Introduction
NGS Establishment in Multidisciplinary Healthcare (NEMHESYS) is an Erasmus+ programme with the purpose of providing qualified staff with the essential technical and bioinformatic knowledge and skills on next-generation sequencing (NGS) to be able to carry out NGS studies and perform some of the most common types of analyses. The clinical application of NGS has become easier with advancements in technologies.1 However, the investment needed to bring NGS into medical practice remains significant, with the scale of knowledge required being unprecedented at most hospitals. In addition, these novel technologies bring new challenges in translating NGS to clinical practice, at both technical and regulatory level, in terms of data management, interpretation of the results, and genetic counseling.2,3 All these aspects justify the consideration of what will be the precise role of NGS in diagnosis, risk assessment, response prediction, and treatment monitoring, today and tomorrow. Thus, to evaluate the implementation of NGS in European healthcare/research centers, a mapping survey was carried out, based on previous NGS mapping studies.4,5
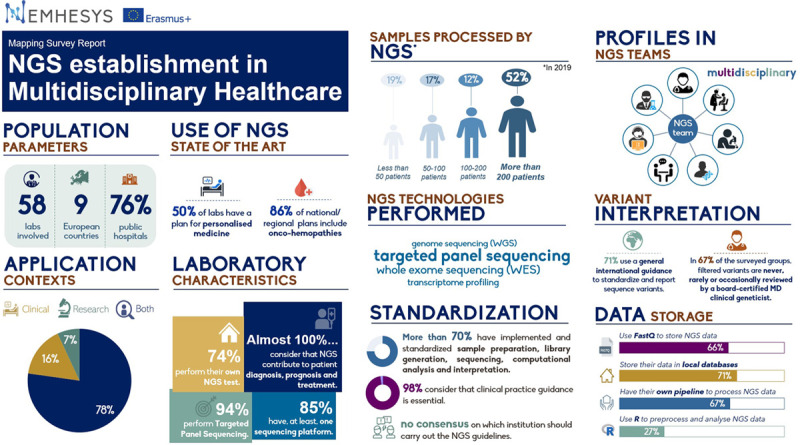
Demographics
The purpose of this survey was to summarize and compare the performance of clinical NGS services in Europe regarding the laboratory’s sequencing, interpretation, and reporting processes, alongside researchers’ interests and limitations. Through this survey, we have gained a better understanding of the use of NGS in Europe as a tool to improve personalized medicine. The 38-question survey, designed in coordination with all partners (University of Salamanca, University of Helsinki, Charité University Medicine Berlin, Queen’s University Belfast, Masaryk University, Artificial Intelligence Techniques, Mnemotix, and IDImás Gestión), was sent by email to various organizations (public hospitals, research centers, private laboratories, and universities) from 9 different countries within Europe between May and July 2020. Questions were formatted as multiple-choice, allowing responders to select one or more options, depending on the item. Finally, questions were grouped topically, generating the following sections: “Demographic Questions,” “State of the Art Use of NGS in your Country,” “Laboratory Characteristics,” “Standardization and Interpretation,” “Data Storage,” “Software Tools,” “Reporting,” and “Open Question.” The report collates the survey results from 58 European university hospitals and other public institutions.
State of the art
The results demonstrated that 78% of respondents use NGS for both research and clinical purposes and that almost 100% of the surveyed researchers consider that NGS can contribute to patient diagnosis, prognosis, and treatment. Interestingly, only 50% of respondents claimed to have a regional/national plan for personalized medicine, highlighting inequalities to patients in the provision of state of the art NGS technologies across Europe. Reimbursement for NGS testing across Europe was received in 66% of cases from government sources, with ~9% not receiving any financial reimbursement.
Laboratory characteristics
Seventy-four percent of surveyed laboratories perform their own NGS tests, while the majority of those outsourcing sent tests to regional/national reference centers. More than half of all respondents reported that they had performed NGS testing on >200 patients in the last year. Targeted panel sequencing was the main interest in 93% of surveyed laboratories, in comparison to 17%, 32%, and 21% of the laboratories performing whole genome sequencing (WGS), whole exome sequencing (WES), or transcriptome profiling, respectively. Ninety percent of centers processed <50 samples in the last year using WGS, WES, or transcriptome profiling, while half of them processed >200 samples by targeted panel sequencing. Thus, centers process considerably fewer samples for WGS, WES, and transcriptome profiling than for targeted panel sequencing
Of those surveyed, 85% have access to at least 1 sequencing platform. The most commonly used sequencing platform is Illumina (92%), with 22% of respondents using Thermo Fisher Scientific.
Standardization and interpretation
Laboratory standardization was surveyed with regards to sample processing/storage, extraction/quantification, and sequencing, alongside computational analysis and interpretation/reporting. The results showed that 81%–91% of wet-lab processes were performed in a standardized manner, while only 66%–78% of computational analyses and interpretation is standardized, indicating that there is a disparity between standardization of wet and dry-lab processes.
The necessity of practice guidance for clinical laboratories seeking to develop NGS assays was surveyed, and it was found that 98% of researchers deemed it an essential requirement; however, there was no consensus on who or which institute should develop these guidelines. General international guidelines, such as the American College of Medical Genetics Best Practice Guidelines for Variant Classification,6 are used by 71% of laboratories for the reporting of sequence variants, while 36% and 47% refer to general national guidelines or disease-specific guidance, respectively.
The profile of those involved with the primary analysis of NGS variants is varied; however, almost 60% of the initial analysis is performed by doctoral-level clinical scientist fellows/molecular genetic pathology resident in training, graduate-level, or PhD-level trained analyst. In 67% of the surveyed groups, filtered variants are never, rarely, or occasionally reviewed by a board-certified MD clinical geneticist. Reported variants are not confirmed by another methodology in 22% of laboratories. Interestingly, 26% of surveyed laboratories do not hold regular meetings to discuss challenging cases or filtered variants from NGS analysis. Of those who hold regular meetings, it was found that the group members were diverse and contributed to a multidisciplinary approach.
Data storage and software tools
The survey asked what format NGS data are stored, and 65.5% of laboratories use FASTQ as their main storage modality, although VCF and BAM files are used to a similar extent (50% and 54%, respectively). These files were stored in local/in-house/LIMS databases in 71% of laboratories, while 20% of those surveyed stored their data in commercial software clouds such as Alamut Visual or BaseSpace Sequence Hub. Nine percent of laboratories use third-party storage such as Amazon and Google. With regards to data processing, 67% of organizations questioned have their own pipeline to process NGS data. Most preprocessing and analysis of NGS data is performed using R (27%), with 35% of respondents not knowing or left the question unanswered. Laboratories were asked which tasks were accomplished using programming, with 38% of respondents using programming to read, preprocess, and visualize the data. Conversely, 27% do not use any programming to accomplish NGS processing tasks, with 25% not knowing, unsure, or did not answer.
Reporting
Laboratories were asked questions referring to the variant reporting process. In 76% of the surveyed organizations, the initial draft of the NGS report is written by a doctoral-level clinical scientist fellow/molecular genetic pathology resident in training graduate level or a PhD-level trained analyst. It was noted that if a second review was performed, there was no consensus of who typically reviews and edits the test report before the sign-out. The professional profile of the NGS report reviewer is varied. Interestingly, 9% of respondents claim they either do not have a second reviewer or that the question is not applicable to them.
Open question
Researchers were asked what the main bottleneck was in their sequencing process, and it was found that there were a variety of issues responsible for slowing down the process. Although most institutions have standardized the main NGS processes (such as sample preparation, library generation, sequencing, computational analysis, or interpretation), most respondents (31%) claimed that a lack of knowledge, training, and exposure to routine analyses and interpretation of NGS resulted in the most critical bottleneck.
Conclusions and future directions
This survey highlights the disparity across Europe with regards to how NGS processes are performed, specifically, the lack of standardization when it comes to bioinformatic analysis. It has emphasized the need to align NGS processing and bioinformatic analysis to an internationally recognized guideline, alongside providing state of the art training to qualify biomedical staff to successfully perform clinical NGS at their own institutions. The patient is the main beneficiary of the application of NGS in health care, and it is imperative that they obtain fast and precise diagnoses and care. To provide this, specialized biomedical personnel equipped with the knowledge and skills are required to give the patient assurance that testing is performed to the highest standard across Europe. It is, however, clear that ensuring the best possible approach to personalized medicine and is a complex but essentially unmet need.
Consequently, the NEMHESYS consortium has generated a masterclass designed to train the most suitable staff based on partners’ experience; this could be clinical scientists, biomedical scientists as well clinicians and trainees. The masterclass will consist of several modules developed by the Higher Education Institution based on their specialization. These modules will be made up of masterclasses, case studies, and workshops, favoring a dynamic and interactive work environment. It will be taught simultaneously via an online platform, with training materials available on the website.
Within the NEMHESYS education program, private companies will run workshops on big data, technological developments, artificial intelligence, entrepreneurship, and innovation, providing participants with practical tasks to strengthen their skills and competencies to solve complex problems in their daily work. Additionally, the simultaneous connection of participants from the different institutions will permit exchange and discussion on these topics. Once educational modules are completed, mobility of the NEMHESYS cohorts among participating institutions will be promoted to complete their training with practical experience.
More information and updates can be found at https://nemhesys.com/
Acknowledgments
We acknowledge all the clinicians and researchers who have participated in the survey. We are also grateful to all members of NEMHESYS consortium (University of Helsinki, Charité University Medicine Berlin, Queen’s University Belfast, Masaryk University, Artificial Intelligence Techniques [Artelnics], Mnemotix, IDImás Gestión, and University of Salamanca).
Sources of funding
This work was supported by Erasmus+ programme (European Commission, Call EAC/A03/2018 NEMHESYS_612639-EPP-1-2019-ES-EPPKA2-KA).
Disclosures
The authors have no conflicts of interest to disclose.
Footnotes
I.S.-G. and K.M.C. have contributed equally to this work. L.B., K.I.M., and J.M.H.R. have jointly supervised this work.
References
- 1.Kumar KR, Cowley MJ, Davis RL. Next-generation sequencing and emerging technologies Semin Thromb Hemost. 2019; 45:661–673 [DOI] [PubMed] [Google Scholar]
- 2.Di Resta C, Galbiati S, Carrera P, et al. Next-generation sequencing approach for the diagnosis of human diseases: open challenges and new opportunities EJIFCC. 2018; 29:4–14 [PMC free article] [PubMed] [Google Scholar]
- 3.Lightbody G, Haberland V, Browne F, et al. Review of applications of high-throughput sequencing in personalized medicine: barriers and facilitators of future progress in research and clinical application Brief Bioinform. 2019; 20:1795–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Daniel JM, McLaughlin HM, Amendola LM, et al. A survey of current practices for genomic sequencing test interpretation and reporting processes in US laboratories Genet Med. 2017; 19:575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gullapalli RR. Evaluation of commercial next-generation sequencing bioinformatics software solutions J Mol Diagn. 2020; 22:147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of medical genetics and genomics and the association for molecular pathology Genet Med. 2015; 17:405–424 [DOI] [PMC free article] [PubMed] [Google Scholar]


