Abstract
Background:
Premature menopause is an independent risk factor for cardiovascular disease in women, but mechanisms underlying this association remain unclear. Clonal hematopoiesis of indeterminate potential (CHIP), the age-related expansion of hematopoietic cells with leukemogenic mutations without detectable malignancy, is associated with accelerated atherosclerosis. Whether premature menopause is associated with CHIP is unknown.
Methods:
We included postmenopausal women from the UK Biobank (N=11,495) aged 40–70 years with whole exome sequences and from the Women’s Health Initiative (WHI, N=8,111) aged 50–79 years with whole genome sequences. Premature menopause was defined as natural or surgical menopause occurring before age 40 years. Co-primary outcomes were the presence of (1) any CHIP and (2) CHIP with variant allele frequency (VAF) >0.1. Logistic regression tested the association of premature menopause with CHIP, adjusted for age, race, the first 10 principal components of ancestry, smoking, diabetes mellitus, and hormone therapy use. Secondary analyses considered natural vs. surgical premature menopause and gene-specific CHIP subtypes. Multivariable-adjusted Cox models tested the association between CHIP and incident coronary artery disease (CAD).
Results:
The sample included 19,606 women, including 418 (2.1%) with natural premature menopause and 887 (4.5%) with surgical premature menopause. Across cohorts, CHIP prevalence in postmenopausal women with vs. without a history of premature menopause was 8.8% vs. 5.5% (P<0.001), respectively. After multivariable adjustment, premature menopause was independently associated with CHIP (all CHIP: OR 1.36, 95% 1.10–1.68, P=0.004; CHIP with VAF >0.1: OR 1.40, 95% CI 1.10–1.79, P=0.007). Associations were larger for natural premature menopause (all CHIP: OR 1.73, 95% CI 1.23–2.44, P=0.001; CHIP with VAF >0.1: OR 1.91, 95% CI 1.30–2.80, P<0.001) but smaller and non-significant for surgical premature menopause. In gene-specific analyses, only DNMT3A CHIP was significantly associated with premature menopause. Among postmenopausal middle-aged women, CHIP was independently associated with incident coronary artery disease (HR associated with all CHIP: 1.36, 95% CI 1.07–1.73, P=0.012; HR associated with CHIP with VAF >0.1: 1.48, 95% CI 1.13–1.94, P=0.005).
Conclusions:
Premature menopause, especially natural premature menopause, is independently associated with CHIP among postmenopausal women. Natural premature menopause may serve as a risk signal for predilection to develop CHIP and CHIP-associated cardiovascular disease.
Keywords: women, menopause, clonal hematopoiesis, hematology, inflammation, coronary artery disease
Introduction
Cardiovascular disease is the leading cause of death among women in the United States and globally,1 and cardiovascular risk increases substantially following menopause.2 The mean age at menopause in Western nations is approximately 51 years, but approximately 10% of women undergo menopause before age 45 years, and at least 1% experience menopause before age 40 years.3, 4 Premature menopause, defined as menopause onset before age 40, is associated with accelerated biologic aging,5, 6 cardiovascular disease, and all-cause mortality,7 with higher cardiovascular risks observed at progressively earlier menopausal age.8, 9 As the association between premature menopause and cardiovascular disease appears partially independent of traditional cardiovascular risk factors,9–11 multi-society cardiovascular guidelines now classify premature menopause as a “risk-enhancing factor” to guide allocation of primary-prevention statin therapy for women otherwise at intermediate risk of atherosclerotic cardiovascular disease (ASCVD).12, 13
Despite the well-described cardiovascular risks associated with premature menopause, the responsible mechanisms remain unclear. Historically, increased cardiovascular risks were attributed to loss of cardioprotective estrogen effects; however, in a recent study, hormone therapy did not lower cardiovascular risks associated with premature menopause,9 and randomized trials of postmenopausal hormone replacement therapy have not specifically targeted women with premature menopause.14 Relevant mechanisms, however, may extend beyond estrogen deficiency. In the largest genome-wide association study of age at natural (i.e., non-surgical) menopause to date, roughly two-thirds of genetic associations mapped to DNA damage repair pathways,15 implying that women with natural premature menopause may be predisposed to accumulate genetic damage; such damage or its downstream consequences may represent one mechanism of accelerated aging in women with natural premature menopause that may heighten cardiovascular disease risk.
Clonal hematopoiesis of indeterminate potential (CHIP) is the expansion of hematopoietic stem cells with somatic mutations in leukemia-associated genes, typically defined as a variant allele prevalence among circulating blood cells (i.e., variant allele frequency, VAF) >2% in the absence of detectable hematologic cytopenia, dysplasia, or malignancy.16, 17 CHIP is a relatively common age-associated phenomenon, affecting 10% of individuals older than 70 years.18–20 CHIP-associated mutations occur most commonly in 1 of 3 genes: DNMT3A and TET2, both of which function as tumor suppressor genes through DNA methylation and demethylation, and ASXL1, a chromatin-binding transcriptional regulator.17 CHIP is associated with elevation in inflammatory cytokines and accelerated atherosclerosis in animal21, 22 and human23, 24 studies. Whether a history of premature menopause is associated with CHIP is currently unknown.
This study tested whether premature menopause overall and stratified by mechanism of menopause (natural and surgical premature menopause) was associated with elevated risk of CHIP among postmenopausal women in the UK Biobank and Women’s Health Initiative (WHI). We further investigated the association of CHIP with incident coronary artery disease (CAD) events in these cohorts.
Methods
Study cohorts
Researchers may apply for UK Biobank data access (https://www.ukbiobank.ac.uk/). WHI data are available to investigators in dbGaP (Accession Number: phs001237.v2.p1). The UK Biobank is a population-based cohort of >500,000 UK adult residents recruited between 2006–2010 and followed prospectively via linkage to national health records.25 At baseline, participants underwent phlebotomy and provided detailed information about medical history, medication use including prior and current hormone therapy, and reproductive history, including history of menarche, menopause, parity, and gynecologic surgery. In the present study, the UK Biobank cohort included postmenopausal women aged 40–70 years at blood draw with complete reproductive history data and available whole exome sequences. As described previously,24 the UK Biobank cohort was restricted to unrelated European ancestry women. Follow-up in the UK Biobank occurred through March 2020 for inpatient diagnosis codes and May 2020 for the death register.
The WHI (NCT00000611) is a prospective, multicenter study of women recruited at 40 centers throughout the United States between 1993 and 1998.26 Participants enrolled in a clinical trial (hormone therapy, calcium/vitamin D supplementation, or dietary modification) or in an observational study. A sub-sample of unrelated WHI women who were 50–79 years old at the time of phlebotomy and underwent whole genome sequencing as part of the National Heart, Lung, and Blood Institute’s Trans-Omics for Precision Medicine (TOPMed) program (total N=11,023) were included in the present analysis.27 To avoid effects of study treatments on outcomes, women in the WHI hormone therapy trial with blood draw ≥2 years after the screening visit (n=486) were excluded. Follow up in WHI occurred through March 2019.
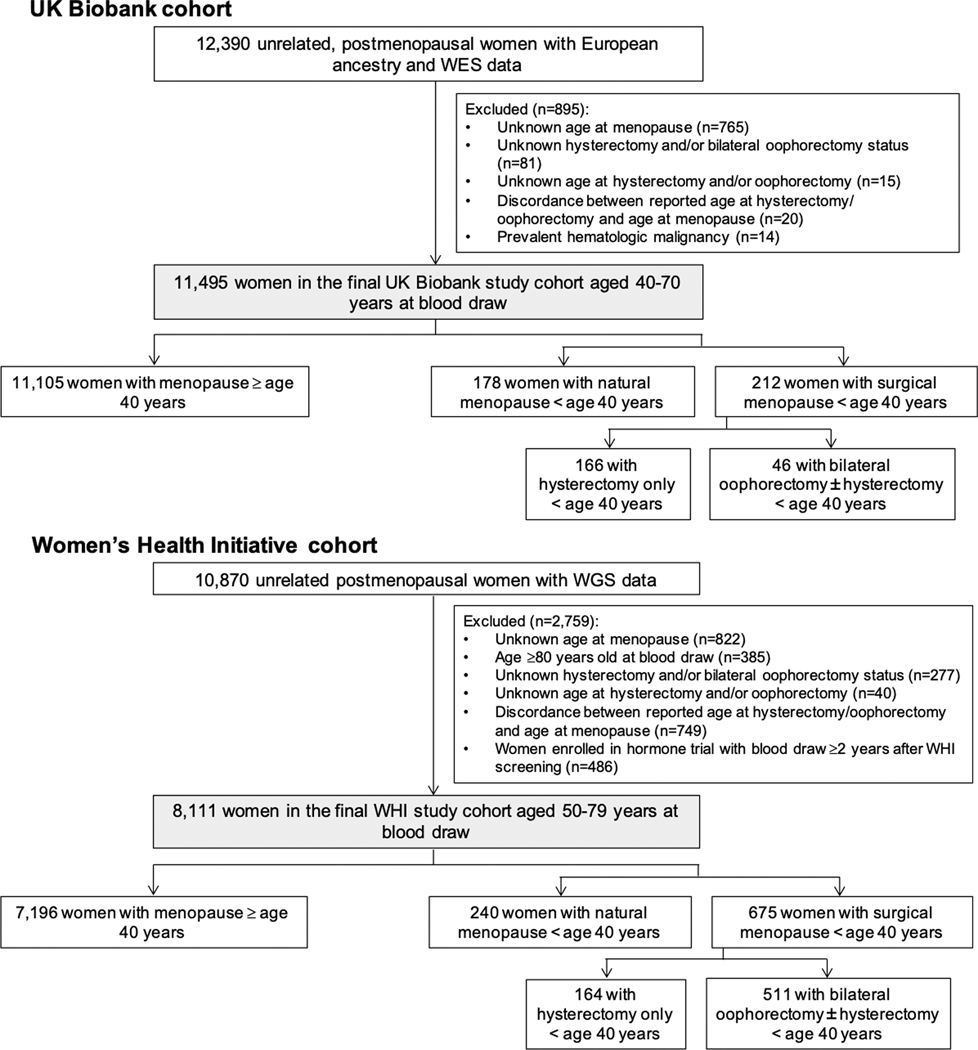
In both cohorts, related women within 3 degree of relatedness were identified using the Kinship-Based Inference for Genome-Wide Association Studies tool28 and excluded. Women with incomplete, unknown, or discordant reproductive history data were also excluded (Figure 1). This research was conducted under UK Biobank application number 7089. All participants provided informed consent. The Massachusetts General Hospital Institutional Review Board approved secondary use of these data.
Figure 1. Creation of the study cohort.
The study cohort included postmenopausal women from the UK Biobank with whole exome sequencing (WES) and from the Women’s Health Initiative (WHI) with whole genome sequencing (WGS).
Exposures
The primary exposure was premature menopause, defined as natural or surgical menopause occurring before age 40 years, for consistency with cardiovascular society guidelines,12, 13 as ascertained by participant self-report. Natural premature menopause was defined as any menopause occurring before age 40 years not resulting from surgery. Natural premature menopause, surgical premature menopause, surgical menopause with bilateral oophorectomy vs. hysterectomy only (to enable comparisons in women with ovarian conservation vs. removal), age at menarche, parity, and nulliparity constituted secondary exposures. Reproductive history, medical conditions, medication use, and health habits were systematically ascertained at study enrollment in both cohorts, and anthropomorphic data and vital signs were measured by study staff.25, 26
Sequencing and CHIP detection
UK Biobank exomes were sequenced from whole blood-derived DNA at the Regeneron Sequencing Center (Tarrytown, NY).29 The MuTect2 software30 analyzed exomes to detect somatic mutations in DNMT3A and TET2, using a pre-defined list of pre-leukemic CHIP driver mutations, in the UK Biobank (Supplemental Table I).24 WHI whole genomes were sequenced from whole blood-derived DNA at the Broad Institute of Harvard and MIT (Cambridge, MA). The MuTect2 software analyzed genomes for the presence of CHIP driver mutations in 74 genes in WHI (Supplemental Table II).31 In both cohorts, common germline variants and sequencing artifacts were excluded as before.24, 31 Compared with whole genome sequencing (>30X sequencing depth), the greater depth of whole exome sequencing (>50X) affords greater sensitivity to detect CHIP clones with VAF ≤0.1.31 In the <1% of women with >1 CHIP mutation, the CHIP gene and VAF used in analyses were assigned based on the CHIP mutation with the largest VAF. Mosaic chromosomal alterations (mCAs) were generated from genotype array data among UK Biobank participants as previously described.32, 33 Mutational signature analysis was performed using the “MutationalPatterns” package in R version 3.6.0.34
Outcomes
The co-primary study outcomes were the presence of (1) any CHIP and (2) CHIP with VAF >0.1, as larger CHIP clones above this threshold have previously been more strongly associated with adverse clinical outcomes.18, 24 The three most commonly mutated CHIP drivers (DNMT3A, TET2, and ASXL1) were each tested as separate secondary outcomes.
Additional models tested the association of CHIP with incident CAD. Incident CAD was defined in the UK Biobank by the appearance of a qualifying International Classification of Diseases (ICD) code in the study record as used previously9, 35 with ICD codes corresponding to acute myocardial infarction and coronary artery revascularization (Supplemental Table III) and in WHI as a composite of fatal and non-fatal myocardial infarction or coronary artery revascularization using standardized physician review, classification, and adjudication as previously described.36
Statistical analysis
Participant characteristics were compared between women with vs. without a history of premature menopause for continuous variables using the Student’s t-test or Wilcoxon rank-sum test, as appropriate, and for categorical variables using Pearson’s chi-squared test or Fisher exact test. Primary analyses tested the association of premature menopause with CHIP, for both UK Biobank and WHI, using multivariable logistic regression models, adjusted for age, the first 10 principal components of ancestry, current or former tobacco use, diabetes mellitus, and a history of current or previous hormone therapy use. Models testing associations among WHI participants were additionally adjusted for race/ethnicity, enrollment in the WHI observational study vs. clinical trial, and whether women were randomized to hormone therapy vs. placebo. To account for diabetes status, models of UK Biobank participants were adjusted for type 2 diabetes mellitus; as diabetes subtypes were not available in WHI, models of WHI participants were adjusted for any treated diabetes mellitus. For primary analyses, models were run separately in each cohort and meta-analyzed with fixed effects models using the “rmeta” package in R 3.6.0. We conducted a variety of sensitivity analyses, including: additionally adjusting for BMI; excluding women with history of cancer, women with history of gynecologic surgery, and women enrolled in the WHI hormone therapy trial; and stratifying by age <70 years (middle age) vs. ≥70 years at blood draw. In exploratory analyses, we tested the association of hormone therapy use with CHIP using logistic regression with the same covariates as the primary analyses, stratified by age at menopause.
Additional models tested the association of CHIP with incident CAD using Cox proportional hazard models, adjusted for age, the first 10 principal components of ancestry, current or former tobacco use, prevalent diabetes mellitus, systolic blood pressure, antihypertensive medication use, cholesterol-lowering medication use, BMI, prior hysterectomy, and a history of prior hormone therapy use; WHI models were further adjusted for race/ethnicity, enrollment in the WHI observational study vs. clinical trial, whether women were randomized to hormone therapy, and an inverse probability weight37 (to account for the non-random selection of women for whole-genome sequencing in WHI). Follow-up began at blood draw, and women with CAD diagnosed prior to blood draw were excluded from incident CAD analyses. Incident CAD models for WHI participants were additionally stratified by age <70 years vs. ≥70 years at blood draw. The proportional hazards assumption was tested using Schoenfeld residuals. As above, primary analyses were performed separately in each cohort and meta-analyzed using fixed effects models.
Given two primary outcomes in prevalent CHIP analyses and two primary exposures in incident CAD analyses, two-sided P<0.05/4 = 0.0125 was considered statistically significant. Findings of secondary analyses should be considered exploratory. Analyses were conducted using R 3.6.0 (R Foundation for Statistical Computing, Vienna, Austria).
Results
Description of the study cohort
The final study sample included 19,606 postmenopausal women across both cohorts, including 418 (2.1%) with natural premature menopause and 887 (4.5%) with surgical premature menopause. The UK Biobank cohort included 11,495 postmenopausal women aged 40–70 years at blood draw with complete reproductive history data and available whole exome sequences (Figure 1). The WHI cohort included 8,111 postmenopausal women with available whole genome sequences from peripheral blood DNA using blood drawn at age 50–79 years. Mean (SD) age at blood draw for whole exome and whole genome sequencing was 60.1 (5.2) years in the UK Biobank and 68.3 (6.6) years in WHI, respectively. The WHI cohort included 6,733 (83.0%) White, 918 (11.3%) Black, 217 (2.7%) Hispanic, and 157 (1.9%) Asian/Pacific Islander women.
In the UK Biobank, 390 women (3.4%) had a history of premature menopause, of whom 178 (1.5%) had natural premature menopause and 212 (1.8%) had surgical premature menopause (46 [0.4%] with bilateral oophorectomy ± hysterectomy vs. 166 [1.4%] with hysterectomy alone). In WHI, 915 women (11.3%) had a history of premature menopause, of whom 240 (3.0%) had natural premature menopause and 675 (8.3%) had surgical premature menopause (511 [6.3%] with bilateral oophorectomy ± hysterectomy vs. 164 [2.0%] with hysterectomy alone). In both cohorts, compared to women without a history of premature menopause, women with a history of premature menopause were 1.2 years younger at blood draw on average, were more likely to be current or former tobacco users, were more likely to be taking antihypertensive medications, and had higher baseline prevalence of diabetes mellitus and CAD (Table 1).
Table 1.
Baseline characteristics of the UK Biobank and Women’s Health Initiative study cohorts.
| UK Biobank (N=11,495) | Women’s Health Initiative (N=8,111) | |||||
|---|---|---|---|---|---|---|
| Menopause at age <40 years (N=390) | Menopause at age ≥40 years (N=11,105) | P-value | Menopause at age <40 years (N=915) | Menopause at age ≥40 years (N=7,196) | P-value | |
| Age at blood draw, years | 58.9 (6.6) | 60.1 (5.2) | <0.001 | 67.3 (6.5) | 68.5 (6.5) | <0.001 |
| Race/ethnicity | 1 | <0.001 | ||||
| • White | 390 (100%) | 11,105 (100%) | 681 (74.4%) | 6,052 (84.1%) | ||
| • Black | 0 (0%) | 0 (0%) | 180 (19.7%) | 738 (10.3%) | ||
| • Hispanic | 0 (0%) | 0 (0%) | 22 (2.4%) | 195 (2.7%) | ||
| • Asian | 0 (0%) | 0 (0%) | 12 (1.3%) | 145 (2.0%) | ||
| • American Indian/Alaskan Native | 0 (0%) | 0 (0%) | 11 (1.2%) | 17 (0.2%) | ||
| • Unknown | 0 (0%) | 0 (0%) | 9 (1.0%) | 49 (0.7%) | ||
| Age at menarche, years | 12.8 (1.8) | 12.9 (1.6) | 0.19 | 12.5 (1.5) | 12.7 (1.3) | 0.008 |
| Age at menopause, years | 35.1 (4.1) | 50.5 (4.2) | <0.001 | 34.3 (3.6) | 49.8 (1.9) | <0.001 |
| Parity (median [IQR]) | 2 [1, 2] | 2 [1, 2] | 0.77 | 3 [2, 4] | 3 [2, 4] | <0.001 |
| Nulliparity | 63 (16.2%) | 1,913 (17.2%) | 0.63 | 38 (4.8%) | 138 (2.1%) | <0.001 |
| History of hysterectomy | 225 (57.7%) | 921 (8.3%) | <0.001 | 770 (84.2%) | 2,402 (33.4%) | <0.001 |
| History of bilateral oophorectomy | 64 (16.4%) | 543 (4.9%) | <0.001 | 498 (54.4%) | 1,428 (19.8%) | <0.001 |
| Ever-use of hormone therapy | 300 (76.9%) | 5,009 (45.2%) | <0.001 | 701 (78.9%) | 4,169 (59.2%) | <0.001 |
| Current use of hormone therapy | 59 (15.1%) | 692 (6.2%) | <0.001 | 365 (41.1%) | 2,238 (31.8%) | <0.001 |
| Duration of prior hormone therapy use (among hormone therapy users) | 9.1 (6.7) | 5.6 (4.5) | <0.001 | 15.3 (12.5) | 8.8 (8.1) | <0.001 |
| Current or former smoking | 203 (52.1%) | 4,578 (41.2%) | <0.001 | 483 (53.1%) | 3,464 (48.8%) | 0.01 |
| Type 2 diabetes mellitus | 15 (3.8%) | 177 (1.6%) | 0.001 | 92 (10.1%) | 452 (6.3%) | <0.001 |
| Coronary artery disease | 19 (4.9%) | 281 (2.5%) | 0.007 | 127 (13.9%) | 728 (10.1%) | <0.001 |
| History of cancer | 70 (18.1%) | 1,197 (10.8%) | <0.001 | 98 (10.8%) | 467 (6.5%) | <0.001 |
| Antihypertensive medication use | 98 (25.1%) | 2,125 (19.1%) | 0.004 | 388 (42.4%) | 2,350 (32.6%) | <0.001 |
| Cholesterol-lowering medication use | 88 (22.6%) | 1,673 (15.1%) | <0.001 | 152 (16.6%) | 1,063 (14.8%) | 0.16 |
| Body-mass index, kg/m2 | 28.5 (6.3) | 26.9 (4.9) | <0.001 | 29.9 (6.3) | 28.5 (6.0) | <0.001 |
| Systolic blood pressure, mmHg | 139.7 (20.1) | 140.5 (20.1) | 0.46 | 133.6 (19.0) | 132.0 (18.3) | 0.02 |
| Low-density lipoprotein cholesterol, mg/dL | 142.8 (34.6) | 144.6 (33.3) | 0.34 | 140.3 (39.4) | 142.9 (37.2) | 0.14 |
| High-density lipoprotein cholesterol, mg/dL | 61.3 (15.3) | 64.0 (14.7) | 0.001 | 55.4 (15.7) | 56.2 (15.3) | 0.25 |
| Triglycerides, mg/dL (median [IQR]) | 134.5 [93.7, 186.5] | 122.0 [89.9, 168.9] | 0.001 | 140.0 [100.0, 195.0] | 135.0 [95.0, 185.0] | 0.17 |
| C-reactive protein, mg/L (median [IQR]) | 2.1 [0.9, 4.1] | 1.4 [0.7, 2.8] | <0.001 | -- | -- | -- |
| Women’s Health Initiative observational study | -- | -- | -- | 399 (43.6%) | 3,222 (44.8%) | 0.53 |
| Any Women’s Health Initiative clinical trial | -- | -- | -- | 516 (56.4%) | 3,974 (55.2%) | 0.53 |
| Women’s Health Initiative hormone trial | -- | -- | -- | 301 (32.9%) | 2,348 (32.6%) | 0.90 |
Prevalence of CHIP
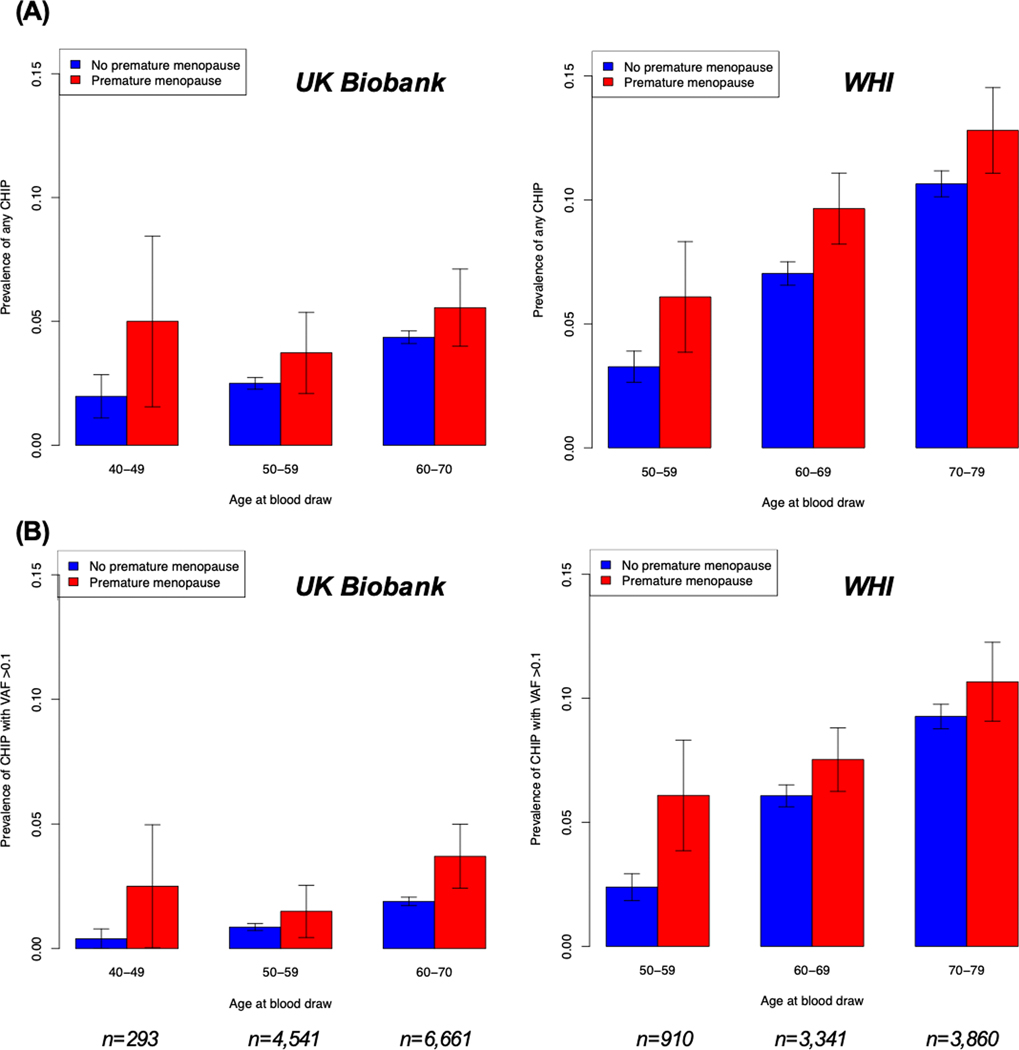
Consistent with the older age of women enrolled and greater number of CHIP drivers ascertained in WHI, CHIP prevalence was greater in WHI compared with the UK Biobank: 698 (8.6%) (including 533 of 698 [76.4%] with DNMT3A or TET2 mutations) in WHI had CHIP vs. 415 (3.6%) in the UK Biobank (P<0.001) (Supplemental Table IV). In the UK Biobank, 172 women with CHIP (41.4% of 415) had large clones (i.e., VAF >0.1). By contrast, 598 women with CHIP (85.7% of 698) in WHI had large CHIP clones, likely because of older age and decreased sensitivity of whole genome sequencing (compared whole exome sequencing) for detecting smaller clones. The most commonly mutated CHIP driver gene was DNMT3A in both cohorts, accounting for 342 (82.4% of 415) CHIP cases in the UK Biobank and 404 (57.9% of 698) CHIP cases in WHI. After DNMT3A, the most common CHIP drivers in WHI were TET2 (129 [18.5%]) ASXL1 (51 [7.3%]), and JAK2 (26 [3.7%]). CHIP prevalence increased progressively with older chronologic age (Figure 2). In WHI, American Indian/Alaskan Native women had 2.56-fold adjusted prevalence of CHIP vs. White women, although this association was not statistically significant (95% CI 0.86–7.67, P=0.09); no other significant associations with race/ethnicity were observed (Supplemental Table V).
Figure 2. Prevalence of (A) any CHIP and (B) CHIP with variant allele frequency >0.1 by age and premature menopause status in the UK Biobank and Women’s Health Initiative.
As expected, CHIP prevalence increased with age. Prevalence of CHIP was higher in women with a history of premature menopause than in women without a history of premature menopause irrespective of age at blood draw. Error bars represent ±1 standard error of the sample proportion. WHI = Women’s Health Initiative; CHIP = clonal hematopoiesis of indeterminate potential; VAF = variant allele frequency.
Premature menopause and clonal hematopoiesis
The overall pooled prevalence of CHIP across cohorts was 8.8% (115/1,305) among women with a history of premature menopause and 5.5% (998/18,301) among women without (P<0.001). Crude CHIP prevalence progressively increased with earlier age at menopause (Supplemental Figure I). CHIP prevalence was greater in women with vs. without premature menopause irrespective of age at blood draw (Figure 2); differences in CHIP prevalence were more pronounced for with CHIP with VAF >0.1 (Supplemental Figure II).
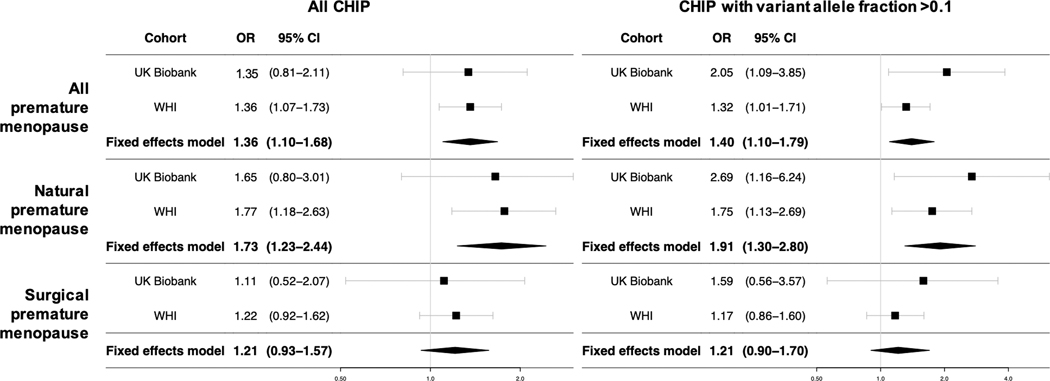
Multivariable-adjusted associations between premature menopause and CHIP were similar in the UK Biobank and WHI (Figure 3). Premature menopause was associated with all CHIP with an odds ratio (OR) of 1.35 (95% CI 0.83–2.17, P=0.22) in the UK Biobank and an OR of 1.36 (95% CI 1.07–1.73, P=0.01) in WHI, yielding a meta-analyzed OR of 1.36 (95% CI 1.10–1.68, P=0.004; P(heterogeneity)=0.97) in our primary analysis. This association appeared to be driven primarily by a stronger association between natural premature menopause and CHIP (meta-analyzed OR 1.73, 95% CI 1.23–2.44, P=0.001; P(heterogeneity)=0.86), compared to the observed relationship between surgical premature menopause and CHIP (meta-analyzed OR 1.21 95% CI 0.93–1.57, P=0.15; P(heterogeneity)=0.81). For CHIP with VAF >0.1, the association with natural premature menopause appeared even stronger (UK Biobank: OR 2.72, 95% CI 1.17–6.29, P=0.02; WHI: OR 1.75, 95% CI 1.13–2.69, P=0.01; meta-analyzed OR 1.91, 95% CI 1.30–2.80. P<0.001; P(heterogeneity)=0.26), whereas there was no evidence of association with surgical premature menopause.
Figure 3. Meta-analysis of associations between premature menopause and CHIP in the UK Biobank and Women’s Health Initiative.
Premature menopause was independently associated with CHIP. Associations were larger for natural premature menopause but smaller and non-significant for surgical premature menopause. Models for both cohorts are adjusted for age, the first 10 principal components of ancestry, ever-smoking, diabetes mellitus, and use of hormone therapy. WHI models are further adjusted for race/ethnicity, enrollment in the WHI observational study vs. clinical trial, and whether women were randomized to hormone therapy vs. placebo. WHI = Women’s Health Initiative; CHIP = clonal hematopoiesis of indeterminate potential; VAF = variant allele frequency.
Among women with a history of surgical premature menopause, associations with CHIP were similar and non-significant among women who underwent bilateral oophorectomy with or without hysterectomy vs. those who underwent hysterectomy alone (Supplemental Table VI). Age at menarche, parity, and nulliparity were not significantly associated with CHIP in either cohort. Associations were not materially changed after further adjustment for body-mass index (BMI, Supplemental Table VII). Sensitivity analyses excluding women with cancer showed results consistent with the primary analyses (Supplemental Table VIII). Sensitivity analyses excluding women with any history of gynecologic surgery and women enrolled in the WHI hormone therapy trial also yielded similar results. When the WHI cohort was stratified by age at blood draw <70 years vs. ≥70 years, associations among women <70 years were stronger and more closely resembled those observed in the UK Biobank cohort (all <70 years old at blood draw), while associations among older women in WHI were attenuated (Supplemental Table IX).
We next compared patterns of somatic mutations among women with CHIP by premature menopause status in the UK Biobank using the Catalogue of Somatic Mutations in Cancer (COSMIC) mutational signatures.38, 39 Among those with CHIP, mutational signatures were similar among women with natural premature menopause, surgical premature menopause, and no premature menopause; the most common mutational signatures across all 3 groups were defective homologous recombination DNA damage repair, age-related spontaneous deamination of 5-methylcytosine, and defective DNA mismatch repair (Supplemental Figure III).39
Having established an association between premature menopause and CHIP, we next assessed whether premature menopause was associated more strongly with specific CHIP genes (Supplemental Table X). Women with natural premature menopause had particular enrichment in DNMT3A CHIP (all DNMT3A CHIP: meta-analyzed OR 1.97, 95% CI 1.34–2.92, P<0.001; P(heterogeneity)=0.78; DNMT3A CHIP with VAF >0.1: meta-analyzed OR 2.28, 95% CI 1.45–3.56, P<0.001; P(heterogeneity)=0.48), but DNMT3A CHIP showed no evidence of association with surgical premature menopause. Natural premature menopause was not associated with TET2 or ASXL1 CHIP. Consistent with known selection from chemotherapy, PPM1D and TP53 CHIP were enriched among women with CHIP with vs. without a history of cancer in WHI (8.8% vs. 3.9%, P=0.06). Among women without prior cancer, no women with premature menopause had TP53 CHIP; no women with natural premature menopause had PPM1D CHIP; and 2 women with surgical premature menopause (3.8% of 53 with CHIP) had PPM1D CHIP, similar to the proportion of women without premature menopause with PPM1D CHIP (17 [3.1%] of 549).
Women with premature menopause and CHIP had higher red blood cell distribution width (RDW) and lower platelet counts compared with other women (Supplemental Table XI). C-reactive protein levels were higher in women with premature menopause irrespective of CHIP status. Among women in the UK Biobank, in an exploratory model adjusted for age, the first 10 principal components of ancestry, and smoking status, premature menopause was associated with 3-fold risk for myeloid dysplasia and neoplasia (hazard ratio [HR] 2.98, 95% CI 1.06–8.38, P=0.04).
Given observed associations between premature menopause and CHIP, we tested whether premature menopause was associated with other clonal somatic phenomena. Mosaic chromosomal alterations (mCAs) were previously ascertained among UK Biobank participants using single nucleotide polymorphism array data.32, 33 Among 147,573 genotyped postmenopausal women in the UK Biobank with complete reproductive history data, premature menopause was associated with 1.3-fold risk of mCA with cell fraction >0.1 (OR 1.31, 95% CI 1.01–1.70, P=0.04), driven by a 2-fold increase in chromosome X mCA with cell fraction >0.1 among women with natural premature menopause (OR 2.07, 95% CI 1.06–4.04, P=0.03; Supplemental Table XII). The proportion of women with mCAs was similar among those with CHIP (3.9%) and without CHIP (3.5%, P=0.57).
Association of hormone therapy use with CHIP
In multivariable-adjusted models, ever-use and current use of hormone therapy at study enrollment were associated with increased odds of CHIP in the UK Biobank (ever-use: OR 1.26, 95% CI 1.03–1.54, P=0.03; current use: OR 1.88, 95% CI 1.33–2.59, P<0.001), but neither was significantly associated with CHIP in WHI (ever-use: OR 1.08, 95% CI 0.91–1.26, P=0.36; current use: OR 0.96, 95% CI 0.81–1.14, P=0.99). Since women with premature menopause are more likely to use hormone therapy than other women, we assessed whether the association with hormone therapy observed in the UK Biobank reflected residual confounding by an indication of premature menopause and whether the association of hormone therapy with CHIP differed by menopausal age. In the UK Biobank, multivariable-adjusted analyses stratified by age at menopause suggested that current or prior use of hormone therapy at study enrollment were associated with increased in risk of CHIP only among women without premature menopause (Supplemental Table XIII), and specifically among women with menopause after age 50 years. WHI participants had no increased or decreased risk of CHIP associated with prior or current hormone therapy use at study enrollment in any age stratum.
Association between CHIP and incident coronary artery disease
Follow-up for incident CAD events (myocardial infarction or coronary revascularization) occurred over a median (interquartile range) 10.0 (9.8–10.2) years in the UK Biobank and 13.1 (6.8–18.8) years in WHI. There were 473 incident CAD cases in the UK Biobank and 1,146 incident CAD cases in WHI.
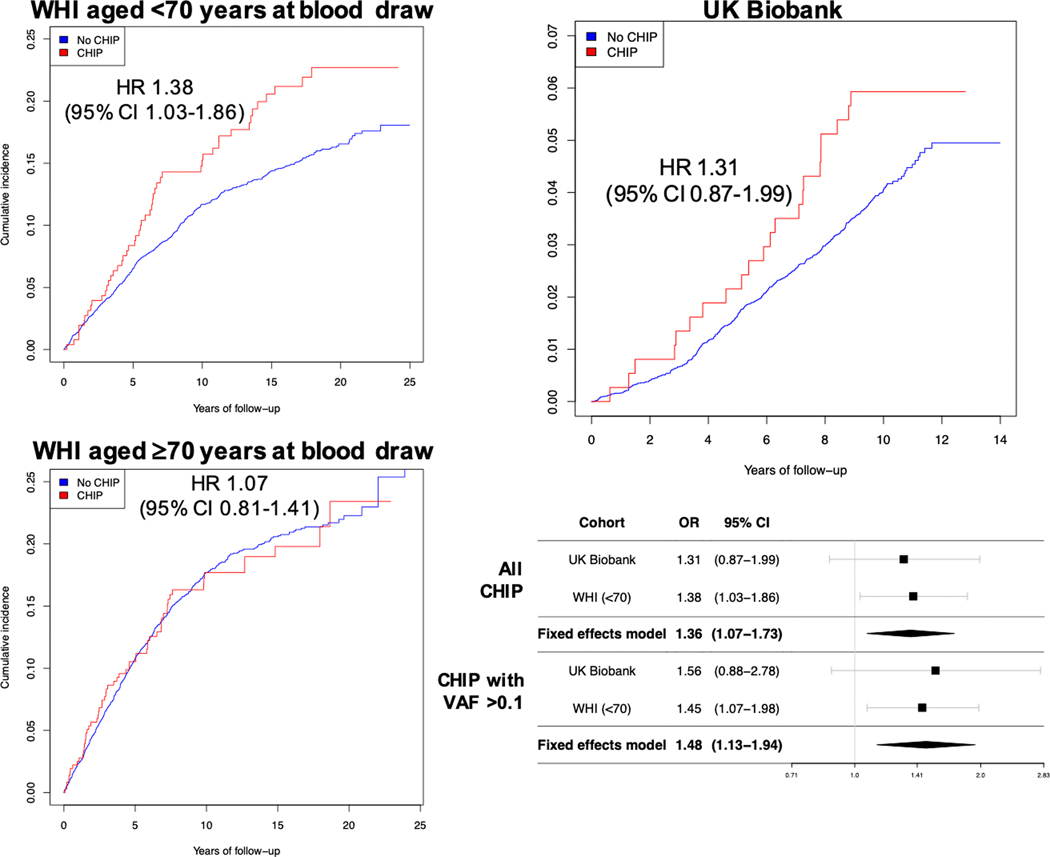
After multivariable adjustment, estimated associations between CHIP and incident CAD were similar among women in the UK Biobank (for all CHIP: HR 1.31, 95% CI 0.87–1.99, P=0.20; for CHIP with VAF >0.1: HR 1.56, 95% CI 0.88–2.78, P=0.13) and among middle-aged women (<70 years old at blood draw) in WHI (for all CHIP: HR 1.38, 95% CI 1.03–1.86, P=0.03; for CHIP with VAF >0.1, HR 1.45, 95% CI 1.07–1.98, P=0.02). By contrast, no association for CHIP with incident CAD was present among women ≥70 years old at blood draw (Supplemental Table XIV; P(interaction) for age <70 vs. ≥70 years *CHIP with VAF >0.1 = 0.06). In meta-analyzed models of women in the UK Biobank and women <70 years old in WHI, all CHIP was associated with an HR of 1.36 (95% CI 1.07–1.73, P=0.012; P(heterogeneity)=0.85) and CHIP with VAF >0.1 with an HR of 1.48 (95% CI 1.13–1.94, P=0.005; P(heterogeneity)=0.83) (Figure 4). The association between CHIP and incident CAD was attenuated for VAF <0.1 (Supplemental Figure IV).
Figure 4. Incidence of coronary artery disease in women with and without CHIP in the UK Biobank and Women’s Health Initiative.
CHIP was independently associated with incident coronary artery disease in meta-analyzed results from postmenopausal women in the UK Biobank (age 40–70 years at blood draw) and postmenopausal women <70 years old at blood draw in the Women’s Health Initiative (WHI). Cox proportional hazard models are adjusted for age, the first 10 principal components of ancestry, current or former tobacco use, prevalent diabetes mellitus, systolic blood pressure, antihypertensive medication use, cholesterol-lowering medication use, body-mass index, prior hysterectomy, and a history of prior hormone therapy use; WHI models are further adjusted for race/ethnicity, enrollment in the WHI observational study vs. clinical trial, whether women were randomized to hormone therapy vs. placebo, and a WHI inverse probability weight. CHIP = clonal hematopoiesis of indeterminate potential; VAF = variant allele frequency; HR = hazard ratio.
In models for incident CAD that were further adjusted for premature menopause status, the association between and incident CAD was unchanged (meta-analyzed HR 1.36, 95% CI 1.07–1.73, P=0.01; P(heterogeneity)=0.84). Across cohorts, cumulative CAD incidence (among women without prevalent CAD) was 8.3% (1,352/16,376) in women without premature menopause or CHIP, 12.5% (132/1,056) in women with premature menopause only, 13.0% (119/916) in women with CHIP only, 15.4% (97/629) in women with CHIP with VAF >0.1 only, 15.5% (16/103) in women with premature menopause and any CHIP, and 17.3% (14/81) in women with premature menopause and CHIP with VAF >0.1 (P<0.001) (Table 2). Compared with women without premature menopause or CHIP, women with both (n=103 without prevalent CAD) had a meta-analyzed adjusted HR for incident CAD of 1.57 (95% CI 0.83–2.97, P=0.17; P(heterogeneity)=0.49), and women with premature menopause and CHIP with VAF >0.1 (n=81) had a meta-analyzed HR of 1.87 (95% CI 0.99–3.53, P=0.053); P(heterogeneity)=0.87). Sensitivity analysis in WHI restricted to women who were <70 years old at blood draw and not enrolled in the hormone trial (n=2,669) confirmed that study treatments did not drive the observed associations with CHIP: all CHIP was associated with an HR of 1.48 (95% CI 1.00–2.18, P=0.05), and CHIP with VAF >0.1 was associated with an HR of 1.54 (95% CI 1.02–2.33, P=0.04). Mosaic chromosomal alterations were not associated with incident CAD (all mCAs: HR 0.84, 95% CI 0.61–1.14, P=0.25; mCAs with cell fraction >0.1: HR 1.04, 95% CI 0.39–2.78, P=0.95). CHIP was associated with all-cause mortality (all CHIP: meta-analyzed HR 1.18, 95% CI 1.06–1.31, P=0.003, P(heterogeneity)=0.29; CHIP with VAF >0.1: HR 1.20, 95% CI 1.07–1.34, P=0.001, P(heterogeneity)=0.31).
Table 2.
Cumulative incidence of coronary artery disease events by premature menopause and clonal hematopoiesis status in the pooled cohort of the UK Biobank and Women’s Health Initiative.
| No CHIP | Any CHIP | CHIP with VAF >0.1 | |
|---|---|---|---|
| No history of premature menopause | 8.3% (1,352/16,376) |
13.0% (119/916) |
15.4% (97/629) |
| History of premature menopause | 12.5% (132/1,056) |
15.5% (16/103) |
17.3% (14/81) |
Cumulative incidences listed are over a median (IQR) follow-up of 10.0 (9.8–11.9) years among women without established coronary artery disease at the time of blood draw. P<0.001 by the Pearson chi-squared test. CHIP = clonal hematopoiesis of indeterminate potential; VAF = variant allele frequency.
Discussion
A history of premature menopause, especially natural premature menopause, was independently associated with increased odds of CHIP, a recently recognized risk factor for cardiovascular disease, in two large cohorts of postmenopausal women with whole exome or genome sequencing of blood DNA. The risks of developing CHIP and specific CHIP driver mutations appeared to differ in women with natural versus surgical premature menopause, implying that postmenopausal reductions in estrogen and other sex steroid hormones alone may not explain the relationship between menopause and CHIP. Premature menopause was more strongly associated with CHIP with VAF >0.1, which were more strongly linked to adverse clinical outcomes in this and prior studies compared with smaller CHIP clones. Furthermore, CHIP was associated with incident CAD in postmenopausal middle-aged women independent of conventional risk factors, extending CHIP’s role as an atherosclerotic cardiovascular disease risk factor to postmenopausal women.
These findings have several key implications for understanding the biological relevance of CHIP in postmenopausal women. First, menopausal age represents a novel risk signal for development of CHIP beyond chronologic age. Independent of established CHIP risk factors, premature menopause was associated with 1.4-fold odds—and natural premature menopause with 1.7-fold odds—of CHIP. Chronologic age, tobacco smoking, and prior exposure to chemotherapy and radiation represent the strongest risk factors for CHIP,18, 40 but our understanding of clinical factors predisposing to CHIP development is otherwise limited. As evidence-based strategies for mitigation of CHIP-associated cardiovascular and cancer risks emerge, and as genomic sequencing becomes increasingly accessible, identification of individuals suitable for CHIP screening will become increasingly clinically relevant.41, 42 History of premature menopause may help identify individuals for CHIP screening and surveillance. Although the present analysis is cross-sectional, prior studies have found very low CHIP prevalence (<0.1%) among individuals than age 40 years18–20—substantially lower than the prevalence of premature menopause in the present cohort and prior cohorts of postmenopausal women8, 9, 43—suggesting that premature menopause likely occurs antecedent to CHIP development. However, the differential association of premature natural versus premature surgical menopause with CHIP lends to the possibility of CHIP occurring prior to premature menopause. Future studies with premenopausal CHIP ascertainment will better clarify this temporal relationship.
Second, the association between premature menopause and CHIP provides insights into potential mechanisms of CHIP development in women. Prior work shows that hematopoietic stem cells possess receptors for sex hormones, including estrogen,44, 45 and that estrogen signaling promotes stem cell renewal46 and exhibits anti-leukemogenic properties.47, 48 We observed that women with natural premature menopause had consistently higher risk of CHIP in both cohorts. However, women with or without bilateral oophorectomy (i.e., removal of the primary source of estrogen) at the time of surgical premature menopause had similar risk of CHIP, suggesting that hormonal deficiency alone does not account for the mechanism of CHIP development. DNA damage repair pathways account for two-thirds of identified genetic determinants of age at natural menopause.15 Genome-wide association studies (GWAS) of age at natural menopause15 have implicated loss-of-function variants in CHEK2, a DNA damage checkpoint kinase.31 Furthermore, genetic variants in TERT, which encodes telomerase, associate strongly with CHIP31, 49 as well as other cancers. Telomeres are key regulators of genomic stability and cellular senescence. TERT variants also link to accelerated epigenetic aging and earlier age at natural menopause.50 Natural premature menopause may reflect a latent predisposition to accumulation of somatic mutations and/or attrition of stem cells in both the ovaries and the bone marrow, in turn enabling selective advantage of CHIP clones. In other words, natural premature menopause may serve as a risk signal for predilection to CHIP development. We also observed enrichment of mosaic chromosomal alterations in women with natural premature menopause, suggesting natural premature menopause may be associated with a variety of clonal somatic phenomena not limited to CHIP and further supporting that natural premature menopause may indicate genomic instability more broadly. Fanconi anemia, a Mendelian syndrome due to defects in DNA damage repair, is characterized by infertility and extremely premature spontaneous menopause,51 accelerated development of hematologic and solid malignancy, and predilection to atherosclerosis.
Notably, we found that exogenous postmenopausal hormone use was associated with increased risk of CHIP among women in the UK Biobank who experienced menopause later in life. Compared with younger women using hormone therapy after early menopause, older women may have accumulated a greater burden of somatic mutations that may be positively selected in the setting of continued estrogen exposure. This premise represents one potential explanation for the “timing hypothesis” of hormone therapy, whereby increased cardiovascular risk associated with exogenous hormone therapy primarily affects women >60 years old and >10 years beyond menopause,52 although this possibility requires further testing in human cohorts and animal models. Our observation of DNMT3A CHIP predilection agrees with prior work demonstrating an inverse relationship between DNMT3A gene expression and cellular activation by estrogen in normal and tumor tissues.53–58
Third, CHIP represents an independent risk factor for incident CAD events among postmenopausal women. An association between CHIP and incident cardiovascular events was recently demonstrated for a composite outcome of myocardial infarction, coronary artery revascularization, stroke, and death among 35,000 male and female individuals in the UK Biobank,24 but whether this association was relevant among women, who have a lower absolute risk of cardiovascular disease, was unclear. Here, we find an association with incident CAD among postmenopausal middle-aged women. Although CHIP is strongly associated with chronologic age, we did not observe an independent association between CHIP and incident CAD among elderly women (≥70 years old); this finding may reflect a greater relative effect of CHIP compared with other conventional risk factors for earlier-onset CAD as suggested by previous cross-sectional analyses,23 exclusion of women with prevalent CAD (which was more common in women with CHIP) from incident disease models, and/or survival bias as is commonly observed in epidemiologic cohort studies. Much mechanistic work linking CHIP to atherothrombotic events has focused on TET2 CHIP and resultant IL-1β/IL-6 pathway activation predisposing to atherosclerosis.21, 23 However, recent studies implicate the same atherogenic inflammatory cytokines with DNMT3A CHIP as well.59, 60 A stratified human genetic analysis recently showed that individuals with CHIP are more likely to derive clinical cardiovascular benefit with IL-1β/IL-6 pathway inhibition.24 Therefore, women with premature menopause may particularly benefit from earlier CHIP screening with the prospect of a novel molecularly-guided preventive strategy. Affirmation of this hypothesis will require prospective randomized controlled trials.
Our study has several limitations. While CHIP was ascertained only in DNMT3A and TET2 in the UK Biobank, these genes constituted the majority (76%) of CHIP drivers in the WHI cohort and in prior analyses.18, 23 The UK Biobank study cohort included European ancestry women only. Although the UK Biobank and WHI differed with respect to age, racial/ethnic composition, the era during which women experienced menopause, associated practices surrounding gynecologic surgery and use of hormone therapy, DNA sequencing methods, and CHIP drivers queried, associations between premature menopause and CHIP—and between CHIP and incident CAD—were nonetheless highly consistent between the cohorts. Age at menopause was ascertained by participant self-report in both cohorts, which may be inaccurate particularly among older women who are further from menopause; however, such misclassification would be expected to minimize observed differences and bias results toward the null. Doses and preparations of hormone therapy used prior to study enrollment were not available; differences in these aspects of hormone therapy use may explain differential associations between hormone therapy and CHIP observed between cohorts. Finally, although multiple prior studies have shown that premature and early menopause independently associate with elevated risk of CAD,7, 9, 61, 62 premature menopause was not independently associated with CAD after covariate adjustment in the present dataset, precluding formal mediation analysis. This is likely due to reduced sample size of women with next-generation sequencing compared to prior larger epidemiologic studies. However, given the observed enrichment for CHIP among women with premature menopause, CHIP-associated CAD risk may be particularly important among some women with premature menopause.
In conclusion, premature menopause, especially natural premature menopause, is independently associated with CHIP among postmenopausal women. CHIP may contribute to the excess cardiovascular risk associated with premature menopause. These findings should stimulate further research to elucidate mechanisms of CHIP development in this population.
Supplementary Material
Clinical Perspective.
What is new?
We tested whether premature menopause (before age 40 years) overall and stratified by mechanism of menopause (natural vs. surgical premature menopause) was associated with clonal hematopoiesis of indeterminate potential (CHIP), the expansion of hematopoietic stem cells with somatic mutations in leukemia-associated genes, among postmenopausal women with next-generation genomic sequencing in the UK Biobank and Women’s Health Initiative.
Premature menopause, especially natural premature menopause, was independently associated with increased odds of CHIP; no significant association with surgical premature menopause was observed.
CHIP was associated with incident CAD events in postmenopausal middle-aged women independent of conventional CAD risk factors.
What are the clinical implications?
Natural premature menopause may represent a risk signal for latent genomic instability and predilection to develop CHIP and CHIP-associated cardiovascular disease.
Women with premature menopause may be particularly well suited to CHIP screening and surveillance and to precision medicine approaches to mitigate CHIP-associated cardiovascular disease risk, such as IL-1β/IL-6 inhibition.
Acknowledgements:
The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201600018C, HHSN268201600001C, HHSN268201600002C, HHSN268201600003C, and HHSN268201600004C. Molecular data for the Trans-Omics in Precision Medicine (TOPMed) program was supported by the National Heart, Lung and Blood Institute (NHLBI). Genome sequencing for NHLBI TOPMed: Women’s Health Initiative (phs001237.v2.p1) was performed at the Broad Institute Genomics Platform (HHSN268201500014C). Core support including centralized genomic read mapping and genotype calling, along with variant quality metrics and filtering were provided by the TOPMed Informatics Research Center (3R01HL-117626–02S1; contract HHSN268201800002I). Core support including phenotype harmonization, data management, sample-identity QC, and general program coordination were provided by the TOPMed Data Coordinating Center (R01HL-120393; U01HL-120393; contract HHSN268201800001I). We gratefully acknowledge the studies and participants who provided biological samples and data for TOPMed.
Sources of Funding
M.C.H. is supported by the National Institutes of Health (T32HL094301–07). J.P.P. is supported by a John S. LaDue Memorial Fellowship in Cardiovascular Research. A.P.R, E.A.W., and P.N. are supported by the National Heart, Lung, and Blood Institute (R01 HL148565). P.N. is additionally supported by grants from the National Heart, Lung, and Blood Institute (R01HL142711, R01HL148565, R01HL148050), Fondation Leducq (TNE-18CVD04), and a Hassenfeld award from the Massachusetts General Hospital.
Dr. Libby receives funding support from the National Heart, Lung, and Blood Institute (1R01HL134892), the American Heart Association (18CSA34080399), the RRM Charitable Fund, and the Simard Fund.
Disclosures
P.N. reports grant support from Amgen, Apple, and Boston Scientific, consulting income from Apple, and spousal employment and equity in Vertex, all unrelated to this work. Dr. Libby is an unpaid consultant to, or involved in clinical trials for Amgen, AstraZeneca, Baim Institute, Beren Therapeutics, Esperion, Therapeutics, Genentech, Kancera, Kowa Pharmaceuticals, Medimmune, Merck, Norvo Nordisk, Merck, Novartis, Pfizer, Sanofi-Regeneron. Dr. Libby is a member of scientific advisory board for Amgen, Corvidia Therapeutics, DalCor Pharmaceuticals, Kowa Pharmaceuticals, Olatec Therapeutics, Medimmune, Novartis, and XBiotech, Inc.
Dr. Libby’s laboratory has received research funding in the last 2 years from Novartis. Dr. Libby is on the Board of Directors of XBiotech, Inc. Dr. Libby has a financial interest in Xbiotech, a company developing therapeutic human antibodies. Dr. Libby’s interests were reviewed and are managed by Brigham and Women’s Hospital and Partners HealthCare in accordance with their conflict of interest policies.
Non-Standard Abbreviations and Acronyms
- CAD
Coronary artery disease
- CHIP
Clonal hematopoiesis of indeterminate potential
- COSMIC
Catalogue of Somatic Mutations in Cancer
- GWAS
Genome-wide association study
- mCA
Mosaic chromosomal alteration
- RDW
Red blood cell distribution width
- TOPMed
National Heart, Lung, and Blood Institute Trans-Omics for Precision Medicine program
- VAF
Variant allele frequency
- WES
Whole exome sequencing
- WGS
Whole genome sequencing
- WHI
Women’s Health Initiative
Footnotes
The other authors declare no competing interests.
References
- 1.Virani SS, Alonso A, Benjamin EJ, Bittencourt MS, Callaway CW, Carson AP, Chamberlain AM, Chang AR, Cheng S, Delling FN, et al. Heart Disease and Stroke Statistics-2020 Update: A Report From the American Heart Association. Circulation. 2020;141:e139–e596. doi: 10.1161/CIR.0000000000000757 [DOI] [PubMed] [Google Scholar]
- 2.Chae CU, Derby CA. The menopausal transition and cardiovascular risk. Obstet Gynecol Clin North Am. 2011;38:477–488. doi: 10.1016/j.ogc.2011.05.005 [DOI] [PubMed] [Google Scholar]
- 3.Zhu D, Chung HF, Pandeya N, Dobson AJ, Kuh D, Crawford SL, Gold EB, Avis NE, Giles GG, Bruinsma F, et al. Body mass index and age at natural menopause: an international pooled analysis of 11 prospective studies. Eur J Epidemiol. 2018;33:699–710. doi: 10.1007/s10654-018-0367-y [DOI] [PubMed] [Google Scholar]
- 4.Shifren JL, Gass ML, NAMS Recommendations for Clinical Care of Midlife Women Working Group. The North American Menopause Society recommendations for clinical care of midlife women. Menopause. 2014;21:1038–1062. doi: 10.1097/GME.0000000000000319 [DOI] [PubMed] [Google Scholar]
- 5.Levine ME, Lu AT, Chen BH, Hernandez DG, Singleton AB, Ferrucci L, Bandinelli S, Salfati E, Manson JE, Quach A, et al. Menopause accelerates biological aging. Proc Natl Acad Sci U S A. 2016;113:9327–9332. doi: 10.1073/pnas.1604558113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canonico M, Artaud F, Tzourio C, Elbaz A. Association of Reproductive History With Motor Function and Disability in Aging Women. J Am Geriatr Soc. 2020;68:585–594. doi: 10.1111/jgs.16257 [DOI] [PubMed] [Google Scholar]
- 7.Muka T, Oliver-Williams C, Kunutsor S, Laven JSE, Fauser BCJM, Chowdhury R, Kavousi M, Franco OH. Association of Age at Onset of Menopause and Time Since Onset of Menopause With Cardiovascular Outcomes, Intermediate Vascular Traits, and All-Cause Mortality: A Systematic Review and Meta-analysis. JAMA Cardiol. 2016;1:767–776. doi: 10.1001/jamacardio.2016.2415 [DOI] [PubMed] [Google Scholar]
- 8.Zhu D, Chung HF, Dobson AJ, Pandeya N, Giles GG, Bruinsma F, Brunner EJ, Kuh D, Hardy R, Avis NE, et al. Age at natural menopause and risk of incident cardiovascular disease: a pooled analysis of individual patient data. Lancet Public Health. 2019;4:e553–e564. doi: 10.1016/S2468-2667(19)30155-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honigberg MC, Zekavat SM, Aragam K, Finneran P, Klarin D, Bhatt DL, Januzzi JL, Scott NS, Natarajan P. Association of Premature Natural and Surgical Menopause With Incident Cardiovascular Disease. JAMA. 2019;322:2411–2421. doi: 10.1001/jama.2019.19191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wellons M, Ouyang P, Schreiner PJ, Herrington DM, Vaidya D. Early menopause predicts future coronary heart disease and stroke: the Multi-Ethnic Study of Atherosclerosis. Menopause. 2012;19:1081–1087. doi: 10.1097/gme.0b013e3182517bd0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ley SH, Li Y, Tobias DK, Manson JE, Rosner B, Hu FB, Rexrode KM. Duration of Reproductive Life Span, Age at Menarche, and Age at Menopause Are Associated With Risk of Cardiovascular Disease in Women. J Am Heart Assoc. 2017;6:e006713. doi: 10.1161/JAHA.117.006713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grundy SM, Stone NJ, Bailey AL, Beam C, Birtcher KK, Blumenthal RS, Braun LT, de Ferranti S, Faiella-Tommasino J, Forman D, et al. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA Guideline on the Management of Blood Cholesterol: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation. 2019;139:e1082–e1143. doi: 10.1161/CIR.0000000000000625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnett DK, Blumenthal RS, Albert MA, Buroker AB, Goldberger ZD, Hahn EJ, Himmelfarb CD, Khera A, Lloyd-Janes D, McEvoy JW, et al. 2019 ACC/AHA Guideline on the Primary Prevention of Cardiovascular Disease: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation. 2019;140:e596–e646. doi: 10.1161/CIR.0000000000000678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boardman HM, Hartley L, Eisinga A, Main C, Roque i Figuls M, Cosp XB, Sanchez RG, Knight B. Hormone therapy for preventing cardiovascular disease in post-menopausal women. Cochrane Database Syst Rev. 2015:CD002229. doi: 10.1002/14651858.CD002229.pub4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Day FR, Ruth KS, Thompson DJ, Lunetta KL, Pervjakova N, Chasman DI, Stolk L, Finucane HK, Sulem P, Bulik-Slluivan B, et al. Large-scale genomic analyses link reproductive aging to hypothalamic signaling, breast cancer susceptibility and BRCA1-mediated DNA repair. Nat Genet. 2015;47:1294–1303. doi: 10.1038/ng.3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, Ebert BL. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126:9–16. doi: 10.1182/blood-2015-03-631747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khetarpal SA, Qamar A, Bick AG, Fuster JJ, Kathiresan S, Jaiswal S, Natarajan P. Clonal Hematopoiesis of Indeterminate Potential Reshapes Age-Related CVD: JACC Review Topic of the Week. J Am Coll Cardiol. 2019;74:578–586. doi: 10.1016/j.jacc.2019.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, Chambert K, Mick E, Neale BM, Fromer M, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, McMichael JF, Schmidt HK, Yellapantula V, Miller CA, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20:1472–1478. doi: 10.1038/nm.3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuster JJ, MacLauchlan S, Zuriaga MA, Polackal MN, Ostriker AC, Chakraborty R, Wu CL, Sano S, Muralidharan S, Ruis C, et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science. 2017;355:842–847. doi: 10.1126/science.aag1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meisel M, Hinterleitner R, Pacis A, Chen Li, Earley ZM, Mayassi T, Pierre JF, Ernest JD, Galipeau HJ, Thuille N, et al. Microbial signals drive pre-leukaemic myeloproliferation in a Tet2-deficient host. Nature. 2018;557:580–584. doi: 10.1038/s41586-018-0125-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, McConkey M, Gupta N, Gabriel S, Ardissino D, et al. Clonal Hematopoiesis and Risk of Atherosclerotic Cardiovascular Disease. N Engl J Med. 2017;377:111–121. doi: 10.1056/NEJMoa1701719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bick AG, Pirruccello JP, Griffin GK, Gupta N, Gabriel S, Saleheen D, Libby P, Kathiresan S, Natarajan P. Genetic Interleukin 6 Signaling Deficiency Attenuates Cardiovascular Risk in Clonal Hematopoiesis. Circulation. 2020;141:124–131. doi: 10.1161/CIRCULATIONAHA.119.044362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stefanick ML, Cochrane BB, Hsia J, Barad DH, Liu JH, Johnson SR. The Women’s Health Initiative postmenopausal hormone trials: overview and baseline characteristics of participants. Ann Epidemiol. 2003;13:S78–S86. doi: 10.1016/s1047-2797(03)00045-0 [DOI] [PubMed] [Google Scholar]
- 27.Taliun D, Harris DN, Kessler MD, Carlson J, Szpiech ZA, Torres R, Gagliano Taliun SA, Corvelo A, Gogarten SM, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. bioRxiv. 2019:563866. doi: 10.1101/563866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Hout CV, Tachmazidou I, Backman JD, Hoffman JX, Ye B, Pandey AK, Gonzaga-Jauregui C, Khalid S, Liu D, Banerjee N, et al. Whole exome sequencing and characterization of coding variation in 49,960 individuals in the UK Biobank. bioRxiv. 2019:572347. doi: 10.1101/572347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjamin D, Sato T, Cibulskis K, Getz G, Stewart C, Lichtenstein L. Calling Somatic SNVs and Indels with Mutect2. bioRxiv. 2019:861054. doi: 10.1101/861054 [DOI] [Google Scholar]
- 31.Bick AG, Weinstock JS, Nandakumar SK, Fulco CP, Bao EL, Zekavat SM, Szeto MD, Liao X, Leventhal MJ, Nasser J, et al. Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature. 2020. [epub ahead of print] 10.1038/s41586-020-2819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loh PR, Genovese G, Handsaker RE, Finucane HK, Reshef YA, Palamara PF, Birman BM, Talkowski ME, Bakhoum SF, McCarroll SA, et al. Insights into clonal haematopoiesis from 8,342 mosaic chromosomal alterations. Nature. 2018;559:350–355. doi: 10.1038/s41586-018-0321-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loh PR, Genovese G, McCarroll SA. Monogenic and polygenic inheritance become instruments for clonal selection. Nature. 2020;584:136–141. doi: 10.1038/s41586-020-2430-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blokzijl F, Janssen R, van Boxtel R, Cuppen E. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med. 2018;10:33. doi: 10.1186/s13073-018-0539-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klarin D, Zhu QM, Emdin CA, Chaffin M, Horner S, McMillan BJ, Leed A, Weale ME, Spencer CCA, Aguet F, et al. Genetic analysis in UK Biobank links insulin resistance and transendothelial migration pathways to coronary artery disease. Nat Genet. 2017;49:1392–1397. doi: 10.1038/ng.3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manson JE, Hsia J, Johnson KC, Rossouw JE, Assaf AR, Lasser NL, Trevisan M, Black HR, Heckbert SR, Detrano R, et al. Estrogen plus progestin and the risk of coronary heart disease. N Engl J Med. 2003;349:523–534. doi: 10.1056/NEJMoa030808 [DOI] [PubMed] [Google Scholar]
- 37.Breslow NE, Amorim G, Pettinger MB, Rossouw J. Using the Whole Cohort in the Analysis of Case-Control Data: Application to the Women’s Health Initiative. Stat Biosci. 2013;5. doi: 10.1007/s12561-013-9080-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maura F, Degasperi A, Nadeu F, Leongamornlert D, Davies H, Moore L, Royo R, Ziccheddu B, Puente XS, Avet-Loiseau H, et al. A practical guide for mutational signature analysis in hematological malignancies. Nat Commun. 2019;10:2969. doi: 10.1038/s41467-019-11037-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Ng AWT, Wu Y, Boot A, Covington KR, Gordenin DA, Bergstrom EN, et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578:94–101. doi: 10.1038/s41586-020-1943-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coombs CC, Zehir A, Devlin SM, Kishtagari A, Syed A, Jonsson P, Hyman DM, Solit DB, Robson ME, Baselga J, et al. Therapy-Related Clonal Hematopoiesis in Patients with Non-hematologic Cancers Is Common and Associated with Adverse Clinical Outcomes. Cell Stem Cell. 2017;21:374–382.e4. doi: 10.1016/j.stem.2017.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gondek LP, DeZern AE. Assessing clonal haematopoiesis: clinical burdens and benefits of diagnosing myelodysplastic syndrome precursor states. Lancet Haematol. 2020;7:e73–e81. doi: 10.1016/S2352-3026(19)30211-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sidlow R, Lin AE, Gupta D, Bolton KE, Steensma DP, Levine RL, Ebert BL, Libby P. The Clinical Challenge of Clonal Hematopoiesis, a Newly Recognized Cardiovascular Risk Factor. JAMA Cardiol. 2020;5:958–961. doi: 10.1001/jamacardio.2020.1271 [DOI] [PubMed] [Google Scholar]
- 43.Velez MP, Alvarado BE, Rosendaal N, de Camara SM, Belanger E, Richardson H, Pirkle CM. Age at natural menopause and physical functioning in postmenopausal women: the Canadian Longitudinal Study on Aging. Menopause. 2019;26:958–965. doi: 10.1097/GME.0000000000001362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mierzejewska K, Borkowska S, Suszynska E, Suszynska M, Poniewierska-Baran A, Maj M, Pedziwaitr D, Adamiak M, Abdel-Latif A, Kakar SS, et al. Hematopoietic stem/progenitor cells express several functional sex hormone receptors-novel evidence for a potential developmental link between hematopoiesis and primordial germ cells. Stem Cells Dev. 2015;24:927–937. doi: 10.1089/scd.2014.0546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abdelbaset-Ismail A, Suszynska M, Borkowska S, Adamiak M, Ratajczak J, Kucia M, Ratajczak MZ. Human haematopoietic stem/progenitor cells express several functional sex hormone receptors. J Cell Mol Med. 2016;20:134–146. doi: 10.1111/jcmm.12712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakada D, Oguro H, Levi BP, Ryan N, Kitano A, Saitoh Y, Takeichi M, Wendt GR, Morrison SJ. Oestrogen increases haematopoietic stem-cell self-renewal in females and during pregnancy. Nature. 2014;505:555–558. doi: 10.1038/nature12932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sánchez-Aguilera A, Arranz L, Martín-Pérez D, Garcia-Garcia A, Stavropoulou V, Kubovcakova L, Isern J, Martin-Salamanca S, Langa X, Skoda RC, et al. Estrogen signaling selectively induces apoptosis of hematopoietic progenitors and myeloid neoplasms without harming steady-state hematopoiesis. Cell Stem Cell. 2014;15:791–804. doi: 10.1016/j.stem.2014.11.002 [DOI] [PubMed] [Google Scholar]
- 48.Shim GJ, Wang L, Andersson S, Nagy N, Levente Kis L, Zhang Q, Makela S, Warner M, Gustafsson JA. Disruption of the estrogen receptor beta gene in mice causes myeloproliferative disease resembling chronic myeloid leukemia with lymphoid blast crisis. Proc Natl Acad Sci U S A. 2003;100:6694–6699. doi: 10.1073/pnas.0731830100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zink F, Stacey SN, Norddahl GL, Frigge ML, Magnusson OT, Jonsdottir I, Thorgeirsson TE, Sigurdsson A, Gudjonsson SA, Gudmundsson J, et al. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood. 2017;130:742–752. doi: 10.1182/blood-2017-02-769869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu AT, Xue L, Salfati EL, Chen BH, Ferrucci L, Levy D, Joehanes R, Murabito JM, Kiel DP, Tsai PC, et al. GWAS of epigenetic aging rates in blood reveals a critical role for TERT. Nat Commun. 2018;9:387. doi: 10.1038/s41467-017-02697-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giri N, Batista DL, Alter BP, Stratakis CA. Endocrine abnormalities in patients with Fanconi anemia. J Clin Endocrinol Metab. 2007;92:2624–2631. doi: 10.1210/jc.2007-0135 [DOI] [PubMed] [Google Scholar]
- 52.Rossouw JE, Prentice RL, Manson JE, Wu L, Barad D, Barnabei VM, Ko M, LaCroix AZ, Margolis KL, Stefanik ML. Postmenopausal hormone therapy and risk of cardiovascular disease by age and years since menopause. JAMA. 2007;297:1465–1477. doi: 10.1001/jama.297.13.1465 [DOI] [PubMed] [Google Scholar]
- 53.Kolodkin MH, Auger AP. Sex difference in the expression of DNA methyltransferase 3a in the rat amygdala during development. J Neuroendocrinol. 2011;23:577–583. doi: 10.1111/j.1365-2826.2011.02147.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamagata Y, Asada H, Tamura I, Lee L, Maekawa R, Taniguchi K, Taketani T, Matsuoka A, Tamura H, Sugino N. DNA methyltransferase expression in the human endometrium: down-regulation by progesterone and estrogen. Hum Reprod. 2009;24:1126–1132. doi: 10.1093/humrep/dep015 [DOI] [PubMed] [Google Scholar]
- 55.Li Y, Hamilton KJ, Lai AY, Lai AY, Burns KA, Li L, Wade PA, Korach KS. Diethylstilbestrol (DES)-stimulated hormonal toxicity is mediated by ERα alteration of target gene methylation patterns and epigenetic modifiers (DNMT3A, MBD2, and HDAC2) in the mouse seminal vesicle. Environ Health Perspect. 2014;122:262–268. doi: 10.1289/ehp.1307351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van der Weijden VA, Flöter VL, Ulbrich SE. Gestational oral low-dose estradiol-17β induces altered DNA methylation of CDKN2D and PSAT1 in embryos and adult offspring. Sci Rep. 2018;8:7494. doi: 10.1038/s41598-018-25831-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nugent BM, Wright CL, Shetty AC, Hodes GE, Lenz KM, Mahurkar A, Russo SJ, Devine SE, McCarthy MM. Brain feminization requires active repression of masculinization via DNA methylation. Nat Neurosci. 2015;18:690–697. doi: 10.1038/nn.3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Du J, Zhou N, Liu H, Jiang F, Wang Y, Hu C, Qi H, Zhong C, Wang X, Li Z. Arsenic induces functional re-expression of estrogen receptor α by demethylation of DNA in estrogen receptor-negative human breast cancer. PLoS One. 2012;7:e35957. doi: 10.1371/journal.pone.0035957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sano S, Oshima K, Wang Y, Katanasaka Y, Sano M, Walsh K. CRISPR-Mediated Gene Editing to Assess the Roles of Tet2 and Dnmt3a in Clonal Hematopoiesis and Cardiovascular Disease. Circ Res. 2018;123:335–341. doi: 10.1161/CIRCRESAHA.118.313225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abplanalp WT, Mas-Peiro S, Cremer S, John D, Dimmeler S, Zeiher AM. Association of Clonal Hematopoiesis of Indeterminate Potential With Inflammatory Gene Expression in Patients With Severe Degenerative Aortic Valve Stenosis or Chronic Postischemic Heart Failure. JAMA Cardiol. 2020;5:1–6. doi: 10.1001/jamacardio.2020.2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Okoth K, Chandan JS, Marshall T, Thangaratinam S, Thomas GN, Nirantharakumar K, Adderley NJ. Association between the reproductive health of young women and cardiovascular disease in later life: umbrella review. BMJ. 2020;371:m3502. doi: 10.1136/bmj.m3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roeters van Lennep JE, Heida KY, Bots ML, Hoek A. Cardiovascular disease risk in women with premature ovarian insufficiency: A systematic review and meta-analysis. Eur J Prev Cardiol. 2016;23:178–186. doi: 10.1177/2047487314556004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.