Abstract
We have recently reported on an experimental model of mitochondrial mistranslation conferred by amino acid exchange V338Y in mitochondrial ribosomal protein MrpS5. Here we used a combination of RNA-seq and metabolic profiling of homozygous transgenic Mrps5V338Y/V338Y mice to analyze the changes associated with the V338Y mutation in postmitotic skeletal muscle. Metabolome analysis demonstrated enhanced levels of age-associated metabolites in the mutant V338Y animals accompanied by increased glycolysis, lipid desaturation and eicosanoid biosynthesis, and alterations of the pentose phosphate pathway. In addition, transcriptome signatures of aged V338Y mutant muscle pointed to elevated inflammation, likely reflecting the increased levels of bioactive lipids. Our findings indicate that mistranslation-mediated impairment of mitochondrial function affects specific bioenergetic processes in muscle in an age-dependent manner.
Keywords: mitochondria, misreading, skeletal muscle, aging, metabolome
INTRODUCTION
A decline in mitochondrial function has been associated with aging and complex age-related changes in metabolism (Sun et al. 2016; Kauppila et al. 2017). Alterations in mitochondrial function including impaired oxidative phosphorylation, increased oxidative damage, reduced activity of metabolic enzymes, and changes in mitochondrial dynamics and biogenesis have all been linked to various aspects of aging (Nunnari and Suomalainen 2012). It is now widely accepted that mitochondria are not mere energy factories, but play central roles in metabolism and signaling (Finkel 2015; Spinelli and Haigis 2018).
The accumulation of somatic mutations in mitochondrial DNA (mtDNA) in post-mitotic tissues is thought to be a key factor in the pathogenesis of age-related diseases (Wallace 2005). mtDNA encodes for genes of oxidative phosphorylation (OXPHOS) and corresponding mutations result in a decline of OXPHOS activity, ultimately impairing cellular bioenergetic capacity (Wallace 1999). Mice that express a mtDNA mutator phenotype, with a threefold to fivefold increase in the levels of random point mutations in mtDNA, display respiratory chain dysfunction and features of accelerated aging (Trifunovic et al. 2004; Kujoth et al. 2005).
Increased error rates in mitochondrial protein synthesis may have similar effects to that of heightened levels of stochastic mtDNA mutations as both would increase the random incorporation of missense mutations. Towards this end, we recently demonstrated that the amino acid exchange V338Y in the mitochondrial ribosomal protein MrpS5 confers mitoribosomal mistranslation (Akbergenov et al. 2018). The identification of amino acid replacement V338Y in mouse mitochondrial ribosomal protein MrpS5 as ribosomal ambiguity mutation (ram) was based on the experimental demonstration that the human homolog V336Y, while not affecting mitochondrial protein synthesis quantitatively, confers incorporation of near-cognate amino acids and stop-codon read-through in in-organello translation assays using mitochondria from human HEK293 cells transfected with mutant V336Y Mrps5 (Akbergenov et al. 2018). Accommodation of mismatched aa-tRNAs by ram mutations increases the natural rate of protein synthesis error frequency, resulting in random missense substitutions in primary amino acid sequences. Due to the physical constraints of the mRNA–tRNA interaction, accommodation of mismatched aa-tRNAs is limited to near-cognate tRNAs, leading to missense amino acid substitutions that are conservative in nature because of the way the genetic code is constructed (Woese 1965; Kurland et al. 1996; Zaher and Green 2009). Homozygous knock-in C57/BL6 mice carrying the Mrps5 V338Y mutation showed impaired oxidative phosphorylation and increased formation of ROS (Akbergenov et al. 2018).
To further dissect the organismal response to increased mitochondrial misreading, we here combined unbiased RNA-seq and metabolome analysis to characterize the response to mitochondrial mistranslation in the skeletal muscle of Mrps5V338Y/V338Y mutant mice. Our choice of tissue was guided by muscle consisting of post-mitotic cells with high energy demands and vulnerability to defects in mitochondrial function (Johnson et al. 2013; Boengler et al. 2017). We show that mitoribosomal misreading results in pronounced age-related metabolite changes in muscle, which are associated with alterations of the pentose phosphate pathway (PPP), increased glycolysis and enhanced lipid biosynthesis. Further, increased levels of bioactive lipids suggest a possible link to the transcriptome signature of enhanced inflammation observed in aged Mrps5V338Y/V338Y mutant mice.
RESULTS
Transcriptomes of skeletal muscle tissue samples from wild-type and Mrps5V338Y/V338Y mice were resolved using RNA-seq. To study the impact of the V338Y mutation on age-related changes, mice were sampled at 3 and 19 mo of age. Considerable transcriptomic changes were detected by differential gene expression analysis of young (3 mo) and old (19 mo) muscle samples (Supplemental Fig. S1). With a threshold P-value <5 × 103 (FDR < 1 × 10−2) there were 3343 and 4232 age-regulated transcripts in the wild-type and V338Y mutant data set, respectively. Further analysis revealed the hallmarks of age-associated transcriptomic alterations for both wild-type and mutant animals—a decrease in mRNA transcripts related to gene annotations associated with mitochondrial function, proteasome degradation, metabolism, and protein translation (Supplemental Fig. S2), for clustering of the experimental groups by BGA see Supplemental Figure S3 (Lee et al. 1999; Su et al. 2015; Shavlakadze et al. 2019).
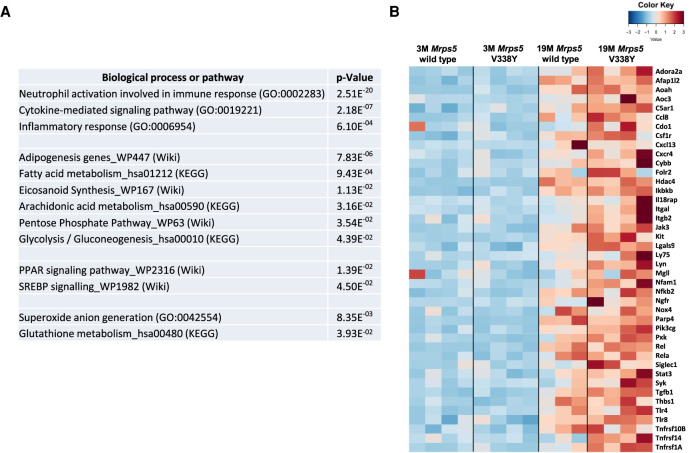
In addition to the characteristic age-associated transcriptome changes, several mutant-specific processes emerged among enriched up-regulated gene transcripts in aged V338Y mice, most prominently multiple terms associated with inflammation and neutrophil activation (Fig. 1A). As evident from the heatmap (Fig. 1B), the expression of inflammation-related transcripts was also elevated in aged wild-type animals, but more prominently in aged V338Y mice. In addition to inflammation, the aged V338Y mutant mice showed enrichment for gene transcripts associated with specific metabolic pathways, that is, adipogenesis, fatty acid (FA) metabolism, eicosanoid synthesis, pentose phosphate shunt, and glycolysis/gluconeogenesis (Fig. 1A).
FIGURE 1.
Muscle transcriptome analysis of mice. (A) Gene enrichment analysis comparing 19 mo MrpS5 WT and MrpS5 V338Y mutant mice; terms and significance for up-regulated gene transcripts, adjusted P-values are shown. (B) Expression heatmap of up-regulated genes enriched in GO term “Inflammatory process” across 3 mo and 19 mo muscle samples.
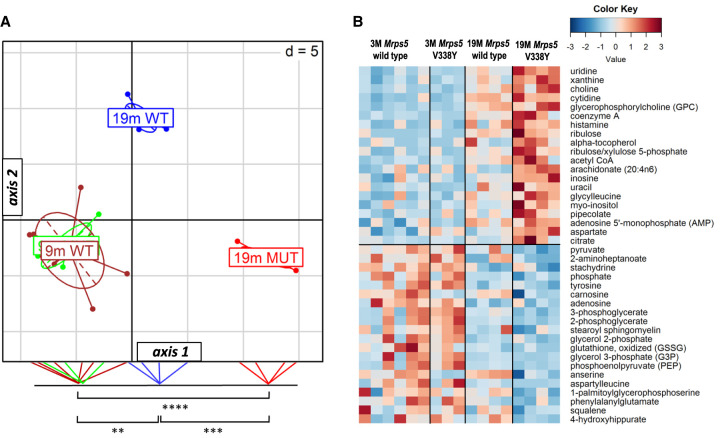
To further characterize the mutant MrpS5 V338Y-associated changes in muscle tissue, we performed metabolic profiling for 9 and 19-mo-old mice. For an initial assessment of differences, we performed a between group analysis (BGA). BGA simplifies a data set by reducing its dimensionality and defining axes that best discriminate the groups. We assigned four groups (9 mo wild-type, 9 mo V338Y, 19 mo wild-type, 19 mo V338Y) and plotted them into two-dimensional space using the first two axes. BGA showed a similar metabolic pattern in wild-type and mutant V338Y mice at 9 mo of age (Fig. 2A). At 19 mo, the metabolic profiles of both genotypes were significantly different from the 9 mo animals. In addition, the 19 mo V338Y mice were also distinct from the age matched wild-type controls (P < 0.0001). The observed metabolic profile indicates that both age and genotype cause the metabolic differences between these two groups. Extraction of the most differentially up- and down-regulated metabolites associated with the horizontal axis (axis 1) included age-associated metabolites described previously (Supplemental Fig. S4A; Houtkooper et al. 2011), suggesting that axis 1 represents aging, as this axis captures the largest fraction of age-related variance. Calculating the BGA on the basis of the published data set and projecting our animals on this model, corroborated our conclusion that the 19 mo Mrps5V338Y/V338Y animals showed enhanced age-associated metabolic changes compared to age-matched wild-type controls (Supplemental Fig. S4B).
FIGURE 2.
Muscle metabolome analysis. (A) Between group analysis (BGA) scatter plot for muscle metabolite profiles (n = 6 for 9-mo-old Mrps5 WT—green; n = 3 for 9-mo-old Mrps5 V338Y—brown; n = 4 for 19-mo-old Mrps5 WT—blue; n = 4 for 19-mo-old Mrps5 V338Y—red). Coordinates of the animals as well as group centers along axis 1 were projected on the bottom line of the plot frame and significant group differences are indicated (Welch's t-test) (**) P = 0.00081, (***) P = 0.000078, (****) P = 0.000002). (B) Heatmap of the top 20 metabolites positively or negatively correlated with horizontal axis (Student's t-test for 19 mo Mrps5 V338Y mice and 19 mo Mrps5 wild-type animals; P < 0.01 for positively correlated metabolites; P < 0.05 for negatively correlated metabolites).
The heatmap of the 40 metabolites with the highest correlation to axis 1 (Fig. 2B) indicates reduced levels of late glycolysis intermediates pyruvate, 3-phosphoglycerate and phosphoenolpyruvate in the 19 mo Mrps5V338Y/V338Y animals, whereas levels of early glycolysis intermediates and lactate remain unchanged (not shown in the heatmap); the same pattern but to a lesser extent was observed in the aged wild-type animals. Transcriptome profiles revealed significant up-regulation (P < 0.05) of hexokinases Hk1 and Hk3 (Hk1/3), phosphofructokinase (Pfk), phosphoglycerate mutase 1 (Pgam1), and Enolase 1 (Eno1) in 19 mo V338Y animals, indicating increased expression of key enzymes in the glycolysis pathway. Further, the pool sizes of ribose-5-phosphate and ribulose-5-phosphate, key metabolites of the pentose phosphate pathway (PPP) were increased in 19 mo V338Y mutant mice and the key enzymes glucose-6-phosphate dehydrogenase X-linked (G6pdx) and transketolase (Tkt) were up-regulated at the transcriptome level, as were 6-phosphogluconolactonase (Pgls) and phosphogluconate dehydrogenase (Pgd) (Supplemental Tables S1, S2), indicative of increased shunting from glycolysis to PPP.
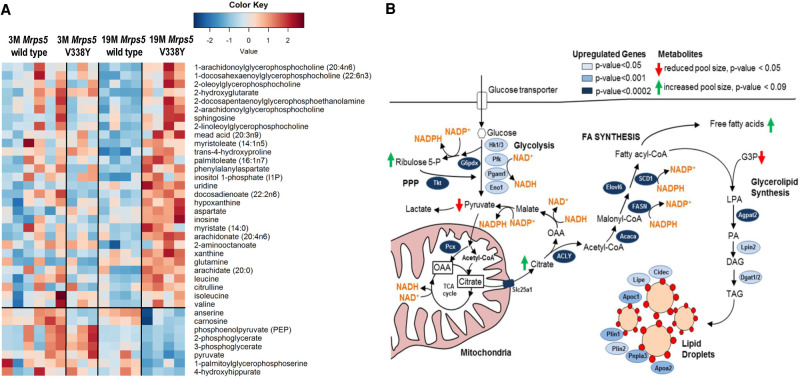
Based on the BGA, we identified the metabolites which were most increased or decreased in 19 mo Mrps5V338Y/V338Y mutant animals and used those to plot a second heatmap (Fig. 3A). The free long chain fatty acids arachidonate, docosadienoate, and arachidate were the most significantly increased metabolites in the mutant animals discriminating between Mrps5 wild-type and V338Y mutant mice at 19 mo. At the transcript level we observed increased expression of enzymes involved in de novo synthesis of lipids and anaplerotic replenishment of the TCA cycle (Fig. 3B). Overall, saturated, monounsaturated, and polyunsaturated free fatty acids (FAs) accumulated in the aged Mrps5V338Y/V338Y mutant animals compared to the 19 mo Mrps5 wild-type mice (Fig. 3A; Supplemental Table S2). Transcriptome data further suggest increased synthesis of triacylglycerides, lipid droplet formation and lipid droplet coating proteins in V338Y mutant mice as compared to 19 mo wild-type animals (Fig. 3B; Supplemental Table S1). Together with elevated levels of arachidonate and dihomo-linoleate, the two key intermediates of eicosanoid metabolism, we found significant enrichment for gene transcripts involved in eicosanoid biosynthesis (e.g., desaturases, arachidonate-lipoxygenases, glutathione peroxidases, prostaglandin E, and thromboxane A synthases; see Supplemental Table S1). In addition, we observed increased levels of lysophospholipids, a side product of arachidonate release from membrane phospholipids (see Supplemental Table S2), together with increased expression of PLA2ε, the main phospholipase in muscle hydrolyzing membrane phospholipids to produce FAs and lysophospholipids (Ohto et al. 2005). The RNA-seq data were corroborated by quantitative RT-PCR for selected key genes involved in glycolysis, PPP, FA synthesis, glycerolipid synthesis, and lipid droplet formation (Supplemental Fig. S5).
FIGURE 3.
Mrps5 V338Y mutation-specific changes in 19-mo-old animals. (A) Heatmap of metabolites associated with the group of 19 mo Mrps5 V338Y animals (Students's t-test for 19 mo Mrps5 V338Y mice and 19 mo Mrps5 wild-type animals; P < 0.05). (B) Model of metabolite pathways, including enzymes and the electron carrying coenzymes, altered in the Mrps5 V338Y animals in contrast to wild-type at 19 mo age (PPP) pentose phosphate pathway, (OAA) oxaloacetate, (G3P) glycerol 3-phosphate, (LPA) lysophosphatidic acid, (PA) phosphatidic acid, (DAG) diacylglycerol, (TAG) triacylglycerol.
DISCUSSION
Translational accuracy of cytoribosomal protein synthesis has been linked to organismal longevity, stress response, and metabolic alterations (Azpurua et al. 2013; Kurylo et al. 2018; McMahon et al. 2019), while little is known about mitochondrial protein synthesis in this respect. We here studied the response to mitochondrial mistranslation in Mrps5V338Y/V338Y mice. Mutation Mrps5 V338Y is a ram mutation, which increases the natural rate of mitochondrial protein synthesis error frequency. We used unbiased RNA-seq to characterize the aging transcriptome in Mrps5 V338Y mutant and Mrps5 wild-type skeletal muscle, complemented by metabolome analysis of muscle from young and old animals. Reflecting the previously reported mutation-mediated increase in reactive oxygen species formation (Akbergenov et al. 2018) we find decreased levels of ROS scavengers anserine and carnosine (Fig. 2B), combined with the transcriptomic signatures of increased superoxide anion generation and glutathione metabolism (Fig. 1A). Presumably, the altered PPP activity observed in the V338Y mutants (Supplemental Tables S1, S2) provides the reducing power to maintain the cellular redox potential and antioxidant activity in response to the mitochondrial production of reactive oxygen species (Zhang et al. 2018).
A common feature of aging is the decline of metabolic systems in general and in particular the link to impaired mitochondrial function (Finkel 2015; Sun et al. 2016). RNA sequencing of muscle confirmed the view of metabolic decline that is thought to underlie part of the aging process. Further metabolomic analyses suggested an enhanced age-associated metabolome profile in the V338Y mutants as compared to the wild-type control mice. This conclusion was supported by analysis of an independent set of metabolites characterized in a previous aging study (see Supplemental Fig. S4B; Shavlakadze et al. 2019). While the mutants essentially showed the same age-associated metabolic changes as the wild-type animals, the effect was significantly more pronounced. In normal aging, these changes are linked to a shortage of NAD+, that is, the main shuttle of electrons to the electron transport chain (Titov et al. 2016; Zhang et al. 2016). Our data indicate that mitochondrial mistranslation aggravates age-related bioenergetic processes in an age-dependent manner. We attribute the observed metabolic shift to a combination of mutation- and age-mediated impairment in mitochondrial function. Reflecting this age-dependency we find no histopathological abnormalities in muscle tissue of 9-mo-old Mrps5 V338Y mice.
Mitochondrial misreading in aged muscle comes along with increased expression of FA synthase, a multifunctional enzyme that catalyzes all seven reactions required for de novo lipid biosynthesis. De novo lipid biosynthesis in MrpS5 mutant cells is initiated by the ATP citrate lyase (Acly) with increased glycolysis and alterations of the pentose phosphate pathway providing the NADPH required for lipogenesis. Acly is a cytosolic enzyme that catalyzes the formation of mitochondria-derived citrate into oxaloacetate and acetyl CoA, which are used as building blocks for lipid synthesis reportedly improving mitochondrial function (Das et al. 2015). Increased expression of genes encoding FA activating enzymes (Acsl5) together with lipid esterification proteins (acylglycerol-3-phosphate acetyltransferase, diacylglycerol acyltransferase) and lipogenesis genes (perilipin 1/2, apolipoprotein CI/AII, lipase) further allows for the coordinate synthesis of lipid droplets in the Mrps5 mutants. Lipid droplet formation, a hallmark of cellular stress, involves a switch in metabolism to glycolysis-dependent ATP generation and lipid biogenesis (Olzmann and Carvalho 2019). In addition to providing an important source of energy, lipid droplets are hypothesized to serve a protective role under conditions of hypoxia and cellular stress by gathering free fatty acids to protect cells against lipotoxicity (Bozza and Viola 2010; Olzmann and Carvalho 2019). Disruption of mitochondrial function by depleting mitochondrial DNA augments the formation of lipid droplets, causing transcriptional activation of FA biosynthesis and metabolic reprogramming to glycolysis (Lee et al. 2013). In addition to increased FA synthesis, impaired mitochondrial respiration has been shown recently to trigger fatty acid desaturation for glycolytic NAD+ recycling (Kim et al. 2019). Our findings of increased glycolysis together with increased expression of fatty acid desaturases (FADS1/2) and heightened levels of unsaturated and polyunsaturated fatty acids would fit well into a scenario of glycolytic NAD+ regeneration by fatty acid desaturation.
While the complex signaling pathways which mediate this metabolic shift in response to mitochondrial misreading are outside the scope of this manuscript, we note that similar metabolic profiles, characterized by increased FA synthesis and expression of genes implicated in lipogenesis together with an enhanced glucose flux through the PPP, have been associated with peroxisome proliferator-activated receptor coactivator 1α (PPARγC-1α) and sterol regulatory element binding protein (SREBP) (Düvel et al. 2010; Summermatter et al. 2010). Interestingly, we find in our mutants a significant enrichment for gene transcripts annotated as PPAR and SREBP signaling pathways, respectively (Fig. 1A).
In addition to the metabolic alterations, V338Y mutant muscle from 19-mo-old mice shows the transcriptomic signature of inflammation. Chronic inflammation is one of the hallmarks of aging and considered to be of multifactorial origin (Ferrucci and Fabbri 2018). In our Mrps5 mutant mice, increased inflammation was accompanied by elevated levels of arachidonic acid, dihomo-linoleate and other ω-6-derived polyunsaturated FAs. The polyunsaturated FAs arachidonate and dihomo-linoleate serve as main substrates for the synthesis of eicosanoids (Buczynski et al. 2009), locally acting bioactive signaling lipids that regulate a diverse set of biological processes and that act as highly potent inflammatory mediators (von Moltke et al. 2012; Lämmermann et al. 2013; Dennis and Norris 2015). The increased levels of the substrates arachidonic and dihomo-linoleic acid together with the enhanced expression of key metabolic enzymes of eicosanoid biosynthesis suggest a possible link of metabolic alterations with the transcriptomic signature of enhanced inflammation observed in the aged Mrps5V338Y/V338Y mutant misreading animals.
MATERIALS AND METHODS
Animals
The transgenic Mrps5V338Y/V338Y mouse strain has been previously described (Akbergenov et al. 2018). Expression levels of Mrps5 mRNA were comparable in mutant and wild-type mice, indicating that the mutation did not lead to deregulation of Mrps5 gene expression. Homozygous mutant mice were able to breed and did not show any gross physiological or morphological phenotype. At 3, 9, and 19 mo of age animals were sacrificed, and muscle tissue was carefully dissected, snap frozen and stored at −80°C.
RNA extraction
For the analysis of quadriceps muscle the following mice were used: four 3-mo-old female animals of Mrps5WT/WT, four 3-mo-old female animals of Mrps5V338Y/V338Y three 19-mo-old female animals of Mrps5WT/WT, and four 19-mo-old animals of Mrps5V338Y/V338Y. RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. The quality of the isolated RNA was assessed using a Qubit (1.0) Fluorometer (Life Technologies) and a Bioanalyzer 2100 (Agilent). Only those samples with a 260 nm/280 nm ratio between 1.8–2.1 and a 28S/18S ratio within 1.5–2 were further processed, all samples used for RNA sequencing had RIN (RNA Integrity Number) ≥ 7.5.
cDNA library preparation and sequencing
RNA sequencing (RNA-seq) was performed at the UZH/ETH Functional Genomics Center Zurich (FGCZ) according to the Illumina RNA sequencing protocol. The TruSeq Stranded mRNA Sample Prep Kit (Illumina) was used in the succeeding steps. Briefly, total RNA samples (100–1000 ng) were ribosome depleted and then reverse-transcribed into double-stranded cDNA with actinomycin added during first-strand synthesis. The cDNA samples were fragmented, end-repaired and polyadenylated. TruSeq adapters containing the index for multiplexing were ligated to the fragmented DNA samples. Fragments containing TruSeq adapters on both ends were selectively enriched with PCR. The quality and quantity of the enriched libraries were validated using Qubit (1.0) Fluorometer and the Caliper GX LabChip GX (Caliper Life Sciences). The product was a smear with an average fragment size of approximately 360 bp. The libraries were normalized to 10 nM in Tris-Cl 10 mM, pH 8.5 with 0.1% Tween 20. The TruSeq SR Cluster Kit v4-cBot-HS (Illumina) was used for cluster generation using 8 pM of pooled normalized libraries on the cBOT. Sequencing was performed on the Illumina HiSeq 2500 single end 126 bp using the TruSeq SBS Kit v4-HS (Illumina).
Transcriptome data analysis
The quality of the reads was assessed using FastQC (Babraham Bioinformatics [http://www.bioinformatics.babraham.ac.uk/projects/fastqc/]) and potential contaminations were evaluated with FastQ Screen (Babraham Bioinformatics [http://www.bioinformatics.babraham.ac.uk/projects/fastq_screen/]) using bowtie2 v. 2.1.0 (Langmead and Salzberg 2012) default parameters. Quantification of gene expression was performed using the RSEM package (version 1.2.18) (Li and Dewey 2011) mapping against the Ensembl 75 annotations derived from the mouse genome assembly GRCm37. Genes not present (<10 counts per gene) in at least 50% of samples from one condition were discarded from further analyses. Differential gene expression analysis between sample groups of interest was performed using the R/bioconductor package edgeR (Robinson et al. 2010). Differences in gene expression levels were calculated as log2 fold changes, resulting P-values were adjusted using Benjamini–Hochberg multiple test correction (Benjamini and Hochberg 1995). An adjusted P-value <0.05 was considered statistically significant; no fold change threshold was applied.
Functional annotation of differentially expressed genes and pathway enrichment analysis were performed using online biological information tool EnrichR (http://amp.pharm.mssm.edu/Enrichr/) (Kuleshov et al. 2016) that integrates several biological databases and provides a comprehensive set of functional annotation information on genes and proteins. The databases KEGG (Kyoto Encyclopedia of Genes and Genomes, http://www.genome.jp/kegg/pathway.html) (Kanehisa et al. 2019), WP (WikiPathways, https://www.wikipathways.org) (Slenter et al. 2018), GO (Gene Ontology enrichment, http://www.geneontology.org) (Harris et al. 2004) from the EnrichR were used for the analysis. The Benjamini–Hochberg false discovery rate (FDR) procedure was applied to pathway analysis as a correction for multiple testing. Pathways or GO terms with FDR corrected P-value (adjusted P-value) <0.05 were considered statistically significant. GO terms with adjusted P-value <0.05 were subjected to the REVIGO web page (Supek et al. 2011) for removal of redundant GO terms from the results.
qRT-PCR
To confirm the results of transcriptome analysis, selected genes were subjected to qRT-PCR (for list of genes and primers see Supplemental Table S3). RNA samples were reverse transcribed into cDNA using the High Capacity RNA-to-DNA Kit (Applied Biosystems). cDNA was analyzed by real-time quantitative PCR (qPCR) using an ABI 7500 Fast Real Time PCR system (Applied Biosystems) and a pair of gene-specific primers for each selected gene. qPCR was performed in triplicates using EvaGreen Mix (Bio&SELL) and 20 ng of cDNA per reaction. The transcript levels of the examined genes were normalized to the geometric mean of four housekeeping genes (GAPDH, Dync1h1, Actb, Rpl41) (Thomas et al. 2014) from the same sample used as an internal reference and the fold change of mutant relative to the WT mice was calculated as 2−ΔΔCT (Pfaffl 2004). Statistical analysis was performed with GraphPad Prism 5.0 software, unpaired Student's t-test was used to estimate significance.
Metabolome analysis
For metabolome analysis of quadriceps muscle, the following mice were compared: six 9-mo-old female animals of Mrps5WT/WT, three 9-mo-old female animals of Mrps5V338Y/V338Y, four 19-mo-old female animals of Mrps5WT/WT animals, and four 19-mo-old female animals of Mrps5V338Y/V338Y (Supplemental Data S1). Metabolome analysis was performed by Metabolon (USA). In brief, samples were prepared by a proprietary series of organic and aqueous extractions in order to remove proteins and to recover the maximum amount of small molecules. The extracted samples were split in equal parts and analyzed via GC–MS or LC–MS/MS. For the LC–MS/MS two equal parts were analyzed in the positive (acidic solvent) and in the negative (basic solvent) ionization mode. Samples for GCMS were bistrimethyl-silyl-trifluoroacetamide derivatized and were run with a 5% diphenyl/95% dimethyl polysiloxane fused silica column.
Metabolome data sets were analyzed by between group analysis (BGA) and random forest (RF) algorithm using the made4 or the RRF package (bioconductor.org) in R, respectively. Normalized MS intensity data were used for RF and BGA analyses, the latter based on Principal Component Analysis (PCA). BGA was applied to a data set consisting of 230 metabolites. Results of the BGA were visualized by a scatter plot for the first two axes of the BGA. The criteria for metabolites being correlated to a group was a maximal angle of 18° (cos18 = 0.95 correlation coefficient) of the metabolite's vector to the vector of the center of the corresponding group. Coordinates were used to calculate significance levels between groups by applying Welch's t-test. A common data set of 88 metabolites derived from the comparison of a published data set (Shavlakadze et al. 2019) and our data set was used to compare age-associated metabolic patterns. The animals of the published data set were used as training data set (3 mo-old animals n = 8, 23-mo-old animals n = 7) and a BGA model was calculated, the animals of the present study were used as test data set and projected on the calculated BGA model. The resulting coordinates of individual mice were plotted. For univariate analyses Welch's t-test and two-way ANOVA were used.
Histopathology
An amount of 4 µm sections of formalin-fixed, paraffin-embedded hindlimb skeletal muscle (M. soleus, M. tibialis anterior) were subjected to hematoxylin-eosin staining. In addition, frozen sections (4 µm) of hindlimb skeletal muscle were subjected to histochemical stains to screen for signs of mitochondrial damage (COX, SDH, NADH, modified Gomori trichrome), abnormal fibre typing and atrophy (ATPases pH 4.2/pH 4.6/pH 9.4), dysregulated autophagic flux (acid phosphatase), pathological accumulation of glycogen (PAS), and neural lipids (ORO). Comparative histopathological analyses of mutant and age-matched wild-type controls at age 9 mo (n = 3 per group) did not reveal any differences. In particular, no histological signs of inflammation (e.g., lymphocytic infiltrates), mitochondrial damage, dysregulated autophagic flux, abnormal accumulation of glycogen, or neural lipids were evident.
DATA DEPOSITION
Transcriptome data are available in the Gene Expression Omnibus (GEO), accession number GSE107520, token utwbackwrzmdxip. Metabolome data have been deposited in EMBL-EBI MetaboLights database and are accessible through accession number MTBLS1594.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank James Moore for editing of the paper and Susanna Salas for expert secretarial assistance.
Author contributions: Concept of study, coordination, and supervision of experiments: E.C.B. Experimental design: E.C.B., D.S., and E.L. Transcriptome studies: D.S. and R.J. Metabolome studies: S.D., D.S., and E.L. Histopathology: L.M.R. and S.F. Writing of the manuscript: E.C.B., E.L., S.D., D.S. with input from R.J. and S.F. All authors analyzed and discussed the data.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.077347.120.
Freely available online through the RNA Open Access option.
REFERENCES
- Akbergenov R, Duscha S, Fritz AK, Juskeviciene R, Oishi N, Schmitt K, Shcherbakov D, Teo Y, Boukari H, Freihofer P, et al. 2018. Mutant MRPS5 affects mitoribosomal accuracy and confers stress-related behavioral alterations. EMBO Rep 19: e46193 10.15252/embr.201846193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpurua J, Ke Z, Chen IX, Zhang Q, Ermolenko DN, Zhang ZD, Gorbunova V, Seluanov A. 2013. Naked mole-rat has increased translational fidelity compared with the mouse, as well as a unique 28S ribosomal RNA cleavage. Proc Natl Acad Sci 110: 17350–17355. 10.1073/pnas.1313473110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 57: 289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- Boengler K, Kosiol M, Mayr M, Schulz R, Rohrbach S. 2017. Mitochondria and ageing: role in heart, skeletal muscle and adipose tissue. J Cachexia Sarcopenia Muscle 8: 349–369. 10.1002/jcsm.12178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozza PT, Viola JP. 2010. Lipid droplets in inflammation and cancer. Prostaglandins Leukot Essent Fatty Acids 82: 243–250. 10.1016/j.plefa.2010.02.005 [DOI] [PubMed] [Google Scholar]
- Buczynski MW, Dumlao DS, Dennis EA. 2009. Thematic review series: proteomics an integrated omics analysis of eicosanoid biology. J Lipid Res 50: 1015–1038. 10.1194/jlr.R900004-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Morvan F, Jourde B, Meier V, Kahle P, Brebbia P, Toussaint G, Glass DJ, Fornaro M. 2015. ATP citrate lyase improves mitochondrial function in skeletal muscle. Cell Metab 21: 868–876. 10.1016/j.cmet.2015.05.006 [DOI] [PubMed] [Google Scholar]
- Dennis EA, Norris PC. 2015. Eicosanoid storm in infection and inflammation. Nat Rev Immunol 15: 511–523. 10.1038/nri3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Düvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, et al. 2010. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell 39: 171–183. 10.1016/j.molcel.2010.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrucci L, Fabbri E. 2018. Inflammageing: chronic inflammation in ageing, cardiovascular disease, and frailty. Nat Rev Cardiol 15: 505–522. 10.1038/s41569-018-0064-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T. 2015. The metabolic regulation of aging. Nat Med 21: 1416–1423. 10.1038/nm.3998 [DOI] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. 2004. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res 32: D258–D261. 10.1093/nar/gkh066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Argmann C, Houten SM, Cantó C, Jeninga EH, Andreux PA, Thomas C, Doenlen R, Schoonjans K, Auwerx J. 2011. The metabolic footprint of aging in mice. Sci Rep 1: 134 10.1038/srep00134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ML, Robinson MM, Nair KS. 2013. Skeletal muscle aging and the mitochondrion. Trends Endocrinol Metab 24: 247–256. 10.1016/j.tem.2012.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. 2019. New approach for understanding genome variations in KEGG. Nucleic Acids Res 47: D590–D595. 10.1093/nar/gky962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppila TES, Kauppila JHK, Larsson NG. 2017. Mammalian mitochondria and aging: an update. Cell Metab 25: 57–71. 10.1016/j.cmet.2016.09.017 [DOI] [PubMed] [Google Scholar]
- Kim W, Deik A, Gonzalez C, Gonzalez ME, Fu F, Ferrari M, Churchhouse CL, Florez JC, Jacobs SBR, Clish CB, et al. 2019. Polyunsaturated fatty acid desaturation is a mechanism for glycolytic NAD. Cell Metab 29: 856–870.e857. 10.1016/j.cmet.2018.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, et al. 2005. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science 309: 481–484. 10.1126/science.1112125 [DOI] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44: W90–W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland CG, Hughes D, Ehrenberg M. 1996. Limitations of translational accuracy. In Escherichia coli and salmonella: cellular and molecular biology (ed. Neidhardt FC, Curtiss R), Vol. 1, pp. 979–1004. ASM Press, Washington DC. [Google Scholar]
- Kurylo CM, Parks MM, Juette MF, Zinshteyn B, Altman RB, Thibado JK, Vincent CT, Blanchard SC. 2018. Endogenous rRNA sequence variation can regulate stress response gene expression and phenotype. Cell Rep 25: 236–248.e236. 10.1016/j.celrep.2018.08.093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lämmermann T, Afonso PV, Angermann BR, Wang JM, Kastenmüller W, Parent CA, Germain RN. 2013. Neutrophil swarms require LTB4 and integrins at sites of cell death in vivo. Nature 498: 371–375. 10.1038/nature12175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, Klopp RG, Weindruch R, Prolla TA. 1999. Gene expression profile of aging and its retardation by caloric restriction. Science 285: 1390–1393. 10.1126/science.285.5432.1390 [DOI] [PubMed] [Google Scholar]
- Lee SJ, Zhang J, Choi AM, Kim HP. 2013. Mitochondrial dysfunction induces formation of lipid droplets as a generalized response to stress. Oxid Med Cell Longev 2013: 327167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12: 323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon M, Contreras A, Holm M, Uechi T, Forester CM, Pang X, Jackson C, Calvert ME, Chen B, Quigley DA, et al. 2019. A single H/ACA small nucleolar RNA mediates tumor suppression downstream of oncogenic RAS. Elife 8: e48847 10.7554/eLife.48847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J, Suomalainen A. 2012. Mitochondria: in sickness and in health. Cell 148: 1145–1159. 10.1016/j.cell.2012.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto T, Uozumi N, Hirabayashi T, Shimizu T. 2005. Identification of novel cytosolic phospholipase A2s, murine cPLA2δ, ɛ, and ζ, which form a gene cluster with cPLA2β. J Biol Chem 280: 24576–24583. 10.1074/jbc.M413711200 [DOI] [PubMed] [Google Scholar]
- Olzmann JA, Carvalho P. 2019. Dynamics and functions of lipid droplets. Nat Rev Mol Cell Biol 20: 137–155. 10.1038/s41580-018-0085-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. 2004. Quantification strategies in real-time PCR. In A-Z of quantitative PCR (ed. Bustin SA), pp. 87–112. International University Line, La Jolla, CA. [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shavlakadze T, Morris M, Fang J, Wang SX, Zhu J, Zhou W, Tse HW, Mondragon-Gonzalez R, Roma G, Glass DJ. 2019. Age-related gene expression signature in rats demonstrate early, late, and linear transcriptional changes from multiple tissues. Cell Rep 28: 3263–3273. 10.1016/j.celrep.2019.08.043 [DOI] [PubMed] [Google Scholar]
- Slenter DN, Kutmon M, Hanspers K, Riutta A, Windsor J, Nunes N, Mélius J, Cirillo E, Coort SL, Digles D, et al. 2018. WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res 46: D661–D667. 10.1093/nar/gkx1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli JB, Haigis MC. 2018. The multifaceted contributions of mitochondria to cellular metabolism. Nat Cell Biol 20: 745–754. 10.1038/s41556-018-0124-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Ekman C, Oskolkov N, Lahti L, Ström K, Brazma A, Groop L, Rung J, Hansson O. 2015. A novel atlas of gene expression in human skeletal muscle reveals molecular changes associated with aging. Skelet Muscle 5: 35 10.1186/s13395-015-0059-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summermatter S, Baum O, Santos G, Hoppeler H, Handschin C. 2010. Peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) promotes skeletal muscle lipid refueling in vivo by activating de novo lipogenesis and the pentose phosphate pathway. J Biol Chem 285: 32793–32800. 10.1074/jbc.M110.145995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Youle RJ, Finkel T. 2016. The mitochondrial basis of aging. Mol Cell 61: 654–666. 10.1016/j.molcel.2016.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6: e21800 10.1371/journal.pone.0021800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KC, Zheng XF, Garces Suarez F, Raftery JM, Quinlan KG, Yang N, North KN, Houweling PJ. 2014. Evidence based selection of commonly used RT-qPCR reference genes for the analysis of mouse skeletal muscle. PLoS One 9: e88653 10.1371/journal.pone.0088653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titov DV, Cracan V, Goodman RP, Peng J, Grabarek Z, Mootha VK. 2016. Complementation of mitochondrial electron transport chain by manipulation of the NAD+/NADH ratio. Science 352: 231–235. 10.1126/science.aad4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly-Y M, Gidlöf S, Oldfors A, Wibom R, et al. 2004. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429: 417–423. 10.1038/nature02517 [DOI] [PubMed] [Google Scholar]
- von Moltke J, Trinidad NJ, Moayeri M, Kintzer AF, Wang SB, van Rooijen N, Brown CR, Krantz BA, Leppla SH, Gronert K, et al. 2012. Rapid induction of inflammatory lipid mediators by the inflammasome in vivo. Nature 490: 107–111. 10.1038/nature11351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. 1999. Mitochondrial diseases in man and mouse. Science 283: 1482–1488. 10.1126/science.283.5407.1482 [DOI] [PubMed] [Google Scholar]
- Wallace DC. 2005. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet 39: 359–407. 10.1146/annurev.genet.39.110304.095751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR. 1965. On the evolution of the genetic code. Proc Natl Acad Sci 54: 1546–1552. 10.1073/pnas.54.6.1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Green R. 2009. Fidelity at the molecular level: lessons from protein synthesis. Cell 136: 746–762. 10.1016/j.cell.2009.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ryu D, Wu Y, Gariani K, Wang X, Luan P, D'Amico D, Ropelle ER, Lutolf MP, Aebersold R, et al. 2016. NAD+ repletion improves mitochondrial and stem cell function and enhances life span in mice. Science 352: 1436–1443. 10.1126/science.aaf2693 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lee JH, Paull TT, Gehrke S, D'Alessandro A, Dou Q, Gladyshev VN, Schroeder EA, Steyl SK, Christian BE, et al. 2018. Mitochondrial redox sensing by the kinase ATM maintains cellular antioxidant capacity. Sci Signal 11: eaaq0702 10.1126/scisignal.aaq0702 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.