Abstract
Introduction
The LumiraDx severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antigen test, which uses a high-sensitivity, microfluidic immunoassay to detect the nucleocapsid protein of SARS-CoV-2, was evaluated for diagnosing acute coronavirus disease 2019 (COVID-19) in adults and children across point-of-care settings (NCT04557046).
Methods
Two paired anterior nasal swabs or two paired nasopharyngeal swabs were collected from each participant. Swabs were tested by the LumiraDx SARS-CoV-2 antigen test and compared with real-time polymerase chain reaction (rt-PCR; Roche cobas 6800 platform). Sensitivity, specificity and likelihood ratios were calculated. Results were stratified on the basis of gender, age, duration of symptoms, and rt-PCR cycle threshold.
Results
Out of the 512 participants, aged 0–90 years, of this prospective validation study, 414 (81%) were symptomatic for COVID-19 and 123 (24%) swabs were positive for SARS-CoV-2 based on rt-PCR testing. Compared with rt-PCR, the 12-min nasal swab test had 97.6% sensitivity and 96.6% specificity, and nasopharyngeal swab had 97.5% sensitivity and 97.7% specificity, within 12 days of symptom onset, representing the period of infectivity. All (100%) samples detected within 33 rt-PCR cycles were also identified using the antigen test. Results were consistent across age and gender. The user error rate of the test system when used by minimally trained operators was 0.7% (95% confidence interval [CI] 0.1–3.7%).
Conclusion
The rapid, high-sensitivity assay using nasopharyngeal or anterior nasal sampling may offer significant improvements for diagnosing acute SARS-CoV-2 infection in clinic- and community-based settings.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40121-021-00413-x.
Keywords: COVID-19, LumiraDx antigen test, Rt-PCR, SARS-CoV-2, Sensitivity
Key Summary Points
| Why carry out this study? |
| There is an urgent need to improve access to point-of-care testing for SARS-CoV-2 during the COVID-19 pandemic. |
| The LumiraDx SARS-CoV-2 antigen test, which uses a high-sensitivity, microfluidic immunoassay to detect the nucleocapsid protein of SARS-CoV-2, was evaluated for diagnosing acute COVID-19 in adults and children across point-of-care settings. |
| What was learned from the study? |
| A 12-min nasal swab test detects 97.6% of COVID-19 infections, compared to gold standard real-time PCR testing, up to 12 days following symptom onset using a microfluidic immunoassay for SARS-CoV-2 nucleocapsid protein. |
| This rapid assay with high sensitivity and anterior nasal sampling offers significant advantages for identification and management of SARS-CoV-2 infection, particularly in clinic- and community-based settings. |
Digital Features
This article is published with digital features, including a summary slide, to facilitate understanding of the article. To view digital features for this article go to https://doi.org/10.6084/m9.figshare.13678606.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in Wuhan, China in December 2019 and rapidly spread across the world [1, 2]. As of November 2020, the World Health Organization (WHO) had reported over 55 million confirmed cases and over 1.3 million deaths [3]. However, as a result of challenges with employing diagnostic testing, the reported numbers may significantly underestimate the global burden of SARS-CoV-2 [4].
Until now, laboratory testing has focused on detecting sequences of the SARS-CoV-2 RNA genome from a nasopharyngeal swab, but this approach has several major limitations. First, laboratory testing remains laborious and expensive, which may limit access for underserved and vulnerable populations. Second, a slow turnaround time for receiving laboratory-based results may delay a person’s ability to self-isolate and prevent transmission [5, 6]. Third, existing diagnostic laboratories have had limited supply of molecular reagents and low- and middle-income countries have limited capacity to scale up nucleic acid testing, to meet the needs of their communities [7]. Whilst laboratory-based testing has the advantage of greater throughput, there remains an urgent need for rapid point-of-care (POC) diagnostic testing of acute SARS-CoV-2 infection in clinic- and community-based settings [8].
SARS-CoV-2 has four major structural proteins, including nucleocapsid (N), spike (S), membrane (M), and small envelope (E). The N protein, which is highly phosphorylated, interacts with SARS-CoV-2 RNA and makes up the viral core and nucleocapsid [9]. The N protein is a highly conserved target of the SARS-CoV-2 nucleocapsid, and therefore allows for reliable detection and quantitation. Our objective was to evaluate a rapid, high-sensitivity immunoassay to detect the N protein of SARS-CoV-2 for use in clinic- and community-based acute coronavirus disease 2019 (COVID-19) testing programs [10].
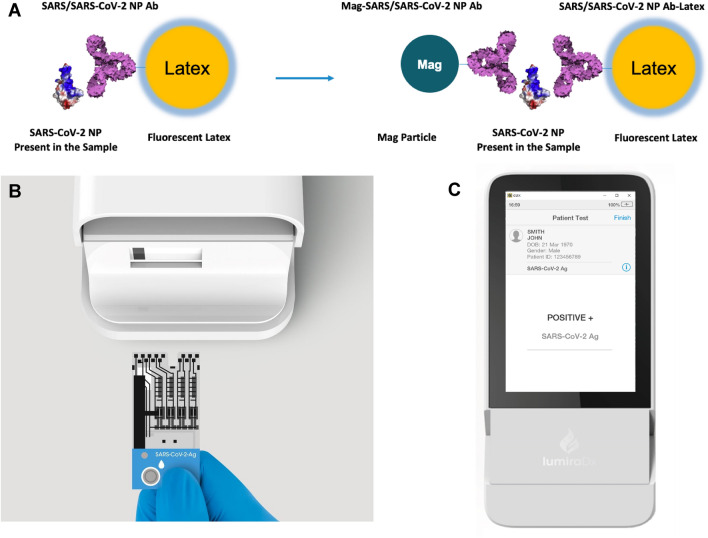
The LumiraDx SARS-CoV-2 antigen test runs on a portable, wall outlet or battery-powered multi-assay point-of-care instrument (Fig. 1) [11] (LumiraDx UK Ltd., Dumyat Business Park, Alloa, FK10 2PB, UK). The assay reagents are dry single-use, disposable, microfluidic test strips that contain specific antibodies to form an immunoassay complex that uses a fluorescent latex signal to detect the N protein of SARS-CoV-2 in a test sample (Fig. 1a). Nasal and nasopharyngeal swab samples are extracted using the extraction buffer and a transfer vial dropper that delivers 20 μL onto a test strip, and runs two simultaneous antigen assays in sub-microliter channels. The test takes 12 min to deliver a positive or negative result after the sample had been added to the test strip and inserted into the instrument. The instrument platform has a touch-screen interface (Fig. 1c), and connects to a cloud server for uploading test data into electronic medical records.
Fig. 1.
LumiraDx SARS-CoV-2 antigen assay. Schematic representation (a), test strip (b), and instrument result screen (c). Images reproduced with permissions from LumiraDx,
copyright 2020
The assay limit of detection (LoD), which was established using reciprocal dilutions of gamma-irradiated SARS-CoV-2, isolate USA WA1/2020, was estimated as 32 median tissue culture infectious dose (TCID50)/mL [11]. The assay cross-reactivity was evaluated by testing a panel of microorganisms that may have high prevalence for people being tested for SARS-CoV-2. 16 viruses, 11 bacteria, and two fungi were tested in the absence or presence of heat-inactivated SARS-CoV-2 at threefold LoD, and no interference was detected when spiking samples with SARS-CoV-2 and other microorganisms [11]. The assay was also evaluated by potential endogenous and exogenous interfering substances, including antiviral medications and over-the-counter cold remedies. Among the 22 substances tested, there was no interference with assay results [11].
A prospective cohort study was conducted to evaluate the LumiraDx SARS-CoV-2 antigen assay among children and adults who presented for COVID-19 testing.
Methods
Clinical Validation Study
A prospective validation study of the LumiraDx SARS-CoV-2 Ag test was conducted among children and adults who presented for COVID-19 testing. The study was conducted at ten sites across the United States (USA) and United Kingdom (UK), and including eight sites in which minimally trained operators collected and tested specimens. The nasal swab sample collection ran first from 26 June to 23 July 2020 in six sites (five in the USA and one in the UK). The nasopharyngeal sample collection was run following the nasal swab study (17 August–28 September 2020), in six US sites of which two of the sites were also in the earlier nasal swab collection in the USA. The clinical study was performed in accordance with the Helsinki Declaration of 1964 and its later amendments, received ethical approval from WCG Institutional Review Board, and all participants, or their parents or guardians for minor patients, provided informed consent (NCT04557046 clinicaltrials.gov). Nasal swab samples were additionally provided by a commercial supplier (MRN Diagnostics, Florida, USA), and also collected from an at-risk population (LumiraDx Stirling, UK), under approved protocols and informed consent. After collecting clinical data, two paired anterior nasal swabs (Copan FLOQ swabs) or two paired nasopharyngeal swabs were collected from each participant. The two swabs were collected simultaneously from both anterior nares, and both nostrils were swabbed using each of the two swabs. Each swab entered one nostril as the first-pass swab, before the swabs were switched to enter the opposite nostril as the second-pass swab. One swab was then placed into 0.7 mL of a proprietary extraction buffer for LumiraDx SARS-CoV-2 antigen test, and the other swab was placed into 3 mL of viral transport media (BD Universal Viral Transport Media, VTM). Swabs in VTM were tested fresh and in agreement with the manufacturer’s instructions by real-time polymerase chain reaction (rt-PCR) using the SARS-CoV-2 assay using a Roche cobas 6800 platform (Roche Molecular Diagnostics, Indianapolis, IN, USA). All buffer specimens for anterior nasal swabs were tested at the clinical site, and then frozen within 1 h of nasal swab collection. They were subsequently retested in a blinded manner. The results from the retested frozen samples were included in the final analysis. Equivalence of fresh and samples frozen, and retested according to the method above, was shown in a separate study. All buffer specimens for nasopharyngeal swabs were tested fresh at the clinical site within 1 h of collection.
Sample Size and Statistical Analysis
On the basis of an anticipated diagnostic sensitivity of 95%, a sample size was targeted to ensure at least 80 positive specimens for the anterior nasal swabs to achieve a 95% confidence interval (CI) from 87% to 98%. For nasopharyngeal swab specimens, 40 positive samples were collected to meet the minimum Emergency Use Authorization (EUA) requirement by the US Food and Drug Administration (FDA). Diagnostic performance was evaluated and results stratified by gender, age, days since symptom onset, and rt-PCR cycle threshold. In addition, sensitivity, specificity, positive likelihood ratios, and 95% CI using a Wilson two-sided analysis [12] were determined.
Operator Usability Study
Eight healthcare workers completed a 12-question Intended Use Operator Questionnaire, which evaluated various metrics of test usability and safety. Each question was assessed on a five-point Likert scale, ranging from 1 = strongly disagree to 5 = strongly agree (Fig. S1 in the supplementary material). The responses of eight test operators, who performed participant tests using the LumiraDx Diagnostic Platform and SARS-CoV-2 Ag test, are summarized in Fig. S1.
Results
Participant Characteristics
Among 512 participants, ages ranged from 0 to 90 years and 287 (56%) were female (Table S1 in the supplementary material). 414 (81%) participants experienced symptoms consistent for COVID-19 with an average duration of 4 days at the time of testing. On the basis of the Roche cobas rt-PCR testing results, 83 nasal swabs were positive for SARS-CoV-2 (prevalence 32.3%), and 40 nasopharyngeal swabs were positive for SARS-CoV-2 (15.7%), giving an overall estimated prevalence of 24% for COVID-19 in this cohort.
SARS-CoV-2 Antigen Assay Clinical Validation
Overall, the LumiraDx SARS-CoV-2 antigen assay had a sensitivity of 97.6% (95% CI 91.6–99.3%) and specificity of 96.6% (95% CI 92.7–98.4%) up to 12 days post symptom onset for nasal swab samples, and sensitivity of 97.5% (95% CI 87.1–99.6%) and specificity of 97.7% (95% CI 94.7–99.0%) for nasopharyngeal swab specimens (Table 1). When restricted to people testing within 10 days of symptom onset, which likely correlates to a period of SARS-CoV-2 viability [13], the diagnostic sensitivity was 98.7% (95% CI 93.0–99.8%) for nasal swabs (Table S2 in the supplementary material). There were no appreciable differences when results were stratified by age or gender.
Table 1.
Diagnostic performance of the LumiraDx SARS-CoV-2 antigen assay nasal and nasopharyngeal swabs compared to rt-PCR for clinical testing
| Anterior nasal swab, n = 257 | Nasopharyngeal swab, n = 255 | |||||
|---|---|---|---|---|---|---|
| Sensitivity, % (CI) | Specificity, % (CI) | LR+ | Sensitivity, % (CI) | Specificity, % (CI) | LR+ | |
| Total cohort | 97.6 (91.6–99.3) | 96.6 (92.7–98.4) | 28.3 | 97.5 (87.1–99.6) | 97.7 (94.7–99.0) | 41.9 |
| Sex | ||||||
| Female | 96.2 (87.2–99.0) | 96.6 (90.6–98.8) | 28.5 | 100 (86.7–100) | 99.2 (95.4–99.9) | 120.0 |
| Male | 100 (88.6–100) | 96.5 (90.1–98.8) | 28.3 | 93.3 (70.2–98.8) | 95.8 (89.7–98.4) | 22.2 |
| Age (years) | ||||||
| ≤ 60 | 97.4 (91.1–99.3) | 96.4 (92.3–98.3) | 26.8 | 97.4 (86.8–99.5) | 97.4 (94.1–98.9) | 38.0 |
| > 60 | 100 (56.6–100) | 100 (70.1–100) | N/A | 100 (20.7–100) | 100 (83.9–100) | N/A |
| rt-PCR Ct threshold (cycles) | ||||||
| ≤ 33 | 100 (94.0–100) | N/A | N/A | 100 (91.0–100) | N/A | N/A |
| > 33 | 60 (23.1–88.2) | N/A | N/A | 0.0 (0.0–79.3) | N/A | N/A |
CI confidence interval, LR likelihood ratio, N/A not available
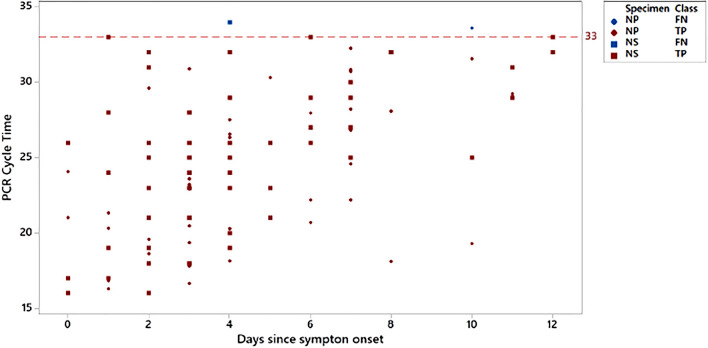
The SARS-CoV-2 antigen assay was highly sensitive up to and including a threshold cycle (Ct) value of 33 cycles (Table 1). As expected, the rt-PCR Ct values increased with more days since the onset of symptoms (Fig. 2). In addition, Ct values of more than 30 cycles were not uncommon shortly after symptom onset, which highlights the need for a high-sensitivity test to identify individuals with a low viral load. Among participants who had an rt-PCR Ct value of less than 33 cycles, the SARS-CoV-2 antigen assay was 100% sensitive for both nasal and nasopharyngeal swab specimen types.
Fig. 2.
Roche cobas SARS-CoV-2 rt-PCR cycle threshold versus days since symptom onset. True positive (TP) and false negative (FN) results using the LumiraDx SARS-CoV-2 antigen test with nasal (NS) or nasopharyngeal (NP) swab specimens. Red circles and squares indicate participants positive by LumiraDx SARS-CoV-2 antigen test. Blue circles and squares indicate participants negative by LumiraDx SARS-CoV-2 antigen test. Dotted line represents Ct 33
SARS-CoV-2 Antigen Assay Usability
During the prospective study, tests were completed by minimally trained healthcare workers who performed field-based testing at various sites including a drive-thru test center, community testing hub, as well as pediatric and family medicine clinics. Usability was assessed by monitoring user errors and obtaining feedback via questionnaire. The user error rate of the test system was recorded at 0.7% (95% CI 0.1–3.7%). Overall, positive responses were obtained for ease of use of the test system, easy to follow instructions, and simple to interpret results (Fig. S1 in the supplementary material).
Discussion
The LumiraDx SARS-CoV-2 antigen assay demonstrated high sensitivity when used to diagnose COVID-19 at the clinical POC. The rapid assay was highly sensitive for people with rt-PCR Ct values of less than 33 cycles within a period of 12 days since the onset of COVID-19 symptoms. These performance characteristics may correlate well with reported infective SARS-CoV-2 viral load and window of infectivity. The assay achieved a lower LoD and higher diagnostic sensitivity than other POC antigen tests and provided fast results (in under 12 min), in a convenient, easy to use POC test format with capacity to transfer data to electronic health records and surveillance systems.
At the outset of the study, EUA for the test was sought from the FDA to cover the following: strip and sample stability, strip and sample freeze–thaw, LoD, analytical specificity, microbial and substance interference, high-dose hook, and POC use [10]. On the basis of the data submitted to the FDA, the LumiraDx SARS-CoV-2 antigen assay with nasal swab received EUA on August 18, 2020 [14]. The performance of the assay was consistent between male and female individuals, and among older and younger adults.
Several other rapid SARS-CoV-2 antigen tests have received EUA from the FDA for use in near-patient settings. Two have limited diagnostic sensitivity (85–88%) when used within 5 days since symptom onset [14]. Another assay demonstrated high diagnostic sensitivity, but was only authorized for the first 7 days since symptom onset [14]. In contrast, this study demonstrated that the LumiraDx SARS-CoV-2 antigen assay had high diagnostic sensitivity, particularly with rt-PCR Ct values of less than 33 cycles, as measured by the Roche cobas SARS-CoV-2 rt-PCR assay, through the first 12 days from the onset of COVID-related symptoms. The high performance of the test is due to the new test technology, using microfluidic and immunofluorescence to precisely measure antigen levels in the picogram per milliliter range.
Recent studies have identified the first 10 days since symptom onset as the likely window of infectivity for the SARS-CoV-2 virus [13, 15, 16]. Several other studies have related infectivity to a low rt-PCR Ct value, which correlates to a high viral load, and/or the ability to culture SARS-CoV-2 virus [17–22]. A study of hospitalized patients with COVID-19 showed that the ability to culture SARS-CoV-2 diminished from day 10 to day 12 since symptom onset [13]. In another study of hospitalized patients, successful isolation of culturable virus correlated with rt-PCR Ct values of less than 33 cycles, while those above this level were considered to be non-infectious [19]. Rt-PCR testing has also been shown to remain positive many days past the window for culturing viable SARS-CoV-2 virus, which suggest that rt-PCR testing may be generating some positive results from people with remnant viral RNA who do not have contagious viral particles [20]. This is further supported by the finding that antigen-based testing, but not rt-PCR testing, correlates with growth of SARS-CoV-2 by viral culture [23].
The recently published WHO guidance [24] suggests minimum performance requirements of at least 80% sensitivity and at least 97% specificity for an antigen-based test compared to a reference assay. This study, together with a recent Scandinavian cooperation for evaluation of near-patient laboratory equipment (SKUP) study using a larger, mixed cohort of symptomatic and asymptomatic patients [25], demonstrates that the LumiraDx SARS-CoV-2 Ag test using nasal swab and nasopharyngeal swab samples meets these requirements.
Conclusions
There is an urgent need to improve access to point-of-care testing for SARS-CoV-2 during the COVID-19 pandemic. Point-of-care testing has the potential to expand access for SARS-CoV-2 testing, and may have imperfect accuracy when compared to laboratory-based testing. This test demonstrated high sensitivity over a wide range of rt-PCR Ct values up to a Ct value of 33 cycles, and over a 12-day infectivity window, making this platform highly suitable for SARS-CoV-2 testing and COVID-19 surveillance programs. This rapid assay with high sensitivity and anterior nasal sampling offers significant advantages for identification and management of SARS-CoV-2 infection, particularly in clinic- and community-based settings.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgments
We thank the children, women and men who participated in this study, the clinical sites for sharing their space, and our research staff and nurses who conducted the study.
Funding
This work was supported by LumiraDx Ltd, including funding of the journal’s Rapid Services fee.
Editorial Support
Editorial support was provided by Maria Haughton, integrated medhealth communication (imc), Cambridge, UK, and was funded by LumiraDx Ltd.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, contributed to the interpretation of the data, critically revised the manuscript for important intellectual content, approved the final version to be published, and agree to be accountable for all aspects of the work.
Prior Presentation
An earlier version of this manuscript was made available on the medRxiv pre-print server on December 14, 2020 (https://www.medrxiv.org/content/10.1101/2020.12.11.20238410v1).
Disclosures
PKD, MA, CC, ABG, MH, MW, RR, SY, EZ and MM have nothing to disclose.
Compliance with Ethics Guidelines
The clinical study received ethical approval from WCG Institutional Review Board and all participants provided informed consent. The study was performed in accordance with the Helsinki Declaration of 1964 and its later amendments. All participants, or their parents or guardians for minor patients, provided informed consent (NCT04557046clinicaltrials.gov).
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- 1.World Health Organization. Health emergencies, coronavirus disease (COVID-19) outbreak. 2020. https://www.euro.who.int/en/health-topics/health-emergencies/coronavirus-covid-19. Accessed 21 Oct 2020.
- 2.Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siordia JA. Epidemiology and clinical features of COVID-19: a review of current literature. J Clin Virol. 2020;127:104357. doi: 10.1016/j.jcv.2020.104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization. Coronavirus disease (COVID-19), situation report–85. 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200414-sitrep-85-covid-19.pdf?sfvrsn=7b8629bb_4. Accessed 21 Oct 2020.
- 5.Emery SL, Erdman DD, Bowen MD, et al. Real-time reverse transcription-polymerase chain reaction assay for SARS-associated coronavirus. Emerg Infect Dis. 2004;10:311. doi: 10.3201/eid1002.030759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Udugama B, Kadhiresan P, Kozlowski HN, et al. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 7.Sheridan C. Fast, portable tests come online to curb coronavirus pandemic. Nat Biotechnol. 2020;38:515–518. doi: 10.1038/d41587-020-00010-2. [DOI] [PubMed] [Google Scholar]
- 8.World Health Organization. A coordinated global research roadmap: 2019 novel coronavirus. 2020. https://www.who.int/blueprint/priority-diseases/key-action/Coronavirus_Roadmap_V9.pdf. Accessed 21 Oct 2020.
- 9.Lai MM. SARS virus: the beginning of the unraveling of a new coronavirus. J Biomed Sci. 2003;10:664–675. doi: 10.1159/000074077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.FDA. EUA antigen template for manufacturers. 2020. https://www.fda.gov/emergency-preparedness-and-response/mcm-legal-regulatory-and-policy-framework/emergency-use-authorization. Accessed 21 Oct 2020.
- 11.LumiraDx. LumiraDx website and SARS-CoV-2 Antigen test EUA Product Insert. 2020. https://www.lumiradx.com/us-en/. Accessed 21 Oct 2020.
- 12.Wilson EB. Probable inference, the law of succession, and statistical inference. J Am Stat Assoc. 1927;22:209–212. doi: 10.1080/01621459.1927.10502953. [DOI] [Google Scholar]
- 13.Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 14.FDA. In vitro diagnostic EUAs. 2020. https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/vitro-diagnostics-euas. Accessed 21 Oct 2020.
- 15.Kujawski SA, Wong KK, Collins JP, et al. First 12 patients with coronavirus disease 2019 (COVID-19) in the United States. medRxiv. 2020. http://medrxiv.org/content/early/2020/03/12/2020.03.09.20032896. Accessed 21 Oct 2020.
- 16.Bullard J, Dust K, Funk D, et al. Predicting infectious SARS-CoV-2 from diagnostic samples. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perera RAPM, Tso E, Tsang OTY, et al. SARS-CoV-2 virus culture and subgenomic RNA for respiratory specimens from patients with mild coronavirus disease. Emerg Infect Dis. 2020;26:2701–2704. doi: 10.3201/eid2611.203219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Kampen JJA, van de Vijver DAMC, Fraaij PLA, et al. Shedding of infectious virus in hospitalized patients with coronavirus disease-2019 (COVID-19): duration and key determinants. medRxiv. 2020. http://medrxiv.org/content/early/2020/06/09/2020.06.08.20125310. Accessed 21 Oct 2020. [DOI] [PMC free article] [PubMed]
- 19.La Scola B, Le Bideau M, et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39:1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singanayagam A, Patel M, Charlett A, et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020;25:2001483. doi: 10.2807/1560-7917.es.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ladhani SN, Chow JY, Janarthanan R, et al. Investigation of SARS-CoV-2 outbreaks in six care homes in London, April 2020. EClinicalMedicine. 2020;26:100533. doi: 10.1016/j.eclinm.2020.100533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gniazdowski V, Morris P, Wohl S, et al. Repeat COVID-19 molecular testing: correlation with recovery of infectious virus, molecular assay cycle thresholds, and analytical sensitivity. medRxiv. 2020 doi: 10.1101/2020.08.05.20168963. [DOI] [Google Scholar]
- 23.Pekosz A, Cooper CK, Parvu V, et al. Antigen-based testing but not real-time PCR correlates with SARS-CoV-2 virus culture. medRxiv. 2020 doi: 10.1101/2020.10.02.20205708. [DOI] [Google Scholar]
- 24.World Health Organization. Antigen-detection in the diagnosis of SARS-CoV-2 infection using rapid immunoassays. 2020. https://www.who.int/publications/i/item/antigen-detection-in-the-diagnosis-of-sars-cov-2infection-using-rapid-immunoassays. Accessed 25 Jan 2021.
- 25.SKUP. Report from the evaluation SKUP/2021/124. LumiraDx SARS-CoV-2 Ag Test (LumiraDx UK Ltd), a system for detection of SARS-CoV-2. www.skup.org. Accessed 25 Jan 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.