Abstract
Methamphetamine (MA) is the largest drug threat across the globe, with health effects including neurotoxicity and cardiovascular disease. Recent studies have begun to link microRNAs (miRNAs) to the processes related to MA use and addiction. Our studies are the first to analyse plasma EVs and their miRNA cargo in humans actively using MA (MA‐ACT) and control participants (CTL). In this cohort we also assessed the effects of tobacco use on plasma EVs. We used vesicle flow cytometry to show that the MA‐ACT group had an increased abundance of EV tetraspanin markers (CD9, CD63, CD81), but not pro‐coagulant, platelet‐, and red blood cell‐derived EVs. We also found that of the 169 plasma EV miRNAs, eight were of interest in MA‐ACT based on multiple statistical criteria. In smokers, we identified 15 miRNAs of interest, two that overlapped with the eight MA‐ACT miRNAs. Three of the MA‐ACT miRNAs significantly correlated with clinical features of MA use and target prediction with these miRNAs identified pathways implicated in MA use, including cardiovascular disease and neuroinflammation. Together our findings indicate that MA use regulates EVs and their miRNA cargo, and support that further studies are warranted to investigate their mechanistic role in addiction, recovery, and recidivism.
Keywords: addiction, extracellular vesicle, methamphetamine, microRNA, plasma, tobacco, vesicle flow cytometry
1. INTRODUCTION
Methamphetamine (MA) is a potent and addictive stimulant that is commonly used for recreational purposes. The 2019 National Survey on Drug Use and Health reported that in 2018 ∼1.9 million people aged 12 and older used MA; 205,000 of those people were new users (SAMSHA, 2019). The 2018 Annual Report of the International Narcotics Control Board of the United Nations reported that despite the surge in opioid‐related issues, amphetamine‐type stimulants, in particular MA, continue to be the largest drug threat across the globe (International Narcotics Control Board, 2019). The long‐term health effects of MA use include cognitive impairments, anxiety, insomnia, and cardiovascular disease (Darke, Duflou, & Kaye, 2017; Huckans, Fuller, Chalker, Adams, & Loftis, 2015; Kevil et al., 2019; Prakash et al., 2017). Aside from the neurotoxic effects and overdose, cardiovascular disease is the leading cause of death for MA users (Darke et al., 2017; Kevil et al., 2019). Compulsive drug use and craving are hallmark behaviours of addiction that are attributed to maladaptive changes in plasticity of dopamine (DA) neurons (Kauer & Malenka, 2007; Koob & Bloom, 1988). MA is a DA transporter substrate (Eshleman, Henningsen, Neve, & Janowsky, 1994), which interferes with DA uptake and leads to increased DA availability in the synapse and interactions with DA receptors. While much is known regarding the physiological effects of MA, there are currently no FDA‐approved treatments for MA use disorder.
Recent studies have focused on emerging roles for microRNAs (miRNAs) in synaptic plasticity and in addiction to drugs of abuse including, cocaine, opiates, nicotine and MA (Smith & Kenny, 2018). MiRNAs are small non‐coding RNAs that regulate post‐transcriptional gene expression by direct effects on target mRNAs (Duchaine & Fabian, 2019; Jonas & Izaurralde, 2015). MA is implicated in regulating protein levels of Dicer1 and Argonaute2 (Ago2), both of which are involved in miRNA silencing complexes (Liu et al., 2019). In addition, miRNA expression levels are altered in response to MA in the rodent ventral tegmental area (VTA) (Bosch, Benton, Macartney‐Coxson, & Kivell, 2015), nucleus accumbens (Sim et al., 2017; Su et al., 2019; Zhu et al., 2015), prefrontal cortex (Du et al., 2016), and hippocampus (Li et al., 2018). Further, miRNAs such as miR‐181a are associated with addiction, and are decreased in plasma from those with MA use disorder (Chandrasekar & Dreyer, 2009; Sun et al., 2020; Zhao et al., 2016). MiRNAs in circulating biofluids are located within extracellular vesicles (EVs) or bound to protein/lipoprotein particles, and they function in cell‐to‐cell communication. Recent studies in cultured cells, rodent models, and humans have begun to examine the effect of stimulants, including MA, cocaine and tobacco on EV biogenesis and release from cells (Carone et al., 2015; Cordazzo et al., 2014; Enjeti, Ariyarajah, D'crus, Seldon, & Lincz, 2017; Nakamura et al., 2019; Nazari, Zahmatkesh, Mortaz, & Hosseinzadeh, 2018; Serban et al., 2016; Trubetckaia, Lane, Qian, Zhou, & Lane, 2019). In regards to MA, plasma endothelial‐derived EVs have been shown to be increased in MA treated rats (Nazari et al., 2018). However, nothing is known about the impact of MA on plasma EVs or their miRNA cargo in humans.
There is great interest in the use of human plasma to identify biomarkers of central nervous system (CNS) diseases. CNS EVs can contribute to cell‐to‐cell communication throughout not only the CNS, but also the peripheral nervous system (Chivet, Hemming, Pernet‐Gallay, Fraboulet, & Sadoul, 2012; Dickens et al., 2017; Zhang & Yang, 2018), under both normal physiological responses and pathological processes (Neven, Nawrot, & Bollati, 2017; Yuyama & Igarashi, 2016). Neurons and other CNS cells package RNA into EVs or lipoprotein complexes, which can be secreted into the extracellular space (Vickers, Palmisano, Shoucri, Shamburek, & Remaley, 2011) where they can serve as biomarkers for CNS diseases (Rao, Benito, & Fischer, 2013). Thus, examining the role of CNS EVs in the setting of drug use disorders, provides novel opportunities to develop biomarkers for recovery, and recidivism in substance use disorders.
We examined the hypothesis that MA use alters EVs and/or their miRNA cargo in humans. Thus, we characterized EVs and their miRNAs in plasma from adult humans with active MA use (MA‐ACT) and in a control group comprised of human adults with no history of MA dependence/use (CTL). Based on our findings we then used the plasma EV miRNAs of interest in MA‐ACT for target prediction and pathway analysis. Our findings show that MA‐ACT users have increased numbers of EVs, and that plasma EV miRNA expression is predominantly decreased in MA‐ACT, compared to CTLs. Further, the expression levels for a subset of the miRNAs of interest for MA‐ACT correlate with clinical features of MA use in the participants. We also report the predicted proteins targeted by the MA‐regulated miRNAs. Together our findings reveal that plasma EVs and their miRNA cargo are altered by MA use in humans, and may serve a role in the mechanisms underlying substance use disorders.
2. MATERIALS AND METHODS
2.1. Ethics statement and participant procedures
The protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki revised in 2013 (World Medical, 2013) and was approved by the Institutional Review Boards at the Veterans Affairs Portland Health Care System and Oregon Health & Science University (OHSU). All participants provided written informed consent and were compensated with grocery store vouchers for completing the following procedures: structured clinical interview; urine drug analysis (MDC‐254, Rapid Exams, Inc., Van Buren, AR); hepatitis C virus and human immunodeficiency virus (HIV) antibody screening (1001‐0181 and 1001‐0079, respectively, OraSure Technologies, Inc., Bethlehem, PA), and blood collection. The structured clinical interview included use of the Mini International Neuropsychiatric Interview (MINI) (Sheehan et al., 1998) and our Methamphetamine Research Center (MARC) Clinical and Lab Study Interview Form.
2.2. Participant characteristics
Plasma samples banked at the MARC were donated by a well‐characterized cohort of MA‐ACT adults (n = 10) that were actively using MA and that met criteria for MA dependence at the time of the study visit and a cohort of control participants with no history of any substance use other than nicotine or caffeine (CTL group, n = 10) (Table 1). All tobacco users self‐reported smoking cigarettes, while one participant also reported smoking cigars. Note: Although the currently accepted terminology is MA use disorder, as per the Diagnostic and Statistical Manual of Mental Disorders‐Fifth Edition (DSM‐5), the diagnostic category was previously termed MA dependence, and this term is used when referring to the research participants described in this paper. Only females were selected for these pilot studies because the groups’ sample sizes were small, and females may be more sensitive to the effects of MA than males (Chang et al., 2005; Winhusen & Lewis, 2013). Inclusion criteria for the MA‐ACT group included: (i) DSM‐IV (American Psychiatric Association 2000) criteria (with confirmation by the MINI (Sheehan et al., 1998)) for MA dependence, (ii) MA use >2 days per week for >1 year, (iii) no dependence (DSM‐IV (American Psychiatric Association, 2000) criteria with confirmation by the MINI (Sheehan et al., 1998)) on other substances, (iv) last use of MA was ≤2 weeks ago, and (v) on the day of the study visits, tests positive on a urine drug analysis for MA, but no other substances. General exclusion criteria included: (i) history of major medical illness or current use of medications that are likely to be associated with serious neurological or immune dysfunction (e.g. stroke, traumatic brain injury, HIV infection, primary psychotic disorder, immunosuppressants, antivirals), (ii) visible intoxication or impaired capacity to understand study risks and benefits or otherwise provide informed consent, and (iii) based on the DSM‐IV (American Psychiatric Association, 2000) with confirmation by the MINI (Sheehan et al., 1998), meets criteria for past or current manic episode, schizophrenia, schizoaffective disorder or other psychotic disorder. Additional exclusion criteria for the CTL group included: (i) meets criteria for lifetime history of dependence on any substance (other than nicotine or caffeine dependence) based on the DSM‐IV (American Psychiatric Association, 2000) with confirmation by the MINI (Sheehan et al., 1998), (ii) heavy alcohol use as defined by the National Institute on Alcohol Abuse and Alcoholism [women: average alcohol use > 7 standard drinks weekly for >1 year, (NIAAA)], (iii) use of marijuana >2 times per month (marijuana is currently legal for recreational and medicinal uses in Oregon), (iv) use of illicit substances, and (v) on the day of the study visits, tests positive on a urine drug analysis for any drug of abuse, including alcohol and marijuana.
TABLE 1.
Participant demographic and clinical characteristics
| CTL (n = 10) | MA‐ACT (n = 10) | Total (n = 20) | |
|---|---|---|---|
| Biological sex, % female | 100 | 100 | 100 |
| Race, % white | 100 | 90 | 95 |
| Age, mean (StDev) | 40.30 (9.92) | 44.90 (7.67) | 42.60 (8.95) |
| Body mass index, mean (SD) | 30.45 (7.86) | 27.39 (5.26) | 28.92 (6.69) |
| Tobacco use, n (%) | 3 (30) | 7 (70) | 10 (50) |
| Infectious disease status | |||
| % seropositive for HCV, HBV, or HIV | 0 | 0 | 0 |
| Noninfectious disease status a , n (%) | |||
| Cancer | 0 | 0 | 0 |
| Diabetes | 0 | 0 | 0 |
| High blood pressure | 0 | 2 (20) | 2 (10) |
| High cholesterol | 3 (30) | 2 (20) | 5 (25) |
| Kidney disease | 0 | 0 | 0 |
| Liver disease | 0 | 0 | 0 |
| Lung disease b | 1 (10) | 1 (10) | 2 (10) |
| Thyroid disease | 1 (10) | 0 | 1 (5) |
| Prescription medications c , n (%) | 3 (30) | 4 (40) | 7 (35) |
Current disease status.
Includes asthma (CTL) and chronic bronchitis (MA‐ACT).
Number of participants that report current prescription medication use. Medications: acyclovir, omeprazole, duloxetine, oxybutynin, simvastatin, levothyroxine, albuterol, hydrochlorothiazide, gabapentin, levonorgestrel, and oral contraception.
2.3. Plasma collection
Non‐fasting blood samples were drawn by venipuncture into 8 ml draw capacity BD Vacutainer CPT cell preparation tubes containing 1 ml of 0.1 M sodium citrate solution (362761, BD Vacutainer CPT, Becton, Dickinson and Company, Franklin Lakes, NJ). Mean (StDev) blood collection times (defined using the 24‐h clock) were as follows: 11:50 (± 2:23 h) for the CTL group, 13:12 (± 1:44 h) for the MA‐ACT group and 12:33 (± 2:07 h) for the total sample. The average time of day for blood collection was reported to demonstrate that our blood collection times did not differ significantly between the groups (t = 1.46, df = 17, P = 0.16) and were not subject to circadian variations. After collection, tubes were stored upright at room temperature until centrifugation within 1 h of collection, then processed according to the manufacturer's instructions. Briefly, blood was centrifuged at 1500 × g for 20 min at room temperature (22–25°C) using GH‐3.8 swinging bucket rotors in an Allegra 6R centrifuge (Beckman Coulter, Indianapolis, IN). Red blood cell (RBC) contamination occurred infrequently. The separated plasma layer was aspirated and aliquoted into polypropylene cryotubes (10‐500‐26, Phenix Research Products, Swedesboro, NJ). The 1.5 ml plasma aliquots were rapidly frozen and stored at −80°C.
2.4. Vesicle flow cytometry
Plasma EVs were analysed by single vesicle flow cytometry (vFC) using fluorescence to estimate vesicle size, concentration, and surface cargo with a commercial flow cytometer (Figure S1, Beckman Coulter CytoFlex, Brea, CA and assay kit [vFC EV Analysis kit, Cellarcus Biosciences, San Diego, CA]). For product details see Supporting Information. Briefly, samples were diluted (plasma 150‐fold, pooled SEC fractions 10‐fold, optimal dilutions determined in preliminary experiments) and stained with a membrane stain (vFRed, Cellarcus Biosciences) plus carboxyfluorescein diacetate succinimidyl ester (40 µM, CFSE, Cellarcus Biosciences). The samples were then incubated with fluorescence‐labeled antibodies against tetraspanins (TS; CD9, CD63, and CD81), or a cocktail comprising a platelet (PLT) surface marker (CD41 BV421), a RBC surface marker (CD235ab PE), and a marker of phosphatidylserine that is exposed on the surface of some EVs [annexin V (AnnV) PECy7], for 1 h at ambient temperature, then diluted and detected using fluorescence triggering (excitation: 488 nm; emission: 690/50 nm). Data analysis was performed using FCS Express (Version 6, De Novo Software, Pasadena, CA). Events were gated with respect to time (to eliminate spurious background that occurs at the start of each sample), vFRed pulse shape (to eliminate short pulse width background events), and violet side scatter (VSSC) vs vFRed fluorescence (to include events with characteristic membrane fluorescence and light scatter) (Figure S2). Membrane fluorescence was calibrated in terms of vesicle size (surface area) using a synthetic vesicle size standard (Lipo100, Cellarcus Biosciences), and diameter calculated assuming a spherical shape (Figure S3). Serial dilutions of a pool of samples was used to establish optimal dilution and demonstrate a lack of coincidence/swarm events (Figure S4). Standardized preparations of platelet (PLT) and red blood cell (RBC) EVs were used as reference samples for surface marker immunofluorescence. Immunofluorescence intensity was calibrated using intensity standard beads (PE Quantibrite, BD Biosciences, San Jose, CA; Quantum MESF FITC, Bangs Laboratories, Inc., Fishers, IN).
2.5. EV isolation by size exclusion chromatography
Plasma EVs were isolated by size exclusion chromatography (SEC) (qEV original/70 nm, Izon Science, Medford, MA), according to the manufacturer's recommendations. Prior to use the qEV columns were brought to room temperature and equilibrated with 0.22 µm filtered PBS. The banked 1.5 ml aliquots of plasma were thawed on ice, divided into 500 µl aliquots and stored at ‐80°C until use. The maximum number of freeze‐thaw cycles was two for each of the samples. A pooled plasma sample that was created with equal amounts of both CTL and MA‐ACT participants was used for isolation of EVs used in NTA, Transmission Electron Microscopy (TEM), Cryo‐Transmission Electron Microscopy (CryoTEM), and immunoblots. Individual plasma samples were used for isolation of EVs used in VFC and miRNA analysis. Immediately prior to EV isolation the plasma was centrifuged at 2000 × g for 10 min at 4°C to remove large particulates. All centrifugation steps in this subsection of the methods were conducted with the Microfuge 22R Centrifuge equipped with an F241.5P fixed angle rotor (Beckman Coulter). 500 µl of individual plasma samples were loaded onto SEC columns and 500 µl fractions (Fxs) collected. Fxs used for NTA, TEM, and CryoTEM were stored at 4°C or on wet ice until processing within 6 h. Fxs used for VFC were aliquoted and stored at ‐80°C until processed. For the pooled void and individual Fxs 7–11 that were concentrated prior to immunoblot analysis, matching volumes were loaded onto individual 0.5 ml Microcon‐30 kDa Centrifugal Filters (MRCF0R030, Millipore Sigma, Burlington, MA) and centrifuged at 14,000 × g for 15 min at 4°C. Concentrated samples were recovered by inverting the centrifugal filter into a clean collection tube and then centrifuged at 1000 × g for 3 min at 4°C, then the concentrated EVs were brought to a final volume of 80 µl with 0.22 µm filtered PBS, stored at ‐80°C, and thawed on ice immediately prior to use. Prior to RNA isolation for miRNA quantitative RT‐PCR (qPCR) we created a 2 ml pool from the individual Fxs 7–10, which was then concentrated with a single 0.5 ml Microcon‐30 kDa Centrifugal at 14,000 × g for 10 min at 4°C per 500 µl of fraction. Concentrated pools were recovered by inverting the centrifugal filter into a clean collection tube and then centrifuged at 1000 × g for 3 min at 4°C then brought to a final volume of 250 µl with 0.22 µm filtered PBS stored at ‐80°C, then thawed on ice immediately prior to use.
2.6. Nanoparticle tracking analysis (NTA) of EVs
Plasma SEC EVs in the void volume (pooled Fxs 1–6) and individual Fxs 7, 8, 9 and 10 were analysed for particle size and concentration by NTA (Malvern Instruments Ltd), as we have previously reported for our cerebrospinal fluid (CSF) studies (Saugstad et al., 2017). Fxs were diluted 1:20 in 0.22 µm filtered PBS, loaded into the NanoSight LM10 chamber, and the microparticles were visualized and acquired by video with a sCMOS camera and analysed using NanoSight NTA Version 3.0 software. Matching capture and analysis settings were maintained for all readings: camera level 14, detection threshold 3, automatic blur size and automatic (8.1–15.9 pix) maximum jump distance. The void volume and each Fx were measured five times with 30 s acquisitions, then averaged to generate the size (nm) and concentration (particles/ml) histograms.
2.7. TEM and CryoTEM evaluation of EVs
Individual plasma SEC Fxs 7–11 were imaged by TEM, and Fx 9 was image by CryoTEM, by the OHSU Multiscale Microscopy Core. For TEM, 5 µl of the SEC preparations were deposited onto glow discharged (120 s 15 mAmp, negative mode) carbon formvar 400 Mesh copper grids (01822‐F, Ted Pella, Inc., Redding, CA) for 3 min, rinsed 15 s in water, wicked on Whatman filter paper 1, stained for 60 s in filtered 1.33 % (w/v) uranyl acetate in water, wicked again, and air dried. Samples were imaged at 120 kV on a FEI Tecnai Spirit TEM system (ThermoFisher Scientific Electron Microscopy, Hillsboro, OR). Images were acquired as 2048 × 2048 pixel, 16‐bit grey scale files using the FEI's TEM Imaging & Analysis interface on an Eagle 2K CCD multiscan camera. For CryoTEM of EVs, 2.5‐3 µl of the SEC suspensions were pipetted onto glow‐discharged (120 s 15 mAmp, negative mode) copper Quantifoil holey carbon support grids (658‐300‐CU, Ted Pella, Inc.) and vitrified on liquid ethane using a Mark IV Vitrobot (ThermoFisher Scientific Electron Microscopy). The conditions utilized for the cryopreservation were 100% humidity, blot force 1, and blotting time 3 s. Low‐dose conditions were used to acquire images on a Talos Arctica (ThermoFisher Scientific Electron Microscopy) at 200 kV with a K2 direct electron detector. CryoTEM images were collected with a defocus range of 2–4 µm.
2.8. Immunoblot evaluation of EV markers in SEC Fxs
To assess the separation of non‐EV proteins from EVs in SEC Fxs we first performed a total protein stain and an albumin immunoblot by loading equal volumes (30 µl) of the void (pooled Fxs 1–6) and individual Fxs 7–15 onto a 10–20% polyacrylamide gel (3450118, Bio‐Rad, Hercules CA). Next, in order to identify the SEC Fxs that contained an enrichment of EV markers, 500 µl of the void volume (pooled Fxs 1–6) and individual Fxs 7–11 were concentrated with Microcon‐30 kDa Centrifugal Filters to a final volume of 80 µl, then protein concentrations were determined using the BCA protein assay (23227, ThermoFisher Scientific, Waltham, MA). Due to undetectable protein amounts in the concentrated samples, we loaded the maximum volume for the void volume (pooled Fxs 1–6) and individual Fxs 7–11 and equal amounts of protein (3 µg) from individual Fxs 12–15. For Alix, TSG101, and Ago2 the samples were separated by SDS‐PAGE through a 10–20% polyacrylamide gel (Bio‐Rad) with a maximum well volume of 30 µl. For CD9, CD63, CD81, and flotillin the samples were separated by SDS‐Page through a NuPAGE 4 to 12% Bis‐Tris polyacrylamide gel (NP0336BOX, ThermoFisher Scientific) with a maximum well volume of 25 µl. For total protein, the gel was stained with GelCode Blue stain reagent (24592, ThermoFisher Scientific) for 1 h then de‐stained with deionized water for 1 h. Gel proteins were transferred to PVDF membranes (1620177, Bio‐Rad), blocked with 5% non‐fat dry milk in 1xTris‐buffered saline with 0.1% Tween20 (TBST) at room temperature for 30 min (Alix, Ago2, TSG101, albumin) or 60 min (CD9, CD63, CD81, flotillin), then incubated at 4°C overnight with primary antibodies diluted 1:1000 in TBST: rabbit anti‐Alix (ab186429, Abcam, Cambridge MA), rabbit anti‐Ago2 (ab186733, Abcam), rabbit anti‐TSG101 (ab125011, Abcam), rabbit anti‐CD9 (EXOAB‐CD9A‐1, SBI, System Biosciences, Palo Alto, CA), rabbit anti‐CD63 (ab134045, Abcam), mouse anti‐CD81 (sc‐166029, Santa Cruz Biotechnology, Inc, Dallas, TX), and rabbit anti‐albumin (4929, Cell Signaling Technology, Danvers, MA). Rabbit anti‐flotillin (ab133497, Abcam) was diluted 1:10,000 in TBST. The immunoblots were washed with TBST and incubated with either donkey anti‐rabbit HRP conjugated (711‐035‐152, Jackson ImmunoResearch laboratories, West Grove, PA) or donkey anti‐mouse HRP‐conjugated (715‐035‐150, Jackson ImmunoResearch) secondary antibodies diluted 1:10,000 in TBST for 1 h at room temperature. Blots were visualized by chemiluminescence (34076, 34080, and 34095, ThermoFisher Scientific) and analysed using Quantity One software (Bio‐Rad) or a ChemiDoc Imaging System (Bio‐Rad).
2.9. EV RNA isolation and miRNA Arrays
EVs were isolated from 500 µl of plasma using SEC, then Fxs 7–10 were pooled and concentrated, and total RNA was isolated using the mirVana PARIS RNA and Native Protein Purification Kit (AM1556, ThermoFisher Scientific), with modification (Burgos et al., 2014) as we have previously reported for CSF (Lusardi et al., 2017). Isolated RNA was then stored at ‐80°C until use. For qPCR, the RNA samples were concentrated (R1013, RNA Clean & Concentrator‐5 Kit, Zymo Research, Irvine, CA) then eluted into 9 µl of RNase/DNase‐free water. Using Megaplex RT Primers Human Pool Set v3.0 (4444750, ThermoFisher Scientific) and MultiScribe Reverse Transcriptase (4311235, ThermoFisher Scientific), 3.2 µl of concentrated RNA was reverse transcribed in a total reaction volume of 7.5 µl. Five microliters of cDNA was pre‐amplified (PreAmp) for 14 cycles using Megaplex PreAmp Primers, Human Pool Set v3.0 (4444748, ThermoFisher Scientific) and TaqMan PreAmp Master Mix (4391128, ThermoFisher Scientific) in a final reaction volume of 25 µl. Reverse transcription and PreAmp reactions were run with a Veriti 96‐Well Thermal Cycler (ThermoFisher Scientific) following the manufacturer's instructions for detection of miRNAs with PreAmp. All cDNA was stored at ‐20°C. Prior to qPCR the PreAmp cDNA was diluted 1:2 in RNase/DNase‐free water and 18 µl was mixed with TaqMan Universal Master Mix II, no UNG (444047, ThermoFisher Scientific). The samples were loaded onto TaqMan low density arrays, specifically the Human MicroRNA Card Set v3.0, a two‐card set (A+B) containing a total of 754 miRNA assays, plus probes for U6 snRNA, RNU44, RNU48 (4444913, ThermoFisher Scientific). The qPCR amplifications and data acquisition were carried out on a QuantStudio 12K Flex Real‐Time PCR System (ThermoFisher Scientific).
2.10. MiRNA expression analysis
MiRNAs were analyzed using relative quantification (∆∆Cq) based on Applied Biosystems recommendations (ThermoFisher, Part Number 4371095 Rev B). Cq values were calculated using automatic baseline and threshold values determined by ExpressionSuite Software v.1.1 (ThermoFisher). The Cq value for each well was reported along with the amplification score (AmpScore) and a Cq confidence (CqConf), which are metrics for the quality of each amplification. Prior to data analysis, amplifications were filtered based on each well's Cq value, AmpScore and CqConf: (i) PCR products with a Cq > 32, or reported as ‘Undetected’, were considered below the detection threshold and assigned a Cq value of ‘32’ and (ii) amplifications with a Cq ≤ 32 and an AmpScore < 1.0 or a CqConf < 0.8 were excluded from analysis. Based on these criteria, miRNAs with a Cq ≤ 32, AmpScore > 1.0 and CqConf > 0.8 were deemed acceptable for further analysis. Next, we excluded miRNAs that were not expressed in at least 80% of the samples within a group from further analysis. We also performed qPCR reactions with water only (no RNA – no template control) and with RNA, but no reverse transcriptase enzymes (no RT), as controls for spurious PCR amplifications (Table S1). MiRNAs that had amplifications with Cq < 32, AmpScore < 1.0 and CqConf < 0.8 in either the water only or no RT group were excluded from any further analysis. For miRNAs that did meet all of the inclusion criteria we then used the following formula for calculating the ∆∆Cq for each miRNA: ∆∆Cq = mean ∆Cq of test samples – mean ∆Cq of CTL samples. Within each sample the ∆Cq for a miRNA was calculated by: ∆Cq = miRNA Cq – mean Cq of endogenous control miRNAs. MiRNAs selected as endogenous control normalizers showed (i) stable good quality expression values in all samples regardless of experimental group and (ii) best endogenous control scores in ExpressionSuite. Because of the different miRNAs expressed in Card A versus Card B, we selected two sets of endogenous controls, one set for analysis of miRNAs assayed on the A card and the other for miRNAs on the B card. The A card endogenous controls were miR‐106a‐5p, miR‐17‐5p, miR‐19a‐3p, miR‐223‐3p, and miR‐93‐5p. The B card endogenous controls were miR‐151‐3p, miR‐22‐5p, miR‐30e‐3p, miR‐151a‐5p, and miR‐340‐3p. For each miRNA the fold change (RQ value) was calculated by 2−ΔΔCq, with RQ > 1 indicates increased miRNA expression in either MA‐ACT or Smoker. Conversely RQ < 1 indicates decreased expression in MA‐ACT or Smoker. The miRNA Microarray data is MIAME compliant and is being submitted to the Gene Expression Omnibus site: https://www.ncbi.nlm.nih.gov/geo/.
2.11. MiRNA target prediction and pathway analysis
We used TargetScan 7.2 (Agarwal, Bell, Nam, & Bartel, 2015) and miRDB (Chen & Wang, 2020; Liu & Wang, 2019) online tools to predict targets of the miRNAs associated with exposure to MA, as these programs are widely used and frequently updated. As pathway analysis is most effective for predictions generated from a limited gene set, predicted targets were excluded if they had a Cumulative Weighted Context Score above ‐0.3 in TargetScan or a target score of below 85 in miRDB. To determine whether a union, an intersection, or an individual target list be used in subsequent pathway analysis we calculated sensitivity, specificity, and precision values using validated miRNA‐target pairs from miRTarBase, an experimentally validated miRNA‐target interactions database (Chou et al., 2018). The values in miRTarBase range from 0 to 1 with high quality results closer to 1 (Fan & Kurgan, 2015; Oliveira et al., 2017). The union of TargetScan and miRDB showed the highest values of sensitivity, specificity, and precision with values of 0.35, 0.83 and 0.98 respectively. The use of a union between the two target prediction algorithm outputs, as opposed to an intersection where only targets predicted by both algorithms are included in analysis, increases the sensitivity of the targets predicted and the chance of predicting a novel target (Fan & Kurgan, 2015; Oliveira et al., 2017). TargetScan predicted 394 targets while miRDB predicted 385 targets. The two target lists from each algorithm overlapped by 65 genes for a total of 714 unique targets in the union between TargetScan and miRDB. Pathway analysis was then performed on the union set of 714 unique targets using Ingenuity Pathway Analysis (IPA; QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis). For this analysis, we excluded cancer‐related tissue and cell lines to avoid knowledge bias towards cancer in IPA.
2.12. Statistical analysis
Data were analysed with GraphPad Prism software v8.4.3 (GraphPad Software, Inc., San Diego, CA). Data are shown as the mean ± standard error mean (SEM). Where possible the individual samples are displayed as symbols. To assess the effects of MA, participants were categorized as either MA‐ACT (n = 10) or CTL (n = 10). To assess the effects of tobacco, data generated from the participants were re‐analysed based on either smoker (n = 10) or non‐smoker (n = 10) status. Given the small number of participants in this study a covariate analysis of MA‐ACT and smoker was not conducted. Thus, conclusions drawn from the analysis assume no interaction between MA‐ACT and smoking. For vFC, the concentration versus diameter histograms were analysed by repeated measures two‐way ANOVA followed by Sidak's multiple comparisons post hoc analysis. We used multiple statistical criteria to identify the miRNAs of interest for either MA‐ACT or smoking. First, for each miRNA that passed quality control we calculated the F‐statistic using a multiple variable linear regression analysis of the ∆Cq, MA‐ACT, and smoker status. The F‐statistic was corrected for sample size then used to rank the miRNA. The top 20% of ranked miRNAs (36 in total) were those that had the largest corrected F‐statistic. Next, the effect size for each miRNA was calculated using Glass's ∆ (∆∆Cq / CTL ∆Cq StDev). Finally, the area under the curve (AUC) for the receiver operating characteristic (ROC) curve was calculated for the top 20% of ranked miRNAs using each participant's ∆Cq, MA‐ACT, and smoker status. From the top ranked miRNAs, those that had a fold change > 1.2 or < ‐1.2, Glass's ∆ large effect size > 0.8 or < ‐0.8 (Cohen, 1988), and AUC ≥ 0.75 were considered to be miRNAs of interest.
2.13. EV‐TRACK
Relevant experimental parameters can be accessed in the EV‐TRACK (evtrack.org) knowledgebase (Deun et al., 2017).
3. RESULTS
3.1. Participant characteristics
The demographic and clinical characteristics for the study participants are reported in Table 1. All 20 participants were female, 95% were white, their average age was 42.60 8.95 SD, and their average body mass index was 28.92 6.69 SD. 50% of the participants also used tobacco.
3.2. TS+ EVs are increased in the plasma of MA‐ACT, but not smokers
To measure EVs in total plasma we performed single vFC, which uses a fluorescent lipid probe, vFRed, combined with sensitive measurement of light scatter, surface immunofluorescence, and intravesicular esterase activity and volume. Staining of diluted plasma revealed a constellation of membranous particles in the diameter range of ∼75 nm to ∼600 nm that exhibit a range of light scatter intensities (Figure 1a‐c). These particles are expected to include vesicles and lipoproteins, which can be in the same size range as EVs, but can be distinguished by their physical properties and molecular composition. Staining diluted plasma with vFRed plus CFSE, a fluorogenic esterase substrate that can stain the EV intravesicular volume (Figure 1d), or with a mixture of fluorescent anti‐TS antibodies (CD9, CD63, CD81), which are expressed on the surface of some EVs (Figure 1e), revealed a population of EVs with intermediate light scatter in the size range of ∼75 to ∼400 nm (Figure 1f). We calculated the concentration of the total number of membrane particles as mean 8.0e10/ml; range 7.0e9‐2.0e11/ml, the concentration of CFSE+ EVs as mean 1.1e10/ml; range 3.0e8‐2.0e10/ml, and the concentration of TS+ EVs as mean 1.3e10/ml; range 5.0e9‐2.0e10/ml. The results show that there was no significant difference in the total concentration of the membrane particle groups between MA‐ACT and CTL, or between smokers and non‐smokers (Figure S5‐6). We also found no difference in the size distributions of total vFRed+ membrane particles and CFSE+ EVs between MA‐ACT and CTL, or smokers and non‐smokers (Figure 1g‐j). However, analysis of the size distributions of TS+ stained plasma revealed a significant increase in the concentration of TS+ EVs at 105 nm in the MA‐ACT relative to CTL (Figure 1k). When participant samples were re‐analysed based on smoking status there was no significant effect on the concentration of TS+ EVs (Figure 1l).
FIGURE 1.
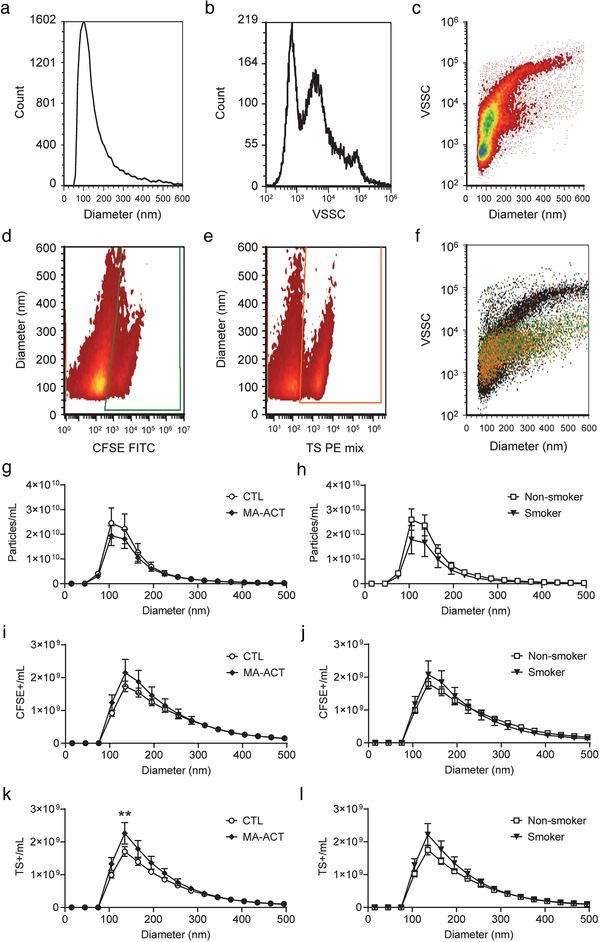
TS+ EVs are increased in the plasma of MA‐ACT, but not smokers. a) Estimated diameter (nm) of all particles, and (b) representative violet side scatter (VSSC) profile of plasma samples stained with vFRed and measured by vesicle flow cytometry (vFC). c) Corresponding diameter versus VSSC distributions of vFRed show three populations of particles differentiated by high, medium and low light scatter. d‐f) Plasma samples stained with vFRed, CFSE FITC and antibodies against CD9, CD63 and CD81 (TS PE mix). Representative diameter versus fluorescence distributions of CFSE FITC (d) or TS PE (e). f) Staining events backgated onto diameter vs VSSC for CFSE FITC (green) and TS PE (orange). G‐L) Analysis of size histograms show the concentration (particles/ml) of all membrane particles (A) and CFSE+ EVs (C) is not significantly different in MA‐ACT (♦, n = 10) versus CTL (○, n = 10). Concentration of TS+ EVs (E) is significantly increased at 105 nm in MA‐ACT (♦, n = 10) versus CTL (○, n = 10). (1B,D,F) Samples re‐categorized for smoking status (non‐smokers, □, n = 10 versus smokers,▼, n = 10) show no significant difference in total particles (B), CFSE+ (D) or TS+ (F) EVs in non‐smokers versus smokers (B). Data are shown as mean ± SEM and analysed by two‐way repeated measures ANOVA followed by Sidak's multiple comparisons post hoc analysis when appropriate; P < 0.01.
RBCs and PLTs are the most abundant cell types in blood and the most likely sources of EVs in plasma. Therefore, we also measured the number of EVs bearing the cell surface markers CD41 (PLT), CD235ab (RBC), and AnnV (phosphatidylserine). EVs bearing the PLT marker CD41 were abundant in plasma (mean 9.0e9/ml; range 4.0e9‐1.4e10/ml) while far fewer EVs with the RBC marker CD235 were detected (8.0e8/ml; range 5.0e8‐1.0e9/ml) (Figure S5‐7). However, we found no statistically significant differences in the total numbers of EVs for each specific marker between MA‐ACT and CTL, or smokers and non‐smokers (Figure S6).
3.3. Characterization of plasma EVs isolated by SEC
To verify that SEC resulted in enriched EV preparations that were depleted of protein contaminants, we examined the SEC column elution fractions using a representative standard containing MA‐ACT and CTL plasma. The NTA profiles showed that pooled Fxs 1–6, which represent the void volume of the SEC column, contain negligible concentrations of particles in plasma that are not discernible from baseline (Figure 2a). In contrast, the individual Fxs 7–10 contain particles ranging in size from ∼40 to 200 nm; Fx 8 had the highest peak EV concentration of 4.0e9 particles/ml. Figure 2 shows representative wide and narrow field of view TEM images of the SEC Fxs. The images revealed that Fx 7 contained a small number of diffuse EVs that are predominantly larger in size (>200 nm, Figure 2b). In line with our NTA results, as the Fx number increased from 8 to 10 so did the concentration of EVs, along with the presence of a greater proportion of smaller EVs (50‐200 nm, Figure 2c‐e). Fxs 7–9 showed negligible levels of background protein, and slightly higher amounts of protein in Fx 10 (Figure 2b‐e), as expected for SEC isolates. TEM images from Fx 11 show small vesicles of ∼50‐80 nm; however, there was a higher degree of background consistent with increasing amounts of proteins that elute later (Figure S8). Wide and narrow view CryoTEM images of Fx 9 confirmed the presence of membrane bound vesicles that are ∼100 nm in diameter (Figure 2f). When each of the SEC Fxs were separated on a protein gel and stained with Coomassie blue, individual Fxs 7–10 showed little evidence of protein, while Fxs 11–15 showed increasing amounts of protein, as expected (Figure 2g). To identify the Fxs that contained EVs versus non‐vesicular protein particles, the SEC Fxs were analysed by immunoblot using antibodies to CD9, CD63, CD81, flotillin, Alix, TSG101, Ago2, and albumin, with each antibody used on an individual immunoblot (Figure 2g). The immunoblots identified bands of the correct size for CD9, CD63, CD81, flotillin, Alix and TSG101 that serve as positive markers for EVs in Fxs 7–10, and only very low levels of albumin, a negative control for EVs, consistent with the MISEV2018 guidelines for EV studies (Théry et al., 2018). Fxs 7–10 also showed bands of the correct size for Ago2. We next used vFC to assess a pool of Fxs 7–10 from individual plasma samples (n = 2 MA‐ACT and n = 2 CTL) that are representative of each group (Figure 2H‐M. We found that the pool of Fxs 7–10 contained CFSE+ (Figure 2k) and TS+ EVs (Figure 2l) that were detergent labile, but with a somewhat smaller and narrower size range, and relatively fewer high‐scattering and low scatter lipoprotein events (Figure 2h‐j, m) compared to total plasma (Figure 1a‐c, f), as might be expected from a size‐fractionated sample, and confirming the effectiveness of the fractionation. Based on these cumulative outcomes, Fxs 7–10 were pooled and the resulting EV samples used for qPCR arrays to measure miRNA expression.
FIGURE 2.

Characterization of plasma EVs isolated by size exclusion chromatography (SEC). a) NTA of plasma SEC fractions (Fx): Fx 1–6 pooled (column void volume), Fx 7, Fx 8, Fx 9, Fx 10. Note that Fxs 7–10 contain particles of the size for expected EVs (∼40–200 nm). Fx 8 has the peak concentration (4 × 109 particles/ml), while Fx 1–6 has a negligible concentration of particles. Representative transmission electron microscopy (TEM) images of SEC Fx 7 (b), Fx 8 (c), Fx 9 (d) and Fx 10 (e) show membrane bound vesicles at a size range of ∼40–200 nm, with most ∼100 nm. Wide field of view (scale bars = 200 nm) and close up views (scale bars = 100 nm) for each TEM image are shown. f) Representative wide field of view and close up CryoTEM image of Fx 9 shows multiple membrane bound vesicles. Scale bars: F = 50nm; F’ = 10 nm. g) Plasma SEC Fxs stained for total protein, and immunoblotted for CD9, CD63, CD81, flotillin, Alix, TSG101, Ago2, and Albumin. Figure represents four separate SEC isolations: SEC isolation 1 = protein stain, Alix, and Ago2; SEC isolation 2 = TSG101 and albumin, SEC isolation 3 = CD9 and flotillin, SEC isolation 4 = CD63 and CD81. h) Estimated diameter (nm) of all particles, and (i) representative violet side scatter (VSSC) profile of a pool of plasma SEC Fxs 7–10 stained with vFRed and measured by vesicle flow cytometry (vFC). j) Corresponding diameter versus VSSC distributions of vFRed show three populations of particles differentiated by high, medium, and low light scatter. K‐M) A pool of plasma SEC Fxs 7–10 stained with vFRed, CFSE FITC and antibodies against CD9, CD63 and CD81 (TS PE mix). Representative diameter versus fluorescence distributions of CFSE FITC (k) or TS PE (l). m) Staining events backgated onto diameter vs VSSC for CFSE FITC (green) and TS PE (orange).
3.4. Plasma EV miRNA expression is altered by MA and smoking
The pooled plasma SEC Fxs (7‐10) were assessed for miRNA expression in MA‐ACT versus CTLs and in smokers versus non‐smokers. We identified 169 miRNAs that passed quality control measures and were expressed in at least 80% of either the MA‐ACT or CTLs (Table S2). A Manhattan plot depicts the 169 miRNAs and their respective fold change in MA‐ACT versus CTL EVs (Figure 3a). These data demonstrate a signature of decreased miRNA expression in MA‐ACT: 19 miRNAs had at least a 1.2 fold increase while 69 miRNAs had at least a 1.2‐fold decrease in MA‐ACT relative to CTL (Figure 3a, red bars). We also observed a similar effect on miRNA expression when samples were re‐analysed for smoking status. There were 175 miRNAs that passed quality control measures and were expressed in at least 80% of either the smoker or non‐smoker samples (Table S3) with a signature of decreased expression in smokers (Figure 3b). Of the 175 miRNAs, 30 had at least a 1.2 fold increase while 77 had at least a 1.2 fold decrease in smokers relative to non‐smokers (Figure 3b, blue bars). Considering that both MA and tobacco are stimulants, and that seven out of 10 MA‐ACT participants also use tobacco, we determined the overlap in miRNAs identified in MA‐ACT versus CTL and smoker versus non‐smoker. Comparison of the miRNAs with a 1.2‐fold increase in either the MA‐ACT or smokers revealed nine miRNAs in common (Figure 3c), while miRNAs with a 1.2 fold decrease in MA‐ACT or smokers revealed 48 miRNAs in common (Figure 3d).
FIGURE 3.

Plasma EV miRNA expression is altered by MA and smoking. MiRNA expression in plasma EVs isolated by SEC was assayed by qPCR in MA‐ACT (n = 10) and CTL (n = 10). a) Manhattan plot shows 169 miRNAs expressed in MA‐ACT and/or CTL plasma EVs; miRNAs with a 1.2 fold increase or decrease in MA‐ACT relative to CTL are depicted in red. b) Recategorizing based on tobacco use shows 175 miRNAs expressed in smokers (n = 10) and/or non‐smokers (n = 10); miRNAs with a 1.2‐fold increase or decrease in smokers relative to non‐smokers are depicted in blue. Venn diagrams show miRNAs that are either 1.2‐fold increased (c) or decreased (d) in MA‐ACT or smokers, and the overlap between miRNAs altered in each group.
3.5. Plasma EV miRNAs differentially expressed by MA and smoking
To identify miRNAs of interest for MA‐ACT we looked at the combined effect of ranking miRNAs based on an F‐statistic, fold change, effect size, and AUC. First, we ranked all miRNAs based on the F‐statistic generated in a multiple variable linear regression that compared the ∆Cq value for each participant to their MA (Table S2) or tobacco (Table S3) status. The top 20% of the ranked miRNAs, 36 in total, brought forward for further analysis identified miRNAs with at least a 1.2 fold increase or decrease in MA‐ACT relative to CTL (Figure 4a, red bars) or smokers relative to non‐smokers (Figure 4b, blue bars). Next, we identified which of the 36 miRNAs had a large effect size > 0.8 or < ‐0.8 in MA‐ACT (Figure 4c, red bars) or smokers (Figure 4d, blue bars). Finally, we used an AUC of the ROC curve ≥ 0.75 to identify the miRNAs that best classify participants based on MA‐ACT (Figure 4e, red bars) or smoking (Figure 4f, blue bars) status. Based on the cumulative criteria of 1.2 fold change in expression, effect size of 0.8, and AUC ≥ 0.75 we identified eight MA‐ACT (Table 2, Figure 5a, red bars) and 15 smoker (Table 3, Figure 5b, blue bars) miRNAs of interest out of the top 36 ranked miRNAs. There was an overlap of two miRNAs in the MA‐ACT and smokers: miR‐301a‐3p and miR‐382‐5p. We next determined if the normalized expression values (ΔCq) for each participant across the MA‐ACT or smoker miRNAs would cluster individuals based on substance use. Linkage analysis with the eight MA‐ACT miRNAs showed that most participants clustered based on their MA status, with the exception of one participant (Figure 5c). In this analysis we designated participants that use tobacco by bold font to show that some participants cluster based on smoking status. In a separate linkage analysis using the 15 miRNAs of interest in smokers, 12 participants clustered based on their smoking status, with the exception of three non‐smokers (Figure 5d). In this analysis MA‐ACT participants are designated in bold font to show minimal clustering of participants based on MA status.
FIGURE 4.

Plasma EV miRNAs differentially expressed by MA and smoking. Identification of miRNAs of interest that are differentially expressed in plasma EVs as a result of MA or smoking. a,b) Manhattan plots of the top 20% of ranked miRNAs based on corrected F‐statistic generated in multiple variable linear regression analysis of each participant's ΔCq, MA‐ACT and smoking status. MiRNAs are shown based on fold change of MA‐ACT versus CTL (a) or smokers versus non‐smokers (4b), with coloured bars representing at least a 1.2‐fold change in expression. c,d) Frequency distribution of the top 20% of ranked miRNAs based on Glass's Δ for MA‐ACT versus CTL (5c) or smokers versus non‐smokers (d). Coloured bars are miRNAs with a large effect size of at least 0.8. e,f) Manhattan plots of area under the curve (AUC) statistic for the top 20% of ranked miRNAs for MA‐ACT versus CTL (4e) or smokers versus non‐smokers (4f), with coloured bars representing an AUC of at least 0.75.
TABLE 2.
MA‐ACT MiRNAs of interest
| miRBase ID | Rank | Fold change | Glass's ∆ | AUC |
|---|---|---|---|---|
| miR‐301a‐3p | 2 | ‐1.36 | 1.41 | 0.770 |
| miR‐590‐3p | 3 | 1.60 | ‐1.02 | 0.820 |
| miR‐125b‐5p | 8 | ‐2.35 | 1.09 | 0.830 |
| miR‐374b‐5p | 13 | ‐1.21 | 1.67 | 0.800 |
| miR‐628‐5p | 18 | ‐1.57 | 1.01 | 0.770 |
| miR‐335‐3p | 25 | 1.29 | ‐0.89 | 0.790 |
| miR‐628‐3p | 29 | ‐1.66 | 0.95 | 0.780 |
| miR‐382‐5p | 33 | ‐2.72 | 1.08 | 0.792 |
FIGURE 5.
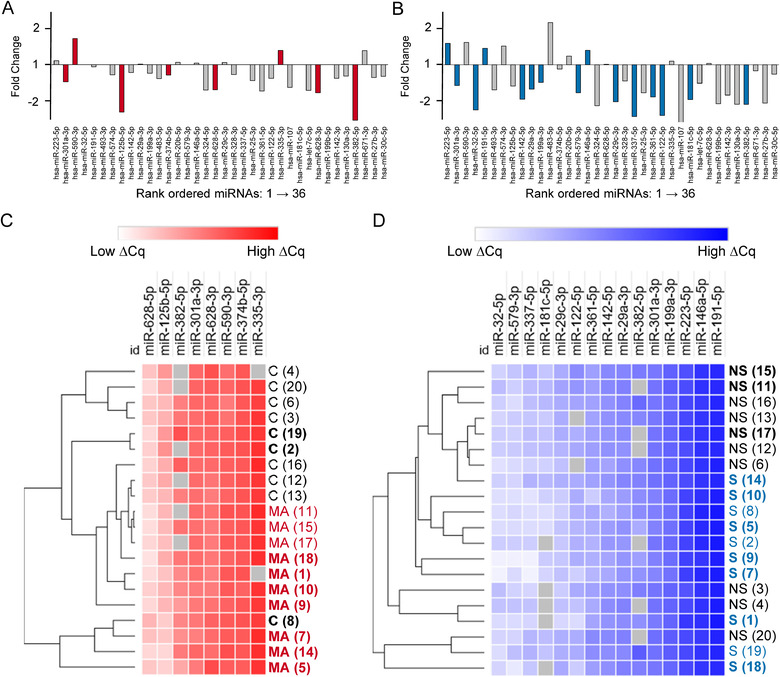
Linkage analysis with miRNA of interest show clustering consistent with MA or smoking. A,B) Manhattan plots of the top 20% of miRNAs in rank order and shown as fold change of MA‐ACT versus CTL (a) or Smokers versus Non‐smokers (b). Coloured bars are miRNAs of interest based on at least a 1.2‐fold change in expression, Glass's Δ of at least 0.8, and AUC of at least 0.75. c) Hierarchal clustering of each participant based on the ΔCq values for 10 miRNAs of interest in MA‐ACT. Participants are labelled based on their MA‐ACT (MA, n = 10, red font) or CTL (c, n = 10, black font). Bold indicates participants that use tobacco. d) Hierarchal clustering of each participant based on the ΔCq values for 16 miRNAs of interest in smoking. Participants are labelled based on their smoking (S, n = 10, blue font) or non‐smoking (NS, n = 10, black font). Bold indicates MA‐ACT participants. Analysis was done using one minus the Pearson correlation and average linkage. The numbers in parenthesis identify corresponding participants in panels c, d.
TABLE 3.
Smoker MiRNAs of interest
| miRBase ID | Rank | Fold change | Glass's ∆ | AUC |
|---|---|---|---|---|
| miR‐223‐5p | 1 | 1.46 | ‐1.68 | 0.870 |
| miR‐301a‐3p | 2 | ‐1.44 | 2.85 | 0.770 |
| miR‐32‐5p | 4 | ‐2.24 | 1.45 | 0.840 |
| miR‐191‐5p | 5 | 1.34 | ‐1.32 | 0.760 |
| miR‐142‐5p | 9 | ‐1.85 | 1.51 | 0.820 |
| miR‐29a‐3p | 10 | ‐1.54 | 1.47 | 0.770 |
| miR‐199a‐3p | 11 | ‐1.37 | 1.98 | 0.780 |
| miR‐579‐3p | 15 | ‐1.65 | 1.21 | 0.770 |
| miR‐146a‐5p | 16 | 1.29 | ‐1.65 | 0.770 |
| miR‐29c‐3p | 19 | ‐1.93 | 1.06 | 0.780 |
| miR‐337‐5p | 21 | ‐2.52 | 1.74 | 0.760 |
| miR‐361‐5p | 23 | ‐1.77 | 1.44 | 0.760 |
| miR‐122‐5p | 24 | ‐2.47 | 1.13 | 0.825 |
| miR‐181c‐5p | 27 | ‐1.86 | 1.46 | 0.750 |
| miR‐382‐5p | 33 | ‐2.03 | 1.22 | 0.800 |
3.6. Expression levels of MA‐ACT miRNAs of interest correlate with clinical features of MA use
We next assessed the correlation between MA‐ACT miRNAs and clinical features of MA use. For each miRNA we correlated each participants miRNA expression levels (ΔCq) to (i) age at first use of MA (age of onset), (ii) percent of lifetime using MA (% lifetime), (iii) how recently MA was used (recency), (iv) frequency of MA use (frequency), (v) quantity of MA use (quantity) and (vi) smoking status (Table 4). We report that three miRNAs correlate with clinical features of MA use: miR‐301a‐3p, miR‐382‐5p, miR‐628‐5p (Table 4). All three miRNAs had a significant correlation with age of onset (Figure 6a‐c) and percent of lifetime using MA (Figure 6d‐f). Lower expression levels of miR‐301a‐3p and miR‐382‐3p correlated with measures of longer lifetime exposure to MA (Figure 6a, b), while higher expression levels correlated to longer exposure for miR‐628‐5p (Figure 6c). Furthermore, miR‐382‐5p has a significant negative correlation with frequency of MA use, with lower expression levels correlated with increased frequency of use (Figure 6g). None of these three miRNAs had a significant correlation with the smoking status. Together these data support that our statistical approach identified miRNAs whose expression levels are linked to MA use, with plasma EV miRNA profiles that closely link with cumulative exposure to MA.
TABLE 4.
MA‐ACT MiRNA correlations with MA use variables
| MA use correlations to ∆Cq: R2 (p‐value) | ||||||
|---|---|---|---|---|---|---|
| miRBase ID | Age of Onset | % Lifetime | Recency | Frequency | Quantity | Smoking |
| miR‐125b‐5p | 0.17 (0.24) | 0.20 (0.19) | 0.0044 (0.86) | 0.069 (0.46) | 0.0010 (0.93) | 0.087 (0.41) |
| miR‐301a‐3p | 0.46 (0.032) | 0.65 (0.0047) | 0.24 (0.15) | 0.19 (0.20) | 0.018 (0.71) | 0.28 (0.11) |
| miR‐335‐3p | 0.052 (0.55) | 0.097 (0.41) | 0.00038 (0.96) | 0.016 (0.74) | 0.051 (0.56) | 0.0078 (0.82) |
| miR‐374b‐5p | 0.32 (0.09) | 0.15 (0.28) | 0.28 (0.12) | 0.041 (0.57) | 0.013 (0.75) | 0.14 (0.29) |
| miR‐382‐5p | 0.66 (0.015) | 0.82 (0.0021) | 0.35 (0.12) | 0.64 (0.017) | 0.0054 (0.86) | 0.23 (0.23) |
| miR‐590‐3p | 0.034 (0.61) | 0.060 (0.50) | 0.11 (0.35) | 0.093 (0.39) | 0.043 (0.57) | 0.018 (0.72) |
| miR‐628‐3p | 0.10 (0.37) | 0.31 (0.09) | 0.090 (0.40) | 0.042 (0.57) | 0.22 (0.17) | 0.078 (0.43) |
| miR‐628‐5p | 0.58 (0.011) | 0.45 (0.033) | 0.24 (0.15) | 0.29 (0.10) | 0.00044 (0.95) | 0.15 (0.28) |
FIGURE 6.

Expression levels of MA‐ACT miRNAs correlate with clinical features of MA use. Correlations of each subjects ΔCq values for miR‐301a‐3p, miR‐382‐5p, and miR‐628‐5p and the age at which the subject started to use MA (a‐c, age of onset), percent of the subjects lifetime spent using MA (d‐f, percent of lifetime), and frequency of MA use (g). All three miRNAs had significant correlations for age of onset and percent of lifetime. MiR‐382‐5p had significant correlation with frequency of use. Data were analysed using Pearson correlations and fitted with a simple linear regression curve.
3.7. Target and pathway predictions for miRNAs affected by MA use
The three miRNAs associated with lifetime exposure to MA (miR‐301a‐3p, miR‐382‐5p, miR‐628) were used for mRNA target prediction with the online analysis tools TargetScan and miRDB. TargetScan predicted 394 targets of the three miRNAs, while miRDB predicted 385 targets of the three miRNAs. The two target lists overlapped by 65 genes for a total of 714 unique targets in the union between TargetScan and miRDB. Ingenuity pathway analysis on these predicted mRNAs identified 144 significant canonical pathways (Figure 7a). The top 20 canonical pathways were commonly related to cardiovascular disease, synaptic plasticity and neuroinflammation (Figure 7b), which are all associated with MA dependency (7).
FIGURE 7.

Target and pathway prediction for miRNA effected by lifetime MA exposure. a) MiRNA target prediction work flow using the three miRNAs effected by lifetime MA exposure. b) Top 20 canonical pathways associated with the predicted mRNA targets. Top pathways are related to neuroinflammation, neurodegeneration, neuronal plasticity, cardiac‐, kidney‐ and liver disease.
4. DISCUSSION
EVs and miRNAs have each been implicated in the pathophysiology of addiction to multiple stimulants (Rao, O'connell, & Finnerty, 2018; Smith & Kenny, 2018). To our knowledge, we are the first to examine the effect of MA on plasma EVs and their miRNA cargo in human study participants with an active dependency on MA. Because of the small number of participants in this pilot study, we focused on females as they may be more sensitive to the effects of MA than males (Chang et al., 2005; Winhusen & Lewis, 2013). In this study, we presented evidence for the enrichment of EVs isolated from plasma by SEC. For the MA‐ACT group, we showed that MA increased the concentration of small TS+ EVs in plasma, and that the predominant signature of the miRNAs was decreased expression. We identified eight miRNAs of interest in the MA‐ACT group. The expression levels of three of these miRNAs correlated with clinical features of MA use, and their predicted mRNA targets and pathways were consistent with the deleterious effects of MA use and addiction. Since nicotine also regulates EV release and miRNA expression (Banerjee, Waters, Camacho, & Minet, 2015; Benedikter, Wouters, Savelkoul, Rohde, & Stassen, 2018; Enjeti et al., 2017; Smith & Kenny, 2018; Stassen et al., 2019), we re‐analysed our data based on participant smoking status. Contrary to MA, there was no change in TS+ EVs in smokers, relative to non‐smokers. However, like MA, a majority of miRNAs affected by smoking were decreased in expression, relative to non‐smokers. Although both MA and smoking had a net effect of decreased plasma EV miRNA expression, our data showed there was minimal overlap in the specific miRNAs of interest for MA use versus smoking. Taken together our findings suggest that MA regulates EVs and their miRNA cargo in females, and implicates a role for EVs in the physiological effects of MA use disorder.
Previous studies have shown that stimulants alter the biogenesis and release of multiple EV subtypes, which in turn may affect cell‐to‐cell communication, and play a role in addiction (Carone et al., 2015; Meng et al., 2020; Nazari et al., 2018). In the mouse VTA, cocaine stimulate the release of EVs that modulate synaptic transmission (Nakamura et al., 2019), an established component to DA neuron plasticity and consequent drug dependency and craving (Kauer & Malenka, 2007). Cocaine also stimulates the release of CD63+ EVs in the mouse VTA, which is dependent on alpha‐synuclein (α‐Syn) (Trubetckaia et al., 2019), a protein that is increased in both the blood and brain of humans that use cocaine and is correlated with increased drug craving (Mash et al., 2008; Qin, Ouyang, Pablo, & Mash, 2005). α‐Syn is also relevant to MA in that (i) α‐Syn protein levels are robustly increased in the brain of MA‐treated mice (Biagioni et al., 2019; Jiang, Li, Zhang, Wang, & Wang, 2014), (ii) MA treated cultured neurons release exosomes that contain α‐Syn, which are taken up by cultured astrocytes to trigger an inflammatory response (Meng et al., 2020), and (iii) a single nucleotide polymorphism in the α‐Syn gene is associated with MA psychosis and dependency in females, but not males (Kobayashi et al., 2004). There is also evidence that humans with a history of MA use are at increased risk of developing Parkinson's disease (Lappin, Darke, & Farrell, 2018), which is associated with propagation of misfolded α‐Syn protein (Hijaz & Volpicelli‐Daley, 2020). In plasma, MA increases plasma microparticles (AnnV+, CD144+, CD31+ and CD41a+) (Nazari et al., 2018); while tobacco use decreases levels of platelet‐derived CD41+ microvesicles (Enjeti et al., 2017). Here we found an effect of MA on increased amounts of TS+ EVs in plasma, but no effect on AnnV+, CD41+ or CD235+ EVs. We also did not find an effect of smoking on any of the plasma EV subclasses that we assayed. Multiple possibilities may account for these differences in outcomes: (i) significant methodological differences in earlier studies, which lacked calibration and important specificity controls, (ii) seven out of 10 of the smokers in our study were also actively using MA, which may confound individual drug effects, and (iii) our study focused on females, while other studies examined both males and females.
To study the effects of MA on EV cargo we isolated EVs by SEC. While our SEC isolation method enriched for markers associated with EVs (CD9, CD63, CD81, flotillin, Alix, and TSG101) while depleting a majority of circulating plasma proteins, we did observe Ago2 staining in the EV fraction immunoblots. Ago proteins have been found in mammalian EVs (Mckenzie et al., 2016; Melo et al., 2014). However, a recent rigorous study employed high‐resolution density gradient fractionation and direct immunoaffinity capture to characterize the nucleic acid and protein constituents of exosomes, and reported that Ago1–4 is not in exosomes, but is secreted from cells in non‐vesicular components and can be present in other small EVs (Jeppesen et al., 2019). Thus, our finding of Ago2 co‐localizing with EV markers suggests that there may be a potential overlap with non‐vesicular particles in this preparation, as SEC is not as stringent as high‐resolution density gradient fractionation for separating particles from vesicles. However, vFC with the SEC pool showed CFSE+ and TS+ EVs, but with a smaller and narrower size range, and relatively fewer high‐scattering and low scatter lipoprotein events, compared to total plasma, as might be expected from a size‐fractionated sample, and confirming the effectiveness of the fractionation.
Plasma is also subject to RBC haemolysis, which could release miRNA contaminants such as miR‐451a, miR‐486‐5p, miR‐92a, and miR‐16 that are associated with RBCs and are increased by 20–30‐fold haemolysed plasma (Pritchard et al., 2012). While SEC isolation followed by concentrating the fractions with a 30‐kDa filter unit may remove free floating miRNA contaminants, we also detected miR‐451a, miR‐486‐5p, miR‐16‐5p, and miR‐92a‐3p in our samples albeit at expression levels that do not reflect RBC haemolysis in individual samples. Thus, we cannot rule out that RBC miRNAs were a part of the plasma EVs. However, none of these RBC markers were identified as miRNAs of interest in either the MA‐ACT or smoker groups in this study.
Our findings show that both MA use and smoking led to a decrease in the expression of most miRNAs in plasma EVs, relative to controls or non‐smokers. This is consistent with previous studies in plasma that show four of six miRNAs (miR‐181a, miR‐15b, miR‐let‐7e, and miR‐let‐7d) were significantly decreased in MA use disorder and negatively correlate with days of drug use in the past month (Zhao et al., 2016). MiR‐181a has been shown to regulate glutamate ionotropic receptor AMPA type subunit 2 (GRIA2), which was increased in humans with MA use disorder (Zhang et al., 2016) and implicated in addictive behaviours (Mead & Stephens, 2003). Also, a single nucleotide polymorphism in miR‐181a (rs10760371) was implicated as a genetic difference between MA users with psychosis and healthy controls (Sun et al., 2020). We did not find any of these four miRNAs in our top eight MA‐regulated miRNAs, which may in part be due to differences in our selection criteria. Our study identified miRNAs based on four statistical measures: (i) top 20% of ranked miRNAs, (ii) at least a 1.2‐fold change in expression, (iii) effect size of at least 0.8, and (iv) AUC of at least 0.75. In comparison, the aforementioned MA use disorder study identified miRNAs based on fold change, involvement with neuroplasticity, neurogenesis, and neural development (Zhao et al., 2016). Based on fold change alone, we did find that the four miRNAs (Zhao et al., 2016) matched our findings with at least a 1.2‐fold decrease in expression of miR‐181a‐5p, miR‐15b‐5p, let‐7e‐5p, and let‐7d‐5p (Table S2). Thus, even using different selection criteria, these same four miRNAs were implicated in MA use in two separate studies.
Rodent models have also been used to investigate the effects of MA on miRNA expression in brain and serum EVs (Bosch et al., 2015; Du et al., 2016; Li et al., 2018; Li et al., 2018; Sim et al., 2017; Zhu et al., 2015,). In the rat VTA, miRNAs previously associated with MA addiction (miR‐124 and miR‐181a) as well as novel miRNAs (miR‐125a‐5p and miR‐145) are decreased in expression following 14 days of abstinence (Bosch et al., 2015). Furthermore, decreases in miR‐125a‐5p and miR‐145 expression are associated with increases in DA transporter (Bosch et al., 2015), and have an established role in regulating VTA DA levels and in neuroprotection (Mijatovic et al., 2007; Tenenbaum & Humbert‐Claude, 2017). While neither miR‐125a‐5p nor miR‐145 were identified as miRNAs of interest in our study, we did find a 1.48 fold decrease in miR‐145, which was also associated with a large effect size (Glass's Δ = 0.85). These data suggest that for some miRNAs, MA induced changes in expression within the brain may be also be detectable in plasma EVs and serve as biomarkers.
MA is also implicated in regulating the expression of miRNA biogenesis genes, which in turn effects miRNA expression levels (Liu et al., 2019; Mulligan et al., 2013). MiRNA biogenesis and processing involves Drosha and Dicer; while miRNA based degradation of mRNAs involves the formation of the RNA‐induced silencing complex (RISC), deadenylation, and decapping (Duchaine & Fabian, 2019; Jonas & Izaurralde, 2015). A large scale association analysis of ∼1,500 phenotypes conducted across the BXD (C57BL/6J x DBA/2J) genetic mouse population identified a significant correlation between behavioural responses to MA and expression levels of Dicer1 and Drosha in the amygdala and hippocampus, respectively (Mulligan et al., 2013). Further, in the nucleus accumbens of MA‐induced locomotor sensitized mice, Ago2 and 15 mature miRNAs were all decreased in expression, while the levels of Dicer1 were increased and levels of precursor miRNAs were either increased or not changed (Liu et al., 2019). These studies imply that MA predominantly effects processing events that lead to production of mature miRNAs. In support of this, we found that miR‐301a‐3p, which correlates with measures of lifetime exposure to MA, is predicted to target proteins associated with miRNA biogenesis (Dicer1), RISC (Ago1, Ago4), mRNA deadenylation (PAN3), and mRNA decapping (DDX6) (data not shown).
Our study investigated plasma EVs and their miRNA cargo to discover new biomarkers that may reflect disease severity and recovery from MA addiction. Identifying predictors of relapse based on a mechanistic understanding of MA use disorder is important for developing relapse prevention strategies. For example, Bayesian cognitive models have been tested to help identify predictive biomarkers of MA relapse (Harlé, Yu, & Paulus, 2019). Other innovative methods have also been used to quantify the severity of MA addiction and to identify biomarkers that may predict response to treatment or risk of relapse—with one potential biomarker being the quantity of MA used (Mendelson, J. Baggott, Flower, & Galloway, 2011). In preclinical studies, the pro‐inflammatory cytokine tumour necrosis factor‐alpha (TNF‐α) has been suggested as a potential diagnostic biomarker for the early stage of MA addiction (Yan, Nitta, Koseki, Yamada, & Nabeshima, 2012). Further, changes in the proliferation and survival of hippocampal neural progenitors and neuronal activation of hippocampal granule cells are able to predict the effects of MA on cognitive performance and relapse to MA seeking (Recinto et al., 2012). Thus, the discovery of objective quantifiable biomarkers for MA use that can be tested in humans is needed.
Here we showed for the first time in humans that a history of MA use increases the concentration of TS+ EVs in plasma and alters EV miRNA expression. We also identified eight miRNAs of interest for MA in plasma EVs. Of note, the expression levels of three of these miRNAs correlated to variables associated with lifetime exposure. These results suggest that the deleterious effects of long‐term exposure to MA may result in abnormal EV biogenesis, including miRNA cargo, which in turn may negatively affect cellular processes. In line with this, target prediction and IPA with these three miRNAs identified pathways related to neuroinflammation, neurodegeneration, or neuronal plasticity. Thus, changes in the miRNA content of EVs may identify novel gene target proteins that address multiple aspects of MA use disorder (diagnosis, prognosis, and treatment). We also identified one miRNA (miR‐382‐5p) that correlated with frequency of MA use, which suggests that plasma EV miRNAs could be used as valuable clinical biomarkers to monitor recover. We do acknowledge the limitations of interpreting our data since MA use behaviour was obtained by self‐report during clinical interview (except recent use, which was confirmed by urine drug analysis). Also, to account for small group sizes and because females may be more sensitive to the effects of MA than males, we chose to limit the study to females (Chang et al., 2005; Harmony, Alderson, Garcia‐Carachure, Bituin, & Crawford, 2020; Winhusen & Lewis, 2013). Female rats are more sensitive to the reinforcing effects of MA (Harmony et al., 2020). Also, prior exposure to nicotine increases MA self‐administration in female rats (Harmony et al., 2020). In contrast, concurrent nicotine exposure decreases MA self‐administration in both males and females (Harmony et al., 2020). Thus, we are now moving forward to carry out large scale validation and cross‐validation studies in both male and female participants that are essential to (i) validate our findings in a new cohort of participants, (ii) investigate putative sex‐dependent effects, and (iii) perform co‐variate analysis to assess potential interactions between MA and smoking. In addition, studies that include MA‐ACT participants in remission will determine the potential of plasma EVs to serve as biomarkers for recovery from MA use disorder. Finally, mechanistic studies that investigate EV subtypes (i.e. neuronal vs. microglial derived EVs) and their targets are necessary to resolve the pathways regulated by circulating EVs and their molecular cargo.
CONFLICTS OF INTEREST
John P. Nolan and Erika Duggan are inventors on patents related to vesicle analysis and have interests in Cellarcus Biosciences. All other authors report no conflicts of interest.
Supporting information
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
ACKNOWLEDGEMENTS
The authors would like to thank the study participants and staff at each of the recruitment sites, including Central City Concern, CODA, Inc., De Paul Treatment Centers, Native American Rehabilitation Association of the Northwest, OutsideIn, Volunteers of America Treatment Centers, OHSU, and the Veterans Affairs Portland Health Care System Mental Health Division and Substance Abuse Treatment Program. The authors are also grateful to Alissa Bazinet, Ph.D., Matthew Arbuckle, B.S., and Bethany Winters, B.S. for their many contributions to this project as Research Assistants and Psychometrists at the MARC. The authors would also like to acknowledge the OHSU Gene Profiling Shared Resource for providing support for the qPCR assays, under the direction of Christina A. Harrington, PhD. Consultation for the analysis of the qPCR data was provided by Jack Wiedrick, M.S. at the OHSU Biostatistics & Design Program. Transmission electron microscopy was performed at the Multiscale Microscopy Core with technical support from the OHSU‐FEI Living Lab and the OHSU Center for Spatial Systems Biomedicine, under the guidance of Claudia S. Lopez, PhD. The work was conducted using facilities at the VA Portland Health Care System and OHSU, Portland, Oregon. Authors JML (Research Scientist), AJ (Senior Research Career Scientist), and MH (Staff Psychologist and Neuropsychologist) acknowledge their appointments at the VA Portland Health Care System, Portland, Oregon. The contents do not represent the views of the Department of Veterans Affairs or the United States Government. This work was supported by the MARC under grant P50 DA018165 and Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, Biomedical Laboratory Research and Development Merit Review Awards [1I01BX002061 (JML), I01BX004934 (AJ)] and Clinical Sciences Research and Development Merit Review Award [1I01CX001592‐01 (MH)].
Sandau US, Duggan E, Shi X, et al. Methamphetamine use alters human plasma extracellular vesicles and their MicroRNA Cargo: An exploratory study. J Extracell Vesicles. 2020;10:e12028 10.1002/jev2.12028
DATA AVAILABILITY STATEMENT
Relevant experimental parameters can be accessed in the EV‐TRACK (evtrack.org) knowledgebase. The vFC data will be available in the Flow Repository https://flowrepository.org/. The miRNA Microarray data is MIAME compliant and will be available on the Gene Expression Omnibus site: https://www.ncbi.nlm.nih.gov/geo/.
REFERENCES
- Agarwal, V. , Bell, G. W. , Nam, J‐Wu , & Bartel, D. P. (2015). Predicting effective microRNA target sites in mammalian mRNAs. Elife, 4, e05005 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association , Diagnostic and statistical manual of mental disorders: DSM‐IV‐TR. (4th ed., text revision. ed). (2000)., Washington, DC: American Psychiatric Association; 10.1002/9780470479216.corpsy0271. [DOI] [Google Scholar]
- Banerjee, A. , Waters, D. , Camacho, O. M. , & Minet, E. (2015). Quantification of plasma microRNAs in a group of healthy smokers, ex‐smokers and non‐smokers and correlation to biomarkers of tobacco exposure. Biomarkers, 20(2), 123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedikter, B. J. , Wouters, E. F. M. , Savelkoul, P. H. M. , Rohde, G. G. U. , & Stassen, F. R. M. (2018). Extracellular vesicles released in response to respiratory exposures: Implications for chronic disease. Journal of Toxicology and Environmental Health. Part B, Critical Reviews, 21(3), 142–160. [DOI] [PubMed] [Google Scholar]
- Biagioni, F. , Ferese, R. , Limanaqi, F. , Madonna, M. , Lenzi, P. , Gambardella, S. , & Fornai, F. (2019). Methamphetamine persistently increases alpha‐synuclein and suppresses gene promoter methylation within striatal neurons. Brain Research, 1719, 157–175. [DOI] [PubMed] [Google Scholar]
- Bosch, P. J. , Benton, M. C. , Macartney‐Coxson, D. , & Kivell, B. M. (2015). mRNA and microRNA analysis reveals modulation of biochemical pathways related to addiction in the ventral tegmental area of methamphetamine self‐administering rats. Bmc Neuroscience [Electronic Resource], 16, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos, K. , Malenica, I. , Metpally, R. , Courtright, A. , Rakela, B. , Beach, T. , … Van Keuren‐Jensen, K. (2014). Profiles of extracellular miRNA in cerebrospinal fluid and serum from patients with Alzheimer's and Parkinson's diseases correlate with disease status and features of pathology. Plos One, 9(5), e94839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carone, C. , Genedani, S. , Leo, G. , Filaferro, M. , Fuxe, K. , & Agnati, L. F. (2015). In vitro effects of cocaine on tunneling nanotube formation and extracellular vesicle release in glioblastoma cell cultures. Journal of Molecular Neuroscience, 55(1), 42–50. [DOI] [PubMed] [Google Scholar]
- Chandrasekar, V. , & Dreyer, J. ‐. L. (2009). microRNAs miR‐124, let‐7d and miR‐181a regulate cocaine‐induced plasticity. Molecular and Cellular Neuroscience, 42(4), 350–362. [DOI] [PubMed] [Google Scholar]
- Chang, L. , Cloak, C. , Patterson, K. , Grob, C. , Miller, E. N. , & Ernst, T. (2005). Enlarged striatum in abstinent methamphetamine abusers: A possible compensatory response. Biological Psychiatry, 57(9), 967–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , & Wang, X. (2020). miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Research, 48(D1), D127–D131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivet, M. , Hemming, F. , Pernet‐Gallay, K. , Fraboulet, S. , & Sadoul, R. (2012). Emerging role of neuronal exosomes in the central nervous system. Front Physiol, 3, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, C.‐H. , Shrestha, S. , Yang, C.‐D. , Chang, N.‐W. , Lin, Y.‐L. , Liao, K.‐W. , … Huang, H.‐D. (2018). miRTarBase update 2018: A resource for experimentally validated microRNA‐target interactions. Nucleic Acids Research, 46, D1, D296–D302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, J. (1988). Statistical Power Analysis for the Behavioral Sciences (2nd ed United States of America: Lawrence Erlbaum; 10.4324/9780203771587. [DOI] [Google Scholar]
- Cordazzo, C. , Petrini, S. , Neri, T. , Lombardi, S. , Carmazzi, Y. , Pedrinelli, R. , … Celi, A. (2014). Rapid shedding of proinflammatory microparticles by human mononuclear cells exposed to cigarette smoke is dependent on Ca2+ mobilization. Inflammation Research, 63(7), 539–547. [DOI] [PubMed] [Google Scholar]
- Darke, S. , Duflou, J. , Kaye, S. Prevalence and nature of cardiovascular disease in methamphetamine‐related death: A national study. Drug and Alcohol Dependence, 179, 174–179. [DOI] [PubMed] [Google Scholar]
- EV‐TRACK Consortium ; Deun, J. V. , Mestdagh, P. , Agostinis, P. , Akay, Ö. , Anand, S. , et al.,(2017). EV‐TRACK: transparent reporting and centralizing knowledge in extracellular vesicle research. Nature Methods, 14(3), 228–232. [DOI] [PubMed] [Google Scholar]
- Dickens, A. M. , Tovar‐Y‐Romo, L. B. , Yoo, S. ‐. W. , Trout, A. L. , Bae, M. , Kanmogne, M. , … Haughey, N. J. (2017). Astrocyte‐shed extracellular vesicles regulate the peripheral leukocyte response to inflammatory brain lesions. Science signaling, eaai7696 10(473) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, H. ‐. Y. , Cao, D‐Ni , Chen, Y. , Wang, Lv , Wu, N. , & Li, J. (2016). Alterations of prefrontal cortical microRNAs in methamphetamine self‐administering rats: From controlled drug intake to escalated drug intake. Neuroscience Letters, 611, 21–27. [DOI] [PubMed] [Google Scholar]
- Duchaine, T. F. , & Fabian, M. R. (2019). Mechanistic insights into MicroRNA‐mediated gene silencing. Cold Spring Harbor perspectives in biology, a032771 11(3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjeti, A. K. , Ariyarajah, A. , D'crus, A. , Seldon, M. , & Lincz, L. F. (2017). Circulating microvesicle number, function and small RNA content vary with age, gender, smoking status, lipid and hormone profiles. Thrombosis Research, 156, 65–72. [DOI] [PubMed] [Google Scholar]
- Eshleman, A. J. , Henningsen, R. A. , Neve, K. A. , & Janowsky, A. (1994). Release of dopamine via the human transporter. Molecular Pharmacology, 45(2), 312–316. [PubMed] [Google Scholar]
- Fan, X. , & Kurgan, L. (2015). Comprehensive overview and assessment of computational prediction of microRNA targets in animals. Briefings in Bioinformatics, 16(5), 780–794. [DOI] [PubMed] [Google Scholar]
- Harlé, K. M. , Yu, A. J. , & Paulus, M. P. (2019). Bayesian computational markers of relapse in methamphetamine dependence. NeuroImage. Clinical, 22, 101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmony, Z. R. , Alderson, E. M. , Garcia‐Carachure, I. , Bituin, L. D. , & Crawford, C. A. (2020). Effects of nicotine exposure on oral methamphetamine self‐administration, extinction, and drug‐primed reinstatement in adolescent male and female rats. Drug and Alcohol Dependence, 209, 107927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijaz, B. A. , & Volpicelli‐Daley, L. A. (2020). Initiation and propagation of alpha‐synuclein aggregation in the nervous system. Molecular Neurodegeneration, 15(1), 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckans, M. , Fuller, B. E. , Chalker, A. L. N. , Adams, M. , & Loftis, J. M. (2015). Plasma inflammatory factors are associated with anxiety, depression, and cognitive problems in adults with and without methamphetamine dependence: An exploratory protein array study. Front Psychiatry, 6, 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Narcotics Control Board. Report of the International Narcotics Control Board for 2018. (2019)., Vienna: United Nations Publication. [Google Scholar]
- Jeppesen, D. K. , Fenix, A. M. , Franklin, J. L. , Higginbotham, J. N. , Zhang, Q. , Zimmerman, L. J. , … Coffey, R. J. (2019). Reassessment of exosome composition. Cell, 177(2), 428–445.e18 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W. , Li, Ji , Zhang, Z. , Wang, H. , & Wang, Z. (2014). Epigenetic upregulation of alpha‐synuclein in the rats exposed to methamphetamine. European Journal of Pharmacology, 745, 243–248. [DOI] [PubMed] [Google Scholar]
- Jonas, S. , & Izaurralde, E. (2015). Towards a molecular understanding of microRNA‐mediated gene silencing. Nature Reviews Genetics, 16(7), 421–433. [DOI] [PubMed] [Google Scholar]
- Kauer, J. A. , & Malenka, R. C. (2007). Synaptic plasticity and addiction. Nature Reviews Neuroscience, 8(11), 844–858. [DOI] [PubMed] [Google Scholar]
- Kevil, C. G. , Goeders, N. E. , Woolard, M. D. , Bhuiyan, Md. S. , Dominic, P. , Kolluru, G. K. , … Orr, A. W. (2019). Methamphetamine use and cardiovascular disease. Arteriosclerosis, Thrombosis, and Vascular Biology, 39(9), 1739–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, H. , Ide, S. , Hasegawa, J. , Ujike, H. , Sekine, Y. , Ozaki, N. , … Soraa, I. (2004). Study of association between alpha‐synuclein gene polymorphism and methamphetamine psychosis/dependence. Annals of the New York Academy of Sciences, 1025, 325–334. [DOI] [PubMed] [Google Scholar]
- Koob, G. , & Bloom, F. (1988). Cellular and molecular mechanisms of drug dependence. Science, 242(4879), 715–723. [DOI] [PubMed] [Google Scholar]
- Lappin, J. M. , Darke, S. , & Farrell, M. (2018). Methamphetamine use and future risk for Parkinson's disease: Evidence and clinical implications. Drug and Alcohol Dependence, 187, 134–140. [DOI] [PubMed] [Google Scholar]
- Li, H. , Li, C. , Zhou, Y. , Luo, C. , Ou, J. , Li, J. , & Mo, Z. (2018). Expression of microRNAs in the serum exosomes of methamphetamine‐dependent rats vs. ketamine‐dependent rats. Experimental and Therapeutic Medicine, 15(4), 3369–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H.‐C. , Lin, Y.‐B. , Li, C. , Luo, C.‐H. , Zhou, Y.‐T. , Ou, J.‐Y. , … Mo, Z.‐X. (2018). Expression of miRNAs in Serum Exosomes versus Hippocampus in Methamphetamine‐Induced Rats and Intervention of Rhynchophylline. Evidence‐Based Complementary and Alternative Medicine : ECAM, 2018, 8025062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D. , Zhu, Li , Ni, T. , Guan, F. ‐. L. , Chen, Y. ‐. J. , Ma, D. ‐. L. , … Chen, T. (2019). Ago2 and Dicer1 are involved in METH‐induced locomotor sensitization in mice via biogenesis of miRNA. Addiction Biology, 24(3), 498–508. [DOI] [PubMed] [Google Scholar]
- Liu, W. , & Wang, X. (2019). Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biology, 20(1), 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusardi, T. A. , Phillips, J. I. , Wiedrick, J. T. , Harrington, C. A. , Lind, B. , Lapidus, J. A. , … Saugstad, J. A. (2017). MicroRNAs in human cerebrospinal fluid as biomarkers for Alzheimer's disease. Journal of Alzheimers Disease, 55(3), 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mash, D. C. , Adi, N. , Duque, L. , Pablo, J. , Kumar, M. , & Ervin, F. R. (2008). Alpha synuclein protein levels are increased in serum from recently abstinent cocaine abusers. Drug and Alcohol Dependence, 94(1‐3), 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mckenzie, A. J. , Hoshino, D. , Hong, N. H. , Cha, D. J. , Franklin, J. L. , Coffey, R. J. , … Weaver, A. M. (2016). KRAS‐MEK signaling controls Ago2 sorting into exosomes. Cell reports, 15(5), 978–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead, A. N. , & Stephens, D. N. (2003). Involvement of AMPA receptor GluR2 subunits in stimulus‐reward learning: Evidence from glutamate receptor gria2 knock‐out mice. Journal of Neuroscience, 23(29), 9500–9507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo, S. A. , Sugimoto, H. , O'connell, J T. , Kato, N. , Villanueva, A. , Vidal, A. , … Kalluri, R. (2014). Cancer exosomes perform cell‐independent microRNA biogenesis and promote tumorigenesis. Cancer Cell, 26(5), 707–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelson, J. , J. Baggott, M. , Flower, K. , & Galloway, G. (2011). Developing biomarkers for methamphetamine addiction. Current Neuropharmacology, 9(1), 100–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, Y. , Ding, J. , Li, C. , Fan, H. , He, Y. , & Qiu, P. (2020). Transfer of pathological alpha‐synuclein from neurons to astrocytes via exosomes causes inflammatory responses after METH exposure. Toxicology Letters, 331, 188–199. [DOI] [PubMed] [Google Scholar]
- Mijatovic, J. , Airavaara, M. , Planken, A. , Auvinen, P. , Raasmaja, A. , Piepponen, T. P. , … Saarma, M. (2007). Constitutive Ret activity in knock‐in multiple endocrine neoplasia type B mice induces profound elevation of brain dopamine concentration via enhanced synthesis and increases the number of TH‐positive cells in the substantia nigra. Journal of Neuroscience, 27(18), 4799–4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan, M. K. , Dubose, C. , Yue, J. , Miles, M. F. , Lu, Lu , & Hamre, K. M. (2013). Expression, covariation, and genetic regulation of miRNA biogenesis genes in brain supports their role in addiction, psychiatric disorders, and disease. Front Genet, 4, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, Y. , Dryanovski, D. I. , Kimura, Y. , Jackson, S. N. , Woods, A. S. , Yasui, Y. , … Lupica, C. R. (2019). Cocaine‐induced endocannabinoid signaling mediated by sigma‐1 receptors and extracellular vesicle secretion. Elife, 8, e47209 10.7554/eLife.47209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazari, A. , Zahmatkesh, M. , Mortaz, E. , & Hosseinzadeh, S. (2018). Effect of methamphetamine exposure on the plasma levels of endothelial‐derived microparticles. Drug and Alcohol Dependence, 186, 219–225. [DOI] [PubMed] [Google Scholar]
- Neven, K. Y. , Nawrot, T. S. , & Bollati, V. (2017). Extracellular Vesicles: How the External and Internal Environment Can Shape Cell‐To‐Cell Communication. Current Environmental Health Reports, 4(1), 30–37. [DOI] [PubMed] [Google Scholar]
- Oliveira, A. C. , Bovolenta, L. A. , Nachtigall, P. G. , Herkenhoff, M. E. , Lemke, N. , & Pinhal, D. (2017). Combining results from distinct MicroRNA target prediction tools enhances the performance of analyses. Frontiers in Genetics, 8, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash, M. D. , Tangalakis, K. , Antonipillai, J. , Stojanovska, L. , Nurgali, K. , & Apostolopoulos, V. (2017). Methamphetamine: Effects on the brain, gut and immune system. Pharmacological Research, 120, 60–67. [DOI] [PubMed] [Google Scholar]
- Pritchard, C. C. , Kroh, E. , Wood, B. , Arroyo, J. D. , Dougherty, K. J. , Miyaji, M. M. , … Tewari, M. (2012). Blood cell origin of circulating microRNAs: A cautionary note for cancer biomarker studies. Cancer prevention research (Philadelphia, Pa.), 5(3), 492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, Y. , Ouyang, Q. , Pablo, J. , & Mash, D. C. (2005). Cocaine abuse elevates alpha‐synuclein and dopamine transporter levels in the human striatum. Neuroreport, 16(13), 1489–1493. [DOI] [PubMed] [Google Scholar]
- Rao, P. S. S. , O'connell, K. , & Finnerty, T. K. (2018). Potential role of extracellular vesicles in the pathophysiology of drug addiction. Molecular Neurobiology, 55(8), 6906–6913. [DOI] [PubMed] [Google Scholar]
- Rao, P. , Benito, E. , & Fischer, A. (2013). MicroRNAs as biomarkers for CNS disease. Frontiers in Molecular Neuroscience, 6, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recinto, P. , Samant, A. R. H. , Chavez, G. , Kim, A. , Yuan, C. J. , Soleiman, M. , … Mandyam, C. D. (2012). Levels of neural progenitors in the hippocampus predict memory impairment and relapse to drug seeking as a function of excessive methamphetamine self‐administration. Neuropsychopharmacology, 37(5), 1275–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAMSHA, 2019. Key Substance Use and Mental Health Indicators in the United States: Results from the 2018 National Survey on Drug Use and Health (HHS Publication No. PEP19‐5068, NSDUH Series H‐54), Rockville, MD, 20857.
- Saugstad, J A. , Lusardi, T A. , Van Keuren‐Jensen, K R. , Phillips, J I. , Lind, B. , Harrington, C A. , … Hochberg, F H. , Analysis of extracellular RNA in cerebrospinal fluid. Journal of Extracellular Vesicles, 2017. 6(1), 1317577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serban, K. A. , Rezania, S. , Petrusca, D. N. , Poirier, C. , Cao, D. , Justice, M. J. , … Petrache, I. (2016). Structural and functional characterization of endothelial microparticles released by cigarette smoke. Scientific Reports, 6, 31596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan, D. V. , Lecrubier, Y. , Sheehan, K. H. , Amorim, P. , Janavs, J. , Weiller, E. , … Dunbar, G. C. (1998). The Mini‐International Neuropsychiatric Interview (M.I.N.I.): The development and validation of a structured diagnostic psychiatric interview for DSM‐IV and ICD‐10. Journal of Clinical Psychiatry, 59(Suppl 20), 22–33;quiz 34–57. [PubMed] [Google Scholar]
- Sim, M. S. , Soga, T. , Pandy, V. , Wu, Y. S. , Parhar, I. S. , & Mohamed, Z. (2017). MicroRNA expression signature of methamphetamine use and addiction in the rat nucleus accumbens. Metabolic Brain Disease, 32(6), 1767–1783. [DOI] [PubMed] [Google Scholar]
- Smith, A. C. W. , & Kenny, P. J. (2018). MicroRNAs regulate synaptic plasticity underlying drug addiction. Genes, Brain, and Behavior, 17(3), e12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stassen, F. R. M. , Van Eijck, P. H. , Savelkoul, P. H. M. , Wouters, E. F. M. , Rohde, G. G. U. , Briedé, J. J. , … Benedikter, B. J. (2019). Cell type‐ and exposure‐specific modulation of CD63/CD81‐positive and tissue factor‐positive extracellular vesicle release in response to respiratory toxicants. Oxidative Medicine and Cellular Longevity, 2019, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, H. , Zhu, Li , Li, J. , Wang, R. , Liu, D. , Han, W. , … Chen, T. (2019). Regulation of microRNA‐29c in the nucleus accumbens modulates methamphetamine ‐induced locomotor sensitization in mice. Neuropharmacology, 148, 160–168. [DOI] [PubMed] [Google Scholar]
- Sun, Q. , Zhao, Y. , Zhang, K. , Su, H. , Chen, T. , Jiang, H. , … Zhao, M. (2020). An association study between methamphetamine use disorder with psychosis and polymorphisms in MiRNA. Neuroscience Letters, 717, 134725. [DOI] [PubMed] [Google Scholar]
- Tenenbaum, L. , & Humbert‐Claude, M. (2017). Glial cell line‐derived neurotrophic factor gene delivery in Parkinson's disease: A delicate balance between neuroprotection, trophic effects, and unwanted compensatory mechanisms. Frontiers in Neuroanatomy, 11, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , … Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 7(1), 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trubetckaia, O. , Lane, A. E. , Qian, L. , Zhou, P. , & Lane, D. A. (2019). Alpha‐synuclein is strategically positioned for afferent modulation of midbrain dopamine neurons and is essential for cocaine preference. Communications Biology, 2, 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers, K. C. , Palmisano, B. T. , Shoucri, B. M. , Shamburek, R. D. , & Remaley, A. T. (2011). MicroRNAs are transported in plasma and delivered to recipient cells by high‐density lipoproteins. Nature Cell Biology, 13(4), 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winhusen, T. , & Lewis, D. (2013). Sex differences in disinhibition and its relationship to physical abuse in a sample of stimulant‐dependent patients. Drug and Alcohol Dependence, 129(1‐2), 158–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Medical, A. (2013). World Medical Association Declaration of Helsinki: Ethical principles for medical research involving human subjects. JAMA, 310(20), 2191–2194. [DOI] [PubMed] [Google Scholar]
- Yan, Y. , Nitta, A. , Koseki, T. , Yamada, K. , & Nabeshima, T. (2012). Dissociable role of tumor necrosis factor alpha gene deletion in methamphetamine self‐administration and cue‐induced relapsing behavior in mice. Psychopharmacology, 221(3), 427–436. [DOI] [PubMed] [Google Scholar]
- Yuyama, K. , & Igarashi, Y. (2016). Physiological and pathological roles of exosomes in the nervous system. Biomolecular Concepts, 7(1), 53–68. [DOI] [PubMed] [Google Scholar]
- Zhang, G. , & Yang, P. (2018). A novel cell‐cell communication mechanism in the nervous system: Exosomes. Journal of Neuroscience Research, 96(1), 45–52. [DOI] [PubMed] [Google Scholar]
- Zhang, K. , Wang, Q. , Jing, X. , Zhao, Y. , Jiang, H. , Du, J. , … Zhao, M. (2016). miR‐181a is a negative regulator of GRIA2 in methamphetamine‐use disorder. Scientific Reports, 6, 35691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Zhang, K. , Jiang, H. , Du, J. , Na, Z. , Hao, W. , … Zhao, M. (2016). Decreased Expression of Plasma MicroRNA in Patients with Methamphetamine (MA) Use Disorder. Journal of neuroimmune pharmacology : The official journal of the Society on NeuroImmune Pharmacology, 11(3), 542–548. [DOI] [PubMed] [Google Scholar]
- Zhu, Li , Zhu, J. , Liu, Y. , Chen, Y. , Li, Y. , Chen, S. , … Chen, T. (2015). Chronic methamphetamine regulates the expression of MicroRNAs and putative target genes in the nucleus accumbens of mice. Journal of Neuroscience Research, 93(10), 1600–1610. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Data Availability Statement
Relevant experimental parameters can be accessed in the EV‐TRACK (evtrack.org) knowledgebase. The vFC data will be available in the Flow Repository https://flowrepository.org/. The miRNA Microarray data is MIAME compliant and will be available on the Gene Expression Omnibus site: https://www.ncbi.nlm.nih.gov/geo/.


