Abstract
Severe acute respiratory syndrome coronavirus (SARS-CoV)-2 induces severe infection, and it is responsible for a worldwide disease outbreak starting in late 2019. Currently, there are no effective medications against coronavirus. In the present study, we utilized a holistic bioinformatics approach to study gene signatures of SARS-CoV- and SARS-CoV-2-infected Calu-3 lung adenocarcinoma cells. Through the Gene Ontology platform, we determined that several cytokine genes were up-regulated after SARS-CoV-2 infection, including TNF, IL6, CSF2, IFNL1, IL-17C, CXCL10, and CXCL11. Differentially regulated pathways were detected by the Kyoto Encyclopedia of Genes and Genomes, gene ontology, and Hallmark platform, including chemokines, cytokines, cytokine receptors, cytokine metabolism, inflammation, immune responses, and cellular responses to the virus. A Venn diagram was utilized to illustrate common overlapping genes from SARS-CoV- and SARS-CoV-2-infected datasets. An Ingenuity pathway analysis discovered an enrichment of tumor necrosis factor- (TNF-) and interleukin (IL)-17-related signaling in a gene set enrichment analysis. Downstream networks were predicted by the Database for Annotation, Visualization, and Integrated Discovery platform also revealed that TNF and TNF receptor 2 signaling elicited leukocyte recruitment, activation, and survival of host cells after coronavirus infection. Our discovery provides essential evidence for transcript regulation and downstream signaling of SARS-CoV and SARS-CoV-2 infection.
Keywords: bioinformatics, coronavirus, interleukin-17, severe acute respiratory syndrome coronavirus, severe acute respiratory syndrome coronavirus-2, tumor necrosis factor
1. Introduction
Coronavirus (CoV) disease 2019 (COVID-19) is causing hundreds of thousands of deaths globally. This infectious disease is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, also named novel CoV, 2019-nCoV). CoVs possess a group of spike-like proteins on their surface and belong to a larger family of enveloped positive-stranded RNA viruses. CoVs induce severe infection on both animals and humans primarily through the respiratory tract.[1–3] SARS-CoV-2, as SARS-CoV responsible for the pandemics in 2003, has caused large-scale infection since December 2019. According to statistical reports from World Health Organization (https://www.who.int/emergencies/diseases/novel-coronavirus-2019), the number of confirmed cases and deaths worldwide caused by the 2019-nCoV/SARS-CoV-2 outbreak have exceeded 54,771,888 and 1,324,249 deaths, respectively, until November 17, 2020. In addition to aerosol transmission, viral transmission also occurs between individuals in close physical contact.[4] Nevertheless, there is currently no effective treatment for COVID-19. It is imperative and urgent to explore effective therapeutic strategies against CoVs.
Low levels of SARS-CoV-2 RNAs are detected in culture supernatants from human lung adenocarcinoma epithelial cells (Calu-3 and A549) at 48 hours after infection. Between these 2 cell lines, Calu-3 is more sensitive to SARS-CoV-2 with a 500-fold higher viral entry rate than the mock-infected control in luciferase activity.[5] Calu-3 is also susceptible to entry driven by SARS-CoV.[6] The reason for susceptibility to these viruses is the expression of angiotensin-converting enzyme 2 (ACE2) membrane receptor. ACE2 is a cellular entry receptor for SARS-CoV-2.[7] Calu-3 cell is an ideal model for investigating and studying SARS-CoV/SARS-CoV-2 infection. Therefore, we selected Calu-3 cells as SARS-CoV-2 infection model for the present study.
High-throughput technology is a robust tool for large-scale studies of functional genomics and biological systems. It facilitates research by providing thousands of gene expression profiles in a single experiment. The advantage of this technology could be applied in studying CoV systematically and comprehensively.[7] Several researchers use RNA sequencing or transcriptomes to study viral infection and search for predictive biomarkers or potential therapies.[8–12] However, a comprehensive approach to distinguish significant differences in gene expressions between SARS-CoV-2- and SARS-CoV-infected Calu-3 cells is still lacking until now. Taken together, we aimed to investigate gene expression profiles in SARS-CoV-/SARS-CoV-2-infected Calu-3 models using a bioinformatics approach in the present study. Regulatory networks were predicted and evaluated for their potential role as therapeutic biomarkers for SARS-CoV-/SARS-CoV-2-infected disease. These valuable evidence-based findings from the present study illustrate the essential roles of novel transcript regulation in SARS-CoV-/SARS-CoV-2-infected diseases.
2. Materials and methods
The National Center for Biotechnology Information GEO database was used to query all publicly available datasets related to SARS-CoV- or SARS-CoV-2-infected human lung adenocarcinoma cell models. The GSE17400 and GSE147507 datasets were downloaded for further analysis, the information of ethics committee or institutional review board were described in the original article.[13,14] The GSE17400 dataset contained SARS-CoV-infected cells and mock-infected controls. Bronchial epithelial cell lines derived from Calu-3 cells were infected by SARV-CoV or mock control for 24 hour. Total RNAs were extracted from infected cells, and purified RNAs were sequenced with an Affymetrix Human Genome U133 Plus 2.0 Array.[13] The GSE147507 dataset included SARS-CoV-2-infected cells and mock-infected controls. Calu-3 cells were infected by the SARS-CoV-2 or a mock control for 24 hour. The sequencing platform was the Illumina NextSeq 500 system.[14] The GSE45042 dataset was used as external validation and it collected data from Human CoV EMC 2012 (HCoV-EMC) or mock infected Calu-3 cells. The high throughput platform was the Agilent-014850 Whole Human Genome Microarray 4x44K G4112F system.[15] The analytical methods are briefly summarized. The biomaRt package v. 2.26.1 (https://bioconductor.org/packages/release/bioc/html/biomaRt.html) was utilized to convert gene IDs to gene symbols using the ENSEMBL database with a dataset named mpfuro_gene_ensembl. The Gene Ontology (GO) Elite and pheatmap (v. 1.0.12) platforms were employed for gene clustering based on messenger (m)RNA expression profiles.[16–18] Signals were processed and normalized with the methodology we previously described.[19–21] All these packages and procedures were included in R studio vers. 1.2.1335 and R vers. 3.6.3 (https://rstudio.com/). The top 10% highly differentially expressed genes in SARS-CoV-/SARS-CoV-2-infected groups relative to mock-infected controls were computed as previously described.[22–24] Both P values < .05 and adjusted false discovery rates were utilized to screen for significantly different genes. The final gene lists were uploaded to the GO Elite platform for constructing biological networks, processes, and diseases using the Database for Annotation, Visualization, and Integrated Discover version. 6.8.[25–28] A gene set enrichment analysis (GSEA) platform was applied to enrich the viral- and immune-related pathways from the list of genes.[29] A P value of < .05 was set as a significant cutoff point of enrichment. The Ingenuity pathway analysis (IPA) was adopted to discover interaction networks and pathways related to SARS-CoV-/SARS-CoV-2-infected models. Furthermore, we also introduced the IPA platform to compare average expression levels of the highest top 10% of genes with significantly expressed differences between SARS-CoV- and SARS-CoV-2-infected models. A P value of < .05 indicated a statistically significant difference.
3. Results
3.1. Gene enrichment analysis of SARS-CoV-/SARS-CoV-2-infected human lung adenocarcinoma models
The scheme of the present study was designed to explore differentially expressed genes in cell models of human lung adenocarcinomas between SARS-CoV- or SARS-CoV-2-infected cells and mock-infected controls. Up-regulated genes from both SARS-CoV- or SARS-CoV-2-infected human lung Calu-3 cells were merged together through a Venn diagram to identify shared genes. These commonly up-regulated genes were analyzed with a series of bioinformatics platforms, inluding the IPA to find protein-protein interactions, GO to search for associated pathways, Database for Annotation, Visualization, and Integrated Discovery to investigate related functions, and GSEA to explore downstream-regulated networks (Fig. 1).
Figure 1.
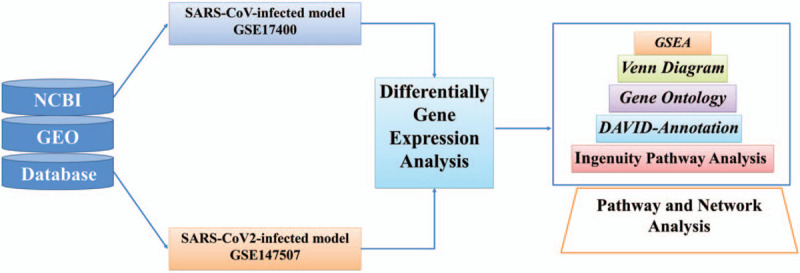
Schematic illustration of the study design. High-throughput data of severe acute respiratory syndrome coronavirus (SARS-CoV-)/SARS-CoV-2-infected human lung adenocarcinoma models were acquired from National Center for Biotechnology Information database with the accession number GSE17400 and GSE147507 datasets. Differentially expressed genes in the top 10% compared to mock-infected controls from (SARS-CoV) and SARS-CoV-2 were merged by the Venn diagram to find shared genes between these 2 groups. Furthermore, gene annotation and gene ontology were done using the Database for Annotation, Visualization, and Integrated Discovery database. Finally, gene set enrichment analysis, and Ingenuity pathway analysis were used for enrichment and downstream pathways analyses. DAVID = the Database for Annotation, Visualization, and Integrated Discovery, GEO = Gene Expression Omnibus, GO = gene ontology, GSEA = gene set enrichment analysis, IPA = Ingenuity pathway analysis, NCBI = National Center for Biotechnology Information.
3.2. GO analysis of SARS-CoV-2-infected human lung adenocarcinoma models
To investigate up-regulated genes in human lung adenocarcinoma cells after infection, we compared sequencing results from SARS-CoV-2-infected cells with mock-infected controls (Fig. 2A). Several cytokine genes were highly expressed in this model of acute infection (24 hour), including IL6 (protein: interleukin (IL)-6), CXCL11 (protein: C-X-C motif chemokine ligand 11 (CXCL11)), CXCL10 (protein: CXCL10), TNF (protein: tumor necrosis factor (TNF)), and IL17C (protein: IL-17C) (Fig. 2B). GO-enriched pathways were categorized into 2 clusters, according to P values. Several immune-related pathways were significantly enriched in SARS-CoV-2-infected cells, including defense response to virus (GO:0051607), immune response (GO:0006955), cytokine-mediated signaling pathway (GO:0019221), etc (Fig. 3 and Supplementary Table 1).
Figure 2.
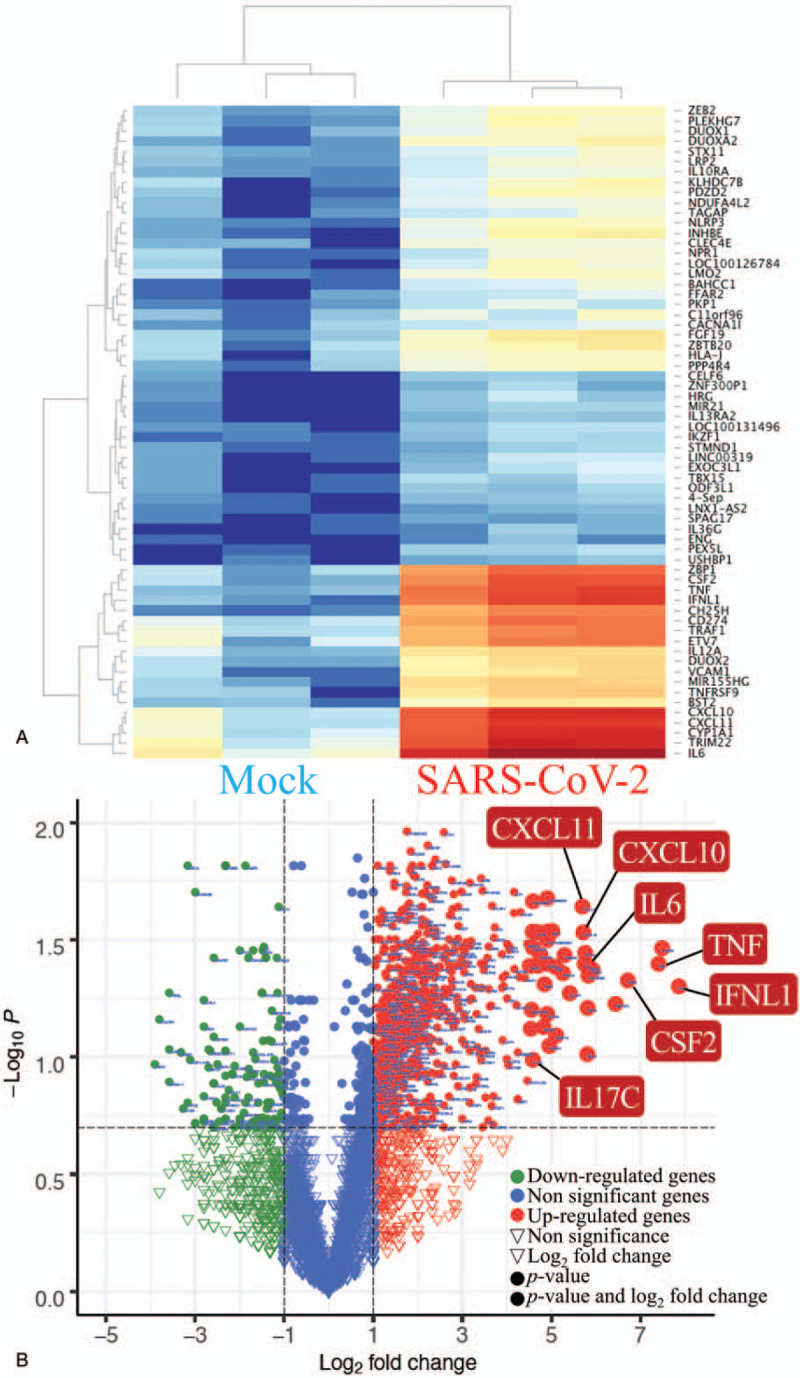
Gene enrichment analysis and heatmap visualization of severe acute respiratory syndrome coronavirus (SARS-CoV)-2-infected human lung adenocarcinoma models from the GSE147507 dataset. (A) Hierarchical clustering heatmap for comparing gene expression between mock and SARS-CoV2. The heatmap could distinctly cluster mock and SARS-CoV-2 groups. SARS-CoV-2-infected cells (n = 3) were compared to mock-infected controls (n = 3). Up-regulated genes are shown in red and downregulated genes in blue on the heatmap. The names of genes are listed from the lower right corner with the lowest P values and to the upper right corner with higher P values. (B) Volcano plot displays the gene expression comparing mock and SARS-CoV-2. Under-expressed genes and overexpressed genes were plotted in the upper left and upper right corner with a threshold of P value > .05 and absolute fold-change larger than 1.5. Most genes play important roles in the immune system such as interleukin (IL)17C, IL6, tumor necrosis factor, CSF2, TFNL1, CXCL11, and CXCL10 were highlighted with a bigger bubble size. The x-axis is log2 of the multiple changes of SARS-CoV-infected cells compared to mock-infected controls. The y-axis is –log10 of P values. A higher Y value means a lower P-value. Critical genes are listed in the figure. CSF2 = colony-stimulating factor 2, CXCL10 = C-X-C motif chemokine ligand 10, CXCL11 = C-X-C motif chemokine ligand 11, IFNL1 = interferon-alpha 1, IL6 = interleukin-6, IL17C, interleukin-17C, TNF = tumor necrosis factor.
Figure 3.
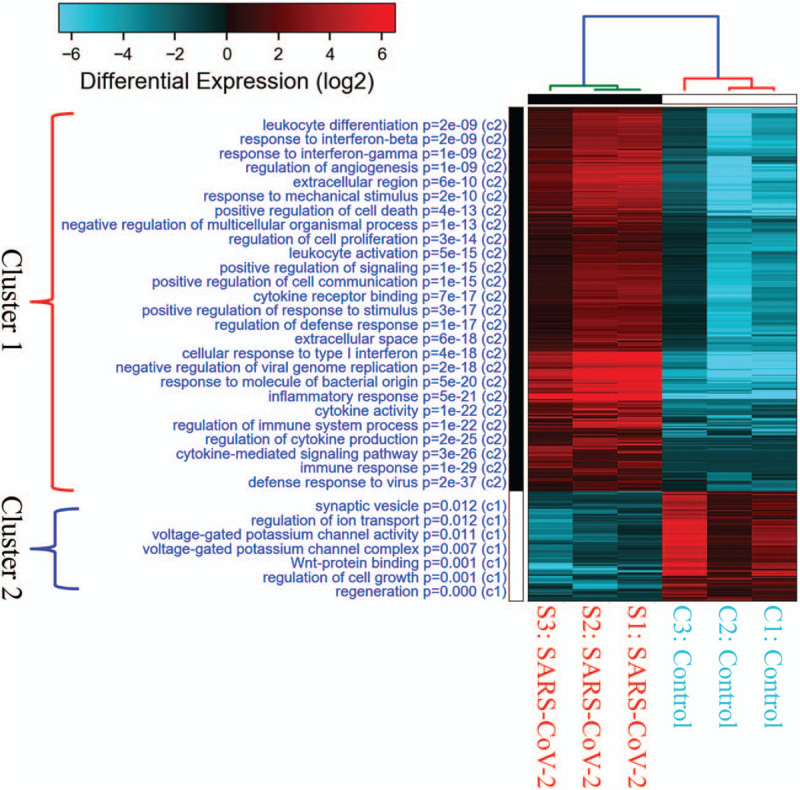
Gene ontology enrichment analysis and heatmap visualization of severe acute respiratory syndrome coronavirus-2-infected human lung adenocarcinoma. SARS-CoV-2-infected 24 hour cells (n = 3) were compared to mock-infected controls (n = 3) from from GSE147507 database, Enriched pathways were grouped into 2 clusters. Pathways in cluster 1 were closely related to SARS-CoV-2 infection with lower P values than those in cluster 2. The highly differentially expressed genes were classified by Gene Onology and expressed as a heatmap. Up-regulated genes are in red, and downregulated ones are in blue.
3.3. GSEA of SARS-CoV-2 infected human lung adenocarcinoma
To verify the significance of the pathways identified in Figure 3 and Supplementary Table 1, up-regulated genes were analyzed with relevant regulatory networks using a GSEA with Kyoto Encyclopedia of Genes and Genomes,[30–32] GO,[33] and the Hallmark platform.[34] Multiple chemokines, cytokines, cytokine receptors, or inflammation-related pathways were identified (Fig. 4A-E, K, L). Highly differentiated expressed cytokines included INF-α, INF-γ, TNF-α, IL-6, and transforming growth factor-β (Fig. 4F, G, H, I, J). Defense responses of host cells to viral infection or biotic stimuli and intracellular machinery for viral reproduction were simultaneously activated, reflecting reactions of the host (Supplementary Fig. 1). Active intracellular signal pathways were also detected, including p53, hypoxia, and signal transducer and activator of transcription signaling (Supplementary Fig. 2A-C). The response of infected cells also mimicked other cells or other diseases; for example, hematopoietic cells, complement system, and type I diabetes (Supplementary Fig. 2D-F).
Figure 4.
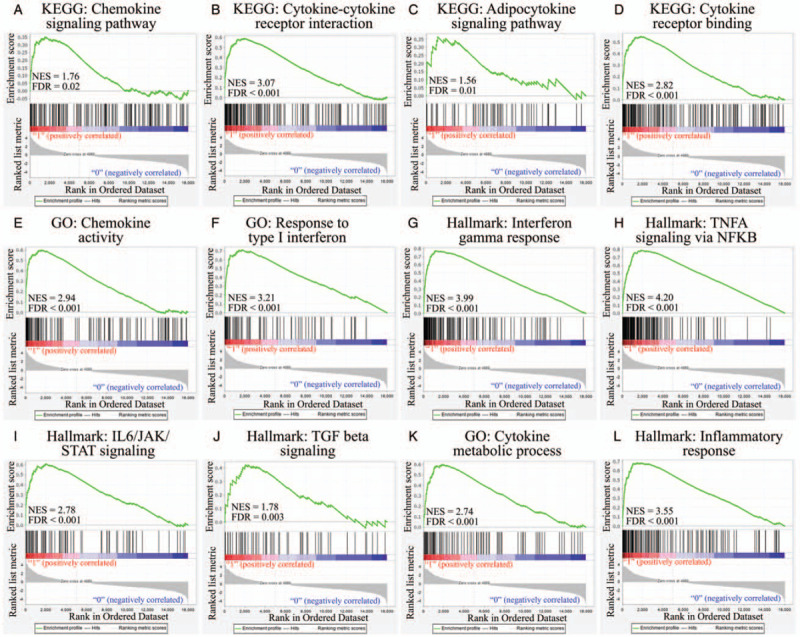
Gene enrichment analysis by Kyoto Encyclopedia of Genes and Genomes, gene ontology, and Hallmark to obtain immune-related networks. Differentially expressed genes in severe acute respiratory syndrome coronavirus (SARS-CoV)-2-infected cells (n = 3) were compared to mock-infected controls (n = 3), and immune-related pathways were selected. (A) Chemokine signaling pathway. (B) Cytokine-cytokine receptor interactions. (C) Adipocytokine signaling pathway. (D) Cytokine receptor binding. (E) Chemokine activity. (F) Response to type I interferon. (G) Iinterferon-γ response. (H) Tumor necrosis factor-α signaling via nuclear factor-κB. (I) Interleukin-6/Janus kinase/signal transducer and activator of transcription signaling. (J) Transforming growth factor-β signaling. (K) Cytokine metabolic process. (L) Inflammatory response. The green line over the upper third of each graph is the enrichment score and a score of > 0 was defined as upregulation. Y-axis was enrichment score to reflect the increased degree of associated genes in SARS-CoV-2-infected model. Y = 1 indicated those genes possitively correlated with SARS-CoV-2 infection. Y = 0 implicated those genes negatively correlated with SARS-CoV-2 infection. We computed the density of genes in dataset to get the normalized enrichment score. The false discovery rate was used to estimate probability value with a given normalized enrichment score. Each solid bar represented the rank of genes in the ordered dataset in the middle third of graph. Red means a positive correlation, and blue means a negative correlation. The distribution of the ranked list along the gene list is shown as the gray part in the lower third of each graph.
3.4. Distinguishing the co-regulated interactions network in SARS-CoV-/SARS-CoV-2-infected models
Next, in order to explore the universal effect between SARS-CoV- and SARS-CoV-2-infected models, up-regulated genes in SARS-CoV-infected cells were compared to mock-infected controls, and 5466 genes were acquired. Highly expressed genes among SARS-CoV-2-infected genes were compared to mock-infected controls, and 2180 genes were obtained. Then, we recognized 441 common genes that overlapped between the 2 gene lists (Fig. 5A). In order to perform external validation and further confirm the significance of these 441 up-regulated genes in Figure 5A, we investigated their expression levels in HCoV-EMC-infected Calu-3 cells in the GSE45042 dataset. Up-regulated genes in HCoV-EMC-infected cells included FNDC3A, NR3C1, GEM, LRIG2, ERBB4, DLG4, IDI2-AS1, DUSP1, BACH1, BACH2 (Supplementary Fig. 3 and Supplementary Table 2). According to the analysis from these datasets, we confirmed that cellular response to different CoVs infection was similar.
Figure 5.
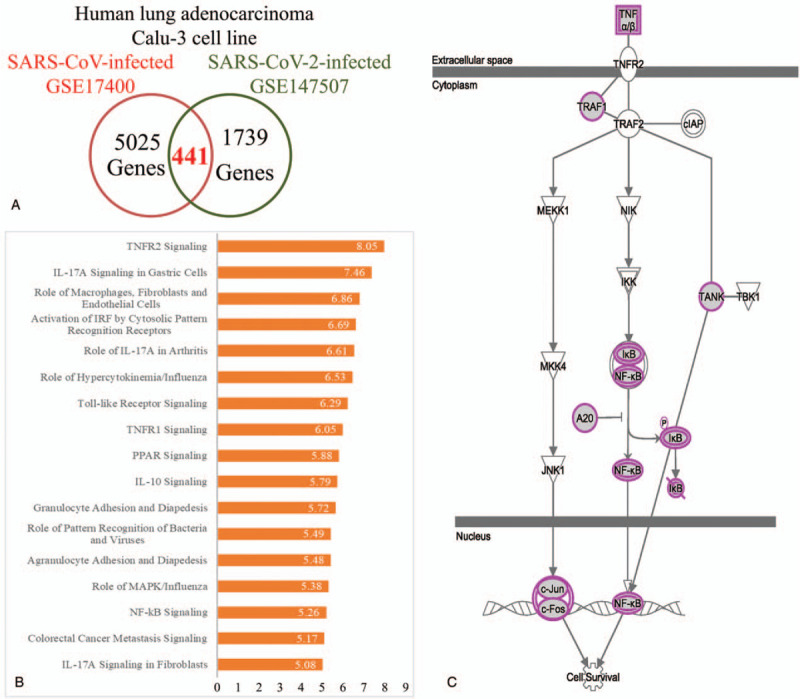
Common gene signatures in severe acute respiratory syndrome coronavirus (SARS-CoV)- and SARS-CoV-2-infected cells. A Venn diagram was used to illustrate common overlapping genes from SARS-CoV- and SARS-CoV-2-infected models, and ngenuity pathway analysis discovered the co-regulated interactions network in these coronavirus infected models. (A) Upregulated genes in SARS-CoV-infected cells (n = 3) compared to mock-infected controls (n = 3) which overlapped with highly expressed genes in SARS-CoV-2 cells (n = 3) compared to mock-infected controls (n = 3). Common genes were explored with a Venn diagram. (B) The Ingenuity pathway analysis platform was used to analyze commonly up-regulated genes of SARS-CoV- and SARS-CoV-2-infected cells. Enriched pathways are shown with the P-value on each bar. (C) Through the Ingenuity pathway analysis platform, downstream tumor necrosis factor-2 recpetor signling was predicted. IL-10 = interleukin-10, IL-17 = interleukin-17, MAPK = mitogen-activated protein kinase, NF-κB = nuclear factor-κB, TNFR1 = tumor necrosis factor receptor 1, TNFR2 = tumor necrosis factor receptor 2.
The IPA platform analyzed these 441 genes from Figure 5A and the most highly enriched pathways were discovered (Fig. 5B). The “tumor necrosis factor receptor 2 (TNFR2) signaling”, “IL-17A signaling in gastric cells”, and “role of macrophages, fibroblasts, and endothelial cells” were the top 3 most highly enriched pathways in both SARS-CoV- and SARS-CoV-2-infected Calu-3 cells. IL-17-associated signal pathways were discovered in 7 of the top 50 pathways and were ranked 2, 5, 17, 24, 34, 40, and 48 (Supplementary Table 3). Furthermore, we predicted these downstream signaling pathways. Activation of TNFR2 promoted phosphorylation of the inhibitor of κB (IκB) protein and released nuclear factor (NF)-κB. Nuclear localization of NF-κB initiates the transcription of targeted genes and supports host cells’ survival (Fig. 5C). Other interacting networks were constructed through analyzing these 441 up-regulated genes on the Kyoto Encyclopedia of Genes and Genomes platform (Supplementary Table 4). There were multiple up-regulated genes associated with TNF signaling pathways, and the corrected P value was 1.36 × 10−9. The interaction between TNF-α and TNF-2R is described in Figure 6. We predicted that TNF-associated signaling pathways in SARS-CoV- and SARS-CoV-2-infected cells were responsible for leukocyte recruitment, leukocyte activation, and survival of host cells. Interactions between cytokines and cytokine receptors were also constructed (Fig. 6). Up-regulated genes in SARS-CoV-2-infected cells were marked with a red star. Increased expressions of chemokines, class I helical cytokines, IL-1-like cytokines, the TNF family, and the TNFR family were detected (Supplementary Fig. 4). Meanwhile, in order to compare the genetic differences between SARS-CoV- and SARS-CoV-2-infected lung adenocarcinoma cells, we also analyzed the genetic signatures in SARS-CoV-infected cell model (Supplementary Fig. 5 and Supplementary Table 5). The enriched GO terms include multicellular organismal process (GO:0032501), anatomical structure development (GO:0048856), and transmembrane receptor protein serine/threonine kinase signaling pathway (GO:0007178). These signatures were completely different from those in SARS-CoV-2-infected cells (Fig. 3).
Figure 6.
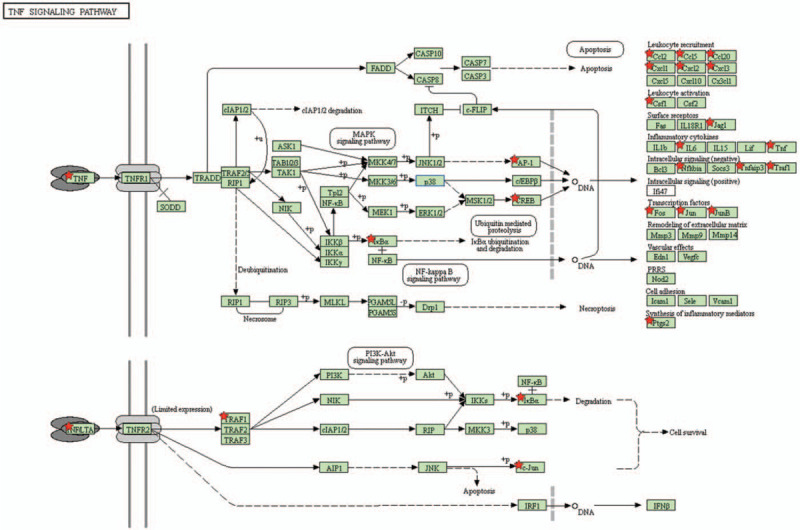
Interaction between tumor necrosis factor and tumor necrosis factor receptor 2 signaling. Up-regulated genes were extracted from the intersection of severe acute respiratory syndrome coronavirus (SARS-CoV)- and SARS-CoV-2-infected cells. These genes were examined by an Ingenuity pathway analysis for interacting networks. Downstream molecules are shown on the right side. Up-regulated genes in SARS-CoV-2-infected cells are marked with a red star, including multiple cytokines and intracellular proteins. CCL = C-C chemokine ligands, CSF = colony-stimulating factor, CXCL = C-X-C motif chemokine ligand, IκB = inhibitor of κB, IL = interleukin, NF-κB = nuclear factor-κB, PTGS2 = prostaglandin-endoperoxide synthase 2, TNFR1 = tumor necrosis factor receptor 1, TNFR2 = tumor necrosis factor receptor, TRAF1 = tumor necrosis factor receptor-associated factor 1.
4. Discussion
Since traditional drug development is time-consuming, drug repurposing, also known as drug repositioning, through bioinformatics is emerging as a crucial experimental approach in drug research and development fields.[35–38] In the present study, we used large amounts of high-throughput data to analyze the genetic signatures of SARS-CoV- and SARS-CoV-2-infected human lung Calu-3 cells compared to mock-infected control groups. By integrating these high-throughput data, we strengthened lines of evidence for certain gene candidates using their genomic as well as transcriptomic datasets. Consequently, multi-omics from public datasets supply a comprehensive analysis for selecting targets. In the present study, we included mRNA expressions from SARS-CoV- and SARS-CoV-2-infected human lung Calu-3 cells. The essential regulatory downstream networks were predicted and evaluated for their potential as therapeutic targets for SARS-CoV- and SARS-CoV-2-infected disease.
Previous studies demonstrated that infection with CoVs induces a cytokine storm and severe inflammation.[39,40] For SARS-CoV-2 infection, several signaling pathways are reported to be potential drug targets for therapy, SARS-CoV-2 relies on ACE2 and TMPRSS2 to enter cells.[6,8] IL family members, including IL-6, IL-1, IL-1β, TNF, and CCL2, are profoundly released by the immune system after SARS-CoV-2 infection[41,42] SARS-CoV was also found to regulate collagen expression via the transforming growth factor-β1 signaling pathway.[43] The response of host cells after SARS-CoV-2 infection induces cytokine-related acute respiratory distress, and cytokine release is associated with worse outcomes. Imbalance of the INF response exacerbates the dysregulation of innate and adaptive immunity after SARS-CoV infection.[44,45] Markedly increased levels of IL-2R, IL-6, IL-10, and TNF-α were detected in patients with serious SARS-CoV-2 infection. Those results are consistent with the results of the present study. Chemokine, cytokine, cytokine receptor, cytokine metabolism, and inflammation-related pathways were enriched (Figs. 3 and 4). Highly expressed genes in acute infection of Calu-3 lung adenocarcinoma cells with SARS-CoV-2 included IL-6 and TNF (Fig. 2). Activation of TNFR2 signaling and TNF downstream signaling was discovered in conjunction with 2 different datasets, SARS-CoV infection and SARS-CoV-2 infection of Calu-3 cells (Fig. 5). These 2 highly pathogenic CoVs have similar structures and genomes. Patients with SARS-CoV and those with SARS-CoV-2 infection also have similar clinical manifestations. Our results confirmed the importance of TNF and the immune system in CoV infection.
In the present study, we also detected several new targeting cytokines. IL-17-associated signaling was discovered in 7 of the top 50 pathways and was ranked 2, 5, 17, 24, 34, 40, and 48 (Supplementary Table 3). The “IL-17A signaling in gastric cells” pathway was enriched in the combined SARS-CoV- and SARS-CoV-2-infected datasets analyzed by the IPA platform (Fig. 5). The upregulation of IL-17C was detected in SARS-CoV-2-infected cells (Fig. 2). Activation of IL-17 signaling was reported in cell-infection models of SARS-CoV and the Middle East respiratory syndrome-CoV. The synergy of IL-17 and IL-6 mediates the cytokine storm after SARS-CoV-2 infection.[46] IL-17 is also overproduced during other viral infections. IL-17C modulates the response of respiratory epithelial cells. It establishes a complex network with other IL-17 family proteins to regulate the innate immune response.[47] Our study further proved the overactivity of IL-17 signaling in host cells after SARS-CoV and SARS-CoV-2 infection. Based on our knowledge, this is the first comprehensive analysis to study correlations of SARS-CoV/SARS-CoV-2 infection with IL-17A signaling. Therefore, IL-17-targeted therapy may provide another opportunity for treating CoV infection.
An inflammatory response is elicited by SARA-CoV and SARS-CoV-2 infection. Treatment with low-dose glucocorticoids can control symptoms of the most-severe patients.[48] The combination of thalidomide and low-dose glucocorticoids is administrated to patients with lung injury and immunological stress caused by COVID-19 pneumonia.[49] Monoclonal anti-IL-6 antibody (tocilizumab) treatment ameliorates serum levels of IL-6 and C-reactive protein; however, overall survival does not improve. A persistent and dramatic increase in serum IL-6 was detected in critically ill patients, and tocilizumab is only effective in a certain type of patient. In the present study, SARS-CoV-2 infection-induced gene expressions of multiple cytokines (Fig. 2) and elicited a complex network of immune responses (Figs. 3 and 4). IL-6 is not the only up-regulated gene. From combining the SARS-CoV and SARS-CoV-2 datasets, TNF and TNFR2 downstream signaling were elevated (Fig. 5). Increased expression of IL-17C and enrichment of multiple IL-17-associated signaling pathways were detected and are illustrated in Fig. 2 and Supplementary Table 3. These pathways could provide potential targets for further treatment of patients with CoV infection.
External validation data in Supplementary Figure 3 and Supplementary Table 2 also indicate that these genes are up-regulated during different coronavirus infection, and these data were consistent with previous studies. Cardiac infection association with SARS-CoV-2 is confirmed via autopsy cases and circular RNA Fndc3b can modulate cardiac repair.[50] NR3C1 and neutrophil axis determines the severity of COVID-19.[51] GEM is a member of the small GTP-binding proteins within the Ras superfamily to increase cellular migration of human T-lymphotropic virus 1-infected cells.[52] LRIG2 is associated with prognosis in relation to Human Papillomavirus-DNA and p16INK4a status.[53] HCV induces a substantial reduction of ErbB3 and ErbB4 expression.[54,55] DUSP1 regulates respiratory Syncytial virus and Sendai virus infection.[56] BACH1 knockdown reduces BZLF1 expression and further Epstein-Barr virus (EBV) infection.[57] BACH2 triggers viral reservoir in T regulatory cells in HIV-1 models.[58] Plasma concentration of CXCL2 and CXCL10 is increased in COVID-19 patients.[59] NFKBIA is a key regulator of immune responsiveness implicated in the SARS and HIV infected model.[60] PTX3 plays a crucial role in coronaviral infection-induced acute lung injury.[61] The elevation of plasma IL-6 in patients is closely correlated with COVID-19 disease.[62] These up-regulated genes in our study provided sufficient evidence for further research of SARS-CoV-2-infected disease.
Collectively, the current study aimed to identify signal pathways related to gene signatures of human lung adenocarcinoma with CoV infection, which could provide potential targets for future treatments. Furthermore, our research revealed regulatory pathways played by several novel genes, which could reduce the gap between bench research and clinical applications. Overactivation of TNF-associated and IL-17-related signaling was explored in the present study. Several cytokine genes were up-regulated in SARS-CoV-2-infected cells, including TNF, IL6, CSF2, IFNL1, IL-17C, CXCL10, and CXCL11. These genes may guide future experimental directions to combat the COVID-19 pandemic.
Acknowledgments
We are thankful to the National Center for High-performance Computing in Taiwan for computer time and facilities (GOV109116 and TRI1091523 to C-Y.W.). This research was supported in part by Higher Education Sprout Project, Ministry of Education to the Headquarters of University Advancement at National Cheng Kung University.
Author contributions
Conceptualization: Li-Chin Cheng, Tzu-Jen Kao, Chih-Yang Wang, Hui-Ping Hsu.
Data curation: Tzu-Jen Kao, Meng-Chi Yen, Chien-Fu Chen, Jui-Hsiang Hung.
Formal analysis: Tzu-Jen Kao, Nam Nhut Phan, Jia-Zhen Jiang.
Methodology: Chung-Chieh Chiao, Zhengda Sun Sun.
Project administration: Chih-Yang Wang, Hui-Ping Hsu.
Writing – original draft: Li-Chin Cheng, Chung-Chieh Chiao, Nam Nhut Phan.
Writing – review & editing: Tzu-Jen Kao, Chih-Yang Wang, Hui-Ping Hsu.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Footnotes
Abbreviations: CoV = coronavirus, COVID 19 = coronavirus disease 2019, CXCL = C-X-C motif chemokine ligand, GO = gene ontology, GSEA = gene set enrichment analysis, HCoV-EMC = Human CoV EMC, IL = interleukin, IPA = Ingenuity pathway analysis, SARS-CoV = Severe acute respiratory syndrome coronavirus, TNF = tumor necrosis factor, TNFR = tumor necrosis factor receptor.
How to cite this article: Cheng L-C, Kao T-J, Phan NN, Chiao C-C, Yen M-C, Chen C-F, Hung J-H, Jiang J-Z, Sun Z, Wang C-Y, Hsu H-P. Novel signaling pathways regulate SARS-CoV and SARS-CoV-2 infectious disease. Medicine. 2021;100:7(e24321).
LC and CW contributed equally to this work.
This study was supported by the Ministry of Science and Technology (MOST) of Taiwan MOST108-2314-B-006-082 to HPH, and MOST109-2320-B-038-009-MY2 to CYW), National Cheng Kung University Hospital (NCKUH11002013 to HPH), Chi Mei Medical Center (CMNCKU B106-F058E and B107-F408E to HPH), Kaohsiung Medical University Hospital (KMUH108-8R72 to MCY), and Taipei Medical University (grant TMU-108-AE1-B16 to CYW).
Computational analyses and data mining were performed using the system provided by Taipei Medical University.
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are publicly available.
Supplemental digital content is available for this article.
References
- [1].Morens DM, Daszak P, Taubenberger JK. Escaping pandora's box - another novel coronavirus. N Engl J Med 2020;382:1293–5. [DOI] [PubMed] [Google Scholar]
- [2].Lam TT, Shum MH, Zhu HC, et al. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature 2020;258:282–5. [DOI] [PubMed] [Google Scholar]
- [3].Wong G, Bi Y-H, Wang Q-H, et al. Zoonotic origins of human coronavirus 2019 (HCoV-19/SARS-CoV-2): why is this work important? Zool Res 2020;41:213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Barton LM, Duval EJ, Stroberg E, et al. COVID-19 Autopsies, Oklahoma, USA. Am J Clin Pathol 2020;153:725–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ou X, Liu Y, Lei X, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun 2020;11:e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020;181:271–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Irigoyen N, Firth AE, Jones JD, et al. High-resolution analysis of coronavirus gene expression by RNA sequencing and ribosome profiling. PLoS Pathog 2016;12:e1–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lukassen S, Chua RL, Trefzer T, et al. SARS-CoV-2 receptor ACE 2 and TMPRSS 2 are primarily expressed in bronchial transient secretory cells. EMBO J 2020;39:e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kim D, Lee J-Y, Yang J-S, et al. The architecture of SARS-CoV-2 transcriptome. Cell 2020;181:914–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Fabozzi G, Oler AJ, Liu P, et al. Strand-specific dual RNA sequencing of bronchial epithelial cells infected with influenza A/H3N2 viruses reveals splicing of gene segment 6 and novel host-virus interactions. J Virol 2018;92: e00518–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yu W-B, Tang G-D, Zhang L, et al. Decoding the evolution and transmissions of the novel pneumonia coronavirus (SARS-CoV-2/HCoV-19) using whole genomic data. Zool Res 2020;41:247–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gong Y-N, Tsao K-C, Hsiao M-J, et al. SARS-CoV-2 genomic surveillance in Taiwan revealed novel ORF8-deletion mutant and clade possibly associated with infections in Middle East. Emerg Microbes Infect 2020;9:1457–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yoshikawa T, Hill TE, Yoshikawa N, et al. Dynamic innate immune responses of human bronchial epithelial cells to severe acute respiratory syndrome-associated coronavirus infection. PLoS One 2010;5:e1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Blanco-Melo D, Nilsson-Payant BE, Liu W-C, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 2020;181:1036–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Menachery VD, Eisfeld AJ, Schäfer A, et al. Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. mBio 2014;5:e01174–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Durinck S, Spellman PT, Birney E, et al. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc 2009;4:1184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004;5:e1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wu C, Ekanem T, Phan N, et al. Gene signatures and prognostic analyses of the Tob/BTG pituitary tumor-transforming gene (PTTG) family in clinical breast cancer patients. Int J Med Sci 2020;17:3112–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Depeille P, Henricks LM, van de Ven RA, et al. RasGRP1 opposes proliferative EGFR-SOS1-Ras signals and restricts intestinal epithelial cell growth. Nat Cell Biol 2015;17:804–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lawson DA, Bhakta NR, Kessenbrock K, et al. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature 2015;526:131–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ou KC, Wang CY, Liu KT, et al. Optimization protein productivity of human interleukin-2 through codon usage, gene copy number and intracellular tRNA concentration in CHO cells. Biochem Biophys Res Commun 2014;454:347–52. [DOI] [PubMed] [Google Scholar]
- [22].Wang CY, Chiao CC, Phan NN, et al. Gene signatures and potential therapeutic targets of amino acid metabolism in estrogen receptor-positive breast cancer. Am J Cancer Res 2020;10:95–113. [PMC free article] [PubMed] [Google Scholar]
- [23].Wang CY, Li CY, Hsu HP, et al. PSMB5 plays a dual role in cancer development and immunosuppression. Am J Cancer Res 2017;7:2103–20. [PMC free article] [PubMed] [Google Scholar]
- [24].Liu H-L, Yeh I-J, Phan NN, et al. Gene signatures of SARS-CoV/SARS-CoV-2-infected ferret lungs in short-and long-term models. Infect Genet Evol 2020;85:e1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zambon AC, Gaj S, Ho I, et al. GO-Elite: a flexible solution for pathway and ontology over-representation. Bioinformatics 2012;28:2209–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Olsson A, Venkatasubramanian M, Chaudhri VK, et al. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature 2016;537:698–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sun Z, Wang C-Y, Lawson DA, et al. Single-cell RNA sequencing reveals gene expression signatures of breast cancer-associated endothelial cells. Oncotarget 2018;9:10945–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- [29].Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci 2019;28:1947–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kanehisa M, Furumichi M, Sato Y, et al. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res 2021;49:D545–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. Nat Genet 2000;25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Liberzon A, Birger C, Thorvaldsdóttir H, et al. The molecular signatures database hallmark gene set collection. Cell Syst 2015;1:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Brown AS, Patel CJ. A standard database for drug repositioning. Sci Data 2017;4:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Li J, Lu Z. A new method for computational drug repositioning using drug pairwise similarity. Proceedings (IEEE Int Conf Bioinformatics Biomed) 2012;2012:1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kim E, Choi A-s, Nam H. Drug repositioning of herbal compounds via a machine-learning approach. BMC Bioinformatics 2019;20:e33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zeng X, Zhu S, Liu X, et al. deepDR: a network-based deep learning approach to in silico drug repositioning. Bioinformatics 2019;35:5191–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Mehta P, McAuley DF, Brown M, et al. Hlh Across Speciality Collaboration UK. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet 2020;395:1033–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Di Gennaro F, Pizzol D, Marotta C, et al. Coronavirus diseases (COVID-19) current status and future perspectives: a narrative review. Int J Environ Res Public Health 2020;17:e1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Conti P, Ronconi G, Caraffa A, et al. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies. J Biol Regul Homeost Agents 2020;34:327–31. [DOI] [PubMed] [Google Scholar]
- [42].Malavolta M, Giacconi R, Brunetti D, et al. Exploring the relevance of senotherapeutics for the current SARS-CoV-2 emergency and similar future global health threats. Cells 2020;9:e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wang CY, Lu CY, Li SW, et al. SARS coronavirus papain-like protease up-regulates the collagen expression through non-Samd TGF-beta1 signaling. Virus Res 2017;235:58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Thiel V, Weber F. Interferon and cytokine responses to SARS-coronavirus infection. Cytokine Growth Factor Rev 2008;19:121–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Qian Z, Travanty EA, Oko L, et al. Innate immune response of human alveolar type ii cells infected with severe acute respiratory syndrome–coronavirus. Am J Respir Cell Mol Biol 2013;48:742–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Cafarotti S. Severe acute respiratory syndrome-coronavirus-2 infection and patients with lung cancer: the potential role of interleukin-17 target therapy. J Thorac Oncol 2020;15:e101–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ryzhakov G, Lai CC, Blazek K, et al. IL-17 boosts proinflammatory outcome of antiviral response in human cells. J Immunol 2011;187:5357–62. [DOI] [PubMed] [Google Scholar]
- [48].Lin L, Lu L, Cao W, et al. Hypothesis for potential pathogenesis of SARS-CoV-2 infection-a review of immune changes in patients with viral pneumonia. Emerg Microbes Infect 2020;9:727–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Chen C, Qi F, Shi K, et al. Thalidomide combined with low-dose glucocorticoid in the treatment of COVID-19 pneumonia. Clin Transl Med 2020;10:e35.1–e35.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Garikipati VNS, Verma SK, Cheng Z, et al. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat Commun 2019;10:e1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Park JH, Lee HK. Re-analysis of single cell transcriptome reveals that the NR3C1-CXCL8-neutrophil axis determines the severity of COVID-19. Front Immunol 2020;11:e1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Chevalier SA, Turpin J, Cachat A, et al. Gem-induced cytoskeleton remodeling increases cellular migration of HTLV-1-infected cells, formation of infected-to-target T-cell conjugates and viral transmission. PLoS Pathog 2014;10:e1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Stefansson K, Oda H, Öfverman C, et al. LRIG1-2 and LMO7 immunoreactivity in vulvar squamous cell carcinoma: association with prognosis in relation to HPV-DNA and p16INK4a status. Oncol Rep 2019;42:142–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Stindt S, Cebula P, Albrecht U, et al. Hepatitis C virus activates a neuregulin-driven circuit to modify surface expression of growth factor receptors of the ErbB family. PLoS One 2016;11:e1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Yun S, Koh J, Nam SK, et al. Clinical significance of overexpression of NRG1 and its receptors, HER3 and HER4, in gastric cancer patients. Gastric Cancer 2018;21:225–36. [DOI] [PubMed] [Google Scholar]
- [56].Robitaille AC, Caron E, Zucchini N, et al. DUSP1 regulates apoptosis and cell migration, but not the JIP1-protected cytokine response, during respiratory syncytial virus and sendai virus infection. Sci Rep 2017;7:e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Keck KM, Moquin SA, He A, et al. Bromodomain and extraterminal inhibitors block the Epstein-Barr virus lytic cycle at two distinct steps. J Biol Chem 2017;292:13284–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Cesana D, Santoni de Sio FR, Rudilosso L, et al. HIV-1-mediated insertional activation of STAT5B and BACH2 trigger viral reservoir in T regulatory cells. Nat Commun 2017;8:e1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Blot M, Jacquier M, Aho Glele LS, et al. CXCL10 could drive longer duration of mechanical ventilation during COVID-19 ARDS. Crit Care 2020;24:e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Moni MA, Liò P. Network-based analysis of comorbidities risk during an infection: SARS and HIV case studies. BMC Bioinformatics 2014;15:e1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Han B, Ma X, Zhang J, et al. Protective effects of long pentraxin PTX3 on lung injury in a severe acute respiratory syndrome model in mice. Lab Invest 2012;92:1285–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Chen X, Zhao B, Qu Y, et al. Detectable serum SARS-CoV-2 viral load (RNAaemia) is closely correlated with drastically elevated interleukin 6 (IL-6) level in critically ill COVID-19 patients. Clin Infect Dis 2020;71:1937–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


