Abstract
Uracil DNA glycosylases are an important class of enzymes that hydrolyze the N-glycosidic bond between the uracil base and the deoxyribose sugar to initiate uracil excision repair. Uracil may arise in DNA either because of its direct incorporation (against A in the template) or because of cytosine deamination. Mycobacteria with G, C rich genomes are inherently at high risk of cytosine deamination. Uracil DNA glycosylase activity is thus important for the survival of mycobacteria. A limitation in evaluating the druggability of this enzyme, however, is the absence of a rapid assay to evaluate catalytic activity that can be scaled for medium to high-throughput screening of inhibitors. Here we report a fluorescence-based method to assay uracil DNA glycosylase activity. A hairpin DNA oligomer with a fluorophore at its 5′ end and a quencher at its 3′ ends was designed incorporating five consecutive U:A base pairs immediately after the first base pair (5′ C:G 3’) at the top of the hairpin stem. Enzyme assays performed using this fluorescent substrate were seen to be highly sensitive thus enabling investigation of the real time kinetics of uracil excision. Here we present data that demonstrate the feasibility of using this assay to screen for inhibitors of Mycobacterium tuberculosis uracil DNA glycosylase. We note that this assay is suitable for high-throughput screening of compound libraries for uracil DNA glycosylase inhibitors.
Highlights
-
•
A novel molecular beacon based fluorescent method to assay uracil DNA glycosylase (UDG) activity has been developed.
-
•
The single step assay is useful to determine real-time kinetics of uracil release.
-
•
The assay is useful for high throughput screening of uracil DNA glycosylase inhibitors.
1. Introduction
Uracils are occasionally found in DNA because of the deamination of the resident cytosines or the direct incorporation of dUMP during replication [[1], [2], [3]]. Uracil DNA glycosylases (UDGs) recognize and excise the uracil base from DNA to initiate the base excision repair pathway. Of the six families of UDGs [4], family I UDGs (Ung), are evolutionarily conserved, and highly specific for excision of uracils from both the single stranded and double stranded DNAs [[4], [5], [6], [7], [8], [9]].
Mycobacterium tuberculosis Ung (MtuUng) plays a crucial role in maintaining the integrity of its genome, which is rendered vulnerable to cytosine deaminations not only because it is G, C rich but also because of the exposure of the pathogen to RNI and ROI discharges by the host macrophages [3,[10], [11], [12]]. Ung removes uracil by cleaving the N-glycosidic bond between the uracil base and the deoxyribose sugar phosphate backbone generating an apurinic/apyrimidinic (AP) site which is further acted upon by the downstream repair enzymes [1,3,[13], [14], [15], [16]]. MtuUng has been reported to be crucial for in vivo growth of M. tuberculosis [17]. The function of MtuUng makes it important to screen for inhibitors for various biochemical studies and growth interventions. With similar objectives, the inhibitors of DNA ligase of Haemophilus influenza, Staphylococcus aureus and Streptococcus pneumoniae; or DNA polymerase III of M. tuberculosis were deemed useful [18,19].
Ung activity assays were originally carried out using 3H uracil containing DNA as a substrate followed by the chromatography of the reaction products on anion exchange columns [20,21]. The free uracil (having no charge) elutes in the flowthrough and is quantified by scintillation counting. While this method provides a direct assay for Ung activity, it is cumbersome especially when a large set of reactions are to be analysed either for determining enzyme kinetics parameters or for setting up inhibitor screens. Subsequently, uracil containing DNA oligomer-based assays were developed [[22], [23], [24]] where the end labelled (radioactively or with a fluorescent probe) DNA oligomers are used in the reactions with Ung. In this assay, excision of uracil leaves behind an AP-site, which is susceptible to cleavage under alkaline conditions. Following the Ung reaction, the samples are treated with alkali and separated on denaturing polyacrylamide gels [25,26]. The presence of a faster migrating product band shows Ung activity. While this assay has been used extensively, the requirement of alkali treatment, as a second step in the assay, makes this method non-usable for real time monitoring of Ung activity. To overcome such limitations and to potentially study real time kinetics of the enzyme, molecular beacon based fluorescence assays were developed [22,27]. The assays employed (FITC)-d (GCACTUAAGAATTCACGCCATGTCGAAATTCTTAAGTGC)-Dabcyl [22] or 5′-6-FAM-CCACTUTTGAATTGACACGCCATGTCGATCAATTCAAAAGTGG-Dabcyl-3’ [27] oligomers and relied on an AP-endonuclease to cleave at the AP-sites. While this method eased the analysis of the reaction products, the use of the AP-endonuclease curtailed its versatility for the real time monitoring of uracil excision as the assay of the Ung activity is limited by the activity of the downstream enzyme (i. e. the AP-endonuclease). To overcome this limitation, subsequent improvements on the molecular beacon assays incorporated many uracils on both strands of the hairpin stem (FITC-GCACUUAAGAAUUCACGCCATGTCGAAAUUCUUAAGUGC)-Dabcyl) [22]. Although such a design avoided the AP-endonuclease step, the authors observed that it resulted in less accurate kinetic parameters of uracil excision [22]. Subsequently, other molecular beacon-based assays were designed, which suffered from the slow kinetics of fluorescence development on account of inefficient melting of the hairpin stem harbouring the fluorophore and the quencher pair [28]. It is worth noting that inefficient stem melting also limits the potential to generate maximal difference in the fluorescence of reacted and unreacted substrate in real time and displays high background.
We had earlier shown that Ung excises uracil residues from DNA oligomers even when they are flanked by AP-sites [29]. The observation was further supported by structural determinations of DNA bound Ung. In these structures, besides making specific contacts with the uracil base, Ung was seen to interact with only the flanking phosphate residues of the sugar-phosphate backbone [30,31]. We had reported that consecutively present uracils could be efficiently excised from DNA [29]. Based on this observation, for rapid melting of the stem region in the oligomer and an improved signal to background ratio, in this study, we designed a substrate (5′-5-FAM-CUUUUUGAGCTTTTGCTCAAAAAG-BHQ-1-3′) wherein five consecutive uracil residues in the stem region of the hairpin were incorporated. Further, we reasoned that given the high turnover of the Ung class of UDGs (~800 per min [1]) excision of consecutive uracils (in contrast to the earlier design where uracils were incorporated at different sites) would least affect the macrolevel kinetic parameters of uracil excision (Km and kcat), which are more likely to be limited by the step of the search of the uracil sites in the substrate. The hairpin oligomer was terminally attached with a commonly used fluorophore, 5-carboxyfluorescein (5-FAM) which is quenched by BHQ-1 [22,32]. Upon treatment with Ung, excision of the consecutive uracils results in unwinding of the stem region of the oligomer to rapidly separate the quencher from the fluorophore yielding an intense fluorescence signal. We demonstrate the sensitivity of this method and its application in determining the efficiency of inhibition of MtuUng by uracil derivatives [33].
2. Materials and methods
2.1. MtuUng purification
The MtuUng expression vector [34] was introduced into E. coli BL21 (DE3) Rosetta by the calcium chloride method of transformation [35]. A loopful of freshly obtained transformants were inoculated into 100 ml Luria–Bertani medium (LB), grown overnight and sub-cultured into 5.2 L LB containing 0.1 mg/ml ampicillin and 0.03 mg/ml chloramphenicol. The cultures were incubated at 24 °C for 36 h. Cells were harvested by centrifugation at 4 °C and resuspended in 15 ml 10 mM Tris–HCl (pH 8) containing 10% (v/v) glycerol, 500 mM NaCl and 1 mM phenyl methyl sulfonyl fluoride (PMSF), disrupted by sonication and spun in an SW-41Ti rotor at 25,000 rpm for 60 min at 4 °C. The supernatant was loaded onto a pre-equilibrated Ni–NTA column (5 ml) and eluted with imidazole gradient (0–500 mM) in the above mentioned buffer using FPLC. Fractions enriched for MtuUng were loaded on to Sephadex-G75 column, and eluted with a buffer containing 10 mM Tris-Cl (pH 8), 10% glycerol, 500 mM NaCl and 2 mM β-mercaptoethanol. The fractions enriched for MtuUng were pooled and loaded onto Ni–NTA column (1 ml), eluted with 280 mM imidazole, dialyzed against 20 mM Tris–HCl (pH 8), 10% glycerol, 2 mM β-mercaptoethanol and 50 mM NaCl, estimated by Bradford's method using bovine serum albumin (BSA) as standard, and stored at −20 °C [36].
2.2. DNA oligomer and Ugi
A hairpin DNA oligomer, containing five consecutive uracils (5′-CUUUUUGAGCTTTTGCTCAAAAAG-3′) and modified at the 5′ and 3′ ends by fluorescein (5-FAM) and black hole quencher (BHQ1) respectively, was obtained from Macrogen, Inc, S. Korea following HPLC purification, resuspended in water (Milli Q), to a concentration of 100 μM. The excitation and emission wavelengths of 5-FAM are 496 nm and 520 nm, respectively, whereas the absorbance maxima of BHQ-1 is 534 nm.
Ugi was used from the lab stock purified [37] from an overexpression construct of PBS1 phage encoded Ugi [38].
2.3. Compounds for inhibitor screening
The uracil derivatives selected for analyzing their potential for MtuUng activity inhibition (Table 1) were obtained from Sigma-Aldrich [33,39] and dissolved in 100% DMSO to prepare 100 mM stock solutions.
Table 1.
List of compounds screened with Dose-dependent inhibition analysis: All the compounds were treated with 0.3 x 10−9 M of MtuUng followed by addition of 30 x 10−9 M of the fluorescence -labelled oligomer. The reaction volume was taken to be 100 μl buffered by 50 mM Tris-Cl (pH 8), 1 mM Na2EDTA and 37.59 nM BSA (n = 3).
| S. No. | Inhibitor | Structure | Docking Score | IC50 in mM (mean ± SEM) |
|---|---|---|---|---|
| 1 | Uracil | 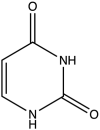 |
−10.268 | 2.05 ± 0.3 |
| 2 | 4-Thio-Uracil | 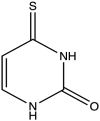 |
−4.806 | 3.57 ± 0.35 |
| 3 | 5- Iodo-Uracil | 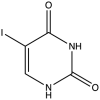 |
−9.103 | 4.92 ± 0.51 |
| 4 | 5-Chloro-Uracil |  |
−8.805 | 8.71 ± 0.70 |
| 5 | 5-Fluoro-Uracil | 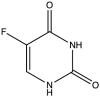 |
−8.963 | 50.18 ± 3.85 |
| 6 | 5-Nitro-Uracil | 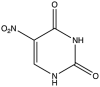 |
−5.054 | 10.77 ± 0.32 |
| 7 | 5-Amino-Uracil | 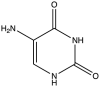 |
−9.636 | 7.18 ± 0.97 |
| 8 | 5-Bromo-Uracil | 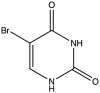 |
−8.733 | 13.59 ± 0.73 |
| 9 | Barbituric acid |  |
−12.03 | 7.29 ± 0.32 |
2.4. Assays for Ung activity and inhibition using the fluorescence assay
The reactions (100 μl) were carried out in a Corning clear-bottom 96 well black plates. The concentrations of the oligomer, MtuUng and Ugi were standardised to 30 nM, 0.3 nM and 2.1 nM, respectively. The reaction was carried out in a buffer containing 50 mM Tris-Cl (pH 8), 1 mM Na2EDTA and 37.6 nM BSA. A control reaction with 80 mM NaOH (in place of MtuUng) was included to monitor melting of the oligomer. After 1.5 mm amplitude of orbital shaking of 10 s, the fluorescence was quantified every 2 min for 40 min in bottom reading mode at 37 °C, with 495 nm as excitation wavelength and 520 nm as emission wavelength using Tecan infinite 200Pro. The excitation and emission bandwidth were 9 nm and 20 nm respectively.
2.5. Gel based method to monitor MtuUng activity on uracil containing substrate
The reactions (5 μl) set up as above, consisted of 6 nM MtuUng, 42 nM Ugi (when required) followed by the addition of 100 nM oligomer. The reactions were incubated at 37 °C for 40 min, treated or not treated with and 0.096 N NaOH at 95 °C for 10 min, mixed with loading buffer (0.5 mM Na2EDTA, 64% formamide) and analysed by electrophoresis on 20% polyacrylamide (19:1 cross-linking) and on Sapphire Biomolecular Imager at 488 nm.
2.6. Graphical analysis
All the graphs were plotted and analysed using GraphPad Prism Version 5.
3. Results and discussion
3.1. Development of fluorescence-based assay to analyse uracil excision by MtuUng
A 24-mer hairpin DNA oligomer containing five consecutive uracil residues in its stem, incorporated immediately after the cytosine at the 5′ end, was used as substrate. The oligomer has a high quantum yield fluorophore, 5-carboxyfluorescein attached with cytosine at the 5′ end which is quenched by black hole quencher, BHQ-1 attached with guanine at the 3’ end, and which efficiently overlaps the emission spectra of the fluorophore with low background signal [40]. The G-C pair at the end ensures stability to the hairpin and thus low background fluorescence. In the presence of MtuUng, the uracil excision from the oligomer generates AP sites and destabilises the duplex, opening it to emit fluorescent signal (Fig. 1A). Quantification of fluorescence was used to measure the enzymatic activity of MtuUng. The temporal increase in fluorescence intensity implies real time uracil removal by the enzyme, which was also confirmed by a similar increase in fluorescence signal due to the melting of the oligomer in the presence of alkaline environment of sodium hydroxide (NaOH), hence ensuring the feasibility of this method [41]. Importantly, we noticed that the kinetics of the rise of the fluorescence in the Ung reaction paralleled that of the NaOH added reaction, suggesting that the opening of the stem was instantaneous and represented real time measurements of the Ung reaction. The inhibition of the enzymatic activity and the lack of the fluorescence rise when Ugi [38,42,43] was pre-incubated with MtuUng (prior to adding the substrate) showed that the fluorescence rise in the MtuUng alone reaction is specific to uracil excision (Fig. 1B). Ugi, an early gene product encoded by Bacillus subtilis phage (PBS1/2), forms an extremely tight complex with Ung [44]. To further validate these observations we examined the reaction contents using native polyacrylamide gel electrophoresis (Fig. 1C). As expected, no fluorescence was observed in the absence of the oligomer (lane 1). The oligomer alone or when treated with MtuUng-Ugi complex, generated only the background fluorescence (compare lane 4 with lane 2), suggesting that inhibition of MtuUng activity by Ugi is total. However, when the oligomer was treated with MtuUng alone (lane 3), bright fluorescence is generated (albeit the gel electrophoresis conditions do lead to cleavage at the unstable AP sites, and the fluorescence is seen to migrate as diffuse band). A band migrating slower than the substrate and faintly fluorescent is also seen in this reaction. This could also be a product band where the uracil(s) have been excised but the stem has not completely melted. When the oligomer was treated with NaOH (at 37 °C) and then electrophoresed, it showed only the background fluorescence suggesting that the strong intramolecular stem-loop structure, required for low background, is restored as soon as the oligomer migrates into the gel. However, when similar to conventional methods, the reactions were treated with NaOH at 95 °C for 10 min to cleave the AP sites (lanes 6–9), the background fluorescence was slightly increased (lanes 6 and 8). The product band (corresponding to FAM-cytosine) showed a sharp and intensely fluorescent product band (lane 7), which migrated faster than the smear generated in the NaOH untreated reaction (lane 3). It may be noted that the presence of the various lengths of sugar moieties to FAM-cytosine (lane 3) would lead to its slower mobility compared to the product band seen in lane 7 [29]. The faint bands seen towards the end of the gel in lanes 6–8 most likely represent the free FAM generated by its cleavage from the oligomer because of treatment under alkaline conditions at high temperature of 95 °C. In fact, in these lanes we see some background bands (migrating between the substrate and product bands) which could be due to damage to the integrity of the stem loop structure of the oligomer when heated at 95 °C in the presence of NaOH. Nonetheless, the analyses in Fig. 1C further support the oligomer design and show that conventional treatment of the oligomer with NaOH at high temperature is not required to generate high signal to noise ratio of the fluorescence for Ung assays. Importantly, the treatment of the oligomer with NaOH at 37 °C does not show any degradation products (lane 5) suggesting that the fluorescence enhancement in the NaOH treated reaction in Fig. 1B, is due to the melting of the oligomer.
Fig. 1.
Fluorescence based real time analysis of uracil excision by MtuUng: [A] Schematic of the molecular beacon used for Ung assays. The action of MtuUng opens the hairpin structure resulting in recovery of fluorescence which was masked by the quencher in the closed state. [B] Real time detection of uracil excision by MtuUng as a measure of increasing fluorescence (purple). The assay was performed with 30 nM oligomeric substrate and 0.3 nM MtuUng. Addition of 2.1 nM Ugi or 80 mM NaOH in the reactions is as indicated. The reactions were carried out in 100 μl volumes in a buffer containing 50 mM Tris-Cl (pH 8), 1 mM Na2EDTA and 37.59 nM BSA, n = 3 and p < 0.0001 with control v/s Ung containing group. [C] Gel electrophoretic analysis of the reactions. Lanes 1–5 depict the products generated in the fluorescence assay (Fig. 1B) at the end of 40 min incubation. Lanes 6–9 show further treatment of the reactions with 0.096 N NaOH and heating at 95 °C. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. Assay of MtuUng activity and determination of kinetic parameters of uracil excision
To check the sensitivity of this assay, we performed an experiment with different concentrations of the substrate against a fixed enzyme concentration which showed direct relationship between fluorescence intensity and substrate concentration (Fig. 2A). The hyperbolic curve of velocity (vo) with respect to the substrate concentration [S] explained the velocity dependence on the substrate concentration (Fig. 2B) [45]. To avoid systemic errors, Km and Vmax (kcat) were calculated from the non-linear regression curve and plotted on double reciprocal graph (1/vo versus 1/[S]) to check goodness of fit (Fig. 2C). The double reciprocal graph yielded a straight line with R2 of 0.97 (the Lineweaver-Burk plot). Km derived from this method (152.3 ± 0.2 nM) is comparable to those determined earlier for MtuUng or other Ung proteins [[46], [47], [48]], and the calculated kcat was 0.38 ± 0.03 s−1. In addition, the results confirm the uracil excision being first order reaction with respect to MtuUng concentration.
Fig. 2.
Determination of kinetic parameters of uracil excision by MtuUng: [A] Background subtracted continuous graph showing linear relationship between product formation and substrate concentration in the presence of MtuUng. The enzyme concentration of 0.3 x 10−9 M was used with the substrate concentrations ranging from 40 x 10−9 to 640 x 10−9 M, n = 3. [B] Graph of the reaction velocities, calculated as the slopes of the data from [A], as a function of the oligomer concentration depicting the Michaelis-Menten trend. [C] The Lineweaver-Burk graph from data in [B] with inverse range of substrate concentration from 40x10−9-640x10−9 M to determine Km and Vmax, n = 3, R2 = 0.97.
3.3. Analysis of inhibitory effect of uracil derivatives on MtuUng activity
The products formed from the MtuUng action on uracil containing DNA are known to act as its inhibitors [1]. The product, uracil, binds to the enzyme and acts as its inhibitor [1,49]. To show the utility of the method and to investigate their efficiencies towards MtuUng, different uracil derivatives were screened to bind to MtuUng in terms of thermodynamic parameters [33] and computational analysis with measure of docking score using Schrödinger Software (Table 1). In addition, barbituric acid was checked owing to its high similarity to the uracil ring and its general applicability in inhibitor design for a variety of targets [[50], [51], [52], [53]]. The selected compounds were analysed for dose dependent inhibition. The graphs of fluorescence produced as a function of inhibitor concentration showed an IC50 (half-maximal inhibitory concentration) value which was taken as a measure of inhibitor efficiency (Table 1). The IC50 of uracil derived from this method is 2.05 ± 0.3 mM (Fig. 3A and B).
Fig. 3.
Dose-dependent inhibition analysis of MtuUng by uracil: [A] Normalised graph illustrating inverse relation of inhibitor (uracil) concentration (1 x 10−3 to 25 x 10−3 M) towards fluorescence production. Uracil was dissolved in DMSO (dimethyl-sulfoxide) and treated against 0.3 x 10−9 M enzyme in the presence of 30 x 10−9 M of substrate (n = 3). [B] The linear time course of graph A is then used to analyse relative product conversion with respect to log concentration of the inhibitor to determine dose-dependent response (IC50) of uracil on MtuUng (n = 3).
3.4. Application of the method in determination of inhibition kinetics
We could further exploit the real time detection of this method by understanding type of inhibition the enzyme undergoes by graphically plotting initial velocity of MtuUng in the presence of the inhibitor as a function of substrate concentration. For instance, one of the uracil derivatives, 5-chloro-uracil was checked by plotting such graphs (Fig. 4A). The Lineweaver-Burk plot of the same data showed conversion point of different inhibitor concentrations above the x-axis and to the left of y-axis, signifying linear mixed inhibition (Fig. 4B) [54]. The data reported earlier [1,[55], [56], [57], [58], [59]] had also suggested the a non-competitive mode of inhibition by uracil. In addition, the inhibitor efficiency was explored by means of Ki (inhibitory constant) [45]. To obtain values of Ki of 5-chloro-uracil, secondary graph was plotted. The slope (Fig. 4A) as a function of inhibitor concentration showed a value of -Ki (intercept on X axis) as 7.036 mM (Fig. 4B).
Fig. 4.
Inhibition kinetics of MtuUng by 5-chloro-uracil: [A] Kinetics of MtuUng inhibition by chloro-uracil as double reciprocal plots in the presence of different inhibitor concentrations. The inhibitors were dissolved in DMSO and used with 0.3 x 10−9 M of the enzyme in the presence of 30 x 10−9 M of the oligomer. The double-reciprocal graph interprets the type of inhibition by chloro-uracil towards MtuUng (n = 3). [B] Secondary plot of slopes from Lineweaver-Burk graph as a function of inhibitor concentration to determine Ki as x-intercept (n = 3).
4. Conclusions
The novelty of this method is defined by a single reaction, real time analysis of an enzyme (Ung) action on DNA. Unlike other methods, requirement of minimal constituents and quick results from the assay ensure speedy and reliable analysis of comparatively complex enzyme kinetics. The one step analysis is supported by the design of substrate since incorporation of multiple consecutive uracils in the oligomer eliminates the need for thermal or pH stimuli to facilitate the opening of the hairpin stem of the DNA oligomer to separate the fluorophore and the quencher. Moreover, the sensitivity of 5 nM difference in substrate shows the method as highly promising for detecting the in vitro efficiency of several biological enzymes acting on DNA damage. The inhibition analysis by this method, endorses high-throughput screening of compounds which can accelerate the process of drug discovery against infectious diseases by targeting their DNA associated proteins.
Declaration of competing interest
The authors declare no conflicts of interest.
Acknowledgments
We thank Dr. D. N. Rao for his critical comments on the manuscript, and Dr. M. Vijayan for his encouragement to carry out this work. This work was supported by funding from Principal Scientific Advisor's Office, Government of India, Department of Biotechnology (DBT), Ministry of Science and Technology, Science and Engineering Research Board (SERB), and Jamsetji Tata Trust. UV is a J. N. Tata Chair Professor and a J.C. Bose Fellow. The authors acknowledge the support of DBT-IISc partnership program, University Grants Commission, New Delhi for the Centre of Advanced Studies, and the DST-FIST level II infrastructure.
References
- 1.Lindahl T. DNA N-glycosidases: properties of uracil-DNA glycosidase from Escherichia coli. J. Biol. Chem. 1977;252(10):3286–3294. [PubMed] [Google Scholar]
- 2.Lindahl T. DNA repair enzymes. Annu. Rev. Biochem. 1982;51(1):61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- 3.Krokan H.E., Drabløs F., Slupphaug G. Uracil in DNA–occurrence, consequences and repair. Oncogene. 2002;21(58):8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- 4.Schormann N., Ricciardi R., Chattopadhyay D. Uracil‐DNA glycosylases—structural and functional perspectives on an essential family of DNA repair enzymes. Protein Sci. 2014;23(12):1667–1685. doi: 10.1002/pro.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savva R. The structural basis of specific base-excision repair by uracil–DNA glycosylase. Nature. 1995;373(6514):487–493. doi: 10.1038/373487a0. [DOI] [PubMed] [Google Scholar]
- 6.Mol C.D. Crystal structure and mutational analysis of human uracil-DNA glycosylase: structural basis for specificity and catalysis. Cell. 1995;80(6):869–878. doi: 10.1016/0092-8674(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 7.Xiao G. Crystal structure of Escherichia coli uracil DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited. Proteins: Structure, Function, and Bioinformatics. 1999;35(1):13–24. [PubMed] [Google Scholar]
- 8.Aravind L., Koonin E.V. The α/β fold uracil DNA glycosylases: a common origin with diverse fates. Genome Biol. 2000;1(4):1–8. doi: 10.1186/gb-2000-1-4-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zharkov D.O., Mechetin G.V., Nevinsky G.A. Uracil-DNA glycosylase: structural, thermodynamic and kinetic aspects of lesion search and recognition. Mutat. Res. Fund Mol. Mech. Mutagen. 2010;685(1–2):11–20. doi: 10.1016/j.mrfmmm.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurthkoti K., Varshney U. Detrimental effects of hypoxia-specific expression of uracil DNA glycosylase (Ung) in Mycobacterium smegmatis. J. Bacteriol. 2010;192(24):6439–6446. doi: 10.1128/JB.00679-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venkatesh J. Importance of uracil DNA glycosylase in Pseudomonas aeruginosa and Mycobacterium smegmatis, G+ C-rich bacteria, in mutation prevention, tolerance to acidified nitrite, and endurance in mouse macrophages. J. Biol. Chem. 2003;278(27):24350–24358. doi: 10.1074/jbc.M302121200. [DOI] [PubMed] [Google Scholar]
- 12.Kumar P., Bharti S.K., Varshney U. Uracil excision repair in Mycobacterium tuberculosis cell-free extracts. Tuberculosis. 2011;91(3):212–218. doi: 10.1016/j.tube.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Kubota Y. Reconstitution of DNA base excision‐repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 1996;15(23):6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 14.Nicholl I.D., Nealon K., Kenny M.K. Reconstitution of human base excision repair with purified proteins. Biochemistry. 1997;36(24):7557–7566. doi: 10.1021/bi962950w. [DOI] [PubMed] [Google Scholar]
- 15.Parikh S.S., Mol C.D., Tainer J.A. Base excision repair enzyme family portrait: integrating the structure and chemistry of an entire DNA repair pathway. Structure. 1997;5(12):1543–1550. doi: 10.1016/s0969-2126(97)00303-1. [DOI] [PubMed] [Google Scholar]
- 16.Raff M. fourth ed. National Center for Biotechnology InformationÕs Bookshelf; 2002. Molecular Biology of the Cell. [Google Scholar]
- 17.Sassetti C.M., Rubin E.J. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. Unit. States Am. 2003;100(22):12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mills S.D. Novel bacterial NAD+-dependent DNA ligase inhibitors with broad-spectrum activity and antibacterial efficacy in vivo. Antimicrobial agents and chemotherapy. 2011;55(3):1088–1096. doi: 10.1128/AAC.01181-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jadaun A., Subbarao N., Dixit A. In silico screening for novel inhibitors of DNA polymerase III alpha subunit of Mycobacterium tuberculosis (mtb DnaE2, H 37 R v) PloS One. 2015;10(3) doi: 10.1371/journal.pone.0119760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krokan H., Urs Wittwer C. Uracil DNA-glycosylase from HeLa cells: general properties, substrate specificity and effect of uracil analogs. Nucleic Acids Res. 1981;9(11):2599–2614. doi: 10.1093/nar/9.11.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindahl T. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc. Natl. Acad. Sci. Unit. States Am. 1974;71(9):3649–3653. doi: 10.1073/pnas.71.9.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maksimenko A. A molecular beacon assay for measuring base excision repair activities. Biochemical and biophysical research communications. 2004;319(1):240–246. doi: 10.1016/j.bbrc.2004.04.179. [DOI] [PubMed] [Google Scholar]
- 23.Delort A.-M. Excision of uracil residues in DNA: mechanism of action of Escherichia coli and Micrococcus luteus uracil-DNA glycosylases. Nucleic Acids Res. 1985;13(2):319–335. doi: 10.1093/nar/13.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bellamy S.R., Baldwin G.S. A kinetic analysis of substrate recognition by uracil-DNA glycosylase from herpes simplex virus type 1. Nucleic Acids Res. 2001;29(18):3857–3863. doi: 10.1093/nar/29.18.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaconas G., van de Sande J.H. Methods in Enzymology. Elsevier; 1980. [10] 5′-32P labeling of RNA and DNA restriction fragments; pp. 75–85. [DOI] [PubMed] [Google Scholar]
- 26.Maxam A.M., Gilbert W. Methods in Enzymology. Elsevier; 1980. [57] Sequencing end-labeled DNA with base-specific chemical cleavages; pp. 499–560. [DOI] [PubMed] [Google Scholar]
- 27.Li J. DNA Repair Molecular Beacon assay: a platform for real-time functional analysis of cellular DNA repair capacity. Oncotarget. 2018;9(60):31719. doi: 10.18632/oncotarget.25859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X. A rapid fluorescence assay for hSMUG1 activity based on modified molecular beacon. Mol. Cell. Probes. 2011;25(5–6):219–221. doi: 10.1016/j.mcp.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 29.Varshney U., van de Sande J.H. Specificities and kinetics of uracil excision from uracil-containing DNA oligomers by Escherichia coli uracil DNA glycosylase. Biochemistry. 1991;30(16):4055–4061. doi: 10.1021/bi00230a033. [DOI] [PubMed] [Google Scholar]
- 30.Kavli B. Excision of cytosine and thymine from DNA by mutants of human uracil‐DNA glycosylase. EMBO J. 1996;15(13):3442–3447. [PMC free article] [PubMed] [Google Scholar]
- 31.Parikh S.S. Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil‐DNA glycosylase with DNA. EMBO J. 1998;17(17):5214–5226. doi: 10.1093/emboj/17.17.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Didenko V.V. DNA probes using fluorescence resonance energy transfer (FRET): designs and applications. Biotechniques. 2001;31(5):1106–1121. doi: 10.2144/01315rv02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arif S. Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015;71(7):1514–1527. doi: 10.1107/S1399004715009311. [DOI] [PubMed] [Google Scholar]
- 34.Singh P. Overexpression, purification, crystallization and preliminary X-ray analysis of uracil N-glycosylase from Mycobacterium tuberculosis in complex with a proteinaceous inhibitor. Acta Crystallogr. F: Structural Biology and Crystallization Communications. 2006;62(12):1231–1234. doi: 10.1107/S1744309106045805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J., Russell D.W. Preparation and transformation of competent E. coli using calcium chloride. Cold Spring Harb. Protoc. 2006;2006(1):prot3932. doi: 10.1101/pdb.prot3932. p. pdb. [DOI] [PubMed] [Google Scholar]
- 36.Sedmak J.J., Grossberg S.E. A rapid, sensitive, and versatile assay for protein using Coomassie brilliant blue G250. Anal. Biochem. 1977;79(1–2):544–552. doi: 10.1016/0003-2697(77)90428-6. [DOI] [PubMed] [Google Scholar]
- 37.Acharya N., Roy S., Varshney U. Mutational analysis of the uracil DNA glycosylase inhibitor protein and its interaction with Escherichia coli uracil DNA glycosylase. Journal of molecular biology. 2002;321(4):579–590. doi: 10.1016/s0022-2836(02)00654-x. [DOI] [PubMed] [Google Scholar]
- 38.Roy S. Use of a coupled transcriptional system for consistent overexpression and purification of UDG–ugi complex and Ugi fromEscherichia coli. Protein Expr. Purif. 1998;13(2):155–162. doi: 10.1006/prep.1998.0878. [DOI] [PubMed] [Google Scholar]
- 39.Pałasz A., Cież D. In search of uracil derivatives as bioactive agents. Uracils and fused uracils: synthesis, biological activity and applications. Eur. J. Med. Chem. 2015;97:582–611. doi: 10.1016/j.ejmech.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 40.Yeung A.T. Evaluation of dual-labeled fluorescent DNA probe purity versus performance in real-time PCR. Biotechniques. 2004;36(2):266–275. doi: 10.2144/04362RR01. [DOI] [PubMed] [Google Scholar]
- 41.Ageno M., Dore E., Frontali C. The alkaline denaturation of DNA. Biophys. J. 1969;9(11):1281–1311. doi: 10.1016/S0006-3495(69)86452-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cone R., Bonura T., Friedberg E. Inhibitor of uracil-DNA glycosylase induced by bacteriophage PBS2. Purification and preliminary characterization. J. Biol. Chem. 1980;255(21):10354–10358. [PubMed] [Google Scholar]
- 43.Warner H., Johnson L., Snustad D. Early events after infection of Escherichia coli by bacteriophage T5. III. Inhibition of uracil-DNA glycosylase activity. J. Virol. 1980;33(1):535–538. doi: 10.1128/jvi.33.1.535-538.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Assefa N.G. Structural and biophysical analysis of interactions between cod and human uracil-DNA N-glycosylase (UNG) and UNG inhibitor (Ugi) Acta Crystallogr. Sect. D Biol. Crystallogr. 2014;70(8):2093–2100. doi: 10.1107/S1399004714011699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Copeland R.A. second ed. John Wiley & Sons; New York, NY: 2000. A Practical Introduction to Structure, Mechanism, and Data Analysis. Enzymes, p. 104. [Google Scholar]
- 46.Handa P., Roy S., Varshney U. The role of leucine 191 of Escherichia coliUracil DNA glycosylase in the formation of a highly stable complex with the substrate mimic, Ugi, and in uracil excision from the synthetic substrates. J. Biol. Chem. 2001;276(20):17324–17331. doi: 10.1074/jbc.M011166200. [DOI] [PubMed] [Google Scholar]
- 47.Purnapatre K., Varshney U. Uracil DNA glycosylase from Mycobacterium smegmatis and its distinct biochemical properties. Eur. J. Biochem. 1998;256(3):580–588. doi: 10.1046/j.1432-1327.1998.2560580.x. [DOI] [PubMed] [Google Scholar]
- 48.Krusong K. A comparative study of uracil-DNA glycosylases from human and herpes simplex virus type 1. J. Biol. Chem. 2006;281(8):4983–4992. doi: 10.1074/jbc.M509137200. [DOI] [PubMed] [Google Scholar]
- 49.Bennett S.E., Schimerlik M., Mosbaugh D. Kinetics of the uracil-DNA glycosylase/inhibitor protein association. Ung interaction with Ugi, nucleic acids, and uracil compounds. J. Biol. Chem. 1993;268(36):26879–26885. [PubMed] [Google Scholar]
- 50.Liao Y.-J. Treatment with a New barbituric acid derivative exerts antiproliferative and antimigratory effects against sorafenib resistance in hepatocellular carcinoma. Molecules. 2020;25(12):2856. doi: 10.3390/molecules25122856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Penthala N.R. 1-Benzyl-2-methyl-3-indolylmethylene barbituric acid derivatives: anti-cancer agents that target nucleophosmin 1 (NPM1) Bioorg. Med. Chem. 2015;23(22):7226–7233. doi: 10.1016/j.bmc.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sundriyal S. New PPARγ ligands based on barbituric acid: virtual screening, synthesis and receptor binding studies. Bioorg. Med. Chem. Lett. 2008;18(18):4959–4962. doi: 10.1016/j.bmcl.2008.08.028. [DOI] [PubMed] [Google Scholar]
- 53.Sokmen B.B. Antibacterial, antiurease, and antioxidant activities of some arylidene barbiturates. Appl. Biochem. Biotechnol. 2013;171(8):2030–2039. doi: 10.1007/s12010-013-0486-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Segel I.H. vol. 115. Wiley; New York: 1975. (Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady State Enzyme Systems). [Google Scholar]
- 55.Blaisdell P., Warner H. Partial purification and characterization of a uracil-DNA glycosylase from wheat germ. J. Biol. Chem. 1983;258(3):1603–1609. [PubMed] [Google Scholar]
- 56.Borle M., Campagnari F., Creissen D. Properties of purified uracil DNA glycosylase from calf thymus. J. Biol. Chem. 1982;257:1208ą1214. [PubMed] [Google Scholar]
- 57.Domena J.D. Purification and properties of mitochondrial uracil-DNA glycosylase from rat liver. Biochemistry. 1988;27(18):6742–6751. doi: 10.1021/bi00418a015. [DOI] [PubMed] [Google Scholar]
- 58.Williams M., Pollack J. A mollicute (mycoplasma) DNA repair enzyme: purification and characterization of uracil-DNA glycosylase. J. Bacteriol. 1990;172(6):2979–2985. doi: 10.1128/jb.172.6.2979-2985.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caradonna S., Cheng Y.-C. Uracil DNA-glycosylase. Purification and properties of this enzyme isolated from blast cells of acute myelocytic leukemia patients. J. Biol. Chem. 1980;255(6):2293–2300. [PubMed] [Google Scholar]






