Abstract
SARS-CoV-2 is mutating and creating divergent variants by altering the composition of essential constituent proteins. Pharmacologically, it is crucial to understand the diverse mechanism of mutations for stable vaccine or anti-viral drug design. Our current study concentrates on all the constituent proteins of 469 SARS-CoV-2 genome samples, derived from Indian patients. However, the study may easily be extended to the samples across the globe.
We perform clustering analysis towards identifying unique variants in each of the SARS-CoV-2 proteins. A total of 536 mutated positions within the coding regions of SARS-CoV-2 proteins are detected among the identified variants from Indian isolates. We quantify mutations by focusing on the unique variants of each SARS-CoV-2 protein. We report the average number of mutation per variant, percentage of mutated positions, synonymous and non-synonymous mutations, mutations occurring in three codon positions and so on. Our study reveals the most susceptible six (06) proteins, which are ORF1ab, Spike (S), Nucleocapsid (N), ORF3a, ORF7a, and ORF8. Several non-synonymous substitutions are observed to be unique in different SARS-CoV-2 proteins. A total of 57 possible deleterious amino acid substitutions are predicted, which may impact on the protein functions. Several mutations show a large decrease in protein stability and are observed in putative functional domains of the proteins that might have some role in disease pathogenesis. We observe a good number of physicochemical property change during above deleterious substitutions.
Abbreviations: SARS, severe acute respiratory syndrome; CoV, coronaviruses; NS, non-synonymous; Syn, synonymous; CP, codon position; Mut, mutation; AA, amino acid; TM, transmembrane domain; NTD, N-terminal domain; CTD, C-terminal domain; HR, heptapeptide repeat
Keywords: COVID-19, Non-synonymous mutations, Codon position, Protein stability, Deleterious substitutions, Functional domain
1. Introduction
Due to the massive outbreak of COVID-19 disease, caused by the highly infectious novel coronavirus- SARS-CoV-2, the world is passing through a difficult situation. There are seven species of human coronaviruses reported so far that causing diseases in humans. Out of them, four species (HCoV-229E, HKU1, NL63 and OC43) causing mild respiratory apparatus infection which can easily be treated. However, three species, termed as beta coronaviruses (SARS-CoV, MERS-CoV, and SARS-CoV-2), are severe in nature, leads to potentially fatal consequences (Andersen et al., 2020). The scientific community trying hard to decipher parthenogenesis mechanism of SARS-CoV-2 and its therapeutic control, in silico, using various computational tools. An exhaustive study is available in (Das et al., 2020a).
Scientists observed a number of variants among novel coronavirus, SARS-CoV-2, reported from different geographical regions (Joshi and Paul, 2020; Sardar et al., 2020; Chang et al., 2020). Most of the evolutionary changes in the genome of viruses occur due to mutation. In some cases, it is due to insertion or deletion in the genome. In the course of evolution, variations bring novelty (Baer, 2008). The small variations might be beneficial or detrimental for the organism (Loewe and Hill, 2010). The mutational study helps in understanding viral transmission, replication efficiency, and magnitude of virulence of the pathogen (Eaaswarkhanth et al., 2020). A minor change in the genome might lead to the variation in functionality of constituent proteins of the organism (Chaudhuri, 2020). Previous studies revealed significant alternation in structural and pathogenic properties due to even single point mutation in virus proteins (André et al., 2019; Sakai et al., 2017). Characterizing mutations in different functional domains of SARS-CoV-2 genome might help in designing potential vaccine (Kaur et al., 2020).
Determining the mutation types (synonymous or non-synonymous) that influence a lot in gene regulation is vital for understanding the role of regulatory variation during evolution (DiMaio and Nathans, 1982; Foy et al., 2003). Studying the mutations at different codon positions is essential, particularly for quantification of synonymous and non-synonymous amino acid substitutions (Plotkin and Kudla, 2011). Though the non-synonymous mutation is primarily crucial (from codon usage bias point of view) as it alters the amino acid, synonymous mutations too have their strong impact (Plotkin and Kudla, 2011; Kristofich et al., 2018; Gustafsson et al., 2004). It is worth to mention that the changes in the physicochemical properties of nucleotides (purine-R or pyrimidine-Y) due to the mutations have remarkable biological significance (Lyons and Lauring, 2017; Sengupta et al., 2018; Guo et al., 2017). It is reported that in the case of codons, various evolutionary constraints at different codon positions occur due to the functional constraints imposed by the genetic code and the physicochemical properties of encoded amino acids (Bofkin and Goldman, 2007; Simmons, 2017; Plotkin and Kudla, 2011). For example, mutations at the 2nd position of a codon directly impact the changes in replaced amino acids (hydrophobic to hydrophilic and vice versa). The change is due to the transversion (A ↔ C or A ↔ T or G ↔ C or G ↔ T) (Haig and Hurst, 1991; Wolfenden et al., 1979; Błażej et al., 2017), although A ↔ G or C ↔ T transition is mostly occurring for single point mutation (Beletskii and Bhagwat, 1996; Błażej et al., 2017). Further, the changes in physicochemical properties of amino acids have a significant functional role (Das et al., 2019; Basak et al., 2017). Hence, understanding the genetic diversity is important that might hint towards the susceptible antigen targets of SARS-CoV-2. It can be used for potential therapeutic and prophylactic interventions in order to prevent this deadly outbreak. Mutations in SARS-CoV-2 proteins may lead to different phenotypic changes, and hence virus can adapt to new hosts and environments. In addition, codon bias study helps in revealing the host-virus interaction mechanism in SARS-CoV-2 (Dilucca et al., 2020; Kurland, 1991; Das et al., 2020b).
A detailed in-silico study on putative mutations in SARS-CoV-2 is of utmost important to understand any significant pattern and its possible impact on the functional and structural characteristics of the virus.
India is the second-largest SARS-CoV-2 infected country in the world. Due to the volume of study, we restricted our current study within Indian isolates only. However, our study can easily be extended to other variants from any part of the world. Although a couple of studies have been carried out to learn various crucial facts about SARS-CoV-2 genome from Indian patient samples (Kaur et al., 2020; Saha et al., 2020; Samaddar et al., 2020), there are certain facts yet to explore. Therefore, in this work, we broadly focused on the mutational study on SARS-CoV-2 genomes, isolated from Indian patient, as discussed in the following section.
2. Material and methods
2.1. Collection of SARS-CoV-2 genome sequences extracted from Indian patients
We collect SARS-CoV-2 genome sequences isolated from Indian patients that are achieved in public repositories. Several protein-coding genes are present in each SARS-CoV-2 genome. SARS-CoV-2 encodes different types of essential proteins: (i) nonstructural proteins - polyprotein (ORF1ab), structural proteins - Spike glycoprotein (S), Envelope (E), Membrane (M) and Nucleocapsid (N), and accessory proteins - ORF3a, ORF6, ORF7a, ORF7b, ORF8, and ORF10 (Kim et al., 2020; Yadav et al., 2020; Ruan et al., 2003; Gordon et al., 2020). A complete topological structure (position) of all SARS-CoV-2 proteins is shown in Table 1. Each of these proteins is highly essential and has diverse functional roles. The first full genome sequence of SARS-CoV-2 virus from India sample was reported during February 2020 (Yadav et al., 2020). We collect sequences from NCBI database1 (Supplementary-1). We find around 469 complete SARS-CoV-2 nucleotide sequences. Protein wise, we extract the coding region from each nucleotide sequence and ignore noisy sequences. The final list of obtained unique sequences is utilized for sub-sequence analysis (Table 2).
Table 1.
Topological structure of all SARS-CoV-2 proteins shown by respective genomic location. For each protein, the range of CDS region and amino acids used in this paper are numbered starting from 1 to length of the nucleotide or protein sequence.
| Gene/protein | Genome location (nucleotide) | Protein length (aa) | Nucleotide location used | Amino acid location used |
|---|---|---|---|---|
| ORF1ab | 266–21,555 | 7096 | 1–21,288 | 1–7096 |
| S | 21,563–25,384 | 1273 | 1–3819 | 1–1273 |
| ORF3a | 25,393–26,220 | 275 | 1–825 | 1–275 |
| E | 26,245–26,472 | 75 | 1–225 | 1–75 |
| M | 26,523–27,191 | 222 | 1–666 | 1–222 |
| ORF6 | 27,202–27,387 | 61 | 1–183 | 1–61 |
| ORF7a | 27,394–27,759 | 121 | 1–363 | 1–121 |
| ORF7b | 27,756–27,887 | 43 | 1–129 | 1–43 |
| ORF8 | 27,894–28,259 | 121 | 1–363 | 1–121 |
| N | 28,274–29,533 | 419 | 1–1257 | 1–419 |
| ORF10 | 29,558–29,674 | 38 | 1–114 | 1–38 |
Table 2.
The number of collected samples from Indian isolates, unique variant, sample to variant ratio in each SARS-CoV-2 protein.
| Protein | # collected samples | # noise free samples | # unique variant | Sample to variant ratio |
|---|---|---|---|---|
| ORF1ab | 462 | 400 | 262 | 1.52 |
| E | 460 | 445 | 3 | 148.33 |
| M | 460 | 457 | 18 | 25.38 |
| N | 463 | 455 | 53 | 8.58 |
| S | 462 | 436 | 90 | 4.84 |
| ORF3a | 459 | 445 | 33 | 13.48 |
| ORF6 | 460 | 459 | 3 | 153.0 |
| ORF7a | 460 | 454 | 11 | 41.27 |
| ORF7b | 456 | 455 | 3 | 151.66 |
| ORF8 | 461 | 451 | 11 | 41.00 |
| ORF10 | 460 | 460 | 3 | 153.33 |
2.2. Workflow design
Protein specific nucleotide sequences are first clustered to extract set of unique sequences (or unique variants). Next, unique sequences (representative of each group) are aligned using multiple sequence alignment. As a reference sequence we use sequence of SARS-CoV-2 proteins from Wuhan-Hu-1 (accession no: NC_045512). We compare every variant with the reference sequence to identify and localize mutations. We consider only single point mutation as a substitution. Observed mutations are then analyzed based on the number of synonymous and non-synonymous substitutions, quantification of nucleotide mutations in three different codon positions (1st/2nd/3rd), type of nucleotide mutations and amino acid substitutions. We then characterize non-synonymous amino acid substitutions and their biological implications using various computational tools.
2.3. Computational tools and techniques used
We use web-based tool PROVEAN2 and I-mutant3 for functional assessment of single point mutation. PROVEAN (Protein Variation Effect Analyzer), a web server, is used to predict any non-synonymous amino acid substitution or indel impacts on the biological function of a protein (Choi et al., 2012). The tool predicts two kinds of substitution effects: deleterious effect and neutral effect on protein function by measuring the combined score of substitution matrix, alignment, the position of substitution with the neighborhood that surrounds the site of variation. The cut-off value of the PROVEAN score is set as −2.5, below which it indicates deleterious substitution, otherwise, neutral. For predicting stability changes due to mutation, we use I-Mutant (Capriotti et al., 2005). The tool is designed based on Support Vector Machine (SVM) that produces Gibbs free energy of unfolding (ΔΔG value in kcal/mol, in terms of increased or decreased stability) for each non-synonymous substitution. The stability predictors value ΔΔG < − 0.5 indicates high decrease in stability, whereas, ΔΔG > 0.5 indicates high increase in stability, and −0.5 < ΔΔG ≤ 0.5 signifies neutral stability.
We use simple Python scripting for rest of the quantitative analysis.
We report the functionally important mutations identified using the above tools, highlighting the various putative functional domains of SARS-CoV-2 proteins. We also study wild type and new amino acid changes in two categories of physicochemical properties, Hydropathy profile (Aftabuddin and Kundu, 2007), and side-chain structure (Das et al., 2016). The categorizations are as follows:
-
•
Hydropathy based classes: The three classes are Hydrophobic (F, M, W, I, V, L, P, A), Hydrophilic (N, C, Q, G, S, T, Y), and Charged (R, D, E, H, K).
-
•
Side-Chain based classes: According to this grouping, twenty (20) amino acids are clustered into eight groups as Acidic (D, E), Basic (R, H, K), Aromatic (F, W, Y), Aliphatic (A, G, I, L, V), Cyclic (P), Sulfur-containing (C, M), Hydroxyl-containing (S, T), and Acidic amide (N, Q).
3. Results and discussion
Our first objective is to find out unique variants by clustering the SARS-CoV-2 gene sequences. We then identify point mutation (as substitution) in each observed variant by comparing it with the reference sequence. Observed mutations occurring at different codon positions are then classified and quantified based on different perspectives as discussed below.
3.1. Clustering of unique variants
The majority of the input genomes are redundant with respect to sequence similarity. We cluster them based on sequence similarity and consider a sequence from each cluster as cluster representative (termed as unique variant). We use a string matching technique to cluster the sequences, where exactly similar sequences are put in a single cluster (Table 2). The cardinality of each cluster indicates the number of similar sequences in that cluster. We report clusters by variant numbering i.e., v1, v2⋯vn; n is the number of clusters or variants for each SARS-CoV-2 protein. The group of similar sequences belonging to a cluster (or variant) for each SARS-CoV-2 protein is reported with accession numbers (Supplementary-1). We draw phylogenetic tree for each SARS-CoV-2 protein taking all distinct variants and report in Supplementary-2. Our analysis shows that distinct variants in ORF1ab, S, N and ORF3a proteins are comparatively higher than other SARS-CoV-2 proteins, signifying that such proteins are highly susceptible.
3.2. Indian vs. world-wide variants
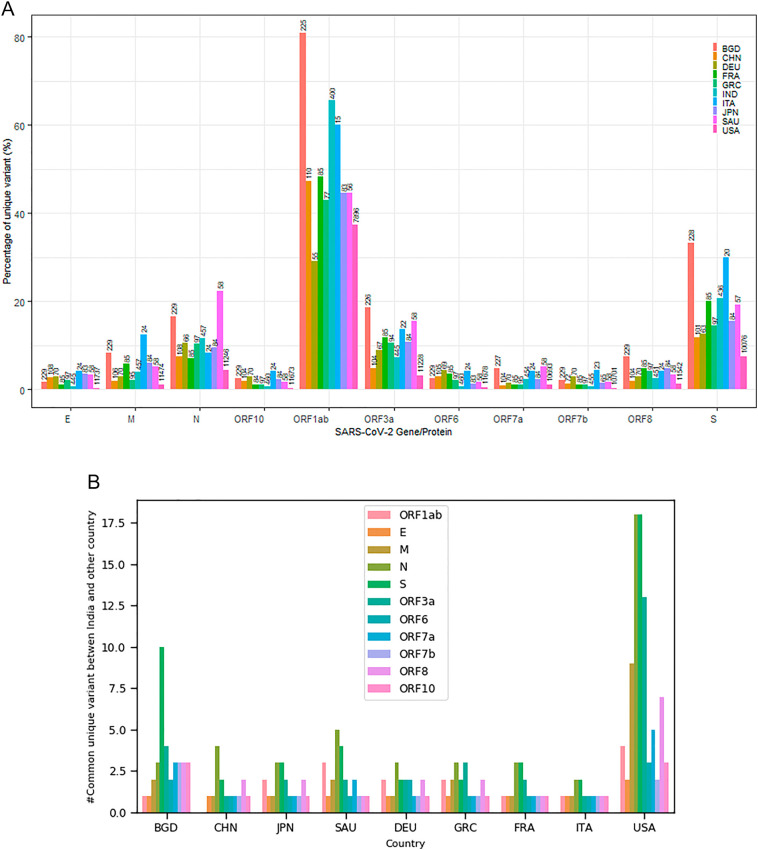
We compare Indian variants with the variants collected from nine (09) major countries such as China (CHN), Bangladesh (BGD), Japan (JPN), Saudi Arabia (SAU), French (FRA), Germany (DEU), Greece (GRC), Italy (ITA), and United States (USA). The protein-specific unique variants observed from all the above countries are reported in Fig. 1(A). We observe a high percentage of unique variants in BGD isolates, followed by Indian isolates. However, the percentage may be an indicator (not conclusive) as the total sample available is non-uniform. We even quantify common variants across nine different countries that are matching with Indian variants is reported in Fig. 1(B). Interestingly, common protein-specific variants are relatively rare while comparing with variants from different countries.
Fig. 1.
Comparison of unique variant among ten different countries. (A) Percentage of unique variant in each SARS-CoV-2 protein. The number at the top of the bar indicates the number of noise-free collected samples; (B) number of common unique variant between India and other country.
3.3. Quantification of observed mutations in SARS-CoV-2 proteins
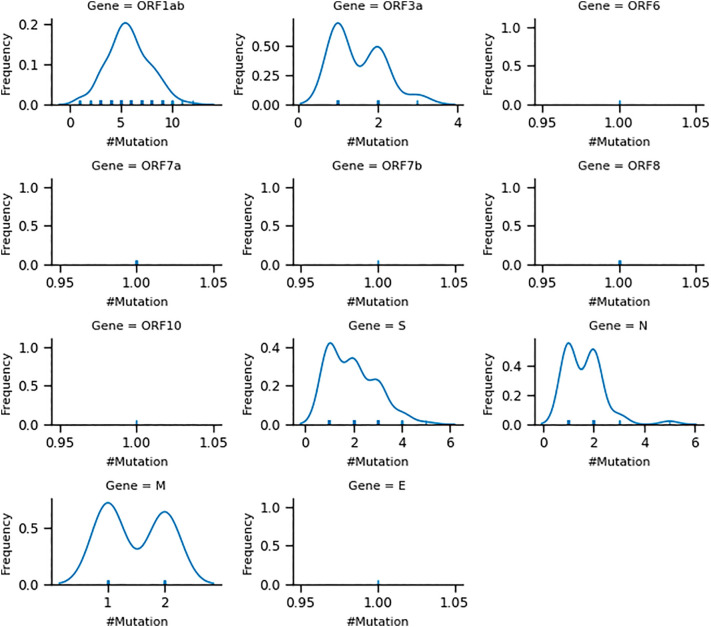
Among the distinct variants in each SARS-CoV-2 protein, we consider a particular variant as a reference sequence (exactly similar to NC_045512) except ORF1ab protein. We then compare other variants for studying nucleotide level substitutions. The frequency distribution of the number of mutations for each protein is shown in Fig. 2. We observe at least one mutation in case of five proteins (ORF1ab, ORF3a, S, N, M). The average number of mutations per variant for such proteins is relatively higher (Fig. 3). In case of other six proteins (ORF6, ORF7a, ORF7b, ORF8, ORF10, and E), we observe only single mutation in each variant. Upon examining mutations in SARS-CoV-2 proteins, we observe several substitutions, the majority of which are associated with a single variant. The protein wise mutations are highlighted and reported for all the variants associated with more than one samples (Supplementary-3). We list mutations considering only M, N, and S proteins (having mutations in more than one sample). In the case of E protein, only a single mutation is observed in all the variants. In case of accessory proteins, mutations in more than one sample are observed in ORF3a, and ORF8. Most of which are from the non-synonymous category and having more than one sample frequency. We discuss below few top variants and mutations observed in our candidate SARS-CoV-2 proteins.
-
•
ORF1ab protein: We observe several mutations in ORF1ab because this protein is a polyprotein that consists of sixteen non-structural proteins. We compare all Indian SARS-CoV-2 ORF1ab protein variants with the reference sequence (NC_045512). We observe mutation in 40 variants that are associated with more than one sample. The top non-synonymous mutation ([C14144T, P4715L]) is observed in 233 variants of total 359 samples. Here, the first numbering in bracket refers to the nucleotide mutation position, whereas the second numbering refers to the amino acid substitution position. The majority of the mutations are synonymous. Several non-synonymous mutations are observed associated with five or more samples, which are [A2027C, Q676P], [C18304T, L6102F], [C18890T, T6297I], [G15814A, V5272I], [G10818T, L3606F], [C6047A, T2016K], [C13466T, A4489V], [G4601T, S1534I], [C9173T, T3058I], [C14161A, L4721I]. Further, several synonymous mutations observed in more than one sample are [C2772T, F924F], [C18613T, L6205L], [C2571T, C857C], [G4035T, V1345V], [A16248G, L5416L], [C15060T, N5020N], [C3369T, N1123N], [C3819T, D1273D], [C8517T, S2839S], [C11355T, F3785F].
-
•
Envelope (E) protein: In the case of E protein, a total 443 (out of 445) variants are observed that are exactly matching with the reference sequence (NC_045512). The only two non-synonymous mutations of single instance are [G184T, V62F] and [G223T, V75F].
-
•
Membrane (M) protein: A total 224 (out of 457) matching samples (with NC_045512) are found in Indian genome. In M protein, significant synonymous mutations are observed. [C213T, Y71Y] observe in 9 variants of total 223 (≈) 50% samples. In addition, one non-synonymous ([C425T, A142V] in 2 variants of total two samples) and one synonymous ([G429T, V143V] in 2 variants of total four samples) are also observed.
-
•
Nucleocapsid (N) protein: A total of 204 (out of 455) matching sequences of N protein are observed in Indian samples. We observe mutations in fifty three (53) variants. Each of them is associated with more than one sample. The mutation in the top variant (v2) is [C581T, S194L], which is found in 19 variants of a total of 158 samples. The other important non-synonymous mutations associated in more than one sample are [C581T, S194L], [C38T, P13L], [G605A, S202N], [G608A, R203K], [G609A, R203K], [G610C, G204R], [C614T, T205I], [G578T, S193I], and one synonymous mutations is [G578T, S193I] observed in 2.
-
•
Spike (S) protein: We observe only 11 samples (out of 436) that are exactly similar to the reference S protein. A total of twenty (20) variants in S protein are found to be associated with mutations in more than one sample. Mutations in each variant show either synonymous or non-synonymous or both the categories. For example, we observe mutations in variant v2 [A1841G, D614G] and [T2367C, Y789Y] that are non-synonymous and synonymous, respectively, and associated with 164 samples. Similarly, the variant (v3) shows two synonymous mutations ([C882T, D294D] and [T2367C, Y789Y]), and two non-synonymous mutations ([G162T, L54F] and [A1841G, D614G]) found in 63 samples. Few findings are consistent with the previously reported results. For example, D614G substitution is observed ≈60% in Indian samples (Saha et al., 2020). In our candidate dataset, we observe D614G substitution in ≈ 93% samples covering 77 variants. The majority of the substitutions are in variant v2, along with a synonymous mutation [T2367C, Y789Y] in the same variant. Few other important mutations are found in five and more samples, which are three non-synonymous mutations ([G162T, L54F], [G1749T, E583D], [G2031T, Q677H]) and three synonymous mutation ([T2367C, Y789Y], [C882T, D294D], [G906T, T302T], [T328C, L110L]).
-
•
ORF3a protein: In ORF3a protein, we observe only 190 samples (out of 445) in Indian SARS-CoV-2 S proteins, which are exactly similar to the reference sequence. We observe mutations in ORF3a protein of eight (08) variants associated with more than one sample. The top variant is v2 with only non-synonymous mutation [G171T, Q57H] in 17 variants of a total of 234 samples. This non-synonymous mutation (Q57H) is found in Ion channels domain and consistence with previous study (Issa et al., 2020), and shows quite higher percentage ((53%)) in Indian SARS-CoV-2 genome as compared to 17.43% a global study reported in (Issa et al., 2020). Although, another mutation G251V is also found 9.71% of the genomes but, we did not observe this mutation in the Indian SARS-CoV-2 candidate genome. The other observed important mutations associated with more than one sample, where six mutations are non-synonymous ([C121T, L41F], [C277T, H93Y], [G67T, A23S], [C452T, T151I], [G463T, D155Y], [C512T, S171L]), and only one synonymous mutation is [C246T, N82N].
-
•
ORF6 protein: In the case of ORF6 protein, 457 (out of 459) sequences of SARS-CoV-2 Indian samples are exactly matching with the reference sequence. Similar to E protein, we observe two mutations, each associated with only one sample, one is synonymous ([C12T, L4L]), and the other is non-synonymous ([G39T, E13D]).
-
•
ORF7a and ORF7b proteins: All the sequences (except two) from the Indian SARS-CoV-2 genome for both the proteins are matched with the reference sequence (NC_045512). Two non-synonymous mutations are observed ORF7a protein associated with two samples each of in a single variant ([C280G, Q94E], [G283A, E95K]). In ORF7b protein, only two non-synonymous are [C92T, S31L] and [G127A, A43T] with an equal number of samples and variants (only 1).
-
•
ORF8 protein: Majority (423 out of 451) of the sequences are similar with the reference sequence. The top two variants are v2 (non-synonymous mutation: [T251C, L84S]) with sample frequency 19, and v3 (synonymous mutation: [G108T, P36P]) with sample frequency 2.
-
•
ORF10 protein: In ORF10 protein, 457 out of 460 sequences from the Indian SARS-CoV-2 genome are similar to the reference sequence. The only non-synonymous mutation is [L37F] and synonymous mutation is [C109T] with sample frequency 2 and 1.
Fig. 2.
Distribution of observed number of mutations (x-axis) and relative frequency of number of variants (y-axis) for each SARS-CoV-2 protein. The five proteins ORF1ab, ORF3a, S, N, M are observed multiple mutations in different variants, whereas in six proteins, ORF6, ORF7a, ORF7b, ORF8, ORF10 and E are found exactly a single mutation in each variant.
Fig. 3.
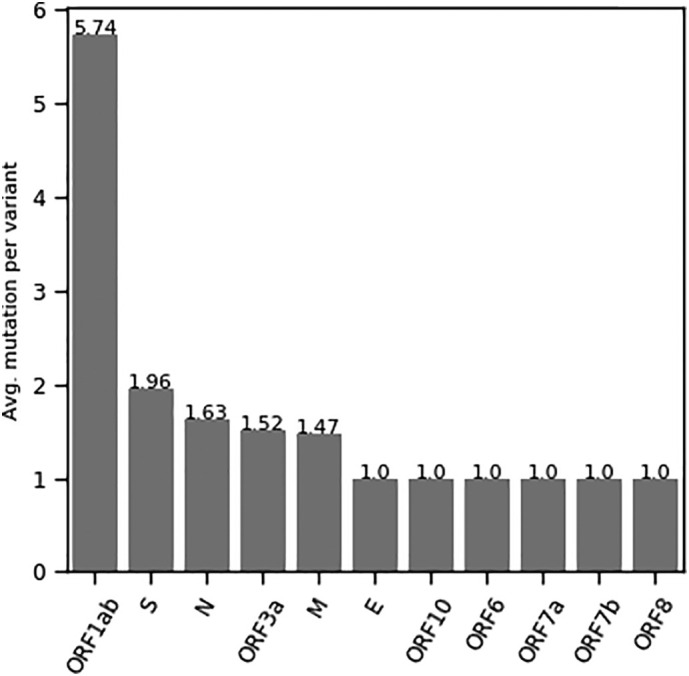
Average number of mutation per variant. Proteins are ranked by avg. mutation, highest (left) to lowest (right).
It can be noted that some of the observed mutations in different variants are common (Supplementary-3). Therefore, with respect to mutation types those variants are highly similar. However, we observe a total of 536 mutated positions located in different SARS-CoV-2 proteins in Indian isolates (Table 3). It is noted that the ORF3a protein shows the highest (≈3.96%) number of mutated locations followed by N protein. We observe a few numbers of mutated locations in E, ORF6, ORF7b and ORF10 proteins.
Table 3.
Number of mutated positions (or locations) in each SARS-CoV-2 protein.
| Gene | Gene length | # mutated position | Mutated position (%) |
|---|---|---|---|
| ORF1ab | 21,291 | 328 | 1.541 |
| E | 228 | 2 | 0.877 |
| M | 669 | 15 | 2.242 |
| N | 1260 | 49 | 3.889 |
| S | 3822 | 83 | 2.172 |
| ORF3a | 828 | 33 | 3.986 |
| ORF6 | 186 | 2 | 1.075 |
| ORF7a | 336 | 10 | 2.976 |
| ORF7b | 132 | 2 | 1.515 |
| ORF8 | 366 | 10 | 2.732 |
| ORF10 | 117 | 2 | 1.709 |
3.4. Characterizing the mutations into synonymous and non-synonymous categories
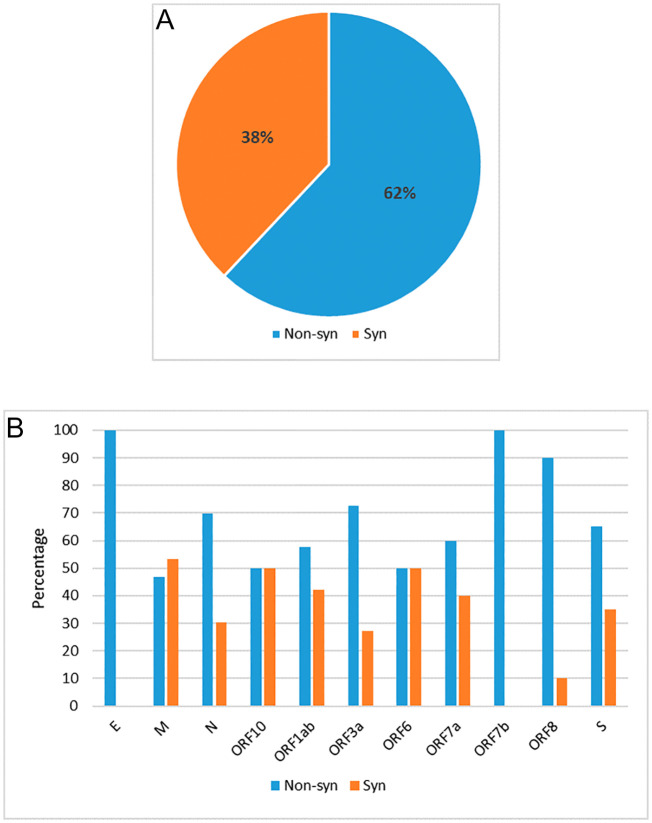
We account for both synonymous and non-synonymous mutations irrespective of any codon positions. Among the observed nucleotide mutations, 541 nucleotide mutations in 536 locations are then characterized in synonymous and non-synonymous categories (see Fig. 4(A)). Overall, percentage of non-synonymous mutation is more (≈62%, count-333) in comparison to synonymous mutations (≈38%, count-208). Observed mutations by the percentage of synonymous and non-synonymous category for all SARS-CoV-2 proteins are shown in Fig. 4(B). Overall, the non-synonymous category percentage is more (except for M protein), where E and ORF7b proteins show 100% non-synonymous mutations.
Fig. 4.
Quantification of synonymous and non-synonymous mutation. (A) Percentage of synonymous vs. non-synonymous mutation type in three codon positions taking all proteins together; (B) percentage of non-synonymous and synonymous mutation type in all SARS-CoV-2 protein.
3.5. Quantifying mutations in three different positions of codon
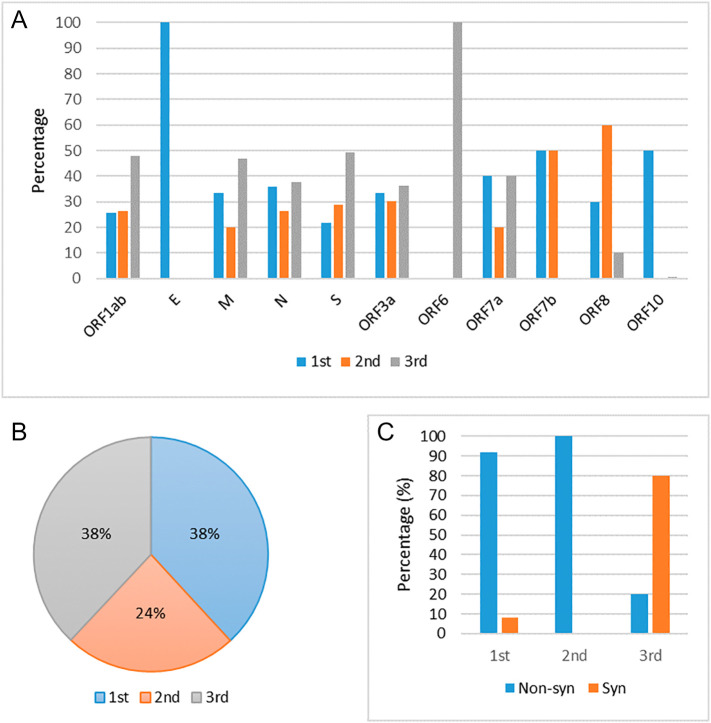
In case of any coding region mutation may occurs at any three different codon positions. Mutations at the third (3rd) position of the codon are almost synonymous that is the least functionally constrained. In contrast, the majority of the mutations at 1st and 2nd positions of the codon are non-synonymous that alter amino acid. The second codon position is the most functionally constrained as any change to the second codon position causes a non-synonymous change in the coding sequence. We observe mutations in all the three codon positions for ORF1ab, M, N, S, ORF3a, and ORF8 genes (Fig. 5(A)). In, and ORF10 genes, mutations are observed in 1st, 3rd, and 1st codon positions, respectively. We observe mutations in 1st and 2nd positions of ORF7b codons. We do not observe any mutation at the 2nd position of codon. It is worth mentioning that most highly mutated genes (ORF1ab, M, N, S, ORF3a) show a higher percentage of mutations at the third position of the codon, i.e., all these are in the synonymous category.
Fig. 5.
Percentage of mutation in SARS-CoV-2 proteins in each codon position (1st, 2nd and 3rd). (A) Protein-wise in each codon position, and (B) aggregate by all proteins in three different codon positions; (C) overall percentage of synonymous vs. non-synonymous mutation taking all codon positions and proteins.
We account overall mutations that are taking place in all SARS-CoV-2 proteins (Fig. 5(B)). Mutations at 1st and 3nd positions of codons are found almost equal (38%), whereas mutations at 2nd position are comparatively less (24%). More than 90% mutations at 1st codon position are non-synonymous, whereas around 80% mutations at 3rd codon position are synonymous (Fig. 5(C)). Protein-wise the mutations at three different codon positions are reported in Table 4 . In case of non-synonymous mutations, the percentage of mutation at 1st codon position is more (≥50%) for E, ORF10, and ORF7b protein, whereas at 2nd codon position, the mutation percentage is more (≥50%). For ORF8 and ORF7b, and ORF6 highest percentage of mutation occurs at the 3rd codon position.
Table 4.
Percentage of synonymous (syn) and non-synonymous (non-syn) mutation in three different codon positions (CP)-1st/2nd/3rd in each of the SARS-CoV-2 protein.
| Protein | CP | Type | Percentage |
|---|---|---|---|
| E | 1st | Non-syn | 100.00 |
| M | 1st | Non-syn | 26.67 |
| M | 1st | Syn | 6.67 |
| N | 1st | Non-syn | 33.96 |
| N | 1st | Syn | 1.89 |
| ORF10 | 1st | Non-syn | 50.00 |
| ORF1ab | 1st | Non-syn | 22.80 |
| ORF1ab | 1st | Syn | 2.74 |
| ORF3a | 1st | Non-syn | 33.33 |
| ORF7a | 1st | Non-syn | 40.00 |
| ORF7b | 1st | Non-syn | 50.00 |
| ORF8 | 1st | Non-syn | 30.00 |
| S | 1st | Non-syn | 20.48 |
| S | 1st | Syn | 1.20 |
| M | 2nd | Non-syn | 20.00 |
| N | 2nd | Non-syn | 26.42 |
| ORF1ab | 2nd | Non-syn | 26.44 |
| ORF3a | 2nd | Non-syn | 30.30 |
| ORF7a | 2nd | Non-syn | 20.00 |
| ORF7b | 2nd | Non-syn | 50.00 |
| ORF8 | 2nd | Non-syn | 60.00 |
| S | 2nd | Non-syn | 28.92 |
| M | 3rd | Syn | 46.67 |
| N | 3rd | Non-syn | 9.43 |
| N | 3rd | Syn | 28.30 |
| ORF10 | 3rd | Syn | 50.00 |
| ORF1ab | 3rd | Non-syn | 8.51 |
| ORF1ab | 3rd | Syn | 39.51 |
| ORF3a | 3rd | Non-syn | 9.09 |
| ORF3a | 3rd | Syn | 27.27 |
| ORF6 | 3rd | Non-syn | 50.00 |
| ORF6 | 3rd | Syn | 50.00 |
| ORF7a | 3rd | Syn | 40.00 |
| ORF8 | 3rd | Syn | 10.00 |
| S | 3rd | Non-syn | 15.66 |
| S | 3rd | Syn | 33.73 |
3.6. Characterizing nucleotide mutation types in non-synonymous category
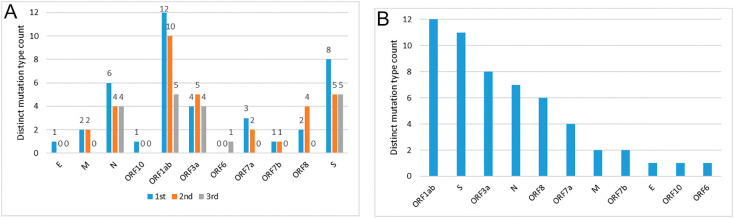
There are twelve possible nucleotide changes that can occur due to nucleotide mutation (see Methods and Materials). Protein-wise observed mutation counts in three different codon positions are shown in Fig. 6 (A). A majority of the nucleotide mutations are observed in the 1st codon position. Considering three codon positions (Fig. 6(A)), all 12 nucleotide mutation are observed in ORF1ab proteins followed by S proteins. ORF3a and N show comparatively fewer number of mutations (8 and 7 respectively). Similarly, in E, ORF10, and ORF6 proteins only a single mutation is observed.
Fig. 6.
(A) Number of distinct mutation type count shown for non-synonymous category in each protein and each codon position; (B) number of distinct mutation type count shown for non-synonymous category by aggregate all codon positions.
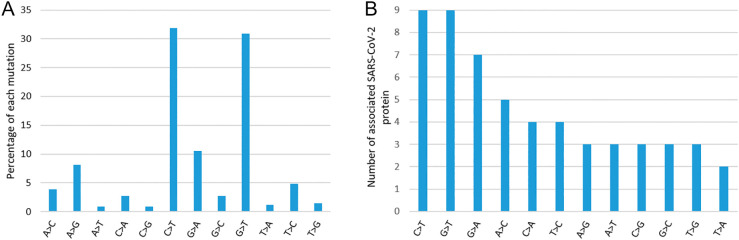
Quantification of nucleotide mutation type shows higher for G > T and C > T, and protein hit count also observer maximum for these two mutation types (Fig. 7(A) and (C)). In terms of percentage (considering all codon positions), the mostly occurring two mutations are C > T (≈32%) and G > T (≈30%). Further, these two mutations (C > T and G > T) are observed in 9 (out of taken 11) SARS-CoV-2 proteins followed by G > A (07) and A > C (05) mutations, respectively. T > A mutation is observed to be rare (only 2). Among the two above mostly occurring mutations, G > T is observed within the top two positions (by percentage) in all three codon positions (Table 5 ), whereas C > T is observed only in 1st and 2nd codon positions. The percentage of abundance of other two important mutations, G > A (in 1st codon position) and A > G (in 2nd codon position), is 17% and 12%, respectively. Further, we observe diversity in different codon positions in individual protein (Table 6 ). For example, G > T is mostly occurring at 2nd codon position (ORFF1ab, S), at the 1st codon position (E, M, N ORF3a), at 3rd codon position (ORF6).
Fig. 7.
The quantification of nucleotide mutation type in non-synonymous category. (A) Percentage of each type of nucleotide mutation; (B) mutation type by associate number of SARS-CoV-2 protein count.
Table 5.
The percentage of nucleotide mutation type for all non-synonymous cases shown for three codon positions independently and arranged by highest to lowest percentage. Mut-type: Mutation type;
| Codon position-1st |
Codon position-2nd |
Codon position-3rd |
|||
|---|---|---|---|---|---|
| Mut-type | Percentage | Mut-type | Percentage | Mut-type | Percentage |
| G>T | 33.09 | C>T | 48.98 | G>T | 68 |
| C>T | 25.00 | G>T | 16.33 | A>C | 6 |
| G>A | 16.91 | A>G | 12.24 | G>A | 6 |
| A>G | 6.62 | T>C | 7.48 | G>C | 6 |
| A>C | 5.88 | G>A | 6.12 | C>A | 4 |
| T>C | 3.68 | C>A | 2.72 | T>A | 4 |
| G>C | 2.94 | A>C | 1.36 | T>G | 4 |
| C>A | 2.21 | G>C | 1.36 | A>T | 2 |
| C>G | 1.47 | T>G | 1.36 | ||
| A>T | 0.74 | A>T | 0.68 | ||
| T>A | 0.74 | C>G | 0.68 | ||
| T>G | 0.74 | T>A | 0.68 | ||
Table 6.
Percentage of nucleotide mutation type for all non-synonymous cases shown by three codon positions for all proteins. CP: codon position; Mut-type: mutation type.
| Protein | CP | Mut-type | Percentage |
|---|---|---|---|
| ORF1ab | 2 | C>T | 23.16 |
| 3 | G>T | 11.58 | |
| 1 | C>T | 10.53 | |
| 1 | G>T | 10.53 | |
| 2 | A>G | 8.42 | |
| 1 | G>A | 7.37 | |
| 2 | T>C | 4.74 | |
| 1 | A>G | 4.21 | |
| 2 | G>A | 2.63 | |
| 2 | G>T | 2.63 | |
| 1 | A>C | 2.11 | |
| 1 | T>C | 2.11 | |
| 2 | C>A | 1.58 | |
| 2 | A>C | 1.05 | |
| 3 | C>A | 1.05 | |
| 3 | G>C | 1.05 | |
| 3 | A>C | 0.53 | |
| 2 | A>T | 0.53 | |
| 1 | C>A | 0.53 | |
| 1 | C>G | 0.53 | |
| 3 | G>A | 0.53 | |
| 1 | G>C | 0.53 | |
| 1 | T>A | 0.53 | |
| 2 | T>A | 0.53 | |
| 1 | T>G | 0.53 | |
| 2 | T>G | 0.53 | |
| E | 1 | G>T | 100.00 |
| M | 1 | C>T | 28.57 |
| 2 | C>T | 28.57 | |
| 1 | G>T | 28.57 | |
| 2 | G>T | 14.29 | |
| N | 2 | C>T | 18.92 |
| 1 | G>T | 18.92 | |
| 1 | G>A | 10.81 | |
| 2 | G>T | 10.81 | |
| 1 | C>T | 8.11 | |
| 2 | G>A | 5.41 | |
| 1 | G>C | 5.41 | |
| 3 | G>T | 5.41 | |
| 3 | A>C | 2.70 | |
| 1 | A>G | 2.70 | |
| 1 | C>A | 2.70 | |
| 3 | G>A | 2.70 | |
| 2 | G>C | 2.70 | |
| 3 | G>C | 2.70 | |
| S | 2 | C>T | 18.52 |
| 2 | G>T | 18.52 | |
| 3 | G>T | 14.81 | |
| 1 | G>T | 12.96 | |
| 1 | C>T | 5.56 | |
| 1 | A>C | 3.70 | |
| 2 | A>G | 3.70 | |
| 3 | T>A | 3.70 | |
| 3 | A>C | 1.85 | |
| 1 | A>T | 1.85 | |
| 1 | C>A | 1.85 | |
| 1 | G>A | 1.85 | |
| 2 | G>A | 1.85 | |
| 3 | G>A | 1.85 | |
| 1 | G>C | 1.85 | |
| 2 | G>C | 1.85 | |
| 1 | T>C | 1.85 | |
| 3 | T>G | 1.85 | |
| ORF3a | 1 | G>T | 20.83 |
| 1 | C>T | 16.67 | |
| 2 | C>T | 16.67 | |
| 2 | G>T | 12.50 | |
| 1 | A>C | 4.17 | |
| 3 | A>T | 4.17 | |
| 2 | C>G | 4.17 | |
| 1 | G>A | 4.17 | |
| 3 | G>T | 4.17 | |
| 2 | T>C | 4.17 | |
| 2 | T>G | 4.17 | |
| 3 | T>G | 4.17 | |
| ORF6 | 3 | G>T | 100.00 |
| ORF7a | 1 | G>A | 33.33 |
| 1 | C>G | 16.67 | |
| 1 | C>T | 16.67 | |
| 2 | C>T | 16.67 | |
| 2 | G>T | 16.67 | |
| ORF8 | 2 | C>T | 33.33 |
| 1 | G>T | 22.22 | |
| 1 | A>C | 11.11 | |
| 2 | C>A | 11.11 | |
| 2 | G>A | 11.11 | |
| 2 | T>C | 11.11 | |
| ORF7b | 2 | C>T | 50.00 |
| 1 | G>A | 50.00 | |
| ORF10 | 1 | C>T | 100.00 |
3.7. Quantification of non-synonymous amino acid substitutions
As highlighted earlier, there are total 380 amino acid substitutions. Out of 333 non-synonymous substitutions, we observe only 86 distinct substitutions in Indian SARS-CoV-2 genome (Table 7 ).
Table 7.
Amino acid substitution type by associated protein and number of mutated locations in that protein.
| Substitution type | Protein (#mutated position) |
|---|---|
| A>D | ORF1ab-(2) |
| A>S | M-(1),N-(2),ORF1ab-(4),ORF3a-(3),ORF8-(1),S-(3) |
| A>T | ORF1ab-(2),ORF7b-(1) |
| A>V | M-(2),N-(2),ORF1ab-(16),ORF3a-(1),ORF8-(2),S-(4) |
| C>F | ORF1ab-(1),S-(3) |
| C>Y | ORF1ab-(1) |
| D>E | ORF1ab-(1) |
| D>G | ORF1ab-(4) |
| D>N | N-(1),ORF1ab-(2) |
| D>Y | N-(3),ORF1ab-(6),ORF3a-(1),S-(3) |
| E>D | ORF1ab-(5),ORF6-(1),S-(2) |
| E>G | ORF1ab-(1) |
| E>K | ORF1ab-(3),ORF7a-(1) |
| E>Q | N-(1),ORF1ab-(1),S-(1) |
| F>L | ORF1ab-(1),S-(1) |
| G>A | S-(1) |
| G>C | N-(1),ORF1ab-(3) |
| G>D | ORF1ab-(3),S-(1) |
| G>E | ORF8-(1) |
| G>R | N-(2),ORF1ab-(1) |
| G>S | N-(1),ORF1ab-(2),S-(1) |
| G>T | N-(2) |
| G>V | ORF1ab-(2),ORF3a-(1),ORF7a-(1),S-(1) |
| G>W | N-(1) |
| H>Q | ORF3a-(1),S-(1) |
| H>R | ORF1ab-(2) |
| H>Y | M-(1),N-(1),ORF1ab-(5),ORF3a-(1),S-(1) |
| I>K | ORF1ab-(1) |
| I>L | ORF1ab-(1),ORF8-(1) |
| I>T | ORF1ab-(4),ORF3a-(1) |
| K>E | ORF1ab-(1) |
| K>N | ORF1ab-(7),ORF3a-(1) |
| K>Q | ORF3a-(1),S-(2) |
| K>R | ORF1ab-(7),S-(1) |
| K>T | ORF1ab-(1) |
| L>F | M-(1),N-(1),ORF10-(1),ORF1ab-(10),ORF3a-(3),ORF7a-(1),S-(2) |
| L>I | ORF1ab-(1) |
| L>P | ORF1ab-(2) |
| L>S | ORF8-(1) |
| L>V | ORF1ab-(1) |
| L>W | ORF3a-(1) |
| M>I | N-(2),ORF1ab-(10),S-(3) |
| N>D | ORF1ab-(2) |
| N>H | ORF1ab-(1) |
| N>K | S-(1) |
| N>L | ORF1ab-(2) |
| N>Y | S-(1) |
| P>A | ORF1ab-(1) |
| P>L | N-(1),ORF1ab-(7),ORF7a-(1),ORF8-(1) |
| P>R | ORF3a-(1) |
| P>S | N-(2),ORF1ab-(5),S-(1) |
| P>T | N-(1) |
| Q>E | ORF7a-(1) |
| Q>H | ORF1ab-(2),S-(4) |
| Q>K | S-(1) |
| Q>P | ORF1ab-(1) |
| Q>R | ORF1ab-(2),S-(1) |
| R>C | ORF1ab-(2) |
| R>G | N-(1) |
| R>I | ORF3a-(1) |
| R>K | N-(2) |
| R>L | M-(1),N-(1),ORF1ab-(1),ORF3a-(1) |
| R>M | S-(1) |
| R>Q | ORF1ab-(1) |
| R>S | N-(1) |
| S>F | ORF1ab-(3),S-(2) |
| S>G | ORF1ab-(2) |
| S>I | N-(3),ORF1ab-(1),S-(3) |
| S>L | N-(1),ORF1ab-(2),ORF3a-(1),ORF7b-(1) |
| S>N | N-(1) |
| S>P | ORF1ab-(2) |
| S>R | ORF1ab-(2) |
| S>T | ORF1ab-(1) |
| T>A | ORF1ab-(3) |
| T>I | N-(3),ORF1ab-(15),ORF3a-(2),S-(4) |
| T>K | ORF1ab-(1) |
| T>M | ORF1ab-(1) |
| T>N | ORF8-(1) |
| V>A | ORF1ab-(3) |
| V>F | E-(2),M-(1),ORF1ab-(5),ORF3a-(1) |
| V>G | ORF1ab-(1) |
| V>I | ORF1ab-(4),ORF3a-(1),ORF7a-(1) |
| V>L | ORF1ab-(2),ORF8-(1),S-(1) |
| W>C | ORF3a-(1) |
| W>L | S-(2) |
| Y>H | ORF1ab-(1),S-(1) |
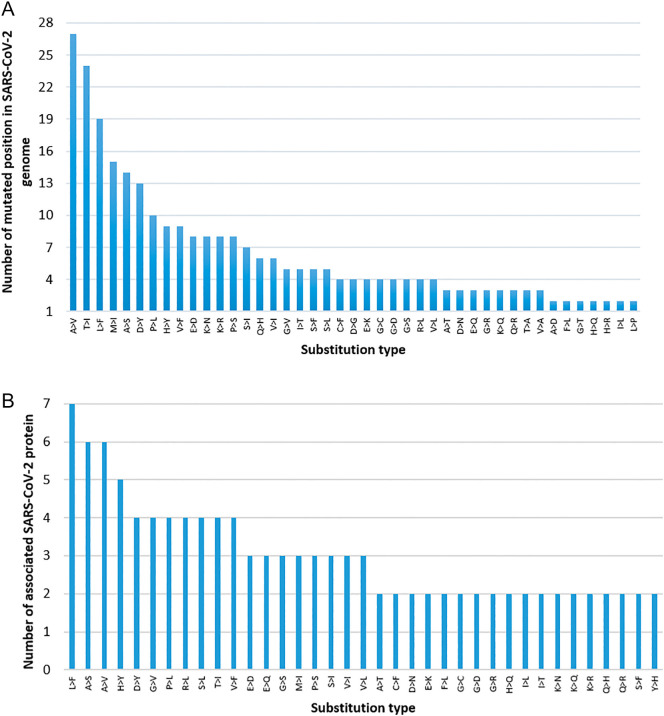
We rank amino acid substitutions by the number of substituted positions (Fig. 8(A)). The top substitutions are A > V, which is observed in 27 locations of six (06) different proteins (Fig. 8(B)). The substitution, L > F is observed with maximum hit, occurring in 07 proteins, M (1), N (1), ORF10 (1), ORF1ab (10), ORF3a (3), ORF7a (1), and S (2). Overall it is observed in nineteen (19) different positions of Indian SARS-CoV-2. Similarly, several other important substitutions with regards to number of substituted positions and associated SARS-CoV-2 proteins can be seen from Fig. 8(A) and (B). Further, there are few substitutions, which are observed uniquely in different SARS-CoV-2 proteins. For example, A > D, C > Y, D > E, D > G are observed in ORF1ab protein, G > A, N > K, N > Y, R > M are observed in Spike (S) protein. Several other unique substitutions with their count and type in each SARS-CoV-2 proteins are reported in Fig. 9 and Table 7, respectively. It is to be noted that the highly mutated four proteins are ORF1ab, S, N, and ORF3a. The number of mutations per variant in SARS-CoV-2 proteins of Indian isolates is shown in Fig. 3.
Fig. 8.
(A) The amino acid substitution type observed with more than two mutated positions in SARS-CoV-2 genome. (B) The amino acid substitution type associated with more than two SARS-CoV-2 proteins.
Fig. 9.
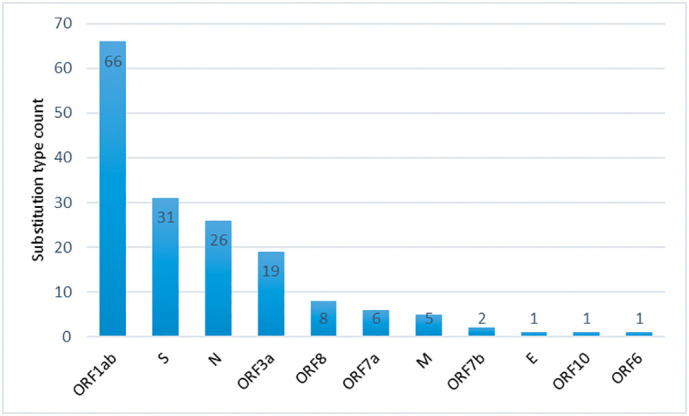
The non-synonymous amino acid substitution type count in each of the SARS-CoV-2 protein.
3.8. Functional assessment of non-synonymous amino acid substitutions
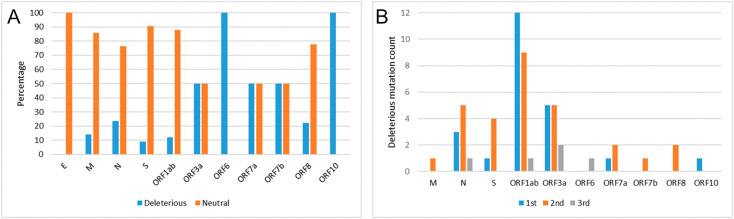
Non-synonymous substitutions are vital as they alter the amino acid that impact on the structural and functional imbalance of the target protein. To understanding the functional alteration during non-synonymous substitutions, we use PROVEAN (Choi and Chan, 2015) to predict mutation type whether deleterious or neutral. We calculate ΔΔG values (Capriotti et al., 2005) for predicting the stability variations (increase or decrease or neutral). We report the PROVEAN and ΔΔG scores in Fig. 10 .
Fig. 10.
The non-synonymous amino acid substitution categorization by percentage of deleterious and neutral mutation type predicted by PROVEAN score. (A) Percentage is shown for SARS-CoV-2 proteins taking all codon positions together; (B) percentage is shown for three codon positions in each of SARS-CoV-2 proteins.
It is to be noted that the deleterious percentage is comparatively low for structural proteins (except E) and high for accessory proteins. We predicted a total of 57 (out of 333 non-synonymous substitution) deleterious substitutions as shown in Table 8, Table 9, Table 10 for ORF1ab, structural, and accessory proteins, respectively. All these substitutions are also listed with NCBI protein accession number (Supplementary-4). While considering codon positions of all the deleterious substitutions, we observe that the deleterious substitutions occurs mostly in 2nd codon position (≈51%) followed by 40% and 9% in 1st and 3rd codon positions, respectively (Fig. 10). Moreover, few neutral mutations with a considerable decrease in stability are observed that might impact on protein structural conformation. For example, we observe D614G mutation occurred in the 2nd codon position, which is neutral with a large decrease in stability. This mutation can potentially decrease the structural stability (Maitra et al., 2020). The change in Asp with Gly at this position resulting in the enhancement of local conformational entropy (Ramakrishnan and Ramachandran, 1965). The most frequently observed non-synonymous mutations, Q57H in ORF3a protein and S194L in N protein, occurred in 3rd and 2nd codon positions respectively. In ORF7b, ORF8, and M proteins, deleterious substitutions occur only in 2nd position, whereas in case of ORF6 and ORF10 it is in 3rd and 1st places, respectively. For all other cases, deleterious substitutions are observed either in any two or all three codon positions.
Table 8.
The non-synonymous amino acid substitutions in ORF1ab protein with the predicted PROVEAN score and ΔΔG prediction value.
| Substitution | PROVEAN score | Type | ΔΔG prediction | RI | Freq. |
|---|---|---|---|---|---|
| G30S | −0.673 | Neutral | −1.15 | 8 | 1 |
| D33N | −0.733 | Neutral | −1.33 | 4 | 2 |
| V38F | −0.553 | Neutral | −1.48 | 9 | 1 |
| G112C | −1.223 | Neutral | −1.08 | 7 | 1 |
| D147E | −1.123 | Neutral | 0.01 | 4 | 1 |
| V169A | 0.027 | Neutral | −1.7 | 8 | 1 |
| G192D | −1.198 | Neutral | −1.23 | 8 | 1 |
| L204F | 0.327 | Neutral | −1.11 | 8 | 2 |
| S212L | 0.097 | Neutral | 0.24 | 1 | 1 |
| T265I | −0.693 | Neutral | −0.67 | 6 | 1 |
| T283I | −0.088 | Neutral | −0.57 | 7 | 2 |
| P309A | −0.135 | Neutral | −1.8 | 8 | 1 |
| P309L | 0.518 | Neutral | −0.72 | 5 | 1 |
| G327D | −1.072 | Neutral | −0.89 | 5 | 1 |
| K338R | −0.685 | Neutral | 0.05 | 2 | 1 |
| A339V | −0.465 | Neutral | 0.07 | 1 | 1 |
| E347D | −0.548 | Neutral | −0.39 | 6 | 1 |
| H417Y | 0.379 | Neutral | 0.26 | 7 | 1 |
| S443P | −0.678 | Neutral | −0.23 | 2 | 1 |
| G519S | −0.633 | Neutral | −1.2 | 8 | 4 |
| Q575R | −0.331 | Neutral | −0.49 | 6 | 3 |
| E633D | −0.233 | Neutral | −0.37 | 7 | 5 |
| E658K | −0.707 | Neutral | −0.44 | 7 | 1 |
| G662R | −1.425 | Neutral | −0.35 | 7 | 1 |
| Q676P | −0.531 | Neutral | −0.58 | 7 | 69 |
| V682L | 0.035 | Neutral | −1.02 | 7 | 1 |
| T882I | −0.691 | Neutral | −0.1 | 2 | 1 |
| P892S | 0.996 | Neutral | −1.35 | 7 | 1 |
| E940D | 0.515 | Neutral | −0.41 | 4 | 1 |
| G989V | 0.35 | Neutral | −0.36 | 5 | 1 |
| D1036G | −0.887 | Neutral | −1.54 | 8 | 1 |
| P1054L | −1.268 | Neutral | −0.44 | 0 | 2 |
| T1055I | −0.496 | Neutral | −0.38 | 2 | 3 |
| E1126D | −0.535 | Neutral | −0.52 | 6 | 1 |
| P1158S | −0.909 | Neutral | −1.75 | 9 | 5 |
| H1160Y | 0.734 | Neutral | 0.11 | 4 | 3 |
| V1211F | −0.667 | Neutral | −0.7 | 5 | 1 |
| E1251K | −0.511 | Neutral | −0.7 | 6 | 1 |
| A1268T | 0.092 | Neutral | −0.78 | 4 | 2 |
| A1283V | −0.232 | Neutral | −0.15 | 2 | 2 |
| A1298V | 0.22 | Neutral | −0.01 | 1 | 1 |
| T1429I | 0.457 | Neutral | −0.5 | 5 | 1 |
| A1432V | 0.864 | Neutral | 0.07 | 2 | 1 |
| S1534I | 0.319 | Neutral | 0.36 | 1 | 7 |
| I1551T | −0.057 | Neutral | −1.98 | 4 | 1 |
| T1573A | −2.402 | Neutral | −1.47 | 9 | 1 |
| M1588I | −0.746 | Neutral | −0.08 | 0 | 2 |
| D1625Y | −2.143 | Neutral | 0.01 | 2 | 2 |
| S1733G | −1.551 | Neutral | −1.07 | 8 | 2 |
| M1769I | −0.349 | Neutral | −0.11 | 5 | 3 |
| A1812D | −0.753 | Neutral | −0.63 | 3 | 6 |
| T1822I | −0.406 | Neutral | 0.1 | 0 | 1 |
| L1853F | −0.808 | Neutral | −1.19 | 6 | 1 |
| T1854A | −0.326 | Neutral | −1.28 | 8 | 1 |
| T1854I | −0.193 | Neutral | −0.25 | 3 | 1 |
| T1874I | −1.364 | Neutral | −0.16 | 3 | 1 |
| D1939G | −0.936 | Neutral | −1.28 | 6 | 1 |
| D1940Y | −0.872 | Neutral | −0.16 | 2 | 1 |
| Q1943H | −0.464 | Neutral | −0.78 | 6 | 1 |
| K1973R | −0.294 | Neutral | −0.37 | 1 | 1 |
| S2015R | −0.501 | Neutral | −0.17 | 0 | 5 |
| T2016K | −0.166 | Neutral | −0.86 | 4 | 10 |
| K2029E | −0.63 | Neutral | −0.5 | 6 | 1 |
| K2029N | −0.431 | Neutral | −0.64 | 2 | 1 |
| P2046L | −1.038 | Neutral | −0.65 | 5 | 3 |
| T2093I | 0.565 | Neutral | 0.1 | 4 | 1 |
| S2103F | −0.372 | Neutral | 0.24 | 6 | 1 |
| L2146P | −1.386 | Neutral | −1.61 | 7 | 1 |
| S2242P | 0.105 | Neutral | −0.06 | 3 | 1 |
| I2307T | −0.03 | Neutral | −2.34 | 8 | 1 |
| L2323V | −0.361 | Neutral | −1.47 | 8 | 1 |
| H2357Y | 0.301 | Neutral | 0.38 | 7 | 3 |
| S2488F | 2.899 | Neutral | −0.05 | 2 | 1 |
| K2511N | −0.966 | Neutral | −0.38 | 0 | 2 |
| H2520R | −0.243 | Neutral | −0.29 | 3 | 2 |
| A2593V | −1.178 | Neutral | −0.24 | 0 | 1 |
| A2732D | −3.463 | Deleterious | −0.68 | 6 | 3 |
| P2739L | −1.595 | Neutral | −0.62 | 4 | 1 |
| H2831Y | 3.17 | Neutral | 0.27 | 6 | 1 |
| A2891V | −0.835 | Neutral | 0 | 3 | 1 |
| D2980G | 0.071 | Neutral | −1.27 | 5 | 1 |
| A2994V | −1.769 | Neutral | −0.05 | 0 | 3 |
| T3058I | 1.463 | Neutral | −0.48 | 4 | 7 |
| G3072C | −5.058 | Deleterious | −1.07 | 6 | 2 |
| M3087I | 0.614 | Neutral | −0.56 | 5 | 3 |
| T3150I | 0.112 | Neutral | −0.43 | 1 | 2 |
| S3158G | −0.785 | Neutral | −1.35 | 7 | 1 |
| L3338F | −3.068 | Deleterious | −1.09 | 6 | 2 |
| K3353R | −1.343 | Neutral | −0.13 | 1 | 1 |
| V3377G | −6.124 | Deleterious | −2.51 | 9 | 1 |
| Q3390R | −0.324 | Neutral | −0.33 | 4 | 1 |
| N3405L | −4.454 | Deleterious | −0.05 | 0 | 5 |
| P3447S | −1.913 | Neutral | −1.7 | 9 | 1 |
| T3453A | −0.882 | Neutral | −0.83 | 7 | 1 |
| V3475F | −2.291 | Neutral | −1.42 | 9 | 1 |
| K3499R | −0.421 | Neutral | −0.25 | 2 | 5 |
| L3606F | −1.432 | Neutral | −1 | 6 | 14 |
| I3618T | −1.397 | Neutral | −1.49 | 7 | 1 |
| M3655I | 0.174 | Neutral | −0.75 | 7 | 1 |
| D3681N | −0.466 | Neutral | −1.11 | 7 | 1 |
| L3711F | −0.348 | Neutral | −1.21 | 7 | 2 |
| I3731T | −0.744 | Neutral | −2.36 | 9 | 2 |
| V3759F | −1.765 | Neutral | −1.52 | 9 | 1 |
| E3909G | −3.759 | Deleterious | −1.17 | 9 | 1 |
| E3962K | −0.041 | Neutral | −0.34 | 3 | 1 |
| S3983F | −2.722 | Deleterious | −0.34 | 7 | 2 |
| R3993C | −6.175 | Deleterious | −0.86 | 5 | 1 |
| R3993L | −5.422 | Deleterious | −0.3 | 7 | 1 |
| K4069T | −2.268 | Neutral | −0.48 | 6 | 1 |
| V4073I | −0.106 | Neutral | −0.55 | 8 | 1 |
| K4081R | −0.921 | Neutral | −0.33 | 7 | 1 |
| M4116I | −0.46 | Neutral | −0.74 | 5 | 1 |
| K4176N | 0.651 | Neutral | −0.36 | 3 | 1 |
| V4181I | −0.046 | Neutral | −0.7 | 7 | 1 |
| A4271V | −3.278 | Deleterious | −0.25 | 1 | 1 |
| A4273V | −3.349 | Deleterious | −0.23 | 1 | 1 |
| K4451N | −0.49 | Neutral | −0.65 | 2 | 2 |
| K4483N | −1.326 | Neutral | −0.42 | 4 | 1 |
| A4487V | 0.357 | Neutral | −0.24 | 2 | 1 |
| A4489V | −2.346 | Neutral | −0.31 | 1 | 10 |
| D4532G | −3.086 | Deleterious | −1.08 | 6 | 1 |
| A4577V | −1.878 | Neutral | −0.17 | 4 | 1 |
| M4588I | −1.074 | Neutral | −0.81 | 8 | 1 |
| I4593L | 0.213 | Neutral | −0.9 | 7 | 1 |
| E4670D | −0.609 | Neutral | −0.59 | 5 | 2 |
| P4715L | −0.446 | Neutral | −0.83 | 6 | 359 |
| L4721I | −1.085 | Neutral | −1.29 | 7 | 7 |
| V4746A | −2.528 | Deleterious | −2.01 | 9 | 1 |
| M4855I | −1.728 | Neutral | −0.81 | 8 | 3 |
| C4856F | −0.483 | Neutral | −0.21 | 3 | 1 |
| L5030F | −2.739 | Deleterious | −0.99 | 7 | 3 |
| T5035I | −0.622 | Neutral | −0.43 | 2 | 1 |
| T5036M | −1.529 | Neutral | −0.29 | 2 | 1 |
| M5060I | −0.117 | Neutral | −0.56 | 7 | 1 |
| A5091S | −1.821 | Neutral | −0.82 | 9 | 2 |
| Q5214H | 1.016 | Neutral | −0.77 | 6 | 2 |
| V5272I | −0.551 | Neutral | −0.17 | 2 | 18 |
| D5285Y | −1.381 | Neutral | 0.27 | 3 | 2 |
| T5300I | −0.542 | Neutral | −0.42 | 4 | 1 |
| S5305L | −2.332 | Neutral | 0.22 | 4 | 1 |
| P5377S | −0.897 | Neutral | −1.73 | 9 | 2 |
| H5488Y | 0.534 | Neutral | 0.19 | 7 | 1 |
| E5492Q | −2.053 | Neutral | −0.7 | 7 | 1 |
| G5530C | −2.742 | Deleterious | −0.82 | 4 | 4 |
| H5569R | 1.004 | Neutral | −0.1 | 5 | 1 |
| V5571F | −1.592 | Neutral | −1.62 | 8 | 2 |
| Y5577H | −0.845 | Neutral | −1.53 | 7 | 2 |
| S5583T | −0.858 | Neutral | −0.66 | 4 | 1 |
| P5624L | −5.36 | Deleterious | −0.63 | 7 | 2 |
| R5766Q | 0.366 | Neutral | −0.91 | 8 | 1 |
| F5823L | −3.989 | Deleterious | −1.17 | 5 | 1 |
| A5926S | 0.351 | Neutral | −1.08 | 10 | 1 |
| N5928H | −0.711 | Neutral | −0.77 | 9 | 1 |
| K5957R | −0.861 | Neutral | −0.09 | 0 | 1 |
| I5970K | −1.93 | Neutral | −2.18 | 9 | 1 |
| M5997I | −0.985 | Neutral | −0.96 | 8 | 1 |
| G6039V | −6.16 | Deleterious | −0.14 | 1 | 1 |
| A6044V | 2.2 | Neutral | 0.14 | 2 | 1 |
| P6065S | 0.176 | Neutral | −1.67 | 8 | 2 |
| L6082F | −0.771 | Neutral | −1.04 | 6 | 1 |
| R6088C | −5.465 | Deleterious | −1.2 | 7 | 3 |
| L6102F | −1.397 | Neutral | −1.05 | 4 | 67 |
| S6180R | −1.897 | Neutral | 0.16 | 4 | 1 |
| A6199S | −2.053 | Neutral | −0.62 | 8 | 1 |
| D6249Y | 0.823 | Neutral | −0.16 | 3 | 1 |
| K6274N | −0.353 | Neutral | −0.18 | 3 | 3 |
| T6297I | −0.448 | Neutral | −0.8 | 3 | 45 |
| N6313D | −3.422 | Deleterious | −0.62 | 7 | 1 |
| P6368L | −6.762 | Deleterious | −0.79 | 6 | 1 |
| V6385L | −0.789 | Neutral | −1.03 | 7 | 3 |
| K6464N | −1.404 | Neutral | −0.42 | 2 | 1 |
| T6500I | −1.557 | Neutral | −0.42 | 5 | 1 |
| A6533V | −0.465 | Neutral | −0.35 | 5 | 3 |
| D6580Y | −0.868 | Neutral | −0.59 | 5 | 1 |
| G6581D | −2.423 | Neutral | −1.12 | 7 | 2 |
| A6589V | −0.154 | Neutral | −0.17 | 3 | 1 |
| V6600A | −2.262 | Neutral | −1.84 | 9 | 4 |
| L6614F | −1.53 | Neutral | −1.28 | 7 | 1 |
| A6623T | 0.442 | Neutral | −0.76 | 6 | 1 |
| V6688I | −0.141 | Neutral | −0.49 | 4 | 1 |
| M6723I | −0.049 | Neutral | −0.85 | 6 | 1 |
| C6742Y | −0.068 | Neutral | −0.18 | 0 | 1 |
| D6900Y | −3.735 | Deleterious | −0.41 | 1 | 1 |
| L6909F | −0.541 | Neutral | −1.04 | 5 | 1 |
| A6914S | −0.017 | Neutral | −1.04 | 5 | 2 |
| A6914V | −0.428 | Neutral | −0.03 | 1 | 1 |
| K6958R | −0.492 | Neutral | −0.17 | 5 | 2 |
| P7034L | 0.713 | Neutral | −0.88 | 5 | 1 |
| N7083D | −1.153 | Neutral | −0.42 | 2 | 1 |
The substitutions with either high PROVEAN score (< − 2.5, type: deleterious) or large increase stability (ΔΔG < − 0.5) or both are shown in bold.
Table 9.
The functional assessment of non-synonymous amino acid substitutions in four structural SARS-CoV-2 proteins (E, M, N, S). The functional assessment of mutation is predicted on utilizing two different measures (PROVEAN score and stability value).
| Protein | Substitution | PROVEAN score | Type | ΔΔG prediction | RI | Freq. |
|---|---|---|---|---|---|---|
| E | V62F | 0 | Neutral | −1.65 | 8 | 1 |
| V75F | −1.414 | Neutral | −1.57 | 9 | 1 | |
| M | A142V | 0.18 | Neutral | 0.25 | 5 | 2 |
| L29F | −1.646 | Neutral | −0.91 | 7 | 1 | |
| A63V | −1.937 | Neutral | 0.14 | 0 | 1 | |
| A69S | −1.991 | Neutral | −0.82 | 9 | 1 | |
| V70F | −1.365 | Neutral | −1.66 | 10 | 1 | |
| R107L | −4.03 | Deleterious | −0.33 | 4 | 1 | |
| H125Y | 0.799 | Neutral | 0.06 | 5 | 1 | |
| N | S194L | −4.272 | Deleterious | −0.38 | 2 | 158 |
| P13L | −1.23 | Neutral | −0.48 | 3 | 23 | |
| S202N | −0.404 | Neutral | −0.78 | 0 | 20 | |
| R203K | −1.604 | Neutral | −0.93 | 6 | 14 | |
| R203K | −1.604 | Neutral | −0.93 | 6 | 14 | |
| G204R | −1.656 | Neutral | −0.52 | 5 | 14 | |
| T205I | −1.562 | Neutral | −0.53 | 3 | 6 | |
| S193I | −2.755 | Deleterious | −0.36 | 2 | 4 | |
| G97S | −1.98 | Neutral | −1.33 | 8 | 3 | |
| A156S | −0.457 | Neutral | −0.83 | 9 | 3 | |
| P6T | −0.223 | Neutral | −1.1 | 8 | 2 | |
| S33I | −1.372 | Neutral | 0.27 | 6 | 2 | |
| S180I | −3.465 | Deleterious | −0.14 | 3 | 2 | |
| M234I | 0.044 | Neutral | −0.03 | 1 | 2 | |
| D22N | −0.541 | Neutral | −1.4 | 6 | 1 | |
| D22N | −0.541 | Neutral | −1.4 | 6 | 1 | |
| E31Q | 0.054 | Neutral | −0.7 | 5 | 1 | |
| G34W | −1.609 | Neutral | −0.13 | 2 | 1 | |
| R92S | −3.718 | Deleterious | −1.23 | 6 | 1 | |
| G120R | −0.733 | Neutral | −0.29 | 1 | 1 | |
| A134V | −2.811 | Deleterious | −0.12 | 2 | 1 | |
| L139F | −0.697 | Neutral | −0.85 | 8 | 1 | |
| D144Y | −1.764 | Neutral | 0.2 | 1 | 1 | |
| A152S | 1.463 | Neutral | −0.92 | 9 | 1 | |
| R191L | −3.269 | Deleterious | −0.58 | 3 | 1 | |
| R203G | −3.247 | Deleterious | −1.6 | 7 | 1 | |
| R203K | −1.604 | Neutral | −0.93 | 6 | 1 | |
| G204R | −1.656 | Neutral | −0.52 | 5 | 1 | |
| G204T | −1.76 | Neutral | −0.96 | 7 | 1 | |
| A218V | 0.171 | Neutral | 0.21 | 1 | 1 | |
| M234I | 0.044 | Neutral | −0.03 | 1 | 1 | |
| G236C | −2.269 | Neutral | −0.27 | 5 | 1 | |
| H300Y | −1.577 | Neutral | 0.46 | 5 | 1 | |
| P302S | −4.043 | Deleterious | −1.3 | 7 | 1 | |
| P344S | −4.031 | Deleterious | −1.46 | 8 | 1 | |
| D348Y | −0.588 | Neutral | −0.41 | 2 | 1 | |
| T362I | −1.722 | Neutral | −0.35 | 3 | 1 | |
| T393I | −0.613 | Neutral | 0.1 | 2 | 1 | |
| S | D614G | 0.598 | Neutral | −0.93 | 3 | 405 |
| L54F | −0.435 | Neutral | −1.14 | 4 | 80 | |
| E583D | −0.819 | Neutral | −0.44 | 3 | 14 | |
| R78M | 0.986 | Neutral | −0.84 | 7 | 12 | |
| T572I | −0.649 | Neutral | 0 | 3 | 10 | |
| Q677H | 0.002 | Neutral | −0.67 | 5 | 5 | |
| L5F | −1.126 | Neutral | −0.98 | 3 | 3 | |
| Q690H | −0.796 | Neutral | −0.86 | 6 | 3 | |
| S12F | −0.65 | Neutral | 0.14 | 2 | 2 | |
| W152L | −0.159 | Neutral | −0.89 | 7 | 2 | |
| S155I | −0.503 | Neutral | 0 | 4 | 2 | |
| M177I | 0.579 | Neutral | −0.61 | 5 | 2 | |
| G181A | 0.396 | Neutral | −0.58 | 1 | 2 | |
| W258L | −1.084 | Neutral | −0.65 | 7 | 2 | |
| A706S | 0.183 | Neutral | −0.85 | 9 | 2 | |
| A879S | −0.361 | Neutral | 0.54 | 7 | 2 | |
| H1083Q | −1.006 | Neutral | −0.34 | 5 | 2 | |
| C1243F | −4.53 | Deleterious | −0.09 | 2 | 2 | |
| F2L | −0.902 | Neutral | −1.22 | 9 | 1 | |
| S13I | −1.187 | Neutral | 0.27 | 0 | 1 | |
| Y28H | −0.443 | Neutral | −1.38 | 4 | 1 | |
| G35V | −2.112 | Neutral | −0.68 | 7 | 1 | |
| T76I | −0.115 | Neutral | −0.72 | 6 | 1 | |
| K97Q | −0.113 | Neutral | −0.92 | 7 | 1 | |
| N148Y | −0.177 | Neutral | 0.1 | 4 | 1 | |
| M153I | 0.244 | Neutral | −0.98 | 6 | 1 | |
| E156D | 0.958 | Neutral | −0.52 | 4 | 1 | |
| S162I | 0.231 | Neutral | 0.02 | 1 | 1 | |
| Q173H | −0.299 | Neutral | −1.02 | 7 | 1 | |
| S255F | −0.423 | Neutral | −0.03 | 3 | 1 | |
| G261S | 0.485 | Neutral | −1.13 | 8 | 1 | |
| A262S | 0.154 | Neutral | −0.64 | 9 | 1 | |
| Q271R | −0.48 | Neutral | −0.27 | 5 | 1 | |
| C301F | −8.689 | Deleterious | 0.2 | 4 | 1 | |
| E471Q | 0.445 | Neutral | −0.59 | 7 | 1 | |
| D574Y | 0.858 | Neutral | 0.36 | 2 | 1 | |
| Q613H | −0.917 | Neutral | −0.86 | 6 | 1 | |
| H655Y | −0.814 | Neutral | 0.08 | 4 | 1 | |
| A688V | 0.498 | Neutral | −0.37 | 5 | 1 | |
| A701V | 0.597 | Neutral | −0.25 | 4 | 1 | |
| M731I | −0.598 | Neutral | −0.25 | 3 | 1 | |
| K795Q | 0.072 | Neutral | −0.61 | 3 | 1 | |
| P809S | 1.024 | Neutral | −1.55 | 8 | 1 | |
| T827I | −0.378 | Neutral | −0.45 | 6 | 1 | |
| A892V | −1.901 | Neutral | 0.2 | 1 | 1 | |
| A930V | −3.727 | Deleterious | −0.2 | 3 | 1 | |
| T1077I | −1.511 | Neutral | −0.13 | 1 | 1 | |
| V1104L | −0.604 | Neutral | −0.7 | 1 | 1 | |
| D1153Y | −3.275 | Deleterious | −1.52 | 2 | 1 | |
| K1181R | −0.522 | Neutral | −0.48 | 7 | 1 | |
| N1187K | −0.467 | Neutral | −0.29 | 4 | 1 | |
| Q1201K | 1.409 | Neutral | −0.29 | 3 | 1 | |
| C1250F | −5.057 | Deleterious | −0.09 | 2 | 1 | |
| D1259Y | 3.924 | Neutral | −0.21 | 3 | 1 |
The substitutions with either high PROVEAN score (< − 2.5, type: deleterious) or large increase stability (ΔΔG < − 0.5) or both are shown in bold.
Table 10.
The functional assessment of non-synonymous amino acid substitutions in six SARS-CoV-2 accessories proteins (ORF3a, ORF6, ORF7a, ORF7b, ORF8, ORF10). The functional assessment of mutation is predicted on utilizing two different measures (PROVEAN score and stability value).
| Protein | Substitution | PROVEAN score | Type | ΔΔG prediction | RI | Freq. |
|---|---|---|---|---|---|---|
| ORF3a | G18V | −1.571 | Neutral | −0.28 | 6 | 1 |
| K21Q | 0.657 | Neutral | −0.47 | 1 | 1 | |
| A23S | −1.638 | Neutral | −0.86 | 9 | 2 | |
| I35T | −2.619 | Deleterious | −2.39 | 9 | 1 | |
| L41F | −2.724 | Deleterious | −1.08 | 7 | 4 | |
| P42R | −5.495 | Deleterious | −0.96 | 7 | 1 | |
| V50I | −0.657 | Neutral | −0.84 | 8 | 1 | |
| L53F | −3.962 | Deleterious | −1.09 | 7 | 1 | |
| A54S | −1.638 | Neutral | −0.6 | 8 | 1 | |
| Q57H | −3.286 | Deleterious | −0.9 | 7 | 234 | |
| K66N | 3.486 | Neutral | −0.16 | 1 | 1 | |
| R68I | −1.562 | Neutral | 0.17 | 3 | 1 | |
| V77F | 2.638 | Neutral | −1.37 | 8 | 1 | |
| L86W | −3.943 | Deleterious | −1.13 | 1 | 1 | |
| H93Y | −3.943 | Deleterious | 0.3 | 6 | 3 | |
| A103V | −2.876 | Deleterious | 0.2 | 5 | 1 | |
| L108F | −3.4 | Deleterious | −1.24 | 6 | 1 | |
| W131C | −7.752 | Deleterious | −1.29 | 8 | 1 | |
| R134L | −1.543 | Neutral | −0.47 | 9 | 1 | |
| A143S | 0.724 | Neutral | −0.95 | 9 | 1 | |
| T151I | −4.886 | Deleterious | −0.29 | 0 | 2 | |
| D155Y | −6.829 | Deleterious | 0.21 | 0 | 2 | |
| S171L | −2.238 | Neutral | −0.22 | 0 | 2 | |
| T175I | 2.562 | Neutral | −0.04 | 4 | 1 | |
| ORF6 | E13D | −2.786 | Deleterious | −0.24 | 4 | 1 |
| ORF7a | Q94E | −1 | Neutral | −0.24 | 2 | 2 |
| E95K | −2.614 | Deleterious | −0.6 | 8 | 2 | |
| G38V | −6.526 | Deleterious | −0.4 | 4 | 1 | |
| P45L | −10 | Deleterious | −0.7 | 4 | 1 | |
| V71I | −0.667 | Neutral | −0.24 | 5 | 1 | |
| L116F | −1.263 | Neutral | −0.85 | 7 | 1 | |
| ORF7b | S31L | −6 | Deleterious | 0.23 | 1 | 1 |
| A43T | 0 | Neutral | −0.44 | 5 | 1 | |
| ORF8 | L84S | 2.333 | Neutral | −2.29 | 8 | 19 |
| G8E | −3.056 | Deleterious | −0.6 | 1 | 1 | |
| T12N | −1.056 | Neutral | −0.71 | 1 | 1 | |
| A14S | 0.833 | Neutral | −0.47 | 6 | 1 | |
| A51V | −1.222 | Neutral | −0.06 | 2 | 1 | |
| V62L | −0.722 | Neutral | −0.8 | 5 | 1 | |
| A65V | 1.222 | Neutral | 0.02 | 1 | 1 | |
| P85L | −8.778 | Deleterious | −0.73 | 7 | 1 | |
| I121L | −0.278 | Neutral | −0.79 | 5 | 1 | |
| ORF10 | L37F | NA | Deleterious | −0.99 | 6 | 1 |
The substitutions with either high PROVEAN score (< − 2.5, type: deleterious) or large increase stability (ΔΔG < − 0.5) or both are shown in bold.
We also predict the stability impact of single point mutations, where most of the substitutions show a large decrease in stability. We find a total of 32 single point deleterious substitutions (7 from structural proteins) out of total 57 with large decrease in stability (ΔΔG < − 0.5). We highlight all the functional domains of all such non-synonymous deleterious substitutions in Table 11 . Additionally, we study the changes in physicochemical properties during such substitutions. A few numbers of substitution leads to change physicochemical property both in hydropathy class and side-chain structural classes (Table 11).
Table 11.
The 57 deleterious amino acid substitutions in different SARS-CoV-2 proteins highlighted with the putative functional domain and physicochemical property changes. The mutations with large decrease stability (ΔΔG < − 0.5) are shown in bold.
| Protein | Substitution | Putative functional domain | Hydropathy change | Chemical property change |
|---|---|---|---|---|
| ORF1ab | A2732D | NSP3 | Hydrophobic to charge | Aliphatic to acidic |
| G3072C | NSP4 | Hydrophilic (unchanged) | Aliphatic to sulfur containing | |
| L3338F | NSP5 (3CLpro) | Hydrophobic (unchanged) | Aliphatic to aromatic | |
| V3377G | NSP5 (3CLpro) | Hydrophobic to hydrophilic | Aliphatic to aliphatic | |
| N3405L | NSP5 (3CLpro) | Hydrophilic to hydrophobic | Acidic amide to aliphatic | |
| E3909G | NSP7 | Charge to hydrophilic | Acidic to aliphatic | |
| S3983F | NSP8 | Hydrophilic to hydrophobic | Hydroxyl containing to aromatic | |
| R3993C | NSP8 | Charge to hydrophilic | Basic to sulfur containing | |
| R3993L | NSP8 | Charge to hydrophilic | Basic to aliphatic | |
| A4271V | NSP10 | Hydrophobic (unchanged) | Aliphatic (unchanged) | |
| A4273V | NSP10 | Hydrophobic (unchanged) | Aliphatic (unchanged) | |
| D4532G | NSP12 (RdRp) | Charge to hydrophilic | Acidic to aliphatic | |
| V4746A | NSP12 (RdRp) | Hydrophobic (unchanged) | Aliphatic (unchanged) | |
| L5030F | NSP12 (RdRp) | Hydrophobic (unchanged) | Aliphatic to aromatic | |
| G5530C | NSP13 (helicase) | Hydrophilic (unchanged) | Aliphatic to sulfur containing | |
| P5624L | NSP13 (helicase) | Hydrophobic (unchanged) | Cyclic to aliphatic | |
| F5823L | NSP13 (helicase) | Hydrophobic (unchanged) | Aromatic to aliphatic | |
| G6039V | NSP14 (exonuclease) | Hydrophilic to hydrophobic | Aliphatic to aliphatic | |
| R6088C | NSP14 (exonuclease) | Charge to hydrophilic | Basic to sulfur containing | |
| N6313D | NSP14 (exonuclease) | Hydrophilic to charge | Acidic amide to acidic | |
| P6368L | NSP14 (exonuclease) | Hydrophobic (unchanged) | Cyclic to aliphatic | |
| D6900Y | NSP16 | Charge to hydrophobic | Acidic to aromatic | |
| M | R107L | Topological domain | Charge to hydrophobic | Basic to aliphatic |
| N | R92S | NTD | Charge to hydrophilic | Basic to hydroxyl containing |
| A134V | NTD | Hydrophobic (unchanged) | Aliphatic (unchanged) | |
| S180I | SR-rich linker | Hydrophilic (unchanged) | Hydroxyl containing to aliphatic | |
| R191L | SR-rich linker | Charge to hydrophobic | Basic to aliphatic | |
| S193I | SR-rich linker | Hydrophilic to hydrophobic | Hydroxyl containing to aliphatic | |
| S194L | SR-rich linker | Hydrophilic to hydrophobic | Hydroxyl containing to aliphatic | |
| R203G | SR-rich linker | Charge to hydrophilic | Basic to aliphatic | |
| P302S | CTD | Hydrophobic to hydrophilic | Cyclic to hydroxyl containing | |
| P344S | CTD | Hydrophobic to hydrophilic | Cyclic to hydroxyl containing | |
| S | C301F | S1 (N-terminal) | Hydrophilic (unchanged) | Sulfur containing to aromatic |
| A930V | S2 (HR-1) | Hydrophobic (unchanged) | Aliphatic (unchanged) | |
| D1153Y | S2 (between HR1 and HR2) | Charge to hydrophilic | Acidic to aromatic | |
| C1243F | S2 (cytoplasm domain) | Hydrophilic to hydrophobic | Sulfur containing to aromatic | |
| C1250F | S2 (cytoplasm domain) | Hydrophilic to hydrophobic | Sulfur containing to aromatic | |
| ORF3a | I35T | Hydrophobic to hydrophilic | Aliphatic to hydroxyl containing | |
| L41F | TM-1 | Hydrophobic (unchanged) | Aliphatic to aromatic | |
| P42R | TM-1 | Hydrophobic to charge | Cyclic to basic | |
| L53F | TM-1 | Hydrophobic (unchanged) | Aliphatic to aromatic | |
| Q57H | TM-1 | Hydrophilic to charge | Acidic amide to basic | |
| L86W | TM-2 | Hydrophobic (unchanged) | Aliphatic to aromatic | |
| H93Y | Ion channels | Charge to hydrophilic | Basic to aromatic | |
| A103V | Ion channels | Hydrophobic (unchanged) | Aliphatic (unchanged) | |
| L108F | Ion channels | Hydrophobic (unchanged) | Aliphatic to aromatic | |
| W131C | Ion channels | Hydrophobic to hydrophilic | Aromatic to sulfur containing | |
| T151I | C-terminal | Hydrophilic to hydrophobic | Hydroxyl containing to aliphatic | |
| D155Y | C-terminal | Charge to hydrophilic | Acidic to aromatic | |
| ORF6 | E13D | Charge (unchanged) | Acidic (unchanged) | |
| ORF7a | G38V | Luminal domain | Hydrophilic (unchanged) | Aliphatic to aliphatic |
| P45L | Luminal domain | Hydrophobic (unchanged) | Cyclic to aliphatic | |
| E95K | Luminal domain | Charge (unchanged) | Acidic to basic | |
| ORF7b | S31L | Hydrophilic to hydrophobic | Hydroxyl containing to aliphatic | |
| ORF8 | G8E | N-terminal (hydrophobic region) | Hydrophilic to charge | Aliphatic to acidic |
| P85L | Hydrophobic (unchanged) | Cyclic to aliphatic | ||
| ORF10 | L37F | Hydrophobic (unchanged) | Aliphatic to aromatic |
The ORF1ab protein consists of several non-structural polyproteins (NSP1-NSP16). A few deleterious substitutions are detected in putative functional domains of those polyproteins. The 3-chymotrypsin-like cysteine protease (3CLpro) and RNA-dependent RNA polymerase (RdRp) regions located in NSP3 and NSP12 polyproteins, respectively. It’s playing a major role in anti viral drug discovery for SARS-CoV-2 and other coronavirus diseases (ul Qamar et al., 2020; Anand et al., 2003; Calligari et al., 2020). So, the mutations detected in these functional domains might impact protein functions and stability. Three deleterious substitutions are also detected both in 3CLpro and RdRp region of ORF1ab polyprotein. The few deleterious substitutions with large decrease in stability changes are detected in other two important functional domains, namely helicase in NSP13 (Chen et al., 2020; Yu et al., 2012), and exonuclease in NSP14 (Yuen et al., 2020). These are also investigated to inhibit coronavirus (Chen et al., 2020; Shannon et al., 2020; Yu et al., 2012).
The Membrane (M) protein is one of the most abundant structural proteins among coronaviruses protein and has an interaction role with other structural proteins (He et al., 2004; Naskalska et al., 2019). A single deleterious substitution is observed in Topological domain (Bianchi et al., 2020).
N protein contains two distinct RNA-binding domains- the N-terminal domain (NTD, 44–179 residues) and the C-terminal domain (CTD, 247–363 residues) (Zeng et al., 2020), responsible for RNA binding and oligomerization, respectively. These two regions are connected by an intrinsically disordered central Ser/Arg (SR)-rich linker (Kang et al., 2020), which is responsible for primary phosphorylation. The study on the Nucleocapsid protein of other coronaviruses, several residues of N-terminal domain, is associated with RNA binding and virus infectivity (Saikatendu et al., 2007; Tan et al., 2006; Grossoehme et al., 2009). Among the observed seven deleterious substitutions in N protein, we observe 2 in NTD, 5 in SR-rich linker, and 2 in CTD functional domain (Kang et al., 2020).
The S1 subunit (residues: 14–685) and the S2 subunit (residues: 686–1273) in Spike protein regions are responsible for receptor binding and membrane fusion, respectively (Huang et al., 2020). The N-terminal domain (residues: 14–305) belongs to the S1 subunit. The S2 subunit consists of several sub-domains, including heptapeptide repeat sequence 1 (HR1) (residues: 912–984), HR2 (residues: 1163–1213), cytoplasm domain (residues: 1237–1273) (Xia et al., 2020). We observe five deleterious substitution in Spike protein, one each in S1 (N-terminal), S2 (HR-1), and S2 subunit in between HR1 and HR2. Two deleterious substitution occur in S2 subunit (Cytoplasm domain) (Huang et al., 2020; Walls et al., 2020).
SARS-CoV has three major transmembrane domains: (i) Transmembrane domain 1 (TM-1) (approx. residues: 34–56), (ii) transmembrane domain 2 (TM-2) (approx. residues: 77–99), and (iii) transmembrane domain 3 (TM-3) (approx. residues: 103–125) available mostly in ORF3a and C-terminal domain with about 160 amino acid residues (Hofman, 1993; Zeng et al., 2004). In connection with approximate residues of SARS-CoV, We observe four mutations in TM-1 and one in TM-2 domains of SARS-CoV-2 ORF3a protein. Four important deleterious substitutions are observed in the Ion channels domain (Domain II, residues: 91–133) (Issa et al., 2020), which is linked to its pro-apoptotic function as observed for other SARS-coronavirus (Chan et al., 2009; Lu et al., 2010). One of the observed mutations is W131C, located in Cysteine rich region (cysteine-rich region overlaps the third membrane-spanning domain) of ORF3a protein (Zeng et al., 2004). This mutation further increases the Cysteine residue in that region that may alter interchain disulfide linkages with the Spike protein of other viral structural proteins. Additionally, two mutations in the C-terminal domain are observed between the last two Cysteine residues (Zeng et al., 2004).
Both the proteins ORF6 and ORF8 do not have any trans-membrane regions, but ORF8 has an hydrophobic signal peptide (residues: 1–15) and chain (residues: 61–121) (Alam et al., 2020). However, they play significant roles in innate immune suppression during viral infection, regulation of molecular functions, virus growth, replication, and host interactions (Alam et al., 2020; Li et al., 2020; Mohammad et al., 2020). A single deleterious substitution (E13D) is found in ORF6 protein and two mutations in ORF8 protein, where one in the Hydrophobic region (G8E) (Alam et al., 2020; Mohammad et al., 2020).
The domain of ORF7a protein of Indian SARS-CoV-2 consists of seven (07) β strands (Alam et al., 2020). A similar result is reported for ORF7a protein of SARS-CoV in (Nelson et al., 2005; Bartlam et al., 2007). It consists of N-terminal signal peptide (residues: 1–15), luminal domain (16–96), transmembrane segment (residues: 97–117), and a 5 residue cytoplasmic tail. Considering the similar organizational domains of SARS-CoV with SARS-CoV-2, three deleterious substitutions are identified in the luminal domain, two of them (G38V and P45L) are located before and after the 3rd β strand.
4. Conclusion
In this study, we thoroughly investigated and characterized mutations observed in Indian SARS-CoV-2 genome. We reported variants and mutations observed in all the SARS-CoV-2 proteins belong to both synonymous and non-synonymous categories. We highlighted position-specific mutations in the codons. Non-synonymous amino acid substitutions are analyzed further to predict the functional stability of the proteins.
Our study reported a total of 536 mutated positions in the coding region of SARS-CoV-2 proteins. The ORF3a happens to be the mostly mutated protein (≈4% of total length), followed by three structural proteins (N, M, S). However, both in ORF3a and N proteins, we observed fewer mutation types compared to ORF1ab and S. The number of variants and mutations per variant observed to be maximum for ORF1ab followed by Spike protein. Interestingly, counts for non-synonymous mutations are higher compared to synonymous mutations (except for M protein). Mutations in E and ORF7b proteins are all non-synonymous.
Our analysis further reveals that most of the deleterious substitutions with decrease in stability occur in the 2nd position (codon) and putative functional domains. Higher quantity of single point mutation, G > T, is observed both in 1st and 3rd positions in the codon, whereas mutation C > T, shows maximum occurrence in 2nd codon position. The conclusion drawn purely based on computational analysis, needs experimental confirmation. Though we restricted our current study on Indian isolates, it may easily be extended to any other strains. Overall analysis might help in better understanding of the possible role in virulence, infectivity, and virus release in SARS-CoV-2. A further comparative study on the significant mutations observed in Indian isolates may be performed with the strains collected from the rest of the world.
The following are the supplementary data related to this article.
Accession numbers of SARS-CoV-2 collected sequences from Indian isolates.
The polygenetic tree for each of the SARS-CoV-2 proteins.
The variant with frequency and respective nucleotide mutation and type, amino acid change, and number of mutations for each of the SARS-CoV-2 proteins.
The observed non-synonymous substitution with the protein accession number in each of the SARS-CoV-2 proteins.
CRediT authorship contribution statement
Jayanta Kumar Das: Conceptualization, Data curation, Methodology, Software, Formal analysis, Visualization, Writing - original draft, review & editing. Antara Sengupta: Methodology, Software, Validation, Visualization, Review & editing. Pabitra Pal Choudhury: Review & editing. Swarup Roy: Conceptualization, Supervision, Visualization, Review & editing.
Declaration of competing interest
The authors declare that they have no competing interests.
Footnotes
https://www.ncbi.nlm.nih.gov/sars-cov-2/ as reported till August 28, 2020.
https://www.ncbi.nlm.nih.gov/sars-cov-2/ as reported till August 28, 2020.
References
- Aftabuddin M., Kundu S. Hydrophobic, hydrophilic, and charged amino acid networks within protein. Biophys. J. 2007;93:225–231. doi: 10.1529/biophysj.106.098004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam I., Kamau A.A., Kulmanov M., Jaremko Ł., Arold S.T., Pain A., Gojobori T., Duarte C.M. Functional pangenome analysis shows key features of e protein are preserved in sars and sars-cov-2. Frontiers in Cellular and Infection Microbiology. 2020;10:405. doi: 10.3389/fcimb.2020.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3clpro) structure: basis for design of anti-sars drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of sars-cov-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- André N.M., Cossic B., Davies E., Miller A.D., Whittaker G.R. Distinct mutation in the feline coronavirus spike protein cleavage activation site in a cat with feline infectious peritonitis-associated meningoencephalomyelitis. J. Feline Med. Surg. Open Rep. 2019;5 doi: 10.1177/2055116919856103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer C.F. Does mutation rate depend on itself. PLoS Biol. 2008;6 doi: 10.1371/journal.pbio.0060052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlam M., Xu Y., Rao Z. Structural proteomics of the sars coronavirus: a model response to emerging infectious diseases. J. Struct. Funct. Genom. 2007;8:85–97. doi: 10.1007/s10969-007-9024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak P., Maitra-Majee S., Das J.K., Mukherjee A., Ghosh Dastidar S., Pal Choudhury P., Lahiri Majumder A. An evolutionary analysis identifies a conserved pentapeptide stretch containing the two essential lysine residues for rice l-myo-inositol 1-phosphate synthase catalytic activity. PLoS One. 2017;12 doi: 10.1371/journal.pone.0185351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beletskii A., Bhagwat A.S. Transcription-induced mutations: increase in c to t mutations in the nontranscribed strand during transcription in Escherichia coli. Proc. Natl. Acad. Sci. 1996;93:13919–13924. doi: 10.1073/pnas.93.24.13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi M., Benvenuto D., Giovanetti M., Angeletti S., Ciccozzi M., Pascarella S. Sars-cov-2 envelope and membrane proteins: structural differences linked to virus characteristics? BioMed Research International. 2020;2020 doi: 10.1155/2020/4389089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Błażej P., Mackiewicz D., Grabińska M., Wnetrzak M., Mackiewicz P. Optimization of amino acid replacement costs by mutational pressure in bacterial genomes. Scientific reports. 2017;7:1–18. doi: 10.1038/s41598-017-01130-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bofkin L., Goldman N. Variation in evolutionary processes at different codon positions. Mol. Biol. Evol. 2007;24:513–521. doi: 10.1093/molbev/msl178. [DOI] [PubMed] [Google Scholar]
- Calligari P., Bobone S., Ricci G., Bocedi A. Molecular investigation of sars–cov-2 proteins and their interactions with antiviral drugs. Viruses. 2020;12:445. doi: 10.3390/v12040445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E., Fariselli P., Casadio R. I-mutant2. 0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33:W306–W310. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.-M., Tsoi H., Chan W.-M., Zhai S., Wong C.-O., Yao X., Chan W.-Y., Tsui S.K.-W., Chan H.Y.E. The ion channel activity of the sars-coronavirus 3a protein is linked to its pro-apoptotic function. Int. J. Biochem. Cell Biol. 2009;41:2232–2239. doi: 10.1016/j.biocel.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T.-J., Yang D.-M., Wang M.-L., Liang K.-H., Tsai P.-H., Chiou S.-H., Lin T.-H., Wang C.-T. Genomic analysis and comparative multiple sequences of sars-cov2. J. Chin. Med. Assoc. 2020;83:537–543. doi: 10.1097/JCMA.0000000000000335. [DOI] [PubMed] [Google Scholar]
- Chaudhuri A. Comparative analysis of non structural protein 1 of sars-cov2 with sars-cov1 and mers-cov: an in silico study. bioRxiv. 2020 doi: 10.1101/2020.06.09.142570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M., Olinares P.D.B., Maruthi K., Eng E.T., Vatandaslar H., Chait B.T., et al. Structural basis for helicase-polymerase coupling in the sars-cov-2 replication-transcription complex. Cell. 2020;182(6):1560–1573. doi: 10.1016/j.cell.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Chan A.P. Provean web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Sims G.E., Murphy S., Miller J.R., Chan A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7 doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das J.K., Das P., Ray K.K., Choudhury P.P., Jana S.S. Mathematical characterization of protein sequences using patterns as chemical group combinations of amino acids. PLoS One. 2016;11 doi: 10.1371/journal.pone.0167651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das J.K., Singh R., Choudhury P.P., Roy B. Computational Intelligence and Big Data Analytics. Springer; 2019. Identifying driver potential in passenger genes using chemical properties of mutated and surrounding amino acids; pp. 107–118. [Google Scholar]
- Das J.K., Tradigo G., Veltri P., Guzzi A.R.S., Pietro H. Data science in unveiling covid-19 pathogenesis and diagnosis: evolutionary origin to drug repurposing. Briefings in Bioinformatics. 2020 doi: 10.1093/bib/bbaa420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das J., Chakrobarty S., Roy S. Impact analysis of sars-cov2 on signaling pathways during covid19 pathogenesis using codon usage assisted host-viral protein interactions. bioRxiv. 2020 doi: 10.1101/2020.07.29.226217. [DOI] [Google Scholar]
- Dilucca M., Forcelloni S., Georgakilas A.G., Giansanti A., Pavlopoulou A. Codon usage and phenotypic divergences of sars-cov-2 genes. Viruses. 2020;12:498. doi: 10.3390/v12050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaio D., Nathans D. Regulatory mutants of simian virus 40: effect of mutations at a t antigen binding site on dna replication and expression of viral genes. J. Mol. Biol. 1982;156:531–548. doi: 10.1016/0022-2836(82)90265-0. [DOI] [PubMed] [Google Scholar]
- Eaaswarkhanth M., Al Madhoun A., Al-Mulla F. Could the d614 g substitution in the sars-cov-2 spike (s) protein be associated with higher covid-19 mortality? Int. J. Infect. Dis. 2020;96:459–460. doi: 10.1016/j.ijid.2020.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foy E., Li K., Wang C., Sumpter R., Ikeda M., Lemon S.M., Gale M. Regulation of interferon regulatory factor-3 by the hepatitis c virus serine protease. Science. 2003;300:1145–1148. doi: 10.1126/science.1082604. [DOI] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A sars-cov-2 protein interaction map reveals targets for drug repurposing. Nature. 2020:1–13. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossoehme N.E., Li L., Keane S.C., Liu P., Dann C.E., III, Leibowitz J.L., Giedroc D.P. Coronavirus n protein n-terminal domain (ntd) specifically binds the transcriptional regulatory sequence (trs) and melts trs-ctrs rna duplexes. J. Mol. Biol. 2009;394:544–557. doi: 10.1016/j.jmb.2009.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C., McDowell I.C., Nodzenski M., Scholtens D.M., Allen A.S., Lowe W.L., Reddy T.E. Transversions have larger regulatory effects than transitions. BMC Genomics. 2017;18:394. doi: 10.1186/s12864-017-3785-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson C., Govindarajan S., Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22:346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Haig D., Hurst L.D. A quantitative measure of error minimization in the genetic code. J. Mol. Evol. 1991;33:412–417. doi: 10.1007/BF02103132. [DOI] [PubMed] [Google Scholar]
- He R., Leeson A., Ballantine M., Andonov A., Baker L., Dobie F., Li Y., Bastien N., Feldmann H., Strocher U., et al. Characterization of protein–protein interactions between the nucleocapsid protein and membrane protein of the sars coronavirus. Virus Res. 2004;105:121–125. doi: 10.1016/j.virusres.2004.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofman K. Tmbase: a database of membrane spanning protein segments. Biol. Chem. Hoppe Seyler. 1993;374:166. [Google Scholar]
- Huang Y., Yang C., Xu X.-f., Xu W., Liu S.-w. Structural and functional properties of sars-cov-2 spike protein: potential antivirus drug development for covid-19. Acta Pharmacol. Sin. 2020:1–9. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa E., Merhi G., Panossian B., Salloum T., Tokajian S. Sars-cov-2 and orf3a: nonsynonymous mutations, functional domains, and viral pathogenesis. Msystems. 2020;5 doi: 10.1128/mSystems.00266-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A., Paul S. Phylogenetic analysis of the novel coronavirus reveals important variants in indian strains. BioRxiv. 2020 doi: 10.1101/2020.04.14.041301. [DOI] [Google Scholar]
- Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Z., Zhou Z., Chen Q., et al. Crystal structure of sars-cov-2 nucleocapsid protein rna binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B. 2020;10(7):1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur N., Singh R., Dar Z., Bijarnia R.K., Dhingra N., Kaur T. Genetic comparison among various coronavirus strains for the identification of potential vaccine targets of sars-cov2. Infection, Genetics and Evolution. 2020;89 doi: 10.1016/j.meegid.2020.104490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Lee J.-Y., Yang J.-S., Kim J.W., Kim V.N., Chang H. The architecture of sars-cov-2 transcriptome. Cell. 2020;181(4):914–921. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristofich J., Morgenthaler A.B., Kinney W.R., Ebmeier C.C., Snyder D.J., Old W.M., Cooper V.S., Copley S.D. Synonymous mutations make dramatic contributions to fitness when growth is limited by a weak-link enzyme. PLoS Genet. 2018;14 doi: 10.1371/journal.pgen.1007615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland C. Codon bias and gene expression. FEBS Lett. 1991;285:165–169. doi: 10.1016/0014-5793(91)80797-7. [DOI] [PubMed] [Google Scholar]
- Li J.-Y., Liao C.-H., Wang Q., Tan Y.-J., Luo R., Qiu Y., Ge X.-Y. The orf6, orf8 and nucleocapsid proteins of sars-cov-2 inhibit type i interferon signaling pathway. Virus Res. 2020;286:198074. doi: 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewe L., Hill W.G. 2010. The Population Genetics of Mutations: Good, Bad and Indifferent. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Xu K., Sun B. Molecular Biology of the SARS-Coronavirus. Springer; 2010. Sars accessory proteins orf3a and 9b and their functional analysis; pp. 167–175. [Google Scholar]
- Lyons D.M., Lauring A.S. Evidence for the selective basis of transition-to-transversion substitution bias in two rna viruses. Mol. Biol. Evol. 2017;34:3205–3215. doi: 10.1093/molbev/msx251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra A., Sarkar M.C., Raheja H., Biswas N.K., Chakraborti S., Singh A.K., Ghosh S., Sarkar S., Patra S., Mondal R.K., et al. Mutations in Sars-Cov-2 viral rna identified in eastern India: possible implications for the ongoing outbreak in India and impact on viral structure and host susceptibility. Journal of Biosciences. 2020;45 doi: 10.1007/s12038-020-00046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad S., Bouchama A., Mohammad Alharbi B., Rashid M., Saleem Khatlani T., Gaber N.S., Malik S.S. Sars-cov-2 orf8 and sars-cov orf8ab: genomic divergence and functional convergence. Pathogens. 2020;9:677. doi: 10.3390/pathogens9090677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naskalska A., Dabrowska A., Szczepanski A., Milewska A., Jasik K.P., Pyrc K. Membrane protein of human coronavirus nl63 is responsible for interaction with the adhesion receptor. J. Virol. 2019;93 doi: 10.1128/JVI.00355-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C.A., Pekosz A., Lee C.A., Diamond M.S., Fremont D.H. Structure and intracellular targeting of the sars-coronavirus orf7a accessory protein. Structure. 2005;13:75–85. doi: 10.1016/j.str.2004.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin J.B., Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat. Rev. Genet. 2011;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan C., Ramachandran G. Stereochemical criteria for polypeptide and protein chain conformations: II. Allowed conformations for a pair of peptide units. Biophys. J. 1965;5:909–933. doi: 10.1016/S0006-3495(65)86759-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y., Wei C.L., Ling A.E., Vega V.B., Thoreau H., Thoe S.Y.S., Chia J.-M., Ng P., Chiu K.P., Lim L., et al. Comparative full-length genome sequence analysis of 14 sars coronavirus isolates and common mutations associated with putative origins of infection. Lancet. 2003;361:1779–1785. doi: 10.1016/S0140-6736(03)13414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha I., Ghosh N., Maity D., Sharma N., Sarkar J.P., Mitra K. Genome-wide analysis of indian sars-cov-2 genomes for the identification of genetic mutation and snp, Infection. Genet. Evol. 2020;85:104457. doi: 10.1016/j.meegid.2020.104457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikatendu K.S., Joseph J.S., Subramanian V., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P. Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the n-terminal domain of n protein. J. Virol. 2007;81:3913–3921. doi: 10.1128/JVI.02236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y., Kawachi K., Terada Y., Omori H., Matsuura Y., Kamitani W. Two-amino acids change in the nsp4 of sars coronavirus abolishes viral replication. Virology. 2017;510:165–174. doi: 10.1016/j.virol.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaddar A., Gadepalli R., Nag V.L., Misra S. The enigma of low covid-19 fatality rate in India. Front. Genet. 2020;11:854. doi: 10.3389/fgene.2020.00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardar R., Satish D., Birla S., Gupta D. Comparative analyses of sar-cov2 genomes from different geographical locations and other coronavirus family genomes reveals unique features potentially consequential to host-virus interaction and pathogenesis. bioRxiv. 2020 doi: 10.1101/2020.03.21.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta A., Choudhury P.P., Manners H.N., Guzzi P.H., Roy S. 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) IEEE; 2018. Chemical characterization of interacting genes in few subnetworks of alzheimer's disease; pp. 2720–2725. [Google Scholar]
- Shannon A., Le N.T.T., Selisko B., Eydoux C., Alvarez K., Guillemot J.-C., Decroly E., Peersen O., Ferron F., Canard B. Remdesivir and sars-cov-2: structural requirements at both nsp12 rdrp and nsp14 exonuclease active-sites. Antivir. Res. 2020;104793 doi: 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons M.P. Relative benefits of amino-acid, codon, degeneracy, dna, and purine-pyrimidine character coding for phylogenetic analyses of exons. J. Syst. Evol. 2017;55:85–109. [Google Scholar]
- Tan Y.W., Fang S., Fan H., Lescar J., Liu D. Amino acid residues critical for rna-binding in the n-terminal domain of the nucleocapsid protein are essential determinants for the infectivity of coronavirus in cultured cells. Nucleic Acids Res. 2006;34:4816–4825. doi: 10.1093/nar/gkl650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ul Qamar M.T., Alqahtani S.M., Alamri M.A., Chen L.-L. Structural basis of sars-cov-2 3clpro and anti-covid-19 drug discovery from medicinal plants. Journal of Pharmaceutical Analysis. 2020;10(4):313–319. doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the sars-cov-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenden R., Cullis P., Southgate C. Water, protein folding, and the genetic code. Science. 1979;206:575–577. doi: 10.1126/science.493962. [DOI] [PubMed] [Google Scholar]
- Xia S., Zhu Y., Liu M., Lan Q., Xu W., Wu Y., Ying T., Liu S., Shi Z., Jiang S., et al. Fusion mechanism of 2019-ncov and fusion inhibitors targeting hr1 domain in spike protein. Cell. Mol. Immunol. 2020:1–3. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav P.D., Potdar V.A., Choudhary M.L., Nyayanit D.A., Agrawal M., Jadhav S.M., Majumdar T.D., Shete-Aich A., Basu A., Abraham P., et al. Full-genome sequences of the first two sars-cov-2 viruses from India. Indian J. Med. Res. 2020;151:200. doi: 10.4103/ijmr.IJMR_663_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M.-S., Lee J., Lee J.M., Kim Y., Chin Y.-W., Jee J.-G., Keum Y.-S., Jeong Y.-J. Identification of myricetin and scutellarein as novel chemical inhibitors of the sars coronavirus helicase, nsp13. Bioorg. Med. Chem. Lett. 2012;22:4049–4054. doi: 10.1016/j.bmcl.2012.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen C.-K., Lam J.-Y., Wong W.-M., Mak L.-F., Wang X., Chu H., Cai J.-P., Jin D.-Y., To K.K.-W., Chan J.F.-W., et al. Sars-cov-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microb. Infect. 2020:1–29. doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng R., Yang R.-F., Shi M.-D., Jiang M.-R., Xie Y.-H., Ruan H.-Q., Jiang X.-S., Shi L., Zhou H., Zhang L., et al. Characterization of the 3a protein of sars-associated coronavirus in infected vero e6 cells and sars patients. J. Mol. Biol. 2004;341:271–279. doi: 10.1016/j.jmb.2004.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W., Liu G., Ma H., Zhao D., Yang Y., Liu M., Mohammed A., Zhao C., Yang Y., Xie J., et al. Biochemical characterization of sars-cov-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020;527(3):618–623. doi: 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Accession numbers of SARS-CoV-2 collected sequences from Indian isolates.
The polygenetic tree for each of the SARS-CoV-2 proteins.
The variant with frequency and respective nucleotide mutation and type, amino acid change, and number of mutations for each of the SARS-CoV-2 proteins.
The observed non-synonymous substitution with the protein accession number in each of the SARS-CoV-2 proteins.