Abstract
Background
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) epidemic has resulted in thousands of infections and deaths worldwide. Several therapies are currently undergoing clinical trials for the treatment of SARS-CoV-2 infection. However, the development of new drugs and the repositioning of existing drugs can only be achieved after the identification of potential therapeutic targets within structures, as this strategy provides the most precise solution for developing treatments for sudden epidemic infectious diseases.
Summary
In the current investigation, crystal and cryo-electron microscopy structures encoded by the SARS-CoV-2 genome were systematically examined for the identification of potential drug targets. These structures include nonstructural proteins (Nsp-9; Nsp-12; and Nsp-15), nucleocapsid (N) proteins, and the main protease (Mpro).
Key Message
The structural information reveals the presence of many potential alternative therapeutic targets, primarily involved in interaction between N protein and Nsp3, forming replication-transcription complexes (RTCs) which might be a potential drug target for effective control of current SARS-CoV-2 pandemic. RTCs consist of 16 nonstructural proteins (Nsp1-16) that play the most essential role in the synthesis of viral RNA. Targeting the physical linkage between the envelope and single-stranded positive RNA, a process facilitated by matrix proteins may provide a good alternative strategy. Our current study provides useful information for the development of new lead compounds against SARS-CoV-2 infections.
Keywords: Nsp-9, Nsp12, Nsp-15, Mpro, Inhibitors, Targets
Introduction
Public health emergencies, in the form of infectious disease outbreaks, epidemics, and pandemics, represent an increasing risk to the world's population. Management requires coordinated responses, across many disciplines and nations, and the capacity to gather proper national and global public health education, infrastructure, and preventive measures. According to the WHO, viral diseases represent a major threat to public health and are continuously emerging. Several viral epidemics have been observed over the past 20 years, and these include severe acute respiratory syndrome coronavirus (SARS-CoV) from 2002 to 2003 and H1N1 influenza in 2009. In 2012, the Middle East respiratory syndrome CoV (MERS-CoV) emerged in Saudi Arabia [1, 2].
Among the infectious agents, HIV emerged as a global threat in the late twentieth century. AIDS, a retrovirus, was first identified in the early 1980s as a disease [3, 4]. HIV-1 was identified as the causative agent, infecting CD4+ T cells. Advances in understanding the molecular biology of HIV together with global effort have led lifesaving screening, surveillance, and antiretroviral therapy [5, 6]. Later on, another viral epidemic in the form of MERS-CoV was found emerged as a threat to global health. A total of 229 MERS-CoV cases, including 70 deaths (30.5%), were recorded in the disease outbreak. The case fatality rate was recorded as 30.5% (70/229). The MERS-CoV occurrence was higher among men than women [7]. Although sufficient research has been conducted to investigate the molecular structure and epidemiology in these previous outbreaks, no significant global health security has been improved nor future possible outbreaks warning has been predicted. In the year 2003, SARS disease caused by a SARS-associated coronavirus emerged. In February 2003, the infectious agent emerged in China and rapidly spread to 4 other countries. WHO, with the assistance of the Global Outbreak Alert and Response Network and international investigation, worked with health system in affected countries to provide clinical support, but the epidemic of SARS spread among 26 countries, which >8,000 cases reported in 2003 (https://www.who.int/ith/diseases/sars/en/). However, no significant improvement has been observed in the form of vaccine to prevent further human loss in future.
Among all viral epidemics, the deadliest 1 emerged in the form of coronavirus disease 2019 (COVID-19), caused by SARS-CoV-2. The primary disease epicenter is currently Wuhan, China, its transmission and spread can be observed globally, where this disease affects >200 countries. This outbreak is affecting the world economy in a drastic manner, as millions of people, including all types of workers, are being quarantined. Currently, no vaccine or drug has been approved to treat COVID-19; however, several treatment options, such as the use of effective antiviral medications, are under consideration [8].
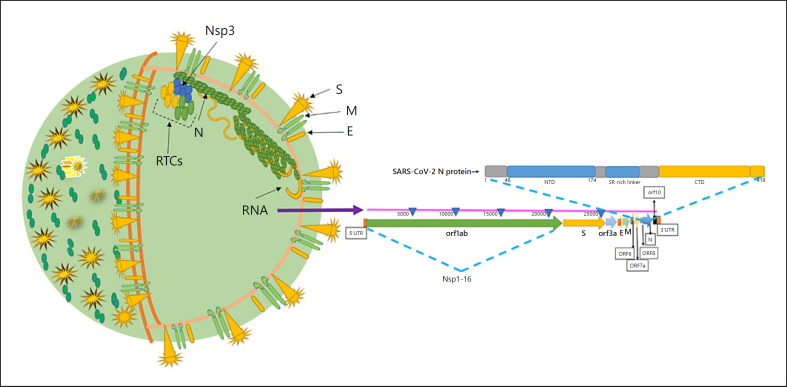
SARS-CoV-2 is a single-stranded positive RNA (+ssRNA) virus of the betacoronavirus family, and which also includes MERS-CoV and SARS-CoV. Coronaviruses are enveloped and nonsegmented positive-sense RNA viruses that belong to the order nidovirales. Among RNA viruses, coronaviruses possess the largest genome (30 kb) that contains structural and accessory genes, ample replicase, and other nonstructural proteins (Nsps) [9, 10, 11, 12]. Two-thirds of the SARS-CoV-2 genome consists of the ORF1a/b region that is translated into 2 polyproteins, pp1a (Nsp1-Nsp11) and pp1ab (Nsp1-Nsp16) [13]. The 4 structural proteins that include envelope (E), matrix (M), nucleocapsid (N) phosphoprotein, and spike (S) function together with the viral RNA and Nsp1-16 to facilitate the replication of the virus within the host cell. The 4 structural proteins in combination with the viral ssRNA genome and the E constitute the complete virion (Fig. 1).
Fig. 1.
Structure of SARS-CoV-2 and its genome organization [40, 92, 93, 94]. N protein is a major facilitator of viral replication within host cells, where it interacts with viral RNA during replication to form the virion after attachment to Nsp3 of the RTCs. RNA also interacts with M proteins via N. RTCs facilitate viral RNA replication. SARS-CoV-2, severe acute respiratory syndrome coronavirus-2; RTC, replication-transcription complex; Nsp, nonstructural protein; S, spike, E, envelope; M, matrix; EM, electron microscopy.
Crystal and cryo-electron microscopy (EM) structure are three-dimensional structures that are determined by X-ray crystallography and nuclear magnetic resonance, and these structures can be representative of proteins and nucleic acids. These structures provide a powerful tool in the elucidation of the three-dimensional structure of a molecule at atomic resolution [14, 15, 16, 17, 18, 19]. They are also favored for proteins that can provide detailed and accurate information for future research in drug design and development. In the present study, we analyzed the structures of 1 structural protein, N, and 3 Nsps that included Nsp-15, Nsp-9, and the main protease ([Mpro] Nsp5) of SARS-CoV-2 that were recently released by the protein data bank to provide for a broader range of possible target identification. All crystal and cryo-EM structures were extracted from protein data banks [20, 21] and were carefully analyzed using PyMOL and Chimera [22, 23]. Structural-related data were also analyzed, and the interacting partners of N, Nsp-15, and Nsp-9 were investigated and compared to SARS-CoVs for potent therapeutic target identification.
Nucleocapsid Protein
Current antiviral drugs have been developed by targeting the S protein. However, the strategy of targeting the S protein does possess some limitations. In certain cases, a number of viruses that possess mutations within the S protein have been observed [24, 25, 26], and these mutant viruses are prone to resist therapy due to an altered manner of attachment to the host-cell receptor that results in changes in binding patterns. Furthermore, in cases involving the Mpro, nonspecific binding may result in antiviral activity and may act on homologs of the cellular protease, ultimately resulting in host-cell toxicity and severe adverse effects. To avoid these limitations, SARS-CoV-2 essential proteins and RNA must be specifically targeted by novel antiviral strategies.
Among the major targets, the N is an important protein that is involved in RNA binding and is essential for RNA activities such as replication. It also plays an integral role in host-cell metabolism and RNA packaging when the most essential viral processes, including replication and transcription, are at their peak, and N protein also modulates the activities of infected cells [27, 28, 29]. The N protein primarily promotes the binding and packing of the RNA ribonucleoprotein complex (N) [30, 31, 32, 33].
To facilitate replication and transcription of the viral genome [28, 34], the N protein also maintains a highly ordered RNA conformation that is suitable for RNA activities Figure 1. Previous studies indicated that N protein is responsible for the regulation of host-cell cycle progression, host-pathogen interactions, actin reorganization, and apoptosis [35, 36]. Additionally, the N protein is extremely immunogenic, induces protective immune responses, and is abundantly expressed during infection [37, 38].
The N-phosphoprotein consists of N-terminal (N-NTD) and C-terminal (CTD) domains. Both of these domains exhibit RNA-binding affinity, while the CTD also binds the M protein to establish the physical linkage between the E and ssRNA. The SARS-CoV N proteins also play regulatory roles in the viral life cycle through the use of host intracellular machinery [30]. The N proteins have been demonstrated as key for incorporating viral RNA into viral progeny particles.
Recent studies have shown that the structure of the N protein includes a right hand-like fold composed of a β-sheet core with an extended central loop. The core region adopts a five-stranded U-shaped right-handed antiparallel β-sheet platform with the topology β4-β2-β3-β1-β5, and this structure is flanked by 2 short α-helices. A prominent feature of the structure is a large extending loop that exists between β2–β3 and forms a long basic β-hairpin (β2′ and β3′) [39, 40].
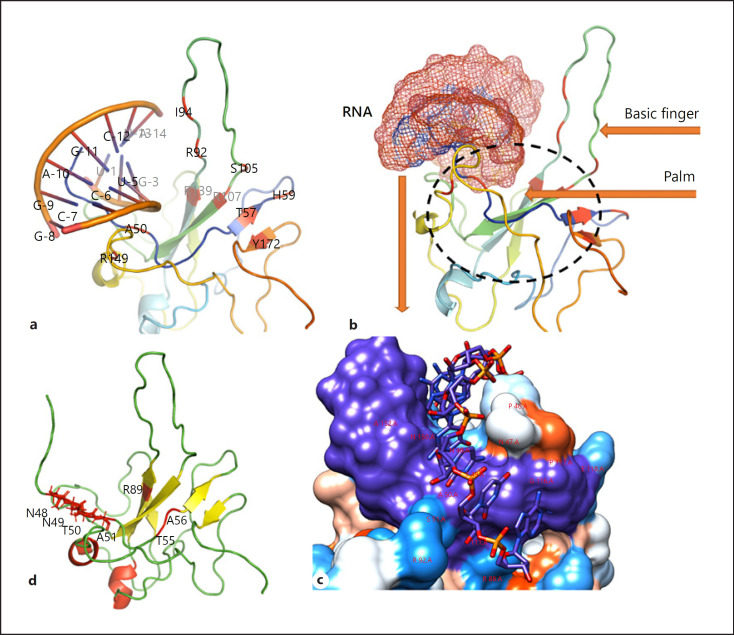
The crystal structure of the RNA [41] and the N protein was docked using HADDOCK [42]. The RNA duplex was found to be bound between the basic finger and the palm of the N-NTD that contains highly positive arginine residues (R92, R107, and R149) that directly contact the RNA. The model predicts several hydrophobic interactions with side chains of residues I94 and L104 that contribute to RNA binding. Residues A50, T57, H59, R89, R92, I94, S105, R107, R149, and Y172 in the surrounding areas may interact with RNA (Fig. 2a). In vitro studies identified several features of the N-NTD, and these revealed the specific targeting of antiviral agents to SARS-CoV-2 [40] at areas that included conserved regions, N-NTD RNA-binding residues, CTD dimerization, and central Ser/Arg (SR)-rich linker regions. The specific roles of each of these regions have been previously described, where N-NTD exhibits RNA-binding affinity, CTD facilitates oligomerization, and SR-rich linkers perform primary phosphorylation roles [43, 44, 45, 46, 47]. Each of these may represent a potential drug target that could be targeted by antiviral inhibitors.
Fig. 2.
Structural organization of N proteins. a RNA-binding location and residues involved in interactions. b Structural components of N proteins (basic finger and palm) that facilitate RNA binding. Surface representation of RNA binding (c) N-terminal tail residues N48, N49, T50, and A51 (d). G phosphate moiety recognition residues T55, A56, and R89. N, nucleocapsid proteins; G, guanosine.
Several residues were identified within the SARS-CoV-2 N-NTD and are involved in the ribonucleotide binding domain functions (Fig. 2). The N-terminal tail residues, N48, N49, T50, and A51 Figure 1d, appear to be more flexible and can open the binding pocket that allows for interaction with SARS-CoV-2 RNA. Residues R89, T55, and A56 are involved in guanosine (G) phosphate moiety recognition [40]. A more recent study [27] reported that the interaction of N proteins with the Nsp3 component of replication-transcription complexes (RTCs), a CoV-RNA synthesis site, plays an essential role in replication (Fig. 1).
In conclusion, inhibitors may be designed to target the RNA-binding site residues. Furthermore, both NTD and CTD are important targets, as they facilitate viral RNA binding and replication. M protein-binding residues may also be a potential target that could be used to break the physical linkage between the E and ssRNA. Inhibition of the N protein-Nsp3 interaction may also block viral replication. In a broader sense, RTCs, which consist of 16 Nsps, play the most essential role in the synthesis of viral RNA and may provide good targets. N proteins may be used as vaccines due to their conservation and immunogenic properties.
Nonstructural Protein 15
The ORFs rep1a and rep1b of SARS-CoV-2 RNA encode 2 large polypeptides (pp1a and pp1ab) [13] that are processed into 3C-like protease and papain-like protease, also called Nsp5 and Nsp3. Processing of pp1a and pp1ab results in 16 viral Nsps [48] that assemble into RTCs. These complexes are involved in multiple functions that range from replication to the processing of polypeptides [49, 50, 51, 52, 53, 54, 55, 56, 57].
Among all the Nsps, Nsp15 is a nidoviral RNA uridylate-specific endoribonuclease (NendoU) that belongs to the EndoU family and possesses a C-terminal catalytic domain. EndoU enzymes exist in all types of organisms and function as RNA endonucleases on phosphodiesters and hydroxyl termini [58]. The prototypic member is XendoU (Xenopus laevis), and it is involved in small nucleolar RNA maturation [59, 60, 61, 62].
NendoU is conserved in several viruses, including coronaviruses, and its function is to respond to the innate immune system [63]. In addition to EndoU activity, evidence in animal models suggests for an immunomodulating property during early viral infection. Nsp15 is playing a leading role in suppressing the IFN-α/β associated innate immune response and avoiding detection of viral mRNA. Nsp15 is a critical component in the life cycle of the coronavirus, is well conserved, and (88% sequence identity) exhibits high similarity (95%) to SARS-CoV-2 and its closest homolog from SARS-CoV [62].
Structural studies have revealed that Nsp15s folds into dimers of trimers, which then form hexamers. A total of ∼345 residues combine into the monomeric unit, and this unit, further, folds into the N-NTD, the middle, and the C-terminal (NendoU) catalytic domain.
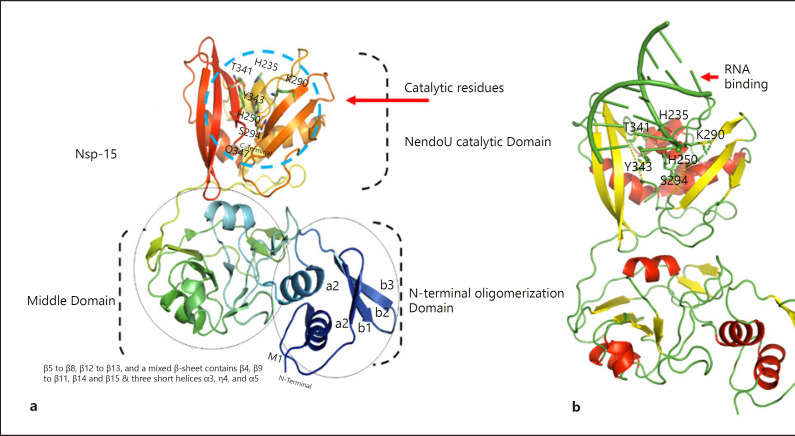
The structure of SARS-CoV-2 Nsp15 contains 1 chloride and 1 magnesium ion, 3 acetate ions, water molecules (346), and 8 molecules of glycerol. The structure of a given monomer possesses 3 distinct domains. The NTD consists of β1, β2, and β3 strands surrounding helices α1 and α2. The middle domain consists of β5–β8, β12–β13, and a mixed β-sheet containing β4, β9–β11, β14, and β15 along with 3 short helices (α3, η4, and α5). The NendoU domain containing the C-terminal catalytic region possesses 2 β-sheets with edges that contain the active sites. Monomers of NendoU join into a double-ring hexamer (Fig. 3. According to a previous report, hexamer creation is critical for proper catalytic activity. Stabilization of the hexamer requires the interaction of oligomers within each subunit domain and NTD oligomerization. The main differences between SARS-CoV-2 and SARS-CoV exist in the middle domains.
Fig. 3.
Domain organization of Nsp15 (a) Catalytic residues are indicated by blue circles (H235, H250, K290, T341, Y343, and S294). b RNA (PDB ID: 4u37) binding to the NendoU catalytic domain. RNA was docked using the HDOCK webserver [95]. Nsp, nonstructural protein.
NendoU Active Site
Nsp15 catalytic activity is facilitated by the NendoU domain. The active site residues, H235, H250, K290, T341, Y343, and S294, are conserved among SARS-CoVs. The main framework of the NendoU CTD and the active site residues (except K290) are well conserved among SARS-CoVs. Residues 235H, 250H, and 290K are involved in catalytic triad formation [64].
NendoU specificity is controlled by S294 and Y343, and these residues correspond to the base recognition roles of F120 and T45 (pyrimidine base B1 sub-site specific site) in RNAse A [65]. Mn2+ dependency has been suggested for nearly all members of the EndoU family with the exception of Nsp11, which did not show such a response [66]. Although the metal-binding site was not detected, it is still essential to note the lack of a protein/RNA complex structure. From the recently published structure of Nsp15 [62], there are 2 subunits in which the presence of electron density was observed at the peak near the active site, and this was uncertainly modeled as Mg2+, despite the poor coordination sphere. The metal ion is correlated by D283, S262, and P263 followed by R258, a side-chain residue in the neighborhood. These 4 amino acids are conserved, and they constitute the metal-binding site in SARS enzymes that are essential for holding and retaining the active site and configuration substrate during catalytic activity. NendoU binds RNA, while large substrates can approach these 6 sites from the site of the hexamer. These sites, together with metal binding, are potential drug targets (Table 1).
Table 1.
Recently released structures, their active sites, and potential drug targets
| Proteins or complex | PDB id | Function | Active site/essential residues | Ref. | Some other drug targets |
|---|---|---|---|---|---|
| N | 6YI3 | RNA binding | Ala50, Thr57, His59, R89, Arg92, Ile94, Ser105, Arg107, Arg149, and Tyr172 | [14, 43] | Nsp3-N interaction site RTCs |
| Nsp9 | 6W9Q | RNA binding | Asn33, Gly100, Met101, Val102, Leu103, Gly104, and Ser105 | [91] | Helical GxxxG interaction motif β2–3- and β3–4-loops are glycine rich and integral for RNA-binding |
| Nsp15 | 6VWW | RNA binding | His235, His250, Lys290, Thr341, Tyr343, and Ser294 | [65] | Ser294 and Tyr343 govern “U” specificity RNA base recognition residues, Phe120 and Thr45 |
| Mpro | 6M03, 6LU7 | Cleaving pp1a and pp1ab | Cys145 and His41 | [83] | Residues involved in dimerization Residues A285 and L286, render enhanced catalytic property Sequence recognition site LQ▼(S, A, and G) |
| RdRp | 7BV1 (apo RdRp complex) 7BV2 (template RNA and Remdesivir bound RdRp complex) |
SARS-CoV-2 RNA synthesis | S759, D760, and D761 | [86] | Nsp7-Nsp8 heterodimer Catalytic active center (residues 759–761) |
SARS-CoV-2, severe acute respiratory syndrome coronavirus-2; RTC, replication-transcription complex; N, nucleocapsid; Mpro, main protease; Nsp, nonstructural protein.
In conclusion, the hexamer clearly represents the active form of Nsp15. Structural comparisons between SARS and MERS-CoVs Nsp15s suggest that druggable compounds targeting Nsp15 might have a good chance of blocking SARS-CoV-2 replication inside the host cells. The CTD of NendoU contains a conserved active site (H235, H250, K290, T341, Y343, and S294), and along with the catalytic triad H235, H250, and K290, it is another potential site that can be targeted by inhibitors. Moreover, S294 and Y343 govern “U” specificity, which corresponds to the base recognition roles of F120 and T45, and the region containing these residues presents another attractive site for druggable compounds.
Nonstructural Protein 9
Nsp9-SARS plays a significant role in viral replication during the infection of human cells [67]. In various coronavirus counts, porcine delta virus (Nsp9 PDCoV), porcine epidemic diarrhea virus (Nsp9 PEDV), avian infectious bronchitis virus (Nsp9 IBV), human coronavirus 229E (Nsp9-HcoV), and SARS-Cov-2 (Nsp9-COV-19), and the homologs of the Nsp9 protein have been recognized. For long oligonucleotides, Nsp9-SARS appears to possess unobtrusive proclivity, thus restricting the idea that function is reliant upon oligomerization state [68, 69]. By means of a conserved α-helical “GxxxG” motif, the Nsp9-SARS dimerizes in solution. Interruption of key buildups within this process weakens RNA-binding [69]. Amongst beta coronaviruses, the order of Nsp9 homologues is preserved, and information regarding their residues can potentially provide useful contrasts among various viruses. Nsp9-COV-19 displays 97% sequence uniqueness to Nsp9-SARS; however, it displays only 44% similarity to Nsp9-HCoV.
The structure of apo-Nsp9-COV-19 displays an infrequent fold similar to other Nsp9 homolog folds that is currently thought to represent the exterior of coronaviruses [69]. From the center of a fold that exists as a small 6-stranded enclosed β-barrel, a sequence of stretched loops ventures outward. The discrete β-strands of the barrel are connected by elongated loops that exist alongside an anticipating N-terminal β-strand and C-terminal α1-helix, and the last 2 components make up the fundamental segments of the dimer interface. From the open face of the barrel, 2 loops extend, and both the β2–3- and β3–4-loops are glycine rich, intricate in RNA-binding, and are positively charged.
In various viruses, the organization of monomers in Nsp9-dimers is well preserved, and this is maintained inside Nsp9-COV-19. The self-connotation of the rationed GxxxG protein-protein restricting motif is the primary component of the inter-subunit interaction that facilitates backbone van der Waals interactions among interfacing copies of the C-terminal α1-helix [70]. These communications were reproduced after a complete helical turn by Gly104 within the separate chains, consequently shaping the molecular basis of the Nsp9-COV-19 dimer interface. When contrasting apo-Nsp9-COV-19 and 3C-Nsp9-COV-19, it is clear that the N-terminal interface strand responsible for separation is in close proximity to L9 (Fig. 4). Within the apo structure, van der Waals interactions occur through the side chains of N33, M101, and S105.
Fig. 4.
a Structure of Nsp9. b β2–3- and β3–4-loops are glycine rich and integral for RNA-binding. β2–3-and β3–4-loops are glycine rich and are involved in RNA-binding. c Secondary structure of Nsp9. Nsp, nonstructural protein.
The function of Nsp9 is important for the proliferation of SARS-CoV. It remains to be determined if Nsp9-COV-19 plays a comparable role in SARS-CoV-2; however, the observed 97% sequence similarity suggests a high level of practical preservation. The CoV-Nsp9 proteins apparently exist as dimers that contain a unique fold that that possesses an unusual α-helical GxxxG interaction motif, and this represents a good antiviral target, as it is required for viral replication. When examining Nsp9 antiviral targets, the side chains of N33, M101, and S105 are significant as it closely mimics the rationed protein-binding motif (33N, 100G, 101M, 102V, 103L, 104G, and 105S). Additionally, the β2–3- and β3–4-loops are glycine rich, integral for RNA-binding, positively charged, and exist as potential hot-spot regions.
Main Protease
One of the most attractive drug targets within SARS-CoV-2 is the Mpro(3CLpro) due to its vital role in processing the polyproteins translated from SARS-CoV-2 RNA. Mpro remains the best-characterized target protease (Mpro, also called 3CLpro) [71, 72, 73, 74, 75] along with papain-like protease(s) [48, 49, 76]. Mpro acts on approximately 11 cleavage locations within polyprotein 1ab. The sequence recognition at most sites consists of LQ▼(S, A, and G) (“▼” shows cleavage site). Inhibitors are likely to be nontoxic to human replication, as human proteases do not share similar cleavage specificity.
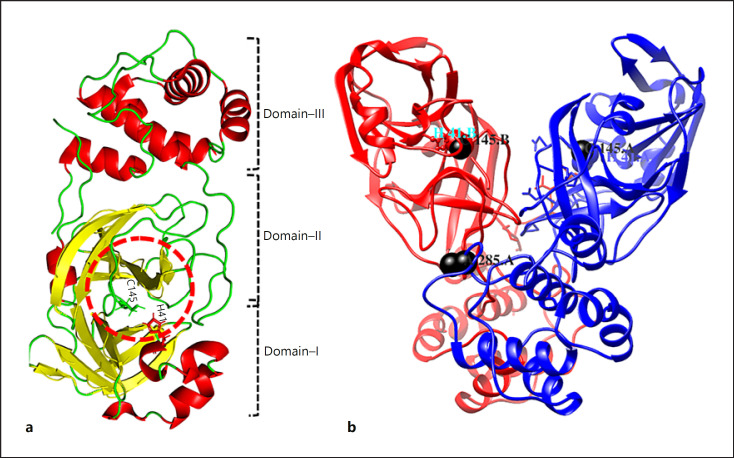
The substrate-binding sites, 3C protease-like residues 10–99 and 100–182 (domains I and II) in picornavirus, are six-stranded antiparallel β-barrels that harbor the substrate between them. Residues 198–303 form domain III that consists of 5 helices that regulate the dimerization of the Mpro between Glu290 and Arg4 of different protomers primarily through salt bridges [77]. Amino-acids C145 and H41 form the catalytic site. The Mpro of SARS-CoV-2 exhibits a tight dimer that creates a contact interface between domain II and the NH2-terminal amino acids (“N-finger”) of molecules A and B, respectively (Figure 5. Catalytic activity depends on the dimerization of the enzyme, as the N-finger interacts with Glu166 to facilitate the S1 pocket shape of the substrate-binding site [78].
Fig. 5.
Domain organization of Mpro. a The 3 domains are shown as domain I (residues 8–101), domain II (residues 102–184), and domain III (residues 201–303). b Dimers. Catalytic residues (C145 and H41) are circled. In SARS-CoV-2, T285 is replaced by A285 (black balls) and Ile286 is replaced by leucine. SARS-CoV-2, severe acute respiratory syndrome coronavirus-2
In CoV-2, residue T285 is substituted by A285, and I286 is substituted by L286 Figure 5 [79]. Substituting S284, T285, and I286 for alanine in Mpro led to a threefold increase in enzymatic activity [80].
The SARS-CoV Mpro (T285, I286) is different from SARS-CoV-2 (A285, L286) due to the catalytic properties conferred by the residue substitutions at 285 and 286. The catalytic properties may be reduced by designing active inhibitors against these locations. Inhibitors are more likely to be toxic if they block the cleavage site (LQ▼[S, A, and G]) specific to SARS-CoV-2, as human proteases do not share a similar cleavage specificity (Table 1). In a more recent study [81], 2 lead compounds have been synthesized (11a and 11b), targeting Mpro, and exhibited good activity as anti-SARS-CoV-2. The crystal structures of Mpro in complex with 11a or 11b demonstrated good binding affinity with C145 of SARS-CoV-2 Mpro. These compounds also exhibited a good pharmacokinetic result in vivo, suggesting that these are potential drug candidates.
RNA-Dependent RNA Polymerase
The polymerase enzymes called RNA-dependent RNA polymerase (RdRp) is playing a key role in corona viral transcription and replication assembly and thus seems as a foremost target for antiviral drug such as remdesivir [82]. Recently, the cryo-EM structure of SARS-CoV-2 RdRp has been released in the apo form (2.8 Å resolution) and in complex (2.5 Å resolution) with a 50-base template-primer RNA and remdesivir [83]. At the central channel of the RdRp, the partial double-stranded RNA template is inserted in Figure 6. This insertion is basically at the first replicated base pair and terminates chain elongation, where remdesivir is covalently incorporated into the primer strand. This structure gives basic bits of knowledge into the component of viral RNA replication and a balanced format for medicate configuration to battle the viral infection.
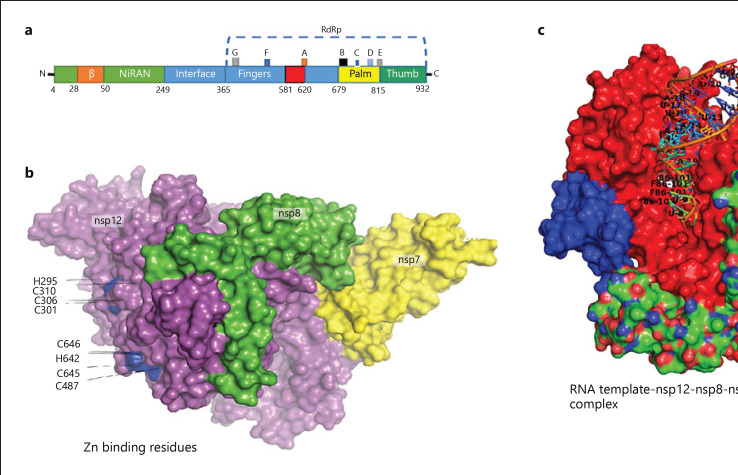
Fig. 6.
Structure assembly of Nsp12-Nsp7-Nsp8 complex in SARS-CoV-2. a Organization of SARS-CoV-2 domains. b Zn binding residues. c Complex structure containing active site residues for RNA template access and remdesivir (F88). SARS-CoV-2, severe acute respiratory syndrome coronavirus-2; Nsp, nonstructural protein.
The catalytic subunit (Nsp12) of an RdRp is the essential constituent of this complex. Nsp12 alone has a little action and its capacities require adornment factors including Nsp7 and Nsp8 [84], which increment RdRp template binding and processability. RdRp is likewise proposed to be the objective of a class of antiviral medications that are nucleotide analogs, including remdesivir [85]. The remdesivir is a prodrug that is converted to the active drug in the triphosphate form (RTP) [86]. The purified Nsp12 demonstrated little activity in binding to a 50-base partial double-stranded template-primer RNA [87]. The binding of Nsp12 to the template-primer RNA is dramatically expanded by the existence of Nsp7 and Nsp8.
Upon addition of adenosine triphosphate, the Nsp12-Nsp7-Nsp8 complex likewise indicated RNA polymerization activity on a poly-U template. By the addition of the active triphosphate form of remdesivir (RTP), this RNA polymerization activity was viably hindered. The apo RdRp complex is composed of unique structure that contains 1 Nsp12, 1 Nsp7, and 2 Nsp8. Unique in relation to the SARS-CoV RdRp structure, the SARS-CoV-2 RdRp structure additionally contains an N-terminal β-hairpin (residues 31–50), with 3 β-strands and 7 helices. An interface domain (residues 251–365) is subsequent the NiRAN domain and is comprised of 3 helices and 5 β-strands, which is associated with the RdRp domain (residues 366–920). The canonical cupped right-handed configuration is displayed by the Nsp12 RdRp domain, in which the finger subdomain (resides 397–581 and residues 621–679) creating a closed circle with the thumb subdomain (residues 819–920). Binding of Nsp7 and Nsp8 stabilizes the closed conformation, with 1 Nsp8 molecule sitting on the top of the finger subdomain and, furthermore, collaborating with the interface domain. The Nsp7-Nsp8 heterodimer further stabilizes the closed conformation of Nsp12, which is packed beside the thumb-finger interface. In the conserved metal-binding motifs, 2 zinc ions, which are also observed in the SARS-CoV RdRp structure have been assigned and are composed by H295, C301, C306, C310, C487, H642, C645, and C646. In keeping up the integrity of the RdRp architecture, these zinc ions likely serve as preserved structural components.
The template-RTP RdRp complex has a unique structure composed of 1 Nsp12, 1 Nsp7, and 1 Nsp8. In the final model, the second Nsp8 was not included as it was largely invisible in the EM map of the template-RTP complex. Furthermore, the template-RTP RdRp structure contains inhibitor remdesivir in its monophosphate form (RMP), and it also contains 14-base RNA in the template strand and 11-base RNA in the primer strand. At the primer strand, the inhibitor (RMP) is covalently linked, as well as 3 magnesium ions and a pyrophosphate that may attend as catalytic ions close to the active site. Although the 2 proteins (Nsp7 or Nsp8) are required for RNA binding by RdRp, surprisingly no RNA interactions are mediated by these proteins. The RMP is located at the 3′ end of the primer strand, which is covalently unified into the primer strand at the +1 location. Supplementary nucleotides interrelate with residues from the back of finger subdomain at the +2 and +3 locations of the template strand. Just a single RMP is assembled into the primer strand regardless of the presence of surplus RTP in complex assembly. Accordingly, remdesivir, in the same way as other nucleotide analog prodrugs, hinders the viral RdRp activity through nonobligate RNA chain termination, a process that necessitates the transformation of the parent medication to the active triphosphate form [88, 89]. The catalytic active center is formed by the SDD sequence (residues 759–761) in motif C. At the catalytic center, both D760 and D761 are engaged in coordination of the 2 magnesium ions. The location of motifs F and G is within the finger subdomain Figure 6a and both interrelate with the template strand RNA and direct this strand into the active site. Motif F, thus, stabilizes the incoming nucleotide in the correct position for catalysis as it can interact with the primer strand RNA with the side chains of K545 and R555 contacting the +1 base. Other than remdesivir, a few nucleotide analog drugs, counting galidesivir, favipiravir, EIDD-2801, and Ribavirin, effectively hinder SARS-CoV-2 replication in cell-based measures [90, 91]. These nucleotide analogs are proposed to repress the viral RdRp as remdesivir with the help of nonobligate RNA chain termination, a process that necessitates the alteration of the parent compound to the triphosphate active form.
Conclusion
Although there are many investigations underway to identify potential drugs and vaccines against the SARS-CoV-2 pandemic, drugs, and vaccines that possess desirable properties may only be designed when accurate target information is available. In the current review, multiple targets have been identified after careful analysis of recently released structures, specific to SARS-CoV-2. Despite identifying several drug targets in recent studies, we suggest that there are many more targets that are conserved and should urgently be considered when designing vaccines and potential inhibitors. Further studies are required to define the role of a number of important enzymes and complexes such as RTCs to allow for improved management of the current pandemic.
Conflict of Interest Statement
The authors declare that they have no competing interests.
Funding Sources
Dong-Qing Wei is supported by the Natural Science Foundation of Henan Province (162300410060), the Ministry of Science and Technology of China (2016YFA0501703), the Science and Technology Commission of Shanghai Municipality (19430750600), the National Natural Science Foundation of China (61503244, 61832019), and the Joint Research Funds for Medical and Engineering and Scientific research at Shanghai Jiao Tong University (YG2017ZD14).
Author Contributions
Conceptualization: D.Q.W. and M.T.K. Data curation: M.T.K., A.A., A.C., and S.C. Formal analysis: M.T.K., S.C., A.A., and M.I. Manuscript: A.A., M.I., M.T.K., A.A., H.A., and A.S.K. Funding acquisition: D.Q.W. Supervision and Approval: D.Q.W.
Acknowledgements
The current study was technically supported by Director PTRL KP, Peshawar, Dr. Sajid Ali, Molecular Biologist, and Dr. Taj Ali, Associate Professor, Kohat University of Science and Technology, Pakistan.
References
- 1.Nur SM, Hasan MA, Amin MA, Hossain M, Sharmin T. Design of potential RNAi (miRNA and siRNA) molecules for Middle East respiratory syndrome coronavirus (MERS-CoV) gene silencing by computational method. Interdiscip Sci. 2015 Sep;7((3)):257–65. doi: 10.1007/s12539-015-0266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nur SM, Hasan MA, Amin MA, Hossain M, Sharmin T. Design of potential RNAi (miRNA and siRNA) molecules for Middle East respiratory syndrome coronavirus (MERS-CoV) gene silencing by computational method. Interdiscip Sci. 2014 Nov; doi: 10.1007/s12539-014-0208-y. [DOI] [PubMed] [Google Scholar]
- 3.Khan U, Mehta R, Arif M, Lakhani O. Pandemics of the past: a narrative review. J Pak Med Assoc. 2020;((0)):1. doi: 10.5455/JPMA.11. [DOI] [PubMed] [Google Scholar]
- 4.Greene WC. A history of AIDS: looking back to see ahead. Eur J Immunol. 2007 Nov;37((Suppl 1)):S94–102. doi: 10.1002/eji.200737441. [DOI] [PubMed] [Google Scholar]
- 5.Fauci AS, Marston HD. Ending the HIV-AIDS pandemic: follow the science. N Engl J Med. 2015 Dec;373((23)):2197–9. doi: 10.1056/NEJMp1502020. [DOI] [PubMed] [Google Scholar]
- 6.Wood E, Montaner JS, Bangsberg DR, Tyndall MW, Strathdee SA, O'Shaughnessy MV, et al. Expanding access to HIV antiretroviral therapy among marginalized populations in the developed world. AIDS. 2003 Nov;17((17)):2419–27. doi: 10.1097/00002030-200311210-00003. [DOI] [PubMed] [Google Scholar]
- 7.Mobaraki K, Ahmadzadeh J. Current epidemiological status of Middle East respiratory syndrome coronavirus in the world from 1.1.2017 to 17.1.2018: a cross-sectional study. BMC Infect Dis. 2019 Apr;19((1)):351. doi: 10.1186/s12879-019-3987-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng J. SARS-CoV-2: an emerging coronavirus that causes a global threat. Int J Biol Sci. 2020 Mar;16((10)):1678–85. doi: 10.7150/ijbs.45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, et al. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020 Apr;5((4)):536–44. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biswas A, Bhattacharjee U, Chakrabarti AK, Tewari DN, Banu H, Dutta S. Emergence of novel coronavirus and COVID-19: whether to stay or die out? Crit Rev Microbiol. 2020 Mar;46((2)):182–93. doi: 10.1080/1040841X.2020.1739001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fung TS, Liu DX. Human coronavirus: host-pathogen interaction. Annu Rev Microbiol. 2019 Sep;73((1)):529–57. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- 12.Cascella M, Rajnik M, Cuomo A, Dulebohn SC, Di Napoli R. StatPearls. Treasure Island, FL: StatPearls Publishing; 2020. Features, evaluation and treatment coronavirus (COVID-19) [PubMed] [Google Scholar]
- 13.Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019 Mar;17((3)):181–92. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aitipamula S, Vangala VR. X-ray crystallography and its role in understanding the physicochemical properties of pharmaceutical cocrystals. J Indian Inst Sci. 2017 Jun;97((2)):227–43. [Google Scholar]
- 15.Picknett TM, Brenner S. X-ray crystallography. In: Brenner S, Miller JH, editors. Encyclopedia of genetics. New York, NY: Academic Press; 2001. p. p.2154. [Google Scholar]
- 16.Rondeau J-M, Schreuder H. Chapter 22: protein crystallography and drug discovery. In: Wermuth CG, Aldous D, Raboisson P, Rognan D, editors. The practice of medicinal chemistry. 4th ed. San Diego, CA: Academic Press; 2015. pp. 511–37. [Google Scholar]
- 17.Smyth MS, Martin JH. X ray crystallography. Mol Pathol. 2000 Feb;53((1)):8–14. doi: 10.1136/mp.53.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garbuzynskiy SO, Melnik BS, Lobanov MY, Finkelstein AV, Galzitskaya OV. Comparison of X-ray and NMR structures: is there a systematic difference in residue contacts between X-ray- and NMR-resolved protein structures? Proteins. 2005 Jul;60((1)):139–47. doi: 10.1002/prot.20491. [DOI] [PubMed] [Google Scholar]
- 19.Berg JM, Tymoczko JL, Stryer L. Three-dimensional protein structure can be determined by NMR spectroscopy and X-ray crystallography. Biochem 5th ed 2002. [Google Scholar]
- 20.Berman HM. The protein data bank: a historical perspective. Acta Crystallogr A. 2008 Jan;64((Pt 1)):88–95. doi: 10.1107/S0108767307035623. [DOI] [PubMed] [Google Scholar]
- 21.Berman HM, Bhat TN, Bourne PE, Feng Z, Gilliland G, Weissig H, et al. The protein data bank and the challenge of structural genomics. Nat Struct Biol. 2000 Jan;7((Suppl. 1)):957–9. doi: 10.1038/80734. [DOI] [PubMed] [Google Scholar]
- 22.DeLano WL. The PyMOL molecular graphics system. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
- 23.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem. 2004 Oct;25((13)):1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 24.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020 Mar;367((6483)):1260–3. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020 Apr;181((2)):281–e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia Y, Shen G, Zhang Y, Huang K-S, Ho H-Y, Hor W-S, et al. Analysis of the mutation dynamics of SARS-CoV-2 reveals the spread history and emergence of RBD mutant with lower ACE2 binding affinity. bioRxiv. 2020 Apr;:034942. [Google Scholar]
- 27.Cong Y, Ulasli M, Schepers H, Mauthe M, V'kovski P, Kriegenburg F, et al. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J Virol. 2020 Jan;94((4)) doi: 10.1128/JVI.01925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stohlman SA, Baric RS, Nelson GN, Soe LH, Welter LM, Deans RJ. Specific interaction between coronavirus leader RNA and nucleocapsid protein. J Virol. 1988 Nov;62((11)):4288–95. doi: 10.1128/jvi.62.11.4288-4295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson GW, Stohlman SA, Tahara SM. High affinity interaction between nucleocapsid protein and leader/intergenic sequence of mouse hepatitis virus RNA. J Gen Virol. 2000 Jan;81((Pt 1)):181–8. doi: 10.1099/0022-1317-81-1-181. [DOI] [PubMed] [Google Scholar]
- 30.Chang CK, Hou MH, Chang CF, Hsiao CD, Huang TH. The SARS coronavirus nucleocapsid protein–forms and functions. Antiviral Res. 2014 Mar;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McBride R, van Zyl M, Fielding BC. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014 Aug;6((8)):2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei DQ, Zhang R, Du QS, Gao WN, Li Y, Gao H, et al. Anti-SARS drug screening by molecular docking. Amino Acids. 2006 Jul;31((1)):73–80. doi: 10.1007/s00726-006-0361-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang R, Wei D-Q, Du Q-S, Chou K-C. Molecular modeling studies of peptide drug candidates against SARS [Internet] 2006 doi: 10.2174/157340606776930736. [DOI] [PubMed] [Google Scholar]
- 34.Tang TK, Wu MP, Chen ST, Hou MH, Hong MH, Pan FM, et al. Biochemical and immunological studies of nucleocapsid proteins of severe acute respiratory syndrome and 229E human coronaviruses. Proteomics. 2005 Mar;5((4)):925–37. doi: 10.1002/pmic.200401204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du L, Zhao G, Lin Y, Chan C, He Y, Jiang S, et al. Priming with rAAV encoding RBD of SARS-CoV S protein and boosting with RBD-specific peptides for T cell epitopes elevated humoral and cellular immune responses against SARS-CoV infection. Vaccine. 2008 Mar;26((13)):1644–51. doi: 10.1016/j.vaccine.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Surjit M, Liu B, Chow VT, Lal SK. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J Biol Chem. 2006 Apr;281((16)):10669–81. doi: 10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahmed SF, Quadeer AA, McKay MR. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020 Feb 25;12((3)):12. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin Y, Shen X, Yang RF, Li YX, Ji YY, He YY, et al. Identification of an epitope of SARS-coronavirus nucleocapsid protein. Cell Res. 2003 Jun;13((3)):141–5. doi: 10.1038/sj.cr.7290158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dinesh DC, Chalupska D, Silhan J, Veverka V, Boura E. Structural basis of RNA recognition by the SARS-CoV-2 nucleocapsid phosphoprotein. bioRxiv. 2020 Apr;:022194. doi: 10.1371/journal.ppat.1009100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang S, Yang M, Hong Z, Zhang L, Huang Z, Chen X, et al. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. bioRxiv. 2020 Mar;:977876. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheng J, Larsen A, Heuberger BD, Blain JC, Szostak JW. Crystal structure studies of RNA duplexes containing s(2)U:A and s(2)U:U base pairs. J Am Chem Soc. 2014 Oct;136((39)):13916–24. doi: 10.1021/ja508015a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003 Feb;125((7)):1731–7. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 43.Lo YS, Lin SY, Wang SM, Wang CT, Chiu YL, Huang TH, et al. Oligomerization of the carboxyl terminal domain of the human coronavirus 229E nucleocapsid protein. FEBS Lett. 2013 Jan;587((2)):120–7. doi: 10.1016/j.febslet.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen IJ, Yuann JM, Chang YM, Lin SY, Zhao J, Perlman S, et al. Crystal structure-based exploration of the important role of Arg106 in the RNA-binding domain of human coronavirus OC43 nucleocapsid protein. Biochim Biophys Acta. 2013 Jun;1834((6)):1054–62. doi: 10.1016/j.bbapap.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang CK, Chen CM, Chiang MH, Hsu YL, Huang TH. Transient oligomerization of the SARS-CoV N protein: implication for virus ribonucleoprotein packaging. PLoS One. 2013 May;8((5)):e65045. doi: 10.1371/journal.pone.0065045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang CK, Sue SC, Yu TH, Hsieh CM, Tsai CK, Chiang YC, et al. Modular organization of SARS coronavirus nucleocapsid protein. J Biomed Sci. 2006 Jan;13((1)):59–72. doi: 10.1007/s11373-005-9035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wootton SK, Rowland RR, Yoo D. Phosphorylation of the porcine reproductive and respiratory syndrome virus nucleocapsid protein. J Virol. 2002 Oct;76((20)):10569–76. doi: 10.1128/JVI.76.20.10569-10576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Báez-Santos YM, St John SE, Mesecar AD. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015 Mar;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014 Sep;281((18)):4085–96. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang H, Yang M, Ding Y, Liu Y, Lou Z, Zhou Z, et al. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc Natl Acad Sci U S A. 2003 Nov;100((23)):13190–5. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anand K, Ziebuhr J, Wadhwani P, Mesters JR, Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003 Jun;300((5626)):1763–7. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 52.Ratia K, Pegan S, Takayama J, Sleeman K, Coughlin M, Baliji S, et al. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc Natl Acad Sci U S A. 2008 Oct;105((42)):16119–24. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006 Apr;117((1)):17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LL, et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol. 2003 Aug;331((5)):991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thiel V, Ivanov KA, Putics A, Hertzig T, Schelle B, Bayer S, et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol. 2003 Sep;84((Pt 9)):2305–15. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 56.Marra MA, Jones SJ, Astell CR, Holt RA, Brooks-Wilson A, Butterfield YS, et al. The genome sequence of the SARS-associated coronavirus. Science. 2003 May;300((5624)):1399–404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 57.Brierley I, Digard P, Inglis SC. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989 May;57((4)):537–47. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ulferts R, Ziebuhr J. Nidovirus ribonucleases: structures and functions in viral replication. RNA Biol. 2011 Apr;8((2)):295–304. doi: 10.4161/rna.8.2.15196. [DOI] [PubMed] [Google Scholar]
- 59.Caffarelli E, Arese M, Santoro B, Fragapane P, Bozzoni I. In vitro study of processing of the intron-encoded U16 small nucleolar RNA in Xenopus laevis. Mol Cell Biol. 1994 May;14((5)):2966–74. doi: 10.1128/mcb.14.5.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Caffarelli E, Losito M, Giorgi C, Fatica A, Bozzoni I. In vivo identification of nuclear factors interacting with the conserved elements of box C/D small nucleolar RNAs. Mol Cell Biol. 1998 Feb;18((2)):1023–8. doi: 10.1128/mcb.18.2.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gioia U, Laneve P, Dlakic M, Arceci M, Bozzoni I, Caffarelli E. Functional characterization of XendoU, the endoribonuclease involved in small nucleolar RNA biosynthesis. J Biol Chem. 2005 May;280((19)):18996–9002. doi: 10.1074/jbc.M501160200. [DOI] [PubMed] [Google Scholar]
- 62.Kim Y, Jedrzejczak R, Maltseva NI, Wilamowski M, Endres M, Godzik A, et al. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci Publ Protein Soc. 2020 Jul;29((7)):1596–605. doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deng X, Hackbart M, Mettelman RC, O‘Brien A, Mielech AM, Yi G, et al. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc Natl Acad Sci U S A. 2017 May 23;114((21)):E4251–60. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ricagno S, Egloff MP, Ulferts R, Coutard B, Nurizzo D, Campanacci V, et al. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc Natl Acad Sci U S A. 2006 Aug;103((32)):11892–7. doi: 10.1073/pnas.0601708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.delCardayré SB, Raines RT. Structural determinants of enzymatic processivity. Biochemistry. 1994 May;33((20)):6031–7. doi: 10.1021/bi00186a001. [DOI] [PubMed] [Google Scholar]
- 66.Nedialkova DD, Ulferts R, van den Born E, Lauber C, Gorbalenya AE, Ziebuhr J, et al. Biochemical characterization of arterivirus nonstructural protein 11 reveals the nidovirus-wide conservation of a replicative endoribonuclease. J Virol. 2009 Jun;83((11)):5671–82. doi: 10.1128/JVI.00261-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frieman M, Yount B, Agnihothram S, Page C, Donaldson E, Roberts A, et al. Molecular determinants of severe acute respiratory syndrome coronavirus pathogenesis and virulence in young and aged mouse models of human disease. J Virol. 2012 Jan;86((2)):884–97. doi: 10.1128/JVI.05957-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Egloff MP, Ferron F, Campanacci V, Longhi S, Rancurel C, Dutartre H, et al. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc Natl Acad Sci U S A. 2004 Mar;101((11)):3792–6. doi: 10.1073/pnas.0307877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sutton G, Fry E, Carter L, Sainsbury S, Walter T, Nettleship J, et al. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure. 2004 Feb;12((2)):341–53. doi: 10.1016/j.str.2004.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu T, Chen C, Li H, Dou Y, Zhou M, Lu D, et al. Structural basis for dimerization and RNA binding of avian infectious bronchitis virus nsp9. Protein Sci. 2017;26((5)):1037–48. doi: 10.1002/pro.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Anand K, Yang H, Bartlam M, Rao Z, Hilgenfeld R. Coronaviruses special emphas first insights concerning S. Basel, Switzerland: Birkhuser Verlag; 2005. Coronavirus main proteinase: target for antiviral drug therapy; pp. p. 173–199. [Google Scholar]
- 72.Xue X, Yu H, Yang H, Xue F, Wu Z, Shen W, et al. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J Virol. 2008 Mar;82((5)):2515–27. doi: 10.1128/JVI.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liang P-H. Characterization and inhibition of SARS-coronavirus main protease. Curr Top Med Chem. 2006;6((4)):361–76. doi: 10.2174/156802606776287090. [DOI] [PubMed] [Google Scholar]
- 74.Pillaiyar T, Manickam M, Namasivayam V, Hayashi Y, Jung SH. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J Med Chem. 2016 Jul 28;59((14)):6595–628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gan YR, Huang H, Huang YD, Rao CM, Zhao Y, Liu JS, et al. Synthesis and activity of an octapeptide inhibitor designed for SARS coronavirus main proteinase. Peptides. 2006 Apr;27((4)):622–5. doi: 10.1016/j.peptides.2005.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Niemeyer D, Mösbauer K, Klein EM, Sieberg A, Mettelman RC, Mielech AM, et al. The papain-like protease determines a virulence trait that varies among members of the SARS-coronavirus species. PLoS Pathog. 2018;14((9)):e1007296. doi: 10.1371/journal.ppat.1007296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shi J, Song J. The catalysis of the SARS 3C-like protease is under extensive regulation by its extra domain. FEBS J. 2006 Mar;273((5)):1035–45. doi: 10.1111/j.1742-4658.2006.05130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Anand K, Palm GJ, Mesters JR, Siddell SG, Ziebuhr J, Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 2002 Jul;21((13)):3213–24. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020 Apr;368((6489)):409–12. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lim L, Shi J, Mu Y, Song J. Dynamically-driven enhancement of the catalytic machinery of the SARS 3C-like protease by the S284-T285-I286/A mutations on the extra domain. PLoS One. 2014;9((7)):e101941. doi: 10.1371/journal.pone.0101941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dai W, Zhang B, Jiang XM, Su H, Li J, Zhao Y, et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020 Jun;368((6497)):1331–5. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao L, et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020 May;368((6492)):779–82. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yin W, Mao C, Luan X, Shen D-D, Shen Q, Su H, et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020 Jun;368((6498)):1499–504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kirchdoerfer RN, Ward AB. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun. 2019 May;10((1)):2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30((3)):269–71. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Siegel D, Hui HC, Doerffler E, Clarke MO, Chun K, Zhang L, et al. Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2, 1-f][triazin-4-amino] adenine C-nucleoside (GS-5734) for the treatment of Ebola and emerging viruses. J Med Chem. 2017 Mar;60((5)):1648–61. doi: 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- 87.Subissi L, Posthuma CC, Collet A, Zevenhoven-Dobbe JC, Gorbalenya AE, Decroly E, et al. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci U S A. 2014 Sep;111((37)):E3900–9. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tchesnokov EP, Feng JY, Porter DP, Götte M. Mechanism of inhibition of Ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses. 2019 Apr;11((4)):326. doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gordon CJ, Tchesnokov EP, Woolner E, Perry JK, Feng JY, Porter DP, et al. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem. 2020 May;295((20)):6785–97. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sheahan TP, Sims AC, Zhou S, Graham RL, Pruijssers AJ, Agostini ML, et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med. 2020 Apr;12((541)):12. doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lu C-C, Chen M-Y, Chang Y-L. Potential therapeutic agents against COVID-19: what we know so far. J Chin Med Assoc. 2020 Apr;83((6)):534–6. doi: 10.1097/JCMA.0000000000000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chan JF, Kok KH, Zhu Z, Chu H, To KK, Yuan S, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020 Jan;9((1)):221–36. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Khailany RA, Safdar M, Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020 Jun;19:100682. doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020 Feb;395((10224)):565–74. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yan Y, Zhang D, Zhou P, Li B, Huang S-Y. HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017 Jul;45((W1)):W365–73. doi: 10.1093/nar/gkx407. [DOI] [PMC free article] [PubMed] [Google Scholar]