Abstract
Emerging data show that microRNA 193a-3p (miR-193a-3p) has a suppressive role in many cancers and is often downregulated in tumors, as compared to surrounding normal tissues. Therefore, mimics of miR-193a-3p could be used as an attractive therapeutic approach in oncology. To better understand and document the molecular mechanism of action of 1B3, a novel synthetic miRNA-193a-3p mimic, RNA sequencing was performed after transfection of 1B3 in six different human tumor cell lines. Genes differentially expressed (DE) in at least three cell lines were mapped by Ingenuity Pathway Analysis (IPA), and interestingly, these results strongly indicated upregulation of the tumor-suppressive phosphatase and tensin homolog (PTEN) pathway, as well as downregulation of many oncogenic growth factor signaling pathways. Importantly, although unsurprisingly, IPA identified miR-193a-3p as a strong upstream regulator of DE genes in an unbiased manner. Furthermore, biological function analysis pointed to an extensive link of 1B3 with cancer, via expected effects on tumor cell survival, proliferation, migration, and cell death. Our data strongly suggest that miR-193a-3p/1B3 is a potent tumor suppressor agent that targets various key oncogenic pathways across cancer types. Therefore, the introduction of 1B3 into tumor cells may represent a promising strategy for cancer treatment.
Keywords: microRNA, miR-193a, PTEN, cancer, RNAi therapeutics, RNA-sequencing
Graphical Abstract
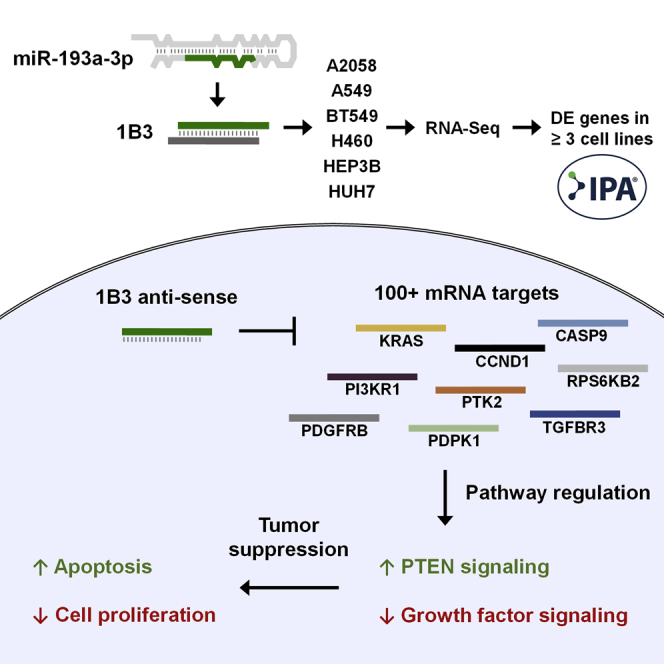
1B3 is a mimic of the naturally occurring miRNA-193a-3p, a gene regulator that has potent tumor suppressor functions. We characterized the effect of 1B3 on gene expression in six human cancer cell lines. 1B3 was shown to strongly activate tumor suppressor pathways while inhibiting oncogenic cell signaling.
Introduction
MicroRNAs (miRNAs) are small noncoding RNA molecules that post-transcriptionally regulate gene expression by controlling the stability and translation of mRNAs.1 Individual miRNAs have multiple mRNA targets, and a single mRNA may be targeted by several miRNAs.2 Given their critical role in a wide range of cellular processes, such as cell proliferation and apoptosis,3 miRNA dysregulation contributes to many hallmarks of cancer.4 Furthermore, miRNAs can modulate tumor-modifying extrinsic factors, such as cancer-immune system interactions, stromal cell interactions, and sensitivity to therapy. Ultimately, it is the balance among these processes that determines whether a miRNA produces a net oncogenic or tumor-suppressive effect. Expression of miRNAs decreases, but does not fully extinguish, expression of their multiple targets, and such a global dampening of biochemical machinery is likely to especially affect transformed cells, while sparing normal cells.5 Introduction of tumor-suppressive miRNAs into tumor cells is therefore an attractive approach for cancer treatment that may result in high efficacy and low drug resistance due to their multi-targeted nature and overlapping downstream effects on cancer cell biology.
Specifically, microRNA (miR)-193a was initially identified in a high-throughput functional lentivirus-based genomics screen of a miRNA library in tumor cells.6 Subsequently, the 3p arm of miR-193a (miR-193a-3p) was demonstrated as the biologically active entity in tumor cell-based assays. A large body of evidence in the literature clearly shows that miR-193a-3p is frequently downregulated in malignant cells, including hepatocellular carcinoma (HCC),7 non-small cell lung cancer (NSCLC),8 and triple negative breast cancer (TNBC).9 Significant evidence points toward a strong tumor-suppressive function of miR-193a-3p. In various cancer types, overexpression of miR-193a-3p in vitro and in vivo increases apoptosis and decreases cell proliferation and migration, leading to reduced tumor growth and metastasis.9, 10, 11, 12, 13 Furthermore, miR-193a-3p was shown to sensitize HCC cells to chemotherapy and radiation treatment.14 Based on compelling evidence that miR-193a-3p plays an undisputed role as a tumor suppressor, we set out to develop 1B3 (Figure 1A), which represents a proprietary, novel, chemically modified miR-193a-3p mimic for therapeutic intervention in oncology. 1B3 has been fully characterized in cell-based assays, showing reduced cell proliferation/survival, cell cycle arrest, induction of apoptosis, increased cell senescence, DNA damage, and inhibition of migration.15 Importantly, 1B3 in a novel lipid nanoparticle-based formulation, INT-1B3, demonstrated marked anti-tumor activity following systemic administration in tumor-bearing mice.
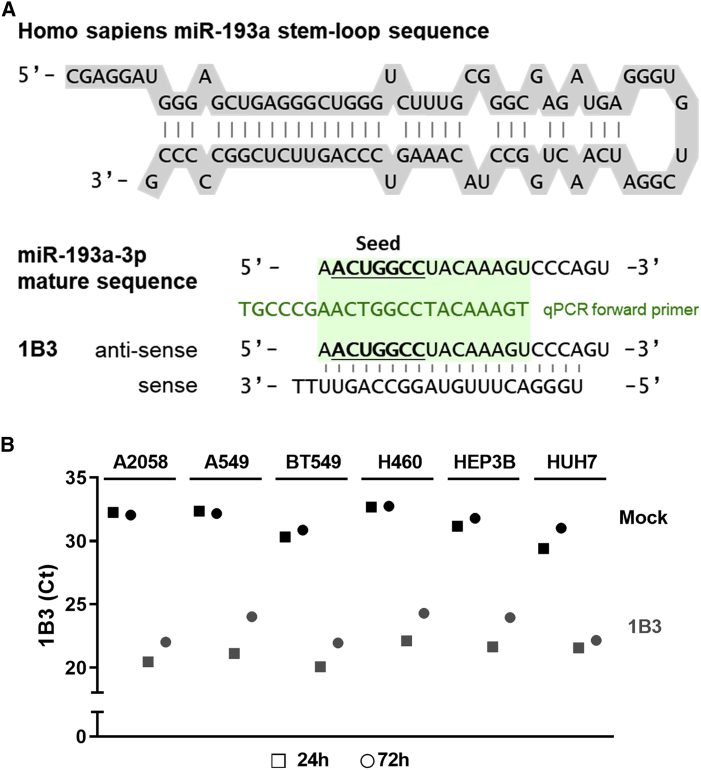
Figure 1.
Nucleotide sequences of precursor miR-193a, miR-193a-3p, and 1B3 and transfection efficiency of 1B3
(A) Nucleotide sequences of precursor miR-193a, miR-193a-3p (according to miRBase), and 1B3. The seed sequence in the guide (anti-sense) strand is underlined. The forward qPCR primer (green) for 1B3 used in stem-loop qRT-PCR also binds to the endogenous mature miR-193a-3p sequence. (B) Six tumor cell lines were transfected with 10 nM 1B3 in the presence of RNAiMAX, and total RNA was harvested after 24 h and 72 h. RNA samples from three independent experiments were pooled. The expression levels of 1B3 in samples submitted for RNA sequencing were analyzed by stem-loop qRT-PCR. Shown are the cycle threshold (Ct) values.
As miRNAs have been shown to be able to modulate expression of several hundreds of genes, the search for targets of miR-193a-3p has, so far, been dictated by prediction algorithms and validation of suspected genes of interest. Among the identified putative targets, KRAS,13 PLAU,16 MCL-1,17 CCND1,5,18 and ERBB411 are the most experimentally validated, and their downregulation by miR-193a-3p has been demonstrated at both mRNA and protein levels. Other published miR-193a-3p targets with important roles in malignant cell behavior include S6K2,19 RAB27B,20 and SRSF2.21 Although focused studies have revealed numerous relevant genes, a comprehensive analysis of miR-193a-3p targets and consequent molecular mechanism of action across cancer types is lacking. We therefore performed transcriptome-wide sequencing of six different human tumor cell lines transfected with 1B3 to identify genes that are directly or indirectly modulated at the mRNA level. Regulated genes were subsequently used in an unbiased analysis (Ingenuity Pathway Analysis [IPA]) to document affected cellular pathways and biological functions.
Results
Transfection of miRNA mimics and controls in different cancer cells
The stem-loop sequence of miR-193a and the naturally occurring miR-193a-3p mature sequence are depicted in Figure 1A. As illustrated, the guide (anti-sense) strand of 1B3 is identical to the 3p strand of the mature miR-193a, which is highly conserved across species. The passenger (sense) strand of the mimic has limited oligonucleotide backbone chemical modifications (2′-O-methyl; as shown in our patent application file22). Besides providing resistance to degradation by endogenous nucleases in vivo,23,24 2′-O-methyl modifications improve anti-sense strand selection. As demonstrated by chemically modified small interfering RNA (siRNA) duplexes, 2′-O-methyl modifications avoid sense strand incorporation into the RNA-induced silencing complex (RISC).25 Anti-sense specificity of 1B3 is further ensured by the 3′ deoxythymidine (dT) overhang of the sense strand26 and the lower 5′ thermodynamic stability of the anti-sense strand.27,28
To perform a comprehensive analysis of the targets of 1B3 and its consequent mode of action, 1B3 was transfected in six different human cancer cell lines, including melanoma (A2058), NSCLC (A549 and H460), TNBC (BT549), and HCC (Hep3B and Huh7) cell lines. Expression of 1B3 was assessed in pooled RNA samples of three independent replicates using stem-loop qRT-PCR . Low endogenous levels (high cycle threshold [Ct]) of miR-193a-3p were detected in all cell lines (Figure 1B). After transfection with 1B3, samples had, on average, a 10-Ct difference compared to mock at 24 h post-transfection, indicative of a ∼1,000-fold higher 1B3 expression as compared to endogenous miR-193a-3p. At 72 h post-transfection, the average difference compared to mock at the same time point was, on average, 8 Ct, pointing at the long-term sustained presence of 1B3 in cells upon transfection and limited degradation. Pooled RNA samples of three independent replicates of each time point were subsequently analyzed by RNA sequencing. Adequate sample pooling has been shown to sufficiently control biological variation and to enable reliable exploratory analysis.29,30
In parallel to exploration of the effects of 1B3 on gene expression, two miRNA controls were also included in the RNA sequencing experiment. 1K1 is a miRNA similar to 1B3, except for three point mutations in the seed sequence leading to a theoretical 1B3 “dead mutant” (Figure S1A). 3A1 is based on a commercially available random sequence miRNA that has been validated to not produce identifiable biological effects on known mRNA functions. Transfection efficiency of these controls was similar among all cell lines (Figure S1B).
Transcriptome-wide analysis of gene expression upon transfection with 1B3
To investigate the transcriptome-wide effect of 1B3, RNA samples were sequenced at two time points after transfection: 24 h to characterize direct targets of 1B3 and initial changes in cell signaling and 72 h to determine consequent differential gene expression and later functional effects following initial 1B3-induced gene-expression modulation. The DESeq analysis was not based on replicates but on a mean-variance relationship by treating the two samples as if they were replicates.31 Single replicate differential gene-expression analysis can therefore only be used to compare between experimental conditions, not between cell lines; hence, all comparisons were done between transfected (1B3) and nontransfected (mock) samples. The estimated expression in the 1B3 sample was divided by the estimated expression in the mock sample to determine the extent of gene down- or upregulation (fold change < 1 or > 1, respectively). Because the p value generated by DESeq is related to the fold change and the estimated gene expression, we chose p <0.05 as an arbitrary cutoff for differential expression. As anticipated from literature data on miRNA properties, at 24 h post-transfection, 1B3 affected the expression of several hundred genes in each tumor cell line, most of which were downregulated, consistent with expected direct effects of the miRNA on target mRNA downregulation (Figure 2A). Furthermore, around 50% of genes downregulated by 1B3 were predicted miR-193a-3p targets.
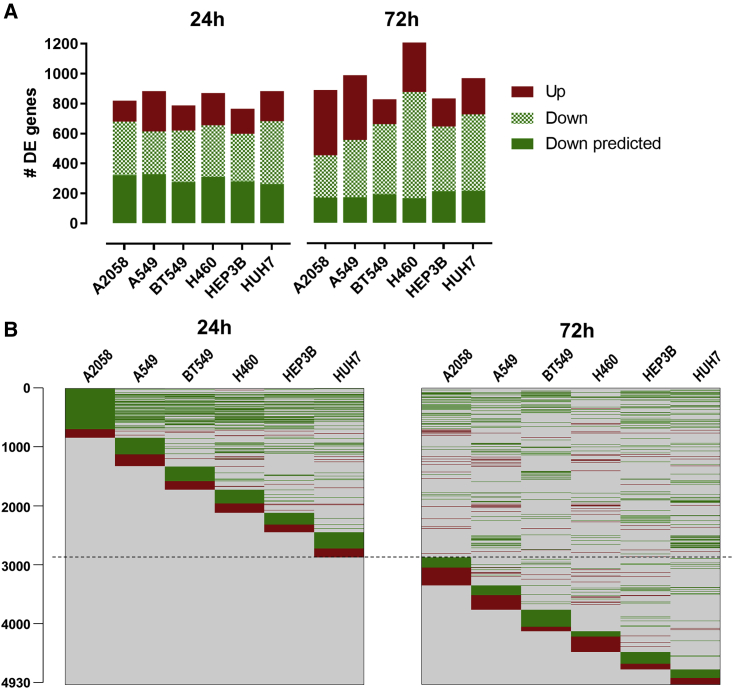
Figure 2.
Gene-expression regulation by 1B3 in individual cell lines
(A) Numbers of differentially expressed (DE) genes that are downregulated (estimated expression 1B3/estimated expression mock < 1) and upregulated (estimated expression 1B3/estimated expression mock > 1) 24 h and 72 h after transfection with 10 nM 1B3 in each cell line are shown as compared to mock-transfection conditions. The number of downregulated genes that are predicted miR-193a-3p targets is also indicated. (B) Heatmaps showing differential gene expression induced by 1B3 per cell line at 24 h and 72 h post-transfection. All DE genes are on the y axis and on the same row across all cell lines and both time points (due to the long list, gene names are not shown). Green means downregulated, and red means upregulated genes. Gray means no observed change in gene expression.
The heatmap presented in Figure 2B visualizes the 1B3-induced gene-expression profile at 24 h and 72 h and the overlap between the tested tumor cell lines, where each row corresponds to a certain gene that is differentially expressed (DE). In line with cell line-specific effects of miRNAs,32 each cell line had a proportion of DE genes that did not overlap with the other cell lines. The DE genes that were common between the cell lines and between 24 h and 72 h were mostly regulated in the same direction. At 72 h post-transfection with 1B3, the pattern of gene expression changed drastically from the profile at 24 h in all cell lines. A greater proportion of genes was upregulated as compared to 24 h, and fewer genes were predicted miR-193a-3p targets, on average 30%, reflecting downstream effects triggered by the initial (24 h) gene-expression modulation (Figure 2A).
DE gene lists from each cell line at 24 h post-transfection were used as input for IPA software, which importantly, identified miR-193a-3p as a very significant upstream regulator in all cell lines (p = 4.4 × 10−6, z = 2.382, on average), based on the observed gene-expression changes in the datasets, independently validating 1B3 as a biologically active miR-193a-3p mimic.
Modulation of gene expression by “negative” miRNA controls
Similarly, gene-expression modulation induced by the negative miRNA controls, 1K1 and 3A1, upon transfection in the same panel of selected tumor cell lines, as compared to mock, was analyzed by RNA sequencing (at 24 h post-transfection). Surprisingly, both 1K1 and 3A1 affected several hundred genes in all tumor cell lines, although only few downregulated genes (<18%) were predicted miR-193a-3p targets (Table S1). The number of overlapping genes between 1K1 and 3A1 was minimal (data not shown), which excludes large general effects on gene-expression modulation due to the transfection procedure or exogenous oligonucleotide introduction. As expected, only a small number of DE genes by 1B3 overlapped with those affected by miRNA controls, 1K1 (<8%) and 3A1 (<7%), as compared to mock (Table S1), highlighting the sequence-specific effect of 1B3 on gene-expression modulation.
Unsurprisingly, DE gene analysis of 1B3 versus either miRNA control (estimated expression in the 1B3 sample divided by the estimated expression in the 1K1 or 3A1 sample) resulted in gene profiles that were not in line with the expected direct effect of 1B3 on mRNA targets, showing only a minority of predicted miR-193a-3p targets and more upregulated than downregulated genes (Figure S1C). Moreover, these gene profiles had, on average, only 22% overlap with the 1B3/mock DE profile (Figure S1D), and therefore, as expected, IPA did not predict miR-193a-3p as a positive upstream regulator when the DE gene-expression profile was based on 1B3 versus 3A1 or 1K1 (data not shown).
Therefore, it was concluded that comparing the gene profile induced by 1B3 to mock transfection, rather than to a negative miRNA control, was more suitable and reliable for correct differential gene-expression analysis.
Cancer gene-expression modulation by 1B3
To further increase stringency and find 1B3 targets that are common across cancer cell types, lists of genes were made that were DE in multiple tumor cell lines at each time point. Narrowing down to genes regulated in more than one cell line may also filter out cell line-specific effects on endogenous miRNA activity, as well as mRNA expression changes that can occur due to competition for intracellular small RNA processing machinery.33 The 24-h samples contained more predicted miR-193a-3p targets common to multiple cell lines than the 72-h samples (Figure 3A). In addition, the more cell lines considered, the larger the proportion of predicted miR-193a-3p targets. 1B3 led to 141 DE genes in all six tumor cell lines at 24 h (Table S2), of which all were downregulated, and 110 genes (78%) were predicted miR-193a-3p targets. Many of these genes, including STMN1,34 CCND1,5 and KRAS,13 have important described roles in cancer, such as regulation of cell proliferation, migration, and apoptosis.
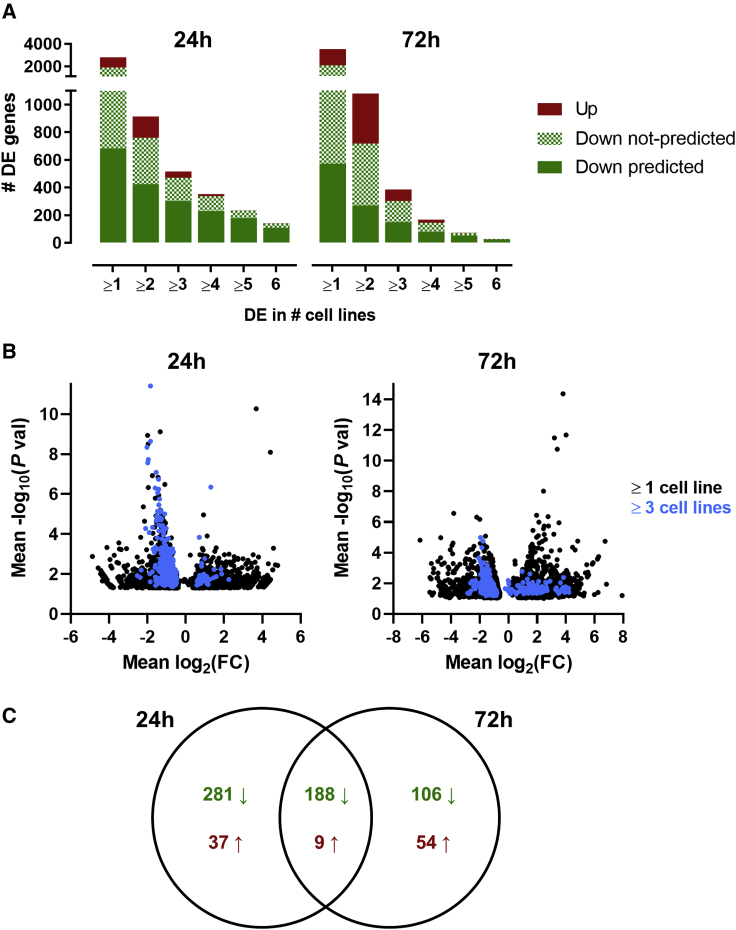
Figure 3.
Gene-expression regulation by 1B3 in multiple cell lines
(A) Numbers of DE genes that are downregulated (estimated expression 1B3/estimated expression mock < 1) and upregulated (estimated expression 1B3/estimated expression mock > 1) 24 h and 72 h after transfection with 10 nM 1B3 in multiple (up to six) tumor cell lines are shown as compared to mock-transfection conditions. The number of downregulated genes that are predicted miR-193a-3p targets is also indicated. (B) Volcano plots show genes DE in at least three cell lines (blue) in the population of all DE genes in at least one cell line (black). (C) Venn diagram demonstrating DE genes common among at least three cell lines. Downward arrow indicates downregulation (average estimated expression 1B3/estimated expression mock < 1), and upward arrow indicates upregulation (average estimated expression 1B3/estimated expression mock > 1).
Because our objective was to further document and refine the mode of action of 1B3 across cancer types, significantly DE genes, common to at least three cell lines at 24 h (Table S3) and 72 h (Table S4), were used as IPA input. This reduced cell-type-specific genes and yielded adequate gene numbers to be used in subsequent pathway analyses. Volcano plots visualize these genes for each time point within the whole population of DE genes (Figure 3B). All genes were DE at least 1.3-fold and were evenly distributed among the different cell lines (Figure S2A). Fewer genes were DE in at least three cell lines at 72 h (357 genes) compared to 24 h (516 genes), likely due to more cell line-specific events secondary to 1B3 target downregulation at the later time point. Almost one-third of DE genes overlapped between the two time points, most of which were downregulated by 1B3 (Figure 3C).
Similar IPA input lists of DE genes common to at least three cell lines (24 h only) were also created for 1K1 (Table S5) and 3A1 (Table S6). These were compared to the IPA input list of 1B3 to rule out the possibility that the DE genes were due to nonspecific effects of each miRNA. This result showed that less than 10% of genes overlapped between 1B3 and 1K1, or between 1B3 and 3A1, and only three genes were DE in at least three cell lines by all three miRNAs (Figure S2B).
1B3 activates the phosphatase and tensin homolog (PTEN) signaling pathway
IPA was used to uncover the significance of the RNA sequencing data and to characterize 1B3-regulated genes within the context of cellular and biological systems. For each time point, a positive or negative expression fold change was assigned to every gene that was DE in at least 3 tumor cell lines, based on its average fold change (estimated expression 1B3/mock). Hence, 515 genes at 24 h and 357 genes at 72 h were consistently down- or upregulated. These gene lists served as input for IPA to find 1B3-regulated mechanisms that are common across cancer types. Data were overlaid onto a global molecular map developed from information contained in the IPA knowledge database.
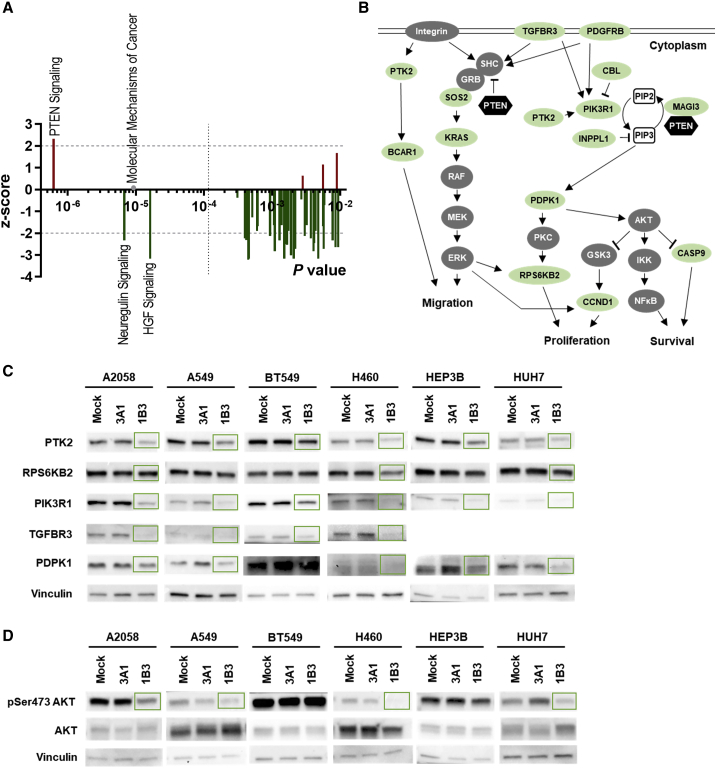
Next, canonical pathways affected by the 1B3-induced differential gene expression were predicted. Statistics were based on the number of genes involved in a defined pathway in the IPA libraries that were up- or downregulated genes in the dataset. By far, the most significantly enriched pathway at 24 h was PTEN signaling (p = 6.2 × 10−7; Figure 4A; Table S7), which was predicted to be activated (z = 2.309). The majority of other significantly regulated pathways were predicted to be inhibited, including neuregulin signaling (p = 6.7 × 10−6, z = −2.333) and hepatocyte growth factor (HGF) signaling (p = 1.6 × 10−5, z = −3.162). The pathway, “molecular mechanisms of cancer,” was identified without directionality (p = 9.1 × 10−6, no Z score). Importantly, PTEN signaling remained significantly activated up to 72 h (p = 4 × 10−4, z = 1.633), alongside multiple inhibited pathways involved in cell cycle control and oncogenesis. PTEN signaling is known to play major roles in suppressing tumors and is frequently inhibited in malignant cells.35 Key genes in this pathway that were downregulated by 1B3 are highlighted in Figure 4B.
Figure 4.
PTEN pathway activation by 1B3
(A) Canonical pathways regulated by 1B3 at 24 h (p < 0.01) according to IPA. Directionality is indicated by Z score (<−2: inhibition; >2: activation). The significance threshold for multiple testing is indicated by a black dotted line. (B) Diagram showing mRNA targets of 1B3 in the PTEN pathway. Genes highlighted in green were downregulated by 1B3 at 24 h in at least three cell lines. (C and D) Indicated human tumor cells were transfected with 10 nM of either 3A1 or 1B3 in the presence of RNAiMAX and lysed after 72 h. Clarified whole-cell lysates were immunoblotted for PTK2, RPS6KB2, PIK3R1, TGFBR3, and PDPK1 (C). TGFBR3 could not be detected in HEP3B and HUH7. Additionally, samples were immunoblotted for pSer473 AKT and AKT total protein (D). Vinculin served as a loading control. Green boxes highlight observed protein downregulation as measured by densitometry.
For confirmation of this result and to gain statistical power, we additionally analyzed the transcriptome data using multiple replicate testing, whereby all six 1B3-transfected cell lines were compared to all corresponding mock-treated cell lines per time point, while controlling for the effect of the cell lines using linear modeling, as implemented in DESeq2.36 In the resulting dataset, 547 genes were significantly DE (adjusted p < 0.05) by at least 1.5-fold compared to mock (477 down, 70 up) at 24 h post-transfection (Figure S3A; Table S8). IPA predicted, identically to the earlier analysis, that PTEN signaling was the top significant pathway and strongly activated (p = 5.5 × 10−6, z = 2.111) in the 1B3-transfected cells (Figure S3B). Also, similarly, the pathway, "molecular mechanisms of cancer," was enriched (p = 8.70964 × 10−5, no Z score), and other identified pathways were all downregulated.
To experimentally demonstrate that 1B3 activates the PTEN pathway and to further validate the results obtained from the RNA sequencing and the IPA, we examined the protein level of selected miR-193a-3p targets involved in the PTEN pathway. Western blotting was performed on lysates of multiple cell lines transfected with 1B3 or 3A1 for 72 h, and results were quantified by densitometry (Table S9). In all tested cell lines, protein downregulation of PTK2 and PIK3R1 was observed in the 1B3 sample compared to mock and 3A1 (Figure 4C). TGFBR3 was also downregulated by 1B3 in cell lines where a constitutive expression level could be observed (A2058, A549, BT549, and H460). RPS6KB2 was reduced by 1B3 in A2058, H460, and HUH7. PDPK1 was downregulated in all cell lines except BT549. In addition to target protein level, we analyzed the activation status of AKT, a critical downstream effector in the PTEN pathway. The phosphorylation level of AKT was decreased by 1B3 in A2058, A549, H460, and HUH7 (Figure 4D). Hence, in line with the RNA sequencing and IPA results, 1B3 was demonstrated to activate the PTEN pathway through direct downregulation and indirect inhibition of key signaling proteins in cell-based assays.
Similar analyses using IPA were performed to identify affected canonical pathways in the dataset of genes that were DE by the miRNA controls compared to mock in at least three cell lines at 24 h (Figures S4A and S4B). 1K1 regulated pathways with much lower significance and directionality as opposed to 1B3. IPA was unable to define meaningful pathway regulation by 3A1 despite significant differential expression of hundreds of genes. Importantly, few pathways regulated by 1K1 and 3A1 were in common with 1B3, and different DE genes participated in these pathways (Table S10). Consistently, IPA identified a very distinct set of pathways for 1B3 when we uploaded the DE gene dataset in which the expression profile of either miRNA control served as baseline for 1B3 (data not shown).
Tumor suppressor functions of 1B3
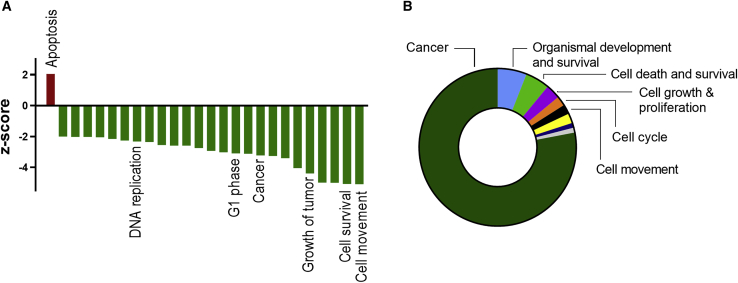
Subsequently, IPA software was used to predict downstream biological effects of the observed gene-expression changes 72 h after transfection. Out of the 100 most significant biological functions that were affected by 1B3, those that were inhibited (z < −2) were related to cell survival, proliferation, migration, or cancer, and those that were activated (z > 2) were related to (tumor) cell death (Figure 5A). Finally, we categorized the top 100 biological functions (all p < 0.00001) regulated by 1B3 and made a ranking based on the number of functions in each category. The great majority of affected biological functions belonged to the category “cancer” (Figure 5B). Categories related to survival, cell proliferation, cell cycle, and cell movement were also prominent.
Figure 5.
Biological functions affected by 1B3
(A) The top 100 biological functions (ranked by p value) affected by 1B3, according to IPA at 72 h, were filtered for functions relevant to tumor cells. Shown are functions with Z score < −2 and > 2. All p values are smaller than 0.00001. (B) Categories of top 100 biological functions (all p < 0.00001) regulated by 1B3 were ranked based on the number of functions in each category.
Discussion
This manuscript is the first to dissect and document the mode of action of 1B3, a novel miR-193a-3p mimic, in a variety of cancer cells via an unbiased transcriptome-wide analysis. We examined the differential gene-expression profiles resulting from transfection with 1B3 in six different human tumor cell lines from various cancer types in comparison to mock-transfection experimental conditions. DE gene lists were then entered into IPA software to characterize affected cellular pathways and biological functions. In accordance with literature data demonstrating that an individual miRNA can recognize several hundreds of mRNAs,37, 38, 39, 40, 41 our data show that 24 h post-transfection with 1B3, 515 genes were DE in at least three cell lines, underlining the multi-targeted feature of miRNAs. At this time point, which is more relevant for identification of direct interactions with the transcriptome, genes were mainly downregulated, consistent with the expected direct primary effect of miRNAs on target mRNA downregulation. 1B3 regulated 141 genes common to all tested tumor cell lines, as well as many cell line-specific genes, in line with known miRNA function.32 At 72 h post-transfection, the pattern of gene-expression modulation drastically changed from the profile at 24 h in all cell lines, and a greater proportion of gene upregulation was observed, reflecting consequent gene-expression modulation triggered by the initial wave of gene downregulation. Many 1B3-regulated genes were components of growth factor signaling pathways and cell cycle machinery, which induce cellular proliferation and tumor progression.
IPA predicted miR-193a-3p as a very significant upstream regulator of the gene-expression changes in all six tested cancer cell lines and confirmed the link between 1B3 and cancer-related biology. Unbiased analysis clearly identified 1B3 as a strong activator of the tumor-suppressive PTEN pathway, which antagonizes the proto-oncogenic phosphoinositide 3-kinase (PI3K)-AKT signaling pathway, thereby governing fundamental cellular processes. The downregulation of multiple PTEN pathway proteins and decrease in AKT phosphorylation because of 1B3 transfection were confirmed by western blotting. PTEN is one of the most commonly lost tumor suppressors in human cancer,42,43 and even small changes in PTEN activity affect susceptibility and prognosis in a range of highly aggressive malignancies. Furthermore, since activation of AKT impairs apoptosis and stimulates cell cycle progression, phosphorylation of Ser473 AKT can predict poor clinical outcome in several cancers.44, 45, 46 Thus, the reactivation of the PTEN signaling pathway and suppression of AKT phosphorylation could potentially represent an effective therapy against cancer.47,48 Very much in line with the mode of action uncovered by this RNA sequencing study, we fully characterized 1B3 as a potent and safe tumor suppressor in cell-based assays as well as animal models.55
Surprisingly, similar evaluation of the control oligonucleotides 1K1 (mutated 1B3) and 3A1 (unrelated miRNA) also revealed extensive mRNA downregulation, despite the absence of a physiological seed sequence. However, target overlap among 1B3, 3A1, and 1K1 was very limited, clearly suggesting distinct molecular mechanisms of action. This also suggests that gene dysregulation by endogenous miRNAs as a side effect of overexpression of exogenous miRNA mimics was minimal.49 A classic way to generate negative controls for small RNAs is to randomize their nucleotides to control for effects that are not sequence-specific but are instead due to the physiochemical properties of the molecules, such as charge.50 CG-rich molecules, for example, have been shown to have nonspecific anti-tumor effects in vivo by inducing an interferon response.51 It is important to note, however, that these negative controls also have their own sequence-specific effects that lead to changes in cellular pathways, as demonstrated by our IPA results, although the direction of regulation was often undetermined. Given the lack of endogenous functions of such miRNAs that do not exist in nature, their effects can be very cell-type dependent and hence, unpredictable. In addition, because of the lack of strong direct targets, the effect of any endogenous miRNA repression due to competition for intracellular small RNA processing machinery33 may be over-represented. Furthermore, we have shown that calculating differential expression of a bona fide miRNA using control oligonucleotide data as baseline, an approach that is often used, may lead to skewed DE gene profiles that are not in line with true miRNA function. Therefore, negative miRNA controls should be used with caution in such studies and possibly even in experimental tumor models, as biological responses to random modifications of gene-expression profiles may have a significant and confusing impact in these settings.
In conclusion, RNA sequencing after transient transfection of a panel of six human tumor cell lines indicated a clear sequence-specific differential effect of 1B3 as compared to either mock-transfection conditions or selected miRNA controls, and IPA demonstrated that 1B3 regulates a wide range of cellular pathways contributing to and/or cooperating with effective tumor suppression. Therefore, based on the significant number of modulated genes and biological links to “hallmarks of cancer,” the introduction of 1B3 into cancer cells represents a promising novel treatment modality in oncology as a single agent or in combination with standard of care.
Materials and methods
Oligonucleotides
The miR-193a-3p mimic, 1B3, and control oligonucleotides, 1K1 (mutated 1B3; point mutations in the seed sequence) and 3A1 (unrelated miRNA; based on Thermo Fisher Scientific; #4464058), were manufactured by BioSpring (Frankfurt am Main, Germany). The nucleotide sequences are as follows (with limited 2′-O-methyl nucleotide modifications on the passenger strand; as shown in our patent application file22):
1B3 passenger (sense) strand: (′5)-UGGGACUUUGUAGGCCAGUUTT-(′3)
1B3 guide (anti-sense) strand: (5′)-AACUGGCCUACAAAGUCCCAGU-(3′)
1K1 passenger (sense) strand: (′5)-UGGGACUUUGUAGCCGACUUTT-(′3)
1K1 guide (anti-sense) strand: (5′)-AAGUCGGCUACAAAGUCCCAGU-(3′)
3A1 passenger (sense) strand: (′5)-UAACGACGCGACGACGUAATT-(′3)
3A1 guide (anti-sense) strand: (5′)-UUACGUCGUCGCGUCGUUATT-(3′)
Cell culture, transfection, and sample preparation
The human cell lines A2058 (melanoma), A549 (NSCLC), BT549 (TNBC), H460 (NSCLC), and Hep3B (HCC) were purchased from ATTC. The human cell line Huh7 (HCC) was obtained from the Japanese Cancer Research Resources Bank (JCRB). All cell lines were cultured at 37°C in a 5% CO2 humidified incubator and in cell culture media recommended by suppliers. For RNA isolation, cells were seeded at 80% confluency into 6-well cell-culture plates, 24 h before mock transfection or Thermo Fisher Scientific Lipofectamine RNAiMAX-assisted transfection with 10 nM of 1B3, 1K1, or 3A1. Transfection media were aspirated at 24 h (mock, 1B3, 3A1, 1K1) and 72 h (mock, 1B3) post-transfection, and culture plates were stored at −80°C. For protein isolation, cells were seeded into 6-well plates at 25% confluency before mock transfection or transfection with either 10 nM of 1B3 or 3A1 using RNAiMAX. Media were aspirated 72 h after transfection, and plates were stored at −80°C. Three independent replicate transfection experiments were performed for each cell line.
RNA isolation
TRIzol Reagent was added onto transfected cells that were stored at −80°C, and total RNAs were isolated according to the instructions of the miRNeasy Mini kit (QIAGEN). The procedure included on-column DNase treatment. RNA concentration was measured on NanodropOne. For RNA sequencing, 150 ng RNA from each independent replicate was pooled.
Protein isolation and western blotting
Radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS, 0.5 mM EDTA), supplemented with protease and phosphatase inhibitor cocktails, was added onto transfected cells stored at −80°C. Lysates were centrifugated at 15,000 × g for 1 h at 4°C and clarified by removing the cell debris pellet. Protein concentration was determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Samples were separated at 120 V by polyacrylamide gel electrophoresis under denaturing conditions (SDS) on Mini-PROTEAN TGX Stain-Free Precast Gels (Bio-Rad). Proteins were transferred at 200 mA for 2 h to polyvinylidene difluoride (PVDF) membranes in transfer buffer (25 mM Tris, 192 mM glycine, 20% methanol, pH 8.3). The membranes were blocked using 5% milk or 5% BSA in Tris-buffered saline with Tween (20 mM Tris, pH 7.6, 137 mM NaCl, 0.1% Tween). Blots were probed with primary and horseradish peroxidase-conjugated secondary antibodies (Table S11). Proteins were detected using enhanced chemiluminescence (ECL) reagents. Membranes were stripped by incubation in stripping buffer (62.5 mM Tris, pH 6.8, 2% SDS, 100 mM 2-mercaptoethanol) for 30 min at 50°C and reprobed as appropriate. Band intensities were quantified by densitometry using ImageJ software.
Stem-loop quantitative real-time PCR
Isolated RNA was reverse transcribed (RT) using stem-loop RT primers (IDT) to amplify specific miRNAs. Subsequently, quantitative real-time PCR was performed using SYBR Green (Bio-Rad). The primer sequences used in this study are provided in Table S11. The forward qPCR primer for 1B3 guide strand also binds to the endogenous mature miR-193a-3p sequence, leading to parallel detection and quantification of both sequences.
RNA sequencing
RNA samples (450 ng) were sent to and processed by GenomeScan BV (Leiden, the Netherlands). Briefly, the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina was used to isolate mRNA from total RNA, followed by mRNA fragmentation and cDNA synthesis. This was used for ligation with the sequencing adapters and PCR amplification of the resulting product. Samples were sequenced using next-generation RNA Sequencing (Illumina HiSeq 4000/NovaSeq 6000). The data processing workflow included raw data quality control, adaptor trimming, and alignment of short reads. The reference GRCh37.75.dna.primary_assembly was used for alignment of the reads for each sample. Based on the mapped locations in the alignment file, the frequency of how often a read was mapped on a transcript was determined (feature counting). The counts were saved to count files, which served as input for downstream RNA sequencing differential expression analysis.
RNA sequencing data analysis
Differential gene-expression analysis was performed on the short-read dataset by GenomeScan BV. The read counts were loaded into the DESeq package version (v.)1.30.0, a statistical package within the R platform v.3.4.4. DESeq was specifically developed to find DE genes between two conditions at the same time point (mock versus transfected with 1B3/3A1/1K1) for RNA sequencing data with small sample size and overdispersion.31 The DESeq algorithm performs an internal normalization to account for sequencing depth and RNA composition. In brief, the raw counts are divided by sample-specific size factors determined by the median ratio of gene counts relative to the geometric mean per gene. To calculate p values in the absence of replicates, DESeq performs a binominal distribution test per gene, whereby the average gene spread is based on the transfected sample and the mock sample. Because the p value is related to the estimated gene expression and the fold change, which represents the gene expression in the transfected sample as compared to the mock sample, genes were selected for subsequent analysis based on the arbitrary cutoff p < 0.05. Furthermore, a minority of genes inf which expression was absent in mock samples (estimated expression = 0) were excluded from further analyses.
An additional analysis of the raw counts was done using the DESeq2 package36 by Single Cell Discoveries BV (Utrecht, the Netherlands). The six cell lines were grouped and considered replicates of each other to compare the gene-expression profiles between 1B3 and mock, while controlling for the cell-line effect using linear modeling. Furthermore, a fold-change threshold of 1.5× was specified. p values were adjusted for multiple testing with the Benjamini-Hochberg method.
Target prediction and pathway analysis
For miR-193a-3p target prediction, three online tools were used: TargetScan 7.2,52 MiRDB,53 and MicroT-CDS/Diana.54 Genes were considered a predicted target if they were listed as the target in at least one of the databases. For pathway analysis, lists of genes that were DE in at least three cell lines were uploaded and analyzed using IPA software55 (https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-ipa, QIAGEN), which computes two statistical measures. The overlap p value is calculated using Fisher’s exact test and compares the significant DE genes in the dataset versus those known to be part of a pathway. The threshold of significance for multiple testing is indicated on the graphs. The Z score provides a prediction of the directional effect. A positive Z score means activation, whereas a negative Z score means inhibition. IPA considers p <0.01 significant and z >2 or <−2 indicative of a clear directionality.
Acknowledgments
The authors thank GenomeScan BV for performing the RNA sequencing and Single Cell Discoveries BV for the statistical analysis of the transcriptome data.
Author contributions
Conceptualization, M.T.J.v.d.B., S.Y., M.F.A., B.J.T., and M.J.; methodology, T.d.G., H.C.d.B., R.M.V., and M.S.; investigation, M.T.J.v.d.B., R.M.V., S.Y., M.F.A., B.J.T., and M.J.; writing – original draft, M.T.J.v.d.B.; writing – review & editing, S.Y. and M.J.; supervision, L.A.H.v.P., R.Q.J.S., and M.J.
Declaration of interests
All authors are employees of InteRNA Technologies BV and hold stock options in the company. R.Q.J.S., L.A.H.v.P., and M.J. are also minority shareholders (<5%) in InteRNA Technologies BV.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.01.020.
Supplemental information
References
- 1.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 3.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ling H., Fabbri M., Calin G.A. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat. Rev. Drug Discov. 2013;12:847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hydbring P., Wang Y., Fassl A., Li X., Matia V., Otto T., Choi Y.J., Sweeney K.E., Suski J.M., Yin H. Cell-Cycle-Targeting MicroRNAs as Therapeutic Tools against Refractory Cancers. Cancer Cell. 2017;31:576–590.e8. doi: 10.1016/j.ccell.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poell J.B., van Haastert R.J., de Gunst T., Schultz I.J., Gommans W.M., Verheul M., Cerisoli F., van Puijenbroek A.A.F.L., van Noort P.I., Prevost G.P. A functional screen identifies specific microRNAs capable of inhibiting human melanoma cell viability. PLoS ONE. 2012;7:e43569. doi: 10.1371/journal.pone.0043569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grossi I., Salvi A., Abeni E., Marchina E., De Petro G. Biological Function of MicroRNA193a-3p in Health and Disease. Int. J. Genomics. 2017;2017:5913195. doi: 10.1155/2017/5913195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao X., Tang R.X., Xie Q.N., Lin J.Y., Shi H.L., Chen G., Li Z.Y. The clinical value of miR-193a-3p in non-small cell lung cancer and its potential molecular mechanism explored in silico using RNA-sequencing and microarray data. FEBS Open Bio. 2018;8:94–109. doi: 10.1002/2211-5463.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu M., Liu Z., Liu Y., Zhou X., Sun F., Liu Y., Li L., Hua S., Zhao Y., Gao H. PTP1B markedly promotes breast cancer progression and is regulated by miR-193a-3p. FEBS J. 2019;286:1136–1153. doi: 10.1111/febs.14724. [DOI] [PubMed] [Google Scholar]
- 10.Khordadmehr M., Shahbazi R., Sadreddini S., Baradaran B. miR-193: A new weapon against cancer. J. Cell. Physiol. 2019;234:16861–16872. doi: 10.1002/jcp.28368. [DOI] [PubMed] [Google Scholar]
- 11.Liang H., Liu M., Yan X., Zhou Y., Wang W., Wang X., Fu Z., Wang N., Zhang S., Wang Y. MiR-193a-3p functions as a tumor suppressor in lung cancer by down-regulating ERBB4. J. Biol. Chem. 2015;290:926–940. doi: 10.1074/jbc.M114.621409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X., Min S., Wu N., Liu H., Wang T., Li W., Shen Y., Zhao C., Wang H., Qian Z. miR-193a-3p inhibition of the Slug activator PAK4 suppresses non-small cell lung cancer aggressiveness via the p53/Slug/L1CAM pathway. Cancer Lett. 2019;447:56–65. doi: 10.1016/j.canlet.2019.01.027. [DOI] [PubMed] [Google Scholar]
- 13.Fan Q., Hu X., Zhang H., Wang S., Zhang H., You C., Zhang C.-Y., Liang H., Chen X., Ba Y. MiR-193a-3p is an Important Tumour Suppressor in Lung Cancer and Directly Targets KRAS. Cell. Physiol. Biochem. 2017;44:1311–1324. doi: 10.1159/000485491. [DOI] [PubMed] [Google Scholar]
- 14.Ma H., Yuan L., Li W., Xu K., Yang L. The LncRNA H19/miR-193a-3p axis modifies the radio-resistance and chemotherapeutic tolerance of hepatocellular carcinoma cells by targeting PSEN1. J. Cell. Biochem. 2018;119:8325–8335. doi: 10.1002/jcb.26883. [DOI] [PubMed] [Google Scholar]
- 15.Telford B.J., Yahyanejad S., de Gunst T., den Boer H.C., Vos R.M., Stegink M. Multi-modal effects of 1B3, a novel synthetic miR-193a-3p mimic, support strong potential for therapeutic intervention in oncology. Oncotarget. 2021 doi: 10.18632/oncotarget.27894. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lv L., Deng H., Li Y., Zhang C., Liu X., Liu Q., Zhang D., Wang L., Pu Y., Zhang H. The DNA methylation-regulated miR-193a-3p dictates the multi-chemoresistance of bladder cancer via repression of SRSF2/PLAU/HIC2 expression. Cell Death Dis. 2014;5:e1402. doi: 10.1038/cddis.2014.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwon J.-E., Kim B.-Y., Kwak S.-Y., Bae I.-H., Han Y.-H. Ionizing radiation-inducible microRNA miR-193a-3p induces apoptosis by directly targeting Mcl-1. Apoptosis. 2013;18:896–909. doi: 10.1007/s10495-013-0841-7. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y., Xu X., Xu X., Li S., Liang Z., Hu Z., Wu J., Zhu Y., Jin X., Wang X. MicroRNA-193a-3p inhibits cell proliferation in prostate cancer by targeting cyclin D1. Oncol. Lett. 2017;14:5121–5128. doi: 10.3892/ol.2017.6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu T., Li J., Yan M., Liu L., Lin H., Zhao F., Sun L., Zhang Y., Cui Y., Zhang F. MicroRNA-193a-3p and -5p suppress the metastasis of human non-small-cell lung cancer by downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. Oncogene. 2015;34:413–423. doi: 10.1038/onc.2013.574. [DOI] [PubMed] [Google Scholar]
- 20.Pu Y., Zhao F., Cai W., Meng X., Li Y., Cai S. MiR-193a-3p and miR-193a-5p suppress the metastasis of human osteosarcoma cells by down-regulating Rab27B and SRR, respectively. Clin. Exp. Metastasis. 2016;33:359–372. doi: 10.1007/s10585-016-9783-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma K., He Y., Zhang H., Fei Q., Niu D., Wang D., Ding X., Xu H., Chen X., Zhu J. DNA methylation-regulated miR-193a-3p dictates resistance of hepatocellular carcinoma to 5-fluorouracil via repression of SRSF2 expression. J. Biol. Chem. 2012;287:5639–5649. doi: 10.1074/jbc.M111.291229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Gunst T., van Pinxteren L.A.H., Janicot M., Schultz I.J., Schaapveld R.Q.J., Yahyanejad S. Anticancer microRNA and lipid formulations thereof. WO/2019/155094. 2019 [Google Scholar]
- 23.Hassler M.R., Turanov A.A., Alterman J.F., Haraszti R.A., Coles A.H., Osborn M.F., Echeverria D., Nikan M., Salomon W.E., Roux L. Comparison of partially and fully chemically-modified siRNA in conjugate-mediated delivery in vivo. Nucleic Acids Res. 2018;46:2185–2196. doi: 10.1093/nar/gky037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Volkov A.A., Kruglova N.S., Meschaninova M.I., Venyaminova A.G., Zenkova M.A., Vlassov V.V., Chernolovskaya E.L. Selective protection of nuclease-sensitive sites in siRNA prolongs silencing effect. Oligonucleotides. 2009;19:191–202. doi: 10.1089/oli.2008.0162. [DOI] [PubMed] [Google Scholar]
- 25.Chen P.Y., Weinmann L., Gaidatzis D., Pei Y., Zavolan M., Tuschl T., Meister G. Strand-specific 5′-O-methylation of siRNA duplexes controls guide strand selection and targeting specificity. RNA. 2008;14:263–274. doi: 10.1261/rna.789808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park J.H., Hong S.W., Yun S., Lee D.K., Shin C. Effect of siRNA with an asymmetric RNA/dTdT overhang on RNA interference activity. Nucleic Acid Ther. 2014;24:364–371. doi: 10.1089/nat.2014.0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khvorova A., Reynolds A., Jayasena S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 28.Lisowiec-Wąchnicka J., Bartyś N., Pasternak A. A systematic study on the influence of thermodynamic asymmetry of 5′-ends of siRNA duplexes in relation to their silencing potency. Sci. Rep. 2019;9:2477. doi: 10.1038/s41598-018-36620-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takele Assefa A., Vandesompele J., Thas O. On the utility of RNA sample pooling to optimize cost and statistical power in RNA sequencing experiments. BMC Genomics. 2020;21:312. doi: 10.1186/s12864-020-6721-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biswas S., Agrawal Y.N., Mucyn T.S., Dangl J.L., Jones C.D. Biological Averaging in RNA-Seq. arXiv. 2013 1309.0670, https:// https://arxiv.org/abs/1309.0670. [Google Scholar]
- 31.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kulkarni V., Naqvi A.R., Uttamani J.R., Nares S. MiRNA-Target interaction reveals cell-specific post-transcriptional regulation in mammalian cell lines. Int. J. Mol. Sci. 2016;17:72. doi: 10.3390/ijms17010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khan A.A., Betel D., Miller M.L., Sander C., Leslie C.S., Marks D.S. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat. Biotechnol. 2009;27:549–555. doi: 10.1038/nbt.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bharambe H.S., Joshi A., Yogi K., Kazi S., Shirsat N.V. Restoration of miR-193a expression is tumor-suppressive in MYC amplified Group 3 medulloblastoma. Acta Neuropathol. Commun. 2020;8:70. doi: 10.1186/s40478-020-00942-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamada K.M., Araki M. Tumor suppressor PTEN: modulator of cell signaling, growth, migration and apoptosis. J. Cell Sci. 2001;114:2375–2382. doi: 10.1242/jcs.114.13.2375. [DOI] [PubMed] [Google Scholar]
- 36.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stark A., Brennecke J., Russell R.B., Cohen S.M. Identification of Drosophila MicroRNA targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farh K.K.-H., Grimson A., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P. The Widespread Impact of Mammalian MicroRNAs on mRNA Repression and Evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 39.Krek A., Grün D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., Rajewsky N. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 40.Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 41.Jackson A.L., Linsley P.S. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat. Rev. Drug Discov. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 42.Leslie N.R., Downes C.P. PTEN function: how normal cells control it and tumour cells lose it. Biochem. J. 2004;382:1–11. doi: 10.1042/BJ20040825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Álvarez-Garcia V., Tawil Y., Wise H.M., Leslie N.R. Mechanisms of PTEN loss in cancer: It’s all about diversity. Semin. Cancer Biol. 2019;59:66–79. doi: 10.1016/j.semcancer.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 44.Tokunaga E., Kimura Y., Oki E., Ueda N., Futatsugi M., Mashino K., Yamamoto M., Ikebe M., Kakeji Y., Baba H., Maehara Y. Akt is frequently activated in HER2/neu-positive breast cancers and associated with poor prognosis among hormone-treated patients. Int. J. Cancer. 2006;118:284–289. doi: 10.1002/ijc.21358. [DOI] [PubMed] [Google Scholar]
- 45.Balsara B.R., Pei J., Mitsuuchi Y., Page R., Klein-Szanto A., Wang H., Unger M., Testa J.R. Frequent activation of AKT in non-small cell lung carcinomas and preneoplastic bronchial lesions. Carcinogenesis. 2004;25:2053–2059. doi: 10.1093/carcin/bgh226. [DOI] [PubMed] [Google Scholar]
- 46.Kreisberg J.I., Malik S.N., Prihoda T.J., Bedolla R.G., Troyer D.A., Kreisberg S., Ghosh P.M. Phosphorylation of Akt (Ser473) is an excellent predictor of poor clinical outcome in prostate cancer. Cancer Res. 2004;64:5232–5236. doi: 10.1158/0008-5472.CAN-04-0272. [DOI] [PubMed] [Google Scholar]
- 47.Lee Y.-R., Chen M., Lee J.D., Zhang J., Lin S.-Y., Fu T.-M., Chen H., Ishikawa T., Chiang S.-Y., Katon J. Reactivation of PTEN tumor suppressor for cancer treatment through inhibition of a MYC-WWP1 inhibitory pathway. Science. 2019;364:eaau0159. doi: 10.1126/science.aau0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moses C., Nugent F., Waryah C.B., Garcia-Bloj B., Harvey A.R., Blancafort P. Activating PTEN Tumor Suppressor Expression with the CRISPR/dCas9 System. Mol. Ther. Nucleic Acids. 2019;14:287–300. doi: 10.1016/j.omtn.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saito T., Sætrom P. Target gene expression levels and competition between transfected and endogenous microRNAs are strong confounding factors in microRNA high-throughput experiments. Silence. 2012;3:3. doi: 10.1186/1758-907X-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaelin W.G., Jr. Common pitfalls in preclinical cancer target validation. Nat. Rev. Cancer. 2017;17:425–440. doi: 10.1038/nrc.2017.32. [DOI] [PubMed] [Google Scholar]
- 51.Brignole C., Pastorino F., Marimpietri D., Pagnan G., Pistorio A., Allen T.M., Pistoia V., Ponzoni M. Immune cell-mediated antitumor activities of GD2-targeted liposomal c-myb antisense oligonucleotides containing CpG motifs. J. Natl. Cancer Inst. 2004;96:1171–1180. doi: 10.1093/jnci/djh221. [DOI] [PubMed] [Google Scholar]
- 52.Agarwal V., Bell G.W., Nam J.-W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong N., Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146–D152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paraskevopoulou M.D., Georgakilas G., Kostoulas N., Vlachos I.S., Vergoulis T., Reczko M., Filippidis C., Dalamagas T., Hatzigeorgiou A.G. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41:W169–W173. doi: 10.1093/nar/gkt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krämer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. https://academic.oup.com/bioinformatics/article/30/4/523/202720 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.