Abstract
Glycyl‐tRNA synthetase 1 (GARS1), a cytosolic enzyme secreted from macrophages, promotes apoptosis in cancer cells. However, the mechanism underlying GARS1 secretion has not been elucidated. Here, we report that GARS1 is secreted through unique extracellular vesicles (EVs) with a hydrodynamic diameter of 20–58 nm (mean diameter: 36.9 nm) and a buoyant density of 1.13–1.17 g/ml. GARS1 was anchored to the surface of these EVs through palmitoylated C390 residue. Proteomic analysis identified 164 proteins that were uniquely enriched in the GARS1‐containing EVs (GARS1‐EVs). Among the identified factors, insulin‐like growth factor II receptor, and vimentin also contributed to the anti‐cancer activity of GARS1‐EVs. This study identified the unique secretory vesicles containing GARS1 and various intracellular factors that are involved in the immunological defence response against tumorigenesis.
Keywords: extracellular vesicles, glycyl‐tRNA synthetase 1, macrophage, palmitoylation, tumour microenvironment
1. INTRODUCTION
Various intracellular proteins that mediate several biological processes in the extracellular space have been identified in the last decade. For example, high mobility group box 1 (HMGB1), a ubiquitously expressed nuclear protein involved in the regulation of gene expression, is secreted to trigger inflammatory responses (Jiang & Pisetsky, 2006, Wang, 1999). Additionally, various heat‐shock proteins (HSPs) are secreted to activate the immune cells (Asea et al., 2000; Basu, Binder, Ramalingam, & Srivastava, 2001). These proteins lack the typical signal peptide associated with secretory proteins. This indicated that these proteins are secreted through the unconventional secretory pathways. For example, the secretion of HSP70 involves exosome, ATP‐binding cassette (ABC) transporter, and lysosomal exocytosis (Mambula & Calderwood, 2006; Mambula, Stevenson, Ogawa, & Calderwood, 2007). HMGB1 is actively secreted through vesicle compartments or secretory lysosomes upon stimulation or passively secreted from the necrotic and damaged cells (Gardella et al., 2002). This indicates that HMGB1 could be secreted from the cells through multiple pathways. Although various secretory intracellular proteins have been identified, the secretory pathways of these proteins have not been elucidated.
The family of aminoacyl‐tRNA synthetases (ARSs) is essential for protein synthesis. Previous studies have reported the secretion of different ARSs and their extracellular functions in diverse cellular processes (Guo & Schimmel, 2013; Son, Park, & Kim, 2014). The immune‐related activities of ARSs are similar to those of previously reported secretory intracellular factors. However, ARSs are unique because they exhibit characteristic intracellular and extracellular functions (Kim, You, & Hwang, 2011; Park, Schimmel, & Kim, 2008; Yao & Fox, 2013). Additionally, the autoantibodies against several ARSs have been detected in the blood of patients with autoimmune diseases, which indicated the pathological role of secreted ARSs (Gelpí, Kanterewicz, Gratacos, Targoff, & Rodriguez‐Sanchez, 1996; Stojanov, Satoh, Hirakata, & Reeves, 1996). Thus, the secretory pathways of ARSs must be elucidated to understand the pathophysiological implications of secreted ARSs. Previously, we had reported that macrophages secrete human glycyl‐tRNA synthetase 1 (GARS1) under stress conditions, which promoted cancer cell death (Park et al., 2012). However, the mechanism underlying GARS1 secretion and the other factors secreted from the macrophages under stress conditions are not known. This study demonstrated that GARS1 was secreted through novel secretory extracellular vesicles (EVs) generated from the macrophages under glucose starvation conditions. The GARS1‐containing EVs (GARS1‐EVs) harboured other intracellular factors that can suppress cancer progression along with GARS1 through multiple mechanisms. The characterization of biophysical properties, protein content, and functions of GARS1‐EVs suggested that they were unique secretory vesicles that are distinct from the known secretory vesicles.
2. MATERIALS AND METHODS
2.1. Cell culture
The RAW 264.7 cells were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum and 50 µg/ml streptomycin/penicillin solution. The H460 cell line was cultured in Rosewell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% fetal bovine serum and 50 µg/ml streptomycin/penicillin solution. The bone marrow derived macrophages (BMDMs) were isolated from the femur and tibia of the C57B/6 mice. The red blood cells in the bone marrow were lysed using ACK lysis buffer (Life Technologies). Next, the bone marrow cells were incubated with murine granulocyte‐macrophage colony‐stimulating factor (Peprotech) for 7 days.
2.2. Reagents
The following primary antibodies were used in this study: anti‐HSP90 (4F10; 1:1000; Santa Cruz; catalog no. sc‐69703), anti‐syntenin‐1 (S‐31; 1:1000; Santa Cruz; catalog no. sc‐100336), anti‐gp96 (9G10; 1:1000; Enzo Life Sciences; catalog no. ADI‐SPA‐850), anti‐GARS1 (1:5000; Abcam; catalog no. ab42905), anti‐IGF2R (EPR6599; 1:50,000; catalog no. ab124767), anti‐vimentin (EPR3776; 1:1000; Abcam; catalog no. ab92547), and anti‐tubulin (TUB 2.1; 1:10,000; Sigma; catalog no. T4026) antibodies. The secondary antibodies used in this study were as follows: goat anti‐rabbit IgG (1:10,000; Invitrogen; catalog no. 31460) and goat anti‐mouse IgG (1:10,000; Invitrogen; catalog no. 31430) antibodies. The secretory pathway inhibitors (brefeldin A (BFA), Y27632, probenecid, sodium azide (NaN3), and methyl‐beta‐cyclodextrin (MβCD)) and 2‐bromopalmitate (2‐BP) were purchased from Sigma and Calbiochem, respectively.
2.3. Immunoblotting
The cells were lysed using 50 mM Tris‐HCl buffer (pH 7.4) containing 150 mM NaCl, 10 mM NaF, 1 mM ethylenediaminetetraacetic acid (EDTA), 1% (v/v) NP‐40, 10% (v/v) glycerol, and protease inhibitor for 30 min. The cell lysate was centrifuged at 18,000 × g for 15 min. The supernatant was mixed with sodium dodecyl sulfate (SDS) sample buffered and subjected to SDS‐polyacrylamide gel electrophoresis (SDS‐PAGE). The target proteins in the supernatant were detected using immunoblotting. The protein band intensities in the immunoblots were quantified using the ImageJ software (LOCI, Taylor et al., 2013). The quantitative values of protein band intensities are shown in Table S1.
2.4. Secretion and isolation of macrophage‐derived GARS1‐EVs
The secretion of GARS1 was induced as described previously (Park et al., 2012). Briefly, the proteins in the conditioned medium were precipitated using 10% trichloroacetic acid (TCA) at 4°C for 12 h. The precipitated proteins were subjected to SDS‐PAGE for immunoblotting. To isolate the GARS1‐EVs, the RAW 264.7 cells were washed twice with serum‐free medium to remove the EVs in the serum and incubated in serum‐free medium in the presence or absence of glucose. The culture medium was harvested and subjected to centrifugation steps based on the MISEV2018 guidelines (Théry et al., 2018). Briefly, the culture medium was centrifuged at 2000 × g for 10 min and 10,000 × g for 30 min to remove cell debris, followed by centrifugation at 100,000 × g in a WX‐1000 Sorvall swing bucket (Thermo) for 90 min to pellet the GARS1‐EVs. The pellet was incubated with trypsin‐EDTA (0.25%, Hyclone) (1:100) in phosphate‐buffered saline (PBS, pH 7.4) at 37°C for 20 min (Fitzgerald et al., 2018).
2.5. Dynamic light scattering
Macrophage‐derived EVs were resuspended in PBS. The hydrodynamic diameters of the EVs were measured a using light‐scattering spectrophotometer (ELS‐Z, Otsuka Electronics). Measurements were performed in the automatic mode after equilibration for 5 min at 20°C. The data were processed using the manufacturer's software in multiple narrow modes.
2.6. Electron microscopy (EM)
For negative staining EM, the isolated vesicles were diluted five fold in PBS. The sample solution (5 µl) was immediate (approximately 5 s) applied to a carbon‐coated grid (Harrick), which was glow‐discharged for 3 min in the air. The grid was negatively stained using 1% uranyl acetate. The same procedure was used for all the negatively stained specimens. For cryo‐EM, 5 µl of sample solution was loaded onto the copper R1.2/1.3 Quantifoil EM grids (SPI Supplies) pretreated with plasma cleaner for 30 s in air. The grids were then blotted and plunged frozen using FEI Vitrobot (FEI) under the following conditions: humidity, 100%; temperature, 4°C; and blot time, 2.5 s. The vitreous ice sample grids were maintained at a temperature of approximately −177°C within the electron microscope using a side‐entry 626 holder (Gatan). For immuno‐EM, the isolated GARS1‐EVs were incubated with the anti‐GARS1 antibody for 6 h, followed by incubation with the secondary antibody conjugated to 6 nm gold vesicles (Jackson Immuno Research Europe). The final mixture was incubated on ice overnight and negatively stained. Negative staining EM and cryo‐EM grids were examined using a Tecnai G2 Spirit Twin transmission electron microscope (FEI) operated at 120 kV. The images were captured using a 4K × 4K Ultrascan 895 CCD (Gatan) at a nominal magnification of 40,000× and under‐focus values ranging from 2 to 5 µm.
2.7. Electron tomography
The immuno‐gold‐labelled GARS1‐EVs localized proximal to the cell membrane were subjected to electron tomography. The images of the sample tilted from −60° to +60° were captured with 2° increments at 50,000× magnification using an eight‐megapixel CCD camera (JEOL). The data were automatically acquired using the recorder module in the TEMography suite (System in Frontier Inc.). Image alignment, reconstruction, 3D volume rendering, and visualization were performed using Composer and Visualizer modules in the TEMography and the IMOD software package (Kremer, Mastronarde, & Mcintosh, 1996).
2.8. Cryo‐fixation and immuno‐gold labelling EM
The cancer cells and macrophages were treated with GARS1‐EVs and cryo‐fixed through rapid contact on a copper block cooled at liquid nitrogen temperature (−196°C) using a multi‐purpose quick freezing system (VFZ‐101). Freeze substitution was performed using anhydrous acetone containing 1% osmium tetroxide and 0.5% glutaraldehyde at −80°C for 72 h. Next, the cell pellet was slowly warmed to room temperature (approximately 20–22°C) with stepwise temperature elevation for 7 h. The cells were washed with anhydrous acetone and embedded in Spurr's resin. The resin was polymerized at 70°C for 8 h. Ultrathin sections (80 nm thickness) were prepared using an Ultracut‐S microtome (Leica). The sections were mounted on the copper grids and stained with uranyl acetate and lead citrate. For the immuno‐gold labelling of GARS1, the frozen cells were freeze‐dried with acetone containing 0.2% uranyl acetate and 0.25% glutaraldehyde at −80°C for 72 h and slowly warmed to 4°C. The cells were washed with anhydrous acetone, infiltrated with Lowicryl HM20 (EMS), and polymerized at 4°C under UV light for 24 h. The ultrathin sections of HM20‐embedded samples were mounted on the Formvar‐coated nickel grids. The sections were blocked using 5% bovine serum albumin (BSA) in PBS containing Tween‐20 (PBS‐T) for 30 min, followed by incubation with anti‐GARS1 antibody solution (1:20) in PBS‐T containing 1% BSA for 1 h at room temperature. Next, the sections were washed thrice with a 1% BSA in PBS‐T and incubated with goat anti‐rabbit IgG antibody conjugated to 6 nm colloidal gold (JIRE) diluted 1:30 in PBS‐T containing 1% BSA for 1 h. Immuno‐gold‐labelled sections were washed 10 times with distilled water. The prepared samples were examined using a JEM‐1400 PLUS Transmission Electron Microscope (JEOL) operated at 120 kV.
2.9. Metabolic labelling palmitoylation assay
The RAW 264.7 cells were pre‐incubated with glucose‐supplemented DMEM for 1 h and labelled with 0.1 mCi/ml [3H] palmitate (PerkinElmer) for 2 h. To examine the GARS1 modifications, the cells were cultured in glucose‐depleted medium containing [3H] palmitate for 2 h. The cells were lysed using radioimmunoprecipitation assay (RIPA) buffer containing 10 mM Tris‐Cl (pH 8.0), 1 mM EDTA, 1% Triton X‐100, 0.1% sodium deoxycholate, 0.1% SDS, and 140 mM NaCl. The lysates were subjected to immunoprecipitation with the anti‐GARS1 antibody. Radiolabel palmitoylated GARS1 was detected using scintillation counting.
2.10. Mass spectrometry
The GARS1‐EVs isolated from the glucose‐supplemented and ‐depleted RAW 264.7 cell culture media were subjected to immunoprecipitation with the anti‐GARS1 antibody. The precipitated EVs were lysed using RIPA buffer after three cycles of freeze/thaw and sonication (10 s). The eluted proteins from the EVs were subjected to SDS‐PAGE and in‐gel trypsin digestion. The tryptic fragments were resolved using reversed‐phase chromatography after each run using an EASY nano‐LC II autosampler (Thermo Scientific) with a reversed‐phase peptide trap EASY‐Column (100 µm inner diameter, 2 cm length) and a reversed‐phase analytical EASY‐Column (75 µm inner diameter, 10 cm length, 3 µm particle sizes, Thermo Scientific). Electrospray ionization was performed using a 30 µm nano‐bore stainless steel online emitter (Thermo Scientific) with the following settings: voltage, 2.6 V; flow rate, 300 nl/min. The chromatography system was coupled online with an LTQ Velos Orbitrap mass spectrometer (Thermo Scientific) equipped with an electron transfer dissociation source. Mass spectrometry (MS) intensity‐based label‐free quantitation was performed using post‐experiment monoisotopic mass refinement (PE‐MMR) as described previously (Shin et al., 2008). In the PE‐MMR analysis, the mass spectral features of a peptide identified using liquid chromatography‐tandem mass spectrometry (LC‐MS/MS) were grouped into a unique mass class (UMC). Peptide abundance for each UMC was calculated as the abundance summation of all UMC mass spectral components. The DTA file was linked to the UMC by matching the UMC mass with the precursor mass in the DTA file to identify the peptide ID for the UMC. When peptide ID was identified in the linked DTA file with a false positive rate of 1% after MS‐GF+ search and target‐decoy analysis, the peptide ID was assigned to the UMC. The UMCs across the replicates were aligned using peptide IDs and normalized elution times. Among the aligned peptides, proteins with more than two non‐redundant peptides and maximum intensity of more than 105 were selected as reliable proteins. Among them, 90 proteins, which had at least one non‐redundant peptide unique to the protein, were identified as the proteins enriched in GARS1‐EVs isolated from the glucose‐depleted culture medium.
2.11. Correlative microscopy
The RAW 264.7 cells were plated on the gold R2/2 Quantifoil finder EM grids (Quantifoil Micro Tools GmbH), which were pre‐coated with 100 µg/ml collagen IV (BD Biosciences) and disinfected under UV light for 2 h. The cells were cultured on the grids overnight and then cultured in the glucose‐depleted medium for 2 h. Next, the cells incubated with the anti‐GARS1 antibody were imaged using a confocal fluorescence microscope. Further, 4 µl of PBS was applied to the cell culture on the gold EM grids for further preparative steps. Rapid verification and cryo‐EM observations were conducted as described above. The fluorescence‐labelled regions were manually identified under EM projection images captured under a series of magnifications (140× to 21,000×) to correlate with confocal fluorescence microscopy images. The low dose (approximately 20 e−/Å2) projection images of the identified region were recorded at under‐focus values ranging from 5 to 8 µm.
2.12. Cell viability assay
The H460 and RAW 264.7 cells (5×103) seeded in 96‐well plates were cultured for 24 h. The cells were washed twice with serum‐free medium and treated with GARS1‐EVs or purified GARS1 in serum‐free medium for 24 h. The cells were then incubated with 0.5 mg/ml of 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyl tetrazolium bromide (MTT; USB) for 4 h. The culture medium was removed, and the MTT formazan crystals were dissolved in 100 µl of dimethyl sulfoxide (DMSO) (Sigma). The absorbance of the mixture was measured at 570 nm using a microplate reader (TECAN). To neutralize the effect of GARS1, the cells were incubated with the anti‐GARS1 antibody for 30 min before treatment with GARS1‐EVs.
2.13. Statistical analysis
All statistical analyses were performed using unpaired two‐tailed Student's t‐test with SPSS statistics software (Version 21.0, IBM Corp). The data are presented as mean ± standard deviation. The differences were considered significant at P < 0.05.
3. RESULTS
3.1. Characterization of GARS1‐EVs derived from macrophages
Previously, we had demonstrated that the co‐cultivation of macrophages with cancer cells promotes the secretion of GARS1 from the macrophages under stress conditions, such as serum or glucose starvation (Park et al., 2012). The major energy source of tumour cells is glucose. Hence, the depletion of glucose is one of the biochemical hallmarks in the cancer microenvironment (Hirayama et al., 2009; Shaw, 2006). The effect of glucose starvation on the secretion of GARS1 from the RAW 264.7 cells was examined. The secretion of GARS1 increased upon glucose starvation without a concomitant increase in its expression (Figure 1a). The mechanism underlying the secretion of GARS1 was examined using the inhibitors of various steps of protein trafficking. The RAW 264.7 cells were treated with brefeldin A (BFA), Y27632, probenecid, and Sodium Azide (NaN3), which inhibit endoplasmic reticulum (ER) and Golgi‐mediated protein secretion (Fujiwara, Oda, Yokota, Takatsuki, & Ikehara, 1988), membrane blebbing (Coleman et al., 2001), ABC transporter (Flieger et al., 2003), and mitochondrial respiratory chain (Gudz, Tserng, & Hoppel, 1997), respectively. Treatment with these inhibitors did not affect the secretion of GARS1 (Figures S1a‐d). In contrast, the RAW 264.7 cells pretreated with methyl‐beta‐cyclodextrin (MβCD), a cholesterol synthesis inhibitor (Rodal et al., 1999), exhibited significantly decreased secretion of GARS1 (Figure 1b). Lipid rafts are rich in cholesterol, which is essential for the formation of EV membrane. The inhibition of GARS1 secretion by MβCD indicated that a membrane‐based structure mediates GARS1 secretion (Brzozowski et al., 2018). The glucose‐supplemented and glucose‐depleted RAW 264.7 cell culture media were subjected to differential centrifugation. GARS1 was detected in the pellets obtained from the culture medium centrifuged at 100,000 × g, which is the speed at which the membrane‐based vesicles sediment (Bobrie, Colombo, Krumeich, Raposo, & Théry, 2012) (Figure 1c). The centrifuged pellet fraction did not contain gp96, which is reported to be secreted through the ER‐Golgi pathway (Peters & Raghavan, 2011) (Figure S1e). Light scattering spectrophotometry and analytical gradient ultracentrifugation revealed that the GARS1‐enriched fraction contained EVs with a hydrodynamic diameter of 20–58 nm (mean diameter of 36.9 nm) (Figure 1d) and a buoyant density of 1.13–1.17 g/ml (Figure 1e). Also, GARS1 was detected in the EVs derived from bone marrow‐derived macrophages (BMDMs) and J774A.1 macrophage cells (Figure 1f and Figure S1f).
FIGURE 1.
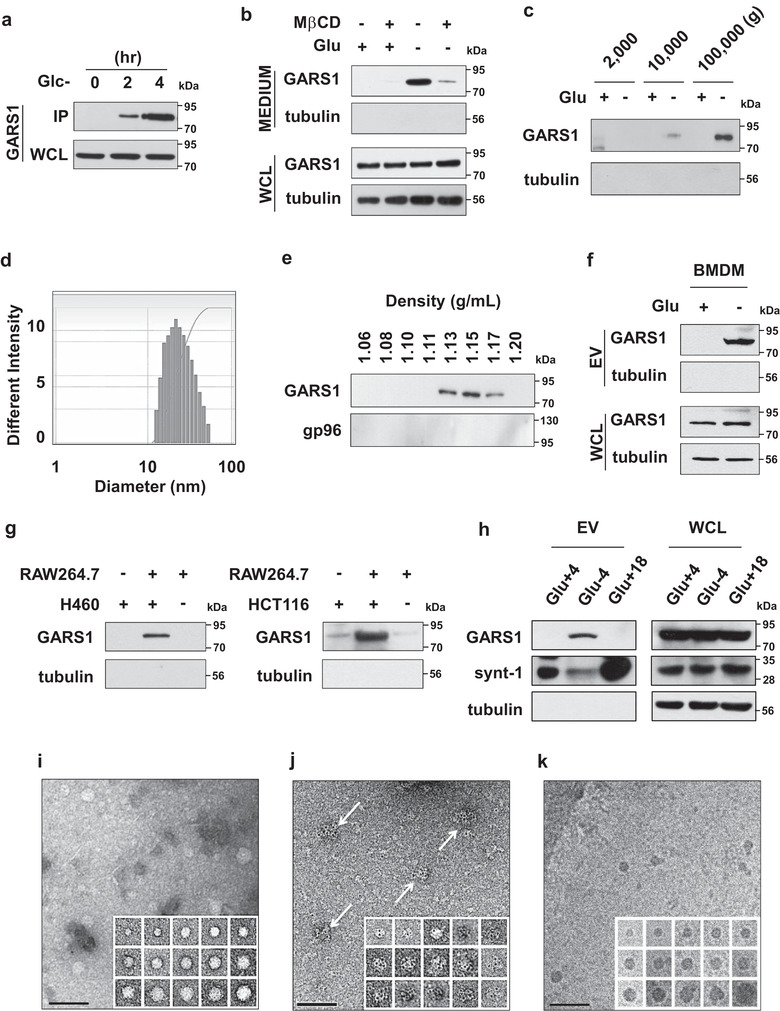
Characteristics of macrophage‐derived GARS1‐containing extracellular vesicles (GARS1‐EVs). (a) RAW 264.7 cells were cultured in the glucose‐depleted medium. The proteins secreted into the culture medium were subjected to immunoprecipitation with the anti‐GARS1 antibody (IP: immunoprecipitation, resuspended with 50 µl of sample buffer). The whole‐cell lysates (WCL) were prepared using 300 µl of lysis buffer. Immunoblotting analysis revealed that GARS1 was detected in the IP sample and 30 µg of WCL at 2 and 4 h. (b) The effect of methyl‐beta‐cyclodextrin (MβCD, 0.1 µM) on the secretion of GARS1 in glucose‐supplemented and glucose‐depleted culture media was evaluated based on the immunoblotting analysis of the secreted proteins precipitated with TCA (trichloroacetic acid). (c) The glucose‐depleted RAW 264.7 cell culture medium was centrifuged at 2000 × g for 10 min to remove the cell debris. The supernatants were centrifuged at 10,000 × g for 30 min. The pellet was removed and the supernatant was centrifuged at 100,000 × g for 90 min. The pellets obtained at 2000 × g, 10,000 × g and 100,000 × g were subjected to immunoblotting with the anti‐GARS1 antibody. (d) The size range of the extracellular vesicles (EVs) was determined using dynamic light scattering spectrophotometry. (e) EVs were loaded on a 10–60% continuous sucrose gradient and subjected to analytical centrifugation to determine the buoyant density. Eight fractions in the gradient were collected and the presence of GARS1 in each fraction was determined using immunoblotting. (f) GARS1 levels in the EVs secreted from the BMDMs (bone marrow derived macrophages) cultured in the presence or absence of glucose were determined using immunoblotting with the anti‐GARS1 antibody. (g) The cancer cells (H460 or HCT116) were loaded in the upper chamber, while the RAW 264.7 cells were loaded in the lower chamber of the transwell plate. The upper and lower chambers were separated by a membrane with a pore size of 0.4 µm. After 12 h of incubation in the serum‐free glucose‐supplemented media, the levels of GARS1 in the EVs obtained from the culture medium of the lower chamber were determined as described previously. (h) The EVs were isolated from the glucose‐supplemented (Glc+4), glucose‐depleted (Glc‐4) after 4 h, and glucose‐supplemented RAW 264.7 cell culture media (Glc+18) after h of incubation. GARS1 in the EVs and WCL were determined using immunoblotting with the anti‐GARS1 antibody. (i) Morphology of GARS1‐EVs was determined using negative staining electron microscopy. (j) Localization of GARS1 was determined using negative staining electron microscopy with the immuno‐gold labelled anti‐GARS1 antibody. (k) GARS1‐EVs were also analyzed using cryo‐electron microscopy. The inset montages display images of individual EVs showing the distribution of different sizes. Scale bar represents 100 nm (i‐k)
The contribution of cancer cells to the secretion of EVs was evaluated. The RAW 264.7 cells were co‐cultured with two cancer cell lines (H460 and HCT116) and two non‐cancerous cell lines (WS‐1 (fibroblasts) and Vero (epithelial cells)). To block the physical interaction, the macrophages and cancer/non‐cancer cells were seeded into the lower and upper chambers of a transwell plate (separated by a membrane with a pore size of 0.4 µm), respectively. GARS1 was detected in the EVs obtained from the macrophage and cancer cell co‐culture media, but not in those obtained from the macrophage and non‐cancerous cell co‐culture media (Figure 1g and Figure S1g). This suggested that the secretion of GARS1‐EVs was triggered by cancer‐derived secreted factors in the cancer microenvironment.
The conditions for GARS1 secretion were compared with those for syntenin‐1 secretion, a known exosome marker (Théry et al., 2001). GARS1 secretion was mainly observed when the RAW 264.7 cells were incubated for 4 h in glucose‐depleted medium, whereas syntenin‐1 was secreted at a high level when the cells were incubated in glucose‐supplemented medium for 18 h (Figure 1h). This suggests that the conditions for the secretion of GARS1 were distinct from those for the secretion of syntenin‐1‐containing exosomes.
The size and morphology of the GARS1‐EVs were examined using negative staining EM. The GARS1‐EVs were globular‐shaped with a diameter of 20–50 nm (Figure 1i). The presence of GARS1 in these EVs was confirmed by staining the EVs with gold particle‐conjugated anti‐GARS1 antibody. The gold spots enriched on the surface of the EVs were GARS1 specific (Figure 1j, arrows, and Figure S2a) . Cryo‐EM analysis also revealed a similar size and morphology of the EVs (Figure 1k). The exact membrane structure of the isolated GARS1‐EVs is unknown. However, the morphology and size of GARS1‐EVs indicated that they were typical lipoprotein EVs (Zhang et al., 2011) and not apoptotic bodies, which have a larger size (1–5 µm) and a distinct morphology (Van Der Pol, Böing, Harrison, Sturk, & Nieuwland, 2012).
The effect of glucose depletion on the total number of GARS1‐EVs was examined. The RAW 264.7 cells were stained with lipophilic fluorescence tracer, DiI (Maier, Oberle, & Hoekstra, 2002). The dye was removed and the cells were incubated in the presence or absence of glucose. The secreted EVs were isolated and the total fluorescence intensity was measured. Glucose starvation increased the total fluorescence intensity and protein content of the EVs (Figure S2b and c, respectively), which suggested that glucose starvation increases the number of GARS1‐EVs. These EVs were confirmed to contain GARS1 (Figure S2d). The cells stained with DiI and 4′,6‐diamidino‐2‐phenylindole (DAPI) did not exhibit any marked difference in the number of cells upon incubation in the presence or absence of glucose (Figure S2e), which indicated that the difference in the fluorescence intensity of the EVs was not due to the cell density. These findings indicated that GARS1 is secreted from macrophages and that it is anchored to the surface of the EVs.
3.2. Lipid modification of GARS1 is required for the attachment to EVs
Next, the isolated EVs were incubated with trypsin. GARS1 was sensitive to trypsin digestion, which suggested that it was exposed on the vesicle surface (Figure 2a). To label the proteins on the surface of EVs, the isolated EVs were treated with membrane‐impermeable EZ‐link Sulfo‐NHS‐LC‐Biotin. The biotinylated proteins were released using detergents and precipitated using streptavidin‐agarose beads, and the biotinylated proteins were subjected to immunoblotting with the anti‐GARS1 antibody. The biotinylated GARS1 levels in the EVs isolated from the glucose‐depleted culture medium were higher than that in the EVs isolated from the glucose‐supplemented culture medium. Additionally, GARS1 was confirmed to be localized on the surface of the EVs (Figure 2b).
FIGURE 2.
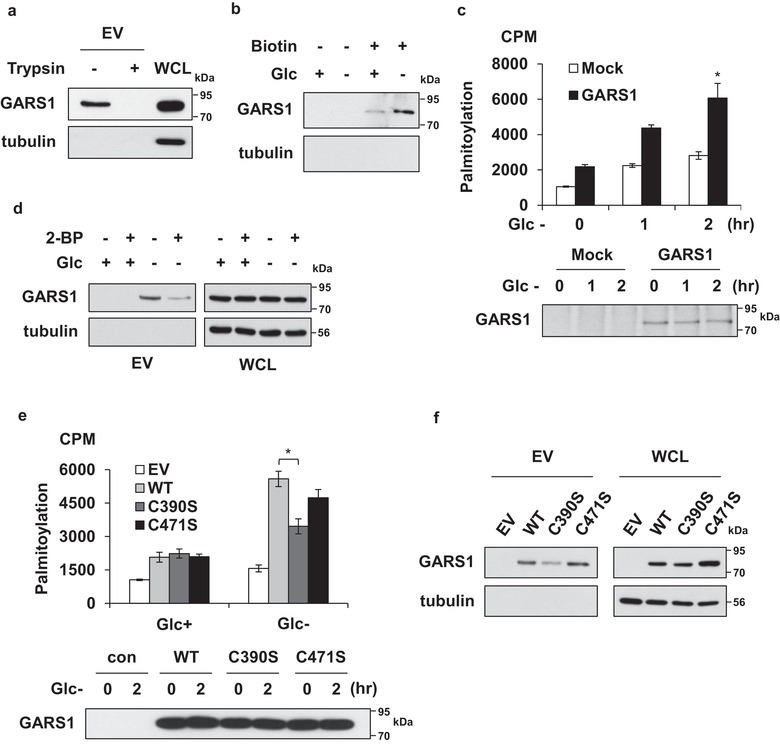
Palmitoylation of GARS1 is required for its secretion through extracellular vesicles (EVs). (a) GARS1‐containing EVs (GARS1‐EVs) were incubated with trypsin to determine the surface exposure of GARS1 on the EVs. (b) GARS1‐EVs from the RAW 264.7 cells cultured in the presence or absence of glucose were incubated with EZ‐link Sulfo‐NHS‐LC‐Biotin and precipitated using streptavidin‐agarose beads. The GARS1 levels in the precipitated EVs were examined using immunoblotting with the anti‐GARS1 antibody. (c) The palmitoylation of GARS1 was analysed using the modified metabolic labelling assay. After [3H]‐palmitic acid labeling, the RAW 264.7 cells were cultured in the glucose‐depleted medium. GARS1 was immunoprecipitated using the anti‐GARS1 antibody. GARS1‐incorporated [3H]‐palmitic acid was detected using a scintillation counter (upper panel). The levels of the immunoprecipitated GARS1 were determined using immunoblotting with the anti‐GARS1 antibody (lower panel). (d) To determine the effect of 2‐bromopalmitate (2‐BP, 2 µM) on the secretion of GARS1, the RAW 264.7 cells were cultured in the glucose‐supplemented and glucose‐depleted medium in the presence or absence of 2‐BP. The GARS1‐EVs were isolated and the presence of GARS1 was determined using immunoblotting. (e) Palmitoylation of the strep‐tagged wild type and mutant (C390S and C471S) GARS1 was analysed using the modified metabolic labelling method (upper panel). The cellular levels of wild type and mutant GARS1 were determined using immunoblotting (lower panel). (f) The effect of a palmitoylation mutation on GARS1 secretion from the RAW 264.7 cells was determined. The levels of wild type and mutant GARS1 in the WCL and isolated EVs were determined using immunoblotting. Data are presented as mean ± standard deviation. *P < 0.01
Next, the attachment of GARS1 to the surface of the secreted vesicles was examined. The amino acid sequence analysis using the TMHMM server (http://www.cbs.dtu.dk/services/TMHMM, data not shown) revealed that GARS1 does not contain potential transmembrane domains. However, the amino acid sequence analysis using the CSS‐Palm 3.0 software suggested the presence of potential palmitoylation sites at the residues C390 and C471. Palmitoylation is a reversible modification that regulates membrane trafficking of proteins (Milligan, Parenti, & Magee, 1995). The alignment of amino acid sequences of GARS1 from nine species demonstrated that C390 (and not C471) were highly conserved, which indicated that C390 was functionally important (Figure S3) (Shen, Wu, Yang, & Gould, 2011). The role of palmitoylation in the secretion of GARS1 was examined using a modified metabolic labelling method. The RAW 264.7 cells were incubated with [3H]‐palmitic acid. GARS1 was immunoprecipitated using the anti‐GARS1 antibody and [3H]‐palmitic acid was detected using a scintillation counter (Figure 2c, upper panel). The immunoprecipitated GARS1 levels were determined using immunoblotting with the anti‐GARS1 antibody (Figure 2c), lower panel). The levels of endogenous palmitoylated GARS1 were upregulated upon glucose starvation. The role of palmitoylation in GARS1 secretion was examined by treating the cells with 2‐bromopalmitate (2‐BP), a palmitoylation inhibitor (Webb, Hermida‐Matsumoto, & Resh, 2000). Treatment with 2‐BP decreased the secretion of GARS1 (Figure 2d). To confirm these results, two mutants of GARS1 were generated in which the cysteines at the 390 or 471 positions were substituted with serine (C390S and C471S, respectively). The effect of these substitutions on palmitoylation was examined. The RAW 264.7 cells expressing the strep‐tagged GARS1 WT (wild type), and C390S and C471S mutants were incubated in the glucose‐depleted medium. The results of the metabolic labelling assay revealed that palmitoylation and secretion of GARS1 in the C390S mutant were lower than those in WT GARS1 and C471S mutant (Figures 2e and f, respectively). This suggested that palmitoylation at C390 is involved in the secretion of GARS1.
Naked GARS1 secreted in the extracellular milieu can also bind to the EVs. Hence, the recruitment of GARS1 to the secretory vesicles in the cytosol was examined using correlative light microscopy and cryo‐EM (Jun et al., 2011). Intracellular GARS1 in the RAW 264.7 cells was examined using immunofluorescence microscopy with the Alexa Fluor 488‐conjugated anti‐GARS1 antibody (indicated by a white arrow in the upper middle panel of Figure S4a). The fluorescent region was subjected to cryo‐EM. The series of magnified images revealed that the fluorescence‐labelled region (white boxes in each panel) contained globular‐shaped vesicles (black arrowheads in the right panel). The morphology and size of these vesicles were consistent with those of vesicles previously observed (Figure 1i–k and Figure S2a). This suggested that GARS1 is recruited and anchored to the surface of the secretory vesicles in the cytosol before secretion. The same method was used to monitor the exosomes labelled with Alexa 594‐conjugated anti‐MHCII antibody (red fluorescence) (Bhatnagar, Shinagawa, Castellino, & Schorey, 2007). The red fluorescence‐labelled exosomes did not co‐localize with the green fluorescence‐labelled GARS1 foci (Figure S4b). The glucose starvation‐induced GARS1 foci did not co‐localize with lysosomes and ER (Figures S4c and d), which indicated that GARS1 transits through a distinct secretory pathway. The role of palmitoylation in the formation of intracellular GARS1 foci during glucose starvation was examined. Treatment with 2‐BP decreased the formation of glucose starvation‐induced GARS1 foci (Figure S4e), which suggested the importance of lipid modification in the intracellular recruitment of GARS1 to EVs.
3.3. Identification of proteins in the GARS1‐EVs
The protein composition of the EVs was examined. The vesicles derived from the macrophages incubated in the presence or absence of glucose were isolated by ultracentrifugation at 100,000 × g. The EVs were purified by subjecting the culture medium to immunoprecipitation with the anti‐GARS1 antibody. The protein components of these EVs were resolved using SDS‐PAGE. The protein bands in the gel were treated with trypsin. The resulting peptides were subjected to LC/MS‐MS analysis using LTQ‐Orbitrap Velos. The MS‐GF+ search identified 184 and 272 proteins (FDR < 0.01) in the EVs obtained from glucose‐supplemented and glucose‐depleted media, respectively. The levels of 73 proteins in the EVs obtained from the glucose‐depleted culture medium were upregulated more than six‐fold when compared with those in the EVs obtained from the glucose‐supplemented culture medium. Additionally, 91 proteins were detected only in the EVs obtained from the glucose‐depleted culture medium (Figure 3a and Table S2). Of these 164 proteins enriched in the glucose‐depleted culture medium, the ExoCarta analysis revealed that 11 proteins overlapped with the 20 representative exosomal marker proteins (Mathivanan & Simpson, 2009) (Figure 3b). The endosomal sorting complexes required for transport proteins (Vps26b and Vps35) were detected in the GARS1‐EVs. However, tetraspanins, such as CD9, CD63, and CD81, which are exosomal marker proteins (Bhatnagar et al., 2007), Alix (PDCD6IP), and Syndecan binding protein (SDCBP) were not detected in the GARS1‐EVs. The comparative analysis of the protein content of GARS1‐EVs and exosomes isolated from different cell types listed in ExoCarta revealed a low degree of overlap (less than 0.12 similarity score) (Figure 3c). This indicated that GARS1‐EVs contain a distinct set of proteins. The proteins associated with apoptotic bodies, such as calreticulin or CD91 (LPR1) were not detected in the GARS1‐EVs, which further confirmed the unique protein composition of GARS1‐EVs (Takemura et al., 2007).
FIGURE 3.
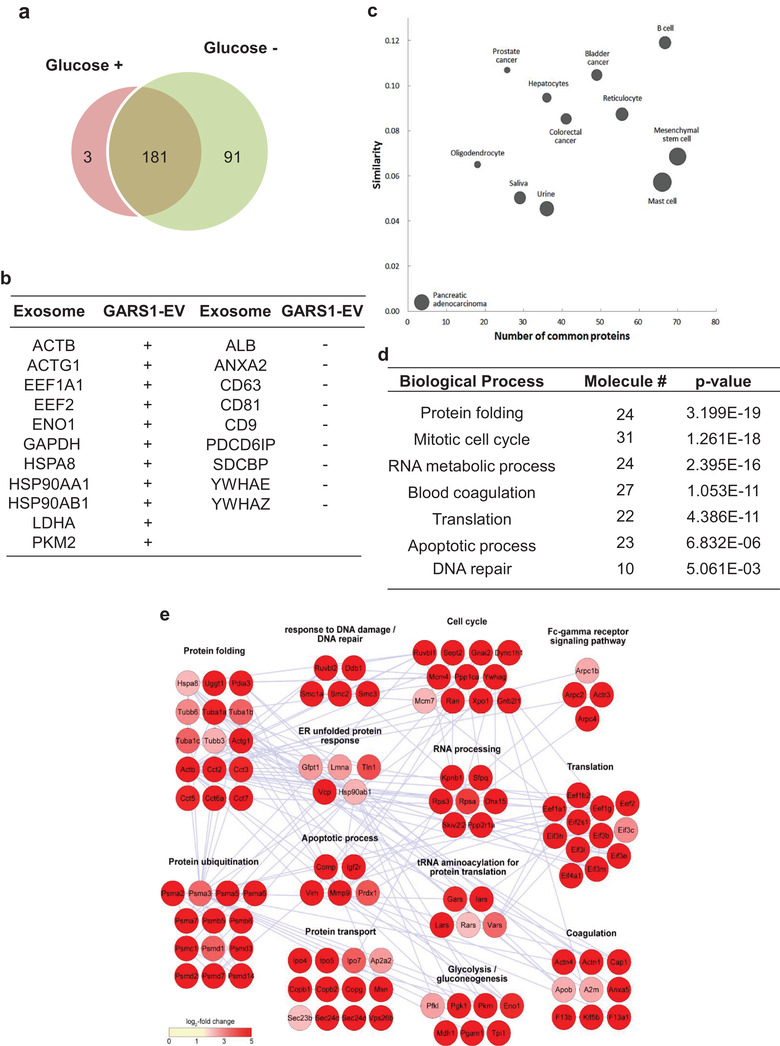
Identification of proteins in GARS1‐EVs. (a) The proteins identified in the GARS1‐EVs isolated from the glucose‐supplemented and glucose‐depleted culture media are displayed in red (3) and green (91) circles, respectively. Among the identified proteins, 181 proteins were commonly identified in both glucose‐supplemented and glucose‐depleted culture media. (b) The proteins enriched in the GARS1‐EVs isolated from the glucose‐depleted culture medium were compared with the top 20 proteins that are frequently found in exosomes (http://exocarta.org/exosome_markers). + and − represent the proteins detected in both exosomes and GARS1‐EVs and only in exosomes, respectively. (c) The proteins enriched in the GARS1‐EVs were further compared with those enriched in the exosomes derived from different cell types shown in Exocarta. The similarity score for each compared pair was calculated using the Jaccard similarity coefficient. The node size represents the average number of proteins identified in each pair. (d) The proteins enriched in the GARS1‐EVs isolated from the glucose‐depleted culture medium were classified according to gene ontology (GO) biological processes. (e) The proteins (164) enriched in the GARS1‐EVs from the glucose‐depleted culture medium were grouped according to their functional processes (P < 0.05). The network model was constructed for the selected proteins
The proteins enriched in the GARS1‐EVs were involved in diverse cellular processes, such as protein folding, cell cycle, RNA metabolism, blood coagulation, apoptosis, and DNA repair (Figure 3d and Table S3). Connectivity mapping of the protein functions using Cytoscape revealed the clusters of protein metabolism (protein folding, ubiquitination, unfolded protein response, transport), cell regulation (cell cycle and apoptosis), and translation (Figure 3e). Interestingly, amino acyl‐tRNA synthetases, such as leucyl‐, isoleucyl‐, arginyl‐ and valyl‐tRNA synthetases were also detected along with GARS1. Although the roles of these factors require experimental validation, they are expected to contribute to the processes of intracellular biogenesis, trafficking, secretion and extracellular functions.
3.4. Effect of GARS1‐EVs on cancer cells
Purified GARS1 was previously reported to promote cancer cell death (Park et al., 2012). In this study, the growth‐inhibiting activity of GARS1‐EVs against cancer cells was evaluated. The uptake of GARS1‐EVs into the H460 cells was examined using EM. Several GARS1‐EVs attached to the membrane surface were detected after 10 min of treatment (Figure 4a). Additionally, EVs at different stages of endocytosis were observed (Figure 4b and c). Immuno‐gold labelling of GARS1, followed by cryo‐fixation EM and 3D electron tomography demonstrated that the endocytosed GARS1‐EVs contained GARS1 (Figure 4d and e, Figure S5a, and Supplementary Movie 1).
FIGURE 4.
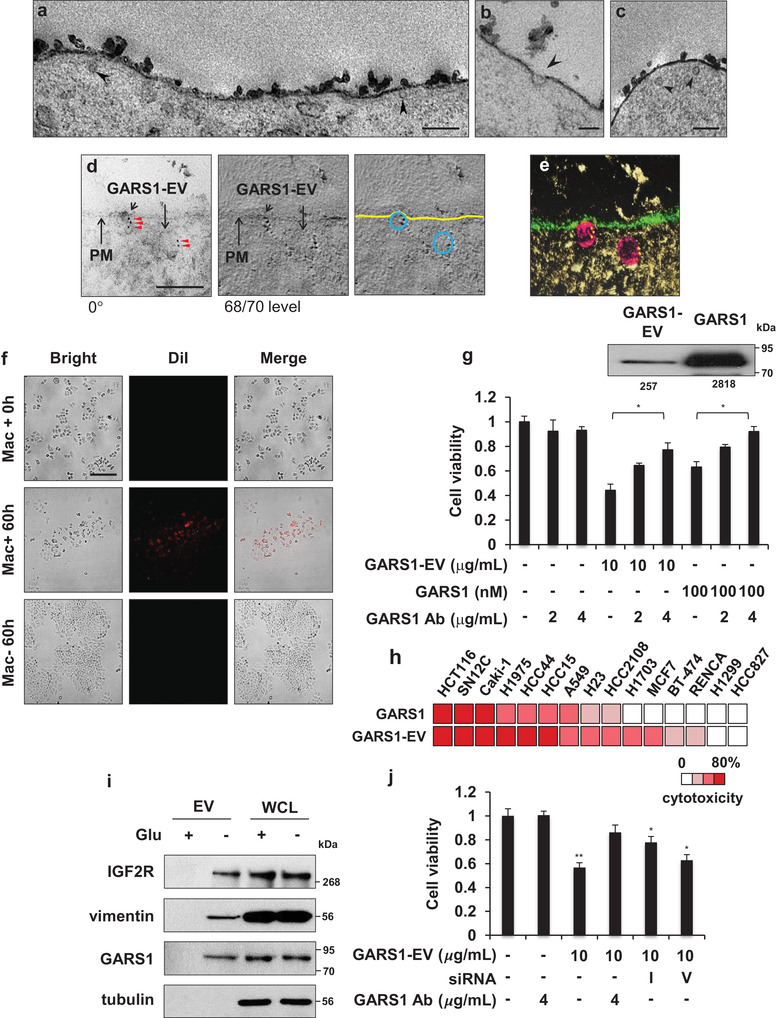
Intracellular uptake and growth‐inhibiting effect of GARS1‐EVs on cancer cells. (a‐c) Electron microscopic images of H460 cell treated with GARS1‐EVs and chemically fixed after 10 min. The images show the plasma membrane with intracellular and extracellular regions and the arrowheads indicate GARS1‐EVs. (d) Images of immuno‐gold labelled GARS1‐EVs endocytosed by the cancer cells. GARS1‐EV‐treated H460 cells were cryo‐fixed and immunostained with the anti‐GARS1 antibody. For tomography, tilted images of the samples were recorded over an angular range from −60° to +60° with 2° increments. Numbers indicate the angle of the tilted sample (left). Tilted images were aligned and used to construct the tomogram. The number indicates the tomographic slice order (center). The plasma membrane (PM) and GARS1‐EVs are indicated in yellow and blue lines, respectively (right). (e) Electron tomography was reconstructed using the collected captured images and visualized using the composer and visualizer modules in TEMography and IMOD software. (f) DiI‐labeled RAW 264.7 and H460 cells were seeded in the upper and lower chambers of transwell (0.4 µm pore size), respectively. After 60 h of co‐culture, the macrophage‐derived DiI‐labeled GARS1‐EVs (red) and cancer cell confluency (bright field) were monitored in the lower chamber using fluorescence microscopy. (g) Cell viability was determined using the 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyl tetrazolium bromide (MTT) assay. The H460 cells were incubated with GARS1‐EVs (10 µg/ml) for 24 h. The purified his‐tagged GARS1 protein (100 nM) was used as a control. To neutralize the GARS1 effect, GARS1‐containing GARS1‐EVs and purified GARS1 were pre‐incubated with the anti‐GARS1 antibody for 30 min before addition to the H460 cell culture medium. Immunoblotting analysis was performed to evaluate the levels of GARS1 in the GARS1‐EVs and compared to naked GARS1 used for experiments. The protein band intensity was analysed using Image J (inset). (h) The comparative analysis of the apoptotic effects of GARS1‐EVs (10 µg/ml) and naked GARS1 (100 nM) on different cancer cells. (i) The presence of IGF2R and vimentin in the GARS1‐EVs was validated using immunoblotting. (j) RAW 246.7 cells were treated with siRNA to suppress the expression levels of IGF2R (I) and vimentin (V). The GARS1‐EVs were then isolated to examine the effect on cancer cell viability. The scale bars indicate 200 nm (a, c), 100 nm (b, d) and 50 µm (f). Data are presented as the mean ± standard deviation. *P < 0.01, **P < 0.001
The growth‐inhibiting activities of GARS1‐EVs against the H460 cancer cells were compared with those of naked GARS1. Treatment with GARS1‐EVs dose‐dependently increased apoptosis in the H460 cells but not in the RAW 264.7 cells (Figure S5b). These results were also confirmed using the terminal TUNEL assay (Figure S5c). The passage of EVs from macrophages to cancer cells was monitored using a transwell chamber. The macrophages whose membranous lipids were labelled with DiI (red fluorescence dye) were placed in the lower chamber, while the cancer cells were placed in the upper chamber. The presence of red fluorescence‐labeled structures in the cancer cells and their ability to promote cancer cell death were evaluated. Red fluorescence‐labelled structures derived from the macrophages were observed in the cancer cells. Additionally, the presence of apoptotic cells was observed at the sites where red fluorescence was detected (Figure 4f).
The anti‐cancer activity of GARS1 in the EVs was examined. The H460 cells were treated with isolated GARS1‐EVs, naked GARS1, and anti‐GARS1 antibody‐treated GARS1‐EVs and naked GARS1. The viability of H460 cells treated with GARS1‐EVs and naked GARS1 was 50% and 40% lower than that of the untreated H460 cells, respectively. Treatment with the anti‐GARS1 antibody‐treated GARS1‐EVs partially inhibited the cytotoxic activity of GARS1‐EVs. In contrast, treatment with anti‐GARS1 antibody‐treated naked GARS1 completely inhibited] the growth‐inhibiting activity of GARS1‐EVs against cancer cells (Figure 4g). This indicated that the GARS1‐EVs contained additional cytotoxic factors. The EVs isolated from the glucose‐supplemented culture medium, which does not contain GARS1, did not inhibit the growth of cancer cells (Figure S5d). Immunoblotting analysis was performed to determine the levels of GARS1 in the EVs and naked GARS1 used for the experiments. The levels of GARS1 in the EVs were eight‐fold lower than those of naked GARS1 (Figure 4g, inset). GARS1‐EVs exhibited potent growth‐inhibiting activity against the cancer cells, which suggested that the EVs contained supplemental cytotoxic factors in addition to GARS1.
The apoptotic activities of naked GARS1 and GARS1‐EVs were comparatively evaluated using different cancer cell lines. Of the 15 tested cancer cell lines, GARS1 and GARS1‐EVs promoted apoptosis in 9 and 13 cancer cell lines, respectively (Figure 4h). This suggested that GARS1‐EVs exhibited a broad‐spectrum growth‐inhibiting activity against the cancer cells. Some components of the GARS1‐EVs were selectively depleted. Among the factors identified in the GARS1‐EVs (Figure 3e), this study focused on the factors involved in apoptotic processes, such as insulin‐like growth factor II receptor (IGF2R) and vimentin (O'Gorman, Costello, Weiss, Firth, & Scott, 1999; Souza et al., 1999; Yang et al., 2005). These factors were detected in the isolated GARS1‐EVs using immunoblotting with specific antibodies (Figure 4i). Each factor was suppressed in the RAW 264.7 cells using specific siRNA and the GARS1‐EVs were isolated as described above. The growth‐inhibiting activities of IGF2R‐depleted and vimentin‐depleted GARS1‐EVs against the cancer cells were examined. Compared with the EVs isolated from the WT macrophages, the EVs isolated from the IGF2R‐suppressed or vimentin‐suppressed macrophages exhibited lower growth‐inhibiting activity against the cancer cells. The IGF2R‐depleted and vimentin‐depleted GARS1‐EVs exhibited varied growth‐inhibitory activities against cancer cells. This suggested that these factors can also partially contribute to the anti‐cancer activity of GARS1‐EVs along with GARS1 (Figure 4j and Figure S5e).
4. DISCUSSION
In this study, the EVs derived from macrophages cultured in the glucose‐depleted medium were demonstrated to harbour GARS1. Additionally, the GARS1‐EVs contained various cellular factors that were involved in immunological defence response against tumorigenesis. In addition to glucose starvation, other stress conditions may induce GARS1‐EV secretion. Macrophages exposed to environmental stresses, such as nutrient deprivation, hypoxia and the signalling factors released from the neighbouring cancer cells recruit various intracellular factors and package them into secretory vesicles to mediate the primary response against cancer cells until the immune surveillance system is activated. In the LPS‐stimulated monocytes, HMGB1 is reported to be phosphorylated by protein kinase C, which is required for the calcium‐dependent secretory pathway (Oh et al., 2009). HMGB1 can also be secreted through vesicle compartments (Gardella et al., 2002), which indicated that it could be released from the cells through multiple pathways. This study identified unique vesicles through which GARS1 is secreted. However, additional secretory pathways for GARS1 cannot be ruled out.
Some membrane‐based vesicles, including exosomes, ectosomes, microvesicles and secretory lysosomes, are reported to mediate the unconventional secretory pathways (Nickel & Seedorf, 2008; Raposo & Stoorvogel, 2013). Currently, the definition and classification of these secreting vesicles remain obscure. However, the biophysical properties of these vesicles are defined based on the size, density, morphology, vesicular contents and biological functions (Théry, Zitvogel, & Amigorena, 2002). The diameter of exosomes, which are enclosed by a lipid bilayer membrane, is in the range of 40–150 nm. The exosomes transport diverse intracellular molecules, including proteins, mRNAs, and microRNAs (Luga et al., 2012; Skog et al., 2008; Valadi et al., 2007). Various cell types, including cancer and immune cells, are reported to release microvesicles to mediate cell‐to‐cell communication (Wen et al., 2017; Yáñez‐Mó et al., 2015). Recently, we reported that lysyl‐tRNA synthetase 1 (KARS1), a tRNA synthetase, is secreted through typical exosomes (Kim et al., 2017). GARS1 is located on the surface of the secreted vesicles, while KARS1 is located within the lumen of the secreted exosomes. This indicates that different aminoacyl‐tRNA synthetases have characteristic signals, secretion pathways, and extracellular activities.
Exosomes secreted from the tumour and tumour‐associated macrophages are reported to promote tumour progression through the delivery of oncogenic miRNAs and oncogenes (Al‐Nedawi et al., 2008; Yang et al., 2011). The intercellular communication between cancer and immune cells plays a crucial role in tumorigenesis (De Visser, Eichten, & Coussens, 2006; Dranoff, 2004). However, the role of secreted vesicles in mediating the interactions between cancer and immune cells is unclear. The findings of this study provided useful insights into the potential roles of macrophage‐derived EVs in the tumour microenvironment. GARS1‐EVs exhibit distinct biophysical characteristics, protein content, and functions when compared with the previously reported membrane‐bound structures.
Although this study identified diverse cellular factors in the GARS1‐EVs, it is unclear if the proteins are carried in a single EV or are differentially packaged into different EVs. The proteins in these vesicles contribute to different functions, such as biogenesis, secretion, and extracellular activities. Among the identified proteins, the depletion of IGF2R and vimentin decreased the anti‐cancer activity of GARS1‐EVs, which suggested that these factors contribute to the anti‐tumorigenic activity of GARS1‐EVs along with GARS1. Previous studies have reported that the overexpression of IGF2R decreased cancer cell and tumour growth, which indicated that IGF2R is a tumour suppressor (O'gorman, Weiss, Hettiaratchi, Firth, & Scott, 2002). Vimentin, a primary constituent of the intermediate filament, was secreted from the activated macrophages. Additionally, vimentin was reported to exhibit antibacterial activity through the enhanced generation of oxidative metabolites (Mor‐Vaknin, Punturieri, Sitwala, & Markovitz, 2003). Vimentin may promote cancer cell death through a similar mechanism.
The exogenous administration of GARS1‐EVs efficiently suppressed tumour growth by promoting apoptosis in the cancer cells, which suggested that the secretion of GARS1‐EVs suppresses tumour growth. The membrane‐bound vesicles, such as exosomes and microvesicles, are a potential tool for the delivery of pharmacologically active molecules and drugs (Jang et al., 2013, Zitvogel et al., 1998). For example, the intravenous injection of exosomes successfully delivered microRNA, which targeted the EGFR of cancer cells (Ohno et al., 2013). Systemic administration of exosomes derived from mesenchymal stromal cells enhanced the recovery of stroke in animal models (Xin et al., 2013). Further studies are needed to establish the functional interaction of GARS1‐EVs with macrophages and to elucidate the physiological and immunological roles of GARS1‐EVs in the tumour microenvironment. The potential application of GARS1‐EVs for cancer immunotherapy is beyond the scope of this study.
AUTHOR CONTRIBUTIONS
Peter C. Goughnour, Min Chul Park, Hyun Suk Jung, and Sunghoon Kim designed the experiments. Peter C. Goughnour, Min Chul Park, Sang Bum Kim, Sangmi Jun, Won Suk Yang, Chihong Song, and Jae Kyung Hyun performed the experiments. Peter C. Goughnour, Min Chul Park, Sangmi Jun, Won Suk Yang, Sehyun Chae., Ji‐Hyun Lee, Byung Gyu Kim, Daehee Hwang, Hyun Suk Jung, and Sunghoon Kim analysed the data. Yong Song Gho provided the materials and technical support for the experiments. Peter C. Goughnour, Min Chul Park, Daehee Hwang, Hyun Suk Jung, and Sunghoon Kim prepared the manuscript. Peter C. Goughnour, Min Chul Park, Hyun Suk Jung, Yong Song Gho, and Sunghoon Kim reviewed the manuscript. Peter C. Goughnour, Min Chul Park, Byung Gyu Kim, Daehee Hwang, Hyun Suk Jung, Yong Song Gho, and Sunghoon Kim evaluated the results and discussed.
CONFLICT OF STATEMENT
The authors report no conflict of interest.
Supporting information
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
ACKNOWLEDGEMENTS
This work was supported by the Global Frontier Project grant (NRF‐M3A6A4‐2010‐0029785) of the National Research Foundation funded by the Ministry of Science and ICT (MSIT) of Korea. Electron Microscopy study (H.S.J.) was supported by the Global Frontier Research grant (NRF‐2015M3A6A4065729) of the National Research Foundation funded by the Ministry of Science and ICT (MSIT) of Korea.
Goughnour PC, Park MC, Kim SB, et al. Extracellular vesicles derived from macrophages display glycyl‐tRNA synthetase 1 and exhibits anti‐cancer activity. J Extracell Vesicles. 2020;10:e12029 10.1002/jev2.12029
Present address: Min Chul Park, CureBio Co., Ltd., Suwon 16229, Korea.
Peter C. Goughnour and Min Chul Park contributed equally to this study.
REFERENCES
- Al‐Nedawi, K. , Meehan, B. , Micallef, J. , Lhotak, V. , May, L. , Guha, A. , & Rak, J. (2008). Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nature Cell Biology, 10(5), 619–624. [DOI] [PubMed] [Google Scholar]
- Asea, A. , Kraeft, S.‐K. , Kurt‐Jones, E. A. , Stevenson, M. A. , Chen, L. B.o , Finberg, R. W. , … Calderwood, S. K. (2000). HSP70 stimulates cytokine production through a CD14‐dependant pathway, demonstrating its dual role as a chaperone and cytokine. Nature Medicine, 6(4), 435–442. [DOI] [PubMed] [Google Scholar]
- Basu, S. , Binder, R. J. , Ramalingam, T. , & Srivastava, P. K. (2001). CD91 is a common receptor for heat shock proteins gp96, hsp90, hsp70, and calreticulin. Immunity, 14(3), 303–313. [DOI] [PubMed] [Google Scholar]
- Bhatnagar, S. , Shinagawa, K. , Castellino, F. J. , & Schorey, J. S. (2007). Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo. Blood, 110(9), 3234–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrie, A. , Colombo, M. , Krumeich, S. , Raposo, G. , & Théry, C. (2012). Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. Journal of Extracellular Vesicles, 1(1), 18397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzozowski, J. S. , Jankowski, H. , Bond, D. R. , Mccague, S. B. , Munro, B. R. , Predebon, M. J. , … Weidenhofer, J. (2018). Lipidomic profiling of extracellular vesicles derived from prostate and prostate cancer cell lines. Lipids in Health Disease, 17(1), 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, M. L. , Sahai, E. A. , Yeo, M. , Bosch, M. , Dewar, A. , & Olson, M. F. (2001). Membrane blebbing during apoptosis results from caspase‐mediated activation of ROCK I. Nature Cell Biology, 3(4), 339–345. [DOI] [PubMed] [Google Scholar]
- De Visser, K. E. , Eichten, A. , & Coussens, L. M. (2006). Paradoxical roles of the immune system during cancer development. Nature Reviews Cancer, 6(1), 24–37. [DOI] [PubMed] [Google Scholar]
- Dranoff, G. (2004). Cytokines in cancer pathogenesis and cancer therapy. Nature Reviews Cancer, 4(1), 11–22. [DOI] [PubMed] [Google Scholar]
- Fitzgerald, W. , Freeman, M. L. , Lederman, M. M. , Vasilieva, E. , Romero, R. , & Margolis, L. (8973). A system of cytokines encapsulated in extracellular vesicles. Science Reports, 8(1), 8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flieger, O. , Engling, A. , Bucala, R. , Lue, H. , Nickel, W. , & Bernhagen, J. (2003). Regulated secretion of macrophage migration inhibitory factor is mediated by a non‐classical pathway involving an ABC transporter. FEBS Letters, 551(1‐3), 78–86. [DOI] [PubMed] [Google Scholar]
- Fujiwara, T. , Oda, K. , Yokota, S. , Takatsuki, A. , & Ikehara, Y. (1988). Brefeldin A causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. Journal of Biological Chemistry, 263(34), 18545–18552. [PubMed] [Google Scholar]
- Gardella, S. , Andrei, C. , Ferrera, D. , Lotti, L. V. , Torrisi, M. R. , Bianchi, M. E. , & Rubartelli, A. (2002). The nuclear protein HMGB1 is secreted by monocytes via a non‐classical, vesicle‐mediated secretory pathway. EMBO Reports, 3(10), 995–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelpí, C. , Kanterewicz, E. , Gratacos, J. , Targoff, I. N. , & Rodriguez‐Sanchez, J. L. (1996). Coexistence of two antisynthetases in a patient with the antisynthetase syndrome. Arthritis and Rheumatism, 39(4), 692–697. [DOI] [PubMed] [Google Scholar]
- Gudz, T. I. , Tserng, K‐Yi , & Hoppel, C. L. (1997). Direct inhibition of mitochondrial respiratory chain complex III by cell‐permeable ceramide. Journal of Biological Chemistry, 272(39), 24154–24158. [DOI] [PubMed] [Google Scholar]
- Guo, M. , & Schimmel, P. (2013). Essential nontranslational functions of tRNA synthetases. Nature Chemical Biology, 9(3), 145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama, A. , Kami, K. , Sugimoto, M. , Sugawara, M. , Toki, N. , Onozuka, H. , … Soga, T. (2009). Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time‐of‐flight mass spectrometry. Cancer Research, 69(11), 4918–4925. [DOI] [PubMed] [Google Scholar]
- Jang, Su C. , Kim, Oh Y. , Yoon, C. M. , Choi, D. ‐. S. , Roh, T.‐Y. , Park, J. , … Gho, Y. S. (2013). Bioinspired exosome‐mimetic nanovesicles for targeted delivery of chemotherapeutics to malignant tumors. ACS Nano, 7(9), 7698–7710. [DOI] [PubMed] [Google Scholar]
- Jiang, W. , & Pisetsky, D. S. (2006). The role of IFN‐alpha and nitric oxide in the release of HMGB1 by RAW 264.7 cells stimulated with polyinosinic‐polycytidylic acid or lipopolysaccharide. Journal of Immunology, 177(5), 3337–3343. [DOI] [PubMed] [Google Scholar]
- Jun, S. , Ke, D. , Debiec, K. , Zhao, G. , Meng, X. , Ambrose, Z. , … Zhang, P. (2011). Direct visualization of HIV‐1 with correlative live‐cell microscopy and cryo‐electron tomography. Structure, 19(11), 1573–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. B. , Kim, H. R. , Park, M. C. , Cho, S. , Goughnour, P. C. , Han, D. , … Kim, S. (2017). Caspase‐8 controls the secretion of inflammatory lysyl‐tRNA synthetase in exosomes from cancer cells. Journal of Cell Biology, 216(7), 2201–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. , You, S. , & Hwang, D. (2011). Aminoacyl‐tRNA synthetases and tumorigenesis: More than housekeeping. Nature Reviews Cancer, 11(10), 708–718. [DOI] [PubMed] [Google Scholar]
- Kremer, J. R. , Mastronarde, D. N. , & Mcintosh, J. R. (1996). Computer visualization of three‐dimensional image data using IMOD. Journal of Structural Biology, 116(1), 71–76. [DOI] [PubMed] [Google Scholar]
- Luga, V. , Zhang, L. , Viloria‐Petit, A. M. , Ogunjimi, A. A. , Inanlou, M. R. , Chiu, E. , … Wrana, J. L. (2012). Exosomes mediate stromal mobilization of autocrine Wnt‐PCP signaling in breast cancer cell migration. Cell, 151(7), 1542–1556. [DOI] [PubMed] [Google Scholar]
- Maier, O. , Oberle, V. , & Hoekstra, D. (2002). Fluorescent lipid probes: Some properties and applications. Chemistry and Physics of Lipids, 116(1‐2), 3–18. [DOI] [PubMed] [Google Scholar]
- Mambula, S. S. , & Calderwood, S. K. (2006). Heat shock protein 70 is secreted from tumor cells by a nonclassical pathway involving lysosomal endosomes. Journal of Immunology, 177(11), 7849–7857. [DOI] [PubMed] [Google Scholar]
- Mambula, S. S. , Stevenson, M. A. , Ogawa, K. , & Calderwood, S. K. (2007). Mechanisms for Hsp70 secretion: Crossing membranes without a leader. Methods, 43(3), 168–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathivanan, S. , & Simpson, R. J. (2009). ExoCarta: A compendium of exosomal proteins and RNA. Proteomics, 9(21), 4997–5000. [DOI] [PubMed] [Google Scholar]
- Milligan, G. , Parenti, M. , & Magee, A. I. (1995). The dynamic role of palmitoylation in signal transduction. Trends in Biochemical Sciences, 20(5), 181–186. [DOI] [PubMed] [Google Scholar]
- Mor‐Vaknin, N. , Punturieri, A. , Sitwala, K. , & Markovitz, D. M. (2003). Vimentin is secreted by activated macrophages. Nature Cell Biology, 5(1), 59–63. [DOI] [PubMed] [Google Scholar]
- Nickel, W. , & Seedorf, M. (2008). Unconventional mechanisms of protein transport to the cell surface of eukaryotic cells. Annual Review of Cell and Developmental Biology, 24, 287–308. [DOI] [PubMed] [Google Scholar]
- O'gorman, D. B. , Weiss, J. , Hettiaratchi, A. , Firth, S. M. , & Scott, C. D. (2002). Insulin‐like growth factor‐II/mannose 6‐phosphate receptor overexpression reduces growth of choriocarcinoma cells in vitro and in vivo. Endocrinology, 143(11), 4287–4294. [DOI] [PubMed] [Google Scholar]
- O'Gorman, D. B. , Costello, M. , Weiss, J. , Firth, S. M. , & Scott, C. D. (1999). Decreased insulin‐like growth factor‐II/mannose 6‐phosphate receptor expression enhances tumorigenicity in JEG‐3 cells. Cancer Research, 59(22), 5692–5694. [PubMed] [Google Scholar]
- Oh, Y. J. , Youn, Ju Ho , Ji, Y. , Lee, S. E. , Lim, K. J. , Choi, Ji E. , & Shin, J. ‐. S. (2009). HMGB1 is phosphorylated by classical protein kinase C and is secreted by a calcium‐dependent mechanism. Journal of Immunology, 182(9), 5800–5809. [DOI] [PubMed] [Google Scholar]
- Ohno, S.‐.I. , Takanashi, M. , Sudo, K. , Ueda, S. , Ishikawa, A. , Matsuyama, N. , … Kuroda, M. (2013). Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Molecular Therapy, 21(1), 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, M. C. , Kang, T. , Jin, D. , Han, J. M. , Kim, S. B. , Park, Y. J. , … Kim, S. (2012). Secreted human glycyl‐tRNA synthetase implicated in defense against ERK‐activated tumorigenesis. Proceedings of National Academy Sciences of the USA., 109(11), E640–E647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S. G. , Schimmel, P. , & Kim, S. (2008). Aminoacyl tRNA synthetases and their connections to disease. Proceedings of the National Academy of Sciences of the USA, 105(32), 11043–11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, L. R. , & Raghavan, M. (2011). Endoplasmic reticulum calcium depletion impacts chaperone secretion, innate immunity, and phagocytic uptake of cells. Journal of Immunology, 187(2), 919–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo, G. , & Stoorvogel, W. (2013). Extracellular vesicles: Exosomes, microvesicles, and friends. Journal of Cell Biology, 200(4), 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal, S. K. , Skretting, G. , Garred, Ø. , Vilhardt, F. , Van Deurs, Bo , & Sandvig, K. (1999). Extraction of cholesterol with methyl‐beta‐cyclodextrin perturbs formation of clathrin‐coated endocytic vesicles. Molecular Biology of the Cell, 10(4), 961–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, R. J. (2006). Glucose metabolism and cancer. Current Opinion in Cell Biology, 18(6), 598–608. [DOI] [PubMed] [Google Scholar]
- Shen, B. , Wu, N. , Yang, Jr‐M. , & Gould, S. J. (2011). Protein targeting to exosomes/microvesicles by plasma membrane anchors. Journal of Biological Chemistry, 286(16), 14383–14395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, B. , Jung, H. ‐. J. , Hyung, S. ‐. W. , Kim, H. , Lee, D. , Lee, C. , … Lee, S.‐W. (2008). Postexperiment monoisotopic mass filtering and refinement (PE‐MMR) of tandem mass spectrometric data increases accuracy of peptide identification in LC/MS/MS. Molecular and Cellular Proteomics, 7(6), 1124–1134. [DOI] [PubMed] [Google Scholar]
- Skog, J. , Würdinger, T. , Van Rijn, S. , Meijer, D. H. , Gainche, L. , Curry, W. T. , … Breakefield, X. O. (2008). Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nature Cell Biology, 10(12), 1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son, S. H. , Park, M. C. , & Kim, S. (2014). Extracellular activities of aminoacyl‐tRNA synthetases: New mediators for cell‐cell communication. Topics in Current Chemistry, 344, 145–166. [DOI] [PubMed] [Google Scholar]
- Souza, R. F. , Wang, S. , Thakar, M. , Smolinski, K. N. , Yin, J. , Zou, T.‐T. , … Meltzer, S. J. (1999). Expression of the wild‐type insulin‐like growth factor II receptor gene suppresses growth and causes death in colorectal carcinoma cells. Oncogene, 18(28), 4063–4068. [DOI] [PubMed] [Google Scholar]
- Stojanov, L. , Satoh, M. , Hirakata, M. , & Reeves, W. H. (1996). Correlation of antisynthetase antibody levels with disease course in a patient with interstitial lung disease and elevated muscle enzymes. Journal of Clinical Rheumatology, 2(2), 89–95. [DOI] [PubMed] [Google Scholar]
- Takemura, Y. , Ouchi, N. , Shibata, R. , Aprahamian, T. , Kirber, M. T. , Summer, R. S. , … Walsh, K. (2007). Adiponectin modulates inflammatory reactions via calreticulin receptor‐dependent clearance of early apoptotic bodies. Journal of Clinical Investigation, 117(2), 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, S.C. , Berkelman, T. , & (2013). A defined methodology for reliable quantification of Western blot data. Molecular Biotechnology, 55, 217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry, C. , Boussac, M. , Véron, P. , Ricciardi‐Castagnoli, P. , Raposo, G. , Garin, J. , & Amigorena, S. (2001). Proteomic analysis of dendritic cell‐derived exosomes: A secreted subcellular compartment distinct from apoptotic vesicles. Journal of Immunology, 166(12), 7309–7318. [DOI] [PubMed] [Google Scholar]
- Théry, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , … Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 7(1), 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry, C. , Zitvogel, L. , & Amigorena, S. (2002). Exosomes: Composition, biogenesis and function. Nature Reviews Immunology, 2(8), 569–579. [DOI] [PubMed] [Google Scholar]
- Valadi, H. , Ekström, K. , Bossios, A. , Sjöstrand, M. , Lee, J. J. , & Lötvall, J. O. (2007). Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology, 9(6), 654–659. [DOI] [PubMed] [Google Scholar]
- Van Der Pol, E. , Böing, A. N. , Harrison, P. , Sturk, A. , & Nieuwland, R. (2012). Classification, functions, and clinical relevance of extracellular vesicles. Pharmacological Reviews, 64(3), 676–705. [DOI] [PubMed] [Google Scholar]
- Wang, H. (1999). HMG‐1 as a late mediator of endotoxin lethality in mice. Science, 285(5425), 248–251. [DOI] [PubMed] [Google Scholar]
- Webb, Y. , Hermida‐Matsumoto, L. , & Resh, M. D. (2000). Inhibition of protein palmitoylation, raft localization, and T cell signaling by 2‐bromopalmitate and polyunsaturated fatty acids. Journal of Biological Chemistry, 275(1), 261–270. [DOI] [PubMed] [Google Scholar]
- Wen, C. , Seeger, R. C. , Fabbri, M. , Wang, L. , Wayne, A. S. , & Jong, A. Y. (2017). Biological roles and potential applications of immune cell‐derived extracellular vesicles. Journal of Extracellular Vesicles, 6(1), 1400370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, H. , Li, Yi , Cui, Y. , Yang, J. J. , Zhang, Z. G. , & Chopp, M. (2013). Systemic administration of exosomes released from mesenchymal stromal cells promote functional recovery and neurovascular plasticity after stroke in rats. Journal of Cerebral Blood Flow and Metabolism, 33(11), 1711–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez‐Mó, M. , Siljander, P. R.‐M. , Andreu, Z. , Bedina Zavec, A. , Borràs, F. E. , Buzas, E. I. , … De Wever, O. (2015). Biological properties of extracellular vesicles and their physiological functions. Journal of Extracellular Vesicles, 4, 27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, M. , Chen, J. , Su, F. , Yu, B. , Su, F. , Lin, L. , … Song, E. (2011). Microvesicles secreted by macrophages shuttle invasion‐potentiating microRNAs into breast cancer cells. Molecular Cancer, 10, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , Wang, J. , Liu, C. , Grizzle, W. E. , Yu, S. , Zhang, S. , … Zhang, H‐Ge (2005). Cleavage of p53‐vimentin complex enhances tumor necrosis factor‐related apoptosis‐inducing ligand‐mediated apoptosis of rheumatoid arthritis synovial fibroblasts. American Journal of Pathology, 167(3), 705–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, P. , & Fox, P. L. (2013). Aminoacyl‐tRNA synthetases in medicine and disease. EMBO Molecular Medicine, 5(3), 332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Song, J. , Cavigiolio, G. , Ishida, B. Y. , Zhang, S. , Kane, J. P. , … Ren, G. (2011). Morphology and structure of lipoproteins revealed by an optimized negative‐staining protocol of electron microscopy. Journal of Lipid Research, 52(1), 175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitvogel, L. , Regnault, A. , Lozier, A. , Wolfers, J. , Flament, C. , Tenza, D. , … Amigorena, S. (1998). Eradication of established murine tumors using a novel cell‐free vaccine: Dendritic cell‐derived exosomes. Nature Medicine, 4(5), 594–600. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.


