Abstract
Minimal residual disease (MRD) is the most powerful prognostic factor in pediatric acute lymphoblastic leukemia (ALL). Real-time quantitative polymerase chain reaction (RQ-PCR) represents the gold standard for molecular MRD assessment and risk-based stratification of front-line treatment. In the protocols of the Italian Association of Pediatric Hematology and Oncology (AIEOP) and the Berlin-Frankfurth-Munschen (BFM) group AIEOP-BFM ALL2009 and ALL2017, B-lineage ALL patients with high RQ-PCR-MRD at day+33 and positive at day+78 are defined slow early responders (SERs). Based on results of the AIEOP-BFM ALL2000 study, these patients are treated as high-risk also when positive MRD signal at day +78 is below the lower limit of quantification of RQ-PCR (“positive not-quantifiable,” POS-NQ). To assess whether droplet digital polymerase chain reaction (ddPCR) could improve patients’ risk definition, we analyzed MRD in 209 pediatric B-lineage ALL cases classified by RQ-PCR as POS-NQ and/or negative (NEG) at days +33 and/or +78 in the AIEOP-BFM ALL2000 trial. ddPCR MRD analysis was performed on 45 samples collected at day +78 from SER patients, who had RQ-PCR MRD ≥ 5.0 × 10–4 at day+33 and POS-NQ at day+78 and were treated as medium risk (MR). The analysis identified 13 of 45 positive quantifiable cases. Most relapses occurred in this patients’ subgroup, while ddPCR NEG or ddPCR-POS-NQ patients had a significantly better outcome (P < 0.001). Overall, in 112 MR cases and 52 standard-risk patients, MRD negativity and POS-NQ were confirmed by the ddPCR analysis except for a minority of cases, for whom no differences in outcome were registered. These data indicate that ddPCR is more accurate than RQ-PCR in the measurement of MRD, particularly in late follow-up time points, and may thus allow improving patients’ stratification in ALL protocols.
Introduction
Acute lymphoblastic leukemia (ALL) is the most frequent cancer in childhood, with peaks of incidence between 2 and 5 years old and with 60% of cases occurring in individuals below 20 years of age.1 The 5-year survival rate for children with ALL has significantly increased over time.1-4 However, relapses still occur in 15-20% of children with ALL1 and are associated with poor outcome.5
ALL is the first neoplasm in which the assessment of early response to therapy by minimal residual disease (MRD) monitoring was proven to be a key prognostic tool for guiding risk-based therapeutic choices. Currently, real-time quantitative polymerase chain reaction (RQ-PCR) of clonotypic immunoglobulin (IG) and T-cell receptor (TR) gene rearrangements is the most widely used molecular method for MRD assessment. About 95% of ALL patients can be investigated by this approach, and sensitivity down to 10–4 can be obtained, depending on the type of patient-specific IG/TR rearrangement and the junctional region sequence analyzed.6,7
To ensure comparable MRD results between different laboratories involved in routine polymerase chain reaction (PCR)–based MRD assessment, rigorous guidelines were established within the EuroMRD consortium.8 Despite the high sensitivity of RQ-PCR method, a nonnegligible fraction of patients with very low MRD levels are classified as positive not-quantifiable (POS-NQ) according to the EuroMRD guidelines. In particular, MRD is defined POS-NQ when the delta cycle threshold of replicates is ≥ 1.5 or when the mean cycle threshold (CT) value of the replicates is greater than the highest CT value of the “quantitative range”.8 In such cases, treatment decision based on MRD risk stratification may be suboptimal.
The reason why a sample results as POS-NQ could reside in lack of reproducibility. However, in most of the cases, in the presence of very low MRD levels, it is difficult to distinguish the PCR amplification signal of very few residual leukemic cells from the nonspecific signal of the regenerating lymphoid population and the donor DNA pool used as negative (NEG) control.
According to the literature,9,10 droplet digital PCR (ddPCR) might be a feasible and attractive alternative method for MRD assessment, with the potential of overcoming some limitations of RQ-PCR. In particular, ddPCR could be more accurate than RQ-PCR since each sample is partitioned in droplets in which the ratio between target DNA molecules and PCR reagents is substantially higher than in RQ-PCR. Each droplet is then analyzed individually, and small changes in fluorescence intensity are more readily detected. Overall, this increases ddPCR amplification efficiency over that of RQ-PCR.11-13 Moreover, ddPCR allows an absolute quantification without the need of a standard curve and is able to provide a reliable quantification of MRD in about 20-30% of RQ-PCR POS-NQ samples, as reported by several groups.9,10,14 The opportunity to analyze follow-up samples in the absence of a standard curve might be crucial for high-risk patients, which might need repeated MRD monitoring before and after hematopoietic cell transplantation15,16
However, no established guidelines for ddPCR MRD analysis and interpretation have been defined so far, and its potential is still under investigation. A major standardization effort is underway within the EuroMRD Consortium (www.euromrd.org) for its future application in standard clinical practice.
In the present study, we measured MRD by ddPCR in pediatric B-lineage ALL cases classified as POS-NQ and/or NEG by RQ-PCR at days +33 and/or +78 within the Italian Association of Pediatric Hematology and Oncology (AIEOP)-Berlin-Frankfurth-Munschen (BFM) ALL 2000 trial to evaluate the potential of ddPCR in improving quantification of low MRD levels and contribute to a better patients’ risk stratification and treatment.
Methods
Study population
A total of 209 pediatric B-lineage ALL patients enrolled in the AIEOP-BFM ALL 2000 trial were included in the study. Patients were stratified in risk categories, and the risk group assignment was based on cytologic and molecular response to treatment and on genetic features of ALL blasts. Patients with either prednisone poor response, or no complete remission (CR) at day +33, or evidence of t(9;22) is the official nomenclature (or breakpoint cluster region-Abelson), or evidence of t(4;11) is the official nomenclature (or mixed lineage leukemia-ALL1 fused gene from chromosome 4), or MRD load ≥ 5 × 10–4 at day +78 were allocated to the high-risk group (MRD-HR). In the absence of high-risk criteria, patients were assigned to the medium risk (MR) group if they had a positive MRD at day +33 and/or day +78, but at a level < 5 × 10–4 at day +78 (MRD-MR) are not classifiable by MRD. When MRD was NEG at both days +33 and +78 with at least 2 markers with a sensitivity of ≥ 10–4, patients were allocated to the standard-risk group (MRD-SR).2,17
We defined 4 tasks, summarized in Figure 1: in the first 2 tasks, we analyzed a series of 124 ALL patients who had positive MRD ≥ 5.0 × 10–4 at day +33, and we focused on day +78 MRD. In task 1, we considered 45 patients who were RQ-PCR POS-NQ at day +78. These patients, classified as MR, in subsequent AIEOP-BFM ALL 2009 and 2017 protocols are defined as slow early responders (SERs) and are instead allocated to high-risk group and treatment. In task 2, we considered 79 MR patients with RQ-PCR NEG at day +78.
Figure 1.
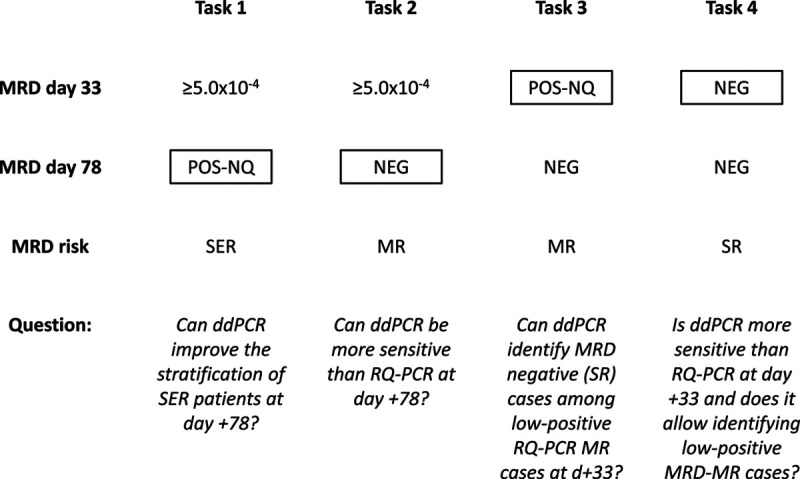
Flowchart of the different tasks. The amplification of IG/TR rearrangements by ddPCR was compared with RQ-PCR to assess if it can improve MRD quantification at clinically critical TP and therefore allows a more precise stratification of patients. MRD results by RQ-PCR are indicated; in boxes are the TP tested by ddPCR in parallel to RQ-PCR, to answer the questions below. ddPCR = droplet digital polymerase chain reaction; IG = immunoglobulin; MR = medium risk; NEG = negative; MRD = minimal residual disease; POS-NQ = positive not-quantifiable; RQ-PCR = real-time quantitative polymerase chain reaction; SER = slow early responder; SR = standard risk; TP = time point; TR = T-cell receptor.
In the remaining 2 tasks, we adopted a case-control design that included 35 ALL patients who relapsed after having MRD POS-NQ (n = 12, MR) or NEG (n = 23, standard risk [SR]) at day +33 by RQ-PCR and a set of 50 (21 MR and 29 SR) matched controls, that is, nonrelapsed patients.
Identification of PCR targets and MRD RQ-PCR analysis
Diagnostic DNA samples were screened by PCR amplification to identify IGH, IGK, TRG, TRD, and TRB rearrangements.18-20 The clonal immune gene rearrangements status was examined and confirmed by homo/heteroduplex analysis. After sequencing, patient-specific primers were designed complementary to the junctional regions of each target identified. Specific and sensitive RQ-PCR assays were developed, and the best performing targets were selected for MRD quantification. MRD RQ-PCR assessment was performed and interpreted according to the EuroMRD guidelines, as previously described.8
ddPCR analysis
The MRD ddPCR analysis was performed as previously reported by Della Starza et al.10 To perform a comparative analysis between ddPCR and RQ-PCR, each sample was tested according to the following criteria:
1.5 μg DNA (500 ng in triplicate, not digested) was used for each follow-up sample;
The undiluted diagnostic DNA sample (or the 10–1 dilution) and the 10–4 dilution were included and performed in 2-fold as positive controls;
As NEG controls, we included the following: (1) the peripheral blood mononuclear cells DNA from a pool of 5 healthy donors, to recognize nonspecific amplification of nonleukemic DNA (background), performed in 6-fold, and (2) a no-template control performed at least in 2-fold.
In addition, 3.0 μg DNA (500 ng in 6-fold, not digested) from follow-up samples were tested for a subset of cases, based on DNA availability, to assess whether a higher sensitivity and/or MRD quantification could be reached when more DNA was used.
All samples were quantified with the following ratio: copies/μL (MRD sample) on copies/μL (diagnosis sample).
Data have been interpreted as it follows10:
Reproducibility: three or six replicates with copies/μL values within the same logarithm were considered as reproducible.
MRD positive quantifiable (POS-Q): a sample was called “positive and quantifiable” if > 3 droplets were observed and the reproducibility rule was achieved. In the presence of positive background, the difference between the lower replicate amplification and the background amplification had to be > 0.5 log.
MRD negative: a sample was considered “negative” if no positive droplets were observed, or if positive droplets were below the background.
MRD POS-NQ: a sample was considered “positive but not-quantifiable” if the reproducibility rule was not achieved, or if the number of positive droplets was ≥ 1 and ≤ 3. In the presence of positive background, the difference between at least one sample amplification and the background amplification had to be > 0.5 log.
MRD quantification: values of replicates were summed up for the calculation.
The distance > 0.5 log between the lower amplification of follow-up sample and the background amplification to consider a sample as positive has been set according to the RQ-PCR EuroMRD guidelines,8 since 0.5 log in ddPCR corresponds to 1.6 CT in RQ-PCR.
Alternative interpretation criteria were also applied21; details and results are reported in Supplemental Digital Content, Supplementary Material (http://links.lww.com/HS/A137).
Statistical analysis
Tasks 1 and 2 considered case series based on available samples at day +78. Event-free survival (EFS) curves were estimated according to Kaplan-Meier with Greenwood standard error, and comparisons performed with the log-rank test. EFS time was calculated from date of diagnosis to date of event and censored if no event occurred. Events considered were resistance, relapse, death, or second malignant neoplasm, whichever occurred first.
In tasks 3 and 4, cases (relapsed patients) were matched to controls (patients relapse free) by risk group (SR and MR) in a 1:2 ratio (task 3) and 1:1 ratio (task 4), according to availability of day +33 samples. Odds ratio (OR) and their corresponding 95% confidence intervals (CIs) were calculated, and χ2 test P values were reported. All analyses were carried out using software package SAS, version 9.4.
Results
Overall, the distribution of IG/TR markers used in the comparison is indicated in Supplemental Digital Content, Supplementary Table 1 (http://links.lww.com/HS/A137). When considering maximal MRD (the highest value of the two IG/TR markers analyzed per TP) (Figure 2), the comparison of MRD results obtained by RQ-PCR and ddPCR showed a concordance rate of 62% (130/209) for patients classified as POS-NQ or NEG by RQ-PCR. When we considered all IG/TR markers/sample (Table 1), a concordance rate of 70.0% (278/397) was observed. The use of ddPCR significantly reduced the proportion of POS-NQ patients compared with RQ-PCR (47/209 [22%] vs 78/209 [37%], P = 0.0013]. As a consequence, ddPCR allowed increasing the proportion of POS-Q samples. In fact, ddPCR detected a quantifiable disease in 17.9% (14/78) of MRD results that were RQ-PCR POS-NQ. Moreover, while only 39 of 292 markers (13.4%) NEG by RQ-PCR were called as POS-Q/POS-NQ by ddPCR (Table 1), ddPCR detected the disease in 19.8% (26/131) of patients who were RQ-PCR NEG, and in 4 of 26 (15.4%), their MRD has also been quantified (4/131 positive, 3.1%) (Figure 2).
Figure 2.
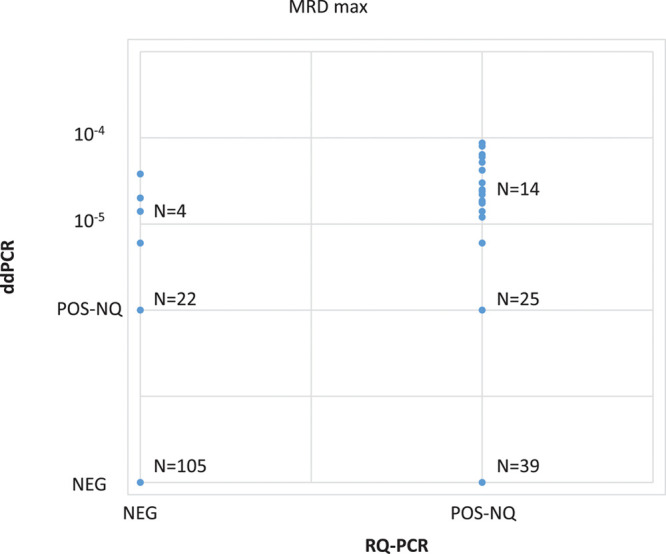
Overall comparison of MRD results performed by RQ-PCR and ddPCR. Analyses were performed in 209 samples classified as POS-NQ and/or NEG by RQ-PCR at days +33 and/or +78. ddPCR = droplet digital polymerase chain reaction; MRD = minimal residual disease; NEG = negative; POS-Q = positive quantifiable; POS-NQ = positive, not-quantifiable; RQ-PCR = real-time quantitative polymerase chain reaction.
Table 1.
Overall Comparison of ddPCR Versus RQ-PCR MRD Results by All IG/TR Markers (1.5 μg DNA Was Used in Both).
| ddPCR | ||||
|---|---|---|---|---|
| RQ-PCR | POS-Q (%) | POS-NQ (%) | NEG (%) | Total (%) |
| POS-NQ | 21 (20) | 25 (23.8) | 59 (56.2) | 105 (100) |
| NEG | 5 (1.7) | 34 (11.6) | 253 (86.7) | 292 (100) |
| Total | 26 (6.5) | 59 (14.9) | 312 (78.6) | 397 (100) |
Concordant values between ddPCR and RQ-PCR are indicated in bold.
ddPCR = droplet digital polymerase chain reaction; IG = immunoglobulin; MRD = minimal residual disease; NEG = negative; POS-NQ = positive not-quantifiable; POS-Q = positive quantifiable; RQ-PCR = real-time quantitative polymerase chain reaction; TR = T-cell receptor.
As expected, by increasing to 3.0 μg the amount of DNA tested by ddPCR, a reduction of NEG cases was observed, though without a significant contribution in discriminating POS-NQs (see Supplemental Digital Content, Supplementary Table 2, http://links.lww.com/HS/A137).
Task 1. Can ddPCR improve the stratification of SER patients at day +78?
We assessed whether ddPCR, compared with RQ-PCR, could improve MRD quantification at day +78 and therefore allow a more precise allocation of the subset of SER patients with B-lineage ALL characterized by RQ-PCR POS-NQ MRD at day +78.
ddPCR performed on 1.5 μg DNA from 45 SER patients with POS-NQ MRD at day +78 revealed that 13 (29%) were POS-Q, 16 (35.5%) were confirmed POS-NQ, and 16 (35.5%) were NEG (Table 2). When 3.0 μg of DNA were used (in 41/45 samples due to material availability), 12 (29%) were POS-Q, 19 (46%) remained POS-NQ, and 10 (24%) were NEG (see Supplemental Digital Content, Supplementary Table 3, http://links.lww.com/HS/A137).
Table 2.
Summary of ddPCR MRD Results by Each Task (1.5 μg DNA Was Used).
| Time Point | RQ-PCR | ddPCR POS-Q, % (n/N) | ddPCR POS-NQ, % (n/N) | ddPCR NEG, % (n/N) | ||
|---|---|---|---|---|---|---|
| Task 1 | POS ≥ 5 × 10–4 +33;POS-NQ +78 | +78 | 45 POS-NQ | 29 (13/45) | 35.5 (16/45) | 35.5 (16/45) |
| Task 2 | POS ≥ 5 × 10–4 +33;NEG +78 | +78 | 79 NEG | 6.3 (5/79) | 21.5 (17/79) | 72.2 (57/79) |
| Task 3 | POS-NQ +33 | +33 | 33 POS-NQ | 6 (2/33) | 27.3 (9/33) | 66.7 (22/33) |
| Task 4 | NEG +33 | +33 | 52 NEG | 0 | 9.6 (5/52) | 90.4 (47/52) |
ddPCR = droplet digital polymerase chain reaction; MRD = minimal residual disease; NEG = negative; POS = positive; POS-NQ = POS not-quantifiable; POS-Q = POS quantifiable; RQ-PCR = real-time quantitative polymerase chain reaction.
The EFS of this subset of SER patients was different by the ddPCR results at day +78 (POS-Q, POS-NQ, or NEG) (Figure 3A). NEG and POS-NQ cases together had a significantly better EFS compared with POS-Q cases (Figure 3B). The use of 3.0 μg of DNA instead of 1.5 μg, although slightly reducing the NEG cases, as expected, did not affect the distribution of events and EFS curves (see Supplemental Digital Content, Supplementary Figure 1, http://links.lww.com/HS/A137).
Figure 3.
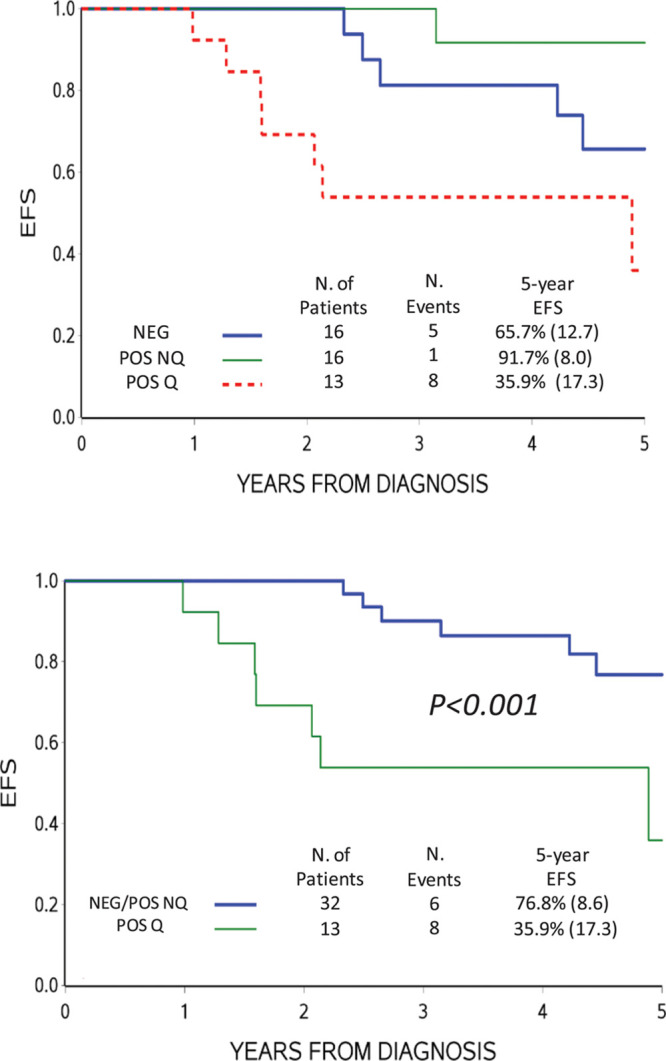
Probability of EFS based on ddPCR MRD. ddPCR MRD results were obtained by analyzing 1.5 μg DNA of patients with high MRD (≥ 5.0 × 10–4) at day +33 and POS-NQ at day +78 by RQ-PCR of IG/TR markers (A). (B), EFS when negative and POS-NQ by ddPCR were cumulatively analyzed. ddPCR = droplet digital polymerase chain reaction; EFS = event-free survival; IG = immunoglobulin; MRD = minimal residual disease; POS-NQ = positive not-quantifiable; RQ-PCR = real-time quantitative polymerase chain reaction; TR = T-cell receptor.
Task 2. Can ddPCR be more sensitive than RQ-PCR at day +78?
The aim of this task was to test whether in the specific subset of MR patients with BCP-ALL rapidly clearing MRD between days +33 and +78, ddPCR could reveal low positivity not detected by RQ-PCR.
Among the 79 patients with high positive (≥ 5.0 × 10–4) MRD at day +33 and NEG MRD at day +78, ddPCR performed on 1.5 μg DNA from day +78 identified 5 (6%) POS-Q, 17 (21%) POS-NQ, and 57 (73%) still NEG (Table 2). When 3.0 μg DNA were used (77/79 samples were available), 9 (12%) patients were POS-Q, 27 (35%) POS-NQ, and 41 (53%) still NEG (see Supplemental Digital Content, Supplementary Table 3, http://links.lww.com/HS/A137).
ddPCR showed higher sensitivity than RQ-PCR in this setting, as it identified several cases with positive MRD values (POS-Q and POS-NQ) in RQ-PCR NEG cases (and even more cases when the amount of tested DNA was increased). However, EFS curves based on ddPCR MRD did not differ significantly, even when 3.0 μg DNA were used (data not shown).
Task 3. Can ddPCR identify MRD NEG (SR) cases among low positive RQ-PCR MR cases at day +33?
The aim of this task was to verify whether ddPCR could discriminate the low positive from NEG MR B-lineage ALL patients better than RQ-PCR.
When ddPCR was applied to 33 RQ-PCR POS-NQ samples at day +33, 2 (6%) were POS-Q, 9 (27%) POS-NQ, and 22 (67%) were NEG (Table 2). When using 3.0 μg of DNA on 33 patients, 1 (3%) was POS-Q, 15 (46%) were POS-NQ, and 17(51%) were NEG (see Supplemental Digital Content, Supplementary Table 3, http://links.lww.com/HS/A137).
Table 3 shows that the prevalence of NEG ddPCR MRD is lower in relapsed patients (7/12, 58.3%) compared with controls (15/21, 71.4%), that is, nonrelapsed patients, but not significantly so (P = 0.44, OR = 0.56, 95% CI, 0.02-1.67).
Table 3.
Summary of ddPCR MRD Results by Patients’ Status (1.5 μg DNA Was Used).
| Relapsed | Alive in CCR | Total | |
|---|---|---|---|
| N (%) | N (%) | N (%) | |
| Task 3 | 12 (0) | 21 (9.5) | 33 (6.1) |
| POS-Q | 0 (41.7) | 2 (19.0) | 2 (27.3) |
| POS-NQ | 5 (58.3) | 4 (71.5) | 9 (66.6) |
| NEG | 7 | 15 | 22 |
| P = 0.44 | |||
| Task 4 | 23 (0) | 29 (0) | 52 (0) |
| POS-Q | 0 (17.4) | 0 (3.4) | 0 (9.6) |
| POS-NQ | 4 (82.6) | 1 (96.6) | 5 (90.4) |
| NEG | 19 | 28 | 47 |
| P = 0.09 |
CCR = continuous complete remission; ddPCR = droplet digital polymerase chain reaction; MRD = minimal residual disease; NEG = negative; POS-NQ = POS not-quantifiable; POS-Q = POS quantifiable.
Task 4. Is ddPCR more sensitive than RQ-PCR at day +33 and does it allow identifying low positive MRD-MR cases?
Finally, we tested whether ddPCR could be more efficient than RQ-PCR in identifying low positive cases among NEG BCP-ALL SR cases at day +33 by RQ-PCR.
ddPCR on 1.5 μg DNA from 52 MRD NEG patients at day +33 showed 5 (10%) POS-NQ and 47 NEG (Table 2), while by using 3.0 μg of 51 sample, 7 (14%) were POS-NQ and 44 (86%) NEG (see Supplemental Digital Content, Supplementary Table 3, http://links.lww.com/HS/A137). Table 3 shows that the prevalence of positive ddPCR MRD is higher in relapsed patients (4/23, 17.4%) compared with that in controls (1/29, 3.4%), that is, nonrelapsed patients, so that ddPCR tends to be more specific and classifies less patients to SR among those who later relapse, yet not significantly so (P = 0.09, OR = 5.9, 95% CI, 0.6-56.9).
The same results interpreted according to alternative guidelines21 are reported in Supplemental Digital Content, Supplementary Material (http://links.lww.com/HS/A137) (see Supplemental Digital Content, Supplementary Tables 4 and 5, http://links.lww.com/HS/A137 and see Supplemental Digital Content, Supplementary Figures 2 and 3, http://links.lww.com/HS/A137).
Discussion
MRD evaluation during and after the induction therapy is the most relevant prognostic factor in pediatric ALL, either in front line and relapse protocols.2,3,5 Although molecular MRD is being applied in clinical protocols since 20 years, there are still some aspects that need to be addressed. By applying the widely accepted EuroMRD methods and guidelines,8 a consistent fraction of patient samples with very low MRD levels cannot be properly quantified and are considered POS-NQ. Since low disease levels are close to the sensitivity limit of the current analytical methods, it is difficult to obtain reproducible results, and this might potentially reflect in a less precise MRD definition for these borderline cases. In the present study, we selected specific and challenging pediatric ALL settings to investigate whether ddPCR could represent an alternative and clinically valuable method compared with the RQ-PCR gold standard. Taking into account all IG/TR markers used, the comparison of MRD results performed by RQ-PCR and ddPCR showed a concordance rate of 70% at the tested time points. The greater accuracy of ddPCR allowed to discriminate very low/POS-NQ samples by RQ-PCR, turning them into POS-Q in 20% (21/105) of cases, or confirming them to be NEG in 56% (59/105). Of note, ddPCR was able to prove a more robust and precise quantification than RQ-PCR for samples with positivity < 10–4, the most challenging cut-off at both clinical and methodologic levels. Importantly, ddPCR MRD data were generated by three different laboratories, and all labs were able to precisely quantify RQ-PCR low positive samples ranging between 10–4 and 10–5, confirming the strength of the ddPCR assay.
From a technical point of view, ddPCR gave concordant positive (quantifiable or not quantifiable) results in RQ-PCR POS-NQ cases, particularly in those with both the markers positive and with 3 amplifications out of three follow-up replicates (91%; 10/11). On the contrary, when MRD was POS-NQ by RQ-PCR with only 1 marker and with 1 or 2 positive amplifications out of 3 replicates, ddPCR resulted NEG in most of the cases (74%; 17/23). This could be due to an inferior sensitivity of ddPCR or to false-positive results by RQ-PCR. Indeed, the use of immune repertoire targets for MRD evaluation, and the clonotypic nature of allele-specific oligonucleotide (ASO) strategies makes RQ-PCR and ddPCR performance variable.
Overall, considering the different experimental settings we tested, ddPCR was convincingly more specific than RQ-PCR when the MRD load was at the limit of sensitivity. Indeed, in the selected subset of SER patients having high disease burden at day +33 and slow kinetics of disease reduction, resulting in MRD-POS-NQ at day +78 by RQ-PCR, most relapses occurred in cases with MRD quantifiable by ddPCR at day +78 (P < 0.001) (Task 1, Figure 3). The outcome of this subset of patients was in fact similar to that of SER patients enrolled in AIEOP-BFM ALL 2000 protocol.2 On the contrary, patients with NEG or POS-NQ MRD results by ddPCR at day +78 had a better outcome compared to patients with MRD high (≥ 5.0 × 10–4) at day +33 and NEG at day +78 (MRD-MR) and similar to that of MR patients enrolled in the same protocol. Based on these results, in next generation MRD-based clinical trials, the high-risk treatment could be given only to patients with quantifiable MRD by ddPCR at day +78, offering them a higher chance of cure and sparing patients with high MRD at day +33 and POS-NQ or NEG MRD at day +78 an unnecessary and more toxic intense protocol.
However, even by applying ddPCR, a certain number of samples remained POS-NQ and showed an outcome similar to NEG cases; such low levels of disease are likely to have a lower impact in determining patients’ outcome, and a larger number of analyzed patients is needed to define the potential impact of ddPCR over RQ-PCR.
In the specular subset of patients with MRD high at day +33 (≥ 5.0 × 10–4) and NEG at day +78, ddPCR did not show a sensitivity higher than RQ-PCR, sufficient to place patients to the high-risk arm (Task 2). Not surprisingly, a small number of cases showed quantifiable disease by ddPCR, but too few to allow any clinical correlation.
Patients with POS-NQ MRD by RQ-PCR at day +33 could represent a biologically different subgroup of cases, with a relatively rapid MRD kinetics compared with those previously discussed who still have a positive MRD at day +78. This could explain why in this subgroup the ability of ddPCR to discriminate POS-Q from POS-NQ and NEG values does not translate into a clinically relevant outcome, being the outcome similar for all patients (Task 3, Table 3).
At this same time point (day +33) and as already shown at day +78, ddPCR was not superior to RQ-PCR in sensitivity and was not able to identify a significant number of positive cases among RQ-PCR NEG patients who could had been classified as MRD-MR (Task 4). Divergent cases were too few to draw any clinical conclusion.
In our study, we tested two different guidelines for interpretation of ddPCR,10,21 also taking advantage of general criteria developed for RQ-PCR, and we obtained similar results (see Supplemental Digital Content, Supplementary Material, http://links.lww.com/HS/A137). Most of the conflicting results between the 2 guidelines regard the discrimination between POS-NQ and NEG MRD results, that, according to the presented data, had similar impact on patients’ outcome.
Overall, our data indicate that ddPCR is as sensitive as RQ-PCR in detecting and quantifying MRD at all the analyzed time points. ddPCR might be slightly more sensitive than RQ-PCR, in particular when the quantitative range of the RQ-PCR assay is < 10–4 and/or other factors reduced the sensitivity of RQ-PCR (ie, background amplification for reduced specificity and/or bone marrow regeneration, assay efficiency). Importantly, this increase in sensitivity does not always translate in a significant prognostic impact, whose cut-off could be different based on time points.
In most MRD-SR patients, ddPCR confirmed the NEG results of RQ-PCR at day +33 and an extremely good kinetics of disease reduction, which was measured independently from the used method. Even testing more DNA (3.0 μg) did not give different results nor substantially modified the risk stratification, at least in these settings.
In contrast, ddPCR can provide a more accurate prognostic stratification for cases defined as MRD-POS-NQ by RQ-PCR and thus allows distinguishing true positive (and quantifiable) cases from NEG, with different clinical outcomes. This substantially reduces the uncertainty of MRD-POS-NQ samples, which at least in the setting analyzed here showed the same outcome as NEG samples, and therefore could be considered as clinically equal.
With regard to a cost comparison between the two techniques, an analysis based on a 96-well plate indicates that the overall cost (consumables and labor, calculated based on hands-on time only) of ddPCR is twice that of RQ-PCR. However, more samples per plate can be run with ddPCR compared with RQ-PCR (29 vs 25 on a 96-well plate), since the diagnostic samples and standard curve are not needed. If testing of a single 500 ng well gives similar results to testing 3 × 500 ng in triplicate, then this would give comparable costs per patient, but with the advantage of sparing precious diagnostic material if using ddPCR. Although the turnaround time for ddPCR is longer than for RQ-PCR (5.5 vs 3.5 h), data interpretation of ddPCR is easier and faster. The intrinsic characteristics of ddPCR to quantify without the need of a standard curve makes this method attractive to spare diagnostic DNA (used to build up standard curves at each MRD evaluation). However, at the moment, the use of ddPCR as a MRD molecular method in clinical protocols is prevented by the lack of published international guidelines for data interpretation, that is a fundamental requirement to ensure reproducibility and to compare MRD data in different clinical protocols; for this reason, the EuroMRD Consortium (www.euromrd.org) is actively working to rapidly achieve this goal.
After the necessarily preliminary stable agreement on standardization and interpretation guidelines, a promising step forward would consist in a parallel prospective testing by ddPCR for samples POS-NQ by RQ-PCR at clinically critical time points in which MRD has proven prognostic significance (ie, end of induction, after high-risk blocks, before and after hematopoietic stem cell transplantation, …). A statistical analysis by the given treatment will validate the findings and define whether ddPCR could contribute to a further improvement of pediatric ALL patients’ stratification and outcome.
Acknowledgments
The authors would like to thank the Associazione Italiana per la Ricerca sul Cancro (AIRC), Milano, Italy: IG 2015 number 17593 to GC, IG 2017 number 20264 to AB and IG 2018 number 21385 to LM), the AIRC Metastases Special Program number 21198 to RF and AB; the Comitato Maria Letizia Verga (Monza); Fondazione Umberto Veronesi (fellowship to FL), Milano, Italy.
Disclosures
The authors have no conflicts of interest to disclose.
Supplementary Material
Footnotes
Supplemental digital content is available for this article.
References
- 1.Pui CH, Pei D, Coustan-Smith E, et al. Clinical utility of sequential minimal residual disease measurements in the context of risk-based therapy in childhood acute lymphoblastic leukaemia: a prospective study. Lancet Oncol. 2015; 16:465–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conter V, Bartram CR, Valsecchi MG, et al. Molecular response to treatment redefines all prognostic factors in children and adolescents with B-cell precursor acute lymphoblastic leukemia: results in 3184 patients of the AIEOP-BFM ALL 2000 study. Blood. 2010; 115:3206–3214 [DOI] [PubMed] [Google Scholar]
- 3.Schrappe M, Valsecchi MG, Bartram CR, et al. Late MRD response determines relapse risk overall and in subsets of childhood T-cell ALL: results of the AIEOP-BFM-ALL 2000 study. Blood. 2011; 118:2077–2084 [DOI] [PubMed] [Google Scholar]
- 4.Pieters R, de Groot-Kruseman H, Van der Velden V, et al. Successful therapy reduction and intensification for childhood acute lymphoblastic leukemia based on minimal residual disease monitoring: study ALL10 from the Dutch childhood oncology group. J Clin Oncol. 2016; 34:2591–2601 [DOI] [PubMed] [Google Scholar]
- 5.Eckert C, Groeneveld-Krentz S, Kirschner-Schwabe R, et al. Improving stratification for children with late bone marrow B-cell acute lymphoblastic leukemia relapses with refined response classification and integration of genetics. J Clin Oncol. 2019; 37:3493–3506 [DOI] [PubMed] [Google Scholar]
- 6.van der Velden VH, Hochhaus A, Cazzaniga G, et al. Detection of minimal residual disease in hematologic malignancies by real-time quantitative PCR: principles, approaches, and laboratory aspects. Leukemia. 2003; 17:1013–1034 [DOI] [PubMed] [Google Scholar]
- 7.Cazzaniga G, Biondi A. Molecular monitoring of childhood acute lymphoblastic leukemia using antigen receptor gene rearrangements and quantitative polymerase chain reaction technology. Haematologica. 2005; 90:382–390 [PubMed] [Google Scholar]
- 8.van der Velden VH, Cazzaniga G, Schrauder A, et al. Analysis of minimal residual disease by Ig/TCR gene rearrangements: guidelines for interpretation of real-time quantitative PCR data. Leukemia. 2007; 21:604–611 [DOI] [PubMed] [Google Scholar]
- 9.Drandi D, Kubiczkova-Besse L, Ferrero S, et al. Minimal residual disease detection by droplet digital PCR in multiple myeloma, mantle cell lymphoma, and follicular lymphoma: a comparison with real-time PCR. J Mol Diagn. 2015; 17:652–660 [DOI] [PubMed] [Google Scholar]
- 10.Della Starza I, Nunes V, Cavalli M, et al. Comparative analysis between RQ-PCR and digital-droplet-PCR of immunoglobulin/T-cell receptor gene rearrangements to monitor minimal residual disease in acute lymphoblastic leukaemia. Br J Haematol. 2016; 174:541–549 [DOI] [PubMed] [Google Scholar]
- 11.Whale AS, Huggett JF, Cowen S, et al. Comparison of microfluidic digital PCR and conventional quantitative PCR for measuring copy number variation. Nucleic Acids Res. 2012; 40:e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hindson CM, Chevillet JR, Briggs HA, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 2013; 10:1003–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vincent ME, Liu W, Haney EB, et al. Microfluidic stochastic confinement enhances analysis of rare cells by isolating cells and creating high density environments for control of diffusible signals. Chem Soc Rev. 2010; 39:974–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavalli M, De Novi LA, Della Starza I, et al. Comparative analysis between RQ-PCR and digital droplet PCR of BCL2/IGH gene rearrangement in the peripheral blood and bone marrow of early stage follicular lymphoma. Br J Haematol. 2017; 177:588–596 [DOI] [PubMed] [Google Scholar]
- 15.Balduzzi A, Di Maio L, Silvestri D, et al. Minimal residual disease before and after transplantation for childhood acute lymphoblastic leukaemia: is there any room for intervention? Br J Haematol. 2014; 164:396–408 [DOI] [PubMed] [Google Scholar]
- 16.Lovisa F, Zecca M, Rossi B, et al. Pre- and post-transplant minimal residual disease predicts relapse occurrence in children with acute lymphoblastic leukaemia. Br J Haematol. 2018; 180:680–693 [DOI] [PubMed] [Google Scholar]
- 17.Möricke A, Zimmermann M, Valsecchi MG, et al. Dexamethasone vs prednisone in induction treatment of pediatric ALL: results of the randomized trial AIEOP-BFM ALL 2000. Blood. 2016; 127:2101–2112 [DOI] [PubMed] [Google Scholar]
- 18.Pongers-Willemse MJ, Seriu T, Stolz F, et al. Primers and protocols for standardized detection of minimal residual disease in acute lymphoblastic leukemia using immunoglobulin and T cell receptor gene rearrangements and TAL1 deletions as PCR targets: report of the BIOMED-1 CONCERTED ACTION: investigation of minimal residual disease in acute leukemia. Leukemia. 1999; 13:110–118 [DOI] [PubMed] [Google Scholar]
- 19.van Dongen JJ, Langerak AW, Brüggemann M, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 concerted action BMH4-CT98-3936. Leukemia. 2003; 17:2257–2317 [DOI] [PubMed] [Google Scholar]
- 20.Szczepanski T, van der Velden VH, Hoogeveen PG, et al. Vdelta2-Jalpha rearrangements are frequent in precursor-B-acute lymphoblastic leukemia but rare in normal lymphoid cells. Blood. 2004; 103:3798–3804 [DOI] [PubMed] [Google Scholar]
- 21.Drandi D, Alcantara M, Benmaad I, et al. Droplet digital PCR quantification of mantle cell lymphoma follow-up samples from four prospective trials of the European MCL Network. HemaSphere. 2020; 4:e347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


