Abstract
Identifying DNA species is crucial for diagnostics. For DNA identification, single-molecule DNA sequence mapping is an alternative to DNA sequencing toward fast point-of-care testing, which traditionally relies on targeting and labeling DNA sequences with fluorescent labels and readout using optical imaging methods. A nanopore is a promising sensor as a complement to optical mapping with advantages of electric measurement suitable for portable devices and potential for high resolution. Here, we demonstrate a high-resolution nanopore-based DNA sequence mapping by labeling specific short sequence motifs with oligodeoxynucleotides (ODNs) using DNA methyltransferase (MTase) and detecting them using nanopores. We successfully detected ODNs down to the size of 11 nucleotides without introducing extra reporters and resolved neighboring sites with a distance of 141 bp (∼48 nm) on a single DNA molecule. To accurately locate the sequence motif positions on DNA, a nanopore data analysis method is proposed by considering DNA velocity change through nanopores and using ensemble statistics to translate the time-dependent signals to the location information. Our platform enables high-resolution detection of small labels on DNA and high-accuracy localization of them for DNA species identification in an all-electrical format. The method presents an alternative to optical techniques relying on fluorescent labels and is promising for miniature-scale integration for diagnostic applications.
Keywords: single-molecule, nanopore sensing, DNA methyltransferase, DNA detection, AdoMet analogue
Nucleic acid analysis including DNA sequencing and testing partial DNA sequence is of vital importance for life science and disease diagnosis. DNA sequencing is the common choice for a detailed analysis of the whole DNA sequence while not necessary for its identification. For DNA identification, testing a specific sequence or a distribution of short sequence motifs is a smart alternative,1,2 suitable for rapid point-of-care testing using portable devices. A variety of biotechnology tools have been used for target DNA sequence detection, for example, PCR, CRISPR-Cas9, and DNA methyltransferases (MTases).3−5 Cas9 can target a specific DNA sequence normally with a length of 20 bp,6,7 requiring a matching guide RNA to be designed for each DNA species. DNA MTases can be used to target a specific sequence with a shorter length (2–8 base pairs) for controlled labeling with a variety of chemical groups.8−10 These short DNA sequences usually appear once or several times in DNA with a length of several kilobase pairs, allowing for a universal method for targeting and labeling different genomes using MTases.
After targeting and labeling the sequence motifs with MTase, the next step is to detect these sites and localize their positions on the DNA. Atomic force microscopes (AFMs) and transmission electron microscopes (TEMs) are direct imaging tools for the characterization of representative molecules if the labels are big enough. However, they are not suitable for diagnostic applications due to the difficulty in sample preparation and the low accessibility of these microscopes. Optical methods have been widely used for DNA mapping by stretching the DNA on a surface or in a micro- or nanoscale confinement and detecting the attached fluorescent labels on the DNA.11−14 Some challenges of the optical mapping—the optical diffraction limit, the thermal fluctuations of DNA, and the bleaching of single fluorophores—limit the resolution for identifying single sequence motifs and the accuracy of localizing their positions. The performance of optical mapping can be improved by using super-resolution imaging methods and reducing DNA fluctuation. For instance, the mapping of surface-stretched DNA using super-resolved structured illumination microscopy (SR-SIM) showed a resolution around 100 nm for identifying microbial species.2 The resolution of optical mapping can also be improved by confining the DNA in nanochannels to overcome the thermal fluctuations, where two labels 676 bp apart were resolved and the resolution was calculated to be ∼100 bp.12 The sample preparation by stretching the DNA and the size and requirement of high-performance optical microscopes prevent their use for point-of-care diagnostics.
A nanopore sensor is an ideal single-molecule tool for DNA detection and suitable for miniature scale integration and multiplexed measurement.15 The measurement principle is that charged DNA can be threaded into a nanopore by applied electric potential and partially block the ionic current flow through the nanopore.16 This change in ionic current translates the 3D structure information on DNA into electrical signals. The detection resolution can outperform optical methods as long as the size and geometry of the nanopore are properly designed or chosen to match the detected object, with the upper limit of identifying single nucleotides.17 Nanopores have been employed to detect not only double-stranded DNA (dsDNA) but also DNA complexes with attached objects including PNA, proteins, and DNA nanostructures.18−25 In a previous study, we detected biotinylated sites on DNA by adding streptavidin to enlarge the signal using ∼15 nm diameter nanopores.26 We introduced an extra-label (streptavidin) to detect the sites, but for higher resolution, the sizes of the labeling object and the nanopore need to be reduced. One option is to use oligodeoxynucleotides (ODNs) for covalent labeling. ODNs offer several advantages over proteins including easy adjustment of size (length) and their stability to conditions which can unfold proteins. A small label combined with a narrow nanopore would significantly increase the resolution (for identifying close labels) and accuracy (due to a narrow signal width). Besides, they can be introduced in one step giving good control of labeling and ensuring high labeling efficiency.
In this study, we use ODNs to label specific DNA sequences by DNA MTase and directly detect the labels using nanopores down to the size of ∼5 nm. Furthermore, a nanopore signal analysis method considering DNA velocity changes during DNA translocation is put forward to increase the accuracy of localizing the sequencing motifs on the DNA. The approach allows high-resolution and high-accuracy mapping of specific short sequence motifs on DNA using nanopores.
Results and Discussion
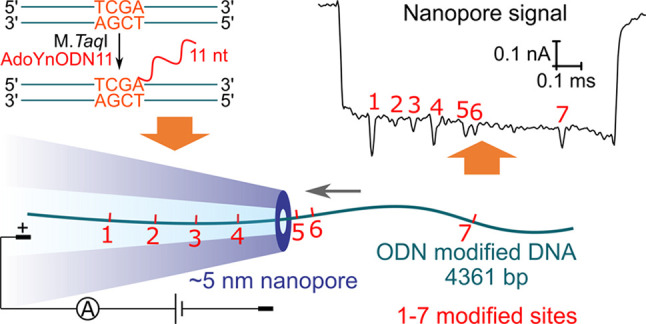
First, we demonstrate the capability of our nanopore to detect DNA with a sufficiently high signal-to-noise (SNR) ratio. Nanopores with inner diameters around 5 nm were fabricated by pulling quartz capillaries. An example of a scanning electron microscope (SEM) image showing the capillary outline is given in Figure S1. Detailed characterization of the nanopores is given in Figures S2 and S3 following a former study.22 The capillary was filled with 4 M LiCl. DNA (8 kbp) was then added with a voltage of 600 mV applied to drive the DNA through the nanopore, as illustrated in Figure 1a. An example of the recorded current trace is shown in Figure 1b, where one DNA translocation caused a downward signal with the zoom-in plot of a representative translocation event shown in the right panel. We analyzed the change of the current ΔI caused by the DNA translocation and the translocation time (usually called event duration). For a single event, ΔI was calculated by dividing the area of the event (commonly defined as the event charge deficit (ECD)) by the event duration. The statistics are shown in Figure S4 with a scatter plot of ΔI as a function of the event duration and the histogram of ΔI for over one hundred events. For this small nanopore, our measurement achieved a high SNR of ∼89 (ΔI = 0.50 nA, Irms = 5.6 pA) at the 600 mV voltage with a 50 kHz low-pass Bessel filter. The high SNR, due to the high signal caused by a relatively narrow nanopore27 and low noise of glass nanopores,28−30 enables a further measurement of closely distributed small labels on DNA.
Figure 1.
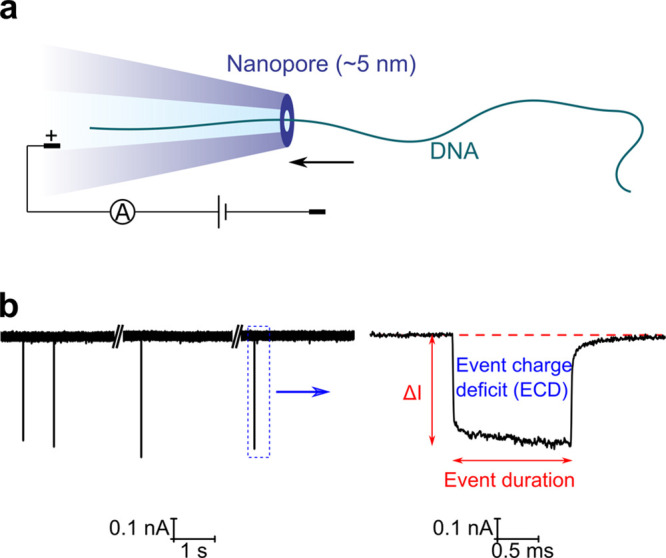
High-resolution nanopore for DNA detection. (a) Schematic of the translocation of a DNA through a ∼5 nm diameter nanopore fabricated by pulling a quartz capillary. (b) Example of the current trace measured at the voltage of 600 mV. The nanopore was filled with 4 M LiCl and then 8 kbp DNA was added. Each spike of more than 0.1 nA represents the translocation of a single DNA molecule with the plot of a zoom-in example in the right panel. See detailed statistics in Figure S4.
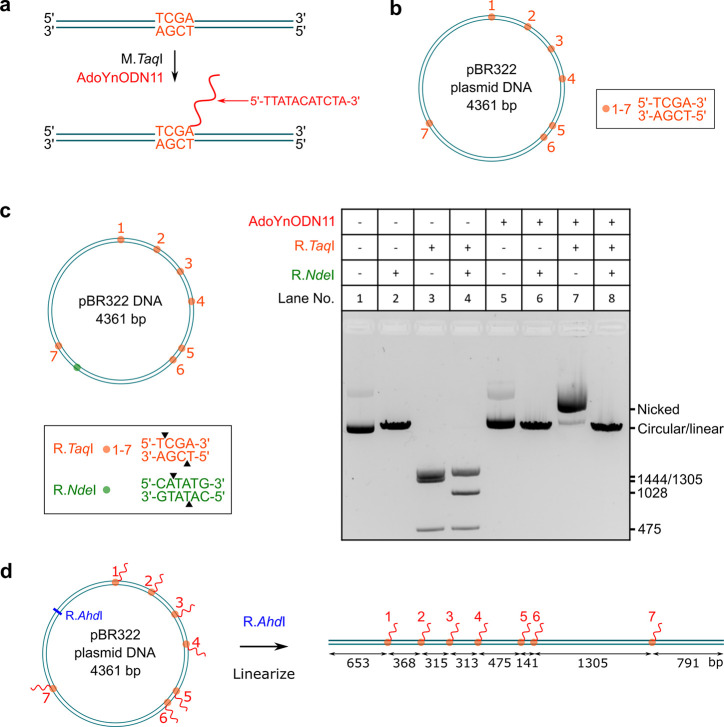
We used pBR322 plasmid DNA as an example to demonstrate the labeling efficiency by DNA MTase. Sites with the specific sequence 5′-TCGA-3′ were targeted on the DNA. As shown in Figure 2a, the sites were covalently labeled with a single-stranded ODN containing 11 nucleotides (nt) using the DNA MTase M.TaqI and AdoYnODN11 cofactor (Figure S5). As a proof of concept, pBR322 DNA was chosen as the plasmid contains seven M.TaqI sites (sites 1–7, Figure 2b) at various distances ideally suited to explore the detection limits of nanopore sensing. To confirm complete labeling, we tested if the modified DNA could be fragmented using the restriction enzyme R.TaqI which targets the same sequence (5′-TCGA-3′) as M.TaqI and cleaves unmodified sites while modified sites are protected. Figure 2c shows the analysis by agarose gel electrophoresis. For the unmodified DNA (lane 3), the DNA was cleaved into the expected fragments while the modified DNA was protected against double-strand cleavage (lane 7). We did see a shift of the band for the modified DNA (compare lanes 5 and 7), which we attributed to nicking of the supercoiled circular DNA by R.TaqI. To double-check nicking, we introduced an extra restriction enzyme R.NdeI with a single site on the pBR322 DNA. With the addition of only R.NdeI, the DNA was linearized no matter if the DNA was modified or not (lanes 6 and 2). With the addition of both (R.TaqI and R.NdeI), there was only one band (lane 8) identical to the single digest with R.NdeI (lanes 2 and 6), showing that modified 5′-TCGA-3′ sites were not cleaved. These results demonstrate nearly a 100% modification efficiency because with R.TaqI the fraction of unmodified DNA would cause fragments, which were not observed in our analysis (lanes 7 and 8).
Figure 2.
Sequence-specific labeling of DNA with an oligodeoxynucleotide (ODN). (a) Modification of the DNA at the sequence 5′-TCGA-3′ using M.TaqI and AdoYnODN11 cofactor. The site was covalently labeled with an ODN containing 11 nucleotides (5′-TTATACATCTA-3′). (b) Distribution of the target sequence (5′-TCGA-3′) sites on pBR322 plasmid DNA. (c) Confirmation of the modification using restriction enzymes. The left panel shows the distribution of the sites of the restriction enzymes. The right panel shows the analysis by agarose gel electrophoresis. (d) Linearization of the pBR322 DNA for nanopore measurement. The labeled DNA was linearized with the restriction endonuclease R.AhdI which cleaves the pBR322 plasmid at a single site.
To prepare the linear sample for nanopore measurement, the modified DNA was linearized with another restriction endonuclease R.AhdI which cleaves pBR322 at a single site (Figure 2d). The restriction site is marked in blue, and locations of the ODN-labeled sites 1–7 on the linearized DNA are shown in orange attached with the ODN labels (red).
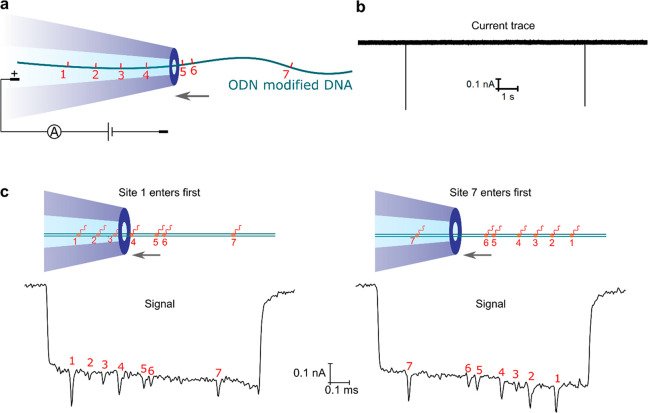
Then we passed the linearized DNA with ODN labels through nanopores with the sample diluted to a concentration of 0.2–2 nM in 4 M LiCl, as illustrated in Figure 3a. An example of a typical current trace at 400 mV is shown in Figure 3b. Figure 3c shows two examples of translocation events. Apart from the current drop caused by the DNA backbone, additional downward spikes caused by the ODN labels can be directly observed. The DNA entered the nanopore in two orientations because the two ends could both be threaded into the nanopore with site 1 or site 7 entering first, respectively. Accordingly, we categorized the nanopore signals in the two orientations by the relative position of the spikes with the corresponding examples listed in Figure 3c. Our measurement using the narrow nanopores achieved a high sensitivity, which was able to capture the signals caused by the ODNs with the size of 11 nt labeled on a double-stranded DNA molecule. More importantly, the signals caused by two close sites with a distance of 141 bp were completely separated, while this distance was not resolved with biotin–streptavidin labels measured with ∼15 nm nanopores in our previous study,26 showing high resolution of our platform in detecting close sequence motifs. With our ∼5 nm nanopores, the resolving distance was demonstrated to be down to 76 bp,22 which could be further improved by using smaller nanopores and smaller labels.
Figure 3.
Detection of the ODN-modified specific sequence motifs on DNA using nanopores. (a) Schematic of the translocation of the labeled DNA through a nanopore. The red marks represent the seven ODN-labeled sites. (b) Recorded current trace at 400 mV with each spike signal representing a single DNA molecule translocation. (c) Examples of the translocation signals. The DNA entered the nanopore in two orientations, with site 1 or site 7 first entering the nanopore, respectively. The additional spikes marked with numbers represent signals caused by the ODN labels on the seven sites.
After demonstrating that
we can detect the ODN labels, we turned
to a thorough analysis of the sequence motif locations on the DNA
from the electrical signals. The results from the two possible orientations
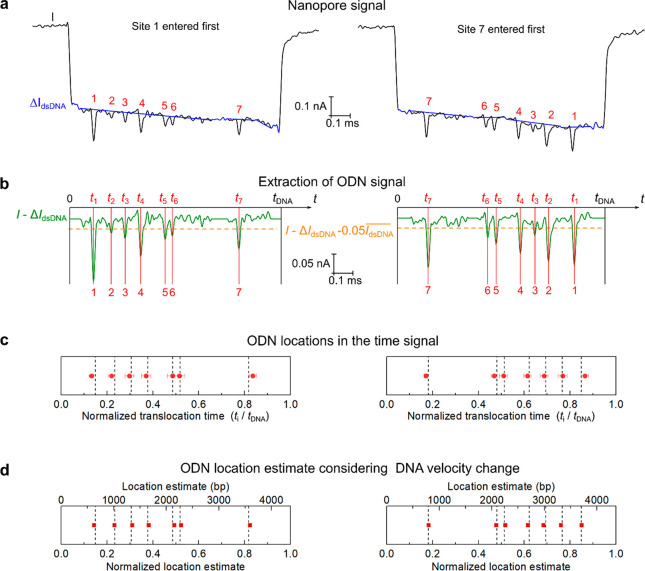
are plotted in the left and right panels in Figure 4, respectively. Figure 4a shows two examples of events. As shown
in Figure 4a,b, the
signals of the ODNs (green) were obtained by subtracting the recorded
current (black) and the fitted dsDNA backbone level (blue). The spikes
in the signals show the positions of the ODN sites. A homemade LabVIEW
algorithm was used for identifying the spikes. In Figure 4b, the spikes with amplitudes
higher than 5% of the dsDNA level (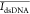 ) were recognized. We calculated the translocation
time of each spike from the beginning of the event and normalized
them to the translocation time of the DNA. Ensemble statistics from
multiple events by averaging the normalized translocations time was
used to reduce the stochastic error caused by single events and plotted
in Figure 4c (values
also listed in Tables S1: the average and
standard deviation were calculated from the 13 events in Figures S6 and S7). If the DNA passed the nanopore
with a constant velocity, the normalized translocation time of each
sequence motif site would equal their normalized location on the DNA
(dashed lines in Figure 4c). However, for both orientations, the experimental values were
lower than the expected locations (indicated by the dashed lines)
at the beginning and higher at the end. The discrepancy between the
expected and observed locations indicates a velocity change of the
DNA during translocation which is in line with previous results.31−33 Using DNA nanotechnology, it was shown that the DNA slows down during
the translocation and speeds up at the very end.33 The changing velocity with higher speed at the beginning
and end explains our results in Figure 4c with the finite differences in expected and measured
locations. The inconstant DNA velocity prevents us from directly translating
the signal in time to the distance information which caused average
localization errors of 0.98% and 0.78% of the DNA length for the two
event orientations (Table S1), corresponding
to 43 and 34 base pairs for the 4361 bp DNA, respectively (with maximum
errors of 1.74% and 1.51% of the DNA length corresponding to 76 and
66 base pairs, respectively).
) were recognized. We calculated the translocation
time of each spike from the beginning of the event and normalized
them to the translocation time of the DNA. Ensemble statistics from
multiple events by averaging the normalized translocations time was
used to reduce the stochastic error caused by single events and plotted
in Figure 4c (values
also listed in Tables S1: the average and
standard deviation were calculated from the 13 events in Figures S6 and S7). If the DNA passed the nanopore
with a constant velocity, the normalized translocation time of each
sequence motif site would equal their normalized location on the DNA
(dashed lines in Figure 4c). However, for both orientations, the experimental values were
lower than the expected locations (indicated by the dashed lines)
at the beginning and higher at the end. The discrepancy between the
expected and observed locations indicates a velocity change of the
DNA during translocation which is in line with previous results.31−33 Using DNA nanotechnology, it was shown that the DNA slows down during
the translocation and speeds up at the very end.33 The changing velocity with higher speed at the beginning
and end explains our results in Figure 4c with the finite differences in expected and measured
locations. The inconstant DNA velocity prevents us from directly translating
the signal in time to the distance information which caused average
localization errors of 0.98% and 0.78% of the DNA length for the two
event orientations (Table S1), corresponding
to 43 and 34 base pairs for the 4361 bp DNA, respectively (with maximum
errors of 1.74% and 1.51% of the DNA length corresponding to 76 and
66 base pairs, respectively).
Figure 4.
Localizing the sequence motifs on the DNA from nanopore signals. (a) Two examples of the nanopore signals with the fitted dsDNA levels (blue). (b) Extraction of the signals of the sequence motifs. The signal (green) was obtained by the subtraction of the recorded current and the fitted dsDNA level. (c) Normalized translocation time of the ODN labels by averaging multiple events. The average and standard deviation were calculated from the 13 events listed in Figures S6 and S7. The dashed lines denote the theoretical values with a constant translocation velocity. (d) Estimate of the locations of the sequence motifs from the ensemble statistics of nanopore signals in (c) by taking the DNA velocity change into account. The dashed lines show the actual values. Detailed analysis is given in Figure S8. Signals with two DNA translocation orientations are shown on the left and right, respectively.
To accurately determine the sequence motif locations from the signals, we took into account the effect of velocity change by using a measured DNA velocity profile (normalized values) from a former study.33 As shown in Figure S8, we used the previously measured velocity change during a DNA translocation to determine the relationship between the length of the DNA that has passed the nanopore (normalized to the DNA length) and the translocation time (normalized to the whole DNA translocation time). From the time positions of the motifs in the signal (normalized to the translocation time) in Figure 4c (also listed in Table S1), we calculated the locations of the motifs using the predetermined relationship between distance and time (Figure S8). The results are shown in Figure 4d and Table 1. In our estimate, the average localization errors were reduced to 0.44% and 0.43% of the DNA length for the two orientations, with the maximum errors of 0.69% and 0.88% of the DNA length, respectively. By introducing DNA with a known length as a reference,26 we can translate the normalized values into distance information (the top X-axis in Figure 4d). Accordingly, the average localization errors were ∼19 bp, with maximum errors of 30 and 38 bp, respectively.
Table 1. Comparison of the Actual Locations and Estimate Locations of the Sequence Motifsa.
| site
1 entered first | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| site | site 1 | site 2 | site 3 | site 4 | site 5 | site 6 | site 7 | avg | max |
| actual location (normalized) | 0.1497 | 0.2341 | 0.3064 | 0.3781 | 0.4870 | 0.5194 | 0.8186 | ||
| estimate location (normalized) | 0.1442 | 0.2324 | 0.3101 | 0.3818 | 0.4939 | 0.5234 | 0.8240 | ||
| error (absolute value) | 0.0055 | 0.0018 | 0.0038 | 0.0036 | 0.0069 | 0.0040 | 0.0053 | 0.0044 | 0.0069 |
| error (in bp) | 24 | 8 | 17 | 16 | 30 | 18 | 23 | 19 | 30 |
| site
7 entered first | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| site | site 7 | site 6 | site 5 | site 4 | site 3 | site 2 | site 1 | avg | max |
| actual location (normalized) | 0.1814 | 0.4806 | 0.5130 | 0.6219 | 0.6936 | 0.7659 | 0.8503 | ||
| estimate location (normalized) | 0.1834 | 0.4776 | 0.5172 | 0.6165 | 0.6848 | 0.7606 | 0.8514 | ||
| error (absolute value) | 0.0020 | 0.0030 | 0.0043 | 0.0054 | 0.0088 | 0.0053 | 0.0011 | 0.0043 | 0.0088 |
| error (in bp) | 9 | 13 | 19 | 23 | 38 | 23 | 5 | 19 | 38 |
The values were corrected according to a translocation velocity profile and normalized to the DNA length. The last row shows the errors in base pairs for the 4361 bp pBR322 DNA.
Our results are superior to those in previous studies using nanopores for locating target sequences on DNA in several aspects.34−36 First, the small label intrinsically enables a high accuracy with a narrow signal, outperforming the larger labels with wide signals.31,34,35 Also, the narrow nanopores and small labels are ideal for separately identifying closely spaced sequence motifs. We compared the result to that from the streptavidin-labeled DNA in our former study (plotted in Figure S9).26 In the former study, sites 5 and 6 were not fully resolved and the wide signal caused by each label prevented an accurate estimate of the sequence motif locations (Table S2). For the ODN-labeled DNA, all of the sequence motifs were resolved with narrow signals caused by ODN labels. The accurate determination of the locations of the specific short sequence motifs combined with the length of the DNA offers a highly reliable and adaptable approach for identifying different DNA species. Depending on prior information on the sample, one can use the length of DNA fragments for velocity calibration, add an additional external calibration standard, or use known labeling sites on the target DNA to determine translocation velocity and direction. A combination of all three approaches should allow for the analysis of any sample that contains target DNA in sufficient concentrations.
Conclusions
In this study, we demonstrate that we can directly detect the ODN-labeled sequence motifs on DNA using narrow nanopores. This assay combining DNA MTase labeling and nanopore sensing could be used as a universal DNA identification method for various genomes with the same labeling and sensing procedure. Using our high SNR nanopores, we were able to detect ODN labels on DNA with a size down to 11 nt. We resolved neighboring labels with a distance of 141 bp. Our platform has shown the ability to achieve a resolution of 76 bp (∼26 nm)22 and could be further improved by optimizing the nanopore and label size, outperforming optical methods.14 We also put forward a nanopore data analysis strategy for accurately translating the nanopore signals into location information on labels on DNA by considering DNA velocity change. This reduces the average localization error down to ∼0.44% of the DNA length. Compared to optical mapping methods allowing for high throughput and long-molecule reading, the advantage of the nanopore approach is that it does not need fluorescent labels. In addition, nanopore spatial resolution could be further improved by optimizing the nanopore size and geometry including further reduction of label size allowing for high-density sequence motifs. The known DNA velocity profile accurately reveals the sequence positions by averaging multiple events over thermal fluctuations. Here, as a proof of concept, we show the detection of a 4361 bp DNA, but this approach has the potential to be scaled for longer molecules as nanopores have shown the ability to read a ∼49 kbp long phage (λ DNA)37 and the methyltransferase enzyme labeling method has also been used for this DNA.1 In the future, high throughput could be achieved by using nanopore arrays and the data analysis could be further assisted by smarter algorithms and employing machine learning.38 More importantly, the detection in an electric format combined with a convenient sample preparation procedure should be ideally suited for easy and rapid point-of-care testing using portable devices.
Methods
Sequence-Specific Labeling with ODN
ODN-labeled DNA was prepared by incubating double-stranded pBR322 plasmid DNA (100 ng/μL, New England BioLabs (NEB), Ipswich, MA), ODN-modified AdoMet analogue AdoYnODN11 (10 μM) and M.TaqI (2.43 μM, 10 equiv of M.TaqI with respect to 5′-TCGA-3′ recognition sequences on the plasmid) in NEB buffer 4 (110 μL, 20 mM Tris–HCl, 50 mM KOAc, 10 mM Mg(OAc)2, 1 mM DTT, pH 7.9) at 65 °C for 1 h. Plasmids were purified using the QIAquick PCR purification kit (QIAGEN, Hilden, Germany) according to the instructions of the manufacturer. Complete labeling was verified by the protection of the modified plasmid against cleavage by the cognate restriction endonuclease R.TaqI. DNA samples were supplemented with R.TaqI (10 Units/μg DNA, New England BioLabs, Ipswich, MA), incubated at 65 °C for 1 h, and analyzed by agarose gel (1%) electrophoresis (0.5 × TBE buffer, 1 h, 6 V/cm, 0.01% GelRed). R.TaqI nicking instead of linearization was verified by cleavage of a second (noncognate) restriction endonuclease (double digest). DNA samples were supplemented with R.TaqI and/or R.NdeI (10 Units/μg DNA each, New England BioLabs, Ipswich, MA), incubated at 37 °C for 2 h, and analyzed by agarose gel (1%) electrophoresis (0.5 × TBE buffer, 1 h, 6 V/cm, 0.01% GelRed).
Nanopore Measurement
The nanopores were fabricated by pulling quartz capillaries with outer and inner diameters of 0.5 mm and 0.2 mm, respectively (Sutter Instrument, USA), by a laser-heated pipet puller (P2000, Sutter Instrument). We fabricated nanopores with inner diameters of ∼5 nm using the one-line program: HEAT = 575, FIL = 0, VEL = 25, DEL = 170, PUL = 225. Detailed description and characterization can be found in a former study.22 The fabricated nanopores were integrated into our homemade PDMS flow cells and filled with 4 M LiCl, 1 × TE (pH 9). DNA was then added with a concentration of 0.2–2 nM diluted in 4 M LiCl. A voltage of 400–600 mV was applied and the ionic current was measured by an Axopatch 200B amplifier (Molecular Devices, USA) with the signal filtered at 50 kHz by an external filter (Frequency Devices). Data was collected by a PCI 6251 data card (National Instruments, USA) at 250 kHz and a homemade LabView algorithm. Data analysis was performed using LabView algorithms.
Acknowledgments
K.C. and U.F.K. acknowledge funding from an ERC Consolidator Grant (Designerpores no. 647144). F.G. and E.W. acknowledge financial support from the German-Israeli Foundation for Scientific Research and Development (I-1196-195.9/2012).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.0c07947.
Characterization of the nanopores and DNA translocation, scheme for the synthesis of AdoYnODN11, examples of the signals of the ODN-labeled pBR322 DNA, and detailed analysis of the sequence motif locations (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Grunwald A.; Dahan M.; Giesbertz A.; Nilsson A.; Nyberg L. K.; Weinhold E.; Ambjornsson T.; Westerlund F.; Ebenstein Y. Bacteriophage Strain Typing by Rapid Single Molecule Analysis. Nucleic Acids Res. 2015, 43, e117 10.1093/nar/gkv563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwens A.; Deen J.; Vitale R.; D’Huys L.; Goyvaerts V.; Descloux A.; Borrenberghs D.; Grussmayer K.; Lukes T.; Camacho R.; Su J.; Ruckebusch C.; Lasser T.; Van De Ville D.; Hofkens J.; Radenovic A.; Janssen K. P. F. Identifying Microbial Species by Single-Molecule DNA Optical Mapping and Resampling Statistics. NAR Genom. Bioinform. 2020, 2, lqz007 10.1093/nargab/lqz007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J. A.; Charpentier E. The New Frontier of Genome Engineering with CRISPR-Cas9. Science 2014, 346, 1258096. 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Goedecke K.; Pignot M.; Goody R. S.; Scheidig A. J.; Weinhold E. Structure of the N6-adenine DNA Methyltransferase M•TaqI in Complex with DNA and a Cofactor Analog. Nat. Struct. Biol. 2001, 8, 121–125. 10.1038/84104. [DOI] [PubMed] [Google Scholar]
- Higuchi R.; Dollinger G.; Walsh P. S.; Griffith R. Simultaneous Amplification and Detection of Specific DNA Sequences. Bio/Technology 1992, 10, 413–417. 10.1038/nbt0492-413. [DOI] [PubMed] [Google Scholar]
- Weckman N. E.; Ermann N.; Gutierrez R.; Chen K.; Graham J.; Tivony R.; Heron A.; Keyser U. F. Multiplexed DNA Identification Using Site Specific dCas9 Barcodes and Nanopore Sensing. ACS Sens. 2019, 4, 2065–2072. 10.1021/acssensors.9b00686. [DOI] [PubMed] [Google Scholar]
- Yang W.; Restrepo-Pérez L.; Bengtson M.; Heerema S. J.; Birnie A.; van der Torre J.; Dekker C. Detection of CRISPR-dCas9 on DNA with Solid-State Nanopores. Nano Lett. 2018, 18, 6469–6474. 10.1021/acs.nanolett.8b02968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottfried A.; Weinhold E. Sequence-Specific Covalent Labelling of DNA. Biochem. Soc. Trans. 2011, 39, 623–628. 10.1042/BST0390623. [DOI] [PubMed] [Google Scholar]
- Hanz G. M.; Jung B.; Giesbertz A.; Juhasz M.; Weinhold E. Sequence-Specific Labeling of Nucleic Acids and Proteins with Methyltransferases and Cofactor Analogues. J. Visualized Exp. 2014, 93, e52014 10.3791/52014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukinavičius G.; Lapienė V.; Staševskij Z.; Dalhoff C.; Weinhold E.; Klimašauskas S. Targeted Labeling of DNA by Methyltransferase-Directed Transfer of Activated Groups (mTAG). J. Am. Chem. Soc. 2007, 129, 2758–2759. 10.1021/ja0691876. [DOI] [PubMed] [Google Scholar]
- Howorka S.; Siwy Z. Nanopores and Nanochannels: From Gene Sequencing to Genome Mapping. ACS Nano 2016, 10, 9768–9771. 10.1021/acsnano.6b07041. [DOI] [PubMed] [Google Scholar]
- Jeffet J.; Kobo A.; Su T.; Grunwald A.; Green O.; Nilsson A. N.; Eisenberg E.; Ambjornsson T.; Westerlund F.; Weinhold E.; Shabat D.; Purohit P. K.; Ebenstein Y. Super-Resolution Genome Mapping in Silicon Nanochannels. ACS Nano 2016, 10, 9823–9830. 10.1021/acsnano.6b05398. [DOI] [PubMed] [Google Scholar]
- Chan E. Y.; Goncalves N. M.; Haeusler R. A.; Hatch A. J.; Larson J. W.; Maletta A. M.; Yantz G. R.; Carstea E. D.; Fuchs M.; Wong G. G. DNA Mapping Using Microfluidic Stretching and Single-Molecule Detection of Fluorescent Site-Specific Tags. Genome Res. 2004, 14, 1137–1146. 10.1101/gr.1635204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely R. K.; Deen J.; Hofkens J. Optical Mapping of DNA: Single-Molecule-Based Methods for Mapping Genomes. Biopolymers 2011, 95, 298–311. 10.1002/bip.21579. [DOI] [PubMed] [Google Scholar]
- Dekker C. Solid-State Nanopores. Nat. Nanotechnol. 2007, 2, 209. 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]
- Li J.; Gershow M.; Stein D.; Brandin E.; Golovchenko J. A. DNA Molecules and Configurations in a Solid-State Nanopore Microscope. Nat. Mater. 2003, 2, 611–615. 10.1038/nmat965. [DOI] [PubMed] [Google Scholar]
- Kasianowicz J. J.; Brandin E.; Branton D.; Deamer D. W. Characterization of Individual Polynucleotide Molecules Using a Membrane Channel. Proc. Natl. Acad. Sci. U. S. A. 1996, 93, 13770–13773. 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer A.; Wanunu M.; Morrison W.; Kuhn H.; Frank-Kamenetskii M.; Meller A. Nanopore Based Sequence Specific Detection of Duplex DNA for Genomic Profiling. Nano Lett. 2010, 10, 738–42. 10.1021/nl100058y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulushev R. D.; Marion S.; Radenovic A. Relevance of the Drag Force during Controlled Translocation of a DNA-Protein Complex through a Glass Nanocapillary. Nano Lett. 2015, 15, 7118–7125. 10.1021/acs.nanolett.5b03264. [DOI] [PubMed] [Google Scholar]
- Plesa C.; Ruitenberg J. W.; Witteveen M. J.; Dekker C. Detection of Individual Proteins Bound along DNA Using Solid-State Nanopores. Nano Lett. 2015, 15, 3153–3158. 10.1021/acs.nanolett.5b00249. [DOI] [PubMed] [Google Scholar]
- Bell N. A.; Keyser U. F. Specific Protein Detection Using Designed DNA Carriers and Nanopores. J. Am. Chem. Soc. 2015, 137, 2035–41. 10.1021/ja512521w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.; Kong J.; Zhu J.; Ermann N.; Predki P.; Keyser U. F. Digital Data Storage Using DNA Nanostructures and Solid-State Nanopores. Nano Lett. 2019, 19, 1210–1215. 10.1021/acs.nanolett.8b04715. [DOI] [PubMed] [Google Scholar]
- Wanunu M.; Sutin J.; Meller A. DNA Profiling Using Solid-State Nanopores: Detection of DNA-Binding Molecules. Nano Lett. 2009, 9, 3498–3502. 10.1021/nl901691v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau C. C.; Radford S. E.; Hewitt E. W.; Actis P. Macromolecular Crowding Enhances the Detection of DNA and Proteins by a Solid-State Nanopore. Nano Lett. 2020, 20, 5553–5561. 10.1021/acs.nanolett.0c02246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh A. Y. Y.; Burgess C. H.; Tanase D. A.; Ferrari G.; McLachlan M. A.; Cass A. E. G.; Albrecht T. Electric Single-Molecule Hybridization Detector for Short DNA Fragments. Anal. Chem. 2018, 90, 14063–14071. 10.1021/acs.analchem.8b04357. [DOI] [PubMed] [Google Scholar]
- Chen K.; Juhasz M.; Gularek F.; Weinhold E.; Tian Y.; Keyser U. F.; Bell N. A. W. Ionic Current-Based Mapping of Short Sequence Motifs in Single DNA Molecules Using Solid-State Nanopores. Nano Lett. 2017, 17, 5199–5205. 10.1021/acs.nanolett.7b01009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.; Li C.; Zhou Y.; Jin Y. Controllable Shrinking of Glass Capillary Nanopores Down to sub-10 nm by Wet-Chemical Silanization for Signal-Enhanced DNA Translocation. ACS Sens. 2017, 2, 1452–1457. 10.1021/acssensors.7b00385. [DOI] [PubMed] [Google Scholar]
- Fragasso A.; Schmid S.; Dekker C. Comparing Current Noise in Biological and Solid-State Nanopores. ACS Nano 2020, 14, 1338–1349. 10.1021/acsnano.9b09353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z.; Duan X.; Li Q.; Wu R.; Wang Y.; Li B. Low-Noise Nanopore Enables in-Situ and Label-Free Tracking of a Trigger-Induced DNA Molecular Machine at the Single-Molecular Level. J. Am. Chem. Soc. 2020, 142, 4481–4492. 10.1021/jacs.0c00029. [DOI] [PubMed] [Google Scholar]
- Chien C.-C.; Shekar S.; Niedzwiecki D. J.; Shepard K. L.; Drndić M. Single-Stranded DNA Translocation Recordings through Solid-State Nanopores on Glass Chips at 10 MHz Measurement Bandwidth. ACS Nano 2019, 13, 10545–10554. 10.1021/acsnano.9b04626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer A.; Rapireddy S.; Ly D. H.; Meller A. Electronic Barcoding of a Viral Gene at the Single-Molecule Level. Nano Lett. 2012, 12, 1722–1728. 10.1021/nl300372a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell N. A. W.; Chen K.; Ghosal S.; Ricci M.; Keyser U. F. Asymmetric Dynamics of DNA Entering and Exiting a Strongly Confining Nanopore. Nat. Commun. 2017, 8, 380. 10.1038/s41467-017-00423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell N. A.; Keyser U. F.. Direct Measurements Reveal Non-Markovian Fluctuations of DNA Threading through a Solid-State Nanopore. arXiv 2016, arXiv:1607.04612 (retrieved 02, 2021-01-02). [Google Scholar]
- Squires A.; Atas E.; Meller A. Nanopore Sensing of Individual Transcription Factors Bound to DNA. Sci. Rep. 2015, 5, 11643. 10.1038/srep11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin T. J.; Shropshire T.; Liu X.; Briggs K.; Huynh C.; Tabard-Cossa V.; Wang H.; Dunbar W. B. Nanopore-Based Target Sequence Detection. PLoS One 2016, 11, e0154426 10.1371/journal.pone.0154426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulushev R. D.; Marion S.; Petrova E.; Davis S. J.; Maerkl S. J.; Radenovic A. Single Molecule Localization and Discrimination of DNA-Protein Complexes by Controlled Translocation Through Nanocapillaries. Nano Lett. 2016, 16, 7882–7890. 10.1021/acs.nanolett.6b04165. [DOI] [PubMed] [Google Scholar]
- Sze J. Y. Y.; Ivanov A. P.; Cass A. E. G.; Edel J. B. Single Molecule Multiplexed Nanopore Protein Screening in Human Serum Using Aptamer Modified DNA Carriers. Nat. Commun. 2017, 8, 1552. 10.1038/s41467-017-01584-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misiunas K.; Ermann N.; Keyser U. F. Quipu Net: Convolutional Neural Network for Single-Molecule Nanopore Sensing. Nano Lett. 2018, 18, 4040–4045. 10.1021/acs.nanolett.8b01709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.