Abstract
Ionizing radiation causes acute radiation syndrome, which leads to hematopoietic, gastrointestinal, and cerebrovascular injuries. We investigated a population of mice that recovered from high-dose radiation to live normal life spans. These “elite-survivors” harbored distinct gut microbiota that developed after radiation and protected against radiation-induced damage and death in both germ-free and conventionally housed recipients. Elevated abundances of members of the bacterial taxa Lachnospiraceae and Enterococcaceae were associated with postradiation restoration of hematopoiesis and gastrointestinal repair. These bacteria were also found to be more abundant in leukemia patients undergoing radiotherapy, who also displayed milder gastrointestinal dysfunction. In our study in mice, metabolomics revealed increased fecal concentrations of microbially derived propionate and tryptophan metabolites in elite-survivors. The administration of these metabolites caused long-term radioprotection, mitigation of hematopoietic and gastrointestinal syndromes, and a reduction in proinflammatory responses.
Graphical Abstract
INTRODUCTION:
The toxicity of high-dose ionizing radiation is associated with the induction of both chronic and acute radiation syndromes that occur after partial or total body radiation and can be further characterized into hematopoietic, gastrointestinal, and cerebrovascular syndromes. The intestine is the major target of radiation and the biggest niche for gut microbiota. Although there are sporadic descriptive studies showing a potential correlation between the gut microbiota and radiation-induced damage, the detailed underpinnings of this relationship remain obscure. In addition, medical intervention to counter-act radiation injury is still a global challenge despite decades of rigorous research.
RATIONALE:
Over the last decade, numerous investigations have demonstrated highly diverse gut microbiota between individuals and significant correlations of gut microbiota with multiple diseases. Gut microbes, as well as microbe-derived metabolites represented by short-chain fatty acids (SCFAs) and tryptophan metabolites, have essential roles in regulating host metabolism and immunity. The imbalance or dysbiosis of a microbial community is associated with potential diseases, risks, or even to the clear onset of clinical symptoms. We have previously corroborated the biological importance of gut microbiota and certain bacteria (e.g., Lachnospiraceae) together with SCFAs in attenuating colitis and obesity. It has also been reported that SCFAs and tryptophan metabolites can reduce proinflammatory cytokines such as tumor necrosis factor-α, interleukin-6, and interferon-γ and promote the anti-inflammatory cytokines, all of which are vital mediators of radiation-induced damage. These findings raise the possibility that the gut microbiota and metabolites play a key role in the regulation of disease susceptibility after radiation challenge.
RESULTS:
We found that a small percentage of mice could survive a high dose of radiation and live a normal life span. These “elite-survivors” harbored a distinct gut microbiome that developed after radiation. Taking advantage of this finding, we used a combination of fecal engraftment and dirty cage sharing to demonstrate that the microbiota from elite-survivors provided substantial radioprotection in both germ-free and conventionally housed recipients, characterized by enhanced survival and ameliorated clinical scores. An unbiased microbiome analysis identified Lachnospiraceae and Enterococcaceae as the most enriched bacteria in elite-survivors. Monoassociation analysis provided direct evidence for the protective role of Lachnospiraceae and Enterococcaceae in promoting hematopoiesis and attenuating gastrointestinal damage. Clinical relevance in humans was supported by an analysis of leukemia patients who were exposed to whole-body radiation. The elevated abundance of Lachnospiraceae and Enterococcaceae was associated with fewer adverse effects in a highly statistically significant fashion. Treatment with SCFAs, especially propionate, rendered mice resistant to radiation, mediated by attenuation of DNA damage and reactive oxygen species release both in hematopoietic and gastrointestinal tissues. Further, an untargeted metabolomics study revealed a realm of metabolites that were affected by radiation and selectively increased in elite-survivors. Among these, two tryptophan pathwaymetabolites,1H-indole-3-carboxaldehyde (I3A) and kynurenic acid (KYNA), provided long-term radioprotection in vivo.
CONCLUSION:
Our findings emphasize a crucial role for the gut microbiota as a master regulator of host defense against radiation, capable of protecting both the hematopoietic and gastrointestinal systems. Lachnospiraceae and Enterococcaceae, together with downstream metabolites represented by propionate and tryptophan pathway members, contribute substantially to radioprotection. This study sheds light on the pivotal role that the microbiota-metabolite axis plays in generating broad protection against radiation and provides promising therapeutic targets to treat the adverse side effects of radiation exposure.
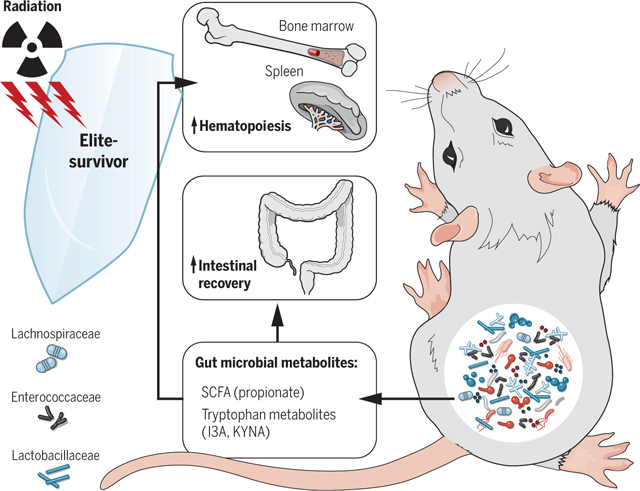
Gut microbiota and metabolites mediate radioprotection. Gut microbes, especially Lachnospiraceae and Enterococcaceae along with bacteria-derived metabolites represented by SCFA (propionate) and tryptophan pathway members (I3A and KYNA), tune host resistance against high doses of radiation by facilitating hematopoiesis and gastrointestinal recovery.
Radiation-induced multiorgan injury represents a sizeable unmet medical need that occurs during cancer radiotherapy and accidental exposure. Substantial federal efforts have been made to develop therapeutic radioprotectants for routine clinical use, but radiation injury remains a long-standing and unresolved problem (1). Acute radiation syndrome or sickness (ARS) occurs after partial or total body exposure to ionizing radiation, and death is frequently attributed to poor hematopoietic recovery and/or death of epithelial cells lining the gastrointestinal (GI) tract. The GI tract is the fastest-renewing adult tissue, making it highly sensitive to radiation. It also harbors a diverse microbial community comprising 10 to 100 trillion microorganisms, raising the possibility that the gut microbiota is involved in responses to radiation-induced damage (2–7). Although there have been some studies showing associations between gut microbiota and radiation-induced damage (6, 8, 9), possible mechanisms are poorly understood.
Our work stemmed from an observation that a small percentage of specific pathogen-free (SPF) C57BL/6 mice survived a high dose of total body radiation and lived a normal life span up to 600 days. These mice have the same genetic background as those that did not survive radiation, thus prompting us to look for differences in the microbiota, which have been implicated in radioprotection (4–9). To better understand the interactions between the gut microbiome and radiation-induced damage, we performed microbiota adoptive transfer experiments as well as monoassociation studies to investigate the function of the gut bacterial community and specific bacterial strains in a radiation model. Both biased and unbiased metabolomics profiling were applied to uncover potential metabolites that play a vital role in radioprotection.
Distinct microbiome in elite-survivors
On administration of a high dose of total body radiation (9.2 Gy), we unexpectedly observed that 5 to 15% of exposed SPF C57BL/6 mice recovered and survived long term (up to 600 days) (Fig. 1A). These mice are referred to as “elite-survivors.” To determine whether the gut microbiota was specific to the elite-survivors, we performed high-throughput gene-sequencing analysis of 16S ribosomal RNA (rRNA) in fecal bacterial DNA isolated from age-matched control mice and elite-survivors at day 290 after radiation. Rarefaction analysis comparing bacterial diversity within individual subjects revealed that elite-survivors harbored a distinct bacterial community relative to that of controls (Fig. 1B). These data were further quantified by UniFrac dissimilarity distance analysis (Fig. 1C) and supported by heatmap analysis of bacterial operational taxonomic units (Fig. 1D; details of the bacterial taxa are listed in table S1). These results led to the speculation that specific gut microbiota may protect against radiation.
Fig. 1. Radiation elite-survivors harbor a distinct gut microbiota compared with age-matched controls.
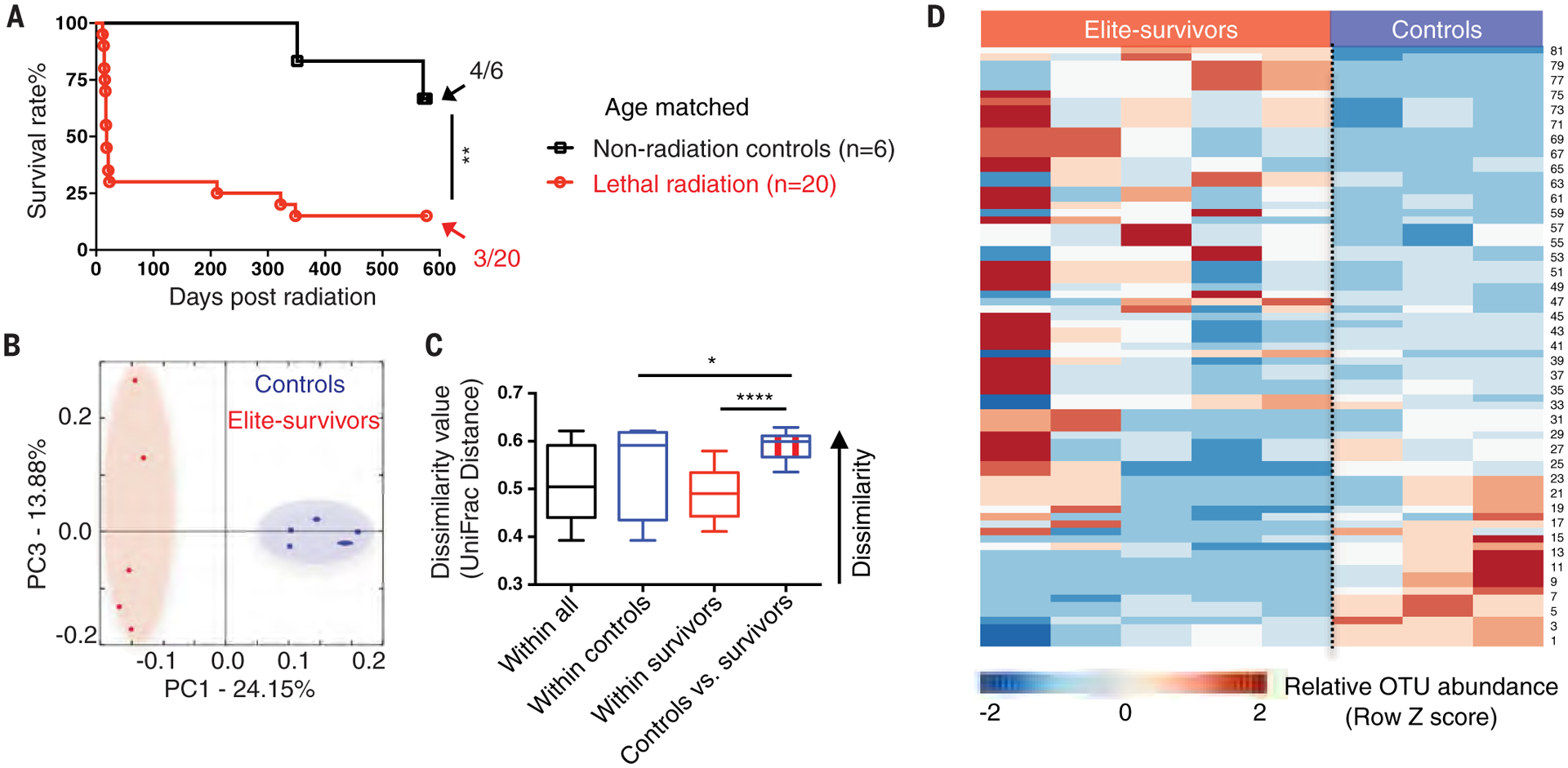
(A) SPF C57BL/6 mice received a high dose of total body radiation and were monitored for >600 days. Fecal samples were collected from age-matched controls and elite-survivors at day 290 after radiation. Fractions indicate the number of mice that survived to the end of the experiment. Data are pooled from two independent experiments. (B and C) PCA plot showing microbial compositional differences (B) as quantified by UniFrac distance (C). (D) Heatmap of sequenced bacterial operational taxonomic unit (OTU) abundances. A detailed bacterial taxa list is provided in table S1. Fecal samples were collected from two independent experiments. Each symbol represents one mouse. Error bars indicate SEM, *P < 0.05, **P < 0.01, ****P < 0.0001 determined by log-rank (Mantel Cox) test (A) and Student’s t test (C).
Radioprotection mediated by microbiota
To investigate more stringently the contribution of microbiota to radioprotection, we performed a “cohousing” assay. Fecal microbiota samples were passively transferred from elite-survivors through a dirty cage-sharing experiment (Fig. 2A). This experimental design was necessary because elite-survivors were old male mice, so a traditional cohousing approach with young male recipients might lead to fighting and injury to the older mice. Instead, dirty cages, which contained fecal material and used bedding from elite-survivors or age-matched control mice, were used to house SPF C57BL/6 mice. Weekly for 8 weeks, recipients were transferred into fresh dirty cages and then exposed to a high dose of total body radiation (8.2 to 9.2 Gy). Nearly 75% of mice housed in dirty cages previously occupied by elite-survivors (ES-Recip) survived high radiation, whereas only 20% of those housed in dirty cages from controls (Ctrl-Recip) survived (Fig. 2B). Clinical score (encompassing the seven body parameters listed in table S2, which were proportional to disease severity) was significantly lower in ES-Recip (Fig. 2C) mice. Histological studies were performed in a double-blinded fashion by a licensed pathologist. Reduced stromal injury, decreased hematopoietic cell death marked by caspase 3, and enhanced proliferation indicated by Ki67 were observed in bone marrow specimens from ES-Recip compared with Ctrl-Recip mice (fig. S1, A to C, upper panels). Splenic architecture was near normal in ES-Recip mice, with the white pulp containing well-developed, lymphocyte-rich follicles and the red pulp exhibiting extramedullary hematopoiesis. Conversely, appreciable atrophy and lymphocyte depletion were observed in Ctrl-Recip spleen samples (fig. S1A, lower panels). A reduction in cleaved caspase 3 staining and immunoblotting and increased Ki67 staining were also observed in spleens from ES-Recip mice (fig. S1, B and C, lower panels, and fig. S1D). These results indicate that the microbiota from elite-survivors mediates the protection of hosts against a high dose of radiation by promoting hematopoietic proliferation and reducing cell death.
Fig. 2. Gut microbiota from elite-survivors protect GF and SPF recipients from radiation-induced death.
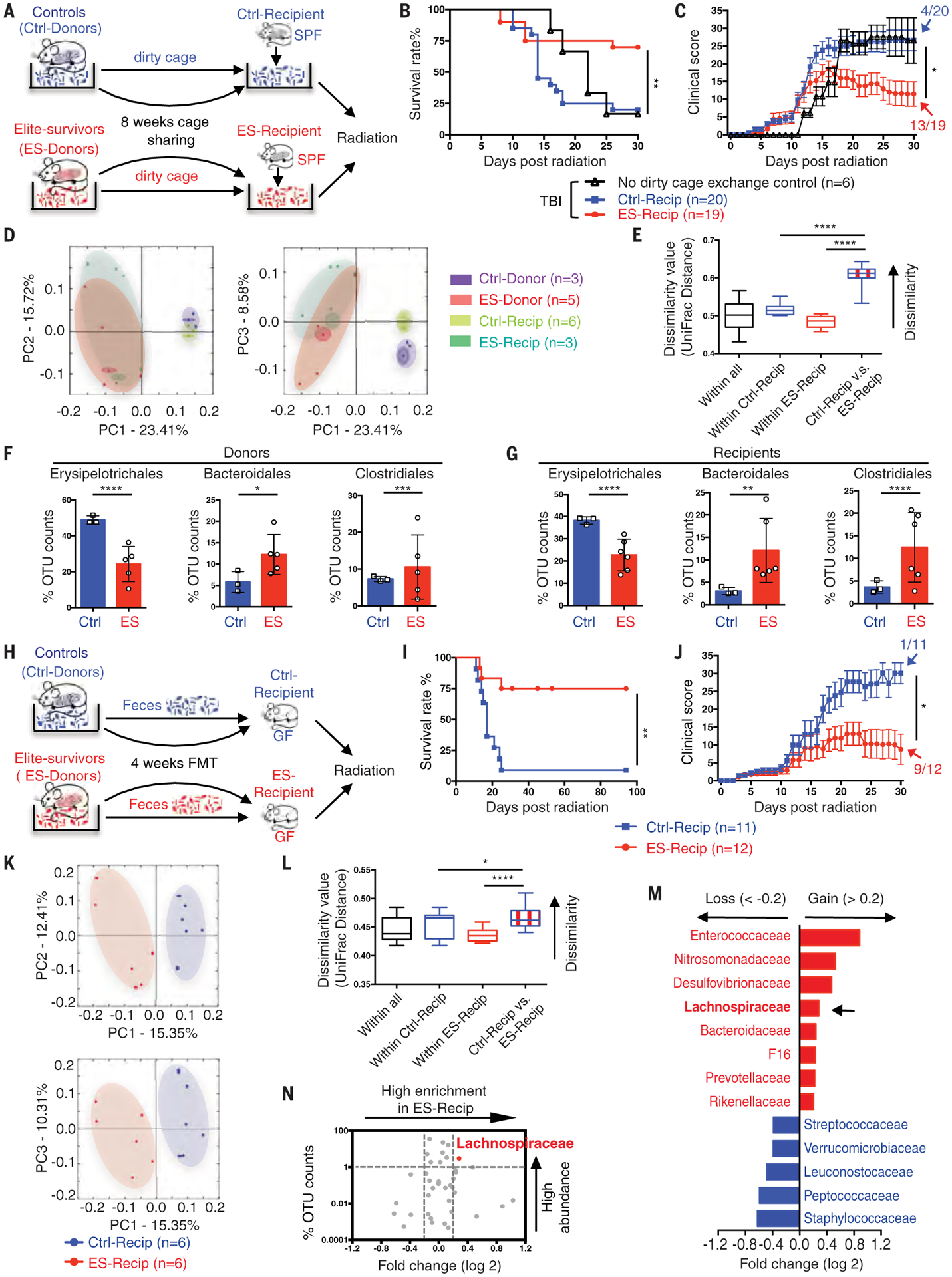
(A) Illustration of dirty cage-sharing experiment. (B and C) Survival rates (B) and clinical scores (C). Significance was found between the Ctrl-Recip and ES-Recip groups. Fecal samples were collected from recipients after 8 weeks of dirty cage sharing. (D and E) PCA plot (D) and UniFrac distance (E) of microbial composition. (F and G) Composite results of substantially changed bacterial groups were identified from donors (F) and recipients (G). (H) Illustration of FMT experiment. (I and J) Survival rates (I) and clinical scores (J). Fecal samples were collected after 4 weeks of FMT from recipients. (K and L) PCA plot (K) and UniFrac distance (L). (M) LDA effect size analysis [LDA significant threshold (log 2) > ±0.2] identified taxonomic biomarkers within recipients. Red bars, enriched within ES-Recip; blue bars, enriched within Ctrl-Recip. (N) Volcano plot displaying relative abundance distribution of microbial OTUs. x axis, log2 relative abundance; y axis, microbial OTU%. Each symbol represents one mouse or bacterial taxa (N). Fractions indicate the number of mice that survived to the end of the experiment [(C) and (J)]. Data are pooled from three [(B) and (C)] or four [(I) and (J)] independent experiments. Error bars indicate SEM, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 determined by log-rank (Mantel Cox) test [(B) and (I)], Mann-Whitney test for area under the curve (AUC) [(C) and (J)], Student’s t test [(E) and (L)], and two-way ANOVA [(F) and (G)].
We profiled bacterial 16S rRNA genes in the feces from both donors and recipients after dirty cage sharing. Principal component analysis (PCA) and UniFrac dissimilarity distance analysis showed differences in microbiome composition in ES-Recip mice (Fig. 2, D and E) compared with Ctrl-Recip mice. Furthermore, the microbiome compositions of the recipients were similar to those of their respective donors, indicating that the dirty cage-sharing strategy was efficient in exchanging gut microbiota from donors to recipients (Fig. 2D). Specifically, a significant decrease in the abundance of members of the order Erysipelotrichales was accompanied by an increase in the abundances of Bacteroidales and Clostridiales in elite-survivors compared with controls. This pattern was recapitulated in ES-Recip mice compared with Ctrl-Recip mice (Fig. 2, F and G).
To confirm the relationship between microbiota and radioprotection, we performed a fecal microbiota transplantation (FMT) experiment in which germ-free (GF) or SPF C57BL/6 mice were reconstituted with fecal materials from elite-survivors or age-matched controls (Fig. 2H and fig. S2A) (10). FMT from elite-survivors resulted in elevated survival rates among GF and SPF mice, which also showed reduced clinical scores compared with that from control mice (Fig. 2, I and J, and fig. S2, B to D), and in an altered microbiome composition as shown by PCA plot and UniFrac dissimilarity distance (Fig. 2, K and L). Bacterial abundances were directly compared between GF ES-Recip and Ctrl-Recip mice. Linear discriminant analysis (LDA) effect size analysis showed that eight taxa were enriched and five were reduced in ES-Recip mice, with an LDA score >0.2 (Fig. 2M). A volcano plot flagged Lachnospiraceae as the most enriched and represented bacteria in ES-Recip mice (Fig. 2N).
We undertook a clinical study to discover whether the bacterial taxa that we identified as radioprotective in our mouse studies might be of relevance to humans. We collected fecal samples from 21 leukemia patients at the start of total body radiation given as a prehematopoietic stem cell transplantation conditioning (detailed clinical patient information is listed in table S3). The incidence and severity of GI side effects were correlated with the extent of diarrhea observed. 16S rRNA sequencing was used to identify the gut microbiome composition. We analyzed the abundance of Lachnospiraceae and Enterococcaceae in all patients (Fig. 3, A and B) during a 30-day observation period. Patients with shorter diarrheal duration (<10 days) had significantly higher abundances of Lachnospiraceae and Enterococcaceae than patients with longer diarrheal duration (>10 days). Lactobacillaceae also showed an increased trend in patients with shorter diarrheal duration, but the value did not reach statistical significance. There was also a statistically significant negative correlation between the abundance of Lachnospiraceae and the occurrence of the GI adverse effects in patients (Fig. 3B). The patient data should be interpreted with caution because these are limited by the small sample size and only show an association of specific bacteria with reduced adverse symptoms; moreover, the study was not designed to show causation.
Fig. 3. Gut microbiota is significantly associated with radioprotection both in patients and mice.
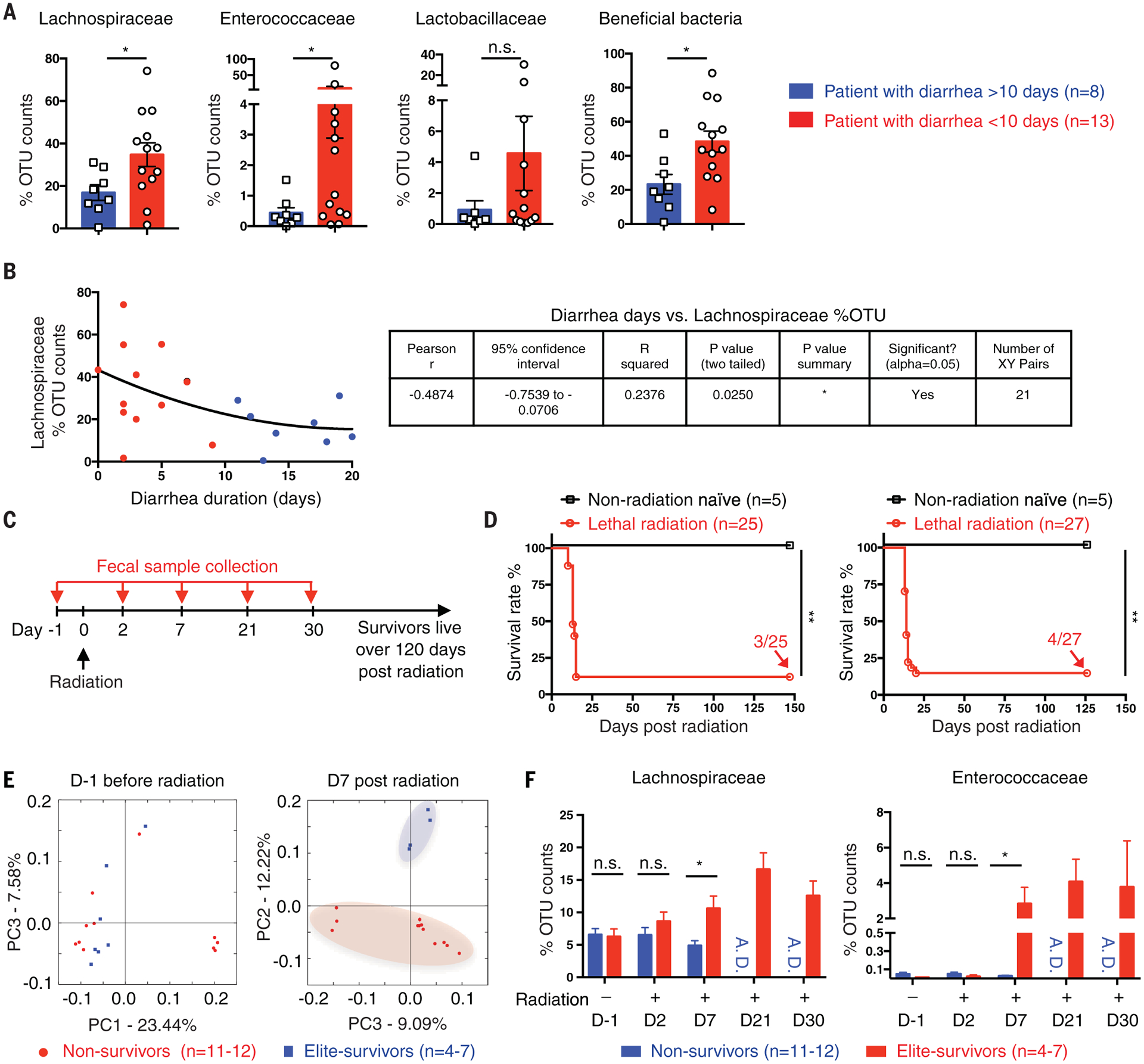
(A and B) Fecal samples were collected from 21 leukemia patients at the start of total body radiation given as a prehematopoietic stem cell transplantation conditioning. The gut microbiome was detected by 16S rRNA sequencing and compared in patients stratified by duration of diarrhea experienced. (A) Relative abundance of Lachnospiraceae, Enterococcaceae, Lactobacillaceae, or a sum of these three beneficial bacteria. (B) Correlation between diarrheal duration and Lachnospiraceae relative abundance. Blue dots, patients with diarrhea > 10 days; red dots, patients with diarrhea <10 days. (C) SPF C57BL/6 mice received a high dose of total body radiation and were monitored for >120 days. Fecal samples were collected from each single mouse at the indicated time points. Mice that survived over day 30 after radiation are referred to as elite-survivors and mice that died are referred to as nonsurvivors. (D) Survival rates (n = 25 in experiment 1 and n = 27 in experiment 2). Fractions indicate the number of mice that survived to the end of the experiment. Fecal 16S rRNA was sequenced. (E) PCA plot. (F) Lachnospiraceae and Enterococcaceae relative abundances. A.D., all mice died. Each symbol represents one patient [(A) and (B)] or one mouse [(C) to (E)]. Error bars indicate SEM, *P < 0.05, **P < 0.01 determined by Mann-Whitney test [(A) and (F)] and log-rank (Mantel Cox) test (D).
Postradiation selection of microbiota in mice
Unlike humans, who have diverse health indicators, the GF and SPF mice used in our study should have similar basal microbiome compositions. To address the question of how elite-survivors harbor a radioprotective microbiota, we investigated whether a distinct microbiota predominated in elite-survivors before radiation or if it was selected after radiation (Fig. 3C). A total of 57 naïve SPF C57BL/6 mice were given a high dose of total body radiation (8.2 Gy), and fecal samples were collected from each mouse before (day −1) and at days 2, 7, 21, and 30 after radiation treatment. Survival was monitored for >120 days (Fig. 3D). Mice that survived >30 days were considered elite-survivors. 16S rRNA sequencing was performed on fecal samples from elite-survivors and those that died after radiation treatment (nonsurvivors). Before radiation (day −1), PCA showed that there was no difference in the bacterial compositions between elite-survivors and nonsurvivors. However, by day 7 after radiation, elite-survivors harbored a bacterial community that was distinct from those that subsequently died (Fig. 3E). Representatives of Lachnospiraceae and Enterococcaceae showed greater abundance in elite-survivors by day 7 after radiation compared with mice that died (Fig. 3F). After 7 days, elite-survivors continued to show a greater abundance of these bacterial taxa, whereas nonsurvivors progressively died before day 21. These data indicate that radiation treatment had a selective effect on Lachnospiraceae and Enterococcaceae taxa in a host “survivor phenotype.”
For monoassociation studies, we selected bacteria that were (i) identifiable to the genus or family level and showed increased abundance in the ES-Recip group (Fig. 2M), (ii) cultivatable for functional studies in vitro and in vivo (11), and (iii) typed strains to ensure reproducibility (11). We tested radioprotection mediated by a mixture of 23 individual strains in the family Lachnospiraceae. We also tested Enterococcus faecalis and Bacteroides fragilis. These were compared with strains that showed reduced abundance in ES-Recip mice from the FMT experiment (Fig. 2M), including Staphylococcus aureus, Staphylococcus saprophyticus, and Streptococcus agalactiae. We also tested Lactobacillus rhamnosus, which has been previously reported to be radioprotective (5, 7). We inoculated SPF C57BL/6 mice with the selected bacteria by oral gavage twice a week for 8 weeks (Fig. 4A). The bacterial growth medium brain heart infusion (BHI) was used as a control. After bacterial reconstitution, recipients received a high dose of total body radiation (8.2 to 8.5 Gy). Mice inoculated with Lachnospiraceae elicited the greatest improvement in survival rate and clinical score (Fig. 4, B to E). Mice given E. faecalis and L. rhamnosus showed a 40 to 60% increase in survival rate (Fig. 4B). Mice gavaged with B. fragilis, S. aureus, S. saprophyticus, or S. agalactiae showed no improvement of outcome compared with the BHI controls (Fig. 4, B and C, and fig. S3).
Fig. 4. Administration of Lachnospiraceae ameliorates radiation-induced syndromes.
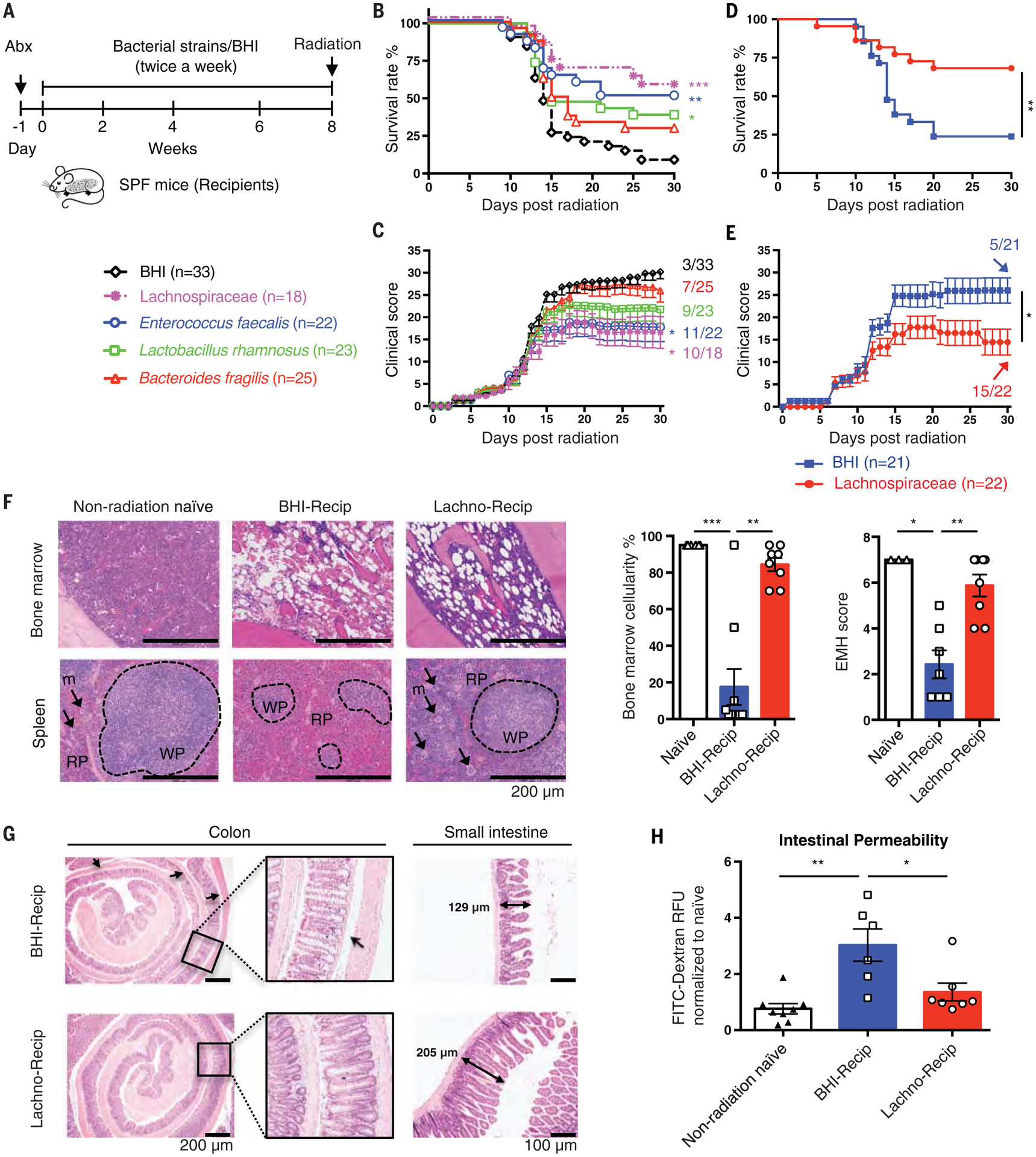
(A) Illustration of transfer experiments with bacteria [“Lachnospiraceae” (a mixture of 23 individual strains in the family Lachnospiraceae), E. faecalis, L. rhamnosus, and B. fragilis] compared with media control (BHI). (B and C) Survival rates (B) and clinical scores (C). Significance is shown between the BHI group and each bacteria group. (D and E) Survival rates (D) and clinical scores (E) from Lachnospiraceae transfer experiments. Fractions indicate the number of mice that survived to the end of the experiment. (F) Femurs and spleens were stained with H&E and quantified for bone marrow cellularity and spleen extramedullary hematopoiesis scores. White pulp (WP), black dashed circles; red pulp (RP), area outside of WP; megakaryocytes (m), black arrows. (G) Colons and small intestines were collected at day 1 after radiation. Arrows indicate gaps between crypt bases and muscularis mucosa. Double-headed arrows indicate the dimensions measured for villi length in small intestines. Representative histology from two independent experiments with seven to eight samples per group is shown. (H) Gut permeability was tested at day 1 after radiation. Fluorescence intensity in sera of each sample was normalized to non-radiation-naïve group average. Data are pooled from three [(B) to (E)] or two [(F) and (H)] independent experiments. Error bars indicate SEM, *P < 0.05, **P < 0.01, ***P < 0.001 determined by log-rank (Mantel Cox) test [(B) and (D)], Mann-Whitney test for AUC [(C) and (E)], and Mann-Whitney test [(F) and (H)].
Recipients of Lachnospiraceae (Lachno-Recip) also showed significantly increased bone marrow cellularity and splenic recovery of the white and red pulp regions (Fig. 4F). Control mice showed more colonic crypt distortion and atrophy, with gaps between the crypt bases and the muscularis mucosa, a common epithelial response to injury (Fig. 4G, indicated by arrows). By contrast, the crypts of mice given Lachnospiraceae remained closely attached to the muscularis mucosa. In control mice given BHI, the intestinal villi were flattened, but this effect was rescued by administration of Lachnospiraceae (Fig. 4G). Fluorescein isothiocyanate (FITC)-dextran assay indicated that after radiation, mice receiving Lachnospiraceae showed reduced gut permeability compared with BHI controls (Fig. 4H).
The radioprotection apparently afforded by Lachnospiraceae raises the possibility that these bacteria may not only be of therapeutic value to patients receiving radiotherapy but may also induce an unintended radioprotective effect to tumor cells. To test whether inoculation of Lachnospiraceae attenuated the antitumor efficacy of radiotherapy, we introduced Lachnospiraceae or BHI into SPF mice for 8 weeks as described above, and then subcutaneously injected B16 (melanoma) or EL4 (lymphoma) cells into the abdomen (fig. S4A). Lachnospiraceae treatment did not affect tumor growth in either tumor model (fig. S4, B and C). To test the effect of Lachnospiraceae on radiotherapy, we transplanted Lachnospiraceae into mice, followed by inoculation of B16 tumor cells. Ten days later, we treated tumor-bearing mice with 10 Gy of localized radiation at the tumor site and monitored survival and tumor growth (fig. S4D). Lachnospiraceae treatment did not negatively affect the efficacy of radiation in reducing tumor growth (fig. S4, E to G).
Metabolite mediators of radioprotection
Members of the Lachnospiraceae are known for their ability to synthesize short-chain fatty acids (SCFAs) by fermentation of dietary polysaccharides (12). SCFAs are important substrates for maintaining intestinal epithelium and for regulating the immune system and inflammatory response (13, 14). We tested whether enhanced SCFA production plays a role in the ameliorative effect of Lachnospiraceae on ARS. Concentrations of three SCFAs (acetate, butyrate, and propionate) were greater in fecal samples from elite-survivors or ES-Recip mice from the dirty cage-sharing experiment compared with controls (fig. S5, A and B). The effects of butyrate approached significance (P = 0.054), showing a clear upward trend, and the levels of propionate and total SCFAs were significantly higher in ES-Recip mice.
SPF C57BL/6 mice were treated with acetate-, butyrate-, or propionate-supplemented water and then given 8.0 to 8.2 Gy of total body radiation (Fig. 5A). The 30-day survival rate was 79% for the propionate-treated group compared with 28% for the control group (Fig. 5, B and C). Acetate and butyrate provided more modest protection (Fig. 5, B and C). To confirm the role of SCFAs in microbiota-mediated radioprotection, we measured the amount of SCFAs produced by several strains of Lachnospiraceae that were either high- or low-SCFA producers (fig. S6, A and B). Subsequent treatment with three high-producer strains protected 100% of mice against radiation, whereas the three low-producer strains only provided 50% protection (fig. S6, C to E).
Fig. 5. Commensal-associated SCFAs suppress radiation-induced death and damage.
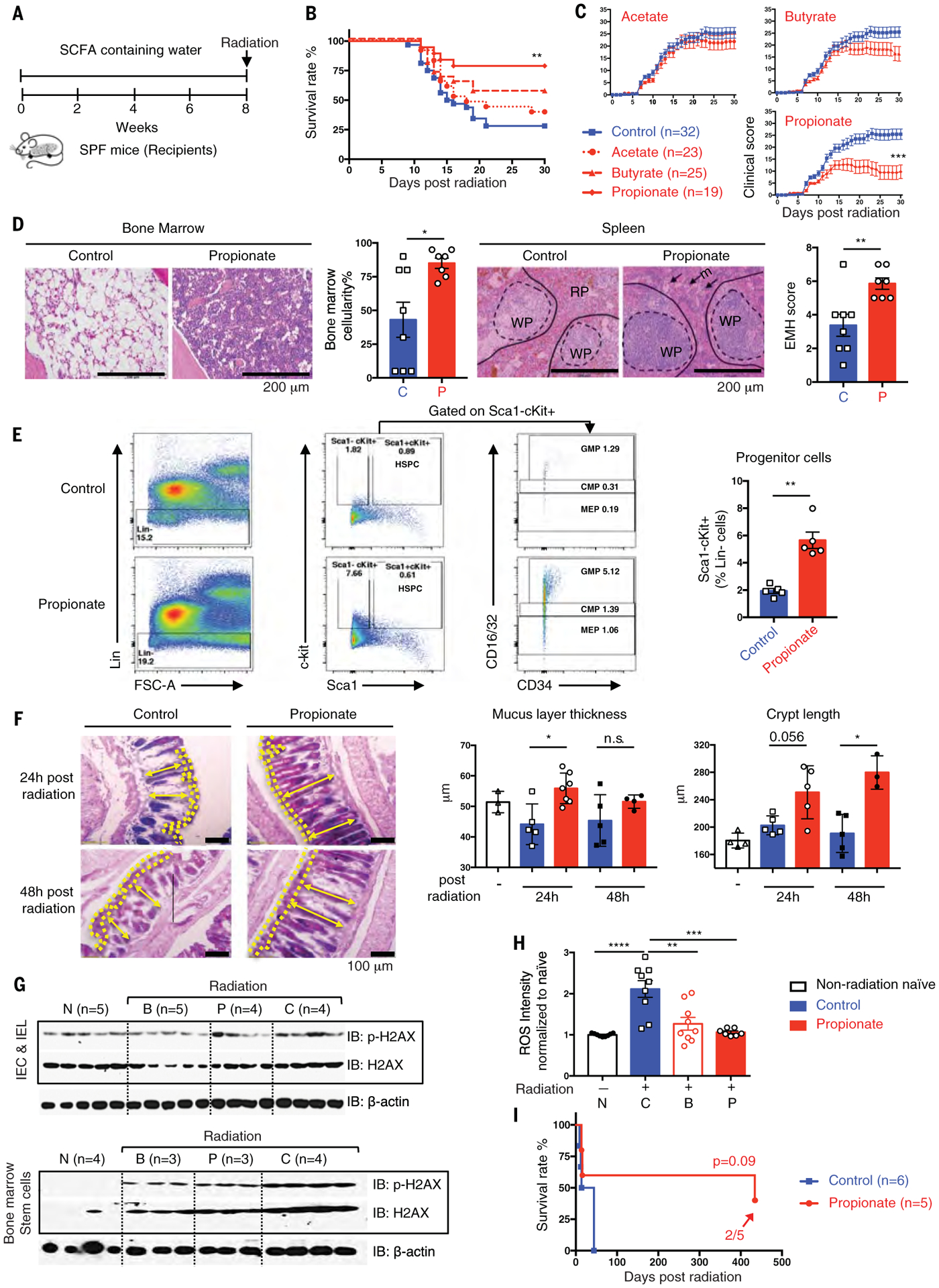
(A) Schematic of SCFA treatment. (B and C) Survival rates (B) and clinical scores (C). (D) Femurs and spleens were stained with H&E and quantified as described in Fig. 4F. (E) Flow cytometric analysis of hematopoietic progenitor cells from bone marrow (CMPs, Lin−Sca1−ckit+CD16/32int; GMPs, Lin−Sca1−ckit+CD16/32hi; MEPs, Lin−Sca1−ckit+CD16/32lo). (F) Representative images of AB/PAS staining in colon. Mucus layer, area between yellow dash lines; crypt length, yellow double-headed arrow. Mucus layer thickness and crypt length were quantified. (G) Intestinal epithelial cells, intestinal intraepithelial lymphocytes, and bone marrow stem cells were isolated (N, non-radiation naïve; B, butyrate; P, propionate; C, control) at 24 hours after radiation. Phosphorylated and total H2AX were detected by Western blot. (H) ROS levels in bone marrow stem cells were detected. Fluorescence intensity of each sample was normalized to non-radiation-naïve group average. (I) Long-term survival in propionate compared with control treated groups were monitored for >400 days after radiation. Fractions indicate the number of mice that survived to the end of the experiment. Data are pooled from three [(B) and (C)] or two [(D) and (H)] independent experiments or represent two independent experiments [(E) to (G)]. Error bars indicate SEM, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 determined by log-rank (Mantel Cox) test [(B) and (I)], Mann-Whitney test for AUC (C), and Mann-Whitney test [(D), (E), (F), and (H)].
Histologic analyses revealed that propionate treatment resulted in elevated bone marrow cellularity and splenic white and red pulp recovery (Fig. 5D). Additionally, propionate attenuated radiation-induced loss of granulocyte-macrophage progenitors (GMPs), common myeloid progenitors (CMPs), and megakaryocyte-erythroid progenitors (MEPs) (Fig. 5E) (15). Furthermore, alcian blue and periodic acid-Schiff (AB/PAS) staining of intracellular mucin glycoproteins within goblet cells (16) showed increased mucus thickness and crypt length in propionate recipients compared with controls (Fig. 5F).
We investigated whether SCFAs mediate radioprotection in mice by alleviating the effects of DNA damage and the release of reactive oxygen species (ROS) generated by ionizing radiation. After SCFA treatment, a high dose of total body radiation (8.4 Gy) was administered and 24 hours later, primary intestinal epithelial cells, intestinal intraepithelial lymphocytes, and bone marrow stem cells were isolated. Levels of DNA damage-related proteins, including γH2AX (Fig. 5G), p53, and 53BP1 (fig. S7), were monitored by Western blot. Phosphorylation of these markers was reduced in butyrate- and propionate-treated groups compared with controls in both GI and hematopoietic tissues. Intracellular ROS levels in bone marrow stem cells (Fig. 5H) were significantly enhanced after radiation, whereas butyrate or propionate reduced the ROS levels by 50 to 60% of that found in non-radiation-naïve mice. Finally, because the presence of Lachnospiraceae provided long-term protection, we also found a protective effect of propionate >400 days (Fig. 5I).
To identify whether metabolites other than SCFAs have radioprotective benefits, an unbiased metabolomics approach was performed on fecal samples from elite-survivors and age-matched control mice (17, 18). Total ion chromatogram metabolomic cloudplot and PCA plot showed very distinct metabolite profiles between elite-survivors and controls (Fig. 6, A and B). From 3787 ion features that were altered between these two groups of mice, we identified 141 individual structures that changed substantially (tables S4 and S5 and fig. S8A). The most highly enriched metabolites from the feces of elite-survivors clustered in the tryptophan metabolic pathway, with a fivefold change seen in level of indole-3-carboxaldehyde (I3A) and an eightfold change in the level of kynurenic acid (KYNA) (Fig. 6, C and D, and fig. S8B). These tryptophan metabolites were tested for radiomitigation in vivo, and both metabolites were found to significantly improve survival and reduce the clinical scores of radiation-treated SPF mice (Fig. 6, E to G). Specifically, both I3A- and KYNA-treated groups showed survival rates of ~75%, with the protection persisting for >200 days after radiation (Fig. 6H).
Fig. 6. Untargeted metabolomics reveals tryptophan metabolites as potent radioprotectants.
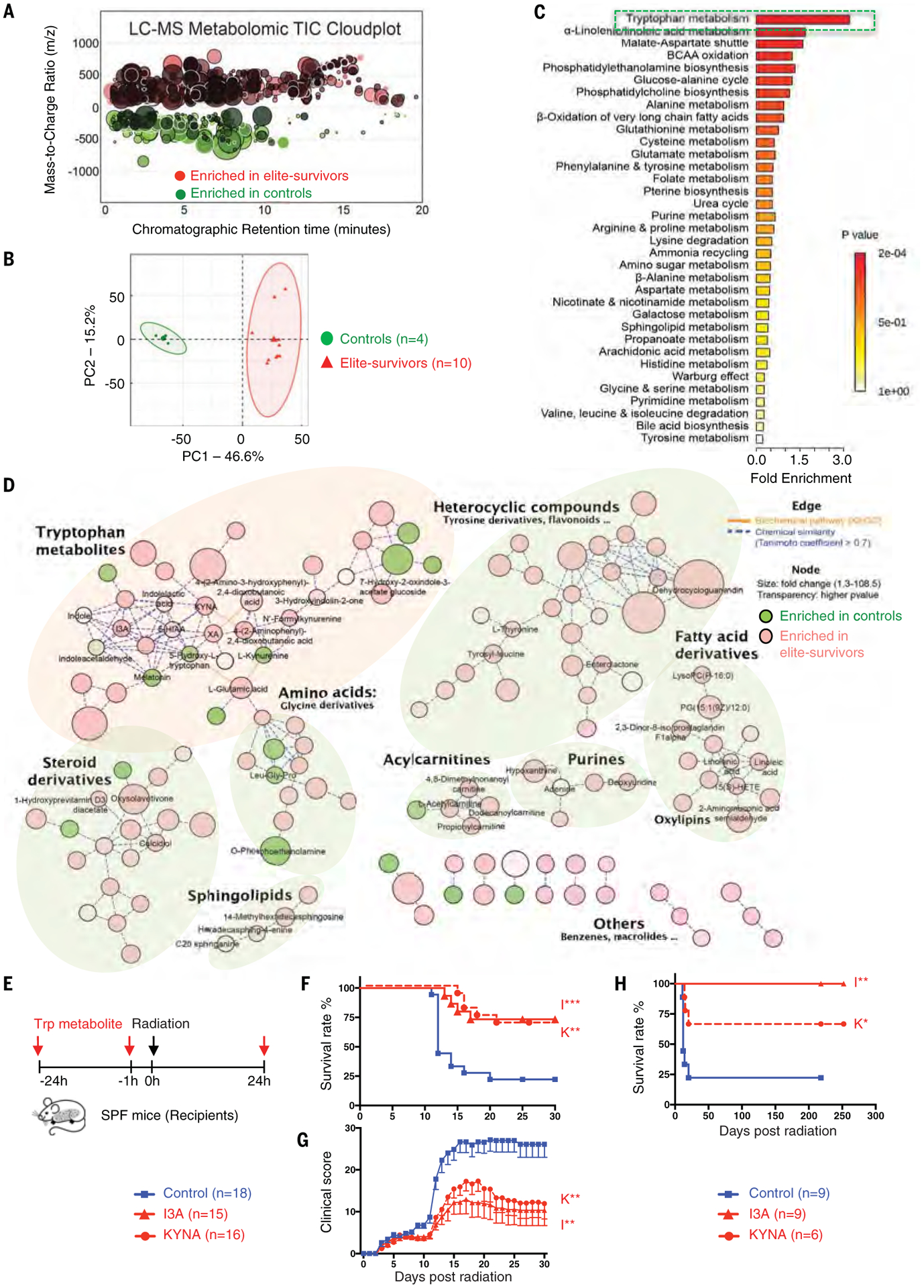
Metabolite profiles were measured in fecal samples of elite-survivors and control mice at day 290 after total body radiation. (A and B) Total ion chromatogram metabolomic cloudplot (P < 0.01) (A) and PCA plot (B) of fecal metabolites. (C) Metabolite set enrichment analysis identified and interpreted metabolites in biochemical contexts. (D) Metabolic network integrated biochemical pathways and chemical relationships of all detected metabolites. Identified metabolites are represented by circular nodes, with lower transparency reflecting lower P values from Welch’s t test. Red nodes, metabolites with higher abundance in elite-survivors; green nodes, those higher in controls. Orange lines connecting metabolites symbolize Kyoto Encyclopedia of Genes and Genomes reactant pair links; green lines symbolize chemical similarity with a Tanimoto coefficient score >0.7. Tryptophan metabolites are highlighted by a pale pink shadow; others are distinguished by pale green shadows. (E) Schematic of tryptophan metabolite treatment. (F and G) Survival rates (F) and clinical scores (G). (H) Long-term survival in tryptophan metabolites compared with control treated groups were monitored for >200 days after radiation. Data are pooled from three [(F) and (G)] or two (H) independent experiments. Error bars indicate SEM, *P < 0.05, **P < 0.01, ***P < 0.001 determined by log-rank (Mantel Cox) test [(F) and (H)] and Mann-Whitney test for AUC (G).
Discussion
We have discovered that a gut microbiome-metabolome network can offer substantial protection against radiation-induced damage in mice. Our studies showed that the presence of Lachnospiraceae and Enterococcaceae was associated with restoration of hematopoiesis and gastrointestinal repair. These two taxa of bacteria were also found to be more abundant in leukemia patients undergoing radiotherapy who showed mild gastrointestinal dysfunction. Propionate, as well as two tryptophan pathway metabolites, I3A and KYNA, were elevated in elite-survivors and provided long-term protection against radiation. This investigation shows that the life-promoting radioprotection of both acute and delayed radiation damage can be provided by intestinal microbiota and their metabolites.
Lachnospiraceae and the microbial metabolite butyrate play an ameliorating role in colitis, food allergy, and graft-versus-host disease (10, 19, 20). SCFAs are metabolized from gut microbial fermentation of indigestible foods in the distal intestine (21), and can directly enter the systemic circulation and act as signaling molecules on peripheral tissues (22). Our work shows that SCFAs can directly promote hematopoiesis and intestinal repair after radiation. Broad inhibition of systemic inflammation, either by reducing the levels of proinflammatory cytokines or by inducing the anti-inflammatory cytokine interleukin-10, mediated by SCFAs may also provide radioprotection (23–26). In addition, this study shows that the tryptophan metabolites I3A and KYNA protect against ARS. Tryptophan metabolites are also known to attenuate proinflammatory cytokine responses (27, 28), and were shown by our untargeted metabolomics approach to be elevated in radiation survivors. Tryptophan metabolites can directly reduce radiation-induced damage and are candidate radioprotective agents. In addition to tryptophan metabolites, we also saw elevations of other metabolite families in the elite-survivor mice. Future studies will be needed to investigate whether these compounds also have radioprotective effects.
Treatment options for the adverse side effects of radiation exposure are limited and expensive, and our study indicates a promising route for further translational investigations.
Materials and Methods
Mice
All experiments used littermate controls or their immediate descendants. Male and female wild-type C57BL/6J mice were obtained from The Jackson Laboratory and house-raised at the University of North Carolina-Chapel Hill (UNC) for at least nine generations. Conventionally raised mice were bred and housed in SPF conditions, and GF animals were generated and housed in the National Gnotobiotic Rodent Resource Center of UNC in AAALAC-accredited facilities. Six- to eight-week-old mice were used in experiments unless otherwise noted. Both male and female mice were used, and both genders responded the same to our bacteria or metabolites. More details are provided in the supplementary materials.
Total body radiation
Nonanesthetized mice were placed in ventilated plastic pie cages and exposed to 8.0 to 9.2 Gy of total body radiation (dose was dependent on sex, age, and SPF/GF). After radiation, mice were housed in sterile autoclaved cages and provided standard chow and water ad libitum unless otherwise noted. Mice were monitored for changes in body weight and other body parameters through 30 days after radiation unless otherwise noted. A clinical score was determined using a cumulative scoring system (table S2) based on weight loss, temperature change, physical appearance, posture, mobility, food consumption, and hydration (29). The last observation carried forward method was used when interpreting the clinical score plots. More details are provided in the supplementary materials.
Elite-survivors
Wild-type C57BL/6J mice received a high dose of total body radiation as described above. Age-matched littermates without radiation treatment were used as controls. After 30 days of monitoring, mice that showed no radiation-induced damage or injury were considered as elite-survivors. At day 290 after radiation, fecal samples were collected for microbiome analyses. Dirty cages from elite-survivors and controls were reserved for the dirty cage-sharing experiment and feces were collected for the FMT experiment.
Dirty cage-sharing experiment
Dirty cages containing feces and used bedding from age-matched controls or elite-survivors were collected every week. Six- to 8-week-old SPF C57BL/6J mice (recipients) were kept in the dirty cages, with mice being transferred to new dirty cages weekly, over a span of 8 weeks. One day after the final dirty cage-housing period, recipients were treated with a high dose of total body radiation. Fecal DNA was collected before radiation for microbiome analysis.
FMT experiment
Feces were collected from controls or elite-survivors. Freshly collected feces were diluted in saline at a ratio of 40 mg feces/ml saline, homogenized, and filtered with a stainless steel sieve (pore size: 0.25 mm). Total bacterial protein concentration was determined by BCA assay to normalize the number of bacterial cells. Fecal mixtures were mixed with 10% autoclaved glycerol, aliquoted, and frozen at −80°C until use. GF or SPF C57BL/6J mice were administered with 200 ml of the fecal mixture by oral gavage twice every week for 4 weeks (GF recipients) or 8 weeks (SPF recipients). Twenty-four hours after the last FMT, recipients were treated with a high dose of total body radiation. Fecal DNA was collected before radiation for microbiome analysis.
Bacteria strain administration
The bacteria taxa termed “Lachnospiraceae” in this study were a mixture of 23 strains that belong to the family Lachnospiraceae as previously reported (11) unless otherwise noted. These bacteria were cultured in an anaerobic chamber in BHI broth supplemented with 5% fetal bovine serum, 0.01% L-cysteine, and 1% corn starch. E. faecalis (ATCC #19433), L. rhamnosus (ATCC #53103), B. fragilis (ATCC #23745), S. aureus (ATCC #25904), S. saprophyticus (ATCC #49907), and S. agalactiae (ATCC #13813) were purchased from ATCC and cultured according to the manufacturer’s instructions. SPF C57BL/6J mice first received antibiotic treatment (20 mg of streptomycin per mouse) by oral gavage. One day later, mice were gavaged with the indicated strains twice a week for 8 weeks. BHI medium was used as a vehicle control. After the last administration, recipients were treated with a high dose of total body radiation.
SCFA treatment
Sodium acetate (Sigma, #S5636), sodium butyrate (Sigma, #303410), or sodium propionate (Sigma, #P1880) (200 mM each) was provided to SPF C57BL/6J mice in drinking water for 8 weeks, and then mice were treated with a high dose of total body radiation. SCFA treatment continued during the monitoring process.
Tryptophan metabolites treatment
I3A (Sigma, #129445) and KYNA (Sigma, #K3375) were dissolved in dimethyl sulfoxide (DMSO). I3A and KYNA (200 mg/kg) were administered by oral gavage into SPF C57BL/6J mice in three doses 1 day before radiation, 1 hour before radiation, and 1 day after radiation. For each gavage, the injection volume was 75 to 100 ml, not to exceed 5 ml/kg (DMSO/body weight).
Histopathology
To assess tissue pathology, spleens, femurs, small intestines, and colons were collected and fixed in 10% neutral-buffered formalin, paraffin embedded, and sectioned (4-μm thickness) at the UNC Lineberger Animal Histopathology Core Facility. Before paraffin embedding, femurs underwent an additional decalcification step in Immunocal (StatLab, McKinney, TX). Slides were stained with hematoxylin and eosin (H&E) and semiquantitatively scored by a board-certified pathologist who was blinded to experimental conditions. Slides of spleen and femur sections were also stained with Ki67 antibody (Cell Signaling Technology, #12202) or cleaved caspase 3 antibody (Cell Signaling Technology, #9661). More details are provided in the supplementary materials.
Clinical patient data
Human stool samples were obtained from 21 leukemia patients at the start of total body radiation given as pretransplantation conditioning before allogeneic hematopoietic stem cell transplantation (see table S3 for detailed information). DNA extraction, polymerase chain reaction amplification of genomic 16S ribosomal RNA V4 and V5 regions, and sequencing were performed as previously described (30).
Tumor inoculation and radiotherapy
Mice were inoculated subcutaneously with 100,000 B16F10 cells or 200,000 EL4 cells in 50% matrigel (Corning, #354234) on the abdomen flank. Tumor volume was measured every 3 days and was calculated using the formula (length × width2) × 0.5. Mouse survival rate was recorded. In accordance with the approved IACUC protocol, mice showing any of the following symptoms were humanely euthanized: (i) weight loss >20%, (ii) ulcerated tumors, or (iii) size reaching 2 cm in any dimension in a single tumor. For radiotherapy, after 10 days of tumor inoculation, mice were anesthetized by isoflurane and immobilized on the operating table by medical tape. Tumor site-localized radiation was performed using an x-ray irradiator with a dose of 10 Gy.
Immunostaining of mucins and measurement of mucus layer and crypt length
Briefly, ~2-cm sections of jejunum and colons were harvested and mounted in CryoMount embedding medium, snap-frozen in liquid nitrogen, and stored at −80°C until use. Four-micrometer-thick sections were cut and mounted onto poly-L-lysine-coated microscope slides, allowed to air dry, and stored at −80°C. Mucus and goblet cell immunostaining was performed using the AB/PAS kit according to the manufacturer’s protocol (Polysciences, #25086). Inner mucus thickness and crypt length were measured in at least 10 different areas of one slide and in more than five mice per group.
Intestine permeability detection
At day 1 after radiation, mice were fasted but allowed water for 4 hours. FITC-dextran (4 kD, Sigma, #46944) was given orally (50 mg/100 g body weight). At 2 hours after gavage, blood was collected and the fluorescence of serum samples was measured using a microplate reader (excitation, 490 nm; emission, 520 nm). Fluorescence intensity of each sample was normalized to the non-radiation-naïve group average.
Bone marrow stem cell, intestinal epithelial cell, and intestinal intraepithelial lymphocyte isolation
Bone marrow stem cells were obtained using a magnetic cell sorting system (MACS, Miltenyi Biotec, #130-091-224) according to the manufacturer’s protocols. Intestinal epithelial cells and intestinal intraepithelial lymphocytes were isolated from fresh colons using a digestion buffer containing 100 μl of DNase (1/50, Sigma, #DN25–1G), 20 ml Collagenase D (1/250, Roche, #11088874103) and 100 μl Dispase (1/50, Stemcell, #07913). More details are provided in the supplementary materials.
Immunoblot analysis
Bone marrow stem cells, intestinal epithelial cells, intestinal intraepithelial lymphocytes or whole spleen tissues were homogenized in radio-immunoprecipitation buffer (Boston BioProducts, #BP-115) containing complete protease inhibitor (Roche, #11697498001) and PhosphoStop (Roche, #4906845001) per the manufacturers’ protocols. Whole-cell lysates were subjected to SDS-polyacrylamide gel electrophoresis and immunoblot analysis. The following primary antibodies were used at a dilution of 1:1000: anti-histone H2AX (Cell Signaling Technology, #7631), anti-phospho-histone H2AX (Cell Signaling Technology, #9718), anti-p53 (Cell Signaling Technology, #2524), anti-phospho-p53 (Cell Signaling Technology, #9284), anti-53BP1 (Invitrogen, #PA1–16566), anti-phospho-53BP1 (Abcam, #ab70323), anti-cleaved caspase 3 (Cell Signaling Technology, #9664), and anti-actin-HRP (C-11) (Santa Cruz Biotechnology, #sc-1615). Goat antirabbit horseradish peroxidase (HRP) (Jackson Immune Research Laboratories, #111-035-144) and goat anti-mouse-IgG HRP (Jackson Immune Research Laboratories, #115-035-146) were used as secondary antibodies. Protein densitometry was quantified with ImageJ software.
ROS detection
Intracellular ROS levels in bone marrow stem cells were detected using the DCFDA Cellular ROS Detection Assay Kit (Abcam, #ab113851) according to the manufacturer’s protocol. The average level of ROS in the non-radiation-naïve group was calculated, and all sample results were normalized to this average. More details are provided in the supplementary materials.
Flow cytometry analysis
Bone marrow was flushed from femurs and then red blood cells were lysed with ammonium chloride-potassium lysing buffer (Lonza, #10–548E). Cells were labeled with phycoerythrin (PE)/Cy7-conjugated anti-CD16/32 clone 93 (BioLegend, #101317), V450 lineage cocktail (anti-CD3ɛ clone 500A2, anti-CD11b clone M1/70, anti B220 clone RA3–6B2, anti-Ly-76 clone TER-119, anti-Ly6G/C clone RB6–8C5, BD Biosciences, #561301), PE-conjugated anti-Sca1 clone E13–161.7 (BD Biosciences, #553336), and FITC-conjugated anti-c-kit clone 2B8 (BD Biosciences, #553354). Labeled cells were fixed in 4% paraformaldehyde (ThermoFisher, #AAJ19943K2) and then analyzed using an LSRII flow cyto-meter (BD Biosciences). The resultant data were gated using FlowJo version 10.4 software (BD Biosciences). Hematopoietic stem and progenitor cells were defined as Lin−Sca1+c-kit+, CMPs as Lin−Sca1−c-kit+CD16/32int, GMPs as Lin−Sca1−c-kit+CD16/32hi, and MEPs as Lin−Sca1−c-kit+CD16/32low.
Fecal DNA extraction, 16S rRNA gene sequencing, and data analysis
Fecal sample preparation and 16S rRNA sequencing were conducted as previously described (10, 31). Fecal DNA samples were amplified by polymerase chain reaction (PCR) using barcoded primer pairs targeting the V1–V2 or V3–V4 region of the 16S rRNA gene. PCR amplicons were sequenced using the Mi-Seq Illumina sequencer. The resulting bacterial sequence fragments were analyzed in QIIME version 1.8.0. More details are provided in the supplementary materials.
SCFAs detection by gas chromatography-mass spectrometry quantitation
Fecal SCFA contents were quantified as described in Zheng et al. (32) with modifications. The instrumental analysis was conducted on a 7820A GC-5977B MSD system (Agilent Technologies, Santa Clara, CA, USA). The relative standard deviations of both intraday and interday precision of analysis were <10%. The limit of quantitation was determined to be 1 ng on the column for acetic acid, 1 ng for propionic acid, and 0.1 ng for butyric acid. More details are provided in the supplementary materials.
Untargeted metabolomics
Untargeted metabolomics of mouse feces was performed on a high-resolution accurate mass spectrometry-based platform coupling to a cutting-edge integrated chemoinformatics pipeline as modified from a previous publication (33). The system consisted of a Vanquish UHPLC and a Thermo Scientific Q-Exactive high resolution/accurate mass spectrometer interfacing with a heated electrospray ionization source and a hybrid quadrupole-orbitrap mass analyzer operated at 70,000 mass resolution (Waltham, MA, USA). Stringent quality assurance and quality control procedures were applied, including timely mass calibration, sample randomization, and intermittent quality control injection.
All MS1 raw data were processed in XCMS (Scripps, La Jolla, CA) for peak picking, peak alignment, gap filling, and sample normalization. Compound identification was implemented in MS-DIAL 2.94 and MS-FINDER 3.12. All identifications were manually curated and validated as available using an in-house metabolomic database established based on authentic chemical standards. Confidence level of annotation was assigned to each structure as required by the Metabolomics Standards Initiative. For pathway analysis and visualization, MetaboAnalyst version 4.0 was used to perform metabolite enrichment analysis and to create a heatmap showing a selection of 50 annotated structures with largest group difference; a MetaMapp approach was used to compute a metabolomic network clustering by biochemical and chemical relationship that was further visualized in CytoScape version 3.7.1. More details are provided in the supplementary materials.
Statistics
All values are presented in the figures as mean ± SEM, with *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; n.s., no significance. The number of animals (n) used in the experiments is indicated in the figures. For all data, one dot or lane represents one mouse or one patient. Linear fitting and normalization were performed in GraphPad Prism software. If not specified in the figure legends, statistical significance between two groups was determined by unpaired, two-tailed Student’s t test, log-rank (Mantel Cox) test, or Mann-Whitney test, and significance between more than two groups was determined using two-way ANOVA in GraphPad Prism with the default parameter based on experience.
Supplementary Material
Funding:
This project was supported by NIAID U19-A1067798, R35-CA232109, T32-CA009156, NMSS CA10068, and P01-DK094779 (J.P.Y.T. and R.B.S.). We thank the following for core and technical support: UNC Flow Cytometry Core Facility, Microbiome Core Facility and National Gnotobiotic Rodent Resource Center [supported by NIH P40-OD010995, P30-DK34987, P30-DK056350, the Crohn’s and Colitis Foundation and the Center for Gastrointestinal Biology and Disease (CGIBD)]. Animal histopathology was performed within the LCCC Animal Histopathology Core Facility at UNC [supported in part by an NCI Center Core Support Grant (CA16086)]. The metabolomics study was performed within the UNC Biomarker Mass Spectrometry Facility and the UNC Center for Environmental Health and Susceptibility (supported by NIEHS ES024950 and P30-ES010126). Clinical data were provided by the Duke Adult Blood and Marrow Transplant Program (supported by R21AG066388 (A.D.S., L.M.B., and N.J.C.), an ASH Scholar Award (A.D.S.), and Duke Pepper Center P30-AG028716-13 Mini#6 (A.D.S.) and Memorial Sloan Kettering Cancer Center [supported by K08HL143189 (J.U.P.), NCI P30 CA008748 (J.U.P.), R01-CA228358 (M.R.M.v.d.B.), R01-CA228308 (M.R.M.v.d.B.), R01-HL147584 (M.R.M.v.d.B.), P30 CA008748 (M.R.M.v.d.B.), Project 2 of P01-CA023766 (M.R.M.v.d.B.), Project 2 of P01-AG052359 (M.R.M.v.d.B.), and U01 AI124275 (M.R.M.v.d.B.)]. Flow cytometry was performed at the Duke Human Vaccine Institute Flow Cytometry Facility in the Immunology Unit of Duke Regional Biocontainment Laboratory (supported by UC6-AI058607).
Footnotes
Competing interests: J.P.Y.T. is a cofounder of IMMvention Therapeutix and GoldCrest Bio. H.G. and W.J.B. hold equity shares in GoldCrest Bio. J.P.Y.T., H.G., and W.J.B. are named as inventors on U.S. Provisional Patent Application Serial No. 62/779,776: “Lachnospiraceae Mitigates Against Radiation-Induced Hematopoietic/Gastrointestinal Injury And Death, And Promotes Cancer Control By Radiation.” M.R.M.v.d.B. has received research support from Seres Therapeutics; has consulted, received honorarium from, or participated in advisory boards for Seres Therapeutics, Novartis, Evelo, Jazz Pharmaceuticals, Therakos, Amgen, Magenta Therapeutics, Merck & Co, Inc., Acute Leukemia Forum (ALF), and DKMS Medical Council (Board); has IP Licensing with Seres Therapeutics, Juno Therapeutics; and has stock options from Smart Immune. J.U.P. reports research funding, intellectual property fees, and travel reimbursement from Seres Therapeutics and consulting fees from DaVolterra.
Data and materials availability: All sequencing data have been deposited to the European Nucleotide Archive (https://www.ebi.ac.uk/ena/browser/home) under the accession numbers PRJEB37999, PRJEB38109, PRJEB38110, PRJEB38111, PRJEB38112, PRJEB38115, PRJEB38116, PRJEB38117, PRJEB38118, and PRJEB38119. All other data generated or analyzed during this study are included in this article and its supplementary materials. All radiotherapy participants provided informed consent before their participation in the studies, which were approved by the institutional review board at Duke University (Pro00050975, Pro00006268, and Pro00078566). The Lachnospiraceae strains were kindly provided by V. B. Young (University of Michigan-Ann Arbor), and the authors do not have the right to disseminate them.
REFERENCES AND NOTES
- 1.Hauer-Jensen M, Denham JW, Andreyev HJ, Radiation enteropathy: Pathogenesis, treatment and prevention. Nat. Rev. Gastroenterol. Hepatol 11, 470–479 (2014). doi: 10.1038/nrgastro.2014.46; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shadad AK, Sullivan FJ, Martin JD, Egan LJ, Gastrointestinal radiation injury: Prevention and treatment. World J. Gastroenterol 19, 199–208 (2013). doi: 10.3748/wjg.v19.i2.199; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI, Human nutrition, the gut microbiome and the immune system. Nature 474, 327–336 (2011). doi: 10.1038/nature10213; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crawford PA, Gordon JI, Microbial regulation of intestinal radiosensitivity. Proc. Natl. Acad. Sci. U.S.A 102, 13254–13259 (2005). doi: 10.1073/pnas.0504830102; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciorba MA et al. , Lactobacillus probiotic protects intestinal epithelium from radiation injury in a TLR-2/cyclo-oxygenase-2-dependent manner. Gut 61, 829–838 (2012). doi: 10.1136/gutjnl-2011-300367; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang A et al. , Gut microbial dysbiosis may predict diarrhea and fatigue in patients undergoing pelvic cancer radiotherapy: A pilot study. PLOS ONE 10, e0126312 (2015). doi: 10.1371/journal.pone.0126312; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riehl TE et al. , Lactobacillus rhamnosus GG protects the intestinal epithelium from radiation injury through release of lipoteichoic acid, macrophage activation and the migration of mesenchymal stem cells. Gut 68, 1003–1013 (2019). doi: 10.1136/gutjnl-2018-316226; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manichanh C et al. , The gut microbiota predispose to the pathophysiology of acute postradiotherapy diarrhea. Am. J. Gastroenterol 103, 1754–1761 (2008). doi: 10.1111/j.1572-0241.2008.01868.x; [DOI] [PubMed] [Google Scholar]
- 9.Kim YS, Kim J, Park SJ, High-throughput 16S rRNA gene sequencing reveals alterations of mouse intestinal microbiota after radiotherapy. Anaerobe 33, 1–7 (2015). doi: 10.1016/j.anaerobe.2015.01.004; [DOI] [PubMed] [Google Scholar]
- 10.Chen L et al. , NLRP12 attenuates colon inflammation by maintaining colonic microbial diversity and promoting protective commensal bacterial growth. Nat. Immunol 18, 541–551 (2017). doi: 10.1038/ni.3690; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reeves AE, Koenigsknecht MJ, Bergin IL, Young VB, Suppression of Clostridium difficile in the gastrointestinal tracts of germfree mice inoculated with a murine isolate from the family Lachnospiraceae. Infect. Immun 80, 3786–3794 (2012). doi: 10.1128/IAI.00647-12; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reichardt N et al. , Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 8, 1323–1335 (2014). doi: 10.1038/ismej.2014.14; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith PM et al. , The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573 (2013). doi: 10.1126/science.1241165; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arpaia N et al. , Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 (2013). doi: 10.1038/nature12726; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim JH et al. , NRF2-mediated Notch pathway activation enhances hematopoietic reconstitution following myelosuppressive radiation. J. Clin. Invest 124, 730–741 (2014). doi: 10.1172/JCI70812; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao X et al. , Chronic stress promotes colitis by disturbing the gut microbiota and triggering immune system response. Proc. Natl. Acad. Sci. U.S.A 115, E2960–E2969 (2018). doi: 10.1073/pnas.1720696115; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rooks MG, Garrett WS, Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol 16, 341–352 (2016). doi: 10.1038/nri.2016.42; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levy M, Blacher E, Elinav E, Microbiome, metabolites and host immunity. Curr. Opin. Microbiol 35, 8–15 (2017). doi: 10.1016/j.mib.2016.10.003; [DOI] [PubMed] [Google Scholar]
- 19.Mathewson ND et al. , Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat. Immunol 17, 505–513 (2016). doi: 10.1038/ni.3400; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feehley T et al. , Healthy infants harbor intestinal bacteria that protect against food allergy. Nat. Med 25, 448–453 (2019). doi: 10.1038/s41591-018-0324-z; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macfarlane GT, Macfarlane S, Bacteria, colonic fermentation, and gastrointestinal health. J. AOAC Int 95, 50–60 (2012). doi: 10.5740/jaoacint.SGE_Macfarlane; [DOI] [PubMed] [Google Scholar]
- 22.Canfora EE, Jocken JW, Blaak EE, Short-chain fatty acids in control of body weight and insulin sensitivity. Nat. Rev. Endocrinol 11, 577–591 (2015). doi: 10.1038/nrendo.2015.128; [DOI] [PubMed] [Google Scholar]
- 23.Maa MC et al. , Butyrate reduced lipopolysaccharide-mediated macrophage migration by suppression of Src enhancement and focal adhesion kinase activity. J. Nutr. Biochem 21, 1186–1192 (2010). doi: 10.1016/j.jnutbio.2009.10.004; [DOI] [PubMed] [Google Scholar]
- 24.Liu L et al. , Butyrate interferes with the differentiation and function of human monocyte-derived dendritic cells. Cell. Immunol 277, 66–73 (2012). doi: 10.1016/j.cellimm.2012.05.011; [DOI] [PubMed] [Google Scholar]
- 25.Chang PV, Hao L, Offermanns S, Medzhitov R, The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. U.S.A 111, 2247–2252 (2014). doi: 10.1073/pnas.1322269111; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trompette A et al. , Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med 20, 159–166 (2014). doi: 10.1038/nm.3444; [DOI] [PubMed] [Google Scholar]
- 27.Wlodarska M et al. , Indoleacrylic acid produced by commensal Peptostreptococcus species suppresses inflammation. Cell Host Microbe 22, 25–37.e6 (2017). doi: 10.1016/j.chom.2017.06.007; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krishnan S et al. , Gut microbiota-derived tryptophan metabolites modulate inflammatory response in hepatocytes and macrophages. Cell Rep. 23, 1099–1111 (2018). doi: 10.1016/j.celrep.2018.03.109; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurkjian CJ et al. , The toll-like receptor 2/6 agonist, FSL-1 lipopeptide, therapeutically mitigates acute radiation syndrome. Sci. Rep 7, 17355 (2017). doi: 10.1038/s41598-017-17729-9; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peled JU et al. , Microbiota as predictor of mortality in allogeneic hematopoietic-cell transplantation. N. Engl. J. Med 382, 822–834 (2020). doi: 10.1056/NEJMoa1900623; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Truax AD et al. , The inhibitory innate immune sensor NLRP12 maintains a threshold against obesity by regulating gut microbiota homeostasis. Cell Host Microbe 24, 364–378.e6 (2018). doi: 10.1016/j.chom.2018.08.009; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng X et al. , A targeted metabolomic protocol for short-chain fatty acids and branched-chain amino acids. Metabolomics 9, 818–827 (2013). doi: 10.1007/s11306-013-0500-6; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai Z et al. , Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nat. Methods 15, 53–56 (2018). doi: 10.1038/nmeth.4512; [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


