Abstract
Introduction:
We characterized the cervical 16S rDNA microbiome of patients in Botswana with high-grade cervical dysplasia and locally advanced cervical cancer.
Methods:
This prospective study included 31 patients: 21 with dysplasia and 10 with cancer. The Shannon diversity index was used to evaluate alpha (intra-sample) diversity, while the UniFrac (weighted and unweighted) and Bray-Curtis distances were employed to evaluate beta (inter-sample) diversity. The relative abundance of microbial taxa was compared among samples using linear discriminant analysis effect size.
Results:
Alpha diversity was significantly higher in patients with cervical cancer than in patients with cervical dysplasia (p<0.05). Beta diversity also differed significantly (weighted UniFrac Bray-Curtis, p<0.01). Neither alpha diversity (p=0.8) nor beta diversity (p=0.19) varied by HIV status. The results of linear discriminant analysis effect size demonstrated that multiple taxa differed significantly between patients with cervical dysplasia vs. cancer. Lachnospira bacteria (in the Clostridia class) were particularly enriched among cervical dysplasia patients, while Proteobacteria (members of the Firmicutes phyla and the Comamonadaceae family) were enriched in patients with cervical cancer.
Discussion:
The results of our study suggest that differences exist in the diversity and composition of the cervical microbiota between patients with cervical dysplasia and patients with cervical cancer in Botswana. Additional studies are warranted to validate these findings and elucidate their clinical significance among women living in sub-Saharan Africa, as well as other regions of the world.
Keywords: Cervical dysplasia, cervical cancer, gynecologic cancer, cervical microbiota, microbiome, HIV, Botswana, sub-Saharan Africa
INTRODUCTION
Cervical cancer is one of the most common malignancies globally and the most common cause of cancer death among African women1. More than half a million new cases of invasive cervical cancer are expected to be diagnosed worldwide in 2020, resulting in over 300,000 deaths2. African women have a far higher risk of cervical cancer than women in regions with ready access to preventative health care screening1. Fourteen percent of the world’s cervical cancer cases and 18% of cervical cancer-related deaths occur in women living in sub-Saharan Africa1,3. The incidence of cervical cancer in sub-Saharan Africa—which includes Botswana, Lesotho, Namibia, South Africa, and Swaziland—is expected to increase by 35% come 20301.
Persistent exposure to human papilloma virus (HPV) is the well-established antecedent to cervical cancer4,5. Women with HIV are at increased risk of HPV infection and thus cervical cancer, despite access to anti-retroviral therapy6. The high regional prevalence of HIV in sub-Saharan countries such as Botswana underscores the importance of cervical cancer prevention in these regions7. Botswana established one of the first nationwide HIV treatment programs8 in Africa, with a corresponding decline in HIV-associated mortality; however, the incidence of cervical cancer in Botswana remains among the highest globally (36.6 per 100,000), with nearly two-thirds of cases occurring in HIV-positive women9.
The microbiome has recently been demonstrated to play a critical role in cancer progression, metastasis, and therapy response10. The female cervix is a microbiome-rich environment, but the effect of this microbiome on cancer development and pathogenesis remains limited and poorly understood11. Given the rising incidence of cervical cancer, understanding the effect of the cervical flora on cancer progression and response (as well as the converse effect of definitive treatments on said flora), represents an unmet need with high potential benefit for vulnerable patient populations. Cervical cancer is uniquely positioned for such an investigation, as it allows direct visualization and contact with the primary tumor at the start of treatment. Here we aim to characterize cervical microbiome of patients with cervical dysplasia and cancer living in Botswana.
To our knowledge, no published studies have specifically explored the cervical dysplasia vs tumor microbiome among women in sub-Saharan Africa. As cervical microbial differences can affect cancer risk and treatment response through several pathways, we characterized the 16S rDNA cervical microbiome of women with dysplasia and locally advanced cervical cancer in Botswana. We hypothesized that the cervical microbiome of cancer patients is distinct from that of dysplasia patients. Furthermore, the longitudinal identification of bacterial strains associated with the cervical microbiome will allow further study of the organisms that stably colonize cervical cancers. The detection of bacterial strains associated with treatment response will then lay groundwork for interventions that alter the tumor microbiota to improve cancer outcomes.
PATIENTS AND METHODS
Participants and Clinical Data
Patients with newly diagnosed, biopsy-proven cervical dysplasia or locally advanced, non-metastatic cervical carcinoma presenting at Princess Marina Hospital were prospectively identified between July 2018 and February 2019. Ineligibility criteria included any history of non-cervical primary cancer and active pregnancy. Comprehensive medical history, including current medication, was assessed via interview with clinical provider or trained study staff. Patient medical records were reviewed to obtain demographic and clinico-pathologic data. Tissue samples were collected prior to initiation of definitive chemoradiation entailing external beam radiation therapy and brachytherapy with concurrent cisplatin for invasive cancer.
The study protocol, including subject recruitment and tissue sampling, was approved by the Institutional Review Boards (IRBs) at the University of Botswana (UBR/RES/IRB/BIO/045), the University of Pennsylvania (830039), and The University of Texas MD Anderson Cancer Center (MDACC 2014–0543). Informed consent was mandatory for study participation and documented by patients in writing.
Sample Collection and DNA Extraction
Cervical samples were collected using a matrix-designed quick-release Isohelix swab. The swabs were placed in 20 μL of protease K and 400 μL of lysis buffer (Isohelix) and stored at −80°C within 1 hour of sample collection. Bacterial genomic DNA was extracted using a MO BIO PowerSoil DNA Isolation Kit (MO BIO Laboratories). Samples were shipped to the US for downstream applications including DNA processing and sequencing.
16S rRNA Gene Sequencing and Sequence Data Processing
16S rRNA gene sequencing of the cervical swabs was performed at the Alkek Center for Metagenomics and Microbiome Research at Baylor College of Medicine (Houston, Texas), using methods adapted from those used for the Human Microbiome Project.12 The 16S rDNA V4 region was amplified by PCR using primers that contained sequencing adapters and single-end barcodes, allowing the pooling and direct sequencing of PCR products. Amplicons were sequenced on the MiSeq platform (Illumina) using the 2×250-bp paired-end protocol, yielding paired-end reads that overlapped almost completely. The sequence reads were de-multiplexed, quality filtered, and subsequently merged using USEARCH version 7.0.1090 (4). 16S rRNA gene sequences were clustered into OTUs at a similarity cut-off value of 97% using the UPARSE algorithm.13 To generate taxonomies, we mapped OTUs to an optimized version of the SILVA rRNA database containing the 16S v4 region. A custom script was used to construct an OTU table from the output files generated, as described above, for downstream analyses of alpha diversity, beta diversity, and phylogenetic trends. Principal coordinates analysis was performed by institution and sample set to ensure that no batch effects were present.
Statistical Analyses
For the microbiome analysis, the rarefaction depth was set at 3561 reads. Alpha (within sample) diversity was examined using the Shannon diversity index, and beta (between sample) diversity was examined using UniFrac (weighted and unweighted) and Bray-Curtis distances. The relative abundance of microbial taxa and genera were compared between samples; we then determined differentially abundant bacterial genera by case status using linear discriminant analysis effect size,14 applying the 1-against-all strategy with a threshold of 4 on the logarithmic linear discriminant analysis score for discriminative features and an α of 0.05 for the factorial Kruskal-Wallis test among classes. linear discriminant analysis effect size was restricted to bacteria that were present in 20% or more of the study population. Observed differences were subjected to paired analysis using two sample Z test for proportions, or Student t test where appropriate.
RESULTS
The 16S rDNA cervical microbiome was characterized among 31 patients: 21 with cervical dysplasia and 10 with cancer. Clinico-pathologic data are summarized in Table 1. Cervical dysplasia patients were classified according to histologic grade of cervical intraepithelial neoplasia ([CIN]1–3). Approximately 58% of study patients (18 of 31) had CIN 3, while 32% (10 of 31) had moderate- or poorly-differentiated squamous cell cancer of the cervix. HPV status was unknown at the time of cervical sampling.
Table 1.
Clinico-pathological features of 31 patients in Botswana with cervical dysplasia or cervical cancer
| Feature | Result |
|---|---|
| Type of cervical lesion, (n=31) | |
| CIN 1 | 0 |
| CIN 2 | 3 |
| CIN 3 | 18 |
| Cervical cancer | 10 |
| HIV status, % | |
| Positive | 77 |
| Negative | 23 |
| Smoking status, % | |
| Smoker | 6 |
| Non-Smoker | 94 |
CIN, cervical intraepithelial neoplasia.
Patient microbiota were initially analyzed with respect to HIV status. Neither α diversity (p=0.8) nor β diversity (p=0.19) varied by HIV status (Figure 1A,B). Figure 1C shows the top 15 most abundant genera for both HIV positive and HIV negative patients.
Figure 1. The cervical microbiota of cervical dysplasia and cervical cancer individuals with and without HIV.
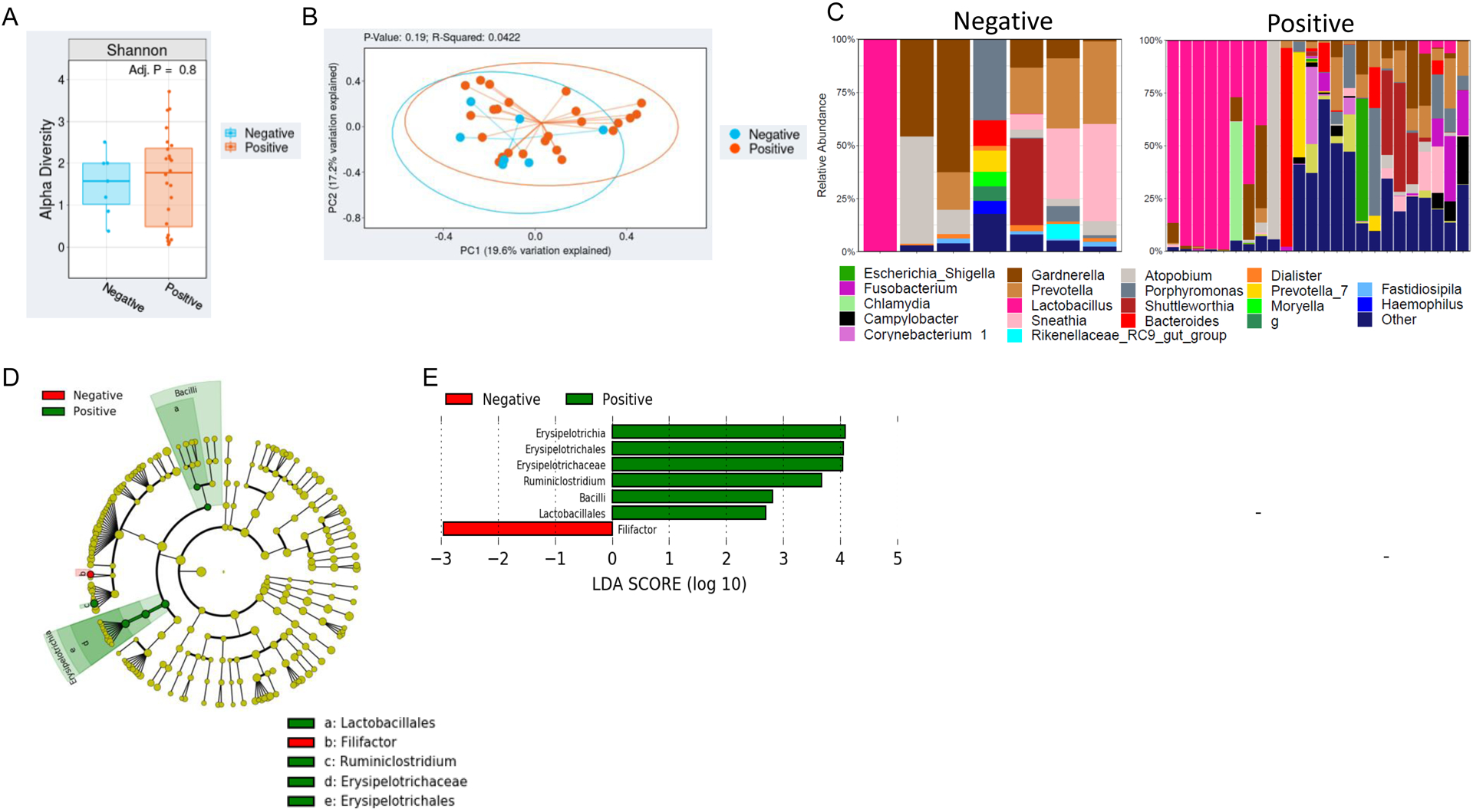
A) Overall alpha diversity, as assessed by Shannon diversity in HIV positive cervical dysplasia and cervical cancer patient’s (n=24) vs negative patients (n=7). B) Beta diversity, as assessed by Bray-Curtis unweighted UniFrac in HIV positive patients vs HIV negative patients. C) Stacked bar plot of the top 10 most abundant genus-level bacteria in HIV positive vs negative patients. Each bar represents a single participant. D,E) LEfSe analysis identified the most differentially abundant taxa between HIV positive and negative patients. D) Cladogram representation of the significantly different taxa features from phylum (inner circle) to genus (outer circle). E) Histogram showing the LDA scores of genera differentially abundant between the two groups. LEfSe was restricted to p < 0.05 for class and subclass analysis and a minimum LDA score of 2.0.
Subsequent analysis was conducted to characterize variations in the cervical microbiome among dysplasia vs. cervical cancer. Clinical and demographic characteristics are displayed in Table 2. Mean age and BMI were similar among cervical dysplasia and cancer patients (mean age, 41.8 vs 50.7 years [p=0.1]; and mean BMI, 26.3 vs. 30.0 kg/m2 [p=0.19], respectively). Significantly higher α diversity, as measured by SDI (p<0.05), was observed in cervical dysplasia patients relative to cervical cancer patients (Figure 2A). Patients with CIN 3 tended to have higher α diversity than those with CIN 2 (Figure 2B). As with α diversity, overall β diversity differed significantly by cancer status (weighted Bray-Curtis Unifrac; p<0.01) (Figure 2C,D). Figure 2E shows the top 15 most abundant genera in cervical dysplasia samples and cervical cancer samples, showing overall higher diversity in the cancer group. The percentage of subjects with a cervical microbiome dominated by Lactobacillus or Gardnerella appears lower in the cervical cancer cohort (1 of 10 patients).
Table 2.
Selected characteristics of 31 patients in Botswana with cervical dysplasia vs. cervical cancer
| Characteristic | Dysplasia (n=21) | Cancer (n=10) | P value* |
|---|---|---|---|
| Mean age (SD), years | 41.8 (7.5) | 50.7 (12) | 0.1 |
| Mean BMI (SD), kg/m2 | 26.3 (6.4) | 30.0 (7.2) | 0.2 |
| HIV status, % | |||
| Positive | 17 (81%) | 7 (70%) | 0.5 |
| Negative | 4 (19%) | 3 (30%) | 0.5 |
| Smoking status, % | |||
| Smoker | 2 (10%) | 0 (0%) | 0.3 |
| Non-Smoker | 19 (90%) | 10 (100%) | 0.3 |
P values were based on a t-test (continuous variables) or z-test (proportions). All tests were 2-sided.
Figure 2. The cervical microbiota in individuals with cervical cancer is significantly different than individuals with cervical dysplasia.
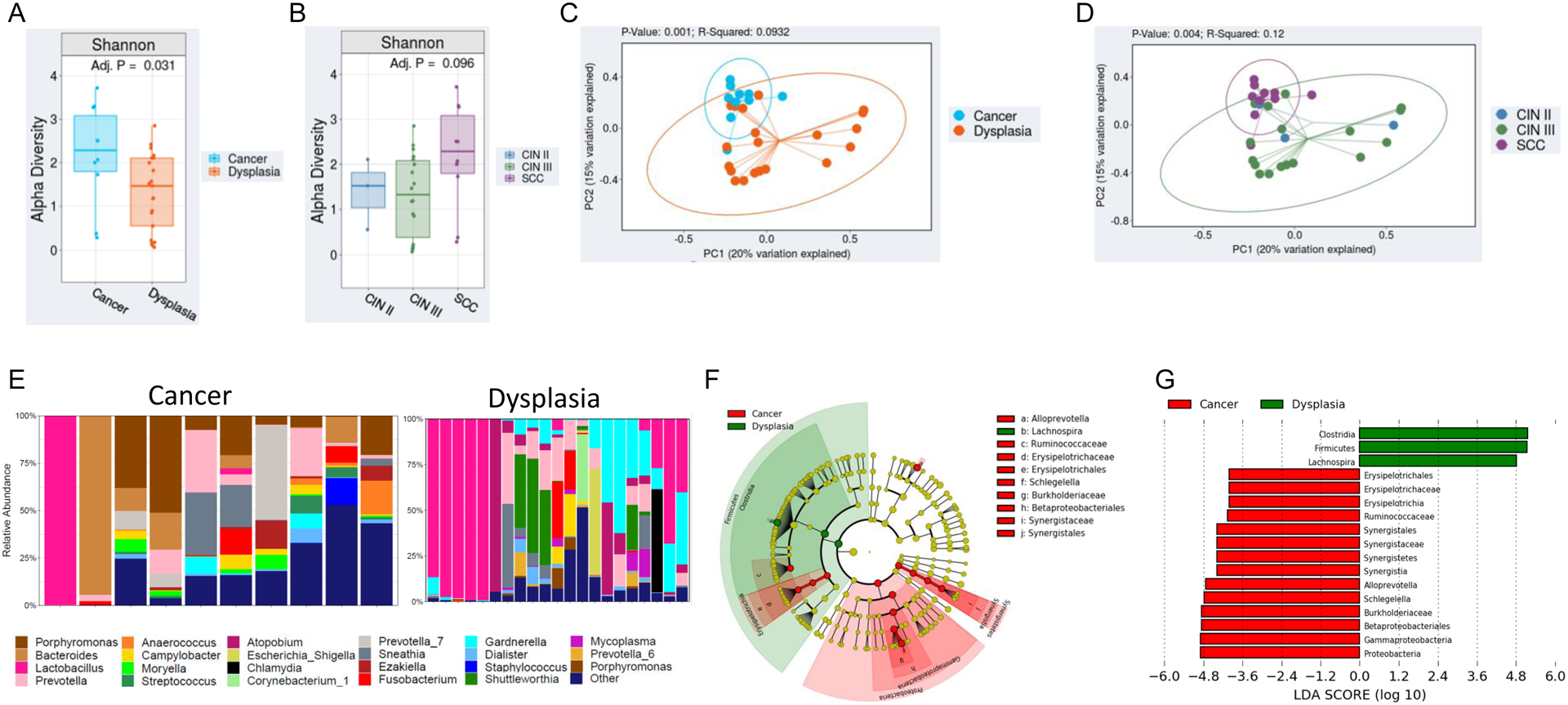
A,B) Overall alpha diversity, as assessed by Shannon diversity in cervical dysplasia (n=21) and cervical cancer patients (n=10). C,D) Beta diversity, as assessed by Bray-Curtis weighted UniFrac in Cervical dysplasia vs cervical cancer patients. E) Stacked bar plot of the top 15 most abundant genus-level bacteria in cervical dysplasia patients vs cervical cancer patients. Each bar represents a single participant. F,G) LEfSe analysis identified the most differentially abundant taxa between cervical dysplasia and cervical cancer patients. F) Cladogram representation of the significantly different taxa features from phylum (inner circle) to genus (outer circle). G) Histogram showing the LDA scores of genera differentially abundant between the two groups. LEfSe was restricted to p < 0.05 for class and subclass analysis and a minimum LDA score of 2.0.
Linear discriminant analysis effect size was used to identify bacterial genera that were differentially enriched among our patient cohort (p<0.05). The genera Ersipelotrichia, Erysipelotrichales, Erysipelotrichaceae, and Ruminiclostridium were enriched in HIV-positive patients, while only Filifactor had higher abundance in HIV-negative patients (Figure 1D,E). Interestingly, the genus Lachnospira (in the Clostridia class of bacteria) was significantly enriched among cervical dysplasia patients; while several Proteobacteria taxa (Betaproteobacteria, Gammaproteobacteria, and Burkholderiaceae) and members of the Firmicutes phyla (Erysiopelotrichaceae and Synergistaceae) and the Comamonadaceae family had higher abundance in cervical cancer patients (p<0.05) (Figure 2F,G).
DISCUSSION
In this novel prospective study, we characterized the cervical microbiome of women with cervical dysplasia and cervical cancer living in Botswana, with the hypothesis that the cervical microbiome would be distinct between these two patient groups. Accordingly, significant differences in cervical α and β diversity were observed among these groups of patients, as well as compositional differences. Interestingly, the overall α and β diversity analysis results did not indicate a difference with regard to HIV status.
Prior microbiome cancer research has focused on exploring the relative abundance of bacteria in the vaginal epithelium, with the assignment of community-state types based on the richness of Lactobacilli species13–15. The presence and abundance of specific Lactobacilli species—for example L. crispatus, L. gasseri, or L. jensenii—is associated with a predisposition towards bacterial vaginosis and other pro-inflammatory states consequently leading to DNA cell damage and potentially carcinogenic changes15,18–20.
However, despite the comparative wealth of data focused on the vaginal microbiome21, the ectocervical microbiome has yet to be well described. Pertinent studies have thus far concentrated on the setting of pregnancy or pelvic inflammatory disease. Previous work with 16S rDNA sequencing in pregnancy suggests that cervical microbiota diversity differs by race22 and that the presence of non-Lactobacillus community state types is associated with a robust cervical inflammatory response in the setting of pre-term, premature membrane rupture23,24. In patients with pelvic inflammatory disease, Wang et al. demonstrated a dominance by Lactobacillus and Gardnerella in the cervical microbiota, suggesting that the abundance of different taxa is associated with both acute and chronic inflammatory states25. In turn, states of polybacterial dysbiosis and chronic local inflammation are thought to encourage the perseverance of HPV, thus ultimately promoting the development of cervical dysplasia and carcinogenesis through persistent HPV exposure15,17,22–25.
Persistent HPV infections are thought to trigger an innate immune response, resulting in the suppression of infected cervicovaginal mucosal cells18,29,30. An altered mucosal microenvironment leads to the growth of anaerobic organisms at the expense of Lactobacillus growth, creating cervicovaginal dysbiosis31. Linear discriminant analysis effect size was designed to detect bacterial taxa that are associated with a specific state32. In our study, linear discriminant analysis effect size identified Clostridia, Firmicutes, and Lachnospira as taxa negatively associated with cervical cancer, while several Proteobacteria were identified as taxa positively-associated with cervical cancer (as compared with dysplasia).
Dysbiosis causes cervicovaginal inflammation and other unfavorable changes in the cervicovaginal mucosal barrier. Worldwide, the most common type of cervicovaginal dysbiosis (defined as a cervicovaginal microbiome not dominated by Lactobacilli) is bacterial vaginosis33. Bacterial vaginosis is characterized by a persistent decrease in Lactobacilli and an increase in fastidious anaerobes29. Globally, the prevalence of bacterial vaginosis is highest among women living in sub-Saharan Africa and in women of sub-Saharan African descent33. Cervicovaginal dysbiotic states, such as bacterial vaginosis, lead to an altered metabolic profile and reduced cervicovaginal barrier function. This dysbiotic state is not only associated with an increased acquisition of HIV, but also with high-risk HPV, cervical dysplasia, and ultimately cervical cancer29,34. The percentage of subjects with their cervical microbiome dominated by Lactobacillus was low in our cohort of patients, although the proportion of dysplasia patients with Lactobacillus-dominated cervical microbiomes was higher than that of cancer patients. The lack of Lactobacilli identified in our cervical dysplasia and cervical cancer patients supports this rationale and suggests that cervicovaginal microbes are important in preventing or enhancing the acquisition and pathogenesis of HPV and HIV. Identifying the microbes that are associated with enhanced pathogenesis and ultimately oncogenesis or tumorigenesis is especially important in susceptible populations such as HIV-positive women in Botswana. Previous studies evaluating the noncancerous cervical microbiome have shown HIV status to be associated with a decrease in microbiome diversity and increase in bacterial richness11,35,36. In our study, although neither α nor β diversity varied by HIV status, HIV status was shown to have a significant effect on the cervical microbiome bacterial composition with the genera Ersipelotrichia, Erysipelotrichales, Erysipelotrichaceae, and Ruminiclostridium being enriched in HIV-positive patients, and only Filifactor being enriched in HIV-negative patients. These results support what has been previously reported in other studies investigating the cervicovaginal microbiome and imply that a patients HIV status exerts some selective pressure on the cervical epithelium microenvironment11,37.
The gut microbiome and its influence on carcinogenesis and prognosis has been well described, most notably in melanoma and colorectal cancer10,38,39. Bullman et al. recently identified colonization by Fusobacterium (and its associated microbiome Bacteriodes, Selenomas, and Prevotella) at both the primary tumor and distant paired metastatic sites of colorectal cancer. This finding suggests that colonized organisms inhabiting the primary tumor could potentially migrate with primary tumor cells to distant locations and manipulate microbiota diversity at metastatic sites, contributing to poor anti-tumor immunity40. Thus, identifying the specific organisms that colonize the tumor microbiota may provide further insight into mechanisms that modulate immune response and potentiate tumor cell growth31. Historically, microbiome cervical cancer research has been limited to mainly Western industrialized populations. We hope that our findings in women in Botswana provide a timely and critical glimpse into this uniquely vulnerable population.
Although the present study yielded intriguing findings, an important limitation is the lack of a control group of “normal” patients without dysplasia or cancer. Additionally, details of menstrual phase, sexual, behavioral and reproductive characteristics, HPV infection and genotypes, antibiotics, synthetic hormones for contraceptive purposes, vaginal douching and presence of other sexually transmitted diseases were not known in our data set. Furthermore our modest sample size remains a potential limitation. Yet despite the relatively small size of this prospective patient cohort, large statistically significant differences were still observed between cervical dysplasia and cancer patients, alluding to the underlying complexity associated with the cervical microbiome in this patient population. Furthermore, this proof-of-concept study demonstrates the feasibility of using 16S rDNA next-generation sequencing to evaluate cervical microbiome differences, even among a unique remote population and within an international collaborative setting.
In conclusion, our study demonstrated hypothesis-generating differences in the cervical microbial profiles of women in Botswana with cervical cancer compared to those with cervical dysplasia. The cervical microbiome of women with cervical dysplasia and cervical cancer did not differ with respect to HIV status. The lack of Lactobacilli in our samples supports the rationale that cervicovaginal dysbiotic states (characterized by a persistent decrease in Lactobacilli) are associated with a higher incidence of HIV, cervical dysplasia, and cervical cancer. These findings help improve comprehension of the essential function of the tumor microbiome in cervical cancer. Additional studies are warranted to validate these findings and to elucidate the biological significance of these observed differences among women living in sub-Saharan Africa.
Highlights:
This prospective study fills the void of data on cervical microbiome in sub-Saharan African women
Among patients in Botswana, cervical microbiome diversity was greater for cancer versus dysplasia
Cervical microbiota of women with cancer are distinct in composition as compared with dysplasia
Acknowledgements
This work was supported in part by the National Institutes of Health through MD Anderson’s Cancer Center Support Grant P30 CA016672 (TTS), the National Institutes of Health T32 grant #5T32 CA101642-14 (TTS), and the Mentored Patient Oriented Career Research Development Award 1-K08CA230170-01A1 (SG). This study was partially funded by the MD Anderson HPV-Related Cancers Moonshot (AK). We gratefully acknowledge the patients who participated in this study.
Role of Funding Sources
The funding sources were not involved in the research hypothesis development, study design, data analysis, or manuscript writing. Data access was limited to the authors of this manuscript.
Footnotes
Conflicts of Interest
The authors report no conflicts of interest, financial or otherwise, related to the subject matter of the article submitted.
REFERENCES
- 1.De Vuyst H, Alemany L, Lacey C, et al. The burden of human papillomavirus infections and related diseases in sub-saharan Africa. Vaccine. 2013;31 Suppl 5:F32–46. doi: 10.1016/j.vaccine.2012.07.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 3.Klein C, Kahesa C, Mwaiselage J, West JT, Wood C, Angeletti PC. How the Cervical Microbiota Contributes to Cervical Cancer Risk in Sub-Saharan Africa. Front Cell Infect Microbiol. 2020;10. doi: 10.3389/fcimb.2020.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hammer A, Rositch A, Qeadan F, Gravitt PE, Blaakaer J. Age-specific prevalence of HPV16/18 genotypes in cervical cancer: A systematic review and meta-analysis. Int J Cancer. 2016;138(12):2795–2803. doi: 10.1002/ijc.29959 [DOI] [PubMed] [Google Scholar]
- 5.Grover S, Bvochora-Nsingo M, Yeager A, et al. Impact of Human Immunodeficiency Virus Infection on Survival and Acute Toxicities From Chemoradiation Therapy for Cervical Cancer Patients in a Limited-Resource Setting. Int J Radiat Oncol Biol Phys. 2018;101(1):201–210. doi: 10.1016/j.ijrobp.2018.01.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dryden-Peterson S, Bvochora-Nsingo M, Suneja G, et al. HIV Infection and Survival Among Women With Cervical Cancer. J Clin Oncol Off J Am Soc Clin Oncol. 2016;34(31):3749–3757. doi: 10.1200/JCO.2016.67.9613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grover S, Raesima M, Bvochora-Nsingo M, et al. Cervical Cancer in Botswana: Current State and Future Steps for Screening and Treatment Programs. Front Oncol. 2015;5:239. doi: 10.3389/fonc.2015.00239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaolathe T, Wirth KE, Holme MP, et al. Botswana’s progress toward achieving the 2020 UNAIDS 90-90-90 antiretroviral therapy and virological suppression goals: a population-based survey. Lancet HIV. 2016;3(5):e221–230. doi: 10.1016/S2352-3018(16)00037-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dryden-Peterson S, Medhin H, Kebabonye-Pusoentsi M, et al. Correction: Cancer Incidence following Expansion of HIV Treatment in Botswana. PloS One. 2015;10(9):e0138742. doi: 10.1371/journal.pone.0138742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell. 2018;33(4):570–580. doi: 10.1016/j.ccell.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein C, Gonzalez D, Samwel K, et al. Relationship between the Cervical Microbiome, HIV Status, and Precancerous Lesions. mBio. 2019;10(1). doi: 10.1128/mBio.02785-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bombardelli C, Ayuso JH, Pelayo RG. A framework for human microbiome research. Nature. 2012;486(7402):215–221. doi: 10.1038/nature11209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edgar RC. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10(10):996–998. doi: 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- 14.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6). doi: 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kyrgiou M, Mitra A, Moscicki A-B. Does the vaginal microbiota play a role in the development of cervical cancer? Transl Res J Lab Clin Med. 2017;179:168–182. doi: 10.1016/j.trsl.2016.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ravel J, Gajer P, Abdo Z, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4680–4687. doi: 10.1073/pnas.1002611107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brotman RM, Shardell MD, Gajer P, et al. Interplay between the temporal dynamics of the vaginal microbiota and human papillomavirus detection. J Infect Dis. 2014;210(11):1723–1733. doi: 10.1093/infdis/jiu330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van de Wijgert JHHM, Borgdorff H, Verhelst R, et al. The vaginal microbiota: what have we learned after a decade of molecular characterization? PloS One. 2014;9(8):e105998. doi: 10.1371/journal.pone.0105998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King CC, Jamieson DJ, Wiener J, et al. Bacterial vaginosis and the natural history of human papillomavirus. Infect Dis Obstet Gynecol. 2011;2011:319460. doi: 10.1155/2011/319460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet Lond Engl. 2001;357(9255):539–545. doi: 10.1016/S0140-6736(00)04046-0 [DOI] [PubMed] [Google Scholar]
- 21.Champer M, Wong AM, Champer J, et al. The role of the vaginal microbiome in gynaecological cancer. BJOG Int J Obstet Gynaecol. 2018;125(3):309–315. doi: 10.1111/1471-0528.14631 [DOI] [PubMed] [Google Scholar]
- 22.Wheeler S, Pryor K, Antczak B, Truong T, Murtha A, Seed P. The relationship of cervical microbiota diversity with race and disparities in preterm birth. J Neonatal-Perinat Med. 2018;11(3):305–310. doi: 10.3233/NPM-17111 [DOI] [PubMed] [Google Scholar]
- 23.Kacerovsky M, Pliskova L, Bolehovska R, et al. Lactobacilli-dominated cervical microbiota in women with preterm prelabor rupture of membranes. Pediatr Res. Published online December 2, 2019. doi: 10.1038/s41390-019-0692-1 [DOI] [PubMed] [Google Scholar]
- 24.Kacerovsky M, Vrbacky F, Kutova R, et al. Cervical microbiota in women with preterm prelabor rupture of membranes. PloS One. 2015;10(5):e0126884. doi: 10.1371/journal.pone.0126884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Zhang Y, Zhang Q, Chen H, Feng Y. Characterization of pelvic and cervical microbiotas from patients with pelvic inflammatory disease. J Med Microbiol. 2018;67(10):1519–1526. doi: 10.1099/jmm.0.000821 [DOI] [PubMed] [Google Scholar]
- 26.Marullo R, Werner E, Zhang H, Chen GZ, Shin DM, Doetsch PW. HPV16 E6 and E7 proteins induce a chronic oxidative stress response via NOX2 that causes genomic instability and increased susceptibility to DNA damage in head and neck cancer cells. Carcinogenesis. 2015;36(11):1397–1406. doi: 10.1093/carcin/bgv126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitra A, MacIntyre DA, Lee YS, et al. Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci Rep. 2015;5:16865. doi: 10.1038/srep16865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balkwill FR, Mantovani A. Cancer-related inflammation: common themes and therapeutic opportunities. Semin Cancer Biol. 2012;22(1):33–40. doi: 10.1016/j.semcancer.2011.12.005 [DOI] [PubMed] [Google Scholar]
- 29.van de Wijgert JHHM, Gill AC, Chikandiwa A, et al. Human papillomavirus infection and cervical dysplasia in HIV-positive women: potential role of the vaginal microbiota. AIDS. 2020;34(1):115–125. doi: 10.1097/QAD.0000000000002381 [DOI] [PubMed] [Google Scholar]
- 30.Karim R, Tummers B, Meyers C, et al. Human papillomavirus (HPV) upregulates the cellular deubiquitinase UCHL1 to suppress the keratinocyte’s innate immune response. PLoS Pathog. 2013;9(5):e1003384. doi: 10.1371/journal.ppat.1003384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ilhan ZE, Łaniewski P, Thomas N, Roe DJ, Chase DM, Herbst-Kralovetz MM. Deciphering the complex interplay between microbiota, HPV, inflammation and cancer through cervicovaginal metabolic profiling. EBioMedicine. 2019;44:675–690. doi: 10.1016/j.ebiom.2019.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brooks JP, Edwards DJ, Blithe DL, et al. Effects of combined oral contraceptives, depot medroxyprogesterone acetate and the levonorgestrel-releasing intrauterine system on the vaginal microbiome. Contraception. 2017;95(4):405–413. doi: 10.1016/j.contraception.2016.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van de Wijgert JHHM, Jespers V. The global health impact of vaginal dysbiosis. Res Microbiol. 2017;168(9–10):859–864. doi: 10.1016/j.resmic.2017.02.003 [DOI] [PubMed] [Google Scholar]
- 34.Borgdorff H, Gautam R, Armstrong SD, et al. Cervicovaginal microbiome dysbiosis is associated with proteome changes related to alterations of the cervicovaginal mucosal barrier. Mucosal Immunol. 2016;9(3):621–633. doi: 10.1038/mi.2015.86 [DOI] [PubMed] [Google Scholar]
- 35.Curty G, Costa RL, Siqueira JD, et al. Analysis of the cervical microbiome and potential biomarkers from postpartum HIV-positive women displaying cervical intraepithelial lesions. Sci Rep. 2017;7(1):17364. doi: 10.1038/s41598-017-17351-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oh HY, Kim B-S, Seo S-S, et al. The association of uterine cervical microbiota with an increased risk for cervical intraepithelial neoplasia in Korea. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2015;21(7):674.e1–9. doi: 10.1016/j.cmi.2015.02.026 [DOI] [PubMed] [Google Scholar]
- 37.Borgdorff H, Tsivtsivadze E, Verhelst R, et al. Lactobacillus-dominated cervicovaginal microbiota associated with reduced HIV/STI prevalence and genital HIV viral load in African women. ISME J. 2014;8(9):1781–1793. doi: 10.1038/ismej.2014.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu S, Rhee K-J, Albesiano E, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15(9):1016–1022. doi: 10.1038/nm.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arthur JC, Perez-Chanona E, Mühlbauer M, et al. Intestinal Inflammation Targets Cancer-Inducing Activity of the Microbiota. Science. 2012;338(6103):120–123. doi: 10.1126/science.1224820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bullman S, Pedamallu CS, Sicinska E, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358(6369):1443–1448. doi: 10.1126/science.aal5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Y, Weng W, Peng J, et al. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-κB, and Up-regulating Expression of MicroRNA-21. Gastroenterology. 2017;152(4):851–866.e24. doi: 10.1053/j.gastro.2016.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195–206. doi: 10.1016/j.chom.2013.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]


