Abstract
Point-of-care (POC) technologies and testing programs hold great potential to significantly improve diagnosis and disease surveillance. POC tests have the intrinsic advantage of being able to be performed near the patient or treatment facility, owing to their portable character. With rapid results often in minutes, these diagnostic platforms have a high positive impact on disease management. POC tests are, in addition, advantageous in situations of a shortage of skilled personnel and restricted availability of laboratory-based analytics. While POC testing programs are widely considered in addressing health care challenges in low-income health systems, the ongoing pandemic of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections could largely benefit from fast, efficient, accurate, and cost-effective point-of-care testing (POCT) devices for limiting COVID-19 spreading. The unrestrained availability of SARS-CoV-2 POC tests is indeed one of the adequate means of better managing the COVID-19 outbreak. A large number of novel and innovative solutions to address this medical need have emerged over the last months. Here, we critically elaborate the role of the surface ligands in the design of biosensors to cope with the current viral outbreak situation. Their notable effect on electrical and electrochemical sensors’ design will be discussed in some given examples.
Graphical abstract
Keywords: Point-of-care (POC) technologies, SARS-CoV-2, Biosensors, Surface ligands, Electrochemistry, Field effect transistors
Introduction
Transmission of communicable disease is an ever-evolving subject, and transmission of pathogens that cause acute respiratory diseases, is no exception. Since the COVID-19 outbreak early December 2019, millions of people have been infected, thousands have died, and the economies of many countries have come to a halt [1]. The main mode of transmission of SARS-CoV-2 like other acute respiratory diseases is through large droplets, but transmission through contact (including hand contamination followed by self-inoculation) and infectious respiratory aerosols of various sizes at the short range may also occur. Many symptoms of COVID-19 are non-specific, and the importance of infection prevention and control measures in health care settings have become evident over the weeks. The types of diagnostic tests being used clinically include, next to chest computed tomography (CT) scans and reverse transcription-polymerase chain reaction (RT-PCR) assays, which identify the virus based on its genetic material. The use of a CT scan is limited to equipped hospitals and trained personnel and it is not specific. RT-PCR is a time-consuming assay, which encounters large possibilities of false-negative results due to the complexity of the test. While multiplex PCR (mPCR) has emerged as an interesting strategy for the rapid detection and precise identification of a large number of respiratory viruses [2, 3] (https://www.sfm-microbiologie.org/wp-content/uploads/2020/09/Rapport_FilmArray_RP2.1plus_30072020-1.pdf; https://www.qiagen.com/ie/products/diagnostics-and-clinical-research/infectious-disease/qiastat-dx-covid-19-ruo/#orderinginformation) and can provide results within 1 h (compared to > 4 h for RT-PCR), this technique is less sensitive. The mPCR film array assay (https://www.sfm-microbiologie.org/wp-content/uploads/2020/09/Rapport_FilmArray_RP2.1plus_30072020-1.pdf) offers the possibility of multiplex testing up to 19 viruses, one of them being SARS-CoV-2. Besides being expensive, it can only test one sample at a time. The mPCR QIAstat-Dx COVID-19 assay is comparable, allowing the detection of 22 respiratory viruses, one being SARS-CoV-2.
Rapid diagnostic tests with high specificity for SARS-CoV-2 antigens can be performed using biosensors [4]. Biosensors are smart analytical diagnostic devices that combine a biological recognition element immobilized on a physical transducer to detect biomarkers within bodily liquids. Their sensing capacity is based on the efficient and selective transformation of a biological reaction event (interaction of a surface-attached ligand with a biomarker in solution) into a measurable and quantifiable detectable signal. Since the advent of the first biosensor, a glucose sensor based on glucose oxidase as a surface ligand [5], continuous efforts have been made to render these sensors more efficient, in terms of sensitivity, and more portable to fully explore their potential for point-of-care applications [6, 7]. These point-of-care devices have been recognized to be the most effective mean to help contain the spread of the virus. In the context of viral sensing, their large advantage over RT-PCR is the timescale and the multiple formats they can provide including, next to antigen assays, which detect part of the SARS-CoV-2 virus, antibody tests that sense the production of antibodies once a person has been infected. However, quicker, eventually cheaper, and less-complicated diagnostic platforms such as biosensors are often accompanied with the drawback of being not as sensitive as RT-PCR assays, which can detect a single molecule of RNA in a microliter of solution. To date, biosensor platforms have not demonstrated the capability to distinguish between infected and non-infected patients.
Since April 2020, there is a flurry of activity on COVID-19 (the disease caused by SARS-CoV-2 virus) diagnostic kits to sublimate hospitals and diagnostic laboratories with alternatives to RT-PCR assays [8–16]. SARS-CoV-2 has 28 different types of proteins, with the structural membrane (M) and envelope (E) proteins being important for assembling the virus structure (Fig. 1a).
Fig. 1.
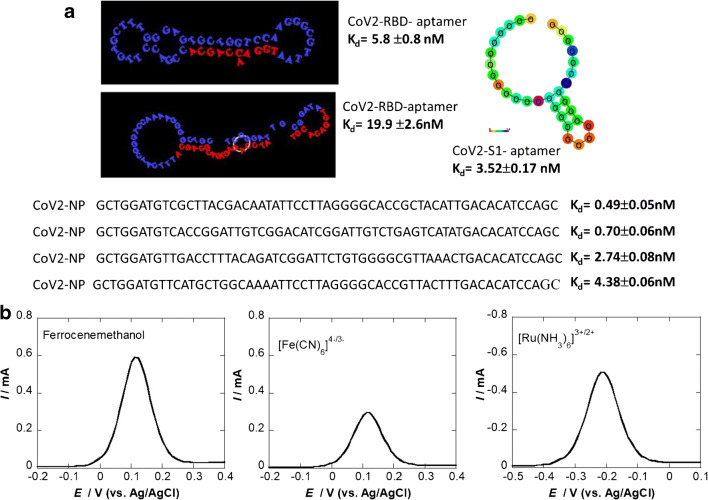
SARS-CoV-2 bioreceptors used in diagnostic devices: a schematic presentation of the SARS-CoV-2 viral particle organization presenting S (spike protein), E (envelope protein), M (membrane protein), and N (nucleocapsid protein); b schematic illustration of a spike protein together with its 3D structure; c single-stranded DNA approach as a surface ligand: genome positions according to GenBank, SARS-CoV-2 NC_045512 together with thiol-cDNA used (reprinted from ref. [11]); d antigen sensing approach based on IgG recognizing the RBD of the virus [14]; e structure of a proposed SARS-CoV-2 aptamer (together with the SARS-CoV-2 S1-protein complex) [17]
The nucleocapsid protein (NP) packages the genomic RNA to form a helical nucleocapsid. The envelope protein together with the M protein and the spike protein (S) is embedded in a lipidic membrane to form the viral outer wall. The spike protein protrudes from the viral membrane, and the viral entry into host cells is mediated by the spike protein that recognizes the host receptor ACE2 by using its receptor-binding domain (RBD) region (Fig. 1b). The RBD region of the S protein is therefore a major target for viral neutralization as well as an excellent target for diagnostics. In addition, the spike protein is repeated some 50–200 times on the viral surface, the exact copy number being still a subject of research [18]. The technologies behind the different SARS-CoV-2 biosensors are based on the linkage of a COVID-19 single-stranded DNA (Fig. 1c), IgG specific to the spike protein (Fig. 1d), or aptamers (Fig. 1e). What is the best choice of a surface ligand for a biosensor in the COVID-19 context? A survey of current diagnostic devices will provide insights into what has been done until now. Some other concepts will be discussed as it is important to widen the horizon of surface anchors of such biosensors in the context of the current epidemic as well as for further ones that may occur in the next future.
DNA, antibodies, aptamers: different recognition elements for SARS-CoV-2
The recognition element is one of the most important parts of a biosensor due to its role in target-analyte recognition [19]. Looking through the literature, the choice between an antibody, an aptamer, and more lately a “simplified antibody” is often very arbitrary and linked to what is commercially available or has been described by others. Considering that access to any ligand is ensured, what would be then the ideal ligand of choice for an electrical, electrochemical, or optical sensor?
End of August, the Abbott’s immunochromatographic antigen assay (COVID-19 Ag) received emergency use authorization and stands out as it does not require instrumentation for reading [20]. This technology is based on using unique antibodies specific to the nucleoprotein (Fig. 1a). In a test performed in a clinical setting [20], it was concluded that for patients with mild respiratory symptoms, the Panbio™ COVID-19 Ag Rapid Test had 99.4% specificity and a sensitivity above 98.2% for nasopharyngeal samples when using Ct values < 33 cycles as cut-off for a RT-qPCR positive test. The Panbio™ COVID-19 Ag assay differs from the BinaxNOW™ COVID-19 Ag CARD, which is a lateral flow immunoassay intended for the qualitative detection of nucleocapsid protein antigen from SARS-CoV-2 in direct nasal swabs. The Roche’s SARS-CoV-2 Rapid Antigen Test (https://www.roche.com/media/releases/med-cor-2020-09-01b.htm) is also a rapid chromatographic immunoassay for nasopharyngeal swab samples. The test has a sensitivity of 96.52% and a specificity of 99.68%, based on 426 samples from two independent study centers.
The accuracy of saliva-based tests [21] still remains a debate, even so some of them got FDA clearance, such as the SalivaDirect developed by researchers at the Yale School of Public Health. The test can run approximately 90 samples in 3 h in a laboratory. EasyCov, a saliva SARS-CoV-2 screening test based on the RT-LAMP technique, developed in the Sys2diag Laboratory of the CHU Montpellier, required 40 min and showed 87% concordance to RT-PCR for COVID-19-positive samples and 99.4% specificity for COVID-19-negative ones.
Next to these commercialized antigen sensors, what is the state-of-the-art for other sensing concepts in the literature and their potential to be taken up into the market? As there are several review articles available by now [4, 8–10, 12, 15, 16], the discussion will be limited to the most adapted surface ligand for electrochemical and electrical transducers. Some of the considerations can be potentially applied to optical sensing, but will not be evoked here. In addition, the focus is on antigen sensing and discussion around serological biosensors, based on the detection of formed COVID-19 antibodies, is excluded. The readers are encouraged to check some interesting articles on this topic, one by Ali et al. using electrochemical impedance spectroscopy (EIS) sensing on graphene-coated 3D micro-electrodes modified with spike S and RBD [22], and the other by Torrente-Rodríguez et al. [23] using a multiplexed electrochemical graphene-based platform for sensitive and simultaneous interrogation of NP viral antigen, S1-IgG, and S1 (IgM isotypes and C-reactive protein).
Electrical and electrochemical COVID-19 diagnosis: what is the best surface ligand choice?
Deoxyribonucleic acid sequencing technology
While the clinical analysis of SARS-CoV-2 is for most people directly related with the deoxyribonucleic acid (DNA) sequencing technology and RNA detection methods (e.g., PCR, loop-mediated isothermal amplification (LAMP)), the only reported biosensor concepts are the thermo-plasmonic-based one by Qiu et al. [11] and the colorimetric-based one by Moitra et al. [24]. Qiu et al. showed that localized heat, generated on gold nanoislands when irradiated with light, can increase the hybridization temperature and discriminate similar gene sequences down to 0.22 pM of the RdRp Covid sequence (Fig. 1c), a 25-based DNA strand (thiol5′-GCATC TCCTG ATGAG GTTCC ACCT) [11]. The assay by Moitra et al. is based on gold nanoparticles capped with thiol-modified antisense oligonucleotides specific to the nucleocapsid protein of SARS-CoV-2. Aggregation of the 4 different Au NPs-DNA particles in the presence of viral RNA can be seen with naked eyes in color change, achieving a detection limit of 18 ng/μL RNA. With a persistence length of about 6.6–8.0 nm (corresponding to 20–25 base pairs [25]), DNA hybridization is inside the strong localized surface plasmon resonance field. While such an approach would be ideally suited for electrochemical sensors, the reports are scarce and lack information [26] (https://www.zimmerpeacocktech.com/2020/04/13/electrochemical-rt-pcr-detection-for-covid-19/). This might be that viral lysing is still required for RNA-based sensors making it overall more complex and time consuming.
Aptamer-based viral sensing
Aptamers are single-stranded DNA or RNA sequences that bind to their targets through shape recognition. Since the first DNA aptamer was selected for human thrombin in 1992, DNA aptamers have been researched and applied in various fields, including for sensing viral biomarkers [27, 28]. These aptamers, consisting of 15 to 70 bases in length, have the same level of target-binding affinity as monoclonal antibodies with dissociation constants (Kd) usually ranging from 0.1 to 50 nM. While DNA and RNA aptamers are functionally similar, they have some differences, notably RNA aptamers being chemically more unstable and can be hydrolyzed. These aptamers are selected via combinatorial libraries of nucleic acid sequences known as the SELEX (systemic evolution of ligands by exponential enrichment) process. While a handful of aptamers for COVID-19 was postulated [29, 30] (Fig. 2a), their use for electrochemical sensing has not been reported so far. Their rather small size, 6–8 nm, for 20–25 base pairs makes them ideal surface ligands for electrochemical sensors (Fig. 2b).
Fig. 2.
SARS-CoV-2 aptamers: a 51-bp aptamer with 3 hair-pined structures selective to RBD [17], 67-bp 2 hairpin sequences [17], 20-bp aptamer with 1 loop (www.basepairbio.com), 58-bp aptamer [30]. b Differential pulse voltammograms (DPVs) of a gold electrode functionalized with a cysteine-modified 20-bp COVID-19 aptamer (10 μg mL−1 for 2 h) using three different electrochemical mediators (1 mM in 0.1 M PBS, pH 7.4). DPV parameters: Estep = 0.01 V; Epulse = 0.06 V, t = 0.02 s, scan rate = 0.06 V/s
A key concept in electrochemical systems is the fact that the kinetics of the heterogeneous electron transfer at modified electrodes is strongly dependent on the thickness of the functionalization layer and the surface coverage. High coverage results in a large decrease in the apparent rate constant for mediator oxidation/reduction, but depends, in addition, on the thickness of the modifying layer [31]. Using ferrocenemethanol, it was demonstrated, for example, that this mediator can permeate to a small extent into a monolayer-modified gold electrode, while the electron transfer at the bilayer is mainly governed by diffusion of the mediator through pinholes with the electron transfer occurring at the free sites on the electrode rather than permeation. In our preliminary study, a thiolated COVID-19 aptamer was grafted onto a gold electrode via self-assembly and the electron transfer kinetics were investigated using three different redox mediators (Fig. 2b). The results indicated that ferrocenemethanol is best suited for further studies on the charge transfer of aptamer-modified electrodes. Using the negatively charged ferro/ferricyanide redox couple [Fe(CN)6]4−/3− revealed that electron transfer is hindered most likely due the repulsive effect between negatively charged DNA aptamers and negatively charged redox probe and the larger size of the redox couple compared to ferrocenemethanol, restricting the diffusion through pinholes. When using the positively charged [Ru(NH3)6]3+/2+ redox meditator, some of the current could be recovered.
Antibody-based viral sensing
In the case of SARS-CoV-2 antigenic tests, antibodies are the most commonly employed surface ligands. Structurally, an antibody is a globular tetramer, consisting of two longer heavy (H) chains (50 kDa, about 450 residues) and two shorter light (L) chains (25 kDa, about 220 residues), connected together by disulfide bridges and stabilized by non-covalent bonds (Fig. 3a). Antibodies have the advantages of greater specificity: interaction of the antibody paratope with the epitope of the SARS-CoV-2 antigen leads to the formation of the antigen-antibody complex. In vivo, the formation of this complex blocks the virus penetration in the cell and results in signalization to the immune system, leading to the antigen destruction. In diagnostic platforms such as antigenic tests, the formation of the antigen-antibody on the surface of the transducer results in a signal change, with quantitative and qualitative information.
Fig. 3.
Antibody-based SARS-CoV-2 antigenic sensors: a Schematic representation of the full-length IgG antibody Fab (fragment antigen binding) element, Fv (fragment variable element), and Fc (fragment crystallizable). b Schematic diagram of the COVID-19 field effect transistor (FET) using graphene as a channel material, where SARS-CoV-2 spike antibodies were covalently immobilized via the 1-pyrenebutyric acid N-hydroxysuccinimide ester linker (reprinted with permission from [14, 32]
The first biosensor based on surface-linked SARS-CoV-2 spike antibodies is the graphene-based field effect transistor (FET) developed by Seo et al. [14] (Fig. 3b). The use of the FET concept, allowing the signal enhancement of surface-binding events with an applied electric field, is a lab-on-a-chip approach with short assay times down to seconds in the best cases and has been applied for viral protein sensing [28, 34]. Selective sensing of SARS-CoV-2 was ensured through anchoring the SARS-CoV-2-specific antibody on the conducting graphene channel of the transistor via the widely used 1-pyrenebutyric acid N-hydroxysuccinimide ester linkage [28]. In a model experiment using the spike protein, a detection limit as low as 100 fg/mL was attained in clinical transport medium. In addition, the sensor performed well in virus culture medium with a detection limit of 1.6 × 101 plaque-forming units (pfu/mL) and a detection limit of 2.42 × 102 copies/mL in nasal swab samples. These results are indeed encouraging as the electrical probing of antibody-protein interactions via FET is a challenging task due to the Debye screening length (Fig. 3c) [32, 33]. The expected size of the antibody is about 13 nm; the Debye length in 10 mM phosphate-buffered saline (PBS; pH 7.4) is about 0.7 nm: The positively charged COVID-19 S1 subunit protein cannot in principle approach the graphene surface close enough to induce a notable field effect under this high saline condition, but only in diluted PBS. Still, the authors were able to perform diagnostics in nasopharyngeal swabs suspended in universal transport medium (UTM), which contains various reagents including BSA (0.5%) and about 10 mM PBS due to charge-related effects. A French biotechnology company, Grapheal, constructed a sensor using a similar approach. While the performance characteristics are not available, using molecular modeling, the detection mechanism is explained (https://www.linkedin.com/pulse/making-grapheals-graphene-based-sars-cov-2-sensor-animation-redon/).
Fabiani et al. [13] proposed an electrochemical immunosensor for SARS-CoV-2 detection in saliva. This assay was conceived for the detection of the S1 protein as well as the nucleocapsid protein using magnetic beads (MB) as support (Fig. 4a). The assay relies on the formulation of COVID-19 antibody–modified MB, which allowed the viral capture and preconcentration of the sample. These beads with their immunological chain labeled with alkaline phosphate enzymes were deposited on a carbon-based working electrode, and the immune complex was revealed by detecting the formation of 1-napthol via differential pulse voltammetry (DPV) using a portable potentiostat. The enzymatic byproduct could be detected at lower potentials (0.1 V) on the electrode compared to the unmodified one. The response of the assay to native virus was tested in a Biosafety Level 3 environment and resulted in a sensitivity for S protein of 6.5 pfu mL−1.
Fig. 4.
Electrochemical antigenic COVID-19 sensors: a magnetic bead–based electrochemical immunosensors for S1 protein detection: change in DPV signal upon viral binding together with selectivity test (reprinted with permission from ref. [13]). b Electrochemical biosensors, eCoVSense, using glass slides coated with fluorine-doped tin oxide and modified with Au NPs serving for COVID-19 antibody conjugation by physisorption or electrostatic interactions; the same was performed on screen-printed carbon electrodes where antibodies were immobilized [36]. c Reagent-less sensing of COVID-19 viral particles based on the change in hydrodynamic diameter in the presence of the spike protein of the viral particle. Sensor architecture based on antibodies tethered to a stranded DNA bearing a ferrocene moiety. Application of a positive potential attracts the negatively charged DNA strand to the surface. Experimental time-dependent current transitions in the presence of the SARS-CoV-2 viral S1 protein (1 n mL−1) and viral particles (107 mL−1) [37])
The research group of Mahari et al. [36] reported eCovSens, a laboratory-based antigenic COVID-19 sensor for detecting the S1 protein of COVID-19 on fluorine-doped tin oxide (FTO) decorated with Au NPs (Fig. 4b) or screen-printed carbon electrodes. A detection limit of 80 fM and 120 fM for the S1 protein on the FTO and SPCE, respectively, was achieved in saliva in 30 s.
An interesting concept was proposed by a Canadian team who reported a reagent-free electrochemical sensing of SARS-CoV-2 viral particles [37] (Fig. 4c). It is based on the linkage of an anti-S1 antibody to a negatively charged DNA linkage that also features a tethered redox probe, ferrocene. When a positive potential is applied, the sensor is linked to the surface and a chronoamperometric signal is detected.
Alternative ligands for viral sensing: some unexplored routes using engineered antibody fragments (Table 1)
Table 1.
Summary of currently reported SARS-CoV-2 electrochemical and electrical diagnostic platforms together with some other ones for comparison
| Target analyte | Surface ligand | Sensor characteristics | Ref. | |
|---|---|---|---|---|
| Electrical SARS-CoV-2 sensors | ||||
| GFET | S1 protein | Anti-S1 antibody |
100 fg mL−1 (S1 protein) 1.6 × 101 pfu mL−1 (cultured virus) 2.42 × 102 copies mL−1 |
[14] |
| Electrochemical SARS-CoV-2 sensors | ||||
| Coupled with magnetic beads |
S1 protein N protein |
Anti-S1 antibody on magnetic beads | 6.5 pfu mL−1 (saliva) | [13] |
| eCovSens | S1 protein | Anti-S1 antibody | 80 fM (S1 protein) | [36] |
| Reagent-less approach | S1 protein | Anti-S1 antibody attached to DNA bearing ferrocene |
1 pg mL−1 (S1 protein) 4 × 103 viral particles mL−1 |
[37] |
| Portable platform | S1 protein | Engineered antibodies |
2.2 × 102 pfu mL−1 (cultured virus) S-PCR: 72% to PCR-positive samples and 92% to PCR-negative ones |
[45] |
| Other selected SARS-CoV-2 sensors | ||||
| Abbott | N protein | Anti-N protein |
S: 99.4% S-PCR: 98.3% Nasopharyngeal samples using Ct values < 33 cycles |
[20] |
| Roche | S1 protein | Anti-S1 antibody |
S: 99.68% S-PCR: 96.52% |
(https://www.roche.com/media/releases/med-cor-2020-09-01b.htm) |
| Plasmonic sensor | SARS-CoV-2 sequence | DNA | 0.22 pM | [11] |
| Colorimetric assay | N protein | DNA | 18 ng/μL viral RNA | [24] |
N protein nucleocapsid protein, S specificity, S-PCR sensitivity vs. PCR, LAMP loop-mediated isothermal amplification, GFET graphene-based field effect transistor
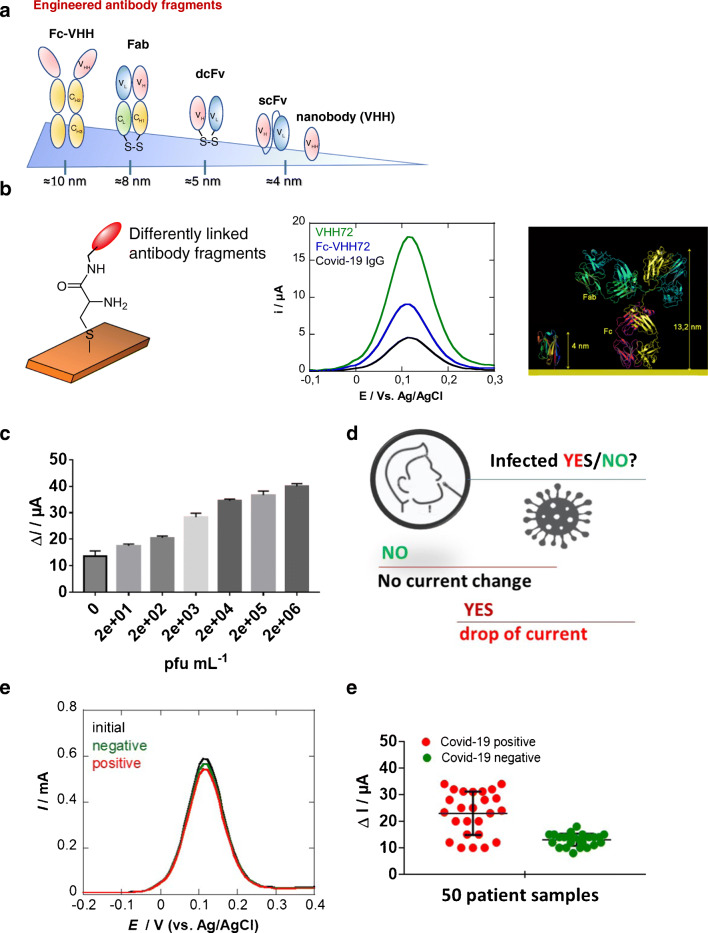
A considerable effort has been put in the last years to design small antibody fragments, such as single-chain variable fragments (ScFv), single-domain antibodies (sdAb), diabodies, and fragment antigen-binding (Fab) fragments (Fig. 5a) [38, 39]. These fragments are mostly obtained as recombinant proteins produced in E. coli or mammalian cells. All of these fragments retain at least one antigen-binding region and are able to recognize the active target. They offer several advantages for diagnostic applications such as low costs and high synthetic yield. Their small distance between the attachment site and the antigen binding site (4 to 10 nm) should make them attractive as surface ligands in electrochemical and electronic configurations (Fig. 5b). Camelids (dromedary, camel, llama, etc.) possess an alternative immune system that produces heavy-chain-only antibodies. Furthermore, besides the conserved Fc, antigen-recognizing moiety is composed of a unique immunoglobulin domain quoted VHH or nanobody. When expressed recombinantly after phage display selection, this VHH domain offers new opportunities for biosensors. Showing a high sequence homology to the variable domains of the heavy chain in humans, but being much smaller in size than a conventional antibody Fab (conventional Fab ≈ 60 kDa, nanobodies ≈ 12–15 kDa) makes them ideal for sensing of antigens. It has been shown that, in contrast with Fabs, nanobodies are able to bind concave epitopes of antigens [39, 40]. Being devoid of Fc domain that may exhibit binding interference from contaminants in the sample, surface-anchored nanobodies interact solely in ligand-receptor interactions in a highly specific manner. Given their excellent solubility, long-term stability at + 4 °C, strong acid-base resistance, and being amendable to genetic manipulation for targeted applications, they seem highly suitable for sensors [35, 43].
Fig. 5.
a Schematic representations of some engineered antibody fragments and their respective height dimension. b Influence of the COVID-19 surface ligand on the current of DPVs recorded on cysteine-modified gold electrodes (2 mM, 2 h) followed by covalent linkage of 10 μg mL−1 VHH-72 (green), VHH-72-Fc (blue), or COVID-19 anti-spike IgG (Abcam, ab273074) via EDC/NHS coupling chemistry (15 mM/15 mM, 1 × PBS, 2 h): mediator: ferrocenemethanol (1 mM in 0.1 M PBS, pH 7.4). DPV parameters: Estep = 0.01 V, Epulse = 0.06 V, t = 0.02 s, scan rate = 0.06 V/s. c Dose-dependent response curve toward SARS-CoV-2-cultured virus. d Concept of the distinction of infection state based on current difference. e DPVs acquired on VHH-72-modified electrodes before (black) and after immersion into nasal swab samples of PCR-categorized COVID-19-positive (red) or COVID-19-negative (green) patient samples. f Results of the preclinical trial on 25 PCR-categorized positive and 25 PCR-categorized negative patient samples [45]
Recently, our group compared the electrochemical response of a gold electrode modified with 2 differently engineered antibody fragments (VHH-72 and Fc-VHH-72) to that of an interface modified with a classical antibody (Fig. 5b) using ferrocenemethanol as the redox mediator. VHH-72 (PDB ID 6WAQ) corresponds to the recently reported SARS-CoV-2 spike–specific nanobody [44], with an equilibrium dissociation constant in the low nM range (~ 39 nM). Larger initial currents are recorded on electrodes modified with VHH-72 compared to the COVID-19 antibody. Fusing a Fc chain to VHH-72 resulted further in an immunoglobulin-like construct (VHH-72-Fc) where the Fab domains are replaced by nanobodies. This bivalent binder exhibited improved equilibrium dissociation constant as reported in [44] and displayed an intermediate current signal between VHH-72 and COVID-19, as expected.
VHH-72-modified electrodes were further investigated for their potential to sense SARS-CoV-2 viral particles in culture medium via binding toward the receptor-binding domain (RBD) of the spike protein (Fig. 5c). Upon immersion of the sensor into a cultured SARS-CoV-2 medium, the electrochemical sensor responded to concentration above 2.2 × 102 pfu/mL with saturation at 1.5 × 105 pfu/mL. These results suggest that the sensor has the potential to be used for COVID-19 diagnosis. Indeed, in a currently ongoing preclinical trial of 25 samples from people tested positive and negative for COVID-19 using RT-PCR, and using a current difference of 15 μA as a cut-off level, the two groups were clearly distinguished (Fig. 5c). Comparison with RT-PCR results in a 72% concordance of PCR-positive samples and a 92% concordance for the RT-PCR-negative ones. We are currently working on the improvement of the concept in believing that binding to the S1 antigen, one of the 4 key proteins (Fig. 1a), minimizes the risk of false-positive samples or identifying someone having already an immune response for past infection to the virus.
Conclusion
The major limitations of most electrochemical detection platforms that restrict their application in the current pandemic crisis are related to their sensitivity, reproducibility, and ultimate upscaling issues. We believe strongly that the success of such diagnostic devices is connected to the choice of the right surface ligand and chemistry, and thus, we chose to focus this critical review on a discussion about surface ligands. Next to oligonucleotides, aptamers, antibodies, and engineered antibody fragments were presented herewith in believing that they might offer significant advantages for viral POC testing approaches for obtaining sensitive diagnostic devices without high costs and improved shelf lives.
While companies and academic research laboratories have presented a panel of different SARS-CoV-2 POC testing devices over the last months, what remains surprising is how long it takes for new sensor technologies to enter the market. All of the discussed sensors are of low technological readiness levels, being mainly based on proven concepts in a research environment, reaching in the best cases TRL 3. This seems even more astonishing in COVID-19 times, where something as complicated as a vaccine can pass clinical trials and regulatory approval in several months.
It is widely recognized that a great number of academic sensors in development never reaches the market. One of the great success stories of electrochemical biosensors is the glucose sensor, which still took several years to reach the market. No doubt, the various efforts invested in virus-oriented, and in particular COVID-19-oriented, electrochemical and electrical sensors might be rewarded in the future. Their fast response time and user-friendly approach make them of great potential for viral sensing. Their real potential has still to be validated, however, before they can be considered a real alternative to standard PCR assays, of which millions of tests are run every day.
Acknowledgments
The unrestricted help of Jean-Pierre Voison from HTDS and his team is acknowledged.
Funding
Financial support is received from the Centre National de la Recherche Scientifique (CNRS), the University of Lille, I-SITE, via the COVID task force and the Hauts-de-France region via ANR Resilience (CorDial-FLU). This work was supported by the French Infrastructure for Integrated Structural Biology (FRISBI) ANR-10-INBS-0005.
Compliance with ethical standards
Conflict of interest
The authors declare that there are no conflicts of interest.
Footnotes
Published in the topical collection celebrating ABCs 20th Anniversary.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fauci AS, Lane C, Redfield R. Covid-19 — navigating the uncharted. New Engl J Med. 2020;382:1268–1269. doi: 10.1056/NEJMe2002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huguenin A, Moutte L, Renois F, Leveque N, Talmud D, Abely M, Nguyen Y, Carrat F, Andreoletti L. Broad respiratory virus detection in infants hospitalized for bronchiolitis by use of a multiplex RT-PCR DNA microarray system. J Med Virol. 2012;84:979–985. doi: 10.1002/jmv.23272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Visseaux B, Le Hingrat Q, Collin G, Bouzid D, Lebourgeois S, Le Pluart D, Deconinck L, Lescure F-X, Lucet J-C, Bouadma L, Timsit J-F, Descamps D, Yazdanpanah Y, Casalino E, Houhou-Fidouh N. Evaluation of the QIAstat-Dx respiratory SARS-CoV-2 panel, the first rapid multiplex PCR commercial assay for SARS-CoV-2 detection. J Clin Microbiol. 2020;58:e00630. doi: 10.1128/JCM.00630-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asif M, Ajmal M, Ashraf G, Muhammad N, Aziz A, Iftikhar T, Wang J, Liu H. The role of biosensors in coronavirus disease-2019 outbreak. Curr Opin Electrochem. 2020;23:174–184. doi: 10.1016/j.coelec.2020.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Updike SJ, Hicks GP. The enzyme electrode. Nature. 1967;214:986–988. doi: 10.1038/214986a0. [DOI] [PubMed] [Google Scholar]
- 6.Wang J. Electrochemical glucose biosensors. Chem Rev. 2008;108:814–825. doi: 10.1021/cr068123a. [DOI] [PubMed] [Google Scholar]
- 7.Juska VB, Pemble ME. A critical review of electrochemical glucose sensing:evolution of biosensor platforms based on advanced nanosystems. Sensors. 2020;20:6013. doi: 10.3390/s20216013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ji T, Liu Z, Wang GQ, Gui X, Khan SA, Lai C, Chen H, Huang S, Xia S, Chen B, Jia H, Chen Y, Zhou Q. Detection of COVID-19: a review of the current literature and future perspectives. Biosens Bioelectron. 2020;166:112455. doi: 10.1016/j.bios.2020.112455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandra P. Miniaturized label-free smartphone assisted electrochemical sensing approach for personalized COVID-19 diagnosis. Sens Int. 2020;1:100019. doi: 10.1016/j.sintl.2020.100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi JR. Development of point-of-care biosensors for COVID-19. Front Chem. 2020;8:517. doi: 10.3389/fchem.2020.00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu G, Gai Z, Tao Y, Schmitt J, Kullak-Ublick GA, Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 12.Yuan X, Yang C, He Q, Chen J, Yu D, Li J, Zhai S, Qin Z, Du K, Chu Z, Qin P. Current and perspective diagnostic techniques for COVID-19. ACS Infect Dis. 2020;6:1998–2016. doi: 10.1021/acsinfecdis.0c00365. [DOI] [PubMed] [Google Scholar]
- 13.Fabiani L, Saroglia M, Galatà G, De Santis R, Fillo S, Luca V, Faggioni G, D’Amore N, Regalbuto E, Salvatori P, Terova G, Moscone D, Lista F, Arduini A. Magnetic beads combined with carbon black-based screen-printed electrodes for COVID-19: a reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva. Biosens Bioelectron. 2021;171:11286. doi: 10.1016/j.bios.2020.112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seo G, Lee G, Kim MJ, Baek S-H, Choi M, Ku KB, Lee C-S, Jun S, Park D, Kim HG, Kim S-J, Lee J-O, Kim BT, Park EC, Kim SI. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 15.Giri B, Pandey S, Shrestha R, Pokharel K, Ligler FS, Neupane BB. Review of analytical performance of COVID-19 detection methods. Anal Bioanal Chem. 2020. 10.1007/s00216-020-02889-x. [DOI] [PMC free article] [PubMed]
- 16.Jayamohan H, Lambert CJ, Sant HJ, Jafek A, Patel D, Feng H, et al. SARS-CoV-2 pandemic: a review of molecular diagnostic tools including sample collection and commercial response with associated advantages and limitations. Anal Bioanal Chem. 2020. 10.1007/s00216-020-02958-1. [DOI] [PMC free article] [PubMed]
- 17.Song Y, Song J, Wei X, Huang M, Sun M, Zhu L, Lin B, Shen H, Zhu Z, Yang C. Discovery of aptamers targeting receptor-binding domain of the SARS-CoV-2 spike glycoprotein. Anal Chem. 2020;92:9895–9990. doi: 10.1021/acs.analchem.0c01394. [DOI] [PubMed] [Google Scholar]
- 18.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Justino CIL, Freitas AC, Pereira R, Duarte AC, Rocha Santos TAP. Recent developments in recognition elements for chemical sensors and biosensors. Trend Anal Chem. 2015;98:2–17. doi: 10.1016/j.trac.2015.03.006. [DOI] [Google Scholar]
- 20.Gremmels H, Winkel BMF, Schuurman R, Rosingh A, Rigter NAM, Rodriguez O, Ubijaan J, Wensing AMJ, Bonten MJM, Hofstra VM. Real-life validation of the Panbio COVID-19 Antigen Rapid Test (Abbott) in community-dwelling subjects with symptoms of potential SARS-CoV-2 infection. medRxiv 2020; 10.1101/2020.10.16.20214189 [DOI] [PMC free article] [PubMed]
- 21.Han P, Ivanovski S. Saliva—friend and foe in the COVID-19 outbreak. Diagnostics. 2020;10:290. doi: 10.3390/diagnostics10050290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ali A, Hu C, Jahan S, Yoan B, Saleh MS, Ju E, Gao S-J, Panat R. Sensing of COVID-19 antibodies in seconds via areosol jet printed three dimensional electrodes. medRxiv 2020. [DOI] [PMC free article] [PubMed]
- 23.Torrente-Rodríguez RM, Lukas H, Tu J, Min J, Yang Y, Xu C, Rossiter HB, Gao W. SARS-CoV-2 RapidPlex: a graphene-based multiplexed telemedicine platform for rapid and low-cost COVID-19 diagnosis and monitoring. Matter. 2020;3:981–1998. doi: 10.1016/j.matt.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moitra P, Alafeef M, Dighe K, Frieman MB, Pan D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano. 2020;14:7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manning GS. The persistence length of DNA is reached from the persistence length of its null isomer through an internal electrostatic stretching force. Biophys J. 2006;91:3607–3616. doi: 10.1529/biophysj.106.089029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tripathy S, Singh SG. Label-free electrochemical detection of DNA hybridization: a method for COVID-19 diagnosis. Trans Indian Natl Acad Eng. 2020;28:1–5. doi: 10.1007/s41403-020-00103-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zou X, Wu J, Gu J, Shen L, Mao L. Application of aptamers in virus detection and antiviral therapy. Front Microbiol. 2019;10:1462. doi: 10.3389/fmicb.2019.01462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aspermair P, Mishyn V, Bintinger J, Happy H, Bagga K, Subramanian P, Knoll W, Boukherroub R, Szunerits S. Reduced graphene oxide-based field effect transistors for the detection of E7 protein of human papillomavirus in saliva. Anal Bioanal Chem. 2020;412:1–9. doi: 10.1007/s00216-019-02168-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torabi R, Ranjbar R, Halaji M, Heiat M. Aptamers, the bivalent agents as probes and therapies for coronavirus infections: a systematic review. Mol Cell Probes. 2020;53:101636. doi: 10.1016/j.mcp.2020.101636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, Fang X, Liu X, Ou H, Zhang H, Wang J, Li Q, Cheng H, Zhang W, Luo Z. Discovery of sandwich type COVID-19 nucleocapsid protein DNA aptamers. Chem Commun. 2020;56:10235. doi: 10.1039/D0CC03993D. [DOI] [PubMed] [Google Scholar]
- 31.Cannes C, Kanoufi F, Bard AJ. Cyclic voltammetry and scanning electrochemical microscopy of ferrocenemethanol at monolayer and bilayer-modified gold electrodes. J Electroanal Chem. 2003;547:83–91. doi: 10.1016/S0022-0728(03)00192-X. [DOI] [Google Scholar]
- 32.Vacic A, Criscione JM, Rajan NK, Stern E, Fahmy TM, Reed MA. Determination of molecular configuration by Debye length modulation. J Am Chem Soc. 2011;133:13886–13889. doi: 10.1021/ja205684a. [DOI] [PubMed] [Google Scholar]
- 33.Chu C-H, Sarangadharan I, Regmi A, Chen Y-W, Hsu C-P, Chang W-H, Lee G-Y, Chyi J-I, Chen C-C, Shiesh S-C, Lee G-B, Wang Y-L. Beyond the Debye length in high ionic strength solution: direct protein detection with field-effect transistors (FETs) in human serum. Sci Rep. 2017;7:5256. doi: 10.1038/s41598-017-05426-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y, Ren R, Pu H, Guo X, Chang J, Zhpu G, et al. Field-effect transistor biosensor for rapid detection of Ebola antigen. Sci Rep. 2017;7:10974. [DOI] [PMC free article] [PubMed]
- 35.Goode J, Dillon D, Miller PA. The development and optimisation of nanobody based electrochemical immunosensors for IgG. Sens Actuators B. 2016;234:478–84.
- 36.Mahari S, Roberts A, Shadeo D, Ghandi S. sCovSens-ultrasensitive novel in-house builg pcinred circuit boared based electrochelical device for rapid deteciton of nCovid-19. bioRxiv 2020; 10.1101/2020.04.24.059204
- 37.Yousefi H, Mahmud A, Chang D, Das J, Gomis S, Chen JB, Wang H, Been T, Yip L, Coomes E, Li Z, Mubareka S, McGeer A, Christie N, Gray-Owen S, Cochrane A, Rini JM, Sargent EH, Kelley SO. Detection of SARS-CoV-2 viral particle using direct, reagent-free electrochemical sensing. ChemRxiv 2020, 10.26434/chemrxiv.13103561.v1 [DOI] [PubMed]
- 38.Richards DA, Maruani A, Chudasama V. Antibody fragments as nanoparticle targetingligands: a step in the right direction. Chem Sci. 2017;8:63. doi: 10.1039/C6SC02403C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu X, Xu Q, Wu Y, Jiang H, Wei W, Zulipikaer YG, et al. Nanobodies derived from Camelids represent versatile biomolecules for biomedical applications. Biomater Sci. 2020;8:3559–73. [DOI] [PubMed]
- 40.Desmyter A, Spinelli S, Roussel A, Cambillau C. Camelid nanobodies: killing two birds with one stone. Curr Opin Struct Biol. 2015;32C:1–8. doi: 10.1016/j.sbi.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Q, Li G, Zhang Y, Zhu M, Wan Y, Shen Y. Highly selective and sensitive electrochemical immunoassay of Cry1C using nanobody and π–π stacked graphene oxide/thionine assembly. Anal Chem. 2016;88:9830–9836. doi: 10.1021/acs.analchem.6b02945. [DOI] [PubMed] [Google Scholar]
- 42.Liu X, Wang D, Chu J, Xu Y, Wang W. Sandwich pair nanobodies, a potential tool for electrochemical immunosensing serum prostate-specific antigen with preferable specificity. J Pharm Biomed Anal. 2018;158:361. doi: 10.1016/j.jpba.2018.06.021. [DOI] [PubMed] [Google Scholar]
- 43.Oloketuyi S, Mazzega E, Zvasnik J, Pungjunun K, Kalcher K, de Marco A, Mehmeti E. Electrochemical immunosensor functionalized with nanobodies for the detection of the toxic microalgae Alexandrium minutum using glassy carbon electrode modified with gold nanoparticles. Biosens Bioelectron. 2020;154:112052. doi: 10.1016/j.bios.2020.112052. [DOI] [PubMed] [Google Scholar]
- 44.Wrapp D, De Vlieger D, Corbett KS, Torres GM, Wang N, Van Breedam W, Roose K, van Schi L, Hoffman M, Puhlmann S, Graham BS, Callewaert N, Schepens B, Saelens X, McLellan JS. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. 2020;181:1004–1015. doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roussel A, Pagneux Q, Mishyn V, Cambillau C, Engelman I, Alidjinou KE, Happy H, Boukherroub R, Devos D, Szunerits S. A fast, cheap and reliable SARS-CoV-2 detection method based on a portable voltammetric device and modified nanobodies. unpublished results.