Abstract
Protein metal-occupancy (metalation) in vivo has been elusive. To address this challenge, the available free energies of metals have recently been determined from the responses of metal sensors. Here, we use these free energy values to develop a metalation-calculator which accounts for inter-metal competition and changing metal-availabilities inside cells. We use the calculator to understand the function and mechanism of GTPase CobW, a predicted CoII-chaperone for vitamin B12. Upon binding nucleotide (GTP) and MgII, CobW assembles a high-affinity site that can obtain CoII or ZnII from the intracellular milieu. In idealised cells with sensors at the mid-points of their responses, competition within the cytosol enables CoII to outcompete ZnII for binding CobW. Thus, CoII is the cognate metal. However, after growth in different [CoII], CoII-occupancy ranges from 10 to 97% which matches CobW-dependent B12 synthesis. The calculator also reveals that related GTPases with comparable ZnII affinities to CobW, preferentially acquire ZnII due to their relatively weaker CoII affinities. The calculator is made available here for use with other proteins.
Subject terms: Proteins, Chemical tools, Metals
The connection between metal binding to proteins and availabilities of different metals in cells has been unclear. Here, the authors report a metalation-calculator that takes into account competition between metals and their variable accessibility, and thereby elucidate in vivo metal occupancies of three different proteins, CobW, YeiR and YjiA.
Introduction
Paradoxically, in vitro, most metalloproteins prefer to bind incorrect metals1,2. A non-cognate metal may bind more tightly to the native site or bind by using a subset of the native ligands, by recruiting additional ligand(s) and/or by distorting the geometry of a binding site. Some enzymes are cambialistic and can function with alternative metals3, but more commonly a non-cognate metal inactivates an enzyme4,5. Correct metalation occurs in vivo because cells control the availability of metals to nascent proteins1,6–8. For example, specialised delivery proteins support metal acquisition by about a third of metalloproteins, (which in turn represent about a third of all proteins and about a half of all enzymes)1,8. However, metal delivery proteins do not ultimately solve the challenge of metalation because now the correct metal must somehow partition onto the delivery protein.
For metalloproteins generally, there is a need to relate metal binding to the intracellular availability of metals. Our recent work provides the basis for such contextualisation9. Cells are thought to assist protein metalation by maintaining availabilities to the opposite of the Irving-Williams series with weaker binding metals such as MgII, MnII and FeII highly available and tighter binding metals such as NiII, ZnII and CuI at low availabilities10–12. We have demonstrated this to be correct by determining the sensitivities of the DNA-binding metal-sensing transcriptional regulators of Salmonella enterica serovar Typhimurium (hereafter Salmonella)9. The sensors trigger expression of genes whose products, for example, import metals that are deficient or export those in excess6,13. A collection of thermodynamic parameters were measured for each sensor and used to calculate the (dynamic range of) buffered intracellular metal concentrations to which each sensor is finely tuned to switch gene expression9,14. For the more competitive metals, detection is so sensitive as to suggest that there is no hydrated metal in the cell9,10. Instead, rapid associative metal-exchange can occur between labile ligands in the crowded cytosol and the binding sites of metalloproteins, making it unhelpful to express metal availabilities as concentrations of the (largely irrelevant and negligible) hydrated species: thus, the chemical potentials of the bound available metals were expressed as free energies ΔG9. It is hypothesised that metal-delivery proteins acquire their metals from these exchangeable, buffered pools. By reference to available ΔG values and by assuming an idealised cell in which the sensors are at the mid-points of their dynamic ranges, the correct metal (CoII) was previously predicted to partition to the known chelatase of the anaerobic cobalamin biosynthetic pathway, CbiK9. There is a need to build upon this approach to account for (1) multiple competing metals and (2) non-idealised (conditional) cell cultures in order to understand the actions of putative metal delivery proteins (such as CobW and related GTPases) and to simplify such calculations for general use.
The G3E GTPase superfamily contains two branches of delivery proteins involved in the assembly of NiII centres (HypB, UreG), one for handling the cobalamin cofactor (MeaB), plus a fourth family, COG0523, investigated here15,16. Though ubiquitous, from bacteria to plants and humans, members of COG0523 have been persistently enigmatic16. Gene context and informatics have linked subgroups of the COG0523 family to at least three different metals: these include Nha3 associated with FeIII-requiring nitrile hydratases17–19, various subgroups (including YeiR, ZigA and ZagA) implicated in ZnII metallostasis16,20–24, and CobW associated with the aerobic biosynthesis of cobalamin (vitamin B12) and hence CoII (ref. 25). Metal insertion into the preformed corrin ring in the aerobic pathway for vitamin B12 biosynthesis appears to be irreversible26,27, highlighting the importance of CoII specificity at this step. Disruption of cobW impairs B12 biosynthesis25, and a role in CoII delivery has been suggested28, but not established. The roles of YeiR and YjiA (two homologues of CobW in Salmonella) are undefined, albeit ZnII has been predicted for YeiR16,20, and CoII, NiII and ZnII shown to bind Escherichia coli (E. coli) YjiA in vitro21. The impact of GTP binding on metal binding remains to be tested for COG0523 GTPases.
Vitamin B12 is an essential nutrient for human health but is neither made nor required by plants29. Prokaryotes present in the ruminant microbiome produce B12 and hence meat and dairy products provide a dietary source30. Vitamin B12 supplements are recommended for those on a vegan diet and its biomanufacture is in demand31. E. coli has significant advantages over currently employed production strains32. Native E. coli does not make vitamin B12 but strains containing functional B12 pathways have been created: the most promising of these use genes of the aerobic pathway, primarily from Rhodobacter capsulatus, and produce high levels of metal-free corrinoids33–35. In R. capsulatus, CoII is inserted into the corrin ring by a cobalt chelatase ATPase (CobNST)36, putatively via CobW28. An understanding of CoII-availability inside engineered E. coli strains (referred to hereafter as E. coli*) is required to optimise CoII supply for the B12 pathway, with relevance to biomanufacturing. High B12 production coupled with similarity between the metal sensors of E. coli and Salmonella also make this system tractable for testing metalation in vivo: the metal sensors of Salmonella having been thermodynamically characterised9.
Here we calculate intracellular metalation to discover which metals partition onto three proposed metal delivery proteins (CobW, YeiR and YjiA). This work makes it widely possible to quantify metal occupancy of proteins and other molecules in vivo based on thermodynamic parameters. The cognate metals of proteins can thus be identified where this was uncertain, and the contributions of additional mechanisms that enable metalation (such as molecular interactions or bespoke growth conditions) exposed. We determine metal affinities of CobW, YeiR and YjiA, and calculate their in vivo metal occupancies (in Salmonella and closely related species), establishing that CobW cannot acquire CoII from the intracellular milieu in the absence of MgIIGTP and revealing ZnII as the preferred metal for MgIIGTP-YeiR and MgIIGTP-YjiA. Predictions of CoII occupancy of MgIIGTP-CobW in CoII-supplemented media are reflected in CobW-dependent production of B12 in E. coli*, establishing the function of CobW in CoII-supply for B12 and further validating an easy-to-use metalation calculator.
Results
Guanine nucleotides create two metal sites in CobW
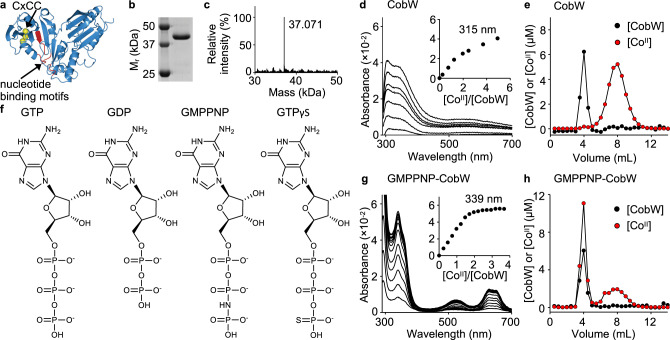
The first objective was to measure the CoII affinities of the form of CobW that acquires metal inside a cell. A modelled structure of CobW (Fig. 1a) showed hypothetical nucleotide-binding sequences adjacent to a putative metal-binding motif, CxCC, and both of these features are conserved in the COG0523 subfamily15,16. To assess the effect of nucleotides on metal binding, CobW was overexpressed and purified (Fig. 1b and Supplementary Fig. 1). The protein mass determined by electrospray ionisation mass spectrometry (ESI-MS) (37,071 Da; Fig. 1c) is consistent with that expected for CobW after cleavage of the N-terminal methionine (37,072.6 Da).
Fig. 1. CoII binding to CobW is enhanced by guanine nucleotides.
a Homology model of CobW (generated with SWISS-MODEL67 using E. coli YjiA PDB entry 1NIJ68 as template; image generated using CCP4 Molecular Graphics software) showing sulfur atoms from conserved CxCC motif (in yellow) and nucleotide-binding (GxxGxGKT, hhhExxG, SKxD*) motifs15,16 (in red). *Ordinarily NKxD but [ST]KxD observed in some COG0523 proteins15. b Purified CobW analysed by SDS-PAGE (full image in Supplementary Fig. 1; n = 1 under these conditions). c ESI-MS analysis (de-convoluted spectra) of purified CobW. d Representative (n = 2) apo-subtracted spectra of CoII-titrated CobW (26.1 µM); feature at 315 nm (inset) shows a non-linear increase. e Representative (n = 2) elution profile following gel-filtration of a mixture of CobW (10 µM) and CoII (30 µM) showing no co-migration of metal (red) with protein (black). Fractions were analysed for protein by Bradford assay and for metal by ICP-MS. f Structures of nucleotides used in this work (generated using ChemDraw software). g As in d for a mixture of CobW (24 µM) and GMPPNP (60 µM); feature at 339 nm (inset) showing a linear increase saturating at 2:1 ratio CoII:CobW (n = 2). h As in e for a mixture of CobW (10 µM), CoII (30 µM) and GMPPNP (30 µM) showing co-migration of 1.8 equivalents CoII per CobW (mean value from peak integration, n = 2 independent experiments). Data replicates are shown in Supplementary Fig. 1.
CoII-titration of CobW alone (26.1 µM) produced a non-linear increase in absorbance at 315 nm (Fig. 1d) but gel-filtration of a mixture of CobW (10 µM) and CoII (30 µM) resulted in their complete separation (Fig. 1e). Taken together, these results suggest only weak interactions between CoII and CobW in the absence of cofactors. In the presence of excess GMPPNP (60 µM), a less readily hydrolysed analogue of GTP (Fig. 1f), CoII-titration of CobW (24 µM) produced an absorbance feature at 339 nm characteristic of ligand-to-metal charge transfer with an extinction coefficient (ε ~2800 M−1 cm−1) indicative of coordination by three cysteine side-chains37 (Fig. 1g). Visible absorbance features (500–700 nm, ε ~300–700 M−1 cm−1) are characteristic of d–d transitions, diagnostic of tetrahedral CoII-coordination geometry (Fig. 1g and Supplementary Fig. 2). Equivalent experiments performed with GTP and an alternate stable analogue, GTPγS, generated indistinguishable spectra (Supplementary Fig. 3a, b). These absorbance features increased linearly, saturating at 2:1 ratio CoII:CobW, and gel filtration of a mixture of CobW (10 µM) and CoII (30 µM) pre-incubated with GMPPNP (30 µM) resulted in co-migration of ~2 equivalents CoII per protein monomer (Fig. 1h). These data show that binding of guanine nucleotides to CobW promotes tight coordination of two metal ions.
Cellular [MgII] generates one CoII site in nucleotide-bound CobW
The uniform absorbance increase observed across both metal-binding events in Fig. 1g could be explained by either the presence of two sequentially filled sites with identical spectroscopic features or two spectrally distinct sites being filled in a pairwise manner. Competition between GMPPNP-CobW and ethylene glycol tetraacetic acid (EGTA) or nitrilotriacetic acid (NTA) for CoII produced sigmoidal binding isotherms indicating positive cooperativity (KD2 < KD1) between the two metal sites (Fig. 2a and Supplementary Fig. 3c, d). Such cooperativity will result in pairwise filling of the two metal sites. Given that GTPases typically bind nucleotides in complex with MgII, we hypothesised that the cognate metal for the first (weak-affinity) site is MgII, and that MgII binding triggers assembly of the second (tight-affinity) metal site in GMPPNP-CobW. CoII titration of CobW (20 µM) with GMPPNP (60 µM) and MgII (2.7 mM, i.e. available idealised intracellular concentration, [MgII]cell9,12) produced identical spectra to that observed without MgII but saturating at 1:1 ratio CoII:CobW (Fig. 2b and Supplementary Fig. 3e). Equivalent experiments performed with GTP and GTPγS also revealed 1:1 CoII:CobW stoichiometry in the presence of [MgII]cell (Supplementary Fig. 3f, g). Thus, binding of MgII and guanine nucleotides preassembles one distinct CoII site in CobW. Occupancy of the first site by MgII was spectroscopically silent in these experiments. The features at 339 nm and at 500–700 nm therefore correspond exclusively to a distinct tetrahedral CoII site and the coordinating sulfhydryl side-chains likely derive (at least in part) from the CxCC motif adjacent to the nucleotide-binding site.
Fig. 2. MgII and the γ-phosphate group of GTP are necessary for high affinity CoII binding.
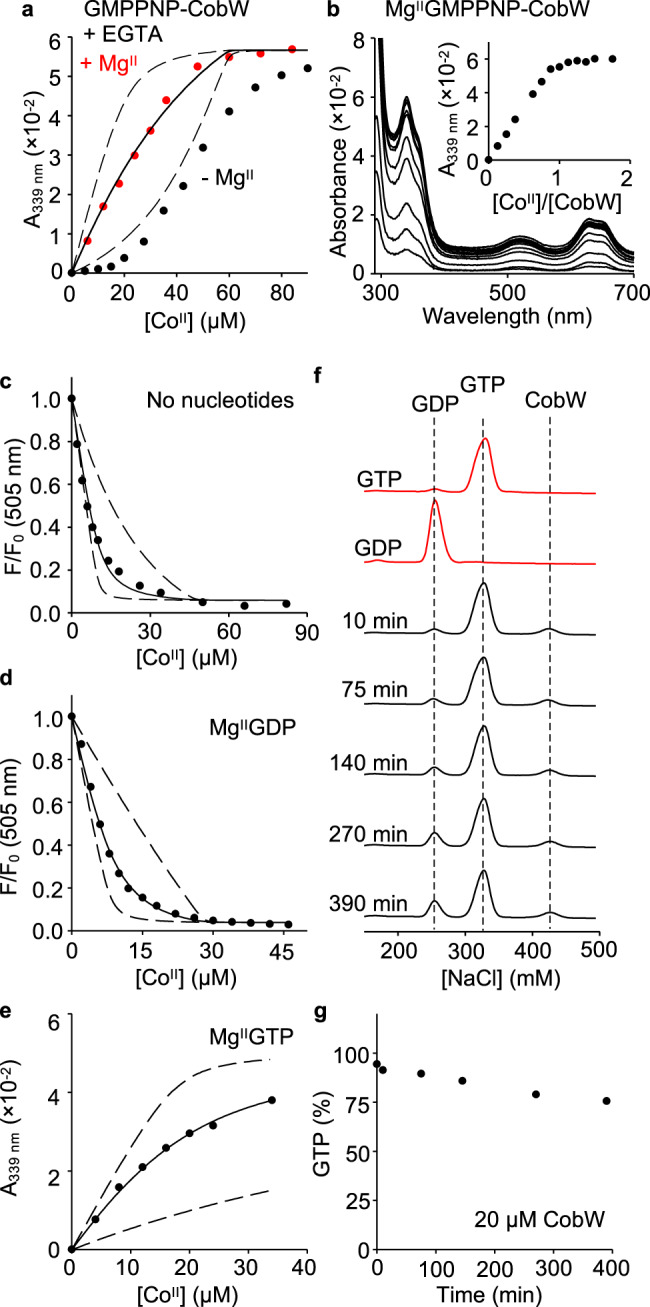
a Absorbance (relative to CoII-free solution) of CoII-titrated CobW (20 µM) with GMPPNP (60 µM) in competition with EGTA (40 µM); titrations in the absence (black) or presence (red) of MgII (2.7 mM, i.e. concentration in a bacterium9,12). Data shown are representative of n = 3 independent experiments (with varying [competitor] and/or identity; see Supplementary Figs. 3c, d and 4a, b). b Absorbance (relative to CoII-free solution) of CoII-titrated CobW (20 µM) with GMPPNP (60 µM) and MgII (2.7 mM) in the absence of competing ligand; feature at 339 nm (inset) showing linear increase saturating at 1:1 ratio CoII:CobW (n = 2; see Supplementary Fig. 3e). c–e Representative KCo(II) quantification for CobW in the absence or presence of nucleotides (n = 3 independent experiments, details in Supplementary Fig. 4 and Supplementary Table 1). c Fluorescence quenching of CoII-titrated fura-2 (10 µM) in competition with CobW alone (37 µM). d Fluorescence quenching of CoII-titrated fura-2 (8.1 µM) in competition with CobW (20 µM) with MgII (2.7 mM) and GDP (200 µM). e Absorbance (relative to CoII-free solution) of CoII-titrated CobW (18 µM) in competition with EGTA (2.0 mM) with MgII (2.7 mM) and GTP (200 µM). Solid traces in a, c, d, e show curve fits of experimental data to a model where CobW binds one molar equivalent CoII per protein monomer. Dashed lines show simulated responses for KCo(II) tenfold tighter or weaker than the fitted value. f Analysis of GTP hydrolysis by anion-exchange chromatography. Control samples of GTP and GDP elute as distinct peaks (red traces) measured by absorbance at 254 nm. Black traces show the extent of hydrolysis of GTP (200 µM) incubated with CobW (20 µM), MgII (2.7 mM) and CoII (18 µM) over time. g Analysis of data from f showing % GTP remaining over time. After 390 min incubation, nucleotides remain primarily (>75 %) unhydrolysed. Equivalent data using 4:1 ratio GTP:CobW is shown in Supplementary Fig. 6.
Due to the tight coordination of CoII to nucleotide-bound forms of CobW (i.e. no measurable dissociation at the micromolar-range protein concentrations required for detection), it was necessary to employ competition assays, whereby CoII is partitioned between the protein and a ligand of well-matched and defined CoII affinity, for reliable quantification of metal-binding affinities38. Competition between MgIIGMPPNP-CobW and EGTA for CoII yielded a binding isotherm consistent with 1:1 stoichiometry for both CoII:protein and CoII:EGTA, and enabled KCo(II) of 2.7 (±0.4) × 10−9 M for MgIIGMPPNP-CobW to be determined (Fig. 2a, Supplementary Fig. 4a, b and Supplementary Tables 1, 2). MgII had negligible impact on the conditional affinity of EGTA for CoII at the concentrations used here (Supplementary Table 3): For this reason, MgII was not incorporated into curve-fitting models. Competition with EGTA revealed a CoII affinity for MgIIGTPγS-CobW (KCo(II) = 1.7 (±0.8) × 10−10 M; Supplementary Fig. 4c–e and Supplementary Tables 1, 2), that was more than tenfold tighter than MgIIGMPPNP-CobW, establishing that the nature of the bound nucleotide exerts an effect on metal binding to CobW.
CoII binds CobW 1000-fold tighter with GTP than GDP
Observed variation in CoII affinities of CobW in association with MgIIGTPγS versus MgIIGMPPNP prompted us to assess the CoII affinities of all three anticipated biological species: nucleotide-free CobW, MgIIGTP-CobW and MgIIGDP-CobW. CoII affinities of CobW and MgIIGDP-CobW were determined via competition with the probe ligand fura-2 (Fig. 2c, d, Supplementary Fig. 4f–i and Supplementary Tables 1, 2), which undergoes fluorescence quenching upon CoII binding39. Fura-2 is too weak to compete effectively with MgIIGTP-CobW (Supplementary Fig. 4j), but high concentrations of EGTA or NTA imposed sufficient competition to enable KCo(II) of 3.0 (±0.8) × 10−11 M to be determined (Fig. 2e, Supplementary Fig. 4k–m and Supplementary Tables 1, 2). GTP concentration was not a limiting factor in these affinity measurements (Supplementary Fig. 5). Under identical conditions used for affinity measurements, we confirmed that CobW-catalysed GTP hydrolysis is sufficiently slow such that nucleotides remain predominantly unhydrolysed over the duration of metal-binding experiments (Fig. 2f, g and Supplementary Fig. 6). MgIIGDP-CobW, despite displaying identical absorbance features indicating the persistence of the cysteine-rich tetrahedral site (Supplementary Fig. 7), has a CoII affinity more than 1000-fold weaker than MgIIGTP-CobW and only marginally tighter than unbound CobW which lacks this site altogether (Supplementary Table 2). GTP also confers higher CoII affinity than either of the tested non-hydrolysable analogues in which the γ-phosphates have been modified (Fig. 1f and Supplementary Table 2). Thus, the presence of an intact nucleotide γ-phosphate is a prerequisite for high-affinity CoII binding.
CuI and ZnII bind MgIIGTP-CobW more tightly than CoII
In view of the challenges associated with correct metal–protein speciation, we sought to determine MgIIGTP-CobW affinities for other first-row transition metals (FeII, NiII, CuI, ZnII). FeII-titration into a mixture of MgIIGTP-CobW (50 µM) and probe ligand 4-(2-thiazolylazo)-resorcinol (Tar) (16 µM) showed FeII being withheld by Tar which revealed a limiting affinity (KFe(II) > 10−6 M) for MgIIGTP-CobW (Fig. 3a, Supplementary Fig. 8 and Supplementary Tables 1, 2). Competition between MgIIGTP-CobW (10 µM) and mag-fura-2 (Mf2; 20 µM) for NiII showed that MgIIGTP-CobW has one NiII site which outcompetes Mf2 (KNi(II) < 10−8 M) in addition to two weaker sites which compete with Mf2 for NiII (KNi(II) ~10−7 M) and are also present in CobW alone (Supplementary Fig. 9a). Competition with Tar allowed the affinity of the tight NiII site in MgIIGTP-CobW to be determined (KNi(II) = 9.8 (±6.5) × 10−10 M; Fig. 3b, Supplementary Fig. 9b, c and Supplementary Tables 1, 2). The conditional β2 value (4.3 (±0.6) × 1015 M−2) for NiIITar2 formation under experimental conditions (pH 7.0, 100 mM NaCl, 400 mM KCl) was independently established by competition with EGTA (Supplementary Fig. 10). Titration of MgIIGTP-CobW (15 µM) and bathocuproine disulfonate (Bcs; 30 µM) with CuI did not reach the expected intensity at saturating metal concentrations (Supplementary Fig. 11a) suggesting the presence of a stable ternary complex, which would preclude accurate affinity determinations40. An equivalent experiment with alternative CuI-probe bicinchoninic acid (Bca) showed that MgIIGTP-CobW has two CuI sites which outcompete Bca and at least three additional weaker CuI sites which effectively compete with the probe (Supplementary Fig. 11b). Effective competition imposed by excess Bca enabled KCu(I) of 2.4 (±0.9) × 10−16 M to be determined (Fig. 3c, Supplementary Figs. 11c, d, 12 and Supplementary Tables 1, 2), assuming only the tightest CuI site can acquire metal at the limiting CuI availabilities employed (e.g. [CuIaq] <3 × 10−16 M in Fig. 3c). ZnII titration into a mixture of quin-2 (10 µM) and MgIIGTP-CobW (10 µM) revealed one high-affinity ZnII site in the protein which was too tight to be quantified by using quin-2 thus showing KZn(II) < 10−12 M (Fig. 3d).
Fig. 3. Binding of MgIIGTP-CobW to FeII, NiII, CuI and ZnII.
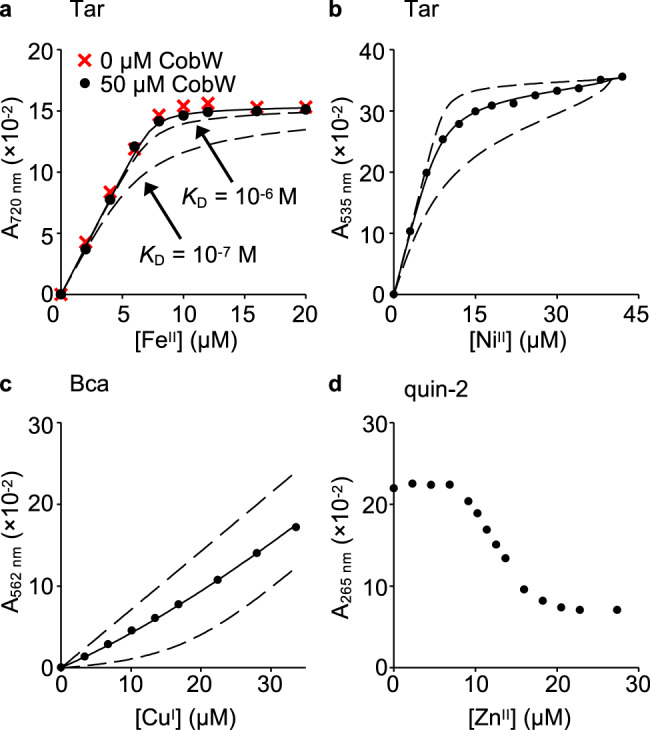
a Absorbance upon FeII titration into a mixture of Tar (16 µM), MgII (2.7 mM) and GTP (500 µM) in the absence (red crosses) or presence (black circles) of CobW (50 µM). Dashed lines show simulated responses for specified KFe(II) of MgIIGTP-CobW, providing limiting KFe(II) ≥ 10−6 M. Control FeII titration into a solution of Tar (16 µM) in buffer only (Supplementary Fig. 8a) confirmed that MgII and GTP did not inhibit stoichiometric FeIITar2 formation. b Absorbance change (relative to NiII-free solution) of NiII-titrated Tar (20 µM) in competition with CobW (30 µM) in the presence of MgII (2.7 mM) and GTP (300 µM). c Absorbance of CuI-titrated Bca (1.0 mM) in competition with CobW (20 µM) in the presence of MgII (2.7 mM) and GTP (200 µM). In a–c, solid traces show curve fits of experimental data to models where CobW binds one molar equivalent of metal per protein monomer. Supplementary Table 2 contains mean ± standard deviation (SD) Kmetal values from n = 3 independent experiments (full details in Supplementary Figs. 8–12 and Supplementary Table 1). In b, c, dashed lines show simulated responses for Kmetal tenfold tighter or weaker than the fitted value. d Absorbance (relative to probe-free solution) upon titration of ZnII into a mixture of quin-2 (10 µM), MgII (2.7 mM), GTP (100 µM) and CobW (10 µM).
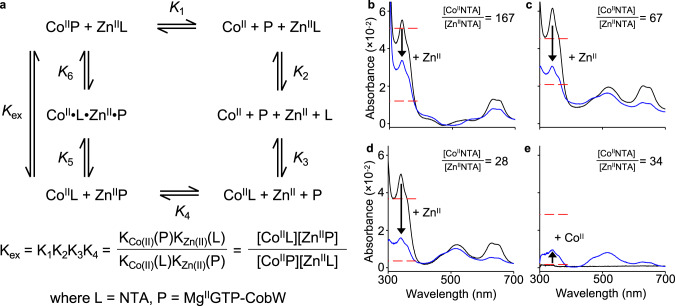
Because of the limiting affinity of quin-2, we employed inter-metal competition, which presumably also occurs within the buffered intracellular milieu, to determine KZn(II) for MgIIGTP-CobW. KZn(II) was determined, relative to the known KCo(II), via competition between the two metals. This approach required an excess of metal ions competing for a limited number of protein metal-sites (i.e. [CoII]tot + [ZnII]tot > [CobW]tot), thus it was essential to include a buffering ligand, in this case NTA, to control the speciation of all CoII and ZnII in solution (i.e. [NTA]tot > [CoII]tot + [ZnII]tot). The measured equilibrium (Kex in Fig. 4a) was the exchange constant for CoII/ZnII exchange between the protein (MgIIGTP-CobW) and buffering ligand (NTA). Equilibrium ratios of [CoIIMgIIGTP-CobW]/[ZnIIMgIIGTP-CobW] were determined (Fig. 4b–e and Supplementary Table 4): absorbance intensity at A339 nm reported specifically on the CoII–protein complex and all remaining protein was ZnII-bound (since MgIIGTP-CobW was metal-saturated under experimental conditions; Supplementary Fig. 13). The concentrations of NTA-bound metals were determined from mass balance relationships (Eqs. (6–8) in “Methods”). Experiments were conducted at multiple relative availabilities of CoII and ZnII and reciprocally (Fig. 4b–e), with consistent results (Supplementary Table 4), to confirm reliability of measured affinities. We thus determined a tight KZn(II) of 1.9 (±0.6) × 10−13 M for MgIIGTP-CobW (Supplementary Table 2).
Fig. 4. MgIIGTP-CobW binds ZnII with sub-picomolar affinity.
a Representation of the equilibrium for exchange of CoII and ZnII between ligand (L = NTA) and protein (P = MgIIGTP-CobW). b–e Absorbance (relative to metal-free solution) of solutions of CobW (17.9–20.4 µM), MgII (2.7 mM), GTP (200 µM) and NTA (0.4–4.0 mM) upon (b–d) first addition of CoII (black trace) then ZnII (blue trace) or (e) the reverse, at equilibrium (n = 1 for each panel). The absorbance peak at 339 nm corresponds to CoII-bound protein. An excess of ligand NTA was used to buffer both metals in each experiment: varying the ratios of ligand-bound metal ions ([CoIINTA]/[ZnIINTA] = 28–167) shifted the ratios of CoII- and ZnII-bound protein as predicted by the equilibrium exchange constant in a. Consistent KZn(II) values for MgIIGTP-CobW were generated at all tested conditions (Supplementary Table 4). Dashed red lines show expected A339 nm peak intensities for KZn(II) of MgIIGTP-CobW tenfold tighter or weaker than calculated values.
GTP not GDP enables CoII acquisition by CobW in cells
In the same manner that Fig. 3 considered competition between a ligand (Tar, Bca or quin-2) and a protein (MgIIGTP-CobW) for metal binding in vitro, metal acquisition by proteins in vivo likewise involves competition with a surplus of cytosolic ligands that buffer metals to different availabilities8,9,14,41,42. Recent work has estimated the buffered availabilities of metals M (where M = MgII, MnII, FeII, CoII, NiII, CuI, ZnII) in a reference bacterium (Salmonella9) expressed here as free energies (ΔG; Fig. 5). The intracellular available ΔG for each metal, ΔGM, is defined as the free energy required for a ligand to become 50% metalated from available and exchangeable intracellular metal (see Supplementary Note 1). Figure 5 and Supplementary Fig. 14 show the intracellular available ΔGM values in an idealised cell (i.e. neither metal deficiency nor excess) defined as the metal availabilities at which the cognate sensors undergoes half of their transcriptional responses. Bars show the changes in available intracellular ΔGM as sensors shift from 10–90% (Fig. 5) or 1–99% (Supplementary Fig. 14) of their dynamic ranges. The percentage occupancy of a protein, P, with metal, M, in vivo is governed by the difference between the free energy for protein metalation, ΔGMP, and the intracellular available ΔGM (Eq. (1)) and can be calculated via Eq. (2) (see Supplementary Note 1):
| 1 |
| 2 |
Fig. 5. MgIIGTP-CobW is predicted to acquire CoII or ZnII in a bacterial cell.
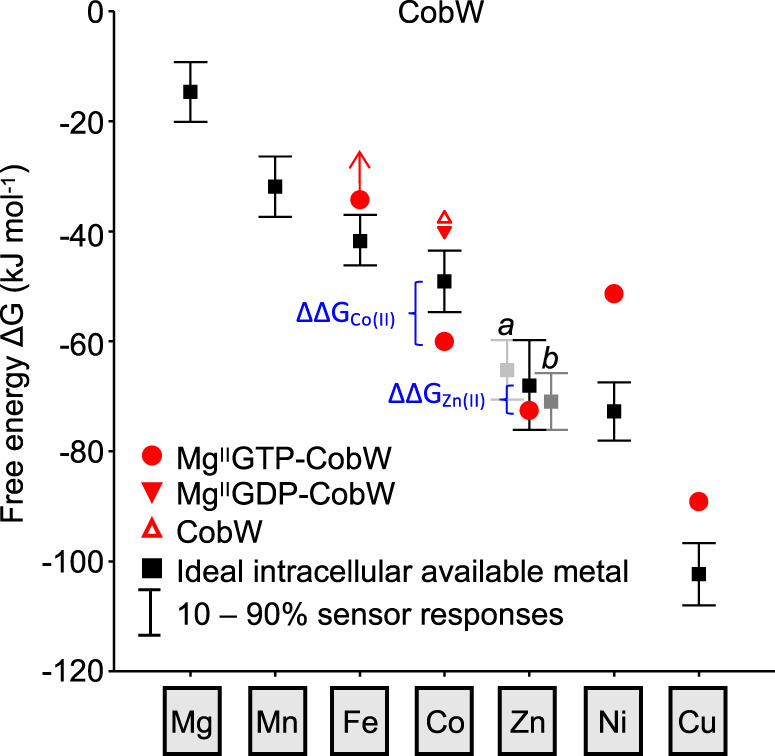
Free-energy change (ΔG) for metal binding to MgIIGTP-CobW (red circles) plotted against the intracellular available free energies for metal binding in a reference bacterial cytosol (values correspond to Salmonella) under idealised conditions (i.e. where each metal sensor undergoes half of its transcriptional response; black squares). Intracellular available ΔGZn(II) is the mean of the values determined from the two ZnII sensors ZntR (a) and Zur (b). Bars show the change in intracellular available ΔG as cognate sensors shifts from 10–90% of their responses. Free energy differences (ΔΔG) which favour acquisition of metals by MgIIGTP-CobW in vivo are indicated in blue. ΔG values for CoII complexes of CobW alone (open red triangle) and MgIIGDP-CobW (closed red triangle) are also shown. For FeII binding to MgIIGTP-CobW, arrow indicates limiting ΔG > −34.2 kJ mol−1.
In an idealised cell, the ΔGCo(II) for CobW and MgIIGDP-CobW were both significantly more positive than intracellular available ΔGCo(II) (ΔΔGCo(II) ≫ 0; Fig. 5) resulting in negligible CoII occupancies of 1.0% and 2.5% for these two protein forms, respectively. Conversely, ΔGCo(II) for MgIIGTP-CobW was significantly more negative than intracellular available ΔGCo(II) (ΔΔGCo(II) ≪ 0), resulting in almost complete protein metalation (99%). Thus, CobW needs MgIIGTP to acquire CoII in a cell.
MgIIGTP-CobW may also acquire ZnII
In addition to CoII, other metals also bound to MgIIGTP-CobW (Figs. 3 and 4). However, ΔΔG for FeII, NiII and CuI was significantly greater than zero (Eq. (1) and Fig. 5), thus preventing acquisition of these metals (Eq. (2) and Table 1). In contrast, ΔΔGZn(II) was <0 with in vivo ZnII occupancy predicted to be 86% (Fig. 5 and Table 1). However, based on Eq. (2), the sum of metal occupancies of MgIIGTP-CobW gave an impossible total metalation >100% (Table 1). Since ΔΔG was <0 for both CoII and ZnII, a more sophisticated approach needs to account for competition between multiple buffered metals in order to predict how much ZnII binds MgIIGTP-CobW in vivo.
Table 1.
Calculated metal occupancies of COG0523 proteins in an idealised cella.
| Metal | MgIIGTP-CobW Eq. 2b | MgIIGTP-CobW Eq. 4c | MgIIGTP-YeiR Eq. 4c | MgIIGTPγS-YjiA Eq. 4c |
|---|---|---|---|---|
| MnII d | <0.8% | <1.8% | ||
| FeII d | <4.6% | <0.1% | <3.0% | < 3.4% |
| CoII | 98.8% | 91.9% | 10.3% | 2.0% |
| ZnII | 86.2% | 6.9% | 24.4% | 22.6% |
| NiII | 0.1% | 0.0% | 0.0% | 0.0% |
| CuI | 0.5% | 0.0% | 0.2% | 0.1% |
| Total | 190.3% | 98.9% | 38.6% | 29.8% |
aBased on metal availabilities in Salmonella under idealised conditions (ref. 9).
bDoes not account for competition between different metals for the same high-affinity site in MgIIGTP-CobW.
cTakes into account competition between multiple intracellular metals for the same site in each protein.
dWhere only limiting metal–protein affinities (strongest KD limit) were determined, calculated occupancy corresponds to a maximum value (denoted by <).
Calculating inter-metal competition in a cell
Figure 4 considered competition between CoII and ZnII for a single metal-binding site in a protein (MgIIGTP-CobW) when the metals were buffered to different availabilities in vitro by an excess of NTA. This can be represented as an available ΔGM (Supplementary Table 4). The observed CoII occupancy was a function of the protein’s affinities for both CoII and ZnII relative to their buffered availabilities in solution (i.e. ΔΔG values), as described by Eq. (3) (see Supplementary Note 1).
| 3 |
By analogy, in a cytoplasm multiple metals, each buffered to different intracellular available ΔGM, compete for a single protein-binding site. We generalised Eq. (3) to account for n different metals (Eq. (4) and Supplementary Note 1).
| 4 |
Thus, we developed a metalation calculator (based on Salmonella, Supplementary Data 1) for determining in vivo metal occupancies of proteins, accounting for multiple inter-metal competitions plus competition from components of the intracellular milieu.
MgIIGTP-CobW selects CoII in idealised (Salmonella) cells
Since ΔΔG was <0 for binding of both CoII and ZnII to MgIIGTP-CobW (Fig. 5), Eq. (4) was next used to predict in vivo metalation in an idealised cell. Between the five metals considered (FeII, CoII, NiII, CuI and ZnII), MgIIGTP-CobW will favour CoII binding in a cell and calculations via Eq. (4) predicted occupancies of 92% and 7%, for CoII and ZnII, respectively (Table 1). Thus, although MgIIGTP-CobW affinities for both CoII and ZnII are tight enough to extract either metal from the cytosolic buffer, CoII will outcompete ZnII, rationalising specificity but only in an intracellular context where there is competition from other cellular components.
Related GE3 GTPase YeiR prefers ZnII in idealised Salmonella
To test the calculator on a second protein, YeiR from Salmonella was overexpressed and purified (Supplementary Fig. 15) in order to determine metal affinities. In view of similarity between YeiR, ZigA22,23 and ZagA24, notably a deduced binding site for ZnII-sensor Zur in the yeiR promoter (Supplementary Fig. 16), occupancy with ZnII might be predicted. Perhaps unexpectedly, MgIIGTP-YeiR showed a similar (slightly weaker) affinity for ZnII relative to MgIIGTP-CobW, the greatest difference in affinity was for CoII (Supplementary Figs. 17–21 and Supplementary Tables 5, 6).
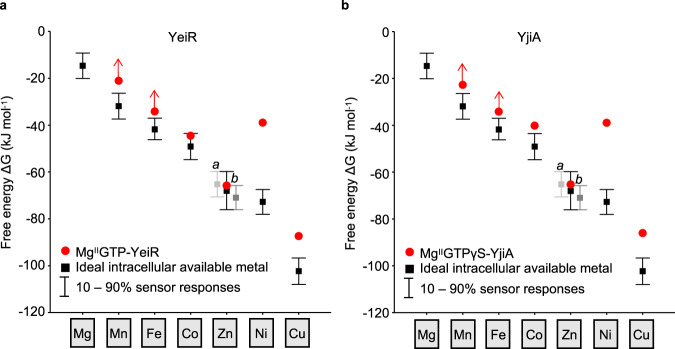
MnII failed to migrate through a gel filtration column with MgIIGTP-YeiR even when the running buffer was supplemented with an additional 20 μM MnCl2, revealing a MnII affinity >2 × 10−4 M (Supplementary Figs. 17a and 18). MgIIGTP-YeiR did not compete with Tar under conditions that imply FeII affinity ≥1 × 10−6 M (Supplementary Fig. 17b). The CoII and NiII affinities of MgIIGTP-YeiR were determined by competition with fura-2 and Mf2, respectively (Supplementary Fig. 17c, d and Supplementary Fig. 19a–d). Data were fit to a 1:1 metal-binding model giving a CoII affinity of 1.5 (±0.7) × 10−8 M and NiII affinity of 1.5 (±0.6) × 10−7 M. Competition for CuI between MgIIGTP-YeiR and Bca identified a CuI affinity of 4.9 (±5.1) × 10−16 M (Supplementary Fig. 17e). Competition with quin-2 was used to determine ZnII affinities of MgIIGTP-YeiR of 3.0 (±1.2) × 10−12 M, and 4.1 (±2.7) × 10−12 M for MgIIGTPγS-YeiR (Supplementary Figs. 17f, 19e–g, 20d and 21h, i). Equation (4) was used to predict in vivo metalation of MgIIGTP-YeiR in an idealised cell. ZnII binding is favoured with calculated occupancies of 24% and 10%, for ZnII and CoII respectively, when sensors are at the mid-points of their dynamic ranges (Fig. 6a and Table 1), and trace amounts of zinc were detected after extensive purification (Supplementary Figs. 22 and 23).
Fig. 6. MgIIGTP-YeiR and MgIIGTP-YjiA preferentially acquire ZnII.
a Free-energy change (ΔG) for metal binding to MgIIGTP-YeiR (red circles) plotted against the intracellular available free energies for metal binding (as described in Fig. 5; black squares and bars). b As a for MgIIGTPγS-YjiA (red circles). Arrows indicate where only a limiting ΔG was determined (thus ΔG > plotted value).
Related GE3 GTPase YjiA prefers ZnII in idealised Salmonella
CoII, NiII and ZnII have all been shown to bind recombinant YjiA in vitro21, and the yjiA promoter contains no deduced recognition sequence for Zur. To test the calculator on a third COG0523 protein, YjiA was overexpressed, purified (Supplementary Fig. 15), and its affinities for metals determined (Supplementary Figs. 24 and 25, Supplementary Tables 5 and 7).
Affinities were determined for MgIIGTPγS-YjiA to avoid nucleotide hydrolysis: this is supported by similar ZnII affinities being measured for MgIIGTP-YeiR and MgIIGTPγS-YeiR (Supplementary Table 5). Mf2 fully outcompeted MgIIGTPγS-YjiA for MnII and simulations indicate a dissociation constant ≥1 × 10−4 M (Supplementary Figs. 24a and 25a–c). MgIIGTPγS-YjiA did not compete with Tar under conditions that imply FeII affinity ≥1 × 10−6 M (Supplementary Figs. 24b and 25d–f). The CoII, NiII and CuI affinities of MgIIGTPγS-YjiA were determined using fura-2, Mf2 and Bca, respectively, as described for YeiR, giving a CoII affinity of 9.1 (±2.0) × 10−8 M, NiII affinity of 1.5 (±0.3) × 10−7 M and CuI affinity of 7.6 (±1.4) × 10−16 M (Supplementary Figs. 24c–e and 25g–l). Competition with quin-2 was used to determine a ZnII affinity for MgIIGTPγS-YjiA of 3.7 (±1.1) × 10−12 M, and this was repeated with MgIIGTP-YjiA generating a near identical value of 3.3 (±2.5) × 10−12 M (Supplementary Figs. 24f and 25m–p). Equation (4) was used to predict in vivo metalation of MgIIGTP-YjiA in an idealised cell. ZnII binding is favoured with occupancies of 23% and 2.0% for ZnII and CoII, respectively (Fig. 6b and Table 1). Notably, the two ZnII sensors show a relatively wide dynamic range for ΔGZn(II), suggesting that ZnII occupancy could increase dependent upon media [ZnII].
MgIIGTP-CobW outcompetes MgIIGTP-YeiR for CoII
Counterintuitively, MgIIGTP-YeiR and MgIIGTP-YjiA were predicted to preferentially bind ZnII in vivo, not due to tighter affinities for ZnII, but rather due to their weaker CoII affinities relative to MgIIGTP-CobW (Table 1, Figs. 5 and 6, and Supplementary Tables 2 and 5). To test relative CoII affinities, MgIIGTP-YeiR was competed against MgIIGTP-CobW (Fig. 7). YeiR and CobW were incubated with CoII in the presence of MgII and GTP, then separated by anion exchange chromatography. The chromatography was also conducted with each protein separately. Individually each protein eluted bound to CoII, but in competition CoII eluted almost exclusively with MgIIGTP-CobW (Fig. 7 and Supplementary Fig. 26), confirming its tighter affinity for CoII.
Fig. 7. MgIIGTP-CobW outcompetes MgIIGTP-YeiR for CoII.
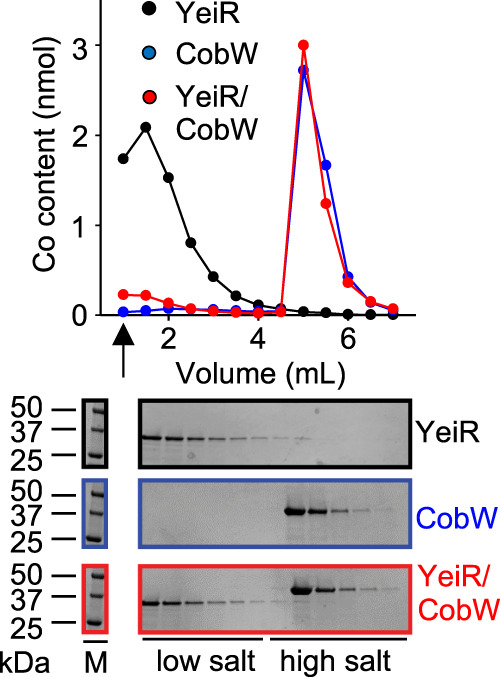
Elution profile of YeiR (10 μM), CobW (10 μM) or both proteins following incubation with GTP (100 μM), MgII (2.7 mM) and CoII (8 μM) resolved by differential elution from an anion exchange column. Fractions were analysed for CoII by ICP-MS and protein by SDS-PAGE (YeiR alone black, CobW alone blue, both proteins red; n = 1). Arrow denotes flow through fractions. Full gel images and SDS-PAGE analysis of flow through fractions shown in Supplementary Fig. 26.
Fine tuning ∆G for metalation in a cell
Calculated free energies for intracellular metalation (ΔGM) in Figs. 5 and 6 are based on an assumption that cellular metal availabilities are fixed at ideal buffered concentrations where every metal sensor undergoes half of its transcriptional response (i.e. normalised fractional DNA occupancy θD = 0.5, see ref. 9). In reality, cellular metal availabilities, and consequently θD of sensors, fluctuate conditionally (e.g. in response to addition of metals or chelators to the growth media). For example, the dynamic response range (defined as θD = 0.99–0.01) of RcnR, the CoII sensor from Salmonella, coincides with an increase in the intracellular available [CoII] from 2.4 × 10−11 to 2.7 × 10−7 M, corresponding to an increase in intracellular available ΔGCo(II) from −60.6 to −37.5 kJ mol−1 (Fig. 8a and Supplementary Table 8).
Fig. 8. Calculations of conditional CoII availabilities in B12-producing E. coli*.
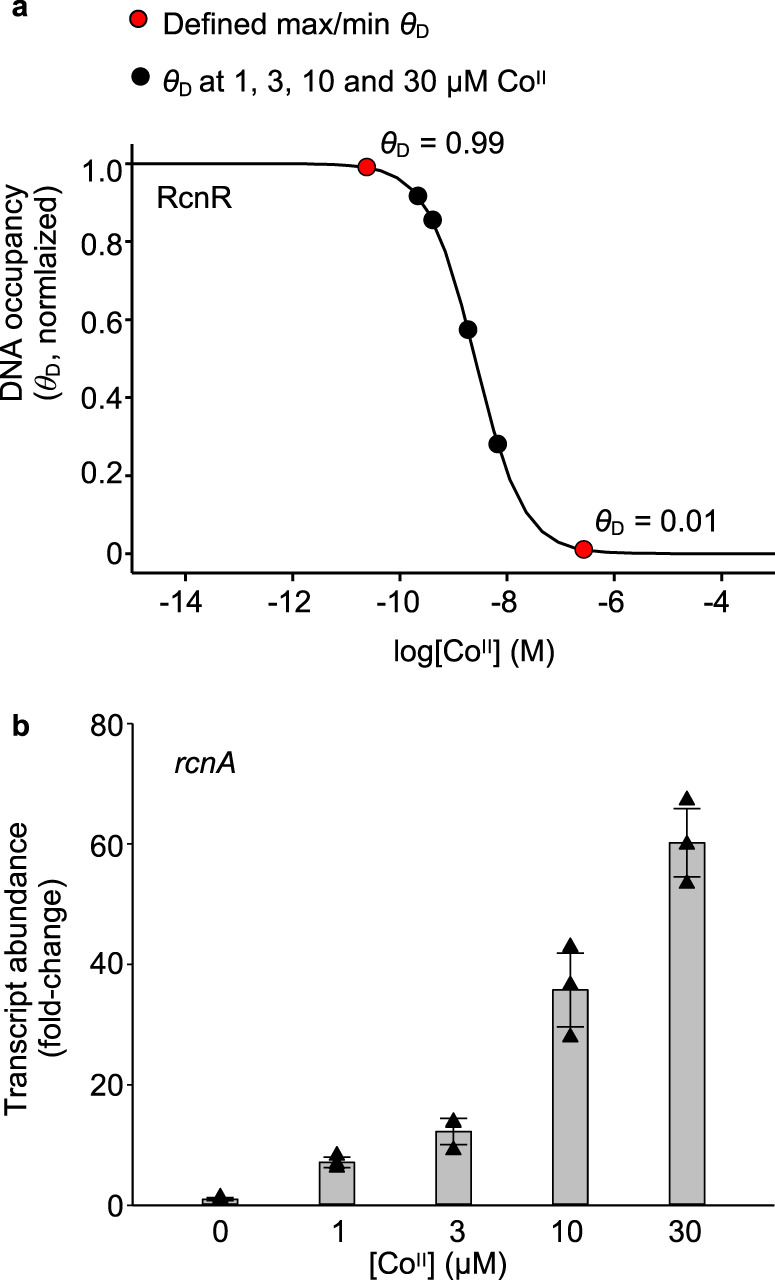
a Calculated relationship between intracellular CoII availability and normalised DNA occupancy (θD) by RcnR. θD of 0 and 1 are the maximum and minimum calculated DNA occupancies. The dynamic range (within which RcnR responds to changing intracellular CoII availability) has been defined as θD of 0.01–0.99 (i.e. 1–99% of RcnR response). The calibrated maximum and minimum fold changes in rcnA transcript abundance (i.e. boundary conditions, see Supplementary Fig. 27) therefore correspond to θD of 0.01 and 0.99 in these calculations (red circles). θD for each growth condition (black circles) was calculated from the qPCR response in b, assuming a linear relationship between change in θD and change in transcript abundance (Eq. (10)). Corresponding CoII availabilities are listed in Supplementary Table 8. b Transcript abundance (relative to untreated control) of the RcnR-regulated gene rcnA following 1 h exposure of E. coli* to increasing [CoII], measured by qPCR. Data are the mean ± SD of n = 3 biologically independent replicates. Triangle shapes represent individual experiments (some data points overlap, experimental values are available in Source Data files).
In order to account for this variation, we developed a method to fine-tune free energy calculations under bespoke culture conditions using quantitative polymerase chain reaction (qPCR) analyses of transcripts regulated by metal sensors. Fine-tuning was performed for CoII in E. coli* which has been engineered to synthesise vitamin B12 (E. coli and Salmonella RcnR share 93% sequence identity and equivalent responses to available CoII were assumed). E. coli* cells were cultured in Luria-Bertani (LB) medium with increasing CoII supplementation. Abundance of the RcnR-regulated rcnA transcript (Fig. 8b) was used to calculate θD of RcnR for each condition (via Eq. (10) in “Methods”) following calibration of the maximum and minimum responses (defined as θD = 0.99 and 0.01 at low and high [CoII], respectively; Supplementary Fig. 27). This enabled the intracellular CoII availabilities, as conditional free energies, to be calculated from the RcnR θD for each condition (Fig. 8a and Supplementary Table 8).
CoII-acquisition by MgIIGTP-CobW predicts B12 synthesis
Does the amount of CoII inserted into B12 follow the predicted metalation of MgIIGTP-CobW? Metal occupancies of MgIIGTP-CobW in E. coli* samples were recalculated (via Eq. (4)) using bespoke intracellular available free energies, ΔGCo(II), for each growth condition (Fig. 8 and Supplementary Table 8). This predicted that in unsupplemented LB media the protein would be predominantly ZnII-bound (10% CoII and 77% ZnII) and that CoII occupancies would increase from 10% to 97% as added [CoII] increased from 0 to 30 µM (Fig. 9a). Since intracellular ZnII availability was also significant in our predictions, we confirmed that our previous estimation of ΔGZn(II) was valid in LB media (Supplementary Fig. 28). Corrin concentrations (presumed to be predominantly B12, noting that intermediates after CoII insertion may also be detected, and that ZnII may competitively inhibit the chelatase complex but not insert into ring-contracted corrins36) were measured in E. coli* strains containing or missing cobW (Fig. 9b and Supplementary Fig. 29), under the growth conditions for which intracellular available ΔGCo(II) was defined (Supplementary Table 8). As the added [CoII] increased so did B12 production in cobW(+), consistent with the predicted loading of MgIIGTP-CobW with CoII (Fig. 9). At higher [CoII], CobW-independent B12 synthesis became evident. As anticipated, total cellular cobalt increases with supplementation, and the amount of cobalt in B12 is <10% of the total cellular cobalt (Supplementary Table 9). The number of additional atoms accumulated per cell exceeds the amount predicted if CoII were not buffered, noting that the internal buffered concentration at 10 μM exogenous CoII is 1.9 nM (Fig. 8 and Supplementary Table 8), and that only 1 atom per cell volume (approximately 1 femtolitre) equates to 1.7 nM. Most importantly, B12 synthesis which is dependent on CobW (Fig. 9b, compare cobW(+) with cobW(−)) matches the trend in predicted metalation of MgIIGTP-CobW (Fig. 9a).
Fig. 9. B12 production follows predicted metalation of MgIIGTP-CobW.
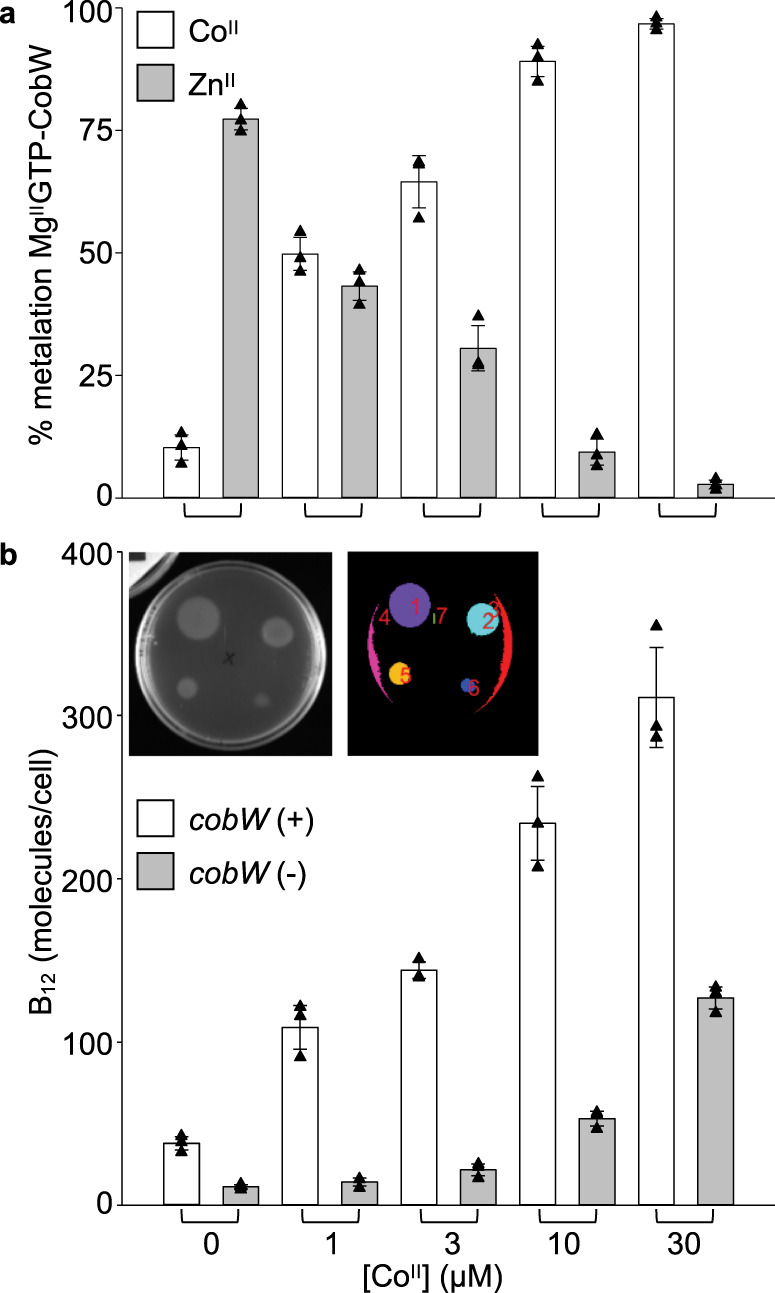
a Predicted metalation of MgIIGTP-CobW with CoII and ZnII (open and grey bars, respectively) in samples treated with defined media [CoII]. Intracellular ΔGCo(II) for each condition was calculated from rcnA expression (Fig. 8 and Supplementary Table 8). b B12 produced by E. coli* strains with and without cobW (open and grey bars, respectively) following 4 h exposure to defined [CoII]. B12 was detected using a Salmonella AR2680 bioassay65 (detects corrins, expected to be predominantly B12; see “Methods”) and quantified by automated analysis of growth areas (Supplementary Fig. 29 and Supplementary Software 1). Inset shows original image and detected areas (each false coloured) for representative (n = 3) bioassay plate of B12 calibration standards. All data are the mean ± SD of n = 3 biologically independent replicates (with errors in a propagated from qPCR data in Fig. 8b). Triangles represent individual experiments (some data points overlap, experimental values are available in Source Data files).
Discussion
Here we relate metal affinities of three putative metallochaperones to a thermodynamic framework, identifying their cognate metals which align with previous speculations16,20,25 (Figs. 5, 6 and Table 1). We establish the connection between CobW and CoII and show how CobW can acquire CoII in a cell (Figs. 1–5 and Table 1). Free-energy calculations reveal that in an idealised cell CoII ions will not flow from the cellular milieu to nucleotide-free CobW (ΔΔGCo(II) > 0). Crucially, CoII will flow from the cellular milieu to the MgIIGTP form of CobW (ΔΔGCo(II) < 0) (Fig. 5, Supplementary Fig. 30a, Table 1 and Supplementary Table 2). Thus, CobW must first bind MgIIGTP in order to acquire CoII inside a cell. In contrast, the product of GTP hydrolysis, MgIIGDP-CobW, will release CoII to the cellular milieu (ΔΔGCo(II) > 0) (Fig. 5, Supplementary Fig. 30b and Supplementary Table 2). Thus, the GTPase activity of CobW will facilitate CoII release, for example to CobNST for insertion into the corrin ring of B12 (Fig. 2f, g and Supplementary Fig. 6). We establish that CobW enhances B12 production when CoII is limiting (Fig. 9b), and Supplementary Fig. 30 illustrates the proposed mechanism. ZnII is identified as the preferred metal for nucleotide-bound forms of YeiR and YjiA: this is due to their weaker affinities for CoII relative to MgIIGTP-CobW (Fig. 7 and Supplementary Table 5), rather than tighter affinities for ZnII. These data illustrate the value of using the metalation calculator provided as Supplementary Data 1, which can now be broadly applied to metal-speciation in the context of intracellular competition.
MgIIGTP-CobW binds ZnII and CuI more tightly than CoII (Figs. 2e, 3, 4 and Supplementary Table 2), and likewise nucleotide-bound forms of YeiR and YjiA bind CuI more tightly than ZnII. Notably, by taking into account intracellular metal availability, ΔΔG for CuI was shown to be greater than zero for all three proteins in idealised cells, and also in conditional cells at either 90% or 99% of the dynamic range of the CuI sensor CueR (Figs. 5, 6 and Supplementary Fig. 14). Thus, these proteins will not acquire CuI. However, for MgIIGTP-CobW ΔΔG for ZnII was below zero in an idealised cell suggesting mis-metalation with ZnII (Fig. 5). Indeed, given that CobW binds ZnII more tightly than many known ZnII proteins14,43, and comparably to YeiR and YjiA (Supplementary Tables 2 and 5), it seems remarkable that ZnII is not the cognate metal. The data in Fig. 4, plus Supplementary Table 4, illustrate how occupancies of MgIIGTP-CobW with CoII versus ZnII change as a function of relative buffered metal availabilities. By reference to intracellular available free energies, the metal with the most negative ΔΔG will have the highest occupancy in vivo (Eq. (4)). In an idealised cell, ΔΔG for CoII is more negative than ΔΔG for ZnII and so the weaker binding metal dominates (Fig. 5 and Supplementary Table 2). In contrast, for nucleotide-bound forms of YeiR and YjiA ΔΔG for ZnII is more negative than ΔΔG for CoII making these deduced ZnII proteins. The previously intractable challenge to understand inter-metal competition in a cell now becomes tractable.
Initial calculations here, and in previous work9, assume an idealised cell in which the metal sensors are at the mid-points of their dynamic ranges (θD = 0.5). Therefore, we have calculated the available ΔGCo(II) in real (conditional) cells from the responses of RcnR (θD) estimated experimentally by qPCR of rcnA (Fig. 8 and Supplementary Fig. 27). As with other metallochaperones10,44, CobW is crucial when the cognate metal is limiting but at elevated CoII, CobW-independent synthesis of B12 occurs (Fig. 9b). CobNST must acquire CoII directly from the cytosol at the higher available ΔGCo(II). Importantly, CobW-dependent B12 synthesis tracked with the calculated CoII occupancy of MgIIGTP-CobW in cells supplemented with different amounts of CoII (Fig. 9). By monitoring the responses of sensors for different metals, it will be possible to define available ΔG, and predict protein occupancies with diverse metals, in different growth conditions. The calculator should be most accurate in Salmonella and closely related species such as E. coli. However, metal availabilities can also be adjusted (and/or simulated) to account for species differences, noting that the dynamic ranges of available ΔG values might be similar even when total cellular metal changes greatly between species.
Spectral features indicate that the CoII site in MgIIGTP-CobW involves thiols, likely derived from the CxCC motif in the GTPase domain, and a tetrahedral geometry (Figs. 1, 2 and Supplementary Fig. 3). However, all COG0523 proteins contain the CxCC motif16 and there is now a quest to understand why CoII affinities are weaker for MgIIGTP-bound YeiR and YjiA (and, hypothetically, ZigA and ZagA), which bind predominantly to ZnII as a result (Fig. 6). Notably a further pair of conserved cysteine residues (C56, C61) in CobWs are absent from the homologues (Supplementary Fig. 31). Intriguingly, NiII binding to MgIIGTP-CobW, MgIIGTP-YeiR and MgIIGTPγS-YjiA does not follow the order of stabilities of metal binding predicted by the Irving-Williams series (Figs. 5 and 6). An appealing explanation is that the allosteric coupling of GTP- and metal-binding imposes a (tetrahedral) geometry on the metal site that would disfavour NiII coordination (the Irving-Williams series applies where there is no steric selection): notably, related G3E GTPases involved in NiII homoeostasis (HypB and UreG) display square planar NiII coordination geometry45,46.
The metalation calculator has identified cognate metals for three members of the COG0523 sub-family of GE3 GTPases (Figs. 5, 6 and Table 1), and this work establishes vitamin B12 as the ultimate CoII-client of CobW (Fig. 9). For YeiR and YjiA, there is now a quest to identify their distinct roles and potential ZnII-requiring client(s). Under-metalation of MgIIGTP-CobW with CoII (and resultant mis-metalation with ZnII, Fig. 9a) could be especially problematic in E. coli* due to the lack of a dedicated CoII import system in this bacterium47. This suggests tantalising opportunities to engineer strains suited to the manufacture of vitamin B12, either via enhanced CoII uptake through engineered CoII-import, or via impaired ZnII accumulation by endogenous ZnII-transport systems. By analogy, with almost a half of enzymes requiring metals, an ability to calculate metalation in vivo should have broad applicability in optimising (or subverting) metalation in biotechnology. The calculator (Supplementary Data 1) can be widely used to understand metalation and mismetalation of proteins that acquire MgII, MnII, FeII, CoII, NiII, CuI or ZnII from the milieu inside living cells.
Methods
Protein expression and purification
The DNA sequence coding CobW was amplified by PCR using primers 1 and 2 (Supplementary Table 10) with genomic DNA from R. capsulatus SB1003 as template. The amplified fragment contained an NdeI restriction site at the 5ʹ end and a SpeI site at the 3ʹ end, allowing it to be cloned into a modified pET-3a vector34. The DNA coding sequence of yeiR (SL1344_2189) from S. enterica serovar Typhimurium strain SL1344 was obtained as a synthetic gene from Eurofins in a pEX plasmid with the T7 promoter and terminator from pET29a flanking yeiR. Additionally, the start codon of the gene (GTG) was changed to the more common ATG (pEXyeiR). The native coding sequence of yjiA (SL1344_4461) was obtained in the same manner (pEXyjiA). The correct sequence of each gene (cobW, yeiR and yjiA) was confirmed by DNA sequencing (DBS Genomics – Durham University).
E. coli BL21(DE3) pLysS, transformed with either pET3a-cobW, pEX-yeiR or pEX-yjiA were cultured in LB medium with antibiotics carbenicillin (50–100 mg L−1) and chloramphenicol (30–34 mg L−1). At mid-log phase, protein expression was induced by addition of 0.4 mM (CobW), 0.5 mM (YjiA) or 1.0 mM (YeiR) IPTG. Cells were cultured (with shaking) for 3–4 h at 37 °C (CobW) or at 20 °C overnight (YeiR and YjiA). Cells were harvested and stored at −20 °C prior to use.
Cells overexpressing CobW were resuspended in 20 mM sodium phosphate pH 7.4, 500 mM NaCl, 5 mM imidazole, 5 mM DTT and 1 mM PMSF for lysis (sonication) and cell debris was pelleted by centrifugation (38,000 × g, 45 min, 4 °C). Lysate was loaded to a 5-mL HisTrap HP column (GE Heathcare) pre-equilibrated in suspension buffer. CobW binds to the HisTrap column courtesy of a natural His-rich region within the protein. The column was washed with suspension buffer (10 column volumes), then eluted with suspension buffer containing 100 mM imidazole. Protein-containing fractions were incubated with excess (≥10-fold) EDTA for ≥1 h before being loaded to a HiLoad 26/600 Superdex 75 size exclusion column equilibrated in 50 mM Tris pH 8.0, 150 mM NaCl, 5 mM DTT and eluted in the same buffer. Peak CobW-containing fractions (determined from SDS-PAGE) were pooled, concentrated to ~0.5 mL (using a Vivaspin® 15 Turbo centrifugal concentrator). Protein identity was confirmed using ESI-MS by Durham University Department of Chemistry Mass Spectrometry Service. ESI-MS data were recorded on a QtoF Premier mass spectrometer coupled to an Acuity UPLC system (Waters). Protein samples were desalted prior to injection using a Waters MassPrep desalting cartridge (2.1 × 10 mm) and eluted with a linear acetonitrile gradient (20–80% v/v; 0.1% formic acid). Spectra were processed using Masslynx 4.1, deconvoluted using MaxEnt 1 and data imported into SigmaPlot software for preparation of figures.
Cells overexpressing YeiR were resuspended in 20 mM sodium phosphate pH 7.4, 100 mM NaCl, 5 mM DTT, 1 mM PMSF for lysis (sonication) and cell debris was pelleted by centrifugation (31,191 × g, 15 min, 8 °C). Soluble lysate was applied to a 5-mL HisTrap column (GE Healthcare) equilibrated with lysis buffer without PMSF. The column was washed with equilibration buffer before elution with equilibration buffer containing 10, 50 and 100 mM imidazole. YeiR eluted in the buffer containing 50 mM imidazole. EDTA was added to the YeiR-containing fraction to a final concentration of 10 mM and stored overnight at 4 °C. The sample was applied to HiLoad 26/600 Superdex 75 (GE Healthcare) equilibrated with 50 mM Tris pH 8, 150 mM NaCl, 5 mM DTT and eluted with the same buffer. Peak fractions were pooled and applied to a 5-mL Q anion exchange column (GE Healthcare) equilibrated with the size exclusion column buffer. Column flow through and wash were collected before eluting the column with size exclusion column buffer with the addition of 1 M NaCl. YeiR displays no affinity for the Q column and elutes in the flow through and wash. The remaining major contaminant elutes with 1 M NaCl. The flow through and wash from the Q column were pooled and concentrated using a Spin-X UF concentrator (Corning, 10 kDa molecular weight cut-off).
Cells overexpressing YjiA were resuspended in 20 mM Tris 7.5, 100 mM NaCl, 5 mM DTT, 1 mM PMSF for lysis (sonication) and cell debris was pelleted by centrifugation (two consecutive 20 min runs, 39,191 × g, 4 °C) before passing clarified supernatant through a 20-µm nylon membrane filter. Soluble lysate was applied to a 5-mL HisTrap column (GE Healthcare) equilibrated with lysis buffer without PMSF, collecting the flow through and a one column volume wash. This step removes a major contaminant which binds to the HisTrap column. Pooled flow through and wash fractions were applied to a 5-mL HiTrap Q-Sepharose fast flow column (GE Healthcare) equilibrated with lysis buffer without PMSF. The column was washed with equilibration buffer then equilibration buffer with 200 mM NaCl before elution of YjiA by application of a 50-mL gradient of 200–600 mM NaCl in equilibration buffer collecting 5 mL fractions. YjiA typically eluted between 10 and 30 mL. Fractions containing the highest concentration of YjiA with the lowest degree of contamination, as judged by SDS-PAGE, were stored overnight (4 °C) with EDTA to a final concentration of 5 mM. Fractions were concentrated to 5 mL using a Spin-X UF concentrator (Corning, 10 kDa molecular weight cut-off) and applied to HiLoad 26/600 Superdex 75 (GE Healthcare) equilibrated with lysis buffer without PMSF and eluted with the same buffer. Fractions were pooled and concentrated to 5–20 mg mL−1 using a Spin-X UF concentrator (Corning, 10 kDa molecular weight cut-off) before storage at −80 °C.
Following purification, CobW, YeiR and YjiA samples were transferred to an anaerobic glovebox (Belle Technology), (0.5–1 mL) applied to a PD-10 Desalting Column prepacked with Sephadex G-25 medium (GE Healthcare) equilibrated with chelex-treated and N2-purged buffer (10 mM HEPES pH 7.0, 100 mM NaCl, 400 mM KCl) and eluted in the same buffer. Proteins were quantified by A280 nm using experimentally determined extinction coefficients (ε = 15,300 M−1 cm−1 for CobW, 52,745 M−1 cm−1 for YeiR, and 27,900 M−1 cm−1 for YjiA) determined by quantitative amino acid analysis (Alta Bioscience Ltd). Samples were confirmed to be of high purity by SDS-PAGE (full gel images are available in the Supplementary Information), and ≥92.5% (YeiR) and ≥95% (CobW, YjiA) metal-free (by inductively coupled plasma-mass spectrometry; ICP-MS). ICP-MS of one sample of YjiA was performed 3 weeks after buffer exchange and >5% zinc detected, but results were consistent with replicates containing <5% zinc. ICP-MS was conducted using Durham University Bio-ICP-MS Facility (PlasmaLab software; Thermo Fisher). Reduced thiol content was determined by reaction with ~10-fold excess of Ellman’s reagent 5,5ʹ-dithio-bis-[2-nitrobenzoic acid] (DTNB; produces one equivalent of chromophore TNB2− per protein thiol, A412 nm = 14,150 M−1 cm−1)48,49. For CobW >5.5 cysteines were found to be reactive with DTNB (expected value = 6), for YeiR 4–5 cysteines (expected value = 5), and for YjiA >4.5 cysteines (expected value = 5).
Preparation of metal stocks
All metal stocks were prepared in ultrapure water from appropriate salts (MgCl2, (NH4)2Fe(SO4)2, CoCl2, NiSO4, NiCl2, CuSO4, ZnCl2, ZnSO4) and quantified by ICP-MS analysis. FeII stocks were prepared by dissolving (NH4)2Fe(SO4)2·6H2O in deoxygenated 0.1% (v/v) HCl in an anaerobic chamber. Reaction with excess ferrozine (Fz; ∼50-fold) confirmed that iron was ≥95% reduced (FeIIFz3 ε562 nm = 27,900 M−1 cm−1)50. Concentrated stocks were diluted daily in deoxygenated ultrapure water to prepare working solutions and confirmed to be ≥90% FeII. Other metal stocks were prepared aerobically as concentrated stocks and diluted to working solutions with deoxygenated ultrapure water in an anaerobic chamber. Cu(I) was generated in situ (from CuSO4) by hydroxylamine (1–10 mM) which quantitatively reduces Cu(II) to Cu(I) in the presence of excess chelator (L = Bca or Bcs) to form CuIL2 complexes51.
Determination of CobW CoII-binding stoichiometries
Metal-binding experiments were conducted in an anaerobic chamber in deoxygenated, chelex-treated 10 mM HEPES pH 7.0, 100 mM NaCl, 400 mM KCl. For stoichiometry determinations, CoII was titrated into a solution of CobW (15–30 µM) together with relevant nucleotides (supplied in ∼10-fold excess of protein concentration for GTP and GDP and ∼3-fold excess for GMPPNP and GTPγS, as specified in figure legends) in the absence or presence of MgII (2.7 mM). Absorbance was recorded using a Lambda 35 UV-visible spectrophotometer (Perkin Elmer; UV-Win lab software). The extinction coefficient of CoIIMgIIGTP-CobW (ε339 nm = 2800 ± 100 M−1 cm−1, average ± s.d of n = 3 independent titrations) was determined from absorbance at saturating metal concentrations (Supplementary Fig. 3g). Extinction coefficients of related complexes CoIIMgIIGMPPNP-CobW, CoIIMgIIGTPγS-CobW, CoII2GTP-CobW, CoII2GMPPNP-CobW and CoII2GTPγS-CobW were similarly determined (Figs. 1, 2 and Supplementary Figs. 1, 3): within experimental error, all produced the same extinction coefficient as for CoIIMgIIGTP-CobW, thus ε339 nm = 2800 M−1 cm−1 was assigned to all species.
Gel-filtration chromatography experiments were performed by incubating CobW (10 µM) and CoII (30 µM) for 30 min with or without cofactor GMPPNP (30 µM) then applying 0.5 mL to a PD-10 Sephadex G-25 gel-filtration column (GE Healthcare). Eluted fractions (0.5 mL) were analysed for cobalt by ICP-MS and for protein by Bradford assay.
Determination of CobW metal affinities via ligand competition
Ligand competition experiments were conducted in an anaerobic chamber in deoxygenated, chelex-treated 10 mM HEPES pH 7.0, 100 mM NaCl, 400 mM KCl, except where high concentrations (≥1 mM) of competing ligand were employed, where 50 mM HEPES was used to maintain buffered pH 7.0. Absorbance was recorded using a Lambda 35 UV-visible spectrophotometer (Perkin Elmer). Fluorescence spectra were recorded using a Cary Eclipse fluorescence spectrophotometer (Agilent; Cary Eclipse scan application software). Affinities were determined at a range of different competing conditions (by varying the competing ligand and/or the protein:ligand ratio) to ensure reliability: details are documented in Supplementary Table 1. Scripts used for data fitting (using Dynafit52) are provided in Supplementary Software 2. The effect of MgII (2.7 mM) on apparent dissociation constants of ligand standards (EGTA, NTA, fura-2, Mf2 and quin-2) was calculated to be insignificant under the conditions of competition experiments (Supplementary Table 3). For probes with undefined MgII affinities (Tar, Bca), control experiments confirmed that addition of MgII (2.7 mM) had negligible effect on competition experiments (Supplementary Figs. 10d and 12). Thus, MgII was not incorporated into the curve-fitting models.
For determination of weaker (KD > 10 nM) CoII binding affinities (CobW and MgIIGDP -CobW), CoCl2 was titrated into a solution of 5-Oxazolecarboxylic acid, 2-(6-(bis(carboxymethyl)amino)-5-(2-(2-(bis(carboxymethyl)amino)-5-methylphenoxy)ethoxy)-2-benzofuranyl)-pentapotassium salt (fura-2; quantified by ε363 nm = 28,000 M−1 cm−1)53 and CobW in the presence or absence of cofactors (MgCl2 and GDP) and fluorescence emission (λex = 360 nm; λmax ~505 nm) was recorded at equilibrium. CoII-dependent fluorescence quenching of fura-2 was used to determine CoII speciation. For determination of CoII binding affinities tighter than 10 nM (MgIIGMPPNP-CobW, MgIIGTPγS-CobW and MgIIGTP-CobW), CoCl2 was titrated into a solution containing CobW, competing ligand (EGTA or NTA), MgCl2 and nucleotide (GMPPNP, GTPγS or GTP). UV-visible absorbance (relative to metal-free solution) was recorded at equilibrium to determine CoII speciation (ε339 nm = 2800 M−1 cm−1 for CoII-bound proteins). Data were fit using Dyanfit52 to models describing 1:1 binding stoichiometry for CoII:protein and 1:1 binding stoichiometry for CoII:ligand (ligand = fura-2, EGTA or NTA). Ligand dissociation constants at pH 7.0: fura-2 KCo(II) = 8.6 × 10−9 M (ref. 39); EGTA KCo(II) = 7.9 × 10−9 M (ref. 38); NTA KCo(II) = 2.2 × 10−8 M (ref. 38).
(NH4)Fe(SO4)2 was titrated into a solution of Tar (16 µM), MgCl2 (2.7 mM) and GTP (500 µM) in the absence or presence of CobW (50 µM) and UV-visible absorbance recorded at equilibrium to define FeII speciation (FeIITar2 ε720 = 19,560 M−1 cm−1 under experimental conditions, Supplementary Fig. 8a). Data were fit in Dynafit52 to a model describing 1:1 binding stoichiometry for FeII:protein and 1:2 binding stoichiometry for FeII:Tar using β2,Fe(II) = 4.0 × 1013 M−2 for Tar at pH 7.0 (ref. 54). Experimental data were compared to simulated fits with defined protein KFe(II) = 10−6 M, 10−7 M, allowing limiting KD ≥ 10−6 M for MgIIGTP-CobW to be determined. Tar stock concentrations were quantified using ε470 nm = 24,800 M−1 cm−1 (reported value at pH 7.0 (ref. 54)) and verified by titration with metal stocks (FeII or NiII, quantified by ICP-MS).
NiSO4 was titrated into a solution of Tar (20 µM), CobW (10–30 µM), MgCl2 (2.7 mM) and GTP (100–300 µM) and UV-visible absorbance recorded at equilibrium to determine NiII speciation (NiIITar2 Δε535 nm = 3.8 (±0.1) × 104 M−1 cm−1 relative to ligand only solution; Supplementary Fig. 10a). Tar stock concentrations were quantified as above. Data were fit using Dynafit52 to a model describing 1:1 stoichiometry NiII:protein and 1:2 stoichiometry NiII:Tar; β2,Ni(II) = 4.3 (±0.6) × 1015 M−2 for Tar at pH 7.0 was independently determined by preparing a series of solutions of NiTar2 ([NiII] = 15 µM, [Tar] = 36 µM) with varying EGTA concentrations (0–400 µM) and measuring UV-visible absorbance at equilibrium (following 1–2 h incubation). EGTA KNi(II) = 5.0 × 10−10 M at pH 7.0 (ref. 38). Data were fit to Eq. (5)51 using Kaleidagraph (Synergy Software).
| 5 |
CuSO4 was titrated into a solution of Bca (1.0 mM), CobW (10–30 µM), MgCl2 (2.7 mM), GTP (100–300 µM) and reductant NH2OH (1.0 mM) which quantitatively reduces CuII to CuI in the presence of a strong CuI ligand (e.g. Bca: β2,Cu(I) = 1.6 × 1017 M−2 (ref. 38)). UV-visible absorbance was recorded at equilibrium to define CuI speciation (CuIBca2 ε562 = 7900 M−1 cm−1 (ref. 38)) and data were fit using Dynafit52 to a model describing 1:1 stoichiometry CuI:protein and 1:2 stoichiometry CuI:Bca.
ZnCl2 was titrated into a solution containing quin-2 (10 µM), CobW (10 µM), MgCl2 (2.7 mM) and GTP (50 µM) and UV-visible absorbance recorded at equilibrium. Quin-2 was quantified using ε261 nm = 37,000 M−1 cm−1 (ref. 55). KZn(II) for MgIIGTP-CobW was beyond the range of this experiment (significantly tighter than quin-2) and only a limiting affinity was determined (KZn(II) < 10−12 M).
ZnII affinity of MgIIGTP-CobW via inter-metal competition
Solutions containing CobW (17.9–20.4 µM), MgCl2 (2.7 mM), GTP (200 µM) and ligand NTA (0.4–4.0 mM) were titrated with CoCl2 (0.3–3.0 mM) and ZnCl2 (15.3–25.5 µM) and UV-visible absorbance was recorded at equilibrium to determine CoII occupancy of CobW (ε339 nm = 2800 M−1 cm−1 for CoIIMgIIGTP-CobW). Details of individual experiments are in Supplementary Table 4. The total concentration of CoII and ZnII in each solution was limiting, such that both metals were buffered by ligand NTA. Metal speciation was determined from the mass balance relationships given in Eqs. (6–8) (cofactors MgIIGTP omitted for clarity). Thus, KZn(II) for MgIIGTP-CobW was calculated from the exchange equilibria (Kex) in Fig. 4a, relative to known KCo(II) for the protein (Supplementary Table 2) and ligand dissociation constants (NTA KZn(II) = 1.18 × 10−8 M, KCo(II) = 2.24 × 10−8 M (ref. 38)). These calculations are valid given that [M]free ≪ [M]tot (M = CoII or ZnII, buffered by excess NTA), the concentration of non-metalated protein is negligible (Supplementary Fig. 13) and potential ternary complexes involving metal, protein and NTA are transient species only with insignificant concentration at thermodynamic equilibrium (varying ratios of buffered metals, [CoIINTA]/[ZnIINTA], were used to confirm consistency of KD values at multiple equilibria; see Fig. 4 and Supplementary Table 4).
| 6 |
| 7 |
| 8 |
Determination of YeiR and YjiA metal stoichiometries and affinities
Investigation of protein–metal interactions and competition experiments to determine metal affinities were performed in 10 mM HEPES pH 7, 100 mM NaCl, 400 mM KCl (chelex treated and N2 purged) with the inclusion of nucleotides and MgCl2 as noted in figure legends. Absorbance was recorded using a Lambda 35 UV-visible spectrophotometer (Perkin Elmer). Fluorescence spectra were recorded using a Cary Eclipse fluorescence spectrophotometer (Agilent). Scripts used for data fitting (using Dynafit52) are provided in Supplementary Software 2.
CoCl2 was titrated into a solution of fura-2 (ε363 nm = 28,000 M−1 cm−1, KCo(II) = 8.6 × 10−9 M (refs. 39,53)) in the presence of YeiR or YjiA and fluorescence emission (510 nm) recorded at equilibrium (λex = 360 nm, 20 °C). Data were fit to a model describing 1:1 CoII:fura-2 and 1:1 CoII:protein binding stoichiometries using Dynafit52.
NiCl2 was titrated into a solution of Mf2 (ε369 nm = 22,000 M−1 cm−1, KNi(II) = 5 × 10−8 M (refs. 53,56)) in the presence of YeiR or YjiA and the absorbance (323–325 and 365–366 nm) recorded at equilibrium. Data were fit (both wavelengths simultaneously) to a model describing 1:1 NiII:Mf2 and 1:1 NiII:protein binding stoichiometries using Dynafit52.
ZnSO4 was titrated into a solution of Mf2 (KZn(II) = 2 × 10−8 M (ref. 57)), PAR (β2 Zn(II) = 2 × 1012 M−2 (ref. 58)) or quin-2 (KZn(II) = 3.7 × 10−12 M (ref. 55)) in the presence of YeiR or YjiA and the absorbance (325 and 366 nm Mf2; 500 nm PAR; 265 or 269 nm quin-2) recorded at equilibrium. Concentrations of PAR and quin-2 stocks were determined by direct titration with ZnSO4. Data were fit to a model describing 1:1 ZnII:quin-2 and 1:1 ZnII:YjiA binding stoichiometries. Zn:YeiR stoichiometries were fit as 1:1, or allowed to be determined in fitting as described in the text using Dynafit52.
CuSO4 was titrated into a solution of Bca (CuIBca2 ε562 nm = 7900 M−1 cm−1, β2 Cu(I) = 1017.2 M−2 (ref. 59)) in the presence and absence of YeiR or YjiA (with inclusion of hydroxylamine) and absorbance (562 nm) recorded at equilibrium. Protein CuI affinity was calculated using Eq. (9), for the tightest binding event. Calculated affinities were simulated using Dynafit52, and overlaid on the data.
| 9 |
(NH4)Fe(SO4)2 was titrated into a solution of Tar (Tar2Fe(II) ε720 nm = 19,000 M−1 cm−1, β2 Fe(II) = 1013.6 M−2 (at pH 7.0) (ref. 54) in the presence and absence of YeiR or YjiA and absorbance (720 nm) recorded at equilibrium. Data were fit to a model describing 1:2 FeII:Tar and 1:1 FeII:protein binding stoichiometries using Dynafit52.
MnCl2 was titrated into a solution of Mf2 (KMn(II) = 6.1 × 10−6 M (ref. 9)) in the presence of YjiA and the absorbance (330 and 365 nm) recorded at equilibrium. Data were fit (both wavelengths simultaneously) to a model describing 1:1 MnII:Mf2 and 1:1 MnII:protein binding stoichiometries using Dynafit52.
Gel filtration chromatography of YeiR was performed by application of 0.5 mL (10 μM) to a PD-10 Desalting Column prepacked with Sephadex G-25 medium equilibrated with buffer supplemented with 2.7 mM MgCl2 with or without 20 μM MnCl2 and eluted with the same buffer. YeiR was incubated with 20 μM MnCl2 for 20 min prior to application to the column. Protein content of collected fractions was assayed by A280 nm and Bradford assay, metal content by ICP-MS.
Inter-protein competition for Co(II)
Experiments were performed in an anaerobic glovebox. YeiR (10 μM) was incubated with GTP (100 μM), MgCl2 (2.7 mM) and CoCl2 (8 μM) in 10 mM HEPES pH 7.0, 40 mM NaCl, 160 mM KCl (chelex treated and N2 purged) for 10 min before addition of CobW (10 μM) (total volume upon CobW addition = 1.1 mL). The mixture was incubated for a further 30 min before application of 1 mL of the incubation reaction to a 1-mL Q anion exchange column (GE Healthcare) equilibrated with 10 mM HEPES pH 7.0, 40 mM NaCl, 160 mM KCl (chelex treated and N2 purged), collecting the flow through. The column was sequentially eluted with equilibration buffer collecting six 0.5 mL fractions followed by 10 mM HEPES pH 7.0, 200 mM NaCl, 800 mM KCl (chelex treated and N2 purged) collecting six 0.5-mL fractions. Fractions were analysed for protein content by SDS-PAGE and for metal content by ICP-MS. Controls were conducted concurrently as above but with YeiR or CobW alone.
GTPase activity assays
CobW (20–50 µM) was incubated with CoCl2 (0.9 equivalents CoII:protein) and GTP (200 µM) in an anaerobic chamber in N2-purged, chelex-treated 10 mM HEPES pH 7.0, 100 mM NaCl, 400 mM KCl. Aliquots of solution taken at various time intervals (0–390 min) were loaded to a 5-mL HiTrap Q HP column (GE Healthcare) equilibrated in buffer (20 mM HEPES pH 7.0, 100 mM NaCl) and eluted with a linear NaCl gradient (100–500 mM NaCl). Nucleotides were detected by UV absorbance (254 nm or 280 nm) and the ratio of GTP:GDP in each sample was calculated by integration of the respective peak areas.
Growth of E. coli* strains
E. coli* strains used in this work are derived from E. coli MG1655 (DE3) engineered to contain a set of B12 biosynthesis genes from R. capsulatus60,61, and Brucella melitensis (B. melitensis)34. Strain ED741 (E. coli* without cobW) is MG1655 with Plac-T7RNAP-PT7-cobAIGJFMKLHBROQJD-bluE-C-bluF-PUB-cbiW-VE-PT7-cobNST while strain ED732 (E. coli* with cobW) is MG1655 with Plac-T7RNAP-PT7-cobAIGJFMKLHBROQJD-bluE-C-bluF-PUB-cbiW-VE-PT7-cobWNST. All R. capsulatus and B. melitensis (cobG, cobR, cobE) genes were cloned individually in pET3a and subcloned together using the link and lock method34. The synthetic operons were transferred into the E. coli genome using CRISPR technology62. Although chromosomally integrated B12 biosynthesis genes are IPTG-inducible under the control of the T7 promoter, in the current experiments IPTG was not added to cell cultures to avoid potential disruptions of cellular metal homoeostasis caused by over-production of metalloproteins.
All cultures and media were prepared in plasticware or acid-washed glassware to minimise trace metal contamination. LB medium was inoculated with overnight culture of E. coli* (OD600 nm = 0.025) and incubated at 37 °C with shaking until OD600 nm reached ~0.2. Aliquots (5 mL or 50 mL) of this culture were treated with sterile CoCl2, H2O, EDTA or ZnCl2 (100× concentrated stocks) to reach final concentrations as specified in figure legends (Figs. 8b, 9b and Supplementary Figs. 27, 28a, b, d and 29c) and incubated under the same conditions for a further 1–4 h. Samples used for RNA extraction were taken 1 h after treatment. Samples for B12 quantification and OD600 nm readings were taken 4 h after treatment to ensure detectable corrinoid production.
Determination of transcript abundance in E. coli*
Aliquots (1 mL) of E. coli* culture from each growth condition were stabilised in RNAProtect Bacteria Reagent (2 mL; Qiagen) and cells pellets were frozen at −80 °C prior to processing. RNA was extracted using an RNeasy Mini Kit (Qiagen) as described by the manufacturer. RNA was quantified by absorbance at 260 nm and treated with DNAse I (2.5 U/μL; Fermentas). cDNA was generated using the ImProm-II Reverse Transcriptase System (Promega) with 300 ng RNA per reaction, and control reactions without reverse transcriptase were conducted in parallel. Transcript abundance was determined using primers 3 and 4 for rcnA, 5 and 6 for zntA, 7 and 8 for znuA, 9 and 10 for rpoD, each pair designed to amplify ~110 bp fragment. Quantitative PCR analysis was carried out in 20 µL reactions using 5 ng of cDNA, 0.8 µM of each appropriate primer and PowerUp SYBR Green Master Mix (Thermo Fisher Scientific). Three technical replicates of each sample (i.e. biological replicate) were analysed using a Rotor-Gene Q 2plex (Qiagen; Rotor-Gene-Q Pure Detection software), plus control reactions without cDNA template for each primer pair. The fold change, relative to the mean of the control condition for each sensor, was calculated using the 2−ΔΔCT method63, with rpoD as the reference gene. Cq values were calculated with LinRegPCR after correcting for amplicon efficiency64.
Intracellular available ∆GCo(II) under bespoke conditions
Intracellular available ∆Gmetals were first calculated from available metal concentrations where the cognate sensor is at 1%, 10%, 50%, 90% and 99% of its response (i.e., θD = 0.01, 0.1, 0.5, 0.9, 0.99; Supplementary Note 1). Available metal concentrations corresponding to these fractional occupancies were determined using known metal affinities, DNA affinities, protein abundances and numbers of DNA binding sites determined for Salmonella sensors9, using excel spreadsheet (Supplementary Dataset 1) and MATLAB code (Supplementary Note 3) available in ref. 9.
Fractional responses (θD) of RcnR at bespoke growth conditions were calculated from transcript abundance of rcnA via Eq. (10):
| 10 |
where fold-changeobs is the observed fold-change in rcnA transcript abundance at the bespoke condition and fold-changemax is the maximum fold-change in rcnA transcript abundance at the calibration limit (corresponding to maximum abundance); all fold-changes were determined relative to the defined control condition (untreated LB) corresponding to minimum rcnA transcript abundance (see Supplementary Fig. 27c). Equation (10) defines maximum and minimum transcript abundances as corresponding to θD of 0.01 and 0.99, respectively (see Fig. 8a), and assumes a linear relationship between change in θD and change in transcript abundance.
The intracellular available [CoII] concentration corresponding to each RcnR θD was calculated using known metal affinity, DNA affinities, protein abundance, number of DNA binding sites determined for Salmonella RcnR9, to calculate the CoII-dependent response of E. coli RcnR (93% sequence identity) using excel spreadsheet (Supplementary Dataset 1) and MATLAB code (Supplementary Note 3) available in ref. 9. The intracellular available ΔGCo(II) for each condition was calculated using Eq. (11), where [CoII] is the intracellular available CoII concentration, R (gas constant) = 8.314 × 10−3 kJ K−1 mol−1 and T (temperature) = 298.15 K (see Supplementary Note 1).
| 11 |
Estimation of intracellular available ∆GZn(II) in LB media
Fractional responses (θD) of Zur and ZntR in LB media were calculated from transcript abundance of znuA and zntA, via Eqs. (10) and (12), respectively:
| 12 |
where fold-changeobs is the observed fold-change in transcript abundance in LB and fold-changemax is the maximum fold-change in transcript abundance at the calibration limit (corresponding to maximum abundance); all fold-changes were determined relative to defined control conditions corresponding to minimum transcript abundance (see Supplementary Fig 28a, b). Equation (12) defines maximum and minimum transcript abundances as corresponding to θD of 0.99 and 0.01, respectively, and assumes a linear relationship between change in θD and change in transcript abundance.
The intracellular available [ZnII] concentration corresponding to each θD was calculated using known metal affinities, DNA affinities, protein abundance, number of DNA binding sites determined for Salmonella homologues, to calculate the ZnII-dependent responses of E. coli ZntR and Zur (both >92% sequence identity to Salmonella) using excel spreadsheet (Supplementary Dataset 1) and MATLAB code (Supplementary Note 3) available in ref. 9. The intracellular available ΔGZn(II) was calculated using Eq. (13), where [ZnII] is the intracellular available ZnII concentration, R (gas constant) = 8.314 × 10−3 kJ K−1 mol−1 and T (temperature) = 298.15 K (see Supplementary Note 1).
| 13 |
Quantification of vitamin B12 in E. coli* cultures
Aliquots (20 mL) of E. coli* culture from each growth condition were taken, and cell pellets frozen at −20 °C. To quantify corrin production (assumed to be predominantly B12, since E. coli* contains genes for the complete pathway), E. coli* pellets were thawed, resuspended in H2O (0.2 mL), boiled for 15 min (95 °C) and centrifuged to remove cell debris. An aliquot (10 µL) of each supernatant was applied to Salmonella typhimurium AR2680 (ΔmetE, ΔcbiB) pre-innoculated bioassay plates65, and incubated at 37 °C overnight. Plates were imaged together with a 1-cm2 reference area on black background (see example in Supplementary Data 2) using a Gel-Doc XR + gel documentation system (BioRad; ImageLab software). Images were analysed in MATLAB using the code in Supplementary Software 1 to determine the growth area (in cm2) of each sample. A calibration curve relating growth areas to B12 concentration was generated using B12 standards (cyanocobalamin; 1–100 nM; quantified by A360 nm = 27,500 M−1 cm−1 at pH 10 (ref. 66)) in parallel with E. coli* lysates, using the same batch of bioassay plates (Supplementary Fig. 29a, b). To determine the number of cells in each sample, solutions of E. coli* at varying cell densities (OD600 nm = 0.2–0.9) were prepared, serially diluted (2000-fold), and the number of cells per mL quantified using a CASY® cell counter. The resulting correlation factor (4.4 ± 0.1 × 108 cells mL−1 OD600 nm−1) was used to convert OD600 nm to cell number (Supplementary Fig. 29c, d).
Metal content of E. coli* cells
Aliquots (20 mL) of E. coli* culture from each growth condition were taken and pellets were washed twice with 0.5 M sorbitol, 200 μM EDTA, 20 mM Tris pH 8.5. Cell pellets were suspended in ultrapure 65% (v/v) HNO3 (0.4 mL) to digest (>24 h), then diluted tenfold in 2.5% HNO3 before metal analysis by ICP-MS.
Statistics and reproducibility
Sample sizes were chosen based on prior experimental experience, and to give consistent results, following convention in the literature for equivalent analyses. Experiments designed to derive quantitative values used to model or test calculations of metalation were performed in triplicate or more (n = 3–5) to enable calculation of SD (listed in Tables or shown as error bars in figures). The number of independent experiments or biologically independent samples is shown in figure legends or footnotes of Tables.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This work was supported by a COFUND European Union/Durham University Junior Research Fellowship under EU grant agreement 609412 (T.R.Y.), a Royal Commission for the Exhibition of 1851 Research Fellowship (T.R.Y.), an UKRI Future Leaders Fellowship MR/T019891/1 (R.J.M.), a US-UK Fulbright Commission award (A.G.), Biotechnology and Biological Sciences Research Council awards BB/S009787/1, BB/J017787/1, BB/R002118/1, BB/S002197/1, BB/S014020/1 and Royal Society award INF\R2\180062. We thank Peter Chivers (Durham University, UK) for constructive scientific discussions.
Source data
Author contributions
T.R.Y. conducted the in vitro metal-binding experiments for CobW, GTP-hydrolysis assays, in vivo gene expression experiments and B12-production experiments. A.W.F. conducted the in vitro metal-binding experiments for YeiR. A.G. conducted the in vitro metal-binding experiments for YjiA. T.R.Y. and M.A.M. developed the experimental protocols for determining metal sensor responses by qPCR. M.A.M. derived equations for the metalation calculator and produced the spreadsheet. R.J.M. and D.O. generated the MATLAB code for analysis of B12 bioassays. E.D. generated the CobW expression plasmid. E.D. and M.J.W. donated the B12-producing E. coli* strains and advised on B12 biochemistry. E.D., M.J.W. and T.R.Y. co-designed the B12-production experiments. T.R.Y. and N.J.R. drafted the manuscript with input from A.W.F. T.R.Y. and N.J.R., in conjunction with A.W.F., A.G., M.A.M. and D.O., interpreted the significance of the data. T.R.Y. and N.J.R. had overall responsibility for the design and management of the project. All authors reviewed the results and edited and approved the final version of the manuscript.
Data availability
All data are available within the article, its Supplementary Information files, plus PDB entry 1NIJ. Source data are provided with this paper.
Code availability
Equation derivations, Excel spreadsheet (with instructions) constituting a metalation calculator, MATLAB code (with instructions) for use in B12 assays and Dynafit scripts are available in Supplementary Note 1, Supplementary Data 1, Supplementary Software 1 and Supplementary Software 2, respectively.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tessa R. Young, Email: tessa.r.young@durham.ac.uk
Nigel J. Robinson, Email: nigel.robinson@durham.ac.uk
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-021-21479-8.
References
- 1.Foster AW, Osman D, Robinson NJ. Metal preferences and metallation. J. Biol. Chem. 2014;289:28095–28103. doi: 10.1074/jbc.R114.588145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Irving H, Williams RJP. Order of stability of metal complexes. Nature. 1948;162:746–747. doi: 10.1038/162746a0. [DOI] [Google Scholar]
- 3.Martin ME, et al. A Streptococcus mutans superoxide dismutase that is active with either manganese or iron as a cofactor. J. Biol. Chem. 1986;261:9361–9367. doi: 10.1016/S0021-9258(18)67663-X. [DOI] [PubMed] [Google Scholar]
- 4.Culotta VC, Yang M, O’Halloran TV. Activation of superoxide dismutases: putting the metal to the pedal. Biochim. Biophys. Acta Mol. Cell Res. 2006;1763:747–758. doi: 10.1016/j.bbamcr.2006.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ranquet C, Ollagnier-de-Choudens S, Loiseau L, Barras F, Fontecave M. Cobalt stress in Escherichia coli: the effect on the iron-sulfur proteins. J. Biol. Chem. 2007;282:30442–30451. doi: 10.1074/jbc.M702519200. [DOI] [PubMed] [Google Scholar]
- 6.Waldron KJ, Robinson NJ. How do bacterial cells ensure that metalloproteins get the correct metal? Nat. Rev. Microbiol. 2009;7:25–35. doi: 10.1038/nrmicro2057. [DOI] [PubMed] [Google Scholar]
- 7.Tottey S, et al. Protein-folding location can regulate manganese-binding versus copper- or zinc-binding. Nature. 2008;455:1138–1142. doi: 10.1038/nature07340. [DOI] [PubMed] [Google Scholar]
- 8.Capdevila DA, Edmonds KA, Giedroc DP. Metallochaperones and metalloregulation in bacteria. Essays Biochem. 2017;61:177–200. doi: 10.1042/EBC20160076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osman D, et al. Bacterial sensors define intracellular free energies for correct enzyme metalation. Nat. Chem. Biol. 2019;15:241–249. doi: 10.1038/s41589-018-0211-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rae TD, et al. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science. 1999;284:805–808. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- 11.Outten CE, Halloran, Thomas V. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 12.Dann CE, et al. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- 13.Ma Z, Jacobsen FE, Giedroc DP. Coordination chemistry of bacterial metal transport and sensing. Chem. Rev. 2009;109:4644–4681. doi: 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osman D, et al. Fine control of metal concentrations is necessary for cells to discern zinc from cobalt. Nat. Commun. 2017;8:1884. doi: 10.1038/s41467-017-02085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 16.Haas CE, et al. A subset of the diverse COG0523 family of putative metal chaperones is linked to zinc homeostasis in all kingdoms of life. BMC Genomics. 2009;10:470–490. doi: 10.1186/1471-2164-10-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nojiri M, et al. Functional expression of nitrile hydratase in Escherichia coli: requirement of a nitrile hydratase activator and post-translational modification of a ligand cysteine. J. Biochem. 1999;125:696–704. doi: 10.1093/oxfordjournals.jbchem.a022339. [DOI] [PubMed] [Google Scholar]
- 18.Lu J, et al. Motif CXCC in nitrile hydratase activator is critical for NHase biogenesis in vivo. FEBS Lett. 2003;553:391–396. doi: 10.1016/S0014-5793(03)01070-6. [DOI] [PubMed] [Google Scholar]
- 19.Gumataotao N, Lankathilaka KPW, Bennett B, Holz RC. The iron-type nitrile hydratase activator protein is a GTPase. Biochem. J. 2017;474:247–258. doi: 10.1042/BCJ20160884. [DOI] [PubMed] [Google Scholar]
- 20.Blaby-Haas CE, Flood JA, Crecy-Lagard VD, Zamble DB. YeiR: a metal-binding GTPase from Escherichia coli involved in metal homeostasis. Metallomics. 2012;4:488–497. doi: 10.1039/c2mt20012k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sydor AM, et al. Metal binding properties of Escherichia coli YjiA, a member of the metal homeostasis-associated COG0523 family of GTPases. Biochemistry. 2013;52:1788–1801. doi: 10.1021/bi301600z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nairn BL, et al. The response of Acinetobacter baumannii to zinc starvation. Cell Host Microbe. 2016;19:826–836. doi: 10.1016/j.chom.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jordan MR, et al. Mechanistic insights into the metal-dependent activation of Zn(II)-dependent metallochaperones. Inorg. Chem. 2019;58:13661–13672. doi: 10.1021/acs.inorgchem.9b01173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandrangsu P, Huang X, Gaballa A, Helmann JD. Bacillus subtilis FolE is sustained by the ZagA zinc metallochaperone and the alarmone ZTP under conditions of zinc deficiency. Mol. Microbiol. 2019;112:751–765. doi: 10.1111/mmi.14314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crouzet J, et al. Nucleotide sequence and genetic analysis of a 13.1-kilobase-pair Pseudomonas denitrificans DNA fragment containing five cob genes and identification of structural genes encoding Cob(I)alamin adenosyltransferase, cobyric acid synthase, and bifunctional cobinamide kinase-cobinamide phosphate guanylyltransferase. J. Bacteriol. 1991;173:6074–6087. doi: 10.1128/JB.173.19.6074-6087.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis NJ, Nussberger R, Kräutler B, Eschenmoser A. 5,15-bisnorcobester: an unexpected mode of formation. Angew. Chem. Int. Ed. Engl. 1983;22:736–737. doi: 10.1002/anie.198307361. [DOI] [Google Scholar]
- 27.Kieninger C, et al. Zinc substitution of cobalt in vitamin B12: zincobyric acid and zincobalamin as luminescent structural B12-mimics. Angew. Chem. Int. Ed. Engl. 2019;58:14568–14572. doi: 10.1002/anie.201908428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heldt D, et al. Aerobic synthesis of vitamin B12: ring contraction and cobalt chelation. Biochem. Soc. Trans. 2005;33:815–819. doi: 10.1042/BST0330815. [DOI] [PubMed] [Google Scholar]
- 29.Roth JR, Lawrence JG, Bobik TA. Cobalamin (coenzyme B12): synthesis and biological significance. Annu. Rev. Microbiol. 1996;50:137–181. doi: 10.1146/annurev.micro.50.1.137. [DOI] [PubMed] [Google Scholar]
- 30.Gille D, Schmid A. Vitamin B12 in meat and dairy products. Nutr. Rev. 2015;73:106–115. doi: 10.1093/nutrit/nuu011. [DOI] [PubMed] [Google Scholar]
- 31.Antony AC. Vegetarianism and vitamin B12 (cobalamin) deficiency. Am. J. Clin. Nutr. 2003;78:3–6. doi: 10.1093/ajcn/78.1.3. [DOI] [PubMed] [Google Scholar]
- 32.Martens JH, Barg H, Warren M, Jahn D. Microbial production of vitamin B12. Appl. Microbiol. Biotechnol. 2002;58:275–285. doi: 10.1007/s00253-001-0902-7. [DOI] [PubMed] [Google Scholar]
- 33.McGoldrick HM, et al. Identification and characterization of a novel vitamin B12 (cobalamin) biosynthetic enzyme (CobZ) from Rhodobacter capsulatus, containing flavin, heme, and Fe-S cofactors. J. Biol. Chem. 2005;280:1086–1094. doi: 10.1074/jbc.M411884200. [DOI] [PubMed] [Google Scholar]
- 34.Deery E, et al. An enzyme-trap approach allows isolation of intermediates in cobalamin biosynthesis. Nat. Chem. Biol. 2012;8:933–940. doi: 10.1038/nchembio.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kieninger C, et al. The hydrogenobyric acid structure reveals the corrin ligand as an entatic state module empowering B12 cofactors for catalysis. Angew. Chem. Int. Ed. Engl. 2019;58:10756–10760. doi: 10.1002/anie.201904713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Debussche L, et al. Assay, purification, and characterization of cobaltochelatase, a unique complex enzyme catalyzing cobalt insertion in hydrogenobyrinic acid a,c-diamide during coenzyme B12 biosynthesis in Pseudomonas denitrificans. J. Bacteriol. 1992;174:7445–7451. doi: 10.1128/JB.174.22.7445-7451.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.VanZile ML, Cosper NJ, Scott RA, Giedroc DP. The zinc metalloregulatory protein Synechococcus PCC7942 SmtB binds a single zinc ion per monomer with high affinity in a tetrahedral coordination geometry. Biochemistry. 2000;39:11818–11829. doi: 10.1021/bi001140o. [DOI] [PubMed] [Google Scholar]
- 38.Xiao Z, Wedd AG. The challenges of determining metal-protein affinities. Nat. Prod. Rep. 2010;27:768–789. doi: 10.1039/b906690j. [DOI] [PubMed] [Google Scholar]
- 39.Kwan C-Y, Putney J. Uptake and intracellular sequestration of divalent cations in resting and methacholine-stimulated mouse lacrimal acinar cells: dissociation by Sr(II) and Ba(II) of agonist-stimulated divalent cation entry from the refilling of the agonist-sensitive intracellular pool. J. Biol. Chem. 1990;265:678–684. doi: 10.1016/S0021-9258(19)40103-8. [DOI] [PubMed] [Google Scholar]
- 40.Young TR, Wedd AG, Xiao Z. Evaluation of Cu(i) binding to the E2 domain of the amyloid precursor protein – a lesson in quantification of metal binding to proteins via ligand competition. Metallomics. 2018;10:108–119. doi: 10.1039/C7MT00291B. [DOI] [PubMed] [Google Scholar]
- 41.Finney LA, O’Halloran TV. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science. 2003;300:931–936. doi: 10.1126/science.1085049. [DOI] [PubMed] [Google Scholar]
- 42.Foster AW, et al. A tight tunable range for Ni(II) sensing and buffering in cells. Nat. Chem. Biol. 2017;13:409–414. doi: 10.1038/nchembio.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hitomi Y, Outten CE, O’Halloran TV. Extreme zinc-binding thermodynamics of the metal sensor/regulator protein, ZntR. J. Am. Chem. Soc. 2001;123:8614–8615. doi: 10.1021/ja016146v. [DOI] [PubMed] [Google Scholar]
- 44.Waugh R, Boxer DH. Pleiotropic hydrogenase mutants of Escherichia coli K12: growth in the presence of nickel can restore hydrogenase activity. Biochimie. 1986;68:157–166. doi: 10.1016/S0300-9084(86)81080-X. [DOI] [PubMed] [Google Scholar]
- 45.Sydor AM, Lebrette H, Ariyakumaran R, Cavazza C, Zamble DB. Relationship between Ni(II) and Zn(II) coordination and nucleotide binding by the Helicobacter pylori [NiFe]-hydrogenase and urease maturation factor HypB. J. Biol. Chem. 2014;289:3828–3841. doi: 10.1074/jbc.M113.502781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuen, M. H., Fong, Y. H., Nim, Y. S., Lau, P. H. & Wong, K. -B. Structural insights into how GTP-dependent conformational changes in a metallochaperone UreG facilitate urease maturation. Proc. Natl. Acad. Sci. USA114, E10890–E10898 (2017). [DOI] [PMC free article] [PubMed]
- 47.Rodionov DA, Hebbeln P, Gelfand MS, Eitinger T. Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: evidence for a novel group of ATP-binding cassette transporters. J. Bacteriol. 2006;188:317–327. doi: 10.1128/JB.188.1.317-327.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ellman GL. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 49.Riddles PW, Blakeley RL, Zerner B. Ellman’s reagent: 5,5′-dithiobis(2-nitrobenzoic acid)—a reexamination. Anal. Biochem. 1979;94:75–81. doi: 10.1016/0003-2697(79)90792-9. [DOI] [PubMed] [Google Scholar]
- 50.Stookey LL. Ferrozine: a new spectrophotometric reagent for iron. Anal. Chem. 1970;42:779–781. doi: 10.1021/ac60289a016. [DOI] [Google Scholar]
- 51.Xiao Z, Gottschlich L, van der Meulen R, Udagedara SR, Wedd AG. Evaluation of quantitative probes for weaker Cu(i) binding sites completes a set of four capable of detecting Cu(i) affinities from nanomolar to attomolar. Metallomics. 2013;5:501–513. doi: 10.1039/c3mt00032j. [DOI] [PubMed] [Google Scholar]
- 52.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal. Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 53.Golynskiy MV, Gunderson WA, Hendrich MP, Cohen SM. Metal binding studies and EPR spectroscopy of the manganese transport regulator MntR. Biochemistry. 2006;45:15359–15372. doi: 10.1021/bi0607406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cortes L, Roberts BR, Wedd AG, Xiao Z. Molecular aspects of a robust assay for ferroxidase function of ceruloplasmin. Inorg. Chem. 2017;56:5275–5284. doi: 10.1021/acs.inorgchem.7b00372. [DOI] [PubMed] [Google Scholar]
- 55.Jefferson JR, Hunt JB, Ginsburg A. Characterization of indo-1 and quin-2 as spectroscopic probes for Zn(II)-protein interactions. Anal. Biochem. 1990;187:328–336. doi: 10.1016/0003-2697(90)90465-L. [DOI] [PubMed] [Google Scholar]
- 56.Reyes-Caballero H, Lee CW, Giedroc DP. Mycobacterium tuberculosis NmtR harbors a nickel sensing site with parallels to Escherichia coli RcnR. Biochemistry. 2011;50:7941–7952. doi: 10.1021/bi200737a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simons TJB. Measurement of free Zn2+ ion concentration with the fluorescent probe mag-fura-2 (furaptra) J. Biochem. Biophys. Methods. 1993;27:25–37. doi: 10.1016/0165-022X(93)90065-V. [DOI] [PubMed] [Google Scholar]
- 58.Hunt JB, Neece SH, Ginsburg A. The use of 4-(2-pyridylazo)resorcinol in studies of zinc release from Escherichia coli aspartate transcarbamoylase. Anal. Biochem. 1985;146:150–157. doi: 10.1016/0003-2697(85)90409-9. [DOI] [PubMed] [Google Scholar]
- 59.Xiao Z, et al. Unification of the copper(I) binding affinities of the metallo-chaperones Atx1, Atox1, and related proteins: DETECTION PROBES AND AFFINITY STANDARDS. J. Biol. Chem. 2011;286:11047–11055. doi: 10.1074/jbc.M110.213074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raux E, Schubert H, Warren M. Biosynthesis of cobalamin (vitamin B12): a bacterial conundrum. Cell. Mol. Life Sci. 2000;57:1880–1893. doi: 10.1007/PL00000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McGoldrick H, Deery E, Warren M, Heathcote P. Cobalamin (vitamin B12) biosynthesis in Rhodobacter capsulatus. Biochem. Soc. Trans. 2002;30:646–648. doi: 10.1042/bst0300646. [DOI] [PubMed] [Google Scholar]
- 62.Jiang Y, et al. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 2015;81:2506. doi: 10.1128/AEM.04023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 64.Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003;339:62–66. doi: 10.1016/S0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 65.Raux E, et al. Salmonella typhimurium cobalamin (vitamin B12) biosynthetic genes: functional studies in S. typhimurium and Escherichia coli. J. Bacteriol. 1996;178:753–767. doi: 10.1128/JB.178.3.753-767.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hill J, Pratt J, Williams R. The chemistry of vitamin B12. Part I: the valency and spectrum of the coenzyme. J. Chem. Soc. 1964;987:5149–5153. doi: 10.1039/jr9640005149. [DOI] [Google Scholar]
- 67.Waterhouse A, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khil PP, et al. Crystal structure of the Escherichia coli YjiA protein suggests a GTP‐dependent regulatory function. Proteins Struct. Funct. Bioinform. 2004;54:371–374. doi: 10.1002/prot.10430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All data are available within the article, its Supplementary Information files, plus PDB entry 1NIJ. Source data are provided with this paper.
Equation derivations, Excel spreadsheet (with instructions) constituting a metalation calculator, MATLAB code (with instructions) for use in B12 assays and Dynafit scripts are available in Supplementary Note 1, Supplementary Data 1, Supplementary Software 1 and Supplementary Software 2, respectively.