Abstract
The evolutionary dynamics of polyploid genomes and consequences of polyploidy have been studied extensively in angiosperms but very rarely in gymnosperms. The gymnospermous genus Ephedra is characterized by a high frequency of polyploidy, and thus provides an ideal system to investigate the evolutionary mode of allopolyploid genomes and test whether subgenome dominance has occurred in gymnosperms. Here, we sequenced transcriptomes of two allotetraploid species of Ephedra and their putative diploid progenitors, identified expressed homeologs, and analyzed alternative splicing and homeolog expression based on PacBio Iso-Seq and Illumina RNA-seq data. We found that the two subgenomes of the allotetraploids had similar numbers of expressed homeologs, similar percentages of homeologs with dominant expression, and approximately equal numbers of isoforms with alternative splicing, showing an unbiased subgenome evolution as in a few polyploid angiosperms, with a divergence of the two subgenomes at ∼8 Ma. In addition, the nuclear DNA content of the allotetraploid species is almost equal to the sum of two putative progenitors, suggesting limited genome restructuring after allotetraploid speciation. The allopolyploid species of Ephedra might have undergone slow diploidization, and the unbiased subgenome evolution implies that the formation of large genomes in gymnosperms could be attributed to even and slow fractionation following polyploidization.
Keywords: transcriptome, genome evolution, unbiased subgenome evolution, allotetraploid, Ephedra, gymnosperm
Significance
The evolutionary dynamics of polyploid genomes and consequences of polyploidy have been very rarely studied in gymnosperms. Our previous study showed that the gymnospermous genus Ephedra is characterized by a high frequency of allotetraploidy, and thus provides an ideal system to explore the evolutionary mode of allopolyploid genomes and investigate whether subgenome dominance has occurred in gymnosperms. Our present study investigated the subgenome evolution in two allotetraploid species of Ephedra and found that, unlike most allopolyploid species of angiosperms, the two Ephedra species showed an unbiased subgenome evolution, indicating a slow diploidization. This finding also implies that the formation of large genomes in gymnosperms could be attributed to even and slow fractionation following polyploidization.
Introduction
A high frequency of polyploidy or whole-genome duplication (WGD) plays an important role in plant evolution and has broad effects on phenotypic diversification, ecological tolerance, and species richness for both autopolyploids and allopolyploids (Otto 2007; Paterson et al. 2010; Fawcett et al. 2013; Weiss-Schneeweiss et al. 2013; Jiao and Paterson 2014; Wendel 2015; Soltis PS and Soltis DE 2016; Rice et al. 2019). Following WGD, generally, the polyploids will revert to a stable status, similar to diploids, by fractionation, the loss of one copy of duplicated genes or cis-regulatory sites (Wendel 2015; Dodsworth et al. 2016; Soltis PS and Soltis DE 2016). In addition, subgenome dominance is commonly associated with polyploid evolution, with the dominant subgenome showing genome-wide high expression levels and more alternative splicing events (Liu et al. 2014; Mei et al. 2017) and retaining more ancestral genes (i.e., biased fractionation) immediately and over the long term (Cheng et al. 2018; Bird et al. 2019; Edger et al. 2019). Subgenome dominance has been observed in many allopolyploids of varying ages, such as Mimulus peregrinus (140 years old, Edger et al. 2017), Arabidopsis suecica (0.02 Ma, Novikova et al. 2017), maize (8 Ma, Schnable et al. 2011), Brassica rapa (15 Ma, Wang et al. 2011), Arabidopsis thaliana (47 Ma, Thomas et al. 2006), and Medicago sativa (58 Ma, Garsmeur et al. 2014), whereas it does not occur in autopolyploids such as Populus trichocarpa (Liu et al. 2017) and Musa acuminata (Garsmeur et al. 2014), and even a few allopolyploids such as soybean (Glycine max) (Zhao et al. 2017), Cucurbita maxima, and Cucurbita moschata (Sun et al. 2017).
In contrast to the high frequency of polyploids documented in angiosperms, polyploidy is exceedingly rare in gymnosperms, although gymnosperms are characterized by large genome sizes, with a mean value of 1 C = 18.35 pg, which is much larger than that of angiosperms (1 C = 5.9 pg) (Leitch and Leitch 2013). Nevertheless, recent studies have suggested that the evolution of gymnosperms was accompanied by several ancient WGD events (e.g., Li et al. 2015; Guan et al. 2016; Roodt et al. 2017; One Thousand Plant Transcriptomes Initiative 2019), and polyploidy is a dominant mode of speciation in Ephedra, a unique genus with 83% of the species being polyploids or having polyploid cytotypes (Wu et al. 2016; Ickert-Bond et al. 2020). Compared with numerous studies of the diploidization process in angiosperm allopolyploids, few studies have investigated the evolutionary dynamics and consequences of polyploidy in gymnosperms (only Juniperus in Farhat et al. 2019, and Ephedra in Ickert-Bond et al. 2015, 2020). Studies of the origin, accumulation, and fate of duplicated functional genes are helpful to unravel the mechanisms underlying genome evolution, including the evolution of large genome size and important pathways in gymnosperms. Moreover, it has been hypothesized that gymnosperms might not have an efficient way to eliminate nonfunctional gene copies, as observed in the accumulation of long terminal repeat retrotransposons (LTR-RTs), leading to the accumulation of degenerate gene copies and gene-like sequences (Prunier et al. 2016). However, this hypothesis needs to be tested empirically.
Ephedra provides an ideal system for investigating evolutionary dynamics of polyploid genomes and consequences of polyploidy in gymnosperms. With the exception of the natural polyploid species in three genera of Cupressaceae (Fitzroya cuprssoides and Sequioa sempervirens, Ahuja 2005; 17.3% of Juniperus species, Farhat et al. 2019) and sporadic polyploids with multiple and aneuploid chromosome numbers in Amentotaxus (Chuang and Hu 1963; Guan et al. 1993; Zhou et al. 2000), Pseudolarix amabilis (Murray 2012), Encephalartos hildebrandtii (Abraham and Mathew 1966), and Gnetum montanum (Ickert-Bond and Renner 2016), all other natural polyploid species of gymnosperms belong to Ephedra, in which 83% of species show tetraploid or very rarely octoploid cytotypes (Khoshoo 1959; Huang et al. 2005; Wu et al. 2016; Ickert-Bond et al. 2020). In particular, these polyploid species originated mainly in the Neogene with a crown age of all extant Ephedra species dated to ∼30 Ma (Ickert-Bond et al. 2009), although the earliest fossil record of the genus was dated to the Early Cretaceous (Yang and Wang 2013). Based on the analyses of two single-copy nuclear genes (LFY and DDB2) and two chloroplast DNA fragments, Wu et al. (2016) inferred that all polyploid species of Ephedra distributed in the Qinghai–Tibetan Plateau (QTP) and neighboring areas, such as Ephedra saxatilis, Ephedra intermedia, and Ephedra sinica, are allotetraploids, and deduced that the high frequency of allopolyploid speciation could be associated with some biological features of Ephedra, such as a shrub habit and vegetative propagation. The allotetraploid E. sinica is a shrub or small erect herbaceous shrub that tends to be clonal, with a vast distribution from northwestern China northward to Mongolia and Russia and eastward to the Gulf of Bohai. Ephedra sinica has been used as a traditional Chinese medicine for over 5,000 years and is still being used in various Ephedra-containing herbal mixtures all over the world (Hagel et al. 2012). This species experienced a WGD event, by tetraploidization after hybridization with one diploid species most closely related to Ephedra przewalskii and Ephedra regeliana as the maternal donor and another diploid species most closely related to Ephedra equisetina–Ephedra minuta–Ephedra monosperma as the paternal donor (Wu et al. 2016). The allotetraploid E. intermedia also has a vast distribution and its putative progenitors are similar to those of E. sinica. Moreover, significant ecological divergence has occurred between the allotetraploid species and their putative progenitors (Wu et al. 2016). Therefore, it is of great interest to investigate how the subgenomes of these allopolyploid species evolved and whether the expression patterns and evolutionary dynamics of their subgenomes are correlated to the genome size evolution and biological features.
Although RNA-seq using short-read sequencing technology has been increasingly used in studying plant transcriptomes, it is still challenging to assemble transcriptomes of allopolyploid species without reference genome sequences because homeologs are difficult to disentangle, particularly when the divergence between subgenomes is low at the sequence level. In contrast, using long-read sequencing technology, full-length isoforms can be directly obtained from sequencing without assembly (Sharon et al. 2013). Recently, Pacific Biosciences (Pacific Biosciences of California Inc., Menlo Park, CA) single-molecule real-time long-read isoform sequencing has performed well in sequencing transcriptomes of cotton, maize, and sorghum, especially accurately predicting alternative splicing and revealing transcriptomic complexity (Abdel-Ghany et al. 2016; Wang et al. 2016, 2018). In the present study, we selected Ephedra sinica, E. intermedia, and their putative progenitors to investigate subgenome evolution in allotetraploid species of Ephedra. First, isoforms of tetraploids were generated on PacBio Iso-Seq, and were used to identify expressed homeologs and explore the patterns of alternative splicing. Then, transcriptome sequencing was conducted on the Illumina HiSeq 2000/2500 platform, and was used to investigate homeolog expression of the two subgenomes. Finally, based on a comprehensive analysis of genome size, expression patterns of subgenomes, and biological attributes, we discussed the mechanisms underlying the evolution of large genomes and the possible correlation between allopolyploid speciation and some biological features in gymnosperms.
Materials and Methods
Plant Sampling and an Outline of Methods
Two allotetraploid species (Ephedra sinica and E. intermedia) and their putative diploid progenitors (E. equisetina, E. minuta, E. monosperma, E. przewalskii, and E. regeliana), and an outgroup species Ephedra rhytidosperma (based on our unpublished research) were sampled. For the two allotetraploid species, a total of 8 samples, representing different tissues (young stems and female strobili) and environments (field and green house), were analyzed. For the five diploid species and the outgroup, each species was represented by only one sample (young stem) collected in the field. All 14 samples were immediately immersed in RNAlater Solution (Life Technologies) after collection. The details of sampling are shown in table 1 and supplementary table S1, Supplementary Material online.
Table 1.
Statistics of Sampled Transcriptomes
| Category | Species | Ploidy Level | Pop. | Sample | Illumina Sequencing and Data Analyses |
PacBio Iso-Seq and Data Analyses |
|||
|---|---|---|---|---|---|---|---|---|---|
| No. of Reads After Filtering | No. of CDS | No. of Polymerase Reads | No. of Consensus Transcripts | No. of High-Quality Consensus Transcripts | |||||
| Putative maternal progenitors | Ephedra equisetina | 2x | ZL | S | 30,714,049 (100 bp) | 30,158 | |||
| Ephedra monosperma | 2x | YX | S | 37,104,728 (100 bp) | 27,102 | ||||
| Ephedra minuta | 2x | MY | S | 29,722,252 (100 bp) | 34,623 | ||||
| Putative paternal progenitors | Ephedra przewalskii | 2x | KLMY | S | 51,820,340 (100 bp) | 28,992 | |||
| Ephedra regeliana | 2x | YS | S | 30,074,428 (100 bp) | 26,883 | ||||
| Polyploids | Ephedra sinica | 4x | XW | S | 21,987,457 (100 bp) | 1,129,502 | 179,191 | 34,517 | |
| F | 23,947,858 (100 bp) | 1,035,706 | 184,925 | 38,655 | |||||
| KB | S | 20,206,814 (150 bp) | 929,043 | 159,591 | 26,623 | ||||
| F | 18,600,852 (150 bp) | 764,474 | 184,929 | 46,439 | |||||
| XL-W | S | 18,902,491 (150 bp) | 586,803 | 123,351 | 26,370 | ||||
| F | 18,724,916 (150 bp) | 566,519 | 131,275 | 28,437 | |||||
| XL-C | S | 20,526,994 (150 bp) | 688,763 | 152,536 | 29,753 | ||||
| Ephedra intermedia | 4x | INT | S | 15,570,339 (150 bp) | 481,241 | 98,103 | 21,950 | ||
| Outgroup | Ephedra rhytidosperma | 2x | RHY | S | 32,633,573 (100 bp) | 32,414 | |||
Note.—XL-C, transplanted from population XL and cultivated in the green house of the Institute of Botany, Chinese Academy of Sciences.
Combining the advantages of Illumina sequencing and PacBio Iso-Seq, our study was conducted as follows: 1) full-length transcriptomes of polyploid samples were sequenced by PacBio Iso-Seq, and transcriptome sequences of diploid samples were obtained from de novo assembly of short reads generated on the Illumina platform; 2) expressed homeolog identification and alternative splicing analysis were performed for the full-length transcriptomes of polyploid samples based on the reference of one-to-one orthologous groups (OGs) identified from transcriptomes of diploid species; 3) single-nucleotide polymorphisms (SNP) analysis was further conducted based on the reads of both diploids and polyploids generated on the Illumina platform to investigate homeolog expression patterns of the two subgenomes of polyploids with the transcriptome of E. regeliana (a putative diploid progenitor) as the reference, due to the lack of a reference genome in Ephedra and the complexity of transcriptomes of polyploid species (supplementary fig. S1, Supplementary Material online).
Illumina Sequencing and Data Analyses
For all samples, total RNA was extracted using the RNAplant Plus Reagent (Tiangen, China). Sequencing libraries were prepared using a NEBNext Ultra RNA Library Prep Kit for Illumina (NEB) and then sequenced on an Illumina HiSeq 2000/X-Ten platform with 100-bp/150-bp paired-end raw reads (supplementary table S1, Supplementary Material online). For the five diploid samples, reads filtering and de novo assembly were performed with Trimmomatic 0.38 (Bolger et al. 2014), Trinity 2.0.6 (Grabherr et al. 2011; Haas et al. 2013), CD-HIT 4.6.8 (Li and Godzik 2006), and TransDecoder 0.36 (https://github.com/TransDecoder, last accessed December 08, 2020), as described in Ran et al. (2018). The completeness of transcripts was evaluated using BUSCO (Benchmarking Universal Single-Copy Orthologs) v4 (Simão et al. 2015) with the Embryophyta (odb10) database. One-to-one OGs were identified by OrthoFinder 2.1.2 (Emms and Kelly 2015), following Liu et al. (2019).
PacBio Iso-Seq and Data Analyses
For the polyploid species, cDNA of each sample was synthesized using the SMARTer PCR cDNA Synthesis Kit (Clontech Laboratories, CA). PCR amplification and size fractionation (1–6 kb, 0.5–6 kb) were conducted using the KAPA HiFi PCR Kits (Kapa Biosystems) and BluePippin Size Selection System (Sage Science), respectively. Libraries were constructed using the SMRTbell Template Prep Kit 1.0 (PacBio) and sequenced on a PacBio Sequel Platform. Sequence data were analyzed using SMRT Link 5.1 (http://www.pacb.com/products-and-services/analytical-software/smrt-analysis/, last accessed December 08, 2020). The raw reads were filtered with the settings of length >100, pass >3, and accuracy >0.75, and then selected nonchimeric reads were classified into nonfull-length reads and full-length reads, which were determined by a length of at least 300 bp and presence of poly(A) tails, 5′ primers and 3′ primers. Further, full-length reads were processed by isoform-level clustering (ICE) to obtain unpolished consensus transcripts. Finally, full-length consensus transcripts were polished using the Quiver software module for error correction and categorized into high-quality consensus transcripts (min_accuracy >0.99, min_pass >2) and low-quality consensus transcripts. The high-quality consensus transcripts were corrected based on the corresponding Illumina RNA-seq data using the software Proovread 2.14.0 (Hackl et al. 2014), and then the coding sequences (CDS) were predicted using TransDecoder 0.36 (https://github.com/TransDecoder, last accessed December 08, 2020), and redundant sequences were removed using CD-HIT 4.6.8 (Li and Godzik 2006). The completeness of high-quality consensus transcripts was evaluated by using BUSCO4 (Benchmarking Universal Single-Copy Orthologs) (Simão et al. 2015) with the Embryophyta (odb10) database.
Identification of Expressed Homeologs
For the allopolyploid samples, the subgenomes derived from the paternal parent and the maternal parents were designated as P subgenome and M subgenome, respectively. Because genic regions of the P and M subgenomes were highly similar, we developed a pipeline to separate these homeologs based on homeologous SNPs matched, respectively, to the sequences of the putative paternal parents E. equisetina–E. minuta–E. monosperma and the maternal parents E. przewalskii–E. regeliana in the alignment of one-to-one OGs using custom MATLAB 2014b scripts snp_based_on_sequence.m (https://github.com/yazhicao/Ephedraanalysis/, last accessed December 08, 2020). Consensus transcripts were aligned to the OGs identified in the five diploid species using BLAST. We removed low quality or short sequences from the alignment and counted the number of SNPs in each consensus transcript that are only shared with one parent, corresponding to maternal (M) sites and paternal (P) sites. Based on the SNPs, after removing recombination sequences, the sequences with at least four M sites were considered as homeologs from subgenome M, and the sequences with at least four P sites were considered as homeologs from subgenome P. We set a minimum of 4 SNPs because of the low site variation in the coding sequences. Only the isoforms that can be unambiguously assigned to the parental species were included in the analysis. The isoforms with the proportion of M sites >0.8 were assigned to subgenome M, and those with the proportion of M sites <0.2 were assigned to subgenome P.
To verify the accuracy of the above pipeline, a phylogenetic approach was also used to identify expressed homeologs. The alignments of the OGs identified in the five diploid species and the orthologous consensus transcripts of polyploid samples were, respectively, used to construct ML trees using RaxML 8.2.11 (Stamatakis 2014) with 100 bootstrap replicates and the GTRGAMMA model. After excluding the trees with bootstrap support values lower than 60% at node E. equisetina–E. minuta–E. monosperma-consensus transcript of polyploids and node E. przewalskii–E. regeliana-consensus transcript of polyploids, statistics of expressed homeologs were performed using Newick utilities 1.7.0 (Junier and Zdobnov 2010).
Gene ontology (GO) annotation of the identified OGs was obtained by Blast2GO program (Conesa et al. 2005) against the Nr annotation, and GO categories were analyzed using the Web Gene Ontology Annotation Plot (WEGO 2.0) (Ye et al. 2018).
To investigate the expression patterns of homeologs, pairwise Pearson correlations were examined between samples and a clustering map was drawn based on the expressed homeologs of each OG. Moreover, to explore the minimal number of consensus transcripts that are required to obtain the maximum number of expressed OGs and OGs that homeolog pairs from both parents are expressed, a series of subdata sets, including 30,000–150,000 sequences with an increment of 30,000 sequences with 3 replicates, were extracted from the consensus transcripts of the samples KB-F and XW-S, respectively. For each subdata set, the identification of expressed homeologs was performed as mentioned earlier. Then, we used the polynomial function of degree 2 in MATLAB 2014b (poly_curve.m) to fit the saturation curve between the logarithmic number of consensus transcripts and the logarithmic number of expressed OGs, and between the logarithmic number of consensus transcripts and the logarithmic number of OGs where both homeologs are expressed, respectively. The two polynomial functions are expressed as:
where x is the number of consensus transcripts, y1 is the number of expressed OGs, and y2 is the number of OGs.
Phylogenetic Analysis and Divergence Time Estimation
Phylogenetic relationships of the polyploids and their putative diploid progenitors were reconstructed using the OGs found in the diploids that have homeologs (high-quality consensus transcripts) in both subgenomes of the polyploids, with E. rhytidosperma as the outgroup. The different samples of polyploids were used, separately, in the identification of homeologs, and then the homologous OGs found in different tissue samples of the same individual were combined in the phylogenetic analysis. For the OGs with multiple high-quality consensus transcripts of the polyploid samples, only one transcript with the maximum length and the least numbers of single-nucleotide insertions/deletions was retained from each subgenome, and then the errors of single-nucleotide insertions/deletions were manually corrected.
Both concatenation and coalescence strategies were used in phylogenetic reconstruction. In the concatenation analysis, all OGs were combined into a concatenated supermatrix using FASConCAT-G 1.02 (Kück and Longo 2014), and a maximum-likelihood (ML) tree was generated by RaxML 8.2.11 (Stamatakis 2014) using the GTRGAMMA model with 100 bootstrap replicates. In the coalescence analysis, the ML tree was generated for each OG using RaxML 8.2.11 with the same parameter settings as above, and then all individual gene trees were used to estimate the species tree in ASTRAL 5.7.3 (Mirarab et al. 2014).
To estimate the divergence times between the putative diploid progenitors and the ages of the polyploids, the OGs of one individual of E. sinica (KB) and E. intermedia were used to determine density distributions of synonymous substitution rates (Ks), considering that the two subgenomes of E. sinica form reciprocal monophyletic groups (see Results). We estimated Ks for sequence pairs using paraAT 2.0 (Zhang et al. 2012). After excluding Ks values <0.001 to avoid spurious frequency peaks, Gaussian mixture models were used to identify significant peaks in the Ks distribution with the best fitting model selected based on Bayesian information criterion scores using script gaussian_analysis.m. According to the phylotranscriptomic study of gymnosperms (Ran et al. 2018), an average mutation rate of 4.8 × 10−9 synonymous substitutions per synonymous site per year for Ephedra was used to estimate approximate ages of the polyploids.
Analysis of Alternative Splicing
Alternative splicing (AS) analysis can also provide evidence for gene expression patterns in different subgenomes. Recent studies have shown that it is feasible to use PacBio sequences to identify AS events by searching for deletions or insertions in the clustering units when reference genomes are unavailable (Ner-Gaon et al. 2007; Zhou et al. 2011; Wu et al. 2014; Liu et al. 2017). Based on the alignments composed of consensus transcripts from all samples of E. sinica or E. intermedia for each one-to-one OG, the longest CDS of the allotetraploids was predicted by the merge of high-quality consensus isoforms’ CDS and validated as the reference. The AS events of each OG were identified with lengths of deletion or insertion >51 bp from the alignment.
SNP Analysis
Due to the lack of a reference genome in Ephedra, filtered clean reads of both diploid and polyploid samples were mapped to E. regeliana (the best reference species based on the phylogenetic analysis, see Results) using BWA-MEM (Li and Durbin 2009), with default parameters. The mapped reads were sorted with SAMtools 1.1 (Li et al. 2009). Variant calling was conducted using HaplotypeCaller and GenotypeGVCFs in Genome Analysis Toolkit GATK 3.6 (McKenna et al. 2010; DePristo et al. 2011). To obtain high-quality SNPs, variant sites were filtered using GATK’s VariantFiltration tool based on the following criteria: quality of depth <2.0, Fisher strand bias (FS) >10.0, mapping quality (MQ) <40.0, depth of coverage (DP) <30.0, ReadPosRankSum <−8.0, and genotype quality (GQ) <20.0. We focused on SNPs that are not shared between two parents, and then classified them into heterozygous sites (SPM) that have fixed differences between two subgenomes for polyploid samples, or homozygous sites (SPP or SMM) that are only shared with maternal or paternal parents, where PP and MM represent paternal and maternal homozygosities, respectively (SNP_analysis.m). The autapomorphic SNPs in either the diploids or one subgenome of the polyploids were excluded from the analysis. Further, genes with at least four informative “heterozygous” sites or four homozygous sites were classified into GPM, GPP, or GMM. For these genes, homeolog-specific expression was measured by calculating the proportion of all reads mapping to the subgenome P using the DepthPerAlleleBySample values found in the VCF file.
Genome Size Estimated by Flow Cytometry
Fresh young branchlets were used in the flow cytometry measurement for each species, mainly following the one-step protocol of Doležel et al. (2007). Vicia faba L. “Inovec” (2 C = 26.90 pg) (Doležel et al. 1992) was selected as the internal standard and Galbraith’s buffer was used as the most appropriate nuclei isolation buffer. The DNA ploidy levels were inferred based on the DNA content measured in E. equisetina (2 C = 16.61 pg), a diploid species with 14 chromosomes (Wu et al. 2016).
Results
Transcriptome Data Collection and Processing
A total of 8 transcriptomes, representing different tissues (young stems and female strobili) and environments (field and green house), were generated from the two allotetraploid species E. sinica and E. intermedia using PacBio Sequel and Illumina platforms. For the full-length transcriptomes, we obtained 16.7–30 G raw data for each sample, including 481,241–1,129,502 polymerase reads with average lengths of 14,752–43,461 bp. The mean number of passes in polymerase reads was almost higher than 15, indicating that the circular consensus sequences have a high accuracy according to the PacBio sequencing study of Eid et al. (2009) (15 passes may yield >99% accuracy). After the clustering step, a total of 98,103–184,929 consensus transcripts were collected and the average lengths were 1,350–2,257 bp, of which 21,950–46,439 were high-quality transcripts (min_accuracy >0.99, min_pass >2) (table 1 and supplementary table S2 and fig. S2, Supplementary Material online). For the Illumina HiSeq data, an average of 42.94 million (M) clean reads were obtained per sample (table 1).
In addition, transcriptome sequences of the five putative progenitors E. przewalskii, E. regeliana, E. equisetina, E. minuta, and E. monosperma were generated on the Illumina platform and used as references. For each sample, we obtained 41,602–69,343 transcripts with an N50 value of 1,464–1,703 bp from de novo assembly. The number of predicted CDS varied from 26,883 to 34,623 (table 1 and supplementary table S1, Supplementary Material online), from which 6,245 one-to-one OGs were obtained, with aligned length ranging from 150 to 6,735 bp. Based on the BUSCO assessment, the completeness of the transcripts from diploids is better than that of the high-quality transcripts from polyploids (supplementary fig. S3, Supplementary Material online).
Characterization of Expressed Homeologs Based on PacBio Iso-Seq Data
The consensus transcripts of 8 polyploid samples were aligned to the 6,245 one-to-one OGs. The expressed homeologs corresponding to the P subgenome (represented by E. equisetina–E. minuta–E. monosperma) and the M subgenome (represented by E. przewalskii–E. regeliana) were identified based on the homeologous SNPs. The OGs with expressed homeolog pairs from both subgenomes were denoted as HMP, and the OGs with expressed homeologs only from subgenome M or subgenome P were denoted as HM or HP. After filtering, 206,314 isoforms (accounting for 47% of the aligned isoforms and 17% of all isoforms, supplementary table S3, Supplementary Material online) were well-classified into 5,402 OGs, with the mean length of isoforms ranging from 974.8 to 1,311.1 bp and the average SNPs per isoform ranging from 13.1 to 17.3. The numbers of transcripts assigned to subgenome M and subgenome P, and the proportions of M sites in the isoforms are shown in supplementary figure S4, Supplementary Material online. Among the 5,402 OGs, 4,893 were annotated to Nr annotation and classified into 3 groups based on GO terms, including 3,523 in “biological process,” 4,773 in “cellular component,” and 3,845 in “molecular function” (supplementary fig. S5, Supplementary Material online).
In E. sinica, we obtained 3,438–4,389 expressed OGs from different samples, with the average number of isoforms ranging from 5.1 to 10.8 (supplementary table S4, Supplementary Material online). For each sample, the HMP expression was detected in most OGs (2,164–3,285, accounting for 62–75%). In contrast, the HM expression and the HP expression occurred in fewer OGs and in approximately equal proportions. Similar homeolog expression patterns were found in E. intermedia, in which HMP, HM, and HP in identified 3,097 OGs accounted for 60%, 19%, and 21%, respectively (table 2). Among all of the samples, the female strobili of an individual of E. sinica from population KB (KB-F) showed the highest number of expressed OGs (4,389), the most consensus transcripts (184,929) with the highest average number of isoforms per OG (10.8), and the highest HMP expression (75%) (table 2 and supplementary table S4, Supplementary Material online). These results indicated that no obvious expression difference occurred between the two subgenomes (M and P) of the allotetraploid species, although this analysis was only based on the presence/absence of expressed homeologs and the expression patterns showed a little difference between tissues. The unbiased homeolog expression in different subgenomes was also revealed by the phylogenetic analysis (supplementary table S5, Supplementary Material online).
Table 2.
Distribution Patterns of Expressed Homeologs
| SampleCategory |
Ephedra sinica
|
Ephedra intermedia
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| XW |
KB |
XL-W |
XL-C |
INT |
|||||||
| S | F | S + F | S | F | S + F | S | F | S + F | S | S | |
| Total | 3,509 | 3,995 | 4,528 | 3,614 | 4,389 | 4,789 | 3,438 | 3,575 | 4,369 | 3,514 | 3,097 |
| HMP | 2,164 (62%) | 2,636 (66%) | 3,364 (74%) | 2,571 (71%) | 3,285 (75%) | 3,833 (80%) | 2,257 (65%) | 2,481 (69%) | 3,364 (77%) | 2,492 (71%) | 1,874 (60%) |
| HM | 614 (17%) | 641 (16%) | 535 (12%) | 480 (13%) | 504 (11%) | 423 (9%) | 575 (17%) | 534 (15%) | 475 (11%) | 470 (13%) | 576 (19%) |
| HP | 731 (21%) | 718 (18%) | 629 (14%) | 563 (16%) | 600 (14%) | 533 (11%) | 606 (18%) | 560 (16%) | 530 (12%) | 552 (16%) | 647 (21%) |
Note.—HMP, OGs with expressed homeolog pairs from both subgenomes; HM, OGs with expressed homeologs only from subgenome M; HP, OGs with expressed homeologs only from subgenome P; S, young stems; F, female strobili.
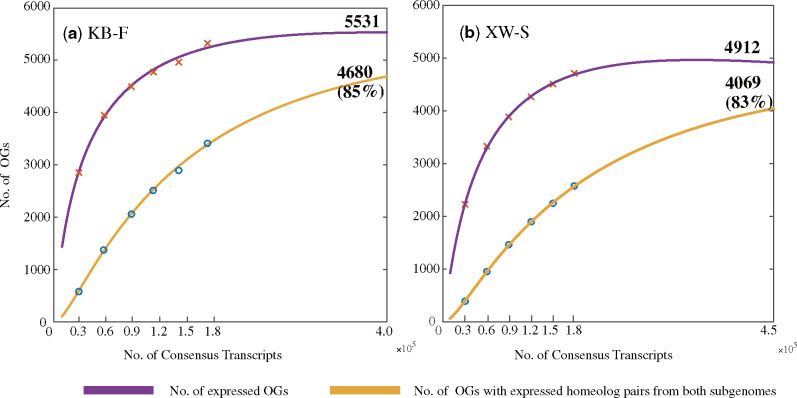
Based on the expression patterns of homeologs, no obvious correlation was found between the polyploid samples, with the pairwise Pearson correlation coefficients ranging from 0.20 to 0.42 (supplementary table S6, Supplementary Material online). The distributions of the expressed homeologs in each OG are shown in supplementary figure S6, Supplementary Material online, which also does not show obvious correlation between samples except that 825 OGs with expressed homeolog pairs from both subgenomes are shared among at least seven samples. In addition, HM and HP mostly (60–81%) occurred in OGs at a low coverage with one to four isoforms (supplementary table S4, Supplementary Material online). To further investigate the maximum number and proportion of expressed OGs and HMP, the saturation curve was used to predict the relationship between the number of consensus transcripts and the number of expressed OGs, and between the number of consensus transcripts and the number of HMP. We found that, in the sample KB-F, the saturation values of expressed OGs and HMP were close to 5,531 and 4,690, respectively, indicating that the proportion of HMP could be close to 85% when the number of consensus transcripts ≥400,000. In the young stem sample of an individual of E. sinica from population XW (XW-S), the proportion of HMP was also up to 83% with 450,000 consensus transcripts (fig. 1).
Fig. 1.
Saturation curves used to predict the maximum numbers of expressed OGs and OGs with expressed homeolog pairs from both subgenomes.
Phylogenetic Reconstruction and Divergence Time Estimation
Among the 6,245 OGs found in the diploid species, 3,953 OGs matched homeologs (high-quality consensus transcripts) in the two subgenomes of the two polyploid species, including 2,627 OGs in XW, 2,802 OGs in KB, 2,231 OGs in XL-W, and 1,033 OGs in XL-C of E. sinica, and 871 OGs in INT of E. intermedia, which were used to infer phylogenetic relationships. The length of the concatenated sequences was 4,666,169 bp, with 179,269 variable sites and 86,671 parsimony-informative sites.
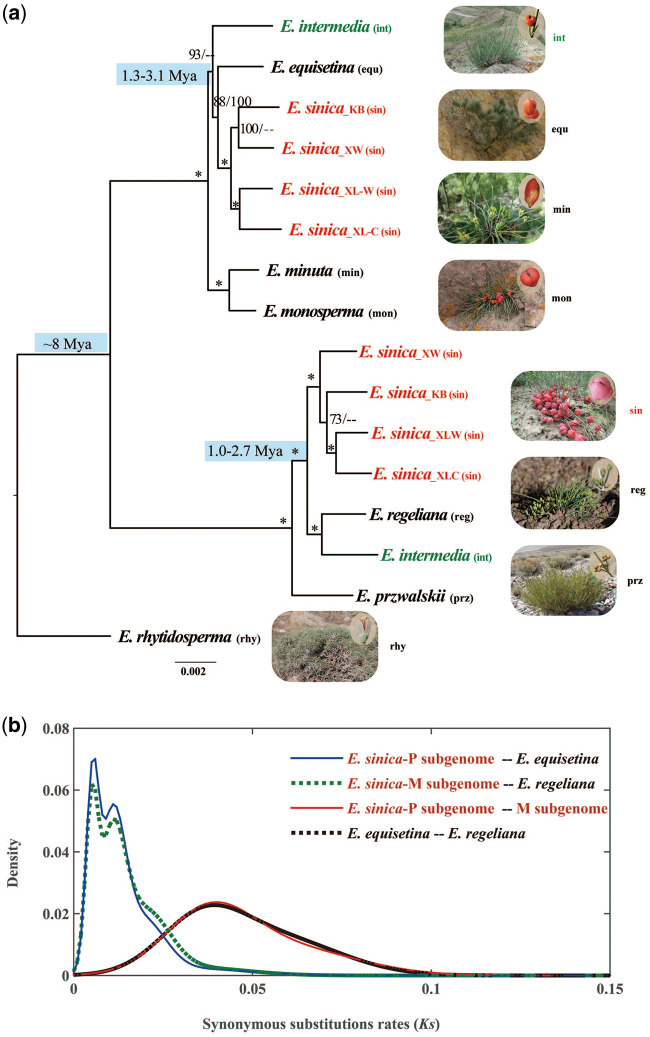
The phylogenies reconstructed based on the concatenation and coalescence methods are largely consistent in topology with high bootstrap support (fig. 2a). Two clades were resolved, one containing E. equisetina, E. minuta, E. monosperma, E. sinica-P subgenome, and E. intermedia-P subgenome, and the other comprising E. przewalskii, E. regeliana, E. sinica-M subgenome, and E. intermedia-M subgenome. In particular, the P subgenomes of the two polyploid species formed one monophyletic subclade with the diploid E. equisetina, and the M subgenomes of them formed another monophyletic subclade (100% bootstrap support) with the diploid E. regeliana. This result suggested that the two polyploid species very likely originated from hybridization with the two diploid species as parents, although the possibility of E. monosperma/E. minuta as the paternal progenitor of E. intermedia cannot be ruled out given the low bootstrap support for a close relationship between E. intermedia and E. equisetina in the coalescent tree.
Fig. 2.
Phylogenetic analysis based on the concatenation method and divergence time estimation. (a) A maximum-likelihood tree showing the phylogenetic relationships among the two subgenomes of two polyploid species and their putative diploid parents. Numbers associated with nodes are bootstrap support (BS) values obtained from the concatenation (left) and coalescent (right) analysis, respectively. An asterisk indicates BS of 100%. Diploids are in black, and polyploids are in color. (b) Density distribution of Ks among the two subgenomes of Ephedra sinica and its putative diploid parents.
Based on the Ks analysis, we estimated the divergence times between E. equisetina and E. regeliana (putative diploid parents), between the two subgenomes of the polyploids, and between each subgenome and the corresponding paternal/maternal progenitor. The density distributions of Ks values are shown in figure 2b and supplementary figure S7, Supplementary Material online, and the Gaussian components are listed in supplementary table S7, Supplementary Material online. The mixture model analysis indicated the presence of peaks at 0.04 for both between the two subgenomes of E. sinica and E. intermedia and between E. equisetina and E. regeliana, and the divergence times between them were estimated to be ∼8 Ma. However, the divergence between each subgenome and the corresponding paternal/maternal progenitor occurred much later, at 1.0∼3.1 Ma (supplementary table S7, Supplementary Material online).
Identification of Alternative Splicing without a Reference Genome
One of the most important features of Iso-Seq is to give access to the direct detection of AS by directly comparing isoforms of the same gene. Based on the clustering isoforms in the 5,204 OGs, we carefully analyzed AS in Ephedra. The reference of each OG was predicted by the merge of high-quality consensus isoforms’ CDS, with an average length of 1,343 bp for E. sinica and 1,292 bp for E. intermedia. For the polyploid samples of E. sinica and E. intermedia, we detected 337–1,343 AS events from 314 to 1,179 isoforms, which occurred in 267–857 OGs, accounting for 8.62–21.45% of all identified OGs. The numbers of isoforms with AS from subgenome M and from subgenome P were approximately equal for all samples (table 3).
Table 3.
Characterization of Alternative Splicing (AS) Events in Ephedra sinica and Ephedra intermedia
| No. | Sample |
Ephedra sinica
|
Ephedra intermedia
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| XW |
KB |
XL-W |
XL-C |
INT |
|||||
| S | F | S | F | S | F | S | S | ||
| Subgenome M | Isoforms (%) | 457 (2.08%) | 576 (2.03%) | 411 (1.61%) | 553 (1.46%) | 208 (0.93%) | 358 (1.44%) | 450 (1.65%) | 160 (0.89%) |
| AS events | 534 | 658 | 450 | 604 | 220 | 387 | 510 | 174 | |
| OGs (%) | 389 (11.09%) | 489 (12.24%) | 363 (10.04%) | 464 (10.57%) | 187 (5.44%) | 312 (8.73%) | 379 (10.79%) | 140 (4.52%) | |
| Subgenome P | Isoforms (%) | 433 (1.97%) | 603 (2.12%) | 465 (1.83%) | 565 (1.49%) | 228 (1.02%) | 351 (1.41%) | 446 (1.64%) | 154 (0.86%) |
| AS events | 484 | 685 | 500 | 620 | 254 | 373 | 499 | 163 | |
| OGs (%) | 365 (10.4%) | 508 (12.72%) | 392 (10.85%) | 475 (10.82%) | 208 (6.05%) | 315 (8.81%) | 390 (11.10%) | 143 (4.62%) | |
| All | Isoforms | 890 | 1,179 | 876 | 1,118 | 436 | 709 | 896 | 314 |
| AS events | 1,018 | 1,343 | 950 | 1,224 | 474 | 760 | 1,009 | 337 | |
| OGs (%) | 644 (18.35%) | 857 (21.45%) | 659 (18.23%) | 803 (18.30%) | 341 (10.00%) | 555 (15.52%) | 668 (19.00%) | 267 (8.62%) | |
| All isoforms | 22,017 | 28,392 | 25,454 | 37,892 | 22,445 | 24,906 | 27,224 | 17,984 | |
| Identified OGs | 3,509 | 3,995 | 3,614 | 4,389 | 3,438 | 3,575 | 3,514 | 3,097 | |
Homeolog Expression Based on SNP Analysis
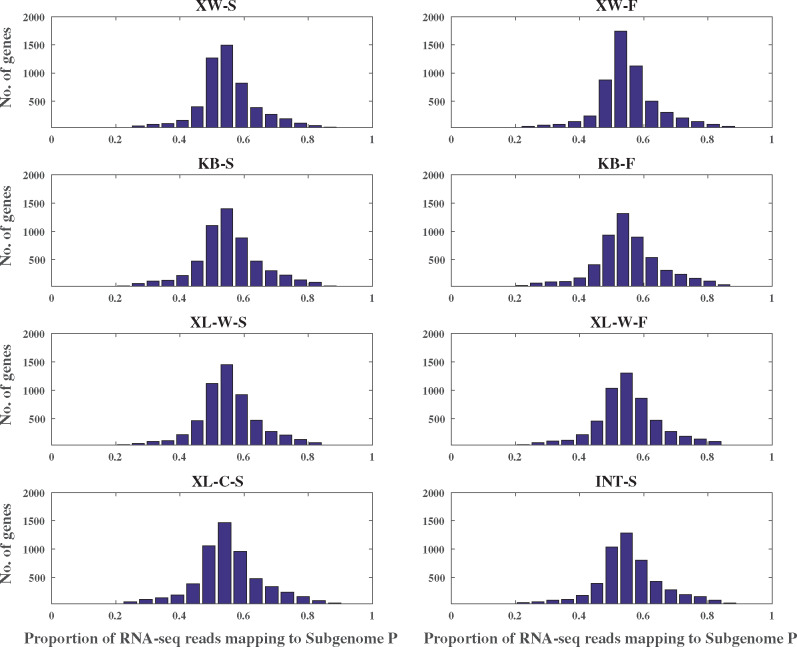
Based on the RNA-seq data, we identified a total of 159,933 SNPs which differed between the putative paternal parent E. equisetina and maternal parent E. regeliana, representing the differences between subgenome P and subgenome M. Of these SNPs, the polyploid samples’ sites were annotated with respect to variants. The most abundant SNPs were SPM sites (80,351–94,151 per sample, accounting for 76–79%). The SPP and SMM sites were far less abundant (SPP: 12,219–14,855 per sample; SMM: 10,721–14,642 per sample). Corresponding to the types of SNPs, 5,279–5,835 GPM, 298–358 GPP, and 265–537 GMM genes were identified per sample. The gene distributions showed that ∼90% of genes in the expressed transcripts had expressed homeolog pairs from both subgenomes across all samples (table 4). If the percentage of an expressed homeolog is higher than 0.6, we defined it as the dominant homeolog. The percentage of homeologs with dominant expression in subgenome M was close to the percentage of homeologs with dominant expression in subgenome P in different samples. A large number of genes (78–83%) showed conserved expression levels (fig. 3). Thus, there appeared to be no differences between subgenomes in the number of expressed genes or the overall expression patterns.
Table 4.
Distributions of SNPs and Genes in Polyploid Samples
| Sample | Category | No. of SNPs |
No. of Genes |
|||||
|---|---|---|---|---|---|---|---|---|
| SPM | SPP | SMM | GPM | GPP | GMM | |||
| Ephedra sinica | XW | S | 87,443 | 13,856 | 14,179 | 5,488 | 341 | 509 |
| F | 90,253 | 14,117 | 14,642 | 5,672 | 342 | 537 | ||
| KB | S | 91,812 | 14,417 | 13,313 | 5,823 | 358 | 453 | |
| F | 87,696 | 13,862 | 13,261 | 5,634 | 354 | 479 | ||
| XL-W | S | 86,696 | 13,529 | 14,156 | 5,725 | 325 | 538 | |
| F | 81,541 | 12,834 | 12,602 | 5,455 | 303 | 456 | ||
| XL-C | S | 94,151 | 14,855 | 10,721 | 5,835 | 363 | 265 | |
| Ephedra intermedia | INT | S | 80,351 | 12,219 | 13,456 | 5,279 | 298 | 452 |
Note.—SPM, heterozygous sites that have fixed differences between two subgenomes; SPP or SMM, sites that only shared with one subgenome; GPM, GPP, and GMM, genes with at least four sites.
Fig. 3.
Transcriptome analyses for homeolog expression.
Genome Size Diversity
Based on the flow cytometry measurement, the genome size of the putative maternal progenitor E. regeliana was 14.86 pg, and that of the putative paternal progenitor E. equisetina was slightly larger. The nuclear DNA contents of the allotetraploids E. sinica and E. intermedia were almost equal to the sum of two putative progenitors (table 5), consistent with the study of Ickert-Bond et al. (2020).
Table 5.
Genome Sizes of the Ephedra Species Estimated by Flow Cytometry
| Species | Holoploid Genome Size (1C-value, pg) | SD | CV% | Inferred Ploidy Level | Monoploid Genome Size (1Cx-value, pg) |
|---|---|---|---|---|---|
| Ephedra minuta | 8.19 | 0.02 | 3.15 | 2x | 8.19 |
| Ephedra equisetina | 8.30 | 0.01 | 2.63 | 2x | 8.30 |
| Ephedra monosperma | 8.54 | 0.01 | 1.85 | 2x | 8.54 |
| Ephedra regeliana | 7.43 | 0.03 | 2.01 | 2x | 7.43 |
| Ephedra przewalskii | 7.65 | 0.01 | 3.53 | 2x | 7.65 |
| Ephedra sinica | 15.42 | 0.03 | 2.46 | 4x | 7.71 |
| Ephedra intermedia | 16.06 | 0.04 | 1.98 | 4x | 8.03 |
Note.—SD, standard deviation; CV, calculated coefficient of variation.
Discussion
Unbiased Subgenome Evolution in Allotetraploid Species of Ephedra
In the past two decades, numerous studies have yielded valuable insights into the evolutionary dynamics of polyploid genomes and consequences of polyploidy in angiosperms, but very rarely in gymnosperms (Bird et al. 2018). Previous studies also indicate that subgenome dominance is often associated with allopolyploid evolution, although unbiased WGD has been reported in a few allopolyploids (Cheng et al. 2018; Liang and Schnable 2018). However, based on the analyses of transcriptome sequences generated from PacBio Iso-Seq and Illumina HiSeq, our present study found unbiased subgenome evolution in two allotetraploid species of Ephedra, a unique gymnosperm genus with 83% of the studied 52 species being polyploids (Ickert-Bond et al. 2020). The two allotetraploid species E. sinica and E. intermedia possibly originated from hybridization with E. regeliana as the maternal parent and E. equisetina as the paternal parent with the divergence time of two subgenomes at 8 Ma, although the paternal progenitor of E. intermedia has not been completely resolved (fig. 2).
This finding is supported by several lines of evidence. First, genes of the putative diploid progenitors are retained in the two subgenomes of the allotetraploids in similar numbers. Expressed homeolog pairs from both subgenomes (HMP) were detected in 60–75% OGs, and the HMP expression can reach 85% as predicted by the saturation curve (fig. 1). In the remaining OGs, the HM expression is also approximately equal to the HP expression in percentage (table 2 and supplementary fig. S6, Supplementary Material online). In addition, the numbers of isoforms with alternative splicing are approximately equal between subgenome M and subgenome P for all samples (table 3). Moreover, the SNP analysis indicates that 76–79% of surveyed sites are heterozygous with fixed differences between two subgenomes and ∼90% of genes show expressed homeolog pairs from both subgenomes across all samples (table 4). These results are similar to the observations in a few allopolyploids of angiosperms such as G. max (Garsmeur et al. 2014), C. maxima and C. moschata (Sun et al. 2017), and Pyrus bretschneideri (Li et al. 2019), which have two ancestral subgenomes with similar gene numbers and show unbiased fractionation. Second, expression dominance was not found between two subgenomes. The percentage of homeologs with dominant expression in subgenome M is close to that of homeologs with dominant expression in subgenome P, and large numbers of genes (78–83%) generally show conserved expression levels (fig. 3). These results strongly contrast with the reports from most allopolyploids of angiosperms such as maize (Swigonová et al. 2004; Schnable et al. 2011) and B. rapa (Wang et al. 2011; Cheng et al. 2016), which show expression dominance and many more events of alternative splicing in one subgenome (Liu et al. 2014; Mei et al. 2017). Finally, the nuclear DNA content of the allotetraploid species is almost equal to the sum of two putative progenitors, suggesting limited genome restructuring after allotetraploid speciation (table 5), as reported in Ickert-Bond et al. (2015, 2020). This characteristic is similar to the modes in G. max (Garsmeur et al. 2014), Capsella bursa-pastoris (Douglas et al. 2015), the allotetraploid Cucurbita species (Sun et al. 2017), and Eragrostis tef (VanBuren et al. 2020), which display karyotype stability after polyploidization.
Implications of the Unbiased Subgenome Evolution for the Formation of Large Genomes in Gymnosperms
For most allopolyploid species of angiosperms, a large fraction of genes from progenitor genomes were lost in the subsequent diploidization process, showing biased fractionation (Soltis PS and Soltis DE 2016; Van de Peer et al. 2017a, 2017b). Subgenome expression dominance is one important mechanism responsible for biased fractionation (Yoo et al. 2014; Cheng et al. 2018). Mechanistically, the unequal gene expression between duplicates may result in differential fitness, leading to biased gene loss with respect to ancestral genomes (Freeling et al. 2012; Bottani et al. 2018; Cheng et al. 2018; Wendel et al. 2018). For example, studies on maize genomes showed that the homeologs in the overfractionated subgenome tend to have lower levels of gene expression, relaxed selection, and higher gene loss, but this evolutionary pattern did not occur in soybean (Pophaly and Tellier 2015; Renny-Byfield et al. 2017; Zhao et al. 2017). In maize, the estimated 85% of originally duplicate gene pairs have become reduced singletons, and the chromosomes (2n = 20) are almost equal to its diploid outgroups sorghum (2n = 20) and rice (2n = 24) (Schnable et al. 2011; Brohammer et al. 2018). In contrast, although the soybean experienced a tetraploidization event (13 Ma, Schmutz et al. 2010) at roughly the same time as maize (11.4 Ma, Gaut and Doebley 1997), soybunderwent slow diploidization, retaining the majority of duplicates and containing 40 chromosomes (2n = 40) that are nearly double the number of chromosomes in the common bean (2n = 22) and pigeon pea (2n = 22) (Du et al. 2012; Zhao et al. 2017).
The unbiased subgenome evolution found in the allotetraploid species of Ephedra leads us to infer that these polyploids might also have undergone slow diploidization with limited genome downsizing. This inference is also supported by previous studies on two conifer genera Sequoia and Juniperus (Scott et al. 2016; Farhat et al. 2019). The unbiased subgenome evolution might also have contributed to the formation of large genomes in gymnosperms considering that at least one round of WGD occurred before the divergence of seed plants (Jiao et al. 2011) and several ancient WGD events occurred in the evolution of gymnosperms (e.g., Li et al. 2015; Guan et al. 2016; Roodt et al. 2017), although some of these WGD events remain controversial (Zwaenepoel and Van de Peer 2019). The accumulation of transposable elements, accounting for 74%, 76.58%, 79%, and 85.9% of the genomes of Pinus taeda (Neale et al. 2014; Wegrzyn et al. 2014), Ginkgo biloba (Guan et al. 2016), Pinus lambertiana (Stevens et al. 2016), and Gnetum montanum (Wan et al. 2018), respectively, could be attributed to slow rates of chromosome rearrangements, as evidenced not only by the cytological stability between diploid and polyploid species in Ephedra (this study, Ickert-Bond et al. 2020), possibly with the formation of disomic inheritance similar to wheat (Yousafzai et al. 2010; Mercier et al. 2015) but also by the moderate genome downsizing following polyploidization in Juniperus (Farhat et al. 2019), and a high degree of synteny between Picea and Pinus (Pavy et al. 2012). In addition, in the large genome of gymnosperms, there is a surprisingly large fraction of gene-like sequences or pseudogenes, in which gene-like sequences represent 2.4% and 2.9% of the Picea abies and Pinus taeda genomes, respectively (Nystedt et al. 2013; Neale et al. 2014). Moreover, a large fraction of gene duplications predated the angiosperm–gymnosperm split. The unbiased subgenome evolution of the allotetraploid species of Ephedra further supports the hypothesis that gymnosperms might lack a mechanism for eliminating redundant gene copies (Nystedt et al. 2013; Warren et al. 2015).
Notably, in Ephedra, 83% of species show tetraploid or very rarely octoploid cytotypes (Ickert-Bond et al. 2020), with a high frequency of allopolyploid speciation reported in Asia (Wu et al. 2016). Why has a high frequency of polyploidy occurred in Ephedra? From the biological view, clonal reproduction is positively associated with polyploidy incidence in angiosperms (Husband et al. 2013; Weiss-Schneeweiss et al. 2013; Freeling 2017; Van Drunen and Husband 2019). The high percentage of polyploid species in Ephedra could be related to a shrub habit and clonal propagation. In contrast to the large trees in conifers, all species of Ephedra are perennial shrubs or sometimes vines or small trees, and have underground rhizomes. The rhizomes of Ephedra can sometimes reach as long as several meters (Pearson 1929), even in rocky slopes (our field investigation), which provide a good mechanism for vegetative propagation (Cutler 1939) and could be helpful to the survival of polyploids. The vegetative propagation also partially contributed to the success of the hexaploid Sequoia sempervirens (Scott et al. 2016) and polyploid Juniperus species (e.g., in Juniperus sabina and Juniperuscommunis; Houle and Babeux 1994; Wesche et al. 2005). The complete genome sequencing of Ephedra species and comparative genomic analyses will further reveal the mechanisms underlying the genome evolution, speciation, and adaptation of the genus.
Supplementary Material
Acknowledgments
This study was supported by Key Research Program of Frontier Sciences, the Chinese Academy of Sciences (Grant No. QYZDJ-SSW-SMC027), National Key Research and Development Program of China (Grant No. 2017YFA0605100), and National Natural Science Foundation of China (Grant Nos. 31330008 and 31170197). We thank Mr Zhen Ma, Dr Ting-Ting Shen, and Dr Wen-Jun Li for their help in sample collection; Ms Wan-Qing Jin for her assistance in transcriptome sequencing; Dr Ren-Hua Peng (Institute of Acousltics, the Chinese Academy of Sciences) and Dr Zhe Cai for their help in the data analysis, especially assisting in writing MATLAB scripts.
Author Contributions
X.-Q.W. designed the research. H.W. and Q.Y. performed the experiments. H.W., J.-H.R., Q.Y., and X.-Q.W contributed to data analysis. H.W. and X.-Q.W. wrote the article.
Data Availability
The sequence data are deposited in GenBank under the BioProject accession PRJNA602052. The sequence alignments are deposited at Dryad under the accession: doi: 10.5061/dryad.bvq83bk5g.
Literature Cited
- Abdel-Ghany SE, et al. 2016. A survey of the sorghum transcriptome using single-molecule long reads. Nat Commun. 7(1):11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham A, Mathew PM.. 1966. Cytology of Encephalartos hildebrandtii A. Br. & Bouche. Ann Bot. 30(2):239–241. [Google Scholar]
- Ahuja MR. 2005. Polyploidy in gymnosperms: revisited. Silvae Genet. 54(1–6):59–69. [Google Scholar]
- Bird K, et al. 2019. Replaying the evolutionary tape to investigate subgenome dominance in allopolyploid Brassica napus. bioRxiv: 814491. [DOI] [PMC free article] [PubMed]
- Bird KA, VanBuren R, Puzey JR, Edger PP.. 2018. The causes and consequences of subgenome dominance in hybrids and recent polyploids. New Phytol. 220(1):87–93. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottani S, Zabet NR, Wendel JF, Veitia RA.. 2018. Gene expression dominance in allopolyploids: hypotheses and models. Trends Plant Sci. 23(5):393–402. [DOI] [PubMed] [Google Scholar]
- Brohammer AB, Kono TJY, Springer NM, McGaugh SE, Hirsch CN.. 2018. The limited role of differential fractionation in genome content variation and function in maize (Zea mays L.) inbred lines. Plant J. 93(1):131–141. [DOI] [PubMed] [Google Scholar]
- Cheng F, et al. 2016. Epigenetic regulation of subgenome dominance following whole genome triplication in Brassica rapa. New Phytol. 211(1):288–299. [DOI] [PubMed] [Google Scholar]
- Cheng F, et al. 2018. Gene retention, fractionation and subgenome differences in polyploid plants. Nat Plants. 4(5):258–268. [DOI] [PubMed] [Google Scholar]
- Chuang TI, Hu WW.. 1963. Study of Amentotaxus argotaenia (Hance) Pilger. Bot Bull Acad Sin. 4:10–14. [Google Scholar]
- Conesa A, et al. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18):3674–3676. [DOI] [PubMed] [Google Scholar]
- Cutler HC. 1939. Monograph of the North American species of the genus Ephedra. Ann Mo Bot Gard. 26(4):373–428. [Google Scholar]
- DePristo MA, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodsworth S, Chase MW, Leitch AR.. 2016. Is post-polyploidization diploidization the key to the evolutionary success of angiosperms? Bot J Linn Soc. 180(1):1–5. [Google Scholar]
- Doležel J, Greilhuber J, Suda J.. 2007. Estimation of nuclear DNA content in plants using flow cytometry. Nat Protoc. 2(9):2233–2244. [DOI] [PubMed] [Google Scholar]
- Doležel J, Sgorbati S, Lucretti S.. 1992. Comparison of three DNA fluorochromes for flow cytometric estimation of nuclear DNA content in plants. Physiol Plant 85(4):625–631. [Google Scholar]
- Douglas GM, et al. 2015. Hybrid origins and the earliest stages of diploidization in the highly successful recent polyploid Capsella bursa-pastoris. Proc Natl Acad Sci U S A. 112(9):2806–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, et al. 2012. Pericentromeric effects shape the patterns of divergence, retention, and expression of duplicated genes in the paleopolyploid soybean. Plant Cell 24(1):21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger PP, et al. 2017. Subgenome dominance in an interspecific hybrid, synthetic allopolyploid, and a 140-year-old naturally established neo-allopolyploid monkeyflower. Plant Cell 29(9):2150–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger PP, et al. 2019. Origin and evolution of the octoploid strawberry genome. Nat Genet. 51(3):541–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid J, et al. 2009. Real-time DNA sequencing from single polymerase molecules. Science 323(5910):133–138. [DOI] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16(1):157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhat P, et al. 2019. Polyploidy in the conifer genus Juniperus: an unexpectedly high rate. Front Plant Sci. 10: 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett JA, Van de Peer Y, Maere S.. 2013. Significance and biological consequences of polyploidization in land plant evolution. In: Leitch IJ, Greilhuber J, Doležel J, Wendel JF, editors. Plant genome diversity 2: physical structure, behaviour and evolution of plant genomes. Wien: Springer. p. 277–293. [Google Scholar]
- Freeling M, et al. 2012. Fractionation mutagenesis and similar consequences of mechanisms removing dispensable or less-expressed DNA in plants. Curr Opin Plant Biol. 15(2):131–139. [DOI] [PubMed] [Google Scholar]
- Freeling M. 2017. Picking up the ball at the K/Pg boundary: the distribution of ancient polyploidies in the plant phylogenetic tree as a spandrel of asexuality with occasional sex. Plant Cell 29(2):202–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsmeur O, et al. 2014. Two evolutionarily distinct classes of paleopolyploidy. Mol Biol Evol. 31(2):448–454. [DOI] [PubMed] [Google Scholar]
- Gaut BS, Doebley JF.. 1997. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc Natl Acad Sci U S A. 94(13):6809–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan QL, Yu ZL, Feng YL.. 1993. Studies of chromosomes in Amentotaxus argotaenia. Acta Bot Yunnan. 15:85–391. [Google Scholar]
- Guan R, et al. 2016. Draft genome of the living fossil Ginkgo biloba. GigaScience 5(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the using the Trinity platform for reference generation and analysis. Nat Protoc. 8(8):1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackl T, Hedrich R, Schultz J, Förster F.. 2014. Proovread: large-scale high accuracy PacBio correction through iterative short read consensus. Bioinformatics 30(21):3004–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagel JM, et al. 2012. Biosynthesis of amphetamine analogs in plants. Trends Plant Sci. 17(7):404–412. [DOI] [PubMed] [Google Scholar]
- Houle G, Babeux P.. 1994. Variations in rooting ability of cuttings and in seed characteristics of five populations of Juniperus communis var. depressa from subarctic Quebec. Can J Bot. 72(4):493–498. [Google Scholar]
- Huang J, Giannasi DE, Price RA.. 2005. Phylogenetic relationships in Ephedra (Ephedraceae) inferred from chloroplast and nuclear DNA sequences. Mol Phylogenet Evol. 35(1):48–59. [DOI] [PubMed] [Google Scholar]
- Husband BC, Baldwin SJ, Suda J.. 2013. The incidence of polyploidy in natural plant populations: major patterns and evolutionary processes. In: Leitch IJ, Greilhuber J, Doležel J, Wendel JF, editors. Plant genome diversity Volume 2, physical structure, behaviour and evolution of plant genomes. Wien: Springer-Verlag. p. 255–276. [Google Scholar]
- Ickert-Bond SM, et al. 2015. Ephedra – the gymnosperm genus with the largest and most diverse genome sizes driven by a high frequency of recently-derived polyploid taxa and a lack of genome downsizing. Edmonton (Canada): Botany 2015, Botanical Society of America Meeting. Available from: http://2015.botanyconference.org/engine/search/index.php?func=detail&aid=862. Accessed December 08, 2020.
- Ickert-Bond SM, et al. 2020. Polyploidy in gymnosperms – insight into the genomic and evolutionary consequences of polyploidy in Ephedra. Mol Phylogenet Evol. 147:106786. [DOI] [PubMed] [Google Scholar]
- Ickert-Bond SM, Renner SS.. 2016. The Gnetales: recent insights on their morphology, reproductive biology, chromosome numbers, biogeography, and divergence times. J Syst Evol. 54(1):1–16. [Google Scholar]
- Ickert-Bond SM, Rydin C, Renner SS.. 2009. A fossil-calibrated relaxed clock for Ephedra indicates an Oligocene age for the divergence of Asian and New World clades and Miocene dispersal into South America. J Syst Evol. 47(5):444–456. [Google Scholar]
- Jiao Y, Paterson AH.. 2014. Polyploidy-associated genome modifications during land plant evolution. Philos Trans R Soc B. 369(1648):20130355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao YN, et al. 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473(7345):97–100. [DOI] [PubMed] [Google Scholar]
- Junier T, Zdobnov EM.. 2010. The Newick utilities: high-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics 26(13):1669–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshoo TN. 1959. Polyploidy in gymnosperms. Evolution 13(1):24–39. [Google Scholar]
- Kück P, , Longo GC. 2014. FASconCAT-G: extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front Zool. 11(1):81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch IJ, Leitch AR.. 2013. Genome size diversity and evolution in land plants. In: Leitch IJ, Greilhuber J, Doležel J, Wendel JF, editors. Plant genome diversity volume 2, physical structure, behaviour and evolution of plant genomes. Wien: Springer-Verlag. p. 307–322. [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, et al. 2019. Unbiased subgenome evolution following a recent whole-genome duplication in pear (Pyrus bretschneideri Rehd.). Hort Res. 6:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A.. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22(13):1658–1659. [DOI] [PubMed] [Google Scholar]
- Li Z, et al. 2015. Early genome duplications in conifers and other seed plants. Sci Adv. 1(10):e1501084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Schnable JC.. 2018. Functional divergence between subgenomes and gene pairs after whole genome duplications. Mol Plant. 11(3):388–397. [DOI] [PubMed] [Google Scholar]
- Liu S, et al. 2014. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun. 5(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Mei W, Soltis PS, Soltis DE, Barbazuk WB.. 2017. Detecting alternatively spliced transcript isoforms from single-molecule long read sequences without a reference genome. Mol Ecol Resour. 17(6):1243–1256. [DOI] [PubMed] [Google Scholar]
- Liu Y, et al. 2017. Two highly similar poplar paleo-subgenomes suggest an autotetraploid ancestor of Salicaceae plants. Front Plant Sci. 8:571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YY, Jin WT, Wei XX, Wang XQ.. 2019. Cryptic speciation in the Chinese white pine (Pinus armandii): Implications for the high species diversity of conifers in the Hengduan Mountains, a global biodiversity hotspot. Mol Phylogenet Evol. 138:114–125. [DOI] [PubMed] [Google Scholar]
- McKenna A, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei W, et al. 2017. A comprehensive analysis of alternative splicing in paleopolyploid maize. Front Plant Sci. 8:694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, et al. 2015. The molecular biology of meiosis in plants. Annu Rev Plant Biol. 66(1):297–327. [DOI] [PubMed] [Google Scholar]
- Mirarab S, et al. 2014. ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics 30(17):i541–i548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray B, Leitch IJ, Bennett MD.. 2012. Gymnosperm DNA C-values database (release 5.0, Dec. 2012). the Royal Botanic Gardens, Kew. Available online at: http://cvalues.science.kew.org/. Accessed December 08, 2020.
- Neale DB, et al. 2014. Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biol. 15(3):R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ner-Gaon H, Leviatan N, Rubin E, Fluhr R.. 2007. Comparative cross-species alternative splicing in plants. Plant Physiol. 144(3):1632–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova PY, et al. 2017. Genome sequencing reveals the origin of the allotetraploid Arabidopsis suecica. Mol Biol Evol. 34(4):957–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystedt B, et al. 2013. The Norway spruce genome sequence and conifer genome evolution. Nature 497(7451):579–584. [DOI] [PubMed] [Google Scholar]
- One Thousand Plant Transcriptomes Initiative. 2019. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574:679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP. 2007. The evolutionary consequences of polyploidy. Cell 131(3):452–462. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Freeling M, Tang H, Wang X.. 2010. Insights from the comparison of plant genome sequences. Annu Rev Plant Biol. 61(1):349–372. [DOI] [PubMed] [Google Scholar]
- Pavy N, et al. 2012. A spruce gene map infers ancient plant genome reshuffling and subsequent slow evolution in the gymnosperm lineage leading to extant conifers. BMC Biol. 10(1):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson HHW. 1929. Gnetales. Cambridge: Cambridge University Press. [Google Scholar]
- Pophaly SD, Tellier A.. 2015. Population level purifying selection and gene expression shape subgenome evolution in maize. Mol Biol Evol. 32(12):3226–3235. [DOI] [PubMed] [Google Scholar]
- Prunier J, Verta JP, MacKay JJ.. 2016. Conifer genomics and adaptation: at the crossroads of genetic diversity and genome function. New Phytol. 209(1):44–62. [DOI] [PubMed] [Google Scholar]
- Ran JH, Shen TT, Wu H, Gong X, Wang XQ.. 2018. Phylogeny and evolutionary history of Pinaceae updated by transcriptomic analysis. Mol Phylogenet Evol. 129:106–116. [DOI] [PubMed] [Google Scholar]
- Renny-Byfield S, Rodgers-Melnick E, Ross-Ibarra J.. 2017. Gene fractionation and function in the ancient subgenomes of maize. Mol Biol Evol. 34(8):1825–1832. [DOI] [PubMed] [Google Scholar]
- Rice A, et al. 2019. The global biogeography of polyploid plants. Nat Ecol Evol. 3(2):265–273. [DOI] [PubMed] [Google Scholar]
- Roodt D, et al. 2017. Evidence for an ancient whole genome duplication in the cycad lineage. PLoS One 12(9):e0184454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, et al. 2010. Genome sequence of the palaeopolyploid soybean. Nature 463(7278):178–183. [DOI] [PubMed] [Google Scholar]
- Schnable JC, Springer NM, Freeling M.. 2011. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc Natl Acad Sci U S A. 108(10):4069–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott AD, Stenz NW, Ingvarsson PK, Baum DA.. 2016. Whole genome duplication in coast redwood (Sequoia sempervirens) and its implications for explaining the rarity of polyploidy in conifers. New Phytol. 211(1):186–193. [DOI] [PubMed] [Google Scholar]
- Sharon D, Tilgner H, Grubert F, Snyder M.. 2013. A single-molecule long-read survey of the human transcriptome. Nat Biotechnol. 31(11):1009–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE.. 2016. Ancient WGD events as drivers of key innovations in angiosperms. Curr Opin Plant Biol. 30:159–165. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens KA, et al. 2016. Sequence of the sugar pine megagenome. Genetics 204(4):1613–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, et al. 2017. Karyotype stability and unbiased fractionation in the paleo-allotetraploid Cucurbita genomes. Mol Plant. 10(10):1293–1306. [DOI] [PubMed] [Google Scholar]
- Swigonová Z, et al. 2004. Close split of sorghum and maize genome progenitors. Genome Res. 14(10A):1916–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BC, Pedersen B, Freeling M.. 2006. Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 16(7):934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Mizrachi E, Marchal K.. 2017a. The evolutionary significance of polyploidy. Nat Rev Genet. 18(7):411–424. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Mizrachi E, Marchal K.. 2017b. Evolutionary associations between polyploidy, clonal reproduction, and perenniality in the angiosperms. New Phytol. 224:1266–1277. [DOI] [PubMed] [Google Scholar]
- Van Drunen WE, , Husband BC. 2019. Evolutionary associations between polyploidy, clonal reproduction, and perenniality in the angiosperms. New Phytol. 224(3):1266–1277. [DOI] [PubMed] [Google Scholar]
- VanBuren R, et al. 2020. Exceptional subgenome stability and functional divergence in the allotetraploid Ethiopian cereal teff. Nat Commun. 11(1):884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan T, et al. 2018. A genome for gnetophytes and early evolution of seed plants. Nat Plants. 4(2):82–89. [DOI] [PubMed] [Google Scholar]
- Wang B, et al. 2016. Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat Commun. 7(1):11708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, et al. 2018. A global survey of alternative splicing in allopolyploid cotton: landscape, complexity and regulation. New Phytol. 217(1):163–178. [DOI] [PubMed] [Google Scholar]
- Wang X, et al. 2011. The genome of the mesopolyploid crop species Brassica rapa. Nat Genet. 43(10):1035–1039. [DOI] [PubMed] [Google Scholar]
- Warren RL, et al. 2015. Improved white spruce (Picea glauca) genome assemblies and annotation of large gene families of conifer defense metabolism. Plant J. 83(2):189–212. [DOI] [PubMed] [Google Scholar]
- Wegrzyn JL, et al. 2014. Unique features of the loblolly pine (Pinus taeda L.) megagenome revealed through sequence annotation. Genetics 196(3):891–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss-Schneeweiss H, Emadzade K, Jang TS, Schneeweiss GM.. 2013. Evolutionary consequences, constraints and potential of polyploidy in plants. Cytogenet Genome Res. 140(2–4):137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF. 2015. The wondrous cycles of polyploidy in plants. Am J Bot. 102(11):1753–1756. [DOI] [PubMed] [Google Scholar]
- Wendel JF, Lisch D, Hu G, Mason AS.. 2018. The long and short of doubling down: polyploidy, epigenetics, and the temporal dynamics of genome fractionation. Curr Opin Genet Dev. 49:1–7. [DOI] [PubMed] [Google Scholar]
- Wesche K, Ronnenberg K, Hensen I.. 2005. Lack of sexual reproduction within mountain steppe populations of the clonal shrub Juniperus sabina L. in semi-arid southern Mongolia. J Arid Environ. 63(2):390–405. [Google Scholar]
- Wu B, Suo F, Lei W, Gu L.. 2014. Comprehensive analysis of alternative splicing in Digitalis purpurea by strand-specific RNA-Seq. PLoS One 9(8):e106001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, et al. 2016. A high frequency of allopolyploid speciation in the gymnospermous genus Ephedra and its possible association with some biological and ecological features. Mol Ecol. 25(5):1192–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang Q.. 2013. The earliest fleshy cone of Ephedra from the early Cretaceous Yixian formation of northeast China. PLoS One 8(1):e53652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, et al. 2018. WEGO 2.0: a web tool for analyzing and plotting GO annotations, 2018 update. Nucleic Acids Res. 46(W1):W71–W75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo MJ, Liu X, Pires JC, Soltis PS, Soltis DE.. 2014. Nonadditive gene expression in polyploids. Annu Rev Genet. 48(1):485–517. [DOI] [PubMed] [Google Scholar]
- Yousafzai KF, Al-Kaff N, Moore G.. 2010. The molecular features of chromosome pairing at meiosis: the polyploid challenge using wheat as a reference. Funct Integr Genomics. 10(2):147–156. [DOI] [PubMed] [Google Scholar]
- Zhang Z, et al. 2012. ParaAT: a parallel tool for constructing multiple protein-coding DNA alignments. Biochem Biophys Res Commun. 419(4):779–781. [DOI] [PubMed] [Google Scholar]
- Zhao M, Zhang B, Lisch D, Ma J.. 2017. Patterns and consequences of subgenome differentiation provide insights into the nature of paleopolyploidy in plants. Plant Cell 29(12):2974–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q-X, Zhi-Jian G, Zhong-Shu Y.. 2000. Karyomorphology and relationships of Amentotaxus Pilg. J Syst Evol. 38:522–527. [Google Scholar]
- Zhou R, Moshgabadi N, Adams KL.. 2011. Extensive changes to alternative splicing patterns following allopolyploidy in natural and resynthesized polyploids. Proc Natl Acad Sci U S A. 108(38):16122–16127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaenepoel A, Van de Peer Y.. 2019. Inference of ancient whole-genome duplications and the evolution of gene duplication and loss rates. Mol Biol Evol. 36(7):1384–1404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence data are deposited in GenBank under the BioProject accession PRJNA602052. The sequence alignments are deposited at Dryad under the accession: doi: 10.5061/dryad.bvq83bk5g.