Abstract
Small RNAs (sRNAs) are essential regulatory molecules, and there are three major sRNA classes in plants: microRNAs (miRNAs), phased small interfering RNAs (phased siRNAs or phasiRNAs), and heterochromatic siRNAs (hc-siRNAs). Excluding miRNAs, the other two classes are not well annotated or available in public databases for most sequenced plant genomes. We performed a comprehensive sRNA annotation of 143 plant species that have fully sequenced genomes and next-generation sequencing sRNA data publicly available. The results are available via an online repository called sRNAanno (www.plantsRNAs.org). Compared with other public plant sRNA databases, we obtained was much more miRNA annotations, which are more complete and reliable because of the consistent and highly stringent criteria used in our miRNA annotations. sRNAanno also provides free access to genomic information for >22,721 PHAS loci and >22 million hc-siRNA loci annotated from these 143 plant species. Both miRNA and PHAS loci can be easily browsed to view their main features, and a collection of archetypal trans-acting siRNA 3 (TAS3) genes were annotated separately for quick access. To facilitate the ease of sRNA annotation, sRNAanno provides free service for sRNA annotations to the community. In summary, the sRNAanno database is a great resource to facilitate genomic and genetic research on plant small RNAs.
Subject terms: Non-coding RNAs, Gene silencing
Introduction
Small RNAs (sRNAs) are essential regulatory molecules in plants. With the rapid development of deep-sequencing technologies and bioinformatics, sRNAs have been characterized in an increasing number of plant species, leading to the generation of large amounts of next-generation sequencing (NGS) data. A large number of raw NGS sRNA data have been deposited in public databases, such as the Sequence Read Archive (SRA), Gene Expression Omnibus (GEO) and European Nucleotide Archive (ENA) databases. MicroRNAs (miRNAs) are the most well-studied class of sRNAs in plants. To date, miRBase is the primary repository and online database for annotated miRNAs1. As a routine practice in the research community, the annotated miRNAs of a species are required to be deposited into miRBase before publication; i.e., author submission is the primary data source of the database. This process makes it hard to maintain a high quality of annotated miRNAs deposited in miRBase because of the variable stringency of the criteria, controlled by the submitting authors who are responsible for miRNA annotations. In other words, rather than the developers or maintainers of miRBase, quality control is more reliant on the authors, reviewers and editors, who likely have a different understanding of the criteria of miRNA annotation. Therefore, the variable reliability of annotated miRNAs in miRBase is of great concern to the community2.
In addition to miRNAs, other types of sRNAs exist in plants, including phased small interfering RNAs (phased siRNAs or phasiRNAs) and heterochromatic siRNAs (hc-siRNAs). phasiRNAs have recently emerged as critical regulatory molecules in nearly all aspects of plant growth and development3. They are widely present in plants—from algae to angiosperms. However, compared to miRNAs, phasiRNAs are much less studied and are not well annotated for most of the plants species whose genome has been fully sequenced4,5. To date, there is no public database of annotation information of plant phasiRNAs, hindering the application of already annotated phasiRNA information. Heterochromatic siRNAs are the most abundant class of sRNAs in plants, and they usually play roles related to DNA methylation, which is a process important for transcriptional regulation. Although we know their functional importance, thorough annotations of hc-siRNA-generating genomic regions (hc-siRNA loci) are lacking for most plant genomes.
In this study, we conducted extensive sRNA annotations of 143 plant species whose genome has been fully sequenced and for which at least one sRNA deep-sequencing data set is available in public databases. The annotations include all three sRNA classes: miRNAs, phasiRNAs, and hc-siRNAs. To achieve high confidence for miRNA annotations, we applied a set of uniform criteria adopted from the recently updated rules2. For phasiRNA annotations, a p value-based approach established by our group was used for annotations of loci, yielding 21-nt phasiRNAs (21-PHAS) or 24-nt phasiRNAs (24-PHAS)6,7. We also developed an algorithm based on sequence repetitiveness for the accurate annotations of loci generating hc-siRNAs, given their primary feature of generation from repetitive genomic regions. In total, we annotated 24,630 miRNA hairpins or precursors, 22,721 PHAS loci (18,239 21-PHAS and 4,482 24-PHAS), and 22,404,950 hc-siRNA loci. All these results have been deposited in an online database of sRNA annotations (sRNAanno) for open access. This database is a great resource for research on plant sRNAs.
Database content
sRNAanno database
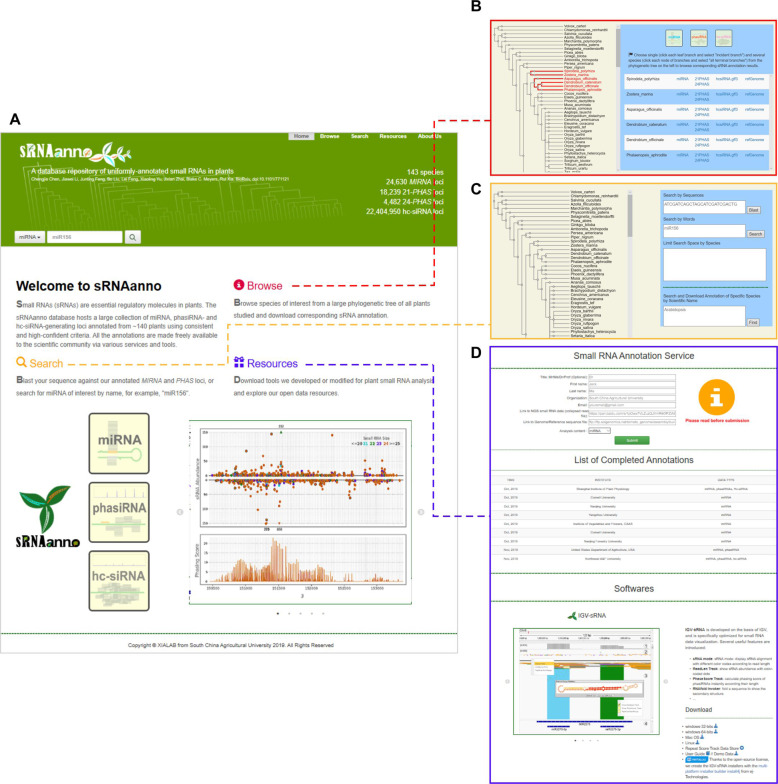
Small RNA annotation of three major sRNA classes was performed for 143 plant species, and an online database (sRNAanno, www.plantsRNAs.org) was constructed to store all the annotation results for easy and quick public access. There are three major functions within sRNAanno:
BROWSE, for browsing annotation results, SEARCH, for searching for certain information, and RESOURCES, for data sharing (Fig. 1A). On the BROWSE page, users can select a single or several species from a large phylogenetic tree and browse or download corresponding small RNA annotation results (Fig. 1B). The SEARCH function includes miRNA searches by either miRNA name or sequence comparison using the BLAST function (Fig. 1C). The RESOURCES page provides quick access to the Small RNA Annotation Service page and other relevant data (Fig. 1D); for instance, the free software IGV-sRNA, which is designed for the exploration of sRNA data, can be download here.
Fig. 1. Overview of the sRNAanno database.
A Screenshots of the HOME page and subpages of the three main functions: BROWSE (B), SEARCH (C), and RESOURCES (D)
miRNA annotations
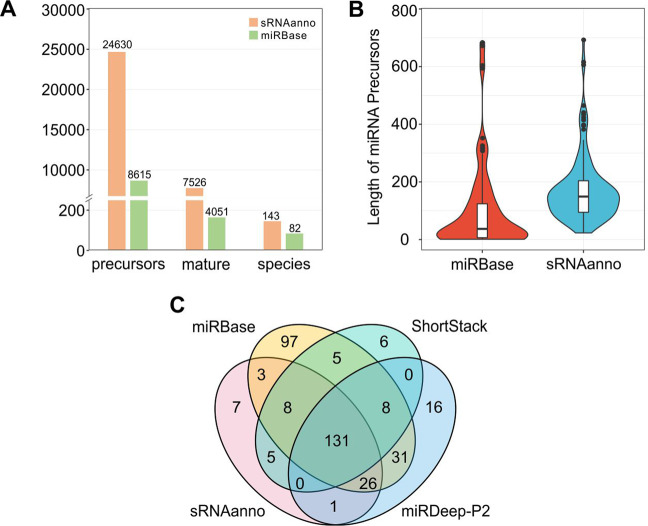
In this study, we aimed to conduct genome-wide annotations of plant miRNAs using a set of uniform and well-established criteria (Fig. S1), as well documented before2,8. To this end, we downloaded the genome sequences for nearly all species for which both their genome and their sRNA NGS data are available from public databases (such as SRA, ENA, and GEO). We found 143 plant species with corresponding sRNA data available. In total, there are 1,606 small RNA sequencing data sets, most of which are generated from well-studied model plant species, such as Arabidopsis and rice. We performed miRNA annotations of all these species and obtained 24,630 annotated hairpin precursors encoding 7,526 unique mature miRNA sequences (Fig. 2A). Compared with other public sRNA databases, like miRbase1, PmiREN9, and “Plant Small RNA Genes”10, sRNAanno employed more small RNA datasets, or covered many more plants, with the annotation of more MIRNA loci. For example, compared with the annotations in the latest release of miRBase (v22), which contains 8,615 annotated hairpin precursors from 82 plant species with 4,051 mature miRNA sequences (Fig. 2A), our annotations yielded more results in terms of not only the number of species annotated (a 1.74-fold increase) but also the number of miRNA precursors (a 2.86-fold increase) (Fig. 2A). In plants, ~24 miRNA families predominate in angiosperms11. To assess the completeness of miRNA annotations of a species, we compared the number of conserved miRNA families in species that have data in both sRNAanno and miRBase. We found that nearly all 45 species analyzed had a more complete list of conserved miRNAs in sRNAanno, while conserved miRNAs in 14 species were obviously incomplete in miRBase (v22) (Fig. S2). Moreover, in terms of the length distribution of MIRNA precursors, the length of most precursors in miRBase is much shorter (<100 bp) than that in sRNAanno (100–200 bp, Fig. 2B), which seems reasonable, as the majority of Arabidopsis MIRNA precursors are 100–200 bp in length12. Therefore, compared with miRBase (v22), sRNAanno has more complete and reliable plant miRNA data. When comparing with miRNAs deposited in miRBase or identified by other tools (miRDeep-P2 and ShortStack) within a species (using Arabidopsis and rice as examples), we found that the majority of miRNAs annotated in sRNAanno were also identified by at least one of the other tools13,14 (Fig. 2C and Fig. S3); only a few of them were unique to sRNAanno. Overall, we contend that our miRNA annotations are of high stringency and high confidence.
Fig. 2. Summary of annotated miRNAs in sRNAanno.
A Comparison of the numbers of plant species and MIRNA loci annotated between miRBase and sRNAanno. B Length of miRNA precursors in miRBase and sRNAanno. C Comparison of the miRNA annotation results of sRNAanno for Arabidopsis with those from three other sources/pipelines: miRBase, ShortStack, and miRDeep-P2
PHAS locus annotation
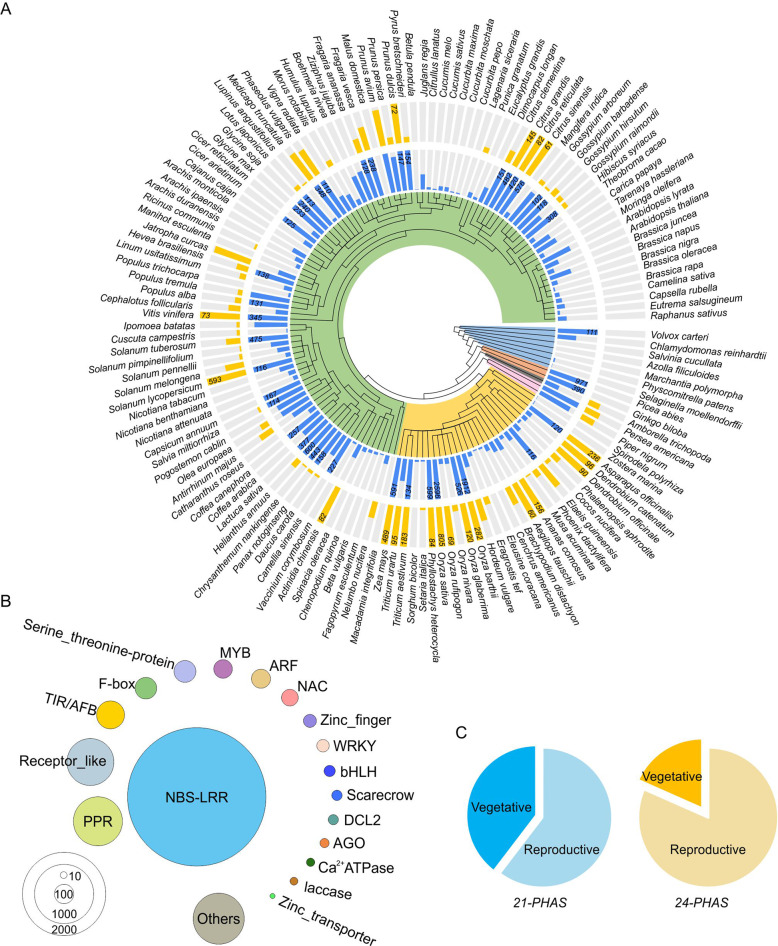
Phased siRNAs (phasiRNAs) are another major class of sRNAs found in plants, and these are universally present in all plants, mainly as members of the trans-acting siRNA (tasiRNA) subgroup15. This group is characterized by the phasing pattern of sRNAs, which exhibit an approximately head-to-tail arrangement. To date, unlike for miRNAs, there is no database of identified or reported PHAS genes or genomic loci, although phasiRNAs have been profiled in a large number of plants3. Therefore, we performed an exhaustive PHAS profiling of the 143 plant species for which at least one sequenced sRNA library exists. We used a well-developed p value-based protocol to perform PHAS analysis7,16 (Fig. S4). The cutoff of the p value was set to 1e-3. For analysis of 24-PHAS loci (generating 24-nt phasiRNA), we added an additional filter to remove repetitive sequences that usually give yield abundant 24-nt hc-siRNAs. In total, we identified 18,239 21-PHAS loci generating 21-nt phasiRNAs and 4,482 24-PHAS loci generating 24-nt phasiRNAs. In general, the number of 21-PHAS loci was substantially greater than that of 24-PHAS loci within a species (Fig. 3A). Both types of PHAS loci are not evenly present across species (Fig. 3A), perhaps because of the intrinsic genomic differences among plant species; for instance, plants from certain families, such as the Brassicaceae (including the model plant Arabidopsis) and Cucurbitaceae, consistently yield fewer PHAS loci than do other species (Fig. 3A). As reported before, 24-PHAS loci are noticeable more widespread in monocots but are dispersed in eudicots7(Fig. 3A). Other factors accounting for this uneven distribution of PHAS loci are likely the sampling for sRNA sequencing (tissue, stage, etc.) and sequencing technology (sequencing platforms, sequencing depth, etc.).
Fig. 3. Summary of annotated PHAS loci in sRNAanno.
A Twenty-one (inner circle, blue, with the maximum value set to 100) and 24 (outer circle, yellow, with the maximum value set to 50) PHAS loci annotated in each species. All the species are ordered according to the phylogenetic tree of APG IV (Angiosperm Phylogeny Group IV). B Functional classification of protein-coding 21-PHAS loci. The circle sizes are scaled by the number of 21 PHAS loci in a given gene group. C Tissue-specific enrichment of 21- and 24-PHAS loci
Protein-coding genes are a rich source of phasiRNAs. After functionally annotating the 21-PHAS loci and assessing their protein-coding capacity, we found that a large number of gene families produce a large number of phasiRNAs, especially for members of the gene families NBS-LRR, PPR, Receptor-like kinase, etc. (Fig. 3B). In particular, many transcription factor-encoding genes produce phasiRNAs, including TIR/AFB, F-box, NAC, MYB, ARF, WRKY, zinc finger, and bHLH genes (Fig. 3B). In terms of noncoding PHAS loci, for both 21-PHAS and 24-PHAS, most were enriched in reproductive tissues (Fig. 3C).
Browsing miRNA and PHAS loci in sRNAanno
All the annotated miRNA and PHAS loci can be easily explored on the BROWSE page in sRNAanno (Fig. 4A). For a given species, the miRNAs are listed in a table, with the main information included, such as chromosomal coordinates, sequences of miRNAs, and miRNAs* (Fig. 4B). For each miRNA, a page of detailed information is also linked, in which a folded secondary structure can be found. For PHAS loci, a summary table is also provided for each species (Fig. 4C). Major features of each PHAS locus, such as chromosomal coordinates, p value, sRNA abundance, maximum phasing score, and data sources, are listed in detail. The sRNA distribution and phasing score of each PHAS locus are displayed on a linked page, with additional information listed, including the sequence of the PHAS locus, SwissProt annotations, and potential Pfam domain structure (Fig. 4C).
Fig. 4. Exploration of miRNA and PHAS loci in sRNAanno.
Demonstrative screenshots of the BROWSE page and linked pages for three main functions: Phylogenetic tree of all species in sRNAanno (A), miRNA list (B), PHAS loci list (C), and TAS gene list (D)
TAS3 is an ancient and archetypal PHAS gene that is widely conserved in all terrestrial plants15. TAS3 has features distinct from most PHAS genes: it is usually targeted at two sites by miR390 and generates one or two tasiARFs (tasiRNAs targeting ARF genes)15,17. The miR390-TAS3-ARF pathway plays essential role in the auxin signaling network. Here, we annotated 232 TAS3 genes from our PHAS analysis results, and all these TAS3 genes are collectively represented in an independent table (Fig. 4D), in which the sequences of miR390 target sites and tasiARF(s) are included. Links to the corresponding page of PHAS features are also provided for convenient browsing.
hc-siRNA locus annotations
hc-siRNAs account for a major part of the plant cellular sRNA population. At present, the genomic loci generating hc-siRNAs have not been thoroughly annotated owing to their large number, variability in sRNA abundances, and complexity of their biogenesis. hc-siRNAs are generated from repeat-related sequence regions, typically transposons, and heterochromatic regions, to direct cis DNA methylation in plants18. As these regions generating hc-siRNA are also important components of genomic information for a species, we annotated hc-siRNA loci for genomes with sRNA data available according to the criteria listed in Fig. S5. Indeed, these hc-siRNA-generating loci are abundant in almost every genome (Fig. S6).
sRNA annotation service
Small RNA data analysis using various bioinformatic software or pipelines relying on programming and command-line environments is challenging and time-consuming for most wet-lab biologists. To facilitate the ease of sRNA annotation, we are providing free service for sRNA annotations in sRNAanno. Users can upload sRNA NGS data and corresponding genome/reference sequence file to an accessible online repository (such as an FTP site) and submit download links to these files to sRNAanno on the RESOURCES page. Upon receiving the information, we will download the data files, perform the sRNA annotations, and return the annotation results to the users by email. For this service, we will maintain high confidence of users’ data and results and will not, under any circumstance, use them or release them to the public without users’ permission.
Discussion
The new database repository of plant small RNAs described here, sRNAanno, is a repository of major types of sRNAs for >140 plant genomes. These extensive annotations were achieved by analyzing ~1,600 sRNA datasets using well-established computation pipelines with reliable and highly stringent criteria. The sRNAanno database includes miRNA annotations of ~64% more plant species than the number within miRBase, the main and most popular miRNA hub, and the number of miRNA annotations in sRNAanno is also much greater than the number in recently published databases, including PmiREN, and “Plant Small RNA Genes”9,10. Moreover, all the miRNAs in sRNAanno were annotated via an identical process with consistent criteria, in contrast to the variable annotation criteria used for the miRNAs in miRBase, whose annotations was conducted by different research groups with various tools19. Generally, we believe that the miRNA annotations in sRNAanno are more reliable than those in miRBase. However, there is no gold standard for annotations of plant miRNAs. Although there are misannotations in miRBase, there may be a certain number of bona fide miRNAs that are possibly missing in sRNAanno. Moreover, miRBase also houses miRNAs from plant species whose genome has not been sequenced or for which no publicly available sRNA data are available (for which we are unable to perform miRNA annotations). Therefore, sRNAanno is a good complement, instead of a substitute, to miRBase.
In addition to miRNAs, sRNAanno also stores information concerning genomic loci generating phasiRNAs or hc-siRNAs. Annotations of these sRNA-generating loci were conducted using high confidence settings according to the widely accepted criteria. In plants, phasiRNAs have emerged as one of the major types of sRNAs, and their targets function in a broad range of biotic and abiotic processes. For instance, phasiRNAs are abundantly produced during the reproductive stage, especially in monocots and their subgroups of grasses15,20. In rice, there are >2,000 PHAS loci generating 21-nt phasiRNAs and ~400 loci generating 24-nt phasiRNAs21. Although phasiRNAs, including well-known tasiRNAs, have been characterized in an increasing number of plant species, there is no public online repository of reported or annotated phasiRNAs to provide convenient and quick access to this information. Similarly, hc-siRNAs are widely present in plant cells and are well known for their connection to DNA methylation or other epigenetic modifications, but the majority of plant genomes lack good annotations of hc-siRNA loci. In this study, we performed broad annotations of phasiRNAs and hc-siRNAs in plants, and the resulting annotations stored in sRNAanno constitute a valuable resource to facilitate genomic and genetic research in plants.
Conclusions
Thorough annotations of miRNAs, phasiRNAs, and hc-siRNAs were conducted for the genome of 143 plant species. All the annotation results are of high quality and confidence and have been deposited in the public database repository sRNAanno (www.plantsRNAs.org) for quick and convenient access. Both miRNA and PHAS loci can be easily browsed to view their main features. All these data and results are valuable resources facilitating research on sRNAs or related areas of plants.
Supplementary information
Acknowledgements
This work was funded by the National Key Research and Developmental Program of China (2018YFD1000104). This work was also supported by awards from the National Natural Science Foundation of China (#31872063), the Special Support Program of Guangdong Province (2019TX05N193), the Guangzhou Science and Technology Key Project (201804020063), the Innovation Team Project of the Department of Education of Guangdong Province (2016KCXTD011), and the Key Areas of Science and Technology Planning Project of Guangdong Province (2018B020202011). The group of J.Z. was supported by the Guangdong Innovative and Entrepreneurial Research Team Program (2016ZT06S172). We thank all members of the Xia lab for their help with this project.
Author contributions
CC and RX designed the study. JL, CC, JF, LF, BL, and XY collected the data and performed the analyses. JZ and BCM helped with the data analysis and database design. JL, CC, and RX constructed the database and wrote the manuscript.
Data availability
All the open-access data (including all the annotated sRNA results) are available through the sRNAanno database (www.plantsRNAs.org)
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
These authors contributed equally: Chengjie Chen, Jiawei Li.
Supplementary information
The online version contains supplementary material available at 10.1038/s41438-021-00480-8.
References
- 1.Kozomara A, Birgaoanu M, Griffiths-Jones S. MiRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Axtell MJ, Meyers BC. Revisiting criteria for plant microRNA annotation in the era of big data. Plant Cell. 2018;30:272–284. doi: 10.1105/tpc.17.00851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fei Q, Xia R, Meyers BC. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell. 2013;25:2400–2415. doi: 10.1105/tpc.113.114652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng Y, et al. Identification of microRNAs, phasiRNAs and their targets in pineapple. Trop. Plant Biol. 2016;9:176–186. doi: 10.1007/s12042-016-9173-4. [DOI] [Google Scholar]
- 5.Feng L, Xia R, Liu Y. Comprehensive characterization of miRNA and PHAS loci in the diploid strawberry (Fragaria vesca) genome. Hortic. Plant J. 2019;5:255–267. doi: 10.1016/j.hpj.2019.11.004. [DOI] [Google Scholar]
- 6.Xia R, Xu J, Arikit S, Meyers BC. Extensive families of miRNAs and PHAS loci in Norway Spruce demonstrate the origins of complex phasiRNA networks in seed plants. Mol. Biol. Evol. 2015;32:2905–2918. doi: 10.1093/molbev/msv164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xia R, et al. 24-nt reproductive phasiRNAs are broadly present in angiosperms. Nat. Commun. 2019;10:627. doi: 10.1038/s41467-019-08543-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyers BC, et al. Criteria for annotation of plant microRNAs. Plant Cell. 2008;20:3186–3190. doi: 10.1105/tpc.108.064311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo Z, et al. PmiREN: a comprehensive encyclopedia of plant miRNAs. Nucleic Acids Res. 2020;48:D1114–D1121. doi: 10.1093/nar/gkz894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lunardon A, et al. Integrated annotations and analyses of small RNA-producing loci from 47 diverse plants. Genome Res. 2020;30:497–513. doi: 10.1101/gr.256750.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C, Zeng Z, Liu Z, Xia R. Small RNAs, emerging regulators critical for the development of horticultural traits. Hortic. Res. 2018;5:63. doi: 10.1038/s41438-018-0072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evers M, Huttner M, Dueck A, Meister G, Engelmann J. C. miRA: Adaptable novel miRNA identification in plants using small RNA sequencing data. BMC Bioinformatics. 2015;16:370. doi: 10.1186/s12859-015-0798-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuang Z, Wang Y, Li L, Yang X. MiRDeep-P2: accurate and fast analysis of the microRNA transcriptome in plants. Bioinformatics. 2019;35:2521–2522. doi: 10.1093/bioinformatics/bty972. [DOI] [PubMed] [Google Scholar]
- 14.Axtell MJ. ShortStack: comprehensive annotation and quantification of small RNA genes. RNA. 2013;19:740–751. doi: 10.1261/rna.035279.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia R, Xu J, Meyers BC. The emergence, evolution, and diversification of the miR390-TAS3-ARF pathway in land plants. Plant Cell. 2017;29:1232–1247. doi: 10.1105/tpc.17.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia R, et al. MicroRNA superfamilies descended from miR390 and their roles in secondary small interfering RNA biogenesis in eudicots. Plant Cell. 2013;25:1555–1572. doi: 10.1105/tpc.113.110957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 18.Axtell MJ. Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 2013;64:137–159. doi: 10.1146/annurev-arplant-050312-120043. [DOI] [PubMed] [Google Scholar]
- 19.Nakano M, et al. Plant MPSS databases: signature-based transcriptional resources for analyses of mRNA and small RNA. Nucleic Acids Res. 2006;34:D731–D735. doi: 10.1093/nar/gkj077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhai J, et al. Spatiotemporally dynamic, cell-type–dependent premeiotic and meiotic phasiRNAs in maize anthers. Proc. Natl Acad. Sci. 2015;112:3146–3151. doi: 10.1073/pnas.1418918112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamim S, et al. Cis-directed cleavage and nonstoichiometric abundances of 21-nucleotide reproductive phased small interfering RNAs in grasses. N. Phytol. 2018;220:865–877. doi: 10.1111/nph.15181. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the open-access data (including all the annotated sRNA results) are available through the sRNAanno database (www.plantsRNAs.org)