Abstract
Fowl adenovirus strains were isolated from the internal organs of 3-wk-old broiler flocks exhibited clinical signs associated with inclusion body hepatitis (IBH). The isolated strains were molecularly characterised and sequencing revealed three distinct clusters. One cluster showed close proximity at the nucleotide level with adenovirus type/species - 6/E, 7/E, 8a/E, and 8b/E. The second cluster contained five reference sequences belonging to the species FAdV-D and E. A third cluster contained one field and four reference sequences belonging to the FAdV-5/B, FAdV-4/C, FAdV-2/D, and FAdV-1/A type/species respectively. The heterogenicity, Relative Synonymous Codon Usage (RSCU), codon composition, and nucleotide frequencies were examined. Statistical analyses, were carried out. The maximum likelihoods for the examined sequences were estimated. The data indicated that correlation between isolated of adenovirus type/species 5/B, and E in Poland have been presented. Indicated adenovirus types and their combinations with locally circulating FAdVs strains could have implications for current detection methods and pathogenicity on infected chickens.
Keywords: Avian adenovirus, Broilers, Eastern Poland, Genome, Molecular analysis, Sequencing
avian adenovirus, broilers, eastern Poland, genome, molecular analysis, sequencing
1. Introduction
The poultry industry in Poland represents one of the largest segments within Polish industry and is constantly growing. This growth provides several opportunities for disease spread, in some cases in the absence of vaccine control, leading to heavy economic losses for poultry producers. Some of these diseases are caused by adenoviruses which are non-enveloped icosahedral viruses belonging to the genus Aviadenovirus in the family Adenoviridae and consist of linear double-stranded DNA ranging in size from 25-46 kbp (Adair and Smyth, 2008a; Benko et al., 2005). Adenoviruses isolated in poultry are mainly fowl adenoviruses with five species FAdV-A-E represented by twelve types FAdV-1-8a-8b-11 (Harrach and Kajan, 2011). Adenovirus strains are distributed worldwide and different types/species have been discovered in various geographic locations (Niczyporuk, 2016; Schachner et. al., 2014). Almost all adenovirus type/species can induce syndromes or diseases including Inclusion Body Hepatitis (IBH) which is associated with intra-nuclear inclusion bodies in the hepatocytes and liver with haemorrhages and has been linked to type/species: FAdV-2/D, -8a/E, -8b/E, and -11/D (Adair and Smyth, 2008a; Zhao et al., 2015; Maartens et al., 2014; Hess, 2013; Grgić et al., 2011; Niczyporuk et al., 2013; Schachner et al., 2014; Schachner et al., 2018). Members of the biologically more distant type belonged to FAdV-1/A and have been shown to induce Gizzard Erosion (GEU) (Hess, 2013). Additionally, epidemiological studies confirmed type/species FAdV-4/C as an agent of Hydropericardium Hepatitis Syndrome (HPS) (Mazaheri et al., 1998; Schonewille et al., 2008; Niu et al., 2018; Schachner et al., 2018) which can have a huge impact on poultry health contributing to economic losses in the poultry industry worldwide. Currently, Poland has large numbers of positive cases of adenovirus infections in poultry flocks. Commercial vaccines against IBH are not available in Poland. Autogenously FAdV broiler vaccines specific for the exact strain in infected flock have been used in some regions where outbreaks of IBH are occurring.
Molecular characterisation of Polish adenovirus strains is ongoing, however, 1/A, 7/E, and 8a/E type/species have been well characterised (Niczyporuk, 2016, 2017, 2018a, 2018b; Matczuk et al., 2017; Niczyporuk and Czekaj, 2018). It is mentioned in the literature that immunosuppression induces Infectious Bursal Disease Virus (IBDV) or Chicken Anaemia Virus (CAV) (Choi et al., 2012; Eregae et al., 2014), and Marek's Disease virus (MD) (Niczyporuk, 2016) can be possible as a cofactor for IBH cases in chickens (Meng et al., 2018; Su et al., 2018; Toro et al., 2000, 2001). However, recent studies have suggested that in some cases immunosuppressant factors may not be needed for induction of the disease based on IBH in chickens (Morshed et al., 2017).
The main objective of this study was to detect and fully characterise adenovirus strains associated with IBH in an affected flocks, and to improve knowledge about its characteristics in Poland which can led to more effective control measures and vaccine development.
2. Materials and methods
2.1. Chicken samples
The samples were collected in March 2020 from one broiler farm located in the Masovia region in the eastern part of Poland (GPS location: latitude 52°1644, geographic longitude 22,2728 52°9′52″ North, 22°16′22″ East). The affected flocks were kept in an all-in all-out management system. Each affected flock contained approximately 20,000 ROSS 308 broiler chickens. Chickens showed low body mass index and there were several casualties. Sick birds (10 birds per flock) were selected from farms and sent to National Veterinary Research Instituite (NVRI) for anatomopathological, histopathological, and virologic examinations. Birds were euthanised by cervical dislocation and anatomopathological examinations were conducted. Internal organs including the liver, spleen, gizzard, and intestines were collected and stored at -20 °C for DNA virus isolation.
2.2. Adenovirus reference strains
The reference strain, which belonged to the type Grp FAdV-1/A, was obtained from a commercial company (Charles River, US) and was used as a positive control in the detection methods such as: Cytopathic effect (CPE) and PCR studies.
The reference strains with Accession numbers according to the GenBank, 1982 (NCBI) database which were used for analysis were as follows: fowl adenovirus E/6 strain CR119, complete genome (KT862808), fowl adenovirus E/8b strain 764, complete genome (KT862811), fowl adenovirus E/7 strain YR36, complete genome (KT862810), fowl adenovirus E/8a strain TR 59 (KT862810), fowl adenovirus E/8a isolate JSN72/08 hexon gene, partial CDS, fowl adenovirus B/5 strain 340, complete genome (KC493646), fowl adenovirus C/4, complete genome KR5 (HE608152), fowl adenovirus A/1 complete genome (AC_000014), fowl adenovirus A/1 61/11z complete genome (KX247012), fowl adenovirus type D/2, complete genome, fowl adenovirus E/8 hexon gene, partial CDS (KT862811), fowl adenovirus D/2 strain 685, complete genome (KT862805), fowl adenovirus D/9, complete genome (AC_000013), fowl adenovirus D/3 strain SR49 (KT862807), and fowl adenovirus D/11 strain 380 (KT862812) were used as a reference sequences during the molecular analysis.
2.3. Adenovirus field strains
Fowl adenovirus field strains have been isolated from the same farm but form two different flocks. Fowl adenovirus strains: 1-G044/20-L-liver-FAdV-E-MT525091, 2-G044/20-S-spleen-FAdV-E-MT525092, 3-G044/20-G-gizzard-FAdV-E-MT525093, 4-G044/20-I-intestinum-FAdV-E-MT525094, have been isolated from one flock, and fowl adenovirus strains: 5-G045/20-L-liver-FAdV-5/B-MT525095, 6-G045/20-S-spleen-FAdV-E-MT525096, 7-G045/20-G-gizzard-FAdV-E-MT525097, and 8-G045/20-I-intestinum-FAdV-E-MT525098 have been isolated from the second flock.
2.4. Chicken embryo fibroblast (CEF) cultures
CEF cultures were prepared from 9-11-day-old specific-pathogen-free (SPF) chicken embryos (Lohman, Germany) according to the standard protocol. Eagle's growth medium (MEM) was used with additions of 10% foetal bovine serum and 1% antibiotic mixture (Antibiotic–Antimycotic, Gibco, U.K.). The maintenance medium consisted of MEM with 1% of an antibiotic-antimycotic mixture. A monolayer of CEF culture was present after 24 h incubation at 37 °C.
2.5. Histopathology
Liver and spleen samples were collected in a 10% formaldehyde solution for histopathological examination. Samples were fixed in 10% neutral-buffered formalin. The samples were then routinely processed, embedded in paraffin blocks and cut on a microtome at 4 μm. The cut sections were stained using haematoxylin-eosin method (H&E) and examined using light microscopy for the presence of histopathological lesions.
2.6. Virus replication on CEF cultures
Homogenates from internal organs of sick chickens were prepared as 1:1 dilutions in Eagle's medium with an addition of 1% antibiotic mixture (Antibiotic–Antimycotic, Gibco, UK) and then were filtrated through the Millipore's 0.45 μm filter (Minisart, Sartorius, Germany). Filtrated homogenates and lyophilisates were used for CEFs infection. CEF cultures were incubated at 37 °C for five to seven days in an atmosphere of 5% CO2. The appearance of cytopathic effects characteristic of FAdV infection were monitored daily under the microscope. The third passage of each strain was preserved at -20 °C for the next step of the study.
2.7. DNA extraction
Total DNA of the reference adenovirus strain grp FAdV-1 and eight field strains were extracted using the DNA Mini Kit (Qiagen, Germany) according to the manufacturer's recommended procedure. DNA was isolated directly from the CEF cultures infected with the field and reference strains (as a positive control). DNA was also extracted from non-infected CEF cultures as a negative control. DNA samples were then stored at -20 °C for the next step of the study.
2.8. Determination of tissue culture infection doses (TCID50)
TCID50 of field and reference strains were determined on 24 well plates (Thermo Scientific, USA) coated with CEF cultures (18–24 h). CEFs were infected with ten-fold dilutions of virus stocks from 10−1.0 to 10−7.0 in triplicate for each dilution and three wells for negative control. The plates were incubated at 37 °C with 85% humidity in an atmosphere of 5% CO2. CPE appearance was observed under a microscope (Zeiss HXP 120, Germany) on daily basis. After six to seven days of incubation results were read according to the Reed and Muench (1938) model and TCID50 was determined.
2.9. PCR for the identification of the hexon gene of adenovirus strain
PCR for the amplification of FAdV, specific to the L1 loop region of the hexon gene, was performed as described previously (Niczyporuk et al., 2013) with the PCR products of 830 bp in size.
2.10. Sequencing and molecular analysis
The PCR products after the reaction of amplification from the Loop L1 HVR1-4 region of the hexon gene were obtained and purified using NucleoSpin Extract II (Marcherey-Nagel, France) then the sequencing step was performed using the Sanger method by the commercial company, GENOMED (Warsaw, Poland).
Sequence analysis was performed by alignment of the nucleotide sequences of the amplified fragments from the loop L1 HVR1-4 hexon gene, with sequences obtained from the Gene Bank database by the Basic Local Alignment Search Tool (BLASTn) search at the National Centre for Biotechnology Information (Altschul et al., 1990; http://www.ncbi.nlm.nih.gov/nucleotide). The nucleotide sequences were aligned using the Clustal W multiple alignment method, and the sequence distances and the phylogenetic trees were calculated according to the International Committee on Taxonomy of Viruses, ICTV classification. .
The evolutionary distances were computed using the maximum composite likelihood method and are in the units of the number of base substitutions per site. The phylogenetic tree has been created with the sum of branch length = 23.59191946 is shown. Overall, the analyses included the 8 field and 15adenovirus reference sequences. Codon positions included were the first, second, third, and noncoding regions. All positions containing gaps and missing data were eliminated. There were a total of 417 positions in the final dataset. Evolutionary analyses the nt and aa sequence identities were calculated using the MegAlign module of the Clustal W algorithm were conducted using MEGA7.0.26(7170509-x86_64) software.
2.11. Tajima's neutrality test
The statistical analysis was performed on 23 nucleotide sequences of examined strains. Codon positions included were 1st+2nd+3rd + Noncoding. All positions containing gaps and missing data were eliminated. The designated abbreviations were as follows: number of sequences (m), total number of sites (n), number of segregating sites (S), Sin (), (), nucleotide diversity (),and the Tajima's test statistic ().
2.12. Ethics approval and consent to participate
Sick birds were euthanised by cervical dislocation and anatomopathological examinations were conducted. All the samples were collected under the permission in accordance with the local license in accordance with institutional guidelines from our local Bioethics Committee. This study did not include live animals in our experiments.
3. Results
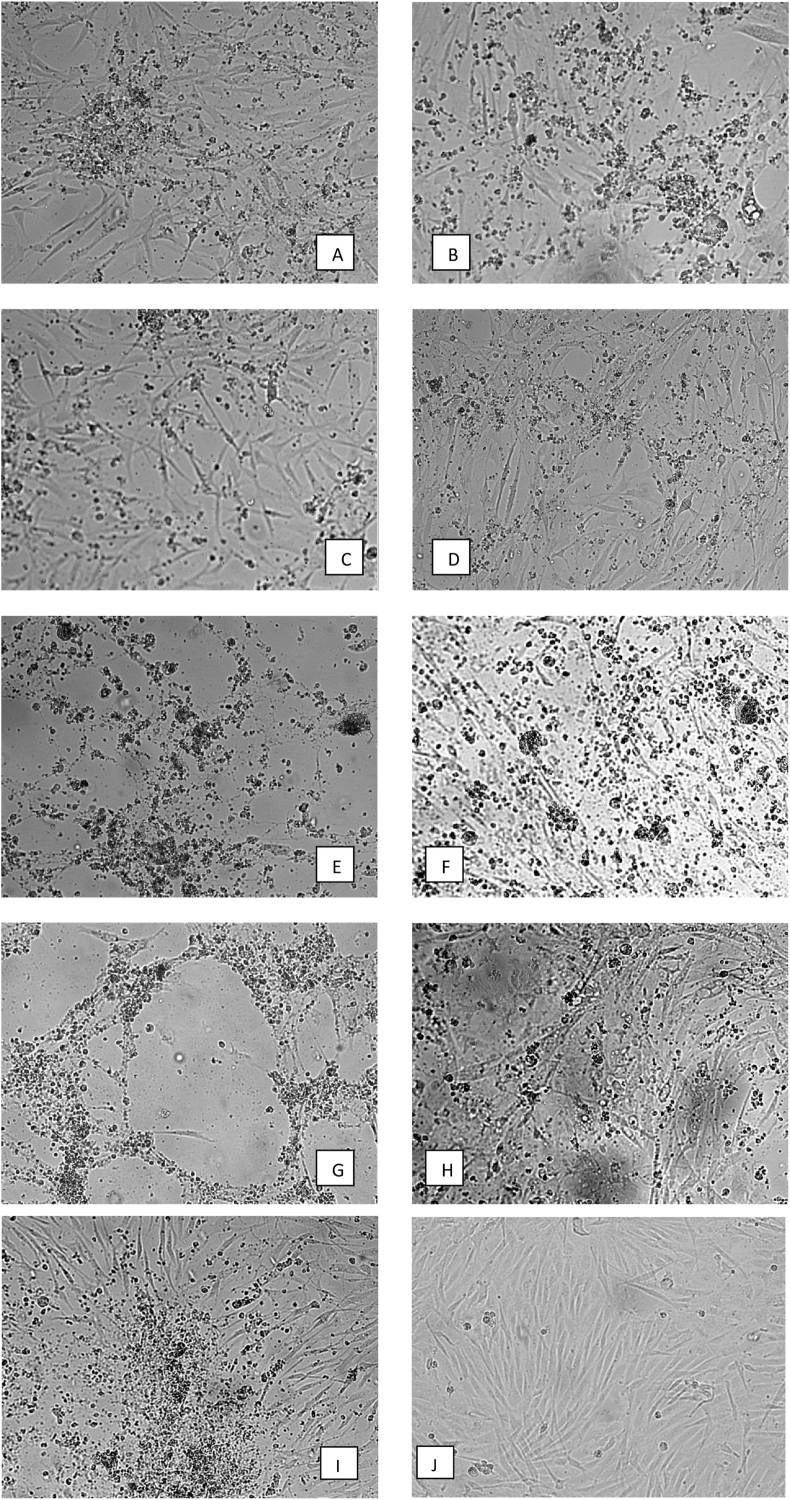
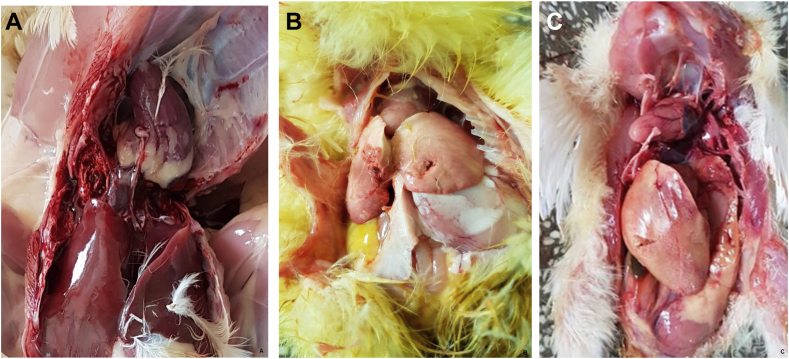
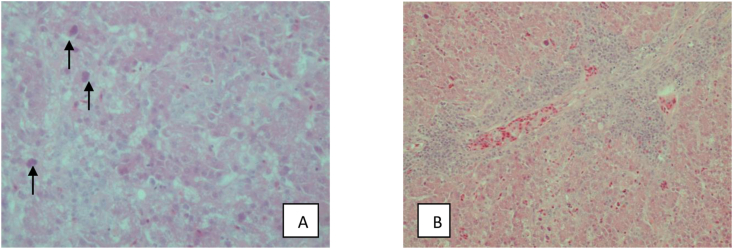
The GPS location of examined flocks have been indicated. Infected chickens were characterised by a rapid disease, diarrhea, lethargy, sudden onset of decreased appetite, increased mortality, and hepatitis. Eight adenovirus strains were successfully cultivated on CEF cultures. The infected cultures cytopathic effects were observed that were consistent with FAdV infection Figure 1(A-H). TCID50 have been determined. The TCID50 which have been calculated make maximum use of the information in the replicate data of the examined virus strains. The eight field isolates all grew to an end-point titer of 104.5TCID50/0.1ml. Positive and negative control have been presented in Figure 1(I-J). Liver and spleen samples collected from the two affected poultry flocks were examined for macroscopic lesions. Liver characteristics for IBH infection with intranuclear inclusion bodies have been observed and presented (Figure 2(A-C). During the histopathological examinations, lesions which could be caused by IBH were noted. Multifocal areas of cytoplasmic vacuolisation of the hepatocytes were indicated (evident, showing reticulated nuclei with pyknosis and karyorrhexis - necrosis). The presence of mononuclear cell infiltrates with abundant lymphoid cells in various degrees of maturation were also noteworthy among the vacuolisation areas. In addition, while observing these areas, some large hepatocyte nuclei with strong basophilic content (intranuclear basophilic inclusion bodies) were found and have been presented (Figure 3A), and lymphoid cell infiltration (inclusion bodies) in spleen were confirmed (Figure 3B).
Figure 1.
Cytopathic effect of eight field adenovirus strains: A – G044/20-L-liver-FAdV-E-MT525091-E, B-G044/20-S-spleen-FAdV-E–MT525092-E, C-G044/20-G-gizzard-FAdV-E- MT525093-E, D-G044/20-I-intestinum-FAdV-E-MT525094-E, E-G045/20-L-liver-FAdV-E-MT525095-5/B, F-G045/20-S-spleen-FAdV-E-MT525096-E, G-G045/20-G-gizzard-FAdV-E-MT525097-E, H-G045/20-I-intestunum-FAdV-E-MT525098-5/E, I – FAdV-5/B Charles River, US. Cytopathic effect of -examined strains IIIp, 96 hpi.(20x magnification) J – Uninfected CEFs, K – negative control.
Figure 2.
A-No pathological changes have been obseved, control tissues -B-C Post-mortem lesions in the liver from two broiler flocks after 21 d being affected by adenoviruses.
Figure 3.
Fatty degeneration and basophilic intranuclear inclusions (arrow) in liver (H&E, bar 2μm) × 40. B – Spleen. Lymphoid cell infiltration, inclusion bodies, HE × 20.
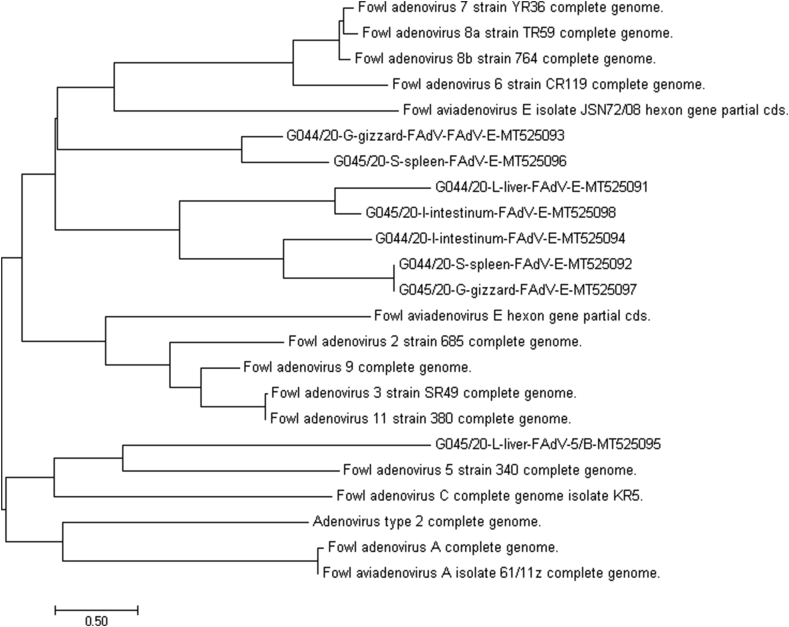
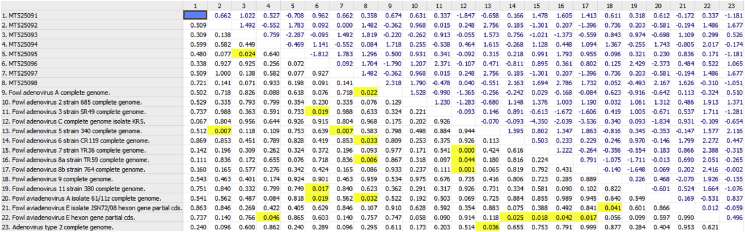
The pair-wise identities of the examined strains hexon gene sequences were 90.55%, as was expected for strains which are representing the same adenovirus species (6, 7, 8a, and 8b/E) and formed the same independent cluster. Strains belonged to this cluster have been indicated as: 3-G044/20-G-gizzard-FAdV-E-MT525093, 6-G045/20-S-spleen-FAdV-E-MT525096, 1-G044/20-L-liver-FAdV-E-MT525091, 8-G045/20-I-intestinum-FAdV-E-MT525098, 4-G044/20-I-intestinum-FAdV-E-MT525094, 2-G044/20-S-spleen-FAdV-E-MT525092, and 7-G045/20-G-gizzard-FAdV-E-MT525097. The most diverse strain was the strain with accession number 5-G045/20-L-liver- FAdV-5/B-MT525095-, which was isolated from the liver of infected chicken, and was closely related with type/species 5/B with 99.12% pair-wise identity of hexon gene and formed a single cluster which is formed in close relation with species A, C, and D. The third cluster consisted of five reference sequences closely related with type/species 2, 3, 9, and 11/D, and species E with 91.12% of identity. Details are presented in Figure 4 with evaluation of pair-wise distance. The phylogenetic tree was based on nt and aa sequence analysis. Among the eight field adenovirus sequences, the nt and aa sequences showed similarities. Sequences obtained from field adenovirus strains shared similarities with the reference strain sequences which are presented in phylogenetic relations/tree in Figure 5.
Figure 4.
Pair wise distance with overall mean distance.
Figure 5.
Phylogenetic tree. The phylogenetic was based on derived amino acid sequences of the hexon gene. Adenovirus strains isolated from chicken flocks are represented using the designations: G-044/20-G-gizzard-FAdV-E-MT525093, G045/20-S-spleen-FAdV-E-MT525096, G044/20-L-liver-FAdV-E-MT525091, G045/20-I-intestinum-FAdV-E-MT525098, G044/20-I-intestinum-FAdV-E-MT525094, G044/20-S-spleen-FAdV-E-MT5250932, G045/20-G-gizzard-FAdV-E-MT525097, G045/20-L-liver-FAdV-5/B-MT525095. The tree was rooted by the 13 reference sequences (Fowl adenovirus 7 strain YR36, Fowl adenovirus 8a strain TR59, Fowl adenovirus 8b strain 764, Fowl adenovirus 6 strain CR119, Fowl adenovirus E, Fowl adenovirus 2 strain 685, Fowl adenovirus 9, Fowl adenovirus 3 strain SR49, Fowl adenovirus 11 strain 380, Fowl adenovirus 5 strain 340, Fowl adenovirus C KR5 strain, Adenovirus type 2, Fowl adenovirus A, and two sequences A-61/11z and Fowl adenovirus E JSN72/08 strains obtained from database GenBank (NCBI).
Each entry shows the probability of substitution (r) from one base (row) to another base (column). For simplicity, the sum of (r) values was made equal to 100. Rates of different transitional substitutions are shown in table in bold and those of transversional substitutions are shown in italics. The nucleotide frequencies were 22.98% (A), 23.30% (T/U), 25.43% (C), and 28.29% (G). The transition/transversion rate ratios were K1 = 3.368 (purines) and k2 = 1.562 (pyrimidines). The overall transition/transversion bias was R = 1.247, where R = (A∗G∗k1 + T∗ C∗K2)/(A + G)∗(T + C). The analysis involved 23 nucleotide sequences. Codon positions included were the first, second, third, and noncoding regions. All positions containing gaps and missing data were eliminated. The results of Maximum Composite Likelihood Estimate of the Pattern of Nucleotide Substitution are presented in Table 1, and heterogeneity of examined adenovirus strain sequences have been indicated in yellow Figure 6.Concerning the data from Tajima's Neutrality Test, there were a total of 417 positions in the final dataset, and the results which have been obtained were presented in Table 2.
Table 1.
Maximum Composite Likelihood Estimate of the Pattern of Nucleotide Substitution indicated different nucleotide sequences of examined strains.
| A | T | C | G | |
|---|---|---|---|---|
| A | - | 5.19 | 5.67 | 21.23 |
| T | 5.12 | - | 8.85 | 6.3 |
| C | 5.12 | 8.11 | - | 6.3 |
| G | 17.25 | 5.19 | 5.67 | - |
Figure 6.
Heterogeneity of examined adenovirus strain sequences.
Table 2.
Tajima's neutrality test.
| m | S | ps | ø | π | D |
|---|---|---|---|---|---|
| 23 | 417 | 1.000000 | 0.270943 | 0.715605 | 6.678223 |
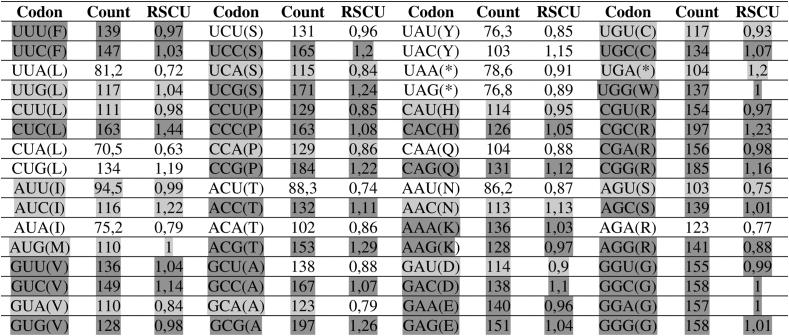
Relative synonymous codon usage (RSCU) which represents the ratio of the observed frequency of codons to the expected frequency given that all the synonymous codons for the same amino acids are used equally. RSCU values have been widely utilised to measure the gene expression level Halder et al., 2017. Analysis of the synonymous codon usage in the hexon gene region revealed differences in this region depending on the strain and adenovirus type/species. The results are presented in Figure 7.
Figure 7.
Analysis of the number of successive codons and relative synonymous codon usage. All frequencies are given in percentages.The color intensity indicates how highly an amino acid is preferred in a particular position amongst each strains Darker shading indicates a higher RSCU value.
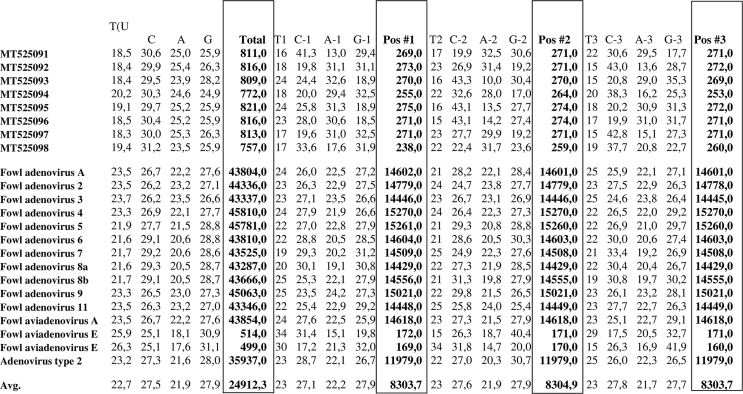
The codon usage in the loop L1 region of the hexon gene sequences from field and reference strains were examined, and it was found that C (cytosine) was the most frequent nucleotide for each examined adenovirus type/species ranging from 29.5% to 31.2% compared with reference strain sequences which ranged from 25.1% to 29.3%. G (guanine) appeared most often in the first position of the codon in all examined sequences, and the percentages were estimated to be between 18.5% to 32.5%. In the second position of the codon, C (cytosine) appeared most and values were estimated between 19.9% and 43.3% whereas these values in the reference strain sequences were between 24.7% and 31.8%. A (adenine) appeared most often (13.0%–32.6%) in the first position of the codon. C (cytosine) was most frequent in the third position of the codon in each examined field strain sequence and the values for cytosine were between 19.9% and 43.0%, and 17.5%–33.4% in the reference sequences. Full results which were obtained for adenovirus field type/species compared to other reference strain sequences are presented in Figure 8.
Figure 8.
Analysis of nucleotide codon composition of the eight examined sequences and 15 sequences of adenovirus strains derived from database GenBank (NCBI). Total-number of nucleotides of tested strain sequences, Pos#1, Pos#2, Pos#3- number of nucleotides in the sequences tested in the first, second, and third codon positions, respectively.
4. Discussion
Characterisation of virus strains were performed by indication of the CPE on CEF cultures, and determination of TCID50. Our investigation included data from two broiler flocks on which two adenovirus species B and E have been confirmed. This manuscript describes a novel fowl adenovirus strain isolated from infected poultry flocks. The hexon gene of isolated and described strains presented differences in the examined sequences, however the examined strains still had a high degree of identity (90.55%).
As part of the analysis a population genetic statistic test (Tajima's) was performed (Tajima, 1989). Analysis results could distinguish between a DNA sequence evolving randomly (neutrally) or evolving under a non-random process. It could also indicate directional selection, balancing selection, demographic expansion, contraction, genetic hitchhiking, or introgression. During the analyses, the obtained DNA sequences indicated mutations with no effect suggesting they were not important (neutral) on the evolution of the strains. However, mutations formed under selection pressure (non-neutral) were identified. For example, mutations which can have an impact on virulence of the examined strains or indicate genetic drift. Detailed data are presented in Table 2.
Fowl adenovirus strains were isolated and described in detail based on nucleotide sequence analyses of the hexon gene loop 1 for the first time in Morocco (Redondo et al., 2018; Abghour et al., 2019). Strains were identified as fowl adenovirus closely related to type 11 and 8a, which represented species E like strains isolated nowadays in Poland. During recent years, adenovirus strains identified as species E types 11 and 8b, and D have been identified, mostly in Iran, with high prevalence and are pathogenic enough to cause IBH in young chickens (Morshed et al., 2017). This same epidemiological situation was observed in Spain where the most redundant adenovirus type/species which are responsible for IBH infections in broilers are strains identified as type/species 8b/E, 7/E, and -11/D, respectively. In previous studies on IBH infections in broilers, the disease also appeared to have been induced by type/species 8b/E, 7/E or 11/D, and have been reported by many research groups (Zhao et al., 2015; Ojkic et al., 2008; Steer et al., 2011; Choi et al., 2012; Kaján et al., 2013; Niczyporuk, 2017). In examining Chinese studies, only 1 of the 10 isolated strains belonged to type 8b/E, whereas others strains have been represented, type-4/C, indicating that in Chinese flocks FAdVs were mainly represented by type/species 4/C which is responsible for HP infection and the phylogenetic analysis indicated that sequences of the hexon regions of the examined strains were clustered into three distinct types: FAdV-4 (79.4%, 123/155), FAdV-8a (13.5%, 21/155) and FAdV-8b (3.9%, 6/155), of which FAdV-4 was the dominant type in China (Chen et al., 2019). The reported FAdV-8b strain is fatal to chicken embryos and possesses horizontal transmission capacity in chickens (Cui et al., 2020).
Based on the growing existing FAdV genome data in the NCBI database, topological switches were investigated in the phylogenetic tree of the hexon gene Loop L1 region HVR1-4, providing evidence for recombination and its systematic occurrence in FAdVs (two type/species in one flock). The discovery of recombination could be interesting, and newly isolated field strains with mutations/evolutions in the hexon Loop L1 HVR1-4 region will be possible. These results suggest that circulating FAdVs, mainly from species FAdV-B and E, can be genetically more diverse than previously indicated in Poland (Niczyporuk, 2016, 2017, 2018a, 2018b).
Eight characteristic sites have been located in the Loop L1 region, and one in the L2 region of the hexon gene to determine whether the complete hexon gene was under positive selection pressure or not. Analysis was performed by Niu et al, 2018) to identify specific codons that underwent evolutionary selection. During the study, 7 positively selected codons in L1 (aa 164, 168, 195, and 243) and L2 (aa 379, 402, and 408) were found. The other 2 sites (aa 140 and 680) were located in regions P1 and P2 (regions not focused on in our study). The results reveal that hexon loops L1 and L2 are hypervariable regions (Niu et al., 2018; Niczyporuk, 2018a, 2018b). Determinations of the relative synonymous codon usage values for the sequences Loop L1 of hexon gene FAdVs type/species strains have been calculated in Poland (Niczyporuk, 2017, 2018a, 2018b), and indicated a significant influence on the evaluation of genetic pressure and adenovirus virulence. Additionally, these indicated significant differences in preference codon presence in RSCU values obtained for Leucine (L) and Valine (V) codons compared to all examined FAdVs type/species can be significant for higher expression of the examined gene. The analysis which was performed by Niczyporuk, 2018a, and 2018b indicated the antigenic properties in sequences of examined adenovirus strains. The indication of relative synonymous codon values and codon mutation designations leads to better understand of the mechanisms of pathogenicity of FAdVs strains. During the molecular/interspecies studies conducted by Niczyporuk, 2017, 2018a, and 2018b, it was indicated that designation/indication of specific conservative sequence GQMT in examined sequences of genomes of FAdVs strains and the constant and value of survive without mutations in sequence existed/determined in adenovirus bird strain sequences indicated that the sequence GQMT in Loop L1 HVR1-4 region of the hexon gene can be responsible for important functions and influence the neutralizing antibody production during infection in birds and can help improve. The Loop L1 region of the hexon gene has been described as a region that varies between different types (Meulemans et al., 2004; Steer et al., 2011; Niczyporuk, 2018a, 2018b) but the types and genotypes overlap only partially (Marek et al., 2010). The present study is concerned with the first recorded presence of FAdV in two species of adenovirus strains B and E in correlation between which have been isolated and analysed on the sequence of loop L1 HVR1-4 of the hexon gene, the most variable region in the adenovirus genome, and to better understanding of the antigenic properties of adenovirus receptors.
Research on the genomic structure of adenoviruses is extremely valuable and constantly improves our understanding. So far it is known that non-pathogenic fowladenoviruses (FAdVs) are amenable for engineering multivalent vaccine platforms due to large stretches of nonessential DNA sequences in their genomes (Corredor et al., 2017).
In studies conducted by (Niu et al, 2018), molecular analysis on selection pressure revealed that the capsid protein of the hexon gene had undergone positive selection, although the evolution of the major capsid protein is steady with only some mutation. However, the future acquisition of additional mutations could lead us to deduce and perform studies on antigenic drift which could cause further outbreaks. Many researchers suggest that future studies on the evolution of adenovirus strains are still needed. Research conducted by (Niu et al, 2018) indicated that polyclonal antibodies, based on hexon regions loops L1 and L2, could be used as a detection of antibody in IHC analysis to elucidate the distribution of the virus in target organs, especially the liver (Niu et al., 2018). Wang, 2019 indicated that phylogenetic analysis of five adenovirus strains belonged to type/species 4/C and indicated an 11 amino-acid deletion in ORF29 which was related to older viral strains and during in vivo studies on chickens indicated lesions characteristic for HP-IBH.
The viral loads were recorded in all the organs after infection and displayed significantly different tissue tropism and cytokine profiles. The full genome of 8b/E was 44,454 nucleotides in length with 58.1% G and C content. No homologous regions to early regions E1, E3, and E4 of mastadenoviruses were identified, and were very similar to the typical organisation of FAdV-E genomes (Huang et al., 2019). Matczuk et al., 2017 indicated than in the type/species 1/A adenovirus strains presented point mutations in the genome sequences of the pathogenic adenovirus strains (1/A-W-15 and 1/A-61/11z) using next generation sequencing. Some non-structural proteins, ORF, ORF1, ORF14, E2b polymerase and IVa 2, as well as structural proteins, hexon IIIa, penton and fibre 2, were related to the genome 1/A reference sequence. Matczuk et al., 2017 also indicated the ORF14 protein of unknown function had accumulated the highest number of amino acid point mutations in 1/A-W-15 and 1/A-61/11z strains, which might suggest a high evolutionary selection pressure on this viral protein, and also indicates the deletion in the codon coding for proline (P) in positions 2 and 53 in the sequence of the adenovirus strain 1/A-61/11z and in position 3 in the sequence of the adenovirus strain 1/A-W-15. The indicated mutation has a significant impact on gene expression in pathogenic 1/A strains.
5. Conclusion
The research presented in the above manuscript is a continuation of the research being conducted on FAdV strains isolated in Poland on type/species 1/A, 7/E, and 8a/E. It can be clearly seen that in the timeframe from 2018-2020, over 94% of isolated strains are strains representing species D and E. Therefore, detailed analyses related to these species are needed to better understand their variability. Following the worldwide literature, it can be deduced that currently these species dominate in Europe, Australia and the Middle East, while in China the species C representing type 4 is definitely still the dominant species.
Declarations
Author contribution statement
Jowita Samanta Niczyporuk: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Wojciech Kozdrun, Hanna Czekaj, Karolina Piekarska, Natalia Stys-Fijol: Contributed reagents, materials, analysis tools or data.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgments
National Veterinary Research Institute, NVRI has sponsored the edition of this manuscript.
References
- Abghour S., Zro K., Mouahid M., Tahiri F., Tarta M., Berrada B., Kichou F. Isolation and characterization of fowl Aviadenovirus serotype 11 from chickens with inclusion body hepatitis in Morocco. PloS One. 2019;12 doi: 10.1371/journal.pone.0227004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adair B.M., Smyth J.A. Group I adenovirus infections. In: Saif Y.M., Fadly A.M., Glisson J.R., McDougald L.R., Nolan L.K., Swayne D.E., editors. Diseases of Poultry. twelfth ed. Iowa State University Press; Iowa, USA: 2008. pp. 252–266. [Google Scholar]
- Altschul S.F. Basic local alignment search tool. J. Molec. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Benko M., Harrach B., Both G., Russel W., Adair B., Adam E., Jong J., Hess M., Johnson M., Kajan A., Kidd A.H., Lehmkuhl H.D., Li Q.G., Mautner V., Pring- Akerblom P., Wadell G. Family Adenoviridae. In: Fauquet C., Mayo M., Maniloff J., Desselberger U., Ball L., editors. Virus Taxonomy. Eighth Report of the International Committee on the Taxonomy of Viruses. Elsevier; New York: 2005. pp. 213–228. [Google Scholar]
- Chen L., Yin L., Zhou Q., Peng P., Du Y., Liu L., Zhang Y., Xue Ch., Cao Y. Epidemiological investigation of fowl adenovirus infections in poultry in China during 2015-2018. BMC Vet. Res. 2019;15:271. doi: 10.1186/s12917-019-1969-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K.S., Kye S.J., Kim J.Y., Jeon W.J., Lee E.K., Park K.Y., Sung H.W. Epidemiological investigation of outbreaks of fowl adenovirus infection in commercial chickens in Korea. Poultry Sci. 2012;91:2502–2506. doi: 10.3382/ps.2012-02296. [DOI] [PubMed] [Google Scholar]
- Corredor J.C., Pei Y., Nagy E. Fowl adenovirus-based vaccine platform. Recomb. Virus. 2017;1581:29–54. doi: 10.1007/978-1-4939-6869-5_3. [DOI] [PubMed] [Google Scholar]
- Cui J., Xu Y., Zhou Z., Xu Q., Wang J., Xiao Y., Li Z., Bi D. Pathogenicity and molecular typing of fowl adenovirus-associated with hepatitis/hydropericardium syndrome in Central China (2015-2018) Front. Vet. Sci. 2020;28:7–19. doi: 10.3389/fvets.2020.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eregae M.E., Dewey C.E., McEwen S.A., Ouckama R., Ojkić D., Guerin M.T. Flock prevalence of exposure to avian adeno-associated virus, chicken anemia virus, fowl adenovirus, and infectious bursal disease virus among Ontario broiler chicken flocks. Avian Dis. 2014 Mar;58(1):71–77. doi: 10.1637/10612-071113-Reg.1. [DOI] [PubMed] [Google Scholar]
- GenBank . National Library of medicine (US), National Center for Biotechnology Information; Bethesda (MD): 1982. http://www.ncbi.nlm.nih.gov/nucleotide [Google Scholar]
- Grgić H., Yang D.H., Nagy E. Pathogenicity and complete genome sequence of a fowl adenovirus serotype 8 isolate. Virus Res. 2011;156(1-2):91–97. doi: 10.1016/j.virusres.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Halder B., Kumar Malakar A., Chakraborty S. Nucleotide composition determines the role of translational efficiency in human genes. Bioinformation. 2017;13(2):46–53. doi: 10.6026/97320630013046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrach B., Kajan G. Aviadenovirus. In: Darai G., Konle S., Simniok J., editors. Aviadenoviridae. Springer; Berlin: 2011. pp. 13–28. 4. Niczyporuk JS. Phylogenetic and geographic analysis of fowl adenovirus field strains isolated from poultry in Poland. Archioves of Virol 2015. [Google Scholar]
- Hess M. Aviadenovirus infections. In: Swayne D.E., Glisson J.R., McDougald L.R., Nolan L.K., Suarez D.L., Nair V., editors. Diseases of Poultry. thirteenth ed. Wiley-Blackwell; Ames: 2013. pp. 290–300. [Google Scholar]
- Huang Q., Ma X., Huang X., Huang Y., Yang S., Zhang L., Cui N., Xu C. Pathogenicity and complete genome sequence of a fowl adenovirus serotype 8b isolate from China. Poultry Sci. 2019;98:573–580. doi: 10.3382/ps/pey425. [DOI] [PubMed] [Google Scholar]
- Kaján G.L., Kecskeméti S., Harrach B., Benkő M. Molecular typing of fowl adenoviruses, isolated in Hungary recently, reveals high diversity. Vet. Microbiol. 2013 Dec 27;167(3- 4):357–363. doi: 10.1016/j.vetmic.2013.09.025. Epub 2013 Oct 1. [DOI] [PubMed] [Google Scholar]
- Maartens L.H., Hilda W., Aitchison J.H., Estelle H. Venter 3 Inclusion body hepatitis associated with an outbreak of fowl adenovirus type 2 and type 8b in broiler flocks in South Africa. J. S. Afr. Vet. Assoc. 2014;85(1):5. doi: 10.4102/jsava.v85i1.1146. Art. #1146. [DOI] [PubMed] [Google Scholar]
- Marek A., Gunes E., Schulz M. Hess. Classification of fowl adenoviruses by use of phylogenetic analysis and high-resolution melting-curve analysis of the hexon L1 gene region. J. Virol. Methods. 2010;170:147–154. doi: 10.1016/j.jviromet.2010.09.019. [DOI] [PubMed] [Google Scholar]
- Matczuk A., Niczyporuk J.S., Kuczkowski M., Wozniakowski G., Nowak M., Wieliczko A. Whole genome sequencing of Fowl aviadenovirus A - a causative agent of gizzard erosion and ulceration, in adult laying hens. Infect. Genet. Evol. 2017;48:47–53. doi: 10.1016/j.meegid.2016.12.008. [DOI] [PubMed] [Google Scholar]
- Mazaheri A., Prusas C., Voss Hess M. Somestrains of serotype 4 fowl adenoviruses cause inclusionbody hepatitis and hydropericardium syndrome in chick- ens. Avian Pathol. 1998;27:269–276. doi: 10.1080/03079459808419335. [DOI] [PubMed] [Google Scholar]
- Meng G., Dong Y., Zhang S., Tian Z., Cui S., Chang P.Zhao. Co-infection of fowl adenovirus with different immunosuppressive viruses in a chicken flock. Poult. Sci. 2018;97:1699–1705. doi: 10.3382/ps/pex414. [DOI] [PubMed] [Google Scholar]
- Meulemans G., Couvreur B., Decaesstecker M., Boschmans M., van den Berg T.P. Phylogenetic analysis of fowl adenoviruses. Avian Pathol. 2004;33:164–170. doi: 10.1080/03079450310001652086. [DOI] [PubMed] [Google Scholar]
- Morshed R., Hosseini H., Langeroudi A.G., Bozorgmehri M.H.F., Charkhkar S. Fowl adenoviruses D and E cause inclusion body hepatitis outbreaks in broiler and broiler breeder pullet flocks. Avian Dis. 2017;61:205–210. doi: 10.1637/11551-120516-Reg.1. [DOI] [PubMed] [Google Scholar]
- Niczyporuk J.S. Phylogenetic and geographic analysis of fowl adenovirus field strains isolated from poultry in Poland. Arch. Virol. Jan. 2016;161:33–42. doi: 10.1007/s00705-015-2635-4. [DOI] [PubMed] [Google Scholar]
- Niczyporuk J.S. Molecular characterisation of fowl adenovirus type 7 isolated from poultry associated with inclusion body hepatitis in Poland. Arch. Virol. 2017;162:1325–1333. doi: 10.1007/s00705-017-3240-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niczyporuk J.S. Deep analysis of Loop L1 HVRs1-4 region of the hexon gene of adenovirus field strains isolated in Poland. PloS One. 2018;27:1–14. doi: 10.1371/journal.pone.0207668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niczyporuk J.S. A complete analysis of Relative Synonymous Codon Usage in HVRs1-4 region in adenovirus genome. Pol. J. Vet. Sci. 2018;21:459–468. doi: 10.24425/122619. [DOI] [PubMed] [Google Scholar]
- Niczyporuk J.S., Czekaj H. A comparative pathogenicity analysis of two adenovirus strains, 1/A and 8a/E, isolated from poultry in Poland. Arch. Virol. 2018;163:3005–3013. doi: 10.1007/s00705-018-3965-9. [DOI] [PubMed] [Google Scholar]
- Niczyporuk J.S., Samorek-Salamonowicz E., Czekaj H. Analysis of adenovirusstrains isolated from poultry in Poland. Bull. Vet. Inst. Pulawy. 2013;57:305–310. [Google Scholar]
- Niu Y., Sun Q., Zhang G., Sun W., Liu X., Xiao Y., Shang Y., Liu S. Epidemiological investigation of outbreaks of fowl adenovirus infections in commercial chickens in China. Transbound Emerg. Dis. 2018 Feb;65(1):e121–e126. doi: 10.1111/tbed.12691. [DOI] [PubMed] [Google Scholar]
- Ojkic D., Martin E., Swinton J., Vaillancourt J.P., Boulianne M., Gomis S. Genotyping of Canadian isolates of fowl adenoviruses Avian. For. Pathol. 2008 Feb;37(1):95–100. doi: 10.1080/03079450701805324. [DOI] [PubMed] [Google Scholar]
- Redondo H., Fragoso J.S., Tahala M.A., Bensassi Y., Gil I., Elbachir E., Rodrıguez M.J., Moreno J.C.A. Characterization of strain of fowl adenoviruses circulating in Morocco. Poult. Sci. Assoc. 2018;97:4057–4062. doi: 10.3382/ps/pey271. [DOI] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 1938;27:493–497. [Google Scholar]
- Schachner A., Marek A., Jaskulska B., Bilic I., Hess M. Recombinant FAdV-4 fiber-2 protein protectschickens against hepatitis-hydropericardium syndrome(HHS) Vaccine. 2014;32:1086–1092. doi: 10.1016/j.vaccine.2013.12.056. [DOI] [PubMed] [Google Scholar]
- Schachner A., Matos M., Grafl B., Hess M. Fowl adenovirus-induced diseases and strategies for their control - a review on the current global situation. Avian Pathol. 2018 Apr;47(2):111–126. doi: 10.1080/03079457.2017.1385724. [DOI] [PubMed] [Google Scholar]
- Schonewille E., Singh A., Goebel T.W., Gerner W., Saalmueller A., Hess M. Fowl adenovirus (FAdV) serotype 4 causes depletion of B and T cells in lymphoid organs in specific pathogen-free chickens following experimental infection. Vet. Immunol. Immunopathol. 2008;121(1-2):130–139. doi: 10.1016/j.vetimm.2007.09.017. [DOI] [PubMed] [Google Scholar]
- Steer P.A., O'Rourke D., Ghorashi S.A., Noormohammadi A.H. Application of high- resolution melting curve analysis for typing of fowl adenoviruses in field cases of inclusion body hepatitis. Aust. Vet. J. 2011;89:184–192. doi: 10.1111/j.1751-0813.2011.00695.x. [DOI] [PubMed] [Google Scholar]
- Su Q., Meng F., Li Y., Zhao P., Zhang Y., Chang S., Zhao P. Newcastle disease virus- attenuated vaccine co-contaminated with fowl adenovirus and chicken infectious anemia virus results in inclusion body hepatitis-hydropericardium syndrome in poultry. Vet. Microbiol. 2018;218:52–59. doi: 10.1016/j.vetmic.2018.03.019. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro H., Gonzalez C., Cerda L., Hess M., Reyes E., Geissea C. Chicken anemia virus and fowl adenoviruses: association to induce the inclusion body hepatitis/hydropericardium syndrome. Avian Dis. 2000;44:51–58. [PubMed] [Google Scholar]
- Toro H., Gonzalez O., Escobar C., Cerda L., Morales M.A., Gonzalez C. Vertical induction of the inclusion body hepatitis/hydropericardium syndrome with fowl adenovirus and chicken anemia virus. Avian Dis. 2001;45:215–222. [PubMed] [Google Scholar]
- Wang Kai, Haiwei Sun, Yunzhang Li, Zhiwei Yang, Jianqiang Ye, Hongjun Chen. Characterization and pathogenicity of fowladenovirus serotype 4 isolated fromeastern China. MC Vet. Res. 2019;15:2–10. doi: 10.1186/s12917-019-2092-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Qi Zhong., Ye Zhao., Yan-xin Hu., Guo-zhong, Zhang Pathogenicity and complete GenomeCharacterization of fowl AdenovirusesIsolated from chickens associated withInclusion body hepatitis andHydropericardium syndrome in China. PloS One. 2015;10(7) doi: 10.1371/journal.pone.0133073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supplementary material/referenced in article.