Abstract
Inflammatory bowel disease (IBD) is an idiopathic intestinal inflammatory disease, including ulcerative colitis (UC) and Crohn’s disease (CD). The abnormality of inflammatory and immune responses in the intestine contributes to the pathogenesis and progression of IBD. Autophagy is a vital catabolic process in cells. Recent studies report that autophagy is highly involved in various kinds of diseases, especially inflammation-related diseases, such as IBD. In this review, the biological characteristics of autophagy and its role in IBD will be described and discussed based on recent literature. In addition, several therapies for IBD through modulating the inflammasome and intestinal microbiota taking advantage of autophagy regulation will be introduced. We aim to bring new insight in the exploration of mechanisms for IBD and development of novel therapeutic strategies against IBD.
Keywords: inflammatory bowel disease, autophagy, inflammasome, apoptosis, mutation
Introduction
Traditionally, it is well acknowledged that the intestinal tract is the largest digesting organ. It mainly functions in absorbing food and nutrition orally taken into the intestinal lumen majorly through the absorptive enterocytes located in the gut lacunae and plica (Choi and Snider, 2019; Ruder et al., 2019; Yamamoto et al., 2020). In the recent decade, an increasing number of studies have revealed that the gut is also regarded as a critical immune organ because of its exposed and challenged with exogenous invasive pathogenic microorganisms in the gut lumen (James et al., 2020; Muller et al., 2020; Zhou et al., 2020). Several self-protective mechanisms are induced with the invasion, including the triggering of the inflammasome formation, neutrophil extracellular traps, pro-inflammatory cytokine secretion and other inflammatory, and immune responses in the gut wall. Those mechanisms largely limit the invasion of pathogenic factors through the gut wall (Bischoff et al., 2014; Tong and Tang, 2017; Ye et al., 2019; Zhou et al., 2020). However, despite of the numerous researches on the gut, there are still unsolved questions or even contradictions on the issues of the development and component of the immune system in the gut and how the intestinal immune system responds to the external and internal stress. So far, a large number of studies have revealed that during the process of intestinal self-defense system, the intestinal mucosal barrier has been widely investigated and recognized as the most important factor in the intestinal defense system. The intestinal mucosal barrier functions in maintaining the gut microbiota homeostasis and peaceful coexistence in organisms (Sanchez de Medina et al., 2014; Jarret et al., 2020). In the following contents, the components, functions, and relations with intestinal diseases will be introduced in detail.
So far, it is revealed that the intestinal mucosal barrier is generally comprised by three layers, including the mucus layer on the surface, epithelial cell layer, and inflammatory and immune cell layer in the submucosa (Zhang et al., 2017; Wang et al., 2018c; Schroeder, 2019). The mucus layer is referred to as a gel-like layer on the surface of the mucosa containing numerous kinds of proteins, carbohydrates, and lipids in mucus. The mucus layer functions in avoiding the direct contacting between epithelial cells and intestinal microbiota (Vita et al., 2019; Pelaseyed and Hansson, 2020). The intestinal epithelial cells in the second layer of intestinal mucosal barrier are mainly including absorptive enterocytes, Paneth cells, goblet cells, micro-fold villus cells, enteroendocrine cells, and tuft cells. The containing of those cells contribute to the absorption of nutrition and secreting mucus and functional proteins (Goll and van Beelen Granlund, 2015; Wu et al., 2019; Yin et al., 2019). Besides, the inflammatory and immune cells in the submucosa, such as macrophages, neutrophils, and lymphocytes, can trigger self-defensive responses through endocytosis, antigen presentation, and secretion of cytokines. Those cells in the third layer fight against the invasion of pathogens (Bui et al., 2018; Luissint et al., 2019; Ho et al., 2020). However, the disturbance of microbiota balance and damage of intestinal mucosal barrier tend to trigger the over-induction of self-defensive processes, including oxidative stress, inflammasome formation, and so on. The overwhelming induction of those self-defensive mechanisms results in several intestinal disorders such as inflammatory bowel disease (IBD; Ramos and Papadakis, 2019; Han et al., 2020; Watanabe et al., 2020). As a result, targeting on the maintaining of intestinal microbiota homeostasis and suppressing the overwhelmingly induced self-defensive inflammatory and immune responses serves as an effective pathway for the treatment of IBD.
Autophagy is regarded as a vital metabolic process. It is for degrading and recycling long-lived or misfolded proteins and useless organelles under stress conditions relying on lysosomes (Dikic and Elazar, 2018; Racanelli et al., 2018). Since initially reported in the mid-1950s, autophagy has been demonstrated to be involved in the pathogenesis and progression of various kinds of diseases (Li and Chen, 2019; Yamaguchi, 2019; Keller et al., 2020; Yim and Mizushima, 2020). In IBD, autophagy has been widely revealed to regulate the onset and development of IBD via immune and inflammatory modulation (Iida et al., 2017; Palmela et al., 2018; Wang et al., 2018b; Hooper et al., 2019). Modern studies indicate that targeting on autophagy might serve as a promising therapeutic strategy for the treatment of IBD. Based on those evidence, here in this paper, the latest literature on autophagy and IBD will be reviewed and the role of autophagy in IBD will be discussed. We aim to provide novel insight in the treatment of IBD taking advantage of autophagy in this review.
Part I: Autophagy in IBD
Biological Characteristics of Autophagy
Autophagy is a catabolic cellular process, through which some protein aggregates and damaged organelles are degraded into metabolic elements via lysosomes for recycling to maintain the homeostasis and vitality of cells (Allen and Baehrecke, 2020; Yang and Klionsky, 2020). The process of autophagy is widely existed in almost all kinds of cells and evolutionarily conserved from yeast to mammals (Bento et al., 2016; Reggiori and Ungermann, 2017; Tyler and Johnson, 2018). According to the mode of cargo delivery and physiological function, autophagy is mainly divided into three classic forms, including macroautophagy, microautophagy, and chaperone-mediated autophagy (Xilouri and Stefanis, 2016; Wang et al., 2018a; Cerri and Blandini, 2019). Macroautophagy is uncovered as a catabolic process featured with sequestration of degrading materials into double-membraned autophagosomes, which are subsequently fused with lysosomes for degradation (Yu et al., 2018; Lieberman and Sulzer, 2020). Microautophagy is a non-selective form of autophagy, through which cytoplasmic degrading materials are engulfed via invagination of the lysosomal/vacuolar membranes (Mijaljica et al., 2011; Li and Hochstrasser, 2020). Chaperone-mediated autophagy is a selective autophagy form relying on the presentation of chaperones via certain target motif in the substrate proteins and lysosomal chaperons (Yang et al., 2019; Dash et al., 2020). Since macroautophagy is so far the most studied form of autophagy and has been reported to be involved in diseases, here in this review, we will mainly discuss the biological features of macroautophagy and its role in IBD (herein referred to as “autophagy”).
The induction of autophagy mainly includes two steps according to our previous reviews (Shao et al., 2016; Wang et al., 2018a). In the first step, substrate materials, such as aggregated proteins, are surrounded by a cup-shaped phagophore with lipid bilayer membrane, which subsequently sequestrated into double-membrane sphere-shaped autophagosomes. The formation of phagophores demands the formation of the autophagy-related gene 1 (ATG1) complex and Class III phosphatidylinositol 3-kinase (PI3K) complex, with Unc-51-like kinase (ULK1, aka Atg1 in yeast), FIP200, ATG13, ATG101 assembly for ATG1 complex and Beclin-1, ATG14, vacuolar protein sorting 15 (VPS15), and VSP34 for PI3K complex (Bento et al., 2016; Wang et al., 2018a). The process of membrane expansion from phagophores to autophagosomes is dependent on the formation of ATG5-ATG12-ATG16L1 complex (Sheng et al., 2018). In the second step, autophagosomes dispose of “coat proteins (LC3-II)” on the surface and fuse with lysosomes with the assistance of ATG3 and ATG7 for the formation of the functional autolysosomes (Nath et al., 2014). On the occurrence of autophagy, more than 30 ATGs are involved. In addition, two classic signaling pathways are vital in the regulation of autophagy process. The Class I PI3K-mammalian target of rapamycin (mTOR) is illustrated as an inhibitory pathway of autophagy via the stimulation of mTOR complex 1 (mTORC1), with AMP-activated protein kinase (AMPK) considered as the classic upstream suppressor (Bork et al., 2020; Kaleagasioglu et al., 2020; Tamaddoni et al., 2020). The other regulatory pathway is regarded as an inductive pathway, which depends on the formation of Class III PI3K-Beclin-1 complex (Tong et al., 2015; Ohashi et al., 2019).
In the recent few years, the role of autophagy in diseases have been widely explored, including cardiovascular diseases such as myocardial infarction (Liu et al., 2020; Shi et al., 2020) and atherosclerosis (Wang et al., 2019; Cao et al., 2020), neuro-degenerative disorders like multiple sclerosis (Shao et al., 2017; Andhavarapu et al., 2019), metabolic diseases including diabetes (Marasco and Linnemann, 2018; Ding et al., 2020) and obesity (Bartelt and Widenmaier, 2020) and inflammatory and immune-related disorders such as IBD (Dalmasso et al., 2019; Tan et al., 2019; Sola Tapias et al., 2020), and arthritis (Vomero et al., 2018). Although the inflammatory and immune suppressive effect of autophagy has been revealed, some researchers also identified autophagy as a form of cellular death. They pointed out that the induction of autophagy in certain conditions might lead to the so-called “autophagic cellular death” (Denton and Kumar, 2019; Ma et al., 2020; Zhang et al., 2020a). Given those evidence, the mechanisms of autophagy in diseases are complicated. In the following contents, the role of autophagy in IBD will be further described and discussed in detail.
Autophagy in the Pathogenesis and Progression of IBD
IBD is recognized as a chronic disorder with pathogenic factors including intestinal microbiota disturbance, mucus barrier damage, gene mutation, and some environmental stimulation (Kelsen and Sullivan, 2017). The incidence of IBD is significantly increased and the age of pathogenesis is becoming younger and younger in the past few decades (Kelsen et al., 2019). IBD comprises two chronic idiopathic inflammatory diseases, namely ulcerative colitis (UC) and Crohn’s disease (CD) (Sairenji et al., 2017). It has been widely reported that autophagy produces an important effect in the onset and development of both UC and CD. In the following contents of this section, the role of autophagy in UC and CD will be discussed, respectively (illustrated in Figure 1).
Figure 1.
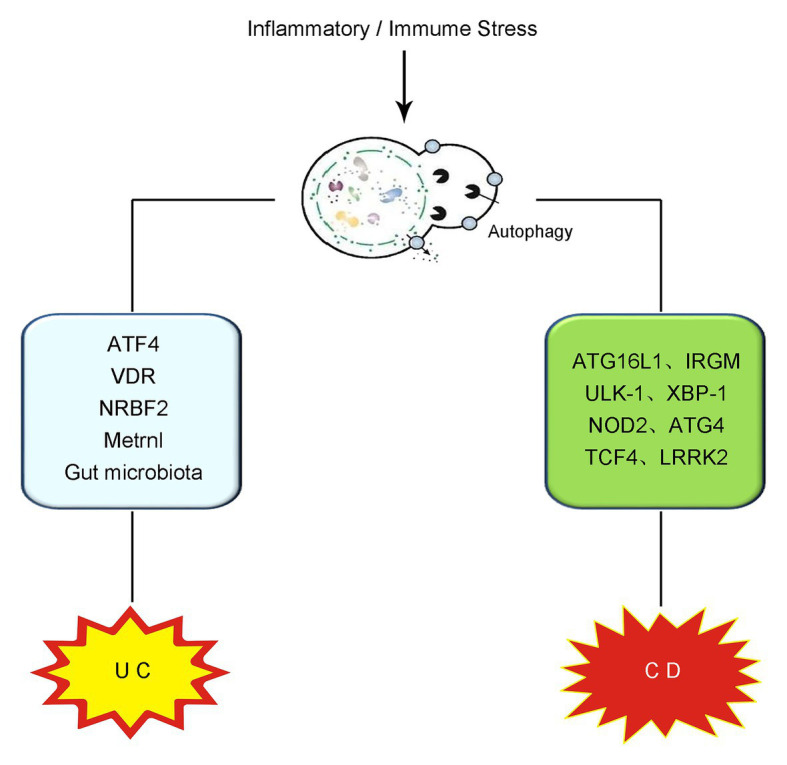
Schematic illustration of the role of autophagy in IBD. The overwhelming inflammatory and immune stress in the gut contributes to the pathogenesis and progression of IBD. On the occurrence of UC, intestinal autophagy is induced and produces effects on UC via the modulation of ATF4, VDR, NRBF2, Metrnl, gut microbiota regulation, and so on. In CD, the mutation or deletion of some autophagy-related genes, including ATG16L1, IRGM, ULK-1, XBP-1, NOD2, ATG4, TCF4, and LRRK2, have been reported to be vital in the onset and development of CD. IBD, inflammatory bowel disease; UC, ulcerative colitis; CD, Crohn’s disease; ATF4, activating transcription factor 4; VDR, vitamin D receptor; NRBF2, nuclear receptor binding factor 2; ATG16L1, autophagy-related protein 16-like protein 1; IRGM, immunity-related GTPase M; ULK-1, Unc-51-like kinase-1; XBP-1, X box-binding protein 1 spliced-1; NOD2, nucleotide binding oligomerization domain containing protein 2; TCF4, transcription factor 4; LRRK2, leucine-rich repeat serine/threonine-protein kinase 2.
Autophagy in UC
UC is a chronic recurrent disorder, affecting all length of the colon and rectum (Ko et al., 2019). So far, it has been revealed that the abnormality of immune system, environment, gut microbiota disturbance, exogenous infection, and certain gene mutations contribute to UC (Feuerstein et al., 2019; Singh et al., 2019; Murphy et al., 2020). However, the specific pathogenesis remains unclarified. UC often spreads from the rectum up to the distal colon and finally involves the inflammatory damage of all the large intestine (Ko et al., 2019; Pravda, 2019). The onset and development of UC often lead to malnutrition syndrome, toxic megacolon, and even colorectal cancer (Burke et al., 1997; Gallinger et al., 2017). In the recent few years, the role of autophagy in UC has been widely explored. In patients with active UC, the level of activating transcription factor 4 (ATF4), an important autophagy-related protein, was significantly decreased in the inflamed intestinal mucosa compared with that in normal mucosa. The findings revealed by that study indicate the reduction of autophagy process on the occurrence of UC (Hu et al., 2019). Recent evidence demonstrated that activating intestinal nuclear receptor vitamin D receptor (VDR) could downregulate intestinal inflammation via inducing autophagy-mediated inflammasome suppression (Bakke and Sun, 2018; Karimi et al., 2019; Law et al., 2019). A low VDR expression and dysfunction of vitamin D/VDR signaling was also observed in UC patients (Bakke and Sun, 2018). In addition, Zhou et al. (2018) revealed the mTOR-dependent autophagy flux deficiency in human intestinal epithelial cells from active UC patients. Those findings suggest the correlation between baseline autophagy impairment and the occurrence of UC. However, under the condition of intestinal inflammation, some kinds of microbiota, such as adherent-invasive Escherichia coli, was shown to adhere to intestinal epithelial cells and escape autophagy clearance via swallowed inside macrophages (Palmela et al., 2018).
In the current studies on UC, dextran sulfate sodium (DSS) is widely applied for the creation of UC animal model, which significantly facilitates the study of UC (Martin et al., 2016; Nunes et al., 2018). In DSS-induced UC models, Wu et al. (2020) reported that nuclear receptor binding factor 2 (NRBF2), a regulatory subunit of the autophagy-related Beclin-1-ATG14-VPS15-VSP34 complex, was necessary for the clearance of apoptotic cells and inflammation suppression during DSS-induced colitis. Studies demonstrated that autophagy contributed to the maintaining and restoration of gut microbiota homeostasis in DSS-induced UC models. Those studies indicate that enhancing the level of autophagy may probably serve as a potential regulator of gut microbiota (Ng, 2015; Xuan et al., 2020; Zha et al., 2020). In addition, it was lately demonstrated by Zhang et al. (2020b) that specific knocking out intestinal Metrnl in epithelial cells produced a detrimental effect UC severity via the reduction of autophagy in epithelial cells in DSS-induced colitis models. The AMPK-mTOR-p70S6K signaling, a classic autophagic pathway, was shown to be involved in this process (Zhang et al., 2020b). In consistent with those findings, recent studies conducted by our lab demonstrated that inducing AMPK-mTOR-p70S6K-mediated autophagy through the activation of certain receptors contributed to the attenuation of UC symptoms and suppression of intestinal inflammation in DSS-induced mice models (Ke et al., 2016; Shao et al., 2019a). All of those studies suggest the alleviative role of autophagy in the onset and development of UC.
However, as we have mentioned above, the induction of autophagy under certain conditions or overwhelming induction of autophagy might produce negative effects on cells via the triggering of autophagic cellular death. In IBD, it has been reported that deficiency of erbin, a protein required for the polarity of epithelial cells, significantly exacerbated the induction of autophagy process and autophagic cell death in DSS-induced mice models (Shen et al., 2018). The damage of intestinal epithelial cells triggered intestinal inflammation and exacerbated the severity of UC symptoms (Shen et al., 2018). Based on those evidence, to safely taking advantage of autophagy in the treatment of UC, further studies are demanded to explore the pathway to properly induce autophagy.
Autophagy in CD
CD is a sort of localized or regional enteritis of unknown etiology associated with pathogenetic factors including impaired immunity, environmental stress, familial inheritance, and certain gene mutations (Harb, 2015; Torres et al., 2017; Kienle, 2018). CD often onsets in the region of perineal blind and spreads to the whole length of the colon and distal ileum in combination of some extraintestinal complications (Kienle, 2018). Compared with UC, CD tends to be more strongly affected by genetic factors. It was previously reported by a European study of twins that the proband concordance in monozygotic twins was 58.3 and 6.3% in CD and UC, respectively (Tysk et al., 1988). However, in dizygotic twins, the proband concordance was reduced to 3.9% in CD and no significance in UC (Tysk et al., 1988). According to several genome-wide association study (GWAS) studies conducted in 2007, the mutation of some autophagy-related genes, including ATG16L1 and immunity-related GTPase M (IRGM), two important autophagy-related proteins, were highly related to the pathogenesis of CD (Tysk et al., 1988; Hampe et al., 2007). Buisson et al. (Rioux et al., 2007) revealed that the polymorphisms of autophagy-related genes, including IRGM, Unc-51-like kinase-1 (ULK-1) and X box-binding protein 1 spliced-1 (XBP-1), were associated with macrophage inability to control the replication of CD-associated adherent-invasive E. coli. They demonstrated that compared with that from UC patients or healthy controls, the survival rate of adherent-invasive E. coli was significantly increased in CD patients with CD-associated polymorphism IRGM (p = 0.05) and decreased in those with polymorphisms CD-associated XBP-1 (p = 0.026) and CD-associated ULK-1 (p = 0.033; Rioux et al., 2007). In addition, some single nucleotide polymorphisms (SNPs) of ATG16L1, such as rs2241880 and rs6754677, have been detected in CD patients and shown to be associated with CD onset (Baradaran Ghavami et al., 2019; Buisson et al., 2019; Kee et al., 2020; Tsianos et al., 2020; Varma et al., 2020). NOD2 is an important autophagy inducer via recruiting ATG16L1 into the cell membrane (Park and Jeen, 2019). In CD patients, nucleotide binding oligomerization domain containing protein 2 (NOD2) mutation has been shown to lead to the intestinal microbiota homeostasis and over-induction of gut inflammatory responses (Frade-Proud’Hon-Clerc et al., 2019; Waldschmitt et al., 2019; Belyayev et al., 2020). Besides NOD2, IRGM, ULK-1, and XBP-1, it was recently reviewed by us that the mutation or deletion of some other autophagy-related genes, including ATG4, transcription factor 4 (TCF4), leucine-rich repeat serine/threonine-protein kinase 2 (LRRK2), and ATG5 in Paneth cells, were related to the pathogenesis of IBD, especially CD (Wang et al., 2018c). Apart from autophagy-gene mutation, Lu et al. (2014) also showed that microRNA106B and microRNA93 could impair the removal of CD-associated bacteria from epithelial cells though the combination of ATG16L1, thus inhibiting the formation of autophagosomes and autophagy-dependent eradication of intracellular bacteria.
Intrarectal administration of 2,4,6-trinitrobenzenesulfonic acid (TNBS) dissolved in ethanol was initially applied by Morris et al. (1989) for the creation of CD animal models. So far, in TNBS-induced CD models, it was reported that the administration of 3-methyladenine (3-MA) for the blockade of autophagy aggravated the CD symptoms and intestinal inflammation, suggesting the protective effect of autophagy on CD (Macias-Ceja et al., 2017). In addition, Gerster et al. (2015) revealed that high-density lipoprotein (HDL) cholesterol-induced autophagy led to the recruitment of phosphorylated IκB kinase to the autophagosome compartment. Such recruitment prevented nuclear factor-κB (NF-κB)-mediated intestinal inflammation in TNBS-induced CD models. However, it has been demonstrated by Banerjee et al. (2019) that the administration of TNBS per se might induce intestinal autophagy toward autophagic cell death, indicating the limitation of TNBS-induced CD models in the study of autophagy.
Part II: Application of Autophagy Regulators in the Treatment of IBD
As discussed above, autophagy is highly involved in the pathogenesis and progression of IBD. So far, some autophagy regulators are available to us in the treatment of IBD, although most of them have not been applied in the clinical practice. Hereafter, the most popular kinds of such agents, including the well-studied inflammasome inhibitors and intestinal microbiota regulators, will be introduced in the following contents (listed in Table 1).
Table 1.
Potential pharmacological mechanisms of autophagy regulators in IBD treatment.
| Autophagy regulators | Potential pharmacological mechanisms | Targeted autophagy pathway | References | |
|---|---|---|---|---|
| Inflammasome inhibitors taking advantage of autophagy | GL-V9 | Inducing AMPK signaling | AMPK signaling | Zhao et al., 2017 |
| Ginsenoside Rd | Triggering p62-driven mitophagy | AMPK/ULK-1 signaling | Liu et al., 2018 | |
| Palmatine | Inducing mitophagy | PINK1/Parkin-mediated signaling | Mai et al., 2019 | |
| Metformin/MCC950 | Modulating HSP90/NLRP3 interaction | AMPK signaling | Saber and El-Kader, 2020 | |
| α7nAChR agonist | Inducing AMPK-mTOR-p70S6K signaling pathway | AMPK-mTOR-p70S6K signaling | Shao et al., 2019a | |
| CB2R agonist | Inducing AMPK-mTOR-p70S6K signaling pathway | AMPK-mTOR-p70S6K signaling | Ke et al., 2016 | |
| Intestinal microbiota regulators taking advantage of autophagy | Vitamin D | VDR-mediated signaling | Acting on ATG16L1 | Sun, 2016; Bakke and Sun, 2018 |
| TREM-1 | Restoration of impaired autophagy activity | Unfolded protein response pathway | Kokten et al., 2018 | |
| Galangin | Modulation of inflammatory reaction and myeloperoxidasactivities | AMPK signaling | Xuan et al., 2020 | |
GL-V9, AMP-activated protein kinase activator; AMPK, AMP-activated protein kinase; PINK1, PTEN-induced putative kinase 1; HSP90, heat shock protein 90; NLRP3, NLR family pyrin domain containing 3; α7nAChR, alpha7 nicotinic acetylcholine receptor; mTOR, mammalian target of rapamycin; CB2R, cannabinoid receptor 2; VDR, vitamin D receptor; TREM-1, triggering receptor expressed on myeloid cells-1; ULK-1, Unc-51-like kinase-1; ATG16L1, autophagy-related protein 16-like protein 1.
Inflammasome Inhibitors Taking Advantage of Autophagy
Inflammasome is recognized as a multi-protein oligomer responsible for activating inflammatory responses (Mariathasan et al., 2004). It belongs to the family of the innate immunity, mainly existing in epithelial cells and most inflammatory and immune cells such as macrophages and dendritic cells in the gut (Lei-Leston et al., 2017; Song and Li, 2018; Shao et al., 2019b). The crosstalk between the inflammasome and autophagy has been well-studied in many disorders (Harris et al., 2017; Deretic and Levine, 2018). So far, several members of inflammasomes have been described, including NLR family pyrin domain containing 1 (NLRP1), NLRP2, NLRP3, NLR family caspase recruitment domain-containing protein 4 (NLRC4), and double-stranded DNA sensors absent in melanoma 2 (AIM2) (Suarez and Buelvas, 2015). Among them, the NLRP3 inflammasome is the most characterized and studied form of inflammasome, which belongs to a member of innate immunity and special form of inflammatory reaction (Huang et al., 2020; Zewinger et al., 2020). The NLRP3 inflammasome comprises three components, including NLRP3 protein, adapter protein apoptosis associated speck-like protein (ASC), and procaspase-1 (Coll et al., 2015). It is mainly produced in inflammatory and immune cells, such as macrophages, NK cells, and lymphocytes (Rathinam et al., 2019). It has been demonstrated that in the recognition of some pathogen-associated molecular patterns (PAMPs) and danger-associated molecular patterns (DAMPs), the reactive components of the NLRP3 inflammasome is accumulated, followed by the formation of the NLRP3 inflammasome complex. The successful formation of the NLRP3 inflammasome induces the transformation of the procaspase-1 to enzymatic caspase-1, which catalyzes the maturation of interleukin (IL)-1β and IL-18 outside cells to stimulate inflammatory reaction cascade (Schroder et al., 2010; Shao et al., 2018; Swanson et al., 2019). So far, it has been revealed by us and other researchers that the over-activation of the NLRP3 inflammasome contributes greatly to the onset and development of IBD (Mangan et al., 2018; Mao et al., 2018; Shao et al., 2019b). According to recent studies, some autophagy regulators have been reported to alleviate IBD via suppressing the NLRP3 inflammasome activation (Ke et al., 2016; Zhao et al., 2017; Liu et al., 2018; Mai et al., 2019; Shao et al., 2019a; Saber and El-Kader, 2020).
For instance, it was demonstrated that a small-molecular AMPK activator (GL-V9) significantly degraded the NLRP3 inflammasome complex in macrophages via the induction of autophagy, thus protecting against colitis (Zhao et al., 2017). Liu et al. (2018) revealed that Ginsenoside Rd (Rd), a well-known tetracyclic triterpenoid derivative, significantly attenuated the severity of colitis in DSS-induced UC models through the induction of p62-driven mitophagy-mediated NLRP3 inflammasome inactivation. Another natural derivative, Palmatine, was also shown to ameliorate DSS-induced colitis through promoting mitophagy-mediated NLRP3 inflammasome suppression (Mai et al., 2019). In addition, Saber and El-Kader (2020) demonstrated that the combined therapy of metformin and MCC950 produced a protective effect on UC and might become a candidate in the future treatment of UC. They showed that metformin/MCC950 attenuated DSS-induced colitis via autophagy-mediated NLRP3 inflammasome inhibition by modulating heat shock protein 90 (HSP90)/NLRP3 interaction. Recently, two studies from us reported the anti-colitis effect of the NLRP3 inflammasome inhibitors taking advantage of autophagy regulation (Ke et al., 2016; Shao et al., 2019a). In one study, we uncovered that the triggering of cholinergic anti-inflammatory pathway by the administration of alpha7 nicotinic acetylcholine receptor (α7nAChR) agonist alleviated DSS-induced colitis. The alleviative process was through autophagy-mediated suppression of the NLRP3 inflammasome-related IL-1β and IL-18 production related to the AMPK-mTOR-p70S6K signaling pathway (Shao et al., 2019a). Similar results were found in another study conducted by us, revealing that cannabinoid receptor 2 (CB2R) agonist contributed to the amelioration of DSS-induced colitis through the induction of AMPK-mTOR-p70S6K-mediated autophagy and inhibition of the NLRP3 inflammasome (Ke et al., 2016).
Intestinal Microbiota Regulators Taking Advantage of Autophagy
Intestinal microbiota is recognized as diverse bacterial colonies planting in the gut. Intestinal microbiota is generally divided into four phyla, namely Bacteroidetes, Firmicutes, Actinobacteria, and Proteobacteria, among which Bacteroidetes and Firmicutes have been shown to be dominant communities in the gut (Frank et al., 2007; Sender et al., 2016; Zhang et al., 2017). It is widely explored that the disturbance of intestinal microbiota homeostasis is highly involved in the onset and development of IBD (Caruso et al., 2020; Lavelle and Sokol, 2020; Neurath, 2020). As a result, restoring the homeostasis of intestinal microbiota provides a potential and effective therapeutic strategy in the treatment of IBD.
Autophagy has been revealed to play an important role in the regulation of intestinal microbiota. For instance, it was shown by Chu et al. (2016) that the mutation of autophagy-related ATG16L1 and NOD2 led to the inactivation of noncanonical autophagy process, resulting in the dysbiosis of gut microbiota in IBD. There is another study on IBD uncovering the effect of autophagy-related gene mutation on intestinal microbiota involved in the induction of the unfolded protein response (UPR; Hooper et al., 2019). In addition, Bel and Hooper (2018) reported the regulatory role of secretory autophagy from Paneth cells in intestinal microbiota in CD patients. They explored the effect of autophagy-related genes on the intestinal defense system. Recently, one of the selective forms of autophagy, named xenophagy, has been largely reported to contribute to modulating gut microbiota in IBD. Xenophagy is recognized as the process of autophagy engulfing intracellular pathogens, because it leads to the elimination of foreign microbes (Sui et al., 2017). It was reported that intestinal epithelial cells with a Crohn’s disease-susceptibility mutation in ATG16L1 exhibited less xenophagy, thus leading to the epithelial barrier dysfunction (Lopes et al., 2018). In addition, Kim et al. (2019) have demonstrated the involvement of some autophagy-related genes including ATG16L1, IRGM, ATG7, and p62 in the maintaining of intestinal homeostasis through the triggering of xenophagy. Those studies indicated that taking advantage of autophagy, especially xenophagy, could facilitate the development of new therapeutic opportunities for IBD.
So far, several therapies are available in the alleviation of IBD taking advantage of autophagy-mediated modulation of intestinal microbiota (Sun, 2016; Bakke and Sun, 2018; Kokten et al., 2018; Xuan et al., 2020). It has been reported that vitamin D promoted intestinal autophagy, thus alleviating IBD (Sun, 2016). The signaling of vitamin D/VDR was shown to be beneficial in the maintaining and restoration of intestinal microbiota homeostasis (Bakke and Sun, 2018). In addition, Kokten et al. (2018) demonstrated that the inhibition of triggering receptor expressed on myeloid cells-1 (TREM-1) contributed to the restoration of impaired autophagy activity, thus positively regulating the intestinal microbiota in colitis mice. Xuan et al. (2020) uncovered that Galangin, a natural flavonoid, could fight against DSS-induced colitis through the promotion of autophagy-mediated regulation of gut microbiota. The modulation of inflammatory reaction and myeloperoxidase activities was shown to be involved in this process.
Conclusion
All in all, recent studies have revealed the role of autophagy in the pathogenesis and progression of IBD. In addition, we are lucky to have several autophagy-related therapies, which have been proven to be effective in the amelioration of IBD. As we have discussed above, several agents taking advantage of autophagy have been demonstrated to be effective in the treatment of IBD. Those agents include some inflammasome inhibitors, especially those targeting on the inflammasome, and some intestinal microbiota regulators. However, although the mechanisms of IBD has been widely investigated, the specific molecular mechanisms of IBD remain unclarified, which largely limits the development of novel treatment for IBD. In addition, because of the complicated effect of autophagy on IBD, to ultimately apply therapeutic strategies taking advantage of autophagy in the clinical practice, further studies are demanded on this issue.
Author Contributions
B-ZS and YY retrieved concerned literatures and wrote the manuscript. J-SZ and J-HZ designed the table and figure. J-PL and KW revised the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by grants from General Hospital of the Chinese People’s Liberation Army (No. 2018ZD-006 and 2018MS-012) and Project of Military Health and Disease Prevention and Control (No. 13BJYZ26).
References
- Allen E. A., Baehrecke E. H. (2020). Autophagy in animal development. Cell Death Differ. 27, 903–918. 10.1038/s41418-020-0497-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andhavarapu S., Mubariz F., Arvas M., Bever C., Jr., Makar T. K. (2019). Interplay between ER stress and autophagy: a possible mechanism in multiple sclerosis pathology. Exp. Mol. Pathol. 108, 183–190. 10.1016/j.yexmp.2019.04.016, PMID: [DOI] [PubMed] [Google Scholar]
- Bakke D., Sun J. (2018). Ancient nuclear receptor VDR with new functions: microbiome and inflammation. Inflamm. Bowel Dis. 24, 1149–1154. 10.1093/ibd/izy092, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S., Ghosh S., Sinha K., Chowdhury S., Sil P. C. (2019). Sulphur dioxide ameliorates colitis related pathophysiology and inflammation. Toxicology 412, 63–78. 10.1016/j.tox.2018.11.010, PMID: [DOI] [PubMed] [Google Scholar]
- Baradaran Ghavami S., Kabiri F., Nourian M., Balaii H., Shahrokh S., Chaleshi V., et al. (2019). Association between variants of the autophagy related gene ATG16L1 in inflammatory bowel diseases and clinical statues. Gastroenterol. Hepatol. Bed. Bench. 12, S94–S100. PMID: [PMC free article] [PubMed] [Google Scholar]
- Bartelt A., Widenmaier S. B. (2020). Proteostasis in thermogenesis and obesity. Biol. Chem. 401, 1019–1030. 10.1515/hsz-2019-0427, PMID: [DOI] [PubMed] [Google Scholar]
- Bel S., Hooper L. V. (2018). Secretory autophagy of lysozyme in Paneth cells. Autophagy 14, 719–721. 10.1080/15548627.2018.1430462, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyayev L., Hawksworth J., Khan K., Kaufman S., Subramanian S., Kroemer A., et al. (2020). Immunologic complications and graft survival in Crohn’s disease and NOD2 mutant non-Crohn’s disease adult recipients following intestine transplantation. Transplant. Direct 6:e556. 10.1097/TXD.0000000000001006, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento C. F., Renna M., Ghislat G., Puri C., Ashkenazi A., Vicinanza M., et al. (2016). Mammalian autophagy: how does it work? Annu. Rev. Biochem. 85, 685–713. 10.1146/annurev-biochem-060815-014556, PMID: [DOI] [PubMed] [Google Scholar]
- Bischoff S. C., Barbara G., Buurman W., Ockhuizen T., Schulzke J. D., Serino M., et al. (2014). Intestinal permeability—a new target for disease prevention and therapy. BMC Gastroenterol. 14:189. 10.1186/s12876-014-0189-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork T., Liang W., Yamahara K., Lee P., Tian Z., Liu S., et al. (2020). Podocytes maintain high basal levels of autophagy independent of mtor signaling. Autophagy 16, 1932–1948. 10.1080/15548627.2019.1705007, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui T. M., Mascarenhas L. A., Sumagin R. (2018). Extracellular vesicles regulate immune responses and cellular function in intestinal inflammation and repair. Tissue Barriers 6:e1431038. 10.1080/21688370.2018.1431038, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buisson A., Douadi C., Ouchchane L., Goutte M., Hugot J. P., Dubois A., et al. (2019). Macrophages inability to mediate adherent-invasive E. coli replication is linked to autophagy in Crohn’s disease patients. Cell 8:1394. 10.3390/cells8111394, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke A., Lichtenstein G. R., Rombeau J. L. (1997). Nutrition and ulcerative colitis. Baillieres Clin. Gastroenterol. 11, 153–174. 10.1016/s0950-3528(97)90059-2, PMID: [DOI] [PubMed] [Google Scholar]
- Cao Q., Du H., Fu X., Duan N., Liu C., Li X. (2020). Artemisinin attenuated atherosclerosis in high-fat diet-fed ApoE−/− mice by promoting macrophage autophagy through the AMPK/mTOR/ULK1 pathway. J. Cardiovasc. Pharmacol. 75, 321–332. 10.1097/FJC.0000000000000794, PMID: [DOI] [PubMed] [Google Scholar]
- Caruso R., Lo B. C., Nunez G. (2020). Host-microbiota interactions in inflammatory bowel disease. Nat. Rev. Immunol. 20, 411–426. 10.4161/gmic.20228, PMID: [DOI] [PubMed] [Google Scholar]
- Cerri S., Blandini F. (2019). Role of autophagy in Parkinson’s disease. Curr. Med. Chem. 26, 3702–3718. 10.2174/0929867325666180226094351, PMID: [DOI] [PubMed] [Google Scholar]
- Choi S., Snider A. J. (2019). Diet, lipids and colon cancer. Int. Rev. Cell Mol. Biol. 347, 105–144. 10.1016/bs.ircmb.2019.07.001, PMID: [DOI] [PubMed] [Google Scholar]
- Chu H., Khosravi A., Kusumawardhani I. P., Kwon A. H., Vasconcelos A. C., Cunha L. D., et al. (2016). Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science 352, 1116–1120. 10.1126/science.aad9948, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll R. C., Robertson A. A., Chae J. J., Higgins S. C., Munoz-Planillo R., Inserra M. C., et al. (2015). A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat. Med. 21, 248–255. 10.1038/nm.3806, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmasso G., Nguyen H. T. T., Fais T., Massier S., Barnich N., Delmas J., et al. (2019). Crohn’s disease-associated adherent-invasive Escherichia coli manipulate host autophagy by impairing SUMOylation. Cell 8:35. 10.3390/cells8010035, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash S., Aydin Y., Wu T. (2020). Integrated stress response in hepatitis C promotes Nrf2-related chaperone-mediated autophagy: a novel mechanism for host-microbe survival and HCC development in liver cirrhosis. Semin. Cell Dev. Biol. 101, 20–35. 10.1016/j.semcdb.2019.07.015, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton D., Kumar S. (2019). Autophagy-dependent cell death. Cell Death Differ. 26, 605–616. 10.1038/s41418-018-0252-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Levine B. (2018). Autophagy balances inflammation in innate immunity. Autophagy 14, 243–251. 10.1080/15548627.2017.1402992, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I., Elazar Z. (2018). Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 19, 349–364. 10.1038/s41580-018-0003-4, PMID: [DOI] [PubMed] [Google Scholar]
- Ding F., Shan C., Li H., Zhang Y., Guo C., Zhou Z., et al. (2020). Simvastatin alleviated diabetes mellitus-induced erectile dysfunction in rats by enhancing AMPK pathway-induced autophagy. Andrology 8, 780–792. 10.1111/andr.12758, PMID: [DOI] [PubMed] [Google Scholar]
- Feuerstein J. D., Moss A. C., Farraye F. A. (2019). Ulcerative Colitis. Mayo Clin. Proc. 94, 1357–1373. 10.1016/j.mayocp.2019.01.018, PMID: [DOI] [PubMed] [Google Scholar]
- Frade-Proud’Hon-Clerc S., Smol T., Frenois F., Sand O., Vaillant E., Dhennin V., et al. (2019). A novel rare missense variation of the NOD2 gene: evidencesof implication in Crohn’s disease. Int. J. Mol. Sci. 20:835. 10.3390/ijms20040835, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D. N., St Amand A. L., Feldman R. A., Boedeker E. C., Harpaz N., Pace N. R. (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U. S. A. 104, 13780–13785. 10.1073/pnas.0706625104, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallinger Z. R., Rumman A., Pivovarov K., Fortinsky K. J., Steinhart A. H., Weizman A. V. (2017). Frequency and variables associated with fasting orders in inpatients with ulcerative colitis: The Audit of Diet Orders-Ulcerative Colitis (ADORE-UC) Study. Inflamm. Bowel Dis. 23, 1790–1795. 10.1097/MIB.0000000000001244, PMID: [DOI] [PubMed] [Google Scholar]
- Gerster R., Eloranta J. J., Hausmann M., Ruiz P. A., Cosin-Roger J., Terhalle A., et al. (2015). Anti-inflammatory function of high-density lipoproteins via autophagy of IkappaB kinase. Cell. Mol. Gastroenterol. Hepatol. 1, 171.e1–187.e1. 10.1016/j.jcmgh.2014.12.006, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll R., van Beelen Granlund A. (2015). Intestinal barrier homeostasis in inflammatory bowel disease. Scand. J. Gastroenterol. 50, 3–12. 10.3109/00365521.2014.971425, PMID: [DOI] [PubMed] [Google Scholar]
- Hampe J., Franke A., Rosenstiel P., Till A., Teuber M., Huse K., et al. (2007). A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 39, 207–211. 10.1038/ng1954, PMID: [DOI] [PubMed] [Google Scholar]
- Han C., Guo L., Sheng Y., Yang Y., Wang J., Gu Y., et al. (2020). FoxO1 regulates TLR4/MyD88/MD2-NF-kappaB inflammatory signalling in mucosal barrier injury of inflammatory bowel disease. J. Cell. Mol. Med. 24, 3712–3723. 10.1111/jcmm.15075, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harb W. J. (2015). Crohn’s disease of the colon, rectum, and anus. Surg. Clin. North Am. 95, 1195–1210, vi. 10.1016/j.suc.2015.07.005, PMID: [DOI] [PubMed] [Google Scholar]
- Harris J., Lang T., Thomas J. P. W., Sukkar M. B., Nabar N. R., Kehrl J. H. (2017). Autophagy and inflammasomes. Mol. Immunol. 86, 10–15. 10.1016/j.molimm.2017.02.013, PMID: [DOI] [PubMed] [Google Scholar]
- Ho J., Chan H., Liang Y., Liu X., Zhang L., Li Q., et al. (2020). Cathelicidin preserves intestinal barrier function in polymicrobial sepsis. Crit. Care 24:47. 10.1186/s13054-020-2754-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper K. M., Barlow P. G., Henderson P., Stevens C. (2019). Interactions between autophagy and the unfolded protein response: implications for inflammatory bowel disease. Inflamm. Bowel Dis. 25, 661–671. 10.1093/ibd/izy380, PMID: [DOI] [PubMed] [Google Scholar]
- Hu X., Deng J., Yu T., Chen S., Ge Y., Zhou Z., et al. (2019). ATF4 deficiency promotes intestinal inflammation in mice by reducing uptake of glutamine and expression of antimicrobial peptides. Gastroenterology 156, 1098–1111. 10.1053/j.gastro.2018.11.033, PMID: [DOI] [PubMed] [Google Scholar]
- Huang Y., Wang H., Hao Y., Lin H., Dong M., Ye J., et al. (2020). Myeloid PTEN promotes chemotherapy-induced NLRP3-inflammasome activation and antitumour immunity. Nat. Cell Biol. 22, 716–727. 10.1038/s41556-020-0510-3, PMID: [DOI] [PubMed] [Google Scholar]
- Iida T., Onodera K., Nakase H. (2017). Role of autophagy in the pathogenesis of inflammatory bowel disease. World J. Gastroenterol. 23, 1944–1953. 10.3748/wjg.v23.i11.1944, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- James K. R., Gomes T., Elmentaite R., Kumar N., Gulliver E. L., King H. W., et al. (2020). Distinct microbial and immune niches of the human colon. Nat. Immunol. 21, 343–353. 10.1038/s41590-020-0602-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarret A., Jackson R., Duizer C., Healy M. E., Zhao J., Rone J. M. (2020). Enteric nervous system-derived IL-18 orchestrates mucosal barrier immunity. Cell 180, 813–814. 10.1016/j.cell.2019.12.016, PMID: [DOI] [PubMed] [Google Scholar]
- Kaleagasioglu F., Ali D. M., Berger M. R. (2020). Multiple facets of autophagy and the emerging role of alkylphosphocholines as autophagy modulators. Front. Pharmacol. 11:547. 10.3389/fphar.2020.00547, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi S., Tabataba-Vakili S., Yari Z., Alborzi F., Hedayati M., Ebrahimi-Daryani N., et al. (2019). The effects of two vitamin D regimens on ulcerative colitis activity index, quality of life and oxidant/anti-oxidant status. Nutr. J. 18:16. 10.1186/s12937-019-0441-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke P., Shao B. Z., Xu Z. Q., Wei W., Han B. Z., Chen X. W., et al. (2016). Activation of cannabinoid receptor 2 ameliorates DSS-induced colitis through inhibiting NLRP3 inflammasome in macrophages. PLoS One 11:e0155076. 10.1371/journal.pone.0155076, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee B. P., Ng J. G., Ng C. C., Hilmi I., Goh K. L., Chua K. H. (2020). Genetic polymorphisms of ATG16L1 and IRGM genes in Malaysian patients with Crohn’s disease. J. Dig. Dis. 21, 29–37. 10.1111/1751-2980.12829, PMID: [DOI] [PubMed] [Google Scholar]
- Keller M. D., Torres V. J., Cadwell K. (2020). Autophagy and microbial pathogenesis. Cell Death Differ. 27, 872–886. 10.1038/s41418-019-0481-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsen J. R., Russo P., Sullivan K. E. (2019). Early-onset inflammatory bowel disease. Immunol. Allergy Clin. N. Am. 39, 63–79. 10.1016/j.iac.2018.08.008, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsen J. R., Sullivan K. E. (2017). Inflammatory bowel disease in primary immunodeficiencies. Curr Allergy Asthma Rep 17:57. 10.1007/s11882-017-0724-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienle P. (2018). Impact of modern drug therapy on surgery: Crohn’s disease. Visc. Med. 34, 422–425. 10.1159/000495127, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Eun H. S., Jo E. K. (2019). Roles of autophagy-related genes in the pathogenesis of inflammatory bowel disease. Cell 8:77. 10.3390/cells8010077, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko C. W., Singh S., Feuerstein J. D., Falck-Ytter C., Falck-Ytter Y., Cross R. K., et al. (2019). AGA clinical practice guidelines on the management of mild-to-moderate ulcerative colitis. Gastroenterology 156, 748–764. 10.1053/j.gastro.2018.12.009, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokten T., Gibot S., Lepage P., D’alessio S., Hablot J., Ndiaye N. C., et al. (2018). TREM-1 inhibition restores impaired autophagy activity and reduces colitis in mice. J. Crohns Colitis 12, 230–244. 10.1093/ecco-jcc/jjx129, PMID: [DOI] [PubMed] [Google Scholar]
- Lavelle A., Sokol H. (2020). Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 17, 223–237. 10.1038/s41575-019-0258-z, PMID: [DOI] [PubMed] [Google Scholar]
- Law A. D., Dutta U., Kochhar R., Vaishnavi C., Kumar S., Noor T., et al. (2019). Vitamin D deficiency in adult patients with ulcerative colitis: prevalence and relationship with disease severity, extent, and duration. Indian J. Gastroenterol. 38, 6–14. 10.1007/s12664-019-00932-z, PMID: [DOI] [PubMed] [Google Scholar]
- Lei-Leston A. C., Murphy A. G., Maloy K. J. (2017). Epithelial cell inflammasomes in intestinal immunity and inflammation. Front. Immunol. 8:1168. 10.3389/fimmu.2017.01168, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Chen Y. (2019). AMPK and autophagy. Adv. Exp. Med. Biol. 1206, 85–108. 10.1007/978-981-15-0602-4_4, PMID: [DOI] [PubMed] [Google Scholar]
- Li J., Hochstrasser M. (2020). Microautophagy regulates proteasome homeostasis. Curr. Genet. 66, 683–668. 10.1007/s00294-020-01059-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman O. J., Sulzer D. (2020). The synaptic autophagy cycle. J. Mol. Biol. 432, 2589–2604. 10.1016/j.jmb.2019.12.028, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Liu S., Qiu X., Yang X., Bao L., Pu F., et al. (2020). Donor MSCs release apoptotic bodies to improve myocardial infarction via autophagy regulation in recipient cells. Autophagy 16, 2140–2155. 10.1080/15548627.2020.1717128, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Wang J., Yang Y., Liu X., Zhu Y., Zou J., et al. (2018). Ginsenoside Rd ameliorates colitis by inducing p62-driven mitophagy-mediated NLRP3 inflammasome inactivation in mice. Biochem. Pharmacol. 155, 366–379. 10.1016/j.bcp.2018.07.010, PMID: [DOI] [PubMed] [Google Scholar]
- Lopes F., Keita A. V., Saxena A., Reyes J. L., Mancini N. L., Al Rajabi A. (2018). ER-stress mobilization of death-associated protein kinase-1-dependent xenophagy counteracts mitochondria stress-induced epithelial barrier dysfunction. J. Biol. Chem. 293, 3073–3087. 10.1074/jbc.RA117.000809, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Chen J., Xu H. G., Zhou X., He Q., Li Y. L., et al. (2014). MIR106B and MIR93 prevent removal of bacteria from epithelial cells by disrupting ATG16L1-mediated autophagy. Gastroenterology 146, 188–199. 10.1053/j.gastro.2013.09.006, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luissint A. C., Williams H. C., Kim W., Flemming S., Azcutia V., Hilgarth R. S., et al. (2019). Macrophage-dependent neutrophil recruitment is impaired under conditions of increased intestinal permeability in JAM-A-deficient mice. Mucosal Immunol. 12, 668–678. 10.1038/s41385-019-0143-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K., Guo J., Wang G., Ni Q., Liu X. (2020). Toll-like receptor 2-mediated autophagy promotes microglial cell death by modulating the microglial M1/M2 phenotype. Inflammation 43, 701–711. 10.1007/s10753-019-01152-5, PMID: [DOI] [PubMed] [Google Scholar]
- Macias-Ceja D. C., Cosin-Roger J., Ortiz-Masia D., Salvador P., Hernandez C., Esplugues J. V., et al. (2017). Stimulation of autophagy prevents intestinal mucosal inflammation and ameliorates murine colitis. Br. J. Pharmacol. 174, 2501–2511. 10.1111/bph.13860, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai C. T., Wu M. M., Wang C. L., Su Z. R., Cheng Y. Y., Zhang X. J. (2019). Palmatine attenuated dextran sulfate sodium (DSS)-induced colitis via promoting mitophagy-mediated NLRP3 inflammasome inactivation. Mol. Immunol. 105, 76–85. 10.1016/j.molimm.2018.10.015, PMID: [DOI] [PubMed] [Google Scholar]
- Mangan M. S. J., Olhava E. J., Roush W. R., Seidel H. M., Glick G. D., Latz E. (2018). Targeting the NLRP3 inflammasome in inflammatory diseases. Nat. Rev. Drug Discov. 17:688. 10.1038/nrd.2018.149, PMID: [DOI] [PubMed] [Google Scholar]
- Mao L., Kitani A., Strober W., Fuss I. J. (2018). The role of NLRP3 and IL-1beta in the pathogenesis of inflammatory bowel disease. Front. Immunol. 9:2566. 10.3389/fimmu.2018.02566, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasco M. R., Linnemann A. K. (2018). β-Cell autophagy in diabetes pathogenesis. Endocrinology 159, 2127–2141. 10.1210/en.2017-03273, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S., Newton K., Monack D. M., Vucic D., French D. M., Lee W. P., et al. (2004). Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430, 213–218. 10.1038/nature02664, PMID: [DOI] [PubMed] [Google Scholar]
- Martin J. C., Beriou G., Josien R. (2016). Dextran sulfate sodium (DSS)-induced acute colitis in the rat. Methods Mol. Biol. 1371, 197–203. 10.1007/978-1-4939-3139-2_12, PMID: [DOI] [PubMed] [Google Scholar]
- Mijaljica D., Prescott M., Devenish R. J. (2011). Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy 7, 673–682. 10.4161/auto.7.7.14733, PMID: [DOI] [PubMed] [Google Scholar]
- Morris G. P., Beck P. L., Herridge M. S., Depew W. T., Szewczuk M. R., Wallace J. L. (1989). Hapten-induced model of chronic inflammation and ulceration in the rat colon. Gastroenterology 96, 795–803. PMID: [PubMed] [Google Scholar]
- Muller P. A., Matheis F., Mucida D. (2020). Gut macrophages: key players in intestinal immunity and tissue physiology. Curr. Opin. Immunol. 62, 54–61. 10.1016/j.coi.2019.11.011, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy B., Kavanagh D. O., Winter D. C. (2020). Modern surgery for ulcerative colitis. Updat. Surg. 72, 325–333. 10.1007/s13304-020-00719-4, PMID: [DOI] [PubMed] [Google Scholar]
- Nath S., Dancourt J., Shteyn V., Puente G., Fong W. M., Nag S., et al. (2014). Lipidation of the LC3/GABARAP family of autophagy proteins relies on a membrane-curvature-sensing domain in Atg3. Nat. Cell Biol. 16, 415–424. 10.1038/ncb2940, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neurath M. F. (2020). Host-microbiota interactions in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 17, 76–77. 10.1038/s41575-019-0248-1, PMID: [DOI] [PubMed] [Google Scholar]
- Ng S. C. (2015). Emerging leadership lecture: inflammatory bowel disease in Asia: emergence of a “Western” disease. J. Gastroenterol. Hepatol. 30, 440–445. 10.1111/jgh.12859, PMID: [DOI] [PubMed] [Google Scholar]
- Nunes N. S., Kim S., Sundby M., Chandran P., Burks S. R., Paz A. H., et al. (2018). Temporal clinical, proteomic, histological and cellular immune responses of dextran sulfate sodium-induced acute colitis. World J. Gastroenterol. 24, 4341–4355. 10.3748/wjg.v24.i38.4341, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi Y., Tremel S., Williams R. L. (2019). VPS34 complexes from a structural perspective. J. Lipid Res. 60, 229–241. 10.1194/jlr.R089490, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmela C., Chevarin C., Xu Z., Torres J., Sevrin G., Hirten R., et al. (2018). Adherent-invasive Escherichia coli in inflammatory bowel disease. Gut 67, 574–587. 10.1136/gutjnl-2017-314903, PMID: [DOI] [PubMed] [Google Scholar]
- Park S. C., Jeen Y. T. (2019). Genetic studies of inflammatory bowel disease-focusing on Asian patients. Cell 8:404. 10.3390/cells8050404, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaseyed T., Hansson G. C. (2020). Membrane mucins of the intestine at a glance. J. Cell Sci. 133:jcs240929. 10.1242/jcs.240929, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pravda J. (2019). Can ulcerative colitis be cured? Discov. Med. 27, 197–200. PMID: [PubMed] [Google Scholar]
- Racanelli A. C., Kikkers S. A., Choi A. M. K., Cloonan S. M. (2018). Autophagy and inflammation in chronic respiratory disease. Autophagy 14, 221–232. 10.1080/15548627.2017.1389823, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos G. P., Papadakis K. A. (2019). Mechanisms of disease: inflammatory bowel diseases. Mayo Clin. Proc. 94, 155–165. 10.1016/j.mayocp.2018.09.013, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam V. A. K., Zhao Y., Shao F. (2019). Innate immunity to intracellular LPS. Nat. Immunol. 20, 527–533. 10.1038/s41590-019-0368-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Ungermann C. (2017). Autophagosome maturation and fusion. J. Mol. Biol. 429, 486–496. 10.1038/s41590-019-0368-3, PMID: [DOI] [PubMed] [Google Scholar]
- Rioux J. D., Xavier R. J., Taylor K. D., Silverberg M. S., Goyette P., Huett A., et al. (2007). Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 39, 596–604. 10.1038/ng2032, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruder B., Atreya R., Becker C. (2019). Tumour necrosis factor alpha in intestinal homeostasis and gut related diseases. Int. J. Mol. Sci. 20:1887. 10.3390/ijms20081887, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saber S., El-Kader E. M. A. (2020). Novel complementary coloprotective effects of metformin and MCC950 by modulating HSP90/NLRP3 interaction and inducing autophagy in rats. Inflammopharmacology. 10.1007/s10787-020-00730-6, PMID: [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Sairenji T., Collins K. L., Evans D. V. (2017). An update on inflammatory bowel disease. Prim. Care 44, 673–692. 10.1016/j.pop.2017.07.010, PMID: [DOI] [PubMed] [Google Scholar]
- Sanchez de Medina F., Romero-Calvo I., Mascaraque C., Martinez-Augustin O. (2014). Intestinal inflammation and mucosal barrier function. Inflamm. Bowel Dis. 20, 2394–2404. 10.1097/MIB.0000000000000204, PMID: [DOI] [PubMed] [Google Scholar]
- Schroder K., Zhou R., Tschopp J. (2010). The NLRP3 inflammasome: a sensor for metabolic danger? Science 327, 296–300. 10.1126/science.1184003, PMID: [DOI] [PubMed] [Google Scholar]
- Schroeder B. O. (2019). Fight them or feed them: how the intestinal mucus layer manages the gut microbiota. Gastroenterol. Rep. 7, 3–12. 10.1093/gastro/goy052, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender R., Fuchs S., Milo R. (2016). Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14:e1002533. 10.1371/journal.pbio.1002533, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao B. Z., Cao Q., Liu C. (2018). Targeting NLRP3 inflammasome in the treatment of CNS diseases. Front. Mol. Neurosci. 11:320. 10.3389/fnmol.2018.00320, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao B. Z., Han B. Z., Zeng Y. X., Su D. F., Liu C. (2016). The roles of macrophage autophagy in atherosclerosis. Acta Pharmacol. Sin. 37, 150–156. 10.1038/aps.2015.87, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao B. Z., Ke P., Xu Z. Q., Wei W., Cheng M. H., Han B. Z., et al. (2017). Autophagy plays an important role in anti-inflammatory mechanisms stimulated by Alpha7 nicotinic acetylcholine receptor. Front. Immunol. 8:553. 10.3389/fimmu.2017.00553, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao B. Z., Wang S. L., Fang J., Li Z. S., Bai Y., Wu K. (2019a). Alpha7 nicotinic acetylcholine receptor alleviates inflammatory bowel disease through induction of AMPK-mTOR-p70S6K-mediated autophagy. Inflammation 42, 1666–1679. 10.1007/s10753-019-01027-9, PMID: [DOI] [PubMed] [Google Scholar]
- Shao B. Z., Wang S. L., Pan P., Yao J., Wu K., Li Z. S., et al. (2019b). Targeting NLRP3 inflammasome in inflammatory bowel disease: putting out the fire of inflammation. Inflammation 42, 1147–1159. 10.1007/s10753-019-01008-y, PMID: [DOI] [PubMed] [Google Scholar]
- Shen T., Li S., Cai L. D., Liu J. L., Wang C. Y., Gan W. J., et al. (2018). Erbin exerts a protective effect against inflammatory bowel disease by suppressing autophagic cell death. Oncotarget 9, 12035–12049. 10.18632/oncotarget.23925, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y., Song Y., Li Z. G., Wang Y. B., Lin H. M., Cheng H. H., et al. (2018). RAB37 interacts directly with ATG5 and promotes autophagosome formation via regulating ATG5-12-16 complex assembly. Cell Death Differ. 25, 918–934. 10.1038/s41418-017-0023-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C. C., Pan L. Y., Peng Z. Y., Li J. G. (2020). MiR-126 regulated myocardial autophagy on myocardial infarction. Eur. Rev. Med. Pharmacol. Sci. 24, 6971–6979. 10.26355/eurrev_202006_21689, PMID: [DOI] [PubMed] [Google Scholar]
- Singh S., Feuerstein J. D., Binion D. G., Tremaine W. J. (2019). AGA technical review on the management of mild-to-moderate ulcerative colitis. Gastroenterology 156, 769.e729–808.e729. 10.1053/j.gastro.2018.12.008, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola Tapias N., Vergnolle N., Denadai-Souza A., Barreau F. (2020). The interplay between genetic risk factors and proteolytic dysregulation in the pathophysiology of inflammatory bowel disease. J. Crohns Colitis 14, 1149–1161. 10.1093/ecco-jcc/jjaa033, PMID: [DOI] [PubMed] [Google Scholar]
- Song N., Li T. (2018). Regulation of NLRP3 inflammasome by phosphorylation. Front. Immunol. 9:2305. 10.3389/fimmu.2018.02305, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez R., Buelvas N. (2015). Inflammasome: activation mechanisms. Investig. Clin. 56, 74–99. PMID: [PubMed] [Google Scholar]
- Sui X., Liang X., Chen L., Guo C., Han W., Pan H., et al. (2017). Bacterial xenophagy and its possible role in cancer: a potential antimicrobial strategy for cancer prevention and treatment. Autophagy 13, 237–247. 10.1080/15548627.2016.1252890, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J. (2016). VDR/vitamin D receptor regulates autophagic activity through ATG16L1. Autophagy 12, 1057–1058. 10.1080/15548627.2015.1072670, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson K. V., Deng M., Ting J. P. (2019). The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 19, 477–489. 10.1038/s41577-019-0165-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaddoni A., Mohammadi E., Sedaghat F., Qujeq D., As’habi A. (2020). The anticancer effects of curcumin via targeting the mammalian target of rapamycin complex 1 (mTORC1) signaling pathway. Pharmacol. Res. 156:104798. 10.1016/j.phrs.2020.104798, PMID: [DOI] [PubMed] [Google Scholar]
- Tan P., Ye Y., Mao J., He L. (2019). Autophagy and immune-related diseases. Adv. Exp. Med. Biol. 1209, 167–179. 10.1007/978-981-15-0606-2_10, PMID: [DOI] [PubMed] [Google Scholar]
- Tong X. P., Chen Y., Zhang S. Y., Xie T., Tian M., Guo M. R., et al. (2015). Key autophagic targets and relevant small-molecule compounds in cancer therapy. Cell Prolif. 48, 7–16. 10.1111/cpr.12154, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y., Tang J. (2017). Candida albicans infection and intestinal immunity. Microbiol. Res. 198, 27–35. 10.1016/j.micres.2017.02.002, PMID: [DOI] [PubMed] [Google Scholar]
- Torres J., Mehandru S., Colombel J. F., Peyrin-Biroulet L. (2017). Crohn’s disease. Lancet 389, 1741–1755. 10.1016/S0140-6736(16)31711-1, PMID: [DOI] [PubMed] [Google Scholar]
- Tsianos V. E., Kostoulas C., Gazouli M., Frillingos S., Georgiou I., Christodoulou D. K., et al. (2020). ATG16L1 T300A polymorphism is associated with Crohn’s disease in a Northwest Greek cohort, but ECM1 T130M and G290S polymorphisms are not associated with ulcerative colitis. Ann. Gastroenterol. 33, 38–44. 10.20524/aog.2019.0434, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler J. K., Johnson J. E. (2018). The role of autophagy in the regulation of yeast life span. Ann. N. Y. Acad. Sci. 1418, 31–43. 10.1111/nyas.13549, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tysk C., Lindberg E., Jarnerot G., Floderus-Myrhed B. (1988). Ulcerative colitis and Crohn’s disease in an unselected population of monozygotic and dizygotic twins. A study of heritability and the influence of smoking. Gut 29, 990–996. 10.1136/gut.29.7.990, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma M., Kadoki M., Lefkovith A., Conway K. L., Gao K., Mohanan V., et al. (2020). Cell type- and stimulation-dependent transcriptional programs regulated by Atg16L1 and its Crohn’s disease risk variant T300A. J. Immunol. 205, 414–424. 10.4049/jimmunol.1900750, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita A. A., Royse E. A., Pullen N. A. (2019). Nanoparticles and danger signals: oral delivery vehicles as potential disruptors of intestinal barrier homeostasis. J. Leukoc. Biol. 106, 95–103. 10.1002/JLB.3MIR1118-414RR, PMID: [DOI] [PubMed] [Google Scholar]
- Vomero M., Barbati C., Colasanti T., Perricone C., Novelli L., Ceccarelli F. (2018). Autophagy and rheumatoid arthritis: current knowledges and future perspectives. Front. Immunol. 9:1577. 10.3389/fimmu.2018.01577, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldschmitt N., Kitamoto S., Secher T., Zacharioudaki V., Boulard O., Floquet E., et al. (2019). The regenerating family member 3 beta instigates IL-17A-mediated neutrophil recruitment downstream of NOD1/2 signalling for controlling colonisation resistance independently of microbiota community structure. Gut 68, 1190–1199. 10.1136/gutjnl-2018-316757, PMID: [DOI] [PubMed] [Google Scholar]
- Wang S., Huang Y., Zhou C., Wu H., Zhao J., Wu L., et al. (2018b). The role of autophagy and related microRNAs in inflammatory bowel disease. Gastroenterol. Res. Pract. 2018:7565076. 10.1155/2018/7565076, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Shao B. Z., Deng Z., Chen S., Yue Z., Miao C. Y. (2018a). Autophagy in ischemic stroke. Prog. Neurobiol. 163–164, 98–117. 10.1016/j.pneurobio.2018.01.001, PMID: [DOI] [PubMed] [Google Scholar]
- Wang S. L., Shao B. Z., Zhao S. B., Fang J., Gu L., Miao C. Y., et al. (2018c). Impact of paneth cell autophagy on inflammatory bowel disease. Front. Immunol. 9:693. 10.3389/fimmu.2018.00693, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Xu W., Liang M., Huang D., Huang K. (2019). CTRP13 inhibits atherosclerosis via autophagy-lysosome-dependent degradation of CD36. FASEB J. 33, 2290–2300. 10.1096/fj.201801267RR, PMID: [DOI] [PubMed] [Google Scholar]
- Watanabe T., Fujiwara Y., Chan F. K. L. (2020). Current knowledge on non-steroidal anti-inflammatory drug-induced small-bowel damage: a comprehensive review. J. Gastroenterol. 55, 481–495. 10.1007/s00535-019-01657-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M. Y., Liu L., Wang E. J., Xiao H. T., Cai C. Z., Wang J., et al. (2020). PI3KC3 complex subunit NRBF2 is required for apoptotic cell clearance to restrict intestinal inflammation. Autophagy 1–16. 10.1080/15548627.2020.1741332, PMID: [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Tang L., Wang B., Sun Q., Zhao P., Li W. (2019). The role of autophagy in maintaining intestinal mucosal barrier. J. Cell. Physiol. 234, 19406–19419. 10.1002/jcp.28722, PMID: [DOI] [PubMed] [Google Scholar]
- Xilouri M., Stefanis L. (2016). Chaperone mediated autophagy in aging: starve to prosper. Ageing Res. Rev. 32, 13–21. 10.1016/j.arr.2016.07.001, PMID: [DOI] [PubMed] [Google Scholar]
- Xuan H., Ou A., Hao S., Shi J., Jin X. (2020). Galangin protects against symptoms of dextran sodium sulfate-induced acute colitis by activating autophagy and modulating the gut microbiota. Nutrients 12:347. 10.3390/nu12020347, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi O. (2019). Autophagy in the heart. Circ. J. 83, 697–704. 10.1253/circj.CJ-18-1065, PMID: [DOI] [PubMed] [Google Scholar]
- Yamamoto A., Ukai H., Morishita M., Katsumi H. (2020). Approaches to improve intestinal and transmucosal absorption of peptide and protein drugs. Pharmacol. Ther. 211:107537. 10.1016/j.pharmthera.2020.107537, PMID: [DOI] [PubMed] [Google Scholar]
- Yang Y., Klionsky D. J. (2020). Autophagy and disease: unanswered questions. Cell Death Differ. 27, 858–871. 10.1038/s41418-019-0480-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Wang R., Zhu L. (2019). Chaperone-mediated autophagy. Adv. Exp. Med. Biol. 1206, 435–452. 10.1007/978-981-15-0602-4_20, PMID: [DOI] [PubMed] [Google Scholar]
- Ye L., Schnepf D., Staeheli P. (2019). Interferon-lambda orchestrates innate and adaptive mucosal immune responses. Nat. Rev. Immunol. 19, 614–625. 10.1038/s41577-019-0182-z, PMID: [DOI] [PubMed] [Google Scholar]
- Yim W. W., Mizushima N. (2020). Lysosome biology in autophagy. Cell Discov. 6:6. 10.1038/s41421-020-0141-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y. B., De Jonge H. R., Wu X., Yin Y. L. (2019). Enteroids for nutritional studies. Mol. Nutr. Food Res. 63:e1801143. 10.1002/mnfr.201801143, PMID: [DOI] [PubMed]
- Yu L., Chen Y., Tooze S. A. (2018). Autophagy pathway: cellular and molecular mechanisms. Autophagy 14, 207–215. 10.1080/15548627.2017.1378838, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zewinger S., Reiser J., Jankowski V., Alansary D., Hahm E., Triem S., et al. (2020). Apolipoprotein C3 induces inflammation and organ damage by alternative inflammasome activation. Nat. Immunol. 21, 30–41. 10.1038/s41590-019-0548-1, PMID: [DOI] [PubMed] [Google Scholar]
- Zha Z., Lv Y., Tang H., Li T., Miao Y., Cheng J., et al. (2020). An orally administered butyrate-releasing xylan derivative reduces inflammation in dextran sulphate sodium-induced murine colitis. Int. J. Biol. Macromol. 156, 1217–1233. 10.1016/j.ijbiomac.2019.11.159, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang S. L., Li Z. Y., Wang D. S., Xu T. Y., Fan M. B., Cheng M. H., et al. (2020b). Aggravated ulcerative colitis caused by intestinal Metrnl deficiency is associated with reduced autophagy in epithelial cells. Acta Pharmacol. Sin. 41, 763–770. 10.1038/s41401-019-0343-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. L., Wang S. N., Miao C. Y. (2017). Influence of microbiota on intestinal immune system in ulcerative colitis and its intervention. Front. Immunol. 8:1674. 10.3389/fimmu.2017.01674, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zhou Y., Xia Q., Chen Y., Li J. (2020a). All-trans-retinal induces autophagic cell death via oxidative stress and the endoplasmic reticulum stress pathway in human retinal pigment epithelial cells. Toxicol. Lett. 322, 77–86. 10.1016/j.toxlet.2020.01.005, PMID: [DOI] [PubMed] [Google Scholar]
- Zhao Y., Guo Q., Zhao K., Zhou Y., Li W., Pan C., et al. (2017). Small molecule GL-V9 protects against colitis-associated colorectal cancer by limiting NLRP3 inflammasome through autophagy. Onco. Targets. Ther. 7:e1375640. 10.1080/2162402X.2017.1375640, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Xu W., Wang J., Yan J., Shi Y., Zhang C., et al. (2018). Boosting mTOR-dependent autophagy via upstream TLR4-MyD88-MAPK signalling and downstream NF-kappaB pathway quenches intestinal inflammation and oxidative stress injury. EBioMedicine 35, 345–360. 10.1016/j.ebiom.2018.08.035, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Yuan Y., Zhang S., Guo C., Li X., Li G., et al. (2020). Intestinal flora and disease mutually shape the regional immune system in the intestinal tract. Front. Immunol. 11:575. 10.3389/fimmu.2020.00575, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]


