Abstract
The human gut microbiota is transmitted from mother to infant through vaginal birth and breastfeeding. Bifidobacterium, a genus that dominates the infants’ gut, is adapted to breast milk in its ability to metabolize human milk oligosaccharides; it is regarded as a mutualist owing to its involvement in the development of the immune system. The composition of microbiota, including the abundance of Bifidobacteria, is highly variable between individuals and some microbial profiles are associated with diseases. However, whether and how birth and feeding practices contribute to such variation remains unclear. To understand how early events affect the establishment of microbiota, we develop a mathematical model of two types of Bifidobacteria and a generic compartment of commensal competitors. We show how early events affect competition between mutualists and commensals and microbe-host-immune interactions to cause long-term alterations in gut microbial profiles. Bifidobacteria associated with breast milk can trigger immune responses with lasting effects on the microbial community structure. Our model shows that, in response to a change in birth environment, competition alone can produce two distinct microbial profiles post-weaning. Adding immune regulation to our competition model allows for variations in microbial profiles in response to different feeding practices. This analysis highlights the importance of microbe–microbe and microbe–host interactions in shaping the gut populations following different birth and feeding modes.
Keywords: microbial population dynamics, infant gut microbes, ordinary differential equations, C-section and formula milk, antibodies, mutualist-commensal competition
1. Introduction
The human gut is a complex ecosystem whose bacterial residents play a crucial role in host health. Interactions between bacteria, including cooperation and competition, are essential in shaping the dynamics of the gut community. While cooperation can promote the colonization and growth of bacterial species, competition stabilizes the gut community by dampening the positive feedback from cooperation [1]. Discrete stable compositions of the adult gut microbiota have been recognized [2], and some are associated with diseases [3]. It is important, therefore, to understand how gut microbiotas are established and the mechanisms that result in different stable microbiota compositions. Early infancy is a key period in gut microbiota development, as microbes are vertically transmitted from mother to infant [4,5]. While considerable debate continues as to whether vertical transmission starts at or before birth [6,7], it is understood that vaginal birth [8] and breast feeding are two important transmission routes as infants collect maternal microbes from the birth canal and breast milk [9,10]. After weaning, the infant gut microbiota stabilizes to the adult form by around the age of 3 years [11,12].
Bifidobacterium, which is highly abundant in the infant’s gut, is believed to exert positive health benefits to the host. The transmission of Bifidobacteria is facilitated by the consumption of breast milk [9,10,13]. Species of Bifidobacteria, namely Bifidobacterium longum subspecies infantis (B. infantis) [14], Bifidobacterium longum subspecies longum (B. longum) [15] and Bifidobacterium bifidum [16], are adapted to the neonatal gut; they can metabolize the human milk oligosaccharides (HMO) that are present in breast milk. Other Bifidobacteria species benefit from the presence of these HMO metabolizing Bifidobacteria through cross-feeding as they use products of HMO degradation [17–19]. The presence of HMO in breast milk provides Bifidobacteria a competitive advantage over other species in the neonatal gut. Vaginal birth is also an important source of Bifidobacteria colonization: vaginally delivered infants share strains of Bifidobacteria with their mothers [8], and gut-derived and vaginal-derived Bifidobacteria strains are indistinguishable [20].
Bifidobacterium is considered an essential genus in the gut as it is involved in the development of the immune system. The abundance of Bifidobacteria varies across individuals [21], and is correlated with faecal immunoglobulin A (IgA) levels in infants [22,23] and children [24]. Bifidobacteria have been shown to stimulate the production of IgA [25,26]. This mediates immune exclusion by preventing pathogen adsorption to the mucosal epithelium [27,28]. Given that the abundance of Bifidobacteria affects the level of IgA and therefore the host’s immune response to other bacteria, Bifidobacteria may have important regulatory effects on neonatal microbial populations.
Early interventions such as caesarean section (C-section) and formula feeding are believed to interrupt the vertical transmission of microbiota [29] and can have significant effects on neonatal gut composition. The microbiota of breastfed infants is characterized by the dominance of Bifidobacteria [11,30,31], while that of infants born through C-section or who receive formula milk are associated with a lower abundance of Bifidobacteria [11,30,32–34] and a high abundance of opportunistic pathogens [35].
While an increasing number of studies reveal correlations between birth mode or feeding practice on one hand and infant gut composition on the other, the empirical evidence for long-term effect of these interventions is mixed. The impact of C-sections appears to last for at least 2 years [36] or 7 years after birth [37]. By contrast, other studies found that the effect of birth mode on the variations in microbiota is no longer evident within a few months after birth [38,39], and that the effect of birth mode was not reflected in adult microbiota [40]. Breastfeeding is reported to have a significant effect on the composition of microbiota until 14 months of age [11], after which the profile of breastfed and formula-fed infants converged [11,41]. However, it has also been found that the microbiome community type in adults is associated with breastfeeding history [42]. Understanding the mechanisms by which different community structures are established, and therefore the effect of early events on their establishment can help to resolve these apparently contradictory results.
Gut populations are shaped by ecological interactions between microbes and by interactions between microbes and the immune system. It is unclear whether and how birth and feeding practices exert long-term effects on the microbiota. Here, we develop mathematical models to investigate how microbe-microbe and microbe–host interactions contribute to the varying abundance of Bifidobacteria post-weaning. We use these models to study the conditions under which birth environment and feeding practices alter the structure of bacterial communities in infants. The models include two Bifidobacteria populations (milk-consuming and fibre-consuming Bifidobacteria) and a commensal population. We also investigate the role of immune regulation on the effects of birth and feeding modes.
2. Methods
(a). Competition model
To study the effect of infant diet and delivery mode on infant microbiota development, we start with a model in which the dynamics are driven by competition between Bifidobacteria and commensals. Species of Bifidobacteria are coarsely divided into two types (B1 and B2) distinguished by their metabolic characteristics. One population of Bifidobacteria (B1) is able to metabolize HMO while the other (B2) is able to metabolize plant polysaccharides and the products of HMO degradation. Figure 1a illustrates our initial model, showing the cross-feeding behaviour between two Bifidobacteria populations and their competition with a generic group of commensal bacteria (B3). This compartment is non-specific in that it groups all species of commensals. We use B1, B2, B3 to refer to the (non-dimensionalized) density of these bacteria.
Figure 1.
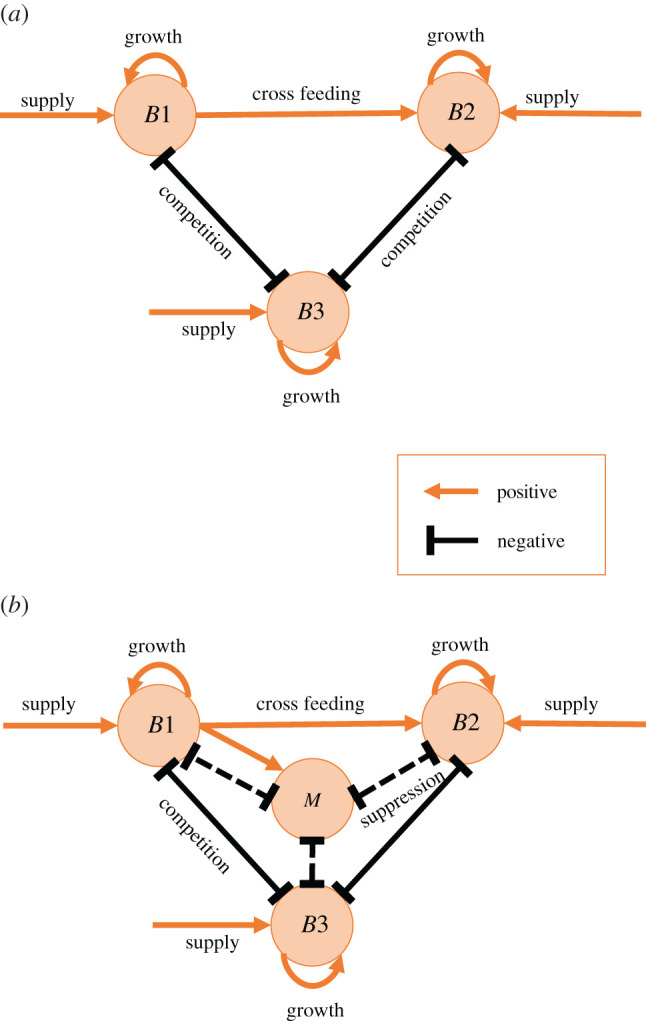
Schematic of the competition model (a) and extended model (b). Orange arrows represent positive effects on the population. Black lines represent negative effects on the population. Fibre consuming Bifidobacteria B2 benefits from cross-feeding by metabolizing the HMO-degradation product of milk consuming Bifidobacteria B1. In the extended model (b), the presence of B1 stimulates the growth of the immune factor (M), which suppresses the growth of all bacterial populations. (Online version in colour.)
We introduce a system of ordinary differential equations (ODEs) to track the abundances of the three bacterial populations, as follows:
| 2.1 |
The strength of competition between Bifidobacteria and commensals is characterized by a competition coefficient α. If α > 1 the competition between Bifidobacteria and commensals is stronger than the competition within each population; if α < 1 the within-population competition is stronger; if α = 1 between-population competition is exactly as strong as within-population competition. The total population is constrained by a carrying capacity parameter K. The growth of the HMO-metabolizing Bifidobacteria B1 population depends on the proportion of milk in the diet, denoted by Z(t); the supply of B1 is proportional to breast-feeding and maternal interaction and is given by fZ Z(t). The proportion of milk Z(t) in the infant’s diet is modelled by a logistic function, which reflects the gradual replacement of milk by solid food during the first 1000 days of life. The weaning process is characterized by the rate of weaning ω and the half-life of milk in infant’s diet h, where half-life is the time required for milk to fall to 50% of an infant’s diet. We define the function Z(t) as
| 2.2 |
Belonging to the same genus, fibre-consuming Bifidobacteria B2 are assumed to have similar competition properties as B1; therefore, the competition between B1 and B2 is considered as equivalent to intrapopulation competition. The presence of B1 is beneficial to the growth of B2 owing to cross-feeding activities described in the introduction and characterized by αc. We assume a constant supply of Bifidobacteria B2 with rate f2 and commensal competitor B3 with rate f3 owing to environmental exposure.
To reduce the number of parameters, we non-dimensionalize the system by defining the following new variables and parameters
We omit the asterisk for notational simplicity and obtain the following non-dimensionalized system
| 2.3 |
| 2.4 |
| 2.5 |
The parameters and their baseline values used in simulation are summarized in table 1.
Table 1.
Summary of (non-dimensionalized) parameters in the model.
| model parameters | |||
|---|---|---|---|
| symbol | description | value | unit |
| r | growth rate of the bacterial populations | 1 | day−1 |
| μb | death rate of Bifidobacteria | 50 | day−1 |
| μc | death rate of commensal competitor | 200 | day−1 |
| αc | effect of cross-feeding between B1 and B2 | 1.7 | — |
| α | competitive effect between bacterial populations | 2 | — |
| fz | supply of B1 associated with milk | 0.01 | day−1 |
| f2 | supply of B2 from the environment | 0.03 | day−1 |
| f3 | supply of B3 from the environment | 0.05 | day−1 |
| ω | rate of weaning | 0.014 | day−1 |
| h | day since birth when milk is half of diet | 500 | day |
| γ | immune effect | 0.0001 | — |
We assume that the supply of bacteria is small relative to the carrying capacity, and that the environmental supply of fibre-consuming Bifidobacteria B2 is higher than the milk-facilitated supply of HMO-consuming Bifidobacteria B1. Because the commensal population B3 includes a broader range of species, we assume that the supply of commensals is higher than that of the Bifidobacteria. Therefore, fz, f2 and f3, which represent the supply of B1, B2 and B3, respectively, are set at 1%, 3% and 5% of the carrying capacity. We focus on the mutualistic interaction between different types of Bifidobacteria [17–19] as the HMO-consuming Bifidobacteria B1 help the other Bifidobacteria population B2 survive through infancy. Therefore, we assume the effect of cross-feeding αc to be higher than the within-population competition (αc > 1) and set αc = 1.7 (we note that αc does not affect the equilibria because it is linked to B1, which is eventually eliminated). Furthermore, to specify high between-population competition (between both Bifidobacteria populations and the generic commensal population), we set the competition coefficient α at 2. We choose h = 500 following the recommendation of the World Health Organisation to introduce solid food at around six months (180 days) and continuous breastfeeding for 2 years (730 days) or more [43].
To study the effects of birth mode on gut bacterial population, we choose two sets of initial conditions representing vaginal birth and C-section; those values are our non-dimensionalized variables and thus represent a fraction of the carrying capacity K. For vaginal birth, the initial colonisation of Bifidobacteria and commensal are set at 0.05 and 0.001, respectively; for C-section, the initial colonization of Bifidobacteria and commensal are set at 0.001 and 0.05. These two conditions qualitatively reflect the higher initial colonization of maternal Bifidobacteria during a vaginal birth and the higher initial colonization of commensals from health care facilities during a C-section [35]. While we choose the default parameters in table 1 based on general empirical observations, we also investigate the sensitivity of our model to variation in a few key parameters.
(b). An extension with immune effects
To investigate the effect of birth and feeding modes under immune regulation as well as competition, we extend the above model by introducing an immune factor M. As highlighted in the introduction, Bifidobacteria stimulates the production of IgA; thus the supply of Bifidobacteria associated with breast milk is likely to trigger immune reaction that suppresses the gut populations. In this modified model, HMO-consuming Bifidobacteria promotes the growth of the immune factor M, and M, in turn, suppresses all bacterial populations. The extended model schematic is given in figure 1b.
The expansion of the immune factor M depends linearly on Bifidobacteria B1 with rate parameter γ. The level of immune reaction is thus determined by the cumulative abundance of Bifidobacteria B1 in the system. The initial condition for M is set at 0.001 for both birth modes. IgA stimulated by Bifidobacteria restricts pathogen and potentially harmful attachment of commensals to the epithlelium [27,28]. Thus, we assume that the immune system favours the establishment of mutualists by making them more resistant to the immune response than commensals. Because we include a wider range of species in the commensal population, we assume that they are more susceptible to this immune clearance. Therefore, denoting death rates of Bifidobacteria and commensals by μb and μc, respectively, we take μc > μb. The extended system is described by the following differential equations:
| 2.6 |
In addition to this model, we considered a generalization in which fibre-consuming Bifidobacteria B2 also contribute to immune stimulation and in which the immune response decays (see electronic supplementary material, figures S10 and S11).
3. Results
(a). Competition model: the effect of birth environment
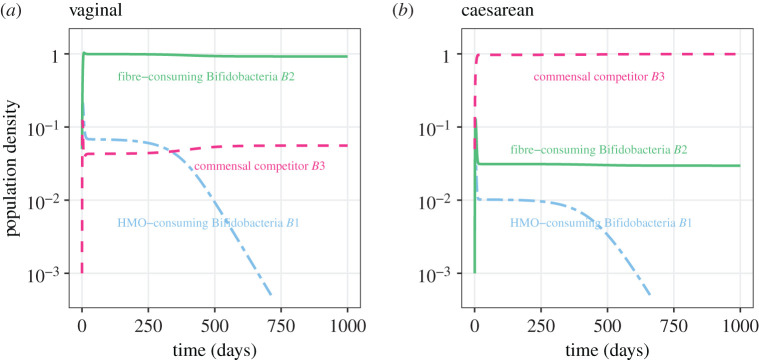
The temporal dynamics of the competition model given by equations (2.3)–(2.5) are shown in figure 2. The populations stabilize at the age of 3 years by the end of weaning as the proportion of milk drops from one to zero (electronic supplementary material, figure S1). During this transitional period Bifidobacteria B1 decreases and is eliminated, leaving the system with two populations. With our baseline parameter values (table 1), the system stabilizes such that under vaginal birth there is a high abundance of Bifidobacteria B2 (figure 2a), while under C-section there is a high abundance of commensal competitor B3 (figure 2b). When the between-population competition is weaker than within-population competition (α = 0.7), the system stabilizes at the same equilibrium regardless of the birth mode (electronic supplementary material, figure S2). We next explore the conditions within which either of these equilibrium patterns arise.
Figure 2.
Longitudinal dynamics of the three bacterial population abundances (on a log scale) using baseline parameter values in table 1 and initial conditions representing vaginal birth (a) and C-section (b). The blue two-dash line represents the infant-type Bifidobacteria B1, the green solid line represents the adult-type Bifidobacteria B2 and the red dashed line represents the commensal competitor B3. The dynamics are simulated for 3 years allowing the system to reach a post-weaning steady state. (Online version in colour.)
We observed a drop of HMO-consuming Bifidobacteria B1 closely following the drop of milk in infant diet Z(t) (dynamics shown in figure 2, and the function Z(t) shown in the electronic supplementary material, figure S1). Studies show that the gut microbiota stabilizes after the cessation of milk [11]. Thus, we make the assumption that B1 reaches its steady state before the other bacterial populations equilibrate, and as Z(t) → 0. We first solve for the steady state of B1 by applying Z(t) = 0 and ; the HMO-consuming Bifidobacteria B1 goes to zero at equilibrium. We approximate the model using a quasi-steady state with and reduce our system to two dimensions (see the electronic supplementary material, equations (S1)–(S2)).
To check the accuracy of our quasi-steady-state approximation against the behaviour of the full model, we compared the numerical solution of the original system with the quasi-steady-state approximation (electronic supplementary material, figure S3) and find a close match under baseline parameters. Under different competition strengths α and weaning schedule h (electronic supplementary material, figures S4 and S5) the approximation shows slight mismatch in the time at which the dominating population switches from commensal B3 to Bifidobacteria B2 (electronic supplementary material, figure S4). However, in general, the approximate equilibria match the full model within the range of parameter values we consider (electronic supplementary material, figures S4 and S5).
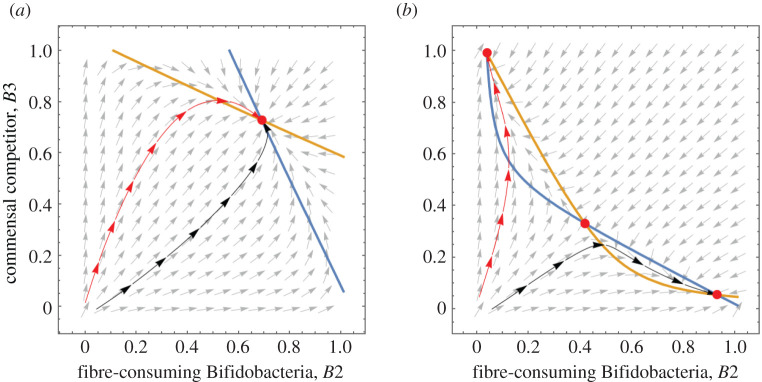
We use this approximate system to explore the long-term equilibria of the fibre-consuming Bifidobacteria B2 and commensal competitor B3 by considering a phase portrait (figure 3). Figure 3a shows the nullclines and trajectories of this approximate system where between-population competition is weaker than within-population competition (α < 1). Two trajectories, with initial conditions representing C-section (red) and vaginal birth (black) meet at a stable node (the red dot) in the positive quadrant. There is only one positive equilibrium, which is consistent with the temporal dynamics of the full model at α = 0.7 (electronic supplementary material, figure S2). This implies a long-term population dynamic independent of birth mode; both birth modes stabilize at the same state.
Figure 3.
Vector plot and nullclines of B2 (blue) and B3 (yellow). Two nullclines meet at a stable equilibria. Black and red lines are the trajectories of the population dynamics following an initial condition representing vaginal birth and C-section, respectively. (a) α = 0.7, other parameters are as specified in table 1. (b) Parameters specified in table 1. (Online version in colour.)
We also find three positive equilibria when between-population competition is higher than within-population competition (α > 1) (electronic supplementary material, table S1). A phase portrait with α = 2 (our baseline value) shows bistability, with two stable nodes and a saddle point (figure 3b). A higher initial Bifidobacteria B2 colonization (vaginal birth) leads to an equilibrium dominated by B2 while higher initial commensal B3 colonization (C-section) leads to an equilibrium dominated by B3.
Whether a trajectory goes to one equilibrium or the other depends on the initial ratio of Bifidobacteria to the commensal competitor. To investigate this dependence on the initial conditions, we further explore the population dynamics by numerically solving the ODEs of the original system to obtain the long-term equilibria at a range of initial conditions (electronic supplementary material, figure S6). As the ratio of initial Bifidobacteria to commensals increases from 0 to 2, the equilibrium switches from one dominated by the commensal to one dominated by Bifidobacteria.
Both our analytic and numerical results imply that birth mode may impose a long term effect on the gut microbiota. We then investigate the long-term effect of feeding practices. Breastfeeding involves a higher level of mother-infant interaction compared with formula feeding, and breast milk carries maternal microbes that are absent in formula milk. Therefore, we assume that the supply (fz) of HMO-metabolizing Bifidobacteria is positively associated with the amount of breast milk given to infants. Different fz values correspond to various feeding practices such as exclusive formula feeding (low fz) and exclusive breastfeeding (high fz). It also reflects the individual differences in the amount of HMO present in breast milk. Under this assumption, we numerically solve the system against a range of milk-associated Bifidobacteria supply fz (electronic supplementary material, figure S7). The model shows a compensatory effect of breastfeeding on C-section delivered infants. Their microbiota composition may eventually resemble that of vagina-delivered infants provided enough Bifidobacteria is supplied (electronic supplementary material, figure S7b). However, the system is generally insensitive to changes in feeding practices as the amount of breast milk has no effect on the population dynamics following vaginal birth (electronic supplementary material, figure S7a).
In this model, variation in response to different birth environment and feeding practices is limited. At low between-species competition (α < 1), the long-term population dynamics is independent of birth environment (figure 3a). At high between-species competition (α > 1), the system is highly constrained to approaching one of two equilibria, where either Bifidobacteria B2 or commensal competitor B3 dominates.
(b). Extended model: effect of breast milk and weaning schedule
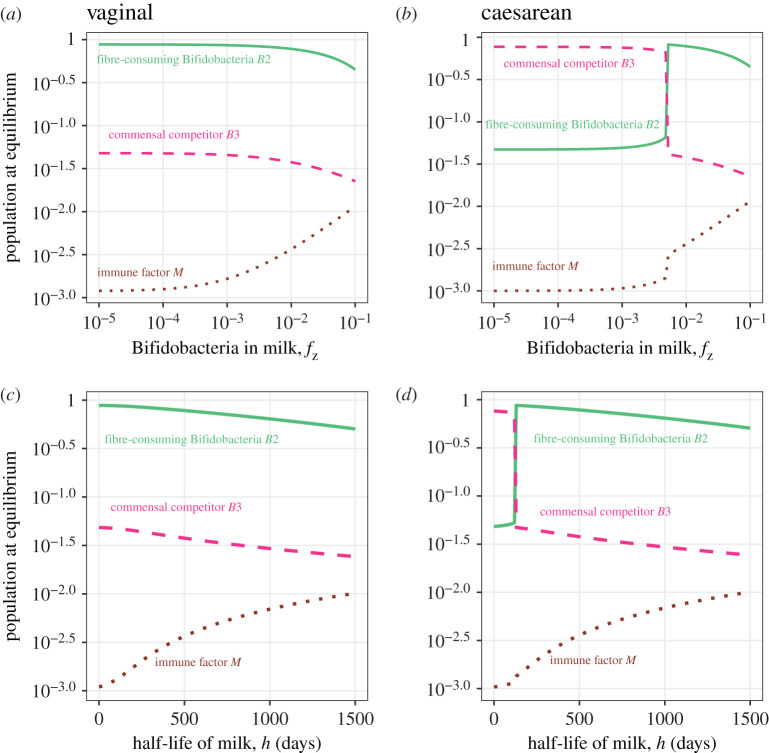
The extended model (equation (2.6)) helps us to gain further insight into the long-term effect of feeding practices on the population dynamics. In figure 4,we show the effect of feeding practices (breast milk fz and half life of milk h) on the equilibria at different initial conditions (which themselves represent birth mode). The dynamics of the bacterial populations in response to feeding practices are influenced by birth mode. Unlike the simple competition model, the population dynamics change continuously with breast milk (rather than abruptly) in this extended model, exhibiting a range of possible outcomes in an individual (figure 4).
Figure 4.
Steady states of the three bacterial populations and the immune factor M in response to feeding practices. The simulations used the baseline parameter values in table 1 and the initial conditions representing vaginal birth (a and c) and C-section (b and d). The green solid line represents the adult-type Bifidobacteria B2, the red dashed line represents the commensal competitor B3 and the brown dotted line represents immune factor M. The y-axes are log-scaled population densities at equilibria. The x-axes are log-scaled Bifidobacteria levels carried by milk fz representing the amount of breast milk in diet (a and b), and the time h when milk takes up of the diet representing the duration of breastfeeding (c and d). (Online version in colour.)
The half-life of milk h has a similar effect on the population dynamics to the input of Bifidobacteria in breast milk fz (figure 4). Vaginal birth promotes the dominance of fibre-consuming Bifidobacteria B2. The increase of breast milk and age of weaning reduces the abundance of both Bifidobacteria B2 and commensals B3, with Bifidobacteria decreasing at a lower rate. C-sections tend to promote the dominance of commensals B3. The abundance of fibre-consuming Bifidobacteria increases with breast milk until a turning point is reached, after which the trajectory resembles that following a vaginal birth. This again indicates the compensatory effect of breast milk on infants delivered by C-section.
The dynamics of B2 and B3 in response to the amount of breast milk and weaning schedule reflect the combined effect of cross-feeding, immune reaction and competition between the populations. Similar to increasing the amount of breast milk fz, a higher half-life of milk h, which models weaning at an older age, increases the total supply of HMO-consuming Bifidobacteria B1 to the system. This can increase the level of cross-feeding between B1 and B2 as well as the immune factor M. While cross-feeding tends to increase the abundance of Bifidobacteria B2 at equilibrium, increasing the immune factor has the opposite effect. As Bifidobacteria is more resistant to the immune clearance, B2 decreases at a slower rate than commensal B3 in response to higher level of breast milk and older weaning age. If Bifidobacteria are subject to the same strength of immune clearance as the commensals, the immune effect together with competition tends to suppress its abundance at stable state (electronic supplementary material, figure S8).
The ratio of Bifidobacteria B2 to commensal B3 at equilibria is sensitive to the changes of their supply from the environment (f2 and f3, respectively). The supply of Bifidobacteria increases the relative abundance of Bifidobacteria while the supply of commensals causes a decrease (electronic supplementary material, figure S9). With a low supply of Bifidobacteria and a high supply of commensals, an increase in breast milk, which facilitates the transmission of maternal Bifidobacteria, can promote the relative abundance of Bifidobacteria. As the supply of Bifidobacteria and commensals increases, the relative abundance of Bifidobacteria at equilibria becomes less sensitive to changes in breast milk. With a high Bifidobacteria supply and low commensal supply, the Bifidobacteria population dominates over the commensals regardless of feeding practices (electronic supplementary material, figure S9).
4. Discussion
We have introduced a simple ecological model to investigate the mechanisms of gut microbiota establishment. The dynamics of the gut populations in our model are driven by competition and can be affected by birth environment (initial colonization). The effects of initial colonization on the competitive outcome between an invasive and a commensal gut population have been studied with chemostat models [44,45] and a recent Lotka–Volterra-type ecological model [46]. The prevalence of competition has been shown in ecological network models [1,47] and human gut community interaction topologies [48,49]. While network models use data-driven inferences to learn microbial interactions, metabolic models reconstruct biochemical reactions to understand those interactions [50–52]. Here, we focused on the population dynamics affected by ecological interaction (competition) and host immune regulation in the context of early interventions. We studied the establishment of two mutualist populations (Bifidobacteria) and a generalized commensal population, and focused on the competition between those populations (Bifidobacteria and commensals) and the immune response triggered by the mutualist (Bifidobacteria). By considering just these two mechanisms, we have shown that immune regulation, along with competition, can produce long-term variation in gut microbial profile following different birth and feeding practices.
Inconsistent observations regarding the long-term effect of birth and feeding practices on microbiota profiles have been made [36,40,42,53]. We offer an explanation for this apparent contradiction. Under our model, it is possible to produce both kinds of outcomes; the impact of early interventions can be either inconsequential or significant depending on the magnitude of the intervention. A change in birth environment can alternatively lead to a long-term alteration in microbial populations, as shown in empirical studies [36,37]. This can occur if the between-population competition is sufficiently strong (higher than within-population competition), and the intervention causes a large enough difference in initial colonization for the populations to switch between two stable states. Following birth, infant feeding continues to shape the microbiota, and we find that its effect depends on birth environment. The gut microbial profile of formula-fed and breast-fed infants born through C-section may diverge [42]; we find a similar divergence by showing that the maternal microbes carried by breast milk can compensate for the loss of beneficial bacteria from C-section. However, if infants receive a large enough number of bacterial cells from vaginal birth, our model suggests that feeding practices early in life may not have a significant impact on the microbial composition, in agreement with findings of empirical studies [40,53].
We illustrate the importance of microbe–microbe and microbe–host interactions in shaping gut populations. Increased strength of competition leads to the divergence of microbial profile following different birth environments. The divergent outcomes are owing to bistability in the dynamics, which was also observed in studies that model commensal-pathogen competition [46] and microbe-immune system interaction [54]. Variation in post-weaning Bifidobacteria abundance in response to different birth and feeding modes cannot be produced by competition alone. Interaction with the immune system facilitates the possibility of this variation in microbiome composition. Early differences in Bifidobacteria levels, driven by birth environment and consumption of breast milk, induce different levels of immune regulation, which in turn have a lasting effect on the microbial structure. While the gut microbiota is influenced by numerous lifestyle factors [55], our work implies that early events can contribute to regional differences observed in individuals from non-industrialized [12,56–59] and industrialized societies [12,60]. The effects of those early events are regulated by microbial competition and immune regulation.
Antibiotic exposure at birth, a common practice in industrialized societies, may also be part of the modern birth environment. This can be a form of early intervention that affects the microbial competition and immune regulation, and therefore the development of microbiota. While not modelled explicitly, antibiotic administration at birth is analogous in our model to restarting the microbial succession with different initial colonization conditions. Early antibiotic exposure has been reported to disrupt early microbial succession and alter the taxonomic composition of the infant gut [61,62]. Our model shows the divergence of microbial populations in response to altered initial colonization, implying the possibility of a long-term shift in microbial configuration owing to antimicrobial exposure. This aligns with other mathematical models that demonstrated the extinction of antimicrobial-sensitive bacteria in the gut after antibiotic treatments [47,63]. However, our results also show that the potential loss of Bifidobacteria resulting from an altered birth environment (initial colonization or, perhaps, antibiotic exposure) can be reversed with a sufficient supply of breast milk. This is consistent with studies showing that continued breastfeeding can compensate for disturbances caused by antibiotics [61].
While we have investigated the effect of early-life interventions on the establishment of infant microbiota, it is clear that the post-weaning diet also plays a key role in shaping gut microbial communities. Microbe acquisition from the environment, for example, has been shown to increase within-host microbiome diversity [64]. The composition of microbiota is shown to be dependent on long-term dietary practice [57,65]. This is reflected in our model where the microbial configuration is sensitive to the change in environmental supply of the bacteria. The microbiota is reported to be responsive to changes in diet [66–68]. However, our model does not capture the full range of possible changes in exposure to environmental microbes that may steer the microbial community to different configurations. In future work, our model can be extended to study the effect of long-term dietary patterns and drastic diet shifts.
Supplementary Material
Acknowledgements
We thank Ruiting Lan, Vanessa Venturi and Catherine Penington for useful discussions.
Data accessibility
All data and results generated in this study can be reproduced using the source code we provide on github https://github.com/xiyanxiongnico/Modelling-the-effect-of-birth-and-feeding-modes-on-the-development-of-human-gut-microbiota.git.
Authors' contributions
X.X. and M.T. conceived of the project, X.X., S.L.L. and M.M.T. developed the mathematical model with input from L.Z. X.X. generated the analytical and numerical results and figures, and drafted the manuscript. All authors critically revised and edited the manuscript. All authors gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by a University Postgraduate Award from UNSW and grant no. DP170101917 from the Australian Research Council.
Reference
- 1.Coyte KZ, Schluter J, Foster KR. 2015. The ecology of the microbiome: networks, competition, and stability. Science 350, 663–666. ( 10.1126/science.aad2602) [DOI] [PubMed] [Google Scholar]
- 2.Arumugam M et al. 2011. Enterotypes of the human gut microbiome. Nature 473, 174–180. ( 10.1038/nature09944) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van de Guchte M, Blottière HM, Doré J. 2018. Humans as holobionts: implications for prevention and therapy. Microbiome 6, 81 ( 10.1186/s40168-018-0466-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferretti P et al. 2018. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 24, 133–145. ( 10.1016/j.chom.2018.06.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yassour M et al. 2018. Strain-level analysis of mother-to-child bacterial transmission during the first few months of life. Cell Host Microbe 24, 146–154. ( 10.1016/j.chom.2018.06.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. 2014. The placenta harbors a unique microbiome. Sci. Transl. Med. 6, 237ra65 ( 10.1126/scitranslmed.3008599) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Goffau MC, Lager S, Sovio U, Gaccioli F, Cook E, Peacock SJ, Parkhill J, Charnock-Jones DS, Smith GC. 2019. Human placenta has no microbiome but can contain potential pathogens. Nature 572, 329–334. ( 10.1038/s41586-019-1451-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makino H et al. 2013. Mother-to-infant transmission of intestinal bifidobacterial strains has an impact on the early development of vaginally delivered infant’s microbiota. PLoS ONE 8, e78331 ( 10.1371/journal.pone.0078331) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duranti S et al. 2017. Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome 5, 66 ( 10.1186/s40168-017-0282-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makino H et al. 2011. Transmission of intestinal Bifidobacterium longum subsp. longum strains from mother to infant determined by multilocus sequencing typing and amplified fragment length polymorphism. Appl. Environ. Microbiol., pages AEM–05346.
- 11.Stewart CJ et al. 2018. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 562, 583–588. ( 10.1038/s41586-018-0617-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yatsunenko T et al. 2012. Human gut microbiome viewed across age and geography. Nature 486, 222–227. ( 10.1038/nature11053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martín R, Jiménez E, Heilig H, Fernández L, Marín ML, Zoetendal EG, Rodríguez JM. 2009. Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR. Appl. Environ. Microbiol. 75, 965–969. ( 10.1128/AEM.02063-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sela D et al. 2008. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl Acad. Sci. USA 105, 18 964–18 969. ( 10.1073/pnas.0809584105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garrido D et al. 2016. A novel gene cluster allows preferential utilization of fucosylated milk oligosaccharides in Bifidobacterium longum subsp. longum SC596. Sci. Rep. 6, 35045 ( 10.1038/srep35045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duranti S et al. 2015. Insights from genomes of representatives of the human gut commensal Bifidobacterium bifidum. Environ. Microbiol. 17, 2515–2531. ( 10.1111/1462-2920.12743) [DOI] [PubMed] [Google Scholar]
- 17.Egan M, Motherway MO, Kilcoyne M, Kane M, Joshi L, Ventura M, van Sinderen D. 2014. Cross-feeding by Bifidobacterium breve ucc2003 during co-cultivation with Bifidobacterium bifidum PRL2010 in a mucin-based medium. BMC Microbiol. 14, 282 ( 10.1186/s12866-014-0282-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turroni F, Özcan E, Milani C, Mancabelli L, Viappiani A, van Sinderen D, Sela D, Ventura M. 2015. Glycan cross-feeding activities between bifidobacteria under in vitro conditions. Front. Microbiol. 6, 1030 ( 10.3389/fmicb.2015.01030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turroni F et al. 2016. Deciphering bifidobacterial-mediated metabolic interactions and their impact on gut microbiota by a multi-omics approach. ISME J. 10, 1656–1668. ( 10.1038/ismej.2015.236) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freitas AC, Hill JE. 2018. Bifidobacteria isolated from vaginal and gut microbiomes are indistinguishable by comparative genomics. PLoS ONE 13, e0196290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Odamaki T, Kato K, Sugahara H, Hashikura N, Takahashi S, Xiao J-z., Abe F, Osawa R. 2016. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol. 16, 90 ( 10.1186/s12866-016-0708-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holscher HD et al. 2012. Bifidobacterium lactis Bb12 enhances intestinal antibody response in formula-fed infants: a randomized, double-blind, controlled trial. J. Parenter. Enteral. Nutr. 36, 106S–117S. ( 10.1177/0148607111430817) [DOI] [PubMed] [Google Scholar]
- 23.Janzon A, Goodrich JK, Koren O, Waters JL, Ley RE, TEDDY Study Group. 2019. Interactions between the gut microbiome and mucosal immunoglobulins A, M, and G in the developing infant gut. Msystems 4, e00612–e00619. ( 10.1128/mSystems.00612-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukushima Y, Kawata Y, Hara H, Terada A, Mitsuoka T. 1998. Effect of a probiotic formula on intestinal immunoglobulin a production in healthy children. Int. J. Food Microbiol. 42, 39–44. ( 10.1016/S0168-1605(98)00056-7) [DOI] [PubMed] [Google Scholar]
- 25.Lundell A-C et al. 2012. Infant B cell memory differentiation and early gut bacterial colonization. J. Immunol. 188, 4315–4322. ( 10.4049/jimmunol.1103223) [DOI] [PubMed] [Google Scholar]
- 26.Yasui H, Nagaoka N, Mike A, Hayakawa K, Ohwaki M. 1992. Detection of bifidobacterium strains that induce large quantities of IgA. Microb. Ecol. Health Dis. 5, 155–162. [Google Scholar]
- 27.Mantis NJ, Forbes SJ. 2010. Secretory IgA: arresting microbial pathogens at epithelial borders. Immunol. Invest. 39, 383–406. ( 10.3109/08820131003622635) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Underdown BJ, Schiff JM. 1986. Immunoglobulin A: strategic defense initiative at the mucosal surface. Annu. Rev. Immunol. 4, 389–417. ( 10.1146/annurev.iy.04.040186.002133) [DOI] [PubMed] [Google Scholar]
- 29.Blaser MJ 2017. The theory of disappearing microbiota and the epidemics of chronic diseases. Nat. Rev. Immunol. 17, 461–463. ( 10.1038/nri.2017.77) [DOI] [PubMed] [Google Scholar]
- 30.Bezirtzoglou E, Tsiotsias A, Welling GW. 2011. Microbiota profile in feces of breast-and formula-fed newborns by using fluorescence in situ hybridization (FISH). Anaerobe 17, 478–482. ( 10.1016/j.anaerobe.2011.03.009) [DOI] [PubMed] [Google Scholar]
- 31.O’Callaghan A, van Sinderen D. 2016. Bifidobacteria and their role as members of the human gut microbiota. Front. Microbiol. 7, 925 ( 10.3389/fmicb.2016.00925) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dogra S et al. 2015. Dynamics of infant gut microbiota are influenced by delivery mode and gestational duration and are associated with subsequent adiposity. MBio 6, e02419–e02414 ( 10.1128/mBio.02419-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fallani M et al. 2011. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology 157, 1385–1392. ( 10.1099/mic.0.042143-0) [DOI] [PubMed] [Google Scholar]
- 34.Hesla HM, Stenius F, Jäderlund L, Nelson R, Engstrand L, Alm J, Dicksved J. 2014. Impact of lifestyle on the gut microbiota of healthy infants and their mothers–the ALADDIN birth cohort. FEMS Microbiol. Ecol. 90, 791–801. ( 10.1111/1574-6941.12434) [DOI] [PubMed] [Google Scholar]
- 35.Shao Y et al. 2019. Stunted microbiota and opportunistic pathogen colonization in Caesarean-section birth. Nature 574, 117–121. ( 10.1038/s41586-019-1560-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, Björkstén B, Engstrand L, Andersson AF. 2014. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut 63, 559–566. ( 10.1136/gutjnl-2012-303249) [DOI] [PubMed] [Google Scholar]
- 37.Salminen S, Gibson G, McCartney A, Isolauri E. 2004. Influence of mode of delivery on gut microbiota composition in seven year old children. Gut 53, 1388–1389. ( 10.1136/gut.2004.041640) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chu DM, Ma J, Prince AL, Antony KM, Seferovic MD, Aagaard KM. 2017. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med. 23, 314–326. ( 10.1038/nm.4272) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hill CJ et al. 2017. Evolution of gut microbiota composition from birth to 24 weeks in the INFANTMET cohort. Microbiome 5, 4 ( 10.1186/s40168-016-0213-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Falony G et al. 2016. Population-level analysis of gut microbiome variation. Science 352, 560–564. ( 10.1126/science.aad3503) [DOI] [PubMed] [Google Scholar]
- 41.Magne F, Hachelaf W, Suau A, Boudraa G, Mangin I, Touhami M, Bouziane-Nedjadi K, Pochart P. 2006. A longitudinal study of infant faecal microbiota during weaning. FEMS Microbiol. Ecol. 58, 563–571. ( 10.1111/j.1574-6941.2006.00182.x) [DOI] [PubMed] [Google Scholar]
- 42.Ding T, Schloss PD. 2014. Dynamics and associations of microbial community types across the human body. Nature 509, 357–360. ( 10.1038/nature13178) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Organization WH et al. 2008. Indicators for assessing infant and young child feeding practices: part 1-definition: conclusions of a consensus meeting held 6–8 November 2007 in Washington DC.
- 44.Ballyk MM, Jones DA, Smith HL. 2001. Microbial competition in reactors with wall attachment. Microb. Ecol. 41, 210–221. ( 10.1007/s002480000005) [DOI] [PubMed] [Google Scholar]
- 45.Freter R, Brickner H, Fekete J, Vickerman MM, Carey KE. 1983. Survival and implantation of Escherichia coli in the intestinal tract. Infect. Immun. 39, 686–703. ( 10.1128/IAI.39.2.686-703.1983) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young G, Ermentrout B, Rubin JE. 2015. A boundary value approach to optimization with an application to salmonella competition. Bull. Math. Biol. 77, 1327–1348. ( 10.1007/s11538-015-0087-3) [DOI] [PubMed] [Google Scholar]
- 47.Stein RR, Bucci V, Toussaint NC, Buffie CG, Rätsch G, Pamer EG, Sander C, Xavier JB. 2013. Ecological modeling from time-series inference: insight into dynamics and stability of intestinal microbiota. PLoS Comput. Biol. 9, e1003388 ( 10.1371/journal.pcbi.1003388) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, Huttenhower C. 2012. Microbial co-occurrence relationships in the human microbiome. PLoS Comput. Biol. 8, e1002606 ( 10.1371/journal.pcbi.1002606) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fisher CK, Mehta P. 2014. Identifying keystone species in the human gut microbiome from metagenomic timeseries using sparse linear regression. PLoS ONE 9, e102451 ( 10.1371/journal.pone.0102451) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heinken A, Thiele I. 2015. Systematic prediction of health-relevant human-microbial co-metabolism through a computational framework. Gut. Microbes 6, 120–130. ( 10.1080/19490976.2015.1023494) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Magnúsdóttir S et al. 2017. Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat. Biotechnol. 35, 81–89. ( 10.1038/nbt.3703) [DOI] [PubMed] [Google Scholar]
- 52.Shoaie S, Karlsson F, Mardinoglu A, Nookaew I, Bordel S, Nielsen J. 2013. Understanding the interactions between bacteria in the human gut through metabolic modeling. Sci. Rep. 3, 2532 ( 10.1038/srep02532) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bergström A, Skov TH, Bahl MI, Roager HM, Christensen LB, Ejlerskov KT, Mølgaard C, Michaelsen KF, Licht TR. 2014. Establishment of intestinal microbiota during early life: a longitudinal, explorative study of a large cohort of Danish infants. Appl. Environ. Microbiol. 80, 2889–2900. ( 10.1128/AEM.00342-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hara A, Iwasa Y. 2019. Coupled dynamics of intestinal microbiome and immune system: a mathematical study. J. Theor. Biol. 464, 9–20. ( 10.1016/j.jtbi.2018.12.021) [DOI] [PubMed] [Google Scholar]
- 55.David LA, Materna AC, Friedman J, Campos-Baptista MI, Blackburn MC, Perrotta A, Erdman SE, Alm EJ. 2014. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 15, R89 ( 10.1186/gb-2014-15-7-r89) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clemente JC et al. 2015. The microbiome of uncontacted Amerindians. Sci. Adv. 1, e1500183 ( 10.1126/sciadv.1500183) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. 2010. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl Aacd. Sci. USA 107, 14 691–14 696. ( 10.1073/pnas.1005963107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martínez I, Stegen JC, Maldonado-Gómez MX, Eren AM, Siba PM, Greenhill AR, Walter J. 2015. The gut microbiota of rural Papua New Guineans: composition, diversity patterns, and ecological processes. Cell Rep. 11, 527–538. ( 10.1016/j.celrep.2015.03.049) [DOI] [PubMed] [Google Scholar]
- 59.Schnorr SL et al. 2014. Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 5, 3654 ( 10.1038/ncomms4654) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huttenhower C et al. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. ( 10.1038/nature11234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Azad MB et al. 2016. Impact of maternal intrapartum antibiotics, method of birth and breastfeeding on gut microbiota during the first year of life: a prospective cohort study. Int. J. Gynecol. Obstetrics 123, 983–993. ( 10.1111/1471-0528.13601) [DOI] [PubMed] [Google Scholar]
- 62.Fouhy F et al. 2012. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob. Agents Chemother. 56, 5811–5820. ( 10.1128/AAC.00789-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bucci V, Bradde S, Biroli G, Xavier JB. 2012. Social interaction, noise and antibiotic-mediated switches in the intestinal microbiota. PLoS Comput. Biol. 8, e1002497 ( 10.1371/journal.pcbi.1002497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zeng Q, Rodrigo A. 2018. Neutral models of short-term microbiome dynamics with host subpopulation structure and migration limitation. Microbiome 6, 80 ( 10.1186/s40168-018-0464-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu GD et al. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108. ( 10.1126/science.1208344) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.David LA et al. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. ( 10.1038/nature12820) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smits SA et al. 2017. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science 357, 802–806. ( 10.1126/science.aan4834) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang C, Zhang M, Pang X, Zhao Y, Wang L, Zhao L. 2012. Structural resilience of the gut microbiota in adult mice under high-fat dietary perturbations. ISME J. 6, 1848–1857. ( 10.1038/ismej.2012.27) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and results generated in this study can be reproduced using the source code we provide on github https://github.com/xiyanxiongnico/Modelling-the-effect-of-birth-and-feeding-modes-on-the-development-of-human-gut-microbiota.git.