Jones et al. determine the role of matrix stiffness in regulating chromatin accessibility in freshly isolated lung fibroblasts. They identify ZNF416 as a key transcriptional regulator controlling fibroblast activation by the mechanical environment with relevance to wound healing and fibrosis.
Abstract
Matrix stiffness is a central regulator of fibroblast function. However, the transcriptional mechanisms linking matrix stiffness to changes in fibroblast phenotype are incompletely understood. Here, we evaluated the effect of matrix stiffness on genome-wide chromatin accessibility in freshly isolated lung fibroblasts using ATAC-seq. We found higher matrix stiffness profoundly increased global chromatin accessibility relative to lower matrix stiffness, and these alterations were in close genomic proximity to known profibrotic gene programs. Motif analysis of these regulated genomic loci identified ZNF416 as a putative mediator of fibroblast stiffness responses. Genome occupancy analysis using ChIP-seq confirmed that ZNF416 occupies a broad range of genes implicated in fibroblast activation and tissue fibrosis, with relatively little overlap in genomic occupancy with other mechanoresponsive and profibrotic transcriptional regulators. Using loss- and gain-of-function studies, we demonstrated that ZNF416 plays a critical role in fibroblast proliferation, extracellular matrix synthesis, and contractile function. Together, these observations identify ZNF416 as novel mechano-activated transcriptional regulator of fibroblast biology.
Introduction
Fibroblasts are tissue-resident mesenchymal cells responsible for maintenance and remodeling of the ECM. In the case of tissue injury or insult, transient fibroblast activation is critical for proper wound repair (Darby et al., 2014). Following injury resolution, activated fibroblasts revert to their quiescent state or undergo apoptosis. Persistent fibroblast activation leads to enhanced ECM deposition and progression of fibrocontractile diseases (Tschumperlin et al., 2018). In the case of idiopathic pulmonary fibrosis, sustained fibroblast activation leads to replacement of functional alveolar tissue architecture with scar tissue, impairing lung function (Gabbiani, 2003; Darby et al., 2014).
Fibroblasts respond to the rigidity of their extracellular surroundings both in vitro and in vivo such that increased local tissue stiffness acts as an amplification feedback loop driving fibrosis progression (White, 2015; Ligresti et al., 2019; Liu et al., 2015; Haak et al., 2019; Balestrini et al., 2012; Liu et al., 2010; Szeto et al., 2016). For example, fibroblasts in a rigid microenvironment display robust transcriptomic changes (Moreno-Vicente et al., 2018; Liu et al., 2010), accompanied by enhanced ECM deposition and remodeling (Liu et al., 2010), amplified proliferation capacity (Mih et al., 2012), and increased contractility (Marinković et al., 2013; Mih et al., 2012) compared with their counterparts in a compliant microenvironment. Importantly, prior work has shown that primary cells can acquire an epigenetic “mechanical memory” after prolonged culture in rigid mechanical environments (Balestrini et al., 2012; Yang et al., 2014; Li et al., 2017; Heo et al., 2015), suggesting that the use of immortalized or serially passaged cells could hamper efforts to identify the full regulatory programs that determine fibroblast transcriptional mechanoresponses relevant to in vivo processes such as wound healing and fibrosis.
External mechanical stimuli are relayed into intracellular biochemical cascades that ultimately converge onto transcriptional regulators (Tschumperlin et al., 2018). While Yes-Assocated Protein (YAP)/Transcriptional Coactivator With PDZ-Binding Motif (TAZ) and Myocardin-related transcription factor A (MRTF-A) are highly studied mechanosensing transcriptional regulators implicated in fibroblast function (Tschumperlin et al., 2018; Liu et al., 2015; Huang et al., 2012), additional transcriptional regulatory mechanisms of mechanosensing likely remain to be identified and could serve as targets for reversing pathogenic fibroblast activation. Epigenomic profiling techniques, such as the assay for transposase-accessible chromatin followed by next-generation sequencing (ATAC-seq) have emerged as a method for identifying transcriptional regulators governing epigenetic/transcriptional responses to stimuli. For example, recent ATAC-seq analyses have identified matrix stiffness–dependent chromatin accessibility changes in an in vitro 3D model of epithelial breast cancer invasion (Stowers et al., 2019). Here, we globally assessed the effect of matrix stiffness on chromatin accessibility in freshly isolated mouse lung fibroblasts and identified ZNF416 as a novel transcriptional regulator of matrix stiffness–dependent fibroblast activation.
Results and discussion
Matrix stiffness increases chromatin accessibility
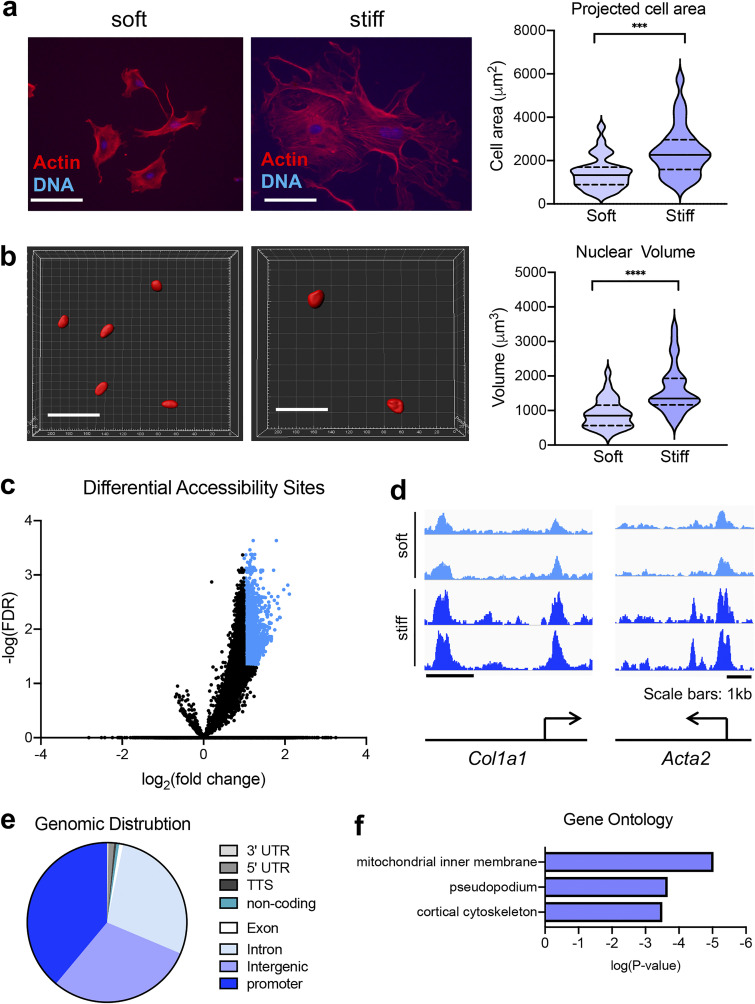
To address the role of matrix rigidity in shaping the chromatin accessibility landscape of “naive” fibroblasts, we used a Col1α1-GFP reporter mouse to isolate lung fibroblasts using FACS (Fig. S1 a). Freshly isolated fibroblasts were placed on identically collagen I–coated soft (0.2 kPa polydimethylsiloxane [PDMS]) or stiff (tissue-culture plastic) substrates for 8 d. Naive fibroblasts were responsive to the rigidity of their substrate as shown by their change in total cell area (Fig. 1 a) and nuclear volume (Fig. 1 b). To address whether matrix rigidity alters chromatin accessibility of naive lung fibroblasts, we used ATAC-seq. Using a false discovery rate (FDR; i.e., adjusted P value) ≤0.05 and a log2 fold-change of ±1, we found that stiff matrices increased accessibility at 2,114 genomic loci compared to soft matrices (Fig. 1 c; GEO accession no. GSE161434). Stiff matrices did not promote any reduction in chromatin accessibility compared with soft matrices (Fig. 1 c; GEO accession no. GSE161434).
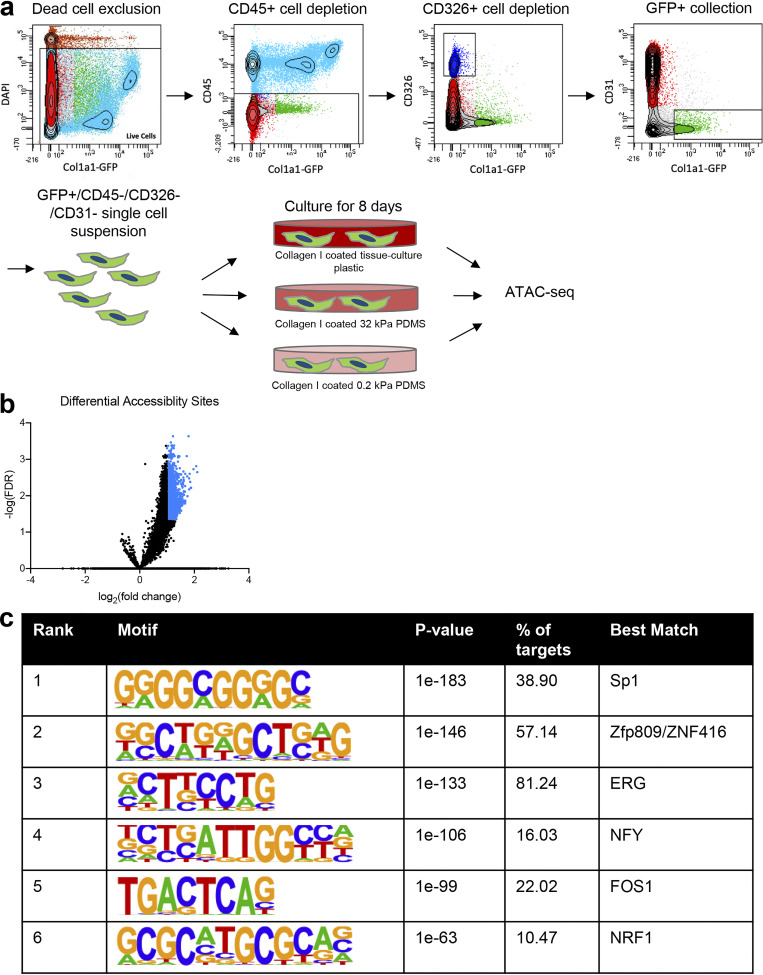
Figure S1.
Approach and validation of matrix stiffness in regulating chromatin accessibility. (a) Schematic displaying approach to isolate mouse lung GFP+ fibroblasts. Single-cell suspensions were first depleted of dead cells, followed by CD45, CD326, and CD31 depletion, and then followed by a GFP+ selection. Cells were then plated on collagen I–coated tissue-culture plastic and 0.2-kPa PDMS for 8 d. Cells were then submitted for ATAC-seq. (b) Volcano plot of DASs of fibroblasts on 0.2-kPa versus 32-kPa PDMS matrices. Each dot represents a single accessibility site. Significantly different accessibility sites colored in blue. (c) De novo DNA motif enrichment analysis of differential chromatin accessibility loci of fibroblasts on 0.2-kPa versus 32-kPa PDMS matrices ranked by P value. “% of targets” indicates the percentage of total input sequences that contain that respective motif.
Figure 1.
Matrix stiffness facilitates increased chromatin accessibility. (a) 20× fluorescent images of FACS-sorted Col1a1-GFP mouse lung fibroblasts accompanied with quantification of cell area on soft versus stiff matrices (soft: n = 32; stiff: n = 27). Solid lines in violin plots represent median. Dotted lines in violin plots represent upper and lower quartiles. Scale bars represent 50 µm. (b) 3D nuclei reconstructed in Imaris accompanied by quantification of nuclear volume on soft versus stiff matrices (soft: n = 32; stiff: n = 30). Solid lines in violin plots represent median. Dotted lines in violin plots represent upper and lower quartiles. Scale bars represent 50 µm. (c) Volcano plot of differential accessibility sites of fibroblasts on soft versus stiff matrices. Each dot represents a single accessibility site. Significantly different accessibility sites colored in blue. (d) Genomic views of differential chromatin accessibility sites in close proximity to known profibrotic genes. Scale bars represent 1 kb. (e) Genomic distribution of differential accessibility sites annotated to their nearest TSS. (f) Ontology analysis of genes annotated to all differential accessibility sites. ***, P < 0.001; ****, P < 0.0001, evaluated by unpaired Student’s t test.
Using Homer, we annotated these 2,114 genomic loci to the nearest transcriptional start site (TSS). This analysis revealed accessibility changes in close proximity to fibroblast activation genes such as Col1a1 and Acta2 (Fig. 1 d). The genomic distribution of these differentially accessible sites (DASs) revealed promoter regions as the dominant regulatory regions responding to matrix stiffness (38.9% of DASs), followed by intergenic/enhancer regions (29.7% of DASs) and introns (28% of DASs; Fig. 1 e). Using the Genomic Regions Enrichment of Annotations Tool (GREAT), we then conducted Gene Ontology analysis of the DASs and identified enrichment of genes involved in “cortical cytoskeleton” (Fig. 1 f). To confirm these results were matrix stiffness driven, we also compared ATAC-seq from 0.2-kPa to 32-kPa PDMS substrates. This comparison yielded a similar number and distribution of DASs (2,111; Fig. S1 b; GEO accession no. GSE161434), verifying the mechanoresponsiveness of chromatin alterations in freshly isolated fibroblasts.
Motif enrichment analysis identifies ZNF416 as a putative regulator of fibroblast activation
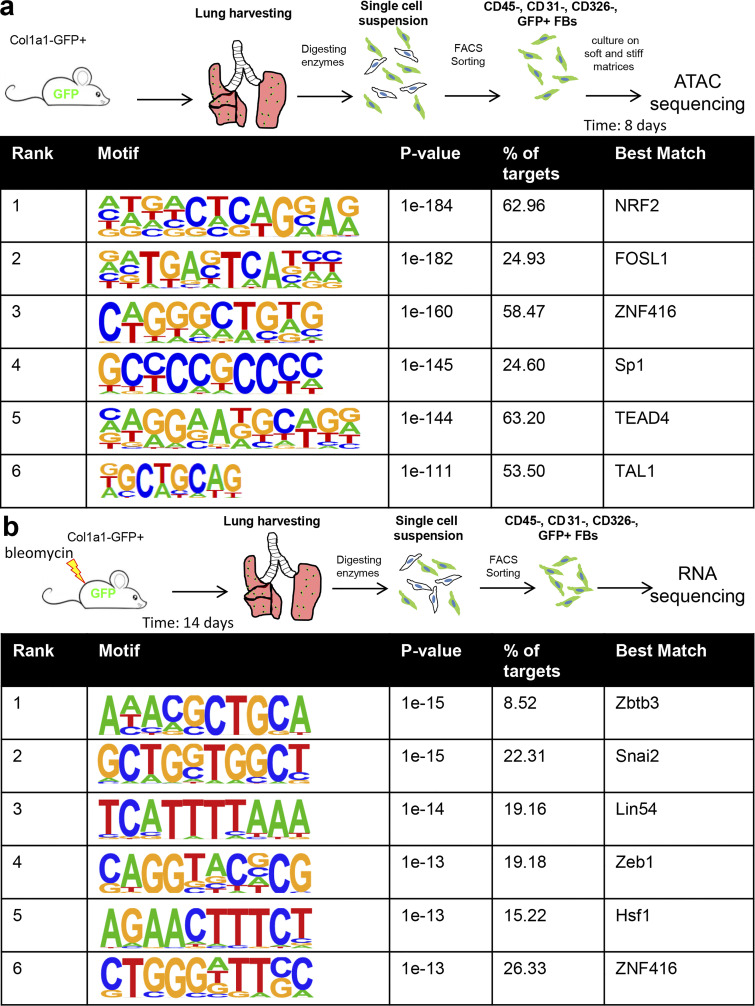
To identify candidate transcription factors associated with matrix stiffness–directed changes in chromatin accessibility, we performed de novo motif analysis on the 2,114 matrix stiffness DASs between 0.2 kPa and plastic using Homer (Fig. 2 a). This analysis identified several known mechanoresponsive factors, including Nrf2 (McSweeney et al., 2016), Sp1 (Stowers et al., 2019), and TEAD4 (Dupont et al., 2011), as well as factors not previously implicated in mechanoresponses (ZNF416 and TAL1). An identical analysis comparing DAS between 0.2 and 32 kPa PDMS matrices confirmed potential roles for Sp1 and ZNF416 (Fig. S1 c).
Figure 2.
Motif analysis identifies ZNF416 as a putative regulator of fibroblast activation in vitro and in vivo. (a) De novo DNA motif enrichment analysis of differential chromatin accessibility loci of fibroblasts on soft versus stiff matrices ranked by P value. Percentage of targets indicates the percentage of total input sequences which contain that respective motif. (b) Schematic of approach to identify differentially expressed genes in mouse fibroblasts following bleomycin challenge to induce experimental lung fibrosis. De novo DNA motif enrichment analysis was done in the promoter region (±1 kb from the TSS) of each differentially expressed gene. P value describes a statistical measure of the enrichment of the DNA motif within the input genomic DNA sequences compared with randomly generated “background” sequences.
To assess whether these transcriptional regulators contribute to fibroblast activation in vivo, we performed motif enrichment analysis scanning promoters (±1 kb from the TSS) of genes differentially expressed in lung fibroblasts following bleomycin-induced experimental lung fibrosis in mice (Fig. 2 b). Interestingly, the top-ranked transcription factor binding motif enriched in all three datasets was ZNF416 (Fig. 2, a–c), suggesting ZNF416 may play an unappreciated role in mechanoregulation of fibroblasts in vitro and in vivo. ZNF416 is a C2H2-type zinc-finger transcription factor and contains a Kruppel-associated box (KRAB) domain and 12 tandem C2H2 zinc-finger domains, suggesting ZNF416 can function as both a transcriptional activator and a repressor (Cassandri et al., 2017). The DNA binding motif for ZNF416 was identified in a study by Najafabadi et al. (2015). However, beyond its binding motif and protein structure, nothing has previously been reported about ZNF416 biological roles or regulation. Based on our primary interest in fibroblast function in human health and disease, as well as the absence of a defined mouse orthologue for ZNF416, we next turned to human lung fibroblasts to delineate the function of ZNF416.
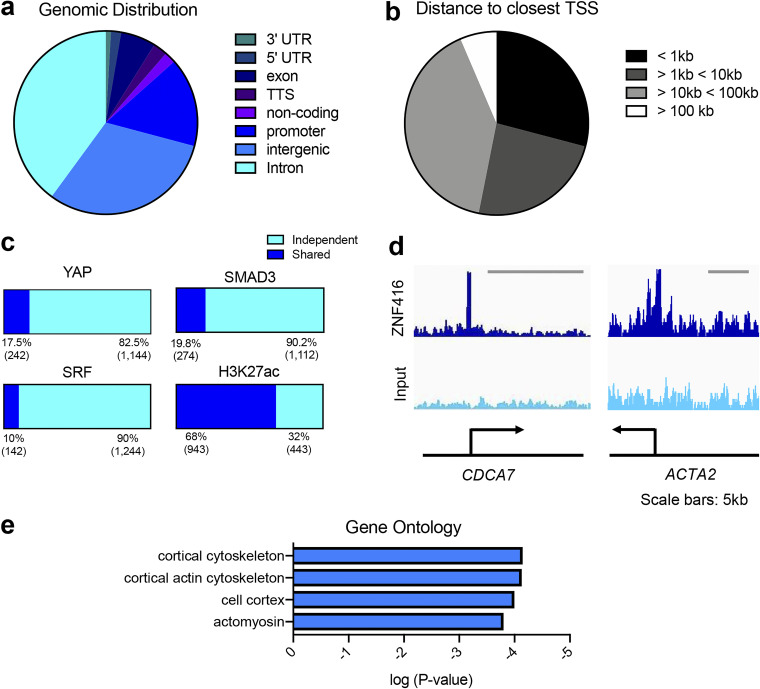
ZNF416 occupies and regulates key genes involved in fibroblast activation
To confirm that ZNF416 directly occupies regulatory genomic loci of genes central to fibroblast function, we performed chromatin immunoprecipitation followed by next-generation sequencing (ChIP-seq) in IMR90 human lung fibroblasts stably expressing ZNF416-FLAG (Fig. S2 d). ChIP-seq for FLAG identified ZNF416 uniquely binds to 1,386 locations in the human genome. We then annotated ZNF416’s binding sites to the nearest TSS using Homer (GEO accession no. GSE161446), revealing ZNF416 predominantly binds to intronic regions (40% of binding sites) and intergenic regions (30% of binding sites), followed by promoter regions (15% of binding sites; Fig. 3 a). Interestingly, ZNF416 largely binds to distal regions of the genome (>1 kb away from the nearest TSS), highlighting a consistency between ZNF416 occupancy and previously reported occupancy of YAP, SMAD3, and SRF (Fig. 3 b; Stein et al., 2015; Zanconato et al., 2015; Ding et al., 2013; Tsankov et al., 2015).
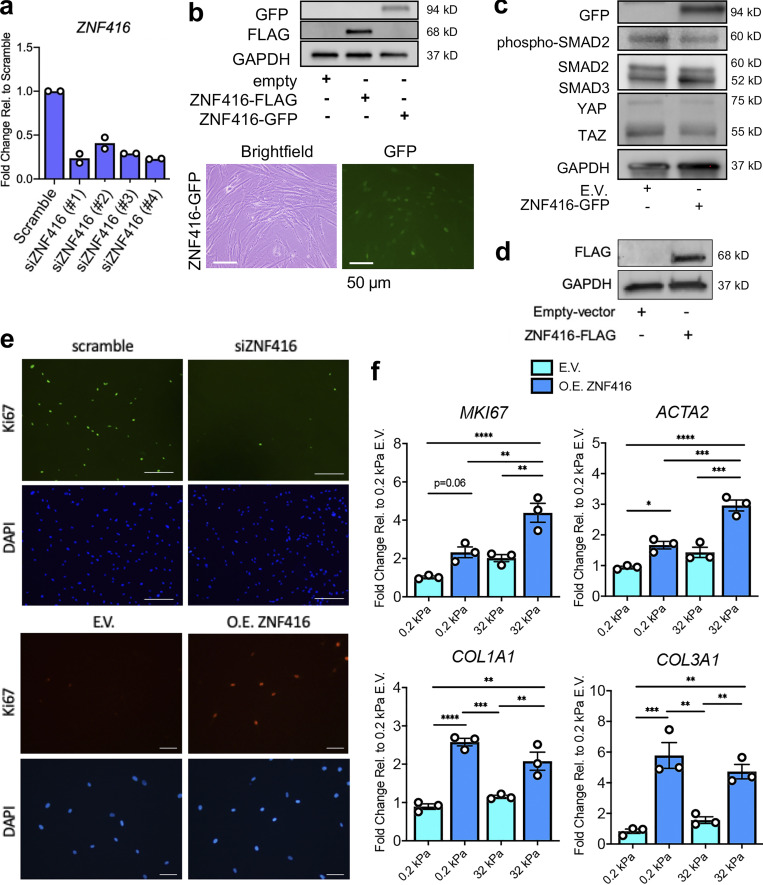
Figure S2.
Evaluation of the role of ZNF416 in fibroblast function. (a) qRT-PCR analysis of primary human lung fibroblasts treated with four individual siRNAs targeting ZNF416 to validate knockdown. Each dot represents an independent biological replicate from a single donor. Data normalized to primary human lung fibroblasts treated with scramble siRNA. (b) Western blot validation for expression of ZNF416-FLAG and ZNF416-GFP in primary human lung fibroblasts. Bright-field and GFP images (10×) of ZNF416-GFP–expressing primary human lung fibroblasts. Scale bars represent 50 µm. (c) Western blot of empty vector controls and ZNF416-GFP–overexpressing primary human lung fibroblasts. (d) Western blot depicting expression of ZNF416-FLAG in IMR90 cells. (e) Representative fluorescence images of Ki67 and DAPI counterstain of scramble and siZNF416-treated lung fibroblasts. Scale bars represent 100 µm. Representative fluorescence images of Ki67 and DAPI counterstain of empty vector and overexpressing (O.E.) ZNF416 lung fibroblasts. Scale bars represent 50 µm. (f) qRT-PCR analysis of empty vector control and ZNF416-overexpressing primary human lung fibroblasts placed on collagen I–coated soft (0.2-kPa PDMS) and stiff (32-kPa PDMS) matrices for 24 h. Experiment completed in biological triplicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, evaluated by one-way ANOVA with Tukey’s correction for multiple comparisons. All error bars represent SEM.
Figure 3.
ZNF416 genomic occupancy is distinct from other profibrotic factors and is associated with wound healing and fibrogenic activation programs. (a) Genomic distribution of ZNF416-FLAG binding sites. (b) Genomic linear distance of ZNF416-FLAG binding sites to its nearest TSS. (c) Comparisons of co-occupancy of ZNF416-FLAG with publicly available YAP, SMAD3, SRF, and H3K27ac ChIP-seq. (d) Genomic view of example ZNF416-FLAG binding proximal to known profibrotic genes. (e) Ontology analysis of genes annotated to ZNF416-FLAG binding sites.
We next sought to determine whether ZNF416 occupancy overlaps with established transcriptional regulators implicated in mechanosignaling and fibrotic pathologies such as YAP, SRF, and SMAD3 (Tschumperlin et al., 2018). To test this, we analyzed publicly available YAP, SMAD3, and SRF ChIP-seq data (GSE61852, GSE38103, and GSE61475, respectively; Stein et al., 2015; Tsankov et al., 2015; Ding et al., 2013) and evaluated the percentage of ZNF416 binding sites that reside within two nucleosomes (300 bp) of YAP, SRF, and SMAD3. Surprisingly, only 17.5% (242) of ZNF416 binding sites were in close proximity to YAP binding sites, 19.8% to SMAD3, and 10% to SRF, suggesting that ZNF416 functions in a fashion that is largely independent of established fibrotic transcriptional regulators (Fig. 3 c). Due to ZNF416 containing both activating and repressive protein domains, we next cross-referenced our ZNF416-FLAG occupancy with publicly available H3K27ac ChIP-seq in IMR90 fibroblasts (GEO accession no. GSE16256; Hawkins et al., 2010). Interestingly, we found that 943 (68%) ZNF416 binding sites were co-occupied by H3K27ac in human fibroblasts (Fig. 3 c), suggesting that ZNF416 predominantly acts as a transcriptional activator.
Annotation of ChIP-seq-identified ZNF416 binding sites to the nearest TSS identified ZNF416 as potentially regulating genes central to fibroblast contractility (e.g., ACTA2), ECM remodeling (e.g., LOXL2), profibrotic growth factor signaling (e.g., PDGFB), and proliferation (e.g., CDCA7; Fig. 3 d; GSE161446). Using GREAT, we then conducted an unbiased ontology analysis of the genomic regions occupied by ZNF416, revealing control of gene programs involved in actomyosin network and cortical cytoskeleton (Fig. 3 e).
ZNF416 is central to fibroblast activation
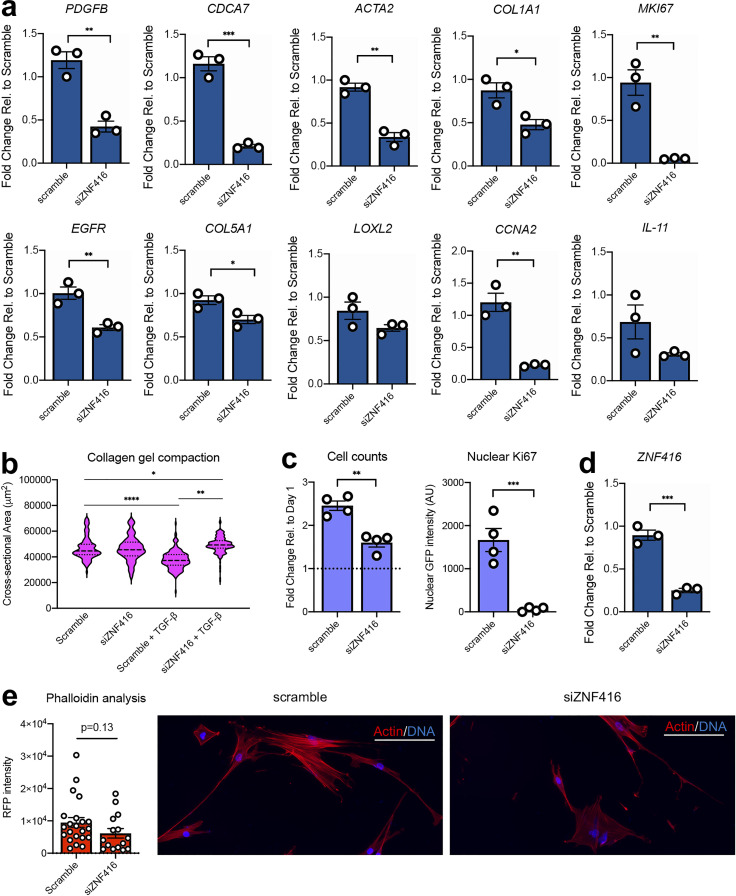
We next conducted gain- and loss-of-function studies to examine the role of ZNF416 in fibroblast function. RNAi-mediated knockdown of ZNF416 in fibroblasts cultured on rigid tissue-culture plastic attenuated baseline transcript levels of ZNF416 target genes identified via ChIP-seq (e.g., PDGFB, EGFR, CDCA7, ACTA2, and COL5A1; Fig. 4 a). More broadly, ZNF416 knockdown reduced transcript levels of ECM-related (e.g., COL1A1) and proliferation associated genes (e.g., MKI67 and CCNA2), suggesting ZNF416 plays a pleiotropic role in regulating fibroblast functions associated with fibrotic pathologies (Fig. 4 a). To examine whether ZNF416 modulates fibroblast cytoskeletal and contractile function, we performed gel compaction assays and phalloidin staining and confirmed that ZNF416 knockdown attenuated TGFβ-mediated fibroblast contraction and matrix remodeling (Fig. 4 b). In the absence of TGFβ, ZNF416 knockdown did not alter collagen compaction and exhibited only a modest trend toward reduced F-actin staining and stress fiber formation (Fig. 4 e). Additionally, we confirmed that knockdown of ZNF416 attenuated fibroblast proliferation when compared with a nontargeting siRNA control (Fig. 4, c and d; and Fig. S2 e).
Figure 4.
Knockdown of ZNF416 attenuates lung fibroblast activation. (a) qRT-PCR of primary human lung fibroblasts treated with siRNA targeting ZNF416 for 72 h. n = 3 independent biological replicates. (b) Violin plot of cross-sectional area of collagen gels containing fibroblasts with siRNA-mediated knockdown of ZNF416 in the presence or absence of TGFβ. Solid lines represent median. Dotted lines represent upper and lower quartiles. (c) Fold change in cell counts of primary human lung fibroblasts between 1 and 3 d in culture following knockdown of ZNF416. Data normalized to the cell counts at day 1 to observe proliferation. Ki67 nuclear intensity, assessed by quantitative immunofluorescence, in primary human lung fibroblasts following ZNF416 knockdown. n = 4 biological replicates. (d) qRT-PCR validation of ZNF416 knockdown. n = 3 independent biological replicates. (e) Phalloidin and DAPI staining with quantification of fibroblasts following siRNA-mediated knockdown of ZNF416. Scale bars represent 100 µm. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 evaluated by unpaired t test (a and c–e) or one-way ANOVA (b). All error bars represent SEM.
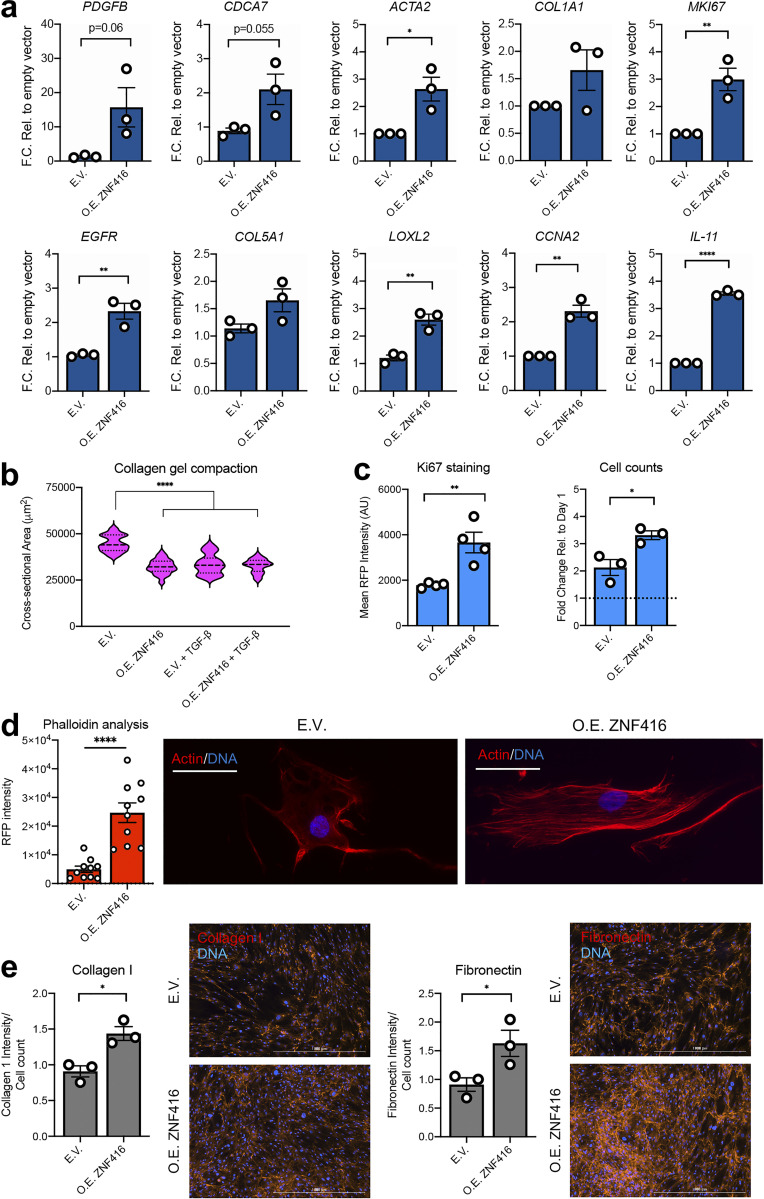
To study gain-of-function of ZNF416, we used a lentiviral approach to generate stable overexpression of ZNF416 in primary human lung fibroblasts (Fig. S2 b). Overexpression of ZNF416 amplified expression of ZNF416 target genes identified via ChIP-seq (e.g., PDGFB, EGFR, CDCA7, ACTA2, and LOXL2; Fig. 5 a). More broadly, ZNF416 gain of function increased transcript levels of proliferation-associated (e.g., MKI67 and CCNA2), soluble profibrotic signaling (IL11), and contractile genes (e.g., ACTA2; Fig. 5 a) when compared with empty vector control fibroblasts. Using collagen compaction assays, we found that overexpression of ZNF416 enhanced contractile and matrix remodeling function when compared with empty vector control fibroblasts (Fig. 5 b) and did so without altering baseline SMAD2 phosphorylation or total SMAD2/3 and YAP/TAZ levels (Fig. S2 c). Overexpression of ZNF416 also increased F-actin staining and stress fiber formation (Fig. 5 d), consistent with increased contractile function (Fig. 5 b). Additionally, we found that overexpression of ZNF416 amplified proliferation when compared with empty vector controls (Fig. 5 c and Fig. S2 e). To test whether overexpression of ZNF416 increased ECM deposition, we probed for collagen I and fibronectin deposition and normalized to cell count; ZNF416 overexpression enhanced deposition of both collagen I and fibronectin (Fig. 5 e). Finally, we studied cells on 0.2- and 32-kPa matrices to test whether gain of function of ZNF416 can override the effect of soft matrices. We found that overexpression of ZNF416 on soft matrices elevated ECM, contractile, and proliferative gene expression to levels close to or higher than empty vector control fibroblasts placed on stiff matrices (Fig. S2 f). Taken together, these data identify ZNF416 as a mechanoregulator of fibroblast biology and demonstrate that ZNF416 function is critical to fibroblast activation.
Figure 5.
Overexpression of ZNF416 drives lung fibroblast proliferative, contractile, and matrix synthetic activation. (a) qPCR of primary human lung fibroblasts stably overexpressing ZNF416 (O.E. ZNF416) or an empty vector control. n = 3 independent biological replicates. (b) Violin plot of overexpressing ZNF416 primary human lung fibroblasts and empty vector controls in the presence or absence of TGFβ. Solid lines represent median. Dotted lines represent upper and lower quartiles. (c) Ki67 nuclear intensity, assessed by quantitative immunofluorescence, in primary human lung fibroblasts following ZNF416 overexpression. n = 4 biological replicates. Fold change in cell counts of primary human lung fibroblasts between 1 and 3 d following overexpressing ZNF416 or empty vector control. Data normalized to the cell counts at day 1 to observe proliferation. n = 3 biological replicates. (d) Phalloidin and DAPI staining with quantification of fibroblasts overexpressing ZNF416 and empty vector. Scale bars represent 50 µm. (e) Deposition of collagen I and fibronectin by primary human lung fibroblasts overexpressing ZNF416 or empty vector control following 3 d of culture. Data normalized to cell counts per image. *, P < 0.05; **, P < 0.01; ****, P < 0.0001, evaluated by unpaired t test (a, c, and d) or one-way ANOVA (b). All error bars represent SEM. E.V., empty vector control; F.C. Rel., fold change relative; O.E., overexpressing.
Matrix stiffness has emerged as a central modulator of fibroblast function both in vitro and in vivo, but the transcriptional and epigenetic mechanisms by which matrix stiffness influences fibroblast function remain incompletely understood. Our work used freshly isolated FACS-sorted (CD45−/CD326−/CD31−/Col1a1−GFP+) fibroblasts to probe the chromatin accessibility changes induced by exposure to matrices of different stiffness. Previous studies that have established transcriptional regulators such as YAP/TAZ and MRTF-A as central mediators of mesenchymal response to mechanical stimuli have used serially passaged or immortalized cell lines. These proteins have been identified to interact with epigenetic regulating enzymes, such as BRD4, p300, and PCAF (Zanconato et al., 2018; Kim et al., 2018; Andrianifahanana et al., 2013), suggesting these mechanoresponsive transcriptional regulators could mediate lasting mechanodriven epigenomic changes in cells. Indeed, mesenchymal cell populations have been shown to acquire a mechanical memory after prolonged exposure to rigid substrates (Killaars et al., 2019; Yang et al., 2014; Heo et al., 2016; Li et al., 2017; Heo et al., 2015). Thus, the prior use of serially passaged cells to examine physiological cell mechanoresponses may have only incompletely identified key factors due to the accumulation of epigenetic mechanical memory in rigid cell culture before stiffness control experiments. Our examination of the effect of matrix stiffness on chromatin accessibility of freshly isolated lung fibroblasts confirmed a role for the YAP/TAZ interacting transcription factor TEAD4 as well as the recently identified matrix stiffness responsive factor Sp1 (Stowers et al., 2019) but also identified ZNF416 as a novel putative mediator of these changes (Fig. 2 a). Importantly, ZNF416 was also implicated in our motif analysis of genes regulated in vivo in fibroblasts during experimental lung fibrosis, highlighting its potential relevance in wound healing and fibrosis. Moreover, comparison of ChIP-seq data from ZNF416 with other known mechanoresponsive and profibrotic transcription factors identified relatively little overlap, suggesting a unique functional role for ZNF416.
ZNF416 contains 12 tandem C2H2 zinc-finger domains that have been shown to interact with activating epigenetic regulating enzymes such as p300 (Brayer et al., 2008), a major histone acetyltransferase. ZNF416 additionally contains a KRAB domain, which has been shown to interact with repressive complexes, such as the nucleosome remodeling and deacetylase complex (Schultz et al., 2001), HP1⍺ (CBX5; Sripathy et al., 2006), and DNA methyltransferases (Quenneville et al., 2012). The diverse functions of ZNF416’s domains indicate ZNF416 has potential to act as either a transcriptional activator or repressor. However, our cross-reference analysis of ZNF416 and H3K27ac ChIP-seq indicates that ZNF416 largely co-occupies sequences associated with H3K27ac (Fig. 3 c), consistent with ZNF416 acting as a transcriptional activator and our observation that ZNF416 overexpression drives fibroblast gene expression and activation. Taken together, our observations identify ZNF416 as a transcriptional activator that promotes fibroblast proliferative, matrix synthetic, and contractile activation.
A key remaining question is whether ZNF416 pioneers alterations in chromatin accessibility, potentially through interactions with epigenetic regulators, or instead is binding to chromatin already opened through independent mechanisms. A recent study observed that mechanical stretch of chromatin was sufficient to induce change in transcription, suggesting mechano-driven changes in chromatin accessibility could be a rapid event (Tajik et al., 2016). Based on the important functional roles identified here for ZNF416, further elucidation of its roles in epigenetic remodeling and the upstream pathways linking matrix stiffness to its transcriptional functions is likely to provide additional insights relevant to wound healing and tissue fibrosis. In conclusion, our work demonstrates that matrix stiffness is a key driver of increased chromatin accessibility in fibroblasts and identifies a novel role for ZNF416 in activation of fibroblast gene programs and functions central to pathological fibroblast activation.
Materials and methods
Primary cell isolation, cell sorting, and cell culture
Primary mouse lung fibroblasts were isolated under a protocol approved by the Mayo Clinic Institutional Animal Care and Use Committee. Col1α1-GFP transgenic mice, generated as previously described (Yata et al., 2003), were kindly provided by Dr. Derek Radisky (Mayo Clinic, Jacksonville, FL). Briefly, 6- to 8-wk-old Col1α1-GFP mice were anesthetized with ketamine/xylazine solution (100 mg/kg and 10 mg/kg, respectively) injected intraperitoneally. The left ventricle of the heart was perfused with ice-cold PBS (Thermo Fisher Scientific) to remove blood content from the lung. Lungs were then immediately harvested and minced in 10-cm Petri dishes and then incubated in digestive solution (DMEM, 0.2 mg/ml Liberase DL, and 100 U/ml DNase I). Samples were digested at 37°C for 45 min. Digestive solution was inactivated with 1x DMEM (Thermo Fisher Scientific) containing 10% FBS (Thermo Fisher Scientific). Cell and tissue suspension was put through a 40-µm filter and centrifuged. Cell pellet was resuspended in red blood cell lysis buffer (BioLegend) for 90 s and then diluted in 3× volume of PBS. Cells were centrifuged and resuspended in 200 µl FACS buffer (1% BSA and 0.5 mM EDTA, pH 7.4 in PBS). The single-cell suspension was then incubated with anti-mouse CD45:PerCp-Cy5.5 (BioLegend; 1:200), anti-mouse CD31-PE (BioLegend; 1:200), anti-mouseCD326-APC (BioLegend; 1:200), and DAPI (Sigma-Aldrich; 1:1,000) antibodies for 30 min on ice. Detailed antibody information is provided in Table S1.
Samples were subjected to FACS using a BD FACS Aria II (BD Biosciences). To isolate the CD45- (hematopoietic), EpCAM− (epithelial), CD31− (endothelial), GFP+ (collagen I–expressing) population, the following isolation strategy was used (Fig. S1): debris exclusion (forward scatter [FSC]-A by side scatter [SSC]-A), doublet exclusion (SSC-W by SSC-H and FSC-W by FSC-H), dead cell exclusion (DAPI by anti-CD31-PE), CD45+ cell exclusion (anti-CD45-PerCP-Cy5.5 by Col1a1-GFP), EpCAM, and CD31+ cell exclusion (anti-CD325-APC by anti-CD31-PE), and isolation of GFP+ cells (APC by GFP). A recent single-cell RNA sequencing (RNA-seq) analysis using the same mouse strain confirmed that the vast majority of GFP+ cells lacking other lineage markers in the lungs are indeed fibroblasts, though a relatively minor population of smooth muscle cells and pericytes is also likely represented (Tsukui et al., 2020). Primary mouse fibroblasts were cultured in DMEM (Thermo Fisher Scientific) supplemented with 10% FBS (Thermo Fisher Scientific) and Anti-Anti (Thermo Fisher Scientific) unless otherwise stated.
Primary human lung fibroblasts were isolated by explant culture from the lungs of subjects who underwent lung transplantation and were kindly provided by Peter Bitterman (University of Minnesota, Minneapolis, MN) under a protocol approved by the University of Minnesota Institutional Review Board. Primary human lung fibroblasts were maintained in DMEM (Thermo Fisher Scientific) supplemented with 10% FBS (Thermo Fisher Scientific) and Anti-Anti (Thermo Fisher Scientific).
ATAC-seq and analysis
FACS-sorted mouse GFP+ lung fibroblasts were seeded (5,000 cells/cm2) onto 0.2-kPa PDMS, 32-kPa commercially available PDMS (Advanced Biomatrix), or tissue-culture plastic. All culture plates were identically coated with collagen I (0.1 mg/ml diluted in PBS; Advanced Biomatrix) for 1 h at room temperature. Cells were cultured for 8 d. Fibroblasts were then trypsinized and counted. 50,000 cells were subjected to Omni ATAC-seq following the published protocol (Corces et al., 2017). The size of library DNA was determined from the amplified and purified library by a Fragment Analyzer (Advanced Analytical Technologies), and the enrichment of accessible regions was determined by the fold difference between positive and negative genomic loci using real-time PCR. The following primer sequences were used: accessibility-positive control locus: AT-P7-F: 5′-GGCTTATCCGGAGCGGAAAT-3′, AT-P7-R: 5′GGCTGGAACAGGTTGTGTTG-3′. Accessibility-negative control locus: AT-P13-F: 5′-TCCCCTTTACTGTTTTCCTCTAC-3′, AT-P13-R: 5′-GGATTGATGAGGAAACAGCCTC-3′. The libraries were sequenced to 51 bp from both ends on an Illumina HiSeq 4000 instrument.
Paired-end reads were mapped to the mm10 genome using BWA (Li and Durbin, 2009). BWA is widely used to map ATAC-seq reads to a reference genome, as BWA is both fast and memory efficient. SAM files were converted to BAM files using Picard SortSam (https://broadinstitute.github.io/picard/command-line-overview.html#SortSam) and sorted by chromosomal coordinates. PCR duplicates were removed using Picard MarkDuplicates (https://broadinstitute.github.io/picard/command-line-overview.html#MarkDuplicates). Pairs of reads with one or both reads uniquely mapped to chromosomes 1–22, X, and Y were retained. Processed BAM files were used to call peaks using MACS2 with the following options “–keep-dup all -q 0.01 -no model” (Zhang et al., 2008). To identify differentially accessible genomic loci, DiffBind was used with the processed BAM files and peak files obtained from MACS2 using an FDR threshold of 0.05 (Stark and Brown, 2011). Differentially accessible loci were then annotated to their closest TSS using Homer (Heinz et al., 2010). Gene ontology was performed using GREAT (Tsukui et al., 2020) on the genomic regions identified in the ATAC-seq analyses using default settings. Motif analysis was completed using the findMotifsGenome.pl command within Homer.
In vivo model of experimental lung fibrosis and motif analysis
To evaluate motif enrichment from fibroblasts activated in vivo, we analyzed an unpublished dataset from previously completed experiments (GSE161322). These animal experiments were performed under protocols approved by the Mayo Clinic Institutional Animal Care and Use Committee. 2-mo-old Col1a1-GFP male mice were administered bleomycin (1.2 U/kg) or PBS intratracheally as described previously (Oh et al., 2018). Mice were sacrificed 14 d after bleomycin exposure to allow lung fibrosis to manifest. Mice were anaesthetized and euthanized, and lung GFP+ fibroblasts were FACS sorted as detailed above (Fig. S1 a). GFP+ lung fibroblasts were sorted into RLT lysis buffer, an RNA lysis buffer supplied in Qiagen RNA kits, and RNA was isolated as detailed below and submitted for sequencing. RNA-seq reads were aligned to the mm10 build of the mouse reference genome using STAR with default parameter setting (Dobin et al., 2013). STAR is an ultrafast aligner developed for mapping RNA-seq reads. STAR addresses major challenges of RNA-seq data mapping, particularly in spliced alignments. Specifically, STAR aligns noncontiguous sequences directly to the reference genome, without the need to align reads to a splice junction database and to split reads/align split reads to a reference genome as most other RNA-seq aligners do. FeatureCounts was used to generate raw counts as well as normalized reads per kilobase of exon per million mapped reads (Liao et al., 2013). To identify differentially expressed genes in bleomycin-treated mice compared with sham, we used an FDR cutoff of 0.05 and a log2 fold change of ±1. The list of 4,124 genes was submitted to Homer for de novo motif enrichment analysis in the promoter region (±1 kb from the TSS). Motifs corresponding to transcription factors not expressed in lung fibroblasts (reads per kilobase of exon per million mapped reads <0.01) were discarded from our analysis.
Microscopy and image analysis
Fibroblasts were fixed with 4% PFA (Sigma-Aldrich) for 15 min followed by permeabilization with 0.1% Triton X-100 (Sigma-Aldrich) in PBS (Thermo Fisher Scientific) for 10 min. Samples were then blocked in 5% BSA (Sigma-Aldrich) in PBS supplemented with 0.1% Tween-20 (Sigma-Aldrich) for 1 h. Samples were then placed in primary lamin A/C antibody (Cell Signaling Technology) diluted in blocking buffer and incubated overnight at 4°C. The following morning, samples were then washed with PBS three times and placed in RFP secondary antibody (Thermo Fisher Scientific) for 1 h. Detailed antibody information is supplied in Table S1. RFP phalloidin (Thermo Fisher Scientific) was used to quantify cell area from freshly isolated fibroblasts on soft and stiff matrices and was done according to manufacturer’s protocol. Samples were then washed with PBS three times, and Prolong Gold Anti-fade mounting media (Thermo Fisher Scientific) was placed on each sample, followed by a small glass coverslip. Samples were then imaged using a z-stack algorithm on a confocal microscope (Zeiss; LSM780) at 40× (NA 1.2) at room temperature using Zen Acquisition software. Z-stack images (0.2 µm per stack) were then reconstructed into 3D images using Imaris (Bitplane) from which nuclear volume was extracted. To measure cell area, confocal RFP phalloidin images were quantified in ImageJ. For evaluation of the gain and loss of function of ZNF416 on the cytoskeleton, RFP phalloidin (Thermo Fisher Scientific) was used on fixed cells in PBS according to manufacturer’s protocol. Images were acquired at room temperature on a Keyence BZ-x810 microscope (Keyence) with the in-microscope camera using 10× (NA 0.3) and 20× (NA 0.75) objectives. Phalloidin intensity is quantified as RFP intensity within cell area (outlined in ImageJ) subtracted by background. Background is defined as an empty well treated identically to cell samples. To measure Ki67 nuclear intensity, cells were prepared for immunofluorescence as described above using a Ki67 antibody (Cell Signaling) and nuclei were outlined using DAPI. Fluorescence intensity was then measured within each outlined nuclei. Images were acquired on a Olympus CKX53 microscope using an Olympus DP22 camera (Olympus Life Sciences). Images were acquired at room temperature using 10× (NA 0.25) and 20× (NA 0.4) objectives using CellSense software (Olympus Life Sciences). Data are reported as average fluorescence intensity within nuclei subtracted by background. Background is defined as an empty well treated with primary and secondary antibodies.
RNAi knockdown and lentiviral-mediated overexpression of ZNF416
RNAi was completed with SMARTpool: ON-TARGETplus siRNA’s (Dharmacon) specific for ZNF416 or scramble nontargeting siRNA as a control. Oligo siRNA sequences are located in Table S1. To confirm on target efficacy of the siRNA pool, we tested individual siRNAs (n = 4) and measured ZNF416 transcript levels (Fig. S2 a). Primary human lung fibroblasts were transfected in 10% FBS in DMEM for 72 h, after which they were used for experiments.
Lentiviral expression plasmids for ZNF416 were purchased from OriGene. Briefly, we used three plasmids: (1) TRC2 pLKO.5-puro empty vector control plasmid (Sigma-Aldrich; SHC201), (2) ZNF416-GFP (OriGene; RG201632), and (3) ZNF416-FLAG (OriGene; RC201632). Using Lipofectamine 3000, HEK293T cells were transfected with appropriate expression vector along with psPAX (Addgene; 12260) and pMD2.G (Addgene; 12259) in Opti-MEM (Thermo Fisher Scientific) in 5% FBS with 1 mM sodium pyruvate (Thermo Fisher Scientific) with no antibiotics. Viral media was collected 24 and 48 h after transfection. To remove potential HEK293T contamination, viral media was spun down at 1,250 rpm for 10 min at 4°C and then passed through a 40-µm PES filter. Viral media with polybrene (Thermo Fisher Scientific; 10 µg/ml) was then added to primary human lung fibroblasts and IMR90s, a commercially available human lung fibroblast cell line. The next day, fibroblasts were washed with PBS and placed in 10% FBS in DMEM with Anti-Anti. Three days after infection, fibroblasts were subjected to puromycin selection (Sigma-Aldrich; 1 µg/ml) for 7 d. Following puromycin selection, surviving cells were then validated for expression of ZNF416-GFP and ZNF416-FLAG by Western blot (Fig. S2 b).
RNA isolation
mRNA was isolated using RNeasy mini kit (Qiagen); the concentration of mRNA was quantified using a Nanodrop spectrophotometer. cDNA was synthesized using the SuperScript VILO kit (Thermo Fisher Scientific). Quantitative real time-PCR (qRT-PCR) was completed using FastStart Essential DNA Green Master (Roche Diagnostics) and analyzed using a LightCycler 96 (Roche Diagnostics). Primers used for qRT-PCR are listed in Table S1.
Protein lysis and Western blotting
For protein analysis, cells were lysed with RIPA buffer (Thermo Fisher Scientific) supplemented with Halt protease and phosphatase inhibitor (Thermo Fisher Scientific). Protein concentration was quantified with the Pierce BCA Protein Assay kit (Thermo Fisher Scientific). Protein samples were loaded onto 4–15% gradient SDS-PAGE gels (Bio-Rad), transferred to 0.2 µm pore-size polyvinylidene difluoride membranes, and blocked for 1 h in blocking buffer (Bio-Rad; 5% milk, 0.1% Tween-20 in 1x TBS). Blots were then placed into primary antibodies overnight at 4°C. The following morning, blots were then incubated with secondary HRP-conjugated antibody for 1 h at room temperature. All antibody information is located in Table S1. Protein bands were observed using Super Signal West Pico Plus (Thermo Fisher Scientific), and images were acquired using a ChemiDoc Imaging System (Bio-Rad).
Collagen gel compaction assays
Compaction of cell-embedded collagen micro tissues was analyzed as follows: fibroblasts (4 × 106 cells/ml) were diluted in rat-tail collagen type I (Corning; 6 mg/ml; Cummins et al., 2019). Polystyrene beads (Thermo Fisher Scientific; 1:50) 1 µm in diameter were added to the solution to visualize the droplet. Emulsions were formed by flow-focusing the collagen/cell solution and oil (Sigma-Aldrich; fluorocarbon oil FC-40) with 2% FluoroSurfactant (RAN Biotechnologies) into a microfluidic PDMS device. Droplet formations were performed at 4°C while the droplets were recovered at 37°C. The fibroblast-containing droplets were incubated at 37°C in medium containing either DMSO (Thermo Fisher Scientific) or 2 ng/ml TGFβ (PeproTech). To quantify the effect of knockdown of ZNF416 on fibroblast compaction, primary human lung fibroblasts were treated with siRNA for 48 h before embedding into collagen microtissues.
ECM deposition
Fibroblasts were plated in 96-well plates (10,000 fibroblasts/well) for 3 d in 2% FBS in DMEM supplemented with ascorbic acid (Thermo Fisher Scientific; 50 µg/ml). Fibroblasts and matrix were then fixed and immunostained as detailed above. Briefly, we used collagen I and fibronectin antibodies to probe for ECM deposition. Antibody information can be found in Table S1. Secondary antibodies were used as detailed above. Images were acquired using a Cytation 5 microscope (BioTek) with the in-microscope camera using a 4× objective (NA 0.13). ECM deposition was quantified by RFP image intensity divided by cell number, calculated by DAPI nuclear stain.
ChIP-seq and analysis
ZNF416-FLAG–expressing IMR90 cells were subjected to ChIP-seq for FLAG as follows: IMR90 cells (n = 2 biological replicates) stably expressing ZNF416-FLAG were cross-linked with 1% formaldehyde (Sigma-Aldrich) for 10 min, followed by quenching with 125 mM glycine (Sigma-Aldrich) for 5 min at room temperature. Fixed cells were washed with 1X TBS (Thermo Fisher Scientific). IMR90 cells were then resuspended in cell lysis buffer (10 mM Tris-HCl, pH 7.5, 10 mM NaCl, and 0.5% NP-40) and incubated on ice for 10 min. The lysates were washed with MNase digestion buffer (20 mM Tris-HCl, pH7.5, 15 mM NaCl, 60 mM KCl, and 1 mM CaCl2) and incubated for 20 min at 37°C in the presence of MNase. After adding the same volume of sonication buffer (100 mM Tris-HCl, pH 8.1, 20 mM EDTA, 200 mM NaCl, 2% Triton X-100, and 0.2% sodium deoxycholate), the lysate was sonicated for 10 min (30 s on/30 s off) in a Bioruptor instrument (Diagenode) and then centrifuged at 15,000 rpm for 10 min. The cleared supernatant equivalent to ∼15 million cells was incubated with 30 µl prewashed anti-FLAG M2 magnetic beads (Sigma-Aldrich; M8823) on a rocker overnight. The beads were extensively washed with ChIP buffer, high-salt buffer, LiCl2 buffer, and TE buffer. Bound chromatin was eluted and reverse-cross-linked at 65°C overnight. DNA was treated with RNase A and Proteinase K (Qiagen) and then purified using a Min-Elute PCR purification kit (Qiagen). Sequencing libraries were prepared from 5 to 10 ng ChIP and input DNA with the ThruPLEX DNA-seq Kit V2 (Rubicon Genomics) and were sequenced to 51 bp from both ends using an Illumina HiSeq 4000 instrument (Illumina).
Paired-end fastq files were aligned to the hg19 build of the human genome using Bowtie2 using the default settings (Langmead and Salzberg, 2012). SAM files were converted to BAM and were sorted by chromosomal coordinates using Picard SortSam. Duplicates were removed using Picard MarkDuplicates. BAM files were then used to call peaks using MACS2 using default settings with a q threshold of 0.05 (Zhang et al., 2008). To generate BigWig files, deepTools BamCoverage was used with default settings and a bin size of 10 bp (Ramírez et al., 2016). Integrative Genome Viewer was used to visualize BigWig files on the hg19 genome track. Homer was used to annotate ZNF416 binding sites to the nearest TSS as described above. Gene ontology of ZNF416 target genes was completed as described above.
To test whether ZNF416-FLAG binding locations are co-occupied by the mechanoresponsive or profibrotic transcription factors YAP, SMAD3, or SRF or H3K27 acetylation (H3K27ac), an active enhancer mark indicating gene activation, publicly available ChIP-seq data were downloaded, including GEO accession no. GSE61852 for YAP (Stein et al., 2015), GEO accession no. GSE38103 for SMAD3 (Ding et al., 2013), GEO accession no. GSE61475 for SRF (Tsankov et al., 2015), and GEO accession no. GSE16256 for H3K27ac (Hawkins et al., 2010). Homer was used to identify co-occupancy using the mergePeaks command within Homer. Regions were considered to be co-occupied if the peak distance between two marks was 300 bp or less.
Statistical analysis and figure preparation
GraphPad Prism 8.0 (GraphPad Software) was used for generating Figs. 1, 2, 3, 4, and 5 and statistical analyses. Data are represented as mean ± SEM. Statistical analysis was done using either an unpaired two-tailed t test when directly comparing two groups or one-way ANOVA with Tukey multiple-comparison adjustment when comparing three or more groups.
Data availability
Raw and analyzed ATAC-seq and ChIP-seq data generated in this study are available through the Gene Expression Omnibus under the GEO accession nos. GSE161434 and 46, respectively. Analyzed ATAC-seq data include TDF genome coverage files and differential accessibility analysis bed files (supplied as .txt) to allow users to analyze these datasets. Analyzed ChIP-seq data include BigWig genome coverage files and peak calling analysis (supplied as .txt). Scripts and code used to analyze these datasets are available from the corresponding author upon request.
Online supplemental material
Fig. S1 shows the flow-sorting strategy used for the selection and isolation of mouse lung fibroblasts and ATAC-seq/motif analysis of mouse lung fibroblasts on soft (0.2 kPa) and stiff (32 kPa) substrates for 8 d. Fig. S2 shows validation of ZNF416 gene modulation and role in fibroblast function. Table S1 lists the antibodies, PCR primers, and siRNA sequences used in this study.
Supplementary Material
lists the antibodies, PCR primers, and siRNA sequences used in this study.
Acknowledgments
We would like to thank the Flow Cytometry and Microscopy cores at Mayo Clinic for their assistance.
Funding support was provided by National Institutes of Health grants T32 HL105355 (D.L. Jones and P.A. Link), DK058185 (J-H. Lee and T. Ordog), DK084567 (J-H. Lee and T. Ordog), HL142596 (G. Ligresti), HL124392 (X. Varelas), HL092961 (D.J. Tschumperlin), and HL133320 (D.J. Tschumperlin).
The authors declare no competing financial interests.
Author contributions: D.L. Jones, H. Yan, T. Ordog, G. Ligresti, X. Varelas, and D.J. Tschumperlin conceived the study, designed the experiments, and interpreted the data. D.L. Jones, J.A. Meridew, M.T. Ducharme, K.L. Lydon, K.M. Choi, N. Caporarello, Q. Tan, P.A. Link, A.M. Diaz Espinosa, Y. Xiong, J-H. Lee, and Z. Ye performed the experiments. D.L. Jones and D.J. Tschumperlin wrote the manuscript. All authors participated in reviewing and editing the manuscript.
References
- Andrianifahanana, M., Wilkes M.C., Gupta S.K., Rahimi R.A., Repellin C.E., Edens M., Wittenberger J., Yin X., Maidl E., Becker J., and Leof E.B.. 2013. Profibrotic TGFβ responses require the cooperative action of PDGF and ErbB receptor tyrosine kinases. FASEB J. 27:4444–4454. 10.1096/fj.12-224907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrini, J.L., Chaudhry S., Sarrazy V., Koehler A., and Hinz B.. 2012. The mechanical memory of lung myofibroblasts. Integr. Biol. 4:410–421. 10.1039/c2ib00149g [DOI] [PubMed] [Google Scholar]
- Brayer, K.J., David A.E., and Segal J.. 2008. Keep Your Fingers Off My DNA: Protein-Protein Interactions Mediated by C2H2 Zinc Finger Domains. Cell Biochem. Biophys. 50:111–131. [DOI] [PubMed] [Google Scholar]
- Cassandri, M., Smirnov A., Novelli F., Pitolli C., Agostini M., Malewicz M., Melino G., and Raschellà G.. 2017. Zinc-finger proteins in health and disease. Cell Death Discov. 3:17071. 10.1038/cddiscovery.2017.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces, M.R., Trevino A.E., Hamilton E.G., Greenside P.G., Sinnott-Armstrong N.A., Vesuna S., Satpathy A.T., Rubin A.J., Montine K.S., Wu B., et al. 2017. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods. 14:959–962. 10.1038/nmeth.4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins, K.A., Crampton A.L., and Wood D.K.. 2019. A High-Throughput Workflow to Study Remodeling of Extracellular Matrix-Based Microtissues. Tissue Eng. Part C Methods. 25:25–36. 10.1089/ten.tec.2018.0290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby, I.A., Laverdet B., Bonté F., and Desmoulière A.. 2014. Fibroblasts and myofibroblasts in wound healing. Clin. Cosmet. Investig. Dermatol. 7:301–311. 10.2147/CCID.S50046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, N., Yu R.T., Subramaniam N., Sherman M.H., Wilson C., Rao R., Leblanc M., Coulter S., He M., Scott C., et al. 2013. A vitamin D receptor/SMAD genomic circuit gates hepatic fibrotic response. Cell. 153:601–613. 10.1016/j.cell.2013.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont, S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S., et al. 2011. Role of YAP/TAZ in mechanotransduction. Nature. 474:179–183. 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- Gabbiani, G. 2003. The myofibroblast in wound healing and fibrocontractive diseases. J. Pathol. 200:500–503. 10.1002/path.1427 [DOI] [PubMed] [Google Scholar]
- Haak, A.J., Kostallari E., Sicard D., Ligresti G., Choi K.M., Caporarello N., Jones D.L., Tan Q., Meridew J., Diaz Espinosa A.M., et al. 2019. Selective YAP/TAZ inhibition in fibroblasts via dopamine receptor D1 agonism reverses fibrosis. Sci. Transl. Med. 11:eaau6296. 10.1126/scitranslmed.aau6296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins, R.D., Hon G.C., Lee L.K., Ngo Q., Lister R., Pelizzola M., Edsall L.E., Kuan S., Luu Y., Klugman S., et al. 2010. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 6:479–491. 10.1016/j.stem.2010.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz, S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., and Glass C.K.. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38:576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo, S.J., Thorpe S.D., Driscoll T.P., Duncan R.L., Lee D.A., and Mauck R.L.. 2015. Biophysical Regulation of Chromatin Architecture Instills a Mechanical Memory in Mesenchymal Stem Cells. Sci. Rep. 5:16895. 10.1038/srep16895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo, S.J., Driscoll T.P., Thorpe S.D., Nerurkar N.L., Baker B.M., Yang M.T., Chen C.S., Lee D.A., and Mauck R.L.. 2016. Differentiation alters stem cell nuclear architecture, mechanics, and mechano-sensitivity. eLife. 5:e18207. 10.7554/eLife.18207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X., Yang N., Fiore V.F., Barker T.H., Sun Y., Morris S.W., Ding Q., Thannickal V.J., and Zhou Y.. 2012. Matrix stiffness-induced myofibroblast differentiation is mediated by intrinsic mechanotransduction. Am. J. Respir. Cell Mol. Biol. 47:340–348. 10.1165/rcmb.2012-0050OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killaars, A.R., Grim J.C., Walker C.J., Hushka E.A., Brown T.E., and Anseth K.S.. 2019. Extended exposure to stiff microenvironments leads to persistent chromatin remodeling in human mesenchymal stem cells. Adv. Sci. (Weinh.). 6:1801483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M.K., Jang J.W., and Bae S.C.. 2018. DNA binding partners of YAP/TAZ. BMB Rep. 51:126–133. 10.5483/BMBRep.2018.51.3.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., and Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C.X., Talele N.P., Boo S., Koehler A., Knee-Walden E., Balestrini J.L., Speight P., Kapus A., and Hinz B.. 2017. MicroRNA-21 preserves the fibrotic mechanical memory of mesenchymal stem cells. Nat. Mater. 16:379–389. 10.1038/nmat4780 [DOI] [PubMed] [Google Scholar]
- Liao, Y., Smyth G.K., and Shi W.. 2013. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41:e108. 10.1093/nar/gkt214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligresti, G., Caporarello N., Meridew J.A., Jones D.L., Tan Q., Choi K.M., Haak A.J., Aravamudhan A., Roden A.C., Prakash Y.S., et al. 2019. CBX5/G9a/H3K9me-mediated gene repression is essential to fibroblast activation during lung fibrosis. JCI Insight. 5:127111. 10.1172/jci.insight.127111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F., Mih J.D., Shea B.S., Kho A.T., Sharif A.S., Tager A.M., and Tschumperlin D.J.. 2010. Feedback amplification of fibrosis through matrix stiffening and COX-2 suppression. J. Cell Biol. 190:693–706. 10.1083/jcb.201004082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F., Lagares D., Choi K.M., Stopfer L., Marinković A., Vrbanac V., Probst C.K., Hiemer S.E., Sisson T.H., Horowitz J.C., et al. 2015. Mechanosignaling through YAP and TAZ drives fibroblast activation and fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 308:L344–L357. 10.1152/ajplung.00300.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinković, A., Liu F., and Tschumperlin D.J.. 2013. Matrices of physiologic stiffness potently inactivate idiopathic pulmonary fibrosis fibroblasts. Am. J. Respir. Cell Mol. Biol. 48:422–430. 10.1165/rcmb.2012-0335OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSweeney, S.R., Warabi E., and Siow R.C.M.. 2016. Nrf2 as an Endothelial Mechanosensitive Transcription Factor: Going With the Flow. Hypertension. 67:20–29. 10.1161/HYPERTENSIONAHA.115.06146 [DOI] [PubMed] [Google Scholar]
- Mih, J.D., Marinkovic A., Liu F., Sharif A.S., and Tschumperlin D.J.. 2012. Matrix stiffness reverses the effect of actomyosin tension on cell proliferation. J. Cell Sci. 125:5974–5983. 10.1242/jcs.108886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Vicente, R., Pavón D.M., Martín-Padura I., Català-Montoro M., Díez-Sánchez A., Quílez-Álvarez A., López J.A., Sánchez-Álvarez M., Vázquez J., Strippoli R., and Del Pozo M.A.. 2018. Caveolin-1 Modulates Mechanotransduction Responses to Substrate Stiffness through Actin-Dependent Control of YAP. Cell Rep. 25:1622–1635.e6. 10.1016/j.celrep.2018.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafabadi, H.S., Mnaimneh S., Schmitges F.W., Garton M., Lam K.N., Yang A., Albu M., Weirauch M.T., Radovani E., Kim P.M., et al. 2015. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat. Biotechnol. 33:555–562. 10.1038/nbt.3128 [DOI] [PubMed] [Google Scholar]
- Oh, R.S., Haak A.J., Smith K.M.J., Ligresti G., Choi K.M., Xie T., Wang S., Walters P.R., Thompson M.A., Freeman M.R., et al. 2018. RNAi screening identifies a mechanosensitive ROCK-JAK2-STAT3 network central to myofibroblast activation. J. Cell Sci. 131:jcs209932. 10.1242/jcs.209932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville, S., Turelli P., Bojkowska K., Raclot C., Offner S., Kapopoulou A., and Trono D.. 2012. The KRAB-ZFP/KAP1 system contributes to the early embryonic establishment of site-specific DNA methylation patterns maintained during development. Cell Rep. 2:766–773. 10.1016/j.celrep.2012.08.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez, F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., and Manke T.. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44(W1):W160-5. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, D.C., Friedman J.R., and Rauscher F.J. III. 2001. Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2α subunit of NuRD. Genes Dev. 15:428–443. 10.1101/gad.869501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripathy, S.P., Stevens J., and Schultz D.C.. 2006. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol. Cell. Biol. 26:8623–8638. 10.1128/MCB.00487-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark, R., and Brown G.. 2011. DiffBind : differential binding analysis of ChIP-Seq peak data. Bioconductor [Google Scholar]
- Stein, C., Bardet A.F., Roma G., Bergling S., Clay I., Ruchti A., Agarinis C., Schmelzle T., Bouwmeester T., Schübeler D., and Bauer A.. 2015. YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers. PLoS Genet. 11:e1005465. 10.1371/journal.pgen.1005465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers, R.S., Shcherbina A., Israeli J., Gruber J.J., Chang J., Nam S., Rabiee A., Teruel M.N., Snyder M.P., Kundaje A., and Chaudhuri O.. 2019. Matrix stiffness induces a tumorigenic phenotype in mammary epithelium through changes in chromatin accessibility. Nat. Biomed. Eng. 3:1009–1019. 10.1038/s41551-019-0420-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto, S.G., Narimatsu M., Lu M., He X., Sidiqi A.M., Tolosa M.F., Chan L., De Freitas K., Bialik J.F., Majumder S., et al. 2016. YAP/TAZ are mechanoregulators of TGF-b-smad signaling and renal fibrogenesis. J. Am. Soc. Nephrol. 27:3117–3128. 10.1681/ASN.2015050499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajik, A., Zhang Y., Wei F., Sun J., Jia Q., Zhou W., Singh R., Khanna N., Belmont A.S., and Wang N.. 2016. Transcription upregulation via force-induced direct stretching of chromatin. Nat. Mater. 15:1287–1296. 10.1038/nmat4729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankov, A.M., Gu H., Akopian V., Ziller M.J., Donaghey J., Amit I., Gnirke A., and Meissner A.. 2015. Transcription factor binding dynamics during human ES cell differentiation. Nature. 518:344–349. 10.1038/nature14233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschumperlin, D.J., Ligresti G., Hilscher M.B., and Shah V.H.. 2018. Mechanosensing and fibrosis. J. Clin. Invest. 128:74–84. 10.1172/JCI93561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukui, T., Sun K.H., Wetter J.B., Wilson-Kanamori J.R., Hazelwood L.A., Henderson N.C., Adams T.S., Schupp J.C., Poli S.D., Rosas I.O., et al. 2020. Collagen-producing lung cell atlas identifies multiple subsets with distinct localization and relevance to fibrosis. Nat. Commun. 11:1920. 10.1038/s41467-020-15647-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, E.S. 2015. Lung extracellular matrix and fibroblast function. Ann. Am. Thorac. Soc. 12:S30–S33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C., Tibbitt M.W., Basta L., and Anseth K.S.. 2014. Mechanical memory and dosing influence stem cell fate. Nat. Mater. 13:645–652. 10.1038/nmat3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yata, Y., Scanga A., Gillan A., Yang L., Reif S., Breindl M., Brenner D.A., and Rippe R.A.. 2003. DNase I-hypersensitive sites enhance α1(I) collagen gene expression in hepatic stellate cells. Hepatology. 37:267–276. 10.1053/jhep.2003.50067 [DOI] [PubMed] [Google Scholar]
- Zanconato, F., Forcato M., Battilana G., Azzolin L., Quaranta E., Bodega B., Rosato A., Bicciato S., Cordenonsi M., and Piccolo S.. 2015. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 17:1218–1227. 10.1038/ncb3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato, F., Battilana G., Forcato M., Filippi L., Azzolin L., Manfrin A., Quaranta E., Di Biagio D., Sigismondo G., Guzzardo V., et al. 2018. Transcriptional addiction in cancer cells is mediated by YAP/TAZ through BRD4. Nat. Med. 24:1599–1610. 10.1038/s41591-018-0158-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., and Liu X.S.. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists the antibodies, PCR primers, and siRNA sequences used in this study.
Data Availability Statement
Raw and analyzed ATAC-seq and ChIP-seq data generated in this study are available through the Gene Expression Omnibus under the GEO accession nos. GSE161434 and 46, respectively. Analyzed ATAC-seq data include TDF genome coverage files and differential accessibility analysis bed files (supplied as .txt) to allow users to analyze these datasets. Analyzed ChIP-seq data include BigWig genome coverage files and peak calling analysis (supplied as .txt). Scripts and code used to analyze these datasets are available from the corresponding author upon request.