Abstract
Sex differences in the brain have prompted many researchers to investigate the underlying molecular actors, such as the glucocorticoid receptor (GR). This nuclear receptor controls gene expression, including microRNAs (miRNAs), in non-neuronal cells. Here, we investigated sex-biased effects of GR on hippocampal miRNA expression and neuronal morphology by generating a neuron-specific GR knockout mouse (Emx1-Nr3c1−/−). The levels of 578 mature miRNAs were assessed using NanoString technology and, in contrast to males, female Emx1-Nr3c1−/− mice showed a substantially higher number of differentially expressed miRNAs, confirming a sex-biased effect of GR ablation. Based on bioinformatic analyses we identified several transcription factors potentially involved in miRNA regulation. Functional enrichment analyses of the miRNA-mRNA interactions revealed pathways related to neuronal arborization and both spine morphology and density in both sexes. Two recognized regulators of dendritic morphology, CAMKII-α and GSK-3β, increased their protein levels by GR ablation in female mice hippocampus, without changes in males. Additionally, sex-specific effects of GR deletion were observed on CA1 neuronal arborization and dendritic spine features. For instance, a reduced density of mushroom spines in apical dendrites was evidenced only in females, while a decreased length in basal dendrites was noted only in males. However, length and arborization of apical dendrites were reduced by GR ablation irrespective of the sex. Overall, our study provides new insights into the sex-biased GR actions, especially in terms of miRNAs expression and neuronal morphology in the hippocampus.
Keywords: Glucocorticoid receptor, Hippocampus, miRNAs, Neuroplasticity, Dendrites, Dendritic spines, Sex
Abbreviations: Glucocorticoids, (GCs); glucocorticoid response elements, (GREs); microRNA, (miRNA); hypothalamic-pituitary-adrenal, (HPA); Transcription factor, (TF)
1. Introduction
Glucocorticoids (GCs) influence diverse physiological functions, including brain maturation, growth, development, metabolism, immune response, and behavior (Uchoa et al., 2014). GCs are released from the adrenal cortex (corticosterone in rodents) by circadian or stress-induced activity of the hypothalamic-pituitary-adrenal (HPA) axis (Hall et al., 2015). Importantly, GCs differentially exert their actions in males and females in some tissues (e.g., heart and liver) (Duma et al., 2010; Quinn et al., 2014; Cruz-Topete et al., 2019), suggesting a complex action of these hormones in a sex-dependent manner.
GCs bind to mineralocorticoid (MR; NR3C2) and glucocorticoid receptors (GR; NR3C1) with high and low affinity, respectively. Both receptors act as ligand-dependent transcription factors that modulate the expression of several genes (Ramamoorthy and Cidlowski, 2016). The GCs-GRs complex binds to specific DNA sequences (glucocorticoid response elements, GREs) in target genes, increasing their expression (Ramamoorthy and Cidlowski, 2016). In contrast, a negative regulation of gene expression occurs through negative GREs, composite GREs, or by a trans-repression mechanism (Ramamoorthy and Cidlowski, 2016). Moreover, GRs may trigger rapid actions upon their activation by interacting with several types of membrane receptors and protein kinases (Panettieri et al., 2019). Notably, the GR may exert various actions through a sophisticated combination of direct gene expression control or a direct cross-talk with ERK1/2–MSK1–Elk-1 signaling to the nucleus (Gutierrez-Mecinas et al., 2011).
The GR is widely expressed in several limbic brain areas and plays a pivotal role in the modulation of the stress response and memory processing (Wirth, 2015). Furthermore, in utero GR silencing in mouse embryos (E15–15.5) established that GR promotes increased spine density and reduced spine size in apical dendrites of cortical neurons evaluated at the juvenile stage (Arango-Lievano et al., 2015). These findings suggest that these receptors participate in neuronal morphology during neurodevelopment. Additionally, evidences have shown that circadian glucocorticoid oscillations produce a balance between synapse formation and pruning after learning through a delicate contribution of both the GR and the MR (Liston et al., 2013; Hall et al., 2015). Thus, GR-dependent actions of GCs -by both genomic and non-genomic mechanisms- may modulate neuronal functioning and structural plasticity (de Kloet et al., 2018). Notably, the GR is highly expressed in the hippocampus, a brain area involved in mood/cognition processes, and the stress response (McEwen et al., 2016). Therefore, GR contributions to structural plasticity in the hippocampus may contribute to promote adaptive behavior.
Interestingly, a bidirectional control between GR and particular miRNAs -key post-transcriptional regulators of gene expression- (Olde Loohuis et al., 2012) has emerged. For instance, both miR-124a and miR-18 target the Nr3c1 3′UTR transcript to downregulate GR expression (Vreugdenhil et al., 2009). On the other hand, evidences in non-neuronal cells indicate that the GR participates in the modulation of miRNA levels (Li et al., 2014; Laxman et al., 2016). Specifically, a synthetic GR agonist induces a broad repression of GC-responsive mature miRNAs in rat primary thymocytes and human lymphoid cell lines (Smith et al., 2010, 2013). However, GR-dependent effects on miRNAs in brain cells have not been described yet. Surprisingly, miRNAs show uneven expression profiles across different brain areas, suggesting region-specific functions (Olsen et al., 2009). Additionally, miRNAs participate in neurodevelopment, synaptic plasticity, and neuronal activity (Olde Loohuis et al., 2012). These important miRNA actions have raised a great interest in deciphering the sex-specific miRNA processes in several tissues, including the brain; differences which may explain the sex-biased prevalence of some diseases (Pak et al., 2013). Recently, a comprehensive analysis of sex differences in human miRNA expression has been conducted with public databases (Cui et al., 2018). This study identified 73 female-biased miRNAs and 163 male-biased miRNAs across four different tissues, including the brain (Cui et al., 2018). Notably, the neonatal brain shows sex-biased expression of miRNAs, an effect which seems to be regulated by a combination of both sex hormones and sex chromosomes (Morgan and Bale, 2012). Furthermore, sex-specific microRNAome variations are detected in the hippocampus after X-ray irradiation, suggesting a sex-biased regulation of miRNA expression (Koturbash et al., 2011).
Although the aforementioned evidences highlight that miRNA expression is sensitive to GR activity in non-neural models, the effect of GR on miRNA regulation in the brain, particularly in the hippocampus, is still unknown. Here, we report a conditional Nr3c1 ablation, through the expression of Cre recombinase under the control of the Emx1 (empty spiracles homeobox 1) promoter, to study GR contributions to both miRNA expression and neuronal morphology in the mouse hippocampus. Besides, since sex influences both GCs actions and miRNAs expression, it is pertinent to evaluate sex contribution to GR-dependent actions on hippocampal miRNAs and pyramidal neurons architecture.
2. Material and methods
2.1. Animals
Mice with a floxed GR/Nr3c1 locus were generated at the National Institute of Environmental Health Sciences (NIEHS) by standard gene-targeting procedures, as previously described (Oakley et al., 2013, 2019). Briefly, the GR locus was modified by inserting loxP sites upstream of exon 3 and downstream of exon 4. Mice carrying the modified GR allele were then derived by blastocyst (albino B6) injection. Homozygous floxed GR (GRloxP/loxP) mice were then mated at NIEHS with mice expressing Cre recombinase under the control of the Emx1 promoter (#005628, The Jackson Laboratory), which restricts Cre recombinase expression to neocortex and hippocampal neurons (Guo et al., 2000). Therefore, the resulting offspring were GRloxP/loxP Emx1-Cre/+ mice (designated Emx1-Nr3c1−/−). The Cre-negative GRloxP/loxP Emx1+/+ littermate mice served as controls. All mice presented a C57BL/6N/J genetic background. Male and female knockouts appeared grossly normal. They were born at the predicted Mendelian ratio and exhibited normal body weights and survival through 12 months of age. Four-to 6-month-old male and female littermates were used for all experiments. All experiments were approved and performed according to the guidelines of the Animal Care and Use Committee of the NIEHS, National Institutes of Health (2015–0004). To ensure that the potential effects of GR ablation would be related to its absence rather than just to a reduction on its levels, we decided to conduct our study only in homozygous GR knockout mice (hereafter called Emx1-Nr3c1−/−). We have used a total of 24 animals segregated in 6 animals per group (female Control, male Control, female Emx1-Nr3c1−/− and male Emx1-Nr3c1−/−). The hippocampi from left hemispheres of all animals were rapidly dissected and immersed in RNAlater™ (Qiagen, California, USA) for subsequent TRIzol RNA and protein extractions. The remaining right hemispheres were used for morphological studies using Golgi Stain (n = 3 for each experimental group) or alternatively, hippocampi were immediately homogenized for protein extraction for GR and MR Western blotting determination (n = 3 for each experimental group).
2.2. RNA extraction and quantitative real-time reverse transcription-polymerase chain reaction (RT-qPCR)
RNAs from one hippocampus of each mouse were isolated and quality-controlled, as we have previously described (Munoz-Llanos et al., 2018). For the RT-qPCR procedure, predesigned TaqMan™ primer/probe sets for Nr3c1 and Nr3c2 mRNAs were used to perform a one-step reaction in the 7900HT-Fast Real-Time PCR System (Applied Biosystems, USA) thermocycler. The thermal profile was defined according to the manufacturer's instructions, and gene expression was calculated based on the 2−ΔΔCt method (Schmittgen and Livak, 2008).
2.3. nCounter® miRNA Expression Assay of miRNAs
For the quantitative determination of mature miRNAs, purified hippocampal RNA was treated with RNase-Free DNase Set (Qiagen, Germany), followed by a clean-up and concentration step of miRNA fraction and small RNAs (<200 nucleotides) by RNeasy MinElute Cleanup Kit (Qiagen, Germany). The sample concentration was determined by NanoDrop, diluted in a range of 40–80 ng/μl, and RNA integrity was determined by Advance Analytics Fragment Analyzer. miRNA levels were determined with the nCounter® miRNA Expression Assay (NanoString Technologies, Seattle, USA), using the mouse miRNA panel v1.5., which detects the levels of 578 mouse mature miRNAs, 33 viral mouse miRNAs, and 4 housekeeping genes. The automated nCounter® platform allows the quantitative measurement of multiple miRNAs within one sample by the hybridization of fluorescent barcodes directly to one specific nucleic acid sequence (Veldman-Jones et al., 2015). The hybridization cartridges were digitized with the nCounter® Digital Analyzer instrument and the raw data (RCC files) were generated from the images with the nSolver™ Analysis Software (v4.0, NanoString Technologies, Inc.). These procedures were conducted at the Molecular Genomics Core Laboratory of the National Institute of Environmental Health Sciences (NIEHS).
2.4. Bioinformatic analyses
Full bioinformatic workflows are described in the supplementary methods. In brief, raw RCC files data were normalized and background-corrected. Differential expression analyses were performed between female and male controls, followed by Emx1-Nr3c1−/− and controls by sex. Probes with an adjusted p-value < 0.05 and |fold change| ≥ 1.3 were considered as differentially expressed.
Putative trancriptional regulators of differentially expressed miRNAs were retrieved from TransmiR 2.0 database (Wang et al., 2010). GR-binding sites were scanned in the promoters of differentially expressed miRNAs using the JASPAR database (Fornes et al., 2020) and FIMO from MEME-Suite (Grant et al., 2011).
Functional enrichment analysis of differentially expressed miRNAs was firstly assessed using the TAM 2.0 webserver (Li et al., 2018). Considering that TAM 2.0 contains only information about human miRNAs, we matched the mouse and human miRNA names and/or the miRNA sequences based on sequence and annotation data from miRBase v22 (Kozomara and Griffiths-Jones, 2011). An extra step was performed using CD-HIT (Li and Godzik, 2006; Fu et al., 2012) to find miRNAs in the human miRNAome, specifically with those mouse miRNAs that did not previously matched. We considered only those miRNAs with an identity greater than or equal to 80%.
Differentially expressed miRNAs in each group were compared against two predicted target mRNA databases: miRDB v6.0 (Chen and Wang, 2020) and miRWalk 3.0 (Sticht et al., 2018). Similarly, the same datasets of miRNAs were used as input for the miRNet platform (Fan et al., 2016), which collects validated interactions from the well-annotated databases miRTarBase v7.0 (Hsu et al., 2011), TarBase v8.0 (Karagkouni et al., 2018) and miRecords (Xiao et al., 2009), to identify validated target mRNAs. Predicted and validated target mRNAs were refined by reported expression in the rodent hippocampus using the Expression Protein Atlas (Petryszak et al., 2016). Data handling and functional enrichment analysis were performed in R v3.6.1 with the R package enrichR v2.1 (Kuleshov et al., 2016), and KEGG 2019 mouse databases (Kanehisa and Goto, 2000). KEGG pathways were retrieved by filtering for statistical significance (p-value < 0.05) and the top 15 pathways with the highest combined score were selected, according to Enrichr recommendations for best ranking (Kuleshov et al., 2016). Data visualization was performed with R package ggplot2 v3.2.1, and circular plots were obtained with R package circlize v0.4.8 (Wickham, 2011; Gu et al., 2014).
2.5. Golgi staining and morphological analyses
One fresh hemisphere was used for Golgi staining, using the FD Rapid GolgiStain™ kit (FD Neuro Technologies, Baltimore, MD, USA), as described (Castaneda et al., 2015; Garcia-Rojo et al., 2017; Aguayo et al., 2018). Full details of dendritic complexity analyses are described within the supplementary methods. Briefly, focal Z-planes were digitally converted into virtual slides using the Hamamatsu NDP slide scanner (Hamamatsu Nanozoomer, Japan). These Z-stack images were then used to create semi-automated 3D reconstructions of each neuron, using the neuTube software (Feng et al., 2015), and saved in SWC format. Using the simple Neurite Tracer plugin for Fiji (Longair et al., 2011), each path of the reconstructed neuron was assigned as being a part of the soma, the apical dendrite, or a basal dendrite. Using the assigned paths and the L-Measure software (Scorcioni et al., 2008), the total dendritic length and the number of branch points were determined for both apical and basal dendrites. The Sholl analysis (Sholl, 1953) was performed within Fiji environment (Ferreira et al., 2014) using a “skeleton” of the constructed image that was overlapped in a series of concentric rings, at an interval of 10 μm centered on the cell body. The number of intersections crossing each concentric ring was determined in basal and apical dendrites.
Dendritic spines analyses were performed as described in prior studies (Castaneda et al., 2015; Garcia-Rojo et al., 2017; Aguayo et al., 2018). Full details of dendritic spine density and morphology analyses are described within the supplementary methods.
2.6. Western blot analysis
A detailed description of Western blot analyses can be found in the supplementary methods. Briefly, for the evaluation of nuclear receptors, hippocampi were homogenized in RIPA buffer (Pierce, Thermo Scientific) containing a cocktail of protease (Co-Ro, Sigma Aldrich) and phosphatase (Calbiochem, Millipore) inhibitors. 15 μg of protein were resolved on 4–20% MiniPROTEAN© TGX™ Stain-Free precast protein gels (BioRad, California, Unites States) and then electroblotted onto 0.2-μm nitrocellulose membranes. After several washes with TBS, membranes were blocked and then incubated overnight at 4 °C with the desired primary antibody (Table 1). Blots were then washed and incubated with goat anti-mouse IRDye800-conjugated (LI-COR Biosciences, Lincoln, NE, USA; cat: 926–32210) or goat anti-rabbit (Cell Signaling Technology, USA; cat. 7074). Membranes were washed, and bands were detected with an Odyssey® CLx Imaging System (Li-Cor Biosciences, Lincoln, NE, USA). Alternatively, we used the protein recovered from hippocampi homogenized in TRIzol for GSK-3β and CaMKII-α, according to a described protocol (Kopec et al., 2017), with some modifications fully described in the supplementary methods.
Table 1.
Primary antibodies and blocking conditions used.
| Antigen | Description of Immunogen | Source, Host Species, Cat. #, RRID |
Concentration used | Primary antibody and blocking solution |
|---|---|---|---|---|
| β-Actin | Synthetic peptide corresponding to aa. 2 to 16 from mouse β-Actin. | Synaptic Systems, rabbit polyclonal, cat. no. 251 003, RRID: AB_11042458 or Merck Millipore, mouse monoclonal, cat. no. MAB1501, RRID: AB_2223041 |
1:10000 1:10000 |
3% non-fat dried milk, TBS-0.1% Tween-20 Odyssey Blocking buffer |
| GR | Amino acids 121–420 of GR of human origin. | Santa Cruz Biotechnology, mouse monoclonal, cat. no. sc-393232, RRID: AB_2687823 |
1:100 | 1% non-fat dried milk, TBS-0.05% Tween-20 |
| MR | Synthetic peptide corresponding to N terminal region (A/B domain) of rat MR | rMR1-18 1D5, mouse monoclonal. (Gomez-Sanchez et al., 2006) | 1:400 | Odyssey Blocking buffer |
| GSK-3β | Synthetic peptide corresponding to the sequence of human GSK-3β | Cell Signaling Technology, rabbit monoclonal, cat. no. 9315, RRID: AB_490890 |
1:1000 | 3% BSA, TBS-0.1% Tween-20 |
| CaMKII-α | Synthetic peptide within Human CaMKII aa 200–300. The exact sequence is proprietary. | Abcam, rabbit monoclonal, cat. no. ab52476, RRID: AB_868641 |
1:3000 | 5% BSA, TBS-0.1% Tween-20 |
2.7. Statistical analyses
Data from molecular and morphological analyses are presented as means ± SEM. Statistical significance was set with a confidence level of 95% (p-value ≤ 0.05). Differences between only two groups (GR and MR protein levels, n = 3) were determined by Welch's t-test. The effects of the sex, genotype, and their interaction (sex × genotype) were determined by two-way ANOVA, using Prism 8 (GraphPad Software Inc, CA, USA). Post-hoc pairwise comparisons were computed with Tukey's test for both molecular and dendritic length and branching analyses, or with Fischer's LSD for dendritic spine density. For the Sholl analysis, the effects of sex, genotype and radial distance from the soma were evaluated by three-way ANOVA, using R v3.6.3 and rstatix packages, v0.4.0 (Kassambara, 2020). For the GSK-3β Western blotting analysis, one female control datapoint was excluded from statistics because the value deviated from the observed central tendency (> mean + 2 SD). Appart from this, no other outlier was identified.
3. Results
3.1. Validation of a conditional GR knockout mice
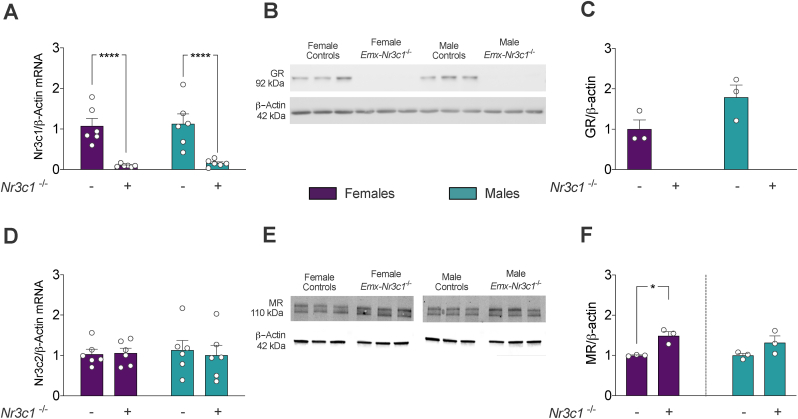
To asses the effect of GR ablation on miRNA expression, we generated conditional GR knockout mice through Cre recombinase expression under the control of the Emx1 gene promoter (Emx1-Nr3c1−/−). The EMX1 is a transcription factor implicated in forebrain development (Muzio and Mallamaci, 2003), whose expression starts around embryonic day 9.5 and is restricted to the dorsal telencephalon (Simeone et al., 1992). The GR/Nr3c1 mRNA levels were significantly reduced by 90% in female and male Emx1-Nr3c1−/− mice compared to controls (genotype: F1,20 = 41.07, p-value < 0.0001), with no significant differences between female and male, both controls and Emx1-Nr3c1−/− mice (Fig. 1A). This was further supported by Western blot analyses of mice hippocampi, where GR protein levels were undetectable on Emx1-Nr3c1−/− female and male mice (Fig. 1B). The GR protein levels tended (p-value = 0.1052) to be similar between female and male controls (Fig. 1B and C).
Fig. 1.
Nr3c1 and Nr3c2 mRNA and protein levels in the hippocampus ofEmx1-Nr3c1−/−mice.(A) GR/Nr3c1 mRNA levels measured by RT-qPCR (n = 6 per group; two-way ANOVA genotype: F1,20 = 47.01, ****p < 0.0001). (B) Western blot analyses of GR and (C) relative quantities of GR in female and male controls (n = 3 per group). (D) MR/Nr3c2 mRNA levels measured by RT-qPCR (n = 6 per group; two-way). (E) Western blot analyses of MR. (F, left) Relative quantities of MR in female Emx1-Nr3c1−/− compared to female controls (n = 3 per group, two-tailed Welch's t-test: t2.036 = 4.558, *p = 0.0434). (F, right) Relative quantities of MR in male Emx1-Nr3c1−/− compared to male controls (n = 3 per group, two-tailed Welch's t-test: t2.4 = 1.837, p = 0.1861). β-actin was used as normalizer across both RT-qPCR and Western blot.
Next, considering the functional interactions between GR and MR, we evaluated whether GR ablation alters MR/Nr3c2 mRNA and protein levels in the hippocampus. MR mRNA levels were similar across female and male mice, both controls and Emx1-Nr3c1−/− (Fig. 1D). Then, MR levels were estimated utilizing a well-characterized antibody (Gomez-Sanchez et al., 2006) that detected two immunoreactive bands in our assays. One band was at the expected molecular mass (~110 kDa), and the other near 130 kDa (Fig. 1E). Since MR is subjected to post-translational modifications (Pascual-Le Tallec and Lombes, 2005) that may explain the double band, we decided to quantify both bands. Despite the low number of biological replicates, MR protein levels tended to be higher in female Emx1-Nr3c1−/− mice compared to their controls (Welch-corrected t(2.036) = 4.558, p-value = 0.0434; Fig. 1F), with no effect in Emx1-Nr3c1−/− males. These findigs corroborate our GR ablation on hippocampal tissue.
3.2. Sex differences on basal miRNA expression on control mice hippocampus
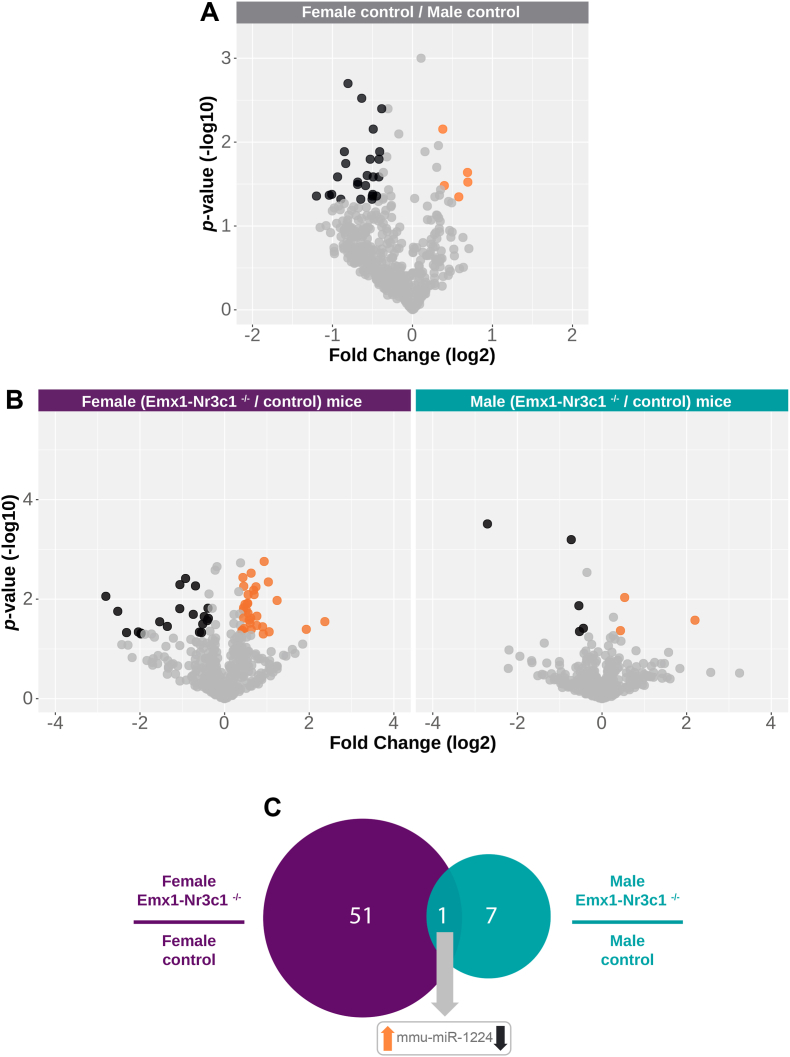
To establish whether there are sex differences on basal miRNAs levels, we used the NanoString technology, which includes probes for 578 mouse mature miRNAs. Six biological replicas were employed to control the reproducibility of our findings (Figure S1). To compare the miRNA expression pattern between female and male controls, we performed a differential expression analysis. This analysis yields differences in the type, number, and direction of miRNAs expression, using a |fold change| ≥ 1.3 and an adjusted p-value < 0.05 as a cutoff parameters. Fig. 2A shows the expression of miRNAs from control mice hippocampi. Females exhibited five highly expressed miRNAs (orange filled circles), and twenty-four lowly expressed miRNAs (black filled circles), compared to males. These data indicate that female and male controls exhibit a similar miRNA profile, where only a reduced number of miRNAs (i.e., 29 miRNAs, corresponding to almost 5% of the total evaluated miRNAs) displays a sex-biased expression. Interestingly, the sex-differentially expressed miRNA genes derive from several chromosomes; nonetheless, six of them are found in the X-chromosome, which exhibited a higher (mmu-miR-2132) or lower (mmu-miR-19 b, −224, −448, −504, -542–5p) expression in females, compared to males (Table S1).
Fig. 2.
Sex differences on hippocampal miRNA expression in control mice and sex-biased effects of GR-ablation. (A) Volcano plot of differentially expressed miRNAs in control mice. The horizontal axis represents the log2 fold change (Female Control/Male Control), and the vertical axis represents the negative log10 of the p-value. miRNAs that significantly decreased their expression are shown in black dots (24 miRNAs), and those that significantly increased their expression are shown in orange (5 miRNAs). (B) Volcano plots of differentially expressed hippocampal miRNAs in female (left) and male (right) Emx1-Nr3c1−/− mice, compared to their respective sex controls. The horizontal axis represents a log2 fold change (Emx1-Nr3c1−/−/Control), and the vertical axis represents a negative log10 of the p-value. miRNAs that decreased their expression are shown in black, and those that increased their expression are shown in orange. Gray dots across volcano plots represent those miRNAs that were outside the cutoff parameters (|fold change| ≥ 1.3; p-value < 0.05). (C) Venn diagram of differentially expressed miRNAs in female (purple) and male (teal) hippocampus of Emx-Nr3c1−/− mice vs controls. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Glucocorticoid receptor knockout induces a sex bias miRNAs expression profile in the mouse hippocampus
In order to establish whether GR ablation triggers a sex-biased miRNA expression profile, first we conducted a differential expression analysis between each Emx1-Nr3c1−/− dataset to its respective sex control group. When female Emx1-Nr3c1−/− mice were contrasted against female controls, 52 miRNAs significantly changed their expression levels (Fig. 2B), of which 20 decreased and 32 increased (Table S2). Interestingly, mmu-miR-219, which was female-biased in control mice, was reduced by GR ablation in females. In contrast, only eight differentially expressed miRNAs were found in male mice (Fig. 2B), of which 5 decreased and 3 increased, compared to male controls (Table S2). We found that mmu-miR-224, which was male-biased in control mice, displayed a reduction in Emx1-Nr3c1−/− males compared to controls.
Next, we compared the obtained datasets of differentially expressed miRNAs to identificate sex-biased effects of GR ablation on miRNA expression (Venn diagram, Fig. 2C). Indeed, while the vast majority of differentially expressed miRNA displayed a sex-biased expression by GR ablation, only one miRNA (mmu-miR-1224) was common in both datasets (Fig. 2C), being up-regulated in females and down-regulated in male mice by GR ablation.
Remarkably, many of the differentially expressed miRNA genes in the female hippocampus are spread across the genome. For instance, many of them derived from autosomic chromosomes and only four derived from the X chromosome (Table S2). Similarly, the differentially expressed miRNAs in the male hippocampus -which were lesser in number- were mapped to several autosomic chromosomes, and only one derive from the X chromosome (Table S2). Chi-square analysis indicated that the distribution of female and male miRNAs genes in autosomic and X chromosomes showed no sex-bias (χ2 1,60 = 0.2098, p-value = 0.6469).
To this extent, our results show that GR ablation triggers a sex-biased miRNA expression profile, which seems to be more sensitive in females -regarding the multiplicity of deregulated miRNAs- than in males. Unexpectedly, only a reduced number of the sex-biased miRNA genes were located in the X chromosome, suggesting that genetic dosage of this chromosome does not influence the sex-biased miRNA expression by GR ablation.
3.4. Search for transcription factors that may regulate miRNA gene expression
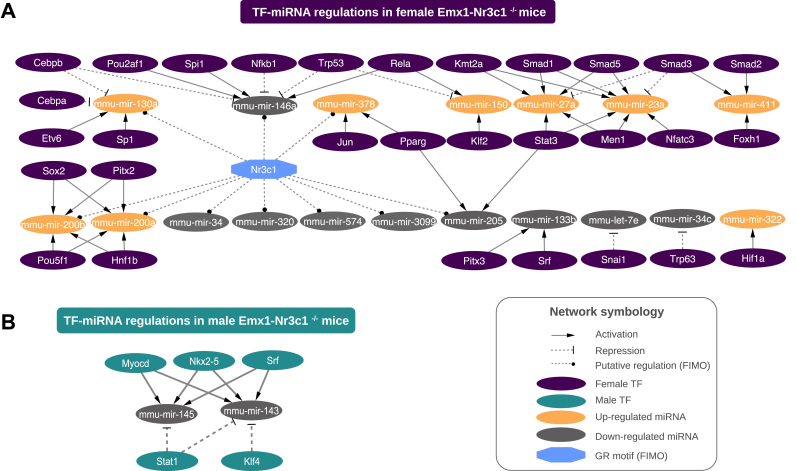
Since the GR-sensitive miRNAs genes derived from multiple genomic locations, common upstream transcription factors (TFs) may explain their co-expression. Therefore, we first searched for putative TF-miRNA regulations utilizing the TransmiR 2.0 curated database. In females, we identified 46 TF-miRNA interactions (Fig. 3A), which included 30 TFs and 14 miRNAs. Several TFs were related to Smad family members. For instance, mmu-miR-411, mmu-miR-27a, and mmu-miR-23a showed a rise in their levels by GR ablation and were found to be putatively responsive to Smad members (Fig. 3A). Also, mmu-miR-200a and mmu-miR-200b -which belong to the miR-200b/200a/429 cluster- are potentially up-regulated by Sox2, Pitx2, Pou5f1 and Hnf1b (Fig. 3A). In addition to Smad regulation, mmu-miR-27a and mmu-miR-23a -which belong to the miR-23a/27a/24-2 cluster- are probably regulated by Stat3, Men1, and Kmt2a (Fig. 3A). Furthermore, we detected five miRNAs that reduced their levels by GR ablation and are putatively repressed by TFs. For instance, mmu-miR-let-7e and mmu-miR-34c are likely repressed by Snai1 and Trp63, respectively. Finally, miR-146a may be negatively regulated by Nfkb1 and Trp53, which are recognized TFs that interact with the GR. In order to gain insight into a putative direct role of GR in miRNA gene expression, we next used the FIMO algorithm to search for GR-binding motifs in promoter regions of miRNA genes sensitive to GR ablation. We identified 10 miRNA genes with a GR-binding motif, and 6 of them (mmu-miR-146, -34, −320, −574, −3099, −205) displayed a reduced expression in female Emx1-Nr3c1−/− mice. The other 4 miRNAs with a GR-binding motif (mmu-miR-378, -130a, and the miR-200a/miR-200b gene cluster) increased their levels upon GR deletion.
Fig. 3.
Putative transcription factor regulations of the sex-biased miRNA genes. Network representation of transcription factors and type of regulation (activation: direct arrows or repression: dashed lines with truncated edge) found in TransmiR 2.0 database for differentially expressed miRNA genes in (A) female Emx1-Nr3c1−/− mice vs controls (purple transcription factors) and (B) male Emx1-Nr3c1−/− mice vs controls (teal transcription factors). Up-regulated miRNAs are shown in orange and down-regulated miRNAs, in gray. FIMO analysis unveiled GR-binding sites in the promoter of several sex-biased miRNA genes and is represented by a dashed line with a dotted-edge. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In males, only the members of the miR-143/145 cluster were annotated in the TransmiR database. Both miRNAs were down-regulated and we identified TFs with both potentially repressive actions (Stat1 and Klf4) and positive actions (Myocd, Nkx 2-5 and SrF) (Fig. 3B). With the FIMO tool, we were unable to find GR-binding motifs in the promoters differentially expressed miRNAs in males.
These results suggest that GR deletion may impact the regulation of miRNA levels through both direct and indirect transcriptional mechanisms, which may impact global gene expression in the hippocampus with a sex-biased effect.
3.5. Functional enrichment analysis of the differentially expressed miRNAs
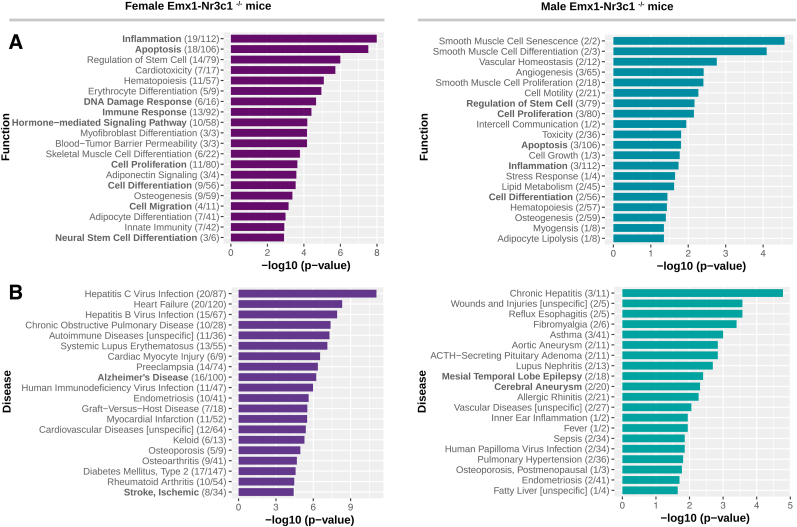
In order to explore the physiological impact of the differentially expressed miRNAs by GR ablation, we used the TAM 2.0 analysis, which is based on text mining to construct miRNA-term annotations from both tabulated information in other databases and those extracted from publications describing miRNAs. The top-20 function terms that were significantly enriched (p-value < 0.05) in female Emx1-Nr3c1−/− mice vs controls are shown in Fig. 4A. Interestingly, several functions are relevant in the brain and hippocampus, e.g., inflammation, apoptosis, DNA damage response, immune response, hormone-mediated signaling, cell proliferation and differentiation, cell migration and neural stem cell differentiation. Additionally, the top-20 significantly enriched miRNA-disease associations of the differentially expressed miRNAs in females mice are shown in Fig. 4B. We identified relevant CNS dysfunctions, such as Alzheimer's disease and stroke/ischemic disease, involving 16 and 8 miRNAs, respectively (Fig. 4B).
Fig. 4.
TAM functional enrichment analysis of the sex-biased miRNA genes. (A) Functional enrichment analysis of the sex-biased miRNA genes of female and male Emx1-Nr3c1−/− mice, using the TAM 2.0 tool (p-value < 0.05). Numbers in parentheses indicate the overlap of the hits over the total annotated miRNA in their respective function. (B) Disease enrichment analysis of female and male miRNA genes in Emx1-Nr3c1−/− animals. Only the top 20 significant terms (p-value < 0.05) are shown. Numbers in parentheses indicate the overlap of the hits over the total annotated miRNA in their respective disease.
A similar analysis was conducted with the differentially expressed miRNAs in male Emx1-Nr3c1−/− vs controls. We found enrichments with 6 miRNAs (mmu-miR-143, -145, −323, −224 and −1224) of the 8 total miRNAs in several functions, suggesting that their variation may be relevant in different processes. These functions -also detected in the female dataset-include regulation of stem cell, cell proliferation and differentiation, apoptosis, and inflammation (Fig. 4A). Additionally, of the top-20 enriched diseases, those relevant to the brain are mesial temporal lobe epilepsy and cerebral aneurysm, both associated to 2 miRNAs (Fig. 4B).
3.6. Pathways constructed with predicted targets of differentially expressed miRNAs
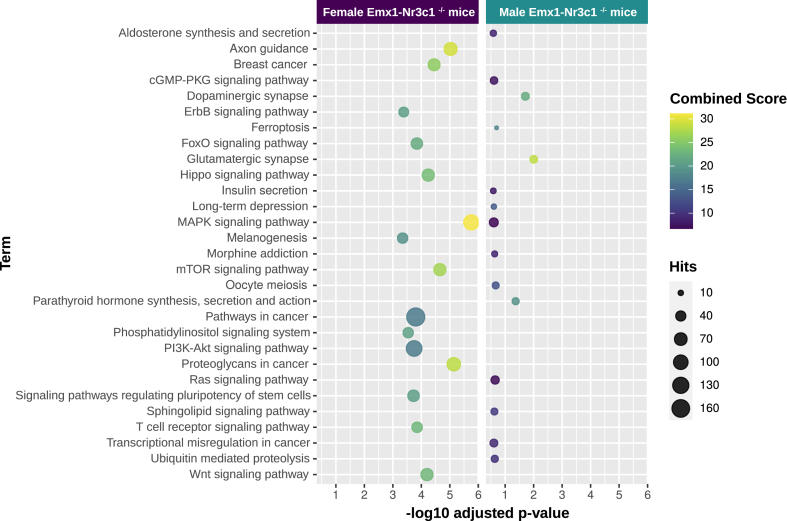
Our TAM analyses yielded many brain-relevant functional and disease-associated processes that may occur by GR ablation in the mouse hippocampus. However, these analyses are limited to text mining relationships and currently annotated interactions described in the literature. To render our functional analyses more complex and less biased, we conducted a functional enrichment analysis with the miRNA-mRNA predicted interactions to evaluate the possible impact of the differentially expressed miRNAs in the hippocampus. By using the predicted target mRNAs of differentially expressed miRNAs in Emx1-Nr3c1−/− mice of each sex, compared to their respective controls, we selected the top 15 KEGG pathways with the highest combined score for each sex dataset (Table S3). Fig. 5 represents them in a bubble plot and the higher combined scores are seen warmer in color (yellow), and the more numerous predicted target mRNA hits, are represented by a bigger bubble. Interestingly, eight of the fifteen pathways identified in female and male enrichment analysis were in common: Axon guidance, Rap1 signaling pathway, MAPK signaling, Phosphatidylinositol signaling system, mTOR pathways, Autophagy, Protein processing in the endoplasmic reticulum and Glutamatergic synapse; pathways which are particularly relevant for synapse formation and neuroplasticity (Kumar et al., 2005; Liu et al., 2006; Switon et al., 2017, Nikoletopoulou and Tavernarakis, 2018).
Fig. 5.
Functional enrichment analysis for the miRNA-mRNA predicted interactions from miRDB. The functional enrichment analysis was performed with predicted target mRNAs of differentially expressed miRNAs in female (left) and male (right) Emx1-Nr3c1−/− mice, using the enrichR package. The enriched terms were then filtered by using Fisher's exact test, p-value < 0.05. The top 15 KEGG terms with the highest combined score are shown for each sex, with dots colored by the combined score (cs = log [p] *z, p = Fisher's exact test p-value, and z = z-score for deviation from expected rank). Hits correspond to the number of targets in the gene set which are potentially regulated by the differentially expressed miRNAs. The predicted target mRNAs were obtained from the miRDB database.
3.7. Pathways constructed with validated targets of differentially expressed miRNAs
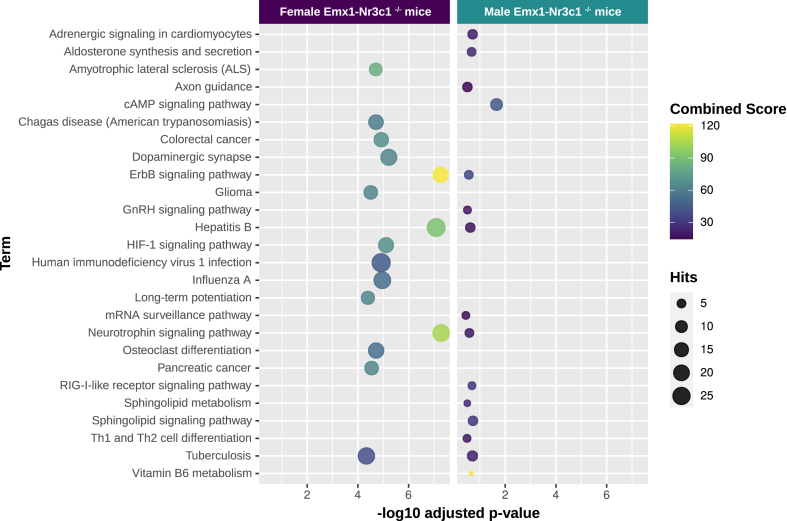
To refine our analysis, we conducted an enrichment analysis with experimentally validated miRNA targets obtained from the miRNet database, which we manually updated with reported expression in the mouse hippocampus. The functional enrichment analysis in females identified 149 significant pathways (p-value < 0.05). As in Fig. 5, the top 15 pathways were selected (Table S4) and are represented in a bubble plot (Fig. 6). We found that eight pathways in the female dataset were related to neuronal function, and four of them were coincident with the prediction analysis: Axon guidance, MAPK signaling pathway, Autophagy, and protein processing in the endoplasmic reticulum. The other pathways were related to cytoskeleton dynamics (focal adhesion and regulation of actin cytoskeleton) and FoxO signaling pathway regulation of cytoskeletal dynamics and organization (McLaughlin and Broihier, 2018). The analysis of the male dataset showed eight pathways that were related to neuronal function, and three of them were also found in the prediction analysis: MAPK, mTOR, and the Sphingolipid signaling pathway (Fig. 6, Table S4). Additionally, the enrichment analysis identified the AMPK signaling pathway, along with PI3K-Akt signaling, Neurotrophin signaling, and ErbB signaling pathways, which are implicated in neuronal morphology (Russo et al., 2009; Shi et al., 2009). Interestingly, three of the top 15 pathways constructed with validated target mRNAs were found both in male and female enrichment analysis: MAPK signaling, Focal adhesions and FoxO signaling. Interestingly, FoxO also converges onto the pathways that mediate morphological and synaptic plasticity in neurons (McLaughlin and Broihier, 2018).
Fig. 6.
Functional enrichment analysis for the miRNA-mRNA validated interactions from miRNet. Functional enrichment analysis was performed with validated target mRNAs of differentially expressed miRNAs in female (left) and male (right) Emx1-Nr3c1−/− mice, using the enrichR package. Enriched terms were filtered by Fisher's exact test, p-value < 0.05. The top 15 KEGG terms are shown for each sex, with dots colored by the combined score (cs = log [p] *z, p = Fisher's exact test p-value, and z = z-score for deviation from expected rank). Hits correspond to the number of validated interactions between differentially expressed miRNAs and target mRNAs. The validated target mRNAs were obtained from the miRNet database.
3.8. Intersectional analysis of pathways constructed with both validated and predicted targets of differentially expressed miRNAs
The analyses of both predicted and validated targets for differentially expressed miRNAs induced by Emx1-directed GR ablation yielded key pathways that control many aspects of neuronal morphology, including dendritic morphogenesis and spine growth (Kumar et al., 2005); events that are crucial during neuronal development and plasticity (Switon et al., 2017). Considering the inexistence of databases with miRNA-mRNAs interactions related to neuronal cytoarchitecture, we searched the literature to identify key proteins involved in morphological plasticity (dendrite formation and arborization, and spine development, shaping, stability, and density) (Koleske, 2013; Forrest et al., 2018) to match this information with target mRNAs contained within the KEGG pathways described above (Table S3, Table S4, and Table S5).
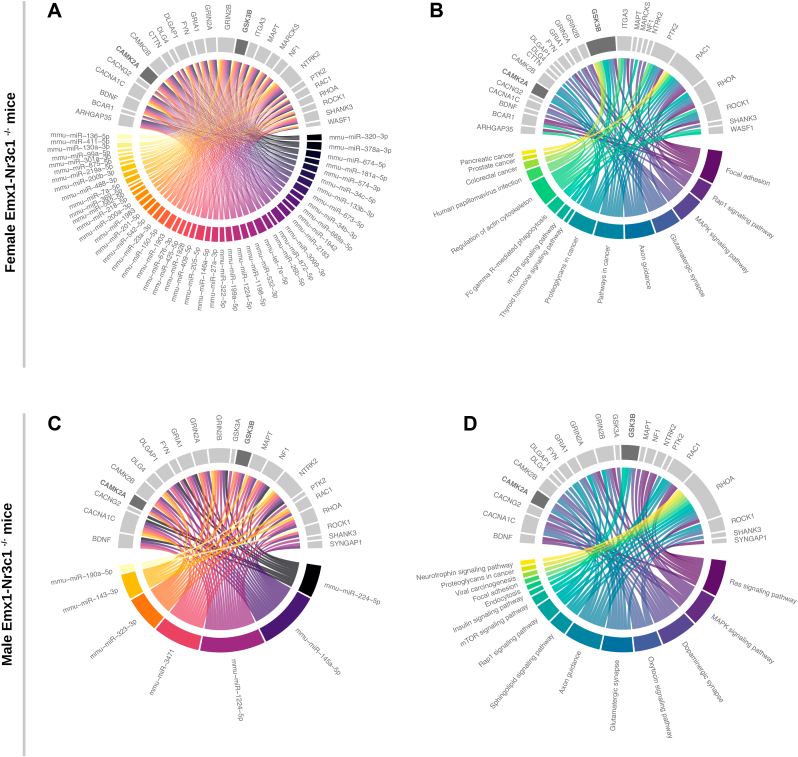
Fig. 7A summarizes both analyses in a chord diagram constructed with the 48 differentially expressed miRNAs in female Emx1-Nr3c1−/− mice (colored arc) and both predicted (26 transcripts) and validated (16 transcripts) miRNA-mRNA interactions (gray arc). Furthermore, we noted that 15 transcripts were common as predicted and validated miRNA targets (Table S5). Then, we matched these gene products (gray arc) with KEGG pathways represented by a colored arc and whose arc length was proportional to the number of transcripts included in the pathway (Fig. 7B). Notably, several of these KEGG pathways were related to the nervous system: axon guidance (8 mRNAs), focal adhesions (7 mRNAs), MAPK signaling (7 mRNAs), regulation of actin cytoskeleton (7 mRNAs), glutamatergic synapse (7 mRNAs), Rap1 signaling pathway (5 mRNAs) and mTOR signaling (2 mRNAs) (Fig. 7B). Alternatively, the length of each gray arc qualitatively represents the number of KEGG pathways in which a particular gene product participates; being Rac1, Gsk3b, RhoA, Rock1, Camk2a, Camk2b and Bcar1 the most represented gene products (Fig. 7B). Similar analysis in male Emx1-Nr3c1−/− mice indicated that 7 differentially expressed miRNAs (mmu-miR-1224–5p, −145a-5p, -323–3p, −3471, -224–5p, -143–3p, and −190a-5) were predicted to interact with 22 transcripts and with one validated target involved in spine and dendrite dynamics (Table S5 and Fig. 7C). The Chord diagram also shows that the more prominent KEGG pathways in function of the number of terms and related to the nervous system were: glutamatergic synapse (10 mRNAs), axon guidance (9 mRNAs), Ras signaling (8 mRNAs), dopaminergic synapse (8 mRNAs), MAPK signaling pathway (6 mRNAs); and with the lower number of terms, the Rap1 (4 mRNAs) and mTOR signaling pathways (2 mRNAs) (Fig. 7C). Moreover, RhoA, Rock1, Gsk3b, Grin2a, Grin2b, Camk2a and Camk2b and Cacna1C -derived transcripts were the most represented in the KEEG pathways (Fig. 7D).
Fig. 7.
mRNAs related to neuronal arborization, spine stability and density, among predicted and validated targets of differentially expressed miRNAs in female and male Emx1-Nr3c1−/−mice. (A) Chord diagram representing the predicted interaction of differentially expressed miRNAs (colored arc) with mRNAs in female Emx1-Nr3c1−/− mice (gray arc). (B) Chord diagram representing the involvement of the mRNAs described above in the KEGG biological pathways enriched in female Emx1-Nr3c1−/− mice, with transcripts involved in spine and dendrite dynamics (gray arc) (Koleske, 2013; Forrest et al., 2018) (Table S5). (C) Chord diagram representing the predicted interaction of differentially expressed miRNAs (colored arc) with mRNAs in male Emx1-Nr3c1−/− mice. (D) Chord diagram representing the involvement of mRNAs described above in the KEEG biological pathways enriched in male Emx1-Nr3c1−/− mice, with transcripts involved in spine and dendrite dynamics (gray arc) [(Koleske, 2013; Forrest et al., 2018), Table S5]. The darker gray arc observed for both GSK3B and CAMK2A transcript denotes a rank >70th percentile of the total number of miRNA-mRNA and genes-pathways interactions.
Overall, these results predict that GR ablation may impact several cellular processes involved in neuronal cytoarchitecture, probably in a sex-biased manner, considering the larger array of miRNAs differentially expressed in females under GR ablation.
3.9. Hippocampal GR deletion triggers a sex-biased effect on GSK-3β and CaMKII-α levels
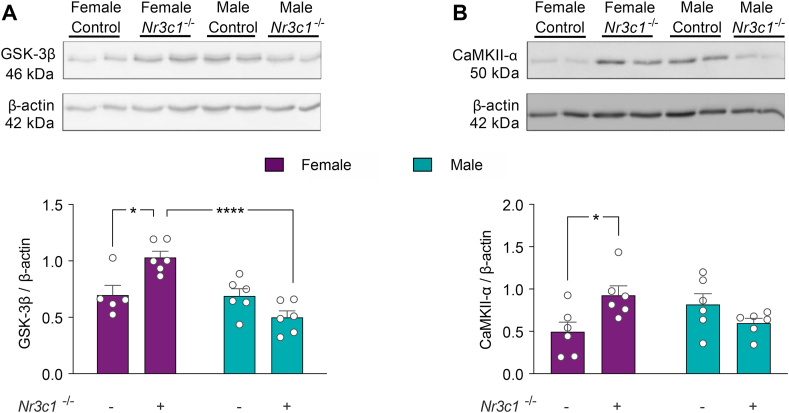
Our bioinformatics analysis indicated many putative and proven miRNA targets related to neuronal morphology. Interestingly, among the miRNA targets, GSK-3β and CAMKII-α displayed a large amount of miRNA-mRNA interactions and pathway hits, being in the upper 70th percentile (Table S5; Fig. 7, dark gray arc). Besides, GSK-3β and CAMKII-α have been described as key neuronal morphology regulators (Koleske, 2013; Forrest et al., 2018). Therefore, we next addressed whether the dysregulations of miRNAs induced by GR ablation were accompanied by changes in GSK-3β and CAMKII-α levels in the hippocampus. Fig. 8A shows a representative Western blot of GSK-3β relative levels in the hippocampus of control and Emx1-Nr3c1−/− animals. Two-way ANOVA indicated a main effect of sex (F1, 19 = 17.17, p-value = 0.0006) and sex × genotype interaction (F1, 19 = 16.19, p-value = 0.0007; Fig. 8A). Tukey's post-hoc test showed no significant effect in males, but a significant increase of GSK-3β levels in female Emx1-Nr3c1−/− mice, compared to both female controls and male Emx1-Nr3c1−/− mice. Interestingly, CaMKII-α levels were increased in female Emx1-Nr3c1−/− mice, compared to controls (sex × genotype: F1,20 = 9.476, p-value = 0.0059; Fig. 8B). These results suggest that Emx1-directed GR deletion in the hippocampus could promote a sex-biased impact on GSK-3β and CaMKII-α levels, which may be related to the sex-biased miRNAs profiles.
Fig. 8.
GSK-3β and CaMKII-α levels are increased in the hippocampus of female Emx1-Nr3c1−/−mice. Representative Western blot analyses and relative immunoreactivity of (A) GSK-3β, (B) CaMKII-α in the hippocampus of control and Emx1-Nr3c1−/− female and male mice. Fifteen μg of protein were loaded for each sample. β-actin was used as a loading control. Data were analyzed by two-way ANOVA with Tukey's post hoc test: **** p-value < 0.0001, * p-value < 0.05. n = 5–6 per group (panel A: main effect of sex F1,19 = 17.11, p-value 0.0006; and sex × genotype F1,19 = 16.19, p-value = 0.0007. Panel B: main effect of sex × genotype F1,20 = 9.476, p-value = 0.0059).
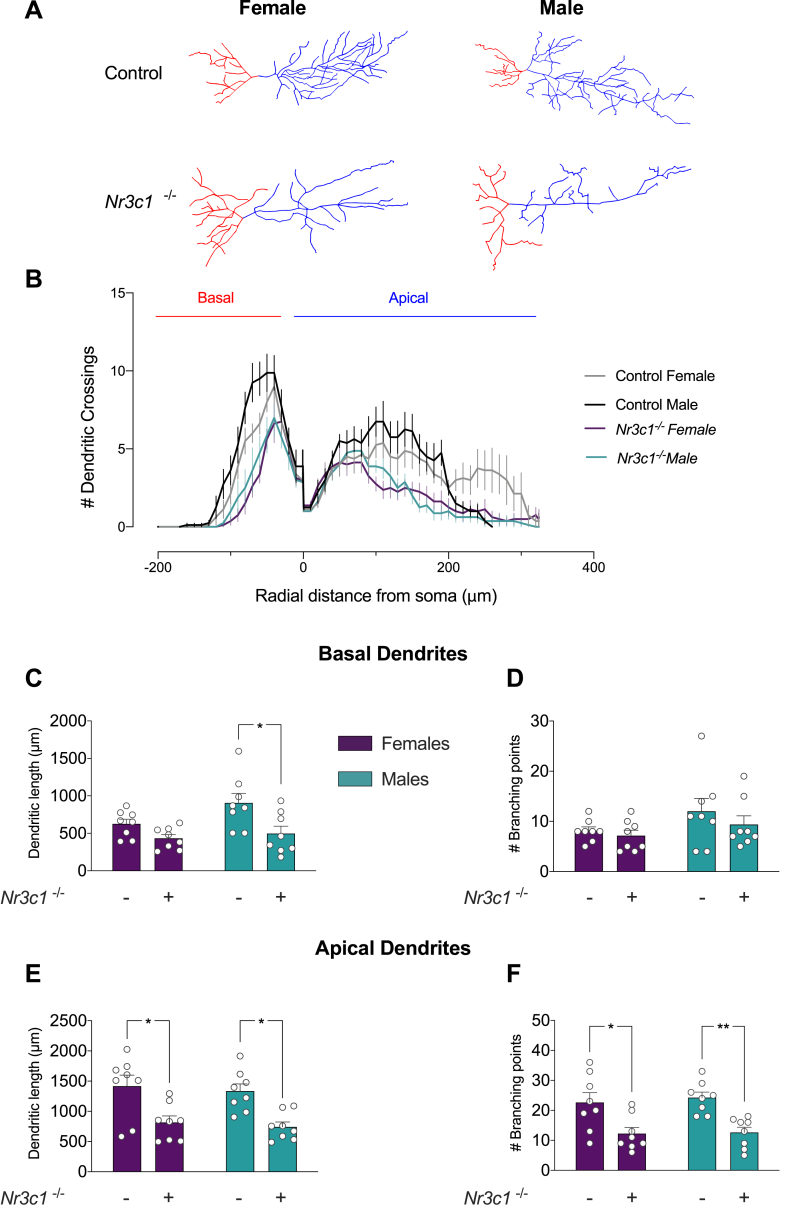
3.10. GR ablation induces a sex-biased effect on the structural morphology of CA1 neuronal dendrites
Our analyses have indicated that both predicted and validated miRNA targets are related to spine formation and dendritic arborization. These observations suggest that GR ablation may have profound effects on neuronal morphology in the hippocampus. Thus, we next evaluated the impact of GR deletion on neuronal morphology in the hippocampus; namely, dendritic arborization and spine morphology and density. We evaluated the arborization of CA1 neurons and dendritic complexity by using tridimensional imaging and neuronal reconstruction of Golgi-impregnated CA1 pyramidal neurons. Fig. 9A shows representative neurons of control and Emx1-Nr3c1−/− mice, and the Sholl analyses of basal and apical dendrites revealed remarkable modifications (Fig. 9B). Three-way ANOVA of the number of intersections in basal dendrites revealed significant main effects of genotype (F1,7 = 19.159, p-value = 3 × 10−3), distance (F21,147 = 86.253, p-value ≈ 0), sex × distance (F21,147 = 1,663, p-value = 4.3 × 10−2), genotype × distance (F21,147 = 6.850, p-value = 2.14 × 10−13), but no sex × genotype × distance interaction (F21,147 = 0.425, p-value = 9.87 × 10−1). These data indicate a main effect of genotype, in which Emx1-Nr3c1−/− mice show lower neuronal complexity, irrespective of sex. The effect of genotype × distance could be easily observed; compared to Emx1-Nr3c1−/− mice, female and male controls exhibited a greater number of intersections at a lesser radial distance in basal dendrites (Fig. 9B). Similar analysis with apical dendrites showed no effect of sex, but significant main effects of genotype (F1,7 = 21.805, p-value = 2 × 10−3), distance (F34,238 = 24.591, p-value = 5.73 × 10−60), sex × distance (F34,238 = 2.243, p-value = 2.39 × 10−4), genotype × distance (F34,238 = 3.881, p-value = 3.29 × 10−10) and sex × genotype × distance interaction (F34,238 = 1.864, p-value = 4 × 10−3). These effects are displayed in distal dendrites as a lower number of intersections in female and male Emx1-Nr3c1−/− mice, compared to their respective controls. Additionally, at higher radii from the soma (over 200 μm), female controls showed increased complexity, in contrast to both male control and female Emx1-Nr3c1−/− mice (Fig. 9B). The variations found in the Sholl analysis may represent effects on dendritic length and/or the number of ramifications from dendrites (i.e., branching points). Two-way ANOVA test of basal dendritic length (Fig. 9C) revealed an effect of genotype (F1,28 = 11.36, p = 0.0022), but no effect of sex (F1,28 = 3.712, p-value = 0.0642), nor sex × genotype interaction (F1,28 = 1.447, p-value = 0.2391). Post-hoc analysis indicated that the Emx1-directed GR ablation triggers a significant reduction in the length of basal dendrites in males, compared to controls (Fig. 9C). Moreover, we did not observe any effect of sex (F1,28 = 3.285, p-value = 0.0807), genotype (F1,28 = 1.15, p-value = 0.2926), or sex × genotype interaction (F1,28 = 0.2312, p = 0.6344) in the number of branching points of basal dendrites (Fig. 9D). In contrast, we only found an effect of genotype (F1,28 = 21.79, p-value < 0.0001) on the length of apical dendrites (Fig. 9E) and the number of branching points of apical dendrites (F1,28 = 22.31, p-value < 0.0001) (Fig. 9F). The post-hoc test indicated that Emx1-Nr3c1−/− mice showed a significant reduction in the length and branching points of apical dendrites compared to controls, irrespective of sex. Altogether, these results indicate that the Emx1-directed GR deletion only generates a sex-biased length reduction in basal dendrites (males, but not females), but evokes both a reduction in branching points and dendritic length in apical dendrites of the hippocampus, irrespective of sex.
Fig. 9.
Emx1-Nr3c1−/−CA1 neurons show a reduction in apical arborization. (A) Representative images and traces from Golgi-labeled pyramidal neurons of the CA1 for each sex and genotype. Red and blue color traces represent basal and apical dendrites, respectively. Neuron reconstruction and analysis were done by the neuTube software and Fiji plugins. (B) Schematic representation of morphometric Sholl analysis. The number of intersections between dendrites (basal and apical) and concentric spheres centered on the soma was determined at various distances from the soma (10 μm increments). Data were analyzed by three-way ANOVA test. (C) Dendritic length and (D) number of branching points of basal dendrites; (E) dendritic length and (F) number of branching points of apical dendrites. Data were analyzed by the two-way ANOVA test with Tukey's post hoc test: * p-value < 0.05, ** p-value < 0.01 (Panel C: main effect of genotype F1,28 = 11.36, p-value = 0.0022. Panel D: no significant effects. Panel E: main effect of genotype F1,28 = 21.79, p-value < 0.0001. Panel F: main effect of genotype F1,28 = 22.31, p-value < 0.0001). n = 8 neurons from 3 mice were analyzed for each condition. Graph values represent mean ± SEM. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
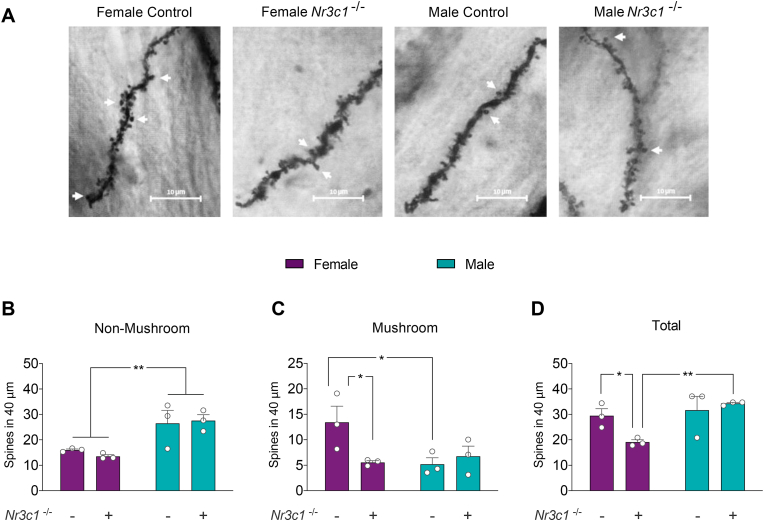
3.11. GR ablation induces a sex-biased effect on CA1 dendritic spine density
Considering that Emx1-directed GR ablation triggers profound alterations in dendritic complexity in a sex-biased manner, we next analyzed whether GR deletion alters the dendritic spine number of secondary apical dendrites from CA1 pyramidal neurons. Fig. 10A shows a secondary dendrite segment used to count the number of spines along a 40 μm distance. In order to refine the dendritic spine analysis, we segregated the spines according to their morphology. The total number of non-mushroom spines counted along 40 μm was analyzed by the two-way ANOVA test and revealed a significant main effect of sex (F1,15 = 5.666, p-value < 0.03), but not of genotype (F1,15 = 1.67, p-value < 0.3). Post-hoc analysis indicated that spine density in female controls was reduced in CA1 dendrites, compared to male controls (p-value = 0.0321). A similar effect was observed when comparing female Emx1-Nr3c1−/− and male Emx1-Nr3c1−/− mice (p-value < 0.0083) (Fig. 10B). We observed no effect of genotype (F1,10 = 2.530, p-value = 0.15) or sex (F1,10 = 2.530, p-value = 0.11), but an effect of sex × genotype interaction (F1,10 = 2.530, p-value = 0.0445) on the number of mushroom spines. Post-hoc analysis revealed that CA1 dendrites from female controls have a higher density of mushroom spines than male controls (p-value < 0.05). Additionally, CA1 dendrites from female Emx1-Nr3c1−/− mice showed a significant reduction in the number of mushroom spines compared to female controls. No similar effect of Emx1-Nr3c1−/− on mature spines was observed in male hippocampi (Fig. 10C). The analysis of the total number of spines along the 40 μm segment revealed no effect of genotype (F1,10 = 0.2458, p-value = 0.2458), but a significant effect of sex (F1,15 = 0.0225, p-value < 0.0225), with no interaction between both factors (F1,10 = 4.448, p-value = 0.068). Post-hoc analysis indicated that the total spine number was significantly reduced in female Emx1-Nr3c1-/-mice, compared to both female controls (p-value < 0.05) and male Emx1-Nr3c1−/− mice (p-value < 0.01) (Fig. 10D).
Fig. 10.
Sex-biased effect on spine morphology and density in CA1 neurons of Emx1-Nr3c1−/−mice. (A) Representative microphotographs of apical secondary dendrites of the hippocampal CA1 region identified by Golgi Stain in female and male controls and Emx1-Nr3c1−/−mice. A total of six dendrites were analyzed in each animal. The white arrows represent mushroom-shaped spines; scale bar represents 10 μm. (B) Non-mushroom spine density represents the number of immature spines along 40 μm (sex: F1,8 = 7.952, p-value = 0.0225). (C) Mushroom spines correspond to mature spines with a head larger than 0.6 μm (sex × genotype: F1,8 = 5.668, p-value = 0.0445). (D) Total spine density corresponds to the sum of immature and mature spines in each experimental condition (sex: F1,8 = 7.952, p-value = 0.0225; sex × genotype: F1,8 = 4.448, p-value = 0.0680). Bar graphs (purple denotes females, and teal, males) represent the mean ± SEM. Female controls (n = 3), male controls (n = 3), female Emx1-Nr3c1−/− (n = 3) and male Emx1-Nr3c1−/− (n = 3). Data were analyzed by two way ANOVA test followed by Fischer's post-hoc test: * p-value < 0.05, ** p-value < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Altogether, the morphological analysis of CA1 neurons from Emx1-Nr3c1−/− mice indicated a reduction in the length in both basal and apical dendrites in males, compared to controls. In contrast, a reduction in the length and branching of apical dendrites was observed in female knockout. Additionally, we observed a sex-biased effect of Emx1-Nr3c1−/− on spine morphology and density; i.e., female mice showed fewer mushroom spines.
4. Discussion
For the first time, here we report the impact of Emx1-directed ablation of mice forebrain GRs on sex differences in miRNAs and hippocampal CA1 neurons morphology. Our main finding is that females showed a higher sensitivity to hippocampal GR ablation in terms of the number of affected miRNAs and changes in dendritic spine density and morphology than males.
An exciting approach to dissect the action of the GR in the brain has been through the genetic disruption of GR in prefrontal cortex, amygdala, pituitary, paraventricular nucleus of the hypothalamus and even in the forebrain (Boyle et al., 2006; Solomon et al., 2012; Arango-Lievano and Jeanneteau, 2016; Scheimann et al., 2019). Considering the ever-rising discovery of sex differences in both nervous system physiology and pathology, there is a need for more studies addressing sex biases of GR actions. For instance, the selective deletion of GR in forebrain cortico-limbic areas induces depression-like behaviors in male but not female mice (Solomon et al., 2012). Moreover, a higher basal and stress-induced corticosterone levels in female than male counterparts has been extensively reported, evidencing differences in HPA axis regulation (Fernandez-Guasti et al., 2012). Notably, the GR in the hippocampus plays a pivotal role in the negative control of the HPA axis response to stress (Franklin et al., 2012). However, the molecular cues by which these sex differences may arise, specially in the hippocampus, have been less studied. Here, we have used Emx1-directed Cre expression, which is active at embryonic day 9.5 (E9.5) (Simeone et al., 1992) and ensues GR ablation of most progenitors early in development (Li et al., 2011). Moreover, Emx1 expression is restricted to pyramidal neurons, both in the cortex (Chan et al., 2001) and the hippocampus (Gorski et al., 2002).
Some studies have shown that the GR is abundantly expressed in the CA1 field and hippocampal dentate gyrus of rodents (Morimoto et al., 1996). Interestingly, the distribution of GR in the hippocampus is dimorphic; i.e., males displayed higher levels in CA1 hippocampal strata (Kitraki et al., 2004) with a nuclear localization under resting conditions compared to females (Brkic et al., 2017). However, we detected similar levels of whole hippocampal GR in females and male controls by Western blot. This discrepancy may be related to the lack of subcellular and hippocampal strata evaluation of our approach. As expected, Emx1-directed ablation of GR reduced both the mRNAs and protein levels of GR to a similar extent in both sexes. On the other hand, it has been described that males and females do not show differences in basal MR immunoreactive hippocampal cells (Kitraki et al., 2004). Nonetheless, we evaluated the impact of GR ablation on hippocampal MR expression, observing a trend to increase in females but not males, while MR transcript levels remained unchanged. These findings may suggest a repressive action of GR on MR expression in the female hippocampus.
GCs exert genomics and non-genomic mechanisms through the GR and the MR, which may influence brain structure and function (Gray et al., 2017). Compelling evidences have shown that part of the GR effects are mediated by controlling the expression of miRNAs in human lymphoid cell lines (Smith et al., 2010, 2013) and bone marrow–derived cells (Li et al., 2014; Laxman et al., 2016). This novel way in which GR can regulate gene expression further opens the spectrum of actors that may be influenced by this receptor, especially in our area of interest: the hippocampus. In mammals, more than 50% of all described miRNAs are expressed in the brain with uneven distribution across different areas, suggesting region-specific functions (Olsen et al., 2009). Conditional neuronal ablation of Dicer -a key enzyme in miRNA maturation- at different embryonic stages causes abnormal hippocampal morphology by both reducing the number of hippocampal progenitors and increasing apoptosis (Li et al., 2011); indicating a pivotal role of miRNAs in neuronal renewal. These evidences led us to evaluate GR contributions to hippocampal miRNA expression.
In the present study we employed the NanoString technology to conduct a comprehensive analysis of miRNA expression in hippocampal tissue. The main advantage of this approach is the direct measurement of miRNAs abundance (Prokopec et al., 2013; Veldman-Jones et al., 2015), that does not require a further validation by another method (Chatterjee et al., 2015). Furthermore, it has a demonstrated high technical reproducibility in both experimental and technical replicates (Knutsen et al., 2013; Kolbert et al., 2013) with high correlation, similar to the RT-qPCR technique (Knutsen et al., 2013). With this analysis, we identified differences in both the identity of the detected miRNAs and the amount of quantitated miRNAs. Between control animals, only 5% of the total examined miRNAs showed a different expression between sexes. Nonetheless, this pattern changed dramatically in Emx1-Nr3c1−/− mice vs controls, showing a sex-biased miRNA profile, with a higher number of dysregulated miRNAs in the female hippocampus than male (52 miRNAs vs 8 miRNAs, respectively). Similar to our results, a report has indicated that miRNA expression in the male hippocampus is less responsive to certain stimuli, e.g., radiation (Koturbash et al., 2011). Besides, a higher impact of GR deletion in females has been reported in other peripheral tissues, such as the heart (Cruz-Topete et al., 2019), suggesting that the sex-biased effects of GR ablation may be replicated in the brain and, specifically in the hippocampus.
Interestingly, a sex-dependent change in miRNA expression in the Bed Nucleus of the Stria Terminalis has been observed following stress (Mavrikaki et al., 2019). Although few studies have shown that brain miRNA expression is sex-biased (Morgan and Bale, 2012; Cui et al., 2018), these have proposed a combination of both sex hormones and sex chromosomal regulation to explain this phenomenon (Morgan and Bale, 2012). Besides these actors, our study provides evidence that GR signaling also contributes to the sex-biased miRNA expression in the hippocampus, that seems to be dissociated from miRNA gene expression on sex chromosomes. Surprisingly, mmu-miR-1224 was the only common differentially expressed miRNA in both sexes induced by GR ablation, but its expression changed in opposite directions. This miRNA acts as a tumor suppressor by targeting Creb1 in malignant gliomas (Qian et al., 2015) and is upregulated in the rat cortex after cerebral ischemia (Hunsberger et al., 2012). Nonetheless, how GR deletion produces this opposite effect in male and female mice remains to be determined.
Notably, in the female Emx1-Nr3c1−/− hippocampus, most miRNAs tended to be up-regulated, in contrast to the down-regulated tendency in males, indicating that the GR may have a negative or positive action on miRNA expression, respectively. Whether this effect is either a result of direct or indirect actions of GR ablation is unknown. At least in lymphocytes, GR activation reduces miRNA levels by repressing key miRNA processing enzymes: Dicer, Drosha, and DGCR8/Pasha (Smith et al., 2010). However, until now, there is no information indicating a sex-biased effect of GR on miRNA machinery. Other possibilities that may explain the sex-biased actions of GCs is the involvement of several co-chaperones that promote or inhibit nuclear GR translocation, which are regulated in a sex-specific way (Bourke et al., 2012). Also, estrogen receptors may interfere with GR actions; an effect described in uterine epithelial cells from human and murine models (Whirledge and Cidlowski, 2013).
Notably, it has been reported that sex-biased miRNAs are associated to clustering expression (Cui et al., 2018). Our study also indicated a sex-biased expression of different clusters; with the miR-200b/200a/429 and miR-23a/27a/24-2 clusters up-regulated in females, and the miR-143/145 cluster down-regulated in males. The molecular actors that may explain the sex-biased miRNA expression -either individually or clustered- could be transcriptional regulators. Firstly, we found only a few differentially expressed miRNA genes hosting a putative GR-binding site in females; while none could be predicted in the male dataset. Remarkably, several transcriptional interactions were predicted to control the differentially expressed miRNA by GR ablation in a sex-biased manner. For instance, various miRNAs may be sensitive to Smad family members in females and, in some non-neuronal tissues, GR may reduce Smad3-controlled gene transcription by protein-protein interactions (Petta et al., 2016). Considering that the GR may interact with different co-activators, enzymes, receptors and also non-coding RNAs (Petta et al., 2016), unveiling how this receptor mediates sex-biased effects on miRNA expression constitutes a big challenge and will allow a better understanding of the complex actions of the GR in the hippocampus.
Regarding the potential downstream effects of GR-sensitive miRNA expression, there is an extensive association between these non-coding RNAs and essential brain processes, such as neurodevelopment, synaptic plasticity, and neuronal activity (Pak et al., 2013; Cui et al., 2018). The TAM 2.0 analysis unveiled significantly enriched functional terms related to CNS functioning and disease in both sexes. Indeed, the differentially expressed miRNAs may be involved in some brain pathologies, such as Alzheimer's disease observed in the female dataset and temporal epilepsy in the male dataset. Of note, in both sexes, we detected terms relevant for hippocampal neurogenesis (e.g., cell differentiation/proliferation), in which GCs have a pivotal role (Odaka et al., 2017). Furthermore, the combined analyses of predicted and validated direct miRNA-mRNA interactions suggests an impact on several targets involved in morphological neuroplasticity, including dendritogenesis and dendritic maturation. Particulary, the former involves growth and extension of primary dendritic shafts, while the latter comprises branching, elongation, spine formation, and pruning (Scott and Luo, 2001). In acute hippocampal slices, a report showed that GR is locally synthesized in dendritic spines and participates in promoting actin polymerization, probably through RhoA/ROCK signaling, which may impact both spine morphology and number (Jafari et al., 2012). Whether these processes may be influenced by GR actions, from a sex-biased perspective, has not been addressed so far.
To conduct a more refined downstream functional analysis, we curated gene-dendritic morphology associations based on literature (Koleske, 2013; Forrest et al., 2018), further confirming the putative involvement of hippocampal transcripts -targeted by differentially expressed miRNA- with neuronal morphology and glutamatergic synapses. We highlight Bdnf and its receptor Ntrk2, Rac1, Rhoa and Rock, all which participate in the maintenance of dendritic spines and dendritic arborization of hippocampal pyramidal neurons (Nakayama et al., 2000; Kowianski et al., 2018). Other overrepresented targets in both sexes were Camk2a and Gsk3b, both crucial serine/threonine kinases associated with structural and functional aspects of synaptic plasticity, including the modulation of dendritic branching (Liu and Murray, 2012) and spine density (Jaworski et al., 2009). Remarkably, we observed a rise in CaMKII-α and GSK-3β expression only in the female hippocampus induced by GR ablation, suggesting that these kinases may be negatively controlled by GR. This regulation may arise by direct GR modulation of gene expression and/or through an indirect mechanism involving a miRNAs. In silico studies (TRRUST and ENCODE) revealed no GR response elements in the promoter of the Gsk3b gene (W. Corrales, Personal Communication, March 2020), in agreement with a report showing that GSK-3β levels are independent of GR activity (Yu et al., 2019). These findings highlight a potential regulation by miRNAs.
Regarding dendritic complexity analysis, we found that adult males exhibited more complex dendritic arbors in CA1 neurons than female controls, in compliance with other studies in juvenile mice (Keil et al., 2017). Sholl analyses in Emx1-Nr3c1−/− mice evidenced a reduction in apical dendritic complexity and length in both sexes. Conversely, GR ablation induced a reduction in basal dendritic length only in males, suggesting a peculiar sex-biased impact of GR on the growth of these dendrites. These alterations may be related to GR-responsive cellular processes, differentially impacting dendritic arborization and length of CA1 hippocampal neurons.
Another fundamental process for the development, maintenance, and plasticity of neuronal circuits, during both neurodevelopment and adulthood, is dendritic spine remodeling (Forrest et al., 2018). We found that total dendritic spine density in secondary dendrites of CA1 neurons was similar in control mice irrespective of sex. However, females showed a higher mature spine density, while males displayed a higher number of non-mushroom spines. These observations suggest that synaptic turn-over and cytoskeleton dynamics may be influenced by sex. Additionally, GR ablation reduced the density of mushroom spines only in the female hippocampus, with no alteration in non-mushroom spines density. Notably, male dendritic spine density and morphology were not affected by GR ablation. Compelling evidences have noted the involvement of GR in dendritic spine dynamics. For instance, an elegant experiment indicated that MR and GR activation during circadian GC oscillations are required for synapse formation and pruning after experience-dependent learning (Liston et al., 2013; Hall et al., 2015). Importantly, another report in mice cortical neurons showed that GR silencing by a specific shRNA reduces spine density, but increases spine size in apical dendrites (Arango-Lievano et al., 2015). Despite no studies have stressed the effect of sex in GR-mediated spine density and shape, our results suggest that males are less responsive to GR influences on this matter than females. This idea, in addition to the magnitude of miRNAs variations, may account for a higher sensitivity or responsiveness to GCs in females, potentially explaining the sex-differential consequences of HPA dysregulation in some pathologies.
In a wider context, the hippocampus and GCs are of particular relevance for the stress response. In fact, either in acute or chronic stress, different transcriptional signatures exist between the female and male brain (Brivio et al., 2020). For instance, a sex-dependent change in miRNA expression in the Bed Nucleus of the Stria Terminalis has been observed following chronic stress (Mavrikaki et al., 2019); however, this study did not explore the contribution of GR to this effects. Besides, many reports have described a variation in miRNAome after chronic stress in the hippocampus of male rodent (Meerson et al., 2010; Castaneda et al., 2015; Munoz-Llanos et al., 2018) but it is urgent to dissect whether these variations are related to corticosteroid receptors and if they are replicable in females. Besides, since stress and the GR may impact RNA subpopulations other than miRNAs, further studies addressing GR ablation effects in a more comprehensive transcriptomic scenario may reveal sex differences at diverse transcriptional levels.
Finally, a current limitation of our model is the lack of a developmental perspective. Since the morphological variations could be installed early during development or occur postnatally in later stages, the GR deletion during these critical periods should be considered. Moreover, we do not know whether GR ablation in cortical neurons may account for part of the observed variations, or whether our results are replicated across other brain areas. Due to circuitry-level, direct or indirect connections between cortical areas and the hippocampus, we cannot conclude whether the molecular and morphological effects of Emx1-directed GR ablation are solely caused by hippocampal GR ablation. Both neuronal miRNAs and dendrite morphology are highly activity-regulated across the brain. Therefore, Emx1-directed GR ablation may cause a complex circuit-level alteration that may impact, directly or indirectly, the hippocampus. Another important issue to consider is the contribution of MR to miRNA expression and neuronal morphology in a context of GR ablation. Thus, future comprehensive studies are needed to shape the molecular basis underlying the sex differences in GR-mediated actions, specially in physiological and pathological scenarios.
5. Conclusion
Altogether, we have found sex-biased changes in the hippocampus in response to GR ablation. Particularly, a higher sensitivity on miRNAs levels, key dendritic remodeling protein levels and dendritic morphology at the hippocampus was observed in female Emx1-Nr3c1−/− mice. Our study provides an important preliminary roadmap for further analysis of GR-mediated transcriptomic signatures involved in sex differences on the brain.
Data Availability disclosure statement
The authors declare that all supporting data and method descriptions are available within the article, or from the corresponding author, upon reasonable request.
Research organism
Mice.
Funding Information
This work was supported by the Intramural Research Program of the NIEHS, National Institutes of Health (JC) and FONDECYT 1190899 (JLF).
CRediT authorship contribution statement
Macarena Tejos-Bravo: Conceptualization, Methodology, Investigation, Writing - original draft, Writing - review & editing. Robert H. Oakley: Conceptualization, Methodology, Writing - review & editing. Shannon D. Whirledge: Conceptualization, Methodology, Investigation, Writing - review & editing. Wladimir A. Corrales: Methodology, Software, Data curation, Writing - original draft, Writing - review & editing. Juan P. Silva: Methodology, Software, Data curation, Writing - original draft, Writing - review & editing. Gonzalo García-Rojo: Methodology, Validation, Writing - original draft. Jorge Toledo: Methodology, Validation, Writing - original draft, Writing - review & editing. Wendy Sanchez: Methodology, Validation, Writing - original draft. Luciano Román-Albasini: Investigation, Formal analysis, Writing - original draft, Writing - review & editing. Esteban Aliaga: Validation, Writing - review & editing. Felipe Aguayo: Validation, Writing - review & editing. Felipe Olave: Validation, Writing - review & editing. Vinicius Maracaja-Coutinho: Supervision, Writing - review & editing. John A. Cidlowski: Resources, Writing - review & editing. Jenny L. Fiedler: Conceptualization, Methodology, Validation, Resources, Writing - review & editing, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that no competing interests exist.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ynstr.2021.100306.
Contributor Information
Vinicius Maracaja-Coutinho, Email: vinicius.maracaja@uchile.cl.
John A. Cidlowski, Email: cidlows1@niehs.nih.gov.
Jenny L. Fiedler, Email: jfiedler@ciq.uchile.cl.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Aguayo F.I., Tejos-Bravo M., Diaz-Veliz G., Pacheco A., Garcia-Rojo G., Corrales W., Olave F.A., Aliaga E., Ulloa J.L., Avalos A.M., Roman-Albasini L., Rojas P.S., Fiedler J.L. Hippocampal memory recovery after acute stress: a behavioral, morphological and molecular study. Front. Mol. Neurosci. 2018;11:283. doi: 10.3389/fnmol.2018.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arango-Lievano M., Jeanneteau F. Timing and crosstalk of glucocorticoid signaling with cytokines, neurotransmitters and growth factors. Pharmacol. Res. 2016;113:1–17. doi: 10.1016/j.phrs.2016.08.005. [DOI] [PubMed] [Google Scholar]
- Arango-Lievano M., Lambert W.M., Bath K.G., Garabedian M.J., Chao M.V., Jeanneteau F. 2015. Neurotrophic-priming of Glucocorticoid Receptor Signaling Is Essential for Neuronal Plasticity to Stress and Antidepressant Treatment. 201509045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourke C.H., Harrell C.S., Neigh G.N. Stress-induced sex differences: adaptations mediated by the glucocorticoid receptor. Horm. Behav. 2012;62:210–218. doi: 10.1016/j.yhbeh.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle M.P., Kolber B.J., Vogt S.K., Wozniak D.F., Muglia L.J. Forebrain glucocorticoid receptors modulate anxiety-associated locomotor activation and adrenal responsiveness. J. Neurosci. : the official journal of the Society for Neuroscience. 2006;26:1971–1978. doi: 10.1523/JNEUROSCI.2173-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brivio E., Lopez J.P., Chen A. Sex differences: transcriptional signatures of stress exposure in male and female brains. Gene Brain Behav. 2020;19 doi: 10.1111/gbb.12643. [DOI] [PubMed] [Google Scholar]
- Brkic Z., Francija E., Petrovic Z., Franic D., Lukic I., Mitic M., Adzic M. Distinct modifications of hippocampal glucocorticoid receptor phosphorylation and FKBPs by lipopolysaccharide in depressive female and male rats. J. Psychopharmacol. 2017;31:1234–1249. doi: 10.1177/0269881117725914. [DOI] [PubMed] [Google Scholar]
- Castaneda P., Munoz M., Garcia-Rojo G., Ulloa J.L., Bravo J.A., Marquez R., Garcia-Perez M.A., Arancibia D., Araneda K., Rojas P.S., Mondaca-Ruff D., Diaz-Veliz G., Mora S., Aliaga E., Fiedler J.L. Association of N-cadherin levels and downstream effectors of Rho GTPases with dendritic spine loss induced by chronic stress in rat hippocampal neurons. J. Neurosci. Res. 2015;93:1476–1491. doi: 10.1002/jnr.23602. [DOI] [PubMed] [Google Scholar]
- Cruz-Topete D., Oakley R.H., Carroll N.G., He B., Myers P.H., Xu X., Watts M.N., Trosclair K., Glasscock E., Dominic P., Cidlowski J.A. Deletion of the cardiomyocyte glucocorticoid receptor leads to sexually dimorphic changes in cardiac gene expression and progression to heart failure. J Am Heart Assoc. 2019;8 doi: 10.1161/JAHA.118.011012. e011012-e011012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C., Yang W., Shi J., Zhou Y., Yang J., Cui Q. Identification and analysis of human sex-biased MicroRNAs. Dev. Reprod. Biol. 2018;16:200–211. doi: 10.1016/j.gpb.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.H., Godinho L.N., Thomaidou D., Tan S.S., Gulisano M., Parnavelas J.G. Emx1 is a marker for pyramidal neurons of the cerebral cortex. Cerebr. Cortex. 2001;11:1191–1198. doi: 10.1093/cercor/11.12.1191. [DOI] [PubMed] [Google Scholar]
- Chatterjee A., Leichter A.L., Fan V., Tsai P., Purcell R.V., Sullivan M.J., Eccles M.R. A cross comparison of technologies for the detection of microRNAs in clinical FFPE samples of hepatoblastoma patients. Sci. Rep. 2015;5:10438. doi: 10.1038/srep10438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48:D127–D131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kloet E.R., Meijer O.C., de Nicola A.F., de Rijk R.H., Joels M. Importance of the brain corticosteroid receptor balance in metaplasticity, cognitive performance and neuro-inflammation. Front. Neuroendocrinol. 2018;49:124–145. doi: 10.1016/j.yfrne.2018.02.003. [DOI] [PubMed] [Google Scholar]
- Duma D., Collins J.B., Chou J.W., Cidlowski J.A. Sexually dimorphic actions of glucocorticoids provide a link to inflammatory diseases with gender differences in prevalence. Sci. Signal. 2010;3 doi: 10.1126/scisignal.2001077. ra74-ra74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Siklenka K., Arora S.K., Ribeiro P., Kimmins S., Xia J. miRNet - dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016;44:W135–W141. doi: 10.1093/nar/gkw288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L., Zhao T., Kim J. 2015. neuTube 1.0: A New Design for Efficient Neuron Reconstruction Software Based on the SWC Format. eNeuro 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Guasti A., Fiedler J.L., Herrera L., Handa R.J. Sex, stress, and mood disorders: at the intersection of adrenal and gonadal hormones. Hormone and metabolic research = Hormon- und Stoffwechselforschung = Hormones et metabolisme. 2012;44:607–618. doi: 10.1055/s-0032-1312592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira T., Ou Y., Li S., Giniger E., van Meyel D.J. Dendrite architecture organized by transcriptional control of the F-actin nucleator Spire. Development. 2014;141:650–660. doi: 10.1242/dev.099655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornes O., Castro-Mondragon J.A., Khan A., van der Lee R., Zhang X., Richmond P.A., Modi B.P., Correard S., Gheorghe M., Baranasic D., Santana-Garcia W., Tan G., Cheneby J., Ballester B., Parcy F., Sandelin A., Lenhard B., Wasserman W.W., Mathelier A. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020;48:D87–D92. doi: 10.1093/nar/gkz1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest M.P., Parnell E., Penzes P. Dendritic structural plasticity and neuropsychiatric disease. Nat. Rev. Neurosci. 2018;19:215–234. doi: 10.1038/nrn.2018.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin T.B., Saab B.J., Mansuy I.M. Neural mechanisms of stress resilience and vulnerability. Neuron. 2012;75:747–761. doi: 10.1016/j.neuron.2012.08.016. [DOI] [PubMed] [Google Scholar]
- Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rojo G., Fresno C., Vilches N., Diaz-Veliz G., Mora S., Aguayo F., Pacheco A., Parra-Fiedler N., Parra C.S., Rojas P.S., Tejos M., Aliaga E., Fiedler J.L. The ROCK inhibitor fasudil prevents chronic restraint stress-induced depressive-like behaviors and dendritic spine loss in rat Hippocampus. Int. J. Neuropsychopharmacol. 2017;20:336–345. doi: 10.1093/ijnp/pyw108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Sanchez C.E., de Rodriguez A.F., Romero D.G., Estess J., Warden M.P., Gomez-Sanchez M.T., Gomez-Sanchez E.P. Development of a panel of monoclonal antibodies against the mineralocorticoid receptor. Endocrinology. 2006;147:1343–1348. doi: 10.1210/en.2005-0860. [DOI] [PubMed] [Google Scholar]
- Gorski J.A., Talley T., Qiu M., Puelles L., Rubenstein J.L., Jones K.R. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J. Neurosci. : the official journal of the Society for Neuroscience. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant C.E., Bailey T.L., Noble W.S. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J.D., Kogan J.F., Marrocco J., McEwen B.S. Genomic and epigenomic mechanisms of glucocorticoids in the brain. Nat. Rev. Endocrinol. 2017;13:661–673. doi: 10.1038/nrendo.2017.97. [DOI] [PubMed] [Google Scholar]
- Gu Z., Gu L., Eils R., Schlesner M., Brors B. Circlize Implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- Guo H., Hong S., Jin X.L., Chen R.S., Avasthi P.P., Tu Y.T., Ivanco T.L., Li Y. Specificity and efficiency of Cre-mediated recombination in Emx1-Cre knock-in mice. Biochem. Biophys. Res. Commun. 2000;273:661–665. doi: 10.1006/bbrc.2000.2870. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Mecinas M., Trollope A.F., Collins A., Morfett H., Hesketh S.A., Kersante F., Reul J.M. Long-lasting behavioral responses to stress involve a direct interaction of glucocorticoid receptors with ERK1/2-MSK1-Elk-1 signaling. Proc. Natl. Acad. Sci. U. S. A. 2011;108:13806–13811. doi: 10.1073/pnas.1104383108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B.S., Moda R.N., Liston C. Glucocorticoid mechanisms of functional connectivity changes in stress-related neuropsychiatric disorders. Neurobiology of stress. 2015;1:174–183. doi: 10.1016/j.ynstr.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S.D., Lin F.M., Wu W.Y., Liang C., Huang W.C., Chan W.L., Tsai W.T., Chen G.Z., Lee C.J., Chiu C.M., Chien C.H., Wu M.C., Huang C.Y., Tsou A.P., Huang H.D. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011;39:D163–D169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunsberger J.G., Fessler E.B., Wang Z., Elkahloun A.G., Chuang D.M. Post-insult valproic acid-regulated microRNAs: potential targets for cerebral ischemia. Am. J. Tourism Res. 2012;4:316–332. [PMC free article] [PubMed] [Google Scholar]
- Jafari M., Seese R.R., Babayan A.H., Gall C.M., Lauterborn J.C. Glucocorticoid receptors are localized to dendritic spines and influence local actin signaling. Mol. Neurobiol. 2012;46:304–315. doi: 10.1007/s12035-012-8288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski J., Kapitein L.C., Gouveia S.M., Dortland B.R., Wulf P.S., Grigoriev I., Camera P., Spangler S.A., Di Stefano P., Demmers J., Krugers H., Defilippi P., Akhmanova A., Hoogenraad C.C. Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron. 2009;61:85–100. doi: 10.1016/j.neuron.2008.11.013. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagkouni D., Paraskevopoulou M.D., Chatzopoulos S., Vlachos I.S., Tastsoglou S., Kanellos I., Papadimitriou D., Kavakiotis I., Maniou S., Skoufos G., Vergoulis T., Dalamagas T., Hatzigeorgiou A.G. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018;46:D239–D245. doi: 10.1093/nar/gkx1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassambara A. Rstatix: pipe-friendly framework for basic statistical tests. R package version 0.4.0. 2020. https://CRANR-projectorg/package=rstatix
- Keil K.P., Sethi S., Wilson M.D., Chen H., Lein P.J. In vivo and in vitro sex differences in the dendritic morphology of developing murine hippocampal and cortical neurons. Sci. Rep. 2017;7:8486. doi: 10.1038/s41598-017-08459-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitraki E., Kremmyda O., Youlatos D., Alexis M.N., Kittas C. Gender-dependent alterations in corticosteroid receptor status and spatial performance following 21 days of restraint stress. Neuroscience. 2004;125:47–55. doi: 10.1016/j.neuroscience.2003.12.024. [DOI] [PubMed] [Google Scholar]
- Knutsen E., Fiskaa T., Ursvik A., Jorgensen T.E., Perander M., Lund E., Seternes O.M., Johansen S.D., Andreassen M. Performance comparison of digital microRNA profiling technologies applied on human breast cancer cell lines. PloS One. 2013;8 doi: 10.1371/journal.pone.0075813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbert C.P., Feddersen R.M., Rakhshan F., Grill D.E., Simon G., Middha S., Jang J.S., Simon V., Schultz D.A., Zschunke M., Lingle W., Carr J.M., Thompson E.A., Oberg A.L., Eckloff B.W., Wieben E.D., Li P., Yang P., Jen J. Multi-platform analysis of microRNA expression measurements in RNA from fresh frozen and FFPE tissues. PloS One. 2013;8 doi: 10.1371/journal.pone.0052517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koleske A.J. Molecular mechanisms of dendrite stability. Nat. Rev. Neurosci. 2013;14:536–550. doi: 10.1038/nrn3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopec A.M., Rivera P.D., Lacagnina M.J., Hanamsagar R., Bilbo S.D. Optimized solubilization of TRIzol-precipitated protein permits Western blotting analysis to maximize data available from brain tissue. J. Neurosci. Methods. 2017;280:64–76. doi: 10.1016/j.jneumeth.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koturbash I., Zemp F., Kolb B., Kovalchuk O. Sex-specific radiation-induced microRNAome responses in the hippocampus, cerebellum and frontal cortex in a mouse model. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2011;722:114–118. doi: 10.1016/j.mrgentox.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Kowianski P., Lietzau G., Czuba E., Waskow M., Steliga A., Morys J. BDNF: a key factor with multipotent impact on brain signaling and synaptic plasticity. Cell. Mol. Neurobiol. 2018;38:579–593. doi: 10.1007/s10571-017-0510-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., McDermott M.G., Monteiro C.D., Gundersen G.W., Ma'ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Zhang M.X., Swank M.W., Kunz J., Wu G.Y. Regulation of dendritic morphogenesis by Ras-PI3K-Akt-mTOR and Ras-MAPK signaling pathways. J. Neurosci. : the official journal of the Society for Neuroscience. 2005;25:11288–11299. doi: 10.1523/JNEUROSCI.2284-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxman N., Rubin C.J., Mallmin H., Nilsson O., Tellgren-Roth C., Kindmark A. Second generation sequencing of microRNA in human bone cells treated with parathyroid hormone or dexamethasone. Bone. 2016;84:181–188. doi: 10.1016/j.bone.2015.12.053. [DOI] [PubMed] [Google Scholar]
- Li J., Han X., Wan Y., Zhang S., Zhao Y., Fan R., Cui Q., Zhou Y. TAM 2.0: tool for MicroRNA set analysis. Nucleic Acids Res. 2018;46:W180–W185. doi: 10.1093/nar/gky509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Bian S., Hong J., Kawase-Koga Y., Zhu E., Zheng Y., Yang L., Sun T. Timing specific requirement of microRNA function is essential for embryonic and postnatal hippocampal development. PloS One. 2011;6 doi: 10.1371/journal.pone.0026000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Li H., Li T., Fan J., Zhao R.C., Weng X. MicroRNA expression profile of dexamethasone-induced human bone marrow-derived mesenchymal stem cells during osteogenic differentiation. J. Cell. Biochem. 2014;115:1683–1691. doi: 10.1002/jcb.24831. [DOI] [PubMed] [Google Scholar]
- Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Liston C., Cichon J.M., Jeanneteau F., Jia Z., Chao M.V., Gan W.B. Circadian glucocorticoid oscillations promote learning-dependent synapse formation and maintenance. Nat. Neurosci. 2013;16:698–705. doi: 10.1038/nn.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B.P., Cafferty W.B., Budel S.O., Strittmatter S.M. Extracellular regulators of axonal growth in the adult central nervous system. Phil. Trans. Roy. Soc. Lond. B Biol. Sci. 2006;361:1593–1610. doi: 10.1098/rstb.2006.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.B., Murray K.D. Neuronal excitability and calcium/calmodulin-dependent protein kinase type II: location, location, location. Epilepsia. 2012;53(Suppl. 1):45–52. doi: 10.1111/j.1528-1167.2012.03474.x. [DOI] [PubMed] [Google Scholar]
- Longair M.H., Baker D.A., Armstrong J.D. Simple Neurite Tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics. 2011;27:2453–2454. doi: 10.1093/bioinformatics/btr390. [DOI] [PubMed] [Google Scholar]
- Mavrikaki M., Pantano L., Potter D., Rogers-Grazado M.A., Anastasiadou E., Slack F.J., Amr S.S., Ressler K.J., Daskalakis N.P., Chartoff E. Sex-dependent changes in miRNA expression in the bed nucleus of the Stria Terminalis following stress. Front. Mol. Neurosci. 2019;12:236. doi: 10.3389/fnmol.2019.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen B.S., Nasca C., Gray J.D. Stress effects on neuronal structure: Hippocampus, amygdala, and prefrontal cortex. Neuropsychopharmacology. 2016;41:3–23. doi: 10.1038/npp.2015.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin C.N., Broihier H.T. Keeping neurons young and foxy: FoxOs promote neuronal plasticity. Trends Genet. : TIG (Trends Genet.) 2018;34:65–78. doi: 10.1016/j.tig.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerson A., Cacheaux L., Goosens K.A., Sapolsky R.M., Soreq H., Kaufer D. Changes in brain MicroRNAs contribute to cholinergic stress reactions. J. Mol. Neurosci. : M. 2010;40:47–55. doi: 10.1007/s12031-009-9252-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan C.P., Bale T.L. Sex differences in microRNA regulation of gene expression: no smoke, just miRs. Biol. Sex Differ. 2012;3:22. doi: 10.1186/2042-6410-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto M., Morita N., Ozawa H., Yokoyama K., Kawata M. Distribution of glucocorticoid receptor immunoreactivity and mRNA in the rat brain: an immunohistochemical and in situ hybridization study. Neurosci. Res. 1996;26:235–269. doi: 10.1016/s0168-0102(96)01105-4. [DOI] [PubMed] [Google Scholar]
- Munoz-Llanos M., Garcia-Perez M.A., Xu X., Tejos-Bravo M., Vidal E.A., Moyano T.C., Gutierrez R.A., Aguayo F.I., Pacheco A., Garcia-Rojo G., Aliaga E., Rojas P.S., Cidlowski J.A., Fiedler J.L. MicroRNA profiling and bioinformatics target analysis in dorsal Hippocampus of chronically stressed rats: relevance to depression pathophysiology. Front. Mol. Neurosci. 2018;11:251. doi: 10.3389/fnmol.2018.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzio L., Mallamaci A. Emx1, emx2 and pax 6 in specification, regionalization and arealization of the cerebral cortex. Cerebr. Cortex. 2003;13:641–647. doi: 10.1093/cercor/13.6.641. [DOI] [PubMed] [Google Scholar]
- Nakayama A.Y., Harms M.B., Luo L. Small GTPases Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J. Neurosci. : the official journal of the Society for Neuroscience. 2000;20:5329–5338. doi: 10.1523/JNEUROSCI.20-14-05329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoletopoulou V., Tavernarakis N. Regulation and roles of Autophagy at synapses. Trends Cell Biol. 2018;28:646–661. doi: 10.1016/j.tcb.2018.03.006. [DOI] [PubMed] [Google Scholar]
- Oakley R.H., Cruz-Topete D., He B., Foley J.F., Myers P.H., Xu X., Gomez-Sanchez C.E., Chambon P., Willis M.S., Cidlowski J.A. Cardiomyocyte glucocorticoid and mineralocorticoid receptors directly and antagonistically regulate heart disease in mice. Sci. Signal. 2019;12 doi: 10.1126/scisignal.aau9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley R.H., Ren R., Cruz-Topete D., Bird G.S., Myers P.H., Boyle M.C., Schneider M.D., Willis M.S., Cidlowski J.A. Essential role of stress hormone signaling in cardiomyocytes for the prevention of heart disease. Proc. Natl. Acad. Sci. U. S. A. 2013;110:17035–17040. doi: 10.1073/pnas.1302546110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odaka H., Adachi N., Numakawa T. Impact of glucocorticoid on neurogenesis. Neural regeneration research. 2017;12:1028–1035. doi: 10.4103/1673-5374.211174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olde Loohuis N.F., Kos A., Martens G.J., Van Bokhoven H., Nadif Kasri N., Aschrafi A. MicroRNA networks direct neuronal development and plasticity. Cell. Mol. Life Sci. : CM. 2012;69:89–102. doi: 10.1007/s00018-011-0788-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen L., Klausen M., Helboe L., Nielsen F.C., Werge T. MicroRNAs show mutually exclusive expression patterns in the brain of adult male rats. PloS One. 2009;4 doi: 10.1371/journal.pone.0007225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak T.R., Rao Y.S., Prins S.A., Mott N.N. An emerging role for microRNAs in sexually dimorphic neurobiological systems. Pflügers Archiv. 2013;465:655–667. doi: 10.1007/s00424-013-1227-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panettieri R.A., Schaafsma D., Amrani Y., Koziol-White C., Ostrom R., Tliba O. Non-genomic effects of glucocorticoids: an updated view. Trends Pharmacol. Sci. 2019;40:38–49. doi: 10.1016/j.tips.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual-Le Tallec L., Lombes M. The mineralocorticoid receptor: a journey exploring its diversity and specificity of action. Mol. Endocrinol. 2005;19:2211–2221. doi: 10.1210/me.2005-0089. [DOI] [PubMed] [Google Scholar]
- Petryszak R., Keays M., Tang Y.A., Fonseca N.A., Barrera E., Burdett T., Fullgrabe A., Fuentes A.M., Jupp S., Koskinen S., Mannion O., Huerta L., Megy K., Snow C., Williams E., Barzine M., Hastings E., Weisser H., Wright J., Jaiswal P., Huber W., Choudhary J., Parkinson H.E., Brazma A. Expression Atlas update--an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res. 2016;44:D746–D752. doi: 10.1093/nar/gkv1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petta I., Dejager L., Ballegeer M., Lievens S., Tavernier J., De Bosscher K., Libert C. The interactome of the glucocorticoid receptor and its influence on the actions of glucocorticoids in combatting inflammatory and infectious diseases. Microbiol. Mol. Biol. Rev. : MMBR (Microbiol. Mol. Biol. Rev.) 2016;80:495–522. doi: 10.1128/MMBR.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokopec S.D., Watson J.D., Waggott D.M., Smith A.B., Wu A.H., Okey A.B., Pohjanvirta R., Boutros P.C. Systematic evaluation of medium-throughput mRNA abundance platforms. RNA. 2013;19:51–62. doi: 10.1261/rna.034710.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J., Li R., Wang Y.Y., Shi Y., Luan W.K., Tao T., Zhang J.X., Xu Y.C., You Y.P. MiR-1224-5p acts as a tumor suppressor by targeting CREB1 in malignant gliomas. Mol. Cell. Biochem. 2015;403:33–41. doi: 10.1007/s11010-015-2334-1. [DOI] [PubMed] [Google Scholar]
- Quinn M., Ramamoorthy S., Cidlowski J.A. Sexually dimorphic actions of glucocorticoids: beyond chromosomes and sex hormones. Ann. N. Y. Acad. Sci. 2014;1317:1–6. doi: 10.1111/nyas.12425. [DOI] [PubMed] [Google Scholar]
- Ramamoorthy S., Cidlowski J.A. Corticosteroids: mechanisms of action in Health and disease. Rheum. Dis. Clin. N. Am. 2016;42:15–31. doi: 10.1016/j.rdc.2015.08.002. vii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo S.J., Mazei-Robison M.S., Ables J.L., Nestler E.J. Neurotrophic factors and structural plasticity in addiction. Neuropharmacology. 2009;56(Suppl. 1):73–82. doi: 10.1016/j.neuropharm.2008.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorcioni R., Polavaram S., Ascoli G.A. L-Measure: a web-accessible tool for the analysis, comparison and search of digital reconstructions of neuronal morphologies. Nat. Protoc. 2008;3:866–876. doi: 10.1038/nprot.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott E.K., Luo L. How do dendrites take their shape? Nat. Neurosci. 2001;4:359–365. doi: 10.1038/86006. [DOI] [PubMed] [Google Scholar]
- Scheimann J.R., Moloney R.D., Mahbod P., Morano R.L., Fitzgerald M., Hoskins O., Packard B.A., Cotella E.M., Hu Y.C., Herman J.P. Conditional deletion of glucocorticoid receptors in rat brain results in sex-specific deficits in fear and coping behaviors. eLife. 2019;8 doi: 10.7554/eLife.44672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Shi Y., Pontrello C.G., DeFea K.A., Reichardt L.F., Ethell I.M. Focal adhesion kinase acts downstream of EphB receptors to maintain mature dendritic spines by regulating cofilin activity. J. Neurosci. : the official journal of the Society for Neuroscience. 2009;29:8129–8142. doi: 10.1523/JNEUROSCI.4681-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sholl D.A. Dendritic organization in the neurons of the visual and motor cortices of the cat. J. Anat. 1953;87:387–406. [PMC free article] [PubMed] [Google Scholar]
- Simeone A., Gulisano M., Acampora D., Stornaiuolo A., Rambaldi M., Boncinelli E. Two vertebrate homeobox genes related to the Drosophila empty spiracles gene are expressed in the embryonic cerebral cortex. EMBO J. 1992;11:2541–2550. doi: 10.1002/j.1460-2075.1992.tb05319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L.K., Shah R.R., Cidlowski J.A. Glucocorticoids modulate MicroRNA expression and processing during lymphocyte apoptosis. J. Biol. Chem. 2010;285:36698–36708. doi: 10.1074/jbc.M110.162123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L.K., Tandon A., Shah R.R., Mav D., Scoltock A.B., Cidlowski J.A. Deep sequencing identification of novel glucocorticoid-responsive miRNAs in apoptotic primary lymphocytes. PloS One. 2013;8 doi: 10.1371/journal.pone.0078316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon M.B., Furay A.R., Jones K., Packard A.E., Packard B.A., Wulsin A.C., Herman J.P. Deletion of forebrain glucocorticoid receptors impairs neuroendocrine stress responses and induces depression-like behavior in males but not females. Neuroscience. 2012;203:135–143. doi: 10.1016/j.neuroscience.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sticht C., De La Torre C., Parveen A., Gretz N. miRWalk: an online resource for prediction of microRNA binding sites. PloS One. 2018;13 doi: 10.1371/journal.pone.0206239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Switon K., Kotulska K., Janusz-Kaminska A., Zmorzynska J., Jaworski J. Molecular neurobiology of mTOR. Neuroscience. 2017;341:112–153. doi: 10.1016/j.neuroscience.2016.11.017. [DOI] [PubMed] [Google Scholar]
- Uchoa E.T., Aguilera G., Herman J.P., Fiedler J.L., Deak T., de Sousa M.B. Novel aspects of glucocorticoid actions. J. Neuroendocrinol. 2014;26:557–572. doi: 10.1111/jne.12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman-Jones M.H., Brant R., Rooney C., Geh C., Emery H., Harbron C.G., Wappett M., Sharpe A., Dymond M., Barrett J.C., Harrington E.A., Marshall G. Evaluating robustness and sensitivity of the NanoString technologies nCounter platform to enable multiplexed gene expression analysis of clinical samples. Canc. Res. 2015;75:2587–2593. doi: 10.1158/0008-5472.CAN-15-0262. [DOI] [PubMed] [Google Scholar]
- Vreugdenhil E., Verissimo C.S., Mariman R., Kamphorst J.T., Barbosa J.S., Zweers T., Champagne D.L., Schouten T., Meijer O.C., de Kloet E.R., Fitzsimons C.P. MicroRNA 18 and 124a down-regulate the glucocorticoid receptor: implications for glucocorticoid responsiveness in the brain. Endocrinology. 2009;150:2220–2228. doi: 10.1210/en.2008-1335. [DOI] [PubMed] [Google Scholar]
- Wang J., Lu M., Qiu C., Cui Q. TransmiR: a transcription factor-microRNA regulation database. Nucleic Acids Res. 2010;38:D119–D122. doi: 10.1093/nar/gkp803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whirledge S., Cidlowski J.A. Estradiol antagonism of glucocorticoid-induced GILZ expression in human uterine epithelial cells and murine uterus. Endocrinology. 2013;154:499–510. doi: 10.1210/en.2012-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. ggplot2. WIREs Comp Stat. 2011;3:180–185. [Google Scholar]
- Wirth M.M. Hormones, stress, and cognition: the effects of glucocorticoids and oxytocin on memory. Adaptive human behavior and physiology. 2015;1:177–201. doi: 10.1007/s40750-014-0010-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Zuo Z., Cai G., Kang S., Gao X., Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105–D110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Guo Y., Zhao Y., Zhou F., Zhao K., Li M., Wen J., He Z., Zhu X., He X. Both insufficient and excessive glucocorticoid receptor-mediated signaling impair neuronal migration. J. Endocrinol. 2019;242:103–114. doi: 10.1530/JOE-19-0207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all supporting data and method descriptions are available within the article, or from the corresponding author, upon reasonable request.