Abstract
Background
The orchid is one of the top ten Chinese flowers and has high ornamental value and elegant color. However, orchids are vulnerable to abiotic stresses during their growth and development, and the molecular mechanism of the abiotic stress response in orchids is unclear. WRKY proteins belong to a transcription factor family that plays important roles in biotic stress, abiotic stress, growth and development in plants, but little is known about the WRKY family in Cymbidium goeringii.
Methods
The specific fragment of the CgWRKY57 gene of C. goeringii was analyzed by bioinformatics. The expression of the CgWRKY57 gene of C. goeringii under 4 °C, 42 °C water and ABA stress as well as different tissues was detected by real-time fluorescence quantitative PCR. CgWRKY57 gene was overexpressed in wild type Arabidopsis thaliana by inflorescence infection method, and the function of transgenic lines under ABA stress was analyzed.
Results
CgWRKY57 was cloned from C. goeringii and found to encode 303 amino acids. The CgWRKY57 protein is an acidic, nonsecreted hydrophilic protein without a signal peptide or transmembrane domain. The CgWRKY57 protein is located to the nucleus and may function intracellularly according to its predicted subcellular localization. A domain analysis and homology comparison showed that the CgWRKY57 protein has a “WRKYGQK” domain and belongs to Group III of the WRKY family, and a phylogenetic analysis demonstrated that CgWRKY57 is closely related to OsWRKY47. CgWRKY57 was expressed in the roots, stems, leaves and floral organs of C. goeringii, and its expression level was highest in the roots according to real-time qPCR analysis. There were significant differences in CgWRKY57 expression under 4 °C, 42 °C ABA and water stress treatments, and its expression changed greatly under ABA stress. The expression of CgWRKY57 in transgenic plants was significantly higher than that in wild type plants under ABA stress, and the root length and germination rate were reduced in transgenic plants compared to wild type plants.
Conclusions
These results indicate that CgWRKY57 overexpression is responsive to ABA stress, and they provide a foundation for future analyses of the biological functions of the WRKY family in C. goeringii.
Keywords: Cymbidium goeringii, WRKY genes, ABA stress, Transgenic
Introduction
WRKY transcription factors (TFs) are vital regulatory proteins and are involved in regulating responses to abiotic stress in plants (Rushton et al., 2010). The first WRKY gene was identified from Ipomoea batatas in 1994 (Ishiguro & Nakamura, 1994). Two WRKY proteins, ABF1 and ABF2, were identified from Avena fatua and their DNA-binding regions had a Zinc-finger motif (Rushton et al., 1995). WRKY proteins have a highly conserved WRKY domain, but these proteins differ in size. The WRKY domain is composed of 60 amino acid residues and has highly conserved “WRKYGQK” sequences in the N-terminal region and zinc finger domains (C-X4-5-C-X22-23-H-X1-H or C-X7-C-X23-H-X1-C) in the C-terminal region (Eulgem et al., 2000). WRKY proteins bind to the cis-acting element W box in the promoters of target genes to regulate stress-related gene expression, allowing plants to achieve stress resistance (Ciolkowski et al., 2008).
Currently, the classification of the WRKY family is based on the genomic characteristics of the Arabidopsis WRKY family (Eulgem et al., 2000). The WRKY family is divided into three groups (I, II and III) according to the number of domains and the types of zinc finger domains. Group I has two WRKY domains and two types of zinc finger domains, while Groups II and III have one WRKY domain per protein. The zinc finger domains of Group II are identical to those of Group I (C-X4-5-C-X22-23-H-X1-H) and different from those of Group III (C-X7-C-X23-H-X1-C) (Chen et al., 2017). In addition, Group II is divided into five subgroups, a, b, c, d, and e, according to sequence similarity. Many studies have shown that WRKY transcription factors play important roles in response to biotic stress and regulation of plant growth and development. However, there have been relatively few studies on their other potential roles, especially biotic stress response (Niu et al., 2012).
More recently, in-depth studies have shown that the WRKY family plays an important role in abiotic stress tolerance in plants (Zhou et al., 2015). Fifty-four OsWRKY genes respond to salt, cold and drought stress in rice (Ramamoorthy et al., 2008). The expressions of 15 TaWRKY genes are reduced by cold, high temperature, NaCl and PEG treatments in Triticum aestivum (Wu et al., 2008). There are 18 AtWRKY genes responded to salt stress and AtWRKY25 or AtWRKY33 overexpression increases salt resistance in A. thaliana (Jiang & Deyholos, 2009). AtWRKY25 also responds to high-temperature stress based on AtWRKY25 mutant and overexpression transgenic lines (Li et al., 2009). There have been reports that AtWRKY25, AtWRKY26 and AtWRKY33 oppositely regulate the cooperation between heat shock proteins and the ethylene-activated signaling pathway in A. thaliana, and this signaling pathway plays a role in the plant heat stress response (Li et al., 2011). The transcription levels of AtWRKY6 and AtWRKY75 are significantly improved under low Pi stress in Arabidopsis (Devaiah, Karthikeyan & Raghothama, 2007; Chen et al., 2009). When HaWRKY76 transgenic lines and wild type Helianthus annuus were subjected to water stress, the transgenic plants showed stronger resistance and increased yield (Raineri, Ribichich & Chan, 2015). The expression of 11 WRKY genes is upregulated in the roots of Actinidia chinensis under water stress, and researchers have speculated that WRKY genes may regulate the transcriptional response to water stress in A. chinensis (Zhang et al., 2015). AtWRKY57 overexpression in Oryza sativa not only improves drought resistance but also tolerance to salt and PEG in rice (Jiang et al., 2016).
Furthermore, a lot of researchers have suggested that the WRKY family plays a key role in the abscisic acid (ABA) response signal network (Qiao, Li & Zhang, 2016). LtWRKY21 from the desert plant Larrea tridentata activates the promoter of the ABA-induced gene, HVA22, and combines VP1 and ABI5 to form a complex acting downstream of ABI1 to control ABA-related gene expression (Chen et al., 2010). The AtWRKY1 knockout mutant displays high sensitivity to ABA and low drought resistance (Qiao, Li & Zhang, 2016). ABO3 induces A. thaliana to respond to ABA and drought stress (Ren et al., 2010). AtWRKY40, AtWRKY18 and AtWRKY60 interact with the ABA receptor CHLH/ABAR, to suppress the inhibition of ABA response genes. Among them, AtWRKY40 is a negative regulator and suppresses the expression of the ABI5 gene (Shang et al., 2010). OsWRKY24 and OsWRKY45 are involved in ABA signal transduction as inhibitory factors, but OsWRKY72 and OsWRKY77 regulate positively ABA inducible promoters in rice (Xie, Ruas & Shen, 2005). CmWRKY10 in transgenic Chrysanthemum morifolium improved tolerance to drought stress compared to wild-type plants and the drought tolerance mechanism was associated with ABA pathway (Jaffar et al., 2016). In Camellia sinensis, CsWRKY2 gene has a higher expression level in leaves. The content of ABA increases when Camellia sinensis is subjected to drought and cold stresses. CsWRKY2 gene plays an important role in plant response to cold and drought stress by regulating the downstream ABA signaling pathway (Wang et al., 2016).
During past years, there are plenty WRKY genes reported in plants including the dicotyledons A. thaliana (74 genes), Glycine max (197 genes), Chaenomeles sinensis (66 genes), Citrullus lanatus (63 genes) and as well as the monocotyledons O. sativa (112) , Sorghum bicolor (68 genes) , Ananas comosus (54 genes) and Zea mays (119) (Rushton et al., 2010; Yang et al., 2018; Tao et al., 2018; Ross, Liu & Shen, 2010; Wei et al., 2012). WRKY superfamily has been one of the biggest families in plants (Zhang & Wang, 2005; Rushton et al., 2010). However, the WRKY family genes of Orchidaceae remain lacking and there are still few reports about the field.
Orchidaceae is one of the largest angiosperm families, representing approximately 10% of all angiosperms, and it includes 700 genera and more than 25,000 species worldwide (Dressler, 1993). The researches mainly focus on the flower development and pigment, mycorrhizal symbiosis, crossbreeding and conservation of Orchidaceae plants. With the development of molecular biology technology, MADS-box and MYB family identification and analysis of their functions have had great achievement. Identification of gene family. WRKY superfamily, one of the biggest families in plants, remains in mystery for Orchidaceae plants. DoWRKY1 was identified from Dendrobium nobile in 2015 and activated the downstream reporter genes, His3 and Ade2 by yeast one-hybrid analysis. Besides, DoWRKY1 gene can improve the synthesis of MeJA and chitosan (Zhao et al., 2015). The expression of DoWRKY5 of D. nobile is upregulated by cold stress treatment and DoWRKY5 can be induced by ABA and source stresses (Jiang et al., 2016). Fifty-two DoWRKY genes were identified by fungi and Dendrobium nobile transcriptomics and genomic analysis. The cis-acting element predictions of DoWRKY promoters show that a promoter includes at least two elements related to stress or hormone response (Wang et al., 2018). When Oncidium was grown under high temperature, the leaves were yellowish and spraying different concentrations of salicylic acid (SA) had a protective effect. OnWRKY1 was cloned from Oncidium and it may be associated with heat resistance (Kong, 2017). Cymbidium goeringii is a ground-living orchid with small flowers in the family Orchidaceae and genus Cymbidium. C. goeringii has high ornamental and economic value because of the elegant color and fragrance of its flower and its beautiful leaves. However, C. goeringii is vulnerable to abiotic stresses, which lead to a decline in ornamental quality and even plant death, during its growth and development. To address the adverse effects of complex abiotic stress on plant growth and development, plants must regulate their gene expression to adapt to abiotic stress conditions. Transcription factors play an important role in this process (Wei et al., 2016). There are few reports on the field of C. goeringii. We identified WRKY family genes from RNA isoform sequencing data of C. goeringii and analyzed several WRKY genes. A CgWRKY gene had higher expression level among them. In this study, the WRKY gene was identified and cloned from C. goeringii and we further explored the response mechanisms of CgWRKY genes to abiotic stress, to lay a foundation for further study of the resistance mechanism and to provide a theoretical basis for genetic breeding of C. goeringii.
Materials and Methods
Plant materials and treatments
The plant material used in the study was C. goeringii cv. Song Mei. ‘Song Mei’ is a well-known and recognized traditional variety, and it has a long history of cultivation in China. The flowers of ‘Song Mei’ are typically plum blossom-shaped, and the outer three petals are round similar to the petals of plum trees. The color is green, the petal has a silkworm-moth shape, and the lip has irregular red dots. The fragrance is mild, and the scape is long with one flower. The leaves are dark green, glossy and beautiful (Sun et al., 2012).
‘Song Mei’ was planted in the orchid germplasm resource conservation nursery of Zhejiang Academy of Agricultural Sciences (30°18′N and 120°11′E) under conditions of 70% relative humidity and 22 °C under natural light. The soil was a mixture of bark block, perlite and peat soil. The growing containers of the orchids are 30 cm high approximately to allow better straight root growth. Several two-year-old plants with strong growth were selected for the following treatments. There were three orchid seedlings per pot. The two year old plants have three or four shoots and pseudobulb per pot approximately.
For the high-temperature stress treatment, plants were cultured in an artificial climate chamber at 42 °C, and the leaves were sampled at 0 h, 2 h, 6 h, 12 h, 24 h and 48 h. For the low-temperature stress treatment, plants were cultured in an artificial climate chamber at 4 °C, and the leaves were sampled at 0 h, 2 h, 6 h, 12 h, 24 h and 48 h. For the water stress treatment, roots were completely immersed in water, and the leaves were sampled at 0 h, 2 h, 6 h, 12 h, 24 h and 48 h. For the ABA stress treatment, leaves were sprayed with 100 µM ABA until dripping, and the leaves were sampled at 0 h, 2 h, 6 h, 12 h, 24 h and 48 h (Ren et al., 2010).
The roots, pseudobulbs and leaves were sampled on April 20th, 2018, with the light intensity of 30 µmol/m2/s approximately. The floral organs including sepals, petals, lips and column, were sampled from two-year-old ‘Song Mei’ plants on October 10th, 2018, with the light intensity of 50 µmol/m2/s approximately.
Each treatment had three groups, and each group included four plants. All samples were cut into approximately 2-cm segments, immediately put into liquid nitrogen and stored at −80 °C.
The transgenic lines used in this study were in the A. thaliana ecotype Col-0 background. The plant materials were cultured in an artificial climate chamber under conditions of 80% relative humidity, 22 °C, light intensity of 150 µmol/m2/s approximately and a 14 h light/10 h dark photoperiod. The T3 transgenic overexpression plants were used for ABA stress treatment. The leaves of A. thaliana were sprayed with 100 µM ABA until dripping and sampled at 0 h, 1 h, 3 h, 6 h and 12 h. All samples were cut into approximately 2-cm segments, immediately put into liquid nitrogen and stored at −80 °C.
WRKY gene cloning
Total RNA was extracted (MiniBEST Plant RNA Extraction kit, Takara, Dalian) from C. goeringii, and first-strand cDNA was synthesized by reverse transcriptase in a volume of 20 µL (PrimeScript RT Master Mix, Takara, Dalian). cDNA was diluted to a suitable concentration for PCR. The primers were designed by Oligo6 software according to the sequence of the CgWRKY gene (Table 1), and the full-length CgWRKY gene was cloned by RT-PCR. The PCR procedure was as follows: 94 °C for 3 min; 32 cycles of 98 °C for 10 s, 56 °C for 15 s, 72 °C for 30 s; and 72 °C for 5 min. The PCR products were detected by 1.5% agarose gel electrophoresis. The amplified fragment was purified and connected to the pEASY-Blunt vector. The vector was then transformed into Trans5 α cells, and the positive clones were tested.
Table 1. Primers used for cloning, RT-qPCR and double enzyme digestion assays.
| Primer name | Primer sequence (5′ → 3′) |
|---|---|
| CgWRKY57-cloning-F | ATGGGATCTCAGGAGGAGG |
| CgWRKY57-cloning-R | CTAACCTTGAAACAGATCGGT |
| CgWRKY57- RT-qPCR-F | CTGCTCTACTCCAAATCGG |
| CgWRKY57- RT- qPCR-R | GGATCAGCCACTCGATTAA |
| CgWRKY57-SmaI-F | CCCGGGATGGGATCTCAGGAGGAGG |
| CgWRKY57-SnaBI-R | TACGTAACCTTGAAACAGATCGGTATG |
| 35S-F | GACGCACAATCCCACTATCC |
Sequence analysis
Online prediction of the physical and chemical properties of the amino acid sequence, including molecular weight and isoelectric point, was conducted by the ExPASy server (http://web.expasy.org/compute_pi/) (Wei et al., 2016). Online prediction of protein subcellular localization was conducted by WOLF PSORT (http://www.genscript.com/psort/wolf_psort.html) (Horton et al., 2007). Online prediction of conserved domains was conducted by SMART (http://smart.embl-heidelberg.de/) (Lvica & Peer, 2018). The multiple sequence alignment was analyzed by Clustal W (http://www.clustal.org/clustal2/) and DNAMAN5.0 software (Lynnon Biosoft, https://www.lynnon.com/dnaman.html) (Larkin et al., 2007). The amino acid sequences of AtWRKY and OsWRKY proteins were downloaded from TAIR 9.0 (http://www.arabidopsis.org/index.jsp) and the Plant Transcription Factor Database (http://planttfdb.cbi.pku.edu.cn/). The phylogenetic tree was constructed by MEGA 7.0 software with the neighbor-joining method (NJ) and a bootstrap value of 1000 (Kumar, Stecher & Tamura, 2016).
Real-time quantitative PCR analysis
RT-qPCR was performed using SYBR Green (Takara) on an ABI qPCR system ABI (StepOne Plus, USA) in a volume of 10 µL. The reaction system included 5 µL of 2 × SYBR Green Premix Ex Taq (Tli RNaseH Plus), 0.2 µL of 50 × ROX Reference Dye, 0.2 µL of forward and reverse primers (10 µM), 1 µL of cDNA template and 3.4 µL of RNase-Free ddH2O. The procedure was as follows: 95 °C for 3 min; and 40 cycles of 95 °C for 3 s and 60 °C for 30 s. The 18S RNA of C. goeringii was used as the reference gene, and the data were analyzed by the 2−ΔΔCt method. All experiments included three independent biological replicates and three technical replicates. The primer sequences used for RT-qPCR are listed in Table 1.
PBI121-CgWRKY57 vector construction and plant transformation
The coding sequence (CDS) of CgWRKY57 was cloned into the PBI121 vector by Sma I/SnaB I double digestion to generate the 35S::CgWRKY57 vector. The primer sequences used for double digestion are listed in Table 1. The recombinant plasmid was introduced into Agrobacterium tumefaciens strain EHA105 and transformed into A. thaliana by the floral dip method. The inflorescences were infected for 60 s and incubated in the dark for 24 h. The resulting plants were screened for resistance until the T3 transgenic plants were obtained.
ABA stress treatment for T3 transgenic plants
Forty-day-old wild-type (WT) Col-0 plants and T3 transgenic plants were cultured in an artificial climate chamber under conditions of 22 °C and 150 µmol/m2/s. The plants were sprayed with 100 µM ABA and incubated for 24 h. The leaves were sampled at 0 h, 1 h, 3 h, 6 h and 12 h, and they were stored at −80 °C.
Determination of germination rate and root length of transgenic plants
Fifty WT Col-0 and T3 transgenic seeds were disinfected and cultured in vitro on culture media with 0 µM ABA, 1 µM ABA and 2 µM ABA, respectively. Three repeated experiments were conducted. After vernalization for 2 days at 4 °C, culture media were placed in an artificial climate chamber under conditions of 80% relative humidity, 22 °C, light intensity of approximately 150 µmol/m2/s, and a 14 h light/10 h dark photoperiod. The in vitro seed germination rate was recorded on the 6th day, and the root length was measured on the 12th day in the 0 µM ABA treatment. The criterion for germination was seeds showing 2 mm long white roots.
Results
CgWRKY57 cloning and analysis of protein physical and chemical properties
The CgWRKY57 sequence was selected from C. goeringii transcriptome data. CgWRKY57 gene was annotated in the Gene Ontology (GO) (Cellular component: nucleus, GO:0005634; Molecular function: transcription factor activity, GO:0003700; sequence-specific DNA binding, GO:0043565), Pfam (PF03106.15) and Clusters of Orthologous Groups (COG) (Functional categories: K: Transcription) databases. Primers were designed to amplify the CgWRKY57 sequence, and the cDNA of C. goeringii was used as a template for PCR. The PCR product was detected by 1% agarose gel electrophoresis, and the amplified fragment was found to match the sequenced fragment (Fig. S1A). The CgWRKY57 ORF is 912 bp in length. The CgWRKY57 protein is acidic, and its molecular weight and isoelectric point were predicted by the ExPASy server. When predicting protein hydrophobicity, if the total average hydrophilicity is positive, the protein is hydrophobic, and larger positive values indicate stronger hydrophobicity. If the total average hydrophobicity is negative, the protein is hydrophilic, and larger negative values indicate stronger hydrophilicity (Dai et al., 2014). Therefore, CgWRKY57 is a hydrophilic protein. Guruprasad assigned a weight value of instability to each of the 400 different dipeptides (DIWV) to compute an instability index and proposed that a protein whose instability index is smaller than 40 is predicted as stable, a value above 40 predicts that the protein may be unstable (Guruprasad, Reddy & Pandit, 1990). CgWRKY57 was predicted to be an unstable protein because its instability index was greater than 40. The CgWRKY57 protein has no signal peptide according to the results of SignalP 4.1 (Fig. S1B). The CgWRKY57 protein is localized outside the membrane without a transmembrane region according to transmembrane helix prediction (Fig. S1C). The CgWRKY57 protein is located to the nucleus and may function intracellularly according to the results of the subcellular localization prediction by PSORT online software (Table 2). The secondary structure of CgWRKY57 was analyzed by SOPMA software and consisted of 62.71% random coil, 19.80% α-helix, 13.86% extended strand and 3.63% β-fold structure (Fig. S1D).
Table 2. Prediction of CgWRKY57 protein subcellular localizations.
| Subcellular structures | CgWRKY57 |
|---|---|
| Nuclear | 11 |
| Mitochondrial matrix | 2 |
| Chloroplast | 1 |
Sequence and phylogenetic tree analysis
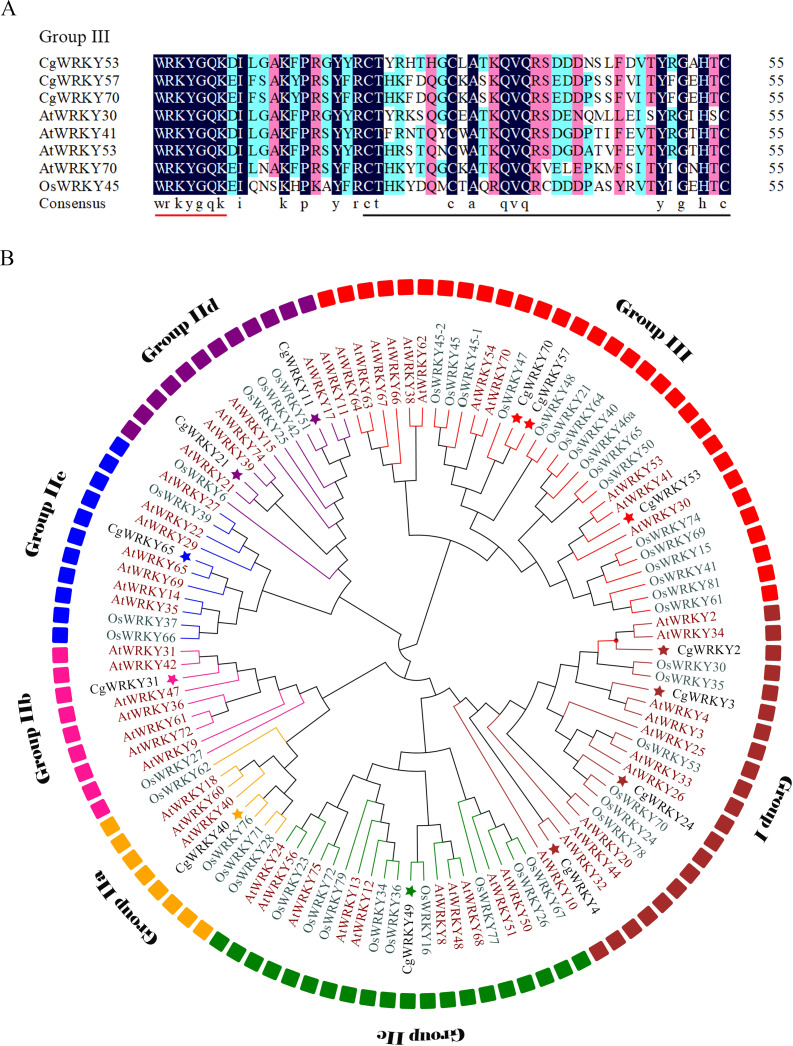
CgWRKY57 has the typical “WRKYGQK” domain and zinc finger domain of the WRKY family according to the SMART online software analysis. A sequence alignment showed that CgWRKY57 has a WRKY domain and a zinc finger domain (C2HC) and that it belongs to Group III (Fig. 1A). To study the phylogenetic relationships among the WRKY proteins of C. goeringii, O. sativa, and A. thaliana, a phylogenetic tree was constructed (Fig. 1B). CgWRKY57 has a close relationship with OsWRKY47, and we hypothesized that they have similar biological functions. Group I genes are reportedly the most ancient WRKY family members, and they may have retained or lost the WRKY domain in the N-terminus. Group II and Group III are the offspring of Group I, and they only have a WRKY domain in the N-terminal (Zhang & Wang, 2005).
Figure 1. Sequence alignment and phylogenetic tree analysis of WRKY domains from C. goeringii, O. sativa, and A. thaliana.
(A) Amino acid sequence alignment of conserved WRKY domains in C. goeringii, O. sativa, and A. thaliana; (B) Phylogenetic tree analysis of CgWRKY57; Black font, C. goeringii; Brown font, A. thaliana; Green font, O. sativa. The phylogenetic tree was constructed by MEGA 7.0 software with the neighbor-joining (NJ) method and a bootstrap value of 1000. The phylogenetic tree was divided into seven subgroups named Groups I, IIA–E, and III.
Phylogenetic tree analysis not only shows the phylogenetic relationships among family members but also provides potential support for functional similarity. WRKY proteins with the same function will group together. For example, AtWRKY54 and AtWRKY70 of Group III are in the same clade, and both regulate leaf senescence (Besseau, Li & Palva, 2012). Similarly, AtWRKY18, AtWRKY40 and AtWRKY60 of Group IIa are in the same branch and negatively regulate the transcription of receptor-like kinase CRK5 (Lu et al., 2016).
Expression patterns of CgWRKY57 in different tissues and under abiotic stresses
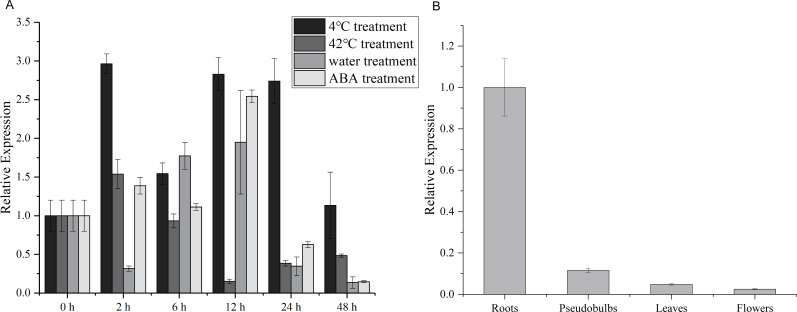
To explore the potential role of the CgWRKY57 gene in response to stress and in four different tissues, RT-qPCR was used to analyze the dynamics of CgWRKY57 under low-temperature, high-temperature, water and ABA stress as well as the expression of CgWRKY57 in four tissues (roots, pseudobulbs, leaves and flowers). The leaves of C. goeringii appeared yellow and aged after 48 h of high temperature or water immersion, and the expression analysis showed that the expression of CgWRKY57 was induced by high-temperature and water stresses (Fig. S2). The results showed that CgWRKY57 was expressed in all the organs and showed different expression levels in different organs (Fig. 2). The expression of CgWRKY57 was highest in roots (P < 0.05), suggesting that CgWRKY57 may be related to root development. The expression of CgWRKY 57 was relatively high at 2 h, 12 h and 24 h after low-temperature treatment; it decreased at 48 h but was still higher than that at 0 h. The expression of CgWRKY57 was enhanced at 2 h after high-temperature treatment but then decreased continuously. The expression of CgWRKY 57 was upregulated during water treatment at 6 h and 12 h and was highest at 12 h, while its expression at 48 h was similar to that at 0 h. After ABA treatment, the expression of CgWRKY 57 showed a downward trend with its highest point at 12 h and its lowest at 48 h. Remarkably, the expression level of CgWRKY57 changed most obviously after ABA stress, and it had a downward trend. Thus, it is predicted that CgWRKY57 responds most strongly to ABA stress.
Figure 2. Expression patterns of CgWRKY57 in different tissues and under abiotic stresses of C. goeringii.
(A) Expression patterns of CgWRKY57 in 4 °C, 42 °C, waterlogging and ABA treatment; (B) Expression patterns of CgWRKY57 in different tissues.
Identification and phenotype observation of transgenic plants overexpressing CgWRKY57
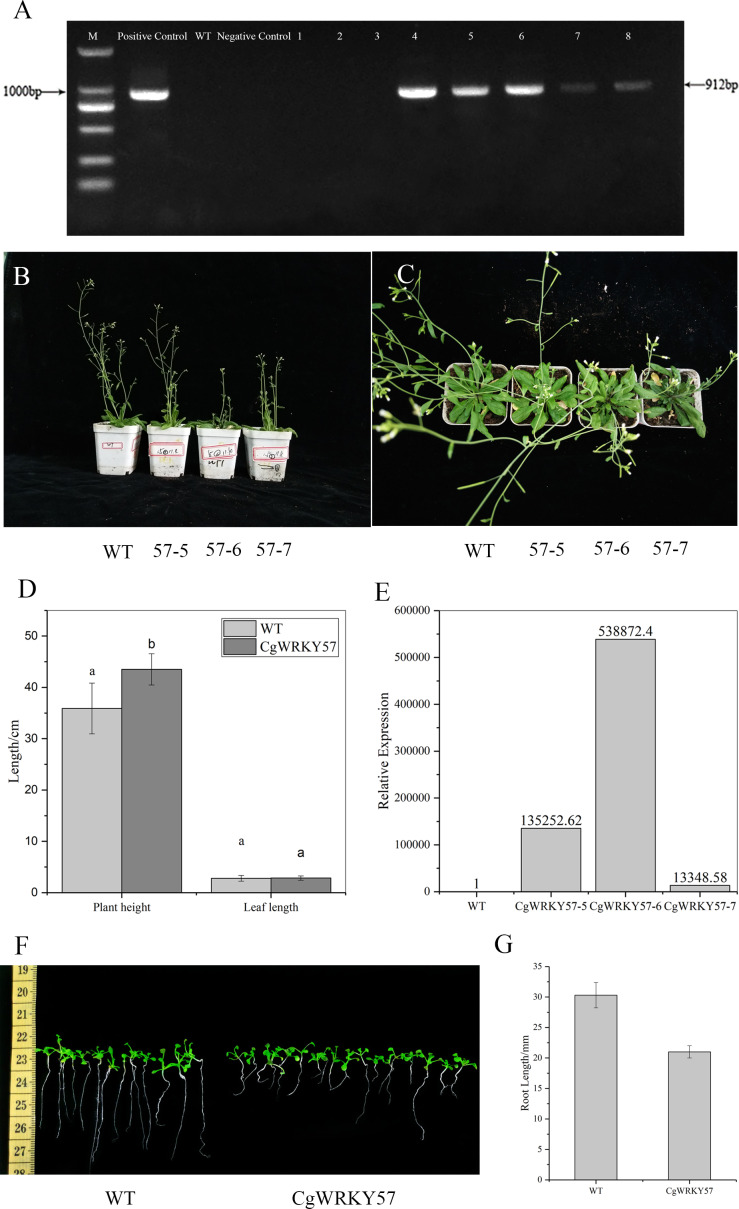
PCR detection was conducted with the 35S forward primer and CgWRKY57 reverse primer using the DNA from T3 transgenic and WT Arabidopsis as well as the PBI121-CgWRKY57 plasmid and water as templates. The results showed that the target product was not amplified from WT Arabidopsis DNA, while T3 transgenic plant DNA allowed amplification. These results indicated that CgWRKY57 was successfully transformed into Arabidopsis (Fig. 3A).
Figure 3. PCR identification and phenotype observation of T3 overexpression CgWRKY57 transgenic plants.
(A) The PCR identification of overexpression CgWRKY57 transgenic plants; (B) side view of overexpression CgWRKY57 transgenic plants and WT plants; (C) top view of overexpression CgWRKY57 transgenic plants and WT plants; (D) the plant height and leaf length of overexpression CgWRKY57 transgenic plants and WT plants; (E) the expression level of overexpression CgWRKY57 transgenic plants and WT plants; (F, G) root length measurements of transgenic plants overexpressing CgWRKY57.
Seven-day-old Arabidopsis seedlings were transplanted into pots and cultured in a light incubator. The phenotype of the transgenic plants was observed after 30 days (Figs. 3B and 3C). Compared to WT Arabidopsis, transgenic plants overexpressing CgWRKY57 grew slowly and were dwarfed. The leaf shape showed no change, but leaf etiolation was obvious (Fig. 3D). The plant height of WT Arabidopsis was about 35.90 cm, but the plant height of overexpressing CgWRKY57 was 43.50 cm (Fig. 3D). The of transgenic plants flowered in 26 days after being transplanted into pots and the inflorescence emerged approximately five days later than that of WT Arabidopsis. Then, we detected the expression level of overexpressing CgWRKY57 and WT Arabidopsis plants (Fig. 3E). The results indicated the CgWRKY57 expression was significantly enhanced in transgenic plants than WT plants.
To measure the root length of transgenic plants overexpressing CgWRKY57, transgenic and WT Arabidopsis were in vitro germinated on MS solid culture medium, and their root lengths were recorded 10 days later. The results showed that the root length of the WT plants was 31 mm and that of the transgenic plants was 20 mm. The roots of the WT Arabidopsis seedlings were long and straight, and those of the transgenic plants were short and curved. The root length of WT Arabidopsis was significantly longer than that of the transgenic plants (Figs. 3F and 3G).
Expression analysis and germination rate of transgenic plants overexpressing CgWRKY57 under ABA stress
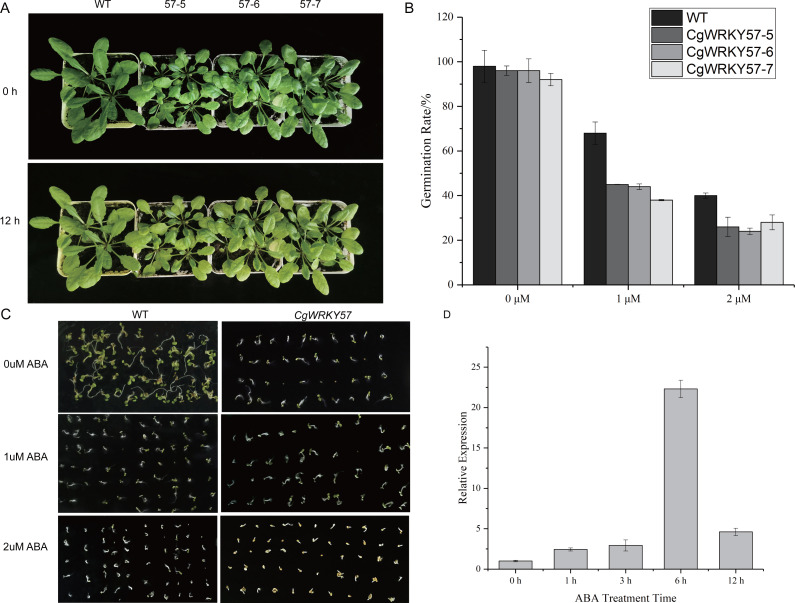
To further study the function of CgWRKY57 under ABA stress, CgWRKY57 transgenic and WT Arabidopsis plants were treated with ABA. At 0 h of ABA treatment, the leaves of the transgenic Arabidopsis plants were yellow, smaller and more wilted than those of the WT plants. At 12 h, the yellow leaves of the transgenic Arabidopsis plants were yellower and weaker than those of the WT plants (Fig. 4A). The expression of CgWRKY57 was detected by RT-qPCR. The results showed that CgWRKY57 was not expressed in WT plants and that its expression in transgenic Arabidopsis was high. The expression level was the lowest at 0 h and increased significantly at 6 h, indicating that CgWRKY57 was responsive to ABA (Fig. 4B).
Figure 4. Phenotype observation and RT-qPCR analysis of transgenic plants overexpressing CgWRKY57 under ABA stress treatment.
(A) Top view of overexpression CgWRKY57 transgenic plants and WT plants under ABA stress treatment; (B) the expression level of overexpression CgWRKY57 transgenic plants under ABA stress treatment; (C, D) germination rates of transgenic plants overexpressing CgWRKY57 under ABA treatment.
Then, we investigated the germination rate of transgenic plants overexpressing CgWRKY57 under ABA stress. CgWRKY57-overexpressing transgenic and WT Arabidopsis were in vitro germinated on culture media with 0 µM ABA, 1 µM ABA and 2 µM ABA. After 7 days, the germination rate was determined. The germination criterion was that the white root length reached 1/2 of the seed length. The germination rates of the transgenic and WT Arabidopsis seeds were both more than 90%. The germination rate of the WT seeds was slightly higher than that of the CgWRKY57 transgenic seeds, but the difference was not significant. Under the 1 µM ABA stress treatment, however, the germination rates of both the WT and transgenic Arabidopsis seeds decreased significantly. The germination rate of WT Arabidopsis seeds was more than 65% and significantly higher than that of the CgWRKY57 transgenic seeds, which was approximately 40%. Under the 2 µM ABA stress treatment, the germination rate of WT Arabidopsis seeds decreased to 40% and that of CgWRKY57 transgenic seeds decreased to approximately 25%. Overall, the germination rate of CgWRKY57 transgenic Arabidopsis seeds decreased more than that of WT seeds compared to the germination rates in the 0 µM ABA treatment, indicating that the germination of CgWRKY57-overexpressing seeds was affected more by ABA (Figs. 4C and 4D).
Discussion
C. goeringii has high ornamental value, but it is vulnerable to abiotic stresses and its ornamental quality will be affected. WRKY transcription factors play important roles in plant growth, development and stress response and have been identified from a variety of plants (Yang et al., 2018). However, there are few reports on the field of C. goeringii. In this study, a CgWRKY protein was obtained and we analyzed the structural characteristics by comparing its conserved WRKY domain to those of previously characterized proteins and constructing a phylogenetic tree. The results indicated that the conserved WRKY domain is indispensable for CgWRKY genes. Many studies have also indicated that the highly conserved WRKY domain is the most prominent feature of the WRKY family (Huang et al., 2012). Many species have variants of the WRKYGQK sequence, such as WRKYGKK, WSKYEQK and WRKYSEK (Mohanta, Park & Bae, 2016). However, the core sequence of the CgWRKY domain contains no mutations, suggesting that the CgWRKY domain is highly conserved and that its amino acid sequence did not undergo a large amount of change during evolution. In addition, the phylogenetic analysis showed that CgWRKY57 belongs to Group III of the WRKY family. The zinc finger domains of different groups of WRKY proteins vary greatly in sequence length and structure, and the zinc finger domain of Group III is C2HC, indicating that the different groups in the WRKY family have undergone different selection pressures and evolution patterns (Bi et al., 2016; Wu, 2005). Phylogenetic analysis not only shows the phylogenetic relationships among members but also provides potential support for functional similarity (Jiang et al., 2014). CgWRKY57 and OsWRKY47 cluster together in a clade, and we hypothesized that they have similar biological functions.
Gene expression patterns provide clues for functional research, and the WRKY family, which is highly expressed in plant tissues, is usually involved in regulating plant growth and development (Yu et al., 2012). TRANSPARENT TESTA GLABRA2 (TTG2) encoding a WRKY TF of Arabidopsis is strongly expressed in trichomes and seed coat throughout their development (Johnson, Kolevski & Smyth, 2002).12 of the 37 members of WRKY TF family in Arabidopsis are found in localized expression domains of roots, indicating a possible specialized role in root cell maturation (Birnbaum et al., 2003). It has been previously reported that SmWRKY2 and SmWRKY24 of Salvia miltiorrhiza are highly expressed in roots with many root hairs, indicating that this gene is related to root morphogenesis (Li et al., 2015). OsWRKY74 is highly expressed in the root, and the Pi content of OsWRKY74-overexpressing plants is significantly higher than that of control plants under phosphate deficiency indicating that OsWRKY74 promotes phosphate absorption by the root (Dai, 2016). As a positive regulator of Pi, AtWRKY75 also affects the development of roots. When AtWRKY75 gene is inhibited, the number of lateral roots and root hairs increases (Devaiah, Karthikeyan & Raghothama, 2007). In the study, a WRKY gene were cloned from C. goeringii, named CgWRKY57. And CgWRKY57 was highly expressed in roots. Above all, these findings suggest that WRKY family is related to the development of roots.
Plants defend against environmental stresses through a complex regulatory network in vivo, and WRKY transcription factors play important roles in this network (Rushton et al., 2010). Plants often encounter water stress, which affects growth and development and reduces oxygen, due to irrigation throughout their life cycles (Voesenek & Bailey-Serres, 2015). Notably, the expression patterns of WRKY genes are similar under different stresses in many studies. The expression of CsWRKY46 in Cucumis sativus is upregulated under cold stress and ABA treatment (Ying et al., 2016), and the expression of SlWRKY3 in Solanum lycopersicum is induced under NaCl, KCl and drought stresses (Hichri et al., 2017). AtWRKY6 plays roles in pathogen defense and aging, and AtWRKY70 is involved in regulating plant growth, drought response and cell death (Robatzek & Somssich, 2010). It has been reported that AtWRKY54 and AtWRKY70 negatively regulate the response to necrotrophic pathogens in Arabidopsis. Increased SA content in the wrky54wrky70 double mutant leads to the accumulation of H2O2 and strengthening of the cell wall, which improves resistance to necrotrophic bacteria in Arabidopsis (Li, Zhong & Palva, 2017). Another study has found that the wrky54wrky70 double mutant shows significantly greater tolerance to osmotic stress, indicating that WRKY70 and WRKY54 are negative regulators of stomatal closure and regulate osmotic stress tolerance in plants (Li et al., 2013). In this study, we analyzed the expression patterns of CgWRKY57 under four stress treatments, including high-temperature, low-temperature, water, and ABA stresses. C. goeringii has fleshy roots, which rot easily under water stress, and this problem can even lead to plant death. The expression of CgWRKY57 under water stress was upregulated at 6 h and 12 h. The expression of AtWRKY22 is also induced rapidly under water stress (Hsu et al., 2013). The AtWRKY22 protein binds to the promoter of TREHALASE1 (TRE1) and inhibits its expression, enhancing water stress resistance by affecting the stomata (Hsu et al., 2013). In addition, AtWRKY22 regulates the expression of MYB15, PUB24 and ACS7, which are related to plant immune responses (Cai et al., 2013). Immune responses in A. thaliana can be caused by water stress, and AtWRKY22 enhances the ability to resist pathogen infection. Both CgWRKY57 and AtWRKY22 belong to the WRKY transcription factor family and have the same WRKY domain. CgWRKY57 belongs to Group III and AtWRKY22 belongs to Group IIe of WRKY family. Thus, we hypothesized that many WRKY family proteins are likely to have similar functions. The expression of CgWRKY57 was downregulated under high-temperature but upregulated under low-temperature stress, indicating that CgWRKY57 was induced by low temperature and inhibited by high temperature. In addition, the degree and timing of CgWRKY57 expression in response to different stresses were significantly different. the expression of CgWRKY57 rapidly reached a peak (at 2 h) under low-temperature treatment, but it did not change at 2 h under water stress, instead increasing rapidly at 6 h and reaching a peak at 12 h. The ABA stress hormone plays a key role in response to abiotic stresses, and WRKY proteins are a key node in the ABA response signal network, acting as activators or inhibitors in this signaling pathway (Qiao, Li & Zhang, 2016; Chi et al., 2013). AtWRKY40 inhibits the expression of ABI5 in response to ABA and negatively regulates the ABA signal transduction pathway by interacting with ABAR, which is a negative regulatory factor (Shang et al., 2010). The expression of CgWRKY57 in C. goeringii was inhibited by ABA treatment, consistent with previous research. Thus, we speculated that CgWRKY57 might also regulate the expression of genes related to ABA stress. These results suggested that CgWRKY57 responded to four abiotic stresses, indicating that CgWRKY57 plays an important role in adaptation to environmental stresses in C. goeringii and a single gene can regulate responses to various stresses. Moreover, the results lay a foundation for further functional study.
WRKY transcription factors constitute one of the largest regulatory protein families in plants and play an important role in response to abiotic stresses. Song et al. transferred OsWRKY72 into Arabidopsis, obtained 35S-OsWRKY72 transgenic Arabidopsis plants and observed their phenotypes. The results showed that the transgenic plants also have abnormal phenotypes, such as leaf senescence and dwarfing, indicating that OsWRKY72 has a strong impact on Arabidopsis morphology (Song et al., 2010). Some studies have shown that AtWRKY53 is significantly upregulated in the early stage of leaf senescence and that its overexpression in transgenic Arabidopsis plants promotes senescence, while silencing of AtWRKY53 delays leaf senescence (Miao et al., 2004). Besseau et al. knocked out AtWRKY54 and AtWRKY70 to obtain an atwrky54-atwrky70 double mutant and found that the leaves of these plants show premature senescence. This result indicated that there might be a synergistic effect between AtWRKY70 and AtWRKY54 in the negative regulation of leaf senescence (Besseau, Li & Palva, 2012). To further study the function of the CgWRKY57 transcription factor, CgWRKY57-overexpressing plants were obtained by the floral dip method. CgWRKY57-overexpressing transgenic plants were dwarfed, and their leaves appeared withered and yellow comparing with WT Arabidopsis, suggesting that overexpression of this gene might positively regulate the plant senescence process. The root length of CgWRKY57 transgenic plants was significantly shorter than that of WT plants, indicating that CgWRKY57 inhibits Arabidopsis root growth.
ABA is a stress hormone with an important role in plant responses to abiotic stress. Previous studies have shown that WRKY proteins positively or negatively regulate ABA signal transduction (Mohanta, Park & Bae, 2016). LtWRKY21 of Larrea tridentata positively regulates the ABA signaling pathway (Zou et al., 2004). Transient expression analysis has shown that OsWRKY24 and OsWRKY45 inhibit ABA signal transduction but that OsWRKY72 and OsWRKY77 positively regulate inducible promoters related to ABA (Xie, Ruas & Shen, 2005). As shown by inserting the atwrky2 T-DNA into the mutant, AtWRKY2 is part of a negative feedback loop in seed germination and later development mediated by ABA (Jiang & Yu, 2009). Another study has shown that AtWRKY18 and AtWRKY60 increased ABA sensitivity, inhibiting seed germination and root growth. Moreover, overexpression of AtWRKY18 and AtWRKY60 increased sensitivity to salt and osmotic stresses (Chen et al., 2010). Transgenic plants overexpressing CgWRKY57 turned yellow under ABA stress and the expression of CgWRKY57 in transgenic plants increased significantly at 6 h of ABA stress. And, the germination rates of both WT and transgenic plants decreased significantly with increasing ABA concentration, and the germination rate of the latter was lower than that of the former. Together, the above results showed that CgWRKY57 has a strong response to ABA and positively regulates the inhibition of ABA. Therefore, it is inferred that CgWRKY57 is sensitive to ABA and may be involved in ABA signal transduction as an inhibitor, but the response mechanism still requires further study.
Conclusion
In this study, the CgWRKY57 gene of C. goeringii was comprehensively analyzed. CgWRKY57 belongs to Group III according to the bioinformatics analysis, and its homology and phylogenetic relationships with rice and Arabidopsis WRKY genes provide valuable clues to its function. The expression patterns of CgWRKY57 in different tissues of C. goeringii and under various abiotic stresses were determined, and the function of CgWRKY57 was characterized in Arabidopsis. These results lay a foundation for further study of the function of CgWRKY57 and provide ideas for future research on breeding C. goeringii for adversity resistance.
Supplemental Information
Funding Statement
This work was supported by the National Natural Science Foundation of China (No. 31801891) and the University Brand Major Construction Foundation of Jiangsu Province (PPZY2015A063). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Yue Chen, Email: earnchen@163.com.
Fengrong Hu, Email: hufengrong2003@sina.com.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Huanhuan Liu performed the experiments, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Lianping Wang performed the experiments, prepared figures and/or tables, and approved the final draft.
Xijun Jing performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Yue Chen and Fengrong Hu conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
DNA Deposition
The following information was supplied regarding the deposition of DNA sequences:
The sequences described here are available in the Supplemental Files and at GenBank: MW228372.
Data Availability
The following information was supplied regarding data availability:
Raw data are available in the Supplemental Files.
References
- Besseau, Li & Palva (2012).Besseau S, Li J, Palva ET. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. Journal of Experimental Botany. 2012;63(7):2667–2679. doi: 10.1093/jxb/err450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi et al. (2016).Bi C, Xu Y, Ye Q, Yin T, Ye N. Genome-wide identification and characterization of WRKY gene family in Salix suchowensis. Peer J. 2016;4(9):e2437. doi: 10.7717/peerj.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum et al. (2003).Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. A gene expression map of the Arabidopsis root. Science. 2003;302(5652):1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- Cai et al. (2013).Cai T, Ye X, Ma Y, Wang X, Chen F, Cheng Z. Over-expression of 1-aminocyclopropane-1-carboxylic acid oxidase genes (ACO) from Vitis vinifera in tomato (Solanum locopersicum) and its effects on ethylene release rates. Journal of Agricultural Biotechnology. 2013;21:1037–1044. doi: 10.3969/j.issn.1674-7968.2013.09.004. [DOI] [Google Scholar]
- Chen et al. (2017).Chen F, Hu Y, Vannozzi A, Wu K, Cai H, Qin Y. The WRKY transcription factor family in model plants and crops. Critical Reviews in Plant Sciences. 2017;36(5–6):311–335. doi: 10.1080/07352689.2018.1441103. [DOI] [Google Scholar]
- Chen et al. (2010).Chen H, Lai Z, Shi J, Xiao Y, Chen Z, Xu X. Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biology. 2010;10:281. doi: 10.1186/1471-2229-10-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen et al. (2009).Chen YF, Li LQ, Xu Q, Kong YH, Wang H, Wu WH. The WRKY6 transcription factor modulates Phosphate1 expression in response to low Pi stress in Arabidopsis. The Plant Cell. 2009;21(11):3554–3566. doi: 10.1105/tpc.108.064980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi et al. (2013).Chi Y, Yang Y, Zhou Y, Zhou J, Fan B, Yu JQ, Chen Z. Protein-protein interactions in the regulation of WRKY transcription factors. Molecular Plant. 2013;6(2):287–300. doi: 10.1093/mp/sst026. [DOI] [PubMed] [Google Scholar]
- Ciolkowski et al. (2008).Ciolkowski I, Wanke D, Birkenbihl RP, Somssich IE. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Molecular Biology. 2008;68(1–2):81–92. doi: 10.1007/s11103-008-9353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai (2016).Dai X. OsWRKY74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice. Journal of Experimental Botany. 2016;67(3):823–826. doi: 10.1093/jxb/erv515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai et al. (2014).Dai MY, Ao T, Xu W, Liu AZ. Sequence and expression analysis of DNA glycosylase in castor bean (Ricinus communis L.) Chinese Journal of Oil Crop Sciences. 2014;36(2):181–188. doi: 10.7505/j.issn.1007-9084.2014.02.007. [DOI] [Google Scholar]
- Devaiah, Karthikeyan & Raghothama (2007).Devaiah BN, Karthikeyan AS, Raghothama KG. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiology. 2007;143(4):1789–1801. doi: 10.1104/pp.106.093971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler (1993).Dressler RL. Phylogeny and classification of the orchid family. Cambridge University Press; Cambridge: 1993. [Google Scholar]
- Eulgem et al. (2000).Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends in Plant Science. 2000;5(5):199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- Guruprasad, Reddy & Pandit (1990).Guruprasad K, Reddy BVB, Pandit MW. Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Engineering. 1990;4(2):155–161. doi: 10.1093/protein/4.2.155. [DOI] [PubMed] [Google Scholar]
- Hichri et al. (2017).Hichri I, Muhovski Y, Žižková E, Dobrev PI, Gharbi E, Franco-Zorrilla JM, Lopez-Vidriero I, Solano R, Clippe A, Errachid A, Motyka V, Lutts S. The Solanum lycopersicum WRKY3 transcription factor SlWRKY3 is involved in salt stress tolerance in tomato. Frontiers in Plant Science. 2017;8:1353. doi: 10.3389/fpls.2017.01343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton et al. (2007).Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Research. 2007;35(1):W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu et al. (2013).Hsu FC, Chou MY, Chou SJ, Li YR, Peng HP, Shih MC. Submergence confers immunity mediated by the WRKY22 transcription factor in Arabidopsis. The Plant Cell. 2013;25:2699–2713. doi: 10.1105/tpc.113.114447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang et al. (2012).Huang S, Gao Y, Liu J, Peng X, Niu X, Fei Z, Cao SQ, Liu YS. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Molecular Genetics and Genomics. 2012;287(6):495–513. doi: 10.1007/s00438-012-0696-6. [DOI] [PubMed] [Google Scholar]
- Ishiguro & Nakamura (1994).Ishiguro S, Nakamura K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and beta-amylase from sweet potato. Molecular and General Genetics. 1994;244(6):563–571. doi: 10.1007/BF00282746. [DOI] [PubMed] [Google Scholar]
- Jaffar et al. (2016).Jaffar MA, Song A, Faheem M, Chen S, Jiang J, Liu C, Fan Q, Chen F. Involvement of CmWRKY10 in drought tolerance of chrysanthemum through the ABA-signaling pathway. International Journal of Molecular Sciences. 2016;17(5):693. doi: 10.3390/ijms17050693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang & Deyholos (2009).Jiang Y, Deyholos MK. Functional characterization of Arabidopsis NaCl-inducible WRKY25, and WRKY33, transcription factors in abiotic stresses. Plant Molecular Biology. 2009;69(1–2):91–105. doi: 10.1007/s11103-008-9408-3. [DOI] [PubMed] [Google Scholar]
- Jiang et al. (2014).Jiang Y, Duan Y, Yin J, Ye S, Zhu J, Zhang F, Lu W, Fan D, Luo K. Genome-wide identification and characterization of the Populus WRKY transcription factor family and analysis of their expression in response to biotic and abiotic stresses. Journal of Experimental Botany. 2014;65(22):6629–6644. doi: 10.1093/jxb/eru381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang et al. (2016).Jiang Y, Qiu Y, Hu Y, Yu D. Heterologous expression of AtWRKY57 confers drought tolerance in Oryza sativa. Frontiers in Plant Science. 2016;7:145. doi: 10.3389/fpls.2016.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang & Yu (2009).Jiang W, Yu D. Arabidopsis WRKY2, transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biology. 2009;9(1):96. doi: 10.1186/1471-2229-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, Kolevski & Smyth (2002).Johnson CS, Kolevski B, Smyth DR. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. The Plant Cell. 2002;14:1359–1375. doi: 10.1105/tpc.001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong (2017).Kong Q. M.S. Thesis. 2017. Physiological response to exogenous salicylic acid on high temperature stress and cloning of WRKY gene in Oncidium. [Google Scholar]
- Kumar, Stecher & Tamura (2016).Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology Evolution. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin et al. (2007).Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Li et al. (2013).Li J, Besseau S, Törönen P, Sipari N, Kollist H, Holm L, Palva ET. Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytologist. 2013;200(2):457–472. doi: 10.1111/nph.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2011).Li S, Fu Q, Chen L, Huang W, Yu D. Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermo tolerance. Planta. 2011;233(6):1237–1252. doi: 10.1007/s00425-011-1375-2. [DOI] [PubMed] [Google Scholar]
- Li et al. (2009).Li SJ, Fu QT, Huang WD, Yu DQ. Functional analysis of an Arabidopsis transcription factor WRKY25 in heat stress. Plant Cell Reports. 2009;28(4):683–693. doi: 10.1007/s00299-008-0666. [DOI] [PubMed] [Google Scholar]
- Li et al. (2015).Li C, Li D, Shao F, Lu S. Molecular cloning and expression analysis of WRKY transcription factor genes in Salvia miltiorrhiza. BMC Genomics. 2015;16:200. doi: 10.1186/s12864-015-1411-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Zhong & Palva (2017).Li J, Zhong R, Palva ET. WRKY70 and its homolog WRKY54 negatively modulate the cell wall-associated defenses to necrotrophic pathogens in Arabidopsis. PLOS ONE. 2017;12(8):e0183731. doi: 10.1371/journal.pone.0183731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu et al. (2016).Lu K, Liang S, Wu Z, Bi C, Yu YT, Ma Y, Wang XF, Zhang DP. Overexpression of an Arabidopsis cysteine-rich receptor-like protein kinase, CRK5, enhances abscisic acid sensitivity and confers drought tolerance. Journal of Experimental Botany. 2016;67:5009–5027. doi: 10.1093/jxb/erw266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lvica & Peer (2018).Lvica L, Peer B. 20 years of the SMART protein domain annotation resource. Nucleic Acids Research. 2016;46(D1):D493–D496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao et al. (2004).Miao Y, Laun T, Zimmermann P, Zentgraf U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Molecular Biology. 2004;55(6):853–867. doi: 10.1007/s11103-005-2142-1. [DOI] [PubMed] [Google Scholar]
- Mohanta, Park & Bae (2016).Mohanta TK, Park YH, Bae H. Novel genomic and evolutionary insight of WRKY transcription factors in plant lineage. Scientific Reports. 2016;6:37309. doi: 10.1038/srep37309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu et al. (2012).Niu CF, Wei W, Zhou QY, Tian AG, Hao YJ, Zhang WK, Ma B, Lin Q, Zhang ZB, Zhang JS, Chen SY. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell & Environment. 2012;35(6):1156–1170. doi: 10.1111/j.1365-3040.2012.02480.x. [DOI] [PubMed] [Google Scholar]
- Qiao, Li & Zhang (2016).Qiao Z, Li CL, Zhang W. WRKY1 regulates stomatal movement in drought-stressed Arabidopsis thaliana. Plant Molecular Biology. 2016;91(1–2):53–65. doi: 10.1007/s11103-016-0441-3. [DOI] [PubMed] [Google Scholar]
- Raineri, Ribichich & Chan (2015).Raineri J, Ribichich KF, Chan RL. The sunflower transcription factor HaWRKY76 confers drought and flood tolerance to Arabidopsis thaliana, plants without yield penalty. Plant Cell Reports. 2015;34(12):2065–2080. doi: 10.1007/s00299-015-1852-3. [DOI] [PubMed] [Google Scholar]
- Ramamoorthy et al. (2008).Ramamoorthy R, Jiang SY, Kumar N, Venkatech PN, Ramachandran S. A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant and Cell Physiology. 2008;49(6):865–879. doi: 10.1093/pcp/pcn061. [DOI] [PubMed] [Google Scholar]
- Ren et al. (2010).Ren X, Chen Z, Liu Y, Zhang H, Zhang M, Liu Q, Hong X, Zhu JK, Gong Z. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant Journal. 2010;63(3):417–429. doi: 10.1111/j.1365-313x.2010.04248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek & Somssich (2010).Robatzek S, Somssich IE. A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant Journal. 2010;28(2):123–133. doi: 10.1046/j.1365-313X.2001.01131.x. [DOI] [PubMed] [Google Scholar]
- Ross, Liu & Shen (2010).Ross CA, Liu Y, Shen QJ. The WRKY gene family in rice (Oryza sativa) Journal of Integrative Plant Biology. 2010;49(6):827–842. doi: 10.1111/j.1744-7909.2007.00504.x. [DOI] [Google Scholar]
- Rushton et al. (1995).Rushton PJ, Macdonald H, Huttly AK, Lazarus CM, Hooley R. Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of alpha-Amy2 genes. Plant Molecular Biology. 1995;29(4):691–702. doi: 10.1007/BF00041160. [DOI] [PubMed] [Google Scholar]
- Rushton et al. (2010).Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends in Plant Science. 2010;15(5):247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Shang et al. (2010).Shang Y, Yan L, Liu ZQ, Cao Z, Mei C, Xin Q, Wu FQ, Wang XF, Du SY, Jiang T, Zhang XF, Zhao R, Sun HL, Liu R, Yu YT, Zhang DP. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. The Plant Cell. 2010;22(6):1909–1935. doi: 10.1105/tpc.110.073874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song et al. (2010).Song Y, Chen L, Zhang L, Yu D. Overexpression of OsWRKY72 gene interferes in the abscisic acid signal and auxin transport pathway of Arabidopsis. Journal of Biosciences. 2010;35(3):459–471. doi: 10.1007/s12038-010-0051-1. [DOI] [PubMed] [Google Scholar]
- Sun et al. (2012).Sun CB, Xiang L, Li B, Qin D, Li X. A new Cymbidium goeringii cultivar ‘Fuwamei’. Acta Horticulturae Sinica. 2012;39(12):2549–2551. [Google Scholar]
- Tao et al. (2018).Tao X, Chen C, Li C, Liu J, Liu C, He Y. Genome-wide investigation of WRKY gene family in pineapple: evolution and expression profiles during development and stress. BMC Genomics. 2018;19(1):490. doi: 10.1186/s12864-018-4880-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voesenek & Bailey-Serres (2015).Voesenek LACJ, Bailey-Serres J. Flood adaptive traits and processes: an overview. New Phytologist. 2015;206:57–73. doi: 10.1111/nph.13209. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2018).Wang T, Song Z, Wei L, Li L. Molecular characterization and expression analysis of WRKY family genes in Dendrobium officinale. Genes & Genomics. 2018;40:265–279. doi: 10.1007/s13258-017-0602-z. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2016).Wang Y, Shu Z, Wang W, Jiang X, Li D, Pan J, Li X. CsWRKY2, a novel WRKY gene from Camellia sinensis, is involved in cold and drought stress responses. Biologia Plantarum. 2016;60(3):1–9. doi: 10.1007/s10535-016-0618-2. [DOI] [Google Scholar]
- Wei et al. (2012).Wei KF, Chen J, Chen YF, Wu LJ, Xie DX. Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Research. 2012;19(2):153–164. doi: 10.1093/dnares/dsr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei et al. (2016).Wei W, Hu Y, Han YT, Zhang K, Zhao FL, Feng JY. The WRKY transcription factors in the diploid woodland strawberry Fragaria vesca: identification and expression analysis under biotic and abiotic stresses. Plant Physiology and Biochemistry. 2016;105:129–144. doi: 10.1016/j.plaphy.2016.04.014. [DOI] [PubMed] [Google Scholar]
- Wu (2005).Wu KL. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Research. 2005;12(1):9–26. doi: 10.1093/dnares/12.1.9. [DOI] [PubMed] [Google Scholar]
- Wu et al. (2008).Wu H, Ni Z, Yao Y, Guo G, Sun Q. Cloning and expression profiles of 15 genes encoding WRKY transcription factor in wheat (Triticum aestivem L.) Progress in Natural Science. 2008;18(6):697–705. doi: 10.1016/j.pnsc.2007.12.006. [DOI] [Google Scholar]
- Xie, Ruas & Shen (2005).Xie Z, Ruas P, Shen QJ. Regulatory networks of the phytohormone abscisic acid. Vitamins & Hormones. 2005;72:235–269. doi: 10.1016/S0083-6729(05)72007-0. [DOI] [PubMed] [Google Scholar]
- Yang et al. (2018).Yang X, Li H, Yang Y, Wang Y, Mo Y, Zhang R, Zhang Y, Ma J, Wei C, Zhang X. Identification and expression analyses of WRKY genes reveal their involvement in growth and abiotic stress response in watermelon (Citrullus lanatus) PLOS ONE. 2018;915-916(1):146–149. doi: 10.1371/journal.pone.0191308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying et al. (2016).Ying Z, Yu H, Yang X, Li Q, Ling J, Wang H, Gu X, Huang S, Jiang W. CsWRKY46, a WRKY transcription factor from cucumber, confers cold resistance in transgenic-plant by regulating a set of cold-stress responsive genes in an ABA-dependent manner. Plant Physiology & Biochemistry. 2016;108:478–487. doi: 10.1016/j.plaphy.2016.08.013. [DOI] [PubMed] [Google Scholar]
- Yu et al. (2012).Yu F, Huaxia Y, Lu W, Cao X, Guo X. GhWRKY15, a member of the WRKY transcription factor family identified from cotton (Gossypium hirsutum L.), is involved in disease resistance and plant development. BMC Plant Biology. 2012;12:144. doi: 10.1186/1471-2229-12-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2015).Zhang J, Huang S, Mo Z, Xuan J, Jia X, Wang G, Guo Z. De novo transcriptome sequencing and comparative analysis of differentially expressed genes in kiwifruit under waterlogging stress. Molecular Breeding. 2015;35(11):208. doi: 10.1016/j.gene.2013.04.066. [DOI] [Google Scholar]
- Zhang & Wang (2005).Zhang Y, Wang L. The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evolutionary Biology. 2005;5(1):1–12. doi: 10.1186/1471-2148-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao et al. (2015).Zhao J, Sun S, Meng C, Jing Q, Fan H, Lin Y, Cai Y. Cloning and expression analysis of transcription factor gene DoWRKY1 in Dendrobium officinale. China Journal of Chinese Materia Medica. 2015;40(14):2807–2813. doi: 10.4268/cjcmm20151423. [DOI] [PubMed] [Google Scholar]
- Zhou et al. (2015).Zhou L, Wang NN, Gong SY, Lu R, Li Y, Li XB. Overexpression of a cotton (Gossypium hirsutum) WRKY gene, GhWRKY34, in Arabidopsis enhances salt-tolerance of the transgenic plants. Plant Physiology & Biochemistry. 2015;96:311–320. doi: 10.1016/j.plaphy.2015.08.016. [DOI] [PubMed] [Google Scholar]
- Zou et al. (2004).Zou XL, Seemann JR, Neuman D, Shen QX. A WRKY gene from creosote bush encodes an activator of the abscisic acid signaling pathway. Journal of Biological Chemistry. 2004;279(53):55770–55779. doi: 10.1074/jbc.m408536200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
Raw data are available in the Supplemental Files.