Abstract
Filamentous fungi possess the capacity to produce a wide array of secondary metabolites with diverse biological activities and structures, such as lovastatin and swainsonine. With the advent of the post-genomic era, increasing amounts of cryptic or uncharacterized secondary metabolite biosynthetic gene clusters are continually being discovered. However, owing to the longstanding lack of versatile, comparatively simple, and highly efficient genetic manipulation techniques, the broader exploration of industrially important secondary metabolites has been hampered thus far. With the emergence of CRISPR/Cas9-based genome editing technology, this dilemma may be alleviated, as this advanced technique has revolutionized genetic research and enabled the exploitation and discovery of new bioactive compounds from filamentous fungi. In this review, we introduce the CRISPR/Cas9 system in detail and summarize the latest applications of CRISPR/Cas9-mediated genome editing in filamentous fungi. We also briefly introduce the specific applications of the CRISPR/Cas9 system and CRISPRa in the improvement of secondary metabolite contents and discovery of novel biologically active compounds in filamentous fungi, with specific examples noted. Additionally, we highlight and discuss some of the challenges and deficiencies of using the CRISPR/Cas9-based genome editing technology in research on the biosynthesis of secondary metabolites as well as future application of CRISPR/Cas9 strategy in filamentous fungi are highlighted and discussed.
Keywords: CRISPR-Cas9, genome editing, secondary metabolites, filamentous fungi, CRISPRa
Introduction
Filamentous fungi have a major impact on many aspects of human diets and health more broadly. Their benefits are derived from various mechanisms underlying the production of enzymes, organic acids, and flavors, as well as, more importantly, antibiotic compounds and therapeutic molecules (Satish et al., 2020). Moreover, they are well known as appealing microbial cell factories that possess the industrial capability to secrete a large repertoire of different bioactive secondary metabolites, such as paclitaxel and swainsonine, which have become important clinical therapeutics (Hautbergue et al., 2018; Keller, 2019; Mosunova et al., 2020). There has been a focus on research into secondary metabolites because some secondary metabolites can be used as anti-cancer drugs and anti-bacterial compounds (Lobanovska and Pilla, 2017). A majority of these secondary metabolites can be classified into three chemical categories: polyketides derived from acyl-CoAs, terpenes produced from acyl-CoAs, and small peptides derived from amino acids (Keller, 2019; Mosunova et al., 2020). Most of the genes involved in the biosynthesis of secondary metabolites are frequently clustered together on chromosomes in biosynthetic gene clusters (SM-BGCs), and some are not expressed under standard laboratory culture conditions (Lin et al., 2013; Jiang et al., 2015; Nah et al., 2017; Keller, 2019; Kjaerbolling et al., 2019). Another characteristic of genes in such clusters is that they are not constitutively expressed, and formerly actively expressed genes can become transcriptionally quiescent upon repeated culturing (Kale et al., 1994; Hoffmeister and Keller, 2007). Although secondary metabolite biosynthetic genes in filamentous fungi are generally found in clusters that provide a convenient genetic locus for manipulation (Hoffmeister and Keller, 2007), the exploitation of new bioactive compounds is hindered by intrinsic difficulties involving complex genetic backgrounds and poor efficiency of gene targeting. With increases in genomic insight and gene mining from high-throughput sequencing data and the advancement of genomics and transcriptomics, there has been an acceleration in the identification and utilization of SM-BGCs. Thus, elucidating the genetic basis and the biosynthetic pathways of secondary metabolites has become comparatively easier.
Filamentous fungi, such as Trichoderma reesei, Aspergillus niger, Aspergillus oryzae, and Aspergillus nidulans, are universally used as model eukaryotic microorganisms to produce industrial secondary metabolites. For example, researchers have utilized rice blast fungus Pyricularia oryzae to yield tenuazonic acid, Aspergillus fumigatus to secrete the natural product trypacidin, Fusarium fujikuroi to produce gibberellic acid, Fusarium heterosporum to synthesize the polyketide equisetin, A. nidulans to secrete microperfuranone, and Penicillium chrysogenum to biosynthesize sorbicillin (Gauthier et al., 2012; Kakule et al., 2015; Yun et al., 2017; Shi et al., 2019). However, while the genetic manipulation of secondary metabolites in filamentous fungi is being explored, there are several factors that limit genetic research on secondary metabolite biosynthetic pathways of filamentous fungi. First, filamentous fungi, like the model organism yeast, have complex genetic backgrounds when compared to prokaryotes (Song et al., 2019). Second, it is difficult to apply genetic manipulation and molecular biology tools in filamentous fungi. Furthermore, low homologous recombination efficiency (generally less than 5%) and a lack of suitable selection markers, and plasmids also hamper the exploitation of novel secondary metabolites from filamentous fungi (Mei et al., 2019).
More recently, a variety of genetic engineering methods, such as RNA interference, heterologous expression, gene-targeting strategies, and zinc finger nuclease (ZFN) and transcription-activator-like effector nuclease (TALEN) – based genome editing have been developed to explore and demonstrate the biosynthetic and regulatory mechanisms in filamentous fungi (Boettcher and McManus, 2015; Wang et al., 2017; Dasgupta et al., 2020). Although the existing approaches can be utilized to edit target genes at the genomic level, these do not meet the needs of industrial secondary metabolite production in filamentous fungi owing their low editing efficiency and cumbersome manipulation (Shi et al., 2017). Thus, the exploitation of new secondary metabolites with potential pharmaceutical applications in filamentous fungi is extremely challenging. Fortunately, the emergence of the clustered regularly interspersed short palindromic repeats (CRISPR)/associated protein (Cas) system in recent years has raised hopes of solving the problem presented by the largely inefficient gene editing tools available for use in filamentous fungi. The current CRISPR/Cas systems were discovered in archaea and bacteria and can be classified into three group based on the different Cas effectors (Cas9, Cas13, and Cas12), which can then be further divided into six types and more than 20 subtypes (Makarova et al., 2018; Li et al., 2019). The type-II CRISPR/Cas system from Streptococcus pyogenes has been widely applied across species, including filamentous fungi, as it is much simpler than other CRISPR systems and has proven to be particularly powerful for use in precise DNA modification (Cong et al., 2013; Deng et al., 2017b). Using specific codon optimization and in vitro RNA transcription, Liu et al. (2015) first adopted the CRISPR/Cas9 system in the filamentous fungus T. reesei and achieved relatively high homologous recombination efficiencies (>93%) when the lengths of the homology arms were 200 bp. In the same year, Nodvig et al. (2015) successfully targeted the yA gene by applying this system in the model fungus A. nidulans and obtained a genome-edited phenotype. Additionally, Fuller et al. (2015) demonstrated that the CRISPR/Cas9 system can be applied to high-efficiency gene disruption in A. fumigatus. These instances illustrate that this powerful system has been widely and effectively applied to industrial filamentous fungi. More details about the CRISPR/Cas9 system and its specific application in the biosynthesis of secondary metabolites by filamentous fungi are reviewed in the following sections.
This review introduces and summarizes the current knowledge and applications of the CRISPR/Cas9 system in filamentous fungi. By detailing several examples, we introduce the specific application of the CRISPR/Cas9 system and CRISPRa for precise gene editing and gene cluster activation, respectively. Additionally, we discuss and summarize the challenges and limitations as well as further prospects of this technology in the production of secondary metabolites by filamentous fungi. Our review lays a solid foundation for the exploration of secondary metabolites in filamentous fungi and will be beneficial to future research on activating silent gene clusters involved in secondary metabolites produced by filamentous fungi.
Principle and Advantages of the CRISPR/CAS9 System
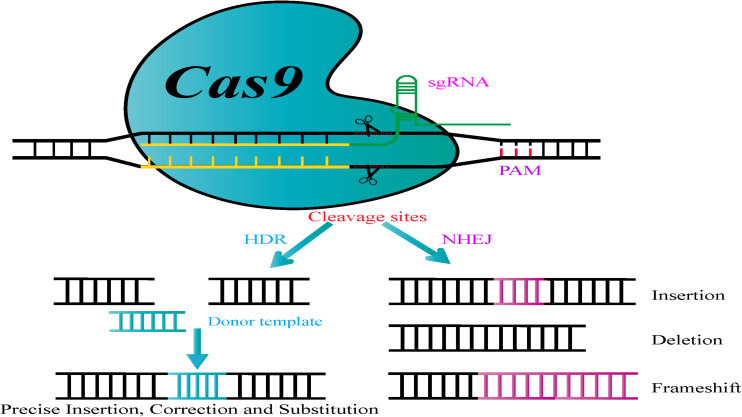
The CRISPR/Cas9 system that has emerged as an advanced technology for genome engineering originated from adaptive immune systems in bacteria as a special defense mechanism against invading viruses and plasmids (Barrangou and Marraffini, 2014; Wang and Coleman, 2019). This genome editing system is composed of two components, a Cas9 nuclease and a guide RNA molecule (gRNA) that targets the nuclease to a specific genomic target site in the genome (Deng et al., 2017b). The single chimeric guide RNA (sgRNA) consisting of a fusion of a CRISPR RNA (crRNA) and a fixed trans-activating crRNA (tracrRNA) are processed by the endogenous bacterial machinery to generate the mature gRNA (Deltcheva et al., 2011). The Cas9 endonuclease is guided to a specific locus by a gRNA, which then forms Watson-Crick base pairs with the target DNA sequence, thereby permitting Cas9 to break the double-stranded DNA at specific sites (Figure 1; Doudna and Charpentier, 2014; Mei et al., 2019). Importantly, to achieve the complementary target-DNA binding and cleavage, Cas9 requires the presence of its major specificity determinant, a well-defined short protospacer adjacent motif (PAM) that is located immediately adjacent to the non-target DNA strand (Mojica et al., 2009; Sternberg et al., 2014). Subsequently, two unique repair mechanisms (Figure 1), non-homologous end-joining (NHEJ) or homology-directed repair (HDR), mend DNA double-strand breaks (DSBs). NHEJ is an error-prone and dominant RNA repair pathway for DSBs via direct ligation of the break ends without using a homologous template. Thus, it can sometimes cause targeted mutations, such as random deletions, insertions, replacement of bases, targeted chromosomal rearrangements, or frameshift mutations at DNA breakage points leading to premature stop codons within the open reading frame (ORF) of the targeted gene (Kim and Kim, 2014; Boettcher and McManus, 2015; Mei et al., 2019). Compared to NHEJ, which is the most common DSB repair mechanism in microorganisms, HDR is a less efficient but high-fidelity pathway. HDR precisely repairs DSBs with the help of a homologous DNA template or exogenous donor fragment, thus having the potential to generate gene modifications by introducing desired nucleotide substitutions or gene insertions (Bortesi and Fischer, 2015; El-Sayed et al., 2017). With the advancement of research and accumulating knowledge on endogenous DNA repair mechanisms, a series of technological tools have been explored to precisely induce DSBs in target genes by using exogenous nucleases. Prior to CRISPR, genome engineering strategies utilizing ZFN or TALEN-based genome editing required the design, generation, and validation of an appropriate protein for a specific DNA locus of interest, thus limiting their widespread application (Doudna and Charpentier, 2014; Boettcher and McManus, 2015; El-Sayed et al., 2017; Leitão et al., 2017). Owing to its high efficiency and the possibility of multi-gene editing, the CRISPR/Cas9 system has rapidly emerged as an extraordinary genome engineering approach has outstripped the performance of earlier technologies (Doudna and Charpentier, 2014; Wang et al., 2017).
FIGURE 1.
Principle of the CRISPR/Cas9 genome engineering tool. Nuclease-induced double-stranded breaks (DSBs) can be repaired by non-homologous end joining (NHEJ) or homology-directed repair (HDR) pathways. Imprecise NHEJ-mediated repair can produce insertion/deletion/frameshift mutations of variable lengths at DSB sites. Homologous recombination (HR)-mediated repair can introduce precise point mutations, by substitutions, insertions, or deletions, depending on the donor DNA template.
Applications of CRISPR/CAS9 in Filamentous Fungi
The versatility and programmability of Cas9 has made the CRISPR/Cas9 genome editing strategy a revolutionary approach in biological research, and it has been considered useful for creating gene deletions, substitutions, and insertions in filamentous fungi. Here, we gather considerable information regarding the application of the CRISPR/Cas9 system in filamentous fungi over recent years (Table 1). Apart from genetic modification, Cas9 can also modulate transcription without editing the genomic sequence by fusing the enzymatically inactive version of Cas9 (dCas9) with transcriptional repression and activation domains (Gilbert et al., 2013, 2014; Chavez et al., 2015). The former approach, fusion of dCas9 with repression domains (e.g., KRAB/Kox1), has been used to negatively regulate the transcription of specific genomic loci (Gilbert et al., 2013; Leitão et al., 2017). This strategy, commonly termed CRISPR inhibition (CRISPRi), decreases the transcription of target DNA loci mainly by blocking transcriptional elongation, impeding transcription factor binding, or interfering with RNA polymerase transcription initiation (Bikard et al., 2013; Larson et al., 2013; Doudna and Charpentier, 2014; Piatek et al., 2015). Several reports have revealed that this approach can be successfully applied to simultaneously repress the transcription of multiple target genes and that it is reversible (Bikard et al., 2013; Gilbert et al., 2013; Zhao et al., 2014). CRISPR activation (CRISPRa), can be achieved through direct fusing dCas9 with activation domains, such as VP64/p65/HSF1 (Bikard et al., 2013; Gilbert et al., 2014; Konermann et al., 2015; Jia et al., 2018; Strezoska et al., 2020). For example, Cheng et al. (2013) achieved robust endogenous gene activation utilizing VP160 transcriptional activation domain in human and mouse cells. With the intention of enhancing transcriptional activation, Chavez et al. (2015) introduced a unique activation strategy that required the fusion of dCas9 to three activation domains, VP64-p65-Rta (VPR) and thus proved its utility in activating endogenous coding and non-coding genes. CRISPRa also has potential in exploring gene expression in filamentous fungi, particularly, for genes that are closely related to secondary metabolite biosynthesis (Wang and Coleman, 2019). For example, Roux et al. (2020) constructed the first CRISPRa system in A. nidulans by using the same method as Chavez et al. (2015) to improve production of the compound microperfuranone and identify a new metabolite of dehydromicroperfuranone. Filamentous fungi have the capacity to produce a diverse spectrum of valuable secondary metabolites, and the genetic potential of secreting important metabolites has not yet been fully utilized for most SM-BGCs. Therefore, there is little doubt that a strategy that combines the CRISPR/Cas9 technology (i.e., CRISPRa) with the activation of expression of secondary metabolite biosynthetic clusters is on the horizon. This would be particularly useful in developing bioactive products or derivatives for biopharmaceuticals.
TABLE 1.
Applications of CRISPR-Cas9 genome engineering tool in filamentous fungi.
| Species | Cas9 expression (selection marker, promoter) | Transformation method | Editing method | Application | Reference |
| Phytophthora sojae | Human-optimized codons Cas9, G418, Ham34 | PMT1 | NHEJ/HDR | Single-gene disruption | Fang and Tyler, 2016 |
| Ustilago maydis | Codon-optimized Cas9, ip, Otef (modified tef1)/hsp70 | PMT | NHEJ | Single/double-gene disruption | Schuster et al., 2016 |
| A. fumigatus | Human codon-optimized Cas9, pyr4/hph, niiA/gpdA | PMT | HDR | Single/double-gene disruption | Zhang et al., 2016 |
| A. oryzae | Codon-optimized Cas9, pyrG, amyB | PMT | NHEJ | Single-gene disruption | Katayama et al., 2016 |
| Penicillium chrysogenum | Human codon-optimized Cas9, amdS, xlnA | PMT | NHEJ/HDR | Single-gene disruption | Pohl et al., 2016 |
| Candida albicans | Codon-optimized Cas9, Nat, ENO1 | Lithium acetate | NHEJ/HDR | Single-gene disruption | Min et al., 2016 |
| A. fumigatus | pFC332, pyrG, TetON | PMT | NHEJ/HDR | Single-gene disruption | Weber et al., 2017 |
| A. niger | pFC332, pyrG/hph, tef1 | PMT | NHEJ/HDR | Single-gene disruption | Kuivanen et al., 2016 |
| Ustilago maydis | B913, ip, Otef (modified tefl)/ hsp70 | PMT | NHEJ | Multiple-gene disruption | Schuster et al., 2018 |
| A. oryzae | Codon-optimized Cas9, niaD/pyrG | PMT | NHEJ | Double-gene disruption | Nakamura et al., 2017 |
| Myceliophthora thermophila | Codon-optimized Cas9, Bar, tef1 | PMT/AMT2 | NHEJ/HDR | Single/multiple-gene disruption | Liu et al., 2017 |
| Talaromyces atroroseus | pFC332, hph, tef1 | PMT | NHEJ | Single-gene disruption | Nielsen et al., 2017 |
| Aspergillus carbonarius | pFC332, hph, tef1 | AMT | NHEJ/HDR | Single-gene disruption | Weyda et al., 2017 |
| A. niger | pFC332, hph, tef1 | PMT | NHEJ | Single-gene disruption | Kuivanen et al., 2017 |
| Ganoderma lucidum | Codon-optimized Cas9, ura3, gpdA | PMT | NHEJ | Single-gene disruption | Qin et al., 2017 |
| Beauveria bassiana | Codon-optimized Cas9, gfp/ura5/bar, gpdA | PMT | NHEJ/HDR | Single/multiple-gene disruption | Chen et al., 2017 |
| Alternaria alternata | pFC332, pyr4/hph, gpdA | PMT | NHEJ | Single-gene disruption | Wenderoth et al., 2017 |
| Shiraia bambusicola | Codon-optimized Cas9, hph, TrpC | PMT | NHEJ | Single-gene disruption | Deng et al., 2017a |
| Shiraia bambusicola | Codon-optimized Cas9, hph, TrpC | PMT | NHEJ/HDR | Single-gene disruption | Deng et al., 2017c |
| Nodulisporium sp. | Codon-optimized Cas9, Bar, TrpC | PMT | NHEJ/HDR | Single-gene disruption | Zheng et al., 2017 |
| Mucor circinelloides | SpCas9, pyr4 | PMT | NHEJ/HDR | Single/double-gene disruption | Nagy et al., 2017 |
| Leptosphaeria maculans | Human codon-optimized Cas9, Ip/G418/hph, act1 | AMT | NHEJ | Single-gene disruption | Idnurm et al., 2017 |
| Sporisorium scitamineum | HCas9, Nat/Hph, gapd | AMT | HDR | Single-gene disruption | Lu et al., 2017 |
| A. niger | pCas9, hyg/pyrG | PMT | NHEJ | Single-gene disruption | Sarkari et al., 2017 |
| A. fumigatus | In vitro-assembled Cas9 RNPs, hyg | PMT | HDR | Single-gene disruption | Al Abdallah et al., 2017 |
| Magnaporthe oryzae | Codon-optimized Cas9, hyg | PMT | NHEJ/HR | Single/double-gene disruption | Foster et al., 2018 |
| M. thermophila | Codon-optimized Cas9, neo, tef1 | PMT | NHEJ | Single-gene disruption | Xu et al., 2018 |
| Rhizopus delemar | Codon-optimized Cas9, pyrF | Electroporation | NHEJ | Single-gene disruption | Bruni et al., 2018 |
| M. thermophila | Codon-optimized Spmae, hyg/bar/neo, tef1/gpdA/TrpC | PMT | HDR | Single/multiple-gene disruption | Gu et al., 2018 |
| A. niger | Codon-optimized Cas9, pyrG/ble, tRNA | PMT | HDR | Single-gene disruption | Song et al., 2018 |
| Penicillium decumbens | Cas9 RNPs, gpdA, ergA | PMT | HDR | Single/multiple-gene disruption | Grijseels et al., 2018 |
| Fusarium graminearum | Codon-optimized Cas9, fludioxonil, gpdA | PMT | NHEJ/HDR | Single-gene disruption | Gardiner and Kazan, 2018 |
| A. niger | pFC332, hph, tef1 | PMT | NHEJ | Single-gene disruption | Kuivanen and Richard, 2018 |
| C. albicans | SpCas9, Nat/Phloxine B | Electroporation | NHEJ | Double-gene disruption | Shapiro et al., 2017 |
| Aspergilli | pFC332, argB/pyrG, tef1 | AMT | HDR | Multiple-gene disruption | Nodvig et al., 2018 |
| Blastomyces dermatitidis | pFC332, hph, tef1 | AMT | NHEJ | Single/double-gene disruption | Kujoth et al., 2018 |
| A. niger | Codon-optimized Cas9, hph/amdS, glaA | PMT | NHEJ/HDR | Single-gene disruption | Zheng et al., 2019 |
| C. albicans | Codon-optimized Cas9, Nat, ENO1 | Electroporation | HDR | Single/double-gene disruption | Vyas et al., 2018 |
| P. sojae | Human-optimized codons Cas9, G418, Ham34 | PMT | NHEJ | Double-gene disruption | Miao et al., 2018 |
| F. oxysporum | pFC332, hph | PMT | NHEJ/HDR | Single-gene disruption | Wang et al., 2018 |
| Ustilaginoidea virens | Codon-optimized Cas9, G418, pdc/cbh1 | AMT/PMT | NHEJ | Multiple-gene disruption | Liang et al., 2018 |
| Cordyceps militaris | Codon-optimized Cas9, 5-FOA/blpR, tef1 | AMT/PMT | NHEJ/HDR | Single-gene disruption | Chen et al., 2018 |
| Cryptococcus neoformans | Codon-optimized Cas9, Ntc, tef1 | Electroporation | HDR | Single-gene disruption | Wang, 2018 |
| Sclerotinia sclerotiorum | Codon-optimized Cas9, hph, tef1 | PMT | NHEJ/HDR | Single-gene disruption | Li et al., 2018 |
| Aspergillus fumigatus | pFC332, pyrG, tef1 | PMT | NHEJ | Single-gene disruption | Matsuda et al., 2018 |
| Ustilago trichophora | Human-optimized codons Cas9, marker-free, U6 | PMT | NHEJ | Single-gene disruption | Huck et al., 2019 |
| Penicillium subrubescens | Codon-optimized Cas9, hph, tef1 | PMT | NHEJ | Single-gene disruption | Salazar-Cerezo et al., 2020 |
| A. niger | pFC332, hph, tef1 | PMT | HDR | Multiple-gene disruption | van Leeuwe et al., 2019 |
| M. thermophila | Codon-optimized Cas12a, neo/bar, U6 | PMT | NHEJ/HDR | Multiple-gene disruption | Liu et al., 2019 |
| A. niger | pFC332, argB/pyrG, tef1 | PMT | HDR | Single-gene disruption | Leynaud-Kieffer et al., 2019 |
| A. oryzae | Codon-optimized Cas9, pyrG, amyB | PMT | HDR | Single/double-gene disruption | Katayama et al., 2019 |
| Alternaria alternata | pFC332, hph, tef1 | PMT | NHEJ | Single-gene disruption | Igbalajobi et al., 2019 |
| A. niger | Codon-optimized Cas9, pyrG | PMT | NHEJ/HDR | Single/multiple-gene disruption | Kuivanen et al., 2019 |
| Mucor circinelloides | SpCas9, pyr4 | PMT | HDR | Multiple-gene disruption | Nagy et al., 2019 |
| Leptosphaeria maculans | Human codon-optimized Cas9, hph, act1 | AMT | NHEJ | Double-gene disruption | Darma et al., 2019 |
| F. fujikuroi | Codon-optimized Cas9, hph, U6/5SrRNA | PMT | HDR | Multiple-gene disruption | Shi et al., 2019 |
| Ashbya gossypii | Human-optimized codons Cas9, G418, tef1 | Electroporation | NHEJ | Single-gene disruption | Jimenez et al., 2019 |
| Duddingtonia flagrans | pFC332, hph, tef1 | PMT | HDR | Single-gene disruption | Youssar et al., 2019 |
| A. oryzae | Aspergillus-optimized codons Cas9, pyrG, U6 | PMT | HDR | Single-gene disruption | Chutrakul et al., 2019 |
| T. reesei | Codon-optimized Cas9, pyr4 | PMT | HDR | Single-gene disruption | Hao and Su, 2019 |
| M. thermophila | Codon-optimized Cas9, bar/neo, tef1 | PMT | HDR | Double-gene disruption | Li et al., 2020a |
| Glarea lozoyensis | Codon-optimized Cas9, neo, 5S rRNA | AMT | NHEJ/HDR | Single/multiple-gene disruption | Wei et al., 2020 |
| Talaromyces pinophilus EMU | Codon-optimized Cas9, hph, gdpA | PMT | NHEJ | Single-gene disruption | Manglekar and Geng, 2020 |
| A. niger | pCsR1, hph, tef1 | PMT | NHEJ/HDR | Single/multiple-gene disruption | Rojas-Sánchez et al., 2020 |
| M. thermophila | Codon-optimized Cas9, neo, trpC | PMT/AMT | HDR | Single/multiple-gene disruption | Li et al., 2020b |
| A. niger | pFC332, pyrG/amdS, PnaII/TPI | PMT | HDR | Single-gene disruption | Dong et al., 2020 |
| T. reesei | Codon-optimized Cas9, ura5, U6 | AMT | NHEJ | Single-gene disruption | Wu et al., 2020 |
| A. niger | pCas9, pyrG/hph, 5S rRNA | PMT | HDR | Double-gene disruption | Zhang et al., 2020 |
1AMT, Agrobacterium tumefaciens-mediated transformation; 2PMT, Protoplast transformation.
Specific Application of CRISPR/CAS9 in Secondary Metabolites Synthesis Pathways
In the past few years, the CRISPR/Cas9 system has been introduced into filamentous fungi to explore the potential of this strategy in modulating production of secondary metabolites. From A. oryzae and T. reesei to A. niger and A. nidulans, CRISPR/Cas9 based systems have become versatile platforms for precise genome editing, and great progress has already been made for production of valuable secondary metabolites. Here, we highlight four examples illustrating the application of CRISPR/Cas9 (gene deletion/substitution/insertion) and CRISPRa (gene cluster activation) systems in filamentous fungi.
Application of CRISPR/Cas9-Based Genome Editing in the Production of Gibberellic Acid by Fusarium fujikuroi
Natural products derived from the secondary metabolism of filamentous fungi have a wide array of applications, especially in the pharmaceutical and agricultural industries. Gibberellic acids (GAs) are a class of natural plant growth hormones that are notably produced in F. fujikuroi and are widely applied to regulate the growth of diverse plant species. Among all GAs, the most biologically active ones are GA1, GA3, GA4, and GA7 (Ullah et al., 2014). Of these, the production and application of GA3 has reached a particularly mature stage, while that of GA4 and GA7 have been hindered by the low efficiency of the existing production methods (Joshi et al., 2018; Qian et al., 2018).
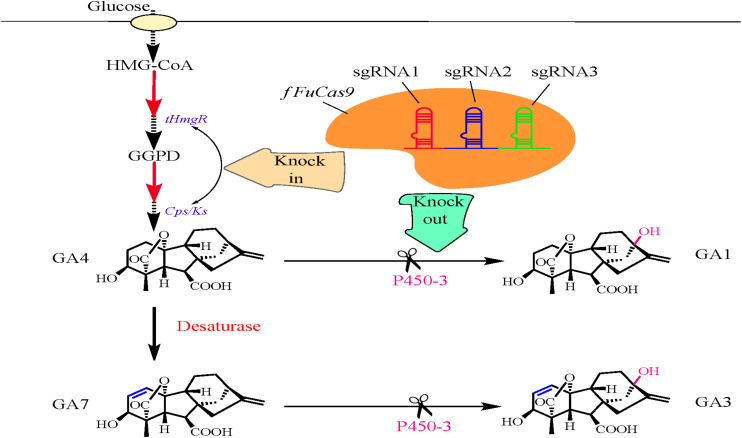
Shi et al. (2019) established an efficient CRISPR/Cas9-based genome editing tool to improve the production of GA4 and GA7 in F. fujikuroi (Figure 2). Initially, a f FuCas9 vector carrying different nuclear localization signals (NLSs) was constructed. The endogenous NLS from histone H2B (HTBNLS) was selected to individually fuse with the f FuCas9 protein, owing to its higher editing efficiency and previous success in Fusarium oxysporum (Wang et al., 2018). The f FuCas9 fusion protein vector along with sgRNAs was assembled in vitro to establish the CRISPR/Cas9 system (pUC-f FuCas9-HTBNLS-hph). Simultaneously, owing to the need for efficient sgRNA transcription, the endogenous 5S rRNA (Ff5SrRNA) was utilized to express the sgRNA designed for the target gene. Subsequently, the Ff5SrRNA-P450-3 sgRNA cassette was synthesized with three sgRNAs and ligated into the EcoRI site of pUC-f FuCas9-HTBNLS-hph, thus yielding the pUC-f FuCas9-HTBNLS-hph-P450-3 vector. This vector was used to disrupt the gene encoding P450-3, thus yielding the final disruption mutant ΔP450-3. Subsequently, sgRNA and f FuCas9 vector were introduced into F. fujikuroi through the modified protoplast-based polyethylene glycol and Ca2+ transformation method (Hwang and Ahn, 2016). Additionally, overexpression of two key genes encoding copalyl diphosphate synthase/kaurene synthase (Cps/Ks) and a truncated HMG-CoA reductase (tHmgR) were also performed separately and simultaneously to explore the possibility of increasing the amount of GA4/GA7 mixture produced based on previous research (Albermann et al., 2013). To overexpress Cps/Ks and tHmgR genes, the f FuCas9 vector and donor vector were introduced by protoplast transformation into the ΔP450-3 disruption mutant. The resulting transformants were subjected to a second round of hygromycin screening and a series of cultivation cycles and finally inoculated into the fermentation medium. The contents of four GAs in the supernatant fraction of the fermentation medium were determined by high-performance liquid chromatography (HPLC). Compared with the control strain (88.38 mg/L), the accumulation of a GA4/GA7 mixture was evidently improved in the ΔP450-3 mutant (410.27 mg/L). Thus, the CRISPR/Cas9-based genome editing system proved to be efficient enough to enhance the levels of secondary metabolites in F. fujikuroi. Thus, overexpression of both Cps/Ks and tHmgR has been demonstrated to be an effective mean of increasing the contents of GA4 and GA7 (by 24.23% to 509.68 mg/L and by 70.14% to 698.03 mg/L, respectively), which is clearly higher than in the ΔP450-3 disruption mutant. Indeed, the combined concentration of GA4 and GA7 reached a higher level (716.37 mg/L) when Cps/Ks and tHmgR were overexpressed simultaneously in the ΔP450-3 mutant. Thus, Shi et al. (2019) successfully demonstrated that CRISPR/Cas9-based genome editing strategies and overexpression approaches are suitable for improving the content of GA4/GA7 mixtures. These results will also greatly facilitate further research on production of other metabolites in F. fujikuroi.
FIGURE 2.
Application of the CRISPR/Cas9-based genome editing tool in Fusarium fujikuroi for improving the production of the gibberellic acids GA4 and GA7.
Functional Reconstitution of Trypacidin Biosynthesis Gene Cluster in A. fumigatus by the CRISPR/Cas9-Based Approach
Aspergillus fumigatus is an important human pathogen responsible for various forms of aspergillosis in humans as well as in animals (Paulussen et al., 2017). It also possesses the capacity to secrete a large repertoire of natural products, some of which are involved in pathogenicity (Scharf et al., 2014). Trypacidin, one of the natural antimicrobial antibiotic compounds produced by A. fumigatus, is a spore-borne product that has been demonstrated to be a potent toxin to human lung cells (Gauthier et al., 2012). This compound may also be a virulence determinant that plays a role in the phagocytosis of different cells such as murine alveolar macrophages and the amoeba Dictyostelium discoideum (Mattern et al., 2015).
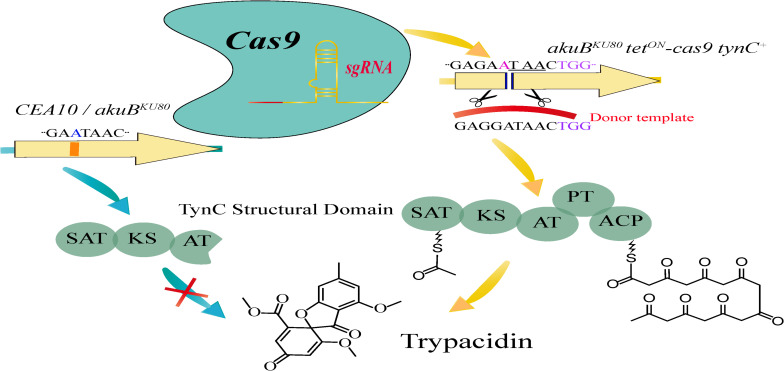
In a previous study, Frisvad et al. (2009) analyzed and detected trypacidin in 30 out 40 A. fumigatus strains of different origins, including the clinical isolate strain Af293. The remaining 10 strains from which they did not isolate trypacidin, included a second clinical isolate and lab strain CEA10 (Frisvad et al., 2009). Based on these results, Weber et al. (2017) explored the mechanisms underlying the difference in trypacidin production and reconstituted the biosynthetic pathway of this compound by advanced genome editing in a non-producing strain. Initially, the area of surrounding a single nucleotide insertion in the polyketide synthase (PKS) coding gene tynC that potentially led to a frameshift and appearance of a premature stop codon in the CEA10 strain genome was sequenced from the tynC alleles of Af293 as well as CEA10 and its descendant strain CEA17 (ΔakuBKU80) (Throckmorton et al., 2016). The sequencing results identified a single adenosine base insertion at position 3881 of tynC in strain CEA10 and ΔakuBKU80. This insertion resulted in a premature stop codon, thus eliminating the predicted acyl carrier protein (ACP) and the product template (PT) domains that are essential for the catalytic function of the PKS. The authors then performed single nucleotide editing by the traditional gene substitution method and detected trypacidin in the complemented strain. However, this is a cumbersome multistep method. Therefore, an alternative strategy using a CRISPR/Cas9-based tool was adopted to reconstitute trypacidin production (Figure 3). Weber et al. (2017) initially constructed a recombinant Cas9 expression cassette and integrated it into strain CEA17 ΔakuB pyrG + (akuBKU80 tetON-cas9). They then developed a plasmid containing a split-marker (Kuck and Hoff, 2010) and gRNA (pJW split-ptrA tynC), which was finally transformed into the same strain. By adding doxycycline to the fungal preculture before transformation, the expression of the Cas9 gene was induced to cause gene editing at the tynC locus. Subsequently, the plasmid pJW split-ptrA tynC coupled with donor DNA fragment were co-transformed into akuBKU80 tetON-cas9 via protoplast-mediated transformation (Weidner et al., 1998). The genomic DNA at the target site of selected positive transformants was sequenced. LC-MS (Mattern et al., 2015) was then applied to analyze stationary phase cultures, and a RNeasy Plant Mini kit was used to extract total RNA extraction. As expected, the single adenosine insertion was eliminated from the tynC locus of the akuBKU80 tetON-cas9 tynC + strain, and the presence of trypacidin as well as the tynC mRNA was detected. Thus, trypacidin was reconstituted through CRISPR/Cas9-mediated deletion of an adenosine insertion in the genome. Compared with the conventional strategy, the CRISPR/Cas9 gene editing system was demonstrated to be highly effective and a powerful tool in researching natural products from biosynthetic genes in A. fumigatus. This approach can also be helpful for the integration of fusion-tags and paves the way to exploit novel natural products derived from other filamentous fungi.
FIGURE 3.
Overview of the CRISPR/Cas9-based tool for functional reconstitution of the Aspergillus fumigatus secondary metabolite (trypacidin) gene cluster.
CRISPR/Cas9-Based Genome Editing in Glarea lozoyensis Produces Pneumocandin B0
Pneumocandins are lipohexapeptides within the echinocandin family, and they potently impede fungal cell wall formation via non-competitive inhibition of β-(1, 3)-glucan synthases (Emri et al., 2013; Robbins et al., 2017). One of these compounds, pneumocandin B0, can be isolated from the industrial filamentous fungus Glarea lozoyensis and is used in the synthesis of the potent antifungal drug caspofungin (Emri et al., 2013; Balkovec et al., 2014). Because it has a strong inhibitory effect on invasive aspergillosis, caspofungin has been approved by the U.S. FDA for treating patients who are refractory or intolerant to standard therapy as well as for the main treatment of certain types of Candida infections (Leonard et al., 2007). Several researchers have explored the possibilities of improving pneumocandin B0 production because of its value in the pharmaceutical industry. However, owing to limitations and a lack of sophistication of traditional genetic tools, generation of a genetically engineered industrial G. lozoyensis strain has been impeded.
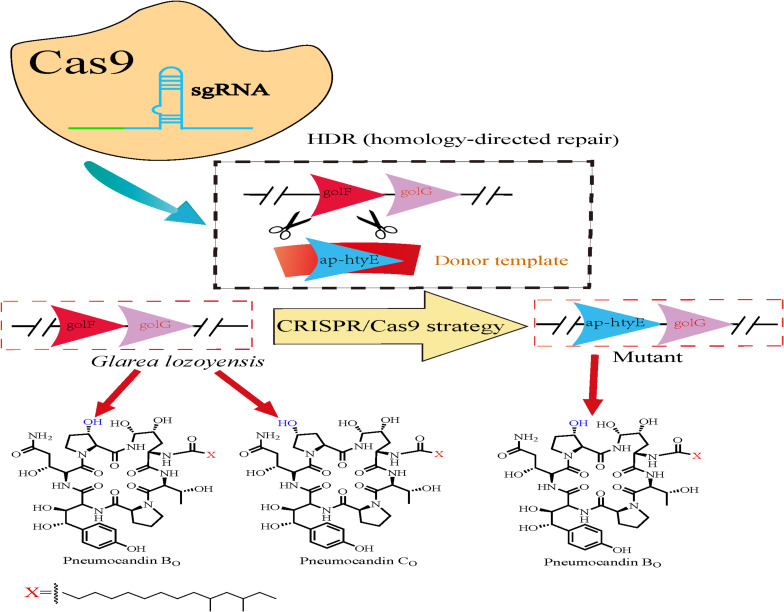
Recently, Wei et al. (2020) utilized an efficient CRISPR/Cas9-based gene editing tool in G. lozoyensis SIPI1208 to significantly enhance the accumulation of pneumocandin B0 (Figure 4). Specifically, this strategy was used to replace GloF with Ap-HtyE (proline hydroxylase, which is responsible for pneumocandin and echinocandin B biosynthesis, separately) in G. lozoyensis using CRISPR/Cas9 system-mediated homology-directed repair (HDR), thus changing the ratio of pneumocandin B0 and pneumocandin C0 products (which constitute a pair of isomers). They initially designed a protospacer sequence targeted to gloF and constructed the pAgG-sgRNA-gloF plasmid to perform gloF gene editing. Thereafter, donor DNA was ligated into the linearized plasmid to construct the final replacement plasmid pAgG-sgRNA-gloF-ap-htyE in order to create a knock-in mutant, ap-htyE. After Agrobacterium tumefaciens-mediated transformation, a series of experimental verifications confirmed that the genomic DNA of ap-htyE was knocked in correctly (Zhang et al., 2003). Based on combined reversed-phase-HPLC and normal phase-HPLC analyses of the fermentation extracts (Osawa et al., 1999), it was found that PC0 was not present in the final fermented product of the knock-in strain, as compared to 33.5% PC0 in the original strain G. lozoyensis. These results illustrated that the ability to produce PC0 was abolished in the gene-edited strain generated using the CRISPR/Cas9 system, thus enabling increased industrial production of PB0 (Wei et al., 2020). In summary, the CRISPR/Cas9-based gene editing method can efficiently manipulate genes in G. lozoyensis and thus enabled for the development of production of other secondary metabolites with similar characteristics.
FIGURE 4.
Schematic representation of gene replacement by the CRISPR/Cas9 strategy. The mutant retained the capability to produce pneumocandin B0, while the production of pneumocandin C0 was abolished.
CRISPR-Mediated Activation of micA Synthetase Gene Increases Microperfuranone Production
The CRISPR-mediated strategy mentioned earlier is not only capable of genetic modification but can also modulate transcription by fusing dCas9 to transcriptional repression and activation domains (i.e., through CRISPRa and CRISPRi). In contrast to the type-II CRISPR/Cas system, CRISPRa and CRISPRi do not require DSBs or donor DNA. The core part of achieving transcriptional activation is the link between the activation domain and dCas9, which then forms a complex with a sgRNA that includes an editable 20-nucleotide sequence complementary to the target site in the gene regulatory region (Chavez et al., 2015; Tak et al., 2017). Furthermore, by concurrently expressing multiple gRNAs, several genes can be simultaneously activated with multiplexed CRISPRa (McCarty et al., 2020). A previous study demonstrated that CRISPRa is a simple and universally applicable technology, by successfully employing it to increase the transcriptional efficiency of Myxococcus xanthus secondary metabolites (Peng et al., 2018). Nevertheless, the application of CRISPRa to the transcriptional regulation of biosynthetic pathways of secondary metabolites in filamentous fungi could be exploited further.
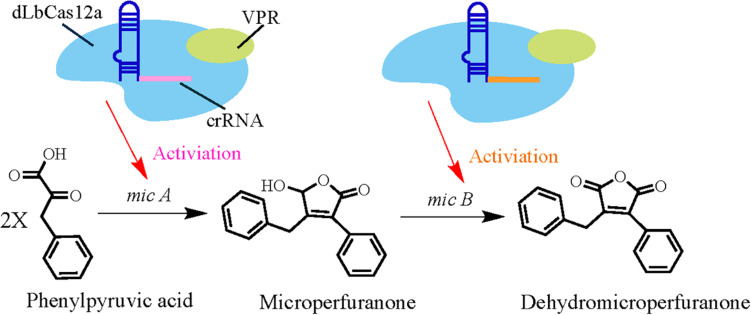
In a recent report, researchers developed a CRISPRa system for A. nidulans for the first time in a filamentous fungus (Roux et al., 2020). They used dCas9 protein fusion to VP64-p65-Rta (VPR, a fusion of three activation domains) and tested whether it leads to strong activation (Chavez et al., 2015). Initially, CRISPR/dLbCas12a-VPR and CRISPR/dSpCas9-VPR-based systems were constructed and tested to exploit their utility. Thereafter, by delivering the four-crRNA array on an AMA1-pyroA vector and the sgRNA in a single AMA1-pyrG vector in two separate systems, they evaluated the multiplexing capability of the dCas12a- and dCas9- driven systems. Ultimately, the CRISPR/dLbCas12a-based CRISPRa system was chosen to probe the activation of biosynthetic genes in A. nidulans for its potential in BCG activation and accelerating the discovery of secondary metabolites (Fonfara et al., 2016). Using this CRISPRa system, Sanson et al. (2018) targeted the micA gene (which is related to microperfuranone biosynthesis) with multiple crRNAs in order to improve the likelihood of achieving strong activation (Figure 5). Following previously designed guidelines, region 119–303 bp and 139–324 bp upstream of the micA transcription start site (Jensen, 2018) were targeted with four-crRNAs to explore the utility of CRISPRa. Then, linearized pCRI001-3 vectors were used to promote homologous recombination followed by polyethylene glycol (PEG)-calcium-based protoplast transformation (Lim et al., 2012). Liquid chromatography coupled with diode array detector and mass spectrometer (LC-DAD-MS) analysis of the media extracts from all CRISPRa transformants showed that the accumulation of microperfuranone was improved when compared with control strains. In addition, the dCas12a-driven system was also utilized to explore simultaneous activation of multiple genes in the mic cluster by adopting the above-mentioned method, but with the addition of two crRNA arrays delivered into the strains. Analysis of A. nidulans culture extracts by LC-DAD-MS indicated that microperfuranone levels decreased in the multiplexed-CRISPRa strains and also revealed a new compound. Subsequent LC-MS/MS and NMR analysis identified this new compound as dehydromicroperfuranone, which is the metabolic product of the mic cluster (Figure 5). Thus, these results demonstrate that CRISPRa is efficient in multi-gene activation of BGCs in A. nidulans and can therefore be used to achieve higher throughput natural production of known and novel bioactive secondary metabolites in filamentous fungi.
FIGURE 5.
Schematic representation of CRISPRa-mediated increases in the production of microperfuranone and discovery of the mic cluster product, dehydromicroperfuranone.
Conclusion and Perspectives
Filamentous fungi, like other eukaryotes, are extensively used in industrial and pharmaceutical production owing to their ability to secrete a plethora of hydrolytic enzymes, polyunsaturated fatty acids, and, more importantly, bioactive small molecules, including antibiotic agents. Furthermore, filamentous fungi are the most compatible heterologous hosts for expression of compounds such as Penicillium citrinum nuclease P1 in A. niger and fungal BGCs in Fusarium graminearum, owing to their lack of a requirement of intron-removal or codon optimization (He et al., 2018; Chen et al., 2019; Nielsen et al., 2019). With advances in genome sequencing and phylogenetic sleuthing, large repertoires of cryptic or silent secondary metabolite biosynthetic gene clusters are being uncovered. However, the difficulties in genetic manipulation have traditionally impeded secondary metabolite molecular studies on filamentous fungi. Therefore, a powerful and comparatively simple genetic technique for overcoming the obstacle of activating these silent secondary metabolite biosynthetic gene clusters is urgently needed. CRISPR/Cas9 is based on a conservative immune defense mechanism found in bacteria and archaea and has been developed as a convenient and flexible technique for genome editing. The CRISPR/Cas9 genome editing technology has shown great promise in revolutionizing the field of fungal research. The CRISPR/Cas9 genome editing system was first introduced into Saccharomyces cerevisiae (DiCarlo et al., 2013). Subsequently, Liu et al. (2015) applied the CRISPR/Cas9 genome editing system to Trichoderma reesei; Matsu-Ura et al. (2015) and Nodvig et al. (2015) applied this system to the model fungal Neurospora crassa and A. nidulans, respectively. Since then, the CRISPR/Cas9 genome editing system has found wide applications in genetic alteration of many filamentous fungi. These studies strongly confirm that the CRISPR/Cas9 gene-editing system will undoubtedly bring great possibilities for the discovery of new secondary metabolites, on account of its straightforward design, high efficiency, and versatility.
Compared with genome editing approaches that utilize protein-guided programmable nucleases such as ZFN and TALEN-based genome editing tools, nucleic acid-guided nucleases such as that primarily utilized in the CRISPR/Cas9 system have several advantages for genetic engineering to produce secondary metabolites. One advantage is that the CRISPR/Cas9 system can potentially edit almost all genes containing a PAM in their target sequences owing to its simplicity and modularity. Only two components are required in this system: a Cas9 endonuclease and sgRNA (Song et al., 2019). Indeed, there remain challenges and limitations to utilize this approach, such as off-target effects and the need to perform precise editing. As research advances, a well-designed gRNA sequence and specific Cas9 variants can be developed to avoid off-target effects, as can control the amount of intracellular Cas9 or enhance Cas9 for higher specificity through protein engineering (Cho et al., 2014; Tong et al., 2019). For example, Pohl et al. (2016) adopted a strategy for assembling the Cas9-gRNA complex in vitro and co-transformed it with a donor DNA into Penicillium chrysogenum, which proved to be effective in reducing off-target effects. With regard to precise editing, the biggest hurdle is the native NHEJ that intensely affects the efficiency of HDR despite the presence of a homologous template (Tong et al., 2019). By employing the inhibitor Scr7, Maruyama et al. (2015) successfully targeted the DNA ligase IV responsible for the NHEJ pathway, causing suppression of the native NHEJ activity, thus improving the efficiency of precise genome editing. Additionally, several advanced strategies like CRISPRi and FokI-dCas9 have been applied to optimize the Cas9 system to perform more precise gene-editing (Guilinger et al., 2014; Kao and Ng, 2017).
The potential of fungal biosynthetic capabilities in generating secondary metabolites seems virtually unlimited. Researchers have continually explored and developed secondary metabolites from filamentous fungi, with CRISPR/Cas9 technologies now accelerating progress. The powerful advantages of the CRISPR/Cas9 system in studies on filamentous fungal secondary metabolites reduce the application of selective markers. Most secondary metabolites are regulated by gene clusters in filamentous fungi. When three or more genes need to be manipulated at the same time, multiple selection markers were required and thus developed for filamentous fungi. The CRISPR/Cas9 system can edit multiple genes at the same time, and it is possible to obtain mutants with multiple site mutations in a single transformation, which greatly improves the efficiency of genome editing in studies on secondary metabolites of filamentous fungi. However, relatively few reports about the “actual applications” of the CRISPR/Cas9 system in the production of secondary metabolites can be obtained. Thus, the application is still in its initial stages, as most of the research focus is on aspects such as assessing the feasibility of CRISPR/Cas9 systems in fungi (Tong et al., 2019). Apart from CRISPR/Cas9 genome editing strategies, alternative methods, such as CRISPRi and CRISPRa, which do not depend on DSBs, were rarely reported in filamentous fungi, especially with respect to secondary metabolite production. So far, only one application of the CRISPRa technique being used to increase the secondary metabolite contents of a filamentous fungus has been reported (Roux et al., 2020). CRISPRi has not been used in filamentous fungi, but has been successfully applied in other fungi (Roman et al., 2019). In addition, prime editing, an emerging and precise genome editing method that expands the scope of CRISPR genome editing, with few byproducts and without requiring DSBs or donor DNA templates, has attracted great attention (Anzalone et al., 2019). Consequently, this method has potential value in research on secondary metabolites from filamentous fungi. Related technologies and applications of the CRISPR/Cas9 possess great potential and will play a greater role in the discovery of new secondary metabolites and engineering of the strains that produce them in the near future.
In summary, CRISPR/Cas9-based genome editing technology still requires development and improvement in genetic modification of secondary metabolites in filamentous fungi, and the general scope of applications can be expanded further. It is premature to declare that the CRISPR/Cas9 technique is accelerating the metabolic engineering of filamentous fungi for secondary metabolites. However, with the development of full genomes available and metabolomics, knowledge of the secondary metabolite biosynthetic gene clusters of filamentous fungi together with exploitation of CRISPR/Cas9 approaches will help overcome current limitations in increasing production of secondary metabolites. Such advances will also promote the discovery of new bioactive compounds.
Author Contributions
GL: conceptualization and writing—original draft preparation. CJ: writing—review and editing and conceptualization. XC and YT: project administration. YD: supervision. BZ: supervision and funding acquisition. BH: writing—review, editing, and funding acquisition. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Thanks to those who made critical review on this manuscript.
Footnotes
Funding. This research was funded by the National Natural Science Foundation of China (31900063), the Natural Science Foundation of Jiangxi Province (Grant Nos. 20202BABL203043 and 20202BAB215003), the Science and Technology Research Project of Jiangxi Provincial Department of Education (GJJ180630), and the Innovation Fund Designated for Graduate Students of Jiangxi Science and Technology Normal University (YC2020-X20).
References
- Al Abdallah Q., Ge W., Fortwendel J. R. (2017). A simple and universal system for gene manipulation in aspergillus fumigatus: in vitro-assembled Cas9-guide RNA ribonucleoproteins coupled with microhomology repair templates. mSphere 2 e417–e446. 10.1128/mSphere.00446-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albermann S., Linnemannstons P., Tudzynski B. (2013). Strategies for strain improvement in Fusarium fujikuroi: overexpression and localization of key enzymes of the isoprenoid pathway and their impact on gibberellin biosynthesis. Appl. Microbiol. Biotechnol. 97 2979–2995. 10.1007/s00253-012-4377-5 [DOI] [PubMed] [Google Scholar]
- Anzalone A. V., Randolph P. B., Davis J. R., Sousa A. A., Koblan L. W., Levy J. M., et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576 149–157. 10.1038/s41586-019-1711-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkovec J. M., Hughes D. L., Masurekar P. S., Sable C. A., Schwartz R. E., Singh S. B. (2014). Discovery and development of first in class antifungal caspofungin (CANCIDAS(R))–a case study. Nat. Prod. Rep. 31 15–34. 10.1039/c3np70070d [DOI] [PubMed] [Google Scholar]
- Barrangou R., Marraffini L. A. (2014). CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol. Cell 54 234–244. 10.1016/j.molcel.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D., Jiang W., Samai P., Hochschild A., Zhang F., Marraffini L. A. (2013). Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 41 7429–7437. 10.1093/nar/gkt520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettcher M., McManus M. T. (2015). Choosing the right tool for the job: RNAi, TALEN, or CRISPR. Mol. Cell 58 575–585. 10.1016/j.molcel.2015.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortesi L., Fischer R. (2015). The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33 41–52. 10.1016/j.biotechadv.2014.12.006 [DOI] [PubMed] [Google Scholar]
- Bruni G. O., Zhong K., Lee S. C., Wang P. (2018). CRISPR-Cas9 induces point mutation in the mucormycosis fungus Rhizopus delemar. Fungal Genet. Biol. 124 1–7. 10.1016/j.fgb.2018.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A., Scheiman J., Vora S., Pruitt B. W., Tuttle M., P R Iyer E., et al. (2015). Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 12 326–328. 10.1038/nmeth.3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B. X., Wei T., Ye Z. W., Yun F., Kang L. Z., Tang H. B., et al. (2018). Efficient CRISPR-Cas9 gene disruption system in edible-medicinal mushroom cordyceps militaris. Front. Microbiol. 9:1157. 10.3389/fmicb.2018.01157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Lai Y., Wang L., Zhai S., Zou G., Zhou Z., et al. (2017). CRISPR/Cas9-mediated efficient genome editing via blastospore-based transformation in entomopathogenic fungus beauveria bassiana. Sci. Rep. 8:45763. 10.1038/srep45763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Wang B., Pan L. (2019). Heterologous expression and characterization of Penicillium citrinum nuclease P1 in Aspergillus niger and its application in the production of nucleotides. Protein Expr. Purif. 156 36–43. 10.1016/j.pep.2018.12.004 [DOI] [PubMed] [Google Scholar]
- Cheng A. W., Wang H., Yang H., Shi L., Katz Y., Theunissen T. W., et al. (2013). Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 23 1163–1171. 10.1038/cr.2013.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. W., Kim S., Kim Y., Kweon J., Kim H. S., Bae S., et al. (2014). Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 24 132–141. 10.1101/gr.162339.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chutrakul C., Panchanawaporn S., Jeennor S., Anantayanon J., Vorapreeda T., Vichai V., et al. (2019). Functional characterization of Novel U6 RNA polymerase III promoters: their implication for CRISPR-Cas9-mediated gene editing in aspergillus oryzae. Curr. Microbiol. 76 1443–1451. 10.1007/s00284-019-01770-0 [DOI] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S. L., Barretto R., Habib N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339 819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darma R., Lutz A., Elliott C. E., Idnurm A. (2019). Identification of a gene cluster for the synthesis of the plant hormone abscisic acid in the plant pathogen Leptosphaeria maculans. Fungal Genet Biol. 130 62–71. 10.1016/j.fgb.2019.04.015 [DOI] [PubMed] [Google Scholar]
- Dasgupta A., Chowdhury N., De R. K. (2020). Metabolic pathway engineering: perspectives and applications. Comput. Methods Programs Biomed. 192:105436. 10.1016/j.cmpb.2020.105436 [DOI] [PubMed] [Google Scholar]
- Deltcheva E., Chylinski K., Sharma C. M., Gonzales K., Chao Y., Pirzada Z. A., et al. (2011). CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471 602–607. 10.1038/nature09886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H., Gao R., Liao X., Cai Y. (2017a). Characterization of a major facilitator superfamily transporter in Shiraia bambusicola. Res. Microbiol. 168 664–672. 10.1016/j.resmic.2017.05.002 [DOI] [PubMed] [Google Scholar]
- Deng H., Gao R., Liao X., Cai Y. (2017b). CRISPR system in filamentous fungi: current achievements and future directions. Gene 627 212–221. 10.1016/j.gene.2017.06.019 [DOI] [PubMed] [Google Scholar]
- Deng H., Gao R., Liao X., Cai Y. (2017c). Genome editing in Shiraia bambusicola using CRISPR-Cas9 system. J. Biotechnol. 259 228–234. 10.1016/j.jbiotec.2017.06.1204 [DOI] [PubMed] [Google Scholar]
- DiCarlo J. E., Norville J. E., Mali P., Rios X., Aach J., Church G. M. (2013). Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 41 4336–4343. 10.1093/nar/gkt135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Yu D., Lin X., Wang B., Pan L. (2020). Improving expression of thermostable trehalase from Myceliophthora sepedonium in Aspergillus niger mediated by the CRISPR/Cas9 tool and its purification, characterization. Protein Expr. Purif. 165:105482. 10.1016/j.pep.2019.105482 [DOI] [PubMed] [Google Scholar]
- Doudna J. A., Charpentier E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096. 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- El-Sayed A. S. A., Abdel-Ghany S. E., Ali G. S. (2017). Genome editing approaches: manipulating of lovastatin and taxol synthesis of filamentous fungi by CRISPR/Cas9 system. Appl. Microbiol. Biotechnol. 101 3953–3976. 10.1007/s00253-017-8263-z [DOI] [PubMed] [Google Scholar]
- Emri T., Majoros L., Toth V., Pocsi I. (2013). Echinocandins: production and applications. Appl. Microbiol. Biotechnol. 97 3267–3284. 10.1007/s00253-013-4761-9 [DOI] [PubMed] [Google Scholar]
- Fang Y., Tyler B. M. (2016). Efficient disruption and replacement of an effector gene in the oomycete Phytophthora sojae using CRISPR/Cas9. Mol. Plant Pathol. 17 127–139. 10.1111/mpp.12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I., Richter H., Bratovic M., Le Rhun A., Charpentier E. (2016). The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532 517–521. 10.1038/nature17945 [DOI] [PubMed] [Google Scholar]
- Foster A. J., Martin-Urdiroz M., Yan X., Wright H. S., Soanes D. M., Talbot N. J., et al. (2018). CRISPR-Cas9 ribonucleoprotein-mediated co-editing and counterselection in the rice blast fungus. Sci. Rep. 8:14355. 10.1038/s41598-018-32702-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad J. C., Rank C., Nielsen K. F., Larsen T. O. (2009). Metabolomics of Aspergillus fumigatus. Med. Mycol. 47 S53–S71. 10.1080/13693780802307720 [DOI] [PubMed] [Google Scholar]
- Fuller K. K., Chen S., Loros J. J., Dunlap J. C. (2015). Development of the CRISPR/Cas9 system for targeted gene disruption in Aspergillus fumigatus. Eukaryot Cell 14 1073–1080. 10.1128/EC.00107-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner D. M., Kazan K. (2018). Selection is required for efficient Cas9-mediated genome editing in Fusarium graminearum. Fungal. Biol. 122 131–137. 10.1016/j.funbio.2017.11.006 [DOI] [PubMed] [Google Scholar]
- Gauthier T., Wang X., Sifuentes Dos Santos J., Fysikopoulos A., Tadrist S., Canlet C., et al. (2012). Trypacidin, a spore-borne toxin from Aspergillus fumigatus, is cytotoxic to lung cells. PLoS One 7:e29906. 10.1371/journal.pone.0029906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L. A., Horlbeck M. A., Adamson B., Villalta J. E., Chen Y., Whitehead E. H., et al. (2014). Genome-Scale CRISPR-mediated control of gene repression and activation. Cell 159 647–661. 10.1016/j.cell.2014.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L. A., Larson M. H., Morsut L., Liu Z., Brar G. A., Torres S. E., et al. (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154 442–451. 10.1016/j.cell.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grijseels S., Pohl C., Nielsen J. C., Wasil Z., Nygard Y., Nielsen J., et al. (2018). Identification of the decumbenone biosynthetic gene cluster in Penicillium decumbens and the importance for production of calbistrin. Fungal. Biol. Biotechnol. 5:18. 10.1186/s40694-018-0063-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S., Li J., Chen B., Sun T., Liu Q., Xiao D., et al. (2018). Metabolic engineering of the thermophilic filamentous fungus Myceliophthora thermophila to produce fumaric acid. Biotechnol. Biofuels 11:323. 10.1186/s13068-018-1319-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilinger J. P., Thompson D. B., Liu D. R. (2014). Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 32 577–582. 10.1038/nbt.2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Z., Su X. (2019). Fast gene disruption in trichoderma reesei using in vitro assembled Cas9/gRNA complex. BMC Biotechnol. 19:2. 10.1186/s12896-018-0498-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hautbergue T., Jamin E. L., Debrauwer L., Puel O., Oswald I. P. (2018). From genomics to metabolomics, moving toward an integrated strategy for the discovery of fungal secondary metabolites. Nat. Prod. Rep. 35 147–173. 10.1039/c7np00032d [DOI] [PubMed] [Google Scholar]
- He Y., Wang B., Chen W., Cox R. J., He J., Chen F. (2018). Recent advances in reconstructing microbial secondary metabolites biosynthesis in Aspergillus spp. Biotechnol. Adv. 36 739–783. 10.1016/j.biotechadv.2018.02.001 [DOI] [PubMed] [Google Scholar]
- Hoffmeister D., Keller N. P. (2007). Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat. Prod. Rep. 24 393–416. 10.1039/b603084j [DOI] [PubMed] [Google Scholar]
- Huck S., Bock J., Girardello J., Gauert M., Pul U. (2019). Marker-free genome editing in Ustilago trichophora with the CRISPR-Cas9 technology. RNA Biol. 16 397–403. 10.1080/15476286.2018.1493329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I. S., Ahn I. P. (2016). Multi-homologous recombination-based gene manipulation in the rice pathogen fusarium fujikuroi. Plant Pathol. J. 32 173–181. 10.5423/PPJ.OA.12.2015.0263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A., Urquhart A. S., Vummadi D. R., Chang S., Van de Wouw A. P., Lopez-Ruiz F. J. (2017). Spontaneous and CRISPR/Cas9-induced mutation of the osmosensor histidine kinase of the canola pathogen Leptosphaeria maculans. Fungal Biol. Biotechnol. 4:12. 10.1186/s40694-017-0043-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igbalajobi O., Yu Z. Z., Fischer R. (2019). Red- and blue-light sensing in the plant pathogen alternaria alternata depends on phytochrome and the white-collar protein LreA. Mbio 10 e319–e371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M. K. (2018). Design principles for nuclease-deficient CRISPR-based transcriptional regulators. FEMS Yeast Res. 18:foy039. 10.1093/femsyr/foy039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., Xu R. G., Ren X., Ewen-Campen B., Rajakumar R., Zirin J., et al. (2018). Next-generation CRISPR/Cas9 transcriptional activation in drosophila using flySAM. Proc. Natl. Acad. Sci. U S A. 115 4719–4724. 10.1073/pnas.1800677115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Wang L., Zhang J., Liu H., Hong B., Tan H., et al. (2015). Identification of novel mureidomycin analogues via rational activation of a cryptic gene cluster in Streptomyces roseosporus NRRL 15998. Sci. Rep. 5:14111. 10.1038/srep14111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez A., Munoz-Fernandez G., Ledesma-Amaro R., Buey R. M., Revuelta J. L. (2019). One-vector CRISPR/Cas9 genome engineering of the industrial fungus Ashbya gossypii. Microb. Biotechnol. 12 1293–1301. 10.1111/1751-7915.13425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi M., Baghel R. S., Fogelman E., Stern R. A., Ginzberg I. (2018). Identification of candidate genes mediating apple fruit-cracking resistance following the application of gibberellic acids 4 + 7 and the cytokinin 6-benzyladenine. Plant Physiol. Biochem. 127 436–445. 10.1016/j.plaphy.2018.04.015 [DOI] [PubMed] [Google Scholar]
- Kakule T. B., Jadulco R. C., Koch M., Janso J. E., Barrows L. R., Schmidt E. W. (2015). Native promoter strategy for high-yielding synthesis and engineering of fungal secondary metabolites. ACS Synth. Biol. 4 625–633. 10.1021/sb500296p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale S. P., Bhatnagar D., Bennett J. W. (1994). Isolation and characterization of morphological variants of Aspergillus parasiticus deficient in secondary metabolite production. Mycological. Res. 98 645–652. 10.1016/s0953-7562(09)80411-5 [DOI] [PubMed] [Google Scholar]
- Kao P. H., Ng I. S. (2017). CRISPRi mediated phosphoenolpyruvate carboxylase regulation to enhance the production of lipid in Chlamydomonas reinhardtii. Bioresour. Technol. 245 1527–1537. 10.1016/j.biortech.2017.04.111 [DOI] [PubMed] [Google Scholar]
- Katayama T., Nakamura H., Zhang Y., Pascal A., Fujii W., Maruyama J. I. (2019). Forced recycling of an AMA1-based genome-editing plasmid allows for efficient multiple gene deletion/integration in the industrial filamentous fungus aspergillus oryzae. Appl. Environ. Microbiol. 85 e1818–e1896. 10.1128/AEM.01896-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama T., Tanaka Y., Okabe T., Nakamura H., Fujii W., Kitamoto K., et al. (2016). Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillus oryzae. Biotechnol. Lett. 38 637–642. 10.1007/s10529-015-2015-x [DOI] [PubMed] [Google Scholar]
- Keller N. P. (2019). Fungal secondary metabolism: regulation, function and drug discovery. Nat. Rev. Microbiol. 17 167–180. 10.1038/s41579-018-0121-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Kim J. S. (2014). A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 15 321–334. 10.1038/nrg3686 [DOI] [PubMed] [Google Scholar]
- Kjaerbolling I., Mortensen U. H., Vesth T., Andersen M. R. (2019). Strategies to establish the link between biosynthetic gene clusters and secondary metabolites. Fungal Genet. Biol. 130 107–121. 10.1016/j.fgb.2019.06.001 [DOI] [PubMed] [Google Scholar]
- Konermann S., Brigham M. D., Trevino A. E., Joung J., Abudayyeh O. O., Barcena C., et al. (2015). Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517 583–588. 10.1038/nature14136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuck U., Hoff B. (2010). New tools for the genetic manipulation of filamentous fungi. Appl. Microbiol. Biotechnol. 86 51–62. 10.1007/s00253-009-2416-7 [DOI] [PubMed] [Google Scholar]
- Kuivanen J., Arvas M., Richard P. (2017). Clustered genes encoding 2-Keto-l-gulonate reductase and l-idonate 5-dehydrogenase in the novel fungal d-glucuronic acid pathway. Front. Microbiol. 8:225. 10.3389/fmicb.2017.00225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuivanen J., Korja V., Holmström S., Richard P. (2019). Development of microtiter plate scale CRISPR/Cas9 transformation method for Aspergillus niger based on in vitro assembled ribonucleoprotein complexes. Fungal Biol. Biotechnol. 6 66–69. 10.1186/s40694-019-0066-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuivanen J., Richard P. (2018). NADPH-dependent 5-keto-D-gluconate reductase is a part of the fungal pathway for D-glucuronate catabolism. FEBS Lett. 592 71–77. 10.1002/1873-3468.12946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuivanen J., Wang Y. M. J., Richard P. (2016). Engineering Aspergillus niger for galactaric acid production: elimination of galactaric acid catabolism by using RNA sequencing and CRISPR/Cas9. Microbial. Cell Fact. 15:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujoth G. C., Sullivan T. D., Merkhofer R., Lee T. J., Wang H., Brandhorst T., et al. (2018). CRISPR/Cas9-mediated gene disruption reveals the importance of zinc metabolism for fitness of the dimorphic fungal pathogen blastomyces dermatitidis. mBio 9 412–418. 10.1128/mBio.00412-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson M. H., Gilbert L. A., Wang X., Lim W. A., Weissman J. S., Qi L. S. (2013). CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protocols 8 2180–2196. 10.1038/nprot.2013.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitão A. L., Costa M. C., Enguita F. J. (2017). Applications of genome editing by programmable nucleases to the metabolic engineering of secondary metabolites. J. Biotechnol. 241 50–60. 10.1016/j.jbiotec.2016.11.009 [DOI] [PubMed] [Google Scholar]
- Leonard W. R., Jr., Belyk K. M., Conlon D. A., Bender D. R., DiMichele L. M., Liu J., et al. (2007). Synthesis of the antifungal beta-1,3-glucan synthase inhibitor CANCIDAS (caspofungin acetate) from pneumocandin B0. J. Org. Chem. 72 2335–2343. 10.1021/jo062008i [DOI] [PubMed] [Google Scholar]
- Leynaud-Kieffer L. M. C., Curran S. C., Kim I., Magnuson J. K., Gladden J. M., Baker S. E., et al. (2019). A new approach to Cas9-based genome editing in Aspergillus niger that is precise, efficient and selectable. PLoS One 14:e0210243. 10.1371/journal.pone.0210243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Liu Q., Li X., Zhang C., Li J., Sun W., et al. (2020a). Construction of a new thermophilic fungus Myceliophthora thermophila platform for enzyme production using a versatile 2A peptide strategy combined with efficient CRISPR-Cas9 system. Biotechnol. Lett. 42 1181–1191. 10.1007/s10529-020-02882-5 [DOI] [PubMed] [Google Scholar]
- Li X., Liu Q., Sun W., He Q., Tian C. (2020b). Improving cellulases production by Myceliophthora thermophila through disruption of protease genes. Biotechnol. Lett. 42 219–229. 10.1007/s10529-019-02777-0 [DOI] [PubMed] [Google Scholar]
- Li J., Zhang Y., Zhang Y., Yu P. L., Pan H., Rollins J. A. (2018). Introduction of large sequence inserts by CRISPR-Cas9 to create pathogenicity mutants in the multinucleate filamentous pathogen sclerotinia sclerotiorum. mBio 9 518–567. 10.1128/mBio.00567-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li S. Y., Wang J., Liu G. (2019). CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol. 37 730–743. 10.1016/j.tibtech.2018.12.005 [DOI] [PubMed] [Google Scholar]
- Liang Y., Han Y., Wang C., Jiang C., Xu J. R. (2018). Targeted deletion of the USTA and UvSLT2 genes efficiently in ustilaginoidea virens with the CRISPR-Cas9 system. Front. Plant Sci. 9:699. 10.3389/fpls.2018.00699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim F. Y., Sanchez J. F., Wang C. C., Keller N. P. (2012). Toward awakening cryptic secondary metabolite gene clusters in filamentous fungi. Methods Enzymol. 517 303–324. 10.1016/B978-0-12-404634-4.00015-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H. C., Chooi Y. H., Dhingra S., Xu W., Calvo A. M., Tang Y. (2013). The fumagillin biosynthetic gene cluster in Aspergillus fumigatus encodes a cryptic terpene cyclase involved in the formation of beta-trans-bergamotene. J. Am. Chem. Soc. 135 4616–4619. 10.1021/ja312503y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Gao R., Li J., Lin L., Zhao J., Sun W., et al. (2017). Development of a genome-editing CRISPR/Cas9 system in thermophilic fungal Myceliophthora species and its application to hyper-cellulase production strain engineering. Biotechnol. Biofuels 10:1. 10.1186/s13068-016-0693-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Zhang Y., Li F., Li J., Sun W., Tian C. (2019). Upgrading of efficient and scalable CRISPR-Cas-mediated technology for genetic engineering in thermophilic fungus Myceliophthora thermophila. Biotechnol. Biofuels 12:293. 10.1186/s13068-019-1637-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Chen L., Jiang Y., Zhou Z., Zou G. (2015). Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov. 1:15007. 10.1038/celldisc.2015.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanovska M., Pilla G. (2017). Penicillin’s discovery and antibiotic resistance: lessons for the future? Yale J. Biol. Med. 90 135–145. [PMC free article] [PubMed] [Google Scholar]
- Lu S., Shen X., Chen B. (2017). Development of an efficient vector system for gene knock-out and near in-cis gene complementation in the sugarcane smut fungus. Sci. Rep. 7:3113. 10.1038/s41598-017-03233-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K. S., Wolf Y. I., Koonin E. V. (2018). Classification and Nomenclature of CRISPR-Cas systems: where from here? CRISPR J. 1 325–336. 10.1089/crispr.2018.0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manglekar R. R., Geng A. (2020). CRISPR-Cas9-mediated seb1 disruption in talaromyces pinophilus EMU for its enhanced cellulase production. Enzyme Microb. Technol. 140:109646. 10.1016/j.enzmictec.2020.109646 [DOI] [PubMed] [Google Scholar]
- Maruyama T., Dougan S. K., Truttmann M. C., Bilate A. M., Ingram J. R., Ploegh H. L. (2015). Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 33 538–542. 10.1038/nbt.3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y., Bai T., Phippen C. B. W., Nodvig C. S., Kjaerbolling I., Vesth T. C., et al. (2018). Novofumigatonin biosynthesis involves a non-heme iron-dependent endoperoxide isomerase for orthoester formation. Nat. Commun. 9:2587. 10.1038/s41467-018-04983-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsu-Ura T., Baek M., Kwon J., Hong C. (2015). Efficient gene editing in neurospora crassa with CRISPR technology. Fungal Biol. Biotechnol. 2:4. 10.1186/s40694-015-0015-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattern D. J., Schoeler H., Weber J., Novohradská S., Kraibooj K., Dahse H.-M., et al. (2015). Identification of the antiphagocytic trypacidin gene cluster in the human-pathogenic fungus Aspergillus fumigatus. Appl. Microbiol.Biotechnol. 99 10151–10161. 10.1007/s00253-015-6898-1 [DOI] [PubMed] [Google Scholar]
- McCarty N. S., Graham A. E., Studena L., Ledesma-Amaro R. (2020). Multiplexed CRISPR technologies for gene editing and transcriptional regulation. Nat. Commun. 11:1281. 10.1038/s41467-020-15053-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y. Z., Zhu Y. L., Huang P. W., Yang Q., Dai C. C. (2019). Strategies for gene disruption and expression in filamentous fungi. Appl. Microbiol. Biotechnol. 103 6041–6059. 10.1007/s00253-019-09953-2 [DOI] [PubMed] [Google Scholar]
- Miao J., Li X., Lin D., Liu X., Tyler B. M. (2018). Oxysterol-binding protein-related protein 2 is not essential for Phytophthora sojae based on CRISPR/Cas9 deletions. Environ. Microbiol. Rep. 10 293–298. 10.1111/1758-2229.12638 [DOI] [PubMed] [Google Scholar]
- Min K., Ichikawa Y., Woolford C. A., Mitchell A. P. (2016). Candida albicans gene deletion with a transient CRISPR-Cas9 system. mSphere 1 e00116–e00130. 10.1126/sciadv.1500248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica F. J. M., Diez-Villasenor C., Garcia-Martinez J., Almendros C. (2009). Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155 733–740. 10.1099/mic.0.023960-0 [DOI] [PubMed] [Google Scholar]
- Mosunova O., Navarro-Muñoz J. C., Collemare J. (2020). “The Biosynthesis of Fungal Secondary Metabolites: From Fundamentals to Biotechnological Applications,” in Reference Module in Life Sciences, (London: Elsevier; ). [Google Scholar]
- Nagy G., Szebenyi C., Csernetics A., Vaz A. G., Toth E. J., Vagvolgyi C., et al. (2017). Development of a plasmid free CRISPR-Cas9 system for the genetic modification of Mucor circinelloides. Sci. Rep. 7:16800. 10.1038/s41598-017-17118-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy G., Vaz A. G., Szebenyi C., Tako M., Toth E. J., Csernetics A., et al. (2019). CRISPR-Cas9-mediated disruption of the HMG-CoA reductase genes of mucor circinelloides and subcellular localization of the encoded enzymes. Fungal Genet. Biol. 129 30–39. 10.1016/j.fgb.2019.04.008 [DOI] [PubMed] [Google Scholar]
- Nah H. J., Pyeon H. R., Kang S. H., Choi S. S., Kim E. S. (2017). Cloning and heterologous expression of a large-sized natural product biosynthetic gene cluster in streptomyces species. Front. Microbiol. 8:394. 10.3389/fmicb.2017.00394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H., Katayama T., Okabe T., Iwashita K., Fujii W., Kitamoto K., et al. (2017). Highly efficient gene targeting in Aspergillus oryzae industrial strains under ligD mutation introduced by genome editing: strain-specific differences in the effects of deleting EcdR, the negative regulator of sclerotia formation. J. General Appl. Microbiol. 63 172–178. 10.2323/jgam.2016.10.002 [DOI] [PubMed] [Google Scholar]
- Nielsen M. L., Isbrandt T., Rasmussen K. B., Thrane U., Hoof J. B., Larsen T. O., et al. (2017). Genes linked to production of secondary metabolites in Talaromyces atroroseus revealed using CRISPR-Cas9. Plos One 12:e169712. 10.1371/journal.pone.0169712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M. R., Wollenberg R. D., Westphal K. R., Sondergaard T. E., Wimmer R., Gardiner D. M., et al. (2019). Heterologous expression of intact biosynthetic gene clusters in Fusarium graminearum. Fungal Genet. Biol. 132:103248. 10.1016/j.fgb.2019.103248 [DOI] [PubMed] [Google Scholar]
- Nodvig C. S., Hoof J. B., Kogle M. E., Jarczynska Z. D., Lehmbeck J., Klitgaard D. K., et al. (2018). Efficient oligo nucleotide mediated CRISPR-Cas9 gene editing in Aspergilli. Fungal Genet. Biol. 115 78–89. 10.1016/j.fgb.2018.01.004 [DOI] [PubMed] [Google Scholar]
- Nodvig C. S., Nielsen J. B., Kogle M. E., Mortensen U. H. (2015). A CRISPR-Cas9 System for genetic engineering of filamentous fungi. PLoS One 10:e0133085. 10.1371/journal.pone.0133085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa A. E., Sitrin R., Lee S. S. (1999). Purification of pneumocandins by preparative silica-gel high-performance liquid chromatography. J. Chromatography A. 831 217–225. 10.1016/s0021-9673(98)00936-4 [DOI] [Google Scholar]
- Paulussen C., Hallsworth J. E., Alvarez-Perez S., Nierman W. C., Hamill P. G., Blain D., et al. (2017). Ecology of aspergillosis: insights into the pathogenic potency of aspergillus fumigatus and some other aspergillus species. Microb. Biotechnol. 10 296–322. 10.1111/1751-7915.12367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng R., Wang Y., Feng W. W., Yue X. J., Chen J. H., Hu X. Z., et al. (2018). CRISPR/dCas9-mediated transcriptional improvement of the biosynthetic gene cluster for the epothilone production in Myxococcus xanthus. Microb. Cell Fact. 17:15. 10.1186/s12934-018-0867-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatek A., Ali Z., Baazim H., Li L., Abulfaraj A., Al-Shareef S., et al. (2015). RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol. J. 13 578–589. 10.1111/pbi.12284 [DOI] [PubMed] [Google Scholar]
- Pohl C., Kiel J. A., Driessen A. J., Bovenberg R. A., Nygard Y. (2016). CRISPR/Cas9 based genome editing of penicillium chrysogenum. ACS Synth. Biol. 5 754–764. 10.1021/acssynbio.6b00082 [DOI] [PubMed] [Google Scholar]
- Qian C., Ren N., Wang J., Xu Q., Chen X., Qi X. (2018). Effects of exogenous application of CPPU, NAA and GA4+7 on parthenocarpy and fruit quality in cucumber (Cucumis sativus L.). Food Chem. 243 410–413. 10.1016/j.foodchem.2017.09.150 [DOI] [PubMed] [Google Scholar]
- Qin H., Xiao H., Zou G., Zhou Z., Zhong J.-J. (2017). CRISPR-Cas9 assisted gene disruption in the higher fungus ganoderma species. Process. Biochem. 56 57–61. 10.1016/j.procbio.2017.02.012 [DOI] [Google Scholar]
- Robbins N., Caplan T., Cowen L. E. (2017). Molecular evolution of antifungal drug resistance. Annu. Rev. Microbiol. 71 753–775. 10.1146/annurev-micro-030117-020345 [DOI] [PubMed] [Google Scholar]
- Rojas-Sánchez U., López-Calleja A. C., Millán-Chiu B. E., Fernández F., Loske A. M., Gómez-Lim M. A. (2020). Enhancing the yield of human erythropoietin in Aspergillus niger by introns and CRISPR-Cas9. Protein Expr. Purif. 168:105570. 10.1016/j.pep.2020.105570 [DOI] [PubMed] [Google Scholar]
- Roman E., Coman I., Prieto D., Alonso-Monge R., Pla J. (2019). Implementation of a CRISPR-based system for gene regulation in candida albicans. mSphere 4 e01–e19. 10.1128/mSphere.00001-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux I., Woodcraft C., Hu J., Wolters R., Gilchrist C. L. M., Chooi Y.-H., et al. (2020). CRISPR-mediated activation of biosynthetic gene clusters for bioactive molecule discovery in filamentous fungi. ACS Synt. Biol. 9 1843–1854. 10.1021/acssynbio.0c00197 [DOI] [PubMed] [Google Scholar]
- Salazar-Cerezo S., Kun R. S., de Vries R. P., Garrigues S. (2020). CRISPR/Cas9 technology enables the development of the filamentous ascomycete fungus Penicillium subrubescens as a new industrial enzyme producer. Enzyme Microb. Technol. 133:109463. 10.1016/j.enzmictec.2019.109463 [DOI] [PubMed] [Google Scholar]
- Sanson K. R., Hanna R. E., Hegde M., Donovan K. F., Strand C., Sullender M. E., et al. (2018). Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat. Commun. 9:5416. 10.1038/s41467-018-07901-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkari P., Marx H., Blumhoff M. L., Mattanovich D., Sauer M., Steiger M. G. (2017). An efficient tool for metabolic pathway construction and gene integration for aspergillus niger. Bioresour. Technol. 245 1327–1333. 10.1016/j.biortech.2017.05.004 [DOI] [PubMed] [Google Scholar]
- Satish L., Shamili S., Muthubharathi B. C., Ceasar S. A., Kushmaro A., Singh V., et al. (2020). ”CRISPR-Cas9 system for fungi genome engineering toward industrial applications,” in Genome Engineering via CRISPR-Cas9 System.). Amsterdam: Elsevier Science, 69–81. [Google Scholar]
- Scharf D. H., Heinekamp T., Brakhage A. A. (2014). Human and plant fungal pathogens: the role of secondary metabolites. PLoS Pathog 10:e1003859. 10.1371/journal.ppat.1003859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M., Schweizer G., Kahmann R. (2018). Comparative analyses of secreted proteins in plant pathogenic smut fungi and related basidiomycetes. Fungal Genet. Biol. 112 21–30. 10.1016/j.fgb.2016.12.003 [DOI] [PubMed] [Google Scholar]
- Schuster M., Schweizer G., Reissmann S., Kahmann R. (2016). Genome editing in Ustilago maydis using the CRISPR-Cas system. Fungal Genet. Biol. 89 3–9. 10.1016/j.fgb.2015.09.001 [DOI] [PubMed] [Google Scholar]
- Shapiro R. S., Chavez A., Porter C. B. M., Hamblin M., Kaas C. S., DiCarlo J. E., et al. (2017). A CRISPR–Cas9-based gene drive platform for genetic interaction analysis in candida albicans. Nat. Microbiol. 3 73–82. 10.1038/s41564-017-0043-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T. Q., Gao J., Wang W. J., Wang K. F., Xu G. Q., Huang H., et al. (2019). CRISPR/Cas9-based genome editing in the filamentous fungus fusarium fujikuroi and its application in strain engineering for gibberellic acid production. ACS Synth. Biol. 8 445–454. 10.1021/acssynbio.8b00478 [DOI] [PubMed] [Google Scholar]
- Shi T.-Q., Liu G.-N., Ji R.-Y., Shi K., Song P., Ren L.-J., et al. (2017). CRISPR/Cas9-based genome editing of the filamentous fungi: the state of the art. Appl. Microbiol. Biotechnol. 101 7435–7443. 10.1007/s00253-017-8497-9 [DOI] [PubMed] [Google Scholar]
- Song L., Ouedraogo J. P., Kolbusz M., Nguyen T. T. M., Tsang A. (2018). Efficient genome editing using tRNA promoter-driven CRISPR/Cas9 gRNA in Aspergillus niger. Plos One 13:e0202868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R., Zhai Q., Sun L., Huang E., Zhang Y., Zhu Y., et al. (2019). CRISPR/Cas9 genome editing technology in filamentous fungi: progress and perspective. Appl. Microbiol. Biotechnol. 103 6919–6932. 10.1007/s00253-019-10007-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg S. H., Redding S., Jinek M., Greene E. C., Doudna J. A. (2014). DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507 62–67. 10.1038/nature13011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strezoska Z., Dickerson S. M., Maksimova E., Chou E., Gross M. M., Hemphill K., et al. (2020). CRISPR-mediated transcriptional activation with synthetic guide RNA. J. Biotechnol. 319 25–35. 10.1016/j.jbiotec.2020.05.005 [DOI] [PubMed] [Google Scholar]
- Tak Y. E., Kleinstiver B. P., Nunez J. K., Hsu J. Y., Horng J. E., Gong J., et al. (2017). Inducible and multiplex gene regulation using CRISPR-Cpf1-based transcription factors. Nat. Methods 14 1163–1166. 10.1038/nmeth.4483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throckmorton K., Lim F. Y., Kontoyiannis D. P., Zheng W., Keller N. P. (2016). Redundant synthesis of a conidial polyketide by two distinct secondary metabolite clusters in aspergillus fumigatus. Environ. Microbiol. 18 246–259. 10.1111/1462-2920.13007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y., Weber T., Lee S. Y. (2019). CRISPR/Cas-based genome engineering in natural product discovery. Nat. Prod. Rep. 36 1262–1280. 10.1039/c8np00089a [DOI] [PubMed] [Google Scholar]
- Ullah I., Khan A. R., Jung B. K., Khan A. L., Lee I.-J., Shin J.-H., et al. (2014). Gibberellins synthesized by the entomopathogenic bacterium, Photorhabdus temperata M1021 as one of the factors of rice plant growth promotion. J. Plant Interact. 9 775–782. 10.1080/17429145.2014.942956 [DOI] [Google Scholar]
- van Leeuwe T. M., Arentshorst M., Ernst T., Alazi E., Punt P. J., Ram A. F. J., et al. (2019). Efficient marker free CRISPR/Cas9 genome editing for functional analysis of gene families in filamentous fungi. Fungal Biol. Biotechnol. 6:13. 10.1186/s40694-019-0076-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas V. K., Bushkin G. G., Bernstein D. A., Getz M. A., Sewastianik M., Barrasa M. I., et al. (2018). New CRISPR mutagenesis strategies reveal variation in repair mechanisms among fungi. mSphere 3 e118–e154. 10.1128/mSphere.00154-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P. (2018). Two distinct approaches for CRISPR-Cas9-mediated gene editing in cryptococcus neoformans and related species. mSphere 3 e208–e218. 10.1128/mSphereDirect.00208-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Cobine P. A., Coleman J. J. (2018). Efficient genome editing in Fusarium oxysporum based on CRISPR/Cas9 ribonucleoprotein complexes. Fungal Genet. Biol. 117 21–29. 10.1016/j.fgb.2018.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Coleman J. J. (2019). Progress and challenges: development and implementation of CRISPR/Cas9 technology in filamentous fungi. Comput. Struct. Biotechnol. J. 17 761–769. 10.1016/j.csbj.2019.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Chen H., Tang X., Zhang H., Chen W., Chen Y. Q. (2017). Molecular tools for gene manipulation in filamentous fungi. Appl. Microbiol. Biotechnol. 101 8063–8075. 10.1007/s00253-017-8486-z [DOI] [PubMed] [Google Scholar]
- Weber J., Valiante V., Nodvig C. S., Mattern D. J., Slotkowski R. A., Mortensen U. H., et al. (2017). Functional reconstitution of a fungal natural product gene cluster by advanced genome editing. ACS Synth. Biol. 6 62–68. 10.1021/acssynbio.6b00203 [DOI] [PubMed] [Google Scholar]
- Wei T. Y., Wu Y. J., Xie Q. P., Tang J. W., Yu Z. T., Yang S. B., et al. (2020). CRISPR/Cas9-based genome editing in the filamentous fungus glarea lozoyensis and its application in manipulating gloF. ACS Synth. Biol. 9 1968–1977. 10.1021/acssynbio.9b00491 [DOI] [PubMed] [Google Scholar]
- Weidner G., d’Enfert C., Koch A., Mol P. C., Brakhage A. A. (1998). Development of a homologous transformation system for the human pathogenic fungus aspergillus fumigatus based on the pyrG gene encoding orotidine 5’-monophosphate decarboxylase. Curr. Genet. 33 378–385. 10.1007/s002940050350 [DOI] [PubMed] [Google Scholar]
- Wenderoth M., Pinecker C., Voss B., Fischer R. (2017). Establishment of CRISPR/Cas9 in alternaria alternata. Fungal Genet. Biol. 101 55–60. 10.1016/j.fgb.2017.03.001 [DOI] [PubMed] [Google Scholar]
- Weyda I., Yang L., Vang J., Ahring B. K., Lübeck M., Lübeck P. S., et al. (2017). A comparison of Agrobacterium-mediated transformation and protoplast-mediated transformation with CRISPR-Cas9 and bipartite gene targeting substrates, as effective gene targeting tools for Aspergillus carbonarius. J. Microbiol. Methods 135 26–34. 10.1016/j.mimet.2017.01.015 [DOI] [PubMed] [Google Scholar]
- Wu C., Chen Y., Qiu Y., Niu X., Zhu N., Chen J., et al. (2020). A simple approach to mediate genome editing in the filamentous fungus Trichoderma reesei by CRISPR/Cas9-coupled in vivo gRNA transcription. Biotechnol. Lett. 42 1203–1210. 10.1007/s10529-020-02887-0 [DOI] [PubMed] [Google Scholar]
- Xu G., Li J., Liu Q., Sun W., Jiang M., Tian C. (2018). Transcriptional analysis of Myceliophthora thermophila on soluble starch and role of regulator AmyR on polysaccharide degradation. Biores. Technol. 265 558–562. 10.1016/j.biortech.2018.05.086 [DOI] [PubMed] [Google Scholar]
- Youssar L., Wernet V., Hensel N., Yu X., Hildebrand H. G., Schreckenberger B., et al. (2019). Intercellular communication is required for trap formation in the nematode-trapping fungus duddingtonia flagrans. PLoS Genet 15:e1008029. 10.1371/journal.pgen.1008029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun C. S., Motoyama T., Osada H. (2017). Regulatory mechanism of mycotoxin tenuazonic acid production in pyricularia oryzae. ACS Chem. Biol. 12 2270–2274. 10.1021/acschembio.7b00353 [DOI] [PubMed] [Google Scholar]
- Zhang A., Lu P., Dahl-Roshak A. M., Paress P. S., Kennedy S., Tkacz J. S., et al. (2003). Efficient disruption of a polyketide synthase gene (pks1) required for melanin synthesis through Agrobacterium-mediated transformation of Glarea lozoyensis. Mol. Genet. Genom. 268 645–655. 10.1007/s00438-002-0780-4 [DOI] [PubMed] [Google Scholar]
- Zhang C., Meng X., Wei X., Lu L. (2016). Highly efficient CRISPR mutagenesis by microhomology-mediated end joining in Aspergillus fumigatus. Fungal. Genet. Biol. 86 47–57. 10.1016/j.fgb.2015.12.007 [DOI] [PubMed] [Google Scholar]
- Zhang L., Zheng X., Cairns T. C., Zhang Z., Wang D., Zheng P., et al. (2020). Disruption or reduced expression of the orotidine-5’-decarboxylase gene pyrG increases citric acid production: a new discovery during recyclable genome editing in Aspergillus niger. Microb. Cell Fact. 19:76. 10.1186/s12934-020-01334-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Dai Z., Liang Y., Yin M., Ma K., He M., et al. (2014). Sequence-specific inhibition of microRNA via CRISPR/CRISPRi system. Sci. Rep. 4:3943. 10.1038/srep03943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Zheng P., Zhang K., Cairns T. C., Meyer V., Sun J., et al. (2019). 5S rRNA promoter for guide RNA expression enabled highly efficient CRISPR/Cas9 genome editing in Aspergillus niger. ACS Synth. Biol. 8 1568–1574. 10.1021/acssynbio.7b00456 [DOI] [PubMed] [Google Scholar]
- Zheng Y. M., Lin F. L., Gao H., Zou G., Zhang J. W., Wang G. Q., et al. (2017). Development of a versatile and conventional technique for gene disruption in filamentous fungi based on CRISPR-Cas9 technology. Sci. Rep. 7:9250. 10.1038/s41598-017-10052-3 [DOI] [PMC free article] [PubMed] [Google Scholar]