Abstract
Objective
To assess the role of STAT4 activation in driving pathogenic follicular helper T (Tfh) cell secretion of the cytokines IL-21 and IFN-γ during murine and human lupus.
Methods
We temporally assessed STAT4-dependent Tfh cell signaling with cytokine production and autoreactive B cell maturation during the course of murine lupus, with further assessment of Tfh cell gene transcription using RNA-seq. STAT4-dependent signaling and cytokine production were also determined in circulating Tfh-like cells in patients with SLE, compared to cells from controls, with correlation to disease activity in the former.
Results
IL-21 and IFN-γ co-producing Tfh cells expanded prior to the detection of potentially pathogenic IgG2c autoantibodies in lupus-prone mice. Tfh cells transcriptionally evolved during the course of disease with acquisition of a STAT4-dependent gene signature. Maintenance of Tfh cell cytokine synthesis was dependent upon STAT4 signaling, driven by type I interferons. Circulating Tfh-like (cTfh) cells from patients with SLE also secreted IL-21 and IFN-γ, with STAT4 phosphorylation enhanced by IFN-β, in association with clinical disease activity.
Conclusion
We identified a role for IFN-I signaling in driving STAT4 activation and production of IL-21 and IFN-γ by Tfh cells in murine and human lupus. Enhanced STAT4 activation in Tfh cells may underlie pathogenic B cell responses in both murine and human lupus. These data indicate that STAT4 guides pathogenic cytokine and immunoglobulin production in SLE, providing a potential therapeutic target to modulate autoimmunity.
Introduction
Systemic autoimmune diseases such as rheumatoid arthritis and systemic lupus erythematosus (SLE, lupus) are characterized by T cell activation driving the production of pathogenic class-switched autoantibodies (1). Activated CD4 T cells in SLE are correlated with autoantibody production and disease activity, suggesting their role in promoting disease (1, 2). An understanding of pathways driving heightened T cell activation in SLE are necessary to identify and define therapeutic targets.
Follicular helper T (Tfh) cells are a subset of CD4 T cells that promote B cell maturation in conventional and systemic autoimmune responses, and autoantibody production in SLE. Tfh cells deliver soluble signals, including IL-21 and IFN-γ, which promote B cell maturation with plasma cell formation and autoantibody production (3–6). Autoantibody production and disease activity in murine lupus are diminished in the absence of IL-21 or IFN-γ (7–9), but the specific effects of these cytokines in disease remain unknown. Conversely, the frequency of Tfh-like cells and amount of IL-21 in circulation are associated with autoantibody production and disease activity in SLE (1, 10). While IL-21 and IFN-γ are pathogenic in lupus, the regulation of these T-dependent cytokines in disease remains unclear.
Signal transducer and activator of transcription 4 (STAT4) and Tbx21 (encoding T-bet) can regulate cytokine production in Tfh cells (11). Tfh cells produce IFN-γ and IL-21 upon type 1 immune challenge, with Bcl6 promoting lineage differentiation and STAT4 and T-bet driving the IFN-γ and STAT4 required for IL-21 production(11). Expression of both these transcription factors declines as the GC response continues, although cytokine production continues with their gene loci remaining accessible (11, 12). STAT4 activation and signaling is driven by type 1 inflammatory cytokines IL-12 and IFN-I (13) in both Th1 and Tfh cells upon viral infection. Aberrant production of IL-12 and IFN-I is a hallmark of lupus, with these cytokines promoting GC persistence and production of pathogenic, inflammatory IgG autoantibodies (14–18). Thus, in chronic autoimmune settings, type 1 inflammatory cytokines may drive STAT4 and T-bet-mediated transcription of IL-21 or IFN-γ in Tfh cells.
STAT4 is strongly associated with disease susceptibility in SLE. The single nucleotide polymorphism (SNP) rs7574865 located within intron 3 in the STAT4 locus has been shown in genome-wide association studies (GWAS) to be a risk allele for SLE (19–21). A separate SNP, rs7574885T, enhances the risk for lupus susceptibility by increasing Stat4 transcription and augmenting responses to IL-12 and IFN-I signaling (21, 22). In MRLlpr/lpr lupus-prone mice, Stat4-specific antisense oligonucleotide therapy decreases the severity of lupus nephritis (23). Yet, the distinct contribution of STAT4 to lupus susceptibility remains unclear.
We hypothesized that STAT4 signaling, in response to ongoing cytokine stimuli in lupus, promotes dysregulated cytokine production by Tfh cells and aberrant B cell maturation. To address this hypothesis, we dissected the phenotype and function of Tfh cells in murine lupus by assessing their transcriptional landscape at differing stages of disease and determined their inflammatory cytokine response and that of human circulating Tfh-like cells in SLE. We found that Tfh cells are chronically expanded in murine lupus with co-production of IL-21 and IFN-γ driving pathogenic IgG2c autoantibody production. Maintenance of Tfh-cell cytokine synthesis was STAT4-dependent, and heightened by IFN-I, with a similar phenotype and cytokine response observed in circulating Tfh-like cells from patients with SLE. Thus, the STAT4 pathway is critical for the maintenance of pathogenic Tfh cell function in both murine and human lupus.
PATIENTS AND METHODS
Human PBMC Isolation and Analysis
Twenty-five SLE patients from ages 18 to 75 years and 19 age-matched healthy controls were enrolled. Disease activity was assessed with the SLE Disease Activity Index 2000 (SLEDAI2K). PBMCs were isolated using Ficoll-Paque density gradient media and rested in DMEM medium with 10% serum overnight. Phosphorylated STAT4 in cTfh cells and CD4+ T memory cells were analyzed as previously described (11). Isolated PBMCs were rested and then stimulated by IFN-β (1000 U/ml) or IL-12 (10 ng/ml) for 20 minutes in DMEM with 1% serum. Stimulated cells were immediately fixed with 4% paraformaldehyde for 15 min, then cold methanol was used to permeabilize the cells for staining by flow cytometry. Antibodies used for flow cytometry are listed (Supplemental Table 1). Informed consent was obtained from human subjects.
Mice
Mice were housed in pathogen-free conditions at the Yale School of Medicine or Rutgers New Jersey Medical School. C57BL/6J (B6), B6.Sle1.Yaa and MRLlpr/lprJ animals were purchased from Jackson Laboratory. All animals were used at 8–28 weeks of age, with approval for procedures given by The Institutional Animal Care and Use Committee of Yale University and Rutgers New Jersey Medical School.
ELISA for Autoantibodies and IFN-β
Anti-chromatin and anti-dsDNA were measured by ELISA using sonicated chicken chromatin and calf thymus dsDNA, respectively. Serum samples were tested at a 1:250 dilution followed by incubation with alkaline phosphatase-labeled goat anti-mouse IgG (1:1000 dilution) (Southern Biotechnology), and development with p-nitrophenylphosphate substrate (Sigma-Aldrich). Optical density at 405 nm was read using a VERSAmax microplate reader (Softmax Pro 3.1 software; Molecular Devices). Sera IFN-β concentrations were measured using the VeriKine-HS Mouse IFN Beta ELISA kit from PBL assay science following manufacture’s protocol.
In Vivo IFNAR-1 Blockade
4-month-old lupus-prone mice were injected twice a week i.p. with either 375 μg of anti-IFNAR1 (24) (MAR1–5A3 mAB, bioXCell) or 1 mg of anti-rat IgG isotype control (Sigma-Aldrich) for 4 weeks.
Flow Cytometry and Cell Sorting
Tissues were homogenized as previously described (11). Antibodies used for flow cytometry staining are listed (Supplemental Table 1). Intracellular staining for cytokines was performed using Cytofix/Cytoperm™ kits (BD Biosciences) following the manufacturer’s protocol. Stained and rinsed cells were analyzed using an LSRII Cytometer (BD Biosciences). For sorting, CD4+ T cells were enriched using a magnetic separation kit (EasySep™, StemCell Technologies) prior to cell surface staining, with specific populations sorted using a FACSAria™ (BD Biosciences).
Microscopy
Spleens were snap frozen in OCT tissue-freezing solution and stored at −80°C. Tissues were cut into 8μm sections and processed as described (25). Reagents used to stain sections are listed (Supplemental Table 2). Images were obtained from a laser-scanning confocal microscope (Model 510 META; Carl Zeiss) at 25x magnification. ImageJ software was used for the measurement of GC and B cell follicle size, distance measurements, and for T cell counting. The latter analyses were performed in a blinded manner.
RNA-Seq and Analysis
Tfh populations were sorted by flow cytometry, with two separate sorts performed on different days, pooling spleens of 3 mice each day. Quality verification, library preparation, and sequencing were performed at the Yale Center for Genome Analysis. Samples were sequenced on an Illumina HiSeq 2500 using 75-bp paired end reads. RNA-seq analysis was performed as described (11). in accordance with the NF-Core RNA-seq guidelines (version 1.4.2). Pathway enrichment analysis was performed using MetaCoretm, version 6.31 build. Enrichment analysis was performed using GSEA software v3.0 (Broad Institute). GSE156578
ATAC-seq
ATAC was performed as described (11). Sorted cells were lysed in buffer (10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, and 0.1% octylphenoxypolyethoxyethanol for 15 min at 4°C and then centrifuged and resuspended with transposase reaction mix (2× tagmented DNA buffer, transposase [Ilumina Nextera], and nuclease-free water) and incubated for 30 min at 37°C. Cells were then PCR amplified in KAPA HiFi 2× mix (Kapa Biosystems) with barcoding primers. Quantitative PCR library amplification test and PCR library amplification were performed as previously described (11). DNA processing and high-throughput sequencing were performed as described in the previous section. Sequenced reads were mapped to the mouse genome (mm10 NCBI Build 38) using the Burrows-Wheeler Aligner version 0.7.9a. Statistically significant differentially accessible regions were then identified (11).
Deposition of data
RNA sequencing and ATAC-seq data have been deposited into the GEO database (accession no GSE156578 and GSE156575)
Statistics
Data were analyzed using the Student’s t-test, unpaired t-test (Mann-Whitney), and Pearson correlation with Prism 8 (GraphPad Software). The number of asterisks represents the degree of significance with respect to p value, with the exact value presented within each figure legend. Pearson correlation coefficient is presented within the graph on each figure.
Results
Tfh cells develop prior to autoantibodies in murine lupus
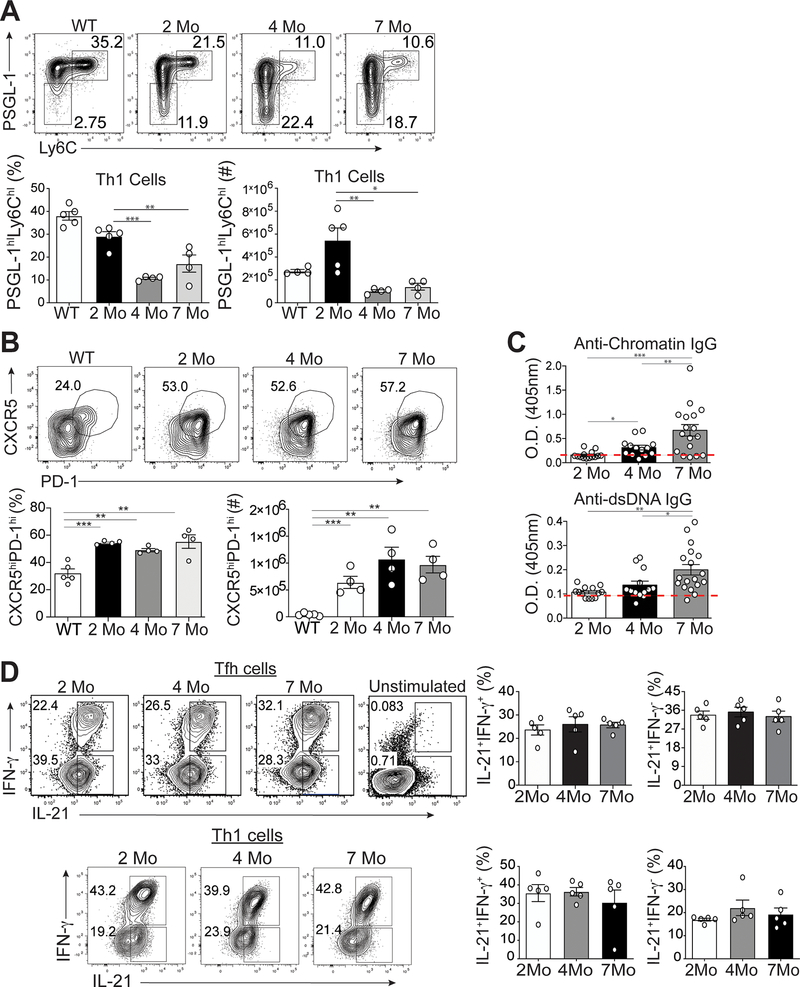
Tfh cells are associated with disease severity in both human and murine lupus (2). While Tfh cells promote autoantibody production in lupus-prone mice (25, 26), the kinetics of their development and expansion during disease progression are unknown. We accordingly analyzed splenic Tfh and Th1 cells over time in two murine lupus models, B6.Sle1.Yaa (aged 2, 4 and 7 months) and MRLlpr/lpr mice (aged 2 and 6 months), asking if they evolve phenotypically and genetically as lupus develops and persists. The numbers and percentages of CD4+CD44hiLy6chiPSGL-1hi Th1 cells of B6.Sle1.Yaa mice were reduced by age 4 months to 2 month-old and B6 control mice (Figure 1A) (11, 27). Conversely, the numbers and percentages of CD4+CD44hiLy6cloPSGL-1loCXCR5hiPD-1hi Tfh cells of B6.Sle1.Yaa mice were similarly increased among all 3 ages compared to control mice (Figure 1B). There were no significant differences in percentages and numbers of splenic CD4+CD44hiLy6cloPSGL-1lo Tfh cells in differently aged MRL lupus-prone mice (Supplemental Figure 1A) (25, 28).
Figure 1.
Temporal formation of Tfh and Th1 cells during different stages of disease. Spleens were harvested from either B6.Sle1.Yaa at 2, 4, and 7 months of age or control 2 months of age wild type B6 mice. (A) Representative flow cytometry plots of Ly6chiPSGL-1hi Th1 cells from B6.Sle1.Yaa mice with percentages and numbers of cells (bottom left and right, respectively) (B) Representative flow cytometry plots of CXCR5hiPD-1hi Tfh cells from B6.Sle1.Yaa mice with percentages and numbers cells (bottom left and right, respectively). (C) Anti-Chromatin IgG (left), and Anti-IgG antibodies (right) in sera of mice B6.Sle1.Yaa at 2, 4, and 7 months of age, with a dashed line representing the average O.D. from 6-month-old wild type B6 mice. (D) Intracellular IL-21 and IFN-γ staining in Tfh cells (top) and Th1 cells (bottom) with percentages (right) of cytokine-positive cells. Cytokine positive cells gates were based on unstimulated Tfh cells (top right). Data are representative of 3 experiments with 3–5 mice per group. ***p < 0.001; **p < 0.01; *p < 0.05 by Student’s t-test. Error bars represent SEM.
To determine if Tfh or Th1 cells were linked with the onset of autoantibody production, we examined anti-chromatin and anti-dsDNA production in B6.Sle1.Yaa mice at ages 2, 4 and 7 months. Although Tfh and Th1 cells were present and expanded in 2-month-old compared to B6 mice, we found no differences in anti-chromatin or anti-dsDNA autoantibodies (Figure 1C). Hence, we will refer to the 2-month time point as the pre-disease stage. While Tfh cells persisted and Th1 cells declined by age 4 months, referred to here as the onset of disease since their autoantibodies were significantly increased compared to those in pre-disease mice; yet were less than those in 6-month-old mice, with the latter referred to as diseased. Together, these data showed that Tfh, and not Th1, cell expansion precedes autoantibody production.
IL-21 and IFN-γ production is maintained as disease progresses
Tfh and Th1 cells co-produce IL-21 and IFN-γ upon acute viral infection of non-autoimmune mice (11). We thus asked whether the temporal onset of autoantibodies was due to varied production of Tfh cell-specific IL-21 and IFN-γ over the course of disease, examining Tfh and Th1 cytokines from B6.Sle1.Yaa mice at 2, 4 and 7 months of age. There were comparable percentages of IL-21+IFN-γ+ and IL-21+IFN-γ-−secreting Tfh and Th1 cells at the three timepoints, as autoantibodies expanded (Figure 1D). Likewise, Tfh cells from 2- and 6-month-old MRL mice maintained cytokine production as disease progressed (Supplemental Figure 1B). These data suggested that IL-21 and IFN-γ-producing Tfh and Th1 cells develop prior to the onset of autoantibodies, yet Tfh, but not Th1, cells are maintained chronologically.
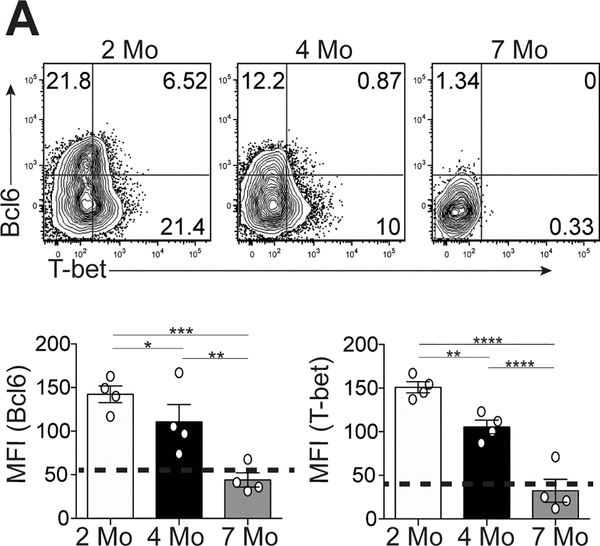
Bcl6 and T-bet expression is temporally decreased in Tfh cells
Given the co-production of Tfh cell cytokines in B6.Sle1.Yaa mice over time (Figure 1D), we examined their transcription factors Bcl6 and T-bet. Tfh cells from 4- and 7-month-old lupus mice had significant decrease in Bcl6 and T-bet expression compared to cells from 2-month-old lupus mice (Figure 2A). Bcl6 and T-bet expression continued to decrease in Tfh cells from 7-month-old compared to cells from 4-month-old lupus mice (Figure 2A). We observed similar decreased expression of Bcl6 and T-bet in Tfh cells from MRL mice, as well as Th1 cells in B6.Sle1.Yaa mice at 6-months compared to 2-months of age (Supplemental Figure 1C and D). Thus, Tfh cells in lupus continually produced pathogenic IL-21 and IFN-γ, despite losing expression of T-bet and Bcl6.
Figure 2.
Temporal expression of T-bet and Bcl6 in Tfh cells in lupus-prone mice. Spleens were harvested from either B6.Sle1.Yaa at 2, 4, and 7 months of age. (A) Representative flow cytometry plots of intracellular Bcl6 and T-bet in splenic Ly6cloPSGL-1loCXCR5hiPD-1hi Tfh cells, with bar graphs that summarize the quantified geometric mean fluorescence intensity of staining (MFI) of Bcl6 (left) or T-bet (right). Data are representative of 3 experiments with 3–5 mice per group. ****p < 0.0001; ***p < 0.001; **p < 0.01; *p < 0.05 by Student’s t-test. Error bars represent SEM.
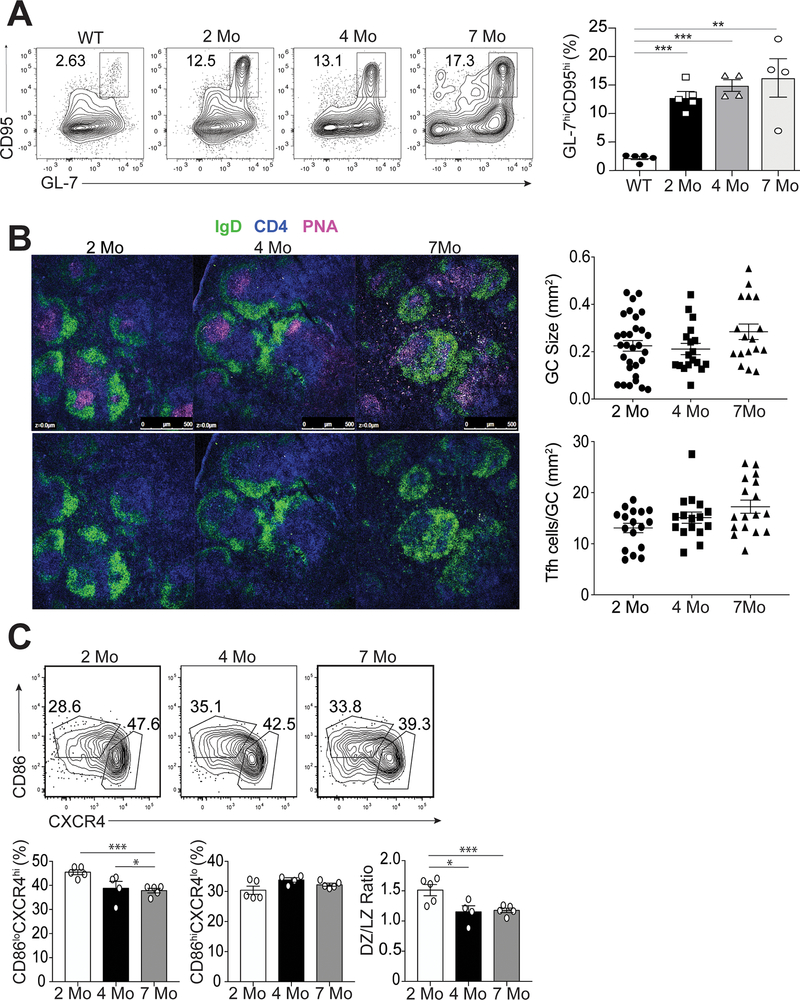
Germinal centers develop prior to the onset of disease
Autoreactive B cells in lupus undergo antigen selection within the germinal center (GC) (29). Since Tfh cells are generated at the pre-disease stage prior to the onset of autoantibodies (Figure 1B and 1C), we assessed temporal generation of GC B cells in 2, 4 and 7 month-old B6.Sle1.Yaa mice. The percentages of splenic B220+IgDloCD95hiGL-7hi GC B cells were similarly increased among all 3 ages compared to cells from non-autoimmune control mice at all three timepoints (Figure 3A). Moreover, the number and size of GCs did not differ over time (Figure 3B) with similar numbers of Tfh cells located within the GCs (Figure 3B). Tfh cell signaling to GC B cells is necessary for dark zone to light zone transition (30). While the total number of GC B cells did not change over time, the percentage of B220+IgDloCD95hiGL-7hiCXCR4hiCD86lo dark zone GC B cells at ages 4 and 7 months significantly decreased compared to that of mice at 2 months of age (Figure 3C). However, the percentage of B220+IgDloCD95hiGL-7hiCXCR4loCD86hi light zone GC B cells remained unchanged throughout (Figure 3C). Thus, Tfh cells persist within the GC, but dysregulation of GC B cell subsets only occurred later during disease.
Figure 3.
Characterization of GC B cells during the progression of disease. Spleens were harvested from B6.Sle1.Yaa at 2, 4, and 7 months of age or control 2 months of age wild type B6 mice. (A) Representative flow cytometry plots of CD4-B220+IgDloCD95hiGL-7hi GC B cells at each time point with cell percentages (right). (B) Splenic B cell follicles of B6.Sle1.Yaa mice stained with anti-IgD (green), anti-CD4 (blue) and PNA (magenta), with GC sizes quantified (top right) and the numbers of T cells per GC size (bottom right). (C) Representative flow cytometry plots of dark zone (CD86-CXCR4+) and light zone (CD86+CXCR4-) GC B cells gated on CD4-B220+IgDloCD95hiGL-7hi GC B cells, with cell percentages and numbers (right). Data are representative of 3 experiments with 3–5 mice per group. ***p < 0.001; *p < 0.05 by Student’s t-test. Error bars represent SEM.
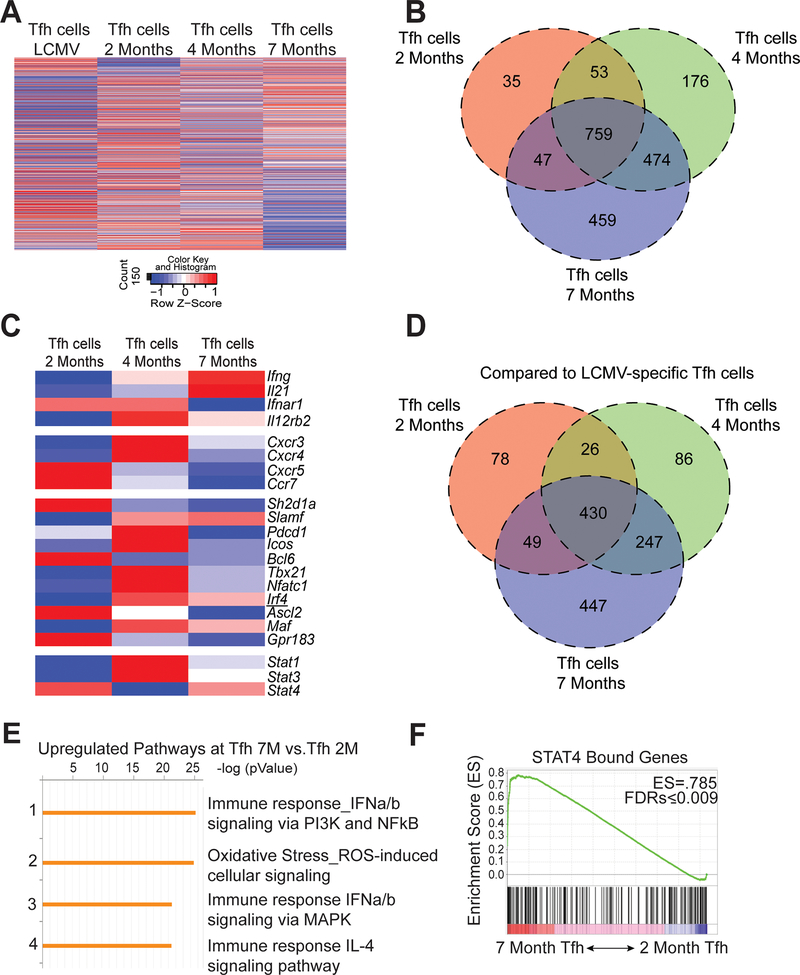
Temporal transcriptomic analysis of Tfh cells in lupus
During disease progression, Tfh cells maintained production of IL-21 and IFN-γ despite diminished transcription factor expression. We next assessed the temporal transcriptional profiles of sorted Tfh cells (CD4+CD44+GITR-PSGL-1loCXCR5hiPD-1hi) from 2, 4- and 7-month-old mice compared to naive CD4 T cells from B6.Sle1.Yaa mice. 732 genes were found to be differentially expressed in Tfh cells from 4-month compared to 2-month-old mice, with 1021 genes differentially expressed in cells from 7- compared to 2-month-old animals (Figure 4A and 4B). Next, we compared the differently expressed genes between the three disease stages in lupus and found that of the 1021 differentially expressed genes in Tfh cells from mice aged 2 and 7 months, 474 overlapped with genes that were already differentially expressed by 4 months of age (Figure 4B). To further analyze the transcriptional differences among the Tfh populations, we examined a curated set of genes previously described to be up- or down-regulated in Tfh cells compared to other CD4+ Th subsets (31). Despite the temporal increase in differentially expressed genes in Tfh cells, the three Tfh populations shared expression of Tfh-cell defining genes including Bcl6, Icos, and Pdcd1 (Figure 4C). Although we observed temporal decline of Bcl6, Tfh-cell defining gene expression was consistent with maintenance of CXCR5 and PD-1 expression (Supplemental Figure 2B). By comparison to Tfh cells from mice with acute LCMV infection, those from pre-diseased 2-month-old lupus-prone mice had the fewest differentially expressed genes compared to those from older lupus-prone mice (Figure 4D), indicating greater divergence in older lupus-prone mice from the physiological response to acute infection. Enrichment analysis of upregulated genes revealed increased expression of genes involved in pathways of type I interferon (IFN-I) signaling (Figure 4E). IFN-I promotes phosphorylation of STAT4, the gene of which is associated with lupus susceptibility and regulates IL-21 and IFN-γ production in acute viral infection of mice (11). Gene set enrichment analysis (GSEA) using a published STAT4 ChIP-seq data set (32) identified enrichment of STAT4-bound genes in Tfh cells from 2-month to 7-month-old lupus mice (Figure 4F). Hence, as disease progressed, Tfh cells developed an enhanced Stat4 gene signature.
Figure 4.
Evolving transcriptional profile of Tfh cells during the progression of disease. Spleens were harvested from B6.Sle1.Yaa at 2, 4, and 7 months of age or from LCMV infected mice 8 days post infection. (A) Heat map of normalized hit counts for differentially expressed genes at 7 or 2 months (rows) in Tfh cells (columns). (B) Venn diagram of differentially expressed genes comparing Tfh cells from 2, 4, and 7 months of age mice to 2-month naïve control. (C) Heatmap of selected T cell-related genes. * FDR-adjusted q < 0.05 comparing Tfh cells at 7 months or 2 months. (D) Venn diagram comparing differentially expressed genes of Tfh cells from 2, 4, and 7 months of age mice to Tfh cells from Day 8 LCMV. (E) Enrichment pathways analysis of the upregulated genes in Tfh cells at 7 months to 2 months, with the top four pathways listed, including the enrichment p value. (F) GSEA revealed a significant enrichment in Stat4-dependent genes at 7-Mo vs. 2-Mo in Tfh cells. Number in top-right corner is the enrichment score. Data are representative of 2 replicates with 3 mice per group.
STAT4 phosphorylation in murine lupus is promoted by IFN-I
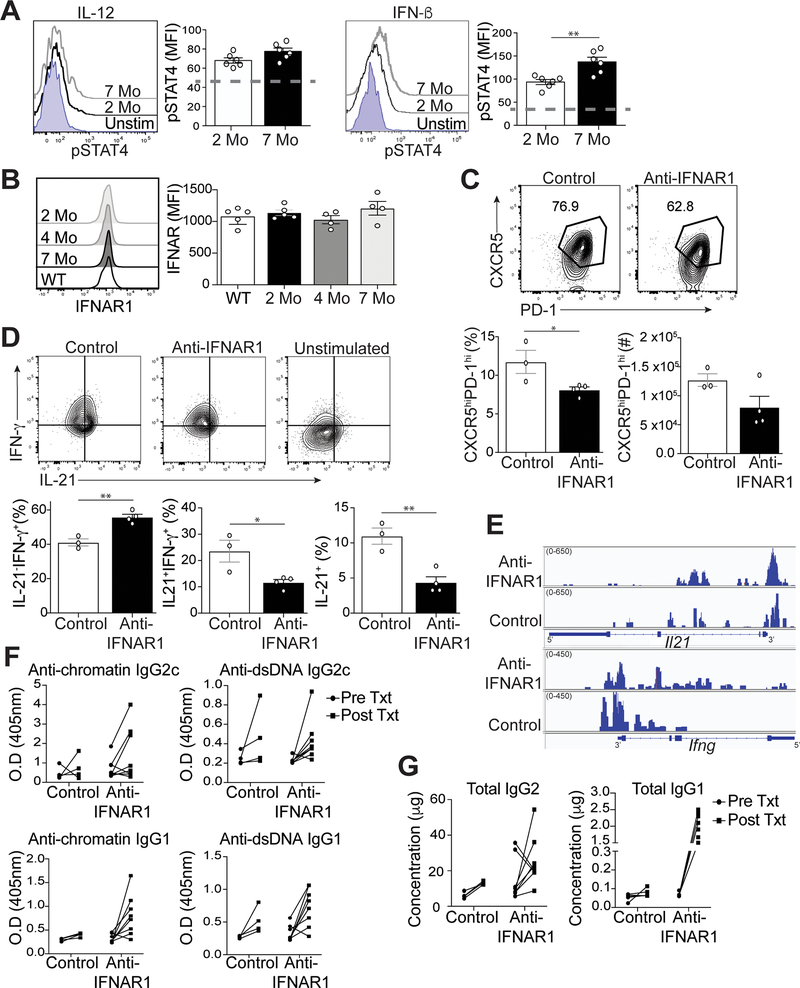
Inflammatory cytokines abundant in lupus, including IL-12 and IFN-I, drive STAT4 activation (14–16). While we found no difference in the temporal expression of Stat4 in Tfh cells from lupus-prone mice (Figure 4C), the expression of IL-12 and IFN-β in B6.Sle1.Yaa mice (33) compared to controls may promote phosphorylation of STAT4 (pSTAT4) in Tfh cells (11). We accordingly measured pSTAT4 in splenic Tfh cells at early and late stages of disease, upon stimulation with IL-12 or IFN-β. In response to IL-12 stimulation, Tfh cells from diseased 7-month-old lupus mice had modestly increased phosphorylated STAT4 compared to Tfh cells from 2-month-old mice (Figure 5A). In response to IFN-β stimulation, pSTAT4 was significantly increased in Tfh cells from 7-month-old compared to 2-month-old lupus mice. We next examined IL-12- or IFN-β-induced STAT4 activation in Th1 cells, finding significantly enhanced pSTAT4 in cells from 7-month-old compared to 2-month-old lupus mice (Supplemental Figure 3A). Thus, IL-12 and IFN-β more readily activated STAT4 in splenic Tfh and Th1 cells from the later disease stage compared to the pre-disease stage, which is consistent with the Tfh cell enhanced Stat4 gene signature (Figure 4F).
Figure 5.
IFN-I regulates Tfh cell-IL-21 and IFN-γ production in lupus. Spleens were harvested from B6.Sle1.Yaa mice at 2 and 7 months of age. (A) Splenocytes were stimulated with IL-12 or IFN-I. Representative flow histogram of pSTAT4 in Tfh cells was determined by intracellular staining (dashed line represents unstimulated control). (B) Representative flow cytometry plots of IFNAR1 on Tfh cells from B6.Sle1.Yaa mice with the MFI of IFNAR1 (right). (C) Splenocytes from 5-month B6.Sle1.Yaa mice treated twice a week for 4 weeks with either anti-IFNAR1 (375ug) or anti-rat IgG (1mg/mL). Representative flow cytometry plots of CXCR5hiPD-1hi Tfh cells from treated B6.Sle1.Yaa mice with percentages and numbers cells (bottom left and right, respectively). (D) Representative flow cytometry plots of intracellular IL-21 and IFN-γ staining in Tfh cells (top) with percentages of (bottom) of cytokine positive cells. Gates were based on unstimulated Tfh cells (top right). (E) Assay for transposase-accessible chromatin using sequencing (ATAC-seq) for Il21 and Ifng loci. (F) Anti-chromatin (left), anti-dsDNA antibodies (right), and (G) total Ig from sera of pre and post-treated mice. Data are representative of 2 experiments with 3–5 mice per group. **p < 0.01; *p < 0.05 by Student’s t-test. Error bars represent SEM.
Since IFN-I promoted increased activation of STAT4 in Tfh cells from lupus-prone mice, we next asked if the amount of IFN-I increased with disease progression, finding no difference in the concentration of circulating IFN-β between 2- and 7-month-old lupus mice (Supplemental Figure 3B). The temporal increase in pSTAT4 was also not due to a difference in the expression of the IFN-I receptor (IFNAR1) on Tfh cells, as its expression on cells from older mice was similar compared to that of 2 and 4 month-old mice (Figure 5B), suggesting that Tfh cells from older mice were more responsive to IFN-I.
To determine if IFN-I signaling was regulating IL-21 and IFN-γ production, we treated 4-month-old B6.Sle1.Yaa mice at the onset of disease for 4 weeks with either anti-IFNAR-1 or control rat IgG antibody. Treatment with anti-IFNAR1 resulted in a decrease in the percentage and numbers of Tfh cells compared to the control group (Figure 5C). Tfh cells from mice treated with anti-IFNAR1 had reduced IFN-γ or IL-21 secretion compared to the controls (Supplemental Figure 3C). Moreover, anti-IFNAR1 treated mice had significantly reduced percentages of IL-21+IFN-γ + and IL-21+IFN-γ- −secreting Tfh cells compared to the control group (Figure 5D).
To determine if the effect of IFNAR1 blockade on Tfh-cell production of IL-21 and IFN-γ was a consequence of altered chromatin accessibility, we used ATAC-seq to characterize their gene loci. Comparison of differentially called peaks in three replicates of Tfh cells from the two revealed no significant differences in chromatin accessibility at the Il21 and Ifng loci (Figure 5E). These findings suggested that in Tfh cells, Il21 and Ifng remain accessible in the absence of IFN-I signaling, yet cytokine production is not maintained.
In concert with the decreased Tfh-cell cytokine production in anti-IFNAR1 treated mice, we also observed enhanced IgG1 switching in autoantibodies and total immunoglobulins, compared to that in control animals (Figure 5F and 5G), albeit without an effect on GC B cell formation (Supplemental Figures 3D and E) or IgG2c antibodies and autoantibodies (Figure 5F). Overall, IFN-I signaling via STAT4 promoted IL-21 and IFN-γ production at onset of disease in lupus prone mice, with this effect enhanced as mice age.
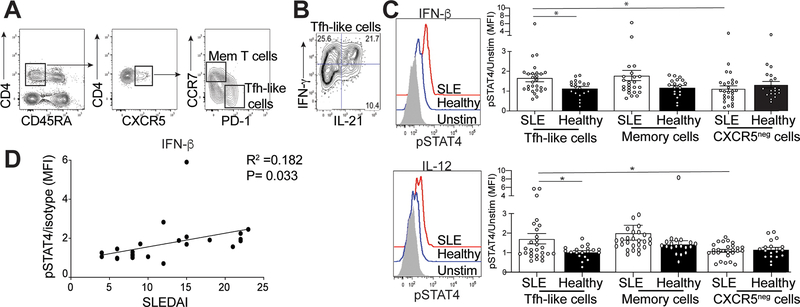
Activation of STAT4 in Tfh-like cells from SLE patients correlates with disease activity
The presence of autoantibodies in human lupus patients precedes clinical disease (34). cTfh (Tfh-like) cells from SLE patients correlate with disease severity and are phenotypically similar to tonsillar Tfh cells, but have reduced Bcl6 and T-bet expression, similar to Tfh cells in lupus-prone mice (2, 35–37). To extend our murine studies to SLE patients, we examined cytokine production in their cTfh-like cells. PBMCs were collected from 25 lupus patients and 19 age- and gender-matched healthy individuals (Supplemental Table 2). IFN-γ and IL-21 production was assessed in CD4+CD45RA-CCR7-PD-1hiCXCR5hi Tfh-like cells and CD4+CD45RA-CCR7+PD-lloCXCR5hi memory T cells (Figure 6A) (36). A substantial portion of both populations co-secreted IL-21 and IFN-γ, mirroring our data from lupus-prone B6.Sle1.Yaa and MRLlpr/lpr mice (Figures 6B, 1D and Supplemental Figure 1C). We next compared pSTAT4 expression in the two T cell populations following IFN-β or IL-12 cytokine stimulation, analogous to our experiments using murine Tfh cells (Figure 5A). We found no difference in pSTAT4 expression in lupus memory T cells compared to healthy controls after stimulation with IFN-β or IL-12 (Figure 6C); however, pSTAT4 expression in lupus cTfh-like cells was significantly increased compared to controls (Figure 6C). We also examined pSTAT4 following IFN-β or IL-12 stimulation of activated CD45RA-CXCR5- cells, finding no differences in pSTAT4 expression in cells from patients compared to healthy controls. However, pSTAT4 expression in lupus cTfh-like cells was increased compared to CD45RA-CXCR5- cells (Figure 6C).
Figure 6.
Activation of STAT4 is correlated with increased disease activity. Isolated mononuclear cells from the peripheral blood from SLE patients or healthy controls were rested in 10% complete DMEM solution overnight, then stimulated by IL-12 of IFN-β. (A) Representative gating strategy to identify memory T cells or Tfh-like cells. (B) Intracellular IL-21 and IFN-γ staining in Tfh-like cells from an SLE patient. (C) Representative flow histogram of pSTAT4 from cells stimulated with either IFN-β (top) or IL-12 (bottom), graphs summarizing the ratio of MFI of stimulated pSTAT4 in Tfh-like (left), memory T (middle) or activated CXCR5- cells (right) to their unstimulated pSTAT4 MFI for each individual sample. (D) Linear regression analysis of the pSTAT4/unstimulated ratio in SLE patients compared to their disease activity measured by SLEDAI. Data are representative of 27 SLE and 19 HC, *p < 0.05 by Mann-Whitney. Error bars represent SEM. Pearson R =0.182, P=0.033
We did not observe a correlation between the percentage of Tfh-like cells and their expression of pSTAT4 in Tfh-like cells from lupus patients (Supplemental Fig 4A). While there was no association of pSTAT4 with complement markers or autoantibodies (Supplemental Figure 4B), increased pSTAT4 expression in Tfh-like cells from lupus patients correlated with increased SLEDAI 2K (Figure 6D). Increased pSTAT4 and co-production of IL-21 and IFN-γ suggested that Tfh-like cells from lupus patients are comparable to those from diseased lupus-prone mice.
Discussion
We define a previously unappreciated role for IFN-I signaling in driving STAT4 activation and production of IL-21 and IFN-γ by Tfh cells in murine and human lupus. We show that in lupus mice, Tfh cells transcriptionally evolve during the course of disease losing their Bcl6 expression. Tfh cells target genes were still expressed, suggesting Bcl6 transcripts remain regulated even as protein expression declines, analogous to what goes on with T-bet(12). As disease progress, Tfh cells acquire an enhanced STAT4 gene signature, consistent with our finding that IFN-I augments STAT4 phosphorylation in Tfh cells at later time points. Blockade of IFNAR1 after the onset of autoantibodies reduced IL-21 and IFN-γ secretion by Tfh cells, indicating that IFN-I signaling maintains the production of these cytokines throughout disease. Similar to our findings in mice, we find that Tfh-like and memory T cells from SLE patients secreted IL-21 and IFN-γ and had increased pSTAT4 correlating with higher disease activity. Collectively, our data suggest that IFN-I enhance STAT4 activation in Tfh cells, driving their aberrant production of IL-21 and IFN-γ, which may underlie pathogenic GC responses in both murine and human lupus.
Our data show that cytokine-secreting-Tfh cells and GCs are generated prior to the onset of autoantibodies in murine lupus. As the development of autoantibodies progressed, we found that Tfh cells continued to produce cytokines and remained within germinal centers but transcriptionally evolved. The pattern of transcription factor expression and cytokine production by Tfh cells from older B6.Sle1.Yaa mice emulated that of Tfh-like cells from SLE patients (37). IFNAR blockade reduced the co-production of these cytokines in Tfh cells but not their development or function as the demonstrated by the increase in IgG1 autoantibodies in the presence of ongoing GCs. While IgG2c autoantibodies were detected, these may be generated by existing plasma cells or those driven to maturation by the fraction of Tfh cells that continued to produce only IFN-γ after treatment. The continued Tfh cell-IFN-γ production may be due to the increased circulating IL-12 as disease progresses in these mice (33). In aggregate, our data suggest that the development of Tfh cells and GCs precedes the onset of autoantibodies and tissue destruction in lupus and that activation of STAT4 by IFN-I sustains aberrant cytokine production.
Previous in vitro studies established that activated STAT4 binds and epigenetically regulates the expression of Il21 and Ifng in Th cells (38–40). We find that both IL-12 and IFN-I are able to activate STAT4, suggesting a role for each in promoting IL-21 and IFN-γ secretion by Tfh cells via STAT4 in lupus. In lupus, we find that although co-production of IL-21 and IFN-γ is maintained in Tfh cells despite progressive loss of T-bet expression, pSTAT4 continues to increase. These findings suggest that cytokine production is driven by IFN-I activation of STAT4 in Tfh cells as diseased progressed. Thus, IFN-I continue to activate STAT4 in Tfh cells maintaining IL-21 and IFN-γ production throughout disease.
Our findings have implications for clinical treatment of SLE. IFN-I-induced genes are expressed in ~ 70% of patients (41). Blocking IFN-I has emerged as a potential treatment for SLE. This idea is supported by the recent TULIP II phase III clinical trial of anifrolumab, a monoclonal anti-IFN-I receptor antibody (42), although the TULIP I trial failed to achieve efficacy in reducing disease activity (43). Nonetheless, JAK-STAT signaling plays an important downstream role from IFN-I in regulating IL-21 and IFN-γ production and their associated biological effects (44). The IFN-I signaling complex can activate STAT4 through Janus kinase 1 (Jak1) and Tyk2 in T cells in humans and in murine models of disease (22, 45). Tofacitinib, an inhibitor of JAK1 and JAK3, is FDA approved for treatment of rheumatoid arthritis. The critical role of JAK1 and JAK3 signaling in host defenses underlie the toxicities associated with its use, which includes infections, cytopenia and alterations to lipid metabolism (46, 47). Modulation of the IL-12/IFN-I -STAT4 signaling pathway may be a preferable way to restrict a set of cytokines and their associated pathologic responses in lupus. In particular, IL-12 and IFN-I pathway inhibitor Tyk2 was developed and employed in preclinical models of SLE. Tyk2 inhibition demonstrated amelioration of disease pathology in (NZB x NZW)F1 lupus-prone mice, highlighting the role of Tyk2-STAT4 signaling in lupus development. Our data support these clinical findings showing that in both murine and human lupus, IL-21 and IFN-γ production by Th cells is downstream of aberrant IL-12/IFN-I-STAT4 signaling. Furthermore, we demonstrated that IL-12 and IFN-I signaling promote increased pSTAT4 in Tfh cells from both murine and human lupus with active disease.
IL-21 and IFN-γ have been identified as compelling therapeutic targets in the treatment of SLE (48, 49). To our knowledge, we show for the first time that the transcriptional regulation of these cytokines in Tfh cells changes over the course of disease. STAT4 is strongly associated with disease susceptibility in SLE, yet the distinct contribution of STAT4 to pathogenesis remained ambiguous (19–21). Here, we identify a potential pathogenic role for STAT4 following activation through IFN-I and IL-12 signaling in Tfh cells, thereby maintaining their IL-21 and IFN-γ production in lupus. Collectively, our findings solidify the STAT4 pathway as a target for the development of a successful therapeutic strategy in SLE.
Supplementary Material
Supplemental Figure 1. Temporal characterization of eTfh cells and their cytokines and transcription factors in MRL lrp/lpr mice. Spleens were harvested from MRLlrp/lpr at 2 and 6 months of age. (A) Representative flow cytometry plots of PSGL-1loLy6Clo eTfh cells from MRLlpr/lpr mice, with percentages of Th1 and Tfh cells (left and right, respectively). (B) Intracellular IL-21 and IFN-γ staining of eTfh cells with percentages of (right) of cytokine positive cells. (C) Representative flow cytometry plots of PSGL-1loLy6Clo eTfh cells from MRL lrp/lpr mice, with the quantified geometric mean fluorescence intensity of staining (MFI) of Bcl6 (left) or T-bet (right). (D) Tbet and Bcl6 in Th1 cells in B6.Sle1.Yaa at 2, 4, and 7 months of age; bar graphs summarize the quantified geometric MFI of Bcl6 (left) or T-bet (right). Data are representative of 2 experiments with 3–5 mice per group. **p < 0.01; Student’s t-test. Error bars represent SEM.
Supplemental Figure 2. Confirmation of differentially expressed genes and downregulated pathways of Tfh cells during the progression of disease. Spleens were harvested from either B6.Sle1.Yaa at 2, 4, and 7 months of age and cDNA was synthesized from sorted Tfh cells. (A) Quantitative RT-PCR analysis of selected T cell-related genes (Cxcr4, Ccr7 and Bcl6), normalized to results obtained for the control gene Hprt. (B) CXCR5 and PD-1 expression in Tfh cells in B6.Sle1.Yaa at 2, 4, and 7 months of age; bar graphs summarize the quantified geometric MFI of CXCR5 (left) or PD-1 (right). (C) Using significantly differentially expressed genes identified by transcriptome analysis of Tfh cells from B6.Sle1.Yaa at 2, 4, and 7 months of age mice. Enrichment pathways analysis of the downregulated genes in Tfh cells at 7 months to 2 months, with the top four pathways listed, including the enrichment p value. Data are representative of 3 experiments with 3–5 mice per group. **p < 0.01; *p< 0.05 by Student’s t-test. Error bars represent SEM.
Supplemental Figure 3. Blockade of IFN-I has no affect in GC B cells in SLE. (A) IFN-β ELISA was performed on the sera from B6.Sle1.Yaa at 2 or 7 months of age. 4-month old B6.Sle1.Yaa mice treated for twice a week for 4 weeks with either anti-IFNAR1 (375ug) or anti-rat IgG (1mg/mL). (B) Representative flow cytometry histogram of IL-21 and IFN-γ in Tfh cells from anti-IFNAR1 or control treated B6.Sle1.Yaa mice compared to unstimulated cells from controls. (C) Representative flow cytometry plots of GL-7hiCD95hi GC B cells following treatment with percentages (left and right, respectively). (D) Splenic B cell follicles of treated B6.Sle1.Yaa mice stained with anti-IgD (green), anti-CD4 (blue) and PNA (magenta), with the numbers of T cells per GC size (left) and GC sizes quantified (right). Data are representative of 2 experiments with 3–5 mice per group.
Supplemental Figure 4. Correlation of pSTAT4 with disease markers in SLE. Isolated mononuclear cells from the peripheral blood from SLE patients were rested in 10% complete DMEM solution overnight, then stimulated by IL-12 of IFN-β. (A) Linear regression analysis of the MFI of pSTAT4 in SLE patients with the percentage of Tfh-like cells (CD4+CD45RA-CCR7-PD-1hiCXCR5hi) determined by flow cytometry. Pearson R =0.0023, P=0.8085. (B) Linear regression analysis of the pSTAT4/unstimulated ratio in SLE patients compared to their complement C4, complement C3 and dsDNA scores. Pearson R =0.0581, P=0.2257 top left, Pearson R =0.0090, P=0.6373 top right, Pearson R =0.1172, P=0.0805 bottom.
ACKNOWLEDGEMENTS
The authors acknowledge members of our departments for critical review of the manuscript. X.D. was supported by NIH 2 T32 AR007107. J.S.W. was supported in part by a National Institutes of Health (NIH) grants K01AR067892-04, P30 AR053495 and R01 AR073912 and the Yale CTSA grant UL1TR0001863 from the National Center for Advancing Translational Science (NCATS), NIH. J.C. and laboratory support came from NIH grants R37 AR40072 and R01 AR074545, and the Lupus Research Alliance. The authors declare no competing financial interests. Correspondence and requests for materials should be addressed to J.S.W. (Jason.weinstein@rutgers.edu).
Footnotes
The authors have declared that no conflict of interest exists.
References
- 1.Choi JY, Ho JH, Pasoto SG, Bunin V, Kim ST, Carrasco S, et al. Circulating follicular helper-like T cells in systemic lupus erythematosus: association with disease activity . Arthritis & rheumatology (Hoboken, NJ). 2015;67(4):988–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simpson N, Gatenby PA, Wilson A, Malik S, Fulcher DA, Tangye SG, et al. Expansion of circulating T cells resembling follicular helper T cells is a fixed phenotype that identifies a subset of severe systemic lupus erythematosus. Arthritis Rheum. 2010;62(1):234–44. [DOI] [PubMed] [Google Scholar]
- 3.Linterman MA, Beaton L, Yu D, Ramiscal RR, Srivastava M, Hogan JJ, et al. IL-21 acts directly on B cells to regulate Bcl-6 expression and germinal center responses. J Exp Med. 2010;207(2):353–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zotos D, Coquet JM, Zhang Y, Light A, D’Costa K, Kallies A, et al. IL-21 regulates germinal center B cell differentiation and proliferation through a B cell-intrinsic mechanism. J Exp Med. 2010;207(2):365–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee SK, Silva DG, Martin JL, Pratama A, Hu X, Chang PP, et al. Interferon-gamma excess leads to pathogenic accumulation of follicular helper T cells and germinal centers. Immunity. 2012;37(5):880–92. [DOI] [PubMed] [Google Scholar]
- 6.Roco JA, Mesin L, Binder SC, Nefzger C, Gonzalez-Figueroa P, Canete PF, et al. Class-Switch Recombination Occurs Infrequently in Germinal Centers. Immunity. 2019;51(2):337–50.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herber D, Brown TP, Liang S, Young DA, Collins M, Dunussi-Joannopoulos K. IL-21 Has a Pathogenic Role in a Lupus-Prone Mouse Model and Its Blockade with IL-21R.Fc Reduces Disease Progression. Journal of immunology (Baltimore, Md : 1950). 2007;178(6):3822–30. [DOI] [PubMed] [Google Scholar]
- 8.Miyake K, Nakashima H, Akahoshi M, Inoue Y, Nagano S, Tanaka Y, et al. Genetically determined interferon-gamma production influences the histological phenotype of lupus nephritis. Rheumatology (Oxford). 2002;41(5):518–24. [DOI] [PubMed] [Google Scholar]
- 9.Peng SL, Moslehi J, Craft J. Roles of interferon-gamma and interleukin-4 in murine lupus. J Clin Invest. 1997;99(8):1936–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, Zhao P, Ma L, Shan Y, Jiang Z, Wang J, et al. Increased interleukin 21 and follicular helper T-like cells and reduced interleukin 10+ B cells in patients with new-onset systemic lupus erythematosus. J Rheumatol. 2014;41(9):1781–92. [DOI] [PubMed] [Google Scholar]
- 11.Weinstein JS, Laidlaw BJ, Lu Y, Wang JK, Schulz VP, Li N, et al. STAT4 and T-bet control follicular helper T cell development in viral infections. J Exp Med. 2018;215(1):337–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang D, Cui K, Mao K, Hu G, Li R, Zheng M, et al. Transient T-bet expression functionally specifies a distinct T follicular helper subset. J Exp Med. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thieu VT, Yu Q, Chang HC, Yeh N, Nguyen ET, Sehra S, et al. Signal transducer and activator of transcription 4 is required for the transcription factor T-bet to promote T helper 1 cell-fate determination. Immunity. 2008;29(5):679–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirou KA, Lee C, George S, Louca K, Peterson MG, Crow MK. Activation of the interferon-alpha pathway identifies a subgroup of systemic lupus erythematosus patients with distinct serologic features and active disease. Arthritis Rheum. 2005;52(5):1491–503. [DOI] [PubMed] [Google Scholar]
- 15.Harigai M, Kawamoto M, Hara M, Kubota T, Kamatani N, Miyasaka N. Excessive production of IFN-gamma in patients with systemic lupus erythematosus and its contribution to induction of B lymphocyte stimulator/B cell-activating factor/TNF ligand superfamily-13B. J Immunol. 2008;181(3):2211–9. [DOI] [PubMed] [Google Scholar]
- 16.Luzina IG, Atamas SP, Storrer CE, daSilva LC, Kelsoe G, Papadimitriou JC, et al. Spontaneous formation of germinal centers in autoimmune mice. J Leukoc Biol. 2001;70(4):578–84. [PubMed] [Google Scholar]
- 17.Munroe ME, Lu R, Zhao YD, Fife DA, Robertson JM, Guthridge JM, et al. Altered type II interferon precedes autoantibody accrual and elevated type I interferon activity prior to systemic lupus erythematosus classification. Ann Rheum Dis. 2016;75(11):2014–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alleva DG, Kaser SB, Beller DI. Intrinsic defects in macrophage IL-12 production associated with immune dysfunction in the MRL/++ and New Zealand Black/White F1 lupus-prone mice and the Leishmania major-susceptible BALB/c strain. J Immunol. 1998;161(12):6878–84. [PubMed] [Google Scholar]
- 19.Remmers EF, Plenge RM, Lee AT, Graham RR, Hom G, Behrens TW, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007;357(10):977–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41(11):1234–7. [DOI] [PubMed] [Google Scholar]
- 21.Alarcon-Riquelme ME, Ziegler JT, Molineros J, Howard TD, Moreno-Estrada A, Sanchez-Rodriguez E, et al. Genome-Wide Association Study in an Amerindian Ancestry Population Reveals Novel Systemic Lupus Erythematosus Risk Loci and the Role of European Admixture . Arthritis & rheumatology (Hoboken, NJ). 2016;68(4):932–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagberg N, Joelsson M, Leonard D, Reid S, Eloranta ML, Mo J, et al. The STAT4 SLE risk allele rs7574865[T] is associated with increased IL-12-induced IFN-gamma production in T cells from patients with SLE. Ann Rheum Dis. 2018;77(7):1070–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh RR, Saxena V, Zang S, Li L, Finkelman FD, Witte DP, et al. Differential contribution of IL-4 and STAT6 vs STAT4 to the development of lupus nephritis. J Immunol. 2003;170(9):4818–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baccala R, Gonzalez-Quintial R, Schreiber RD, Lawson BR, Kono DH, Theofilopoulos AN. Anti-IFN-alpha/beta receptor antibody treatment ameliorates disease in lupus-predisposed mice. J Immunol. 2012;189(12):5976–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Odegard JM, Marks BR, DiPlacido LD, Poholek AC, Kono DH, Dong C, et al. ICOS-dependent extrafollicular helper T cells elicit IgG production via IL-21 in systemic autoimmunity. J Exp Med. 2008;205(12):2873–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mittereder N, Kuta E, Bhat G, Dacosta K, Cheng LI, Herbst R, et al. Loss of Immune Tolerance Is Controlled by ICOS in Sle1 Mice. J Immunol. 2016;197(2):491–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marshall HD, Chandele A, Jung YW, Meng H, Poholek AC, Parish IA, et al. Differential Expression of Ly6C and T-bet Distinguish Effector and Memory Th1 CD4+ Cell Properties during Viral Infection. Immunity. 2011;35(4):633–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.William J, Euler C, Christensen S, Shlomchik MJ. Evolution of autoantibody responses via somatic hypermutation outside of germinal centers. Science. 2002;297(5589):2066–70. [DOI] [PubMed] [Google Scholar]
- 29.Degn SE, van der Poel CE, Firl DJ, Ayoglu B, Al Qureshah FA, Bajic G, et al. Clonal Evolution of Autoreactive Germinal Centers. Cell. 2017;170(5):913–26.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo W, Weisel F, Shlomchik MJ. B Cell Receptor and CD40 Signaling Are Rewired for Synergistic Induction of the c-Myc Transcription Factor in Germinal Center B Cells. Immunity. 2018;48(2):313–26.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weinstein JS, Herman EI, Lainez B, Licona-Limon P, Esplugues E, Flavell R, et al. TFH cells progressively differentiate to regulate the germinal center response. Nat Immunol. 2016;17(10):1197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho SS, Bacon CM, Sudarshan C, Rees RC, Finbloom D, Pine R, et al. Activation of STAT4 by IL-12 and IFN-alpha: evidence for the involvement of ligand-induced tyrosine and serine phosphorylation. J Immunol. 1996;157(11):4781–9. [PubMed] [Google Scholar]
- 33.Fairhurst AM, Hwang SH, Wang A, Tian XH, Boudreaux C, Zhou XJ, et al. Yaa autoimmune phenotypes are conferred by overexpression of TLR7. European Journal of Immunology. 2008;38(7):1971–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arbuckle MR, McClain MT, Rubertone MV, Scofield RH, Dennis GJ, James JA, et al. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med. 2003;349(16):1526–33. [DOI] [PubMed] [Google Scholar]
- 35.Yang JH, Zhang J, Cai Q, Zhao DB, Wang J, Guo PE, et al. Expression and function of inducible costimulator on peripheral blood T cells in patients with systemic lupus erythematosus. Rheumatology (Oxford). 2005;44(10):1245–54. [DOI] [PubMed] [Google Scholar]
- 36.He J, Tsai LM, Leong YA, Hu X, Ma CS, Chevalier N, et al. Circulating precursor CCR7(lo)PD-1(hi) CXCR5(+) CD4(+) T cells indicate Tfh cell activity and promote antibody responses upon antigen reexposure. Immunity. 2013;39(4):770–81. [DOI] [PubMed] [Google Scholar]
- 37.Morita R, Schmitt N, Bentebibel S-E, Ranganathan R, Bourdery L, Zurawski G, et al. Human Blood CXCR5+CD4+ T Cells Are Counterparts of T Follicular Cells and Contain Specific Subsets that Differentially Support Antibody Secretion. Immunity. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitt N, Morita R, Bourdery L, Bentebibel SE, Zurawski SM, Banchereau J, et al. Human dendritic cells induce the differentiation of interleukin-21-producing T follicular helper-like cells through interleukin-12. Immunity. 2009;31(1):158–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei L, Vahedi G, Sun HW, Watford WT, Takatori H, Ramos HL, et al. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity. 2010;32(6):840–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakayamada S, Kanno Y, Takahashi H, Jankovic D, Lu KT, Johnson TA, et al. Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity. 2011;35(6):919–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiche L, Jourde-Chiche N, Whalen E, Presnell S, Gersuk V, Dang K, et al. Modular transcriptional repertoire analyses of adults with systemic lupus erythematosus reveal distinct type I and type II interferon signatures. Arthritis & rheumatology (Hoboken, NJ). 2014;66(6):1583–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morand EF, Furie R, Tanaka Y, Bruce IN, Askanase AD, Richez C, et al. Trial of Anifrolumab in Active Systemic Lupus Erythematosus. N Engl J Med. 2020;382(3):211–21. [DOI] [PubMed] [Google Scholar]
- 43.Furie RA, Morand EF, Bruce IN, Manzi S, Kalunian KC, Vital EM, et al. Type I interferon inhibitor anifrolumab in active systemic lupus erythematosus (TULIP-1): a randomised, controlled, phase 3 trial. The Lancet Rheumatology. 2019;1(4):e208–e19. [DOI] [PubMed] [Google Scholar]
- 44.O’Shea JJ, Plenge R. JAK and STAT signaling molecules in immunoregulation and immune-mediated disease. Immunity. 2012;36(4):542–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen KB, Watford WT, Salomon R, Hofmann SR, Pien GC, Morinobu A, et al. Critical role for STAT4 activation by type 1 interferons in the interferon-gamma response to viral infection. Science. 2002;297(5589):2063–6. [DOI] [PubMed] [Google Scholar]
- 46.Yuan K, Chen J, Xu A. Tofacitinib versus methotrexate in rheumatoid arthritis. N Engl J Med. 2014;371(12):1163–4. [DOI] [PubMed] [Google Scholar]
- 47.Schwartz DM, Kanno Y, Villarino A, Ward M, Gadina M, O’Shea JJ. JAK inhibition as a therapeutic strategy for immune and inflammatory diseases. Nature reviews Drug discovery. 2017;17(1):78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spolski R, Leonard WJ. Interleukin-21: a double-edged sword with therapeutic potential. Nat Rev Drug Discov. 2014;13(5):379–95. [DOI] [PubMed] [Google Scholar]
- 49.Ronnblom L, Elkon KB. Cytokines as therapeutic targets in SLE . Nat Rev Rheumatol. 2010;6(6):339–47. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Temporal characterization of eTfh cells and their cytokines and transcription factors in MRL lrp/lpr mice. Spleens were harvested from MRLlrp/lpr at 2 and 6 months of age. (A) Representative flow cytometry plots of PSGL-1loLy6Clo eTfh cells from MRLlpr/lpr mice, with percentages of Th1 and Tfh cells (left and right, respectively). (B) Intracellular IL-21 and IFN-γ staining of eTfh cells with percentages of (right) of cytokine positive cells. (C) Representative flow cytometry plots of PSGL-1loLy6Clo eTfh cells from MRL lrp/lpr mice, with the quantified geometric mean fluorescence intensity of staining (MFI) of Bcl6 (left) or T-bet (right). (D) Tbet and Bcl6 in Th1 cells in B6.Sle1.Yaa at 2, 4, and 7 months of age; bar graphs summarize the quantified geometric MFI of Bcl6 (left) or T-bet (right). Data are representative of 2 experiments with 3–5 mice per group. **p < 0.01; Student’s t-test. Error bars represent SEM.
Supplemental Figure 2. Confirmation of differentially expressed genes and downregulated pathways of Tfh cells during the progression of disease. Spleens were harvested from either B6.Sle1.Yaa at 2, 4, and 7 months of age and cDNA was synthesized from sorted Tfh cells. (A) Quantitative RT-PCR analysis of selected T cell-related genes (Cxcr4, Ccr7 and Bcl6), normalized to results obtained for the control gene Hprt. (B) CXCR5 and PD-1 expression in Tfh cells in B6.Sle1.Yaa at 2, 4, and 7 months of age; bar graphs summarize the quantified geometric MFI of CXCR5 (left) or PD-1 (right). (C) Using significantly differentially expressed genes identified by transcriptome analysis of Tfh cells from B6.Sle1.Yaa at 2, 4, and 7 months of age mice. Enrichment pathways analysis of the downregulated genes in Tfh cells at 7 months to 2 months, with the top four pathways listed, including the enrichment p value. Data are representative of 3 experiments with 3–5 mice per group. **p < 0.01; *p< 0.05 by Student’s t-test. Error bars represent SEM.
Supplemental Figure 3. Blockade of IFN-I has no affect in GC B cells in SLE. (A) IFN-β ELISA was performed on the sera from B6.Sle1.Yaa at 2 or 7 months of age. 4-month old B6.Sle1.Yaa mice treated for twice a week for 4 weeks with either anti-IFNAR1 (375ug) or anti-rat IgG (1mg/mL). (B) Representative flow cytometry histogram of IL-21 and IFN-γ in Tfh cells from anti-IFNAR1 or control treated B6.Sle1.Yaa mice compared to unstimulated cells from controls. (C) Representative flow cytometry plots of GL-7hiCD95hi GC B cells following treatment with percentages (left and right, respectively). (D) Splenic B cell follicles of treated B6.Sle1.Yaa mice stained with anti-IgD (green), anti-CD4 (blue) and PNA (magenta), with the numbers of T cells per GC size (left) and GC sizes quantified (right). Data are representative of 2 experiments with 3–5 mice per group.
Supplemental Figure 4. Correlation of pSTAT4 with disease markers in SLE. Isolated mononuclear cells from the peripheral blood from SLE patients were rested in 10% complete DMEM solution overnight, then stimulated by IL-12 of IFN-β. (A) Linear regression analysis of the MFI of pSTAT4 in SLE patients with the percentage of Tfh-like cells (CD4+CD45RA-CCR7-PD-1hiCXCR5hi) determined by flow cytometry. Pearson R =0.0023, P=0.8085. (B) Linear regression analysis of the pSTAT4/unstimulated ratio in SLE patients compared to their complement C4, complement C3 and dsDNA scores. Pearson R =0.0581, P=0.2257 top left, Pearson R =0.0090, P=0.6373 top right, Pearson R =0.1172, P=0.0805 bottom.