Abstract
Non-small cell lung cancer (NSCLC) is the most common form of cancer, resulting in cancer-related deaths worldwide. Exosomes, a subclass of extracellular vesicles, are produced and secreted from various types of cells, including cancer cells. Cancer-derived exosomes can deliver nucleic acids, proteins, and lipids to provide a favorable microenvironment that supports tumor growth through enhancing cell proliferation and metastasis. Our results showed that miR-224-5p was upregulated in NSCLC patient tissues and cell lines, with a tumor-promoting phenotype. Meanwhile, exosome-derived miR-224-5p induced cell proliferation and metastasis in NSCLC and human lung cells. Moreover, we characterized the androgen receptor (AR) as a direct target of miR-224-5p. Tumor xenograft assay experiments revealed that overexpression of miR-224-5p drove NSCLC tumor growth via the suppression of AR and the mediation of epithelial-mesenchymal transition (EMT). Collectively, our results suggest that miR-224-5p-enriched exosomes promote tumorigenesis by directly targeting AR in NSCLC, which may provide novel potential therapeutic and preventive targets for NSCLC.
Keywords: miR-224-5p, AR, Exosome, NSCLC, Tumorigenesis
Graphical Abstract
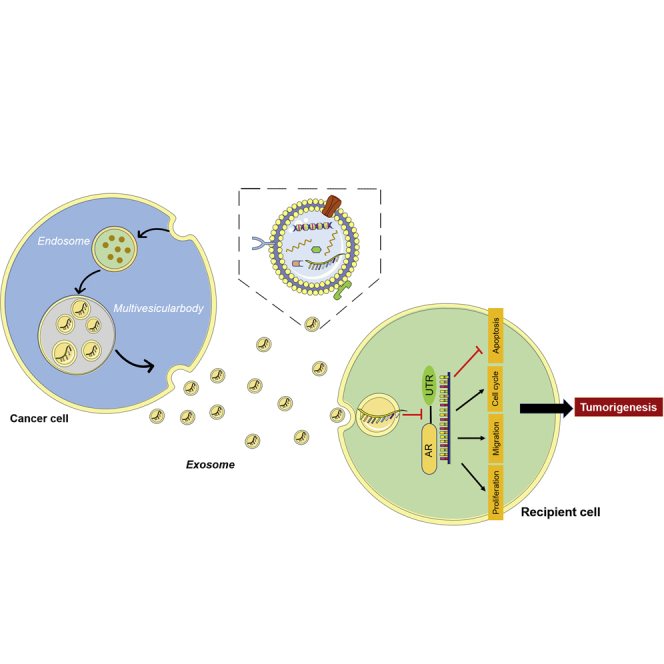
Exosomes and microRNAs play important roles in tumorigenesis. In non-small lung cancer, the cancer-derived exosome miR-224-5p increased significantly. With exosomes transporting, miR-224-5p promoted the proliferation and metastasis ability of their receptor cells via targeting androgenic receptor. Consequently, exosomal miR-224-5p may have a promising prospect in lung cancer therapy.
Introduction
Lung cancer is the most common malignant cancer and the leading cause of cancer-related deaths worldwide.1,2 More than 85% of lung cancer cases are classified as non-small cell lung cancer (NSCLC), with the predicted 5-year survival rate remaining below 15%.2,3 There are many factors associated with the high incidence and mortality of NSCLC, such as late diagnosis, smoking history,4 age differences,5 and sex.6
Exosomes are extracellular vesicles with a size range of 30–150 nm in diameter that are secreted from almost all types of cells, including cancer cells.7 Cancer-derived exosomes can deliver nucleic acids,8 proteins,9 and lipids10 to neighboring or distant cells and subsequently modulate recipient cells. Recently, high levels of microRNAs (miRNAs) have been identified in cancer-derived exosomes, which provide a favorable microenvironment that contribute to tumorigenesis,11 tumor metastasis,8,9 angiogenesis,12 chemoresistance,13 and immune escape.14 Among miRNAs, miR-224-5p has been reported to participate in promoting NSCLC progression and enhance tumor invasion and metastasis.15,16 Meanwhile, the high expression of exosomal miR-224-5p can promote the proliferation and invasion of liver cancer cells, which may serve as a promising non-invasive diagnostic biomarker and therapy monitor of hepatocellular carcinoma.17 However, how cancer-derived exosomal miR-224-5p is involved in NSCLC tumorigenesis has not been elucidated clearly.
Steroid hormones and their receptors are linked to the survival rate of lung cancer patients.18, 19, 20 The androgen receptor (AR) belongs to the steroid hormone receptor family as a nuclear transcription factor. Upon the binding of its androgen ligand, AR protein translocates to the nucleus where it stimulates or suppresses transcription of androgen-responsive genes.21,22 Therefore, the status of AR signaling has become increasingly important, for it is used as both a prognostic marker and potential therapeutic target in prostate cancer,23 bladder cancer,24 and breast cancer.22,25 However, the role of AR signals in the progression of sex-unrelated cancers presents a complex situation and evidence remains contradictory. Male NSCLC patients exposed to androgen pathway manipulation (APM) were significantly associated with a better survival rate.26 Alternatively, the overall survival rate of female NSCLC patients with higher levels of AR was significantly better than that in NSCLC patients with no AR exprsssion.27 Whether AR participates in the adverse outcomes and progression of NSCLC needs to be further investigated.
In this study our results showed that miR-224-5p-enriched exosomes can be transmitted to adjacent and remote cells, promoting human lung cells or NSCLC cell proliferation and migration by targeting AR.
Results
miR-224-5p is upregulated in NSCLC
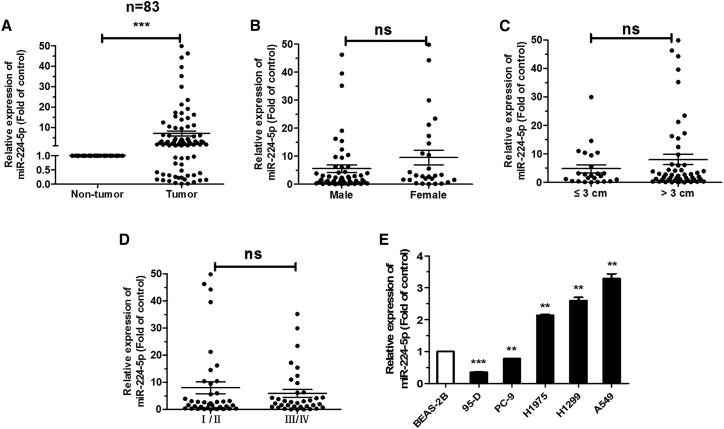
In order to investigate whether miR-224-5p plays a crucial role in the progression of NSCLC, 83 pairs of NSCLC patient tissues were collected for detecting the expression of miR-224-5p. Compared with corresponding para-carcinoma tissues, the results indicated that miR-224-5p was upregulated in NSCLC tissues (Figure 1A). However, the levels of miR-224-5p were not related to sex, tumor size, and pathological stage in these patient tissues (Figures 1B–1D). Meanwhile, miR-224-5p was observably much higher in H1975, H1299, and A549 cells, and it is lower in PC-9 and 95-D cells, with BEAS-2B cells as a control (Figure 1E). Collectively, these results indicated that elevated miR-224-5p might be relevant to the development of NSCLC.
Figure 1.
miR-224-5p is upregulated in NSCLC tissues and cell lines
(A) qRT-PCR analysis of miR-224-5p expression in tumor and corresponding para-carcinoma tissues from 83 NSCLC patients. (B–D) qRT-PCR analysis of miR-224-5p expression related to tumors based on sex (B), size (C), and pathological stage (D). (E) qRT-PCR analysis of miR-224-5p expression in human lung epithelial cell line BEAS-2B, and NSCLC cell lines 95-D, PC-9, H1975, H1299, and A549. U6 was used as an internal control. Error bars represent the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
miR-224-5p promotes cell proliferation and migration of NSCLC
We first constructed stable overexpression of miR-224-5p by lentivirus infection with pLenti-miR-224 or pLenti in H1299 and A549 cells. As shown by qRT-PCR analysis, miR-224-5p increased in these two stably transfected cell lines (Figure 2A). Meanwhile, an inhibitor was transfected into H1299 and A549 cells to inhibit the expression of miR-224-5p, and the results validated that transfection of the miR-224-5p inhibitor did reduce its level compared with a negative control (NC) inhibitor (Figure 2B).
Figure 2.
miR-224-5p promotes cell proliferation and migration of NSCLC
(A and B) qRT-PCR analysis of miR-224-5p expression in H1299 and A549 cells, stably transfected with pLenti/pLenti-miR-224 by lentivirus infection (A) or transiently transfected with NC/miR-224-5p inhibitor. U6 was used as an internal control. (C and D) Cell proliferation ability of H1299 and A549 cells stably transfected with pLenti/pLenti-miR-224 (C), and transiently transfected with NC/miR-224-5p inhibitor (D), as measured by a Cell Counting Kit-8 (CCK-8) assay. (E–H) Representative images (E and G) and quantification of colony formation in H1299 and A549 cells transfected with pLenti/pLenti-miR-224 (F), and transiently transfected with NC/miR-224-5p inhibitor (H). (I–K) Representative images (I) and quantification of cell migration in H1299 and A549 cells stably transfected with pLenti/pLenti-miR-224. Scale bars, 100 μm. (J), and transiently transfected with NC/miR-224-5p inhibitor (K), as measured by wound-healing assays. Error bars represent the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
To investigate the effect of miR-224-5p on NSCLC cell growth, Cell Counting Kit-8 (CCK-8) and colony formation assays were utilized. Overexpression of miR-224-5p exhibited a significant facilitation of cell proliferation (Figures 2C, 2E, and 2F). The same trend was also found in 95-D cells with lower expression of miR-224-5p (Figures S1A–S1C). Similarly, inhibition of miR-224-5p dramatically decreased cell proliferative ability in H1299 and A549 cells (Figures 2D, 2G, and 2H). In addition, we examined the effect of miR-224-5p on NSCLC migration ability by a wound-healing assay. As predicted, miR-224-5p overexpression led to an increase and miR-224-5p inhibition led to a reduction in the migration rate of NSCLC cells (Figures 2I–2K). Cells that had migrated from the serum-free medium in the top chamber to the lower chamber in 24 h were imaged and analyzed by a transwell assay (Figures S2A and S2B). These results indicate that miR-224-5p contributes to NSCLC cell proliferation and migration ability in vitro.
miR-224-5p accelerates cell-cycle progression and inhibits apoptosis
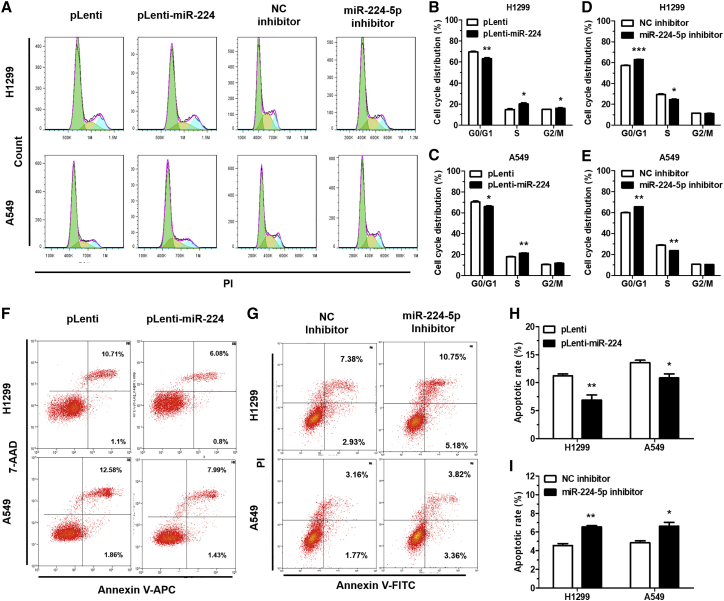
To investigate the effect of miR-224-5p on cell-cycle progression and apoptosis, we performed flow cytometry analysis after miR-224-5p overexpression or inhibition in NSCLC cells. With regard to cell-cycle progression, miR-224-5p overexpression or miR-224-5p inhibition in NSCLC cells could significantly facilitate or arrest the conversion from the G0/G1 phase to the S phase (Figures 3A–3E). Furthermore, we used flow cytometry to evaluate the apoptosis rate in NSCLC cells. The results showed that upregulation of miR-224-5p decreased NSCLC cell apoptosis (Figures 3F and 3H; Figures S3A and S3B). Meanwhile, an increased apoptosis rate was found in the group transfected with miR-224-5p inhibitor as compared with NC inhibitor (Figures 3G and 3I). Taken together, our results demonstrated that miR-224-5p could accelerate cell-cycle progression and inhibit apoptosis of NSCLC cells.
Figure 3.
miR-224-5p accelerates cell-cycle progression and inhibits apoptosis
(A–E) Representative results (A) and quantification of the cell-cycle distributions in H1299 and A549 cells stably transfected with pLenti/pLenti-miR-224 (B and C), and transiently transfected with NC/miR-224-5p inhibitor (D and E). Green, yellow, and blue represent the populations of cells in the G0/G1, S, and G2/M phases, respectively. (F–I) Representative results (F and G) and quantification of the rate of apoptosis in H1299 and A549 cells, stably transfected with pLenti/pLenti-miR-224 (H), and transiently transfected with NC/miR-224-5p inhibitor (I). Cell-cycle distribution and rate of apoptosis were detected by flow cytometry. Error bars represent the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
miR-224-5p-enriched exosomes facilitate tumorigenesis of NSCLC cells in vitro
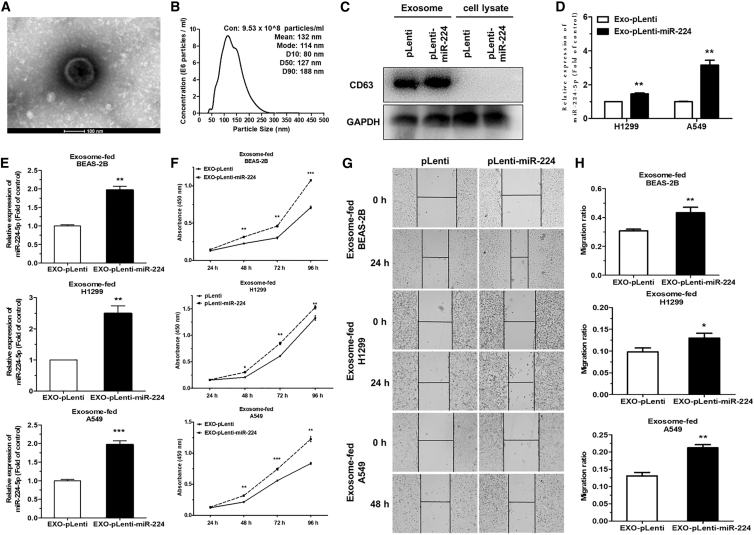
Recent studies have suggested that miRNAs can be packaged into tumor-derived exosomes and delivered to neighboring or distal cells, which functionally modulate the level of target mRNAs to alter target cell fate.8,28,29 Herein, we isolated and purified extracellular exosomes from the culture medium of H1299 and A549 cells stably transfected with pLenti-miR-224 or pLenti (EXO-pLenti-miR-224/pLenti). Then, we observed a canonical cup-shaped morphology of the isolated exosomes by transmission electron microscopy (TEM) (Figure 4A). Simultaneously, NanoSight particle tracking analysis (NTA) further confirmed the concentration and size of the exosomes (Figure 4B). The specific exosomal markers CD63 and Tsg101 were only identified in the exosomes by western blot assay compared with cell lysates (Figure 4C; Figure S4A).
Figure 4.
miR-224-5p-enriched exosomes facilitate tumorigenesis of NSCLC cells in vitro
(A and B) Transmission electron microscopy (TEM) (A) and NanoSight particle tracking analysis (NTA) (B) identified the morphology, size distributions, and concentration of isolated exosomes collected from A549 cell culture medium. Scale bar, 100 nm. (C) Western blot analysis of exosomal marker CD63 in A549-EXO-pLenti/pLenti-miR-224 or A549 cell lysates. GAPDH was used for as an internal control. (D) qRT-PCR analysis of miR-224-5p expression in EXO-pLenti or EXO-pLenti-miR-224 isolated from miR-224-5p-overexpressing H1299 and A549 cells. (E and F) qRT-PCR analysis of miR-224-5p expression (E) and cell proliferation ability (F) in BEAS-2B, H1299, and A549 cells fed with EXO-pLenti or EXO-pLenti-miR-224. U6 was used as an internal control. (G and H) Representative images (G) and quantification (H) of cell migration in BEAS-2B, H1299, and A549 cells fed with EXO-pLenti or EXO-pLenti-miR-224, as measured by wound-healing assays. Scale bars, 100 μm. Error bars represent the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Since miR-224-5p facilitates NSCLC cell growth and migration, we further investigated whether miR-224-5p might contribute to this effect through tumor-derived exosomes. First, we detected that miR-224-5p was obviously enriched in EXO-pLenti-miR-224 derived from H1299 and A549 cells, compared to EXO-pLenti (Figure 4D). Then, exosome feeding assays found that pure BEAS-2B, H1299, and A549 cells had elevated miR-224-5p levels following incubation with A549-EXO-pLenti-miR-224 (Figure 4E), suggesting that miR-224-5p-enriched exosomes could increase the level of miR-224-5p in receptive cells. A549-EXO-pLenti-miR-224 remarkably promoted the proliferation and migration ability of pure BEAS-2B, H1299, and A549 cells compared to those fed with A549-EXO-pLenti (Figures 4F–4H).
Taken together, these data suggest that miR-224-5p can be enriched in tumor-derived exosomes and then delivered to neighboring cells, functionally facilitating recipient cell proliferation and migration in vitro.
AR is a direct target of miR-224-5p
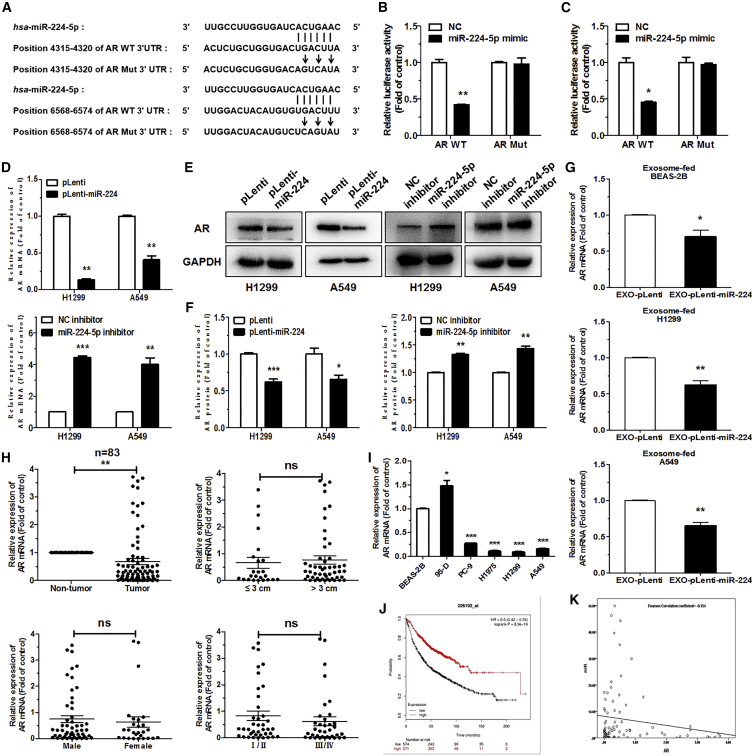
To explore the molecular mechanism of miR-224-5p’s contribution to the tumorigenesis of NSCLC, we identified AR as a potential target for miR-224-5p with two different binding sites by TargetScan and miRWalk 2.0. The wild-type (WT) and corresponding mutant (Mut) binding sites of miR-224-5p with the AR 3′ untranslated region (UTR) are shown in Figure 5A. Then, a dual-luciferase reporter assay was performed to prove whether AR is a direct target of miR-224-5p. In brief, pGL3-AR-WT-3′ UTR-1/2 or pGL3-AR-Mut-3′ UTR-1/2, along with pRL and miR-224-5p mimic or NC mimic, were co-transfected into HEK293T cells. The results showed that compared to transfection with pGL3-AR-Mut-3′ UTR-1/2, the relative luciferase activity was strikingly reduced by about 50% in the cells transfected with pGL3-AR-WT-3′ UTR-1/2, suggesting that the two-stage binding sites of AR 3′ UTR are specifically inhibited by miR-224-5p mimic (Figures 5B and 5C). qRT-PCR and western blot analysis of AR indicated that the level of AR mRNA and protein was strikingly decreased in miR-224-5p-overexpression cells and increased in miR-224-5p-inhibition cells (Figures 5D–5F). Moreover, following being fed with A549-EXO-pLenti-miR-224, the expressions of AR mRNA were reduced in BEAS-2B, H1299, and A549 cells (Figure 5G). Western blot detected the changes in the level of AR protein (Figure S5A). These results support that AR is a direct target of miR-224-5p, and exosome-enriched miR-224-5p can directly target AR after exosomes were taken up by adjacent cells.
Figure 5.
AR is a direct target of miR-224-5p
(A) The two-stage predicted wild-type (WT) and corresponding mutant (Mut) binding sites of the AR 3′ UTR with miR-224-5p. The vertical lines data indicate the binding sites between miR-224-5p and the AR WT 3′ UTR, and arrows indicate mutagenic nucleotides of the AR Mut 3′ UTR. (B and C) Relative firefly luciferase expression in HEK293T cells co-transfected with pGL3-AR-WT-3′ UTR-1/Mut-3′ UTR-1 (B) or pGL3-AR-WT-3′ UTR-2/Mut-3′ UTR-2 (C), including pRL and NC/miR-224-5p mimic. Renilla luciferase expression was used for as an internal control. (D–F) qRT-PCR analysis (D), western blot analysis (E), and quantification (F) of AR mRNA or protein expression in H1299 and A549 cells stably transfected with pLenti/pLenti-miR-224 or transiently transfected with NC/miR-224-5p inhibitor. (G) qRT-PCR analysis of AR mRNA expression in BEAS-2B, H1299, and A549 cells fed with EXO-pLenti or EXO-pLenti-miR-224. (H) qRT-PCR analysis of AR mRNA expression and relationship with tumor size, sex, and pathological stage in 83 pairs of NSCLC patient tumor and corresponding para-carcinoma tissues. (I) qRT-PCR analysis of AR mRNA expression in human lung epithelial cell line BEAS-2B, and in NSCLC cell lines 95-D, PC-9, H1975, H1299, and A549. 18S rRNA and GAPDH were used as internal controls for qRT-PCR and western blot analysis, respectively. (J) Kaplan-Meier curve of overall survival of 1,926 lung cancer patients from a Kaplan-Meier plotter online database. (K) The negative correlation between miR-224-5p and AR, according to the Pearson correlation coefficient. Error bars represent the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
To examine the expression of AR in NSCLC tumors, we evaluated the mRNA expression of AR in the 83 pairs of primary NSCLC tumors and corresponding para-carcinoma tissues, which were identical to that of miR-224-5p detection. The level of AR mRNA was downregulated in tumor tissues. However, there was no significant relationship between the level of AR mRNA and tumor size, sex, or pathological stage in these patient tissues (Figure 5H). Additionally, compared with BEAS-2B cells, the levels of AR mRNA in H1299, A549, PC-9, and H1975 cells were much lower. The level of AR was higher in 95-D cells, which was contrary to the expression of miR-224-5p (Figure 5I). Based on the Kaplan-Meier plotter online database, we generated a Kaplan-Meier overall survival curve of NSCLC patients. Kaplan-Meier analysis revealed that AR overexpression was correlated with longer overall survival in 1,145 NSCLC cases (Figure 5J). Furthermore, the Pearson correlation coefficient indicated a negative correlation between relative AR expression and miR-224-5p in 83 pairs of NSCLC patient samples (Figure 5K). Collectively, these results suggest that AR is downregulated in NSCLC tissues and cell lines, and it might be a novel potential diagnostic biomarker for NSCLC.
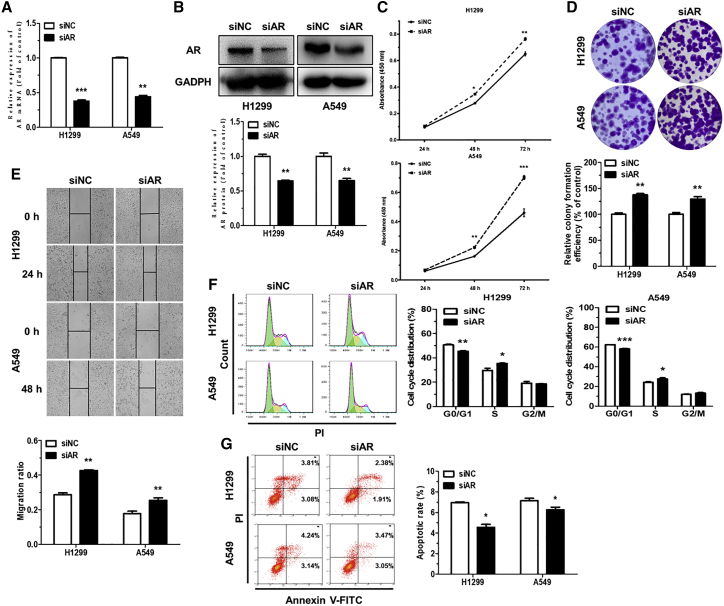
AR functions to suppress NSCLC
To investigate the biological function of AR in NSCLC cells, AR was inhibited by transfecting with AR small interfering RNA (siRNA) (siAR). The downregulated levels of AR mRNA and protein were subsequently detected in transfected H1299 and A549 cells, suggesting efficient knockdown of AR (Figures 6A and 6B). CCK-8, colony formation, and wound-healing assays demonstrated that siAR significantly promoted H1299 and A549 cell proliferation, colony formation, and migration ability, compared to siRNA NC (siNC) (Figures 6C–6E). Next, we further analyzed the effects of siAR on cell-cycle progression and apoptosis. The results of flow cytometry analysis showed that siAR significantly facilitated the conversion of cell cycle from the G0/G1 phase to the S phase and decreased the rate of apoptosis in H1299 and A549 cells (Figures 6F and 6G). Taken together, these data demonstrated that AR knockdown exhibits a similar phenotype as miR-224-5p overexpression in NSCLC cells. It also showed that miR-224-5p promotes tumorigenesis of NSCLC by directly downregulating AR.
Figure 6.
AR functions to suppress NSCLC
(A and B) qRT-PCR (A) and western blot analysis (B) of AR mRNA and protein expression in H1299 and A549 cells transiently transfected with siNC or siAR. 18S rRNA and GAPDH were used as internal controls for qRT-PCR and western blot analysis, respectively. (C–E) Representative images and quantification of cell proliferation (C), colony formation (D), and migration ability (E) of H1299 and A549 cells transiently transfected with siAR. Scale bars, 100 μm. (F and G) Representative results and quantification of the cell-cycle distributions (F) and rate of apoptosis (G) in H1299 and A549 cells transiently transfected with siNC or siAR. Green, yellow, and blue represent the populations of cells in the G0/G1, S, and G2/M phases, respectively. Cell-cycle distribution and rate of apoptosis were detected by flow cytometry. Error bars represent the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
miR-224-5p drives tumor growth and metastases in vivo
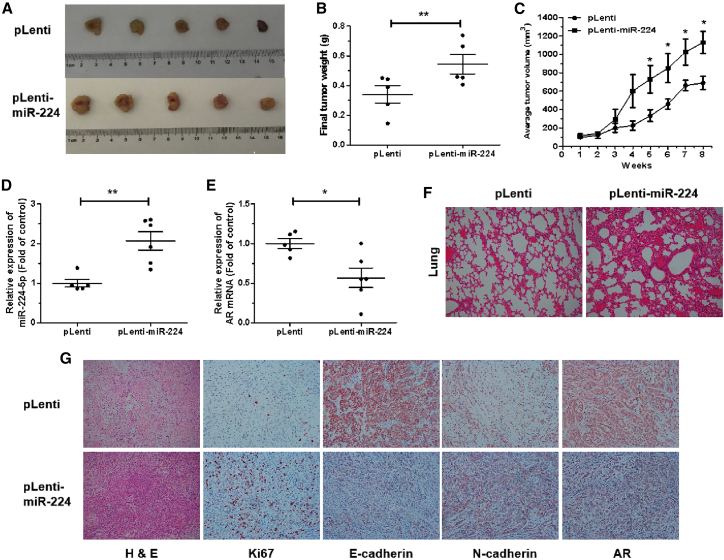
To further examine the effect of miR-224-5p on NSCLC tumor growth and metastases in vivo, we established a subcutaneous xenograft and metastatic model. Nude mice were injected subcutaneously and intravenously with miR-224-5p-overexpression (pLenti-miR-224) or control (pLenti) A549 cells, then randomly divided into two groups (n = 10). Xenograft tumors in the above nude mice were measured every week and were subsequently excised and weighed. The xenograft tumors in the pLenti-miR-224 group significantly facilitated tumor growth and were of heavier weight and larger size at 8 weeks after implantation compared with the pLenti group (Figures 7A–7C). Then, there were significant upregulations of miR-224-5p and downregulations of AR mRNA levels in xenograft tumors from the pLenti-miR-224 group compared with pLenti group (Figures 7D and 7E).
Figure 7.
miR-224-5p drives tumor growth in vivo
(A and B) Representative images (A) and weights (B) of subcutaneous xenograft tumors from nude mice 8 weeks after injection with pLenti or pLenti-miR-224 A549 cells. (C) Tumor growth curves with volume were measured weekly after injection. (D and E) qRT-PCR analysis of miR-224-5p (D) and AR mRNA expression (E) in subcutaneous xenograft tumors. (F) Histopathology of metastases in the lungs of two groups of nude mice stained with hematoxylin and eosin (original magnification, ×100). (G) Representative images of tumor tissues stained with hematoxylin and eosin solution and representative images of immunohistochemistry staining for Ki67, E-cadherin, N-cadherin, and AR expression in tumor tissues (original magnification, ×100). Error bars represent the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Subsequently, the bilateral lungs and xenograft tumors of these two groups were fabricated into sections to observe corresponding pathological changes. Hematoxylin and eosin staining in the bilateral lungs of the pLenti-miR-224 group showed larger and more destructive or necrotic regions compared with pLenti group, which suggested that miR-224-5p promoted the lung colonization of A549 cells (Figure 7F). Moreover, immunohistochemistry in xenograft tumors of the pLenti-miR-224 group showed higher levels of the cell proliferation marker Ki67 and epithelial-mesenchymal transition (EMT) marker N-cadherin, along with lower levels of E-cadherin and AR, compared with the pLenti group (Figure 7G). Taken together, these results indicate that miR-224-5p can drive tumor growth and lung colonization in vivo via the suppression of AR and the mediation of EMT.
Discussion
Our results demonstrated that miR-224-5p was upregulated in NSCLC patient tissues and cell lines. AR was confirmed to be a direct target of miR-224-5p. Furthermore, we have identified that miR-224-5p can be enriched in tumor-derived exosomes, then transmitted to adjacent cells, promoting cell proliferation and metastasis through directly inhibiting AR in vitro. Meanwhile, our results also indicated that miR-224-5p could drive tumor growth in vivo.
Numerous epidemiological studies suggested that the steroid hormones and their receptors are associated with the incidence and clinical outcomes of lung cancer.18,30 Estrogen receptor (ER) has been reported to be likely to modulate epidermal growth factor receptor (EGFR), Notch1, and GSK3β/β-catenin signaling pathways to promote NSCLC progression.30 In addition, ERβ was sensitive to chemotherapy by regulating DNA damage response31 and correlated with resistance of EGFR tyrosine kinase inhibitors (EGFR-TKIs) in NSCLC,32 supporting the potential predictive and therapeutic value of the steroid hormones receptor in clinical management of NSCLC.
AR signaling has a crucial role in the progression of multiple hormone-related cancers. However, the expression patterns and underlying molecular mechanism of AR in NSCLC are poorly understood and remain controversial.26,27 In our study, we found that AR was downregulated in NSCLC tissues but there were no significant difference between tumor size, sex, and staging. Meanwhile, NSCLC patients with a high level of AR had a good survival rate. Further experiments indicated that AR could restrain cell proliferation and migration ability, arrest cell-cycle progression, and increase cell apoptosis in NSCLC cells.
Usually, AR works as an oncogene in sex-related tumors by stimulating or suppressing transcription of androgen-responsive genes, upon binding of its androgen ligand.21 Moreover, the AR is also regulated through non-ligand-dependent pathways. In breast cancer, AR activity can be activated by signal transduction in an extracellular signal-regulated kinase (ERK)-dependent or -independent manner.33 In prostate cancer, AR interacts with Src kinase and phosphatidylinositol 3-kinase (PI3K) and then activates mitogen-activated protein kinase (MAPK) and Akt pathways.34 Nevertheless, after being inhibited by upregulated miR-224-5p, how AR participates in regulation of cell phenotype needs further exploration, which will offer perspective into the implications for therapies targeting the AR in NSCLC.
Exosomes are extracellular vesicles that can deliver nucleic acids, proteins, and lipids to neighboring or distant cells. They originate from the endosomal pathway by the formation of intracellular multivesicular bodies.35 Exosomes have recently become the focus of intensive scientific research as novel mediators of intercellular communication. Several studies have recently established that miRNAs or long non-coding RNAs (lncRNAs) secreted by tumor cells through exosomes are involved in the communication between the tumor and its surrounding microenvironment. Fang et al.8 found that tumor-derived exosomal miR-1247-3p controls lung metastasis of high-metastatic hepatocellular carcinoma, demonstrating an intercellular crosstalk between tumor cells and fibroblasts. Exosomes also selectively package an amount of lncRNAs, which are known to regulate the tumor microenvironment or modulate gene expression by acting as competitive endogenous RNA.34,36
Previous studies from our laboratory have confirmed that miR-18a-5p,37 miR-183-5p,38 miR-199a-5p,39 and miR-146a-5p40 have a diversity effect on the progression of NSCLC. These findings revealed that the dysregulation of miRNAs might be associated with the tumorigenesis of lung cancer. Herein, our data indicate that miR-224-5p was packed into exosomes and transmitted intercellular signals. Then, tumor-derived exosomal miR-224-5p was absorbed into adjacent cells, facilitating cell proliferation and migration ability of NSCLC or normal human lung epithelial cells. In fact, exosomes are currently being evaluated as possible methods to target drug delivery, owing to their ability to shield miRNAs, lncRNAs, peptides, and synthetic drugs from degradation. In addition, miR-224-5p could evade recognition by the immune system and be accurately delivered to recipient cells or tissues.41 Further exploration on the detailed molecular mechanism between exosomes and tumorigenesis will make it possible to utilize exosome-derived miR-224-5p as a diagnostic and therapy target for precision medicine.
Overall, our study shows that miR-224-5p-enriched exosomes promote tumorigenesis of NSCLC by directly targeting AR, perhaps providing novel potential targets for prevention and treatment of NSCLC.
Materials and methods
Clinical tissue samples and cell lines
The Shanghai Chest Hospital affiliated with Shanghai Jiao Tong University provided us with 83 pairs (carcinoma and corresponding para-carcinoma) of human NSCLC samples. The study was approved by the Ethics Committee of Shanghai Chest Hospital. Details of all samples used in this study are presented in Table S1.
The human NSCLC cell lines 95-D, PC-9, H1975, H1299, and A549 were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). The BEAS-2B (human lung epithelial cells) and HEK293T (human embryonic kidney 293T) cell lines were obtained from the Cell Bank, China Academy of Sciences (Shanghai, China).
Cell culture
95-D, PC-9, A549, BEAS-2B, and HEK293T cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Corning Cellgro, Manassas, VA, USA). H1975 and H1299 cells were cultured in RPMI 1640 medium (Corning Cellgro). All media were supplemented with 10% fetal bovine serum (FBS, Gibco, Gaithersburg, MD, USA) and the antibiotic mixture of 100 μg/mL streptomycin and 100 U/mL penicillin (HyClone, Logan, UT, USA) at 37°C in a 5% CO2 humidified atmosphere.
Total RNA extraction, reverse transcription, and qRT-PCR
Clinical tissue samples were ground with liquid nitrogen before total RNA was extracted. Total RNA was isolated with TRIzol Up reagent (TransGen Biotech, Beijing, China). The PrimeScript RT reagent kit and QuantiMir cDNA kit (Takara, Dalian, China) were used to construct reverse transcription of mRNAs and miRNAs. 18S rRNA and U6 snRNA were used as the endogenous controls for mRNAs and miRNAs. Then, qRT-PCR was performed with SYBR Premix Ex Taq (Takara). Results were processed by the relative quantification (2−ΔΔCt) method for relative quantification of mRNAs and miRNAs. All primer sequences are shown in Table S2.
Lentivirus construction and infection
In short, the primary (pri-)miR-224 sequence was cloned into the pLenti lentiviral vector (Invitrogen, Carlsbad, CA, USA) with BamHI and XhoI (Takara), named pLenti-miR-224. pSPAX2 and pMD2G as auxiliary vectors for lentiviral assembly were co-transfected with pLenti-miR-224 or pLenti into HEK293T cells. The virus particles were collected after incubation for 48 and 72 h. Subsequently, they were centrifuged together at 4,000 rpm for 5 min at 4°C and filtered with a 0.45-μm filter (Millipore, Billerica, MA, USA). The viral particles of pLenti-miR-224 or pLenti infected H1299 and A549 cells. Then, the infected cells were sorted for green fluorescence intensity via a flow cytometer (Beckman Coulter, CA, USA) to generate the miR-224-5p overexpression cell lines.
Transient transfection
According to the manufacturer’s instructions for Invitrogen Lipofectamine 2000 (Thermo Fisher Scientific, Life Technologies, New York, NY, USA), H1299 and A549 cells were transiently transfected with 50 nM miR-224-5p mimic/inhibitor, 100 nM siAR, or the corresponding NC (NC mimic/inhibitor and siNC) (RiboBio, Guangzhou, China). The level of RNA or protein was measured using qRT-PCR or western blot at 48 h after transfection, and the subsequent experiments were completed within 72 h.
Cell proliferation assay
Briefly, the cells were plated into a 96-well plate (Corning Life Sciences) at a density of 2,000 cells per well. After cultivating every 24 h, to each well was added 5 μL of CCK-8 kit solution (Dojindo, Tokyo, Japan) with 95 μL of serum-free medium, then incubated in the dark for 1.5–2 h. Finally, the absorbance of every well was measured at 450 nm using a multi-function enzyme-linked analyzer (BioTek, Winooski, VT, USA).
Colony formation assay
The cells were plated into a six-well plate (Corning Life Sciences) at a density of 500 cells per well and continuously cultured for about 2 weeks. After that, the colonies were successively fixed with absolute ethanol for 15 min and stained with crystal violet (0.5% w/v) for 30 min. Finally, the colonies per well were counted and imaged.
Cell migration assay
The cells were plated into a 12-well plate (Corning Life Sciences) at a density of 2 × 105 cells per well and continuously cultured until 100% density. Then, by using a sterile pipette tip, single-scratch wounds were made in the center of each well. Cell debris was washed clearly with phosphate-buffered saline (PBS). After incubating with serum-free medium for 24 or 48 h, the cell migration distance was measured and imaged.
A cell transwell assay was performed using 24-well transwells (6.5-mm pore size, Corning Life Sciences, Manassas, VA, USA). 8,000 cells were added to the upper chamber of each well in 100 μL of FBS-free medium, and 500 μL of medium with 10% FBS was added to the lower part of the chamber. Cells were fixed, stained, and imaged 24 h after incubation, and then the cell numbers were recorded.
Apoptosis analysis
An annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit (BD Pharmingen, San Diego, CA, USA) and an annexin V-allophycocyanin (APC)/7-aminoactinomycin D (7-AAD) apoptosis detection kit (KeyGEN, Nanjing, China) were used to measure the level of cell apoptosis, according to the manufacturers’ instructions. Apoptotic cells were analyzed using flow cytometer (Beckman Coulter).
Cell-cycle analysis
In brief, 2 × 105 cells were collected to fix in 75% ethanol overnight at −20°C. Then, the cells were collected by centrifugation, washed once with PBS, resuspended with 250 μL of RNase A (100 ng/mL; Sigma-Aldrich) for 20 min, and stained with propidium iodide (PI) (50 ng/mL; Sigma-Aldrich) for 15 min in the dark. Next, the processing cells were measured for cell-cycle distribution via a flow cytometer (Beckman Coulter).
Protein extraction and western blot
Total protein was extracted from the cells or exosome using Radioimmunoprecipitation (RIPA) lysis buffer (CWBIO, Beijing, China) and normalized with a Protein BCA Assay Kit (Bio-Rad). Equal amounts of protein were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride membrane (PVDF; Millipore). Then, the PVDF was blocked in 5% non-fat powdered milk for 1.5 h at room temperature, followed incubated with antibodies against AR (rabbit monoclonal antibody; 5153S; Cell Signaling Technology, Danvers, MA, USA), CD63 (rabbit monoclonal antibody; ab134045; Abcam, California, USA), Tsg101 (mouse monoclonal antibody; sc-7964; Santa Cruz Biotechnology, USA) and GAPDH (rabbit monoclonal antibody; 5174; Cell Signaling Technology, Danvers, MA, USA) overnight at 4°C, respectively.
Next, after washing with Tris-buffered saline Tween 20 (TBST) 3-4 times, the PVDF were incubated with a goat-anti-rabbit secondary antibody (1:10000, Cell Signaling Technology) conjugated to horseradish peroxidase (HRP) for 1.5 h at room temperature, followed being washed with TBST 3-4 times. Finally, protein bands were detected with a chemiluminescent horseradish peroxidase substrate (Millipore) and exposed using an E-Gel Imager (Tanon, Shanghai, China).
Exosome isolation and characterization
Briefly, we first planted equivalent cells into 10-cm plates (Corning Life Sciences) to ensure that the cell density reached about 80%. We changed the culture medium without FBS after repeated rinsing twice with PBS. After 48 h, cell culture media were collected and centrifuged at 2,000 × g for 20 min to remove cellular debris. Subsequently, supernatants were successively centrifuged at 5,000 × g for 15 min and at 12,000 × g for 30 min. Thereafter, supernatants were filtered using 0.22-μm filters (Millipore) and then centrifuged at 5,000 × g for 10 min with an ultrafiltration tube (Ultracel-100 regenerated cellulose membrane, 15-mL sample volume; UFC910024; Millipore). The superficial fluid of the ultrafiltration tube contained exosomes. All centrifugation steps were performed at 4°C, and isolated exosomes were stored at −80°C. The representative marker of exosomes, CD63, was assayed by western blot assay. Furthermore, the morphology, concentration, and size distribution of exosomes were detected by TEM or NTA, respectively.
Effect of miR-224-5p-enriched exosomes on NSCLC
The exosomes (100 μg/mL) and serum-free medium were incubated with NSCLC and seeded onto 7-cm plates for 24 h until the cells were grown to an appropriate density.
The expression of miR-224-5p in NSCLC after co-incubation was measured by qRT-PCR. The influence of exosomes on proliferation and migration of NSCLC was analyzed as before.
Dual-luciferase reporter assay
The 3′ UTR of AR containing two-stage binding sites of miR-224-5p was cloned downstream of the firefly luciferase reporter gene in the pGL3 miReport vector (Promega, Madison, WI, USA) and defined as pGL3-AR-WT-3′ UTR-1/2. Then, the pGL3-AR-Mut-3′ UTR-1/2 vectors were also constructed by interval site-directed mutagenesis with the corresponding mutated primer pairs. All of the above-mentioned recombinant vectors were confirmed by DNA sequencing (Sangon Biotech, Shanghai, China). Primer sequences are listed in Table S2.
HEK293T cells were plated in 24-well plates (Corning Life Sciences) until the cell density reached about 50%. Subsequently, the cells were transiently co-transfected with a final concentration of 50 nM miR-224-5p mimic or NC mimic (RiboBio), 200 ng of luciferase reporter vectors pGL3-AR-WT-3′ UTR-1/2 or pGL3-AR-Mut-3′ UTR-1/2, and 15 ng of the Renilla luciferase expression vector (pRL; Promega, Madison, WI, USA) as the control for checking co-transfection efficiency. Following the manufacturer’s instructions for an Orion II microplate illuminometer (Titertek-Berthold, South San Francisco, CA, USA), the luciferase activities of the cells were detected after 48 h post-transfection. The relative firefly luciferase activities were normalized to Renilla luciferase activities and expressed as the fold change in luciferase activity.
Subcutaneous tumor xenograft assay and metastatic assay
The 6-week-old female nude mice were purchased from the SLRC Laboratory Animal Center (Shanghai, China) and maintained in a specific pathogen-free condition. Ten nude mice were randomly assigned into two groups (five mice per group). 5 × 106 A549 cells stably transfected with pLenti or pLenti-miR-224 (resuspended in 100 μL of pure DMEM medium) were injected subcutaneously into the right flank of two groups nude mice. To investigate the metastatic ability, two groups were simultaneously injected with 2.5 × 106 A549 cells (resuspended in 200 μL of pure DMEM medium) via the tail vein. Tumor sizes were measured weekly with a Vernier caliper, and tumor volumes were calculated using the following formula: volume = (length × width × width)/2. Mice were sacrificed until 8 weeks post-injection, and then the xenograft tumors and bilateral lung tissues were excised. Ten tumor tissues and bilateral lung tissues were subjected to serial sectioning, hematoxylin and eosin staining, or immunohistochemistry. All experimental protocols were approved by the Institutional Animal Care and Use Committee of Shanghai University (Shanghai, China).
Immunohistochemistry
Primary tumor tissues were fixed with 10% formalin, embedded in paraffin, and cut into slices of about 4-μm thickness. Following deparaffinization and dehydration by xylene and graded alcohols, the slices were subsequently rehydrated with demineralized water. Then, the slices were placed in boiled antigen retrieval buffer for 5 min and incubated in 3% H2O2 solution for 25 min at room temperature in the dark. Next, the slices were treated with primary antibodies against Ki67 (1:200; Servicebio, Wuhan, China), E-cadherin, N-cadherin, and AR (1:500 to 1:200, Cell Signaling Technology), followed by application of goat anti-rabbit horseradish peroxidase (HRP)-conjugated antibodies, staining with a 3,3′-diaminobenzidine reaction solution, and imaging using a digitalized microscope camera.
Statistical analysis
Data are expressed as the mean ± SEM. Differences between two experimental groups were evaluated using a double-sided t test with statistical significance set as p < 0.05. GraphPad Prism 5 software was used to make graph presentations. Flow cytometry data were analyzed using FlowJo 7.6.2. Every experiment was performed at least three times with independent biological replicates.
Acknowledgments
We acknowledge Linda (Ningjing) Ma (Mount Holyoke College) for critical reading of the manuscript. This work was supported by Shanghai Chest Hospital (no. YJXT20190208). This work was also supported by the Scientific Research Project Foundation of Shanghai (no. 20S11901300) and the National Natural Science Foundation of China (no. 82073285). All laboratory members are acknowledged for stimulating discussions.
Author contributions
Z.M., W.F., and J.Z. conceived the experiments; J.Z., H.W., Q.S., and X.L. developed methodology; J.Z., H.W., X.L., Z.W., and X.W. performed the experiments; J.Z., H.W., X.L., and Z.W. analyzed and interpreted the data; J.Z. wrote, and H.W., Z.M., and W.F. edited, the manuscript. All authors read and approved the contents of the manuscript and its publication.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.01.028.
Contributor Information
Wentao Fang, Email: vwtfang@hotmail.com.
Zhongliang Ma, Email: zlma@shu.edu.cn.
Supplemental information
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chen Z., Fillmore C.M., Hammerman P.S., Kim C.F., Wong K.K. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat. Rev. Cancer. 2014;14:535–546. doi: 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herbst R.S., Morgensztern D., Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 4.Reck M., Blais N., Juhasz E., Gorbunova V., Jones C.M., Urban L., Orlov S., Barlesi F., Kio E., Keilholz U. Smoking history predicts sensitivity to PARP inhibitor veliparib in patients with advanced non-small cell lung cancer. J. Thorac. Oncol. 2017;12:1098–1108. doi: 10.1016/j.jtho.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 5.Hansen O., Schytte T., Nielsen M., Brink C. Age dependent prognosis in concurrent chemo-radiation of locally advanced NSCLC. Acta Oncol. 2015;54:333–339. doi: 10.3109/0284186X.2014.958529. [DOI] [PubMed] [Google Scholar]
- 6.Harichand-Herdt S., Ramalingam S.S. Gender-associated differences in lung cancer: clinical characteristics and treatment outcomes in women. Semin. Oncol. 2009;36:572–580. doi: 10.1053/j.seminoncol.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Ruivo C.F., Adem B., Silva M., Melo S.A. The biology of cancer exosomes: insights and new perspectives. Cancer Res. 2017;77:6480–6488. doi: 10.1158/0008-5472.CAN-17-0994. [DOI] [PubMed] [Google Scholar]
- 8.Fang T., Lv H., Lv G., Li T., Wang C., Han Q., Yu L., Su B., Guo L., Huang S. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat. Commun. 2018;9:191. doi: 10.1038/s41467-017-02583-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H., Deng T., Liu R., Bai M., Zhou L., Wang X., Li S., Wang X., Yang H., Li J. Exosome-delivered EGFR regulates liver microenvironment to promote gastric cancer liver metastasis. Nat. Commun. 2017;8:15016. doi: 10.1038/ncomms15016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skotland T., Sandvig K., Llorente A. Lipids in exosomes: current knowledge and the way forward. Prog. Lipid Res. 2017;66:30–41. doi: 10.1016/j.plipres.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Melo S.A., Sugimoto H., O’Connell J.T., Kato N., Villanueva A., Vidal A., Qiu L., Vitkin E., Perelman L.T., Melo C.A. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng Z., Li Y., Pan Y., Lan X., Song F., Sun J., Zhou K., Liu X., Ren X., Wang F. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat. Commun. 2018;9:5395. doi: 10.1038/s41467-018-07810-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng Z., Rong Y., Teng Y., Zhuang X., Samykutty A., Mu J., Zhang L., Cao P., Yan J., Miller D., Zhang H.G. Exosomes miR-126a released from MDSC induced by DOX treatment promotes lung metastasis. Oncogene. 2017;36:639–651. doi: 10.1038/onc.2016.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song Y., Dou H., Li X., Zhao X., Li Y., Liu D., Ji J., Liu F., Ding L., Ni Y., Hou Y. Exosomal miR-146a contributes to the enhanced therapeutic efficacy of interleukin-1β-primed mesenchymal stem cells against sepsis. Stem Cells. 2017;35:1208–1221. doi: 10.1002/stem.2564. [DOI] [PubMed] [Google Scholar]
- 15.Cui R., Meng W., Sun H.L., Kim T., Ye Z., Fassan M., Jeon Y.J., Li B., Vicentini C., Peng Y. MicroRNA-224 promotes tumor progression in nonsmall cell lung cancer. Proc. Natl. Acad. Sci. USA. 2015;112:E4288–E4297. doi: 10.1073/pnas.1502068112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li S., Zhang J., Zhao Y., Wang F., Chen Y., Fei X. miR-224 enhances invasion and metastasis by targeting HOXD10 in non-small cell lung cancer cells. Oncol. Lett. 2018;15:7069–7075. doi: 10.3892/ol.2018.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui Y., Xu H.F., Liu M.Y., Xu Y.J., He J.C., Zhou Y., Cang S.D. Mechanism of exosomal microRNA-224 in development of hepatocellular carcinoma and its diagnostic and prognostic value. World J. Gastroenterol. 2019;25:1890–1898. doi: 10.3748/wjg.v25.i15.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng T.D., Darke A.K., Redman M.W., Zirpoli G.R., Davis W., Payne Ondracek R., Bshara W., Omilian A.R., Kratzke R., Reid M.E. Smoking, sex, and non-small cell lung cancer: steroid hormone receptors in tumor tissue (S0424) J. Natl. Cancer Inst. 2018;110:734–742. doi: 10.1093/jnci/djx260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skjefstad K., Grindstad T., Khanehkenari M.R., Richardsen E., Donnem T., Kilvaer T., Andersen S., Bremnes R.M., Busund L.T., Al-Saad S. Prognostic relevance of estrogen receptor α, β and aromatase expression in non-small cell lung cancer. Steroids. 2016;113:5–13. doi: 10.1016/j.steroids.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 20.Rades D., Setter C., Dahl O., Schild S.E., Noack F. The prognostic impact of tumor cell expression of estrogen receptor-α, progesterone receptor, and androgen receptor in patients irradiated for nonsmall cell lung cancer. Cancer. 2012;118:157–163. doi: 10.1002/cncr.26282. [DOI] [PubMed] [Google Scholar]
- 21.Dai C., Heemers H., Sharifi N. Androgen signaling in prostate cancer. Cold Spring Harb. Perspect. Med. 2017;7:a030452. doi: 10.1101/cshperspect.a030452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kono M., Fujii T., Lim B., Karuturi M.S., Tripathy D., Ueno N.T. Androgen receptor function and androgen receptor-targeted therapies in breast cancer: a review. JAMA Oncol. 2017;3:1266–1273. doi: 10.1001/jamaoncol.2016.4975. [DOI] [PubMed] [Google Scholar]
- 23.Fletcher C.E., Sulpice E., Combe S., Shibakawa A., Leach D.A., Hamilton M.P., Chrysostomou S.L., Sharp A., Welti J., Yuan W. Androgen receptor-modulatory microRNAs provide insight into therapy resistance and therapeutic targets in advanced prostate cancer. Oncogene. 2019;38:5700–5724. doi: 10.1038/s41388-019-0823-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li P., Chen J., Miyamoto H. Androgen receptor signaling in bladder cancer. Cancers (Basel) 2017;9:20. doi: 10.3390/cancers9020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kensler K.H., Poole E.M., Heng Y.J., Collins L.C., Glass B., Beck A.H., Hazra A., Rosner B.A., Eliassen A.H., Hankinson S.E. Androgen receptor expression and breast cancer survival: results from the Nurses’ Health Studies. J. Natl. Cancer Inst. 2019;111:700–708. doi: 10.1093/jnci/djy173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harlos C., Musto G., Lambert P., Ahmed R., Pitz M.W. Androgen pathway manipulation and survival in patients with lung cancer. Horm. Cancer. 2015;6:120–127. doi: 10.1007/s12672-015-0218-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berardi R., Morgese F., Santinelli A., Onofri A., Biscotti T., Brunelli A., Caramanti M., Savini A., De Lisa M., Ballatore Z. Hormonal receptors in lung adenocarcinoma: expression and difference in outcome by sex. Oncotarget. 2016;7:82648–82657. doi: 10.18632/oncotarget.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu T., Zhang X., Du L., Wang Y., Liu X., Tian H., Wang L., Li P., Zhao Y., Duan W. Exosome-transmitted miR-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Mol. Cancer. 2019;18:43. doi: 10.1186/s12943-019-0981-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Qin X., Guo H., Wang X., Zhu X., Yan M., Wang X., Xu Q., Shi J., Lu E., Chen W., Zhang J. Exosomal miR-196a derived from cancer-associated fibroblasts confers cisplatin resistance in head and neck cancer through targeting CDKN1B and ING5. Genome Biol. 2019;20:12. doi: 10.1186/s13059-018-1604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao X., Cai Y., Wang Z., He W., Cao S., Xu R., Chen H. Estrogen receptors promote NSCLC progression by modulating the membrane receptor signaling network: a systems biology perspective. J. Transl. Med. 2019;17:308. doi: 10.1186/s12967-019-2056-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nikolos F., Thomas C., Bado I., Gustafsson J.A. ERβ sensitizes NSCLC to chemotherapy by regulating DNA damage response. Mol. Cancer Res. 2018;16:233–242. doi: 10.1158/1541-7786.MCR-17-0201. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z., Li Z., Ding X., Shen Z., Liu Z., An T., Duan J., Zhong J., Wu M., Zhao J. ERβ localization influenced outcomes of EGFR-TKI treatment in NSCLC patients with EGFR mutations. Sci. Rep. 2015;5:11392. doi: 10.1038/srep11392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerratana L., Basile D., Buono G., De Placido S., Giuliano M., Minichillo S., Coinu A., Martorana F., De Santo I., Del Mastro L. Androgen receptor in triple negative breast cancer: a potential target for the targetless subtype. Cancer Treat. Rev. 2018;68:102–110. doi: 10.1016/j.ctrv.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 34.Sun Z., Yang S., Zhou Q., Wang G., Song J., Li Z., Zhang Z., Xu J., Xia K., Chang Y. Emerging role of exosome-derived long non-coding RNAs in tumor microenvironment. Mol. Cancer. 2018;17:82. doi: 10.1186/s12943-018-0831-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalluri R., LeBleu V.S. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977. doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qu L., Ding J., Chen C., Wu Z.J., Liu B., Gao Y., Chen W., Liu F., Sun W., Li X.F. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29:653–668. doi: 10.1016/j.ccell.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 37.Liang C., Zhang X., Wang H.M., Liu X.M., Zhang X.J., Zheng B., Qian G.R., Ma Z.L. MicroRNA-18a-5p functions as an oncogene by directly targeting IRF2 in lung cancer. Cell Death Dis. 2017;8:e2764. doi: 10.1038/cddis.2017.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H., Ma Z., Liu X., Zhang C., Hu Y., Ding L., Qi P., Wang J., Lu S., Li Y. miR-183-5p is required for non-small cell lung cancer progression by repressing PTEN. Biomed. Pharmacother. 2019;111:1103–1111. doi: 10.1016/j.biopha.2018.12.115. [DOI] [PubMed] [Google Scholar]
- 39.Li Y., Wang D., Li X., Shao Y., He Y., Yu H., Ma Z. miR-199a-5p suppresses non-small cell lung cancer via targeting MAP3K11. J. Cancer. 2019;10:2472–2479. doi: 10.7150/jca.29426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qi P., Li Y., Liu X., Jafari F.A., Zhang X., Sun Q., Ma Z. Cryptotanshinone suppresses non-small cell lung cancer via microRNA-146a-5p/EGFR axis. Int. J. Biol. Sci. 2019;15:1072–1079. doi: 10.7150/ijbs.31277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barile L., Vassalli G. Exosomes: therapy delivery tools and biomarkers of diseases. Pharmacol. Ther. 2017;174:63–78. doi: 10.1016/j.pharmthera.2017.02.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.