Abstract
Super-enhancers (SEs) mediate high transcription levels of target genes. Previous studies have shown that SEs recruit transcription complexes and generate enhancer RNAs (eRNAs). We characterized transcription at the human and murine β-globin locus control region (LCR) SE. We found that the human LCR is capable of recruiting transcription complexes independently from linked globin genes in transgenic mice. Furthermore, LCR hypersensitive site 2 (HS2) initiates the formation of bidirectional transcripts in transgenic mice and in the endogenous β-globin gene locus in murine erythroleukemia (MEL) cells. HS2 3′eRNA is relatively unstable and remains in close proximity to the globin gene locus. Reducing the abundance of HS2 3′eRNA leads to a reduction in β-globin gene transcription and compromises RNA polymerase II (Pol II) recruitment at the promoter. The Integrator complex has been shown to terminate eRNA transcription. We demonstrate that Integrator interacts downstream of LCR HS2. Inducible ablation of Integrator function in MEL or differentiating primary human CD34+ cells causes a decrease in expression of the adult β-globin gene and accumulation of Pol II and eRNA at the LCR. The data suggest that transcription complexes are assembled at the LCR and transferred to the globin genes by mechanisms that involve Integrator mediated release of Pol II and eRNA from the LCR.
INTRODUCTION
Enhancers are cis-regulatory DNA elements usually 200–400 bp long, that recruit specific combinations of transcription factors and activate transcription initiation or elongation at target gene promoters (1,2). Many enhancers also initiate transcription of eRNAs, and genome-wide studies have shown that recruitment of RNA polymerase II (Pol II) occurs at a large number of enhancers (3,4). Importantly, association of Pol II at enhancers and transcription of eRNA often precedes activation of target genes during cell differentiation (5). Super-enhancers (SEs) are extended cis-regulatory elements that associate with extremely high levels of Mediator and Pol II (6,7). Locus control regions (LCRs) are SEs that have been demonstrated to provide position-independent expression of linked genes in transgenic mice through dominant chromatin opening activities (8,9). Like other SEs, LCRs associated with the human and murine β-globin gene loci consist of multiple DNase I hypersensitive sites (HSs) that function together to mediate extremely high-level expression of the cis-linked globin genes (10–12).
The Tuan and Proudfoot laboratories were the first to document non-coding transcription originating within HS sites of the human LCR (13,14). Subsequent reports demonstrated that LCR HS sites harbor promoter-like activity (15,16). While Tuan et al. proposed a facilitated tracking mechanism by which the enhancer is delivered to the globin gene promoters by a transcription mediated process (13), others hypothesized that LCR recruited transcription complexes are transferred to the globin gene promoters by looping mechanisms (17–19). Evidence for both mechanisms exist. For example, in support of a tracking mechanism investigators have shown that placing an insulator between the LCR and the globin genes caused accumulation of Pol II at the respective locations and reduced expression of globin genes (20,21). On the other hand, the LCR is positioned in relatively close proximity to the adult β-globin gene promoter in differentiated cells expressing the gene (22). In erythroid cells lacking the transcription factor NF-E2, adult β-globin expression was reduced and Pol II accumulated at the LCR (17). Moreover, in vitro studies demonstrated that Pol II is transferred from an immobilized LCR to a β-globin gene template in a process stimulated by NF-E2 (18). Deletion of the murine LCR reduced recruitment of Pol II at the adult globin gene promoter by about 50%; however, the remaining Pol II recruited to the globin gene was not transcriptionally competent (23). These data suggest that the LCR regulates both recruitment of Pol II and transcription elongation at the β-globin gene.
The functional role of transcription initiation at enhancers or that of eRNAs is not completely understood (24,25). Some studies have shown that eRNAs participate in the transcription activation function of enhancers (26). Other studies demonstrated that the process of transcription itself contributes to enhancer function (27). Mounting evidence suggests that eRNAs play a role in orchestrating gene expression in cis or trans (24,25). Knockdown of eRNAs by RNA interference mechanisms or by drug mediated transcription termination decreased target gene expression (28,29). eRNAs are relatively short in length and it was shown that early termination of enhancer mediated transcription requires the Integrator complex, which also terminates transcription of small nuclear RNAs (snRNAs) (30). Inhibition of Integrator complex function led to prolonged transcription at enhancers and a reduction in enhancer function (30).
The mammalian β-type globin genes are expressed exclusively and at extremely high levels in erythroid cells in a developmental stage-specific manner (9). Those globin genes close to the LCR are expressed during the embryonic and fetal stages, whereas the more distant globin genes are expressed during the adult stages of hematopoiesis. The stage-specific expression is mediated primarily by gene proximal regulatory elements; however, the order of the genes with respect to the LCR also contributes to developmental regulation (31,32).
Here, we show that Pol II is recruited to the human β-globin LCR in transgenic mice and initiates generation of eRNAs independently from the linked human β-globin genes. RNA fluorescence in situ hybridization (RNA-FISH) combined with DNA-FISH revealed that eRNA generated from HS2 remains associated with the β-globin gene locus. Depletion of the eRNA 3′ to HS2 (HS2 3′eRNA) in MEL cells caused a reduction in adult β-globin gene transcription and an accumulation of Pol II at LCR element HS2. Ablation of Integrator subunit 11 (INTs11) in MEL or in differentiating human CD34+ cells reduced globin expression and increased the levels of LCR HS2 and HS3 associated eRNA. Furthermore, depletion of Integrator complex function led to accumulation of Pol II at LCR elements HS2 and HS3 and a reduction of Pol II recruitment to the β-globin gene. These data support a model of Pol II transfer from the LCR to the globin genes and implicate eRNA and Integrator in this process.
MATERIALS AND METHODS
Generation and characterization of human LCR transgenic mice
Enzymes ClaI and NotI were used to cut out the human β-globin locus control region from a plasmid (pRS316/LCR) previously described (16). The products were run on a 1% agarose gel and the 15kbp band containing the LCR was extracted from the gel and purified using the QIAquick Gel Extraction Kit (Qiagen, 28706). The DNA was diluted to concentrations of 1 and 0.5 ng/μl in injection buffer and injections were performed on fertilized FVB oocytes as described previously (33). Eggs were implanted into the uterus of pseudo-pregnant mothers and after birth, DNA was isolated and subjected to PCR for genotyping as described previously (33).
The integration sites of the transgenes were determined by inverse PCR technique. For this, 3 μg transgenic mouse genomic DNA was digested using SacI and then run on a 1% agarose gel to verify digestion. 1 μg of this digested DNA was used in a 400 ul intramolecular ligation reaction and then the DNA was purified using phenol-chloroform/isoamyl alcohol extraction and ethanol precipitation. PCR reactions using inverse PCR primers complementary to sequences near the 3′ and the 5′ ends of the inserted LCR were performed and the products were run on 1% agarose gels. Resulting bands were purified using the QIAquick Gel Extraction kit and were ligated with the pGEM-T vector (Promega, A3600). These plasmids were then transformed into Stbl2 competent cells (Invitrogen, 10268-019) and selected using Ampicillin. Colonies were picked and selected by colony PCR (using the same inverse PCR primers) for positive clones. Minipreps (QIAprep Spin Miniprep Kit, 27106) were performed and the samples were sent for Sanger Sequencing. The primers used in the PCR are listed in the Supplementary Table.
Real-time quantitative PCR for copy number determination was carried out as previously described with minor modifications (34). Three to four mice from each transgenic line were analyzed on at least two separate occasions with reactions prepared in triplicate. The 2−ΔΔCt method was used for analysis and results were confirmed using the standard curve method (35). Mouse HS2 primers were used to amplify a reference sequence and DNA from a β-YAC line was used as a one-copy calibrator.
Cell culture and stable transfection
MEL cells were grown in DMEM medium (Cellgro, Manassas, VA) supplemented with 10% (vol/vol) fetal bovine serum (FBS; Sigma) and 1% (vol/vol) penicillin–streptomycin (P–S; Cellgro) in the presence of 5% CO2 at 37°C. The cell density was maintained at 106 cells/ml. MEL cells were transfected with the TripZ vector containing two different shRNAs (shRNA-1 and shRNA-2) against INTs11 and two shRNAs against 3′HS2eRNA [3′HS2(A) and 3′HS2(B)] or scrambled control shRNAs. Single cell clones were generated using an established protocol developed by John A Ryan at Corning (https://www.corning.com/catalog/cls/documents/protocols/Single_cell_cloning_protocol.pdf). The plate was kept at 37°C for 7–9 days to allow single cells to grow into colonies. The colonies were picked and seeded into 96-well plates. The 96-well plates were cultured for about 4 days until the cells reached confluency. Clones were then scaled up to 24-well plates and finally adapted for culture in flasks. The sequences of shRNAs are listed in the Supplementary Table.
The pTRIPZ vector (for the INTs11kd cell line) or pGIPZ vector (for the 3′HS2 eRNA KD cell lines) containing shRNA sequences were transfected into MEL cells via the Trans-Lentiviral packaging kit (Thermo Scientific). Lenti-X 293T cells (Clontech) were passed at ∼70% confluency for 2 days to ensure rapid growth. In six-well cell culture plates (Costar), 1.2 × 106 Lenti-X 293T cells in two mLs of standard DMEM cell culture media [Dulbecco's modified Eagle's medium (Cellgro), 10% (vol/vol) fetal bovine serum (FBS) and 1% (vol/vol) penicillin–streptomycin (Cellgro)] were incubated overnight at 37°C in the presence of 5% CO2. The transfection mix was created by adding 4.3 μl of Trans-Lentiviral packaging mix with 6 μg of pTRIPZ/pGIPZ-shRNA vector and bringing the solution to 135 μl with sterile water. To each transfection mix 15 μl of CaCl2 reagent was added (catalog number TLP5911), and the solution was gently mixed while adding 150 μl 2× HEPES-buffered saline solution (catalog number TLP5910). The completed transfection mix was incubated at room temperature for 3 min before being added in a drop wise manner to the Lenti-X 293T cells. Cells were incubated for 12 h at 37°C in the presence of 5% CO2. The media with transfection mix was exchanged for Dulbecco's modified Eagle's medium with 5% (vol/vol) fetal bovine serum (FBS) and 1% (vol/vol) penicillin–streptomycin. The Lenti-X 293T cells were then incubated with the reduced serum media at 37°C in the presence of 5% CO2 for 48 h. The viral particles were released into the media over a period of 48 h. The media with viral particles was cleared of detached cells and debris by centrifugation at 3600 rpm for 5 min. A mixture of 1 ml virus containing media, 1 ml of standard DMEM cell culture media, and 8 μg/ml polybrene (Sigma) was used to resuspend 4 × 105 MEL cells. MEL cells were incubated with virus for 48 h before the media was refreshed. The transfected MEL cells were then subjected to selection with 1 μg/ml puromycin for 7 days. The transfected MEL cells were maintained with standard DMEM cell culture media containing 0.25 μg/ml puromycin. Single cell clones for each shRNA stably transfected cell line were generated as described above. For induction of differentiation, MEL cells were grown in 2% (vol/vol) dimethyl sulfoxide (DMSO) for up to 72 h. For Integrator shRNA expression, transfected MEL cells were treated with both 1 μg/ml Doxycycline (Sigma, D9891) and 2% final volume DMSO (Sigma, D2650) for 24 and 48 h.
The Umbilical Cord Blood (UCB) CD34+ cells were obtained from STEMCELL Technologies. A total of 2 × 105 cells were expanded in Stem Span Serum Free Expansion Medium II (STEMCELL Technologies, Catalog 09605) supplemented with StemSpan™ Erythroid Expansion Supplement (100×) (Catalog No. 02691) and 1% penicillin–streptomycin. The cells were allowed to expand for 9 days with frequent media changes. On the ninth day, the media was changed and supplemented with 3% human serum (Sigma Aldrich Catalog No. H4522), 2 U/ml human recombinant erythropoietin (EPO; STEMCELL Technologies Catalog No. 78007) for inducing differentiation. The cells were subjected to siRNA treatment 24 h after inducing with EPO. Accell non-targeting pool control siRNA (control siRNA) (Accell Catalog No. D-001910-01) and Accell Human INTs11 (54973) siRNA (INTs11 kd siRNA) SMARTpool (Accell Catalog No. E-013798) were obtained from GE Dharmacon. A total of 50 000 human UCB CD34+ cells were suspended in Accell Delivery Medium (Dharmacon Catalog B-005000) with 2 units/ml human recombinant EPO, 1% penicillin–streptomycin, and seeded in a 96-well plate. The siRNAs were maintained at a final concentration of 1 μmol/l according to the manufacturer's protocol. The cells were transferred to basic growth medium 72 h after transfection. Aliquots of cells were collected at 72 h and at 6 days for downstream experiments.
Western blotting
Western blotting procedures were performed using a previously published protocol (36). Briefly, cells were collected and lysed in radioimmunoprecipitation assay (RIPA) buffer (10 mmol/liter Tris–HCl, pH 8.0, 1 mmol/liter EDTA, 0.5 mmol/l EGTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 140 mmol/l NaCl, 1× protease inhibitor). The supernatant containing the proteins was loaded onto 4–15% (wt/vol) SDS-PAGE gels (Bio-Rad, Hercules, CA). After electrophoresis the proteins were transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad). The PVDF membranes were incubated with anti-INTs11 (A301-274A; Bethyl Laboratories Inc.), anti-GAPDH (catalog number CST5174; Cell Signaling, Beverly, MA), or anti-β-actin (catalog number sc-47778, Santa Cruz, Biotechnology, Santa Cruz, CA) antibodies. The membranes were washed and incubated with secondary goat anti-rabbit IgG-HRP (catalog number sc-2004) or goat anti-mouse IgG-HRP (catalog number sc-2005; Santa Cruz Biotechnology, Santa Cruz, CA). Antibodies were detected using an enhanced chemiluminescence (ECL) reagent (Millipore, Danvers, MA) and visualized with X-ray films (Kodak, Rochester, NY).
RNA extraction and gene expression analysis
Total RNA was extracted using the RNeasy kit (Qiagen, Hilden, Germany) following the manufacturer's protocol. RNA was reverse transcribed into cDNA using the IScript cDNA synthesis kit (Bio-Rad). The cDNAs were subjected to qPCR in a 10-μl reaction mix with SYBR green and 1.5 μmol/l of each forward and reverse primer. Gene expression was analyzed by the normalization of expression to that of GAPDH/ß-Actin using the ΔCT method. The primer sequences used for RT-qPCR are listed in the Supplementary Table.
Chromatin immunoprecipitation
The chromatin immunoprecipitation (ChIP) protocol used in this study was described previously (36). Briefly, 5 × 106 cells were collected and cross-linked with 1% (vol/vol) formaldehyde at room temperature for 10 min. The reaction was quenched with 125 mmol/l glycine, and the cells were incubated in cell lysis buffer (5 mmol/l PIPES [1,4-piperazinediethanesulfonic acid], pH 8.0, 85 mmol/l KCl, 0.5% NP-40 [nonyl octylphenoxypolyethoxylethanol], 1× protease inhibitor) and nucleus lysis buffer (50 mmol/l Tris, pH 8.0, 10 mmol/l EDTA, 0.32% SDS, 1× protease inhibitor). Chromatin was fragmented to average 300-bp fragments on ice using a Bioruptor disruptor (Diagenode, Denville, NJ). For each immunoprecipitation reaction, chromatin from 2 × 105 cells was incubated with 2.5 μg antibody [anti-INTS11 (A301-274A; Bethyl Laboratories Inc.,), anti-GAPDH (CST5174; Cell Signaling), anti-RNA polymerase II, clone CTD4H8 (Millipore, 05-623), anti-RNA polymerase II CTD repeat YSPTSPS (phospho S5) antibody (4H8 ab5408; Abcam), or rabbit polyclonal anti-RNA polymerase II CTD repeat YSPTSPS (phospho S2) ab5095; Abcam] plus protein A/G magnetic beads (Pierce) rotating at 4°C overnight. The chromatin-antibody-bead complexes were subjected to washing, eluting, and reverse cross-linking at 65°C overnight. The DNA was purified using a PCR purification kit (Qiagen) and subjected to qPCR as described earlier. The sequences of the primers used in the ChIP assays are listed in the Supplementary Table.
Chromatin-associated RNA isolation
The chromatin associated RNA was isolated along with the cytoplasmic and nucleoplasmic fraction following a published protocol (37). Briefly, 105–106 cells were homogenized by lysis with cell lysis buffer (10 mM Tris pH 7.4, 150 mM NaCl, 0.15% Igepal CA-630). The lysate was centrifuged through a 24% sucrose cushion (sucrose buffer, 10 mM Tris pH 7.4, 150 mM NaCl, 24% sucrose), which yields a pellet with purified nuclei and the supernatant representing the cytoplasmic fraction. The sedimented nuclei were gently resuspended in glycerol buffer (20 mM Tris pH 7.4, 75 mM NaCl, 0.5 mM EDTA, 50% glycerol) followed by rapid lysis in nuclear lysis buffer (10 mM Tris pH 7.4, 1 M Urea, 0.3 M NaCl, 7.5 mM MgCl2, 0.2 mM EDTA, 1% Igepal CA-630). After a brief centrifugation at 13 000g for 2 min, the supernatant represented the nucleoplasm and the pellet contained the chromatin associated RNA. RNA from all fractions was isolated by Trizol extraction (15596026, Invitrogen). The RNA was reverse transcribed into cDNA using the IScript cDNA synthesis kit (Bio-Rad). The cDNAs were subjected to qPCR in a 10-μl reaction mix with SYBR green and 1.5 μmol/l of each forward and reverse primer. Gene expression was analyzed by the normalization of expression to that of the chromatin fraction of each sample using the ΔCT method. The primer sequences used for RT-qPCR are listed in the Supplementary Table.
DNA and RNA fluorescence in situ hybridization
Mouse Fetal Liver cells/MEL cells were fixed on glass slides and incubated in methanol at –20°C for 10 min, dried and then incubated in 300 μl of 10% formaldehyde and 2× SSC for 8 min. A probe cocktail was prepared by mixing 20 ng of probe DNA (ds Beta major probe labelled with Digoxigenin using DIG Nick Translation Mix by Sigma) and 3 mg of salmon sperm DNA. DNA was lyophilized in a heating block, resuspended in 7 μl of deionized formamide and mixed well (at 15–25°C for at least 30 min). Probe was denatured and pre-labelled with Cot-1 DNA (85°C for 5 min, 37°C for 20 min, 12°C hold). Fixed chromosome specimens were denatured by immersing the slides in denaturation buffer (70% formamide, 2× SSC [pH 7.0]) at 82°C for 2 min. 10 μl of the denatured probe was applied onto an 18 × 18-mm glass coverslip (prewarmed to 37°C) and the slide was lowered onto the probe mix to avoid trapping air bubbles. Rubber cement was applied to seal around the coverslip. The slides with samples were hybridized in a humidified chamber in a hot-air oven at 37°C for 4 h. The slides were washed with 2× SSC followed by washing in wash buffer (70% formamide, 2× SSC (pH 7), preheated to 42°C). 200 μl of detecting solution (4% dried milk powder, 4× SSC) was added to the hybridized sample and incubated at 25°C for 20 min. 50μl of detecting solution containing Anti Digoxigenin Alexa 488 antibody (1:1000) was added to a coverslip and incubated for 2 h at RT. The slides were then washed in wash buffer (containing 2× SSC, 0.1% Tween 20 and DAPI to stain chromatin), three times at 42°C. The slides were mounted, and images were analyzed and documented using a Leica TCS SP5 confocal microscope. The sequences of the probes used in DNA FISH are listed in the Supplementary Table. For immunofluorescence microcopy MEL cells were incubated with the Pol II-S2P primary and FITC-conjugated secondary antibody (Santa Cruz, sc2777).
The RNA FISH probes were prepared and labelled by nick translation using a nick translation kit. (Catalog No. 11745824910, Roche). The hybridization solution was prepared by mixing 50% formamide, 2× SSC, dextran sulfate, yeast t-RNA and nick-translated probe and was incubated at 3°C for 10 min. The probe was denatured by heating at 90°C for 5–10 min. The probe was chilled on ice immediately. Fixed MEL cells were hybridized with the denatured 3′HS2 LCR probe overnight. The probe was detected using a 5-layer Ab system (biotin-StAv) for 12 h (2× signal amplification) (1st antibody StAv Alexa 488 (1:1000) 2 h at RT, wash with 3× PBS for 5 min, second Ab Biotinylated Anti Streptavidin (1:200) 2 h at RT, wash with 3× PBS for 5 min, first antibody StAv Alexa 488 (1:1000) 2 h at RT, wash with 3× PBS for 5 min, second Ab Biotinylated Anti Streptavidin (1:200) 2 h at RT, wash with 3× PBS for 5 min, first antibody StAv Alexa 488 (1:1000) 4°C overnight). Post-hybridization wash was performed using 2× SSC for 5 min, followed by 1× SSC for 5 min and another 1× SSC wash for 5 min at 37°C. DNA was counterstained with DAPI. In control experiments cells were treated with RNase. The slides were mounted, and images were analyzed and documented using a Leica TCS SP5 confocal microscope. The sequence of the probe used in RNA FISH is listed in the Supplementary Table.
Statistics
The experiments were repeated at least three times unless stated otherwise. In experiments involving qPCR, each sample from the three repeats was analyzed by PCR three times. Error bars represent the standard error of the mean (SEM). Statistical significance (P value) was determined using Student's t-test.
RESULTS
Recruitment of Pol II to human LCR HS2 in transgenic mouse fetal liver
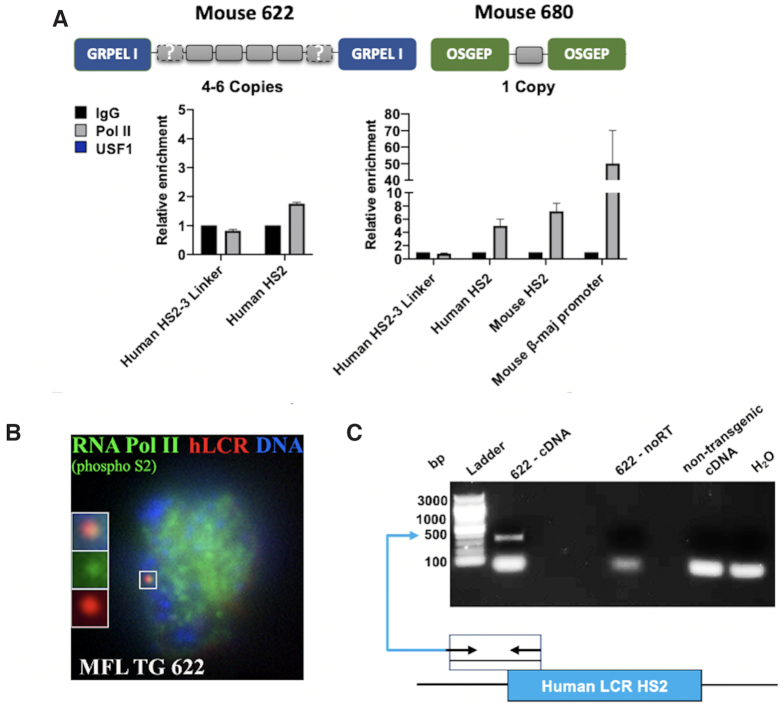
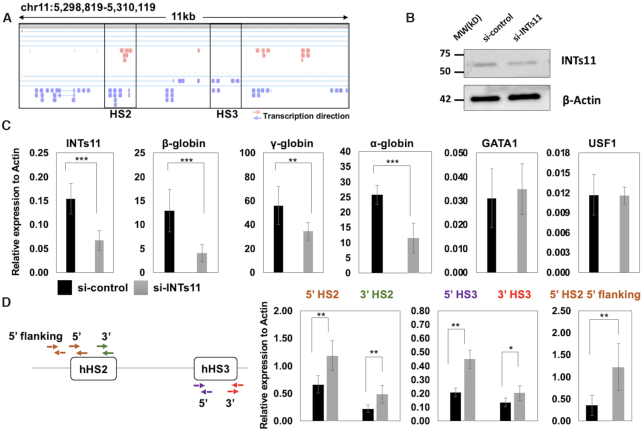
Previous data have shown that LCR HSs associate with transcription complexes (16,17). To determine if the LCR is able to recruit transcription complexes in the absence of linked globin genes we generated transgenic mice with the human LCR (Figure 1A). The fragment encompassed all of the human β-globin LCR HSs but none of the human globin genes. We generated two transgenic lines harboring one or up to six copies of the human LCR (Figure 1A). The copy number was determined by quantitative PCR using a human β-globin yeast artificial chromosome (YAC) transgenic mouse as a one copy calibrator (32). We used inverse PCR to map the integration sites of the transgenes. In the two transgenic lines, the human LCR integrated into an intron of protein-coding genes. In line 622, four to six copies of the transgenic LCR integrated into the last intron of the GrpE-like I gene (GRPELI), which encodes a protein of the mitochondrial PAM (presequence translocase-associated motor) complex (38). In line 680, one copy of the transgenic LCR integrated into an intron of the OSGEP gene, which encodes a putative O-sialoglycoprotein endopeptidase (39).
Figure 1.
Recruitment of Pol II to human LCR HS2 in transgenic mice. (A) Two transgenic mouse lines were generated in which the human LCR integrated with four to six copies or one copy into introns of the GRPEL1 (line 622) or OSGEP (line 680) genes, respectively. Fetal liver (12.5 dpc) was isolated from the transgenic mice and subjected to ChIP followed by qPCR using antibodies specific for Pol II and control IgG, and primers specific for mouse or human HS2, for a region in between human HS2 and HS3 (HS2-3 Linker), or for the mouse β-maj-globin promoter. Error bars reflect SEM from three independent experiments. (B) Association of the transgenic human LCR with Pol II foci in mouse fetal liver cells (12.5 dpc). Fetal liver cells were isolated from transgenic line 622 and subjected to immunofluorescence using antibodies specific for Pol II-S2P (green) and a fluorescently labeled probe specific for the transgenic human LCR (red; DNA in blue). The magnified insert shows association between the transgenic LCR and Pol II-S2P (five out of six nuclei showed association of the transgenic LCR with transcription foci). (C) Analysis of HS2 5′eRNA. RNA was isolated from fetal liver of transgenic line 622 (12.5 dpc) and subjected to PCR analysis with or without reverse transcriptase (noRT). Results from analyzing DNA from wild-type, non-transgenic mice, and from a negative H2O control are also shown. The diagram at the bottom shows the location of primers used for the RT-PCR analysis.
We next examined the interaction of Pol II with the transgenic human LCR and with the endogenous mouse LCR using chromatin immunoprecipitation (ChIP) (Figure 1A). We used an antibody that does not discriminate between the different phosphorylated forms of Pol II (S5P or S2P). The data show that Pol II interacted with human HS2 in the two transgenic lines examined. We also examined association with a region flanking the human LCR HSs 2 and 3 (Human HS2–3 Linker). Pol II did not interact with the transgenic HS flanking region, ruling out the possibility that interaction of Pol II is detected because the transgene integrated into an active gene locus. The binding of Pol II at HS2 in line 680 was as efficient as the binding of Pol II at the endogenous LCR HS elements, and as expected, less efficient compared to binding of Pol II at the endogenous mouse β-maj-globin promoter. This is relevant as line 680 harbored one copy of the human LCR.
We next examined association of the transgenic LCR with transcription foci in mouse fetal liver cells. We isolated fetal liver cells from transgenic line 622 and subjected the cells to immunofluorescence microscopy using antibodies specific for the serine 2 phosphorylated form of Pol II (Pol II-S2P) and a fluorescently labeled DNA probe corresponding to the human LCR (Figure 1B). Pol II-S2P represents the transcription elongating form of Pol II (40). The data show that the transgenic LCR associates with Pol II-S2P foci, suggesting that the LCR is in close proximity to transcription domains. It is possible that transcription of the GRPELI gene contributes to associations with transcription foci. To examine if transgenic LCR HSs initiate formation of eRNA, we subjected fetal liver RNA from transgenic line 622 to reverse transcriptase-PCR analysis (Figure 1C). In this experiment, we analyzed HS2 antisense eRNA at the 5′end, to distinguish the signal from the potential sense transcription of the GRPELI gene. We did indeed detect eRNA at the transgenic HS2 5′region. This transcript was absent in the non-transgenic control. Together, the data demonstrate that the transgenic human LCR is capable of independently recruiting Pol II transcription complexes in the absence of linked globin genes.
Stability, location and function of HS2 3′eRNA in MEL cells
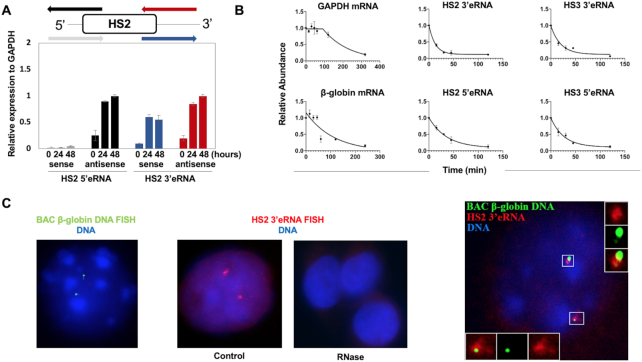
To further analyze the importance of eRNAs with respect to globin gene regulation we examined their expression, stability and localization in differentiating murine erythroleukemia (MEL) cells (Figure 2). We focused on characterizing transcripts around LCR HS2, which has been shown to display the strongest enhancer activity. Examination of sense and antisense transcription 5′ and 3′ of LCR HS2 revealed that both sense eRNA 3′ of HS2 and antisense eRNA transcription 5′ and 3′ of HS2 increased 24 and 48 h after induction of differentiation (Figure 2A). We did not detect upregulation of a sense transcript 5′ of HS2. These data are largely consistent with previous reports which also demonstrated that formation of eRNAs at the LCR precedes the transcription of the β-globin gene (41).
Figure 2.
Formation, stability and localization of LCR associated eRNA. (A) Transcription of 5′ and 3′ HS2 eRNAs during differentiation of MEL cells. Diagram on top showing primers used to assess HS2 5′ and 3′ antisense and sense transcription. MEL cells were subjected to differentiation for 24 and 48 h with 2% (final v/v) DMSO. RNA was isolated, subjected to reverse transcription using strand-specific primers, and analyzed by qPCR. (B) Measurements of GAPDH, β-maj-globin RNA as well as HS2 and HS5 5′ and 3′ eRNA half-life. RNA was isolated from differentiated MEL cells incubated with actinomycin D for up to 8 h and subjected to RT-qPCR using primers specific for GAPDH, β-maj-globin, and HS2 or HS3 5′ and 3′ eRNAs. Error bars reflect SEM from three independent experiments. (C) Differentiated MEL cells were subjected to β-globin gene locus DNA-FISH (left, representative nucleus) or HS2 3′eRNA-FISH (middle-left, representative nucleus; middle-right: RNAse treatment to show specificity of RNA signal, three representative nuclei) using fluorescently labeled probes specific for the murine β-globin gene locus (green) or HS2 3′eRNA (red; DNA: blue). Right: combination of HS2 3′eRNA and β-globin gene locus DNA FISH. HS2 3′eRNA (red) is associated with the β-globin gene locus (green; DNA: blue; four out of four nuclei showed association between the globin gene locus on HS2 3′eRNA).
Previous studies have shown that eRNAs are unstable (24). We measured the stability of HS2 and HS3 5′ and 3′eRNAs as well as GAPDH and βmaj-globin RNA by RT-qPCR of MEL cells treated with the transcription inhibitor actinomycin D (Figure 2B). Actinomycin D intercalates into DNA and prevents unwinding of DNA thus inhibiting RNA polymerase activity (42). The transcripts of the β-maj-globin and GAPDH genes revealed stabilities with a half-life (t1/2) of 98 and 78 min, respectively. In contrast, HS2 3′eRNA revealed a significant shorter half-life of about 9 min. The half-life of the other LCR eRNAs was ∼14 (HS3 3′eRNA), ∼18 (HS3 5′eRNA) and 25 (HS2 5′eRNA) min.
We next performed combined RNA and DNA FISH to examine the location of HS2 3′eRNA with respect to the β-globin gene locus (Figure 2C). We used a bacterial artificial chromosome (BAC) probe for the murine β-globin gene locus and probes specific for HS2 3′eRNA. The data show that most cells revealed two signals for the β-globin gene locus (green) in induced MEL cells (representative nucleus, Figure 2C, left). Cells induced to differentiate for 48 hours revealed two RNA foci (red) as well (representative nucleus, Figure 2C, middle-left), suggesting that a large fraction of the eRNA is concentrated at a specific nuclear location. The HS2 3′eRNA foci were not visible in cells treated with RNase, confirming the specificity of the signals (three representative nuclei, Figure 2C, middle-right). Combination of RNA and DNA FISH revealed that the HS2 3′eRNA is present in cloud-like foci and located in close proximity to and partially overlapping with the β-globin gene locus (representative nucleus, Figure 2C, right).
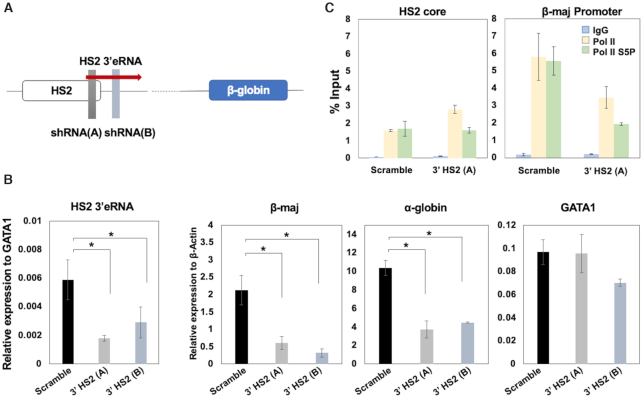
eRNA molecules per se have been shown to contribute to the activation of gene expression by enhancers, whereas other studies demonstrated that it is the process of eRNA transcription that is crucial for gene regulation (24,27). We reduced the abundance of the HS2 3′eRNA by shRNA mediated knockdown in MEL cells (Figure 3). We generated two single cell clones expressing different shRNAs against the HS2 3′eRNA (3′HS2 A and B; Figure 3A). Stably transfected cells expressing the 3′HS2 shRNAs revealed a significant reduction in the HS2 3′eRNA compared to clones expressing scrambled shRNA (Figure 3B). The reduction in HS2 eRNA resulted in a decrease in adult β-globin gene expression. Expression of the GATA1 gene, which encodes for an essential erythropoietic transcription factor, was not affected in the 3′HS2 (A) KD cells, and was only mildly reduced in the 3′HS2 (B) KD cells (43). However, we also detected a decrease in expression of the adult α-globin gene, albeit not to the extent observed for the β-globin gene. We next assessed the binding of Pol II using ChIP at HS2 and the adult β-globin promoter in cells expressing the scrambled shRNA and in cells expressing the shRNA directed against HS2 3′eRNA (Figure 3C). We used two different antibodies; antibodies specific for the serine 5 phosphorylated form (Pol II S5P) and antibodies that do not distinguish between the differentially phosphorylated forms of Pol II (Pol II). Figure 3C shows that reducing the abundance of the HS2 3′eRNA caused a reduction in the association of Pol II and Pol II-S5P with the β-maj-globin gene promoter, consistent with reduced expression of the gene. In contrast, the levels of Pol II, but not Pol II-S5P, were increased at LCR HS2 in cells expressing HS2 3′shRNA. These data show that the HS2 3′eRNA plays an active role in the regulation of adult β-globin gene expression.
Figure 3.
Changes of transcription and Pol II association in MEL cells expressing shRNA against HS2 3′eRNA. (A) Diagram showing targeting of HS2 3′eRNA by two shRNAs. (B) Depletion of HS2 3′eRNA reduces β-maj-globin gene expression. MEL cells stably expressing shRNA against HS2 3′eRNA (clones A and B) or scrambled control shRNA, were subjected to RT-qPCR using primers specific for HS2 3′eRNA or the β-maj-globin, α-globin, and GATA1 genes. HS2 3′eRNA expression was normalized to GATA1 RNA because of the high expression of β-actin. (C) Changes in Pol II accumulation in the β-globin gene locus in cells expressing HS2 3′eRNA shRNA. MEL cells stably expressing shRNA against HS2 3′eRNA (clone A) or scrambled shRNA were subjected to ChIP-qPCR using antibodies specific for Pol II, Pol II-S5P, and control IgG, and primers specific for the HS2 core and the β-maj-globin gene. Error bars reflect SEM from three independent experiments. (*P < 0.05)
Role of Integrator in LCR mediated control of globin gene expression
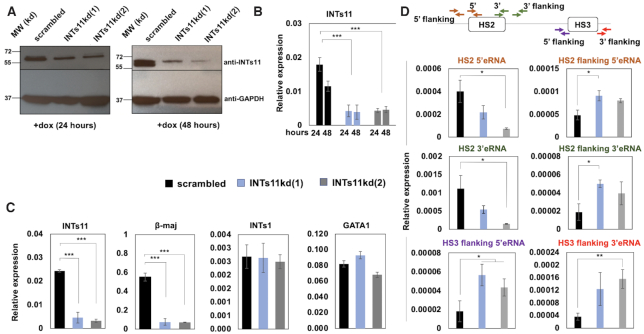
Recently, it was shown that many enhancers recruit the Integrator complex which mediates termination of eRNA transcription (30). Integrator is a component of the transcription termination machinery implicated in the processing of splicing associated small nuclear RNAs (snRNAs) (44). The involvement of Integrator in the early transcription termination of eRNA suggests that the release of Pol II from enhancers could aide in the transfer of Pol II to promoters. We used doxycycline (dox) inducible shRNA mediated knockdown of Integrator subunit 11 (INTs11) in differentiating MEL cells to examine the function of Integrator in LCR mediated recruitment of Pol II to the adult β-globin gene (Figure 4). INTs11 is the catalytic subunit of the Integrator complex exhibiting endonuclease activity (44). Single MEL cell clones expressing two different shRNAs against INTs11 [INTs11kd(1) and INTs11kd(2)] showed reduced levels of INTs11 protein (Figure 4A) and RNA (Figure 4B) compared to MEL cells expressing scrambled shRNA. Gene expression analysis demonstrated that INTs11 and β-maj-globin expression was reduced in differentiated MEL cells with inducible INTs11 KD compared to cells expressing scrambled control shRNA (Figure 4C). In contrast, we did not observe a change in expression of INTs1, the largest subunit of integrator, or GATA1 genes. We also observed a reduction in expression of HS2 5′ and 3′eRNA that are expressed immediately downstream of the HS2 core region (Figure 4D). Using primers located further away from the core, we observed an increase in the levels of eRNAs both further upstream and downstream of HS2 (HS2 flanking 5′ and 3′eRNA). The same was true for outer eRNA associated with HS3, which increased both at the 5′ and 3′ end (HS3 flanking 5′ and 3′eRNA).
Figure 4.
Transcription changes in MEL cells stably expressing shRNA against INTs11. (A) MEL cells stably expressing shRNA against INTs11 (clones 1 and 2) or scrambled control were subjected to western blotting experiments using antibodies specific for INTs11 or GAPDH. (B) MEL cells stably expressing shRNA against INTs11 (clones 1 and 2) or scrambled control were subjected to RT-qPCR using primers specific for INTs11. Expression was normalized to that of GAPDH. Error bars reflect SEM from three independent experiments (***, P < 0.001). (C, D) MEL cells expressing shRNA against INTs11 [INTs11kd(1) and INTs11kd(2)] or scrambled control shRNA (scrambled) were subjected to RT-qPCR using primers specific for the INTs11, β-maj-globin, INTs1, GATA1 genes (C) as well as primers specific for HS2 and HS3 eRNAs (D). The diagram on top of panel D illustrates the location of primers used in the analysis of LCR HS2 and HS3 associated eRNA. Error bars reflect SEM from three independent experiments (***P < 0.001; **P < 0.01; *P < 0.05).
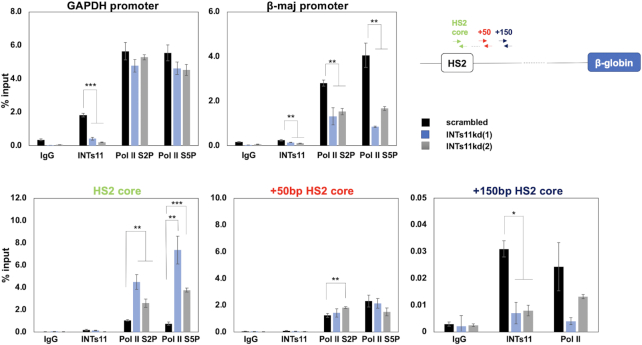
We next analyzed the occupancy of Pol II in the globin gene locus as well as at the GAPDH gene in the INTs11 KD and control differentiated MEL cells using ChIP (Figure 5). We used antibodies specific for the serine 5 (Pol II S5P) and serine 2 (Pol II S2P) phosphorylated forms of Pol II. Pol II-S2P represents the elongating form of Pol II (40). We also used antibodies specific for INTs11. The binding of Pol II-S5P or Pol II-S2P was not altered at the GAPDH promoter in INTs11 KD versus control cells, while the interaction of INTs11 was reduced in INTs11 KD cells. We observed little binding of Integrator subunit 11 at the adult β-maj-globin gene promoter. INTs11 KD reduced association of both forms of Pol II with the β-globin gene consistent with reduced expression levels. Interestingly, we observed an increase in the association of Pol II-S2P and Pol II-S5P with the HS2 core region in INTs11 KD cells. There was little binding of INTs11 at the HS2 core. However, INTs11 associated with a region further downstream of HS2 (+150 bp). The association of INTs11 with the HS2 downstream region (+150 bp HS2 core) was reduced in INTs11 KD cells, as was the association of Pol II (Pol II-S2Pand Pol II-S5P). We did not observe a reduction in the association of Pol II in the region at +50 bp downstream of the HS2 core.
Figure 5.
Increased association of Pol II with the LCR HS2 core region in INTs11 depleted MEL cells. MEL cells stably expressing shRNA against INTs11 (clones 1 and 2) or scrambled control were subjected ChIP-qPCR using antibodies specific for Pol II-S2P, Pol II-S5P and INTs11, and primers specific for GAPDH, β-maj-globin, and different regions downstream of LCR HS2 as indicated. Error bars reflect SEM from three independent experiments (***P < 0.001; **P < 0.01; *P < 0.05).
Lai et al., showed that in the absence of Integrator subunit 11, eRNAs remained bound to RNA Pol II and their primary transcripts accumulated at chromatin (30). To examine if Integrator KD causes alterations in the association of eRNA with chromatin, we isolated cytoplasmic, nuclear, and chromatin associated RNA from INTs11 KD and scrambled control MEL cells differentiated for 48 hours (Figure 6). The control experiments confirmed the successful separation of the RNAs showing that intronic GAPDH RNA is chromatin associated, whereas intron-less GAPDH RNA is located mostly in the cytoplasm. Furthermore, the presence of 45S precursor ribosomal RNA (pre-rRNA) in the chromatin fraction was consistent with previous findings (Figure 6A) (37). We observed an increase in the chromatin associated fraction of HS2 5′ and 3′eRNA in INTs11 KD cells while at the same time there was a decrease of β-maj-globin mRNA in the cytoplasm. Consistent with the observation that INTs11 KD did not affect GAPDH gene expression, the relative abundance of GAPDH RNA in the three fractions did not change after INTs11 KD (Figure 6B). The data were normalized to the chromatin fractions and the presence of RNA species in each fraction has been calculated as a percentage as described by Conrad and Orum (37).
Figure 6.
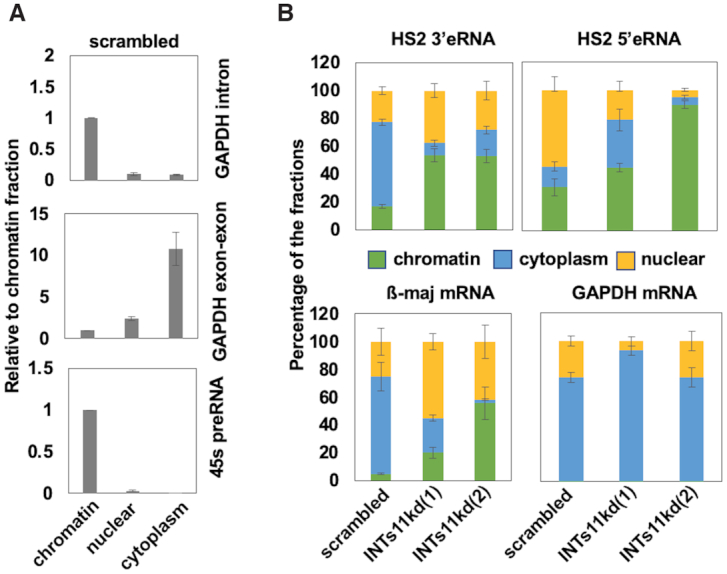
Increased association of HS2 3′eRNA with chromatin in INTs11 depleted cells. (A) Cytoplasmic, nuclear, and chromatin associated RNA was isolated from differentiated MEL cells and subjected to RT-PCR using primers specific for a GAPDH intron, GAPDH exon-exon junctions, and 45Spre-rRNA. (B) Cytoplasmic, nuclear, and chromatin associated RNA was isolated from differentiated MEL cells stably expressing shRNA against INTs11 (clones 1 and 2) or scrambled control and subjected to RT-qPCR using primers specific for HS2 3′eRNA, HS2 5′eRNA, β-maj-globin, and GAPDH. The data were normalized to the chromatin fraction and calculated as a percentage as described by Conrad and Orum (37). Error bars reflect SEM from three independent experiments (**P < 0.01).
To validate our findings with respect to Integrator function in the β-globin gene locus, we reduced Integrator function in differentiating primary human erythroid cells. Figure 7A documents the presence of HS2 and HS3 5′ and 3′ eRNA in differentiated CD34+ cells, according to data from the ENCODE project (45). The CD34+ cells were derived from umbilical cord and transcribe both the adult β- and fetal γ-globin genes upon erythroid differentiation (Figure 7C). We subjected the differentiated cells to siRNA mediated knock-down of INTs11 (si-INTs11) or to a control siRNA (si-control). The data show that RNA and protein expression of INTs11 was reduced in siINTs11 treated cells compared to the si-control treated cells (Figure 7 B and C). We observed a decrease in expression of the α- and β-globin genes, and to a lesser extent expression of γ-globin. Consistent with our observations in MEL cells, we detected an increase in HS2 and HS3 associated 5′ and 3′ eRNA in si-INTs11 treated cells compared to si-control treated cells (Figure 7D). This increase in eRNA was also observed in a region further upstream (5) of HS2 (HS2 5′flanking).
Figure 7.
Reduced globin gene expression and increased LCR HS2 and HS3 eRNA levels in differentiating CD34+ cells deficient for INTs11. (A) RNA sequence reads in the human LCR HS2 and HS3 genomic region. Data were obtained from CD34+ blood progenitor cells from the ENCODE database (ENCSR000CUA,(45). (B) Western blot analysis of INTs11 and β-actin in differentiating CD34+ cells treated with INTs11 siRNA (si-INTs11) or nontargeting control siRNA (si-control) for 72 h. (C) RT-qPCR expression analysis of INTs11, β-globin, γ-globin, α-globin, GATA1 and USF1 genes in differentiating CD34+ cells treated with si-INTs11 or si-control for 72 h. (D) RT-qPCR expression analysis of LCR HS2 and HS3 5′ and 3′ eRNAs in differentiating CD34+ cells treated with si-INTs11 or si-control for 72 hours. The diagram on the left illustrates position of primers used to amply different regions flanking LCR HS2 and HS3. Error bars reflect SEM from three independent experiments (***P < 0.001; **P < 0.01; *P < 0.05). Expression in (C) and (D) was normalized to that of β-actin (Actin).
DISCUSSION
Based on the current and previous data, we propose that the β-globin LCR provides a highly accessible recruitment site for coactivators and Pol II (19,46). A fraction of Pol II is assembled into transcription complexes and generates eRNAs (Figure 8). Integrator terminates transcription at enhancers and releases Pol II and eRNA. The eRNAs together with Pol II, as well as co-activators such as Mediator and Brd4 generate a transcription domain that transiently interacts with the globin promoters to load Pol II onto the genes. Disruption of Integrator function prevents formation of the LCR transcription domain due to the lack of release of Pol II and eRNA. Recently, it was found that Integrator not only terminates transcription but also dephosphorylates the C-terminal domain of Pol II (47). This seems to be an important finding with respect to the Integrator mediated Pol II transfer model, as it is the unphosphorylated form of Pol II that is recruited to Pol II promoters.
Figure 8.
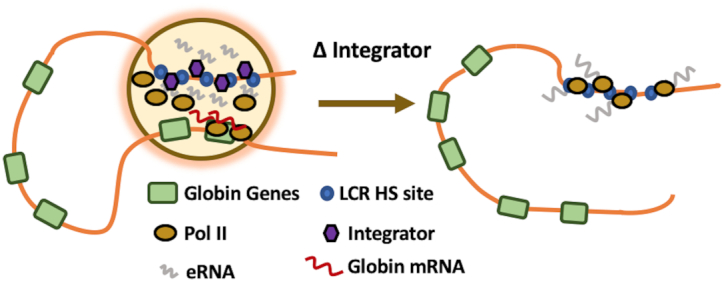
Model of LCR mediated regulation of adult β-globin gene expression focusing on the role of Integrator. The LCR associates with erythroid-specific and basal transcription factors, which recruit co-activators, including Mediator, and Pol II. A fraction of Pol II initiates transcription of eRNA. Integrator terminates transcription at the LCR and releases Pol II and eRNA which contributes to the formation of a transcription domain with high concentration of Pol II (yellow circle).The β-globin gene transiently associates with the transcription domain and Pol II is transferred to strong basal promoter elements at the β-globin gene. Disruption of Integrator function (shown on the right) prevents release of Pol II and eRNA from the LCR and, as a consequence, prevents the loading of Pol II to the globin gene promoter.
SEs recruit a large number of transcription factors, co-activators, as well as Pol II and as a consequence initiate the formation of bidirectional eRNAs (6,7). We studied the role of eRNA and Pol II recruitment at the human and mouse β-globin LCR. Consistent with previous studies, we found that the transgenic human LCR recruits transcription complexes and associates with transcription foci in fetal liver cells independently from the presence of the β-type globin genes (Figure 1) (48). This observation underscores the notion that the association of Pol II at the LCR is not a reflection of interactions with linked transcriptionally active globin genes. Previous studies have shown that during the process of hematopoiesis, many enhancers recruit Pol II before Pol II is detectable at target genes, suggesting that Pol II recruitment to enhancers is involved in priming these regulatory DNA-elements (5). This is consistent with observations that elevated eRNA transcription at the LCR precedes activation of the adult β-globin gene after induction of erythroid differentiation (41).
The ablation of HS2 3′eRNA transcripts by shRNA caused a decrease in the expression of the adult β-globin gene up to 4-fold but did not affect expression of the GATA1 gene (Figure 4). Surprisingly, reduction of HS2 3′eRNA using two different shRNAs also decreased expression of the adult α-globin genes by about 2-fold. We cannot exclude the possibility that the two shRNAs have off-target effects. However, the two shRNAs contain different sequences and we consider it unlikely that they both would reduce α-globin as a consequence of off-target effects. The α- and β-globin gene loci have been shown to associate with the same nuclear speckles suggesting some form of coordinated regulation (49). However, removal of the β-globin LCR in mice had no effect on expression of the α-globin genes (50). It is possible that compensatory mechanisms in mice maintain α-globin expression in the absence of the LCR.
The exact mechanisms by which eRNAs contribute to enhancer function is unclear. It was recently shown that super-enhancers establish domains of a high concentration of Mediator and unphosphorylated Pol II (46,51). RNA could play a role in the formation of SE associated transcription domains. SE associated eRNAs have also been shown to be involved in mediating proximity between SEs and target genes (52).To further examine a potential role of LCR eRNA in β-globin gene regulation, we analyzed the location of HS2 3′eRNA in relation to the β-globin gene locus (Figure 2). We found that HS2 3′eRNA remains largely associated with the β-globin gene locus. The association of the HS2 3′eRNA with the globin gene locus does not necessarily reflect a specific function, it could rather be a consequence of the relatively short half-life, similar to intron containing pre-mRNA. However, we found that the eRNAs associated with LCR HSs exhibit a half-life of 9 min and longer. Furthermore, despite a relative short half-life, eRNAs are constantly being produced and could thus function in the process of gene regulation by contributing to the formation of a transcription domain that concentrates Pol II or by mediating proximity between enhancers and promoters. The turn-over of the eRNAs would allow rapid changes in SE configuration. The observation that KD of HS2 3′eRNA reduced expression of the adult β-globin gene clearly suggests a functional role of at least a subset of LCR associated eRNAs.
The Integrator terminates transcription of snRNAs as well as eRNAs and attenuates transcription at protein coding genes possibly by removing unproductive transcription complexes (30,44,53). Ablation of Integrator function led to reduced transcription and enhancer-promoter looping at epidermal growth factor (EGF) induced genes (30). That study not only implicated the Integrator complex in enhancer function but also suggested that eRNAs released from enhancers contribute to the communication between enhancers and promoters. We conditionally reduced expression of INTs11, the catalytic subunit of Integrator, in differentiating MEL cells and primary human CD34+ cells and examined the consequence on eRNA formation, Pol II recruitment, and β-globin gene transcription. The data demonstrate that Integrator is required for the release of eRNA and Pol II from the β-globin LCR (Figures 4–7). The accumulation of Pol II at the LCR and the decreased recruitment to the β-globin gene in the INTs11 KD cells suggests that Integrator is involved in the transfer of Pol II from the LCR to the β-globin gene promoter. In addition, Integrator mediated release of eRNA could contribute to the formation of transcription domains that concentrate Pol II (54).
Using RNA proximity ligation, Cai et al. (52) demonstrated that eRNA frequently associates with promoter upstream transcripts (PROMPTS) and RNA binding proteins like hnRNPK. Ablation of a MYC SE associated noncoding RNA reduced MYC transcription. The authors suggest that hnRNPK oligomerization, orchestrated by eRNA, contributes to the delivery of Pol II from the SE to the target gene promoter. Recently, it was shown that noncoding chromatin associated RNAs, including eRNAs, are enriched for 5′splice sites and interact with U1snRNP, likely via transcribing Pol II (55). It is possible that HS2 3′eRNA remains at the globin locus first via retention by U1snRNPs and subsequently, after release by Integrator, participates in transcription domain formation and/or communication between the LCR and the genes. This will be the subject of future investigations.
We and others provided evidence for a Pol II transfer mechanism in the β-globin gene locus (19). For example, Johnson et al. showed accumulation of Pol II at the LCR and reduced β-globin expression in MEL cells lacking transcription factor NF-E2 (17). Moreover, we demonstrated that Pol II recruited to an immobilized LCR template is transferred to the β-globin gene in a process depending on the β-globin TATA-box and stimulated by NF-E2 (18). Finally, targeting a synthetic DNA-binding protein to the β-globin downstream promoter inhibited transcription elongation and accumulated Pol II at the LCR (56). These data together with the data presented here support a Pol II transfer model.
There are some observations in our study that appear to be incongruent. For example, we found that Pol II levels increased in the HS2 core but not in a region downstream of the core in INTs11 KD MEL cells. In contrast, an increase in eRNA was mostly observed downstream of the HS2 core region in these cells. It is possible that the lack of termination of transcription downstream of HS2 leads to an accumulation of Pol II at the site of initiation. Moreover, it has been reported that INTs13 regulates early steps in the formation of enhancers (57). Thus, Integrator could play a role in the initiation of transcription at SEs as well.
In summary, the data demonstrate that Integrator releases Pol II and eRNAs from the LCR and thereby contributes to the loading of Pol II to the globin gene promoters.
Supplementary Material
ACKNOWLEDGEMENTS
We thank our colleagues in the Bungert and Xie laboratories for discussions and encouragement. We thank Wara Rattanaphong for technical assistance. We thank Drs Mike Kilberg, Rolf Renne, and Jixiu Shan (UF) for helpful discussions and comments on the manuscript.
Contributor Information
Aishwarya Gurumurthy, Department of Biochemistry and Molecular Biology, Center for Epigenetics, Genetics Institute, UF Health Cancer Center, Powell-Gene Therapy Center, Gainesville, FL 32610, USA.
David T Yu, Department of Biochemistry and Molecular Biology, Center for Epigenetics, Genetics Institute, UF Health Cancer Center, Powell-Gene Therapy Center, Gainesville, FL 32610, USA.
Jared R Stees, Department of Biochemistry and Molecular Biology, Center for Epigenetics, Genetics Institute, UF Health Cancer Center, Powell-Gene Therapy Center, Gainesville, FL 32610, USA.
Pamela Chamales, Department of Biochemistry and Molecular Biology, Center for Epigenetics, Genetics Institute, UF Health Cancer Center, Powell-Gene Therapy Center, Gainesville, FL 32610, USA.
Ekaterina Gavrilova, Department of Biochemistry and Molecular Biology, Center for Epigenetics, Genetics Institute, UF Health Cancer Center, Powell-Gene Therapy Center, Gainesville, FL 32610, USA.
Paul Wassel, Department of Biochemistry and Molecular Biology, Center for Epigenetics, Genetics Institute, UF Health Cancer Center, Powell-Gene Therapy Center, Gainesville, FL 32610, USA.
Lu Li, Department of Biochemistry and Molecular Biology, Center for Epigenetics, Genetics Institute, UF Health Cancer Center, Powell-Gene Therapy Center, Gainesville, FL 32610, USA.
Daniel Stribling, Department of Biochemistry and Molecular Biology, Center for Epigenetics, Genetics Institute, UF Health Cancer Center, Powell-Gene Therapy Center, Gainesville, FL 32610, USA; Department of Molecular Genetics and Microbiology, Gainesville, FL 32610, USA.
Jinyang Chen, Department of Statistics, University of Georgia, Athens, GA 30602, USA.
Marissa Brackett, Department of Biochemistry and Molecular Biology, Center for Epigenetics, Genetics Institute, UF Health Cancer Center, Powell-Gene Therapy Center, Gainesville, FL 32610, USA.
Alexander M Ishov, Department of Anatomy and Cell Biology, UF Health Cancer Center, University of Florida, Gainesville, FL, 32610, USA.
Mingyi Xie, Department of Biochemistry and Molecular Biology, Center for Epigenetics, Genetics Institute, UF Health Cancer Center, Powell-Gene Therapy Center, Gainesville, FL 32610, USA.
Jörg Bungert, Department of Biochemistry and Molecular Biology, Center for Epigenetics, Genetics Institute, UF Health Cancer Center, Powell-Gene Therapy Center, Gainesville, FL 32610, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
American Society of Hematology (to J.B.); National Institutes of Health [R56 DK111439, R01 DK052356 to J.B., R00-CA190886, R35-GM128753 to M.X., R01DE026707, R21CA198820 to A.M.I.]. Funding for open access charge: American Society of Hematology.
Conflict of interest statement. None declared.
REFERENCES
- 1. Andersson R., Sandelin A.. Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet. 2020; 21:71–87. [DOI] [PubMed] [Google Scholar]
- 2. Gasperini M., Tome J.M., Shendure J.. Towards a comprehensive catalogue of validated and target-linked human enhancers. Nat. Rev. Genet. 2020; 21:292–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim T.K., Hemberg M., Gray J.M., Costa A.M., Bear D.M., Wu J., Harmin D.A., Laptewicz M., Barbara-Haley K., Kuersten S.et al.. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010; 465:182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Denisenko E., Guler R., Mhlanga M.M., Suzuki H., Brombacher F., Schmeier S.. Genome-wide profiling of transcribed enhancers during macrophage activation. Epigenet. Chromatin. 2017; 10:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arner E., Daub C.O., Vitting-Seerup K., Andersson R., Lilje B., Drabløs F., Lennartsson A., Rönnerblad M., Hrydziuszko O., Vitezic M.et al.. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science. 2015; 347:1010–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., Young R.A.. Super-enhancers in the control of cell identity and disease. Cell. 2013; 155:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A.. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013; 153:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grosveld F., van Assendelft G.B., Greaves D.R., Kollias G.. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987; 51:975–985. [DOI] [PubMed] [Google Scholar]
- 9. Stamatoyannopoulos G. Control of globin gene expression during development and erythroid differentiation. Exp. Hematol. 2005; 33:259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tuan D., Solomon W., Li Q., London I.M.. The “beta-like-globin” gene domain in human erythroid cells. Proc. Natl. Acad. Sci. U.S.A. 1985; 82:6384–6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Forrester W.C., Takegawa S., Papayannopoulou T., Stamatoyannopoulos G., Groudine M.. Evidence for a locus activation region: the formation of developmentally stable hypersensitive sites in globin-expressing hybrids. Nucleic Acids Res. 1987; 15:10159–10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li Q., Peterson K.R., Fang X., Stamatoyannopoulos G.. Locus control regions. Blood. 2002; 100:3077–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tuan D., Kong S., Hu K.. Transcription of the hypersensitive site HS2 enhancer in erythroid cells. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:11219–11223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ashe H.L., Monks J., Wijgerde M., Fraser P., Proudfoot N.J.. Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev. 1997; 11:2494–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Routledge S.J., Proudfoot N.J.. Definition of transcriptional promoters in the human beta globin locus control region. J. Mol. Biol. 2002; 323:601–611. [DOI] [PubMed] [Google Scholar]
- 16. Leach K.M., Nightingale K., Igarashi K., Levings P.P., Engel J.D., Becker P.B., Bungert J.. Reconstitution of human beta-globin locus control region hypersensitive sites in the absence of chromatin assembly. Mol. Cell. Biol. 2001; 21:2629–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johnson K.D., Christensen H.M., Zhao B., Bresnick E.H.. Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol. Cell. 2001; 8:465–471. [DOI] [PubMed] [Google Scholar]
- 18. Vieira K.F., Levings P.P., Hill M.A., Crusselle V.J., Kang S.H., Engel J.D., Bungert J.. Recruitment of transcription complexes to the beta-globin gene locus in vivo and in vitro. J. Biol. Chem. 2004; 279:50350–50357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Levings P.P., Bungert J.. The human beta-globin locus control region. Eur. J. Biochem. 2002; 269:1589–1599. [DOI] [PubMed] [Google Scholar]
- 20. Zhu X., Ling J., Zhang L., Pi W., Wu M., Tuan D.. A facilitated tracking and transcription mechanism of long-range enhancer function. Nucleic Acids Res. 2007; 35:5532–5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhao H., Dean A.. An insulator blocks spreading of histone acetylation and interferes with RNA polymerase II transfer between an enhancer and gene. Nucleic Acids Res. 2004; 32:4903–4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Allahyar A., Vermeulen C., Bouwman B.A.M., Krijger P.H.L., Verstegen M.J.A.M., Geeven G., van Kranenburg M., Pieterse M., Straver R., Haarhuis J.H.I.et al.. Enhancer hubs and loop collisions identified from single-allele topologies. Nat. Genet. 2018; 50:1151–1160. [DOI] [PubMed] [Google Scholar]
- 23. Sawado T., Halow J., Bender M.A., Groudine M.. The beta-globin locus control region (LCR) functions primarily by enhancing the transition from transcription initiation to elongation. Genes Dev. 2003; 17:1009–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arnold P.R., Wells A.D., Li X.C.. Diversity and emerging roles of enhancer RNA in regulation of gene expression and cell fate. Front. Cell Dev. Biol. 2019; 7:377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tippens N.D., Vihervaara A., Lis J.T.. Enhancer transcription: what, where, when, and why. Genes Dev. 2018; 32:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lam M.T., Cho H., Lesch H.P., Gosselin D., Heinz S., Tanaka-Oishi Y., Benner C., Kaikkonen M.U., Kim A.S., Kosaka M.et al.. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013; 498:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fitz J., Neumann T., Steininger M., Wiedemann E.M., Garcia A.C., Athanasiadis A., Schoeberl U.E., Pavri R.. Spt5-mediated enhancer transcription directly couples enhancer activation with physical promoter interaction. Nat. Genet. 2020; 52:505–515. [DOI] [PubMed] [Google Scholar]
- 28. Hsieh C.L., Fei T., Chen Y., Li T., Gao Y., Wang X., Sun T., Sweeney C.J., Lee G.S., Chen S.et al.. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:7319–7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li W., Notani D., Ma Q., Tanasa B., Nunez E., Chen A.Y., Merkurjev D., Zhang J., Ohgi K., Song X.et al.. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013; 498:516–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lai F., Gardini A., Zhang A., Shiekhattar R.. Integrator mediates the biogenesis of enhancer RNAs. Nature. 2015; 525:399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Peterson K.R., Stamatoyannopoulos G.. Role of gene order in developmental control of human gamma- and beta-globin gene expression. Mol. Cell. Biol. 1993; 13:4836–4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tanimoto K., Liu Q., Bungert J., Engel J.D.. Effects of altered gene order or orientation of the locus control region on human beta-globin gene expression in mice. Nature. 1999; 398:344–348. [DOI] [PubMed] [Google Scholar]
- 33. Bungert J., Davé U., Lim K.C., Lieuw K.H., Shavit J.A., Liu Q., Engel J.D.. Synergistic regulation of human beta-globin gene switching by locus control region elements HS3 and HS4. Genes Dev. 1995; 9:3083–3096. [DOI] [PubMed] [Google Scholar]
- 34. Ballester M., Castelló A., Ibáñez E., Sánchez A., Folch J.M.. Real-time quantitative PCR-based system for determining transgene copy number in transgenic animals. BioTechniques. 2004; 37:610–613. [DOI] [PubMed] [Google Scholar]
- 35. Livak K.J., Schmittgen T.D.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 36. Hossain M.A., Shen Y., Knudson I., Thakur S., Stees J.R., Qiu Y., Pace B.S., Peterson K.R., Bungert J.. Activation of fetal γ-globin gene expression via direct protein delivery of synthetic zinc-finger DNA-binding domains. Mol. Ther. Nucleic Acids. 2016; 5:e378. [DOI] [PubMed] [Google Scholar]
- 37. Conrad T., Ørom U.A.. Cellular fractionation and isolation of chromatin-associated RNA. Methods Mol. Biol. 2017; 1468:1–9. [DOI] [PubMed] [Google Scholar]
- 38. Srivastava S., Savanur M.A., Sinha D., Birje A., R V., Saha P.P., D'Silva P.. Regulation of mitochondrial protein import by the nucleotide exchange factors GrpEL1 and GrpEL2 in human cells. J. Biol. Chem. 2017; 292:18075–18090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Seki Y., Ikeda S., Kiyohara H., Ayabe H., Seki T., Matsui H.. Sequencing analysis of a putative human O-sialoglycoprotein endopeptidase gene (OSGEP) and analysis of a bidirectional promoter between the OSGEP and APEX genes. Gene. 2002; 285:101–108. [DOI] [PubMed] [Google Scholar]
- 40. Zaborowska J., Egloff S., Murphy S.. The pol II CTD: new twists in the tail. Nat. Struct. Mol. Biol. 2016; 23:771–777. [DOI] [PubMed] [Google Scholar]
- 41. Kim Y.W., Lee S., Yun J., Kim A.. Chromatin looping and eRNA transcription precede the transcriptional activation of gene in the β-globin locus. Biosci. Rep. 2015; 35:e00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ayupe A.C., Reis E.M.. Evaluating the stability of mRNAs and noncoding RNAs. Methods Mol. Biol. 2017; 1468:139–153. [DOI] [PubMed] [Google Scholar]
- 43. Katsumura K.R., Bresnick E.H., Group G.F.M.. The GATA factor revolution in hematology. Blood. 2017; 129:2092–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rienzo M., Casamassimi A.. Integrator complex and transcription regulation: Recent findings and pathophysiology. Biochim. Biophys. Acta. 2016; 1859:1269–1280. [DOI] [PubMed] [Google Scholar]
- 45. Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sabari B.R., Dall’Agnese A., Boija A., Klein I.A., Coffey E.L., Shrinivas K., Abraham B.J., Hannett N.M., Zamudio A.V., Manteiga J.C.et al.. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018; 361:eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huang K.L., Jee D., Stein C.B., Elrod N.D., Henriques T., Mascibroda L.G., Baillat D., Russell W.K., Adelman K., Wagner E.J.. Integrator recruits protein phosphatase 2A to prevent pause release and facilitate transcription termination. Mol. Cell. 2020; 80:345–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Noordermeer D., de Wit E., Klous P., van de Werken H., Simonis M., Lopez-Jones M., Eussen B., de Klein A., Singer R.H., de Laat W.. Variegated gene expression caused by cell-specific long-range DNA interactions. Nat. Cell Biol. 2011; 13:944–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Brown J.M., Leach J., Reittie J.E., Atzberger A., Lee-Prudhoe J., Wood W.G., Higgs D.R., Iborra F.J., Buckle V.J.. Coregulated human globin genes are frequently in spatial proximity when active. J. Cell Biol. 2006; 172:177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bender M.A., Ragoczy T., Lee J., Byron R., Telling A., Dean A., Groudine M.. The hypersensitive sites of the murine β-globin locus control region act independently to affect nuclear localization and transcriptional elongation. Blood. 2012; 119:3820–3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gurumurthy A., Shen Y., Gunn E.M., Bungert J.. Phase separation and transcription regulation: are super-enhancers and locus control regions primary sites of transcription complex assembly. Bioessays. 2019; 41:e1800164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cai Z., Cao C., Ji L., Ye R., Wang D., Xia C., Wang S., Du Z., Hu N., Yu X.et al.. RIC-seq for global in situ profiling of RNA–RNA spatial interactions. Nature. 2020; 582:432–437. [DOI] [PubMed] [Google Scholar]
- 53. Elrod N.D., Henriques T., Huang K.L., Tatomer D.C., Wilusz J.E., Wagner E.J., Adelman K.. The integrator complex attenuates promoter-proximal transcription at Protein-Coding genes. Mol. Cell. 2019; 76:738–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hnisz D., Shrinivas K., Young R.A., Chakraborty A.K., Sharp P.A.. A phase separation model for transcriptional control. Cell. 2017; 169:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yin Y., Lu J.Y., Zhang X., Shao W., Xu Y., Li P., Hong Y., Cui L., Shan G., Tian B.et al.. U1 snRNP regulates chromatin retention of noncoding RNAs. Nature. 2020; 580:147–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Barrow J.J., Li Y., Hossain M., Huang S., Bungert J.. Dissecting the function of the adult β-globin downstream promoter region using an artificial zinc finger DNA-binding domain. Nucleic Acids Res. 2014; 42:4363–4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Barbieri E., Trizzino M., Welsh S.A., Owens T.A., Calabretta B., Carroll M., Sarma K., Gardini A.. Targeted enhancer activation by a subunit of the integrator complex. Mol. Cell. 2018; 71:103–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.