Abstract
SARS-CoV-2 is a novel coronavirus, spread among humans, and to date, more than 100 million of laboratory-confirmed cases have been reported worldwide. The virus demonstrates 96% similarity to a coronavirus from a horseshoe bat and most probably emerged from a spill over from bats or wild animal(s) to humans. Currently, two variants are circulating in the UK and South Africa and spread to many countries around the world. The impact of mutations on virus replication, virulence and transmissibility should be monitored carefully. Current data suggest recurrent infection with SARS-CoV-2 correlated to the level of neutralising antibodies and with sustained memory responses following infection. Recently, remdesivir was FDA approved for treatment of COVID-19, however many potential antivirals are currently in different clinical trials. Clinical data and experimental studies indicated that licenced vaccines are helpful in controlling the disease. However, the current vaccines should be evaluated against the emerging variants of SARS-CoV-2.
Keywords: 2019-nCoV, Bats, Betacoronaviruses, Coronavirus disease, COVID-19, COVID-19 vaccine, COVID-19 antivirals, Interspecies transmission, Viral zoonosis
1. Human coronaviruses (HCoVs)
Human coronaviruses belong to the order Nidovirales, suborder Cornidovirineae, family Coronaviridae, and subfamily Orthocoronavirinae, the latter containing four genera designated alpha, beta, gamma and delta CoVs (ICTV, 2019). Human coronaviruses encompass viruses that affect both the upper and lower respiratory tract. Coronaviruses that lead to upper respiratory tract infections (URTIs) are responsible for 10%–30% of common cold infections in humans and include the following: HCoV-229E (Alphacoronavirus, subgenus Duvinacovirus), HCoV-NL63 (Alphacoronavirus, subgenus Setracovirus), HCoV-OC43, and HCoV–KHU1, with the latter two being related to Betacoronavirus subgenus Embecovirus. Currently, three coronaviruses induce severe lower respiratory tract infections (LRTIs): SARS-CoV and SARS-CoV-2 (Betacoronavirus subgenus, Sarbecovirus) as well as MERS-CoV (Betacoronavirus subgenus, Merbecovirus) (ICTV, 2019).
1.1. Virus structure and genome organization
The virus particle is spherical in shape and wrapped in a viral envelope obtained from the host cell Golgi apparatus and endoplasmic reticulum during budding of the progeny viruses. The coronavirus genome is a positive polarity, linear-stranded RNA that is 26–32 kb in length (Fehr and Perlman, 2015). The viral genome encodes structural, accessory and non-structural proteins. Like other coronaviruses, SARS-CoV-2 contains 6 major open-reading frames (ORFs)—ORF1a/b, S, E, M and N—in addition to other accessory ORFs 3, 6, 7a, 7b, 8, and 9b. It is organised as 5′-leader-UTR–ORF1ab-S-ORF3a-ORF4(E)-ORF5(M)-ORF6-ORF7a-ORF7b-ORF8-ORF9(N)-ORF10-3′UTR-poly (A) tail (Zhu et al., 2020c). The S protein is a club-like peplomer projection that mediates both virus entry (via binding with host cell receptors) and virus fusion with the host cell membrane. The M protein is the most abundant structural transmembrane protein, which is important for virus assembly and morphology. The E protein facilitates both virus assembly and release as well as possessing ion-channel activity. The N protein encapsidates the viral RNA genome, while the non-structural proteins (NSP 1–16)—encoded by the replicase gene that constitutes two-thirds of the viral genome (20 kb)—play diverse roles in virus replication and transcription (Fehr and Perlman, 2015).
1.2. SARS-CoV-2 origin and evolution
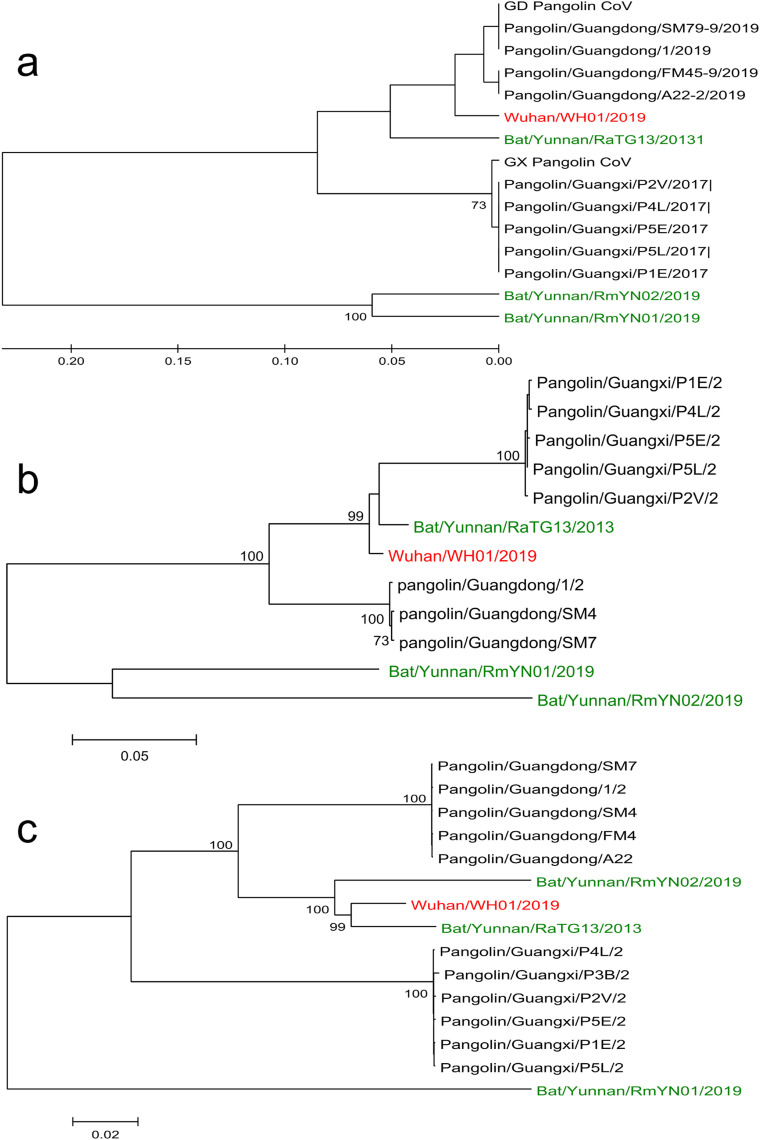
The original infection was traced to a seafood wholesale market in Wuhan city (Zhu et al., 2020c). A novel coronavirus that showed high similarity to a bat betacoronavirus called RaTG13 was identified from the diseased subjects and the virus was successfully isolated in both Vero and Huh7 cells (Zhou et al., 2020b; Zhu et al., 2020c). The receptor binding domain (RBD) of the S gene showed 93.1% similarity to RaTG13, but was divergent from SARS related-CoVs (75% identity) (Zhou et al., 2020b). These have three short insertions in the N-terminal domain and mutations in the receptor-binding motif (Zhou et al., 2020b) (Fig. 1 ). Similar to SARS-CoV and MERS-CoV viruses, bats are the most likely reservoir host for SARS-CoV-2. Interestingly, a recent virus dataset of Malayan pangolins (PRJNA573298 52) from two samples—SRR10168377 20 and SRR10168378 12 (Liu et al., 2019)—were found to share 97% amino acid identity to the RBD (Wong et al., 2020). This finding was augmented by the successful isolation of pangolin coronavirus from archival samples in the Vero-E6 cell line. The high similarity of multiple lineages of coronavirus from pangolins suggests they could be a possible intermediate host of SARS-CoV-2 (Lam et al., 2020; Xiao et al., 2020b). The RBD of the human SARS-CoV-2 and the closely-related bat virus is assumed to have been obtained by a recombination event from a pangolin virus (Fig. 2 ) involving the prior jumping of the virus to humans from a not-yet known species.
Fig. 1.
Deduced amino acids of the SARS-CoV-2 receptor binding domain in comparison to SARS-CoV-2 like bat and pangolin viruses. Amino acid residues that were found critical for ACE-2 binding are shaded in grey.
Fig. 2.
Phylogenetic tree of SARS-CoV-2 with SARS-CoV-2 similar viruses from bats and pangolins. a) Receptor binding domain b) Full-length spike protein (S) c) Full-length genome. The phylogenetic tree was constructed using MEGA 5.2. Maximum likelihood with 1000 bootstrap replications using Jones-Taylor-Thornton (JTT) model was used for amino acids and Tamura-Nei model with uniform rates for nucleotides. ML heuristic method with the Nearest-Neighbor-Interchange was used as tree interface. Bat viruses are in green colour, pangolin viruses in black colour while human SARS-CoV-2 virus is in red colour.
The S protein of SARS-CoV-2 can be proteolytically cleaved by TMPRSS2 and furin-like enzymes into S1 and S2 subunits (Hoffmann et al., 2020). S1 of the SARS-CoV-2 virus efficiently utilises human ACE2 receptors for virus entry while the S2 subunit mediates the fusion of the viral envelope with the cell membrane (Hoffmann et al., 2020). The affinity of SARS-CoV-2 to the ACE2 receptor was found to be higher than for SARS-CoV due to the presence of amino acid substitution in SARS-CoV-2, including L455, GVEG (482–485), F486, Q493, S494 and N501N. The same amino acid substitutions were found in CoV-pangolin/GD/2020, a pangolin coronavirus detected in Guangdong (Shang et al., 2020) (Fig. 1). This finding renders the CoV-pangolin/GD/2020 (or other pangolin coronaviruses with similar affinity to the ACE2 receptor) able to infect humans directly without further adaptation; therefore, pangolins could be a potential intermediate host for betacoronaviruses (Abdel-Moneim and Abdelwhab, 2020).
A furin polybasic cleavage site was identified in the SARS-CoV-2 spike, rendering it different from SARS-CoV (which possesses a monobasic S1/S2 cleavage site). Interestingly, a betacoronvirus (RmYN02) that showed lower homology to SARS-CoV-2 than detected in RaTG13 demonstrated a multiple polybasic amino acid insertion (Zhou et al., 2020a). Polybasic cleavage sites are probably acquired through adaptation in humans. However, this scenario assumes a sufficiently extended period between the first spill over of the virus from the animal host and the adaptation among humans that probably resulted in the acquisition of the polybasic cleavage site (Andersen et al., 2020). The presence of polybasic cleavage site was assumed to increase virus transmissibility and virulence and to expand its cell and tissue tropism.
1.3. Clades and lineages of the SARS-CoV-2
The Global Initiative on Sharing All Influenza Data (GISAID) database clustered SARS-CoV-2 viruses based on the full genome sequences into G, GH, GR, GV, L, O, S, and V clades. The L is the original clade from which S clade emerges due to ORF8 L84S amino acid substitution in addition to a point silent mutation C8782T. The V clade shows G251V amino acid substitution in the ORF3a with co-existence of L37F amino acid substitution in the NSP6. The G clade harbours spike D614G amino acid substitution, while the GR clade contains spike D614G and nucleocapsid G204R amino acid substitutions. Meanwhile, the GH clade contains spike D614G plus NS3-Q57H amino acid substitutions. The O clade contains viral sequences that do not cope with any of above-mentioned criteria (Table 1 ). Currently, G, GH, GV and GR are the major clades and account for more than 91% of all global SARS-CoV-2 sequenced genomes (GISAID sequence database).
Table 1.
GISAID clusters of the SARS-CoV-2 viruses based on the full genome sequences.
| GISAID Clade | Nucleotide changes | Amino acid substitutions |
|
|---|---|---|---|
| ORF | Substitution | ||
| L [Original strain] | C241/C3037/A23403/C8782/G11083/G26144/T28144 | ||
| S [from L clade] | C8782T/T28144C | NS8 | L84S |
| G | C241T/C3037T/A23403G | Spike | D614G |
| GH | C241T/C3037T/A23403G/G25563T | Spike | D614G |
| NS3 | Q57H | ||
| GR | C241T/C3037T/A23403G/G28882A | Spike | D614G |
| Nucleoprotein | G204R | ||
| GV | C241T/C3037T/A23403G/C22227T | Spike | D614G |
| A222V | |||
| V | G11083T/G26144T | NS3 | G251V |
| NSP6 | L37F | ||
| O | Others | ||
The GISAID clades are further divided into two major lineages: A and B. A numerical value of the descendants was given to sub-lineages with a maximum of three sublevels (ex. A.1.1.1). This nomenclature is known as the Phylogenetic Assignment of Named Global Outbreak Lineages (Pangolin), which is supported by a freely-available website (https://pangolin.cog-uk.io/) (Rambaut et al., 2020a).
Nextstrain [https://nextstrain.org/sars-cov-2/] open source tracking introduced a new nomenclature system, in which a novel clade is introduced when the frequency of circulation reaches approximately 20% for worldwide strains (Hadfield et al., 2018). Five clades were identified as being globally widespread: 19A (the root clade that spread in China then to the rest of Asia), 19B (the ancestor of the December 2019 Wuhan strain), and clades 20A, 20B and 20C. Clade 20A emerged due to D614G amino acid substitution in the spike glycoprotein during the European outbreak in February 2020. The latter spread globally during late February, from which clades 20B and 20C emerged in Europe and North America, respectively. All three clades (20A, 20B and 20C) belong to the GISAID G clade and harbour the D614G mutation in the spike (Korber et al., 2020). Two variants of concern emerged. The Nextstrain classification was modified to include labels correspond to an emerging variant and the prominent spike mutation and currently new clades 20D, 20E, 20F, 20G, 20H/501Y.V2, and 20I/501Y.V1 have been added (Bedford et al., 2021) as shown in Table 2 .
Table 2.
The origin and distribution of SARS-CoV-2 clades based on Nextsrain clustering.
| Clade | Root clade/ | Main characteristics | Region |
|---|---|---|---|
| 19A | The root clade that | China then to the rest of Asia | |
| 19B, | The ancestor Wuhan strain (December 2019) | Wuhan strain | |
| 20A | 19A | Spike: D614G | Europe |
| 20B | 20A | Spike: D614G | Europe |
| 20C | 20A | Spike: D614G | North America |
| 20D, | 20B | ORF1a: 1246I and ORF1a 3278S | South America, Southern Europe, and South Africa |
| 20E | 20A | Spike: 222V, N: 220V, ORF10: 30L, and ORF14: 67F | Europe |
| 20F | 20B | ORF1a: 300F and Spike: 477N | Australia |
| 20G | 20C | ORF1b: 1653D, ORF3a: 172V, N 67S and N 199L, | United States |
| 20H/501Y.V2 | 20C | Spike: 80A, 215G, 484K, 501Y, and 701V | South Africa |
| 20I/501Y.V1 | 20B | Spike (501Y, 570D, 681H) ORF8(27*) | United Kingdom |
The SARS-CoV-2, VUI 202012/01 variant under investigation (which has been detected in the UK) belongs to 20B, clade GR, lineage B.1.1.7. and will latterly be referred to as the Variant of Concern (VOC). It has spread to many countries around the world, in Africa (Gambia, Mayotte and Nigeria), Europe (to almost all countries), Asia (Bangaladesh, Hong Kong, India, Iran, Israel, Japan, Jordan, Malaysia, Oman, Pakistan, Singapore, South Korea, Thiland, United Arab Emirates, and Vietnam), Latin America (Argentina, Brazil, Ecuador, Peru, Trinidad and Tobago), North America (Canada Jamica, Mexico, St Lucia, USA) and Australasia (Australia and New Zealand). It possesses mutations in different viral proteins including the spike protein. In the spike protein, it harbours important mutations in the S1 (deletion 69–70, deletion 144, (RBD: N501Y), A570D, D614G), S1/S2 (cleavage site: P681H), and S2 (T716I, S982A, D1118) (Rambaut et al., 2020b). Another VOC also emerged in South Africa that possesses 80A, 215G, 484K, 501Y, and 701V amino acid residues in the spike protein. It spread to Africa (Botswana, Ghana, Kenya Mayotte, Mozambique) Asia, (Bangaladish, Israel, Japan, South Korea, and United Arab of Emirates), Australia (Asutralia and New Zealand), Europe (Austria, Belgium, Denmark, Filand, France, Germany, Irland, Italy, the Netherlands, Norway, Portugal, Spain, Sweden, Switzerland, Turkey, and United Kingdom), North and Central America (Canada, USA, Panama). These mutations developed unexpectedly, and the possible explanations for this are as follows: i) a prolonged infection among patients with reduced immunocompetence (Choi et al., 2020; McCarthy et al., 2020), ii) increased mutation rate as a consequence of immune pressure during reinfection, iii) adaptation of the virus in a susceptible animal then transferring infection back to humans, as was detected in mink in Denmark (deletion 69–70 and Y453F) (Laussauniere et al., 2020).
Some mutations can provide a selective advantage for the virus, including increased transmissibility, increased receptor binding affinity or providing the virus with the capacity to evade the host immune responsiveness or by simply altering neutralising epitopes. An earlier D614G variant increased the viral cellular infectivity but did not alter the clinical outcome of the disease (Volz et al., 2020). An attenuated SARS-CoV-2 phenotype was hypothesised based on 382 nucleotide deletions in SARS-CoV-2 Singapore strains (Su et al., 2020). Such deletions resulted in the removal of the transcription regulatory sequence (TRS) of the ORF8 with subsequent assumption of reduced fitness of virus replication (Muth et al., 2018). Interestingly, a similar deletion phenotype was detected in SARS-CoV at the end of the 2003 outbreak (Chinese-SARS-Molecular-Epidemiology-Consortium, 2004). In addition, an isolate with 81 nucleotide deletions with subsequent deletions of 27 amino acids in the ORF7a was also reported in Arizona, USA (Holland et al., 2020); however, such variants have since disappeared.
The D614G amino acid substitution was found in a B-cell epitope (Koyama et al., 2020). The majority of the SARS-CoV-2 strains with D614G mutation also demonstrated P4715L and P323L amino acid substitutions in ORF1ab and RdRp, respectively. Spike D614G mutation in G, GR, and GH clades is suggested to be linked to higher viral loads (Bhattacharyya et al., 2020; Korber et al., 2020), while rapidly-spreading strains were linked to the presence of the Nsp12 P322L (Pachetti et al., 2020). The GR clade is more prevalent among patients with severe disease manifestations (Korber et al., 2020). Conversely, Grubaugh et al. doubt the possibility of a clear role for the D614G mutation based on currently available data, and that extensive experimental and epidemiological studies should be conducted to determine the impact of D614G on the current epidemic (Grubaugh et al., 2020).
Currently, there is no information on the impact of VUI 202012/01 mutations on the virus binding to the target cells, its transmissibility, disease severity, and the possible impact on virus neutralisation by antibodies from vaccinated and/or infected patients.
It is assumed that N501Y increases the binding affinity to ACE2 (Starr et al., 2020), and it has been found to increase both virulence and infectivity in experimentally-infected mice (Gu et al., 2020). Although each of N501Y and P681H have been detected previously in SARS-CoV-2, it is the first time these have concurrently been present in the same virus. The VUI 202012/01 virus strain possesses the ORF8 Q27stop mutation that shortens the ORF8 protein with subsequent loss of function. However, ORF8 deletion was found to have only a slight effect on virus replication (Gamage et al., 2020).
It is well known that protection against coronaviruses depends on the development of neutralising antibodies against the antigenic epitopes of the spike protein. Phylogenetic analysis of the spike protein of different clades and lineages shows only minor variations with no similar clustering of different strains that belong to different clades. Many strains show different amino acid substitutions in the spike glycoprotein, including: L18F, A222V, N439K, G476S, S477N, T478I, V483A, E484Q, D614G and E780Q. Thus, the significance of such substitution needs to be investigated (Fig. 3 ).
Fig. 3.
Phylogentic tree of the full-length spike protein from different clades and lineages of the SARS-CoV-2. Maximum likelihood with 1000 bootstrap replications using Jones-Taylor-Thornton (JTT) model with uniform rates. ML heuristic method with the Nearest-Neighbor-Interchange was used as tree interface. Strains of the same clade are presented with the same colour. Amino acid substitutions except D614G were represented by grey highlight. Most of the sequences in the tree showed D614G substitution so not highlighted. Strains related to variant 1, that emerged in UK in 2020, were highlighted in cyan while strains related to variant 2, that emerged in South Africa in 2020, were highlighted in yellow.
1.4. Recombination as a potential tool of emergence for SARS-CoV-2 (and possibly others)
Coronaviruses are subjected to high-frequency recombination events. Such recombination was responsible for virus evolution as reported in SARS-CoV (Hon et al., 2008), MERS-CoV (Corman et al., 2014) and was also speculated to occur due to a recombination event of the RBD from Malayan pangolin coronavirus (Wong et al., 2020). Most SARSr-CoV viruses do not use ACE2; however, there is a possibility of acquiring such ability either by cumulative mutations or natural recombination with subsequent emergence of new SARSr-CoVs able to infect humans (Fan et al., 2019). Accordingly, experimental chimeric SARS coronaviruses were successfully synthesised. This was achieved using a bat-SCoV genome and with the SARS-CoV receptor binding domain (Becker et al., 2008) BtCoV HKU5 containing the SARS-CoV spike (S) glycoprotein (BtCoV HKU5-SE) (Agnihothram et al., 2014) in addition to a chimeric virus of murine adapted SARS-CoV backbone containing SHC014 spike bat coronavirus (Menachery et al., 2015). The latter was found to replicate efficiently without prior adaptation in both mice and human airway cultures. Such findings confirmed the possibility of natural recombination in the emergence of potential pathogens to humans and denoted the potential risk of constructing chimeric viruses based on natural circulating betacoronavirus strains since increased virulence to humans could not be excluded (Menachery et al., 2015).
2. Possible causes of SARS-CoV-2 reinfection
All patients exposed to SARS-CoV-2 possess detectable IgG+ RBD-specific plasma antibodies and neutralising plasma for ~3 months (Isho et al., 2020; Marklund et al., 2020; Wajnberg et al., 2020). Seroconversion begins after 7 days in 50% of symptomatic patients and after 14 days in all patients (Lan et al., 2020; Wölfel et al., 2020a, 2020b). However, some mildly infected patients do not show detectable IgG (Lou et al., 2020; Okba et al., 2020). This conflicting finding was explained by how low the sensitivity of the used technique was and the difference in the targeted antigen (Long et al., 2020; Marklund et al., 2020; Wajnberg et al., 2020; Xiao et al., 2020a). This was confirmed by the fact that all patients with undetectable IgG showed evidence of the presence of neutralising antibodies using neutralisation assay (Marklund et al., 2020).
The COVID-19 severity is correlated to the IgG antibody titers since antibody titers were found to be higher in severe cases in comparison to mild ones (Ma et al., 2020; Zhao et al., 2020). Mildly infected COVID-19 patients demonstrated specific immune memory cells that exhibited protective antiviral functions after 3 months of exposure (Rodda et al., 2020). IgM + memory B-cells dominated in the first 20 days followed by a gradual increase in IgG1+ memory B cells (Hartley et al., 2020). The latter cells express CD27 and positively correlate with the T follicular helper (Tfh) cell number that suggest high robustness (Hartley et al., 2020). In addition, memory B cells showed upregulation of CD80, CD180 and TACI that assumed activation upon re-exposure to the antigen (Berkowska et al., 2011, 2015). It was assumed that B cell memory is durable for up to 8 months, which would probably be protective upon reinfection. Accordingly, re-infection is proposed to be milder than the first exposure (Hartley et al., 2020). Increased neutralising antibodies, IgG+ classical memory B cells, Tfh, CD4+ memory T-cells, and IFN-γ CD8+ T cells were detected. The IgA+ against the receptor binding domain sharply declined after 3 months, which denotes that short-lived plasmablasts IgA (Rodda et al., 2020), SARS-CoV-2-specific memory B cells, and predominant IgG + B cells with lower frequencies of cells expressing IgM and IgA were detected (Dan et al., 2020; Hartley et al., 2020; Juno et al., 2020; Rodda et al., 2020). Based on such findings, memory B cells are expected to respond rapidly to SARS-CoV-2 re-exposure, thus generating neutralising antibodies that will provide protection (or at least guard against a severe form of the disease).
In-vitro stimulation of CD4+ memory T cells from SARS-CoV-2 recovered individuals resulted in a rapid expression of Th1- and Th17- cytokines and upregulation of both ICOS and CD40L on CXCR5+ cells (Rodda et al., 2020).
It has been found that the SARS-CoV-2 antibody level declines over time (Gudbjartsson et al., 2020; Ni et al., 2020), which might reflect a retrenchment of immune responsiveness. Meanwhile, mild COVID-19 induces an expanded population of memory B cells and CD4+ memory T cells. However, different levels of severity of COVID-19 could result in different levels of immune memory and subsequent immune protection (Rodda et al., 2020). Rare cases of SARS-CoV-2 reinfection were detected in different countries. It is known that protection correlates with homologous reinfection and antibody titers. To date, no major change in the spike protein was detected based on the GISAID sequence database. Thus, SARS-CoV-2 re-infection was suggested to be milder (Abu-Raddad et al., 2020; To et al., 2020). Three previously-exposed individuals with neutralising antibodies did not get sick when exposed to reinfection (Addetia et al., 2020). Moreover, in an overnight camp, a previous seropositive attendee was not infected, while non-previously infected persons tested positive (Pray et al., 2020). However, severe infections were also detected in some cases following re-infection (Selvaraj et al., 2020; Tillett et al., 2020; To et al., 2020). A severe form of the disease in re-infected cases is unclear. It could be due to a huge viral load in the second attack of infection (Guallar et al., 2020), infection by a more virulent strain, or the presence of poorly neutralising or non-neutralising antibodies being a progenitor for antibody-dependent enhancement (ADE) (Du et al., 2016; Karthik et al., 2020; Khandia et al., 2018; Wang et al., 2014). This phenomenon was documented in both SARS-CoV and MERS-CoV (Du et al., 2016; Wang et al., 2014), where SARS-CoV was found to be able to infect immune cells that lack the ACE-2 receptor through antibody dependent entry (Jaume et al., 2011; Wang et al., 2014). Further, ADE was detected in the presence of diluted anti-spike protein antibodies, but not with anti-nucleoprotein antibodies (Wang et al., 2014). Interestingly, mAbs targeting the SARS-CoV spike epitopes (other than RBD epitopes) can lead to ADE (Wang et al., 2016). Similarly, non-neutralising antibodies against MERS-CoV led to ADE experimentally in rabbits (Houser et al., 2017). Although ADE was recorded in related viruses of SARS-CoV-2, using an animal model for human infection does not accurately reflect immunopathogenesis in humans. To date, there is no evidence that ADE occurs or even induces a serious effect in COVID-19 patients. In addition, the pathogenesis of a model virus strain in animals does not fully reflect human infection because most viruses are highly species-specific (Arvin et al., 2020). Plasma therapy using a passive transfer of antibodies from the recovered patients was successfully used in the treatment of COVID-19 patients and proved to increase the survival rate and reduce the disease severity as reviewed in (Alghamdi and Abdel-Moneim, 2020), in which it was also reported with both MERS-CoV and SARS-CoV infections (Cheng et al., 2005; Ko et al., 2018; Mair-Jenkins et al., 2015).
3. Potential antivirals
3.1. FDA approved COVID-19 treatment
Remdesivir is an adenosine nucleoside analogue that inhibits viral replication (Wang et al., 2020) probably by binding to RdRp (Eweas et al., 2021; Wu et al., 2020), M and nsp14 (Eweas et al., 2021). It is of interest that M and S as well as M and N interactions are critical for the assembly of viral proteins (Siu et al., 2008). Remdesivir also possesses a high affinity to TMPRSS2 and ACE-2 (Eweas et al., 2021) with the subsequent effect of blocking cellular receptors necessary for viral entry in addition to inhibiting TMPRSS2-induced membrane fusion. Moreover, it is assumed to inhibit both SARS-CoV-2 Mpro and PLpro (Deshpande et al., 2020; Eweas et al., 2021). On 22nd October 2020, the FDA approved Veklury (remdesivir) as the first drug for COVID-19: it has been approved for the treatment of COVID-19 among hospitalised patients over the age of 12 years (≥40 kg). The FDA provided an Emergency Use Authorization (EUA) of Veklury for the treatment of hospitalised paediatric patients weighing ≥3.5 kg (FDA, 2020b). An initial dose of 200 mg followed by a daily dose of 100 mg for 4 days is the current recommended regimen for treatment of children (>12 years old and ≥40 kg). A single dose of 5 mg/kg in the first day, followed by daily dose of 2.5 mg/kg is the recommended regimen for treatment of children (≥3.5 kg to <40 kg). The drug is given by intravenous infusion for a period of between 30 and 120 min. This treatment could be extended for an additional 5 days if there is no improvement in the treated patients. Liver injury, nausea, rash, and allergic hypersensitive reactions are among the detected side effects of Veklury (FDA, 2020b). An NIH-sponsored clinical trial revealed that remdesivir shortened the time of recovery (Beigel et al., 2020), however, a WHO SOLIDARITY trial showed that remdesivir showed neither a significant reduction in the duration of hospitalization or mortality (Pan et al., 2021).
3.2. Other potential antiviral drugs
On 16th February, 2020, favipiravir (T-705), a viral RNA polymerase inhibitor, was approved for marketing in Zhejiang Province, China (National-Health-Commission-of-the-People's-Republic-of-China, 2020). To date, SARS-CoV-2 has been tested against five FDA-approved drugs: chloroquine phosphate (an old antimalarial drug), ribavirin, penciclovir, nitazoxanide and nafamostat, in addition to two viral RNA polymerase inhibitors: remdesivir (GS-5734) and favipiravir. Both remdesivir and chloroquine were found to be more effective than the other drugs following in vitro evaluation (Wang et al., 2020).
Ivermectin, an antiparasitic drug, was found to possess antiviral activity against dengue virus and was also found to possess potent in-vitro anti-viral activity against SARS-CoV-2 (Caly et al., 2020). It is assumed to play a role in preventing viral entry since it interacts with both the SARS-CoV-2 S protein and the human ACE-2 receptor (Eweas et al., 2021; Lehrer and Rheinstein, 2020) as well as TMPRSS2 (Eweas et al., 2021; Glowacka et al., 2011). It also binds efficiently to SARS-CoV-2 nsp14, N and M proteins with a potential role in alleviating the efficiency of virus replication and assembly (Eweas et al., 2021) and N nuclear import that is mediated by IMPα/β1 (Caly et al., 2020; Rowland et al., 2005; Tay et al., 2013; Timani et al., 2005; Wagstaff et al., 2012; Yang et al., 2020). It is likely that it binds to SARS-CoV-2 Mpro and PLpro with subsequent inhibition of post-translational cleavage of viral polyproteins (Eweas et al., 2021).
Chloroquine and hydroxychloroquine interfere with virus entry (by blocking the terminal glycosylation of ACE-2), and inhibit the post-entry mechanism through alkalinisation of the endosome pH (Vincent et al., 2005). They also inhibit the biosynthesis of sialic acid (Vincent et al., 2005). Growing evidence alleviates the potential use of hydroxychoroquine as an effective therapy in the treatment of COVID-19 patients. Multiple clinical trials have demonstrated that hydroxychoroquine is of no value when used in moderate doses as either a pre- or post-exposure prophylaxis (Barnabas et al., 2020; Boulware et al., 2020; Rajasingham et al., 2020). In addition, many retrospective observational studies deny its value in COVID-19 treatment (Geleris et al., 2020; Magagnoli et al., 2020; Rosenberg et al., 2020) and also in some prospective clinical trials (Cavalcanti et al., 2020; Horby et al., 2020). However, more than 100 clinical trials conducted on the efficacy of chloroquine and hydroxychloroquine are still ongoing (Chen et al., 2020); thus, it may be too early to determine conclusively the value of such drugs.
Camostat mesylate (a serine protease inhibitor) inhibits TMPRSS2 and may be a candidate drug for treating SARS-CoV-2 since it was proved to inhibit SARS-CoV in BALB/c mice (Zhou et al., 2015). In addition, bromhexine hydrochloride (a mucolytic cough suppressant) is an inhibitor of TMPRSS2 and could also be used for treatment, since it was found to be effective for both influenza viruses and coronaviruses (Lucas et al., 2014; Shen et al., 2017). More recently, a severe COVID-19 patient recovered following treatment with a soluble recombinant human ACE-2. After treatment, the virus disappeared from the patient and there was a significant decline of different cytokines (Zoufaly et al., 2020). EIDD-2801 is a ribonucleoside analogue N4-hydroxycytidine (NHC) that possesses potential antiviral activity against several viruses (Ehteshami et al., 2017; Urakova et al., 2018; Yoon et al., 2018) was found to possess a significant antiviral activity against SARS-CoV-2 (Cox et al., 2021). Interestingly, two-day administration of EIDD-2801 significantly reduced in vivo virus titres. In addition, it also succeeded to prevent against the disease development when used as a pre-exposure prophylactic therapy (Wahl et al., 2021).
4. Immunotherapy
4.1. Convalescent plasma therapy
Immunotherapy using convalescent plasma from recovered patients has demonstrated some success in treating SARS-CoV-2 infected patients earning EUA by the US FDA. In some of these studies, normalisation of body temperature and improvement in clinical and laboratory parameters were demonstrated after receiving convalescent plasma. The treatment was used together with other supportive antiviral therapies (Duan et al., 2020; Shen et al., 2020). Many studies successfully used COVID-19 convalescent plasma in different quantities and variable neutralisation titers. In one study, a convalescent plasma (400 ml, a neutralisation titer >40) was successful in restoring normal temperature and improving PaO2/FiO2 after 3 days post-treatment. It also helps to reduce vial load and inflammatory biomarkers (Shen et al., 2020). Another study reported an improvement of the clinical findings and laboratory biomarkers following treatment with 200 ml convalescent serum (neutralisation titer ≥640) (Duan et al., 2020). After receiving 500 ml of convalescent plasma, the health of critical COVID-19 patients improved post-treatment (Ahn et al., 2020). At least 200 ml of convalescent plasma infusion was required to improve the clinical findings and it was of great benefit in patients with late disease course (Ye et al., 2020). Convalescent plasma was also successful in reducing SARS-CoV-2 viral load in steroid treated patients (Ahn et al., 2020; Shen et al., 2020). More than 170 clinical trials on the effectiveness and safety of using plasma therapy in COVID-19 patients are currently underway. Early February 2021 the US FDA revised its EUA based on the review of the evidence and decided to restrict the use of convalescent plasma use in COVID-19 patients to the use of high titer COVID-19 convalescent plasma for the treatment of hospitalised patients with COVID-19, early in the disease course (FDA, 2020b).
4.2. Monoclonal antibody therapy
On 21st November 2020, the FDA provided an EUA for using a combination of casirivimab and imdevimab, which are monoclonal antibody therapies (MABs) against the SARS-CoV-2 S protein. It is administered by intravenous infusion for mild-to-moderate (but not severe) COVID-19 cases (12 years of age or ≥ 40 kg) and to patients at risk of developing severe COVID-19, including patients ≥65 years old or those suffering from chronic diseases. It was proven that the combined MAB therapy improved the clinical outcomes within 28 days post-treatment. It is not recommended for COVID-19 patients who are hospitalised and in need of high-flow oxygen or mechanical ventilation. Treatment among such patients might be accompanied by deterioration of the disease conditions for those patients (FDA, 2020a).
5. Vaccines
To date, five vaccines have been licenced, as follows: i) two RNA vaccines: BNT162b2 (Pfizer and BioNTech) and mRNA-1273 (Moderna); ii) two inactivated vaccines (SinoPharm and Bharat Biotech); and iii) a non-replicating adenovirus vector vaccine (AstraZenca). The BNT162b2 from Pfizer and BioNTech was the first authorised candidate vaccine (mRNA in lipid nanoparticles) with 95% efficacy (Polack et al., 2020), then the SinoPharm inactivated vaccine, followed by Moderna (mRNA-1273) that demonstrated 94.1% vaccine efficacy (Baden et al., 2020), and more recently the AstraZenca adenovirus vaccine that demonstrated 90% vaccine efficiency together with the COVAXIN from Bharat Biotech (the first indigenous vaccine). The released vaccines are given in a two-dose regimen with the maximum protection achieved after the booster dose.
In addition, dozens of candidate vaccines are in different preclinical and clinical phases. They include different types of vaccines including RNA vaccines, DNA vaccines, non-replicating viral vector vaccines, replicating viral vector vaccines, inactivated vaccines, live attenuated vaccines and subunit vaccines. To date, there are 56 in clinical and 172 in preclinical phases (VAC-LSTM, 2020).
The inactivated vaccines are propagated in the cell culture (Vero cells) and are then chemically inactivated by β-propiolactone and adjuvated with aluminium hydroxide (or other adjuvants). The non-replicating adenovirus vector vaccines include: i) ChAdOx1-S (Oxford and AstraZeneca) that uses the chimpanzee adenovirus as a backbone to express the spike protein of the SARS-CoV-2 (van Doremalen et al., 2020); ii) the CanSino Biological Inc. that uses human adenovirus 5 (Ad5) (Zhu et al., 2020a, 2020b); and iii) the non-replicating adenovirus recombinant vaccine containing both Ad26 and Ad5, which was developed by the Gamaleya Research Institute. The presence of the humoral antibody against human adenoviruses among humans may be a challenge against this type of vaccine, especially when two doses of the vaccine are used. However, this can be overcome using the initial dose from one vector and boosting with different one.
The COVID-19 mRNA vaccine is encapcidated into a lipid nanoparticle (LNP) (Mulligan et al., 2020; Walsh et al., 2020). The mRNA-1273 was developed by Moderna/NIAID (Jackson et al., 2020) and BNT162b1 and BNT162b2 were developed by BioNTech/Fosun Pharma/Pfizer and are among the potential RNA vaccines (Anderson et al., 2020). Another effective vaccine is the subunit vaccine containing recombinant purified spike protein of the SARS-CoV-2 co-formulated with Matrix-M1 adjuvant (Novavax) (VAC-LSTM, 2020). In terms of immunogenicity, adjuvanted, protein-based vaccines are followed by mRNA vaccines then ChAdOx1-based vaccines, and AdV5-based vaccines then the inactivated seem to rank the lowest. Post-vaccine side effects are highest for vectored vaccines and lowest for inactivated and protein-based vaccines and mRNA vaccines (Krammer, 2020).
A live attenuated vaccine is considered among the best for human viruses with many successful models including for poliovirus, measles, rubella, mumps, and yellow fever. The fact that it simulates natural infection without causing the disease causes it to induce mucosal, humoral and cell-mediated immune responsiveness against the virus. However, this type of vaccine requires a long time for development and to ensure vaccine efficacy and safety. Currently, three SARS-CoV-2 live attenuated vaccines (two from India and one from Turkey) currently exist. Two are in pre-clinical testing while Codagenix/Serum institute of India is in clinical phase I (VAC-LSTM, 2020).
6. Challenges
Due to the urgent need for a potent vaccine against COVID-19, an acceleration in advancing vaccine development exists. However, the adoption of novel human vaccines without previous licensed models may lead to uncertainties regarding long-term safety issues. Meanwhile, most vaccine trials have not been conducted on the most vulnerable subjects, which includes the elderly, pregnant women, and children. The role of different vaccines on the autoinflammatory responses such as paediatric inflammatory multisystem syndrome temporally associated with SARS-CoV-2 (PIMS-TS) has not been examined (Koirala et al., 2020).
Like ADE, vaccine-associated enhanced disease (VAED) has been detected in preclinical trials of SARS-CoV and MERS-CoV vaccines when associated with low neutralising antibodies (Graham, 2020; Haynes et al., 2020). However, this was not detected with the SARS-CoV-2 and mRNA-1273 vaccine (Baden et al., 2020), although, monitoring of the possible VAED risk needs to be carefully monitored. However, an important pending issue is the longevity of the neutralising antibodies and protective memory immune responsiveness. It is apt to mention that neutralising antibodies persisted for 3 months after the second booster dose of vaccine (Widge et al., 2020).
Challenge to the virus in the presence of vaccine pressure could help increasing the rate of virus mutations to evade neutralisation; therefore, continuous analysis of virus escape mutation and possible cross reaction are needed. Like other non-replicating vaccines, SARS-CoV-2 virus infection is assumed to replicate at the porta of entry of vaccinated subjects, especially in the absence of evidence of developing local immunity. This assumption is important for the possible role of vaccinated subjects transmitting infection to healthy ones.
The World Health Organization (WHO) estimates that 2 billion doses of an effective vaccine will be available by end of 2021; however, since the world population is 7.7 billion, there will not be enough vaccines to accommodate the world's entire population. A priority list of populations who are at highest risk of infection/spread and high fatality in need of vaccination has been developed by the WHO, and healthcare workers, the elderly population above 60 years of age and people younger than 60 years who have comorbidities have been given the highest priority in early vaccination campaigns.
7. Lessons to be learnt
Lessons from the SARS-CoV outbreak in 2002–2003 were not heeded, the most important being to never allow free will to challenge nature. Although, there is no report of foodborne transmission of SARS-CoV/SARS-CoV-2, transmission is assumed to be introduced by close animal-to-human contact or by the virus being inoculated through skin injuries that probably occur during butchering of such animals for food consumption. It is speculated that consumption of exotic live animals helped in the emergence of SARS-CoV in 2002 and SARS-CoV-2 in 2019. Unfortunately, in China, some African countries and possibly others, consumption of live animals including bats and wild animals is widely practiced, which may increase the possibility of the virus being transmitted to humans as previously reviewed (Fan et al., 2019). This requires a reorientation of food practices in such countries. The emergence of new zoonotic pathogens not only affects the persons involved in such practices but tend to spread very rapidly in local communities and internationally. The patterns of virus evolution and natural selection cannot be predicted, and the host immune responsiveness vary widely among viruses in the same genus. Whether the immune pressure in the absence of sufficient herd immunity will increase the mutation rate or evolution of immune-escape variants is not known. The successful and rapid development of mRNA based vaccines against SARS-CoV-2 will be helpful to control other viruses (e.g. Ebola, Dengue).
8. Conclusion and challenges
Although nobody can predict the future course of the SARS-CoV-2, nonetheless, some facts about this emerging virus are now clear. In the spirit of strong implementation of the One Health concept, the restriction and even the ban of both live wild animal markets and experimental induction of recombination studies on SARSr-CoV are highly recommended. Although remdesivir is the current FDA approved antiviral drug for treatment COVID-19, however, a doubt raised about its significance in reducing the duration of hospitalization or mortality rate. This finding, although requires more confirmation, highlights the need of developing potential antivirals against the SARS-CoV-2.
A second wave of the disease has now been recorded worldwide, with a tremendous increase in the number of COVID-19 patients over recent months, which reflects the long-term battle with COVID-19. Upregulation of both memory B cells and T follicular helper (Tfh) cells upon re-exposure to the SARS-CoV-2 were recorded. Accordingly, re-infection is proposed to be milder than the first exposure. Meanwhile, the sharp decline of IgA+ after 3 months might be responsible for the re-infection in previously infected patients and probably vaccinees. Such finding may necessitate the need for development SARS-CoV-2 vaccines that stimulate the mucosal immunity. The currently-released vaccines are assumed be helpful in controlling the disease since there are no major antigenic variations in the S protein. However, some of the current vaccines may not be as effective against some of the emerging variants of SARS-CoV-2. Like other coronaviruses, both immune and vaccination pressure mutations are expected. This fact necessitates a continuous monitoring of the virus evolution with possible emergence of new variants.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Abdel-Moneim A.S., Abdelwhab E.M. Evidence for SARS-CoV-2 infection of animal hosts. Pathogens. 2020;9:529. doi: 10.3390/pathogens9070529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Raddad L.J., Chemaitelly H., Ayoub H.H., Al Kanaani Z., Al Khal A., Al Kuwari E., Butt A.A., Coyle P., Jeremijenko A., Kaleeckal A.H. Assessment of the risk of SARS-CoV-2 reinfection in an intense re-exposure setting. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1846. ciaa1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addetia A., Crawford K.H.D., Dingens A., Zhu H., Roychoudhury P., Huang M.L., Jerome K.R., Bloom J.D., Greninger A.L. Neutralizing antibodies correlate with protection from SARS-CoV-2 in humans during a fishery vessel outbreak with a high attack rate. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.02107-20. e02107-02120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agnihothram S., Yount B.L., Jr., Donaldson E.F., Huynh J., Menachery V.D., Gralinski L.E., Graham R.L., Becker M.M., Tomar S., Scobey T.D., Osswald H.L., Whitmore A., Gopal R., Ghosh A.K., Mesecar A., Zambon M., Heise M., Denison M.R., Baric R.S. A mouse model for Betacoronavirus subgroup 2c using a bat coronavirus strain HKU5 variant. mBio. 2014;5 doi: 10.1128/mBio.00047-14. e00047-e00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J.Y., Sohn Y., Lee S.H., Cho Y., Hyun J.H., Baek Y.J., Jeong S.J., Kim J.H., Ku N.S., Yeom J.S., Roh J., Ahn M.Y., Chin B.S., Kim Y.S., Lee H., Yong D., Kim H.O., Kim S., Choi J.Y. Use of convalescent plasma therapy in two COVID-19 patients with acute respiratory distress syndrome in Korea. J. Kor. Med. Sci. 2020;35:e149. doi: 10.3346/jkms.2020.35.e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alghamdi A.N., Abdel-Moneim A.S. Convalescent plasma: a potential life-saving therapy for coronavirus disease 2019 (COVID-19) Front Publ. Health. 2020;8:437. doi: 10.3389/fpubh.2020.00437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A.B., Flach B., Lin B.C., Doria-Rose N.A., O'Dell S., Schmidt S.D., Corbett K.S., Swanson P.A., 2nd, Padilla M., Neuzil K.M., Bennett H., Leav B., Makowski M., Albert J., Cross K., Edara V.V., Floyd K., Suthar M.S., Martinez D.R., Baric R., Buchanan W., Luke C.J., Phadke V.K., Rostad C.A., Ledgerwood J.E., Graham B.S., Beigel J.H. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N. Engl. J. Med. 2020;383:2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvin A.M., Fink K., Schmid M.A., Cathcart A., Spreafico R., Havenar-Daughton C., Lanzavecchia A., Corti D., Virgin H.W. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature. 2020;584:353–363. doi: 10.1038/s41586-020-2538-8. [DOI] [PubMed] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., McGettigan J., Kehtan S., Segall N., Solis J., Brosz A., Fierro C., Schwartz H., Neuzil K., Corey L., Gilbert P., Janes H., Follmann D., Marovich M., Mascola J., Polakowski L., Ledgerwood J., Graham B.S., Bennett H., Pajon R., Knightly C., Leav B., Deng W., Zhou H., Han S., Ivarsson M., Miller J., Zaks T. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnabas R.V., Brown E., Bershteyn A., Miller R.S., Wener M., Celum C., Wald A., Chu H., Wesche D., Baeten J.M. Efficacy of hydroxychloroquine for post-exposure prophylaxis to prevent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection among adults exposed to coronavirus disease (COVID-19): a structured summary of a study protocol for a randomised controlled trial. Trials. 2020;21:475. doi: 10.1186/s13063-020-04446-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M.M., Graham R.L., Donaldson E.F., Rockx B., Sims A.C., Sheahan T., Pickles R.J., Corti D., Johnston R.E., Baric R.S., Denison M.R. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc. Natl. Acad. Sci. U.S.A. 2008;105:19944–19949. doi: 10.1073/pnas.0808116105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford T., Hodcroft E.B., Neher R.A. 2021. Updated Nextstain SARS-CoV-2 Clade Naming Strategy. [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M.D., Ruiz-Palacios G.M., Benfield T., Fätkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C. Remdesivir for the treatment of covid-19 - final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowska M.A., Driessen G.J., Bikos V., Grosserichter-Wagener C., Stamatopoulos K., Cerutti A., He B., Biermann K., Lange J.F., Van Der Burg M. Human memory B cells originate from three distinct germinal center-dependent and-independent maturation pathways. Blood. 2011;118:2150–2158. doi: 10.1182/blood-2011-04-345579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowska M.A., Schickel J.-N., Grosserichter-Wagener C., De Ridder D., Ng Y.S., Van Dongen J.J., Meffre E., Van Zelm M.C. Circulating human CD27− IgA+ memory B cells recognize bacteria with polyreactive Igs. J. Immunol. 2015;195:1417–1426. doi: 10.4049/jimmunol.1402708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya C., Das C., Ghosh A., Singh A.K., Mukherjee S., Majumder P.P., Basu A., Biswas N.K. Global spread of SARS-CoV-2 subtype with spike protein mutation D614G is shaped by human genomic variations that regulate expression of TMPRSS2 and MX1 genes. bioRxiv. 2020 doi: 10.1101/2020.1105.1104.075911. [DOI] [Google Scholar]
- Boulware D.R., Pullen M.F., Bangdiwala A.S., Pastick K.A., Lofgren S.M., Okafor E.C., Skipper C.P., Nascene A.A., Nicol M.R., Abassi M., Engen N.W., Cheng M.P., LaBar D., Lother S.A., MacKenzie L.J., Drobot G., Marten N., Zarychanski R., Kelly L.E., Schwartz I.S., McDonald E.G., Rajasingham R., Lee T.C., Hullsiek K.H. A randomized trial of hydroxychloroquine as postexposure prophylaxis for Covid-19. N. Engl. J. Med. 2020;383:517–525. doi: 10.1056/NEJMoa2016638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020:104787. doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalcanti A.B., Zampieri F.G., Rosa R.G., Azevedo L.C.P., Veiga V.C., Avezum A., Damiani L.P., Marcadenti A., Kawano-Dourado L., Lisboa T., Junqueira D.L.M., de Barros E.S.P.G.M., Tramujas L., Abreu-Silva E.O., Laranjeira L.N., Soares A.T., Echenique L.S., Pereira A.J., Freitas F.G.R., Gebara O.C.E., Dantas V.C.S., Furtado R.H.M., Milan E.P., Golin N.A., Cardoso F.F., Maia I.S., Hoffmann Filho C.R., Kormann A.P.M., Amazonas R.B., Bocchi de Oliveira M.F., Serpa-Neto A., Falavigna M., Lopes R.D., Machado F.R., Berwanger O. Hydroxychloroquine with or without azithromycin in mild-to-moderate covid-19. N. Engl. J. Med. 2020;383:e119. doi: 10.1056/NEJMoa2019014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Shen T., Zhong L., Liu Z., Dong X., Huang T., Wang Q., Xiao H. Research progress of chloroquine and hydroxychloroquine on the COVID-19 and their potential risks in clinic use. Front. Pharmacol. 2020;11:1167. doi: 10.3389/fphar.2020.01167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Wong R., Soo Y.O., Wong W.S., Lee C.K., Ng M.H., Chan P., Wong K.C., Leung C.B., Cheng G. Use of convalescent plasma therapy in SARS patients in Hong Kong. Eur. J. Clin. Microbiol. Infect. Dis. 2005;24:44–46. doi: 10.1007/s10096-004-1271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinese-Sars-Molecular-Epidemiology-Consortium Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- Choi B., Choudhary M.C., Regan J., Sparks J.A., Padera R.F., Qiu X., Solomon I.H., Kuo H.H., Boucau J., Bowman K., Adhikari U.D., Winkler M.L., Mueller A.A., Hsu T.Y., Desjardins M., Baden L.R., Chan B.T., Walker B.D., Lichterfeld M., Brigl M., Kwon D.S., Kanjilal S., Richardson E.T., Jonsson A.H., Alter G., Barczak A.K., Hanage W.P., Yu X.G., Gaiha G.D., Seaman M.S., Cernadas M., Li J.Z. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N. Engl. J. Med. 2020;383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Ithete N.L., Richards L.R., Schoeman M.C., Preiser W., Drosten C., Drexler J.F. Rooting the phylogenetic tree of middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J. Virol. 2014;88:11297–11303. doi: 10.1128/JVI.01498-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R.M., Wolf J.D., Plemper R.K. Therapeutically administered ribonucleoside analogue MK-4482/EIDD-2801 blocks SARS-CoV-2 transmission in ferrets. Nat. Microbiol. 2021;6:11–18. doi: 10.1038/s41564-020-00835-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J.M., Mateus J., Kato Y., Hastie K.M., Faliti C., Ramirez S.I., Frazier A., Esther D.Y., Grifoni A., Rawlings S.A. Immunological memory to SARS-CoV-2 assessed for greater than six months after infection. BioRxiv. 2020 doi: 10.1101/2020.1111.1115.383323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande R.R., Tiwari A.P., Nyayanit N., Modak M. In silico molecular docking analysis for repurposing therapeutics against multiple proteins from SARS-CoV-2. Eur. J. Pharmacol. 2020:173430. doi: 10.1016/j.ejphar.2020.173430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Tai W., Zhou Y., Jiang S. Vaccines for the prevention against the threat of MERS-CoV. Expert Rev. Vaccines. 2016;15:1123–1134. doi: 10.1586/14760584.2016.1167603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K., Liu B., Li C., Zhang H., Yu T., Qu J., Zhou M., Chen L., Meng S., Hu Y., Peng C., Yuan M., Huang J., Wang Z., Yu J., Gao X., Wang D., Yu X., Li L., Zhang J., Wu X., Li B., Xu Y., Chen W., Peng Y., Hu Y., Lin L., Liu X., Huang S., Zhou Z., Zhang L., Wang Y., Zhang Z., Deng K., Xia Z., Gong Q., Zhang W., Zheng X., Liu Y., Yang H., Zhou D., Yu D., Hou J., Shi Z., Chen S., Chen Z., Zhang X., Yang X. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. U. S. A. 2020;117:9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehteshami M., Tao S., Zandi K., Hsiao H.M., Jiang Y., Hammond E., Amblard F., Russell O.O., Merits A., Schinazi R.F. Characterization of β-d-N(4)-hydroxycytidine as a novel inhibitor of chikungunya virus. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.02395-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eweas A.F., Alhossary A.A., Abdel-Moneim A.S. Molecular docking reveals that ivermectin and remdesivir are potential repurposing drugs against the SARS-CoV-2. Front. Microbiol. 2021 doi: 10.3389/fmicb.2020.592908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Zhao K., Shi Z.L., Zhou P. 2019. Bat Coronaviruses in China. Viruses 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA . FDA; USA: 2020. Coronavirus (COVID-19) Update: FDA Authorizes Monoclonal Antibodies for Treatment of COVID-19. [Google Scholar]
- FDA . FDA; USA: 2020. Fact sheet for healthcare providers emergency use authorization (EUA) of veklury® (remdesivir) for hospitalized pediatric patients weighing 3.5 kg to less than 40 kgorhospitalized pediatric patients less than 12 years of age weighing at least 3.5 kg.https://www.fda.gov/media/137566/download [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamage A.M., Tan K.S., Chan W.O.Y., Liu J., Tan C.W., Ong Y.K., Thong M., Andiappan A.K., Anderson D.E., Wang Y., Wang L.F. Infection of human Nasal Epithelial Cells with SARS-CoV-2 and a 382-nt deletion isolate lacking ORF8 reveals similar viral kinetics and host transcriptional profiles. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1009130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geleris J., Sun Y., Platt J., Zucker J., Baldwin M., Hripcsak G., Labella A., Manson D.K., Kubin C., Barr R.G., Sobieszczyk M.E., Schluger N.W. Observational study of hydroxychloroquine in hospitalized patients with covid-19. N. Engl. J. Med. 2020;382:2411–2418. doi: 10.1056/NEJMoa2012410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka I., Bertram S., Müller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K., Niemeyer D., Schneider H., Drosten C., Pöhlmann S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham B.S. Rapid COVID-19 vaccine development. Science. 2020;368:945–946. doi: 10.1126/science.abb8923. [DOI] [PubMed] [Google Scholar]
- Grubaugh N.D., Hanage W.P., Rasmussen A.L. Making sense of mutation: what D614G means for the COVID-19 pandemic remains unclear. Cell. 2020;182:794–795. doi: 10.1016/j.cell.2020.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.Q., Wang Y., Teng Y., Zhao Z., Cui Y., Li Y., Li X.F., Li J., Zhang N.N., Yang X., Chen S., Guo Y., Zhao G., Wang X., Luo D.Y., Wang H., Yang X., Li Y., Han G., He Y., Zhou X., Geng S., Sheng X., Jiang S., Sun S., Qin C.F., Zhou Y. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guallar M.P., Meiriño R., Donat-Vargas C., Corral O., Jouvé N., Soriano V. Inoculum at the time of SARS-CoV-2 exposure and risk of disease severity. Int. J. Infect. Dis. 2020;97:290–292. doi: 10.1016/j.ijid.2020.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson D.F., Norddahl G.L., Melsted P., Gunnarsdottir K., Holm H., Eythorsson E., Arnthorsson A.O., Helgason D., Bjarnadottir K., Ingvarsson R.F., Thorsteinsdottir B., Kristjansdottir S., Birgisdottir K., Kristinsdottir A.M., Sigurdsson M.I., Arnadottir G.A., Ivarsdottir E.V., Andresdottir M., Jonsson F., Agustsdottir A.B., Berglund J., Eiriksdottir B., Fridriksdottir R., Gardarsdottir E.E., Gottfredsson M., Gretarsdottir O.S., Gudmundsdottir S., Gudmundsson K.R., Gunnarsdottir T.R., Gylfason A., Helgason A., Jensson B.O., Jonasdottir A., Jonsson H., Kristjansson T., Kristinsson K.G., Magnusdottir D.N., Magnusson O.T., Olafsdottir L.B., Rognvaldsson S., le Roux L., Sigmundsdottir G., Sigurdsson A., Sveinbjornsson G., Sveinsdottir K.E., Sveinsdottir M., Thorarensen E.A., Thorbjornsson B., Thordardottir M., Saemundsdottir J., Kristjansson S.H., Josefsdottir K.S., Masson G., Georgsson G., Kristjansson M., Moller A., Palsson R., Gudnason T., Thorsteinsdottir U., Jonsdottir I., Sulem P., Stefansson K. Humoral immune response to SARS-CoV-2 in Iceland. N. Engl. J. Med. 2020;383:1724–1734. doi: 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley G.E., Edwards E.S., Aui P.M., Varese N., Stojanovic S., McMahon J., Peleg A.Y., Boo I., Drummer H.E., Hogarth P.M. Rapid generation of durable B cell memory to SARS-CoV-2 spike and nucleocapsid proteins in COVID-19 and convalescence. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abf8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes B.F., Corey L., Fernandes P., Gilbert P.B., Hotez P.J., Rao S., Santos M.R., Schuitemaker H., Watson M., Arvin A. Prospects for a safe COVID-19 vaccine. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abe0948. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Krüger N., Müller M., Drosten C., Pöhlmann S. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. bioRxiv. 2020 2020.2001.2031.929042. [Google Scholar]
- Holland L.A., Kaelin E.A., Maqsood R., Estifanos B., Wu L.I., Varsani A., Halden R.U., Hogue B.G., Scotch M., Lim E.S. An 81 nucleotide deletion in SARS-CoV-2 ORF7a identified from sentinel surveillance in Arizona (Jan-Mar 2020) J. Virol. 2020 doi: 10.1128/JVI.00711-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon C.C., Lam T.Y., Shi Z.L., Drummond A.J., Yip C.W., Zeng F., Lam P.Y., Leung F.C. Evidence of the recombinant origin of a bat severe acute respiratory syndrome (SARS)-like coronavirus and its implications on the direct ancestor of SARS coronavirus. J. Virol. 2008;82:1819–1826. doi: 10.1128/JVI.01926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horby P., Mafham M., Linsell L., Bell J.L., Staplin N., Emberson J.R., Wiselka M., Ustianowski A., Elmahi E., Prudon B., Whitehouse T., Felton T., Williams J., Faccenda J., Underwood J., Baillie J.K., Chappell L.C., Faust S.N., Jaki T., Jeffery K., Lim W.S., Montgomery A., Rowan K., Tarning J., Watson J.A., White N.J., Juszczak E., Haynes R., Landray M.J. Effect of hydroxychloroquine in hospitalized patients with Covid-19. N. Engl. J. Med. 2020;383:2030–2040. doi: 10.1056/NEJMoa2022926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houser K.V., Broadbent A.J., Gretebeck L., Vogel L., Lamirande E.W., Sutton T., Bock K.W., Minai M., Orandle M., Moore I.N., Subbarao K. Enhanced inflammation in New Zealand white rabbits when MERS-CoV reinfection occurs in the absence of neutralizing antibody. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICTV Coronaviridae. 2019. https://talk.ictvonline.org/ictv-reports/ictv_9th_report/positive-sense-rna-viruses-2011/w/posrna_viruses/222/coronaviridae Available online at: (last accessed 26-01-2020)
- Isho B., Abe K.T., Zuo M., Jamal A.J., Rathod B., Wang J.H., Li Z., Chao G., Rojas O.L., Bang Y.M. Evidence for sustained mucosal and systemic antibody responses to SARS-CoV-2 antigens in COVID-19 patients. MedRxiv. 2020 doi: 10.1101/2020.1108.1101.20166553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A., Flach B., Doria-Rose N.A., Corbett K.S., Morabito K.M., O'Dell S., Schmidt S.D., Swanson P.A., 2nd, Padilla M., Mascola J.R., Neuzil K.M., Bennett H., Sun W., Peters E., Makowski M., Albert J., Cross K., Buchanan W., Pikaart-Tautges R., Ledgerwood J.E., Graham B.S., Beigel J.H. An mRNA vaccine against SARS-CoV-2 - preliminary report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaume M., Yip M.S., Cheung C.Y., Leung H.L., Li P.H., Kien F., Dutry I., Callendret B., Escriou N., Altmeyer R., Nal B., Daëron M., Bruzzone R., Peiris J.S. Anti-severe acute respiratory syndrome coronavirus spike antibodies trigger infection of human immune cells via a pH- and cysteine protease-independent FcγR pathway. J. Virol. 2011;85:10582–10597. doi: 10.1128/JVI.00671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juno J.A., Tan H.X., Lee W.S., Reynaldi A., Kelly H.G., Wragg K., Esterbauer R., Kent H.E., Batten C.J., Mordant F.L., Gherardin N.A., Pymm P., Dietrich M.H., Scott N.E., Tham W.H., Godfrey D.I., Subbarao K., Davenport M.P., Kent S.J., Wheatley A.K. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat. Med. 2020;26:1428–1434. doi: 10.1038/s41591-020-0995-0. [DOI] [PubMed] [Google Scholar]
- Karthik K., Senthilkumar T.M.A., Udhayavel S., Raj G.D. Role of antibody-dependent enhancement (ADE) in the virulence of SARS-CoV-2 and its mitigation strategies for the development of vaccines and immunotherapies to counter COVID-19. Hum. Vaccines Immunother. 2020:1–6. doi: 10.1080/21645515.2020.1796425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandia R., Munjal A., Dhama K., Karthik K., Tiwari R., Malik Y.S., Singh R.K., Chaicumpa W. Modulation of dengue/zika virus pathogenicity by antibody-dependent enhancement and strategies to protect against enhancement in zika virus infection. Front. Immunol. 2018;9:597. doi: 10.3389/fimmu.2018.00597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J.H., Seok H., Cho S.Y., Ha Y.E., Baek J.Y., Kim S.H., Kim Y.J., Park J.K., Chung C.R., Kang E.S., Cho D., Müller M.A., Drosten C., Kang C.I., Chung D.R., Song J.H., Peck K.R. Challenges of convalescent plasma infusion therapy in Middle East respiratory coronavirus infection: a single centre experience. Antivir. Ther. 2018;23:617–622. doi: 10.3851/IMP3243. [DOI] [PubMed] [Google Scholar]
- Koirala A., Joo Y.J., Khatami A., Chiu C., Britton P.N. Vaccines for COVID-19: the current state of play. Paediatr. Respir. Rev. 2020;35:43–49. doi: 10.1016/j.prrv.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. e819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Weeraratne D., Snowdon J.L., Parida L. Emergence of drift variants that may affect COVID-19 vaccine development and antibody treatment. Pathogens. 2020;9:324. doi: 10.3390/pathogens9050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- Lam T.T., Shum M.H., Zhu H.C., Tong Y.G., Ni X.B., Liao Y.S., Wei W., Cheung W.Y., Li W.J., Li L.F., Leung G.M., Holmes E.C., Hu Y.L., Guan Y. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Lan L., Xu D., Ye G., Xia C., Wang S., Li Y., Xu H. Positive RT-PCR test results in patients recovered from COVID-19. J. Am. Med. Assoc. 2020;323:1502–1503. doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laussauniere R., Fonager J., Rasmussen M., Frische A., Strandh C.P., Rasmussen T.B., Bøtner A., Fomsgaard A. Statens Serum Institut; Copenhagen: 2020. Preliminary Rapporton SARS-CoV-2 Spike Mutations Arising in Danish Mink, Their Spread to Humans and Neutralization data:SARS-CoV-2 Spike Mutations Arising in Danish Minkand Their Spread to Humans, 17 Dec. 2020. [Google Scholar]
- Lehrer S., Rheinstein P.H. vol. 34. 2020. Ivermectin docks to the SARS-CoV-2 spike receptor-binding domain attached to ACE2; pp. 3023–3026. (Vivo). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Chen W., Chen J.P. Viral metagenomics revealed Sendai virus and coronavirus infection of Malayan pangolins (Manis javanica) Viruses. 2019;11:979. doi: 10.3390/v11110979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K., Liao P., Qiu J.F., Lin Y., Cai X.F., Wang D.Q., Hu Y., Ren J.H., Tang N., Xu Y.Y., Yu L.H., Mo Z., Gong F., Zhang X.L., Tian W.G., Hu L., Zhang X.X., Xiang J.L., Du H.X., Liu H.W., Lang C.H., Luo X.H., Wu S.B., Cui X.P., Zhou Z., Zhu M.M., Wang J., Xue C.J., Li X.F., Wang L., Li Z.J., Wang K., Niu C.C., Yang Q.J., Tang X.J., Zhang Y., Liu X.M., Li J.J., Zhang D.C., Zhang F., Liu P., Yuan J., Li Q., Hu J.L., Chen J., Huang A.L. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Lou B., Li T.D., Zheng S.F., Su Y.Y., Li Z.Y., Liu W., Yu F., Ge S.X., Zou Q.D., Yuan Q., Lin S., Hong C.M., Yao X.Y., Zhang X.J., Wu D.H., Zhou G.L., Hou W.H., Li T.T., Zhang Y.L., Zhang S.Y., Fan J., Zhang J., Xia N.S., Chen Y. Serology characteristics of SARS-CoV-2 infection after exposure and post-symptom onset. Eur. Respir. J. 2020;56 doi: 10.1183/13993003.00763-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas J.M., Heinlein C., Kim T., Hernandez S.A., Malik M.S., True L.D., Morrissey C., Corey E., Montgomery B., Mostaghel E., Clegg N., Coleman I., Brown C.M., Schneider E.L., Craik C., Simon J.A., Bedalov A., Nelson P.S. The androgen-regulated protease TMPRSS2 activates a proteolytic cascade involving components of the tumor microenvironment and promotes prostate cancer metastasis. Canc. Discov. 2014;4:1310–1325. doi: 10.1158/2159-8290.CD-13-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Zeng W., He H., Zhao D., Jiang D., Zhou P., Cheng L., Li Y., Ma X., Jin T. Serum IgA, IgM, and IgG responses in COVID-19. Cell. Mol. Immunol. 2020;17:773–775. doi: 10.1038/s41423-020-0474-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magagnoli J., Narendran S., Pereira F., Cummings T.H., Hardin J.W., Sutton S.S., Ambati J. Outcomes of hydroxychloroquine usage in United States veterans hospitalized with COVID-19. Med (N Y) 2020;1:114–127. doi: 10.1016/j.medj.2020.06.001. e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair-Jenkins J., Saavedra-Campos M., Baillie J.K., Cleary P., Khaw F.M., Lim W.S., Makki S., Rooney K.D., Nguyen-Van-Tam J.S., Beck C.R. The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta-analysis. J. Infect. Dis. 2015;211:80–90. doi: 10.1093/infdis/jiu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marklund E., Leach S., Axelsson H., Nyström K., Norder H., Bemark M., Angeletti D., Lundgren A., Nilsson S., Andersson L.M., Yilmaz A., Lindh M., Liljeqvist J., Gisslén M. Serum-IgG responses to SARS-CoV-2 after mild and severe COVID-19 infection and analysis of IgG non-responders. PloS One. 2020;15 doi: 10.1371/journal.pone.0241104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy K.R., Rennick L.J., Nambulli S., Robinson-McCarthy L.R., Bain W.G., Haidar G., al e. Natural deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. bioRxiv. 2020 doi: 10.1126/science.abf6950. 2020.2011.2019.38991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Debbink K., Agnihothram S., Gralinski L.E., Plante J.A., Graham R.L., Scobey T., Ge X.Y., Donaldson E.F., Randell S.H., Lanzavecchia A., Marasco W.A., Shi Z.L., Baric R.S. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015;21:1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Raabe V., Bailey R., Swanson K.A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P.Y., Türeci Ö., Tompkins K.R., Walsh E.E., Frenck R., Falsey A.R., Dormitzer P.R., Gruber W.C., Şahin U., Jansen K.U. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586:589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- Muth D., Corman V.M., Roth H., Binger T., Dijkman R., Gottula L.T., Gloza-Rausch F., Balboni A., Battilani M., Rihtarič D. Attenuation of replication by a 29 nucleotide deletion in SARS-coronavirus acquired during the early stages of human-to-human transmission. Sci. Rep. 2018;8:1–11. doi: 10.1038/s41598-018-33487-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National-Health-Commission-of-the-People's-Republic-of-China . 2020. First Antiviral Drug Approved to Fight Coronavirus. [Google Scholar]
- Ni L., Ye F., Cheng M.L., Feng Y., Deng Y.Q., Zhao H., Wei P., Ge J., Gou M., Li X., Sun L., Cao T., Wang P., Zhou C., Zhang R., Liang P., Guo H., Wang X., Qin C.F., Chen F., Dong C. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52:971–977. doi: 10.1016/j.immuni.2020.04.023. e973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okba N.M.A., Müller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., de Bruin E., Chandler F.D., Yazdanpanah Y., Le Hingrat Q., Descamps D., Houhou-Fidouh N., Reusken C., Bosch B.J., Drosten C., Koopmans M.P.G., Haagmans B.L. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg. Infect. Dis. 2020;26:1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., Masciovecchio C., Angeletti S., Ciccozzi M., Gallo R.C. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020;18:1–9. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H., Peto R., Henao-Restrepo A.M., Preziosi M.P., Sathiyamoorthy V., Abdool Karim Q., Alejandria M.M., Hernández García C., Kieny M.P., Malekzadeh R., Murthy S., Reddy K.S., Roses Periago M., Abi Hanna P., Ader F., Al-Bader A.M., Alhasawi A., Allum E., Alotaibi A., Alvarez-Moreno C.A., Appadoo S., Asiri A., Aukrust P., Barratt-Due A., Bellani S., Branca M., Cappel-Porter H.B.C., Cerrato N., Chow T.S., Como N., Eustace J., García P.J., Godbole S., Gotuzzo E., Griskevicius L., Hamra R., Hassan M., Hassany M., Hutton D., Irmansyah I., Jancoriene L., Kirwan J., Kumar S., Lennon P., Lopardo G., Lydon P., Magrini N., Maguire T., Manevska S., Manuel O., McGinty S., Medina M.T., Mesa Rubio M.L., Miranda-Montoya M.C., Nel J., Nunes E.P., Perola M., Portolés A., Rasmin M.R., Raza A., Rees H., Reges P.P.S., Rogers C.A., Salami K., Salvadori M.I., Sinani N., Sterne J.A.C., Stevanovikj M., Tacconelli E., Tikkinen K.A.O., Trelle S., Zaid H., Røttingen J.A., Swaminathan S. Repurposed antiviral drugs for covid-19 - interim WHO solidarity trial results. N. Engl. J. Med. 2021;384:497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck R.W., Jr., Hammitt L.L., Türeci Ö., Nell H., Schaefer A., Ünal S., Tresnan D.B., Mather S., Dormitzer P.R., Şahin U., Jansen K.U., Gruber W.C. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pray I.W., Gibbons-Burgener S.N., Rosenberg A.Z., Cole D., Borenstein S., Bateman A., Pevzner E., Westergaard R.P. COVID-19 outbreak at an overnight summer school retreat - Wisconsin, july-august 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:1600–1604. doi: 10.15585/mmwr.mm6943a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasingham R., Bangdiwala A.S., Nicol M.R., Skipper C.P., Pastick K.A., Axelrod M.L., Pullen M.F., Nascene A.A., Williams D.A., Engen N.W., Okafor E.C., Rini B.I., Mayer I.A., McDonald E.G., Lee T.C., Li P., MacKenzie L.J., Balko J.M., Dunlop S.J., Hullsiek K.H., Boulware D.R., Lofgren S.M. Hydroxychloroquine as pre-exposure prophylaxis for COVID-19 in healthcare workers: a randomized trial. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1571. ciaa1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., O'Toole Á., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Loman N., Pybus O., Barclay W., Barrett J., Carabelli A., Connor T., Peacock T., Robertson D.L., Volz E. Consortium, C.-g.U. 2020. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. [Google Scholar]
- Rodda L.B., Netland J., Shehata L., Pruner K.B., Morawski P.A., Thouvenel C.D., Takehara K.K., Eggenberger J., Hemann E.A., Waterman H.R., Fahning M.L., Chen Y., Hale M., Rathe J., Stokes C., Wrenn S., Fiala B., Carter L., Hamerman J.A., King N.P., Gale M., Jr., Campbell D.J., Rawlings D.J., Pepper M. Functional SARS-CoV-2-specific immune memory persists after mild COVID-19. Cell. 2020;S0092–8674:31565–31568. doi: 10.1016/j.cell.2020.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E.S., Dufort E.M., Udo T., Wilberschied L.A., Kumar J., Tesoriero J., Weinberg P., Kirkwood J., Muse A., DeHovitz J., Blog D.S., Hutton B., Holtgrave D.R., Zucker H.A. Association of treatment with hydroxychloroquine or azithromycin with in-hospital mortality in patients with COVID-19 in New York state. Jama. 2020;323:2493–2502. doi: 10.1001/jama.2020.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland R.R., Chauhan V., Fang Y., Pekosz A., Kerrigan M., Burton M.D. Intracellular localization of the severe acute respiratory syndrome coronavirus nucleocapsid protein: absence of nucleolar accumulation during infection and after expression as a recombinant protein in vero cells. J. Virol. 2005;79:11507–11512. doi: 10.1128/JVI.79.17.11507-11512.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj V., Herman K., Dapaah-Afriyie K. Severe, symptomatic reinfection in a patient with COVID-19. R. I. Med. J. 2020;103(2013):24–26. [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., Wang F., Li D., Yang M., Xing L., Wei J., Xiao H., Yang Y., Qu J., Qing L., Chen L., Xu Z., Peng L., Li Y., Zheng H., Chen F., Huang K., Jiang Y., Liu D., Zhang Z., Liu Y., Liu L. Treatment of 5 critically Ill patients with COVID-19 with convalescent plasma. Jama. 2020;323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L.W., Mao H.J., Wu Y.L., Tanaka Y., Zhang W. TMPRSS2: a potential target for treatment of influenza virus and coronavirus infections. Biochimie. 2017;142:1–10. doi: 10.1016/j.biochi.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu Y., Teoh K., Lo J., Chan C., Kien F., Escriou N., Tsao S., Nicholls J., Altmeyer R., Peiris J. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 2008;82:11318–11330. doi: 10.1128/JVI.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., King N.P., Veesler D., Bloom J.D. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310. doi: 10.1016/j.cell.2020.08.012. e1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y.C.F., Anderson D.E., Young B.E., Zhu F., Linster M., Kalimuddin S., Low J.G.H., Yan Z., Jayakumar J., Sun L., Yan G.Z., Mendenhall I.H., Leo Y.-S., Lye D.C., Wang L.-F., Smith G.J.D. 2020. Discovery and Genomic Characterization of a 382-nucleotide Deletion in ORF7b and ORF8 during the Early Evolution of SARS-CoV-2 mBio 11. e01610-01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay M., Fraser J.E., Chan W., Moreland N.J., Rathore A.P., Wang C., Vasudevan S.G., Jans D.A. Nuclear localization of dengue virus (DENV) 1–4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin. Antivir. Res. 2013;99:301–306. doi: 10.1016/j.antiviral.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Tillett R.L., Sevinsky J.R., Hartley P.D., Kerwin H., Crawford N., Gorzalski A., Laverdure C., Verma S.C., Rossetto C.C., Jackson D., Farrell M.J., Van Hooser S., Pandori M. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect. Dis. 2020;21:52–58. doi: 10.1016/S1473-3099(20)30764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timani K.A., Liao Q., Ye L., Zeng Y., Liu J., Zheng Y., Ye L., Yang X., Lingbao K., Gao J. Nuclear/nucleolar localization properties of C-terminal nucleocapsid protein of SARS coronavirus. Virus Res. 2005;114:23–34. doi: 10.1016/j.virusres.2005.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K., Hung I.F., Ip J.D., Chu A.W., Chan W.M., Tam A.R., Fong C.H., Yuan S., Tsoi H.W., Ng A.C., Lee L.L., Wan P., Tso E., To W.K., Tsang D., Chan K.H., Huang J.D., Kok K.H., Cheng V.C., Yuen K.Y. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1275. ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urakova N., Kuznetsova V., Crossman D.K., Sokratian A., Guthrie D.B., Kolykhalov A.A., Lockwood M.A., Natchus M.G., Crowley M.R., Painter G.R., Frolova E.I., Frolov I. β-d-N (4)-hydroxycytidine is a potent anti-alphavirus compound that induces a high level of mutations in the viral genome. J. Virol. 2018;92 doi: 10.1128/JVI.01965-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAC-LSTM . Vaccine Centre at the London School of Hygiene & Tropical Medicine; 2020. COVID-19 Vaccine Development Pipeline. [Google Scholar]
- van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R., Avanzato V., Bushmaker T., Flaxman A., Ulaszewska M., Feldmann F., Allen E.R., Sharpe H., Schulz J., Holbrook M., Okumura A., Meade-White K., Pérez-Pérez L., Bissett C., Gilbride C., Williamson B.N., Rosenke R., Long D., Ishwarbhai A., Kailath R., Rose L., Morris S., Powers C., Lovaglio J., Hanley P.W., Scott D., Saturday G., de Wit E., Gilbert S.C., Munster V.J. ChAdOx1 nCoV-19 vaccination prevents SARS-CoV-2 pneumonia in rhesus macaques. bioRxiv. 2020 doi: 10.1101/2020.1105.1113.093195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent M.J., Bergeron E., Benjannet S., Erickson B.R., Rollin P.E., Ksiazek T.G., Seidah N.G., Nichol S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005;2:69. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E., Hill V., McCrone J.T., Price A., Jorgensen D., O'Toole Á., Southgate J., Johnson R., Jackson B., Nascimento F.F., Rey S.M., Nicholls S.M., Colquhoun R.M., da Silva Filipe A., Shepherd J., Pascall D.J., Shah R., Jesudason N., Li K., Jarrett R., Pacchiarini N., Bull M., Geidelberg L., Siveroni I., Goodfellow I., Loman N.J., Pybus O.G., Robertson D.L., Thomson E.C., Rambaut A., Connor T.R. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2020;S0092–8674:31537–31543. doi: 10.1016/j.cell.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff K.M., Sivakumaran H., Heaton S.M., Harrich D., Jans D.A. Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 2012;443:851–856. doi: 10.1042/BJ20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl A., Gralinski L.E., Johnson C.E., Yao W., Kovarova M., Dinnon K.H., 3rd, Liu H., Madden V.J., Krzystek H.M., De C., White K.K., Gully K., Schäfer A., Zaman T., Leist S.R., Grant P.O., Bluemling G.R., Kolykhalov A.A., Natchus M.G., Askin F.B., Painter G., Browne E.P., Jones C.D., Pickles R.J., Baric R.S., Garcia J.V. SARS-CoV-2 infection is effectively treated and prevented by EIDD-2801. Nature. 2021 doi: 10.1038/s41586-021-03312-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajnberg A., Amanat F., Firpo A., Altman D.R., Bailey M.J., Mansour M., McMahon M., Meade P., Mendu D.R., Muellers K., Stadlbauer D., Stone K., Strohmeier S., Simon V., Aberg J., Reich D.L., Krammer F., Cordon-Cardo C. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science. 2020;370:1227–1230. doi: 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh E.E., Frenck R.W., Jr., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R., Swanson K.A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P.Y., Türeci Ö., Tompkins K.R., Lyke K.E., Raabe V., Dormitzer P.R., Jansen K.U., Şahin U., Gruber W.C. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang L., Kuwahara K., Li L., Liu Z., Li T., Zhu H., Liu J., Xu Y., Xie J., Morioka H., Sakaguchi N., Qin C., Liu G. Immunodominant SARS coronavirus epitopes in humans elicited both enhancing and neutralizing effects on infection in non-human primates. ACS Infect. Dis. 2016;2:361–376. doi: 10.1021/acsinfecdis.6b00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.F., Tseng S.P., Yen C.H., Yang J.Y., Tsao C.H., Shen C.W., Chen K.H., Liu F.T., Liu W.T., Chen Y.M., Huang J.C. Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem. Biophys. Res. Commun. 2014;451:208–214. doi: 10.1016/j.bbrc.2014.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widge A.T., Rouphael N.G., Jackson L.A., Anderson E.J., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A.B., Flach B., Lin B.C., Doria-Rose N.A., O'Dell S., Schmidt S.D., Neuzil K.M., Bennett H., Leav B., Makowski M., Albert J., Cross K., Edara V.V., Floyd K., Suthar M.S., Buchanan W., Luke C.J., Ledgerwood J.E., Mascola J.R., Graham B.S., Beigel J.H. Durability of responses after SARS-CoV-2 mRNA-1273 vaccination. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2032195. NEJMc2032195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Kelly T.C.J., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized cases of coronavirus disease 2019. 2020. Preprint. [DOI]
- Wong M.C., Javornik Cregeen S.J., Ajami N.J., Petrosino J.F. 2020. Evidence of Recombination in Coronaviruses Implicating Pangolin Origins of nCoV-2019. Preprint.https://www.biorxiv.org/content/10.1101/2020.02.07.939207v1 [Google Scholar]
- Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A.T., Gao C., Zhang S. Profile of specific antibodies to SARS-CoV-2: the first report. J. Infect. 2020;81:147–178. doi: 10.1016/j.jinf.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K., Zhai J., Feng Y., Zhou N., Zhang X., Zou J.J., Li N., Guo Y., Li X., Shen X., Zhang Z., Shu F., Huang W., Li Y., Zhang Z., Chen R.A., Wu Y.J., Peng S.M., Huang M., Xie W.J., Cai Q.H., Hou F.H., Chen W., Xiao L., Shen Y. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583:286–289. doi: 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- Yang S.N., Atkinson S.C., Wang C., Lee A., Bogoyevitch M.A., Borg N.A., Jans D.A. Antiviral research; 2020. The Broad Spectrum Antiviral Ivermectin Targets the Host Nuclear Transport Importin α/β1 Heterodimer; p. 104760. [DOI] [PubMed] [Google Scholar]
- Ye M., Fu D., Ren Y., Wang F., Wang D., Zhang F., Xia X., Lv T. Treatment with convalescent plasma for COVID-19 patients in Wuhan, China. J. Med. Virol. 2020;92:1890–1901. doi: 10.1002/jmv.25882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J.J., Toots M., Lee S., Lee M.E., Ludeke B., Luczo J.M., Ganti K., Cox R.M., Sticher Z.M., Edpuganti V., Mitchell D.G., Lockwood M.A., Kolykhalov A.A., Greninger A.L., Moore M.L., Painter G.R., Lowen A.C., Tompkins S.M., Fearns R., Natchus M.G., Plemper R.K. Orally efficacious broad-spectrum ribonucleoside analog inhibitor of influenza and respiratory syncytial viruses. Antimicrob. Agents Chemother. 2018;62 doi: 10.1128/AAC.00766-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., Wang X., Yuan J., Li T., Li J., Qian S., Hong C., Wang F., Liu Y., Wang Z., He Q., Li Z., He B., Zhang T., Fu Y., Ge S., Liu L., Zhang J., Xia N., Zhang Z. Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease 2019. Clin. Infect. Dis. 2020;71:2027–2034. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Chen X., Hu T., Li J., Song H., Liu Y., Wang P., Liu D., Yang J., Holmes E.C., Hughes A.C., Bi Y., Shi W. A novel bat coronavirus reveals natural insertions at the S1/S2 cleavage site of the spike protein and a possible recombinant origin of HCoV-19. bioRxiv. 2020 doi: 10.1101/2020.1103.1102.974139. [DOI] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Vedantham P., Lu K., Agudelo J., Carrion R., Jr., Nunneley J.W., Barnard D., Pöhlmann S., McKerrow J.H., Renslo A.R., Simmons G. Protease inhibitors targeting coronavirus and filovirus entry. Antivir. Res. 2015;116:76–84. doi: 10.1016/j.antiviral.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F.-C., Li Y.-H., Guan X.-H., Hou L.-H., Wang W.-J., Li J.-X., Wu S.-P., Wang B.-S., Wang Z., Wang L., Jia S.-Y., Jiang H.-D., Wang L., Jiang T., Hu Y., Gou J.-B., Xu S.-B., Xu J.-J., Wang X.-W., Wang W., Chen W. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. 2020;395:1845–1854. doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F.C., Guan X.H., Li Y.H., Huang J.Y., Jiang T., Hou L.H., Li J.X., Yang B.F., Wang L., Wang W.J., Wu S.P., Wang Z., Wu X.H., Xu J.J., Zhang Z., Jia S.Y., Wang B.S., Hu Y., Liu J.J., Zhang J., Qian X.A., Li Q., Pan H.X., Jiang H.D., Deng P., Gou J.B., Wang X.W., Wang X.H., Chen W. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020;396:479–488. doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus I., Research T. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoufaly A., Poglitsch M., Aberle J.H., Hoepler W., Seitz T., Traugott M., Grieb A., Pawelka E., Laferl H., Wenisch C., Neuhold S., Haider D., Stiasny K., Bergthaler A., Puchhammer-Stoeckl E., Mirazimi A., Montserrat N., Zhang H., Slutsky A.S., Penninger J.M. Human recombinant soluble ACE2 in severe COVID-19. Lancet Respir. Med. 2020;8:1154–1158. doi: 10.1016/S2213-2600(20)30418-5. [DOI] [PMC free article] [PubMed] [Google Scholar]