Abstract
The development of genome-editing technologies in 1970s has discerned a new beginning in the field of science. Out of different genome-editing approaches such as Zing-finger nucleases, TALENs, and meganucleases, clustered regularly interspaced short palindromic repeats-CRISPR-associated protein 9 (CRISPR/Cas9) is a recent and versatile technology that has the ability of making changes to the genome of different organisms with high specificity. Cancer is a complex process that is characterized by multiple genetic and epigenetic changes resulting in abnormal cell growth and proliferation. As cancer is one of the leading causes of deaths worldwide, a large number of studies are done to understand the molecular mechanisms underlying the development of cancer. Because of its high efficiency and specificity, CRISPR/Cas9 has emerged as a novel and powerful tool in the field of cancer research. CRISPR/Cas9 has the potential to accelerate cancer research by dissecting tumorigenesis process, generating animal and cellular models, and identify drug targets for chemotherapeutic approaches. However, despite having tremendous potential, there are certain challenges associated with CRISPR/Cas9 such as safe delivery to the target, potential off-target effects and its efficacy which needs to be addressed prior to its clinical application. In this review, we give a gist of different genome-editing technologies with a special focus on CRISPR/Cas9 development, its mechanism of action and its applications, especially in different type of cancers. We also highlight the importance of CRISPR/Cas9 in generating animal models of different cancers. Finally, we present an overview of the clinical trials and discuss the challenges associated with translating CRISPR/Cas9 in clinical use.
Keywords: Genome editing, Cancer, CRISPR/Cas9, Genetics, Nucleic acids, Animal models
Background
The establishment of recombinant DNA technology has a significant role in the field of research and development (Carroll 2017). Recombinant DNA technology has helped the researchers to modify the DNA of any organism to understand the biology behind any mechanism, and thus exploiting the novel methods for medicine and diagnosis development (Hsu et al. 2014). Recent developments in genome-editing technologies are being exploited as a new resource in the biological fields where researchers could directly delete, insert, and modify a DNA segments in the genetic material of the cell or an organism, to functionally characterize the biological role of any genomic region at systemic level and identify pathogenic mutations (Lanigan et al. 2020). In the current scenario, genome-editing technologies are being extensively used in almost all the areas of biological research including pharmaceuticals, agriculture, crop enhancement, pest management therapeutics, drug development, etc. (Sander and Joung 2014).
The major application of genome-editing technology includes genomic modification which includes gene inactivation, new sequence insertion, and/or correction of mutated sequences with accurate nucleotide sequences (Li et al. 2020a). The potential to perform it with efficiency in eukaryotes carries enormous applications in biological science. Various methods have been developed for the editing of the genomes, but the most efficient one is the targeted genome modification by the designer nucleases (Corrigan-Curay et al. 2015). Since the eukaryotic genome contains millions of bases, and to manipulate or correct the bases, the genome-editing technologies which are very specific could herald the purpose through emerging nuclease-based editing technologies (Li et al. 2020a). The most commonly used and promising nuclease-based technologies are zinc finger nucleases (ZFN), meganucleases, transcription activator-like effector nucleases (TALENs), and the most recently developed, clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated system (Cas) (Gaj et al. 2013; Zhang 2014; Kim and Kim 2014). Conventional genome editing is done using homologous recombination approaches where a vector containing a desirable DNA construct (homologous to targeted genomic sequence) is introduced in a cell, and through the process of homologous recombination, the targeted DNA is replaced by the introduced DNA (Hirotsune et al. 2020). Such technologies have been implicated well in developing transgenic mice from embryonic stem cells with desirable gene modifications/expression patterns and ‘knock-in’ targeted genes (Rocha-Martins et al. 2015). However, this approach of generating genetically engineered organisms is a slow and expensive mechanism as it would take around 2–3 years to generate a single mutant mouse which will cost more than $100,000 (Lampreht Tratar et al. 2018). In addition, this technology is relatively less efficient in terms of precise editing of DNA sequences due to lowered frequency of recombination events and generation of large number of false positives. To reduce false positives, genome-editing nucleases were introduced (Sander and Joung 2014). Genome-editing nucleases have the ability to recognize and cut at specific sequences resulting into double-stranded breaks (DSB) (Gersbach 2014; Doudna 2015). These breaks are then repaired either by non-homologous end-joining (NHEJ) or homology-directed repair (HDR) methods (Brinkman et al. 2018). NHEJ method of DNA repair is said to be error-prone which produces small insertions or deletions (indel), resulting in gene knockout. On the other hand, HDR method provides a genome-editing approach which is precise and can thus be used for delivering DNA with similar sequences in the target loci (Zhang et al. 2019a).
Nuclease-based tools for genome editing
Zinc finger nucleases (ZFN): the pioneer one
ZFNs are derived from the largest family of metallo-proteins, zinc proteins (Klug 2010). Due to their unique structure and functional applications, they play a major role in regulating transcription and translation in both prokaryotes and eukaryotes (Carroll 2011; Urnov et al. 2010). ZFNs contain two domains: a nuclease domain and a DNA-binding zinc finger protein domain (Davis and Stokoe 2010). The zinc finger domain with Cys2-His2 is abundantly found in the eukaryotes as the DNA-binding motifs, which may include 3–6 finger-like projections held together by the Zn+2, and two of each cysteine and histidine amino acids (Gupta et al. 2014). Each finger-like projection is made up of ~ 30 amino acids, which are folded into ββα configurations. These zinc finger nucleases have the ability to recognize around 9–18 base pairs and the specificity is provided by the recognition helix of the nucleases composed of six amino acids (Durai et al. 2005). The nuclease domain of the ZFN is formed by the C-terminal of the restriction endonuclease, FokI. However, this genomic tool requires the dimerization of two ZFN monomers, i.e., two sets of fingers, to be activated and to produce a nick or a double-stranded break (DSB) in the DNA (Mani et al. 2005). Some of the various programs and online tools for designing ZFN are: genome-wide target scanner for nuclease off-sites, the Segal Laboratory software site, Zinc Finger Tools ZiFiT Targeter software (Sander et al. 2010), and ZFN target site algorithm for identifying sites compatible with zinc fingers (Schierling et al. 2012). The specificity can be altered by mutagenesis, which allows the ZFN to be programmable nucleases, in a fast and convenient method. Lack of DNA target activity and cytotoxic effects of off-targeting are some of the possible disadvantages in using ZFN (Khan 2019). At times when off-targeting is extensive, the number of double-stranded breaks produced may be more than the DNA repair capacity of the cells and, therefore, results in cell death due to cytotoxicity. Hence, the disadvantages of ZFNs include the lack of DNA targeting activity or cytotoxicity due to off-target effects (Miller et al. 2007). Thus, it is still a challenge to construct a ZFN with low cytotoxic effects, high specificity, and high targeting activity. ZFNs have reached the phase I clinical trials for treating HIV (human immunodeficiency virus) infected patients by knocking out the HIV co-receptor CCR5 (chemokine receptor 5) gene (Ashmore-Harris and Fruhwirth 2020; Perez et al. 2008).
Transcription activator-like effector nucleases (TALENs): second generation
The TALENs are derived from transcription activator-like effectors, encoded by Xanthomonas, a plant pathogenic bacterium (Bloom et al. 2015; Boch and Bonas 2010). The TALENs from Ralstonia, another plant pathogenic bacterium, are also engineered to bind to the DNA sequences and have a structural similarity to that of ZFNs (Joung and Sander 2013) The DNA-binding domain includes arrays of single protein modules of ~ 34 amino acids long, each of which recognizes a single base pair of the DNA in the major groove, thus providing the specificity to the level of single nucleotide. These modules have identical amino acids except at the positions 12 and 13 (Deng et al. 2012). The amino acids at these positions are called ‘repeat variable di-residue’ (RVD), which are responsible for determining the nucleotide specificity of the programmable nuclease (Boch et al. 2009). All four different nucleotides are recognized by four different RVDs: Guanine, Adenine, Cytosine, and Thymine by Asn–Asn, Asn–Ile, His–Asp, and Asn–Gly, respectively. The TALENs can be constructed to recognize 13–20 base pairs along with FokI nuclease domain, in the C-terminal (Miller et al. 2011). As different RVD modules recognize different nucleotides, it is comparatively easier to design the TALENS to recognize specific DNA sequences (Bogdanove and Voytas 2011). E-TALEN (Heigwer et al. 2013), Genome Engineering Resources (http://www.genome-engineering.org/), scoring algorithm for predicting its activity (http://baolab.bme.gatech.edu/Research/BioinformaticTools/TAL_targeter.html), ToolGen TALEN Designer (http://www.toolgen.co.kr/talen_designer/), and ZiFiT Targeter (Sander et al. 2010) software are some of the online tools available for constructing a new TALENs. They can be designed in such a way that it can target any given DNA sequences (Kim and Kim 2014). The transcription activator-like effective (TALE) arrays can be assembled rapidly using various strategies such as high-throughput Golden Gate molecular assembly and ligation-independent cloning techniques (Holkers et al. 2013). There are some limitations of TALENs as it cannot be used for producing nicks and thus can create only DSB in the DNA (Joung and Sander 2013). It is also a highly laborious and expensive process to construct individual TALE protein modules. Another limitation of this strategy is the requirement of a Thymine base (recognized by the TALEN) at the 5′ end of the target sequence (Wood et al. 2011). Conventional TALENs are also considered to be incapable of cleaving DNA which contains methylated cytosines (Sakuma et al. 2013).
Mega nucleases
Mega nucleases (homing endonucleases) are natural proteins derived from microbial mobile genetic elements, with the ability to recognize DNA sequences of > 12 bp in length (Silva et al. 2011). Homing endonucleases can be divided into five families based on their sequence and structures: LAGLIDADG, GIY-YIG, HNH, His-Cys box, and PD-(D/E) XK (Chevalier and Stoddard 2001). LAGLIDADG endonucleases are used as molecular tools as genetic endonucleases and the essential element for their enzymatic activity are defined by ‘LAGLIDADG’ sequence motif (Jacoby et al. 2012). The endonucleases may have one or two such sequence motifs followed by ~ 75–200 amino acids. These endodeoxyribonuclease target the recognition site with high specificity and generate a DSB having overhangs, and protein-coding sequence is inserted within the genome by the process of homologous recombination (Maeder and Gersbach 2016). Initially, I-CreI and I-SceI, single and double motif protein, respectively, were used for editing the genome (Chen et al. 2009; Rosen et al. 2006). In both the cases, enzymes obtain a αββαββα fold and recognize a DNA sequence with 14–40 bp in length (Grizot et al. 2009). The recognition and enzymatic properties of the enzymes are intertwined which make it difficult to design and engineer these nucleases (Nomura et al. 2008). The main advantage with meganucleases is that they produce DSB with 3′ overhang, which has more recombination characteristic to stimulate HDR, when compared to the 5′ overhangs produced by the FokI restriction endonucleases (Guha and Edgell 2017). Since they are also the smallest class of modified nucleases, they can be delivered more easily and efficiently (Galetto et al. 2009).
CRISPR–Cas9: recent and versatile tool in genome editing
Another recently developed gene-editing technology, clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated proteins (Cas), is a well-known adaptive and heritable immune system of archaea and bacteria (Barrangou and Marraffini 2014; Rath et al. 2015; Ishino et al. 1987). CRISPR–Cas technology has various advantages compared to the nuclease-based genome-editing technologies (Adli 2018; Ran et al. 2013). Nuclease-based genome-editing technologies involve protein–DNA interaction, whereas CRISPR–Cas technology uses Watson–Crick base pairing for recognizing the target sequence. The various advantages of the CRISPR–Cas system include its high efficiency, specificity, ability to target multiple genes, and being cost-effective (Chen et al. 2017; Kleinstiver et al. 2016). Due to such advantages, the CRISPR–Cas system has triggered a value addition by bringing dramatic changes in the field of therapeutics especially in cancer therapy (Liu et al. 2019a). Furthermore, there are various applications of this technology to this field in combination with immunotherapy, such as the production of therapeutic cells or antibodies (Xia et al. 2019), and providing immunity to prokaryotic species in opposition to viral-mediated infections and bacterial transformations (Price et al. 2016).
The CRISPR–Cas system was first discovered in E.coli, but it has been shown to be present in a large population of prokaryotes (Ishino et al. 2018; Jansen et al. 2002). The CRISPR–Cas system is a sequence-specific adaptive immune system in bacteria and archea which provide resistance against phages, viruses, and other genetic materials (Horvath and Barrangou 2010; Haft et al. 2005). CRISPR primarily prevents bacteriophages infection and plasmid conjugation (Barrangou et al. 2007). In addition to this, several studies have shown that CRISPR can act as a barrier against all mechanisms of Horizontal gene transfer (HGT) which is a main source of genetic variation in prokaryotes (Garneau et al. 2010). Therefore, along with their function in antiviral immunity, CRISPR plays an important role in maintenance of genome integrity (Shabbir et al. 2016). When a foreign pathogenic organism attacks the bacteria, the invading foreign DNA or RNA is recognized and inserted into its own genome, forming a CRISPR locus (Loureiro and Da Silva 2019). This CRISPR locus comprises two main elements: a series of repeat sequences interspaced by variable sequences and a clustered set of CRISPR-associated (Cas) genes (Karimi et al. 2018; Bolotin et al. 2005). These elements of the CRISPR–Cas system provide a three-step defense response against any invading foreign organisms (Hille et al. 2018). The three steps include; Adaptation, CRISPR Expression, and Interference or immunity. (i) Adaptation: Certain regions of foreign DNA or RNA elements from the invading virus or other pathogenic organisms such as bacteria and archaea are selected based on protospacer-adjacent motifs (PAM) and are incorporated into their genome, thus forming protospacers (Karginov and Hannon 2010; Mojica et al. 2000). Cas9 proteins identify different PAM sequences originated from different bacteria; for instance, Streptococcus pyogenes Cas9 recognizes ‘NGG’ and weaker ‘NAG’ PAM sequences (Geng et al. 2016). These new protospacers act as sequence-specific memory against the pathogenic organisms (Brouns et al. 2008). (ii) Expression: The CRISPR regions are transcribed to form pre-CRISPR-RNA (pre-crRNA) which then matures into CRISPR-RNA (crRNA) by RNaseIII (Charpentier et al. 2015). The crRNA is composed of two sequences: a sequence complementary to the foreign gene sequence of 20-nucleotide length and the sequence which is complementary to the trans-activating CRISPR-RNA (tracrRNA), a binding scaffold to the Cas nuclease (Deltcheva et al. 2011) (iii) Interference: crRNA guides the Cas9 protein to the PAM and produces a double-stranded break (DSB) in the foreign DNA and thus provides defense against the pathogenic organism by blocking the propagation of foreign DNA (Lone et al. 2018; Cong and Zhang 2015). In the CRISPR system, tracrRNA base pairs with crRNA and forms a functional single-guide RNA (sgRNA). The sgRNA of the CRISPR–Cas9 system is designed as a single strand, so that the guide sequence is present at the 5′ end and the RNA duplex at the 3′ end (Anders et al. 2014). The guide sequence is complementary to the foreign DNA and, hence, directs the binding with the target gene sequence, whereas the RNA duplex binds with the Cas9. Hence, the sgRNA recognizes the sequence at which the Cas9 acts as endonucleases and catalyzes the cleavage of 3–4 nucleotides upstream of the PAM (Ma et al. 2015; Wu et al. 2014) (Table 1, Fig. 1).
Table 1.
Comparison of the various genome-editing tools
| Features | Zinc Fingers | TALENs | Meganucleases | CRISPR/Cas9 |
|---|---|---|---|---|
| Origin | Eukaryotic transcription factors, metallo-proteins | Transcription activator-like effectors from plant pathogenic bacteria | Microbial mobile genetic elements | Bacterial adaptive immune system (Gupta and Musunuru 2014) |
| Recognition method | Protein:DNA | Protein:DNA | Protein:DNA | RNA:DNA |
| Size or length | 20–30 amino acids per finger | 33–35 amino acids per repeat | 75–200 amino acids | Approximately 1400 amino acids for SpCas9/dCas9 |
| Specificity | Low and higher frequency of off-target binding | When compared to other genome-editing tools, TALENS have high and comparatively lower frequency of off-target binding | High lower frequency of off-target binding | Highest specificity and the reports vary about the frequency of off-target binding (Kim and Kim 2014) |
| Design | Simple | Slightly complex | Complex | Simpler (Guha et al. 2017) |
| Multiplexing potentiality | Must be custom made and built for each new target | Must be custom made and built for each new target repetitive structures could cause cloning problems | Must be custom made and built for each new target | Allows simultaneous editing of multiple target sites |
Fig. 1.
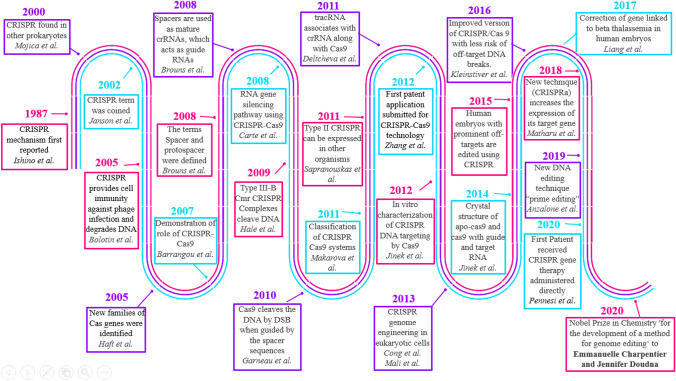
Schematic representation of timelines of CRISPR development which highlights the important discoveries related to CRISPR technology from 1987 to current year. CRISPR technology was first reported in 1987 in Osaka University, whereas the term CRISPR–Cas9 was first coined in 2002. In 2012, first patent for CRISPR–Cas9 technology was submitted, and in 2015, first report of human genes edited by CRISPR came out which fueled the controversy about ethical issues related to gene-editing technologies. In the same year, US scientists used CRISPR/Cas9 for making genetically modified mosquitoes, to prevent them carrying malaria parasite. In 2018, first CRISPR–Cas9 clinical trial was launched. In 2020, first patient received gene therapy where CRISPR was administered directly into the body and in the same year Emmanuelle Charpentier and Jennifer Doudna won the Nobel Prize in chemistry for CRISPR technology
Different types of Cas systems
Another functional form of CRISPR/Cas system composed of a small molecular weight protein known as Casφ (approx. 70 Kda protein) and CRISPR array is specifically involved in encoding large-sized bacteriophage genomes (Pausch et al. 2020). The Casφ protein utilizes a similar active site for both CRISPR-RNA (crRNA) and crRNA-guided DNA target molecules for cleaving external DNA molecules. This compact system is found stable in multicellular eukaryotic species with highly efficient target prediction abilities in contrast to other CRISPR/Cas systems and is also applicable in significant gene-editing functions and DNA recognition (Pausch et al. 2020). CasX is another new protein which has been identified from metagenomic analysis of bacteria (Yang and Patel 2019). CasX protein has a RuvC domain at C-terminus and lacks HNH domain. It works in the same way as Cas9 as it employs both crRNA and tracrRNA which leads to dual RNA-guided DNA cleavage, but compared to Cas9, it has few advantages. The size being the most important factor which allows it to get inside the cells much easier and since it has been isolated from bacteria, the chances of triggering an immune response are humans is less which has always been a concern of Cas9 (Liu et al. 2019b). Similarly, CRISPR–CasY is another newly identified functional form which has some distinct features compared to other CRISPR systems. In CRISPR–CasY, most of the CRISPR arrays contain 17–19 nucleotides spacers which are comparatively shorter from other reported systems. Recent studies have shown that CRISPR–CasY exhibit RNA-guided DNA cleavage activity, but whether they use tracrRNA for cleavage is still not known (Burstein et al. 2017).
Structure and classification of CRISPR system
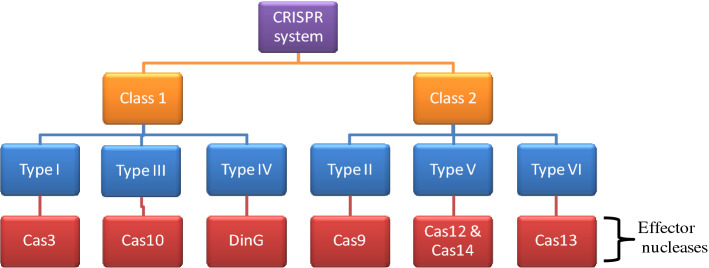
Due to peculiar structural and functional diversification between the CRISPR/Cas systems, Cas gene arrangement, and features of crRNA-cas effector complexes, these systems have been well categorized into 6 major types (types I–VI) which are supplemented by a broader classification as Class 1 and Class 2 (Koonin et al. 2017; Makarova et al. 2011). Class 1 system includes type I, III, and IV, and is characterized by the presence of a multiprotein effector complex known as CRISPR-associated complex for antiviral defense (Cascade) (Hidalgo-Cantabrana et al. 2019). Class 1 system utilizes Cas3, Cas10, and DinG as nucleases (Pickar-Oliver et al. 2019). Also, they are more commonly present in genomes of bacteria and archaea; where the type I system comprises seven subtypes (I–A to I–F plus I–U) as the most abundant and widespread in nature. Although type I is the most abundant, yet only few models have been characterized with most of the studies focusing on type I–E from Escherichia coli (Zheng et al. 2020b). Endogenous type I systems which are naturally present in bacteria and archaea can be repurposed for targeted genome editing or transcriptional approaches which would help in alteration of genome or transcriptome of microbiome of industrial and economical importance. Class 2 system comprises type II, V, and VI, and is associated with Cas9, Cas12–Cas14, and Cas13, respectively (Chylinski et al. 2014) (Fig. 2).
Fig. 2.
A simple schematic representation of CRISPR system and associated nucleases. The CRISPR/Cas system can be divided into two classes as Class1 and Class2. Class1 CRISPR/Cas system utilize multi-Cas protein complex, whereas Class2 CRISPR/Cas system employ single Cas protein. Furthermore, these two classes are subdivided into six types based on the presence of specific genes. Class1 includes types I, III, and IV, whereas Class2 includes types II, V, and VI
Types I–III systems associated with Cas3, Cas9, and Cas10, respectively, are the most studied one, whereas the remaining IV–VI has been recently identified (Makarova et al. 2011). Surprisingly, Cas3, Cas10, and Cas12 are present in both bacteria and archaea, whereas Cas9 and Cas13 are present exclusively in bacteria and Cas14 is uniquely present in archaea (Harrington et al. 2018). RNase III, which is present only in bacteria (and not in archea), plays a crucial role in maturation of pre-crRNA into crRNA, and this explains why type II systems are found only in bacteria (Mir et al. 2018; Durand et al. 2012). Out of these, Cas3, Cas9, and Cas10 are the most commonly occurring caspases and can easily be identified from microbial and human environments. However, within every type, there are several subtypes which display different functional features utilized by prokaryotic species to protect themselves from viral attack as well as from other mobile genomic substances. For instance, class 1 systems’ (type I, III, and IV) effector complexes are composed of diverse cas proteins in close coalition with crRNA, while class 2 systems (type II, V, and VI) harbor only one effector cas protein having nuclease property (Shmakov et al. 2017; Liu and Doudna 2020). In addition, the nucleic acid substance identification and destruction by different crRNA-cas effector complexes vary among disparate CRISPR/Cas systems (Koonin et al. 2017). For instance, type I, II, and V targets DNA molecules, and type III system assists cleavage of both DNA and RNA molecules, while type VI system specifically targets RNA molecules. The effector molecule functioning via DNA targeting system have been dependent on prediction of small sequences, known as PAM which are situated near the target sequences in invading DNA and are responsible for degradation (Leenay et al. 2016). Contrastingly, the type III and VI system targeting RNA molecules does not depend upon PAM sequences as these systems are known to be regulated by flanking sequences surrounding the protospacer segments of target RNA molecules (Gleditzsch et al. 2019). Cas13 protein has ribonuclease activity and can bind to single-stranded RNA and cleave the target (Granados-Riveron and Aquino-Jarquin 2018). There are 4 Cas13 proteins identified so far; Cas13a, Cas13b, Cas13c, and Cas13d. Recently, there are two RNA base editing systems have been developed; one is REPAIR which allows A-to-I (G) replacement and the other one is RESCUE system which allows C-to-U replacement (Fry et al. 2020). RNA editing is more specific and efficient compared to DNA editing. Also, it makes temporary genetic edits/changes which are reversible avoiding the potential ethical issues (Fukuda et al. 2017). The capability of type II and V systems to efficiently form transgenic crRNAs, which degrade specific DNA sequences has been utilized in a variety of applications such as gene modification, controlling genetic expression, and DNA checking in populations. Due to its simple structure and ease of use, class 2 CRISPR–Cas system is popular among the other tools used for the genome-editing approaches for various applications in the field of medicine, agriculture, and biotechnology (Manghwar et al. 2019). In contrast, the CRISPR-based RNA targeting system has shown their wide applications in the development of engineered RNA as well as in identifying prognosis for virus-mediated, bacteria-mediated, and human-associated disorders (Strutt et al. 2018; Sampson et al. 2013). Among the other types of nucleases in class 2, type II CRISPR–Cas9 is the most routinely used tool and is commonly known as CRISPR (Doudna and Charpentier 2014). The Cas9 proteins have two domains RuvC and HNH. RuvC is further subdivided into three subdomains, RuvC I, which is near the N-terminal of the protein and RuvC II/III, which flanks the HNH domain of the protein (Mali et al. 2013). The RuvC domain of the Cas9 protein cleaves the non-complementary regions of the DNA, whereas the HNS domain cleaves the complementary regions (Chen et al. 2014). This cleavage results in the formation of DSBs which is further repaired by NHEJ or HDR pathway. However, HDR pathway/gene-knocking requires a de novo DNA template and is considered to be less effective than the NHEJ pathway/gene-knockout (Rodgers and Mcvey 2016). CRISPR along with Cas9 from Francisella novicida (Fn) can target and degrade mRNA. FnCas9 forms a complex with its tracrRNA and a novel and small CRISPR/Cas-associated RNA (termed scaRNA) instead of the crRNA. The detailed molecular mechanism of FnCas9 is still not clear, but there are studies, which will definitely improve its further application in targeting endogenous RNAs (Burmistrz et al. 2020). A simplified representation of CRISPR technology is described in Fig. 3.
Fig. 3.
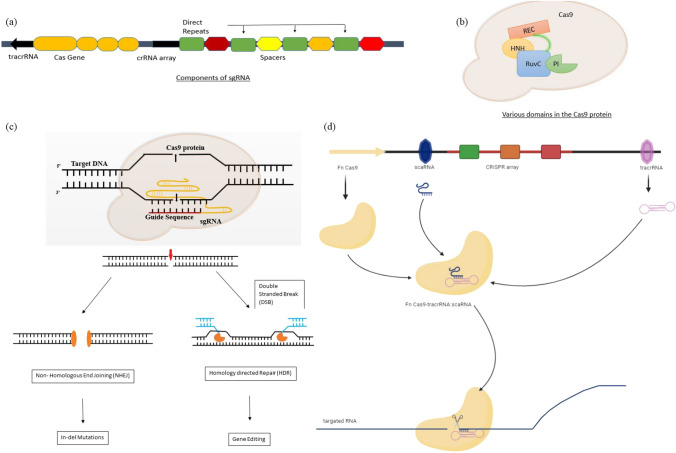
Simplified mechanism of the CRISPR technology (modified from “Delivery Strategies of the CRISPR–Cas9 Gene-Editing System for Therapeutic Applications” (Liu et al. 2017a). a Single guide RNA (sgRNA structure) consists of CRISPR-RNA (crRNA) and trans-activating CRISPR-RNA (tracrRNA). The crRNA and tracrRNA forms a complex and acts as a guide RNA for the Cas9 enzyme. b Cas9 is a dual RNA-guided DNA endonuclease enzyme in Streptococcus pyrogenes. There are two nuclease domains, RuvC (which cleaves the non-target DNA strand) and HNH nuclease domain (that cleaves the target strand of DNA). Target DNA must contain a PAM-motif which is recognized by PAM-interacting domain (PI) of Cas9. Cas9 also have a recognition lobe (REC). Both REC and Nuclease lobe folds to give a positive charge that can accommodate the negative charged sgRNA:target DNA heteroduplex. c CRISPR/Cas9 induces double-stranded breaks which can be repaired either by the non-homologous end-joining DNA repair pathway (NHEJ) or the homology-directed repair (HDR) pathway. In NHEJ process, the two broken ends of DNA are ligated without a template donor which causes insertion and deletion (indel) mutations. This repair process is error-prone and can result in frameshift or loss-of-function and finally gene disruption. In case of HDR, it requires almost identical DNA template to repair the breaks that result in to precise insertion or edition ultimately leading to full correction of the DNA cleavage. d RNA targeting by CRISPR–FnCas9. Cas9 from Francisella novicida (Fn) can target and degrade mRNA. FnCas9 forms a complex with its tracrRNA and a novel and small CRISPR/Cas-associated RNA (termed a scaRNA) instead of the crRNA. The exact mechanism is not very clear yet
Delivery of CRISPR/Cas9 in cells
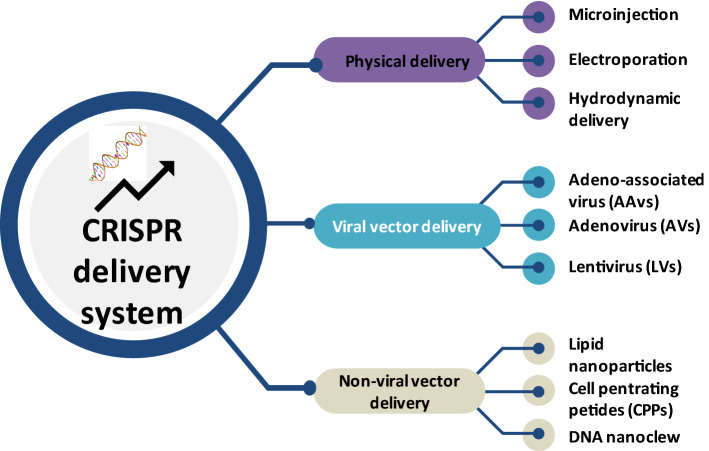
For successful implementation of CRISPR/Cas9 in vivo, a proper and effective delivery of Cas9 and sgRNA to the target cell is required (Wilbie et al. 2019). The main criterion for this approach is that it should cause low toxicity and avoid potential off-target genome editing (Lino et al. 2018). Initially, it was achieved in mammalian cells, by plasmid based expression of Cas9 and sgRNA. Similar approach was adapted for delivery into model organisms such as mice, but the outcome of editing efficiency was poor. Therefore, to improve the efficiency of in vivo delivery, different viral and non-viral methods have been adapted (Yin et al. 2016; Li et al. 2015a). Non-viral methods include lipid nanoparticles, gold nanoparticles, cell penetrating peptides, etc., but these methods are not being widely used; however, it is becoming a burgeoning area of research (Li et al. 2018). Viral methods include adeno-associated virus (AAV), Adenovirus (AV), and lentivirus (LV) vehicles which act as the prominent method of delivery vectors. AAV is a single-stranded DNA virus that has been extensively used for gene delivery because of its non-toxic nature (Lee et al. 2017; Xu et al. 2019a). As such, AAV is not known to induce innate or adaptive immune response or any associated toxicity which makes it a great choice for this purpose (Chew et al. 2016). There are different approaches for CRISPR/Cas9 delivery using AAV vectors which include packaging of SpCas9 (Streptococcus pyrogenes) and sgRNA onto one plasmid vector and then delivery via one AAV article (Luther et al. 2018). The only drawback of this method is that AAV allows approximately 4.5–5 kb of packaged genomic material, whereas the size of Cas9 and sgRNA is roughly around 4.2 kb; this makes the packaging little challenging, and also, it does not allow addition of other elements such as fluorescent tags, reporters, etc. There has been different modification to this approach for example by packaging Cas9 and sgRNA into two separate AAV particles which allows an increase in the overall size of the construct, but this has its own pitfalls which include decrease in efficiency in terms of delivery as well as target DNA cutting (Sun et al. 2003; Ronzitti et al. 2020). Another approach is using a different version of Cas9 from S. aureus rather than S. pyrogenes, which is less in size, but has the same editing ability, so that Cas9 and sgRNA expression cassette can be packed into a single AAV genome-editing vector (Lino et al. 2018). The method of delivering CRISPR/Cas9 using LV and AV is pretty much similar to each other where backbone virus in LV is a provirus of HIV, and in AV, it is different known serotypes of known AVs. Both LV and AV can infect dividing and non-dividing cells, but interestingly, AV does not integrate into the genome. This is very advantageous for reducing off-target effects. LV and AV have a better advantage compared to AAV in terms of their size which ultimately affects the size of the construct (Kabadi et al. 2014; Cheng et al. 2014). All these viral delivery methods can be used for in vitro, in vivo, and ex vivo applications which makes this as the most suitable choice for CRISPR delivery (van Haasteren et al. 2020). A brief summary of all the delivery methods of CRISPR is represented in Fig. 4.
Fig. 4.
A pictorial representation of different types of CRISPR delivery systems. The effective delivery of CRISPR is one of the most challenging steps in the genome-editing process. The main mode of delivery is either by viral (adenoassociated, adenoviral, or lentiviral) or by non-viral methods (lipid nanoparticles, gold nanoparticles, cell penetrating peptides, etc.). Non-viral methods have lesser advantage over viral vectors especially in case of gene knock-ins. Viral vectors are the most prominent one, but it induces off-targets and immune response which needs to be improved
Applications of CRISPR
Applications of CRISPR in agriculture
Till date, many crops have been genetically modified using CRISPR/Cas9 such as rice, wheat, maize, soybean, cotton, lettuce, grapes, potato, etc. Gene knockouts are the most frequently used application to produce null alleles (Jaganathan et al. 2018). LAZY1 gene was knocked out in rice using CRISPR/Cas9, to produce a phenotype which could result in increased crop yield (Miao et al. 2013). Similarly the GN1a, DEP1, and GS3 genes of the rice cultivar ZH11 (Zhonghua11) were modified using the CRISPR/Cas9, resulting in phenotypes with enhanced grain number dense erect particles and larger grain size (Zhang et al. 2019e; Arora and Narula 2017). CRISPR/Cas9 can also be used to improve nutritional profiles of the crops, to improve resistance to biotic stresses, to improve shelf life, and also to create herbicide resistant crops (Zhou et al. 2020). In 2014, Haun et al. (2014) have used TALENs technology to improve oleic acid content in Soybean by targeting FAD2 gene. Similarly, some other teams studied the mutation efficiency of CRISPR/Cas9 for evaluating exogenous and endogenous genes in hairy roots of soybean (Jacobs et al. 2015; Li et al. 2015b). This was further supported by Du et al. (2016) who compared TALENs and CRISPR/Cas9 efficiency for targeting phytoene desaturase genes, and concluded that CRISPR is much more efficient than TALEN in targeting these alleles. Whereas in rice, this technique was used to produce targeted mutations in SBEIIb, thereby resulting in the production of higher proportions of amylopectin which enhanced the nutritional value of starch (Sun et al. 2017). Zhang et al. (2017) produced wheat plants resistant to powdery mildew by targeted modification of three homologs of EDR1. Also, SlMLO1 was edited to produce powdery mildew-resistant tomatoes (Nekrasov et al. 2017). When the eIF4E gene of the cucumber was disrupted, broad virus-resistant plants were generated as they were resistant against Ipomovirus, Zucchini yellow mosaic virus, and Papaya ringspot mosaic virus-W (Chandrasekaran et al. 2016). Although the performance of cas9 genes in plants and breeding varieties has not been to the expectations, there is a greater scope and promise of applying these variants in wide cultivars (Zhang et al. 2019d). Table 2 summarizes the more recent and advance application of CRISPR in agriculture.
Table 2.
Applications of CRISPR in different fields
| Field | Applications | References |
|---|---|---|
| Agriculture | Knockout of two homologs of the BnaMAX1, which resulted in the increase of yield in rapeseed | Zheng et al. (2020a) |
| CRISPR-mediated knockout of phytochrome C in maize resulted in regulation of flowering time and height of the plant | Li et al. (2020c) | |
| Semi-dwarf rice lines lacking any residual transgene-DNA and off-target effects were generated through CRISPR/Cas9-guided mutagenesis of the OsGA20ox2 gene in a high yielding Basmati rice line | Nawaz et al. (2020) | |
| Complete reproductive sterility in the poplar sterile apetala (PopSAP) via the CRISPR/Cas9 | Azeez and Busov (2020) | |
| Knockout of OsGhd7 via CRISPR/Cas9 resulted in rice varieties with early flowering and early maturity | Wang et al. (2020) | |
| CRISPR/Cas9 editing of SlHyPRP1 resulted in salt stress-tolerant events in cultivated tomato | Tran et al. (2020) | |
| CRISPR/Cas9 editing of OsROS1 gene resulted in pollen and embryo sac defects in the rice | Xu et al. (2020) | |
| Genome-editing via CRISPR/Cas9 resulted in the modification of MaGA20ox2 gene which created semi-dwarf banana | Shao et al. (2020) | |
| OsPYL9 was mutagenized through CRISPR/Cas9 enhanced Drought Tolerance and Grain Yield in Rice (Oryza sativa L.) | Usman et al. (2020) | |
| Gene therapy | The targeting of IVS1-110G>A mutation using Cas9 ribonucleoprotein (RNP) and the IVS2-654C>T mutation by Cas12a/Cpf1 RNP in primary CD34+ hematopoietic stem and progenitor cells (HSPCs) from β-thalassemia patients | Xu et al. (2019b) |
| Cas9:sgRNA ribonucleoprotein (RNP)-mediated cleavage within a GATA1 binding site at the + 58 BCL11A erythroid enhancer has resulted in the induction of γ fetal globulin in Sickle cell anemia patients | Wu et al. (2019) | |
| Development of EDIT-101, a candidate genome-editing therapeutic using CRISPR/Cas9, to remove the aberrant splice donor created by the IVS26 mutation in the CEP290 gene and restore normal CEP290 expression in the Lebercongenital amaurosis type 10 | Maeder et al. (2019) | |
| CRISPR/Cas9 system was used in the mdx mouse model of DMD to remove the mutated exon 23 from the dystrophin gene | Nelson et al. (2016) | |
| CRISPR/Cas9 endonucleases coupled with paired guide RNAs flanking the mutated Dmd exon23 were used in excision of intervening DNA and restored the Dmd reading frame in myofibers, cardiomyocytes, and muscle stem cells after local or systemic delivery | Tabebordbar et al. (2016) | |
| It has been demonstrated that in a mouse model of tyrosinaemia, hydrodynamic tail-vein injection of plasmid DNA encoding the adenine base editor (ABE) and a single-guide RNA (sgRNA), can correct an A>G splice-site mutation | Song et al. (2020) | |
| AAV delivery of CRISPR can effectively correct Z-AAT mutation in the liver of a transgenic mouse model of Alpha1-Antitrypsin Deficiency | Song et al. (2018) | |
| Cell and animal disease models | A human muscle cell model of Duchenne muscular dystrophy created through CRISPR/Cas9 by targeted removal of DMD exons 51–57 | Shimo et al. (2018) |
| CRISPR/Cas9 technology was used to introduce a heterozygous nonsense mutation in the PAX6 gene of LSCs, which is found in Aniridia-Related Keratopathy patients | Roux et al. (2018) | |
| CRISPR/Cas9-mediated knockout of Abcd1 and Abcd2 genes in BV-2 cells resulted in microglial models for X-linked Adrenoleukodystrophy | Raas et al. (2019) | |
| Development of DMD mouse model was achieved by deleting exons 8–34 of the X-linked mouse Dmd gene using CRISPR/Cas9 genome editing, which led to a reading frame shift and the absence of functional dystrophin production | Egorova et al. (2019) | |
| First CRISPR/Cas9-induced Lep and Lepr knockout (KO) mouse models for diabetics and obesity were generated using CRISPR/Cas9 technique by specifically targeting Lep or Lepr in C57BL/6J embryos, which resulted in phenotypic such as an increase in body weight, hyperglycemia, and hepatic steatosis | Roh et al. (2018) | |
| A new tau knockout strain (tauΔex1) of Alzheimer’s was generated by CRISPR/Cas9-mediated genome-editing of intron-1/exon 1 of Mapt in C57Bl/6J mice | Tan et al. (2018) | |
| A knock-in (KI) pig model of Huntington Disease, which endogenously expresses full-length mutant huntingtin (HTT) was developed using CRISPR/Cas9 and somatic nuclear transfer technology | Yan et al. (2018) |
Applications of CRISPR in medicine
One of the most early and important applications of CRISPR in the medical field is disease modeling. Nakamura and colleagues (2015) produced a rat model for Duchenne muscular dystrophy (DMD) by targeting two exons of the DMD gene in the rat using CRISPR/Cas9. This genome-editing technology can be used to make multigenic disease models more easily than the conventional transgenic techniques. Similarly, various somatic mutations can be induced in adult mice simultaneously or sequentially, resulting in oncogenic mutations and ultimately cancer. Xue and colleagues (2014) were successful in inducing both gain- and loss-of-function mutations in the liver cells of the mouse which resulted in hepatocarcinogenesis (Maddalo et al. 2014). CRISPR/Cas9 can also be used for interrogation of functions of genes in health and disease and identification of genes involved in resistance to adverse conditions, i.e., toxins or drugs. Another most crucial application of CRISPR/Cas9 in medicinal research is its potential for gene therapy. It was demonstrated that using CRISPR/Cas9 HIV-1 gene can be mutated which resulted in its decreased expression in the human T cells (De Masi et al. 2020). The potentiality of this technique against numerous infections such as hepatitis B virus and human papillomavirus are being explored along with their ability to correct genetic mutations (Kennedy et al. 2014, 2015). Recently, various studies reported that CRISPR/Cas9 components delivered into mice corrected a genetic mutation in the DMD mouse models. Some of the studies reported the efficacy of the CRISPR/Cas9-mediated HDR for in vivo gene therapy in the mouse models of human hereditary liver disease (Yang et al. 2016; Yin et al. 2016). Despite all these advances in the use of CRISPR/Cas9 in gene therapy, there are some ethical and safety concerns. As the changes caused by CRISPR in human genome are permanent and inheritable, there are still some uncertainty for using it in clinical settings (Nelson et al. 2016). It is also possible that they may produce unpredictable and uncontrollable off-target effects to the genome. The use of CRISPR in gene therapy and in animal models is summarized in Table 2.
CRISPR and cancer
Cancer is a complex disease which is characterized by multiple changes in genetic and epigenetic alterations in tumor suppressors and oncogenes (Garraway and Lander 2013). Therefore, it is essential to look for experimental approaches to modify the genome of normal as well as cancer cells and identify different genes and pathways involved in the process (Stratton et al. 2009). Different genome-editing approaches have made it possible to study the function of different genes in cancer initiation and progression by modifying specific DNA sequences in vivo as well as in vitro (Barman et al. 2020). Also, these technologies have helped to understand the function of a particular gene (either an oncogene or tumor suppressor) in cancer (Shen et al. 2018). The use of CRISPR/Cas-9 in research laboratories has significantly increased because of its simplicity and efficiency (Liu et al. 2019a). Moreover, its successful implementation in mammalian cells by different pioneer groups in the fields has strengthened this genome-editing approach which also dealt with many limitations of other methods. There has been an enormous increase in the research using CRISPR-mediated efficient gene modification in a variety of cells and organisms strengthening this approach for further use (Sánchez-Rivera and Jacks 2015; Moses et al. 2018). Several research laboratories have generated in vitro and in vivo knockout models to study the molecular mechanisms underlying different pathways in different cancers (Ng et al. 2020). In addition, CRISPR technology is used for identification of potential therapeutic approaches in different cancers mainly in solid tumors such as breast, lung, brain, bone, liver, prostate, and colorectal cancer (Hazafa et al. 2020).
Lung cancer
Lung cancer is one of the leading causes of cancer-related deaths worldwide where non-small cell lung cancer (NSCLC) subtypes account for more than 90% of all lung cancers (Devarakonda et al. 2015). Different studies have used CRISPR/Cas9 for effectively targeting different genes involved in the initiation and progression of lung cancer (Nair et al. 2020). For example, Cheung et al. have used the CRISPR technology to target mutant versions of EGFR gene, elimination of which resulted in reduced cell proliferation both in vitro and in vivo (Cheung et al. 2018). Similarly, Koo et al. deleted EGFR in a NSCLC cell line which resulted in death of cancer cells and reduction in tumor size in vivo (Koo et al. 2017). There are many more similar results that highlight the importance of CRISPR technology, especially for targeting EGFR which takes us forward in cancer therapy (Xiao-Jie et al. 2015). Elumalai et al. have successfully knocked out the tumor-suppressor phosphatase and tensin homolog (PTEN) gene via CRISPR/Cas9 genome-editing approach in NSCLC which resulted in increased cancer growth by promoting Akt pathway (Perumal et al. 2019). PTEN inactivation further contributed to epithelial-to-mesenchymal transition by regulating β-catenin translocation. This study reported the mechanism of EMT transition via PTEN, which was previously unknown, with the help of CRISPR technology. Recent studies by Lu et al. (2020b) have used CRISPR-edited T cells for the treatment of NSCLC by targeting PD-1 gene which showed promising results in the first-in-human phase I clinical trial (NCT02793856). This study demonstrated safety and feasibility of CRISPR-edited T cells for the treatment of lung cancer with minimal off-target effects. This can be a breakthrough study as this will lead to more clinical trials for the treatment of lung cancer (Lu et al. 2020b).
Breast cancer
Breast cancer is another most common cancer and one of the leading causes of cancer-related deaths in women worldwide (Momenimovahed and Salehiniya 2019). Breast cancer can be divided into four molecular subtypes based on the expression of progesterone receptor (PR), estrogen receptor (ER), and HER2 (Voduc et al. 2010). The most common among them is ER-positive which shows maximum resistance to therapies (Waks and Winer 2019; Rani et al. 2019). Recent advancements in CRISPR technology have led to the identification of different potential therapeutic targets in different breast cancer subtypes (Yang et al. 2018). Currently, many research laboratories are exploring therapeutic targets in breast cancer by studying multiple proteins with oncogenic effects (Lima et al. 2019). Ebright and colleagues used CRISPR-based genome screening to identify genes which are responsible for metastasis. They reported over-expression of RPL15, a component of a large ribosomal unit, to be a major factor responsible for the metastatic growth. By the advent of CRISPR-based screening, they identified multiple genes which are well-established oncogenes or genes involved in cancer hallmark pathways (Ebright et al. 2020). A recent study by Selinas et al. has used CRISPR technology to study the role of FASN gene (involved in ERα signaling) in the development of breast cancer. Using CRISPR, they generated different clones with frameshift mutations in FASN gene and observed different stages of cancer development. They concluded that FASN knockout resulted in decreased proliferation and migration of breast cancer cells (Gonzalez-Salinas et al. 2020). Triple negative breast cancer (TNBC) is a subtype of breast cancer which is characterized by the loss of estrogen receptor, human epidermal growth factor receptor, and progesterone receptor (Telli 2016). There are not many therapeutic options available for this type of cancer and prognosis of TNBC remains the poorest among all other types of breast cancer (Wahba and El-Hadaad 2015). Not many studies have been done to investigate the therapeutic approach of CRISPR in TNBC. Guo et al. have synthesized a noncationic tumor-targeted nanolipogel system (tNLG) to knockout Lipocalin 2 (Lcn2), a breast cancer-promoting gene, in vitro and in vivo. This approach resulted in significant decrease in tumor growth and highlighted the importance of CRISPR genome editing for therapeutic applications in TNBC (Guo et al. 2019). Almost 70–80% of BRCA1 mutations lead to the development of TNBC (Brianese et al. 2018). The poly(ADP‐ribose) polymerase 1 (PARP1) gene is the synthetic lethal pair of BRCA1 and can be targeted for TNBC treatment (Faraoni and Graziani 2018). There are some clinical studies which are using PARP1 inhibitors such as olaparib (AZD‐2281) and veliparib (ABT‐888), and are currently under investigation (Lord and Ashworth 2017). Chemotherapy has also been used along with PARP1 inhibitors, but not much improvement was seen. Because of all these inconsistent data, CRISPR can be used to expedite the drug testing in preclinical studies. Mintz et al. (2020) used CRIPSR approach for generating PARP1-deficient TNBC cell lines (with and without BRCA1 mutation). The therapeutic efficiency of different drugs was checked both in 2D and 3D tumor-chip models. They reported that TNBC cells (with both BRCA1 and PARP1 mutations) were more sensitive to chemotherapeutic breast cancer drugs docetaxel, doxorubicin, and gemcitabine in 2D culture, but the result was not that promising for 3D tumors. Further investigation is required to understand the exact mechanism of difference in both the systems, so that better therapy approaches for treating BRCA1 mutant TNBC can be developed (Mintz et al. 2020).
Colorectal cancer
Colorectal cancer is the fourth cause of cancer-related deaths worldwide where adenocarcinomas are the most prominent one (Compton 2003). The well-known driver mutations in colorectal cancers are KRAS and BRAF (Yokota 2012; Yau et al. 2017). Using CRISPR/Cas9 genome-wide screening of KRAS wild type and mutant xenografts, different genes, which act as either an oncogene or tumor suppressor, have been screened out, but clinical studies are needed to confirm their efficacy (Yau et al. 2017). A very recent study by Wan et al. (2020) have identified a new strategy for the construction of supramolecular vectors which facilitate in vivo delivery of CRISPR/Cas9 in the form of ribonucleoprotein (RNP) and inhibit tumor growth and metastasis in experimental mouse models by targeting mutant KRAS (Gao et al. 2020). This study provides a new effective therapeutic strategy for treatment of colorectal cancer using CRISPR approach. Similarly, Ryu et al. (2020) have developed a nanoliposomal particle, which contains Cas9 protein and a single-guide RNA (sgRNA) to specifically target KRAS mutation in colorectal cancer. Takeda et al. have used CRISPR/Cas9 to functionally characterize colorectal cancer driver genes in mouse intestinal tumor organoids and human colorectal cancer-derived organoids. This study resulted in the identification of Arid2, Acvr2a, and Acvr1b as tumor suppressors. This system can also be used in other organoid system to identify novel driver cancer genes (Takeda et al. 2019). Li et al. have used CRISPR/Cas9 approach to generate CD133 knockout colon cancer cells to study its role. CD133 is a cancer stem cell marker and plays a very crucial role in proliferation and invasion in colon cancer (Li et al. 2019b). Izumi et al. have generated xenograft mouse models using CRISPR/Cas9 approach with stable knockdown of TIAM1, a gene involved in Wnt signaling pathways and is over-expressed in colorectal cancer. By studying the role of TIAM1 in colon cancer cells, it was identified as a therapeutic target to reverse drug resistance (Izumi et al. 2019). Environmental and genetic factors play an important role in the colon cancer development. Haiwen li et al. have used CRISPR approach to identify the genetic factors involved in the regulation of oxidative stress. Galectin 2 (Gal2) protein, encoded by LGALS2, is known to be down-regulated in colon cancers. With the help of some in vitro and mice studies, this group showed the therapeutic potential of Gal2 in colon cancer. CRISPR has been increasingly used in genome-wide screening to study various signaling pathways in different disease conditions, and altogether, all these studies have provided a new dimension to this approach (Li et al. 2020b).
Prostate cancer
There are different studies showing the importance of the CRISPR/Cas9 system in Prostate cancer. Kawamura et al. (2015) established NANOG1 and NANOGP8-knockout PCa cell lines and studied the function of these genes in the same cell lines. Several studies have suggested NANOG1, a transcription factor, to be involved in the process of malignancy using RNAi-based approaches. NANOGP8 is a pseudogene which encodes full-length NANOG1 protein. RNAi can have off-target effects due to high similarity between NANOG1 and NANOGP8 mRNA. CRISPR/Cas9 proved to be a boon for this study as it resulted in the proper knockout of NANOG1 and NANOGP8 in PCa cell lines. Multiple experimental approaches further suggested the role of these two genes in the malignant potential of PCa (Kawamura et al. 2015). Another recent interesting study by Kounatidou et al. (2019) have used CRISPR technology to generate PCa cell lines which have lost expression of Androgen Receptors of Full Length (AR-FL), but have all endogenous AR-Vs. One of the major problems in treating PCa is the development of resistance to androgen receptor (AR)-targeted therapies. A very recent article by Warner et al. (2020) highlighted the importance of ERβ in PCa. Complete knockout of ERβ was not successful in the previous studies as the knockout techniques only deleted the DNA-binding domain of ERβ which is not required for ERβ signaling. With the help of CRISPR/Cas9 technology, the ERβ gene was completely deleted (Warner et al. 2020). In another study, Ye et al. (2017) have deleted GPRC6A, a G-protein coupled receptor known to be involved in the progression of PCa, using CRISPR/Cas9 and found out that GPRC6A editing could reduce androgen biosynthesis by regulating enzyme expression involved in the process. A very novel approach to target tumors in vivo by chimera delivery system consisting of RNA aptamers-liposome-CRISPR/Cas9 was developed by Zhen and team (2017). This could act as a potential therapeutic approach for treating PCa. However, this has its own challenge which includes isolation of aptamers which itself can be a herculean task (Esposito et al. 2018). In a very recent study by Jiang et al., CRISPR was used to knockout PCa associated miRNAs for functional validation. Different miRNAs such as miR-205, miR-221, miR-455-3p, miR-222, miR-224, miR-505, miR-23b, miR-30c, miR-1225-5p, and miR-663a were knocked out in PCa cell line LNCaP, and their effects were studied. This study, to the best of author’s knowledge, first time reported that miR-663a and miR 1225 5p may be involved with the progression of prostate cancer, suggesting their potential as candidate biomarker (Jiang et al. 2020). Another recent study by Rushworth et al. (2020) have used whole-genome CRISPR screening to identify genes and related pathways which is responsible for sensitization of prostate cancer cells against taxane treatment. Through this study, they reported that suppression of transcription elongation factor A-like 1 (Tceal1) enhances taxanes (docetaxel) efficacy. Park et al. used CRISPR to produce NKX3.1 knockout mice. NKX3.1 is a well-known tumor-suppressor gene which is known to act as an androgen-regulated transcription factor. This study provided evidence that CRISPR-mediated knock-down of NKX3.1 gene leads to PIN lesions and alteration in different cancer pathways (Park et al. 2020). Chakraborty et al. have used CRISPR for concomitant deletions of BRCA1 and RB1 which induces EMT transition and leads to aggressive prostate cancer progression. This is the first study which shows that co-loss of two genes induces a distinct phenomenon in prostate cancer which is associated with worse prognosis (Chakraborty et al. 2020).
Bone cancer
Osteosarcoma is the most common type of malignant bone cancer affecting children and adults (Yan et al. 2016). Earlier to the 1970′s, osteosarcoma was seldom treatable, even through certain surgical treatments. Since then, the combined effect of surgical treatment and use of chemotherapeutic agents such as doxorubicin, cisplatin, and methotrexate have been used worldwide and shown effective survival rate among the population. Unfortunately, none of the tyrosine kinase inhibitors like imatinib and sorafenib are proven to be better diagnostic therapies than the conventional methods (Lodish 2013); however, few of the tyrosine kinase inhibitors such as vascular endothelial growth factor, platelet-derived growth factors, and IGF1 get over-expressed during osteosarcoma (Li et al. 2019c). In addition to this, many osteosarcoma cells have shown dependency on CDK11 for growth (Feng et al. 2015). Thus, a clear understanding of biological processes underlying osteosarcoma and an urgent requirement of novel chemotherapeutic agents are needed. In recent years, the CRISPR/Cas9 system of bacterial host defense mechanisms has proven to be a better genome-editing tool in cancer diagnosis. Feng et al. have given evidence for utilizing the CRISPR/Cas9 system as a robust genome-editing tool for determining direct effect of CDK11 gene in osteosarcoma cell line by efficiently silencing it. The CDK11 inhibition is connected with decreased cellular proliferation and viability in osteosarcoma cell lines, and thus, CDK11 knockout could be used as a powerful prognostic marker for diagnosing osteosarcoma (Feng et al. 2015). Furthermore, Liao et al. (2017) have proved that higher level of PD-L1 expression in osteosarcoma patients could also be disrupted by CRISPR/Cas9 strategy and PD-L1 knockout can be used as a promising therapeutic approach for osteosarcoma diagnosis. Another recent report for osteosarcoma diagnosis was established by Wu et al. (2020b) where they reported the development of kinase library for CRISPR/Cas9 which can be used as a promising tool for genomic screening in osteosarcoma as well as for drug discovery in any other kind of cancer.
Ovarian cancer
Ovarian cancer is one of the cancer-associated causes of mortality in female individuals (Permuth-Wey and Sellers 2009). It is of extreme importance to understand the molecular mechanism behind tumor development and determine certain oncogenic markers for inhibiting its malignancy. The over-expression of OC-2 gene has been proven to be highly regulating tumor progression in ovarian cancer (Khaider et al. 2012; Wu et al. 2020a). Lu et al. (2020a) have reported a CRISPR/Cas9-based genome-editing tool which directly inhibits the OC-2 expression in ovarian cancer cell lines and, thus, finally results in down regulation of pre-tumor growth factors such as FGF2, vascular endothelial growth factor, and human growth factors. Furthermore, utilizing CRISPR/Cas9 editing tool for targeting altered DNA methyltransferase I might be used as a potential therapeutic agent for ovarian cancer diagnosis (He et al. 2018). In addition, CRISPR/Cas9 system-mediated induction of mutations in genes which are found altered in high-grade-serous ovarian cancer showed the dual origin for these high-grade tumors which could also be used to design biomarkers for studying their molecular mechanisms (Govindarajan et al. 2020; Lõhmussaar et al. 2020).
Hepatocellular cancer
Hepatocellular cancer is the second most leading cancer-related death in humans worldwide (Savitha et al. 2017). Despite several advancements in interpreting various processes regulating the hepatocellular carcinoma progression through tumor-associated genes such as TP53, the underlying mechanisms driving the tumor progression are rarely understood. Thus, it is important to discover carcinoma progression mechanisms with an aim to improve diagnosis for individuals suffering from hepatocellular cancer. The nuclear receptor-binding SET domain proteins (NSD) are known to be involved in the tumorigenesis process (Bennett et al. 2017), while in reference to hepatocellular carcinoma, the molecular mechanisms of these proteins are rarely discovered. The CRISPR/Cas9-mediated genome-editing tool is an efficient approach in inducing variations with single guideRNA or producing knockout of genetic fragments with multiple guideRNAs in human cells to better understand the biological mechanism underlying any disorder (Rodríguez-Rodríguez et al. 2019). For example, CRISPR/Cas9 system influences alterations in Pten and P53 genes which has been reported to assist hepatocellular cancer development in transgenic mice (Liu et al. 2017c). In addition, genome editing of CXC chemokine receptor 4 through CRISPR/Cas9 system has been reported to reduce cellular proliferation and progression of hepatocellular carcinoma in living cells (Wang et al. 2017). Another study reported that CRISPR/Cas9 system mediates knockout of NSD-1 which assists in suppression of hepatocellular carcinoma progression and transfer in cells, suggesting that NSD1 could be treated as a potential regiment for its diagnosis (Zhang et al. 2019b).
Advancements of CRISPR/Cas9 technique on mouse cancer models
As cancer genomes are characterized with complex prospects of mutations and several kinds of genomic alterations (Bailey et al. 2018). One of the major limitations in interpreting the cancer genome is to extract the genetic information about mutations that are controlling the tumor evolution process (Vogelstein et al. 2013). Genetic screening is one of the most substantial tools for ascertaining pathogenic mutations in tumor progression (Hanahan and Weinberg 2000, 2011). In the past, RNA interference and open-reading frame expression studies have been extensively used for predicting cancerous genes in oncogenic mouse models (Livshits and Lowe 2013). Currently, the Cas9 nuclease enzyme from the bacterial type II CRISPR system has been exploited majorly in eukaryotic species (Cong et al. 2013). The CRISPR system is used to introduce genetic alterations in less than 4 weeks of duration, thus providing an efficient platform for functionally annotating the cancer genome. In particular, there are currently two interrelated approaches available for building mouse models through genome editing: germline and somatic. The germline mouse models could be generated by inducing cancer-derived variations into mice embryonic stem cells, thereby generating germline-transmitting alleles and permitting their maintenance through animal husbandry. The CRISPR/Cas9 system can successfully develop double-stranded breaks into mouse embryos and thus generating genetically modified mice which are briefly not resting on their suitable embryonic stem cells (Amitai and Sorek 2016; Inui et al. 2014). In contrast, the somatic cancer mouse models reiterate the somatic capability of tumor progression and divert the embryonic damage caused due to knockout of particular genes in mice (Lampreht Tratar et al. 2018). The brief applications of CRISPR technology in mouse models and cancer research are shown in Table 3.
Table 3.
Generation of different animal models using CRISPR approach to study about various cancers
| Cancer type | Animal model | References |
|---|---|---|
| Non-small cell lung cancers (NSCLC) | Mouse model of Eml4-Alk-driven lung cancer was generated using the CRISPR/Cas9 technology | Maddalo et al. (2014), Blasco et al. (2014) |
| Liver cancer | Direct mutation of tumor-suppressor genes such as PTEN and p53 and oncogenes in the mouse liver using the CRISPR/Cas system, may lead to the rapid development of the animal models | Xue et al. (2014) |
| Lung adenocarcinoma | KRAS, p53, and LKB1genes were mutated for lung adenocarcinoma for developing a mouse model | Platt et al. (2014) |
| Lung adenocarcinoma | Cre-dependent somatic activation of oncogenic K-Ras (G12D) along with CRISPR/Cas9-mediated genome editing of tumor-suppressor genes resulted in lung adenocarcinomas mouse models | Sanchez-Rivera et al. (2014) |
| Pancreatic ductal adenocarcinoma | CRISPR/Cas9-mediated gene inactivation of Lkb1in combination with oncogenic K-Ras expression in mice pancreas | Chiou et al. (2015) |
| Alveolar rhabdomyosarcoma (A-RMS) | In vivo mimicking of reciprocal translocation t(2;13)(q36.1;q14.1) in mice creates a pathognomonic PAX3-FOXO1 fusion gene | Lagutina et al. (2015) |
| Brain tumor | Deleting single (Ptch1) or multiple genes (Trp53, Pten, Nf1) in the mouse brain, using CRISPR/Cas9, results in the development of medulloblastoma and glioblastoma, respectively | Zuckermann et al. (2015) |
| Pancreatic cancer | Multiplex genetic engineering of multiple genes in the pancreatic cells of mice | Maresch et al. (2016) |
| Breast cancer | PYCR1 knockout in an in vivo mouse disease model | Loayza-Puch et al. (2016) |
| Lobular breast carcinoma | Generation of mice models by targeting the ILC-initiating cells and induce specific gene disruption of PTEN with the CRISPR/Cas9 gene-editing tool | Annunziato et al. (2016) |
| Ovarian cancer | CRISPR/Cas9-mediated Trp53 and BRCA2 Knockout to generate improved murine models of Ovarian High-Grade Serous Carcinoma | Walton et al. (2016) |
| Retinoblastoma | Knockout of rb1 and rbl1 leads to retinoblastoma development in Xenopus tropicalis | Naert et al. (2016) |
| Metastatic renal cell carcinoma (mRCC) | Knock out of Von Hippel Lindau (VHL), a tumor suppressor in the RENCA model leads to morphologic and molecular changes indicative of epithelial-mesenchymal transition (EMT) phenotype, which in turn drives increased metastasis to the lungs | Schokrpur et al. (2016) |
| Colorectal cancer (CRC) | Tumor formation is induced editing of the tumor-suppressor genes such as Apc and in colon epithelial cells and later by transplantation of Apc-edited colon organoids | Roper et al. (2017) |
| Ovarian cancer | CRISPR/Cas9 gene editing was used to generate derivatives with deletions in Brca1, PTEN and Nf1 to produce murine models of ovarian high-grade-serous carcinoma (HGSC) | Walton et al. (2017) |
| Brain tumor | For precise modeling of human tumors through the somatic deletion of tumor-suppressor genes, such as Trp53, Cdkn2a, and Pten in the neural stem cells, a series of mouse strains were generated by combined approach of RCAS-TVA and CRISPR/Cas-9 | Oldrini et al. (2018) |
| Retinoblastoma | Rb1 knockout in Xenopus tropicalis using CRISPR/Cas9 tool | Naert and Vleminckx (2018) |
| Leukemia | CRISPR/Cas9 gene editing is used to induce MLL chromosomal translocation t(9;11) in human hematopoietic stem and progenitor cells | Jeong et al. (2019) |
| Brain tumors | Accurate cell targeting of genetic recombination, Sleeping beauty and piggyBac transposons and CRISPR/Cas9 for generating genetic knockout and functional screens | Noorani (2019) |
| Hepatocellular carcinoma (HCC) | Specific knock-out of Androgen Receptor (AR) gene in the liver of zebrafish via the CRISPR/Cas9 system | Li et al. (2019a) |
| Leukemia | An experimental pipeline, based on mosaic genome editing by CRISPR/Cas9, to generate high-performing leukemia models in Xenopus tropicalis | Dimitrakopoulou et al. (2019) |
| Small cell lung cancer (SCLC) | Using CRISPR/Cas9 genome-editing tool, a murine model of SCLC is rapidly modeled for the loss of the gene p107, which significantly accelerated the tumor progression | Ng et al. (2020) |
| Breast cancer | Generation of a knock-in mouse with Cre-conditional expression of a cytidine base editor | Annunziato et al. (2020) |
CRISPR in cancer clinical trial
The somatic genome editing with CRISPR/Cas9 approach has enhanced the therapeutic applications of genes and immunotherapy in cancer biology (Sayin and Papagiannakopoulos 2017). For instance, in vivo modeling of repetitive ERCC3 variation contributes to medium risk of breast tumor in ERCC3-deficient cells, thus permitting to visualize effects of ERCC3 malformed-repair phenotype (Vijai et al. 2016). Furthermore, genome-editing process has also enabled in vivo sensitive biomarkers prediction for smaller biomolecules, for, e.g., detecting SLFN11 which is sensitive to PARP inhibitors as a predictive biomarker for lung cancer (Lok et al. 2017). Moreover, examination of mutations of EGFR tyrosine kinase inhibitors in EGFR-mutant non-small cell lung cancer has led to detection of pathogenic variations and thus resulting in better targeted resistance therapies (Leonetti et al. 2019). The genome editing through CRISPR/Cas9 knockout has allowed the progression of mutations and detection of candidate risk genes connected with p53 and KRAS proto-oncogenes in mouse embryonic fibroblasts, assisting in comprehension of primary mice sarcomas (Huang et al. 2019). In another study, total knockout of ORF57 was performed in HEK293/Bac36 cells and simultaneously expressed in Cas9 protein along with two guide RNAs connected with cell selection from an isolated vector (BeltCappellino et al. 2019). In further clinical trials, investigators have examined the role of FRK oncogene in lung cancer cell lines, depicting FRK knockout leading to cellular colony formation cancer cell proliferation (Zhang et al. 2020). In a recent report, ex vivo genome editing for cancer treatments is performed through chimeric antigen receptor T-cell (CAR-T) therapy and PD-1-induced immunotherapy utilizing CRISPR/Cas9 editing approach to destroy the tumors (Stadtmauer et al. 2020). In this clinical trial NCT03399448, three patients were administered CRIPSR-mediated T cells where two patients were suffering from multiple myeloma and one patient was suffering from liposarcoma. All the patients were heavily pretreated both by chemotherapeutic regimens and transplants. Since the trial, 1 patient has died and there is disease progression in rest of the two patients (Stadtmauer et al. 2020). In addition, CRISPR/Cas9 systems are also used to produce in vivo chromosomal abnormalities such as deletions, duplications, insertions, and inversions (Cheong et al. 2018). The results of the first-in-human phase I clinical trial of CRISPR/Cas9 PD-1-edited T cells in patients with advanced non-small-cell lung cancer (NCT02793856) were reported to be safe, feasible, and effective (Lu et al. 2020b). In another clinical trial, genome-wide CRISPR screen was used to identify the reduction in gemcitabine-induced apoptosis in the Gall Bladder cancer cells, due to the loss of ELP5, which results in the poor survival rate of the patients with lower ELP5, hnRNPQ, or P53 expression after the gemcitabine chemotherapy (Xu et al. 2019c). All the above-cited instances support the efficacy of CRISPR/Cas9 system in both in vivo and in vitro studies and their implementation in gene therapy fields. However, the approach has not been used extensively; the increased reported studies will probably promote its understanding in wide applications in the medical field and contribute toward better understanding in disease etiology (Doudna 2020).
CRISPR/Cas9 and long non-coding RNAs
Long non-coding RNAs constitute a large family of non-coding endogenous RNAs with > 200 nt, which are considered to play a major role in regulation of biological processes (Mercer et al. 2009). There is also evidence, suggesting that lncRNA plays a pivotal role in the development of a number of human cancers (Jiang et al. 2019). These are also identified as potential biomarkers for various diseases. While numerous lncRNAs are characterized and studied, the biological functions of most of the lncRNAs are yet to be discovered (Bhan et al. 2017). Understanding each of the lncRNA and their role in regulation is critical for various fields in biological research such as developmental biology, genetics, evolution, etc. Genetic modification is one of the essential approaches to study the function of the lncRNAs and other genes, which has been accelerated by the usage of CRISPR/Cas9 gene-editing technology (Goyal et al. 2017). Three common strategies are used to knock down the lncRNAs: (i) RNA degradation by RNA interference (RNAi; Elbashir et al. 2001); (ii) degradation of the RNAs by RNase H, which is activated by antisense oligonucleotides (ASOs) (Bennett and Swayze 2010); (iii) CRISPR/Cas9 technology (Liu et al. 2017b). Similarly, as the CRISPR/Cas9 is seen as the revolutionary genome-editing tool for all areas of molecular biology, it can also be used in the lncRNA research to delete lncRNA genes to introduce RNA-destabilizing elements into their locus. Each of these methods has their own advantages and disadvantages, and the success of the editing process is also influenced by the sub-cellular localization of the lncRNAs. Some lncRNAs are located in the nucleus and are involved in the regulation of transcription and RNA processing, whereas other lncRNAs are located in the cytoplasm, where they target the protein localization, mRNA stability, and translation (Zhang et al. 2019c). Some lncRNAs are found to be equally present in both nucleus and cytoplasm. RNAi is one of the commonly used methods for the knockdown of the lncRNAs within the nucleus. It utilizes the RNAi-induced silencing complex (RISC), a multiprotein molecule with siRNA to target a specific RNA for degradation. While the ASOs are used for editing the lncRNAs present in the cytoplasm. They are used to target and bind to the RNA which then employs endogenous RNase H1 enzyme which readily cleaves the RNA in the heteroduplex RNA:DNA. It has also been reported that only 38% of the lncRNA loci are safely amenable to apply CRISPR technology, whereas the remaining lncRNA loci are at risk of deregulating the adjacent genes as most of these lncRNAs are derived from bidirectional promoters or they overlap with promoters or regions of sense or antisense genes. There will be some cases where lncRNAs will not be knocked down either by RNAi or by ASOs which could be due to their sub-cellular localization and inaccessibility to RNase H or RNAi machinery. In that situation, CRISPR technology will play a very important role, and currently, there are many researches which have successfully implemented CRISPR/Cas9 genome-editing technology to knock out lncRNAs (Horlbeck et al. 2020). One such study by Singh et al. (2016) involved deletion of BC200, a lncRNA which is known to be involved in protein synthesis in ER-positive breast tumors, in vitro and in vivo which resulted in the reduced cell growth through expression of a proapoptotic protein (Richard and Eichhorn 2018). This also opened up new avenues for therapeutic approaches for treatment of breast cancer.
Challenges in using CRISPR technology in cancer
The CRISPR–Cas9 system has facilitated specific genome-targeted editing processes and has been comprehensively applied to cancer treatment therapies in a broader way, opening new possibilities for cancer management. However, there remain a few challenges concerning the efficiency and precision which needs to be concerted for clinical and diagnostic applications, such as gene-editing ability, plausible non-specific insertion, potential off-targets effects, and delivery mechanisms. Hence, it is important to consider crucial factors that might influence the clinical outcome of CRISPR–Cas9 system-mediated genomic manipulation in cancer.
Genome-editing efficiency is the major factor for the utility of the CRISPR technology, because as of now, the system can result in significant level of changes in the non-targeted sites (Zhang et al. 2015). These undesirable mutations may be silent or may have deleterious effects. The various mechanisms have already been discussed. Another important challenge came across the development of CRISPR–Cas9 system is its successful delivery to the target cells in vivo for its implementation in clinical therapeutics. Adeno Associated Virus (AAV) associated Cas9 transportation occurs well in a laboratory system, but some limitations still emerge in clinical and therapeutic settings. For example, expression of some important genes could be altered when transgene is introduced into the target genomic region. Some of the physical delivery methods including Electroporation have been widely used for ribonucleoproteins delivery which has been successfully employed in Cas9–sgRNA transportation into cells in vitro. Other methods for Cas9–sgRNA delivery include microinjections and liposome-mediated transfection systems. Since, Cas9–sgRNA complex is anionic, thus cationic lipid transfection reagent can be used to transport this complex which further results in around 80% genetic alteration in cells with enhanced editing accuracy (Lino et al. 2018). Moreover, the most suitable carriers could be non-viral-mediated in vivo delivery of nucleases in RNA or protein which revokes the transcriptional and translational processes. In contrast to virus-mediated and DNA/RNA vectors, the nanoparticle-based delivery mechanisms might have potential advantages, leading to precise dose duration, minimized risk of off-target cleavage effects, and reduced immunogenic and cytotoxic effects (Wilbie et al. 2019). These entire advantages support a safe and highly efficient genome modification process in vivo. However, nanoparticle-mediated ribonucleoprotein harbors a few challenges in the form of difficult packing into smaller particles to maintain intact biological function and to further avoid their degradation prior to entering nuclei of cells. Hence, the non-viral delivery reagent should be biologically compatible, non-tumorigenic, and non-cytotoxic, and also have the capability to transport Cas9–sgRNA complex to the cell nuclei for efficient genome-editing mechanism (Chen et al. 2020).
Discussion
In this review, we have attempted to summarize the role of CRISPR/Cas9 in different fields, but mainly focusing on cancers such as lung cancer, bone cancer, prostate cancer, liver cancer, etc. We have also made a comparison among CRISPR/Cas9 and other established genome-editing technologies. Out of all the other established technologies, CRISPR is the most advanced and highly efficient technique. Because of its high efficiency, CRIPSR holds a great potential for therapeutic applications. There are still some limitations for CRISPR which include proper delivery to the target cells and to minimize off-target effects. There has been continuous modification for better implementation of CRISPR technology in humans.
Conclusions
CRISPR-mediated genome editing has a great potential in the field of cancer therapeutics, but for its successful implementation, there are certain issues which need to be resolved; for example, standardization of delivery methods to minimize off-target effects and to reduce toxicity to other cells, etc. Despite all these challenges, CRISPR/Cas9 has given an invaluable contribution in the field of gene therapy and played an important role in cancer treatment. Also, CRISPR/Cas9 is able to manipulate non-coding regions of the genome which gives a valuable source for understanding the poorly characterized part of the cancer genome. Another major application of CRISPR technology is the generation of chimeric antigen receptor (CAR) T cells that can recognize specific antigens on cancer cells. The research in CAR-T cells has been increasing tremendously, and for this purpose, CRISPR/Cas9 can play an indispensable role.
Acknowledgements
The authors gratefully acknowledge Dr. Prashanth N Suravajhala for his critical comments and suggestions. JNS acknowledges Department of Biotechnology, GOI for Ramalingaswamy re-entry fellowship awarded to him. NS gratefully acknowledges Department of Science and technology, GOI for Women Scientist fellowship (WOS-A) award.
Abbreviations
- AAV
Adeno-associated virus
- CRISPR
Clustered regularly interspaced short palindromic repeats
- Cas
CRISPR-associated (protein)
- ZFN
Zinc finger nucleases
- TALENs
Transcription activator-like effector nucleases
- crRNA
CRISPR-RNA
- scaRNA
Small, CRISPR–Cas-associated RNA
- sgRNA
Single guide RNA
- tracrRNA
Trans-activating CRISPR-RNA
- PAM
Protospacer-Associated Motif
- REPAIR
RNA editing for programmable A-to-I replacement
- RNP
Ribonucleoprotein
- NHEJ
Non-homologous end-joining
- HDR
Homology Direct repair
- DSBs
Double-stranded breaks
- TCR
T-cell receptors
- HGT
Horizontal gene transfer
Authors’ contributions
NaS, SG, and NiS conceived the manuscript. NaS and SG wrote the first draft with NiS. AG prepared the figures. BM helped with all the tables in the manuscript. JNS helped in the revision of the manuscript. NiS proofread the manuscript before uploading and all others agreed to the changes in the manuscript. All authors contributed to the article and approved the submitted version.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Narmadhaa Siva and Sonal Gupta have contributed equally to this work
References
- Adli M. The CRISPR tool kit for genome editing and beyond. Nat Commun. 2018 doi: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amitai G, Sorek R. CRISPR–Cas adaptation: insights into the mechanism of action. Nat Rev Microbiol. 2016 doi: 10.1038/nrmicro.2015.14. [DOI] [PubMed] [Google Scholar]
- Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014 doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziato S, Kas SM, Nethe M, et al. Modeling invasive lobular breast carcinoma by CRISPR/Cas9-mediated somatic genome editing of the mammary gland. Genes Dev. 2016 doi: 10.1101/gad.279190.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziato S, Lutz C, Henneman L, et al. In situ CRISPR–Cas9 base editing for the development of genetically engineered mouse models of breast cancer. EMBO J. 2020 doi: 10.15252/embj.2019102169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora L, Narula A. Gene editing and crop improvement using CRISPR–cas9 system. Front Plant Sci. 2017;8:193. doi: 10.3389/fpls.2017.01932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashmore-Harris C, Fruhwirth GO. The clinical potential of gene editing as a tool to engineer cell-based therapeutics. Clin Transl Med. 2020 doi: 10.1186/s40169-020-0268-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azeez A, Busov V. CRISPR/Cas9-mediated single and biallelic knockout of poplar STERILE APETALA (PopSAP) leads to complete reproductive sterility. Plant Biotechnol J. 2020 doi: 10.1111/pbi.13451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MH, Tokheim C, Porta-Pardo E, et al. Comprehensive characterization of cancer driver genes and mutations. Cell. 2018 doi: 10.1016/j.cell.2018.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barman NC, Khan NM, Islam M, et al. CRISPR–Cas9: a promising genome editing therapeutic tool for alzheimer’s disease—a narrative review. Neurol Ther. 2020;9:419–434. doi: 10.1007/s40120-020-00218-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Marraffini LA. CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol Cell. 2014;54(2):234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science (80−) 2007 doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- BeltCappellino A, Majerciak V, Lobanov A, et al. CRISPR/Cas9-mediated knockout and in situ inversion of the ORF57 gene from all copies of the Kaposi’s sarcoma-associated herpesvirus genome in BCBL-1 cells. J Virol. 2019 doi: 10.1128/jvi.00628-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010 doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- Bennett RL, Swaroop A, Troche C, Licht JD. The role of nuclear receptor-binding SET domain family histone lysine methyltransferases in cancer. Cold Spring Harb Perspect Med. 2017;7(6):a026708. doi: 10.1101/cshperspect.a026708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco RB, Karaca E, Ambrogio C, et al. Simple and rapid invivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Rep. 2014 doi: 10.1016/j.celrep.2014.10.051. [DOI] [PubMed] [Google Scholar]
- Bloom K, Mussolino C, Arbuthnot P. Transcription activator-like effector (TALE) nucleases and repressor TALEs for antiviral gene therapy. Curr Stem Cell Rep. 2015;1:1–8. doi: 10.1007/s40778-014-0008-7. [DOI] [Google Scholar]
- Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol. 2010 doi: 10.1146/annurev-phyto-080508-081936. [DOI] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science (80−) 2009 doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333(6051):1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Dusko Ehrlich S. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005 doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- Brianese RC, Nakamura KDdM, de Almeida FGdSR, et al. BRCA1 deficiency is a recurrent event in early-onset triple-negative breast cancer: a comprehensive analysis of germline mutations and somatic promoter methylation. Breast Cancer Res Treat. 2018 doi: 10.1007/s10549-017-4552-6. [DOI] [PubMed] [Google Scholar]
- Brinkman EK, Chen T, de Haas M, et al. Kinetics and fidelity of the repair of Cas9-induced double-strand DNA breaks. Mol Cell. 2018 doi: 10.1016/j.molcel.2018.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJJ, Jore MM, Lundgren M, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science (80−) 2008 doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmistrz M, Krakowski K, Krawczyk-Balska A. RNA-targeting CRISPR–Cas systems and their applications. Int J Mol Sci. 2020;21(3):1122. doi: 10.3390/ijms21031122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein D, Harrington LB, Strutt SC, et al. New CRISPR–Cas systems from uncultivated microbes. Nature. 2017 doi: 10.1038/nature21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D. Genome editing: past, present, and future. Yale J Biol Med. 2017;90(4):653–659. [PMC free article] [PubMed] [Google Scholar]
- Chakraborty G, Armenia J, Mazzu YZ, et al. Significance of BRCA2 and RB1 co-loss in aggressive prostate cancer progression. Clin Cancer Res. 2020 doi: 10.1158/1078-0432.CCR-19-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran J, Brumin M, Wolf D, et al. Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol Plant Pathol. 2016 doi: 10.1111/mpp.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier E, Richter H, van der Oost J, White MF. Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity. FEMS Microbiol Rev. 2015;39(3):428–441. doi: 10.1093/femsre/fuv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Wen F, Sun N, Zhao H. Directed evolution of homing endonuclease I-SceI with altered sequence specificity. Protein Eng Des Sel. 2009 doi: 10.1093/protein/gzp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Choi J, Bailey S. Cut site selection by the two nuclease domains of the Cas9 RNA-guided endonuclease. J Biol Chem. 2014 doi: 10.1074/jbc.M113.539726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JS, Dagdas YS, Kleinstiver BP, et al. Enhanced proofreading governs CRISPR–Cas9 targeting accuracy. Nature. 2017 doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Alphonse M, Liu Q. Strategies for nonviral nanoparticle-based delivery of CRISPR/Cas9 therapeutics. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2020;12(3):e1609. doi: 10.1002/wnan.1609. [DOI] [PubMed] [Google Scholar]
- Cheng R, Peng J, Yan Y, et al. Efficient gene editing in adult mouse livers via adenoviral delivery of CRISPR/Cas9. FEBS Lett. 2014 doi: 10.1016/j.febslet.2014.09.008. [DOI] [PubMed] [Google Scholar]
- Cheong TC, Blasco RB, Chiarle R (2018) The CRISPR/Cas9 system as a tool to engineer chromosomal translocation in vivo. In: Advances in experimental medicine and biology [DOI] [PubMed]
- Cheung AHK, Chow C, Zhang J, et al. Specific targeting of point mutations in EGFR L858R-positive lung cancer by CRISPR/Cas9. Lab Investig. 2018 doi: 10.1038/s41374-018-0056-1. [DOI] [PubMed] [Google Scholar]
- Chevalier BS, Stoddard BL. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 2001 doi: 10.1093/nar/29.18.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew WL, Tabebordbar M, Cheng JKW, et al. A multifunctional AAV-CRISPR–Cas9 and its host response. Nat Methods. 2016 doi: 10.1038/nmeth.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou SH, Winters IP, Wang J, et al. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes Dev. 2015 doi: 10.1101/gad.264861.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chylinski K, Makarova KS, Charpentier E, Koonin EV. Classification and evolution of type II CRISPR–Cas systems. Nucleic Acids Res. 2014;42(10):6091–6105. doi: 10.1093/nar/gku241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton CC. Colorectal carcinoma: diagnostic, prognostic, and molecular features. Mod Pathol. 2003;16:376–388. doi: 10.1097/01.MP.0000062859.46942.93. [DOI] [PubMed] [Google Scholar]
- Cong L, Zhang F. Genome engineering using CRISPR–Cas9 system. Methods Mol Biol. 2015 doi: 10.1007/978-1-4939-1862-1_10. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science (80−) 2013 doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan-Curay J, O’Reilly M, Kohn DB, et al (2015) Genome editing technologies: defining a path to clinic. In: Molecular therapy. pp 796–806 [DOI] [PMC free article] [PubMed]
- Davis D, Stokoe D. Zinc finger nucleases as tools to understand and treat human diseases. BMC Med. 2010;8:42. doi: 10.1186/1741-7015-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Masi C, Spitalieri P, Murdocca M, et al. Application of CRISPR/Cas9 to human-induced pluripotent stem cells: From gene editing to drug discovery. Hum Genomics. 2020;14:25. doi: 10.1186/s40246-020-00276-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011 doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng D, Yan C, Pan X, et al. Structural basis for sequence-specific recognition of DNA by TAL effectors. Science (80−) 2012 doi: 10.1126/science.1215670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devarakonda S, Morgensztern D, Govindan R. Genomic alterations in lung adenocarcinoma. Lancet Oncol. 2015;16(7):e342–351. doi: 10.1016/S1470-2045(15)00077-7. [DOI] [PubMed] [Google Scholar]
- Dimitrakopoulou D, Tulkens D, Van Vlierberghe P, Vleminckx K (2019) Xenopus tropicalis: joining the Armada in the fight against blood cancer. Front Physiol 10:48. 10.3389/fphys.2019.00048. Erratum in: Front Physiol 10:210 [DOI] [PMC free article] [PubMed]
- Doudna J. Genome-editing revolution: my whirlwind year with CRISPR. Nature. 2015;528(7583):469–471. doi: 10.1038/528469a. [DOI] [PubMed] [Google Scholar]
- Doudna JA. The promise and challenge of therapeutic genome editing. Nature. 2020;578:229–236. doi: 10.1038/s41586-020-1978-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science (80−) 2014 doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Du H, Zeng X, Zhao M, et al. Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J Biotechnol. 2016 doi: 10.1016/j.jbiotec.2015.11.005. [DOI] [PubMed] [Google Scholar]
- Durai S, Mani M, Kandavelou K, et al. Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005 doi: 10.1093/nar/gki912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand S, Gilet L, Condon C. The essential function of B. subtilis RNase III is to silence foreign toxin genes. PLoS Genet. 2012 doi: 10.1371/journal.pgen.1003181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebright RY, Lee S, Wittner BS, et al. Deregulation of ribosomal protein expression and translation promotes breast cancer metastasis. Science (80−) 2020 doi: 10.1126/science.aay0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egorova TV, Zotova ED, Reshetov DA, et al. CRISPR/Cas9-generated mouse model of Duchenne muscular dystrophy recapitulating a newly identified large 430 kb deletion in the human DMD gene. DMM Dis Model Mech. 2019 doi: 10.1242/dmm.037655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001 doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Esposito CL, Catuogno S, Condorelli G, Ungaro P, de Franciscis V. Aptamer chimeras for therapeutic delivery: the challenging perspectives. Genes (Basel) 2018;9(11):529. doi: 10.3390/genes9110529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraoni I, Graziani G. Role of BRCA mutations in cancer treatment with poly(ADP-ribose) polymerase (PARP) inhibitors. Cancers (Basel) 2018;10:487. doi: 10.3390/cancers10120487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Sassi S, Shen JK, et al. Targeting Cdk11 in osteosarcoma cells using the CRISPR-cas9 system. J Orthop Res. 2015 doi: 10.1002/jor.22745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry LE, Peddle CF, Barnard AR, McClements ME, MacLaren RE. RNA editing as a therapeutic approach for retinal gene therapy requiring long coding sequences. Int J Mol Sci. 2020;21(3):777. doi: 10.3390/ijms21030777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M, Umeno H, Nose K, et al. Construction of a guide-RNA for site-directed RNA mutagenesis utilising intracellular A-To-I RNA editing. Sci Rep. 2017 doi: 10.1038/srep41478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31(7):397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galetto R, Duchateau P, Pâques F. Targeted approaches for gene therapy and the emergence of engineered meganucleases. Expert Opin Biol Ther. 2009;9(10):1289–1303. doi: 10.1517/14712590903213669. [DOI] [PubMed] [Google Scholar]
- Gao Q, Ouyang W, Kang B, et al. Selective targeting of the oncogenic KRAS G12S mutant allele by CRISPR/Cas9 induces efficient tumor regression. Theranostics. 2020 doi: 10.7150/thno.42325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis MÈ, Villion M, et al. The CRISPR/cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010 doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153(1):17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Geng Y, Deng Z, Sun Y. An insight into the protospacer adjacent motif of Streptococcus pyogenes Cas9 with artificially stimulated RNA-guided-Cas9 DNA cleavage flexibility. RSC Adv. 2016 doi: 10.1039/c6ra02774a. [DOI] [Google Scholar]
- Gersbach CA. Genome engineering: The next genomic revolution. Nat Methods. 2014 doi: 10.1038/nmeth.3113. [DOI] [PubMed] [Google Scholar]
- Gleditzsch D, Pausch P, Müller-Esparza H, Özcan A, Guo X, Bange G, Randau L. PAM identification by CRISPR–Cas effector complexes: diversified mechanisms and structures. RNA Biol. 2019;16(4):504–517. doi: 10.1080/15476286.2018.1504546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Salinas F, Rojo R, Martinez-Amador C, et al. Transcriptomic and cellular analyses of CRISPR/Cas9-mediated edition of FASN show inhibition of aggressive characteristics in breast cancer cells. Biochem Biophys Res Commun. 2020 doi: 10.1016/j.bbrc.2020.05.172. [DOI] [PubMed] [Google Scholar]
- Govindarajan M, Wohlmuth C, Waas M, Bernardini MQ, Kislinger T, et al. High-throughput approaches for precision medicine in high-grade serous ovarian cancer. J Hematol Oncol. 2020;13(1):134. doi: 10.1186/s13045-020-00971-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal A, Myacheva K, Groß M, et al. Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkw883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granados-Riveron JT, Aquino-Jarquin G. CRISPR–Cas13 precision transcriptome engineering in cancer. Cancer Res. 2018;78:4107–4113. doi: 10.1158/0008-5472.CAN-18-0785. [DOI] [PubMed] [Google Scholar]
- Grizot S, Epinat JC, Thomas S, et al. Generation of redesigned homing endonucleases comprising DNA-binding domains derived from two different scaffolds. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha TK, Edgell DR. Applications of alternative nucleases in the age of CRISPR/Cas9. Int J Mol Sci. 2017;18(12):2565. doi: 10.3390/ijms18122565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha TK, Wai A, Hausner G. Programmable genome editing tools and their regulation for efficient genome engineering. Comput Struct Biotechnol J. 2017;15:146–160. doi: 10.1016/j.csbj.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Yang J, Huang J, et al. Therapeutic genome editing of triple-negative breast tumors using a noncationic and deformable nanolipogel. Proc Natl Acad Sci USA. 2019 doi: 10.1073/pnas.1904697116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RM, Musunuru K. Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR–Cas9. J Clin Invest. 2014;124(10):4154–4161. doi: 10.1172/JCI72992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Christensen RG, Bell HA, et al. An improved predictive recognition model for Cys2-His 2 zinc finger proteins. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005 doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Harrington LB, Burstein D, Chen JS, et al. Programmed DNA destruction by miniature CRISPR–Cas14 enzymes. Science (80−) 2018 doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haun W, Coffman A, Clasen BM, et al. Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol J. 2014 doi: 10.1111/pbi.12201. [DOI] [PubMed] [Google Scholar]
- Hazafa A, Mumtaz M, Farooq MF, Bilal S, Chaudhry SN, Firdous M, Naeem H, Ullah MO, Yameen M, Mukhtiar MS, Zafar F, et al. CRISPR/Cas9: a powerful genome editing technique for the treatment of cancer cells with present challenges and future directions. Life Sci. 2020;263:118525. doi: 10.1016/j.lfs.2020.118525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He ZY, Zhang YG, Yang YH, et al. In vivo ovarian cancer gene therapy using CRISPR-Cas9. Hum Gene Ther. 2018 doi: 10.1089/hum.2017.209. [DOI] [PubMed] [Google Scholar]
- Heigwer F, Kerr G, Walther N, et al. E-TALEN: a web tool to design TALENs for genome engineering. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo-Cantabrana C, Goh YJ, Barrangou R. Characterization and repurposing of Type I and Type II CRISPR–Cas systems in bacteria. J Mol Biol. 2019;431(1):21–33. doi: 10.1016/j.jmb.2018.09.013. [DOI] [PubMed] [Google Scholar]
- Hille F, Richter H, Wong SP, Bratovič M, Ressel S, Charpentier E, et al. The biology of CRISPR-Cas: backward and forward. Cell. 2018;172(6):1239–1259. doi: 10.1016/j.cell.2017.11.032. [DOI] [PubMed] [Google Scholar]
- Hirotsune S, Kiyonari H, Jin M, et al. Enhanced homologous recombination by the modulation of targeting vector ends. Sci Rep. 2020 doi: 10.1038/s41598-020-58893-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holkers M, Maggio I, Liu J, et al. Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res. 2013 doi: 10.1093/nar/gks1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlbeck MA, Liu SJ, Chang HY, Lim DA, Weissman JS. Fitness effects of CRISPR/Cas9-targeting of long noncoding RNA genes. Nat Biotechnol. 2020;38(5):573–576. doi: 10.1038/s41587-020-0428-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327(5962):167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR–Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Chen M, Xu ES, et al. Genome-wide CRISPR screen to identify genes that suppress transformation in the presence of endogenous Kras G12D. Sci Rep. 2019 doi: 10.1038/s41598-019-53572-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inui M, Miyado M, Igarashi M, et al. Rapid generation of mouse models with defined point mutations by the CRISPR/Cas9 system. Sci Rep. 2014 doi: 10.1038/srep05396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino Y, Shinagawa H, Makino K, et al. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isoenzyme conversion in Escherichiacoli, and identification of the gene product. J Bacteriol. 1987 doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino Y, Krupovic M, Forterre P. History of CRISPR–Cas from encounter with a mysterious repeated sequence to genome editing technology. J Bacteriol. 2018 doi: 10.1128/JB.00580-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi D, Toden S, Ureta E, et al. TIAM1 promotes chemoresistance and tumor invasiveness in colorectal cancer. Cell Death Dis. 2019 doi: 10.1038/s41419-019-1493-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs TB, LaFayette PR, Schmitz RJ, Parrott WA. Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 2015 doi: 10.1186/s12896-015-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby K, Metzger M, Shen BW, et al. Expanding LAGLIDADG endonuclease scaffold diversity by rapidly surveying evolutionary sequence space. Nucleic Acids Res. 2012 doi: 10.1093/nar/gkr1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaganathan D, Ramasamy K, Sellamuthu G, Jayabalan S, Venkataraman G. CRISPR for crop improvement: an update review. Front Plant Sci. 2018;9:985. doi: 10.3389/fpls.2018.00985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Van Embden JDA, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002 doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- Jeong J, Jager A, Domizi P, et al. High-efficiency CRISPR induction of t(9;11) chromosomal translocations and acute leukemias in human blood stem cells. Blood Adv. 2019 doi: 10.1182/bloodadvances.2019000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M-C, Ni J-J, Cui W-Y, et al. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am J Cancer Res. 2019;9:1354. [PMC free article] [PubMed] [Google Scholar]
- Jiang FN, Liang YX, Wei W, et al. Functional classification of prostate cancer-associated miRNAs through CRISPR/Cas9-mediated gene knockout. Mol Med Rep. 2020 doi: 10.3892/mmr.2020.11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14(1):49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabadi AM, Ousterout DG, Hilton IB, Gersbach CA. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov FV, Hannon GJ. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol Cell. 2010;37(1):7–19. doi: 10.1016/j.molcel.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi Z, Ahmadi A, Najafi A, Ranjbar R. Bacterial CRISPR regions: general features and their potential for epidemiological molecular typing studies. Open Microbiol J. 2018 doi: 10.2174/1874285801812010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura N, Nimura K, Nagano H, et al. CRISPR/Cas9-mediated gene knockout of NANOG and NANOGP8 decreases the malignant potential of prostate cancer cells. Oncotarget. 2015 doi: 10.18632/oncotarget.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Kornepati AVR, Goldstein M, et al. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease. J Virol. 2014 doi: 10.1128/jvi.01879-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Bassit LC, Mueller H, et al. Suppression of hepatitis B virus DNA accumulation in chronically infected cells using a bacterial CRISPR/Cas RNA-guided DNA endonuclease. Virology. 2015 doi: 10.1016/j.virol.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaider NG, Lane D, Matte I, et al. Targeted ovarian cancer treatment: the TRAILs of resistance. Am J Cancer Res. 2012;2:75. [PMC free article] [PubMed] [Google Scholar]
- Khan SH. Genome-editing technologies: concept, pros, and cons of various genome-editing techniques and bioethical concerns for clinical application. Mol Ther Nucleic Acids. 2019;16:326–334. doi: 10.1016/j.omtn.2019.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kim JS. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016 doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu Rev Biochem. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- Koo T, Yoon AR, Cho HY, et al. Selective disruption of an oncogenic mutant allele by CRISPR/Cas9 induces efficient tumor regression. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR–Cas systems. Curr Opin Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kounatidou E, Nakjang S, McCracken SRC, et al. A novel CRISPR-engineered prostate cancer cell line defines the AR-V transcriptome and identifies PARP inhibitor sensitivities. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagutina IV, Valentine V, Picchione F, et al. Modeling of the human alveolar rhabdomyosarcoma Pax3-Foxo1 chromosome translocation in mouse myoblasts using CRISPR–Cas9 nuclease. PLoS Genet. 2015 doi: 10.1371/journal.pgen.1004951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampreht Tratar U, Horvat S, Cemazar M. Transgenic mouse models in cancer research. Front Oncol. 2018;8:268. doi: 10.3389/fonc.2018.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanigan TM, Kopera HC, Saunders TL. Principles of genetic engineering. Genes (Basel) 2020;11(3):291. doi: 10.3390/genes11030291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Bishop ES, Zhang R, Yu X, Farina EM, Yan S, Zhao C, Zheng Z, Shu Y, Wu X, Lei J, Li Y, Zhang W, Yang C, Wu K, Wu Y, Ho S, Athiviraham A, Lee MJ, Wolf JM, Reid RR, He TC. Adenovirus-mediated gene delivery: potential applications for gene and cell-based therapies in the new era of personalized medicine. Genes Dis. 2017;4(2):43–63. doi: 10.1016/j.gendis.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenay RT, Maksimchuk KR, Slotkowski RA, et al. Identifying and visualizing functional PAM diversity across CRISPR–Cas systems. Mol Cell. 2016 doi: 10.1016/j.molcel.2016.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonetti A, Sharma S, Minari R, Perego P, Giovannetti E, Tiseo M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br J Cancer. 2019;121(9):725–737. doi: 10.1038/s41416-019-0573-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, He ZY, Wei XW, Gao GP, Wei YQ. Challenges in CRISPR/CAS9 delivery: potential roles of nonviral vectors. Hum Gene Ther. 2015;26(7):452–462. doi: 10.1089/hum.2015.069. [DOI] [PubMed] [Google Scholar]
- Li Z, Bin LZ, Xing A, et al. Cas9-guide RNA directed genome editing in soybean. Plant Physiol. 2015 doi: 10.1104/pp.15.00783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Hu S, Chen X. Non-viral delivery systems for CRISPR/Cas9-based genome editing: challenges and opportunities. Biomaterials. 2018;171:207–218. doi: 10.1016/j.biomaterials.2018.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li Y, Lu JW, et al. Liver-specific androgen receptor knockout attenuates early liver tumor development in zebrafish. Sci Rep. 2019 doi: 10.1038/s41598-019-46378-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Cho MY, Lee S, et al. CRISPR-Cas9 mediated CD133 knockout inhibits colon cancer invasion through reduced epithelial-mesenchymal transition. PLoS ONE. 2019 doi: 10.1371/journal.pone.0220860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YS, Liu Q, He HB, Luo W. The possible role of insulin-like growth factor-1 in osteosarcoma. Curr Probl Cancer. 2019;43(3):228–235. doi: 10.1016/j.currproblcancer.2018.08.008. [DOI] [PubMed] [Google Scholar]
- Li H, Yang Y, Hong W, et al. Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduct Target Ther. 2020;5:1–23. doi: 10.1038/s41392-019-0089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhao L, Lau YS, et al. Genome-wide CRISPR screen identifies LGALS2 as an oxidative stress-responsive gene with an inhibitory function on colon tumor growth. Oncogene. 2020 doi: 10.1038/s41388-020-01523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Wu G, Zhao Y, et al. CRISPR/Cas9-mediated knockout and overexpression studies reveal a role of maize phytochrome C in regulating flowering time and plant height. Plant Biotechnol J. 2020 doi: 10.1111/pbi.13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Chen L, Feng Y, et al. Targeting programmed cell death ligand 1 by CRISPR/Cas9 in osteosarcoma cells. Oncotarget. 2017 doi: 10.18632/oncotarget.16326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima ZS, Ghadamzadeh M, Arashloo FT, et al. Recent advances of therapeutic targets based on the molecular signature in breast cancer: genetic mutations and implications for current treatment paradigms. J Hematol Oncol. 2019;12:38. doi: 10.1186/s13045-019-0725-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lino CA, Harper JC, Carney JP, Timlin JA. Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 2018;25(1):1234–1257. doi: 10.1080/10717544.2018.1474964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TY, Doudna JA. Chemistry of Class 1 CRISPR-Cas effectors: binding, editing, and regulation. J Biol Chem. 2020;295(42):14473–14487. doi: 10.1074/jbc.REV120.007034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Zhang L, Liu H, Cheng K. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J Control Release. 2017;266:17–26. doi: 10.1016/j.jconrel.2017.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SJ, Horlbeck MA, Cho SW, et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science (80−) 2017 doi: 10.1126/science.aah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Qi X, Zeng Z, et al. CRISPR/Cas9-mediated p53 and Pten dual mutation accelerates hepatocarcinogenesis in adult hepatitis B virus transgenic mice. Sci Rep. 2017 doi: 10.1038/s41598-017-03070-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Saber A, Haisma HJ. CRISPR/Cas9: a powerful tool for identification of new targets for cancer treatment. Drug Discov Today. 2019;24(4):955–970. doi: 10.1016/j.drudis.2019.02.011. [DOI] [PubMed] [Google Scholar]
- Liu JJ, Orlova N, Oakes BL, et al. CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature. 2019 doi: 10.1038/s41586-019-0908-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livshits G, Lowe SW. Accelerating cancer modeling with RNAi and nongermline genetically engineered mouse models. Cold Spring Harb Protoc. 2013 doi: 10.1101/pdb.top069856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loayza-Puch F, Rooijers K, Buil LCM, et al. Tumour-specific proline vulnerability uncovered by differential ribosome codon reading. Nature. 2016 doi: 10.1038/nature16982. [DOI] [PubMed] [Google Scholar]
- Lodish MB. Clinical review: kinase inhibitors: adverse effects related to the endocrine system. J Clin Endocrinol Metab. 2013;98(4):1333–1342. doi: 10.1210/jc.2012-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lõhmussaar K, Kopper O, Korving J, et al. Assessing the origin of high-grade serous ovarian cancer using CRISPR-modification of mouse organoids. Nat Commun. 2020 doi: 10.1038/s41467-020-16432-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok BH, Gardner EE, Schneeberger VE, et al. PARP Inhibitor activity correlates with slfn11 expression and demonstrates synergy with temozolomide in small cell lung cancer. Clin Cancer Res. 2017 doi: 10.1158/1078-0432.CCR-16-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lone BA, Karna SKL, Ahmad F, et al (2018) CRISPR/Cas9 system: a bacterial tailor for genomic engineering. Genet Res Int [DOI] [PMC free article] [PubMed]
- Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science. 2017;355(6330):1152–1158. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro A, Da Silva GJ. Crispr-cas: Converting a bacterial defence mechanism into a state-of-the-art genetic manipulation tool. Antibiotics. 2019 doi: 10.3390/antibiotics8010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Zhang L, Zhu W, et al. CRISPR/Cas9-mediated OC-2 editing inhibits the tumor growth and angiogenesis of ovarian cancer. Front Oncol. 2020 doi: 10.3389/fonc.2020.01529. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lu Y, Xue J, Deng T, et al. Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat Med. 2020 doi: 10.1038/s41591-020-0840-5. [DOI] [PubMed] [Google Scholar]
- Luther DC, Lee YW, Nagaraj H, Scaletti F, Rotello VM. Delivery approaches for CRISPR/Cas9 therapeutics in vivo: advances and challenges. Expert Opin Drug Deliv. 2018;15(9):905–913. doi: 10.1080/17425247.2018.1517746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma E, Harrington LB, O’Connell MR, et al. Single-stranded DNA cleavage by divergent CRISPR–Cas9 enzymes. Mol Cell. 2015 doi: 10.1016/j.molcel.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddalo D, Manchado E, Concepcion CP, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014 doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Gersbach CA. Genome-editing technologies for gene and cell therapy. Mol Ther. 2016;24:430–446. doi: 10.1038/mt.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Stefanidakis M, Wilson CJ, et al. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat Med. 2019 doi: 10.1038/s41591-018-0327-9. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, et al. Evolution and classification of the CRISPR–Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science (80−) 2013 doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manghwar H, Lindsey K, Zhang X, Jin S. CRISPR/Cas system: recent advances and future prospects for genome editing. Trends Plant Sci. 2019;24(12):1102–1125. doi: 10.1016/j.tplants.2019.09.006. [DOI] [PubMed] [Google Scholar]
- Mani M, Smith J, Kandavelou K, et al. Binding of two zinc finger nuclease monomers to two specific sites is required for effective double-strand DNA cleavage. Biochem Biophys Res Commun. 2005 doi: 10.1016/j.bbrc.2005.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresch R, Mueller S, Veltkamp C, et al. Multiplexed pancreatic genome engineering and cancer induction by transfection-based CRISPR/Cas9 delivery in mice. Nat Commun. 2016 doi: 10.1038/ncomms10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Miao J, Guo D, Zhang J, Huang Q, Qin G, Zhang X, Wan J, Gu H, Qu LJ. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 2013;23(10):1233–1236. doi: 10.1038/cr.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007 doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011 doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Mintz RL, Lao YH, Chi CW, et al. CRISPR/Cas9-mediated mutagenesis to validate the synergy between PARP1 inhibition and chemotherapy in BRCA1-mutated breast cancer cells. Bioeng Transl Med. 2020 doi: 10.1002/btm2.10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir A, Edraki A, Lee J, Sontheimer EJ. Type II-C CRISPR–Cas9 biology, mechanism, and application. ACS Chem Biol. 2018;13:357–365. doi: 10.1021/acschembio.7b00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Díez-Villaseñor C, Soria E, Juez G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea. Bacteria and mitochondria. Mol Microbiol. 2000;36(1):244–246. doi: 10.1046/j.1365-2958.2000.01838.x. [DOI] [PubMed] [Google Scholar]
- Momenimovahed Z, Salehiniya H (2019) Epidemiological characteristics of and risk factors for breast cancer in the world. Breast Cancer Targets Ther. [DOI] [PMC free article] [PubMed]
- Moses C, Garcia-Bloj B, Harvey AR, Blancafort P. Hallmarks of cancer: the CRISPR generation. Eur J Cancer. 2018;93:10–18. doi: 10.1016/j.ejca.2018.01.002. [DOI] [PubMed] [Google Scholar]
- Naert T, Vleminckx K (2018) CRISPR/cas9-mediated knockout of RB1 in xenopus tropicalis. In: Methods in molecular biology [DOI] [PubMed]
- Naert T, Colpaert R, Van Nieuwenhuysen T, et al. CRISPR/Cas9 mediated knockout of rb1 and rbl1 leads to rapid and penetrant retinoblastoma development in Xenopus tropicalis. Sci Rep. 2016 doi: 10.1038/srep35264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair J, Nair A, Veerappan S, Sen D. Translatable gene therapy for lung cancer using CRISPR CAS9—an exploratory review. Cancer Gene Ther. 2020;27(3–4):116–124. doi: 10.1038/s41417-019-0116-8. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Fujii W, Tsuboi M, et al. Generation of muscular dystrophy model rats with a CRISPR/Cas system. Sci Rep. 2015 doi: 10.1038/srep05635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz G, Usman B, Zhao N, et al. Crispr/cas9 directed mutagenesis of osga20ox2 in high yielding basmati rice (Oryzasativa L.) line and comparative proteome profiling of unveiled changes triggered by mutations. Int J Mol Sci. 2020 doi: 10.3390/ijms21176170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V, Wang C, Win J, et al. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci Rep. 2017 doi: 10.1038/s41598-017-00578-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CE, Hakim CH, Ousterout DG, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science (80−) 2016 doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SR, Rideout WM, Akama-Garren EH, et al. CRISPR-mediated modeling and functional validation of candidate tumor suppressor genes in small cell lung cancer. Proc Natl Acad Sci USA. 2020 doi: 10.1073/pnas.1821893117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura N, Nomura Y, Sussman D, et al. Recognition of a common rDNA target site in archaea and eukarya by analogous LAGLIDADG and His-Cys box homing endonucleases. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noorani I. Genetically engineered mouse models of gliomas: technological developments for translational discoveries. Cancers (Basel) 2019;11:1335. doi: 10.3390/cancers11091335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldrini B, Curiel-García Á, Marques C, et al. Somatic genome editing with the RCAS-TVA-CRISPR–Cas9 system for precision tumor modeling. Nat Commun. 2018 doi: 10.1038/s41467-018-03731-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JJ, Kim JE, Jeon Y, et al. Deletion of NKX3.1 via CRISPR/Cas9 induces prostatic intraepithelial neoplasia in C57BL/6 mice. Technol Cancer Res Treat. 2020 doi: 10.1177/1533033820964425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pausch P, Al-Shayeb B, Bisom-Rapp E, et al. Crispr-casf from huge phages is a hypercompact genome editor. Science (80−) 2020 doi: 10.1126/science.abb1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez EE, Wang J, Miller JC, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008 doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Permuth-Wey J, Sellers TA (2009) Epidemiology of ovarian cancer. Methods Mol Biol [DOI] [PubMed]
- Perumal E, So Youn K, Sun S, et al. PTEN inactivation induces epithelial-mesenchymal transition and metastasis by intranuclear translocation of β-catenin and snail/slug in non-small cell lung carcinoma cells. Lung Cancer. 2019 doi: 10.1016/j.lungcan.2019.01.013. [DOI] [PubMed] [Google Scholar]
- Pickar-Oliver A, Black JB, Lewis MM, et al. Targeted transcriptional modulation with type I CRISPR–Cas systems in human cells. Nat Biotechnol. 2019 doi: 10.1038/s41587-019-0235-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RJ, Chen S, Zhou Y, et al. CRISPR–Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014 doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AA, Grakoui A, Weiss DS. Harnessing the prokaryotic adaptive immune system as a eukaryotic antiviral defense. Trends Microbiol. 2016;24(4):294–306. doi: 10.1016/j.tim.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raas Q, Gondcaille C, Hamon Y, et al. CRISPR/Cas9-mediated knockout of Abcd1 and Abcd2 genes in BV-2 cells: novel microglial models for X-linked adrenoleukodystrophy. Biochim Biophys Acta Mol Cell Biol Lipids. 2019 doi: 10.1016/j.bbalip.2019.02.006. [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013 doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani A, Stebbing J, Giamas G, Murphy J. Endocrine resistance in hormone receptor positive breast cancer—from mechanism to therapy. Front Endocrinol (Lausanne) 2019;10:245. doi: 10.3389/fendo.2019.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath D, Amlinger L, Rath A, Lundgren M. The CRISPR–Cas immune system: biology, mechanisms and applications. Biochimie. 2015;117:119–128. doi: 10.1016/j.biochi.2015.03.025. [DOI] [PubMed] [Google Scholar]
- Richard JLC, Eichhorn PJA. Deciphering the roles of lncRNAs in breast development and disease. Oncotarget. 2018;9(28):20179–20212. doi: 10.18632/oncotarget.24591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha-Martins M, Cavalheiro GR, Matos-Rodrigues GE, Martins RAP. From gene targeting to genome editing: transgenic animals applications and beyond. An Acad Bras Cienc. 2015 doi: 10.1590/0001-3765201520140710. [DOI] [PubMed] [Google Scholar]
- Rodgers K, Mcvey M. Error-Prone repair of DNA double-strand breaks. J Cell Physiol. 2016;231(1):15–24. doi: 10.1002/jcp.25053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Rodríguez DR, Ramírez-Solís R, Garza-Elizondo MA, Garza-Rodríguez ML, Barrera-Saldaña HA. Genome editing: a perspective on the application of CRISPR/Cas9 to study human diseases (Review) Int J Mol Med. 2019;43(4):1559–1574. doi: 10.3892/ijmm.2019.4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh JI, Lee J, Park SU, et al. CRISPR–Cas9-mediated generation of obese and diabetic mouse models. Exp Anim. 2018 doi: 10.1538/expanim.17-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronzitti G, Gross DA, Mingozzi F. Human immune responses to adeno-associated virus (AAV) vectors. Front Immunol. 2020;11:670. doi: 10.3389/fimmu.2020.00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper J, Tammela T, Cetinbas NM, et al. In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat Biotechnol. 2017 doi: 10.1038/nbt.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen LE, Morrison HA, Masri S, et al. Homing endonuclease I-CreI derivatives with novel DNA target specificities. Nucleic Acids Res. 2006 doi: 10.1093/nar/gkl645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux LN, Petit I, Domart R, et al. Modeling of aniridia-related keratopathy by CRISPR/Cas9 genome editing of human limbal epithelial cells and rescue by recombinant PAX6 protein. Stem Cells. 2018 doi: 10.1002/stem.2858. [DOI] [PubMed] [Google Scholar]
- Rushworth LK, Harle V, Repiscak P, et al. In vivo CRISPR/Cas9 knockout screen: TCEAL1 silencing enhances docetaxel efficacy in prostate cancer. Life Sci Alliance. 2020 doi: 10.26508/LSA.202000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JY, Choi YJ, Won EJ, et al. Gene editing particle system as a therapeutic approach for drug-resistant colorectal cancer. Nano Res. 2020 doi: 10.1007/s12274-020-2773-1. [DOI] [Google Scholar]
- Sakuma T, Hosoi S, Woltjen K, et al. Efficient TALEN construction and evaluation methods for human cell and animal applications. Genes Cells. 2013 doi: 10.1111/gtc.12037. [DOI] [PubMed] [Google Scholar]
- Sampson TR, Saroj SD, Llewellyn AC, et al. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013 doi: 10.1038/nature12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Rivera FJ, Papagiannakopoulos T, Romero R, et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature. 2014 doi: 10.1038/nature13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Rivera FJ, Jacks T. Applications of the CRISPR–Cas9 system in cancer biology. Nat Rev Cancer. 2015 doi: 10.1038/nrc3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Joung JK. CRISPR–Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32(4):347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Maeder ML, Reyon D, et al. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitha G, Vishnupriya V, Krishnamohan S. Hepatocellular carcinoma—a review. J Pharm Sci Res. 2017;9:1276–1280. [Google Scholar]
- Sayin VI, Papagiannakopoulos T. Application of CRISPR-mediated genome engineering in cancer research. Cancer Lett. 2017;387:10–17. doi: 10.1016/j.canlet.2016.03.029. [DOI] [PubMed] [Google Scholar]
- Schierling B, Dannemann N, Gabsalilow L, et al. A novel zinc-finger nuclease platform with a sequence-specific cleavage module. Nucleic Acids Res. 2012 doi: 10.1093/nar/gkr1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schokrpur S, Hu J, Moughon DL, et al. CRISPR-mediated VHL knockout generates an improved model for metastatic renal cell carcinoma. Sci Rep. 2016 doi: 10.1038/srep29032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabbir MAB, Hao H, Shabbir MZ, Hussain HI, Iqbal Z, Ahmed S, Sattar A, Iqbal M, Li J, Yuan Z. Survival and evolution of CRISPR–Cas system in prokaryotes and its applications. Front Immunol. 2016;7:375. doi: 10.3389/fimmu.2016.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao X, Wu S, Dou T, et al. Using CRISPR/Cas9 genome editing system to create MaGA20ox2 gene-modified semi-dwarf banana. Plant Biotechnol J. 2020 doi: 10.1111/pbi.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Shi Q, Wang W. Double agents: genes with both oncogenic and tumor-suppressor functions. Oncogenesis. 2018 doi: 10.1038/s41389-018-0034-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimo T, Hosoki K, Nakatsuji Y, et al. A novel human muscle cell model of Duchenne muscular dystrophy created by CRISPR/Cas9 and evaluation of antisense-mediated exon skipping. J Hum Genet. 2018 doi: 10.1038/s10038-017-0400-0. [DOI] [PubMed] [Google Scholar]
- Shmakov S, Smargon A, Scott D, et al. Diversity and evolution of class 2 CRISPR–Cas systems. Nat Rev Microbiol. 2017 doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva G, Poirot L, Galetto R, et al. Meganucleases and other tools for targeted genome engineering: perspectives and challenges for gene therapy. Curr Gene Ther. 2011;11:11–27. doi: 10.2174/156652311794520111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Gupta SC, Peng WX, et al. Regulation of alternative splicing of Bcl-x by BC200 contributes to breast cancer pathogenesis. Cell Death Dis. 2016 doi: 10.1038/cddis.2016.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CQ, Wang D, Jiang T, et al. In vivo genome editing partially restores alpha1-antitrypsin in a murine model of AAT deficiency. Hum Gene Ther. 2018 doi: 10.1089/hum.2017.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CQ, Jiang T, Richter M, et al. Adenine base editing in an adult mouse model of tyrosinaemia. Nat Biomed Eng. 2020 doi: 10.1038/s41551-019-0357-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtmauer EA, Fraietta JA, Davis MM, et al. CRISPR-engineered T cells in patients with refractory cancer. Science (80−) 2020 doi: 10.1126/science.aba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt SC, Torrez RM, Kaya E, et al. RNA-dependent RNA targeting by CRISPR–Cas9. Elife. 2018 doi: 10.7554/eLife.32724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JY, Anand-Jawa V, Chatterjee S, Wong KK. Immune responses to adeno-associated virus and its recombinant vectors. Gene Ther. 2003;10(11):964–976. doi: 10.1038/sj.gt.3302039. [DOI] [PubMed] [Google Scholar]
- Sun Y, Jiao G, Liu Z, et al. Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front Plant Sci. 2017 doi: 10.3389/fpls.2017.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabebordbar M, Zhu K, Cheng JKW, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science (80−) 2016 doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda H, Kataoka S, Nakayama M, et al. CRISPR-Cas9-mediated gene knockout in intestinal tumor organoids provides functional validation for colorectal cancer driver genes. Proc Natl Acad Sci USA. 2019 doi: 10.1073/pnas.1904714116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan DCS, Yao S, Ittner A, et al. Generation of a new tau knockout (tau Δex1) line using crispr/cas9 genome editing in mice. J Alzheimer’s Dis. 2018 doi: 10.3233/JAD-171058. [DOI] [PubMed] [Google Scholar]
- Telli ML (2016) Triple-negative breast cancer. In: Molecular pathology of breast cancer
- Tran MT, Doan DTH, Kim J, et al. CRISPR/Cas9-based precise excision of SlHyPRP1 domain(s) to obtain salt stress-tolerant tomato. Plant Cell Rep. 2020 doi: 10.1007/s00299-020-02622-z. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11(9):636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- Usman B, Nawaz G, Zhao N, et al. Precise editing of the ospyl9 gene by rna-guided cas9 nuclease confers enhanced drought tolerance and grain yield in rice (Oryza sativa l.) by regulating circadian rhythm and abiotic stress responsive proteins. Int J Mol Sci. 2020 doi: 10.3390/ijms21217854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Haasteren J, Li J, Scheideler OJ, Murthy N, Schaffer DV. The delivery challenge: fulfilling the promise of therapeutic genome editing. Nat Biotechnol. 2020;38(7):845–855. doi: 10.1038/s41587-020-0565-5. [DOI] [PubMed] [Google Scholar]
- Vijai J, Topka S, Villano D, et al. A recurrent ERCC3 truncating mutation confers moderate risk for breast cancer. Cancer Discov. 2016 doi: 10.1158/2159-8290.CD-16-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voduc KD, Cheang MCU, Tyldesley S, et al. Breast cancer subtypes and the risk of local and regional relapse. J Clin Oncol. 2010 doi: 10.1200/JCO.2009.24.9284. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba HA, El-Hadaad HA. Current approaches in treatment of triple-negative breast cancer. Cancer Biol Med. 2015;12(2):106–116. doi: 10.7497/j.issn.2095-3941.2015.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waks AG, Winer EP. Breast cancer treatment: a review. JAMA. 2019;321(3):288–300. doi: 10.1001/jama.2018.19323. [DOI] [PubMed] [Google Scholar]
- Walton J, Blagih J, Ennis D, et al. CRISPR/Cas9-mediated Trp53 and Brca2 knockout to generate improved murine models of ovarian high-grade serous carcinoma. Cancer Res. 2016 doi: 10.1158/0008-5472.CAN-16-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton JB, Farquharson M, Mason S, et al. CRISPR/Cas9-derived models of ovarian high grade serous carcinoma targeting Brca1, Pten and Nf1, and correlation with platinum sensitivity. Sci Rep. 2017 doi: 10.1038/s41598-017-17119-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan T, Chen Y, Pan Q, et al. Genome editing of mutant KRAS through supramolecular polymer-mediated delivery of Cas9 ribonucleoprotein for colorectal cancer therapy. J Control Release. 2020 doi: 10.1016/j.jconrel.2020.03.015. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang W, Ding Y, et al. CRISPR/Cas9-mediated genome engineering of CXCR4 decreases the malignancy of hepatocellular carcinoma cells in vitro and in vivo. Oncol Rep. 2017 doi: 10.3892/or.2017.5601. [DOI] [PubMed] [Google Scholar]
- Wang G, Wang C, Lu G, et al. Knockouts of a late flowering gene via CRISPR–Cas9 confer early maturity in rice at multiple field locations. Plant Mol Biol. 2020 doi: 10.1007/s11103-020-01031-w. [DOI] [PubMed] [Google Scholar]
- Warner M, Wu WF, Montanholi L, et al. Ventral prostate and mammary gland phenotype in mice with complete deletion of the ERβ gene. Proc Natl Acad Sci USA. 2020 doi: 10.1073/pnas.1920478117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbie D, Walther J, Mastrobattista E. Delivery aspects of CRISPR/Cas for in vivo genome editing. Acc Chem Res. 2019 doi: 10.1021/acs.accounts.9b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Meyer BJ. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333(6040):307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Kriz AJ, Sharp PA. Target specificity of the CRISPR-Cas9 system. Quant Biol. 2014;2(2):59–70. doi: 10.1007/s40484-014-0030-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zeng J, Roscoe BP, et al. Highly efficient therapeutic gene editing of human hematopoietic stem cells. Nat Med. 2019 doi: 10.1038/s41591-019-0401-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Zhang L, Yu Y, et al. miR-6086 inhibits ovarian cancer angiogenesis by downregulating the OC2/VEGFA/EGFL6 axis. Cell Death Dis. 2020 doi: 10.1038/s41419-020-2501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhou L, Wang Z, et al. Systematic screening for potential therapeutic targets in osteosarcoma through a kinome-wide CRISPR-Cas9 library. Cancer Biol Med. 2020 doi: 10.20892/j.issn.2095-3941.2020.0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia AL, He QF, Wang JC, et al. Applications and advances of CRISPR-Cas9 in cancer immunotherapy. Genet: J. Med; 2019. [DOI] [PubMed] [Google Scholar]
- Xiao-Jie L, Hui-Ying X, Zun-Ping K, et al. CRISPR–Cas9: a new and promising player in gene therapy. J Med Genet. 2015;52:289–296. doi: 10.1136/jmedgenet-2014-102968. [DOI] [PubMed] [Google Scholar]
- Xu CL, Ruan MZC, Mahajan VB, Tsang SH (2019a) Viral delivery systems for CRISPR. Viruses 11(1):28. 10.3390/v11010028 [DOI] [PMC free article] [PubMed]
- Xu S, Luk K, Yao Q, et al. Editing aberrant splice sites efficiently restores b-globin expression in b-thalassemia. Blood. 2019 doi: 10.1182/blood-2019-01-895094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Zhan M, Jiang C, et al. Genome-wide CRISPR screen identifies ELP5 as a determinant of gemcitabine sensitivity in gallbladder cancer. Nat Commun. 2019 doi: 10.1038/s41467-019-13420-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wang F, Chen Z, et al. CRISPR/Cas9-targeted mutagenesis of the OsROS1 gene induces pollen and embryo sac defects in rice. Plant Biotechnol J. 2020 doi: 10.1111/pbi.13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Chen S, Yin H, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014 doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan GN, Lv YF, Guo QN. Advances in osteosarcoma stem cell research and opportunities for novel therapeutic targets. Cancer Lett. 2016;370:268–274. doi: 10.1016/j.canlet.2015.11.003. [DOI] [PubMed] [Google Scholar]
- Yan S, Tu Z, Liu Z, et al. A huntingtin knockin pig model recapitulates features of selective neurodegeneration in Huntington’s disease. Cell. 2018 doi: 10.1016/j.cell.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Patel DJ. CasX: a new and small CRISPR gene-editing protein. Cell Res. 2019 doi: 10.1038/s41422-019-0165-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang L, Bell P, et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol. 2016 doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Jaeger M, Walker A, Wei D, Leiker K, Weitao T. Break breast cancer addiction by CRISPR/Cas9 genome editing. J Cancer. 2018;9(2):219–231. doi: 10.7150/jca.22554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau EH, Kummetha IR, Lichinchi G, et al. Genome-wide CRISPR screen for essential cell growth mediators in mutant KRAS colorectal cancers. Cancer Res. 2017 doi: 10.1158/0008-5472.CAN-17-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R, Pi M, Cox JV, et al. CRISPR/Cas9 targeting of GPRC6A suppresses prostate cancer tumorigenesis in a human xenograft model. J Exp Clin Cancer Res. 2017 doi: 10.1186/s13046-017-0561-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Song CQ, Dorkin JR, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016 doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T. Are KRAS/BRAF mutations potent prognostic and/or predictive biomarkers in colorectal cancers? Anticancer Agents Med Chem. 2012 doi: 10.2174/187152012799014968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Genome editing with ZFN, TALEN and CRISPR/Cas systems: the applications and future prospects. Adv Genet Eng. 2014 doi: 10.4172/2169-0111.1000e108. [DOI] [Google Scholar]
- Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids. 2015;4(11):e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Bai Y, Wu G, et al. Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 2017 doi: 10.1111/tpj.13599. [DOI] [PubMed] [Google Scholar]
- Zhang HX, Zhang Y, Yin H. Genome Editing with mRNA Encoding ZFN, TALEN, and Cas9. Mol Ther. 2019;27:735–746. doi: 10.1016/j.ymthe.2019.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Zhang F, Chen Q, et al. CRISPR/Cas9-mediated knockout of NSD1 suppresses the hepatocellular carcinoma development via the NSD1/H3/Wnt10b signaling pathway. J Exp Clin Cancer Res. 2019 doi: 10.1186/s13046-019-1462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang W, Zhu W, Dong J, Cheng Y, Yin Z, Shen F. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int J Mol Sci. 2019;20(22):5573. doi: 10.3390/ijms20225573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Malzahn AA, Sretenovic S, Qi Y. The emerging and uncultivated potential of CRISPR technology in plant science. Nat Plants. 2019;5(8):778–794. doi: 10.1038/s41477-019-0461-5. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Massel K, Godwin ID, Gao C. Correction to: Applications and potential of genome editing in crop improvement. Genome Biol. 2019 doi: 10.1186/s13059-019-1622-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yang Y, Chai L, et al. FRK plays an oncogenic role in non-small cell lung cancer by enhancing the stemness phenotype via induction of metabolic reprogramming. Int J Cancer. 2020 doi: 10.1002/ijc.32530. [DOI] [PubMed] [Google Scholar]
- Zhen S, Takahashi Y, Narita S, et al. Targeted delivery of CRISPR/Cas9 to prostate cancer by modified gRNA using a flexible aptamer-cationic liposome. Oncotarget. 2017 doi: 10.18632/oncotarget.14072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Zhang L, Tang M, et al. Knockout of two BnaMAX1 homologs by CRISPR/Cas9-targeted mutagenesis improves plant architecture and increases yield in rapeseed (Brassica napus L.) Plant Biotechnol J. 2020 doi: 10.1111/pbi.13228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Li J, Wang B, Han J, Hao Y, Wang S, Ma X, Yang S, Ma L, Yi L, Peng W. Endogenous type I CRISPR–Cas: from foreign DNA defense to prokaryotic engineering. Front Bioeng Biotechnol. 2020;8:62. doi: 10.3389/fbioe.2020.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Li D, Wang G, et al (2020) Application and future perspective of CRISPR/Cas9 genome editing in fruit crops. J. Integr. Plant Biol. [DOI] [PMC free article] [PubMed]
- Zuckermann M, Hovestadt V, Knobbe-Thomsen CB, et al. Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat Commun. 2015 doi: 10.1038/ncomms8391. [DOI] [PMC free article] [PubMed] [Google Scholar]