Abstract
Background
Malignant pleural mesothelioma (MPM) is a rare and aggressive neoplasia affecting the lung mesothelium. Immune checkpoint inhibitors (ICI) in MPM have not been extremely successful, likely due to poor identification of suitable candidate patients for the therapy. We aimed to identify cellular immune fractions associated with clinical outcome and classify patients with MPM based on their immune contexture. For each defined group, we sought for molecular specificities that could help further define our MPM classification at the genomic and transcriptomic level, as well as identify differential therapeutic strategies based on transcriptional signatures predictive of drug response.
Methods
The abundance of 20 immune cell fractions in 516 MPM samples from 7 gene expression datasets was inferred using gene set variation analysis. Identification of clinically relevant fractions was performed with Cox proportional-hazards models adjusted for age, stage, sex, and tumor histology. Immune-based groups were defined based on the identified fractions.
Results
T-helper 2 (TH2) and cytotoxic T (TC) cells were found to be consistently associated with overall survival. Three immune clusters (IG) were subsequently defined based on TH2 and TC immune infiltration levels: IG1 (54.5%) was characterized by high TH2 and low TC levels, IG2 (37%) had either low or high levels of both fractions, and IG3 (8.5%) was defined by low TH2 and high TC levels. IG1 and IG3 groups were associated with worse and better overall survival, respectively. While no differential genomic alterations were identified among immune groups, at the transcriptional level, IG1 samples showed upregulation of proliferation signatures, while IG3 samples presented upregulation of immune and inflammation-related pathways. Finally, the integration of gene expression with functional signatures of drug response showed that IG3 patients might be more likely to respond to ICI.
Conclusions
This study identifies a novel immune-based signature with potential clinical relevance based on TH2 and TC levels, unveiling a fraction of patients with MPM with better prognosis and who might benefit from immune-based therapies. Molecular specificities of the different groups might be used to tailor specific potential therapies in the future.
Keywords: lung neoplasms, gene expression profiling, tumor microenvironment, computational biology, tumor biomarkers
Background
Malignant pleural mesothelioma (MPM) is a rare and highly aggressive neoplasia arising in the pleural cavity, and its incidence is increasing worldwide. MPM tumors are classified into three distinct histological subtypes: epithelioid, biphasic, and sarcomatoid. Median overall survival in patients with MPM is approximately 12 months, although patients with epithelioid tumors usually have better prognosis than individuals with biphasic or sarcomatoid MPM.1 Furthermore, most patients are diagnosed in advanced stages, which still aggravates the burden caused by the disease. The use of surgery in patients with MPM is very limited, and current standard of care treatment for patients with MPM is chemotherapy with platinum combined with pemetrexed, which has been shown to improve overall survival and quality of life.2 However, benefits from chemotherapy are generally modest and prognosis remains dismal with 5-year survival rates lower than 5%.3 Furthermore, there is no standard second-line treatment when tumor progresses to front-line chemotherapy.
Immunotherapy based on monoclonal antibodies against programmed cell death protein 1 (PD-1) and programmed death-ligand 1 (PD-L1) have been tested in clinical trials in MPM. Several nonrandomized phase I/II trials testing single-agent immune checkpoint inhibitors (ICI) showed antitumor activity (9%–29%) and median progression-free survival ranging from 2.8 to 6.2 months.4 Preliminary results from phase II clinical trials combining in second-line inhibitors of cytotoxic T-lymphocyte-associated antigen 4 (CTLA4) plus anti-PD1/PD-L1, such as ipilimumab and nivolumab or tremelimumab and durvalumab, showed promising efficacy results and also significant toxicity.5 In those clinical trials, PD-L1 expression had limited value to predict benefit from ICI. Moreover, tumor mutational burden (TMB) which is generally low in MPM6 is not likely to become a predictive marker in this tumor. Therefore, it is essential to further study the role of the different components of the immune system in the evolution and prognosis of MPM, as well as the identification of additional biomarkers of immunotherapy response beyond PD-L1 immunohistochemical expression.
In this study, our aim is to capture the landscape of the immune microenvironment in a large collection of 516 MPM gene expression profiles comprised in seven different publicly available datasets.6–12 Once the immune contexture is determined for every tumor, we will identify those immune cell fractions with potential prognostic ability and classify MPM samples based on these previously identified clinically relevant immune cell types. We will subsequently use the defined MPM groups to identify genomic and transcriptomic group specificities and evaluate functional signatures of drug response to suggest possible therapeutic strategies.
Methods
Data collection and processing
A systematic search to identify publicly available datasets was performed in January 2018 using the query “pleural mesothelioma” and filtering to keep only human samples and expression profiling experiments in multiple open access repositories. We also filtered datasets with less than 30 samples and kept experiments that covered most of the transcriptome. Raw data were downloaded whenever possible and subsequently processed accordingly as summarized hereafter: for whole-exome sequencing data, Broad’s Genome Analysis Toolkit best practices were followed, RNA-sequencing data were processed according to an in-house pipeline based on ENCODE best practices, and for expression arrays, robust multiarray average algorithm was used to normalize the data. A detailed description of data identification and data processing is available in online supplemental extended methods.
jitc-2020-001601supp002.pdf (58.1KB, pdf)
Association of immune fractions with overall survival
The immune infiltrate composition of 20 different cell types was deconvoluted from the obtained gene expression profiles using gene set variation analysis (GSVA) method.13 Briefly, for each sample and immune cellular fraction, we obtained a score between [−1,+1], with extreme values close to 1 or −1 indicating a relative strong presence or absence of a fraction in a specific sample, respectively. Similarly, GSVA scores close to 0 correspond to an unbiased distribution (ie, centered or randomly distributed) of the genes across the whole gene expression profile. A more detailed explanation on how this algorithm works or why its application is more robust to batch effect than other deconvolution methods can be found in Hänzelmann et al13 and Tamborero et al,14 respectively.
Gene signatures of the different immune fractions were obtained from a previous study15 in which signatures for a total of 24 different immune cell types were generated, including innate immune cells (eg, dendritic cells, eosinophils, mast cells, macrophages, natural killer cells, neutrophils) and adaptive immune cells (eg, B cells, T cells). Since GSVA can only deconvolute multigene signatures, we replaced two single-gene signatures (plasmacytoid dendritic cells and regulatory T cells) with their corresponding multigene signatures from another study.16 Additionally, when more specific categories were available for a cellular fraction (eg, NK CD56dim and NK CD56bright), general categories (eg, NK cells) were removed to avoid signature redundancy.
Once relative abundances for each immune fraction and sample were obtained, Cox proportional-hazards models adjusted for sex, stage, age, and histology were fitted using the largest dataset with available survival information as the discovery dataset,6 and setting the optimal cut point that maximized survival differences for each immune fraction using R packages survival (V.3.1.7) and survMisc (V.0.5.5). Using the GSVA cut-points defined in the discovery dataset, Cox proportional-hazards models adjusted for age, stage, sex, and histology were fitted in two additional validation datasets.7 10 After verifying there was no association of neoadjuvant therapy status with clinical outcome in Bueno et al6 (p=0.2, data not shown), we did not adjust for neoadjuvant therapy status due to lack of information in Bott et al10 and lack of diversity in Hmeljak et al7 (all patients are naïve) in order to have equivalent models to test the association of clinical outcome with the immune fractions. Fractions associated to clinical outcome were determined as those significant (p<0.05) after false discovery rate (FDR) adjustment in the discovery dataset and at least one validation dataset, and with a consistent HR across the three datasets.
Group characterization
Clinicopathological variables
Association with clinicopathological variables (age, sex, stage, tumor histology, asbestos exposure, and neoadjuvant therapy) was done using compareGroups package for R (V.4.2.0). For categorical variables, a χ² test was performed, and continuous variables were tested using Kruskal-Wallis test. Moreover, a multivariate analysis was performed using a multinomial logistic regression model.
Immune checkpoints and T cell exhaustion status
To assess immune checkpoints status and T cell exhaustion status in the different immune groups, multiple sets of genes were obtained from previous studies, including lists of immune checkpoint activators, ICI,17 and T cell exhaustion and effector markers.18 For each gene, we performed a linear model adjusted for sex, age, stage, and histology in each dataset, where the immune group was codified as a numeric variable and the gene expression was the dependent variable. Datasets with less than three samples in any immune group were discarded. The weighted Z method from the survcomp package (V.1.36.0) was used to combine the p-values obtained for the different datasets. Bonferroni adjusted p values lower than 0.05 were considered as significant. β values were combined using the weighted mean.
Genomic alterations
Both Bueno et al6 and Hmeljak et al7 datasets had somatic variant data available to evaluate the mutational burden. The total number of variants per sample was calculated excluding non-exonic and synonymous mutations. Using somatic single nucleotide variants, mutational signatures were inferred using the R package deconstructSigs (V.1.8.0). Neoepitope binding affinity was also derived from somatic single nucleotide variants. More details on these analyses are available in online supplemental extended methods.
Copy number alteration burden was evaluated in the samples from the Hmeljak et al dataset. Fisher tests were computed to assess the differences between immune groups in terms of number of altered/lost/gained genes. R package copynumber (V.1.24.0) was used to plot the genome overview of altered samples.
Transcriptomic analysis
The association of the expression levels of individual genes with the immune groups was done as described in the immune checkpoint evaluation section. For pathway analysis, a preranked gene set enrichment analysis (GSEA) was run with genes ranked using the product of the slope and the negative log10(p value) from the linear model used to assess transcriptomic status at the single-gene level. Gene sets tested included hallmarks and canonical pathways (KEGG, Biocarta, Reactome) from MSigDB V.6.1.19
Assessment of response to therapy
Transcriptional signatures of sensitivity or resistance to specific treatments were used to compute GSVA scores for each sample. The tested signatures were: pemetrexed resistance,20 cisplatin resistance,21 PARP inhibitors sensitivity,22 and palbociclib resistance,23 as well as multiple signatures predictive of benefit to immunotherapy found after a systematic search literature. A total of seven previously published studies with publicly available transcriptomic signatures were found.24–30 Signatures with less than 10 genes were filtered out and linear models were fitted to obtain the slope of the signature correlation with the immune groups. Analysis of variance was used to test for significance of the models adjusted for histology and dataset.
Results
Profiling of immune fractions in a large cohort of MPM tumors leads to clinically relevant immune cell populations
In order to identify clinically relevant immune-cell fractions in MPM tumors, we collected data from seven MPM gene expression datasets that fulfilled our selection criteria.6–12 Overall, we collected 516 MPM gene expression profiles, with available survival information and covariates for 300 patients (58%). A summary of the clinicopathological characteristics of the samples in each dataset is available in online supplemental table S1.
jitc-2020-001601supp003.pdf (96KB, pdf)
For each sample included in the analysis, we quantified the abundance of 20 immune-cell populations by applying GSVA13 on a previously established set of immune-specific gene signatures (online supplemental table S2).15 16
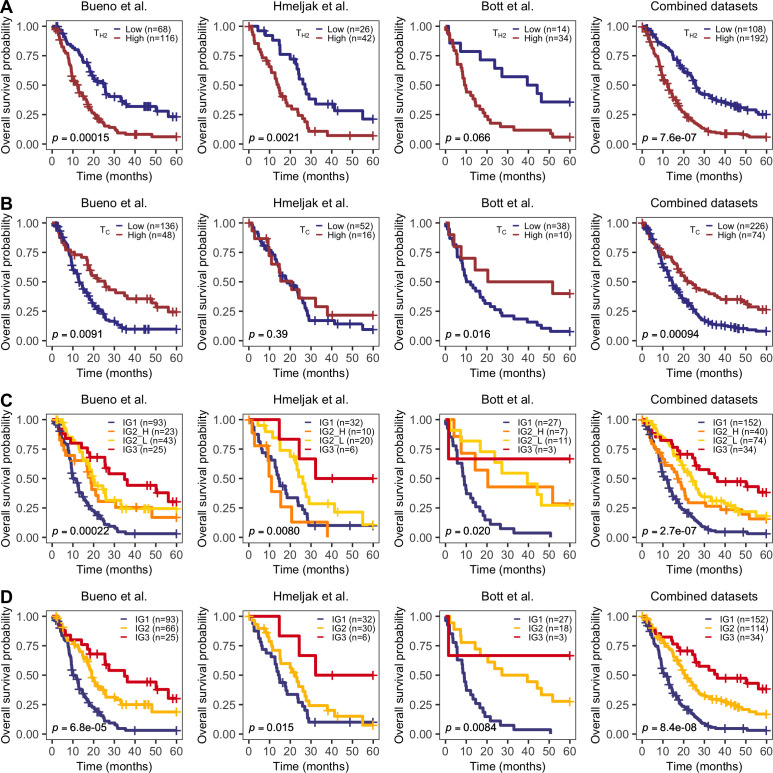
Once the abundance of all immune fractions was quantified in the full cohort of samples, we aimed to assess whether there existed any individual fractions associated with overall patient survival using Cox proportional-hazards regression models adjusted for potential confounders (ie, age, sex, tumor stage, histology, and dataset). Thus, for each cell type, we identified the cut point that maximized the survival differences between high versus low using the largest available dataset (Bueno et al6). The cut points obtained in the discovery dataset were further validated in the two additional cohorts with available survival information (Hmeljak et al7 and Bott et al10). Out of the 20 studied immune cell types, T-helper 2 (TH2) and cytotoxic T cells (TC) showed a consistent association with patient survival across datasets based on our criteria (Cox p<0.05 in at least two of the three datasets, and the remaining dataset showing a consistent trend, although not necessarily significant). In our analysis, higher levels of TH2 cells (GSVA score >−0.16) were associated with worse outcome (HRBueno=2.1, p=0.00015; HRHmeljak=2.6, p=0.0021; HRBott=2.3, p=0.066; HRCombined=2.1, p=7.6·10-7) (figure 1A), while higher levels of TC (GSVA score >0.40) were associated with better prognosis (HRBueno=0.57, p=0.0091; HRHmeljak=0.73, p=0.39; HRBott=0.29, p=0.016; HRCombined=0.59, p=0.00094) (figure 1B). Survival analysis results for the complete set of immune fractions can be found in online supplemental figure S1. When individuals were stratified in four groups based on the combination of the abundance (ie, low versus high) of both TH2 and TC cells, patients with MPM with low abundance of TH2 and high TC infiltration consistently showed the best overall survival when compared with patients with high levels of TH2 and low levels of TC (HRBueno=0.34, p=0.00022; HRHmeljak=0.20, p=0.0080; HRBott=0.12, p=0.020; HRCombined=0.31, p=2.7×10−7) (figure 1C). Additionally, patients with either both high (IG2-H) or low abundance levels of both cell types (IG2-L) showed intermediate prognosis, suggesting some type of compensatory effect between these two immune cell types. Moreover, we do not observe statistically significant differences in survival when comparing IG2-H and IG2-L in any of the datasets. In Hmeljak et al cohort where although IG2-H patients (n=10) have a similar survival to IG1 patients, the HR and 95% CI between IG2-L and IG2-H (HRIG2-L=0.39 (0.15 to 1.02)) is not statistically significant, with IG3 being the only group that shows an improved survival in that cohort. Therefore, we decided to merge these two groups into a single category to create a final classification of three immune-based groups: TH2High-TCLow (IG1, worse prognosis, 54.5% of the patients), TH2High-TCHigh or TH2Low-TCLow (IG2, intermediate prognosis, 37% of the patients, HRCombined=0.52), TH2Low-TCHigh (IG3, better prognosis, 8.5% of the patients, HRCombined=0.32) (figure 1D).
Figure 1.
Overall survival by significant immune fractions. Kaplan-Meier curves of T-helper 2 cells (A), cytotoxic T cells (B), the combination of both fractions in four groups (C), and in three groups (D). HR and p values come from a Cox proportional-hazards model adjusted for age, sex, stage, and histology.
jitc-2020-001601supp001.pdf (10.1MB, pdf)
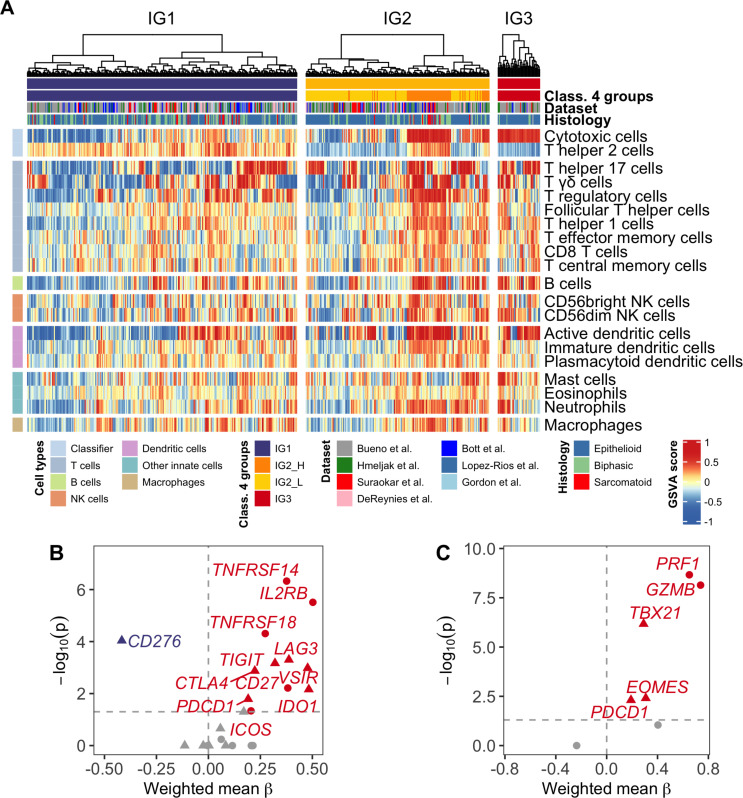
Once the immune-based MPM groups were defined, we aimed to identify any potential differences in the remaining immune fractions among them. Groups IG1 and IG2 comprised a mixture of samples with both strongly and less immune-enriched tumors, while on average, group IG3 showed significant higher abundance for 13 of the 20 immune cell fractions (figure 2A, online supplemental figure S2). These results suggest that the combination of these two immune fractions interplays with the remaining immune cell types and contributes to identify a subset of highly immune-infiltrated MPM tumors with improved prognosis, as already reported in previous publications.14
Figure 2.
Overview of MPM groups at the immune level. (A) Relative abundances of the 20 immune fractions among 516 MPM tumors. Samples are stratified in each group and clustered using Ward’s method and Euclidean distance. Class 4 groups: classification in four groups as reported in figure 1C. (B) Volcano plot of immune checkpoints inhibitors (triangle) and activators (dot). Effect size (β) correlates with immune groups: negative β values correspond to decreasing expression from IG1 to IG3, while positive values indicate increasing expression from IG1 to IG3. Grey-colored dots are not significant. (C) Volcano plot of T-cell exhaustion (triangle) and effector (dot) markers. Effect size (β) correlates with immune groups. Grey-colored dots are not significant. MPM, malignant pleural mesothelioma.
Regarding tumors in IG2 (figure 2A), we observe that samples from the cluster on the left-side are mainly from IG2-L, although 6.06% (n=6/105) is composed by IG2-H samples. On the right-side, consisting mainly of IG2-H samples, 41.0% of the samples (n=25/86) are from IG2-L, suggesting that samples with similar survival rates can have a heterogeneous landscape regarding immune cell abundance.
In order to explore potential additional clinical correlates of the three identified MPM immune groups, we analyzed their association with available clinical covariates: age, sex, tumor stage at the time of diagnosis, tumor histology, asbestos exposure, and neoadjuvant therapy. Out of all the variables explored, we identified a significant association with age and histology (table 1). Patients’ median age in group IG1 is 66 years, compared with 63.7 years and 63.2 years for groups IG2 and IG3, respectively (p=0.021). Regarding tumor histology, IG3 showed a strong enrichment in epithelioid tumors (90.70% in IG3 vs 62.04% in IG1 and 73.68% in IG2, p=0.001).
Table 1.
Summary of clinicopathological variables by immune group
| N | IG1 | IG2 | IG3 | P value | |
| N=281 | N=191 | N=44 | |||
| Age, median (range) | 385 | 66.00 (28.00–86.00) | 63.70 (30.20–84.50) | 63.20 (18.80–76.00) | 0.021 |
| Sex, N (%) | 385 | 0.115 | |||
| Male | 166 (83.42%) | 120 (81.63%) | 27 (69.23%) | ||
| Female | 33 (16.58%) | 27 (18.37%) | 12 (30.77%) | ||
| Stage, N (%) | 348 | 0.888 | |||
| Stage I | 10 (5.32%) | 9 (7.26%) | 3 (8.33%) | ||
| Stage II | 33 (17.55%) | 21 (16.94%) | 7 (19.44%) | ||
| Stage III | 108 (57.45%) | 68 (54.84%) | 22 (61.11%) | ||
| Stage IV | 37 (19.68%) | 26 (20.97%) | 4 (11.11%) | ||
| Histology, N (%) | 507 | 0.001 | |||
| Epithelioid | 170 (62.04%) | 140 (73.68%) | 39 (90.70%) | ||
| Biphasic | 84 (30.66%) | 39 (20.53%) | 4 (9.30%) | ||
| Sarcomatoid | 20 (7.30%) | 11 (5.79%) | 0 (0.00%) | ||
| Asbestos exposure, N (%) | 351 | 0.068 | |||
| No | 41 (22.53%) | 42 (31.82%) | 15 (40.54%) | ||
| Possible | 1 (0.55%) | 2 (1.52%) | 1 (2.70%) | ||
| Yes | 140 (76.92%) | 88 (66.67%) | 21 (56.76%) | ||
| Neoadjuvant therapy, N (%) | 325 | 0.11 | |||
| No | 163 (93.68%) | 101 (87.07%) | 30 (85.71%) | ||
| Yes | 11 (6.32%) | 15 (12.93%) | 5 (14.29%) |
A multivariate analysis with the observations with complete data for all the covariates (n=231) confirmed the association with histology and age, and the lack of association of our classification with sex and stage (online supplemental table S3). A slight increase of asbestos exposure in IG2 as well as a marginal association with neoadjuvant therapy in IG2 was also observed.
To further exclude a potential confounding role of the tumor histology on the differential survival among the immune groups, we evaluated whether the immune-based classification still was associated with patient survival within both epithelioid and non-epithelioid tumors. In epithelioid tumors, we observed improved survival in IG2 and IG3 patients compared with IG1 (HRIG2=0.47, HRIG3=0.24, p=3.3×10−8) (online supplemental figure S3A). For non-epithelioid tumors, the very small number of IG3 patients (n=3) prevented us from obtaining reliable results for this subgroup, but we still observed a trend towards better survival in IG2 patients compared with IG1 (HRIG2=0.60, p=0.074) (online supplemental figure S3B).
Finally, a comparison with previously published classifications6 7 9 was done. While the immune-based classification was not associated to De Reyniès et al9 classification, it was associated to Bueno et al6 classification, where IG3 is enriched in samples from Epithelioid subtype, and Hmeljak et al7 classification, where IG3 is enriched in S1 samples which is also enriched in epithelioid histology and has the best prognosis.
Immune groups correlate with most immune checkpoint markers and markers of effector and exhausted T cells
To further evaluate the potential clinical relevance of the identified immune groups, we evaluated the expression of a wide selection of ICI and activators. We observed a significant increasing expression pattern across MPM immune groups for most of the evaluated checkpoint markers (figure 2B). TNFRSF14, IL2RB, TNFRSF18 (GITR), CD27, and ICOS were among the strongest positively associated activator markers with the immune groups. Regarding ICI, LAG3, TIGIT, VSIR (VISTA), IDO1, CTLA4, and PDCD1 (PD-1) were the strongest correlated markers. In opposition, CD276 (B7-H3) showed negative correlation with the immune groups, being its expression higher in IG1 tumors than tumors in IG2 and IG3 groups. Detailed expression of CD274 (PD-L1), PDCD1 (PD-1), CTLA4, CD276 (B7-H3), and VSIR (VISTA) in each dataset is available in online supplemental figure S4A, showing the distribution of tumors in each group. An overall higher expression of VSIR with respect to the other immune checkpoints is observed, as previously reported in other publications.7 31 Importantly, immune groups still show significant differences in prognosis independently of the expression level of any of these five immune checkpoints (online supplemental figure S4B). Additionally, we also evaluated markers of effector and exhausted T cells correlated with the identified MPM immune groups (figure 2C). In our analysis, all evaluated markers—excluding CD44—showed a positive correlation with the immune group classification, with a majority being significantly enriched in IG3 (PRF1, GZB, TBX21, EOMES, PDCD1).
Genomic characterization shows no differences in genomic instability among immune groups
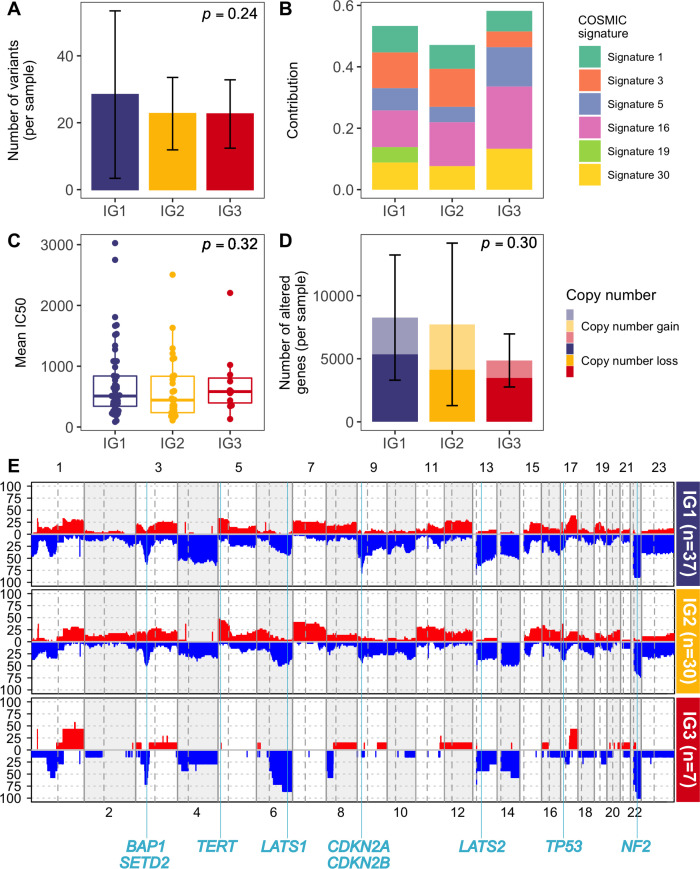
Once we identified the different immune-based groups in MPM tumors and established their potential clinical relevance, we wanted to characterize the different groups at the genomic level to identify any potential differential genomic patterns. These analyses were performed using Bueno et al6 and Hmeljak et al7 datasets.
Using whole-exome data from Bueno et al6 and Hmeljak et al7 datasets, we assessed if immune groups were different in terms of TMB, mutational signatures,32 or antigen immunogenicity (figure 3A–C). Although we did not find statistically significant differences among immune groups for any of the previous analyses, we did observe a slight decrease of mutational Signature 3 in IG3 tumors compared with IG1 and IG2 patients. Signature 3 is associated with homologous recombination deficiency and BRCA1/2 deficiency.32 Additionally, at the single-gene level, no gene was found to be preferentially mutated in any group (data not shown).
Figure 3.
Genomic characterization. (A) Tumor mutational burden among immune groups. Mean number of variants per sample. Error bars indicate SD. (B) Distribution of COSMIC mutational signatures among each immune group. (C) Immunogenicity as the mean of IC50 affinity of mutations among immune groups. (D) Copy number burden among immune groups. Mean number of altered genes per sample. Error bars indicate SD. Lighter colors show copy number gains while darker ones depict copy number losses. (E) Genomic overview showing the percentage of samples with copy number alterations among each immune group. Landmark MPM genes are depicted according to their genomic location. MPM, malignant pleural mesothelioma.
Copy number alterations were assessed for Hmeljak et al dataset.7 There were no significant differences between immune groups in the overall number of altered genes per sample, although tumors in IG3 tend to have fewer copy number alterations overall compared with the other groups as shown in figure 3D. Figure 3E shows the overall percentage of altered samples across the genome for each immune group. Regarding MPM landmark genes, genes like BAP1, SETD2, LATS2, TP53, and NF2 did not show differences between groups. CDKN2A is altered in 81% of IG1 patients, 47% of IG2 patients, and 14% of IG3 patients, but it does not reach statistical significance after multiple testing correction, probably due to small sample size. Online supplemental figure S5 shows expression of frequent tumor suppressor genes inactivated in MPM.3 For the case of CDKN2A, we see an increasing expression IG1<IG2<IG3, which is in agreement with the pattern observed at the genomic level.
We also evaluated differences in methylation data at the single-gene level and no significant results were found after adjusting for multiple testing (data not shown).
Transcriptional cell cycle/immune system activity trade-off across immune groups
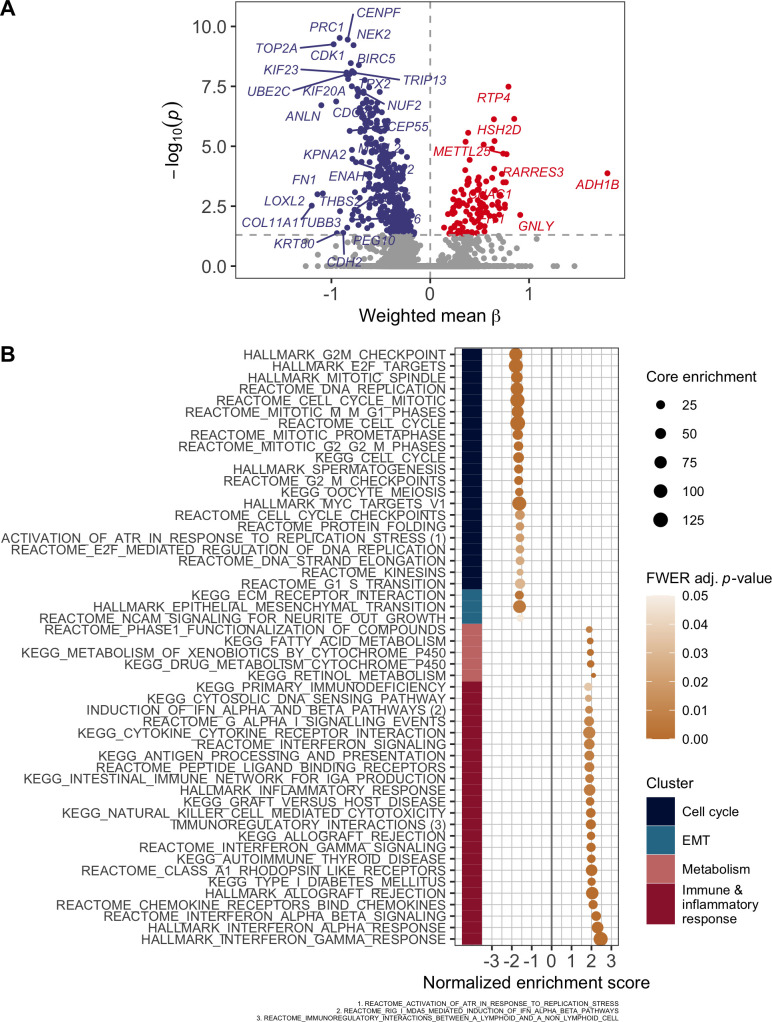
Next, we sought for gene expression differences among the three immune groups, to identify those genes with an increasing or decreasing pattern among the three groups. The results are depicted in figure 4A, which shows a high number of genes with decreasing expression from IG1 to IG3 tumors related to cell proliferation such as CENPF (centromere protein also known as mitosin), BIRC5 (otherwise known as survivin, preventing apoptotic cell death), or kinesins like KIF23, involved in cell division. On the other hand, genes positively correlated with the immune groups include immune system related genes such as HSH2D, a target of T-cell activation signaling pathways, and GNLY, which encodes for a protein present in cytotoxic granules of cytotoxic T lymphocytes.
Figure 4.
Transcriptomic characterization. (A) Volcano plot of gene expression. Effect size (β) correlates with immune groups. (B) Significant pathways from preranked GSEA. Positive normalized enrichment scores correlate with an increasing expression of pathway from IG1 to IG3, while negative ones depict higher pathway expression for IG1 tumors versus IG3 tumors. Pathways are clustered using Ward’s method and an OR-based distance. GSEA, gene set enrichment analysis.
In order to capture more precisely the enriched pathways and biological functions of the genes identified in the previous analysis, we performed a preranked GSEA using a set of transcriptional signatures covering a wide range of pathways, and the genes ranked according to their linear association with the immune groups. The results are shown in figure 4B and we observed that signatures more positively correlated with the immune group (ie, more expressed in IG3 tumors) were mostly related to immune system and inflammatory response processes. Contrarily, signatures negatively correlated with the immune groups mostly relate to cell cycle and epithelial mesenchymal transition, suggesting a trade-off between these molecular processes and the immune system.
Finally, since IG1 tumors appeared to be driven by cell cycle deregulation, we assessed the potential confounder effect among cell proliferation and immune groups. Therefore, we tested the association between the surrogate cell proliferation marker MKI67 and the immune groups. Notably, regardless of MKI67 expression levels, the association of the improving survival pattern across immune groups is upheld (online supplemental figure S6), meaning that our classification is not confounded with cell proliferation.
Potential therapeutic strategies
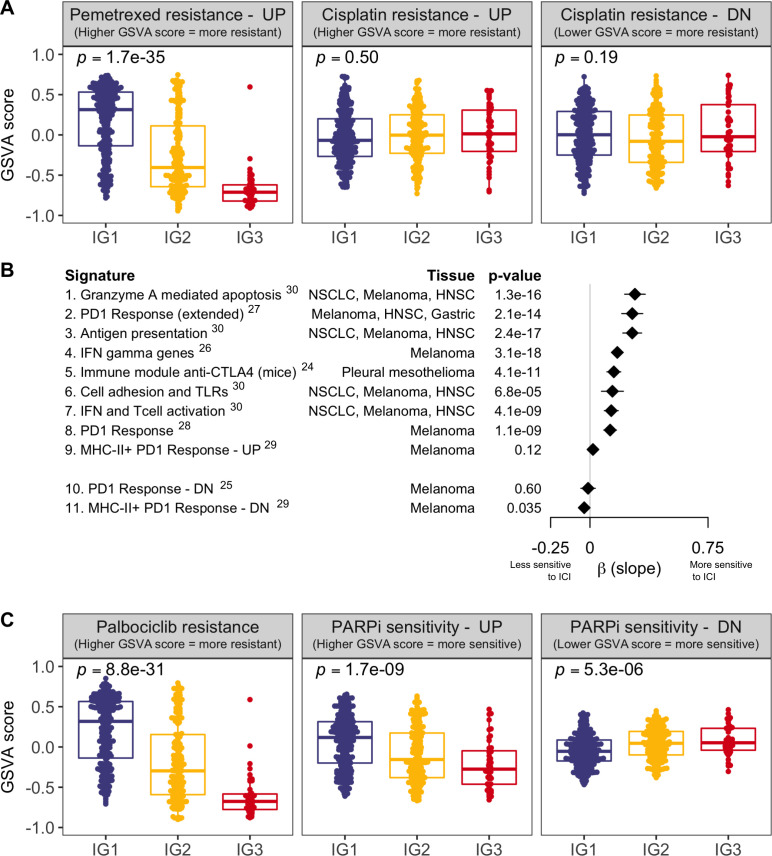
To further evaluate the potential clinical benefit of the immune-based MPM classification, we decided to assess the behavior of different treatment-response signatures among the three groups. We first wanted to identify whether there were any differences in the response to the currently available standard chemotherapy treatment based on the combination of cisplatin and pemetrexed. We obtained two expression-based signatures derived from non-small cell lung cancer that predicted resistance to pemetrexed20 and to cisplatin.21 Figure 5A displays GSVA scores for the two signatures across the three MPM immune groups. Overall, the results of the analysis suggested that on average, MPM tumors in IG1 might be more resistant to pemetrexed, while IG3 tumors show a trend to lower resistance. On the other hand, cisplatin resistance analysis showed no differences between immune groups.
Figure 5.
Assessment of gene expression signatures predictive of benefit or resistance to multiple treatments. (A) Enrichment assessment for signatures of resistance to currently established first-line chemotherapy in patients with MPM (pemetrexed—left panel, cisplatin—mid and right panels). Each MPM tumor is represented with a dot, and GSVA scores (y-axis) indicate upregulation or downregulation of the signature in each patient (as indicated in each plot title). Signature of resistance to cisplatin is divided into two subsignatures of upregulated and downregulated genes in cisplatin resistant cells. (B) Assessment of response to multiple ICI therapies. Each line represents a different ICI predictive signature. Signatures 1–9 are upregulated in ICI responders, while signatures 10 and 11 are downregulated in ICI responders, based on the literature. For each signature, we represent the β coefficient and the 95% CI of the linear model of the GSVA scores across the three immune groups. Positive β values indicate increasing GSVA scores (IG1<IG2<IG3) and negative β values indicate decreasing GSVA scores (IG1>IG2>IG3). Superscripts in immunotherapy signatures correspond to bibliographic references (C) Analogously to panel A, assessment for signatures of resistance or sensitivity to targeted therapy (palbociclib—left panel, PARP inhibitors—mid and right panels). Signature of sensitivity to PARP inhibitors is divided into two subsignatures of upregulated and downregulated genes in PARP inhibitors sensitive cells. GSVA, gene set variation analysis; HNSC, head and neck squamous carcinoma; ICI, immune checkpoint inhibitor; IFN, interferon; MHC, major histocompatibility complex; NSCLC, non-small cell lung carcinoma; PARP, poly ADP ribose polymerase; PD1, programmed cell death protein 1; TLR, toll-like receptor; UP/DN, up/down-regulated gene signature in either sensitive or resistant cells for the specified treatment.
Next, we tested a large set of gene-expression signatures predictive of benefit from ICI treatment24–30 to formally evaluate if MPM tumors in IG3 could benefit from an immune-based therapeutic approach (figure 5B). The overlap between these signatures and the classifier is almost non-existent (three shared genes at most). We observed that all signatures were positively correlated with the immune groups (ie, higher GSVA scores in IG3 for upregulated signatures (signatures 1 to 9) and lower GSVA scores in IG3 for downregulated signatures (signatures 10 and 11)), with IG3 tumors presenting a significant association for all but two of the signatures (signatures 9 and 10).
Given the increased activation of cell cycle and DNA repair processes in the IG1 group of MPM tumors, we wanted to explore whether any differences in sensitivity to cyclin-dependent kinase inhibitors and to drugs that inhibit DNA repair among the three groups. To do that, we collected a signature of resistance to palbociclib derived from breast cancer23 and another computationally-derived signature predicting sensitivity to DNA repair inhibition with PARP inhibitors22 (figure 5C). Despite a higher cycling nature of tumors in group IG1, they appear to be more resistant to palbociclib than tumors in other groups, based on this predictive signature. Strikingly, tumors in IG1 showed increased sensitivity to PARP inhibitors compared with the other groups.
Discussion
In this study, we performed a comprehensive analysis of publicly available transcriptome data from more than 500 MPM tumors in order to characterize the immune landscape of this disease. By the identification of immune cells associated with clinical outcome, we classified MPM tumors according to their T-helper 2 and cytotoxic T cell abundance levels. This classification stratifies patients into three groups that represent different immune infiltration patterns and are associated with distinct survival outcomes. The group with the shortest overall survival, IG1, represents more than 50% of the analyzed tumors, while IG3, with better prognosis, accounts for only 8.5% of the tumors. We observed an increasing pattern of abundance for most immune fractions across the three immune groups. Interestingly, the survival benefit of this classification is consistent in both epithelioid and non-epithelioid tumors. We further characterized these three MPM immune groups at the genomic and transcriptomic levels and identified potential therapeutic strategies using predictive signatures and large-scale pharmacogenomics data.
Different immune-related MPM phenotypes have already been described in recent studies using comprehensive approaches.33–37 These studies use different techniques such as Nanostring technology,34 mass cytometry,33 or RNA-sequencing.36 The diversity of obtained results suggests that the composition and role of the tumor microenvironment in MPM is remarkably complex and controversial, and thus further studies will contribute to elucidate this question.
In our work, MPM tumors are classified based on TH2 and TC cells abundance, as previously stated. TH2 are CD4+ T cells which are induced by the presence of interleukin 4 (IL4) via STAT6 signaling and regulate humoral immune responses and responses to extracellular pathogens. TH2 cells secrete IL4, IL5, IL10, and IL13 which promote immune suppression by inhibiting T-helper 1 cytokine production. Additionally, a recent study identified higher levels of IL5 and IL13 after exposure to asbestos in mesothelial cells.38 The role of TH2 cells in cancer has been found to be dual and context-dependent. While they can generate antitumor immunity by recruiting specific populations of innate immune cells, they have also been found to increase tumorigenicity in certain experimental models.39 Consistent with our results, higher levels of TH2 cells have already been associated with poor prognosis in multiple cancer types.40–42 Finally, in the analysis performed by Hmeljak et al,7 this signature has already been associated to the group with the worst prognosis.
On the other hand, CD8+ TC cells are essential actors of the effector function of adaptive cellular immune response.43 Along with natural killer (NK) cells, TC cells are ultimately responsible for targeting and attacking cancer cells by secreting cytotoxins (eg, granzymes and perforins) which reach the tumor cell cytoplasm and trigger a caspase-mediated apoptotic process.44 Additionally, in agreement with our study, a recent pan-cancer study observed that patients with tumors with higher TC infiltration tended to have better survival than patients with less TC-infiltrated tumors.14
While most immune checkpoints correlate with the defined immune groups, CD276 (B7-H3) shows an opposite pattern of expression, with decreasing expression from IG1 to IG3. CD276 is a member of the B7 family of immunoregulatory proteins and is overexpressed in distinct tumor types. It has been shown that CD276 can promote tumor proliferation, angiogenesis and metastasis and is associated with shorter survival time.45 This result ties in with the survival pattern among the groups as well as with results from functional analysis in which cell cycle shows a decreasing pattern from IG1 to IG3. Similarly, CD44 is the only T-cell exhaustion marker that shows negative correlation with the immune groups. This marker has been associated with metastasis and low survival rate in multiple cancer types, and chemoresistance in prostate and head and neck squamous cell cancer types.46 In MPM, CD44 has been shown to promote invasiveness when interacting with hyaluronan.47
Regarding molecular characteristics, immune groups were not significantly correlated with specific genomic alterations in terms of gene mutations or copy number alterations, and although there seems to be a pattern of decreasing genomic instability among the groups, there is no statistically significant evidence to support this observation. In terms of copy number, no global differences were found among MPM groups, possibly due to the smaller sample size (only one dataset with available data). However, genomic regions like 9p21, harboring landmark genes in MPM as CDKN2A or type I interferon gene cluster, were likely to be lost in the IG1 compared with the other two groups. We would require larger copy number data in order to further validate this apparent lack of differences in genomic alterations among the three immune groups given that previous studies have reported that tumors with more aneuploidy overexpress cell proliferation markers and underexpress immune infiltration markers48 which are the main characteristics of IG1 tumors.
Transcriptomic characterization of these groups by gene enrichment analysis revealed a trade-off between immune response activity and cell proliferation mechanisms, showing higher activation of cell cycle in IG1 tumors, in agreement with genomic characteristics. This trade-off has already been described in other studies, reinforcing the hypothesis that cell cycle regulators and genomic instability could also have an impact on immune checkpoints.14 49 Besides, IFN activation has also been associated with better clinical outcomes in other studies.30
Finally, the lack of effective therapeutic strategies beyond chemotherapy in MPM calls for assessment of potential options and tailoring for each subset of patients. Currently, several phase III clinical trials are assessing the role of ICI in combination with chemotherapy in the first-line setting of patients with advanced MPM. The results of the CheckMate-743 trial evaluating nivolumab plus ipilimumab versus platinum-based chemotherapy in previously untreated MPM50 showed the survival benefit from ICI treatment. This trial also reports that ICI treatment performs better than chemotherapy in non-epithelioid samples, and this might seem contradictory with our results. In fact, the survival benefit of ICI treatment was observed, regardless of histology (in non-epithelioid samples chemotherapy performs worse, thus giving an impression of higher performance for ICI treatment), and PD-L1 ≥1% was also predictive of benefit, which is higher expressed in IG3 tumors from our classification. In this sense, the treatment landscape of advanced MPM is likely to evolve and predictive markers of ICI benefit are needed. In our work, we observed that patients belonging to IG3 may benefit from pemetrexed and immunotherapy. IG3 molecular characteristics have been associated with non-progressive disease after treatment with ICI in another study.30 Also, no group showed any strong predisposition to benefit from chemotherapy with cisplatin treatment, although we were unable to assess this predisposition using a signature that included both therapies used in standard of care.
Moreover, our data might suggest that tumors in IG1 could be more sensitive to PARP inhibitors. However, the reasons for this increased sensitivity remain unclear, since we do not observe differences at the genomic level among the three immune groups.
It is important to stress that the presented classification in three groups is based on clinical outcome and this can generate some heterogeneity at the molecular level, as shown in IG2 group immune cell abundances. These results need further validation in a prospective cohort in order to confirm these findings. Moreover, further research in prospective cohorts could contribute to the identification of the most suitable therapeutic strategy for patients in each immune group. It is also important to point out that this study focused on gene expression signatures as proxies since there is no information regarding immunotherapy in large pharmacogenomic assays and also that even though pemetrexed and cisplatin are commonly administered in combination; we assessed their effect as single therapies due to the lack of signatures for the combined treatment.
To sum up, this study identifies a novel signature with potential clinical relevance based on TH2 and TC levels and unveils a small fraction of patients with MPM that show better prognosis. These groups have different molecular characteristics, mainly at the transcriptomic level. These differences might be useful to tailor potential therapies for each group in the future. Further research is needed towards the identification of a signature valid for formalin-fixed, paraffin-embedded (FFPE) samples, and validation of these results is warranted in an independent and prospective cohort of patients with MPM preferentially treated with ICI.
Acknowledgments
We thank CERCA Programme/Generalitat de Catalunya for institutional support. We thank all the researchers who kindly put their research data for public use. Bueno et al data were accessed through a data transfer agreement with Genentech (DAT-015429). We thank the reviewers for their time and for contributing to improve the manuscript.
Footnotes
Twitter: @NadalErnest, @xsole1977
Contributors: XS and EN conceived and designed the study. AA and DC performed the computational and statistical analysis. XS, EN, AA, DC, SHP, and EA contributed to the interpretation of the results. XS, EN, AA, and DC drafted the manuscript. ALD, RM, RP, RL, IE, RR, SP, and VM provided critical revisions of the article. All authors read and approved the final manuscript.
Funding: This work is supported by the Carlos III National Health Institute funded by FEDER funds—a way to build Europe—(PI14/01109; PI18/00920); the Government of Catalonia (2017SGR448). XS is supported by RTI2018-102134-A-I00 grant funded by Spanish Ministry of Science and Innovation. EN received support from the SLT006/17/00127 grant, funded by the Department of Health of the Generalitat de Catalunya by the call “Acció instrumental d’intensificació de professionals de la salut”. This study has been funded by Sociedad Española de Oncología Médica (SEOM) through “Proyectos de Investigación para Grupo Emergente”.
Competing interests: EN participated in advisory boards from Bristol Myers Squibb, Merck Sharp & Dohme, Lilly, Roche, Pfizer, Takeda, Boehringer Ingelheim, Amgen, and AstraZeneca. VM is consultant to Bioiberica S.A.U. and Grupo Ferrer S.A., received research funds from Universal DX and is coinvestigator in grants with Aniling.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data sharing not applicable as no datasets were generated and/or analyzed for this study. Data are available under the accession IDs provided by the researchers who kindly put their research data for public use.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
References
- 1.Meyerhoff RR, Yang C-FJ, Speicher PJ, et al. Impact of mesothelioma histologic subtype on outcomes in the surveillance, epidemiology, and end results database. J Surg Res 2015;196:23–32. 10.1016/j.jss.2015.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kindler HL, Ismaila N, Armato SG, et al. Treatment of malignant pleural mesothelioma: American Society of clinical oncology clinical practice guideline. J Clin Oncol 2018;36:1343–73. 10.1200/JCO.2017.76.6394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yap TA, Aerts JG, Popat S, et al. Novel insights into mesothelioma biology and implications for therapy. Nat Rev Cancer 2017;17:475–88. 10.1038/nrc.2017.42 [DOI] [PubMed] [Google Scholar]
- 4.McCambridge AJ, Napolitano A, Mansfield AS, et al. Progress in the Management of Malignant Pleural Mesothelioma in 2017. J Thorac Oncol 2018;13:606–23. 10.1016/j.jtho.2018.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scherpereel A, Wallyn F, Albelda SM, et al. Novel therapies for malignant pleural mesothelioma. Lancet Oncol 2018;19:e161–72. 10.1016/S1470-2045(18)30100-1 [DOI] [PubMed] [Google Scholar]
- 6.Bueno R, Stawiski EW, Goldstein LD, et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat Genet 2016;48:407–16. 10.1038/ng.3520 [DOI] [PubMed] [Google Scholar]
- 7.Hmeljak J, Sanchez-Vega F, Hoadley KA, et al. Integrative molecular characterization of malignant pleural mesothelioma. Cancer Discov 2018;8:1548–65. 10.1158/2159-8290.CD-18-0804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suraokar MB, Nunez MI, Diao L, et al. Expression profiling stratifies mesothelioma tumors and signifies deregulation of spindle checkpoint pathway and microtubule network with therapeutic implications. Ann Oncol 2014;25:1184–92. 10.1093/annonc/mdu127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Reyniès A, Jaurand M-C, Renier A, et al. Molecular classification of malignant pleural mesothelioma: identification of a poor prognosis subgroup linked to the epithelial-to-mesenchymal transition. Clin Cancer Res 2014;20:1323–34. 10.1158/1078-0432.CCR-13-2429 [DOI] [PubMed] [Google Scholar]
- 10.Bott M, Brevet M, Taylor BS, et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet 2011;43:668–72. 10.1038/ng.855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.López-Ríos F, Chuai S, Flores R, et al. Global gene expression profiling of pleural mesotheliomas: overexpression of Aurora kinases and P16/CDKN2A deletion as prognostic factors and critical evaluation of microarray-based prognostic prediction. Cancer Res 2006;66:2970–9. 10.1158/0008-5472.CAN-05-3907 [DOI] [PubMed] [Google Scholar]
- 12.Gordon GJ, Rockwell GN, Jensen RV, et al. Identification of novel candidate oncogenes and tumor suppressors in malignant pleural mesothelioma using large-scale transcriptional profiling. Am J Pathol 2005;166:1827–40. 10.1016/S0002-9440(10)62492-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics 2013;14:7. 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamborero D, Rubio-Perez C, Muiños F, et al. A pan-cancer landscape of interactions between solid tumors and infiltrating immune cell populations. Clin Cancer Res 2018;24:3717–28. 10.1158/1078-0432.CCR-17-3509 [DOI] [PubMed] [Google Scholar]
- 15.Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39:782–95. 10.1016/j.immuni.2013.10.003 [DOI] [PubMed] [Google Scholar]
- 16.Charoentong P, Finotello F, Angelova M, et al. Pan-Cancer Immunogenomic analyses reveal Genotype-Immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep 2017;18:248–62. 10.1016/j.celrep.2016.12.019 [DOI] [PubMed] [Google Scholar]
- 17.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012;12:252–64. 10.1038/nrc3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 2015;15:486–99. 10.1038/nri3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hou J, Lambers M, den Hamer B, et al. Expression profiling-based subtyping identifies novel non-small cell lung cancer subgroups and implicates putative resistance to pemetrexed therapy. J Thorac Oncol 2012;7:105–14. 10.1097/JTO.0b013e3182352a45 [DOI] [PubMed] [Google Scholar]
- 21.Whiteside MA, Chen D-T, Desmond RA, et al. A novel time-course cDNA microarray analysis method identifies genes associated with the development of cisplatin resistance. Oncogene 2004;23:744–52. 10.1038/sj.onc.1207164 [DOI] [PubMed] [Google Scholar]
- 22.McGrail DJ, Lin CC-J, Garnett J, et al. Improved prediction of PARP inhibitor response and identification of synergizing agents through use of a novel gene expression signature generation algorithm. NPJ Syst Biol Appl 2017;3:8. 10.1038/s41540-017-0011-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malorni L, Piazza S, Ciani Y, et al. A gene expression signature of retinoblastoma loss-of-function is a predictive biomarker of resistance to palbociclib in breast cancer cell lines and is prognostic in patients with ER positive early breast cancer. Oncotarget 2016;7:68012–22. 10.18632/oncotarget.12010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lesterhuis WJ, Rinaldi C, Jones A, et al. Network analysis of immunotherapy-induced regressing tumours identifies novel synergistic drug combinations. Sci Rep 2015;5:12298. 10.1038/srep12298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hugo W, Zaretsky JM, Sun L, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 2016;165:35–44. 10.1016/j.cell.2016.02.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao J, Shi LZ, Zhao H, et al. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell 2016;167:397–404. 10.1016/j.cell.2016.08.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ayers M, Lunceford J, Nebozhyn M, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930–40. 10.1172/JCI91190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen P-L, Roh W, Reuben A, et al. Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov 2016;6:827–37. 10.1158/2159-8290.CD-15-1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson DB, Estrada MV, Salgado R, et al. Melanoma-Specific MHC-II expression represents a tumour-autonomous phenotype and predicts response to anti-PD-1/PD-L1 therapy. Nat Commun 2016;7:10582. 10.1038/ncomms10582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prat A, Navarro A, Paré L, et al. Immune-Related gene expression profiling after PD-1 blockade in non-small cell lung carcinoma, head and neck squamous cell carcinoma, and melanoma. Cancer Res 2017;77:3540–50. 10.1158/0008-5472.CAN-16-3556 [DOI] [PubMed] [Google Scholar]
- 31.Muller S, Victoria Lai W, Adusumilli PS, et al. V-Domain Ig-containing suppressor of T-cell activation (vista), a potentially targetable immune checkpoint molecule, is highly expressed in epithelioid malignant pleural mesothelioma. Mod Pathol 2020;33:303–11. 10.1038/s41379-019-0364-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415–21. 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee H-S, Jang H-J, Choi JM, et al. Comprehensive immunoproteogenomic analyses of malignant pleural mesothelioma. JCI Insight 2018;3. 10.1172/jci.insight.98575. [Epub ahead of print: 05 04 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patil NS, Righi L, Koeppen H, et al. Molecular and histopathological characterization of the tumor immune microenvironment in advanced stage of malignant pleural mesothelioma. J Thorac Oncol 2018;13:124–33. 10.1016/j.jtho.2017.09.1968 [DOI] [PubMed] [Google Scholar]
- 35.Blum Y, Meiller C, Quetel L, et al. Dissecting heterogeneity in malignant pleural mesothelioma through histo-molecular gradients for clinical applications. Nat Commun 2019;10:1333. 10.1038/s41467-019-09307-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alcala N, Mangiante L, Le-Stang N, et al. Redefining malignant pleural mesothelioma types as a continuum uncovers immune-vascular interactions. EBioMedicine 2019;48:191–202. 10.1016/j.ebiom.2019.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sneddon S, Rive CM, Ma S, et al. Identification of a CD8+ T-cell response to a predicted neoantigen in malignant mesothelioma. Oncoimmunology 2020;9:1684713. 10.1080/2162402X.2019.1684713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maki Y, Nishimura Y, Toyooka S, et al. The proliferative effects of asbestos-exposed peripheral blood mononuclear cells on mesothelial cells. Oncol Lett 2016;11:3308–16. 10.3892/ol.2016.4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liudahl SM, Coussens LM. To help or to harm. In: Immunology. Elsevier 2018:97–116. [Google Scholar]
- 40.De Monte L, Reni M, Tassi E, et al. Intratumor T helper type 2 cell infiltrate correlates with cancer-associated fibroblast thymic stromal lymphopoietin production and reduced survival in pancreatic cancer. J Exp Med 2011;208:469–78. 10.1084/jem.20101876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kusuda T, Shigemasa K, Arihiro K, et al. Relative expression levels of Th1 and Th2 cytokine mRNA are independent prognostic factors in patients with ovarian cancer. Oncol Rep 2005;13:1153–8. 10.3892/or.13.6.1153 [DOI] [PubMed] [Google Scholar]
- 42.Nevala WK, Vachon CM, Leontovich AA, et al. Evidence of systemic Th2-driven chronic inflammation in patients with metastatic melanoma. Clin Cancer Res 2009;15:1931–9. 10.1158/1078-0432.CCR-08-1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andersen MH, Schrama D, Thor Straten P, et al. Cytotoxic T cells. J Invest Dermatol 2006;126:32–41. 10.1038/sj.jid.5700001 [DOI] [PubMed] [Google Scholar]
- 44.Martínez-Lostao L, Anel A, Pardo J. How do cytotoxic lymphocytes kill cancer cells? Clin Cancer Res 2015;21:5047–56. 10.1158/1078-0432.CCR-15-0685 [DOI] [PubMed] [Google Scholar]
- 45.Seaman S, Zhu Z, Saha S, et al. Eradication of tumors through simultaneous ablation of CD276/B7-H3-Positive tumor cells and tumor vasculature. Cancer Cell 2017;31:501–15. 10.1016/j.ccell.2017.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen C, Zhao S, Karnad A, et al. The biology and role of CD44 in cancer progression: therapeutic implications. J Hematol Oncol 2018;11:64. 10.1186/s13045-018-0605-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cortes-Dericks L, Schmid RA. Cd44 and its ligand hyaluronan as potential biomarkers in malignant pleural mesothelioma: evidence and perspectives. Respir Res 2017;18:58. 10.1186/s12931-017-0546-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davoli T, Uno H, Wooten EC, et al. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 2017;355. 10.1126/science.aaf8399. [Epub ahead of print: 20 01 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wellenstein MD, de Visser KE. Cancer-Cell-Intrinsic mechanisms shaping the tumor immune landscape. Immunity 2018;48:399–416. 10.1016/j.immuni.2018.03.004 [DOI] [PubMed] [Google Scholar]
- 50.Baas P, Scherpereel A, Nowak A, et al. ID:2908 first-line nivolumab + ipilimumab vs chemotherapy in unresectable malignant pleural mesothelioma: CheckMate 743. Journal of Thoracic Oncology 2020;15:e42. 10.1016/j.jtho.2020.08.00432093860 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-001601supp002.pdf (58.1KB, pdf)
jitc-2020-001601supp003.pdf (96KB, pdf)
jitc-2020-001601supp001.pdf (10.1MB, pdf)