ABSTRACT
Spindle orientation is important in multiple developmental processes as it determines cell fate and function. The orientation of the spindle depends on the assembly of a proper astral microtubule network. Here, we report that the spindle assembly factor TPX2 regulates astral microtubules. TPX2 in the spindle pole area is activated by GM130 (GOLGA2) on Golgi membranes to promote astral microtubule growth. GM130 relieves TPX2 inhibition by competing for importin α1 (KPNA2) binding. Mitotic phosphorylation of importin α at serine 62 (S62) by CDK1 switches its substrate preference from TPX2 to GM130, thereby enabling competition-based activation. Importin α S62A mutation impedes local TPX2 activation and compromises astral microtubule formation, ultimately resulting in misoriented spindles. Blocking the GM130–importin α–TPX2 pathway impairs astral microtubule growth. Our results reveal a novel role for TPX2 in the organization of astral microtubules. Furthermore, we show that the substrate preference of the important mitotic modulator importin α is regulated by CDK1-mediated phosphorylation.
KEY WORDS: Mitosis, Astral microtubules, Spindle orientation, TPX2, Importin α, GM130, CDK1, Phosphorylation, Golgi
Summary: TPX2 regulates astral microtubules and spindle orientation. TPX2 is locally activated via the Golgi protein GM130 by sequestering importin α from TPX2, a process enabled by importin α phosphorylation at S62.
INTRODUCTION
Cell proliferation requires accurate partitioning of all vital cellular constituents during cell division. In eukaryotic cells, the microtubule-based spindle machinery delivers a precise set of chromosomes into each daughter cell. Spindle microtubules are further responsible for the correct positioning and orientation of the spindle to define the cell division plane, which plays a critical role in cell fate decisions, morphogenesis and maintenance of tissue organization (Bergstralh et al., 2017). Consequently, aberrant spindle orientation causes defects in various physiological processes such as gastrulation, neuronal differentiation, epithelial self-renewal and tissue stratification (Gong et al., 2004; Lechler and Fuchs, 2005; Fish et al., 2006; Cabernard and Doe, 2009). Determination and maintenance of spindle orientation largely rely on the mitotic microtubule network and its connections to the cell cortex.
Assembly of the mitotic spindle is tightly controlled in space and time through regulated microtubule nucleation and polymerization (Prosser and Pelletier, 2017). In higher eukaryotes, mitotic microtubule arrays predominantly originate at the centrosomes, which form the spindle poles. Astral microtubules emanate from the spindle poles and extend to the cell cortex. This spindle–cortex connection is required to generate the pulling forces that dictate positioning and orientation of the spindle. A reduction of astral microtubules weakens the connections between the spindle and cell cortex, often leading to spindle oscillation and misorientation (Petry, 2016). Although astral microtubules are important in spindle positioning, it is the microtubule filaments connected to kinetochores that are directly engaged in the segregation of chromosomes into the daughter cells. Kinetochore microtubules are bundles of parallel microtubule filaments known as K-fibers that originate at the spindle poles and attach to the chromosomes via kinetochores. Proper attachment of all K-fibers in the bipolar spindle is required to silence the spindle assembly checkpoint and allow progression into anaphase (Prosser and Pelletier, 2017).
In addition to centrosomes, mitotic microtubules are also nucleated and elongated around the kinetochores and chromatin (O'Connell and Khodjakov, 2007). This key step of spindle formation ensures that all chromosomes are captured by the kinetochore microtubules and it is regulated by the small GTPase Ran. Upon loading with GTP (via its chromatin-localized guanine-nucleotide-exchange factor RCC1), RanGTP destabilizes ternary complexes of importins α/β and classical nuclear localization sequence (NLS)-containing proteins, including several essential spindle assembly factors, and promotes their dissociation (Carazo-Salas et al., 1999). Through this mechanism, RanGTP relieves importin α-mediated inhibition of these spindle assembly factors to promote microtubule nucleation, stabilization and spindle organization (Kalab and Heald, 2008). One of the key spindle assembly factors is the microtubule nucleator TPX2. Once released from importin α inhibition, TPX2 activates and stabilizes the Aurora A kinase (AURKA) by shielding it from dephosphorylation (Bayliss et al., 2003). The Aurora A–TPX2 complex binds to microtubules, further enhancing microtubule polymerization at the chromosomes (Kufer et al., 2002; Anderson et al., 2007).
In addition to functioning at chromatin, TPX2 also nucleates microtubules at mitotic Golgi membranes through activation by the Golgi-resident membrane protein GM130 (also known as GOLGA2; Wei et al., 2015). During mitosis, the mammalian Golgi apparatus is disassembled into a collection of vesicles that cluster and localize in the spindle pole area (Shima et al., 1998). These mitotic Golgi clusters are subsequently delivered by the spindle into the daughter cells where the membranes fuse to re-establish a contiguous Golgi structure (Wei and Seemann, 2017; Mascanzoni et al., 2019). Mitotic disassembly of the Golgi is initiated by CDK1-mediated phosphorylation of GM130 (Lowe et al., 2000), which exposes its NLS motif that sequesters importin α1 (KPNA2; hereafter referred to as importin α) from TPX2 by direct competition. TPX2 is thereby activated to locally drive microtubule nucleation at mitotic Golgi membranes. The nascent microtubules can then be captured by GM130 and couple Golgi membranes to the spindle (Wei et al., 2015). However, it remains unclear how TPX2 is preferentially liberated from importin α by the NLS motif of GM130, given the high abundance of NLS-containing proteins that are released upon nuclear envelope breakdown at mitotic entry. Furthermore, how importin α regulates this event spatiotemporally so that the process is coordinated with other microtubule nucleation activities to form a functional spindle remains elusive. Moreover, the exact role of Golgi-based microtubule nucleation in shaping the mitotic microtubule network has not been characterized.
During mitosis, importin α silences several essential spindle assembly factors (such as TPX2) in the cytoplasm by binding to their NLS. It is unclear how importin α preferentially recognizes these spindle assembly factors in the presence of other abundant NLS-containing proteins that are released upon nuclear breakdown. Previous studies have shown that post-translational modifications of importin α, such as phosphorylation by casein kinase 2, can modulate its activity (Hachet et al., 2004; Brownlee and Heald, 2019). Furthermore, proteomics analyses have revealed mitosis-specific phosphorylation of importin α1 (KPNA2) at residue serine 62 (S62; Nousiainen et al., 2006; Cantin et al., 2008; Dephoure et al., 2008; Ly et al., 2017), while a recent study chose threonine 9 (T9) and S62 in silico as potential phosphorylation sites and investigated the role of their phosphorylation in importin α function (Guo et al., 2019). Overexpression of GFP-tagged T9A and S62A (T9A/S62A) mutant importin α increases mutant protein association with the spindle assembly factors NUMA1, TPX2 and KIF1C, an observation that correlates with shortened spindles, a reduced microtubule mass and a delay in mitotic progression (Guo et al., 2019). However, it remains unclear whether importin α is phosphorylated by CDK1 in vivo and whether the induced spindle defects are directly caused by the differential binding of the importin α T9A/S62A mutant to spindle assembly factors.
Here, we describe a pathway responsible for driving astral microtubule formation and spindle orientation. We show that TPX2 promotes astral microtubule growth around the spindle poles through Aurora A kinase. TPX2 becomes locally activated by GM130, which sequesters importin α from TPX2 on mitotic Golgi membranes around the spindle poles. This sequestration-activation process is triggered by CDK1-mediated phosphorylation of importin α at S62. Mitotic phosphorylation of importin α weakens its interaction with TPX2 but enhances its binding to GM130, thereby enabling competition-based activation of TPX2. This Golgi-based TPX2 activation is required for astral microtubule growth from the spindle poles, since suppressing importin α phosphorylation results in diminished astral microtubules and misorientation of the spindle.
RESULTS
Importin α is phosphorylated at serine 62 by CDK1 in mitosis
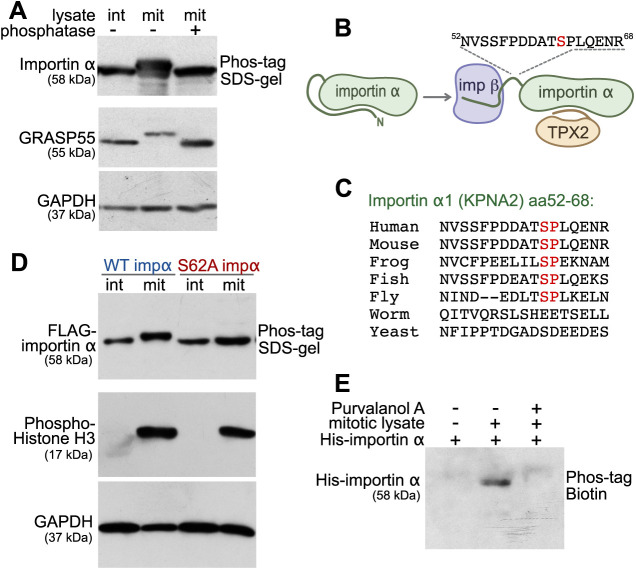
To examine whether importin α is phosphorylated during mitosis, we analyzed lysates from interphase and mitotic HeLa cells on a Phos-tag acrylamide gel, through which phosphorylated proteins migrate more slowly (Fig. 1A). Upon immunoblotting, we observed that the band corresponding to importin α from mitotic lysate was shifted upward relative to that from interphase lysate, as was the positive control GRASP55 (also known as GORASP2; Xiang and Wang, 2010). To further establish that the decreased mobility of mitotic importin α was induced by phosphorylation, we pretreated the mitotic lysate with λ-phosphatase before analyzing the band shift. Phosphatase treatment abolished the band shift of both GRASP55 and importin α, demonstrating that importin α is indeed mitotically phosphorylated. Next, we applied mass spectrometry to identify mitosis-specific phosphorylation sites of importin α. To this end, we generated a HeLa cell line that stably expresses FLAG-tagged importin α in a doxycycline-inducible manner, and then isolated tagged importin α from the interphase and mitotic cell lysates. Consistent with previous proteomics data (Nousiainen et al., 2006; Cantin et al., 2008; Dephoure et al., 2008; Ly et al., 2017), our mass spectrometry analysis identified a mitosis-specific peptide comprised of residues 52–68 of importin α among which S62 was phosphorylated (Fig. 1B; Fig. S1A). The S62 site is consistent with the well-defined [Ser/Thr]-Pro consensus motif of the master mitotic kinase CDK1. Although T9 of human importin α, but not of other species, also matches the CDK1 consensus motif (Guo et al., 2019), T9 phosphorylation was not identified in our approach (Fig. S1A) or in previous proteomics studies (Nousiainen et al., 2006; Cantin et al., 2008; Dephoure et al., 2008; Ly et al., 2017).
Fig. 1.
Importin α is mitotically phosphorylated at serine 62 by CDK1. (A) Importin α is phosphorylated during mitosis. Cell lysates from interphase (int) and mitotic (mit) HeLa cells, as well as λ phosphatase-treated mitotic lysate, were separated on a 10% SDS gel containing 25 μM Phos-tag acrylamide and immunoblotted for importin α. GRASP55 and GADPH were separated by regular SDS–PAGE. (B) Schematic illustration of the location of the mitotically-phosphorylated peptide identified by mass spectrometry. The identified peptide (residues 52–68) harbors the mitosis-specific phosphorylation site of importin α1 (KPNA2) and is located between the importin β (imp β)-binding domain and the NLS-binding domain. (C) Within the identified peptide sequence, residue serine 62 is conserved among higher eukaryotes. The Ser-Pro sequence matches a CDK1 phosphorylation consensus motif. (D) Importin α (impα) is phosphorylated at serine 62. Interphase or mitotic cell lysates from cells expressing FLAG-tagged WT importin α or importin α S62A were separated on a Phos-tag acrylamide gel and immunoblotted for FLAG. Phospho-histone H3 and GAPDH were separated by SDS–PAGE. (E) Importin α phosphorylation is blocked by the CDK1 inhibitor purvalanol A. Beads coated with recombinant full-length His-tagged importin α were incubated with mitotic HeLa extract in the presence or absence of purvalanol A. The beads were then washed by high-salt buffer (PBS, 1M NaCl), and importin α was eluted by boiling in SDS sample buffer. Phosphorylation was detected by blotting with Phos-tag biotin and streptavidin–HRP. Data in A,D,E are representative of three experiments.
Importin α binds its substrates and forms a ternary complex with importin β. In the absence of NLS-containing substrates, the autoinhibitory N-terminal domain of importin α (IBB domain) folds back into its NLS-binding groove (Fig. 1B). Importin β can bind to the IBB domain of importin α to open up its NLS-binding region. The phosphorylated fragment we identified (residues 52–68) is situated between the IBB domain and the substrate-binding domain. This importin α peptide (NVSSFPDDATSPLQENR) contains several serine and threonine residues as potential phosphorylation sites, but harbors only one [Ser/Thr]-Pro consensus motif for CDK1 at S62. Since this Ser-Pro motif is highly conserved (Fig. 1C), we reasoned that S62 is mitotically phosphorylated by CDK1. To validate this supposition, we generated stable HeLa cell lines that inducibly express FLAG-tagged wild-type importin α (WT) or the importin α S62A mutant protein, and then analyzed their interphase and mitotic lysates on a Phos-tag gel (Fig. 1D). We found that the mobility of WT importin α from mitotic lysate was reduced compared to that of interphase lysate. In contrast, the band shift of importin α was abolished in the mutant harboring the single-point S62A mutation. Together with our mass spectrometry data, this result indicates that residues other than S62, such as T9, are not phosphorylated during mitosis. Next, we determined whether mitotic phosphorylation of importin α is indeed triggered by CDK1. We incubated recombinant importin α with mitotic extract in the presence or absence of the CDK1 inhibitor purvalanol A. Phosphorylation was then detected by blotting with a biotinylated Phos-tag combined with streptavidin-conjugated HRP (Fig. 1E). Phos-tag biotin specifically labeled the phosphorylated importin α in mitotic extracts, which was abolished upon treatment of extracts with the Cdk1 inhibitor purvalanol A. Taken together, these results show that importin α is phosphorylated at serine 62 by CDK1 in mitosis.
Impaired importin α phosphorylation causes mitotic defects
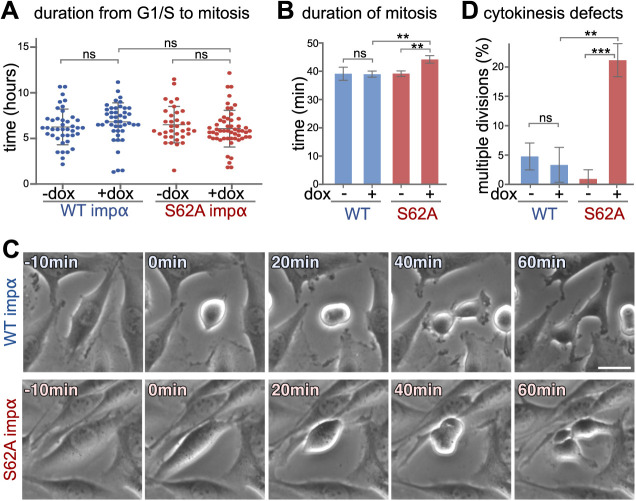
We then sought to determine the effects of importin α phosphorylation on mitotic progression. To rule out potential chronic effects of expressing phospho-deficient mutant importin α, we generated stable normal rat kidney (NRK) cell lines that express human FLAG-tagged wild-type importin α (hereafter referred to as ‘WT-expressing’) or mutant importin α S62A (hereafter referred to as ‘S62A-expressing’) upon doxycycline treatment. These importin α-expressing cells were synchronized and released from a thymidine G1-S block, and then their progression into M phase was followed by phase-contrast time-lapse microcopy. Average time to mitotic entry of WT- and S62A-expressing cells was comparable, indicating that ectopic expression of importin α does not impact interphase progression or the G2-M transition (Fig. 2A). However, the time from cell rounding to cytokinesis onset (as a measure of mitosis duration) was prolonged by 13% (or 5 min) in S62A-expressing cells compared to WT-expressing cells (Fig. 2B). Moreover, whereas WT-expressing cells divided normally, 21.2% of mutant cells with phospho-deficient importin α S62A exhibited cytokinesis defects, often generating more than two daughter cells (Fig. 2C,D).
Fig. 2.
Importin α S62A induces mitotic defects. (A–D) NRK cells non-induced or induced using doxycycline (dox) to express WT importin α (impα) or importin α S62A were synchronized at the G1-S transition by means of thymidine treatment for 16 h. The cells were then released from the thymidine block and cell cycle progression was monitored by time-lapse phase-contrast microscopy. (A) Importin α expression does not affect mitotic entry. The duration between release from G1-S arrest to mitotic entry is plotted. n>90. ns indicates P=0.075 for WT +dox versus S62A +dox, P=0.211 for WT −dox versus WT +dox and P=0.316 for S62A −dox versus S26A +dox (unpaired, two-tailed Student's t-test). (B) Importin α S62A-expressing cells exhibit prolonged mitotic duration. n>90. **P=0.0065 for WT +dox versus S62A +dox and **P=0.0062 for S62A −dox versus S26A +dox; ns, P=0.93 for WT −dox versus WT +dox (unpaired, two-tailed Student's t-test). (C,D) NRK cells expressing importin α S62A exhibit cytokinesis defects. Still images from a representative time-lapse movie at the indicated time points after mitotic entry (0 min) are shown in C. n>90. ***P=0.0004; **P=0.0017; ns P=0.545 (unpaired, two-tailed Student's t-test). Data are presented as mean±s.d. Scale bar: 20 µm.
Mitotic phosphorylation of importin α controls spindle orientation
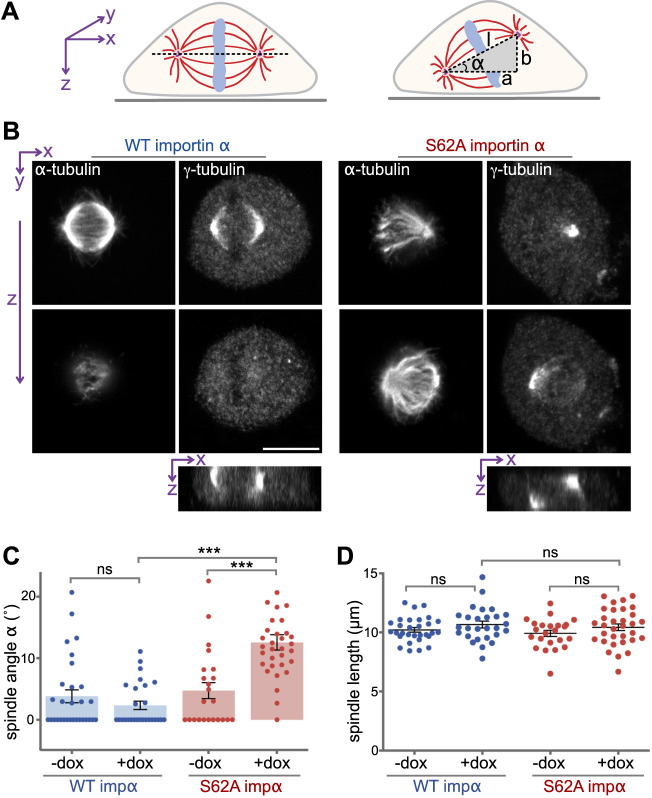
To further investigate the effects of mutant importin α S62A on mitosis, we analyzed the 3D orientation of metaphase spindles using confocal microscopy. Cells arrested in metaphase were double-stained for α-tubulin and γ-tubulin to label spindle microtubules and mark the spindle poles, respectively. We then measured spindle length and calculated spindle orientation along the z-axis (Fig. 3A). We found that average distances between the two spindle poles did not differ significantly between WT- and S62A-expressing cells (Fig. 3B,D), indicating that the overall structure of the spindle was not affected (Baudoin and Cimini, 2018). Notably, in WT-expressing cells, both spindle poles were detected within the same z-section and were aligned parallel to the substratum. In contrast, the spindle poles of S62A-expressing cells were located in different z-planes, revealing that the spindle axis was tilted relative to the coverslip surface. The angle between the coverslip surface and the plane in which the two spindle poles resided showed an aberrant 13° tilt of the spindle axis in S62A-expressing cells (Fig. 3C). These results indicate that expression of phospho-deficient importin α S62A alters the orientation of the mitotic spindle and affects cell division.
Fig. 3.
Phosphorylation of importin α controls spindle orientation. (A) Schematic illustration of spindle orientation in tissue culture cells. The axis of a normal spindle is oriented parallel to the coverslip (left), whereas the axis of a misoriented spindle is tilted (right). The angle α of spindle tilt was calculated as: α=arctan (b/a). Spindle length (l) was calculated as the 3D distance between the spindle poles (l2=a2+b2). (B) Importin α S62A causes misorientation of the spindle. NRK cells expressing WT importin α or importin α S62A were arrested at metaphase using MG132 and then dual-stained for α-tubulin and γ-tubulin to label spindle microtubules and the spindle poles, respectively. Z-sections were captured by confocal microscopy. Orthogonal sections through the z-stacks along the x-axis (bottom panels) indicate the relative spindle pole localization on the z-axis. Scale bar: 10 μm. (C) Importin α (impα) S62A results in tilted spindles. The spindle tilt angle α was calculated as shown in A for cells with or without doxycyclin (dox) treatment. n>30. ***P<0.0001; ns, WT −dox versus WT +dox P=0.2478 (Student's t-test). (D) Importin α S62A expression does not alter spindle length. The spindle length l was determined as depicted in A. n>30. ns for WT +dox versus S62A +dox indicates P=0.5676, WT −dox versus WT +dox P=0.185 and S62A −dox versus S62 +dox P=0.196 (Student's t-test). Data are presented as mean±s.e.m.
Mutant importin α S62A inhibits astral microtubule growth
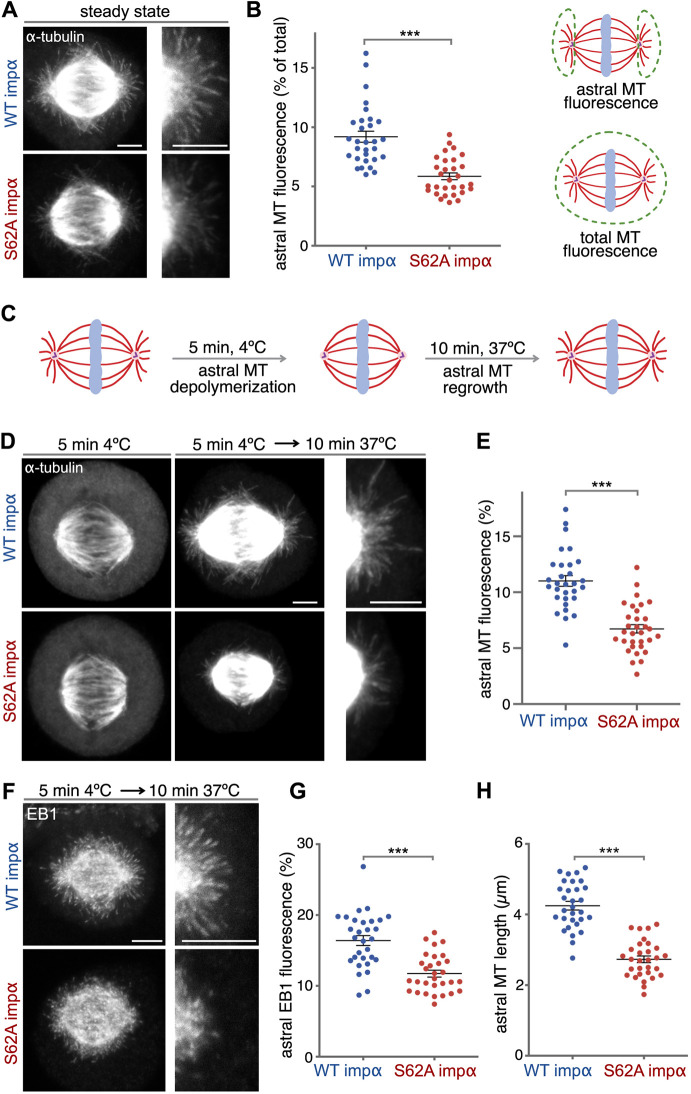
Proper orientation of the spindle depends on the astral microtubules that connect the spindle to the cell cortex (Lu and Johnston, 2013). To decipher whether astral microtubules in cells expressing mutant importin α S62A were compromised, we arrested the cells in metaphase using MG132 and then investigated the microtubule networks by confocal microscopy. To better visualize astral microtubules, which are less abundant and present weaker signal compared to interpolar and kinetochore microtubules, we increased image exposure. We found that microtubule mass in the astral area of S62A-expressing cells at steady state was reduced, as indicated by less dense and shorter filaments relative to those in WT-expressing cells (Fig. 4A). Furthermore, the relative fluorescence intensity of α-tubulin staining in the astral area of S62A-expressing cells was diminished by 30% compared to that in WT-expressing cells (reduced from 9% to 6%, Fig. 4B).
Fig. 4.
Importin α S62A inhibits microtubule growth. (A) Importin α (impα) S62A compromises astral microtubules. NRK cells expressing WT importin α or importin α S62A were arrested at metaphase using MG132 for 1 h and stained for α-tubulin. Maximum intensity projections of confocal z-sections are shown. Enlargements of the astral microtubule area are shown in the panels on the right. (B) Quantitation of the experiment shown in A. Astral microtubule (MT) fluorescence was measured as the percentage of astral microtubule intensity relative to the total microtubule intensity, as shown in the schematic. n>30. ***P<0.0001 (Student's t-test). (C) Schematic illustration of the cold-induced astral microtubule regrowth assay. Astral microtubules were depolymerized by 5 min of cold treatment, and then allowed to regrow by shifting the cells back to 37°C for 10 min. (D) Importin α S62A inhibits astral microtubule regrowth upon cold treatment. NRK cells arrested at metaphase using MG132 were placed on ice for 5 min to depolymerize astral microtubules but not spindle filaments. After 10 min regrowth at 37°C, cells were fixed and stained for α-tubulin. Confocal microscopy images represent maximum intensity projections. Enlargements of the astral microtubule area are shown in the panels on the right. (E) Astral microtubule fluorescence was calculated as in B. n>30. ***P<0.0001 (Student's t-test). (F) Importin α S62A inhibits EB1 localization to the astral area. EB1 was stained following the procedure described in C,D. Maximum intensity projections are shown. Enlargements of the astral microtubule area are shown in the panels on the right. (G) EB1 fluorescence was calculated as for astral microtubule intensity. n>30. ***P<0.0001 (Student's t-test). (H) Importin α S62A shortens astral microtubule length. The lengths of astral microtubules were determined as the distance between the tip of an EB1 signal comet and the spindle pole. For each cell, the average length of the three longest filaments from each pole was used to represent nascent microtubule length. n>30. ***P=0.019 (Student's t-test). Data in B,E,G,H are presented as mean±s.e.m. Scale bars: 5 µm.
To determine whether the S62A mutation affects initiation of astral microtubules, we performed a microtubule regrowth assay upon cold-induced depolymerization (Fig. 4C). Cells arrested in metaphase were placed on ice for 5 min, resulting in full depolymerization of astral microtubules (Fig. 4D). The cells were then shifted to 37°C for 10 min to allow microtubule regrowth and stained for α-tubulin. This assay showed that the astral microtubule mass in WT-expressing cells was restored to steady-state conditions upon temperature shift, whereas astral microtubule reformation was reduced by 25% in S62A-expressing cells (Fig. 4E).
To further test whether the diminished astral microtubule mass in S62A-expressing cells was due to diminished microtubule growth, we immunostained the cells for EB1 (also known as MAPRE1) after the microtubule regrowth assay (Fig. 4F). EB1 preferentially localizes at the tips of growing microtubules and functions as a marker for growing microtubule filaments. Consistent with our tubulin staining results (Fig. 4E), EB1 signal intensity in the astral microtubule region of S62A-expressing cells was reduced by 29% (from 16.4% to 11.7% of the EB1 signal in the astral area; Fig. 4G). Furthermore, the EB1 comets did not emanate as far from the spindle poles of S62A-expressing cells (mean=2.7 µm) as they did in WT-expressing cells (mean=4.2 μm) (Fig. 4H). These results demonstrate that expression of mutant importin α S62A impairs astral microtubule growth and dynamics, resulting in spindle misorientation.
TPX2 is required for astral microtubule formation
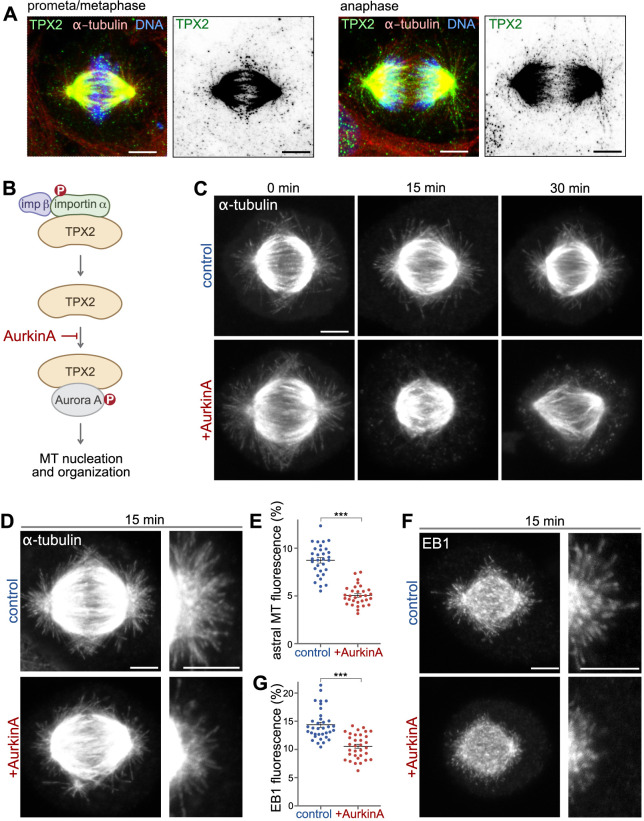
During mitosis, importin α silences the activity of the spindle assembly factor TPX2 in the cytoplasm. Upon release from importin α inhibition, TPX2 localizes to spindle microtubules and activates Aurora A, an essential kinase for spindle assembly (Garrido and Vernos, 2016). TPX2 was also found on astral microtubules in Caenorhabditis elegans and upon overexpression in human RPE-1 cells (Mangal et al., 2018; Naso et al., 2020). We also detected endogenous TPX2 on astral and spindle microtubules (Fig. 5A), suggesting that TPX2 might regulate astral microtubule assembly. Previously, we demonstrated that mitotic Golgi membranes can initiate localized microtubule assembly by liberating TPX2 from importin α inhibition (Wei et al., 2015). Given that mitotic Golgi membranes accumulate around the spindle poles (Wei and Seemann, 2009b) and TPX2 localizes to astral microtubules (Fig. 5A), we wondered whether TPX2 binding to Aurora A drives astral microtubule nucleation. To test this possibility, we took advantage of AurkinA, a small-molecule inhibitor that specifically blocks the interaction of TPX2 with Aurora A (Fig. 5B) (Janeček et al., 2016). We first tested the overall efficacy of the inhibitor on spindle microtubules. Cells arrested in metaphase were treated with 200 µM AurkinA for 0, 15 or 30 min, and then were immunostained for tubulin (Fig. 5C). We found that prolonged AurkinA treatment (for 30 min) weakened the microtubule filament network, whereas 15 min AurkinA incubation mainly impacted astral microtubules compared to spindle filaments (Fig. 5D,E). Furthermore, the 15 min AurkinA treatment reduced the lengths of astral microtubules, as well as EB1 signal intensity and density, in the astral microtubule area (Fig. 5F,G). These results indicate that TPX2 binding and activation of Aurora A kinase are required for astral microtubule nucleation.
Fig. 5.
TPX2 is required for astral microtubule formation. (A) Endogenous TPX2 localizes to astral microtubules and spindle microtubules. SV589 cells in prometa/metaphase and anaphase were stained for TPX2 (green), α-tubulin (red) and DNA (blue). Panels on the right show the TPX2 signal in inverted grayscale. (B) AurkinA is a small molecule inhibitor that specifically blocks the interaction of TPX2 with Aurora A kinase (MT, microtubule). (C) AurkinA alters spindle organization. NRK cells arrested at metaphase using MG132 for 1 h were treated with AurkinA for 0, 15 or 30 min and then fixed and stained for α-tubulin. (D) AurkinA compromises astral microtubules. After 15 min of AurkinA treatment, NRK cells were fixed and stained for α-tubulin. Enlargements of the astral microtubule area are shown in the panels on the right. (E) Quantitation of the experiment shown in D. n>30, ***P<0.0001 (Student's t-test). (F) AurkinA inhibits astral microtubule growth. After 15 min of Aurkin A treatment, NRK cells were fixed and stained for EB1. Enlargements of the astral microtubule area are shown in the panels on the right. (G) Quantitation of the experiment shown in F. n>30. ***P<0.0001 (Student's t-test). In E,G, data are presented as mean±s.e.m. Maximum intensity projections of confocal z-sections are shown. Scale bars: 5 µm.
GM130 regulates astral microtubule growth via phosphorylated importin α
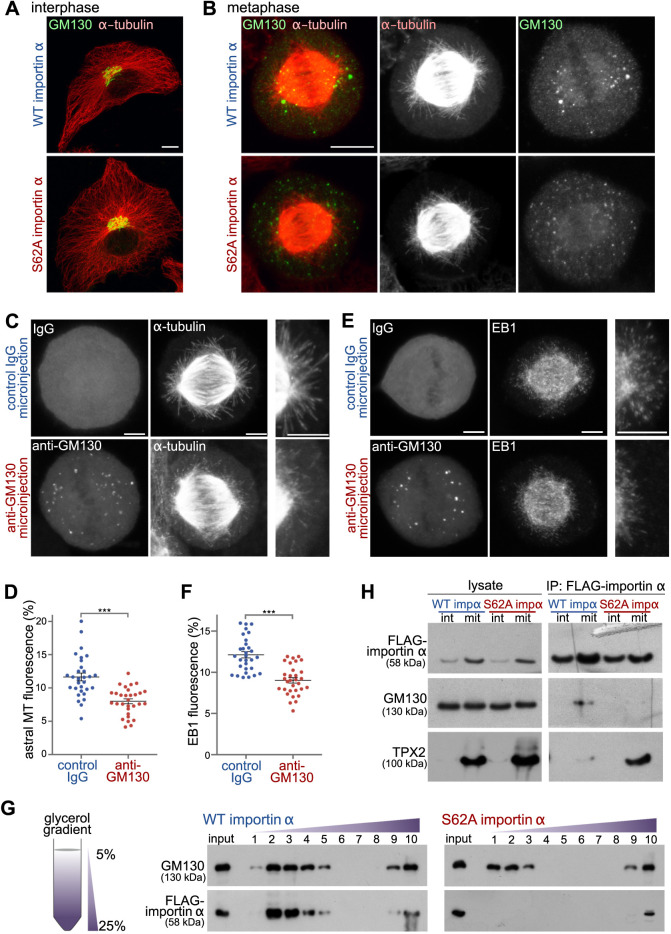
Upon mitotic entry, the Golgi ribbon is disassembled into a collection of vesicles and clusters of membranes (termed mitotic Golgi clusters; Shima et al., 1997). By metaphase, these mitotic Golgi clusters are concentrated around the two spindle poles in the vicinity of astral microtubule filaments (Jokitalo et al., 2001). Competition for importin α binding by GM130 on Golgi membranes activates the microtubule initiation role of TPX2 (Wei et al., 2015). Therefore, we tested whether the mitotic Golgi clusters can initiate astral microtubule formation through TPX2 and if, in turn, the astral microtubules are important for proper localization of mitotic Golgi clusters. We found that expression of WT or S62A-mutant importin α in interphase NRK cells did not affect the morphology or localization of the Golgi ribbon in the perinuclear region of those cells (Fig. 6A). However, during mitosis, we observed that expression of importin α S62A not only diminished astral microtubule mass (as shown in Fig. 4), but also caused increased dispersal of mitotic Golgi clusters (Fig. 6B).
Fig. 6.
GM130 regulates astral microtubule growth via phosphorylated importin α. (A) Importin α S62A does not alter Golgi structure or the microtubule network during interphase. Interphase NRK cells expressing WT importin α or importin α S62A were fixed and stained for α-tubulin and the Golgi membrane protein GM130. Maximum intensity projections are shown. Scale bar: 10 µm. (B) Importin α S62A delocalizes mitotic Golgi clusters from the spindle pole area. NRK cells expressing WT importin α or importin α S62A were arrested at metaphase using MG132, and then fixed and stained for GM130 and α-tubulin. Maximum intensity projections are shown. Scale bar: 10 µm. (C,E) Blocking the GM130-importin α interaction inhibits astral microtubule growth and delocalizes Golgi clusters from the spindle poles. NRK cells were arrested at metaphase for 1 h and microinjected with inhibitory antibodies against GM130 or with control IgG. Injected cells were incubated for another 30 min at 37°C, followed by cold treatment for 5 min. They were then allowed to regrow microtubules for 10 min, before fixation and staining for α-tubulin (C), EB1 (E) and injected IgG. Maximum intensity projections are shown. Enlargements of the astral microtubule area are shown in the panels on the right. Scale bars: 5 µm. (D,F) Quantitation of astral microtubule (MT) fluorescence (D) and astral EB1 fluorescence (F) from the experiments shown in C and E, respectively. Data are presented as mean±s.e.m. n>30. ***P<0.0001 (Student's t-test). (G) Importin α S62A does not associate with mitotic Golgi membranes. Post-chromosomal supernatant of mitotic HeLa cells expressing FLAG-tagged WT importin α or importin α S62A was centrifuged through a linear 5–25% glycerol gradient to separate membrane components by size (left). Ten fractions were collected from the top. The membranes from each fraction were collected by centrifugation and then analyzed by western blotting of the indicated proteins (right). (H) Phosphorylation of importin α shifts its binding preference from TPX2 to GM130. Interphase (int) or mitotic (mit) HeLa cells expressing FLAG-tagged WT importin α or importin α S62A were lysed and incubated with anti-FLAG antibody-conjugated beads (IP, immunoprecipitation). Beads were then washed with lysis buffer and analyzed by western blotting for the indicated proteins. Data in G and H are representative of three experiments.
We then sought to acutely interfere with the microtubule initiation capacity of the mitotic Golgi clusters. The microtubule initiation activity of GM130 can be blocked by microinjection of inhibitory anti-GM130 antibodies (Wei et al., 2015). We microinjected affinity-purified anti-GM130 antibodies or control IgG into MG132-arrested metaphase NRK cells. Then, 30 min after the injection, we depolymerized astral microtubules by cold treatment, before shifting the cells to 37°C for 10 min to allow filament regrowth (Fig. 6C). The cells were then stained for the injected antibodies (to discriminate cells), as well as for tubulin and EB1. Blocking GM130 activity phenocopied the effects of AurkinA treatment (Fig. 5) by attenuating astral microtubules (Fig. 6C,D), as well as their growth dynamics as revealed by reduced EB1 labeling (Fig. 6E,F). Consistent with the effect of mutant importin α S62A expression, we further noted from the staining patterns of injected GM130 antibodies that Golgi clusters were less concentrated at the spindle pole area (Fig. 6C,E). These results suggest that the GM130–TPX2 pathway activates astral microtubule assembly and is enabled by importin α phosphorylation.
Importin α is recruited to Golgi membranes during mitosis via GM130 (Wei et al., 2015). If mitotic phosphorylation of importin α is required for binding to GM130, then importin α S62A should not associate with mitotic Golgi membranes. We tested this possibility using velocity gradient centrifugation to separate smaller mitotic Golgi vesicles from larger membranes (Jesch et al., 2001; Seemann et al., 2002). HeLa cells arrested in mitosis by means of the Eg5 kinesin (KIF11) inhibitor S-trityl-L-cysteine were collected by shake-off and mechanically lysed using a ball bearing homogenizer. The post-chromosomal supernatant was then centrifuged through a linear 5–25% glycerol gradient, and the membranes of collected fractions were immunoblotted for GM130 and FLAG-tagged importin α (Fig. 6G). Consistent with our previous report (Wei et al., 2015), WT importin α co-migrated with GM130 in the top fractions of the gradient, demonstrating that importin α had been recruited to mitotic Golgi membranes. In contrast, phospho-deficient importin α S62A did not associate with mitotic Golgi membranes in the top fractions of the gradient. We also observed that the GM130-containing membrane fractions were shifted upwards to the lighter membranes in the gradient. Some of the importin α remained in the bottom fraction of the gradient comprising the bulk of the cellular membranes, consistent with previous reports indicating that importin α also associates with the nuclear envelope and the plasma membrane during mitosis (Hachet et al., 2004; Brownlee and Heald, 2019).
To further assess the phosphorylation dependency of the importin α–substrate interaction, we pulled down FLAG-tagged WT or S62A importin α from mitotic lysates and probed for association of endogenous GM130 and TPX2 by means of immunoblotting (Fig. 6H). The results showed that TPX2 binds much more strongly to importin α S62A than to phosphorylated WT importin α. Conversely, binding of GM130 to phosphorylated WT importin α was greatly enhanced relative to binding of GM130 to phospho-deficient importin α S62A. These results provide evidence that phosphorylation of importin α shifts its substrate preference from TPX2 to GM130, allowing GM130 on mitotic Golgi membranes to sequester importin α from TPX2 and activate Aurora A kinase to regulate astral microtubules and spindle orientation.
DISCUSSION
Here, we report that TPX2 activation regulates astral microtubule dynamics and is essential for correct spindle orientation (Fig. 7). Upon mitotic entry, phosphorylation of importin α at S62 by CDK1 induces a switch in substrate preference from TPX2 to GM130 and thereby enables GM130 to activate TPX2 through direct competition. Phospho-deficient importin α S62A mutant protein prevents GM130 from activating TPX2, reducing astral microtubule mass and causing spindle misorientation. Furthermore, the characteristic clustering of mitotic Golgi membranes at the spindle poles is astral microtubule-dependent and requires TPX2 activation.
Fig. 7.
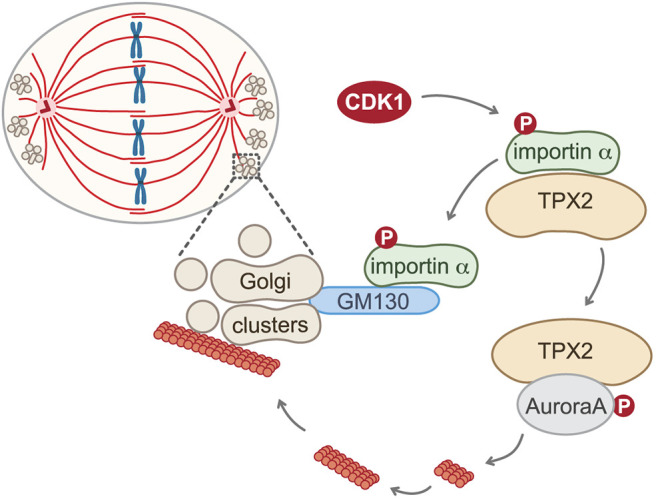
Model of the TPX2-mediated astral microtubule assembly pathway. Phosphorylation (P) of importin α at S62 by CDK1 switches its substrate preference from TPX2 to GM130. This switch enables GM130 on the mitotic Golgi clusters at the spindle poles to locally sequester importin α from TPX2 through direct competition. TPX2 is thereby relieved from inhibition, allowing it to bind to and activate Aurora A kinase to drive astral microtubule assembly and facilitate spindle orientation.
Spindle orientation is important in determining cell fate and function. For example, in mammalian skin epidermal cells, the orientation of the spindle defines the division plane and, consequently, cell fate (Barr and Gruneberg, 2017). Embryonic epidermal progenitor cells initially proliferate in a single layer by dividing parallel to the basement membrane. During stratification, the cells may also divide perpendicular to the basement membrane, generating one daughter cell that remains proliferative in the basal layer and another that is destined for terminal differentiation. Therefore, spindle orientation must be tightly controlled, as defects in spindle orientation trigger unbalanced growth and differentiation, outcomes linked to hyperproliferation and cancer (Ragkousi and Gibson, 2014; Asare et al., 2017). Similarly, aberrant spindle orientation due to impairment of astral microtubule–cortex connectivity in Drosophila alters the balance between neural progenitor expansion versus differentiation, leading to defects in neuroblast homeostasis (Cabernard and Doe, 2009). Moreover, knockdown of ASPM (abnormal spindle-like microcephaly-associated), which directly contributes to the growth of astral microtubules, causes abnormal spindle orientation in neuroepithelial progenitor cells, and mutation of ASPM is the most common defect observed in human autosomal recessive primary microcephaly (Fish et al., 2006; Gai et al., 2016). Interestingly, similar defects have been reported for GM130 mutations in vivo. Knockdown of gm130 in zebrafish leads to microcephaly (Shamseldin et al., 2016). The Purkinje cells of GM130-deficient mice exhibit neurodegeneration (Liu et al., 2017), and a patient with a homozygous loss-of-function mutation of GM130 developed neuromuscular disorder including progressive microcephaly (Shamseldin et al., 2016). These phenotypes have been broadly assigned to defective secretory pathways in which GM130 functions as a vesicle tethering factor during interphase. However, our present work shows that GM130 regulates spindle orientation, suggesting that deficiencies in GM130 mitotic function might further contribute to these neuronal disorders through impaired cell division and development.
Ran-dependent TPX2 activation takes place at the chromosomes. An analysis of RanGTP distribution in mitotic cells detected a low concentration of RanGTP in the astral area where Ran-mediated activation of TPX2 is unlikely to happen (Kaláb et al., 2006). Therefore, initiation and growth of astral microtubules has often been attributed solely to the centrosomes and microtubule-stabilizing proteins. Our findings have identified TPX2 as a key molecule in promoting astral microtubule growth. The direct competition-based activation of TPX2 on Golgi clusters does not depend on RanGTP and it facilitates microtubule organization in a distinct spatiotemporal fashion. This competition-based activation of TPX2 is enabled through S62 phosphorylation of importin α, which weakens the interaction of importin α with TPX2 and simultaneously enhances its binding to GM130. Similarly, it was recently reported that phosphorylation of importin α might also help in the RanGTP-dependent release of spindle assembly factors and their loading onto the mitotic spindle (Guo et al., 2019). However, the dominant-negative effect of S62A importin α on microtubule formation we observed here seems to be restricted to astral microtubules. A universal effect on microtubule formation and spindle assembly would require that the canonical Ran-dependent pathway activating TPX2 in the chromosomal area is equally regulated and affected by importin α. In other words, RanGTP generated around the chromatin would fail to dissociate the ternary complex of TPX–importin α–importin β, which should phenocopy TPX2 RNAi treatment, which blocks cells in mitosis with asters that fail to establish a bipolar spindle (Garrett et al., 2002; Gruss et al., 2002). However, upon importin α S62A expression, we found that bipolar spindles still assembled, the spindle length was not affected and the cells still progressed through the metaphase checkpoint. These observations suggest that TPX2 silenced by importin α S62A can still be relieved and activated by RanGTP around the chromatin, at least sufficiently to generate spindle microtubules that satisfy requirements for the spindle assembly checkpoint.
Our results also indicate that phosphorylation-deficient importin α S62A inhibits TPX2 from being activated by GM130 at the spindle poles, leading to a diminished astral microtubule mass that, in turn, misorients the spindle. Because importin α modulates several mitotic regulators, other mitotic events could also be affected by importin α S62A, as indicated by the cytokinesis defects we report here. Indeed, it was recently revealed that overexpression of an importin α T9A/S62A mutant protein increases its association with the spindle assembly factors NUMA1, TPX2 and KIF1C, and that this correlates with shortened spindles, a reduced microtubule mass and a delay in mitotic progression (Guo et al., 2019). Whether the observed spindle defects were directly caused by differential binding of mutant importin α T9A/S62A to spindle assembly factors awaits further investigation. We also noticed a slight delay in the duration of mitosis upon importin α S62A expression (Fig. 2B), but we did not observe shortened spindles or a noticeable reduction in spindle microtubule mass. The additional phenotypes reported by Guo et al. (2019) may be attributable to the double mutation (T9A/S62A) of importin α employed in that study, with the combination of T9A and S62A mutation potentially affecting protein–protein interactions or causing additional functional changes in importin α. We did not detect any such spindle defects in our experiments using the single S62A mutant. Instead, we found that spindle length was unaffected (as measured in Fig. 3D) and that spindle microtubules formed normally. Moreover, we detected a specific effect on astral microtubule formation as well as spindle orientation.
The spindle misorientation induced by importin α S62A likely mainly arises from compromised astral microtubule dynamics. We did not record spindle oscillation, which has often been reported for cells with spindle–cell cortex junction defects (Kotak and Gönczy, 2014; Petry, 2016). The centrosomal protein ASPM targets the citron kinase CITK (also known as CIT) to the spindle poles, where it specifically regulates astral microtubule dynamics (Gai et al., 2016). In ASPM-knockdown cells, spindle length is unchanged and spindle oscillation is not observed. However, the metaphase-to-anaphase transition is delayed by five minutes in such cells, and the spindle is misoriented (Gai et al., 2016). Expression of importin α S62A essentially phenocopied the results of ASPM knockdown. Therefore, it is reasonable to conclude that importin α regulates astral microtubule growth by promoting astral microtubule nucleation from the Golgi clusters at the pole area. Moreover, our acute inhibition of the GM130–TPX2 pathway diminished astral microtubule mass, prompting us to conclude that the GM130–TPX2 pathway is an important element of the mechanism controlling astral microtubules.
In humans there are seven importin α proteins, which are classified into three subfamilies: α1 and α8 (KPNA7); α3 (KPNA4) and α4 (KPNA3); and α5 (KPNA1), α6 (KPNA5) and α7 (KPNA6) (Miyamoto et al., 2016). The CDK1 consensus motif at S62 is restricted to importin α1 and α6 but is absent from the other importin α proteins (Fig. S1B). In contrast to importin α1, importin α6 is only expressed in testis (Köhler et al., 1997) and its phosphorylation in mitotic tissue culture cells has not been reported in proteomics studies (Dephoure et al., 2008; Ly et al., 2017; Nousiainen et al., 2006; Olsen et al., 2006). Thus, CDK1-mediated phosphorylation appears to specifically regulate importin α1, representing the most conserved isoform and the key regulator of mitotic spindle assembly factors (Pumroy and Cingolani, 2015; Guo et al., 2019).
Apart from the Golgi, other membrane components have been reported to influence astral microtubule dynamics (Barr and Gruneberg, 2017). Recycling endosomes can promote microtubule growth in mitotic extracts, and knockdown of the recycling endosomal protein RAB11 results in compromised astral microtubules and rotating spindles (Hehnly and Doxsey, 2014). Based on those findings, it has been proposed that the poleward movement of recycling endosomes transports microtubule-nucleating material like γ-tubulin and GCP4 (TUBGCP4) protein to the centrosomes and facilitates the polymerization of astral microtubules. Peroxisomes also play a role in orienting the spindle. Knockdown of peroxisomal protein PEX11B results in diminished cell cortex localization of NUMA1, contributing to spindle oscillation and misorientation (Asare et al., 2017). Here, we report mitotic Golgi clusters as being membranes that coordinate spindle orientation. We showed in a previous study that the association between mitotic Golgi membranes and the spindle is required for a single Golgi ribbon to reform upon mitotic exit (Wei and Seemann, 2009b). In the current study, we have shown that the association of mitotic Golgi clusters with astral microtubules depends on the Golgi-resident membrane protein GM130. When we blocked GM130-triggered TPX2 activation by microinjecting inhibitory antibodies or expressed importin α S62A, the Golgi clusters were delocalized from the spindle poles and dispersed. Although it is still not clear how mitotic Golgi clusters are organized in vivo, our results suggest that the astral microtubule network plays an integral role in maintaining the morphology and localization of the mitotic Golgi clusters at the spindle poles.
Post-translational modifications of importin-family proteins affect their binding affinity to substrates, influencing the speed of cargo transport into the nucleus during interphase. Importin α is repurposed as a spindle modulator during mitosis, but it had not been clear how it specifically recognizes spindle assembly factors at this time given the amount of NLS-containing proteins that are released into the cytoplasm upon nuclear envelope breakdown (Giesecke and Stewart, 2010). Our study shows that CDK1-mediated and mitotic-specific phosphorylation of importin α at S62 shifts its binding preference from TPX2 to GM130. Since importin α regulates other proteins that play important roles in mitosis, phosphorylation of importin α potentially also modulates their activities. In addition to tilted spindles in metaphase, we observed cytokinesis deficiencies in S62A importin α-expressing cells (Fig. 2), which might be caused by other clients of importin α that act in different steps of mitosis. Indeed, it was recently reported that overexpression of a GFP-tagged T9A/S62A double mutant of importin α results in decreased binding to the spindle assembly factors TPX2, NUMA1 and KIFC1, which is correlated with various spindle defects (Guo et al., 2019). However, the contribution of T9 and/or S62 to the underlying mechanism needs to be further dissected. Furthermore, another recent report based on the crystal structure of non-phosphorylated importin α complexed with the N-terminal fragment of GM130 indicates that, unlike other NLS sequences, GM130 competes with TPX2 for the minor binding site of the NLS-binding pockets in importin α, thereby preventing other NLS-containing cargos from activating TPX2 by competitive inhibition (Chang et al., 2019). A more thorough study on phosphorylation-induced conformational change should help further establish the exact mechanism by which importin α is regulated and repurposed throughout the cell cycle.
MATERIALS AND METHODS
Cell culture and drug treatments
HEK293T (ATCC), HeLa (ATCC), normal rat kidney (NRK) (ATCC) and SV589 (SV40 immortalized human fibroblasts; Yamamoto et al., 1984) cells were cultured at 37°C and 5% CO2 in complete growth medium [Dulbecco's modified Eagle's medium (Mediatech) supplemented with 10% cosmic calf serum (HyClone), 100 units/ml penicillin and 100 µg/ml streptomycin]. Cells were routinely monitored for contamination by phase-contrast microscopy and by staining with Hoechst. NRK cells were synchronized at the G1-S phase transition by 16 h treatment with 2 mM thymidine (Chem-Impex). Then, we washed out the thymidine and added 24 µM 2′-deoxycytidine (Chem-Impex). Five hours after release from thymidine, we added 10 µM MG132 (Boston Biotech) for 1 h to arrest cells in metaphase. HeLa cells and HeLa cell lines expressing FLAG-tagged WT or S62A importin α were synchronized in mitosis by treatment with 20 µM S-trityl-L-cysteine (Acros) for 16 h. The mitotic cells were then collected by shake-off, followed by centrifugation. We employed 200 µM AurkinA (Aobious) in complete culture medium to block the association of TPX2 with Aurora A kinase.
Plasmids and cell lines
The cDNA of human importin α1 (KPNA2) from pGEX TEV-KPNA2 (received from Yuh Min Chook, UT Southwestern Dallas, USA) was used to generate the S62A mutation by quick-change PCR mutagenesis. WT importin α1 or the S62A point mutant was then cloned into pET30a (Novagen) for His–importin α recombinant protein purification. Importin α with an N-terminal FLAG tag was cloned into pLVX-TetOne-Puro (Clontech) to generate stable cell lines. pLVX-FLAG-KPNA2 WT or S62A was co-transfected with psPAX and pVSVG (Addgene) into HEK293T cells to produce lentivirus, which was then used to transduce NRK and HeLa cells. Two days later, we added 5 μg/ml puromycin (RPI) and stable clones were selected for 7 days. Individual colonies were picked, and single clones were isolated by limited dilution. Expression of FLAG–importin α WT or S62A was induced for 16 h by addition of 1 µg/ml doxycycline (Sigma).
Antibodies
For western blotting, we used mouse monoclonal antibodies against importin α1 (BD Transduction Lab, cat. #610485, 25 ng/ml) and GAPDH (GA1R, Invitrogen, cat. #MA5-15738, 20 ng/ml), as well as rabbit polyclonal antibodies against phospho-histone H3 (Ser10) (Millipore, cat. #06-570, 25 ng/ml), TPX2 (Proteintech, cat. #11741-1-AP, 150 ng/ml), FLAG-tag (Sigma, cat. #F7425, 100 ng/ml), and GRASP55 (serum 1:5000; Wei and Seemann, 2009b). For immunofluorescence, we used mouse monoclonal antibodies against EB1 (BD Transduction Lab, cat. #61534, 1 μg/ml), α-tubulin (TAT1; supernatant 1:200; Woods et al., 1989), GM130 (NN2C10, 5 µg/ml; used in Fig. 6A,B; Seemann et al., 2000) and TPX2 (BioLegend, cat. #628001, 5 µg/ml); rabbit polyclonal antibodies against γ-tubulin (Sigma, cat. # T3559, 1:2000); as well as rat monoclonal antibody against α-tubulin (Chemicon, cat. #MAB1860, 5 µg/ml). For microinjection, we used affinity-purified rabbit anti-GM130 (Wei et al., 2015) and rabbit IgG (Sigma) as control. Secondary antibodies were as follows: Alexa Fluor 488- or Alexa Fluor 594-conjugated to highly cross-adsorbed goat anti-mouse or goat anti-rabbit IgG (Invitrogen), Alexa Fluor 568-conjugated to goat anti-rat IgG (Invitrogen), HRP-conjugated goat anti-rabbit (Jackson ImmunoResearch) and HRP-conjugated goat anti-mouse IgG (Invitrogen).
Immunofluorescence
For the astral microtubule regrowth assay, we replaced the medium with 4°C complete growth medium for 5 min to depolymerize astral microtubules and then changed it back to complete growth medium at 37°C and continued incubating the cells at 37°C for 10 min to allow microtubule regrowth. To stain for aster microtubules, the cells were fixed for 20 min at 37°C in microtubule-stabilizing fixative (3.7% formaldehyde, 0.1% glutaraldehyde, 60 mM PIPES, 25 mM HEPES, 2 mM MgCl2 and 10 mM EGTA, pH 6.9). The reaction was quenched for 5 min with sodium borohydride in 20 mM Tris-HCl pH 7.4 and 150 mM NaCl, washed three times with TBS-Tx (20 mM Tris-HCl pH 7.4, 150 mM NaCl and 0.1% Triton X-100), and blocked with 1 mg/ml BSA in TBS-Tx at room temperature for 30 min. For all other immunofluorescence stainings, the cells were fixed and permeabilized for 15 min in methanol at −20°C, then incubated with respective antibodies at 37°C for 30 min, followed by Alexa Fluor-conjugated secondary antibodies. DNA was stained for 10 min at room temperature with 1 µg/ml Hoechst 33342 (Invitrogen) in phosphate-buffered saline (PBS) and cells were embedded in Mowiol (Calbiochem) mounting solution (Wei and Seemann, 2009a).
Microscopy and quantitation
Confocal microscopy was performed using a Zeiss LSM780 inverted microscope in combination with a Plan-Apochromat 63×/1.4 objective. Z-sections were captured at 0.5 µm intervals, and maximum intensity projections are presented. Spindle angle was calculated using the formula α=arctan (b/a), where b=0.5 μm multiplied by the number of z sections between the spindle poles and a=the distance between the projections of the two spindle poles on the x–y plane. Spindle length (l) was calculated as l2=a2+b2.
Relative fluorescence of astral microtubules was measured in ImageJ (as depicted in Fig. 4B; NIH, Bethesda, MD). A line was drawn through the spindle pole perpendicular to the spindle pole-to-pole axis. The fluorescence signal of tubulin staining outside the interpolar spindle area was measured as the astral microtubule fluorescence. The percentage of the astral microtubule intensity was calculated as the fluorescence signal of the astral microtubules/total microtubule fluorescence signal of the cell. Astral microtubule length was determined by averaging the distance between the tip of the EB1 ‘comet’ and the pole for the three longest filaments from each of the two asters.
Phase-contrast time-lapse microscopy was performed using an Axiovert 200M microscope (Zeiss) with an LD-A-PLAN 20×/0.3 Ph1 objective (Zeiss) and a Retiga 2000R CCD camera (QImaging) using MetaMorph 7.1.3 software (Molecular Devices). NRK cells expressing WT importin α or importin α S62A released from thymidine blocking were changed to CO2-independent medium (Invitrogen), 10% cosmic calf serum (HyClone), 2 mM GlutaMax (Invitrogen) and 100 units/ml penicillin and 100 μg/ml streptomycin. Phase-contrast images were captured at 10 min intervals for 12 h at 37°C, and cell cycle progression was analyzed using MetaMorph 7.1.3 (Molecular Devices) and ImageJ. The duration from G1-S phase to mitosis was determined as being from 30 min after thymidine washout until rounding of the cells. The duration of mitosis was measured as the time from rounding of the cells to onset of cytokinesis.
Image analysis was performed using ImageJ 2.0. Statistical analyses were conducted using Prism 8.3 software (GraphPad). Data shown are from three or more independent experiments. Statistical significance was assessed by unpaired two-tailed Student's t-tests.
Microinjection
NRK cells grown on glass coverslips were microinjected using a FemtoJet system (Eppendorf) and a micromanipulator 5171 (Eppendorf) attached to an Eclipse TE300 inverted microscope (Nikon). The cells were switched to complete growth medium supplemented with 50 mM HEPES-KOH pH 7.4 before injections. The cells were microinjected with 2 mg/ml affinity-purified rabbit anti-GM130 or 2 mg/ml control IgG in H/K buffer (20 mM HEPES-KOH pH 7.4, 50 mM KOAc). Injected cells were incubated at 37°C for 30 min and then subjected to an astral regrowth assay. Cells were then fixed in −20°C methanol for EB1 staining or in microtubule-stabilizing fixative to stain for α-tubulin.
Co-immunoprecipitation
Interphase or mitotic HeLa cells expressing FLAG–importin α WT or S62A mutants were lysed for 20 min at 4°C in lysis buffer [10 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% TritonX-100, cOmplete protease inhibitor cocktail (Roche), 1 mM DTT, 10 mM β-glycerophosphate, 1 mM Na3VO4 and 10 mM NaF]. Lysates were cleared by centrifugation at 16,000 g for 10 min at 4°C and then incubated with 10 µl anti-FLAG M2-Sepharose beads (Sigma) for 60 min at 4°C. Beads were then washed three times with lysis buffer and boiled in SDS sample buffer to elute proteins for western blotting analysis. For mass spectrometry, FLAG–importin α purified from mitotic and interphase HeLa cells was eluted from the beads using 300 µg/ml 3×FLAG-tag peptide (Apexbio). The 58-kDa bands were cut from the gel, digested with trypsin and analyzed by using a short reverse-phase LC-MS/MS on a Orbitrap Fusion Lumos Mass Spectrometer (Thermo Fisher Scientific). Proteins and posttranslational modifications were identified using Proteome Discoverer 2.4 software (Thermo Fisher Scientific).
Subcellular fractionation
Mitotic HeLa cells expressing WT importin α or importin α S62A were washed with PBS followed by homogenization buffer (20 mM HEPES pH 7.4, 50 mM NaCl). The cells were then mechanically lysed using a ball bearing homogenizer with a clearance of 12 μm (Isobiotec) in homogenization buffer containing cOmplete protease inhibitor cocktail (Roche), 1 mM DTT, 10 mM β-glycerophosphate, 1 mM Na3VO4 and 10 mM NaF. The post-chromosomal supernatant (PCS) was collected by centrifugation for 5 min at 1000 g at 4°C, loaded onto a continuous gradient of 5–25% (v/v) glycerol in 20 mM HEPES-KOH pH 7.4, and centrifuged for 30 min in a SW55Ti rotor (Beckman) at 35,000 rpm (149,000 g) at 4°C. Ten 0.5 ml fractions were collected from the top of the gradient, diluted with 1 ml of 20 mM HEPES-KOH pH 7.4, and membranes were collected by centrifugation for 30 min at 100,000 g at 4°C. The supernatant was discarded, and the membrane pellets were resuspended in SDS sample buffer for further analysis by western blotting.
Protein purification
BL21(DE3) competent E. coli were transformed with pET30a-importin α (WT or S62A). Single clones were inoculated into 20 ml LB medium and grown overnight at 37°C. The 20 ml cultures were inoculated into 1000 ml LB to allow the bacteria to grow to OD600=0.6–0.8. Protein expression was induced by addition of 0.2 mM IPTG and incubation for 18 h at 16°C. Bacteria were collected by centrifugation, resuspended in high salt buffer [HSB; PBS supplemented with 150 mM NaCl, cOmplete protease inhibitor cocktail (Roche), 1 mM DTT and 1 mM PMSF] and lysed using a French press. Nickel-NTA agarose beads (Qiagen) were used to purify His–importin α (1 ml slurry for 1000 ml culture). The beads were then washed with HSB and directly used for in vitro phosphorylation (5 μl slurry per experiment).
In vitro phosphorylation assay
Mitotic HeLa cells were lysed for 20 min at 4°C in lysis buffer [10 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% TritonX-100, cOmplete protease inhibitor cocktail (Roche), 1 mM DTT, 10 mM β-glycerophosphate, 1 mM Na3VO4 and 10 mM NaF], and then the lysates were cleared by centrifugation at 16,000 g for 10 min at 4°C. Nickel beads coated with recombinant His-tagged importin α were incubated at 30°C for 30 min with pre-cleared mitotic lysate supplemented with 2 mM ATP pH 7.2. Beads were then washed three times with HBS, eluted by boiling in SDS sample buffer, and analyzed by Phos-tag biotin blotting. The CDK1 inhibitor purvalanol A (LC Labs) was used at a concentration of 0.5 mM.
Phos-tag gel and Phos-tag biotin
To detect importin α phosphorylation via in vitro phosphorylation assay, protein samples were separated by SDS–PAGE on a 10% gel and transferred to PVDF membrane. The PVDF membrane was then washed with TBST for 20 min before incubation with Phos-tag biotin solution [TBST supplemented with 6 µM ZnCl2, 2 µM Phos-tag Biotin (ApexBio) and 2 ng/ml streptavidin–HRP (Jackson ImmunoResearch)]. After 30 min incubation, the blot was developed by ECL detection.
To detect importin α phosphorylation on Phos-tag gels, mitotic or interphase cells were lysed for 20 min at 4°C in lysis buffer (without EDTA) and cleared by centrifugation at 16,000 g for 10 min at 4°C. For phosphatase treatment, 40 µl of the cleared lysates were incubated with 1 µl Lambda Protein Phosphatase (λ phosphatase) (New England Biolabs), 5 µl 10× Buffer for Protein MetalloPhosphatases (New England Biolabs), and 5 µl 10 mM MnCl2 for 30 min at 30°C. Proteins were separated on a 10% SDS gel containing 20 μM Phos-tag acrylamide (ApexBio) and 100 µM MnCl2. The gel was then washed for 1 h in transfer buffer containing 1 mM EDTA to remove the manganese. Proteins were subsequently transferred to PVDF membrane for immunoblotting.
Supplementary Material
Acknowledgements
We thank Gianni Guizzunti for insightful discussions and suggestions, the Proteomics Core Facility for mass spectrometry analysis and the Light Microscopy Facility at the University of Texas Southwestern Medical Center for imaging support.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: H.G., J.-H.W., J.S.; Methodology: H.G., Y.Z., J.-H.W., J.S.; Validation: H.G., Y.Z., J.S.; Formal analysis: H.G., Y.Z., J.S.; Investigation: H.G., J.-H.W., Y.Z., J.S.; Resources: H.G., J.-H.W. J.S.; Data curation: H.G., Y.Z., J.S.; Writing - original draft: H.G., J.S.; Writing - review & editing: H.G., J.-H.W., J.S.; Visualization: H.G., Y.Z., J.S.; Supervision: J.S.; Project administration: J.S.; Funding acquisition: J.S.
Funding
J.-H.W. is supported by an Academia Sinica Career Development Award (AS-CDA-109-L02). This work was supported by grants from the National Institutes of Health (GM096070 to J.S.) and the Welch Foundation (I-1910 to J.S.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.258356.supplemental
References
- Anderson, K., Yang, J., Koretke, K., Nurse, K., Calamari, A., Kirkpatrick, R. B., Patrick, D., Silva, D., Tummino, P. J., Copeland, R. A.et al. (2007). Binding of TPX2 to Aurora A alters substrate and inhibitor interactions. Biochemistry 46, 10287-10295. 10.1021/bi7011355 [DOI] [PubMed] [Google Scholar]
- Asare, A., Levorse, J. and Fuchs, E (2017). Coupling organelle inheritance with mitosis to balance growth and differentiation. Science 355, eaah4701 10.1126/science.aah4701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr, F. A. and Gruneberg, U (2017). Organelle inheritance—what players have skin in the game? Science 355, 458-459. 10.1126/science.aam6381 [DOI] [PubMed] [Google Scholar]
- Baudoin, N. C. and Cimini, D (2018). A guide to classifying mitotic stages and mitotic defects in fixed cells. Chromosoma 127, 215-227. 10.1007/s00412-018-0660-2 [DOI] [PubMed] [Google Scholar]
- Bayliss, R., Sardon, T., Vernos, I. and Conti, E (2003). Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol. Cell 12, 851-862. 10.1016/S1097-2765(03)00392-7 [DOI] [PubMed] [Google Scholar]
- Bergstralh, D. T., Dawney, N. S. and St Johnston, D (2017). Spindle orientation: a question of complex positioning. Development 144, 1137-1145. 10.1242/dev.140764 [DOI] [PubMed] [Google Scholar]
- Brownlee, C. and Heald, R (2019). Importin α partitioning to the plasma membrane regulates intracellular scaling. Cell 176, 805-815.e8. 10.1016/j.cell.2018.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabernard, C. and Doe, C. Q (2009). Apical/basal spindle orientation is required for neuroblast homeostasis and neuronal differentiation in Drosophila. Dev. Cell 17, 134-141. 10.1016/j.devcel.2009.06.009 [DOI] [PubMed] [Google Scholar]
- Cantin, G. T., Yi, W., Lu, B., Park, S. K., Xu, T., Lee, J.-D. and Yates, J. R (2008). Combining protein-based IMAC, peptide-based IMAC, and MudPIT for efficient phosphoproteomic analysis. J. Proteome Res. 7, 1346-1351. 10.1021/pr0705441 [DOI] [PubMed] [Google Scholar]
- Carazo-Salas, R. E., Guarguaglini, G., Gruss, O. J., Segref, A., Karsenti, E. and Mattaj, I. W (1999). Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature 400, 178-181. 10.1038/22133 [DOI] [PubMed] [Google Scholar]
- Chang, C.-C., Chen, C.-J., Grauffel, C., Pien, Y.-C., Lim, C., Tsai, S.-Y. and Hsia, K.-C (2019). Ran pathway-independent regulation of mitotic Golgi disassembly by Importin-α. Nat. Commun. 10, 4307 10.1038/s41467-019-12207-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure, N., Zhou, C., Villén, J., Beausoleil, S. A., Bakalarski, C. E., Elledge, S. J. and Gygi, S. P (2008). A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA 105, 10762-10767. 10.1073/pnas.0805139105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish, J. L., Kosodo, Y., Enard, W., Pääbo, S. and Huttner, W. B (2006). Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc. Natl. Acad. Sci. USA 103, 10438-10443. 10.1073/pnas.0604066103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai, M., Bianchi, F. T., Vagnoni, C., Vernì, F., Bonaccorsi, S., Pasquero, S., Berto, G. E., Sgrò, F., Chiotto, A. M., Annaratone, L.et al. (2016). ASPM and CITK regulate spindle orientation by affecting the dynamics of astral microtubules. EMBO Rep. 17, 1396-1409. 10.15252/embr.201541823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett, S., Auer, K., Compton, D. A. and Kapoor, T. M (2002). hTPX2 is required for normal spindle morphology and centrosome integrity during vertebrate cell division. Curr. Biol. 12, 2055-2059. 10.1016/S0960-9822(02)01277-0 [DOI] [PubMed] [Google Scholar]
- Garrido, G. and Vernos, I (2016). Non-centrosomal TPX2-dependent regulation of the aurora a kinase: functional implications for healthy and pathological cell division. Front. Oncol. 6, 88 10.3389/fonc.2016.00088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesecke, A. and Stewart, M (2010). Novel binding of the mitotic regulator TPX2 (target protein for Xenopus kinesin-like protein 2) to importin-alpha. J. Biol. Chem. 285, 17628-17635. 10.1074/jbc.M110.102343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, Y., Mo, C. and Fraser, S. E (2004). Planar cell polarity signalling controls cell division orientation during zebrafish gastrulation. Nature 430, 689-693. 10.1038/nature02796 [DOI] [PubMed] [Google Scholar]
- Gruss, O. J., Wittmann, M., Yokoyama, H., Pepperkok, R., Kufer, T., Silljé, H., Karsenti, E., Mattaj, I. W. and Vernos, I (2002). Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat. Cell Biol. 4, 871-879. 10.1038/ncb870 [DOI] [PubMed] [Google Scholar]
- Guo, L., Mohd, K. S., Ren, H., Xin, G., Jiang, Q., Clarke, P. R. and Zhang, C (2019). Phosphorylation of importin-α1 by CDK1-cyclin B1 controls mitotic spindle assembly. J. Cell Sci. 132, jcs232314 10.1242/jcs.232314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet, V., Köcher, T., Wilm, M. and Mattaj, I. W (2004). Importin alpha associates with membranes and participates in nuclear envelope assembly in vitro. EMBO J. 23, 1526-1535. 10.1038/sj.emboj.7600154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehnly, H. and Doxsey, S (2014). Rab11 endosomes contribute to mitotic spindle organization and orientation. Dev. Cell 28, 497-507. 10.1016/j.devcel.2014.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeček, M., Rossmann, M., Sharma, P., Emery, A., Huggins, D. J., Stockwell, S. R., Stokes, J. E., Tan, Y. S., Almeida, E. G., Hardwick, B.et al. (2016). Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2. Sci. Rep. 6, 28528 10.1038/srep28528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesch, S. A., Mehta, A. J., Velliste, M., Murphy, R. F. and Linstedt, A. D (2001). Mitotic Golgi is in a dynamic equilibrium between clustered and free vesicles independent of the ER. Traffic 2, 873-884. 10.1034/j.1600-0854.2001.21203.x [DOI] [PubMed] [Google Scholar]
- Jokitalo, E., Cabrera-Poch, N., Warren, G. and Shima, D. T (2001). Golgi clusters and vesicles mediate mitotic inheritance independently of the endoplasmic reticulum. J. Cell Biol. 154, 317-330. 10.1083/jcb.200104073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab, P. and Heald, R (2008). The RanGTP gradient - a GPS for the mitotic spindle. J. Cell. Sci. 121, 1577-1586. 10.1242/jcs.005959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaláb, P., Pralle, A., Isacoff, E. Y., Heald, R. and Weis, K (2006). Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature 440, 697-701. 10.1038/nature04589 [DOI] [PubMed] [Google Scholar]
- Köhler, M., Ansieau, S., Prehn, S., Leutz, A., Haller, H. and Hartmann, E (1997). Cloning of two novel human importin-alpha subunits and analysis of the expression pattern of the importin-alpha protein family. FEBS Lett. 417, 104-108. 10.1016/S0014-5793(97)01265-9 [DOI] [PubMed] [Google Scholar]
- Kotak, S. and Gönczy, P (2014). NuMA phosphorylation dictates dynein-dependent spindle positioning. Cell Cycle 13, 177-178. 10.4161/cc.27040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufer, T. A., Silljé, H. H. W., Körner, R., Gruss, O. J., Meraldi, P. and Nigg, E. A (2002). Human TPX2 is required for targeting Aurora-A kinase to the spindle. J. Cell Biol. 158, 617-623. 10.1083/jcb.200204155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler, T. and Fuchs, E (2005). Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature 437, 275-280. 10.1038/nature03922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C., Mei, M., Li, Q., Roboti, P., Pang, Q., Ying, Z., Gao, F., Lowe, M. and Bao, S (2017). Loss of the golgin GM130 causes Golgi disruption, Purkinje neuron loss, and ataxia in mice. Proc. Natl. Acad. Sci. USA 114, 346-351. 10.1073/pnas.1608576114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, M., Gonatas, N. K. and Warren, G (2000). The mitotic phosphorylation cycle of the cis-Golgi matrix protein GM130. J. Cell Biol. 149, 341-356. 10.1083/jcb.149.2.341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, M. S. and Johnston, C. A (2013). Molecular pathways regulating mitotic spindle orientation in animal cells. Development 140, 1843-1856. 10.1242/dev.087627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly, T., Whigham, A., Clarke, R., Brenes-Murillo, A. J., Estes, B., Madhessian, D., Lundberg, E., Wadsworth, P. and Lamond, A. I (2017). Proteomic analysis of cell cycle progression in asynchronous cultures, including mitotic subphases, using PRIMMUS. Elife 6, e27574 10.7554/eLife.27574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangal, S., Sacher, J., Kim, T., Osório, D. S., Motegi, F., Carvalho, A. X., Oegema, K. and Zanin, E (2018). TPXL-1 activates Aurora A to clear contractile ring components from the polar cortex during cytokinesis. J. Cell Biol. 217, 837-848. 10.1083/jcb.201706021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascanzoni, F., Ayala, I. and Colanzi, A (2019). Organelle inheritance control of mitotic entry and progression: implications for tissue homeostasis and disease. Front. Cell Dev. Biol. 7, 133 10.3389/fcell.2019.00133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, Y., Yamada, K. and Yoneda, Y (2016). Importin α: a key molecule in nuclear transport and non-transport functions. J. Biochem. 160, 69-75. 10.1093/jb/mvw036 [DOI] [PubMed] [Google Scholar]
- Naso, F. D., Sterbini, V., Crecca, E., Asteriti, I. A., Russo, A. D., Giubettini, M., Cundari, E., Lindon, C., Rosa, A. and Guarguaglini, G (2020). Excess TPX2 interferes with microtubule disassembly and nuclei reformation at mitotic exit. Cells 9,374 10.3390/cells9020374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousiainen, M., Silljé, H. H. W., Sauer, G., Nigg, E. A. and Körner, R (2006). Phosphoproteome analysis of the human mitotic spindle. Proc. Natl. Acad. Sci. USA 103, 5391-5396. 10.1073/pnas.0507066103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell, C. B. and Khodjakov, A. L (2007). Cooperative mechanisms of mitotic spindle formation. J. Cell. Sci. 120, 1717-1722. 10.1242/jcs.03442 [DOI] [PubMed] [Google Scholar]
- Olsen, J. V., Blagoev, B., Gnad, F., Macek, B., Kumar, C., Mortensen, P. and Mann, M (2006). Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635-648. 10.1016/j.cell.2006.09.026 [DOI] [PubMed] [Google Scholar]
- Petry, S (2016). Mechanisms of mitotic spindle assembly. Annu. Rev. Biochem. 85, 659-683. 10.1146/annurev-biochem-060815-014528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser, S. L. and Pelletier, L (2017). Mitotic spindle assembly in animal cells: a fine balancing act. Nat. Rev. Mol. Cell Biol. 18, 187-201. 10.1038/nrm.2016.162 [DOI] [PubMed] [Google Scholar]
- Pumroy, R. A. and Cingolani, G (2015). Diversification of importin-α isoforms in cellular trafficking and disease states. Biochem. J. 466, 13-28. 10.1042/BJ20141186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragkousi, K. and Gibson, M. C (2014). Cell division and the maintenance of epithelial order. J. Cell Biol. 207, 181-188. 10.1083/jcb.201408044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann, J., Jokitalo, E., Pypaert, M. and Warren, G (2000). Matrix proteins can generate the higher order architecture of the Golgi apparatus. Nature 407, 1022-1026. 10.1038/35039538 [DOI] [PubMed] [Google Scholar]
- Seemann, J., Pypaert, M., Taguchi, T., Malsam, J. and Warren, G (2002). Partitioning of the matrix fraction of the Golgi apparatus during mitosis in animal cells. Science 295, 848-851. 10.1126/science.1068064 [DOI] [PubMed] [Google Scholar]
- Shamseldin, H. E., Bennett, A. H., Alfadhel, M., Gupta, V. and Alkuraya, F. S (2016). GOLGA2, encoding a master regulator of golgi apparatus, is mutated in a patient with a neuromuscular disorder. Hum. Genet. 135, 245-251. 10.1007/s00439-015-1632-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima, D. T., Haldar, K., Pepperkok, R., Watson, R. and Warren, G (1997). Partitioning of the Golgi apparatus during mitosis in living HeLa cells. J. Cell Biol 137, 1211-1228. 10.1083/jcb.137.6.1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima, D. T., Cabrera-Poch, N., Pepperkok, R. and Warren, G (1998). An ordered inheritance strategy for the Golgi apparatus: visualization of mitotic disassembly reveals a role for the mitotic spindle. J. Cell Biol. 141, 955-966. 10.1083/jcb.141.4.955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, J.-H. and Seemann, J (2017). Golgi ribbon disassembly during mitosis, differentiation and disease progression. Curr. Opin. Cell Biol. 47, 43-51. 10.1016/j.ceb.2017.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, J.-H. and Seemann, J (2009a). Induction of asymmetrical cell division to analyze spindle-dependent organelle partitioning using correlative microscopy techniques. Nat. Protoc. 4, 1653-1662. 10.1038/nprot.2009.160 [DOI] [PubMed] [Google Scholar]
- Wei, J.-H. and Seemann, J (2009b). The mitotic spindle mediates inheritance of the Golgi ribbon structure. J. Cell Biol. 184, 391-397. 10.1083/jcb.200809090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, J.-H., Zhang, Z. C., Wynn, R. M. and Seemann, J (2015). GM130 Regulates golgi-derived spindle assembly by activating TPX2 and capturing microtubules. Cell 162, 287-299. 10.1016/j.cell.2015.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods, A., Sherwin, T., Sasse, R., MacRae, T. H., Baines, A. J. and Gull, K (1989). Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell. Sci. 93, 491-500. [DOI] [PubMed] [Google Scholar]
- Xiang, Y. and Wang, Y (2010). GRASP55 and GRASP65 play complementary and essential roles in Golgi cisternal stacking. J. Cell Biol. 188, 237-251. 10.1083/jcb.200907132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, T., Davis, C. G., Brown, M. S., Schneider, W. J., Casey, M. L., Goldstein, J. L. and Russell, D. W (1984). The human LDL receptor: a cysteine-rich protein with multiple Alu sequences in its mRNA. Cell 39, 27-38. 10.1016/0092-8674(84)90188-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.