORj | PERk | ORl | PCIMBAm | HRCIMBAn |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| chr11:46773616-47773616 | 1 | 74 | 11p11.2 | rs60882887 | 11 | 47475675 | RAPSN, CELF1 | Intergenic | A | G | 0.14 | 0.95 | 2.20e−10 | 0.82 | 3.20e−06 | 0.82 | 7.00e−01 | 0.99 |
| chr17:39141815-40141815 | 1 | 2 | 17q21.2 | rs5820435 | 17 | 39961558 | LEPREL4 | Intronic | A | C | 0.45 | 0.51 | 1.10e−11 | 0.82 | 2.80e−05 | 0.85 | 9.10e−01 | 1.00 |
| 2 | 2 | 17q21.2 | rs7222250 | 17 | 39938469 | JUP | Intronic | C | T | 0.44 | 0.66 | 5.50e−14 | 1.23 | 3.90e−07 | 1.20 | 8.70e−01 | 1.00 | |
| 3 | 6 | 17q21.2 | rs9901834 | 17 | 39926811 | JUP | Intronic | A | G | 0.10 | 0.55 | 7.20e−10 | 0.72 | 3.90e−06 | 0.72 | 7.40e−01 | 1.02 | |
| 4 | 3 | 17q21.2 | rs58117746 | 17 | 39305775 | KRTAP4-5 | Intronic | TGGCAGCAGCTGGGGC | T | 0.39 | 0.60 | 5.50e−09 | 1.17 | 4.60e−04 | 1.13 | 2.20e−02 | 1.06 | |
| 5 | 13 | 17q21.2 | rs2239711 | 17 | 39633317 | KRT35 | Intronic | A | G | 0.29 | 0.93 | 4.90e−11 | 0.85 | 2.90e−04 | 0.88 | 5.00e−01 | 0.98 | |
| 6 | 4 | 17q21.2 | rs10708222 | 17 | 40137437 | DNAJC7 | Intronic | T | TA | 0.17 | 0.60 | 8.40e−07 | 1.18 | 6.10e−04 | 1.17 | 2.28e−01 | 0.95 | |
| 7 | 4 | 17q21.2 | rs41283425 | 17 | 39925713 | JUP | Intronic | T | C | 0.06 | 0.54 | 4.30e−07 | 0.73 | 1.30e−05 | 0.69 | 4.82e−01 | 0.95 | |
| 8 | 15 | 17q21.2 | rs56291217 | 17 | 39858199 | JUP | Intronic | AT | A | 0.44 | 0.76 | 6.70e−08 | 0.88 | 1.20e−06 | 0.85 | 4.06e−01 | 1.03 | |
| 9 | 1 | 17q21.2 | rs111637825 | 17 | 40134782 | DNAJC7 | Intronic | A | G | 0.06 | 0.89 | 3.60e−07 | 0.74 | 3.50e−04 | 0.75 | 4.47e−01 | 0.96 |
N = 67,469 breast cancer cases (60,212 BCAC cases and 7,257 BRCA1 mutation carrier cases).
aSignificant region in the main analysis used to look for credible causal variants (CCV).
bSignal number (the first one corresponds to the CCV set without any adjustment and the following are those with adjustment on each most significant SNP of the previous signals).
cNumber of credible causal variants at each signal (SNP with p-value at 2 order of magnitude of the most significant one).
dThe most significant SNP after adjustment on the most significant SNPs of the previous signals (except for these of the signal 1).
eChromosome.
fBuild 37 position.
gFrequency of the allele for which effect is estimated in BCAC cases (OncoArray dataset).
hImputation accuracy.
ip-value in the case-only analysis after adjustment on the most significant SNPs of the previous signals (except for these of the signal 1).
jPer allele odds ratio estimated in the case-only analysis. OR values were computed from a two sided logistic regression using a 1df lrtest adjusted for age at BC diagnosis, country, the first four principal components and the most significant SNPs of the previous signals (except for these of the signal 1).
kP-value in the case-only analysis restricted to ER-negative BCAC cases and after adjustment with the most significant SNP of the previous signals (except for these of the signal 1).
lPer allele odds ratio estimated in the case-only analysis restricted to ER-negative BCAC cases and after adjustment with the most significant SNP of the previous signals (except for these of the signal 1).
mp-value found in CIMBA cohort analysis.
n Per allele hazard ratio estimated in CIMBA cohort analysis.
We used INQUISIT35,54 to prioritize target genes by intersecting each CCV with publicly available annotation data from breast cells and tissues (see “Methods”). The results for BRCA1 mutation carriers are summarized in Supplementary Data 7. For BRCA1 mutation carriers, we predicted 38 unique target genes for six of the 10 independent signals. Seven target genes in two regions (MTCH2, MADD, PSMC3, RP11-750H9.5, SLC39A13, SPI1, and EIF1) were predicted with high confidence (designated Level 1, scoring range between Level 1 [highest confidence] to Level 3 [lowest confidence]). All seven Level 1 genes were predicted to be distally regulated by CCVs.
Similarly, sets of CCVs were sought from the four regions found in the previous step-wise analyses to be associated with risk in BRCA2 mutation carriers. A total of 17 signals were found. One signal composed of 78 CCVs was found in the region located at 2p14 (Table 6). One CCV was genotyped and the others were imputed with r2 between 0.95 and 0.99 (Supplementary Data 8). Twelve signals were found from the two regions previously found in 13q13.1 which contained from one to 46 CCVs. The analysis in the region of rs79183898 in 13q13.1 found three signals out of the 12, which are located in 13q12.3 (with top SNPs: rs71434801, rs77197167, rs114300732). Finally, four signals in the previously identified region located in 13q13.2 containing from three to 40 CCVs were found. Among all CCVs, 11 are genotyped and the imputed ones have an r2 higher than 0.58 (Table 6 and Supplementary Data 8).
Table 6.
List of most significant SNPs in the CCV analysis for BRCA2 mutation carriers.
| Fine mapping regiona | Signalb | #CCVc | Location | SNP named | Chre | Positionf | Nearest gene | Localisation | Estimated effect allele | Referent allele | Frequency g | r² h | Pi | ORj | PCIMBAk | HRCIMBAl |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| chr2:67099466-68099466 | 1 | 78 | 2p14 | rs12470785 | 2 | 67634003 | ETAA1 | Intronic | G | A | 0.30 | 0.98 | 4.20e−11 | 0.85 | 7.70e−05 | 0.89 |
| chr13:31015494-32515494 | 1 | 8 | 13q13.1 | rs79183898 | 13 | 32221794 | B3GALTL, RXFP2 | Intergenic | A | T | 0.07 | 0.84 | 1.10e−10 | 1.33 | 3.60e−01 | 1.04 |
| 2 | 23 | 13q12.3 | rs71434801 | 13 | 31249461 | USPL1, ALOX5AP | Intergenic | G | C | 0.13 | 0.76 | 3.40e−08 | 1.22 | 8.40e−01 | 0.99 | |
| 3 | 35 | 13q12.3 | rs77197167 | 13 | 31693513 | WDR95P, HSPH1 | Intergenic | C | T | 0.09 | 0.76 | 1.80e−07 | 1.25 | 4.00e−01 | 1.04 | |
| 4 | 7 | 13q12.3 | rs114300732 | 13 | 31662987 | WDR95P | Intronic | T | C | 0.07 | 0.90 | 1.70e−08 | 0.67 | 8.80e−02 | 1.09 | |
| 5 | 12 | 13q13.1 | 13:32231513:CAA:C | 13 | 32231513 | B3GALTL, RXFP2 | Intergenic | CAA | C | 0.25 | 0.92 | 8.40e−07 | 0.86 | 1.70e−02 | 1.08 | |
| 6 | 6 | 13q13.1 | rs1623189 | 13 | 32232683 | B3GALTL, RXFP2 | Intergenic | G | T | 0.26 | 0.95 | 1.30e−31 | 2.70 | 6.60e−01 | 1.01 | |
| chr13:33395975-34395975 | 1 | 1 | 13q13.1 | rs736596 | 13 | 33776506 | STARD13 | Intronic | T | G | 0.09 | 0.66 | 1.20e−12 | 1.37 | 2.50e−01 | 0.95 |
| 2 | 1 | 13q13.1 | rs77889880 | 13 | 33776161 | STARD13 | Intronic | T | A | 0.10 | 0.80 | 3.00e−21 | 0.51 | 1.90e−02 | 1.12 | |
| 3 | 1 | 13q13.1 | rs67776313 | 13 | 33934343 | RP11-141M1.3 | Intronic | A | AT | 0.33 | 0.70 | 7.70e−12 | 0.81 | 4.60e−01 | 0.98 | |
| 4 | 42 | 13q13.1 | rs71196514 | 13 | 33800572 | STARD13 | Intronic | C | CT | 0.38 | 0.67 | 1.00e−07 | 0.86 | 6.20e−01 | 1.01 | |
| 5 | 52 | 13q13.1 | rs2555605 | 13 | 33833810 | STARD13 | Intronic | C | T | 0.36 | 1.00 | 4.60e−08 | 0.87 | 2.00e−01 | 1.03 | |
| 6 | 46 | 13q13.1 | rs74796280 | 13 | 33700860 | STARD13 | Intronic | C | A | 0.06 | 0.96 | 4.70e−07 | 0.77 | 3.10e−02 | 0.89 | |
| chr13:34793902-35793902 | 1 | 18 | 13q13.2 | rs4943263 | 13 | 35376357 | RP11-266E6.3, RP11-307O13.1 | intergenic | T | C | 0.27 | 0.99 | 6.30e−11 | 1.18 | 9.80e−01 | 1.00 |
| 2 | 3 | 13q13.2 | rs2202781 | 13 | 35292372 | RP11-266E6.3, RP11-307O13.1 | Intergenic | G | A | 0.24 | 0.93 | 3.10e−11 | 1.20 | 6.00e−01 | 0.98 | |
| 3 | 40 | 13q13.2 | rs55675572 | 13 | 35315594 | RP11-266E6.3, RP11-307O13.1 | intergenic | A | T | 0.40 | 0.77 | 5.60e−08 | 0.86 | 7.50e−01 | 0.99 | |
| 4 | 21 | 13q13.2 | rs17755120 | 13 | 35270340 | RP11-266E6.3, RP11-307O13.1 | intergenic | T | A | 0.20 | 0.98 | 6.30e−07 | 0.76 | 4.80e−01 | 0.98 |
N = 62,822 breast cancer cases (57,725 BCAC cases and 5,097 BRCA2 mutation carrier cases).
aSignificant region in the main analysis used to look for credible causal variants (CCV).
bSignal number (the first one corresponds to the CCV set without any adjustment and the following are those with adjustment on each most significant SNP of the previous signals).
cNumber of credible causal variants at each signal (SNP with p-value at 2 order of magnitude of the most significant one).
dThe most significant SNP after adjustment on the most significant SNPs of the previous signals (except for these of the signal 1).
eChromosome.
fBuild 37 position.
gFrequency of the allele for which effect is estimated in BCAC cases (OncoArray dataset).
hImputation accuracy.
ip-value in the case-only analysis after adjustment on the most significant SNPs of the previous signals (except for these of the signal 1).
jPer allele odds ratio estimated in the case-only analysis. OR values were computed from a two sided logistic regression using a 1df lrtest adjusted for age at BC diagnosis, country, the first four principal components and the most significant SNPs of the previous signals (except for these of the signal 1).
kp-value found in CIMBA cohort analysis.
lPer allele hazard ratio estimated in CIMBA cohort analysis.
For BRCA2 mutation carriers, we predicted 24 unique target genes for 10 of the 17 independent signals, including one high confidence target gene, STARD13 at chr13:33395975-34395975. STARD13 was also predicted to be targeted by three independent signals. All results are presented in Supplementary Data 9.
Discussion
To identify novel genetic modifiers of BC risk for BRCA1 and BRCA2 mutation carriers and to further clarify the effects of known BC susceptibility SNPs on BC risk for carriers, a novel case-only analysis strategy was used based on GWAS data from unselected BC cases in BCAC and mutation carriers with BC from CIMBA. This strategy provides increased statistical power for detecting new associations and for clarifying the risk associations of known BC susceptibility SNPs in mutation carriers55.
Of the 179 known BC susceptibility SNPs identified through GWAS in the general population5–35, only 10 showed evidence of interaction with BRCA1 or BRCA2 mutation carrier status after taking the tumour ER-status into account. None of these 10 SNPs was among the fifty SNPs previously shown to be associated with BC for mutation carriers5,6,15,18,20,39–48. However, 82% of all 179 known susceptibility SNPs showed a predicted OR point estimate for mutation carriers closer to 1 than that estimated in the general population. The effect sizes in the general population may be somewhat exaggerated as the BCAC dataset used here contributed to the discovery of most of the loci, although this effect is likely to be small as most loci are highly significant and the effects have been replicated in independent datasets7. Taken together, these results suggest that, while most SNPs associated with risk in the general population are associated with risk for mutation carriers, the average effect sizes for mutation carriers are smaller. These findings are in line with previous results by Kuchenbaecker et al.49 and suggest that a PRS built using data from the general population will have a smaller effect size for BRCA1/2 mutation carriers.
For 10 SNPs, an interaction was observed with BRCA1 or BRCA2 mutation carrier status, suggesting that these SNPs have different effect sizes in BRCA1 or BRCA2 mutation carriers compared to the general population (seven for BRCA1 mutation carriers and three for BRCA2 mutation carriers). Specifically, for seven SNPs the confidence intervals were consistent with no effect on BC risk for mutation carriers, one SNP was associated with a larger OR for mutation carriers compared to the general population and two were associated in the opposite direction to that observed in the general population. However, distinguishing between a smaller effect size for mutation carriers compared to the general population OR estimates and no association for mutation carriers is very challenging since, even with the large sample size here, it is not possible to estimate precisely the effect sizes for individual variants. Larger sample sizes will be required for this purpose. Determining the precise effects of the SNPs in BRCA1 and BRCA2 mutation carriers will provide insights for understanding the biological basis of cancer development associated with BRCA1 and BRCA2 mutations.
We also identified eight novel conditionally independent common SNPs associated with BC risk (four for BRCA1 mutation carriers, four for BRCA2 mutation carriers). These have not been reported in previous association studies5,6,15,18,20,39–47. The case-only OR estimates for these SNPs varied from 0.85 to 1.37 for BRCA2 mutation carriers and from 0.78 to 1.22 for BRCA1 mutation carriers. For five of these SNPs the estimated ORs from the case-only analysis results were in the same direction as the estimated HRs from previously reported GWAS using cohort analyses restricted in BRCA1 and BRCA2 mutation carriers in CIMBA56. Two of these five SNPs also demonstrated some evidence of association in mutation carriers (p = 2.2 × 10−2 for rs58117746 for BRCA1 mutation carriers; and p = 7.7 × 10−5 for rs12470785 in ETAA1 for BRCA2 mutation carriers; Tables 3 and 4). For the remaining three variants, rs5820435 and rs11079012 at 17q21.2 and rs736596 at 13q13.1, the associations in BRCA1 or BRCA2 mutation carriers in the CIMBA data were not consistent with the observed interactions and might be artefactual. One possibility is that the associations with SNPs on 17q and 13q in BRCA1 and BRCA2 carriers respectively, reflect confounding due to linkage disequilibrium (LD) with specific mutations. Although we excluded variants with evidence of association in the control only analyses, it is possible that residual confounding due to specific mutations was still present.
Seven genes at a locus at 11p11.2 marked by rs60882887, were predicted with high confidence as targets, including MADD, SP11 and EIF1 which have previously been reported to be associated with BC biology57–59. However, no likely target genes were predicted at the 17q21.2 region. The lack of target gene predictions may be due to reliance on breast cell line data which does not represent the in vivo tissue of interest or due to the fact that the target transcripts are not annotated.
Only one gene, STARD13, was predicted as a potential target of SNPs at 13q13.1. This tumour suppressor gene has been previously implicated in metastasis, cell proliferation and development of BC60. However, rs736596, localized at 13q13.1, showed no association in CIMBA analyses and the association observed in our case-only analysis showed heterogeneity by country.
At the 2p14 locus, INQUISIT-predicted target genes included ETAA1 with lower confidence. The OR estimates obtained in the case-only analysis for the SNPs located in this gene were consistent with the HR estimated in previously reported CIMBA analyses56. Moreover, around one hundred correlated SNPs, were associated with BRCA2 mutation carrier status at p < 10−8, including the genotyped SNP chr2_67654113_C_T.
The validity of the case-only analysis as evidence of interaction relies on the assumption of independence between the mutation status and the SNPs under investigation61. Therefore, based on the control-only analyses, we excluded ~2,000 SNPs which were associated with BRCA1 or BRCA2 mutation carrier status and also showed an association with risk in the case-only analyses (Supplementary Fig. 5). While most of these associations are probably spurious, due to (intra- or inter-chromosomal) LD with BRCA1 or BRCA2 mutations, it is possible that some may reflect true associations and that the higher frequency in unaffected BRCA1/2 may be because they are relatives of BC cases. These associations may warrant further evaluation using other study designs. A recent publication using data from the Framingham Heart Study suggested that interchromosomal LD can be caused by bio-genetic mechanisms possibly associated with favourable or unfavourable epistatic evolution62. SNPs for which no association with mutation carrier status was found at the significance level of 10−8 were assumed to be independent of the mutation status. However, this does not necessarily rule out residual LD between the novel SNPs on chromosomes 13 and 17 and BRCA1 or BRCA2 mutations. Therefore, the OR estimates for these SNPs might be biased and may further explain the lack of evidence of association in the CIMBA only analyses.
Our findings highlight the importance of imputation in GWAS. The imputed genome-wide genotype data used in the main case-only association analyses were based on carrying out the imputation separately for the BC cases from BCAC and CIMBA. We found that 28 out of the 33 regions associated with BRCA1 or BRCA2 mutation carrier status were no longer associated with risk after re-imputing all samples together. By re-imputing all the data together we ensured that the associations observed for the remaining regions are robust to potential differences in the imputation accuracy between the BCAC and CIMBA samples.
Under our analytical strategy, only the regions for which evidence of associated with BC risk was observed were re-imputed using all BCAC and CIMBA samples combined. This re-imputation was not done at genome-wide level due to computational constraints and this may have led to false-negative associations being excluded for further evaluation as potential novel modifiers. Future analyses should aim to analyse the genome-wide associations after the genome-wide re-imputation across the combined BCAC and CIMBA dataset. However, our approach using joint one-step imputation should have ensured that associations we report (all of which are common SNPs with imputation scores > 0.5) are not driven by inaccuracies in imputation.
Due to the recruitment of participants in CIMBA studies primarily through genetic counselling, the mean age at diagnosis of mutation carriers was 16 years younger than the BC cases participating in BCAC. Although all analyses were adjusted for age, the observed associations might be related to the ageing process instead of interactions with mutation carrier status. Another source of bias could be related to the fact that there are 1.5 times more prevalent cases among CIMBA (68.1%) than BCAC (42.3%) with a delay between diagnosis and study recruitment of 6.83 years and 2.07 years respectively. An observed association might be due to a differential survival between CIMBA and BCAC cases. However, none of the identified SNPs has been found to be associated with BC survival63.
The majority (92.5%) of cases and controls in BCAC were not tested for BRCA1/2 mutations at the time of enrolment, potentially leading to some attenuation in the interaction OR (as some BCAC cases will be carriers). However, most BCAC studies were population-based case-control studies and the proportion of cases and controls that carry pathogenic BRCA1/2 mutations will be small (<5%), hence any attenuation is likely to be negligible.
Despite heterogeneity in the interaction ORs by country for some SNPs, results were generally robust to the exclusion of each country sequentially except, for two SNPs (rs79183898 and rs736596) found associated with BRCA2 mutation carrier status; for these, the association seemed to be driven by data from the USA. For the other SNPs, the observed heterogeneity may be due to random error, given the relatively small sample sizes of each country. However, if these differences are real, future PRS for BRCA1 and BRCA2 carriers should consider the country-specific differences.
This is the first analysis of genetic modifiers of BC risk that investigated the differences in the association of common genetic variants with BC risk in the general population and in women with BRCA1 or BRCA2 mutations. The inclusion of unselected BC cases resulted in increased sample size and hence a gain in statistical power for identifying novel SNPs. These represent the largest currently available datasets, but it is important to replicate these observations in independent samples. This should be possible through the ongoing CONFLUENCE (https://dceg.cancer.gov/research/cancer-types/breast-cancer/confluence-project) large-scale genotyping experiment. More detailed fine mapping and functional analysis will be required to elucidate the role of the novel variants identified in BC development for BRCA1 and BRCA2 mutation carriers. Our findings should contribute to the improved performance of BC PRS for absolute risk prediction for BRCA1 and BRCA2 mutation carriers, which will help inform decisions on the best timing for risk-reducing surgery, risk reduction medication, or the start of surveillance.
Methods
Study sample
We used data from two international consortia, BCAC64 and CIMBA56. BCAC included data from 108 studies of BC from 33 countries in North America, Europe and Australia, the majority (88%) of which were case-control studies. The majority of BCAC cases/controls were not tested for BRCA1/2 mutations at the time of enrolment. However, most studies were population-based, hence the proportion of cases and controls that carry pathogenic BRCA1/2 mutations will be small. CIMBA participants were women with pathogenic mutations in BRCA1 or BRCA2. All participants were at least 18 years old. The majority of mutation carriers were recruited through cancer genetics clinics and enroled into national or regional studies. Data were available on 30,500 BRCA1 mutation carriers and 20,500 BRCA2 mutation carriers from 77 studies in 32 countries. A total of 188,320 BC cases and 161,669 controls were available from both consortia. All studies provided information on disease status, age at diagnosis or at interview. Oestrogen receptor status was available for 72% of BCAC cases and 71% of CIMBA cases. All subjects provided written informed consent and participated in studies with protocols approved by ethics committees at each participating institution.
Sample selection
BCAC cases were women diagnosed with BC7. To define disease status in CIMBA participants, women were censored at the first of the following events: age at BC diagnosis, age at ovarian cancer diagnosis, other cancer, bilateral prophylactic mastectomy or age at study recruitment. Subjects censored at a BC diagnosis were considered as cases.
A control-only analysis was carried out to test the independence between the SNPs and the BRCA1 and BRCA2 mutation carrier status. In BCAC, controls were defined as individuals unaffected by BC at study recruitment35. In CIMBA, participants were considered as controls if they were unaffected at recruitment.
Only women of European ancestry were included. To minimise the chance of observing spurious associations due to differences in the distribution of BC cases in the population by tumour characteristics (defined as unselected BC cases), 3,478 BCAC cases from four studies were excluded because they were included in clinical trials based on breast tumour characteristics as HER-2 receptor status (see Supplementary Table 2). Because all the analyses were adjusted for country, to ensure that the number of subjects in each country stratum was large enough, we excluded the CIMBA data from any country for which there were less than ten BC cases with BRCA1 or BRCA2 mutation. Consequently, data from Poland and Russia were excluded from the BRCA2 analyses (Supplementary Table 3). Finally, duplicate subjects between BCAC and CIMBA were excluded from the BCAC data (114 and 80 subjects from the BRCA1 and BRCA2 case-only analyses, respectively; eight subjects from control-only analyses).
A total of 60,212 BCAC cases and 7,257 BRCA1 mutation carrier cases were available for the BRCA1 case-only analyses and 57,725 BCAC cases and 5097 BRCA2 mutation carrier cases were available for the BRCA2 case-only analyses (Fig. 1). A total of 45,881 BCAC controls and 5,750 BRCA1 mutation carrier controls were available for the BRCA1 control-only analyses and 43,549 BCAC controls and 4,456 BRCA2 mutation carrier controls for the BRCA2 control-only analyses (Fig. 2).
Genotype data
All the study samples were genotyped using the OncoArray Illumina beadchip65. The array includes a backbone of ~260,000 SNPs that provide genome-wide coverage of most common variants, together with markers of interest for breast and other cancers identified through GWAS, fine-mapping of known susceptibility regions, and other approaches65.
A standard genotype quality control process was followed for both the BCAC and CIMBA samples which have been described in detail elsewhere35,48. Briefly, this involved excluding SNPs located on chromosome Y; SNPs with call rates <95%; SNPs with MAF < 0.05 and call rate <98%; monomorphic SNPs; and SNPs for which evidence of departure from Hardy-Weinberg equilibrium was observed (P < 10−7 based on a country-stratified test).
Genotypes for ~21 million SNPs were imputed for all subjects using the 1000 Genomes Phase III data (released October 2014) as reference panel, as described previously66. Briefly, the number of reference haplotypes used as templates when imputing missing genotypes was fixed to 800 (-k_hap = 800). A two-stage imputation approach was used: phasing with SHAPEIT67,68 and imputation with IMPUTE269 using 5 Mb non-overlapping intervals. Genotypes were imputed for all SNPs that were found polymorphic (MAF > 0.1%) in either European or Asian populations.
The genome-wide imputation process described above was carried out separately for the BCAC and CIMBA samples. However, this may potentially lead to spurious associations if there are differences in the quality of the imputation (measured using the imputation accuracy r² metric70) for a given SNP between the two datasets. To address this, a stringent approach was employed which involved including only SNPs for which the difference in r² between the BCAC and CIMBA SNP imputations (Δr²) was minimal relative to their r² values. SNPs with r² > 0.9 in both BCAC and CIMBA were kept in the analyses only if Δr² < 0.05; SNPs with 0.8 < r² ≤ 0.9 in both BCAC and CIMBA were kept if Δr² < 0.02 and, SNPs with 0.5 < r² ≤ 0.8 in both BCAC and CIMBA were kept if Δr² < 0.01. All SNPs with r² < 0.5 in either CIMBA or BCAC were excluded. Only SNPs with a MAF > 0.01 in BCAC cases were included.
Consequently, 9,072,535 SNPs were included in the BRCA1 analyses (402,336 genotyped and 8,670,199 imputed SNPs) and 9,047,403 SNPs in the BRCA2 analyses (402,397 genotyped and 8,645,006 imputed SNPs).
Case-only and control-only analyses
The comparison of SNP frequency between CIMBA cases and BCAC cases (case-only analyses), or between unaffected CIMBA subjects and BCAC controls (control-only analyses), was performed using logistic regression adjusted for age at BC diagnosis in the case-only analyses and for age at interview for BCAC controls or at censure for CIMBA unaffected subjects in the control-only analyses, as well as for country and principal components (PCs) to account for population structure. Separate analyses were carried out for BRCA1 and BRCA2 mutation carriers. To define the number of PC for inclusion in the models, the principal component analysis was carried out using 35,858 uncorrelated genotyped SNPs on the OncoArray and purpose-written software (http://ccge.medschl.cam.ac.uk/software/pccalc/). The inflation statistic was calculated and converted to an equivalent statistic for a study of 1,000 subjects for each outcome (λ1,000) by adjusting for effective study size:
where n and m are the numbers of BCAC and CIMBA subjects respectively. The models were adjusted with the first four PCs (λ1,000 with and without PCs in the model = 1.03 and 1.21, respectively) since additional PCs did not result in further reduction in the inflation of the test statistics.
Strategy for determining significant associations
The analytical process is summarised in Figs. 3 and 4. A fundamental assumption when using a case-only design in this context is that the SNPs and mutation carrier status are independent61. To confirm independence, SNPs likely to be in linkage disequilibrium (LD) with BRCA1 or BRCA2 mutations, i.e., those located in or within 500 kb of either gene, were excluded. However, LD also exists between variants at long-distance on the same chromosome or even on a different chromosome (interchromosomal LD)62,71. Therefore, control-only analyses were performed to further exclude SNPs associated with mutation carrier status in unaffected women72, using a stringent statistical significance level of 10−8).
After excluding SNPs in LD or in interchromosomal LD with BRCA1 or BRCA2 mutations, case-only analyses were performed to assess the association between SNPs and BRCA1 or BRCA2 mutation carrier status. We considered two categories of SNPs depending on whether they had been previously found to be associated with BC in published BCAC studies35,48. For known BC susceptibility SNPs (Fig. 3) we used a significance threshold of 2.7 × 10−4 (applying Bonferroni correction to 179 tests) and for potential novel SNP modifier (Fig. 4) a stringent significance threshold of 10−8 was used.
Because BRCA1mutation-associated tumours are more often ER-negative than those in the general population73, a subsequent case-only analysis was performed restricting the BCAC cases to those with ER-negative disease. We used this strategy for two reasons. First, we wished to exclude associations driven by differences in the tumour ER-status distributions between BRCA1 carriers and BCAC cases. Therefore, in the BRCA1 analysis, SNPs were considered to be associated with mutation carrier status only if they were also associated in the ER-negative case-only analysis at a prior defined significance threshold of 10−7 for novel SNP modifiers (Fig. 4) and of 0.05 for the established BC susceptibility SNPs after a pre-selection at P < 2.7 × 10−4 in the BRCA1 overall case-only analysis (Fig. 3). The second reason we applied this strategy was to identify novel SNP modifiers specific to BRCA1/ER-negative tumours that had not been detected in the overall analysis; for this we applied a significance threshold of 10−8.
To confirm that potentially novel associations in the case-only analysis were not driven by differences in the imputation accuracy between the CIMBA and BCAC data, each of the regions defined as ±500 kb around the associated SNP, were re-imputed for the combined CIMBA and BCAC samples. The more accurate one-stage imputation was carried out, using IMPUTE2 without pre-phasing. Associations with all the SNPs in the re-imputed regions were then re-evaluated using the control-only and case-only analytical approaches described above. Finally, we used a step-wise regression analysis using a significance threshold of 10−8 in order to determine whether associations with SNPs in the same region are independent and to define the conditionally independent SNPs (top SNPs).
Among the 179 established BC susceptibility SNPs, 107 were genotyped and 71 were imputed. As previously, although none of these 71 SNPs were excluded based on their Δr², to exclude potentially spurious associations, regions around these 71 SNPs were re-imputed using the one-stage imputation applied to BCAC and CIMBA data combined, and before performing the control-only and case-only analyses.
Determining the magnitude of association
For the potentially novel SNP modifiers the risk ratio of BC applicable to mutation carriers was assumed to be equal to the OR estimate from the case-only analysis (with the hypothesis that their relative risk equals 1 in the general population, given that none of them was found to be associated with BC in BCAC)55.
For the known BC susceptibility SNPs, a significant association in the case-only analysis implies that the magnitude of association is different for BRCA1 or BRCA2 mutation carriers than for the general population. Therefore, the risk ratio of BC for mutation carriers was computed as the product of OR × ORBCAC where OR was obtained from the case-only analysis, and ORBCAC was the odds ratio of association obtained from either Michailidou et al.35 for the SNPs associated with overall BC risk and from Milne et al.48 for the SNPs associated with ER-negative BC.
For all associated SNPs in case-only analyses, heterogeneity by country was assessed using likelihood ratio tests that compared models with and without an SNP by country interaction term. When the heterogeneity test was significant at P < 0.05, a leave-one-out analysis was performed, by excluding each country in turn to assess the influence of a data from a specific country on the overall association.
Credible causal variants
For each novel region, we defined sets of credible causal variants (CCVs) to use in the prediction of the likely target genes. For this purpose, we defined a first set of CCVs including the top SNP of the region of interest and the SNPs with p-values of association within two orders of magnitude of the top SNP association. Then, we sequentially performed logistic regression analyses using all other SNPs in the region, adjusted for the top SNP. We defined a second set of CCVs which included the most significant SNP after adjusting for the top SNP and the SNPs with p-values within two orders of magnitude of the most significant SNP association. This was repeated (conditioning on the previously found most significant SNPs) to define additional sets of CCVs as long as at least one p-value remained <10−6.
eQTL analysis
Data from BC tumours and adjacent normal breast tissue were accessed from The Cancer Genome Atlas74 (TCGA). Germline SNP genotypes (Affymetrix 6.0 arrays) from individuals of European ancestry were processed and imputed to the 1000 Genomes reference panel (October 2014)35. Tumour tissue copy number was estimated from the Affymetrix 6.0 and called using the GISTIC2 algorithm75. Complete genotype, RNA-seq and copy number data were available for 679 genetically European patients (78 with adjacent normal tissue). Further, RNA-seq for normal breast tissue and imputed germline genotype data were available from 80 females from the GTEx Consortium76. Genes with a median expression level of 0 RPKM across samples were removed, and RPKM values of each gene were log2 transformed. Expression values of samples were quantile normalized. Genetic variants were evaluated for association with the expression of genes located within ±2 Mb of the lead variant at each risk region using linear regression models, adjusting for ESR1 expression. Tumour tissue was also adjusted for copy number variation77. eQTL analyses were performed using the MatrixEQTL program in R78.
INQUISIT analyses
Candidate target genes were evaluated by assessing each CCV’s potential impact on regulatory or coding features using a computational pipeline, INtegrated expression QUantitative trait and In SIlico prediction of GWAS Targets (INQUISIT)35,54. Briefly, genes were considered as potential targets of candidate causal variants through effects on: (1) distal gene regulation, (2) proximal regulation, or (3) a gene’s coding sequence. We intersected CCV positions with multiple sources of genomic information chromatin interaction analysis by paired-end tag sequencing (ChIA-PET79) in MCF7 cells and genome-wide chromosome conformation capture (Hi-C) in HMECs. We used breast cell line computational enhancer–promoter correlations (PreSTIGE80, IM-PET81, FANTOM582) breast cell super-enhancer83, breast tissue-specific expression variants (eQTL) from multiple independent studies (TCGA (normal breast and breast tumour) and GTEx breast—see eQTL methods), transcription factor and histone modification chromatin immunoprecipitation followed by sequencing (ChIP-seq) from the ENCODE and Roadmap Epigenomics Projects together with the genomic features found to be significantly enriched for all known breast cancer CCVs54, gene expression RNA-seq from several breast cancer lines and normal samples (ENCODE) and topologically associated domain (TAD) boundaries from T47D cells (ENCODE84). To assess the impact of intragenic variants, we evaluated their potential to alter primary protein coding sequence and splicing using Ensembl Variant Effect Predictor85 using MaxEntScan and dbscSNV modules for splicing alterations based on ada and rf scores. Nonsense and missense changes were assessed with the REVEL ensemble algorithm, with CCVs displaying REVEL scores >0.5 deemed deleterious.
Each target gene prediction category (distal, promoter or coding) was scored according to different criteria. Genes predicted to be distally regulated targets of CCVs were awarded points based on physical links (for example ChIA-PET), computational prediction methods, or eQTL associations. All CCVs were considered as potentially involved in distal regulation. Intersection of a putative distal enhancer with genomic features found to be significantly enriched54 were further upweighted. Multiple independent interactions were awarded an additional point. CCVs in gene proximal regulatory regions were intersected with histone ChIP-Seq peaks characteristic of promoters and assigned to the overlapping transcription start sites (defined as −1.0 kb – +0.1 kb). Further points were awarded to such genes if there was evidence for eQTL association, while a lack of expression resulted in down-weighting as potential targets. Potential coding changes including missense, nonsense and predicted splicing alterations resulted in addition of one point to the encoded gene for each type of change, while lack of expression reduced the score. We added an additional point for predicted target genes that were also breast cancer drivers (278 genes35,54). For each category, scores potentially ranged from 0 to 8 (distal); 0 to 4 (promoter) or 0 to 3 (coding). We converted these scores into ‘confidence levels’: Level 1 (highest confidence) when distal score > 4, promoter score ≥3 or coding score >1; Level 2 when distal score ≤ 4 and ≥1, promoter score = 1 or = 2, coding score = 1; and Level 3 when distal score <1 and >0, promoter score <1 and >0, and coding <1 and >0. For genes with multiple scores (for example, predicted as targets from multiple independent risk signals or predicted to be impacted in several categories), we recorded the highest score.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
BCAC acknowledgements. We thank all the individuals who took part in these studies and all the researchers, clinicians, technicians and administrative staff who have enabled this work to be carried out. ABCFS thank Maggie Angelakos, Judi Maskiell, Gillian Dite. ABCS thanks the Blood bank Sanquin, The Netherlands. ABCTB Investigators: Christine Clarke, Deborah Marsh, Rodney Scott, Robert Baxter, Desmond Yip, Jane Carpenter, Alison Davis, Nirmala Pathmanathan, Peter Simpson, J. Dinny Graham, Mythily Sachchithananthan. Samples are made available to researchers on a non-exclusive basis. BBCS thanks Eileen Williams, Elaine Ryder-Mills, Kara Sargus. BCEES thanks Allyson Thomson, Christobel Saunders, Terry Slevin, BreastScreen Western Australia, Elizabeth Wylie, Rachel Lloyd. The BCINIS study would not have been possible without the contributions of Dr. K. Landsman, Dr. N. Gronich, Dr. A. Flugelman, Dr. W. Saliba, Dr. E. Liani, Dr. I. Cohen, Dr. S. Kalet, Dr. V. Friedman, Dr. O. Barnet of the NICCC in Haifa, and all the contributing family medicine, surgery, pathology and oncology teams in all medical institutes in Northern Israel. The BREOGAN study would not have been possible without the contributions of the following: Manuela Gago-Dominguez, Jose Esteban Castelao, Angel Carracedo, Victor Muñoz Garzón, Alejandro Novo Domínguez, Maria Elena Martinez, Sara Miranda Ponte, Carmen Redondo Marey, Maite Peña Fernández, Manuel Enguix Castelo, Maria Torres, Manuel Calaza (BREOGAN), José Antúnez, Máximo Fraga and the staff of the Department of Pathology and Biobank of the University Hospital Complex of Santiago-CHUS, Instituto de Investigación Sanitaria de Santiago, IDIS, Xerencia de Xestion Integrada de Santiago-SERGAS; Joaquín González-Carreró and the staff of the Department of Pathology and Biobank of University Hospital Complex of Vigo, Instituto de Investigacion Biomedica Galicia Sur, SERGAS, Vigo, Spain. BSUCH thanks Peter Bugert, Medical Faculty Mannheim. CBCS thanks study participants, co-investigators, collaborators and staff of the Canadian Breast Cancer Study, and project coordinators Agnes Lai and Celine Morissette. CCGP thanks Styliani Apostolaki, Anna Margiolaki, Georgios Nintos, Maria Perraki, Georgia Saloustrou, Georgia Sevastaki, Konstantinos Pompodakis. CGPS thanks staff and participants of the Copenhagen General Population Study. For the excellent technical assistance: Dorthe Uldall Andersen, Maria Birna Arnadottir, Anne Bank, Dorthe Kjeldgård Hansen. The Danish Cancer Biobank is acknowledged for providing infrastructure for the collection of blood samples for the cases. CNIO-BCS thanks Guillermo Pita, Charo Alonso, Nuria Álvarez, Pilar Zamora, Primitiva Menendez, the Human Genotyping-CEGEN Unit (CNIO). The CTS Steering Committee includes Leslie Bernstein, Susan Neuhausen, James Lacey, Sophia Wang, Huiyan Ma, and Jessica Clague DeHart at the Beckman Research Institute of City of Hope, Dennis Deapen, Rich Pinder, and Eunjung Lee at the University of Southern California, Pam Horn-Ross, Peggy Reynolds, Christina Clarke Dur and David Nelson at the Cancer Prevention Institute of California, Hoda Anton-Culver, Argyrios Ziogas, and Hannah Park at the University of California Irvine, and Fred Schumacher at Case Western University. DIETCOMPLYF thanks the patients, nurses and clinical staff involved in the study. The DietCompLyf study was funded by the charity Against Breast Cancer (Registered Charity Number 1121258) and the NCRN. We thank the participants and the investigators of EPIC (European Prospective Investigation into Cancer and Nutrition). ESTHER thanks Hartwig Ziegler, Sonja Wolf, Volker Hermann, Christa Stegmaier, Katja Butterbach. GC-HBOC thanks Stefanie Engert, Heide Hellebrand, Sandra Kröber and LIFE - Leipzig Research Centre for Civilization Diseases (Markus Loeffler, Joachim Thiery, Matthias Nüchter, Ronny Baber). The GENICA Network: Dr. Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart, and University of Tübingen, Germany [HB, Wing-Yee Lo], German Cancer Consortium (DKTK) and German Cancer Research Center (DKFZ) Partner Site Tübingen [[HB], gefördert durch die Deutsche Forschungsgemeinschaft (DFG) im Rahmen der Exzellenzstrategie des Bundes und der Länder - EXC 2180 - 390900677 [HB], Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany [YDK, Christian Baisch], Institute of Pathology, University of Bonn, Germany [Hans-Peter Fischer], Molecular Genetics of Breast Cancer, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, Germany [Ute Hamann], Institute for Prevention and Occupational Medicine of the German Social Accident Insurance, Institute of the Ruhr University Bochum (IPA), Bochum, Germany [Thomas Brüning, Beate Pesch, Sylvia Rabstein, Anne Lotz]; and Institute of Occupational Medicine and Maritime Medicine, University Medical Center Hamburg-Eppendorf, Germany [Volker Harth]. HABCS thanks Michael Bremer. HEBCS thanks Kirsimari Aaltonen, Irja Erkkilä. HUBCS thanks Shamil Gantsev. KARMA and SASBAC thank the Swedish Medical Research Counsel. KBCP thanks Eija Myöhänen, Helena Kemiläinen. kConFab/AOCS wish to thank Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow-Up Study (which has received funding from the NHMRC, the National Breast Cancer Foundation, Cancer Australia, and the National Institute of Health (USA)) for their contributions to this resource, and the many families who contribute to kConFab. LMBC thanks Gilian Peuteman, Thomas Van Brussel, EvyVanderheyden and Kathleen Corthouts. MARIE thanks Petra Seibold, Dieter Flesch-Janys, Judith Heinz, Nadia Obi, Alina Vrieling, Sabine Behrens, Ursula Eilber, Muhabbet Celik, Til Olchers and Stefan Nickels. MBCSG (Milan Breast Cancer Study Group): Mariarosaria Calvello, Davide Bondavalli, Aliana Guerrieri Gonzaga, Monica Marabelli, Irene Feroce, and the personnel of the Cogentech Cancer Genetic Test Laboratory. The MCCS was made possible by the contribution of many people, including the original investigators, the teams that recruited the participants and continue working on follow-up, and the many thousands of Melbourne residents who continue to participate in the study. We thank the coordinators, the research staff and especially the MMHS participants for their continued collaboration on research studies in breast cancer. MSKCC thanks Marina Corines, Lauren Jacobs. MTLGEBCS would like to thank Martine Tranchant (CHU de Québec – Université Laval Research Center), Marie-France Valois, Annie Turgeon and Lea Heguy (McGill University Health Center, Royal Victoria Hospital; McGill University) for DNA extraction, sample management and skilful technical assistance. J.S. is Chair holder of the Canada Research Chair in Oncogenetics. NBHS and SBCGS thank study participants and research staff for their contributions and commitment to the studies. For NHS and NHS2 the study protocol was approved by the institutional review boards of the Brigham and Women’s Hospital and Harvard T.H. Chan School of Public Health, and those of participating registries as required. We would like to thank the participants and staff of the NHS and NHS2 for their valuable contributions as well as the following state cancer registries for their help: A.L., A.Z., A.R., C.A., C.O., C.T., D.E., F.L., G.A., I.D., I.L., I.N., I.A., K.Y., L.A., M.E., M.D., M.A., M.I., N.E., N.H., N.J., N.Y., N.C., N.D., O.H., O.K., O.R., P.A., R.I., S.C., T.N., T.X., V.A., W.A., and W.Y. The authors assume full responsibility for analyses and interpretation of these data. OFBCR thanks Teresa Selander, Nayana Weerasooriya. ORIGO thanks E. Krol-Warmerdam, and J. Blom for patient accrual, administering questionnaires, and managing clinical information. PBCS thanks Louise Brinton, Mark Sherman, Neonila Szeszenia-Dabrowska, Beata Peplonska, Witold Zatonski, Pei Chao, Michael Stagner. The ethical approval for the POSH study is MREC /00/6/69, UKCRN ID: 1137. We thank staff in the Experimental Cancer Medicine Centre (ECMC) supported Faculty of Medicine Tissue Bank and the Faculty of Medicine DNA Banking resource. RBCS thanks Jannet Blom, Saskia Pelders, Annette Heemskerk and the Erasmus MC Family Cancer Clinic. We thank the SEARCH and EPIC teams. SKKDKFZS thanks all study participants, clinicians, family doctors, researchers and technicians for their contributions and commitment to this study. SZBCS thanks Ewa Putresza. UCIBCS thanks Irene Masunaka. UKBGS thanks Breast Cancer Now and the Institute of Cancer Research for support and funding of the Breakthrough Generations Study, and the study participants, study staff, and the doctors, nurses and other health care providers and health information sources who have contributed to the study. We acknowledge NHS funding to the Royal Marsden/ICR NIHR Biomedical Research Centre. We acknowledge funding to the Manchester NIHR Biomedical Research Centre (IS-BRC-1215-20007). The authors thank the WHI investigators and staff for their dedication and the study participants for making the program possible. CIMBA acknowledgments. All the families and clinicians who contribute to the studies; Catherine M. Phelan for her contribution to CIMBA until she passed away on 22 September 2017; Sue Healey, in particular taking on the task of mutation classification with the late Olga Sinilnikova; Maggie Angelakos, Judi Maskiell, Gillian Dite, Helen Tsimiklis; members and participants in the New York site of the Breast Cancer Family Registry; members and participants in the Ontario Familial Breast Cancer Registry; Vilius Rudaitis and Laimonas Griškevičius; Drs Janis Eglitis, Anna Krilova and Aivars Stengrevics; Yuan Chun Ding and Linda Steele for their work in participant enrollment and biospecimen and data management; Bent Ejlertsen and Anne-Marie Gerdes for the recruitment and genetic counseling of participants; Alicia Barroso, Rosario Alonso and Guillermo Pita; all the individuals and the researchers who took part in CONSIT TEAM (Consorzio Italiano Tumori Ereditari Alla Mammella), in particular: Bernard Peissel, Dario Zimbalatti, Daniela Zaffaroni, Alessandra Viel, Giuseppe Giannini Liliana Varesco, Viviana Gismondi, Maria Grazia Tibiletti, Daniela Furlan, Antonella Savarese, Aline Martayan, Stefania Tommasi, Brunella Pilato and the personnel of the Cogentech Cancer Genetic Test Laboratory, Milan, Italy. Ms. JoEllen Weaver and Dr. Betsy Bove; FPGMX: members of the Cancer Genetics group (IDIS): Marta Santamariña, Miguel Aguado and Olivia Ríos; IFE - Leipzig Research Centre for Civilization Diseases (Markus Loeffler, Joachim Thiery, Matthias Nüchter, Ronny Baber); We thank all participants, clinicians, family doctors, researchers, and technicians for their contributions and commitment to the DKFZ study and the collaborating groups in Lahore, Pakistan (Noor Muhammad, Sidra Gull, Seerat Bajwa, Faiz Ali Khan, Humaira Naeemi, Saima Faisal, Asif Loya, Mohammed Aasim Yusuf) and Bogota, Colombia (Ignacio Briceno, Fabian Gil). Genetic Modifiers of Cancer Risk in BRCA1/2 Mutation Carriers (GEMO) study is a study from the National Cancer Genetics Network UNICANCER Genetic Group, France. We wish to pay a tribute to Olga M. Sinilnikova, who with Dominique Stoppa-Lyonnet initiated and coordinated GEMO until she sadly passed away on the 30th June 2014. The team in Lyon (Olga Sinilnikova, Mélanie Léoné, Laure Barjhoux, Carole Verny-Pierre, Sylvie Mazoyer, Francesca Damiola, Valérie Sornin) managed the GEMO samples until the biological resource centre was transferred to Paris in December 2015 (Noura Mebirouk, Fabienne Lesueur, Dominique Stoppa-Lyonnet). We want to thank all the GEMO collaborating groups for their contribution to this study: Coordinating Centre, Service de Génétique, Institut Curie, Paris, France: Muriel Belotti, Ophélie Bertrand, Anne-Marie Birot, Bruno Buecher, Sandrine Caputo, Anaïs Dupré, Emmanuelle Fourme, Marion Gauthier-Villars, Lisa Golmard, Claude Houdayer, Marine Le Mentec, Virginie Moncoutier, Antoine de Pauw, Claire Saule, Dominique Stoppa-Lyonnet, and Inserm U900, Institut Curie, Paris, France: Fabienne Lesueur, Noura Mebirouk. Contributing Centres: Unité Mixte de Génétique Constitutionnelle des Cancers Fréquents, Hospices Civils de Lyon - Centre Léon Bérard, Lyon, France: Nadia Boutry-Kryza, Alain Calender, Sophie Giraud, Mélanie Léone. Institut Gustave Roussy, Villejuif, France: Brigitte Bressac-de-Paillerets, Olivier Caron, Marine Guillaud-Bataille. Centre Jean Perrin, Clermont–Ferrand, France: Yves-Jean Bignon, Nancy Uhrhammer. Centre Léon Bérard, Lyon, France: Valérie Bonadona, Christine Lasset. Centre François Baclesse, Caen, France: Pascaline Berthet, Laurent Castera, Dominique Vaur. Institut Paoli Calmettes, Marseille, France: Violaine Bourdon, Catherine Noguès, Tetsuro Noguchi, Cornel Popovici, Audrey Remenieras, Hagay Sobol. CHU Arnaud-de-Villeneuve, Montpellier, France: Isabelle Coupier, Pascal Pujol. Centre Oscar Lambret, Lille, France: Claude Adenis, Aurélie Dumont, Françoise Révillion. Centre Paul Strauss, Strasbourg, France: Danièle Muller. Institut Bergonié, Bordeaux, France: Emmanuelle Barouk-Simonet, Françoise Bonnet, Virginie Bubien, Michel Longy, Nicolas Sevenet, Institut Claudius Regaud, Toulouse, France: Laurence Gladieff, Rosine Guimbaud, Viviane Feillel, Christine Toulas. CHU Grenoble, France: Hélène Dreyfus, Christine Dominique Leroux, Magalie Peysselon, Rebischung. CHU Dijon, France: Amandine Baurand, Geoffrey Bertolone, Fanny Coron, Laurence Faivre, Caroline Jacquot, Sarab Lizard. CHU St-Etienne, France: Caroline Kientz, Marine Lebrun, Fabienne Prieur. Hôtel Dieu Centre Hospitalier, Chambéry, France: Sandra Fert Ferrer. Centre Antoine Lacassagne, Nice, France: Véronique Mari. CHU Limoges, France: Laurence Vénat-Bouvet. CHU Nantes, France: Stéphane Bézieau, Capucine Delnatte. CHU Bretonneau, Tours and Centre Hospitalier de Bourges France: Isabelle Mortemousque. Groupe Hospitalier Pitié-Salpétrière, Paris, France: Chrystelle Colas, Florence Coulet, Florent Soubrier, Mathilde Warcoin. CHU Vandoeuvre-les-Nancy, France: Myriam Bronner, Johanna Sokolowska. CHU Besançon, France: Marie-Agnès Collonge-Rame, Alexandre Damette. CHU Poitiers, Centre Hospitalier d’Angoulême and Centre Hospitalier de Niort, France: Paul Gesta. Centre Hospitalier de La Rochelle: Hakima Lallaoui. CHU Nîmes Carémeau, France: Jean Chiesa. CHI Poissy, France: Denise Molina-Gomes. CHU Angers, France: Olivier Ingster; Ilse Coene en Brecht Crombez; Ilse Coene and Brecht Crombez; Alicia Tosar and Paula Diaque; Drs.Sofia Khan, Taru A. Muranen, Carl Blomqvist, Irja Erkkilä and Virpi Palola; The Hereditary Breast and Ovarian Cancer Research Group Netherlands (HEBON) consists of the following Collaborating Centers: Coordinating center: Netherlands Cancer Institute, Amsterdam, NL: M.A. Rookus, F.B.L. Hogervorst, F.E. van Leeuwen, S. Verhoef, M.K. Schmidt, N.S. Russell, D.J. Jenner; Erasmus Medical Center, Rotterdam, NL: J.M. Collée, A.M.W. van den Ouweland, M.J. Hooning, C. Seynaeve, C.H.M. van Deurzen, I.M. Obdeijn; Leiden University Medical Center, NL: C.J. van Asperen, J.T. Wijnen, R.A.E.M. Tollenaar, P. Devilee, T.C.T.E.F. van Cronenburg; Radboud University Nijmegen Medical Center, NL: C.M. Kets, A.R. Mensenkamp; University Medical Center Utrecht, NL: M.G.E.M. Ausems, R.B. van der Luijt, C.C. van der Pol; Amsterdam Medical Center, NL: C.M. Aalfs, T.A.M. van Os; VU University Medical Center, Amsterdam, NL: J.J.P. Gille, Q. Waisfisz, H.E.J. Meijers-Heijboer; University Hospital Maastricht, NL: E.B. Gómez-Garcia, M.J. Blok; University Medical Center Groningen, NL: J.C. Oosterwijk, A.H. van der Hout, M.J. Mourits, G.H. de Bock; The Netherlands Foundation for the detection of hereditary tumours, Leiden, NL: H.F. Vasen; The Netherlands Comprehensive Cancer Organization (IKNL): S. Siesling, J.Verloop; the ICO Hereditary Cancer Program team led by Dr. Gabriel Capella; the ICO Hereditary Cancer Program team led by Dr. Gabriel Capella; Dr Martine Dumont for sample management and skillful assistance; Ana Peixoto, Catarina Santos and Pedro Pinto; members of the Center of Molecular Diagnosis, Oncogenetics Department and Molecular Oncology Research Center of Barretos Cancer Hospital; Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow-Up Study (which has received funding from the NHMRC, the National Breast Cancer Foundation, Cancer Australia, and the National Institute of Health (USA)) for their contributions to this resource, and the many families who contribute to kConFab; the investigators of the Australia New Zealand NRG Oncology group; members and participants in the Ontario Cancer Genetics Network; Leigha Senter, Kevin Sweet, Caroline Craven, Julia Cooper, Amber Aielts, and Michelle O’Conor; HVH: acknowledgments to the Cellex Foundation for providing research facilities and equipment. Dr Juliette Coignard was supported by a fellowship of INCa Institut National du Cancer N°2015-181, la Ligue Nationale contre le Cancer IP/SC-15229 and Olga Sinilnikova’s fellowship (2016). BCAC Funding. BCAC is funded by Cancer Research UK [C1287/A16563, C1287/A10118], the European Union’s Horizon 2020 Research and Innovation Programme (grant numbers 634935 and 633784 for BRIDGES and B-CAST respectively), and by the European Community´s Seventh Framework Programme under grant agreement number 223175 (grant number HEALTH-F2-2009-223175) (COGS). The EU Horizon 2020 Research and Innovation Programme funding source had no role in study design, data collection, data analysis, data interpretation or writing of the report. Genotyping of the OncoArray was funded by the NIH Grant U19 CA148065, and Cancer UK Grant C1287/A16563 and the PERSPECTIVE project supported by the Government of Canada through Genome Canada and the Canadian Institutes of Health Research (grant GPH-129344) and, the Ministère de l’Économie, Science et Innovation du Québec through Genome Québec and the PSRSIIRI-701 grant, and the Quebec Breast Cancer Foundation. The Australian Breast Cancer Family Study (ABCFS) was supported by grant UM1 CA164920 from the National Cancer Institute (USA). The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the USA Government or the BCFR. The ABCFS was also supported by the National Health and Medical Research Council of Australia, the New South Wales Cancer Council, the Victorian Health Promotion Foundation (Australia) and the Victorian Breast Cancer Research Consortium. J.L.H. is a National Health and Medical Research Council (NHMRC) Senior Principal Research Fellow. M.C.S. is a NHMRC Senior Research Fellow. The ABCS study was supported by the Dutch Cancer Society [grants NKI 2007-3839; 2009 4363]. The Australian Breast Cancer Tissue Bank (ABCTB) was supported by the National Health and Medical Research Council of Australia, The Cancer Institute NSW and the National Breast Cancer Foundation. The work of the BBCC was partly funded by ELAN-Fond of the University Hospital of Erlangen. The BBCS is funded by Cancer Research UK and Breast Cancer Now and acknowledges NHS funding to the NIHR Biomedical Research Centre, and the National Cancer Research Network (NCRN). The BCEES was funded by the National Health and Medical Research Council, Australia and the Cancer Council Western Australia and acknowledges funding from the National Breast Cancer Foundation (JS). For the BCFR-NY, BCFR-PA, BCFR-UT this work was supported by grant UM1 CA164920 from the National Cancer Institute. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the BCFR. The BREast Oncology GAlician Network (BREOGAN) is funded by Acción Estratégica de Salud del Instituto de Salud Carlos III FIS PI12/02125/Cofinanciado FEDER; Acción Estratégica de Salud del Instituto de Salud Carlos III FIS Intrasalud (PI13/01136); Programa Grupos Emergentes, Cancer Genetics Unit, Instituto de Investigacion Biomedica Galicia Sur. Xerencia de Xestion Integrada de Vigo-SERGAS, Instituto de Salud Carlos III, Spain; Grant 10CSA012E, Consellería de Industria Programa Sectorial de Investigación Aplicada, PEME I + D e I + D Suma del Plan Gallego de Investigación, Desarrollo e Innovación Tecnológica de la Consellería de Industria de la Xunta de Galicia, Spain; Grant EC11-192. Fomento de la Investigación Clínica Independiente, Ministerio de Sanidad, Servicios Sociales e Igualdad, Spain; and Grant FEDER-Innterconecta. Ministerio de Economia y Competitividad, Xunta de Galicia, Spain. The BSUCH study was supported by the Dietmar-Hopp Foundation, the Helmholtz Society and the German Cancer Research Center (DKFZ). CBCS is funded by the Canadian Cancer Society (grant # 313404) and the Canadian Institutes of Health Research. CCGP is supported by funding from the University of Crete. The CECILE study was supported by Fondation de France, Institut National du Cancer (INCa), Ligue Nationale contre le Cancer, Agence Nationale de Sécurité Sanitaire, de l’Alimentation, de l’Environnement et du Travail (ANSES), Agence Nationale de la Recherche (ANR). The CGPS was supported by the Chief Physician Johan Boserup and Lise Boserup Fund, the Danish Medical Research Council, and Herlev and Gentofte Hospital. The CNIO-BCS was supported by the Instituto de Salud Carlos III, the Red Temática de Investigación Cooperativa en Cáncer and grants from the Asociación Española Contra el Cáncer and the Fondo de Investigación Sanitario (PI11/00923 and PI12/00070). The CTS was initially supported by the California Breast Cancer Act of 1993 and the California Breast Cancer Research Fund (contract 97-10500) and is currently funded through the National Institutes of Health (R01 CA77398, UM1 CA164917, and U01 CA199277). Collection of cancer incidence data was supported by the California Department of Public Health as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885. The University of Westminster curates the DietCompLyf database funded by Against Breast Cancer Registered Charity No. 1121258 and the NCRN. The coordination of EPIC is financially supported by the European Commission (DG-SANCO) and the International Agency for Research on Cancer. The national cohorts are supported by: Ligue Contre le Cancer, Institut Gustave Roussy, Mutuelle Générale de l’Education Nationale, Institut National de la Santé et de la Recherche Médicale (INSERM) (France); German Cancer Aid, German Cancer Research Center (DKFZ), Federal Ministry of Education and Research (BMBF) (Germany); the Hellenic Health Foundation, the Stavros Niarchos Foundation (Greece); Associazione Italiana per la Ricerca sul Cancro-AIRC-Italy and National Research Council (Italy); Dutch Ministry of Public Health, Welfare and Sports (VWS), Netherlands Cancer Registry (NKR), LK Research Funds, Dutch Prevention Funds, Dutch ZON (Zorg Onderzoek Nederland), World Cancer Research Fund (WCRF), Statistics Netherlands (The Netherlands); Health Research Fund (FIS), PI13/00061 to Granada, PI13/01162 to EPIC-Murcia, Regional Governments of Andalucía, Asturias, Basque Country, Murcia and Navarra, ISCIII RETIC (RD06/0020) (Spain); Cancer Research UK (14136 to EPIC-Norfolk; C570/A16491 and C8221/A19170 to EPIC-Oxford), Medical Research Council (1000143 to EPIC-Norfolk, MR/M012190/1 to EPIC-Oxford) (United Kingdom). The ESTHER study was supported by a grant from the Baden Württemberg Ministry of Science, Research and Arts. Additional cases were recruited in the context of the VERDI study, which was supported by a grant from the German Cancer Aid (Deutsche Krebshilfe). The GC-HBOC (German Consortium of Hereditary Breast and Ovarian Cancer) is supported by the German Cancer Aid (grant no 110837, coordinator: Rita K. Schmutzler, Cologne). This work was also funded by the European Regional Development Fund and Free State of Saxony, Germany (LIFE - Leipzig Research Centre for Civilization Diseases, project numbers 713-241202, 713-241202, 14505/2470, 14575/2470). The GENICA was funded by the Federal Ministry of Education and Research (BMBF) Germany grants 01KW9975/5, 01KW9976/8, 01KW9977/0 and 01KW0114, the Robert Bosch Foundation, Stuttgart, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, the Institute for Prevention and Occupational Medicine of the German Social Accident Insurance, Institute of the Ruhr University Bochum (IPA), Bochum, as well as the Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany. The GESBC was supported by the Deutsche Krebshilfe e. V. [70492] and the German Cancer Research Center (DKFZ). The HABCS study was supported by the Claudia von Schilling Foundation for Breast Cancer Research, by the Lower Saxonian Cancer Society, and by the Rudolf Bartling Foundation. The HEBCS was financially supported by the Helsinki UniversityHospital Research Fund, the Finnish Cancer Society, and the Sigrid Juselius Foundation. The HUBCS was supported by a grant from the German Federal Ministry of Research and Education (RUS08/017), and by the Russian Foundation for Basic Research and the Federal Agency for Scientific Organizations for support the Bioresource collections and RFBR grants 14-04-97088, 17-29-06014 and 17-44-020498. Financial support for KARBAC was provided through the regional agreement on medical training and clinical research (ALF) between Stockholm County Council and Karolinska Institutet, the Swedish Cancer Society, The Gustav V Jubilee foundation and Bert von Kantzows foundation. The KARMA study was supported by Märit and Hans Rausings Initiative Against Breast Cancer. The KBCP was financially supported by the special Government Funding (EVO) of Kuopio University Hospital grants, Cancer Fund of North Savo, the Finnish Cancer Organizations, and by the strategic funding of the University of Eastern Finland. kConFab is supported by a grant from the National Breast Cancer Foundation, and previously by the National Health and Medical Research Council (NHMRC), the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, and the Cancer Foundation of Western Australia. Financial support for the AOCS was provided by the United States Army Medical Research and Materiel Command [DAMD17-01-1-0729], Cancer Council Victoria, Queensland Cancer Fund, Cancer Council New South Wales, Cancer Council South Australia, The Cancer Foundation of Western Australia, Cancer Council Tasmania and the National Health and Medical Research Council of Australia (NHMRC; 400413, 400281, 199600). G.C.T. and P.W. are supported by the NHMRC. RB was a Cancer Institute NSW Clinical Research Fellow. LMBC is supported by the ‘Stichting tegen Kanker’. The MARIE study was supported by the Deutsche Krebshilfe e.V. [70-2892-BR I, 106332, 108253, 108419, 110826, 110828], the Hamburg Cancer Society, the German Cancer Research Center (DKFZ) and the Federal Ministry of Education and Research (BMBF) Germany [01KH0402]. MBCSG is supported by grants from the Italian Association for Cancer Research (AIRC; IG2014 no.15547) to P. Radice. The MCBCS was supported by the NIH grants CA192393, CA116167, CA176785 an NIH Specialized Program of Research Excellence (SPORE) in Breast Cancer [CA116201], and the Breast Cancer Research Foundation and a generous gift from the David F. and Margaret T. Grohne Family Foundation. The Melbourne Collaborative Cohort Study (MCCS) cohort recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further augmented by Australian National Health and Medical Research Council grants 209057, 396414 and 1074383 and by infrastructure provided by Cancer Council Victoria. Cases and their vital status were ascertained through the Victorian Cancer Registry and the Australian Institute of Health and Welfare, including the National Death Index and the Australian Cancer Database. The MEC was support by NIH grants CA63464, CA54281, CA098758, CA132839 and CA164973. The MISS study is supported by funding from ERC-2011-294576 Advanced grant, Swedish Cancer Society, Swedish Research Council, Local hospital funds, Berta Kamprad Foundation, Gunnar Nilsson. The MMHS study was supported by NIH grants CA97396, CA128931, CA116201, CA140286 and CA177150. MSKCC is supported by grants from the Breast Cancer Research Foundation and Robert and Kate Niehaus Clinical Cancer Genetics Initiative. The work of MTLGEBCS was supported by the Quebec Breast Cancer Foundation, the Canadian Institutes of Health Research for the CIHR Team in Familial Risks of Breast Cancer program – grant # CRN-87521 and the Ministry of Economic Development, Innovation and Export Trade – grant # PSR-SIIRI-701. The NBHS was supported by NIH grant R01CA100374. Biological sample preparation was conducted the Survey and Biospecimen Shared Resource, which is supported by P30 CA68485. The Northern California Breast Cancer Family Registry (NC-BCFR) and Ontario Familial Breast Cancer Registry (OFBCR) were supported by grant UM1 CA164920 from the National Cancer Institute (USA). The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the USA Government or the BCFR. The Carolina Breast Cancer Study was funded by Komen Foundation, the National Cancer Institute (P50 CA058223, U54 CA156733, U01 CA179715), and the North Carolina University Cancer Research Fund. The NHS was supported by NIH grants P01 CA87969, UM1 CA186107, and U19 CA148065. The NHS2 was supported by NIH grants UM1 CA176726 and U19 CA148065. The ORIGO study was supported by the Dutch Cancer Society (RUL 1997-1505) and the Biobanking and Biomolecular Resources Research Infrastructure (BBMRI-NL CP16). The PBCS was funded by Intramural Research Funds of the National Cancer Institute, Department of Health and Human Services, USA. Genotyping for PLCO was supported by the Intramural Research Program of the National Institutes of Health, NCI, Division of Cancer Epidemiology and Genetics. The PLCO is supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics and supported by contracts from the Division of Cancer Prevention, National Cancer Institute, National Institutes of Health. The POSH study is funded by Cancer Research UK (grants C1275/A11699, C1275/C22524, C1275/A19187, C1275/A15956 and Breast Cancer Campaign 2010PR62, 2013PR044. The RBCS was funded by the Dutch Cancer Society (DDHK 2004-3124, DDHK 2009-4318. SEARCH is funded by Cancer Research UK [C490/A10124, C490/A16561] and supported by the UK National Institute for Health Research Biomedical Research Centre at the University of Cambridge. The University of Cambridge has received salary support for PDPP from the NHS in the East of England through the Clinical Academic Reserve. The Sister Study (SISTER) is supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (Z01-ES044005 and Z01-ES049033). The Two Sister Study (2SISTER) was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (Z01-ES044005 and Z01-ES102245), and, also by a grant from Susan G. Komen for the Cure, grant FAS0703856. SKKDKFZS is supported by the DKFZ. The SMC is funded by the Swedish Cancer Foundation and the Swedish Research Council (VR 2017-00644) grant for the Swedish Infrastructure for Medical Population-based Life-course Environmental Research (SIMPLER). The SZBCS and IHCC were supported by Grant PBZ_KBN_122/P05/2004 and the program of the Minister of Science and Higher Education under the name Regional Initiative of Excellence in 2019–2022 project number 002/RID/2018/19 amount of financing 12 000 000 PLN. The TNBCC was supported by: a Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA116201), a grant from the Breast Cancer Research Foundation, a generous gift from the David F. and Margaret T. Grohne Family Foundation. The UCIBCS component of this research was supported by the NIH [CA58860, CA92044] and the Lon V Smith Foundation [LVS39420]. The UKBGS is funded by Breast Cancer Now and the Institute of Cancer Research (ICR), London. The UKOPS study was funded by The Eve Appeal (The Oak Foundation) and supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre. CIMBA Funding. CIMBA: The CIMBA data management and data analysis were supported by Cancer Research – UK grants C12292/A20861, C12292/A11174. GCT and ABS are NHMRC Research Fellows. iCOGS: the European Community’s Seventh Framework Programme under grant agreement no 223175 (HEALTH-F2-2009-223175) (COGS), Cancer Research UK (C1287/A10118, C1287/A 10710, C12292/A11174, C1281/A12014, C5047/A8384, C5047/A15007, C5047/A10692, C8197/A16565), the National Institutes of Health (CA128978) and Post-Cancer GWAS initiative (1U19 CA148537, 1U19 CA148065 and 1U19 CA148112 - the GAME-ON initiative), the Department of Defence (W81XWH-10-1-0341), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer (CRN-87521), and the Ministry of Economic Development, Innovation and Export Trade (PSR-SIIRI-701), Komen Foundation for the Cure, the Breast Cancer Research Foundation, and the Ovarian Cancer Research Fund. The PERSPECTIVE project was supported by the Government of Canada through Genome Canada and the Canadian Institutes of Health Research, the Ministry of Economy, Science and Innovation through Genome Québec, and The Quebec Breast Cancer Foundation. BCFR: UM1 CA164920 from the National Cancer Institute. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the BCFR. BIDMC: Breast Cancer Research Foundation. CNIO: Spanish Ministry of Health PI16/00440 supported by FEDER funds, the Spanish Ministry of Economy and Competitiveness (MINECO) SAF2014-57680-R and the Spanish Research Network on Rare diseases (CIBERER). COH-CCGCRN: Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under grant number R25CA112486, and RC4CA153828 (PI: J. Weitzel) from the National Cancer Institute and the Office of the Director, National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. CONSIT TEAM: Funds from Italian citizens who allocated the 5 × 1000 share of their tax payment in support of the Fondazione IRCCS Istituto Nazionale Tumori, according to Italian laws (INT-Institutional strategic projects ‘5 × 1000’) to S. Manoukian. Associazione Italiana Ricerca sul Cancro (AIRC; IG2015 no.16732) to P. Peterlongo. DEMOKRITOS: European Union (European Social Fund – ESF) and Greek national funds through the Operational Program Education and Lifelong Learning of the National Strategic Reference Framework (NSRF) - Research Funding Program of the General Secretariat for Research & Technology: SYN11_10_19 NBCA. Investing in knowledge society through the European Social Fund. DKFZ: German Cancer Research Center. EMBRACE: Cancer Research UK Grants C1287/A10118 and C1287/A11990. D. Gareth Evans and Fiona Lalloo are supported by an NIHR grant to the Biomedical Research Centre, Manchester. The Investigators at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust are supported by an NIHR grant to the Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust. Ros Eeles and Elizabeth Bancroft are supported by Cancer Research UK Grant C5047/A8385. Ros Eeles is also supported by NIHR support to the Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust. FCCC: A.K.G. was in part funded by the NCI (R01 CA214545), The University of Kansas Cancer Center Support Grant (P30 CA168524), The Kansas Institute for Precision Medicine (P20 GM130423), and the Kansas Bioscience Authority Eminent Scholar Program. A.K.G. is the Chancellors Distinguished Chair in Biomedical Sciences Professorship. A.Vega is supported by the Spanish Health Research Foundation, Instituto de Salud Carlos III (ISCIII), partially supported by FEDER funds through Research Activity Intensification Program (contract grant numbers: INT15/00070, INT16/00154, INT17/00133), and through Centro de Investigación Biomédica en Red de Enferemdades Raras CIBERER (ACCI 2016: ER17P1AC7112/2018); Autonomous Government of Galicia (Consolidation and structuring program: IN607B), and by the Fundación Mutua Madrileña (call 2018). GC-HBOC: German Cancer Aid (grant no 110837, Rita K. Schmutzler) and the European Regional Development Fund and Free State of Saxony, Germany (LIFE - Leipzig Research Centre for Civilization Diseases, project numbers 713-241202, 713-241202, 14505/2470, 14575/2470). GEMO: Ligue Nationale Contre le Cancer; the Association Le cancer du sein, parlons-en! Award, the Canadian Institutes of Health Research for the CIHR Team in Familial Risks of Breast Cancer program and the French National Institute of Cancer (INCa grants 2013-1-BCB-01-ICH-1 and SHS-E-SP 18-015). GEORGETOWN: the Non-Therapeutic Subject Registry Shared Resource at Georgetown University (NIH/NCI grant P30-CA051008), the Fisher Center for Hereditary Cancer and Clinical Genomics Research, and Swing Fore the Cure. G-FAST: Bruce Poppe is a senior clinical investigator of FWO. Mattias Van Heetvelde obtained funding from IWT. HCSC: Spanish Ministry of Health PI15/00059, PI16/01292, and CB-161200301 CIBERONC from ISCIII (Spain), partially supported by European Regional Development FEDER funds. HEBCS: Helsinki University Hospital Research Fund, the Finnish Cancer Society and the Sigrid Juselius Foundation. HEBON: the Dutch Cancer Society grants NKI1998-1854, NKI2004-3088, NKI2007-3756, the Netherlands Organization of Scientific Research grant NWO 91109024, the Pink Ribbon grants 110005 and 2014-187.WO76, the BBMRI grant NWO 184.021.007/CP46 and the Transcan grant JTC 2012 Cancer 12-054. HEBON thanks the registration teams of Dutch Cancer Registry (IKNL; S. Siesling, J. Verloop) and the Dutch Pathology database (PALGA; L. Overbeek) for part of the data collection. ICO: The authors would like to particularly acknowledge the support of the Asociación Española Contra el Cáncer (AECC), the Instituto de Salud Carlos III (organismo adscrito al Ministerio de Economía y Competitividad) and Fondo Europeo de Desarrollo Regional (FEDER), una manera de hacer Europa (PI10/01422, PI13/00285, PIE13/00022, PI15/00854, PI16/00563 and CIBERONC) and the Institut Català de la Salut and Autonomous Government of Catalonia (2009SGR290, 2014SGR338 and PERIS Project MedPerCan). INHERIT: Canadian Institutes of Health Research for the CIHR Team in Familial Risks of Breast Cancer program – grant # CRN-87521 and the Ministry of Economic Development, Innovation and Export Trade – grant # PSR-SIIRI-701. IOVHBOCS: Ministero della Salute and 5 × 1000 Istituto Oncologico Veneto grant. kConFab: The National Breast Cancer Foundation, and previously by the National Health and Medical Research Council (NHMRC), the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, and the Cancer Foundation of Western Australia. MAYO: NIH grants CA116167, CA192393 and CA176785, an NCI Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA116201),and a grant from the Breast Cancer Research Foundation. MCGILL: Jewish General Hospital Weekend to End Breast Cancer, Quebec Ministry of Economic Development, Innovation and Export Trade. Marc Tischkowitz is supported by the funded by the European Union Seventh Framework Program (2007Y2013)/European Research Council (Grant No. 310018). MSKCC: the Breast Cancer Research Foundation, the Robert and Kate Niehaus Clinical Cancer Genetics Initiative, the Andrew Sabin Research Fund and a Cancer Center Support Grant/Core Grant (P30 CA008748). NCI: the Intramural Research Program of the US National Cancer Institute, NIH, and by support services contracts NO2-CP-11019-50, N02-CP-21013-63 and N02-CP-65504 with Westat, Inc, Rockville, MD. NNPIO: the Russian Foundation for Basic Research (grants 17-00-00171, 18-515-45012 and 19-515-25001). NRG Oncology: U10 CA180868, NRG SDMC grant U10 CA180822, NRG Administrative Office and the NRG Tissue Bank (CA 27469), the NRG Statistical and Data Center (CA 37517) and the Intramural Research Program, NCI. OSUCCG: Ohio State University Comprehensive Cancer Center. PBCS: Italian Association of Cancer Research (AIRC) [IG 2013 N.14477] and Tuscany Institute for Tumours (ITT) grant 2014-2015-2016. SMC: the Israeli Cancer Association. SWE-BRCA: the Swedish Cancer Society. UCHICAGO: NCI Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA125183), R01 CA142996, 1U01CA161032 and by the Ralph and Marion Falk Medical Research Trust, the Entertainment Industry Fund National Women’s Cancer Research Alliance and the Breast Cancer research Foundation. UCSF: UCSF Cancer Risk Program and Helen Diller Family Comprehensive Cancer Center. UPENN: Breast Cancer Research Foundation; Susan G. Komen Foundation for the cure, Basser Research Center for BRCA. UPITT/MWH: Hackers for Hope Pittsburgh. VFCTG: Victorian Cancer Agency, Cancer Australia, National Breast Cancer Foundation. WCP: Dr Karlan is funded by the American Cancer Society Early Detection Professorship (SIOP-06-258-01-COUN) and the National Center for Advancing Translational Sciences (NCATS), Grant UL1TR000124. HVH: Supported by the Carlos III National Health Institute funded by FEDER funds – a way to build Europe – PI16/11363. MT Parsons is supported by a grant from Newcastle University. Kelly-Anne Phillips is an Australian National Breast Cancer Foundation Fellow.
Source data
Author contributions
A.C.A., D.F.E. and N.A. conceived the study design. J.C., N.A. and A.C.A. drafted the initial manuscript, while the complete writing group consisted of J.C., N.A., A.C.A., G.C.T. and D.F.E. J.C. performed the statistical analyses and J.B. and T.A.O. the INQUISIT predictions. M.L. and J.P.T. contributed to bioinformatics analyses. D.R.B. contributed to statistics analyses. J.D., L.M., G.L. and M.K.B. performed the data management. M.A.A., S.A., T.A., K.A., I.L.A., H.A-.C., V.A., N. Arnold, K.J.A., B.K.A., A.A., J.A., C.B., H.B., M.B., L.B., K.B., C. Blomqvist, D.B., S.E.B., B.B., A.B., H.B., H. Brenner, B. Burwinkel, S.S.B., T.C., M.A.C., D.C., B.D.C., J.E.C., J.C-.C., S.J.C., F.J.C., W.K.C., K.B.M.C., C.L.C., J.M.C., D.M.C., K.C., M.B.D., P.D., O.D., Y.C.D., S.M.D., T.D., I.D-.S-.S., A.M.D., M.D., D.M.E., A.H.E., C.E., M.E., D.G.E., P.A.F., H.F., F.F., E.F., L.F., D.F., M.G-.D., S.M.G., J.G., V.G-.B., M.G-.C., J.A.G-.S., M.M.G., S.A.G., A.G., V.G., G.G.G., A.K.G., M.S.G., D.E.G., A.G-.N., M.H.G., P.G., L.H., E.H., C.A.H., N.H., P.H., U.H., P.A.H., S.N.H., W.H., F.B.L.H., A.H., J.L.H., D.J.H., P.J.H., D.J. Hunter, E.N.I., A.J., A. Jakubowska, P.A.J., U.B.J., E.M.J., M.E.J., R.K., P.M.K., B.Y.K., R. Keeman, E.K., J.I.K., Y-.D.K., V-.M.K., P.K., A.W.K., Y.L., D.L., L.L.M., J.L., F.L., T.L., A.L-.F., J.T.L., C.L., A.M., S.M., S. Margolin, J.W.M.M., N.M., A. Meindl, A. Miller, R.L.M., M.M., K.L.N., S.L.N., H.N., F.C.N., K.M.O’.B., O.I.O., J.E.O., H.O., A.O., K.O., L.O., T-.W.P-.S., M.T.P., I.S.P., B.P., P.P., J.P., P.D.P.P., K-.A.P., E.C.P., B. Poppe, N.P., M.A.P., K.P., S.K.P., P.R., J.R., M.U.R., G.R., H.S.R., M.R., A.R., M. Rossing, E.S., D.P.S., R.S., M.T.S., M.K.S., G.S., C.S., P.S., P. Soucy, M.C.S., J.J.S., Z.S., J.S., J. Stone, R.K.S., D.S-.L., A.S., R.M.T., W.J.T., J.A.T., M.B.T., A.T., D.L.T., M.T., M. Tischkowitz, A.E.T., D.T., A.H.T., T.T., N.T., C.M.V., A.V., J.V., Q.W., B.W., C.R.W., J.N.W., C.W., A.W., S.Y., X.R.Y., D.Y., W.Z., A.Z., K.K.Z., KConFab Investigators, HEBON Investigators, ABCTB Investigators, GEMO Study Collaborators, EMBRACE Collaborators provided DNA samples and/or phenotypic data. All authors read and approved the final manuscript. The funders had no role in the design of the study, the collection, analysis, or interpretation of the data, the writing of the manuscript, or the decision to submit the manuscript for publication.
Data availability
Among BCAC data used in this study, data from 2SISTER, BREOGAN, CGPS, CPSII, EPIC, MEC, NBHS, MCCS, NHS, NHS2, PBCS, PLCO, SEARCH, SISTER, SMC, WAABCS and WHI are available in the dbGaP database under accession code phs001265.v1.p1. Among CIMBA data used in this study, data from KCONFAB, KUMC, MAYO, MSKCC, MUV, NCI, NNPIO, NORTHSHORE, OSU CCG, PBCS, SMC, SWE-BRCA, UCHICAGO, UCSF, UPENN, UPITT, UTMDACC, VFCTG and WCP studies are available in the dbGaP database under accession code phs001321.v1.p1. The complete dataset is not publicly available due to restraints imposed by the ethical committees of individual studies. Requests for the complete data can be made to the corresponding author or the Data Access Coordinating Committees (DACCs) of BCAC (BCAC@medschl.cam.ac.uk) and CIMBA (ljm26@medschl.cam.ac.uk). BCAC DACC approval is required to access data from the following studies ABCFS, ABCS, ABCTB, BBCC, BBCS, BCEES, BCFR-NY, BCFR-PA, BCFR-UTAH, BCINIS, BSUCH, CBCS, CECILE, CGPS, CTS, DIETCOMPLYF, ESTHER, GC-HBOC, GENICA, GEPARSIXTO, GESBC, HABCS, HCSC, HEBCS, HUBCS, KARBAC, KBCP, LMBC, MARIE, MBCSG, MCBCS, MISS, MMHS, MSKCC, MTLGEBCS, NC-BCFR, OFBCR, ORIGO, PBCS, pKARMA, POSH, PREFACE, RBCS, SKKDKFZS, SUCCESSB, SUCCESSC, SZBCS, TNBCC, UCIBCS, UKBGS and UKOPS (see Supplementary Table 2—for a list of all studies). CIMBA DACC approval is required to access data from studies BCFR-ON, BRICOH, CONSIT TEAM, DKFZ, EMBRACE, FPGMX, G-FAST, GC-HBOC, GEMO, HEBCS, HEBON, IHCC, INHERIT, IOVHBOCS, MCGILL, NRG_ONCOLOGY, OUH and UKGRFOCR (see Supplementary Table 1—for a list of all CIMBA studies). Case-control summary results from CIMBA and BCAC consortia are publicly available and can be downloaded at http://cimba.ccge.medschl.cam.ac.uk/oncoarray-complete-summary-results/ and at http://bcac.ccge.medschl.cam.ac.uk/bcacdata/oncoarray/oncoarray-and-combined-summary-result/gwas-summary-associations-breast-cancer-risk-2020/). The top 10 000 SNPs from the current BCAC-CIMBA case-only study can be found at http://cimba.ccge.medschl.cam.ac.uk/projects/BCAC-CIMBA_Case-only_analysis. The remaining data are available within the Article, Supplementary Information or available from the authors upon request. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks Mark Iles, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised this work: Douglas F. Easton, Nadine Andrieu, Antonis C. Antoniou.
Lists of authors and their affiliations appear at the end of the paper.
Change history
5/14/2021
A Correction to this paper has been published: 10.1038/s41467-021-23162-4
Contributor Information
Nadine Andrieu, Email: nadine.andrieu@curie.fr.
Antonis C. Antoniou, Email: aca20@medschl.cam.ac.uk
GEMO Study Collaborators:
Ophélie Bertrand, Sandrine Caputo, Anaïs Dupré, Marine Le Mentec, Muriel Belotti, Anne-Marie Birot, Bruno Buecher, Emmanuelle Fourme, Marion Gauthier-Villars, Lisa Golmard, Claude Houdayer, Virginie Moncoutier, Antoine de Pauw, Claire Saule, Olga Sinilnikova, Sylvie Mazoyer, Francesca Damiola, Laure Barjhoux, Carole Verny-Pierre, Mélanie Léone, Nadia Boutry-Kryza, Alain Calender, Sophie Giraud, Olivier Caron, Marine Guillaud-Bataille, Brigitte Bressac-de-Paillerets, Yves- Jean Bignon, Nancy Uhrhammer, Christine Lasset, Valérie Bonadona, Pascaline Berthet, Dominique Vaur, Laurent Castera, Tetsuro Noguchi, Cornel Popovici, Hagay Sobol, Violaine Bourdon, Tetsuro Noguchi, Audrey Remenieras, Catherine Noguès, Isabelle Coupier, Pascal Pujol, Aurélie Dumont, Françoise Révillion, Claude Adenis, Danièle Muller, Emmanuelle Barouk-Simonet, Françoise Bonnet, Virginie Bubien, Nicolas Sevenet, Michel Longy, Christine Toulas, Rosine Guimbaud, Laurence Gladieff, Viviane Feillel, Dominique Leroux, Hélène Dreyfus, Christine Rebischung, Magalie Peysselon, Fanny Coron, Laurence Faivre, Amandine Baurand, Caroline Jacquot, Geoffrey Bertolone, Sarab Lizard, Fabienne Prieur, Marine Lebrun, Caroline Kientz, Sandra Fert Ferrer, Véronique Mari, Laurence Vénat-Bouvet, Capucine Delnatte, Stéphane Bézieau, Isabelle Mortemousque, Florence Coulet, Chrystelle Colas, Florent Soubrier, Mathilde Warcoin, Johanna Sokolowska, Myriam Bronner, Marie-Agnès Collonge-Rame, Alexandre Damette, Paul Gesta, Hakima Lallaoui, Jean Chiesa, Denise Molina-Gomes, and Olivier Ingster
EMBRACE Collaborators:
Helen Gregory, Zosia Miedzybrodzka, Patrick J. Morrison, Kai-ren Ong, Alan Donaldson, Mark T. Rogers, M. John Kennedy, Mary E. Porteous, Carole Brewer, Rosemarie Davidson, Louise Izatt, Angela Brady, Julian Barwell, Julian Adlard, Claire Foo, Fiona Lalloo, Lucy E. Side, Jacqueline Eason, Alex Henderson, Lisa Walker, Rosalind A. Eeles, Jackie Cook, Katie Snape, Diana Eccles, Alex Murray, and Emma McCann
KConFab Investigators:
Stephen Fox, Ian Campbell, Amanda Spurdle, Penny Webb, Anna de Fazio, Margaret Tassell, Judy Kirk, Geoff Lindeman, Melanie Price, Melissa Southey, Roger Milne, Sid Deb, and David Bowtell
HEBON Investigators:
Annemieke H. van der Hout, Ans M. W. van den Ouweland, Arjen R. Mensenkamp, Carolien H. M. van Deurzen, Carolien M. Kets, Caroline Seynaeve, Christi J. van Asperen, Cora M. Aalfs, Encarna B. Gómez Garcia, Flora E. van Leeuwen, G. H. de Bock, Hanne E. J. Meijers-Heijboer, Inge M. Obdeijn, J. Margriet Collée, J. J. P. Gille, Jan C. Oosterwijk, Juul T. Wijnen, Lizet E. van der Kolk, Maartje J. Hooning, Margreet G. E. M. Ausems, Marian J. E. Mourits, Marinus J. Blok, Matti A. Rookus, Muriel A. Adank, Rob B. van der Luijt, T. C. T. E. F. van Cronenburg, Carmen C. van der Pol, Nicola S. Russell, Sabine Siesling, Lucy Overbeek, R. Wijnands, and Judith L. de Lange
ABCTB Investigators:
Christine Clarke, Dinny Graham, Mythily Sachchithananthan, Deborah Marsh, Rodney Scott, Robert Baxter, Desmond Yip, Jane Carpenter, Alison Davis, Nirmala Pathmanathan, and Peter Simpson
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-020-20496-3.
References
- 1.Ferlay J, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Pharoah PDP, Day NE, Duffy S, Easton DF, Ponder BAJ. Family history and the risk of breast cancer: a systematic review and meta-analysis. Int. J. Cancer. 1997;71:800–809. doi: 10.1002/(sici)1097-0215(19970529)71:5<800::aid-ijc18>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 3.Nelson HD, et al. Risk assessment, genetic counseling, and genetic testing for BRCA-related cancer in women: a systematic review to update the U.S. Preventive Services Task Force recommendation. Ann. Intern. Med. 2014;160:255–266. doi: 10.7326/M13-1684. [DOI] [PubMed] [Google Scholar]
- 4.Kuchenbaecker KB, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA. 2017;317:2402–2416. doi: 10.1001/jama.2017.7112. [DOI] [PubMed] [Google Scholar]
- 5.Antoniou AC, et al. Common variants in LSP1, 2q35 and 8q24 and breast cancer risk for BRCA1 and BRCA2 mutation carriers. Hum. Mol. Genet. 2009;18:4442–4456. doi: 10.1093/hmg/ddp372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antoniou AC, et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor–negative breast cancer in the general population. Nat. Genet. 2010;42:885–892. doi: 10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michailidou K, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet. 2013;45:353–361. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas G, et al. A multi-stage genome-wide association in breast cancer identifies two novel risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat. Genet. 2009;41:579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finucane HK, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Closas M, et al. Genome-wide association studies identify four ER negative–specific breast cancer risk loci. Nat. Genet. 2013;45:392–398e2. doi: 10.1038/ng.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Couch, F. J. et al. Identification of four novel susceptibility loci for oestrogen receptor negative breast cancer. Nat. Commun. 7, 11375 (2016). [DOI] [PMC free article] [PubMed]
- 12.Lin W-Y, et al. Identification and characterization of novel associations in the CASP8/ALS2CR12 region on chromosome 2 with breast cancer risk. Hum. Mol. Genet. 2015;24:285–298. doi: 10.1093/hmg/ddu431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milne RL, et al. Common non-synonymous SNPs associated with breast cancer susceptibility: findings from the Breast Cancer Association Consortium. Hum. Mol. Genet. 2014;23:6096–6111. doi: 10.1093/hmg/ddu311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haiman CA, et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor–negative breast cancer. Nat. Genet. 2011;43:1210–1214. doi: 10.1038/ng.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bojesen SE, et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat. Genet. 2013;45:371–384.e2. doi: 10.1038/ng.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghoussaini M, et al. Evidence that the 5p12 variant rs10941679 confers susceptibility to estrogen-receptor-positive breast cancer through FGF10 and MRPS30 regulation. Am. J. Hum. Genet. 2016;99:903–911. doi: 10.1016/j.ajhg.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glubb DM, et al. Fine-scale mapping of the 5q11.2 breast cancer locus reveals at least three independent risk variants regulating MAP3K1. Am. J. Hum. Genet. 2015;96:5–20. doi: 10.1016/j.ajhg.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaudet, M. M. et al. Identification of a BRCA2-Specific Modifier Locus at 6p24 Related to Breast Cancer Risk. PLoS Genet. 9, e1003173 (2013). [DOI] [PMC free article] [PubMed]
- 19.Siddiq A, et al. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum. Mol. Genet. 2012;21:5373–5384. doi: 10.1093/hmg/dds381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunning AM, et al. Breast cancer risk variants at 6q25 display different phenotype associations and regulate ESR1, RMND1 and CCDC170. Nat. Genet. 2016;48:374–386. doi: 10.1038/ng.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sawyer E, et al. Genetic predisposition to in situ and invasive lobular carcinoma of the breast. PLoS Genet. 2014;10:e1004285. doi: 10.1371/journal.pgen.1004285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Easton DF, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turnbull C, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orr N, et al. Fine-mapping identifies two additional breast cancer susceptibility loci at 9q31.2. Hum. Mol. Genet. 2015;24:2966–2984. doi: 10.1093/hmg/ddv035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darabi H, et al. Polymorphisms in a putative enhancer at the 10q21.2 breast cancer risk locus regulate NRBF2 expression. Am. J. Hum. Genet. 2015;97:22–34. doi: 10.1016/j.ajhg.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer KB, et al. Fine-scale mapping of the fgfr2 breast cancer risk locus: putative functional variants differentially bind FOXA1 and E2F1. Am. J. Hum. Genet. 2013;93:1046–1060. doi: 10.1016/j.ajhg.2013.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.French JD, et al. Functional variants at the 11q13 risk locus for breast cancer regulate cyclin D1 expression through long-range enhancers. Am. J. Hum. Genet. 2013;92:489–503. doi: 10.1016/j.ajhg.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeng, C. et al. Identification of independent association signals and putative functional variants for breast cancer risk through fine-scale mapping of the 12p11 locus. Breast Cancer Res.18, 64 (2016). [DOI] [PMC free article] [PubMed]
- 29.Ghoussaini M, et al. Genome-wide association analysis identifies three new breast cancer susceptibility loci. Nat. Genet. 2012;44:312–318. doi: 10.1038/ng.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Udler MS, et al. Fine scale mapping of the breast cancer 16q12 locus. Hum. Mol. Genet. 2010;19:2507–2515. doi: 10.1093/hmg/ddq122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darabi H, et al. Fine scale mapping of the 17q22 breast cancer locus using dense SNPs, genotyped within the Collaborative Oncological Gene-Environment Study (COGs) Sci. Rep. 2016;6:32512. doi: 10.1038/srep32512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long J, et al. Genome-Wide Association Study in East Asians Identifies Novel Susceptibility Loci for Breast Cancer. PLOS Genet. 2012;8:e1002532. doi: 10.1371/journal.pgen.1002532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai Q, et al. Genome-wide association analysis in East Asians identifies breast cancer susceptibility loci at 1q32.1, 5q14.3 and 15q26.1. Nat. Genet. 2014;46:886–890. doi: 10.1038/ng.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Long J, et al. A Common Deletion in the APOBEC3 Genes and Breast Cancer Risk. J. Natl Cancer Inst. 2013;105:573–579. doi: 10.1093/jnci/djt018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michailidou K, et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551:92–94. doi: 10.1038/nature24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mavaddat, N. et al. Prediction of breast cancer risk based on profiling with common genetic variants. J. Natl. Cancer Inst. 107, djv036 (2015). [DOI] [PMC free article] [PubMed]
- 37.Pashayan, N., Morris, S., Gilbert, F. J. & Pharoah, P. D. P. Cost-effectiveness and benefit-to-harm ratio of risk-stratified screening for breast cancer: a life-table model. JAMA Oncol. 10.1001/jamaoncol.2018.1901 (2018). [DOI] [PMC free article] [PubMed]
- 38.Mavaddat N, et al. Polygenic risk scores for prediction of breast cancer and breast cancer subtypes. Am. J. Hum. Genet. 2019;104:21–34. doi: 10.1016/j.ajhg.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Antoniou AC, et al. RAD51 135G→C modifies breast cancer risk among BRCA2 mutation carriers: results from a combined analysis of 19 studies. Am. J. Hum. Genet. 2007;81:1186–1200. doi: 10.1086/522611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcia-Closas M, et al. Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet. 2008;4:e1000054. doi: 10.1371/journal.pgen.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Antoniou AC, et al. Common breast cancer-predisposition alleles are associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers. Am. J. Hum. Genet. 2008;82:937–948. doi: 10.1016/j.ajhg.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silva LD, Lakhani SR. Pathology of hereditary breast cancer. Mod. Pathol. 2010;23:S46–S51. doi: 10.1038/modpathol.2010.37. [DOI] [PubMed] [Google Scholar]
- 43.Antoniou AC, et al. Common alleles at 6q25.1 and 1p11.2 are associated with breast cancer risk for BRCA1 and BRCA2 mutation carriers. Hum. Mol. Genet. 2011;20:3304–3321. doi: 10.1093/hmg/ddr226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Antoniou AC, et al. Common variants at 12p11, 12q24, 9p21, 9q31.2 and in ZNF365 are associated with breast cancer risk for BRCA1 and/or BRCA2 mutation carriers. Breast Cancer Res. 2012;14:R33. doi: 10.1186/bcr3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Couch, F. J. et al. Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS Genet. 9, e1003212 (2013). [DOI] [PMC free article] [PubMed]
- 46.Kuchenbaecker, K. B. et al. Associations of common breast cancer susceptibility alleles with risk of breast cancer subtypes in BRCA1 and BRCA2 mutation carriers. Breast Cancer Res.16, 3416 (2014). [DOI] [PMC free article] [PubMed]
- 47.Lawrenson, K. et al. Functional mechanisms underlying pleiotropic risk alleles at the 19p13.1 breast–ovarian cancer susceptibility locus. Nat. Commun. 7, 12675 (2016). [DOI] [PMC free article] [PubMed]
- 48.Milne RL, et al. Identification of ten variants associated with risk of estrogen-receptor-negative breast cancer. Nat. Genet. 2017;49:1767–1778. doi: 10.1038/ng.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuchenbaecker, K. B. et al. Evaluation of polygenic risk scores for breast and ovarian cancer risk prediction in BRCA1 and BRCA2 mutation carriers. J. Natl. Cancer Inst. 109, djw302 (2017). [DOI] [PMC free article] [PubMed]
- 50.Chenevix-Trench G, et al. An international initiative to identify genetic modifiers of cancer risk in BRCA1 and BRCA2 mutation carriers: the Consortium of Investigators of Modifiers of BRCA1 and BRCA2 (CIMBA) Breast Cancer Res. 2007;9:104. doi: 10.1186/bcr1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pierce BL, Ahsan H. Case-only genome-wide interaction study of disease risk, prognosis and treatment. Genet. Epidemiol. 2010;34:7–15. doi: 10.1002/gepi.20427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ottman R. Gene–environment interaction: definitions and study designs. Prev. Med. 1996;25:764–770. doi: 10.1006/pmed.1996.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andrieu N, Goldstein AM. Epidemiologic and genetic approaches in the study of gene-environment interaction: an overview of available methods. Epidemiol. Rev. 1998;20:137–147. doi: 10.1093/oxfordjournals.epirev.a017976. [DOI] [PubMed] [Google Scholar]
- 54.Fachal L, et al. Fine-mapping of 150 breast cancer risk regions identifies 191 likely target genes. Nature genetics. 2020;52:56–73. doi: 10.1038/s41588-019-0537-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whittemore AS. Assessing environmental modifiers of disease risk associated with rare mutations. Hum. Hered. 2007;63:134–143. doi: 10.1159/000099185. [DOI] [PubMed] [Google Scholar]
- 56.CIMBA - Consortium of Investigators of Modifiers of BRCA1/2 —. http://cimba.ccge.medschl.cam.ac.uk/ (2017).
- 57.Turner A, et al. MADD knock-down enhances doxorubicin and TRAIL induced apoptosis in breast cancer cells. PLoS ONE. 2013;8:e56817. doi: 10.1371/journal.pone.0056817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng T, Wang A, Hu D, Wang Y. Molecular mechanisms of breast cancer metastasis by gene expression profile analysis. Mol. Med. Rep. 2017;16:4671–4677. doi: 10.3892/mmr.2017.7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharma, D. K., Bressler, K., Patel, H., Balasingam, N. & Thakor, N. Role of Eukaryotic initiation factors during cellular stress and cancer progression. J. Nucleic Acids2016, 8235121 (2016). [DOI] [PMC free article] [PubMed]
- 60.HANNA S, et al. StarD13 is a tumor suppressor in breast cancer that regulates cell motility and invasion. Int. J. Oncol. 2014;44:1499–1511. doi: 10.3892/ijo.2014.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Piegorsch WW, Weinberg CR, Taylor JA. Non-hierarchical logistic models and case-only designs for assessing susceptibility in population-based case-control studies. Stat. Med. 1994;13:153–162. doi: 10.1002/sim.4780130206. [DOI] [PubMed] [Google Scholar]
- 62.Kulminski AM. Complex phenotypes and phenomenon of genome-wide inter-chromosomal linkage disequilibrium in the human genome. Exp. Gerontol. 2011;46:979–986. doi: 10.1016/j.exger.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 63.Escala-Garcia M, et al. Genome-wide association study of germline variants and breast cancer-specific mortality. Br. J. Cancer. 2019;120:647–657. doi: 10.1038/s41416-019-0393-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.BCAC - The Breast Cancer Association Consortium —. http://bcac.ccge.medschl.cam.ac.uk/ (2017).
- 65.Amos CI, et al. The OncoArray Consortium: A Network for Understanding the Genetic Architecture of Common Cancers. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 2017;26:126–135. doi: 10.1158/1055-9965.EPI-16-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Consortium T. 1000 G. P. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Delaneau O, Marchini J, Zagury J-F. A linear complexity phasing method for thousands of genomes. Nat. Methods. 2011;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 68.O’Connell J, et al. A General approach for haplotype phasing across the full spectrum of relatedness. PLOS Genet. 2014;10:e1004234. doi: 10.1371/journal.pgen.1004234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Howie, B. N., Donnelly, P. & Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009). [DOI] [PMC free article] [PubMed]
- 70.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hohenlohe PA, Bassham S, Currey M, Cresko WA. Extensive linkage disequilibrium and parallel adaptive divergence across threespine stickleback genomes. Philos. Trans. R. Soc. B Biol. Sci. 2012;367:395–408. doi: 10.1098/rstb.2011.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Umbach DM, Weinberg CR. Designing and analysing case-control studies to exploit independence of genotype and exposure. Stat. Med. 1997;16:1731–1743. doi: 10.1002/(sici)1097-0258(19970815)16:15<1731::aid-sim595>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 73.Spurdle, A. B. et al. Refined histopathological predictors of BRCA1 and BRCA2mutation status: a large-scale analysis of breast cancer characteristics from the BCAC, CIMBA, and ENIGMA consortia. Breast Cancer Res.16, 3419 (2014). [DOI] [PMC free article] [PubMed]
- 74.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature, 490, 61–70 (2012). [DOI] [PMC free article] [PubMed]
- 75.Mermel CH, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Q, et al. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell. 2013;152:633–641. doi: 10.1016/j.cell.2012.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinforma. Oxf. Engl. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fullwood MJ, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Corradin O, et al. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 2014;24:1–13. doi: 10.1101/gr.164079.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.He B, Chen C, Teng L, Tan K. Global view of enhancer–promoter interactome in human cells. Proc. Natl Acad. Sci. USA. 2014;111:E2191–E2199. doi: 10.1073/pnas.1320308111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dixon JR, et al. Integrative detection and analysis of structural variation in cancer genomes. Nat. Genet. 2018;50:1388–1398. doi: 10.1038/s41588-018-0195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McLaren W, et al. The ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Among BCAC data used in this study, data from 2SISTER, BREOGAN, CGPS, CPSII, EPIC, MEC, NBHS, MCCS, NHS, NHS2, PBCS, PLCO, SEARCH, SISTER, SMC, WAABCS and WHI are available in the dbGaP database under accession code phs001265.v1.p1. Among CIMBA data used in this study, data from KCONFAB, KUMC, MAYO, MSKCC, MUV, NCI, NNPIO, NORTHSHORE, OSU CCG, PBCS, SMC, SWE-BRCA, UCHICAGO, UCSF, UPENN, UPITT, UTMDACC, VFCTG and WCP studies are available in the dbGaP database under accession code phs001321.v1.p1. The complete dataset is not publicly available due to restraints imposed by the ethical committees of individual studies. Requests for the complete data can be made to the corresponding author or the Data Access Coordinating Committees (DACCs) of BCAC (BCAC@medschl.cam.ac.uk) and CIMBA (ljm26@medschl.cam.ac.uk). BCAC DACC approval is required to access data from the following studies ABCFS, ABCS, ABCTB, BBCC, BBCS, BCEES, BCFR-NY, BCFR-PA, BCFR-UTAH, BCINIS, BSUCH, CBCS, CECILE, CGPS, CTS, DIETCOMPLYF, ESTHER, GC-HBOC, GENICA, GEPARSIXTO, GESBC, HABCS, HCSC, HEBCS, HUBCS, KARBAC, KBCP, LMBC, MARIE, MBCSG, MCBCS, MISS, MMHS, MSKCC, MTLGEBCS, NC-BCFR, OFBCR, ORIGO, PBCS, pKARMA, POSH, PREFACE, RBCS, SKKDKFZS, SUCCESSB, SUCCESSC, SZBCS, TNBCC, UCIBCS, UKBGS and UKOPS (see Supplementary Table 2—for a list of all studies). CIMBA DACC approval is required to access data from studies BCFR-ON, BRICOH, CONSIT TEAM, DKFZ, EMBRACE, FPGMX, G-FAST, GC-HBOC, GEMO, HEBCS, HEBON, IHCC, INHERIT, IOVHBOCS, MCGILL, NRG_ONCOLOGY, OUH and UKGRFOCR (see Supplementary Table 1—for a list of all CIMBA studies). Case-control summary results from CIMBA and BCAC consortia are publicly available and can be downloaded at http://cimba.ccge.medschl.cam.ac.uk/oncoarray-complete-summary-results/ and at http://bcac.ccge.medschl.cam.ac.uk/bcacdata/oncoarray/oncoarray-and-combined-summary-result/gwas-summary-associations-breast-cancer-risk-2020/). The top 10 000 SNPs from the current BCAC-CIMBA case-only study can be found at http://cimba.ccge.medschl.cam.ac.uk/projects/BCAC-CIMBA_Case-only_analysis. The remaining data are available within the Article, Supplementary Information or available from the authors upon request. Source data are provided with this paper.