Abstract
Oral squamous cell carcinomas (OSCCs) are the most common cancers of the oral cavity, but the molecular mechanisms driving OSCC carcinogenesis remain unclear. Our group previously established a murine OSCC model based on a 10-week carcinogen [4-nitroquinoline 1-oxide (4-NQO)] treatment. Here we used K14CreERTAM;Rosa26LacZ mice to perform lineage tracing to delineate the mutational profiles in clonal cell populations resulting from single, long-lived epithelial stem cells, here called LacZ+ stem cell clones (LSCCs). Using laser-capture microdissection, we examined mutational changes in LSCCs immediately after the 10-week 4-NQO treatment and >17 weeks after 4-NQO treatment. We found a 1.8-fold ±0.4 (P = 0.009) increase in single-nucleotide variants and insertions/deletions (indels) in tumor compared with pre-neoplastic LSCCs. The percentages of indels and of loss of heterozygosity events were 1.3-fold±0.3 (P = 0.02) and 2.2-fold±0.7 (P = 0.08) higher in pre-neoplastic compared with tumor LSCCs. Mutations in cell adhesion- and development-associated genes occurred in 83% of the tumor LSCCs. Frequently mutated genes in tumor LSCCs were involved in planar cell polarity (Celsr1, Fat4) or development (Notch1). Chromosomal amplifications in 50% of the tumor LSCCs occurred in epidermal growth factor receptor, phosphoinositide 3-kinase and cell adhesion pathways. All pre-neoplastic and tumor LSCCs were characterized by key smoking-associated changes also observed in human OSCC, C>A and G>T. DeconstructSigs analysis identified smoking and head and neck cancer as the most frequent mutational signatures in pre-neoplastic and tumor LSCCs. Thus, this model recapitulates a smoking-associated mutational profile also observed in humans and illustrates the role of LSCCs in early carcinogenesis and OSCCs.
Introduction
Oral squamous cell carcinoma (OSCC), a subtype of head and neck squamous cell carcinoma (HNSCC), is the most common form of oral cancer (1) and is characterized by high invasive capacity to neighboring tissues, metastatic potential to distant organs (2) and an overall 5-year survival of lower than 50% (3). Tobacco use is a well-established risk factor that predisposes to OSCCs (4) that are associated with a high mutagenic potential (5). Recent estimates of oral cavity and pharyngeal cancer indicate 51 540 new cases in 2018 in the USA, with 30% of these cancers in the tongue (6). Many current therapies for HNSCC target epidermal growth factor receptor (Egfr) and the phosphoinositide 3-kinase (Pi3k) pathway because of their frequent overexpression in these tumors (7). Lifelong smokers with tongue tumors usually exhibit a higher number of mutations than younger and occasional or mild smokers (8), indicating that the combination of exposure to the carcinogenic effects of cigarette smoking over a longer period and aging (9) plays a major role in carcinogenesis.
The typical mutational profile of smokers with OSCC shows recurrent C:G>A:T transversions found at 5′-GCX sites (10), G>T transversions (11), an increase in C>A and A>T substitutions (8) and frequent TP53 and NOTCH1 mutations (12). However, despite these established mutational signatures that include tumors of the head and neck area, there are some patients with a history of smoking whose tongue tumors do not fully align with the previously established mutational signatures (8), suggesting that carcinogenesis in different regions of the head and neck exposed to tobacco may result from different molecular mechanisms.
Our group has developed and characterized a mouse model of oral carcinogenesis by using tamoxifen-treated transgenic K14CreErTAM;Rosa26LacZ mice treated with the carcinogen 4-nitroquinoline-1-oxide (4-NQO), a surrogate of tobacco. 4-NQO-associated changes share similarities with tobacco-associated tissue changes; for example, an increase in oxidative stress markers (13), a gradual increase in Bcl2 and Bax proteins (14), decreased expression of tumor suppressors, such as p16 and p53 (15), and changes in cellular adhesion protein dynamics are seen with 4-NQO treatment (16). By using this model we can follow the fates of permanently marked, long-lived LacZ+ stem cells of the tongue epithelium. These LacZ+ stem cells are representative of all long-lived epithelial stem cells in the basal layer (17). Using this lineage tracing approach, our group demonstrated that the LacZ+ stem cells and their progeny (LSCCs) can be followed during the pre-neoplastic stages and during the formation of OSCCs. We also showed that some cells in these tumors originated from these long-lived stem cells in the basal layer of the tongue epithelium (17). Using this 4-NQO murine model, we demonstrated that these LacZ+ stem cells in the basal layer of the epithelium, which normally divide asymmetrically (18), undergo some symmetric cell divisions following 4-NQO treatment (17).
4-NQO, like tobacco-associated carcinogens, forms DNA adducts preferentially on G and A sites (19). However, there are limited data on the impact of 4-NQO mutagenesis at a genome-wide level (19). Moreover, it is not clear how 4-NQO-induced mutagenesis leads to frank tumors in some cell types, but not others. Furthermore, we do not have a clear understanding of which mutations are needed to develop a tumor as opposed to a pre-neoplastic lesion.
Here, we delineated the mutational profiles in clones of cells resulting from single, long-lived tongue epithelial stem cells in pre-neoplastic tissues and in OSCCs. Our data define the mutational composition of OSCCs in this model and provide information about why only some stem-cell clones with mutations develop into frank OSCC (20).
Materials and methods
Mice
We crossed double-positive K14-CreERTAM (cat. no 56822) transgenic mice with ROSA26 floxed STOP-LacZ (cat. no 003474) (Jackson Laboratory, Bar Harbor, ME) double-positive transgenic mice to obtain K14CreERTAM;ROSA26 mice (17). As previously reported by our group (16,17), we treated 6-week-old K14CreERTAM;ROSA26 female mice with tamoxifen (4 mg/mouse/day) by intraperitoneal injection for two consecutive days, causing the transient activation of the CreER recombinase in the K14-positive cells of the basal tongue epithelium and the subsequent permanent expression of LacZ in these long-lived stem cells and their progeny (LSCCs, LacZ+ stem cell clones) (17). Thus, following the long-term fates of the LSCCs of the tongue epithelium will provide new information about the role of these cells in tongue tumor development and progression.
Ethical statement
The experimental protocols and animal handling were approved by the Institutional Animal Care and Use Committee (IACUC) of Weill Cornell Medical College.
X-Gal staining and treatments
To identify the LacZ+ cells of the tongue epithelium, we used X-Gal, with some modifications from a previously published method (17) (Figure 1). For details about X-Gal staining and treatments, see the Supplementary Methods, available at Carcinogenesis Online.
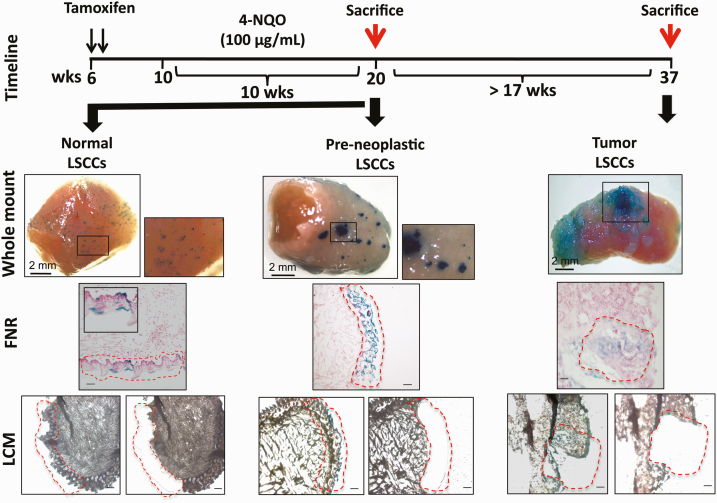
Figure 1.
Timeline of 4-NQO treatment in the tamoxifen-treated K14CreER;Rosa26LacZ mice, and experimental design. Two intraperitoneal injections on two consecutive days in 6-week-old mice transiently activated the truncated K14 promoter-driven expression of CreER in only a small percentage of the long-lived stem cells and their clonal progeny (LSCCs), resulting in permanent LacZ expression in these stem cells and all of their progeny. We visualize these stem cell clones by x-Gal staining. Four weeks after the 2-day tamoxifen treatment, 12 mice received the carcinogen 4-NQO (100 μg/mL) in the drinking water for 10 weeks, whereas six mice received water with no carcinogen (pre-neoplastic, age-matched controls). After 10 weeks of 4-NQO treatment, we sacrificed six mice (red arrow referring to the control and pre-neoplastic LSCCs) and returned to regular water without 4-NQO the remaining 6 mice; these mice developed tongue squamous cell carcinomas after ≈17 weeks and were then sacrificed (red arrow referring to the tumor LSCCs). For each time point, the figure shows an example of a whole-mount tongue after x-Gal staining; the x-Gal+ tongue sections counterstained with fast nuclear red (FNR); and representative consecutive tongue sections on which we performed LCM. The red dashed areas highlight the x-Gal+ LSCCs before and after LCM. Whole-mount images, scale bar = 2 mm; in all other images, the scale bar = 50 μm.
Laser capture microdissection
For precise isolation of the X-Gal-positive cell clones of the tongue epithelium, we performed laser-capture microdissection (LCM), which takes advantage of the cutting power of UV lasers and the capturing power of the infrared lasers to isolate small cell populations from a tissue (21). For further details, see Supplementary Methods, available at Carcinogenesis Online.
Genomic DNA isolation
After collection of the cells of interest in the Arcturus Cap Sure Macro LCM Caps (#LCM0211, Life Technologies), we digested the cells in 15-μl digestion buffer and 10-μl Proteinase K provided by the QIAmp DNA Micro Kit (# 56304, Qiagen) for 4 h at 56°C. We extracted genomic DNA (gDNA) from LCM samples using the Qiagen kit following the manufacturer’s protocol. We quantified total gDNA by Qbit Quantitation Platform (Invitrogen Life Sciences) and assessed the quality by using the Bioanalyzer.
We performed exome sequencing using HiSeq4000 (Illumina) using paired-end 100 bp reads. The NCBI BioProject accession numbers for the exome sequencing are PRJNA528979 and PRJNA605291. For detailed information about Library Preparation and Whole-Exome Sequencing, Functional Categories and Mutational Signature Analysis, see Supplementary Methods, available at Carcinogenesis Online.
Copy number alterations
We performed copy number alteration (CNA) analysis in pre-neoplastic and tumor LSCCs using VarScan2 (22). For details, see Supplementary Methods, available at Carcinogenesis Online.
Statistical analysis
We represented bar plots as mean ± standard deviation (SD). For two-group comparisons, we used Student’s t-test, whereas for multiple-group comparisons, we used one-way ANOVA. P < 0.05 was considered statistically significant.
Results
We used a lineage tracing model to characterize mutations in long-lived epithelial stem cells and their clonal progeny
The mutational changes accumulating over time in OSCC are not fully understood, and this lack of understanding limits our ability to find new, targeted therapeutic strategies to reduce the burden of this malignancy. Therefore, we took advantage of the 4-NQO carcinogenesis model (23), established by our laboratory and used a lineage tracing approach in K14CreErTAM;Rosa26LacZ mice (17) to delineate the mutational profiles of cell clones, each clone derived from a single, long-lived LacZ+ stem cell (here called LSCC, LacZ+ stem cell clones), at defined times during tumor development.
We dissected the LSCCs by LCM (24) in cohorts of mice either treated for 10 weeks with the carcinogen 4-NQO or not (control tongues) (Figure 1). X-Gal whole-mount staining, which marks the LacZ+ cells of the control tongues, showed a typical dot-like, blue pattern across the entire tongue dorsal surface (Figure 1), indicating X-Gal staining of individual, long-lived stem cells and their progeny, LSCCs, that arise primarily from asymmetric division of these stem cells (17). In mice that have not been treated with carcinogen, we observed blue staining extending from the K14+ basal layer to the top keratinized layers (Figure 1) (17,18). We performed LCM of LSCCs, each derived from one stem cell, in six control mice, and isolated gDNA for exome sequencing.
Next, we focused on 12 mice treated with 4-NQO for 10 weeks, as outlined in Methods and Figure 1. X-Gal staining at the end of 10 weeks of 4-NQO treatment (six mice) consistently showed larger, blue-stained epithelial regions with areas ranging from 0.12 to 0.4 mm2. The expanded blue areas are consistent with a model of stem cell proliferation by symmetric cell division during 4-NQO treatment (Figure 1) (17,18). Based on our histological evaluations of the tissue sections at this time point, the tongue epithelia at the 10th and last week of 4-NQO treatment showed scattered areas of hyperplasia, necrosis and foci of mild inflammation, but no frank tumors (Supplementary Figure 2, available at Carcinogenesis Online). We did not detect dysplastic areas; the basal epithelial stem cells showed regular margins and nuclei did not indicate the presence of tumors. Thus, the tongues analyzed at the end of the 10-week 4-NQO treatment are informative with respect to the mutations that may precede tumor formation. We refer to these samples as pre-neoplastic (PN) samples. We collected the blue x-Gal+ areas from 15 to 20 consecutive tongue sections per mouse by LCM (Figure 1) and isolated gDNA for exome sequencing.
By 17–23 weeks following the end of the 10-week 4-NQO treatment (Figure 1), the mice showed large tumors on the tongue surface. Histopathologic evaluation confirmed the presence of squamous cell carcinomas in all six mice; the LSCC samples (T1, T2, T3 and T6) had invasive foci, whereas two samples (T4 and T5) did not (Supplementary Figure 2, available at Carcinogenesis Online). We dissected each LSCC by LCM (Figure 1) from five to eight consecutive sections and retrieved gDNA for exome sequencing of the tumor LSCCs.
We characterized the pre-neoplastic- and tumor-associated mutations and insertions/deletions (indels) in each LSCC by exome sequencing
To examine the single-nucleotide variants (SNVs) and indels present in each of the LSCCs, we performed whole exome sequencing of LSCCs of different sizes in the 12 tongues from mice treated with the carcinogen 4-NQO for 10 weeks and from LSCCs in tongues of six mice of same genetic background used as controls (Figure 1).
After genome alignment, we identified somatic mutations in pre-neoplastic (n = 6) and tumor LSCCs (n = 6). In the pre-neoplastic LSCCs, we achieved an average of 48× coverage (range: 31× to 60×) on captured exome regions, and 61.9% bases (range: 49.2%–69.2%) had more than 20× coverage (Supplementary Table 1, available at Carcinogenesis Online). In the tumor LSCCs, we achieved an average of 55× coverage (range: 46× to 68×) on captured exome regions, and 73.5% bases (range: 61.5%–81.9%) had more than 20× coverage (Supplementary Table 1, available at Carcinogenesis Online). Based on these data, the numbers of the captured exome regions and their coverage in the pre-neoplastic LSCCs were comparable to those from tumor LSCCs, allowing us to determine and compare the mutational profiles from both groups.
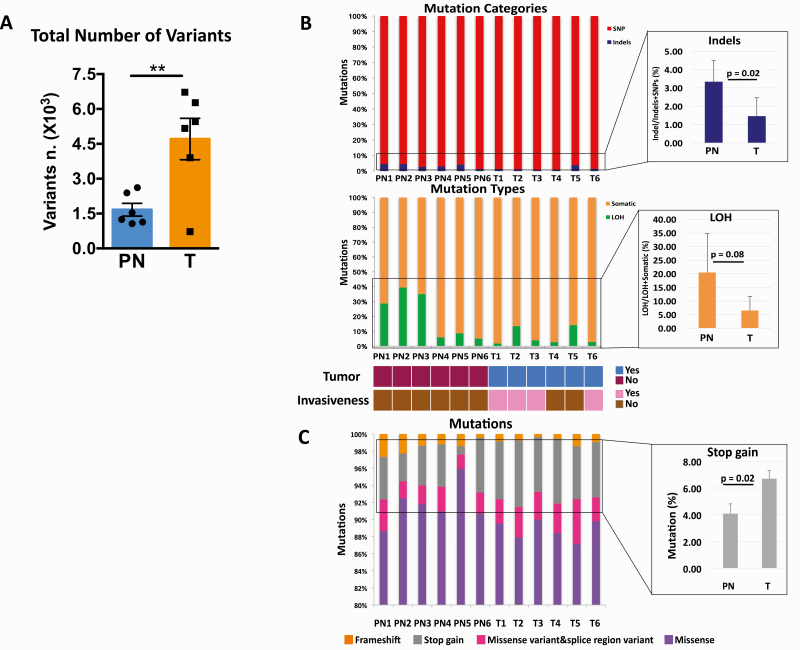
The LSCCs in pre-neoplastic samples showed an average of 1,664 ± 614 SNVs and indels, whereas the LSCCs in tumors showed an average of 4,707 ± 1992 SNVs and indels, indicating a 1.8-fold (± 0.4; P = 0.009) increase in the total number of SNVs and indels compared with the LSCCs in pre-neoplastic tissues (Figure 2A and B; Supplementary Table 1, available at Carcinogenesis Online). The LSCCs from the tumors with histopathological classifications indicating invasive foci (T1, T2, T3 and T6) (Figure 2B; Supplementary Table 1, available at Carcinogenesis Online) had the highest number of SNVs and indels. These data indicate that the tumor LSCCs accumulated a higher number of mutations compared with the pre-neoplastic LSCCs and that within tumor LSCCs, the numbers of mutations increase with tumor invasiveness.
Figure 2.
Exome sequencing of LCM-isolated LSCCs. The total numbers of single-nucleotide variants (SNVs) and insertions/deletions (indels) after exome sequencing (A). Mutation categories that indicate single nucleotide variants (SNVs) and insertions/deletions (Indels), as well as mutation types that show somatic and loss of heterozygosity (LOH) events in the LSCCs from pre-neoplastic (PN; n = 6) and tumor samples (T; n = 6). The enlarged blue bar plot shows in detail the different numbers of indels between PN and T expressed as mean ± standard deviation (SD). The enlarged orange bar plot shows in detail the different numbers of LOH and their averaged differences between PN and T, expressed as mean ± standard deviation (SD). The color-coded diagram below the bar plots provides information about the presence or absence of tumor and tumor invasiveness (B). Main classes of mutations identified in PN and T depicting the proportion of missense, stop gain and frameshift mutations in PN and T. The enlarged gray bar plot shows the averaged differences between PN and T expressed as mean ± standard deviation (SD) (C). We calculated the statistical significance between averaged PN and T with a t-test in which *P < 0.05 and **P < 0.001.
The percentages of indels in the LSCCs in pre-neoplastic samples were increased by 1.3-fold (± 0.3; P = 0.02) compared with the LSCCs from the tumors (Figure 2B). We also discovered that the LSCCs in pre-neoplastic samples showed a 2.2-fold (± 0.7; P = 0.08) higher loss of heterozygosity (LOH) events compared with LSCCs from the tumors (Figure 2B). Thus, we identified distinct mutational changes in pre-neoplastic LSCCs compared with tumor LSCCs. These results suggest that the mutational compositions of the tumor LSCCs are characterized by a higher proportion of SNVs compared with LOHs and indels, which accumulate at earlier stages of carcinogenesis.
Using the variant annotation tool SnpEff (25), we classified mutations into high, moderate, low and modifier categories based on their impact on the corresponding proteins. A higher impact predicts a more disruptive effect on the corresponding protein (Supplementary Methods, available at Carcinogenesis Online). Among the SNVs and indels with high and moderate impact (Supplementary Tables 1 and 2, available at Carcinogenesis Online), the most abundant mutations in both pre-neoplastic and tumor LSCCs were missense SNVs, followed by stop gain and frameshift mutations (Figure 2C).
The mutated genes in LSCCs from OSCCs are more frequently associated with adhesion and development than the LSCCs from pre-neoplastic lesions
To find potential enrichment indicative of selection across pre-neoplastic and tumor LSCCs, we examined the high- and moderate-impact SNVs and indels with WebGestalt (www.webgestalt.org) (26) (Supplementary Methods, available at Carcinogenesis Online). We focused on the top 10 functional categories in all samples.
Before analyzing the functional categories in pre-neoplastic and tumor LSCCs, we ensured that our approach to align the tumor samples to the pre-neoplastic age-matched controls was appropriate and that the differences in the genomes of 20-week-old healthy mice (age-matched with the pre-neoplastic samples) and 37-week-old healthy mice (age-matched with the tumor samples) were not affecting the functional category analysis by Webgestalt. We aligned one representative tumor sample (T6) to both the pre-neoplastic age-matched controls and the tumor age-matched controls and compared the first 10 functional categories. The comparison of the two alignments resulted in almost identical functional categories (Supplementary Figure 3, available at Carcinogenesis Online), indicating that there were no substantial differences among control LSCCs from mice of different ages.
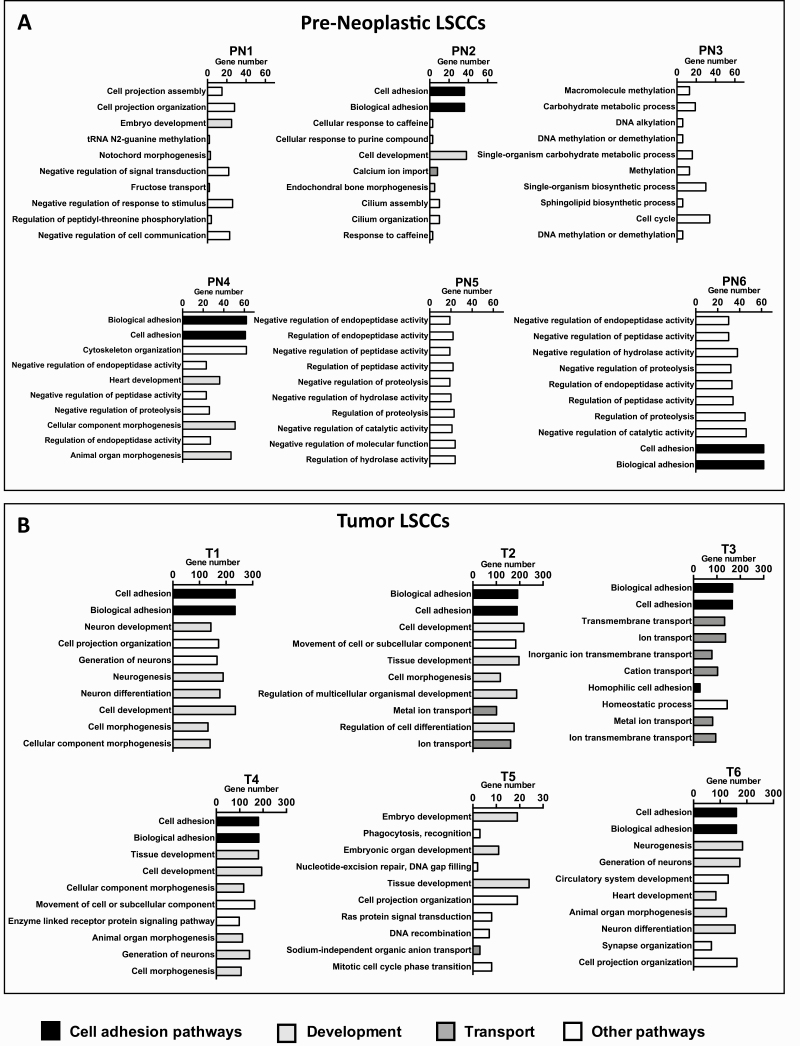
Most of the functional categories among the pre-neoplastic LSCCs lacked a common mutational profile, with the exception of cell-adhesion-related functional categories present in 50% of these samples (Figure 3A). The majority of the tumor LSCCs (83% in T1, T2, T3, T4 and T6) had mutations in genes associated with cell adhesion and biological adhesion in the top two categories (Figure 3B). In contrast, LSCC T5 did not show the cell adhesion-related category, indicating that this LSCC had a different mutational composition (Figure 3B). The second highest functional category, in 83% of the tumor LSCCs (T1, T2, T4, T5 and T6), was development-related (Figure 3B). The only tumor LSCC that did not display any of the development-associated categories was T3, which instead showed 6 of 10 functional categories related to transport, including transmembrane transport, ion transport and cation transport (Figure 3B). These data indicate that, unlike the pre-neoplastic LSCCs, the tumor LSCCs exhibit many mutations in cell adhesion- and development-related pathways.
Figure 3.
Functional categories of the mutated genes with high- and moderate-impact mutations. The image shows the functional categories of the mutated genes resulting from the WebGestalt analysis (Supplementary Methods). Functional categories of the pre-neoplastic LSCCs (PN; n = 6) (A) and the tumor LSCCs (T; n = 6) analyzed (B).
Mutated genes involved in cell adhesion and development
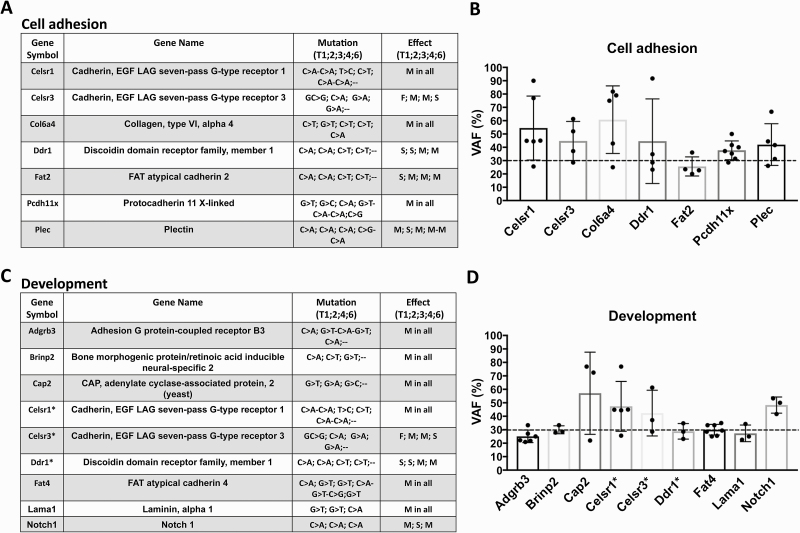
To determine whether the categories cell adhesion and development contained recurrent SNVs and indels indicative of specific pathways with potential roles in OSCC carcinogenesis, we next analyzed the mutated genes found in the majority of the tumor LSCCs in greater detail (Figure 4). We found that in the 83% of the tumor LSCCs that showed cell adhesion-related functional categories, the SNVs in the genes Celsr1, Celsr3, Col6a4, Ddr1, Fat2, Pcdh11x and Plec were predominantly C>A substitutions that caused the majority of missense and stop gain mutations (Figure 4A).
Figure 4.
Cell adhesion and development frequently mutated genes in the tumor LSCCs. Cell adhesion-associated genes and relative mutation substitutions, as well as the predicted effect associated with each mutation (M = missense; F = frameshift; S = stop codon gain) found in 83% of the tumor LSCCs (A). For each of the cell adhesion-associated genes, the figure shows the variant allele frequency (VAF) % of all substitutions found as mean ± standard deviation (SD) (B). Development-associated genes and relative substitutions with predicted effect associated with each mutation found in 75% of the LSCCs from the tumor samples (C). For each of the development-associated genes, the figure shows the variant allele frequency (VAF) % of all substitutions found as mean ± standard deviation (SD) (D). Each dot in the graphs represents one substitution, as indicated in the tables. The dashed line in the bar plots indicates the 30% VAF cutoff.
We then analyzed the mutated genes found in at least 75% of the tumor LSCCs that showed development-associated functional categories and identified six genes, Adgrb3 (BAI3), Brinp2, Cap2, Fat4, Lama1 and Notch1, that showed a predominance of C>A substitutions (Figure 4C). We also identified three genes, Celsr1, Celsr3 and Ddr1, that overlapped with the previous category, cell adhesion (Figure 4A and C).
Because Celsr1, Celsr3 and Ddr1 were frequently mutated in our analyses, we interrogated the Human Protein Atlas database (http://www.proteinatlas.org) to determine whether the mRNA expression levels of these genes had prognostic value in human HNSCCs (27). Among 499 patients with HNSCC, we found no prognostic value for Celsr1 (P = 0.18). However, patients with HNSCC overexpressing Celsr3 showed increased survival rates (P = 0.000002), whereas patients overexpressing Ddr1 exhibited decreased survival rates (P = 0.008) (Supplementary Figure 4, available at Carcinogenesis Online). Because OSCCs are one subtype of HNSCC, these data potentially implicate these genes in oral carcinogenesis.
There is growing evidence that the variant allele frequency (VAF) percentage, the ratio between the mutated reads and the total number of reads in a given locus, may provide a better indication of the potential roles of some mutations in various tumor types (28). The expected VAF % for a heterozygous variant should be close to 50% (29). Therefore, to evaluate the potential relevance of the mutated genes in the tumor LSCCs, we tracked the VAFs % of the mutated genes belonging to the top functional categories, cell adhesion (Figure 4A) and development (Figure 4C). The majority of the mutations found in these functional categories showed a VAF > 30% (Figure 4B and D), indicating the presence of such mutations in the majority of the LSCCs from tumors. In some mutated genes, such as Pcdh11x, involved in cell adhesion, and Notch1, involved in development and frequently mutated in human oral carcinomas (20,30,31), all VAFs were >30% (Figure 4B and D). Notch1 showed mutations in exons 32 (T1), 26 (T2) and 20 (T4) (Supplementary Table 2, available at Carcinogenesis Online). The Notch1 mutation in exon 32, located in an intracellular Ankyrin domain (32), is a missense variant with a VAF of 53.33%, whereas the mutation in exon 26 is a stop gain in a heterodimerization-N terminal domain with a VAF of 41.67%. The Notch1 mutation found in exon 32, located in an epidermal growth factor-like domain (EGF like) in the extracellular region of Notch1 (32), is a missense mutation with a VAF of 50%. In human head and neck cancer, Notch1 mutations occur mainly in the EGF-like domain (33). Notch mutations in human HNSCC are generally inactivating mutations (12). The presence of a mutated Notch1 in tumor LSCCs suggests that the 4-NQO model recapitulates some aspects of human oral carcinogenesis. Thus, defining the mutations occurring in these LSCCs can elucidate the potential contribution of each mutation in OSCC carcinogenesis.
We took the same approach to analyze the LSCCs from pre-neoplastic samples. Because there were no common functional categories, we did not detect any predominant mutations in these LSCCs. However, the pre-neoplastic LSCC sample 2 (PN2) displayed cell adhesion-related functional categories and a missense mutation in a gene, Lama1, that also exhibited mutations in 50% of the tumor LSCCs (Figure 4C and D). Lama1 showed an LOH in chromosome 17, at position 67795272 (A>G; VAF = 95%), indicating that the pre-neoplastic LSCCs had lost one allele compared with the control samples (Supplementary Table 2–4, available at Carcinogenesis Online). Thus, despite the lack of common functional categories within the LSCCs from pre-neoplastic tissues, one LSCC sample shared the top functional category, cell adhesion, with tumor LSCCs.
To complement the WebGestalt analysis, we also manually surveyed the data to find potentially relevant genes that did not map in any of the WebGestalt categories. With this approach, we found that 67% of the pre-neoplastic LSCCs and 83% of the tumor LSCCs exhibited mutations in obscurin (Obscn) (Supplementary Figure 5 and Supplementary Table 5, available at Carcinogenesis Online), a protein involved in cell morphology and signaling pathways (34).
Somatic CNAs involve the Pi3k, Egfr and cell adhesion pathways
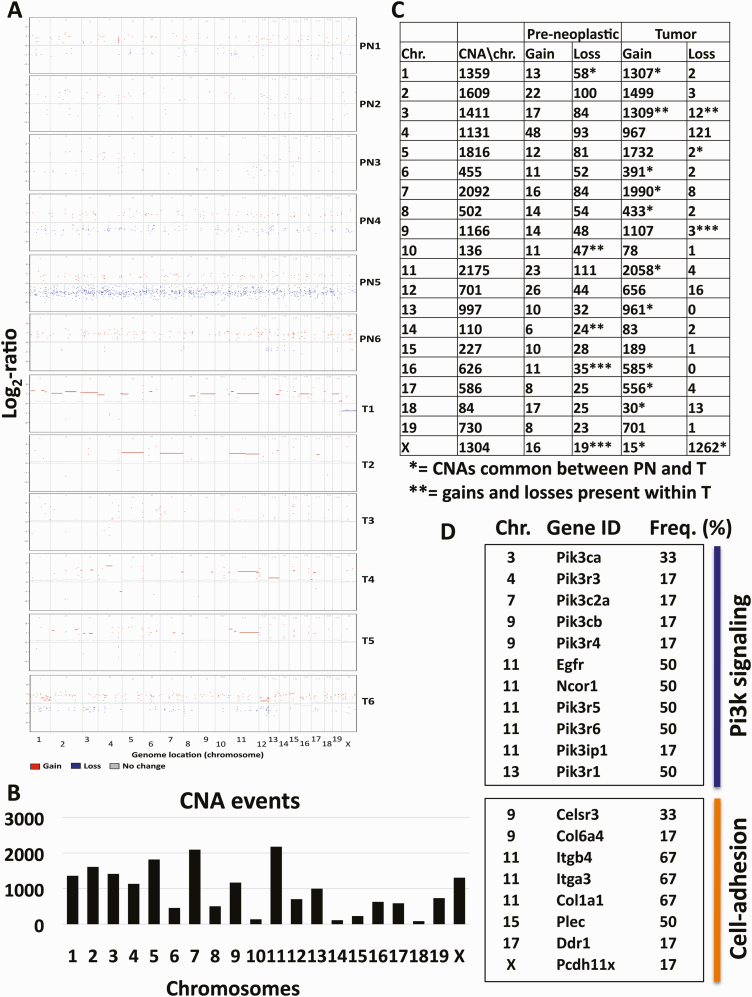
Genome-wide analyses of multiple cancers, including over 300 HNSCC human cases, showed that CNAs play a major role in HNSCC carcinogenesis (35). Thus, we characterized CNAs in our mouse model of OSCC carcinogenesis. We identified CNAs, including amplifications and losses, indicating chromosomal rearrangements, in pre-neoplastic LSCCs (Figure 5). We detected >100-fold more CNAs in tumor when compared with pre-neoplastic LSCCs (Figure 5; Supplementary Table 5, available at Carcinogenesis Online). In addition, all tumor LSCCs except T3 and T6 (67%) showed amplifications in most exons analyzed on chromosome 11 (Figure 5A), whereas we observed no common chromosomal areas with amplifications in the pre-neoplastic LSCCs. These results indicate that the great majority of chromosomal changes occur in the later stages of carcinogenesis, that such changes are amplifications, and that chromosome 11 includes amplifications common to the majority of tumor LSCCs (Figure 5).
Figure 5.
Somatic CNA analysis in pre-neoplastic and tumor LSCCs. The image shows distribution of CNAs in pre-neoplastic (PN) and tumor (T) LSCCs across 56 Mb of the exome-capture regions per chromosome. Each point is expressed as a log2-ratio value with a cutoff of log2-ratio > 0.25. Red dots indicate amplifications, blue dots losses and gray dots no changes compared with matched control samples (A). Representation of CNA events for each mouse chromosome (B). Total numbers of CNA events throughout the exome of the mouse genome (C). Selected genes showing copy number amplifications in the tumor LSCCs organized by chromosome (Chr.), Gene ID, and Frequency (%) (D).
Egfr, Ncor1, Pik3r5, Pik3r6 and Pik3ip1, some of the genes that show CNAs in human HNSCC (7), are located on mouse chromosome 11. We detected amplifications in these genes, as well as in other key genes of the Pi3k pathway, such as Pik3ca (Chr. 3), Pik3r3 (Chr. 4), Pik3c2a (Chr. 7), Pik3cb and Pik3r4 (Chr. 9) and Pik3r1 (Chr. 13) (Figure 5D; Supplementary Tables 5 and 6, available at Carcinogenesis Online). This CNA analysis also uncovered similarities between our murine OSCC carcinogenesis model and human HNSCC.
Several genes in the cell adhesion pathway (Itgb4, Itga3 and Col1a1) located on chromosome 11 displayed amplifications in 67% of the tumor LSCCs. We also identified amplifications in genes, such as Celsr3 and Col6a4 (Chr. 9), Plec (Chr.15), Ddr1 (Chr. 17) and Pcdh11x (Chr. X), that we previously highlighted because of the presence of SNVs and indels with VAFs > 30% in the majority of the tumor LSCCs (Figure 4). These findings indicate that the cell adhesion-associated genes are altered not only by SNVs and indels, but also by chromosomal rearrangements, and the data underscore the importance of the cell adhesion pathway in this OSCC model.
4-NQO treatment results in C>A and G>T transversions in LSCCs from both pre-neoplastic and tumor tissues
4-NQO confers a high mutagenic potential, with a preference for binding guanine (G), and mimics smoking- and tobacco consumption-associated carcinogenesis (36). Because C>A is one of the most common transversions in humans with a history of smoking and therefore is also present in a high proportion of human head and neck cancers (HNSCC) (8), we performed a comprehensive query of the mouse genome to determine whether 4-NQO showed a preference for inducing C>A substitutions. We found that the tumor LSCCs showed an average of 1045.8 ± 494.4 C>A substitutions (transversions) and an average of 1080.6 ± 512 G>T transversions (Supplementary Figure 6, available at Carcinogenesis Online). If compared with lower abundance base changes, such as A>C (67.6 ± 34.5), C>A and G>T substitutions were increased by 14-fold (± 0.5; P < 0.0001). Interestingly, we found that the pre-neoplastic LSCCs displayed the same type of mutational pattern as the tumor LSCCs, with a predominance of C>A and G>T, but with increases of 3.8-fold (± 0.5; P < 0.0001) compared with A>C, which we used as a representative transversion with a low 4-NQO effect for comparison (Supplementary Figure 6, available at Carcinogenesis Online). This indicates that 4-NQO induces the same type of mutational substitutions observed in human HNSCC and that increases in such substitutions start in LSCCs from pre-neoplastic samples, providing new information about early carcinogenesis.
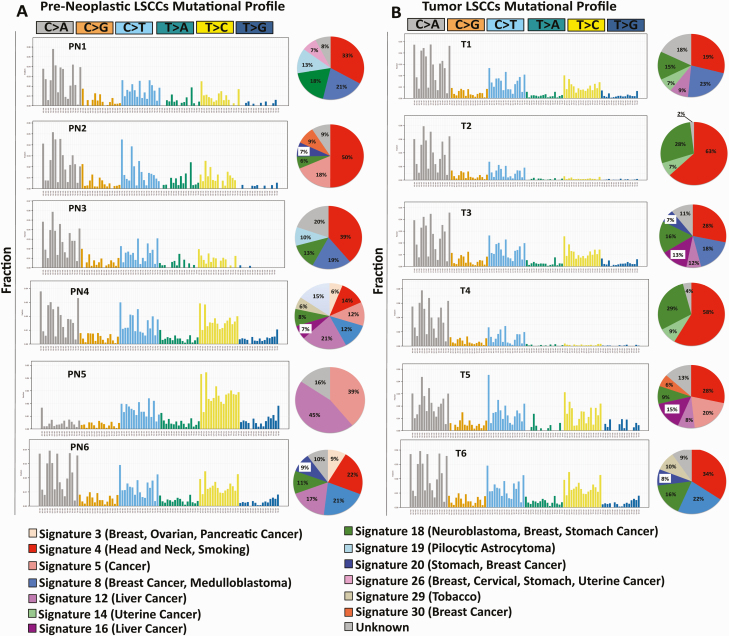
4-NQO confers a mutational signature typical of HNSCC and smoking in LSCCs from both pre-neoplastic and tumor tissues
To examine the translational significance of the 4-NQO murine carcinogenesis model for OSCC and HNSCC, we analyzed the composition of 30 canonical COSMIC signatures (37–39) in our samples using deconstructSigs (Supplementary Methods, available at Carcinogenesis Online). We found that the most frequent mutational signatures in the LSCCs from pre-neoplastic and tumor samples were Signature 4 and Signature 18 (Figure 6A and B) (39). Signature 4 is associated with HNSCC and smoking, based on the COSMIC database from which the signatures originated (38–40), suggesting that genomic alterations in the 4-NQO OSCC carcinogenesis model also occur in humans. Signature 18 is associated with neuroblastoma, breast and stomach cancers, but unlike Signature 4, for which smoking is an established etiologic factor, there is no known etiology associated with Signature 18. Both signatures are characterized by a prevalence of C>A substitutions.
Figure 6.
DeconstructSigs trinucleotide signatures in the pre-neoplastic and tumor LSCCs. The mutational signatures in the pre-neoplastic and tumor LSCCs. As explained in detail in Supplementary Methods, available at Carcinogenesis Online, this analysis infers the composition of 30 canonical COSMIC signatures based on the 96-substitution combination, indicated on the x-axis, whereas on the y-axis, we indicated the fraction of each substitution combination. Next to each signature, the pie diagram shows the frequency of each represented mutational signature, color-coded. The color-coded legend describes the number and name of each mutational signature according to the 30 canonical COSMIC signatures.
Discussion
Lineage tracing and exome sequencing of the LSCCs
We used a lineage-tracing approach by permanently labeling LacZ+ epithelial stem cells in the tongue in a mouse model of OSCC carcinogenesis that utilizes 4-NQO as a carcinogen. We examined the mutational changes in these stem cells and their progeny (LSCCs) at early (pre-neoplastic) and late (tumor) stages of carcinogenesis (Figure 1) by exome sequencing. We found that the LSCCs from tumors exhibited an increased number of SNVs compared with the LSCCs from the pre-neoplastic samples (Figure 2A). We also observed a higher LOH proportion in pre-neoplastic LSCCs compared with tumor LSCCs (Figure 2B), suggesting that accumulation of LOH events characterizes early carcinogenesis and precedes the tumors. Human leukoplakias, benign lesions on the tongue that often precede OSCCs (41), showed an enrichment of LOH events occurring at the chromosomal microsatellites 8p and 9p (41). These chromosomal changes correlated with early development of head and neck cancer (9p), a poor prognosis and greater aggressiveness of this type of tumor (8p) (41).
We noted that the numbers of SNVs and indels were similar in pre-neoplastic LSCCs with different x-Gal+ areas. Sample PN1 had the smallest area analyzed, 0.12 mm2, whereas LSCC samples PN2 and PN3 had larger x-Gal+ areas of 0.4 and 0.36 mm2, respectively. These differences in LSCC sizes did not correlate with distinct mutational signatures.
Functional analyses of the mutated genes
Examination of the functional categories of the mutated genes showed cell adhesion and development in 83% of the tumor LSCCs, whereas we found that cell adhesion was the only recurrent functional category in 50% of the pre-neoplastic LSCCs (Figure 3). These results highlight several key points potentially relevant in OSCC. We found Celsr1, Celsr3 and Ddr1 in the cell adhesion and development functional categories (Figure 4), suggesting that mutations in these genes may alter key signaling pathways leading to OSCC development. Mutations in genes involved in orientation and cell polarization (42) may contribute to OSCC by altering the orientation of the dividing stem cells of the tongue epithelium, as summarized in a model proposed by our group (17). To assess the prognostic value of mutations in these three genes, we surveyed the Human Protein Atlas and found that although Celsr1 overexpression had no prognostic value, Celsr3 mRNA overexpression suggested a better prognosis and Ddr1 overexpression a worse prognosis in patients with HNSCC (Supplementary Figure 4, available at Carcinogenesis Online). Celsr1 and Fat4, both defined as atypical cadherins (43,44), play a central role in controlling planar cell polarity (43). Functionally, Celsr1 may interact with the Wnt signaling pathway in development, but its role is not fully understood (43,44). Likewise, Fat4 controls aspects of planar cell polarity (45). Loss of Fat4 in mice caused disorganization in oriented cell division during kidney development (46). Furthermore, Fat4 knockdown in BGC-823 and HGC-27 gastric cancer cells resulted in increased cell proliferation and epithelial-to-mesenchymal transition (47). Additionally, Fat4 exhibited reduced expression and was associated with a poor prognosis in human gastric cancer specimens (48). Fat4 was frequently mutated in gingivo-buccal OSCCs in an Indian cohort (49), with a 96% predominance of tobacco users, underscoring the relevance of our 4-NQO carcinogenesis model to the mutagenic effects of tobacco and smoking-related OSCC in humans.
CNA analysis in the 4-NQO OSCC carcinogenesis model reveals similarities with human oral cancers
In contrast to SNVs and indels, CNAs affect the functions of broad chromosomal areas and shape the genomic profiles of HNSCC (35). From CNA analysis in our mouse OSCC model, we show that CNA events are interspersed across the mouse chromosomes even in pre-neoplastic LSCCs; that tumor LSCCs exhibited over 100-fold more CNA events, a vast majority of which were amplifications, than pre-neoplastic LSCCs; and that some tumor LSCCs have amplifications in common areas of chromosome 11, areas that include the phosphoinositide 3-kinase pathway genes, the epidermal growth factor receptor gene and cell adhesion-related genes (Figure 5).
Despite the identification of CNA events in pre-neoplastic LSCCs, we did not identify common patterns among pre-neoplastic LSCCs or CNAs in genes already described in other types of cancers. We therefore suggest that CNAs occur in later stages of carcinogenesis or in frank tumors. Our findings show almost no gene losses in the tumor LSCCs, which are instead characterized by gene amplifications.
We found amplification events in the Egfr gene in 50% and in the Pi3k family genes in 17%–50% of the tumor LSCCs (Figure 5), similar to the percentages of these genomic amplifications observed in human HNSCC (50,51). In fact, a recent comprehensive genomic profiling of 243 HPV (human papilloma virus)-negative HNSCC showed Egfr amplifications in 15% of the patients and PIK3CA amplifications in 34%, indicating that amplifications of these genes represent a hallmark of HNSCC (51). Moreover, Egfr protein is overexpressed in 90% of human HNSCC, and Egfr is one of the genes targeted by drugs currently available for HNSCC (7).
Mutational signatures of pre-neoplastic and tumor LSCCs
Our findings showed a predominant smoking-associated signature (13), characterized by C>A substitutions, in both pre-neoplastic and tumor LSCCs (Supplementary Figure 6, available at Carcinogenesis Online). This indicates that many mutational changes caused by 4-NQO were present in pre-neoplastic LSCCs.
One of our main goals was to determine whether tongue epithelia with no evidence of tumors, such as the pre-neoplastic LSCCs, contained mutated genes also found in tumor LSCCs. We discovered that Obscn was mutated in 67% of the pre-neoplastic and 83% of the tumor LSCCs (Supplementary Figure 5, available at Carcinogenesis Online). Human tongue SCCs (31) and several SCC cell lines, such as FaDu, SCC4, SCC15 and SCC25 (52), have Obscn mutations. However, the effects of mutated Obscn in long-term stem cells of the tongue are still undefined. In breast cancers, Obscn loss resulted in increased cell survival (34), implicating Obscn as a tumor suppressor gene. That Obscn mutations are found in pre-neoplastic LSCCs suggests that mutations in Obscn drive OSCC development.
Mutational signature analysis with deconstructSig
To understand better the translational power of the 4-NQO OSCC carcinogenesis model, we utilized deconstructSigs in pre-neoplastic and tumor LSCCs and deconvoluted the composition of 30 COSMIC (Signatures of Mutational Processes in Human Cancer) mutational signatures (39,40). The most frequent mutational signature found in both pre-neoplastic and tumor LSCCs was Signature 4 (Figure 6). Signature 4 is associated with smoking-related C>A mutations (5), head and neck cancer, small cell lung carcinoma, lung squamous carcinoma, liver cancer and esophageal cancer.
Mutated genes common to human head and neck cancer
Among genes with the highest mutation frequency in head and neck SCCs shown in the COSMIC database, p53 (32%; 3124 patients), CDKN2A (11%; 1391 patients) and Notch1 (9%; 863 patients) are the top three. We found that Notch1 was mutated in 75% of tumor LSCCs. Notch1 exhibited a key substitution associated with smoking, C>A, and was characterized by a VAF % higher than 30% in all cases. Recent research conducted in Asian individuals demonstrated that some Notch1 mutations are present in healthy individuals that may have acquired such mutations as early as in infancy (20). However, the authors showed that the strongest factors that led to esophageal tumors were aging, heavy smoking and drinking, which caused further accumulation of Notch1 mutations associated with this tumor (20). These recent data support our findings in which Notch1 is mutated in the 4-NQO carcinogenesis mouse model. We did not detect mutations in two highly mutated genes, according to the COSMIC database, p53 (32%) and Cdkn2a (11%). This could mean that p53 and CDKN2A mutations occur later in OSCC development. It is known that adding a mutation in p53 to the 4-NQO treatment enhances tumorigenesis (53).
In conclusion, we combined a carcinogenesis model with lineage tracing technology to advance our understanding of the roles of mutations in individual long-lived stem cells of the tongue epithelium in OSCC development.
Supplementary material
Supplementary data are available at Carcinogenesis online.
Supplementary Figure 1. The diagram shows the workflow of the experiments, starting from the tongue isolation and followed by fixation in ethanol (ETOH), x-Gal+ staining, and optimal cutting temperature (OCT) embedding of the tongues. After cryostat-sectioning, one slide including the x-Gal+ area is stained with fast nuclear red (FNR) and is used as a guide slide, whereas a consecutive slide is stained with hematoxylin and eosin (H&E) and is used for histopathologic evaluation. The remaining sequential sections (5–20) are placed on LCM-specific polyethylene naphthalate (PEN) membrane slides for genomic DNA (gDNA) isolation and exome sequencing.
Supplementary Figure 2. Representative Hematoxylin and Eosin (H&E) tongue sections of 4-NQO-treated and non-treated mice. Representative control and pre-neoplastic tongue sections, as well as noninvasive and invasive squamous cell carcinomas (SCCs) from tongues taken >17–23 weeks after the end of the 4-NQO treatment. Bar = 100 μm.
Supplementary Figure 3. Functional categories of the mutated genes with high and moderate impact mutations in tumor sample T6. The Image shows the functional categories of the mutated genes resulting from T6 alignment to pre-neoplastic age-matched controls and tumor age-matched controls, as explained in Supplementary Methods. The red arrows indicate the shared functional categories between the two analyses.
Supplementary Figure 4. Kaplan Meier curves for the genes Celsr1. Celrs3, and Ddr1. The figure shows the survival rates associated with overexpression of Celsr1. Celrs3, and Ddr1 transcripts in HNSCC patients, according to the Human Protein Atlas database (http://www.proteinatlas.org).
Supplementary Figure 5. Mutations affecting Obscurin in pre-neoplastic and tumor LSCCs. The Obscurin (Obscn) gene, located on mouse chromosome 11, showed mutations in all pre-neoplastic and 80% of tumor LSCCs. For each mutation, the table indicates the chromosome location, the type of substitution, the category and type of mutation, the amino acid change resulting from each mutation, the variance allele frequency (VAF %), and the mutation involved.
Supplementary Figure 6. Distribution of base-pair substitutions in pre-neoplastic and tumor LSCCs. The image shows the transition and transversion substitutions in the pre-neoplastic (top panel) and tumor (lower panel) LSCCs. Each dot is the number of a given substitution (listed in the x-axis) for each sample. The substitutions are represented as average ± standard deviation (SD). We calculated the statistical differences among the substitutions with a one-way ANOVA test. The asterisks indicate statistical significance of C>A substitutions (p < 0.0001 = ****), whereas the hashtags indicate statistical significance of G>T substitutions (p < 0.0001 = ####).
Funding
This research was supported in part by National Institutes of Health (T32 CA062948 to MM and R01 CA205258 to LJG) and Weill Cornell Medical College funds.
Conflict of Interest Statement:
None declared.
Acknowledgements
We thank the Gudas lab for insightful discussions, and Dr Olivier Elemento and Dr John Wagner for critical comments on the manuscript. We thank Dr Bing He, Department of Pathology and Laboratory Medicine and Translational Research Core lab, Weill Cornell Medicine, for providing access to the laser capture microdissection microscope. These data files have been uploaded to the NCBI BioProject (accession numbers PRJNA528979 and PRJNA605291); the data files are embargoed until publication.
Glossary
Abbreviations
- 4-NQO
4-nitroquinoline 1-oxide
- CNA
copy number alteration
- Egfr
epidermal growth factor receptor
- gDNA
genomic DNA
- HNSCC
head and neck squamous cell carcinoma
- LCM
laser-capture microdissection
- LSCC
LacZ+ stem cell clones
- OSCC
oral squamous cell carcinoma
- Pi3k
phosphoinositide 3-kinase
- SNVs
single-nucleotide variants
- VAF
variant allele frequency
References
- 1. Jemal, A. et al. (2008) Cancer statistics, 2008. CA. Cancer J. Clin., 58, 71–96. [DOI] [PubMed] [Google Scholar]
- 2. Leemans, C.R. et al. (2011) The molecular biology of head and neck cancer. Nat. Rev. Cancer, 11, 9–22. [DOI] [PubMed] [Google Scholar]
- 3. Zhang, H. et al. (2013) Survival outcomes of patients with advanced oral cavity squamous cell carcinoma treated with multimodal therapy: a multi-institutional analysis. J. Otolaryngol. Head Neck Surg., 42, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Scully, C. et al. (2009) Oral squamous cell carcinoma overview. Oral Oncol., 45, 301–308. [DOI] [PubMed] [Google Scholar]
- 5. Pfeifer, G.P. et al. (2002) Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene, 21, 7435–7451. [DOI] [PubMed] [Google Scholar]
- 6. Siegel, R.L. et al. (2018) Cancer statistics, 2018. CA. Cancer J. Clin., 68, 7–30. [DOI] [PubMed] [Google Scholar]
- 7. Xu, M.J. et al. (2017) EGFR-targeted therapies in the post-genomic era. Cancer Metastasis Rev., 36, 463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pickering, C.R. et al. (2014) Squamous cell carcinoma of the oral tongue in young non-smokers is genomically similar to tumors in older smokers. Clin. Cancer Res., 20, 3842–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vijg, J. et al. (2013) Genome instability and aging. Annu. Rev. Physiol., 75, 645–668. [DOI] [PubMed] [Google Scholar]
- 10. Network C.G.A.R. et al. (2017) Integrated genomic characterization of oesophageal carcinoma. Nature, 541, 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stransky, N. et al. (2011) The mutational landscape of head and neck squamous cell carcinoma. Science, 333, 1157–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Agrawal, N. et al. (2011) Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science, 333, 1154–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kanojia, D. et al. (2006) 4-Nitroquinoline-1-oxide induced experimental oral carcinogenesis. Oral Oncol., 42, 655–667. [DOI] [PubMed] [Google Scholar]
- 14. Nishimura, A (1999) Changes in Bcl-2 and Bax expression in rat tongue during 4-nitroquinoline 1-oxide-induced carcinogenesis. J. Dent. Res., 78, 1264–1269. [DOI] [PubMed] [Google Scholar]
- 15. Nakahara, Y. et al. (2004) High frequency methylation of p16INK4A gene during 4-nitroquinoline 1-oxide-induced rat tongue carcinogenesis. Oncol. Rep., 12, 101–106. [PubMed] [Google Scholar]
- 16. Osei-Sarfo, K. et al. (2013) The molecular features of tongue epithelium treated with the carcinogen 4-nitroquinoline-1-oxide and alcohol as a model for HNSCC. Carcinogenesis, 34, 2673–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tang, X.H. et al. (2013) Basal stem cells contribute to squamous cell carcinomas in the oral cavity. Carcinogenesis, 34, 1158–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jones, K.B. et al. (2013) Oral epithelial stem cells in tissue maintenance and disease: the first steps in a long journey. Int. J. Oral Sci., 5, 121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Downes, D.J. et al. (2014) Characterization of the mutagenic spectrum of 4-nitroquinoline 1-oxide (4-NQO) in Aspergillus nidulans by whole genome sequencing. G3 (Bethesda), 4, 2483–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yokoyama, A. et al. (2019) Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature, 565, 312–317. [DOI] [PubMed] [Google Scholar]
- 21. Simone, N.L. et al. (1998) Laser-capture microdissection: opening the microscopic frontier to molecular analysis. Trends Genet., 14, 272–276. [DOI] [PubMed] [Google Scholar]
- 22. Koboldt, D.C. et al. (2012) VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res., 22, 568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tang, X.H. et al. (2004) Oral cavity and esophageal carcinogenesis modeled in carcinogen-treated mice. Clin. Cancer Res., 10(1 Pt 1), 301–313. [DOI] [PubMed] [Google Scholar]
- 24. Ornstein, D.K. et al. (2000) Characterization of intracellular prostate-specific antigen from laser capture microdissected benign and malignant prostatic epithelium. Clin. Cancer Res., 6, 353–356. [PubMed] [Google Scholar]
- 25. Cingolani, P. et al. (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin), 6, 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang, J. et al. (2013) WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res., 41, W77–W83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Uhlen, M. et al. (2017) A pathology atlas of the human cancer transcriptome. Science, 357, 1–11. [DOI] [PubMed] [Google Scholar]
- 28. Sallman, D.A. et al. (2016) Integrating mutation variant allele frequency into clinical practice in myeloid malignancies. Hematol. Oncol. Stem Cell Ther., 9, 89–95. [DOI] [PubMed] [Google Scholar]
- 29. Metzeler, K.H. et al. ; AMLCG Study Group (2016) Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood, 128, 686–698. [DOI] [PubMed] [Google Scholar]
- 30. Zhong, R. et al. (2015) Notch1 activation or loss promotes HPV-induced oral tumorigenesis. Cancer Res., 75, 3958–3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Krishnan, N. et al. (2015) Integrated analysis of oral tongue squamous cell carcinoma identifies key variants and pathways linked to risk habits, HPV, clinical parameters and tumor recurrence. F1000Res., 4, 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nakagaki, T. et al. (2017) Profiling cancer-related gene mutations in oral squamous cell carcinoma from Japanese patients by targeted amplicon sequencing. Oncotarget, 8, 59113–59122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Izumchenko, E. et al. (2015) Notch1 mutations are drivers of oral tumorigenesis. Cancer Prev. Res. (Phila.), 8, 277–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Perry, N.A. et al. (2012) Loss of giant obscurins promotes breast epithelial cell survival through apoptotic resistance. FASEB J., 26, 2764–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ciriello, G. et al. (2013) Emerging landscape of oncogenic signatures across human cancers. Nat. Genet., 45, 1127–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. El-Bayoumy, K. et al. (2017) Carcinogenesis of the oral cavity: environmental causes and potential prevention by black raspberry. Chem. Res. Toxicol., 30, 126–144. [DOI] [PubMed] [Google Scholar]
- 37. Nik-Zainal, S. et al. ; Breast Cancer Working Group of the International Cancer Genome Consortium (2012) Mutational processes molding the genomes of 21 breast cancers. Cell, 149, 979–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Alexandrov, L.B. et al. (2014) Mutational signatures: the patterns of somatic mutations hidden in cancer genomes. Curr. Opin. Genet. Dev., 24, 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rosenthal, R. et al. (2016) DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol., 17, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Alexandrov, L.B. et al. ; Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain (2013) Signatures of mutational processes in human cancer. Nature, 500, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. el-Naggar, A.K. et al. (1995) Sequential loss of heterozygosity at microsatellite motifs in preinvasive and invasive head and neck squamous carcinoma. Cancer Res., 55, 2656–2659. [PubMed] [Google Scholar]
- 42. Butler, M.T. et al. (2018) Spatial and temporal analysis of PCP protein dynamics during neural tube closure. Elife, 7, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Goffinet, A.M. et al. (2017) Seven pass Cadherins CELSR1-3. Semin. Cell Dev. Biol., 69, 102–110. [DOI] [PubMed] [Google Scholar]
- 44. Wang, Y. et al. (2016) Patterning of papillae on the mouse tongue: a system for the quantitative assessment of planar cell polarity signaling. Dev. Biol., 419, 298–310. [DOI] [PubMed] [Google Scholar]
- 45. Rock, R. et al. (2005) Expression of mouse dchs1, fjx1, and fat-j suggests conservation of the planar cell polarity pathway identified in Drosophila. Dev. Dyn., 234, 747–755. [DOI] [PubMed] [Google Scholar]
- 46. Saburi, S. et al. (2008) Loss of Fat4 disrupts PCP signaling and oriented cell division and leads to cystic kidney disease. Nat. Genet., 40, 1010–1015. [DOI] [PubMed] [Google Scholar]
- 47. Cai, J. et al. (2015) FAT4 functions as a tumour suppressor in gastric cancer by modulating Wnt/β-catenin signalling. Br. J. Cancer, 113, 1720–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jiang, X. et al. (2018) Low FAT4 expression is associated with a poor prognosis in gastric cancer patients. Oncotarget, 9, 5137–5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. India Project Team of the International Cancer Genome Consortium (2013) Mutational landscape of gingivo-buccal oral squamous cell carcinoma reveals new recurrently-mutated genes and molecular subgroups. Nat. Commun., 4, 2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Temam, S. et al. (2007) Epidermal growth factor receptor copy number alterations correlate with poor clinical outcome in patients with head and neck squamous cancer. J. Clin. Oncol., 25, 2164–2170. [DOI] [PubMed] [Google Scholar]
- 51. Cancer Genome Atlas Network (2015) Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature, 517, 576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nichols, A.C. et al. (2012) Frequent mutations in TP53 and CDKN2A found by next-generation sequencing of head and neck cancer cell lines. Arch. Otolaryngol. Head. Neck Surg., 138, 732–739. [DOI] [PubMed] [Google Scholar]
- 53. Zhang, Z. et al. (2006) p53 Transgenic mice are highly susceptible to 4-nitroquinoline-1-oxide-induced oral cancer. Mol. Cancer Res., 4, 401–410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.