This work shows that GATA transcription factors transduce the BMP signaling and, with SOX17 and TFAP2C, induce the human germ-cell fate, delineating the mechanism for human germ-cell specification.
Abstract
The in vitro reconstitution of human germ-cell development provides a robust framework for clarifying key underlying mechanisms. Here, we explored transcription factors (TFs) that engender the germ-cell fate in their pluripotent precursors. Unexpectedly, SOX17, TFAP2C, and BLIMP1, which act under the BMP signaling and are indispensable for human primordial germ-cell-like cell (hPGCLC) specification, failed to induce hPGCLCs. In contrast, GATA3 or GATA2, immediate BMP effectors, combined with SOX17 and TFAP2C, generated hPGCLCs. GATA3/GATA2 knockouts dose-dependently impaired BMP-induced hPGCLC specification, whereas GATA3/GATA2 expression remained unaffected in SOX17, TFAP2C, or BLIMP1 knockouts. In cynomolgus monkeys, a key model for human development, GATA3, SOX17, and TFAP2C were co-expressed exclusively in early PGCs. Crucially, the TF-induced hPGCLCs acquired a hallmark of bona fide hPGCs to undergo epigenetic reprogramming and mature into oogonia/gonocytes in xenogeneic reconstituted ovaries. By uncovering a TF circuitry driving the germ line program, our study provides a paradigm for TF-based human gametogenesis.
Introduction
Germ cells are the carriers of genetic as well as epigenetic information into new individuals, and thus serve as an enduring link between generations. Accordingly, they bear a capacity to replicate genetic information with high fidelity (1, 2, 3). On the other hand, they also create genetic and epigenetic diversity through meiotic recombination and epigenetic reprogramming/programming, respectively, providing a driving force for evolution (4, 5). Anomalies in such processes often lead to diseased states, including infertility and genetic/epigenetic disorders of offspring. Therefore, investigations into the mechanism of germ-cell development not only promote our understanding of fundamental principles of heredity and evolution but also provide salient information regarding the etiology of critical diseases.
Despite such importance, investigations into human germ-cell development have been limited because of the difficulties in accessing relevant experimental materials and ethical restrictions. Notably, recent advancements in the in vitro reconstitution of human germ-cell development using human pluripotent stem cells (hPSCs), including embryonic stem cells (hESCs) and induced pluripotent stem cells (hiPSCs), have created novel opportunities for such studies, permitting investigations into the mechanisms of human germ-cell development as an emerging frontier in reproductive biology/medicine (6, 7). Accordingly, hPSCs are induced into cells bearing properties similar to human primordial germ cells (hPGCs) (8, 9), the founding population of the human germ-cell lineage that eventually gives rise to either spermatozoa or oocytes. The induced hPGC-like cells (hPGCLCs) are further differentiated into oogonia/early oocyte-like cells with appropriate epigenetic reprogramming in a reconstituted ovary culture (10, 11), or into pro-spermatogonia-like cells in a reconstituted testis culture (12). Although further reconstitution of human germ-cell development remains a key challenge, these advances recapitulate a period of more than 10 wk of human germ-cell development, leading to a number of key findings with regard to the mechanism of human germ-cell development in general, and germ-cell specification in particular (8, 9, 13, 14, 15, 16, 17).
In the case of germ-cell specification, humans as well as non-human primates such as cynomolgus monkeys (Macaca fascicularis), use transcriptional and signaling programs evolutionarily distinct from those in mice, which have long been a paradigm for mammalian development (8, 9, 13, 18, 19). Specifically, in humans, WNT signaling induces EOMES, which, together with bone morphogenetic protein 4 (BMP4) signaling, induces SOX17 as one of the most upstream transcription factors (TFs) for hPGC(LC) specification (13). SOX17 is essential for the expression of key downstream genes, including BLIMP1, and for activating other germ-cell specification programs (8, 13). TFAP2C also serves as a key upstream TF that functions in parallel and in an interdependent fashion with SOX17 and is critical for the repression of somatic programs (13, 15). Such programs for germ-cell specification appear to be relatively well conserved in cynomolgus monkeys (18, 19). In contrast, in mice, Sox17 has no role in germ-cell specification (20), and BMP4 signaling activates endogenous WNT signaling that in turn induces T (T has no role in humans (13)), which up-regulates Blimp1 and Prdm14, two of the most upstream TFs for germ-cell specification (21, 22, 23). Blimp1, Prdm14, and Tfap2c are essential and sufficient for the global control of downstream programs, including by reactivating pluripotency programs, repressing somatic programs, and initiating epigenetic reprogramming (22, 23, 24, 25). These findings demonstrate that the TFs and TF hierarchies involved in conferring the germ-cell fate in humans are distinct from those in mice, highlighting the importance of further promoting human germ-cell biology.
In regard to the mechanism of human germ-cell specification, a fundamental question remains to be answered: That is, which TFs or TF combinations are sufficient to give rise to the germ-cell fate in their precursors? The answer to this question could help establish a foundation for TF-based human gametogenesis. In mice, three TFs (Blimp1, Prdm14, and Tfac2c), and to a lesser extent, two TFs (Blimp1 and Tfap2c; Prdm14 and Tfap2c) or a single TF (Prdm14), are sufficient to confer the germ-cell fate to their precursors, and such TF-induced mouse PGCLCs (mPGCLCs) contribute to spermatogenesis (25). We therefore set out to define the TFs that replace the BMP4 signaling and are sufficient to establish the identity of hPGCs on their precursors. Unexpectedly, we found that three TFs that are essential for hPGCLC specification—that is, SOX17, TFAP2C, and BLIMP1—are nonetheless not sufficient, and in contrast, the GATA family of TFs, combined with SOX17 and TFAP2C, drives the hPGCLC program.
Results
SOX17, TFAP2C, and BLIMP1 are not sufficient to generate hPGCLCs
For hPGCLC induction, hiPSCs are first induced into incipient mesoderm-like cells (iMeLCs) by stimulating with activin A and a WNT signal activator (CHIR99021) for 2 d, and iMeLCs are then induced into hPGCLCs by stimulating with bone morphogenetic protein 4, together with proliferation/survival factors, including stem cell factor (SCF), EGF, and leukemia inhibitory factor (LIF), under a floating aggregate condition (9, 13, 26). hPGCLCs that express key genes such as SOX17, TFAP2C, BLIMP1, and NANOS3 are induced as early as day 2 of induction (d2 hPGCLCs), show a progressive maturation, and persist at least until around d10 (9, 13, 26).
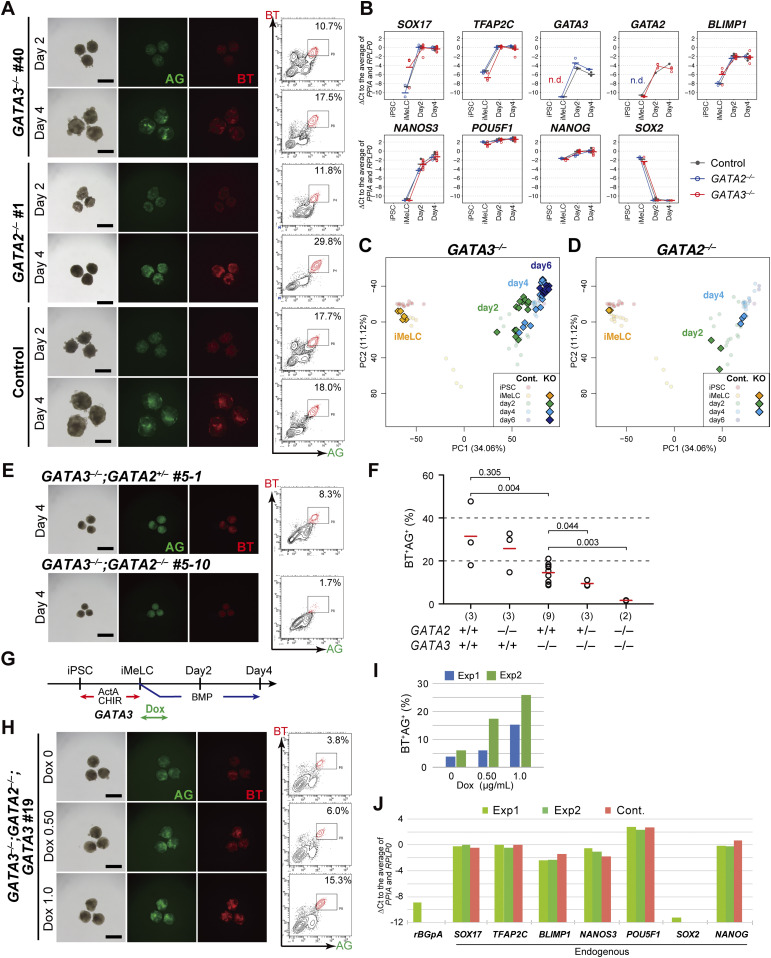
We set out to identify TFs that are sufficient to confer the germ-cell fate on iMeLCs in the absence of BMP signaling. At the outset, we evaluated whether SOX17, TFAP2C, or BLIMP1, three TFs essential for hPGCLC specification (8, 9, 13), would be sufficient to induce the germ-cell fate when expressed either singly or in one of various combinations. For this purpose, hiPSCs bearing the BLIMP1-2A-tdTomato (BT) and TFAP2C-2A-EGFP (AG) alleles (585B1 BTAG (XY)) (9) were transfected with piggyBac-based vectors expressing (i) the reverse tetracycline trans-activator (rtTA) under a constitutively active promoter and (ii) the genes of interest (SOX17, TFAP2C, SOX17/TFAP2C, SOX17/BLIMP1, TFAP2C/BLIMP1, or SOX17/TFAP2C/BLIMP1) under the control of tetracycline regulatory elements with transcription termination by the rabbit β-globin poly A sequence (rBGpA), so that the genes of interest exhibited timed expression in a doxycycline (Dox)-dependent manner and could be distinguished from the endogenous ones by the presence of rBGpA (Fig 1A). For each transfectant, we selected two clones that exhibited transgene expression levels in hiPSCs comparable with the corresponding endogenous gene expression levels in hPGCLCs (Figs 1B and S1A). The expression of the transgenes in a clone expressing SOX17/TFAP2C/BLIMP1 was confirmed with Western blotting (Fig S1B). All the hiPSC clones selected exhibited undifferentiated morphology.
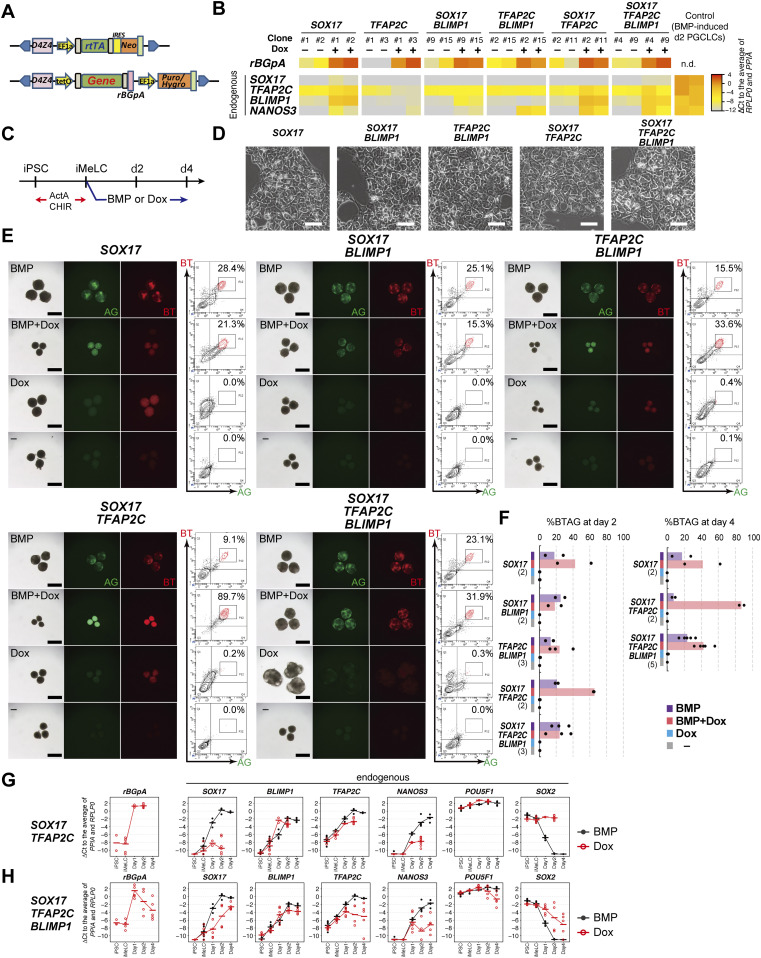
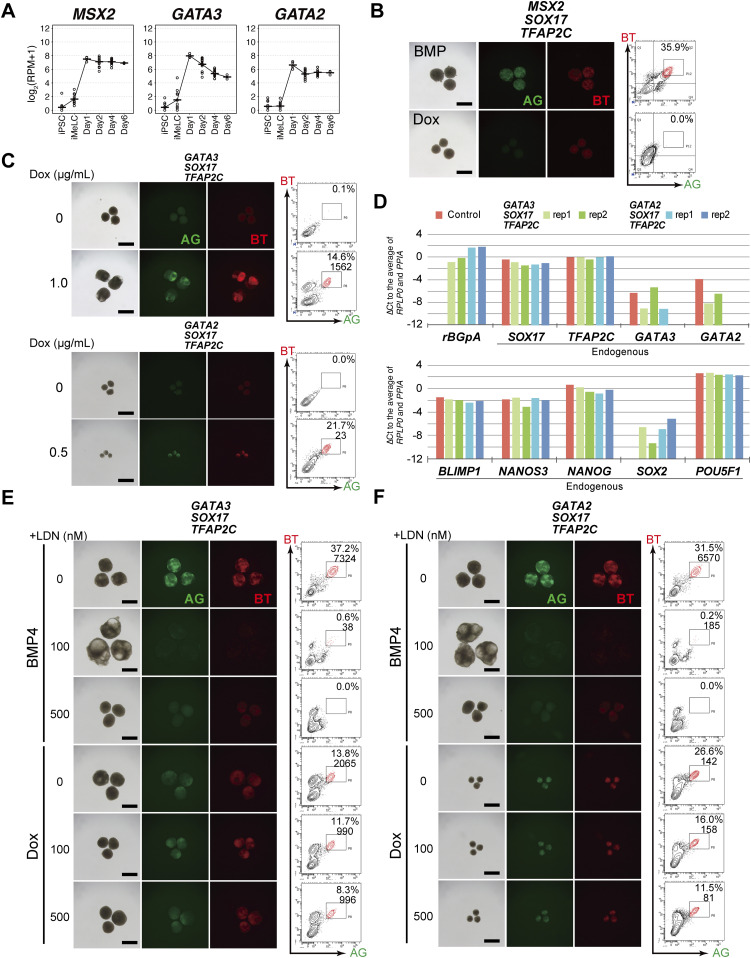
Figure 1. SOX17, TFAP2C, and BLIMP1 do not create human primordial germ-cell-like cells (hPGCLCs).
(A) A schematic of the vectors used for Dox-inducible expression. The coding sequence of each gene was cloned in the designated position. D4Z4: D4Z4 macrosatellite repeat insulator; EF1α: promoter sequence of human EEF1A1; rtTA: reverse tetracycline trans-activator; IRES: internal ribosome entry site; Neo: Neomycin resistance gene; tetO: Tet operator sequence; rBGpA: rabbit β-globin polyadenylation signal; Puro/Hygro: resistance gene for puromycin/hygromycin. (B) Heat map representation of the expression levels of the indicated genes in the designated hiPSC clones stimulated with (+) or without (−) Dox (1.0 μg/ml) for 24 h. Two clones were examined for each transgene combination. To quantify the expression levels of the transgenes or endogenous genes by qRT-PCR, primer pairs for the rBGpA or 3′ untranslated regions were used, respectively, and the ΔCt was calculated from the average Ct value of two housekeeping genes, RPLP0 and PPIA (set as 0). (C) The protocol for hPGCLC induction. iMeLC aggregates were induced for hPGCLC fate by bone morphogenetic protein 4 (BMP4) or Dox (1.0 μg/ml) in the presence of stem cell factor, EGF, and leukemia inhibitory factor. ActA: activin A; CHIR: CHIR99021. (D) Phase-contrast images of iMeLCs in the designated clones. No apparent morphological differences were seen among the clones. Representative images of at least two independent experiments are shown (shown in Fig 1F). Bar: 50 μm. (E) Bright-field and fluorescence (TFAP2C-EGFP [AG] and BLIMP1-tdTomato [BT]) images, and FACS analyses for BTAG expression in floating aggregates of the indicated transgene-expressing clones at day 4 of the indicated stimulation. (−): induction only with stem cell factor, EGF and leukemia inhibitory factor. Representative images of at least two independent experiments are shown (see Fig 1F). Bars, 200 μm. (F) Percentage of BT+AG+ cells of the indicated transgene-expressing clones with the indicated stimulations at day 2 (left) and day 4 (right). Dots represent values for each experiment and the bars represent their averages. The numbers of inductions performed are shown in parenthesis. (G, H) Expression dynamics of rBGpA (transgenes) and the indicated endogenous genes in the SOX17/TFAP2C (G) and SOX17/TFAP2C/BLIMP1 (H) clones induced by BMP4 (black) or Dox (red). d1: whole aggregates; d2/d4: BT+AG+ cells for induction by BMP4, BT+ cells for induction by Dox. For each gene, the ΔCt was calculated from the average Ct value of two housekeeping genes, RPLP0 and PPIA (set as 0). (G, H) Three independent experiments with two SOX17/TFAP2C clones (G) and three SOX17/TFAP2C/BLIMP1 clones (H) were performed.
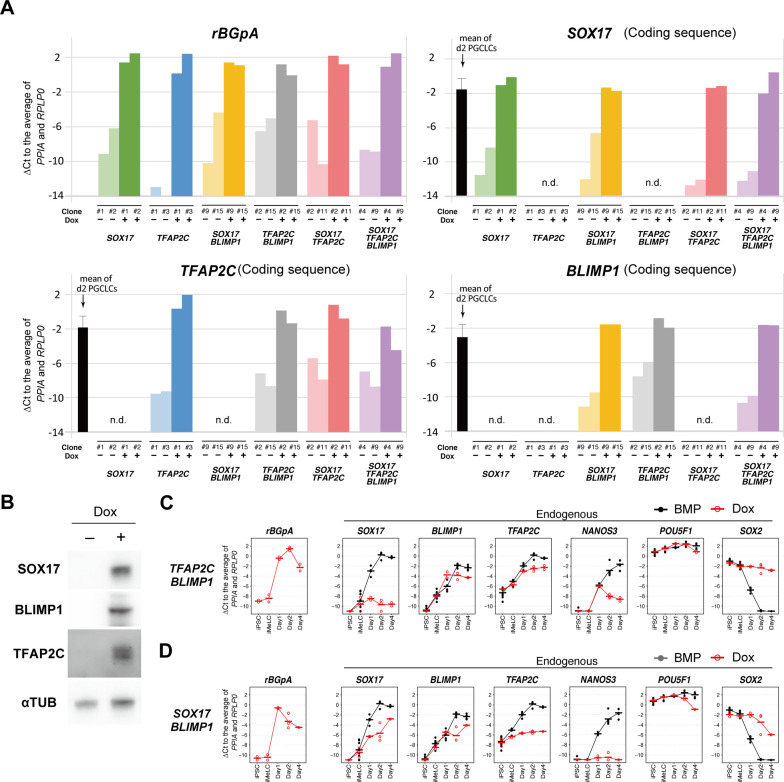
Figure S1. SOX17, TFAP2C, and BLIMP1 do not create human primordial germ-cell-like cells.
(A) The transgene expression levels in the designated hiPSC clones stimulated with (+) or without (−) Dox (1.0 μg/ml) for 24 h. Two clones were examined for each transgene combination. To quantify the transgene expression levels by qRT-PCR, primer pairs for the rabbit β-globin polyA sequence (rBGpA) or for the transgene coding sequences were used, and the ΔCt was calculated from the average Ct value of two housekeeping genes, RPLP0 and PPIA (set as 0). The expression levels of SOX17, TFAP2C and BLIMP1 in d2 human primordial germ-cell–like cells (BT+AG+ cells) measured by primer pairs for detecting their 3′ untranslated regions are shown as references. n.d., not determined. (B) Western blot analysis of the expression of SOX17, BLIMP1, and TFAP2C in the SOX17/TFAP2C/BLIMP1 hiPSC clone without (−) or with (+) 1.0 μg/ml of Dox for 24 h. α-TUBULIN was used as loading control. (C, D) Expression dynamics of rBGpA (transgenes) and the indicated endogenous genes in the TFAP2C/BLIMP1 (C) and SOX17/BLIMP1 (D) clones induced by bone morphogenetic protein 4 (black) or Dox (red). d1: whole aggregates; d2/d4: BT+AG+ cells for induction by bone morphogenetic protein 4, BT+ cells for induction by Dox. For each gene, the ΔCt was calculated from the average Ct value of two housekeeping genes, RPLP0 and PPIA (set as 0). The bars indicate the mean value of each time point. (C, D) Two independent experiments for the TFAP2C/BLIMP1 (C) and SOX17/BLIMP1 (D) clones were performed.
Source data are available for this figure.
We first examined the effects of the transgene expression in hiPSCs cultured with Dox for 24 h. Quantitative Real Time-PCR (qRT-PCR) for the endogenous key loci (SOX17, TFAP2C, BLIMP1, and NANOS3) showed that no clones up-regulated SOX17, whereas the SOX17, SOX17/TFAP2C, and SOX17/TFAP2C/BLIMP1 clones up-regulated TFAP2C mildly and BLIMP1 to an extent comparable with that in d2 hPGCLCs (Fig 1B). The TFAP2C/BLIMP1, SOX17/TFAP2C, and SOX17/TFAP2C/BLIMP1 clones up-regulated endogenous NANOS3 to a level similar to that in d2 hPGCLCs (Fig 1B). Because the TFAP2C clones had no impact on all these genes (TFAP2C appeared to repress endogenous TFAP2C) (Fig 1B), we excluded them from the subsequent analyses.
We next analyzed whether the expression of these genes in iMeLCs might induce the germ-cell fate (Fig 1C). The iMeLCs induced by activin A and CHIR99021 from all the clones bore a morphology indistinguishable from that of the parental clone (Fig 1D). Upon d4 of induction by BMP4 or BMP4 and Dox, iMeLC aggregates from all the clones exhibited a distinct cluster of BT-positive (BT+) and AG-positive (AG+) cells, as revealed by observation under a fluorescence dissection microscope or FACS (Fig 1E). We noted that the SOX17/TFAP2C clones stimulated by BMP4 and Dox, although forming small aggregates, differentiated into BT+AG+ cells at a very high efficiency (∼90%), whereas the other clones formed BT+AG+ cells with an efficiency of ∼20–30% (Fig 1E and F). This might be because SOX17 and TFAP2C expression could be a rate-limiting event for hPGCLC specification, and the Dox-induced expression of SOX17 and TFAP2C would create a state highly competent for BMP-induced hPGCLC specification. In addition, the iMeLC aggregates of the SOX17, SOX17/TFAP2C, and TFAP2C/BLIMP1 clones became smaller when stimulated with BMP4 and Dox, which might have been due to a subtle but significant difference in the expression levels of SOX17, TFAP2C, or BLIMP1 (e.g., Blimp1/BLIMP1 is known to induce cell-cycle arrest in various contexts (27, 28)).
In contrast, with Dox stimulation alone, no iMeLC aggregates showed BT+AG+ cells (Fig 1E and F). Upon stimulation with Dox, the SOX17 clones induced weak BT+ cells, the SOX17/BLIMP1 clones showed no BTAG positivity, the TFAP2C/BLIMP1 clones generated small aggregates with weak BTAG positivity (less than the threshold levels), the SOX17/TFAP2C clones induced weak BT+ cells, and the SOX17/TFAP2C/BLIMP1 clones induced weak BT+ or AG+ cells, with the generation of very minor BT+AG+ cells (<1%) (Fig 1E and F). Because very few/no BT+AG+ cells were induced under any conditions, we characterized the properties of BT+ cells induced by the transgenes by qRT-PCR. All such cells failed to show a proper up-regulation of endogenous SOX17, TFAP2C, and NANOS3 and down-regulation of SOX2, despite their substantial transgene expression (Figs 1G and H and S1C and D).
Taken together, these facts lead us to conclude that no combination of SOX17, TFAP2C, and BLIMP1 is sufficient to induce appropriate endogenous SOX17/TFAP2C expression and other hPGCLC properties, including NANOS3 expression and SOX2 repression, in iMeLCs. Thus, the three TFs are not sufficient to replace the BMP4 signaling for hPGCLC induction. On the other hand, the finding that all the clones generated BT+AG+ cells upon BMP4 and Dox provision indicates that the expression of SOX17/TFAP2C/BLIMP1 transgenes does not interfere with the hPGC specification program.
Exploration of relevant TFs by transcriptome analysis
To explore the effects of the TF expression more globally and to identify a relevant TF(s) that might be required for hPGCLC generation, we compared the transcriptome dynamics associated with the TF expression to those of the parental clone induced by BMP4 by RNA-sequencing (RNA-seq) (13, 29) (see Table S1 for the RNA-seq samples analyzed in this study).
Table S1 List of cell types used for transcriptome analysis. (41.6KB, xlsx)
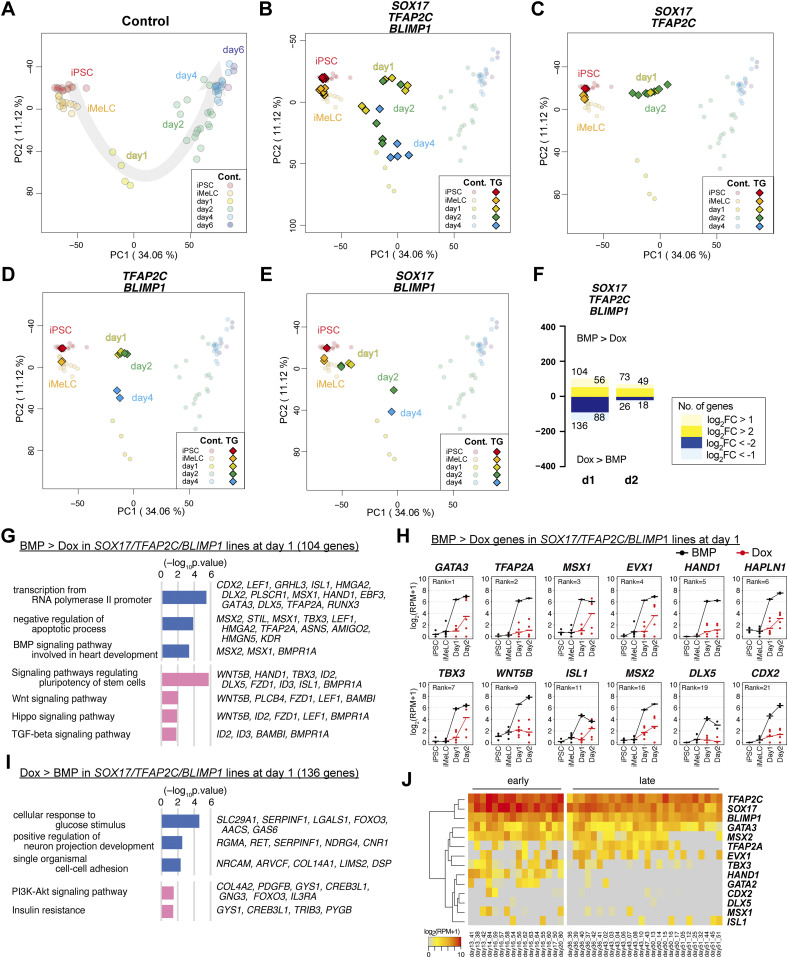
In agreement with the previous report (13), principal component analysis (PCA) revealed that parental cells exhibited characteristic cell–property transitions during hPGCLC induction: on the PC1/2 plane, they transitioned along a V-shaped trajectory, with hiPSCs and iMeLCs plotted on the top-left corner, d1 iMeLC aggregates progressing diagonally toward the bottom, and d2/d4 BT+AG+ cells progressing up toward the top-right corner (Fig 2A). In contrast, whereas hiPSCs/iMeLCs of all the TF-expressing clones (no Dox provision) were plotted at positions similar to those of parental hiPSCs/iMeLCs, upon Dox provision on iMeLC aggregates (i.e., TF expression), all the clones shifted their positions in parallel along the PC1 coordinate with a retention or some down-regulation of their PC2 scores, but none of the clones acquired a state similar to d4 hPGCLCs (Fig 2B–E). We examined the expression of 481 genes that characterize the hPGCLC specification process (13), which revealed that the d2 TF-expressing cells failed to show a property similar to those of d2/d4 BT+AG+ cells (Fig S2A). Most notably, they lacked sufficient expression of the genes specifying hPGCLC properties (Fig S2A).
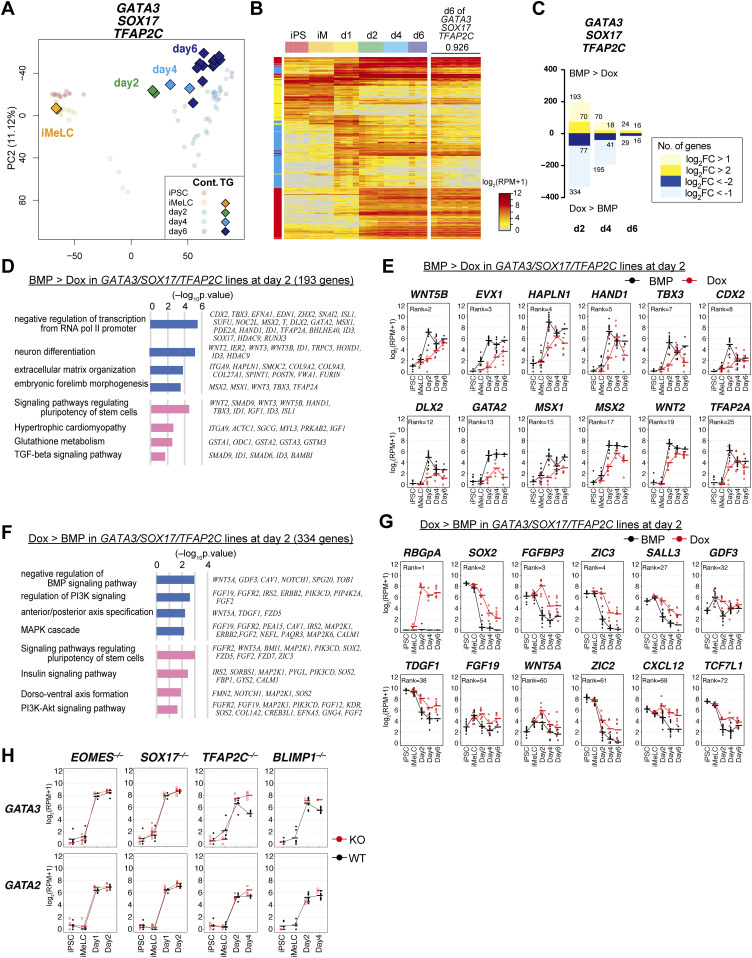
Figure 2. Transcriptome analysis of the effects of the transcription factor expression.
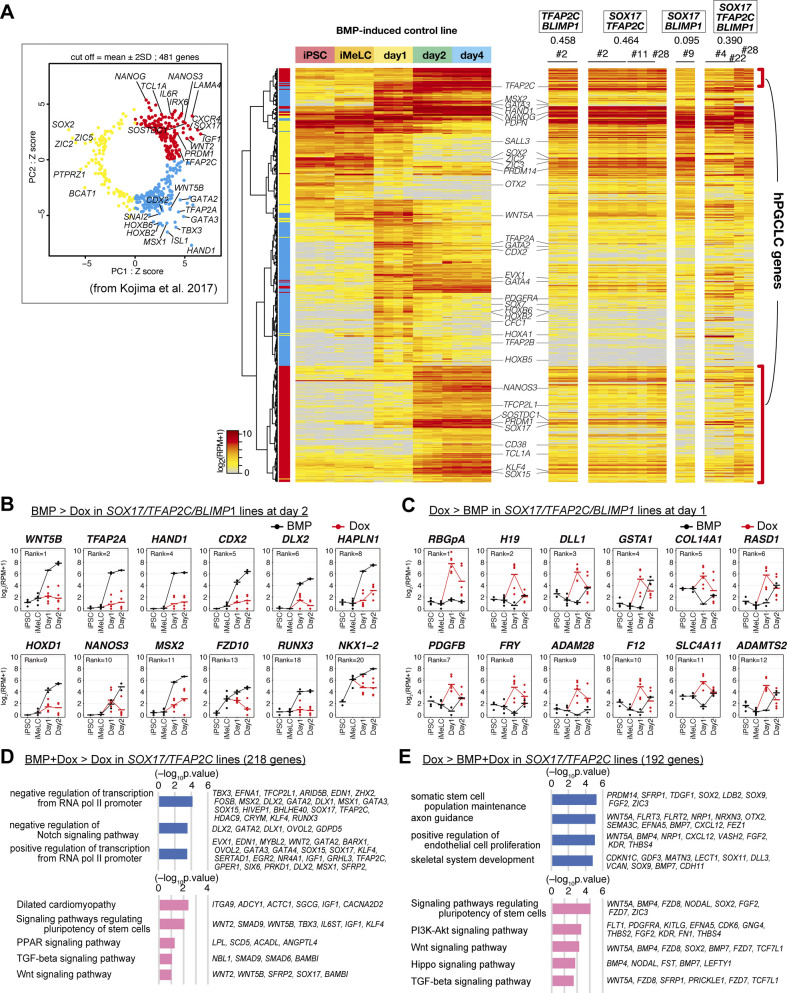
(A, B, C, D, E) Principal component analysis (PCA) of the effects of the transcription factor expression (see the Materials and Methods section for details). (A) The PCA plots of the cells (hiPSCs, iMeLCs, d1 whole aggregates, d2/d4/d6 BT+AG+ cells) derived from the parental clone (585B1 BTAG). The developmental progression is indicated by an arrow. (B, C, D, E) The PCA plots of the SOX17/TFAP2C/BLIMP1 (B), SOX17/TFAP2C (C), TFAP2C/BLIMP1 (D), and SOX17/BLIMP1 (E) clones (squares) are overlaid with those of the parental clone (circles with pale color). See Table S1 for the samples analyzed. The color coding is as indicated. (F) The numbers of the differentially expressed genes at d1/d2 between bone morphogenetic protein (BMP)– and Dox-stimulated cells of the SOX17/TFAP2C/BLIMP1 clones (P < 0.01 by Tukey–Kramer test, log2[RPM + 1] > 4 in cells with higher expression, log2[fold change: FC] >1 [up, pale yellow; down, pale blue] or 2 [up, yellow; down, blue]). d1: iMeLC whole aggregates; d2: BT+AG+ and BT+ cells for BMP- and Dox-stimulated cells, respectively. Note that the numbers of differentially expressed genes were smaller at d2, because the gene expression of the BT+ cells of the Dox-induced SOX17/TFAP2C/BLIMP1 clones was somewhat variable. (G, I) Gene ontology terms (blue) and KEGG pathways (pink) enriched in differentially expressed genes between BMP- and Dox-stimulated d1 SOX17/TFAP2C/BLIMP1 clone aggregates. (G, I) Representative genes up-regulated in BMP- (G) or Dox- (I) stimulations and P-values are shown. (F, H) Expression dynamics of the genes up-regulated at d1 (F) in BMP-stimulated (black) compared with Dox-stimulated (red) SOX17/TFAP2C/BLIMP1 clone-derived cells. The ranks of the genes ordered by the fold changes between BMP and Dox stimulation are shown. Note that TFAP2A, HAND1, HAPLN1, MSX2, and CDX2 were highly up-regulated in BMP-stimulated cells also at d2 (Fig S2B). See Table S1 for the samples analyzed. (E, H, J) Heat map representation of the expression of the genes in (H) in cynomolgus monkey fetal germ cells (early: embryonic day (E) 13-E17; late: E36-E51) (9, 18, 30, 31).
Figure S2. Transcriptome analysis of the effects of the transcription factor expression.
(A) (left) Principal component analysis of the expression of the 481 genes characterizing human primordial germ-cell–like cell specification shown in reference 13. Red (first quadrant): genes for PGCLC specification; cyan (fourth quadrant): genes for endoderm/mesoderm specification; yellow (second and third quadrants): genes for pluripotency (right). Heat map representation of the expression of the 481 genes in the parental hiPSCs, iMeLCs, bone morphogenetic protein (BMP)4-induced d1 whole aggregates, and d2/d4 BT+AG+ cells, and in TFAP2C/BLIMP1, SOX17/TFAP2C, SOX17/BLIMP1, SOX17/TFAP2C/BLIMP1 clone-derived, Dox-induced d2 BT+ cells. Representative genes are annotated, and the correlation coefficient values of each induction with the d4 BT+AG+ cells are shown. The color coding is as indicated. (B, C) Expression dynamics of the genes up-regulated in BMP-stimulated (black, BT+AG+) compared with Dox-stimulated (red, BT+) d2 cells (B) (Note that TFAP2A, HAND1, CDX2, HAPLN1, and MSX2 were highly up-regulated also at d1 [Fig 2H]) or those up-regulated in Dox-stimulated (red) compared with BMP-stimulated (black) SOX17/TFAP2C/BLIMP1 clone-derived d1 iMeLC aggregates (C). The bars indicate the mean value of each time point, and the ranks of the genes ordered by the fold changes between BMP- and Dox-stimulation are shown. See Table S1 for the samples analyzed. (D, E) Gene ontology terms (blue) and KEGG pathways (pink) enriched in differentially expressed genes between BMP and Dox- and Dox-stimulated SOX17/TFAP2C clone-derived cells at d2 (BMP and Dox: BT+AG+; Dox: BT+). (D, E) Representative genes up-regulated in BMP and Dox- (D) or Dox- (E) stimulations and P-values are shown.
With a focus on the SOX17/TFAP2C/BLIMP1 clones, we determined the differentially expressed genes (DEGs) between BMP4- and Dox-stimulated cells at d1 (whole iMeLC aggregates) and d2 (BMP4: BT+AG+ cells; Dox: BT+ cells) of the respective stimulations. Compared with the Dox-stimulated cells, the BMP4-stimulated cells up-regulated 136 and 26 genes and down-regulated 104 and 73 genes at d1 and d2, respectively (Fig 2F and Table S2) (note that the numbers of DEGs were smaller at d2 because of a variability in gene expression of the BT+ cells of the Dox-induced SOX17/TFAP2C/BLIMP1 clones [Fig 2B]). The genes up-regulated in BMP4-stimulated cells at d1 and/or d2 were enriched with those for “transcription from RNA polymerase II promoter” (Gene Ontology [GO] functional terms), “signaling pathways regulating pluripotency of stem cells,” and “WNT/HIPPO/TGF-β signaling pathways” (Kyoto Encyclopedia of Genes and Genomes [KEGG] pathway) (Fig 2G and Table S2), and included known BMP targets such as GATA3, TFAP2A, MSX1, EVX1, HAND1, TBX3, MSX2, and CDX2 (Figs 2G and H and S2B and Table S2) (Note that TFAP2A, HAND1, HAPLN1, MSX2, and CDX2 were highly up-regulated at both d1 [Fig 2H] and d2 [Fig S2B]). The genes up-regulated in Dox-stimulated cells at d1 and/or d2 were enriched with those for “cellular response to glucose stimulus” (GO terms) and “PI3K-AKT signaling pathway” (KEGG pathways) (Fig 2I and Table S2), and included H19, DLL1, GSTA1, COL14A1, TCF7L1, HHEX, NRP1, CHODL, and LEFTY1 (Figs 2I and S2C and Table S2), some of which are characteristic for anterior epiblast/endoderm in gastrulating mouse embryos (32, 33, 34). The DEGs between the SOX17/TFAP2C clone-derived BMP4/Dox- and Dox-stimulated cells at d2 (BMP4/Dox: BT+AG+ cells; Dox: BT+ cells) included similar genes (Fig S2D and E and Table S2). These findings raised a possibility that, unlike in mice (25), a TF(s) recognized as a canonical BMP target(s) may play a key role in conferring the germ-cell fate on iMeLCs.
Table S2 List of differentially expressed genes between relevant cell types generated in this study. (143.3KB, xlsx)
To explore the relevance of such TFs in an in vivo context, we examined their expression in the single-cell transcriptome of PGCs (cyPGCs) of cynomolgus monkeys, a primate model for human development (18, 30). Among the TFs examined, GATA3 and MSX2 were expressed at relatively high levels in cyPGCs from embryonic day (E) 13 to E17 (early cyPGCs) as well as in cy germ cells from E36 to E51 (oogonia/gonocytes), and whereas TBX3 and HAND1 were expressed in early cyPGCs (at lower levels than GATA3 and MSX2), their expression was sporadic/repressed in oogonia/gonocytes (Fig 2J). The other TFs showed more sporadic/no expression in early cy germ cells (Fig 2J). Accordingly, we decided to focus on exploring the function of GATA3 and MSX2. They were indeed expressed at high levels upon induction of hPGCLCs by BMP4 (Fig 3A), and in the iMeLC aggregates of the SOX17/TFAP2C and SOX17/TFAP2C/BLIMP1 clones stimulated by BMP4 and Dox (Fig S3A).
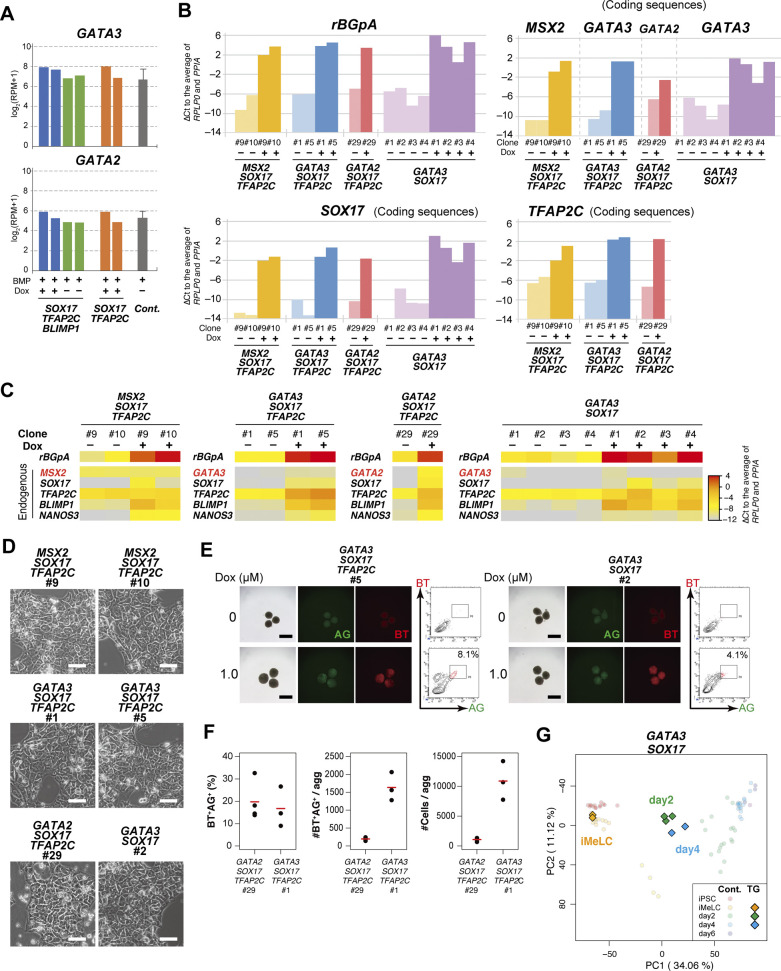
Figure 3. GATA3/2, SOX17, and TFAP2C generate human primordial germ-cell-like cells.
(A) Expression dynamics of MSX2, GATA3, and GATA2 during bone morphogenetic protein (BMP)-driven human primordial germ-cell–like cell induction from the parental hiPSCs. Log2(RPM + 1) values are plotted and the bars indicate the mean value of each time point. See Table S1 for the samples analyzed. (B) Bright-field and fluorescence (AG/BT) images, and FACS analyses for BTAG expression in floating aggregates of the MSX2/SOX17/TFAP2C clone induced by BMP4 or Dox at d4. Representative images of three independent experiments are shown. Bars, 200 μm. (C) Bright-field and fluorescence (AG/BT) images, and FACS analyses for BTAG expression in floating aggregates of the GATA3/SOX17/TFAP2C clone (top) and GATA2/SOX17/TFAP2C clone (bottom) induced with or without Dox 1.0 μg/ml at d4. Representative images for 10 (GATA3/SOX17/TFAP2C) and six (GATA2/SOX17/TFAP2C) experiments are shown. Bars, 200 μm. (D) Expression of rBGpA (transgenes) and the indicated endogenous genes in BMP-induced parental clone-derived and Dox-induced GATA3/SOX17/TFAP2C clone- and GATA2/SOX17/TFAP2C clone-derived d4 BT+AG+ cells. Two replicates from independent experiments were analyzed. For each gene, the ΔCt was calculated from the average Ct value of two housekeeping genes, RPLP0 and PPIA (set as 0). (E, F) Bright-field and fluorescence (AG/BT) images, and FACS analyses for BTAG expression at d4 in floating aggregates of the GATA3/SOX17/TFAP2C (E) and GATA2/SOX17/TFAP2C (F) clones induced by BMP4 or Dox with 0, 100, 500 nM of LDN193189. Representative images of at least two independent experiments are shown. Bars, 200 μm.
Figure S3. GATA3/2, SOX17, and TFAP2C generate human primordial germ-cell-like cells.
(A) Expression of GATA3 and GATA2 at d2 in bone morphogenetic protein (BMP) and Dox- or BMP-treated BT+ cells of the SOX17/TFAP2C/BLIMP1 clones, BMP and Dox-treated BT+ cells of the SOX17/TFAP2C clones, and BMP-treated BT+AG+ PGCLCs of the parental line. The transcriptome data were used and the values are shown in log2(RPM + 1). See Table S1 for the samples analyzed. (B) The transgene expression levels in the designated hiPSC clones stimulated with (+) or without (−) Dox (1.0 μg/ml) for 24 h. Two clones for MSX2/SOX17/TFAP2C and GATA3/SOX17/TFAP2C, one clone for GATA2/SOX17/TFAP2C, and four clones for GATA3/SOX17 expression were examined. To quantify the transgene expression levels by qRT-PCR, primer pairs for rabbit rBGpA or for the transgene coding sequences were used, and the ΔCt was calculated from the average Ct value of two housekeeping genes, RPLP0 and PPIA (set as 0). (C) Heat map representation of the expression levels of the indicated genes in the designated hiPSC clones stimulated with (+) or without (−) Dox (1.0 μg/ml) for 24 h. To quantify the expression levels of the transgenes or endogenous genes, primer pairs for rBGpA or the 3′ untranslated regions were used, respectively, and the ΔCt was calculated from the average Ct value of two housekeeping genes, RPLP0 and PPIA (set as 0). The color coding is as indicated. (D) Phase-contrast images of iMeLCs of the designated clones. No apparent morphological differences were seen among the clones. Representative images of at least two independent experiments are shown. Bars, 50 μm. (E) Bright-field and fluorescence (AG/BT) images, and FACS analyses for BTAG expression in d2/d4 floating aggregates of the GATA3/SOX17/TFAP2C (left) and GATA3/SOX17 (right) clones induced by 0 (top) or 1.0 μg/ml (bottom) of Dox. Representative data for six (GATA3/SOX17/TFAP2C) and two (GATA3/SOX17) experiments are shown. Bars, 200 μm. (F) BT+AG+ cell induction efficiency (%) (left), BT+AG+ cell numbers per aggregate (middle), and total cell numbers per aggregate (right) of the GATA2/SOX17/TFAP2C and GATA3/SOX17/TFAP2C clones. Mean values (n = 3 for each line) are shown as red bars. (G) The principal component analysis plots of the cells (iMeLCs, Dox-induced d2/d4 BT+ cells) derived from the GATA3/SOX17 clone (squares), overlaid with the indicated cells derived from the parental clone (circles with pale color). See Table S1 for the samples analyzed. The color coding is as indicated.
GATA TFs, SOX17, and TFAP2C drive hPGCLC induction
GATA3 is a member of the evolutionarily conserved GATA family of TFs, which bears six paralogs in vertebrates and plays key roles in the specification of a wide range of cell types in three germ layers as well as in extraembryonic tissues (see reference 35 for review; see the Discussion section for the roles of GATA3 in relevant contexts), but its role in germ-cell development has not been reported. MSX2 is a member of the evolutionarily conserved homeodomain TFs, which bear three and two paralogs in mice and humans, respectively, and play important functions during neural tube, tooth, and limb development (see reference 36 for review). In mice, Msx1 and Msx2 double mutants show a defect in meiotic prophase in female embryonic germ cells (37).
We decided to examine the effect of GATA3 or MSX2 expression combined with the expression of SOX17 and TFAP2C, for the following reasons: (1) both SOX17 and TFAP2C are required for hPGCLC specification (8, 13); (2) SOX17 and TFAP2C did not activate each other (Fig 1B and E–H); (3) BLIMP1 was activated by SOX17 (Fig 1B and E) (13); (4) nearly all the cells in the SOX17/TFAP2C clone-derived iMeLC aggregates became BT+AG+ in response to BMP4 and Dox (Fig 1E and F). We isolated two independent clones expressing MSX2, SOX17, and TFAP2C, verified the transgene expression in hiPSCs by qRT-PCR (Fig S3B and C), and examined whether their expression in iMeLC aggregates (Fig S3D) induces the germ-cell fate; however, we found that MSX2, SOX17, and TFAP2C expression failed to induce BT+AG+ cells (Fig 3B).
Next, therefore, we isolated two independent clones expressing GATA3, SOX17, and TFAP2C, and verified the transgene expression in hiPSCs by qRT-PCR (Fig S3B). We found that GATA3, SOX17, and TFAP2C expression up-regulated endogenous SOX17 to a moderate extent and endogenous TFAP2C to a substantial extent (Fig S3C). We then expressed these genes in iMeLC aggregates; remarkably, we observed a progressive induction of distinct populations of BT+AG+ cells by d4 of the transgene induction (Figs 3C and S3D and E). We then isolated induced BT+AG+ cells at d4, and examined their expression of key genes by qRT-PCR. As shown in Fig 3D, they up-regulated endogenous SOX17, TFAP2C, BLIMP1, and NANOS3 to levels indistinguishable from those in BT+AG+ cells induced by BMP4, and repressed SOX2 to a great extent.
We next examined whether other GATA TFs might also induce the BT+AG+ cells in iMeLCs. We decided to explore the function of GATA2 because (1) GATA2 is also up-regulated upon hPGCLC induction, although at a lower level than GATA3 (Fig 3A), and is expressed in early cyPGCs (GATA2 is detectable in at least 6 of 16 early cy germ cells) (Fig 2J); (2) GATA2 shows the highest structural similarity to GATA3 among the GATA TFs (35, 38); and (3) GATA3 and GATA2 show a compensatory function and bear overlapping genome-wide binding profiles in other relevant contexts (35, 39, 40, 41). Accordingly, we isolated a clone expressing GATA2, SOX17, and TFAP2C, verified the transgene expression in hiPSCs, and found that GATA2, SOX17, and TFAP2C expression up-regulated both endogenous SOX17 and TFAP2C to a substantial extent (Fig S3C). Consistent with this result, the expression of GATA2, SOX17, and TFAP2C in iMeLCs robustly induced BT+AG+ cells with an expression profile of key genes similar to that in BT+AG+ cells induced by GATA3, SOX17, and TFAP2C or BMP4 (Fig 3C and D). Interestingly, however, the sizes/cell numbers of the iMeLC aggregates induced with GATA2, SOX17, and TFAP2C were smaller than those induced with GATA3, SOX17, and TFAP2C or BMP4 (Figs 3C and S3F), suggesting that GATA3 and GATA2 play overlapping but distinct functions in iMeLC aggregates.
A recent report has shown that in hESCs, BMP signaling activates GATA3, which in turn up-regulates BMP4, thereby creating a feedforward loop for persistent activation of the BMP signaling pathway (42). To exclude the possibility that GATA3/GATA2, SOX17, and TFAP2C expression activates the BMP signaling that in turn induces the BT+AG+ cells, we induced GATA3/GATA2, SOX17, and TFAP2C in iMeLCs in the presence of LDN193189, a potent inhibitor of the key receptor for BMP signaling, ALK2/3 (43, 44). As shown in Fig 3E and F, whereas the provision of LDN193189 blocked the induction of BT+AG+ cells by BMP4, it had little, if any, effect on the BT+AG+-cell induction by GATA3/GATA2, SOX17, and TFAP2C. Next, we explored the possibility that if GATA3 up-regulates BMP4 to a substantial extent, then the expression of GATA3 with SOX17, but without TFAP2C, may also induce the BT+AG+ cells. For this purpose, we isolated a number of clones expressing GATA3 and SOX17, verified the transgene expression (Fig S3B and C), and induced them in iMeLC aggregates (Fig S3D). Although the GATA3/SOX17-derived iMeLC aggregates up-regulated BT and activated AG to some extent, they did not form a distinct population of BT+AG+ cells (Fig S3E). Thus, GATA3/2, SOX17, and TFAP2C directly and cell-autonomously drive the hPGCLC program.
We determined the transcriptomes of the GATA3/SOX17/TFAP2C and GATA3/SOX17 clones during the induction of BT+AG+/BT+ cells. PCA revealed that whereas the iMeLCs from the GATA3/SOX17/TFAP2C clones were nearly identical to wild-type iMeLCs, upon induction of the transgenes by Dox, they progressed directly toward the hPGCLC fate, bypassing the d1 iMeLC-aggregate state, and by d6 of induction, they acquired a transcriptome close to that of the d4/d6 hPGCLCs induced by BMP4 (Fig 4A). Accordingly, regarding the expression of 481 genes that characterize the hPGCLC specification process (13) (Fig S2A), d6 BT+AG+ cells from the GATA3/SOX17/TFAP2C clone exhibited high similarity/correlation to d2/d4/d6 wild-type BT+AG+ cells induced by BMP4 (Fig 4B). In contrast, although d2/d4 BT+ cells induced from the GATA3/SOX17 clone appeared to take a similar pathway until d2, they failed to progress further by d4 (Fig S3G).
Figure 4. The transcription factor-induced BT+AG+ cells directly acquire human primordial germ-cell-like cell (hPGCLC) transcriptome.
(A) The principal component analysis plots of the cells (iMeLCs, Dox-induced d2/d4/d6 BT+AG+ cells) derived from the GATA3/SOX17/TFAP2C clone (squares), overlaid with the indicated cells derived from the parental clone (circles with pale color). See Table S1 for the samples analyzed. The color coding is as indicated. (B) Heat map representation (color coding as indicated) of the expression of the 481 genes characterizing hPGCLC specification (13) (Fig S2) in the parental hiPSCs, iMeLCs, d1 whole aggregates, and d2/d4/d6 BT+AG+ cells and in Dox-induced, GATA3/SOX17/TFAP2C–derived d6 BT+AG+ cells. The correlation coefficient (0.926) between bone morphogenetic protein (BMP)– and Dox-induced d6 BT+AG+ cells is shown. The color coding in the left column is as follows: red, genes for PGCLC specification; cyan, genes for endoderm/mesoderm specification; yellow, genes for pluripotency. See Fig S2 for details. (C) The numbers of differentially expressed genes between BMP- and Dox-induced cells of the GATA3/SOX17/TFAP2C clone in d2/d4/d6 BT+AG+ cells (P < 0.01 by Tukey–Kramer test, log2[RPM + 1] > 4 in cells with higher expression, log2[fold change: FC] > 1 [up, pale yellow; down, pale blue] or 2 [up, yellow; down, blue]). (D, F) Gene ontology terms (blue) and KEGG pathways (pink) enriched in genes up-regulated in BMP-induced (D) and in Dox-induced (F) GATA3/SOX17/TFAP2C clone-derived d2 BT+AG+ cells. (E, G) Expression dynamics of the genes up-regulated in BMP-induced (E, the parental clone) or Dox-induced (G, the GATA3/SOX17/TFAP2C clone) d2 BT+AG+ cells during the respective induction processes (BMP: black; Dox: red). The bars indicate the mean value of each time point, and the rank of the gene ordered by the fold change is shown. See Table S1 for the samples analyzed. (H) Expression dynamics of GATA3 and GATA2 during BMP-induced hPGCLC induction from the parental (gray) and the indicated knockout hiPSCs (red) (d1: whole iMeLC aggregates; d2: EOMES−/−: whole aggregates; SOX17−/−: whole aggregates; TFAP2C−/−: BT+ cells; BLIMP1−/−: AG+ cells) (13). The bars indicate the mean value of each time point of each genotype. See Table S1 for the samples analyzed.
The numbers of DEGs between BT+AG+ cells from the GATA3/SOX17/TFAP2C clone induced by Dox and from the parental cells induced by BMP4 were the largest at d2 (527), and decreased thereafter (d4: 265; d6: 53) (Fig 4C and Table S2). The genes up-regulated in BMP4-induced cells at day 2 (334 genes) were enriched with those for “negative regulation of transcription from RNA pol II promoter,” “embryonic forelimb morphogenesis” (GO terms), and “TGF-β signaling pathway” (KEGG pathway), and included key BMP targets, such as ID1, ID3, CDX2, TBX3, MSX1, MSX2, HAND1, and TFAP2A (Fig 4D and E), suggesting that these BMP effectors are dispensable for hPGCLC specification. In contrast, the genes up-regulated in GATA3/SOX17/TFAP2C-induced cells at day 2 (193 genes) were enriched with those for “negative regulation of BMP signaling pathway,” “anterior/posterior axis specification” (GO terms), and “PI3K-Akt signaling pathway” (KEGG pathway), and included FGF2, FGF12, FGF19, FGFR2, MAP2K1, MAP2K6, IRS2, and SOS2 (Fig 4F and G). The genes up-regulated with high fold-changes included epiblast/ectoderm genes such as SOX2, ZIC3, and SALL3, which were repressed in slower kinetics by the transgene expression (Fig 4G).
We addressed whether GATA3 and GATA2 expression were affected by other TFs relevant for hPGCLC specification. As shown in Fig 4H, in any of the knockout clones for EOMES, SOX17, TFAP2C, and BLIMP1 induced for the germ-cell fate by BMP4 (13), GATA3 and GATA2 up-regulation was un-affected, indicating that their expression is independent from these TF pathways. We conclude that among the BMP effectors, the GATA TFs are the key that, together with SOX17 and TFAP2C, is sufficient to drive the transcriptional program for hPGCLC specification.
Critical requirements of the GATA TF paralogs for hPGCLC specification
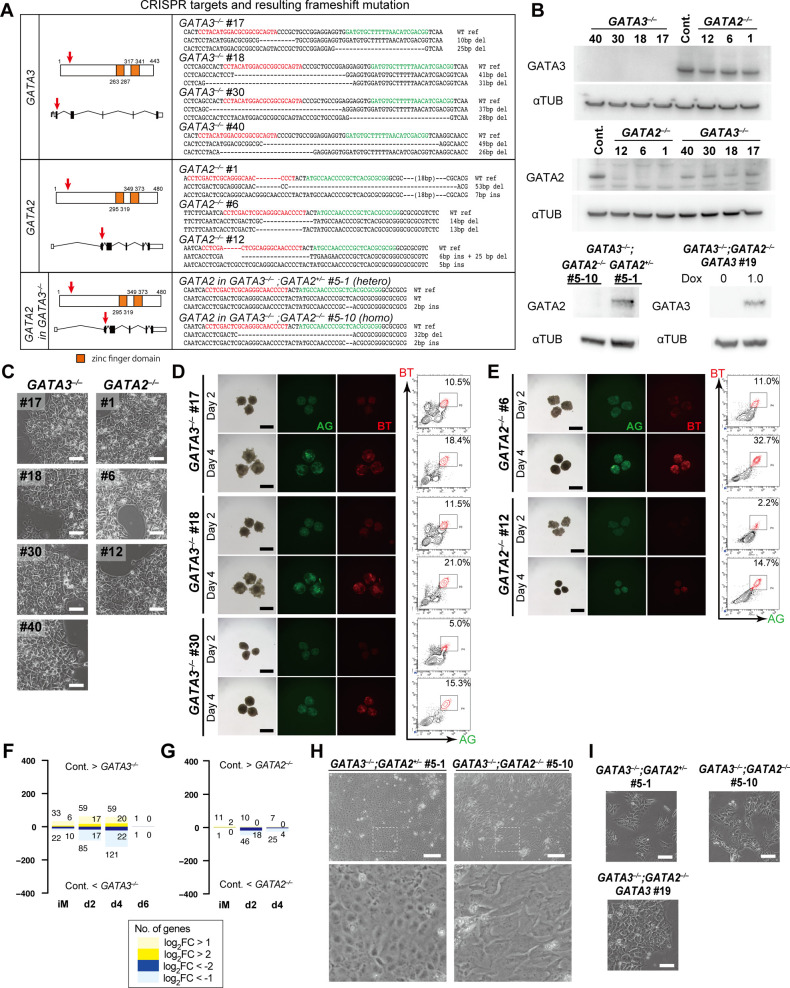
We next explored whether GATA TFs are essential for hPGCLC induction. Using CRISPR/Cas9 technology (45), we targeted GATA3 or GATA2 loci in parental 585B1 BTAG hiPSCs, and isolated four and three clones bearing frameshift mutations in both alleles of GATA3 or GATA2, respectively (GATA3 or GATA2 homozygous knockout−/− clones) (Fig S4A). The lack of GATA3 or GATA2 expression in these clones was verified by Western blot analyses following the differentiation of these clones into TE-like cells (46) (Fig S4B).
Figure S4. Dose-dependent function of GATA transcription factors in human primordial germ-cell-like cell specification.
(A) The CRISPR targets (left) and resulting mutations (right) for GATA3 (top), GATA2 (middle), and GATA2 in the GATA3−/− #40 clone (bottom). The same gRNA was used to create GATA2 mutants. (left) The amino acid sequences and the genomic structures (the exons: black boxes; introns: folded lines; un-translated regions: white boxes) of GATA3 and GATA2 are indicated, with the position of zinc finger DNA binding domain (orange) and the CRISPR target sites (red arrows). (right) The DNA sequences of both alleles for the indicated knockout lines are shown. Red and green letters indicate the positions of the gRNAs including the PAM sequences. del, deletion; ins, insertion. (B) Western blot analysis of GATA3 and GATA2 expression in TE-like cells induced from the GATA3−/−, GATA2−/−, GATA3−/−:GATA2+/−, and GATA3−/−:GATA2−/− clones, and in the GATA3−/−:GATA2−/−: GATA3 (rescued) clones cultured with or without 1.0 μg/ml of Dox for 24 h. α-TUBULIN was used as a loading control. (C) Phase-contrast images of iMeLCs in the designated clones. No apparent morphological differences were seen among the clones. Representative images of four (GATA3−/−) and two (GATA2−/−) independent experiments are shown. Bars, 50 μm. (D, E) Bright-field and fluorescence (AG/BT) images, and FACS analyses for BTAG expression in d2/d4 floating aggregates of the GATA3−/−#17, #18, #30 (D) or GATA2−/− #6, #12 (E) clones induced by bone morphogenetic protein 4. Representative images of four (D) and two (E) independent experiments are shown. Bars, 200 μm. (F, G) The numbers of differentially expressed genes between the parental and GATA3−/− (E) or GATA2−/− (F) clones in iMeLCs (iM), d2/d4/d6 BT+AG+ cells (P < 0.01 by Tukey–Kramer test, log2[RPM + 1] > 4 in cells with higher expression, log2[fold change: FC] > 1 [up, pale yellow; down, pale blue] or 2 [up, yellow; down, blue]). (H) Phase-contrast images of the TE-like cells induced from the designated clones. The GATA3−/−:GATA2+/− #5-1 clone exhibited a cobblestone-like epithelial morphology similar to TE, whereas the GATA3−/−:GATA2−/− #5-10 clone failed to show TE-like differentiation and exhibited a mesenchymal morphology. Bars, 200 μm. (I) Phase-contrast images of iMeLCs in the designated clones. No apparent morphological differences were seen among the clones. Representative images of at least two independent experiments are shown (indicated in Fig 5F). Bars, 50 μm.
Source data are available for this figure.
We induced these clones into iMeLCs (Fig S4C), and then into hPGCLCs by BMP4. Unexpectedly, all the GATA3−/− or GATA2−/− clones were induced into BT+AG+ cells in a manner similar to the parental clone (Figs 5A and S4D and E). We isolated total RNAs from iMeLCs and d2/d4/d6 BT+AG+ cells induced from all the clones, analyzed the expression of key genes by qRT-PCR, and found that the GATA3−/− or GATA2−/− clones expressed relevant genes for hPGCLC specification in an apparently normal fashion (Fig 5B). We performed an RNA-seq analysis, which revealed that GATA3−/− and GATA2−/− cells differentiated into BT+AG+ cells in a manner equivalent to the parental clone (Fig 5C and D), and exhibited small numbers of DEGs compared with the parental counterparts (Fig S4F and G). However, we noted that the BT+AG+-cell induction efficiencies at d4 of the GATA3−/− clones (∼14.5%) were significantly lower than those of the control (∼31.4%) or of the GATA2−/− clones (∼25.7%) (Fig 5F), raising the possibility that GATA3 and GATA2 have a compensatory function, with GATA3 playing the more dominant role, during hPGCLC induction.
Figure 5. Dose-dependent function of GATA transcription factors in human primordial germ-cell-like cell specification.
(A) Bright-field and fluorescence (AG/BT) images, and FACS analyses for BTAG expression in floating aggregates of the GATA3−/−, GATA2−/−, and parental clones induced by bone morphogenetic protein 4 (BMP4) at d2/d4. Representative images of at least two independent experiments are shown (indicated in Fig 5F). Bars, 200 μm. (B) Expression dynamics of the indicated genes during human primordial germ-cell–like cell induction (iMeLCs, d2/d4 BT+AG+ cells) by BMP4 from the parental (black), GATA3−/− (red), and GATA2−/− (blue) clones. For each gene, the ΔCt was calculated from the average Ct value of two housekeeping genes, RPLP0 and PPIA (set as 0). The bars indicate the mean value of each time point of each genotype. Replicate numbers: GATA2−/−: 3; GATA3−/−: 4 for iMeLCs and 8 for d2/d4 BT+AG+ cells; parental clone: 2. (C, D) The principal component analysis plots of the cells (iMeLCs, d2/d4/d6 BT+AG+ cells) derived from the GATA3−/− (C) and GATA2−/− (D) clones (squares), overlaid with the indicated cells derived from the parental clone (circles with pale colors). See Table S1 for the samples analyzed. The color coding is as indicated. (E) Bright-field and fluorescence (AG/BT) images, and FACS analyses for BTAG expression in floating aggregates of the GATA3−/−; GATA2+/− and GATA3−/−; GATA2−/− clones induced by BMP4 at d4. Representative images of at least two independent experiments are shown (indicated in Fig 5F). Bars, 200 μm. (F) The percentages of BT+AG+ cell induction from the indicated genotypes at d4. The replicate numbers and the P-values (t test) are as indicated. The inductions were performed side by side. Typically, the efficiency for BT+AG+ cell induction from parental hiPSCs varies to this extent (20%∼60%) (9, 13, 26). (G) A scheme for GATA3 expression in the GATA3−/−; GATA2−/−; GATA3 clone. (H) Bright-field and fluorescence (AG/BT) images, and FACS analyses for BTAG expression in floating aggregates of the GATA3−/−; GATA2−/−; GATA3 clone upon induction with BMP4 and 0, 0.5, and 1.0 μg/ml of Dox at d4. Representative images of at least two independent experiments are shown (indicated in Fig 5F). Bars, 200 μm. (I) The percentages of BT+AG+ cell induction at d4 (two replicates) from the GATA3−/−; GATA2−/−; GATA3 clone induced with BMP4 and 0, 0.5, and 1.0 μg/ml of Dox treatment. (J) Expression of the indicated genes in d4 BT+AG+ cells induced from the parental (with BMP4, red) or GATA3−/−; GATA2−/−; GATA3 (with BMP4 and Dox, green) clones (two replicates). For each gene, the ΔCt was calculated from the average Ct value of two housekeeping genes, RPLP0 and PPIA (set as 0).
To investigate this possibility, we knocked out the GATA2 alleles in the GATA3−/− clone, and obtained one line with the GATA3−/−; GATA2+/− genotype and one line with the GATA3−/−; GATA2−/− genotype (Fig S4A). Upon the differentiation of these clones into TE-like cells (46), the GATA3−/−; GATA2+/− cells formed an epithelial-sheet structure with a typical TE-like cobblestone morphology, but the GATA3−/−; GATA2−/− cells failed to show such differentiation and exhibited a mesenchyme-like appearance (Fig S4H). Consistently, we confirmed that the GATA3−/−; GATA2−/− cells, but not the GATA3−/−; GATA2+/− cells, lost the expression of GATA2 proteins (Fig S4B). We then induced these cells into iMeLCs (Fig S4I) and successively to hPGCLCs, which revealed that the GATA3−/−; GATA2+/− cells still formed the BT+AG+ cells, but at a further reduced efficiency (∼9.5%), whereas the GATA3−/−; GATA2−/− cells barely differentiated into such a state (∼1.6%) (Fig 5E and F).
To exclude the possibility that the differentiation failure of the GATA3−/−; GATA2−/− clone was due to a clonal effect, we performed a rescue experiment. We introduced the Dox-inducible GATA3 expression system into the GATA3−/−; GATA2−/− clone and isolated a line that showed an appropriate GATA3 expression in hiPSCs. We induced this line into iMeLCs (Fig S4I), and stimulated the iMeLC aggregates with BMP4 and Dox. Although we found that a continuous stimulation of the iMeLC aggregates with BMP4 and Dox led to major cell death for an unknown reason, the timed stimulation of Dox (∼32 h) resulted in the induction of BT+AG+ cells in a Dox-dose dependent manner (Fig 5G–I), and the induced BT+AG+ cells expressed key genes for hPGCLCs in an appropriate fashion (Fig 5J). Thus, we conclude that the GATA TF paralogs, GATA3 and GATA2, show a dose-dependent requirement for hPGCLC specification. Considering that GATA3 was expressed at a higher level than GATA2 during hPGCLC induction (Fig 3A) and upon cyPGC specification (Fig 2I) and that GATA3 knockouts, but not GATA2 knockouts, exhibited a significant decrease in hPGCLC induction efficiency (Fig 5F), we propose that GATA3 plays a major role in hPGCLC induction.
GATA3 expression in post-implantation primate embryos
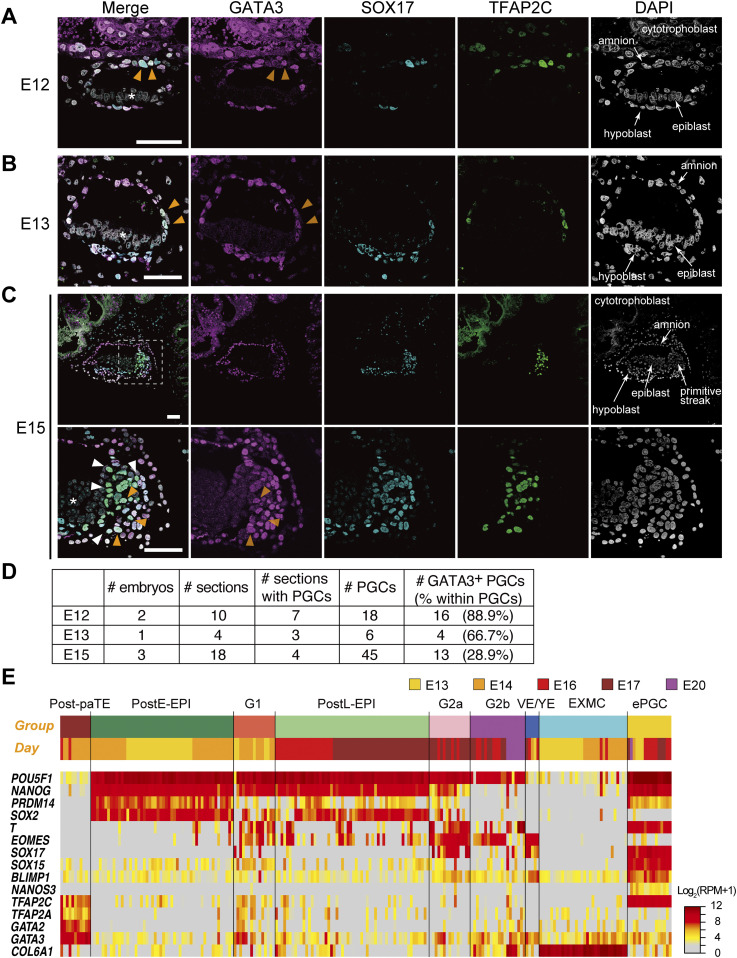
To explore the spatial relationship of GATA3, SOX17, and TFAP2C expression in a developmental context, we examined their expression during PGC specification in the early post-implantation embryos of cynomolgus monkeys. By immunofluorescence analysis, we detected SOX17+ and TFAP2C+ cyPGCs in the dorsal amnion at E12, in the posterior amnion at E13, and near the primitive streak region between the epiblast and hypoblast at E15 (Fig 6A–D). During this period, SOX17 was also expressed in the hypoblast, and TFAP2C was also expressed diffusely in the amnion and cytotrophoblast/syncytiotrophoblast at E12 but became negative in the amnion thereafter (Fig 6A–C). We found that GATA3 was expressed strongly in the cytotrophoblast/syncytiotrophoblast and also in the amnion and the hypoblast, and importantly, in the SOX17+/TFAP2C+ cyPGCs (Fig 6A–D). Along with the embryonic development, we noted a gradual decrease in the ratio of GATA3+ cyPGCs (Fig 6D). Notably, the epiblast did not express any of these TFs.
Figure 6. GATA3 expression in post-implantation cynomolgus monkey embryos.
(A, B, C) Immunofluorescence analysis of GATA3 (magenta), SOX17 (cyan), and TFAP2C (green) expression (merged with DAPI) in cynomolgus monkey embryos at E12 (A), E13 (B) and E15 (C). For E15 (C), the boxed area (top) is magnified (bottom). Orange arrowheads indicate triple-positive PGCs, whereas white arrowheads indicate SOX17/TFAP2C+ but GATA3− cells. Note that there was no signal in the epiblast (asterisk). (D) Representative images of the samples shown in (D) are presented. Bars, 50 μm. (D) The numbers of embryos (E12, E13, and E15), total sections, and sections with PGCs (SOX17+TFAP2C+ cells), PGCs, and GATA3+ PGCs analyzed/detected in this study. (E) Heat map representation of the expression of key genes in the indicated cell types of the post-implantation cy embryos (30). The colored bars on top indicate cell types (top) and embryonic days (bottom), respectively. The color coding is as indicated. Post-paTE, PostE-EPI, post-implantation early epiblast; G1, gastrulating cells, group 1; postL-EPI, post-implantation late epiblast; G2a/G2b, gastrulating cells, group 2a/2b; VE/YE, visceral endoderm/yolk sac endoderm; EXMC, extraembryonic mesenchyme; ePGC, early PGC.
In addition, we re-analyzed GATA3/GATA2 expression in the single-cell transcriptome of cy post-implantation embryos (E13–E20) (30). In addition to early cyPGCs (Fig 6A–D), GATA3 was expressed in the TE, extra-embryonic mesenchyme, visceral endoderm/yolk-sac endoderm, and gastrulating cells, and weakly/sporadically in the epiblast (Fig 6E). GATA2 was expressed strongly in the TE, but was weak/sporadic in the other cell types, including early cyPGCs (Figs 2J and 6E). These findings delineate the spatial relationship of the expression of GATA3, SOX17, and TFAP2C during primate development, demonstrating that cyPGCs, but not other relevant cell types, co-express these TFs.
TF-induced hPGCLCs are competent for epigenetic reprogramming and differentiation into oogonia/gonocytes
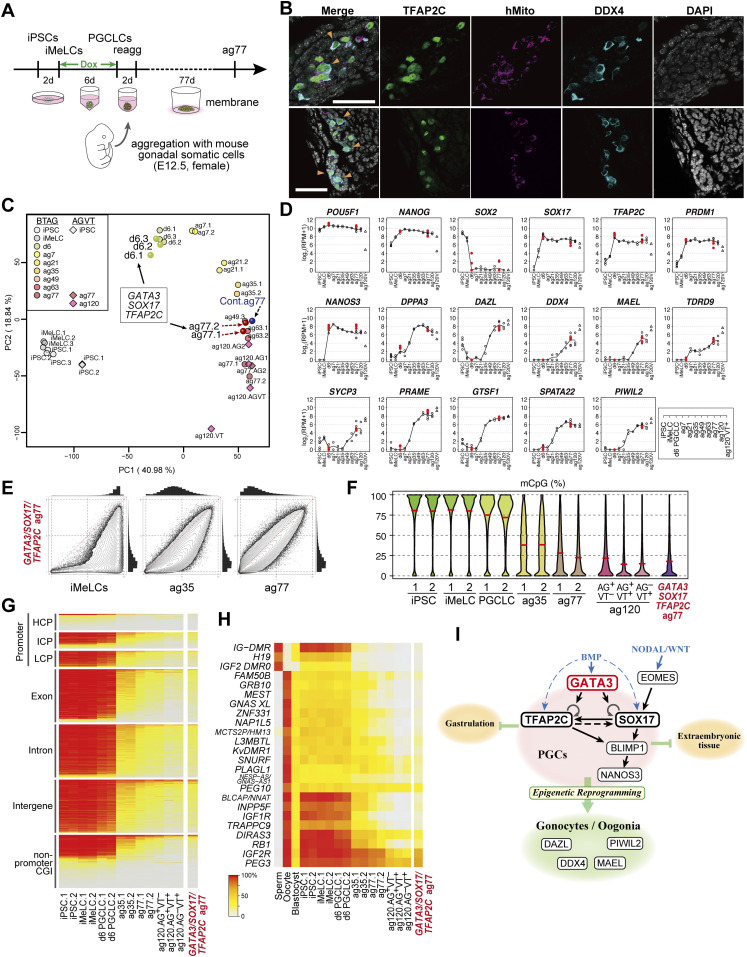
To evaluate the functional property of the BT+AG+ cells induced by GATA3, SOX17, and TFAP2C as human germ cells, we explored whether they are competent to undergo epigenetic reprogramming and to differentiate into oogonia/gonocytes, which are female/male gonadal germ cells before overt sexual differentiation and exhibit similar gene-expression and epigenetic profiles (47, 48, 49). Both female and male hiPSCs differentiate into oogonia/gonocytes (10). Accordingly, for this purpose, we isolated the Dox-induced GATA3/SOX17/TFAP2C clone-derived d6 BT+AG+ cells, and aggregated them with mouse embryonic ovarian somatic cells at E12.5 to form xenogeneic reconstituted ovaries (xrOvaries) in vitro (10, 11) (Fig 7A). As a control, we generated xrOvaries using the BMP4-induced parental clone-derived d6 hPGCLCs. We isolated xrOvaries at d77 of aggregation culture (ag77) and evaluated them with immunofluorescence analyses. The analyses revealed that multiple clusters of GATA3/SOX17/TFAP2C clone-derived AG+/human mitochondrial-antigen+ cells persisted in xrOvaries, and many of them expressed DDX4, a key marker of oogonia/gonocytes (Fig 7B).
Figure 7. Transcription factor-induced BT+AG+ cells differentiate into oogonia/gonocytes in xrOvaries.
(A) A scheme for xrOvary culture (10, 11) with Dox-induced GATA3/SOX17/TFAP2C clone-derived d6 BT+AG+ cells. (B) Immunofluorescence analysis of TFAP2C (green), human mitochondria antigen (magenta), and DDX4 (cyan) expression (merged with DAPI) on aggregation day (ag) 77 xrOvaries. In two independent experiments, 28 and 23 TFAP2C/DDX4-expressing cells/7 sections, respectively, were detected. Bars, 50 μm. (C) The principal component analysis plots of the transcriptome of the GATA3/SOX17/TFAP2C clone-derived d6 BT+AG+ cells, d6ag77 BT+AG+ cells, and the parent clone-derived bone morphogenetic protein-induced d6ag77 BT+AG+ cells (see Table S1) with the relevant cell types during in vitro oogonia/gonocyte differentiation reported in reference 10, in which 585B1 BTAG hiPSCs (XY) and 1390G3 AGVT (AG; DDX4 [also known as VASA]-tdTomato [VT]) hiPSCs (XX) were used as starting materials. Numbers following “ag” indicate the culture days in xrOvaries. For the AGVT cells, ag77 and 120 AG+VT− (AG), AG+VT+ (AGVT) or AG−VT+ (VT) were used for analysis. (D) Expression dynamics of the key genes in GATA3/SOX17/TFAP2C clone-derived d6 BT+AG+ cells, and d6ag77 BT+AG+ cells (n = 2, red circles) (see Table S1), overlaid with those in the relevant cell types during the in vitro oogonia/gonocyte differentiation reported in reference 10. (E) Scatter-plot comparisons, combined with histogram representations (top and right of scatter plots), of the genome-wide 5 mC levels (genome-wide 2-kb windows) between the indicated cell types. (F) Violin-plot representation of the genome-wide 5 mC levels determined by whole-genome bisulfite sequence analysis in the cell types indicated. The mean levels are indicated by red bars. (G, H) Heat map representation showing the 5 mC levels in the indicated genomic elements on the autosomes (G) and in the differentially methylated regions of the indicated imprinted genes (H) in the indicated cells. HCP/ICP/LCP, high/intermediate/low-CpG promoters. The color coding is as indicated. (I) A model of the transcription factor circuitry driving human primordial germ-cell like cell specification.
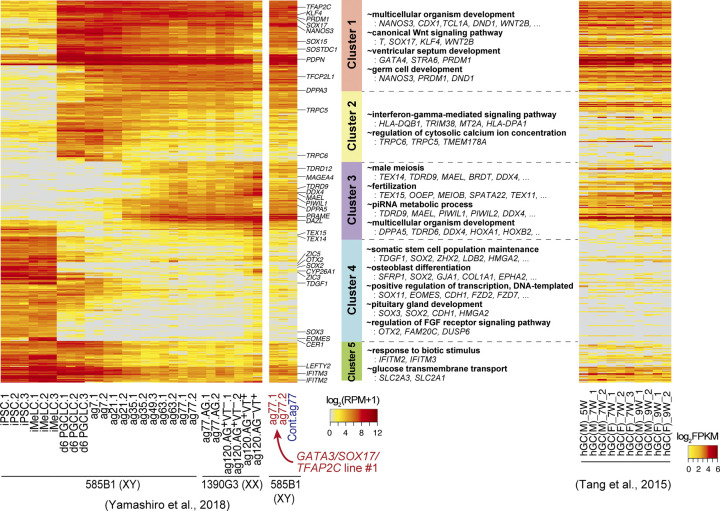
To further characterize the GATA3/SOX17/TFAP2C clone-derived BT+AG+ cells at ag77, we isolated them by FACS, determined their transcriptome by an RNA-sequence, and analyzed their properties. PCA revealed that both the GATA3/SOX17/TFAP2C clone-derived cells and the parental d6 hPGCLC-derived cells (generated in the present study) differentiated as human germ cells in an appropriate manner, acquiring the transcriptome property of oogonia/gonocytes (Fig 7C). Accordingly, the GATA3/SOX17/TFAP2C clone-derived cells expressed key markers of oogonia/gonocytes, including DPPA3, DAZL, DDX4, and MAEL, at high levels (Fig 7D), and with regard to the expression of 451 genes that characterize the oogonia/gonocyte differentiation process from hiPSCs (10), the GATA3/SOX17/TFAP2C clone-derived cells exhibited profiles equivalent to those of the oogonia/gonocytes (Fig S5). The finding that both the GATA3/SOX17/TFAP2C clone-derived cells and the control cells generated in this study were plotted at positions closest to the previous ag63 cells rather than ag77 cells in the PCA (Fig 7C) would be attributable to experimental variation.
Figure S5. Transcription factor-induced BT+AG+ cells differentiate into oogonia/gonocytes in xrOvaries.
Heat map representation of the expression of the 453 genes (the cluster 1–5 genes) characterizing the in vitro oogonia/gonocyte differentiation process reported in reference 10 in the GATA3/SOX17/TFAP2C-derived d6ag77 BT+AG+ cells and the parent clone-derived bone morphogenetic protein–induced d6ag77 BT+AG+ cells, in comparison with those in the relevant cell types in reference 10 and in human fetal germ cells reported in reference 47, as indicated. Representative genes are annotated, and gene ontology-term enrichments in each cluster and key genes are listed. ag, aggregation culture days; VT: DDX4 (also known as VASA)-tdTomato; hGC, human germ cells; M, male; F, female; W, developmental week. Numbers following the indications of cellular states denote replicate numbers. The color coding is as indicated.
Epigenetic reprogramming, including genome-wide DNA demethylation, is a hallmark and exclusive event during germ-cell development (4, 6, 7). To examine whether the GATA3/SOX17/TFAP2C clone-derived BT+AG+ cells at ag77 undergo epigenetic reprogramming, we determined their genome-wide DNA methylation profile by whole-genome bisulfite sequence (WGBS) analysis. Importantly, we found that the genome-wide DNA methylation properties of the GATA3/SOX17/TFAP2C clone-derived cells at ag77 were similar to those of oogonia/gonocytes reported previously (10,47,50) with regard to both their distribution profiles (Fig 7E) and total levels (reduced to as low as ∼17.5%) (Fig 7F). Accordingly, the GATA3/SOX17/TFAP2C clone-derived cells erased their DNA methylation throughout their genomic regions, including promoters, exons, introns, intergenic regions, and non-promoter CpG islands (CGIs) (Fig 7G), as well as parental imprint control regions (ICRs) (Fig 7H). Collectively, these findings demonstrate that the BT+AG+ cells induced by GATA3, SOX17, and TFAP2C are equivalent to hPGCLCs in their capacity to undergo epigenetic reprogramming and to differentiate into oogonia/gonocytes, and thus are considered to bear key properties of bona fide hPGCs.
Discussion
We have identified core TFs, GATA3/2, SOX17, and TFAP2C, which suffice for the reconstitution of the human germ-cell fate, providing a step forward for delineating the mechanism of human germ-cell specification and a foundation for the TF-based human gametogenesis (Fig 7I). Our finding that SOX17, TFAP2C, and BLIMP1 expression in iMeLCs was not sufficient to induce hPGCLCs was unexpected, given that in mice, the expression of three TFs, Blimp1, Prdm14, and Tfac2c, and with a lesser efficiency, two TFs (Blimp1 and Tfap2c; Prdm14 and Tfap2c) or even a single TF (Prdm14), was sufficient to confer the germ-cell fate on their precursors (25). Notably, none of these TFs are known as direct effectors of the BMP signaling in mice; TFAP2C and BLIMP1 are shared by humans; and PRDM14 is expressed in both hiPSCs and iMeLCs (9, 16). Thus, the mechanism of human germ-cell specification is distinct from that in mice not only with regard to the TFs themselves and their hierarchies of actions (13), but also in terms of how the key TFs regulate each other to drive the downstream PGC pathways.
We showed that SOX17 induces BLIMP1 in both hiPSCs and iMeLC aggregates (Fig 1B and E), and the TFAP2C/BLIMP1, SOX17/TFAP2C, SOX17/TFAP2C/BLIMP1 clones induce NANOS3, a marker for hPGCLCs (Fig 1B). Combined with the finding that the BLIMP1−/− clones failed to up-regulate NANOS3 (9, 13), these data indicate that NANOS3 is most likely a downstream target of BLIMP1. Thus, SOX17 induces BLIMP1, which in turn induces NANOS3; however, this is evidently insufficient for hPGCLC specification (Fig 1E–H). The addition of TFAP2C expression to the SOX17/BLIMP1/NANOS3 pathway also failed to activate the hPGCLC specification program (Fig 1E), indicating that although both SOX17 and TFAP2C are essential and interdependent for hPGC(LC) specification (13), the two pathways do not activate each other to elicit the hPGC(LC) specification program.
We identified two GATA TFs—GATA3 and GATA2, with GATA3 playing the more dominant role—as key BMP effectors that, together with SOX17 and TFAP2C, drive the hPGC(LC) specification program (Figs 3C–F and 7I). Importantly, GATA3 and SOX17 expression in iMeLCs was not sufficient to induce the hPGCLC program (Fig S3E), suggesting that the expression of three TFs, GATA3/2, SOX17, and TFAP2C, is a minimal requirement in replacing the BMP signaling and conferring the germ-cell fate on iMeLCs. This would in turn suggest that in humans, the BMP signaling plays a key role in activating not only GATA3/2, but also, directly or indirectly, SOX17 and TFAP2C, because BMP-induced GATA3/2 per se was not sufficient to activate SOX17 and TFAP2C. The mechanisms by which the three TFs control each other as well as the downstream pathways for hPGCLC specification require further investigation. In this regard, it is interesting to note that GATA3 is required only transiently for hPGCLC induction (Fig 5G–J), and that GATA TFs are known to act as “pioneering factors” that open heterochromatin regions and make them accessible to other TFs (51, 52, 53). Accordingly, it has been reported that hPGCLCs and human fetal germ cells bear open chromatin regions enriched in the binding motifs for OCT4, SOX/TFAP/KLF families, and GATA families as well (15), and that the expression of GATA3 as well as TFAP2A precedes that of SOX17 and TFAP2C at a single-cell level (54), supporting the idea that GATA TFs may operate as “pioneering factors” for other TFs such as SOX17 and TFAP2C to drive germ-cell specification and thereafter to maintain germ-cell identity (Fig 7I).
A number of studies have shown that in response to BMP4, hPSCs differentiate into TE-like cells with the expression of key TFs, including GATA3, GATA2, TFAP2A, and TFAP2C (39, 46, 55, 56), and thus the differentiation of hPSCs into TE-like cells involves signals and TFs that are also involved in hPGCLC specification (this study) (54). These TFs are also associated with the TE development in mice (40, 57, 58), and as in the case of hPGCLC specification, GATA3 and GATA2 play a compensatory function for TE-like cell specification in humans (39) and TE development in mice (40). A characteristic gene involved in hPGC(LC) differentiation is SOX17, the activation of which requires stimulation of hPSCs by WNT signaling that activates EOMES expression (i.e., iMeLC induction), before stimulation by BMP signaling (13). In contrast, TE-like cell differentiation requires direct stimulation of hPSCs by BMP signaling (39, 55, 56) and such differentiation has been shown to be significantly promoted by inhibiting endogenous WNT activity elicited by BMP (59). Thus, evidently, a prior activation of the WNT pathway in hPSCs is a key to the differential cell-fate specification between the germ-cell fate and TE-like cell fate upon BMP stimulation.
Notably, GATA3, GATA2, TFAP2A, and TFAP2C are also expressed in the amnion in cynomolgus monkeys (Fig 6A–C) (18) and most likely in humans (60, 61), indicating that relatively close lineage relationships exist among TEs, amnion and PGCs in primates. Indeed, during primate development, TEs are specified from the inner cell mass (ICM) cells of the pre-implantation blastocysts (around embryonic day [E] 4 in humans and E5 in cy monkeys) 30, 62, 63, 64, and subsequently, the amnion is differentiated from ICM/epiblasts around the peri-implantation stage (around E7 in humans and E11 in cy monkeys) 18, 30, 62, 63, 64 and the PGCs are most likely originated in the nascent amnion (unknown for humans and E11 in cy monkeys) (18, 54). Thus, TEs, amnion and PGCs are the lineages that arise successively from the ICM/epiblast during the relatively short period of early development. On the other hand, the transcriptome of h/cyPSCs is highly similar to those of post-implantation early (E12/13) or late (E16/17) epiblast cells, and is substantially different from those of ICM cells or pre-implantation epiblast (30), making it difficult to naturally reconcile the observation that hPSCs bear a capacity to differentiate into TE-like cells; further investigations will be needed to account for this apparent paradox. The mechanism that segregates the germ-cell fate from the amnion fate, which also responds to WNT signaling (18), also remains an open question, and an understanding of this mechanism may lead to a more efficient induction of the germ-cell fate from hPSCs.
Crucially, we showed that the TF-induced BT+AG+ cells, when cultured in xrOvaries, underwent epigenetic reprogramming and differentiated into oogonia/gonocytes (Fig 7), demonstrating that the TF-induced BT+AG+ cells bear one of the key functions of bona fide hPGCs. Unlike mPGCLC specification, which is directly coupled with epigenetic reprogramming (65, 66, 67, 68), hPGCLC specification itself does not appear to be sufficient to elicit the epigenetic reprogramming: further signaling/environmental cues, including those provided by xrOvaries, are necessary to activate such key processes (10, 50). Upon mPGCLC specification, Blimp1, Prdm14, and Tfap2c repress the expression of genes such as Dnmt3a/b and Uhrf1, and create a cellular state with little, if any, de novo and maintenance DNA methyltransferase (DNMT) activities (50, 65, 66, 67, 68), and this leads to a replication-coupled passive genome-wide DNA demethylation upon mPGCLC proliferation (67, 68). In contrast, the mechanism of epigenetic reprogramming, including genome-wide DNA demethylation, in humans is unclear, and may involve a divergence from that in mice. The identification of the TFs sufficient to create the hPGCLC state (this study), coupled with the development of a method for hPGCLC expansion (50), will be instrumental in clarifying the mechanism of epigenetic reprogramming in human germ cells.
The mechanisms of germ-cell specification in metazoans are classified largely into two modes, “epigenesis” and “preformation” (69, 70). The former, as in mammals, involves a strategy to induce the germ-cell fate into pluripotent precursors by signaling molecules and is evolutionarily ancestral, whereas the latter, as in flies and frogs, involves “preformed” germ plasm in oocytes for germ-cell specification and has been acquired in diverse metazoan lineages as a result of convergent evolution (69, 70). Notably, in the “epigenesis” mode, BMP has been identified as an evolutionarily conserved key signal in species as diverse as gryllus (71), axolotl (72), and mammals, including mice (73, 74), rabbits (75), pigs (76), monkeys (18, 19), and humans (8, 9). On the other hand, there has been a lack of knowledge as to the mechanism of action, including via direct effectors, of the BMP signaling for PGC specification in these species. In future investigations, it would be useful to investigate whether GATA TFs—which are widely evolutionarily conserved—play a similar role in diverse species, including mice.
Materials and Methods
Reagents and tools table
| Reagent/resource | Reference or source | Identifier or catalog number |
|---|---|---|
| Experimental models | ||
| BTAG (BLIMP1-tdTomato and TFAP2C-eGFP knockin reporters in the 585B1 hiPSCs) | Sasaki et al (2015). | N/A |
| BTAG; SOX17 OE#1 | This study | N/A |
| BTAG; SOX17 OE#2 | This study | N/A |
| BTAG; TFAP2C OE#1 | This study | N/A |
| BTAG; TFAP2C OE#3 | This study | N/A |
| BTAG; SOX17+BLIMP1 OE#9 | This study | N/A |
| BTAG; SOX17+BLIMP1 OE#15 | This study | N/A |
| BTAG; TFAP2C+BLIMP1 OE#2 | This study | N/A |
| BTAG; TFAP2C+BLIMP1 OE#15 | This study | N/A |
| BTAG; SOX17+TFAP2C OE#2 | This study | N/A |
| BTAG; SOX17+TFAP2C OE#11 | This study | N/A |
| BTAG; SOX17+TFAP2C OE#28 | This study | N/A |
| BTAG; SOX17+TFAP2C+BLIMP1 OE#4 | This study | N/A |
| BTAG; SOX17+TFAP2C+BLIMP1 OE#9 | This study | N/A |
| BTAG; SOX17+TFAP2C+BLIMP1 OE#22 | This study | N/A |
| BTAG; SOX17+TFAP2C+BLIMP1 OE#28 | This study | N/A |
| BTAG; SOX17+TFAP2C+BLIMP1 OE#31 | This study | N/A |
| BTAG; SOX17+TFAP2C+MSX2 OE#7 | This study | N/A |
| BTAG; SOX17+TFAP2C+MSX2 OE#9 | This study | N/A |
| BTAG; GATA3+SOX17+TFAP2C OE#1 | This study | N/A |
| BTAG; GATA3+SOX17+TFAP2C OE#5 | This study | N/A |
| BTAG; GATA2+SOX17+TFAP2C OE#29 | This study | N/A |
| BTAG; GATA2−/− #1 | This study | N/A |
| BTAG; GATA2−/− #6 | This study | N/A |
| BTAG; GATA2−/− #12 | This study | N/A |
| BTAG; GATA3−/− #17 | This study | N/A |
| BTAG; GATA3−/− #18 | This study | N/A |
| BTAG; GATA3−/− #30 | This study | N/A |
| BTAG; GATA3−/− #40 | This study | N/A |
| BTAG; HAND1−/− #6 | This study | N/A |
| BTAG; GATA3−/−; GATA2+/− #5-1 | This study | N/A |
| BTAG; GATA3−/−; GATA2−/− #5-10 | This study | N/A |
| BTAG; GATA3−/−; GATA2−/−; GATA3 OE #19 | This study | N/A |
| Recombinant DNA | ||
| pX335-U6-Chimeric BB-CBh-hSpCas9n (D10A) | Addgene | Cat. no. 42335 |
| Antibodies | ||
| Goat anti-SOX17 | R&D Systems | AF1924; RRID: AB_355060 |
| Mouse anti-TFAP2C | Santa Cruz | sc-12762; RRID: AB_667770 |
| Mouse anti-BLIMP1 | R&D Systems | MAB36081; RRID: AB_10718104 |
| Mouse anti-GATA3 | BIOCARE | ACR405A; RRID: AB_10895444 |
| Rabbit anti-GATA3 | Cell Signaling | CST5852S; RRID:AB_10835690 |
| Rabbit anti-GATA2 | Novus | NBP82581; RRID:AB_11026191 |
| Rabbit anti-GATA2 | Santa Cruz | sc9008; RRID:AB_2294456 |
| Mouse anti-human mitochondria | Merck Millipore | MAB1273; RRID:AB_94052 |
| Goat anti-DDX4 | R&D Systems | AF2030; RRID:AB_2277369 |
| Mouse IgG – HRP conjugated | Sigma-Aldrich | A5906; RRID: AB_258264 |
| Mouse anti-α Tubulin | Sigma-Aldrich | T9026; RRID: AB_477593 |
| Mouse IgG – HRP conjugated | Sigma-Aldrich | A5906; RRID: AB_258264 |
| Rabbit IgG – HRP conjugated | Sigma-Aldrich | A6154; RRID: AB_258284 |
| Goat IgG – HRP conjugated | Sigma-Aldrich | A5420; RRID: AB_258242 |
| SSEA1 (CD15) microbeads for human and mouse | Miltenyi Biotec | 130-094+530 |
| CD31 microbeads for mouse | Miltenyi Biotec | 130-097-418 |
| Oligonucleotides and sequence-based reagents | ||
| qRT-PCR primers | This study | Table S3 |
| Chemicals, enzymes and other reagents | ||
| StemFit AK03N | Ajinomoto | N/A |
| iMatrix-511 | Nippi | |
| Puromycin | Thermo Fisher Scientific | A1113803 |
| G418, Geneticin | Thermo Fisher Scientific | #10131035 |
| Hygromycin B | Thermo Fisher Scientific | #10131035 |
| Doxycycline | Takara-Clontech | Z1311N |
| Fibronectin | Millipore | FC010 |
| GMEM | Thermo Fisher Scientific | #11710035 |
| Knockout serum replacement | Thermo Fisher Scientific | A3181502 |
| Activin A | Peprotech | 120-14E |
| CHIR99021 | TOCRIS | #4423 |
| Y27632 | FujiFilm | 030-24021 |
| BMP4 | R&D Systems | 314-BP |
| SCF | R&D Systems | 255-SC |
| EGF | R&D Systems | 236-EG |
| LIF | Millipore | LIF1010 |
| LDN193189 | StemGent | 04-0074 |
| Glutamax | Thermo Fisher Scientific | 35050-061 |
| HEPES | Thermo Fisher Scientific | 15630-106 |
| α-Minimum Essential Medium | Thermo Fisher Scientific | 32571-036 |
| L-ascorbic acid | Sigma-Aldrich | A4403 |
| Software | ||
| FACSDiva Software | BD Biosciences | N/A |
| DAVID (v6.8; GO analysis) | https://david.ncifcrf.gov/ | N/A |
| FV10-ASW | Olympus | N/A |
| R (v3.6.0; PCA, DEG, and graphs) | https://www.R-project.org | N/A |
| Bowtie2 v2.2.7 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml | N/A |
| TopHat v2.1.0 | https://ccb.jhu.edu/software/tophat/index.shtml | N/A |
| HTSeq v0.9.1 | https://htseq.readthedocs.io/en/master/overview.html | N/A |
| ImageJ/Fiji | Fiji.sc | N/A |
| Trim_galore v0.6.3 | https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ | N/A |
| cutadapt v118 | http://cutadapt.readthedocs.io/en/stable/guide.html | N/A |
| Bismark v0.22.1 | https://www.bioinformatics.babraham.ac.uk/projects/bismark/ | N/A |
| SAMtools v1.9 | http://samtools.source-forge.net | N/A |
| Other | ||
| pGEM-T Easy Kit | Promega | A3600 |
| Gateway LR Clonase Enzyme Mix | Thermo Fisher Scientific | #11791043 |
| v-bottom 96-well plate | Greiner | #651970 |
| RNeasy Micro Kit | QIAGEN | #74004 |
| NucleoSpin RNA XS | Macherey-Nagel | #740902 |
| Qubit RNA HS assay kit | Thermo Fisher Scientific | Q32855 |
| PowerSYBR Green PCR Master Mix | Thermo Fisher Scientific | #4367659 |
| Qubit dsDNA HS assay kit | Thermo Fisher Scientific | Q32851 |
| Protein Quantification Assay | Macherey-Nagel | #740967 |
| ECL Western Blotting Detection Reagent | GE Healthcare Life Sciences | RPN2106 |
| EZ DNA Methylation-Gold Kit | Zymogen | D5005 |
| DP72 | Olympus | N/A |
| FV1000-IX81 confocal microscope system | Olympus | N/A |
| CFX384 Touch Real-Time PCR detection system | Bio-Rad Laboratories | N/A |
| NextSeq500/550 | Illumina | N/A |
| Hiseq2500 | Illumina | N/A |
Animal care and use
All animal experiments were performed under the ethical guidelines of Kyoto University and Shiga University of Medical Science. Pregnant ICR female mice were purchased from Japan SLC. Experimental procedures using cynomolgus monkeys were approved by the Animal Care and Use Committee of the Shiga University of Medical Science.
Human iPSC culture
All the experiments on the induction of hPGCLCs from hiPSCs and genome editing were approved by the Institutional Review Board of Kyoto University and were performed according to the guidelines of the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan.
The 585B1 BTAG hiPSCs (46, XY) (9) were maintained in StemFit AK03N medium (Ajinomoto) on cell culture plates coated with iMatrix-511 (Nippi) (77). The medium was changed every other day. For the passage or the induction of differentiation, the cells were treated with a 1 to 1 mixture of TrypLE Select (Life Technologies) and 0.5 mM EDTA/PBS to dissociate into single cells, and 10 μM of a ROCK inhibitor (Y-27632; Wako Pure Chemical Industries) was added for 24 h after plating.
Generation of TF-expression lines
The vectors for the Doxycycline-induced expression were constructed based on the Gateway System (Thermo Fisher Scientific) as described previously (13). The full-length cDNA sequences of SOX17, TFAP2C, BLIMP1, MSX2, GATA3, and GATA2 were PCR amplified from d2 hPGCLCs derived from the 585B1 BTAG hiPSC line. Nucleotide sequences for the epitope tags with linkers, 3×FLAG-G4S, V5-G4S, and 2×TY1-G4S, were added to the 5-prime ends of SOX17, TFAP2C, and BLIMP1/MSX2/GATA3/GATA2, respectively. Primers used for the construction are shown in Table S3. The PCR products were cloned between the BamHI and XhoI sites of the pENTR1a vector and were subsequently recombined into the destination vector with LR clonase. In the destination vector, the transgenes were cloned under the TetO promoter repeat region and followed by the rabbit β-globin poly A (rBGpA) sequence. In the region downstream of rβGpA, the puromycin-resistant gene driven by the EF1α promoter was cloned for SOX17 and TFAP2C, neomycin for rtTA, and hygromycin for the rest of the genes.
Table S3 Primers used in this study. (12.1KB, xlsx)
Transfection was performed with the electroporator NEPA21 type II (Nepagene). Half a million hiPSCs were transfected with 500 ng of the piggybac transposase expression vector and 1 μg of each transgene expression vector, except for TFAP2C, which was added at 1.5 μg, then resuspended in 100 μl of OptiMEM (Thermo Fisher Scientific). Selection antibiotics (200 μg/ml geneticin [G418], 10 μg/ml puromycin, and 800 μg/ml hygromycin [all from Thermo Fisher Scientific]) were added 2 d after the transfection and maintained until the surviving colonies were picked up at 12–14 d. The induction of the transgenes with 1.0 μg/ml doxycycline (Takara-Clontech) for the selected hiPSC clones was assessed at 24 h of culture.
Generation of knockout lines
pX335-U6-Chimeric BB-CBh-hSpCas9n (D10A) was a gift of Feng Zhang (plasmid #42335; Addgene) (45), and the eGFP sequence was replaced with the mCherry sequence bearing a silent mutation (G432A) for convenience in clone selection. For one recombination site, a pair of gRNA sequences nicking each strand of genomic DNA were designed using a CRISPR design website (crispr.mit.edu/: now renovated). Two oligo DNAs bearing the gRNA sequence and compatible ends for the BbsI-digested overhang sequence were phosphorylated, annealed and ligated into the BbsI site of the pX335 vector. 2 μg each of the CRISPR vectors in 100 μl of OptiMEM were transfected into 5 × 105 585B1 BTAG hiPSCs using NEPA21 type II (Nepagene). The cells were cultured in AK03N with 10 μM Y27632 for 24 h, then maintained in AK03N alone for the next 24 h, and the cells with high mCherry expression (≈top 0.5%) were plated onto 96-well plates precoated with iMatrix at a single cell per well with the Automatic Cell Deposition Unit of the FACS Aria III (BD Biosciences). The cells were then cultured in AK03N with 10 μM Y27632 for 72 h and subsequently cultured in AK03N alone. 10–14 d after plating, proliferating colonies were collected: half of the cells were frozen in Stem CellBanker (Zenoaq) and the remaining half were pelleted and lysed for genotyping.
Genotype
To extract the genomic DNA, the cells were lysed in 40 μl of KOD Plus Neo buffer (TOYOBO) supplemented with 0.5% NP40 and 0.8 mg/ml Proteinase K (TakaraBio) at 55°C for 3 h, followed by a proteinase inactivation step at 95°C for 10 min. PCR amplification at the CRISPR recombination site was performed from 1 μl of the cell lysate with KOD Plus Neo (TOYOBO) and the primers listed in Table S3. To sequence each allele separately, the amplicon was A-tailed with Taq polymerase (Greiner), cloned into pGEM-T easy (Promega), and transformed into the DH5α Escherichia coli strain, followed by plating onto LB plates with a blue-white selection. White colonies were picked for direct PCR with KOD FX Neo (TOYOBO) using the primers M13-RV and M13-M4. The amplified fragments were sequenced by Eurofins Genomics with the M13-FW primer and searched for insertions and/or deletions.
Western blot analysis
Cell pellets were lysed in Bolt Sample Buffer Reducing Agent and the protein concentrations were measured by protein quantification assay (Macharey-Nagel). Bolt 10% Bis-Tris Plus Gels were used for SDS–PAGE with 1 μg of samples per lane and subsequently transferred to Polyvinylidene difluoride (PVDF) membrane (0.22 μm pore) with an iBlot2 Dry Blotting System. All the reagents and devices were from Thermo Fisher Scientific if not specified and all the experiments were performed following the manufacturer’s instructions. The transferred membrane was washed in PBST (PBS with 0.1% Tween20), blocked with the blocking solution (5% skim milk in PBST) for 20 min at room temperature with continuous rocking and incubated overnight at 4°C with primary antibodies diluted at 1:1,000 in blocking solution. After washing three times × 5 min with PBST, the membrane was incubated with the HRP-conjugated secondary antibodies diluted at 1:1,000 in blocking solution for 1–3 h at room temperature. After washing three times × 5 min with PBST, the chemiluminescent reaction was induced with Amersham ECL Western Blotting Detection Reagent (GE Healthcare Life Sciences) and the signal was detected with Fusion Solo S (Vilber). All the antibodies used are listed in the Reagents and Tools table.
hPGCLC induction
The induction of hPGCLCs via iMeLCs was performed as described previously (9, 13, 26). For the induction of iMeLCs, hiPSCs were plated at a density of 5 × 104 cells/cm2 onto a fibronectin (FC010; Millipore)-coated plate. Either 24-well, 12-well or 6-well plates were used according to the number of cells required. The cells were cultured in GK15 medium (GMEM with 15% KSR, 0.1 mM NEAA, 2 mM L-glutamine, 1 mM sodium pyruvate, penicillin-streptomycin, and 0.1 mM 2-mercaptoethanol) supplemented with 50 ng/ml activin A (R&D Systems), 3 μM CHIR99021 (TOCRIS), and 10 μM of Y-27632 (Wako Pure Chemical Industries) for 44–48 h. Then the cells were dissociated into single cells with TrypLE Select and aggregated in a low-cell-binding V-bottom 96-well plate (Greiner) at 5,000 cells per well in 100 μl of GK15 medium supplemented with 200 ng/ml BMP4 (R&D Systems), 100 ng/ml SCF (R&D Systems), 50 ng/ml EGF (R&D Systems), 1,000 U/ml LIF (Millipore), and 10 μM of Y-27632 to be induced into hPGCLCs. For the transgene-mediated induction, BMP was replaced with doxycycline (Dox) at 1.0 μg/ml except in the case of GATA2 overexpression, for which 0.5 μg/ml Dox was used. The medium was not changed until the analysis up to the sixth day of induction except in the case of the GATA3 rescue experiment. For the GATA3 rescue experiment, 100 μl of medium containing both Dox and BMP4 was applied first, and then, after 32 h, 90 μl of the medium was aspirated and replaced with the same amount of medium containing BMP4, SCF, EGF and LIF, and the culture was continued for the remaining days. The images of the aggregates were taken under an M205C stereo microscope (Leica Microsystems) equipped with a DP72 CCD camera and DP2-BSW software (Olympus).
FACS
For analysis of the cellular contents of the aggregates, the aggregates were collected on the designated days of induction, washed once in PBS, and dissociated with 0.25% Trypsin–EDTA for 10–15 min at 37°C with gentle pipetting every 5 min. Trypsin was neutralized with a 5× volume of 10% FBS in DMEM, and the resuspended cells were processed with FACS Aria III system (BD Biosciences) and analyzed with FACS Diva software.
The method for selecting CRISPR-mediated knockout clones is described in the section “Generation of knockout lines.”
cDNA amplification, qRT-PCR and RNA-seq analysis
Total RNA was extracted from the frozen cell pellets using RNeasy kits (QIAGEN) or NucleoSpin RNA kits (Macherey-Nagel) following the manufacturers’ instructions. The amount of RNA was measured with Qubit 2.0 (Thermo Fisher Scientific) and the cDNAs were synthesized through amplification of their 3′ ends starting from 1 ng of total RNA as described previously (29). The RNA sample was mixed with ERCC (External RNA Controls Consortium; Thermo Fisher Scientific) spike RNA and then reverse transcribed with V1-(dT)24 primer using SuperScript III for 5 min at 50°C. SuperScript III was immediately inactivated at 70°C for 10 min, and the excess primer was digested with Exonuclease I (TakaraBio) for 30 min at 37°C followed by heat inactivation for 25 min at 80°C. Then the poly A tail was added to the cDNA with Terminal Deoxynucleotidyl Transferase (TakaraBio) for 15 min at 37°C and heat inactivated for 10 min at 70°C. Subsequently, PCR amplification was done using ExTaq HS polymerase (TakaraBio); the first cycle was run with V3-(dT)24 primer alone, followed by 14 cycles using both V1-(dT)24 and V3-(dT24). The PCR product was then purified and the primer dimers were removed by adding a 0.6× volume of AMPure XP (Agencourt), washed with 80% ethanol two times, and eluted with 50 μl 5 mM Tris–HCl (pH 8.0) on a magnetic stand. In some cases, an AxyPrep MAG PCR Clean Up Kit (Corning) was used in place of AMPure XP; the two provided comparable results.
qRT-PCR was performed with PowerSYBR Green PCR Master Mix (Thermo Fisher Scientific) on a CFX384 Real-Time PCR Detection System (Bio-Rad) using the primers listed in Table S3. The quality of the amplified cDNA was assessed according to the Ct values by qPCR of the ERCC spike RNA and the housekeeping genes (PPIA and RPLP0).
The cDNA library was prepared as described previously (78). 5 ng aliquots of quality-controlled cDNA samples were further amplified by PCR using ExTaq HS (TakaraBio) with the N-V3 (dT)24 and V1 (dT)24 primers for four cycles, purified with three rounds of binding, washing and eluting steps with AMPureXP, and then fragmented with a Covaris E220 sonicator. The fragmented products were end-polished with T4 DNA polymerase (NEB) and T4 polynucleotide kinase (NEB) for 30 min at 20°C. The products were then purified again with a 0.7× volume of AMPureXP, followed by addition of a 0.9× volume of AMPureXP to the supernatant, and a final washing and elution. To attach adaptor sequences, the cDNA solution was treated first with Rd2SP-V1(dT)20 primer using ExTaqHS, followed by addition of Rd1SP-adaptor with T4 DNA ligase (NEB), and purified with a 0.8× volume of AMPureXP. The adapter attached cDNA was then PCR amplified using Nextera XT Index 1 (N7XX) and Index 2 (S5XX) Primers (Illumina) with ExTaqHS for 10 cycles and purified by two washings with a 0.9× volume of AMPureXP.
The quality and quantity of the resultant library DNAs were evaluated by the LabChip GX (Perkin Elmer), the Qubit dsDNA HS assay kit, and the Taqman-qPCR assay using Thunderbird Probe qPCR mix (TOYOBO) and a TaqMan probe (Ac04364396; Applied Biosystems). The sequence data were acquired using NextSeq 500 (Illumina). Conversion of the sequence read data into expression levels was performed as described previously (29, 78). The reads were first processed with cutadapt-1.3 (79) to trim the V1 and V3 adaptor sequences and poly-A sequences. The trimmed reads longer than 30 bp were then mapped onto the GRCh38.p2 genome using Tophat v2.1.0/Bowtie2 v2.2.7, with the “-no-coverage-search” option (80). The expression levels (reads per million-mapped reads: RPM) were calculated from these mapped reads using the HTSeq v0.9.1 with default settings, and the GRCh38.p2 reference gene annotations were modified, where necessary, so that the transcript termination sites were extended up to 10 kb downstream.
Data analysis of the RNA-seq
All statistical analyses were performed on R (ver 3.6.0) with stringr, gplots, and prcomp packages. The expression data were first converted into log2(RPM + 1) values, the genes with maximum log2(RPM + 1) < 4 (equivalent to 10–20 copies per cell) in all the samples were excluded, and the distribution of the expression levels was assessed with boxplots to confirm the quality of the samples. For hPGCLC induction, 180 samples were added to the 230 samples used in the former study with 12,909 genes (13), and for xenogeneic reconstituted ovary series, three iMeLC samples and three ag77 samples were newly collected and analyzed with 30 samples collected in the previous study (10) with 12,737 genes.
All the heat maps were drawn with the heatmap.2() function, the correlation coefficient between BMP-induced and overexpressed samples was calculated with the cor() function with “Pearson correlation,” and the PCA was performed with the prcomp() function. DEGs among SOX17/TFAP2C/BLIMP1 samples were defined as P < 0.01 by one-way ANOVA among all time points and conditions of SOX17/TFAP2C/BLIMP1 series, P < 0.01 in Tukey–Kramer post hoc test for multiple comparisons, log2(RPM + 1) > 4 in the higher expression group, and more than onefold change of the mean expression values. GO analysis was performed on the DAVID website (https://david.ncifcrf.gov). The GO terms were extracted from the Biological Process (GOTERM_BP_DIRECT) and the pathway terms from KEGG_PATHWAY (Kyoto Encyclopedia of Genes and Genomes) (http://www.genome.jp/kegg/) (81).
Single-cell transcriptome data of cynomolgus monkey embryos was retrieved from the Gene Expression Omnibus database under accession numbers GSE67259, GSE74767 and GSE76267 (30, 31). The following cells were extracted: post-implantation parietal trophectoderm (designated Post-paTE), post-implantation early epiblast (PostE-EPI), post-implantation late epiblast (PostL-EPI), gastrulating cells group 1 (G1), gastrulating cells group 2a (G2a), gastrulating cells group 2b (G2b), visceral endoderm or yolk sac endoderm (VE/YE), extraembryonic mesenchyme (EXMC), early PGCs (ePGC), and late PGCs (lPGC). The heat maps were drawn with the heatmap.2() function on R (ver 3.6.0). Note that SOX17 and GATA2 were named LOC101925698 and LOC101865311, respectively, in these datasets.
Immunofluorescence of iMeLC aggregates and cynomolgus monkey embryos
Cynomolgus monkey embryos were isolated as described previously (18, 30). The samples were formalin-fixed, paraffin-embedded, and stored at 4°C. The sectioned samples were first deparaffinized and hydrated followed by antigen retrieval with HistoVT One (Nacalai) at 90°C for 35 min. After washing in PBS, the samples were permeabilized and blocked with blocking solution (5% donkey serum, 0.2% Tween 20, in PBS) for 2 h at ambient temperature. The samples were then treated with primary antibodies in the blocking solution overnight at 4°C, washed three times with PBS, incubated with secondary antibodies at room temperature for 1 h, washed three times with PBS, mounted in VECTASHIELD Mounting Medium with DAPI (Vector Laboratories), and imaged under an Olympus FV1000 confocal microscope.
For the immunofluorescence analysis of reconstituted xenogeneic ovaries, the harvested reaggregates were fixed with 4% paraformaldehyde at 4°C for 2 h, followed by two-step cryoprotection with 10% and 30% sucrose dissolved in PBS at 4°C for 1 h and overnight, respectively, and finally, freezing in Optimal Cutting Temperature (OCT) compound (Sakura Finetek). The sectioned samples were washed with PBS to remove the OCT compound, and then permeabilized and blocked using the protocol described above.
Xenogeneic reconstituted ovary (xrOvaries) culture
d6 hPGCLCs were further differentiated by aggregation with mouse female gonadal somatic cells at embryonic day (E) 12.5, which we termed xrOvaries as described previously (10, 11). Pregnant ICR females were euthanized by cervical dislocation and the E12.5 embryos were dissected in DMEM containing 10% FBS (Hyclone), 2 mM GlutaMax, 10 mM HEPES, and 100 U/ml penicillin/streptomycin. Fetal ovaries were dissected out with tungsten needles and dissociated into single cells with 0.25% Trypsin, the endogenous mouse PGCs were removed by MACS with anti-CD31 and anti-SSEA1 antibodies (Miltenyi), and the remaining fetal ovarian somatic cells were aliquoted and frozen until use. Thawed somatic cells (75,000 cells/well) were mixed with d6 hPGCLCs (5,000 cells/well) and cultured in a Lipidure-coated U-bottom 96-well plate with GK15 medium containing 10 μM Y27632 (Tocris) to form floating cellular aggregates. After 2 d under this condition, the xrOvaries were transferred onto Transwell-COL membrane inserts (Corning) with a mouth pipette and maintained as an air-liquid interface culture in α-MEM with 10% FBS, 55 μM 2-mercaptoethanol, 150 μM l-ascorbic acid (Sigma-Aldrich), and 100 U/ml penicillin/streptomycin. The xrOvaries were cultured at 37°C, 5%CO2 in air and half of the medium was replaced every 3 d until harvest. All the reagents were from Thermo Fisher Scientific unless otherwise specified.
WGBS
Genomic DNA was prepared from 5,000 cells, then resuspended in lysis buffer containing 0.1% SDS and 1 μg/μl Proteinase K in DNase Free Water (GIBCO) and incubated at 37°C for 60 min; the Proteinase K was then heat inactivated at 98°C for 15 min. This lysate was spiked with 150 pg of unmethylated λ phage DNA (Promega), based on the estimation that the amount of genomic DNA was 6 pg per cell and the phage DNA accounted for 0.5% of the input genomic DNA. Bisulfite conversion and library construction were performed by the post-bisulfite adaptor tagging (PBAT) method (82). The detailed protocol of the PBAT method has been published and is freely available at the website of the International Human Epigenome Consortium (http://www.crest-ihec.jp/english/epigenome/index.html). All the reagents used were the same as described in the protocol, except that Phusion Hot Start II DNA Polymerase (Thermo Fisher Scientific) and an AxyPrep MAG PCR Clean-Up Kit (Corning) were substituted for the Phusion Hot Start High-Fidelity DNA polymerase and Agencourt AMPure XP, respectively. Deep sequencing was performed on an Illumina Hiseq 2500 system to generate 101-nucleotide single-end sequence reads, and cluster generation and sequencing were implemented in single-read mode using a TruSeq SR Cluster Kit v3-cBot-HS and TruSeq SBS Kit v3-HS (Illumina) following the manufacturer's instructions.
WGBS data processing
The WGBS data were first processed with Trim_Galore v0.6.3 (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/)/cutadapt v1.18 (http://cutadapt.readthedocs.io/en/stable/guide.html) with the “--clip_R1 4,” “--trim1” and “-a AGATCGGAAGAGC” options to trim away low-quality bases (four bases from the 5′ ends, one base from the 3′ends and bases with quality score <20) and the adapter sequences. The qualified reads were then mapped onto the human genome (GRCh38.p2) using Bismark v0.22.1 with the “--pbat” option, and the cytosine/methyl-cytosine count at every CpG site on the genome was determined by the bismark_methylation_extractor program included in the package.
For the genome-wide analysis, all the CpG sites with read depth ≥4 were used for the following analyses. The percent methylations of individual CpG sites were plotted in the Violin plot using the vioplot package. For the scatterplots, the average percent methylations of the CpG sites in 2-kb non-overlapping bins that carried four or more CpG sites were used. The scatterplots were overlaid with contour plots to enhance the visibility of the plot density, and a histogram in each scatterplot shows the frequency of CpG methylation at 5% intervals within the samples.
To compare the ratio of DNA methylation in particular regions, the genomic DNA sequence was grouped into the following regions. Promoters were defined as the region between 900-bp upstream and 400-bp downstream of the transcription start sites, and the promoters with high CpG (HCP), intermediate CpG (ICP), and low CpG (LCP) were classified according to the previous report (83). Data for the CpG islands (84) and human imprint loci (85) were downloaded from public repositories and converted to GRCh38 format with LiftOver. No statistical analysis was performed on WGBS data.
Data and Code Availability
All the sequencing data have been deposited in the Gene Expression Omnibus database under accession number GSE154691 (RNA-seq: GSE154688; WGBS: GSE154690) and the R script is available on request.
Supplementary Material
Acknowledgements
We thank the members of our laboratory for their helpful input on this study. We are grateful to Y Nagai, N Konishi, E Tsutsumi, and M Kawasaki of the Saitou Laboratory, and J Asahira and M Kabata of the Yamamoto Laboratory for their technical assistance. This work was supported in part by Grants-in-Aid for Scientific Research from Japan Society for the Promotion of Science (JSPS) (15K08267 and 20K06654) to Y Kojima and by a Grant-in-Aid for Specially Promoted Research from JSPS (17H06098), an Exploratory Research for Advanced Technology Grant from the Japan Science and Technology Agency (JST-ERATO) (JPMJER1104), a Grant from Human Science Frontier Program (RGP0057/2018), and grants from the Pythias Fund and Open Philanthropy Project to M Saitou.
Author Contributions
Y Kojima: conceptualization, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, project administration, and writing—original draft, review, and editing.
C Yamashiro: resources, investigation, and methodology.
Y Murase: resources, investigation, and methodology.
Y Yabuta: resources, formal analysis, investigation, and methodology.
I Okamoto: resources, investigation, and methodology.
C Iwatani: resources, investigation, and methodology.
H Tsuchiya: resources, investigation, and methodology.
M Nakaya: resources, investigation, and methodology.
T Tsukiyama: resources, investigation, and methodology.
T Nakamura: resources, investigation, and methodology.
T Yamamoto: resources, investigation, and methodology.
M Saitou: conceptualization, formal analysis, supervision, funding acquisition, investigation, methodology, project administration, and writing—original draft, review, and editing.
Conflict of Interest Statement
The authors declare that they have no conflict of interest.
References
- 1.Lynch M (2010) Evolution of the mutation rate. Trends Genet 26: 345–352. 10.1016/j.tig.2010.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Segurel L, Wyman MJ, Przeworski M (2014) Determinants of mutation rate variation in the human germline. Annu Rev Genomics Hum Genet 15: 47–70. 10.1146/annurev-genom-031714-125740 [DOI] [PubMed] [Google Scholar]
- 3.Milholland B, Dong X, Zhang L, Hao X, Suh Y, Vijg J (2017) Differences between germline and somatic mutation rates in humans and mice. Nat Commun 8: 15183. 10.1038/ncomms15183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee HJ, Hore TA, Reik W (2014) Reprogramming the methylome: Erasing memory and creating diversity. Cell Stem Cell 14: 710–719. 10.1016/j.stem.2014.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baudat F, Imai Y, de Massy B (2013) Meiotic recombination in mammals: Localization and regulation. Nat Rev Genet 14: 794–806. 10.1038/nrg3573 [DOI] [PubMed] [Google Scholar]
- 6.Saitou M, Miyauchi H (2016) Gametogenesis from pluripotent stem cells. Cell Stem Cell 18: 721–735. 10.1016/j.stem.2016.05.001 [DOI] [PubMed] [Google Scholar]
- 7.Tang WW, Kobayashi T, Irie N, Dietmann S, Surani MA (2016) Specification and epigenetic programming of the human germ line. Nat Rev Genet 17: 585–600. 10.1038/nrg.2016.88 [DOI] [PubMed] [Google Scholar]
- 8.Irie N, Weinberger L, Tang WW, Kobayashi T, Viukov S, Manor YS, Dietmann S, Hanna JH, Surani MA (2015) SOX17 is a critical specifier of human primordial germ cell fate. Cell 160: 253–268. 10.1016/j.cell.2014.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sasaki K, Yokobayashi S, Nakamura T, Okamoto I, Yabuta Y, Kurimoto K, Ohta H, Moritoki Y, Iwatani C, Tsuchiya H, et al. (2015) Robust in vitro induction of human germ cell fate from pluripotent stem cells. Cell Stem Cell 17: 178–194. 10.1016/j.stem.2015.06.014 [DOI] [PubMed] [Google Scholar]
- 10.Yamashiro C, Sasaki K, Yabuta Y, Kojima Y, Nakamura T, Okamoto I, Yokobayashi S, Murase Y, Ishikura Y, Shirane K, et al. (2018) Generation of human oogonia from induced pluripotent stem cells in vitro. Science 36: 356–360. 10.1126/science.aat1674 [DOI] [PubMed] [Google Scholar]
- 11.Yamashiro C, Sasaki K, Yokobayashi S, Kojima Y, Saitou M (2020) Generation of human oogonia from induced pluripotent stem cells in culture. Nat Protoc 15: 1560–1583. 10.1038/s41596-020-0297-5 [DOI] [PubMed] [Google Scholar]
- 12.Hwang YS, Suzuki S, Seita Y, Ito J, Sakata Y, Aso H, Sato K, Hermann BP, Sasaki K (2020) Reconstitution of prospermatogonial specification in vitro from human induced pluripotent stem cells. Nat Commun 11: 5656. 10.1038/s41467-020-19350-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kojima Y, Sasaki K, Yokobayashi S, Sakai Y, Nakamura T, Yabuta Y, Nakaki F, Nagaoka S, Woltjen K, Hotta A, et al. (2017) Evolutionarily distinctive transcriptional and signaling programs drive human germ cell lineage specification from pluripotent stem cells. Cell Stem Cell 21: 517–532.e5. 10.1016/j.stem.2017.09.005 [DOI] [PubMed] [Google Scholar]
- 14.Chen D, Liu W, Lukianchikov A, Hancock GV, Zimmerman J, Lowe MG, Kim R, Galic Z, Irie N, Surani MA, et al. (2017) Germline competency of human embryonic stem cells depends on eomesodermin. Biol Reprod 97: 850–861. 10.1093/biolre/iox138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen D, Liu W, Zimmerman J, Pastor WA, Kim R, Hosohama L, Ho J, Aslanyan M, Gell JJ, Jacobsen SE, et al. (2018) The TFAP2C-regulated OCT4 naive enhancer is involved in human germline formation. Cell Rep 25: 3591–3602.e5. 10.1016/j.celrep.2018.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sybirna A, Tang WWC, Pierson Smela M, Dietmann S, Gruhn WH, Brosh R, Surani MA (2020) A critical role of PRDM14 in human primordial germ cell fate revealed by inducible degrons. Nat Commun 11: 1282. 10.1038/s41467-020-15042-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierson Smela M, Sybirna A, Wong FCK, Surani MA (2019) Testing the role of SOX15 in human primordial germ cell fate. Wellcome Open Res 4: 122. 10.12688/wellcomeopenres.15381.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasaki K, Nakamura T, Okamoto I, Yabuta Y, Iwatani C, Tsuchiya H, Seita Y, Nakamura S, Shiraki N, Takakuwa T, et al. (2016) The germ cell fate of cynomolgus monkeys is specified in the nascent amnion. Dev Cell 39: 169–185. 10.1016/j.devcel.2016.09.007 [DOI] [PubMed] [Google Scholar]
- 19.Sakai Y, Nakamura T, Okamoto I, Gyobu-Motani S, Ohta H, Yabuta Y, Tsukiyama T, Iwatani C, Tsuchiya H, Ema M, et al. (2020) Induction of the germ cell fate from pluripotent stem cells in cynomolgus monkeys. Biol Reprod 102: 620–638. 10.1093/biolre/ioz205 [DOI] [PubMed] [Google Scholar]
- 20.Kanai-Azuma M, Kanai Y, Gad JM, Tajima Y, Taya C, Kurohmaru M, Sanai Y, Yonekawa H, Yazaki K, Tam PP, et al. (2002) Depletion of definitive gut endoderm in Sox17-null mutant mice. Development 129: 2367–2379. [DOI] [PubMed] [Google Scholar]
- 21.Aramaki S, Hayashi K, Kurimoto K, Ohta H, Yabuta Y, Iwanari H, Mochizuki Y, Hamakubo T, Kato Y, Shirahige K, et al. (2013) A mesodermal factor, T, specifies mouse germ cell fate by directly activating germline determinants. Dev Cell 27: 516–529. 10.1016/j.devcel.2013.11.001 [DOI] [PubMed] [Google Scholar]
- 22.Ohinata Y, Payer B, O’Carroll D, Ancelin K, Ono Y, Sano M, Barton SC, Obukhanych T, Nussenzweig M, Tarakhovsky A, et al. (2005) Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 436: 207–213. 10.1038/nature03813 [DOI] [PubMed] [Google Scholar]
- 23.Yamaji M, Seki Y, Kurimoto K, Yabuta Y, Yuasa M, Shigeta M, Yamanaka K, Ohinata Y, Saitou M (2008) Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat Genet 40: 1016–1022. 10.1038/ng.186 [DOI] [PubMed] [Google Scholar]
- 24.Weber S, Eckert D, Nettersheim D, Gillis AJ, Schäfer S, Kuckenberg P, Ehlermann J, Werling U, Biermann K, Looijenga LH, et al. (2010) Critical function of AP-2 gamma/TCFAP2C in mouse embryonic germ cell maintenance. Biol Reprod 82: 214–223. 10.1095/biolreprod.109.078717 [DOI] [PubMed] [Google Scholar]
- 25.Nakaki F, Hayashi K, Ohta H, Kurimoto K, Yabuta Y, Saitou M (2013) Induction of mouse germ-cell fate by transcription factors in vitro. Nature 501: 222–226. 10.1038/nature12417 [DOI] [PubMed] [Google Scholar]
- 26.Yokobayashi S, Okita K, Nakagawa M, Nakamura T, Yabuta Y, Yamamoto T, Saitou M (2017) Clonal variation of human induced pluripotent stem cells for induction into the germ cell fate. Biol Reprod 96: 1154–1166. 10.1093/biolre/iox038 [DOI] [PubMed] [Google Scholar]
- 27.Martins G, Calame K (2008) Regulation and functions of Blimp-1 in T and B lymphocytes. Annu Rev Immunol 26: 133–169. 10.1146/annurev.immunol.26.021607.090241 [DOI] [PubMed] [Google Scholar]
- 28.Bikoff EK, Morgan MA, Robertson EJ (2009) An expanding job description for Blimp-1/PRDM1. Curr Opin Genet Dev 19: 379–385. 10.1016/j.gde.2009.05.005 [DOI] [PubMed] [Google Scholar]
- 29.Nakamura T, Yabuta Y, Okamoto I, Aramaki S, Yokobayashi S, Kurimoto K, Sekiguchi K, Nakagawa M, Yamamoto T, Saitou M (2015) SC3-seq: A method for highly parallel and quantitative measurement of single-cell gene expression. Nucleic Acids Res 43: e60. 10.1093/nar/gkv134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura T, Okamoto I, Sasaki K, Yabuta Y, Iwatani C, Tsuchiya H, Seita Y, Nakamura S, Yamamoto T, Saitou M (2016) A developmental coordinate of pluripotency among mice, monkeys and humans. Nature 537: 57–62. 10.1038/nature19096 [DOI] [PubMed] [Google Scholar]
- 31.Nakamura T, Yabuta Y, Okamoto I, Sasaki K, Iwatani C, Tsuchiya H, Saitou M (2017) Single-cell transcriptome of early embryos and cultured embryonic stem cells of cynomolgus monkeys. Sci Data 4: 170067. 10.1038/sdata.2017.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas PQ, Brown A, Beddington RS (1998) Hex: A homeobox gene revealing peri-implantation asymmetry in the mouse embryo and an early transient marker of endothelial cell precursors. Development 125: 85–94. [DOI] [PubMed] [Google Scholar]
- 33.Brennan J, Lu CC, Norris DP, Rodriguez TA, Beddington RS, Robertson EJ (2001) Nodal signalling in the epiblast patterns the early mouse embryo. Nature 411: 965–969. 10.1038/35082103 [DOI] [PubMed] [Google Scholar]
- 34.Merrill BJ, Pasolli HA, Polak L, Rendl M, García-García MJ, Anderson KV, Fuchs E (2004) Tcf3: A transcriptional regulator of axis induction in the early embryo. Development 131: 263–274. 10.1242/dev.00935 [DOI] [PubMed] [Google Scholar]
- 35.Tremblay M, Sanchez-Ferras O, Bouchard M (2018) GATA transcription factors in development and disease. Development 145: dev164384. 10.1242/dev.164384 [DOI] [PubMed] [Google Scholar]
- 36.Ramos C, Robert B (2005) msh/Msx gene family in neural development. Trends Genet 21: 624–632. 10.1016/j.tig.2005.09.001 [DOI] [PubMed] [Google Scholar]
- 37.Le Bouffant R, Souquet B, Duval N, Duquenne C, Hervé R, Frydman N, Robert B, Habert R, Livera G (2011) Msx1 and Msx2 promote meiosis initiation. Development 138: 5393–5402. 10.1242/dev.068452 [DOI] [PubMed] [Google Scholar]
- 38.Lowry JA, Atchley WR (2000) Molecular evolution of the GATA family of transcription factors: Conservation within the DNA-binding domain. J Mol Evol 50: 103–115. 10.1007/s002399910012 [DOI] [PubMed] [Google Scholar]
- 39.Krendl C, Shaposhnikov D, Rishko V, Ori C, Ziegenhain C, Sass S, Simon L, Müller NS, Straub T, Brooks KE, et al. (2017) GATA2/3-TFAP2A/C transcription factor network couples human pluripotent stem cell differentiation to trophectoderm with repression of pluripotency. Proc Natl Acad Sci U S A 114: E9579–E9588. 10.1073/pnas.1708341114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Home P, Kumar RP, Ganguly A, Saha B, Milano-Foster J, Bhattacharya B, Ray S, Gunewardena S, Paul A, Camper SA, et al. (2017) Genetic redundancy of GATA factors in the extraembryonic trophoblast lineage ensures the progression of preimplantation and postimplantation mammalian development. Development 144: 876–888. 10.1242/dev.145318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Craven SE, Lim KC, Ye W, Engel JD, de Sauvage F, Rosenthal A (2004) Gata2 specifies serotonergic neurons downstream of sonic hedgehog. Development 131: 1165–1173. 10.1242/dev.01024 [DOI] [PubMed] [Google Scholar]
- 42.Gunne-Braden A, Sullivan A, Gharibi B, Sheriff RSM, Maity A, Wang YF, Edwards A, Jiang M, Howell M, Goldstone R, et al. (2020) GATA3 mediates a fast, irreversible commitment to BMP4-driven differentiation in human embryonic stem cells. Cell Stem Cell 26: 693–706.e9. 10.1016/j.stem.2020.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cuny GD, Yu PB, Laha JK, Xing X, Liu JF, Lai CS, Deng DY, Sachidanandan C, Bloch KD, Peterson RT (2008) Structure-activity relationship study of bone morphogenetic protein (BMP) signaling inhibitors. Bioorg Med Chem Lett 18: 4388–4392. 10.1016/j.bmcl.2008.06.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu PB, Deng DY, Lai CS, Hong CC, Cuny GD, Bouxsein ML, Hong DW, McManus PM, Katagiri T, Sachidanandan C, et al. (2008) BMP type I receptor inhibition reduces heterotopic [corrected] ossification. Nat Med 14: 1363–1369. 10.1038/nm.1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Moretto-Zita M, Soncin F, Wakeland A, Wolfe L, Leon-Garcia S, Pandian R, Pizzo D, Cui L, Nazor K, et al. (2013) BMP4-directed trophoblast differentiation of human embryonic stem cells is mediated through a DeltaNp63+ cytotrophoblast stem cell state. Development 140: 3965–3976. 10.1242/dev.092155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang WW, Dietmann S, Irie N, Leitch HG, Floros VI, Bradshaw CR, Hackett JA, Chinnery PF, Surani MA, et al. (2015) A unique gene regulatory network resets the human germline epigenome for development. Cell 161: 1453–1467. 10.1016/j.cell.2015.04.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo F, Yan L, Guo H, Li L, Hu B, Zhao Y, Yong J, Hu Y, Wang X, Wei Y, et al. (2015) The transcriptome and DNA methylome landscapes of human primordial germ cells. Cell 161: 1437–1452. 10.1016/j.cell.2015.05.015 [DOI] [PubMed] [Google Scholar]
- 49.Li L, Dong J, Yan L, Yong J, Liu X, Hu Y, Fan X, Wu X, Guo H, Wang X, et al. (2017) Single-cell RNA-seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell 20: 858–873.e4. 10.1016/j.stem.2017.03.007 [DOI] [PubMed] [Google Scholar]
- 50.Murase Y, Yabuta Y, Ohta H, Yamashiro C, Nakamura T, Yamamoto T, Saitou M (2020) Long-term expansion with germline potential of human primordial germ cell-like cells in vitro. EMBO J 39: e104929. 10.15252/embj.2020104929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS (2002) Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell 9: 279–289. 10.1016/s1097-2765(02)00459-8 [DOI] [PubMed] [Google Scholar]
- 52.Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, Brown M (2007) Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res 67: 6477–6483. 10.1158/0008-5472.can-07-0746 [DOI] [PubMed] [Google Scholar]
- 53.Kong SL, Li G, Loh SL, Sung WK, Liu ET (2011) Cellular reprogramming by the conjoint action of ERalpha, FOXA1, and GATA3 to a ligand-inducible growth state. Mol Syst Biol 7: 526. 10.1038/msb.2011.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen D, Sun N, Hou L, Kim R, Faith J, Aslanyan M, Tao Y, Zheng Y, Fu J, Liu W, et al. (2019) Human primordial germ cells are specified from lineage-primed progenitors. Cell Rep 29: 4568–4582.e5. 10.1016/j.celrep.2019.11.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu RH, Chen X, Li DS, Li R, Addicks GC, Glennon C, Zwaka TP, Thomson JA (2002) BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol 20: 1261–1264. 10.1038/nbt761 [DOI] [PubMed] [Google Scholar]
- 56.Drukker M, Tang C, Ardehali R, Rinkevich Y, Seita J, Lee AS, Mosley AR, Weissman IL, Soen Y (2012) Isolation of primitive endoderm, mesoderm, vascular endothelial and trophoblast progenitors from human pluripotent stem cells. Nat Biotechnol 30: 531–542. 10.1038/nbt.2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Auman HJ, Nottoli T, Lakiza O, Winger Q, Donaldson S, Williams T (2002) Transcription factor AP-2gamma is essential in the extra-embryonic lineages for early postimplantation development. Development 129: 2733–2747. [DOI] [PubMed] [Google Scholar]
- 58.Ralston A, Cox BJ, Nishioka N, Sasaki H, Chea E, Rugg-Gunn P, Guo G, Robson P, Draper JS, Rossant J (2010) Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development 137: 395–403. 10.1242/dev.038828 [DOI] [PubMed] [Google Scholar]
- 59.Kurek D, Neagu A, Tastemel M, Tüysüz N, Lehmann J, van de Werken HJG, Philipsen S, van der Linden R, Maas A, van IJcken WFJ, et al. (2015) Endogenous WNT signals mediate BMP-induced and spontaneous differentiation of epiblast stem cells and human embryonic stem cells. Stem Cel Rep 4: 114–128. 10.1016/j.stemcr.2014.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shao Y, Taniguchi K, Gurdziel K, Townshend RF, Xue X, Yong KMA, Sang J, Spence JR, Gumucio DL, Fu J (2017) Self-organized amniogenesis by human pluripotent stem cells in a biomimetic implantation-like niche. Nat Mater 16: 419–425. 10.1038/nmat4829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng Y, Xue X, Shao Y, Wang S, Esfahani SN, Li Z, Muncie JM, Lakins JN, Weaver VM, Gumucio DL, et al. (2019) Controlled modelling of human epiblast and amnion development using stem cells. Nature 573: 421–425. 10.1038/s41586-019-1535-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O’Rahilly R, Müller F (1987) Developmental Stages in Human Embryos Including a Revision of Streeter’s “Horizons” and a Survey of the Carnegie Collection. Washington, DC: Carnegie Institution. [Google Scholar]
- 63.Luckett WP (1975) The development of primordial and definitive amniotic cavities in early Rhesus monkey and human embryos. Am J Anat 144: 149–167. 10.1002/aja.1001440204 [DOI] [PubMed] [Google Scholar]
- 64.Heuser Streeter (1941) Development of the macaque embryo Contributions to Embryology. pp: 15–55. Washington DC: Carnegie Institution. [Google Scholar]
- 65.Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M (2011) Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146: 519–532. 10.1016/j.cell.2011.06.052 [DOI] [PubMed] [Google Scholar]
- 66.Kurimoto K, Yabuta Y, Hayashi K, Ohta H, Kiyonari H, Mitani T, Moritoki Y, Kohri K, Kimura H, Yamamoto T, et al. (2015) Quantitative dynamics of chromatin remodeling during germ cell specification from mouse embryonic stem cells. Cell Stem Cell 16: 517–532. 10.1016/j.stem.2015.03.002 [DOI] [PubMed] [Google Scholar]
- 67.Shirane K, Kurimoto K, Yabuta Y, Yamaji M, Satoh J, Ito S, Watanabe A, Hayashi K, Saitou M, Sasaki H (2016) Global landscape and regulatory principles of DNA methylation reprogramming for germ cell specification by mouse pluripotent stem cells. Dev Cell 39: 87–103. 10.1016/j.devcel.2016.08.008 [DOI] [PubMed] [Google Scholar]
- 68.Ohta H, Kurimoto K, Okamoto I, Nakamura T, Yabuta Y, Miyauchi H, Yamamoto T, Okuno Y, Hagiwara M, Shirane K, et al. (2017) In vitro expansion of mouse primordial germ cell-like cells recapitulates an epigenetic blank slate. EMBO J 36: 1888–1907. 10.15252/embj.201695862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Extavour CG, Akam M (2003) Mechanisms of germ cell specification across the metazoans: Epigenesis and preformation. Development 130: 5869–5884. 10.1242/dev.00804 [DOI] [PubMed] [Google Scholar]
- 70.Johnson AD, Richardson E, Bachvarova RF, Crother BI (2011) Evolution of the germ line-soma relationship in vertebrate embryos. Reproduction 141: 291–300. 10.1530/rep-10-0474 [DOI] [PubMed] [Google Scholar]
- 71.Donoughe S, Nakamura T, Ewen-Campen B, Green DA 2nd, Henderson L, Extavour CG (2014) BMP signaling is required for the generation of primordial germ cells in an insect. Proc Natl Acad Sci U S A 111: 4133–4138. 10.1073/pnas.1400525111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chatfield J, O’Reilly MA, Bachvarova RF, Ferjentsik Z, Redwood C, Walmsley M, Patient R, Loose M, Johnson AD, et al. (2014) Stochastic specification of primordial germ cells from mesoderm precursors in axolotl embryos. Development 141: 2429–2440. 10.1242/dev.105346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lawson KA, Dunn NR, Roelen BA, Zeinstra LM, Davis AM, Wright CV, Korving JP, Hogan BL (1999) Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev 13: 424–436. 10.1101/gad.13.4.424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ohinata Y, Ohta H, Shigeta M, Yamanaka K, Wakayama T, Saitou M (2009) A signaling principle for the specification of the germ cell lineage in mice. Cell 137: 571–584. 10.1016/j.cell.2009.03.014 [DOI] [PubMed] [Google Scholar]
- 75.Hopf C, Viebahn C, Puschel B (2011) BMP signals and the transcriptional repressor BLIMP1 during germline segregation in the mammalian embryo. Dev Genes Evol 221: 209–223. 10.1007/s00427-011-0373-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kobayashi T, Zhang H, Tang WWC, Irie N, Withey S, Klisch D, Sybirna A, Dietmann S, Contreras DA, Webb R, et al. (2017) Principles of early human development and germ cell program from conserved model systems. Nature 546: 416–420. 10.1038/nature22812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nakagawa M, Taniguchi Y, Senda S, Takizawa N, Ichisaka T, Asano K, Morizane A, Doi D, Takahashi J, Nishizawa M, et al. (2014) A novel efficient feeder-free culture system for the derivation of human induced pluripotent stem cells. Sci Rep 4: 3594. 10.1038/srep03594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ishikura Y, Yabuta Y, Ohta H, Hayashi K, Nakamura T, Okamoto I, Yamamoto T, Kurimoto K, Shirane K, Sasaki H, et al. (2016) In Vitro derivation and propagation of spermatogonial stem cell activity from mouse pluripotent stem cells. Cell Rep 17: 2789–2804. 10.1016/j.celrep.2016.11.026 [DOI] [PubMed] [Google Scholar]
- 79.Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17: 10. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 80.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang da W, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Miura F, Enomoto Y, Dairiki R, Ito T (2012) Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res 40: e136. 10.1093/nar/gks454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Borgel J, Guibert S, Li Y, Chiba H, Schübeler D, Sasaki H, Forné T, Weber M (2010) Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet 42: 1093–1100. 10.1038/ng.708 [DOI] [PubMed] [Google Scholar]
- 84.Illingworth RS, Gruenewald-Schneider U, Webb S, Kerr AR, James KD, Turner DJ, Smith C, Harrison DJ, Andrews R, Bird AP (2010) Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet 6: e1001134. 10.1371/journal.pgen.1001134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Court F, Tayama C, Romanelli V, Martin-Trujillo A, Iglesias-Platas I, Okamura K, Sugahara N, Simón C, Moore H, Harness JV, et al. (2014) Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome Res 24: 554–569. 10.1101/gr.164913.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Source Data for Figure S1LSA-2020-00974_SdataFS1_FS4.pdf (14MB, pdf)
Table S1 List of cell types used for transcriptome analysis. (41.6KB, xlsx)
Table S2 List of differentially expressed genes between relevant cell types generated in this study. (143.3KB, xlsx)
Source Data for Figure S4LSA-2020-00974_SdataFS1_FS4.pdf (14MB, pdf)
Table S3 Primers used in this study. (12.1KB, xlsx)
Data Availability Statement
All the sequencing data have been deposited in the Gene Expression Omnibus database under accession number GSE154691 (RNA-seq: GSE154688; WGBS: GSE154690) and the R script is available on request.