Abstract
Small interfering RNAs (siRNAs) therapeutically induce RNA interference (RNAi) of disease-causing genes, but they also silence hundreds of seed-matched off-targets as behaving similar to microRNAs (miRNAs). miRNAs control the pathophysiology of tumors, wherein their accessible binding sites can be sequenced by Argonaute crosslinking immunoprecipitation (AGO CLIP). Herein, based on AGO CLIP, we develop potent anticancer siRNAs utilizing miRNA-like activity (mi/siRNAs). The mi/siRNAs contain seed sequences (positions 2–7) of tumor-suppressive miRNAs while maintaining perfect sequence complementarity to the AGO-accessible tumor target sites. Initially, host miRNA interactions with human papillomavirus 18 (HPV18) were identified in cervical cancer by AGO CLIP, revealing tumor-suppressive activity of miR-1/206 and miR-218. Based on the AGO-miRNA binding sites, mi/siRNAs were designed to target E6 and E7 (E6/E7) transcript with seed sequences of miR-1/206 (206/E7) and miR-218 (218/E7). Synergistic anticancer activity of 206/E7 and 218/E7 was functionally validated and confirmed via RNA sequencing and in vivo xenograft models (206/E7). Other mi/siRNA sequences were additionally designed for cervical, ovarian, and breast cancer, and available as an online tool (http://ago.korea.ac.kr/misiRNA); some of the mi/siRNAs were validated for their augmented anticancer activity (206/EphA2 and 206/Her2). mi/siRNAs could coordinate miRNA-like activity with robust siRNA function, demonstrating the potential of AGO CLIP analysis for RNAi therapeutics.
Keywords: siRNA, miRNA, AGO CLIP, cervical cancer, ovarian cancer, breast cancer
Graphical Abstract
The gene silencing activity of siRNAs depends on target accessibility of Argonaute (AGO) and has a problem to induce miRNA-like repression of seed-matched off-targets. Based on AGO CLIP sequencing analyses, Gu and colleagues developed a robust strategy to design anticancer siRNAs (mi/siRNAs), which utilize AGO-accessible tumor target sites to augment tumor-suppressive miRNA-like activity.
Introduction
MicroRNAs (miRNAs) are small (about 22 nt), endogenous, non-coding RNAs that induce RNA interference (RNAi).1 miRNAs impart RNA-induced silencing complexes (RISCs) to suppress target transcripts by loading onto Argonaute (AGO) proteins. Guided by miRNA sequences, the AGO-miRNA complexes in RISCs recognize hundreds of target mRNAs through binding to at least 6 consecutive bp in the seed region (positions 2–8), thereby reducing their stability and/or translation.2 Spatiotemporal regulation of specific miRNAs is crucial to properly control diverse physiological functions; thus, any perturbation in miRNA expression could impair physiological homeostasis, thereby prompting various pathogeneses, including neurological disorders, cardiovascular diseases, and many types of cancer.3
Dysregulation of miRNA expression in human malignancies has been observed and is known to contribute to the hallmarks of cancer, ranging from tumor initiation to progression.4 Indeed, profiles of miRNA expression significantly correlate with cancer type, stage, and other clinical information. Therefore, miRNAs should be studied as biomarkers for cancer diagnosis and prognosis.5 miRNAs have been reported to function as either oncogenes (so-called oncomiRs; e.g., miR-17/926 and miR-217) or tumor suppressors (e.g., miR-346 and let-78), depending on the circumstances.9
The biological roles of miRNAs depend on the integrated functions of the target mRNAs; therefore, it is crucial to understand miRNA- target interactions in various pathogenesis. For this reason, high-throughput sequencing of RNAs isolated by crosslinking immunoprecipitation (HITS-CLIP, also called CLIP-esq)10 has been used for AGO (AGO HITS-CLIP, AGO CLIP-seq, or AGO CLIP),11 along with bioinformatics analysis12 to map AGO-associated miRNA targets, including noncanonical sites13,14 and new recognition sites caused by miRNA oxidation.15 AGO CLIP has been applied to various tumor cells, revealing critical miRNA-target interactions and their integrated functions in tumorigenesis.16, 17, 18
In contrast to miRNA, small interfering RNA (siRNA) typically induces RNAi for a single target gene, thereby triggering cleavage of its mRNA with perfect sequence complementarity.19 Due to this simple procedure for suppressing an intended target, siRNAs have been widely used to silence disease-related genes.20 However, by sharing the same RNAi effectors, siRNA loaded on AGO can also function as miRNA, repressing hundreds of off-targets by pairing with the seed region.21 The miRNA-like off-target effect is potent, often leading to inadvertent side effects and toxicity; therefore, for their use as siRNAs, miRNA-like off-targets must be evaluated and suitably controlled.22 To this end, chemical modifications such as abasic pivot substitution23,24 have been developed to avoid miRNA-like off-targeting and maintain on-target activity.25
Although miRNA-like activity is inevitable in siRNA, it can be advantageous when arranged in the seed sequence to kill cancer cells by targeting multiple survival genes.26 This is founded on the basis that the specific base compositions of the 6-mer seeds can be universally toxic by targeting 3ʹ untranslated regions (UTRs) of genes critical for cell survival in a miRNA-like fashion.27 A miRNA-siRNA chimera, a miR-200a seed containing siRNA against AKT1, has been empirically synthesized, thus proving its conceptual multifunctionality in the suppression of cancer.28 However, owing to its dependency on the prediction of miRNA target sites, this method guarantees neither miRNA-like activity nor effectiveness of siRNA functioning, which limits its use. Furthermore, the variable feasibility of siRNA targeting and the susceptibility of tumor-suppressive miRNAs have not been fully considered; therefore, concerns about differential AGO binding and miRNA activity must be addressed.
Therefore, in this study, we designed siRNA sequences from AGO CLIP results wherein AGO-miRNA target sites were confirmed to be susceptible to RNAi, and to have perfect complementary sequences to the seed-paired sites that could naturally determine the function as both siRNA and miRNA (i.e., mi/siRNA). As cervical cancer is known to be initiated by human papillomavirus (HPV) infections,29 the AGO binding sites in the critical oncogenes of human papillomavirus 18 (HPV18), E6, and E7 were initially identified for putative host antitumor miRNAs, miR-1/206,30, 31, 32, 33 and miR-218,34,35 thus enabling the delineation of mi/siRNAs (206/E7 and 218/E7) based on AGO CLIP.11 Then, the mi/siRNAs were further applied to putative oncogenes and tumor-suppressing miRNAs for ovarian and breast cancers and offered as a comprehensive web resource. By utilizing AGO CLIP, mi/siRNA design can be a general means for the development of RNAi medicine, where the miRNA-like off-target effect can be beneficially adjusted to culminate in synergistic roles with on-target repression.
Results
AGO CLIP analysis of HPV18 for cervical cancer
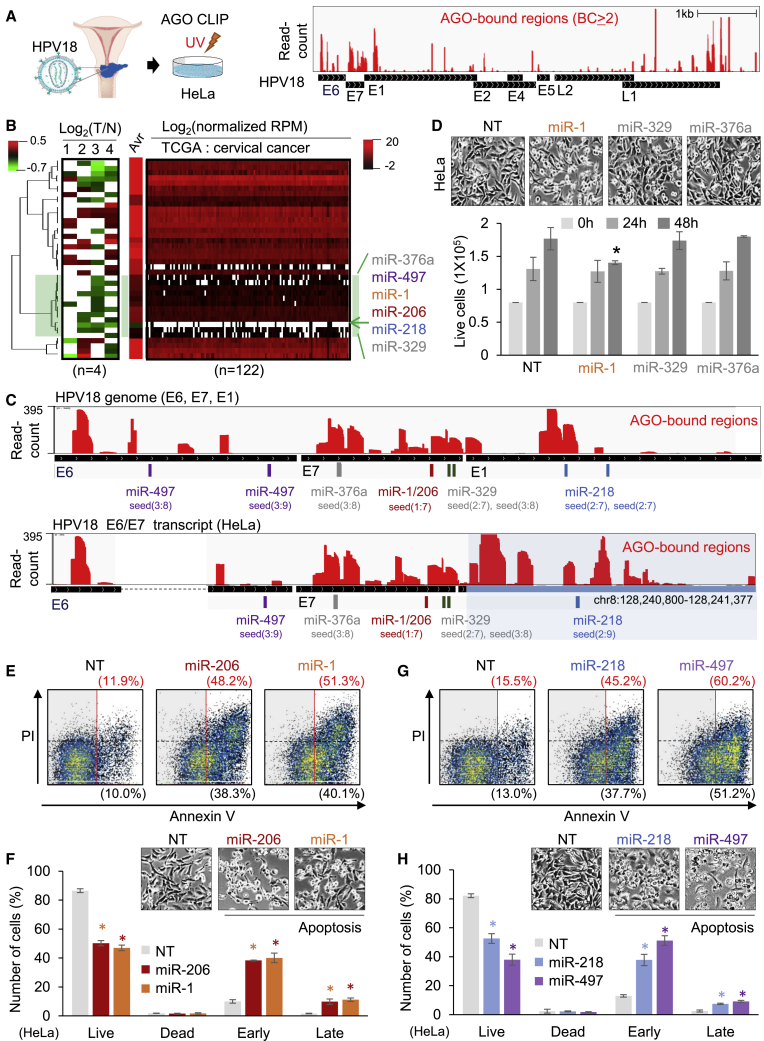
By analyzing the AGO CLIP results in HeLa cells,11 we initially attempted to assess global AGO interactions in HPV18-infected cervical cancer cells where some AGO-target interactions could lead to tumor suppression mediated by host miRNAs (Figure 1A). Since expression of high-risk HPV18 genes is crucial for the initiation and progression of cervical cancer,29 the HPV18 genome was the focus of our analyses (Figure S1A, left panel). We observed that HPV18 transcripts were associated with AGO11 (biological complexity [BC] ≥ 2; Figure 1A, right panel). To further distinguish AGO-miRNA interactions from antitumor activity, putative tumor-suppressor miRNAs were selected by their downregulation during tumorigenesis. In the results of the miRNA expression analyses, tumor-suppressing miRNAs (miR-1,30,33 miR-206,31,32 miR-218,34,35 miR-329,38 miR-376a,39 and miR-49740,41) were identified as a downregulated cluster in different cervical cancer cells (n = 4; Figure 1B, left panel) and patients (The Cancer Genome Atlas [TCGA]; n = 122; Figure 1B, right panel). Among them, miR-1 and miR-206 shared the same seed sequence (positions 2–8) as the miR-1/206 family and exhibited the most decreased expression in cervical cancers (Figure 1B).
Figure 1.
AGO CLIP analyses identified putative anticancer miRNAs associated with HPV18 in cervical cancer
(A) AGO CLIP performed in HeLa cells, a cell line derived from a cervical cancer patient with HPV18 infection (left panel). AGO-bound regions on which AGO CLIP reads were reproducibly mapped between biological replicates (biological complexity [BC] ≥ 2, CLIPick12) are represented on the HPV18 reference genome (GenBank: NC_001357; right panel). Detailed mapping statistics can be found in Figure S1A. (B) Cluster analyses of miRNA expression in different cervical cancer cells: (1) cervical cancer versus normal cervical squamous epithelium,36 (2) tumor/normal pairs of human cervical tissue,37 (3) CaSki and SiHa cervical cancer cells relative to normal cervical cells,34 and (4) HeLa cervical cancer cells relative to normal cervical cells34 (left panel) in parallel with cervical cancer patients (TCGA, n = 122; right panel). Heatmap shows the log2 ratio for tumor (T) versus normal (N); white indicates absence of value; Avr, average log2 (normalized RPM) in TCGA data; green shading indicates the most repressed miRNA cluster; green arrow denotes miR-1; only miRNAs known to suppress tumors are shown. Of note, TCGA data were normalized by median RPM of all expressed miRNAs due to the lack of normal cervix data. (C) AGO-bound regions (BC ≥ 2, CLIPick12) mapped in HPV18 transcripts, including E6 and E7, following annotation of the reference genome (upper panel) or E6/E7 transcript expressed in HeLa cells (GenBank: M20324; lower panel), of which the 3ʹ end of E7 was fused with the host genome (blue shading; hg19, chr8:128,240,800–128,241,377). Seed sites of miRNAs in the identified cluster (green shading in B) were mapped with indication of their positions in seeds. Base pairs between the identified target sites and the cognate miRNAs are indicated in Table S3. (D) Bright-field images of HeLa cells 48 h after transfection of non-targeting control (NT), miR-1, miR-329, or miR-376a (upper panel) with quantitation of live cell numbers (p < 0.01, relative to NT; lower panel). More images, including miR-206 transfection, are shown in Figure S1B. (E) Cell death assays of miR-1- or miR-206-transfected HeLa cells measured by flow cytometry with propidium iodide (PI) and annexin V staining, including total apoptosis (%, red, upper value) and early apoptosis (%, black, lower value). (F) Quantitation of cell death in (E) analyzed for live (PI−, annexin V−), dead (PI+, annexin V−), early (PI−, annexin V+), and late apoptosis (PI+, annexin V+); bright-field images (upper panel). (G and H) Same cell death assays as performed in (E) and (F) except for mir-218 and miR-497. Images of miR-218- or miR-497-transfected HeLa cells are shown in Figure S1C. All p values are from two-sided t tests; ∗p < 0.05; n ≥ 3, repeated with biologically independent samples. Graphs show mean and error bars show SD unless otherwise indicated.
Since AGO-miRNA interactions were found in one common transcript that covered both critical oncogenes, E6 and E7 (E6/E7; Figure 1A, right panel), seed matches of the identified miRNAs were mapped onto their encoded regions in the HPV18 genome (Figure 1C, upper panel). HeLa cells are known to express a variant E6/E7 transcript caused by a different orientation of HPV18 genomic integration.42,43 Therefore, we also mapped AGO CLIP reads on this HeLa-specific E6/E7 transcript (Figure S1A, right panel), where the difference was caused not only by the genomic integration that fused the 3ʹ end of E7 with the host genome sequence (8q24.21), but also by alternative splicing that skipped a part of E642,43 (Figure 1C, lower panel). Regardless of the variants, every miRNA in the list of the putative tumor-suppressing cluster (Figure 1B) was mapped on E6/E7 transcripts, complexed with AGO (miR-376a, miR-1/206, and miR-329 sites in E7; miR-218 site at the 3ʹ end; Figure 1C), except for miR-497, of which the target site in E6 had no overlap with AGO CLIP reads.
Next, we checked their tumor-suppressive functions and observed a significant reduction of HeLa cells with transfection of miR-1 or miR-206, and no effect from miR-329 and miR-376a (Figure 1D; Figure S1B). After confirming the induction of apoptotic cell death by miR-1 (4.3-fold increase) and miR-206 (4.1-fold increase; Figures 1E and 1F), we further elucidated that miR-218 and miR-497 also triggered apoptosis (2.9- and 3.9-fold increases, respectively; Figures 1G and 1H). These findings are consistent with the observed reduction in living cells (Figure S1C). Taken together with the AGO CLIP analyses, we were able to delineate putative tumor-suppressing miRNAs (miR-1/206, miR-218, and miR-497) that target the E6/E7 transcript of HPV18 in cervical cancer cells.
Tumor-suppressive activity of HPV18 E6/E7-targeted miRNAs
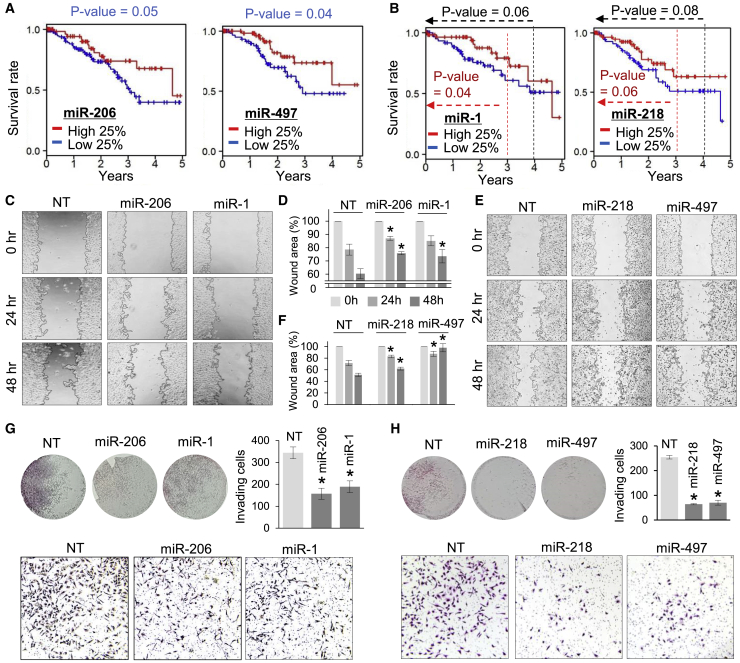
To investigate the clinical relevance of HPV18 E6/E7-targeted miRNAs, survival rates of cervical cancer patients were initially examined in the published results of split-half analyses depending on miRNA expression.44 Among them, miR-206 (p = 0.02) and miR-497 (p = 0.006) showed significant increases in survival rates within 5 years (Figure S2A), but the others (miR-1, miR-218, miR-376a, and miR-329) showed no significant differences (p = 0.725, 0.65, 0.28, and 0.86, respectively; Figures S2B and S2C). Therefore, we decided to further scrutinize TCGA data by performing split-quartile analyses (high of 25% versus low of 25%) to compare more distinct differences of miRNA expression in various periods (within 3, 4, and 5 years). Concordantly, miR-206 (p = 0.05; Figure 2A, left panel) and miR-497 (p = 0.04; Figure 2A, right panel) were found to increase survival rates within 5 years. However, although marginal, we observed similar effects from other miRNAs: cervical cancer patients expressing miR-1 (p = 0.04 and 0.06) or miR-218 (p = 0.06 and 0.08) survived more often within 3 and 4 years, respectively (Figure 2B). No significant effects resulting from miR-376a or miR-329 expression were detected (Figure S2D).
Figure 2.
Tumor-suppressive activity of the putative anticancer miRNAs identified in AGO-bound regions of HPV18
(A) Survival analysis of cervical cancer patients (from TCGA) depending on abundance of miR-206 (left panel) and miR-497 (right panel), represented as Kaplan-Meier plots. Red line indicates patients in upper quartile (high 25%); blue line indicates patients in lower quartile (low 25%); p value, log-rank tests; 5-year survival rate. (B) Same survival analyses as in (A) except for miR-1 (left panel) and miR-218 (right panel) within various time periods. p values, Wilcoxon signed-rank tests (two-sided); black indicates 4-year survival rate; red indicates 3-year survival rate. (C and D) Wound-healing assays of miR-206- or miR-1-transfected HeLa cells compared with NT-transfected cells (C). Quantitation of the image results (as shown in C) was conducted using ImageJ (D). More images are shown in Figure S2D. (E and F) Same wound-healing assays as in (C) and (D) except for miR-218 and miR-497. More images are shown in Figure S2E. (G) Invasion assays of miR-206- or miR-1-transfected HeLa cells using Matrigel-based transwell. Hematoxylin and eosin (H&E)-stained Matrigel inserts are represented as whole (upper left panel) or zoomed-in images (lower panel); quantitation of results are shown in the upper right panel. (H) Same invasion assays as performed in (G) except for miR-218 and miR-497. All p values were from two-sided t tests; ∗p < 0.05; n ≥ 3, repeated with biologically independent samples. Graphs show mean and error bars show SD unless otherwise indicated.
The extent of the metastatic potential of cancers is known to contribute toward patient survival. Thus, the miRNAs that negatively affected cell survival (i.e., miR-1, miR-206, miR-218, and miR-497) were evaluated for their effects on migration and invasion of cervical cancer cells. It was found that expression of either miR-206 or miR-1 significantly reduced migration of HeLa cells in wound-healing assays (Figures 2C and 2D; Figure S2D). Similarly, expression of miR-218 or miR-497 also inhibited cell migration and induced cell death during wound healing (Figures 2E and 2F; Figure S2E). In invasion assays, every miRNA that showed inhibition of cell migration (miR-206, miR-1, miR-218, and miR-497) significantly attenuated the invasive activity of HeLa cells (Figures 2G and 2H). Overall, we found evidence that expression of some HPV18 E6/E7-targeted miRNAs (miR-206, miR-1, miR-218, and miR-497) increased survival rates of cervical cancer patients and could function as tumor suppressors that inhibited metastatic potential of cervical cancer cells.
AGO CLIP-based design of antitumor mi/siRNAs against E6/E7
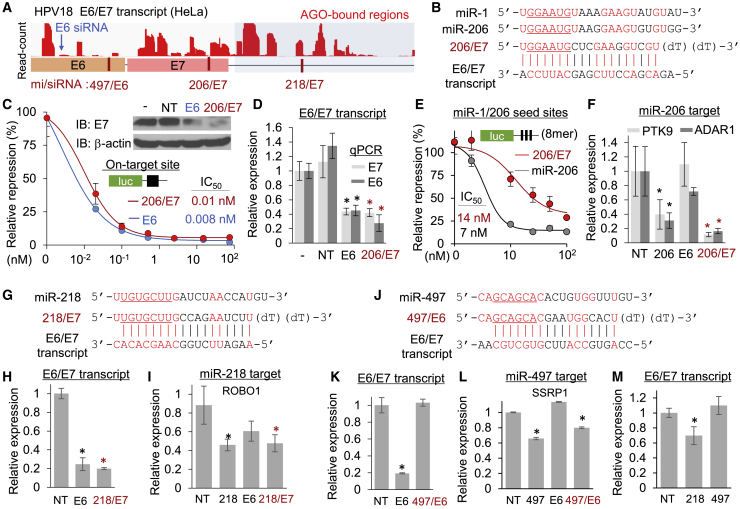
The HPV18 E6/E7 transcript was found to be associated with host miRNAs in cervical cancer cells (HeLa; Figure 3A). Of the target sites in AGO binding regions, the miR-1/206 seed site in E7 could dictate the composition of mi/siRNA sequences containing the miR-1/206 seed (6-mer, positions 2–7), while the entire sequence remained perfectly complementary to function as E7 siRNA (206/E7; Figure 3B). As postulated from the AGO-accessible target region, 206/E7 could trigger RNAi as efficiently as an optimized E6 siRNA, which has been developed for therapeutic purposes.45 This was supported by measuring the half-maximal inhibitory concentration (IC50) with luciferase reporters (0.01 nM versus 0.008 nM; Figure 3C). Similarly, high efficacy of 206/E7 for on-target repression was also verified at the level of E7 proteins (Figure 3C, upper right panel) and E6/E7 transcripts (Figure 3D). 206/E7 exerted miRNA-like activity, repressing the luciferase reporters with miR-1/206 8-mer seed sites (positions 1–8; IC50 = 14 nM; maximal inhibition rate [Imax] = 74%), albeit less efficiently than miR-206 (IC50 = 7 nM, Imax = 85%; Figure 3E), presumably due to a mismatch at position 8 (Figure 3B). In comparison, introduction of 206/E7 to HeLa cells sufficiently downregulated two previously identified miR-1/206 targets (Figure 3F), protein tyrosine kinase 9 (PTK9, also called TWF1) and RNA-specific adenosine deaminase 1 (ADAR1).46,47
Figure 3.
AGO CLIP-based design of anticancer mi/siRNAs suppressing cervical cancer with HPV18 infections
(A) AGO-bound regions (BC ≥ 2) of E6/E7 transcript in HeLa cells (GenBank: M20324) represented with target sites of putative antitumor mi/siRNAs, based on target sites of tumor-suppressing miRNAs identified in Figures 1 and 2. (B) Sequences of miR-1, miR-206, and 206/E7 (a mi/siRNA designed based on the miR-1/206 site) derived from the AGO-associated region of the E6/E7 transcript. (C) Luciferase reporter assays with the cognate on-target site for estimating IC50 of 206/E7-mediated repression under different concentrations, compared with transfection of E6 siRNA (E6). Repression of E7 protein in HeLa cells was also examined by immunoblotting with β-actin as a control, relative to NT transfection (upper right inset). −, non-transfected HeLa cells. (D) On-target repression measured by qRT-PCR for E6 and E7 mRNA in HeLa cells; relative expression was normalized by GAPDH mRNA. Notably, suppression of E6 and E7 mRNAs was observed from both E7 and 206/E7. (E) miRNA-like activity of 206/E7 was measured by luciferase reporters with miR-206 seed sites (three 8-mer sites, positions 1–8) compared with miR-206, as performed in (C). (F) Silencing of miR-206 targets by 206/E7 transfection in HeLa cells, examined by qRT-PCR of the miR-206 targets, PTK9, and ADAR1. “206,” miR-206. Of note, comparable suppression was shown in the presence of 206/E7 as much as miR-206. (G) Sequences of miR-218 and 218/E7 where mi/siRNAs were delineated from the AGO-miR-218 site in the E6/E7 transcript (E6/E7). (H and I) qRT-PCR measurement of on-target E7 mRNA (H) and miR-218 target, ROBO1 mRNA (I), in HeLa cells. “218,” miR-218. (J) Sequences of miR-497 and 497/E6, mi/siRNAs derived from the predicted miR-497 site in the E6/E7 transcript. Notably, the corresponding miR-497 site had no overlap with any AGO CLIP reads. (K and L) qRT-PCR experiments for on-target E7 mRNA (K) and miR-497 target, SSRP1 mRNA (L). “497,” miR-497. Intriguingly, 497/E6 transfection only silenced SSRP1 but had no effect on E7 mRNA in HeLa cells. (M) Level of E6/E7 transcript in the presence of miR-218 or miR-497 in HeLa cells, quantitated by qRT-PCR. All p values were from two-sided t tests, relative to NT; ∗p < 0.05; n ≥ 3, repeated with biologically independent samples. Graphs show mean and error bars show SD unless otherwise indicated.
By harboring the target site in the coding region of E7, 206/E7 could treat most of the HPV18 variants caused by viral genome rearrangement. Indeed, the integration of HPV often generated truncated E6/E7 transcripts fused with different host genome sequences. To assess the possibility of personalized designs for mi/siRNA sequences depending on AGO CLIP analysis, AGO-miRNA sites in the fusion region of the 3ʹ end of the E6/E7 transcript derived from the host genome (Figure 3A) were used to design 218/E7 (Figure 3G). Transfection of 218/E7 silenced the E6/E7 transcript as efficiently as E6 siRNA in HeLa cells (Figure 3H). Similarly, 218/E7 also exerted miRNA-like activity, as it repressed the known miR-218 target roundabout homolog 1 (ROBO1)48 to the same extent as miR-218 (Figure 3I).
Activity of mi/siRNA was also examined depending on AGO accessibility. In the case of the miR-497 site in E6 (497/E6; Figure 3J), there were no AGO CLIP reads associated with this region (Figure 3A). Consistent with low AGO accessibility, 497/E6 showed a negligible effect on silencing of the E6/E7 transcript despite the perfect sequence complementarity (Figure 3K). In contrast, 497/E6 could still mediate miR-497-like repression for other target sites and efficiently suppress a known miR-497 target, structure-specific recognition protein 1 (SSRP1),41 as much as was shown in miR-497 expression (Figure 3L). However, miR-497 expression was unable to reduce the E6/E7 transcript as observed in 497/E6, unlike AGO-associated miR-218 (Figure 3M), indicating the importance of AGO accessibility. Altogether, we could demonstrate the value of designing antitumor mi/siRNAs based on AGO CLIP by targeting HPV18 E6/E7 transcripts with tumor-suppressing miRNA-like activity in cervical cancer.
Transcriptome-wide analyses of mi/siRNAs targeting E6/E7
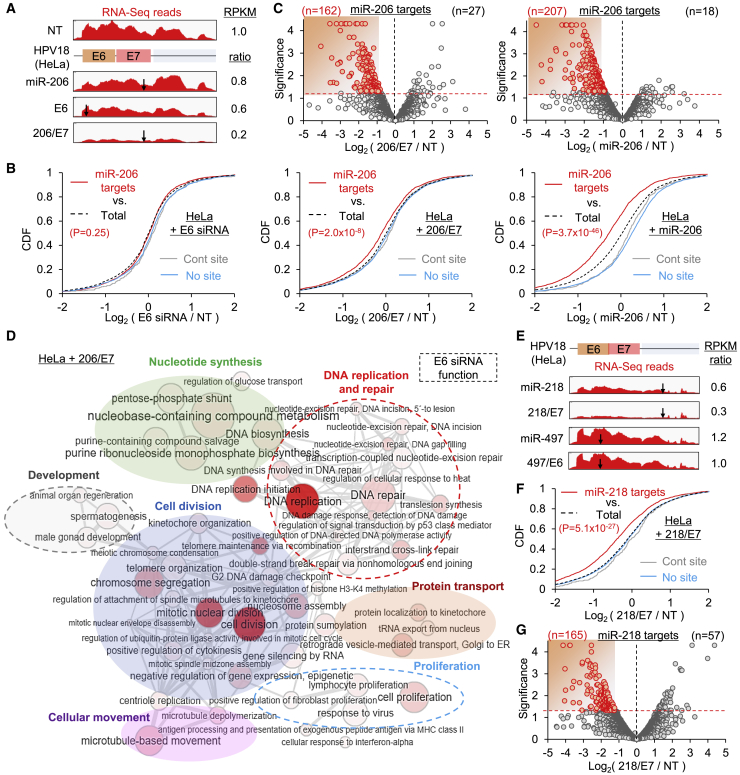
To evaluate the general activity of mi/siRNAs, RNA sequencing (RNA-seq) analyses were conducted after the transfection of 206/E7, miR-206, and E6 siRNA into HeLa cells (Figure S3A). Relative to the non-targeting control (NT), 206/E7 induced the most drastic repression of its on-target E6/E7 transcript compared to miR-206 and E6 siRNA (reads per kilobase transcript per million mapped reads [RPKM] ratio = 0.2, 0.8, and 0.6, respectively; Figure 4A). In contrast to the lack of cleavage activity by miR-206, which showed limited potency in silencing the E6/E7 transcript (only 20% reduction, Figure 4A), 206/E7 could induce the cleavage of an on-target site, as confirmed by in vitro Ago2 cleavage assays and cleaved patterns of RNA-seq reads (Figure S3B). To further examine whether 206/E7 could show miR-206-like activity, cumulative distribution function (CDF) analyses were conducted. These results showed that the putative miR-206 targets that contained the 6-mer seed sites in the 3ʹ UTR were significantly downregulated relative to the total transcripts (p = 2.0 × 10−8, Kolmogorov-Smirnov [KS] test; Figure 4B, middle panel). However, such repression of miR-206 targets was not seen in cells transfected with the E6 siRNA but was observed in miR-206-transfected cells (p = 3.7 × 10−46, KS test; relative to total transcripts; Figure 4B). Notably, E6 siRNA inadvertently repressed its own off-targets through the seed sequences (E6 seed sites in the 3ʹ UTRs; Figure S3C), implicating widespread miRNA-like off-target activity and highlighting the risks in using siRNAs. Differentially expressed gene (DEG) analysis also verified that the putative miR-206 targets were prominently downregulated by 206/E7 (n = 162 versus 27; downregulation versus upregulation, respectively; Figure 4C, left panel) as well as by miR-206 (n = 207 versus 18; downregulation versus upregulation, respectively; Figure 4C, right panel).
Figure 4.
Transcriptome-wide validation of the anticancer mi/siRNAs 206/E7 and 218/E7
(A) RNA-seq reads from NT-, miR-206-, E6-, and 206/E7-transfected HeLa cells, mapped on the HeLa-specific E6/E7 transcript. E6, E6 siRNA; reads per kilobase of transcript per million mapped reads (RPKM) ratio are shown relative to NT; arrows denote cognate target sites. (B) CDF analyses of putative miR-206 targets that contain seed sites in 3ʹ UTRs, depending on their fold changes (log2 ratio) in E6 (left panel), 206/E7 (middle panel), and miR-206 (right panel), relative to NT transfection. RPKM values are from Cufflinks; p values are from KS tests, relative to total transcripts (Total); “Cont site” indicates transcripts with non-nucleation bulge sites in 3ʹ UTRs, previously used as negative control;13 “No site” indicates transcripts with no miR-206 seed site in 3ʹ UTRs. (C) Volcano plot analyses of the putative miR-206 targets for 206/E7 (left panel) and miR-206 transfection (right panel). Significance, −log10(p value); downregulated DEG (p < 0.05; Cuffdiff), highlighted in red. (D) GO analysis results of the downregulated DEG in 206/E7-transfected HeLa cells (red dots in Figure S3D), represented as networks of overrepresented biological process terms (EASE score < 0.2, DAVID; node size and color intensity inversely correlate with p value). Clusters of terms functionally enriched in E6 siRNA transfection (Figure S4B) are highlighted with dotted circles. Of note, the rest of the clustered networks were derived from miR-206 function (Figures S3E and S4C), although there were some overlaps in the function of regulating development. (E) RNA-seq reads on E6/E7 transcript as analyzed in (A) except for miR-218, 218/E7, miR-497, and 497/E6 expression. The RPKM ratio is relative to 497/E6. (F and G) CDF (F) and volcano plot (G) analyses as performed in (B) and (C) except for putative miR-218 targets in 218/E7-transfected HeLa cells. Red dots indicated downregulated DEGs. All p values are from two-sided KS tests, relative to total, unless otherwise indicated.
We further investigated the functional consequences of silencing 206/E7 targets by focusing on downregulated DEGs (Figure S3D). Gene Ontology (GO) analysis found that the downregulated DEGs were involved in several biological process networks, such as DNA replication and repair, proliferation, development, cell division, nucleotide synthesis, protein transport, and cellular movement (Figure 4D). miR-206 targets were significantly enriched in the function of cell division, nucleotide synthesis, protein transport, and cellular movement (Figure S3E), where related pathways regulating P53, retinoblastoma (RB), and the cell cycle were also arranged with on-target activity against E6/E7 (Figure S3F). Supporting this observation, E6 siRNA showed downregulation of transcripts (Figure S4A) functioning in DNA replication and repair, development, and proliferation, and related cell signaling pathways (Figure S4B), which was consistent with previous reports.29 The genes downregulated by miR-206 expression also showed a functional enrichment for miR-206 targets, similar to that observed in 206/E7-expressing cells (e.g., protein transport, development, cellular movement, and related signal transduction; Figure S4C). We also confirmed miR-206-like repression by 206/E7 expression in hierarchical clustering analyses (Figure S4D). All of these results strongly supported the idea that the effects of 206/E7 mi/siRNA ultimately culminates in robust antitumor activity by combining different tumor-suppressive phenotypes, including prohibition of DNA replication and repair (E6 siRNA), induction of tumor cell death, inhibition of proliferation (miR-206 and E6 siRNA), and suppression of tumor cell migration and invasion (miR-206).
Expanding to other mi/siRNAs, RNA-seq analyses were also performed for 218/E7 and 497/E6 (Figure S3A). On-target activity of 218/E7 was verified to reduce the E6/E7 transcript more effectively than miR-218 (RPKM ratio of 0.6 versus 0.3 for miR-218 and miR-218/E7, respectively; Figure 4E) and showed cleavage of target transcripts in RNA-seq reads (Figure S4E). Intriguingly, neither 497/E6 nor miR-497 mediated the repression of the E6/E7 transcript (RPKM ratio of 1.2 and 1.0 for miR-497 and 497/E6, respectively; Figure 4E) with no distinct patterns of target cleavage in RNA-seq reads (Figure S4F). The limited potency of the miR-497 site in E6 might be explained by its location; that is, there were no neighboring AGO-bound regions (Figure 3A). In contrast, both miR-497 and 497/E6 could exert miRNA-like activity in other locations, suppressing most of the putative miR-497 targets in CDF analyses (Figures S4G and S4H). Of note, the level of repression was marginal in 497/E6, possibly because it contained a relatively weak off-set 6-mer seed sequence (positions 3–8; Figure 3J).
In addition, 218/E7 expression significantly silenced putative miR-218 targets in CDF (p = 5.1 × 10−27, KS test, relative to total; Figure 4F) and volcano plot analyses (n = 165 versus 57 for downregulation and upregulation, respectively; Figure 4G) as shown in the miR-218-transfected HeLa cells (Figures S5A and S5B). The putative miR-218 targets, which harbored miR-218 seed sites in 3ʹ UTRs, were significantly downregulated by 218/E7 and enriched in the functions found by miR-218 expression, including cell division, trafficking and movement, stress response, homeostasis, and related signaling pathways (Figures S5C and S5D). 218/E7 also induced siRNA-mediated repression of E6/E7, showing its downregulated targets functionally enriched in DNA replication and repair (Figure S5E), the same functional terms derived from E6 siRNA expression (Figure S4B). Collectively, 218/E7 is thought to affect transcriptome-wide repression as a dual player of miR-218 and E6 siRNA, leading to efficient suppression of cervical cancer by suppressing DNA replication and repair (E6 siRNA), inducing apoptotic cell death, inhibiting proliferation (miR-218 and E6 siRNA), and preventing tumor cell migration and invasion (miR-218).
Suppression of cervical cancer cells by 206/E7 and 218/E7
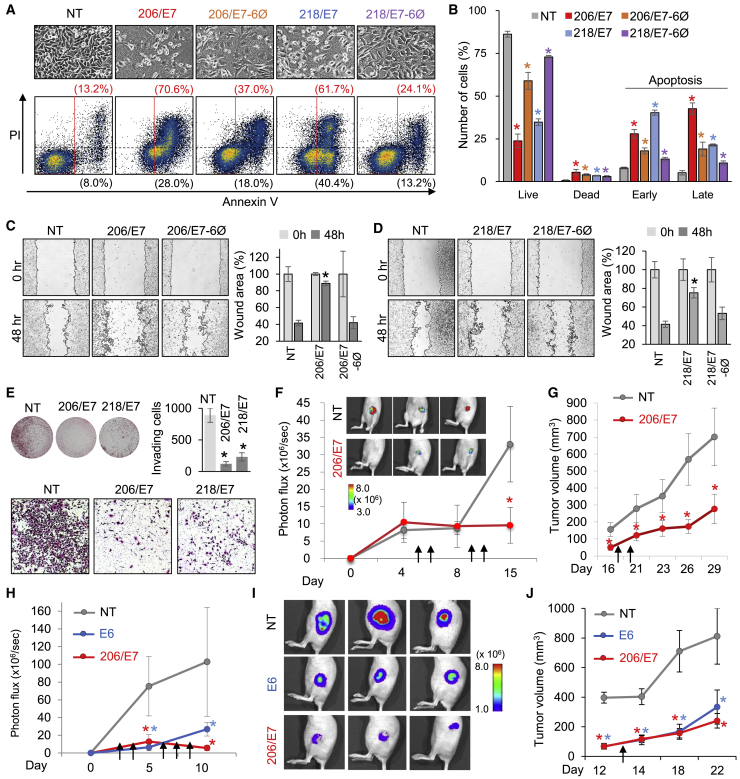
Expected antitumor activity from mi/siRNAs was experimentally examined for 206/E7 and 218/E7. Introduction of 206/E7 into HeLa cells dramatically suppressed tumor growth and showed a marked increase in cell death (Figure S6A) with DNA fragmentation (Figure S6B). To qualify the contribution of miRNA-like activity, we modified the same mi/siRNA sequences with the previously reported abasic pivot substitution (dSpacer at position 6; 6Ø),23,24 of which application to 206/E7 (206/E7-6Ø) was validated to eliminate only miR-206 activity while maintaining on-target activity against E7 (Figure S6C). It was clearly shown that 206/E7 substantially contributed to caspase-independent cell death, whereas 218/E7-induced cell death was caspase-dependent apoptosis (Figure S6D). We found that the considerable increase of apoptotic cell death by 206/E7 (5.3-fold) or 218/E7 (4.7-fold) expression was synergistically mediated by miRNA-like activity because 206/E7-6Ø and 218/E7-6Ø only marginally enhanced apoptosis (2.8- and 1.8-fold, respectively; Figures 5A and 5B). Furthermore, 206/E7 and 218/E7 showed inhibition of tumor cell migration in wound-healing assays, but this largely depended on miRNA-like activity (Figures 5C and 5D; Figures S6E and S6F). Additionally, 206/E7 and 218/E7 also efficiently suppressed tumor cell invasion, as evaluated by Matrigel-based transwell assays (Figure 5E).
Figure 5.
Functional validation of 206/E7 or 218/E7 as synergistic tumor-suppressing treatments
(A and B) Cell death analysis of NT-, 206/E7-, 206/E7-6Ø-, 218/E7-, or 218/E7-6Ø-transfected HeLa cells (images, upper panel), estimated by flow cytometry with PI and annexin V staining (A, lower panel); quantitation results are as calculated in Figure 1F (B). (C and D) Wound-healing assays for 206/E7 (C) and 218/E7 (D) in HeLa cells, compared with the presence of 6Ø modification. Images are shown in the left panels; quantitation results (right panels) are as measured in Figure 2D. More images used for the analyses are shown in Figures S6E and S6F. (E) Invasion assays of 206/E7- and 218/E7-transfected HeLa cells with Matrigel-based transwells. H&E-stained Matrigel insert (upper left panel), zoomed-in images (lower panel), and quantitation results (upper right panel) are shown. (F) Measurement of HeLa-luc cell size in xenograft mouse model after the serial injection of 206/E7 (as complexed with PEI; indicated as arrows), estimated based on bioluminescence images (photon flux; upper inset). Arrows denote time points of the injection: serial RNA injection started 5 days after implantation (1 × 106 HeLa-Luc cells). (G) Tumor volume measured by caliper 16 days after xenograft in (F). Arrows denote time points of additional injections; red line indicates 206/E7; black line indicates NT. (H–J) The same xenograft mouse model experiments with HeLa-luc cells except for the implantation of 5-fold more cells (i.e., 5 × 106 HeLa-luc cells) to compare the effects of 206/E7 and E6 siRNA, of which injection was started at an earlier time (2 days after implantation) and quantitated (H) based on bioluminescence images (I, after 10 days); tumor volume was measured after 12 days using calipers (J). Of note, there was no significant difference in tumor size between 206/E7 and the control NT treatment early on (F; up to day 8), which was likely due to the low number of HeLa cells used (1 × 106) relative to the other experiments (H–J), which used 5-fold as many HeLa cells (5 × 106). Difference in tumor size between E6 and 206/E7 treatments became unnoticeable 4 days (day 12) after the serial siRNA injections (days 2, 4, 6, 7, and 8). Subsequent 206/E7 treatment (day 13) again showed superior tumor-suppressive activity over E6 siRNA (J; day 22). All p values were from two-sided t tests, relative to NT; ∗p < 0.05; n ≥ 3, repeated with biologically independent samples. Graphs show mean and error bars show SD unless otherwise indicated.
The effectiveness of 206/E7 in vivo was then tested by using a xenograft mouse model implanted with HeLa cells (1 × 106 cells). Local administration of 206/E7 complexed with polyethylenimine (PEI) potently alleviated tumor growth within 15 days of subcutaneous injection (greater than 3-fold reduction relative to control; Figure 5F), detected by bioluminescence imaging and, subsequently, by measuring tumor volume at 29 days (Figure 5G). To compare such tumor-suppressive effects between 206/E7 and E6 siRNA in larger xenograft tumors, we increased the number of HeLa cells to 5 × 106, followed by immediate treatment with siRNAs 2 days after implantation (Figures 5H–5J). It was observed that 206/E7 also efficiently restrained tumor growth (Figures 5H and 5I) and later attenuated the increase in tumor volume more significantly than E6 siRNA (Figure 5J). E6 is known to downregulate P21 mRNA by destabilizing P53 proteins, thereby regulating cancer proliferation and apoptosis.29 Since there was no significant difference between 206/E7 and E6 siRNA in terms of derepressing P21 mRNA (Figure S6G), superior antitumor activity of 206/E7 over E6 siRNA was likely brought on by additional miR-206-like activity. Overall, these results strongly suggested robust antitumor activity of 206/E7 in cervical cancer.
AGO CLIP-based strategy of designing mi/siRNAs for cancer treatment
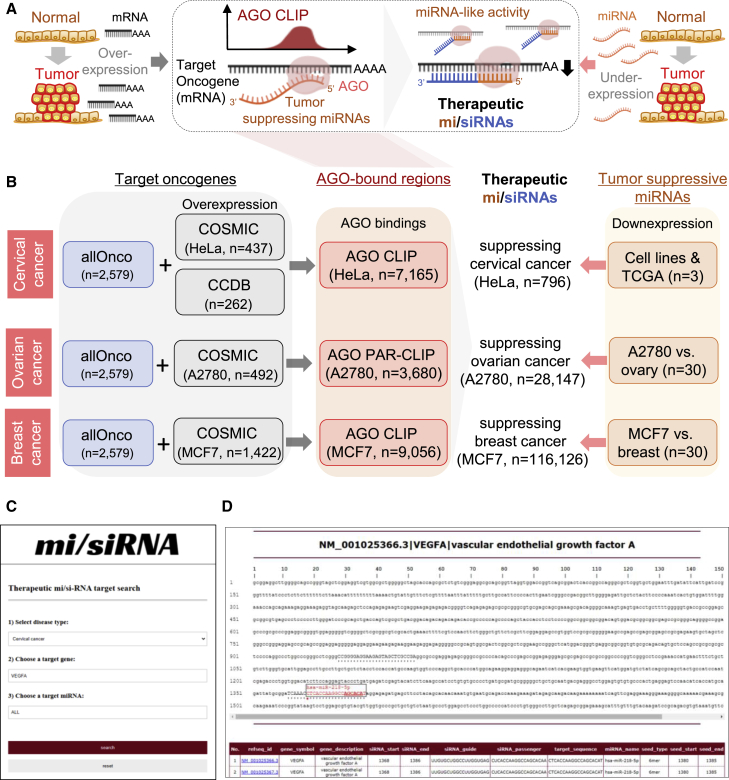
The strategy of designing antitumor mi/siRNAs could be applied to any type of cancer once AGO CLIP data become available. As initially illustrated for cervical cancer, tumor-promoting oncogenes, which could be characterized by their overexpression in tumors, needed to be identified to map their AGO-bound regions as targets (Figure 6A, left panel). Then, after selecting tumor-suppressing miRNAs based on their downregulation in tumors, their binding sites in AGO-associated oncogenes could be identified (Figure 6A, right panel). By adopting complementary sequences to these AGO-miRNA sites in the target oncogenes, mi/siRNA sequences could be determined, simultaneously eliciting the target gene-silencing and tumor-suppressive miRNA-like activity (Figure 6A).
Figure 6.
Strategy of designing antitumor mi/siRNAs applied to cervical, ovarian, and breast cancers
(A) Overview of designing therapeutic mi/siRNAs based on AGO CLIP analysis for cancer treatment, conferring dual roles as siRNA-targeting oncogenes (overexpressed in tumor) and as tumor-suppressive miRNAs (underexpressed in tumor). (B) Antitumor mi/siRNAs delineated for cervical, ovarian, and breast cancers based on AGO CLIP results in HeLa cells,11 A2780,16 and MCF7.17,18 For selection of target oncogenes, a combination of allOnco (http://www.bushmanlab.org/links/genelists) and COSMIC databases was used. For cervical cancer, the list of amplified or overexpressed genes in the cervical cancer gene database (https://webs.iiitd.edu.in/raghava/ccdb/) was also used. Tumor-suppressive miRNAs were selected based on miRNA expression profiles derived from TCGA, the gene expression omnibus, and the microRNA (http://www.microrna.org) databases (details in Materials and methods). (C and D) The mi/siRNA web server (http://ago.korea.ac.kr/misiRNA) with searchable functionality for cervical, ovarian, and breast cancers (C) and an example of output results (D) are shown. Red asterisk indicates start position of the mi/siRNA target site; a black dot indicates AGO-bound regions. Of note, searches in the mi/siRNA web server could be started by defining a type of cancer (cervical, ovarian, or breast cancer), followed by autocompletion of the selected target oncogenes and tumor-suppressing miRNAs. Indication of AGO-bound regions (black dot, D) with detailed information from RefSeq and miRBase links could guide users to narrow down their choices.
Expanding this methodology to cervical, ovarian, and breast cancers, the pool of target oncogenes was initially chosen by combining the allOnco database, a comprehensive list of cancer-related genes (n = 2,579), and a list of overexpressed genes derived from the Catalog of Somatic Mutations in Cancer (COSMIC) database49 for the corresponding cancer cells (HeLa, A2780, and MCF7) for which AGO CLIP had been performed11,16, 17, 18 (Figure 6B). By analyzing the AGO-bound regions with a list of tumor-suppressive miRNAs selected by their downregulation in the matched cancer cell, antitumor mi/siRNA sequences were delineated for cervical (HeLa, n = 796), ovarian (A2780, n = 28,147), and breast cancers (MCF7, n = 116,126). To facilitate their use in other therapeutic studies, we have established a mi/siRNA webserver (http://ago.korea.ac.kr/misiRNA) including these sequences and added search functionality (Figures 6C and 6D).
Validation of mi/siRNAs designed for ovarian and breast cancers
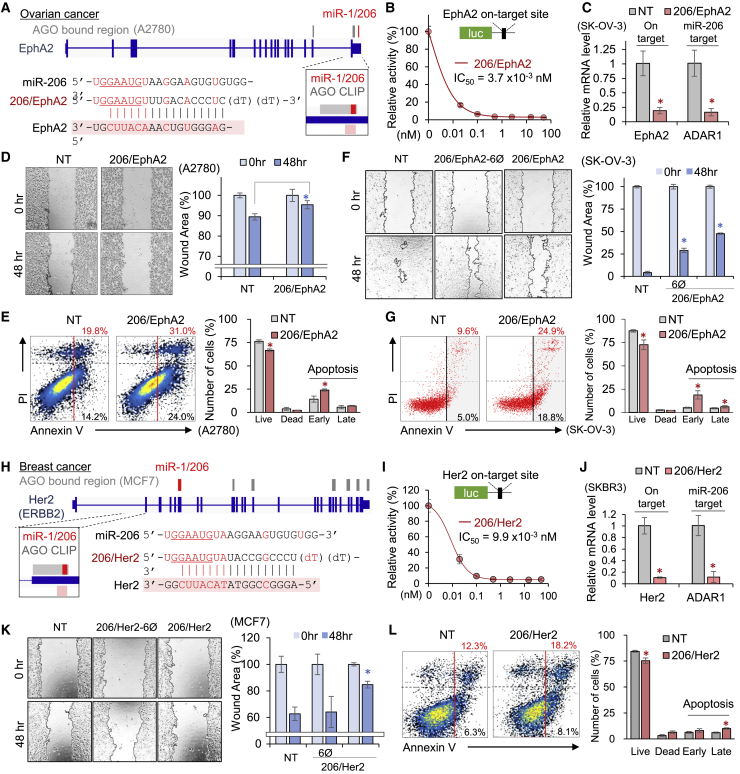
We verified the therapeutic potency of mi/siRNAs by applying a treatment of epithelial ovarian cancer and Her2+ (ERBB2+) breast cancer (Figure 7). First, the miR-1/206 target site in the AGO-bound region16 of epithelial cell receptor protein tyrosine kinase (EphA2), which is frequently overexpressed in ovarian cancer and involved in metastatic progression,50 was utilized to design mi/siRNA (206/EphA2; Figure 7A) and subjected to validation. The results showed that 206/EphA2 efficiently elicited on-target activity (IC50 = 3.7 × 10−3 nM), which was monitored by a luciferase reporter with a cognate EphA2 site (Figure 7B). It was also confirmed that 206/EphA2 could silence both EphA2 and a miR-206 target, ADAR1, in an ovarian cancer cell line (SK-OV-3; Figure 7C). As expected, 206/EphA2 significantly inhibited migration in a wound-healing assay (Figure 7D; Figure S7A) and induced apoptosis of A2780 ovarian cancer cells (Figure 7E). We also confirmed that these potent effects of 206/EphA2 were synergistically brought on by miR-206-like activity because 206/EphA2-6Ø showed attenuation of its inhibitory activity on tumor migration (Figure 7F; Figure S7B). Expression of 206/EphA2 also significantly induced apoptosis in SK-OV-3 ovarian cancer cells (Figure 7G).
Figure 7.
Validation of antitumor mi/siRNAs for ovarian and breast cancer
(A) 206/EphA2, a mi/siRNA designed to target EphA2 with miR-1/206 activity based on the AGO CLIP results of ovarian cancer cell A2780;16 AGO-bound regions in EphA2 (gray line, upper panel) with indication of miR-1/206 seed site (red line) are shown. Sequences of miR-206, 206/EphA2, and its target site in EphA2 (pink shading) are indicated in the lower panel. (B) Efficiency of on-target repression mediated by 206/EphA2; IC50 from luciferase reporter assays as performed in Figure 3C. (C) Repressive activity of 206/EphA2 on EphA2 (on-target) and ADAR1 (miR-206 target), measured by qRT-PCR. (D) Effect of 206/EphA2 on migration activity of A2780; wound-healing assays (left panel) and quantitative results (right panel) as measured in Figure 2D. More images are shown in Figure S7A. (E) Cell death assays of 206/EphA2-transfected A2780; flow cytometry (left panel) and its quantitative results (right panel), as conducted in Figures 1E and 1F. Relatively low responses from 206/EphA2 could be explained by low expression of EphA2 in A2780. (F and G) The same wound-healing and cell death assays as performed in (D) and (E) except that 206/EphA2-6Ø was introduced into another ovarian cancer cell line, SK-OV-3. More images are shown in Figure S7B. (H) 206/Her2, a mi/siRNA designed to target Her2 (ERBB2) with miR-1/206 activity, derived from the AGO CLIP results of the breast cancer cell line, MCF7. (I) On-target repressive efficacy of 206/Her2 as estimated in (B). (J) Repression of Her2 (on-target) and ADAR1 (miR-206 target), quantitated by qRT-PCR in SKBR3. The same experiment performed in another breast cancer cell line, BT474, is shown in Figure S7C. (K and L) Wound-healing (K) and cell death assay (L) as performed in (D) and (E) except with 206/Her2 and 206/Her2-6Ø in MCF7. More wound-healing results are shown in Figure S7D. All p values are from two-sided t tests. ∗p < 0.05, relative to NT; n ≥ 3; repeated with biologically independent samples. Graphs show mean and error bars show SD unless otherwise indicated.
Focusing on a critical oncogene in breast cancer, Her2 (ERBB2),51 a miR-1/206 target site was defined within the AGO-associated region in the Her2 transcript (MCF7)17,18 to generate the antitumor mi/siRNA, 206/Her2 (Figure 7H). This 206/Her2 was able to efficiently trigger on-target repression through the cognate Her2 target site (IC50 = 9.9 × 10−3 nM; Figure 7I) in luciferase reporter assays. Then, 206/Her2 was validated to act similar to miR-206, a finding that was supported by observing effective repression of the miR-206 target, ADAR1, regardless of the cell line being tested (Figure 7J; Figure S7C). Functionally, 206/Her2 inhibited migration of MCF7 cells but lost this ability when miRNA-like activity was eliminated by 6Ø modification (Figure 7K; Figure S7D). Also, 206/Her2 expression promoted cell death in MCF7 cells (Figure 7L), and this effect was dependent on miR-206-like activity because no significant changes were observed from 206/Her2-6Ø (Figure S7E). Taken together, all of these findings support the development of mi/siRNAs for cancer treatment.
Discussion
In this study, the strategy of designing anticancer mi/siRNAs, that is, oncogene-targeting siRNAs with tumor-suppressing miRNA-like activities, was initially applied to cervical cancer HeLa cells. Since persistent HPV18 infection initiated cervical tumorigenesis,29 we primarily used the HPV18 E6/E7 transcript as a target for siRNA activity to demonstrate the validity of therapeutic mi/siRNAs. To induce RNAi, any sequence in the transcript can be targeted by siRNA if it is capable of perfect annealing.22 However, not all perfectly complementary sequences are equally effective; thus, siRNA sequences were generally screened by the gene-walking method that could comprehensively identify the most potent sequence for therapeutic purposes. Bioinformatics that considers the reported features of potent siRNA can assist in this rigorous approach,25 but its application is often limited. Since siRNAs are also AGO-dependent, AGO-bound regions analyzed by CLIP were the most efficiently targeted by RNAi. Indeed, AGO CLIP revealed that such differential AGO-miRNA binding is affected by target site accessibility determined by the secondary structure of RNA and recruitment of RNA-binding proteins.52 Consistent with this notion, we confirmed that all mi/siRNAs designed from AGO-associated regions (206/E7, 218/E7, 206/EphA2, and 206/Her2) were potently suppressed on-target, except one that was designed on a non-AGO-associated region, 497/E6, which showed negligible on-target repression despite its complete sequence match.
Since miRNA-target interactions are predominantly mediated by the seed region (positions 2–8), the miRNA family that contains the same seed sequences represses a similar set of targets and controls the same major phenotypes.1,2 Furthermore, depending on the seed sequence, miRNA and siRNA have shown the same transcriptome profile46 and functional outcome, founding the conceptual basis of multifunctional mi/siRNAs. AGO CLIP analyses can naturally design mi/siRNA by adopting the complementary sequence of AGO-accessible target sites that contain the seed sequence of miRNA and exert miRNA-like activity within perfectly matched siRNA sequences. Therefore, to use this mi/siRNA strategy for a specific type of cancer, AGO CLIP data must be available.
Regarding seed sequences of siRNAs, specific compositions of 6mers have been known to exert toxicity in a miRNA-like fashion.26,27 Thus, such toxicity could be used to design siRNAs that kill tumor cells. Intriguingly, it appears that at least three of the four tumor-suppressive miRNAs studied here (miR-1, miR-206, and miR-497) carry seeds that are at least in part toxic through the 6-mer toxicity because their seed sequences start with one or two Gs.26,27 Involvement of such 6-mer seed toxicity has been supported by multiple GO analyses, showing that one key feature of the 6-mer seed toxicity was downregulation of genes involved in proliferation, cell division, and DNA repair, as observed in 206/E7 expression (Figure 4D; Figure S3E). Therefore, mi/siRNAs in this study could likely be explained by at least three overlapping activities: (1) targeting of an oncogene by an siRNA, (2) cross-reaction with a network of genes targeted by a tumor-suppressive miRNA that carries the same seed sequence, and (3) activation of 6-mer seed toxicity by targeting essential survival genes that carry these seed matches, based on the observation that mi/siRNAs somewhat induce toxicity, resulting in a combination of caspase-dependent apoptosis and caspase-independent cell death (Figure S6D).
Although miRNAs can affect cancer initiation and progression both positively and negatively, tumor-suppressive activity seems to prevail in miRNAs. Indeed, most miRNAs are reported to be downregulated in cancer;5 for example, global deregulation of miRNA caused by defects in biogenesis promoted tumor development.53 AGO-miRNA interactions on oncogenes are prevalently expected to be tumor-suppressive, and so they were selected by the feature of their downregulation during tumorigenesis in the miRNA expression profile. By adapting bioinformatics analysis CLIPick,12 AGO CLIP has been demonstrated to detect noncanonical target sites from lowly expressed miRNAs,12, 13, 14 while being sensitive enough to uncover new targets sites recognized by transient miRNA modification.15 Therefore, even for downregulated tumor-suppressive miRNAs, AGO CLIP might be able to map miRNA target interactions regardless of time points in the tumor development process. In lieu of this, cancer would be the appropriate target disease to use AGO CLIP and design miRNA-like activity for therapeutic siRNA.
In the therapeutic application of conventional siRNAs, the miRNA-like off-target effect should be evaluated and controlled since it often causes serious side effects.22 However, mi/siRNA is free from the issue of miRNA-like off-targeting and instead intentionally utilizes such miRNA-like activity for an additive therapeutic effect. Although mi/siRNA could be replaced by combined use of miRNA and siRNA, use of multiple RNAs could exacerbate the problem of innate immune response, toxicity, and RNA delivery. Furthermore, individual use of siRNA could cause unintended off-target effects, while using only miRNA showed limited potency of on-target repression due to the lack of cleavage activity (Figures S3B, S4D, and S4E). Indeed, therapeutically developed E6 siRNA45 showed comparable effectiveness with 206/E7 in reducing the tumor sizes in xenograft mouse models (Figures 5H–5J), although E6 siRNA also induced potentially inadvertent off-target repression in the transcriptome (Figure S3C).
Designing mi/siRNAs with a similar activity as highly expressed miRNAs in normal tissues could avoid causing detrimental changes in non-targeted cells because of their negligible contribution to preexisting miRNAs. However, the issues remain to be further investigated in follow-up studies to establish the safety. Moreover, depending on the selection of miRNA activity, mi/siRNA could confer slightly different functions tailored to various stages of tumor progression. For example, both 206/EphA2 and 206/Her2 in this study were only marginally effective in inducing apoptosis (13.3% and 5.9% increase, respectively; Figures 7E, 7F, and 7L), but they rather synergistically inhibited metastatic potential of ovarian and breast cancer cells (Figures 7D, 7F, and 7K), implicating their therapeutic potency for treating advanced cancers.
RNAi induced by mi/siRNAs can also be personalized, easily rearranged based on AGO CLIP results, and altered for sequence variation, as illustrated in the matched disease sample. Such flexibility in RNAi may enable us to deal with heterogeneity in cancer patients for whom non-uniform distribution of mutation in oncogenes impedes generalized cancer therapy.3 With the help of AGO CLIP, mi/siRNAs could serve as a personalized or general strategy, as they can be made applicable to a wide range of RNAi therapeutics by silencing a disease-causing target gene with synergistic miRNA-like activity.
Materials and methods
Bioinformatics and statistical analysis
For bioinformatics analysis, we mainly used Python scripts (available on our mi/siRNA website: http://ago.korea.ac.kr/misiRNA) with Biopython (v1.76; https://biopython.org/). Mapping of AGO CLIP sequencing reads was conducted using NovoAlign (v3.07.01; http://www.novocraft.com/products/novoalign/). RNA-seq analysis was performed using StringTie (v2.1.1; https://ccb.jhu.edu/software/stringtie/) or Cufflinks (v2.2.1; http://cole-trapnell-lab.github.io/cufflinks/), which includes Cuffdiff (http://cole-trapnell-lab.github.io/cufflinks/cuffdiff/). Hierarchical clustering analyses were performed in the Cluster program (v3.0; http://www.eisenlab.org/software.html) and visualized as heatmaps using the Java TreeView program (http://jtreeview.sourceforge.net/).
The standard laboratory practice randomization procedure was used for cell line and mouse groups. The investigators were not blinded to allocation during experiments and outcome assessment. All of the statistical tests, including KS tests (two-sided), t tests (unpaired, two-tailed), Wilcoxon signed-rank tests (two-sided), and log-rank tests (two-sided) were performed in Scipy (v0.13; https://www.scipy.org/), Bioconductor (https://www.bioconductor.org/), or Excel. Unless stated otherwise, values represent mean ± SD and statistical significance was set at p < 0.05 (relative to control, equal variance) and repeated with biologically independent samples (n ≥ 3).
AGO CLIP analysis on HPV18
AGO CLIP results, performed in HeLa cells with two different antibodies (2A8 and 7G1-1),11 were retrieved from control sets (http://ago.rockefeller.edu/rawdata.php) and mapped on an HPV18 reference genome (GenBank: NC_001357) or an HPV18 E6/E7 transcript identified in HeLa cells (GenBank: M20324) using NovoAlign (v3.07.01) with the same parameters used previously12 (Figure S1A). They were further analyzed to identify the AGO-bound regions using the CLIPick program12 (http://clip.korea.ac.kr/clipick) with the function of selecting reads in reproducible clusters (CLIPick.reproducible_peaks), where reads were overlapped from experiments using two different AGO antibodies (BC ≥ 2). Read counts were compiled as mapped reads (bedGraph) using BedTools (v2.29.2; https://bedtools.readthedocs.io) and visualized by Integrative Genomics Viewer (IGV) (v2.8.10; https://software.broadinstitute.org/software/igv/). To identify the target sites of given miRNAs within the AGO-bound regions, 6-mer matches to seed sequences at positions 2–8 were searched in the region of the compiled reads.
miRNA profile analysis in cervical cancer
miRNA expression data36 in normal cervical squamous epithelium (n = 7) were compared with tumor tissues that were derived from cervical intraepithelial neoplasia (n = 18), squamous cell carcinoma (n = 10), or adenocarcinoma (n = 9) using GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) (GEO: GSE30656). Based on the false discovery rate (FDR), 97 miRNAs were selected as differentially expressed (FDR < 0.05) and calculated as the log ratio of fold changes (log2[tumor/normal]; set 1). miRNA profiles from 29 tumor/normal pairs of human cervical tissue samples (GEO: GSE20592) were analyzed,37 and 134 miRNAs were selected as significantly differentiated (FDR < 0.01). The tumor/normal ratio in each pair of samples was normalized, and the median log2 values were calculated (set 2). The mean fold changes of miRNA abundance with significant changes (q value = 0) were retrieved from a previous study34 on cervical cancer cell lines, CaSki, SiHa (set 3; HPV16 integration), and HeLa (set 4; HPV18 integration), relative to the normal cervixes. Sets 1–4 were analyzed by hierarchical clustering, performed in the Cluster 3.0 program. Based on the order of the hierarchical clustering results, small RNA sequencing data from cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) in TCGA (n = 122) were analyzed to calculate the log2 ratio of reads per million reads (RPM), then normalized by the median RPM of all expressed miRNAs.
Small RNA synthesis
siRNA, miRNA, and mi/siRNA were synthesized by custom RNA synthesis services from Bioneer (Korea) and TriLink BioTechnologies (USA). The quality of synthesized RNAs and their modification were monitored, reported, and confirmed by the company. As a control RNA, non-targeting miRNA (NT), derived from a cel-miR-67 sequence (C. elegans-specific miRNA), was synthesized as siRNA with a two-thymidine deoxynucleotide (dT) overhang and further modified to contain abasic pivot (abasic deoxynucleotide, dSpacer [6Ø]) in both strands to prevent seed-mediated miRNA-like repression, as previously reported.23,24 Duplex miRNAs (miR-1, miR-206, miR-329, and miR-376a) were synthesized by following the human miRNA annotation in miRBase (http://www.mirbase.org/) with consideration of the 2-nt overhang (especially miR-1; details in Table S1). For RNA-seq analysis, miR-218 and miR-497 sequences were also synthesized based on annotations in miRBase, but their passenger strands contained 2ʹ-O methylation (2ʹOMe) at positions 1 and 2 in the form of siRNA,54 preventing seed-mediated repression from the passenger strand (Table S1). The sequence of E6 siRNA was taken from a previous study.45 mi/siRNAs designed to contain the miR-206 seed sequence were synthesized as indicated in Figures 3B, 7A, and 7H. The sequences of the other mi/siRNAs (218/E7 and 497/E6), represented in Figures 3G, and 3J, also contained 2ʹOMe in the passenger strands. The details of the siRNA or mi/siRNA sequences are represented in Table S2. When we modified the mi/siRNA with abasic pivot substitution, a nucleotide at position 6 was synthesized with dSpacer (6Ø),23,24 resulting in elimination of miRNA-like activity while maintaining the on-target activity as siRNA.
Cell culture and transfection
The human cervical cancer cell line HeLa (Korean Cell Line Bank) was grown in Dulbecco’s modified Eagle’s medium (DMEM; HyClone, USA). The human breast cancer cell lines MCF-7, SKBR3, and BT-474 (Korean Cell Line Bank), the ovarian cancer cell lines SK-OV-3 (Korean Cell Line Bank) and A2780 (a generous gift from Dr. J.H. Choi) were grown in RPMI 1640 (HyClone, USA). All media were supplemented with 10% fetal bovine serum (FBS; Gibco, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin (Welgene, Korea) and incubated at 37°C with 5% CO2. Transfection of 50 nM siRNA, miRNA, or mi/siRNA was performed using Lipofectamine RNAiMAX (Invitrogen, USA), according to the manufacturer’s protocol, unless otherwise indicated. For cotransfection of RNA with plasmid vectors, Lipofectamine 3000 (Invitrogen, USA) was used according to the manufacturer’s instructions. For seeding cells before transfection, the accurate number of cells was quantitated using a Countess II automated cell counter (Invitrogen, USA). The cells were harvested 24 h (qRT-PCR and luciferase reporter assays), 48 h (wound-healing assays), and 72 h (cell death analyses) after transfection, unless otherwise indicated.
Cell death analysis
For quantification of live cells, dead cells were stained with 0.4% trypan blue solution (Gibco, USA) and counted for live cells by a hemocytometer three times. Images of the cells were acquired and analyzed using an inverted light microscope (Leica DMi8). For the quantitative measurements of apoptotic cell death, an annexin V apoptosis detection kit (eBioscience, USA) was used following the manufacturer’s protocol. Briefly, after washing with phosphate-buffered saline (PBS; Biosesang, Korea), 0.4–1 × 105 cells were resuspended in 200 μL of binding buffer containing 5 μL of annexin V and incubated for 10 min at room temperature. After washing with binding buffer, cells were resuspended in 190 μL of binding buffer. Ten microliters of 20 μg/mL propidium iodide (PI) was added and cells were analyzed using an Attune NxT flow cytometer (Thermo Fisher Scientific, USA). In the case of 206/EphA2-transfected SK-OV-3 cells, apoptotic cell death analysis was conducted using a Muse cell analyzer (Luminex, USA) with a Muse annexin V & dead cell kit (Luminex, USA) according to the manufacturer’s protocol. To inhibit caspase activity, 20 μM Z-VAD-fmk (Selleck Chemicals, USA) was treated from the transfection.
Survival analysis
The results shown herein were based on data generated by TCGA Research Network (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga). The clinical data within the dataset of CESC (n = 122) were downloaded as matches to the expression profiles of miRNAs. Thus, some patients were excluded, such as those with missing miRNA expression levels, those without follow-up meetings, and those without complete clinical information. For Kaplan-Meier analysis, the R program was used with the packages “survival” and “KMsurv” applied to compare the survival rates of patients with given miRNA expressions in the upper quartile versus the lower quartile, unless otherwise indicated. Wilcoxon signed-rank tests (two-sided) or log-rank tests (two-sided) were applied to check statistical significance.
Wound-healing assay
For the wound-healing assay, cells were cultured in 24-well plates (1 × 105 cells per well) for 24 h to achieve 90% confluence. A vertical or horizontal wound was created by using a SPLScar scratcher (SPL, Korea). The wounded cells were washed once with DMEM or RPMI 1640 with 10% FBS and allowed to grow in the medium for 24–48 h. Images of cells were taken with a Leica DMi8 inverted light microscope and quantitatively analyzed using ImageJ 1.51s software (https://imagej.nih.gov/) installed with a macro (edge-detection/thresholding method) for automated analysis of scratch-wound assays.
Invasion assay
HeLa cells were plated on a transwell of BioCoat Matrigel invasion chambers (8.0-μm polyester [PET] membrane; Corning Life Sciences, USA) in a 24-well plate (50 × 105 cells per well) and reverse transfected using RNAiMax (Invitrogen, USA) with 50 nM RNA duplexes, according to the manufacturer’s protocol. After 24 h, the cells in the transwell were changed to grow in the media without FBS. The lower chamber contained media with 10% FBS to serve as chemoattractant. After 48 h, non-invading cells were removed with cotton swabs. Then, invading cells in Matrigel-precoated PET membranes were visualized by hematoxylin and eosin (H&E) staining: the membranes were fixed with 100% methanol (Biosesang, Korea) for 5 min after the PBS (Biosesang, Korea) wash. After a quick wash with distilled water, the membranes were stained with hematoxylin (ScyTek Laboratories, USA) for 5 min, again washed with distilled water, and stained with eosin (Sigma-Aldrich, USA) for 1 min. The membranes were mounted on a glass slide after double washing with distilled water. Then, the stained cells were examined and quantitated using a Leica DMi8 inverted light microscope.
Construction of luciferase reporters
To measure the efficiency of RNAi-mediated repression, psiCheck-2 vector (Promega, USA) was used to construct luciferase reporters, wherein on-target sites were inserted into the 3ʹ UTR of synthetic Renilla luciferase. Accordingly, synthetic duplex DNA oligonucleotides (Bionics or Bioneer, Korea) containing various on-target sites (underlined) were cloned into the psiCheck-2 plasmid via XhoI and NotI sites, as indicated: E6, forward, 5ʹ-TCGAGAATAACCTGTGTATATTGCAGC-3ʹ, reverse, 5ʹ-GGCCGCTGCAATATACACAGGTTATTC-3ʹ; 206/E7, forward, 5ʹ-TCGAGCGACCTTCGAGCATTCCGGC-3ʹ, reverse, 5ʹ-GGCCGCCGGAATGCTCGAAGGTCGC-3ʹ; 206/EphA2, forward, 5ʹ-TCGAGGAGGGTGTCAAACATTCCAGC-3ʹ, reverse, 5ʹ-GGCCGCTGGAATGTTTGACACCCTCC-3ʹ; and 206/Her2, forward, 5ʹ-TCGAGAGGGCCGGTATACATTCCAGC-3ʹ, reverse, 5ʹ-GGCCGCTGGAATGTATACCGGCCCTC-3ʹ. To measure the efficacy of seed-mediated miRNA activity, seed sites (positions 2–8) were tandemly repeated (n = 3 or 5) and cloned into psiCheck-2: miR-1/206 seed, forward, 5ʹ-TCGAGACATTCCAACATTCCAACATTCCAGC-3ʹ or 5ʹ-TCGAGACATTCCAACATTCCAACATTCCAACATTCCAACATTCCAGC-3ʹ, reverse, 5ʹ-GGCCGCTGGAATGTTGGAATGTTGGAATGTC-3ʹ or 5ʹ-GGCCGCTGGAATGTTGGAATGTTGGAATGTTGGAATGTTGGAATGTC-3ʹ.
Luciferase reporter assay
Luciferase reporter assays were conducted as previously described.23 In short, psiCheck-2 plasmids (Promega, USA) were cotransfected with different amounts of duplex RNAs (up to 75 nM) using Lipofectamine 2000 or 3000 (Invitrogen, USA). Twenty-four hours after transfection, relative activity (Renilla luciferase activity normalized to firefly luciferase) was measured using a Dual-Luciferase reporter assay system (Promega, USA) with the GloMax-multi detection system (Promega, USA) with six replicates, following the manufacturer’s protocol. IC50 was calculated by performing nonlinear least-squares fitting for the sigmoid function using Scipy (scipy.optimize.curve_fit()). In cases where least-squares fitting failed to fit the function, an approximate IC50 was estimated from the regression line.
Immunoblotting
Cell lysates were prepared using 1× radioimmunoprecipitation assay (RIPA) lysis buffer (Dynebio, Korea), supplemented with a protease inhibitor (Roche, Switzerland), cleared by centrifugation at 12,000 × g, separated by SDS-PAGE, and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, USA). Primary antibodies were incubated overnight at 4°C in 1× Tris-buffered saline with Tween 20 (TBST) with 5% skim milk or 5% BSA, E7 (8E2, 1:1,000; Abcam, UK), and β-actin (8H10D10, 1:1,000; Cell Signaling Technology, USA).
Quantitative RT-PCR analysis
Large RNAs (>200 nt) were isolated using an RNeasy mini kit (QIAGEN, Germany) through on-column DNA digestion with an RNase-free DNase set (QIAGEN, Germany). RT was performed by SuperScript III RT (Invitrogen, USA) with anchored oligo(dT) primers. qPCR was conducted using QuantiFast SYBR Green PCR master mix (QIAGEN, Germany) or Power SYBR Green PCR master mix (Thermo Fisher Scientific, USA) and custom primers for HPV18 transcripts (E6 and E7) and other human mRNAs (PTK9, ADAR1, SSRP1, ROBO1, EphA2, Her2, and GAPDH), using Rotor-Gene Q (QIAGEN, Germany). The sequences of the primers are listed in Table S4. All of the reactions were run in triplicate with the standard two-step cycling protocol. Relative quantification was calculated by the ΔCT method, using GAPDH as a control.
In vitro Ago2 cleavage assay
Slicing activity of the mi/siRNA was assessed by performing an in vitro Ago2 cleavage assay as previously described.23 Briefly, the synthesized guide strands of NT, 206/E7, and miR-206 were complexed with recombinant human Ago2 proteins (Sino Biological, China) in vitro by incubating them at 37°C for 1 h (20 mM Tris-HCl, 150 mM NaCl, and 1 mM MgCl2). The substrate containing the HPV18 206/E7 site (5ʹ-UUUUUUUUUUUUUUUUUUUAGACGACCUUCGAGCAUUCCGUUUUUUUUUUUUUUUUUUUU3ʹ) was also synthesized, and the further 5′ end was labeled with [γ-32P]-ATP by T4 PNK (New England Biolabs [NEB], USA) and purified by a G25 column (GE Healthcare, USA). Subsequently, the labeled substrate was added to the Ago2 guide strand complex and incubated at 37°C for 1 h. The reaction was stopped by boiling with denaturation buffer (NEB, USA) for 5 min and resolved on a 15% denaturing polyacrylamide gel.
RNA-seq analysis
RNA-seq libraries were constructed from the large RNA using the strand-displacement stop/ligation method. From 1.5 μg of large RNA (extracted with an RNeasy mini kit; QIAGEN, Germany), polyadenylated mRNA was extracted using 10 μL of Dynabead oligo(dT)25 (Invitrogen, USA) according to the manufacturer’s protocol. Then, 10 ng of the purified mRNA was subjected to RNA-seq library preparation using a CORALL total RNA-seq library prep kit (Lexogen, Austria), as instructed by the manufacturer. After construction of cDNA, the libraries were amplified by PCR with the lowest optimal cycle determined by qPCR. The quality of the amplified library was checked using a Fragment Analyzer (Agilent, USA), which estimated the size distribution and quantities. After cross-checking the concentration of libraries using Qubit RNA HS assay kit (Thermo Fisher Scientific, USA), the prepared multiplexed libraries were precisely pooled and sequenced by MiniSeq system (Illumina) as 75 single-end reads (SE75). All sequence data were deposited into the Sequence Read Archive database (HeLa, SRP267427; A549, SRP270828).
The demultiplexed sequencing reads were aligned to the human genome (hg19) using STAR (star–outSAMtype BAM SortedByCoordinate–outSAMattrIHstart 0–outFilterType BySJout–outFilterMultimapNmax 20–outFilterIntronMotifs RemoveNoncanonicalUnannotated–outMultimapperOrder Random–alignSJoverhangMin 8–alignSJDBoverhangMin 1) under a supply of RefSeq gene annotations. The first 12 nt from the starter side of the reads were trimmed before mapping, as instructed by the manufacturer.
The abundances of the transcripts were quantified as RPKM using Cufflinks (cufflinks -b -G–compatible-hits-norm–library-type fr-secondstrand) or StringTie (stringtie–conservative–fr -e -G). The statistical significance of differential expression (determined by p value) was analyzed by Cuffdiff (Cuffdiff FDR = 0.1 -b–compatible-hits-norm–library-type fr-secondstrand) with a supply of mapping results in groups with the same conditions as in our experiments. Only the values with a valid status were selected and further analyzed for DEGs with statistical significance.
Cumulative distribution function and volcano plot analyses
To check miRNA-like target repression depending on transfection of a given duplex RNA, the CDF depending on fold change (log2 ratio) was analyzed with RPKM values derived from Cufflinks, Cuffdiff, or StringTie. Putative miRNA targets were selected by those containing cognate seed sites in the 3ʹ UTR (annotated by RefSeq, downloaded from the UCSC genome browser), wherein 7-mer matches to positions 2–8 were generally searched unless otherwise indicated. “No site” indicated transcripts containing no cognate 6-mer sites from seed (positions 2–8) in their mRNA sequences. “Cont site” denoted a subset of “No site” transcripts that also contained control sites in the 3ʹ UTR, wherein the nucleotide that aligned with the pivot (position 6) was substituted with the same nucleotide to be impaired.13 Total mRNAs were selected only for the expressed ones (Cufflinks, RPKM ≥ 0.1; Cuffdiff and StringTie, RPKM > 0). KS testing was performed using Scipy (scypy.stats.ks_2samp()), relative to total mRNAs or control site. The volcano plot was created by calculating the fold change and significance (−log10[p value]), and cutoffs were used to select DEGs.
GO analysis
GO analysis was conducted by using DAVID (6.8; https://david.abcc.ncifcrf.gov/) with a supply of downregulated DEGs (selected by volcano plot analysis) under the background of expressed transcripts in a matched control set (selected as RPKM > 0 and log2[volume] > 2, where volume is the average RPKM across different conditions) with default parameters unless otherwise indicated. The GO analysis results (EASE score of <0.15 or 0.2) were visualized using REVIGO (http://revigo.irb.hr/)55 in the network of biological process terms. An interconnected graph was also analyzed, and highly similar GO terms were linked as representing the degree of similarity with width. Pathway analysis was also separately performed by focusing on BIOCARTA and KEGG pathways (https://www.genome.jp/kegg/).
DNA fragmentation assay with flow cytometry
HeLa cells (1 × 105 cells) were harvested 48 h after transfection and resuspended in 200 μL of PBS containing 20 mM EDTA (Sigma-Aldrich, USA). For fixation, 700 μL of ethanol (Biosesang, Korea) was treated for 1 h at 4°C. Then, 10 μL of 1 μg/μL PI (Sigma-Aldrich, USA) was added after treatment with 5 μL of RNase A (0.2 μg/μL; Sigma-Aldrich, USA) for 30 min at 37°C. The cells were analyzed by BD FACSCalibur (BD Biosciences, USA).
Xenograft mouse model and siRNA treatment
Xenograft production was attempted for 6-week-old nude mice (nu/nu on BALB/c background; Orient, a branch of Charles River Laboratories, Korea). Suspension of 1 × 106 or 5 × 106 HeLa-luc cells (Caliper Life Sciences, USA) in 100 μL of sterile PBS supplemented with 50% thawed Matrigel (BD Biosciences, USA; for high-integrity in vivo) was injected subcutaneously into the flank on both sides. Fifteen micrograms of duplex RNAs were complexed with in vivo-jetPEI (Polyplus Transfection, France) and injected intratumorally according to the manufacturer’s protocol. Tumor growth was monitored by in vivo imaging for short-term effects and caliper measurement for long-term effects. In vivo bioluminescent monitoring was performed with an in vivo imaging system (IVIS) Spectrum imaging system (Caliper Life Sciences, USA). All bioluminescent images were taken 15 min after intraperitoneal injection of d-luciferin (150 mg/kg). Images and bioluminescent signals were selected and quantified through the instrument using the Living Image software (Caliper Life Sciences, USA). Tumors were measured using a digital caliper and tumor volume was calculated using the following formula: tumor volume (mm3) = length × width × height. All experimental procedures were reviewed and approved by the Institutional Animal Care and Use Committee of Samsung Biomedical Research Institute (SBRI). SBRI has been accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International and abides by the guidelines of the Institute of Laboratory Animal Resources.
Construction of mi/siRNA web server for cervical, ovarian, and breast cancer
The mi/siRNA web server was implemented using the Django framework (https://www.djangoproject.com/) in Python, and MySQL and was set up in a 12-core server. The web interface was implemented using HTML, CSS, and JavaScript components, where the JavaScript components utilized the JQuery library (https://jquery.com/). Within the server, the user is assisted in formulating a query by automatic completion of gene names (official gene symbols) or miRNA names, and requests are processed on the server side by the Django framework. All information was computed in advance for every mi/siRNA sequence and target site to speed up queries and stored as pre-built MySQL tables.
For designing mi/siRNAs, target oncogenes were initially selected by comparing genes to the allOnco database (http://www.bushmanlab.org/links/genelists) of overexpressed genes in cancer cell lines (HeLa, A2780, and MCF7; retrieved from the COSMIC database49). A list of amplified or overexpressed genes in the CCDB database (https://webs.iiitd.edu.in/raghava/ccdb/) was additionally included for HeLa. AGO-bound regions were determined by analyzing the AGO CLIP data derived from cervical (HeLa),11 ovarian (A2780; GEO: GSE129076), and breast cancer cells (MCF7; GEO: GSE57855 and GSE78059). For HeLa cells, the mapped reads from the control sets (“Ago CLIP tags in HeLa/-miR-124”; http://ago.rockefeller.edu/) were analyzed by CLIPick, where peak width was determined by adjusting 90% of clusters (default = 0.95) with the option of skipping background simulation (details of modified protocols on http://clip.korea.ac.kr/clipick/), yielding the AGO-bound regions in hg19 (converted from hg18 by LiftOver; https://genome.ucsc.edu/cgi-bin/hgLiftOver). To retrieve the AGO-bound regions in A2780, the peak positions determined in every replicate (bed files) by a previous study16 were merged using bedtools after extension of peak width (±70 nt). For MCF7, AGO-bound regions were inferred from the clusters determined by two independent CLIP experiments17,18 after extending the cluster width (±80 nt) and then merged into BedGraph using bedtools. Finally, AGO-bound regions in putative oncogenes were identified in HeLa (n = 7,165), A2780 (n = 3,680), or MCF7 (n = 9,056).
To select the tumor-suppressive miRNAs for HeLa cells, we used the validated miRNAs from this study (miR-1/206, miR-497, and miR-218). In the case of ovarian cancer, the miRNA expression profiles in A2780 (control set, n = 3; GEO: GSE129076) were retrieved as average RPM values in replicates after adding the pseudocount of 0.1, compared with one in normal ovaries (obtained from http://www.microrna.org; log2 ratio), and used to select the top 30 downregulated miRNAs in tumors as putative tumor suppressors. Similarly, the miRNA profile in MCF7 was obtained from http://www.microrna.org before being compared to expression in normal breast tissue (average, n = 103) and used to choose the top 30 downregulated miRNAs. Finally, all of the selected miRNAs were considered to map their seed sites in the AGO-bound regions of the putative oncogenes, ultimately designing mi/siRNAs for cervical (HeLa, n = 796), ovarian (A2780, n = 28,147), and breast cancer (MCF7, n = 116,126). The designed mi/siRNAs can be explored on the mi/siRNA web server: http://ago.korea.ac.kr/misiRNA.
Acknowledgments
We thank members of the Chi Laboratory for discussion, and H. Seok and J. Park for critical review of the manuscript. We are grateful to the late Y.S. Lee for his help on this work. We also thank Samsung Medical Center for the bioluminescence images, J.H. Choi for providing the A2780 cell line, and K.K. Kim for initial help with bioinformatics analyses. This work was supported by the Korean Health Technology R&D Project, Ministry of Health and Welfare, Republic of Korea (HI15C3137) and by the Korea University Research Grant.
Author contributions
D.G. and S.E. performed the major experiments. D.G., S.H.A., J.H., E.-S.J., and S.W.C. analyzed the data. D.G., S.E., H.-S.L., and J.H. performed the molecular, biochemical, and cell biology experiments. S.H.A., Y.K., Y.K.C., E.I., and S.W.C. performed the bioinformatics analyses. S.H.A. analyzed AGO CLIP sequencing data and constructed the mi/siRNA web server. Y.K.C. and E.I. analyzed cancer data. S.H.A. and Y.K. analyzed RNA-seq data. D.H.L. performed the mouse experiments. E.-S.J. and S.W.C. conceived, designed, and supervised the research. E.-S.J. and S.W.C. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2021.01.018.
Supplemental information
References
- 1.Seok H., Ham J., Jang E.S., Chi S.W. MicroRNA target recognition: insights from transcriptome-wide non-canonical interactions. Mol. Cells. 2016;39:375–381. doi: 10.14348/molcells.2016.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel D.P. Metazoan microRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 4.Di Leva G., Garofalo M., Croce C.M. MicroRNAs in cancer. Annu. Rev. Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 6.He L., He X., Lim L.P., de Stanchina E., Xuan Z., Liang Y., Xue W., Zender L., Magnus J., Ridzon D. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medina P.P., Nolde M., Slack F.J. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 8.Yu F., Yao H., Zhu P., Zhang X., Pan Q., Gong C., Huang Y., Hu X., Su F., Lieberman J., Song E. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 9.Svoronos A.A., Engelman D.M., Slack F.J. OncomiR or tumor suppressor? The duplicity of MicroRNAs in cancer. Cancer Res. 2016;76:3666–3670. doi: 10.1158/0008-5472.CAN-16-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Licatalosi D.D., Mele A., Fak J.J., Ule J., Kayikci M., Chi S.W., Clark T.A., Schweitzer A.C., Blume J.E., Wang X. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chi S.W., Zang J.B., Mele A., Darnell R.B. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park S., Ahn S.H., Cho E.S., Cho Y.K., Jang E.S., Chi S.W. CLIPick: a sensitive peak caller for expression-based deconvolution of HITS-CLIP signals. Nucleic Acids Res. 2018;46:11153–11168. doi: 10.1093/nar/gky917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chi S.W., Hannon G.J., Darnell R.B. An alternative mode of microRNA target recognition. Nat. Struct. Mol. Biol. 2012;19:321–327. doi: 10.1038/nsmb.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim K.K., Ham J., Chi S.W. miRTCat: a comprehensive map of human and mouse microRNA target sites including non-canonical nucleation bulges. Bioinformatics. 2013;29:1898–1899. doi: 10.1093/bioinformatics/btt296. [DOI] [PubMed] [Google Scholar]
- 15.Seok H., Lee H., Lee S., Ahn S.H., Lee H.S., Kim G.D., Peak J., Park J., Cho Y.K., Jeong Y. Position-specific oxidation of miR-1 encodes cardiac hypertrophy. Nature. 2020;584:279–285. doi: 10.1038/s41586-020-2586-0. [DOI] [PubMed] [Google Scholar]
- 16.Muys B.R., Sousa J.F., Plaça J.R., de Araújo L.F., Sarshad A.A., Anastasakis D.G., Wang X., Li X.L., de Molfetta G.A., Ramão A. miR-450a acts as a tumor suppressor in ovarian cancer by regulating energy metabolism. Cancer Res. 2019;79:3294–3305. doi: 10.1158/0008-5472.CAN-19-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pillai M.M., Gillen A.E., Yamamoto T.M., Kline E., Brown J., Flory K., Hesselberth J.R., Kabos P. HITS-CLIP reveals key regulators of nuclear receptor signaling in breast cancer. Breast Cancer Res. Treat. 2014;146:85–97. doi: 10.1007/s10549-014-3004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillen A.E., Yamamoto T.M., Kline E., Hesselberth J.R., Kabos P. Improvements to the HITS-CLIP protocol eliminate widespread mispriming artifacts. BMC Genomics. 2016;17:338. doi: 10.1186/s12864-016-2675-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson R.C., Doudna J.A. Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wittrup A., Lieberman J. Knocking down disease: a progress report on siRNA therapeutics. Nat. Rev. Genet. 2015;16:543–552. doi: 10.1038/nrg3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson A.L., Linsley P.S. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat. Rev. Drug Discov. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 22.Seok H., Lee H., Jang E.S., Chi S.W. Evaluation and control of miRNA-like off-target repression for RNA interference. Cell. Mol. Life Sci. 2018;75:797–814. doi: 10.1007/s00018-017-2656-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee H.S., Seok H., Lee D.H., Ham J., Lee W., Youm E.M., Yoo J.S., Lee Y.S., Jang E.S., Chi S.W. Abasic pivot substitution harnesses target specificity of RNA interference. Nat. Commun. 2015;6:10154. doi: 10.1038/ncomms10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seok H., Jang E.S., Chi S.W. Rationally designed siRNAs without miRNA-like off-target repression. BMB Rep. 2016;49:135–136. doi: 10.5483/BMBRep.2016.49.3.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park J., Ahn S.H., Cho K.M., Gu D., Jang E.S., Chi S.W. siAbasic: a comprehensive database for potent siRNA-6Ø sequences without off-target effects. Database (Oxford) 2018;2018:bay109. doi: 10.1093/database/bay109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Putzbach W., Gao Q.Q., Patel M., van Dongen S., Haluck-Kangas A., Sarshad A.A., Bartom E.T., Kim K.A., Scholtens D.M., Hafner M. Many si/shRNAs can kill cancer cells by targeting multiple survival genes through an off-target mechanism. eLife. 2017;6:e29702. doi: 10.7554/eLife.29702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao Q.Q., Putzbach W.E., Murmann A.E., Chen S., Sarshad A.A., Peter J.M., Bartom E.T., Hafner M., Peter M.E. 6mer seed toxicity in tumor suppressive microRNAs. Nat. Commun. 2018;9:4504. doi: 10.1038/s41467-018-06526-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Z., Liu W., Wang Y., Gao Z., Gao G., Wang X. Rational design of microRNA-siRNA chimeras for multifunctional target suppression. RNA. 2013;19:1745–1754. doi: 10.1261/rna.039677.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crosbie E.J., Einstein M.H., Franceschi S., Kitchener H.C. Human papillomavirus and cervical cancer. Lancet. 2013;382:889–899. doi: 10.1016/S0140-6736(13)60022-7. [DOI] [PubMed] [Google Scholar]
- 30.Stope M.B., Hettenbach D., Kaul A., Paditz M., Diesing K., Burchardt M., Zygmunt M., Mustea A., Koensgen D. The tumor suppressor microRNA-1 exhibits restricted inhibition of proliferation of ovarian cancer cells. Anticancer Res. 2016;36:3329–3334. [PubMed] [Google Scholar]
- 31.Tavazoie S.F., Alarcón C., Oskarsson T., Padua D., Wang Q., Bos P.D., Gerald W.L., Massagué J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song G., Zhang Y., Wang L. MicroRNA-206 targets notch3, activates apoptosis, and inhibits tumor cell migration and focus formation. J. Biol. Chem. 2009;284:31921–31927. doi: 10.1074/jbc.M109.046862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu T., Chang Y.F., Xiao Z., Mao R., Tong J., Chen B., Liu G.C., Hong Y., Chen H.L., Kong S.Y. miR-1 inhibits progression of high-risk papillomavirus-associated human cervical cancer by targeting G6PD. Oncotarget. 2016;7:86103–86116. doi: 10.18632/oncotarget.13344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez I., Gardiner A.S., Board K.F., Monzon F.A., Edwards R.P., Khan S.A. Human papillomavirus type 16 reduces the expression of microRNA-218 in cervical carcinoma cells. Oncogene. 2008;27:2575–2582. doi: 10.1038/sj.onc.1210919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi Z.M., Wang L., Shen H., Jiang C.F., Ge X., Li D.M., Wen Y.Y., Sun H.R., Pan M.H., Li W. Downregulation of miR-218 contributes to epithelial-mesenchymal transition and tumor metastasis in lung cancer by targeting Slug/ZEB2 signaling. Oncogene. 2017;36:2577–2588. doi: 10.1038/onc.2016.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilting S.M., Snijders P.J., Verlaat W., Jaspers A., van de Wiel M.A., van Wieringen W.N., Meijer G.A., Kenter G.G., Yi Y., le Sage C. Altered microRNA expression associated with chromosomal changes contributes to cervical carcinogenesis. Oncogene. 2013;32:106–116. doi: 10.1038/onc.2012.20. [DOI] [PubMed] [Google Scholar]
- 37.Witten D., Tibshirani R., Gu S.G., Fire A., Lui W.O. Ultra-high throughput sequencing-based small RNA discovery and discrete statistical biomarker analysis in a collection of cervical tumours and matched controls. BMC Biol. 2010;8:58. doi: 10.1186/1741-7007-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sannigrahi M.K., Sharma R., Singh V., Panda N.K., Rattan V., Khullar M. Role of host miRNA hsa-miR-139-3p in HPV-16-induced carcinomas. Clin. Cancer Res. 2017;23:3884–3895. doi: 10.1158/1078-0432.CCR-16-2936. [DOI] [PubMed] [Google Scholar]
- 39.Zheng Y., Yin L., Chen H., Yang S., Pan C., Lu S., Miao M., Jiao B. miR-376a suppresses proliferation and induces apoptosis in hepatocellular carcinoma. FEBS Lett. 2012;586:2396–2403. doi: 10.1016/j.febslet.2012.05.054. [DOI] [PubMed] [Google Scholar]
- 40.Luo M., Shen D., Zhou X., Chen X., Wang W. MicroRNA-497 is a potential prognostic marker in human cervical cancer and functions as a tumor suppressor by targeting the insulin-like growth factor 1 receptor. Surgery. 2013;153:836–847. doi: 10.1016/j.surg.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 41.Ding Q., He K., Luo T., Deng Y., Wang H., Liu H., Zhang J., Chen K., Xiao J., Duan X. SSRP1 contributes to the malignancy of hepatocellular carcinoma and is negatively regulated by miR-497. Mol. Ther. 2016;24:903–914. doi: 10.1038/mt.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adey A., Burton J.N., Kitzman J.O., Hiatt J.B., Lewis A.P., Martin B.K., Qiu R., Lee C., Shendure J. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature. 2013;500:207–211. doi: 10.1038/nature12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu L., Zhang X., Zhao Z., Wang L., Li B., Li G., Dean M., Yu Q., Wang Y., Lin X. Full-length single-cell RNA-seq applied to a viral human cancer: applications to HPV expression and splicing analysis in HeLa S3 cells. Gigascience. 2015;4:51. doi: 10.1186/s13742-015-0091-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chung I.F., Chang S.J., Chen C.Y., Liu S.H., Li C.Y., Chan C.H., Shih C.C., Cheng W.C. YM500v3: a database for small RNA sequencing in human cancer research. Nucleic Acids Res. 2017;45(D1):D925–D931. doi: 10.1093/nar/gkw1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujii T., Saito M., Iwasaki E., Ochiya T., Takei Y., Hayashi S., Ono A., Hirao N., Nakamura M., Kubushiro K. Intratumor injection of small interfering RNA-targeting human papillomavirus 18 E6 and E7 successfully inhibits the growth of cervical cancer. Int. J. Oncol. 2006;29:541–548. [PubMed] [Google Scholar]
- 46.Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 47.Hsieh C.L., Liu H., Huang Y., Kang L., Chen H.W., Chen Y.T., Wee Y.R., Chen S.J., Tan B.C. ADAR1 deaminase contributes to scheduled skeletal myogenesis progression via stage-specific functions. Cell Death Differ. 2014;21:707–719. doi: 10.1038/cdd.2013.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tie J., Pan Y., Zhao L., Wu K., Liu J., Sun S., Guo X., Wang B., Gang Y., Zhang Y. miR-218 inhibits invasion and metastasis of gastric cancer by targeting the Robo1 receptor. PLoS Genet. 2010;6:e1000879. doi: 10.1371/journal.pgen.1000879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., Boutselakis H., Cole C.G., Creatore C., Dawson E. COSMIC: the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2019;47(D1):D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Landen C.N., Kinch M.S., Sood A.K. EphA2 as a target for ovarian cancer therapy. Expert Opin. Ther. Targets. 2005;9:1179–1187. doi: 10.1517/14728222.9.6.1179. [DOI] [PubMed] [Google Scholar]
- 51.Oh D.Y., Bang Y.J. HER2-targeted therapies—a role beyond breast cancer. Nat. Rev. Clin. Oncol. 2020;17:33–48. doi: 10.1038/s41571-019-0268-3. [DOI] [PubMed] [Google Scholar]
- 52.Xue Y., Ouyang K., Huang J., Zhou Y., Ouyang H., Li H., Wang G., Wu Q., Wei C., Bi Y. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013;152:82–96. doi: 10.1016/j.cell.2012.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar M.S., Lu J., Mercer K.L., Golub T.R., Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 54.Jackson A.L., Burchard J., Leake D., Reynolds A., Schelter J., Guo J., Johnson J.M., Lim L., Karpilow J., Nichols K. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Supek F., Bošnjak M., Škunca N., Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.