Abstract
Background
Analyses of gut microbiome composition in livestock species have shown its potential to contribute to the regulation of complex phenotypes. However, little is known about the host genetic control over the gut microbial communities. In pigs, previous studies are based on classical “single-gene-single-trait” approaches and have evaluated the role of host genome controlling gut prokaryote and eukaryote communities separately.
Results
In order to determine the ability of the host genome to control the diversity and composition of microbial communities in healthy pigs, we undertook genome-wide association studies (GWAS) for 39 microbial phenotypes that included 2 diversity indexes, and the relative abundance of 31 bacterial and six commensal protist genera in 390 pigs genotyped for 70 K SNPs. The GWAS results were processed through a 3-step analytical pipeline comprised of (1) association weight matrix; (2) regulatory impact factor; and (3) partial correlation and information theory. The inferred gene regulatory network comprised 3561 genes (within a 5 kb distance from a relevant SNP–P < 0.05) and 738,913 connections (SNP-to-SNP co-associations). Our findings highlight the complexity and polygenic nature of the pig gut microbial ecosystem. Prominent within the network were 5 regulators, PRDM15, STAT1, ssc-mir-371, SOX9 and RUNX2 which gathered 942, 607, 588, 284 and 273 connections, respectively. PRDM15 modulates the transcription of upstream regulators of WNT and MAPK-ERK signaling to safeguard naive pluripotency and regulates the production of Th1- and Th2-type immune response. The signal transducer STAT1 has long been associated with immune processes and was recently identified as a potential regulator of vaccine response to porcine reproductive and respiratory syndrome. The list of regulators was enriched for immune-related pathways, and the list of predicted targets includes candidate genes previously reported as associated with microbiota profile in pigs, mice and human, such as SLIT3, SLC39A8, NOS1, IL1R2, DAB1, TOX3, SPP1, THSD7B, ELF2, PIANP, A2ML1, and IFNAR1. Moreover, we show the existence of host-genetic variants jointly associated with the relative abundance of butyrate producer bacteria and host performance.
Conclusions
Taken together, our results identified regulators, candidate genes, and mechanisms linked with microbiome modulation by the host. They further highlight the value of the proposed analytical pipeline to exploit pleiotropy and the crosstalk between bacteria and protists as significant contributors to host-microbiome interactions and identify genetic markers and candidate genes that can be incorporated in breeding program to improve host-performance and microbial traits.
Video Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s40168-020-00994-8.
Keywords: Microbiota, Pig, Gene network, Regulators, Protist, Bacteria
Background
The gut microbiota is a diverse ecosystem predominantly dominated by bacteria, but other microorganisms such as fungi and protists are also present. Influenced by the host immunity and environmental factors such as age, diet, and geography, the gut microbiota is known to be involved in many physiological functions as well as in disease pathogenesis (see for instance the recent reviews of Richard and Sokol, 2019 [1]; Torp Austvoll et al. 2020 [2]).
Lagging behind humans and model organisms, studies of the gut microbiome in livestock species have dramatically increased over the last decade thanks to a decreased metagenome next-generation sequencing cost. In the particular case of the pig, these studies have provided valuable information into the gut microbiota compositional changes, as well as associations between the porcine gut microbial communities and production traits [3–8]. In contrast, little is known about the host genetic control over the gut microbial communities in pigs. Previous studies based on classical “single-gene-single-trait” approaches and have independently evaluated the role of host-genetic genome controlling pig gut prokaryote [9–11] or eukaryote communities [5]. Therefore, ignoring the crosstalk between bacterial and protist communities, but also the contribution of gene-by-gene interactions shaping the pig gut microbial ecosystem. In their review, Aluthge et al. (2019) [12] identified the lack of sophisticated computational and bioinformatics approaches as the major bottleneck to look beyond compositional changes and instead understand the functional causative role of the pig gut microbiome.
To address this void, we propose an association weight matrix approach (AWM) [13, 14] to generate a gene co-association network where nodes are SNP mapped to or nearby gene coding regions, and found to be associated with the diversity and abundance of 31 bacterial and 6 commensal protist genera in the gut of 390 pigs genotyped for 70 K SNPs. In building and interpreting the network, we place emphasis on gene regulators responsible for the crosstalk between the host bacterial and protists communities.
Methods
Animal care and experimental procedures were carried out following national and institutional guidelines for the Good Experimental Practices and were approved by the IRTA Ethical Committee.
Animals, sample collection, and gut microbiome phenotypes
The pigs employed in this study are a subset of those reported in Ramayo-Caldas et al. (2020) [5]. In brief, we used a total of 405 weaned piglets (204 males and 201 females) distributed in seven batches. Samples were collected in a commercial farm at 60 ± 8 days of age. Fecal DNA was extracted with the DNeasy PowerSoil Kit (QIAGEN, Hilden, Germany), following manufacturer’s instructions. The 16S rRNA gene fragment was amplified using the primers V3_F357_N: 5′-CCTACGGGNGGCWGCAG-3′ and V4_R805: 5′-GACTACHVGGGTATCTAATCC-3′. Protist-specific primers F-566: 5′-CAGCAGCCGCGGTAATTCC-3′ and R-1200: 5′-CCCGTGTTGAGTCAAATTAAGC-3′ were used to amplify the 18S rRNA gene fragment. Amplicons were paired-end (2 × 250 nt) sequenced on an Illumina NovaSeq (Illumina, San Diego, CA, USA) at the University of Illinois Keck Center. Sequences were analyzed with QIIME2 [15] and processed into amplicon sequences variants (ASVs) at 99% of identity. Samples with less than 10,000 reads were excluded and ASVs present in less than three samples and representing less than 0.005% of the total counts were discarded. ASVs were classified to the lowest possible taxonomic level based on SILVA v123 database for 18S rRNA genes, and GreenGenes Database for Bacteria [16]. Bacteria and protist alpha diversity were evaluated with the Shannon index (Shannon, 1948); before the estimation of diversity indexes, samples were rarefied at 10,000 reads of depth. Finally, ASVs were aggregated at genera level, and only those genus present in more than 60% of the samples were considered in posteriors data analysis. In total, we captured 39 microbiome phenotypes that included 2 diversity indexes, and the relative abundance of 31 bacterial and 6 commensal protist genera. (Supplementary Table 1).
Genotypes and genome-wide association studies (GWAS)
The Porcine 70 K GGP Porcine HD Array (Illumina, San Diego, CA) was used to genotype 390 out of 405 animals. We excluded single nucleotide polymorphisms (SNPs) with minor allele frequencies < 5%, rates of missing genotypes above 10%, as well as SNPs that did not map to the porcine reference genome (Sscrofa11.1 assembly). Then, to identify SNPs from the host genome associated with the alpha diversity as well as bacteria and protists relative abundances, a series of 39 GWAS were performed between 42,562 SNPs and the alpha diversity or the centered log ratio (clr) transformed genera abundance. For the GWAS, we used the GCTA software [17] using the following model at each SNP:
where yijk corresponds to the microbiome phenotype under scrutiny of the i-th individual animal of sex j in the k-th batch; sexj and bk correspond to the systematic effects of j-th sex (2 levels) and k-th batch (7 levels), respectively; ui is the random additive genetic effect of the i-th individual, collectively distributed as u ~ N(0, G) where is the additive genetic variance and G is the genomic relationship matrix calculated using the filtered autosomal SNPs based on the methodology of Yang et al. (2011) [17]; sli is the genotype (coded as 0,1,2) for the l-th SNP of the i-th individual, and al is the allele substitution effect of the l-th SNP on the microbiome phenotype being analyzed. Following [18] and with equivalent original derivations from [19], FDR was calculated as
Where P is the P value tested, A is the number of SNP that were significant at the P value tested, and T is the total number of SNP tested. For further analyses and in order to allow for direct comparison across phenotypes, estimated SNP effects were standardized by dividing them by the standard deviation of all SNP effects.
Association weight matrix, regulatory impact factors, and gene co-association networks
We used the AWM methodology [13, 14] in combination with the regulatory impact factors (RIF) algorithm [20] to identify the SNP, anchored to genes, to be included in the co-association gene network. Genes in the AWM are tagged by SNP that were found to be either associated with the key phenotype (alpha diversity), pleiotropic, or key regulators. In detail, the AWM was built in accordance with the following steps:
Initial search of SNP in genes (SNP-gene)
Using a population of 20 European pig breeds, Muñoz et al. (2019) [21] reported linkage disequilibrium of r2 > 0.2 for SNP pairs separated by 0.05 Mb. Therefore, we selected SNP located in the coding region or within 5 kb of an annotated gene based on the Sscrofa11.1 reference genome assembly.
SNP-genes associated with key phenotypes
Using the alpha diversity indexes for bacteria and protists as key phenotypes, we selected those SNP-Gene associated with alpha diversity and namely diversity-associated SNPs.
SNP-genes with pleiotropic potential
We captured the average number of phenotypes to which the diversity-associated SNPs were associated with, namely NA, and selected the remaining SNP-Genes associated (P < 0.05) to more than NA phenotypes and referred to as pleiotropic SNPs.
SNP-genes with regulatory potential
We used the RIF algorithm [20] to identify key regulatory SNP. Details are as follows.
To undertake the RIF analysis, we considered the diversity-associated SNPs and the pleiotropic SNP as potential targets of all the SNP-Genes annotated as either transcription factors (TF) or microRNA genes (miRNA). The RIF algorithm is designed to detect loci with high regulatory potential in a set of loci, TFs, and microRNA genes in our case, while contrasting two biological conditions or groups, such as bacteria and protists in our case. The analysis makes use of two metrics: RIF1 and RIF2. While RIF1 prioritizes regulators that are consistently the most differentially co-associated with the highly associated potential target genes, RIF2 highlights regulators with the most altered abilities to predict the association of potential target genes.
To identify significant gene–gene interactions, we used the partial correlation and information theory (PCIT) algorithm [22] which calculates pairwise correlations between loci while accounting for the influence of a third locus. Unlike likelihood-based approaches, which invoke a parametric distribution (e.g., normal) assumed to hold under the null hypothesis and then a nominal P value (e.g., 5%) used to ascertain significance, PCIT is an information theoretic approach. Its threshold is an informative metric; in this case, the partial correlation after exploring all trios in judging the significance of a given correlation, which might then become a connection when inferring a network. It thereby tests all possible 3-way combinations in a dataset and only keeps correlations between loci if they are significant and independent of the expression of another locus, whereas no hard threshold is set for the correlation strength. The significance threshold for each combination of loci depends on the average ratio of partial to direct correlations. Gene interactions were predicted using correlation analysis of the SNP effects across pairwise rows of the AWM. Hence, the AWM-predicted gene interactions are based on significant co-association between SNP. In the network, every node represents a gene (or SNP), whereas every edge connecting two nodes represents a significant gene–gene interaction (based on SNP–SNP co-association). Finally, the Cytoscape software [23] was used to visualize the gene network and the CentiScaPe plugin [24] was used to calculate specific node centrality values and network topology parameters.
Transcription factor binding motifs search
To further investigate the alleged role of the most relevant TFs, i.e., the PR/Set domain 15 (PRDM15) and the signal transducer and activator of transcription 1 (STAT1), a TF-binding motif search was implemented in the promoter region of the predicted target genes (i.e., genes with significant associations according to the AWM approach for PRDM15 and STAT1. To this end, we established putative promoter regions 1 kb upstream from the start of the coding region of each target gene and downloaded the corresponding sequences in the Sscrofa11.1 porcine assembly according to Ensembl repositories [25], by means of the BioMart tool (https://www.ensembl.org/biomart/martview/). Transcription factor binding motifs for PRDM15 and STAT1 genes were retrieved from JASPAR database v.2020 [26] represented as position frequency matrices (PFMs). The FIMO algorithm [27] within MEME Suite [28] was subsequently employed for scanning individual matches between PFMs and retrieved promoter sequences. The Scrofa11.1 assembly was used for building a zero-order Markov model and integrated in FIMO calculations in order to correct for possible biases in nucleotide proportions. Motif occurrences with estimated P value below 10− 4 were considered significant and retained for further analyses.
MicroRNA-binding sites search and structural inference
Following the rationale for detecting putative functional interactions among the set of key regulators identified by the AWM approach, we aimed at determining whether miRNA-mRNA significant co-associations found for relevant miRNA regulator genes were suggestive of an interaction between the miRNA and their co-associated mRNA transcripts. In this way, the ssc-miR-371 gene was among the top regulatory factors according to RIF metrics. The mRNA genes harboring SNPs significantly co-associated with the polymorphism within miR-371 (rs320008166, n.59 T > C) gene were retrieved and their 3′-UTR sequences according to the Sscrofa11.1 assembly annotation in Ensembl repositories [25] were downloaded by means of the BioMart tool (https://www.ensembl.org/biomart/martview/). In order to obtain a putative list of targeted mRNAs by ssc-miR-371, the seed region of the miRNA (2nd to 8th 5′ nucleotides of the mature miRNA) was reverse complemented and miRNA binding sites comprising perfect sequence matches between the seed and the retrieved 3′-UTRs (7mer-m8 sites) were assessed by using the locate tool from the SeqKit toolkit [29]. The potential structural consequences of rs320008166 (n.59 T > C) in the hairpin organization of ssc-miR-371 precursor transcript was assessed with the RNAfold software [30].
Results
Gene-tailored association between microbial traits
Table 1 lists the number of significant SNP and false discovery rate (FDR) at three nominal P value thresholds (P value < 0.05, 0.01, and 0.001) across the 39 phenotypes that were subjected to GWAS. The results highlight the trade-off that exists between significant SNP (the higher the better) and the FDR (the lower the better) as P values become more stringent. On average across the 39 phenotypes, the number of significant SNP (average FDR in brackets) at P values < 0.05, 0.01, and 0.001 was 4015.3 (FDR = 50.6%), 272.8 (FDR = 16.5%), and 87.6 (FDR = 5.8%), respectively. At the most stringent P value threshold of < 0.001, the highest number of significant SNP (N = 381; FDR = 1.1%) was obtained for the abundance of Faecalibacterium. On the other extreme, the lowest number of significant SNP (N = 25; FDR = 17.0%) was obtained for the abundance of Catenibacterium. Details of the genome map position and strength of the statistical association of the most significant SNP in each of the 39 phenotypes are given in Table 2. For each SNP–phenotype pair, the distance to and identity of the nearest gene is also listed in Table 2. With five instances, Chromosome 8 harbored the highest number of most associated SNP including one in the coding region of SORCS2 (SNP rs320095924) and one in the coding region of TRIM2 (rs329143797).
Table 1.
Number of significant SNP (N) and false discovery rate (FDR, %) at three nominal P value thresholds and across the 39 phenotypes
| Phenotype | P value < 0.05 | P value < 0.01 | P value < 0.001 | |||
|---|---|---|---|---|---|---|
| N | FDR, % | N | FDR, % | N | FDR, % | |
| Anaerovibrio | 3836 | 53.13 | 355 | 11.90 | 105 | 4.04 |
| Blautia | 3803 | 53.64 | 232 | 18.26 | 88 | 4.82 |
| Bulleidia | 4114 | 49.18 | 196 | 21.63 | 77 | 5.51 |
| Butyricicoccus | 3830 | 53.22 | 255 | 16.60 | 92 | 4.61 |
| Campylobacter | 4088 | 49.53 | 271 | 15.62 | 69 | 6.15 |
| Catenibacterium | 4407 | 45.56 | 164 | 25.87 | 25 | 17.01 |
| Clostridium | 4024 | 50.40 | 211 | 20.09 | 67 | 6.34 |
| Collinsella | 3969 | 51.17 | 290 | 14.59 | 73 | 5.82 |
| Coprococcus | 4152 | 48.68 | 269 | 15.73 | 68 | 6.24 |
| Desulfovibrio | 3797 | 53.73 | 203 | 20.88 | 70 | 6.07 |
| Dorea | 4170 | 48.45 | 226 | 18.75 | 63 | 6.74 |
| Faecalibacterium | 3644 | 56.21 | 750 | 5.58 | 387 | 1.08 |
| Fibrobacter | 4032 | 50.29 | 225 | 18.83 | 66 | 6.43 |
| Gemmiger | 4077 | 49.68 | 238 | 17.80 | 43 | 9.88 |
| Lachnospira | 4192 | 48.17 | 248 | 17.07 | 64 | 6.64 |
| Lactobacillus | 3966 | 51.21 | 291 | 14.54 | 103 | 4.12 |
| Megasphaera | 4027 | 50.36 | 234 | 18.10 | 69 | 6.15 |
| Mitsuokella | 4201 | 48.06 | 211 | 20.09 | 58 | 7.32 |
| Oscillospira | 4161 | 48.57 | 198 | 21.41 | 68 | 6.24 |
| Parabacteroides | 4206 | 47.99 | 211 | 20.09 | 45 | 9.44 |
| Peptococcus | 4254 | 47.39 | 319 | 13.25 | 95 | 4.47 |
| Phascolarctobacterium | 3921 | 51.86 | 278 | 15.22 | 129 | 3.28 |
| Prevotella | 3952 | 51.41 | 267 | 15.85 | 66 | 6.43 |
| RFN20 | 3946 | 51.50 | 299 | 14.14 | 101 | 4.20 |
| Roseburia | 4025 | 50.39 | 264 | 16.03 | 59 | 7.20 |
| Ruminococcus | 4015 | 50.53 | 265 | 15.97 | 97 | 4.37 |
| Sphaerochaeta | 4143 | 48.80 | 213 | 19.90 | 68 | 6.24 |
| Streptococcus | 4060 | 49.91 | 260 | 16.28 | 73 | 5.82 |
| Succinivibrio | 4071 | 49.76 | 314 | 13.46 | 99 | 4.28 |
| Sutterella | 4016 | 50.51 | 248 | 17.07 | 81 | 5.24 |
| Treponema | 3966 | 51.21 | 307 | 13.77 | 102 | 4.16 |
| AlphaBACT | 3960 | 51.30 | 314 | 13.46 | 89 | 4.77 |
| Entamoeba | 3849 | 52.92 | 335 | 12.61 | 117 | 3.62 |
| Hypotrichomonas | 3897 | 52.21 | 230 | 18.42 | 67 | 6.34 |
| Trichomitus | 3944 | 51.52 | 307 | 13.77 | 126 | 3.36 |
| Tetratrichomonas | 3998 | 50.76 | 266 | 15.91 | 64 | 6.64 |
| Neobalantidium | 3828 | 53.24 | 297 | 14.24 | 139 | 3.05 |
| Blastocystis | 3859 | 52.77 | 320 | 13.21 | 78 | 5.44 |
| AlphaPROTO | 4195 | 48.13 | 259 | 16.34 | 67 | 6.34 |
Table 2.
Genome map position and strength of the association for most significant SNP in each of the 39 phenotypes
| Phenotype | SNP | Chr | Bp | Candidate gene | Distance to gene (Bp) | Effect | P value |
|---|---|---|---|---|---|---|---|
| Anaerovibrio | rs320095924 | 8 | 3,650,253 | SORCS2 | 0 | − 0.364 | 8.89E−09 |
| Blautia | rs331690240 | 23 | 14,234,646 | ENSSSCG00000050465 | 0 | 0.475 | 2.14E−21 |
| Bulleidia | rs340048252 | 11 | 69,840,940 | NALCN | 0 | 1.086 | 5.29E−17 |
| Butyricicoccus | rs81361511 | 2 | 96,004,482 | ENSSSCG00000041042 | 71,388 | − 0.882 | 2.43E−15 |
| Campylobacter | rs81472036 | 18 | 15,768,346 | EXOC4 | 0 | − 0.708 | 6.09E−07 |
| Catenibacterium | rs81323962 | 3 | 74,840,151 | ETAA1 | 103,387 | − 0.567 | 8.52E−08 |
| Clostridium | rs81251808 | 4 | 60,750,431 | HNF4G | 113,341 | − 0.325 | 3.72E−09 |
| Collinsella | rs81430153 | 11 | 18,229,756 | PHF11 | 0 | 0.771 | 4.32E−07 |
| Coprococcus | rs81344777 | 3 | 4,420,359 | RBAK | 2425 | − 0.223 | 1.26E−07 |
| Desulfovibrio | rs331690240 | 23 | 14,234,646 | ENSSSCG00000050465 | 0 | 1.052 | 3.81E−14 |
| Dorea | rs81248474 | 6 | 8,320,740 | ENSSSCG00000042592 | 0 | − 0.199 | 1.34E−09 |
| Faecalibacterium | rs81288412 | 13 | 121,545,470 | ENSSSCG00000050978 | 3524 | 0.237 | 1.07E−17 |
| Fibrobacter | rs343652685 | 1 | 174,156,779 | ENSSSCG00000047027 | 156,250 | 1.092 | 3.99E−09 |
| Gemmiger | rs80980706 | 17 | 58,651,427 | VAPB | 6509 | 0.269 | 1.37E−08 |
| Lachnospira | rs329723588 | 15 | 137,647,629 | SCLY | 0 | − 0.545 | 1.84E−08 |
| Lactobacillus | rs81235044 | 4 | 121,603,127 | ENSSSCG00000046641 | 100,786 | − 0.743 | 2.77E−09 |
| Megasphaera | rs81371842 | 3 | 65,644,946 | ENSSSCG00000042171 | 273,629 | − 1.085 | 1.23E−08 |
| Mitsuokella | rs81358080 | 2 | 41,479,089 | ENSSSCG00000047282 | 5730 | − 1.094 | 3.68E−08 |
| Oscillospira | rs81344398 | 6 | 34,452,081 | ENSSSCG00000042964 | 722 | − 0.223 | 4.09E−10 |
| Parabacteroides | rs81242782 | 5 | 71,843,440 | LRRK2 | 0 | − 0.763 | 2.83E−06 |
| Peptococcus | rs81253718 | 2 | 43,124,329 | ENSSSCG00000045584 | 48,027 | − 0.907 | 1.44E−09 |
| Phascolarctobacterium | rs344728746 | 15 | 6,453,282 | ENSSSCG00000042287 | 110,243 | − 0.289 | 2.32E−09 |
| Prevotella | rs329143797 | 8 | 75,777,330 | TRIM2 | 0 | 0.178 | 1.56E−06 |
| RFN20 | rs81345563 | 2 | 44,851,717 | ENSSSCG00000033786 | 38,407 | − 0.571 | 7.63E−12 |
| Roseburia | rs81222575 | 8 | 19,050,584 | ENSSSCG00000041382 | 5304 | − 0.375 | 1.11E−08 |
| Ruminococcus | rs339681838 | 14 | 35,548,936 | FBXW8 | 0 | 0.133 | 1.26E−07 |
| Sphaerochaeta | rs81222575 | 8 | 19,050,584 | ENSSSCG00000041382 | 5304 | 0.776 | 3.58E−08 |
| Streptococcus | rs80787622 | 11 | 24,468,975 | TNFSF11 | 0 | 1.503 | 2.43E−12 |
| Succinivibrio | rs345030123 | 6 | 267,415 | FANCA | 0 | 0.967 | 3.82E−09 |
| Sutterella | rs322099448 | 8 | 74,295,221 | RBM46 | 16,663 | 0.826 | 4.34E−12 |
| Treponema | rs340048252 | 11 | 69,840,940 | NALCN | 0 | − 0.850 | 2.80E−10 |
| AlphaBACT | rs331027938 | 7 | 118,897,310 | ENSSSCG00000044157 | 12,891 | 0.141 | 1.10E−07 |
| Entamoeba | rs81323991 | 12 | 12,377,085 | ENSSSCG00000043117 | 7568 | 1.175 | 1.71E−08 |
| Hypotrichomonas | rs334181413 | 18 | 4,567,215 | ENSSSCG00000016426 | 0 | − 1.404 | 9.73E−09 |
| Trichomitus | rs81382396 | 4 | 10,874,687 | ENSSSCG00000047619 | 0 | 1.257 | 5.98E−07 |
| Tetratrichomonas | rs81280147 | 9 | 99,713,062 | CD36 | 0 | − 0.960 | 1.57E−08 |
| Neobalantidium | rs81323206 | 6 | 81,951,114 | IFNLR1 | 37,030 | − 1.948 | 7.05E−09 |
| Blastocystis | rs320737267 | 17 | 46,864,439 | TTPAL | 10,153 | 1.030 | 2.81E−12 |
| AlphaPROTO | rs80782515 | 17 | 28,489,220 | RALGAPA2 | 0 | 0.373 | 9.83E−11 |
The results of GWAS served as the basis for the AWM, the gene co-association network, and the regulatory impact factor approaches. In a first step, for all 39 phenotypes, estimated SNP effects were standardized by dividing them by the standard deviation of all SNP effects. After applying the analytical pipeline described in the “Methods” sections, the resulting AWM was comprised of 3561 SNP anchored to individual genes of which 121 were annotated as TF and 7 microRNA genes. In addition, there were 47 key regulators according to the RIF analyses including 3 microRNA genes. Notably, 10 of the 47 key regulators did not have a significant association (P < 0.05) with any of the phenotypes and their relevance would have been overlooked by GWAS alone. For the remaining 3551 genes, the number of associated phenotypes ranged from 1 to 13 and averaged 3.79. A total of 84.08% of the SNPs mapped within genes and the 15.92% were located upstream/downstream of annotated genes. Correlations between microbial traits were calculated using AWM columns (standardized SNP effects across bacteria and protists diversity and abundance) and were visualized as a hierarchical tree cluster, in which strong positive and negative correlations are displayed as proximity and distance, respectively (Fig. 1).
Fig. 1.
SNP co-association correlation: heatmap of the correlation matrix across 32 bacteria (prefix “B_”) and 7 protist (prefix “P_”) phenotypes based on the 3561 SNP-Genes included in the AWM
Gene co-association network linked to microbial phenotypes
Supplementary Fig. 1 shows an overview of the PCIT-inferred gene co-association network for 3561 SNP-Genes included in the AWM procedure, that connected by 738,913 edges of which 374,116 were positive and 364,797 were negative. In the network, node color indicates the phenotype with the strongest association (Supplementary Fig. 1). The network is characterized by a large central module where most of the bacteria vs. protists crosstalk takes place, surrounded by lots of smaller modules mostly phenotype specific. Reflecting the pleiotropic nature of the AWM, while the key phenotypes to capture SNP-Genes were the bacteria and protist alpha diversities, these only represent a minority of the genes: 241 for bacteria alpha diversity (dark blue nodes) and 231 for protist alpha diversity (bright red nodes). In contrast, other bacteria abundance was captured by 2568 genes (light blue nodes), while other protist abundance phenotypes were captured by 521 genes (orange nodes).
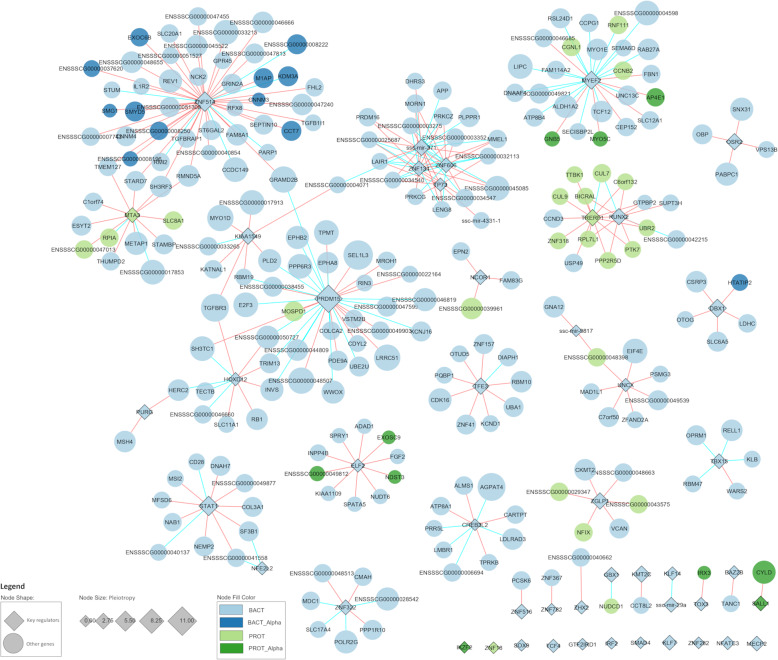
Key regulators in the network
The RIF analyses identified 47 key regulators among genes selected in the AWM procedure, and these are listed in Table 3, and Fig. 2 resumes the gene co-association sub-network comprising the 47 regulators identified by RIF. Prominent among the 47 key regulators were PRDM15, STAT1, ssc-mir-371, SOX9, and RUNX2 which gathered 942, 607, 588, 284, and 273 connections, respectively (Table 3). PRDM15 was associated with 10 traits and was the regulator showing the highest pleotropic value (Table 1). PRDM15 is a zinc-finger sequence-specific chromatin factor that modulates the transcription of upstream regulators of WNT and MAPK-ERK signaling to safeguard naive pluripotency and regulates the production of Th1-and Th2-type immune response [31]. Similarly, the microRNA ssc-mir-371 has been reported to play an important role in pluripotent regulation in pigs [32]. On the other hand, mice with depleted gut microbiome have been found to develop liver damage and bone loss through the mediation of SOX9 [33] and RUNX2 [34], respectively. The signal transducer STAT1 was associated with five microbial traits (Table 3). STAT1 has long been associated with immune processes and was recently identified as a potential regulator of vaccine response to porcine reproductive and respiratory syndrome [35]. It should be noted that between 32 and 22.5% of the predicted target genes had at least one TF-binding site for STAT1 and PRDM15, respectively (Supplementary Table 2). Regarding the microRNA ssc-mir-371, a total of 155 binding sites were identified (Supplementary Table 3), comprising a total of 71 different mRNA genes (12% of the initial 588 co-associated genes). The search for background random miRNA-binding sites in the reverse complemented 3′-UTRs gave a total of 115 different binding sites (Supplementary Table 3), comprising 48 different mRNA genes, i.e., the expected number of miRNA 7mer-m8 binding sites for ssc-miR-371 seed was increased by 1.48 folds compared with background random miRNA-mRNA interactions. When we assessed the putative structural consequences of the presence of the rs320008166 (n.59 T > C) mutated allele, a reduction in the minimum free energy (MFE) of the folding of the precursor miRNA hairpin was highlighted. More specifically, the presence of the alternative C allele at position 59th of the precursor region of the miRNA implied the stabilization of a G:U wobble pairing in the wild-type miRNA sequence, introducing a stable canonical Watson-Crick G:C pairing. While the miRNA hairpin carrying the T allele had a MFE = − 35.44 kcal/mol, the presence of the T allele implied an estimated MFE = − 37.74 kcal/mol. Overall, in agreement with AWM original publication [13], here we provide a promoter sequence in silico validation of some of the predicted TF-target genes and also for the first time predicted miRNA target genes.
Table 3.
List of 47 key regulators revealed by the regulatory impact factors (RIF) analyses and their RIF1 and RIF2 scores, standardized association to the bacteria (Alpha_B) and protist (Alpha_P) diversity, phenotype of strongest association, pleiotropy, and number of connections in the PCIT-inferred network
| Regulator | RIF1 | RIF2 | Alpha_B | Alpha_P | Top_Association | Pleio | Conn |
|---|---|---|---|---|---|---|---|
| PRDM15 | 0.648 | 2.806 | 0.239 | 0.277 | B_Treponema | 10 | 942 |
| HOXD12 | 1.342 | 2.536 | − 0.667 | − 0.624 | B_Sphaerochaeta | 1 | 910 |
| ZNF514 | 0.045 | 2.607 | − 2.938 | 0.310 | B_Sphaerochaeta | 5 | 889 |
| KIAA1549 | 1.270 | 2.715 | − 1.701 | − 1.465 | B_RFN20 | 3 | 812 |
| KLF7 | − 5.625 | 0.204 | − 0.603 | 1.249 | B_Bulleidia | 2 | 807 |
| CREB3L2 | − 2.411 | 0.511 | − 0.694 | 0.818 | B_Sphaerochaeta | 0 | 676 |
| TFE3 | − 0.127 | 2.144 | 1.045 | − 0.216 | B_Phascolarctobacterium | 3 | 627 |
| TBX15 | 2.506 | 1.414 | − 0.373 | − 1.229 | B_Anaerovibrio | 2 | 617 |
| STAT1 | − 1.464 | 2.285 | − 1.043 | 0.370 | B_Catenibacterium | 5 | 607 |
| PURG | 1.542 | 2.173 | − 1.320 | 0.338 | B_Sphaerochaeta | 2 | 595 |
| ssc-mir-371 | − 2.848 | 1.073 | 1.181 | − 0.690 | B_Roseburia | 3 | 588 |
| MTA3 | 0.390 | 2.097 | − 0.952 | − 0.107 | P_Hypotrichomonas | 2 | 556 |
| OSR2 | − 2.512 | − 0.194 | 0.270 | 0.005 | B_Lachnospira | 3 | 473 |
| DBX1 | − 2.047 | − 0.129 | − 2.207 | − 1.577 | B_Peptococcus | 4 | 468 |
| UNCX | − 3.085 | − 0.194 | − 0.051 | 1.258 | B_Desulfovibrio | 1 | 426 |
| MYEF2 | − 0.754 | − 2.657 | 1.215 | 2.430 | B_Fibrobacter | 4 | 391 |
| GBX1 | − 0.730 | − 2.439 | − 0.245 | − 1.460 | B_Campylobacter | 2 | 380 |
| ZNF134 | − 2.419 | 0.128 | − 0.572 | 1.686 | B_Lactobacillus | 2 | 370 |
| ZNF606 | − 2.419 | 0.128 | − 0.572 | 1.686 | B_Lactobacillus | 2 | 370 |
| SOX9 | − 2.524 | − 0.404 | − 0.498 | 0.096 | B_Anaerovibrio | 0 | 284 |
| KMT2C | − 0.311 | − 2.260 | − 0.628 | − 0.811 | B_Catenibacterium | 3 | 281 |
| RUNX2 | − 3.631 | − 1.878 | − 0.206 | − 1.776 | B_Collinsella | 3 | 273 |
| ELF2 | − 1.740 | − 2.300 | − 0.739 | 1.688 | B_Mitsuokella | 1 | 245 |
| ZNF322 | − 2.079 | − 0.792 | − 0.233 | 0.809 | B_Collinsella | 2 | 229 |
| IRF2 | − 0.764 | − 2.656 | − 0.697 | − 1.697 | B_Peptococcus | 0 | 209 |
| GTF2IRD1 | 2.117 | 0.660 | − 1.421 | 0.132 | B_Lachnospira | 0 | 206 |
| ZNF516 | 2.099 | 0.150 | 2.010 | 0.690 | B_Fibrobacter | 2 | 195 |
| NFE2L2 | − 2.077 | − 0.227 | 0.705 | − 0.051 | B_Coprococcus | 1 | 183 |
| NCOR1 | − 1.999 | − 0.455 | − 1.007 | 1.063 | B_Coprococcus | 2 | 165 |
| KLF14 | − 2.287 | − 2.255 | − 0.327 | − 1.073 | B_Sutterella | 1 | 161 |
| ZGLP1 | 2.029 | − 0.491 | 0.577 | 0.729 | B_Butyricicoccus | 3 | 161 |
| TCF4 | − 2.497 | − 1.382 | 0.280 | − 0.333 | B_Clostridium | 1 | 150 |
| TP73 | − 2.788 | − 0.275 | 0.633 | − 1.082 | B_Campylobacter | 1 | 146 |
| ZNF782 | − 2.831 | − 0.500 | 0.382 | − 1.811 | B_Desulfovibrio | 0 | 132 |
| SALL1 | − 1.133 | − 2.716 | − 0.833 | − 2.125 | P_AlphaPROTO | 2 | 131 |
| TOX3 | − 2.335 | − 2.043 | 0.365 | 1.641 | B_Lactobacillus | 1 | 129 |
| TRERF1 | − 2.359 | − 1.043 | − 0.286 | − 2.005 | P_Neobalantidium | 3 | 128 |
| MECP2 | − 2.059 | − 0.864 | 0.536 | − 0.591 | B_Dorea | 2 | 118 |
| SMAD4 | − 2.460 | − 2.206 | 0.431 | 1.586 | B_Catenibacterium | 0 | 118 |
| IKZF2 | − 1.694 | − 2.864 | − 0.578 | − 1.337 | P_AlphaPROTO | 0 | 117 |
| ssc-mir-9817 | − 1.841 | − 2.175 | 2.358 | 2.135 | B_RFN20 | 0 | 116 |
| ZHX2 | − 2.028 | − 1.124 | 0.266 | − 1.654 | B_Desulfovibrio | 1 | 109 |
| ZNF282 | − 1.657 | − 2.258 | 0.954 | 1.573 | B_Ruminococcus | 0 | 85 |
| BAZ2B | − 2.287 | − 0.786 | − 1.879 | 0.484 | B_Clostridium | 1 | 76 |
| ssc-mir-29a | − 2.067 | − 2.748 | − 0.568 | 0.811 | B_Lachnospira | 2 | 62 |
| NFATC3 | − 2.132 | − 2.525 | 0.270 | 0.442 | B_Desulfovibrio | 1 | 24 |
| ZNF18 | − 1.353 | − 2.134 | − 0.490 | − 0.929 | P_Neobalantidium | 0 | 18 |
Fig. 2.
Gene co-association sub-network: PCIT-inferred gene co-association sub-network comprising the 47 key regulators identified by RIF (diamonds) and their first neighbors (ellipses) presenting strong significant correlations (> 0.7). Node color is mapped to the phenotype of the strongest association: dark blue for bacteria alpha diversity (BACT_Alpha), light blue for other bacteria abundance phenotypes (BACT), dark green for protist alpha diversity (PROT_Alpha), and light green for other protist abundance phenotypes (PROT). Pink and cyan edges indicate positive and negative correlations, respectively, and node size indicates the amount of pleiotropy
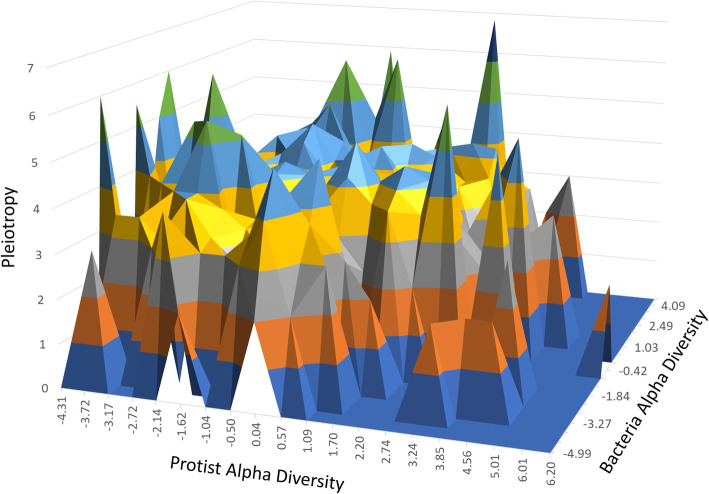
A closer inspection of values in Table 3 reveals some fascinating relationships. Pleiotropy (measured by the number of significantly associated phenotypes) and connectivity (number of first neighbors in the co-association network) are significantly correlated (r = 0.571; P value < 0.0001), suggesting that both metrics are indicators of the pluripotential capacity of the regulators. This finding is of relevance because while pleiotropy was computed from the number of significant phenotypes in the GWAS, the number of connections is a feature of the connectivity in a co-association network. Two vastly different concepts which, when pointing to the same outcome, underscore the relevance of, in this case, regulators. Similarly, while RIF1 and RIF2 scores are moderately correlated (r = 0.421; P value < 0.01), only RIF2 is significantly correlated with pleiotropy (r = 0.471; P value < 0.001) and more significantly with connectivity (r = 0.806; P value < 0.0001). This relationship, to our knowledge, never before documented, indicates the ability of RIF2 scores to prioritize regulators that, in our case study, have an uncanny ability to address the ‘bacteria vs. protists’ crosstalk by differentiating between genes associated with bacterial traits from those associated with protists phenotypes, i.e., the contrast used in developing RIF. To further explore ‘bacteria vs. protists’ crosstalk, Fig. 3 shows the 3-way relationship between a gene’s association with alpha diversity in bacteria, alpha diversity in protists, and its pleiotropy across the 3561 SNP-Genes included in the AWM. While alpha diversities indexes were the key phenotypes used to capture genes for the AWM, a further step aimed at identifying genes with pleiotropic potential (see “Methods”). The plateau of the surface revealed by Fig. 3 indicates that indeed SNP-Genes with near-zero non-significant association with alpha diversity phenotypes were still significantly associated across a large number of other microbial abundance phenotypes which reflect their potential pleiotropic effect.
Fig. 3.
Microbiome diversity and pleiotropy. Surface plot of the 3-way relationship between the SNP association with alpha diversity in protists (width), bacteria (depth) and its pleiotropy (height), measured as the number of significantly associated (adjusted P value < 0.05) protist and bacteria genera
Discussions
In this study, we propose a system biology approach to identify candidate genes, regulators, and biological pathways associated with 39 microbial traits. The cluster distribution based on the estimated additive value mirrored the distinct community composition types (enterotype clusters) as well as the known co-occurrence patterns of the pig gut microbiota [4]. For instance, pig gut enterotypes driver taxon Prevotella and Mitsuokella clustered together and distantly of a second cluster that includes Treponema and Ruminococcus (Fig. 1). We also noted that butyrate producer genera such as Faecalibacterium, Dorea, Blautia, Butyrococcus, and Coprococcus tend to cluster closely, which suggest a common directionality of the additive values, and perhaps a common genetic control for this groups of taxa. Notwithstanding the fact that targeting of 16S rRNA variable regions with short-read sequencing platforms cannot achieve the taxonomic resolution at the species level, our results add confidence and suggest the usefulness of the proposed analytical framework to recover key ecological properties of gut microbial ecosystem.
The gene co-association network (Supplementary Fig. 1) revealed the identity of predicted target genes and the higher complexity and polygenic nature of the diversity and composition of the pig gut microbial ecosystem. It is worth noting that literature mining confirms the association between the pig host-genome and the relative abundance of six bacterial genus reported by Crespo et al. 2019 (Supplementary Table 4). Furthermore, our findings also confirm association of 27 of the 68 genes recently reported by [36] as linked with the alpha diversity and the relative abundance of member of the swine gut microbiota (Supplementary Table 5). Remarkably, PRDM15 was among the genes commonly identify by [36]. As previously mentioned, PRDM15 was associated with the highest number of traits (Table 3) and the regulator showing the highest pleotropic value, despite differences between studies of genetic background, age, diets, and other environmental factors. Confirmed associations includes 11 of the 17 QTLs reported by Crespo et al. 2019 (Supplementary Table 4), as well as QTLs reported by [36] associated with members of Clostridium, Succinivibrio, Bacteroides, Prevotella, Blautia, Turicibacter, Treponema, Mobiluncus, and Oscillibacter genera. Therefore, our results suggest that in contrast to host genome-microbiota association performed in humans and mouse [37–39], several QTLs reported in swine can be replicated which open the possibility to identify genetic markers and candidate genes that can be incorporated in genetic breeding program to improve microbial traits.
We also noted that the list of target-genes contains a total of 200 candidate genes previously reported as linked to microbial traits in mouse or human studies (Supplementary Table 6). Among them, it is worth highlighting the following: (1) SLIT3, reported in the UK Twins Dataset [40] as associated with unclassified Clostridiaceae [41], linked to MetaCyc pathways involved in plant-derived steroid degradation, and whose expression is upregulated in colon crypts during the conventionalization of germ-free mice [42]; (2) SLC39A8, a gene with a pleiotropic missense variant related with Crohn’s disease and the composition of human gut microbiome [43]; and (3) NOS1, for which a pleiotropic association with body fatness and gut microbiota composition in mice had been shown [44]. We also identified other genes that had been shown to be related to β-diversity (CSMD1, ZFAT, FRMPD1, CLEC16A, IL1R2, BANK1, PRKAG2, LHFPL3, ST5, and NXN) and gut bacterial abundance (TOX3, NAPG, DLEC1, COL19A, DDM, and IFNAR1) in mice [45–47]. Genes reported by Allison et al. (2019) [48] as differentially expressed in primary human colonic epithelial cells (NEBL, ASAP3, ABLIM1, CUEDC2, PRRC2C, DENND1A, LAMC1, MAL2, ITGB1, CAST, A2ML1, IL7R, PCDH7, NFATC2IP, SORCS2, and DNM3) were also found in our study. Furthermore, genes linked to the functional profiles of the human gut ecosystem (SORCS2, LRRC32, and ARAP1) were among the predicted target genes in our network. As previously mentioned, SORCS2 was reported as linked to a plant-derived steroid degradation pathway. Meanwhile, genetic variants located in ARAP1 were linked to the bile acid metabolism, and LRRC32 was associated to the profile of ‘cell–cell signaling’ GO term [41]. It should be noted that many of these genes, including the key regulators, would have been missed by traditional single-trait GWAS which highly the usefulness of the proposed analytical pipeline to identify novels regulators and candidate genes linked with diversity and the composition of pig gut microbial ecosystem.
Host-genome markers associated with butyrate producing bacteria and piglets body weight
The identification of host-genetic markers linked with the relative abundance of butyrate producer bacteria such as Faecalibacterium, Dorea, Blautia, Butyrococcus, and Coprococcus (Table 2) prompted us to investigate whether these associations can be expanded in terms of overall pig’s wellbeing, and using the piglets’ body weight as the proxy for health and productivity. Butyrate is an important energy source for intestinal epithelial with anti-inflammatory potential that influences cell differentiation and strengthens the epithelial defense barrier [49, 50]. In fact, the beneficial effect of butyrate on swine growth and intestinal integrity have been documented [51, 52]. Therefore, we focused on the identification of SNPs with pleiotropic effect associated with the relative abundance of butyrate producer bacteria and host performance. After correcting for the systematic effects of sex (2 levels) and batch (7 levels), we found one significant SNP located at 96,004,482 bp of SSC2 (rs81361511) linked with the relative abundance of members of Butyricicoccus (P = 2.43E− 15) and piglets body weight (P = 0.026) (Table 4), as well as two SNPs associated with the relative abundance of Coprococcus (rs81344777, P = 1.26E− 07) and Faecalibacterium (rs81288412, P = 1.07E− 17) that were suggestively associated with piglets body weight. It is noteworthy to highlight that in these three cases, the allelic effects were in the same direction: the same allele affects the relative abundance of Butyricicoccus, Coprococcus, Faecalibacterium and piglets body weight. In agreement with [36], our findings suggest the existence of host-genetic variants jointly associated with the relative abundance of beneficial bacterial and host performance. However, larger studies including experimental validations and alternative source of information at host (additional phenotypic traits) and microbial (whole-metagenome, meta-transcriptomics) level are needed to fully characterize the role of the host genome-associated microbial communities in swine production performance, welfare, and health.
Table 4.
SNPs with pleiotropic effect associated with the relative abundance of butyrate producer bacteria and piglets body weight
| SNP | Associated microbial trait | Body weight (60 days) | |
|---|---|---|---|
| Effect | P value | ||
| rs81361511 | Butyricicoccus | 1.014 | 0.026 |
| rs81344777 | Coprococcus | 0.789 | 0.078 |
| rs81288412 | Faecalibacterium | − 0.787 | 0.082 |
Host-genome microbial interactions are partially modulated by the host immune system
The functional analysis from the list of regulators reveals overrepresentation of immune-related pathways. As many as 64% (16 out of 25) of the pathways reported as overrepresented by IPA relate to the host immune response (Table 5). Of note, this list includes pathways related with the bidirectional host-microbial crosstalk such as ERK/MAPK signaling [53], Th1 and Th2 activation pathway [54], TGF-β signaling [55], Wnt/β-catenin signaling [56], glucocorticoid receptor signaling [57], VDR/RXR activation [58], IL-22 signaling [59], and aryl hydrocarbon receptor [60].
Table 5.
List of pathways significantly enriched by the list of regulators
| Ingenuity canonical pathways | −log(P value) | Ratio | Genes |
|---|---|---|---|
| Osteoarthritis pathway | 3.13 | 0.019 | RUNX2,SMAD4,SOX9,TCF4 |
| TGF-β signaling | 3.07 | 0.031 | RUNX2,SMAD4,TFE3 |
| Role of osteoblasts, osteoclasts, and chondrocytes in rheumatoid arthritis | 3.07 | 0.018 | NFATC3,RUNX2,SMAD4,TCF4 |
| Glucocorticoid receptor signaling | 2.4 | 0.012 | NCOR1,NFATC3,SMAD4,STAT1 |
| VDR/RXR activation | 2 | 0.022 | NCOR1,RUNX2 |
| BMP signaling pathway | 1.93 | 0.024 | RUNX2,SMAD4 |
| Colorectal cancer metastasis signaling | 1.89 | 0.012 | SMAD4,STAT1,TCF4 |
| Regulation of IL-2 expression in activated and anergic T lymphocytes | 1.89 | 0.023 | NFATC3,SMAD4 |
| Senescencepathway | 1.79 | 0.011 | ELF2,NFATC3,SMAD4 |
| Mouse embryonic stem cell pluripotency | 1.77 | 0.019 | SMAD4,TCF4 |
| Pancreatic adenocarcinoma signaling | 1.72 | 0.018 | SMAD4,STAT1 |
| Neuroinflammation signaling pathway | 1.69 | 0.010 | NFATC3,NFE2L2,STAT1 |
| Th1 pathway | 1.64 | 0.016 | NFATC3,STAT1 |
| Human embryonic stem cell pluripotency | 1.55 | 0.015 | SMAD4,TCF4 |
| Aryl hydrocarbon receptor signaling | 1.5 | 0.014 | NFE2L2,TP73 |
| Factors promoting cardiogenesis in vertebrates | 1.47 | 0.013 | SMAD4,TCF4 |
| HOTAIR regulatory pathway | 1.41 | 0.012 | KMT2C,TCF4 |
| Protein kinase A signaling | 1.38 | 0.008 | NFATC3,SMAD4,TCF4 |
| Th1 and Th2 activation pathway | 1.36 | 0.013 | NFATC3,STAT1 |
| Wnt/β-catenin signaling | 1.35 | 0.012 | SOX9,TCF4 |
| T cell exhaustion signaling pathway | 1.34 | 0.011 | NFATC3,STAT1 |
| IL-22 signaling | 1.34 | 0.042 | STAT1 |
| Role of JAK family kinases in IL-6-type cytokine signaling | 1.32 | 0.040 | STAT1 |
| ERK/MAPK signaling | 1.3 | 0.010 | ELF2,STAT1 |
| Hepatic fibrosis/hepatic stellate cell activation | 1.3 | 0.011 | SMAD4,STAT1 |
The list of regulator includes other TFs-related with host immune system such as TFE3, NCOR1, SMAD4, NFE2L2, KLF7, NFATC3, TCF4, IRF2, and IKZF2. TFE3 cooperates with TFEB in the regulation of the innate immune response and macrophages activation [61], while NCOR1 plays an essential role controlling positive and negative selection of thymocytes during T cell development [62]. SMAD4 regulates IL-2 expression [63] and is essential for T cell proliferation [64]. Interestingly, according to String database [65], experimental data confirm the protein-by-protein interaction between SMAD4 and previously mentioned regulators RUNX2, SOX9, TEF3, and NCOR1. Finally, the list of regulators also includes TFs related to biological process like, inflammatory response (NFE2L2, KLF7) [66, 67], hematopoiesis (NFATC3 and IKZF2), cell-mediated immune response (IKZF2, IRF2, NCOR1, NFATC3, RUNX2, STAT1, TCF4), humoral immune response (IRF2, NFATC3, RUNX2, STAT1, TCF4), and the modulation of human B-cell differentiation (KLF14, KLF7, MTA3, STAT1) [68]. Therefore, in concordance with recent findings in human, mice, and pigs [5, 39, 69], our confirm that host-genome microbial interactions are mainly shaping by the host immune system.
Conclusions
In the present study, we built and explored a SNP-gene co-association network comprising 3561 genes related to the diversity and abundance of 31 bacterial and 6 commensal protist genera in pigs gut microbiota. Besides identifying genes associated with alpha diversity in both bacteria and protist, our analytical approach takes advantage of the genetic contribution to related microbial traits and revealed genetic variants with pleiotropic effects on pig gut microbiota profile. We also identify SNPs with pleiotropic effect associated with the relative abundance of butyrate producer bacteria (Faecalibacterium, Butyrococcus, and Coprococcus) and host performance. Placing emphasis on regulatory elements, a total of 47 regulators that enriched for immune-related pathways were identified. Among them, five regulators resulted prominent within the network: PRDM15, STAT1, ssc-mir-371, SOX9, and RUNX2. The list of predicted targets included 200 candidate genes previously reported as associated with microbiota profile in mice and human, such as SLIT3, SLC39A8, NOS1, IL1R2, DAB1, TOX3, SPP1, THSD7B, ELF2, PIANP, A2ML1, and IFNAR1. Taken together, our results highlight the value of the proposed analytical pipeline to exploit pleiotropy and the crosstalk between bacteria and protists as significant contributors to host-microbiome interactions.
Supplementary Information
Additional file 1: Supplementary Table 1. Microbial traits employed in the study.
Additional file 2: Supplementary Fig. 1. Overview of the PCIT-inferred gene co-association network for 3561 SNP-Genes included in the AWM procedure.
Additional file 3: Supplementary Table 2. List of predicted target genes with at least one TF binding site for STAT1 and PRDM15.
Additional file 4: Supplementary Table 3. List of predicted target genes with at least one TF binding site for microRNA ssc-mir-371.
Additional file 5: Supplementary Table 4. Overlappings QTLs confirmed in our study from the reported by Crespo et al., 2019.
Additional file 6: Supplementary Table 5. Overlapping QTLs confirmed in our study from the reported by Bergamaschi et al. 2020.
Additional file 7: Supplementary Table 6. List of candidate genes previously reported as linked to microbial traits in mouse or human studies.
Acknowledgements
The authors warmly thank all technical staff from Selección Batallé S.A for providing the animal material and their collaboration during the sampling.
Ethics approval and consent to participate.
Animal care and experimental procedures were carried out following national and institutional guidelines for the Good Experimental Practices and were approved by the IRTA Ethical Committee. Consent to participate is not applicable in this study.
Abbreviations
- QIIME
Quantitative insights into microbial ecology
- clr
Centered log ratio transformation
- GWAS
Genome-wide association studies
- AWM
Association weight matrix
- PCIT
Partial Correlation coefficient with Information Theory
- RIF
Regulatory impact factors
- TF
Transcription factors
- miRNA
MicroRNA
Authors’ contributions
AR, RQ, and YRC designed the study. MB carried out DNA extraction. MB, RQ, TD, and YRC performed the sampling. AR, EMS, and YRC analyzed the data. AR, MB, PA, EMS, RQ, and YRC interpreted the results and wrote the manuscript. All the authors read and approved the final version of the manuscript.
Funding
YRC was funded by Marie Skłodowska-Curie grant (P-Sphere) agreement No. 6655919 (EU). MB is recipient of a Ramon y Cajal post-doctoral fellowship (RYC-2013–12573) from the Spanish Ministry of Economy and Competitiveness. EMS was funded with a PhD fellowship FPU15/01733 awarded by the Spanish Ministry of Education and Culture. Part of the research presented in this publication was funded by Grants AGL2016–75432-R, AGL2017–88849-R awarded by the Spanish Ministry of Economy and Competitiveness. The authors belong to Consolidated Research Group TERRA (AGAUR, 2017 SGR 1719).
Availability of data and materials
The raw sequencing data employed in this article has been submitted to the NCBI’s sequence read archive (https://www.ncbi.nlm.nih.gov/sra); BioProject: PRJNA608629.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Antonio Reverter, Email: toni.rreverter-Gomez@csiro.au.
Maria Ballester, Email: maria.ballester@irta.cat.
Pamela A. Alexandre, Email: pamela.alexandre@csiro.au
Emilio Mármol-Sánchez, Email: emilio.marmol@cragenomica.es.
Antoni Dalmau, Email: antoni.dalmau@irta.cat.
Raquel Quintanilla, Email: raquel.quintanilla@irta.cat.
Yuliaxis Ramayo-Caldas, Email: yuliaxis.ramayo@irta.cat.
References
- 1.Richard ML, Sokol H. The gut mycobiota: insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol. 2019;16:331–345. doi: 10.1038/s41575-019-0121-2. [DOI] [PubMed] [Google Scholar]
- 2.Torp Austvoll C, Gallo V, Montag D. Health impact of the Anthropocene: the complex relationship between gut microbiota, epigenetics, and human health, using obesity as an example. Glob Health Epidemiol Genomics. 2020;5:e2. doi: 10.1017/gheg.2020.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X, Tsai T, Deng F, Wei X, Chai J, Knapp J, et al. Longitudinal investigation of the swine gut microbiome from birth to market reveals stage and growth performance associated bacteria. Microbiome. 2019;7:109. doi: 10.1186/s40168-019-0721-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramayo-Caldas Y, Mach N, Lepage P, Levenez F, Denis C, Lemonnier G, et al. Phylogenetic network analysis applied to pig gut microbiota identifies an ecosystem structure linked with growth traits. ISME J. 2016;10:2973–2977. doi: 10.1038/ismej.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramayo-Caldas Y, Prenafeta-Boldú F, Zingaretti LM, Gonzalez-Rodriguez O, Dalmau A, Quintanilla R, et al. Gut eukaryotic communities in pigs: diversity, composition and host genetics contribution. Anim Microbiome. 2020;2:18. doi: 10.1186/s42523-020-00038-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCormack UM, Curião T, Buzoianu SG, Prieto ML, Ryan T, Varley P, et al. Exploring a possible link between the intestinal microbiota and feed efficiency in pigs. Appl Environ Microbiol. 2017;83:e00380–e00317. doi: 10.1128/AEM.00380-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang H, Huang X, Fang S, Xin W, Huang L, Chen C. Uncovering the composition of microbial community structure and metagenomics among three gut locations in pigs with distinct fatness. Sci Rep. 2016;6:27427. doi: 10.1038/srep27427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He M, Fang S, Huang X, Zhao Y, Ke S, Yang H, et al. Evaluating the Contribution of Gut Microbiota to the Variation of Porcine Fatness with the Cecum and Fecal Samples. Front Microbiol. 2016;07 Available from: http://journal.frontiersin.org/article/10.3389/fmicb.2016.02108/full. [DOI] [PMC free article] [PubMed]
- 9.Chen C, Huang X, Fang S, Yang H, He M, Zhao Y, et al. Contribution of Host Genetics to the Variation of Microbial Composition of Cecum Lumen and Feces in Pigs. Front Microbiol. 2018;9 Available from: https://www.frontiersin.org/article/10.3389/fmicb.2018.02626/full. [DOI] [PMC free article] [PubMed]
- 10.Camarinha-Silva A, Maushammer M, Wellmann R, Vital M, Preuss S, Bennewitz J. Host Genome Influence on Gut Microbial Composition and Microbial Prediction of Complex Traits in Pigs. Genetics. 2017;206:1637–1644. doi: 10.1534/genetics.117.200782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crespo-Piazuelo D, Migura-Garcia L, Estellé J, Criado-Mesas L, Revilla M, Castelló A, et al. Association between the pig genome and its gut microbiota composition. Sci Rep. 2019;9:8791. doi: 10.1038/s41598-019-45066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aluthge ND, Van Sambeek DM, Carney-Hinkle EE, Li YS, Fernando SC, Burkey TE. BOARD INVITED REVIEW: The pig microbiota and the potential for harnessing the power of the microbiome to improve growth and health1. J Anim Sci. 2019;97:3741–3757. doi: 10.1093/jas/skz208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fortes MRS, Reverter A, Zhang Y, Collis E, Nagaraj SH, Jonsson NN, et al. Association weight matrix for the genetic dissection of puberty in beef cattle. Proc Natl Acad Sci. 2010;107:13642–13647. doi: 10.1073/pnas.1002044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reverter A, Fortes MRS. Association Weight Matrix: A Network-Based Approach Towards Functional Genome-Wide Association. Studies. 2013:437–47 Available from: http://link.springer.com/10.1007/978-1-62703-447-0_20. [DOI] [PubMed]
- 15.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A Tool for Genome-wide Complex Trait Analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolormaa S, Pryce JE, Reverter A, Zhang Y, Barendse W, Kemper K, et al. A Multi-Trait, Meta-analysis for Detecting Pleiotropic Polymorphisms for Stature, Fatness and Reproduction in Beef Cattle. PLoS Genet. 2014;10:e1004198. doi: 10.1371/journal.pgen.1004198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stephens M. False discovery rates: a new deal. Biostatistics. 2017;18:275–294. doi: 10.1093/biostatistics/kxw041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reverter A, Hudson NJ, Nagaraj SH, Pérez-Enciso M, Dalrymple BP. Regulatory impact factors: Unraveling the transcriptional regulation of complex traits from expression data. Bioinformatics. 2010;26:896–904. doi: 10.1093/bioinformatics/btq051. [DOI] [PubMed] [Google Scholar]
- 21.Muñoz M, Bozzi R, García-Casco J, Núñez Y, Ribani A, Franci O, et al. Genomic diversity, linkage disequilibrium and selection signatures in European local pig breeds assessed with a high density SNP chip. Sci Rep. 2019;9:13546. doi: 10.1038/s41598-019-49830-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reverter A, Chan EKF. Combining partial correlation and an information theory approach to the reversed engineering of gene co-expression networks. Bioinformatics. 2008;24:2491–2497. doi: 10.1093/bioinformatics/btn482. [DOI] [PubMed] [Google Scholar]
- 23.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scardoni G, Petterlini M, Laudanna C. Analyzing biological network parameters with CentiScaPe. Bioinformatics. 2009;25:2857–2859. doi: 10.1093/bioinformatics/btp517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yates AD, Achuthan P, Akanni W, Allen J, Allen J, Alvarez-Jarreta J, et al. Ensembl 2020. Nucleic Acids Res. 2019;48:D682–D688. doi: 10.1093/nar/gkz966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fornes O, Castro-Mondragon JA, Khan A, van der Lee R, Zhang X, Richmond PA, et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2019;48:D87–D92. doi: 10.1093/nar/gkz1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen W, Le S, Li Y, Hu F. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLoS One. 2016;11:e0163962. doi: 10.1371/journal.pone.0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lorenz R, Bernhart SH, Höner zu Siederdissen C, Tafer H, Flamm C, Stadler PF, et al. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mzoughi S, Zhang J, Hequet D, Teo SX, Fang H, Xing QR, et al. PRDM15 safeguards naive pluripotency by transcriptionally regulating WNT and MAPK–ERK signaling. Nat Genet. 2017;49:1354–1363. doi: 10.1038/ng.3922. [DOI] [PubMed] [Google Scholar]
- 32.Zhang W, Zhong L, Wang J, Han J. Distinct MicroRNA Expression Signatures of Porcine Induced Pluripotent Stem Cells under Mouse and Human ESC Culture Conditions. PLoS One. 2016;11:e0158655. doi: 10.1371/journal.pone.0158655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang F, Sun N, Li L, Zhu W, Xiu J, Shen Y, et al. Hepatic progenitor cell activation is induced by the depletion of the gut microbiome in mice. Microbiologyopen. 2019;8 Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/mbo3.873. [DOI] [PMC free article] [PubMed]
- 34.Tavakoli S, Xiao L. Depletion of Intestinal Microbiome Partially Rescues Bone Loss in Sickle Cell Disease Male Mice. Sci Rep. 2019;9:8659. doi: 10.1038/s41598-019-45270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Islam MA, Neuhoff C, Aqter Rony S, Große-Brinkhaus C, Uddin MJ, Hölker M, et al. PBMCs transcriptome profiles identified breed-specific transcriptome signatures for PRRSV vaccination in German Landrace and Pietrain pigs. PLoS One. 2019;14:e0222513. doi: 10.1371/journal.pone.0222513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bergamaschi M, Maltecca C, Schillebeeckx C, McNulty NP, Schwab C, Shull C, et al. Heritability and genome-wide association of swine gut microbiome features with growth and fatness parameters. Sci Rep. 2020;10:10134. doi: 10.1038/s41598-020-66791-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 38.Mikkelsen PB, Toubro S, Astrup A. Effect of fat-reduced diets on 24-h energy expenditure: comparisons between animal protein, vegetable protein, and carbohydrate. Am J Clin Nutr. 2000;72:1135–1141. doi: 10.1093/ajcn/72.5.1135. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Chen L, Zhao N, Xu X, Xu Y, Zhu B. Of genes and microbes: solving the intricacies in host genomes. Protein Cell. 2018;9:446–461. doi: 10.1007/s13238-018-0532-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, et al. Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host Microbe. 2016;19:731–743. doi: 10.1016/j.chom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bonder MJ, Kurilshikov A, Tigchelaar EF, Mujagic Z, Imhann F, Vila AV, et al. The effect of host genetics on the gut microbiome. Nat Genet. 2016;48:1407–1412. doi: 10.1038/ng.3663. [DOI] [PubMed] [Google Scholar]
- 42.Sommer F, Nookaew I, Sommer N, Fogelstrand P, Bäckhed F. Site-specific programming of the host epithelial transcriptome by the gut microbiota. Genome Biol. 2015;16:62. doi: 10.1186/s13059-015-0614-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li D, Achkar J-P, Haritunians T, Jacobs JP, Hui KY, D’Amato M, et al. A Pleiotropic Missense Variant in SLC39A8 Is Associated With Crohn’s Disease and Human Gut Microbiome Composition. Gastroenterology. 2016;151:724–732. doi: 10.1053/j.gastro.2016.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leamy LJ, Kelly SA, Nietfeldt J, Legge RM, Ma F, Hua K, et al. Host genetics and diet, but not immunoglobulin A expression, converge to shape compositional features of the gut microbiome in an advanced intercross population of mice. Genome Biol. 2014;15:552. doi: 10.1186/s13059-014-0552-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J, Thingholm LB, Skiecevičienė J, Rausch P, Kummen M, Hov JR, et al. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat Genet. 2016;48:1396–1406. doi: 10.1038/ng.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tschurtschenthaler M, Wang J, Fricke C, Fritz TMJ, Niederreiter L, Adolph TE, et al. Type I interferon signalling in the intestinal epithelium affects Paneth cells, microbial ecology and epithelial regeneration. Gut. 2014;63:1921–1931. doi: 10.1136/gutjnl-2013-305863. [DOI] [PubMed] [Google Scholar]
- 47.Wang J, Kalyan S, Steck N, Turner LM, Harr B, Künzel S, et al. Analysis of intestinal microbiota in hybrid house mice reveals evolutionary divergence in a vertebrate hologenome. Nat Commun. 2015;6:6440. doi: 10.1038/ncomms7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richards AL, Muehlbauer AL, Alazizi A, Burns MB, Findley A, Messina F, et al. Gut microbiota has a widespread and modifiable effect on host gene regulation. mSystems. 2019;4:e00323–e00318. doi: 10.1128/mSystems.00323-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer R-J. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2007;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 50.Guilloteau P, Martin L, Eeckhaut V, Ducatelle R, Zabielski R, Van Immerseel F. From the gut to the peripheral tissues: the multiple effects of butyrate. Nutr Res Rev 2010;23:366–384. Available from: https://www.cambridge.org/core/product/identifier/S0954422410000247/type/journal_article [DOI] [PubMed]
- 51.Le Gall M, Gallois M, Sève B, Louveau I, Holst JJ, Oswald IP, et al. Comparative effect of orally administered sodium butyrate before or after weaning on growth and several indices of gastrointestinal biology of piglets. Br J Nutr. 2009;102:1285–1296. doi: 10.1017/S0007114509990213. [DOI] [PubMed] [Google Scholar]
- 52.Kotunia A, Woliński J, Laubitz D, Jurkowska M, Romé V, Guilloteau P, et al. Effect of sodium butyrate on the small intestine development in neonatal piglets fed [correction of feed] by artificial sow. J Physiol Pharmacol. 2004;55(Suppl 2):59–68. [PubMed] [Google Scholar]
- 53.Soares-Silva M, Diniz FF, Gomes GN, Bahia D. The Mitogen-Activated Protein Kinase (MAPK) Pathway: Role in Immune Evasion by Trypanosomatids. Front Microbiol. 2016;7 Available from: http://journal.frontiersin.org/Article/10.3389/fmicb.2016.00183/abstract. [DOI] [PMC free article] [PubMed]
- 54.Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 55.Bauché D, Marie JC. Transforming growth factor β: a master regulator of the gut microbiota and immune cell interactions. Clin Transl Immunol. 2017;6:e136. doi: 10.1038/cti.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silva-García O, Valdez-Alarcón JJ, Baizabal-Aguirre VM. Wnt/β-Catenin Signaling as a Molecular Target by Pathogenic Bacteria. Front Immunol. 2019;10:2135. doi: 10.3389/fimmu.2019.02135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luo Y, Zeng B, Zeng L, Du X, Li B, Huo R, et al. Gut microbiota regulates mouse behaviors through glucocorticoid receptor pathway genes in the hippocampus. Transl Psychiatry. 2018;8:187. doi: 10.1038/s41398-018-0240-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bakke D, Chatterjee I, Agrawal A, Dai Y, Sun J. Regulation of Microbiota by Vitamin D Receptor: A Nuclear Weapon in Metabolic Diseases. Nucl Recept Res. 2018;5 Available from: http://www.kenzpub.com/journals/nurr/2018/101377/. [DOI] [PMC free article] [PubMed]
- 59.Fatkhullina AR, Peshkova IO, Dzutsev A, Aghayev T, McCulloch JA, Thovarai V, et al. An Interleukin-23-Interleukin-22 Axis Regulates Intestinal Microbial Homeostasis to Protect from Diet-Induced Atherosclerosis. Immunity. 2018;49:943–957.e9. doi: 10.1016/j.immuni.2018.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Korecka A, Dona A, Lahiri S, Tett AJ, Al-Asmakh M, Braniste V, et al. Bidirectional communication between the Aryl hydrocarbon Receptor (AhR) and the microbiome tunes host metabolism. npj Biofilms Microbiomes. 2016;2:16014. doi: 10.1038/npjbiofilms.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pastore N, Brady OA, Diab HI, Martina JA, Sun L, Huynh T, et al. TFEB and TFE3 cooperate in the regulation of the innate immune response in activated macrophages. Autophagy. 2016;12:1240–1258. doi: 10.1080/15548627.2016.1179405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Müller L, Hainberger D, Stolz V, Ellmeier W. NCOR1—a new player on the field of T cell development. J Leukoc Biol. 2018;104:1061–1068. doi: 10.1002/JLB.1RI0418-168R. [DOI] [PubMed] [Google Scholar]
- 63.Kim H-P, Kim B-G, Letterio J, Leonard WJ. Smad-dependent Cooperative Regulation of Interleukin 2 Receptor α Chain Gene Expression by T Cell Receptor and Transforming Growth Factor-β. J Biol Chem. 2005;280:34042–34047. doi: 10.1074/jbc.M505833200. [DOI] [PubMed] [Google Scholar]
- 64.Gu A-D, Zhang S, Wang Y, Xiong H, Curtis TA, Wan YY. A Critical Role for Transcription Factor Smad4 in T Cell Function that Is Independent of Transforming Growth Factor β Receptor Signaling. Immunity. 2015;42:68–79. doi: 10.1016/j.immuni.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Itoh K, Mochizuki M, Ishii Y, Ishii T, Shibata T, Kawamoto Y, et al. Transcription Factor Nrf2 Regulates Inflammation by Mediating the Effect of 15-Deoxy-Δ12,14-Prostaglandin J2. Mol Cell Biol. 2004;24:36–45. doi: 10.1128/MCB.24.1.36-45.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang M, Wang C, Wu J, Ha X, Deng Y, Zhang X, et al. The Effect and Mechanism of KLF7 in the TLR4/NF- κ B/IL-6 Inflammatory Signal Pathway of Adipocytes. Mediat Inflamm. 2018;2018:1–12. doi: 10.1155/2018/1756494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmidlin H, Diehl SA, Blom B. New insights into the regulation of human B-cell differentiation. Trends Immunol. 2009;30:277–285. doi: 10.1016/j.it.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khan AA, Yurkovetskiy L, O’Grady K, Pickard JM, de Pooter R, Antonopoulos DA, et al. Polymorphic Immune Mechanisms Regulate Commensal Repertoire. Cell Rep. 2019;29:541–550.e4. doi: 10.1016/j.celrep.2019.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Table 1. Microbial traits employed in the study.
Additional file 2: Supplementary Fig. 1. Overview of the PCIT-inferred gene co-association network for 3561 SNP-Genes included in the AWM procedure.
Additional file 3: Supplementary Table 2. List of predicted target genes with at least one TF binding site for STAT1 and PRDM15.
Additional file 4: Supplementary Table 3. List of predicted target genes with at least one TF binding site for microRNA ssc-mir-371.
Additional file 5: Supplementary Table 4. Overlappings QTLs confirmed in our study from the reported by Crespo et al., 2019.
Additional file 6: Supplementary Table 5. Overlapping QTLs confirmed in our study from the reported by Bergamaschi et al. 2020.
Additional file 7: Supplementary Table 6. List of candidate genes previously reported as linked to microbial traits in mouse or human studies.
Data Availability Statement
The raw sequencing data employed in this article has been submitted to the NCBI’s sequence read archive (https://www.ncbi.nlm.nih.gov/sra); BioProject: PRJNA608629.