Abstract
WNK kinases auto-activate by autophosphorylation. Crystallography of the kinase domain of WNK1 phosphophorylated on the primary activating site (pWNK1) and in the presence of AMP-PNP reveals a well-ordered but inactive configuration. This new pWNK1 structure features specific and unique interactions of the phosphoserine, less hydration, and smaller cavities as compared with unphophorylated WNK1 (uWNK1). Since WNKs are activated by osmotic stress in cells, we addressed whether the structure was influenced directly by osmotic pressure. pWNK1 crystals formed in PEG3350 were soaked in the osmolyte sucrose. Suc-WNK1 crystals maintained x-ray diffraction, but the lattice constants and pWNK1 structure changed. Differences occurred in the activation loop and helix C, common switch loci in kinase activation. Based on these structural changes, we tested for effects on in vitro activity of two WNKs, pWNK1 and pWNK3. The osmolyte PEG400 enhanced ATPase activity. The data presented suggest multistage activation of WNKs.
Keywords: WNK1, WNK3, kinase, phosphorylation, osmotic pressure, structure, crystallography, cavities, water, PEG400, sucrose, osmolyte, crowding agent, intermediate, multistage activation
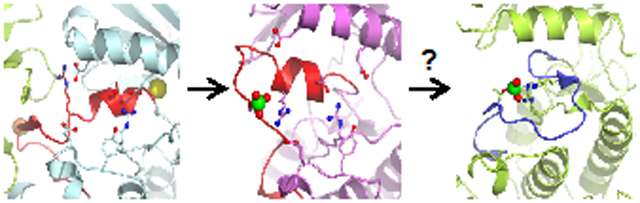
Graphical Abstract
Introduction
With No Lysine (WNK) kinases are large intracellular kinases named for their unique constellation of catalytic residues 1. WNK kinases 1 and 4 have been linked genetically to a familial form of hypertension, Gordon’s syndrome 2. This disease is treated with diuretics targeting the cation chloride cotransporters Na+Cl− and Na+K+2Cl− (NCCs and NKCCs), placing WNKs in the pathway leading to cotransporter activation. Cotransporter regulation is known to involve phosphorylation-dependent chloride inhibition and osmotic stress activation 3, 4, potentially involving WNKs. We demonstrated that both WNK1 activity and autophosphorylation is inhibited by chloride 5. WNK3 chloride sensitivity has also been reported 6. Further, we determined the structure of the unphosphorylated kinase domain of WNK1 (uWNK1) which revealed chloride bound in the active site. Several reports show that WNKs are activated by osmotic stress in cells 7-9. As a consequence, our laboratory as well as several other researchers, have hypothesized that WNKs are at the top of the cascade leading to NKCC and NCC activation. This hypothesis casts WNKs as the long-anticipated protein kinases regulated by chloride and osmotic stress to control cell volume 10, 11. We have recently reported that uWNK1 and uWNK3 autophosphorylation is stimulated by osmotic pressure (submitted for publication).
Here we focus on pWNK1 and pWNK3. pWNK1 is a drug target for hypertension 2, 12 and triple negative breast cancer 13. pWNK3 has been implicated as a drug target for stroke 14. The kinase domains of pWNK1 and pWNK3 are 90% identical. We have obtained crystals of pWNK1 and report a new 2.0 Å structure of pWNK1 in the presence of AMP-PNP. pWNK3 expresses well, shows greater thermal stability than pWNK1, and was used for in vitro assays. The structure of pWNK1 reveals a well-ordered configuration of the activation loop and activating phosphorylation site. However, the observed configuration does not resemble other active kinases according to numerous structural cues. Thus, conformational change is likely required for our pWNK1 conformer to become active. Comparisons are presented here to protein kinase A (PKA, PDB file 1ATP) and a recently deposited structure of doubly phosphorylated pWNK3 (PDB file 5O26, personal communication, Alex Bullock).
Crowding agents and osmolytes put a direct demand on solvent, changing the water vapor pressure of a solution 15-17. In cells, osmotic stress applied externally promotes water efflux and internal macromolecular crowding. Many cellular proteins, including kinases, are influenced by externally applied osmolytes or crowding agents. Polyethylene glycols and ficolls change both the structure and activity of enzymes, in one case inducing an ATPase activity 18. However, there are few reports concerning direct osmotic pressure effects on known cellular osmosensors 19-22. To test whether osmolytes affect the structure of pWNK1, pWNK1 crystals were soaked in sucrose. Significant conformational changes, disorder, reduction in cavity size, and loss of water were observed. Based upon our pWNK1-sucrose (suc-pWNK1) structure, we examined whether sucrose or the stronger osmolyte PEG400 23 affect the kinase activity of pWNK1 in vitro. We found that crowding agents functioned to induce ATPase activity in both pWNK1 and pWNK3, suggestive of influence on the protein conformation.
Experimental Methods
Reagents
BioUltra PEG400 (Sigma) was aliquoted and stored at −80°C and used fresh or within a year. Sucrose and SYPRO-Orange dye were from Sigma-Aldrich Inc, and crystallization screens were from Hampton Research (Viejo CA).
Protein reagents
Protein reagents were expressed and purified. A pHis expression vector 24 harboring the coding sequence for the pWNK1 kinase domain (amino acids 209-483) was constructed and transformed into the E. coli strain Rosetta DE3 25. pWNK1 purification used Ni-NTA affinity and gel filtration, with details available in 5, 25. The purification of pWNK1 included an additional cation-exchange (Mono-S, GE Healthcare) chromatography step to separate the phosphorylated and unphosphorylated forms. Purified protein was buffer exchanged into 50 mM HEPES pH 7.4 and 150 mM NaCl. Serine 382 of pWNK1 was confirmed by mass-spectrometry to be fully phosphorylated.
The coding sequence for the kinase domain of pWNK3 (amino acids 118-409) was optimized and synthesized by Genscript Inc. (New Jersey) and cloned into pET29B. pWNK3 was transformed into E. coli strain Bl21 DE3, grown in TB media at 37 °C to an OD of about 1.2, then protein expression was induced by the addition of IPTG to 0.5 mM and grown overnight at 18°C. pWNK3 was purified using Ni-NTA affinity chromatography followed by anion exchange (MonoQ, GE Healthcare). Purified protein was buffer exchanged into 50 mM HEPES pH 7.4 and 150 mM NaCl. pWNK3 phosphorylation on Ser308 was confirmed by mass-spectrometry.
A construct of glutathione-S-transferase-OSR1(314-344) (gOSR1) encompassing the phosphorylation site S325 was a gift of Clinton Taylor and Melanie Cobb 26. Protein expression of gOSR1 was carried out in the E. coli strain Bl21 DE3. Purification utilized a glutathione sepharose column (GE Healthcare); the gOSR1 fractions were buffer-exchanged into 20mM HEPES, pH 7.4, 50 mM NaCl, and 5% glycerol.
The active kinase proteins used as PEG400 controls, MAP2K6 (MEK6/DD) and TAO2, were expressed and purified as previously reported 27, 28.
Kinase-Glo® Assays
Kinase Glo® reactions were dispensed in 96 well plates using an Oxford 8 channel pipette. A 50 μL reaction contained 40 mM HEPES (pH 7.4), 10 mM MgCl2, 4 μM pWNK, 40 μM gOSR1, 100 μM ATP. The final chloride concentration was maintained at 150 mM. Reactions were stopped at specific time points by addition of 30 μL of Kinase-Glo® Reagent (Promega Inc). The plate was centrifuged for 2 min. at 800 rpm and incubated for 10 min. at room temperature. Luciferase generated luminescence was read on a CLARIOstar plate reader (BMG Labtech, Ortenberg, GER) and the data processed using MARS data analysis software (BMG Labtech, Ortenberg, GER).
32P Assays
Reaction conditions were similar to those used for Kinase Glo®, with exceptions. Reactions were initiated by the addition of pWNK1 or pWNK3 (50 nM final concentration) to reaction solutions containing 50 μM gOSR1 and 5-10 μCi of [γ−32P] ATP in addition to 100 μM cold ATP. Assay termination was accomplished using protocols described by Racker 29. Reactions were stopped at various time points by spotting 25 μL aliquots onto 3MM filter paper followed by immediate immersion in 10% trichloroacetic acid and 10 mM pyrophosphate (termination solution). Washing with fresh termination solution to remove unincorporated radioactivity was followed by filter paper drying. 32P-incorporation was measured by autoradiography using a phosphor screen (Kodak) and Typhoon 9200 imager (General Electric). Additional aliquots of the reaction mixtures were diluted then spotted on unwashed filter paper to establish the ratio of CPM to cold ATP concentration. ImageQuant software (General Electric) was used to quantify the autoradiograms.
Differential scanning fluorimetry (DSF)
Thermal melt temperatures (Tm’s) were measured with SYPRO Orange dye in an RT-PCR instrument 30. Kinases (5 μM) were incubated with 50 mM HEPES (pH 7.4), 2.5× SYPRO Orange dye diluted from manufactures stock (Sigma-Aldrich Inc), water, and the specified crowding agent or osmolyte to make a 25-μl reaction in a 96-well clear-bottom plate. The plate was centrifuged to remove bubbles. The temperature in each well was cycled from an initial 4 to 80°C using 0.5°C increments in a Bio-Rad CFX96 RT-PCR instrument. The SYPRO Orange fluorescence intensity was measured using the CFX96 fluorescein amidite channel. Tm is taken at the lowest point of –d(RFU)/dT during DSF from 0 to 80°C, where RFU is relative fluorescence units. All reactions were conducted in triplicate.
Crystallization and X-Ray Data.
Crystals of pWNK1 (WNK1/209-483, pS382) were obtained by hanging drop vapor diffusion using 9 mg/ml protein in 50 mM NaCl, 50 mM HEPES (pH 8.0), 1 mM EDTA, 2 mM dithiothreitol, 10 mM MgCl2, and 5 mM AMP-PNP. Crystallization conditions were found using Hampton PEG/Ion 2 Screen (Condition 4 and 39). Condition 39 was further optimized to 43 mM citric acid, 57 mM bis-tris propane, and 14% PEG 3350 at pH 6.0. Crystals were grown at 16°C, and cryoprotected in 20% PEG400. X-ray data were collected at the APS beamline 19-ID, yielding diffraction to 1.8Å. Integration and scaling were performed with the HKL-3000 software suite 31. The data collection and refinement statistics are given in Table S1 for reflections out to 2.0 Å where the CC1/2 was above 0.5 32.
Crystallography of pWNK1 in 0.6 M sucrose.
Crystals of pWNK1 grown as stated above were soaked in 0.6 M sucrose for 24 hours by replacing the crystallization solution with 43 mM citric acid, 57 mM bis-tris propane, 14% PEG 3350 and 600 mM sucrose. After soaking, no cracking of the crystals was observed. The crystals were then flash-frozen in liquid nitrogen. Data to 2Å was collected at the 19-ID APS beamline and processed as above. The data collection and refinement statistics are given in Table S1.
Structure and refinement.
The structure of pWNK1 was determined by molecular replacement using MOLREP in CCP4 33 using the uWNK1 structure 25 (PDB 6CN9) as a starting model. The model was built in COOT 34 based on the ∣2Fo-Fc∣ maps. Final restrained refinement against 1.8 Å x-ray data using REFMAC5 in the CCP4 suite for the pWNK1 and including TLS yielded an R-factor and R-free of 0.15 and 0.23, respectively. There are two similar but non-identical subunits in the asymmetric unit. The refined structure is deposited in the RCSB Protein Data Bank (PDB file is 5W7T).
Crystals of pWNK1 soaked with sucrose (suc-pWNK1) exhibited higher symmetry than the pWNK1 crystals, as a pseudo-twofold axis in pWNK1 became a true twofold in suc-pWNK in space group C2. The starting model for molecular replacement was the pWNK1 (PDB 5W7T). Model building and refinement of the structure (as described above) yielded a R-factor/R-free of 0.21 / 0.24.
pWNK1 cavity analysis.
To obtain a semi-quantitative estimate of the cavity volumes in pWNK, the POCASA software was used (altair.sci.hokudai.ac.jp/g6/service/pocasa) 35. POCASA defines cavities using a simple grid-scanning algorithm to identify points surrounded by protein. The recommended probe radius of 2 Å and grid size of 1Å was used in POCASA to compare cavities in pWNK1, suc-pWNK1 and other protein kinases of comparable size. The control kinases were CDK2, PDB file 1QZM, 785 Å3, and IRAK, PDB file 2OIB 750 Å3.
Mass spectrometry of polydisperse PEG400.
Ethylene glycol polymers were separated by an Agilent 1100 series LC system (Agilent Technologies, Palo Alto, CA) with an RP-C18 microbore HPLC column (Phenomenex Aeris WIDEPORE 150 2.1 mm, 3- m particle size, 200Å pore diameter). A ThermoFinnigan LCQ Deca-XP mass spectrometer was used to observe ethylene glycol oligomers.
RESULTS
Crystal structure of pWNK1 in comparison with fully active kinases
Crystals of pWNK1 that were obtained in the presence of AMP-PNP produced reflections to 2.0 Å (Table S1). Two copies of pWNK1 are present in the asymmetric unit, although pWNK1 behaves as a monomer by gel filtration (data not shown). The two copies are similar but not identical, and superimpose with an RMSD of 0.38 Å. The activation loop and pS382 are clearly visible in the electron density (Figure S1A). Both monomers have strong electron density for AMP-PNP, and weaker density for two putative magnesium ions (range 1.6-2.4σ) (Figure S1B). The citrate in the crystallization conditions may have reduced the [Mg+2] thereby attenuating the observed Mg electron densities. pS382 adopts a different position than in the S382 in the unphosphorylated form (PDB file 6CN9)25 where S382 is buried in the dimer interface (Figure S1C). The activation loop of pWNK1 is folded into a helix (helix AL for activation loop, hAL). This helix has been observed in several other protein kinases and is an indication that the structure is not in an active configuration 36. The hAL is similar but not identical to that of uWNK1 (Figure S1C).
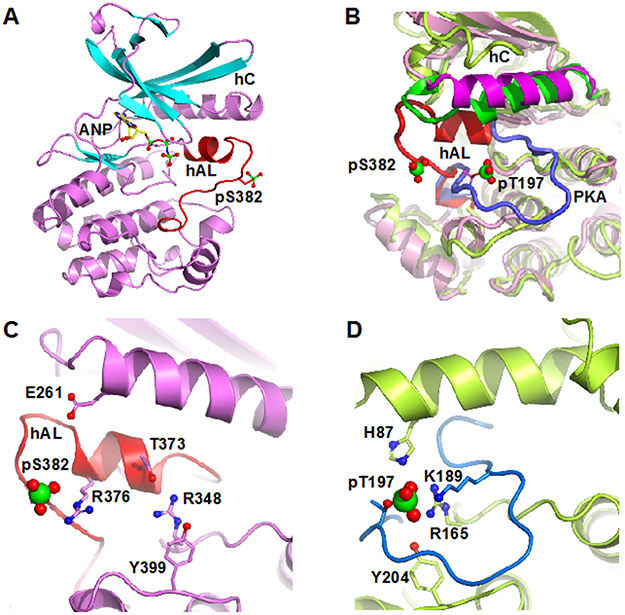
To compare the observed pWNK1 structure with an active kinase, we used phosphorylated protein kinase A (PKA, PDB file 1ATP) as a canonical example. As shown in the overlay in Figure 1B, the activation loop of pWNK1 (red) makes contacts mainly to the N-terminal domain, in contrast to the PKA activation loop (blue) that makes contacts to its C-terminal domain. Figure 1B also highlights the difference in position of hC in pWNK1 in comparison with PKA. In pWNK1, hC is displaced from the body of the kinase, another typical feature of inactive kinases 37. The position of AMP-PNP and the putative Mg++ ions also differs from that of Mn-ATP in PKA (Figure S1D), another indication that the observed configuration is inactive.
Figure 1.
Structure of pWNK1. (A) Overall structure of pWNK1. Helices are pink and β-strands are cyan, AMP-PNP (ANP) and the pS382 are shown in sticks. (B) Overlay of the structure of pWNK1 (PDB file 5W7T) with PKA (PDB file 1ATP). pWNK is magenta and its activation loop red; PKA is green, and it activation loop blue. (C) Unique conformation of the pWNK1 activation loop and unique interactions of pS382 with R376 in hAL, rather than R348 from the catalytic loop. (D) Interactions of the phosphorylation site in PKA from the same perspective as (C) showing canonical interactions with basic residues from the catalytic loop (R165), helix C (H87) and the activation loop (K189). Diagrams drawn in PyMOL 34
Figure 1 C and D contrast the activation loop configuration of pWNK1 and PKA. The pS382 interactions are distinct from those of PKA, forming an ion pair with R376 in the activation loop rather than with its expected ligand, R348 (in the catalytic loop sequence HRD). pWNK R348 corresponds to PKA R165. In pWNK1, R348 instead forms a hydrogen bond with T373 in hAL (Figure 1C, D). R348 further makes an aromatic stacking interaction with Y399. Y399 is homologous to Y204 of PKA (Figure 1D), a ligand to the phosphoserine/phosphothreonine. The recently deposited structure of pWNK3 shows that residues corresponding to R348 and Y399 (pWNK3 R274 and Y325) interact with the phosphoserine (PDB file 5O26).
Cavities and water
Structural comparisons of uWNK1 with pWNK1 indicate that water and cavities play a role in the conformational equilibrium between an unphophorylated, inactive conformer (uWNK1) and a phosphorylated conformer (pWNK1, present structure, PDB file 5W7T) (submitted for publication). We examined both cavities and water to assess their potential involvement in structural change to a fully active configuration. pWNK1 cavity volumes were calculated and found to be ~770Å3, in between the 950Å3 per monomer in uWNK1 (PDB file 6CN9) (Table S2) and cavities calculated for pWNK3 of 563Å3 (PDB file 5O26). Model-building was used to define ~410 water molecules per monomer in pWNK1. This is slightly more than ~350 molecules expected for proteins of comparable size 38.
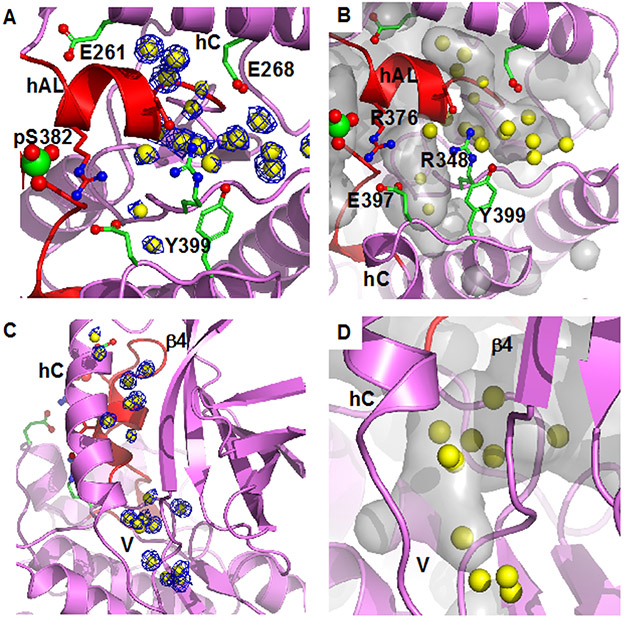
Well-ordered water in pWNK1 is present at the N-terminus of hAL and is associated with the unique hAL configuration. This cluster of waters, shown with electron density (Figure 2A), is partially buried in the cavities shown in Figure 2B. Another set of waters is in the V-shaped linker connecting hC and β−strand 4 and extends into the back of the active site (Figure 2C, electron density, Figure 2D, overlay with calculated cavities). Similar waters are conserved in Subunit B (not shown), but not in unphosphorylated uWNK1 (PDB file 6CN9). Deposited coordinates of active pWNK3 (PDB file 5O26) indicated that comparable waters near hAL (Figure 2 A,B are not observed). Also, few of the waters near the V-shaped linker, Figure 2C, remain in pWNK3. These contrasts suggest a potential role for bound water in the conversion of pWNK1 to a fully active configuration.
Figure 2.
Bound water in pWNK1 associated with the inactive configuration of hAL. (A) Waters in pWNK1 near helix AL. Cartoon rendering of pWNK1 in magenta (main cartoon) and red (activation loop). Waters are shown in yellow; electron density contoured at 0.8 σ in blue. (B) Cavities are depicted in gray in the same orientation as (A). (C) Waters between hC and β4 and extending from the V-shaped linker (V) into the back of the active site (below and β4 in the orientation shown). (D) Closeup of the cavity near the V-shaped linker and active site. (Surfaces calculated in PyMOL using Cavity_cull default settings).
The osmolyte sucrose induces changes in the structure of pWNK1 but not activity.
Since pWNK1 is not in an active configuration, we addressed whether the osmolyte sucrose could alter the configuration of pWNK1. Sucrose-soaked pWNK1 (suc-pWNK1) crystals maintained diffraction to 2Å resolution (Table S1). Sucrose induced significant changes in the structure. Overlays gave an 0.69 Å RMSD change in cα positions (Figure 3A). Conformational changes were observed from β-strand 3 to helix C and the loop between β4 and β5. Increased disorder and conformational changes also occur in hAL and the rest of the activation loop, although hAL is still present. Y399, an anticipated ligand to the phosphoserine (Figure 1C,D), also shows disorder (Figure 3A). However, suc-pWNK1 is still not identical to active kinases.
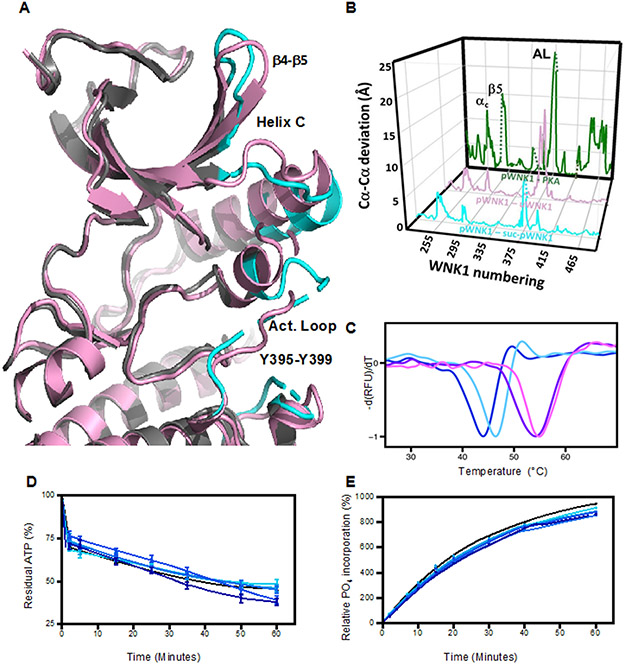
Figure 3.
Effects of sucrose on pWNK1 and pWNK3. (A) Overlay of the AMP-PNP complexed pWNK1 structure in the presence (magenta) and absence (gray) of sucrose (0.6 M). Activation loop, helix C and β4-β5 of suc-pWNK1 are shown in cyan. (B) Placement of conformational changes along the sequence (WNK1 numbering) between pWNK1 and PKA (green), pWNK1 and uWNK1 (pink), and pWNK1 and suc-pWNK1 with 0.6 M sucrose (cyan). (C) DSF melt temperatures (Tm) of pWNK1 in the absence (blue) and presence (cyan) of 0.6 M sucrose and pWNK3 in the absence (purple) and presence (violet) of 0.6 M sucrose. Note that the DSF conditions differ from the crystallographic study in A. * Trace is the negative first derivative of fluorescence from Sypro orange. Note that sucrose stabilizes pWNK1 and pWNK3. (D) Progress curves for the activity of pWNK1 on gOSR1 in increasing sucrose concentrations (0 mM, 50 mM, 100 mM, 200 mM, 400 mM, and 600 mM sucrose shown in colors shifting from black to cyan) tracking the disappearance of ATP using Kinase-Glo®. (E) Progress curves as in (D) following the incorporation of 32P into total protein (4 μM pWNK1 and 40 μM gOSR1) using [γ-32]P ATP with increasing sucrose concentrations (same concentrations and coloring as in (D)). Representative data from three replicates shown in (C). Progress curves in (D) reflect 5 simultaneous experiments, each having 3 independent replicates. Error bars are standard error. Panel E shows 5 simultaneous experiments, with autoradiography shown in Figure S2A. * “Additional DSF experiments with Mg+2 and AMP-PNP added as controls could not be conducted at submission on account of the COVID-19 lockdown.”
The 0.6 M sucrose diminished the size of the largest cavity (Table S2). The size of the remaining cavities could not be estimated due to disorder. Suc-pWNK1 has less bound water by a factor of two (Table S1). The waters shown in Figure 2 near hAL and the V-shaped linker remained. In general, disorder and other factors, such as B-factor, affect the number of waters reported in crystal structures 38. Nevertheless, since the resolution of the pWNK1 and suc-pWNK structures is comparable, we think this reduction in bound water is meaningful.
Conformational changes among pWNK and PKA structures were analyzed as a function of sequence in Figure 3B (pWNK1 vs sucrose-pWNK in cyan, uWNK1 vs pWNK1, in red, and pWNK1 vs PKA in green). Sucrose-induced structural changes (cyan) occur in the same locations as changes seen between pWNK1 and uWNK1 (red). Interestingly, the comparison of pWNK1 to PKA (green) also show changes in the same locations. Thus we speculate that the sucrose-induced effects may mimic activation.
Because soaking in sucrose induced activation loop rearrangement, we asked if sucrose affects pWNK1 stability and activity. Differential Scanning Fluorimetry (DSF) of 5 μM pWNK1 in the presence or absence of 0.6 M sucrose showed that the sucrose increased pWNK1 melt temperature (Tm) by 3°C and by 1.5°C for pWNK3, which is consistent with sucrose-induced structural changes (Figure 3C). We also asked if sucrose affects in vitro pWNK activity using pWNK1. A series of progress curves with increasing sucrose concentrations was measured. Kinase-Glo® tracking ATP consumption in the presence of gOSR1 (Figure 3D) and [γ −32P] measurements tracking phosphate incorporation, also in the presence of gOSR1 (Figure 3E, autoradiography in Figure S2A), both showed little sucrose-induced phosphotransfer.
Biochemical effects of PEG400 on pWNK1 and pWNK3
We have shown that PEG400, a stronger osmolyte than sucrose 22, 23, affect the activity of the unphosphorylated forms of WNK1 and WNK3 (data submitted for publication). We tested whether PEG400 could change the stability or activity of pWNK1 and pWNK3, and found different effects on stability and enhanced ATP consumption. pWNK3 has a higher basal stability than pWNK1, and both are destabilized by 15% PEG400, −3°C for pWNK1 and −1.5°C for pWNK3 (Figure 4A). Multiple simultaneous progress curves for ATP depletion as a function of PEG400 were measured for pWNK1 and pWNK3 with the substrate gOSR1. For both pWNK1 (Figure 4B) and pWNK3 (Figure 4C), ATP consumption increases with PEG400 concentration. Progress curves using 15% PEG400 were triplicated (Figure 4D, E). PEG400 increased pWNK1 ATP consumption by about 30% over 60 min. (Figure 2D).
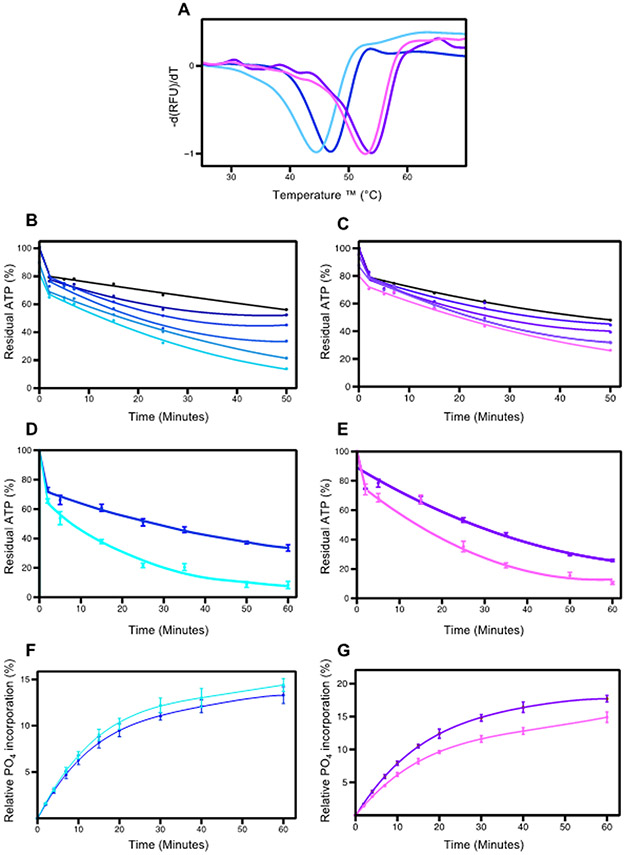
Figure 4.
Effects of PEG400 on pWNK1 and pWNK3. (A) DSF Melt temperatures (Tm) of pWNK1 (blue/cyan) and pWNK3 (purple/violet) with and without 15% PEG400. Note the reduced thermal stability in PEG400. (B) Progress curves for the activity of pWNK1 on gOSR1 in increasing PEG400 concentrations (0 mM, 50 mM, 100 mM, 200 mM, 400 mM, 600 mM in shades of blue from black to cyan) tracking the disappearance of ATP using Kinase-Glo®. (C) Progress curves for pWNK3 activity, PEG400 concentrations as in (B) with colors shifting from black to violet with increasing sucrose. (D) Progress curves for the activity of pWNK1 on gOSR1 with (cyan) and without (blue) 15%PEG400, tracking the disappearance of ATP using Kinase-Glo®. (E) Progress curves for the activity of pWNK3 on gOSR1 with (violet) and without (purple) 15%PEG400, as in (D). (F) Progress curves for the activity of pWNK1 on gOSR1 with (cyan) and without (blue) 15%PEG400, following the incorporation of 32P into total protein. Autoradiography in Figure S2B. (G) Progress curves for the activity of pWNK3 on gOSR1 with (violet) and without (purple) 15%PEG400, as in (F) Autoradiography in Figure S2C. Representative data from three replicates shown in (A). Panels B and C represent 5 simultaneous experiments. Panels D, E, F, and G reflect data from 3 experimental replicates. Error bars are standard error.
PEG400 increased pWNK3 ATP consumption by about 20% over the same time frame. In comparison, two active protein kinases in the MAPK pathway, MEK6/DD and TAO2, were evaluated for PEG400 effects on activity toward p38a and MBP, respectively; no increases in ATP consumption were observed (Figure S3).
To check whether the ATP depletion presented in Figures 4 B-E correspond to protein phosphorylation, autoradiography was used to monitor 32P incorporation into gOSR1 (Figures 4F and G, autoradiography in S2A and S2B). Contrary to our expectations, for pWNK1, the 32P incorporation into gOSR1 did not show a significant change in the presence of PEG400. For pWNK3, the 32P incorporation did not increase, but actually was reduced, suggesting that PEG400 inhibited activity toward gOSR1. PEG400 did not affect blotting efficiency of either pWNK1 or gOSR1 (data not shown). The ATP depletion and lack of 32P incorporation into protein raised the question of whether PEG400 was being phosphorylated. However, mass spectrometric analysis of the initial PEG400 reagent and PEG400 from a pWNK1 phosphorylation assay revealed identical spectra in both samples (Figure S4). These results imply that PEG400 may induce ATP hydrolysis by pWNK1 and pWNK3, but does not appear to induce downstream protein phosphorylation of gOSR1.”
DISCUSSION:
Our high-resolution structure of pWNK1 shows that while phosphorylated, pWNK1 reveals hallmarks of inactive kinases, including a refolded activation loop, displaced helix C and the presence of helix AL. Well-resolved electron density of the phosphoserine pS382 and the activation loop, as well as specific and unique interactions of conserved residues, provides evidence that the crystallographically observed pWNK1 conformer is likely stable in solution. The present pWNK1 structure thus may represent an intermediate state in WNK kinase activation. Work from our lab, and others, show multistage activation of protein kinases. We have observed these effects in the MAPKs ERK2 39, 40 and p38 MAP kinase 41, 42, and in the MAP2K MEK6 43. The Alex Bullock group has deposited structures of mono- and di- phosphorylated WNK3 (PDB files 5O23 and 5O26). PDB file 5O26 reveals a fully active configuration; PDB file 5O23, like the present pWNK1 structure, is not fully active. These structures represent a mounting body of evidence for multistage kinase activation. The functions of these intermediates may be to confer nuanced regulation of activity or pathway specificity.
We have presented data elsewhere suggesting a possible trans-autophosphorylation mechanism for WNK3 (in review) which is supported by the earlier work of Thastrup et al 2012. The kinetics of WNK multi-stage activation requires additional investigation.
Since WNK kinases are activated by osmotic stress in cells, and we have recently shown that uWNK1 (unphosphorylated) is activated by PEG400, we questioned whether pWNKs are affected by osmolytes or crowding agents. We expected osmolytes would enhance pWNK activity and observed increased ATPase activity, but not increased substrate phosphorylation. However, the suc-pWNK1 structure confirms that the pWNK1 structure is altered by an osmolyte. Interestingly, the location of sucrose-induced changes align with the differences in structure between pWNKs and fully active kinases, nudging the conformation more towards a canonicaly active state. The suc-pWNK1 structure also showed smaller cavities and reduced amounts of bound water, suggestive of a direct demand on solvent by sucrose. Thus, a potential mechanism for osmolyte-induced changes is an equilibrium between water-rich and water-poor conformers (see graphical abstract). We observed similar reduction in water when comparing uWNK1 with pWNK1 (submitted for publication). We look forward to more structural data on the effect of osmolytes, mimics on cellular osmotic stress, on other osmoregulated signaling proteins.
CONCLUSIONS
We have used crystallography and biochemical studies to investigate a novel phosphorylation intermediate in WNK1 activation. We observed more bound water in cavities not typically found in canonical kinase structures. This led us to explore the effects of osmotic stress on WNK1 structure and activation. In the presence of sucrose, pWNK1 structure exhibited distinct conformational changes, disorder, reduction in cavity size, and loss of water. Adding the crowding agent PEG400 to WNKs induced ATPase activity in both pWNK1 and pWNK3. We posit that WNKs are the expected protein kinase osmosensor regulating NKCC and NCC activation, making WNKs key players in renal function.
Supplementary Material
Table S1 Data statistics and refinement for pWNK1 and suc-pWNK1.
Table S2 Comparison of cavities volumes for uWNK1, pWNK1, and suc-pWNK1.
Figure S1 Electron density for ANP-PNP, pS382, and comparison to uWNK1.
Figure S2 WNK assay audioradiography
Figure S3 PEG400 effects on control kinases
Figure S4 Mass spectrometry of PEG400
ACKNOWLEDGMENTS
We thank Melanie Cobb and Clint Taylor for the gift of gOSR1 expression plasmid. Results shown in this report are derived in part from work performed at Argonne National Laboratory, Structural Biology Center (SBC) at the Advanced Photon Source. SBC is operated by the U. S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. We thank Zhe Chen and Diana Tomchick for help with organizing APS data collection.
Funding Sources
This research was supported by the American Heart Association (14GRNT20500035 (EJG) and 16CSA28530002 (EJG), NIH DK110358 (EJG) and the Welch Foundation (I1128, EJG).
ABBREVIATIONS
- WNK
With No Lysine(K) kinase
- pWNK1
the kinase domain of WNK1 phosphorylated as expressed in bacteria
- pWNK3
the kinase domain of WNK3 phosphorylated as expressed in bacteria
- uWNK1
the kinase domain of WNK1 dephosphorylated with phosphatases
- OSR1
oxidative stress responsive-1 kinase
- cdk2
cyclin dependent protein kinase 2
- IRAK
interlukin-1 associated kinase
- AMP-PNP
adenylyl-imido-diphosphate
- NCC
Na+Cl− NCC cotransporters
- NKCCs
Na+K+2Cl− cotransporter
Footnotes
Accession Codes
rat WNK1, NP_001002823.2; human WNK3, NP_065973.2; OSR1, O95747.1; rat MEK6, AAH87004.1; rat TAO2, NP_073193.1
pWNK1, PDB file 5W7T; uWNK1/SA, PDB file 6CN9; Suc-pWNK1 coordinates to be deposited
The atomic coordinates and structure factors for the structure described in this study have been deposited to the RCSB PDB (www.rcsb.org) with accession number 5W7T.
REFERENCES
- [1].Xu B, English JM, Wilsbacher JL, Stippec S, Goldsmith EJ, and Cobb MH (2000) WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II, J Biol Chem 275, 16795–16801. [DOI] [PubMed] [Google Scholar]
- [2].Wilson FH, Disse-Nicodeme S, Choate KA, Ishikawa K, Nelson-Williams C, Desitter I, Gunel M, Milford DV, Lipkin GW, Achard JM, Feely MP, Dussol B, Berland Y, Unwin RJ, Mayan H, Simon DB, Farfel Z, Jeunemaitre X, and Lifton RP (2001) Human hypertension caused by mutations in WNK kinases, Science 293, 1107–1112. [DOI] [PubMed] [Google Scholar]
- [3].Lytle C, and Forbush B 3rd. (1996) Regulatory phosphorylation of the secretory Na-K-Cl cotransporter: modulation by cytoplasmic Cl, Am J Physiol 270, C437–448. [DOI] [PubMed] [Google Scholar]
- [4].Lytle C, and Forbush B 3rd. (1992) Na-K-Cl cotransport in the shark rectal gland. II. Regulation in isolated tubules, Am J Physiol 262, C1009–1017. [DOI] [PubMed] [Google Scholar]
- [5].Piala AT, Moon TM, Akella R, He H, Cobb MH, and Goldsmith EJ (2014) Chloride sensing by WNK1 involves inhibition of autophosphorylation, Sci Signal 7, ra41, 1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Terker AS, Zhang C, Erspamer KJ, Gamba G, Yang CL, and Ellison DH (2016) Unique chloride-sensing properties of WNK4 permit the distal nephron to modulate potassium homeostasis, Kidney Int 89, 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lenertz LY, Lee BH, Min X, Xu BE, Wedin K, Earnest S, Goldsmith EJ, and Cobb MH (2005) Properties of WNK1 and implications for other family members, J Biol Chem 280, 26653–26658. [DOI] [PubMed] [Google Scholar]
- [8].Zagorska A, Pozo-Guisado E, Boudeau J, Vitari AC, Rafiqi FH, Thastrup J, Deak M, Campbell DG, Morrice NA, Prescott AR, and Alessi DR (2007) Regulation of activity and localization of the WNK1 protein kinase by hyperosmotic stress, Journal of Cell Biology 176, 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Moriguchi T, Urushiyama S, Hisamoto N, Iemura S, Uchida S, Natsume T, Matsumoto K, and Shibuya H (2005) WNK1 regulates phosphorylation of cation-chloride-coupled cotransporters via the STE20-related kinases, SPAK and OSR1, J Biol Chem 280, 42685–42693. [DOI] [PubMed] [Google Scholar]
- [10].Shekarabi M, Zhang J, Khanna AR, Ellison DH, Delpire E, and Kahle KT (2017) WNK Kinase Signaling in Ion Homeostasis and Human Disease, Cell Metab 25, 285–299. [DOI] [PubMed] [Google Scholar]
- [11].de Los Heros P, Pacheco-Alvarez D, and Gamba G (2018) Role of WNK Kinases in the Modulation of Cell Volume, Curr Top Membr 81, 207–235. [DOI] [PubMed] [Google Scholar]
- [12].Yamada K, Park HM, Rigel DF, DiPetrillo K, Whalen EJ, Anisowicz A, Beil M, Berstler J, Brocklehurst CE, Burdick DA, Caplan SL, Capparelli MP, Chen G, Chen W, Dale B, Deng L, Fu F, Hamamatsu N, Harasaki K, Herr T, Hoffmann P, Hu QY, Huang WJ, Idamakanti N, Imase H, Iwaki Y, Jain M, Jeyaseelan J, Kato M, Kaushik VK, Kohls D, Kunjathoor V, LaSala D, Lee J, Liu J, Luo Y, Ma F, Mo R, Mowbray S, Mogi M, Ossola F, Pandey P, Patel SJ, Raghavan S, Salem B, Shanado YH, Trakshel GM, Turner G, Wakai H, Wang C, Weldon S, Wielicki JB, Xie X, Xu L, Yagi YI, Yasoshima K, Yin J, Yowe D, Zhang JH, Zheng G, and Monovich L (2016) Small-molecule WNK inhibition regulates cardiovascular and renal function, Nat Chem Biol 12, 896–898. [DOI] [PubMed] [Google Scholar]
- [13].Chen L, Jenjaroenpun P, Pillai AM, Ivshina AV, Ow GS, Efthimios M, Zhiqun T, Tan TZ, Lee SC, Rogers K, Ward JM, Mori S, Adams DJ, Jenkins NA, Copeland NG, Ban KH, Kuznetsov VA, and Thiery JP (2017) Transposon insertional mutagenesis in mice identifies human breast cancer susceptibility genes and signatures for stratification, Proc Natl Acad Sci U S A 114, E2215–E2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Begum G, Yuan H, Kahle KT, Li L, Wang S, Shi Y, Shmukler BE, Yang S-S, Lin S-H, Alper SL, and others. (2015) Inhibition of WNK3 Kinase Signaling Reduces Brain Damage and Accelerates Neurological Recovery After Stroke, Stroke 46, 1956−-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ross PD, and Minton AP (1977) Analysis of non-ideal behavior in concentrated hemoglobin solutions, J Mol Biol 112, 437–452. [DOI] [PubMed] [Google Scholar]
- [16].Zhou HX, Rivas G, and Minton AP (2008) Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences, Annu Rev Biophys 37, 375–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Parsegian VA, Rand RP, and Rau DC (1994) Macromolecules and water: probing with osmotic stress., Methods in enzymology 259, 43–94. [DOI] [PubMed] [Google Scholar]
- [18].Akabayov SR, Akabayov B, Richardson CC, and Wagner G (2013) Molecular crowding enhanced ATPase activity of the RNA helicase eIF4A correlates with compaction of its quaternary structure and association with eIF4G, J Am Chem Soc 135, 10040–10047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhou X, Naguro I, Ichijo H, and Watanabe K (2016) Mitogen-activated protein kinases as key players in osmotic stress signaling, Biochim Biophys Acta 1860, 2037–2052. [DOI] [PubMed] [Google Scholar]
- [20].Foo YH, Gao Y, Zhang H, and Kenney LJ (2015) Cytoplasmic sensing by the inner membrane histidine kinase EnvZ, Progress in biophysics and molecular biology 118, 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang LC, Morgan LK, Godakumbura P, Kenney LJ, and Anand GS (2012) The inner membrane histidine kinase EnvZ senses osmolality via helix-coil transitions in the cytoplasm, EMBO J 31, 2648–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Mikles DC, Bhat V, Schuchardt BJ, McDonald CB, and Farooq A (2015) Effect of osmolytes on the binding of EGR1 transcription factor to DNA, Biopolymers 103, 74–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Money NP (1989) Osmotic Pressure of Aqueous Polyethylene Glycols : Relationship between Molecular Weight and Vapor Pressure Deficit, Plant Physiol 91, 766–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sheffield P, Garrard S, and Derewenda Z (1999) Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors, Protein expression and purification 15, 34–39. [DOI] [PubMed] [Google Scholar]
- [25].Min XS, Lee BH, Cobb MH, and Goldsmith EJ (2004) Crystal structure of the kinase domain of WNK1, a kinase that causes a hereditary form of hypertension, Structure 12, 1303–1311. [DOI] [PubMed] [Google Scholar]
- [26].Taylor C. A. t., An SW, Kankanamalage SG, Stippec S, Earnest S, Trivedi AT, Yang JZ, Mirzaei H, Huang CL, and Cobb MH (2018) OSR1 regulates a subset of inward rectifier potassium channels via a binding motif variant, Proc Natl Acad Sci U S A 115, 3840–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Humphreys JM, Piala AT, Akella R, He H, and Goldsmith EJ (2013) Precisely ordered phosphorylation reactions in the p38 mitogen-activated protein (MAP) kinase cascade, Journal of Biological Chemistry 288, 23322–23330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhou T, Raman M, Gao Y, Earnest S, Chen Z, Machius M, Cobb MH, and Goldsmith EJ (2004) Crystal structure of the TAO2 kinase domain: activation and specificity of a Ste20p MAP3K, Structure 12, 1891–1900. [DOI] [PubMed] [Google Scholar]
- [29].Racker E (1991) Use of synthetic amino acid polymers for assay of protein-tyrosine and protein-serine kinases, Methods Enzymol 200, 107–111. [DOI] [PubMed] [Google Scholar]
- [30].Pantoliano MW, Petrella EC, Kwasnoski JD, Lobanov VS, Myslik J, Graf E, Carver T, Asel E, Springer BA, Lane P, and Salemme FR (2001) High-density miniaturized thermal shift assays as a general strategy for drug discovery, J Biomol Screen 6, 429–440. [DOI] [PubMed] [Google Scholar]
- [31].Minor W, Cymborowski M, Otwinowski Z, and Chruszcz M (2006) HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes, Acta Crystallogr D Biol Crystallogr 62, 859–866. [DOI] [PubMed] [Google Scholar]
- [32].Evans PR, and Murshudov GN (2013) How good are my data and what is the resolution?, Acta Crystallogr D Biol Crystallogr 69, 1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Cowtan K, Emsley P, and Wilson KS (2011) From crystal to structure with CCP4, Acta Crystallogr D Biol Crystallogr 67, 233–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Emsley P, and Cowtan K (2004) Coot: model-building tools for molecular graphics, Acta Crystallogr D Biol Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- [35].Yu J, Zhou Y, Tanaka I, and Yao M (2010) Roll: a new algorithm for the detection of protein pockets and cavities with a rolling probe sphere, Bioinformatics 26, 46–52. [DOI] [PubMed] [Google Scholar]
- [36].Jura N, Zhang X, Endres NF, Seeliger MA, Schindler T, and Kuriyan J (2011) Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms, Molecular cell 42, 9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Huse M, and Kuriyan J (2002) The conformational plasticity of protein kinases, Cell 109, 275–282. [DOI] [PubMed] [Google Scholar]
- [38].Carugo O (2017) Protein hydration: Investigation of globular protein crystal structures, Int J Biol Macromol 99, 160–165. [DOI] [PubMed] [Google Scholar]
- [39].Zhou T, Sun L, Humphreys J, and Goldsmith EJ (2006) Docking interactions induce exposure of activation loop in the MAP kinase ERK2, Structure 14, 1011–1019. [DOI] [PubMed] [Google Scholar]
- [40].Taylor C. A. t., Cormier KW, Keenan SE, Earnest S, Stippec S, Wichaidit C, Juang YC, Wang J, Shvartsman SY, Goldsmith EJ, and Cobb MH (2019) Functional divergence caused by mutations in an energetic hotspot in ERK2, Proc Natl Acad Sci U S A 116, 15514–15523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Akella R, Min X, Wu Q, Gardner KH, and Goldsmith EJ (2010) The third conformation of p38alpha MAP kinase observed in phosphorylated p38alpha and in solution, Structure 18, 1571–1578. [DOI] [PubMed] [Google Scholar]
- [42].Chang CI, Xu BE, Akella R, Cobb MH, and Goldsmith EJ (2002) Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b, Mol Cell 9, 1241–1249. [DOI] [PubMed] [Google Scholar]
- [43].Min X, Akella R, He H, Humphreys JM, Tsutakawa SE, Lee SJ, Tainer JA, Cobb MH, and Goldsmith EJ (2009) The structure of the MAP2K MEK6 reveals an autoinhibitory dimer, Structure 17, 96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Data statistics and refinement for pWNK1 and suc-pWNK1.
Table S2 Comparison of cavities volumes for uWNK1, pWNK1, and suc-pWNK1.
Figure S1 Electron density for ANP-PNP, pS382, and comparison to uWNK1.
Figure S2 WNK assay audioradiography
Figure S3 PEG400 effects on control kinases
Figure S4 Mass spectrometry of PEG400