Abstract
Endometrial stromal sarcomas (ESS) are a heterogeneous group of rare mesenchymal cancers. Considerable knowledge has been gained in recent years about the molecular characteristics of these cancers, which helps to classify them in a more meaningful manner leading to improved diagnosis, prognostication, and treatment. According to this classification, ESS is now grouped as low‐ or high‐grade. ESS may have overlapping clinical presentation, morphology, and immunohistochemical profile. Their genetic characteristics allow subdivision of many of them depending on which pathogenetically important fusion genes they carry, but clearly much more needs to be unraveled in this regard. We here provide an overview of the molecular pathogenetic knowledge gained so far on low‐ and high‐grade ESS.
Keywords: chromosomal aberrations, endometrial stromal sarcoma, fusion gene, high‐grade ESS, low‐grade ESS
1. INTRODUCTION
According to the latest World Health Organization (WHO) classification of tumors, Endometrial Stromal Sarcomas (ESS) belongs to the overall category of Endometrial Stromal and related Tumors (EST). The spectrum runs from the completely benign, that is endometrial stromal nodules (ESN) showing well‐circumscribed margins with cells resembling those of proliferative‐phase endometrial stroma, to malignant, high‐grade ESS (HG‐ESS), which show destructive growth with invasion of surrounding myometrium, to the highly aggressive, undifferentiated uterine sarcomas (UUS) showing high‐grade cytological features, but no specific type of differentiation. Thus, ESS are malignant tumors composed of cells resembling stromal cells of proliferative‐phase endometrium, with a tendency toward infiltrative growth into the myometrium and/or lymphovascular spaces. The HG variants show round cell morphology that may be associated with a low‐grade spindle cell component, which is frequently fibromyxoid. 1 Though most of these tumors originate from the uterus, a subset arises in extrauterine locations, such as, the ovary or peritoneum, often in association with endometriosis. 2 , 3
ESS are rare, accounting for 7% to 25% of all uterine mesenchymal tumors or 1% of all malignancies arising in the uterus. They are the second most common uterine malignant mesenchymal tumors after leiomyosarcoma. 1 , 2 The morphologic features, clinical behavior, and genetic aberration pattern identified in ESS allowed for separation into two categories: high and low grade. However, the complexity and heterogeneity of these tumors extend far beyond this diagnostic grouping.
2. CHROMOSOMAL ABERRATIONS AND THEIR MOLECULAR PRODUCTS
Different types of chromosomal aberrations have been described in ESS, with the most common being translocations involving two different chromosomes. Regardless of whether the translocation is balanced or unbalanced, the molecular product of such rearrangements is usually a so‐called fusion gene. This is a hybrid formed from two previously independent genes. It has been known for more than 30 years that gene fusions play an important role in tumorigenesis. 4 , 5 Oncogenic fusions may lead to an abnormal gene product brought about by fusing elements from the two fusion partners. Alternatively, a proto‐oncogene may be fused to a strong promoter leading to its upregulation. Oncogenic fusion transcripts may also be caused by trans‐splicing or read‐through events. Identification of an activated fusion gene improves diagnostic precision as well as prognostication, while at the same time providing pathogenetic information about the tumor. 6
Though different fusion genes have been identified in ESS, generally it seems that the presence of one fusion gene excludes the presence of another in the same tumor.
2.1. LG‐ESS
Since 1988, when the first cytogenetic abnormalities were reported in ESS, 7 many characteristic chromosomal rearrangements have been described in this group of tumors. The most distinctive cytogenetic hallmark of LG‐ESS is the 7;17‐translocation (Figure 1), first described by Sreekantaiah et al in 1991. 8 The aberration leads at the molecular level to fusion of two zinc finger genes, JAZF1 (from 7p15) and SUZ12 (previously known as JJAZ1; from 17q21; Figure 1). 9 Many other chromosomal changes have also been described and their pattern of occurrence is clearly nonrandom (Table 1). The molecular product behind each translocation has been identified for most of them. Ever improving methodological tools have facilitated the discoveries, especially the introduction of deep sequencing technologies allowing rapid screening of tumor genomes and transcriptomes. 10 The second most common rearrangement involves chromosome band 6p21 and the PHF1 gene. 11 PHF1 may recombine with several partners, not least JAZF1 trough an unbalanced 6;7‐translocation. 11 Other partners are EPC1 through a 6;10‐rearrangement 11 ; MEAF6 through a t(1;6)(p34;p21) 12 ; BRD8 via t(5;6)(q31;p21) 13 ; EPC2 through a 2;6‐rearrangement 14 ; and recently a MBTD1/PHF1 was also reported. 15 A study by D'Angelo et al. 16 showed that tumors bearing PHF1 fusions, independently of which partner gene is involved, typically present sex cord‐like differentiation, leading the authors to suggest that rearrangements of this gene preferentially induce such differentiation.
FIGURE 1.
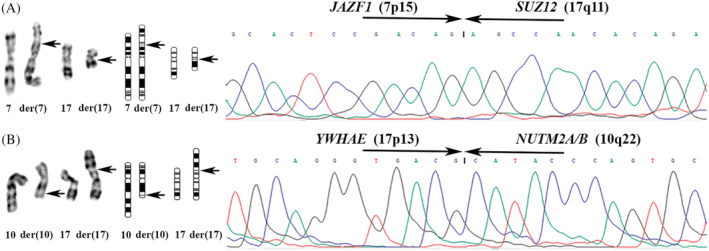
Partial karyogram and chromatogram of the hallmarks for ESS. A, LG‐ESS: partial karyogram showing the t(7;17)(p15;q11) (left), the ideograms for the rearranged chomosomes (center), and sequence chromatogram for the JAZF1/SUZ12 fusion gene (right). B, HG‐ESS: partial karyogram showing the t(10;17)(q22;p13) (left), the ideograms for the rearranged chomosomes (center), and the sequence chromatogram for the YWHAE/NUTM2A/B fusion gene (right). Arrows point at breakpoints
TABLE 1.
Overview of chromosomal rearrangements and their respective fusion genes detected in low‐grade and high‐grade endometrial stromal sarcomas, and their occurrence in other types of neoplasms
| ESS type | Chromosomal rearrangement | Fusion transcript and/or molecular aberration | Other neoplasm a |
|---|---|---|---|
| Low‐grade | t(7;17)(p15;q11) | JAZF1/SUZ12 | ESN |
| Low‐grade | t(6;7)(p21;p15) | JAZF1/PHF1 | cardiac ossifying sarcoma |
| Low‐grade | t(6;10)(p21;p11) | EPC1/PHF1 | OFM |
| Low‐grade | t(1;6)(p34;p21) | MEAF6/PHF1 | ESN, OFM |
| Low‐grade | t(X;17)(p11; q21) | MBTD1/EZHIP | |
| Low‐grade | t(5;6)(q13;p21) | BRD8/PHF1 | |
| Low‐grade | ins(6;2)(p21;q23q23) | EPC2/PHF1 | |
| Low‐grade | t(6;17)(p21;q21) putative b | MBTD1/PHF1 | |
| Low‐grade | t(X;7)(q26;p15) putative | JAZF1/BCORL1 | adenosarcoma |
| Low‐grade | t(1;17)(p34;q11) putative | MEAF6/SUZ12 | |
| Low‐grade | t(10;17)(p11;q11) putative | EPC1/SUZ12 | |
| Low‐grade | t(X;10)(p11;p11) putative | EPC1/BCOR | |
| High‐grade | t(10;17)(q22;p13) | YWHAE/NUTM2A/B | LMS, angiosarcoma, CCSK, SRBCS, URCS, PMMTI, NRCS, |
| High‐grade | t(X;22)(p11; q13) | ZC3H7B/BCOR | OFM |
| High‐grade | t(X;3)(p11;q28) putative | LPP/BCOR | |
| High‐grade | BCOR ITD | CCSK, PMMTI, RCS |
CCSK, clear cell sarcoma of the kidney; ESN, endometrial stromal nodule; OFM, ossifying fibromyxoid tumor; LMS, leiomyosarcoma; NRCS, neonatal round cell sarcoma; PMMTI, primitive myxoid mesenchymal tumor of infancy; RCS, round cell sarcoma; SRBCS, small round blue cell sarcoma; URCS, undifferentiated round cell sarcoma.
Putative, the fusion was found by sequencing analysis and the chromosomal rearrangement designed by default.
A less frequent chromosomal rearrangement is the t(X;17)(p11;q21) leading to the MBTD1/EZHIP (previously known as CXorf67) fusion. 17 Variants of the JAZF1/SUZ12 were recently identified in which JAZF1 recombines with BCORL1 18 and SUZ12 with MEAF6. 19
Another two novel chimeric fusions were reported by Dickson et al., 20 EPC1/SUZ12 and EPC1/BCOR. The identification of these transcripts underlines the promiscuous nature of EPC1, but also obfuscates the molecular distinction between high grade and low grade ESS. Both tumors were described as clinically aggressive and with morphological features compatible with HG‐ESS. 20 The biological potential associated with these fusions remains to be fully characterized. Most likely, also other fusion gene products will emerge. Until a sufficient number of cases is studied and the clinical parameters correlated, it is likely to remain challenging to classify the observed molecular events as being fully specific for LG‐ or HG‐ESS.
The above‐mentioned fusion genes have so far not been seen in leiomyomas, leiomyosarcomas, and uterine tumor resembling ovarian sex cord stromal tumors (UTROSCT), all of which may on occasion be differential diagnoses. Nevertheless, none of the fusions is fully pathognomonic for LG‐ESS as they have all been found also in other neoplasias. JAZF1/SUZ12 fusion is detected in 65% to 75% of ESN. 9 , 21 , 22 , 23 , 24 , 25 , 26 , 27 This chimera has been found more frequently in classic LG‐ESS than in LG‐ESS exhibiting variant features. 28 Recently, an ESN with MEAF6/PHF1 was reported, providing further support for a continuum between these two tumor entities. 29 Interestingly, the ESN showed focal peripheral ossification, a rare feature of ESN and/or LG‐ESS but a hallmark of ossifying fibromyxoid tumors with which they may share also other molecular events such as PHF1 fusions, that is, EP400/PHF1, MEAF6/PHF1, and EPC1/PHF1. It was therefore hypothesized that the MEAF6/PHF1 could be associated with metaplastic bone formation. 30 The aforementioned fusions occur in ossifying fibromyxoid tumors of soft parts, irrespective of whether the tumor is diagnosed as typical, atypical, or malignant, whereas JAZF1/PHF1 has been found in cardiac ossifying sarcomas. 31 , 32 , 33 , 34 Furthermore, two novel fusions, CREBBP/BCORL1 and KDM2A/WWTR1 have been reported in ossifying fibromyxoid tumors 35 showing additional overlap with LG‐ESS as the genes CREBBP and KDM2B were previously found in a chimera in the latter tumor as well. 36 Recently, the JAZF1/BCORL1 fusion was identified in an adenosarcoma arising in the uterus. 37
2.2. HG‐ESS
The cytogenetic hallmark of HG‐ESS is the balanced t(10;17)(q22;p13) translocation simultaneously reported in 2003 by two groups 38 , 39 (Figure 1). The gene product it leads to was identified by Lee et al. 40 as an in‐frame fusion between the YWHAE and NUTM2A/B genes (previously known as FAM22A/B; Figure 1). The fusion seems to be specific for HG‐ESS as it was never identified in other gynecological tumors or neoplastic lesions, such as, uterine adenosarcomas, carcinosarcomas, leiomyosarcomas, leiomyomas, and polypoid endometriosis. 40 Kubo et al. 41 reported a low frequency of YWHAE and NUTM2A/B rearrangements in epithelioid leiomyosarcoma; admittedly, the immunostaining data of that study were suggestive of an unusual ESS. Splitting of probes for the YWHAE, FAM22A, and FAM22B genes has been reported in a uterine angiosarcoma. 42 Despite the fact that no fusion transcript involving the mentioned genes was discovered, the authors suggested that abnormalities of them may contribute to development of uterine angiosarcoma in much the same manner as they do in ESS. 42 Of further note in the context is the fact that no such rearrangement was identified in 21 angiosarcomas of extrauterine soft tissue. 40 However, the very same chromosomal translocation has been reported in clear cell sarcomas of the kidney by different groups 43 , 44 , 45 and shown to correspond to a YWHAE/NUTM2A/B fusion. 46 Kao et al. 47 identified it also in small round blue cell sarcomas of soft tissue, undifferentiated round cell sarcoma, and primitive myxoid mesenchymal tumor of infancy. 47 , 48 Recently, the first neonatal case of a round cell sarcoma bearing this chimera was described in a tumor with aggressive clinical behavior. 49 Sciallis et al. 50 studied 17 HG‐ESS and outlined three morphological patterns in this tumor type: YWHAE rearrangements were identified only in tumors showing high‐grade round cells with brisk mitotic activity and necrosis, not in all examined tumors.
A subset of tumors within the HG‐ESS category has lately been described as having aggressive behavior and mimicking myxoid leiomyosarcoma morphologically. These tumors show the ZC3H7B/BCOR fusion first reported by Panagopoulos et al. 51 in two ESS showing a (X;22)(p11;q13) chromosomal translocation, a rearrangement later confirmed in several tumors by other investigators. 52 , 53 , 54 , 55 Most of the clinical data published on patients diagnosed with HG‐ESS showed stage 3 disease or the patient had a recurrence. However, ESS harboring a ZC3H7B/BCOR fusion may be clinically as well as morphologically heterogeneous. 54 , 55 , 56 A unique case of ZC3H7B/BCOR‐positive HG‐ESS was identified at an early stage when an endocervical polypoid mass from the lower uterine segment was examined. 56 Though macroscopically the tumor presented as a polypoid mass descending into the cervical canal in a myoma nascens‐like fashion, the histomorphologic and immunohistochemical profiles were suggestive of HG‐ESS.
Recently, additional variant partners for BCOR‐fusions, including L3MBTI2, EP300, NUTM2G, RALGPS1, MAP7D2, RGAG1, ING3, NUGGC, KMT2D, and CREBBP, were identified in a series of 40 uterine sarcomas. 57 However, until a sufficient number of tumors showing these fusions are collected and their clinical‐pathological parameters examined in detail and reported, it is difficult to correlate meaningfully the genetic and morphologic features.
In addition to fusion genes, other types of molecular aberrations have also been found in HG‐ESS. Chiang et al. 52 reported the first HG‐ESS with BCOR internal tandem duplication (ITD), the same aberration previously found in clear cell sarcoma of the kidney (CCSK) 58 , 59 and primitive myxoid mesenchymal tumor of infancy. 60 This adds to the growing body of histologic, immunophenotypic, and genetic evidence unifying these tumors pathogenetically with CCSK as well as soft tissue round cell sarcomas. 47 , 52 , 59 The latter tumor type also shows alternative gene fusions involving BCOR, such as, BCOR/CCNB3, BCOR/MAML3, and KMT2D/BCOR. 60 , 61 BCOR gene aberrations have further been found in a tumor of the sinonasal cavity (a sarcoma) and in pediatric glioma, resulting in CIITA/BCOR and EP300/BCOR fusion, respectively. 62 , 63 Truncating mutations or gene deletions occurring in BCOR have also been identified in acute myeloid leukemia, retinoblastoma, diffuse glioma, and medulloblastoma. 64 , 65 , 66 The detection of aberrations of this gene already plays a key role in the diagnosis of some malignancies, for example, high‐grade neuroepithelial tumor of the central nervous system with BCOR gene alteration. 67 BCOR ITD has been reported in a limited number of HG‐ESS; however, this aberration seems to characterize a younger group of patients and be associated with a slightly more favorable clinical course. 68 The identification of the same BCOR genetic aberrations in so many different tumor types suggests a central pathogenetic role of this gene. In all likelihood, the type of stem cell hit by the tumorigenic event, the differentiation pattern it is already locked onto, decides the phenotypic differences observed.
The gene's orientation in the different chimeric fusions is intriguing, whether it is 3′ or 5′. Furthermore, some but not all ZC3H7B/BCOR positive HG‐ESS are characterized by the presence of also the reciprocal transcript 51 , 52 , 56 whose role in tumorigenesis and/or progression is still unknown. Additionally, a new BCOR‐rearranged HG‐ESS was recently reported by Kommoss et al. 69 in which RNAseq identified a fusion between BCOR and LPP. The genic event behind the transcript resulted in overexpression of the C‐terminal, truncated BCOR protein but without generation of the chimera, that is, no coding part of LPP was involved. The functional consequences of this aberration are unclear.
BCOR immunochemical staining has proved to be a highly sensitive marker for HG‐ESS bearing YWHAE/NUTM2 and YWHAE‐rearrangements, be it with classical or unusual morphology; it was found positive in half of the tumors showing BCOR‐rearrangements as well as in tumors showing BCOR ITD. 47 , 52 Overexpression of BCOR mRNA has been described in tumors with YWHAE/NUTM2 fusions. 48 BCOR immunohistochemistry can be used diagnostically to separate all the above‐mentioned tumors (with BCOR genetic rearrangement) from their histological mimics.
As for the LG‐ESS, genetic aberrations found in HG‐ESS have been found also in other tumor types. Of particular interest are the data reported by Cotzia et al. 70 who detected rearrangements of YWHAE, BCOR, and PHF1 in a series of tumors previously classified as UUS based on morphologic and immunohistochemical features. Consequently and logically, the authors suggested that many UUS may represent misdiagnosed HG‐ESS. 70 Stewart et al. 71 identified a YWHAE deletion in a vagina wall metastasis from a monomorphic undifferentiated sarcoma, as the tumor was classified at that time. Also, two tumors with morphologic feature of LG‐ESS have had YWHAE rearrangements: a YWHAE/NUTM2A fusion was identified in a tumor confined within the endometrium, 72 whereas deletion of a 3′ probe for YWHAE was shown in an LG‐ESS and in its recurrence in a case showing progression from LG‐ to HG‐ESS. 73 Antonescu et al. 31 identified ZC3H7B/BCOR in ossifying fibromyxoid tumors showing molecular overlap with ESS.
ESS showing both LG‐ and HG‐features are rare, as are tumors initially diagnosed as LG‐ but then developing metastatic HG‐ESS. 50 , 74 , 75 , 76 These tumors harbor gene fusions that are typically associated with LG‐ESS. 77
The identification so far of four chimeric transcripts in HG‐ESS—YWHAE/NUTM2A/B, ZC3H7B/BCOR, EPC1/SUZ12, and EPC1/BCOR‐is evidence of genetic heterogeneity also within this tumor subgroup. As highlighted above, EPC1 is involved in LG‐ESS‐related fusions. The molecular heterogeneity even within specific pathologic entities, that is, LG‐ and HG‐ESS, as well as the fact that different entities may show the same fusion, makes the diagnosis of these tumors challenging. Some tumors may exhibit morphologic aspects of a specific entity without having any known molecular signature of it, or they show sex‐cord differentiation and/or myxoid morphology reflecting phenotypic overlapping among subgroups. A tumor classification that combines both morphologic and genetic tumor features is necessary to improve diagnostic precision as well as prognostication, while simultaneously providing pathogenetic information about the neoplastic process.
3. PATHOGENETIC CONSEQUENCES OF ESS‐SPECIFIC GENETIC REARRANGEMENTS
In later years, much effort has gone into the identification of molecular mechanisms behind ESS‐specific genetic rearrangements with the goal of unraveling how they contribute to tumorigenesis and, eventually, how this knowledge can lead to novel therapeutic approaches. A striking general feature of the molecular genetics of ESS is that many of the genes involved in chromosomal rearrangements are associated with chromatin modification, that is, JAZF1/SUZ12, JAZF1/PHF1, EPC1/PHF1, MEAF6/PHF1, MBTD1/EZHIP, BRD8/PHF1, EPC2/PHF1, MBTD1/PHF1, JAZF1/BCORL1, MEAF6/SUZ12, EPC1/SUZ12, EPC1/BCOR, and ZC3H7B/BCOR.
LG‐ESS harbor chromosomal rearrangements of genes, such as, SUZ12, PHF1, EZHIP, MBTD1, EPC1, and EPC2 (the latter is a paralog) whose protein products are associated with chromatin remodeling complexes, the NuA4 acetyltransferase complex and the Polycomb group of Protein (PcG; mainly the Polycomb Repressive Complex 2 subunits). It was recently demonstrated that the 5′ partner gene of the fusion codes for a component of the NuA4 acetyltransferase (N‐terminal on the chimeric protein) whereas the 3′ gene codes for a PcG subunit (C‐terminal on the chimeric protein). 78 , 79 The JAZF1/SUZ12 chimera was first demonstrated to inhibit apoptosis and induce proliferation rates above normal in both benign and malignant uterine tumors, although only in the malignant form was suppression of the wild type/unrearranged SUZ12 allele identified. This led to the hypothesis that genetic progression from a benign precursor to sarcoma lay behind the suppression of the unrearranged SUZ12 allele, starting with increased cell survival but followed by accelerated cellular proliferation upon exclusion of the second allele. 80 A similar mechanism was seen for ESS bearing JAZF1/PHF1 fusion with simultaneous silencing of the normal PHF1 allele. 80 The JAZF1/SUZ12 protein has been shown to be an essential component of the Polycomb Repressive Complex 2 (PRC2), a major player in epigenetic silencing responsible for methylation of lysines 9 and 27 of histone 3 (H3K9 and H3K27). JAZF1/SUZ12 destabilizes the PRC2 components leading to a decrease of methyltransferase activity, especially on H3K27, and therefore activates chromatin and/or genes normally repressed by PRC2. 81 Analyses of the gene expression profiles of LG‐ESS have shown overexpression of genes directly regulated by SUZ12 and activation of genes implicated in the Wnt signaling pathway, 82 confirming that different chromosomal rearrangements may lead to similar gene expression profiles. 36 , 82 It has therefore been suggested that LG‐ESS chimeric proteins disrupt the repressive function of the PRC2 complex; possibly these chimeras contribute to overexpression of Wnt ligands with subsequent activation of the Wnt signaling pathway and formation of an active β‐catenin/Lef1 transcriptional complex. 81 , 82 The latter would also explain why there is nuclear expression of β‐catenin in 60% of LG‐ESS. 83 , 84 , 85
BCOR (BCL‐6 interacting corepressor, mapping on Xp11) and BCORL1 (BCL‐6 corepressor‐like 1), like other genes rearranged in EST, are transcriptional corepressors. Specifically, BCOR is part of the PRC complex and promotes transcriptional repression by covalent modification of histone deacetylases and the polycomb repressive complex 1. 86 BCOR has a number of functions within normal tissue and its alteration can result in developmental disorders and a variety of hematologic and solid malignancies. 48 , 58 , 59 , 87 , 88 , 89 , 90
Lately, methylation profiles for different uterine tumors have been determined 91 showing different methylation clusters correlating with established diagnostic entities. The data obtained highlighted that the LG‐ESS pattern differed from that of HG‐ESS, and that, within the latter, distinct subgrouping of YWHAE‐ and BCOR‐rearranged tumors was possible. 91 The copy number‐profile was investigated by the same group in a series of uterine tumors that included LG‐ESS, HG‐ESS, UTROSCT, uterine leiomyomas, and uterine leiomyosarcomas. 69 The authors identified amplification of the MDM2 gene from chromosomal band 12q15 only in BCOR‐rearranged HG‐ESS. 69 Previously, a study by Shoolmeester et al. 92 showed MDM2 amplification in an LG‐ESS with JAZF1‐rearrangement and in a UUS. Since such amplifications have not been identified in other mesenchymal uterine tumors, it is intriguing that Cotzia et al. 70 suggested that UUS are unrecognized HG‐ESS. The discovery of MDM2 amplification opens up for potential use of targeted therapy in a subset of HG‐ESS. 69
Lin et al. 57 recently investigated the genomic profile of 40 uterine sarcomas harboring BCOR alterations. The analyzed tumors were found to be stable at the microsatellite level; however, some of them showed homozygous deletion of CDKN2A which codes for an inhibitor of CDK4 and CDKN2B. Furthermore, a similar copy number profile was identified for the CDK4, MDM2, and FRS2 genes (all located at 12q14.1) in uterine sarcomas bearing BCOR‐fusions, but not in tumors with BCOR ITD. It seems that alteration of CDK4 pathway members contributes to the pathogenesis of BCOR‐rearranged tumors, something that may have therapeutic implications. 57
Histone acetyltransferases (HAT) of the MYST family are known to be involved in vital cellular processes, such as, gene transcription, detection and repair of DNA damage, and DNA replication. They carry out a significant proportion of all nuclear acetylation, and their anomalous activity, or anomalous activity of complexes associated with them (these enzymes work in multisubunit protein complexes), can lead to different anomalies from cell death to uncontrolled growth, the latter leading to cancer formation. 93 There are different HATs in the MYST family, many of which are known to be involved in different types of cancer, for example, MOZ and MORF in acute myeloid leukemia.
Of all chimeric proteins associated with ESS, YWHAE/NUTM2A/B is the only one that does not undergo epigenetic modification. The gene for YWHAE (14‐3‐3 ε) belongs to a broad family of proteins responsible for mediating signal transduction. 40 FAM22A/B was renamed NUTM2A/B due to its sequence homology with NUT (NUTM1), which is notable in NUT midline carcinoma. 94
The issue whether a linear tumor progression exists among the different EST was investigated by means of array based Comparative Genomic Hybridization (aCGH). 95 The fact that no chromosomal aberrations were common to the ESN, LG‐ESS, and UES/UUS investigated led the authors to conclude that this proposition was unlikely. However, an increasing number of aberrations were registered from ESN to UES, correlating well with histological grading and worsening clinical behavior. 95
ACKNOWLEDGMENTS
The authors wish to thank the Norwegian Radium Hospital Foundation and the Anders Jahre's foundation through UNIFOR (University of Oslo) for their support.
Micci F, Heim S, Panagopoulos I. Molecular pathogenesis and prognostication of "low‐grade'' and "high‐grade" endometrial stromal sarcoma. Genes Chromosomes Cancer. 2021;60:160–167. 10.1002/gcc.22907
Funding information Anders Jahre's foundation through UNIFOR (University of Oslo); The Norwegian Radium Hospital Foundation
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- 1. Kurman RJ, Carcangiu ML, Herrington CS, Young RH. WHO Classification of Tumors of Female Reproductive Organs. Lyon, France: IARC; 2014. [Google Scholar]
- 2. Hoang L, Chiang S, Lee CH. Endometrial stromal sarcomas and related neoplasms: new developments and diagnostic considerations. Pathology. 2018;50(2):162‐177. [DOI] [PubMed] [Google Scholar]
- 3. Oliva E, Egger JF, Young RH. Primary endometrioid stromal sarcoma of the ovary: a clinicopathologic study of 27 cases with morphologic and behavioral features similar to those of uterine low‐grade endometrial stromal sarcoma. Am J Surg Pathol. 2014;38(3):305‐315. [DOI] [PubMed] [Google Scholar]
- 4. Edwards PA. Fusion genes and chromosome translocations in the common epithelial cancers. J Pathol. 2010;220(2):244‐254. [DOI] [PubMed] [Google Scholar]
- 5. Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7(4):233‐245. [DOI] [PubMed] [Google Scholar]
- 6. Heim S, Mitelman F. Cancer Cytogenetics—Chromosomal and Molecular Genetic Aberrations of Tumor Cells. 4th ed. New Jersey, NJ: Wiley Blackwell; 2015. [Google Scholar]
- 7. Dal Cin P, Talcott J, Abrams J, Li FP, Sandberg AA. Ins(10;19) in an endometrial stromal sarcoma. Cancer Genet Cytogenet. 1988;36(1):1‐5. [DOI] [PubMed] [Google Scholar]
- 8. Sreekantaiah C, Li FP, Weidner N, Sandberg AA. An endometrial stromal sarcoma with clonal cytogenetic abnormalities. Cancer Genet Cytogenet. 1991;55(2):163‐166. [DOI] [PubMed] [Google Scholar]
- 9. Koontz JI, Soreng AL, Nucci M, et al. Frequent fusion of the JAZF1 and JJAZ1 genes in endometrial stromal tumors. Proc Natl Acad Sci U S A. 2001;98(11):6348‐6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maher CA, Kumar‐Sinha C, Cao X, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458(7234):97‐101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Micci F, Panagopoulos I, Bjerkehagen B, Heim S. Consistent rearrangement of chromosomal band 6p21 with generation of fusion genes JAZF1/PHF1 and EPC1/PHF1 in endometrial stromal sarcoma. Cancer Res. 2006;66(1):107‐112. [DOI] [PubMed] [Google Scholar]
- 12. Panagopoulos I, Micci F, Thorsen J, et al. Novel fusion of MYST/Esa1‐associated factor 6 and PHF1 in endometrial stromal sarcoma. PLoS One. 2012;7(6):e39354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Micci F, Brunetti M, Dal Cin P, et al. Fusion of the genes BRD8 and PHF1 in endometrial stromal sarcoma. Genes, Chromosomes Cancer. 2017;56(12):841‐845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brunetti M, Gorunova L, Davidson B, Heim S, Panagopoulos I, Micci F. Identification of an EPC2‐PHF1 fusion transcript in low‐grade endometrial stromal sarcoma. Oncotarget. 2018;9(27):19203‐19208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Han L, Liu YJ, Ricciotti RW, Mantilla JG. A novel MBTD1‐PHF1 gene fusion in endometrial stromal sarcoma: a case report and literature review. Genes, Chromosomes Cancer. 2020;59(7):428‐432. [DOI] [PubMed] [Google Scholar]
- 16. D'Angelo E, Ali RH, Espinosa I, et al. Endometrial stromal sarcomas with sex cord differentiation are associated with PHF1 rearrangement. Am J Surg Pathol. 2013;37(4):514‐521. [DOI] [PubMed] [Google Scholar]
- 17. Dewaele B, Przybyl J, Quattrone A, et al. Identification of a novel, recurrent MBTD1‐CXorf67 fusion in low‐grade endometrial stromal sarcoma. Int J Cancer. 2014;134(5):1112‐1122. [DOI] [PubMed] [Google Scholar]
- 18. Allen AJ, Ali SM, Gowen K, Elvin JA, Pejovic T. A recurrent endometrial stromal sarcoma harbors the novel fusion JAZF1‐BCORL1. Gynecol Oncol Rep. 2017;20:51‐53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Makise N, Sekimizu M, Kobayashi E, et al. Low‐grade endometrial stromal sarcoma with a novel MEAF6‐SUZ12 fusion. Virchows Arch. 2019;475(4):527‐531. [DOI] [PubMed] [Google Scholar]
- 20. Dickson BC, Lum A, Swanson D, et al. Novel EPC1 gene fusions in endometrial stromal sarcoma. Genes, Chromosomes Cancer. 2018;57(11):598‐603. [DOI] [PubMed] [Google Scholar]
- 21. Chiang S, Ali R, Melnyk N, et al. Frequency of known gene rearrangements in endometrial stromal tumors. Am J Surg Pathol. 2011;35(9):1364‐1372. [DOI] [PubMed] [Google Scholar]
- 22. Chiang S, Oliva E. Cytogenetic and molecular aberrations in endometrial stromal tumors. Hum Pathol. 2011;42(5):609‐617. [DOI] [PubMed] [Google Scholar]
- 23. Conklin CM, Longacre TA. Endometrial stromal tumors: the new WHO classification. Adv Anat Pathol. 2014;21(6):383‐393. [DOI] [PubMed] [Google Scholar]
- 24. Huang HY, Ladanyi M, Soslow RA. Molecular detection of JAZF1‐JJAZ1 gene fusion in endometrial stromal neoplasms with classic and variant histology: evidence for genetic heterogeneity. Am J Surg Pathol. 2004;28(2):224‐232. [DOI] [PubMed] [Google Scholar]
- 25. Nucci MR, Harburger D, Koontz J, Dal Cin P, Sklar J. Molecular analysis of the JAZF1‐JJAZ1 gene fusion by RT‐PCR and fluorescence in situ hybridization in endometrial stromal neoplasms. Am J Surg Pathol. 2007;31(1):65‐70. [DOI] [PubMed] [Google Scholar]
- 26. Oliva E, de Leval L, Soslow RA, Herens C. High frequency of JAZF1‐JJAZ1 gene fusion in endometrial stromal tumors with smooth muscle differentiation by interphase FISH detection. Am J Surg Pathol. 2007;31(8):1277‐1284. [DOI] [PubMed] [Google Scholar]
- 27. Sato K, Ueda Y, Sugaya J, Ozaki M, Hisaoka M, Katsuda S. Extrauterine endometrial stromal sarcoma with JAZF1/JJAZ1 fusion confirmed by RT‐PCR and interphase FISH presenting as an inguinal tumor. Virchows Arch. 2007;450(3):349‐353. [DOI] [PubMed] [Google Scholar]
- 28. Chiang S, Oliva E. Recent developments in uterine mesenchymal neoplasms. Histopathology. 2013;62(1):124‐137. [DOI] [PubMed] [Google Scholar]
- 29. Nomura Y, Tamura D, Horie M, et al. Detection of MEAF6‐PHF1 translocation in an endometrial stromal nodule. Genes, Chromosomes Cancer. 2020;59:702‐708. [DOI] [PubMed] [Google Scholar]
- 30. Chiyoda T, Tsuda H, Tanaka H, et al. Expression profiles of carcinosarcoma of the uterine corpus‐are these similar to carcinoma or sarcoma? Genes, Chromosomes Cancer. 2012;51(3):229‐239. [DOI] [PubMed] [Google Scholar]
- 31. Antonescu CR, Sung YS, Chen CL, et al. Novel ZC3H7B‐BCOR, MEAF6‐PHF1, and EPC1‐PHF1 fusions in ossifying fibromyxoid tumors—molecular characterization shows genetic overlap with endometrial stromal sarcoma. Genes, Chromosomes Cancer. 2014;53(2):183‐193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gebre‐Medhin S, Nord KH, Moller E, et al. Recurrent rearrangement of the PHF1 gene in ossifying fibromyxoid tumors. Am J Pathol. 2012;181(3):1069‐1077. [DOI] [PubMed] [Google Scholar]
- 33. Graham RP, Weiss SW, Sukov WR, et al. PHF1 rearrangements in ossifying fibromyxoid tumors of soft parts: a fluorescence in situ hybridization study of 41 cases with emphasis on the malignant variant. Am J Surg Pathol. 2013;37(11):1751‐1755. [DOI] [PubMed] [Google Scholar]
- 34. Schoolmeester JK, Sukov WR, Maleszewski JJ, Bedroske PP, Folpe AL, Hodge JC. JAZF1 rearrangement in a mesenchymal tumor of nonendometrial stromal origin: report of an unusual ossifying sarcoma of the heart demonstrating JAZF1/PHF1 fusion. Am J Surg Pathol. 2013;37(6):938‐942. [DOI] [PubMed] [Google Scholar]
- 35. Kao YC, Sung YS, Zhang L, Chen CL, Huang SC, Antonescu CR. Expanding the molecular signature of ossifying fibromyxoid tumors with two novel gene fusions: CREBBP‐BCORL1 and KDM2A‐WWTR1. Genes, Chromosomes Cancer. 2017;56(1):42‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Micci F, Gorunova L, Agostini A, et al. Cytogenetic and molecular profile of endometrial stromal sarcoma. Genes, Chromosomes Cancer. 2016;55(11):834‐846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Muthukumarana V, Fix DJ, Stolnicu S, et al. BCOR expression in Mullerian Adenosarcoma: a potential diagnostic pitfall. Am J Surg Pathol. 2020;44(6):765‐770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Leunen K, Amant F, biec‐Rychter M, et al. Endometrial stromal sarcoma presenting as postpartum haemorrhage: report of a case with a sole t(10;17)(q22;p13) translocation. Gynecol Oncol. 2003;91(1):265‐271. [DOI] [PubMed] [Google Scholar]
- 39. Micci F, Walter CU, Teixeira MR, et al. Cytogenetic and molecular genetic analyses of endometrial stromal sarcoma: nonrandom involvement of chromosome arms 6p and 7p and confirmation of JAZF1/JJAZ1 gene fusion in t(7;17). Cancer Genet Cytogenet. 2003;144(2):119‐124. [DOI] [PubMed] [Google Scholar]
- 40. Lee CH, Ou WB, Marino‐Enriquez A, et al. 14‐3‐3 fusion oncogenes in high‐grade endometrial stromal sarcoma. Proc Natl Acad Sci U S A. 2012;109(3):929‐934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kubo T, Sugita S, Wada R, et al. Uterine epithelioid leiomyosarcoma with c‐kit expression and YWHAE gene rearrangement: a case report of a diagnostic pitfall of uterine sarcoma. Diagn Pathol. 2017;12(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Suzuki S, Tanioka F, Minato H, Ayhan A, Kasami M, Sugimura H. Breakages at YWHAE, FAM22A, and FAM22B loci in uterine angiosarcoma: a case report with immunohistochemical and genetic analysis. Pathol Res Pract. 2014;210(2):130‐134. [DOI] [PubMed] [Google Scholar]
- 43. O'Meara E, Stack D, Lee CH, et al. Characterization of the chromosomal translocation t(10;17)(q22;p13) in clear cell sarcoma of kidney. J Pathol. 2012;227(1):72‐80. [DOI] [PubMed] [Google Scholar]
- 44. Punnett HH, Halligan GE, Zaeri N, Karmazin N. Translocation 10;17 in clear cell sarcoma of the kidney. A First Report Cancer Genet Cytogenet. 1989;41(1):123‐128. [DOI] [PubMed] [Google Scholar]
- 45. Rakheja D, Weinberg AG, Tomlinson GE, Partridge K, Schneider NR. Translocation (10;17)(q22;p13): a recurring translocation in clear cell sarcoma of kidney. Cancer Genet Cytogenet. 2004;154(2):175‐179. [DOI] [PubMed] [Google Scholar]
- 46. Fehr A, Hansson MC, Kindblom LG, Stenman G. YWHAE‐FAM22 gene fusion in clear cell sarcoma of the kidney. J Pathol. 2012;227(4):e5‐e7. [DOI] [PubMed] [Google Scholar]
- 47. Kao YC, Sung YS, Zhang L, et al. Recurrent BCOR internal tandem duplication and YWHAE‐NUTM2B fusions in soft tissue undifferentiated round cell sarcoma of infancy: overlapping genetic features with clear cell sarcoma of kidney. Am J Surg Pathol. 2016a;40(8):1009‐1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kao YC, Sung YS, Zhang L, et al. BCOR overexpression is a highly sensitive marker in round cell sarcomas with BCOR genetic abnormalities. Am J Surg Pathol. 2016b;40(12):1670‐1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Guizard M, Karanian M, Dijoud F, et al. Neonatal soft tissue sarcoma with YWHAE‐NUTM2B fusion. Case Rep Oncol. 2019;12(2):631‐638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sciallis AP, Bedroske PP, Schoolmeester JK, et al. High‐grade endometrial stromal sarcomas: a clinicopathologic study of a group of tumors with heterogenous morphologic and genetic features. Am J Surg Pathol. 2014;38(9):1161‐1172. [DOI] [PubMed] [Google Scholar]
- 51. Panagopoulos I, Thorsen J, Gorunova L, et al. Fusion of the ZC3H7B and BCOR genes in endometrial stromal sarcomas carrying an X;22‐translocation. Genes, Chromosomes Cancer. 2013;52(7):610‐618. [DOI] [PubMed] [Google Scholar]
- 52. Chiang S, Lee CH, Stewart CJR, et al. BCOR is a robust diagnostic immunohistochemical marker of genetically diverse high‐grade endometrial stromal sarcoma, including tumors exhibiting variant morphology. Mod Pathol. 2017;30(9):1251‐1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hoang LN, Aneja A, Conlon N, et al. Novel high‐grade endometrial stromal sarcoma: a morphologic mimicker of Myxoid Leiomyosarcoma. Am J Surg Pathol. 2017;41(1):12‐24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lewis N, Soslow RA, Delair DF, Park KJ, Murali R, Hollmann TJ, Davidson B, Micci F, Panagopoulos I, Hoang LN, Arias‐Stella JA, 3rd, Oliva E, Young RH, Hensley ML, Leitao MM, Jr. , Hameed M, Benayed R, Ladanyi M, Frosina D, Jungbluth AA, Antonescu CR, Chiang S. 2018. ZC3H7B‐BCOR high‐grade endometrial stromal sarcomas: a report of 17 cases of a newly defined entity. Mod Pathol 31(4):674–684. [DOI] [PubMed] [Google Scholar]
- 55. Mansor S, Kuick CH, Lim SL, et al. ZC3H7B‐BCOR‐rearranged endometrial stromal sarcomas: a distinct subset merits its own classification? Int J Gynecol Pathol. 2019;38(5):420‐425. [DOI] [PubMed] [Google Scholar]
- 56. Ondič O, Bednářová B, Ptáková N, et al. ZC3H7B‐BCOR high‐grade endometrial stromal sarcoma may present as myoma nascens with cytoplasmic signet ring cell change. Virchows Arch. 2020;476(4):615‐619. [DOI] [PubMed] [Google Scholar]
- 57. Lin DI, Hemmerich A, Edgerly C, et al. Genomic profiling of BCOR‐rearranged uterine sarcomas reveals novel gene fusion partners, frequent CDK4 amplification and CDKN2A loss. Gynecol Oncol. 2020;157(2):357‐366. [DOI] [PubMed] [Google Scholar]
- 58. Roy A, Kumar V, Zorman B, et al. Recurrent internal tandem duplications of BCOR in clear cell sarcoma of the kidney. Nat Commun. 2015;6:8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ueno‐Yokohata H, Okita H, Nakasato K, et al. Consistent in‐frame internal tandem duplications of BCOR characterize clear cell sarcoma of the kidney. Nat Genet. 2015;47(8):861‐863. [DOI] [PubMed] [Google Scholar]
- 60. Kao YC, Owosho AA, Sung YS, et al. BCOR‐CCNB3 fusion positive sarcomas: a Clinicopathologic and molecular analysis of 36 cases with comparison to morphologic Spectrum and clinical behavior of other round cell sarcomas. Am J Surg Pathol. 2018;42(5):604‐615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pierron G, Tirode F, Lucchesi C, et al. A new subtype of bone sarcoma defined by BCOR‐CCNB3 gene fusion. Nat Genet. 2012;44(4):461‐466. [DOI] [PubMed] [Google Scholar]
- 62. Torre M, Meredith DM, Dubuc A, et al. Recurrent EP300‐BCOR fusions in pediatric gliomas with distinct clinicopathologic features. J Neuropathol Exp Neurol. 2019;78(4):305‐314. [DOI] [PubMed] [Google Scholar]
- 63. Yoshida Y, Nobusawa S, Nakata S, et al. CNS high‐grade neuroepithelial tumor with BCOR internal tandem duplication: a comparison with its counterparts in the kidney and soft tissue. Brain Pathol. 2018;28(5):710‐720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kline CN, Joseph NM, Grenert JP, et al. Targeted next‐generation sequencing of pediatric neuro‐oncology patients improves diagnosis, identifies pathogenic germline mutations. Neuro Oncol. 2017;19(5):699‐709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mackay A, Burford A, Carvalho D, et al. Integrated molecular meta‐analysis of 1,000 pediatric high‐grade and diffuse intrinsic pontine glioma. Cancer Cell. 2017;32(4):520‐537.e525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Santiago T, Clay MR, Allen SJ, Orr BA. Recurrent BCOR internal tandem duplication and BCOR or BCL6 expression distinguish primitive myxoid mesenchymal tumor of infancy from congenital infantile fibrosarcoma. Mod Pathol. 2017;30(6):884‐891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sturm D, Orr BA, Toprak UH, et al. New brain tumor entities emerge from molecular classification of CNS‐PNETs. Cell. 2016;164(5):1060‐1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mariño‐Enriquez A, Lauria A, Przybyl J, et al. BCOR internal tandem duplication in high‐grade uterine sarcomas. Am J Surg Pathol. 2018;42(3):335‐341. [DOI] [PubMed] [Google Scholar]
- 69. Kommoss FK, Chang KT, Stichel D, et al. Endometrial stromal sarcomas with BCOR‐rearrangement harbor MDM2 amplifications. J Pathol Clin Res. 2020;6(3):178‐184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cotzia P, Benayed R, Mullaney K, et al. Undifferentiated uterine sarcomas represent under‐recognized high‐grade endometrial stromal sarcomas. Am J Surg Pathol. 2019;43(5):662‐669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Stewart CJ, Leung YC, Murch A, Peverall J. Evaluation of fluorescence in‐situ hybridization in monomorphic endometrial stromal neoplasms and their histological mimics: a review of 49 cases. Histopathology. 2014;65(4):473‐482. [DOI] [PubMed] [Google Scholar]
- 72. Attygalle AD, Vroobel K, Wren D, et al. An unusual case of YWHAE‐NUTM2A/B endometrial stromal sarcoma with confinement to the endometrium and lack of high‐grade morphology. Int J Gynecol Pathol. 2017;36(2):165‐171. [DOI] [PubMed] [Google Scholar]
- 73. Aisagbonhi O, Harrison B, Zhao L, Osgood R, Chebib I, Oliva E. YWHAE rearrangement in a purely conventional low‐grade endometrial stromal sarcoma that transformed over time to high‐grade sarcoma: importance of molecular testing. Int J Gynecol Pathol. 2018;37(5):441‐447. [DOI] [PubMed] [Google Scholar]
- 74. Amant F, Woestenborghs H, Vandenbroucke V, et al. Transition of endometrial stromal sarcoma into high‐grade sarcoma. Gynecol Oncol. 2006;103(3):1137‐1140. [DOI] [PubMed] [Google Scholar]
- 75. Kurihara S, Oda Y, Ohishi Y, et al. Endometrial stromal sarcomas and related high‐grade sarcomas: immunohistochemical and molecular genetic study of 31 cases. Am J Surg Pathol. 2008;32(8):1228‐1238. [DOI] [PubMed] [Google Scholar]
- 76. Ohta Y, Suzuki T, Omatsu M, et al. Transition from low‐grade endometrial stromal sarcoma to high‐grade endometrial stromal sarcoma. Int J Gynecol Pathol. 2010;29(4):374‐377. [DOI] [PubMed] [Google Scholar]
- 77. Zou Y, Turashvili G, Soslow RA, et al. High‐grade transformation of low‐grade endometrial stromal sarcomas lacking YWHAE and BCOR genetic abnormalities. Mod Pathol. 2020;33(9):1861‐1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hübner JM, Müller T, Papageorgiou DN, et al. EZHIP/CXorf67 mimics K27M mutated oncohistones and functions as an intrinsic inhibitor of PRC2 function in aggressive posterior fossa ependymoma. Neuro Oncol. 2019;21(7):878‐889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Piunti A, Smith ER, Morgan MAJ, et al. CATACOMB: an endogenous inducible gene that antagonizes H3K27 methylation activity of Polycomb repressive complex 2 via an H3K27M‐like mechanism. Sci Adv. 2019;5(7):eaax2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Li H, Ma X, Wang J, Koontz J, Nucci M, Sklar J. Effects of rearrangement and allelic exclusion of JJAZ1/SUZ12 on cell proliferation and survival. Proc Natl Acad Sci U S A. 2007;104(50):20001‐20006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ma X, Wang J, Wang J, et al. The JAZF1‐SUZ12 fusion protein disrupts PRC2 complexes and impairs chromatin repression during human endometrial stromal tumorogenesis. Oncotarget. 2017;8(3):4062‐4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Przybyl J, Kidzinski L, Hastie T, Debiec‐Rychter M, Nusse R, van de Rijn M. Gene expression profiling of low‐grade endometrial stromal sarcoma indicates fusion protein‐mediated activation of the Wnt signaling pathway. Gynecol Oncol. 2018;149(2):388‐393. [DOI] [PubMed] [Google Scholar]
- 83. Jung CK, Jung JH, Lee A, et al. Diagnostic use of nuclear beta‐catenin expression for the assessment of endometrial stromal tumors. Mod Pathol. 2008;21(6):756‐763. [DOI] [PubMed] [Google Scholar]
- 84. Kildal W, Pradhan M, Abeler VM, Kristensen GB, Danielsen HE. Beta‐catenin expression in uterine sarcomas and its relation to clinicopathological parameters. Eur J Cancer. 2009;45(13):2412‐2417. [DOI] [PubMed] [Google Scholar]
- 85. Kurihara S, Oda Y, Ohishi Y, et al. Coincident expression of beta‐catenin and cyclin D1 in endometrial stromal tumors and related high‐grade sarcomas. Mod Pathol. 2010;23(2):225‐234. [DOI] [PubMed] [Google Scholar]
- 86. Yamamoto Y, Abe A, Emi N. Clarifying the impact of polycomb complex component disruption in human cancers. Mol Cancer Res. 2014;12(4):479‐484. [DOI] [PubMed] [Google Scholar]
- 87. Astolfi A, Fiore M, Melchionda F, Indio V, Bertuccio SN, Pession A. BCOR involvement in cancer. Epigenomics. 2019;11(7):835‐855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Astolfi A, Melchionda F, Perotti D, et al. Whole transcriptome sequencing identifies BCOR internal tandem duplication as a common feature of clear cell sarcoma of the kidney. Oncotarget. 2015;6(38):40934‐40939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Karlsson J, Valind A, Gisselsson D. BCOR internal tandem duplication and YWHAE‐NUTM2B/E fusion are mutually exclusive events in clear cell sarcoma of the kidney. Genes, Chromosomes Cancer. 2016;55(2):120‐123. [DOI] [PubMed] [Google Scholar]
- 90. Specht K, Zhang L, Sung YS, et al. Novel BCOR‐MAML3 and ZC3H7B‐BCOR gene fusions in undifferentiated small blue round cell sarcomas. Am J Surg Pathol. 2016;40(4):433‐442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kommoss FKF, Stichel D, Schrimpf D, et al. DNA methylation‐based profiling of uterine neoplasms: a novel tool to improve gynecologic cancer diagnostics. J Cancer Res Clin Oncol. 2020;146(1):97‐104. [DOI] [PubMed] [Google Scholar]
- 92. Schoolmeester JK, Sciallis AP, Greipp PT, et al. Analysis of MDM2 amplification in 43 endometrial stromal tumors: a potential diagnostic pitfall. Int J Gynecol Pathol. 2015;34(6):576‐583. [DOI] [PubMed] [Google Scholar]
- 93. Avvakumov N, Cote J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene. 2007;26(37):5395‐5407. [DOI] [PubMed] [Google Scholar]
- 94. Lee CH, Nucci MR. Endometrial stromal sarcoma—the new genetic paradigm. Histopathology. 2015;67(1):1‐19. [DOI] [PubMed] [Google Scholar]
- 95. Flicker K, Smolle E, Haybaeck J, Moinfar F. Genomic characterization of endometrial stromal sarcomas with array comparative genomic hybridization. Exp Mol Pathol. 2015;98(3):367‐374. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


