Abstract
A novel genus, Anastomitrabeculia, is introduced herein for a distinct species, Anastomitrabeculia didymospora, collected as a saprobe on dead bamboo culms from a freshwater stream in Thailand. Anastomitrabeculia is distinct in its trabeculate pseudoparaphyses and ascospores with longitudinally striate wall ornamentation. A new family, Anastomitrabeculiaceae, is introduced to accommodate Anastomitrabeculia. Anastomitrabeculiaceae forms an independent lineage basal to Halojulellaceae in Pleosporales and it is closely related to Neohendersoniaceae based on phylogenetic analyses of a combined LSU, SSU and TEF1α dataset. In addition, divergence time estimates provide further support for the establishment of Anastomitrabeculiaceae. The family diverged around 84 million years ago (MYA) during the Cretaceous period, which supports the establishment of the new family. The crown and stem age of Anastomitrabeculiaceae was also compared to morphologically similar pleosporalean families.
Keywords: BEAST, Dothideomycetes, Pleosporales, Poaceae, taxonomy, three new taxa, trabeculate pseudoparaphyses
1. Introduction
Pleosporales is the largest order within Dothideomycetes (Ascomycota) [1]. The taxonomic and phylogenetic relationships of families and genera within this order are well documented [1,2,3,4,5,6,7]. Pleosporales comprises two suborders, Massarineae and Pleosporineae [1]. Pleosporineae includes economically important plant pathogens and Massarineae includes mainly saprobes from terrestrial or aquatic environments [1,3]. Zhang et al. [1] revised 174 genera and accepted 26 families in Pleosporales. The suborder Massarineae was resurrected to accommodate five families, the Lentitheciaceae, Massarinaceae, Montagnulaceae (Didymosphaeriaceae), Morosphaeriaceae and Trematosphaeriaceae [1]. Hyde et al. [2] correlated morphology with phylogenetic evidence and accepted 41 families in this order. Tanaka et al. [3] introduced two new families, Parabambusicolaceae and Sulcatisporaceae, accepting 12 families in Massarineae. The family Longipedicellataceae was introduced, and the divergence time in Pleosporales was estimated with emphasis on Massarineae [4]. The crown age of Pleosporales was dated to 211 MYA and Massarineae was dated to 130 MYA [4]. Species boundaries in Cucurbitariaceae were revised [5] and the family, Lentimurisporaceae, was introduced in Pleosporales [6].
Species in this order are abundant and occur in terrestrial, marine and freshwater habitats [7,8,9]. The species can be epiphytes, endophytes or parasites of living leaves or stems, hyperparasites on fungi or insects, lichenized, or saprobes of dead plant stems, leaves or bark [7,8,9]. Currently, about 400 genera in 64 families are known in Pleosporales [1,2,7,10,11,12,13], with numerous coelomycetous and hyphomycetous taxa as their asexual morphs [1,13,14,15].
Several pleosporalean taxa are pathogens associated with a broad range of hosts including bamboo. Bamboo (Poaceae) comprises over 115 genera with around 1500 species [16,17,18], can be found in diverse climates [17], and are widely distributed in various forest types in Thailand [18,19]. It has been estimated that around 1100 fungal species belonging to over 200 genera have been described or recorded worldwide on bamboo and most of these bamboo-associated fungi are ascomycetes [20,21].
Divergence time estimates using molecular clock methodologies have been widely used in fungal taxonomy [4,11,22,23,24,25,26,27]. Several studies have applied molecular dating to provide additional evidence for higher taxa ranking in Pleosporales [4,6,7,11]. In this study, we introduce a novel bambusicolous species, Anastomitrabeculia didymospora within Anastomitrabeculia, which is accommodated in a new family, Anastomitrabeculiaceae, based on morphology, multi-loci phylogeny and divergence times estimates.
2. Materials and Methods
2.1. Sample Collection, Isolation and Identification
Dead bamboo culms were collected from a freshwater stream from Krabi province, Thailand, in 2015. The samples were incubated in plastic boxes with sterile and moist tissue at 25–30 °C for 3 days. Pure fungal colonies were obtained using single-spore isolation [28]. Germinating spores were transferred aseptically to potato dextrose agar (PDA) and malt extract agar (MEA) (Difco™). The cultures were incubated at 25 °C with frequent observations. Fungal characters were observed using a stereo microscope (Zeiss SteREO Discovery v. 8) fitted with an Axio Cam ERc5S and a Leica DM2500 compound microscope attached with a Leica MC190 HD camera. All microscopic measurements were carried out using Tarosoft (R) Image Frame Work program and the images were processed with Adobe Photoshop CS6 version 13.0 software (Adobe Systems, San Jose, CA, USA). The type specimens were deposited in the Mae Fah Luang University (MFLU) Herbarium, Chiang Rai, Thailand, and pure cultures were deposited at the Mae Fah Luang University Culture Collection (MFLUCC). The new taxon was linked with Facesoffungi numbers (FoF) [29] and Index Fungorum (Index Fungorum 2020, http://www.indexfungorum.org/, accessed on 2 December 2020) and established based on guidelines recommended by Jeewon and Hyde [30].
2.2. DNA Extraction, PCR Amplification and DNA Sequencing
DNA extraction, PCR amplification, DNA sequencing and phylogenetic analysis were carried out as detailed in Dissanayake et al. [31]. Total genomic DNA was extracted from fresh mycelium with a Biospin Fungus Genomic DNA Extraction Kit (BioFlux®) (Hangzhou, P.R. China) following the manufacturer’s protocol. The nuclear ribosomal large subunit 28S rRNA gene (LSU) [32], the nuclear ribosomal small subunit 18S rRNA gene (SSU) [33] and the translation elongation factor 1-alpha gene (TEF1α) [34] were amplified using primers (LSU: LROR/LR5, SSU: NS1/NS4 and TEF1α: 983F/2218R). Polymerase chain reaction (PCR) was performed using PCR mixtures containing 5–10 ng DNA, 1X PCR buffer, 0.8 units Taq polymerase, 0.3 μM of each primer, 0.2 mM dNTP and 1.5 mM MgCl2. PCR conditions were set at an initial denaturation for 3 min at 94 °C, followed by 40 cycles of 45 s of denaturation at 94 °C, annealing for 50 s at 56 °C for LSU, SSU and 52 °C for TEF1α and extension for 1 min at 72 °C, with a final extension of 10 min at 72 °C. All the PCR products were visualised on 1% Agarose gels with added 6 μL of 4S green dyes, per each 100 mL. Successful PCR products were purified and sequencing was performed by Shanghai Sangon Biological Engineering Technology & Services Co. (Shanghai, P.R. China). All sequences generated in this study were submitted to GenBank (Table 1) and the ITS region of Anastomitrabeculia didymospora was deposited with the accession number MW413900 (MFLUCC 16-0412) and MW413897 (MFLUCC 16-0417).
Table 1.
DNA sequences and GenBank numbers used for the phylogenetic analyses in this study. The ex-type strains are in bold and the new taxon introduced in this study is indicated in blue.
| Taxon | Strain Number | GenBank Accession Numbers | ||
|---|---|---|---|---|
| LSU | SSU | TEF1α | ||
| Acrocalymma aquatica | MFLUCC 11-0208 | JX276952 | JX276953 | - |
| Acrocalymma fici | CBS 317.76 | KP170712 | - | - |
| Acrocalymma medicaginis | CPC 24340 | KP170713 | - | - |
| Acrocalymma medicaginis | CPC 24341 | KP170714 | - | - |
| Acrocalymma medicaginis | CPC 24345 | KP170718 | - | - |
| Acrocalymma pterocarpi | MFLUCC 17-0926 | MK347949 | MK347840 | - |
| Aigialus grandis | BCC 20000 | GU479775 | GU479739 | GU479839 |
| Aigialus mangrovis | BCC 33563 | GU479776 | GU479741 | GU479840 |
| Aigialus parvus | BCC 18403 | GU479778 | GU479743 | GU479842 |
| Aigialus rhizophorae | BCC 33572 | GU479780 | GU479745 | GU479844 |
| Aliquandostipite khaoyaiensis | CBS 118232 | GU301796 | - | GU349048 |
| Amniculicola immersa | CBS 123083 | FJ795498 | GU456295 | GU456273 |
| Amniculicola lignicola | CBS 123094 | EF493861 | EF493863 | - |
| Amniculicola parva | CBS 123092 | GU301797 | GU296134 | GU349065 |
| Amorosia littoralis | NN 6654 | AM292055 | AM292056 | - |
| Anastomitrabeculia didymospora | MFLUCC 16-0412 | MW412978 | MW412977 | MW411338 |
| Anastomitrabeculia didymospora | MFLUCC 16-0417 | MW413899 | MW413898 | MW411339 |
| Angustimassarina populi | MFLUCC 13-0034 | KP888642 | KP899128 | KR075164 |
| Angustimassarina quercicola | MFLUCC 14-0506 | KP888638 | KP899124 | KR075169 |
| Anteaglonium abbreviatum | ANM 925a | GQ221877 | - | - |
| Anteaglonium globosum | SMH 5283 | GQ221911 | - | GQ221919 |
| Anteaglonium parvulum | MFLUCC 14-0821 | KU922915 | KU922916 | - |
| Antealophiotrema brunneosporum | CBS 123095 | LC194340 | LC194298 | LC194382 |
| Aquasubmersa japonica | HHUF 30468 | LC061586 | LC061581 | - |
| Aquasubmersa japonica | HHUF 30469 | LC061587 | LC061582 | - |
| Aquasubmersa mircensis | MFLUCC 11-0401 | JX276955 | JX276956 | - |
| Arthonia dispersa | UPSC 2583 | AY571381 | AY571379 | - |
| Ascocratera manglicola | BCC 09270 | GU479782 | GU479747 | GU479846 |
| Ascocylindrica marina | MD6011 | KT252905 | KT252907 | - |
| Ascocylindrica marina | MD6012 | KT252906 | - | - |
| Ascocylindrica marina | MF416 | MK007123 | MK007124 | - |
| Bahusandhika indica | GUFCC 18001 | KF460274 | - | - |
| Bambusicola massarinia | MFLUCC 11-0389 | JX442037 | JX442041 | - |
| Berkleasmium micronesicum | BCC 8141 | DQ280272 | DQ280268 | - |
| Berkleasmium nigroapicale | BCC 8220 | DQ280273 | DQ280269 | - |
| Bimuria novae-zelandiae | CBS 107.79 | AY016356 | AY016338 | DQ471087 |
| Botryosphaeria dothidea | CBS 115476 | AY928047 | EU673173 | AY236898 |
| Brevicollum hyalosporum | MAFF 243400 | LC271239 | LC271236 | LC271245 |
| Brevicollum hyalosporum | MFLUCC 17-0071 | MG602200 | MG602202 | MG739516 |
| Brevicollum hyalosporum | PUFNI 17628 | MH918671 | - | - |
| Brevicollum versicolor | HHUF 30591 | LC271240 | LC271237 | LC271246 |
| Capnodium salicinum | CBS 131.34 | DQ678050 | DQ677997 | - |
| Cladosporium cladosporioides | CBS 170.54 | DQ678057 | DQ678004 | - |
| Clematidis italica | MFLUCC 15-0084 | KU842381 | KU842382 | - |
| Corynespora cassiicola | CBS 100822 | GU301808 | GU296144 | GU349052 |
| Corynespora smithii | CABI 5649b | GU323201 | - | GU349018 |
| Crassiparies quadrisporus | HHUF 30590 | LC271241 | LC271238 | LC271248 |
| Crassiparies quadrisporus | HHUF 30409 | LC100025 | LC100017 | - |
| Crassiperidium octosporum | KT 2144 | LC373108 | LC373084 | LC373120 |
| Crassiperidium octosporum | KT 2894 | LC373109 | LC373085 | LC373121 |
| Crassiperidium octosporum | KT 3008 | LC373110 | LC373086 | LC373122 |
| Crassiperidium octosporum | KT 3029 | LC373111 | LC373087 | LC373123 |
| Crassiperidium octosporum | KT 3046 | LC373112 | LC373088 | LC373124 |
| Crassiperidium octosporum | KT 3188 | LC373113 | LC373089 | LC373125 |
| Crassiperidium octosporum | KT 3468 | LC373114 | LC373090 | LC373126 |
| Crassiperidium octosporum | KT 3604 | LC373115 | LC373091 | LC373127 |
| Crassiperidium octosporum | KT 3605 | LC373116 | LC373092 | LC373128 |
| Crassiperidium octosporum | MM 9 | LC373117 | LC373093 | LC373129 |
| Crassiperidium quadrisporum | KT 27981 | LC373118 | LC373094 | LC373130 |
| Crassiperidium quadrisporum | KT 27982 | LC373119 | LC373095 | LC373131 |
| Cryptoclypeus oxysporus | HHUF 30507 | LC194345 | LC194303 | LC194390 |
| Cryptocoryneum akitaense | MAFF 245365 | LC194348 | LC194306 | LC096136 |
| Cryptocoryneum japonicum | MAFF 245370 | LC194356 | LC194314 | LC096144 |
| Cryptocoryneum longicondensatum | MAFF 245374 | LC194360 | LC194318 | LC096148 |
| Cyclothyriella rubronotata | CBS 141486 | KX650544 | KX650507 | KX650519 |
| Cyclothyriella rubronotata | CBS 121892 | KX650541 | - | KX650516 |
| Cyclothyriella rubronotata | CBS 385.39 | MH867543 | - | - |
| Cyclothyriella rubronotata | CBS 419 85 | GU301875 | - | GU349002 |
| Delitschia didyma | UME 31411 | DQ384090 | AF242264 | - |
| Delitschia winteri | CBS 225.62 | DQ678077 | DQ678026 | DQ677922 |
| Dendrographa decolorans | Ertz 5003 | AY548815 | AY548809 | - |
| Dendrographa leucophaea f. minor | AF279382 | AF279381 | - | |
| Dendryphion europaeum | CPC 22943 | KJ869203 | - | - |
| Dendryphion europaeum | CPC 23231 | NG_059120 | - | - |
| Dendryphion nanum | MFLUCC 16-0975 | MG208132 | - | MG207983 |
| Didymosphaeria rubi-ulmifolii | MFLUCC 14-0023 | KJ436586 | KJ436588 | - |
| Dissoconium aciculare | CBS 204.89 | GU214419 | GU214523 | - |
| Ernakulamia cochinensis | PRC 3992 | LT964670 | - | - |
| Flavomyces fulophazii | CBS 135761 | KP184040 | KP184082 | - |
| Fuscostagonospora cytisi | MFLUCC 16-0622 | KY770978 | KY770977 | KY770979 |
| Fuscostagonospora sasae | CBS 139687 | AB807548 | AB797258 | - |
| Fusculina eucalyptorum | CBS 145083 | MK047499 | - | - |
| Gordonomyces mucovaginatus | CBS 127273 | JN712552 | ||
| Halojulella avicenniae | JK 5326A | GU479790 | GU479756 | - |
| Halojulella avicenniae | BCC 20173 | GU371822 | GU371830 | GU371815 |
| Halojulella avicenniae | PUFD542 | MK026757 | MK026754 | - |
| Halojulella avicenniae | BCC 18422 | GU371823 | GU371831 | GU371816 |
| Halojulella avicenniae | BCC28357 | KC555567 | KC555565 | - |
| Halojulella avicenniae | GR00584 | KC555568 | KC555566 | - |
| Halotthia posidoniae | BBH 22481 | GU479786 | GU479752 | - |
| Helminthosporium aquaticum | MFLUCC 15-0357 | KU697306 | KU697310 | - |
| Helminthosporium velutinum | MAFF 243854 | AB807530 | AB797240 | - |
| Helminthosporium velutinum | MFLUCC 13-0243 | KU697305 | - | - |
| Helminthosporium velutinum | MFLUCC 15-0423 | KU697304 | - | - |
| Hermatomyces iriomotensis | HHUF 30518 | LC194367 | LC194325 | LC194394 |
| Hermatomyces tectonae | MFLUCC 14-1140 | KU764695 | KU712465 | KU872757 |
| Hermatomyces thailandica | MFLUCC 14-1143 | KU764692 | KU712468 | KU872754 |
| Hobus wogradensis | TI | KX650546 | KX650508 | KX650521 |
| Hysterium angustatum | CBS 236.34 | FJ161180 | GU397359 | FJ161096 |
| Hysterium angustatum | MFLUCC 16-0623 | MH535893 | MH535885 | MH535878 |
| Jahnula seychellensis | SS2113 | EF175665 | EF175643 | - |
| Latorua caligans | CBS 576.65 | KR873266 | - | - |
| Latorua grootfonteinensis | CBS 369.72 | KR873267 | - | - |
| Lentimurispora urniformis | MFLUCC 18-0497 | MH179144 | MH179160 | MH188055 |
| Leptosphaeria doliolum | CBS 505.75 | GQ387576 | GQ387515 | GU349069 |
| Leptoxyphium cacuminum | MFLUCC 10-0049 | JN832602 | JN832587 | - |
| Leucaenicola phraeana | MFLUCC 18-0472 | MK348003 | MK347892 | - |
| Lignosphaeria fusispora | MFLUCC 11-0377 | KP888646 | - | - |
| Lignosphaeria thailandica | MFLUCC 11-0376 | KP888645 | - | - |
| Lindgomyces ingoldianus | ATCC 200398 | AB521736 | AB521719 | - |
| Longiostiolum tectonae | MFLUCC 12 0562 | KU764700 | KU712459 | - |
| Lophiotrema eburnoides | HHUF 30079 | LC001707 | LC001706 | - |
| Lophiotrema nucula | CBS 627.86 | GU301837 | GU296167 | GU349073 |
| Macrodiplodiopsis desmazieri | CBS 140062 | KR873272 | - | - |
| Magnicamarosporium diospyricola | MFLUCC 16-0419 | KY554212 | KY554211 | KY554209 |
| Massarina eburnea | CBS 473.64 | GU301840 | GU296170 | - |
| Massariosphaeria phaeospora | CBS 611.86 | GU301843 | GU296173 | - |
| Mauritiana rhizophorae | BCC 28866 | GU371824 | GU371832 | GU371817 |
| Medicopsis romeroi | CBS 122784 | EU754208 | EU754109 | KF015679 |
| Medicopsis romeroi | CBS 252.60 | EU754207 | EU754108 | KF015678 |
| Medicopsis romeroi | CBS 132878 | KF015622 | KF015648 | KF015682 |
| Murispora rubicunda | IFRD 2017 | FJ795507 | GU456308 | GU456289 |
| Neoastrosphaeriella krabiensis | MFLUCC 11-0025 | JN846729 | JN846739 | - |
| Neohendersonia kickxii | CBS 112403 | KX820266 | - | - |
| Neohendersonia kickxii | CBS 122938 | KX820268 | - | - |
| Neohendersonia kickxii | CBS 114276 | KX820267 | - | - |
| Neohendersonia kickxii | CPC 24865 | KX820270 | - | - |
| Neohendersonia kickxii | CBS 122941 | KX820269 | - | - |
| Neomassaria fabacearum | MFLUCC 16-1875 | KX524145 | KX524147 | KX524149 |
| Neomassaria formosana | NTUCC 17-007 | MH714756 | MH714759 | MH714762 |
| Neomassarina chromolaenae | MFLUCC 17-1480 | MT214466 | MT214419 | MT235785 |
| Neomassarina pandanicola | MFLUCC 16-0270 | MG298945 | - | MG298947 |
| Neomassarina thailandica | MFLUCC 10-0552 | KX672157 | KX672160 | KX672163 |
| Neomassarina thailandica | MFLUCC 17-1432 | MT214467 | MT214420 | MT235786 |
| Neotorula aquatica | MFLUCC 150342 | KU500576 | KU500583 | - |
| Neotorula submersa | KUMCC 15-0280 | KX789217 | - | - |
| Occultibambusa bambusae | MFLUCC 13-0855 | KU863112 | KU872116 | - |
| Occultibambusa pustula | MFLUCC 11-0502 | KU863115 | KU872118 | - |
| Ohleria modesta | MGC | KX650562 | - | KX650533 |
| Ohleria modesta | CBS 141480 | KX650563 | KX650513 | KX650534 |
| Paradictyoarthrinium diffractum | MFLUCC 13-0466 | KP744498 | KP753960 | - |
| Paradictyoarthrinium diffractum | MFLUCC 12-0557 | KP744497 | - | - |
| Paradictyoarthrinium hydei | MFLUCC 13-0465 | MG747497 | - | - |
| Paradictyoarthrinium tectonicola | MFLUCC 13-0465 | KP744500 | KP753961 | - |
| Paradictyoarthrinium tectonicola | MFLUCC 12-0556 | KP744499 | - | - |
| Periconia thailandica | MFLUCC 17-0065 | KY753888 | KY753889 | - |
| Phaeoseptum aquaticum | CBS 123113 | JN644072 | - | - |
| Phaeoseptum terricola | MFLUCC 10-0102 | MH105779 | MH105780 | MH105781 |
| Phyllosticta capitalensis | CBS 226.77 | KF206289 | KF766300 | - |
| Piedraia hortae | CBS 480.64 | GU214466 | - | - |
| Polyplosphaeria fusca | CBS 125425 | AB524607 | AB524466 | AB524822 |
| Preussia lignicola | CBS 363.69 | DQ384098 | DQ384087 | - |
| Preussia lignicola | CBS 264.69 | GU301872 | GU296197 | GU349027 |
| Pseudoastrosphaeriella bambusae | MFLUCC 11-0205 | KT955475 | KT955455 | KT955437 |
| Pseudoastrosphaeriella longicolla | MFLUCC 11-0171 | KT955476 | KT955456 | KT955438 |
| Pseudoastrosphaeriella thailandensis | MFLUCC 10-0553 | KT955477 | KT955456 | KT955439 |
| Pseudolophiotrema elymicola | HHUF 28984 | LC194381 | LC194339 | LC194418 |
| Pseudomassariosphaeria bromicola | MFLUCC 15-0031 | KT305994 | KT305996 | KT305999 |
| Pseudotetraploa curviappendiculata | CBS 125426 | AB524610 | AB524469 | AB524825 |
| Quadricrura septentrionalis | CBS 125428 | AB524617 | AB524476 | AB524832 |
| Racodium rupestre | L346 | EU048583 | EU048575 | - |
| Racodium rupestre | L424 | EU048582 | EU048577 | - |
| Ramusculicola thailandica | MFLUCC 13-0284 | KP888647 | KP899131 | KR075167 |
| Rimora mangrovei | JK 5246A | GU301868 | GU296193 | |
| Roccella fuciformis | Tehler 8171 | FJ638979 | - | - |
| Rostriconidium aquaticum | KUMCC 15-0297 | MG208144 | - | MG207995 |
| Rostriconidium aquaticum | MFLUCC 16-1113 | MG208143 | - | MG207994 |
| Salsuginea ramicola | KT 2597.1 | GU479800 | GU479767 | GU479861 |
| Salsuginea ramicola | CBS 125781 | MH877872 | - | - |
| Scorias spongiosa | CBS 325.33 | MH866910 | GU214696 | - |
| Seriascoma didymospora | MFLUCC 11-0179 | KU863116 | KU872119 | - |
| Sigarispora arundinis | JCM 13550 | AB618998 | AB618679 | LC001737 |
| Sigarispora ravennica | MFLUCC 14-0005 | KP698414 | KP698415 | - |
| Splanchnonema platani | CBS 222.37 | KR909316 | KR909318 | KR909319 |
| Sporidesmioides thailandica | KUMCC 16-0012 | KX437758 | KX437760 | KX437767 |
| Sporidesmioides thailandica | MFLUCC 13-0840 | NG_059703 | NG_061242 | KX437766 |
| Sporormia fimetaria | UPS:Dissing Gr.81.194 | GQ203729 | - | - |
| Sporormiella minima | CBS 52450 | DQ468046 | - | DQ468003 |
| Stagonospora pseudocaricis | CBS 135132 | KF251762 | KF251259 | KF252741 |
| Stemphylium vesicarium | CBS 191.86 | DQ247804 | DQ247812 | DQ471090 |
| Stemphylium vesicarium | CBS 714.68 | DQ678049 | DQ767648 | DQ677888 |
| Sulcatispora acerina | KT2982 | LC014610 | LC014605 | LC014615 |
| Sulcosporium thailandicum | MFLUCC 12-0004 | KT426563 | KT426564 | - |
| Teichospora quercus | CBS 143396 | MH107966 | - | MH108030 |
| Tetraplosphaeria sasicola | KT 563 | AB524631 | AB524490 | AB524838 |
| Torula gaodangensis | MFLUCC 17-0234 | NG_059827 | NG_063641 | - |
| Torula herbarum | CBS 111855 | KF443386 | KF443391 | KF443403 |
| Triplosphaeria maxima | MAFF 239682 | AB524637 | AB524496 | - |
| Tubeufia chiangmaiensis | MFLUCC 11-0514 | KF301538 | KF301543 | KF301557 |
| Tubeufia javanica | MFLUCC 12-0545 | KJ880036 | KJ880035 | KJ880037 |
| Vargamyces aquaticus | CBS 639.63 | KY853539 | - | - |
| Vargamyces aquaticus | HKUCC 10830 | DQ408575 | - | - |
| Versicolorisporium triseptatum | HHUF 28815 | AB330081 | AB524501 | - |
| Westerdykella dispersa | CBS 297.56 | MH869191 | - | - |
| Westerdykella ornata | CBS 379.55 | GU301880 | GU296208 | GU349021 |
| Xenomassariosphaeria rosae | MFLUCC 15-0179 | MG829092 | MG829192 | - |
2.3. Phylogenetic Analysis
The sequence data were assembled using BioEdit v. 7.2.5 [35] and subjected to a BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to find the closest matches with taxa in Pleosporales. Reference sequence data of this order and some representatives of other orders of Dothideomycetes were downloaded from previously published studies [1,6,36,37,38,39]. The sequences were automatically aligned using default settings in MAFFT v. 7 (http://mafft.cbrc.jp/alignment/server/) [40]. A combined dataset of three gene regions (LSU, SSU and TEF1α) was prepared and manually adjusted using BioEdit and AliView [41]. Phylogenetic analyses of the combined dataset were performed using maximum likelihood, maximum parsimony and Bayesian inference method. Maximum likelihood analyses (ML), including 1000 bootstrap pseudoreplicates, were performed at the CIPRES web portal [42] using RAxML v. 8.2.12 [43]. Maximum parsimony analysis was conducted using PAUP v.4.0b 10 [44] with the heuristic search option and number of replications 1000 each. The Tree Length (TL), Consistency Indices (CI), Retention Indices (RI), Rescaled Consistency Indices (RC) and Homoplasy Index (HI) were documented.
The best model for different genes partition was determined in JModelTest version 2.1.10 [45] for posterior probability (PP). The general time reversible (GTR) model with a discrete gamma distribution plus invariant site (GTR+I+G) substitution model was used for the combined dataset. Posterior probabilities [46] were estimated by Markov Chain Monte Carlo sampling (MCMC) in MrBayes v. 3.2.6 [47]. Four simultaneous Markov chains were run for 10 million generations and trees were sampled every 1000th generation, thus resulting in 10,000 trees. The suitable burn-in phase was determined by inspecting traces in Tracer version 1.7 [48]. The first 10% of generated trees representing the burn-in phase of the analyses were discarded, while the remaining trees were used to calculate posterior probabilities (PP) in the majority rule consensus tree. The phylograms were visualized with FigTree v1.4.0 program [49] and edited using Adobe Illustrator CS6 v15.0 (Adobe Systems, USA).
2.4. Fossil Calibration and Divergence Time Estimates
Divergence times were estimated with BEAST 2.6.2 [50] based on the methodology described in Phukhamsakda et al. [4]. The aligned sequence dataset (LSU, SSU and TEF1α) used for the phylogenetic analyses were loaded into BEAUTI 2.6.2 to prepare the XML file. Nucleotide substitution models were determined using JModelTest version 2.1.10. The GTR+I+G nucleotide substitution model was applied to LSU and TEF1α partitions. The symmetrical (SYM) model with a discrete gamma distribution plus invariant site (SYM+I+G) substitution model was applied to the SSU partition. The data partitions were set with unlinked substitution, linked clock model and linked tree. An uncorrelated relaxed clock model with lognormal distribution was used. The Yule speciation process, which assumes a constant rate of speciation divergence, was used as the tree prior [51]. The analysis was performed in BEAST 2.6.2 for 100 million generations, sampling every 1000 generations. The effective sample size (ESS) was analysed with Tracer version 1.7 to check that the values were greater than 200, as recommended by Drummond et al. [52]. The first 20% trees were discarded as the burn-in phase and the remaining trees were combined in LogCombiner 2.6.2. The maximum clade credibility was calculated in TreeAnnotator v 2.6.2. The phylograms were visualized with FigTree v.1.4.0 program.
To estimate the divergence time for Anastomitrabeculiaceae, the fossil Metacapnodium succinum (Metacapnodiaceae) was used to set the crown age of Capnodiales using a normal distribution, mean of 100 MYA, SD of 150 MYA, giving 95% credibility interval of 346 MYA [4,23,53,54]. The fossil Margaretbarromyces dictyosporus was used to calibrate the crown age of Aigialus (Aigialaceae) using a gamma distribution, with an offset of 35 MYA, a shape of 1.0, scale of 25, providing 95% credibility interval of 110 MYA [4,55,56,57]. The split between Arthoniomycetes (outgroup) and Dothideomycetes was used as the secondary calibration using a normal distribution, mean of 300 MYA, SD of 50 MYA, giving 95% credibility interval of 382 MYA [22,36,53,54].
3. Results
3.1. Phylogenetic Analyses
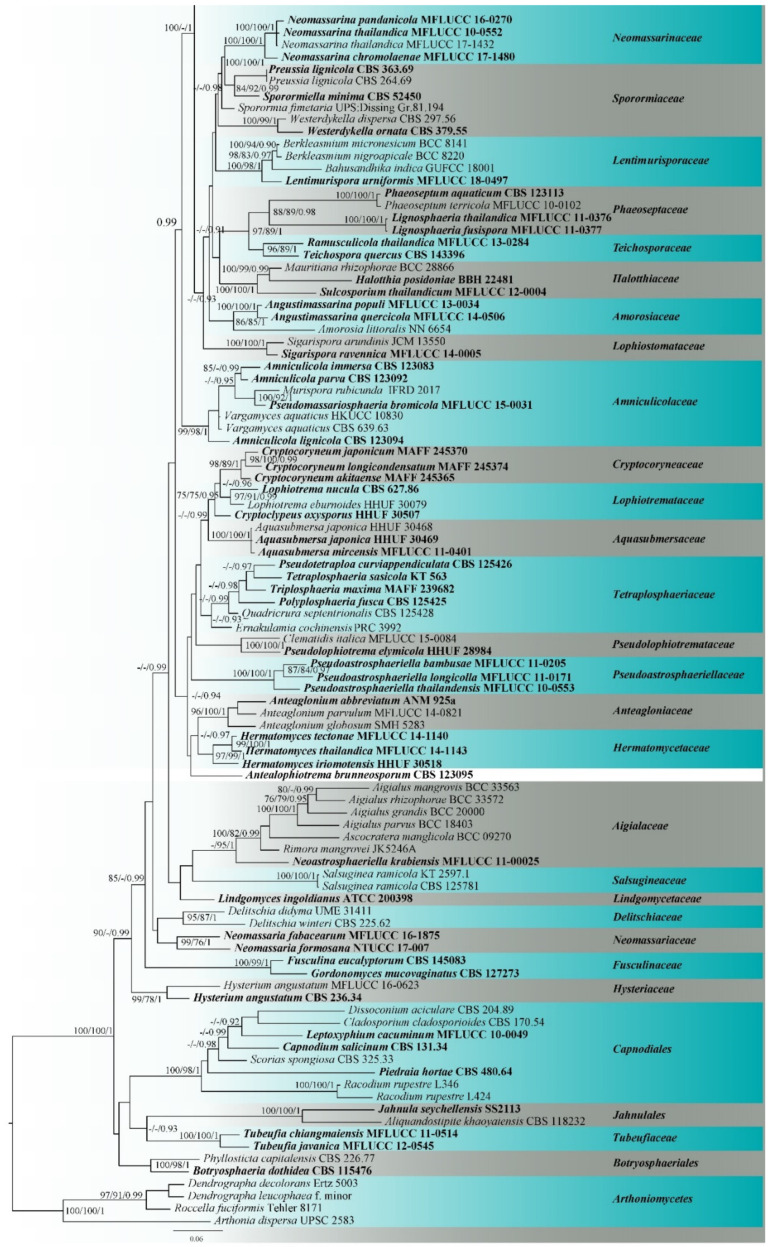
The combined gene alignment comprised 196 strains and 2800 characters (LSU: 860 characters, SSU: 1039 characters and TEF1α: 901 characters). Among the 2800 characters, there were 1492 conserved sites (53%), 364 variable sites (13%) and 944 parsimony informative sites (34%). The parsimony analysis of the data matrix yielded one most parsimonious tree out of 1000 (CI = 0.265, RI = 0.659, RC = 0.175, HI = 0.735, Tree Length = 7606). Based on BLAST search in the NCBI GenBank of the LSU gene, the newly generated taxon MFLUCC 16-0412 and MFLUCC 16-0417 show 95% similarity to Crassiperidium quadrisporum (KT 27981 and KT 27982). The topology of the phylogenetic tree based on the LSU gene was generally congruent with the overall topology of the tree based on the combined dataset. Phylogenetic trees generated from maximum likelihood, maximum parsimony and Bayesian analysis of the combined dataset resulted in similar topologies with some exception. The position of Cyclothyriellaceae and Longiostiolaceae differed between the three methods. The best scoring RAxML tree had a final likelihood value of −40,523.297855 (Figure 1). The new taxon formed an independent lineage basal to the Halojulellaceae with strong Bayesian inference support and moderate support from maximum likelihood (0.99 PP/65% MLBT). A new genus Anastomitrabeculia is therefore introduced within Anastomitrabeculiaceae to accommodate the new species.
Figure 1.
The best scoring RAxML tree based on a combined LSU, SSU and TEF1α dataset. RAxML bootstrap support and maximum parsimony values ≥60% (BT), as well as Bayesian posterior probabilities ≥0.90 (BYPP) are shown, respectively, near the nodes. The ex-type strains are in bold and the scale bar indicates 0.06 changes per site. The tree is rooted with species of Arthoniomycetes and the new taxon is indicated in blue.
3.2. Fossil Calibration and Divergence Time Estimates
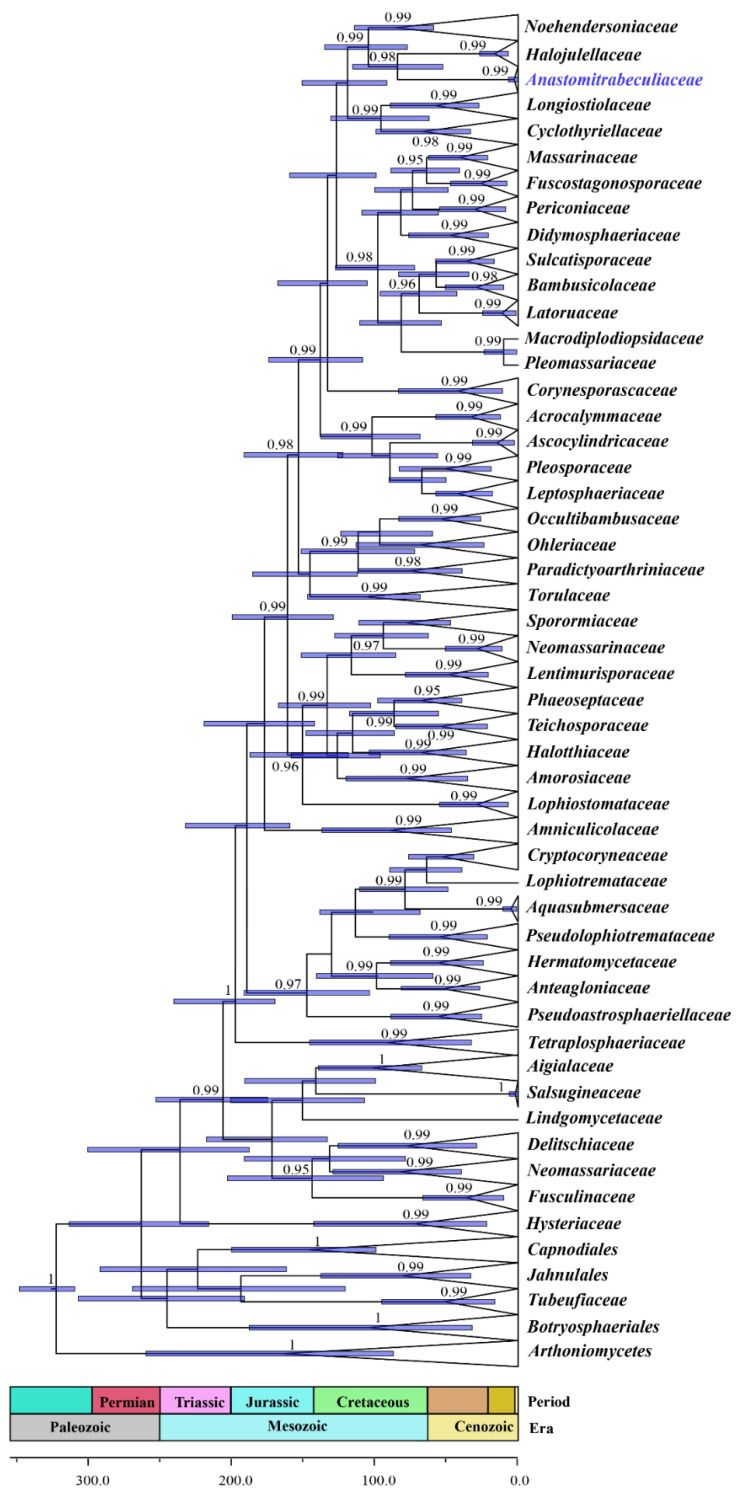
The topology of the maximum clade credibility (MCC) tree (Figure 2) was congruent with the tree obtained from the Bayesian inference analysis and the maximum likelihood analysis. The divergence times of the dating analysis are listed in Table 2. The crown age of Dothideomycetes is estimated at 263 MYA during the Permian period based on the MCC tree. The split of Arthoniomycetes and Dothideomycetes occurred around 323 MYA during the Carboniferous period. The crown age of Pleosporales is estimated at 206 MYA, and Hysteriales diverged from Pleosporales approximately 236 MYA during the Triassic period. The crown age of Anastomitrabeculiaceae is estimated at around 2.6 MYA, and it diverged from Halojulellaceae at around 84 (52–116) MYA. Anastomitrabeculiaceae formed an independent lineage with close relationship to Halojulellaceae with strong posterior probability in the MCC tree (0.99 BYPP). The divergence time of Anastomitrabeculiaceae was compared to Pleosporalean families with trabeculate pseudoparaphyses, cylindrical asci and ascospores with a sheath (Table 3). The divergence time of Anastomitrabeculiaceae was also compared to Didymosphaeriaceae as they are morphologically similar by having trabeculate pseudoparaphyses and cylindrical asci.
Figure 2.
Maximum clade credibility (MCC) tree of families in Dothideomycetes using BEAST. Numbers at nodes indicate posterior probabilities (PP) for node support. Bars correspond to the 95% highest posterior density (HPD) intervals. Posterior probabilities greater than 0.95 are given near the nodes. The new taxon is indicated in blue. Geological time scales are given at the base together with scale in million years ago (MYA) [58].
Table 2.
Divergence time estimates obtained from BEAST analysis. The median and the 95% Highest Posterior Density are provided in million years ago (MYA). The geological time scales are given based on the median node age.
| Nodes | Node Age | Geological Time Period |
|---|---|---|
| Arthoniomycetes–Dothideomycetes | 323 (310–349) | Carboniferous |
| Dothideomycetes crown group | 263 (216–313) | Permian |
| Hysteriales–Pleosporales | 236 (188–300) | Triassic |
| Pleosporales crown group | 206 (171–254) | Triassic |
| Capnodiales crown group | 147 (99–200) | Jurassic |
| Anastomitrabeculiaceae stem group | 84 (52–116) | Cretaceous |
| Aigialaceae–Aigialus sp. | 37 (18–56) | Eocene |
| Anastomitrabeculiaceae crown group | 2.6 (0.19–6.61) | Neogene |
Table 3.
Divergence time estimates obtained from BEAST analysis for families with similar morphology to Anastomitrabeculiaceae. The crown age and the stem age are provided in million years ago (MYA).
| Families | Crown Age | Stem Age |
|---|---|---|
| Aigialaceae | 102 | 141 |
| Amniculicolaceae | 90 | 177 |
| Anastomitrabeculiaceae | 2.6 | 84 |
| Anteagloniaceae | 52 | 98 |
| Bambusicolaceae | 29 | 57 |
| Cyclothyriellaceae | 66 | 95 |
| Delitschiaceae | 78 | 131 |
| Didymosphaeriaceae | 47 | 81 |
| Fuscostagonosporaceae | 26 | 63 |
| Lindgomycetaceae | 31 | 92 |
| Neomassariaceae | 82 | 131 |
| Pseudoastrosphaeriellaceae | 56 | 147 |
| Tetraplosphaeriaceae | 91 | 189 |
3.3. Taxonomy
Anastomitrabeculiaceae Bhunjun, Phukhams and K.D. Hyde, fam. nov.
Index Fungorum number: IF556817, Facesoffungi number: FoF 09521.
Etymology: Referring to the name of the type genus.
Saprobic on dead bamboo culms submerged in freshwater. Sexual morph: Ascomata immersed under a clypeus to semi-immersed, gregarious, uniloculate, globose to subglobose, carbonaceous, black. Ostiole central, apex well developed. Peridium multi-layered, sub-carbonaceous or coriaceous, with dark brown to hyaline cells arranged in a textura angularis. Hamathecium composed of numerous, filamentous, trabeculate pseudoparaphyses, septate, anastomosing between the asci and at the apex. Asci bitunicate, fissitunicate, broad cylindrical to cylindrical-clavate, bulbous pedicel, with an ocular chamber. Ascospores biseriate, broadly fusiform, septate, smooth-walled, with wall ornamentation, surrounded by mucilaginous sheath.
Note: Anastomitrabeculiaceae is introduced to include Anastomitrabeculia, which is reported as a saprobe on bamboo culms. Anastomitrabeculiaceae is characterised by semi-immersed, coriaceous or carbonaceous ascomata with septate, trabeculate pseudoparaphyses and hyaline ascospores with longitudinally striate wall ornamentation, surrounded by mucilaginous sheath. Anastomitrabeculiaceae formed a well-supported independent lineage closely related to Halojulellaceae, but Halojulellaceae differs by its cellular pseudoparaphyses and golden-brown ascospores.
Type genus: Anastomitrabeculia Bhunjun, Phukhams and K.D. Hyde.
Anastomitrabeculia Bhunjun, Phukhams. and K.D. Hyde, gen. nov.
Index Fungorum number: IF556560, Facesoffungi number: FoF 09522.
Etymology: Referring to the trabeculate pseudoparaphyses anastomosing between the asci and at the apex.
Colonies on natural substrate umbonate at the centre, circular, black shiny dots are visible on the host surface. Ascomata on surface of the host, immersed under a clypeus, gregarious, uniloculate, subglobose, carbonaceous. Ostiole orange pigment near ostiole. Peridium comprising multilayers of brown to hyaline cells of textura angularis, inner layers composed of thin, hyaline cells. Asci 8–spored, bitunicate, fissitunicate, broad cylindrical to cylindrical-clavate, with a bulbous pedicellate, rounded at the apex, with an ocular chamber. Ascospores biseriate, broadly fusiform, tapering towards the ends, hyaline, with guttules in each cell, constricted at the septa, with longitudinally striate wall ornamentation, surrounded by mucilaginous sheath.
Note: Anastomitrabeculia is established as a monotypic genus. It is characterised by the presence of carbonaceous ascomata, with orange pigment near ostiole and ascospores with longitudinally striate wall ornamentation. Anastomitrabeculia is morphologically similar to members of Pleosporales in having perithecioid ascomata, bitunicate asci and hyaline ascospores.
Type species: Anastomitrabeculia didymospora Bhunjun, Phukhams and K.D. Hyde.
Anastomitrabeculia didymospora Bhunjun, Phukhams and K.D. Hyde, sp. nov.
Index Fungorum number: IF556559; Facesoffungi number: FoF 09523 Figure 3.
Etymology: Referring to the didymosporous ascospores.
Holotype–MFLU 20-0694.
Figure 3.
Anastomitrabeculia didymospora (MFLU 20-0694, holotype). (a) Ascomata on bamboo. (b) Close-up of ascomata. (c) Vertical section of ascoma. (d) Ostiolar canal. (e) Peridium layer. (f) Trabeculate pseudoparaphyses. (g–i) Asci. (j) Pedicel. (k–o) Ascospores showing mucilaginous sheath. (p) Culture characteristics on PDA from above and below (9 cm diameter petri dish). Scale bar: (b) = 500 µm, (c) = 200 µm, (d–i) = 50 µm, (j–o) = 10 µm.
Saprobic on dead bamboo culms submerged in freshwater. Sexual morph: Ascomata 430–460 μm high, 435–575 μm diam., immersed under a clypeus to semi-immersed, gregarious, uniloculate, globose to subglobose, carbonaceous, rough, black, ostiolate. Ostiole 160 μm high, 270 μm diam., central, apex well developed, papillate, with pore-like opening, with periphyses filling the ostiolar canal, dark brown to black, orange pigment near ostiole. Peridium 6–18 μm wide, comprising 3–5 layers of brown to hyaline cells of textura angularis, inner layers composed of thin, hyaline cells. Hamathecium of dense, long, 0.8–1.25 µm wide ( = 1 μm, n = 50), filiform, filamentous, trabeculate pseudoparaphyses, septate, branched, embedded in a gelatinous matrix, anastomosing between the asci and at the apex. Asci 125–160 × 15–20 μm ( = 145 × 17 μm, n = 20), 8–spored, bitunicate, fissitunicate, broad cylindrical to cylindrical-clavate, with bulbous pedicellate, rounded at the apex, with an ocular chamber. Ascospores 18–28 × 7–10 μm ( = 22.5× 9 μm, n = 20), biseriate, broadly fusiform, tapering towards the ends, hyaline, 1-septate at the centre, constricted at the septum, cell above septate enlarged, straight, smooth-walled, with longitudinally striate wall ornamentation, surrounded by mucilaginous sheath. Asexual morph: Undetermined.
Culture characters: Ascospores germinating on MEA and PDA within 24 h with germ tubes developing from basal cells. Colonies on MEA and PDA umbonate at the centre, circular, friable, reaching 20 mm diameter after four weeks of incubation at 25 °C. Culture on MEA with white aerial mycelium, dark brown at the centre and paler towards the edge from above and below. Culture on PDA dark brown from above and below.
Material examined: THAILAND, Krabi province (8.1° N, 98.9° E), on dead bamboo culms, 15 December 2015, C. Phukhamsakda, KR001 (MFLU 20-0694, holotype), ibid, 18 December 2015 (MFLU 20-0695, paratype); ex-type living culture MFLUCC 16-0412; ex-paratype living culture, MFLUCC 16-0417.
4. Discussion
In this study, we introduce a new species, genus and family for a collection of Pleosporales found on bamboo. The introduction of new taxa, even at the family level, is not surprising, considering that about 93% of fungi remain unknown to science despite ca. 2000 species described every year [59,60]. Pleosporalean species can occur in terrestrial, marine and freshwater habitats [7,8,9]. Several studies have reported new pleosporalean taxa from freshwater or marine habitats or from bambusicolous hosts [1,3]. Pleosporales have unique characters such as perithecioid ascomata typically with a papilla and bitunicate, generally fissitunicate asci, bearing mostly septate ascospores of different colours and shapes, with or without a gelatinous sheath [7]. The morphology of Anastomitrabeculiaceae is similar to members of the Pleosporales based on the presence of pseudoparaphyses, perithecioid ascomata, bitunicate asci and hyaline ascospores. Anastomitrabeculiaceae is characterised by semi-immersed to superficial ascomata, trabeculate pseudoparaphyses, cylindrical asci and ascospores with longitudinally striate wall ornamentation, surrounded by mucilaginous sheath. The newly discovered species formed a well-supported independent lineage basal to the Halojulellaceae based on phylogenetic analyses of the combined dataset (0.99 PP/65% MLBT). Halojulellaceae differs by its cellular pseudoparaphyses and golden brown ascospores [2]. The new taxon is also phylogenetically closely related to Neohendersoniaceae, which differs by its cellular pseudoparaphyses and smooth-walled ascospore [61]. A novel genus Anastomitrabeculia is therefore introduced to accommodate one new species, Anastomitrabeculia didymospora. A new family, Anastomitrabeculiaceae, is also introduced to accommodate this independent lineage.
Several pleosporalean families such as Aigialaceae, Amniculicolaceae, Anteagloniaceae, Astrosphaeriellaceae, Bambusicolaceae, Biatriosporaceae, Caryosporaceae, Cyclothyriellaceae, Delitschiaceae, Didymosphaeriaceae, Fuscostagonosporaceae, Lindgomycetaceae, Melanommataceae, Neomassariaceae, Pseudoastrosphaeriellaceae, Striatiguttulaceae and Tetraplosphaeriaceae share similar characters to Anastomitrabeculiaceae in having trabeculate pseudoparaphyses, cylindrical asci and ascospores with a sheath [7]. The nature of pseudoparaphyses is often overlooked, but they have taxonomic relevance at the genus and possibly family levels [7], but not at the ordinal level [62]. These families differ from Anastomitrabeculiaceae mainly by their ascospores, for example, Aigialaceae and Amniculicolaceae have brown and muriform ascospores [7]. Anteagloniaceae differs by having a peridium composed of dark brown cells of textura epidermoidea, cellular or trabeculate pseudoparaphyses and small, uniseriate ascospores [2]. Astrosphaeriellaceae differs by its brown, sub-fusiform to fusiform, obclavate to ellipsoidal, or limoniform ascospores [63] and Biatriosporaceae differs by its immersed ascomata and fusiform, dark brown ascospores [2]. Caryosporaceae differs by its broad-fusiform, ovoid or ellipsoid, brown ascospores [64]. Bambusicolaceae species have also been isolated from dead bamboo culms, but they differ from Anastomitrabeculiaceae by their cellular pseudoparaphyses and multi-seriate, smooth-walled ascospores [2]. Cyclothyriellaceae differs by its uniseriate, ellipsoid to fusiform, brown ascospores with several eusepta [65]. Fuscostagonosporaceae differs in having globose to subglobose ascomata, fissitunicate asci with long stipes and narrowly fusiform ascospores [66]. Anastomitrabeculiaceae shares several characters with Didymosphaeriaceae in having immersed ascomata formed under a clypeus, trabeculate pseudoparaphyses and cylindrical asci. Didymosphaeriaceae and Melanommataceae differ in having cellular or trabeculate pseudoparaphyses and brown, multi-septate, muriform ascospores [7]. Lindgomycetaceae differs by the presence of cellular or trabeculate pseudoparaphyses and brown, multi-septate ascospores with bipolar mucilaginous appendages [7]. Neomassariaceae differs by its immersed ascomata and ellipsoid ascospores. Pseudoastrosphaeriellaceae differs by its brown to reddish-brown ascospores with longitudinal ridges towards the ends and Striatiguttulaceae differs in having brown, ellipsoid ascospores with paler end cells. Tetraplosphaeriaceae differs by its immersed ascomata and slightly curved, pale brown ascospores [7].
Divergence time estimate has been widely used as supporting evidence to clarify taxonomic status of extant or novel families in fungal taxonomy [4,6,23,24,26,27,67]. In this study, the MCC tree was congruent with the topology of the trees generated from Bayesian inference analysis and maximum likelihood analyses. The divergence time estimates for the crown age of Dothideomycetes (263 MYA), the split of Dothideomycetes and Arthoniomycetes (323 MYA), the crown age of Pleosporales (206 MYA) and the divergence of Hysteriales from Pleosporales (236 MYA) are similar to previous studies [4,7,11]. Hyde et al. [27] recommended that the divergence times of families should be between 50 and 150 MYA. The stem age is usually preferred to the crown age in taxa ranking as it is not affected by the sample size of the clade [27]. Based on the MCC tree, Anastomitrabeculiaceae and Halojulellaceae share the stem age of 84 MYA which supports the establishment of Anastomitrabeculiaceae.
The divergence time of Anastomitrabeculiaceae was also compared to Pleosporalean families with trabeculate pseudoparaphyses, cylindrical asci and ascospores with a sheath (Table 3). Cyclothyriellaceae has an estimated crown age of 66 MYA and it diverged at 95 MYA. Fuscostagonosporaceae has a crown age of approximately 26 MYA and it diverged around 63 MYA. Bambusicolaceae, which was also isolated from dead bamboo culms, has a crown age of 29 MYA and a stem age of about 57 MYA. The stem age of Anastomitrabeculiaceae lies within the range of divergence times of those with similar morphology, but the crown age of Anastomitrabeculiaceae (2.6 MYA) is much earlier compared to these families. Bambusicolaceae was introduced by Hyde at al. [2] to include three bambusicolous taxa, and it currently has 15 species [7]. Fuscostagonosporaceae was introduced by Hyde at al. [66] to accommodate one bambusicolous taxon and it currently has four species [7]. Ariyawansa et al. [64] introduced the pleosporalean family, Caryosporaceae, which is morphologically similar to Astrosphaeriellaceae and Trematosphaeriaceae [7]. Based on Liu et al. [11], the stem age of Caryosporaceae (85 MYA) is similar to Trematosphaeriaceae (88 MYA) compared to Astrosphaeriellaceae (113 MYA), but the crown age of Caryosporaceae (2 MYA) is much earlier compared to Astrosphaeriellaceae (55 MYA) and Trematosphaeriaceae (65 MYA). Astrosphaeriellaceae currently has 111 species, and Trematosphaeriaceae has 103 species, whereas Caryosporaceae has ten species [7]. Compared to their morphologically similar families, the early crown of Anastomitrabeculiaceae and Caryosporaceae could be due to their smaller sample size. Therefore, further collections are needed for an accurate estimation of the crown age as it is affected by the sample size of the clade [27]. This could also be due to rapid speciation of pleosporalean fungal species given their high adaptation capabilities.
The estimated crown age of Pleosporales (206 MYA) lies within the early Triassic period. The origin of monocotyledons is estimated within the late Cretaceous period (around 145 MYA) [68]. This period is associated with the diversification of pleosporalean families, which continued during the early Cretaceous period when there was a major diversification and radiation of angiosperms, which favoured further diversification of Pleosporalean families to adapt to various hosts [69].
Hosts and their symbionts can speciate in parallel, which relates to a high level of congruence between the phylogeny of the hosts and their symbionts [70,71]. Therefore, studies focusing on divergence time is important for a better understanding of host–pathogen interaction as well as co-evolutionary interactions [72]. This study uses a polyphasic approach based on morphology, multi-locus phylogenetic analyses and divergence time estimates. By implementing a polyphasic approach, we provide strong evidence for introducing the new family based on congruent results supporting the establishment of a new family.
Acknowledgments
K.D.H. thanks Chiang Mai University for the award of visiting Professor. R.J. thanks the University of Mauritius for support and the MRC funded project MRC/RUN/1705. We would like to thank Shaun Pennycook from Manaaki Whenua, Landcare Research, New Zealand, for nomenclatural advice.
Author Contributions
Conceptualization, C.S.B., C.P.; methodology, C.S.B., C.P.; resources, K.D.H.; writing—original draft preparation, C.S.B.; writing—review and editing, C.S.B., C.P., R.J., I.P. and K.D.H.; supervision, K.D.H.; funding acquisition, K.D.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Thailand Research Fund, grant RDG6130001 entitled “Impact of climate change on fungal diversity and biogeography in the Greater Mekong Subregion”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All sequences generated in this study were submitted to GenBank.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhang Y., Crous P.W., Schoch C.L., Hyde K.D. Pleosporales. Fungal Divers. 2012;53:1–221. doi: 10.1007/s13225-011-0117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hyde K.D., Jones E.B.G., Liu J.K., Ariyawansa H., Boehm E., Boonmee S., Braun U., Chomnunti P., Crous P.W., Dai D.Q., et al. Families of Dothideomycetes. Fungal Divers. 2013;63:1–313. doi: 10.1007/s13225-013-0263-4. [DOI] [Google Scholar]
- 3.Tanaka K., Hirayama K., Yonezawa H., Sato G., Toriyabe A., Kudo H., Hashimot A., Matsumura M., Harada Y., Kurihara Y., et al. Revision of the Massarineae (Pleosporales, Dothideomycetes) Stud. Mycol. 2015;82:75–136. doi: 10.1016/j.simyco.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phukhamsakda C., Hongsanan S., Ryberg M., Ariyawansa H.A., Chomnunti P., Bahkali A.H., Hyde K.D. The evolution of Massarineae with Longipedicellataceae fam. nov. Mycosphere. 2016;7:1713–1731. doi: 10.5943/mycosphere/7/11/7. [DOI] [Google Scholar]
- 5.Jaklitsch W.M., Checa J., Blanco M.N., Olariaga I., Tello S., Voglmayr H. A preliminary account of the Cucurbitariaceae. Stud. Mycol. 2018;90:71–118. doi: 10.1016/j.simyco.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu N.G., Lin C.G., Liu J.K., Samarakoon M.C., Hongsanan S., Bhat D.J., Hyde K.D., McKenzie E.H., Jumpathong J. Lentimurisporaceae, a new Pleosporalean family with divergence times estimates. Cryptogam. Mycol. 2018;39:259–283. doi: 10.7872/crym/v39.iss2.2018.259. [DOI] [Google Scholar]
- 7.Hongsanan S., Hyde K.D., Phookamsak R., Wanasinghe D.N., Mckenzie E., Sarma V.V., Boonmee S., Lücking R., Pem D., Bhat D.J., et al. Refined families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere. 2020;11:1553–2107. doi: 10.5943/mycosphere/11/1/13. [DOI] [Google Scholar]
- 8.Ramesh C. Loculoascomycetes from India. Front. Fungal Divers. India. 2003:457–479. [Google Scholar]
- 9.Kruys Å., Eriksson O.E., Wedin M. Phylogenetic relationships of coprophilous Pleosporales (Dothideomycetes, Ascomycota), and the classification of some bitunicate taxa of unknown position. Mycol. Res. 2006;110:527–536. doi: 10.1016/j.mycres.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y., Schoch C.L., Fournier J., Crous P.W., De Gruyter J., Woudenberg J.H.C., Hirayama K., Tanaka K., Pointing S.B., Spatafora J.W., et al. Multi-locus phylogeny of Pleosporales: A taxonomic, ecological and evolutionary re-evaluation. Stud. Mycol. 2009;64:85–102. doi: 10.3114/sim.2009.64.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J.K., Hyde K.D., Jeewon R., Phillips A.J., Maharachchikumbura S.S., Ryberg M., Liu Z.Y., Zhao Q. Ranking higher taxa using divergence times: A case study in Dothideomycetes. Fungal Divers. 2017;84:75–99. doi: 10.1007/s13225-017-0385-1. [DOI] [Google Scholar]
- 12.Wijayawardene N.N., Hyde K.D., Lumbsch H.T., Liu J.K., Maharachchikumbura S.S.N., Ekanayaka A.H., Tian Q., Phookamsak R. Outline of Ascomycota: 2017. Fungal Divers. 2018;88:167–263. doi: 10.1007/s13225-018-0394-8. [DOI] [Google Scholar]
- 13.Wijayawardene N.N., Hyde K.D., Al-Ani L.K.T., Tedersoo L., Haelewaters D., Rajeshkumar K.C., Zhao R.L., Aptroot A., Leontyev D.V., Saxena R.K., et al. Outline of Fungi and fungus-like taxa. Mycosphere. 2020;11:1060–1456. doi: 10.5943/mycosphere/11/1/8. [DOI] [Google Scholar]
- 14.Wijayawardene N.N., Hyde K.D., Wanasinghe D.N., Papizadeh M., Goonasekara I.D., Camporesi E., Bhat D.J., Mckenzie E.H.C., Phillips A.J.L., Diederich P., et al. Taxonomy and phylogeny of dematiaceous coelomycetes. Fungal Divers. 2016;77:1–316. doi: 10.1007/s13225-016-0360-2. [DOI] [Google Scholar]
- 15.Wijayawardene N.N., Hyde K.D., Rajeshkumar K.C., Hawksworth D.L., Madrid H., Kirk P.M., Braun U., Singh R.V., Crous P.W., Kukwa M., et al. Notes for genera: Ascomycota. Fungal Divers. 2017;86:1–594. doi: 10.1007/s13225-017-0386-0. [DOI] [Google Scholar]
- 16.Gratani L., Crescente M.F., Varone L., Fabrini G., Digiulio E. Growth pattern and photosynthetic activity of different bamboo species growing in the Botanical Garden of Rome. Flora-Morphol. Distrib. Funct. Ecol. Plants. 2008;203:77–84. doi: 10.1016/j.flora.2007.11.002. [DOI] [Google Scholar]
- 17.Kelchner S.A., Group B.P. Higher level phylogenetic relationships within the bamboos (Poaceae: Bambusoideae) based on five plastid markers. Mol. Phylogenet. Evol. 2013;67:404–413. doi: 10.1016/j.ympev.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Dai D.Q., Phookamsak R., Wijayawardene N.N., Li W.J., Bhat D.J., Xu J.C., Taylor J.E., Hyde K.D., Chukeatirote E. Bambusicolous fungi. Fungal Divers. 2016;82:1–105. doi: 10.1007/s13225-016-0367-8. [DOI] [Google Scholar]
- 19.Kirschner R., Yang Z.L., Zhao Q., Feng B. Ovipoculum album, a new anamorph with gelatinous cupulate bulbilliferous conidiomata from China and with affinities to the Auriculariales (Basidiomycota) Fungal Divers. 2009;43:55–65. doi: 10.1007/s13225-010-0038-0. [DOI] [Google Scholar]
- 20.Hyde K.D., Zhou D., McKenzie E., Ho W., Dalisay T. Vertical distribution of saprobic fungi on bamboo culms. Fungal Divers. 2002;11:109–118. [Google Scholar]
- 21.Hyde K.D., Zhou D., Dalisay T. Bambusicolous fungi: A review. Fungal Divers. 2002;9:1–14. [Google Scholar]
- 22.Prieto M., Wedin M. Dating the diversification of the major lineages of Ascomycota (Fungi) PLoS ONE. 2013;8:e65576. doi: 10.1371/journal.pone.0065576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hongsanan S., Sánchez-Ramírez S., Crous P.W., Ariyawansa H.A., Zhao R.L., Hyde K.D. The evolution of fungal epiphytes. Mycosphere. 2016;7:1690–1712. doi: 10.5943/mycosphere/7/11/6. [DOI] [Google Scholar]
- 24.Hongsanan S., Maharachchikumbura S.S.N., Hyde K.D., Samarakoon M.C., Jeewon R., Zhao Q., Al-Sadi A.M., Bahkali A.H. An updated phylogeny of Sordariomycetes based on phylogenetic and molecular clock evidence. Fungal Divers. 2017;84:25–41. doi: 10.1007/s13225-017-0384-2. [DOI] [Google Scholar]
- 25.Samarakoon M.C., Hyde K.D., Promputtha I., Hongsanan S., Ariyawansa H.A., Maharachchikumbura S.S.N., Daranagama D.A., Stadler M., Mapook A. Evolution of Xylariomycetidae (Ascomycota: Sordariomycetes) Mycosphere. 2016;7:1746–1761. doi: 10.5943/mycosphere/7/11/9. [DOI] [Google Scholar]
- 26.Samarakoon M.C., Hyde K.D., Hongsanan S., McKenzie E.H., Ariyawansa H.A., Promputtha I., Zeng X.Y., Tian Q., Liu J.K. Divergence time calibrations for ancient lineages of Ascomycota classification based on a modern review of estimations. Fungal Divers. 2019;96:285–346. doi: 10.1007/s13225-019-00423-8. [DOI] [Google Scholar]
- 27.Hyde K.D., Maharachchikumbura S.S., Hongsanan S., Samarakoon M.C., Lücking R., Pem D., Harishchandra D., Jeewon R., Zhao R.L., Xu J.C., et al. The ranking of fungi: A tribute to David L. Hawksworth on his 70th birthday. Fungal Divers. 2017;84:1–23. doi: 10.1007/s13225-017-0383-3. [DOI] [Google Scholar]
- 28.Senanayake I.C., Rathnayaka A.R., Marasinghe D.S., Calabon M.S., Gentekaki E., Lee H.B., Hurdeal V.G., Pem D., Dissanayake L.S., Wijesinghe S.N., et al. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere. 2020;11:2678–2754. doi: 10.5943/mycosphere/11/1/20. [DOI] [Google Scholar]
- 29.Jayasiri S.C., Hyde K.D., Ariyawansa H.A., Bhat J., Buyck B., Cai L., Dai Y.C., Abd-Elsalam K.A., Ertz D., Hidayat I., et al. The faces of fungi database: Fungal names linked with morphology, molecular and human attributes. Fungal Divers. 2015;74:3–18. doi: 10.1007/s13225-015-0351-8. [DOI] [Google Scholar]
- 30.Jeewon R., Hyde K.D. Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere. 2016;7:1669–1677. doi: 10.5943/mycosphere/7/11/4. [DOI] [Google Scholar]
- 31.Dissanayake A.J., Bhunjun C.S., Maharachchikumbura S.S.N., Liu J.K. Applied aspects of methods to infer phylogenetic relationships amongst fungi. Mycosphere. 2020;11:2652–2676. doi: 10.5943/mycosphere/11/1/18. [DOI] [Google Scholar]
- 32.Vilgalys R., Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990;172:4238–4246. doi: 10.1128/JB.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White T.J., Bruns T., Lee S., Taylor J. PCR Protocols. Elsevier; Amsterdam, The Netherlands: 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics; pp. 315–322. [DOI] [Google Scholar]
- 34.Rehner S.A., Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences, evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 2005;97:84–98. doi: 10.3852/mycologia.97.1.84. [DOI] [PubMed] [Google Scholar]
- 35.Hall T.A. Nucleic Acids Symposium Series. Volume 41. Information Retrieval Ltd.; London, UK: 1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT; pp. 95–98. [Google Scholar]
- 36.Pinnoi A., Jeewon R., Sakayaroj J., Hyde K.D., Jones E.B.G. Berkleasmium crunisia sp. nov. and its phylogenetic affinities to the Pleosporales based on 18S and 28S rDNA sequence analyses. Mycologia. 2007;99:378–384. doi: 10.1080/15572536.2007.11832562. [DOI] [PubMed] [Google Scholar]
- 37.Beimforde C., Feldberg K., Nylinder S., Rikkinen J., Tuovila H., Dörfelt H., Gube M., Jackson D.J., Reitner J., Seyfullah L.J., et al. Estimating the Phanerozoic history of the Ascomycota lineages: Combining fossil and molecular data. Mol. Phylogenet. Evol. 2014;78:386–398. doi: 10.1016/j.ympev.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 38.Pratibha J., Prabhugaonkar A., Hyde K.D., Bhat D.J. Phylogenetic placement of Bahusandhika, Cancellidium and Pseudoepicoccum (asexual Ascomycota) Phytotaxa. 2014;176:68–80. doi: 10.11646/phytotaxa.176.1.9. [DOI] [Google Scholar]
- 39.Thambugala K.M., Hyde K.D., Tanaka K., Tian Q., Wanasinghe D.N., Ariyawansa H.A., Jayasiri S.C., Boonmee S., Camporesi E., Hashimoto A., et al. Towards a natural classification and backbone tree for Lophiostomataceae, Floricolaceae, and Amorosiaceae fam. nov. Fungal Divers. 2015;74:199–266. doi: 10.1007/s13225-015-0348-3. [DOI] [Google Scholar]
- 40.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larsson A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 2014;30:3276–3278. doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller M.A., Pfeiffer W., Schwartz T. Creating the CIPRES science gateway for inference of large phylogenetic trees; Proceedings of the 2010 Gateway Computing Environments Workshop (GCE); New Orleans, LA, USA. 14 November 2010; pp. 1–8. [Google Scholar]
- 43.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swofford D.L. PAUP: Phylogenetic Analysis Using Parsimony, Version 4.0 b10. Sinauer Associates; Sunderland, UK: 2002. [Google Scholar]
- 45.Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhaxybayeva O., Gogarten J.P. Bootstrap, Bayesian probability and maximum likelihood mapping: Exploring new tools for comparative genome analyses. BMC Genom. 2002;3:4. doi: 10.1186/1471-2164-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rambaut A., Drummond A.J., Xie D., Baele G., Suchard M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018;67:901. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rambaut A. FigTree v1.4. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh. [(accessed on 15 August 2020)]; Available online: http://tree.bio.ed.ac.uk/software/figtree/
- 50.Bouckaert R., Vaughan T.G., Barido-Sottani J., Duchêne S., Fourment M., Gavryushkina A., Heled J., Jones G., Kühnert D., De Maio N., et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019;15:e1006650. doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gernhard T., Hartmann K., Steel M. Stochastic properties of generalised Yule models, with biodiversity applications. J. Math. Biol. 2008;253:769–778. doi: 10.1007/s00285-008-0186-y. [DOI] [PubMed] [Google Scholar]
- 52.Drummond A.J., Suchard M.A., Xie D., Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gueidan C., Ruibal C., De Hoog G.S., Schneider H. Rock-inhabiting fungi originated during periods of dry climate in the late Devonian and middle Triassic. Fungal Biol. 2011;115:987–996. doi: 10.1016/j.funbio.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 54.Pérez-Ortega S., Garrido-Benavent I., Grube M., Olmo R., de los Ríos A. Hidden diversity of marine borderline lichens and a new order of fungi: Collemopsidiales (Dothideomyceta) Fungal Divers. 2016;80:285–300. doi: 10.1007/s13225-016-0361-1. [DOI] [Google Scholar]
- 55.Mindell R.A., Stockey R.A., Beard G., Currah R.S. Margaretbarromyces dictyosporus gen. sp. nov.: A permineralized corticolous ascomycete from the Eocene of Vancouver Island, British Columbia. Mycol. Res. 2007;111:680–684. doi: 10.1016/j.mycres.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 56.Berbee M., Taylor J.W. Dating the molecular clock in fungi-how close are we? Fungal Biol. Rev. 2010;24:1–16. doi: 10.1016/j.fbr.2010.03.001. [DOI] [Google Scholar]
- 57.Taylor T.N., Krings M., Taylor E.L. Fossil fungi. Academic Press; San Diego, CA, USA: 2015. Ascomycota. [Google Scholar]
- 58.Cohen K.M., Finney S.C., Gibbard P.L., Fan J.X. The ICS International Chronostratigraphic Chart. Episodes. 2013;36:199–204. doi: 10.18814/epiiugs/2013/v36i3/002. [DOI] [Google Scholar]
- 59.Willis K.J. State of the World’s Fungi 2018. Royal Botanic Gardens, Kew; London, UK: 2018. [Google Scholar]
- 60.Hyde K.D., Jeewon R., Chen Y.J., Bhunjun C.S., Calabon M.S., Jiang H.B., Lin C.G., Norphanphoun C., Sysouphanthong P., Pem D., et al. The numbers of fungi: Is the descriptive curve flattening? Fungal Divers. 2020;103:219–271. doi: 10.1007/s13225-020-00458-2. [DOI] [Google Scholar]
- 61.Giraldo A., Crous P.W., Schumacher R.K., Cheewangkoon R., Ghobad-Nejhad M., Langer E. The Genera of Fungi—G3: Aleurocystis, Blastacervulus, Clypeophysalospora, Licrostroma, Neohendersonia and Spumatoria. Mycol. Prog. 2017;16:325–348. doi: 10.1007/s11557-017-1270-8. [DOI] [Google Scholar]
- 62.Liew E.C.Y., Aptroot A., Hyde K.D. Phylogenetic significance of the pseudoparaphyses in Loculoascomycete taxonomy. Mol. Phylogenet. Evol. 2000;16:392–402. doi: 10.1006/mpev.2000.0801. [DOI] [PubMed] [Google Scholar]
- 63.Phookamsak R., Norphanphoun C., Tanaka K., Dai D.Q., Luo Z.L., Liu J.K., Su H.Y., Bhat D.J., Bahkali A.H., Mortimer P.E., et al. Towards a natural classification of Astrosphaeriella-like species; introducing Astrosphaeriellaceae and Pseudoastrosphaeriellaceae fam. nov. and Astrosphaeriellopsis, gen. nov. Fungal Divers. 2015;74:143–197. doi: 10.1007/s13225-015-0352-7. [DOI] [Google Scholar]
- 64.Ariyawansa H.A., Hyde K.D., Jayasiri S.C., Buyck B., Chethana K.W.T., Dai D.Q., Dai Y.C., Daranagama D.A., Jayawardena R.S., Lücking R., et al. Fungal diversity notes 111–252: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2015;75:27–274. doi: 10.1007/s13225-015-0346-5. [DOI] [Google Scholar]
- 65.Jaklitsch W.M., Voglmayr H. Hidden diversity in Thyridaria and a new circumscription of the Thyridariaceae. Stud. Mycol. 2016;85:35–64. doi: 10.1016/j.simyco.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hyde K.D., Norphanphoun C., Abreu V.P., Bazzicalupo A., Chethana K.W.T., Clericuzio M., Dayarathne M.C., Dissanayake A.J., Ekanayaka A.H., He M.Q., et al. Fungal diversity notes 603–708: Taxonomic and phylogenetic notes on genera and species. Fungal Divers. 2017;87:1–235. doi: 10.1007/s13225-017-0391-3. [DOI] [Google Scholar]
- 67.Zhang S.N., Hyde K.D., Jones E.B.G., Jeewon R., Cheewangkoon R., Liu J.K. Striatiguttulaceae, a new pleosporalean family to accommodate Longicorpus and Striatiguttula gen. nov. from palms. MycoKeys. 2019;49:99–129. doi: 10.3897/mycokeys.49.30886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chaw S.M., Chang C.C., Chen H.L., Li W.H. Dating the Monocot–Dicot divergence and the origin of core Eudicots using whole chloroplast genomes. J. Mol. Evol. 2004;58:424–441. doi: 10.1007/s00239-003-2564-9. [DOI] [PubMed] [Google Scholar]
- 69.Phillips A.J., Hyde K.D., Alves A., Liu J.K. Families in Botryosphaeriales: A phylogenetic, morphological and evolutionary perspective. Fungal Divers. 2019;94:1–22. doi: 10.1007/s13225-018-0416-6. [DOI] [Google Scholar]
- 70.De Vienne D.M., Refrégier G., López-Villavicencio M., Tellier A., Hood M.E., Giraud T. Cospeciation vs host-shift speciation: Methods for testing, evidence from natural associations and relation to coevolution. New Phytol. 2013;198:347–385. doi: 10.1111/nph.12150. [DOI] [PubMed] [Google Scholar]
- 71.Phukhamsakda C., McKenzie E., Phillips A.J.L., Jones E.B.G., Bhat D.J., Marc S., Bhunjun C.S., Wanasinghe D.N., Thongbai B., Camporesi E., et al. Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Divers. 2020;102:1–203. doi: 10.1007/s13225-020-00448-4. [DOI] [Google Scholar]
- 72.Zeng X.Y., Jeewon R., Hongsanan S., Hyde K.D., Wen T.C. Unravelling evolutionary relationships between epifoliar Meliolaceae and angiosperms. J. Syst. Evol. 2020 doi: 10.1111/jse.12643. in press. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All sequences generated in this study were submitted to GenBank.