Abstract
Coronaviruses, such as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) responsible for the coronavirus disease 2019 (COVID-19) pandemic, present a significant threat to human health by inflicting a wide variety of health complications and even death. While conventional therapeutics often involve administering small molecules to fight viral infections, small non-coding RNA sequences, known as microRNAs (miRNAs/miR-), may present a novel antiviral strategy. We can take advantage of their ability to modulate host–virus interactions through mediating RNA degradation or translational inhibition. Investigations into miRNA and SARS-CoV-2 interactions can reveal novel therapeutic approaches against this virus. The viral genomes of SARS-CoV-2, severe acute respiratory syndrome coronavirus (SARS-CoV), and Middle East respiratory syndrome coronavirus (MERS-CoV) were searched using the Nucleotide Basic Local Alignment Search Tool (BLASTn) for highly similar sequences, to identify potential binding sites for miRNAs hypothesized to play a role in SARS-CoV-2 infection. miRNAs that target angiotensin-converting enzyme 2 (ACE2), the receptor used by SARS-CoV-2 and SARS-CoV for host cell entry, were also predicted. Several relevant miRNAs were identified, and their potential roles in regulating SARS-CoV-2 infections were further assessed. Current treatment options for SARS-CoV-2 are limited and have not generated sufficient evidence on safety and efficacy for treating COVID-19. Therefore, by investigating the interactions between miRNAs and SARS-CoV-2, miRNA-based antiviral therapies, including miRNA mimics and inhibitors, may be developed as an alternative strategy to fight COVID-19.
Key Points
| MicroRNAs (miRNAs) regulate host–virus interactions through direct interactions with the viral genome or by altering the host’s cellular microenvironment. |
| RNA and miRNA-based antiviral therapeutics are evolving and represent a promising therapeutic option. |
| In this study, we utilized available computational and miRNA target prediction tools and databases to identify key miRNAs that may have a role in modulating severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. |
SARS-CoV-2 and the COVID-19 Pandemic
The newly emerged human coronavirus (HCoV), named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is the etiologic agent responsible for the ongoing coronavirus disease 2019 (COVID-19) pandemic and has infected ~ 100 million people and caused ~ 2 million deaths worldwide at the time of submission [1, 2]. While some COVID-19 patients remain asymptomatic or present with mild flu-like symptoms, others develop severe respiratory distress, cardiac complications, renal failure, septic shock, and other long-term health complications [3]. Despite global efforts to control the spread of the virus, many countries are now facing a second rise in COVID-19 cases with uncontrolled SARS-CoV-2 spreading in populations, leading to a need for effective antiviral treatments and vaccine developments [2].
Coronaviruses are enveloped single-stranded RNA viruses and are divided into four genera, Alpha-, Beta-, Gamma- and Deltacoronavirus, with Alpha- and Betacoronavirus being the only genera infecting humans [4, 5]. HCoVs originate from animal hosts, and SARS-CoV-2 is now the third highly pathogenic Betacoronavirus to cross the species barrier, along with the previously identified severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) [4–7]. SARS-CoV-2 presents high sequence homology with SARS-CoV (around 80%) and similar cell tropism in the lower respiratory tract, infecting pulmonary epithelial alveolar type II cells [8, 9]. Notably, both SARS-CoV-2 and SARS-CoV use the angiotensin-converting enzyme 2 (ACE2) as their functional receptor and gain access to the cell cytoplasm after the specific interaction of their Spike glycoprotein with ACE2 and subsequent viral membrane–host membrane fusion in the endosomal compartment [10]. In addition to the cell receptor, several membrane proteins have been shown to facilitate SARS-CoV-2 cell entry such as the transmembrane protease serine 2 (TMPRSS2), the lysosomal cathepsins B/L, and neuropilin-1 [10–12]. Moreover, SARS-CoV-2 acquired a furin cleavage site between the S1 and S2 subunits of its Spike protein, leading to the proteolytic pre-activation of the glycoprotein, a feature necessary for viral entry, and could explain the high pathogenicity of the virus given the ubiquitous expression of the furin protease combined with the large distribution of ACE2 outside of the lungs [10].
Following entry, the viral genome is released into the cell cytoplasm to start the replicative cycle. SARS-CoV-2 possesses a large single-stranded, positive-sense RNA genome (29.9 kb), organized in 11 open reading frames (ORF) surrounded by a 5′ and 3′ untranslated region (UTR) and coding for 16 non-structural, four structural, and six accessory proteins [13, 14]. The viral replication machinery comprises several viral proteins (replicase, helicase, RNA-dependent RNA polymerase complex, and endoribonuclease) that are synthesized as large polyproteins called PP1a and PP1ab, encoded by ORF1a and ORF1b, and cleaved into individual proteins by the viral proteases PLpro and 3CLpro [15].
Current therapeutic strategies to treat COVID-19 patients rely on the management of the disease’s most severe symptoms and on the administration of the antiviral drug remdesivir in the most severe cases [16]. Among the current efforts to fight COVID-19, some notable approaches include finding entry and replication inhibitors and repurposing existing antiviral drugs [17]. To date, over 2000 COVID-19 clinical trials have been registered worldwide, ranging from small molecule approaches (including drugs such as lopinavir-ritonavir) to convalescent plasma and stem cell therapies [18]. As of January 2021, several COVID-19 vaccines have been authorized for use by different regulatory bodies around the world. However, despite the development of effective vaccines, continued investigations on all therapeutic approaches remains crucial, especially in the context of emerging SARS-CoV-2 variants and potential vaccine-resistant strains. Herein, we evaluate microRNA (miRNA/miR-)-based approaches for their potential use therapeutically against SARS-CoV-2 and emerging coronavirus variants of concern for global human health.
MicroRNAs as an Antiviral Therapeutic Approach
Another relevant SARS-CoV-2 antiviral strategy could come from the host’s natural antiviral response. Host cells can develop different antiviral strategies against RNA viruses, and one effective strategy is RNA interference. RNA interference includes a group of RNA molecules known as miRNA, which are non-coding RNA molecules, typically 20–25 nucleotides in length, that regulate gene expression, cell signaling, and the host cell environment [19]. Acting as post-transcriptional gene regulators, miRNAs base-pair with the 3′-UTR of complementary mRNA sequences to mediate their degradation or inhibit their translation [20]. Non-canonical miRNA–mRNA interactions, such as those that interact with the 5′-UTR or coding regions of target genes, have also been reported to be functional [21]. Given that approximately 2000 human miRNAs have been identified (miRBase 22), and they have been predicted to regulate over 60% of all human protein-coding genes [22], miRNAs serve as a plausible avenue for therapeutic investigation, especially for the modulation of proteins that cannot be targeted by other small molecules [23]. There are two general approaches for developing miRNA-based therapeutics (Fig. 1). The first one is based on the development of miRNA antagonists or inhibitors to increase or rescue the expression of specific proteins that are downregulated. The second approach consists of the use of miRNA mimics to downregulate the expression of proteins abnormally induced. In recent years, several miRNA-based treatments have showed therapeutic promise in clinical trials [24, 25]. For example, the experimental drug known as miravirsen, developed by Santaris Pharma A/S, was the first miRNA-based drug that successfully completed a phase 2 clinical trial. Serving as an miR-122 antagonist, this treatment for hepatitis C virus (HCV) infection reduces viral RNA levels by sequestering miR-122 away from the viral genome, where it is normally used by the virus to enhance its propagation [26, 27].
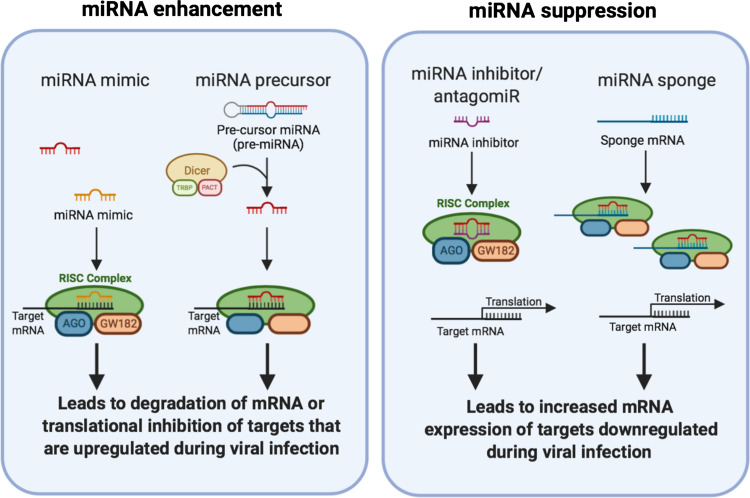
Figure 1.
Potential therapeutic applications of miRNAs. Through miRNA enhancement, mRNA expression and translation that is upregulated during viral infection can be repressed. In contrast, through miRNA suppression, mRNA expression and translation that is downregulated during viral infection can be restored. miRNA microRNA, RISC RNA-induced silencing complex, AGO argonaute, TRBP trans-activation response RNA-binding protein, PACT protein kinase RNA activator
It has also been reported that miRNAs have a role in interrogating host–virus interactions during viral infection. Whether it be through direct interactions with the viral genome or by inducing antiviral cellular responses in the host, miRNAs serve as critical regulators of viral pathogenesis (Fig. 2). More specifically, viruses have been shown to create specific microenvironments that facilitate the viral life cycle through alterations in host miRNA expression [19, 28–30]. This cross-talk between host miRNAs and viruses can alter the host’s transcriptome and indirectly regulate viral infections through modulation of cellular host pathways, resulting in overall pro- or antiviral effects. For example, one study found that elevated miR-155 expression in Epstein-Barr virus (EBV)-infected lymphocytes contributed to an overall proviral cellular state by simultaneously downregulating nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) signaling, which is important for cytokine production and cell survival, and suppressing innate immune responses to viral infection [31]. In fact, miRNAs have been shown to regulate the host’s innate immune response to viral infections by a variety of mechanisms, including by altering the lipid microenvironment, promoting cellular signaling pathways that lead to the expression of interferon (IFN)-inducible antiviral genes, or by enhancing the expression of immune response regulators [32–34]. Moreover, another study identified miR-24, miR-124, and miR-744 as broad-spectrum antiviral miRNAs against influenza A virus (IAV) and respiratory syncytial virus (RSV) [35]. By targeting the p38 mitogen-activated protein kinase (MAPK) signaling pathway, these miRNAs regulate the production of cytokines involved in the immune response, as well as viral entry and propagation, contributing to an overall antiviral phenotype [36, 37]. With respect to HCV infection, IFN-induced antiviral miRNAs have been shown to target viral RNA and inhibit its replication [38], while other miRNAs, such as miR-135a, have been found to enhance viral replication through antagonism of cellular antiviral pathways [39]. Evidently, as established by many reports, miRNA-virus interactions are extensive and complex and possess the ability to enhance or suppress viral replication.
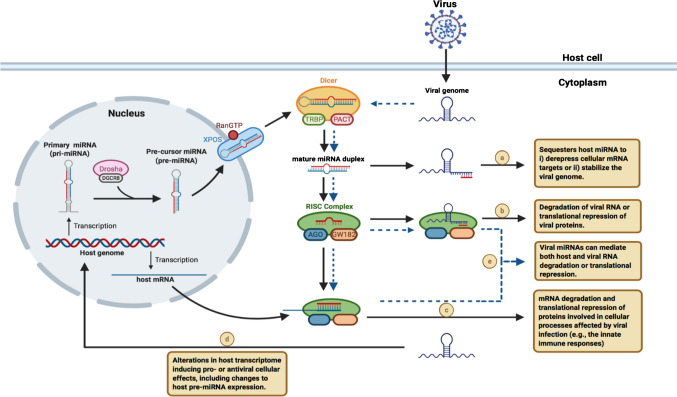
Figure 2.
Potential interactions between miRNAs and viral infection. Host-derived miRNA transcripts can serve as substrates for the endonuclease Dicer to produce mature miRNA, which can complex with the RISC. The miRNAs can be sequestered by the viral genome to derepress cellular targets or stabilize the genome for replication (a), induce degradation or translational inhibition of viral RNA (b), or host mRNA to modulate cellular pathways relevant for the viral infection (c). The virus can also interact directly with the host genome to alter the transcriptome and modulate miRNA expression to induce pro- or antiviral cellular effects (d). Some viruses can also encode viral miRNAs that can target both host and viral RNA (e). miRNA microRNA, RISC RNA-induced silencing complex
Host miRNAs can also interact directly with the viral genome to regulate viral infections. In order to enhance viral replication, some viruses use host-derived miRNAs to stabilize their genome and prevent degradation [40, 41]. One example of this includes the aforementioned proviral miR-122, which directly binds to the 5′-UTR of HCV and effectively stabilizes the RNA to promote viral replication. Interestingly, the HCV genome can also act as a “sponge” to sequester miR-122 away from its cellular targets, thereby providing another mechanism to enhance its infective potential [42]. Other viruses, such as some herpesviruses, also use this miRNA sequestering method to derepress cellular targets as a viral strategy to modulate the host cell [43]. In contrast, it has also been suggested that cellular miRNAs can interact with the viral genome to inhibit replication. For example, in human immunodeficiency virus 1 (HIV-1) infection, a cluster of cellular miRNAs was found to induce viral latency through interactions with the 3′ ends of viral RNA [44]. In addition to host-derived miRNAs, viral-encoded miRNAs (or viral miRNAs) have also been identified in several viruses, including the human EBV [45]. By exploiting the host’s cellular miRNA machinery, viral miRNAs can be expressed to modulate both host and viral RNA targets, with some even helping the virus to evade the host’s immune response [46, 47]. Overall, by making the host cell more amenable to viral infection or through direct interactions with the virus, miRNAs serve an important role in regulating viral infections and can be explored for developing novel antiviral therapeutics.
Evidently, miRNAs can regulate a wide variety of viral infections, including respiratory infections such as HCoVs. For example, a study conducted by Mallick et al. found that the SARS-CoV viral proteins N and S downregulated miR-223 and miR-98, respectively, in order to modulate host cell differentiation and induce pro-inflammatory responses such as increased inflammatory chemokine activation [48]. While interactions between miRNAs and SARS-CoV have been established, the ability of miRNAs to regulate SARS-CoV-2 is yet to be fully understood. With that said, several groups have recently conducted computational investigations on miRNA targets in the SARS-CoV-2 genome [49–52]; however, further experimental validation is still required to validate these predictions and determine their downstream cellular effects. This gap in knowledge, coupled with the limited treatment options for SARS-CoV-2, highlights the importance for further investigation on miRNA and SARS-CoV-2 interactions. Here, we used the Nucleotide Basic Local Alignment Search Tool (BLASTn) for highly similar sequences, to identify potential miRNA binding sites in the viral genomes of SARS-CoV-2, SARS-CoV, and MERS-CoV. We identified miRNAs with 100% seed site complementarity to the viral genome and then conducted further literature investigations on their potential roles in SARS-CoV-2 infection.
While studies on the interactions between host miRNAs and viral genomes are vital, they merely reflect an initial step in the development of miRNA-based antiviral therapies. Despite the increasing advancements in miRNA technologies, there remain challenges, namely the unstable nature of RNA molecules and the requirement for targeted delivery based on its site of action [53], that complicate the direct translatability of this approach for the current COVID-19 pandemic. However, with the emergence of mRNA-based vaccines being used as a frontline method for SARS-CoV-2 management, other RNA-based therapeutics are expected to also emerge as alternatives to disease management. Thus, miRNA approaches undoubtedly present a promising therapeutic avenue for future antiviral applications.
Identification of MicroRNAs with Potential Roles in SARS-CoV-2 Infection
Based on the established potential for miRNAs to influence viral infections (either with pro- or antiviral effects), we sought to investigate the interactions between human miRNAs and the SARS-CoV-2 genome in order to gain insight and direct the development of miRNA-based therapeutics (Fig. 3a). In our approach, we opted to look at the most commonly expressed miRNAs in tissue and cell types often targeted by the virus, as a way to identify miRNAs that may modify the host’s cellular microenvironment to be more favorable for viral infection. Here, we used the NCBI Nucleotide database to obtain the viral genome sequences for SARS-CoV-2 (MN908947.3) [54], SARS-CoV (NC_004718.3) [55], and MERS-CoV (NC_019843.3) [56]. Using the mimiRNA database [57], we focused on the 20-30 most significantly expressed miRNAs in A549 lung epithelial cells, heart, hepatoma (liver), and small intestine tissues as a starting point for our analysis; however, further computational analysis needs to be done to screen through all the miRNAs found in these tissue types. From these results, the top 12 miRNAs that were present in all cell types and highly abundant in lung epithelial A549 cells were selected for further investigation. Additional miRNAs were identified through a literature review of miRNAs shown to have roles in regulating metabolism, the immune response, and other viral infections (Table 1) [28, 32, 35, 40, 46, 48, 58–65].
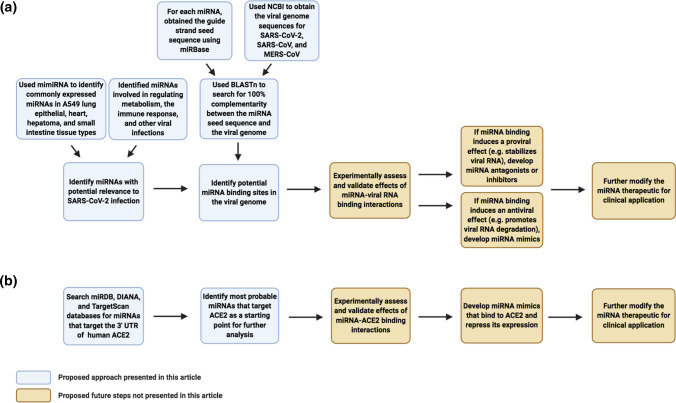
Figure 3.
Overview of the proposed miRNA-based antiviral therapeutic approach against SARS-CoV-2 infection. Flowcharts outlining the strategies used to identify miRNAs that bind to the SARS-CoV-2 viral genome (a) or the ACE2 receptor (b) for the development of miRNA-based therapeutics are presented. ACE angiotensin-converting enzyme, BLASTn Nucleotide Basic Local Alignment Search Tool, MERS-CoV Middle East respiratory syndrome coronavirus, miRNA/miR- microRNA, SARS-CoV severe acute respiratory syndrome coronavirus, SARS-CoV-2 severe acute respiratory syndrome coronavirus 2, UTR untranslated region
Table 1.
Selection of miRNAs with potential relevance to SARS-CoV-2 infection
| hsa-miRNA | Potential interest related to SARS-CoV-2 infection |
|---|---|
| miR-16 | Abundant in A549 cells, suppresses viral replication in EV71 infection, regulation of apoptosis [58] |
| miR-29 | Abundant in A549 cells |
| miR-30 | Abundant in A549 cells, upregulated in RSV exosomes [59] |
| miR-186 | Indirect regulation of HIV-1 infection [60] |
| miR-130 | Downregulated in HCV infection [40] |
| miR-27 | Abundant in A549 cells, role in MCMV and HCV infection [40] |
| miR-595 | Repressed in RSV infected A549 cells [61] |
| miR-182 | Upregulated in RSV exosomes [59], miR-183 cluster regulation of innate antiviral response [32] |
| miR-199 | Proviral functions in HCV infection [46] |
| miR-20 | Abundant in A549 cells |
| miR-93 | Potential target site in VSV genome [40] |
| miR-218 | Role in NF-κB signaling pathway [46] |
| miR-23 | Abundant in A549 cells, upregulated in RSV exosomes [59] |
| miR-203 | Upregulated IAV infection, inhibits viral replication [62] |
| miR-320 | Upregulated in HCV infection, proviral functions [46], upregulated in RSV exosomes [59] |
| miR-135 | Role in immune response [63] |
| miR-19 | Role in type I interferon signaling pathway, antiviral effects [46], upregulated in RSV exosomes [59] |
| miR-122 | Role in type I interferon signaling pathway, antiviral effects [46] |
| miR-520 | Role in immune response [62], induced in RSV infected A549 cells [61] |
| miR-21 | Abundant in A549 cells, NF-κB signaling pathway [46], upregulated in RSV exosomes [59] |
| miR-26 | Role in immune response [63] |
| miR-125 | Abundant in A549 cells, regulation of apoptosis [46] |
| miR-92 | Abundant in A549 cells |
| miR-155 | Role in type I interferon signaling pathway, antiviral effects [46] |
| miR-183 | miR-183 cluster regulation of innate antiviral response [32] |
| miR-224 | Tumor suppressive functions [64] |
| miR-200 | Upregulated in H5N1 infection, targets the 3′-UTR of ACE2 [65] |
| miR-24 | Upregulated in RSV exosomes [59], abundant in A549 cells, antiviral in IAV and RSV infection [35] |
| miR-198 | Repressed in RSV infected A549 cells [61] |
| let-7 | Abundant in A549 cells, involved in NF-κB signaling and inflammation [46] |
| miR-223 | Downregulated in RSV exosomes [58], role in SARS-CoV infection [48] |
| miR-98 | Role in immune response [62], role in SARS-CoV infection [48] |
| miR-337 | Induced in RSV infected A549 cells [61] |
| miR-146 | Role in NF-κB signaling pathway, proviral functions [46], downregulated in RSV exosomes [59] |
| miR-185 | Regulation of host metabolic pathways and HCV infection [28] |
| miR-744 | Antiviral functions against RSV and influenza viruses [46] |
| miR-192 | Abundant in A549 cells |
| miR-127 | Upregulated in M2 macrophages, downregulated by inflammation [64] |
| miR-187 | Anti-inflammatory effects [64] |
Human (hsa) miRNAs that were identified to be highly abundant in lung epithelial A549 cells and/or to have roles in metabolism, the immune response, or other viral infections are presented. miRNAs are listed in decreasing order of the total number of binding sites predicted in the SARS-CoV-2 genome
ACE angiotensin-converting enzyme, EV71 enterovirus 71, H5N1 highly pathogenic Asian avian influenza A virus, HCV hepatitis C virus, HIV-1 human immunodeficiency virus 1, IAV influenza A virus, MCMV murine cytomegalovirus, miRNA/miR- microRNA, RSV respiratory syncytial virus, SARS-CoV severe acute respiratory syndrome coronavirus, SARS-CoV-2 severe acute respiratory syndrome coronavirus 2, UTR untranslated region, VSV vesicular stomatitis virus
Prediction of Binding Sites
Guide strand sequences for the human (hsa) miRNAs identified for investigation were obtained from miRBase [66–71]. For each miRNA, the seed site (nucleotides 2–8) and its corresponding complementary sequence were determined. BLASTn for highly similar sequences was used to search for matches between the viral genome sequences (SARS-CoV-2, SARS-CoV, and MERS-CoV) and the complementary miRNA seed sequences. Potential miRNA binding sites were predicted at locations with 100% complementarity. Imperfect seed site complementarity was not considered in this analysis. Binding sites predicted in the SARS-CoV genome were cross-referenced with the ViTa database using SARS-CoV Strain TWH [72]. For each potential binding site, the location and corresponding region in the viral genome was identified.
We also used miRDB [73], DIANA microT-CDS [74], and TargetScan [75] prediction software to predict miRNAs that target the 3′-UTR of the ACE2 receptor in humans (NM_021804.3) in order to explore whether miRNAs might be good targets for treatment of SARS-CoV-2 infection [76]. Searches included both highly and poorly conserved sites, as well as 6mer, 7mer, 8mer, and 9mer binding sites. The top hits generated here will provide an ideal starting point for further miRNA-ACE2 expression analysis which in turn, can help direct the development of antiviral miRNA therapies (Fig. 3b).
Potential Roles and Binding Sites of MicroRNAs in the SARS-CoV-2, SARS-CoV, and MERS-CoV Genomes
Among the 39 miRNAs investigated, 29 were predicted to have binding sites within the SARS-CoV-2 genome based on seed site complementarity (Table 2, supplementary Table S1—see the electronic supplementary material). A large number of these binding sites were predicted in the ORF1ab, as it is the largest region of the genome, spanning about 21 kb in length, and encodes for numerous viral proteins. No miRNA binding sites were predicted in the SARS-CoV-2 and SARS-CoV UTRs. This suggests that the miRNAs investigated in this study may not directly bind to the viral genome to stabilize or promote viral RNA degradation; however, the binding sites in the coding regions may still be functional. Since only one genomic sequence was used to assess each virus, variations in the viral genomes that may allow for additional miRNA binding sites were not captured. Also, the fact that miRNA binding sites in the viral genomes were defined as locations with 100% seed site complementarity is quite restrictive as miRNAs are known to be able to bind to sites imperfectly [77]. This leads us to believe that other potential binding sites may not have been represented. Furthermore, these cellular miRNAs, both with or without binding sites in the viral genome, may still have a role in regulating inflammatory or signaling pathways in the host, as seen with other viral infections (Table 1). In terms of the MERS-CoV genome, miR-92 was predicted to target the 3′-UTR, while miR-130 and miR-199 were predicted to target the 5′-UTR. While host miRNA-mediated regulation of MERS-CoV is not well explored, it is interesting to note that miR-199a has been previously identified to regulate TMPRSS2 expression in the liver, stomach, and uterine corpus, as this protease is critical for SARS-CoV-2, SARS-CoV, and MERS-CoV entry into cells [78]. Therefore, upon further analysis and experimental validation to determine whether these are true miRNA binding sites, the roles of these miRNAs in modulating MERS-CoV infection, along with other related viruses, can be explored.
Table 2.
Potential miRNA binding sites in the SARS-CoV-2, SARS-CoV, and MERS-CoV genomes
| hsa-miRNA | Seed site sequence (5′–3′) | SARS-CoV-2 | SARS-CoV | MERS-CoV | |||
|---|---|---|---|---|---|---|---|
| Number of potential binding sites | Location in genome (number of sites in the region) | Number of potential binding sites | Location in genome (number of sites in the region) | Number of potential binding sites | Location in genome (number of sites in the region) | ||
| miR-16 | AGCAGCA | 15 | ORF1ab (7), S (2), ORF3a, M, N (4) | 21 | ORF1ab (13), S (3), sars3a, M, sars7a, N (2) | 14 | ORF1ab (10), S, N (3) |
| miR-29 | AGCACCA | 10 | ORF1ab (8), S, N | 9 | ORF1ab (7), S, M | 11 | ORF1ab (10), S |
| miR-30 | GUAAACA | 10 | ORF1ab (8), M, ORF8 | 7 | ORF1ab (7) | 3 | ORF1ab (2), S |
| miR-186 | AAAGAAU | 7 | ORF1ab (5), S, ORF7a | 4 | ORF1ab (2), S, N | 6 | ORF1ab (6) |
| miR-130 | AGUGCAA | 6 | ORF1ab (3), S, ORF3a, ORF7a | 4 | ORF1ab (3), S, M | 5 | 5'-UTR, ORF1ab (3), ORF5 |
| miR-27 | UCACAGU | 6 | ORF1ab (4), S, N | 2 | ORF1ab, N | 2 | ORF1ab (2) |
| miR-595 | AAGUGUG | 5 | ORF1ab (4), S | 3 | ORF1ab, S, sars7a | 2 | ORF1ab |
| miR-182 | UUGGCAA | 4 | S (3), N | 4 | ORF1ab (3), N | 5 | ORF1ab (4), S |
| miR-199 | CCAGUGU | 4 | ORF1ab (2), S, ORF3a | 5 | ORF1ab (4), sars3a/3b | 3 | 5'-UTR, N, N/ORF8b |
| miR-20 | AAAGUGC | 4 | ORF1ab (3), S | 4 | ORF1ab (3), S | 4 | ORF1ab (3), S |
| miR-93 | AAAGUGC | 4 | ORF1ab (3), S | 4 | ORF1ab (3), S | 4 | ORF1ab (3), S |
| miR-218 | UGUGCUU | 4 | ORF1ab, S (2), ORF3a | 3 | ORF1ab, sars3a, N | 2 | ORF1ab (2) |
| miR-23 | UCACAUU | 4 | ORF1ab (4) | 2 | ORF1ab (2) | 2 | ORF1ab, S |
| miR-203 | UGAAAUG | 3 | ORF1ab, ORF7a, ORF8 | 7 | ORF1ab (2), S (3), sars3a, sars7a | 1 | ORF5 |
| miR-320 | AAAGCUG | 3 | ORF1ab (3) | 4 | ORF1ab (4) | 3 | ORF1ab (3) |
| miR-135 | AUGGCUU | 3 | ORF1ab (3) | 1 | ORF1ab | 0 | none |
| miR-19 | GUGCAAA | 2 | S, ORF7a | 4 | ORF1ab (2), S, sars7a | 4 | ORF1ab (2), S, ORF5 |
| miR-122 | GGAGUGU | 2 | ORF1ab (2) | 3 | ORF1ab (2), M | 4 | ORF1ab (3), S |
| miR-520 | UCCAGAG | 2 | ORF1ab (2) | 2 | ORF1ab (2) | 2 | ORF1ab, S |
| miR-21 | AGCUUAU | 2 | ORF1ab (2) | 2 | ORF1ab (2) | 1 | ORF1ab |
| miR-26 | UCAAGUA | 2 | ORF1ab (2) | 3 | ORF1ab (3) | 0 | none |
| miR-125 | CCCUGAG | 2 | ORF1ab, S | 0 | none | 0 | none |
| miR-92 | AUUGCAC | 1 | S | 2 | ORF1ab, S | 6 | ORF1ab (4), E, |
| miR-155 | UAAUGCU | 1 | ORF1ab | 5 | ORF1ab (3), S (2) | 1 | ORF5 |
| miR-183 | AUGGCAC | 1 | N | 4 | ORF1ab (3), S | 2 | ORF1ab (2) |
| miR-224 | CAAGUCA | 1 | ORF7a | 4 | ORF1ab (3), sars8a | 1 | ORF1ab |
| miR-200 | AAUACUG | 1 | ORF1ab | 0 | none | 2 | ORF1ab, S |
| miR-24 | GGCUCAG | 1 | S | 1 | S | 1 | S |
| miR-198 | GUCCAGA | 1 | ORF1ab | 1 | ORF1ab | 0 | none |
| let-7 | GAGGUAG | 0 | none | 3 | ORF1ab (3) | 2 | ORF1ab, ORF4a/b |
| miR-223 | GUCAGUU | 0 | none | 2 | ORF1ab, N | 3 | ORF1ab (2), ORF5 |
| miR-98 | GAGGUAG | 0 | none | 3 | ORF1ab (3) | 2 | ORF1ab, ORF4a/b |
| miR-337 | UCCUAUA | 0 | none | 2 | ORF1ab, S | 2 | 3′-UTR |
| miR-146 | GAGAACU | 0 | none | 1 | S | 2 | ORF1ab (2) |
| miR-185 | GGAGAGA | 0 | none | 0 | none | 2 | ORF1ab, ORF1ab/S |
| miR-744 | GCCCCGC | 0 | none | 0 | none | 0 | none |
| miR-192 | UGACCUA | 0 | none | 1 | ORF1ab | 0 | none |
| miR-127 | CGGAUCC | 0 | none | 0 | none | 0 | none |
| miR-187 | CGUGUCU | 0 | none | 0 | none | 0 | none |
Binding sites in the genome were predicted at locations with 100% complementarity to the hsa-miRNA seed site sequence using BLASTn. The locations of binding sites include coding regions [ORFs, spike glycoprotein (S), matrix protein (M), small envelope protein (E), and nucleocapsid protein (N)] and non-coding regions (5′- and 3′-UTR). miRNAs are listed in decreasing order of the total number of binding sites predicted in the SARS-CoV-2 genome
BLASTn Nucleotide Basic Local Alignment Search Tool, MERS-CoV Middle East respiratory syndrome coronavirus, miRNA/miR- microRNA, ORF open reading frame, SARS-CoV severe acute respiratory syndrome coronavirus, SARS-CoV-2 severe acute respiratory syndrome coronavirus 2, UTR untranslated region
The relative number and locations of potential miRNA binding sites in the genome can also provide some insight into the roles these miRNAs may play during viral infections. From our analysis, miR-16 was found to have the highest total number of binding sites in all three viral genomes, with 21 binding sites in the SAR-CoV genome, 15 binding sites in the SARS-CoV-2 genome, and 14 binding sites in the MERS-CoV genome (Table 2, Fig. 4). The fact that the high number of miR-16 binding sites is common among all three HCoVs could indicate that these viruses have evolved proviral mechanisms that use this miRNA to enhance their propagation, as previously observed in HCV [79]. Similarly, miR-29 and miR-30 were also found to have a large number of potential binding sites, with ten binding sites each predicted in the SARS-CoV-2 genome. In terms of SARS-CoV-2 infection, we further identified miR-200 and miR-24 to be of particular interest as they have been shown to be linked to the regulation of ACE2 and Furin, respectively (Fig. 5).
Figure 4.
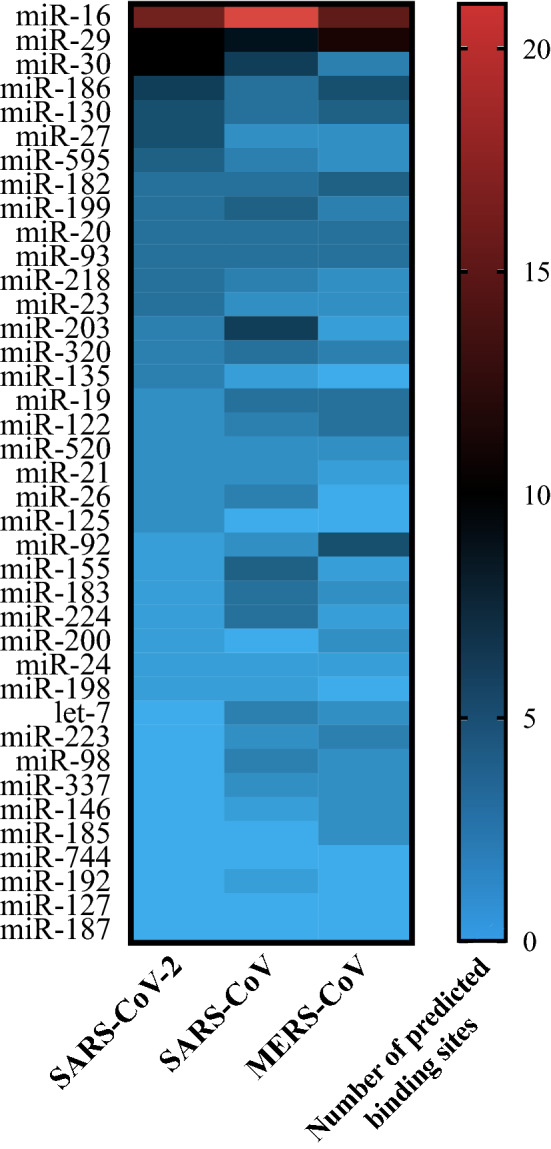
Number of predicted miRNA binding sites in the SARS-CoV-2, SARS-CoV, and MERS-CoV genomes. A heat map indicating the number of binding sites predicted in each viral genome is presented (values ranging from 0 to 21). Binding sites were predicted at locations with 100% complementarity to the hsa-miRNA seed site sequence using BLASTn. miRNAs are listed in decreasing order of the total number of binding sites predicted in the SARS-CoV-2 genome. BLASTn Nucleotide Basic Local Alignment Search Tool, MERS-CoV Middle East respiratory syndrome coronavirus, miRNA/miR- microRNA, SARS-CoV severe acute respiratory syndrome coronavirus, SARS-CoV-2 severe acute respiratory syndrome coronavirus 2
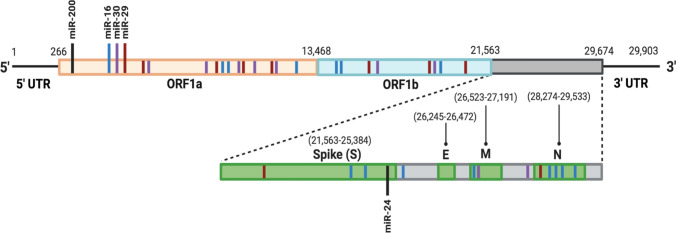
Figure 5.
Schematic representation of predicted miRNA binding sites in the SARS-CoV-2 genome. The location of potential binding sites in the SARS-CoV-2 genome for select miRNA of interest, including the top three most abundant miRNAs in lung epithelial A549 cells (miR-16, miR-29, and miR-30), miR-24, and miR-200 are presented. miRNA/miR- microRNA, ORF open reading frame, SARS-CoV-2 severe acute respiratory syndrome coronavirus 2, UTR untranslated region
MicroRNAs Regulating the Expression of the ACE2 Receptor
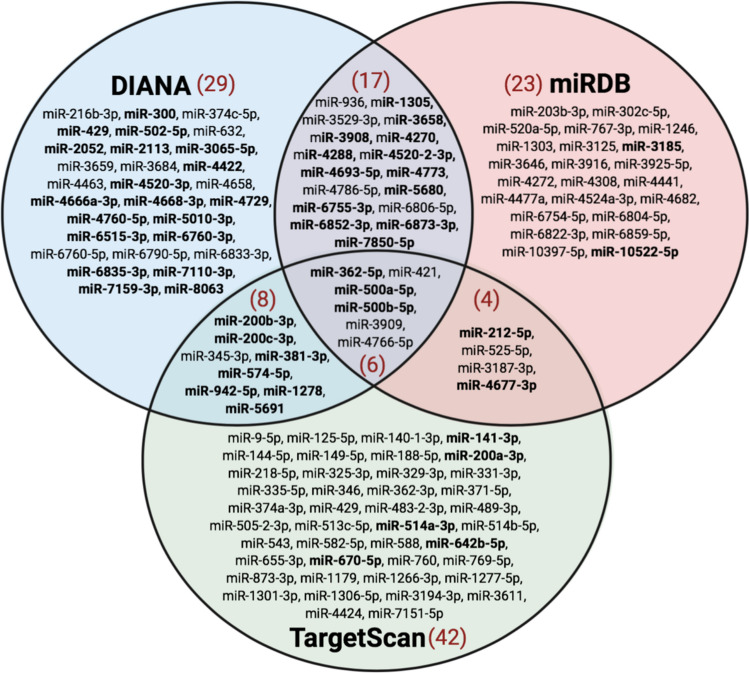
In addition to miRNAs that directly interact with the viral genome, host miRNAs that target ACE2 can also be involved in the regulation of SARS-CoV-2 infection. As ACE2 is the viral receptor that facilitates SARS-CoV-2 entry into cells, an understanding of which miRNAs modulate its expression can provide insight for prospective antiviral miRNA-based therapeutics. In this study, we identified numerous miRNAs that target the 3′-UTR of ACE2 in humans (Fig. 6). A total of six miRNAs (miR-362-5p, miR-421, miR-500a-5p, miR-500b-5p, miR-3909, and miR-4766-5p) were predicted by all three online miRNA prediction tools. Interestingly, miR-421 has also been previously identified as a potential ACE2 regulator [80], making this a plausible target for further therapeutic investigation against SARS-CoV-2.
Figure 6.
miRNAs predicted to bind to the 3′-UTR of ACE2. The miRDB, DIANA microT-CDS, and TargetScan databases were used to predict miRNAs that bind to the 3′-UTR of ACE2. Searches included both highly and poorly conserved sites, as well as 6mer, 7mer, 8mer, and 9mer binding sites. miRNAs in bold were predicted to have more than one miRNA binding site. Numbers in red indicate the total number of miRNAs predicted in each group. ACE2 angiotensin-converting enzyme 2, miRNA/miR- microRNA, UTR untranslated region
Abundant MicroRNAs Whose Normal Function may be Modulated by SARS-CoV-2
In most cases, miRNAs regulate target gene expression by binding to the 3′-UTR of mRNAs; however, viral genomes are significantly larger than normal mRNA, so their regulation and interactions with miRNAs can be more complicated. It has been shown that miRNA binding sites in the 5′- and 3′-UTRs and coding regions can all be functional [21, 81]. While no binding sites were predicted in the 3′-UTR of the SARS-CoV-2 genome in our analysis, we identified three miRNAs with predicted binding sites in SARS-CoV-2 coding regions (miR-16, miR-200, and miR-24) and will further discuss their putative functions in the context of SARS-CoV-2 infection.
Functions and Pathways of miR-16 that may be Perturbed During SARS-CoV-2 Infection
The high number of miR-16 binding sites predicted in all three HCoV genomes suggests that the viral RNA may act as a “sponge” to sequester miR-16, thereby decreasing its abundance in infected cells leading to a derepression of its targets in the host [82]. As determined from previous studies, one function of miR-16 is to induce apoptosis by downregulating survival factor BCL2 [83]. Apoptosis has been indicated as an important part of the innate antiviral immune response in some viral infections. For example, in influenza-infected mouse cells, apoptosis was found to be induced with the apoptotic cells subsequently being targeted by macrophages and neutrophils for phagocytosis [84]. By downregulating miR-16, viruses such as SARS-CoV-2 may evade this innate immune mechanism by suppressing host cell apoptosis.
The suppression of miR-16 has also been shown to increase mitochondrial reactive oxygen species (ROS) production and toll-like receptor 4 (TLR4) expression [85, 86]. This is of particular interest as both ROS production and TLR4 expression can have negative effects on lung cells and even cause lung damage, something which is pertinent in respiratory viruses such as SARS-CoV-2 and SARS-CoV. ROS production triggers a reaction through the TLR4-TRIF signaling pathway that ultimately increases cytokine production in lung macrophages causing inflammation and damage in the lungs [86]. The redox imbalance resulting from SARS-CoV infection and increased ROS production have also been shown to induce transforming growth factor beta 1 (TGF-β1) expression, a profibrogenic cytokine that further increases ROS production, and has been linked to the development of damaged and scarred lung tissue, commonly referred to as pulmonary fibrosis [87, 88]. Due to the similarities and high homology between SARS-CoV-2 and SARS-CoV, it is not surprising that these phenomena, namely the increased cytokine production and pulmonary fibrosis, are also evident in some severe COVID-19 cases [89–91]. Taken together, these observations suggest that the viral suppression of miR-16 may contribute to lung damage in severe COVID-19 cases as a result of increased production of ROS and TLR4. Therefore, upregulation of miR-16, perhaps in the form of an miRNA mimic, may be a plausible option to combat the detrimental symptoms of COVID-19. It will be interesting to see if further studies show that the predicted binding sites are indeed functional and if sponging of miR-16 by SARS-CoV-2, SARS-CoV, and MERS-CoV occurs in vivo. Finally, it is important to note that miR-16 is highly conserved among mammals, indicating that miR-16 may be involved in a mechanism of SARS-CoV-2 regulation that exists in humans as well as other mammals, such as bats, which can be infected by homologous bat CoVs [92]. If these HCoVs have indeed evolved a mechanism that takes advantage of the regulatory potential of miR-16 in their host, developing therapeutics that antagonize this interaction, such as miRNA inhibitors to prevent miR-16 interactions with the SARS-CoV-2 genome, is another plausible antiviral strategy.
Potential Effects of SARS-CoV-2 Interactions with miR-200
miR-200b and miR-200c were predicted by DIANA and TargetScan to bind to a highly conserved region of the ACE2 3′-UTR (Fig. 6), suggesting that these miRNAs play an important role in the regulation of ACE2. This is particularly important in terms of SARS-CoV-2 infection as the ACE2 receptor is used by both SARS-CoV-2 and SARS-CoV for attachment and entry into host cells. ACE2 is also considered to be a negative regulator of lung fibrosis as it is responsible for cleaving angiotensin II (Ang II), a peptide hormone that has been linked to the development of lung fibrosis through the activation of TGF-β [88]. Previous studies have shown that the upregulation of miR-200c-3p during avian influenza A virus (H5N1) infection induces a decrease in ACE2 levels and can consequently cause lung damage [65]. Similar to H5N1 infection, SARS-CoV-2 and SARS-CoV have been shown to induce a decrease in ACE2 expression; however, it is unclear if this is due to the binding of the S protein to the receptor or the result of upregulated miR-200b/c expression [88, 93]. As such, more in vitro and in vivo research would be required to determine the expression profile of miR-200b/c during SARS-CoV and SARS-CoV-2 infection. Furthermore, ACE2 has been identified as an important regulator of heart function, and it has been suggested that Ang II accumulation may lead to heart disease [94, 95]. Therefore, further research on miR-200b/c regulation of ACE2 may provide useful insight on heart-related risk factors and complications that occur in some SARS-CoV-2-infected patients, such as acute coronary syndrome, myocarditis, arrhythmias, and, in severe cases, cardiac arrest [96].
Functions and Pathways of miR-24 that are Susceptible to Modulation by SARS-CoV-2
The predicted miR-24 binding site in the viral S protein appears to be conserved between the SARS-CoV-2, SARS-CoV, and MERS-CoV genomes. miR-24 has previously been shown to have a notable role in the regulation of furin, a serine protease that is important for the proteolytic activation and host-cell entry of some respiratory viruses, such as SARS-CoV-2, MERS-CoV, and H5N1 [97–99]. Often abundant in several tissue types, including the lungs, many enveloped respiratory viruses make use of furin to cleave and modify surface glycoproteins involved in membrane binding and fusion, thereby promoting the fusogenic potential of the virus, which can ultimately lead to effects on cell tropism and viral pathogenicity [98, 99]. As there is a furin-like cleavage site (FCS) in the SARS-CoV-2 S protein that is not present in SARS-CoV, there is reason to believe that furin may contribute to SARS-CoV-2 infection and pathogenesis [100, 101]. In fact, this FCS may be responsible for the high membrane fusion activity that is specific to SARS-CoV-2 [98, 100]. Furin has been shown to activate TGF-β1, which, as discussed previously, may be related to the development of pulmonary fibrosis in severe COVID-19 cases [101]. Therefore, the potential downregulation of miR-24 during SARS-CoV-2 infection may lead to both an increase in furin, resulting in higher infectivity, along with an increase in TGF-β1 that can eventually lead to lung damage.
On the contrary, one study found that the FCS and activation by furin may not be crucial for SARS-CoV-2 infection. It was determined that the treatment of infected samples with a high concentration of human airway trypsin-like proteases (HATs) restored the fusogenic capacity of SARS-CoV-2 without FCS, indicating that the FCS and presence of furin may not be completely necessary for SARS-CoV-2 entry into host cells [100]. Instead, this study suggests that the overall structure of the S protein may be responsible for the high fusogenic activity of SARS-CoV-2, not the presence of the FCS; however, more in vivo evidence would be necessary to support this hypothesis and dismiss the possible role of furin in SARS-CoV-2 infection [100]. Regardless, it is evident that the interaction between miR-24 and furin during SARS-COV-2 infection may provide another avenue for antiviral development.
Conclusion and Future Directions
In conclusion, miRNAs represent novel and emerging targets for therapeutic intervention, including against SARS-CoV-2. The cellular targets of miRNAs can be suppressed by adding miRNA mimics or can be upregulated with the use of anti-miRs, of which there are several chemical classes. RNA viruses can interact with and suppress the function of endogenous miRNAs, regulate miRNAs, or even produce their own miRNAs under certain circumstances. Counteracting these evolved interactions has been shown to be antiviral for other viruses such as HCV, where antagomirs have been used successfully in clinical trials. Given the similarities and size of the SARS-CoV-2 genome, it is reasonable to hypothesize that such strategies will also be effective in developing new therapeutic strategies.
Upon closer examination, several miRNAs were found to have potential binding sites in the SARS-CoV-2, SARS-CoV, and MERS-CoV genomes. Among them, we identified three that showed promise in modulating SARS-CoV-2 infection, namely miR-16, miR-200, and miR-24. Many human miRNAs may see altered function during acute infection owing to a sponge effect of the SARS-CoV-2 genome and resulting derepression of mRNA targets. It is interesting to consider that some of these interactions may have evolved as part of virus tropism as a respiratory virus, and thus selection of virus strains could have occurred to subvert miRNA signaling in host cells.
Current and future work is centered on obtaining additional evidence to determine if SARS-CoV-2 indeed relies on specific miRNAs and through what mechanism. A starting point will be to validate the predicted binding sites of specific miRNAs in vivo and to determine whether or not these binding sites are functionally important to the SARS-CoV-2 life cycle. This will likely involve examining additional characteristics such as RNA secondary structure, miRNA abundance, and other cellular interactions. Profiling changes in miRNA expression during viral infection (i.e., which miRNAs are up- or downregulated) as well as extending our miRNA screening analysis to identify miRNAs that target the UTR regions will also be important.
Information on the cellular downstream effects resulting from altered miRNA expression can also be useful to understand the complications and symptoms associated with the viral infection. It is also important to investigate the conservation of miRNA binding sites and regulatory mechanisms among other mammals as CoV infections have been observed in mammals other than humans. As some CoV can cross species barriers, understanding the miRNA interactions and regulations in these non-human genomes may serve to improve our understanding of potential emerging HCoV such as SARS-COV-2.
In terms of translating miRNA targets into therapies, it is interesting to note that miRNAs can be delivered through injection or directly into the lungs using aerosolization technologies [102, 103]. Both mimics and inhibitors can be delivered in either of these formats. Furthermore, such strategies can be used in conjunction with small interfering RNA (siRNA) technology to directly target the viral genome, leading to powerful all RNA-based multicomponent therapies. Other dependencies on and susceptibilities to miRNA function may also emerge as antiviral strategies against SARS-CoV-2 [104]. Finally, small molecule modulators of miRNA function may be used to recapitulate miRNA function [105]. However, the latter cannot be effective without us first understanding the roles of miRNAs as therapeutic targets for intervention and treatment of SARS-CoV-2.
Acknowledgements
All figures were designed with Biorender.
Declarations
Funding
C.H. is supported by NSERC CREATE. N.A. is supported by an NSERC Postgraduate Scholarship-Doctoral. This work is supported by funding from a Natural Sciences and Engineering Research Council (NSERC) Grant (210719) and a Canadian Institutes of Health Research (CIHR) Grant (221125).
Conflict of interest
The authors, Christine Hum, Julia Loiselle, Nadine Ahmed, Tyler Shaw, Caroline Toudic, and John Paul Pezacki, have no conflicts of interest to declare.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and materials
The data generated or analyzed during this study are included within the article and its supplementary information files.
Code availability
Not applicable.
References
- 1.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML. Author correction: a new coronavirus associated with human respiratory disease in China. Nature. 2020;580(7803):2202–2203. doi: 10.1038/s41586-020-2202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui J, Li F, Shi LZ. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhong NS, Zheng BJ, Li YM, Poon LLM, Xie ZH, Chan KH, Li PH, Tan SY, Chang Q, Xie JP, Liu XQ. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet. 2003;362(9393):1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J of Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 7.Zaki AM, Van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J of Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 8.Chan JFW, Kok KH, Zhu Z, Chu H, To KKW, Yuan S, Yuen KY. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg ing Microbes Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi F, Qian S, Zhang S, Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem Biophys Res Commun. 2020;526(1):135–140. doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cantuti-Castelvetri L, Ojha R, Pedro LD, Djannatian M, Franz J, Kuivanen S, van der Meer F, Kallio K, Kaya T, Anastasina M, Smura T. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370(6518):856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu X, Chen P, Wang J, Feng J, Zhou H, Li X, Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63(3):457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181(4):914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92(4):418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartenian E, Nandakumar D, Lari A, Ly M, Tucker JM, Glaunsinger BA. The molecular virology of coronaviruses. J Biol Chem. 2020;295(37):12910–12934. doi: 10.1074/jbc.REV120.013930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eastman RT, Roth JS, Brimacombe KR, Simeonov A, Shen M, Patnaik S, Hall MD. Remdesivir: a review of its discovery and development leading to emergency use authorization for treatment of COVID-19. ACS Cent Sci. 2020;6(5):672–683. doi: 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O’Meara MJ, Rezelj VV, Guo JZ, Swaney DL, Tummino TA. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorlund K, Dron L, Park J, Hsu G, Forrest JI, Mills EJ. A real-time dashboard of clinical trials for COVID-19. Lancet Dig Health. 2020;2(6):e286–e287. doi: 10.1016/S2589-7500(20)30086-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singaravelu R, Chen R, Lyn RK, Jones DM, O’Hara S, Rouleau Y, Pezacki JP. Hepatitis C virus induced up-regulation of microRNA-27: A novel mechanism for hepatic steatosis. Hepatology. 2014;59(1):98–108. doi: 10.1002/hep.26634. [DOI] [PubMed] [Google Scholar]
- 20.Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16(7):421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 21.Broughton JP, Lovci MT, Huang JL, Yeo GW, Pasquinelli AE. Pairing beyond the seed supports microRNA targeting specificity. Mol Cell. 2016;64(2):320–333. doi: 10.1016/j.molcel.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedman RC, Farh KKH, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt MF. Drug target miRNAs: chances and challenges. Trends Biotechnol. 2014;32(11):578–585. doi: 10.1016/j.tibtech.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 24.Gallant-Behm CL, Piper J, Lynch JM, Seto AG, Hong SJ, Mustoe TA, Maari C, Pestano LA, Dalby CM, Jackson AL, Rubin P. A microRNA-29 Mimic (Remlarsen) represses extracellular matrix expression and fibroplasia in the skin. J Investig Dermatol. 2019;139(5):1073–1081. doi: 10.1016/j.jid.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 25.Seto AG, Beatty X, Lynch JM, Hermreck M, Tetzlaff M, Duvic M, Jackson AL. Cobomarsen, an oligonucleotide inhibitor of miR-155, co-ordinately regulates multiple survival pathways to reduce cellular proliferation and survival in cutaneous T-cell lymphoma. Br J Haematol. 2018;183(3):428–444. doi: 10.1111/bjh.15547. [DOI] [PubMed] [Google Scholar]
- 26.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, Van Der Meer AJ, Patick AK, Chen A, Zhou Y, Persson R. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368(18):1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 27.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309(5740):1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 28.Singaravelu R, O’Hara S, Jones DM, Chen R, Taylor NG, Srinivasan P, Pezacki JP. MicroRNAs regulate the immunometabolic response to viral infection in the liver. Nat Chem Biol. 2015;11(12):988–993. doi: 10.1038/nchembio.1940. [DOI] [PubMed] [Google Scholar]
- 29.Motsch N, Pfuhl T, Mrazek J, Barth S, Grässer FA. Epstein–Barr virus-encoded latent membrane protein 1 (LMP1) induces the expression of the cellular microRNA miR-146a. RNA Biol. 2007;4(3):131–137. doi: 10.4161/rna.4.3.5206. [DOI] [PubMed] [Google Scholar]
- 30.Miller S, Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol. 2008;6(5):363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin Q, McBride J, Fewell C, Lacey M, Wang X, Lin Z, Cameron J, Flemington EK. MicroRNA-155 is an Epstein–Barr virus-induced gene that modulates Epstein-Barr virus-regulated gene expression pathways. J Virol. 2008;82(11):5295–5306. doi: 10.1128/JVI.02380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singaravelu R, Ahmed N, Quan C, Srinivasan P, Ablenas CJ, Roy DG, Pezacki JP. A conserved miRNA-183 cluster regulates the innate antiviral response. J Biol Chem. 2019;294(51):19785–19794. doi: 10.1074/jbc.RA119.010858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su C, Hou Z, Zhang C, Tian Z, Zhang J. Ectopic expression of microRNA-155 enhances innate antiviral immunity against HBV infection in human hepatoma cells. Virol J. 2011;8(1):354. doi: 10.1186/1743-422X-8-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riess M, Fuchs NV, Idica A, Hamdorf M, Flory E, Pedersen IM, König R. Interferons induce expression of SAMHD1 in monocytes through down-regulation of miR-181a and miR-30a. J Biol Chem. 2017;292(1):264–277. doi: 10.1074/jbc.M116.752584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCaskill JL, Ressel S, Alber A, Redford J, Power UF, Schwarze J, Dutia BM, Buck AH. Broad-spectrum inhibition of respiratory virus infection by microRNA mimics targeting p38 MAPK signaling. Mol Ther Nucleic Acids. 2017;7:256–266. doi: 10.1016/j.omtn.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luig C, Köther K, Dudek SE, Gaestel M, Hiscott J, Wixler V, Ludwig S. MAP kinase-activated protein kinases 2 and 3 are required for influenza A virus propagation and act via inhibition of PKR. FASEB J. 2010;24(10):4068–4077. doi: 10.1096/fj.10-158766. [DOI] [PubMed] [Google Scholar]
- 37.Marchant D, Singhera GK, Utokaparch S, Hackett TL, Boyd JH, Luo Z, Si X, Dorscheid DR, McManus BM, Hegele RG. Toll-like receptor 4-mediated activation of p38 mitogen-activated protein kinase is a determinant of respiratory virus entry and tropism. J Virol. 2010;84(21):11359–11373. doi: 10.1128/JVI.00804-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449(7164):919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sodroski C, Lowey B, Hertz L, Jake Liang T, Li Q. MicroRNA-135a modulates hepatitis C virus genome replication through downregulation of host antiviral factors. Virologica Sinica. 2019;34(2):197–210. doi: 10.1007/s12250-018-0055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Girardi E, L.pez P, Pfefer S. On the importance of host Micro-RNAs during viral infection. Front Genet. 2018;9:439. doi: 10.3389/fgene.2018.00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheel TK, Luna JM, Liniger M, Nishiuchi E, Rozen-Gagnon K, Shlomai A, Auray G, Gerber M, Fak J, Keller I, Bruggmann R. A broad RNA virus survey reveals both miRNA dependence and functional sequestration. Cell Host Microbe. 2016;19(3):409–423. doi: 10.1016/j.chom.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luna JM, Scheel TK, Danino T, Shaw KS, Mele A, Fak JJ, Nishiuchi E, Takacs CN, Catanese MT, de Jong YP, Jacobson IM. Hepatitis C virus RNA functionally sequesters miR-122. Cell. 2015;160(6):1099–1110. doi: 10.1016/j.cell.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Libri V, Helwak A, Miesen P, Santhakumar D, Borger JG, Kudla G, Grey F, Tollervey D, Buck AH. Murine cytomegalovirus encodes a miR-27 inhibitor disguised as a target. Proc Natl Acad Sci. 2012;109(1):279–284. doi: 10.1073/pnas.1114204109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, Huang W, Squires K, Verlinghieri G, Zhang H. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4 + T lymphocytes. Nat Med. 2007;13(10):1241–1247. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- 45.Pfeffer S, Zavolan M, Grässer FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. Identification of virus-encoded microRNAs. Science. 2004;304(5671):734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 46.Barbu MG, Condrat CE, Thompson DC, Bugnar OL, Cretoiu D, Toader OD, Suciu N, Voinea SC. MicroRNA involvement in signaling pathways during viral infection. Front Cell Dev Biol. 2020;8:143. doi: 10.3389/fcell.2020.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mishra R, Kumar A, Ingle H, Kumar H. The interplay between viral-derived miRNAs and host immunity during infection. Front Immunol. 2020;10:3079. doi: 10.3389/fimmu.2019.03079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mallick B, Ghosh Z, Chakrabarti J. MicroRNome analysis unravels the molecular basis of SARS infection in bronchoalveolar stem cells. PLoS ONE. 2009;4(11):e7837. doi: 10.1371/journal.pone.0007837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arisan ED, Dart A, Grant GH, Arisan S, Cuhadaroglu S, Lange S, Uysal-Onganer P. The prediction of miRNAs in SARS-CoV-2 genomes: Hsa-miR databases identify 7 key miRs linked to host responses and virus pathogenicity-related KEGG pathways significant for comorbidities. Viruses. 2020;12(6):614. doi: 10.3390/v12060614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bartoszewski R, Dabrowski M, Jakiela B, Matalon S, Harrod KS, Sanak M, Collawn JF. SARS-CoV-2 may regulate cellular responses through depletion of specific host miRNAs. Am J Physiol Lung Cell Mol Physiol. 2020;319(3):L444–L455. doi: 10.1152/ajplung.00252.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duygu M, Demirci S, Adan A. Computational analysis of microRNA-mediated interactions in SARS-CoV-2 infection. PeerJ. 2020;8:e9369. doi: 10.7717/peerj.9369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khan MAAK, Sany MRU, Islam MS, Mehebub MS, Islam AB. Epigenetic regulator miRNA pattern differences among SARS-CoV, SARS-CoV-2, and SARS-CoV-2 world-wide isolates delineated the mystery behind the epic pathogenicity and distinct clinical characteristics of pandemic COVID-19. Front Genet. 2020;11:765. doi: 10.3389/fgene.2020.00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bonneau E, Neveu B, Kostantin E, Tsongalis GJ, De Guire V. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. Ejifcc. 2019;30(2):114. [PMC free article] [PubMed] [Google Scholar]
- 54.Nucleotide [Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]. Accession No. MN908947.3, Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1, complete genome. https://www.ncbi.nlm.nih.gov/nuccore/MN908947. Cited 1 Sept 2020.
- 55.Nucleotide [Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]. Accession No. NC_004718.3, SARS coronavirus Tor2, complete genome. https://www.ncbi.nlm.nih.gov/nuccore/NC_004718.3. Cited 1 Sept 2020.
- 56.Nucleotide [Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]. Accession No. NC_019843.3, Middle East respiratory syndrome-related coronavirus isolate HCoV-EMC/2012, complete genom. https://www.ncbi.nlm.nih.gov/nuccore/NC_019843.3?report=genbank. Cited 1 Sept 2020.
- 57.Ritchie W, Flamant S, Rasko JE. mimiRNA: a microRNA expression profiler and classification resource designed to identify functional correlations between microRNAs and their targets. Bioinformatics. 2010;26(2):223–227. doi: 10.1093/bioinformatics/btp649. [DOI] [PubMed] [Google Scholar]
- 58.Zheng C, Zheng Z, Sun J, Zhang Y, Wei C, Ke X, Liu Y, Deng L, Wang H. MiR-16-5p mediates a positive feedback loop in EV71-induced apoptosis and suppresses virus replication. Sci Rep. 2017;7(1):16422. doi: 10.1038/s41598-017-16616-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chahar HS, Corsello T, Kudlicki AS, Komaravelli N, Casola A. Respiratory syncytial virus infection changes cargo composition of exosome released from airway epithelial cells. Sci Rep. 2018;8(1):387. doi: 10.1038/s41598-017-18672-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Modai S, Farberov L, Herzig E, Isakov O, Hizi A, Shomron N. HIV-1 infection increases microRNAs that inhibit Dicer1, HRB and HIV-EP2, thereby reducing viral replication. PLoS ONE. 2019;14(1):e0211111. doi: 10.1371/journal.pone.0211111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bakre A, Mitchell P, Coleman JK, Jones LP, Saavedra G, Teng M, Tompkins SM, Tripp RA. Respiratory syncytial virus modifies microRNAs regulating host genes that affect virus replication. J Gen Virol. 2012;93(Pt 11):2346–2356. doi: 10.1099/vir.0.044255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang S, Li J, Li J, Yang Y, Kang X, Li Y, Wu X, Zhu Q, Zhou Y, Hu Y. Up-regulation of microRNA-203 in influenza A virus infection inhibits viral replication by targeting DR1. Sci Rep. 2018;8:6797. doi: 10.1038/s41598-018-25073-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pandey RK, Sundar S, Prajapati VK. Diferential expression of miRNA regulates T cell diferentiation and plasticity during visceral leishmaniasis infection. Front Microbiol. 2016;7:206. doi: 10.3389/fmicb.2016.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Curtale G. MiRNAs at the crossroads between innate immunity and cancer: focus on macrophages. Cells. 2018;7(2):12. doi: 10.3390/cells7020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Q, Du J, Yu X, Xu J, Huang F, Li X, Zhang C, Li X, Chang J, Shang D, Zhao Y. miRNA-200c-3p is crucial in acute respiratory distress syndrome. Cell Discov. 2017;3(1):1–17. doi: 10.1038/s41421-019-0132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(suppl_1):D152–7. [DOI] [PMC free article] [PubMed]
- 67.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(D1):D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47(D1):D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36(suppl_1):D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(suppl_1):D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32(suppl_1):D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hsu PW, Lin LZ, Hsu SD, Hsu JB, Huang HD. ViTa: prediction of host microRNAs targets on viruses. Nucleic Acids Res. 2007;35(Database issue):D381–D385. doi: 10.1093/nar/gkl1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48(D1):D127–D131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C, Dalamagas T, Hatzigeorgiou AG. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41(Web Server issue):W169–W173. doi: 10.1093/nar/gkt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nucleotide [Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]. Accession No. NM_021804.3, Homo sapiens angiotensin I converting enzyme 2 (ACE2), transcript variant 2, mRNA. https://www.ncbi.nlm.nih.gov/nuccore/NM_021804.3/. Cited 1 Sept 2020.
- 77.Martin HC, Wani S, Steptoe AL, Krishnan K, Nones K, Nourbakhsh E, Vlassov A, Grimmond SM, Cloonan N. Imperfect centered miRNA binding sites are common and can mediate repression of target mRNAs. Genome Biol. 2014;15(3):1–22. doi: 10.1186/gb-2014-15-3-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nersisyan S, Shkurnikov M, Turchinovich A, Knyazev E, Tonevitsky A. Integrative analysis of miRNA and mRNA sequencing data reveals potential regulatory mechanisms of ACE2 and TMPRSS2. PLoS ONE. 2020;15(7):e0235987. doi: 10.1371/journal.pone.0235987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu B, Wei XX, Wang TB, Zhou YC, Liu AM, Zhang GW. Increased miR-16 expression induced by hepatitis C virus infection promotes liver fibrosis through downregulation of hepatocyte growth factor and Smad7. Arch Virol. 2015;160(8):2043–2050. doi: 10.1007/s00705-015-2474-3. [DOI] [PubMed] [Google Scholar]
- 80.Lambert DW, Lambert LA, Clarke NE, Hooper NM, Porter KE, Turner AJ. Angiotensin-converting enzyme 2 is subject to post-transcriptional regulation by miR-421. Clin Sci. 2014;127(4):243–249. doi: 10.1042/CS20130420. [DOI] [PubMed] [Google Scholar]
- 81.Nasheri N, Singaravelu R, Goodmurphy M, Lyn RK, Pezacki JP. Competing roles of microRNA-122 recognition elements in hepatitis C virus RNA. Virology. 2011;410(2):336–344. doi: 10.1016/j.virol.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 82.Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Curr Biol. 2010;20(19):R858–R861. doi: 10.1016/j.cub.2010.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102(39):13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nainu F, Shiratsuchi A, Nakanishi Y. Induction of apoptosis and subsequent phagocytosis of virus-infected cells as an antiviral mechanism. Front Immunol. 2017;8:1220. doi: 10.3389/fimmu.2017.01220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moon HG, Yang J, Zheng Y, Jin Y. miR-15a/16 regulates macrophage phagocytosis after bacterial infection. J Immunol. 2014;193(9):4558–4567. doi: 10.4049/jimmunol.1401372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Imai Y, Kuba K, Neely GG, Yaghubian-Malhami R, Perkmann T, van Loo G, Ermolaeva M, Veldhuizen R, Leung YH, Wang H, Liu H, Sun Y, Pasparakis M, Kopf M, Mech C, Bavari S, Peiris JS, Slutsky AS, Akira S, Hultqvist M, Penninger JM. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell. 2008;133(2):235–249. doi: 10.1016/j.cell.2008.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu RM, Desai LP. Reciprocal regulation of TGF-β and reactive oxygen species: a perverse cycle for fibrosis. Redox Biol. 2015;6:565–577. doi: 10.1016/j.redox.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zuo W, Zhao X, Chen YG. SARS coronavirus and lung fibrosis. In: Lal S, editor. Molecular Biology of the SARS-Coronavirus. Springer, Berlin: Heidelberg; 2010. pp. 247–258. [Google Scholar]
- 89.Spagnolo P, Balestro E, Aliberti S, Cocconcelli E, Biondini D, Casa GD, Sverzellati N, Maher TM. Pulmonary fibrosis secondary to COVID-19: a call to arms? Lancet Respir Med. 2020;8(8):750–752. doi: 10.1016/S2213-2600(20)30222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu J, Xu X, Jiang L. SARS-CoV-2 induces transcriptional signatures in human lung epithelial cells that promote lung fibrosis. Respir Res. 2020;21(182). [DOI] [PMC free article] [PubMed]
- 91.Shi Y, Wang Y, Shao C, Huang J, Gan J, Huang X, Bucci E, Piacentini M, Ippolito G, Melino G. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27(5):1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yue J, Tigyi G. Conservation of miR-15a/16-1 and miR-15b/16-2 clusters. Mamm Genome. 2010;21(1–2):88–94. doi: 10.1007/s00335-009-9240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kai H, Kai M. Interactions of coronaviruses with ACE2, angiotensin II, and RAS inhibitors-lessons from available evidence and insights into COVID-19. Hypertens Res. 2020;43(7):648–654. doi: 10.1038/s41440-020-0455-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Crackower MA, Sarao R, Oudit GY, Yagil C, Kozieradzki I, Scanga SE, Oliveira-dos-Santos AJ, da Costa J, Zhang L, Pei Y, Scholey J, Ferrario CM, Manoukian AS, Chappell MC, Backx PH, Yagil Y, Penninger JM. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002;417(6891):822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- 95.Oudit GY, Kassiri Z, Patel MP, Chappell M, Butany J, Backx PH, Tsushima RG, Scholey JW, Khokha R, Penninger JM. Angiotensin II-mediated oxidative stress and inflammation mediate the age-dependent cardiomyopathy in ACE2 null mice. Cardiovasc Res. 2007;75(1):29–39. doi: 10.1016/j.cardiores.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 96.Kang Y, Chen T, Mui D, Ferrari V, Jagasia D, Scherrer-Crosbie M, Chen Y, Han Y. Cardiovascular manifestations and treatment considerations in COVID-19. Heart Br Card Soc. 2020;106(15):1132–1141. doi: 10.1136/heartjnl-2020-317056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Loveday EK, Diederich S, Pasick J, Jean F. Human microRNA-24 modulates highly pathogenic avian-origin H5N1 influenza A virus infection in A549 cells by targeting secretory pathway furin. J Gen Virol. 2015;96(Pt 1):30–39. doi: 10.1099/vir.0.068585-0. [DOI] [PubMed] [Google Scholar]
- 98.Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir Res. 2020;176(104742). [DOI] [PMC free article] [PubMed]
- 99.Millet JK, Whittaker GR. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc Natl Acad Sci. 2014;111(42):15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xia S, Lan Q, Su S, Wang X, Xu W, Liu Z, Zhu Y, Wang Q, Lu L, Jiang S. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal Transd Targeted Ther. 2020;5(1):92. doi: 10.1038/s41392-020-0184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen Z, Lu S, Xu M, Liu P, Ren R, Ma W. Role of miR-24, furin, and transforming growth factor-β1 signal pathway in fibrosis after cardiac infarction. Med Sci Monit. 2017;23:65–70. doi: 10.12659/MSM.898641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lam JKW, Liang W, Chan HK. Pulmonary delivery of therapeutic siRNA. Adv Drug Deliv Rev. 2012;64(1):1–15. doi: 10.1016/j.addr.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schlosser K, Taha M, Stewart DJ. Systematic assessment of strategies for lung-targeted delivery of microRNA mimics. Theranostics. 2018;8(5):1213–1226. doi: 10.7150/thno.22912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Powdrill MH, Desrochers GF, Singaravelu R, Pezacki JP. The role of microRNAs in metabolic interactions between viruses and their hosts. Curr Opin Virol. 2016;19:71–76. doi: 10.1016/j.coviro.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 105.Filip r, Desrochers GF, Lefebvre DM, Reed A, Singaravelu R, Cravatt BF, Pezacki JP. Functional profiling of microRNA targets using activity-based protein profiling: linking enzyme activity to microRNA-185 altered lipid metabolism. Cell Chem Biol. 2021;28(2):202–212. doi: 10.1016/j.chembiol.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data generated or analyzed during this study are included within the article and its supplementary information files.