Abstract
Capturing the folding dynamics of large, functionally important RNAs has relied primarily on global measurements of structure or on per-nucleotide chemical probing. These approaches infer, but do not directly measure, through-space structural interactions. Here we introduce trimethyloxonium (TMO) as a chemical probe for RNA. TMO alkylates RNA at high levels in seconds, and thereby enables time-resolved, single-molecule, through-space probing of RNA folding using the RING-MaP correlated chemical probing framework. Time-resolved correlations in the RNase P RNA – a functional RNA with a complex structure stabilized by multiple noncanonical interactions – revealed that a long-range tertiary interaction guides native RNA folding for both secondary and tertiary structure. This unanticipated non-hierarchical folding mechanism was directly validated by examining the consequences of concise disruption of the through-space interaction. Single-molecule, time-resolved RNA structure probing with TMO is poised to reveal a wide range of dynamic RNA folding processes and principles of RNA folding.
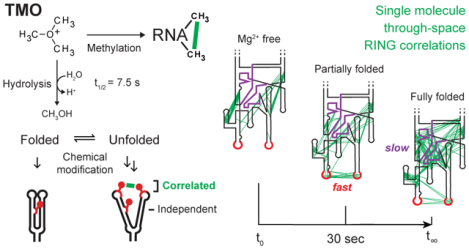
Graphical Abstract
Chemical probing is widely used to monitor RNA folding reactions. Broadly, chemical probing involves reacting an RNA with a small molecule that is sensitive to the underlying nucleic acid structure1–4. Chemical probing has revealed numerous features of RNA biology and allows complex RNA structures to be modeled with good to outstanding accuracy4–7. However, most chemical probing strategies merely infer RNA structure from observed reactivity, but do not measure RNA structure directly.
RNA interaction groups analyzed by mutational profiling (RING-MaP) technologies directly detect through-space structural communication in RNA. In RING-MaP, RNAs are modified at high per-nucleotide rates such that multiple chemical adducts occur in the same RNA strand and are comprehensively detected by a processive relaxed fidelity reverse transcription (MaP) reaction8–10. RING-MaP measures reagent-induced sequential unfolding of RNA molecules; however, every molecule is perturbed in a unique manner and perturbations average out over a population of molecules8,10. Modification of one nucleotide in a dynamically exposed base pair or tertiary interaction can block local refolding, increasing the probability of subsequent modification of the pairing partner (Figure 1A). Through-space structural communication is detected as the subset of interdependent or correlated modification events in the same molecular strand of RNA (Figure 1A). RING-MaP chemical probing can be used to measure multiple features of RNA structure, depending on how correlation data are interpreted. RING-MaP has been used directly detect RNA duplexes (termed PAIR-MaP)9,10, to define multiple conformational states in an RNA ensemble8, and to reveal tertiary interactions and through-space structural communication8,11,12 (Figure 1B). In this study, we focus on the latter, examining through-space RINGs, in a time-resolved way. To date, RING-MaP correlated chemical probing has been carried out using dimethyl sulfate (DMS).
Figure 1.
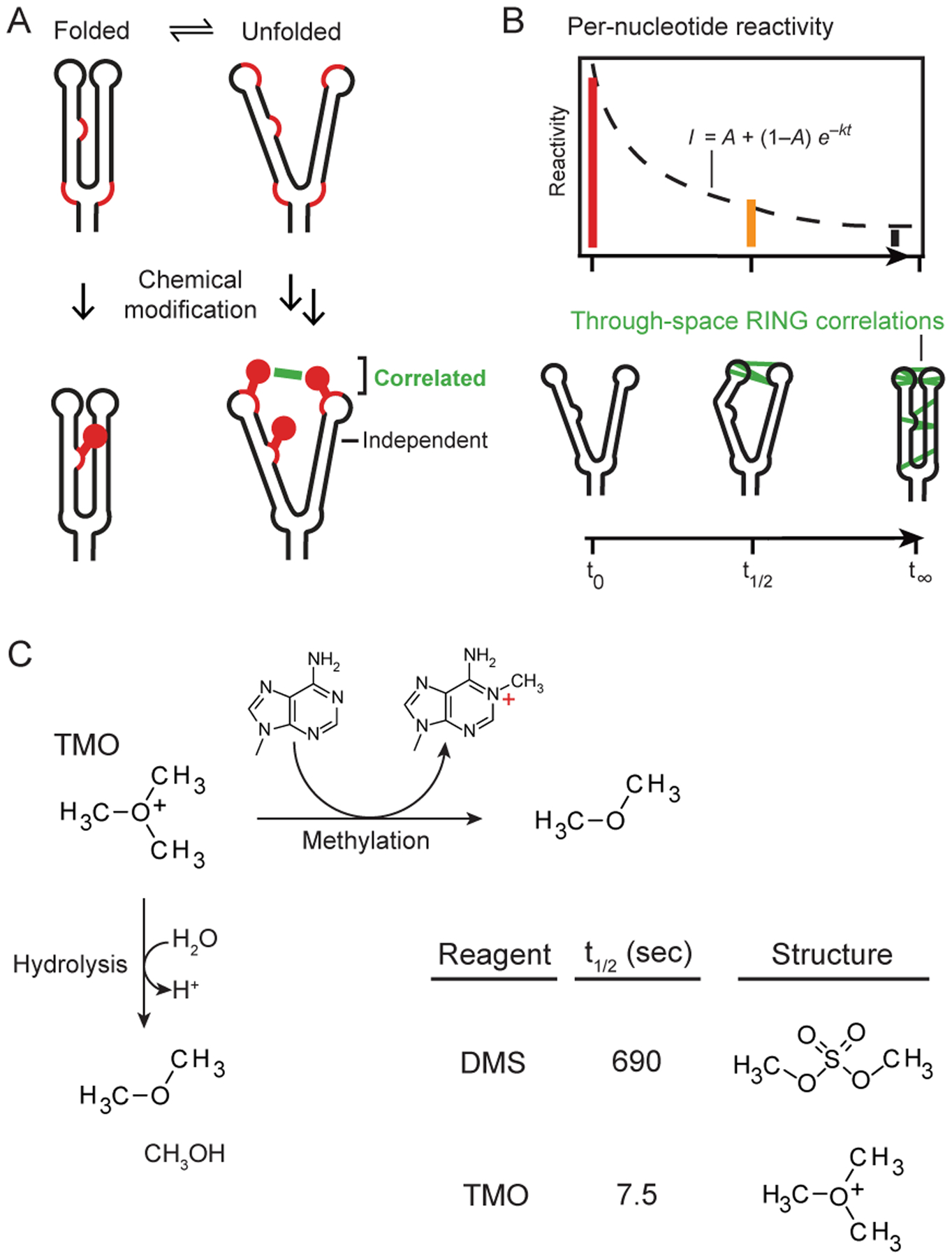
Correlated probing and TMO reactivity. (A) Mechanism of correlated chemical probing. Fluctuations in base-pairing or higher-order structure enable chemical modification, such that one modification event promotes reaction with a second site (correlated adducts). In contrast, modifications at low-structure sites occur independently. Reactive sites emphasized with red backbone. (B) Strategies for analyzing time-resolved probing data. (top) Conventional time series of per-nucleotide reactivity changes and (bottom) pairwise through-space RING correlations. (C) RNA methylation and hydrolysis reactions for TMO.
DMS is widely used to examine interrelationships between RNA structure and cellular function, including in cells1,13. DMS has high reactivity with unpaired adenosine and cytidine nucleotides, and our laboratory has developed reaction conditions that now enable DMS to modify uridine and guanosine at structurally informative levels (Supporting Methods and ref. 10). DMS reacts slowly with RNA, with a reaction half-life of ~12 min (Figure 1C).
We sought to identify a chemical probe with the advantages of DMS but much faster reactivity. Trimethyloxonium tetrafluoroborate (TMO)14 is highly soluble, reacts through RNA alkylation and self-quenching hydrolysis, and has half-life in buffered solution of 7.5 sec (Figure 1C, Supporting Figure 1). TMO thus reacts 90 times more rapidly than DMS, does not require an explicit quenching step, and probes RNA folding on the seconds time scale.
We compared TMO and DMS reactivities by probing the structure of the B. stearothermophilus ribonuclease P (RNase P) catalytic domain, an RNA with a complex secondary and tertiary structure15,16. TMO and DMS both achieve the high level of reactivity required for correlated chemical probing (Figure 2A). TMO, like DMS, measures RNA structure in accurate and useful ways, as shown by multiple experiments.
Figure 2.
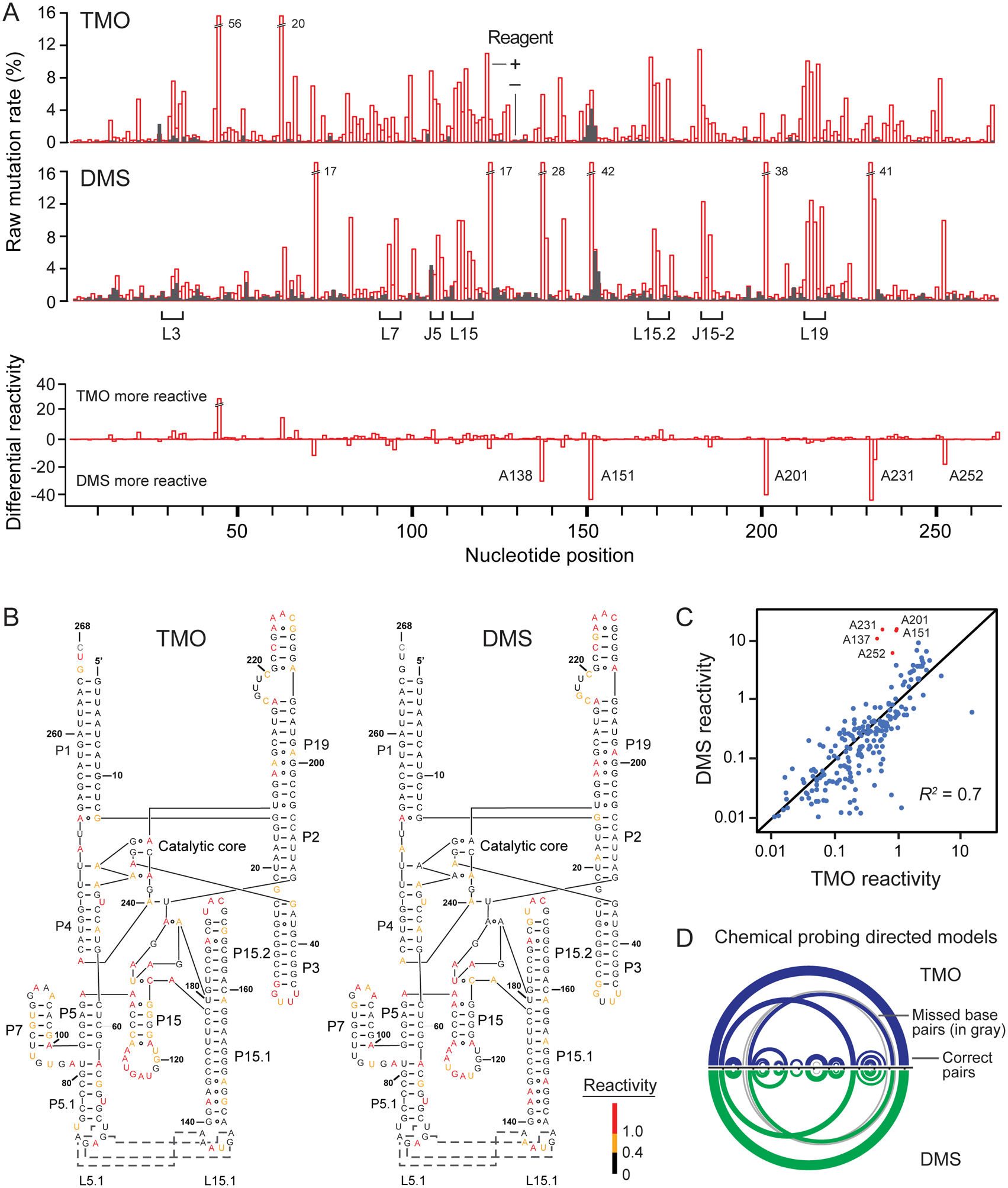
Comparison of TMO and DMS reactivities. (A) Raw reactivity rates (top) for the RNase P catalytic domain RNA. RNA adducts were identified using MaP8,10; median modifications per strand were 5–8. Loop (L) and junction (J) landmarks are emphasized; off scale rates are labeled. Differential reactivity (bottom) occurs preferentially at sites of noncanonical base pairing (labeled nucleotides). (B) Reactivities superimposed on the RNase P secondary structure. Red, yellow, and black nucleotides indicate high, medium, and low reactivities, respectively. Dashes and circles connecting nucleotides show Watson-Crick and non-canonical base pairing, respectively. (C) Correlation between TMO and DMS reactivity. Points indicate individual nucleotides. Positions with preferential reactivity toward DMS (red) correspond to Hoogsteen-paired adenosine nucleotides, and were excluded from R2 calculation. (D) Chemical probing-directed secondary structure models for the RNase P catalytic domain based on TMO and DMS reactivities, modeled using the PAIR-MaP framework10. Arcs indicate base pairs; gray arcs indicate missed base pairs relative to the accepted structure.
First, superimposition of TMO and DMS reactivities on the secondary structure of RNase P (Figure 2B) and a second model RNA (Supporting Figure 2) reveals that both reagents react with all four RNA nucleotides and preferentially with unpaired RNA regions. Second, TMO reactivities correlate strongly with those for DMS (Figure 2C) and, third, discriminate strongly and similarly between paired and unpaired positions at all four ribonucleotides (Supporting Figure 3). A few nucleotides show differences, reflective of distinct probe structure and charge features (Figure 1C, 2A); specifically, adenosine residues that form Hoogsteen base pairs react preferentially toward DMS (Figure 2C).
Fourth, per-nucleotide reactivity data direct accurate models of the RNase P using both DMS and TMO, which requires both good detection of unpaired sites and maintenance of the overall RNA structure4,10. When reactivities are converted to pseudo-free energy change restraints, DMS reactivity-informed modeling of both RNase P and other model RNA structures recovered both long- and short-range helices, and a pseudoknot, as expected10; TMO reactivity data yield secondary structure models of comparable accuracy (Figure 2D, Supporting Figure 2).
Fifth and finally, TMO reactions performed at 40 and 100 mM reagent are highly correlated (R2 = 0.96; Supporting Figure 4A). Taken together with accurate structure modeling (Figure 2, Supporting Figure 2), these data emphasize that high level TMO reactivity measures RNA structure accurately and does not disrupt the underlying RNA structure.
Given this extensive validation, we used TMO to follow time-dependent RNA folding processes in two ways: (i) Time-series per-nucleotide reactivity data to reveal folding kinetics and (ii) pair-wise correlation data to map through-space RNA interaction networks (Figure 1B).
We performed time-resolved probing experiments with TMO, in replicate (Supporting Figure 4), to characterize the folding landscape of RNase P. RNA folding was initiated by adding Mg2+ (to 10 mM) to RNase P (5 μM, pH 8) at 37 °C. Aliquots were removed at intervals 5 to 1200 seconds after Mg2+ addition, and were added to one-tenth volume of TMO (to 100 mM final). TMO reactivity profiles were used to generate data-directed structural models of the RNase P RNA as a function of time. Roughly 40% of nucleotides in the RNA showed time-dependent behavior.
RNase P architecture is comprised of canonical base pairing in helices P1–P19, noncanonical pairing and stacking throughout the catalytic core, and a long-range tertiary pairing between loops L5.1 and L15.1 (Figure 3A)15. Prior to Mg2+addition, helices P2, P5, and P15 were reactive and thus not formed. P2, in particular, forms a pseudoknot critical for tertiary folding of the RNA. Nucleotides in the catalytic core and in the L5.1-L15.1 loop-loop interaction were also reactive and thus not formed. (detailed reactivities shown in Supporting Figure 5). Time-resolved TMO probing revealed that nucleotides in the P2, P5 and P15 helices refolded relatively slowly upon Mg2+ addition, with nucleotides in the catalytic core showing even slower reactivity changes (Figure 3B). Conversely and intriguingly, loops L5.1 and L15.1 were relatively rapidly protected from TMO reaction, suggestive of a critical role for this element in the folding landscape.
Figure 3.
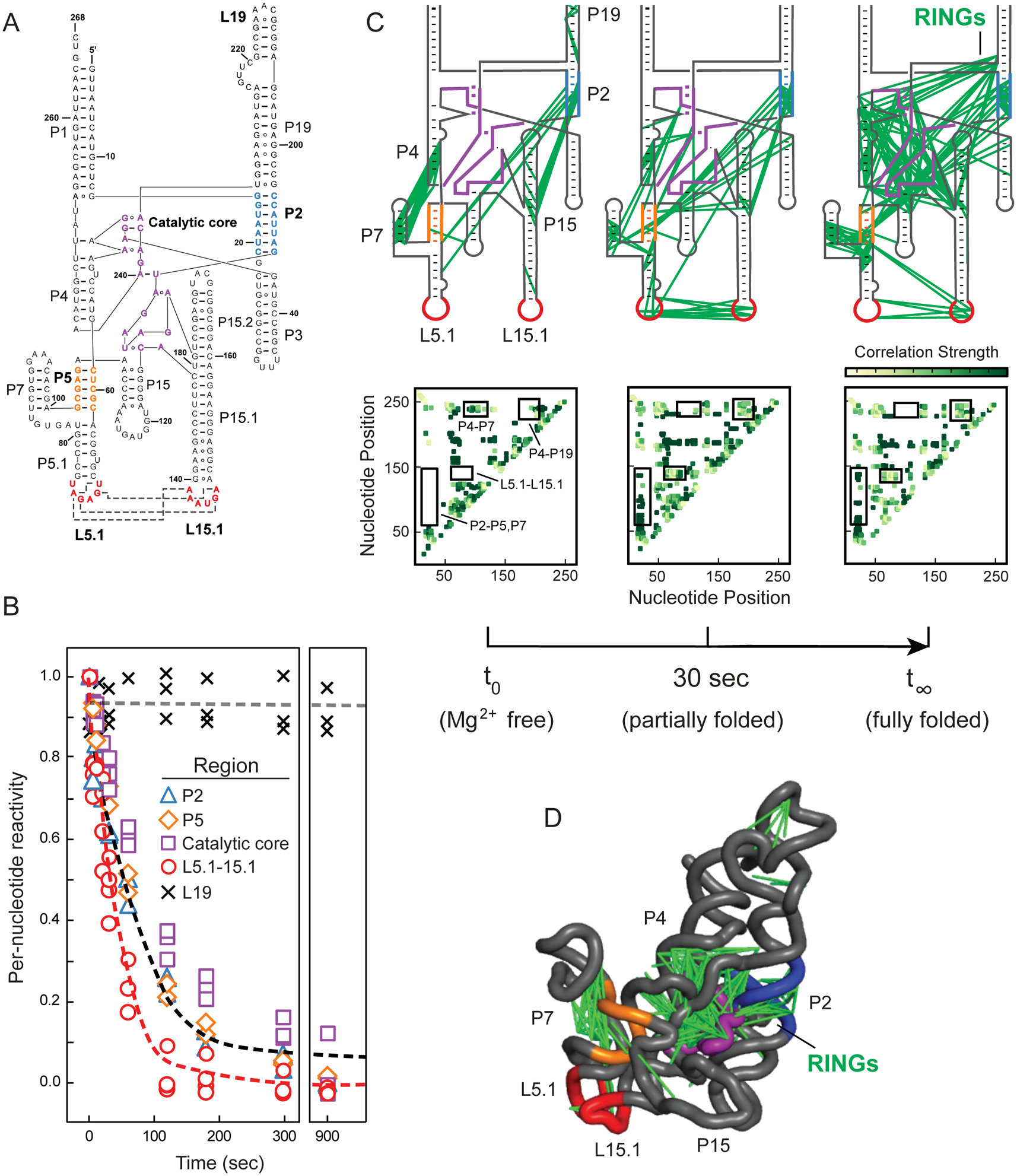
Time-resolved folding of the RNase P RNA. (A) Secondary structure. Regions undergoing folding transitions upon Mg2+ addition are emphasized in color (in all panels). (B) Time-dependent reactivity profiles. Individual per-nucleotide reactivities are shown as points. Red and black lines show best fits to averaged reactivities for nucleotides that form the L5.1-L15.1 interaction and for all other folding motifs. Rate constants are 0.030 and 0.011 sec–1, respectively. Representative unchanging region is shown with × and gray line. (C) Pairwise through-space RING-MaP correlations as a function of folding time. Inter-nucleotide correlations are shown with green lines superimposed on secondary structure; heatmaps of same data are shown below each panel, with key regions boxed and labeled. (D) Three-dimensional structure of RNase P RNA15, with superimposed RING correlations.
Global structural transitions of the RNase P catalytic domain have been monitored previously by their circular dichroism profile. These studies revealed that the RNA folds through a, currently uncharacterized, meta-stable intermediate to form the equilibrium structure17. Time-resolved TMO reactivity data suggested that P2, P5, the catalytic core, and the L5.1-L15.1 interaction might all contribute to formation of intermediates in the RNase P RNA folding pathway.
We next analyzed these same TMO data based on pairwise RING correlations (Figure 1A, 1B). Extensive changes in through-space structural communication occur as the RNase P RNA folds (Figure 3C, green lines and heatmap intensities). Interactions are relatively sparse in the absence of Mg2+, consistent with the role of Mg2+ in stabilizing tertiary RNA structures. Correlations linking P4 and P7, and P2 and P15.2 occurred in the absence of Mg2+ but disappeared in the final structure, indicative of misfolding in the Mg2+-free structure. Correlations between the L5.1 and L15.1 loops appeared rapidly upon addition of Mg2+, and then decreased gradually as the RNA folded (Figure 3C, Supporting Figure 6). The decrease in correlation density in L5.1-L15.1 after 60 seconds likely reflects that fewer pairwise modifications are observed as nucleotides in these stable non-canonical interactions become unreactive. In sum, visualization of through-space folding (Figure 3C, 3D, Supporting Figure 6) reveals rapid formation of the tertiary L5.1-L15.1 loop-loop interaction followed by slower folding of the P2 pseudoknot and catalytic core.
Our study thus far suggested that the L5.1-L15.1 tertiary interaction guides native RNase P RNA folding. We made a concise 2-nucleotide mutation that extends the P15.1 helix and disrupts non-canonical tertiary base pairing between L5.1 and L15.1 (Figure 4A). TMO probing showed that the native sequence and L15.1 mutant RNAs formed the same misfolded starting structure in the absence of Mg2+. Upon Mg2+addition, TMO reactivities suggested that neither the L5.1-L15.1 tertiary interaction nor the P2–P4 pseudoknot formed fully in the mutant, as these nucleotides were more reactive than for the native sequence RNA (Figure 4B). RING correlations then directly showed that neither the L5.1-L15.1 interaction nor the P2 pseudoknot formed in the mutant (Figure 4C, Supporting Figure 7). Indeed, the final mutant structure resembles that observed for early folding stages of the native RNA. The L5.1-L15.1 loop-loop tertiary interaction thus coordinates RNA folding at both base-pairing and tertiary-structure levels.
Figure 4.
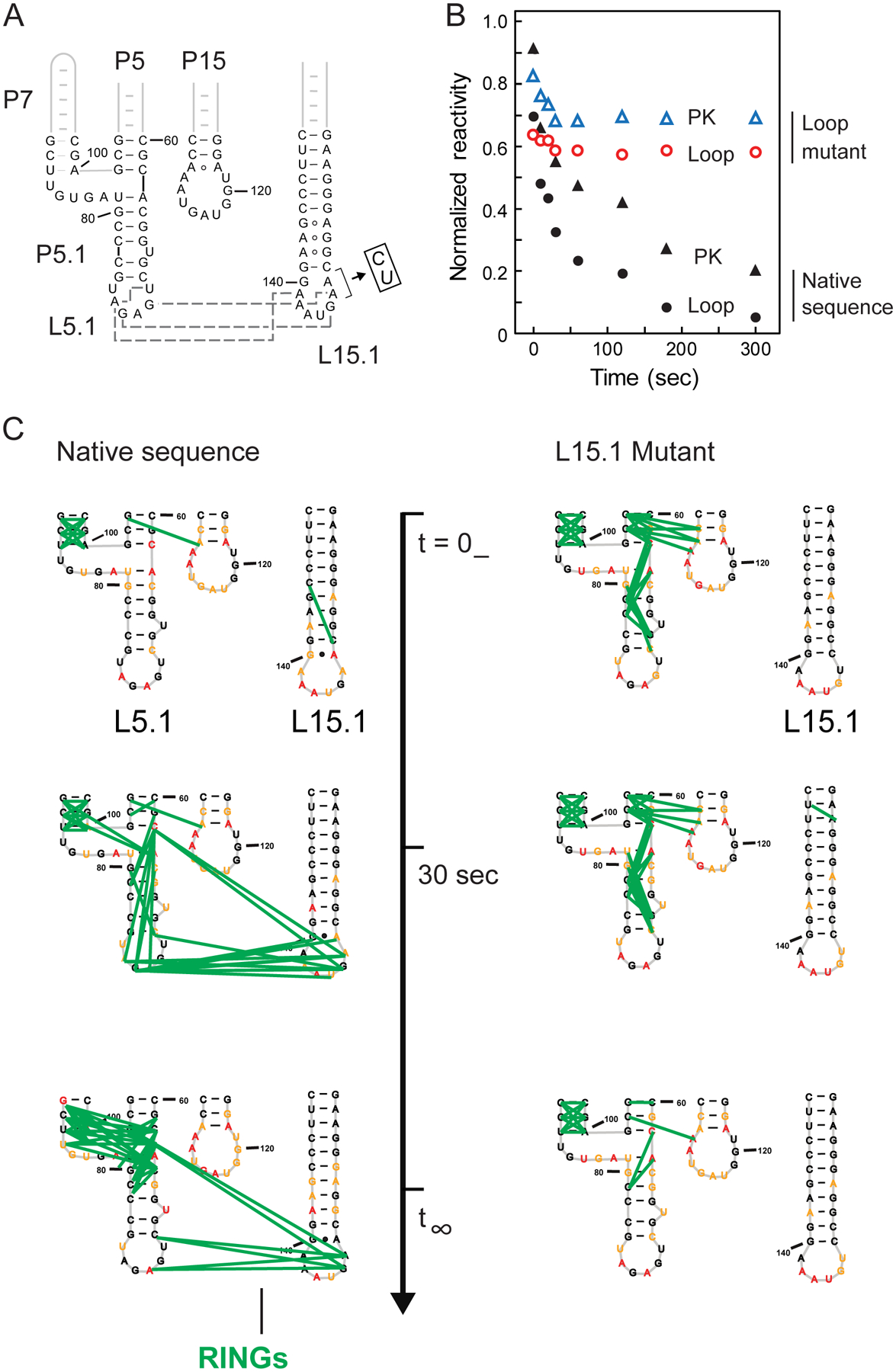
Role of L5.1-L15.1 tertiary interaction on RNase P RNA folding. (A) Mutant that disrupts L5.1-L15.1 interaction. Tertiary contacts (in native sequence) are shown with gray dashed lines. (B) Time-dependent reactivities in the pseudoknot (PK) and L5.1 and L15.1 loops for native-sequence and mutant RNAs. (C) Time-dependent through-space correlations for native-sequence and mutant RNAs (green lines); complete correlation maps are shown in Supporting Figure 7.
RNA folding is broadly modeled as hierarchical, meaning that base-paired secondary structures form independently of and prior to higher-order tertiary structures18. Within this framework, structural coupling is also observed such that disruption of a tertiary structure can destabilize secondary structure19,20,21.
Single-molecule correlated probing directly visualizes time evolution of dozens of through-space interactions (Figures 3, 4; Supporting Figures 6, 7). Unanticipatedly, our study shows that RNase P folding is clearly not hierarchical. First, non-native interactions occur at early folding time points but then disappear, consistent with reorganization of both secondary and tertiary structure. Second, early formation of the long-range L5.1-L15.1 tertiary interaction drives native folding of both secondary and tertiary structures.
Time-resolved, correlated chemical probing experiments provide a powerful approach to characterize RNA folding landscapes. TMO has rapid, readily controlled, alkylation reaction kinetics and high reactivity rates, allowing for layering of single-molecule pairwise correlation data on per-nucleotide reactivities to reveal RNA folding intermediates and pathways directly. This study moves chemical probing beyond merely inferring dynamics of RNA structure formation, to enable simultaneous and direct measurement of multiple complex folding features for functionally important RNAs.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the US NIH (R35 GM122532 to K.M.W.).
Footnotes
Supporting Information
Detailed experimental methods, two tables, and seven figures. The Supporting Information is available free of charge on the ACS Publications website.
K.M.W. is an advisor to and holds equity in Ribometrix.
REFERENCES
- (1).Peattie DA; Gilbert W Chemical probes for higher order structure in RNA. Proc. Natl. Acad. Sci. USA 1980, 77, 4679–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Brenowitz M; Chance M; Dhavan G; Takamoto K Probing the structural dynamics of nucleic acids by quantitative time-resolved and equilibrium hydroxyl radical ‘footprinting’. Curr. Opin. Struc. Biol 2002, 12, 648–653. [DOI] [PubMed] [Google Scholar]
- (3).Mortimer S; Weeks KM C2’-endo nucleotides as molecular timers suggested by the folding of an RNA domain. Proc. Natl. Acad. Sci. U. S. A 2009, 106, 15622–15627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Weeks KM Advances in RNA structure analysis by chemical probing. Curr. Opin. Struct. Biol 2010, 20, 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Smola MJ; Rice GM; Busan S; Siegfried NA; Weeks KM Selective 2’-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat. Protoc 2015, 10, 1643–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Mustoe AM; Corley M; Laederach A; Weeks KM Messenger RNA Structure Regulates Translation Initiation: A Mechanism Exploited from Bacteria to Humans. Biochemistry 2018, 57, 3537–3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Boerneke MA; Ehrhardt JE; Weeks KM Physical and Functional Analysis of Viral RNA Genomes by SHAPE. Annu. Rev. Virol 2019, 6, 93–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Homan PJ; Favorov OV; Lavender CA; Kursun O; Ge X; Busan S; Dokholyan NV; Weeks KM Single-molecule correlated chemical probing of RNA. Proc. Natl. Acad. Sci. U. S. A 2014, 111, 13858–13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Krokhotin A; Mustoe AM; Weeks KM; Dokholyan NV Direct identification of base-paired RNA nucleotides by correlated chemical probing. RNA 2017, 23, 6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Mustoe AM; Lama NN; Irving PS; Olson SW; Weeks KM RNA base-pairing complexity in living cells visualized by correlated chemical probing. Proc. Natl. Acad. Sci. U. S. A 2019, 116, 24574–24582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Sengupta A; Rice GM; Weeks KM Single-molecule correlated chemical probing reveals large-scale structural communication in the ribosome and the mechanism of the antibiotic spectinomycin in living cells. PLOS Biol 2019, 17, e3000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Dethoff EA; Boerneke MA; Gokhale NS; Muhire BM; Martin DP; Sacco MT; McFadden MJ; Weinstein JB; Messer WB; Horner SM; Weeks KM Pervasive tertiary structure in the dengue virus RNA genome. Proc. Natl. Acad. Sci. U. S. A 2018, 115, 11513–11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Wells SE; Hughes JMX; Ares M Use of dimethylsulfate to probe RNA structure in vivo. Methods Enzymol 2000, 318, 479–493. [DOI] [PubMed] [Google Scholar]
- (14).Stahl I; Seapy DG Trimethyloxonium Tetrafluoroborate. In Encyclopedia of Reagents for Organic Synthesis; John Wiley & Sons, Ltd: Chichester, UK, 2008. [Google Scholar]
- (15).Kazantsev AV; Krivenko AA; Pace NR Mapping metal-binding sites in the catalytic domain of bacterial RNase P RNA. RNA 2009, 15, 266–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Kazantsev AV; Rambo RP; Karimpour S; Santalucia J; Tainer JA; Pace NR Solution structure of RNase P RNA. RNA 2011, 17, 1159–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Fang XW; Golden BL; Littrell K; Shelton V; Thiyagarajan P; Pan T; Sosnick TR The thermodynamic origin of the stability of a thermophilic ribozyme. Proc. Natl. Acad. Sci. U. S. A 2001, 98, 4355–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Brion P; Westhof E Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct 1997, 26, 113–137. [DOI] [PubMed] [Google Scholar]
- (19).Woodson SA Compact Intermediates in RNA Folding. Annu. Rev. Biophys 2010, 39, 61–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Tian S; Das R RNA structure through multidimensional chemical mapping. Quarterly Rev. Biophys 2016, 49, e7. [DOI] [PubMed] [Google Scholar]
- (21).Herschlag D; Bonilla S; Bisaria N The story of RNA folding, as told in epochs. Cold Spring Harb. Perspect. Biol 2018, 10, a032433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


