Abstract
The protozoal pathogen Tritrichomonas foetus infects the colon of domestic cats and is a major cause of chronic colitis and diarrhea. Treatment failure is common, but antibiotics may improve clinical signs in a subset of cats, leading researchers to question involvement of the colonic microbiota in disease pathogenesis. Studies performed in women with venereal Trichomonas vaginalis infections have revealed that dysbiosis of host microbiota contributes to pathogenicity with similar findings also found in mice with intestinal Tritrichomonas musculis The aim of this study was to characterize differences in the fecal microbiota of cats with and without naturally occurring T. foetus infection and in a group of kittens prior to and after experimentally induced infection. Archived fecal DNA from cats undergoing testing for T. foetus infection (n = 89) and experimentally infected kittens (n = 4; at pre-, 2 weeks, and 9 weeks post-infection) were analyzed by sequencing of 16S rRNA genes. Amongst the naturally infected population, the genera Megamonas and Helicobacter were significantly increased in prevalence and abundance in cats testing positive for T. foetus infection. In the group of four experimentally infected kittens, fecal samples post-infection had significantly lower abundance of genus Dialister and Megamonas and greater abundance of the class Betaproteobacteria and family Succinivibrionaceae. We hypothesize that T. foetus promotes dysbiosis by competition for fermentable substrates used by these bacteria and that metabolic byproducts may contribute to the pathogenesis of colonic inflammation and diarrhea. Future studies are warranted for the measurement of fecal concentrations of microbial and protozoal metabolites in cats with T. foetus infection for the identification of potential therapeutic targets.
Introduction
Tritrichomonas foetus is a common cause of chronic colitis and diarrhea in domestic cats worldwide [1–6]. Infections caused by this protozoan are notoriously difficult to treat as many isolates are resistant to current therapies and relapse is common [7–9]. Trichomonads are obligate parasites or commensals of anaerobic mucosa-lined body cavities such as the gastrointestinal and reproductive tract. These protozoa cause disease by a variety of mechanisms including disruption of the mucosal microenvironment [10, 11], adhesion and degradation of the mucosal barrier [12], induction of epithelial apoptosis [13, 14], and evasion of the host immune system [15, 16]. In cats, T. foetus infection is transmitted via the fecal-oral route and restricted in final residence to the colon where the organisms live intermixed with colonic bacteria in close proximity to the mucosa [2, 17]. Presence of the infection results in lymphocytic, plasmacytic, and neutrophilic colitis and chronic, oftentimes lifelong, diarrhea [18].
Studies in women with the venereal protozoan Trichomonas vaginalis pinpoint dysbiotic vaginal microbiota as mediating not only pathogenic effects of infection but also predisposition to infection [19–22]. The dysbiotic bacteria serve as pathobionts that enhance pathogenicity by promoting adhesion of T. vaginalis to cervical cells [21] and increasing paracellular permeability [20]. In laboratory mice, colonization of the intestine by commensal Tritrichomonas musculis is influenced by competitive and cooperative relationships with members of the intestinal microbiota for metabolic byproducts [23]. Trichomonads are reliant on acquisition of micronutrients from the host environment and may also prey upon host bacteria via phagocytosis [24]. Currently, no studies have investigated the impact of T. foetus infection on the colonic microbiota of cats. This relationship is particularly interesting as treatment with antibiotics may improve clinical signs of diarrhea in some cats with T. foetus infection while precipitating a relapse of diarrhea in others [1, 3, 9].
Our objective in this study was to characterize the impact of naturally occurring and experimentally introduced T. foetus infection on the colonic microbiota of cats using Illumina-based 16S rRNA gene sequencing of archived fecal DNA. We hypothesized that cats with T. foetus infection would have a dysbiosis of specific members of the fecal microbiota whose characteristics could provide insights into disease pathogenesis and targets for treatment.
Methods
Fecal DNA samples
Cats having PCR testing for diagnosis of naturally occurring T. foetus infection
Archived DNA was identified for fecal samples submitted for T. foetus PCR testing to the North Carolina State University Intestinal Pathogens Research Laboratory between the years of 2012 to 2017. DNA extractions were performed using a Zymo Fecal/Soil DNA extraction kit (Zymo Research, Irvine CA) as previously described [25]. All DNA samples were stored at -20°C prior to use in the current study. A complete description of the parent population from which these samples were selected was recently published [26]. Inclusion criteria for this study were samples originally accompanied by a submission form in which the treatment history, or indication of lack of treatment, was provided. Additional patient information extracted from the submission forms included breed, age, life stage [27], sex, method of fecal collection (voided, loop, colonic flush), and indication for and results of T. foetus PCR testing. As previously described [5], loop collections are performed by inserting a fecal loop device through the anus into the colon to collect a fecal sample. A colonic flush is performed by inserting an 8 to 10 French red rubber catheter through the anus and instilling saline into the proximal colon to create a fecal slurry. The slurry material is collected and the sediment is used as the fecal sample. Samples were excluded from the study if no treatment history was provided or if the treatment history reported any prior administration of antimicrobial or anthelminthic drugs.
Kittens experimentally infected with feline T. foetus
Archived DNA was identified for fecal samples collected from kittens prior to and after experimental infection with T. foetus in 2006 [28]. DNA extractions were performed using a QIAamp DNA Stool minikit (Qiagen) with minor modifications as previously described [29]. All DNA samples were stored at -20°C prior to use in the current study. For the experimental infection, four specific pathogen free 7-week old sexually intact male (n = 2) and female (n = 2) domestic shorthair kittens were purchased from the same vendor (Liberty Laboratories, Liberty, NY). Each kitten was housed in a separate cage in the same laboratory animal resources facility and received the same dry food ad libitum (Hill’s Science Diet Feline Growth) throughout the study. After an acclimation period of seven weeks, and at an age of 14 weeks, kittens were infected with a single isolate of T. foetus that was cultivated from feces of a cat with diarrhea [28]. Each kitten was inoculated with 2 x 106 T. foetus by orogastric intubation. Included in this study are DNA samples extracted from feces collected from each kitten using a fecal loop prior to infection and at 2 and 9 weeks post-infection. The study was approved by the North Carolina State University Institutional Animal Care and Use Committee #03-095B. Due to inability to later eradicate the induced infection, cats were euthanized by IV injection of pentobarbital at the conclusion of the original study.
Bacterial 16S rRNA gene amplification and sequencing
Extracted DNA was quantified via PicoGreen analysis and used for bacterial 16S rRNA gene amplicon sequencing. Total DNA (12.5 ng) was amplified using universal primers targeting the V4 region of the bacterial 16S rRNA gene [30]. The sequences of the primers were: 515F - 5′ TCGTCGGCAGCGTCA GATGTGTATAAGAGACAGGTGCCAGCMGCCGCGGTAA 3′ and 806R - 5′GTCTCGTGGGCTCGGAGATGT GTATAAGAGACAGGGACTACHVGGGTWTCTAAT 3′. Overhang adapters were appended to the 5′ end of each primer sequence for compatibility with the Illumina sequencing platform. Master mixes contained 12.5 ng of total DNA, 0.2 μM of each primer and 2× KAPA HiFi HotStart ReadyMix (KAPA Biosystems, Wilmington, MA). The thermal profile for the amplification of each sample had an initial denaturing step at 95°C for 3 minutes, followed by 25 cycles of denaturing of 95°C for 30 seconds, annealing at 55°C for 30 seconds for 16S rRNA and a 30 second extension at 72°C, a 5 minute extension at 72°C and a final hold at 4°C. Each 16S amplicon was purified using the AMPure XP reagent (Beckman Coulter, Indianapolis, IN). In the next step each sample was amplified using a limited cycle PCR program, adding Illumina sequencing adapters and dual index barcodes (index 1(i7) and index 2(i5)) (Illumina, San Diego, CA) to the amplicon target. The thermal profile for the amplification of each sample had an initial denaturing step at 95°C for 3 minutes, followed by a denaturing cycle of 95°C for 30 seconds, annealing at 55°C for 30 seconds and a 30 second extension at 72°C (8 cycles), a 5 minute extension at 72°C and a final hold at 4°C. The final libraries were again purified using the AMPure XP reagent (Beckman Coulter), quantified with Quant-iT™ PicoGreen® dsDNA Reagent (Molecular Probes, Thermo Fisher Scientific, Waltham, MA), and normalized prior to equimolar pooling. The DNA library pool was then denatured with NaOH, diluted with hybridization buffer and heat denatured before loading on the MiSeq reagent cartridge MiSeq instrument (Illumina). Automated cluster generation and paired–end sequencing with dual reads were performed according to the manufacturer’s instructions.
Sequencing data analysis
Sequencing output from the Illumina MiSeq platform were converted to fastq format and demultiplexed using Illumina Bcl2Fastq 2.18.0.12. The resulting paired-end reads were processed using QIIME 2 [31] 2018.11. Index and linker primer sequences were trimmed using the QIIME 2 invocation of cutadapt. The resulting paired-end reads were processed with DADA2 through QIIME 2 including merging paired ends, quality filtering, error correction, and chimera detection. Amplicon sequencing units from DADA2 were assigned taxonomic identifiers with respect to Green Genes release 13_08.
Alpha diversity estimates were calculated within QIIME 2 using Evenness (Shannon) index and observed species number metrics at a rarefaction depth of 5,000 reads. Pairwise significance was tested using a Kruskal-Wallis ANOVA with Benjamini-Hochberg corrected q-values calculated as implemented in QIIME 2. Beta diversity estimates were calculated within QIIME 2 using weighted and unweighted Unifrac distances as well as Bray-Curtis dissimilarity between samples at a subsampling depth of 5,000 reads. Results were summarized, visualized through principal coordinate analysis in Emperor, and significance was estimated by PERMANOVA with Benjamini-Hochberg corrected q-values calculated as implemented in QIIME 2. Differentially abundant microbial taxa were identified by Analysis of Composition of Microbiomes (ANCOM) in QIIME 2. Relative abundance data were normalized by sample library size and taxa were removed if they were present in less than 10% of all samples or if lower than 0.01% average abundance [32]. Differences in abundance were tested for significance using a Kruskal-Wallis One-Way Analysis of Variance on Ranks. When significant, pairwise comparisons between groups were performed using a Dunn’s test. Generated p-values were corrected for multiple testing using the Benjamini-Hochberg procedure [33] at a false discovery rate of 0.15. For selected taxa, odds ratio and 95% confidence intervals for association with infection status were calculated using a Chi-square Test with Yates Continuity Correction. Statistical examination of abundance data were conducted using Systat Software (SigmaPlot 12.0, San Jose, CA). Results were represented graphically using GraphPad Software (Prism version 7.03, San Diego, CA).
Results
Cats with naturally occurring T. foetus infection
From a total of 1,717 archived DNA samples extracted from feces submitted for T. foetus PCR testing, 89 samples met criteria for inclusion in the study. Sixteen breeds were represented including domestic (46 cats), Bengal (11), Abyssinian (7), and Ragdoll (6) most commonly. Mean and median ages of cats were 3.7 and 1.7 years respectively (range 3 months– 16 years). The majority of cats were classified by life stage as junior (28 cats), followed by kitten (20 cats), prime (19 cats), mature (14 cats), senior (2 cats), geriatric (1 cat), or unknown age (2 cats). Sexes included male castrated (35 cats), female spayed (24 cats), female intact (16 cats), and male intact (14 cats). Test results for T. foetus infection, clinical signs, method of fecal sample collection, and treatment history of cats from which DNA samples were included in the study are shown in Table 1.
Table 1. Clinical data pertaining to 89 cats that underwent PCR testing for T. foetus infection and had archived fecal DNA used in this study.
| Clinical Variable | PCR test result for T. foetus infection No. (%) | |
|---|---|---|
| Positive | Negative | |
| PCR test results | 17 (19) | 72 (81) |
| Clinical signs | ||
| Diarrhea | 12 (13) | 54 (61) |
| No diarrhea | 5 (6) | 16 (18) |
| Not reported | 0 (0) | 2(2) |
| Method of fecal collection | ||
| Voided | 8 (9) | 33 (37) |
| Fecal loop | 4 (4) | 16 (18) |
| Colon flush | 4(4) | 21 (24) |
| Not reported | 1 (1) | 2 (2) |
| Treatment history† | ||
| Prescription diet | 5 (6) | 5 (6) |
| Probiotic | 3 (3) | 11 (12) |
| Steroid | 1 (1) | 10 (11) |
| Supplement | 0 (0) | 4 (4) |
| No treatment | 9 (10) | 49 (55) |
Specialty diets included unspecified (6 cats), Hills Prescription Diet I/D (2), Purina EN (1), or a duck-based diet (1). Probiotics used included Purina Fortiflora (10), unspecified (3), or NutraMax Proviable (1). Specific steroids used were prednisolone (10) or Depomedrol (1). Supplements included one each of Diagel, Benefiber, or Clay.
†Some cats received more than 1 listed treatment.
Major phyla in fecal microbiota of cats undergoing testing for T. foetus infection include Firmicutes, Bacteroidetes, Proteobacteria, Fusobacteria, and Actinobacteria
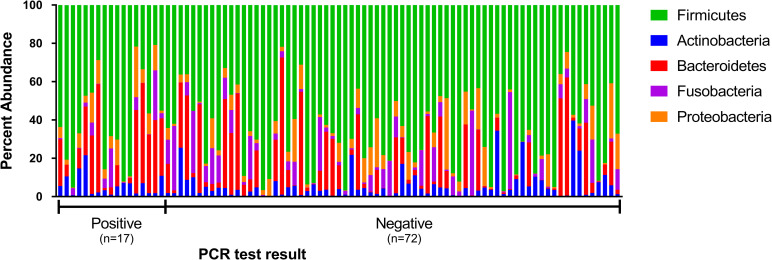
The sequence analysis of all 89 cats yielded 9,691,382 quality sequences (mean ± SD = 108,892 ± 21,089). The major phyla represented in the fecal microbiota of cats tested for T. foetus infection were Firmicutes, Bacteroidetes, Proteobacteria, Fusobacteria, and Actinobacteria (Fig 1).
Fig 1. Percent abundance of the major phyla observed in feces from 89 individual cats undergoing testing for T. foetus infection.
Each sample is designated as having been obtained from a cat testing negative versus positive for T. foetus infection as determined by results of fecal PCR testing.
There were no significant differences in alpha diversity measures of evenness (Shannon), diversity (Faith), or observed number of taxa between cats testing positive versus negative for T. foetus infection. Both weighted and unweighted measures of beta diversity identified small but significant differences in phylogenetic composition of the microbiota between cats testing positive versus negative for naturally occurring T. foetus infection (PERMANOVA, q < 0.05) (Fig 2).
Fig 2. Beta diversity of microbiota from 89 cats tested for naturally occurring infection with T. foetus.
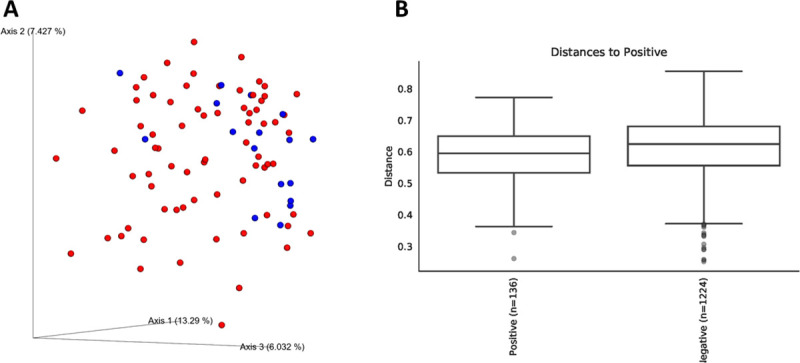
Panel A: Bray-Curtis principal coordinates analysis plot showing clustering of microbial communities from feces of cats testing positive (blue circles) versus negative (red circles) for T. foetus infection. Axis 1, 13.29%; Axis 2, 7.427%; Axis 3, 6.032%. Panel B: Unweighted Unifrac distances between microbial communities from feces testing positive versus negative for T. foetus infection. PERMANOVA q = 0.033.
Cats testing positive for T. foetus infection have more frequent membership and higher abundance of genus Megamonas and Helicobacter
Abundance of specific taxa observed in the feces of cats testing positive versus negative for naturally occurring T. foetus infection are shown in Table 2.
Table 2. Relative abundance of genus level taxa (percent of total sequences) in the fecal microbiota of cats undergoing PCR testing for naturally occurring T. foetus infection.
| Phylum | Family | Genus | Median % abundance [range min to max] | Kruskal-Wallis ANOVA p-value | |
|---|---|---|---|---|---|
| PCR Positive (n = 17 cats) | PCR Negative (n = 72 cats) | ||||
| Actinobacteria | 5.19 [0.78–21.59] | 3.63 [0.04–39.61] | 0.484 | ||
| Bifidobacteriaceae | Bifidobacterium | 0.03 [0.0–5.79] | 0.02 [0.0–31.05] | 0.688 | |
| Coriobacteriaceae | Uncl. Coriobacteriaceae | 0.09 [0.0–0.75] | 0.04 [0.0–1.21] | 0.342 | |
| Adlercreutzia | 0.05 [0.0–0.12] | 0.02 [0.0–0.21] | 0.141 | ||
| Collinsella | 3.05 [0.24–18.62] | 2. 80 [0.0–12.24] | 0.262 | ||
| Slackia | 0.05 [0.0–0.35] | 0.08 [0.0–1.56] | 0.431 | ||
| Bacteroidetes | 24.40 [0.05–56.30] | 4.48 [0.01–71.26] | 0.115 | ||
| Bacteroidaceae | Bacteroides | 0.63 [0.0–29.94] | 0.96 [0.0–43.68] | 0.774 | |
| Prevotellaceae | Prevotella | 9.71 [0.02–50.82] | 1.27 [0.0–47.79] | 0.075 | |
| [Paraprevotellaceae] | [Prevotella] | 0.10 [0.0–7.75] | 0.02 [0.0–23.98] | 0.885 | |
| Firmicutes | 63.66 [20.84–95.03] | 65.32 [21.71–97.00] | 0.342 | ||
| Enterococcaceae | Enterococcus | 0.06 [0.0–12.57] | 0.08 [0.0–47.67] | 0.7 | |
| Turicibacteraceae | Turicibacter | 0.0 [0.0–3.51] | 0.01 [0.0–48.24] | 0.142 | |
| Streptococcaceae | Streptococcus | 0.01 [0.0–10.00] | 0.02 [0.0–49.65] | 0.179 | |
| Uncl. Clostridiales | Uncl. Clostridiales | 0.07 [0.0–0.62] | 0.21 [0.0–5.64] | 0.071 | |
| Clostridiaceae | Clostridium | 5.55 [0.86–57.11] | 7.26 [0.04–59.92] | 0.888 | |
| Lachnospiraceae | Uncl. Lachnospiraceae | 3.58 [0.93–40.76] | 3.59 [0.01–25.73] | 0.822 | |
| Blautia | 5.50 [2.01–27.11] | 8.11 [0.0–38.68] | 0.806 | ||
| Clostridium | 0.02 [0.0–8.11] | 0.04 [0.0–5.48] | 0.707 | ||
| Coprococcus | 0.13 [0.0–2.24] | 0.19 [0.0–7.99] | 0.378 | ||
| Dorea | 0.75 [0.0–3.31] | 0.61 [0.0–9.00] | 0.896 | ||
| Roseburia | 0.29 [0.0–2.92] | 0.06 [0.0–18.42] | 0.772 | ||
| [Ruminococcus] | 1.52 [0.19–9.28] | 1.40 [0.0–9.57] | 0.719 | ||
| Peptococcaceae | Peptococcus | 0.43 [0.0–4.29] | 0.93 [0.0–19.38] | 0.241 | |
| Peptostreptococcaceae | Uncl. Peptostreptococcaceae | 0.52 [0.0–15.03] | 0.37 [0.0–83.14] | 0.637 | |
| Ruminococcaceae | Uncl. Ruminococcaceae | 0.70 [0.0–2.28] | 1.05 [0.0–12.31] | 0.407 | |
| Faecalibacterium | 0.13 [0.0–2.43] | 0.15 [0.0–9.64] | 0.851 | ||
| Oscillospira | 0.13 [0.0–1.87] | 0.28 [0.0–1.90] | 0.43 | ||
| Ruminococcus | 0.03 [0.0–0.27] | 0.03 [0.0–6.41] | 0.975 | ||
| Veillonellaceae | Dialister | 0.16 [0.0–3.03] | 0.03 [0.0–4.11] | 0.327 | |
| Megamonas | 1.52 [0.0–17.31] | 0.03 [0.0–7.39] | 0.001*** † | ||
| Megasphaera | 1.37 [0.0–14.27] | 0.11 [0.0–15.54] | 0.116 | ||
| Phascolarctobacterium | 0.30 [0.0–3.15] | 0.03 [0.0–7.32] | 0.42 | ||
| [Mogibacteriaceae] | Uncl. [Mogibacteriaceae] | 0.22 [0.0–6.54] | 0.09 [0.0–2.55] | 0.325 | |
| Erysipelotrichaceae | Bulleidia | 0.04 [0.0–4.07] | 0.09 [0.0–9.59] | 0.974 | |
| Catenibacterium | 0.44 [0.0–7.25] | 0.09 [0.0–5.46] | 0.121 | ||
| [Eubacterium] | 0.83 [0.02–8.04] | 0.60 [0.0–11.56] | 0.855 | ||
| Fusobacteria | 0.59 [0.01–25.95] | 2.08 [0.0–50.63] | 0.251 | ||
| Uncl. Fusobacteriaceae | Uncl. Fusobacteriaceae | 0.59 [0.0–24.24] | 1.52 [0.0–28.00] | 0.507 | |
| Fusobacteriaceae | Fusobacterium | 0.0 [0.0–0.91] | 0.03 [0.0–6.99] | 0.334 | |
| Proteobacteria | 6.40 [0.33–26.37] | 3.18 [0.0–37.26] | 0.306 | ||
| Alcaligenaceae | Sutterella | 0.34 [0.0–2.96] | 0.59 [0.0–5.88] | 0.389 | |
| Campylobacteraceae | Campylobacter | 0.06 [0.0–3.96] | 0.01 [0.0–15.88] | 0.050* | |
| Succinivibrionaceae | Anaerobiospirillum | 1.62 [0.0–11.27] | 0.01 [0.0–23.51] | 0.046* | |
| Enterobacteriaceae | Uncl. Enterobacteriaceae | 0.01 [0.0–2.34] | 0.05 [0.0–22.12] | 0.264 | |
| Helicobacteraceae | Helicobacter | 0.09 [0.0–5.13] | 0.0 [0.0–5.25] | 0.003**† | |
*p<0.05 and
***p<0.001.
†Benjamini-Hochberg corrected p-value <0.15.
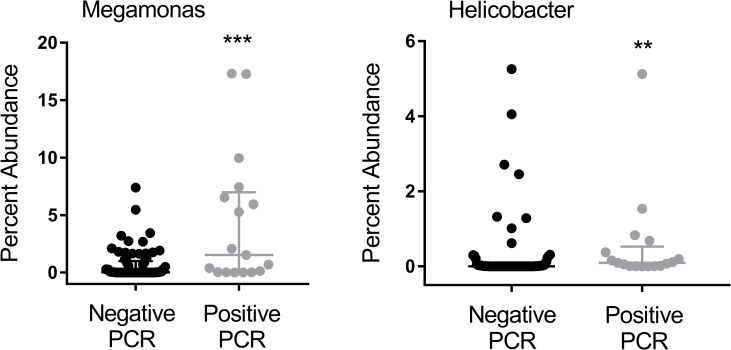
After accounting for the impact of multiple statistical comparisons, cats testing positive for T. foetus infection had a significantly greater abundance of members of the genus Megamonas and Helicobacter compared to cats that tested negative for T. foetus infection (Fig 3).
Fig 3. Percent abundance of Megamonas and Helicobacter in feces of cats testing PCR negative (n = 72 cats) or PCR positive (n = 17 cats) for naturally occurring T. foetus infection.
Data points represent individual cats. Bars represent median and interquartile range. Mann-Whitney Rank Sum Test ***p = 0.001 (BH-corrected p = 0.043). **p = 0.003 (BH-corrected p = 0.06). BH = Benjamini-Hochberg.
Cats testing positive for T. foetus infection were 10.8 times more likely to have Megamonas (95%CI = 1.356 to 85.9, p = 0.016, Chi-square test) and 7.9 times more likely to have Helicobacter present in the microbial community (95%CI = 1.689 to 37.210, p = 0.007, Chi-square test) compared to cats testing negative for T. foetus infection.
Method of fecal sample collection significantly influence the abundance of select taxa in feces of cats tested for T. foetus infection
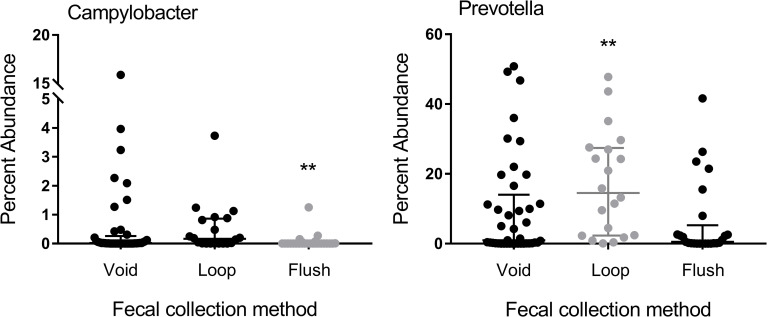
When examining the impact of available metadata in the cats undergoing testing for T. foetus infection, no significant differences in alpha or beta diversity were observed based on life stage, breed, sex, method of fecal sample collection, clinical signs of diarrhea, or treatment history. However, method of fecal sample collection had a significant impact on relative abundance of members of the genus Prevotella and Campylobacter (Fig 4).
Fig 4. Percent abundance of Campylobacter and Prevotella in feces of cats having the sample collected after voiding (n = 41 cats), using a fecal loop (n = 20 cats), or by means of colonic flush (n = 25 cats).
Data points represent individual cats. Bars represent median and interquartile range. Kruskal-Wallis One Way ANOVA on Ranks **p<0.01 (BH-corrected p = 0.129). BH = Benjamini- Hochberg.
Campylobacter was less abundant in feces collected by colonic flush compared to samples collected after voiding or using a fecal loop. Fecal samples collected using a loop recovered a greater percent abundance of Prevotella compared to samples collected after voiding or by colonic flush.
Cats with experimentally induced T. foetus infection
Following orogastric infection, all four specific pathogen free kittens were confirmed positive for T. foetus by means of fecal DNA PCR testing at both the 2 week and 9 week post-infection time points. None of the kittens developed clinical signs of diarrhea at any time point following infection.
The sequence analysis yielded 1,408,793 quality sequences for the 12 analyzed fecal samples (mean ± SD = 117,399 ± 26,660). Among the measures of alpha diversity, significant differences were observed between pre- and post-infection fecal samples in the evenness of species abundances (Shannon alpha diversity; Kruskal-Wallis q = 0.04) but not in the number of different taxa observed.
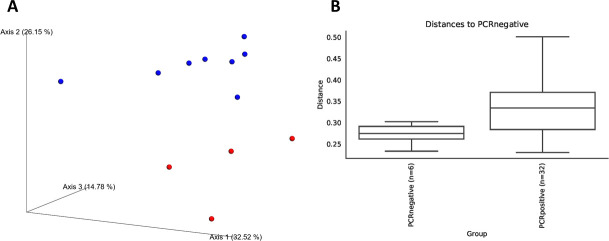
Weighted measures of beta diversity identified significant differences in phylogenetic composition of the fecal microbiota between pre- and post-infection samples from cats with experimentally induced T. foetus infection (PERMANOVA, q < 0.01) (Fig 5).
Fig 5. Bray-Curtis beta diversity of microbiota from 4 kittens prior to (PCR negative) and at 2 time points after (PCR positive) experimental infection with feline T. foetus.
Panel A: Principal coordinates analysis plot showing clustering of microbial communities from feces of cats prior to (red circles) and after (blue circles) experimental T. foetus infection. Axis 1, 35.52%; Axis 2, 26.15%; Axis 3, 14.78%. Panel B: Weighted Unifrac distances between microbial communities from feces of cats prior to (PCR negative) and after (PCR positive) experimental T. foetus infection. Bray-Curtis PERMANOVA q = 0.003.
Experimental infection with T. foetus shifts the abundance of specific taxa in the feline fecal microbiome
Abundance of specific taxa observed in the feces of kittens prior to and at two time points after undergoing experimental infection with T. foetus are shown in Table 3.
Table 3. Relative abundance of genus level taxa (percent of total sequences) in the fecal microbiota of 4 cats prior to and following experimentally induced T. foetus infection.
| Median % abundance [Range min to max] |
Kruskal-Wallis ANOVA |
|||||
|---|---|---|---|---|---|---|
| Phylum | Family | Genus | Pre-infection | 2 weeks post-infection | 9 weeks post-infection | |
| p-value | ||||||
| Actinobacteria | 1.45 [0.25–4.72] | 0.54 [0.18–1.21] | 0.39 [0.01–2.84] | 0.706 | ||
| Bifidobacteriaceae | Bifidobacterium | 0.01 [0.0–0.03] | 0.00 [0.0–0.0] | 0.00 [0.0–0.01] | 0.136 | |
| Coriobacteriaceae | Adlercreutzia | 0.05 [0.01–0.1] | 0.04 [0.01–0.11] | 0.06 [0.0–0.12] | 1.000 | |
| Collinsella | 1.33 [0.23–4.65] | 0.48 [0.13–1.03] | 0.33 [0.01–2.55] | 0.706 | ||
| Bacteroidetes | 60.16 [51.79–61.82] | 59.53 [53.31–64.53] | 56.81 [50.80–66.33] | 0.815 | ||
| Bacteroidaceae | Bacteroides | 10.45 [9.64–16.54] | 16.21 [15.17–17.05] | 16.23 [14.61–23.30] | 0.136 | |
| Porphyromonadaceae | Parabacteroides | 1.31 [0.52–2.23] | 2.14 [2.02–2.45] | 1.60 [1.38–2.17] | 0.197 | |
| Prevotellaceae | Prevotella | 29.69 [25.78–33.00] | 21.08 [18.70–27.07] | 25.26 [9.18–27.40] | 0.044* | |
| Rikenellaceae | Uncl. Rikenellaceae | 0.00 [0.0–0.03] | 0.08 [0.0–0.18] | 0.06 [0.0–0.14] | 0.173 | |
| Alistipes | 0.18 [0.01-.33] | 0.36 [0.08–0.95] | 0.09 [0.05–0.92] | 0.540 | ||
| S24-7 | Uncl. S24-7 | 0.77 [0.61–0.94] | 1.72 [0.88–2.75] | 0.90 [0.40–1.01] | 0.074 | |
| [Odoribacteraceae] | Odoribacter | 0.43 [0.16–1.04] | 0.77 [0.62–0.95] | 0.59 [0.22–1.08] | 0.592 | |
| [Paraprevotellacea] | Paraprevotella | 0.07 [0.04–0.13] | 0.10 [0.04–0.17] | 0.11 [0.08–0.23] | 0.397 | |
| [Prevotella] | 13.01 [9.36–21.52] | 15.10 [13.51–19.13] | 14.77 [9.37–19.92] | 0.770 | ||
| Firmicutes | 28.96 [27.83–36.96] | 22.60 [20.25–26.23] | 22.01 [18.24–30.92] | 0.086 | ||
| Lactobacillaceae | Lactobacillus | 0.00 [0.0–0.01] | 0.00 [0.0–0.01] | 0.00 [0.0–0.03] | 0.941 | |
| Leuconostocaceae | Leuconostoc | 0.00 [0.0–0.0] | 0.00 [0.0–0.01] | 0.00 [0.0–0.01] | 0.630 | |
| Streptococcaceae | Lactococcus | 0.01 [0.0–0.02] | 0.01 [0.0–0.02] | 0.03 [0.01–0.03] | 0.040* | |
| Streptococcus | 0.00 [0.0–0.06] | 0.00 [0.0–0.21] | 0.01 [0.0–0.03] | 0.968 | ||
| Uncl. Clostridiales | Uncl. Clostridiales | 0.73 [0.60–1.35] | 1.57 [0.57–1.98] | 1.27 [0.71–2.06] | 0.436 | |
| Clostridiaceae | Candidatus Arthromitus | 0.16 [0.0–0.46] | 0.13 [0.01–0.65] | 0.03 [0.01–0.13] | 0.480 | |
| Clostridium | 0.36 [0.15–0.89] | 0.26 [0.09–0.46] | 0.16 [0.07–0.54] | 0.452 | ||
| Lachnospiraceae | Uncl. Lachnospiraceae | 1.94 [0.75–2.58] | 2.77 [1.79–3.21] | 2.42 [2.34–4.76] | 0.252 | |
| Blautia | 0.39 [0.20–0.72] | 0.34 [0.19–0.43] | 0.35 [0.12–1.00] | 0.913 | ||
| Clostridium | 0.07 [0.0–0.15] | 0.02 [0.02–0.03] | 0.01 [0.0–0.04] | 0.397 | ||
| Coprococcus | 0.03 [0.0–0.03] | 0.04 [0.01–0.04] | 0.03 [0.01–0.19] | 0.540 | ||
| Dorea | 0.10 [0.04–0.32] | 0.05 [0.01–0.12] | 0.02 [0.01–0.08] | 0.234 | ||
| Lachnospira | 0.00 [0.0–0.16] | 0.43 [0.0–0.69] | 0.60 [0.07–1.11] | 0.074 | ||
| Roseburia | 0.54 [0.17–0.83] | 0.36 [0.01–0.47] | 0.77 [0.13–4.21] | 0.592 | ||
| [Ruminococcus] | 0.12 [0.09–0.27] | 0.07 [0.06–0.14] | 0.09 [0.07–0.45] | 0.277 | ||
| Peptococcaceae | Peptococcus | 0.16 [0.02–0.25] | 0.08 [0.03–0.18] | 0.07 [0.03–0.16] | 0.706 | |
| Peptostreptococcaceae | Uncl. Peptostreptococcaceae | 0.69 [0.12–6.23] | 0.59 [0.07–1.55] | 0.08 [0.0–0.52] | 0.252 | |
| Ruminococcaceae | Uncl. Ruminococcaceae | 1.75 [0.65–2.70] | 1.47 [1.14–2.82] | 1.28 [0.69–3.71] | 0.968 | |
| Butyricicoccus | 0.01 [0.01–0.01] | 0.01 [0.0–0.01] | 0.01 [0.01–0.03] | 0.592 | ||
| Faecalibacterium | 2.68 [1.83–5.03] | 3.36 [2.26–4.40] | 2.87 [0.65–3.75] | 0.840 | ||
| Oscillospira | 1.14 [0.40–1.54] | 1.03 [0.71–1.37] | 0.94 [0.61–1.34] | 0.941 | ||
| Ruminococcus | 0.13 [0.02–0.21] | 0.22 [0.19–0.27] | 0.21 [0.16–0.47] | 0.074 | ||
| Veillonellaceae | Acidaminococcus | 0.75 [0.08–2.66] | 0.33 [0.07–0.92] | 0.09 [0.01–0.23] | 0.145 | |
| Dialister | 10.54 [8.42–11.66] | 6.17 [5.06–7.30] | 7.09 [6.72–7.69] | 0.002**† | ||
| Megamonas | 0.83 [0.72–2.14] | 0.18 [0.18–0.20] | 0.10 [0.04–0.46] | 0.005**† | ||
| Megasphaera | 2.36 [1.49–3.65] | 0.77 [0.47–1.92] | 0.56 [0.26–0.68] | 0.016* | ||
| Mitsuokella | 0.05 [0.0–0.82] | 0.00 [0.0–0.15] | 0.00 [0.0–0.01] | 0.706 | ||
| [Mogibacteriaceae] | Uncl. [Mogibacteriaceae] | 0.79 [0.24–1.17] | 0.41 [0.15–0.62] | 0.34 [0.14–0.83] | 0.252 | |
| Erysipelotrichaceae | Uncl. Erysipelotrichaceae | 0.05 [0.0–0.09] | 0.00 [0.0–0.0] | 0.03 [0.0–0.14] | 0.131 | |
| Allobaculum | 0.01 [0.0–0.03] | 0.07 [0.0–0.25] | 0.04 [0.02–0.18] | 0.370 | ||
| Bulleidia | 0.00 [0.0–0.10] | 0.00 [0.0–0.19] | 0.00 [0.0–0.49] | 0.941 | ||
| Catenibacterium | 0.41 [0.13–0.72] | 0.46 [0.18–0.62] | 0.38 [0.12–1.63] | 0.968 | ||
| Holdemania | 0.02 [0.0–0.02] | 0.02 [0.02–0.04] | 0.02 [0.01–0.94] | 0.540 | ||
| [Eubacterium] | 0.20 [0.09–0.31] | 0.21 [0.17–0.30] | 0.18 [0.10–0.50] | 0.913 | ||
| Fusobacteria | 2.59 [1.68–3.34] | 5.07 [2.06–6.78] | 5.22 [4.34–8.00] | 0.074 | ||
| Uncl. Fusobacteriaceae | Uncl. Fusobacteriaceae | 1.80 [1.06–2.65] | 4.08 [1.91–5.40] | 3.85 [3.25–5.72] | 0.026* | |
| Fusobacteriaceae | Uncl. Fusobacteriaceae | 0.66 [0.62–0.94] | 0.99 [0.15–1.37] | 1.50 [0.82–2.27] | 0.063 | |
| Proteobacteria | 6.60 [4.69–6.72] | 12.51 [10.62–13.91] | 11.64 [9.42–16.49] | 0.011* | ||
| Betaproteobacteria (class) | 0.01 [0.01–0.03] | 0.19 [0.10–0.32] | 0.12 [0.04–0.16] | 0.002**† | ||
| Burkholderiales (order) | 0.74 [0.58–0.98] | 2.53 [1.29–2.95] | 2.04 [0.59–2.62] | 0.055 | ||
| Alcaligenaceae | Sutterella | 2.19 [1.61–2.69] | 3.31 [2.53–4.91] | 3.79 [1.72–7.07] | 0.348 | |
| Methylophilaceae | Methylotenera | 0.00 [0.0–0.0] | 0.00 [0.0–0.0] | 0.00 [0.0–0.0] | 1.000 | |
| Desulfovibrionaceae | Uncl. Desulfovibrionaceae | 0.15 [0.13–0.21] | 0.30 [0.19–0.43] | 0.23 [0.12–0.32] | 0.122 | |
| Desulfovibrio | 0.89 [0.22–1.68] | 1.14 [0.65–1.55] | 1.31 [0.73–1.44] | 0.840 | ||
| Campylobacteraceae | Campylobacter | 1.70 [1.13–2.07] | 4.92 [1.45–6.64] | 4.02 [2.37–6.68] | 0.080 | |
| Succinivibrionaceae | Uncl. Succinivibrionaceae | 0.03 [0.01–0.05] | 0.16 [0.14-.18] | 0.30 [0.14–0.57] | 0.007**† | |
| Succinivibrio | 0.37 [0.19–0.80] | 0.24 [0.11–0.28] | 0.16 [0.08–0.36] | 0.234 | ||
| Enterobacteriaceae | Uncl. Enterobacteriaceae | 0.02 [0.0–0.04] | 0.01 [0.0–0.09] | 0.04 [0.02–0.08] | 0.252 | |
| Pseudomonadaceae | Pseudomonas | 0.00 [0.0–0.0] | 0.00 [0.0–0.0] | 0.00 [0.0–0.0] | 1.000 | |
*p<0.05 and
**p<0.01.
†Benjamini-Hochberg corrected p-value <0.15.
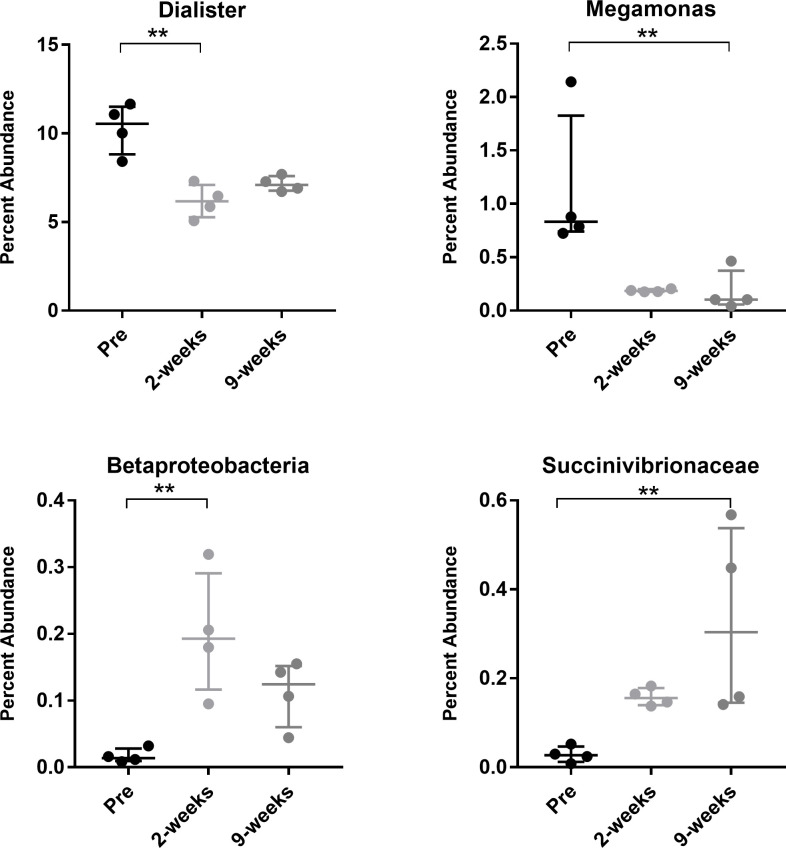
After accounting for the impact of multiple statistical comparisons, four taxa were identified as significantly impacted by introduction of T. foetus infection. Following experimental infection with T. foetus, the fecal microbiota of kittens had a significantly lower abundance of members of the genus Dialister and Megamonas and significantly greater abundance of members of the class Betaproteobacteria and family Succinivibrionaceae compared to kittens prior to experimental infection (Figs 6 and 7).
Fig 6. Differentially abundant microbial taxa identified by Analysis of Composition of Microbiomes (ANCOM).
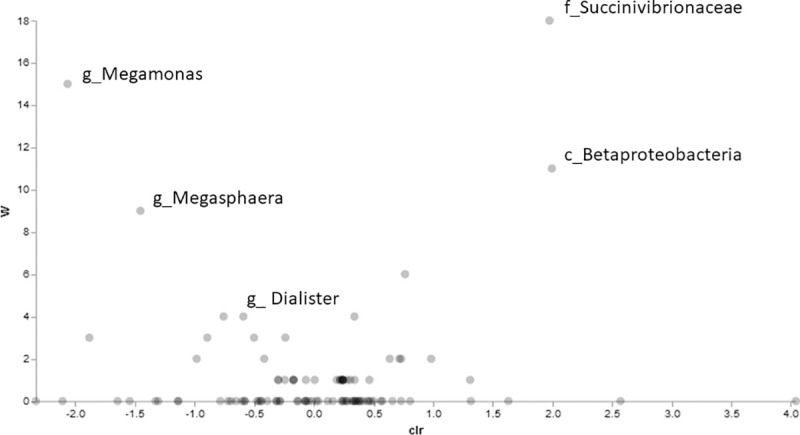
Volcano plot of differential abundance comparing kittens prior to versus after experimental infection with T. foetus. Clr (x-axis) is a measure of the effect size difference for a particular species between the two conditions. The W-statistic (y-axis) is the strength of the ANCOM test for the tested number of species.
Fig 7. Percent abundance of Dialister, Megamonas, Betaproteobacteria, and Succinivibrionaceae in feces of 4 kittens prior to (Pre) and at 2 time points after (2 weeks and 9 weeks) experimental infection with feline T. foetus.
Data points represent individual cats. Bars represent median and interquartile range. **p<0.01 (Benjamini-Hochberg-corrected p<0.15).
Discussion
To better understand the role of the colonic microbiota in the pathogenesis of T. foetus infection in cats, we characterized the fecal microbiota of 89 cats undergoing testing for natural infection. Each fecal sample was submitted for diagnostic T. foetus PCR testing based on clinical signs such as diarrhea, history of exposure, or for purposes of screening for the infection. Overall, 19% of cats in this population were diagnosed with T. foetus while the other 81% had any number of other potential causes for their clinical signs. Based on 16S rRNA gene sequencing, T. foetus infection did not alter the predominant bacterial phyla present in feces which were similar in identity to prior descriptions of the fecal microbiota in cats [34–38]. These phyla were represented mainly by Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, and Fusobacteria. The presence of T. foetus also did not affect the overall number or diversity of different taxa present in the microbial community. However, T. foetus had a significant impact on community membership that could be attributed to increases in both the presence and relative abundance of the genus Megamonas (phylum Firmicutes, family Veillonellaceae) and Helicobacter (phylum Proteobacteria). A key attribute of this study was our ability to extend this population level observation to assessment of the specific impact of experimentally introduced T. foetus infection in a group of purpose-bred kittens that were controlled for age, sex, diet, and background microbiome. In these kittens, introduction of T. foetus resulted in a decrease in relative abundance of the genus Megamonas and Dialister (phylum Firmicutes, family Veillonellaceae) and significantly increased abundance of members of the class Betaproteobacteria and family Succinivibrionaceae (phylum Proteobacteria).
It is remarkable that changes in the family Viellonellaceae and the specific genus Megamonas were specifically and significantly impacted in both naturally occurring and experimentally induced T. foetus infection. Both Megamonas and Dialister produce large amounts of short chain fatty acids such as propionate and lactate which are thought to possess anti-inflammatory properties and provide a large portion of the energy supply for colonocytes [39]. A higher abundance of Megamonas in the microbial community is a distinguishing characteristic of healthy cats [40]. Decreases in these taxa, as observed in kittens with experimentally induced T. foetus infection, is commonly associated with disease states such as IBD [40–42]. Low levels of short chain fatty acids may lead to colonocyte autophagy which could contribute to the pathogenic effects of T. foetus or somehow benefit its survival.
It is likely that T. foetus promotes dysbiosis in part by competition for fermentable substrates. In mice, the presence of fermentable fiber is a requirement for successful intestinal colonization with the commensal trichomonad Tritrichomonas musculis [43]. Specific colonic bacterial species also compete for this fiber source. Studies have identified that the availability of fermentable fiber can affect the abundance of Megamonas and other members of the family Veillonellaceae [44–48]. For example, cats fed a chicken-based extruded diet had agreater abundance of Megamonas species compared to those fed raw whole chicks [45]. Similarly, administration of fermentable prebiotics containing fructooligosaccharides or inulin results in a higher abundance of Veillonellaceae in the feces of cats [46] and specifically the genus Megamonas in dogs [47]. By consuming available fiber, T. foetus may indirectly decrease the abundance of Megamonas and Dialister via competition for available nutrients.
The major fermentation products of T. foetus are succinate, acetate, and molecular hydrogen [49]. Succinate is also a key substrate or product of metabolism by the Veillonellaceae (Megamonas and Dialister) and Succinovibrionaceae [50]. Succinate is an important pro-inflammatory signaling molecule [50] that may be of interest as a mediator of disease pathogenesis in T. foetus infection. In mice, colonization with T. musculis increases the concentration of intestinal succinate which stimulates tuft cell succinate receptors leading to activation of a type 2 immune response [43, 51]. Activation of the host epithelial inflammasome by T. musculis exacerbates the development of T- cell- driven colitis [52]. Succinate increases in the intestinal lumen of mice and humans with inflammatory bowel disease and is correlated with disease activity [53]. A decrease in succinate-consuming bacterial strains is reported in people with IBD [54]. Accordingly, it is worth considering that metabolic products of T. foetus or the dysbiotic microbiota contribute to the pathogenesis of colonic inflammation and diarrhea in cats with T. foetus infection. If this is the case, then manipulation of dietary fermentation substrates, addition of competing probiotics, or administration of select metabolites might influence T. foetus survival, secondary microbial dysbiosis, or pathogenic effects.
In contrast to experimentally infected kittens, cats with naturally occurring T. foetus infection had a significant increase in abundance of Megamonas as well as Helicobacter. This is an interesting finding as a recent study comparing the microbiome of normal cats versus those with chronic and acute diarrhea found a significant increase in Helicobacter and Megamonas in the normal cats as compared to those with diarrhea [40]. This finding in our naturally infected cats could indicate resolving dysbiosis with a more chronic T. foetus infection as compared to the acute stages of experimental infection. Other explanations could include use of a single strain of T. foetus for the experimental infection that resulted in a unique effect, differences in age between the two populations of cats, underlying differences in microbiome ecology between the purpose-bred kittens and an outbred population of cats, or the use of different DNA extraction methods for naturally infected cats vs. experimentally infected kittens. It is also possible that cats with a higher abundance of Megamonas or Helicobacter are more susceptible to infection by T. foetus via changes to the colonic microenvironment. A similar phenomenon has been described in women where changes in the abundance and specific identity of vaginal Lactobacillus species are linked to increased risk for development of T. vaginalis infection [22].
Inherent in the outbred population of cats included in this study were numerous variables having potential impact on the microbiota including differences in age, breed, sex, clinical signs of diarrhea, and treatments administered [34, 37, 40, 55, 56]. Additional influences that were unknown for these cats include differences in diet, environment, comorbidities, or co-infections [35, 36, 38, 56–58]. While these variables have a demonstrated impact on the fecal microbiota in studies controlling for their effects, their impact in this large, cross-sectional study population was not found to be significant. We did however observe a significant impact of fecal collection method on composition of the microbiome. These results likely reflect preferential sampling of microbiota from different microbial niches. In the present study, Prevotella was observed in greater abundance in fecal samples collected with a loop than by the other methods. In people, Prevotella is a predominant taxa in the rectum where it is observed in higher abundance in samples of the rectal mucosa compared to feces [59]. Samples collected with a loop were more likely to disrupt the mucosal surface microbiota thereby increasing recovery of Prevotella. A significantly lower abundance of Campylobacter was observed in samples collected using the flush method in our study. In pigs, Campylobacter is significantly more abundant in the mucosa than in lumen content [60]. Accordingly, samples collected by flush were more likely to capture non-mucosa-associated bacteria. These observations emphasize both the impact of sample collection method on results of microbiome analysis as well as support the power of our study to detect expected differences between these methods. All cats in the study had a treatment history provided and those reporting use of antimicrobials were excluded. Reports of other treatments were too few in number to enable a robust examination of treatment effects on microbiota composition.
In this first study of the fecal microbiota in cats with and without T. foetus infection, specific changes in the abundance of members of the Veillonellaceae and Succinivibrionaceae suggest altered fermentative metabolism in the colon of cats with T. foetus infection. These findings support additional investigations into the composition and functional impacts of the fecal metabolome on colonic inflammation and diarrhea in cats with T. foetus infection.
Acknowledgments
The authors thank Dr. Benjamin Callahan for his input on this study.
Data Availability
All sequence files are available from the NCBI Sequence Read Archive database (accession numbers PRJNA692480 and PRJNA692463) https://www.ncbi.nlm.nih.gov/bioproject/PRJNA692463https://www.ncbi.nlm.nih.gov/bioproject/PRJNA692480.
Funding Statement
These studies were supported by the Paul and Lea Levine Foundation (JLG) and the North Carolina Veterinary Medical Foundation’s Support for T. foetus Research and Innovation in Veterinary Education fund (NCVMF-STRIVE)(JLG). The UNC Microbiome Core is supported in part by P30 DK034987 Center for Gastrointestinal Biology and Disease (CGIBD) and P30 DK056350 UNC Nutrition Obesity Research Center (NORC). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gookin JL, Breitschwerdt EB, Levy MG, Gager RB, Benrud JG. Diarrhea associated with trichomonosis in cats. Journal of the American Veterinary Medical Association. 1999;215(10):1450–4. . [PubMed] [Google Scholar]
- 2.Yaeger MJ, Gookin JL. Histologic features associated with Tritrichomonas foetus-induced colitis in domestic cats. Veterinary pathology. 2005;42(6):797–804. 10.1354/vp.42-6-797 . [DOI] [PubMed] [Google Scholar]
- 3.Gookin JL, Stebbins ME, Hunt E, Burlone K, Fulton M, Hochel R, et al. Prevalence of and risk factors for feline Tritrichomonas foetus and giardia infection. Journal of clinical microbiology. 2004;42(6):2707–10. 10.1128/JCM.42.6.2707-2710.2004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gunn-Moore DA, McCann TM, Reed N, Simpson KE, Tennant B. Prevalence of Tritrichomonas foetus infection in cats with diarrhoea in the UK. Journal of feline medicine and surgery. 2007;9(3):214–8. 10.1016/j.jfms.2007.01.003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gookin JL, Hanrahan K, Levy MG. The conundrum of feline Trichomonosis. Journal of feline medicine and surgery. 2017;19(3):261–74. 10.1177/1098612X17693499 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao C, Koster LS. Tritrichomonas foetus infection, a cause of chronic diarrhea in the domestic cat. Vet Res. 2015;46:35 10.1186/s13567-015-0169-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gookin JL, Stauffer SH, Dybas D, Cannon DH. Documentation of in vivo and in vitro aerobic resistance of feline Tritrichomonas foetus isolates to ronidazole. Journal of veterinary internal medicine. 2010;24(4):1003–7. 10.1111/j.1939-1676.2010.0534.x . [DOI] [PubMed] [Google Scholar]
- 8.Kather EJ, Marks SL, Kass PH. Determination of the in vitro susceptibility of feline tritrichomonas foetus to 5 antimicrobial agents. Journal of veterinary internal medicine. 2007;21(5):966–70. 10.1892/0891-6640(2007)21[966:dotivs]2.0.co;2 . [DOI] [PubMed] [Google Scholar]
- 9.Foster DM, Gookin JL, Poore MF, Stebbins ME, Levy MG. Outcome of cats with diarrhea and Tritrichomonas foetus infection. Journal of the American Veterinary Medical Association. 2004;225(6):888–92. 10.2460/javma.2004.225.888 . [DOI] [PubMed] [Google Scholar]
- 10.Figueroa-Angulo EE, Rendon-Gandarilla FJ, Puente-Rivera J, Calla-Choque JS, Cardenas-Guerra RE, Ortega-Lopez J, et al. The effects of environmental factors on the virulence of Trichomonas vaginalis. Microbes Infect. 2012;14(15):1411–27. 10.1016/j.micinf.2012.09.004 . [DOI] [PubMed] [Google Scholar]
- 11.Tolbert MK, Gookin JL. Mechanisms of Tritrichomonas foetus Pathogenicity in Cats with Insights from Venereal Trichomonosis. Journal of veterinary internal medicine. 2016;30(2):516–26. 10.1111/jvim.13920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.da Costa RF, de Souza W, Benchimol M, Alderete JF, Morgado-Diaz JA. Trichomonas vaginalis perturbs the junctional complex in epithelial cells. Cell research. 2005;15(9):704–16. 10.1038/sj.cr.7290340 . [DOI] [PubMed] [Google Scholar]
- 13.Singh BN, Lucas JJ, Hayes GR, Kumar I, Beach DH, Frajblat M, et al. Tritrichomonas foetus induces apoptotic cell death in bovine vaginal epithelial cells. Infect Immun. 2004;72(7):4151–8. 10.1128/IAI.72.7.4151-4158.2004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sommer U, Costello CE, Hayes GR, Beach DH, Gilbert RO, Lucas JJ, et al. Identification of Trichomonas vaginalis cysteine proteases that induce apoptosis in human vaginal epithelial cells. The Journal of biological chemistry. 2005;280(25):23853–60. 10.1074/jbc.M501752200 . [DOI] [PubMed] [Google Scholar]
- 15.Ahn MH, Song HO, Ryu JS. Trichomonas vaginalis-induced neutrophil apoptosis causes anti-inflammatory cytokine production by human monocyte-derived macrophages. Parasite Immunol. 2008;30(8):410–6. 10.1111/j.1365-3024.2008.01037.x . [DOI] [PubMed] [Google Scholar]
- 16.Chang JH, Ryang YS, Morio T, Lee SK, Chang EJ. Trichomonas vaginalis inhibits proinflammatory cytokine production in macrophages by suppressing NF-kappaB activation. Mol Cells. 2004;18(2):177–85. . [PubMed] [Google Scholar]
- 17.Gookin JL, Levy MG, Law JM, Papich MG, Poore MF, Breitschwerdt EB. Experimental infection of cats with Tritrichomonas foetus. American journal of veterinary research. 2001;62(11):1690–7. 10.2460/ajvr.2001.62.1690 . [DOI] [PubMed] [Google Scholar]
- 18.Gookin JL, Stone MR, Yaeger MJ, Meyerholz DK, Moisan P. Fluorescence in situ hybridization for identification of Tritrichomonas foetus in formalin-fixed and paraffin-embedded histological specimens of intestinal trichomoniasis. Vet Parasitol. 2010;172(1–2):139–43. 10.1016/j.vetpar.2010.04.014 . [DOI] [PubMed] [Google Scholar]
- 19.Brotman RM, Bradford LL, Conrad M, Gajer P, Ault K, Peralta L, et al. Association between Trichomonas vaginalis and vaginal bacterial community composition among reproductive-age women. Sexually transmitted diseases. 2012;39(10):807–12. 10.1097/OLQ.0b013e3182631c79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinderfeld AS, Phukan N, Bar AK, Roberton AM, Simoes-Barbosa A. Cooperative Interactions between Trichomonas vaginalis and Associated Bacteria Enhance Paracellular Permeability of the Cervicovaginal Epithelium by Dysregulating Tight Junctions. Infect Immun. 2019;87(5). 10.1128/IAI.00141-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinderfeld AS, Simoes-Barbosa A. Vaginal dysbiotic bacteria act as pathobionts of the protozoal pathogen Trichomonas vaginalis. Microbial pathogenesis. 2020;138:103820 10.1016/j.micpath.2019.103820 . [DOI] [PubMed] [Google Scholar]
- 22.Jarrett OD, Srinivasan S, Richardson BA, Fiedler T, Wallis JM, Kinuthia J, et al. Specific Vaginal Bacteria Are Associated With an Increased Risk of Trichomonas vaginalis Acquisition in Women. The Journal of infectious diseases. 2019;220(9):1503–10. 10.1093/infdis/jiz354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei Y, Gao J, Kou Y, Meng L, Zheng X, Liang M, et al. Commensal Bacteria Impact a Protozoan’s Integration into the Murine Gut Microbiota in a Dietary Nutrient-Dependent Manner. Applied and environmental microbiology. 2020;86(11). 10.1128/AEM.00303-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Midlej V, Benchimol M. Trichomonas vaginalis kills and eats—evidence for phagocytic activity as a cytopathic effect. Parasitology. 2010;137(1):65–76. 10.1017/S0031182009991041 . [DOI] [PubMed] [Google Scholar]
- 25.Stauffer SH, Birkenheuer AJ, Levy MG, Marr H, Gookin JL. Evaluation of four DNA extraction methods for the detection of Tritrichomonas foetus in feline stool specimens by polymerase chain reaction. Journal of veterinary diagnostic investigation: official publication of the American Association of Veterinary Laboratory Diagnosticians, Inc. 2008;20(5):639–41. 10.1177/104063870802000518 . [DOI] [PubMed] [Google Scholar]
- 26.Hedgespeth BA, Stauffer SH, Robertson JB, Gookin JL. Association of fecal sample collection technique and treatment history with Tritrichomonas foetus polymerase chain reaction test results in 1717 cats. Journal of veterinary internal medicine. 2020;34(2):734–41. 10.1111/jvim.15727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogt AH, Rodan I, Brown M, Brown S, Buffington CA, Forman MJ, et al. AAFP-AAHA: Feline life stage guidelines. J Am Anim Hosp Assoc. 2010;46(1):70–85. 10.5326/0460070 . [DOI] [PubMed] [Google Scholar]
- 28.Gookin JL, Stauffer SH, Coccaro MR, Poore MF, Levy MG, Papich MG. Efficacy of tinidazole for treatment of cats experimentally infected with Tritrichomonas foetus. American journal of veterinary research. 2007;68(10):1085–8. 10.2460/ajvr.68.10.1085 . [DOI] [PubMed] [Google Scholar]
- 29.Gookin JL, Birkenheuer AJ, Breitschwerdt EB, Levy MG. Single-tube nested PCR for detection of tritrichomonas foetus in feline feces. Journal of clinical microbiology. 2002;40(11):4126–30. 10.1128/jcm.40.11.4126-4130.2002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences of the United States of America. 2011;108 Suppl 1:4516–22. Epub 2010/06/11. 10.1073/pnas.1000080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature biotechnology. 2019;37(8):852–7. Epub 2019/07/26. 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bourgon R, Gentleman R, Huber W. Independent filtering increases detection power for high-throughput experiments. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(21):9546–51. 10.1073/pnas.0914005107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological). 1995;57(1):289–300. [Google Scholar]
- 34.Garcia-Mazcorro JF, Lanerie DJ, Dowd SE, Paddock CG, Grutzner N, Steiner JM, et al. Effect of a multi-species synbiotic formulation on fecal bacterial microbiota of healthy cats and dogs as evaluated by pyrosequencing. FEMS microbiology ecology. 2011;78(3):542–54. 10.1111/j.1574-6941.2011.01185.x . [DOI] [PubMed] [Google Scholar]
- 35.Hooda S, Vester Boler BM, Kerr KR, Dowd SE, Swanson KS. The gut microbiome of kittens is affected by dietary protein:carbohydrate ratio and associated with blood metabolite and hormone concentrations. The British journal of nutrition. 2013;109(9):1637–46. 10.1017/S0007114512003479 . [DOI] [PubMed] [Google Scholar]
- 36.Jia J, Frantz N, Khoo C, Gibson GR, Rastall RA, McCartney AL. Investigation of the faecal microbiota of kittens: monitoring bacterial succession and effect of diet. FEMS microbiology ecology. 2011;78(2):395–404. 10.1111/j.1574-6941.2011.01172.x . [DOI] [PubMed] [Google Scholar]
- 37.Deusch O, O’Flynn C, Colyer A, Swanson KS, Allaway D, Morris P. A Longitudinal Study of the Feline Faecal Microbiome Identifies Changes into Early Adulthood Irrespective of Sexual Development. PloS one. 2015;10(12):e0144881 10.1371/journal.pone.0144881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bermingham EN, Young W, Kittelmann S, Kerr KR, Swanson KS, Roy NC, et al. Dietary format alters fecal bacterial populations in the domestic cat (Felis catus). MicrobiologyOpen. 2013;2(1):173–81. 10.1002/mbo3.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roediger WE. Utilization of nutrients by isolated epithelial cells of the rat colon. Gastroenterology. 1982;83(2):424–9. . [PubMed] [Google Scholar]
- 40.Suchodolski JS, Foster ML, Sohail MU, Leutenegger C, Queen EV, Steiner JM, et al. The fecal microbiome in cats with diarrhea. PloS one. 2015;10(5):e0127378 10.1371/journal.pone.0127378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suchodolski JS, Dowd SE, Wilke V, Steiner JM, Jergens AE. 16S rRNA gene pyrosequencing reveals bacterial dysbiosis in the duodenum of dogs with idiopathic inflammatory bowel disease. PloS one. 2012;7(6):e39333 10.1371/journal.pone.0039333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Minamoto Y, Otoni CC, Steelman SM, Buyukleblebici O, Steiner JM, Jergens AE, et al. Alteration of the fecal microbiota and serum metabolite profiles in dogs with idiopathic inflammatory bowel disease. Gut microbes. 2015;6(1):33–47. 10.1080/19490976.2014.997612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider C, O’Leary CE, von Moltke J, Liang HE, Ang QY, Turnbaugh PJ, et al. A Metabolite-Triggered Tuft Cell-ILC2 Circuit Drives Small Intestinal Remodeling. Cell. 2018;174(2):271–84 e14. 10.1016/j.cell.2018.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandri M, Dal Monego S, Conte G, Sgorlon S, Stefanon B. Raw meat based diet influences faecal microbiome and end products of fermentation in healthy dogs. BMC veterinary research. 2017;13(1):65 10.1186/s12917-017-0981-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kerr KR, Dowd SE, Swanson KS. Faecal microbiota of domestic cats fed raw whole chicks v. an extruded chicken-based diet. Journal of nutritional science. 2014;3:e22 10.1017/jns.2014.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcia-Mazcorro JF, Barcenas-Walls JR, Suchodolski JS, Steiner JM. Molecular assessment of the fecal microbiota in healthy cats and dogs before and during supplementation with fructo-oligosaccharides (FOS) and inulin using high-throughput 454-pyrosequencing. PeerJ. 2017;5:e3184 10.7717/peerj.3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beloshapka AN, Dowd SE, Suchodolski JS, Steiner JM, Duclos L, Swanson KS. Fecal microbial communities of healthy adult dogs fed raw meat-based diets with or without inulin or yeast cell wall extracts as assessed by 454 pyrosequencing. FEMS microbiology ecology. 2013;84(3):532–41. 10.1111/1574-6941.12081 . [DOI] [PubMed] [Google Scholar]
- 48.Jackson MI, Jewell DE. Balance of saccharolysis and proteolysis underpins improvements in stool quality induced by adding a fiber bundle containing bound polyphenols to either hydrolyzed meat or grain-rich foods. Gut microbes. 2019;10(3):298–320. 10.1080/19490976.2018.1526580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lindmark DG, Eckenrode BL, Halberg LA, Dinbergs ID. Carbohydrate, energy and hydrogenosomal metabolism of Tritrichomonas foetus and Trichomonas vaginalis. J Protozool. 1989;36(2):214–6. 10.1111/j.1550-7408.1989.tb01076.x . [DOI] [PubMed] [Google Scholar]
- 50.Fernandez-Veledo S, Vendrell J. Gut microbiota-derived succinate: Friend or foe in human metabolic diseases? Reviews in endocrine & metabolic disorders. 2019;20(4):439–47. 10.1007/s11154-019-09513-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nadjsombati MS, McGinty JW, Lyons-Cohen MR, Jaffe JB, DiPeso L, Schneider C, et al. Detection of Succinate by Intestinal Tuft Cells Triggers a Type 2 Innate Immune Circuit. Immunity. 2018;49(1):33–41 e7. 10.1016/j.immuni.2018.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chudnovskiy A, Mortha A, Kana V, Kennard A, Ramirez JD, Rahman A, et al. Host-Protozoan Interactions Protect from Mucosal Infections through Activation of the Inflammasome. Cell. 2016;167(2):444–56 e14. 10.1016/j.cell.2016.08.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Macias-Ceja DC, Ortiz-Masia D, Salvador P, Gisbert-Ferrandiz L, Hernandez C, Hausmann M, et al. Succinate receptor mediates intestinal inflammation and fibrosis. Mucosal Immunol. 2019;12(1):178–87. 10.1038/s41385-018-0087-3 . [DOI] [PubMed] [Google Scholar]
- 54.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome biology. 2012;13(9):R79 10.1186/gb-2012-13-9-r79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jia J, Frantz N, Khoo C, Gibson GR, Rastall RA, McCartney AL. Investigation of the faecal microbiota of geriatric cats. Letters in applied microbiology. 2011;53(3):288–93. 10.1111/j.1472-765X.2011.03105.x . [DOI] [PubMed] [Google Scholar]
- 56.Bermingham EN, Young W, Butowski CF, Moon CD, Maclean PH, Rosendale D, et al. The Fecal Microbiota in the Domestic Cat (Felis catus) Is Influenced by Interactions Between Age and Diet; A Five Year Longitudinal Study. Front Microbiol. 2018;9:1231 10.3389/fmicb.2018.01231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marsilio S, Pilla R, Sarawichitr B, Chow B, Hill SL, Ackermann MR, et al. Characterization of the fecal microbiome in cats with inflammatory bowel disease or alimentary small cell lymphoma. Sci Rep. 2019;9(1):19208 10.1038/s41598-019-55691-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Young W, Moon CD, Thomas DG, Cave NJ, Bermingham EN. Pre- and post-weaning diet alters the faecal metagenome in the cat with differences in vitamin and carbohydrate metabolism gene abundances. Sci Rep. 2016;6:34668 10.1038/srep34668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li G, Yang M, Zhou K, Zhang L, Tian L, Lv S, et al. Diversity of Duodenal and Rectal Microbiota in Biopsy Tissues and Luminal Contents in Healthy Volunteers. Journal of microbiology and biotechnology. 2015;25(7):1136–45. 10.4014/jmb.1412.12047 . [DOI] [PubMed] [Google Scholar]
- 60.Adhikari B, Kim SW, Kwon YM. Characterization of Microbiota Associated with Digesta and Mucosa in Different Regions of Gastrointestinal Tract of Nursery Pigs. International journal of molecular sciences. 2019;20(7). 10.3390/ijms20071630 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All sequence files are available from the NCBI Sequence Read Archive database (accession numbers PRJNA692480 and PRJNA692463) https://www.ncbi.nlm.nih.gov/bioproject/PRJNA692463https://www.ncbi.nlm.nih.gov/bioproject/PRJNA692480.