Abstract
Human African Trypanosomiasis (HAT) is a disease of major economic importance in Sub-Saharan Africa. The HAT is caused by Trypanosoma brucei rhodesiense (Tbr) parasite in eastern and southern Africa, with suramin as drug of choice for treatment of early stage of the disease. Suramin treatment failures has been observed among HAT patients in Tbr foci in Uganda. In this study, we assessed Tbr parasite strains isolated from HAT patients responsive (Tbr EATRO-232) and non-responsive (Tbr EATRO-734) to suramin treatment in Busoga, Uganda for 1) putative role of suramin resistance in the treatment failure 2) correlation of suramin resistance with Tbr pathogenicity and 3) proteomic pathways underpinning the potential suramin resistance phenotype in vivo. We first assessed suramin response in each isolate by infecting male Swiss white mice followed by treatment using a series of suramin doses. We then assessed relative pathogenicity of the two Tbr isolates by assessing changes pathogenicity indices (prepatent period, survival and mortality). We finally isolated proteins from mice infected by the isolates, and assessed their proteomic profiles using mass spectrometry. We established putative resistance to 2.5 mg/kg suramin in the parasite Tbr EATRO-734. We established that Tbr EATRO-734 proliferated slower and has significantly enriched pathways associated with detoxification and metabolism of energy and drugs relative to Tbr EATRO-232. The Tbr EATRO-734 also has more abundantly expressed mitochondrion proteins and enzymes than Tbr EATRO-232. The suramin treatment failure may be linked to the relatively higher resistance to suramin in Tbr EATRO-734 than Tbr EATRO-232, among other host and parasite specific factors. However, the Tbr EATRO-734 appears to be less pathogenic than Tbr EATRO-232, as evidenced by its lower rate of parasitaemia. The Tbr EATRO-734 putatively surmount suramin challenges through induction of energy metabolism pathways. These cellular and molecular processes may be involved in suramin resistance in Tbr.
Keywords: Suramin; Drug resistance; Drug sensitive, Trypanosoma brucei rhodesience
Graphical abstract
Highlights
-
•
Tbr EATRO-734 and Tbr EATRO-232 are resistant and susceptible to suramin respectively.
-
•
Tbr EATRO-734 highly expressed mitochondrion proteins/enzymes than Tbr EATRO-232.
-
•
Tbr EATRO-734 surmounts suramin challenges by induction of energy metabolism.
-
•
VSG proteins that may confer suramin resistance were identified in Tbr EATRO-734.
-
•
Proteins involved in trypanosome sensitive to suramin were induced in Tbr EATRO-232.
1. Introduction
African trypanosomiases (AT) are diseases of humans (known as Human African Trypanosomiasis (HAT)/sleeping sickness) and their domestic animals (known as African Animal Trypanosomiasis (AAT)/nagana) with devastating medical and economic consequences for Africa. The AT are caused by single-celled trypanosome protozoan parasites transmitted by infected tsetse flies (Glossina spp.). The HAT is specifically caused by Trypanosoma brucei gambiense (Tbg) and Trypanosoma brucei rhodesiense (Tbr), while AAT is caused by Trypanosoma brucei brucei (Tbb), Trypanosome vivax (Tv) and Trypanosome congolense (Tc). The Tbr and Tbb are genetically similar, differing only in phenotype of human infectivity (host range expansion) in Tbr. The human infectivity in Tbr is due to the presence of Tbr-specific serum-resistance associated (SRA) protein (Xong et al., 1998). The SRA gene is widely disturbed and readily exchanged among lineages of T. brucei in eastern Africa, potentially providing Tbr with extensive gene pool with which to respond to selective pressures, including drugs (Balmer et al., 2011). The Tbg causes the chronic form of HAT in central and West Africa while Tbr causes the acute form of HAT in East Africa. There are no HAT vaccines and treatment is costly, and with adverse side effects (Brun et al., 2010). Typical Tbr transmission cycle involves wild and domestic animals. However, intensified human or animal to human Tbr transmission may occur during epidemics. On the other hand, Tbg transmission cycle is mostly from human to human, with limited involvement of animals.
Tsetse fly vector transmit Tbr and Tbg to human through bites, hereby the parasites multiply at the bite site (forming chancre). The parasites then migrate to the lymphatic fluid, blood and other body tissues causing first stage (early or haemo-lymphatic stage) of HAT. The parasites then cross blood brain barrier (BBB) into central nervous system (CNS) where they cause second stage of HAT (late or meningo-encephalitic stage). This stage is characterized by neurological symptoms that include disturbance of sleep cycle (from which HAT derived its name). The acute Tbr infections evolve rapidly in a matter of weeks or months, with the chronic Tbg infections lasting many months or even years. Both infections are fatal if untreated. Treatment of sleeping sickness is stage specific where pentamidine and suramin drugs are used for treatment of the first stage infections by Tbg and Tbr respectively. The second stage infections by Tbg and Tbr are treated with eflornithine and melarsoprol drugs respectively. A special combination of eflornithine and nifurtimox (NECT) drugs is also available for treatment of the second stage of Tbg infections (Babokhov et al., 2013). Various trials for new, modified or re-adapted drug compounds have recently been undertaken (Bisser et al., 2007; Vodnala et al., 2009; Trunz et al., 2011; Jacobs et al., 2011; Rodgers et al., 2011; Pohlig et al., 2016).
The mode of action of suramin against Tbr in largely unknown, but the drug is thought to be internalised by the parasite through receptor-mediated low-density lipoprotein (LDL), variant surface glycoprotein (VSG) and invariant surface glycoprotein (ISG) 75 (Vansterkenburg et al., 1993; Wiedemar et al., 2018; Alsford et al., 2012). The drug appears to elicit its anti-trypanocidal effects through inhibition of glycolysis (glycosomal) enzymes, a source of energy for bloodstream form (BSF) of the parasites (Vansterkenburg et al., 1993; Wang, 1995). The activity of suramin appears to be synergized by import of ornithine and its metabolism (Alsford et al., 2012; Macedo et al., 2017).
Cases of suramin treatment failure/relapses have been reported and potentially attributed to misdiagnosis of second stage of HAT (against which suramin is ineffective), insufficient dosage compliance and/or suramin resistance. Suramin resistance can be experimentally induced (Apted, 1980). However, physiological and molecular process that underpin this resistance phenotype are poorly understood in Tbr. In Tbb, (the Tbr variant), suramin resistance phenotype appears to be stage specific, confined to bloodstream form (BSF) parasites without progression to procylic (PC) stage, which are less sensitive to suramin (Alsford et al., 2013a). These observations suggest that suramin affects BSF-specific biological processes. Other studies link the resistance to reduced cellular uptake through endocytic pathway (Alsford et al., 2012, 2013a) and VSG antigenic variation, where expression of a particular VSG (VSGSur) appear to impart resistance phenotype to Tbb (Wiedemar et al., 2018).
In this study, we interrogated one Tbr strain isolated from HAT patient that did not respond to suramin treatment (treatment failure) (Tbr EATRO-734 isolate) against another stain (Tbr EATRO-232 isolate) from a patient that responded to the treatment (cured) in Busoga, Uganda in 1964 and 1959 respectively (Murilla et al., 2014). From this interrogation, we sought to establish putative 1) role of suramin resistance in the treatment failure 2) correlation of suramin resistance with Tbr pathogenicity and 3) proteomic pathways underpinning the potential suramin resistance phenotype in vivo using murine model.
2. Materials and methods
2.1. Test trypanosome isolate parasites
We utilized Tbr parasites isolate (Tbr EATRO 232) from HAT patient who responded to suramin (Germanin®, Bayer schering pharma, Germany), and compared this to isolate (Tbr EATRO 734) from a patient who did not respond to suramin treatment in Busoga Uganda in 1959 and 1964 respectively. We also utilized a third isolate (Tbr EATRO-2285) that was established as valid Tbr subspecies (Gibson et al., 2002). This isolate served as a positive control for validation of Tbr EATRO 734 and Tbr EATRO 232 subspecies status. We have summarized biological and historical data on Tbr EATRO 734 and Tbr EATRO 232 strains in Table 1. We obtained each parasites strain (Tbr EATRO 734, Tbr EATRO 232 or Tbr EATRO-2285) as stabilates from Biotechnology Research Institute of the Kenya Agricultural and Livestock Research Organization (BioRI-KALRO) cryo-bank, Muguga, Kenya (Murilla et al., 2014).
Table 1.
Biological and historical data of selected Trypanosoma brucei rhodesiense strains.
| Strain | Year of Isolation | Region of Isolation | Type of Isolate | Comment |
|---|---|---|---|---|
| EATRO 734 EATRO 232 |
1964 1959 |
Busoga, Uganda Busoga, Uganda |
Pleomorphic Pleomorphic |
Eight passages since isolation Four passages since isolation |
2.2. Test vertebrate animals
We separately expanded the parasites from the individual stabilates using 25–30 g male Swiss white mice (6–8 weeks old) provided by BioRI-KALRO, Muguga, Kenya. We utilized the mice to establish putative 1) role of suramin resistance to the treatment failure 2) differential pathogenicity of the isolates. Our proteomic assessment of the molecular pathways potentially associated with the resistance phenotype required greater parasitaemia (harvesting) that could not be provided by mice. We therefore utilized rats (bigger size hence more parasites than mice) instead of mice for that assessment. We consequently expanded the parasites using 180–250 g male Wistar rats (4–6 weeks old). We sourced for these rats from School of Veterinary Medicine, University of Nairobi, Kenya. We housed both rodents (mice and rats) in standard (30.80 × 30.80 × 18.72 cm) plastic cages (Thoren Caging Systems, inc., Hazleton, PA, USA) with wood shavings as bedding material. We maintained the rodents on commercial Unga® pellets (Unga® Kenya Ltd, Nairobi, Kenya) according to the manufacturer's instructions, and provided them with water ad libitum. We allowed the rodents to acclimatize for two weeks in their new environment before we involved them in any of our experiments. We initiated our experiments by concurrently collecting standard pre-inoculation baseline data on body weight and packed cell volume (PCV) twice a week and screened the rodents for ecto- and endo-parasites. We cleared the parasites off by administrating ivermectin drug (Ivermectin®, Anupco, Suffolk, England) according to established procedures (Soll, 1989). We strictly adhered to procedures and protocols as outlined in The Guide for the Care and Use of Laboratory Animals in our use of the rodents for our experiments (Institute for Laboratory Animal Research, 2011). Our procedures and protocols were reviewed and approved by Institutional Animal Care and Use Committee (IACUC) of BioRI-KALRO (Ref. No. C/BioRI/4/325/II/20).
2.3. Validation of species status of the Tbr isolates and expansion in donor mice
Both isolates (Tbr EATRO 734 and Tbr EATRO 232) were initially classified as Tbr (and not Tbg) based on their human infectivity and isolation from Busoga, Uganda, a known Tbr foci. However, Uganda has geographically separated Tbg and Tbr zones (Picozzi et al., 2005). We thus reasoned that the patients could have possibly been infected in either zone by respective resident parasites (Tbg or Tbr) and travelled to Busoga where the infections were diagnosed. We therefore validated the Tbr status of these isolates. We thus validated the taxonomic status of the isolates using Polymerase Chain Reaction (PCR) to detect Tbr specific/diagnostic SRA gene (Gibson et al., 2002) in the respective parasite isolate genome. To achieve this, we first thawed the respective cryopreserved parasite isolate (Tbr EATRO 734, Tbr EATRO 232 or Tbr EATRO-2285) stabilates and visually confirmed viability of the parasites by observing their motility (as evidence of viability) under wet film light microscope (Leitz, Wetzlar, Germany) at ×40 magnification. We then separately suspended the viable parasites from each stabilate in Phosphate Buffered Saline (PBS) with 1% glucose (44 mM NaCl, 57 mM Na2HPO4, 3 mM KH2PO4, 55 mM glucose) (PSG) pH 8.0, and extracted genomic DNA from respective stabilate using Qiagen DNeasy blood and tissue kit (Qiagen Inc, Valencia, CA, USA) according to manufacturer's instructions. We then separately amplified the DNA via PCR using Tbr-SRA specific 5′GACAACAAGTACCTTGGCGC3′ forward and 5′TACTGTTGTTGTACCG CCGC3′ reverse primers (Gibson et al., 2002). For the amplification, we used 1 μl of each DNA template in 10 μl PCR mixture containing 1x PCR buffer, 2.5 mM MgCl2, 10 mM of each of the four dNTPs, 10 μM of each primer and 1 unit of Taq polymerase (Promega, Madison, MO, USA) in the buffer (provided by the manufacturer which contained no MgCl2). Our first PCR cycle included an initial denaturation step at 95 °C for 3 min, followed by 35 cycles each at 95 °C for 45 s, 56 °C for 30 s, 72 °C for 1 min and a final extension step at 72 °C for 4 min. We specifically amplified the DNA from Tbr EATRO-2285, a known Tbr species (Gibson et al., 2002) to serve as a positive control. We also included no-sample negative control (tripled distilled water used for re-suspension of extracted genomic DNA) in our PCR regime. We loaded and resolved the PCR products on 3% agarose gel in Tris Borate EDTA (TBE) buffer with ethidium bromide (Sambrook et al., 1989). We also run 100 bp DNA ladder molecular weight marker (Promega, Madison, MO, USA) to confirm expected (460bp) molecular weight of the amplification products.
2.4. Expansion of the Tbr isolates in donor mice
We received the parasites in cryo-preserved form in single capillary tube vial stabilates. These vials typically have limited parasites that were insufficient for our analyses. We therefore expanded these parasite populations by separately infecting mice with the different isolates and propagated the parasites for our downstream analyses. To achieve this, we first, suppressed immunity of the mice using cyclophosphamide drug (Sigma-Aldrich, Laborchemikalien, GmbH, Germany) (Renoux and Renoux, 1980) at 300 mg/kg/day, for three consecutive days to increase the odds of their infection by the parasites, and then separately inoculated them with the respective parasite isolates/strain (Kagira et al., 2005). We administered the cyclophosphamide drug intraperitoneally (ip) on four donor mice as previously described by (Antoine-Moussiaux et al., 2008). We then separately ip injected the trypanosomes diluted to 1 × 104 parasites in a 200 μl volume of PBS (Turner, 1990) into each of the donor mice (two donor mice per isolate) and monitored individual mice daily for development of parasitaemia. We monitored the parasitaemia by collecting blood using tail snip (Parasuraman et al., 2010), examined the parasites under microscopy (Van Meirvenne, 1999) and scored the parasitaemia using improved Neubauer chamber method (Herbert and Lumsden, 1976). We euthanized the donor mice at peak parasitaemia (~1.0 × 108 trypanosomes), using carbon dioxide according to American Veterinary Medical Association (AVMA) guidelines for the euthanasia of animals. We then collected the parasites through cardiac puncture in tubes containing 10% EDTA and quantified the parasites using improved Neubauer chamber method (Herbert and Lumsden, 1976). We subsequently diluted the parasites (in blood) to 1.0 × 104 trypanosomes/ml with PSG pH 8.0 solution for downstream suramin resistance/sensitivity, pathogenicity and proteomic profile assessments.
2.5. Validation of suramin resistance or sensitivity in T. b. rhodesiense-isolates
We evaluated sensitivity of individual Tbr isolates to suramin using established guidelines for testing drug resistance in trypanosomes (Eisler et al., 2001). Briefly, we separately ip inoculated Tbr EATRO-734 isolate into four groups of mice consisting of six experimental mice per group. We then ip administered 2.5, 5.0 and 10.0 mg/kg of suramin (Germanin®, Bayer schering pharma, Germany) to the first, second and third groups respectively, 24 h post parasite inoculation. We chose these doses based on the previous finding that showed a minimum of 2.0 mg/kg of suramin was required to clear apparent suramin-resistant trypanosomes as opposed to 1.5 mg/kg required for treatment of their wild-type counterparts (Scott et al., 1996; Thomas et al., 2018). We did not treat the inoculated fourth group. This group served to provide baseline control data on performance of the parasites (parasitaemia) in the absence of treatment. We concurrently performed similar experiments with Tbr EATRO-232 on mice. We included an additional independent group of six mice that were neither infected with any of the parasites, nor treated with suramin. This group served as a negative control for both isolates. We subsequently monitored changes in mice parasitaemia daily during the first week, three times a week during the second week and twice a week thereafter as previously documented (Herbert and Lumsden, 1976; Kagira and Maina, 2007) for up to 60 days post inoculation (dpi). We, thereafter, euthanized surviving mice by placing them in a chamber containing CO2 according to AVMA and BioRI-KALRO IACUC guidelines for the euthanasia of animals. We employed the guidelines on standardized tests in mice for detection of drug resistance trypanosomes (Eisler et al., 2001) to establish inclusion/exclusion criteria for assessment of our Tbr resistance/sensitivity to suramin. Based on these criteria, we considered trypanosome isolate suramin-sensitive if at least five out of the six treated mice are cured (they remain aparasitaemic until the end of the 60-day observation period) and suramin-resistant if fewer than five mice were cured.
2.6. Assessment of pathogenicity of suramin sensitive/resistant Tbr isolates
We sought to determined differences in pathogenicity between the two isolates (Tbr EATRO-734 and Tbr EATRO-232) in mice. To achieve this, we inoculated the mice with respective isolates and subsequently monitored parasite pre-patent period (pp) (day of first appearance of the parasite in the peripheral blood) and parasitaemia profiles of the isolates. We also concurrently monitored changes in PCV and body weight of the mice due to the parasite infections. We performed these by ip inoculation of two groups (consisting of ten mice each), with the 1 × 104 of Tbr EATRO-734 or Tbr EATRO-232 trypanosomes in 200 μl of PSG (pH = 8.0) per mouse. We then monitored (daily) establishment of the parasites in peripheral blood of the mice to establish 1) respective pp (days) and 2) subsequently changes in parasitaemia every other day for 60 days. We achieved these using a combination of tail snip, microcopy and parasitaemia scoring we have as detailed above. We recorded PCV weekly using micro-haematocrit method (McInroy, 1954) where we collected blood from the tail vein into heparinized capillary tubes as previously described (Parasuraman et al., 2010). We also recorded weight and survival of individual mouse weekly and daily respectively. We considered a mouse at extremis (at the point of death) and withdrew it from the study if the PCV declined by 25% or more and/or had high (at least 1 × 109 trypanosomes/ml) terminal parasitaemia for at least two consecutive days (Kamidi et al., 2018). We similarly euthanized mice surviving beyond 60 days post infection as per BioRI-KALRO IACUC guidelines. We categorized survival time for these mice as censored data.
2.7. Parasites isolation and protein preparation
We sequentially 1) propagated the Tbr EATRO-734 or Tbr EATRO-232 isolates we expanded in mice above in rats to raise sufficient parasite numbers, 2) harvested and purified the parasites from the rats 3) isolated and prepared their proteome for subsequent high throughput proteomic analyses. More specifically, we propagated the parasites by ip inoculating three rats (representing three independent replicates) with 1 × 104 of the mice-expanded Tbr EATRO-734 of Tbr EATRO-232 isolate suspended in PSG (pH 8.0). We then harvested the parasites of either strain from the infected rats at their peak parasitaemia (~1.0 × 108 trypanosomes/ml) in the respective rats. We achieved this by euthanizing the rats following the methods as we have described above and immediately drawing their infected blood into syringe containing EDTA (anti-coagulant) using established cardiac puncture technique (Parasuraman et al., 2010). We then purified the harvested parasites from the blood components using blood DEAE-cellulose 52 column chromatography as previously described (Lanham and Godfrey, 1970). We then scored the parasitaemia of the purified parasites using improved Neubauer chamber method (Herbert and Lumsden, 1976), and then pelleted the isolated trypanosomes by centrifugation at 1400g for 10 min at 4 °C. We finally isolated their proteome by first washing the pellet once with PBS and then lysing the parasites in the pellet by bead-overtaxing for 10 min in cell lysis buffer RLT (Qiagen, Hilden, Germany) according to the manufacturer's instructions. We then extracted the proteome from the lysate using AllPrep DNA/RNA/Protein Mini Kit (Qiagen, Hilden, Germany) following manufacturer's instructions. We quantified total protein in respective extracts using Bradford assay (Bio-Rad, Hercules, CA, and Hercules, CA, USA) following the manufacturer's instructions.
We prepared the isolated protein for the high throughput proteomics analysis by first reducing 30 μg of protein from each of the three-independent biological replicates in each isolate with 10 mM tris (2-carboxyethyl) phosphine (TCEP, Sigma-Aldrich, USA) at 55 °C for 1 h. We then alkylated the reduced proteins with 18 mM iodoacetamide (Sigma-Aldrich, St. Louis, MO, USA) for 30 min at room temperature away from light. We subsequently precipitated the alkylated protein in six volumes of pre-chilled (−20 °C) acetone (Sigma-Aldrich, St. Louis, MO, USA) overnight at −20 °C, and then pelleted the protein by centrifuging the suspension at 8000×g for 10 min at 4 °C. We then re-suspended the pellet in 100 μl of 50 mM triethylammonium bicarbonate (Sigma-Aldrich, St. Louis, MO, USA) and subsequently digested the protein by adding trypsin (Sigma-Aldrich, St. Louis, MO, USA) at a trypsin-protein sample ratio of 1:10. We then incubated the trypsin-protein mixture overnight at 37 °C with shaking. We subsequently labelled the resulting peptides using the Tandem Mass Tag™ (TMT™) 10-plex mass tag kit (Thermo Scientific, Waltham, MA USA) using 6 channels (TMT10-126, TMT10-127C, TMT10-128C, TMT10-129C, TMT10-130 N and TMT10-131) according to manufacturer's instructions. We then combined the labelled peptides into a single pool and desalted the peptides using P10 C18 pipette ZipTips (Millipore, Burlington, MA, USA) according to the manufacturer's instructions. We eluted resultant peptides and then dried them using Speedvac concentrator (Thermo Scientific, Waltham, MA, USA). We then re-suspended the dried peptides in 15 μl loading solvent (97.05% H2O, 2% acetonitrile, 0.05% formic acid) for mass spectrometry analysis.
2.8. Mass spectrometry (MS) analysis of the proteome
We employed MS analytical technique to measure the mass-to-charge ratio of the individual peptides to help separate the peptides into a mass spectrum aid in identification if the individual peptides. We achieved this by first loading 8 μl of the peptides per replicate on to a 75 μm × 2 cm C18 trap column (Thermo Scientific) and separated on a 75 μm × 25 cm C18 reverse-phase analytical column (Thermo Scientific, Waltham, MA, USA) heated at 40 °C, using Dionex Ultimate 3000 nano-flow ultra-high-pressure liquid chromatography system (Thermo Scientific, Waltham, MA, USA). We then eluted peptides with mobile phase consisting of 80% acetonitrile and 0.1% formic acid at a gradient of 4–30% over 180 min at a flow rate of 0.25 μl/min. We measured the mass of the eluted peptides using a Q Exactive Orbitrap MS (Thermo Scientific, Waltham, MA, USA). The MS was coupled to the chromatography system via a nano-electrospray ion source (Thermo Scientific, Waltham, MA, USA). In this process, our ms^1 settings consisted of a resolution of 70,000, AGC target of 3e6, maximum IT of 120 ms and scan range of 400–1800 m/z. Our ms^2 settings consisted of a resolution of 17,000, AGC target of 5e4, maximum IT of 120 ms and isolation window of 1.6 m/z. We obtained the MS data by data dependent acquisition. In this procedure, we selected our top 12 most intense precursor ions in positive mode for ms^2 HCD fragmentation, which we subsequently excluded for the next 45 s following fragmentation event. We set charge exclusion to ignore peptide spectrum matches that were 1) unassigned, 2) singly charged and 3) those with ≥ +8 charges (Mirzaei et al., 2017).
2.9. Protein identification
We identified putative function(s) of the respective peptides by searching for their match in among the Annotated Tbb genes. To achieve this, we first processed our raw MS peptides in MaxQuant version 1.6.0.1 (Cox and Mann, 2008), and searched the output Andromeda search engine against TriTrypDB-9.0_TbruceiTREU927_ Annotated Proteins FASTA data. We downloaded this data from TriTrypDB (Aslett et al., 2010) on August 1, 2017. In this process, we set cysteine carbamidomethylation and TMT-10plex labelled N-terminus and lysine as a fixed modification. We also set N-terminal acetylation and methionine oxidations as variable modifications. We used both types of modifications for protein quantification. We set False Discovery Rate (FDR) to 0.01 for both proteins and peptide-spectrum matches. We determined the FDR by searching a FASTA protein database-comprising target and reversed target sequences (decoy) derived from Tbb, by switching the amino-carboxyl orientation of amino acids of protein to generate random sequences. We set enzyme specificity as C-terminal to arginine and lysine with trypsin as the protease. We allowed a maximum of two missed cleavages in the database search. We performed peptide identification with initial precursor mass deviation of up to 7 ppm and fragment mass deviation of up to 20 ppm. We only accepted peptides that were at least seven amino acids long and at most 4600 Da. We specified TmT10plex under isobaric labels for reporter ion MS^2 and set reporter mass tolerance at 0.01Da. We extracted the 10plex reporter ion intensity matrix from the Maxquant protein group matrix file and used the information for downstream comparative proteome analyses. We deposited the mass spectrometry proteomics data to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD021560.
2.10. Identification of differentially expressed proteins
We determined differentially expressed peptides between the Tbr EATRO-734 and Tbr EATRO-232 strains by comparing the quantities of peptides from respective strains using differential expression quantification mass spectrometry (DEqMS) software (Zhu et al., 2020). We selected global differentially expressed peptide by adopting a regime that minimizes type I statistical errors by accepting a peptide as differentially expressed only if it had at least 1.5-fold change (FC) and FDR corrected p < 0.05. We considered the FC as a ratio of individual peptide quantities in the Tbr EATRO-734 relative to their counterparts in the Tbr EATRO-232 strains. We graphically represented the differentially expressed peptides between the parasite strains on volcano plot through in R software (R Core Team, 2016). We determined the suramin resistant or sensitive enriched gene ontology (GO) terms from the global differential expression data through the algorithms in TriTrypDB (Aslett et al., 2010) at 1% p-value (https://tritrypdb.org/tritrypdb/showApplication.do) (Aslett et al., 2010). We summarized the generated GO terms using REVIGO (Supek et al., 2011) at a cut-off (small, 0.53) that gives 99% chance of semantic similarity measure within the cluster.
2.11. Statistical analyses
We evaluated differences in pre-patent periods and effect of trypanosome isolates on peak parasitaemia using t-test with parasite species as factors. We separated significantly different means using Tukey's HSD post hoc analysis (Brillinger, 1984). We established differences in rates of increase in parasitaemia in either isolate following inoculations by comparing effective median times (ET50) for each isolate using Probit analysis (Finney, 1971). We analyzed temporal differences in PCV and body weights caused by the isolates by linear regression analysis of the changes of individual mice against time (days). We assessed the impact of different isolates on survivorship of mice using Kaplan-Meier method to determine survival distribution function with log-rank (Mantel-Cox) and Gehan-Breslow-Wilcoxon test (Machin and Cheung YB, 2006). We conducted all the analyses using GraphPad Prism version 7.00 for Mac (GraphPad Software, La Jolla CA, USA).
3. Results
3.1. Comparative pathogenicity of Tbr EATRO-232 and Tbr EATRO734 in Swiss white mice
We successfully PCR amplified the SRA gene in the two isolates, and the positive control (Fig. S1). The presence of the diagnostic 460bp band of SRA gene (De Greef et al., 1989; De Greef and Hamers, 1994) effectively confirmed the Tbr EATRO-232 and Tbr EATRO-734 isolates as the Tbr subspecies. Our analysis of the phenotypic responses of either isolates to 2.5, 5 or 10 mg/kg) of suramin dosages revealed that all these doses cleared the suramin Tbr EATRO-232 isolate infections in all mice. The Tbr EATRO-734 isolate parasites were cleared in all except mice treated with 2.5 mg/kg of suramin, confirming that the phenotype status of the Tbr EATRO-232 and Tbr EATRO-734 isolates as suramin sensitive and resistance stains respectively to about 2.5 mg/kg of suramin according to established criteria (Kagira and Maina, 2007). Having confirmed the phenotype status of the two isolates, we will thus hereinafter refer to Tbr EATRO-232 and Tbr EATRO-734 isolates as suramin sensitive and resistant strains respectively.
When we evaluated the prepatent period of these isolates in murine model, our results showed no significant differences in the prepatent period (days) between suramin sensitive (5.5 ± 0.167) and resistant (5.9 ± 0.277) strains (two-tailed t-test, t DF = 18 = 1.238, P = 0.2317) (Fig. S2A). Our analyses of the parasitaemia profile in mice revealed similar parasitaemia profile between the two isolates/strains. Our median time (in days) for parasitaemia to reach 50% of the peak parasitaemia (LT50) for suramin resistant and sensitive strains were 5.21 ± 0.23 (CI 2.04–14.27) and 4.10 ± 0.26 (CI 1.33–12.72) days respectively (Fig. S2B).
Our survival analyses of mice infected with either of the isolates revealed that mice infected with suramin sensitive strain survived marginally longer (25.4 ± 1.118 days) than those infected with suramin resistant strain (23.9 ± 1.84 days) (two-tailed t-test, t df = 18 = 0.697, P = 0.4949) (Fig. S2C). Our Kaplan–Meier analysis of survivorship of mice cohorts infected by either parasite isolate revealed similar mortality/survivorship pattern between the cohorts (Log rank P = 0.8 and Wilcoxon P = 0.5) (Fig. S2C). Our regression analyses of changes in weight revealed significant difference between control and infected groups, where the control group gained weight faster than mice infected with suramin sensitive or resistant strains (Table 2, Fig. S2D). Our analysis of PCV in the control (uninfected) mice show that the mice generally maintained their pre-infection PCV levels throughout the 60 days. We however, observed significant decrease in PCV levels in the infected groups compared to the control (Fig. S2E).
Table 2.
Linear regression analysis for changes in body weight of individual mice against time (days).
| Treatment | Slope (β ± SE) | 95% CI | χ2 | Slope deviation from Zero |
|---|---|---|---|---|
| Control | 0.075 ± 0.007 | 0.061–0.089 | 0.502 | F1,16 = 133.9, P < 0.001 |
| Tbr EATRO 232 | −0.032 ± 0.032 | −0.107–0.043 | 0.860 | F1,7 = 1.045, P < 0.341 |
| Tbr EATRO 734 | −0.175 ± 0.051 | −0.296–−0.054 | 1.391 | F1,7 = 11.62, P < 0.011 |
Tbr –T. b. rhodesiense.
3.2. Differential proteome analysis of Tbr EATRO-232 and Tbr EATRO-734
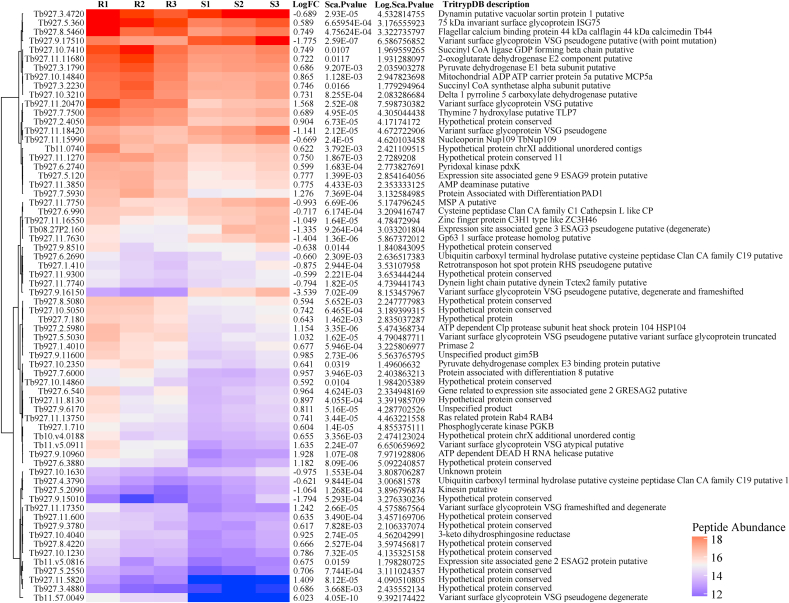
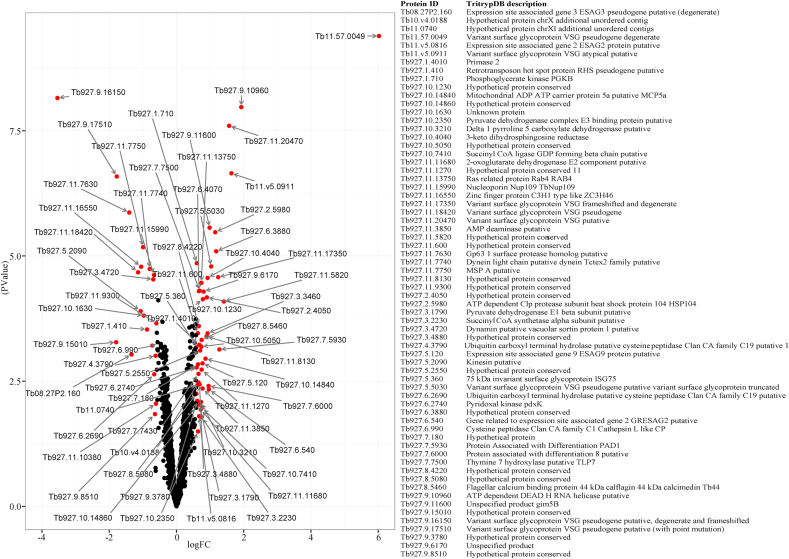
From our proteomic analyses, we identified 67 differentially expressed proteins between suramin-resistant and sensitive isolates/strains, most of which (71.6%) were significantly abundant in the suramin resistant isolate (Fig. 1a, Fig. 1ba and b). Among the significantly abundant proteins in the suramin resistant isolate included six variant surface glycoproteins (VSGs), two proteins associated with differentiation (PADs; PAD1 and PAD8), two expression site associated genes (ESAGs, 2 and 9), heat shock protein 104, several mitochondrial related proteins and 17 hypothetical proteins (Fig. 1a, Fig. 1ba and b). The ISG75 associated with suramin metabolism by acting as the drug receptor (Alsford et al., 2012) was also up regulated in the suramin resistant strain. On the other hand, some of the proteins significantly abundant in the suramin sensitive isolate relative to suramin resitant isolate included the following; lysosomal protease Cathepsin L, two ubiquitin carboxyl-terminal hydrolase enzymes, three VSGs (designated as pseudo proteins), two major surface metalloprotease (MSPs), kinesin, zinc finger protein and three uncharacterized hypothetical proteins (Fig. 1a, Fig. 1ba and b).
Fig. 1a.
Heatmap of differentially expressed proteins between T. brucei rhodesiense EATRO 734 and T. brucei rhodesiense EATRO 232 isolates. R1, R2, R3 and S1, S2, S3 are replicates for the suramin resistant (EATRO-734 and sensitive (EATRO-232) isolates respectively. Red – Highly expressed proteins and Blue – Lowly expressed proteins.
Fig. 1b.
A Volcano plot showing differentially expressed proteins between T. brucei rhodesiense EATRO 734 and T. brucei rhodesiense EATRO 232 isolates. Red dots indicate differentially expressed proteins with an FC of ≥1.5 and false detection rate (FDR) corrected P-value of <0.05 between the isolates. The x-axis displays magnitude of fold-changes and y-axis the statistical significance (-log10 of P-value). Points having FC of <1.5 on an FDR corrected P-value of <0.05 are shown in black, and indicate proteins with non-significance change between different developmental states.
We obtained a global snapshot of molecular mechanisms that underlie the resistant and sensitive phenotypes of the two parasite isolates (Table 3) after subjecting differentially expressed proteins between the two isolates to gene ontology (GO) analyses. From these analyses, cell and cellular components that included those involved in detoxification or oxidation (peroxisome) and energy (pyruvate dehydrogenase complex and proton-transporting two sector ATPase complex) were enriched in the suramin resistant isolate. We did not identify enriched cell and cellular components in the suramin sensitive isolate. We identified the following enriched molecular functions in the suramin resistant isolate; proteins/enzyme involved in oxidation-reduction processes (3-dehydrosphinganine reductase activity and 3-chloroallyl aldehyde dehydrogenase activity), ATP production (dihydrolipoyllsine-residue-acetyltransferase, S-acyltransferase, and 1-pyrroline-5-carboxylate dehydrogenase activities), pyridoxal kinase activity and catabolism. Our molecular functions enriched in suramin sensitive strain included catabolic enzymatic activities. We identified enrichment of drug metabolism, ion transport and energy production biological processes in the suramin-resistant isolate. We similarly established enrichment of proteins linked to protein degradation and cell adhesion in the suramin sensitive isolates.
Table 3.
Gene ontology enrichment analysis of differentially expressed proteins in Trypanosoma brucei rhodesiense suramin resistant (Tbr EATRO-734) and susceptible (Tbr EATRO-232) isolates.
|
Tbr EATRO-734 (Resistance) Isolate | |||||
|---|---|---|---|---|---|
| GO-Category | Term ID | Term Description | Frequency | log10 p-value | Uniqueness |
| Cellular Component | GO:0005623 | Cell | 53.55% | −2.6461 | 0.972 |
| GO:0016020 | Membrane | 61.59% | −5.1657 | 0.977 | |
| GO:0016469 | Proton-transporting two-sector ATPase complex | 0.66% | −4.6575 | 0.767 | |
| GO:0031975 | Envelope | 2.32% | −4.6648 | 0.826 | |
| GO:0043226 | Organelle | 20.79% | −3.1882 | 0.953 | |
| GO:0044425 | Membrane part | 57.39% | −2.9842 | 0.927 | |
| GO:0031967 | Organelle envelope | 1.26% | −4.6648 | 0.422 | |
| GO:0005886 | Plasma membrane | 10.51% | −2.7293 | 0.782 | |
| GO:0071944 | Cell periphery | 11.58% | −2.3157 | 0.796 | |
| GO:0044444 | Cytoplasmic part | 12.66% | −4.5961 | 0.634 | |
| GO:0044464 | Cell part | 52.39% | −2.6461 | 0.765 | |
| GO:0045254 | Pyruvate dehydrogenase complex | 0.03% | −2.0724 | 0.673 | |
| GO:0005777 | Peroxisome | 0.22% | −2.2605 | 0.568 | |
| GO:0042579 | Microbody | 0.22% | −2.2605 | 0.59 | |
| GO:0005622 | Intracellular | 41.18% | −2.2369 | 0.761 | |
| GO:0005737 | Cytoplasm | 26.02% | −2.7724 | 0.653 | |
| GO:0005739 | Mitochondrion | 2.16% | −3.8191 | 0.505 | |
| GO:0044424 | Intracellular part | 35.65% | −2.2637 | 0.683 | |
| Molecular function | GO:0003824 | Catalytic activity | 65.83% | −2.9153 | 0.939 |
| GO:0008478 | Pyridoxal kinase activity | 0.02% | −2.3725 | 0.725 | |
| GO:0015078 | Hydrogen ion transmembrane transporter activity | 0.93% | −2.0714 | 0.828 | |
| GO:0033293 | Monocarboxylic acid binding | 0.19% | −2.0724 | 0.722 | |
| GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity | 0.02% | −2.3725 | 0.755 | |
| GO:0016417 | S-acyltransferase activity | 0.14% | −2.0724 | 0.572 | |
| GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity | 0.00% | −2.3725 | 0.762 | |
| GO:0047560 | 3-dehydrosphinganine reductase activity | 0.00% | −2.3725 | 0.762 | |
| GO:0004742 | Dihydrolipoyllysine-residue acetyltransferase activity | 0.02% | −2.0724 | 0.5 | |
| Biological processes | GO:0017144 | Drug metabolic process | 0.06% | −3.3703 | 0.883 |
| GO:0034220 | Ion transmembrane transport | 3.53% | −3.2422 | 0.553 | |
| GO:0006091 | Generation of precursor metabolites and energy | 1.94% | −2.6952 | 0.866 | |
| GO:0042823 | Pyridoxal phosphate biosynthetic process | 0.17% | −2.3725 | 0.519 | |
| GO:0044281 | Small molecule metabolic process | 15.14% | −2.4195 | 0.719 | |
| GO:0006562 | Proline catabolic process | 0.03% | −2.3725 | 0.61 | |
| GO:0019752 | Carboxylic acid metabolic process | 8.83% | −2.585 | 0.566 | |
| GO:0006811 | Ion transport | 5.34% | −3.4857 | 0.688 | |
| GO:0006850 | Mitochondrial pyruvate transport | 0.02% | −2.0724 | 0.547 | |
| Tbr EATRO-232 (Susceptible) Isolate | |||||
| GO-Category | Term ID | Term Description | Frequency | log10 p-value | Uniqueness |
| Molecular Function | GO:0003824 | Catalytic activity | 65.83% | −2.9333 | 0.958 |
| GO:0070011 | Peptidase activity, acting on L-amino acid peptides | 3.58% | −5.2555 | 0.247 | |
| GO:0016787 | Hydrolase activity | 22.29% | −6.2877 | 0.816 | |
| GO:0101005 | Ubiquitinyl hydrolase activity | 0.18% | −3.3175 | 0.547 | |
| GO:0003774 | Motor activity | 0.40% | −2.0131 | 0.424 | |
| GO:0008233 | Peptidase activity | 4.05% | −5.1587 | 0.438 | |
| GO:0016817 | Hydrolase activity, acting on acid anhydrides | 7.22% | −2.7046 | 0.412 | |
| GO:0003777 | Microtubule motor activity | 0.22% | −2.0604 | 0.445 | |
| Biological Processes | GO:0007155 | Cell adhesion | 0.54% | −3.157 | 0.696 |
| GO:0016579 | Protein deubiquitination | 0.20% | −3.3629 | 0.323 | |
| GO:0022610 | Biological adhesion | 0.55% | −3.1212 | 0.696 | |
| GO:0000291 | Nuclear-transcribed mRNA catabolic process, exonucleolytic | 0.01% | −2.7771 | 0.593 | |
| GO:0006508 | Proteolysis | 5.22% | −4.9456 | 0.445 | |
| GO:0070647 | Protein modification by small protein conjugation or removal | 0.82% | −2.9914 | 0.378 | |
Collectively, these data suggest that the suramin-resistant isolate can produce energy (mitochondrial ATP via Krebs cycle) and concurrently withstand oxidative stress from their toxic environment.
4. Discussion
There has been no formal report of suramin resistance in clinical HAT. However, emergence of resistance in animal trypanosomiasis (Babokhov et al., 2013) and Tbr lab isolates of human origin (Kibona et al., 2006) have been reported. We utilized Tbr parasite strains previously isolated from suramin responsive and non-responsive HAT patient to establish if potential rare suramin resistance was potentially responsible for the treatment failure. We also sought to establish if this rare resistance could potentially induce differential clinical manifestations. We were finally interested in determining molecular process that potentially underpin the serum resistance/susceptivity phenotypes in these isolates. Our suramin dose-response evaluation of the isolates revealed that the isolate from the patient with treatment failure was not susceptible to about 2.5 mg/kg of suramin that the isolate from the suramin responsive patient was susceptible to. These findings pointed to potential suramin resistance phenotype in the isolate derived from the patient with suramin treatment failure, based on established criteria (Kagira and Maina, 2007). Treatment failure could also be attributed to misdiagnosis of second stage for HAT against which suramin is ineffective, insufficient dosage compliance and a plethora of host specific factors. Further studies can help delineate proportional contributions of these individual factors, including the rare suramin resistance phenotype potentially contribute to the treatment failure.
In our assessment of relationship between the putative rare resistance and potential differential clinical manifestations by the parasite, our results revealed that the resistance phenotype did not affect the pre-patent period and survivorship in the mouse model. However, this phenotype marginally delayed development of parasitaemia, suggesting a reduction in rate of parasite proliferation in the mouse model due to the resistance phenotype. These observations support similar observations in Tbb, a Tbr variant, where suramin resistance reduced rate of Tbb proliferation (parasitaemia) (Wiedemar et al., 2018). The reduced parasitaemia suggest reduced pathogenicity of the resistance phenotype and vice versa, a notion that is supported by previous studies that established direct correlation between parasitaemia and pathogenesis/virulence in T.bb and Trypanasoma congolense (Murray and Morrison, 1979). These observations concur with other studies that suggest trypanosomes that proliferate faster have higher parasitaemia and greater virulence (Turner, 1990). We observed a gradual and significant gain/loss of weight in the control mice than the infected groups with both isolates. Previous studies have shown weight loss as a common feature in trypanosome infected animals (Toth et al., 1994; Nishimura et al., 2001). We potentially attribute the reduction in body weight to decreased food intake in trypanosome infected mice. This suggestion is indicative of potential interference with body weight control by the hypothalamus (Darsaud et al., 2003). Our results also indicated a significant decrease in PCV among infected mice, suggesting a state of anemia in the infected group, which is in agreement with previous studies (Murray and Morrison, 1979; Ndung'u et al., 2008; Sharma et al., 2000).
Global assessment of molecular process potentially underpinning the potential rare suramin resistance revealed significant induction of more proteins in the resistant than susceptible isolate. This enhanced protein production can potentially be linked additional molecular process that prime the parasite to surmount subsequent drug challenge, which typically occur at a biological cost to the parasite. Consequently, the pronounced induction may be due to the molecular process associated with the significant enrichment of GO terms in the suramin resistant strain. These terms were linked to energy production, drug metabolism and detoxification, which potentially tie with the associated phenotype. Mechanism underpinning suramin trypanocidal activity is poorly understood, suramin inhibit activity of cytosolic pyruvate kinase and all seven-glycosome compartmentalized enzymes, including phosphoglycerate kinase that function in glycolysis, selectively interfering with energy production (Michels et al., 2006; Willson et al., 1993). Our findings revealed more abundant expression of phosphoglycerate kinase by suramin resistant relative to the susceptible one, suggesting the potential sustenance of glycolytic pathway by this strain despite the drug pressure.
Besides inhibition of glycolytic process, suramin reduce the overall cellular ATP levels and partially activates mitochondrial Krebs’ cycle (Zoltner et al., 2020). In our analysis, most of the proteins associated with energy in suramin resistant strain are mitochondrial related. These proteins include pyruvate dehydrogenase (PDH) E1 and E3 always present in mitochondrion inner membrane. These two enzymes are part of three enzymes complex that form PDH-complex. The PDH-complex catalyze oxidative decarboxylation of pyruvate to acetyl-CoA and links aerobic glycolysis to Krebs cycle. The 2-oxoglutarate dehydrogenase (α-ketoglutarate dehydrogenase; α-KD), succinyl coenzyme A synthetase (SCoAS) Delta-1-pyrroline-5-carboxylate dehydrogenase Krebs cycle enzymes were significantly abundant in the resistant isolate. ScoAS degrade proline and glutamate to succinate (Weelden et al., 2003). Delta-1-pyrroline-5-carboxylate dehydrogenase converts γ-glutamate semialdehyde into glutamate (Mantilla et al., 2017). These enzymes are essential for energy production in PC trypanosomes (Weelden et al., 2003). The α-KD is also important in BSF trypanosomes growth (Sykes et al., 2015).
Mitochondrion in BSF parasites are typically reduced to simple, tubular, acristate like organelle without respiratory cytochromes and functional Krebs cycle (Matthews, 2005). The α-KD was recently found to be localizes in the glycosome of these parasites (Sykes et al., 2015). In trypanosome, SCoAS function in ATP-generating reaction that converts acetyl-CoA from threonine metabolism to acetate (Rivière et al., 2004). Enhanced expression of Delta-1-pyrroline-5-carboxylate dehydrogenase is an indication of accelerated enzymatic activity. This suggests increased proline catabolic process in the resistant strain, which is a feature of PC metabolism (Mantilla et al., 2017). Another mitochondrion inner membrane carrier protein identified in the resistant Tbr is mitochondrial carrier family protein (TbMCP5). This is an ADP/ATP carrier protein essential for procyclic trypanosome growth, particularly when the parasites depend on proline for energy production (Peña-Diaz et al., 2012). The TbMCP5 enables exchange of ADP into the mitochondrion with ATP, which is released into the cell to provide energy for cell function. This was surprising, as the resistant strain was not subjected to suramin drug pressure during the study and BSF T. brucei are known to predominantly rely on glycolysis for energy while in mammalian host bloodstream (Van Hellemond et al., 2005; Hannaert and Michels, 2003; Creek et al., 2015). However, this factor could have been carried over from the treatment failure regime where the parasite was subjected to suramin selection pressure. Enrichment of mitochondria in the suramin resistant strain suggests ATP production via Krebs cycle using acetate obtained from glucose. That process is essential for trypanosome viability in mammal host (Mazet et al., 2013). Mitochondrion is generally well developed in PC trypanosomes (Matthews, 2005) and is essential for energy metabolism in this parasite stage (Weelden et al., 2003). This suggests that the BSF of this parasite isolate exhibit characteristic of insect stage trypanosomes. Role of mitochondria in ATP generation in suramin resistant BSF requires further investigation.
Enrichment of GO terms associated with drug metabolism and detoxification in the suramin-resistant Tbr EATRO-734 suggest that this parasite can potentially handle toxic xenobiotic substances (like suramin). Enhanced mitochondrial (respiratory) activity can also result in production of deleterious reactive oxygen species in the parasite. Activation of detoxification pathway may thus concomitantly be a protective response mechanism against lethal oxidative stress in trypanosome. Enrichment of GO term, cell and associated components in suramin-resistant strain suggest normal parasite multiplication process in the mammalian host. The enhanced expression primase 2, a mitochondrial protein in the suramin-resistant isolate further suggests that these parasite isolates are undergoing cell division since these protein is involved in DNA replication and cell growth. Previous studies showed that T. brucei genome encode for two mitochondrial primases (PRI1 and PRI2) both of which are essential for trypanosome growth and kinetoplast minicircle DNA replication in these parasites (Hines and Ray, 2010, 2011). Suramin inhibits trypanosome replication (Alsford et al., 2012) by impeding cytokinesis process (Gibson et al., 2002).
Evaluation of individual proteins identified six VSGs to be significantly abundant in the suramin-resistant isolate suggesting aggressive evasion of host immune system (Horn and Duraisingh, 2014; Mugnier et al., 2016; McCulloch et al., 2017). In addition, VSGs are involved in resistance of trypanosome to suramin. A recent in vitro study identified a specific VSG gene, VSGsur to be consistently up regulated in suramin-resistant BSF Tbr suggesting their involvement in drug resistance (Wiedemar et al., 2018). It was later shown that VSGsur causes suramin resistance in Tbr BSF by reducing specific receptor-mediated endocytosis pathways (Wiedemar et al., 2019). However, none of the VSGs abundantly expressed in our suramin resistance in Tbr isolate was related to VSGsur, suggesting that VSGsur may be strain or isolate specific. This begs for further functional studies of VSGs from different trypanosomes isolates/strains beyond immune evasion in conferring parasite resistance to suramin. Heat shock protein 104 (Hsp104) was also significantly enriched in the suramin resistant isolate. The Hsp104 is a hexameric member of AAA + family of ATPases that uses energy from ATP hydrolysis to dissolve disordered protein aggregates (Glover and Lindquist, 1998; Shorter and Lindquist, 2004; Shorter, 2008). The Hsp104 is highly conserved in prokaryotic and eukaryotic organisms (Shorter, 2011; Torrente and Shorter, 2013). The protein is essential for cell viability under challenging conditions when proteins aggregate more readily (Sanchez and Lindquist, 1990; Sanchez et al., 1992). Suramin inhibit ATPase and disaggregase activity of Hsp104 (Torrente et al., 2014), reducing functionality of this protein. Variants of Hsp104 containing mutations in inactivating sensor-1 in the nucleotide-binding domain (NBD) 1 or 2 are more resistant to suramin compared to the wild type (Torrente et al., 2014). The presence of similar mutation in the Hsp104 expressed in our suramin resistant isolate that may confer suramin resistance phenotype in this parasite isolate deserves investigation. On the other hand, cells also induce heat shock proteins when exposed to different environmentally stressful conditions (Fuqua et al., 1994; Miller and Fort, 2018). Enhanced expression of Hsp104 in our study could thus be an indication of effective response of our suramin resistant isolate to lethal oxidative stress.
The overexpression of ISG75 in the suramin resistant compared sensitive isolates suggests that the resistant isolate can to take-up suramin drug through endocytosis. The physicochemical characteristics of suramin does not allow it to gain entry into the trypanosomes by diffusion through the plasma membrane, thus requires an active process for uptake (Alsford et al., 2013b; Zoltner et al., 2016). Previous study via genome-wide screening for loss-of function in an RNAi library of BSF Tbb revealed that the ISG75 plays a critical role in suramin internalization through receptor-mediated endocytosis by acting as a receptor for the drug (Alsford et al., 2012, 2013b; Zoltner et al., 2015, 2016). For suramin to achieve its anti-trypanosome effect upon entry into the parasite, several trypanosome endosomal proteins, lysosomal components are required (Alsford et al., 2012; Zoltner et al., 2015). These proteins were downregulated in the suramin-resistant isolate. These molecules actively degrade ISG75-suramin complex and subsequently release of suramin into the lysosome lumen (Alsford et al., 2012; Quintana et al., 2018; de Koning, 2020). As such, the upregulation of ISG75 in the suramin resistant isolate is an indication that this parasite isolate can sufficiently take up suramin. The ISG75 expression level correlate with suramin accumulation in the parasite (Zoltner et al., 2020). We thus hypothesize that, 1) this parasite isolate may remain resistant to suramin due to inadequate expression of supporting internal lysosomal components required for the downstream effectiveness of drug action or 2) the possibility of the presence of mutation in ISG75 expressed by suramin resistance isolate that interfere with suramin binding efficiency hence lowering drug uptake cannot be underestimated. Finally, our study also showed enhanced expression of two proteins associated with differentiation (PAD1 and PAD8). The PAD1 is highly expressed by stumpy BSF trypanosomes and signal trypanosome differentiation from BSF to PC (Dean et al., 2009). Role of PAD8 is unknown. Expression of PADs is an indication that these parasites undergo normal developmental process.
In the suramin susceptible isolate parasite, our results indicated an enrichment of Gp63-1 and MSP-A major surface metalloproteases. In Leishmania, Gp63 is essential for survival of the parasite in the mammalian hosts by protecting them against complement-mediated lysis (Brittingham et al., 1995). The observed expression of Gp63 in our suramin-sensitive Tbr isolate could be due to normal induction of parasite defense mechanism against the host immune response since our study was done in-vivo. Though MSP-A is highly expressed in the blood stream Tbb (LaCount et al., 2003), its function is still unknown. Important proteins enriched in the suramin sensitive relative to the resistant isolates were lysosomal protease cathepsin L (Tb927.6.960) and two homologs of deubiquitylating (DUB) enzymes (Tb927.9.14470, TbUsp7 and Tb927.11.12240, TbVdu1), and ubiquitin carboxyl-terminal hydrolase (Tb927.4.3790, Tb927.6.2690) previously shown to be critical in suramin sensitive trypanosomes (Alsford et al., 2012; Zoltner et al., 2015). Ubiquitylation stabilizes ISG75 to enable it to appropriately bind and internalize suramin (Leung et al., 2011) for subsequent lysosomal release promoting endosomal targeting and degradation (Zoltner et al., 2015). The TbUsp7 and TbVdu1 enzymes, then act on the ubiquitylated ISG75, mediating the removal of ubiquitin (Zoltner et al., 2015). Two homologs of these enzymes are up regulated in our suramin sensitive isolate. This suggest that these enzymes may perform similar functions as their relatives in vivo. We also observed significant enhanced expression of kinesin (cell motility) and zinc finger (mRNA binding) proteins in suramin sensitive relative to the resistant isolates. These proteins required for normal functioning of the parasite.
5. Conclusion
In conclusion, our results reveal potential role of suramin resistance in the treatment failure of the Tbr HAT patient. The results also indicate the potential role of the putative rare suramin resistance phenotype in reducing the Tbr parasitaemia. We also established that drug resistance initiates physiological changes in the parasite and multiple pathways in the parasite undergo alterations to accommodate the resistant state. The enrichment and overexpression of mitochondrial related proteins and/or enzymes in the suramin resistant isolate BSF indicate that this parasite isolate partially activates and utilizes mitochondrial ATP-generating activity. Over-expressed proteins in suramin resistant isolate are altered by suramin in the PC parasites compared to BSF trypanosomes (Zoltner et al., 2020). As expected, the expression of some of the parasite proteins associated with suramin sensitivity were upregulated in the suramin sensitive isolate. Functional studies are needed on the identified pathways using parasites generated under drug pressure to confirm their role in suramin resistance. This will provide insight on development of efficacious HAT drugs.
Declaration of competing interest
The authors declare that no competing interests exist.
Acknowledgements
We are grateful to Purity Gitonga, Samuel Guya, John Ndichu and Jane Hanya for their help in trypanosome sample preparation and laboratory analysis.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpddr.2021.02.001.
Contributor Information
Rosemary Bateta, Email: batetarw@gmail.com.
Paul O. Mireji, Email: mireji.paul@gmail.com.
Note
Supplementary data associated with this manuscript.
Authors’ contributions
Conceptualization and study design: RB, POM, KN. Sample generation: CNM, KN, Sample processing: CNM, KN, MKR and JKN. Data generation: CNM, RB, KN, MKR and JKN. Data analysis: CNM, RB, MKR, POM, CMM, MOA and EOA. First manuscript draft: CNM, RB, POM and VOA. Manuscript review: MKR, JKN, KN, CMM, MOA, EOA and BNO. Coordination and overseeing of the study: RB and POM. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Animal experiments were carried out in strict accordance with recommendations in Institutional Animal Care and Use Committee (IACUC) of Biotechnology Research Institute of Kenya Agricultural and Livestock Research Organization (BioRI-KALRO), Kikuyu, Kenya (Ref. No. C/BioRI/4/325/II/20).
Availability of data and material
Proteomic data are available via ProteomeXchange with identifier PXD021560 and all other data is presented in the main text and supplementary material.
Funding
This work was supported by the Foundation for the National Institutes of Health (https://fnih.org) grant number U01 AI115648.
Author approval
All authors read and approved the final manuscript to be submitted for publication.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Alsford S., Eckert S., Baker N., Glover L., Sanchez-Flores A., Leung K.F. High-throughput decoding of antitrypanosomal drug efficacy and resistance. Nature. 2012;482(7384):232–236. doi: 10.1038/nature10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsford S., Kelly J.M., Baker N., Horn D. Genetic dissection of drug resistance in trypanosomes. Parasitology. 2013 Oct;140(12):1478–1491. doi: 10.1017/S003118201300022X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsford S., Field M.C., Horn D. Receptor-mediated endocytosis for drug delivery in African trypanosomes: fulfilling Paul Ehrlich's vision of chemotherapy. Trends Parasitol. 2013;29(5):207–212. doi: 10.1016/j.pt.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Antoine-Moussiaux N., Saerens D., Desmecht D. Flow cytometric enumeration of parasitaemia and haematologic changes in trypanosoma-infected mice. Acta Trop. 2008 Aug;107(2):139–144. doi: 10.1016/j.actatropica.2008.05.010. [DOI] [PubMed] [Google Scholar]
- Apted F.I.C. Present status of chemotherapy and chemoprophylaxis of human trypanosomiasis in the Eastern hemisphere. Pharmacol. Ther. 1980;11:391–413. doi: 10.1016/0163-7258(80)90035-2. [DOI] [PubMed] [Google Scholar]
- Aslett M., Aurrecoechea C., Berriman M., Brestelli J., Brunk B.P., Carrington M. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010;38(1):457–462. doi: 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babokhov P., Sanyaolu A.O., Oyibo W.A., Fagbenro-Beyioku A.F., Iriemenam N.C. A current analysis of chemotherapy strategies for the treatment of human African trypanosomiasis. Pathog. Glob. Health. 2013;107(5):242–252. doi: 10.1179/2047773213Y.0000000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer O., Beadell J.S., Gibson W., Caccone A. Phylogeography and taxonomy of Trypanosoma brucei. PLoS Neglected Trop. Dis. 2011;5(2):1–11. doi: 10.1371/journal.pntd.0000961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisser S., N'Siesi F., Lejon V., Preux P., Van Nieuwenhove S., Miaka Mia Bilenge C. Equivalence trial of melarsoprol and nifurtimox monotherapy and combination therapy for the treatment of second‐stage Trypanosoma brucei gambiense sleeping sickness. J. Infect. Dis. 2007 Feb;195(3):322–329. doi: 10.1086/510534. [DOI] [PubMed] [Google Scholar]
- Brillinger D. The Collected Works of John W. Tukey: Philosophy and Principles of Data Analysis 1965-1986 Volume IV - CRC. Press Book; 1984. The collected works of John W. Tukey”. [Google Scholar]
- Brittingham A., Morrison C.1., McMaster W.R., McGwire B.S., Chang K.-P., Mosse D.M. Role of the HRM role of the HRM. J. Immunol. 1995;1(7):1–35. [PubMed] [Google Scholar]
- Brun R., Blum J., Chappuis F., Burri C. Human african trypanosomiasis. Lancet. 2010;375(9709):148–159. doi: 10.1016/S0140-6736(09)60829-1. [DOI] [PubMed] [Google Scholar]
- Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008 Dec;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Creek D.J., Mazet M., Achcar F., Anderson J., Kim D.H., Kamour R. Probing the metabolic network in bloodstream-form Trypanosoma brucei using untargeted metabolomics with stable isotope labelled glucose. PLoS Pathog. 2015;11(3):1–25. doi: 10.1371/journal.ppat.1004689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsaud A., Bourdon L., Chevrier C., Keita M., Bouteille B., Queyroy A. Clinical follow-up in the rat experimental model of African trypanosomiasis. Exp. Biol. Med. 2003 Dec;228(11):1355–1362. doi: 10.1177/153537020322801114. [DOI] [PubMed] [Google Scholar]
- Dean S., Marchetti R., Kirk K., Matthews K.R. A surface transporter family conveys the trypanosome differentiation signal. Nature. 2009;459(7244):213–217. doi: 10.1038/nature07997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisler M.C., Brandt J., Bauer B., Clausen P.H., Delespaux V., Holmes P.H. Standardised tests in mice and cattle for the detection of drug resistance in tsetse-transmitted trypanosomes of African domestic cattle. Vet. Parasitol. 2001;97(3):171–183. doi: 10.1016/s0304-4017(01)00415-0. [DOI] [PubMed] [Google Scholar]
- Finney D.J. Cambridge University Press; London: 1971. Probit Analysis. [Google Scholar]
- Fuqua S.A.W., Oesterreich S., Hilsenbeck S.G., Von Hoff D.D., Eckardt J., Osborne C.K. Heat shock proteins and drug resistance. Breast Canc. Res. Treat. 1994;32(1):67–71. doi: 10.1007/BF00666207. [DOI] [PubMed] [Google Scholar]
- Gibson W., Backhouse T., Griffiths A. The human serum resistance associated gene is ubiquitous and conserved in Trypanosoma brucei rhodesiense throughout East Africa. Infect. Genet. Evol. 2002 May;1(3):207–214. doi: 10.1016/s1567-1348(02)00028-x. [DOI] [PubMed] [Google Scholar]
- Glover J.R., Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94(1):73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- De Greef C., Hamers R. The serum resistance-associated (SRA) gene of Trypanosoma brucei rhodesiense encodes a variant surface glycoprotein-like protein. Mol. Biochem. Parasitol. 1994 Dec;68(2):277–284. doi: 10.1016/0166-6851(94)90172-4. [DOI] [PubMed] [Google Scholar]
- De Greef C., Imberechts H., Matthyssens G., Van Meirvenne N., Hamers R. A gene expressed only in serum-resistant variants of Trypanosoma brucei rhodesiense. Mol. Biochem. Parasitol. 1989 Sep;36(2):169–176. doi: 10.1016/0166-6851(89)90189-8. [DOI] [PubMed] [Google Scholar]
- Hannaert Bringaud, Michels Kinetoplastid biology and disease. Kinetoplastid Biol. Dis. 2003;11(2):1–10. doi: 10.1186/1475-9292-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hellemond J.J., Bakker B.M., Tielens A.G.M. vol. 50. Elsevier Masson SAS; 2005. Energy metabolism and its compartmentation in Trypanosoma brucei; pp. 199–226. (Advances in Microbial Physiology). [DOI] [PubMed] [Google Scholar]
- Herbert W.J., Lumsden W.H.R. Trypanosoma brucei: a rapid “matching” method for estimating the host's parasitemia. Exp. Parasitol. 1976;40(3):427–431. doi: 10.1016/0014-4894(76)90110-7. [DOI] [PubMed] [Google Scholar]
- Hines J.C., Ray D.S. A mitochondrial DNA primase is essential for cell growth and kinetoplast DNA replication in Trypanosoma brucei. Mol. Cell Biol. 2010;30(6):1319–1328. doi: 10.1128/MCB.01231-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines J.C., Ray D.S. A second mitochondrial DNA primase is essential for cell growth and kinetoplast minicircle DNA replication in Trypanosoma brucei. Eukaryot. Cell. 2011;10(3):445–454. doi: 10.1128/EC.00308-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn D., Duraisingh M.T. Antiparasitic chemotherapy: from genomes to mechanisms. Annu. Rev. Pharmacol. Toxicol. 2014;54(1):71–94. doi: 10.1146/annurev-pharmtox-011613-135915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs R.T., Plattner J.J., Nare B., Wring S.A., Chen D., Freund Y. Benzoxaboroles: a new class of potential drugs for human African trypanosomiasis. Future Med. Chem. 2011 Aug;3(10):1259–1278. doi: 10.4155/fmc.11.80. [DOI] [PubMed] [Google Scholar]
- Kagira J.M., Maina N. Occurrence of multiple drug resistance in Trypanosoma brucei rhodesiense isolated from sleeping sickness patients. Onderstepoort J. Vet. Res. 2007 Mar;74(1):17–22. doi: 10.4102/ojvr.v74i1.135. [DOI] [PubMed] [Google Scholar]
- Kagira J.M., Maina N.W., Thuita J.K., Ngotho M., Hau J. Influence of cyclophosphamide on the haematological profile of laboratory bred African soft-furred rats (Mastomys natalensis) Scand. J. Lab. Anim. Sci. 2005;32(3):153–158. [Google Scholar]
- Kamidi C.M., Auma J., Mireji P.O., Ndungu K., Bateta R., Kurgat R. Differential virulence of camel Trypanosoma evansi isolates in mice. Parasitology. 2018;145(9):1235–1242. doi: 10.1017/S0031182017002359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibona S.N., Matemba L., Kaboya J.S., Lubega G.W. Drug-resistance of Trypanosoma b . rhodesiense isolates from Tanzania. Trop. Med. Int. Health. 2006;11(2):144–155. doi: 10.1111/j.1365-3156.2005.01545.x. [DOI] [PubMed] [Google Scholar]
- de Koning H.P. The drugs of sleeping sickness: their mechanisms of action and resistance, and a brief history. Trav. Med. Infect. Dis. 2020;5(1):1–23. doi: 10.3390/tropicalmed5010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCount D.J., Gruszynski A.E., Grandgenett P.M., Bangs J.D., Donelson J.E. Expression and function of the Trypanosoma brucei major surface protease (GP63) genes. J. Biol. Chem. 2003;278(27):24658–24664. doi: 10.1074/jbc.M301451200. [DOI] [PubMed] [Google Scholar]
- Lanham S.M., Godfrey D.G. Isolation of salivarian trypanosomes from man and other mammals using DEAE-cellulose. Exp. Parasitol. 1970 Dec;28(3):521–534. doi: 10.1016/0014-4894(70)90120-7. [DOI] [PubMed] [Google Scholar]
- Leung K.F., Riley F.S., Carrington M., Field M.C. Ubiquitylation and developmental regulation of invariant surface protein expression in trypanosomes. Eukaryot. Cell. 2011;10(7):916–931. doi: 10.1128/EC.05012-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macedo J.P., Currier R.B., Wirdnam C., Horn D., Alsford S., Rentsch D. Ornithine uptake and the modulation of drug sensitivity in Trypanosoma brucei. Faseb. J. 2017;31(15):1–12. doi: 10.1096/fj.201700311R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machin D., Cheung Yb P.M. second ed. John Wiley & Sons, Ltd; West Sus sex: 2006. Survival Analysis – A Practical Approach; p. 278. [Google Scholar]
- Mantilla B.S., Marchese L., Dyer N.A., Casas-sa A., Lehane M.J., Ejeh N. Proline metabolism is essential for Trypanosoma brucei brucei survival in the tsetse vector. PLoS Pathog. 2017;13(1):1–29. doi: 10.1371/journal.ppat.1006158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K.R. The developmental cell biology of Trypanosoma brucei. J. Cell Sci. 2005;118:283–290. doi: 10.1242/jcs.01649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazet M., Morand P., Biran M., Bouyssou G., Courtois P., Daulouède S. Revisiting the central metabolism of the bloodstream forms of Trypanosoma brucei: production of acetate in the mitochondrion is essential for parasite viability. PLoS Neglected Trop. Dis. 2013;7(12) doi: 10.1371/journal.pntd.0002587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch R., Cobbold C.A., Figueiredo L., Jackson A., Morrison L.J., Mugnier M.R. Emerging challenges in understanding trypanosome antigenic variation. Emerg Top Life Sci. 2017;1(6):585–592. doi: 10.1042/ETLS20170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInroy R.A. A micro-haematocrit for determining the packed cell volume and haemoglobin concentration on capillary blood. J. Clin. Pathol. 1954 Feb;7(1):32–36. doi: 10.1136/jcp.7.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meirvenne N. Progress in Human African Trypanosomiasis, Sleeping Sickness. Springer Paris; Paris: 1999. Biological diagnosis of human African trypanosomiasis; pp. 235–252. [Google Scholar]
- Michels P.A.M., Bringaud F., Herman M., Hannaert V. Metabolic functions of glycosomes in trypanosomatids. Biochim. Biophys. Acta Mol. Cell Res. 2006;1763(12):1463–1477. doi: 10.1016/j.bbamcr.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Miller D.J., Fort P.E. Heat shock proteins regulatory role in neurodevelopment. Front. Neurosci. 2018;12(821):1–15. doi: 10.3389/fnins.2018.00821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaei M., Gupta V.B., Chick J.M., Greco T.M., Wu Y., Chitranshi N. Age-related neurodegenerative disease associated pathways identified in retinal and vitreous proteome from human glaucoma eyes. Sci. Rep. 2017;7(1):12685. doi: 10.1038/s41598-017-12858-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugnier M.R., Stebbins C.E., Papavasiliou F.N. Masters of disguise: antigenic variation and the VSG coat in Trypanosoma brucei. PLoS Pathog. 2016;12(9):1–6. doi: 10.1371/journal.ppat.1005784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murilla G.A., Ndung’u K., Thuita J.K., Gitonga P.K., Kahiga D.T., Auma J.E. Kenya trypanosomiasis research Institute cryobank for human and animal trypanosome isolates to support research: opportunities and challenges. Tschudi C, editor. PLoS Neglected Trop. Dis. 2014 May;8(5) doi: 10.1371/journal.pntd.0002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M., Morrison W.I. Non-specific induction of increased resistance in mice to Trypanosoma congolense and Trypanosoma brucei by immunostimulants. Parasitology. 1979 Dec;79(3):349–366. doi: 10.1017/s0031182000053750. [DOI] [PubMed] [Google Scholar]
- Ndung’u K., Ngotho M., Kinyua J., Kagira J., Guya S., Ndung’u J. Pathogenicity of bloodstream and cerebrospinal fluid forms of Trypanosoma brucei rhodesiense in Swiss White Mice. Afr. J. Health Sci. 2008 Nov;15(1):34–41. [Google Scholar]
- Nishimura K., Araki N., Ohnishi Y., Kozaki S. Effects of dietary polyamine deficiency on Trypanosoma gambiense infection in rats. Exp. Parasitol. 2001 Feb;97(2):95–101. doi: 10.1006/expr.2000.4588. [DOI] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2016. A Language and Environment for Statistical Computing. [Google Scholar]
- Parasuraman S., Raveendran R., Kesavan R. Blood sample collection in small laboratory animals. J. Pharmacol. Pharmacother. 2010 Jul;1(2):87–93. doi: 10.4103/0976-500X.72350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Diaz P., Pelosi L., Ebikeme C., Colasante C., Gao F., Bringaud F. Functional characterization of TbMCP5, a conserved and essential ADP/ATP carrier present in the mitochondrion of the human pathogen Trypanosoma brucei. J. Biol. Chem. 2012;287(50):41861–41874. doi: 10.1074/jbc.M112.404699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47(D1):D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlig G., Bernhard S.C., Blum J., Burri C., Mpanya A., Lubaki J.P.F. Efficacy and safety of pafuramidine versus pentamidine maleate for treatment of first stage sleeping sickness in a randomized, comparator-controlled, international phase 3 clinical trial. Franco-paredes C, editor. PLoS Neglected Trop. Dis. 2016 Feb;10(2) doi: 10.1371/journal.pntd.0004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana J.F., Del Pino R.C., Yamada K., Zhang N., Field M.C. Adaptation and therapeutic exploitation of the plasma membrane of African trypanosomes. Genes. 2018;9(7):1–16. doi: 10.3390/genes9070368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renoux G., Renoux M. The effects of sodium diethyldithiocarbamate, azathioprine, cyclophosphamide, or hydrocortisone acetate administered alone or in association for 4 weeks on the immune responses of BALB/C mice. Clin. Immunol. Immunopathol. 1980 Jan;15(1):23–32. doi: 10.1016/0090-1229(80)90017-3. [DOI] [PubMed] [Google Scholar]
- Rivière L., Van Weelden S.W.H., Glass P., Vegh P., Coustou V., Biran M. Acetyl:succinate CoA-transferase in procyclic Trypanosoma brucei. Gene identification and role in carbohydrate metabolism. J. Biol. Chem. 2004;279(44):45337–45346. doi: 10.1074/jbc.M407513200. [DOI] [PubMed] [Google Scholar]
- Rodgers J., Jones A., Gibaud S., Bradley B., McCabe C., Barrett M.P. Melarsoprol cyclodextrin inclusion complexes as promising oral candidates for the treatment of human african trypanosomiasis. Raper J, editor. PLoS Neglected Trop. Dis. 2011 Sep;5(9):e1308. doi: 10.1371/journal.pntd.0001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E., Maniatis T. Cold Spring Harbor Laboratory Press; New York.: 1989. Cloning: A Laboratory Manual. Spring; p. 1989. [Google Scholar]
- Sanchez Y., Lindquist S.L. HSP104 required for induced thermotolerance. Science (80- ) 1990;248(4959):1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- Sanchez Y., Taulien J., Borkovich K.A., Lindquist S. Hsp104 is required for tolerance to many forms of stress. EMBO J. 1992;11(6):2357–2364. doi: 10.1002/j.1460-2075.1992.tb05295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A.G., Tait A., Turner C.M. Characterisation of cloned lines of Trypanosoma brucei expressing stable resistance to MelCy and suramin. Acta Trop. 1996 Feb;60(4):251–262. doi: 10.1016/0001-706x(96)00131-3. [DOI] [PubMed] [Google Scholar]
- Sharma D.K., Chauhan Saxena VK., Agrawal R.D. Haematological changes in experimental trypanosomiasis in Barbari goats. Small Rumin. Res. 2000 Oct;38(2):145–149. [Google Scholar]
- Shorter J. Hsp104: a weapon to combat diverse neurodegenerative disorders. Neurosignals. 2008;16(1):63–74. doi: 10.1159/000109760. [DOI] [PubMed] [Google Scholar]
- Shorter J. The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PloS One. 2011;6(10) doi: 10.1371/journal.pone.0026319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J., Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science (80- ) 2004;304(5678):1793–1797. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- Soll M. Use of ivermectin in laboratory and exotic mammals and in birds. Fish, and Reptiles. 1989:260–286. [Google Scholar]
- Supek F., Bošnjak M., Škunca N., Šmuc T. Revigo summarizes and visualizes long lists of gene ontology terms. PloS One. 2011;6(7) doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes S., Szempruch A., Hajduk S. The Krebs cycle enzyme ␣ -ketoglutarate decarboxylase is an essential glycosomal protein in bloodstream african trypanosomes. Eukaryot. Cell. 2015;14(3):206–215. doi: 10.1128/EC.00214-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J., Baker N., Hutchinson S., Dominicus C., Trenaman A., Glover L. Insights into antitrypanosomal drug mode-of-action from cytology-based profiling. PLoS Neglected Trop. Dis. 2018;12(11) doi: 10.1371/journal.pntd.0006980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrente M.P., Shorter J. The metazoan protein disaggregase and amyloid depolymerase system: Hsp110, Hsp70, Hsp40, and small heat shock proteins. Prion. 2013;7(6):457–463. doi: 10.4161/pri.27531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrente M.P., Castellano L.M., Shorter J. Suramin inhibits Hsp104 ATPase and disaggregase activity. PloS One. 2014;9(10):1–12. doi: 10.1371/journal.pone.0110115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth L.A., Tolley E.A., Broady R., Blakely B., Krueger J.M. Sleep during experimental trypanosomiasis in rabbits. Proc Soc Exp Biol Med. 1994 Feb;205(2):174–181. doi: 10.3181/00379727-205-43694. [DOI] [PubMed] [Google Scholar]
- Trunz B.B., Jędrysiak R., Tweats D., Brun R., Kaiser M., Suwiński J. 1-Aryl-4-nitro-1H-imidazoles, a new promising series for the treatment of human African trypanosomiasis. Eur. J. Med. Chem. 2011 May;46(5):1524–1535. doi: 10.1016/j.ejmech.2011.01.071. [DOI] [PubMed] [Google Scholar]
- Turner C.M. The use of experimental artefacts in African trypanosome research. Parasitol. Today. 1990 Jan;6(1):14–17. doi: 10.1016/0169-4758(90)90385-h. [DOI] [PubMed] [Google Scholar]
- Vansterkenburg E.L.M., Coppens I., Wilting J., Bos O.J.M., Fischer M.J.E., Janssen L.H.M. The uptake of the trypanocidal drug suramin in combination with low-density lipoproteins by Trypanosoma brucei and its possible mode of action. Acta Trop. 1993;54:237–250. doi: 10.1016/0001-706x(93)90096-t. [DOI] [PubMed] [Google Scholar]
- Vodnala S.K., Ferella M., Lundén-Miguel H., Betha E., van Reet N., Amin D.N. Preclinical assessment of the treatment of second-stage african trypanosomiasis with cordycepin and deoxycoformycin. Ndung'u J, editor. PLoS Neglected Trop. Dis. 2009 Aug;3(8):e495. doi: 10.1371/journal.pntd.0000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.C. Molecular mechanisms and therapeutic treatment of African trypanosomiasis. Annu. Rev. Pharmacol. Toxicol. 1995;35:93–127. doi: 10.1146/annurev.pa.35.040195.000521. [DOI] [PubMed] [Google Scholar]
- Weelden SWH Van, Fast B., Vogt A., Meer Van Der, Saas J., Hellemond JJ Van. Procyclic trypanosoma brucei do not use Krebs cycle activity for energy generation *. J. Biol. Chem. 2003;278(15):12854–12863. doi: 10.1074/jbc.M213190200. [DOI] [PubMed] [Google Scholar]
- Wiedemar N., Graf F.E., Zwyer M., Ndomba E., Kunz Renggli C., Cal M. Beyond immune escape: a variant surface glycoprotein causes suramin resistance in Trypanosoma brucei. Mol. Microbiol. 2018 Jan;107(1):57–67. doi: 10.1111/mmi.13854. [DOI] [PubMed] [Google Scholar]
- Wiedemar N., Zwyer M., Zoltner M., Cal M., Field M.C., Mäser P. Expression of a specific variant surface glycoprotein has a major impact on suramin sensitivity and endocytosis in Trypanosoma brucei. FASEB BioAdvances. 2019;1(10):595–608. doi: 10.1096/fba.2019-00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willson M., Callens M., Kuntz D.A., Perié J., Opperdoes F.R. Synthesis and activity of inhibitors highly specific for the glycolytic enzymes from Trypanosoma brucei. Mol. Biochem. Parasitol. 1993;59(2):201–210. doi: 10.1016/0166-6851(93)90218-m. [DOI] [PubMed] [Google Scholar]
- Xong H Van, Vanhamme L., Chamekh M., Chimfwembe C.E., Abbeele J Van Den, Pays A. A VSG expression site – associated gene confers resistance to human serum in Trypanosoma rhodesiense prince leopold Institute of tropical medicine. Cell. 1998;95:839–846. doi: 10.1016/s0092-8674(00)81706-7. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Orre L.M., Zhou Tran Y., Mermelekas G., Johansson H.J., Malyutina A. DEqMS: a method for accurate variance estimation in differential protein expression analysis. Mol. Cell. Proteomics. 2020;19:1047–1057. doi: 10.1074/mcp.TIR119.001646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoltner M., Leung K.F., Alsford S., Horn D., Field M.C. Modulation of the surface proteome through multiple ubiquitylation pathways in african trypanosomes. PLoS Pathog. 2015;11(10):1–26. doi: 10.1371/journal.ppat.1005236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoltner M., Horn D., de Koning H.P., Field M.C. Exploiting the Achilles' heel of membrane trafficking in trypanosomes. Curr. Opin. Microbiol. 2016;34:97–103. doi: 10.1016/j.mib.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoltner M., Campagnaro G.D., Taleva G., Burrell A., Cerone M., Leung K.F. Suramin exposure alters cellular metabolism and mitochondrial energy production in African trypanosomes. J. Biol. Chem. 2020;295(24):8331–8347. doi: 10.1074/jbc.RA120.012355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.