Abstract
Mitochondrial abnormalities have been noted in lupus, but the causes and consequences remain obscure. Autophagy-related genes ATG5, ATG7, and IRGM have been previously implicated in autoimmune disease. We reasoned that failure to clear defective mitochondria via mitophagy might be a foundational driver in autoimmunity by licensing mitochondrial (mt)DNA-dependent induction of type I interferon (IFN-I). Here, we show that mice lacking the GTPase IRGM1 (IRGM homologue) exhibited a type I interferonopathy with autoimmune features. Irgm1 deletion impaired execution of mitophagy with cell-specific consequences. In fibroblasts, mtDNA soiling of the cytosol induced cyclic GMP-AMP synthase (cGAS)–Stimulator of Interferon Genes (STING)-dependent IFN-I, whereas in macrophages, lysosomal TLR7 was activated. In vivo, Irgm1−/− tissues exhibited mosaic dependency upon nucleic acid receptors. Whereas salivary and lacrimal gland autoimmune pathology were abolished and lung pathology was attenuated by cGAS and STING deletion, pancreatic pathology remained unchanged. These findings reveal fundamental connections between mitochondrial quality control and tissue-selective autoimmune disease.
Introduction
Transient, signal-dependent induction of type I interferon (IFN-I) is critical for host defense. Spontaneous and sustained IFN induction, however, is a hallmark of and causal factor in lupus, Sjogren’s Syndrome (SS), and other autoimmune diseases (ADs)1, 2. Whereas the proximal cause(s) of IFN-I in AD remain obscure, monogenic type I interferonopathies have recently taught us that aberrant (e.g., cytosolic) host nucleic acids can induce chronic IFN-I via activation of the double-stranded (ds)DNA receptor, cyclic GMP-AMP synthase (cGAS), and its downstream adaptor, stimulator of IFN genes (STING)2, 3. Soiling of the cytosol with mitochondrial (mt)DNA induces cGAS–STING-dependent IFN-I in experimental settings, including genetically enforced mitochondrial stress4 and apoptosis with caspase inhibition5. Recently, increased cytosolic mtDNA has also been documented in human and murine lupus leukocytes and linked to blocked autophagic flux arising from lysosomal alkalinization6, 7, 8. Given that mitochondrial abnormalities have long been noted in lupus9, we reasoned that failure to clear defective mitochondria via autophagy (i.e., mitophagy) and consequent mtDNA-dependent IFN-I induction might conceivably be a foundational event in the pathogenesis of human AD.
IRGM1 is one of a ~20-member family of dynamin-like immunity-related GTPases (IRGs) in mice10. IRGs have been categorized into two groups based on the amino acid sequence of their GTP-binding site: (i) GKS ‘effector’ proteins traffic to pathogen-containing vacuoles during infection, where they cooperatively initiate antimicrobial membranolysis; whereas (ii) ‘GMS’ regulatory proteins such as IRGM1 prevent off-target (host-directed) GKS activation by binding to organellar endomembranes (Golgi, mitochondria, ER, lysosomes), where they act as GKS-suppressive guanine dissociation inhibitors10, 11, 12. Irgm1−/− mice display defective cell-autonomous host defense against a wide array of intracellular pathogens that is thought to derive in part from defective autophagy (reviewed in10). IRGM1 has been shown to be required for lysosomal degradation of autophagosomes (APs) and their cargo, likely by preventing GKS protein-mediated lysosomal dysfunction11, 13, 14. Indeed, multiple parenchymal and immune cell types from Irgm1−/− mice exhibit abnormal accumulation of APs13, 14.
We previously reported that Irgm1−/− mice exhibit a mucosal-selective autoimmune disorder reminiscent of SS as well as upregulation of IFN-stimulated genes (ISGs), suggesting spontaneous IFN-I induction15. Intriguingly, genetic association studies have recently implicated autophagy-related genes ATG5, ATG7, and IRGM (human orthologue for Irgm1) in lupus and other ADs16, 17, 18. Given this, we hypothesized that defective mitophagic clearance of mtDNA and consequent activation of the cGAS–STING axis might be a cross-cutting mechanism in human AD, and that the Irgm1−/− mouse might provide a window into key steps in this failed pathway.
Results
IRGM1 deficiency triggers type I interferonopathy
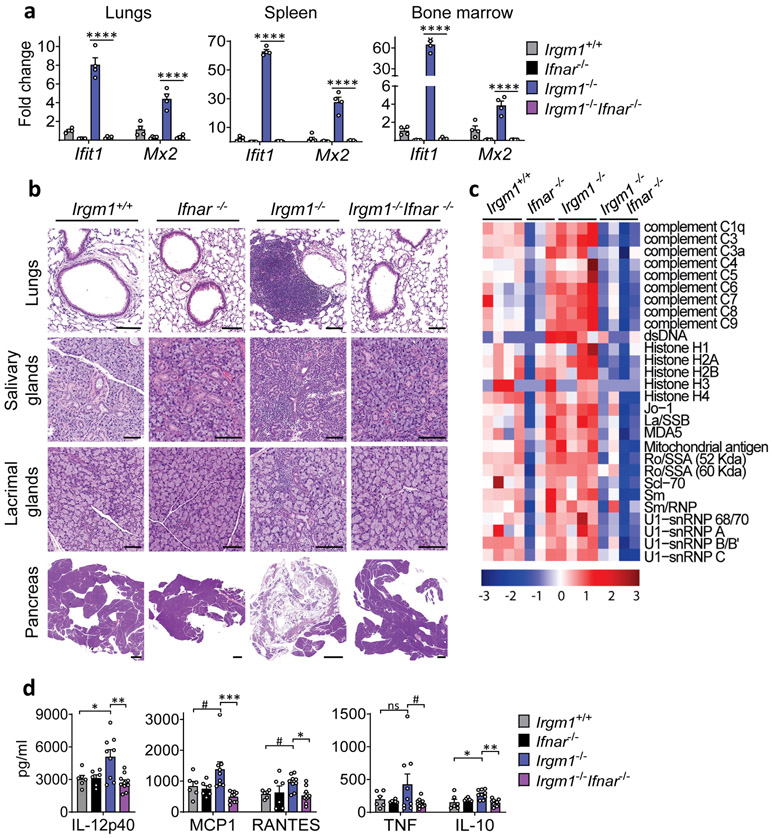
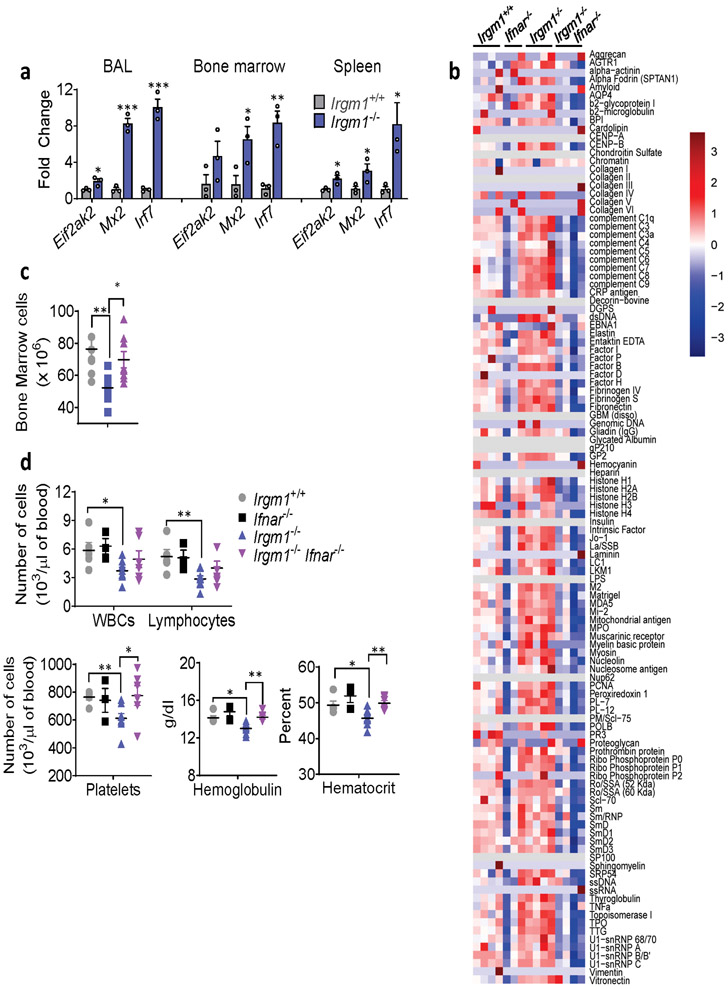
Naïve Irgm1−/− mice display lymphocytic infiltration of the lung, salivary gland, lacrimal gland, and exocrine pancreas and elevated serum autoantibodies and cytokines15, a pattern highly reminiscent of SS, the third most common human AD19. IFN-I plays a central role in SS pathogenesis1, 19. To test the requirement for IFN-I in the Irgm1−/− phenotype, we disrupted IFN-I signaling in Irgm1−/− mice by crossing them with mice deleted for the IFN-I receptor (IFNAR). In Irgm1−/−Ifnar−/− mice, the upregulation of ISGs (Ifit1, Mx2, Eif2ak2, Irf7) in the lungs, spleen, and bone marrow of Irgm1−/− mice was abolished (Fig. 1a, Extended Data Fig. 1a), confirming these genes as a specific IFN-I signature. Histopathology in all four target tissues of Irgm1−/− mice was abolished in Irgm1−/−Ifnar−/− mice and the elevated serum autoantibodies and cytokines were also normalized (Fig. 1b-d; Extended Data Fig. 1b). Irgm1−/− mice have been reported to have hematopoietic stem cell abnormalities and multilineage blood cell deficits15, 20. The bone marrow hypocellularity of Irgm1−/− mice was corrected in Irgm1−/−Ifnar−/− mice, as were systemic anemia and thrombocytopenia, but not lymphocytopenia (Extended Data Fig. 1c,d). Collectively, these findings demonstrate that IRGM1 deficiency provokes a type I interferonopathy with multi-organ involvement and autoimmune features.
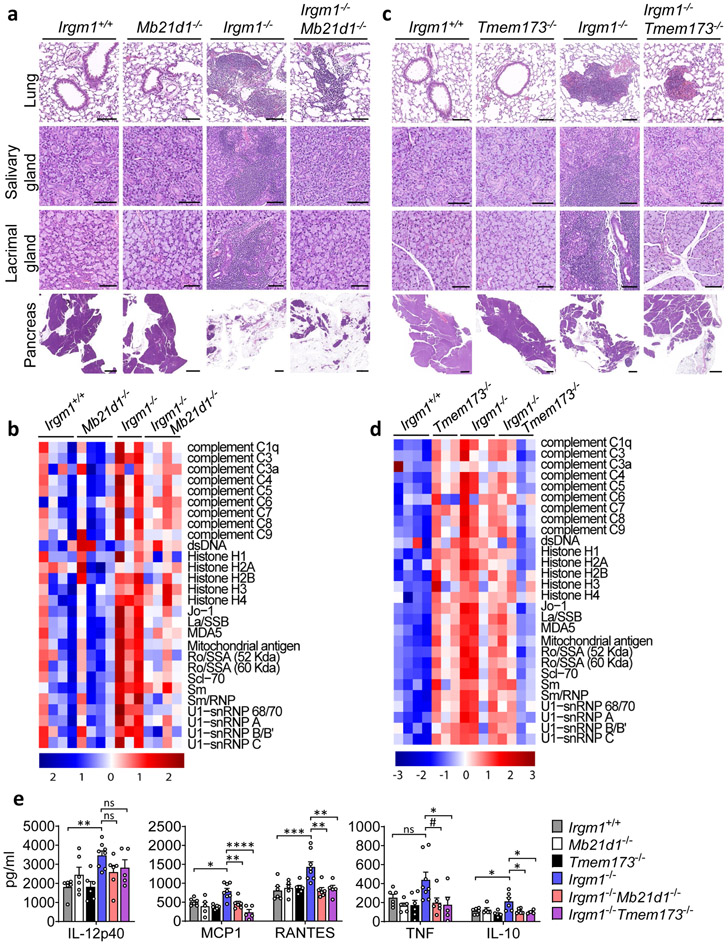
Figure 1: IRGM1 deficiency induces type I interferonopathy.
a, Expression of interferon-stimulated genes (ISG) Ifit1 and Mx2 in lungs, spleen and bone marrow of wild type, Ifnar−/−, Irgm1−/− and Irgm1−/−Ifnar−/− animals (n=4/genotype). b, Representative H&E-stained sections of lungs, salivary glands (submandibular), lacrimal glands, and pancreas from indicated genotypes (n = 2-7/genotype) (scale bars, 100 μm, except pancreas [1000 μm]). c, Autoantibodies against indicated antigens, measured in serum of animals (n=3-4 mice/genotype). d, Cytokines and chemokines measured in serum (n=6 [Irgm1+/+, Ifnar−/−]; n=9 [Irgm1−/−]; n=10 [Irgm1−/−Ifnar−/−]). Data are mean +/− s.e.m. #P = 0.1, *P < 0.05, **P < 0.01, and ****P < 0.0001 (one-way ANOVA with Tukey’s adjustment).
Aberrant mtDNA induces type I IFN in Irgm1-null cells
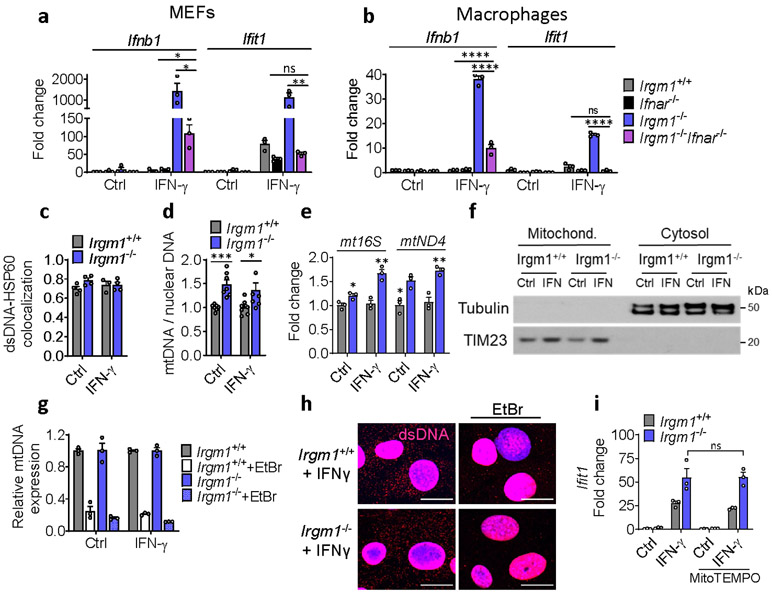
To define the cellular mechanism of IFN-I induction, we examined Irgm1−/− murine embryonic fibroblasts (MEFs) both in the unstimulated state and following overnight priming with IFN-γ. IRGs are robustly upregulated by IFN-γ, provoking autophagic deficits in Irgm1−/− cells, likely due to unregulated GKS effector protein activity10, 11, 13, 14. Ex vivo treatment of Irgm1-null cells with IFN-γ, a widely used experimental paradigm11, 12, 13, 14, 21, may not be artificial given that naïve Irgm1−/− mice have elevated serum IFN-γ and their autophagic defects are corrected by IFN-γ receptor deletion15, 22. We found that Irgm1−/− MEFs displayed marked upregulation of Ifnb1 and ISGs upon overnight priming with IFN-γ (Fig. 2a). ISG induction was abolished in Irgm1−/−Ifnar−/− MEFs, confirming that it was caused by autocrine/paracrine IFN-I, and Ifnb1 induction was also attenuated, suggesting an amplification loop (Extended Data Fig. 2a). Similar findings were noted in Irgm1−/− bone marrow-derived macrophages (BMDMs) (Fig. 2b, Extended Data Fig. 2b), indicating that multiple Irgm1−/− cell types exhibit an IFN-I response.
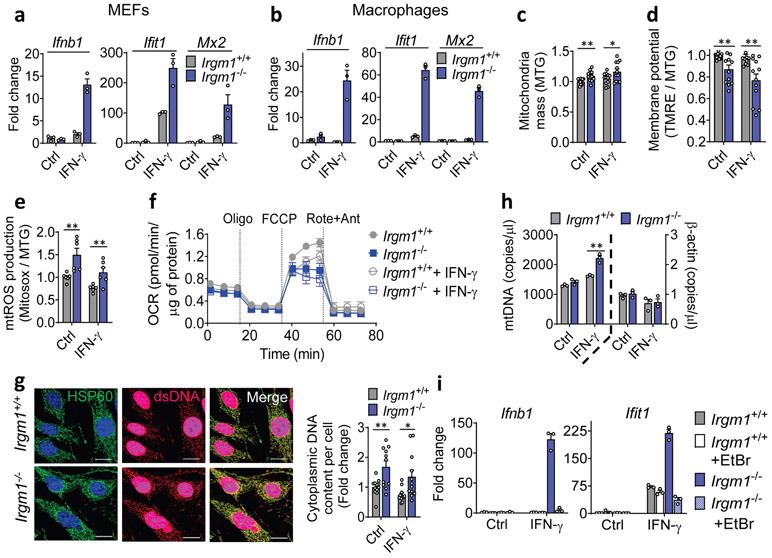
Figure 2: Mitochondrial DNA induces type I interferon response in Irgm1-null cells.
a, b, Interferon-β (Ifnb1) and interferon-stimulated genes (Ifit1, Mx2) expression in mouse embryonic fibroblasts (MEFs) (a) and bone marrow-derived macrophages (BMDMs) (b) left untreated or treated with IFN-γ (20 ng/ml, 16h) (n=3). c, Mitochondrial mass in MEFs as measured by MitoTracker Green (MTG) staining in flow cytometry. Data are pooled from four independent experiments (n=12). d, Mitochondrial membrane potential as measured by tetramethylrhodamine, ethyl ester (TMRE [a cell-permeant, cationic, fluorescent dye sequestered by polarized mitochondria]) and normalized to MitoTracker Green (MTG) fluorescence. Data are pooled from four independent experiments (n=12). e, Mitochondrial ROS (mtROS) production measured by MitoSox and normalized by MTG. Data are pooled from two independent experiments (n=6). f, Oxygen consumption rate (OCR) analyzed by Seahorse assay (Oligo=Oligomycin; FCCP = Carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone; Rote+Ant=Rotenone + Antimycin A) (n=15). g, Immunofluorescence staining of double-stranded (ds)DNA and HSP60 (a mitochondrial matrix marker) in MEFs (scale bar, 20 μm). HSP60 intensity was generally comparable between genotypes across replicate experiments. Cytoplasmic (ds)DNA signal was quantified by subtracting nuclear DNA signal (right). Data are pooled from three independent experiments (n=12). h, Cytosolic fractions of MEFs assayed for mitochondrial (mt)DNA (ND1) and nuclear DNA (β-actin) by digital droplet PCR (n=3). i, Gene expression in MEFs with or without mtDNA depletion by ethidium bromide (EtBr), followed by buffer or IFN-γ treatment (n=3). a, b, f, h, and i are representative of three independent experiments. c, d, and e are expressed as fold change of mean fluorescence intensity (MFI) or its ratio. Data are mean +/− s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001 (Two-tailed unpaired t-test).
The finding of IFN-I induction in primed Irgm1−/− cells suggested that an endogenous interferonogenic stimulus (e.g., nucleic acids, or mtDNA in particular)4, 5 might be responsible. Multiple mitochondrial abnormalities were detected even in non-primed Irgm1−/− MEFs. Specifically, Irgm1−/− MEFs had an increased mass of depolarized mitochondria, elevated mitochondrial reactive oxygen species (mtROS), and reduced spare respiratory capacity, together suggesting accumulation of dysfunctional mitochondria (Fig. 2c-f). Irgm1−/− MEFs also exhibited a significant increase in extranuclear dsDNA signal colocalizing with the mitochondrial matrix protein, HSP60 (Fig. 2g, Extended Data Fig. 2c), suggesting increased mtDNA nucleoids. Increased mtDNA copy number, indexed to nuclear DNA, was also detected in Irgm1−/− cells, as was an increase in RNA transcripts deriving from mitochondrially encoded genes (Extended Data Fig. 2d-e).
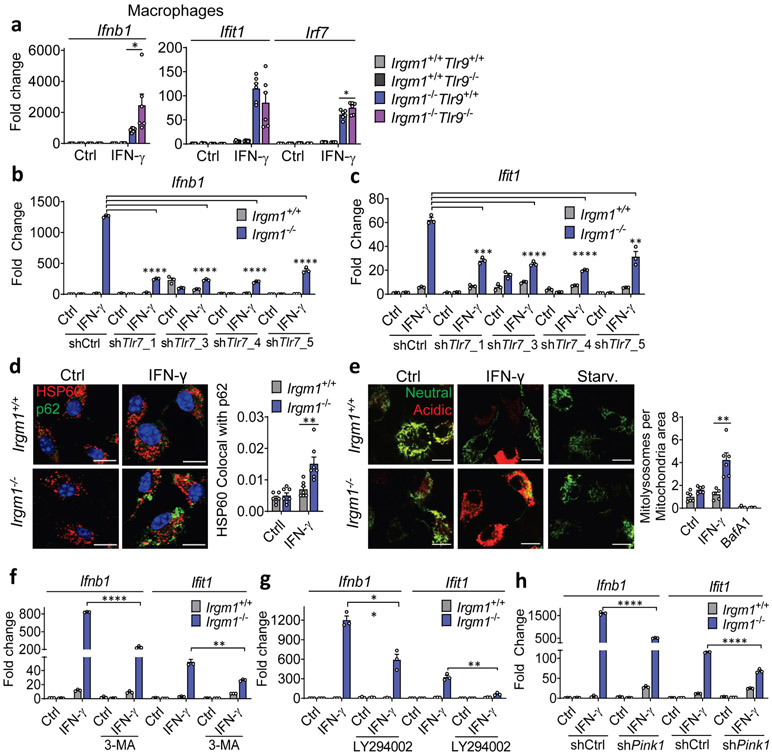
mtDNA that accesses the cytosol in increased amounts can induce IFN-I through activation of the dsDNA receptor cGAS4, 5. Whereas Irgm1+/+ and Irgm1−/− MEFs had equivalent cytosolic mtDNA in the non-stimulated state as assessed by digital droplet PCR, Irgm1−/− MEFs exhibited a significant increase following IFN-γ treatment (Fig. 2h, Extended Data Fig. 2f). In the cytosolic extracts, mtDNA copy number was >3 orders of magnitude higher than copy number for Actb, and the latter was equivalent between genotypes, together arguing against confounding by nuclear-integrated mitochondrial sequences23, and against mislocalized nuclear DNA as a cause for the IFN-I phenotype. To test whether mtDNA was playing a causal role in the IFN-I response, we depleted mtDNA by cell culture in ethidium bromide5, 24. mtDNA depletion, confirmed by PCR and microscopy (Extended Data Fig. 2g,h), abolished the abnormal induction of Ifnb1 and Ifit1 in Irgm1−/− MEFs (Fig. 2i). Although excess mtROS have been reported in some settings to play a role in mtDNA escape into the cytosol24, quenching mtROS in Irgm1−/− MEFs with mitoTEMPO did not reduce the IFN-I signature (Extended Data Fig. 2i). Taken together, these findings suggest that Irgm1−/− MEFs accumulate dysfunctional mitochondria in the steady state, which are provoked by IFN-γ to release mtDNA into the cytosol through a mtROS-independent mechanism, thereby inducing IFN-I, followed by autocrine/paracrine induction of ISGs.
Irgm1-null fibroblasts induce cGAS–STING–IFN-I pathway
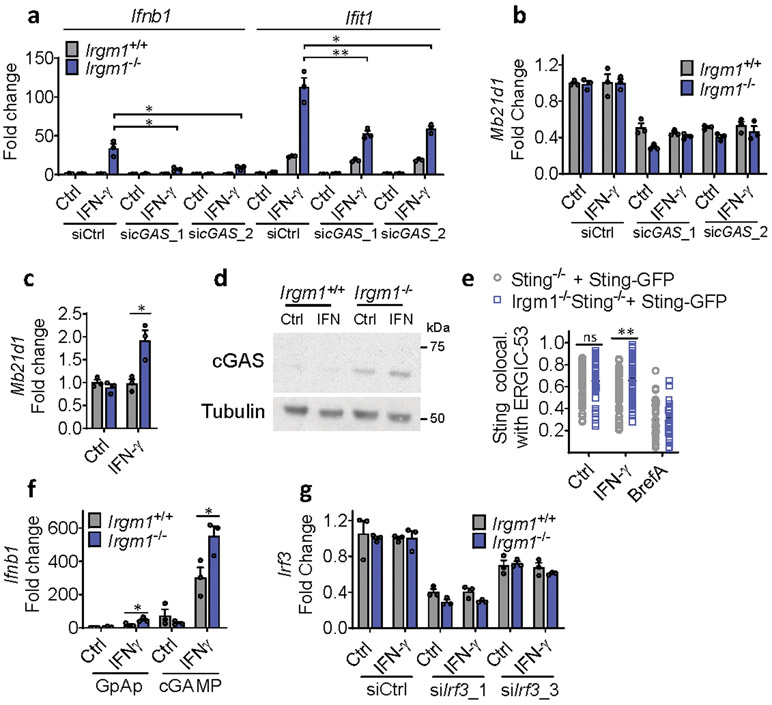
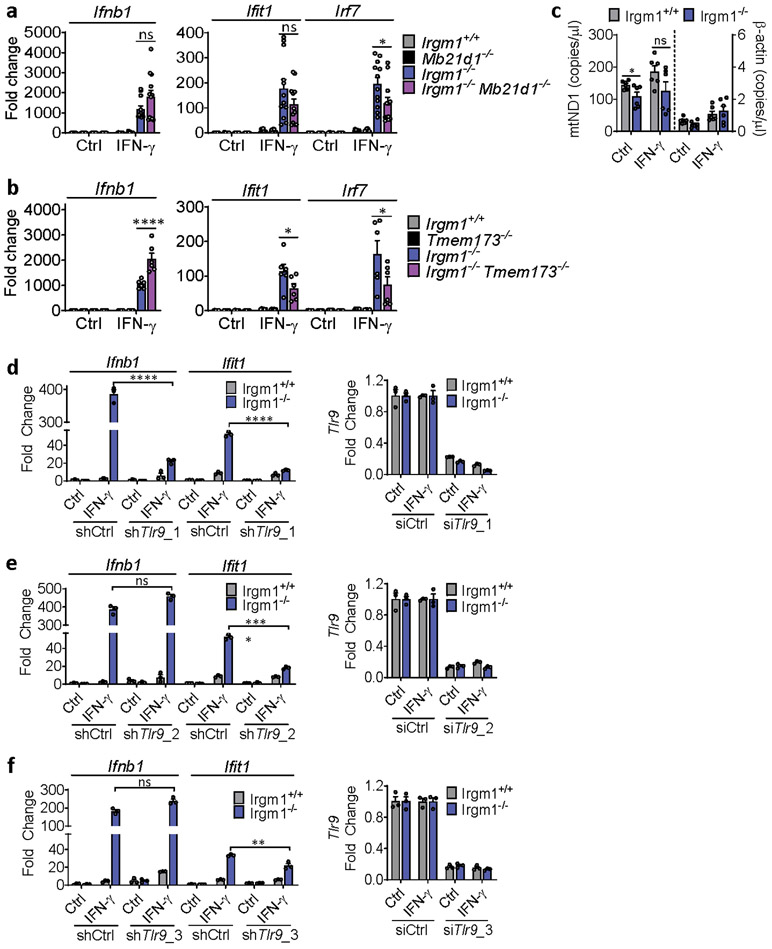
Upon ligation by dsDNA, including mtDNA, cGAS synthesizes the dinucleotide second messenger, 2′3′-cGAMP, which then activates STING, an ER-resident adaptor, inducing its trafficking to the ER-Golgi intermediate complex (ERGIC), where TANK-binding kinase (TBK)1 phosphorylates the IFN-I-inducing transcription factor Interferon Regulatory Factor (IRF)34. To test for a role of cGAS in the IFN-I induction in Irgm1−/− cells, we crossed Irgm1−/− mice with cGAS (Mb21d1)-deficient mice. Notably, the Ifnb1 and Ifit1 upregulation in Irgm1−/− MEFs was abolished in Irgm1−/−Mb21d1−/−MEFs (Fig. 3a), confirming an absolute requirement for cGAS. siRNA-mediated partial (~60%) silencing of cGAS in Irgm1−/− MEFs also caused a comparable partial reduction in IFN-γ-induced Ifnb1 and Ifit1 expression (Extended Data Fig. 3a,b). Mb21d1 is itself an ISG, supplying positive feedback to IFN-I induction by dsDNA25. Consistent with this, Mb21d1 mRNA and cGAS protein were elevated in IFN-γ-primed Irgm1−/− MEFs (Extended Data Fig. 3c,d).
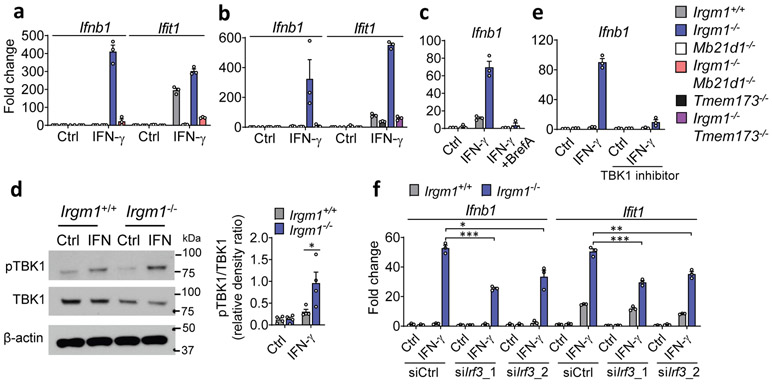
Figure 3: Type I interferon induction in Irgm1-null fibroblasts is cGAS/STING-dependent.
a,b, Interferon-β (Ifnb1) and interferon-stimulated gene (Ifit1) expression in murine embryonic fibroblasts (MEFs) of indicated genotypes, treated with or without IFN-γ (16h) (n=3). c, Ifnb1 expression in MEFs co-treated with brefeldin A (2 μg/ml) and IFN-γ (n=3). d, Phosphorylation of TBK1 (S172), total TBK1, and β-actin (loading control) detected by Western blot in cells untreated or treated with IFN-γ. At right, densitometry of four independent experiments is shown. e, Ifnb1 expression in MEFs pretreated with TBK1 inhibitor (MRT-67307, 10 μM, 2h) and then IFN-γ (16h) (n=3). f, Ifnb1 and Ifit1 expression in MEFs transfected with two different siRNAs against Irf3 or control siRNA and then treated as shown (n=3). b, d, and e are representative of three independent experiments. a, c, and f are representative of two independent experiments. Data are mean +/− s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001 (Two-tailed unpaired t-test).
IFN-γ priming caused translocation of GFP-STING to the ERGIC in Irgm1−/− MEFs that was suppressed with the ER trafficking inhibitor brefeldin A (Extended Data Fig. 3e), suggesting activation of STING downstream of cGAS. Transfected 2′3′-cGAMP caused more robust Ifnb1 upregulation in IFN-γ-treated Irgm1−/− than Irgm1+/+ MEFs (Extended Data Fig. 3f), suggesting that STING was primed for a stronger response to ligand. To test whether STING (encoded by Tmem173) is required for the IFN-I phenotype of Irgm1−/− cells, we crossed Irgm1−/− and Tmem173−/− mice. The Ifnb1 and Ifit1 upregulation observed in Irgm1−/− MEFs was abolished in Irgm1−/−Tmem173−/−MEFs (Fig. 3b), revealing a requirement for STING. Consistent with this, brefeldin A also abolished Infb1 induction (Fig. 3c). We found increased TBK1 phosphorylation in Irgm1−/− MEFs following IFN-γ, as well as abrogation of increased Ifnb1 expression in Irgm1−/− MEFs upon pretreatment with the TBK1 inhibitor MRT-67307 (Fig. 3d-e). RNAi knockdown also indicated a role for IRF3 (Fig. 3f, Extended Data Fig. 3g). Together, these findings suggest that mtDNA released into the cytosol in IFN-γ-primed Irgm1−/− MEFs activates the cGAS-STING-TBK1-IRF3 axis to induce IFN-I and ISGs.
Impaired mitophagy induces IFN in Irgm1−/− fibroblasts
The increased mass of dysfunctional mitochondria in Irgm1−/− MEFs was suggestive of defective mitophagy. During mitophagy, autophagy adaptor proteins decorate depolarized mitochondria that have been ubiquitinated by the Pink1-Parkin pathway, processively recruiting microtubule-associated protein 1A/1B light chain 3 (‘LC3’) to enclose the mitochondrion in an AP, which then fuses with degradative acidified lysosomes26.
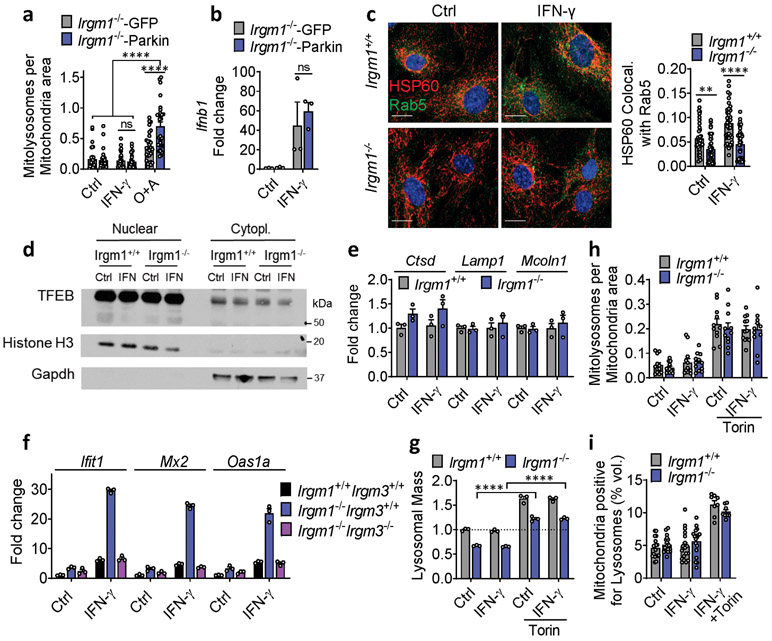
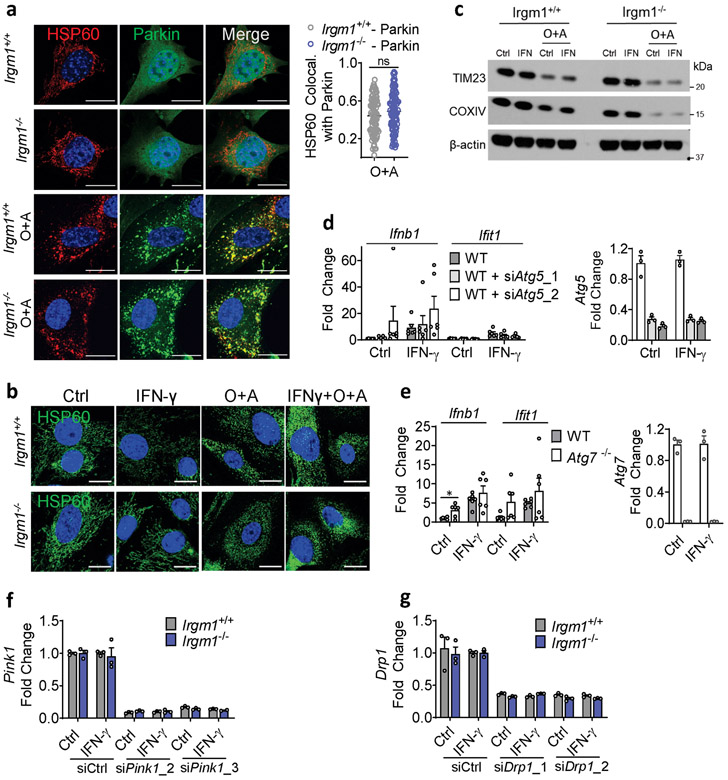
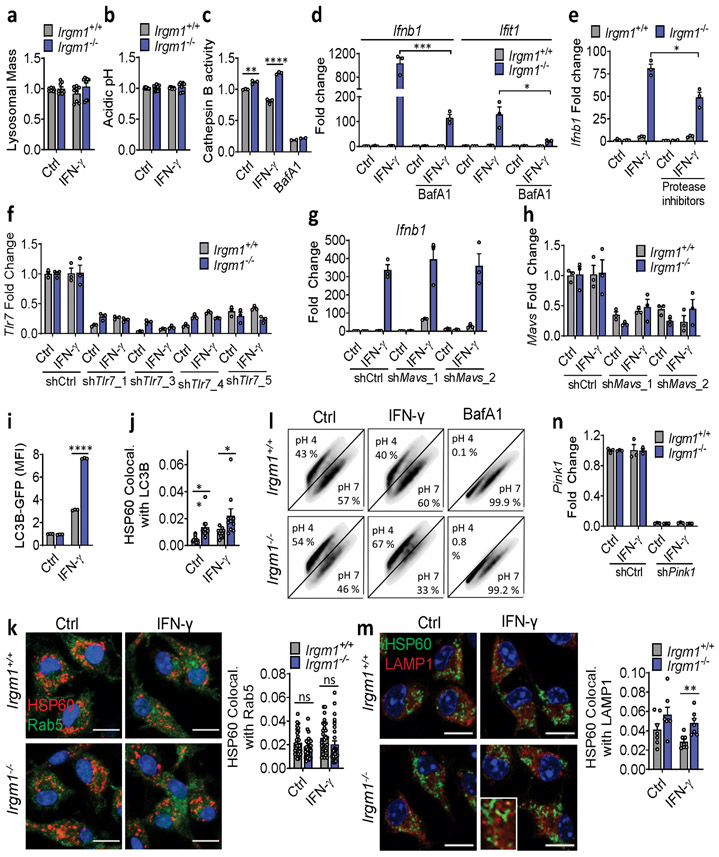
To assay mitophagy, we transfected MEFs with mt-mKeima, a mitochondrial-localizing construct that fluoresces differentially at pH 7.0 (cytosol) vs. pH 4.0 (acidified lysosome), allowing measurement of mitochondrial delivery to lysosomes27. Irgm1+/+ and Irgm1−/− MEFs showed no increase over baseline in mitolysosome signal after IFN-γ, whereas both displayed increased signal after starvation, a potent inducer of macroautophagy28 (Fig. 4a), and after the mitochondrial depolarizing dual treatment oligomycin plus antimycin29 (not depicted). Although MEFs express low amounts of PARKIN30, this did not appear to be limiting for mitophagy under IFN-γ treatment conditions, as PARKIN overexpression neither boosted mt-mKeima signal in Irgm1−/− MEFs nor rescued increased Ifnb1 and Ifit1 expression (Extended Data Fig. 4a,b). Collectively, these findings suggest that Irgm1−/− MEFs are mitophagy-competent at least under high-stress conditions, but that, despite their accumulation of depolarized mitochondria, they exhibit at best very low levels of mitophagic flux. Reduced Rab5-HSP60 colocalization was observed in Irgm1−/− MEFs in both the unstimulated and IFN-γ-stimulated state (Extended Data Fig. 4c). This result suggests that a deficit in endosomal transfer may contribute, at least in part, to defective mitochondrial quality control in Irgm1−/− MEFs.
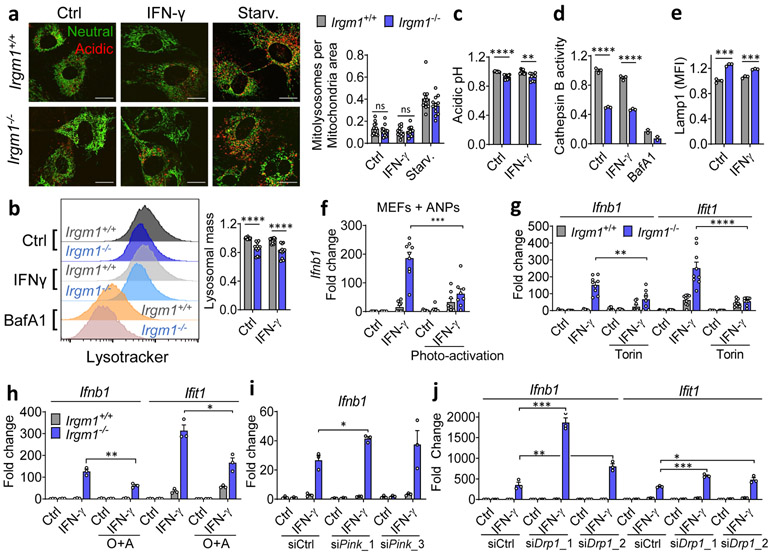
Figure 4: Mitophagic flux deficit drives type I IFN response in Irgm1-null fibroblasts.
a, Mitochondria (mt)-mKeima-expressing murine embryonic fibroblasts (MEFs) (representative images, left) analyzed for pixel area in red channel (acidic mitolysosomes), normalized to signal in green channel (total mitochondria) (scale bar, 20 μm). MEFs were untreated, treated with IFN-γ, or starved (Starv.) by incubation in HBSS buffer. Quantification at right (n=12). b, Lysosomal mass assessed by LysoTracker fluorescence in flow cytometry. Representative histogram at left, quantification at right (n=12). c, Relative acidic pH measured by ratiometric Lysosensor yellow/blue dye (n=9). d, Cathepsin activity measured by cleavage of Magic Red substrate (n=3). Bafilomycin A1 (BafA1, 100 nM) was used as negative control in (b) and (d). e, LAMP1 fluorescence measured by flow cytometry (MFI, mean fluorescence intensity) (n=3). f, Interferon-β (Ifnb1) expression in MEFs preloaded with photoactivatable acidic nanoparticles (aNPs), treated with IFN-γ and then exposed to UV light (n=9). g,h, Ifnb1 and Ifit1 expression in MEFs co-treated with IFN-γ and 1 μM Torin1 (n=9) (g) or Oligomycin + Antimycin (O+A, 10 μM each) (n=3) (h). i, j, Ifnb1 and Ifit1 expression in MEFs transfected with two different siRNAs against Pink1 (n=3) (i) or Drp1 (n=3) (j) or control siRNA followed by treatment as shown. b, c, d and e are expressed as fold change of MFI. a, h and j are representative of three independent experiments. c, e, and i are representative of two independent experiments. b, d, f and g are pooled from at least three independent experiments. Data are mean +/− s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (Two-tailed unpaired t-test).
mt-mKeima acidification (and mitophagy) require fusion of mitochondria-bearing APs (mitoAPs) with acidified lysosomes. We found that naïve and IFN-γ-primed Irgm1−/− MEFs had reduced signal for both LysoTracker, an indicator of acidified organelles, and LysoSensor, an indicator of low lysosomal pH (Fig. 4b,c). Proteolytic activity of the acid-dependent lysosomal protease cathepsin B was also reduced (Fig. 4d), collectively confirming acidifying and degradative insufficiency of Irgm1−/− MEF lysosomes and suggesting a potential explanation for defective mitophagic flux.
We observed equivalent nuclear localization in Irgm1+/+ and Irgm1−/− MEFs of transcription factor EB (TFEB), and equivalent expression of members of the Coordinated Lysosomal Expression and Regulation (CLEAR) network of TFEB target genes31 (Extended Data Fig. 4d,e), collectively suggesting no change in lysosomal biogenesis in Irgm1-null cells. Instead, consistent with a prior report that lysosomal mistargeting of GKS effector proteins impairs lysosomal function in Irgm1-null cells13, we found that Irgm1−/− MEFs had increased signal intensity for LAMP1, an endolysosomal marker (Fig. 4e). Taken together with the reduction in LysoTracker signal, this suggests that a block in lysosomal maturation in Irgm1−/− MEFs compromises fusion of mitoAPs with degradative lysosomes, impairing mitophagy.
Several of the defects of Irgm1−/− cells, including lysosomal dysfunction, have been shown to be dependent on the alternate IRG protein, IRGM3. Loss of IRGM3 in Irgm1−/− cells rescues their autophagic flux defects and has been proposed to do so by redirecting GKS effector proteins away from lysosomes10, 11, 13, 22. We previously reported that Irgm3 deletion normalizes the histopathologic findings in Irgm1−/− mice15. Here, we found that Irgm1−/−Irgm3−/− MEFs also have normalized ISG expression (Extended Data Fig. 4f). This finding suggests that IRGM3-dependent GKS effector protein mistargeting to lysosomes with resultant lysosomal dysfunction may be responsible for IFN-I induction in Irgm1−/− cells.
We next sought to test whether correcting lysosomal function could rescue IFN-I induction in Irgm1−/− MEFs. Treatment of Irgm1−/− MEFs with photoactivatable acidifying nanoparticles (ANPs) that restore lysosomal acidification after internalization32 led to a marked reduction in Ifnb1 expression (Fig. 4f). Torin, a mTOR inhibitor that triggers lysosomal biogenesis33, increased LysoTracker signal and mitochondrial delivery to acidified lysosomes in both Irgm1+/+ and Irgm1−/− MEFs (Extended Data Fig. 4g-i). Under these conditions, it normalized both Ifnb1 and Ifit1 in Irgm1−/− MEFs (Fig. 4g). Finally, to test whether chemically enforced mitophagy could rescue the IFN-I phenotype, we used the potent mitophagy inducer oligomycin plus antimycin29. In both Irgm1+/+ and Irgm1−/− MEFs treated with oligomycin plus antimycin, YFP-tagged Parkin colocalized with HSP60, mitochondria become more punctate, and mitochondrial markers TIM23 and COXIV were reduced (Extended Data Fig. 5a-c), collectively confirming successful execution of mitophagy. In parallel, Ifnb1 and Ifit1 were downregulated in IFN-γ-primed Irgm1−/− MEFs (Fig. 4h). Thus, chemically enforcing clearance of mitochondria in Irgm1−/− MEFs through any of several means rescues mtDNA-dependent activation of the cGAS-STING pathway. Conversely, we found that, although silencing of ATG5 did not provoke an IFN-I response in wild-type MEFs, ATG7-null MEFs exhibited spontaneous induction of Ifnb1 (Extended Data Fig. 5d,e), corroborating that defective autophagic clearance of mitochondria may be sufficient in some contexts to induce IFN-I in MEFs.
RNAi-mediated knockdown of the mitophagic kinase PINK1 and of the pro-mitophagy mitochondrial fission protein DRP126 both augmented expression of Ifnb1 and/or Ifit1 in IFN-γ-primed Irgm1−/− MEFs (Fig. 4i,j; Extended Data Fig. 5f,g), suggesting that residual native PINK1–PARKIN-dependent mitophagy does play a small role in dampening mtDNA-dependent IFN-I induction in Irgm1−/− MEFs. Taken together with our observation that reacidification of the Irgm1−/− lysosomal lumen rescues IFN-I induction, these findings suggest that low-level mitophagic flux is operative in Irgm1−/− cells but insufficient to prevent mtDNA-dependent activation of cGAS.
Irgm1−/− pathology is cGAS- and STING-dependent
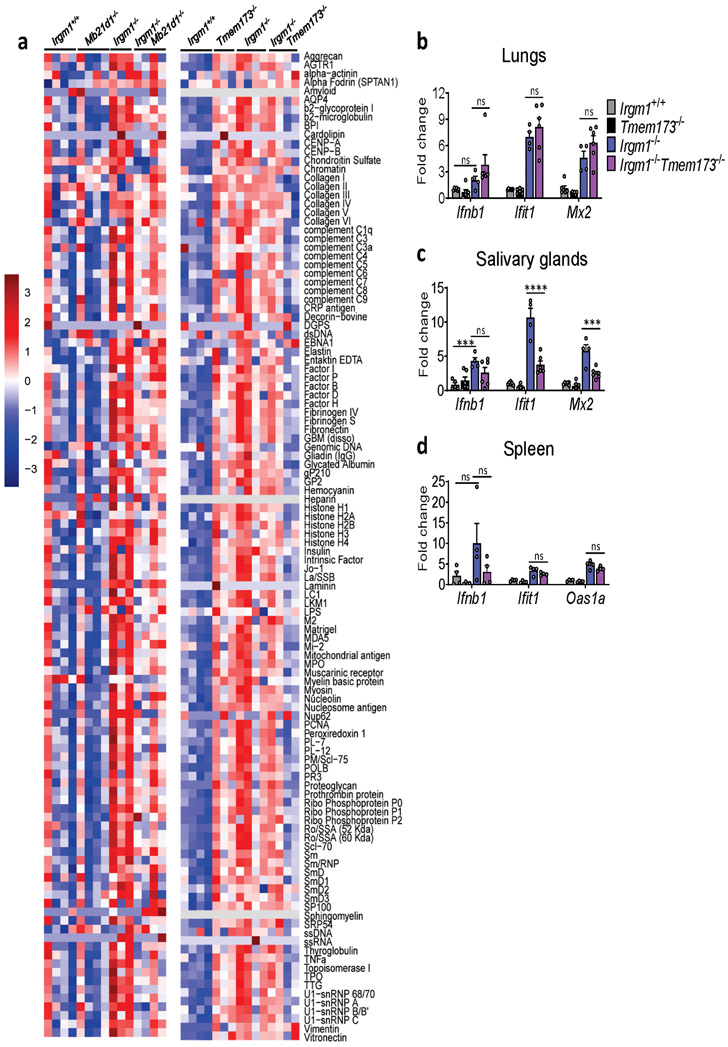
Having established that Irgm1−/− mice have a type I interferonopathy and that IFN-I induction in Irgm1−/− MEFs is cGAS- and STING-dependent, we next sought to confirm whether the autoimmune pathology of Irgm1−/− mice is cGAS- and STING-dependent. Irgm1−/−Mb21d1−/− mice exhibited partial, tissue-selective rescue of the histopathologic changes noted in Irgm1−/− mice (Fig. 5a), indicating a role for cGAS. Specifically, pathology was abolished in the salivary and lacrimal glands of all double knockout mice, but not in the pancreas, which showed extensive acinar cell atrophy with variable adipocyte replacement and lymphocytic infiltration typical of Irgm1−/− mice. By contrast, the lungs of Irgm1−/−Mb21d1−/− mice showed partial rescue. Three of six Irgm1−/−Mb21d1−/− mice exhibited minimal changes in the lungs (peribronchovascular lymphocytic foci) compared to Irgm1−/− mice, whereas the other three mice exhibited lesions comparable in extent and severity to those of Irgm1−/− mice. Confirming a role for cGAS in the humoral autoimmunity of Irgm1−/− mice, Irgm1−/−Mb21d1−/− mice had normalization of a wide array of serum autoantibodies (Fig. 5b, Extended Data Fig. 6a).
Figure 5: Tissue-selective involvement of cGAS/STING in autoimmune pathology of Irgm1-null mice.
a,c, Representative H&E sections of lungs, salivary glands (submandibular), lacrimal glands, and pancreas from indicated genotypes (n=3-6/genotype) (scale bars, 100 μm, except pancreas [1000 μm]). b,d, Autoantibodies against indicated antigens, measured in serum of animals (n=3-4 mice/genotype). e, Cytokine levels in serum of animals (n=5 [Mb21d1−/−, Irgm1−/−Tmem173−/−]; n=6 [Irgm1+/+, Tmem173−/−, Irgm1−/−Mb21d1−/−]; n=8 [Irgm1−/−]). Data are mean +/− s.e.m. #P = 0.08, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (one-way ANOVA with Tukey’s adjustment).
Irgm1−/−Tmem173−/− mice exhibited a pattern of pathological rescue similar, but not identical to that of Irgm1−/−Mb21d1−/− mice, suggesting that cGAS and STING are dissociable in the Irgm1−/− autoimmune syndrome (Fig. 5c). As with Irgm1−/−Mb21d1−/− mice, salivary and lacrimal glands were rescued whereas the pancreas was not. In Irgm1−/−Tmem173−/− mice, however, lung lesions were comparable in extent and severity to those observed in Irgm1−/− mice, indicating no rescue. Suggesting that STING is required for the spontaneous IFN-I phenotype of the Irgm1−/− salivary gland but not lung, ISG expression was markedly attenuated in the salivary glands but not lungs of Irgm1−/−Tmem173−/− mice (Extended Data Fig. 6b,c). STING ablation also did not rescue elevated ISG expression in Irgm1−/− spleen (Extended Data Fig. 6d), a tissue without overt histopathologic changes. Unlike the case for Mb21d1−/− mice, we found that Tmem173−/− mice had elevated serum autoantibodies and Irgm1−/−Tmem173−/− mice displayed inconsistent differences in autoantibodies compared to Irgm1−/− mice (Fig. 5d, Extended Data Fig. 6a). Regardless of their slight differences in histopathology and more marked differences in autoantibody profile, Irgm1−/−Mb21d1−/− and Irgm1−/−Tmem173−/− mice both displayed normalization of the serum cytokines that were elevated in Irgm1−/− mice (Fig. 5e). Taken together, these findings suggest that the autoimmune pathogenesis of Irgm1−/− mice is partially dependent upon cGAS and STING, with some organs (e.g., lung) displaying histopathology that is IFNAR-dependent but STING-independent.
TLR7-dependent IFN-I in Irgm1-null macrophages
Given that lung is a macrophage-rich tissue and macrophages are developmentally distant from fibroblasts, we next focused our attention on the mechanism of IFN-I induction in Irgm1−/− macrophages. We found that IFN-γ-primed Irgm1−/−Mb21d1−/− BMDMs had no significant rescue of Ifnb1 or Ifit1, but had reduced Irf7 expression (Extended Data Fig. 7a). By contrast, Irgm1−/−Tmem173−/− BMDMs had substantial rescue of ISGs induction but augmentation of Ifnb1 upregulation beyond that observed in Irgm1−/−Tmem173+/+ cells (Extended Data Fig. 7b). Unlike Irgm1−/− MEFs, there was no increase in cytosolic mtDNA in Irgm1−/− BMDMs (Extended Data Fig. 7c). Collectively, these findings indicated that, unlike the case for Irgm1−/− MEFs, cGAS detection of mtDNA does not play a role in the IFN-I phenotype of Irgm1−/− BMDMs and that STING may have an indirect regulatory role that is dissociated from cGAS. This suggested potential involvement of a distinct innate immune receptor pathway in Irgm1−/− BMDMs.
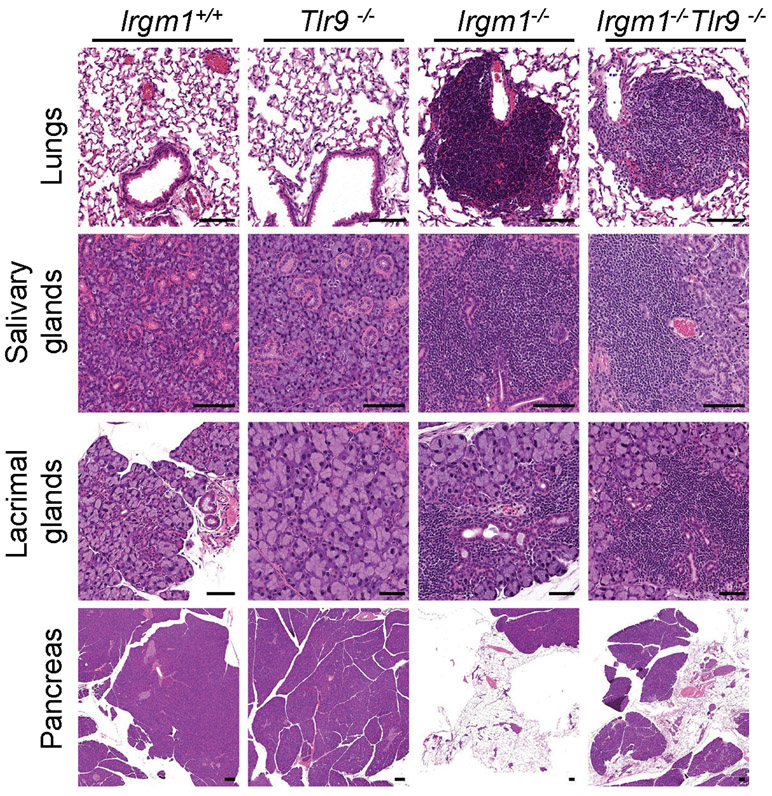
TLR9, an endolysosomal ssDNA receptor, has been shown to induce IFN-I in response to mtDNA through both cell-autonomous (i.e., mitochondrial trafficking to lysosomes) and transcellular (i.e., internalization of extracellular mtDNA) pathways34, 35. MEFs reportedly have negligible TLR9 activity36, but BMDMs display robust TLR9 responses. We found that, whereas three lentiviral shRNA TLR9 silencing constructs attenuated Ifit1 expression in IFN-γ-primed Irgm1−/− BMDMs, only one of the three attenuated Ifnb1 (Extended Data Fig. 7d-f). This suggested a nonspecific effect. Supporting that, genetic deletion of Tlr9 in Irgm1−/− macrophages (i.e., Irgm1−/−Tlr9−/− BMDMs) induced a nonsignificant reduction in Ifit1 and a significant further increase in Ifnb1 and Irf7 (Fig. 6a). Further corroborating that TLR9 does not mediate the IFN-I response in Irgm1−/− macrophages, we found that there was no rescue of histopathology in the lungs or other tissues of Irgm1−/−Tlr9−/− mice (Extended Data Fig. 8).
Figure 6: Type I IFN induction in Irgm1-null macrophages is TLR7-dependent.
a, BMDMs from the indicated strains were treated as shown and then analyzed for expression of Ifnb1, Ifit1, and Irf7 by qPCR (n=6). Results derive from cultures from two animals. * p ≤ 0.05 (one-way ANOVA with Tukey’s multiple comparison test). b,c, BMDMs underwent Tlr7 silencing with four different shRNA constructs (or control), were treated as shown, and then analyzed for expression of Ifnb1 (b) and Ifit1 (c) (n=3). Results are representative of two independent experiments. d, BMDMs were stained with HSP60 and p62 and analyzed for colocalization (Mander’s coefficient) (n=7) (scale bar, 10 μm). e, Mitochondria-mKeima-expressing BMDMs (representative images, left) were analyzed for pixel area in red channel (mitolysosomes) and normalized by signals in green channel (total mitochondria) (scale bar, 10 μm). Bafilomycin (BafA) treatment is used as negative control (n=6). f-h, Ifnb1 and Ifit1 expression by qPCR in BMDM treated with 3-methyladenine (3-MA, 5 mM) (n=3) (f) or LY94002 (10 μM) (g) or silenced for Pink1 (n=3) (h) prior to being left untreated or treated with IFN-γ. a and d are representative of three independent experiments. b, c, e, f, g, and h are representative of two independent experiments. Data are mean +/− s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (Two-tailed unpaired t-test).
Unlike Irgm1−/− MEFs, Irgm1−/− BMDMs had intact or elevated lysosomal function, as indicated by normal signal for LysoTracker and LysoSensor, and increased cathepsin B activity (Extended Data Fig. 9a-c). Similar to TLR9, TLR7, an endolysosomal ssRNA receptor, is dependent upon acidification of the endolysosome because of its requirement for processing by acidic cathepsins36, 37. Given the absence of detectable cytosolic mtDNA and the finding of enhanced cathepsin activity, we speculated that TLR7 might be mediating Ifnb1 induction in Irgm1−/− macrophages. It has recently been reported that TLR7 is activated by segments of mitochondrial 16S rRNA38. Consistent with a requirement for acid protease-dependent processing within the lysosome for IFN-I induction in Irgm1−/− BMDMs, we found that the lysosomal V-ATPase inhibitor bafilomycin A abrogated cathepsin activity and substantially attenuated induction of Ifnb1 and Ifit1 in IFN-γ-primed Irgm1−/− BMDMs (Extended Data Fig. 9c,d). Dual treatment with E64 and pepstatin A, cathepsin inhibitors that block endosomal TLR maturation36, also attenuated Ifnb1 and Ifit1 induction (Extended Data Fig. 9e), whereas they had no effect in Irgm1−/− MEFs (not depicted). Finally, TLR7 silencing with four independent lentiviral shRNAs reduced Ifnb1 and Ifit1 expression in IFN-γ-primed Irgm1−/− BMDMs (Fig. 6b,c; Extended Data Fig. 9f), confirming a requirement for TLR7 in their IFN-I phenotype. By contrast, silencing of mitochondrial antiviral signaling protein, an adaptor for the cytosolic RNA-sensing RIG-I-Like Receptors, had no effect on Ifnb1 (Extended Data Fig. 9g,h).
To better define the mechanism of accumulation of mitochondrial cargo in lysosomes, we crossed Irgm1−/− and wild-type mice with LC3-GFP transgenic mice39. Treatment with saponin to release soluble LC3-I39 revealed that IFN-γ-primed Irgm1−/− BMDMs had abnormal accumulation of LC3-II signal by flow cytometry (Extended Data Fig. 9i). This is consistent with past reports from our group and others that Irgm1−/− cells have deficient degradative clearance of APs due to failure to fuse with and/or be degraded by lysosomes13, 14. Suggesting abnormal accumulation of mitoAPs, we found increased colocalization of HSP60 with both p62 (Fig. 6d) and LC3 (Extended Data Fig. 9j) in IFN-γ-primed Irgm1−/− BMDMs. Taken together with our finding of intact degradative lysosomal function, the accumulation of mitoAPs suggested defective fusion of mitoAPs with lysosomes (i.e., a blockade in mitophagic flux). Contrary to the case for Irgm1−/− MEFs, there was no change in Rab5-HSP60 colocalization in Irgm1−/− BMDMs (Extended Data Fig. 9k), suggesting no alteration in endosomal transfer of mitochondria to the lysosome.
To assay mitochondrial delivery to acidified lysosomes, we transfected BMDMs with mt-mKeima. Unexpectedly, Irgm1−/− BMDMs had increased mitolysosomes after IFN-γ (Fig. 6e, Extended Data Fig. 9l). Corroborating increased mitochondrial accumulation in late endosomes and/or lysosomes of Irgm1−/− BMDMs, we also observed increased HSP60-LAMP1 colocalization (Extended Data Fig. 9m). 3-methyladenine28 and LY294002, phosphatidylinositol-3-kinase inhibitors that block autophagy, both reduced Ifnb1 and Ifit1 expression in IFN-γ-primed Irgm1−/− BMDMs (Fig. 6f,g), suggesting that the upstream signaling of autophagy initiation is required for delivery of mitochondria to TLR7 in Irgm1−/− BMDMs. Moreover, opposite to the case in MEFs, we found that, in Irgm1−/− BMDMs, lentiviral silencing of PINK126 also downregulated Ifnb1 and Ifit1 (Fig. 6h, Extended Data Fig. 9n). Collectively, these findings suggest that, in Irgm1−/− BMDMs, mitochondria are delivered through a PINK1- and LC3-dependent pathway to an endolysosomal compartment that harbors TLR7.
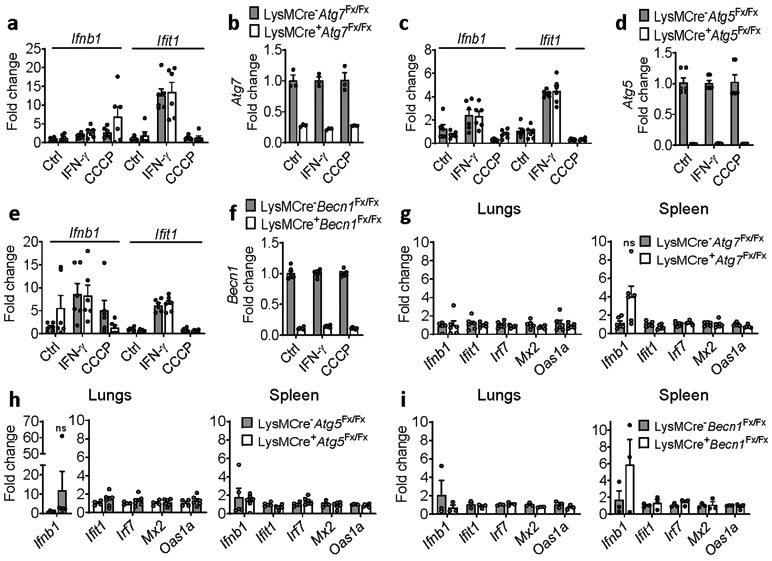
BMDMs deficient in any of the core autophagy proteins ATG5, ATG7, or BECLIN1 did not exhibit induction of Ifnb1 or Ifit1 either spontaneously or after challenge with IFN-γ or the mitochondrial stressor CCCP; nor did we detect induction of Ifnb1 or ISGs in the lungs or spleen of mice with myeloid-specific deletion of these genes (Extended Data Fig. 10). Given that ATG7-null MEFs exhibit spontaneous Ifnb1 induction, these findings further underline the fundamental difference in mitophagic flux deficits between Irgm1−/− MEFs and BMDMs. Taken together with prior work documenting defective degradation of APs in Irgm1−/− cells13, 14, we thus propose a cell-autonomous model in which frustrated autophagic flux to degradative lysosomes in Irgm1−/− macrophages promotes increased delivery of mitoAPs to a TLR7-expressing subcellular compartment, leading to IFN-I induction by mtRNA species.
Discussion
Hereditary interferonopathies have recently revealed that multiple safeguards are in place to prevent inappropriate immune activation by endogenous nucleic acids. The recent findings that mtDNA may accumulate in lupus due to failure of mitochondria to be delivered to40, or degraded in6, 7, 8, lysosomes, and that polymorphisms in autophagy-related genes increase risk for AD16, 17, 18 together suggested to us that defective mitophagy might be a cross-cutting mechanism underlying human ADs. Here, complementing recent genetic association studies that have implicated the autophagy-related gene IRGM in human AD16, 17, 18, we identify the murine homologue Irgm1 as a master suppressor of mitochondrial nucleic acid-induced autoinflammation.
Of interest, we provide evidence that Irgm1 deficiency provokes a mosaic pattern of activation of different nucleic acid sensors in different tissues, inducing a systemic IFN-I-dependent autoimmune syndrome reminiscent of SS. This finding may suggest that caution is warranted in developing therapeutics for systemic AD that target single innate immune receptors, such as cGAS41, on the basis of pathways validated in select tissues such as blood. Although STING is activated by cGAS-derived cGAMP, we found that STING deletion, unlike cGAS deletion, failed to rescue autoantibody levels in Irgm1−/− mice. A prior report that Tmem173−/− mice have increased susceptibility to TLR-dependent autoimmunity42 may offer insight on this dissociation.
In Irgm1−/− MEFs, silencing of PARKIN, PINK1, and DRP1 further, albeit marginally, increased IFN-I, suggesting that mitophagy is active – as would be expected given the mitochondrial depolarization – and also somewhat successful at dampening mtDNA-induced responses. Direct reacidification of lysosomes with ANPs rescued mtDNA-dependent IFN-I induction, indicating that lysosomal dysfunction and attendant mitophagic deficiency is, however, a root cause of cGAS activation in these cells. As lysosomal alkalinization can compromise AP-lysosome fusion43, our studies do not distinguish between defects in AP-lysosome fusion and intralysosomal degradation, both of which have been reported in Irgm1−/− MEFs13, 14. Nonetheless, the equivalent mitochondrial-lysosome colocalization we observed between Irgm1+/+ and Irgm1−/− MEFs, in the face of increased mitochondrial mass in the latter cell, implies a deficit in fusion.44
Remarkably, Irgm1 deficiency induced very different responses in macrophages. Irgm1−/− macrophages exhibited a marked increase in mitoAPs and in mitochondrial markers within acidified lysosomes. This suggests a deficit in mitochondrial degradation despite overtly intact lysosomal function and/or accelerated delivery of mitochondria to lysosomes beyond their degradative capacity. The ISG induction phenotype of Irgm1−/− BMDMs was partially STING-dependent. Although this may suggest, as previously reported in DNAse-deficient cells34, that nondegraded host DNA can escape the lysosome and access cytosolic dsDNA receptors, we were unable to detect increased mtDNA in macrophage cytosol preparations. Unlike MEFs, which are endosomal TLR-incompetent36, BMDMs from Irgm1−/− mice exhibited TLR7-dependent IFN-I induction, implying a role for endosomally delivered mtRNA. Mitochondria contain several TLR7-active molecules, including mRNA and ncRNAs such as miRNA45, 46.
Our findings that inhibition of autophagy (3-MA), mitophagy (Pink1 silencing), and lysosomal function (cathepsin inhibitors, bafilomycin) all suppressed rather than augmented IFN-I in Irgm1−/− macrophages further underlines that TLR7, and not cGAS, serves as the primary detector for defective disposal of mitochondrial nucleic acid in these cells. We speculate that in Irgm1−/− macrophages, mtRNA is delivered to a TLR7+ endolysosomal compartment via a mechanism requiring canonical mitophagic mechanisms, where its persistence induces immune activation. Future studies are warranted to determine whether excess mitoAPs in Irgm1−/− macrophages access TLR7 via direct fusion with an endolysosomal (amphisomal) compartment, as well as whether proximal inhibition of autophagy initiation (e.g., PI3-kinase inhibitors) attenuates lung disease in Irgm1−/− animals.
Important differences have been identified between murine IRGM1 and human IRGM. The latter is truncated and functions as an adaptor for the signaling that initiates autophagy47, a role that appears not to be shared by the murine homologue21. Both proteins, however, appear to play an important role in delivery of APs to degradative lysosomes14, 48, suggesting that key events in IRGM1-deficient murine cells may be shared by IRGM-deficient human cells. Parkin-deficient humans, like Parkin-null mice, exhibit elevated serum cytokines, including TNF, that are thought to arise from mtDNA-induced cGAS–STING activation49. Intriguingly, similar to this and parallel to our findings in Irgm1−/− mice, human subjects with IRGM deficiency (due to a polymorphic ~20kb deletion upstream of IRGM) have elevated TNF expression in blood50.
Here, we identify IRGM1 as a master suppressor of autoinflammation that prevents spontaneous IFN-I induction by ensuring mitochondrial quality control. These findings suggest that mtDNA and mtRNA may serve as an alarm system for failures in autophagic flux and that mitophagy – classically considered a housekeeping process – warrants much closer examination for its potentially fundamental roles in the programming of immune function.
Methods
Animals.
Irgm1−/− mice have been described before and were backcrossed more than 8 generations onto the C57BL/6 background51. Tmem173−/− mice were a generous gift from G. Barber (University of Miami) and were backcrossed more than 8 generations onto C57BL/6. Ifnar−/− (stock #032045) and Mb21d1−/− (stock #026554) mice were from Jackson Laboratory and were 8 and 7 generations backcrossed to C57BL/6, respectively. Tlr9−/− mice, a generous gift from C. Bosio (NIAID/NIH), have been described before52 and were 12 generations backcrossed to C57BL/6. LC3-GFP mice39 and LysM-Cre Atg5fx/fx, LysM-Cre Atg7fx/fx and LysM-Cre Becn1fx/fx mice53 were as described. Age- and gender-matched colony-mate controls were used for all experiments. Male and female animals were used in accordance with the Animal Welfare Act and the U.S. Public Health Service Policy on Humane Care and Use of Laboratory Animals. Housing conditions were 72+/−2 °F, humidity 40-60%, and dark/light schedule of 12/12 hours. Experiments were reviewed by the Animal Care and Use Committee of the NIEHS.
Cell Culture and reagents.
MEFs were isolated from embryos at day 12.5-14.5 and maintained in culture for maximum 6 passages. BMDMs were differentiated from bone-marrow cells by using L929-supernatant (10%) enriched media for 6 days. Cells were grown in DMEM with 10% FBS (Atlanta Biological) and Penicillin/Streptomycin (Sigma). Interferon-γ (BioLegend #575306), Torin1 (EMD Millipore #475991), Brefeldin A (BD Biosciences #555029), MRT67307 (Sigma #CAS1190378-57-4), LY294002 (Cayman #70920), Bafilomycin A1 (Santa Cruz #sc-201550), E64d (Sigma #E8640), Pepstatin A (Sigma #P5318), 3-Methyladenine (3MA) (Sigma #M9281), MitoTEMPO (Sigma #SML0737), Oligomycin A (Sigma #75351), Antimycin A (Sigma #A8674), and Ethidium bromide (Bio-Rad #161-0433) were purchased.
Plasmids, transduction and transfection.
pCHAC-mt-mKeima-IRES-MCS2 was a generous gift from R. Youle (NINDS/NIH). pMRX-STING-GFP was a generous gift from N. Yan (UTSW). pBMN-YFP-Parkin (#59416) and pBMN-I-GFP (#1736) were bought from Addgene. Cells were transduced with these plasmids for 48 h and sorted for GFP-positive cells (except for mKeima transduced cells) and further incubated for 24-48 h before using for experiments. FACS sorting was done using a BD FACSAria II. The Supplementary Information shows representative flow cytometry gating for GFP vector-transduced and YFP-Parkin-transduced cells. For knockdown experiments, lentiviral scrambled pLKO.1 shRNA and shRNA against TLR9 (Sigma #TRCN0000360485, TRCN0000360556, TRCN0000066085), PINK1 (Sigma #TRCN0000026743, TRCN0000026727), MAVS (Sigma # TRCN0000446155, TRCN0000124770) and TLR7 (Sigma #TRCN0000360477, TRCN0000360478, TRCN0000360550, and TRCN0000065983) were used to transduce BMDMs at day 3 of differentiation in a 10-cm dish, selected with puromycin after 48h and seeded onto 12-well plate. For RNA interference, primary MEFs were transfected in a 12-well plate with 12.5 pmol targeted siRNA and negative control siRNA (Integrated DNA Technologies) using Lipofectamine RNAiMAX (Invitrogen) in ratios recommended by manufacturer. 2′3′-cGAMP (Invivogen #tlrl-nacga23) and 2′5′-GpAp (Invivogen #tlrl-nagpap) were also transfected using the same protocol.
Quantitative-PCR.
Total RNA was isolated from cells and tissues using RNeasy kits (Qiagen), converted to cDNA using cDNA Reverse transcription kit (Applied Biosystems) or iScript™ cDNA synthesis kit (Bio-Rad #1708891) and used for quantitative PCR in Real-Time PCR System (Applied Biosystems). Tissues were lysed using stainless steel beads (5 mm) in TissueLyser II (Qiagen). Validated TaqMan® Gene Expression Assays (Invitrogen) with TaqMan™ Universal PCR Master Mix (Invitrogen) was used to quantitate expression for all the genes. For relative quantification of mitochondrial DNA (mtDNA), whole cell DNA was isolated using DNeasy Blood and tissue kit (Qiagen) and mtDloop1 (Fwd 5′-AATCTACCATCCTCCGTGAAACC-3′, Rev 5′-TCAGTTTAGCTACCCCCAAGTTTAA-3′) and housekeeping Tert (Fwd 5′-CTAGCTCATGTGTCAAGACCCTCTT-3′, Rev 5′-GCCAGCACGTTTCTCTCGTT-3′)4 were amplified using Power SYBR Green Master Mix (Invitrogen). 2−ΔΔCt method was used to analyze the gene expression fold change after normalization with GAPDH or HPRT. QuantStudio Software (ThermoFisher Scientific) was used.
Confocal Microscopy.
Cells grown in 8-chamber slide or glass-bottom dish (MatTek) were fixed using 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, blocked by 3% BSA solution, stained with HSP60 (1:100, Santa Cruz, sc-1052), p62 (1:500, Enzo, #BML-PW9860), LAMP1 (1:50, DSHB 1D4B), LC3B (1:2000, Novus #NB600-1384), Rab5 (1:100, Abcam ab109534 for BMDM and Cell Signaling #3547 for MEF) and dsDNA (1:400, Abcam #ab27156) for 1 h at 22°C followed by secondary antibody (Alexa Fluors, Invitrogen) and mounted with cover slip (1.5 thickness, 22 × 50 or 12 mm diameter) on ProLong Diamond Antifade Mountant with DAPI (Invitrogen). Cells were imaged in a Zeiss LSM 780 inverted confocal microscope with 63× 1.4 NA oil-immersion objective and Zen (Zeiss) and Image J software was used. At least 2 × 2 tile scan of 4-12 random fields were taken for all experiments with fixed cells. Images were analyzed by using ImageJ software plugins – Analyze Particles for cytoplasmic DNA after subtracting Nuclear signal and JaCOP for colocalization to determine Mander’s coefficient. For YFP-Parkin and GFP-Sting overexpressed cells, YFP+ or GFP+ sorted cells (at least 20-30 cells) were analyzed during each experiment by ImageJ as described above. For live cell imaging of mitophagy, mt-mKeima transduced cells were re-seeded in glass bottom dish (MatTek P35GC-1.5-14-C). To evaluate mitophagy in Parkin-overexpressed MEFs, YFP+ cells were first sorted and then transduced with mt-mKeima. Control cells were incubated with HBSS or Bafilomycin (5 h) or Torin1 (8 or 14 h) before imaging. At least 8 images were taken as 1024 × 1024 pixels frame in a 37°C incubation chamber with 40×1.2 W objective and analyzed as described before46. Briefly, to quantify the extent of mitochondrial delivery to lysosomes (Mitolysosomes), pixel area (calculated in ImageJ) from 594 nm excitation (acidic) was normalized with pixel area from 458 nm excitation (neutral). Emission filters of 597-695 was used. For live imaging of MitoTracker and Lysotracker, after co-staining in serum-free media, cells were washed with warm (37°C) PBS and kept in complete media during imaging with 63× 1.4 NA oil-immersion objective. Eight slices were taken in the z plane. Surfaces of mitochondria and lysosomes were constructed in Imaris software. To determine the volume (voxels) of mitochondria-containing lysosomes, the lysosomal channel was masked by setting the inside voxel to 1 and the intensity sum of the new masked channel (i.e. number of lysosomal masked voxels on mitochondria channel) was divided by the total voxels of mitochondria.
Immunoblotting.
Cell lysates were run on a 4-12% gradient Bis Tris gel (Invitrogen) and transferred to a PVDF membrane using standard methods. The membrane was probed with Tubulin (Sigma #T6047; 1:1000), β-actin (Sigma #A3854; 1:14000), TIM23 (BD Biosciences #611222; 1:1000), TBK1 (Abcam #ab40676; 1:1000), phospho-TBK1 S172 (Cell Signaling #5483; 1:800), cGAS (Cell Signaling #31659; 1:800), TFEB (Abcam #ab2636; 1:800), Histone H3 (Abcam #ab1791; 1:1000). Nuclear fractions were isolated using NE-PER™ Nuclear and Cytoplasmic Extraction Reagents (ThermoFisher #7883). Membranes were then washed and exposed (60 min) to 1:2000 HRP-conjugated secondary antibody (GE Healthcare) in 5% milk. After further washes, signal was detected with HyGLO quick spray (Thomas Scientific), followed by film exposure.
Mitochondria and Lysosome measurements.
Tetramethylrhodamine ethyl ester (TMRE) (Invitrogen #T669) and MitoTracker green (Invitrogen #M7514) was both used at 100 nM in serum-free DMEM for 30 min at 37°C. MitoSox red (Invitrogen # M36008) was used at 5 μM in HBSS for 15 min at 37°C. LysoTracker deep red (Invitrogen #L12492) was used at 50 nM for 30 min. To detect cathepsin B activity, Magic red (Immunochemistry Tech. #937) was incubated with cells for 45 min (manufacturer’s protocol was followed). After staining, cells were washed and analyzed by flow cytometry. LysoSensor yellow/blue DND (Invitrogen #L7545) was used 2 μM for 5 min at 37°C. The ratiometric dye was excited by UV laser (355 nm) and fluorescence intensities through 585/42 (545 LP) and 450/50 filters were used to calculate relative pH by flow cytometry. LAMP1 was stained (Invitrogen #53-1071-82, 1:100) after permeabilizing the cells (BD Perm/Wash buffer) and analyzed by flow cytometry (BD LSR Fortessa instrument; BD FACSDiva and FlowJo software for analysis). LC3-GFP was measured after permeabilizing the cells with 0.05% saponin. Representative flow cytometry gating and histograms for LysoTracker Deep Red and Magic Red (cathepsin B activity) are shown in the Supplementary Information.
Nanoparticle treatment.
MEFs (3 × 104) seeded for 8 h were treated with 50 μg photoactivatable acidic nanoparticles32 for 14 h. After washing, cells were treated with IFN-γ for 8 h, exposed to UV 365 nm light (UVP UVLMS-38 EL Series UV Lamp) for 5 min to activate the nanoparticles and lysed after 6 h for RNA isolation.
Autoantibody profiling using autoantigen microarrays.
Autoantibody reactivities against a panel of 124 autoantigens were measured using an autoantigen microarray platform developed by University of Texas Southwestern Medical Center (https://microarray.swmed.edu/products/category/protein-array/) and analyzed by Genepix Pro 7.0 software (Molecular Devices).
Seahorse assay.
MEFs were seeded on Seahorse cell culture plate at 5400 cells per well. Oxygen consumption rate (OCR) was measured by using Seahorse XF Cell Mito Stress Test kit (#103015-100) with serial injection of Oligomycin (2 μM), FCCP (2 μM) and Rotenone/Antimycin A (0.5 μM). Each well was then washed and lysed with 50 μl lysis buffer and BCA protein assay (Thermo Scientific) was performed to determine the protein content and used to normalize the OCR measurements. Seahorse XF96 Analyzer and Wave softwares were used for analysis.
Cytosolic fraction.
Cells grown in 10-cm dish were harvested, washed, and used for Mitochondria/Cytosol Fractionation Kit (Biovision, K256) to isolate mitochondria-enriched and cytosolic fractions following manufacturer’s protocol with slight modifications. Approximately 1 × 106 MEFs and 5 × 106 BMDMs were resuspended in 2 ml of 1× Cytosolic extraction buffer in a 4 ml glass tube compatible with Potter-Elvehjem PTFE pestle (Sigma P7859-1EA). Maximum 6 samples were handled at a time. Cole Palmer mixer (Fisher S111) was used to homogenize samples at 600 rpm with 11-12 strokes on ice. The suspension was centrifuged at 700g for 10 min in 4°C, and the supernatant was again centrifuged at 7000g for 10 min in 4°C. The supernatant from this spin was collected as cytosolic fraction, and the pellet was further washed with 1 ml cytosolic extraction buffer and centrifuged for mitochondria-enriched pellets. Total DNA was isolated from 300-350 μl of cytosolic fraction by using QIAquick Nucleotide Removal Kit (Qiagen) and eluted in 30 μl. mtDNA was quantified using ddPCR copy number assay for ND1 (Bio-rad, Assay ID #dMmuCNS343824284) and β-actin (Bio-Rad, Assay ID #dMmuCNS292036842) in QX200™ Droplet Digital™ PCR System.
mtDNA depletion.
MEFs (0.5 × 105 cells) grown in a 6-well plate were treated with 400 ng/ml ethidium bromide, which was refreshed every 48 h with media. At day 6, MEFs were harvested and seeded for experiments.
Cytokine analysis.
Cytokines were quantified by multiplex assay (Bio-Plex; Bio-Rad Laboratories). For serum IL-10, three values were below the limit of detection (Irgm1−/− [n=2]; Irgm1 Tmem173−/− [n=1]) and were excluded from analysis (Fig. 5e).
Histopathologic analysis.
Tissues were fixed in 10% neutral buffered formalin, trimmed, processed for paraffin, embedding, sectioned (5 μm), and stained with H&E. The slides were scanned using an Aperio slide scanner (Leica Biosystems) and images were captured using Aperio’s ImageScope. Tissues were evaluated for pathology by a board-certified veterinary pathologist.
Statistical analysis.
Analysis was performed using GraphPad Prism software. Data are represented as mean ± SEM. Two-tailed Student’s t test was applied for comparisons of 2 groups and one-way ANOVA for comparisons of >2 groups. For all tests, P < 0.05 was considered significant.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1. Role for type I IFN in disease phenotypes of Irgm1−/− mouse.
a, Expression of interferon-stimulated genes in bronchoalveolar lavage (BAL) cells, bone marrow cells, and spleen of wild type and Irgm1−/− animals (n=3/genotype). b, Autoantibodies against full array of 124 antigens, measured in serum of animals (n=3-4 mice/genotype; each column is an independent mouse). c, Total count of bone marrow cells in wild type (n=8), Irgm1−/−(n=8), and Irgm1−/−Ifnar−/− (n=9) mice. d, Total leukocyte count (WBC), lymphocyte count, platelet count, hemoglobin concentration, and hematocrit in peripheral blood of wild-type (n=5), Ifnar−/− (n=3), Irgm1−/−(n=8), and Irgm1−/−Ifnar−/− (n=6) mice. Data are mean +/− s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001. Two-tailed unpaired t-test (a, d) and one-way ANOVA with Tukey’s adjustment (c).
Extended Data Fig. 2. Mitochondrial abnormalities in Irgm1−/− MEFs.
a,b, Interferon-β (Ifnb1) and interferon-stimulated gene (Ifit1) expression in mouse embryonic fibroblasts (MEFs) (n=3) (a) and bone marrow-derived macrophages (n=3) (b) treated with or without IFN-γ (20 ng/ml, 16h). c, Colocalization of dsDNA and HSP60 immunostaining expressed as Mander’s coefficient (n=4 for all conditions, except n=3 for Irgm1+/++IFN-γ). d, Mitochondrial gene Dloop1 quantified by qPCR in total DNA isolates of MEFs, normalized to nuclear gene Tert (n=7). e, Expression of mtDNA-encoded genes mt16S and mtND4 quantified by qPCR in MEFs (n=3). f, Mitochondrial fractions evaluated for purity by immunoblotting for mitochondrial protein TIM23 and cytosolic protein tubulin. g,h, qPCR of mtDloop1 (n=3) (g) and immunostaining for cytoplasmic dsDNA (h) confirming mitochondrial DNA depletion by EtBr (scale bar, 20 μm). i, Ifit1 expression in MEFs treated with mitochondria-specific antioxidant MitoTempo (50 μM) prior to IFN-γ (n=3). a, b and e-i are representative of at least two independent experiments. d is combination of two independent experiments. Data are mean +/− s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001. Two-tailed unpaired t-test.
Extended Data Fig. 3. cGAS/STING/IRF3 axis in type I IFN response of Irgm1−/− MEFs.
a, Interferon-β (Ifnb1) and Ifit1 expression in murine embryonic fibroblasts (MEFs) transfected with two different siRNAs against cGAS or control siRNA and then treated as shown (n=3). b, Mb21d1 qPCR confirmation of siRNA silencing (n=3). c,d, Mb21d1 expression measured by qPCR (n=3) (c) and cGAS by Western blot (d) in MEFs. e, Sting−/− (i.e., Tmem173−/−; n=53 for control, n=62 for IFN-γ and n=44 for Brefeldin A [BrefA] conditions) and Irgm1−/−Sting−/− (n=49 for control, n=70 for IFN-γ and n=36 for BrefA conditions) MEFs transduced with Sting-GFP were analyzed for colocalization (Mander’s coefficient) of GFP with ER-Golgi intermediate complex (ERGIC)-53. Brefeldin A (2 μg/ml) treatment was used as negative control. f, Ifnb1 expression in MEFs treated with IFN-γ and then transfected with 2 μg/ml cyclic-GAMP or linear GpAp negative control (n=3). g, qPCR confirmation of Irf3 silencing by siRNA (n=3). a-d and f-g are representative of at least two independent experiments. e is combination of two independent experiments. Data are mean +/− s.e.m. *P < 0.05, **P < 0.01. Two-tailed unpaired t-test.
Extended Data Fig. 4. Deficient mitophagic flux in Irgm1−/− MEFs.
a, GFP-Parkin-expressing Irgm1−/− MEFs transduced with mt-mKeima and analyzed for mitolysosome signals. GFP vector served as transduction control. Oligomycin and Antimycin A (O+A; 10 μM each for 5-6h) were used as positive control for mitophagy (n=30 for Irgm1−/−-GFP, Irgm1−/−-Parkin treated with IFN-γ or O+A, n=28 for Irgm1−/−-GFP treated with O+A, n=31 for Irgm1−/−-GFP treated with IFN-γ and n=27 for Irgm1−/−-Parkin control conditions). b, PARKIN-expressing (and GFP control) MEFs assessed for Ifnb1 expression by qPCR (n=3). c, MEFs treated with or without IFN-γ were stained for mitochondria (HSP60) and endosomes (Rab5) and analyzed for colocalization (Mander’s coefficient shown at right) (n=38 fields) (scale bar, 20 μm). d, Nuclear and cytoplasmic fractions were isolated and stained for TFEB. Nuclear Histone H3 and cytoplasmic GAPDH serve as fraction markers. e, Expression of lysosomal biogenesis genes in MEFs (n=3). f, Immortalized MEFs of indicated genotypes were assessed by qPCR for expression of ISGs (Ifit1, Mx2, and Oas1a) (n=3). g, LysoTracker fluorescence fold change in MEFs treated with Torin1 (1 μM, 8h) (n=3). h, Mitophagy measured in mt-mKeima-expressing MEFs treated with Torin1 (n=11). i, Mitochondrial volume colocalizing with lysosomes analyzed by live-imaging of MitoTracker and LysoTracker-stained MEFs (n=18 for all conditions, except n=8 for IFN-γ +Torin). Surface rendering was performed in Imaris software for volumetric analysis. a, d, f and g are representative of two independent experiments. b, e and h are representative of three independent experiments. c and i are combination of three independent experiments. Data are mean +/− s.e.m. *P < 0.05, **P < 0.01, ****P < 0.0001, ns=not significant.Two-tailed unpaired t-test.
Extended Data Fig. 5. Mitophagy controls in Irgm1−/− MEFs.
a, YFP-Parkin expressing MEFs treated with Oligomycin and Antimycin A (O+A, 10 μM each for 6h), stained for HSP60 and analyzed for colocalization (Mander’s coefficient; right panel) of HSP60 with Parkin (n=85 for Irgm1+/+-Parkin and n=100 for Irgm1−/−-Parkin cells) (scale bar, 20 μm). b,c, MEFs treated with O+A analyzed for mitochondrial fragmentation by HSP60 staining (b) and for mitochondrial protein expression (TIM23, COXIV, actin control) by Western blot (c) (scale bar, 20 μm). d, WT MEFs underwent Atg5 silencing with two siRNAs (or control siRNA), were untreated or treated with IFN-γ, and then analyzed by qPCR for Ifnb1, Ifit1, and Atg5 (n=6). e, WT and Atg7−/− MEFs were treated as shown and then analyzed by qPCR for Ifnb1, Ifit1, and Atg7 (n=6). f,g, Silencing efficiency for siRNAs against Pink1 (n=3) (f) and Drp1 (n=3) (g). b, c, f, and g are representative of at least two independent experiments. a, d, and e are combinations of two independent experiments. Data are mean +/− s.e.m. ***P < 0.001, ns = not significant. Two-tailed unpaired t-test.
Extended Data Fig. 6. Tissue-selective role of STING in autoimmune pathology of Irgm1-null mice.
a, Autoantibodies against full array of 124 antigens, measured in serum of animals (n=3-4 mice/genotype; each column is an independent mouse). b-d, Expression of interferon-stimulated genes in lungs (n=6 for all genotypes, except n=4 for Irgm1−/−), salivary glands (n=6 for all genotypes, except n=4 for Irgm1−/−), and spleen of indicated genotypes (n=4 for all genotypes). Data are mean +/− s.e.m. ***P < 0.001, ns = not significant. One-way ANOVA with Tukey’s adjustment.
Extended Data Fig. 7. Effects of cGAS, STING, and TLR9 silencing on type I IFN response of Irgm1−/− macrophages.
a,b, BMDMs of the indicated genotypes were treated as shown and then evaluated for expression of Ifnb1 and interferon-stimulated genes by qPCR. For (a) n=12, and (b) n=6. c, Cytosolic fractions of BMDMs were assayed for mitochondrial (mt)DNA (ND1) and nuclear DNA (β-actin) by digital droplet PCR (n=3). d-f, WT and Irgm1−/− BMDMs were treated with three different lentiviral shRNAs targeting TLR9 (or control shRNA), treated as shown, and then analyzed by qPCR for Ifnb1 and Ifit1 (left) and Tlr9 (right) (n=3). a is a combination of three independent experiments. b and c are combinations of two independent experiments. d and f are representative of three independent experiments. e is a representative of two independent experiments. Data are mean +/− s.e.m. ** p ≤ 0.01, ****p ≤ 0.0001, ns = not significant. Two-tailed unpaired t-test.
Extended Data Fig. 8. No impact of TLR9 deletion on histopathology of the Irgm1−/− mouse.
Representative H&E-stained sections of lungs, salivary glands (submandibular), lacrimal glands, and pancreas from the indicated genotypes (n=5-7/genotype) (all scale bars are 100 μm, except for lacrimal glands of Tlr9−/−, Irgm1−/−, and Irgm1−/−Tlr9−/− [50 μm]).
Extended Data Fig. 9. Role of lysosome and mitochondrial cargo in type I IFN response of Irgm1−/− macrophages.
a, Lysosomal mass analyzed by LysoTracker staining (n=9). b, Relative acidic pH measured by ratiometric Lysosensor yellow/blue dye (n=9). c, Cathepsin B activity assessed by Magic red substrate (n=3). Bafilomycin (BafA1) is used as negative control. d,e, qPCR for indicated targets in BMDM pretreated prior to IFN-γ with BafA1 (100 nM, 2h) (d) or co-treated with protease inhibitors (20 μM E64d, 50 μM pepstatin A) and IFN-γ (e) (n=3). f-h, BMDM silenced for Tlr7 using four different lentiviral shRNAs (or control shRNA) and analyzed for Tlr7 expression (f); silenced for Mavs using two different shRNAs and analyzed for Ifnb1 (g) or Mavs expression (h) (n=3). i, Mean fluorescence intensity (MFI) of LC3-GFP transgenic WT and Irgm1−/− BMDM after washing with 0.05% saponin (n=3). Release of LC3-I was confirmed by microscopy showing only punctate LC3-II signal (not shown). j, BMDM stained for mitochondria (HSP60) and endogenous LC3, expressed as Mander’s coefficient of colocalization (n=10). k, BMDM analyzed for colocalization of HSP60 and endosome (Rab5) (n=30). l, mt-mKeima-expressing BMDM analyzed for mitophagy by flow cytometry using ratiometric measurements at 488 (pH 7) and 561 nm (pH 4) lasers with 610/20 nm emission and 600 nm long pass filters. m, BMDM analyzed for HSP60 and LAMP1 colocalization (n=7). n, Evaluation of knockdown of Pink1 in BMDM (n=3). a and k are combination of three independent experiments. b is combination of two independent experiments. c, d, i, j, l, and m are representative of three independent experiments. e-h and n are representative of two independent experiments. All scale bars are 10 μm. Data are mean +/− s.e.m. #P = 0.06, *P < 0.05, **P < 0.01, ****P < 0.0001, ns = not significant. One-way ANOVA or two-tailed unpaired t-test.
Extended Data Fig. 10. Deletion of ATG5, ATG7, and BECLIN1 does not induce type I IFN.
BMDMs from mice with myeloid-specific deficiency (LysM-Cre-targeted deletion) of Atg7 (a-b), Atg5, (c-d), and Beclin1 (e-f) were treated as shown and analyzed for expression of Ifnb1, Ifit1, and the respective deleted gene targets. Results are a combination of BMDM cultures from two animals (n=6), except for (b) where n=3. CCCP = carbonyl cyanide m-chlorophenyl hydrazine. g-i, Lungs and spleen from naïve mice from the three strains were harvested and analyzed by qPCR for the targets shown. n=6, 58 week-old females for (g), n=4 for LysMCre−Atg5Fx/Fx lungs, n=5 for LysMCre−Atg5Fx/Fx spleen and n=6 for LysMCre+Atg5Fx/Fx lungs and spleens, 9-14 week-old females for (h), and N=3, 9-14 week-old females for (i). Data are mean +/− s.e.m, ns = not significant. Two-tailed unpaired t-test.
Supplementary Material
Acknowledgments
The authors thank P. West, D. Sliter, F. Zhao, and J. Santos for helpful discussions; N. Yan (UTSW) and R. Youle (NINDS/NIH) for reagents; G. Barber (University of Miami) for Tmem173−/− mice; C. Bosio (NIAID/NIH) for Tlr9−/− mice; L. Perrow for assistance with breeding; D. King for blood cell count analysis; the NIEHS histology core laboratory for assistance with processing, sectioning and staining of tissues; K. Gerrish, B. Elgart and N. Clausen of the NIEHS Molecular Genomics Core; N. Martin and D. Chen of the NIEHS Viral Vector Core; C.J. Tucker, A.K. Janoshazi, and E. Scappini of the NIEHS Fluorescence Microscopy and Imaging Center; and C. Bortner and M. Sifre of the NIEHS Flow Cytometry Core Facility. This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (Z01 ES102005 [M.B.F] and ZIA ES103286 [J.Ma]); and by AI135398, AI145929, and AI148243 (G.A.T.); R21AG063373 (M.W.G.); and R21AG060456 (O.S.).
Footnotes
The authors have declared that no conflict of interest exists.
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. Uncropped versions of the immunoblots are archived in the Source Data available online.
References
- 1.Thorlacius GE, Wahren-Herlenius M & Ronnblom L An update on the role of type I interferons in systemic lupus erythematosus and Sjogren's syndrome. Curr Opin Rheumatol 30, 471–481 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Crow YJ & Manel N Aicardi-Goutieres syndrome and the type I interferonopathies. Nature reviews. Immunology 15, 429–440 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Rodero MP & Crow YJ Type I interferon-mediated monogenic autoinflammation: The type I interferonopathies, a conceptual overview. J Exp Med 213, 2527–2538 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.West AP et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520, 553–557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rongvaux A et al. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell 159, 1563–1577 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gkirtzimanaki K et al. IFNalpha Impairs Autophagic Degradation of mtDNA Promoting Autoreactivity of SLE Monocytes in a STING-Dependent Fashion. Cell Rep 25, 921–933 e925 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monteith AJ et al. Defects in lysosomal maturation facilitate the activation of innate sensors in systemic lupus erythematosus. Proceedings of the National Academy of Sciences of the United States of America 113, E2142–2151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caza TN et al. HRES-1/Rab4-mediated depletion of Drp1 impairs mitochondrial homeostasis and represents a target for treatment in SLE. Ann Rheum Dis 73, 1888–1897 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perl A, Gergely P Jr. & Banki K Mitochondrial dysfunction in T cells of patients with systemic lupus erythematosus. Int Rev Immunol 23, 293–313 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Pilla-Moffett D, Barber MF, Taylor GA & Coers J Interferon-Inducible GTPases in Host Resistance, Inflammation and Disease. J Mol Biol 428, 3495–3513 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haldar AK et al. IRG and GBP host resistance factors target aberrant, "non-self" vacuoles characterized by the missing of "self" IRGM proteins. PLoS Pathog 9, e1003414 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao YO, Konen-Waisman S, Taylor GA, Martens S & Howard JC Localisation and mislocalisation of the interferon-inducible immunity-related GTPase, Irgm1 (LRG-47) in mouse cells. PLoS One 5, e8648 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maric-Biresev J et al. Loss of the interferon-gamma-inducible regulatory immunity-related GTPase (IRG), Irgm1, causes activation of effector IRG proteins on lysosomes, damaging lysosomal function and predicting the dramatic susceptibility of Irgm1-deficient mice to infection. BMC biology 14, 33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Traver MK et al. Immunity-related GTPase M (IRGM) proteins influence the localization of guanylate-binding protein 2 (GBP2) by modulating macroautophagy. The Journal of biological chemistry 286, 30471–30480 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Azzam KM et al. Irgm1 coordinately regulates autoimmunity and host defense at select mucosal surfaces. JCI Insight 2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou XJ et al. Genetic association of PRDM1-ATG5 intergenic region and autophagy with systemic lupus erythematosus in a Chinese population. Ann Rheum Dis 70, 1330–1337 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Xia Q et al. Autophagy-related IRGM genes confer susceptibility to ankylosing spondylitis in a Chinese female population: a case-control study. Genes Immun 18, 42–47 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Yao QM et al. Polymorphisms in Autophagy-Related Gene IRGM Are Associated with Susceptibility to Autoimmune Thyroid Diseases. Biomed Res Int 2018, 7959707 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nocturne G & Mariette X Advances in understanding the pathogenesis of primary Sjogren's syndrome. Nature reviews. Rheumatology 9, 544–556 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Feng CG, Weksberg DC, Taylor GA, Sher A & Goodell MA The p47 GTPase Lrg-47 (Irgm1) links host defense and hematopoietic stem cell proliferation. Cell Stem Cell 2, 83–89 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuzawa T et al. IFN-gamma elicits macrophage autophagy via the p38 MAPK signaling pathway. Journal of immunology 189, 813–818 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King KY et al. Irgm1 protects hematopoietic stem cells by negative regulation of IFN signaling. Blood 118, 1525–1533 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.West AP & Shadel GS Mitochondrial DNA in innate immune responses and inflammatory pathology. Nature reviews. Immunology (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakahira K et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nature immunology 12, 222–230 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma F et al. Positive feedback regulation of type I IFN production by the IFN-inducible DNA sensor cGAS. Journal of immunology 194, 1545–1554 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamacher-Brady A & Brady NR Mitophagy programs: mechanisms and physiological implications of mitochondrial targeting by autophagy. Cell Mol Life Sci 73, 775–795 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katayama H, Kogure T, Mizushima N, Yoshimori T & Miyawaki A A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem Biol 18, 1042–1052 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Singh SB, Davis AS, Taylor GA & Deretic V Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science 313, 1438–1441 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Georgakopoulos ND, Wells G & Campanella M The pharmacological regulation of cellular mitophagy. Nat Chem Biol 13, 136–146 (2017). [DOI] [PubMed] [Google Scholar]
- 30.McWilliams TG et al. Phosphorylation of Parkin at serine 65 is essential for its activation in vivo. Open Biol 8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Malta C, Cinque L & Settembre C Transcriptional Regulation of Autophagy: Mechanisms and Diseases. Front Cell Dev Biol 7, 114 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trudeau KM et al. Lysosome acidification by photoactivated nanoparticles restores autophagy under lipotoxicity. J Cell Biol 214, 25–34 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He L, Weber KJ, Diwan A & Schilling JD Inhibition of mTOR reduces lipotoxic cell death in primary macrophages through an autophagy-independent mechanism. J Leukoc Biol 100, 1113–1124 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oka T et al. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature 485, 251–255 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez-Nuevo A et al. Mitochondrial DNA and TLR9 drive muscle inflammation upon Opa1 deficiency. EMBO J 37 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ewald SE et al. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature 456, 658–662 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ewald SE et al. Nucleic acid recognition by Toll-like receptors is coupled to stepwise processing by cathepsins and asparagine endopeptidase. J Exp Med 208, 643–651 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kruger A et al. Human TLR8 senses UR/URR motifs in bacterial and mitochondrial RNA. EMBO reports 16, 1656–1663 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez J et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nature cell biology 17, 893–906 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Caielli S et al. Oxidized mitochondrial nucleoids released by neutrophils drive type I interferon production in human lupus. J Exp Med 213, 697–713 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lama L et al. Development of human cGAS-specific small-molecule inhibitors for repression of dsDNA-triggered interferon expression. Nat Commun 10, 2261 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma S et al. Suppression of systemic autoimmunity by the innate immune adaptor STING. Proceedings of the National Academy of Sciences of the United States of America 112, E710–717 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi B et al. SNAPIN is critical for lysosomal acidification and autophagosome maturation in macrophages. Autophagy 13, 285–301 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vitner EB et al. Induction of the type I interferon response in neurological forms of Gaucher disease. J Neuroinflammation 13, 104 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borralho PM, Rodrigues CM & Steer CJ microRNAs in Mitochondria: An Unexplored Niche. Adv Exp Med Biol 887, 31–51 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Gao S et al. Two novel lncRNAs discovered in human mitochondrial DNA using PacBio full-length transcriptome data. Mitochondrion 38, 41–47 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Chauhan S, Mandell MA & Deretic V IRGM governs the core autophagy machinery to conduct antimicrobial defense. Mol Cell 58, 507–521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar S et al. Mechanism of Stx17 recruitment to autophagosomes via IRGM and mammalian Atg8 proteins. J Cell Biol 217, 997–1013 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sliter DA et al. Parkin and PINK1 mitigate STING-induced inflammation. Nature 561, 258–262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Ajayi TA et al. Crohn's disease IRGM risk alleles are associated with altered gene expression in human tissues. American journal of physiology. Gastrointestinal and liver physiology 316, G95–G105 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collazo CM et al. Inactivation of LRG-47 and IRG-47 reveals a family of interferon gamma-inducible genes with essential, pathogen-specific roles in resistance to infection. J Exp Med 194, 181–188 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hemmi H et al. A Toll-like receptor recognizes bacterial DNA. Nature 408, 740–745 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Martinez J et al. Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature 533, 115–119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. Uncropped versions of the immunoblots are archived in the Source Data available online.