Abstract
The prognosis of patients with human papillomavirus (HPV)-negative head and neck squamous cell carcinoma (HNSCC) is poorer than those with HPV-positive HNSCC. The present study aimed to identify novel and specific biomarkers of HPV-negative HNSCC using bioinformatics analysis and associated experiments. The gene expression profiles of HPV-negative HNSCC tissues and corresponding clinical data were downloaded from The Cancer Genome Atlas database and used in a weighted gene co-expression network analysis. Genes in clinically significant co-expression modules were used to construct a protein-protein interaction (PPI) network. The genes demonstrating a high degree score in the PPI network and a high correlation with tumor grade were considered hub genes. The diagnostic value of the hub genes associated with HPV-negative and HPV-positive HNSCC was analyzed using differential expression gene (DEG) analysis, immunohistochemical (IHC) staining and a receiver operating characteristic (ROC) curve analysis. Seven genes [Serrate RNA effector molecule (SRRT), checkpoint kinase 2 (CHEK2), small nuclear ribonucleoprotein polypeptide E (SNRPE), proteasome 26S subunit ATPase 2 (PSMC2), origin recognition complex subunit 5 (ORC5), S100 calcium binding protein A7 and keratinocyte differentiation associated protein (KRTDAP)] were demonstrated to be hub genes in clinically significant co-expression modules. DEG, IHC and ROC curve analyses revealed that SRRT, CHEK2 and SNRPE were significantly upregulated in HPV-negative and HPV-positive HNSCC tissues compared with in adjacent tissues, and these genes demonstrated a high diagnostic value for distinguishing HNSCC tissues. However, PSMC2, ORC5 and KRTDAP were the only differentially expressed genes identified in HPV-negative HNSCC tissues, and these genes demonstrated a high diagnostic value for HPV-negative HNSCC. PSMC2, ORC5 and KRTDAP may therefore serve as novel and specific biomarkers for HPV-negative HNSCC, potentially improving the diagnosis and treatment of patients with HPV-negative HNSCC.
Keywords: human papillomavirus-negative head and neck squamous cell carcinoma, weighted gene co-expression network analysis, specific, biomarker
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common malignancy worldwide (1). There are ~650,000 new cases of HNSCC diagnosed every year, with >350,000 associated deaths occurring in a year (2). Several risk factors involved in the development of HNSCC have been identified, including alcohol, tobacco use and human papillomavirus (HPV) infection, the latter of which is a major independent risk factor for HNSCC (3). Compared with HPV-positive HNSCC, HPV-negative HNSCC is more aggressive and is associated with a higher rate of drug resistance (4). Similarly, patients with HPV-negative HNSCC demonstrate lower overall survival rates than those with HPV-positive HNSCC (5). The identification of novel and specific biomarkers for HPV-negative HNSCC may therefore help to improve the understanding of the mechanism that underlies the progression of HPV-negative HNSCC, as well as to improve the diagnosis and treatment of the disease.
With the progression of high-throughput sequencing technology and bioinformatics methods, additional oncogenes involved in the development of HNSCC have been identified (6,7). Weighted gene co-expression network analysis (WGCNA) is a bioinformatics method that involves calculating the associations between gene co-expression modules and clinical traits using a weighted soft threshold (8). Recently, with the use of WGCNA, a series of biomarkers for HNSCC have been identified. For example, Song et al (9) identified 16 genes involved in the immune and inflammatory response that promoted the development of HNSCC. Similarly, using WGCNA, Zhang et al (10) identified 12 genes associated with perineural invasion in patients with HNSCC. However, the molecular mechanism of each HNSCC subtype is different, and the feasibility and specificity of the majority of biomarkers obtained from previous studies are limited.
The current study combined WGCNA, differential expression gene (DEG) analysis and experimental verification to identify specific biomarkers for HPV-negative HNSCC.
Materials and methods
Clinical specimens
A total of 62 pairs of primary HNSCC tissues and adjacent non-tumor tissues (1-2 cm distance from the tumor tissues) were collected from patients at The Affiliated Hospital of Guizhou Medical University (Guiyang, China). None of the enrolled patients underwent treatment (such as chemotherapy or radiation) prior to tissue collection. HPV infection status had been determined prior to study commencement by performing in situ hybridization (ISH), as previously described (11), and P16 staining (Fig. S1). HPV-positive HNSCC was diagnosed based on ISH or P16 (also named p16INK4a) positive results, while HPV-negative HNSCC was defined based on negative ISH and P16 results. A total of 35 patients with HPV-positive HNSCC (26 tissues were ISH and P16 positive, 9 tissues were ISH positive only and none of the tissues were P16 positive only) and 27 patients with HPV-negative HNSCC were enrolled in the present study. The basic clinical characteristics (American Joint Committee on Cancer) (12) of the patients with HPV-negative and HPV-positive HNSCC are presented in Table I. The current study was approved by the Human Trait Ethics Committee of Guizhou Medical University and performed in accordance with the Declaration of Helsinki. All patients who provided samples signed written informed consent.
Table I.
Basic clinical characteristics of patients with HPV-negative (n=27) and HPV-positive (n=35) HNSCC.
| Clinicopathological feature | HPV-negative HNSCC | HPV-positive HNSCC | P-value |
|---|---|---|---|
| Mean age ± SD, years | 71.4±6.7 | 72.4±7.3 | >0.05 |
| Sex, n (%) | >0.05 | ||
| Male | 12 (44.4) | 17 (48.5) | |
| Female | 15 (55.6) | 18 (51.5) | |
| Tumor grade (Broders), n (%) | >0.05 | ||
| Grade I | 12 (44.4) | 16 (45.7) | |
| Grade II | 13 (48.1) | 16 (45.7) | |
| Grade III | 2 (7.5) | 3 (9.6) | |
| Metastasis at diagnosis, n (%) | >0.05 | ||
| Yes | 2 (7.5) | 4 (11.4) | |
| No | 25 (92.5) | 31 (88.6) | |
| Tumor subsite, n (%) | >0.05 | ||
| Oral tongue | 10 (37.0) | 13 (37.1) | |
| Buccal mucosa | 2 (7.4) | 2 (5.7) | |
| Larynx | 7 (26.0) | 9 (25.7) | |
| Oral cavity | 4 (14.8) | 5 (14.2) | |
| Floor of mouth | 4 (14.8) | 6 (17.1) |
HPV, human papillomavirus; HNSCC, head and neck squamous cell carcinoma.
Data processing
For constructing the WGCNA, the gene expression profiles of 54 HPV-negative HNSCC tissues (ISH and P16 negative stain), along with their corresponding clinical characteristics (Table SI), were downloaded from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). After normalization, outliers were assessed using the hierarchical cluster algorithm with a cut-off of 140. A fragments per kilobase of transcript per million mapped reads (FPKM) value of <0.5 was used as the threshold for removing the genes with low expression. This was necessary as a gene must be expressed at a minimal level before it is likely to be translated into a protein or be considered biologically important (13). Consequently, 15,703 genes expressed in the 54 HPV-negative HNSCC tissues, along with the corresponding clinical characteristics data, were used in the WGCNA. For DEGs analysis, the gene expression prolife in TCGA and Gene Expression Omnibus datasets [GSE117973 (14), GSE85446 (15) and GSE112026 (16); https://www.ncbi.nlm.nih.gov/gds] containing 103 HPV-positive HNSCC and 165 HPV-negative HNSCC was downloaded. After merge and batch normalization using the sva package (version: 3.11; Bioconductor; https://www.bioconductor.org/packages/release/bioc/html/sva.htl), the merge gene expression profile were used for DEGs analysis. |Log fold-change (FC)| >0.5 and adjusted P<0.05 were set as cut-off values to consider genes differentially expressed between HPV-positive and HPV-negative HNSCC.
WGCNA
The WGCNA R package (version 1.69; https://cran.r-project.org/web/packages/WGCNA/index.html) was used to conduct the WGCNA. All gene pairs were analyzed using Pearson's correlation analysis, the results of which were used to construct a matrix of similarity. Subsequently, to produce a scale-free co-expression network, the matrix of similarity was constructed using a soft power of β=7. The adjacency matrix was then translated into a topological overlap matrix (TOM). Furthermore, median linkage hierarchical clustering was analyzed using the TOM-based dissimilarity measure with a minimum size of 50.
Identification of clinically significant modules
Following WGCNA, the Pearson's correlation analyses between various module eigengenes and the clinical characteristics of patients were assessed. These traits included node (N) stage, tumor (T) stage, tumor grade, recurrence, perineural invasion, tobacco use history and alcohol consumption history. Modules were considered clinically significant if they were correlated with two clinical characteristics (r>0.3 and P<0.05). In the clinically significant modules, gene significance (GS) was quantified using associations between the individual genes and the clinical characteristics of interest, along with the module membership (MM), which was itself determined using the correlation between the module eigengenes and the gene expression profiles. If P<0.05 for the correlation between GS and MM in the clinically significant modules, the modules were subjected to further analysis.
Construction of a protein-protein interaction (PPI) network
The online Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; http://string-db.org) was used to construct the PPI network. Nodes without connections were removed. The minimum required interaction score was set at 0.7 (high confidence). Node and edge information was exported into a text file, which was subsequently imported into Cytoscape software (version 6.1; http://cytoscape.org/). In the PPI network, nodes represented the protein information encoded by the gene of interest. Lines indicated the interactions between two proteins. The degree score was calculated using the Cytohub plug-in (version 1.0; http://github.com/cytoscape/appstore) and was set as the criterion used to calculate the interaction between a single protein and other proteins. Finally, proteins encoded by genes with a degree score in the top 10% were visualized using Cytoscape, and the genes coding for proteins with a degree score in the top 1% were considered candidate hub genes.
Pearson's correlation analysis between the candidate hub genes and tumor grade
Pearson's correlation analysis between the expression of candidate hub genes and tumor grade was performed using SPSS version 20.0 (IBM Corp.). P<0.05 was set as the cut-off indicating a significant correlation. The genes significantly correlated with tumor grade were considered hub genes.
Immunohistochemical (IHC) staining
All 27 HPV-negative and 35 HPV-positive HNSCC tissues were fixed in 4% paraformaldehyde (Boster Biological Technology) at room temperature. Subsequently, they were embedded into paraffin and cut into 4-µm-thick sections. After heating at 65°C, the sections were deparaffinized using xylene and rehydrated in a graded ethanol series (100, 80, 60 and 40%). After conducting antigen retrieval using sodium citrate (0.01 mol/l; Boster Biological Technology), endogenous peroxidase activity was blocked using 3% H2O2. Subsequently, 5% bovine serum albumin (Wuhan Servicebio Technology Co., Ltd.) was added at room temperature for 1 h. The sections were then incubated for 16 h at 4°C with the following primary antibodies: p16INK4a (1:200; cat. no. A11651; ABclonal Biotech Co., Ltd.), Serrate RNA effector molecule (SRRT; 1:200; cat. no. A8219; ABclonal Biotech Co., Ltd.), checkpoint kinase 2 (CHEK2; 1:200; cat. no. A19543; ABclonal Biotech Co., Ltd.), small nuclear ribonucleoprotein polypeptide E (SNRPE; 1:200; cat. no. A5488; ABclonal Biotech Co., Ltd.), proteasome 26S subunit ATPase 2 (PSMC2; 1:400; cat. no. 14905-1-AP; ProteinTech Group, Inc.), origin recognition complex subunit 5 (ORC5; 1:400; cat no. 11542-1-AP, ProteinTech Group, Inc.), S100 calcium binding protein A7 (S100A7; 1:200; cat no. 13061-1-AP, ProteinTech Group Inc.) and keratinocyte differentiation associated protein (KRTDAP; 1:1,000; cat. no. ab204583; Abcam). Subsequently, the sections were incubated for 2 h at room temperature with anti-mouse (cat. no. BM3895) and anti-rabbit (cat. no. BM3894) horseradish peroxidase-conjugated goat secondary antibodies (both 1:500; Boster Biological Technology). Samples were then stained with 3,3′-diaminobenzidine and hematoxylin at room temperature for 1 min, after which a light orthophoto microscope (magnification, ×200 and ×400) was used to obtain images. Finally, the protein expression levels of the target genes were evaluated based on the sum of the intensity score (0, no staining; 1, weakly positive; 2, moderately positive; and 3, strongly positive) and the score for the proportion of positive cells (0, <1; 1, 1–33; 2, 34–66; and 3, 67–100%) using Image-Pro Plus software (version 6.0; Media Cybernetics, Inc.). Total scores of 0–2, 3–4 and 5–6 indicated low, moderate and high expression, respectively. The difference in the protein expression of tissues was determined using Fisher's exact test combined with Bonferroni's correction test based on their expression levels. P<0.05 was considered to indicate a statistically significant difference.
Verification of the diagnostic value of hub genes
The diagnostic value of hub genes for HPV-negative and HPV-positive HNSCC was analyzed using a receiver operating characteristic (ROC) curve analysis using the IHC-based protein levels. The protein expression scores of the HNSCC and normal tissues were imported into SPSS version 20.0 (IBM Corp.), after which ROC curve analysis was performed. An area under the curve (AUC) value of >0.7 was considered to indicate a high diagnostic value.
Results
WGCNA
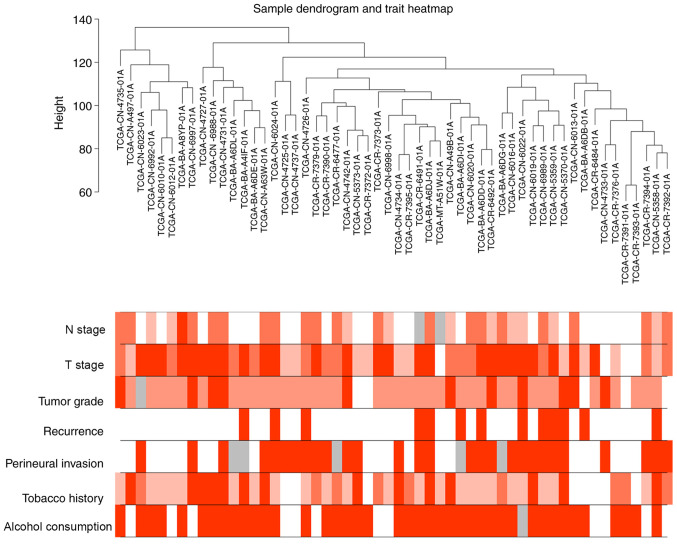
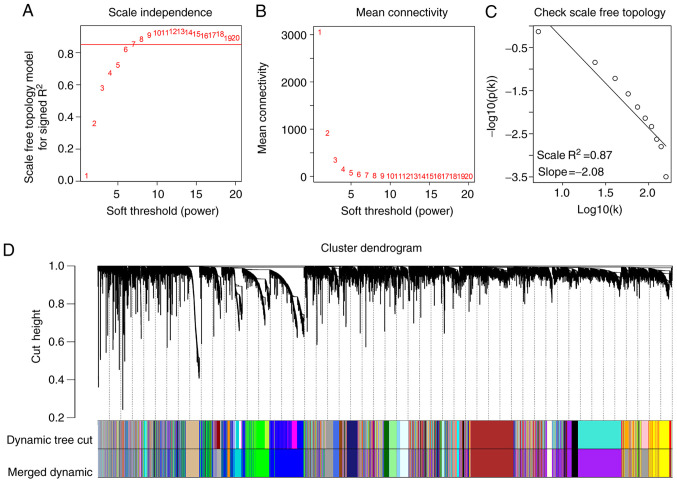
The gene expression profiles of 54 HPV-negative HNSCC tissues (with negative ISH and P16 staining) were downloaded from the TCGA database, as well as corresponding clinical characteristic data. The sample dendrogram revealed that there were no outliers; similarly, the trait heatmap showed that clinical data of most patients in TCGA database were completely documented (Fig. 1). Therefore, after removing the genes with low expression, the gene expression profiles of all 54 HPV-negative HNSCC tissues were used in the WGCNA. When the soft power β was set as 7, the scale independence of the topology network reached >0.85 (Fig. 2A) with the mean connectivity close to 0 (Fig. 2B). Therefore, a soft power of β=7 was selected as the soft threshold for performing subsequent analyses. The results revealed that when β=7, the topological overlap matrix was able to meet the scale-free topology criterion with R2=0.87 (Fig. 2C). The results identified 27 gene co-expression modules (paleturquoise, cyan, salmon, sienna3, violet, royalblue, saddlebrown, darkmagenta, brown, black, purple, lightyellow, darkolivegreen, grey60, yellow, darkgreen, skyblue, lightcyan, lightgreen, tan, darkturquoise, darkorange, blue, green, steelblue, darkgrey and midnightblue), while the genes that were not co-expressed were clustered in the grey module (Fig. 2D). The 27 gene co-expression modules were then used for further analysis.
Figure 1.
Sample dendrogram of 54 human papillomavirus-negative head and neck squamous cell carcinoma tissues and the corresponding clinical characteristics of patients, including N stage, T stage, tumor grade, recurrence, perineural invasion, tobacco use history and alcohol consumption history. Red, white and grey indicate high, low and missing values, respectively. N, node; T, tumor.
Figure 2.
Weighted gene co-expression network analysis. (A) Analysis of the scale-free fit index for various β soft-thresholding powers. (B) Analysis of the mean connectivity for various soft-thresholding powers. (C) Scale-free topology when β=7. (D) Dendrogram of all genes clustered based on a dissimilarity measure (1-topological overlap matrix). There were 27 gene co-expression modules in total (paleturquoise, cyan, salmon, sienna3, violet, royalblue, saddlebrown, darkmagenta, brown, black, purple, lightyellow, darkolivegreen, grey60, yellow, darkgreen, skyblue, lightcyan, lightgreen, tan, darkturquoise, darkorange, blue, green, steelblue, darkgrey and midnightblue). Genes without co-expression were clustered into the grey module.
Identification of clinically significant modules
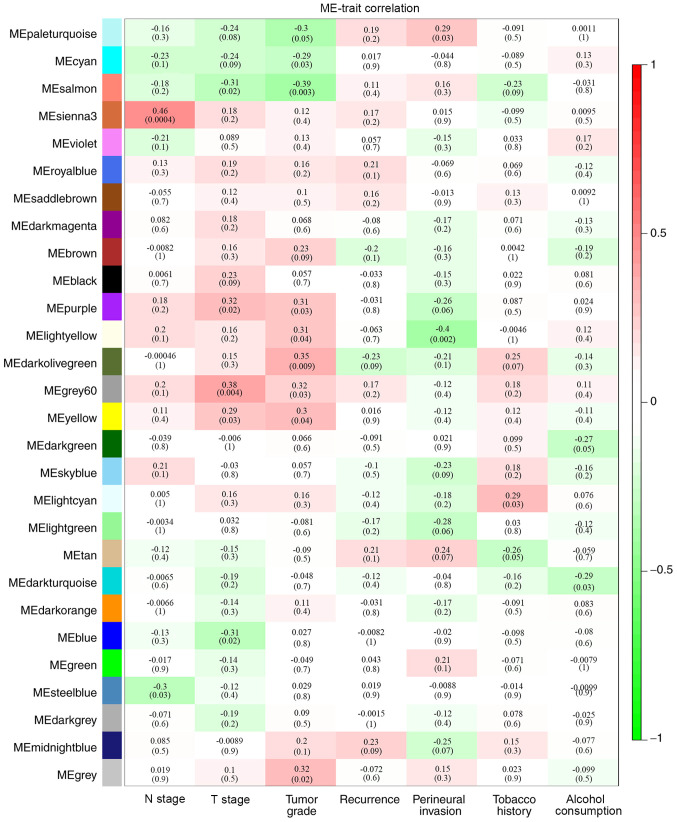
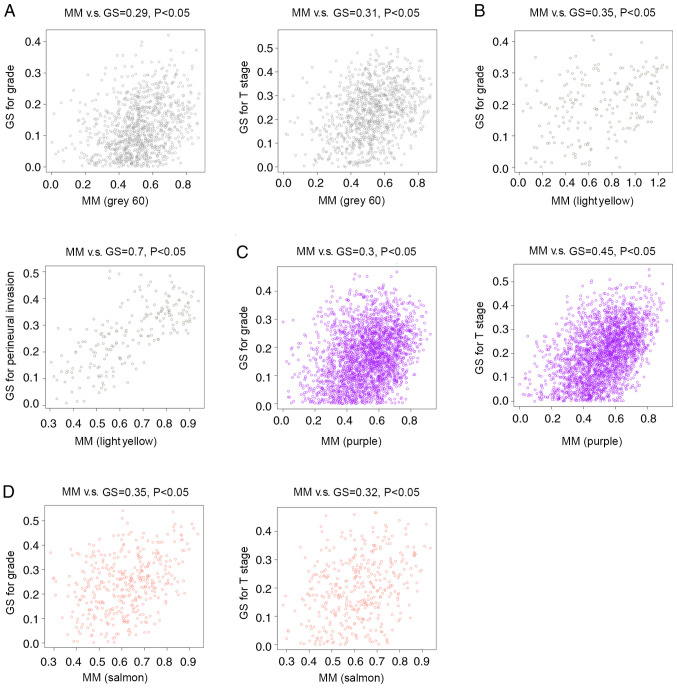
By analyzing the correlation between the gene co-expression modules and clinical characteristics, it was determined that four gene co-expression modules were simultaneously and significantly correlated with two clinical characteristics in patients with HPV-negative HNSCC. The gene of the grey60 module was positively correlated with T stage (R=0.38; P=0.004) and tumor grade (R=0.32; P=0.03). Genes of the lightyellow module were positively correlated with tumor grade (R=0.31; P=0.04) and negatively correlated with perineural invasion (R=−0.4; P=0.002). Additionally, genes in the purple module were positively correlated with T stage (R=0.32; P=0.02) and tumor grade (R=0.31; P=0.03), while genes of the salmon module were negatively correlated with T stage (R=−0.31; P=0.02) and tumor stage (R=−0.39; P=0.003) (Fig. 3). Correlations between GS and MM were subsequently calculated in the four aforementioned modules. The MM of the grey60 module was correlated with the GS for tumor grade (correlation=0.29; P<0.05) and T stage (correlation=0.31; P<0.05) (Fig. 4A). The MM of the lightyellow module was correlated with the GS for tumor grade (correlation=0.35; P<0.05) and perineural invasion (correlation=0.7; P<0.05) (Fig. 4B). The MM of the purple module was correlated with the GS for tumor grade (correlation=0.3; P<0.05) and T stage (correlation=0.45; P<0.05) (Fig. 4C), while the MM of the salmon module was correlated with the GS for tumor grade (correlation=0.35; P<0.05) and T stage (correlation=0.32; P<0.05) (Fig. 4D). Therefore, these four modules (grey60, lightyellow, purple and salmon modules) were set as key gene modules and were analyzed further.
Figure 3.
Identification of clinically significant modules based on the correlation of module eigengenes with clinical characteristics. Each cell contains the corresponding correlation coefficient and P-value (in brackets). The colors indicate the strength of correlation according to the color legend. ME, module; T, tumor; N, node.
Figure 4.
Analysis of GS and MM in the clinically significant modules. (A) MM of the grey60 module was correlated with GS for tumor grade and T stage. (B) MM in the lightyellow module was correlated with GS for tumor grade and perineural invasion. (C) MM of the purple module was correlated with GS for tumor grade and T stage. (D) MM of the salmon module was correlated with GS for tumor grade and T stage. GS, gene significance; MM, module membership; cor, correlation; T, tumor; N, node.
Construction of the PPI network and selection of candidate hub genes
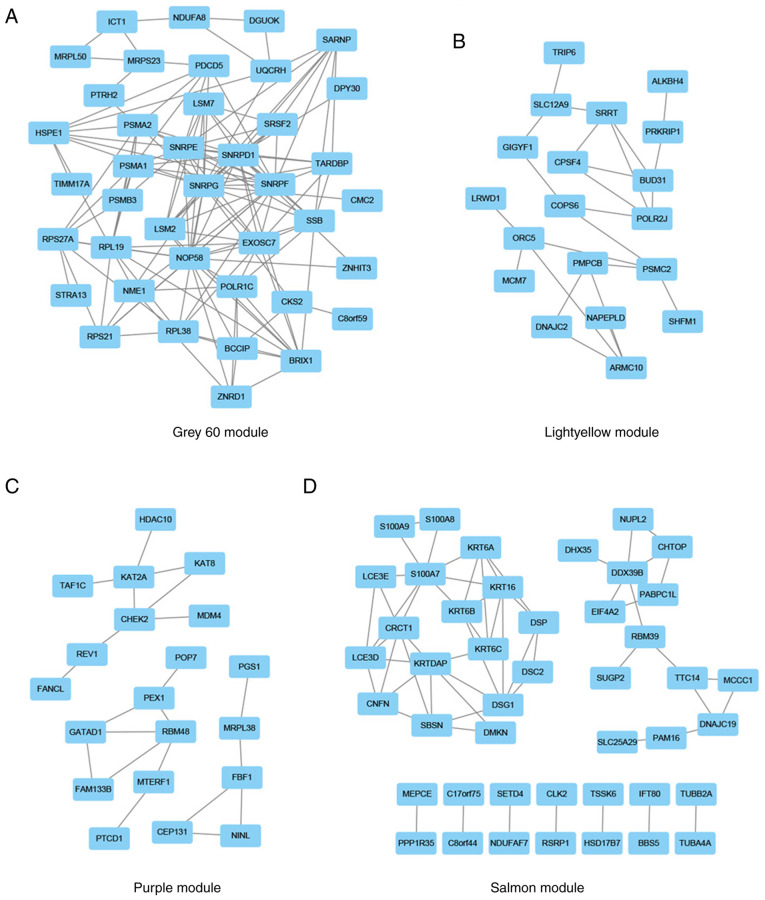
The lightyellow, grey60, purple and salmon modules included 201, 997, 2,538 and 389 genes, respectively. After importing these genes into the STRING database and removing the nodes without connections, genes with a degree score in the top 10% in the grey60 (Fig. 5A), lightyellow (Fig. 5B), purple (Fig. 5C) and salmon (Fig. 5D) modules were visualized using Cytoscape. The genes with a degree score in the top 1% in each module are presented in Table II. The resultant 17 genes were considered candidate hub genes and may serve key roles in the development of HPV-negative HNSCC.
Figure 5.
PPI network. Genes with a degree score in the top 10% in the (A) grey60, (B) light yellow, (C) purple and (D) salmon modules were used to construct the PPI network. Nodes and edges indicate the genes and their interactions, respectively. PPI, protein-protein interaction.
Table II.
Genes with a degree score in the top 1% in each module.
| Module | Gene | Degree score |
|---|---|---|
| Grey60 module | SNRPG | 20 |
| SNRPF | 18 | |
| SNRPE | 16 | |
| SNRPD1 | 16 | |
| NOP58 | 16 | |
| Salmon module | S100A7 | 8 |
| KRTDAP | 7 | |
| Purple module | DDX39B | 5 |
| CHEK2 | 4 | |
| KAT2A | 4 | |
| Lightyellow module | ORC5 | 4 |
| PSMC2 | 4 | |
| SRRT | 4 | |
| BUD31 | 4 | |
| POLR2J | 4 | |
| ARMC10 | 4 | |
| RBM48 | 4 |
SRRT, serrate RNA effector molecule; CHEK2, checkpoint kinase 2; SNRPE, small nuclear ribonucleoprotein polypeptide E; PSMC2, proteasome 26S subunit ATPase 2; ORC5, origin recognition complex subunit 5; S100A7, S100 calcium binding protein A7; KRTDAP, keratinocyte differentiation associated protein.
Pearson's correlation analysis between the candidate hub genes and tumor grade
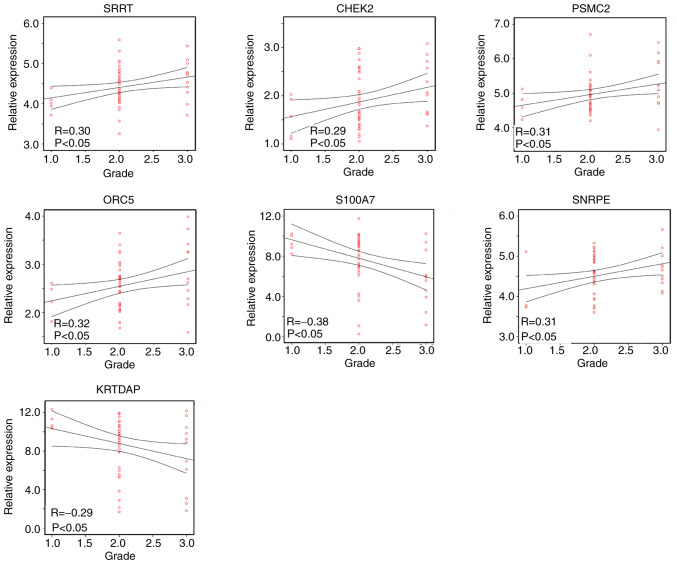
As the four gene modules were all correlated with tumor grade, the correlation between candidate hub gene expression and the tumor grade of HPV-negative HNSCC was further analyzed using Pearson's correlation analysis. The results revealed that SRRT (R=0.30; P<0.05), CHEK2 (R=0.29; P<0.05), SNRPE (R=0.31; P<0.05), PSMC2 (R=0.31; P<0.05) and ORC5 (R=0.32; P<0.05) were positively correlated with the tumor grade of HPV-negative HNSCC tissue, while S100A7 (R=−0.38; P<0.05) and KRTDAP (R=−0.29; P<0.05) were negatively correlated with tumor grade (Fig. 6). Therefore, SRRT, CHEK2, SNRPE, PSMC2, ORC5, S100A7 and KRTDAP were identified as hub genes in HPV-negative HNSCC.
Figure 6.
Pearson's correlation analysis between hub gene expression and tumor grade in human papillomavirus-negative head and neck squamous cell carcinoma. SRRT, serrate RNA effector molecule; CHEK2, checkpoint kinase 2; SNRPE, small nuclear ribonucleoprotein polypeptide E; PSMC2, proteasome 26S subunit ATPase 2; ORC5, origin recognition complex subunit 5; S100A7, S100 calcium binding protein A7; KRTDAP, keratinocyte differentiation associated protein.
Identification of the expression levels of hub genes in HPV-negative and HPV-positive HNSCC
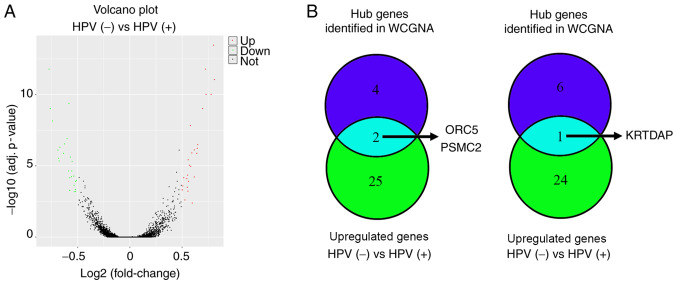
To analyze the expression levels of hub genes in HPV-negative and HPV-positive HNSCC, the merged gene data expression profile of 103 HPV-positive HNSCC and 165 HPV-negative HNSCC samples in TCGA and GEO databases (GSE117973, GSE85446 and GSE112026) was used to perform DEGs analysis. The results revealed that there were 27 upregulated genes and 25 downregulated genes in HPV-negative HNSCC compared with in HPV-positive HNSCC (Fig. 7A; Table III). Through an intersection analysis, it was found that the hub genes ORC5 and PSMC2 were upregulated in HPV-negative HNSCC tissues compared with in HPV-positive HNSCC tissues, while KRTDAP was downregulated (Fig. 7B).
Figure 7.
Differential expression gene analysis for HPV-negative and HPV-positive HNSCC. (A) Volcano plot of the differentially expressed genes between HPV-negative and HPV-positive HNSCC. (B) Intersection analysis for the differentially expressed genes (HPV-negative HNSCC versus HPV-positive HNSCC) and hub genes identified from WGCNA. HPV, human papillomavirus; HNSCC, head and neck squamous cell carcinoma; WGCNA, weighted gene co-expression network analysis; PSMC2, proteasome 26S subunit ATPase 2; ORC5, origin recognition complex subunit 5; KRTDAP, keratinocyte differentiation associated protein.
Table III.
Detailed information of genes differently expressed in HPV-negative and HPV-positive head and neck squamous cell carcinoma according to the different expression analysis.
| Gene | Log (fold-change) |
|---|---|
| DSG1 | 0.814689016 |
| SPRR2G | 0.807843405 |
| KLK5 | 0.78303004 |
| KRT1 | 0.735022158 |
| PSMC2 | 0.729090901 |
| MMP10 | 0.701782465 |
| ORC5 | 0.655262803 |
| SPRR2B | 0.650839232 |
| LCE3E | 0.646742549 |
| DSC1 | 0.622678824 |
| KLK7 | 0.622523674 |
| DEFB103B | 0.602302594 |
| PTHLH | 0.598655999 |
| MMP13 | 0.588604647 |
| MMP3 | 0.584023444 |
| FAM25A | 0.576520859 |
| ASPRV1 | 0.566257594 |
| LCE3D | 0.564179626 |
| CCNA1 | 0.561757208 |
| KLK8 | 0.558113758 |
| CPA4 | 0.555893249 |
| IGFL1 | 0.551152103 |
| CDSN | 0.532585037 |
| KLK6 | 0.52189567 |
| CDA | 0.512697702 |
| KRT75 | 0.507135684 |
| WFDC12 | 0.506385839 |
| CYP4X1 | −0.50678768 |
| SYNGR3 | −0.514548202 |
| RIBC2 | −0.514701295 |
| RNF212 | −0.517145557 |
| ABCA3 | −0.528349222 |
| TMPRSS2 | −0.530093548 |
| TMSB15A | −0.53215901 |
| KRTDAP | −0.54853466 |
| MEI1 | −0.551064139 |
| GABRP | −0.554796624 |
| CDKN2C | −0.565098291 |
| YBX2 | −0.569455778 |
| TCP11 | −0.570473276 |
| KRT19 | −0.578420257 |
| SMC1B | −0.580791027 |
| PODXL2 | −0.59651466 |
| PLAC8 | −0.620386125 |
| FAM3B | −0.630548652 |
| STAG3 | −0.665598925 |
| TAF7L | −0.672735588 |
| ZNF541 | −0.677964463 |
| KCNS1 | −0.683860665 |
| SYCP2 | −0.735795983 |
| NEFH | −0.757761211 |
| CDKN2A | −0.768176467 |
HPV, human papillomavirus; PSMC2, proteasome 26S subunit ATPase 2; ORC5, origin recognition complex subunit 5; KRTDAP, keratinocyte differentiation associated protein.
Verification of the protein expression levels of hub genes in HPV-negative and HPV-positive HNSCC tissues
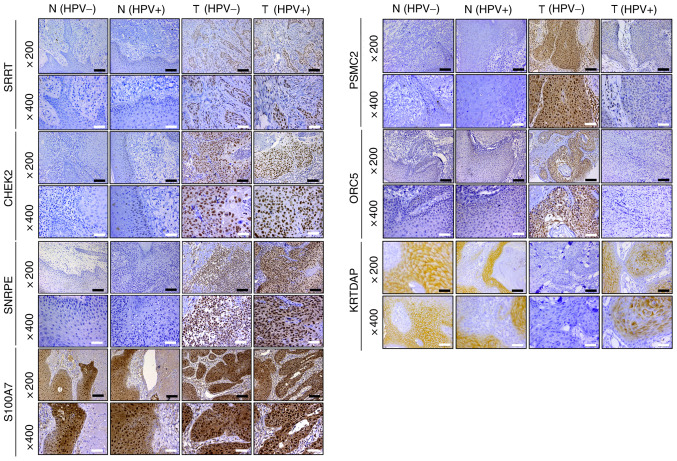
To verify the bioinformatics results, IHC staining of 27 HPV-negative and 35 HPV-positive HNSCC tissues was performed, alongside their corresponding adjacent tissues. The results revealed that the protein expression levels of SRRT, CHEK2 and SNRPE were increased in HPV-negative HNSCC and HPV-positive HNSCC tissues compared with in their adjacent counterparts; however, there was no significant difference between HPV-negative and HPV-positive HNSCC tissues. S100A7 expression was not significantly different among HPV-negative HNSCC, HPV-positive HNSCC and adjacent tissues. Furthermore, it was demonstrated that PSMC2 and ORC5 expression was significantly increased, while KRTDAP expression was significantly decreased in HPV-negative HNSCC tissues compared with in adjacent and HPV-positive HNSCC tissues (Table IV; Figs. 8 and S2).
Table IV.
Detailed immunohistochemical scoring of SRRT, CHEK2, SNRPE, PSMC2, ORC5, S100A7 and KRTDAP expression in HPV− HNSCC (n=27), HPV+ HNSCC (n=35) and corresponding adjacent tissues.
| Gene | Tissues | Low expression (0–2), n | Medium expression (3–4), n | High expression (5–6), n |
|---|---|---|---|---|
| SRRT | Tumor (HPV+)a | 5 | 4 | 26 |
| Adjacent (HPV+) | 27 | 6 | 2 | |
| Tumor (HPV−)b | 2 | 3 | 22 | |
| Adjacent (HPV−) | 18 | 6 | 3 | |
| CHEK2 | Tumor (HPV+)a | 3 | 3 | 29 |
| Adjacent (HPV+) | 28 | 4 | 3 | |
| Tumor (HPV−)b | 5 | 4 | 18 | |
| Adjacent (HPV−) | 18 | 7 | 2 | |
| PSMC2 | Tumor (HPV+) | 15 | 13 | 7 |
| Adjacent (HPV+) | 22 | 7 | 6 | |
| Tumor (HPV−)b,c | 2 | 3 | 22 | |
| Adjacent (HPV−) | 20 | 5 | 2 | |
| OCR5 | Tumor (HPV+) | 19 | 10 | 6 |
| Adjacent (HPV+) | 24 | 9 | 2 | |
| Tumor (HPV−)b,c | 2 | 7 | 18 | |
| Adjacent (HPV−) | 19 | 5 | 3 | |
| S100A7 | Tumor (HPV+) | 6 | 5 | 24 |
| Adjacent (HPV+) | 4 | 4 | 27 | |
| Tumor (HPV−) | 2 | 4 | 21 | |
| Adjacent (HPV−) | 4 | 5 | 18 | |
| SNRPE | Tumor (HPV+)a | 3 | 4 | 28 |
| Adjacent (HPV+) | 26 | 3 | 6 | |
| Tumor (HPV−)b | 4 | 2 | 21 | |
| Adjacent (HPV−) | 17 | 6 | 4 | |
| KRTDAP | Tumor (HPV+) | 9 | 6 | 20 |
| Adjacent (HPV+) | 8 | 8 | 19 | |
| Tumor (HPV−)b,c | 20 | 4 | 3 | |
| Adjacent (HPV−) | 4 | 7 | 16 |
P<0.05 vs. HPV+ adjacent tissues
P<0.05 vs. HPV− adjacent tissues
P<0.05 vs. HPV+ HNSCC. SRRT, serrate RNA effector molecule; CHEK2, checkpoint kinase 2; SNRPE, small nuclear ribonucleoprotein polypeptide E; PSMC2, proteasome 26S subunit ATPase 2; ORC5, origin recognition complex subunit 5; S100A7, S100 calcium binding protein A7; KRTDAP, keratinocyte differentiation associated protein; HPV, human papillomavirus; HNSCC, head and neck squamous cell carcinoma.
Figure 8.
Immunohistochemistry was used to detect the protein expression levels of SRRT, CHEK2, SNRPE, PSMC2, ORC5, S100A7 and KRTDAP in HPV-negative HNSCC, HPV-positive HNSCC and adjacent tissues (magnification, ×200 and ×400). Black scale bar, 100 µm; white scale bar, 50 µm. SRRT, serrate RNA effector molecule; CHEK2, checkpoint kinase 2; SNRPE, small nuclear ribonucleoprotein polypeptide E; PSMC2, proteasome 26S subunit ATPase 2; ORC5, origin recognition complex subunit 5; S100A7, S100 calcium binding protein A7; KRTDAP, keratinocyte differentiation associated protein; HPV, human papillomavirus; HNSCC, head and neck squamous cell carcinoma; N, normal tissues; T, tumor tissues.
Verification of the diagnostic value of hub genes for HPV-negative and HPV-positive HNSCC
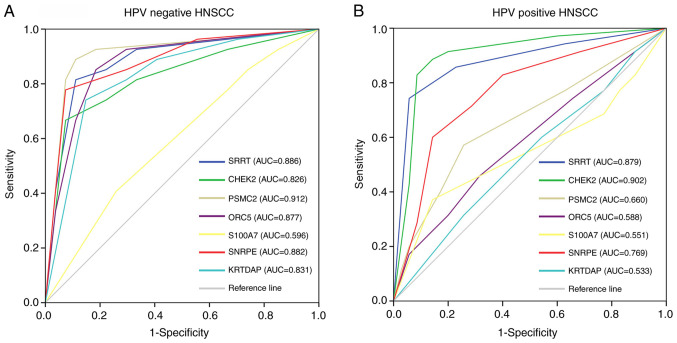
Using protein level data obtained by IHC staining, ROC curve analysis was performed. The results revealed that SRRT, CHEK2, SNRPE, PSMC2, ORC5 and KRTDAP exhibited high diagnostic value for distinguishing between HPV-negative HNSCC and adjacent tissues, while S100A7 did not exert this effect (Fig. 9A). However, only SRRT, CHEK2 and SNRPE had high diagnostic value for distinguishing between HPV-positive HNSCC and adjacent tissues (Fig. 9B). Thus, SRRT, CHEK2 and SNRPE may be used as common biomarkers for both HPV-negative and HPV-positive HNSCC, while PSMC2, ORC5 and KRTDAP may be specific biomarkers for HPV-negative HNSCC only.
Figure 9.
Receiver operating characteristic curve analysis was used to determine the diagnostic value of SRRT, CHEK2, SNRPE, PSMC2, ORC5, S100A7 and KRTDAP for (A) HPV-negative and (B) HPV-positive HNSCC based on protein level data obtained by immunohistochemical staining. AUC >0.7 indicated genes with high diagnostic value for distinguishing between HNSCC and adjacent tissues. SRRT, serrate RNA effector molecule; CHEK2, checkpoint kinase 2; SNRPE, small nuclear ribonucleoprotein polypeptide E; PSMC2, proteasome 26S subunit ATPase 2; ORC5, origin recognition complex subunit 5; S100A7, S100 calcium binding protein A7; KRTDAP, keratinocyte differentiation associated protein; HPV, human papillomavirus; HNSCC, head and neck squamous cell carcinoma; AUC, area under the curve.
Discussion
There are numerous risk factors involved in the pathogenesis of HNSCC, including HPV infection. Although the majority of HNSCC cases involve HPV infection, certain patients develop HNSCC without it (17,18). The molecular mechanisms underlying these two subtypes are different (19). Furthermore, HPV-negative HNSCC is more aggressive than HPV-positive HNSCC (20,21). Therefore, the identification of novel and specific biomarkers for HPV-negative HNSCC may improve the understanding of the specific molecular mechanism associated with HPV-negative HNSCC, which may improve the diagnosis and treatment of patients.
The current study identified four gene co-expression modules associated with clinical characteristics of patients via WGCNA. The results revealed 17 genes in these gene co-expression modules that had a high degree score (top 1% of genes in each module) in the PPI network. Additionally, seven of these genes (SRRT, CHEK2, SNRPE, PSMC2, ORC5, S100A7 and KRTDAP) were correlated with tumor grade in HPV-negative HNSCC. DEG analysis demonstrated that PSMC2 and ORC5 expression was higher in HPV-negative HNSCC compared with in HPV-positive HNSCC, while KRTDAP expression was lower. Furthermore, using IHC staining and ROC curve analysis, it was revealed that SRRT, CHEK2, SNRPE, PSMC2, ORC5 and KRTDAP were differentially expressed in HPV-negative HNSCC tissues compared with in adjacent tissues and had high diagnostic values for distinguishing between HPV-negative HNSCC tissues and adjacent tissues. However, after performing additional IHC staining and ROC curve analysis, it was demonstrated that SRRT, CHEK2 and SNRPE were differentially expressed in HPV-positive HNSCC tissues compared with in adjacent tissues, exhibiting a high diagnostic value for distinguishing between HPV-positive HNSCC and adjacent tissues. Thus, while SRRT, CHEK2 and SNRPE may be used as common biomarkers for both HPV-negative and HPV-positive HNSCC, PSMC2, ORC5 and KRTDAP may be specific to HPV-negative HNSCC only.
SRRT (also named Ars2) was first isolated from a hamster cell line and serves a key role in sodium arsenite resistance (22). Furthermore, it has been demonstrated that SRRT is involved in the biosynthesis of certain microRNAs (23). However, research on the role of SRRT in the development of cancer is conflicting. It has been demonstrated that SRRT serves as an oncogene in glioblastoma and promotes the proliferation and migration of LN-229 and U87 glioblastoma cells via the MAPK/ERK signaling pathway (24). Additionally, SRRT expression is upregulated in cholangiocarcinoma and associated with poor outcomes (25). However, pediatric acute lymphoblastic leukemia involving low SRRT expression has poor chemotherapy outcomes (26). CHEK2 was first reported as an important breast cancer susceptibility gene; it serves a key role in regulating cell apoptosis, cell cycle and DNA repair (27). It increases the stability of P53 and induces cell cycle arrest in the G1 phase (28). However, CHEK2 mutations are very common in several types of cancer, including breast cancer (29), colorectal cancer (30) and oral squamous cell carcinoma (31). Additionally, CHEK2 promotes the progression of various types of cancer, including hepatocellular carcinoma (32) and colorectal cancer (33). SNRPE is a member of a large family of polypeptides that are conserved in eukaryotes and archaebacteria (34). Previous studies have revealed that it is involved in RNA processing and mRNA degradation (35,36). Furthermore, a previous study has revealed that SNRPE is highly expressed in high-grade prostate cancer, promoting cell proliferation (37). In the present study, it was demonstrated that SRRT, CHEK2 and SNRPE were in clinically significant gene modules, had high degree scores in the PPI network, were associated with tumor grade and were highly expressed in HPV-negative and HPV-positive HNSCC tissues. Furthermore, it was revealed that SRRT, CHEK2 and SNRPE had a high diagnostic value in distinguishing HNSCC and adjacent tissues. Therefore, to the best of our knowledge, the present study was the first to demonstrate that SRRT, CHEK2 and SNRPE may be used as common biomarkers for both HPV-negative and HPV-positive HNSCC. Targeting SRRT, CHEK2 and SNRPE may aid the therapy of patients with HPV-negative and HPV-positive HNSCC. However, the mechanism of SRRT, CHEK2 and SNRPE in the development of HNSCC requires further study.
PSMC2 is a novel gene on chromosome 7q22.1-q22.3 that encodes a member of the 19S proteasome (38). Recently, PSMC2 was revealed as an oncogene in several types of cancer, including pancreatic cancer (39) and osteosarcoma (40). Similarly, high PSMC2 expression predicts a poor prognosis in patients with osteosarcoma (41). ORC5 is a key member of the origin recognition complex that binds to replication origins, initiating replication itself (42). Additionally, it induces large-scale chromatin decondensation and regulates the cell cycle and proliferation (43). KRTDAP acts as a soluble regulator of keratinocyte differentiation and serves a key role in embryonic skin morphogenesis (44). Research has revealed that KRTDAP mRNA is upregulated in HPV-positive HNSCC tissues compared with in adjacent tissues (45). In the present study, it was demonstrated that PSMC2 and ORC5 expression was significantly increased in HPV-negative HNSCC tissues compared with in corresponding adjacent and HPV-positive HNSCC tissues. Additionally, KRTDAP was significantly decreased in HPV-negative HNSCC tissues compared with in corresponding adjacent and HPV-positive HNSCC tissues. Similarly, it was revealed that PSMC2, ORC5 and KRTDAP had a high diagnostic value in distinguishing HPV-negative HNSCC and adjacent tissues. However, it could not distinguish between HPV-positive HNSCC and adjacent tissues. Therefore, it was hypothesized that PSMC2, ORC5 and KRTDAP may be used as specific biomarkers for patients with HPV-negative HNSCC only. It is well known that HPV-negative HNSCC is more aggressive than HPV-positive HNSCC, so the downregulation of KRTDAP and upregulation of PSMC2 and ORC5 may be key for HPV-negative HNSCC cells to become aggressive. Targeting ORC5, PSMC and KRTDAP may therefore help the individualized treatment of patients with HPV-negative HNSCC. However, additional experiments should be performed to verify this hypothesis.
In conclusion, based on WGCNA, PPI network, Pearson's correlation, DEG, IHC and ROC curve analyses, SRRT, CHEK2 and SNRPE were revealed to be common biomarkers for both HPV-negative and HPV-positive HNSCC, while PSMC2, ORC5 and KRTDAP may be potential specific biomarkers for HPV-negative HNSCC. The results may improve the understanding of HNSCC, as well as providing potential biomarkers and targets for the diagnosis and treatment of patients with this disease.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
The current study was supported by Guizhou Big Data Health Management Industry Technology Innovation Strategic Alliance [grant no. Qianketong (2016)193].
Funding
The current study was supported by Guizhou Big Data Health Management Industry Technology Innovation Strategic Alliance [grant no. Qianketong (2016)193].
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request, and the GSE117973, GSE85446 and GSE112026 datasets analyzed during the current study are available in the Gene Expression Omnibus repository (https://www.ncbi.nlm.nih.gov/gds).
Authors' contributions
YS, YC and ZZ were responsible for the experimental design, analysis and interpretation of data. DR, YY and BW performed the experiments. YS and YC were responsible for confirming the authenticity of the data. All authors took part in drafting the article and revising it critically for important intellectual content, agreed to be accountable for all aspects of the work, and read and approved the final manuscript.
Ethics approval and consent to participate
The current study was approved by the Human Trait Ethics Committee of Guizhou Medical University (Guiyang, China) and performed in accordance with the Declaration of Helsinki. All patients from which samples were obtained provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Johnson DE, Burtness B, Leemans CR, Lui VWY, Bauman JE, Grandis JR. Head and neck squamous cell carcinoma. Nat Rev Dis Primers. 2020;6:92. doi: 10.1038/s41572-020-00224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dhull AK, Atri R, Dhankhar R, Chauhan AK, Kaushal V. Major risk factors in head and neck cancer: A retrospective analysis of 12-year experiences. World J Oncol. 2018;9:80–84. doi: 10.14740/wjon1104w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plzák J, Bouček J, Bandúrová V, Kolář M, Hradilová M, Szabo P, Lacina L, Chovanec M, Smetana K., Jr The head and neck squamous cell carcinoma microenvironment as a potential target for cancer therapy. Cancers (Basel) 2019;11:440. doi: 10.3390/cancers11040440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabatini ME, Chiocca S. Human papillomavirus as a driver of head and neck cancers. Br J Cancer. 2020;122:306–314. doi: 10.1038/s41416-019-0602-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Viros Porcuna D, Pollan Guisasola C, Viña Soria C, Cirauqui Cirauqui B, Pardo Muñoz L, Collurá F, Mesia Nin R. Transoral robotic surgery for squamous cell carcinoma of the oropharynx in a primarily human papillomavirus-negative patient population. Clin Transl Oncol. 2020;22:1303–1311. doi: 10.1007/s12094-019-02256-y. [DOI] [PubMed] [Google Scholar]
- 6.Shen Y, Liu J, Zhang L, Dong S, Zhang J, Liu Y, Zhou H, Dong W. Identification of potential biomarkers and survival analysis for head and neck squamous cell carcinoma using bioinformatics strategy: A study based on TCGA and GEO datasets. Biomed Res Int. 2019;2019:7376034. doi: 10.1155/2019/7376034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang B, Fu L, Xu S, Xiao J, Li Z, Liu Y. A nomogram based on a gene signature for predicting the prognosis of patients with head and neck squamous cell carcinoma. Int J Biol Markers. 2019;34:309–317. doi: 10.1177/1724600819865745. [DOI] [PubMed] [Google Scholar]
- 8.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song Y, Pan Y, Liu J. The relevance between the immune response-related gene module and clinical traits in head and neck squamous cell carcinoma. Cancer Manag Res. 2019;11:7455–7472. doi: 10.2147/CMAR.S201177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z, Liu R, Jin R, Fan Y, Li T, Shuai Y, Li X, Wang X, Luo J. Integrating clinical and genetic analysis of perineural invasion in head and neck squamous cell carcinoma. Front Oncol. 2019;9:434. doi: 10.3389/fonc.2019.00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chi J, Preeshagul IR, Sheikh-Fayyaz S, Teckie S, Kohn N, Ziemba Y, Laser A, Frank D, Ghaly M, Kamdar D, et al. Evaluating of HPV-DNA ISH as an adjunct to p16 testing in oropharyngeal cancer. Future Sci OA. 2020;6:FSO606. doi: 10.2144/fsoa-2020-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lydiatt WM, Patel SG, O'Sullivan B, Brandwein MS, Ridge JA, Migliacci JC, Loomis AM, Shah JP. Head and neck cancers-major changes in the American joint committee on cancer eighth edition cancer staging manual. CA Cancer J Clin. 2017;67:122–137. doi: 10.3322/caac.21389. [DOI] [PubMed] [Google Scholar]
- 13.Bi N, Sun Y, Lei S, Zeng Z, Zhang Y, Sun C, Yu C. Identification of 40S ribosomal protein S8 as a novel biomarker for alcohol-associated hepatocellular carcinoma using weighted gene co-expression network analysis. Oncol Rep. 2020;44:611–627. doi: 10.3892/or.2020.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng B, Shen Y, Pastor Hostench X, Bieg M, Plath M, Ishaque N, Eils R, Freier K, Weichert W, Zaoui K, Hess J. Integrative analysis of multi-omics data identified EGFR and PTGS2 as key nodes in a gene regulatory network related to immune phenotypes in head and neck cancer. Clin Cancer Res. 2020;26:3616–3628. doi: 10.1158/1078-0432.CCR-19-3997. [DOI] [PubMed] [Google Scholar]
- 15.Mes SW, Te Beest D, Poli T, Rossi S, Scheckenbach K, van Wieringen WN, Brink A, Bertani N, Lanfranco D, Silini EM, et al. Prognostic modeling of oral cancer by gene profiles and clinicopathological co-variables. Oncotarget. 2017;8:59312–59323. doi: 10.18632/oncotarget.19576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelley DZ, Flam EL, Izumchenko E, Danilova LV, Wulf HA, Guo T, Singman DA, Afsari B, Skaist AM, Considine M, et al. Integrated analysis of whole-genome ChIP-Seq and RNA-Seq data of primary head and neck tumor samples associates HPV integration sites with open chromatin marks. Cancer Res. 2017;77:6538–6550. doi: 10.1158/0008-5472.CAN-17-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jethwa AR, Khariwala SS. Tobacco-related carcinogenesis in head and neck cancer. Cancer Metastasis Rev. 2017;36:411–423. doi: 10.1007/s10555-017-9689-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paver EC, Currie AM, Gupta R, Dahlstrom JE. Human papilloma virus related squamous cell carcinomas of the head and neck: Diagnosis, clinical implications and detection of HPV. Pathology. 2020;52:179–191. doi: 10.1016/j.pathol.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 19.Koneva LA, Zhang Y, Virani S, Hall PB, McHugh JB, Chepeha DB, Wolf GT, Carey TE, Rozek LS, Sartor MA. HPV Integration in HNSCC correlates with survival outcomes, immune response signatures, and candidate drivers. Mol Cancer Res. 2018;16:90–102. doi: 10.1158/1541-7786.MCR-17-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qian X, Nguyen DT, Dong Y, Sinikovic B, Kaufmann AM, Myers JN, Albers AE, Graviss EA. Prognostic score predicts survival in HPV-negative head and neck squamous cell cancer patients. Int J Biol Sci. 2019;15:1336–1344. doi: 10.7150/ijbs.33329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vossen DM, Verhagen CVM, van der Heijden M, Essers PBM, Bartelink H, Verheij M, Wessels LFA, van den Brekel MWM, Vens C. Genetic factors associated with a poor outcome in head and neck cancer patients receiving definitive chemoradiotherapy. Cancers (Basel) 2019;11:445. doi: 10.3390/cancers11040445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elahi S, Egan SM, Holling GA, Kandefer RL, Nemeth MJ, Olejniczak SH. The RNA binding protein Ars2 supports hematopoiesis at multiple levels. Exp Hematol. 2018;64:45–58.e9. doi: 10.1016/j.exphem.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gruber JJ, Olejniczak SH, Yong J, La Rocca G, Dreyfuss G, Thompson CB. Ars2 promotes proper replication-dependent histone mRNA 3′ end formation. Mol Cell. 2012;45:87–98. doi: 10.1016/j.molcel.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ke XX, Pang Y, Chen K, Zhang D, Wang F, Zhu S, Mao J, Hu X, Zhang G, Cui H. Knockdown of arsenic resistance protein 2 inhibits human glioblastoma cell proliferation through the MAPK/ERK pathway. Oncol Rep. 2018;40:3313–3322. doi: 10.3892/or.2018.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He Q, Cai L, Shuai L, Li D, Wang C, Liu Y, Li X, Li Z, Wang S. Ars2 is overexpressed in human cholangiocarcinomas and its depletion increases PTEN and PDCD4 by decreasing microRNA-21. Mol Carcinog. 2013;52:286–296. doi: 10.1002/mc.21859. [DOI] [PubMed] [Google Scholar]
- 26.Cui L, Gao C, Zhang RD, Jiao Y, Li WJ, Zhao XX, Liu SG, Yue ZX, Zheng HY, Deng GR, et al. Low expressions of ARS2 and CASP8AP2 predict relapse and poor prognosis in pediatric acute lymphoblastic leukemia patients treated on China CCLG-ALL 2008 protocol. Leuk Res. 2015;39:115–123. doi: 10.1016/j.leukres.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Ansari N, Shahrabi S, Khosravi A, Shirzad R, Rezaeean H. Prognostic significance of CHEK2 mutation in progression of breast cancer. Lab Med. 2019;50:e36–e41. doi: 10.1093/labmed/lmz009. [DOI] [PubMed] [Google Scholar]
- 28.Luo Q, Guo H, Kuang P, Cui H, Deng H, Liu H, Lu Y, Wei Q, Chen L, Fang J, et al. Sodium fluoride arrests renal G2/M phase cell-cycle progression by activating ATM-Chk2-P53/Cdc25C signaling pathway in mice. Cell Physiol Biochem. 2018;51:2421–2433. doi: 10.1159/000495899. [DOI] [PubMed] [Google Scholar]
- 29.Apostolou P, Papasotiriou I. Current perspectives on CHEK2 mutations in breast cancer. Breast Cancer (Dove Med Press) 2017;9:331–335. doi: 10.2147/BCTT.S111394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang W, Guo M, Xia X, Zhang C, Zeng Y, Wu S. XRRA1 targets ATM/CHK1/2-mediated DNA repair in colorectal cancer. Biomed Res Int. 2017;2017:5718968. doi: 10.1155/2017/5718968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoon AJ, Shen J, Santella RM, Zegarelli DJ, Chen R, Weinstein IB. Activated checkpoint kinase 2 expression and risk for oral squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16:2768–2772. doi: 10.1158/1055-9965.EPI-07-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carloni V, Lulli M, Madiai S, Mello T, Hall A, Luong TV, Pinzani M, Rombouts K, Galli A. CHK2 overexpression and mislocalisation within mitotic structures enhances chromosomal instability and hepatocellular carcinoma progression. Gut. 2018;67:348–361. doi: 10.1136/gutjnl-2016-313114. [DOI] [PubMed] [Google Scholar]
- 33.Advani SJ, Camargo MF, Seguin L, Mielgo A, Anand S, Hicks AM, Aguilera J, Franovic A, Weis SM, Cheresh DA. Kinase-independent role for CRAF-driving tumour radioresistance via CHK2. Nat Commun. 2015;6:8154. doi: 10.1038/ncomms9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quidville V, Alsafadi S, Goubar A, Commo F, Scott V, Pioche-Durieu C, Girault I, Baconnais S, Le Cam E, Lazar V, et al. Targeting the deregulated spliceosome core machinery in cancer cells triggers mTOR blockade and autophagy. Cancer Res. 2013;73:2247–2258. doi: 10.1158/0008-5472.CAN-12-2501. [DOI] [PubMed] [Google Scholar]
- 35.Valles I, Pajares MJ, Segura V, Guruceaga E, Gomez-Roman J, Blanco D, Tamura A, Montuenga LM, Pio R. Identification of novel deregulated RNA metabolism-related genes in non-small cell lung cancer. PLoS One. 2012;7:e42086. doi: 10.1371/journal.pone.0042086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen T, Zhang B, Ziegenhals T, Prusty AB, Fröhler S, Grimm C, Hu Y, Schaefke B, Fang L, Zhang M, et al. A missense mutation in SNRPE linked to non-syndromal microcephaly interferes with U snRNP assembly and pre-mRNA splicing. PLoS Genet. 2019;15:e1008460. doi: 10.1371/journal.pgen.1008460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anchi T, Tamura K, Furihata M, Satake H, Sakoda H, Kawada C, Kamei M, Shimamoto T, Fukuhara H, Fukata S, et al. SNRPE is involved in cell proliferation and progression of high-grade prostate cancer through the regulation of androgen receptor expression. Oncol Lett. 2012;3:264–268. doi: 10.3892/ol.2011.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He J, Xing J, Yang X, Zhang C, Zhang Y, Wang H, Xu X, Wang H, Cao Y, Xu H, et al. Silencing of proteasome 26S subunit ATPase 2 regulates colorectal cancer cell proliferation, apoptosis, and migration. Chemotherapy. 2019;64:146–154. doi: 10.1159/000502224. [DOI] [PubMed] [Google Scholar]
- 39.Qin J, Wang W, An F, Huang W, Ding J. PSMC2 is up-regulated in pancreatic cancer and promotes cancer cell proliferation and inhibits apoptosis. J Cancer. 2019;10:4939–4946. doi: 10.7150/jca.27616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song M, Wang Y, Zhang Z, Wang S. PSMC2 is up-regulated in osteosarcoma and regulates osteosarcoma cell proliferation, apoptosis and migration. Oncotarget. 2017;8:933–953. doi: 10.18632/oncotarget.13511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li GW, Yan X. Lower miR-630 expression predicts poor prognosis of osteosarcoma and promotes cell proliferation, migration and invasion by targeting PSMC2. Eur Rev Med Pharmacol Sci. 2019;23:1915–1925. doi: 10.26355/eurrev_201903_17229. [DOI] [PubMed] [Google Scholar]
- 42.Shibata E, Dutta A. A human cancer cell line initiates DNA replication normally in the absence of ORC5 and ORC2 proteins. J Biol Chem. 2020;295:16949–16959. doi: 10.1074/jbc.RA120.015450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giri S, Chakraborty A, Sathyan KM, Prasanth KV, Prasanth SG. Orc5 induces large-scale chromatin decondensation in a GCN5-dependent manner. J Cell Sci. 2016;129:417–429. doi: 10.1242/jcs.178889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oomizu S, Sahuc F, Asahina K, Inamatsu M, Matsuzaki T, Sasaki M, Obara M, Yoshizato K. Kdap, a novel gene associated with the stratification of the epithelium. Gene. 2000;256:19–27. doi: 10.1016/S0378-1119(00)00357-7. [DOI] [PubMed] [Google Scholar]
- 45.Thibodeau BJ, Geddes TJ, Fortier LE, Ahmed S, Pruetz BL, Wobb J, Chen P, Wilson GD, Akervall JA. Gene expression characterization of HPV positive head and neck cancer to predict response to chemoradiation. Head Neck Pathol. 2015;9:345–353. doi: 10.1007/s12105-014-0597-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request, and the GSE117973, GSE85446 and GSE112026 datasets analyzed during the current study are available in the Gene Expression Omnibus repository (https://www.ncbi.nlm.nih.gov/gds).